Abstract
Antibodies and other new antibody-like formats have emerged as one of the most rapidly growing classes of biotherapeutic proteins. Understanding the structural features that drive antibody function and, consequently, their molecular recognition is critical for engineering antibodies. Here, we present the structural architecture of conventional IgG antibodies alongside other formats. We emphasize the importance of considering antibodies as conformational ensembles in solution instead of focusing on single-static structures because their functions and properties are strongly governed by their dynamic nature. Thus, in this review, we provide an overview of the unique structural and dynamic characteristics of antibodies with respect to their antigen recognition, biophysical properties, and effector functions. We highlight the numerous technical advances in antibody structure prediction and design, enabled by the vast number of experimentally determined high-quality structures recorded with cryo-EM, NMR, and X-ray crystallography. Lastly, we assess antibody and vaccine design strategies in the context of structure and dynamics.
Keywords: antibody, structure prediction, antibody structure determination, molecular dynamics, X-ray crystallography, NMR, cryo-EM, vaccine design, special formats
1. Antibody Structure Characteristics
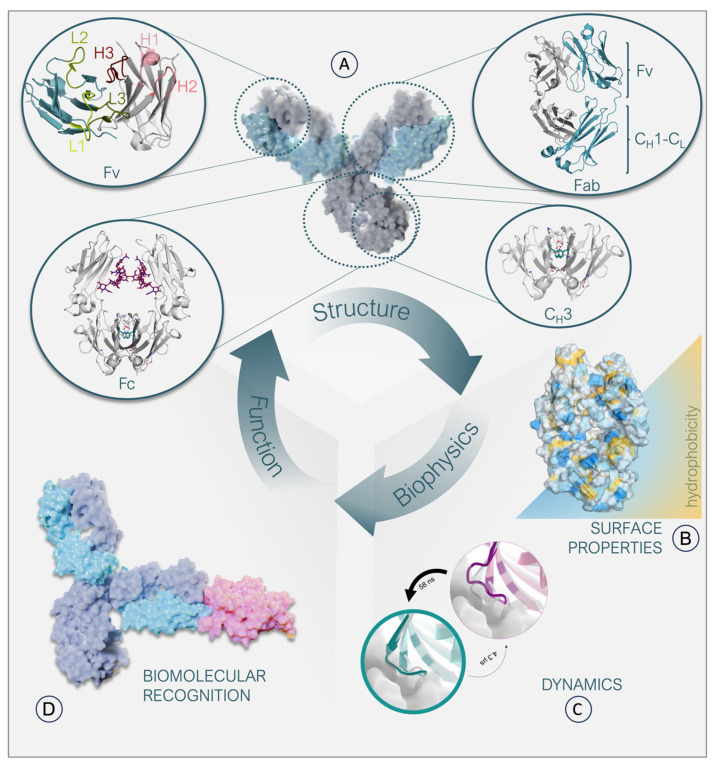
Antibodies and related formats have become enormously important as biological drugs in recent decades, with more than 120 therapeutic antibodies approved by the US Food and Drug Administration (FDA) for use in humans [1,2]. The vast majority of approved therapeutic antibodies share the canonical IgG architecture, i.e., a symmetrical Y-shaped structure (Figure 1A,D). The modular anatomy of an antibody has inspired numerous engineering efforts to optimize and fine-tune the respective biophysical properties and increase their therapeutic potential. The classic IgG structure is composed of two identical heavy chains as well as two identical light chains [3]. The stem of the antibody is built of two analogously paired heavy chains that form the crystallizable fragment region (Fc), comprising the CH3-CH3 and the CH2-CH2 dimers. The CH3 domains are directly involved in interactions with cell surface receptors, such as the neonatal receptor [4,5]. The CH2-CH2 dimer differs from the other interfaces as it contains glycan interactions in the interface instead of forming direct protein-protein contacts (Figure 1A). The arms of the “Y” are heterodimers by nature, each consisting of a paired heavy and light chain. As the arms are responsible for antigen binding, they are called antigen binding fragments (Fab). The Fab can be further subdivided into a constant and variable domain (Fv). The variable fragment comprises six hypervariable loops, which are mainly involved in antigen recognition, called the complementarity determining region (CDR) loops (Figure 1A) [3]. The highest diversity in length, sequence, and structure is concentrated on these CDR loops, which pose challenges in predicting antibody structures, their developability as tools or therapeutics, and in elucidating the antibody-antigen recognition mechanism. While less prominent, sequence changes in the framework region (FR) can also have significant consequences for biophysical properties and antigen binding (Figure 1B) [6,7,8,9,10,11]. For example, the relative orientation of the variable domains (VH-VL) influences the shape of the antigen binding site, deemed the paratope (Figure 1A,D) [12,13].
Figure 1.
Relationship between structure, biophysical properties, and function of an IgG antibody. (A): Surface representation of an antibody together with structural depictions of its individual domains. The heavy chain is colored gray, and the light chain is colored light blue. The CH2-CH2 interface glycan interactions are colored pink. Amino acids constituting the hydrophobic core in the CH3-CH3 interface are colored light green. The Fv contains the CDR loops, which are highlighted in shades of green and red, respectively. (B): Representation of a Fab surface, color-coded based on its hydrophobicity. Hydrophilic patches are colored blue, while hydrophobic patches are colored yellow. (C): Depiction of two distinct loop states, with the more probable state colored in cyan and the less probable state in purple. The thickness of the circles indicates the probability of each state. The transition times between the two states, which can occur at the nanosecond or microsecond level, are indicated by the thickness of the arrows. (D): Surface representation of an antibody-antigen complex. The heavy chains of the antibody are colored dark blue, and the light chains are colored light blue. A model antigen is represented in pink.
2. The Ig-Fold
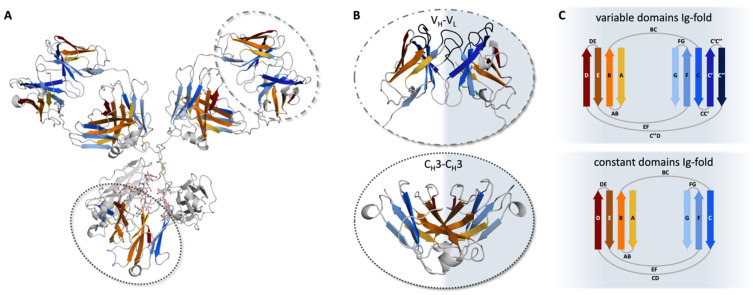
The individual antibody domains are characterized by the evolutionarily conserved immunoglobulin fold (Ig-fold; Figure 2A) [14]. This fold is common among various proteins covering diverse biological functions [15,16]. Each Ig-like domain of an antibody contains approximately 100–110 amino acids, forming two anti-parallel β-sheets stabilized by a buried intramolecular disulfide bridge [14]. The Ig-folds between constant and variable domains vary. Both include a similar β-sheet consisting of four β-strands (A, B, D, and E). The second β-sheet varies with the constant domain consisting of three β-strands (C, F, and G), while the variable domain features five strands (C, C′, C″, F, and G; Figure 2B,C). While the second β-sheet builds up the VH-VL interface, the pairing of the constant domains (CH1–CL and CH3–CH3) is facilitated by the first β-sheet (Figure 2C) [3,17,18,19]. Due to the assembly of the four chains, four to five interfaces are formed depending on the isotype of the heavy chain (Figure 2A) [20]. Generally, the chains belonging to the human IgG structure dimerize, forming either heterodimers, e.g., VH–VL and CH1–CL, or homodimers (CH2–CH2 and CH3–CH3).
Figure 2.
Pairing preferences of the Ig-folds in antibodies. (A) Crystal structure of an IgG antibody (PDB: 8TP9) [21] in cartoon representation. In orange and blue, the two beta-sheets of the Ig-fold are represented, respectively. The gray-shaded regions illustrate loops, or CH2-CH2 domains. VH-VL and CH3-CH3 domains are highlighted by dashed and dotted circles, respectively. (B) VH-VL (top) and CH3-CH3 (bottom) dimers are shown as cartoons. The CDR loops in the Fv are shown in gray, while the framework is colored based on the respective sheets, according to (A). (C) Schematic depiction of the Ig-fold. Variable domains (top) consist of two β-sheets, one with four (D, E, B, and A) and the other with five strands (C″, C′, C, F, and G). The constant domains (bottom) are also made up of two β-sheets, again one with four (D, E, B, and A) and the other with three strands (C, F, and G).
3. Antibody Structure Prediction
Acquiring high-resolution structures of an antibody of interest can be a laborious and resource-intensive task. Therefore, antibody structure prediction can significantly inform and expedite the antibody engineering process [22]. Accurate prediction of protein structures from their amino acid sequences is a critical challenge in computational chemistry [23,24,25]. Precise models could greatly enhance our comprehension of the physical and biological properties of existing therapeutic antibodies and facilitate the search for superior binders, thus positively influencing the development of future therapeutics [3,26].
The structure of most parts of an antibody, including the constant regions and the framework of the variable regions, is highly conserved [27]. Therefore, it is rather straightforward to model. However, the CDR loops in IgG-like antibodies exhibit significant structural diversity [28,29]. Where five of the six CDR loops can be represented by a set of canonical cluster conformations [30,31,32], the CDR-H3 loop has proven to be difficult to model accurately [29,33]. Furthermore, the multimeric structure of antibodies, where multiple non-covalently bound domains interact to form the complete antibody, makes it paramount to include the orientation of these domains [33,34,35,36]. For these reasons, the main conundrum in antibody structure prediction is that the antigen binding site, often the primary region of interest, is also the hardest to model [37]. Over the years, various methods have been developed to tackle this problem [26,38,39,40,41].
Template-based approaches, such as homology modeling, use the structural information from previously measured protein structures to build a model for a given sequence. Although these methods yield good results for most parts of the antibody, including the five canonical CDR loops, they are limited by the availability of structural data, particularly for antibody-like constructs such as bispecific antibodies or nanobodies. Hybrid methods, such as Rosetta antibody [42] or ABodyBuilder [43], instead include ab initio prediction methods to generate better loop ensembles, fill the gaps of missing data, and improve the modeling of uncertain regions. Recent, exhaustive reviews offer an overview of the vast array of available methods [26,38,39,40,41].
For several years, artificial intelligence-driven methods have advanced in predicting protein structures from their amino acid sequences. However, available protein structure prediction tools, such as AlphaFold2 [44,45], are less reliable for predicting the structures of CDR loops and multimeric proteins like antibodies [46]. Recent models tailored to describe CDR loops, such as DeepAB [47], IgFold [48], or ABlooper [49], have shown promising results in predicting the structures of CDR loops with greater accuracy. As highlighted by recent reviews, this is a rapidly evolving field of study set to revolutionize antibody in silico design [40].
Future tools may be further improved by incorporating information on the dynamic nature of the protein, as a description through a single structure may not capture all relevant properties. Especially the CDR loops are known to be notoriously flexible, and property prediction might therefore suffer from insufficient sampling of their structural ensemble. In addition, while most applications aim to model antibodies close to the crystal structure, these might be distorted compared to the structure in solution, which can be especially problematic for paratope predictions [50,51]. Including information on the accessible structural ensemble in solution through experimental methods such as NMR spectroscopy or cryo-EM as well as through computational methods such as molecular dynamics simulations may provide valuable insights.
It is imperative to exercise caution and thoroughly scrutinize the generated models while employing any modeling approach. In a late comparison of various antibody modeling tools, unphysical inaccuracies were observed, which may affect the antibody’s predicted properties. [52] Notably, a comprehensive comparison of the available antibody modeling tools, including the new artificial intelligence-driven methods, is missing. As the last Antibody Modeling Assessment took place over 8 years ago [22,53,54], it might be time for another large-scale blind study assessing the current state and helping address the existing challenges in antibody 3D structure modeling.
4. Antibody Dynamics
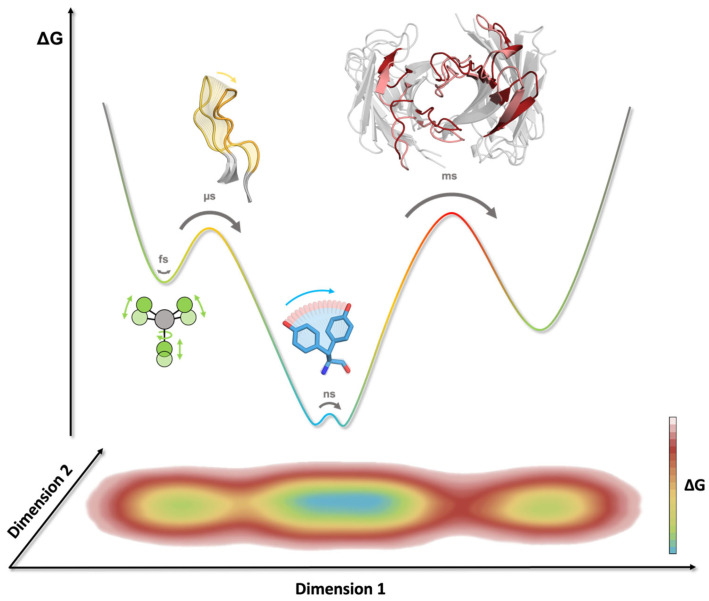
Protein dynamics are a central driving force in all biological processes. Ultimately, protein functions are governed by their dynamic nature, and the same applies for antibodies (Figure 1C) [55,56]. Molecular dynamics simulations allow the characterization of functionally relevant structural rearrangements in atomistic detail as a function of time [57]. The high dimensionality of the conformational space of an antibody leads to a large and rugged free energy surface [55]. The depth of an energetic minimum reflects the probability of a specific conformational state. To transition from one state to another, the system needs to overcome the energetic barrier that separates them. Therefore, the height of this barrier determines the probability of a transition, i.e., the timescale for a conformational change to occur [50]. Thus, resulting from the multidimensionality of the energy landscape representing an antibody, different rearrangements can occur on different timescales. While subtle movements, such as bond vibrations, side-chain fluctuations, and variations in the interdomain orientations, are separated by low energy barriers and can therefore be observed on the nano-to-microsecond timescale, large loop rearrangements in the binding site can take milliseconds or even longer (Figure 3) [50,56].
Figure 3.
Timescales of conformational rearrangements in antibodies. The energy barriers correspond to the timescale required to overcome and assess different conformational states. A 1D representation of the free energy landscape (top) and 2D projection highlight the multidimensional character of certain movements and reaction coordinates. Bond vibration, rotation, and stretching can be observed on the femtosecond (fs) timescale (indicated in green), while sidechain motions can occur within the nanosecond (ns) timescale (highlighted in blue). CDR loop rearrangements take place on the micro- to millisecond (µs/ms) timescale (colored in yellow), whereas transitions between different paratope states can occur on the millisecond (ms) timescale (indicated in red).
Because the antibody scaffold can recognize and accommodate diverse antigens with distinct motifs, Pauling and Landsteiner proposed in the 1940s that antibodies follow the concept of conformational diversity. In other words, a single antibody can adopt various conformations and potentially recognize multiple antigens, thereby directly increasing the size of the antibody repertoire [58,59]. This idea of having an ensemble of pre-existing antibody conformations in solution out of which the functional ones are selected and expanded is in line with the conformational selection paradigm. In agreement with these hypotheses, the antigen binding site exists as multiple interconverting paratope states, revealing correlated CDR loop rearrangements that can result in shifted interdomain orientations [11,13,60,61]. The dominant antibody paratope state in solution has been frequently pointed out to coincide with the binding competent conformation, while unbound crystal structures can be distorted by crystal packing effects [28,62]. Additionally, it has been demonstrated that antibody-antigen docking can profit from incorporating different antibody paratope states and their respective probabilities [63]. Another critical aspect influencing and co-determining the conformations and properties of the antigen binding site are specific antibody framework residues. The importance of non-CDR loop amino acids should not be omitted, since up to 22% of residues that can interact with antigens fall outside the traditionally defined CDR loops [6,7,8,9,10,64,65]. Various studies have structurally and dynamically characterized the role of framework mutations on the CDR loops and the relative VH–VL interdomain orientations and showed that a single-point mutation in the framework can influence antibody affinity and specificity. In particular, residue H71 located in the HV4 loop (Chothia nomenclature) determines the canonical conformation of the CDR-H2 loop and consequently affects the shape of the paratope [6,7,8,9]. Even the low-population states that occur more frequently near hydrophobic surfaces can play an important role in understanding processes such as aggregation or chemical modifications. In fact, these interactions with hydrophobic surfaces can result in a population shift towards more hydrophobic conformations, which are more likely to aggregate. Thus, accounting for the high conformational diversity of antibodies by considering them as ensembles in solution can advance the antibody development process, as the identification of potential liabilities and optimization of biophysical properties, such as hydrophobicity and electrostatics, can be facilitated (Figure 1B) [66,67,68].
Other critical aspects in antibody dynamics and engineering are the interdomain orientations (VH-VL, CH1-CL) and the elbow angle [69,70,71]. The majority of the interdomain movements have been reported to occur in the low nanosecond timescale (<10 ns). Additionally, for the VH-VL, it has been shown that the slow components of the motions (>10 ns) correlate with CDR-loop rearrangements [12,13,50,72]. In line with the surprisingly fast VH-VL interdomain dynamics, we also find fluctuations in the same timescale for the elbow angle, which increase the variability of the whole Fab, enhancing the bivalent binding of antigens [13,73]. The elbow angle is defined as the angle between the pseudo-2-fold axes relating VH to VL and CH1 to CL. Previous studies have demonstrated that naturally occurring forces as well as antibody engineering approaches influence the dynamics of elbow and interface angles [6,13,60,72,73,74]. For example, varying one single so-called Vernier-zone residue (the region of the framework anchoring the CDR loops) or using a different combination of germline heavy and light chains can alter not only the CDR loop conformations but also the VH-VL interface orientations [6,60]. Additionally, the type of the light chain has been determined to affect elbow and interface orientations, as λ-light chain antibodies reveal shifts and higher diversities in possible elbow VH-VL and CH1-CL angles [13,71]. Furthermore, maintaining a specific orientation of the variable domains has been shown to be important during humanization processes to preserve the antigen-binding properties [13,75].
5. Antibody-Antigen Recognition and Structure Guided Vaccine Design
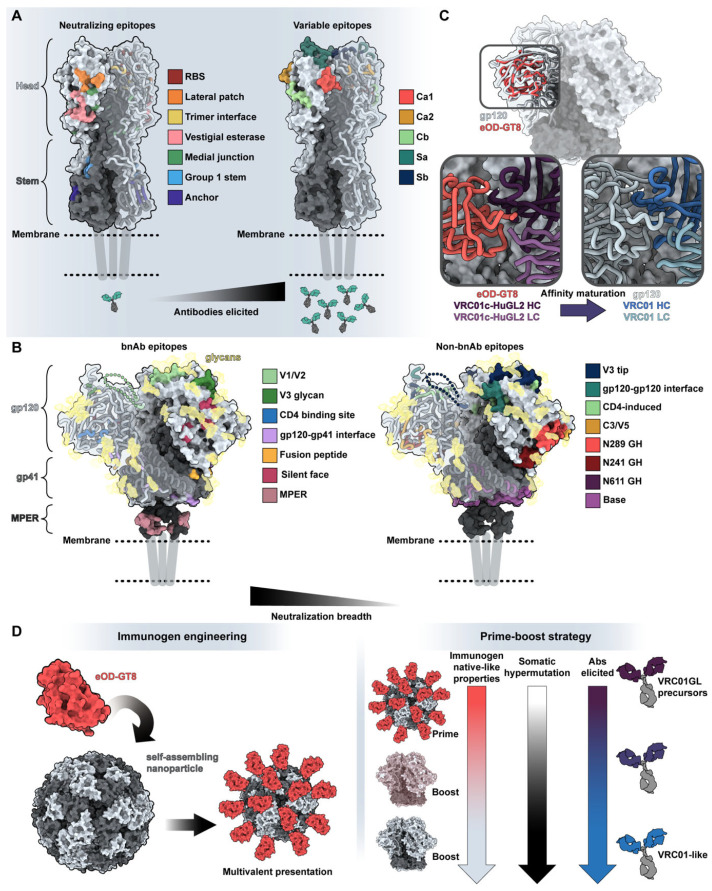
Understanding the structure of antibody-antigen interfaces, molecular dynamic constraints, and consequences of antibody maturation are crucial for the development of next-generation vaccines. The established method of vaccine development, which involves introducing native antigens to the immune system through inactivated or weakened pathogens, proves inefficient for developing protective immunity against challenging pathogens like influenza viruses and the human immunodeficiency virus (HIV). Due to genomic heterogeneity, high evolution and mutation rates, and competing immune responses of varying quality (Figure 4A,B), wild-type immunogens from these pathogens are insufficient at eliciting broadly protective antibody responses.
In a process termed “rational vaccine design” or “structural vaccinology”, high-resolution structural information about antibody–antigen interactions is leveraged to design immune-focusing vaccines [76,77,78]. Pathogens that escape immune protection utilize variable epitopes on their surface proteins to distract antibody responses away from conserved and neutralizing sites. By leveraging crucial interactions of broadly neutralizing or protective antibodies as well as identifying interactions that allow pathogens to overcome these responses, tailored immunogens can train the immune system to home in on the most vulnerable sites of the pathogen.
For example, the hemagglutinin (HA) surface protein of influenza viruses has high sequence variation in the head region but considerable conservation in the stem region [79,80]. The head is significantly more immunogenic, drawing away antibody responses from the conserved stem (Figure 4A). To elicit protective antibody responses, a vaccine will need to redirect immune responses to the stem region. One successful strategy involves focusing antibody responses to the stem region through chimeric HA vaccination in human subjects: by successive immunization of HAs with the same stem region but exotic head regions, broadly-reactive stem antibody responses were recalled and matured [81,82]. Another effective approach used nanoparticles displaying just the stem region to induce broadly reactive stem antibodies [83,84,85,86].
Sometimes, additional specificity to particular epitopes and even targeting of germline genes are needed to elicit protective antibody responses. Due to high mutation rates and immunodominance in the Envelope glycoprotein (Env) of HIV (Figure 4B), only highly specific lineages of antibodies will provide protection. Iterative vaccinology and structural comparison of antibodies to Env (Figure 4C) led to the design of epitope-specific immunogens that guide specific precursors of protective antibodies, a method known as “germline targeting” [87,88,89]. Immunization strategies often include priming with germline-targeting immunogens, followed by boosts by immunogens with increasing structural/sequence similarity to native Env epitopes. This strategy intends to increase the somatic hypermutation found in the B-cell repertoire and the neutralizing behavior and breadth of elicited antibodies with each successive boost.
Indeed, in a landmark clinical trial, an immunogen (eOD-GT8, Figure 4D) displaying a single epitope on a scaffold with a masked antigenic surface resulted in the engagement of broadly neutralizing antibody (bnAb) precursor B cells [90]. Through sequential boosting, the desired antibody lineage increased in somatic hypermutation and affinity.
Obtaining high-resolution structures of broadly protective antibodies in complex with antigens is crucial for advancing the development of next-generation vaccines. Research into the interplay between antibody structure, function, dynamics, and immunogenetics will not only inform iterative vaccine design but also be leveraged to engineer novel antibody therapeutics.
Figure 4.
Structure guides vaccine design. (A) Neutralizing and variable epitopes of influenza’s H1 hemagglutinin protein (PDB: 7T3D) [91]. Neutralizing epitopes on the left are colored based on a distance of <5 Å between residues of known antibodies and H1 HA (RBS with CH65, PDB: 5UGY [92]; lateral patch with 2B05, PDB: 7T3D [91]; trimer interface with FluA-20, PDB: 6OC3 [93]; medial junction PDB: 8TP9 stem with CR6261, PDB: 3GBN [94]; anchor with 222-1C06, PDB: 7T3D [91]; vestigial esterase domain with H5-specific antibody H5M9, PDB: 4MHH [95]). Immunodominant head epitopes associated with high variability and narrow neutralization breadth are shown on the right [96]. (B) Epitopes associated with broadly neutralizing versus limited neutralization breadth on HIV-1’s Env glycoprotein in the prefusion (surface representation of BG505-SOSIP, PDB: 6VFL) [97] and CD4-bound (ribbon representation of B41 SOSIP.664 bound to soluble CD4, PDB: 5VN3) [98] states. Broadly neutralizing epitopes on the left are colored based on a distance of <5 Å between residues of known antibodies and Env (V1/V2 with PG9, PDB: 7T77 [88]; V3 with 10-1074, PDB: 7UCG [99]; CD4 binding site with 3BNC117, PDB: 4JPV [100]; gp120-gp41 interface with 8ANC195, PDB: 5CJX [101]; fusion peptide with ACS202, PDB: 6NC2 [102]; silent face with SF12, PDB: 6OKP [103]; and MPER with 10E8, PDB: 4G6F [104], and 6VPX [105]). Epitopes that generate antibodies without neutralizing breadth are depicted on the right. For simplicity, glycans are depicted as transparent gold surfaces and are not highlighted based on engagement by antibodies. (C) Comparison of germline-targeting immunogen eOD-GT8 (PDB: 5IES) [106] with BG505-SOSIP (PDB: 6VFL) [97]. Insets indicate the differences in antigen engagement by VRC01-class precursor antibody VRC01-cHuGL2 (PDB: 5IES; left) [106] and bnAb VRC01 in complex with gp120 (PDB: 3NGB; right) [107]. (D) General overview of germline-targeting immunization from immunogen design (left) to overall prime-boost strategies (right). Notably, germline-targeting immunogen eOD-GT8 was designed to bind with high affinity and breadth to germline-reverted antibodies and displayed on a multivalent nanoparticle, the 60-mer lumazine synthase (PDB: 1NQU) [108], to increase immune trafficking and engagement of rare B cells [109].
6. State-of-the-Art Experimental Structure Determination
Structural characterization of protective antibodies bound to their cognate antigen is one of the major research goals of rational vaccine design and vaccinology efforts [110,111]. High-resolution structures are key to understanding the molecular basis of antibody-antigen recognition and thereby complementing other biochemical and biophysical characterizations. These empirical structures are essential in providing unparalleled insights into the mechanisms underlying neutralization and the interactions that contribute to its breadth and potency [112]. The three major structural techniques—NMR spectroscopy, X-ray crystallography, and cryo-electron microscopy (cryo-EM)—remain the most common and widely used methods for epitope mapping at an atomic level. Of these three techniques, X-ray crystallography is the most well established, providing structural details at atomic resolution to elucidate antibody recognition [113]. Up to now, the majority of structures available in the Protein Data Bank (PDB) [114] are crystal structures, emphasizing the tremendous contributions of X-ray crystallography to the field [115]. Crystallography has enabled invaluable structural insights to assess antibody–antigen recognition, characterize antibody affinity maturation, and laid the foundation for antibody structure prediction [116]. A pre-requisite for high-resolution crystal structures are high-quality crystals that diffract, which often requires significant amounts of time and resources. Moreover, structures obtained in the crystalline state may not accurately capture the dynamic behavior of a complex due to crystal packing constraints.
NMR spectroscopy has been extensively used for the biochemical characterization of protein–protein interfaces [117], including antibody–antigen complexes [118]. In a classic NMR epitope mapping experiment, the antigen is isotopically labeled. Upon mixing the antigen with the antibody of interest, NMR signals will shift from residues in contact with the paratope relative to the signals of the free antigen, thus providing information on the epitope [119]. Key advantages of NMR are its ability to provide dynamic information on antibody-antigen binding and that the samples are studied in solution at near physiological conditions. Unlike X-ray crystallography, NMR spectroscopy can provide an ensemble representation of the protein or complex of interest, which takes into account its inherent flexibility and dynamic behavior in solution [120]. By analyzing the chemical shifts and nuclear Overhauser effects (NOEs) of a protein or complex in solution, NMR data can reveal information about its secondary and tertiary structure as well as its conformational ensemble. This information is critical for understanding the function and stability of proteins and can inform protein engineering efforts. Still, NMR has relatively low throughput for two central reasons: (1) samples require isotopic enrichment, and (2) users need extensive and time-consuming training for successful data collection and processing. Furthermore, NMR spectroscopy is limited to studying only small-sized complexes due to molecular tumbling constraints [121]. This size constraint often necessitates the use of specialized NMR techniques, such as TROSY, to enable the study of larger molecular complexes.
Cryo-EM is fast emerging as a relatively high-throughput structural technique that can study proteins in physiologically relevant solutions to and investigate proteins and complexes between ~100 and 1000 kDa. Moreover, cryo-EM has fewer sample requirements for successful structure determination than X-ray or NMR [122]. Largely owing to the “resolution revolution” and further advancements in data collection technologies—which brought the cryo-EM field from “blobology” to high resolution—have facilitated the relatively routine acquisition of 3–4 Å resolution structures, and in many cases even higher [123,124].
Further, the dynamic behavior, on a global or domain-level scale, of antibody complexes can be readily and often routinely assessed using cryo-EM. Moreover, implementing a data processing technique called focused classification can provide high-resolution details of epitope-paratope interfaces. In addition to determining high-resolution structures, the single-particle nature of EM allows the processing of heterogeneous samples, which has been leveraged to perform negative stain electron microscopy polyclonal epitope mapping (nsEMPEM). This method uses antigens as bait in Fab-digested or full IgG serum or plasma samples to purify polyclonal antigen-antibody complexes from vaccinated or infected animals or human participants. After imaging and processing, specific epitopes targeted by a polyclonal antibody response can be determined [125], thereby enabling a relatively complete mapping of the immunogenic landscape of an antigen as well as allowing the temporal tracking of an individual’s immune response by sampling over time. These data can be expanded using cryo-EMPEM to obtain high-resolution data on polyclonal antibody responses. When combined with B-cell repertoire data, cryo-EMPEM can potentially identify and discover novel antibodies by predicting antibody sequences from cryoEM density maps [126]. CryoEMPEM does require intensive sample preparation as well as extensive computational resources for data processing.
7. Antibody Engineering—Design of Special Formats/Interface Characterization
7.1. The Role and Organization of Constant Domains
In the mid-twentieth century, it was common belief that the variable and constant domains of antibodies act independently [127]. This was supported by genetic studies, which demonstrated that an immunoglobulin is encoded by different gene segments: V(D)J genes carry variable region information while the C segment encodes the constant domain [128]. However, this hypothesis was soon challenged, and later studies proved that the constant domains can affect the affinity and specificity of an antibody towards its antigen [129,130]. For instance, according to the model proposed by Huber et al., the antigen binding causes a stiffening of the antibody molecule, with consequent folding of the hinge peptide, thus facilitating the CH1-CH2 interaction [131]. Further, the constant regions determine the Ig isotype, which plays a central role in defining the binding affinity to the antigen due to conformational changes of the CH1 domains [127,132,133]. Therefore, Ig-like constant domains are of interest since they bind to effector cells, which activate the immune response, and they contribute to binding specificity [20,127,134,135].
As the name “constant domains” already suggests, these domains are evolutionarily more conserved compared to the variable domains. In addition to disulfide-bridged cysteines, residues forming a hydrophobic core within the two sheets are highly conserved, as they contribute to the stabilization of the fold [136,137]. Despite the Ig-like constant domains sharing a similar fold and structure, they differ in their interface properties and dimer formation mechanisms. In fact, the CH3-CH3 interface is dominated by electrostatic interactions, whereas hydrophobic contacts are mainly responsible for stabilizing the CH1-CL interface [72,138]. Both of these interfaces show surprisingly fast interdomain movements, though the CH1-CL dimer exhibits higher variability in the interface angle compared to the CH3-CH3 dimer [13,72]. Only the CH2-CH2 domains do not directly interact with each other but are instead spatially separated by oligosaccharides. The glycans anchored on the Asn297 residue located on both CH2 domains are fundamental to holding the two domains apart and maintaining the characteristic horseshoe shape of the Fc fragment, which is the favorite conformation for interacting with the Fc receptor (Figure 1A) [139,140].
7.2. Engineering Techniques Led to New Antibody Formats
Nowadays, monoclonal antibodies (mAbs) are clinically significant molecules, and the hybridoma technology is well established for their industrial production [141,142,143]. Currently, ~175 antibody therapeutics are under regulatory review or on the market, and 13 have been approved in either the USA or Europe in 2022 alone, including four bispecific antibodies [1]. The early financial and clinical success of antibodies increased the curiosity of various pharmaceutical companies and research groups. Endeavours to advance the characterization of these biomolecules and engineer innovative and better-performing biotherapeutics followed soon. Antibody engineering can meet diverse needs, including, avoiding immunogenic responses and rejection in vivo, reducing the molecular weight to facilitate the antibody’s penetration in the cell, improving antibody developability, addressing novel therapeutic needs, promoting large-scale production, and decreasing costs. Here, we will provide a general overview of the most common engineering techniques and describe how new antibody formats accommodate pharmaceutical demands.
7.3. Humanization
Several engineering studies attempted to address the strong immunogenic reaction and the undesired anti-drug antibody (ADA) response caused by repeated administration of mAbs [144]. The origin of the ADA response is not fully understood, but experts hypothesize it may be caused by the murine nature of the mAbs [145]. Therefore, several studies aimed to develop humanized antibodies and avoid the human anti-mouse antibody response (HAMA) [146,147]. To achieve this goal, transgenic mice featuring a human immunoglobulin gene repertoire are used [148,149,150]. Generally, humanization describes the engineering effort of making a non-human antibody more human-like, i.e., by reducing as much of the murine content as possible while sustaining affinity and specificity [151,152]. There are several strategies to achieve antibody humanization. A well-established technique is the production of chimeric antibodies, where the non-human constant domains are exchanged with human constant domains and the non-human and antigen-specific variable domains are unaltered [153]. Another common method to obtain humanized Fabs is CDR grafting, which consists of grafting non-human CDR loops onto a human Fab framework [154,155,156,157,158]. In order to avoid decreased affinity, some non-human residues in the framework region are retained; these are the Vernier-zone residues, which can affect the CDR conformation and the binding affinity [7,159].
7.4. Affinity Maturation
Affinity maturation describes a crucial and complex process in the adaptive immune system where antibodies with higher affinity are produced in response to an antigen [160]. Thereby, B-cells adapt their receptors towards the epitope through multiple rounds of somatic hypermutation events followed by selection occasions in germinal centers. This process can increase the affinity of a naïve B-cell receptor (BCR) towards a target antigen up to a thousand-fold. Engineering strategies aim to mimic the affinity maturation process to modulate antibody affinity and specificity [160,161]. Interestingly, residues that are mutated during this process are not solely located in the CDR loops but can also be part of the framework region [162], showcasing the high complexity of predicting relevant amino acids that determine/govern affinity. Various studies have succeeded in improving antibody affinity by using combinatorial libraries [163] or computational design [164,165,166,167,168]. Additionally, high-resolution structural characterization enables the elucidation of the structural consequences of affinity maturation and the identification of key residues contributing to this increase in affinity and specificity [169,170]. Based on these empirical structures, long-timescale MD simulations have been used to provide a mechanistic understanding of the structural and dynamic changes in different stages of affinity maturation. It has been demonstrated that affinity maturation does not only result in a rigidification of the antigen-binding site but also restricts the flexibility of the elbow and interface angles in the Fab [13,169]. The observed rigidification corresponds to a reduction of energy minima, shifting the probability towards the binding competent conformation and favoring the lock-and-key binding mechanism [171,172]. Conversely, germline antibodies follow the conformational selection paradigm, as they adopt multiple weakly populated conformational states that can recognize different antigens [173,174,175]. Upon binding to a specific antigen, the respective state probabilities subsequently shift towards the binding competent state.
7.5. Bispecific Antibodies
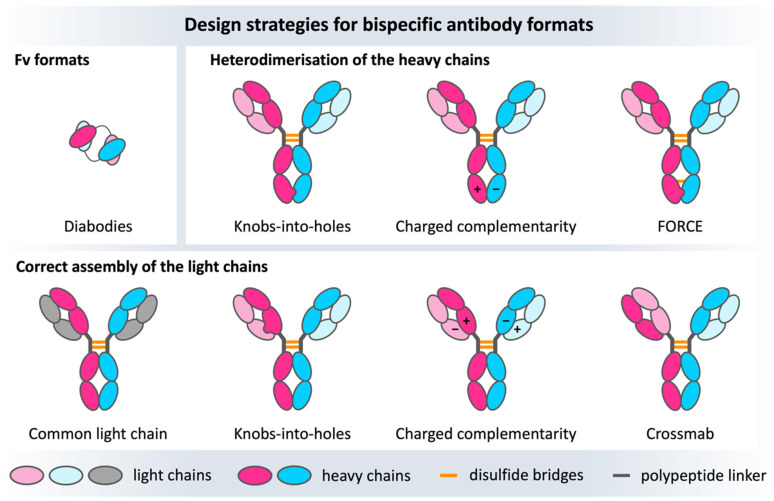
Bispecific antibodies are the products of tremendous advances in antibody engineering, resulting in a molecule that can simultaneously recognize two different epitopes and/or bring two targets in close proximity. The most prominent bispecific antibody designs are full-size and IgG-like, in which the CH3-CH3 domains are engineered to favor heavy chain heterodimerization over homodimerization (Figure 5). The idea behind this antibody format is that the resulting IgG-like molecule is formed by two different heavy chains and two different light chains and consequently offers distinct antigen binding sites. The main challenge in engineering and producing bispecific IgG-like antibodies is the high number of pairing possibilities resulting from two different heavy and light chains. The potential combinations can lead to several unwanted side products [176,177,178]. Nowadays, there are several design strategies that modify domain interfaces by introducing point mutations to ensure the desired heterodimeric interface is formed (Figure 5). The first engineering steps focus on optimizing the association of the heterodimeric heavy chains, which can be achieved by establishing charge interactions or increasing steric complementarity. Charge complementarity can be realized by inserting opposite charged residues in the CH3-CH3 interface [179,180,181], while steric complementarity has been obtained with the knobs-into-holes (KiH) technology. In the KiH approach, a bulky tryptophan residue is introduced on one chain to fit into a designed hydrophobic pocket on the other chain, resulting from three mutations [182,183]. Starting from the KiH method, the expression yields of the heterodimers improved thanks to the format chain exchange (FORCE) technology, in which the final product is further stabilized by a disulfide bridge [184]. Apart from optimizing the heavy chain heterodimerization, the correct light chain pairing must be ensured to obtain two functional and distinct antigen binding sites. The most obvious solution is using a common light chain; however, this strategy restricts the accessible combinations/paratopes [185,186,187]. Alternatively, the CH1-CL interface can be engineered following the same strategies used for the CH3-CH3 interface, resulting in the correct heavy-light chain pairing [188]. With the more recent Crossmab technology developed by Roche, correct association of the light chain with the respective counterpart can be enabled by exchanging heavy and light chain domains within one Fab, thereby retaining antigen-binding affinity while preventing light chain mispairings [189].
Figure 5.
Overview of some of the numerous bispecific design strategies for bispecific antibodies. The heavy chains are colored bright pink/blue, the light chains are depicted in light blue/pink and gray. The upper row shows Fv formats focusing on engineering and optimizing the VH-VL interface on the left, as well as heavy chain heterodimerization approaches (CH3-CH3 complementarity), whereas the lower row represents the light chain heterodimerization strategies concerning mainly the CH1-CL interface.
These tremendous antibody engineering successes are reflected in the numerous bispecific antibodies that are already approved on the market or are currently in late-stage development [1].
7.6. Small Size Antibody Formats
Natural IgGs have a weight of ~150 kDa, and their large size often represents a challenge for their production and administration. Therefore, smaller molecules that retain the binding affinity and specificity of the original antibody but have more favorable expression and pharmacokinetic properties have been pursued. Well-established examples of these small antibody formats are Fabs or single-chain variable fragments (scFv), the latter of which consists of a variable fragment of a heavy chain and one of a light chain, joined by a flexible peptide linker. The length of the linker (~15 residues) is fundamental to obtaining the correct folding, and its amino acid composition can be engineered to increase the affinity and specificity of the scFv [190,191]. Another antibody format is the “diabody”. Diabodies are bivalent antibodies constructed by connecting two Fv domains via a short peptide linker consisting of three to twelve residues. This linker is too short to allow mispairings of the two Fvs, and, therefore, the domains form two binding sites [192]. The two sites of a diabody can be either identical, resulting in a monospecific biologic, or distinct, resulting in bispecificity [193,194].
8. Special Formats from Natural Sources
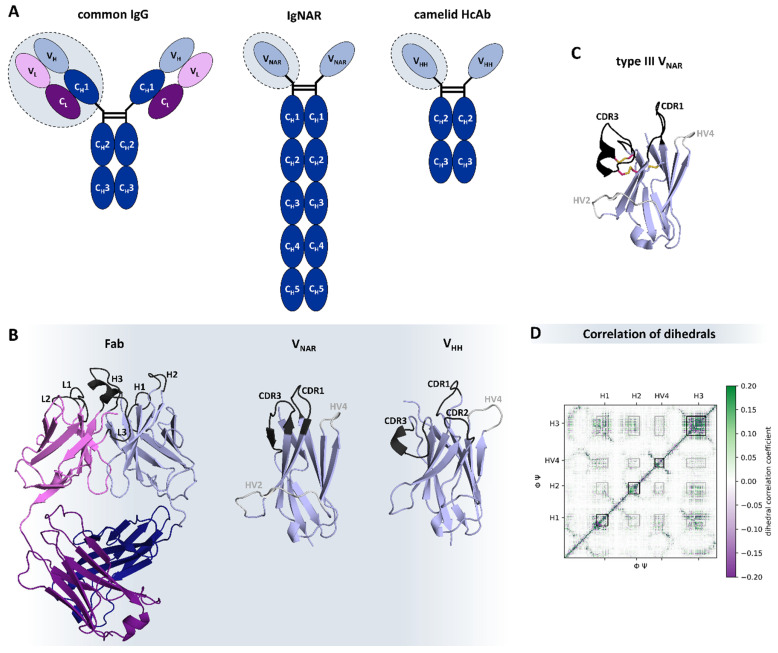
In contrast to the conventional IgG isotype, there are different types of naturally occurring antibody formats found in species like cartilaginous fish, e.g., sharks, and Camelidae, e.g., camelids. These special formats, also known as heavy-chain only antibodies (HcAb), are deprived of the light chains and therefore characterized by the assembly of only two heavy chains [195,196]. While the Immunoglobulin New Antigen Receptor (IgNAR) found in cartilaginous fish is comprised of one variable (VNAR) and five constant domains, the Camelidae variant has one variable (VHH, or nanobody) and two constant domains (Figure 6A) [197,198]. There are further structural differences, especially in the variable domains, which represent the functional and structural analogues of the Fab in conventional antibodies and are critical for antigen-binding (Figure 6B). First, the VNAR and the VHH are much smaller than a whole Fab (consisting of 4 domains) and, hence, are able to wend into tissue in a fast and specific way [199,200]. In this context, it has been shown that VNARs can serve as shuttles for larger proteins to cross the blood-brain barrier [201,202]. Both single-domain antibodies have a characteristically prolonged CDR3 loop that has higher variability in sequence and structure than the CDR-H3 in conventional IgGs [203,204]. The consequently convex-shaped paratope is able to bind into cavities and cryptic epitopes [205,206]. However, the high conformational flexibility of HcAbs can result in a high entropic cost upon antigen binding, which led to the evolution of additional cysteine bonds to rigidify and stabilize the binding site (Figure 6C) [207]. Nevertheless, it is still unclear what precise role rigidity plays in the antigen binding process [208]. The overall high stability of HcAbs arises from the challenging conditions they evolved in, e.g., the high urea concentration in the blood of sharks, which has a denaturizing effect [197,209]. Furthermore, the increased number of solvent-exposed hydrophilic residues in the HcAbs contributes to their good solubility in aqueous solutions and is required to prevent the formation of a heavy-light chain interface, which is mainly mediated by a hydrophobic core [72,210]. For VNARs, this characteristic property cannot be assigned to specific residues, while for nanobodies, this is mainly dependent on four residues in framework 2 (F42, E49, R50, and G52 in IMGT numbering) [198]. Furthermore, VNARs are missing the CDR2 loop, which is replaced by a hypervariable region with hydrophilic character that forms a belt-like conformation, from which side chains point into the alleged VH-VL interface [211]. The hypervariable region 4-loop (HV4-loop) is of special importance for all HcAbs since it influences antigen-binding directly and indirectly by stabilizing distinct paratope conformations [170], as it was previously demonstrated for T-cell receptors [212]. In addition, all loops collude with each other and support the hypothesis of a complementary evolution (Figure 6D) [200].
The greatest difference between nanobodies and VNARs lies in their sequences. VNARs evolved around 500 million years ago and therefore show high sequence dissimilarities to vertebrates yet astonishingly high structural resemblance to κ-light chain variable domains [197,213,214,215]. VHHs, on the other hand, are more related to other vertebrate heavy chain variable domains since they evolved only 40 million years ago [216,217]. Although they share a low sequence similarity, the structural likeliness of the HcAbs to other heavy chain variable domains is remarkable, which highlights their coherent evolution [218,219]. This insight facilitates the engineering efforts of nanobodies and reduces the risk of immunogenicity in comparison to VNARs [220,221]. In fact, various studies have focused on designing and optimizing these single-domain antibodies to exploit their advantageous features for future biotherapeutics.
In addition to their unique sequence and structural features, understanding the consequences of affinity maturation and point mutations on their binding properties, i.e., affinity and specificity, is of utmost importance to enable modulation and fine-tuning of their function [222,223]. Indeed, structural and dynamic characterizations of nanobodies have, on the one hand, revealed residues that play a crucial role in antigen recognition and, on the other, identified that an increase in affinity and specificity can be accompanied by a rigidification of the antigen binding site [170]. Apart from the investigation of the antigen binding properties, there are ongoing studies about the humanization strategies of HcAbs, which are complicated for shark VNARs due to their low sequence similarity [213,221,224]. Nevertheless, there are both nanobodiesy and VNAR drug candidates in clinical trials [225,226,227]. Considering the beneficial features and improvements in designing and engineering HcAbs, predicting their structure, in particular the conformations of CDR loops, still remains challenging. The extended CDR3 loop conformation is especially difficult to predict accurately due to the high diversity in length, sequence, and structure. Additionally, the resulting high conformational variability suggests that one single static structure might not be sufficient to capture the high conformational diversity of this loop [228,229]. However, in addition to a structural and dynamic characterization of these novel antibody formats, various tools are emerging to address these challenges based on deep-learning models [44,230,231].
Figure 6.
Natural occurring heavy-chain only antibodies in comparison to common IgG isotypes: (A) Differences in antibodies in humans, sharks, and camelids as shown by their schematic structures. The heavy chains are colored blue, while the light chains are purple. The variable domains are shown in lighter shades. (B) Structural differences of the antigen binding relevant sections: Fab (PDB: 6MAM) [232], VNAR (type 4, PDB: 4HGK) [213], and VHH (PDB: 2P49) [233], respectively. The CDR loops are colored black, and the hypervariable regions are gray. In (C), the disulfide bridges of a type III VNAR (PDB: 1SQ2) [196] are highlighted. Depending on the number of additional disulfide bridges and distinct conserved residues, shark VNARs can be grouped into four different types [196,199,213]. The pink-colored residues point out the additional bonds in this exemplary system. (D) Exemplary correlation of the backbone torsions of a nanobody simulation. Green depicts a positive correlation coefficient and purple a negative one. Besides the diagonal, which represents the correlation of adjacent residues, a clear influence of the different binding relevant regions (CDR-loops: H1, H2, and H3, as well as HV4) is highlighted, describing the correlated movements involved in shaping the paratope.
9. Conclusions
The numerous high-quality antibody structures available in the PDB (originating from X-ray crystallography, cryo-EM, and NMR spectroscopy) provide an atomic view of the affinity maturation process, influence engineering strategies, and advance the understanding of antigen binding mechanisms. In addition, these structures, in combination with neural networks, substantially improve state-of-the-art antibody structure prediction tools. These models can, however, still suffer from structural and physical inaccuracies and should be carefully reviewed. The CDR loops especially remain challenging to accurately predict due to their high conformational diversity. Thus, capturing the high variability of the CDR loops is another critical aspect for informing antibody engineering efforts and improving structure prediction algorithms. In particular, processes such as aggregation and stability are strongly governed by dynamics, highlighting the importance of considering antibodies as conformational ensembles in solution to guide rational antibody design. The tremendous advances in antibody engineering, focusing on the shape or charge complementarity of the constant domain interfaces, led to various novel formats, even further enhancing the applicability of antibodies as biotherapeutics. Finally, single-domain antibodies, originating from sharks and camelids, represent a highly potent alternative to conventional IgG antibodies, providing numerous applications and design opportunities.
Acknowledgments
We acknowledge CHRONOS for awarding us access to Piz Daint at CSCS, Switzerland. We acknowledge the EuroHPC Joint Undertaking for awarding us access to Karolina at IT4Innovations, Czech Republic.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
N.D.P. has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement number 847476. The views and opinions expressed herein do not necessarily reflect those of the European Commission. This work was supported by the Austrian Science Fund (FWF) via grant P34518. This work was supported by the Austrian Academy of Sciences APART-MINT postdoctoral fellowship to M.L.F.Q. H.R.P. was supported by a David C. Fairchild Endowed Fellowship, and the Achievement Rewards for College Scientists Foundation. J.H, J.F. and A.B.W. were supported by the Bill and Melinda Gates Foundation through grant INV-004923 (A.B.W.).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Kaplon H., Crescioli S., Chenoweth A., Visweswaraiah J., Reichert J.M. Antibodies to Watch in 2023. mAbs. 2023;15:2153410. doi: 10.1080/19420862.2022.2153410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplon H., Chenoweth A., Crescioli S., Reichert J.M. Antibodies to Watch in 2022. mAbs. 2022;14:2014296. doi: 10.1080/19420862.2021.2014296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiu M.L., Goulet D.R., Teplyakov A., Gilliland G.L. Antibody Structure and Function: The Basis for Engineering Therapeutics. Antibodies. 2019;8:55. doi: 10.3390/antib8040055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pyzik M., Rath T., Lencer W.I., Baker K., Blumberg R.S. FcRn: The Architect Behind the Immune and Nonimmune Functions of IgG and Albumin. J. Immunol. 2015;194:4595–4603. doi: 10.4049/jimmunol.1403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ying T., Ju T.W., Wang Y., Prabakaran P., Dimitrov D.S. Interactions of IgG1 CH2 and CH3 Domains with FcRn. Front. Immunol. 2014;5:146. doi: 10.3389/fimmu.2014.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernández-Quintero M.L., Kroell K.B., Hofer F., Riccabona J.R., Liedl K.R. Mutation of Framework Residue H71 Results in Different Antibody Paratope States in Solution. Front. Immunol. 2021;12:243. doi: 10.3389/fimmu.2021.630034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foote J., Winter G. Antibody Framework Residues Affecting the Conformation of the Hypervariable Loops. J. Mol. Biol. 1992;224:487–499. doi: 10.1016/0022-2836(92)91010-M. [DOI] [PubMed] [Google Scholar]
- 8.Krauss J., Arndt M.A.E., Zhu Z., Newton D.L., Vu B.K., Choudhry V., Darbha R., Ji X., Courtenay-Luck N.S., Deonarain M.P., et al. Impact of Antibody Framework Residue VH-71 on the Stability of a Humanised Anti-MUC1 scFv and Derived Immunoenzyme. Br. J. Cancer. 2004;90:1863–1870. doi: 10.1038/sj.bjc.6601759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tramontano A., Chothia C., Lesk A.M. Framework Residue 71 Is a Major Determinant of the Position and Conformation of the Second Hypervariable Region in the VH Domains of Immunoglobulins. J. Mol. Biol. 1990;215:175–182. doi: 10.1016/S0022-2836(05)80102-0. [DOI] [PubMed] [Google Scholar]
- 10.Xiang J., Sha Y., Jia Z., Prasad L., Delbaere L.T.J. Framework Residues 71 and 93 of the Chimeric B72.3 Antibody Are Major Determinants of the Conformation of Heavy-Chain Hypervariable Loops. J. Mol. Biol. 1995;253:385–390. doi: 10.1006/jmbi.1995.0560. [DOI] [PubMed] [Google Scholar]
- 11.Rappazzo C.G., Fernández-Quintero M.L., Mayer A., Wu N.C., Greiff V., Guthmiller J.J. Defining and Studying B Cell Receptor and TCR Interactions. J. Immunol. 2023;211:311–322. doi: 10.4049/jimmunol.2300136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernández-Quintero M.L., Hoerschinger V.J., Lamp L.M., Bujotzek A., Georges G., Liedl K.R. VH -VL Interdomain Dynamics Observed by Computer Simulations and NMR. Proteins. 2020;88:830–839. doi: 10.1002/prot.25872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernández-Quintero M.L., Kroell K.B., Heiss M.C., Loeffler J.R., Quoika P.K., Waibl F., Bujotzek A., Moessner E., Georges G., Liedl K.R. Surprisingly Fast Interface and Elbow Angle Dynamics of Antigen-Binding Fragments. Front. Mol. Biosci. 2020;7:609088. doi: 10.3389/fmolb.2020.609088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teichmann S.A., Chothia C. Immunoglobulin Superfamily Proteins in Caenorhabditis Elegans. J. Mol. Biol. 2000;296:1367–1383. doi: 10.1006/jmbi.1999.3497. [DOI] [PubMed] [Google Scholar]
- 15.Youkharibache P. Topological and Structural Plasticity of the Single Ig Fold and the Double Ig Fold Present in CD19. Biomolecules. 2021;11:1290. doi: 10.3390/biom11091290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lesk A.M., Chothia C. Evolution of Proteins Formed by Beta-Sheets. II. The Core of the Immunoglobulin Domains. J. Mol. Biol. 1982;160:325–342. doi: 10.1016/0022-2836(82)90179-6. [DOI] [PubMed] [Google Scholar]
- 17.Brinkmann U., Kontermann R.E. The Making of Bispecific Antibodies. mAbs. 2017;9:182–212. doi: 10.1080/19420862.2016.1268307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merchant A.M., Zhu Z., Yuan J.Q., Goddard A., Adams C.W., Presta L.G., Carter P. An Efficient Route to Human Bispecific IgG. Nat. Biotechnol. 1998;16:677–681. doi: 10.1038/nbt0798-677. [DOI] [PubMed] [Google Scholar]
- 19.Labrijn A.F., Janmaat M.L., Reichert J.M., Parren P.W.H.I. Bispecific Antibodies: A Mechanistic Review of the Pipeline. Nat. Rev. Drug Discov. 2019;18:585–608. doi: 10.1038/s41573-019-0028-1. [DOI] [PubMed] [Google Scholar]
- 20.Schroeder H.W., Cavacini L. Structure and Function of Immunoglobulins. J. Allergy Clin. Immunol. 2010;125:S41–S52. doi: 10.1016/j.jaci.2009.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris L.J., Skaletsky E., McPherson A. Crystallographic Structure of an Intact IgG1 Monoclonal Antibody. J. Mol. Biol. 1998;275:861–872. doi: 10.1006/jmbi.1997.1508. [DOI] [PubMed] [Google Scholar]
- 22.Almagro J.C., Teplyakov A., Luo J., Sweet R.W., Kodangattil S., Hernandez-Guzman F., Gilliland G.L. Second Antibody Modeling Assessment (AMA-II) Proteins Struct. Funct. Bioinform. 2014;82:1553–1562. doi: 10.1002/prot.24567. [DOI] [PubMed] [Google Scholar]
- 23.Dill K.A., MacCallum J.L. The Protein-Folding Problem, 50 Years On. Science. 2012;338:1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- 24.Dill K.A., Ozkan S.B., Shell M.S., Weikl T.R. The Protein Folding Problem. Annu. Rev. Biophys. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Lazikani B., Jung J., Xiang Z., Honig B. Protein Structure Prediction. Curr. Opin. Chem. Biol. 2001;5:51–56. doi: 10.1016/S1367-5931(00)00164-2. [DOI] [PubMed] [Google Scholar]
- 26.Khetan R., Curtis R., Deane C.M., Hadsund J.T., Kar U., Krawczyk K., Kuroda D., Robinson S.A., Sormanni P., Tsumoto K., et al. Current Advances in Biopharmaceutical Informatics: Guidelines, Impact and Challenges in the Computational Developability Assessment of Antibody Therapeutics. mAbs. 2022;14:2020082. doi: 10.1080/19420862.2021.2020082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W., Singh S., Zeng D.L., King K., Nema S. Antibody Structure, Instability, and Formulation. J. Pharm. Sci. 2007;96:1–26. doi: 10.1002/jps.20727. [DOI] [PubMed] [Google Scholar]
- 28.Fernández-Quintero M.L., Kraml J., Georges G., Liedl K.R. CDR-H3 Loop Ensemble in Solution—Conformational Selection upon Antibody Binding. mAbs. 2019;11:1077–1088. doi: 10.1080/19420862.2019.1618676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weitzner B.D., Dunbrack R.L.J., Gray J.J. The Origin of CDR H3 Structural Diversity. Structure. 2015;23:302–311. doi: 10.1016/j.str.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chothia C., Lesk A.M. Canonical Structures for the Hypervariable Regions of Immunoglobulins. J. Mol. Biol. 1987;196:901–917. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- 31.North B., Lehmann A., Dunbrack R.L.J. A New Clustering of Antibody CDR Loop Conformations. J. Mol. Biol. 2011;406:228–256. doi: 10.1016/j.jmb.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morea V., Lesk A.M., Tramontano A. Antibody Modeling: Implications for Engineering and Design. Methods. 2000;20:267–279. doi: 10.1006/meth.1999.0921. [DOI] [PubMed] [Google Scholar]
- 33.Weitzner B.D., Kuroda D., Marze N., Xu J., Gray J.J. Blind Prediction Performance of RosettaAntibody 3.0: Grafting, Relaxation, Kinematic Loop Modeling, and Full CDR Optimization. Proteins. 2014;82:1611–1623. doi: 10.1002/prot.24534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bujotzek A., Dunbar J., Lipsmeier F., Schäfer W., Antes I., Deane C.M., Georges G. Prediction of VH-VL Domain Orientation for Antibody Variable Domain Modeling. Proteins. 2015;83:681–695. doi: 10.1002/prot.24756. [DOI] [PubMed] [Google Scholar]
- 35.Abhinandan K.R., Martin A.C.R. Analysis and Prediction of VH/VL Packing in Antibodies. Protein Eng. Des. Sel. PEDS. 2010;23:689–697. doi: 10.1093/protein/gzq043. [DOI] [PubMed] [Google Scholar]
- 36.Marze N.A., Lyskov S., Gray J.J. Improved Prediction of Antibody VL-VH Orientation. Protein Eng. Des. Sel. PEDS. 2016;29:409–418. doi: 10.1093/protein/gzw013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marks C., Deane C.M. Antibody H3 Structure Prediction. Comput. Struct. Biotechnol. J. 2017;15:222–231. doi: 10.1016/j.csbj.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sormanni P., Aprile F.A., Vendruscolo M. Third Generation Antibody Discovery Methods: In Silico Rational Design. Chem. Soc. Rev. 2018;47:9137–9157. doi: 10.1039/C8CS00523K. [DOI] [PubMed] [Google Scholar]
- 39.Wilman W., Wróbel S., Bielska W., Deszynski P., Dudzic P., Jaszczyszyn I., Kaniewski J., Młokosiewicz J., Rouyan A., Satława T., et al. Machine-Designed Biotherapeutics: Opportunities, Feasibility and Advantages of Deep Learning in Computational Antibody Discovery. Brief. Bioinform. 2022;23:bbac267. doi: 10.1093/bib/bbac267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hummer A.M., Abanades B., Deane C.M. Advances in Computational Structure-Based Antibody Design. Curr. Opin. Struct. Biol. 2022;74:102379. doi: 10.1016/j.sbi.2022.102379. [DOI] [PubMed] [Google Scholar]
- 41.Norman R.A., Ambrosetti F., Bonvin A.M.J.J., Colwell L.J., Kelm S., Kumar S., Krawczyk K. Computational Approaches to Therapeutic Antibody Design: Established Methods and Emerging Trends. Brief. Bioinform. 2020;21:1549–1567. doi: 10.1093/bib/bbz095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weitzner B.D., Jeliazkov J.R., Lyskov S., Marze N., Kuroda D., Frick R., Adolf-Bryfogle J., Biswas N., Dunbrack R.L.J., Gray J.J. Modeling and Docking of Antibody Structures with Rosetta. Nat. Protoc. 2017;12:401–416. doi: 10.1038/nprot.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leem J., Dunbar J., Georges G., Shi J., Deane C.M. ABodyBuilder: Automated Antibody Structure Prediction with Data-Driven Accuracy Estimation. mAbs. 2016;8:1259–1268. doi: 10.1080/19420862.2016.1205773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A., et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2021;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans R., O’Neill M., Pritzel A., Antropova N., Senior A., Green T., Žídek A., Bates R., Blackwell S., Yim J., et al. Protein Complex Prediction with AlphaFold-Multimer. bioRxiv. 2022 doi: 10.1101/2021.10.04.463034. [DOI] [Google Scholar]
- 47.Ruffolo J.A., Sulam J., Gray J.J. Antibody Structure Prediction Using Interpretable Deep Learning. Patterns. 2022;3:100406. doi: 10.1016/j.patter.2021.100406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruffolo J.A., Chu L.-S., Mahajan S.P., Gray J.J. Fast, Accurate Antibody Structure Prediction from Deep Learning on Massive Set of Natural Antibodies. Nat. Commun. 2023;14:2389. doi: 10.1038/s41467-023-38063-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abanades B., Georges G., Bujotzek A., Deane C.M. ABlooper: Fast Accurate Antibody CDR Loop Structure Prediction with Accuracy Estimation. Bioinformatics. 2022;38:1877–1880. doi: 10.1093/bioinformatics/btac016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernández-Quintero M.L., Georges G., Varga J.M., Liedl K.R. Ensembles in Solution as a New Paradigm for Antibody Structure Prediction and Design. mAbs. 2021;13:1923122. doi: 10.1080/19420862.2021.1923122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishigami H., Kamiya N., Nakamura H. Revisiting Antibody Modeling Assessment for CDR-H3 Loop. Protein Eng. Des. Sel. PEDS. 2016;29:477–484. doi: 10.1093/protein/gzw028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernández-Quintero M.L., Kokot J., Waibl F., Fischer A.-L.M., Quoika P.K., Deane C.M., Liedl K.R. Challenges in Antibody Structure Prediction. mAbs. 2023;15:2175319. doi: 10.1080/19420862.2023.2175319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teplyakov A., Luo J., Obmolova G., Malia T.J., Sweet R., Stanfield R.L., Kodangattil S., Almagro J.C., Gilliland G.L. Antibody Modeling Assessment II. Structures and Models. Proteins. 2014;82:1563–1582. doi: 10.1002/prot.24554. [DOI] [PubMed] [Google Scholar]
- 54.Almagro J.C., Beavers M.P., Hernandez-Guzman F., Maier J., Shaulsky J., Butenhof K., Labute P., Thorsteinson N., Kelly K., Teplyakov A., et al. Antibody Modeling Assessment. Proteins. 2011;79:3050–3066. doi: 10.1002/prot.23130. [DOI] [PubMed] [Google Scholar]
- 55.Kern D. From Structure to Mechanism: Skiing the Energy Landscape. Nat. Methods. 2021;18:435–436. doi: 10.1038/s41592-021-01140-4. [DOI] [PubMed] [Google Scholar]
- 56.Henzler-Wildman K., Kern D. Dynamic Personalities of Proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 57.Karplus M., Petsko G.A. Molecular Dynamics Simulations in Biology. Nature. 1990;347:631–639. doi: 10.1038/347631a0. [DOI] [PubMed] [Google Scholar]
- 58.Landsteiner K., Van Der Scheer J. On the specificity of serological reactions with simple chemical compounds (inhibition reactions) J. Exp. Med. 1931;54:295–305. doi: 10.1084/jem.54.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pauling L. A Theory of the Structure and Process of Formation of Antibodies *. J. Am. Chem. Soc. 1940;62:2643–2657. doi: 10.1021/ja01867a018. [DOI] [Google Scholar]
- 60.Fernández-Quintero M.L., Kroell K.B., Bacher L.M., Loeffler J.R., Quoika P.K., Georges G., Bujotzek A., Kettenberger H., Liedl K.R. Germline-Dependent Antibody Paratope States and Pairing Specific VH-VL Interface Dynamics. Front. Immunol. 2021;12:675655. doi: 10.3389/fimmu.2021.675655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernández-Quintero M.L., Pomarici N.D., Math B.A., Kroell K.B., Waibl F., Bujotzek A., Georges G., Liedl K.R. Antibodies Exhibit Multiple Paratope States Influencing VH–VL Domain Orientations. Commun. Biol. 2020;3:589. doi: 10.1038/s42003-020-01319-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fernández-Quintero M.L., Heiss M.C., Pomarici N.D., Math B.A., Liedl K.R. Antibody CDR Loops as Ensembles in Solution vs. Canonical Clusters from X-Ray Structures. mAbs. 2020;12:1744328. doi: 10.1080/19420862.2020.1744328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fernández-Quintero M.L., Vangone A., Loeffler J.R., Seidler C.A., Georges G., Liedl K.R. Paratope States in Solution Improve Structure Prediction and Docking. Structure. 2022;30:430–440.e3. doi: 10.1016/j.str.2021.11.001. [DOI] [PubMed] [Google Scholar]
- 64.Kunik V., Ashkenazi S., Ofran Y. Paratome: An Online Tool for Systematic Identification of Antigen-Binding Regions in Antibodies Based on Sequence or Structure. Nucleic Acids Res. 2012;40:W521–W524. doi: 10.1093/nar/gks480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacCallum R.M., Martin A.C.R., Thornton J.M. Antibody-Antigen Interactions: Contact Analysis and Binding Site Topography. J. Mol. Biol. 1996;262:732–745. doi: 10.1006/jmbi.1996.0548. [DOI] [PubMed] [Google Scholar]
- 66.Waibl F., Fernández-Quintero M.L., Kamenik A.S., Kraml J., Hofer F., Kettenberger H., Georges G., Liedl K.R. Conformational Ensembles of Antibodies Determine Their Hydrophobicity. Biophys. J. 2021;120:143–157. doi: 10.1016/j.bpj.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waibl F., Fernández-Quintero M.L., Wedl F.S., Kettenberger H., Georges G., Liedl K.R. Comparison of Hydrophobicity Scales for Predicting Biophysical Properties of Antibodies. Front. Mol. Biosci. 2022;9:960194. doi: 10.3389/fmolb.2022.960194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waibl F., Pomarici N.D., Hoerschinger V.J., Loeffler J.R., Deane C.M., Georges G., Kettenberger H., Fernández-Quintero M.L., Liedl K.R. PEP-Patch: Electrostatics in Protein-Protein Recognition, Specificity and Antibody Developability. bioRxiv. 2023 doi: 10.1101/2023.07.14.547811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dunbar J., Fuchs A., Shi J., Deane C.M. ABangle: Characterising the VH-VL Orientation in Antibodies. Protein Eng. Des. Sel. 2013;26:611–620. doi: 10.1093/protein/gzt020. [DOI] [PubMed] [Google Scholar]
- 70.Hoerschinger V.J., Fernández-Quintero M.L., Waibl F., Kraml J., Bujotzek A., Georges G., Liedl K.R. OCD.Py-Characterizing Immunoglobulin Inter-Domain Orientations. bioRxiv. 2021 doi: 10.1101/2021.03.15.435379. [DOI] [Google Scholar]
- 71.Stanfield R.L., Zemla A., Wilson I.A., Rupp B. Antibody Elbow Angles Are Influenced by Their Light Chain Class. J. Mol. Biol. 2006;357:1566–1574. doi: 10.1016/j.jmb.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 72.Fernández-Quintero M.L., Quoika P.K., Wedl F.S., Seidler C.A., Kroell K.B., Loeffler J.R., Pomarici N.D., Hoerschinger V.J., Bujotzek A., Georges G., et al. Comparing Antibody Interfaces to Inform Rational Design of New Antibody Formats. Front. Mol. Biosci. 2022;9:812750. doi: 10.3389/fmolb.2022.812750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sotriffer C.A., Liedl K.R., Linthicum D.S., Rode B.M., Varga J.M. Ligand-Induced Domain Movement in an Antibody Fab: Molecular Dynamics Studies Confirm the Unique Domain Movement Observed Experimentally for Fab NC6.8 upon Complexation and Reveal Its Segmental flexibility. J. Mol. Biol. 1998;278:301–306. doi: 10.1006/jmbi.1998.1684. [DOI] [PubMed] [Google Scholar]
- 74.Fernández-Quintero M.L., Kroell K.B., Grunewald L.J., Fischer A.-L.M., Riccabona J.R., Liedl K.R. CDR Loop Interactions Can Determine Heavy and Light Chain Pairing Preferences in Bispecific Antibodies. mAbs. 2022;14:2024118. doi: 10.1080/19420862.2021.2024118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bujotzek A., Lipsmeier F., Harris S.F., Benz J., Kuglstatter A., Georges G. VH-VL Orientation Prediction for Antibody Humanization Candidate Selection: A Case Study. mAbs. 2016;8:288–305. doi: 10.1080/19420862.2015.1117720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Burton D.R. Scaffolding to Build a Rational Vaccine Design Strategy. Proc. Natl. Acad. Sci. USA. 2010;107:17859–17860. doi: 10.1073/pnas.1012923107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ward A.B., Wilson I.A. Innovations in Structure-Based Antigen Design and Immune Monitoring for next Generation Vaccines. Curr. Opin. Immunol. 2020;65:50–56. doi: 10.1016/j.coi.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kulp D.W., Schief W.R. Advances in Structure-Based Vaccine Design. Curr. Opin. Virol. 2013;3:322–331. doi: 10.1016/j.coviro.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krammer F. The Human Antibody Response to Influenza A Virus Infection and Vaccination. Nat. Rev. Immunol. 2019;19:383–397. doi: 10.1038/s41577-019-0143-6. [DOI] [PubMed] [Google Scholar]
- 80.Kirkpatrick E., Qiu X., Wilson P.C., Bahl J., Krammer F. The Influenza Virus Hemagglutinin Head Evolves Faster than the Stalk Domain. Sci. Rep. 2018;8:10432. doi: 10.1038/s41598-018-28706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nachbagauer R., Feser J., Naficy A., Bernstein D.I., Guptill J., Walter E.B., Berlanda-Scorza F., Stadlbauer D., Wilson P.C., Aydillo T., et al. A Chimeric Hemagglutinin-Based Universal Influenza Virus Vaccine Approach Induces Broad and Long-Lasting Immunity in a Randomized, Placebo-Controlled Phase I Trial. Nat. Med. 2021;27:106–114. doi: 10.1038/s41591-020-1118-7. [DOI] [PubMed] [Google Scholar]
- 82.Bernstein D.I., Guptill J., Naficy A., Nachbagauer R., Berlanda-Scorza F., Feser J., Wilson P.C., Solórzano A., Van der Wielen M., Walter E.B., et al. Immunogenicity of Chimeric Haemagglutinin-Based, Universal Influenza Virus Vaccine Candidates: Interim Results of a Randomised, Placebo-Controlled, Phase 1 Clinical Trial. Lancet Infect. Dis. 2020;20:80–91. doi: 10.1016/S1473-3099(19)30393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yassine H.M., Boyington J.C., McTamney P.M., Wei C.-J., Kanekiyo M., Kong W.-P., Gallagher J.R., Wang L., Zhang Y., Joyce M.G., et al. Hemagglutinin-Stem Nanoparticles Generate Heterosubtypic Influenza Protection. Nat. Med. 2015;21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- 84.Kanekiyo M., Wei C.-J., Yassine H.M., McTamney P.M., Boyington J.C., Whittle J.R.R., Rao S.S., Kong W.-P., Wang L., Nabel G.J. Self-Assembling Influenza Nanoparticle Vaccines Elicit Broadly Neutralizing H1N1 Antibodies. Nature. 2013;499:102–106. doi: 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boyoglu-Barnum S., Ellis D., Gillespie R.A., Hutchinson G.B., Park Y.-J., Moin S.M., Acton O., Ravichandran R., Murphy M., Pettie D., et al. Elicitation of Broadly Protective Immunity to Influenza by Multivalent Hemagglutinin Nanoparticle Vaccines. bioRxiv. 2020 doi: 10.1101/2020.05.30.125179. [DOI] [Google Scholar]
- 86.Kanekiyo M., Joyce M.G., Gillespie R.A., Gallagher J.R., Andrews S.F., Yassine H.M., Wheatley A.K., Fisher B.E., Ambrozak D.R., Creanga A., et al. Mosaic Nanoparticle Display of Diverse Influenza Virus Hemagglutinins Elicits Broad B Cell Responses. Nat. Immunol. 2019;20:362–372. doi: 10.1038/s41590-018-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Steichen J.M., Lin Y.-C., Havenar-Daughton C., Pecetta S., Ozorowski G., Willis J.R., Toy L., Sok D., Liguori A., Kratochvil S., et al. A Generalized HIV Vaccine Design Strategy for Priming of Broadly Neutralizing Antibody Responses. Science. 2019;366:eaax4380. doi: 10.1126/science.aax4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Willis J.R., Berndsen Z.T., Ma K.M., Steichen J.M., Schiffner T., Landais E., Liguori A., Kalyuzhniy O., Allen J.D., Baboo S., et al. Human Immunoglobulin Repertoire Analysis Guides Design of Vaccine Priming Immunogens Targeting HIV V2-Apex Broadly Neutralizing Antibody Precursors. Immunity. 2022;55:2149–2167.e9. doi: 10.1016/j.immuni.2022.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Melzi E., Willis J.R., Ma K.M., Lin Y.-C., Kratochvil S., Berndsen Z.T., Landais E.A., Kalyuzhniy O., Nair U., Warner J., et al. Membrane-Bound mRNA Immunogens Lower the Threshold to Activate HIV Env V2 Apex-Directed Broadly Neutralizing B Cell Precursors in Humanized Mice. Immunity. 2022;55:2168–2186.e6. doi: 10.1016/j.immuni.2022.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leggat D.J., Cohen K.W., Willis J.R., Fulp W.J., deCamp A.C., Kalyuzhniy O., Cottrell C.A., Menis S., Finak G., Ballweber-Fleming L., et al. Vaccination Induces HIV Broadly Neutralizing Antibody Precursors in Humans. Science. 2022;378:eadd6502. doi: 10.1126/science.add6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guthmiller J.J., Han J., Utset H.A., Li L., Lan L.Y.-L., Henry C., Stamper C.T., McMahon M., O’Dell G., Fernández-Quintero M.L., et al. Broadly Neutralizing Antibodies Target a Haemagglutinin Anchor Epitope. Nature. 2022;602:314–320. doi: 10.1038/s41586-021-04356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Whittle J.R.R., Zhang R., Khurana S., King L.R., Manischewitz J., Golding H., Dormitzer P.R., Haynes B.F., Walter E.B., Moody M.A., et al. Broadly Neutralizing Human Antibody That Recognizes the Receptor-Binding Pocket of Influenza Virus Hemagglutinin. Proc. Natl. Acad. Sci. USA. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bangaru S., Lang S., Schotsaert M., Vanderven H.A., Zhu X., Kose N., Bombardi R., Finn J.A., Kent S.J., Gilchuk P., et al. A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface. Cell. 2019;177:1136–1152.e18. doi: 10.1016/j.cell.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ekiert D.C., Bhabha G., Elsliger M.-A., Friesen R.H.E., Jongeneelen M., Throsby M., Goudsmit J., Wilson I.A. Antibody Recognition of a Highly Conserved Influenza Virus Epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rowan-Nash A.D., Korry B.J., Mylonakis E., Belenky P. Cross-Domain and Viral Interactions in the Microbiome. Microbiol. Mol. Biol. Rev. MMBR. 2019;83:e00044-18. doi: 10.1128/MMBR.00044-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu N.C., Wilson I.A. Influenza Hemagglutinin Structures and Antibody Recognition. Cold Spring Harb. Perspect. Med. 2020;10:a038778. doi: 10.1101/cshperspect.a038778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Antanasijevic A., Ueda G., Brouwer P.J.M., Copps J., Huang D., Allen J.D., Cottrell C.A., Yasmeen A., Sewall L.M., Bontjer I., et al. Structural and Functional Evaluation of de Novo-Designed, Two-Component Nanoparticle Carriers for HIV Env Trimer Immunogens. PLoS Pathog. 2020;16:e1008665. doi: 10.1371/journal.ppat.1008665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ozorowski G., Pallesen J., de Val N., Lyumkis D., Cottrell C.A., Torres J.L., Copps J., Stanfield R.L., Cupo A., Pugach P., et al. Open and Closed Structures Reveal Allostery and Pliability in the HIV-1 Envelope Spike. Nature. 2017;547:360–363. doi: 10.1038/nature23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barnes C.O., Schoofs T., Gnanapragasam P.N.P., Golijanin J., Huey-Tubman K.E., Gruell H., Schommers P., Suh-Toma N., Lee Y.E., Cetrulo Lorenzi J.C., et al. A Naturally Arising Broad and Potent CD4-Binding Site Antibody with Low Somatic Mutation. Sci. Adv. 2022;8:eabp8155. doi: 10.1126/sciadv.abp8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Klein F., Diskin R., Scheid J.F., Gaebler C., Mouquet H., Georgiev I.S., Pancera M., Zhou T., Incesu R.-B., Fu B.Z., et al. Somatic Mutations of the Immunoglobulin Framework Are Generally Required for Broad and Potent HIV-1 Neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Scharf L., Wang H., Gao H., Chen S., McDowall A.W., Bjorkman P.J. Broadly Neutralizing Antibody 8ANC195 Recognizes Closed and Open States of HIV-1 Env. Cell. 2015;162:1379–1390. doi: 10.1016/j.cell.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yuan M., Cottrell C.A., Ozorowski G., van Gils M.J., Kumar S., Wu N.C., Sarkar A., Torres J.L., de Val N., Copps J., et al. Conformational Plasticity in the HIV-1 Fusion Peptide Facilitates Recognition by Broadly Neutralizing Antibodies. Cell Host Microbe. 2019;25:873–883.e5. doi: 10.1016/j.chom.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schoofs T., Barnes C.O., Suh-Toma N., Golijanin J., Schommers P., Gruell H., West A.P.J., Bach F., Lee Y.E., Nogueira L., et al. Broad and Potent Neutralizing Antibodies Recognize the Silent Face of the HIV Envelope. Immunity. 2019;50:1513–1529.e9. doi: 10.1016/j.immuni.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huang J., Ofek G., Laub L., Louder M.K., Doria-Rose N.A., Longo N.S., Imamichi H., Bailer R.T., Chakrabarti B., Sharma S.K., et al. Broad and Potent Neutralization of HIV-1 by a Gp41-Specific Human Antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rantalainen K., Berndsen Z.T., Antanasijevic A., Schiffner T., Zhang X., Lee W.-H., Torres J.L., Zhang L., Irimia A., Copps J., et al. HIV-1 Envelope and MPER Antibody Structures in Lipid Assemblies. Cell Rep. 2020;31:107583. doi: 10.1016/j.celrep.2020.107583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jardine J.G., Kulp D.W., Havenar-Daughton C., Sarkar A., Briney B., Sok D., Sesterhenn F., Ereño-Orbea J., Kalyuzhniy O., Deresa I., et al. HIV-1 Broadly Neutralizing Antibody Precursor B Cells Revealed by Germline-Targeting Immunogen. Science. 2016;351:1458–1463. doi: 10.1126/science.aad9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou T., Georgiev I., Wu X., Yang Z.-Y., Dai K., Finzi A., Kwon Y.D., Scheid J.F., Shi W., Xu L., et al. Structural Basis for Broad and Potent Neutralization of HIV-1 by Antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang X., Meining W., Cushman M., Haase I., Fischer M., Bacher A., Ladenstein R. A Structure-Based Model of the Reaction Catalyzed by Lumazine Synthase from Aquifex Aeolicus. J. Mol. Biol. 2003;328:167–182. doi: 10.1016/S0022-2836(03)00186-4. [DOI] [PubMed] [Google Scholar]
- 109.Bennett N.R., Zwick D.B., Courtney A.H., Kiessling L.L. Multivalent Antigens for Promoting B and T Cell Activation. ACS Chem. Biol. 2015;10:1817–1824. doi: 10.1021/acschembio.5b00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.De Gregorio E., Rappuoli R. From Empiricism to Rational Design: A Personal Perspective of the Evolution of Vaccine Development. Nat. Rev. Immunol. 2014;14:505–514. doi: 10.1038/nri3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rappuoli R., Bottomley M.J., D’Oro U., Finco O., De Gregorio E. Reverse Vaccinology 2.0: Human Immunology Instructs Vaccine Antigen Design. J. Exp. Med. 2016;213:469–481. doi: 10.1084/jem.20151960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee P.S., Wilson I.A. Structural Characterization of Viral Epitopes Recognized by Broadly Cross-Reactive Antibodies. Curr. Top. Microbiol. Immunol. 2015;386:323–341. doi: 10.1007/82_2014_413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wilson I.A., Stanfield R.L. Antibody-Antigen Interactions: New Structures and New Conformational Changes. Curr. Opin. Struct. Biol. 1994;4:857–867. doi: 10.1016/0959-440X(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 114.Bernstein F.C., Koetzle T.F., Williams G.J.B., Meyer E.F., Brice M.D., Rodgers J.R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: A Computer-Based Archival File for Macromolecular Structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/S0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 115.Wlodawer A., Minor W., Dauter Z., Jaskolski M. Protein Crystallography for Aspiring Crystallographers or How to Avoid Pitfalls and Traps in Macromolecular Structure Determination. FEBS J. 2013;280:5705–5736. doi: 10.1111/febs.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li Y., Li H., Yang F., Smith-Gill S.J., Mariuzza R.A. X-Ray Snapshots of the Maturation of an Antibody Response to a Protein Antigen. Nat. Struct. Biol. 2003;10:482–488. doi: 10.1038/nsb930. [DOI] [PubMed] [Google Scholar]
- 117.Purslow J.A., Khatiwada B., Bayro M.J., Venditti V. NMR Methods for Structural Characterization of Protein-Protein Complexes. Front. Mol. Biosci. 2020;7:9. doi: 10.3389/fmolb.2020.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rosen O., Anglister J. Epitope Mapping of Antibody-Antigen Complexes by Nuclear Magnetic Resonance Spectroscopy. Methods Mol. Biol. 2009;524:37–57. doi: 10.1007/978-1-59745-450-6_3. [DOI] [PubMed] [Google Scholar]
- 119.Bardelli M., Livoti E., Simonelli L., Pedotti M., Moraes A., Valente A.P., Varani L. Epitope Mapping by Solution NMR Spectroscopy. J. Mol. Recognition JMR. 2015;28:393–400. doi: 10.1002/jmr.2454. [DOI] [PubMed] [Google Scholar]
- 120.Sapienza P.J., Lee A.L. Using NMR to Study Fast Dynamics in Proteins: Methods and Applications. Curr. Opin. Pharmacol. 2010;10:723–730. doi: 10.1016/j.coph.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Krishnan V., Rupp B. Encyclopedia of Life Sciences. John Wiley & Sons, Ltd.; Hoboken, NJ, USA: 2012. Macromolecular Structure Determination: Comparison of X-ray Crystallography and NMR Spectroscopy. [DOI] [Google Scholar]
- 122.Shoemaker S.C., Ando N. X-Rays in the Cryo-Electron Microscopy Era: Structural Biology’s Dynamic Future. Biochemistry. 2018;57:277–285. doi: 10.1021/acs.biochem.7b01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bai X., McMullan G., Scheres S.H.W. How Cryo-EM Is Revolutionizing Structural Biology. Trends Biochem. Sci. 2015;40:49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 124.Cheng Y. Single-Particle Cryo-EM at Crystallographic Resolution. Cell. 2015;161:450–457. doi: 10.1016/j.cell.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bianchi M., Turner H.L., Nogal B., Cottrell C.A., Oyen D., Pauthner M., Bastidas R., Nedellec R., McCoy L.E., Wilson I.A., et al. Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization. Immunity. 2018;49:288–300.e8. doi: 10.1016/j.immuni.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Antanasijevic A., Bowman C.A., Kirchdoerfer R.N., Cottrell C.A., Ozorowski G., Upadhyay A.A., Cirelli K.M., Carnathan D.G., Enemuo C.A., Sewall L.M., et al. From Structure to Sequence: Antibody Discovery Using cryoEM. Sci. Adv. 2022;8:eabk2039. doi: 10.1126/sciadv.abk2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Janda A., Bowen A., Greenspan N.S., Casadevall A. Ig Constant Region Effects on Variable Region Structure and Function. Front. Microbiol. 2016;7:22. doi: 10.3389/fmicb.2016.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tonegawa S. Somatic Generation of Antibody Diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 129.Cooper L.J., Shikhman A.R., Glass D.D., Kangisser D., Cunningham M.W., Greenspan N.S. Role of Heavy Chain Constant Domains in Antibody-Antigen Interaction. Apparent Specificity Differences among Streptococcal IgG Antibodies Expressing Identical Variable Domains. J. Immunol. 1993;150:2231–2242. doi: 10.4049/jimmunol.150.6.2231. [DOI] [PubMed] [Google Scholar]
- 130.Cooper L.J., Robertson D., Granzow R., Greenspan N.S. Variable Domain-Identical Antibodies Exhibit IgG Subclass-Related Differences in Affinity and Kinetic Constants as Determined by Surface Plasmon Resonance. Mol. Immunol. 1994;31:577–584. doi: 10.1016/0161-5890(94)90165-1. [DOI] [PubMed] [Google Scholar]
- 131.Huber R., Deisenhofer J., Colman P.M., Matsushima M., Palm W. Crystallographic Structure Studies of an IgG Molecule and an Fc Fragment. Nature. 1976;264:415–420. doi: 10.1038/264415a0. [DOI] [PubMed] [Google Scholar]
- 132.Pritsch O., Magnac C., Dumas G., Bouvet J.P., Alzari P., Dighiero G. Can Isotype Switch Modulate Antigen-Binding Affinity and Influence Clonal Selection? Eur. J. Immunol. 2000;30:3387–3395. doi: 10.1002/1521-4141(2000012)30:12<3387::AID-IMMU3387>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 133.Sheriff S., Jeffrey P.D., Bajorath J. Comparison of CH1 Domains in Different Classes of Murine Antibodies. J. Mol. Biol. 1996;263:385–389. doi: 10.1006/jmbi.1996.0582. [DOI] [PubMed] [Google Scholar]
- 134.Normansell D.E. Human Immunoglobulin Subclasses. Diagn. Clin. Immunol. 1987;5:115–128. [PubMed] [Google Scholar]
- 135.Torres M., Casadevall A. The Immunoglobulin Constant Region Contributes to Affinity and Specificity. Trends Immunol. 2008;29:91–97. doi: 10.1016/j.it.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 136.Williams A.F., Barclay A.N. The Immunoglobulin Superfamily—Domains for Cell Surface Recognition. Annu. Rev. Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 137.Proctor E.A., Kota P., Demarest S.J., Caravella J.A., Dokholyan N.V. Highly Covarying Residues Have a Functional Role in Antibody Constant Domains. Proteins Struct. Funct. Bioinform. 2013;81:884–895. doi: 10.1002/prot.24247. [DOI] [PubMed] [Google Scholar]
- 138.Pomarici N.D., Fernández-Quintero M.L., Quoika P.K., Waibl F., Bujotzek A., Georges G., Liedl K.R. Bispecific Antibodies—Effects of Point Mutations on CH3-CH3 Interface Stability. Protein Eng. Des. Sel. 2022;35:gzac012. doi: 10.1093/protein/gzac012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Krapp S., Mimura Y., Jefferis R., Huber R., Sondermann P. Structural Analysis of Human IgG-Fc Glycoforms Reveals a Correlation Between Glycosylation and Structural Integrity. J. Mol. Biol. 2003;325:979–989. doi: 10.1016/S0022-2836(02)01250-0. [DOI] [PubMed] [Google Scholar]
- 140.Radaev S., Motyka S., Fridman W.-H., Sautes-Fridman C., Sun P.D. The Structure of a Human Type III Fcγ Receptor in Complex with Fc*. J. Biol. Chem. 2001;276:16469–16477. doi: 10.1074/jbc.M100350200. [DOI] [PubMed] [Google Scholar]
- 141.Köhler G., Milstein C. Continuous Cultures of Fused Cells Secreting Antibody of Predefined Specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 142.Zhang C. Hybridoma Technology for the Generation of Monoclonal Antibodies. In: Proetzel G., Ebersbach H., editors. Antibody Methods and Protocols. Humana Press; Totowa, NJ, USA: 2012. pp. 117–135. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 143.ul Haque Saeed A.F., ul Haque Saeed A.F. Advances in Monoclonal Antibodies Production and Cancer Therapy. MOJ Immunol. 2016;3:99. doi: 10.15406/moji.2016.03.00099. [DOI] [Google Scholar]
- 144.Vaisman-Mentesh A., Rosenstein S., Yavzori M., Dror Y., Fudim E., Ungar B., Kopylov U., Picard O., Kigel A., Ben-Horin S., et al. Molecular Landscape of Anti-Drug Antibodies Reveals the Mechanism of the Immune Response Following Treatment With TNFα Antagonists. Front. Immunol. 2019;10:2921. doi: 10.3389/fimmu.2019.02921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mosch R., Guchelaar H.-J. Immunogenicity of Monoclonal Antibodies and the Potential Use of HLA Haplotypes to Predict Vulnerable Patients. Front. Immunol. 2022;13:885672. doi: 10.3389/fimmu.2022.885672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lynch D.H., Yang X.-D. Therapeutic Potential of ABX-EGF: A Fully Human Anti-Epidermal Growth Factor Receptor Monoclonal Antibody for Cancer Treatment. Semin. Oncol. 2002;29((Suppl. S4)):47–50. doi: 10.1053/sonc.2002.31522. [DOI] [PubMed] [Google Scholar]
- 147.Nelson A.L., Dhimolea E., Reichert J.M. Development Trends for Human Monoclonal Antibody Therapeutics. Nat. Rev. Drug Discov. 2010;9:767–774. doi: 10.1038/nrd3229. [DOI] [PubMed] [Google Scholar]
- 148.Lonberg N., Taylor L.D., Harding F.A., Trounstine M., Higgins K.M., Schramm S.R., Kuo C.-C., Mashayekh R., Wymore K., McCabe J.G., et al. Antigen-Specific Human Antibodies from Mice Comprising Four Distinct Genetic Modifications. Nature. 1994;368:856–859. doi: 10.1038/368856a0. [DOI] [PubMed] [Google Scholar]
- 149.Green L.L. Antibody Engineering via Genetic Engineering of the Mouse: XenoMouse Strains Are a Vehicle for the Facile Generation of Therapeutic Human Monoclonal Antibodies. J. Immunol. Methods. 1999;231:11–23. doi: 10.1016/S0022-1759(99)00137-4. [DOI] [PubMed] [Google Scholar]
- 150.Taylor L.D., Carmack C.E., Schramm S.R., Mashayekh R., Higgins K.M., Kuo C.C., Woodhouse C., Kay R.M., Lonberg N. A Transgenic Mouse That Expresses a Diversity of Human Sequence Heavy and Light Chain Immunoglobulins. Nucleic Acids Res. 1992;20:6287–6295. doi: 10.1093/nar/20.23.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hwang W.Y.K., Foote J. Immunogenicity of Engineered Antibodies. Methods. 2005;36:3–10. doi: 10.1016/j.ymeth.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 152.Shankar G., Shores E., Wagner C., Mire-Sluis A. Scientific and Regulatory Considerations on the Immunogenicity of Biologics. Trends Biotechnol. 2006;24:274–280. doi: 10.1016/j.tibtech.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 153.Morrison S.L., Johnson M.J., Herzenberg L.A., Oi V.T. Chimeric Human Antibody Molecules: Mouse Antigen-Binding Domains with Human Constant Region Domains. Proc. Natl. Acad. Sci. USA. 1984;81:6851–6855. doi: 10.1073/pnas.81.21.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jones P.T., Dear P.H., Foote J., Neuberger M.S., Winter G. Replacing the Complementarity-Determining Regions in a Human Antibody with Those from a Mouse. Nature. 1986;321:522–525. doi: 10.1038/321522a0. [DOI] [PubMed] [Google Scholar]
- 155.Pelat T., Bedouelle H., Rees A.R., Crennell S.J., Lefranc M.-P., Thullier P. Germline Humanization of a Non-Human Primate Antibody That Neutralizes the Anthrax Toxin, by in Vitro and in Silico Engineering. J. Mol. Biol. 2008;384:1400–1407. doi: 10.1016/j.jmb.2008.10.033. [DOI] [PubMed] [Google Scholar]
- 156.Robert R., Streltsov V.A., Newman J., Pearce L.A., Wark K.L., Dolezal O. Germline Humanization of a Murine Aβ Antibody and Crystal Structure of the Humanized Recombinant Fab Fragment. Protein Sci. 2010;19:299–308. doi: 10.1002/pro.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cheung N.-K.V., Guo H., Hu J., Tassev D.V., Cheung I.Y. Humanizing Murine IgG3 Anti-GD2 Antibody m3F8 Substantially Improves Antibody-Dependent Cell-Mediated Cytotoxicity While Retaining Targeting in Vivo. OncoImmunology. 2012;1:477–486. doi: 10.4161/onci.19864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kim J.H., Hong H.J. Humanization by CDR Grafting and Specificity-Determining Residue Grafting. In: Chames P., editor. Antibody Engineering: Methods and Protocols. 2nd ed. Humana Press; Totowa, NJ, USA: 2012. pp. 237–245. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 159.Makabe K., Nakanishi T., Tsumoto K., Tanaka Y., Kondo H., Umetsu M., Sone Y., Asano R., Kumagai I. Thermodynamic Consequences of Mutations in Vernier Zone Residues of a Humanized Anti-Human Epidermal Growth Factor Receptor Murine Antibody, 528*. J. Biol. Chem. 2008;283:1156–1166. doi: 10.1074/jbc.M706190200. [DOI] [PubMed] [Google Scholar]
- 160.Doria-Rose N.A., Joyce M.G. Strategies to Guide the Antibody Affinity Maturation Process. Curr. Opin. Virol. 2015;11:137–147. doi: 10.1016/j.coviro.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ducancel F., Muller B.H. Molecular Engineering of Antibodies for Therapeutic and Diagnostic Purposes. mAbs. 2012;4:445–457. doi: 10.4161/mabs.20776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Boder E.T., Midelfort K.S., Wittrup K.D. Directed Evolution of Antibody Fragments with Monovalent Femtomolar Antigen-Binding Affinity. Proc. Natl. Acad. Sci. USA. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rajpal A., Beyaz N., Haber L., Cappuccilli G., Yee H., Bhatt R.R., Takeuchi T., Lerner R.A., Crea R. A General Method for Greatly Improving the Affinity of Antibodies by Using Combinatorial Libraries. Proc. Natl. Acad. Sci. USA. 2005;102:8466–8471. doi: 10.1073/pnas.0503543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lippow S.M., Wittrup K.D., Tidor B. Computational Design of Antibody Affinity Improvement beyond in Vivo Maturation. Nat. Biotechnol. 2007;25:1171–1176. doi: 10.1038/nbt1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Clark L.A., Boriack-Sjodin P.A., Eldredge J., Fitch C., Friedman B., Hanf K.J.M., Jarpe M., Liparoto S.F., Li Y., Lugovskoy A., et al. Affinity Enhancement of an in Vivo Matured Therapeutic Antibody Using Structure-Based Computational Design. Protein Sci. 2006;15:949–960. doi: 10.1110/ps.052030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Akiba H., Tamura H., Caaveiro J.M.M., Tsumoto K. Computer-Guided Library Generation Applied to the Optimization of Single-Domain Antibodies. Protein Eng. Des. Sel. 2019;32:423–431. doi: 10.1093/protein/gzaa006. [DOI] [PubMed] [Google Scholar]
- 167.Chiba S., Okuno Y., Ohta M. Structure-Based Affinity Maturation of Antibody Based on Double-Point Mutations. In: Tsumoto K., Kuroda D., editors. Computer-Aided Antibody Design. Springer; New York, NY, USA: 2023. pp. 323–331. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 168.Sulea T., Deprez C., Corbeil C.R., Purisima E.O. Optimizing Antibody–Antigen Binding Affinities with the ADAPT Platform. In: Tsumoto K., Kuroda D., editors. Computer-Aided Antibody Design. Springer; New York, NY, USA: 2023. pp. 361–374. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 169.Fernández-Quintero M.L., Loeffler J.R., Bacher L.M., Waibl F., Seidler C.A., Liedl K.R. Local and Global Rigidification Upon Antibody Affinity Maturation. Front. Mol. Biosci. 2020;7:182. doi: 10.3389/fmolb.2020.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fernández-Quintero M.L., Seidler C.A., Quoika P.K., Liedl K.R. Shark Antibody Variable Domains Rigidify Upon Affinity Maturation—Understanding the Potential of Shark Immunoglobulins as Therapeutics. Front. Mol. Biosci. 2021;8:639166. doi: 10.3389/fmolb.2021.639166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fischer E. Einfluss Der Configuration Auf Die Wirkung Der Enzyme. Ber. Dtsch. Chem. Ges. 1894;27:2985–2993. doi: 10.1002/cber.18940270364. [DOI] [Google Scholar]
- 172.Braden B.C., Dall’Acqua W., Eisenstein E., Fields B.A., Goldbaum F.A., Malchiodi E.L., Mariuzza R.A., Schwarz F.P., Ysern X., Poljak R.J. Protein Motion and Lock and Key Complementarity in Antigen-Antibody Reactions. Pharm. Acta Helv. 1995;69:225–230. doi: 10.1016/0031-6865(94)00046-X. [DOI] [PubMed] [Google Scholar]
- 173.Ma B., Kumar S., Tsai C.-J., Nussinov R. Folding Funnels and Binding Mechanisms. Protein Eng. Des. Sel. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- 174.Csermely P., Palotai R., Nussinov R. Induced Fit, Conformational Selection and Independent Dynamic Segments: An Extended View of Binding Events. Trends Biochem. Sci. 2010;35:539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang W., Ye W., Yu Q., Jiang C., Zhang J., Luo R., Chen H.-F. Conformational Selection and Induced Fit in Specific Antibody and Antigen Recognition: SPE7 as a Case Study. J. Phys. Chem. B. 2013;117:4912–4923. doi: 10.1021/jp4010967. [DOI] [PubMed] [Google Scholar]
- 176.Klein C., Sustmann C., Thomas M., Stubenrauch K., Croasdale R., Schanzer J., Brinkmann U., Kettenberger H., Regula J.T., Schaefer W. Progress in Overcoming the Chain Association Issue in Bispecific Heterodimeric IgG Antibodies. mAbs. 2012;4:653–663. doi: 10.4161/mabs.21379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Krah S., Sellmann C., Rhiel L., Schröter C., Dickgiesser S., Beck J., Zielonka S., Toleikis L., Hock B., Kolmar H., et al. Engineering Bispecific Antibodies with Defined Chain Pairing. New Biotechnol. 2017;39:167–173. doi: 10.1016/j.nbt.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 178.Krah S., Kolmar H., Becker S., Zielonka S. Engineering IgG-Like Bispecific Antibodies—An Overview. Antibodies. 2018;7:28. doi: 10.3390/antib7030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Choi H.-J., Seok S.-H., Kim Y.-J., Seo M.-D., Kim Y.-S. Crystal Structures of Immunoglobulin Fc Heterodimers Reveal the Molecular Basis for Heterodimer Formation. Mol. Immunol. 2015;65:377–383. doi: 10.1016/j.molimm.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 180.De Nardis C., Hendriks L.J.A., Poirier E., Arvinte T., Gros P., Bakker A.B.H., de Kruif J. A New Approach for Generating Bispecific Antibodies Based on a Common Light Chain Format and the Stable Architecture of Human Immunoglobulin G1. J. Biol. Chem. 2017;292:14706–14717. doi: 10.1074/jbc.M117.793497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gunasekaran K., Pentony M., Shen M., Garrett L., Forte C., Woodward A., Ng S.B., Born T., Retter M., Manchulenko K., et al. Enhancing Antibody Fc Heterodimer Formation through Electrostatic Steering Effects. J. Biol. Chem. 2010;285:19637–19646. doi: 10.1074/jbc.M110.117382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ridgway J.B., Presta L.G., Carter P. “Knobs-into-Holes” Engineering of Antibody CH3 Domains for Heavy Chain Heterodimerization. Protein Eng. 1996;9:617–621. doi: 10.1093/protein/9.7.617. [DOI] [PubMed] [Google Scholar]
- 183.Elliott J.M., Ultsch M., Lee J., Tong R., Takeda K., Spiess C., Eigenbrot C., Scheer J.M. Antiparallel Conformation of Knob and Hole Aglycosylated Half-Antibody Homodimers Is Mediated by a CH2-CH3 Hydrophobic Interaction. J. Mol. Biol. 2014;426:1947–1957. doi: 10.1016/j.jmb.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 184.Dengl S., Mayer K., Bormann F., Duerr H., Hoffmann E., Nussbaum B., Tischler M., Wagner M., Kuglstatter A., Leibrock L., et al. Format Chain Exchange (FORCE) for High-Throughput Generation of Bispecific Antibodies in Combinatorial Binder-Format Matrices. Nat. Commun. 2020;11:4974. doi: 10.1038/s41467-020-18477-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Krah S., Schröter C., Eller C., Rhiel L., Rasche N., Beck J., Sellmann C., Günther R., Toleikis L., Hock B., et al. Generation of Human Bispecific Common Light Chain Antibodies by Combining Animal Immunization and Yeast Display. Protein Eng. Des. Sel. 2017;30:291–301. doi: 10.1093/protein/gzw077. [DOI] [PubMed] [Google Scholar]
- 186.Van Blarcom T., Lindquist K., Melton Z., Cheung W.L., Wagstrom C., McDonough D., Valle Oseguera C., Ding S., Rossi A., Potluri S., et al. Productive Common Light Chain Libraries Yield Diverse Panels of High Affinity Bispecific Antibodies. mAbs. 2018;10:256–268. doi: 10.1080/19420862.2017.1406570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shiraiwa H., Narita A., Kamata-Sakurai M., Ishiguro T., Sano Y., Hironiwa N., Tsushima T., Segawa H., Tsunenari T., Ikeda Y., et al. Engineering a Bispecific Antibody with a Common Light Chain: Identification and Optimization of an Anti-CD3 Epsilon and Anti-GPC3 Bispecific Antibody, ERY974. Methods. 2019;154:10–20. doi: 10.1016/j.ymeth.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 188.Bönisch M., Sellmann C., Maresch D., Halbig C., Becker S., Toleikis L., Hock B., Rüker F. Novel CH1:CL Interfaces That Enhance Correct Light Chain Pairing in Heterodimeric Bispecific Antibodies. Protein Eng. Des. Sel. 2017;30:685–696. doi: 10.1093/protein/gzx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Schaefer W., Regula J.T., Bähner M., Schanzer J., Croasdale R., Dürr H., Gassner C., Georges G., Kettenberger H., Imhof-Jung S., et al. Immunoglobulin Domain Crossover as a Generic Approach for the Production of Bispecific IgG Antibodies. Proc. Natl. Acad. Sci. USA. 2011;108:11187–11192. doi: 10.1073/pnas.1019002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Shen Z., Yan H., Zhang Y., Mernaugh R.L., Zeng X. Engineering Peptide Linkers for scFv Immunosensors. Anal. Chem. 2008;80:1910–1917. doi: 10.1021/ac7018624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yusakul G., Sakamoto S., Pongkitwitoon B., Tanaka H., Morimoto S. Effect of Linker Length between Variable Domains of Single Chain Variable Fragment Antibody against Daidzin on Its Reactivity. Biosci. Biotechnol. Biochem. 2016;80:1306–1312. doi: 10.1080/09168451.2016.1156482. [DOI] [PubMed] [Google Scholar]
- 192.Holliger P., Prospero T., Winter G. “Diabodies”: Small Bivalent and Bispecific Antibody Fragments. Proc. Natl. Acad. Sci. USA. 1993;90:6444–6448. doi: 10.1073/pnas.90.14.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kwon N.-Y., Kim Y., Lee J.-O. Structural Diversity and Flexibility of Diabodies. Methods. 2019;154:136–142. doi: 10.1016/j.ymeth.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 194.Math B.A., Waibl F., Lamp L.M., Fernández-Quintero M.L., Liedl K.R. Cross-linking Disulfide Bonds Govern Solution Structures of Diabodies. Proteins. 2023;91:1316–1328. doi: 10.1002/prot.26509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Harmsen M., de Haard H. Properties, Production, and Applications of Camelid Single-Domain Antibody Fragments. Appl. Microbiol. Biotechnol. 2007;77:13–22. doi: 10.1007/s00253-007-1142-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Stanfield R.L., Dooley H., Flajnik M.F., Wilson I.A. Crystal Structure of a Shark Single-Domain Antibody V Region in Complex with Lysozyme. Science. 2004;305:1770–1773. doi: 10.1126/science.1101148. [DOI] [PubMed] [Google Scholar]
- 197.Dooley H., Flajnik M.F. Antibody Repertoire Development in Cartilaginous Fish. Dev. Comp. Immunol. 2006;30:43–56. doi: 10.1016/j.dci.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 198.Muyldermans S. Nanobodies: Natural Single-Domain Antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 199.Matz H., Dooley H. Shark IgNAR-Derived Binding Domains as Potential Diagnostic and Therapeutic Agents. Dev. Comp. Immunol. 2019;90:100–107. doi: 10.1016/j.dci.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 200.Gaudreault F., Corbeil C.R., Purisima E.O., Sulea T. Coevolved Canonical Loops Conformations of Single-Domain Antibodies: A Tale of Three Pockets Playing Musical Chairs. Front. Immunol. 2022;13:884132. doi: 10.3389/fimmu.2022.884132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sehlin D., Stocki P., Gustavsson T., Hultqvist G., Walsh F.S., Rutkowski J.L., Syvänen S. Brain Delivery of Biologics Using a Cross-Species Reactive Transferrin Receptor 1 VNAR Shuttle. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020;34:13272–13283. doi: 10.1096/fj.202000610RR. [DOI] [PubMed] [Google Scholar]
- 202.Clarke E., Stocki P., Sinclair E.H., Gauhar A., Fletcher E.J.R., Krawczun-Rygmaczewska A., Duty S., Walsh F.S., Doherty P., Rutkowski J.L. A Single Domain Shark Antibody Targeting the Transferrin Receptor 1 Delivers a TrkB Agonist Antibody to the Brain and Provides Full Neuroprotection in a Mouse Model of Parkinson’s Disease. Pharmaceutics. 2022;14:1335. doi: 10.3390/pharmaceutics14071335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Muyldermans S., Atarhouch T., Saldanha J., Barbosa J.A.R.G., Hamers R. Sequence and Structure of VH Domain from Naturally Occurring Camel Heavy Chain Immunoglobulins Lacking Light Chains. Protein Eng. Des. Sel. 1994;7:1129–1135. doi: 10.1093/protein/7.9.1129. [DOI] [PubMed] [Google Scholar]
- 204.De Genst E., Silence K., Decanniere K., Conrath K., Loris R., Kinne J., Muyldermans S., Wyns L. Molecular Basis for the Preferential Cleft Recognition by Dromedary Heavy-Chain Antibodies. Proc. Natl. Acad. Sci. USA. 2006;103:4586–4591. doi: 10.1073/pnas.0505379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zavrtanik U., Lukan J., Loris R., Lah J., Hadži S. Structural Basis of Epitope Recognition by Heavy-Chain Camelid Antibodies. J. Mol. Biol. 2018;430:4369–4386. doi: 10.1016/j.jmb.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 206.Muyldermans S., Cambillau C., Wyns L. Recognition of Antigens by Single-Domain Antibody Fragments: The Superfluous Luxury of Paired Domains. Trends Biochem. Sci. 2001;26:230–235. doi: 10.1016/S0968-0004(01)01790-X. [DOI] [PubMed] [Google Scholar]
- 207.Govaert J., Pellis M., Deschacht N., Vincke C., Conrath K., Muyldermans S., Saerens D. Dual Beneficial Effect of Interloop Disulfide Bond for Single Domain Antibody Fragments*. J. Biol. Chem. 2011;287:1970–1979. doi: 10.1074/jbc.M111.242818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Löhr T., Sormanni P., Vendruscolo M. Conformational Entropy as a Potential Liability of Computationally Designed Antibodies. Biomolecules. 2022;12:718. doi: 10.3390/biom12050718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ewert S., Cambillau C., Conrath K., Plückthun A. Biophysical Properties of Camelid V(HH) Domains Compared to Those of Human V(H)3 Domains. Biochemistry. 2002;41:3628–3636. doi: 10.1021/bi011239a. [DOI] [PubMed] [Google Scholar]
- 210.Teplyakov A., Zhao Y., Malia T.J., Obmolova G., Gilliland G.L. IgG2 Fc Structure and the Dynamic Features of the IgG CH2-CH3 Interface. Mol. Immunol. 2013;56:131–139. doi: 10.1016/j.molimm.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 211.Fernández-Quintero M.L., Fischer A.-L.M., Kokot J., Waibl F., Seidler C.A., Liedl K.R. The Influence of Antibody Humanization on Shark Variable Domain (VNAR) Binding Site Ensembles. Front. Immunol. 2022;13:953917. doi: 10.3389/fimmu.2022.953917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Fernández-Quintero M.L., Seidler C.A., Liedl K.R. T-Cell Receptor Variable β Domains Rigidify During Affinity Maturation. Sci. Rep. 2020;10:4472. doi: 10.1038/s41598-020-61433-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kovalenko O., Olland A., Piché-Nicholas N., Godbole A., King D., Svenson K., Calabro V., Müller M., Barelle C., Somers W., et al. Atypical Antigen Recognition Mode of a Shark IgNAR Variable Domain Characterized by Humanization and Structural Analysis. J. Biol. Chem. 2013;288:17408–17419. doi: 10.1074/jbc.M112.435289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Brazeau M.D., Friedman M. The Origin and Early Phylogenetic History of Jawed Vertebrates. Nature. 2015;520:490–497. doi: 10.1038/nature14438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Flajnik M.F. A Cold-Blooded View of Adaptive Immunity. Nat. Rev. Immunol. 2018;18:438–453. doi: 10.1038/s41577-018-0003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Nguyen V.K., Hamers R., Wyns L., Muyldermans S. Camel Heavy-Chain Antibodies: Diverse Germline V(H)H and Specific Mechanisms Enlarge the Antigen-Binding Repertoire. EMBO J. 2000;19:921–930. doi: 10.1093/emboj/19.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Burger P.A. The History of Old World Camelids in the Light of Molecular Genetics. Trop. Anim. Health Prod. 2016;48:905–913. doi: 10.1007/s11250-016-1032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Flajnik M.F., Deschacht N., Muyldermans S. A Case of Convergence: Why Did a Simple Alternative to Canonical Antibodies Arise in Sharks and Camels? PLoS Biol. 2011;9:e1001120. doi: 10.1371/journal.pbio.1001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Nguyen V.K., Su C., Muyldermans S., van der Loo W. Heavy-Chain Antibodies in Camelidae; a Case of Evolutionary Innovation. Immunogenetics. 2002;54:39–47. doi: 10.1007/s00251-002-0433-0. [DOI] [PubMed] [Google Scholar]
- 220.Klarenbeek A., Mazouari K.E., Desmyter A., Blanchetot C., Hultberg A., de Jonge N., Roovers R.C., Cambillau C., Spinelli S., Del-Favero J., et al. Camelid Ig V Genes Reveal Significant Human Homology Not Seen in Therapeutic Target Genes, Providing for a Powerful Therapeutic Antibody Platform. mAbs. 2015;7:693–706. doi: 10.1080/19420862.2015.1046648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Rossotti M.A., Bélanger K., Henry K.A., Tanha J. Immunogenicity and Humanization of Single-Domain Antibodies. FEBS J. 2022;289:4304–4327. doi: 10.1111/febs.15809. [DOI] [PubMed] [Google Scholar]
- 222.Stanfield R.L., Dooley H., Verdino P., Flajnik M.F., Wilson I.A. Maturation of Shark Single-Domain (IgNAR) Antibodies: Evidence for Induced-Fit Binding. J. Mol. Biol. 2007;367:358–372. doi: 10.1016/j.jmb.2006.12.045. [DOI] [PubMed] [Google Scholar]
- 223.Hoey R.J., Eom H., Horn J.R. Structure and Development of Single Domain Antibodies as Modules for Therapeutics and Diagnostics. Exp. Biol. Med. 2019;244:1568–1576. doi: 10.1177/1535370219881129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sulea T. Humanization of Camelid Single-Domain Antibodies. Methods Mol. Biol. 2022;2446:299–312. doi: 10.1007/978-1-0716-2075-5_14. [DOI] [PubMed] [Google Scholar]
- 225.Steeland S., Vandenbroucke R.E., Libert C. Nanobodies as Therapeutics: Big Opportunities for Small Antibodies. Drug Discov. Today. 2016;21:1076–1113. doi: 10.1016/j.drudis.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 226.Steven J., Müller M.R., Carvalho M.F., Ubah O.C., Kovaleva M., Donohoe G., Baddeley T., Cornock D., Saunders K., Porter A.J., et al. In Vitro Maturation of a Humanized Shark VNAR Domain to Improve Its Biophysical Properties to Facilitate Clinical Development. Front. Immunol. 2017;8:1361. doi: 10.3389/fimmu.2017.01361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kinoshita S., Nakakido M., Mori C., Kuroda D., Caaveiro J.M.M., Tsumoto K. Molecular Basis for Thermal Stability and Affinity in a VHH: Contribution of the Framework Region and Its Influence in the Conformation of the CDR3. Protein Sci. A Publ. Protein Soc. 2022;31:e4450. doi: 10.1002/pro.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Fernández-Quintero M.L., DeRose E.F., Gabel S.A., Mueller G.A., Liedl K.R. Nanobody Paratope Ensembles in Solution Characterized by MD Simulations and NMR. Int. J. Mol. Sci. 2022;23:5419. doi: 10.3390/ijms23105419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Regep C., Georges G., Shi J., Popovic B., Deane C.M. The H3 Loop of Antibodies Shows Unique Structural Characteristics. Proteins. 2017;85:1311–1318. doi: 10.1002/prot.25291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Abanades B., Wong W.K., Boyles F., Georges G., Bujotzek A., Deane C.M. ImmuneBuilder: Deep-Learning Models for Predicting the Structures of Immune Proteins. bioRxiv. 2022 doi: 10.1038/s42003-023-04927-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Cohen T., Halfon M., Schneidman-Duhovny D. NanoNet: Rapid and Accurate End-to-End Nanobody Modeling by Deep Learning. Front. Immunol. 2022;13:958584. doi: 10.3389/fimmu.2022.958584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.West B.R., Wec A.Z., Moyer C.L., Fusco M.L., Ilinykh P.A., Huang K., Wirchnianski A.S., James R.M., Herbert A.S., Hui S., et al. Structural Basis of Broad Ebolavirus Neutralization by a Human Survivor Antibody. Nat. Struct. Mol. Biol. 2019;26:204–212. doi: 10.1038/s41594-019-0191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Koide A., Tereshko V., Uysal S., Margalef K., Kossiakoff A.A., Koide S. Exploring the Capacity of Minimalist Protein Interfaces: Interface Energetics and Affinity Maturation to Picomolar KD of a Single-Domain Antibody with a Flat Paratope. J. Mol. Biol. 2007;373:941–953. doi: 10.1016/j.jmb.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.