Abstract
Purpose:
Radiation-induced sarcomas (RIS) have a poor prognosis and lack effective treatments. Its genome and tumor microenvironment are not well characterized and need further exploration.
Experimental Design:
Here, we performed whole-exome sequencing (WES) and mRNA sequencing (mRNA-seq) on patients with RIS and primary sarcomas (WES samples 46 vs. 48, mRNA-seq samples 16 vs. 8, mainly in head and neck), investigated the antitumor effect of programmed cell death protein 1 (PD-1) blockade in RIS patient-derived xenograft models, and analyzed clinical data of patients with RIS treated with chemotherapy alone or combined with an anti–PD-1 antibody.
Results:
Compared with primary sarcomas, RIS manifested different patterns of copy-number variations, a significantly higher number of predicted strong MHC-binding neoantigens, and significantly increased immune cell infiltration. Clinical data showed that the combinatorial use of chemotherapy and PD-1 blockade achieved a higher objective response rate (36.67% vs. 8.00%; P = 0.003), longer overall survival (31.9 months vs. 14.8 months; P = 0.014), and longer progression-free survival (4.7 months vs. 9.5 months; P = 0.032) in patients with RIS compared with single chemotherapy.
Conclusions:
Elevated genomic instability and higher immune cell infiltrations were found in RIS than in primary sarcomas. Moreover, higher efficacy of chemotherapy plus PD-1 blockade was observed in animal experiments and clinical practice. This evidence indicated the promising application of immune checkpoint inhibitors in the treatment of RIS.
Translational Relevance.
In this study, a comprehensive and integrative analysis for the genomic, transcriptomic, and immunologic profile of radiation-induced sarcomas (RIS) was performed, revealing that this tumor type harbored a heterogenous genomic variation in comparison with primary sarcomas. It is first proposed that immune checkpoint blockade is a promising therapeutic approach for RIS especially in the head and neck, and the combination of traditional chemotherapy could even enhance the clinical efficacy. Our findings may help to identify diagnostic and prognostic markers as well as molecular targets for the diagnosis and treatment of RIS and provide strong evidence for the application of programmed cell death protein 1 blockade in clinical practice of RIS treatment.
Introduction
Radiotherapy is one of the major therapeutic approaches for cancer treatment beyond surgery and chemotherapy (1, 2). For head and neck malignancies, radiotherapy is a cornerstone of treatments and improves the long-term survival rate of patients (3). However, the longer survival brought by radiotherapy is accompanied with an increased risk of secondary malignancies, mainly radiation-induced carcinomas and radiation-induced sarcomas (RIS; refs. 4–7). In recent decades, increased cases of RIS have been reported with different histologic subtypes including angiosarcoma, fibrosarcoma, osteosarcoma, etc. (7, 8). The incidence of RIS ranges from 0.04% to 0.78% among radiation-treated patients, and up to 0.3% among those received head and neck radiotherapy (9, 10). RIS account for up to 5% of all sarcoma cases (5). The most commonly used diagnostic criteria for RIS include: (i) the tumor develops within the previously irradiated area, (ii) the tumor differs histologically from the primary tumors, (iii) no evidence of any secondary tumor at the time of radiotherapy, and (iv) the tumor often develops 3 years or more after radiotherapy (11, 12).
RIS are highly aggressive and have poorer prognosis compared with primary sarcomas (13), and the main reasons include delayed diagnosis, surgical limit due to tumor size and location, reduced benefit from radiotherapy, and low sensitivity to chemotherapy (14). Moreover, studies have revealed that RIS show a higher local recurrence rate and shorter disease-specific survival than primary sarcomas (15–17). Considering the aggressiveness and poor prognosis of RIS, genomic and transcriptomic features of RIS, which greatly facilitate novel treatment development, need to be characterized.
Radiation has been reported to induce double-stranded DNA damage, cause genomic instability, and further influence the transcriptome of affected cells. The frequent amplification of gene MYC (MYC proto-oncogene) and FLT4 (fms-related receptor tyrosine kinase 4) in radiation-induced angiosarcoma after breast cancer has been reported (18, 19). Whole-exome sequencing (WES) has revealed TP53 mutations in pediatric cancer survivors who developed RIS (20). The gene RB1 (tumor suppressor retinoblastoma 1) and a panel of proteins related to senescence-associated secretory phenotypes have been found to influence the tumorigenesis of radiation-induced osteosarcomas (RIOS) in the Rb +/− mouse model (21). A transcriptomic analysis of limited samples of RIS and primary sarcomas identifies a signature of 135 genes clustered in chronic oxidative stress pathway that discriminates RIS from primary sarcomas (22). However, comprehensive knowledge about genomic features and immune characteristics of RIS is yet to be achieved, and investigations with larger sample numbers of RIS are urgently needed. In clinical practice, previous studies reported that immune checkpoint inhibitors (ICI) showed potential clinical benefit for radiation-induced angiosarcomas in two individual cases as early insight for our research (23, 24). Most studies on RIS have focused on certain aspects, and there is yet to be a study that extensively connects genomic landscape and clinical outcomes to provide in-depth view in how they differentiate RIS from primary sarcomas and in how these factors impact the oncogenesis, tumor microenvironment, and treatment responses of RIS.
Nasopharyngeal carcinoma (NPC) is a common malignancy in southern China, and radiotherapy is one of the most important standard treatments for NPC (25). Patients with NPC receiving radiotherapy are at a risk of developing RIS. In this study, we integratively perform WES and mRNA sequencing (mRNA-seq) to investigate the genomic features, transcriptomic landscape, and tumor microenvironment characteristics of RIS developed in patients with NPC after radiotherapy, and provided clinical data of the largest current cohort for antiprogrammed cell death protein 1 (anti-PD-1) treatment in RIS. We provide new insights into the molecular basis of RIS and show that RIS are distinguished from primary sarcomas, which may help to identify diagnostic and prognostic markers as well as molecular targets for the treatment, especially anti–PD-1 treatment, of RIS.
Materials and Methods
Study population and tumor specimens
A total of 144 patients diagnosed with RIS and 123 patients diagnosed with primary sarcomas at the Sun Yat-sen University Cancer Center (SYSUCC) from January 2000 to October 2022 were identified. The inclusion criteria were listed in Supplementary Fig. S1. All the patients were followed up every 3 months until October 1, 2022. This study was approved by the Institutional Review Board (IRB) of the SYSUCC. Ethical consent was obtained from the Ethics Committee of the SYSUCC. The clinicopathologic characteristics of patients were collected and analyzed by specialists to reconfirm the clinical diagnosis.
Out of the total 144 patients diagnosed with RIS, 46 patients had both tumor samples and matched blood specimens available for WES, while 16 patients had cryopreserved tumor tissue available for mRNA-seq (transcriptome sequencing). Of the 123 primary sarcoma samples, 48 were for WES, and eight for mRNA-seq. The cryopreserved tumor tissue was from the SYSUCC Bio-bank. All tumor specimens were examined with hematoxylin and eosin staining and then reviewed by an expert pathologist to ensure that tumor content within the specimen was above 20%. Representative formalin-fixed, paraffin-embedded (FFPE) sections from each sample were retrieved for DNA extraction, and the matched blood samples were also retrieved to extract DNA as control.
Whole transcriptome analysis
Sample processing and sequencing
Total RNA was extracted from cryopreserved tissues using OMEGA DNA/RNA/Protein extraction kit, and the quality assessment of RNA was carried out by Mingma Technologies Co., Ltd. An aliquot of 250 ng of purified RNA was used with DV200 scores >30% based on Illumina's TruSeq Stranded mRNA protocol. The normalized, pooled libraries were loaded onto the Novaseq 6000 platforms for a goal of 6G data per sample (HiSeq2500). To filter the raw data, fastp (https://github.com/OpenGene/fastp) was used to process the raw pair-end fastq files, after which subread (version 2.0.1) was used for alignment to reference genome hg19 and gene expression quantification. We used RSEM (version 1.3.0) to estimate gene and gene isoform expression levels by aligning individual samples back to the transcriptome using Bowtie (version 1.2.1.1) (26).
Differentially expressed genes analysis
The gene expression matrices rendered were further filtered to remove low expression genes (zero count among less than two-thirds of all samples). About one-third of the genes in each qualified expression matrix had 10 or more counts and was prepared for differential gene expression analysis. The samples were grouped into the RIS group (patients with RIS) and the SARC group (patients with primary sarcomas). The analysis was carried out using the DESeq2 package (version 1.12.3) in the R programming environment (version 4.0.3). The differentially expressed genes (DEG) were determined by two conditions: Padj. value < 0.05 and absolute value of log2 (fold change) > 2. To further investigate the enrichment of DEG in a certain clustering pattern, enrichment analysis was performed using the R package ClusterProfiler (https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html) in gene ontology (GO), Reactome, and Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The packages pheatmap 1.0.1 and ggplot2 3.3.2 were used for visualization.
Principal component analysis
Principal component analysis (PCA) was performed to verify the differences of clustering characteristics between the two groups. The top 5,000 genes with the most significant difference in expression were selected to analyze with the R software. The R packages FactoMineR 2.4 and factoextra were used for visualization. The overview of PCA was plotted in figures.
Immune profile analysis
To quantify the relative infiltration of immune cells in the tumor microenvironment, we selected 28 immune cell types whose feature gene panels were described in previous publications (27, 28). The single-sample gene set enrichment analysis (ssGSEA) was introduced to calculate an enrichment score for the relative abundance of each immune cell type (29). One-sided Wilcox rank-sum test was used, and a P value < 0.05 indicated significant difference. The antitumor immunity score was calculated using a previous reported model including 12 cell types [ActCD4, ActCD8, TcmCD4, TcmCD8, TemCD4, TemCD8, Th1, Th17, ActDC, CD56briNK, natural killer (NK), and NKT; ref. 30]. The proliferation module score was calculated involving eight cell types [regulatory T (Treg), Th2, CD56dimNK, imDC, TAM, myeloid-derived suppressor cell (MDSC), neutrophil, and pDC]. The relationships between antitumor immunity score, proliferation module score, and the expression levels of some immune-related genes [programmed death-ligand 1 (PD-L1), PD-1, CTLA-4, IDO1, TDO2] were analyzed using Student t test and Pearson's correlation analysis. P < 0.05 indicated a significant correlation.
Whole-exome analysis
Sample processing and sequencing
Tumor DNA of FFPE samples was extracted with the Maxwell 16 FFPE Plus LEV DNA Purification Kit (Promega-AS1135), and blood DNA was purified using the OMEGA Blood DNA Kit following the manufacturer's instructions. The quality assessment of DNA was performed by Mingma Technologies Co., Ltd. Briefly, samples were sequenced using the Agilent exome enrichment kit (Sure Select V5; with > 50% of baits above 100×coverage). Base calls and intensities from the Illumina NovaSeq 6000 were processed into fastq files.
Mutational analysis
After using Fastp to filter the paired sequencing data, Sentieon 201711 TNscope standard pipeline was applied to call somatic single-nucleotide variants (SNV) and insertion-deletions (INS/DELs) in the reference of University of California Santa Cruz's hg19 GRCh37 human genome. On the basis of paired alignment files (tumor samples and matched blood samples), somatic SNV and small INS/DELs with default parameters were identified. Mutations were progressively selected for further investigation. Somatic SNV were further filtered to remove potential false positives based on the parameters recommended by the standard protocol of Sentieon; only high confidence variants with an allele fraction greater than 0.05 or a coverage at least 5× were remained. Then, rare variants with frequencies less than 0.005 were selected from all databases (EXAC, ESP6500, DBSNP, and 1000G). The Annovar was used for mutation annotation; only mutations within the gene coding sequence regions of the genes were remained (31). Besides, we rearranged mutated genes by signaling pathways. Then, R package maftools 2.10.0 was applied to visualize the data.
Tumor mutation burden analysis
Tumor mutation burden (TMB) was calculated on the basis of the number of somatic mutations, coding mutations, base substitution mutations, and indel mutations per megabase of genome examined (32). To reduce noise, synonymous mutations in the exon region were included in calculation, while mutations in regions of rearrangements, fusions, copy-number variations (CNV) and noncoding alterations were excluded.
CNV analysis
The CNV identification was performed using CNVKit (33). Matched bam (tumor-normal) files from samples served as the input for this analysis using default parameters. The results of CNVkit analysis were exported to the standard SEG format and were analyzed using GISTIC 2.0 for visualization. We then filtered these CNV using genes listed in the Catalogue of Somatic Mutations in Cancer (COSMIC) Cancer Gene Census. The copy-number gain and amplification are respectively defined as an increase in the number of gene copies of one (gain) or more (amplification) sections of the DNA. The copy-number loss and deletion are respectively defined as the decrease in the number of gene copies of one (loss) or more (deletion) sections of the DNA.
Microsatellite instability and mismatch repair genes
To assess the microsatellite instability (MSI) status computationally, the bam file obtained from the WES analysis pipeline was retrieved as input for MSI analysis using MANTIS (https://github.com/OSU-SRLab/MANTIS), following the recommended parameters for WES data (34). The reference microsatellite region was called using the compiled RepeatFinder within the MANTIS based on hg19 genome, and was further filtered according to the Bethesda panel (35). The MSI status was calculated using the Step-Wise Difference (DIF) method. Samples with a DIF score over 0.4 were considered instable. IHC was performed to detect the expression levels of mismatch repair (MMR) family proteins (MLH1, PMS2, MSH2, and MSH6).
Mutational signatures identification and clustering
The R packages nonnegative matrix factorization and MutationalPatterns were used to discover the mutational patterns of RIS (36–38). The resulting signature contribution proportions were further used to cluster samples by using all known COSMIC signatures (https://cancer.sanger.ac.uk/cosmic/signatures).
Neoantigen analysis
Tumor neoantigen analysis was performed using the NeoPredPipe neoantigen discovery pipeline (https://github.com/MathOnco/NeoPredPipe) as described in previous studies (39, 40). Briefly, the bam files from WES were used as input for HLA typing by Polysolver (http://software.broadinstitute.org/cancer/cga/polysolver; refs. 39, 41). Then, the filtered VCF files and HLA types of samples served as input for neoantigen calling using default parameters, after which the output neoantigen file was used as the source file for neoantigen recognition potential calculation to predict the possibility of recognition of each neoantigen peptide (40).
Multiplex immunofluorescence staining
We performed multiplex immunofluorescence (mIF) staining by the seven-color Fluorescence Kit (Panovue, Beijing, China) according to the manufacturer's protocol. Manual mIF staining was performed at 4-μmol/L sequential histologic tumor sections from representative FFPE samples. The stained slides were scanned with a Vectra Polaris multispectral imaging platform and analyzed by HALO digital pathologic platform. The immunofluorescence markers used consisted of PD-L1 (clone E1L3N, dilution 1:100; #13684, Cell Signaling Technology), PD-1 (clone D4W2J, dilution 1:250; #86163, CST), CD3 (ZM-0417, dilution 1:100; ZSJQB Co., Ltd.) and CD8 (ZA-0508, dilution 1:100; ZSJQB Co., Ltd.), Foxp3 (clone D2W8E; dilution 1:100; #98377, CST), CD56 (clone E7X9M, dilution 1:250; #99746, CST), CD68 (ZM-0060, dilution 1:100; ZSJQB Co., Ltd.), CD11c (clone D3V1E, dilution 1:250; #45581, CST), and Ki67 (ZM-0167, dilution 1:200; ZSJQB Co., Ltd.). The results were calculated using positively stained cells and tissue area ratio (cells/mm2) based on t test.
IHC staining
MMR status was assessed by IHC staining with the antibodies for MLH1 (dilution 1:250; ab92312, Abcam), PMS2 (dilution 1:200; ab110638, Abcam), MSH2 (dilution 1:200; abs135777, Absin), and MSH6 (dilution 1:500; #12988, CST) as previously described (42). Normal colonic epithelium was used as external control. Quality check was done using confirmative positive and negative samples. The interpretation of IHC was carried out independently by two pathologists. Lymphocytes, stromal cells, and blood vessels showed positive nuclear staining (internal control). In the presence of retained internal control, negative staining in tumor cells indicated MMR deficiency.
Establishment of patient-derived xenograft model and animal experiments
All protocols for animal experiments were approved by the Institutional Animal Care and Use Committee of Sun Yat-Sen University, China. Female 5-week-old NOD/SCID gamma (NSG) mice #T001475 purchased from GuangDong GemPharmatech Co., Ltd. (Guangdong, China) were used for the establishment of patient-derived xenograft (PDX) models for human RIS. All human tumor samples were obtained from the donor patients of the SYSUCC from 2018 to 2022. Informed written consent was acquired from the patients. Tissues used in this study and the protocols for experiments were approved by the IRBs of SYSUCC.
Fresh tumor tissues were sliced into fragments (diameter: ∼3 mm) under sterile conditions. Under inhalation anesthesia, fragments were subcutaneously transplanted into the right flanks of mice. The first transplantation directly from a donor patient to a host mouse is termed G0, and subsequent generations were numbered consecutively (G1, G2, and G3, etc.). Tumors were harvested when they reached a volume of 1,000 to 1,500 mm3, sliced and regrafted into new host mice. All experiments on the PDX were performed on G3–5 generations.
The third-generation xenograft of a radiation-induced fibrosarcoma (RIFS) was individually transplanted subcutaneously into the right flanks of 24 mice. The mice would not be ready for subsequent experiments until the tumors reached about 50 to 100 mm3. Human peripheral blood mononuclear cells (PBMC) were isolated from the corresponding patient via Ficoll Hypaque centrifugation, stimulated with precoated anti-CD3 antibody (1 μg/mL, clone OKT; catalog No. 317302, BioLegend) for 1 day under standard culture conditions, and then injected into the PDX-bearing mice intravenous (i.v., 5 × 106 PBMCs per mouse). Subsequently, the mice were randomized into four groups and treated with PBS (vehicle control), anti–PD-1 antibody (anti-human PD-1 Ab, Clone J116, catalog No. BE0188, BioXcell, 10 mg/kg, i.v., every 3 days), doxorubicin (doxorubicin hydrochloride liposome, CSPC Pharmaceutical Group Co., Ltd., 2 mg/kg, i.p., every 3 days), or anti–PD-1 antibody plus doxorubicin. Body weight and tumor size were measured every 3 days. Tumor volume was calculated using the formula length × width2/2. At the end of the experiment, the mice were sacrificed, and the subcutaneous tumors were dissected and stained with Ki67.
Drug treatment and survival analysis for patients
All the cases in survival analysis were defined as advanced sarcomas. Advanced sarcomas included those that were unresectable by surgery or locally advanced, or sarcomas with metastases. All the patients received standard doxorubicin-based chemotherapy every 3 weeks (mainly CAV/IE, AI and MAID; refs. 43, 44). CAV/IE refers to the regimen of cyclophosphamide, doxorubicin (adriamycin, ADM), and vincristine (CAV), and ifosfamide and etoposide (IE), with alternating cycles of CAV and IE every 3 weeks (45). The anti–PD-1 antibody was named toripalimab (Shanghai Junshi Biosciences Co., Ltd.) and administered at a dosage of 240 mg every 3 weeks (46). The retrospective analysis was approved by the IRB of the SYSUCC. The overall survival of advanced patients was defined as the time interval between the date of the start of chemotherapy and the date of death. The progression-free survival (PFS) was defined as the time interval between the date of commencement of chemotherapy and the date of progression or death. Adverse events (AE) were graded according to the Common Terminology Criteria for Adverse Events Version 5.0.
Quantification of cytokines in patient blood samples by ELISA
Peripheral blood samples from patients with RIS with chemotherapy plus immunotherapy were collected pretreatment and four to six cycles posttreatment. The quantification of eight cytokines (IFNγ, IFNβ, TNFα, TNFβ, IL2, IL6, IL16 and IL10) was detected in blood plasma using ELISA. Samples were analyzed as individual values for each patient. In brief, ELISA plates were coated with capture antibody overnight at 4°C. The plates were then washed with wash buffer (0.05% tween-20 diluted in 1x PBS) and blocked with 1% BSA-PBS) for 1 hour at room temperature. The plates were then washed, and samples and standards were added for a further 2 hours of incubation at room temperature. Secondary antibodies, followed by streptavidin-horseradish peroxidase and substrate solution (1x TMB) were then used, and the reaction was terminated by the addition of 2N H2SO4. The results were analyzed with a spectrophotometer (ELISA reader) at 450-nm and 570-nm wavelength. The final data were calculated by subtracting 570 nm from 450 nm.
Detection of PBMCs in patient blood samples by flow cytometry
The PBMCs were collected from patient blood using human lymphocyte separation medium. A total of 0.5×106 to 1×106 PBMCs were washed by 1x PBS and incubated with the indicated antibodies for 30 minutes at 4°C. For intracellular staining, cells were washed, fixed and permeabilized using the transcription factor staining buffer set (eBiosciences, #00–5523–00). Antibodies for intracellular staining were incubated with cells for 20 minutes at 4°C. Cells were washed, resuspended in 1x PBS, and acquired on the cytoFLEX LX Flow Cytometer. Data were analyzed using FlowJo V.10. The antibodies that were used are listed as follows: CD3 (PerCP/Cyanine5.5, clone UCHT1, catalog No. 300430, BioLegend), CD4 (APC/Cyanine7, clone SK3, catalog No. 317418, BioLegend), CD56 (PE-Cy7, catalog No. 985912, BD Biosciences), PD-1 (Brilliant Violet 421, catalog No. 367421, BioLegend), PD-L1 (APC, catalog No. 329707, BioLegend), CD68 (APC, catalog No. 333809, BioLegend), CD86 (FITC, catalog No. 555657, BD Biosciences) and Foxp3 (Brilliant Violet 421, catalog No. 320123, BioLegend), CD163 (APC/Cyanine7, catalog No. 333621, BioLegend), HLA-DR (Brilliant Violet 650, catalog No. 307649, BioLegend), CD11c (PerCP/Cyanine5.5, catalog No. 980610, BioLegend), and Fixable Viability Dye eFluor 506 (eBiosciences, #65–0866–18).
Statistical analysis
We used R software (version 4.0.3, http://www.R-project.org), GraphPad Prism (V.6.0) software, and SPSS (V.19.0) software to analyze and visualize the data. Nonparametric tests such as the Wilcoxon rank-sum test (comparison between two groups) and the Kruskal-Wallis test (comparison between more than two groups) were used for continuous variables unless otherwise noted χ2 test was used for categorical variables. Statistical significance was defined as P < 0.05.
Data availability statement
The raw sequence data have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA004110). The general information of data are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human, and the data files are available upon reasonable academic request.
Ethics approval and consent to participate
This study has been performed in accordance with the research protocol approved by IRB of the SYSUCC (reference No. B2022–071–01). All patients provided written informed consent to participate in this study. The research activities in this study conformed to the principles of the Declaration of Helsinki.
Results
Clinical characteristics of the subjects
A total of 144 patients diagnosed with RIS at the SYSUCC were identified (Supplementary Fig. S1). Most of these patients (118/144, 81.9%) had a disease history of NPCs. Major histologic subtypes of RIS in head and neck were fibrosarcomas and osteosarcomas, followed by undifferentiated pleomorphic sarcomas (UPS) or malignant fibrous histiocytoma (MFH). Twenty-four of 144 (16.7%) samples were defined as RIS without specific subtypes (Supplementary Table S1). All patients received radiotherapy before the diagnosis of RIS, with radiation doses ranging from 36 Gy to 80 Gy. The latency between the initial radiotherapy and the diagnosis of RIS ranged from 2 to 37 years (mean, 9.87 years). These RIS mainly had the French Federation Natinale des centres de Lutte Cotre le Cancer (FNCLCC) grade of G2 and G3.
We also identified 123 patients with primary sarcomas that mainly included fibrosarcoma, osteosarcoma, UPS/MFH as control, named the SARC group. Most fibrosarcomas (36/45, 80.0%) and osteosarcomas (21/29, 72.4%) in this group were located in head and neck. As shown in Supplementary Table S1, compared with the SARC group, the mean age of patients at diagnosis in the RIS group was older (49.0 vs. 38.9; P < 0.001). There was a significant difference in the FNCLCC grade between RIS and SARC groups (P < 0.001).
RIS and primary sarcomas have different gene expression profiles
WES and mRNA-seq were performed on available tumor samples and matched blood samples. In total, 46 WES and 16 mRNA-seq were obtained from the RIS cohort and 48 WES and eight mRNA-seq were obtained from the SARC group. The clinical information for samples with sequencing was shown in Supplementary Table S1. Most of the tumor samples for WES and all the samples for mRNA-seq in two groups were from the head and neck area. The RIS group for mRNA-seq contained 10 RIFSs and six RIOS, and the SARC group contained three fibrosarcomas and five osteosarcomas.
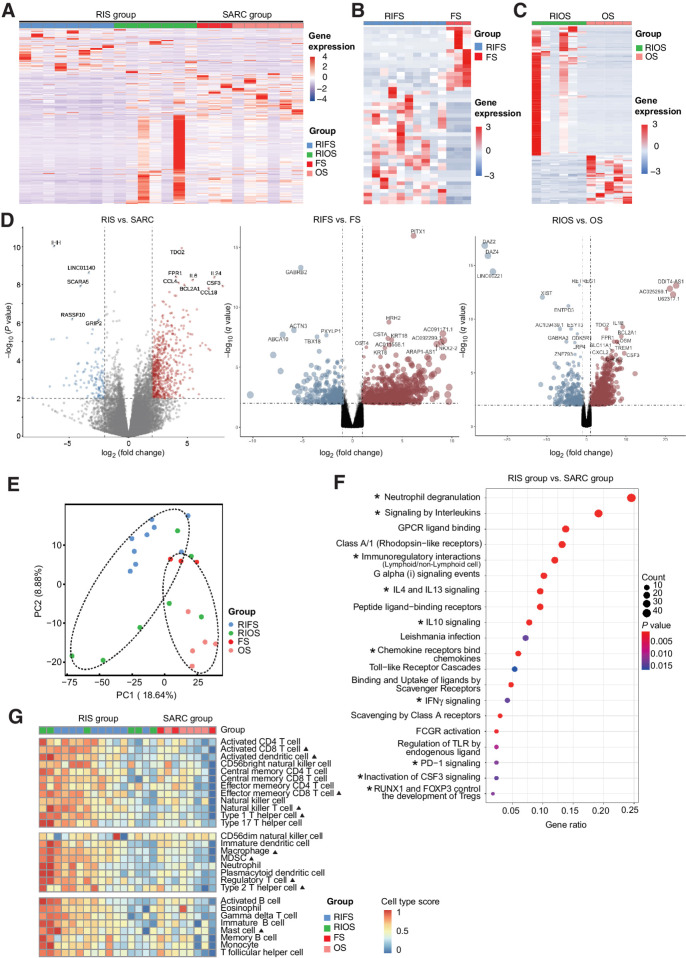
Transcriptome analysis between the RIS group and the SARC group revealed 977 differentially expressed genes (DEG; 756 upregulated and 221 downregulated; Padj. value < 0.05; Supplementary Table S2). Unsupervised hierarchical clustering in the heat map distinguished the RIS group from the SARC group by their gene expression profile (Fig. 1A) and also distinguished RIFS samples from fibrosarcoma samples and RIOS samples from osteosarcoma samples (Fig. 1B and C) in subtype comparison. Compared with the SARC group, expression levels of TDO2 (tryptophan 2,3-dioxygenase), FPR1 (formyl peptide receptor 1), CSF3 (colony-stimulating factor 3), IL6, IL24, CCL4 (C-C motif chemokine ligand 4), BCL2A1 (BCL2 related protein A1), and CCL18 (C-C motif chemokine ligand 18) were higher, while expression levels of IHH (indian hedgehog), LINC01140 (long noncoding RNA 01140), SCARA5 (scavenger receptor class A member 5), RASSF10 (Ras association domain family member 10), and GRIP2 (glutamate receptor-interacting protein 2) were lower in the RIS group (Fig. 1D). In comparison with primary sarcomas (fibrosarcomas and osteosarcomas), certain genes were significantly upregulated/downregulated in RIFS or RIOS samples shown in the volcano plots. A relatively small set of 18 genes was found coexistent in the DEG list for RIS versus SARC, RIFS versus fibrosarcoma, and RIOS versus osteosarcoma, including HRH2, CCL18, TDO2, CXCL3, LRG1, IL6, etc. (Supplementary Fig. S2A). In addition, the two subtypes of the RIS group, RIFS and RIOS, show certain differences in gene expression. The RIOS subgroup showed a transitional pattern between the RIFS subgroup and the SARC group (Fig. 1A). The RIFS samples had a higher expression level of genes LINC00221 (long intergenic nonprotein coding RNA 221), PTGER3 (prostaglandin E receptor 3), CALB2 (calbindin 2), etc. (Supplementary Fig. S2B). The PCAs showed the correlations among all RNA samples in Fig. 1E.
Figure 1.
Gene expression profiles of RIS and primary sarcomas. A, The unsupervised hierarchical clustering separating the RIS group and the SARC group according to their gene expression profiles. B and C, Heat maps showing differences in gene expression profiles between RIFS and fibrosarcoma subtypes (B) and between RIOS and osteosarcoma subtypes (C). D, Volcano plot showing upregulated and downregulated genes in the RIS group versus the SARC group, the RIFS group versus the fibrosarcoma group, and the RIOS group versus the osteosarcoma group. E, PCA separating samples of the RIS group and the SARC group. F, Twenty significantly upregulated pathways in the RIS group compared with the SARC group identified by functional annotations of DEG to the Reactome database. GPCR, G protein–coupled receptor; FCGR, Fc fragment of IgG receptor. *, immune-related pathways. G, Relative abundances of activated immune cells and immunosuppression-related cells in RIS and in primary sarcomas. ▴, P < 0.05 (RIS vs. SARC). FS, fibrosarcoma; OS, osteosarcoma; RIFS, radiation-induced fibrosarcoma; RIOS, radiation-induced osteosarcoma; RIS, radiation-induced sarcoma; SARC, primary sarcoma.
To investigate the functions of DEG, functional enrichment based on the Reactome pathway database was analyzed. Twenty significantly upregulated pathways in RIS were found (Fig. 1F), including neutrophil degranulation, signaling by interleukins, G protein–coupled receptor ligand binding, class A/1 (rhodopsin-like receptors), immunoregulatory interactions between lymphoid/nonlymphoid cells, G alpha (i) signaling events, IL4 signaling, IL13 signaling, IL10 signaling and peptide ligand−binding receptors. We also performed GO enrichment analyses of DEG and found a set of upregulated GO terms in RIS, including chemotaxis of various kinds of immune cells and cytokine activity (Supplementary Fig. S2C).
These results demonstrated that the gene expression patterns of RIS and primary sarcomas were different, especially in the identical subtypes between two groups. A few gene fusion and alternative splicing events were also identified in both groups (Supplementary Table S2). In summary, expression profiling results suggested that RIS might harbor some special transcriptomic alterations important for their tumorigenesis. Notably, expressions of immune-related genes and pathways were upregulated in RIS relative to primary sarcomas.
Immune infiltration and PD-1 expression level are higher in RIS than in primary sarcomas
Because the expressions of immune-related genes were higher in RIS than in primary sarcomas, we then investigated the differences in the tumor immune microenvironment between these two groups. The signaling pathway activation and relative immune cell infiltration were quantitatively measured by ssGSEA and CIBERSORTx (47). The results showed that NK T cells, activated CD8+ T cells, activated dendritic cells (DC), macrophages, effector memory CD8 T cells, Treg cells, MDSC, mast cells, and type 1/2 T helper cells were significantly more abundant in RIS than in primary sarcomas (Fig. 1G). In subtype comparison, CD56 bright NK cells and Tregs were significantly more abundant in RIFS than in fibrosarcomas, and mast cells were significantly more abundant in RIOS than in osteosarcoma (Supplementary Fig. S2D). Overall, most of the 28 types of immune cells were at higher levels in RIS than in primary sarcomas. These tumor-infiltrating immune cells might have important roles in the antitumor response.
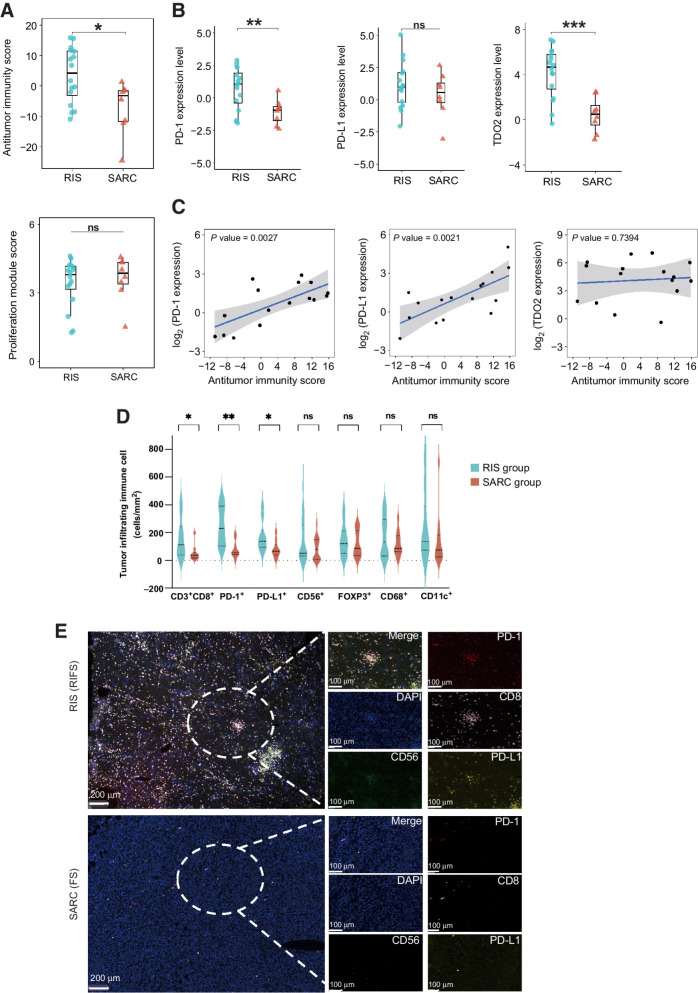
We subsequently calculated the antitumor immunity score and the proliferation module score of the two groups (Supplementary Table S2) and found that RIS had significantly higher antitumor immunity scores than primary sarcomas (P = 0.01422) but had similar proliferation module scores as primary sarcomas (P > 0.05; Fig. 2A). To understand the immune escape mechanisms of RIS, we analyzed expression levels of some immune checkpoints and immune-related genes, including PD-L1, PD-1, CTLA-4, IDO1, and TDO2. We observed that RIS expressed significantly higher levels of PD-1 and TDO2 (P < 0.003) and a slightly higher PD-L1 level (Fig. 2B). There was no difference in expression levels of CTLA-4 and IDO1 between the two groups (Supplementary Fig. S3A). The correlation analysis showed that PD-1 and PD-L1 expression levels were both positively correlated with antitumor immunity scores among RIS samples (Fig. 2C). Samples with higher antitumor immunity scores expressed higher mRNA levels of PD-L1. These results suggested that although RIS had a higher degree of immune infiltration, tumor cells might enhance immune evasion by increasing expressions of immune checkpoints PD-1 and PD-L1.
Figure 2.
The antitumor immunity of RIS. A, The antitumor immunity score (top) and the proliferation module score (bottom) of the RIS group and the SARC group. Box plot: the line inside the box indicates median, the top and the bottom borders of the box indicate upper and lower quartile. B, Expression levels of PD-1, PD-L1, and TDO2 in the RIS group and the SARC group in box plots. C, Correlations between expression levels of PD-1, PD-L1, and TDO2 and the antitumor immunity score in RIS. D, The summary of the density of tumor-infiltrating immune cells (cells/mm2) by mIF in the RIS group and the SARC group (15 specimens in each group). Solid lines indicate median and dotted lines indicate quartiles. E, Representative images of mIF staining showing immune infiltration in an RIS and a primary sarcoma. ***, P < 0.001; **, P < 0.01; *, P < 0.05.
To verify these findings, we performed mIF staining in 15 RIS samples and 15 primary sarcoma samples from exome/mRNA-seq specimens and calculated the cell density (cells/mm2) using the HALO platform. Consistent with the literature, primary sarcomas exhibited minimal infiltration of immune cells (48). Compared with primary sarcomas, RIS had a higher density of CD3+CD8+ T cells and higher immunofluorescence staining intensities of PD-1 and PD-L1 (Fig. 2D and E). Above all, the overview of the intratumoral immune landscape demonstrated that RIS had a higher level of tumor-infiltrating lymphocytes, especially CD8+ T cells, and can be categorized as inflamed tumors (49). Subgroup analysis revealed that the antitumor immunity score was slightly higher in RIFS and RIOS than in fibrosarcomas and osteosarcomas. However, neither the antitumor immunity score nor the proliferation module score was statistically different between RIOS and RIFSs, or RIFSs and fibrosarcomas, or RIOS and osteosarcomas (Supplementary Fig. S3B and S3C), which might be due to the limited sample size in subgroup analysis.
RIS and primary sarcomas have different gene mutations
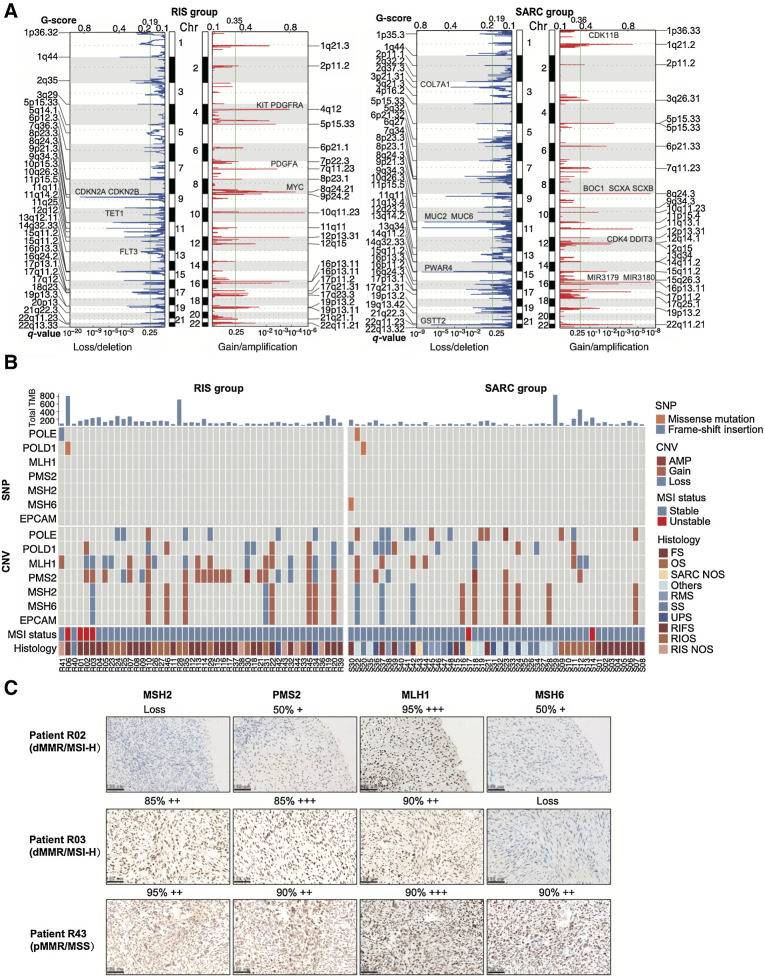
Most nonsilent somatic mutations identified in both RIS and SARC groups were single-nucleotide missense mutations; the majority were the C > T transition (Supplementary Fig. S3D). Figure 3A summarizes mutated genes in important pathways in RIS and SARC groups. The top 10 mutated genes in RIS were mucin families (MUC4, MUC3A, MUC16, MUC6), TP53, ODF1, PABPC3, AHNAK2, MUC17, and CDC27. TP53 was mutated in 13 of 46 analyzed RIS (28.3%), ranked only after genes in the mucin family. Mutation rates of FAT1 (10.9% vs. 0%; P = 0.025), FCGBP (15.2% vs. 0%; P = 0.005), and KCNS1 (15.2% vs. 0%; P = 0.005) were significantly higher in the RIS group than in the SARC group, while the mutation rates of TP53 (28.3% vs. 12.5%; P = 0.074) did not show statistical differences. In subtype analysis, the RIS group had 26 RIFS samples and 12 RIOS samples, while the SARC group had 14 fibrosarcoma samples and six osteosarcoma samples. The mutation rates of genes BCLAF1 (7.7% vs. 50.0%; P = 0.004), TRPM6 (0% vs. 21.4%; P = 0.037), LILRB1 (0% vs. 21.4%; P = 0.037), and CCDC30 (0% vs. 21.4%; P = 0.037) were significantly lower in RIFSs than fibrosarcomas, and the mutation rates of genes TMEM63A (0% vs. 21.4%; P = 0.037) and BCL11B (0% vs. 21.4%; P = 0.037) were significantly lower in RIOS than in osteosarcomas (Supplementary Fig. S3E).
Figure 3.
TMB and CNV in RIS compared with those in primary sarcomas. A, Heat maps for mutated genes in important pathways in the RIS group and the SARC group. NOS, not otherwise specified. B and C, COSMIC mutational signature analysis and nucleotide change type analysis (C) of RIS and primary sarcomas. D, TMB in RIS and primary sarcomas, including comparison between subtypes. The numbers in the plot indicate mean value. E, The predicted total tumor neoantigen burden (left) and the predicted strong binding neoantigen counts (right) in the RIS group and the SARC group. The numbers in the plot indicate mean value. Error bars indicate mean ± SD. ****, P < 0.0001.
We then analyzed mutational signatures in RIS and SARCs based on COSMIC mutational signatures and discovered signatures similar to COSMIC signatures 1 and 5 in both groups, but RIS showed a unique COSMIC signature 3 (Fig. 3B and C). Signature 1 is considered to be related to increased rate of spontaneous deamination of 5-methylcytosine and signature 3 is closely associated with DNA double-strand break (DSB) repair by homologous recombination (HR). These findings indicated potential mechanisms for the genome mutations in RIS and might provide clues for future investigations.
RIS have a higher burden of predicted strong MHC-binding neoantigens
TMB has been demonstrated to be an indicator of response to ICI treatment (50). We therefore analyzed the TMB of RIS and primary sarcomas of our cohorts. There was no significant difference in TMB between the RIS group and the SARC group (3.097 muts/MB vs. 2.737 muts/MB; P = 0.358). The TMB was slightly higher in RIS samples than SARC samples and slightly higher in RIFSs than fibrosarcomas, but the subtype comparison did not show significant differences (Fig. 3D). This finding could be ascribed to the relatively small sample size of different subtypes.
Results of tumor neoantigen inferred from the WES data showed that the number of total neoantigens of the RIS group was slightly, but not significantly, higher than that of the SARC group (mean, 255 vs. 241; P = 0.701; Fig. 3E). However, the mean count of neoantigens predicted to have a strong binding capability with MHC molecules was significantly higher in RIS than in primary sarcomas (mean, 185 vs. 96; P < 0.001). These results indicated a greater potential for tumor cells in RIS being recognized by activated immune cells during immunotherapy.
RIS have a different pattern of CNV and a higher frequency of being MSI-high than primary sarcomas
Patterns of CNV were also different between the RIS group and the SARC group (Fig. 4A). The frequency of copy-number loss was higher than that of copy-number gain in RIS samples, indicating that radiation could potentially induce more deletions in the tumor genome, consistent with previous reports (51). Among 57 chromosomal fragments with somatic CNV in RIS, 34 (59.6%) were loss or deletion, and the most frequent events were the loss of chromosome regions 9q21.3, 10q26.3, 11q11, 1q44, and 11p15.5 and the amplification of 10q11.23, 8q24.21, 4q12, 12p13.31, and 7p11.23. After filtered by genes listed in the COSMIC Cancer Gene Census for drug target discovery, the top five genes in RIS all had more frequent copy-number gains than SARCs including HEY1 (65.2% vs. 41.7%; P = 0.022), NCOA2 (65.2% vs. 43.8%; P = 0.037), PREX2 (63.0% vs. 43.8%; P = 0.038), ARHGAP5 (50.0% vs. 25.0%; P = 0.012), and BAZ1A (52.2% vs. 29.2%; P = 0.023; Supplementary Fig. S4A). In addition, we found more frequent copy-number gains of gene MYC (65.2% vs. 31.3%; P = 0.001), PDGFA (54.3% vs. 18.8%; P = 0.0003), PDGFRA (56.5% vs. 16.7%; P < 0.0001), and KIT (56.5% vs. 18.8%; P = 0.0002) in RIS. The top genes in RIS with copy-number losses more than SARCs included TET1 (60.9% vs. 27.1%; P = 0.001), CDKN2A (71.7% vs. 27.1%; P = 0.0001), CDKN2B (71.7% vs. 27.1%; P = 0.0001), and FLT3 (69.6% vs. 27.1%; P = 0.0001; Fig. 4A). The SARC group had a CNV pattern of different segments, with amplification of 8q24.3, 16p13.11, 1q21.2, 11q13.1, and 22q11.21 and loss of 15q11.2, 11q11, 11p15.5, 22q11.23, and 9p21.3. Subtype analysis also revealed different top genes with CNV between RIFSs and fibrosarcomas, RIOS, and osteosarcomas (Supplementary Fig. S4B and S4C).
Figure 4.
Genome instability and mutational signatures of RIS and primary sarcomas. A, Copy-number analysis of the RIS group and the SARC group. B, Bars showing TMB (top), heat maps summarizing alterations of POLE, POLD1, MLH1, PMS2, MSH2, MSH6, and EPCAM (middle) and predicted MSI status (bottom) in two groups. C, Representative images of IHC staining for MMR proteins in two MMR-deficient (dMMR)/MSI-H RIS and one MMR-proficient (pMMR)/MSS RIS.
We further analyzed the WES data to determine the microsatellite stability status. Figure 4B summarizes alterations in the main MMR genes MLH1, PMS2, MSH2, MSH6, and EPCAM, POLE, and POLD1 in the RIS and SARC groups. The majority of the 46 analyzed RIS were microsatellite stable (MSS); there were some copy-number alterations but no singe-nucleotide changes in MMR genes. Four RIS samples had higher MSI scores and were identified as MSI-high (MSI-H, 4/46, 8.70%) in the RIS group, and two samples were identified as MSI-H (2/48, 4.17%) in the SARC group (P = 0.430). Consistently, two RIS samples with the MSI-H status showed loss of nuclear staining for protein MSH2 and MSH6, respectively, whereas an MSS RIS sample showed positive nuclear staining for all MMR proteins (Fig. 4C).
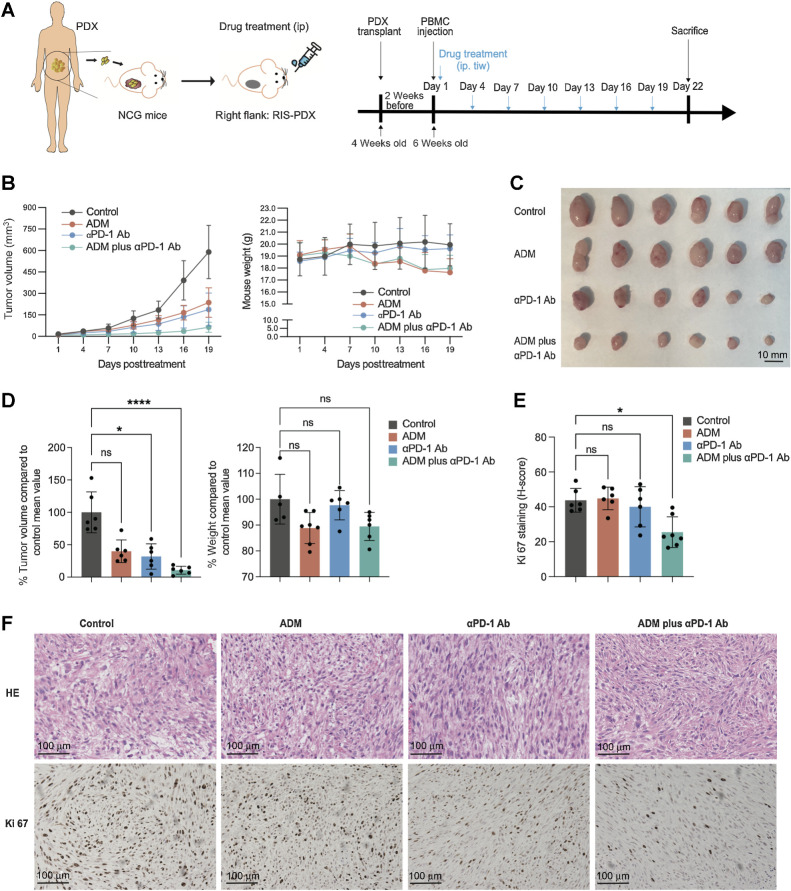
PD-1 inhibition shows promising antitumor effects in RIS PDX mouse model
On the basis of the findings previously described, we hypothesized that combining ICIs with standard chemotherapy might achieve better controls of RIS than chemotherapy alone. To verify this hypothesis, we first established a PDX model of RIS in NSG mice using a primary RIS sample obtained from a patient diagnosed with fibrosarcoma after radiotherapy for NPC. Twenty-four NSG mice were transplanted with the third-generation PDX, transfused with lymphocytes obtained from the corresponding patient, and divided into four treatment groups: the control group (PBS), the doxorubicin (ADM) group, the anti–PD-1 antibody (αPD-1 Ab) group, and the doxorubicin plus anti–PD-1 antibody group (Fig. 5A). As shown in Fig. 5B–D, tumors in the control group mice grew gradually, whereas those in mice of any other groups grew slower. Notably, mice in the ADM plus αPD-1 Ab group had the lowest tumor volume, which was significantly lower (P < 0.03) than that in the control group. There was an insignificant decrease in mouse weight observed in the ADM group and the combination group compared with the control group (P > 0.05; Fig. 5B and D). Tumor samples from these mice were collected and stained for Ki67, and the IHC score (H-score) was calculated using the HALO platform. Among these four groups, the combination group had the lowest H-score of Ki67, which was significantly lower than that of the control group (P = 0.030), whereas the anti–PD-1 group had a slightly lower, but without statistical significance, H-score than the control group (Fig. 5E and F).
Figure 5.
The RIS PDX mouse model responds better to the combination of anti–PD-1 antibody and chemotherapy than to chemotherapy alone. A, Diagrams for the establishment of the PDX mouse model and for the treatment and analysis of the model. B, The curves for tumor growth and mouse weight. C, Images of tumors at the end of the indicated treatment. D, Endpoint tumor volume and mouse weight. E and F, Quantification of Ki-67 (E) and representative images of HE staining and IHC staining for Ki-67 (F) of tumors at the end of indicated treatments. Scale bar = 100 μm. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; one-way ANOVA. Error bars indicate mean ± SD.
These findings suggested that the combination of anti–PD-1 therapy with chemotherapy potentially had better efficacy compared with the chemotherapy alone.
Patients with RIS have good response to anti–PD-1 antibody combined with chemotherapy
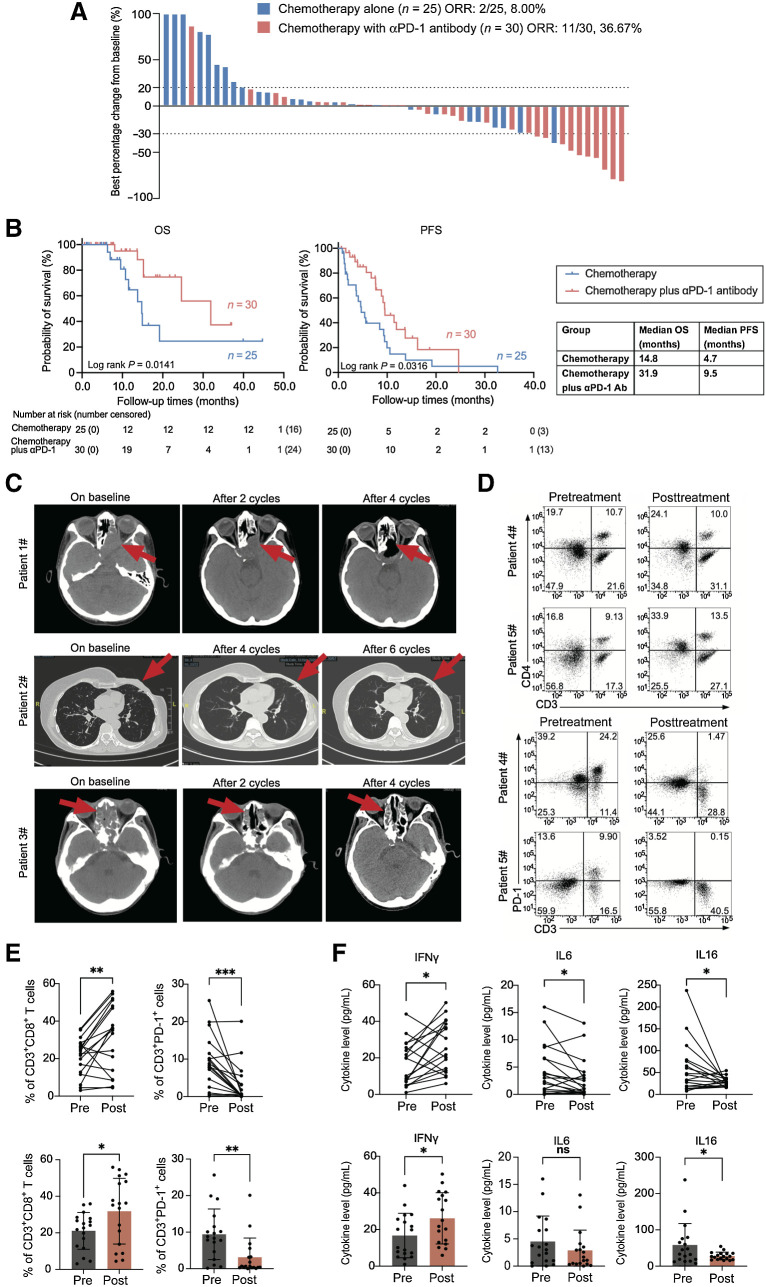
We then investigated the efficacy of the combination of an anti–PD-1 antibody with standard chemotherapy in patients with RIS in retrospective analysis. From January 2015 to October 2022, 30 patients diagnosed with advanced RIS were treated with toripalimab, an anti–PD-1 antibody, and doxorubicin-based chemotherapy every 3 weeks at our hospital (SYSUCC). In addition, we collected 25 patients with RIS treated with doxorubicin-based chemotherapy alone during this time period at our hospital and assessed the drug efficacy and patient outcome. All patients had evaluable imaging for efficacy. The overall objective response rate was 36.67% (11/30) for patients treated with the combination and was 8.00% (2/25) among patients treated with chemotherapy alone (P = 0.024; Supplementary Table S3; Fig. 6A). The addition of anti–PD-1 antibody improved the disease control rate from 64.00% (16/25), the median overall survival (mOS) from 14.8 months, and the median PFS (mPFS) from 4.7 months in the chemotherapy group to 96.67% (29/30; P = 0.003), 31.9 months (P = 0.014) and 9.5 months (P = 0.032), respectively, in the combination treatment group (Fig. 6B). Typical CT imaging in Fig. 6C showed tumor shrinkage in representative patients.
Figure 6.
Tumor and antitumor immunity response in patients with RIS treated with anti–PD-1 antibody in combination with chemotherapy. A, The best percentage changes of all the RIS volume in patients treated with chemotherapy alone or the combination of chemotherapy and anti–PD-1 antibody. Dashed lines indicate RECIST criteria for progressive disease (+20%) or partial response (–30%). B, OS and PFS of patients with RIS treated with the combination of chemotherapy and anti–PD-1 antibody or chemotherapy alone. C, CT imaging of representative patients with RIS treated with anti–PD-1 antibody combined with chemotherapy. Each row represents one patient. Red arrows indicate the tumor lesions. D, Typical dot plots for flow cytometry detection of immune cells in patient blood pre- and posttreatment. E, Percentages of CD3+CD8+ T cells and PD-1+CD3+ cells in pre- and posttreatment peripheral blood from 18 patients with RIS treated with the combination therapy. F, Pre- and posttreatment plasma levels of IFNγ, IL6, and IL16 in 18 patients in the combination treatment group. Paired t test, ***, P < 0.001; **, P < 0.01; *, P < 0.05. Error bars indicate mean ± SD.
Most of the AEs observed in the chemotherapy plus immunotherapy group were grade 1 or 2 (Supplementary Table S4). The most common treatment-related AEs were anemia (26, 86.7%), leukopenia (16, 53.3%), and hypoalbuminemia (9, 30.0%). No patients had any AEs of grade 4 or 5 and no treatment-related deaths occurred. The potential immune-related AEs were hypothyroidism (7, 23.3%).
Pre- and posttreatment blood samples from 18 patients with RIS treated with chemotherapy plus anti–PD-1 antibody were collected. Flow cytometry analysis of the PBMC showed that the percentage of CD3+CD8+ T cells was significantly higher, whereas the expression level of PD-1 in CD3+ cells decreased after anti–PD-1 treatment. Representative dot plots were shown (Fig. 6D and E). However, percentages of macrophages (total, M1, M2, or M1/M2 macrophage), NK cells, NKT cells, DC cells, and Treg cells were not significantly different (Supplementary Fig. S4D). Also, we detected the plasma level of cytokines (IFNγ, IFNβ, TNFα, TNFβ, IL2, IL6, etc.) from peripheral blood by ELISA. The results showed that after four to six cycles of anti–PD-1 treatment, the level of IFNγ was significantly upregulated, whereas levels of IL6 and IL16 were downregulated (Fig. 6F). Levels of other cytokines and ratios of IFNγ/IL10, TNFα/IL10, and IL6/IL10 were similar in pre- and posttreatment plasmas (Supplementary Fig. S4E).
Discussion
Our results together show that RIS have significantly higher immune cell infiltration, higher mRNA level of PD-1, higher predicted strong MHC-binding neoantigen load level, higher MSI-H rate, and slightly higher TMB than primary sarcomas and indicate that PD-1 blockade is a promising treatment approach for RIS.
Radiotherapy is an important cancer therapeutic approach; however, it poses a potential risk of causing secondary tumors. It is generally believed that ionizing radiation induces DNA damage of cells directly and affects the microenvironment of normal tissues to promote the cytokine production and transformation (52, 53). RIS is one of the most common secondary tumors after radiotherapy. It has been reported that RIS have distinct clinical characteristics from primary sarcomas and are more aggressive than primary sarcomas (54). However, the comprehensive information about transcriptomic and genomic characteristics and tumor microenvironment features of RIS remain largely unknown, which limits our understanding of this important clinical entity. In this study, we performed WES and mRNA-seq of human RIS and primary sarcomas to comprehensively characterize the mutational landscape and the expression profile of RIS, focusing on RIS developed after radiotherapy of NPCs in the head and neck area.
Our genomic analyses reveal that some genetic alterations occur more frequently in RIS than in primary sarcomas. Exposure to ionizing radiation involves activation of p53-dependent pathways (55). TP53, known as “the guardian of the genome,” has been shown to be the most commonly mutated gene in RIS, with a reported mutation rate of 58% (56). Consistent with the literature, our results show TP53 among most frequently mutated genes in RIS (13/46, 28.3%). A previous study reported that RIS and primary sarcomas share similar genomic alterations, including gains of chromosomes 1, 9, 14 and losses of chromosomes 10, 13, 16 at high frequencies. Our study shows more copy-number losses than gains in RIS and the loss of chromosome 9q as the most frequent event in RIS. The copy-number gain of MYC and copy-number losses of CDKN2A and CDKN2B have been reported (57–59). In addition, we find more frequent copy-number gains of HEY1, NCOA2, PREX2, ARHGAP5, BAZ1A, PDGFA, PDGFRA, and KIT, and copy-number losses of TET1 and FLT3 in RIS than SARCs. Further studies are needed to understand functional impacts of these CNV in RIS. Moreover, the number of predicted MHC-strong binding neoantigens is significantly higher and the TMB is trended toward higher in RIS than in primary sarcomas. TMB has been identified as an indicator for ICI treatment (60), and the predicted neoantigens also indicate a greater potential for tumor cell recognition by activated immune cells during immunotherapy (40). Furthermore, the predicted MSI-H rate among our RIS is 8.70%, which is higher than that of our SARC group (4.17%) and higher than that reported for primary sarcomas (2.8%; ref. 61). Also, we discovered a unique COSMIC signature 3 related to DNA DSB repair by HR in RIS. These results suggested that RIS exhibit a greater abundance of genomic alterations and higher genomic instability compared to primary sarcomas, which may be partly due to the DNA repair deficiency caused by radiation and needs further exploration.
By mRNA-seq, we identified 977 genes that are differentially expressed between RIS and primary sarcomas, found a small set of 18 genes coexistent in the DEG list for RIS versus SARC, RIFS versus fibrosarcoma, and RIOS versus osteosarcoma, and revealed a variety of pathways that are significantly upregulated in RIS. Notably, most of these pathways are related to tumor immunology. A previous study identified DEG between primary sarcomas and RIS and determined that the main gene ontologies related to RIS included response to oxygenated compounds and organic matter, positive regulation of lipid storage, and inflammatory response (57). DEG found in our study are different from those DEG; we speculate that such difference may be partly due to different pathologic subtypes and locations of RIS analyzed in these two studies. The previous study (57) focuses on angiosarcomas, leiomyosarcomas, and UPS, whereas most RIS in our study are fibrosarcomas, osteosarcomas, and UPS/MFH.
Consistent with our DEG findings, our subsequent analysis of the tumor immune microenvironment shows more abundant infiltration of immune cells in RIS than in primary sarcomas. We further found that RIS have significantly higher antitumor immunity scores than primary sarcomas but have similar proliferation module scores as primary sarcomas, suggesting that a major factor contributing to RIS development and progression is their ability to evade the immune surveillance. PD-1 and TDO2 levels are significantly higher, and the PD-L1 level is slightly higher in RIS than in primary sarcomas. The antitumor immunity score in RIS positively correlates with the expression levels of PD-1 and PD-L1. We thus can suggest that tumor cells may enhance their immune evasion by increasing the expression of PD-L1. These findings together with the abundance of intra-tumoral CD8+ T cells suggest the potential benefit of inhibiting PD-1 in RIS treatment.
Although ICIs exhibit poor efficacy in treating primary sarcomas, our comprehensive characterizations of RIS provide molecular evidence supporting the application of ICIs for RIS treatment. The potential of ICIs in treating RIS is experimentally supported by our preclinical and clinical studies. In the PDX model of RIS, the tumor volumes in mice treated with an anti–PD-1 and with the combination of an anti–PD-1 and chemotherapy are significantly lower than that of mice in the control group. Importantly, in retrospective analysis, the combination of chemotherapy and an anti–PD-1 antibody achieves better OS, PFS, and overall response rate than chemotherapy alone in patients with RIS. The plasma level of IFNγ is significantly upregulated, whereas plasma levels of IL6 and IL16 are downregulated together with a significant increase in the percentage of CD3 CD8+ T cells in the peripheral blood in patients with RIS treated with the combination.
However, the study still has some limitations. Due to the relatively small sample size of each subtype, the differentially revealed mutation or expression pattern may need further exploration in a larger size. The rarity of the disease and the lack of abundant clinical cases with sequencing samples limited our coherent sequencing-to-treatment research, which would require a larger cohort to achieve this purpose in the future.
In conclusion, we examined RIS by integrating WES and mRNA-seq, and our results clearly distinguish RIS from primary sarcomas and reveal multiple genetic alterations that may contribute to the development and progression of RIS, which deserve further investigation.
To our knowledge, this is the first study identifying higher levels of immune cell infiltration by transcriptome sequencing in RIS than in primary sarcomas. Furthermore, we reveal that inhibiting PD-1 may demonstrate good efficacy against RIS in both PDX mouse models and clinical patients. This is currently the largest cohort of the anti–PD-1 application in the treatment of patients with RIS. Our findings provide new insights into the genomic basis of RIS and may help to identify diagnostic and prognostic markers as well as molecular targets for the treatment of RIS.
Supplementary Material
The flowchart showing patient screening and identification, and analysis steps for mRNA-seq and WES data. FS: fibrosarcoma; OS: osteosarcoma.
RNA-seq analysis between RIS and RIS groups and some subtypes.(A) Venn plot showing differentially expressed genes (DEG) of FS and OS subtype comparison between RIS and SARC groups.(B) Volcano plot showing up-regulated and down-regulated genes in the RIFS group versus the RIOS group.(C) GO pathway enrichment of mRNA-seq between the RIS group and SARC group, and between the RIFS and RIOS subtypes.(D) Relative abundances of activated immune cells and immunosuppression-related cells in subtypes of RIS and primary sarcomas.
Comparison between RIS group and SARC group in mRNA-seq and WES data.(A) Expression levels of IDO and CTLA-4 in the RIS group and SARC group in mRNA-seq.(B) The anti-tumor immunity score and the proliferation module score of the RIFS group and the RIOS group.(C) The comparison of the anti-tumor immunity score and the proliferation module score between subtypes RIFS vs FS and between RIOS vs OS.(D) Mutation summary of WES data in the RIS and SARC groups.(E) Top 50 mutated genes in RIFS, FS, RIOS and OS subtype groups.
The landscape of copy number variants and results for blood detection.༈A༉Top genes in copy number changes in RIS and SARC groups filtered by COSMIC gene list.༈B༉Top genes in copy number changes in RIFS and FS subtypes.༈C༉Top genes in copy number changes in RIOS and OS subtypes.(D and E༉Flow cytometry (D) and ELISA (E) results of immune cells and cytokines in peripheral blood from patients.
Clinical characteristics of patients in RIS group and SARC group as a whole, or for WES, or for mRNA- seq.
Differentially expressed genes (DEG), possible fusion genes and alternative splicing events, and the anti-tumor immunity scores and proliferation module scores of each sample.
Clinical characteristics of patients treated with chemotherapy alone and chemotherapy plus anti-PD-1 antibody in RIS group.
Treatment-related adverse events of 30 RIS patients treated with chemotherapy plus anti-PD-1 antibody.
Acknowledgments
This study was supported by grants from the National Key Research and Development Program of China (no. 2021YFC2400601) and National Natural Science Foundation of China (no. 82072958, no. 82272699). We would like to express our gratitude for the support provided by the National Supercomputer Center in Guangzhou during our utilization of Tianhe-2 for processing high-performance sequencing data. This manuscript was edited at Life Science Editors.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This article is featured in Highlights of This Issue, p. 2739
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
D.-C. Hong: Data curation, formal analysis, validation, writing–original draft, writing–review and editing. J. Yang: Data curation, formal analysis, investigation, methodology, writing–review and editing. C. Sun: Software, formal analysis, visualization, methodology. Y.-T. Liu: Software, formal analysis, visualization, methodology. L.-J. Shen: Data curation, supervision, methodology, writing–review and editing. B.-S. Xu: Data curation, validation, investigation, writing–review and editing. Y. Que: Supervision, investigation, project administration, writing–review and editing. X. Xia: Conceptualization, resources, data curation, supervision, validation. X. Zhang: Conceptualization, resources, supervision, project administration, writing–review and editing.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5–29. [DOI] [PubMed] [Google Scholar]
- 2. Ramanathan RC, A'Hern R, Fisher C, Thomas JM. Prognostic index for extremity soft-tissue sarcomas with isolated local recurrence. Ann Surg Oncol 2001;8:278–89. [DOI] [PubMed] [Google Scholar]
- 3. Chow LQM. Head and neck cancer. N Engl J Med 2020;382:60–72. [DOI] [PubMed] [Google Scholar]
- 4. Liu YP, Zheng CC, Huang YN, He ML, Xu WW, Li B. Molecular mechanisms of chemo- and radiotherapy resistance and the potential implications for cancer treatment. MedComm 2021;2:315–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mito JK, Mitra D, Doyle LA. Radiation-associated sarcomas: an update on clinical, histologic, and molecular features. Surgical pathology clinics 2019;12:139–48. [DOI] [PubMed] [Google Scholar]
- 6. Plichta JK, Hughes K. Radiation-induced angiosarcoma after breast-cancer treatment. N Engl J Med 2017;376:367. [DOI] [PubMed] [Google Scholar]
- 7. Veiga LHS, Vo JB, Curtis RE, Mille MM, Lee C, Ramin C, et al. Treatment-related thoracic soft-tissue sarcomas in US breast cancer survivors: a retrospective cohort study. Lancet Oncol 2022;23:1451–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wei Z, Xie Y, Xu J, Luo Y, Chen F, Yang Y, et al. Radiation-induced sarcoma of head and neck: 50 years of experience at a single institution in an endemic area of nasopharyngeal carcinoma in China. Med Oncol 2012;29:670–6. [DOI] [PubMed] [Google Scholar]
- 9. Lou J, Jiang L, Dai X, Wang H, Yang J, Guo L, et al. Radiation-induced sarcoma of the head and neck following radiotherapy for nasopharyngeal carcinoma: a single institutional experience and literature review. Front Oncol 2020;10:526360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Snow A, Ring A, Struycken L, Mack W, Koç M, Lang JE. Incidence of radiation-induced sarcoma attributable to radiotherapy in adults: a retrospective cohort study in the SEER cancer registries across 17 primary tumor sites. Cancer Epidemiol 2021;70:101857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murray EM, Werner D, Greeff EA, Taylor DA. Postradiation sarcomas: 20 cases and a literature review. Int J Radiat Oncol Biol Phys 1999;45:951–61. [DOI] [PubMed] [Google Scholar]
- 12. Goy E, Tomezak M, Facchin C, Martin N, Bouchaert E, Benoit J, et al. The out-of-field dose in radiation therapy induces delayed tumorigenesis by senescence evasion. Elife 2022;11:e67190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang P, Zhao L, Zhu YJ, Qiu B, Guo SP, Li Y, et al. Prognosis of fibrosarcoma in patients with and without a history of radiation for nasopharyngeal carcinoma. Ann Surg Oncol 2017;24:434–40. [DOI] [PubMed] [Google Scholar]
- 14. Bjerkehagen B, Småstuen MC, Hall KS, Skjeldal S, Smeland S, Fosså SD. Why do patients with radiation-induced sarcomas have a poor sarcoma-related survival? Br J Cancer 2012;106:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gladdy RA, Qin LX, Moraco N, Edgar MA, Antonescu CR, Alektiar KM, et al. Do radiation-associated soft-tissue sarcomas have the same prognosis as sporadic soft-tissue sarcomas? J Clin Oncol 2010;28:2064–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dineen SP, Roland CL, Feig R, May C, Zhou S, Demicco E, et al. Radiation-associated undifferentiated pleomorphic sarcoma is associated with worse clinical outcomes than sporadic lesions. Ann Surg Oncol 2015;22:3913–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yeang MS, Tay K, Ong WS, Thiagarajan A, Tan DS, Ha TC, et al. Outcomes and prognostic factors of postirradiation and de novo sarcomas of the head and neck: a histologically matched case–control study. Ann Surg Oncol 2013;20:3066–75. [DOI] [PubMed] [Google Scholar]
- 18. Kadouri L, Sagi M, Goldberg Y, Lerer I, Hamburger T, Peretz T. Genetic predisposition to radiation-induced sarcoma: possible role for BRCA and p53 mutations. Breast Cancer Res Treat 2013;140:207–11. [DOI] [PubMed] [Google Scholar]
- 19. Kokkali S, Moreno JD, Klijanienko J, Theocharis S. Clinical and molecular insights of radiation-induced breast sarcomas: is there hope on the horizon for effective treatment of this aggressive disease? Int J Mol Sci 2022;23:4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sherborne AL, Lavergne V, Yu K, Lee L, Davidson PR, Mazor T, et al. Somatic and germline TP53 alterations in second malignant neoplasms from pediatric cancer survivors. Clin Cancer Res 2017;23:1852–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kansara M, Leong HS, Lin DM, Popkiss S, Pang P, Garsed DW, et al. Immune response to RB1-regulated senescence limits radiation-induced osteosarcoma formation. J Clin Invest 2013;123:5351–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hadj-Hamou NS, Ugolin N, Ory C, Britzen-Laurent N, Sastre-Garau X, Chevillard S, et al. A transcriptome signature distinguished sporadic from postradiotherapy radiation-induced sarcomas. Carcinogenesis 2011;32:929–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Florou V, Rosenberg AE, Wieder E, Komanduri KV, Kolonias D, Uduman M, et al. Angiosarcoma patients treated with immune checkpoint inhibitors: a case series of seven patients from a single institution. J Immunother Cancer 2019;7:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ju T, Foster D, Titan A, Najjar S, Bean GR, Ganjoo K, et al. Skin angiography assisted mastectomy in secondary breast angiosarcoma: complete clinical response after neoadjuvant immunotherapy. Breast J 2021;27:723–5. [DOI] [PubMed] [Google Scholar]
- 25. Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet 2019;394:64–80. [DOI] [PubMed] [Google Scholar]
- 26. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nirmal AJ, Regan T, Shih BB, Hume DA, Sims AH, Freeman TC. Immune cell gene signatures for profiling the microenvironment of solid tumors. Cancer Immunol Res 2018;6:1388–400. [DOI] [PubMed] [Google Scholar]
- 28. Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer immunogenomic analyses reveal genotype–immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep 2017;18:248–62. [DOI] [PubMed] [Google Scholar]
- 29. Hackl H, Charoentong P, Finotello F, Trajanoski Z. Computational genomics tools for dissecting tumor-immune cell interactions. Nat Rev Genet 2016;17:441–58. [DOI] [PubMed] [Google Scholar]
- 30. Jia Q, Wu W, Wang Y, Alexander PB, Sun C, Gong Z, et al. Local mutational diversity drives intratumoral immune heterogeneity in non–small cell lung cancer. Nat Commun 2018;9:5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy-number detection and visualization from targeted DNA sequencing. PLoS Comput Biol 2016;12:e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bonneville R, Krook MA, Kautto EA, Miya J, Wing MR, Chen HZ, et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol 2017;2017:PO.17.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, et al. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998;58:5248–57. [PubMed] [Google Scholar]
- 36. Cao P, Hu S, Kong K, Han P, Yue J, Deng Y, et al. Genomic landscape of ground glass opacities (GGOs) in East Asians. J Thorac Dis 2021;13:2393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinf 2010;11:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Blokzijl F, Janssen R, van Boxtel R, Cuppen E. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med 2018;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schenck RO, Lakatos E, Gatenbee C, Graham TA, Anderson ARA. NeoPredPipe: high-throughput neoantigen prediction and recognition potential pipeline. BMC Bioinf 2019;20:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Łuksza M, Riaz N, Makarov V, Balachandran VP, Hellmann MD, Solovyov A, et al. A neoantigen fitness model predicts tumor response to checkpoint blockade immunotherapy. Nature 2017;551:517–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shukla SA, Rooney MS, Rajasagi M, Tiao G, Dixon PM, Lawrence MS, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol 2015;33:1152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Indraccolo S, Lombardi G, Fassan M, Pasqualini L, Giunco S, Marcato R, et al. Genetic, epigenetic, and immunologic profiling of MMR-deficient relapsed glioblastoma. Clinical Cancer Res 2019;25:1828–37. [DOI] [PubMed] [Google Scholar]
- 43. Bui BN, Chevallier B, Chevreau C, Krakowski I, Peny AM, Thyss A, et al. Efficacy of lenograstim on hematologic tolerance to MAID chemotherapy in patients with advanced soft-tissue sarcoma and consequences on treatment dose-intensity. J Clin Oncol 1995;13:2629–36. [DOI] [PubMed] [Google Scholar]
- 44. Xiao W, Liang Y, Que Y, Li J, Peng R, Xu B, et al. Comparison of the MAID (AI) and CAV/IE regimens with the predictive value of cyclic AMP-responsive element-binding protein 3 like protein 1 (CREB3L1) in palliative chemotherapy for advanced soft-tissue sarcoma patients. J Cancer 2019;10:3517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arndt CA, Nascimento AG, Schroeder G, Schomberg PJ, Neglia JP, Sencer SF, et al. Treatment of intermediate risk rhabdomyosarcoma and undifferentiated sarcoma with alternating cycles of vincristine/doxorubicin/cyclophosphamide and etoposide/ifosfamide. Eur J Cancer 1998;34:1224–9. [DOI] [PubMed] [Google Scholar]
- 46. Yang J, Dong L, Yang S, Han X, Han Y, Jiang S. Safety and clinical efficacy of toripalimab, a PD-1 mAb, in patients with advanced or recurrent malignancies in a phase I study. Eur J Cancer 2020;130:182–92. [DOI] [PubMed] [Google Scholar]
- 47. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smolle MA, Herbsthofer L, Granegger B, Goda M, Brcic I, Bergovec M, et al. T-regulatory cells predict clinical outcome in soft-tissue sarcoma patients: a clinicopathologic study. Br J Cancer 2021;125:717–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hegde PS, Karanikas V, Evers S. The where, the when, and the how of immune monitoring for cancer immunotherapies in the era of checkpoint inhibition. Clin Cancer Res 2016;22:1865–74. [DOI] [PubMed] [Google Scholar]
- 50. Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Morton LM, Karyadi DM, Stewart C, Bogdanova TI, Dawson ET, Steinberg MK, et al. Radiation-related genomic profile of papillary thyroid carcinoma after the Chernobyl accident. Science 2021;372:eabg2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nguyen DH, Oketch-Rabah HA, Illa-Bochaca I, Geyer FC, Reis-Filho JS, Mao JH, et al. Radiation acts on the microenvironment to affect breast carcinogenesis by distinct mechanisms that decrease cancer latency and affect tumor type. Cancer Cell 2011;19:640–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rose LY, Halliwill KD, Adams CJ, Iyer V, Riva L, Mamunur R, et al. Mutational signatures in tumors induced by high and low energy radiation in Trp53 deficient mice. Nat Commun 2020;11:394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Riad S, Biau D, Holt GE, Werier J, Turcotte RE, Ferguson PC, et al. The clinical and functional outcome for patients with radiation-induced soft-tissue sarcoma. Cancer 2012;118:2682–92. [DOI] [PubMed] [Google Scholar]
- 55. Szołtysek K, Janus P, Zając G, Stokowy T, Walaszczyk A, Widłak W, et al. RRAD, IL4I1, CDKN1A, and SERPINE1 genes are potentially co-regulated by NF-κB and p53 transcription factors in cells exposed to high doses of ionizing radiation. BMC Genomics 2018;19:813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gonin-Laurent N, Hadj-Hamou NS, Vogt N, Houdayer C, Gauthiers-Villars M, Dehainault C, et al. RB1 and TP53 pathways in radiation-induced sarcomas. Oncogene 2007;26:6106–12. [DOI] [PubMed] [Google Scholar]
- 57. Lesluyes T, Baud J, Pérot G, Charon-Barra C, You A, Valo I, et al. Genomic and transcriptomic comparison of postradiation versus sporadic sarcomas. Mod Pathol 2019;32:1786–94. [DOI] [PubMed] [Google Scholar]
- 58. Guan J, Luo Z, Xiao Z, Xie Y, Lin L. Treatment of consistent BRAF/HRAS gene mutation and MYC amplification radiation-induced abdominal wall angiosarcoma with low-dose apatinib: a case report. BMC Cancer 2019;19:1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sturm SA, Strauss PG, Adolph S, Hameister H, Erfle V. Amplification and rearrangement of c-myc in radiation-induced murine osteosarcomas. Cancer Res 1990;50:4146–53. [PubMed] [Google Scholar]
- 60. Sha D, Jin Z, Budczies J, Kluck K, Stenzinger A, Sinicrope FA. Tumor mutational burden as a predictive biomarker in solid tumors. Cancer Discov 2020;10:1808–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gan M, Zhang C, Qiu L, Wang Y, Bao H, Yu R, et al. Molecular landscape and therapeutic alterations in Asian soft-tissue sarcoma patients. Cancer Med 2022;11:4070–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The flowchart showing patient screening and identification, and analysis steps for mRNA-seq and WES data. FS: fibrosarcoma; OS: osteosarcoma.
RNA-seq analysis between RIS and RIS groups and some subtypes.(A) Venn plot showing differentially expressed genes (DEG) of FS and OS subtype comparison between RIS and SARC groups.(B) Volcano plot showing up-regulated and down-regulated genes in the RIFS group versus the RIOS group.(C) GO pathway enrichment of mRNA-seq between the RIS group and SARC group, and between the RIFS and RIOS subtypes.(D) Relative abundances of activated immune cells and immunosuppression-related cells in subtypes of RIS and primary sarcomas.
Comparison between RIS group and SARC group in mRNA-seq and WES data.(A) Expression levels of IDO and CTLA-4 in the RIS group and SARC group in mRNA-seq.(B) The anti-tumor immunity score and the proliferation module score of the RIFS group and the RIOS group.(C) The comparison of the anti-tumor immunity score and the proliferation module score between subtypes RIFS vs FS and between RIOS vs OS.(D) Mutation summary of WES data in the RIS and SARC groups.(E) Top 50 mutated genes in RIFS, FS, RIOS and OS subtype groups.
The landscape of copy number variants and results for blood detection.༈A༉Top genes in copy number changes in RIS and SARC groups filtered by COSMIC gene list.༈B༉Top genes in copy number changes in RIFS and FS subtypes.༈C༉Top genes in copy number changes in RIOS and OS subtypes.(D and E༉Flow cytometry (D) and ELISA (E) results of immune cells and cytokines in peripheral blood from patients.
Clinical characteristics of patients in RIS group and SARC group as a whole, or for WES, or for mRNA- seq.
Differentially expressed genes (DEG), possible fusion genes and alternative splicing events, and the anti-tumor immunity scores and proliferation module scores of each sample.
Clinical characteristics of patients treated with chemotherapy alone and chemotherapy plus anti-PD-1 antibody in RIS group.
Treatment-related adverse events of 30 RIS patients treated with chemotherapy plus anti-PD-1 antibody.
Data Availability Statement
The raw sequence data have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA004110). The general information of data are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human, and the data files are available upon reasonable academic request.