Abstract
Adult humans harbor at least as many microbial cells as eukaryotic ones. The largest compartment of this diverse microbial population, the gut microbiota, encompasses the collection of bacteria, archaea, viruses, and eukaryotic organisms that populate the gastrointestinal tract, and represents a complex and dynamic ecosystem that has been increasingly implicated in health and disease. The gut microbiota carries ∼100-to-150-times more genes than the human genome and is intimately involved in development, homeostasis, and disease. Of the several microbial metabolites that have been studied, short-chain fatty acids emerge as a group of molecules that shape gene expression in several types of eukaryotic cells by multiple mechanisms, which include DNA methylation changes, histone post-translational modifications, and microRNA-mediated gene silencing. Butyric acid, one of the most extensively studied short-chain fatty acids, reaches higher concentrations in the colonic lumen, where it provides a source of energy for healthy colonocytes, and its concentrations decrease towards the bottom of the colonic crypts, where stem cells reside. The lower butyric acid concentration in the colonic crypts allows undifferentiated cells, such as stem cells, to progress through the cell cycle, pointing towards the importance of the crypts in providing them with a protective niche. In cancerous colonocytes, which metabolize relatively little butyric acid and mostly rely on glycolysis, butyric acid preferentially acts as a histone deacetylase inhibitor, leading to decreased cell proliferation and increased apoptosis. A better understanding of the interface between the gut microbiota metabolites and epigenetic changes in eukaryotic cells promises to unravel in more detail processes that occur physiologically and as part of disease, help develop novel biomarkers, and identify new therapeutic modalities.
Size and regulatory role of sRNAs in V. cholerae. Functionally characterized sRNAs from V. cholerae have been grouped according to their size and regulatory functions. The sRNAs are shown as boxes and were color-coded regarding their physiological roles.
The human microbiota and its composition in adults
Microbial symbionts and their animal hosts have a history of co-evolution that goes back at least 500 million years (Cho and Blaser 2012, Christian et al. 2015). Various studies estimated the ratio between eukaryotic and bacterial cells in the human body to be 1:10 (Savage 1977, Bull and Plummer 2014), 1:2–3 (Gilbert 2015), or closer to 1:1 (Sender et al. 2016, Sender et al. 2016). Until recently, the importance of the human microbiota, previously called the normal flora (Cho and Blaser 2012), was largely neglected (Riccio and Rossano 2020), but the topic has received increasing attention, as shown by the exponential growth in the number of papers on the gut microbiota published between 2010 and 2022 (Riccio and Rossano 2020).
The composition of the human microbiota varies by anatomical location and across individuals for the same location, and some of the most extensively studied sites include the skin, the colon, the vagina, the oral cavity, the lung, the stomach, and the hair (Cho and Blaser 2012, Mathieu et al. 2018, Lousada et al. 2021, Hou et al. 2022). The collection of microbial genomes encoded by these bacteria, known as the human microbiome, was referred to as our second genome (Grice and Segre 2012, Lemm 2018).
The microbiota that colonizes the gastrointestinal tract, also known as the gut microbiota, comprises 1013–1014 resident microorganisms that include bacteria, archaea, viruses, fungi, and protozoa (Gill et al. 2006, Martín et al. 2014, Thursby and Juge 2017, Gibiino et al. 2021), and represents a highly diverse and dynamic ecosystem with critical roles in human health (Rodríguez et al. 2015). Five bacterial phyla dominate the healthy gut microbiota in adults, and each microorganism encodes unique metabolic functions (Bezek et al. 2020, Ferraris et al. 2020). Most microorganisms that comprise the gut microbiota are in the colon (Jandhyala et al. 2015, Gagnière et al. 2016, Sender et al. 2016, Dieterich et al. 2018, Gibiino et al. 2021), which is the best studied compartment and one of the most densely populated microbial habitats known on Earth (Kelsen and Wu 2012, Rinninella et al. 2019).
As compared to the 24 000 protein-coding genes that are present in the human genome, the gut microbiome collectively contains at least 100-to-150-times more (Hooper and Gordon 2001, Gill et al. 2006, Karlsson et al. 2013), or >3 million genes, which are involved in a multitude of metabolic pathways (Ursell et al. 2012, Valdes et al. 2018, Rinninella et al. 2019).
The gut microbiota and its link to health and disease
The gut microbiota, relevantly referred to as foreign to us while at the same time being part of us (Riccio and Rossano 2020), plays critical functions in the biology of the host (Guinane and Cotter 2013), and the two exist in a state of mutual symbiosis (Durack and Lynch 2019). Its microbial members interact closely with intestinal epithelial cells, which are located at the interface with underlying host cells. These interactions result in a bidirectional communication between host cells and resident microorganisms as well as microbiota-derived metabolites. Among the several groups of metabolites that are synthesized by the gut microbiota, the most extensively studied ones include short- and branched-chain fatty acids, phenolic derivatives, polyamines, tryptophan, and bile acids (Yang and Kweon 2016, Wilson and Nicholson 2017, Agus et al. 2021).
The taxonomical abundance of the gut microbiota and its composition are shaped by factors that include microbial interactions, lifestyle, medication use, social networks, environmental factors, diseases, host genetic factors, and age (Goodrich et al. 2014, Blaser 2016, Lane et al. 2019, Madison and Kiecolt-Glaser 2019, Amato et al. 2021, Herzog et al. 2021, Kaur et al. 2022). Twin studies suggest that environmental factors, such as diet, outweigh the contribution of genetic factors to the gut microbiota composition and function (Rothschild et al. 2018).
The human gut microbiota serves multiple functions in the host. Some of these include its contribution to the structure and function of the intestinal barrier, including that of the mucus layer, which at the same time provides nutrients for the microorganisms (Pickard et al. 2017, Paone and Cani 2020); the breakdown of endogenous mucus and the fermentation of non-digestible dietary fibers (Valdes et al. 2018), a process that harvests energy and also supports the production of short-chain fatty acids (SCFAs); the metabolism of bile acids (Ramírez-Pérez et al. 2017, Valdes et al. 2018); the development of the immune system and protection against autoimmune diseases (Wu and Wu 2012, Rosser and Mauri 2016, Schluter et al. 2020); the synthesis of lipopolysaccharides, several essential vitamins, such as biotin, folate, and vitamin K, and amino acids (Cummings and Macfarlane 1997, Fan and Pedersen 2021); the breakdown of carcinogens (Morotomi and Mutai 1986, Kho and Lal 2018), xenobiotic compounds, and drugs (Nakov and Velikova 2020); the production of molecules that inhibit potentially pathogenic bacteria (Cipe et al. 2015); and the development and functioning of the enteric nervous system (Ochoa-Repáraz and Kasper 2016, De Vadder et al. 2018, Geng et al. 2022).
The microbiota modulates multiple gut-organ axes, such as the gut-brain, gut-lung, gut-skin, gut-bone, and gut-heart axis (Enaud et al. 2020, Afzaal et al. 2022, Bulanda and Wypych 2022), and contributes to the development and functioning of the nervous, endocrine, and immune systems (Clarke et al. 2014, Obata and Pachnis 2016, Foster et al. 2021). Imbalances in the gut microbiota and its metabolites have been implicated not only in gastrointestinal pathologies, but also in extra-intestinal, systemic diseases (Guinane and Cotter 2013, Bull and Plummer 2014, Carding et al. 2015, Tang et al. 2015, Li et al. 2017). Animal and human studies have established links between perturbations in the intestinal microbiota and various medical conditions, including inflammatory bowel disease (Zheng and Wen 2021), Crohn's disease (Manichanh et al. 2006), obesity (Iatcu et al. 2021), metabolic syndrome (Iatcu et al. 2021) and metabolic diseases (Woting and Blaut 2016), type 2 diabetes (Han and Lin 2014, Gurung et al. 2020), cardiovascular disease (Novakovic et al. 2020), depression (Winter et al. 2018, Limbana et al. 2020, Liu et al. 2020), occupational sleep apnea-induced hypertension (Mashaqi and Gozal 2019), Alzheimer's disease (Kowalski and Mulak 2019, Liu et al. 2020), Parkinson's disease (Sampson et al. 2016, Kang et al. 2021, Wang et al. 2021), autism spectrum disorder (Rosenfeld 2015), and cancer (Singh et al. 2017).
Brief description of epigenetic changes
Among the mechanisms that underlie the communication between the gut microbiota and host cells, the ability of microbial metabolites to epigenetically modulate eukaryotic gene expression has received considerable attention, particularly in recent years (Sharma et al. 2019, Woo and Alenghat 2022). The term epigenetics was coined by Conrad Waddington in 1942, as a refinement to the concept of epigenetic landscape that he introduced earlier (Waddington 1940), to refer to interactions between genes and the environment that shape phenotypes during development (Waddington 1942, Deichmann 2016, Tronick and Hunter 2016). Over the past 80 years, the term itself has developed, along with advances in molecular biology (Choudhuri 2011, Felsenfeld 2014).
Epigenetic changes involve potentially heritable alterations in gene expression without changes in the DNA sequence itself and, in addition to development, are implicated in regulating stem cell potential, tissue homeostasis, the response to environmental factors, and disease pathogenesis (Johnstone and Baylin 2010, Hamilton 2011, Beerman and Rossi 2015, Allis and Jenuwein 2016, Zoghbi and Beaudet 2016, Kang et al. 2019). Three major categories of epigenetic changes were described in eukaryotes, and they include DNA methylation, histone post-translational modifications, and RNA interference (RNAi), a process in which microRNAs, a class of non-coding RNA species, inhibit messenger RNA (mRNA) translation (Stephens et al. 2013, Du et al. 2015, Fessele and Wright 2018, Zhang et al. 2019, Aure et al. 2021, Liu et al. 2022). Proteins and protein complexes that epigenetically modify DNA and histones were classified into epigenetic writers, readers, and erasers (Biswas and Rao 2018). Writers include enzymes that deposit epigenetic modifications; readers recognize and bind the covalent epigenetic modifications; and erasers remove the epigenetic marks (Torres and Fujimori 2015, Biswas and Rao 2018).
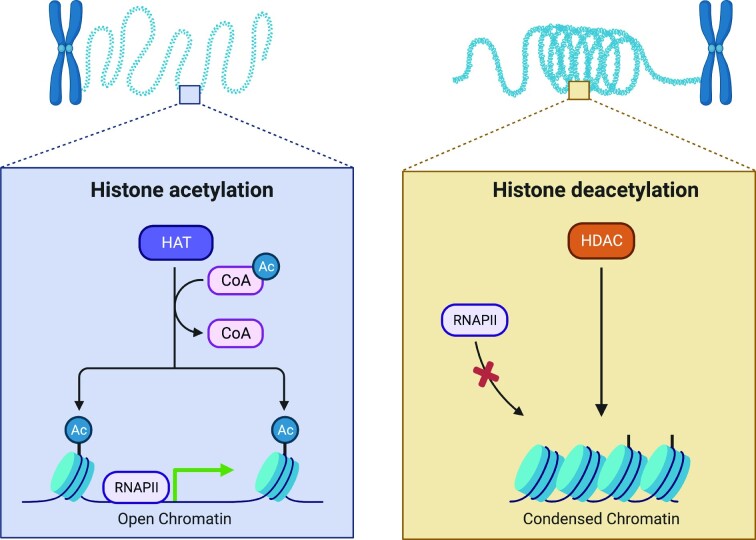
DNA methylation, mediated by DNA methyltransferases (DNMTs), involves the covalent attachment of a methyl group to the C-5 position of cytosine in the DNA, leading to the formation of 5-methylcytosine (5-mC) (Jin et al. 2011, Schmitz et al. 2019). Most instances of cytosine methylation were described when cytosine is part of the CpG dinucleotide, but non-CpG methylation, which refers to cytosine methylation at CpA, CpC, and CpT sites, was also described (He and Ecker 2015, Jang et al. 2017). In human somatic cells, >98% of the DNA methylation occurs in the CpG context, and non-CpG methylation is much more abundant in embryonic stem cells (Lister et al. 2009, Jin et al. 2011). Histone post-translational modifications involve the covalent attachment of functional groups to amino acids, mostly on the N-terminal histone tails, and include acetylation, methylation, phosphorylation, ubiquitylation, SUMOylation, glycosylation, and ADP-ribosylation (Bowman and Poirier 2015, Ramazi et al. 2020). Histone acetyltransferases (HATs), sometimes also referred to as lysine acetyltransferases (KATs), comprise a superfamily of enzymes that attach acetyl groups to the ɛ-amino group of lysine residues on both histone and non-histone proteins, and histone deacetylases (HDACs) (Fig. 1) catalyze the removal of the acetyl groups (Hodawadekar and Marmorstein 2007, Berndsen and Denu 2008, Gong and Miller 2013, Marmorstein and Zhou 2014, Wang et al. 2014). An example of acetyl-lysine readers are the bromodomain-containing proteins (Wu et al. 2019). Acetylation transforms condensed chromatin into a more relaxed structure, making it more accessible and facilitating gene expression, and deacetylation leads to chromatin compaction, which makes genes less accessible for transcription (Liu and Xu 2004, Berndsen and Denu 2008, Verza et al. 2020). Finally, microRNAs (miRNAs), a class of small non-coding RNAs, are 19–24 nucleotides long single-stranded RNA molecules that regulate gene expression by causing gene silencing or, occasionally, by translational activation (Chuang and Jones 2007, Truesdell et al. 2012, Bhaskaran and Mohan 2014, O'Brien et al. 2018). Extensive cross-talk was described among different types of epigenetic changes; for example, DNA methylation influences and is influenced by histone post-translational modifications (Miller and Grant 2013, Rose and Klose 2014, Weinberg et al. 2019), and microRNAs regulate DNMTs and HDACs (Bourassa and Ratan 2014, Lopez-Bertoni et al. 2015).
Figure 1.
Histone acetylation and deacetylation. The acetylation of histone lysine residues involves the covalent attachment of an acetyl group to the ε-position of the lysine side chain, which creates an open chromatin structure that is more permissible for gene transcription. Lysine deacetylation involves the removal of the acetyl group, leading to a more compacted chromatin structure that is less accessible for active gene expression. Created with BioRender.com.
SCFAs in the large intestine
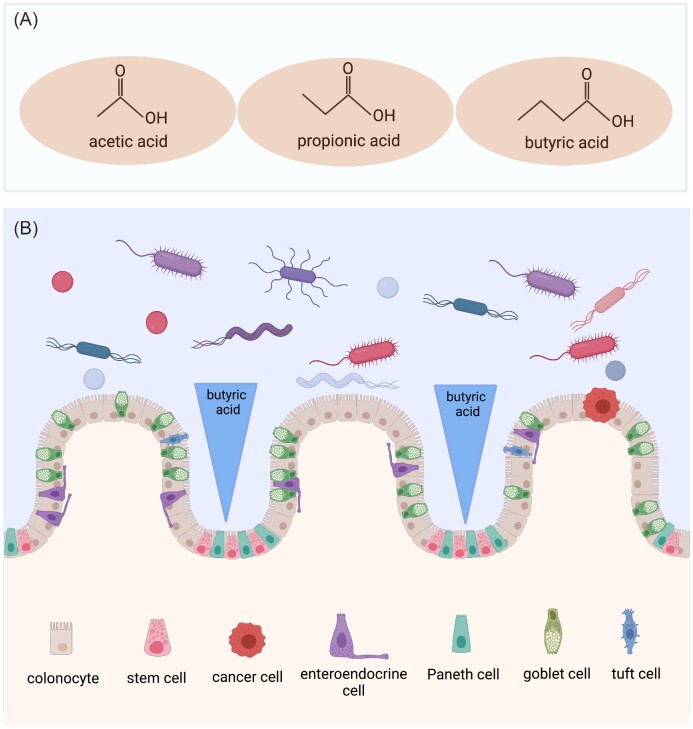
Based on their carbon chain length, fatty acids are classified as short- (<6C), medium- (6–12C), and long-chain (>12C) (Nogal et al. 2021). Short-chain fatty acids (SCFAs) are organic linear carboxylic acids that harbor an aliphatic tail of two to six carbon atoms (Tan et al. 2014, He et al. 2020) and are produced in the colon as a result of the anaerobic fermentation of dietary fiber by the gut microbiota (Parada Venegas et al. 2019). Acetic acid (C2), propionic acid (C3), and butyric acid (C4) are the most abundant SCFAs in the human large intestine (≥95%) (Fig. 2A) and have received particular attention due to their ability to modulate multiple metabolic pathways relevant to human health (Morrison and Preston 2016, Boets et al. 2017, Sun et al. 2017, Overby and Ferguson 2021, Portincasa et al. 2022). Other SCFAs, such as isovalerate and isobutyrate, are present in only trace amounts (Swer et al. 2022).
Figure 2.
Short-chain fatty acids in the large intestine have important local and systemic biological functions. (A) Acetic, propionic, and butyric acid are the three most abundant SCFAs in the large intestine, where they are synthesized by the gut microbiota from dietary fiber. (B) Schematic representation of the butyrate paradox. Butyric acid creates a colonic lumen-to-crypt decreasing concentration gradient, which promotes the growth or decreases the proliferation of healthy colonocytes, but selectively inhibits undifferentiated cells, such as cancer cells and stem cells, by mechanisms that include epigenetic changes. Created with BioRender.com.
Different intestinal microbes produce different amounts of SCFAs (Macfarlane and Macfarlane 2003, He et al. 2020). In the human large intestine, members of the Bacteroidetes synthesize mainly acetate and propionate, and members of the Firmicutes mostly produce butyrate (Parada Venegas et al. 2019). The amount and the rate of SCFA production depends on the host species, the composition of the microbiota, the fermentation substrate, and the transit time in the large intestine (Wong et al. 2006). Small amounts of SCFAs originate in the diet (Shimizu et al. 2019, He et al. 2020), can form by the fermentation of amino acids (Nogal et al. 2021), and are also synthesized in the liver (Tan et al. 2014). Their levels are very low, but measurable, in germ-free animals (Høverstad and Midtvedt 1986). SCFAs enter colonocytes through the apical membrane by passive diffusion and active transport that is mediated by H+-dependent monocarboxylate transporters (MCTs), previously referred to as solute carrier family (SLC) transporters (Fredericks et al. 2020, Deleu et al. 2021, Portincasa et al. 2022). Colonocytes derive 60%–70% of their energetical needs from oxidizing SCFAs (Roediger 1982, den Besten et al. 2013), with butyrate providing their main energy source, and SCFAs that are not used for their energetic needs are transported across their basolateral membrane into the portal vein and, through the peripheral blood, to various organs, where they can be used for metabolic processes or signaling (Sun et al. 2017, He et al. 2020, Thomas and Denu 2021).
The highest SCFA concentration in the human gastrointestinal tract is in the colon (Parada Venegas et al. 2019), where the molar ratio of acetate to propionate to butyrate is about 3:1:1 (Chambers et al. 2018, Deleu et al. 2021, Nogal et al. 2021), but this ratio changes in the peripheral veins to 91:5:4 (Cummings et al. 1987). Acetate, more abundant than butyrate and propionate, is the most abundant SCFA in the distal gut and in the systemic circulation (Qin and Wade 2018, Rahman et al. 2023). The concentration of SCFAs is about 5-times higher in the portal vein than in the peripheral venous blood, suggesting that the gut is their primary source (Cummings et al. 1987). In addition to serving as an energy source for the cells of the colon and ileum (Yao et al. 2022), SCFAs influence microbial composition (Overby and Ferguson 2021), pH (Overby and Ferguson 2021), the integrity of the intestinal barrier (Overby and Ferguson 2021), glucose and lipid metabolism (Morrison and Preston 2016, Nogal et al. 2021), appetite (Morrison and Preston 2016, Blaak et al. 2020), are involved in mucus production (Blaak et al. 2020), and regulate inflammation (He et al. 2020) and the immune response (Kim et al. 2014, Blaak et al. 2020, Yao et al. 2022).
SCFAs impact histone acetylation
Microbial-derived SCFAs are one of the best understood mediators of the microbiota-host interactions (Fellows and Varga-Weisz 2020) and, along with other metabolites, emerge as important participants to the microbiota-gut-brain axis (Swer et al. 2022). In 1978, Sealy and Chalkley reported, in rat hepatoma cells that were treated with SCFAs such as sodium butyrate or acetic, isobutyric, or propionic acid, a global increase in histone acetylation as a result of the noncompetitive and reversible inhibition of HDAC activity (Sealy and Chalkley 1978). The same year, Boffa et al. reported that in HeLa cells, sodium butyrate causes an increase in histone acetylation by inhibiting HDAC activity (Boffa et al. 1978) and Reeves and Candido found that sodium butyrate suppressed histone deacetylation in Friend erythroleukemic cells in culture (Reeves and Candido 1978).
SCFAs generated in the gut mediate at least part of their local and systemic biological effects by signaling through G protein-coupled receptors (GPCRs), including GPR41 (FFAR3), GPR43 (FFAR2), and GPR109A (HCAR2), inhibiting HDACs, and their influence on cellular energy metabolism (Canani et al. 2011, Vinolo et al. 2011, Tan et al. 2014, Dalile et al. 2019, Gasaly et al. 2021, Nogal et al. 2021). The main HDAC inhibitor is butyric acid, which inhibits mostly HDAC enzymes from classes I, IIa, and IV (Davie 2003, Licciardi et al. 2010, Fock and Parnova 2023). While the mechanisms that explain the ability of SCFAs to inhibit HDACs are incompletely understood, it was proposed that they may act directly on the HDAC or indirectly, through GPCR activation (He et al. 2020). Butyrate, by being metabolized to acetyl-CoA, also increases histone acetylation (Donohoe et al. 2012). An analysis that conducted deep profiling of histone modifications using mass spectrometry and chromatin immunoprecipitation sequencing found reduced histone H4 mono-, di-, and tri-acetylation at gene bodies across the genome of cecal and colonic epithelial cells. Isotope tracing studies using labeled fermentable fiber confirmed that the isotope becomes incorporated into acetylated H4 and H3, supporting the view that butyrate generated from fiber, under the influence of the microbiota, provides carbon sources for histone acetylation (Lund et al. 2022). Propionate and butyrate also activate the p300 acetyltransferase, which recently emerged as a previously unknown mechanism (Thomas and Denu 2021).
Butyrate, the most studied SCFA (Vinolo et al. 2011), is the most potent, and acetate is the least potent HDAC inhibitor (Vinolo et al. 2011). The ability of SCFAs to inhibit HDAC activity was shown in the gut epithelium (Li et al. 2017, Han et al. 2018, Mirzaei et al. 2021), the associated immune cells (Chang et al. 2014, Kim et al. 2016, Schulthess et al. 2019, Sanchez et al. 2020, Kibbie et al. 2021, Ney et al. 2023), skeletal (Gao et al. 2009) and vascular smooth muscle cells (Mathew et al. 2010, Mathew et al. 2019, Zhong et al. 2022), endothelial cells (Li et al. 2018), and cardiomyocytes (Zhang et al. 2019, Umei et al. 2020), and was documented at several sites in the body, including the central nervous system (Ziemka-Nalecz et al. 2017, Reddy et al. 2018, Jaworska et al. 2019, Silva et al. 2020), the kidneys (Liu et al. 2021), and the lungs (Folkerts et al. 2020, Karoor et al. 2021, Yip et al. 2021). In addition to being an HDAC inhibitor, butyrate was linked to DNA methylation changes in eukaryotic cells (de Haan et al. 1986, Wippermann et al. 2017, Wang et al. 2022, Xie et al. 2022) and was shown to signal through microRNAs. For example, butyrate decreased the expression of several microRNA clusters in human colon cancer cells (Hu et al. 2011, Hu et al. 2015), and in mouse and human B cells, butyrate and propionate upregulated microRNAs that targeted the 3’-untranslated region of Aicda/AICDA and Prdm1/PRDM1 mRNAs and dose-dependently inhibited their translation as a result of their HDAC inhibitory activity (Sanchez et al. 2020).
The butyrate paradox
The butyric acid that is generated in the colon by microbial fermentation establishes two concentration gradients (Donohoe et al. 2012, van Deuren et al. 2022, van Deuren et al. 2022). A proximal-to-distal concentration gradient occurs because most butyrate is produced in the proximal colon, and the butyrate that is not used by colonocytes for their metabolic requirements travels distally as a result of peristaltic movements (Donohoe et al. 2012, Liu et al. 2018). A second gradient is generated along the lumen-to-crypt axis (Fig. 2B), with larger butyrate concentrations in the colonic lumen and lower concentrations at the base of the crypts of Lieberkühn (Bultman 2016). Butyrate concentrations are about 5 mM in the colonic lumen of mice (Louis and Flint 2007, Kaiko et al. 2016, Vemula and Jala 2016, Linder and Mostoslavsky 2017) and 10–70 mM in the colonic lumen of humans (Cummings et al. 1987, Vemula and Jala 2016, Ota and Sakuraba 2022), but were estimated to be 50–800 μM in the mouse colonic crypts (Donohoe et al. 2012).
The effects of butyrate on the colonic epithelium are complex and depend on its concentration and on the state of cellular differentiation. While butyrate promotes the growth of healthy colonocytes, or decreases their proliferation, depending on its concentration (Lupton 2004, Comalada et al. 2006, Canani et al. 2011, Donohoe et al. 2012, Li et al. 2018, Hajjar et al. 2021), it selectively inhibits undifferentiated cells, such as cancer cells (Barnard and Warwick 1993, Archer et al. 1998, Gonçalves and Martel 2013) and stem cells (Kaiko et al. 2016, Singh et al. 2016). In various studies, butyrate inhibited the proliferation of colorectal cancer cells and cell lines in a dose- and time-dependent manner, caused cell cycle arrest mostly in the G1 phase, increased differentiation, and induced apoptosis (Siavoshian et al. 1997, Orchel et al. 2005, Hong et al. 2015, Ryu et al. 2018, Chen et al. 2019, Klepinina et al. 2021, Salvi and Cowles 2021, Xi et al. 2021). This poorly understood duality, which has become known as the butyrate paradox (Donohoe et al. 2012, Gasaly et al. 2021, Salvi and Cowles 2021), is explained by the fact that healthy colonocytes primarily break down butyrate by β-oxidation and use it as a source of energy, but undifferentiated colonocytes, due to the Warburg effect, preferentially use glucose over butyrate for their energetic needs and, as a result, butyrate accumulates and epigenetically induces gene expression changes (Donohoe et al. 2012, Han et al. 2018, Jung et al. 2021, Salvi and Cowles 2021).
In a study that unveiled fundamental differences in energy metabolism between healthy and malignant colonocytes, Donohoe et al. proposed a model to explain the ability of the lumen-to-crypt butyrate gradient to shape gene expression in the colon epithelium in a dose-dependent and cell type-specific manner. At the low concentrations of butyrate (∼0.5 mM in mice) that exist near the base of the colonic crypts, most of the butyrate is metabolized and contributes to histone acetylation by a mechanism that involves the formation of acetyl-CoA and histone acetyl transferases, and supports the proliferation of mitotically active colonocytes. Transcriptome profiling revealed that target genes upregulated by this mechanism are enriched for functions related to cell proliferation. Healthy colonocytes preferentially use butyrate as an energy source and metabolize it in the mitochondria by β-oxidation. The higher butyrate concentrations (∼5 mM in mice) that exist near the lumen exceed the concentration at which butyrate can be efficiently metabolized, which is approximately 1–2 mM. As a result, butyrate accumulates in the nucleus of colonocytes, acts as an HDAC inhibitor, and decreases cellular proliferation. Transcriptome profiling showed that the targets of this mechanism are enriched for apoptotic genes, supporting the ability of the higher butyrate levels to promote colonocyte apoptosis and exfoliation. In cancerous colonocytes, which due to the Warburg effect rely on glycolysis but metabolize relatively little butyrate, the HDAC inhibition mechanism predominates and leads to decreased cell proliferation and increased apoptosis (Donohoe et al. 2012).
In studies that sought to further interrogate the cellular and molecular intricacies of the butyrate paradox, Kaiko et al. showed that in zebrafish, which do not have colonic crypts (Chen et al. 2012, Flores et al. 2020, Tavakoli et al. 2022) but their intestinal stem cells are exposed to the intestinal lumen, and also lack intestinal bacteria that synthesize butyrate, a marked inhibition of intestinal proliferation occurs when the mucosa is exposed to butyrate (Kaiko et al. 2016). This suggested that the placement of intestinal stem cells and progenitor cells in colonic crypts (Fig. 2B) protects them from the growth inhibitory effects of butyrate, which could otherwise suppress their division. The metabolization of butyrate into acetyl-CoA in the neighboring colonocytes protects stem cells in the crypts from the effects of butyrate (Kaiko et al. 2016), a phenomenon that was relevantly referred to as a metabolic sinkhole (Singh et al. 2016). Due to the ability to protect stem cells from the anti-proliferative effects of butyrate (Salvi and Cowles 2021), the colonic crypts emerge as their natural gatekeepers (Vemula and Jala 2016). In intestinal stem cells and progenitor cells, butyrate increased H3K27 and H3K9 acetylation in a manner dependent, at least in part, on the Forkhead box O3 (Foxo3) transcription factor, which bound the promoters of Cdkn1a, Cdkn1c, and Gadd45b, negative cell cycle regulators that are involved in cell cycle arrest, suppressing cellular proliferation (Kaiko et al. 2016). In a human colorectal cancer cell line, butyrate activated pyruvate kinase M2, the pyruvate kinase isoform expressed in cancer cells and stem cells, and induced a metabolic phenotype that inhibited cellular proliferation (Li et al. 2018).
Relative butyrate levels were shown to increase about 4-fold in infants between 6 and 12 months of life (Nilsen et al. 2020). Based on the fact that butyrate levels in the intestine increase as the number of stem cells and the intestinal crypt length increase early during postnatal life (Midtvedt and Midtvedt 1992), Gasaly et al. hypothesized that colonization with butyrate-producing intestinal microorganisms occurs later during infancy so that butyrate would not interfere with stem cell function and epithelial remodeling during the early postnatal life (Gasaly et al. 2021).
Epigenetic effects of SCFAs on cells of the immune system
Accumulating evidence indicates that metabolites derived from the gut microbiota, including SCFAs, modulate gene expression in cells of the innate and adaptive immune system, and some of these effects involve epigenetic mechanisms. In vitro, sodium butyrate inhibited the proliferation of mouse malignant mast cells by inducing cell cycle arrest in G1, increased caspase 3-dependent apoptosis, and increased H3K9 acetylation at the IL-6 and TNF-α promoters, decreasing cytokine production by acting as an HDAC inhibitor (Zhang et al. 2016). Butyrate and propionate blocked the formation of dendritic cells from bone marrow stem cells, and this was dependent on their HDAC inhibitory activity (Singh et al. 2010). Propionate and butyrate decreased the production of TNF-α, CINC-2αβ (cytokine-induced neutrophil chemoattractant-2) and nitric oxide (NO) by lipopolysaccharide-stimulated neutrophils and they also inhibited HDAC and the activation of NF-κB, and in a rat model, the administration of tributyrin, a pro-drug of butyrate, decreased the migration of neutrophils to the peritoneum after the intraperitoneal administration of glycogen (Vinolo et al. 2011).
Butyrate increased the antimicrobial activity of monocytes that were differentiating into macrophages against several Gram-negative and Gram-positive pathogens through a mechanism that involved HDAC3 inhibition. Macrophages differentiated in the presence of butyrate showed a significant decrease in H3K27 tri-methylation, which is associated with chromatin repression, and an increase in H3K27 acetylation, which is associated with more open chromatin. These effects were relevant in vivo, as shown by the higher antimicrobial activity of colonic macrophages, and a lower dissemination of pathogens from the large intestine to peripheral organs in mice who received butyrate in the drinking water, as compared to control animals (Schulthess et al. 2019).
In vitro, butyrate decreased the production of proinflammatory molecules such as NO, IL-6, and IL-12 in bone marrow-derived macrophages and in macrophages from the lamina propria of the colon, and showed similar effects on colon lamina propria macrophages when orally administered to mice, and these effects occurred through the inhibition of HDACs (Chang et al. 2014). Butyrate promoted the production of the anti-inflammatory cytokine IL-22 in vitro in CD4+ T cells and innate lymphoid cells, and in vivo in mice who received it orally in drinking water, by G-protein receptor 41 (GPR41)-mediated signaling and HDAC inhibition. This occurred by its ability to increase the binding of hypoxia-inducible factor 1α (HIF1α) to the hypoxia response element (HRE) of the Il22 promoter through histone modification. As part of this effect, butyrate increased H3K9 acetylation and suppressed H3K9 trimethylation at the HRE site of the Il22 promoter (Yang et al. 2020). In another study, butyrate modulated the function of Th17 lymphocytes depending on their state of differentiation: in naïve CD4+ T cells undergoing differentiation to Th17 cells, it downregulated the RORγt transcription factor and decreased IL-17 production, but in already differentiated Th17 cells it induced RORγt expression and IL-17 secretion, an effect that was mediated by histone H4 acetylation near the RORγT proximal promoter (Sałkowska et al. 2017). Butyrate, and to a lesser extent acetate and propionate, decreased the human gut lamina propria CD4+ T cell activation and proliferation in vitro by increasing H3K9 acetylation and lowered the production of inflammatory cytokines such as IL-17 and IFNγ (Kibbie et al. 2021). Butyrate increased the expression of Treg-associated FoxP3 in a concentration-dependent manner and enhanced the differentiation of T cells into regulatory T (Treg) cells over inflammatory T helper (Th) cells, including Th17 cells (McBride et al. 2022). Mice that were fed butyrylated high-amylose maize starch, which was formed by the treatment of starch with butyric anhydride, showed an increased differentiation of Treg cells from the colon lamina propria and in the number of IL-10 producing Treg cells as compared to animals fed a control diet. At least one of the mechanisms explaining this effect involved the epigenetic upregulation of the Foxp3 gene, as shown by an increased histone H3 acetylation in the promoter and conserved noncoding sequences of the gene (Furusawa et al. 2013).
Intestinal SCFAs epigenetically change gene expression at extraintestinal sites
Animal and human studies implicated gut dysbiosis in the development of neuropsychiatric conditions (Kowalski and Mulak 2019, Li et al. 2021, Romano et al. 2021, Chen et al. 2022), and manipulating the gut microbiota decreased neuroinflammation and/or improved certain cognitive or pathological features (Sampson et al. 2016, Bonfili et al. 2021, Qian et al. 2022, Wang et al. 2022), implicating the gut microbiota in the regulation of the gut-brain axis. As part of this connection, microbial metabolites such as SCFAs have received increasing attention (Silva et al. 2020, Begum et al. 2022, O'Riordan et al. 2022).
In 1973, Oldendorf reported that 14C-SCFAs injected in the carotid artery of rats cross the blood-brain barrier (BBB), and the relative order of crossing was butyrate (highest), propionate, and acetate (Oldendorf 1973). This finding, and the bioactive properties of SCFAs in the brain, were subsequently confirmed by other studies, and while initially it was believed that SCFA levels are much lower in the brain than in the plasma, the brain levels of butyrate and propionate are higher than originally thought (Wishart et al. 2018, Dalile et al. 2019, Silva et al. 2020, Wenzel et al. 2020, Colombo et al. 2021, Fillier et al. 2022, Fock and Parnova 2023). SCFAs emerge as a potential link between the gut microbiota and the pathology of several neurodegenerative diseases. In human neuroblastoma-derived and rat mesencephalon-derived cell lines, sodium butyrate partially prevented the apoptotic cell death caused by the mitochondrial toxin 1-methyl-4-phenylpyridinium, and this was accompanied by a significant increase in histone H3 acetylation (Kidd and Schneider 2010). In another study, sodium butyrate increased brain-derived neurotrophic factor (BDNF) and glial cell line-derived neurotrophic factor (GDNF) transcription in primary cortical astrocyte cultures, along with an increase in histone H3 acetylation at the GDNF promoter (Wu et al. 2008). Butyrate inhibited the aggregation of β-amyloid 1–40 and 1–42 monomers into fibrils in vitro by interfering with protein-protein interactions (Ho et al. 2018), and showed benefits in several mouse models of Alzheimer's disease (Cao et al. 2018). In a mouse model of early Alzheimer's disease, the oral administration of sodium butyrate decreased β-amyloid levels and improved cognitive memory (Fernando et al. 2020), and in a presenilin-1 and presenilin-2 conditional double knockout mouse model it significantly increased neurogenesis in the subgranular zone of the dentate gyrus, restored contextual memory, and reversed the dysregulated histone acetylation in the hippocampus and the cortex (Cao et al. 2018). A cross-sectional study of elderly individuals found that brain amyloid deposition and endothelial dysfunction were positively correlated with blood acetate levels and negatively correlated with butyrate levels (Marizzoni et al. 2020). In a mouse model of Parkinson's disease, butyrate prevented the DNA damage caused by α-synuclein, possibly by upregulating DNA repair genes, and in cell lines it rescued the decrease in histone H3 acetylation that was mediated by α-synuclein (Paiva et al. 2017). In a rat model of 6-hydroxydopamine-inducecd experimental Parkinson's disease, intraperitoneal sodium butyrate attenuated the motor deficits, increased dopamine levels in the striatum, and lowered oxidative stress, along with increasing striatal global histone H3 acetylation levels (Sharma et al. 2015).
The microbiota additionally regulates the gut-brain axis through several metabolites that preserve the integrity of the BBB, including SCFAs, by mechanisms that include epigenetic changes (Fock and Parnova 2023). Germ-free mice have an increased BBB permeability starting with the intrauterine life and continuing into adulthood, as compared to pathogen-free mice harboring a normal gut microbiota, significantly lower levels of occludin and claudin-5 in the tight junctions from the frontal cortex, striatum, and hippocampus, and fewer intact tight junctions in the striatum. The colonization of germ-free mice with pathogen-free gut microbiota, or the administration of bacteria making SCFAs, such as Clostridium tyrobutyricum, which produces mainly butyrate, or Bacteroides thetaiotaomicron, which produces mostly acetate and propionate, decreased the permeability of the BBB, an effect that was also achieved by the administration of sodium butyrate by oral gavage. Sodium butyrate increased the expression of occludin in the frontal cortex and the hippocampus, and monocolonization of germ-free mice with C. tyrobutyricum, or the administration of sodium butyrate, increased histone H4 acetylation in extracts from the frontal cortex (Braniste et al. 2014).
In vitro, SCFAs downregulated the production of pro-inflammatory molecules by downregulating nuclear factor-κB (NF-κB) (Liu et al. 2012), and in a rat model of transient focal cerebral ischemia, valproic acid, an HDAC inhibitor, reduced the degradation of tight junction proteins and the nuclear translocation of NF-κB, effects that were mimicked by sodium butyrate (Wang et al. 2011). In a rat model of middle cerebral artery occlusion, the administration of a selective HDAC3 inhibitor early after the occlusion decreased cerebral edema and BBB leakage, and these effects were at least in part mediated by upregulating tight junction proteins and decreasing NF-kB-mediated inflammation (Lu et al. 2023). In a mouse model of type 2 diabetes, which showed increased hippocampal and cortical Hdac3 levels and activity, inhibition of Hdac3 significantly upregulated several tight junction and adherens junction proteins and improved BBB permeability. The miR-200a/Keap1/Nrf2 was required for this effect, pointing towards the possibility that the epigenetic modification of Nrf2 could explain the protection that SCFAs confer to the integrity of the BBB (Zhao et al. 2019).
Several studies support the benefits of butyrate in ischemic stroke. The first study to show that sodium butyrate causes epigenetically mediated selective gene expression changes in microglia in ischemic stroke was a mouse model of cerebral artery occlusion, which found that intraperitoneally administered sodium butyrate downregulated pro-inflammatory mediators, such as TNF-α and STAT1, and upregulated the anti-inflammatory mediator IL-10, due to its ability to modulate H3K9 acetylation levels (Patnala et al. 2017). In a rat model of ischemic stroke, sodium butyrate reduced the size of the injury and suppressed neurological deficits, likely by several mechanisms, including suppression of inflammation, and the benefits appeared to be explained by the HDAC inhibitor-induced apoptosis of microglia and monocytes/macrophages, which decreased neuroinflammation (Kim et al. 2007). In another study that investigated a rat model of permanent brain ischemia, the subcutaneous injection of butyrate stimulated neurogenesis in the subventricular zone and hippocampal dentate gyrus, two neurogenic regions of the brain, and this was correlated with increased acetylated histone H3 levels in these regions, and upregulated the levels of BDNF, phospho-CREB, and GFAP in several regions of the brain (Kim et al. 2009).
SCFAs can induce epigenetic changes in various immune cells from multiple body compartments (Yip et al. 2021). Germ-free mice exhibited structural and functional defects in microglia, and some of the genes that were dysregulated, such as Hdac1, Sirt2, and Mll3, encode well-known epigenetic regulators. These phenotypes were mimicked by the deficiency of the FFAR2 receptor for SCFAs and were reversed by SCFA supplementation (Erny et al. 2015). Mice fed with inulin had increased levels of all three SCFAs in the cecum and increased levels of butyric and propionic acid in the hepatic portal vein, and in ex vivo experiments, their microglia secreted significantly less tumor necrosis factor α (TNF-α) in the presence of lipopolysaccharide as compared to mice fed a control diet. This suggested that part of the SCFAs generated in the large intestine may regulate microglial activation. In vitro, butyrate and acetate inhibited the inflammatory response of microglia stimulated with lipopolysaccharide, most likely through an epigenetic mechanism that involved inhibition of HDAC and NF-κB activity (Caetano-Silva et al. 2023).
Propionate and butyrate affected the adhesion of human eosinophils to endothelial cells in vitro, impaired their viability, and activated apoptosis, and butyrate impaired their ability to migrate, effects that were accompanied by increased histone H3 acetylation (Theiler et al. 2019).When administered in the drinking water or intranasally to mice, butyrate regulated the function of type 2 innate lymphoid cells in the lungs and attenuated airway hyperreactivity and inflammation, effects that occurred through HDAC inhibition and were accompanied by increased histone H3 lysine 9 and 14 acetylation (Thio et al. 2018). A high fiber diet or acetate administered to mice protected against allergic airway disease in a manner that required HDAC9 inhibition and was dependent on Treg cells, and maternal high fiber diet or acetate also conferred protection to the offspring when allergic airway disease was induced. These effects were mediated in utero and were not dependent on the transfer of microbiota to the fetus. In both adult mice and the offspring, there was an increased acetylation of histone H4 and histone H3K9 at the Foxp3 promoter, an increased Foxp3 expression in the lungs, and an increase in the number and function of Treg cells in the lungs (Thorburn et al. 2015).
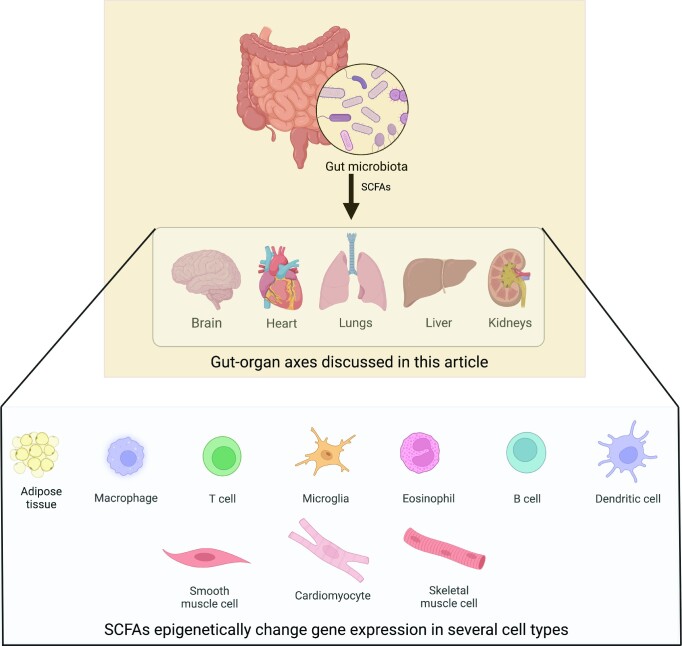
SCFAs epigenetically regulate multiple gut-organ axes and various cell types in the body (Fig. 3). A study on mice found that the gut microbiota regulates global histone acetylation and methylation in several host tissues, including colon, liver, and white adipose tissue, in a manner that is shaped by diet, and the oral administration of SCFAs to germ-free animals recapitulated some of the chromatin modification states associated with gut microbiota colonization (Krautkramer et al. 2016). In mouse adipocytes, butyrate and propionate stimulated lipolysis in vitro, an effect that was mimicked by the HDAC inhibitor trichostatin A (Rumberger et al. 2014), and in rats with type 2 diabetes mellitus fed a high-fat diet and administered streptozotocin, intraperitoneal sodium butyrate significantly reduced plasma glucose, insulin resistance, and liver steatosis, effects that occurred through HDAC inhibition and were accompanied by histone H3 hyperacetylation in liver tissue (Khan and Jena 2016). In another study, the oral administration of sodium butyrate to mice fed a high fat diet alleviated obesity, improved glucose tolerance, restored plasma insulin and leptin levels, and significantly reduced lipid deposition in skeletal muscle. These effects appeared to mainly occur through HDAC inhibition, and H3K9 acetylation was increased at the promoters of Adipor1 and Adipor2, which encode adiponectin receptors, and Ucp2 (uncoupling protein 2) and Ucp3 (uncoupling protein 3), which are involved in mitochondrial thermogenesis and β-oxidation (Hong et al. 2016).
Figure 3.
Short-chain fatty acids and some of their target organs and cells. SCFAs generated in the large intestine influence several gut-organ axes and epigenetically modulate gene expression in multiple cell types in the body. Created with BioRender.com.
In rat neonatal cardiomyocytes, butyrate increased histone H3 acetylation as a result of HDAC inhibition and suppressed endothelin-1-induced cardiomyocyte hypertrophy (Umei et al. 2020). Sodium butyrate attenuated angiotensin II-induced cardiac hypertrophy in rats and, in vitro, this was shown to require the inhibition of the COX2/PGE2 pathway in an HDAC5- and HDAC6-dependent manner (Zhang et al. 2019). Butyrate lowered LDL cholesterol levels in HepG2 cells through HDAC inhibition (Bridgeman et al. 2022), a significant finding considering that the liver can extract butyrate that is absorbed from the large intestine (Guilloteau et al. 2010, Blaak et al. 2020). In an observational study of 92 consecutive patients, the levels of propionate and butyrate in blood and fecal samples negatively correlated with the vascular calcification score of the aorta, and sodium propionate administered orally or rectally to rats ameliorated calcium deposition in the ascending aorta and decreased plasma levels of inflammatory cytokines (Yan et al. 2022). This is a relevant finding, considering that butyrate conferred atheroprotective functions in vitro by inhibiting the proliferation of vascular smooth muscle cells as a result of epigenetic changes in histone and non-histone proteins and through additional mechanisms (Mathew et al. 2010, Cantoni et al. 2013, Aguilar et al. 2014). In a genome-wide association meta-analysis, several single nucleotide polymorphisms at the HDAC9 locus were associated with the atherosclerotic calcification of the abdominal aorta, an association that is intriguing, considering that mice deficient in Hdac9 exhibited reduced aortic calcification and improved survival, and in human aortic smooth muscle cells, HDAC9 overexpression increased mRNA levels of RUNX2, a master regulator of the osteogenic phenotype, and led to calcification and reduced contractility (Malhotra et al. 2019).
In the first study to show that dietary modification of the gut microbiota can prevent experimental acute kidney injury, the administration of a high fiber diet to mice with folic acid nephropathy, or supplementation with acetate, butyrate, or propionate in the drinking water, decreased the expression of several pro-inflammatory cytokines and chemokines and protected against the development of acute and chronic kidney injury. In this study, the high fiber diet or the administration of acetate or propionate in the drinking water significantly downregulated the kidney tissue levels of HDAC4, which has immunomodulatory functions, and HDAC10, which contributes to DNA repair, autophagy, and cancer progression, as compared to controls (Liu et al.2021).
Crotonylation and HDAC-dependent gene expression regulation
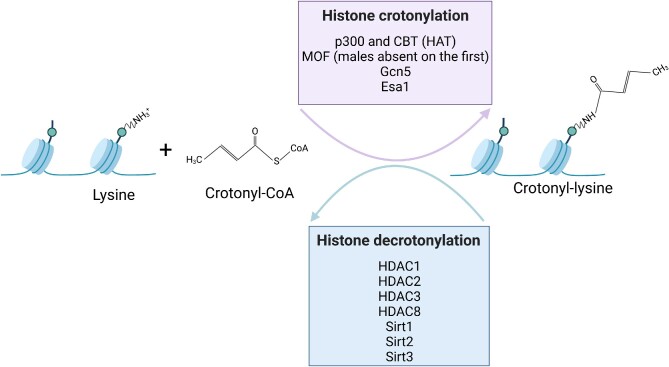
Besides acetylation, crotonylation has emerged as a new epigenetic change involved in the communication between the gut microbiota and the host (Tan et al. 2011, Fellows and Varga-Weisz 2020). Crotonylation is an evolutionarily conserved histone post-translational modification, present from yeast to humans, which involves the reversible covalent attachment of a crotonyl group to the ε-amino group of the lysine side chain (Liu et al. 2018, Wang et al. 2021), and was recently also described on serine residues (Liao et al. 2020). Crotonylation was described on histone H1 and all the core histone proteins (Tan et al. 2011) as well as on non-histone proteins (Xu et al. 2017), and is involved in many cellular functions (Wei et al. 2017). X-ray crystallography showed that crotonylation on histone H3 weakens the hydrogen bonds between the histones and DNA and decreases their interaction, opening the chromatin structure (Suzuki et al. 2016). HATs have histone crotonyltransferase (HCT) activities, and HDAC1, HDAC2, and HDAC3 are the major histone decrotonylases (Sabari et al. 2015, Wei et al. 2017, Fellows et al. 2018), but no crotonyl-specific writers have yet been identified (Jiang et al. 2021) (Fig. 4). In cell-free assays, histone crotonylation catalyzed by the HAT p300 stimulated gene transcription to a greater degree than the one catalyzed by the p300 histone acetylation (Sabari et al. 2015). A study of histone crotonylation in several mouse tissues found that the colon and the brain had the highest levels of H3K18 crotonylation, and while several lysine residues were crotonylated in the small intestine, the H3K18 crotonylation mark was the most abundant one. In the colon epithelium, H3K18 crotonylation was associated with transcription start sites. Antibiotic treatment of the mice led to a decrease in the SCFAs in the colon and serum, and a decrease in histone H3K18 and H4K4 crotonylation in the colon. Butyrate promoted histone crotonylation in human colon carcinoma cells and in gut organoids (Fellows et al. 2018). Histone crotonylation is linked to cell cycle progression (Fellows et al. 2018), it is required for various processes, including spermatogenesis (Liu et al. 2017), the renewal of mouse embryonic stem cells (Wei et al. 2017), and the regulation of telomeres (Fu et al. 2018), and was shown to be dysregulated in several cancer types (Wan et al. 2019). In a rat model of neonatal hypoxic-ischemic brain damage, sodium butyrate decreased the damage in the cerebral cortex and the hippocampus, ameliorated behavioral function, and improved H3K9 crotonylation at several neurotrophic genes where it was downregulated during the hypoxic-ischemic encephalopathy, such as Bdnf, Manf, Ogdh, and Cdnf (He et al. 2022).
Figure 4.
Histone lysine crotonylation, an evolutionarily conserved post-translational epigenetic modification, involves the reversible covalent attachment of a crotonyl group to the ε-amino group of the lysine side chain, and histone lysine decrotonylation involves the removal of the crotonyl group. Several histone acetyltransferases also have histone crotonyltransferase activity, and several histone deacetylases have histone decrotonylation activities as well. Based on information from references (Madsen and Olsen 2012, Bao et al. 2014, Sabari et al. 2015, Liu et al. 2017, Wei et al. 2017, Xu et al. 2017, Fellows et al. 2018, Kollenstart et al. 2019). Created with BioRender.com.
Conclusions and Perspectives
During the past several decades, an increasing number of studies have underscored the importance of the gut microbiota in shaping host physiology, and connected intestinal dysbiosis with various diseases that affect organs within and beyond the gastrointestinal tract. Consequently, significant attention was dedicated to identifying the microbial metabolites that orchestrate these connections. SCFAs are emerging as key molecules that explain the ability of the gut microbiota to shape the function of multiple types of eukaryotic cells from different organs and body sites. In addition to their effects on the cells lining the gastrointestinal tract, where their effects depend on their concentration and the cellular state of differentiation, SCFAs modulate gene expression in cells of the innate and adaptive immune system, adipocytes, and skeletal, cardiac and vascular smooth muscle cells, and shape the function of organs that include the brain, the lungs, and the kidneys. The mechanisms involved in these effects include epigenetic changes, and the ability of SCFAs to increase histone acetylation, and to act as histone deacetylase inhibitors, is emerging as an important topic that will support efforts to better understand physiological processes, interrogate disease pathogenesis, and develop novel biomarkers and therapeutic approaches.
Contributor Information
Richard A Stein, Department of Chemical and Biomolecular Engineering, NYU Tandon School of Engineering, 6 MetroTech Center, Brooklyn, NY 11201, USA.
Leise Riber, Department of Plant & Environmental Sciences, University of Copenhagen, Thorvaldsensvej 40, DK-1871 Frederiksberg, Denmark.
Conflict of interest
None declared.
References
- Afzaal M, Saeed F, Shah YAet al. Human gut microbiota in health and disease: unveiling the relationship. Front Microbiol. 2022;13:999001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar EC, Leonel AJ, Teixeira LGet al. Butyrate impairs atherogenesis by reducing plaque inflammation and vulnerability and decreasing nfκb activation. Nutr Metab Cardiovasc Dis. 2014;24:606–13. [DOI] [PubMed] [Google Scholar]
- Agus A, Clément K, Sokol H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut. 2021;70:1174–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. [DOI] [PubMed] [Google Scholar]
- Amato KR, Arrieta MC, Azad MBet al. The human gut microbiome and health inequities. Proc Natl Acad Sci USA. 2021;118: e2017947118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer S, Meng S, Wu Jet al. Butyrate inhibits colon carcinoma cell growth through two distinct pathways. Surgery. 1998;124:248–53. [PubMed] [Google Scholar]
- Aure MR, Fleischer T, Bjørklund Set al. Crosstalk between microRNA expression and DNA methylation drives the hormone-dependent phenotype of breast cancer. Genome Med. 2021;13:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Wang Y, Li Xet al. Identification of ‘erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. Elife. 2014;3:e02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard JA, Warwick G. Butyrate rapidly induces growth inhibition and differentiation in HT-29 cells. Cell Growth Differ. 1993;4:495–501. [PubMed] [Google Scholar]
- Beerman I, Rossi DJ. Epigenetic control of stem cell potential during homeostasis, aging, and disease. Cell Stem Cell. 2015;16:613–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum N, Mandhare A, Tryphena KPet al. Epigenetics in depression and gut-brain axis: a molecular crosstalk. Front Aging Neurosci. 2022;14:1048333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen CE, Denu JM. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr Opin Struct Biol. 2008;18:682–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezek K, Petelin A, Pražnikar Jet al. Obesity measures and dietary parameters as predictors of gut microbiota phyla in healthy individuals. Nutrients. 2020;12: 2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskaran M, Mohan M. MicroRNAs: history, biogenesis, and their evolving role in animal development and disease. Vet Pathol. 2014;51:759–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Rao CM. Epigenetic tools (The Writers, The Readers and The Erasers) and their implications in cancer therapy. Eur J Pharmacol. 2018;837:8–24. [DOI] [PubMed] [Google Scholar]
- Blaak EE, Canfora EE, Theis Set al. Short chain fatty acids in human gut and metabolic health. Benef Microbes. 2020;11:411–55. [DOI] [PubMed] [Google Scholar]
- Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352:544–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boets E, Gomand SV, Deroover Let al. Systemic availability and metabolism of colonic-derived short-chain fatty acids in healthy subjects: a stable isotope study. J Physiol. 2017;595:541–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffa LC, Vidali G, Mann RSet al. Suppression of histone deacetylation in vivo and in vitro by sodium butyrate. J Biol Chem. 1978;253:3364–6. [PubMed] [Google Scholar]
- Bonfili L, Cecarini V, Gogoi Oet al. Microbiota modulation as preventative and therapeutic approach in Alzheimer's disease. FEBS J. 2021;288:2836–55. [DOI] [PubMed] [Google Scholar]
- Bourassa MW, Ratan RR. The interplay between microRNAs and histone deacetylases in neurological diseases. Neurochem Int. 2014;77:33–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GD, Poirier MG. Post-translational modifications of histones that influence nucleosome dynamics. Chem Rev. 2015;115:2274–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braniste V, Al-Asmakh M, Kowal Cet al. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med. 2014;6:263ra158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgeman S, Woo HC, Newsholme Pet al. Butyrate lowers cellular cholesterol through HDAC inhibition and impaired SREBP-2 signalling. Int J Mol Sci. 2022;23:15506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulanda E, Wypych TP. Bypassing the gut-lung axis via microbial metabolites: implications for chronic Respiratory diseases. Front Microbiol. 2022;13:857418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull MJ, Plummer NT. Part 1: the Human gut microbiome in health and disease. Integr Med (Encinitas). 2014;13:17–22. [PMC free article] [PubMed] [Google Scholar]
- Bultman SJ. Butyrate consumption of differentiated colonocytes in the upper crypt promotes homeostatic proliferation of stem and progenitor cells near the crypt base. Transl Cancer Res. 2016;5:S526–s8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Silva ME, Rund L, Hutchinson NTet al. Inhibition of inflammatory microglia by dietary fiber and short-chain fatty acids. Sci Rep. 2023;13:2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canani RB, Costanzo MD, Leone Let al. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J Gastroenterol. 2011;17:1519–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantoni S, Galletti M, Zambelli Fet al. Sodium butyrate inhibits platelet-derived growth factor-induced proliferation and migration in pulmonary artery smooth muscle cells through Akt inhibition. Febs j. 2013;280:2042–55. [DOI] [PubMed] [Google Scholar]
- Cao T, Zhou X, Zheng Xet al. Histone deacetylase inhibitor alleviates the neurodegenerative phenotypes and Histone dysregulation in presenilins-deficient mice. Front Aging Neurosci. 2018;10:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carding S, Verbeke K, Vipond DTet al. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis. 2015;26:26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers ES, Preston T, Frost Get al. Role of gut microbiota-generated short-chain fatty acids in metabolic and cardiovascular health. Curr Nutr Rep. 2018;7:198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PV, Hao L, Offermanns Set al. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA. 2014;111:2247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Liao J, Xia Yet al. Gut microbiota regulate Alzheimer's disease pathologies and cognitive disorders via PUFA-associated neuroinflammation. Gut. 2022;71:2233–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhao KN, Vitetta L. Effects of intestinal microbial⁻elaborated butyrate on oncogenic signaling pathways. Nutrients. 2019;11: 1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Lu YF, Li ICet al. Zebrafish Agr2 is required for terminal differentiation of intestinal goblet cells. PLoS One. 2012;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri S. From Waddington's epigenetic landscape to small noncoding RNA: some important milestones in the history of epigenetics research. Toxicol Mech Methods. 2011;21:252–74. [DOI] [PubMed] [Google Scholar]
- Christian N, Whitaker BK, Clay K. Microbiomes: unifying animal and plant systems through the lens of community ecology theory. Front Microbiol. 2015;6:869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007;61:24r–9r. [DOI] [PubMed] [Google Scholar]
- Cipe G, Idiz UO, Firat Det al. Relationship between intestinal microbiota and colorectal cancer. World J Gastrointest Oncol. 2015;7:233–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke G, Stilling RM, Kennedy PJet al. Minireview: gut microbiota: the neglected endocrine organ. Mol Endocrinol. 2014;28:1221–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo AV, Sadler RK, Llovera Get al. Microbiota-derived short chain fatty acids modulate microglia and promote aβ plaque deposition. Elife. 2021;10: e59826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comalada M, Bailón E, de Haro Oet al. The effects of short-chain fatty acids on colon epithelial proliferation and survival depend on the cellular phenotype. J Cancer Res Clin Oncol. 2006;132:487–97. [DOI] [PubMed] [Google Scholar]
- Cummings JH, Macfarlane GT. Role of intestinal bacteria in nutrient metabolism. Clin Nutr. 1997;16:3–11. [DOI] [PubMed] [Google Scholar]
- Cummings JH, Pomare EW, Branch WJet al. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalile B, Van Oudenhove L, Vervliet Bet al. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat Rev Gastroenterol Hepatol. 2019;16:461–78. [DOI] [PubMed] [Google Scholar]
- Davie JR Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133:2485S–93.S. [DOI] [PubMed] [Google Scholar]
- de Haan JB, Gevers W, Parker MI. Effects of sodium butyrate on the synthesis and methylation of DNA in normal cells and their transformed counterparts. Cancer Res. 1986;46:713–6. [PubMed] [Google Scholar]
- De Vadder F, Grasset E, Mannerås Holm Let al. Gut microbiota regulates maturation of the adult enteric nervous system via enteric serotonin networks. Proc Natl Acad Sci USA. 2018;115:6458–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deichmann U. Epigenetics: the origins and evolution of a fashionable topic. Dev Biol. 2016;416:249–54. [DOI] [PubMed] [Google Scholar]
- Deleu S, Machiels K, Raes Jet al. Short chain fatty acids and its producing organisms: an overlooked therapy for IBD?. EBioMedicine. 2021;66:103293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Besten G, van Eunen K, Groen AKet al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich W, Schink M, Zopf Y. Microbiota in the gastrointestinal tract. Med Sci (Basel). 2018;6: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe DR, Collins LB, Wali Aet al. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell. 2012;48:612–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe DR, Wali A, Brylawski BPet al. Microbial regulation of glucose metabolism and cell-cycle progression in mammalian colonocytes. PLoS One. 2012;7:e46589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Johnson LM, Jacobsen SEet al. DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol. 2015;16:519–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durack J, Lynch SV. The gut microbiome: relationships with disease and opportunities for therapy. J Exp Med. 2019;216:20–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enaud R, Prevel R, Ciarlo Eet al. The gut-lung axis in health and Respiratory diseases: a place for inter-organ and inter-Kingdom crosstalks. Front Cell Infect Microbiol. 2020;10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erny D, Hrabě de Angelis AL, Jaitin Det al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci. 2015;18:965–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19:55–71. [DOI] [PubMed] [Google Scholar]
- Fellows R, Denizot J, Stellato Cet al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat Commun. 2018;9:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellows R, Varga-Weisz P. Chromatin dynamics and histone modifications in intestinal microbiota-host crosstalk. Mol Metab. 2020;38:100925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G. A brief history of epigenetics. Cold Spring Harb Perspect Biol. 2014;6: a018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando W, Martins IJ, Morici Met al. Sodium butyrate reduces brain amyloid-β levels and improves cognitive memory performance in an Alzheimer's disease transgenic mouse model at an early disease stage. J Alzheimers Dis. 2020;74:91–9. [DOI] [PubMed] [Google Scholar]
- Ferraris C, Elli M, Tagliabue A. Gut microbiota for health: how can diet maintain A healthy Gut microbiota?. Nutrients. 2020;12: 3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessele KL, Wright F. Primer in Genetics and Genomics, article 6: basics of epigenetic control. Biol Res Nurs. 2018;20:103–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillier TA, Shah S, Doody KMet al. Brief exposure of neuronal cells to levels of scfas observed in human systemic circulation impair lipid metabolism resulting in apoptosis. Sci Rep. 2022;12:14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores EM, Nguyen AT, Odem MAet al. The zebrafish as a model for gastrointestinal tract-microbe interactions. Cell Microbiol. 2020;22:e13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fock E, Parnova R. Mechanisms of blood-brain barrier protection by microbiota-derived short-chain fatty acids. Cells. 2023;12: 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkerts J, Redegeld F, Folkerts Get al. Butyrate inhibits human mast cell activation via epigenetic regulation of fcεri-mediated signaling. Allergy. 2020;75:1966–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JA, Baker GB, Dursun SM. The relationship between the gut microbiome-immune system-brain axis and major depressive disorder. Front Neurol. 2021;12:721126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericks E, Theunissen R, Roux S. Short chain fatty acids and monocarboxylate transporters in irritable bowel syndrome. Turk J Gastroenterol. 2020;31:840–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Tian CL, Ye Xet al. Dynamics of telomere rejuvenation during chemical induction to pluripotent stem cells. Stem Cell Reports. 2018;11:70–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda Set al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–50. [DOI] [PubMed] [Google Scholar]
- Gagnière J, Raisch J, Veziant Jet al. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol. 2016;22:501–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Yin J, Zhang Jet al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58:1509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasaly N, Hermoso MA, Gotteland M. Butyrate and the fine-tuning of colonic homeostasis: implication for inflammatory bowel diseases. Int J Mol Sci. 2021;22: 3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng ZH, Zhu Y, Li QLet al. Enteric Nervous system: the bridge between the gut microbiota and neurological disorders. Front Aging Neurosci. 2022;14:810483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibiino G, De Siena M, Sbrancia Met al. Dietary habits and gut microbiota in healthy adults: focusing on the right diet. A systematic review. Int J Mol Sci. 2021;22:6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA. Our unique microbial identity. Genome Biol. 2015;16:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RTet al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves P, Martel F. Butyrate and colorectal cancer: the role of butyrate transport. Curr Drug Metab. 2013;14:994–1008. [DOI] [PubMed] [Google Scholar]
- Gong F, Miller KM. Mammalian DNA repair: hATs and hdacs make their mark through histone acetylation. Mutat Res. 2013;750:23–30. [DOI] [PubMed] [Google Scholar]
- Goodrich JK, Waters JL, Poole ACet al. Human genetics shape the gut microbiome. Cell. 2014;159:789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Segre JA. The human microbiome: our second genome. Annu Rev Genomics Hum Genet. 2012;13:151–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilloteau P, Martin L, Eeckhaut Vet al. From the gut to the peripheral tissues: the multiple effects of butyrate. Nutr Res Rev. 2010;23:366–84. [DOI] [PubMed] [Google Scholar]
- Guinane CM, Cotter PD. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Therap Adv Gastroenterol. 2013;6:295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung M, Li Z, You Het al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020;51:102590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar R, Richard CS, Santos MM. The role of butyrate in surgical and oncological outcomes in colorectal cancer. Am J Physiol Gastrointest Liver Physiol. 2021;320:G601–g8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton JP. Epigenetics: principles and practice. Dig Dis. 2011;29:130–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A, Bennett N, Ahmed Bet al. Butyrate decreases its own oxidation in colorectal cancer cells through inhibition of histone deacetylases. Oncotarget. 2018;9:27280–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JL, Lin HL. Intestinal microbiota and type 2 diabetes: from mechanism insights to therapeutic perspective. World J Gastroenterol. 2014;20:17737–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Zhang P, Shen Let al. Short-chain fatty acids and their association with signalling pathways in inflammation, glucose and lipid metabolism. Int J Mol Sci. 2020;21: 6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang T, Zeng Yet al. Sodium butyrate mediates histone crotonylation and alleviated neonatal rats hypoxic-ischemic brain injury through gut-brain axis. Front Microbiol. 2022;13:993146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Ecker JR. Non-CG methylation in the Human genome. Annu Rev Genomics Hum Genet. 2015;16:55–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog EL, Wäfler M, Keller Iet al. The importance of age in compositional and functional profiling of the human intestinal microbiome. PLoS One. 2021;16:e0258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ono K, Tsuji Met al. Protective roles of intestinal microbiota derived short chain fatty acids in Alzheimer's disease-type beta-amyloid neuropathological mechanisms. Expert Rev Neurother. 2018;18:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodawadekar SC, Marmorstein R. Chemistry of acetyl transfer by histone modifying enzymes: structure, mechanism and implications for effector design. Oncogene. 2007;26:5528–40. [DOI] [PubMed] [Google Scholar]
- Hong J, Jia Y, Pan Set al. Butyrate alleviates high fat diet-induced obesity through activation of adiponectin-mediated pathway and stimulation of mitochondrial function in the skeletal muscle of mice. Oncotarget. 2016;7:56071–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong MY, Turner ND, Murphy MEet al. In vivo regulation of colonic cell proliferation, differentiation, apoptosis, and P27Kip1 by dietary fish oil and butyrate in rats. Cancer Prev Res (Phila). 2015;8:1076–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–8. [DOI] [PubMed] [Google Scholar]
- Hou K, Wu ZX, Chen XYet al. Microbiota in health and diseases. Signal Transduct Target Ther. 2022;7:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høverstad T, Midtvedt T. Short-chain fatty acids in germfree mice and rats. J Nutr. 1986;116:1772–6. [DOI] [PubMed] [Google Scholar]
- Hu S, Dong TS, Dalal SRet al. The microbe-derived short chain fatty acid butyrate targets miRNA-dependent p21 gene expression in human colon cancer. PLoS One. 2011;6:e16221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Liu L, Chang EBet al. Butyrate inhibits pro-proliferative miR-92a by diminishing c-Myc-induced miR-17-92a cluster transcription in human colon cancer cells. Mol Cancer. 2015;14:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iatcu CO, Steen A, Covasa M. Gut microbiota and complications of type-2 diabetes. Nutrients. 2021;14:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandhyala SM, Talukdar R, Subramanyam Cet al. Role of the normal gut microbiota. World J Gastroenterol. 2015;21:8787–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang HS, Shin WJ, Lee JEet al. CpG and non-CpG methylation in epigenetic gene regulation and brain function. Genes (Basel). 2017;8: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworska J, Zalewska T, Sypecka Jet al. Effect of the HDAC inhibitor, sodium butyrate, on neurogenesis in a rat model of neonatal hypoxia-ischemia: potential mechanism of action. Mol Neurobiol. 2019;56:6341–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G, Li C, Lu Met al. Protein lysine crotonylation: past, present, perspective. Cell Death Dis. 2021;12:703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy?. Genes Cancer. 2011;2:607–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone SE, Baylin SB. Stress and the epigenetic landscape: a link to the pathobiology of human diseases?. Nat Rev Genet. 2010;11:806–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung DH, Yong JH, Hwang Wet al. An efficient system for intestinal on-site butyrate production using novel microbiome-derived esterases. J Biol Eng. 2021;15:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiko GE, Ryu SH, Koues OIet al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell. 2016;165:1708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Chovatiya G, Tumbar T. Epigenetic control in skin development, homeostasis and injury repair. Exp Dermatol. 2019;28:453–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Kang X, Zhang Het al. Gut microbiota and Parkinson's disease: implications for faecal microbiota transplantation therapy. ASN Neuro. 2021;13:17590914211016217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson F, Tremaroli V, Nielsen Jet al. Assessing the human gut microbiota in metabolic diseases. Diabetes. 2013;62:3341–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karoor V, Strassheim D, Sullivan Tet al. The short-chain fatty acid butyrate attenuates pulmonary vascular remodeling and inflammation in hypoxia-induced pulmonary hypertension. Int J Mol Sci. 2021;22: 9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Ali SA, Yan F. Interactions between the gut microbiota-derived functional factors and intestinal epithelial cells - implication in the microbiota-host mutualism. Front Immunol. 2022;13:1006081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsen JR, Wu GD. The gut microbiota, environment and diseases of modern society. Gut Microbes. 2012;3:374–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Jena G. Sodium butyrate reduces insulin-resistance, fat accumulation and dyslipidemia in type-2 diabetic rat: a comparative study with metformin. Chem Biol Interact. 2016;254:124–34. [DOI] [PubMed] [Google Scholar]
- Kho ZY, Lal SK. The Human gut microbiome - A potential controller of wellness and disease. Front Microbiol. 2018;9:1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibbie JJ, Dillon SM, Thompson TAet al. Butyrate directly decreases human gut lamina propria CD4 T cell function through histone deacetylase (HDAC) inhibition and GPR43 signaling. Immunobiology. 2021;226:152126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd SK, Schneider JS. Protection of dopaminergic cells from MPP+-mediated toxicity by histone deacetylase inhibition. Brain Res. 2010;1354:172–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Park J, Kim M. Gut microbiota-derived short-chain fatty acids, T cells, and inflammation. Immune Netw. 2014;14:277–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Leeds P, Chuang DM. The HDAC inhibitor, sodium butyrate, stimulates neurogenesis in the ischemic brain. J Neurochem. 2009;110:1226–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Rowe M, Ren Met al. Histone deacetylase inhibitors exhibit anti-inflammatory and neuroprotective effects in a rat permanent ischemic model of stroke: multiple mechanisms of action. J Pharmacol Exp Ther. 2007;321:892–901. [DOI] [PubMed] [Google Scholar]
- Kim M, Qie Y, Park Jet al. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe. 2016;20:202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepinina L, Klepinin A, Truu Let al. Colon cancer cell differentiation by sodium butyrate modulates metabolic plasticity of caco-2 cells via alteration of phosphotransfer network. PLoS One. 2021;16:e0245348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollenstart L, de Groot AJL, Janssen GMCet al. Gcn5 and Esa1 function as histone crotonyltransferases to regulate crotonylation-dependent transcription. J Biol Chem. 2019;294:20122–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski K, Mulak A. Brain-gut-microbiota axis in Alzheimer's disease. J Neurogastroenterol Motil. 2019;25:48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautkramer KA, Kreznar JH, Romano KAet al. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell. 2016;64:982–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane AA, McGuire MK, McGuire MAet al. Household composition and the infant fecal microbiome: the INSPIRE study. Am J Phys Anthropol. 2019;169:526–39. [DOI] [PubMed] [Google Scholar]
- Lemm CL. The second genome. Lancet Gastroenterol Hepatol. 2018;3:535. [Google Scholar]
- Li J, Zhao F, Wang Yet al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 2017;5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, van Esch B, Henricks PAJet al. The anti-inflammatory effects of short chain fatty acids on lipopolysaccharide- or tumor necrosis factor α-stimulated endothelial cells via activation of GPR41/43 and inhibition of hdacs. Front Pharmacol. 2018;9:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Cao L, Tian Yet al. Butyrate suppresses the proliferation of colorectal cancer cells via targeting pyruvate kinase M2 and metabolic reprogramming. Mol Cell Proteomics. 2018;17:1531–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Ding C, Meng Tet al. Butyrate suppresses motility of colorectal cancer cells via deactivating Akt/ERK signaling in histone deacetylase dependent manner. J Pharmacol Sci. 2017;135:148–55. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang R, Li Qet al. Gut microbiota and Alzheimer's disease: pathophysiology and therapeutic perspectives. J Alzheimers Dis. 2021;83:963–76. [DOI] [PubMed] [Google Scholar]
- Liao P, Bhattarai N, Cao Bet al. Crotonylation at serine 46 impairs p53 activity. Biochem Biophys Res Commun. 2020;524:730–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licciardi PV, Wong SS, Tang MLet al. Epigenome targeting by probiotic metabolites. Gut Pathog. 2010;2:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbana T, Khan F, Eskander N. Gut microbiome and depression: how microbes affect the way we think. Cureus. 2020;12:e9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder SJ, Mostoslavsky R. Chapter 15 - interaction between cellular metabolic states and chromatin dynamics. In: Göndör A, ed. Chromatin Regulation and Dynamics. Boston: Academic Press; 2017:373–98. [Google Scholar]
- Lister R, Pelizzola M, Dowen RHet al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Xu D. Inhibition of histone deacetylases. Methods Mol Biol. 2004;287:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang J, He Tet al. Butyrate: a double-edged sword for health?. Adv Nutr. 2018;9:21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Gao J, Zhu Met al. Gut microbiota and dysbiosis in Alzheimer's disease: implications for pathogenesis and treatment. Mol Neurobiol. 2020;57:5026–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Guo R, Liu Fet al. Gut microbiota regulates depression-like behavior in rats through the neuroendocrine-immune-mitochondrial pathway. Neuropsychiatr Dis Treat. 2020;16:859–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Xue C, Fang Yet al. Global involvement of lysine crotonylation in protein modification and transcription regulation in rice. Mol Cell Proteomics. 2018;17:1922–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Yu H, Liu Yet al. Chromodomain protein CDYL acts as a crotonyl-CoA hydratase to regulate histone crotonylation and spermatogenesis. Mol Cell. 2017;67:853–66.e5. [DOI] [PubMed] [Google Scholar]
- Liu T, Li J, Liu Yet al. Short-chain fatty acids suppress lipopolysaccharide-induced production of nitric oxide and proinflammatory cytokines through inhibition of NF-κb pathway in RAW264.7 cells. Inflammation. 2012;35:1676–84. [DOI] [PubMed] [Google Scholar]
- Liu X, Wei W, Liu Yet al. MOF as an evolutionarily conserved histone crotonyltransferase and transcriptional activation by histone acetyltransferase-deficient and crotonyltransferase-competent CBP/p300. Cell Discovery. 2017;3:17016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Leng P, Liu Yet al. Crosstalk between methylation and ncRNAs in breast cancer: therapeutic and diagnostic implications. Int J Mol Sci. 2022;23: 15759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li YJ, Loh YWet al. Fiber derived microbial metabolites prevent acute kidney injury through G-protein coupled receptors and HDAC inhibition. Front Cell Dev Biol. 2021;9:648639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bertoni H, Lal B, Li Aet al. DNMT-dependent suppression of microRNA regulates the induction of GBM tumor-propagating phenotype by Oct4 and Sox2. Oncogene. 2015;34:3994–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, Flint HJ. Development of a semiquantitative degenerate real-time pcr-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl Environ Microbiol. 2007;73:2009–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lousada MB, Lachnit T, Edelkamp Jet al. Exploring the human hair follicle microbiome. Br J Dermatol. 2021;184:802–15. [DOI] [PubMed] [Google Scholar]
- Lu H, Ashiqueali R, Lin CIet al. Histone deacetylase 3 inhibition decreases cerebral edema and protects the blood-brain barrier after stroke. Mol Neurobiol. 2023;60:235–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund PJ, Gates LA, Leboeuf Met al. Stable isotope tracing <em>in vivo</em>reveals a metabolic bridge linking the microbiota to host histone acetylation. Cell Rep. 2022;41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupton JR Microbial degradation products influence colon cancer risk: the butyrate controversy. J Nutr. 2004;134:479–82. [DOI] [PubMed] [Google Scholar]
- Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62:67–72. [DOI] [PubMed] [Google Scholar]
- Madison A, Kiecolt-Glaser JK. Stress, depression, diet, and the gut microbiota: human-bacteria interactions at the core of psychoneuroimmunology and nutrition. Curr Opin Behav Sci. 2019;28:105–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen AS, Olsen CA. Profiling of substrates for zinc-dependent lysine deacylase enzymes: HDAC3 exhibits decrotonylase activity in vitro. Angew Chem Int Ed Engl. 2012;51:9083–7. [DOI] [PubMed] [Google Scholar]
- Malhotra R, Mauer AC, Lino Cardenas CLet al. HDAC9 is implicated in atherosclerotic aortic calcification and affects vascular smooth muscle cell phenotype. Nat Genet. 2019;51:1580–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud Eet al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marizzoni M, Cattaneo A, Mirabelli Pet al. Short-chain fatty acids and lipopolysaccharide as mediators between gut dysbiosis and amyloid pathology in Alzheimer's disease. J Alzheimers Dis. 2020;78:683–97. [DOI] [PubMed] [Google Scholar]
- Marmorstein R, Zhou MM. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol. 2014;6:a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín R, Miquel S, Ulmer Jet al. Gut ecosystem: how microbes help us. Benef Microbes. 2014;5:219–33. [DOI] [PubMed] [Google Scholar]
- Mashaqi S, Gozal D. Obstructive sleep apnea and systemic hypertension: gut dysbiosis as the mediator?. J Clin Sleep Med. 2019;15:1517–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew OP, Ranganna K, Mathew Jet al. Cellular effects of butyrate on vascular smooth muscle cells are mediated through disparate actions on dual targets, histone deacetylase (HDAC) activity and PI3K/Akt signaling network. Int J Mol Sci. 2019;20:: 2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew OP, Ranganna K, Yatsu FM. Butyrate, an HDAC inhibitor, stimulates interplay between different posttranslational modifications of histone H3 and differently alters G1-specific cell cycle proteins in vascular smooth muscle cells. Biomed Pharmacother. 2010;64:733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu E, Escribano-Vazquez U, Descamps Det al. Paradigms of lung microbiota functions in health and disease, particularly, in asthma. Front Physiol. 2018;9:1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride DA, Dorn NC, Yao Met al. Short-chain fatty acid-mediated epigenetic modulation of inflammatory T cells in vitro. Drug Deliv Transl Res. 2022. 13: 1912–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midtvedt AC, Midtvedt T. Production of short chain fatty acids by the intestinal microflora during the first 2 years of human life. J Pediatr Gastroenterol Nutr. 1992;15:395–403. [DOI] [PubMed] [Google Scholar]
- Miller JL, Grant PA. The role of DNA methylation and histone modifications in transcriptional regulation in humans. Subcell Biochem. 2013;61:289–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei R, Afaghi A, Babakhani Set al. Role of microbiota-derived short-chain fatty acids in cancer development and prevention. Biomed Pharmacother. 2021;139:111619. [DOI] [PubMed] [Google Scholar]
- Morotomi M, Mutai M. In vitro binding of potent mutagenic pyrolysates to intestinal bacteria. J Natl Cancer Inst. 1986;77:195–201. [PubMed] [Google Scholar]
- Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakov R, Velikova T. Chemical metabolism of xenobiotics by gut microbiota. Curr Drug Metab. 2020;21:260–9. [DOI] [PubMed] [Google Scholar]
- Ney L-M, Wipplinger M, Grossmann Met al. Short chain fatty acids: key regulators of the local and systemic immune response in inflammatory diseases and infections. Open Biology. 2023;13:230014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen M, Madelen Saunders C, Leena Angell Iet al. Butyrate levels in the transition from an infant- to an adult-like gut microbiota correlate with bacterial networks associated with Eubacterium rectale and Ruminococcus gnavus. Genes (Basel). 2020;11: 1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogal A, Valdes AM, Menni C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes. 2021;13:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakovic M, Rout A, Kingsley Tet al. Role of gut microbiota in cardiovascular diseases. World J Cardiol. 2020;12:110–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien J, Hayder H, Zayed Yet al. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne). 2018;9:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Riordan KJ, Collins MK, Moloney GMet al. Short chain fatty acids: microbial metabolites for gut-brain axis signalling. Mol Cell Endocrinol. 2022;546:111572. [DOI] [PubMed] [Google Scholar]
- Obata Y, Pachnis V. The effect of microbiota and the immune system on the development and organization of the enteric nervous system. Gastroenterology. 2016;151:836–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Repáraz J, Kasper LH. The second brain: is the gut microbiota a link between obesity and central nervous system disorders?. Curr Obes Rep. 2016;5:51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldendorf WH. Carrier-mediated blood-brain barrier transport of short-chain monocarboxylic organic acids. Am J Physiol. 1973;224:1450–3. [DOI] [PubMed] [Google Scholar]
- Orchel A, Dzierzewicz Z, Parfiniewicz Bet al. Butyrate-induced differentiation of colon cancer cells is PKC and JNK dependent. Dig Dis Sci. 2005;50:490–8. [DOI] [PubMed] [Google Scholar]
- Ota S, Sakuraba H. Uptake and advanced therapy of butyrate in inflammatory bowel disease. Immuno. 2022;2:692–702. [Google Scholar]
- Overby HB, Ferguson JF. Gut microbiota-derived short-chain fatty acids facilitate microbiota:host cross talk and modulate obesity and hypertension. Curr Hypertens Rep. 2021;23:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva I, Pinho R, Pavlou MAet al. Sodium butyrate rescues dopaminergic cells from alpha-synuclein-induced transcriptional deregulation and DNA damage. Hum Mol Genet. 2017;26:2231–46. [DOI] [PubMed] [Google Scholar]
- Paone P, Cani PD. Mucus barrier, mucins and gut microbiota: the expected slimy partners?. Gut. 2020;69:2232–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada Venegas D, De la Fuente MK, Landskron Get al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. 2019;10:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnala R, Arumugam TV, Gupta Net al. HDAC inhibitor sodium butyrate-mediated epigenetic regulation enhances neuroprotective function of microglia during ischemic stroke. Mol Neurobiol. 2017;54:6391–411. [DOI] [PubMed] [Google Scholar]
- Pickard JM, Zeng MY, Caruso Ret al. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 2017;279:70–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portincasa P, Bonfrate L, Vacca Met al. Gut microbiota and short chain fatty acids: implications in glucose homeostasis. Int J Mol Sci. 2022;23: 1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian XH, Xie RY, Liu XLet al. Mechanisms of short-chain fatty acids derived from gut microbiota in Alzheimer's disease. Aging Dis. 2022;13:1252–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Wade PA. Crosstalk between the microbiome and epigenome: messages from bugs. J Biochem. 2018;163:105–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Trone K, Kelly Cet al. All Fiber is not Fiber. Curr Gastroenterol Rep. 2023;25:1–12. [DOI] [PubMed] [Google Scholar]
- Ramazi S, Allahverdi A, Zahiri J. Evaluation of post-translational modifications in histone proteins: a review on histone modification defects in developmental and neurological disorders. J Biosci. 2020;45: 135. [PubMed] [Google Scholar]
- Ramírez-Pérez O, Cruz-Ramón V, Chinchilla-López Pet al. The role of the gut microbiota in bile acid metabolism. Ann Hepatol. 2017;16:s15–20. [DOI] [PubMed] [Google Scholar]
- Reddy SD, Clossen BL, Reddy DS. Epigenetic histone deacetylation inhibition prevents the development and persistence of temporal lobe epilepsy. J Pharmacol Exp Ther. 2018;364:97–109. [DOI] [PubMed] [Google Scholar]
- Reeves R, Candido EPM. Turnover of histone acetyl groups in cultured cells is inhibited by sodium butyrate. FEBS Lett. 1978;91:117–20. [DOI] [PubMed] [Google Scholar]
- Riccio P, Rossano R. The human gut microbiota is neither an organ nor a commensal. FEBS Lett. 2020;594:3262–71. [DOI] [PubMed] [Google Scholar]
- Rinninella E, Raoul P, Cintoni Met al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez JM, Murphy K, Stanton Cet al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis. 2015;26:26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roediger WE. Utilization of nutrients by isolated epithelial cells of the rat colon. Gastroenterology. 1982;83:424–9. [PubMed] [Google Scholar]
- Romano S, Savva GM, Bedarf JRet al. Meta-analysis of the Parkinson's disease gut microbiome suggests alterations linked to intestinal inflammation. NPJ Parkinsons Dis. 2021;7:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose NR, Klose RJ. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta. 2014;1839:1362–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld CS. Microbiome disturbances and Autism spectrum disorders. Drug Metab Dispos. 2015;43:1557–71. [DOI] [PubMed] [Google Scholar]
- Rosser EC, Mauri C. A clinical update on the significance of the gut microbiota in systemic autoimmunity. J Autoimmun. 2016;74:85–93. [DOI] [PubMed] [Google Scholar]
- Rothschild D, Weissbrod O, Barkan Eet al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–5. [DOI] [PubMed] [Google Scholar]
- Rumberger JM, Arch JR, Green A. Butyrate and other short-chain fatty acids increase the rate of lipolysis in 3T3-L1 adipocytes. PeerJ. 2014;2:e611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu SH, Kaiko GE, Stappenbeck TS. Cellular differentiation: potential insight into butyrate paradox?. Mol Cell Oncol. 2018;5:e1212685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Tang Z, Huang Het al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol Cell. 2015;58:203–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sałkowska A, Karaś K, Walczak-Drzewiecka Aet al. Differentiation stage-specific effect of histone deacetylase inhibitors on the expression of RORγT in human lymphocytes. J Leukoc Biol. 2017;102:1487–95. [DOI] [PubMed] [Google Scholar]
- Salvi PS, Cowles RA. Butyrate and the intestinal epithelium: modulation of proliferation and inflammation in homeostasis and disease. Cells. 2021;10: 1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Debelius JW, Thron Tet al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's Disease. Cell. 2016;167:1469–80.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez HN, Moroney JB, Gan Het al. B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids. Nat Commun. 2020;11:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–33. [DOI] [PubMed] [Google Scholar]
- Schluter J, Peled JU, Taylor BPet al. The gut microbiota is associated with immune cell dynamics in humans. Nature. 2020;588:303–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Lewis ZA, Goll MG. DNA methylation: shared and divergent features across eukaryotes. Trends Genet. 2019;35:818–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulthess J, Pandey S, Capitani Met al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity. 2019;50:432–45.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sealy L, Chalkley R. The effect of sodium butyrate on histone modification. Cell. 1978;14:115–21. [DOI] [PubMed] [Google Scholar]
- Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164:337–40. [DOI] [PubMed] [Google Scholar]
- Sender R, Fuchs S, Milo R. Revised estimates for the number of Human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Li Y, Stoll MLet al. The epigenetic connection between the gut microbiome in obesity and diabetes. Front Genet. 2019;10:1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Taliyan R, Singh S. Beneficial effects of sodium butyrate in 6-OHDA induced neurotoxicity and behavioral abnormalities: modulation of histone deacetylase activity. Behav Brain Res. 2015;291:306–14. [DOI] [PubMed] [Google Scholar]
- Shimizu H, Masujima Y, Ushiroda Cet al. Dietary short-chain fatty acid intake improves the hepatic metabolic condition via FFAR3. Sci Rep. 2019;9:16574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siavoshian S, Blottiere HM, Cherbut Cet al. Butyrate stimulates cyclin D and p21 and inhibits cyclin-dependent kinase 2 expression in HT-29 colonic epithelial cells. Biochem Biophys Res Commun. 1997;232:169–72. [DOI] [PubMed] [Google Scholar]
- Silva YP, Bernardi A, Frozza RL. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front Endocrinol (Lausanne). 2020;11:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Thangaraju M, Prasad PDet al. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J Biol Chem. 2010;285:27601–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Chang HW, Yan Det al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Yeoh BS, Vijay-Kumar M. Feed your gut with caution!. Transl Cancer Res. 2016;5:S507–s13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens KE, Miaskowski CA, Levine JDet al. Epigenetic regulation and measurement of epigenetic changes. Biol Res Nurs. 2013;15:373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Wu W, Liu Zet al. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J Gastroenterol. 2017;52:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Horikoshi N, Kato Det al. Crystal structure of the nucleosome containing histone H3 with crotonylated lysine 122. Biochem Biophys Res Commun. 2016;469:483–9. [DOI] [PubMed] [Google Scholar]
- Swer NM, Venkidesh BS, Murali TSet al. Gut microbiota-derived metabolites and their importance in neurological disorders. Mol Biol Rep. 2022. 50: 1663–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, McKenzie C, Potamitis Met al. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91–119. [DOI] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee Set al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WH, Wang Z, Kennedy DJet al. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ Res. 2015;116:448–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavakoli S, Zhu S, Matsudaira P. Cell clusters containing intestinal stem cells line, the zebrafish intestine intervillus pocket. Iscience. 2022;25:104280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theiler A, Bärnthaler T, Platzer Wet al. Butyrate ameliorates allergic airway inflammation by limiting eosinophil trafficking and survival. J Allergy Clin Immunol. 2019;144:764–76. [DOI] [PubMed] [Google Scholar]
- Thio CL-P, Chi P-Y, Lai AC-Yet al. Regulation of type 2 innate lymphoid cell–dependent airway hyperreactivity by butyrate. J Allergy Clin Immunol. 2018;142:1867–83.e12. [DOI] [PubMed] [Google Scholar]
- Thomas SP, Denu JM. Short-chain fatty acids activate acetyltransferase p300. Elife. 2021;10: e72171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn AN, McKenzie CI, Shen Set al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. 2015;6:7320. [DOI] [PubMed] [Google Scholar]
- Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. 2017;474:1823–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres IO, Fujimori DG. Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr Opin Struct Biol. 2015;35:68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronick E, Hunter RG. Waddington, Dynamic Systems, and epigenetics. Front Behav Neurosci. 2016;10:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truesdell SS, Mortensen RD, Seo Met al. MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci Rep. 2012;2:842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umei M, Akazawa H, Saga-Kamo Aet al. Protective action of the microbial metabolite butyrate against cardiomyocyte hypertrophy. Eur Heart J. 2020;41: ehaa946.3666. [Google Scholar]
- Ursell LK, Metcalf JL, Parfrey LWet al. Defining the human microbiome. Nutr Rev. 2012;70 Suppl 1:S38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes AM, Walter J, Segal Eet al. Role of the gut microbiota in nutrition and health. BMJ. 2018;361:k2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deuren T, Blaak EE, Canfora EE. Butyrate to combat obesity and obesity-associated metabolic disorders: current status and future implications for therapeutic use. Obes Rev. 2022;23:e13498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deuren T, Blaak EE, Canfora EE. Butyrate to combat obesity and obesity-associated metabolic disorders: current status and future implications for therapeutic use. Obes Rev. 2022;23:e13498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemula PK, Jala VR. Colonic crypts are natural gatekeepers of microbial metabolites to protect stem cells. Transl Cancer Res. 2016;5:S536–s9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verza FA, Das U, Fachin ALet al. Roles of histone deacetylases and inhibitors in anticancer therapy. Cancers (Basel). 2020;12: 1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinolo MA, Rodrigues HG, Hatanaka Eet al. Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils. J Nutr Biochem. 2011;22:849–55. [DOI] [PubMed] [Google Scholar]
- Vinolo MA, Rodrigues HG, Nachbar RTet al. Regulation of inflammation by short chain fatty acids. Nutrients. 2011;3:858–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. Organisers and genes. Organisers and Genes. 1940; 102. [Google Scholar]
- Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- Wan J, Liu H, Ming L. Lysine crotonylation is involved in hepatocellular carcinoma progression. Biomed Pharmacother. 2019;111:976–82. [DOI] [PubMed] [Google Scholar]
- Wang L, Shannar AAF, Wu Ret al. Butyrate drives metabolic rewiring and epigenetic reprogramming in Human colon cancer cells. Mol Nutr Food Res. 2022;66:e2200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhao Z, Zhao Let al. Lactobacillus plantarum DP189 reduces α-SYN aggravation in MPTP-induced Parkinson's Disease mice via regulating oxidative damage, inflammation, and gut microbiota disorder. J Agric Food Chem. 2022;70:1163–73. [DOI] [PubMed] [Google Scholar]
- Wang Q, Luo Y, Ray Chaudhuri Ket al. The role of gut dysbiosis in Parkinson's disease: mechanistic insights and therapeutic options. Brain. 2021;144:2571–93. [DOI] [PubMed] [Google Scholar]
- Wang S, Mu G, Qiu Bet al. The function and related diseases of protein crotonylation. Int J Biol Sci. 2021;17:3441–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Leng Y, Tsai LKet al. Valproic acid attenuates blood-brain barrier disruption in a rat model of transient focal cerebral ischemia: the roles of HDAC and MMP-9 inhibition. J Cereb Blood Flow Metab. 2011;31:52–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Qin G, Zhao TC. HDAC4: mechanism of regulation and biological functions. Epigenomics. 2014;6:139–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Liu X, Chen Jet al. Class I histone deacetylases are major histone decrotonylases: evidence for critical and broad function of histone crotonylation in transcription. Cell Res. 2017;27:898–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Mao A, Tang Bet al. Large-scale identification of protein crotonylation reveals its role in multiple cellular functions. J Proteome Res. 2017;16:1743–52. [DOI] [PubMed] [Google Scholar]
- Weinberg DN, Papillon-Cavanagh S, Chen Het al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature. 2019;573:281–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel TJ, Gates EJ, Ranger ALet al. Short-chain fatty acids (SCFAs) alone or in combination regulate select immune functions of microglia-like cells. Mol Cell Neurosci. 2020;105:103493. [DOI] [PubMed] [Google Scholar]
- Wilson ID, Nicholson JK. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl Res. 2017;179:204–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G, Hart RA, Charlesworth RPGet al. Gut microbiome and depression: what we know and what we need to know. Rev Neurosci. 2018;29:629–43. [DOI] [PubMed] [Google Scholar]
- Wippermann A, Rupp O, Brinkrolf Ket al. Integrative analysis of DNA methylation and gene expression in butyrate-treated CHO cells. J Biotechnol. 2017;257:150–61. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Feunang YD, Marcu Aet al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608-d17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JM, de Souza R, Kendall CWet al. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40:235–43. [DOI] [PubMed] [Google Scholar]
- Woo V, Alenghat T. Epigenetic regulation by gut microbiota. Gut Microbes. 2022;14:2022407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woting A, Blaut M. The intestinal microbiota in metabolic disease. Nutrients. 2016;8:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012;3:4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Heidenreich D, Zhou Set al. A chemical toolbox for the study of bromodomains and epigenetic signaling. Nat Commun. 2019;10:1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Chen PS, Dallas Set al. Histone deacetylase inhibitors up-regulate astrocyte GDNF and BDNF gene transcription and protect dopaminergic neurons. Int J Neuropsychopharmacol. 2008;11:1123–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Jing Z, Wei Wet al. Inhibitory effect of sodium butyrate on colorectal cancer cells and construction of the related molecular network. BMC Cancer. 2021;21:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A, Ensink E, Li Pet al. Bacterial butyrate in Parkinson's Disease is linked to epigenetic changes and depressive symptoms. Mov Disord. 2022;37:1644–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Wan J, Zhan Jet al. Global profiling of crotonylation on non-histone proteins. Cell Res. 2017;27:946–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Pan Y, Shao Wet al. Beneficial effect of the short-chain fatty acid propionate on vascular calcification through intestinal microbiota remodelling. Microbiome. 2022;10:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Kweon MN. The gut microbiota: a key regulator of metabolic diseases. BMB Rep. 2016;49:536–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Yu T, Huang Xet al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat Commun. 2020;11:4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Cai X, Fei Wet al. The role of short-chain fatty acids in immunity, inflammation and metabolism. Crit Rev Food Sci Nutr. 2022;62:1–12. [DOI] [PubMed] [Google Scholar]
- Yip W, Hughes MR, Li Yet al. Butyrate shapes immune cell fate and function in allergic asthma. Front Immunol. 2021;12:628453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Du M, Yang Qet al. Butyrate suppresses murine mast cell proliferation and cytokine production through inhibiting histone deacetylase. J Nutr Biochem. 2016;27:299–306. [DOI] [PubMed] [Google Scholar]
- Zhang L, Deng M, Lu Aet al. Sodium butyrate attenuates angiotensin II-induced cardiac hypertrophy by inhibiting COX2/PGE2 pathway via a HDAC5/HDAC6-dependent mechanism. J Cell Mol Med. 2019;23:8139–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Tian S, Pei Met al. Crosstalk between histone modification and DNA methylation orchestrates the epigenetic regulation of the costimulatory factors, tim–3 and galectin–9, in cervical cancer. Oncol Rep. 2019;42:2655–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Zhang F, Yu Zet al. HDAC3 inhibition prevents blood-brain barrier permeability through Nrf2 activation in type 2 diabetes male mice. J Neuroinflammation. 2019;16:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Wen XL. Gut microbiota and inflammatory bowel disease: the current status and perspectives. World J Clin Cases. 2021;9:321–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Yu H, Chen Jet al. The short-chain fatty acid butyrate accelerates vascular calcification via regulation of histone deacetylases and NF-κb signaling. Vascul Pharmacol. 2022;146:107096. [DOI] [PubMed] [Google Scholar]
- Ziemka-Nalecz M, Jaworska J, Sypecka Jet al. Sodium butyrate, a histone deacetylase inhibitor, exhibits neuroprotective/neurogenic effects in a rat model of neonatal hypoxia-ischemia. Mol Neurobiol. 2017;54:5300–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY, Beaudet AL. Epigenetics and Human disease. Cold Spring Harb Perspect Biol. 2016;8:a019497. [DOI] [PMC free article] [PubMed] [Google Scholar]