Abstract
Mitochondrial toxicity drives several adverse health outcomes. Current high-throughput screening assays for chemically induced mitochondrial toxicity typically measure changes to mitochondrial structure and may not detect known mitochondrial toxicants. We adapted a respirometric screening assay (RSA) measuring mitochondrial function to screen ToxCast chemicals in HepG2 cells using a tiered testing strategy. Of 1042 chemicals initially screened at a singlemaximal concentration, 243 actives were identified and rescreened at 7 concentrations. Concentration-response data for 3 respiration phases confirmed activity and indicated a mechanism for 193 mitochondrial toxicants: 149 electron transport chain inhibitors (ETCi), 15 uncouplers and 29 adenosine triphosphate synthase inhibitors. Subsequently, an electron flow assay was used to identify the target complex for 84 of the 149 ETCi. Sixty reference chemicals were used to compare the RSA to existing ToxCast and Tox21 mitochondrial toxicity assays. The RSA was most predictive (accuracy = 90%) of mitochondrial toxicity. The Tox21 mitochondrial membrane potential assay was also highly predictive (accuracy = 87%) of bioactivity but underestimated the potency of well-known ETCi and provided no mechanistic information. The tiered RSA approach accurately identifies and characterizes mitochondrial toxicants acting through diverse mechanisms and at a throughput sufficient to screen large chemical inventories. The electron flow assay provides additional confirmation and detailed mechanistic understanding for ETCi, the most common type of mitochondrial toxicants among ToxCast chemicals. The mitochondrial toxicity screening approach described herein may inform hazard assessment and the in vitro bioactive concentrations used to derive relevant doses for screening level chemical assessment using new approach methodologies.
Keywords: mitochondria, mechanism, electron transport chain, uncoupling, oxidative phosphorylation, respiration, high-throughput screening, Seahorse, ToxCast, new approach methodologies
Mitochondria are organelles in eukaryotic cells responsible for producing cellular adenosine triphosphate (ATP) through oxidative phosphorylation. Many cellular signaling pathways, including those regulating proliferation, apoptosis, and anabolic biosynthesis processes, are regulated by mitochondria (Meyer and Chan, 2017). The role of mitochondrial toxicity in adverse health outcomes has been the subject of extensive study. Incurable genetic diseases of the mitochondria affect many different organ systems (Nunnari and Suomalainen, 2012). Mitochondrial dysfunction is also implicated in the etiologies of neurodegenerative diseases such as Parkinson’s and Alzheimer’s, several types of cancer, diabetes, and metabolic syndrome (Brandon et al., 2006; Coskun et al., 2012; Nunnari and Suomalainen, 2012; Rolo and Palmeira, 2006; Wallace, 2005). The Organisation for Economic and Cooperative Development also recognizes the linkage between impaired mitochondrial function and parkinsonian motor deficits in their adverse outcome pathway (AOP) development workplan (https://aopwiki.org/aops/3, last accessed May 7, 2020); support for this AOP is largely based on observations of in vitro and in vivo effects resultant to rotenone or 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine exposure. Mitochondrial perturbation is often implicated with drug-induced liver injury as hepatotoxic drugs can elicit mitochondrial toxicity through diverse mechanisms (Pessayre et al., 2012). As such, drugs or their reactive metabolites have been identified that disrupt several mitochondrial processes including respiration, energy homeostasis, membrane integrity, mtDNA replication, ROS production, mitochondrial transport pathways, and protein synthesis (Begriche et al., 2011). These off-target effects leading to potentially serious health complications have compelled the U.S. Food and Drug Administration to issue market withdrawals (eg, troglitazone, phenformin) or, more commonly, black box warnings for several classes of drugs, including flutamide, diclofenac, and ketoconazole (Dykens and Will, 2007). Furthermore, mitochondrial toxicity is recognized as a mechanism of acute toxicity in vivo (Bhhatarai et al., 2015; Hamm et al., 2017), and as such mitochondrial toxicity may be an important screening node for regulatory toxicology and development of screening level chemical assessments.
Several environmental chemicals are mitochondrial toxicants with known mechanisms of toxicity. Rotenone, a broadspectrum pesticide, blocks the mitochondrial electron transport chain (ETC) at complex I by inhibiting electron transfer to ubiquinone. Complex III of the ETC is inhibited by the piscicide antimycin A and the strobilurin class of fungicides (Sierotzki, 2015). ETC complex IV is inhibited by cyanide leading to catastrophic loss of ATP and cell death (Alonso et al., 2003). The pesticide, industrial-use chemical and banned weight loss drug 2,4-dinitrophenol increases the proton permeability of the inner mitochondrial membrane uncoupling respiration from ATP synthesis (Loomis and Lipmann, 1948). In addition, mitochondrial DNA is susceptible to damage by chemicals and ultraviolet radiation due to a lack of DNA repair (Meyer et al., 2018). Other environmental chemicals like heavy metals can induce oxidative stress in mitochondria leading to neurotoxicity (Farina et al., 2013).
As interest in mitochondrial toxicity has increased, so has the demand for newer and more rapid technologies for measuring mitochondrial disruption (reviewed in Meyer et al., 2018; Wallace, 2018; Wills, 2017). Current high-throughput screening methods to assess mitochondrial toxicity include the measurement of mitochondrial membrane potential (Attene-Ramos et al., 2013, 2015; Sakamuru et al., 2012); evaluation of mitochondrial morphology through high-content imaging (Leonard et al., 2015) or flow cytometry (Saunders et al., 2013); forced dependence on oxidative phosphorylation for ATP biosynthesis using alternative sugars (eg, galactose; Delp et al., 2019; Marroquin et al., 2007); and, respirometry measuring oxygen consumption (Ferrick et al., 2008; Wills et al., 2013, 2015). Methods using fluorescent dye probes to measure mitochondrial membrane potential or morphology have the highest sample throughput and predominate the suite of mitochondrial toxicity assays in both the ToxCast and Tox21 programs (Collins et al., 2008; Dix et al., 2007). The limitations of fluorescent dye probe methodologies to comprehensively identify mitochondrial toxicants are well known (Wills, 2017). Dye-based endpoints measure the morphological integrity of the mitochondria or changes in charge across mitochondrial membranes but are unable to directly measure inhibition of the ETC or loss of the proton gradient. In contrast, respirometric assays assess mitochondria function instead of structural changes and detect chemicals that disrupt mitochondria through diverse mechanisms including ETC inhibition, uncoupling, and ATP synthase inhibition (Beeson et al., 2010).
Due to the need to for streamlined cell culture practices in high-throughput screening, ToxCast/Tox21 HTS assays typically use immortalized or tumor cells cultured in high-glucose medium, which together drive these cells to generate ATP via glycolysis instead of oxidative phosphorylation (Warburg and Crabtree Effects; Marroquin et al., 2007). This reduced reliance upon mitochondria for energy production renders these cells more resistant to mitochondrial insult, and as such the chemical concentrations at which mitochondrial toxicity is observed may be higher than the chemical concentrations that elicit specific mitochondrial effects in medium that resembles a more physiologically relevant glucose level. To test this hypothesis and explore an approach to more specifically evaluate mitochondrial toxicity, we adapted a respirometric assay method (Wills et al., 2013, 2015) to screen ToxCast Phase I and II chemicals using a tiered workflow to maximize sample throughput, identify mitochondrial toxicants, and assign putative mechanisms of action.
MATERIALS AND METHODS
Data analysis.
Data were analyzed using R (Version 3.2.2) and RStudio (Version 0.99.467) and plotted using the ggplot2 package. Statistical methods employed for each assay and testing tier are described below. All source files and code are made available at: https://datadryad.org/stash/dataset/doi:10.5061/dryad.zkh189367 (last accessed May 7, 2020).
Control and reagent chemicals.
Antimycin A (AA), carbonyl cyanide-p-(trifluoromethoxy) phenylhydrazone (FCCP), dimethyl sulfoxide (DMSO), 2,4-dinitrophenol (DNP; DTXSID0020523), D-glucose, fenpyroximate (FENP; DTXSID7032557), D-malic acid, myxothiazol (MYXO), rotenone (ROT; DTXSID6021248), sodium ascorbate, sodium pyruvate, and N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) were purchased from Sigma-Aldrich (St. Louis, Missouri). The Distributed Structure-Searchable Toxicity (DSSTox) database identifiers are used to identify probe and test substances (Grulke et al., 2019).
Test chemicals.
The ToxCast Phase I (v2) and II chemical libraries were provided as 1051 blinded chemical samples drawn from a set of 1042 unique chemicals, the difference owing to 9 test chemicals represented as 18 internally duplicated samples used to measure intra-assay reproducibility. The chemical library, quality control (QC) analysis, and structure data format files are available at: http://www.epa.gov/NCCT/toxcast/chemicals.html (last accessed May 7, 2020). Stock solutions of all test chemicals were provided in DMSO at a target concentration of 20 mM (Evotec, Inc; Princeton, New Jersey) in 384PP Echo-qualified source plates (LabCyte Inc; San Jose, California), with any deviation from this top stock concentration based on maximum solubility in DMSO. Compound plates containing all control and test compounds were prepared using 4% DMSO and 96% assay medium (described below) as the diluent, sealed and stored at —80°C, away from light. All experiments were conducted within 10 weeks of compound plate preparation.
Cell culture.
HepG2 human hepatocellular carcinoma cells were purchased from American Type Culture Collection (ATCC; Manassas, Virginia). HepG2 cells were cultured in a humidified 37°C atmosphere containing 5% CO2 in a complete growth medium comprised high-glucose Dulbecco’s Modified Eagle Medium with l-glutamine and sodium pyruvate (Life Technologies; Grand Island, New York) supplemented with 10% qualified fetal bovine serum (Life Technologies), penicillin-streptomycin (100 U/ml–100 μg/ml final concentration; HyClone; Logan, Utah), and 5 mM HEPES (Sigma-Aldrich). HepG2 cells were plated onto Seahorse XF96 cell culture microplates (Agilent Technologies; Santa Clara, California) coated with rat tail collagen type I (Corning, Inc; Corning, New York) at a density of 2.7 × 104 cells per well in 80 μl of complete growth medium for 1 h at room temperature to facilitate uniform seeding. Complete growth medium was added to a final volume of 220 ml and cells were incubated overnight (16–18 h) before testing.
Respirometric screening assay.
A tiered screening approach was employed for the respirometric screening assay (RSA), with a Tier 1 single-concentration screening at a maximal target concentration of 100 μM followed by a Tier 2 multiconcentration screening using 7 concentrations from 125 nM to 100 μM. Tier 1 screening was intended to capture any activity that would affect oxygen consumption rate (OCR), including mitochondrial toxicity and/or other nonmitochondrial modes-of-action. Tier 2 screening was designed to identify a no-effect concentration and provide support for a proposed mode of action for observed effects on OCR, attempting to distinguish selective mitochondrial toxicity from other nonmitochondrial modes-of-action.
Seeded Seahorse XF96 cell microplates (assay plates) were twice aspirated to 20 μl and washed with 155 μl per well of assay medium comprised Seahorse XF base medium (Agilent Technologies) supplemented with 10 mM d-glucose, 1 mM sodium pyruvate, and 2 mM l-glutamine (Life Technologies). Cells were replenished with 155 μl per well of assay medium after a final aspiration to 20 μl (175 μl final volume) and equilibrated for 1 h at 37°C in a humidified atmosphere void of CO2. Assay plates were loaded onto a Seahorse XFe96 Analyzer precalibrated with a hydrated sensor cartridge. Prior to equilibration, the sensor cartridge ports were loaded with 25 μl of controls or test chemicals in port A (provided as ×8 concentrations), FCCP in port B (250 nM final), and rotenone/antimycin A in port C (both 1 μM final). The final target concentrations of controls and test chemicals are listed in Table 1. Measurements were made using 6-min cycles comprised 3 min of mixing followed by 3 min of OCR measurement. A total of 12 OCR measurements were made per experiment, 3 measurements in each of 4 sequential phases: preinjection phase (Figure 2; 6–18 min); basal respiration phase following injection of test compound or control (24–36 min); maximal respiration phase following FCCP injection (42–54 min); and inhibited respiration phase following injection of ROT and AA (60–72 min). Three biological replicates of technical singles were run for each experiment. The electron transport chain inhibitor (ETCi) fenpyroximate was used as the positive control for decreased basal respiration and the uncoupler 2,4-dintirophenol was used as the positive control for increased basal respiration. Both controls were present on each assay plate in 6 titrated concentrations with technical singles of the 5 lowest concentrations and technical triplicates of the maximally effective concentration. All OCR values for each well were normalized (rval) to the final preinjection phase OCR measurement (18 min) of that well to correct for intraplate variability. Bioactivity was determined using only the rval derived from the final measurement in each respiration phase (36, 54, and 72 min for basal, maximal, and inhibited phases, respectively). A median DMSO response (bval) was calculated for each assay plate. A zero-centered fold change response (fc) was calculated using the formulas:
Table 1.
Control and Sample Concentrations
| Testing Tier | Sample | Sample Type | Target Concentrations (μM) |
|---|---|---|---|
|
| |||
| Single concentration | Fenpyroximate | Control (ETCi) | 0.25, 0.625, 0.875, 1.13, 1.38, 2.75 |
| Single concentration | 2,4-Dinitrophenol | Control (uncoupler) | 0.938, 2.19, 4.38, 9.06, 19.1, 40 |
| Single concentration | Test chemical | Blinded sample | 100 |
| Multiconcentration | Fenpyroximate | Control (ETCi) | 0.25, 0.625, 0.875, 1.13, 1.38, 2.75 |
| Multiconcentration | 2,4-Dinitrophenol | Control (uncoupler) | 0.938, 2.19, 4.38, 9.06, 19.1, 40 |
| Multiconcentration | Test chemical | Blinded sample | 0.125, 0.312, 1, 3.19, 10, 31.6, 100 |
| Electron flow | Fenpyroximate | Control (complex I) | 0.5, 1.13, 1.75, 2.5, 3.25, 4 |
| Electron flow | Myxothiazol | Control (complex III) | 0.5, 0.938, 1.34, 1.73, 2.11, 2.5 |
| Electron flow | Test chemical | Blinded sample | 31.6, 64.6, 100 |
Figure 2.
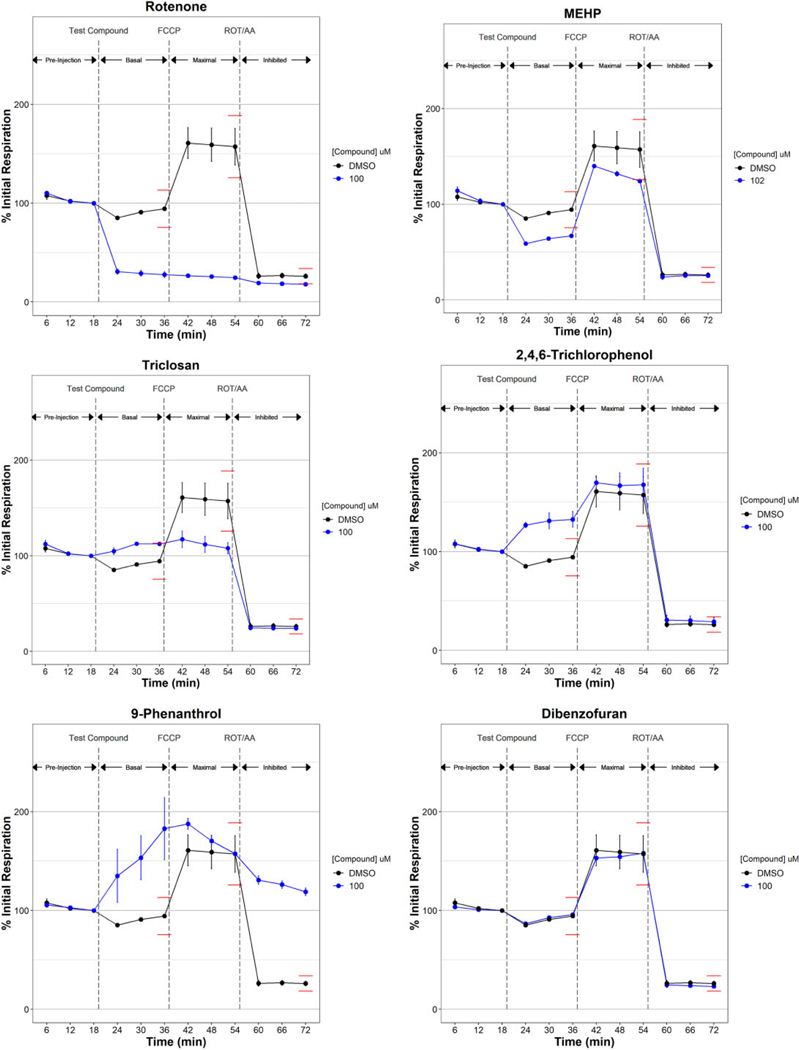
Tier 1 respirometric screening assay temporal response examples. Oxygen consumption was measured in HepG2 cells for 3 3-min cycles during a preinjection phase to establish and initial respiration rate used to normalize data across 3 respiration phases: basal respiration following injection of controls or test compound, maximal respiration following the injection of the uncoupler carbonyl cyanide-p-(trifluoromethoxy) phenylhydrazone (FCCP), and inhibited respiration following the injection of electron transport chain inhibitors rotenone (ROT) and antimycin A (AA). Median % initial respiration ± mad for triplicate experiments are plotted. Global dimethyl sulfoxide (DMSO) controls are plotted in black. Test chemicals are plotted in blue. Global 20% (for basal and maximal phases) and 30% (inhibited phase) thresholds are plotted by red lines and approximate the thresholds used, which were calculated independently for each sample plate. Supplementary Table 2 lists all 1042 ToxCast chemicals tested in this study with the maximal test concentration used and respirometric screening assay activity determination. Abbreviation: MEHP, mono(2-ethylhexyl) phthalate.
Active chemical samples were defined as those with fc responses deviating ≥ 20% from DMSO controls for basal respiration which represents 9.7 times the median absolute deviation (mad), ≥ 20% for maximal respiration (1.7*mad) and ≥ 30% for inhibited respiration (3.5*mad). Only active Tier 1 chemicals were retested (Tier 2) in the same manner as Tier 1 but using 7 titrated concentrations (see Table 1) for each chemical. The Tier 1 activity thresholds were also used for Tier 2: ≥ 20% for basal respiration (6.9*mad), ≥ 20% for maximal respiration (1.9*mad), and ≥ 30% for inhibited respiration (3.5*mad). Dose-response curves were fit separately for fc.up and fc.down for each respiration phase using the ToxCast data pipeline R package tcpl version 2.0.1 (Filer et al., 2017; https://cran.r-project.org/web/packages/tcpl/index.html, last accessed May 7, 2020). Area under fitted curve (AUF) was calculated for each sample that exceeded the activity thresholds using the integrate function in base R and with a uniform concentration (x-axis) range spanning the lowest and highest concentrations across all tested chemical samples.
Assignment of mitochondrial inhibition mechanism.
Chemical samples tested in multiconcentration (Tier 2) were assigned a putative mitochondrial inhibition mechanism as follows: (1) samples eliciting an fc.up ≥ 30% in the inhibited phase were binned as potential false positive redox-cycling chemicals; (2) samples eliciting an fc.up ≥ 20% at any tested concentration in the basal phase and not already identified as redox-cyclers were binned as uncouplers; (3) samples eliciting an fc.down ≥ 20% in the maximal phase and not already identified as uncouplers were binned as ETCi; and (4) samples eliciting an fc.down ≥ 20% in the basal phase and not already identified as uncouplers or ETCi were binned as ATP synthase inhibitors.
Electron flow assay.
Seeded Seahorse XF96 cell microplates (assay plates) were washed twice with 175 μl per well of ×1 MAS buffer comprised 220 mM d-mannitol, 70 mM sucrose, 10 mM potassium phosphate monobasic, 5 mM magnesium chloride, 2 mM HEPES, and 1 mM EGTA. Cells were replenished with 175 μl per well of ×1 MAS supplemented with 10 mM sodium pyruvate, 2 mM malic acid,1 μM FCCP, and 2.5 nM XF Plasma Membrane Permeabilizer (Agilent Technologies) and immediately loaded onto a Seahorse XFe96 Analyzer precalibrated with a hydrated sensor cartridge. Sensor cartridge ports were loaded (25 μl per port) as follows: Port A, controls or test chemicals (provided as ×8 concentrations); Port B, 18 μM rotenone and 90 mM succinate (2 μM and 10 mM final, respectively); Port C, 100 mM ascorbate, 1 mM TMPD, and 10 μM AA (10 mM, 100 μM, and 1 μM final, respectively). Each test chemical was assayed at 3 titrated concentrations. The final target concentrations of controls and test chemicals are listed in Table 1. Measurements were made using 3-min cycles comprised 30 s of mixing followed a 30 s wait then by 2 min of OCR measurement. A total of 12 OCR measurements were made per experiment, 3 measurements in each of 4 sequential phases: preinjection phase (Figure 6; 3–6 min); basal respiration phase following injection of test compound or control (9–24 min); complex I inhibition bypass phase following injection of rotenone/succinate (27–30 min); and complex III inhibition bypass phase following injection of ascorbate/TMPD/ AA (33–36 min). Three biological replicates of technical singles were run for each experiment. The ETCi fenpyroximate and myxothiazol (MYXO) were used as the positive controls for complex I and III inhibition, respectively. Both controls were present on each assay plate in 6 titrated concentrations with technical singles of the 5 lowest concentrations and technical triplicates of the maximally effective concentration. All OCR values for each well were normalized (rval) to the final preinjection phase OCR measurement (6 min) of that well to correct for intra plate variability. Bioactivity was determined using only the final OCR measurement in each postinjection phase: 24 min for the basal phase, 30 min for the complex I bypass phase, and 36 min for the complex III bypass phase. Percent biological response (pc.resp) was calculated using the formula:
where bval is the median DMSO response and pval is the minimum FENP or MYXO response calculated for each assay plate. Active chemical samples were defined as those with pc.resp responses deviating ≥ 20% from DMSO controls for basal respiration (5.9*mad), ≥ 20% for complex I bypass respiration (3.6*mad), and ≥ 40% for complex III bypass respiration (2.2*mad).
Figure 6.
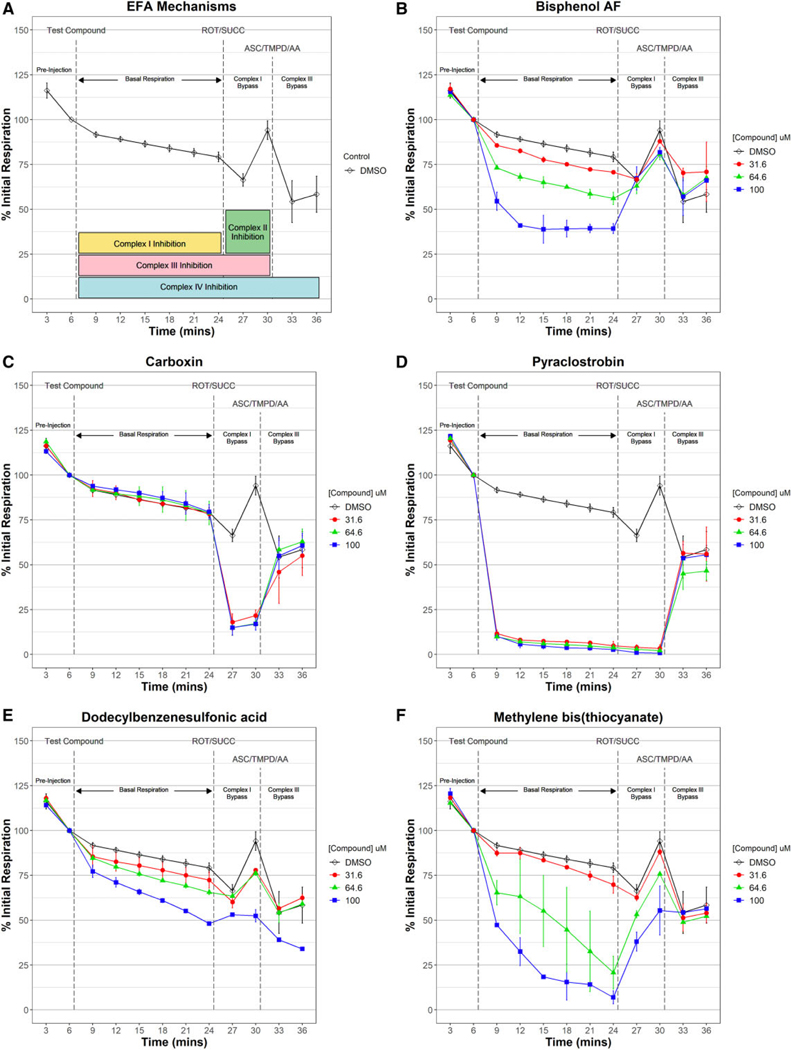
Electron flow assay to determine complex(es) targeted by electron transport chain (ETC) inhibitors. Oxygen consumption was measured in permeabilized and fully uncoupled HepG2 cells supplied with pyruvate and malate during a preinjection phase used to normalize data across 3 respiration phases: basal respiration following injection of controls or test ETC inhibitor, complex I bypass respiration following the injection of the complex I inhibitor rotenone (ROT) and complex II substrate succinate (SUCC), and complex III bypass respiration following the injection of complex III inhibitor antimycin A (AA) and complex IV substrates ascorbate (ASC) and N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD). Median % initial respiration ± mad for triplicate experiments are plotted. Global dimethyl sulfoxide (DMSO) controls are plotted in black. All ETCi were tested at 3 titrated concentrations. The temporal dose-response patterns for all 149 ETC inhibitors tested in the electron flow assay (EFA) are provided in Supplementary Figure 3. Chemicals that inhibited respiration during the basal phase only but recovered with succinate (A, yellow) like bisphenol B (B) were classified as complex I inhibitors. Those that inhibited respiration during the complex I bypass phase only but recovered with ASC/TPMD (A, green) like carboxin (C) were classified as complex II inhibitors. Chemicals that inhibited respiration during both basal and complex I bypass phases but recovered with ASC/ TPMD (A, pink) like pyraclostrobin (D) were classified as complex III inhibitors. Inhibition of respiration during all 3 phases (A, blue) as observed with dodecylbenzene-sulfonic acid (E) was classified as complex IV inhibitors. Chemicals that inhibited respiration during the basal phase and only partially recovered with succinate and fully with ASC/TPMD like methylene bis(thiocyanate) (F) were classified as mixed complex I/III inhibitors.
Assignment of ETC inhibition mechanism.
An ETC inhibition mechanism was assigned to each tested chemical sample using electron flow assay (EFA) data independently for each of the 3 titrated test concentrations as follows:
| Mechanism | By Concentration |
|---|---|
|
| |
| Inactive | Inactive in all 3 phases |
| Complex I inhibitor | Active in the basal phase but inactive in the complex I bypass and complex III bypass phases |
| Complex II inhibitor | Active in the complex III phase but inactive in the basal and complex III bypass phases |
| Complex III inhibitor | Active in the basal and complex I bypass phases but inactive in the complex III bypass phase |
| Mixed complex I/III inhibitor | Complex III inhibitors eliciting a basal phase pc.resp ≥ 2 * complex I bypass pc.resp |
| Complex IV inhibitor | Active in all 3 phases |
Final ETC inhibition mechanism determinations were derived for each chemical from the assignment across the 3 titrated test concentrations as follows:
| Mechanism Assignment | By Chemical |
|---|---|
|
| |
| Inactive | Inactive at the highest tested concentration |
| Consensus mechanism | Mechanisms for all tested concentrations agree; mechanisms for 2 highest test concentrations agree and lower test concentration was inactive; mechanisms for highest and lowest concentrations agree; mechanisms for 2 lowest concentrations agree; consensus for the 2 highest test concentrations that conflicted with that for the lowest test concentration |
| Highest concentration mechanism | Only highest tested concentration was active |
| Mixed mechanism | No consensus for the 2 highest test concentration and the lowest test concentration was inactive |
| Undetermined | Any remaining conditions |
Reference chemical selection for assay comparison.
An open literature search identified 30 mitochondrial-disrupting chemicals spanning a diversity of mechanisms from among the 1042 unique chemicals tested in this study (Table 2). Thirty presumptive inactive chemicals were randomly selected from a pool comprised the remaining test chemicals that met 4 criteria: (1) active in 5% or fewer of all ToxCast assay (mitochondrial toxicity assays included); (2) no PubMed abstracts linking chemical name or CAS registry number (casn) with “mitochondria” as search terms using Abstract Sifter (Baker et al., 2017); (3) 10 or fewer abstracts linking chemical name or casn with any of the following additional search terms: “electron transport”; “respiration”; “uncouple”; “complex I” or “NADH ubiquinone oxidoreductase” or “Type I NADH dehydrogenase”; “complex II” or “succinate dehydrogenase” or “succinate coenzyme Q reductase”; “complex III” or “coenzyme Q cytochrome c oxidoreductase” or “cytochrome bc1 complex”; “complex IV” or “cytochrome c oxidase”; “complex V” or “ATP synthase” or “F1F0”; and, “mitochondrial membrane” or “electrochemical gradient” or “proton gradient”; and (4) no pharmaceutical ToxCast designation such as “CP-” or “PD-.” The reduced stringency between no abstracts specifically linking a chemical to “mitochondria” versus 10 or fewer mentioning mitochondria-related terms is owing to the ambiguity in terms such “respiration” and “uncouple.” The exclusion of chemicals active in more than 5% of ToxCast assays aims to eliminate promiscuous or reactive chemicals that may elicit a nonspecific response in cell-based mitochondrial toxicity assays. The exclusion of ToxCast chemicals with a pharmaceutical designation is due to a lack of published reports on chemical with unknown mitochondrial activities.
Table 2.
Reference Chemicals Used to Assess Interassay Performance
| Chemical | Activity | % Inactive ToxCast | Reference | Mitochondria Abstracts | Total Abstracts |
|---|---|---|---|---|---|
|
| |||||
| Rotenone | Active | 65.4 | Degli Esposti (1998) and Metcalf and Horowitz (2014) | 3261 | 8924 |
| 2,4-Dinitrophenol | Active | 81.9 | Knobeloch et al. (1990) | 689 | 1621 |
| Quercetin | Active | 78.6 | Bohmont et al. (1987) | 393 | 806 |
| Sodium azide | Active | 92.5 | Wallace and Starkov (2000) | 317 | 1048 |
| Clofibrate | Active | 97.2 | Mehta et al. (2008) | 284 | 367 |
| Tamoxifen citrate | Active | 48.5 | Mehta et al. (2008) | 249 | 500 |
| Indomethacin | Active | 94.0 | Mehta et al. (2008) | 183 | 895 |
| Genistein | Active | 78.7 | Bohmont et al. (1987) | 138 | 367 |
| Mercuric chloride | Active | 50.2 | Knobeloch et al. (1990) | 105 | 223 |
| Diethylstilbestrol | Active | 62.7 | Wallace and Starkov (2000) and Knobeloch et al. (1990) | 103 | 184 |
| Diclofenac sodium | Active | 90.9 | Mehta et al. (2008) | 80 | 236 |
| Fenofibrate | Active | 83.5 | Mehta et al. (2008) | 76 | 128 |
| Pentachlorophenol | Active | 69.9 | Knobeloch et al. (1990) | 74 | 223 |
| Troglitazone | Active | 71.9 | Mehta et al. (2008) | 65 | 145 |
| Ketoconazole | Active | 58.5 | Rodriguez and Acosta (1996) | 64 | 123 |
| Carboxin | Active | 92.4 | Stammler et al. (2015) | 26 | 146 |
| Flutamide | Active | 77.9 | Mehta et al. (2008) | 18 | 47 |
| Azoxystrobin | Active | 80.0 | Balba (2007) and Sierotzki (2015) | 17 | 113 |
| 2-Methyl-4,6-dinitrophenol | Active | 73.7 | Metcalf and Horowitz (2014) and Knobeloch et al. (1990) | 17 | 59 |
| Fenpyroximate (Z,E) | Active | 73.0 | Degli Esposti (1998) | 15 | 53 |
| Pyridaben | Active | 76.5 | Degli Esposti (1998) and Metcalf and Horowitz (2014) | 15 | 78 |
| Pyraclostrobin | Active | 70.1 | Balba (2007) and Sierotzki (2015) | 9 | 66 |
| Dinoseb | Active | 72.9 | Metcalf and Horowitz (2014) | 6 | 27 |
| Tebufenpyrad | Active | 67.3 | Degli Esposti (1998) and Metcalf and Horowitz (2014) | 6 | 24 |
| Trifloxystrobin | Active | 78.4 | Balba (2007) and Sierotzki (2015) | 5 | 29 |
| Famoxadone | Active | 72.3 | Sierotzki (2015) | 5 | 41 |
| Nilutamide | Active | 89.5 | Berson et al. (1994) | 2 | 6 |
| Boscalid | Active | 92.6 | Stammler et al. (2015) | 2 | 99 |
| Picoxystrobin | Active | 71.8 | Balba (2007) and Sierotzki (2015) | 1 | 4 |
| Fluoxastrobin | Active | 75.7 | Balba (2007) and Sierotzki (2015) | 0 | 3 |
| 1,2,4,5-Tetrachlorobenzene | Inactive | 98.9 | NA | 0 | 2 |
| 1,3-Diisopropylbenzene | Inactive | 99.5 | NA | 0 | 0 |
| 1,5,9-Cyclododecatriene | Inactive | 98.2 | NA | 0 | 1 |
| 2,3,6-Trimethylphenol | Inactive | 98.3 | NA | 0 | 3 |
| 2-Anisidine | Inactive | 97.9 | NA | 0 | 1 |
| 2-Butoxyethanol | Inactive | 98.6 | NA | 0 | 4 |
| 2-Methyl-5-nitroaniline | Inactive | 95.7 | NA | 0 | 0 |
| 3,4-Dimethylphenol | Inactive | 98.0 | NA | 0 | 0 |
| 3-Bromo-1-propanol | Inactive | 98.9 | NA | 0 | 0 |
| 4-(2-Methylbutan-2-yl)cyclohexanol | Inactive | 95.7 | NA | 0 | 0 |
| 4-Methyl-2-pentanol | Inactive | 98.6 | NA | 0 | 1 |
| Asulam | Inactive | 99.3 | NA | 0 | 2 |
| Bentazone | Inactive | 97.6 | NA | 0 | 9 |
| Bis(2-ethylhexyl) decanedioate | Inactive | 96.9 | NA | 0 | 10 |
| Chlorendic acid | Inactive | 98.9 | NA | 0 | 0 |
| Chlorthal-dimethyl | Inactive | 95.3 | NA | 0 | 0 |
| Cyanazine | Inactive | 95.5 | NA | 0 | 6 |
| Di(propylene glycol) dibenzoate | Inactive | 96.1 | NA | 0 | 0 |
| Dibromoacetonitrile | Inactive | 97.6 | NA | 0 | 1 |
| Diethylene glycol dibenzoate | Inactive | 96.6 | NA | 0 | 0 |
| Dipropyl 2,5-pyridinedicarboxylate | Inactive | 97.5 | NA | 0 | 0 |
| Flufenpyr-ethyl | Inactive | 97.3 | NA | 0 | 0 |
| Fosthiazate | Inactive | 95.5 | NA | 0 | 0 |
| Geranyl acetate | Inactive | 98.6 | NA | 0 | 2 |
| Monobenzyl phthalate | Inactive | 97.4 | NA | 0 | 0 |
| N-Ethylaniline | Inactive | 99.0 | NA | 0 | 1 |
| Pymetrozine | Inactive | 98.3 | NA | 0 | 1 |
| Sodium xylenesulfonate | Inactive | 98.9 | NA | 0 | 0 |
| tert-Butylbenzene | Inactive | 99.0 | NA | 0 | 0 |
| Tributylamine | Inactive | 99.2 | NA | 0 | 2 |
The 60 chemicals were used to evaluate the performance of the RSA and preceding ToxCast/Tox21 mitochondrial toxicity assays are listed in Table 2. Binary classifier statistics were calculated using the R caret package (v6.0–84). RSA-active chemicals were defined as ETCi, uncouplers, or ATP synthase inhibitors based on mechanistic assignment from Tier 2 concentration-response patterns. All other test chemicals were defined as inactive in the RSA. Active chemicals in the ToxCast/ Tox21 mitochondrial toxicity assays (ie, Apredica and Tox21_MMP) were defined as those having an active hit call (invitrodb v3.1; https://doi.org/10.23645/epacomptox.6062623.v3, last accessed May 7, 2020) in either direction (ie, up/down) and at any time point (combined assay endpoint id) for any sample id. Chemicals that tested negative across all assay endpoints for each assay technology were classified as inactive.
RESULTS
Single-concentration Respirometric Screening
A RSA using intact HepG2 cells was used to assess 1042 unique test chemicals (Figure 1; Tier 1) at a single, maximal target concentration of 100 μM (solubility permitting). OCRs were measured in 3 sequential respiration phases (Figure 2): (1) basal respiration following injection of test chemical or controls, (2) maximal respiration following injection of the uncoupler FCCP, and (3) inhibited respiration following injection of ETCi rotenone and antimycin A. Assay performance was evaluated using 5 QC metrics calculated using DMSO (vehicle), fenpyroximate (FENP; positive control for ETC inhibition), and 2,4-dinitrophenol (DNP; positive control for uncoupling) controls (Supplementary Table 1). Plates were accepted after exceeding at least 4 QC criteria. The RSA performed reliably across all 51 assay plates. Vehicle control rCV values ranged from 0.29% to 3.51% with a median of 1.41%, all well below the 5% acceptance criterion. FENP and DNP controls were evaluated by half-maximal activity concentration (AC50) and rZ scores. The median FENP AC50 was 1.07 μM and values ranged from 0.85 to 1.43 μM, all well within the 1 μM ±. 0.5 log-unit acceptance criterion. FENP rZ scores ranged from 0.68 to 0.97 with a median of 0.88, all above the 0.5 acceptance criterion. The median DNP AC50 was 10.1 μM and values ranged from 2.52 to 19.8 μM, all within the 7.5 μM ± 0.5 log-unit acceptance criterion. DNP rZ scores ranged from −0.04 to 0.97 with a median of 0.76; 44 of 51 assay plates exceeded the 0.5 acceptance criterion. Uncouplers such as DNP typically exhibit bell-shaped (inverted-u) dose-responses, ie, increased respiration at lower concentrations and decreased respiration at concentrations exceeding its maximally effective uncoupling concentration (Wallace and Starkov, 2000). The maximally effective response for uncouplers such DNP is therefore more variable than that for ETCi such as FENP, thus QC failure is more likely from DNP rZ’ which incorporates both the magnitude and variability of uncoupling into account. Decreased basal and maximal respiration responses of 9 internally duplicated chemical samples are shown in Supplementary Figure 1. Duplicated chemicals produced similar responses with minimal intra-sample variability. The results typify the high intra- and inter-assay reproducibility of the RSA.
Figure 1.
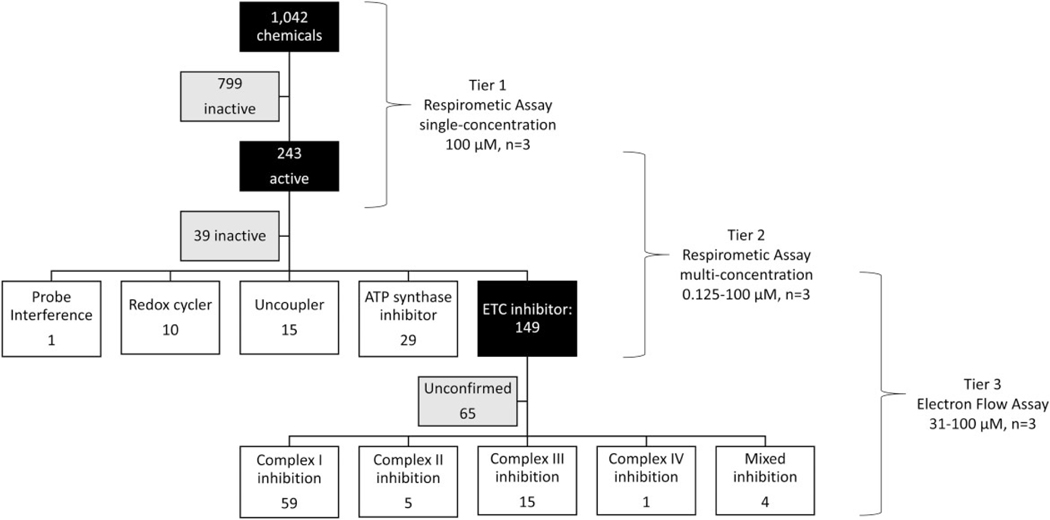
Tiered testing strategy to identify and characterize mitochondrial toxicants. A respirometric screening assay (RSA) was used to test 1042 ToxCast Phase I/II chemicals at singlemaximal concentration resulting in 249 active and 799 inactive test chemicals (Tier 1). The 249 Tier 1 actives were rescreened in the RSA at 7 titrated concentrations (Tier 2). Tier 2 data was fit using the ToxCast pipeline and actives from specific respiratory phased used to mechanistic classify each chemical. A total of 193 mitochondrial toxicants were identified using Tier 2 data. The 149 electron transport chain (ETC) inhibitors identified in Tier 2 were evaluated in an electron flow assay (EFA) at 3 titrated concentrations. EFA data used to identify the targeted ETC complex(es) for 84 chemicals. Abbreviation: ATP, adenosine triphosphate.
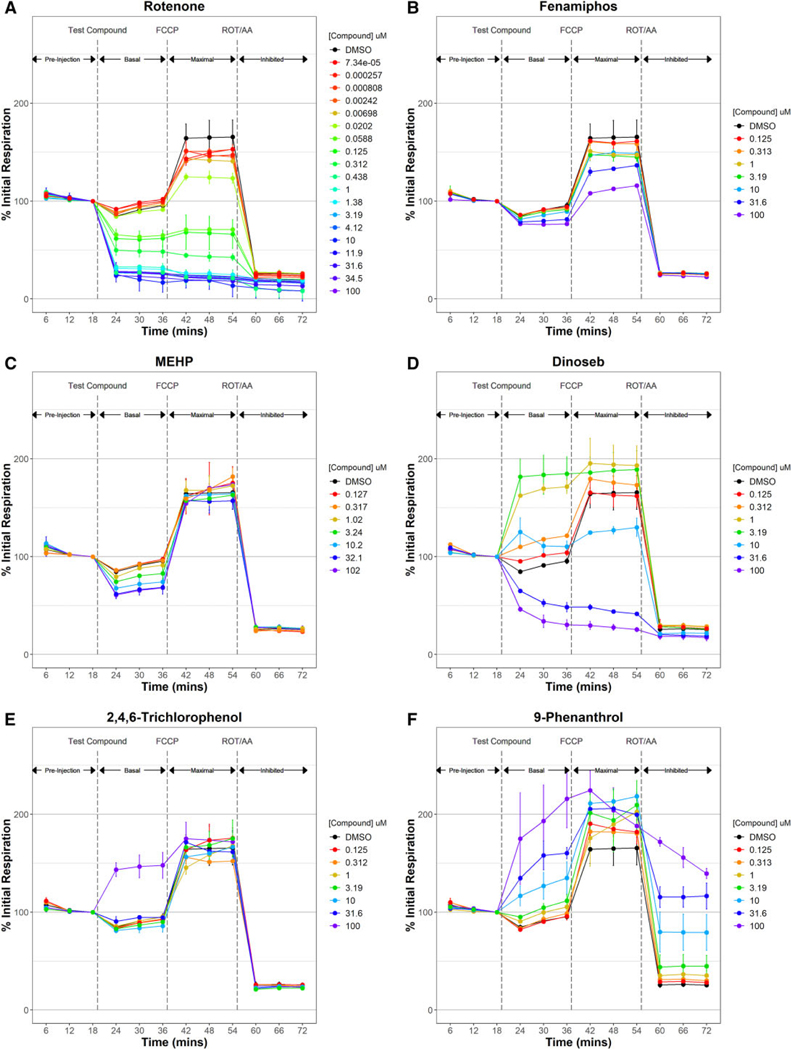
Figure 2 highlights some of the varied temporal response observed in the RSA. Most (54%) of the 243 active chemicals decreased both basal and maximal phase respiration, as typified by the known ETCi rotenone (Figure 2A). An equal number of active chemicals decreased either basal or maximal respiration (48 chemicals each) like mono(2-ethylhexyl) phthalate (MEHP) and triclosan, respectively (Figs. 2B and 2C). Fifteen active chemicals increased basal respiration such as 2,4,6-trichlorophenol (Figure 2D); however, 5 of those chemicals, like 9-phenanthrol (Figure 2E), also increased inhibited phase (nonmitochondrial) respiration. Most tested chemicals (799 of 1042) were inactive like dibenzofuran (Figure 2F), exhibiting a response pattern that coincided with the DMSO vehicle control response.
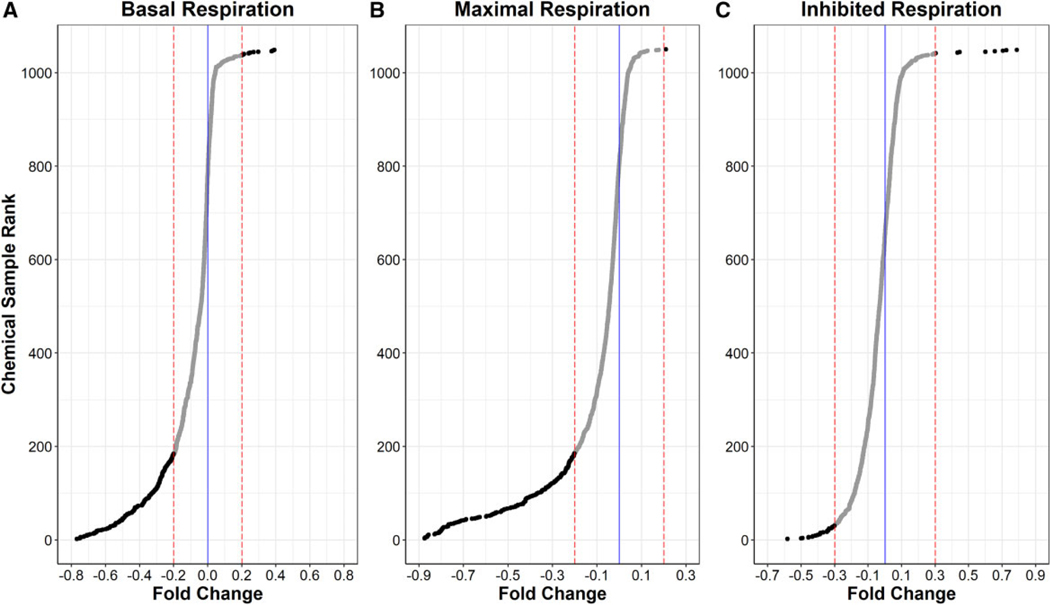
Figure 3 illustrates the wide spectrum of responses and number of actives observed in each of the 3 respiration phases. Most of the tested chemical samples clustered around the vehicle control response (solid blue lines). Of the 1042 unique chemicals tested, 193 (18.5%) altered basal respiration ≥ 20% (Figure 3A dotted red lines), the threshold used to define activity in the basal and maximal respiration phases. Of the 193 basal respiration actives, 179 chemicals significantly decreased respiration whereas only 14 increased respiration. A similar response rate was observed in the maximal respiration phase (Figure 3B) where 181 chemicals (17.4%) tested active, of which 179 chemicals decreased maximal respiration by ≥ 20% and only 2 chemicals increased maximal respiration above the upper threshold. For the inhibited respiration phase (Figure 3C), a 0.3-fold threshold was used to define 38 total actives (3.6%), of which 28 decreased and 10 increased inhibited respiration. A total of 243 unique chemicals (249 samples) tested active in 1 or more respiration phase. Supplementary Table 2 lists the names, test concentrations, and activity determinations for all 1042 unique chemicals assessed in this study.
Figure 3.
Ranked median Tier 1 respirometric screening assay responses of 1042 test chemicals. Median fold changes for triplicate experiments are ranked from lowest (decreased respiration) to highest (increased respiration) for each of 1042 test chemicals for basal respiration (A), maximal respiration (B), and inhibited respiration (C). Tier 1 responses were normalized and zero-centered to DMSO control (blue line). Red dotted lined mark the thresholds used to define active chemicals (black dots) in each respiration phase.
Multiple-concentration Respirometric Screening and Mechanistic Classification
The 243 unique active chemicals identified in single-concentration or Tier 1 testing were rescreened in the RSA (Figure 1; Tier 2) at 7 titrated concentrations spanning 125 nM– 100 μM (solubility permitting). Dose-response curves were fit for 6 assay endpoints: each of the 3 respiration phases in each direction (ie, increased or decreased respiration). Again, the RSA performed reliably across 90 assay plates, and all plate data were accepted after exceeding at least 4 QC criteria. Vehicle control rCV values ranged from 0.11% to 3.42% with a median of 1.50%. The median FENP AC50 was 1.15 μM and values ranged from 0.81 to 2.11 μM. FENP rZ scores ranged from 0.35 to 0.96 with a median of 0.87; 88 of 90 assay plates exceeded the 0.5 acceptance criterion. The median DNP AC50 was 13.8 μM and values ranged from 7.58 to 21.2 μM. DNP rZ scores ranged from −0.12 to 0.99 with a median of 0.80; 87 of 90 assay plates exceeded the 0.5 acceptance criterion.
Temporal-dose-response plots for all 243 active chemicals controls are provided in Supplementary Figure 2. Figure 4 typifies the diversity of multiconcentration response patterns observed. Most of the Tier 2 test chemicals (57%) significantly decreased both basal and maximal respiration in a dose-dependent manner as illustrated with rotenone (Figure 4A). A smaller number of chemicals (10.3% and 11.9%, respectively) decreased maximal respiration with no significant decrease in basal respiration such as fenamiphos (Figure 4B) or vice versa like MEHP (Figure 4C). Eighteen test chemicals (7.4%) increased basal respiration. Some of the more potent chemicals in this group, such as dinoseb (Figure 4D) increased basal respiration in a dose-dependent manner up to maximally effective concentration, but inhibited basal respiration at higher test concentrations, which is typical for uncouplers. 2,4,6-Trichlorophenol (Figure 4E) increased basal respiration but only at the highest test concentration. We identified 11 false positive chemicals exhibiting 1 of 2 distinct response patterns. We identified 10 compounds like 9-phenanthrol (Figure 4F) that increased basal phase respiration, but also increased inhibited phase respiration indicating an increase in nonmitochondrial oxygen consumption likely from redox cycling and not from uncoupling. Redox-cycling chemicals such as 1,2-naphthoquinone have been shown to increased cellular respiration in the presence of rotenone and antimycin A using a similar respirometric method (Lavrich et al., 2018). Following the identification of assay false positives, mechanism of mitochondrial disruption for the remaining RSA-active chemicals can be derived with sequential analysis of Tier 2 data.
Figure 4.
Tier 2 respirometric screening assay temporal dose-response patterns. Two hundred and forty-nine Tier 1 active chemicals were retested in the respirometric screening assay at 7 titrated concentrations to derive potency estimates and assign a putative mechanism of action. Rotenone (A) and fenamiphos (B) are examples of potent and weak electron transport chain inhibitors decreasing maximal phase respiration. mono(2-ethylhexyl) phthalate (MEHP) (C) exemplifies the response pattern of an adenosine triphosphate synthase inhibitor where basal phase respiration is inhibited but not maximal respiration. Dinoseb (D) and 2,4,6-trichlorophenol (E) are examples of potent and weak uncouplers which increase basal phase respiration. Redox-cycling chemicals like 9-phenanthrol (F) increase inhibited phase respiration indicating nonmitochondrial oxygen consumption. Gentian violet (G) elicited a decrease in signal below that of blank (no cell) controls, indicating probe interference. Mechanisms of mitochondrial toxicity (H) were assigned from Tier 2 data using a step-wise classification scheme involving only 4 of the 6 Tier 2 assay endpoints (colored boxes). The temporal dose-response patterns for the 243 Tier 2 test chemicals are provided in Supplementary Figure 2. Abbreviations: AA, antimycin A; DMSO, dimethyl sulfoxide; ETC, electron transport chain; FCCP, carbonyl cyanide-p-(trifluoromethoxy) phenylhydrazone; ROT, rotenone.
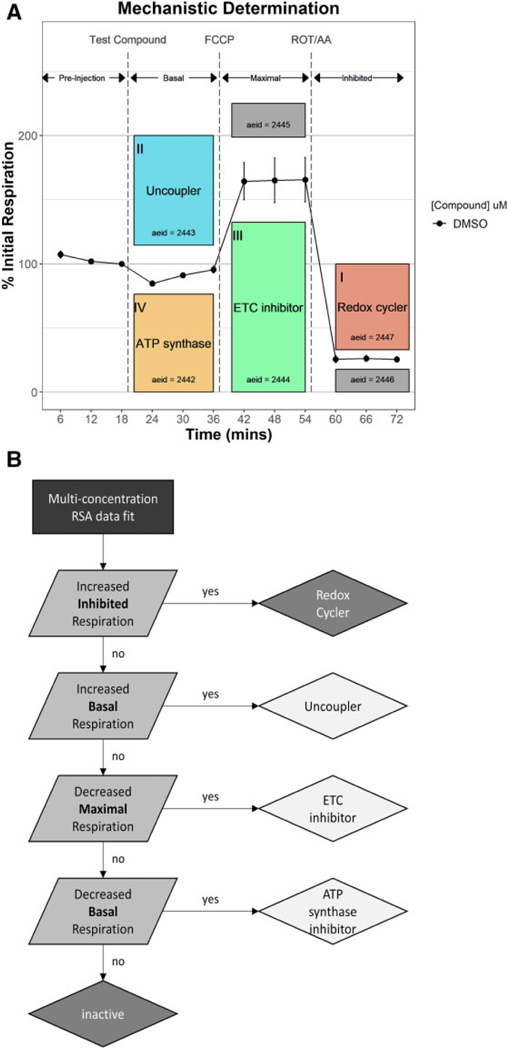
Bioactivity patterns across 4 of 6 assay endpoints (Figure 5A; colored boxes) were used to establish a rule set (Figure 5B) to assign a putative mitochondrial toxicity mechanism as well as identify potential false positives. First, the 10 chemicals that increased inhibited phase respiration (I) were flagged as probable redox-cycling false positives and excluded from mechanistic analysis. Fifteen uncouplers were then identified from the remaining test chemicals that increased basal phase respiration at 1 or more concentrations (II). Decreased maximal phase respiration (III) was used to identify 149 putative ETCi. Test chemicals that decreased basal phase respiration without decreasing maximal phase respiration (IV) were assigned as putative ATP synthase inhibitors, of which 29 were identified. The remaining 39 Tier 2 test chemicals (16.0%) retested inactive across all assay endpoints. Increased maximal phase and decreased inhibited phase respiration (gray boxes) were not used to assign mechanism. Supplementary Table 2 shows the activity calls and putative mechanism assignments for the 243 chemicals tested in Tier 2.
Figure 5.
Mechanistic assignment using Tier 2 data. Mechanisms of mitochondrial toxicity were assigned using 4 of the 6 Tier 2 assay endpoints (A; colored boxes). A rule set (B) was established that sequentially classified active chemicals as redox cyclers (false positives), uncouplers, electron transport chain inhibitors, or adenosine triphosphate (ATP) synthase inhibitors based a Tier 2 bioactivity response patterns. Abbreviations: AA, antimycin A; DMSO, dimethyl sulfoxide; FCCP, carbonyl cyanide-p-(trifluoromethoxy) phenylhydrazone; ROT, rotenone.
Table 3 lists the top 10 chemicals for each of the 3 mitochondrial disruption mechanisms assigned using Tier 2 data (Figure 1), ranked by the AUF dose-response curves, a single metric that incorporates both potency and efficacy, calculated for only that assay endpoint used to make each mechanistic determination.
Table 3.
Top 10 Active Chemicals by Mechanism
| Chemical | Mechanism | AUF | AC50 (mM) |
|---|---|---|---|
|
| |||
| Rotenone | ETC inhibitor | 2.813 | 0.0572 |
| Tebufenpyrad | ETC inhibitor | 2.074 | 0.483 |
| Pyridaben | ETC inhibitor | 1.954 | 0.586 |
| Fenpyroximate (Z,E) | ETC inhibitor | 1.898 | 0.769 |
| Picoxystrobin | ETC inhibitor | 1.561 | 1.28 |
| Pyraclostrobin | ETC inhibitor | 1.556 | 1.62 |
| Azoxystrobin | ETC inhibitor | 1.229 | 6.35 |
| Difenoconazole | ETC inhibitor | 1.221 | 7.33 |
| Tributyltin chloride | ETC inhibitor | 1.214 | 2.94 |
| FR150011 | ETC inhibitor | 1.176 | 4.53 |
| Niclosamide | Uncoupler | 2.562 | 0.229 |
| Dinoseb | Uncoupler | 2.392 | 0.572 |
| Cyazofamid | Uncoupler | 2.334 | 2.28 |
| 2-Methyl-4,6-dinitrophenol | Uncoupler | 2.277 | 5.83 |
| Pentachlorophenol | Uncoupler | 1.488 | 2.22 |
| 2,4-Dinitrophenol | Uncoupler | 1.185 | 33.6 |
| Bromoxynil | Uncoupler | 0.990 | 7.61 |
| CP-100829 | Uncoupler | 0.869 | 13.2 |
| 2,4,5-Trichlorophenol | Uncoupler | 0.628 | 23.8 |
| Fluazinam | Uncoupler | 0.424 | 14.3 |
| PharmaGSID_48510 | ATP synthase inhibitor | 0.718 | 1.33 |
| MEHP | ATP synthase inhibitor | 0.430 | 3.72 |
| 4,4′,4′′-Ethane-1,1,1-triyltriphenol | ATP synthase inhibitor | 0.349 | 17.9 |
| 3,5,3′-Triiodothyronine | ATP synthase inhibitor | 0.334 | 7.46 |
| Indomethacin | ATP synthase inhibitor | 0.326 | 13.2 |
| Decanoic acid | ATP synthase inhibitor | 0.277 | 27.5 |
| 2,4-Dichlorophenoxybutyric acid | ATP synthase inhibitor | 0.246 | 35.4 |
| Chlorothalonil | ATP synthase inhibitor | 0.220 | 22 |
| Propylparaben | ATP synthase inhibitor | 0.209 | 39.2 |
| UK-156819 | ATP synthase inhibitor | 0.208 | 35.3 |
EFA and Assignment of ETC Inhibition Mechanisms
An EFA was used to monitor the effects of 149 putative ETC inhibitors on ETC complexes (Figure 1; Tier 3) at 3 titrated concentrations ranging 31–100 μM (solubility permitting). Permeabilized HepG2 cells with fully uncoupled mitochondria following FCCP exposure were supplied pyruvate and malate to provide the reducing equivalents required for complex I-dependent respiration. OCRs were measured in 3 sequential respiration phases (Figure 6): basal, complex I bypass and complex III bypass. Assay performance was assessed using DMSO (vehicle), fenpyroximate (FENP; positive control for complex I inhibition), and myxothiazol (MYXO; positive control for complex III inhibition) controls. Active compounds were identified as those that decreased basal or complex I bypass respiration by ≥ 20% or complex III bypass respiration by ≥ 40% compared with DMSO control values. The temporal dose-response patterns for the 149 ETCi tested in the EFA are provided in Supplementary Figure 3.
Figure 6A illustrates the EFA response patterns used to derive a consensus electron transport inhibition mechanism for the 149 ETCi identified in Tier 2. Most (70.2%) of the EFA-active test chemicals were determined to be complex I inhibitors. Several well-known complex I inhibitors such as rotenone fully inhibited basal respiration, even at the lowest tested concentration, but respiration was restored with succinate, a substrate that allows electrons to enter though complex II (complex I bypass). Most of the complex I inhibitors identified in this study exhibited dose-dependent and partial inhibition of basal respiration at the highest test concentration, as shown with bisphenol AF (Figure 6B). Five complex II inhibitors were also identified. Complex II inhibition is exemplified by carboxin (Figure 6C), where no test compound effects are observed during basal respiration, but manifest only after complex I is inhibited by rotenone injection. Complex I delivers electrons directly to complex III, thus during basal respiration when pyruvate and malate (complex I substrates) are the only electron donors present, respiration is unimpeded by complex II inhibition. Complex II also delivers electrons directly to complex III independently of complex I status. Thus, complex II inhibition is only observed once complex I is inhibited by rotenone and succinate (oxidizable substrate for complex II) added. The complex II inhibition response pattern is distinct from that elicited by complex III inhibitors such a pyraclostrobin (Figure 6D) where both basal (complex I-mediated) and complex I bypass (complex II-mediated) respiration are impeded and only relieved by the addition of TPMD and ascorbate. Fifteen (17.8%) of the EFA-active chemicals were determined to complex III inhibitors. Only 1 chemical, dodecylbenzenesulforic acid (Figure 6E), was determined to be a complex IV inhibitor marked by an inability to respond to succinate or TMPD/ascorbate. Four chemicals (4.8% of EFA actives) were shown to have a mixed inhibition mechanism. One of these, methylene bis(thiocyanate), is shown in Figure 6F. Methylene bis(thiocyanate) clearly inhibits basal respiration in a dose-dependent manner. Respiration is partially recovered with succinate, then fully recovers with TMPD/ascorbate. That respiration responds even partially to succinate is indicative of complex I inhibition, but the incomplete nature of the recovery suggests additional inhibition at complex II and/ or III. Of the 149 putative ETC inhibitors identified in Tier 2 testing, 65 (43.6%) failed to inhibit respiration in any phase of the EFA and therefore the respiratory complex inhibited by these chemicals in the RFA could not be identified. Supplementary Table 2 shows the activity calls and targeted respiratory complexes for the 149 chemicals tested in Tier 3.
Table 4 lists the 84 EFA-active chemicals and ETC inhibition mechanism.
Table 4.
ETC Complexes Inhibited by 84 EFA-active Chemicals
| Chemical | ETCi Mechanism |
|---|---|
|
| |
| 2-(Thiocyanomethylthio)benzothiazole | Complex I inhibition |
| 2,2-Bis(4-hydroxyphenyl)-1,1,1-trichloroethane | Complex I inhibition |
| 2,4,7,9-Tetramethyl-5-decyne-4,7-diol | Complex I inhibition |
| 2,5-Di-tert-butylbenzene-1,4-diol | Complex I inhibition |
| 3,3′,5,5′-Tetrabromobisphenol A | Complex I inhibition |
| 4,4′-Sulfonylbis[2-(prop-2-en-1-yl)phenol] | Complex I inhibition |
| 4-Cumylphenol | Complex I inhibition |
| 4-Hexylresorcinol | Complex I inhibition |
| 5HPP-33 | Complex I inhibition |
| Acifluorfen | Complex I inhibition |
| Allethrin | Complex I inhibition |
| AVE5638 | Complex I inhibition |
| Besonprodil | Complex I inhibition |
| Bisphenol AF | Complex I inhibition |
| Bisphenol B | Complex I inhibition |
| Butylparaben | Complex I inhibition |
| CJ-013610 | Complex I inhibition |
| CJ-013790 | Complex I inhibition |
| Clorophene | Complex I inhibition |
| Corticosterone | Complex I inhibition |
| CP-105696 | Complex I inhibition |
| CP-457677 | Complex I inhibition |
| CP-544439 | Complex I inhibition |
| Cyclanilide | Complex I inhibition |
| Didecyldimethylammonium chloride | Complex I inhibition |
| Diethylstilbestrol | Complex I inhibition |
| Difenoconazole | Complex I inhibition |
| Dodecyltrimethylammonium chloride | Complex I inhibition |
| Elzasonan | Complex I inhibition |
| Ethofumesate | Complex I inhibition |
| Fenhexamid | Complex I inhibition |
| Fenpyroximate (Z,E) | Complex I inhibition |
| Flumioxazin | Complex I inhibition |
| Forchlorfenuron | Complex I inhibition |
| Hexaconazole | Complex I inhibition |
| Imazalil | Complex I inhibition |
| Isazofos | Complex I inhibition |
| Ketoconazole | Complex I inhibition |
| meso-Hexestrol | Complex I inhibition |
| MK-968 | Complex I inhibition |
| Nilutamide | Complex I inhibition |
| Octyl gallate | Complex I inhibition |
| PD 0343701 | Complex I inhibition |
| Phenolphthalein | Complex I inhibition |
| Prallethrin | Complex I inhibition |
| Propetamphos | Complex I inhibition |
| Pyridaben | Complex I inhibition |
| Rotenone | Complex I inhibition |
| SAR 150640 | Complex I inhibition |
| S-Bioallethrin | Complex I inhibition |
| SR58611 | Complex I inhibition |
| SSR150106 | Complex I inhibition |
| Tebufenpyrad | Complex I inhibition |
| Tetraconazole | Complex I inhibition |
| Tetramethrin | Complex I inhibition |
| Triflumizole | Complex I inhibition |
| Triphenyltin hydroxide | Complex I inhibition |
| Troglitazone | Complex I inhibition |
| Zoxamide | Complex I inhibition |
| Carboxin | Complex II inhibition |
| Chlorpyrifos oxon | Complex II inhibition |
| Chlorpyrifos-methyl | Complex II inhibition |
| MK-274 | Complex II inhibition |
| PharmaGSID_48514 | Complex II inhibition |
| 3-Iodo-2-propynyl-N-butylcarbamate | Complex III inhibition |
| Azoxystrobin | Complex III inhibition |
| CP-085958 | Complex III inhibition |
| Docusate sodium | Complex III inhibition |
| Fenamidone | Complex III inhibition |
| Fluoxastrobin | Complex III inhibition |
| FR150011 | Complex III inhibition |
| Mercuric chloride | Complex III inhibition |
| Milbemectin (mixture of 70% Milbemycin A4, | Complex III inhibition |
| 30% Milbemycin A3) | |
| Octhilinone | Complex III inhibition |
| Phenylmercuric acetate | Complex III inhibition |
| Picoxystrobin | Complex III inhibition |
| Pyraclostrobin | Complex III inhibition |
| Tributyltin chloride | Complex III inhibition |
| Trifloxystrobin | Complex III inhibition |
| Dodecylbenzenesulfonic acid | Complex IV inhibition |
| Famoxadone | Mixed inhibition |
| Methylene bis(thiocyanate) | Mixed inhibition |
| Tiratricol | Mixed inhibition |
| Tributyltin methacrylate | Mixed inhibition |
Comparison of Respirometric and ToxCast Mitochondrial Toxicity Assays
Sixty reference chemicals (Table 2) were selected from the 1042 chemicals tested in this study using an open literature search for both known mitochondrial toxicants and presumptive inactives. These reference chemicals were used to compare the predictivity of the RSA to preceding ToxCast mitochondrial toxicity assays, namely the Tox21 MMP and Apredica HepG2 assays. The results are listed in Table 5.
Table 5.
Binary Classification of ToxCast Mitochondrial Toxicity Assay Performance Using 60 Reference Chemicals
| Assay Technology | Reference Chemicals Tested | TP | TN | FP | FN | Sensitivity | Specificity | BA | MCC |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| NCCT RSA | 60 | 24 | 30 | 0 | 6 | 0.800 | 1.000 | 0.900 | 0.816 |
| TOX21 MMP ratio | 60 | 23 | 29 | 1 | 7 | 0.767 | 0.967 | 0.867 | 0.748 |
| Apredica MitoMembPot | 58 | 18 | 27 | 2 | 11 | 0.621 | 0.931 | 0.776 | 0.580 |
| Apredica MitoMass | 58 | 11 | 26 | 3 | 18 | 0.379 | 0.897 | 0.638 | 0.322 |
The RSA was the most predictive with a balanced accuracy (BA) of 0.9 and a Matthew’s correlation coefficient (MCC) of 0.82. The RSA correctly identified all the presumptive negative chemicals as inactive, but failed to correctly detect boscalid, clofibrate, diclofenac, flutamide, genistein, or quercetin as active. The Tox21 MMP assay was tested on all 60 reference compounds (Attene-Ramos et al., 2013) and was also highly predictive with a BA of 0.87 and MCC of 0.75. Tox21 MMP correctly identified all presumptive negatives except bentazone and all positive reference chemicals except azoxystrobin, boscalid, carboxin, clofibrate, fenofibrate, nilutamide, and sodium azide. The Apredica HepG2 mitochondrial membrane potential and mitochondrial mass assays were tested on 58 of the 60 reference chemicals and were less predictive with MCC values of 0.58 and 0.32, respectively even when combining all assay endpoints (time and direction) to bolster active calls.
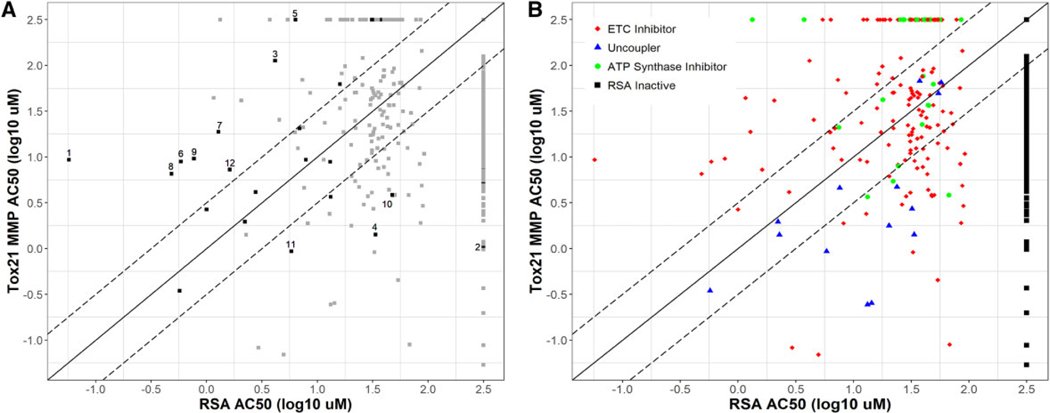
Figure 7 shows a comparison of potency estimates from the RSA and Tox21 MMP assays. Thirteen of the 28 (46%) detected positive reference chemicals (Figure 7A) had an RSA AC50 value (x-axis) more than 0.5 log units (> 3-fold) lower than that for the Tox21 MMP assay (y-axis). These chemicals included some of the most well-studied mitochondrial toxicants such as rotenone, fenpyroximate, pyridaben, tebufenpyrad, and 3 strobilurin compounds and all 13 were ETCi. Eight of the 28 (29%) positive reference compounds had Tox21 MMP AC50 values more than 0.5 log units (> 3-fold) lower than that calculated for the RSA. These included 2 dinitrophenol uncouplers, as well as other chemicals such as mercuric chloride and quercetin. The AC50 values for 7 of the 28 (25%) detected positive reference chemicals were within 0.5 log units (approximately 0.3- to 3-fold; between dashed lines) between the 2 assays, indicating similar potency estimates. Thirteen (87%) of the 15 uncouplers (Figure 7B; blue) identified in this study had a lower AC50 value in the Tox21 MMP assay. Of the 149 ETCi (Figure 7B; red), 91 (61%) had lower AC50 values in the RSA, with 60 (40%) having an RSA AC50 value more than 0.5 log units (> 3-fold) lower than that for the Tox21 MMP assay. The Tox21 MMP assay detected only 11 (38%) of the ATP synthase inhibitors (Figure 7B; green) identified in the RSA.
Figure 7.
Comparison of potency estimates from respirometric screening assay (RSA) and Tox21 MMP assays. Comparison of potency (log AC50 values) estimates derived from the RSA (x-axis) and Tox21 MMP assay (y-axis). Chemicals tested as inactive in the RSA are plotted along the right and those tested as inactive in the Tox21 MMP assay are plotted along the top. The solid black diagonal line denoted equal potency across the 2 assays and the dashed black diagonal lines half-log potency differences between the 2 assays. A, The 60 reference chemicals used to evaluate assay performance are represented as black dots. Gray dots represent the 984 other chemicals tested in this study. Examples are highlighted and ordered by absolute difference in log AC50 values: (1) rotenone, (2) quercetin, (3) fluoxastrobin, (4) 2,4-dintirophenol (5) azoxystrobin, (6) pyridaben, (7) picoxystrobin, (8) tebufenpyrad, (9) fenpyroximate, (10) mercuric chloride, (11) 2-methyl-4,6,-dinitrophenol, and (12) pyraclostrobin. B, Test chemicals (1042) plotted by RSA-assigned mechanisms. Chemicals tested as inactive in the RSA (black dots) are plotted along the right.
DISCUSSION
The RSA approach used in screening was adapted from a previous pilot study that used a respirometric technique to screen and analyze 676 ToxCast Phase II chemicals (Wills et al., 2015), with some intentional changes worth noting, namely: different cell lines, which may have different respiration profiles; use of an exposure paradigm that includes measurement of basal respiration before addition of chemical treatment; and, an experimental design that enabled the identification of ATP synthase inhibitors (more details on the comparison of Wills et al. and our methodology are provided in Supplementary File A). Importantly, this study used human HepG2 tumor cells, the same cell line used in previous ToxCast and Tox21 mitochondrial toxicity screens. Eliminating the confounding issue of cell type in this study facilitated a comparison of the RSA to other ToxCast and Tox21 assay technologies that used HepG2 cells. Although commonly used for high-throughput toxicity programs like ToxCast and Tox21, tumor cells such as HepG2 lack many important xenobiotic metabolism enzymes and transporters present in primary cell models like those used by Wills et al. The absence of xenobiotic metabolism and/or active transport of test chemicals can significantly impact mitochondrial toxicities observed using tumor cells compared with primary cells.
The added value of the RSA screening approach for high-throughput chemical safety evaluation was enabled through a performance evaluation using 60 reference chemicals identified from an open literature search. The 30 active reference chemicals included uncouplers, ATP synthase inhibitors as well as inhibitors of all 4 ETC complexes. Identification of 30 inactive chemicals proved challenging since published studies rarely highlight negative results. We used an indirect approach to randomly select 30 presumptive inactive references from a pool of chemicals meeting 4 criteria, as described in the Materials and Methods section. Although the RSA best predicted the mitochondrial toxicities of the 60 reference chemicals (MCC = 0.90), the Tox21 MMP assay also performed well (0.75). The ToxCast Apredica HepG2 mitochondrial membrane potential and mitochondrial mass assays were not nearly as predictive (0.58 and 0.32, respectively). For several well-known mitochondrial toxicants, there were stark differences between the potencies estimated from the RSA compared with the Tox21 MMP. Rotenone, fenpyroximate, pyridaben, tebufenpyrad, and 3 of the strobilurin pesticides (all ETCi) had RSA AC50 values at least 10-fold lower than those derived from Tox21 MMP data. In contrast, 5 reference chemicals were more potent in the Tox21 MMP assay. All 3 reference uncouplers, 2,4-dinitrophenol, 2-methyl-4,6-dinitrophenol, and dinoseb, were more potent (ie, active at lower concentrations) in the Tox21 MMP assay than the RSA.
The tiered approach used in this study enabled the identification of false positives that have may have been misclassified in previous Tox21 and ToxCast assays. Gentian violet and similar compounds such as methyl violet and fluorescein known to interfere with fluorescent signal but are among the 50 most potent actives in the Tox21 MMP. Eight of the 10 redox-cycling chemicals flagged as false positives in the RSA are listed as active in the Tox21 MMP. Given that the Tox21 MMP has higher throughput than the RSA with comparable active/inactive detection of the reference chemicals, an expedient strategy to screen large chemical inventories would be to use the Tox21 MMP to first rapidly identify actives and then confirm and characterize those actives using the RSA. Because the sources of assay interference between the Tox21 MMP assay and the RSA appear to be unique, this strategy would correctly identify only 19 (63%) of 30 active reference chemicals significantly decreasing sensitivity and predictivity (MCC = 0.68). Alternatively, the Tox21 MMP and a tiered RSA could both be used to screen the entire inventory in parallel and activity in either or both assays then be determinative of mitochondrial toxicity. This alternative strategy would be more resource- and time-intensive but leverages each assay to compensate for the insufficiencies of the other and would correctly identify 93% of the reference positives and 97% the reference negatives, significantly improving predictivity (MCC = 0.90). The combined use of membrane potential and respiration for quantifying the effects of mitochondrial toxicants has been previously recommended (Brand and Nicholls, 2011).
No single assay endpoint from the RSA should be used in isolation to derive a definitive list of mitochondrial toxicants. A proposed data-driven mechanistic assignment required 4 assay endpoints considered sequentially (Figure 4H). We found many more ETCi than uncouplers and ATP synthase inhibitors combined, in agreement with Wills et al. (2015). The ability to determine an underlying mechanism of mitochondrial toxicity is a major feature of the respirometric screening strategy when compared with single-endpoint screening technologies. Understanding the mitochondrial toxicity mechanism of a chemical is critical for predicting the constellation of adverse outcomes that may manifest from exposure. Acute poisoning from ETCi and ATP synthase inhibitors cause metabolic acidosis and hyperpnea leading to convulsions and cardio-respiratory failure, despite normal oxygen levels (Wallace and Starkov, 2000). Uncouplers induce excessive oxygen consumption by maximizing respiration, leading to hypoxia, and cyanosis in addition to metabolic acidosis which results in uncontrolled thermogenesis and hyperthermia.
We identified 15 uncouplers (1.4% of tested chemicals) in this study using multiconcentration RSA testing. Wills et al. identified 5 uncouplers (0.7%) among the 676 chemicals tested. Three of those uncouplers, 2-methyl-4,6-dinitrolphenol, pentachlorophenol, and perfluorooctanesulfonamide were confirmed as uncouplers in this study. The other 2, didecyldimethylammonium chloride and tributyltin chloride were determined to be ETC inhibitors in this study and their mechanisms (complex I and III inhibitors, respectively) confirmed in the EFA. Ten of the 15 uncouplers identified here were nitro- or halogen-substituted phenols, the uncoupling activity of which has been known for decades (Stockdale and Selwyn, 1971). Recently, both perfluorinated sulfonamides and perfluorinated carboxylic acids were listed as structural alerts for protonophoric activity and mitochondrial uncoupling (Enoch et al., 2018). Of the 14 perfluorinated compounds tested in this study, only perfluorooctanesulfonamide (PFOSA) was active in the RSA and was classified an uncoupler. PFOSA was also among the 5 uncouplers identified by Wills et al. and was the only perfluorinated chemical listed as active in that study. Although lacking any of the predictive structural features, fluazinam has long been known to be an uncoupler (Guo et al., 1991). The only seemingly mischaracterized uncoupler in this study was cyazofamid which has been reported to be a complex III inhibitor (Li et al., 2014). Cyazofamid clearly increased basal phase respiration at concentrations ranging from 1 to 10 μM (Supplementary Figure 2) and decreased maximal phase respiration at 100 μM. To eliminate the possibility of chemical degradation or impurity in the blinded sample, we independently procured cyazofamid, prepared a new DMSO solution and retested in the RSA. The results from the newly sourced cyazofamid were indistinguishable from the those obtained using the blinded sample (data not shown). The reasons for this disparity our results from the published reports are unclear. A mechanistic determination using only the 100 mM response pattern from RSA testing would have misclassified cyazofamid as an ETCi, so it is possible cyazofamid was previously tested at concentrations exceeding its effective uncoupling concentration range. Sulfonamides comprise 4 out of 20 structural alerts for mitochondrial uncoupling (Enoch et al., 2018) and cyazofamid harbors a prominent sulfonamide group. Another possibility then is that the cyazofamid parent is a complex III inhibitor but a cyazofamid derivative resulting from degradation or modification in HepG2 cells that retains the sulfonamide moiety is a potent uncoupler.
The EFA was designed to confirm the activity and identify the respiratory complex(es) target by each of the 149 ETCi identified in Tier 2 testing. Permeabilization of cells grants mitochondrial access to substrates like succinate and ascorbate that are not readily absorbed by cells. These substrates facilitate electron transport at downstream complexes, bypassing chemically induced inhibition of upstream complexes. A set of 6 heuristics (described in the Materials and Methods section) was applied to EFA data to identify ETC complexes inhibited by each tested chemical. The first 3 heuristics are straightforward and classify more than 90% of the test chemicals. The remaining 3 rules were developed to adjudicate disagreement between response patterns by concentration, likely owing to promiscuous activity at higher test concentrations. EFA analysis identified ETC complex targets for 84 (56%) of the 149 ETCi identified in Tier 2 testing. Most of the EFA-active chemicals (70%) were complex I inhibitors, unsurprising given the relative size of complex I and the number of known complex I inhibitors compared with those for complexes II, III, and IV. Among the complex I inhibitors identified in this study were well-known mitochondrial toxicants such as rotenone, fenpyroximate, pyridaben, and tebufenpyrad, as well as several bisphenols and conazoles. Five complex II inhibitors were identified, including carboxin which has been extensively studied and 2 metabolites of chlorpyrifos: -methyl and -oxon. Others have observed decreased ATP synthesis in chlorpyrifos-treated cells but attributed this to possible complex I inhibition (Chen et al., 2017). Fifteen complex III inhibitors were identified including 5 strobilurin pesticides and 3 heavy or organo-metal compounds. We identified only a single complex IV inhibitor, dodecylbenzenesulforic acid; this study is the first to report this activity for this chemical. At least one other known complex IV inhibitor, sodium azide, was binned as an ETCi using Tier 2 data from intact cells but failed to inhibit respiration in the EFA. Four chemicals exhibited a mixed complex I/III inhibition response pattern characterized by partial respiration recovery with succinate and complete recovery with TMPD/ascorbate. One of these compounds, tributyltin methacrylate is highly related another chemical classified as a complex III inhibitor, tributyltin chloride. Interestingly, Wills et al. classified tributyltin as an uncoupler using rabbit renal proximal tubule cells.
Sixty-five (44%) of the ETCi identified in Tier 2 failed to inhibit respiration in the EFA and remain unconfirmed. Reduced potencies were observed for both fenpyroximate control and FCCP reagent during optimization of the EFA protocol. Complex I inhibition in the EFA required 4 μM fenpyroximate compared with approximately 1 μM in the RSA. The concentration of FCCP required to induce maximal phase respiration increased from 0.25 μM in the RSA to 2 μM in EFA. The reasons why reagents and test chemicals are less potent and efficacious in the EFA compared with the RSA are not clear, but 1 possible explanation is that many of these chemicals preferentially partition to the cellular compartment in the RSA so that the nominal test concentration underestimates the intracellular chemical concentration. In the EFA, where cells are permeabilized prior to test compound addition, cell partitioning is not possible and thus the nominal test concentration is more likely to accurately reflect the concentration encountered by mitochondria. Analysis of the decreased maximal respiration phase AUFs of EFA-active and -inactive chemicals reveals that EFA-active chemical had higher AUFs (0.39 ± 0.30) than EFA-inactives (0.19 ± 0.08) indicating the EFA-active chemicals were more potent and efficacious in the RSA compared with EFA-inactives.
We identified 29 chemicals that decreased basal respiration with no significant decrease in maximal or inhibited respiration which is typical of ATP synthase inhibitors such as oligomycin in the RSA. This is more than we would have expected prior to screening and may have warranted inclusion of a third positive control like oligomycin if only to clearly demonstrate the ATP synthase inhibition phenotype in the RSA. This number may also reflect misclassification of some chemicals due to a disparity in activity thresholds used for basal versus maximal respiration phases. Additional confirmation using isolated mitochondria activated with ADP would be needed to determine how many of the putative ATP synthase are correctly classified.
It is important to note that some of the mitochondrial-disrupting chemicals identified in this study have superseding toxicities unrelated to mitochondrial dysfunction. Organophosphates such as fenamiphos and propetamphos were also determined to be ETCi in this report but are known to be highly or moderately neurotoxic due to acetylcholinesterase inhibition (www.epa.gov/sites/production/files/documents/rmpp_6thed_ch5_organophosphates.pdf, last accessed May 7, 2020). Several known endocrine-active chemicals, such as 17β-estradiol, diethylstilbestrol, and testosterone propionate, were also found to be ETCi in this study. Like any other high-throughput screening effort, the results presented herein report the effects of test chemicals on a singular toxicity endpoint. Using data of this type to predict in vivo adverse outcomes can be confounded by other factors such superseding toxicities, toxicokinetics, and toxicodynamics. The primary aim of screening assays is to identify all tests chemicals that elicit a particular in vitro toxicity, specifically mitochondrial toxicity. We have shown here that the RSA can be used to efficiently screen a large chemical inventory for mitochondrial toxicants, derive sensitive potency estimates, and effectively assign putative mechanisms of mitochondrial disruption.
Supplementary Material
ACKNOWLEDGMENTS
This study was carried out as part of the EPA High Throughput Testing project within the Chemical Safety for Sustainability (CSS) Program. We thank Dr Brian Chorley, Dr Stephanie Padilla, and Dr James Samet for critical review of the manuscript.
FUNDING
H.B.L worked under the National Student Services Contract managed by Oak Ridge Associated Universities under the Student Services Contracting Authority on behalf of the U.S. Environmental Protection Agency.
Footnotes
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
This work is written by US Government employees and is in the public domain in the US.
Disclaimer: The views expressed in this paper are those of the authors and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
REFERENCES
- Alonso JR, Cardellach F, López S, Casademont J, and Miró O. (2003). Carbon monoxide specifically inhibits cytochrome c oxidase of human mitochondrial respiratory chain. Pharmacol. Toxicol 93, 142–146. [DOI] [PubMed] [Google Scholar]
- Attene-Ramos MS, Huang R, Michael S, Witt KL, Richard A, Tice RR, Simeonov A, Austin CP, and Xia M. (2015). Profiling of the Tox21 chemical collection for mitochondrial function to identify compounds that acutely decrease mitochondrial membrane potential. Environ. Health Perspect 123, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attene-Ramos MS, Huang R, Sakamuru S, Witt KL, Beeson GC, Shou L, Schnellmann RG, Beeson CC, Tice RR, Austin CP, et al. (2013). Systematic study of mitochondrial toxicity of environmental chemicals using quantitative high throughput screening. Chem. Res. Toxicol 26, 1323–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N, Knudsen T, and Williams A. (2017). Abstract Sifter: A comprehensive front-end system to PubMed. F1000Research 6, 2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balba H. (2007). Review of strobilurin fungicide chemicals. J. Environ. Sci. Health B 42, 441–451. [DOI] [PubMed] [Google Scholar]
- Beeson CC, Beeson GC, and Schnellmann RG (2010). A high throughput respirometric assay for mitochondrial biogenesis and toxicity. Anal. Biochem 404, 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begriche K, Massart J, Robin MA, Borgne-Sanchez A, and Fromenty B. (2011). Drug-induced toxicity on mitochondria and lipid metabolism: Mechanistic diversity and deleterious consequences for the liver. J. Hepatol 54, 773–794. [DOI] [PubMed] [Google Scholar]
- Berson A, Schmets L, Fisch C, Fau D, Wolf C, Fromenty B, Deschamps D, and Pessayre D. (1994). Inhibition by nilutamide of the mitochondrial respiratory chain and ATP formation. Possible contribution to the adverse effects of this antiandrogen. J. Pharmacol. Exp. Ther 270, 167–176. [PubMed] [Google Scholar]
- Bhhatarai B, Wilson DM, Bartels MJ, Chaudhuri S, Price PS, and Carney EW (2015). Acute toxicity prediction in multiple species by leveraging mechanistic ToxCast mitochondrial inhibition data and simulation of oral bioavailability. Toxicol. Sci 147, 386–396. [DOI] [PubMed] [Google Scholar]
- Bohmont C, Aaronson LM, Mann K, and Pardini RS (1987). Inhibition of mitochondrial NADH oxidase, succinoxidase, and ATPase by naturally occurring flavonoids. J. Nat. Prod 50, 427–433. [DOI] [PubMed] [Google Scholar]
- Brand MD, and Nicholls DG (2011). Assessing mitochondrial dysfunction in cells. Biochem. J 435, 297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon M, Baldi P, and Wallace DC (2006). Mitochondrial mutations in cancer. Oncogene 25, 4647–4662. [DOI] [PubMed] [Google Scholar]
- Chen T, Tan J, Wan Z, Zou Y, Afewerky HK, Zhang Z, and Zhang T. (2017). Effects of commonly used pesticides in China on the mitochondria and ubiquitin-proteasome system in Parkinson’s disease. Int. J. Mol. Sci 18, 2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Gray GM, and Bucher JR (2008). Transforming environmental health protection. Science 319, 906–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun P, Wyrembak J, Schriner SE, Chen HW, Marciniack C, Laferla F, and Wallace DC (2012). A mitochondrial etiology of Alzheimer and Parkinson disease. Biochim. Biophys. Acta 1820, 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degli Esposti M. (1998). Inhibitors of NADH-ubiquinone reductase: An overview. Biochim. Biophys. Acta 1364, 222–235. [DOI] [PubMed] [Google Scholar]
- Delp J, Funke M, Rudolf F, Cediel A, Hougaard Bennekou S, van der Stel W, Carta G, Jennings P, Toma C, Gardner I, et al. (2019). Development of a neurotoxicity assay that is tuned to detect mitochondrial toxicants. Arch. Toxicol 93, 1585–1608. [DOI] [PubMed] [Google Scholar]
- Dix DJ, Houck KA, Martin MT, Richard AM, Setzer RW, and Kavlock RJ (2007). The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol. Sci 95, 5–12. [DOI] [PubMed] [Google Scholar]
- Dykens JA, and Will Y. (2007). The significance of mitochondrial toxicity testing in drug development. Drug Discov. Today 12, 777–785. [DOI] [PubMed] [Google Scholar]
- Enoch SJ, Schultz TW, Popova IG, Vasilev KG, and Mekenyan OG (2018). Development of a decision tree for mitochondrial dysfunction: Uncoupling of oxidative phosphorylation. Chem. Res. Toxicol 31, 814–820. [DOI] [PubMed] [Google Scholar]
- Farina M, Avila DS, da Rocha JB, and Aschner M. (2013). Metals, oxidative stress and neurodegeneration: A focus on iron, manganese and mercury. Neurochem. Int 62, 575–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrick DA, Neilson A, and Beeson C. (2008). Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov. Today 13, 268–274. [DOI] [PubMed] [Google Scholar]
- Filer DL, Kothiya P, Setzer RW, Judson RS, and Martin MT (2017). tcpl: The ToxCast pipeline for high-throughput screening data. Bioinformatics 33, 618–620. [DOI] [PubMed] [Google Scholar]
- Grulke CM, Williams AJ, Thillanadarajah I, and Richard AM (2019). EPA’s DSSTox database: History of development of a curated chemistry resource supporting computational toxicology research. Comput. Toxicol 12, 100096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Miyoshi H, Komyoji T, Haga T, and Fujita T. (1991). Uncoupling activity of a newly developed fungicide, fluazinam [3-chloro-N-(3-chloro-2,6-dinitro-4-trifluoromethyl-phenyl)-5-trifluoromethyl-2-pyridinamine]. Biochim. Biophys. Acta 1056, 89–92. [Google Scholar]
- Hamm J, Sullivan K, Clippinger AJ, Strickland J, Bell S, Bhhatarai B, Blaauboer B, Casey W, Dorman D, Forsby A, et al. (2017). Alternative approaches for identifying acute systemic toxicity: Moving from research to regulatory testing. Toxicol. In Vitro 41, 245–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobeloch LM, Blondin GA, Read HW, and Harkin JM (1990). Assessment of chemical toxicity using mammalian mitochondrial electron transport particles. Arch. Environ. Contam. Toxicol 19, 828–835. [DOI] [PubMed] [Google Scholar]
- Lavrich KS, Corteselli EM, Wages PA, Bromberg PA, Simmons SO, Gibbs-Flournoy EA, and Samet JM (2018). Investigating mitochondrial dysfunction in human lung cells exposed to redox-active PM components. Toxicol. Appl. Pharmacol 342, 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AP, Cameron RB, Speiser JL, Wolf BJ, Peterson YK, Schnellmann RG, Beeson CC, and Rohrer B. (2015). Quantitative analysis of mitochondrial morphology and membrane potential in living cells using high-content imaging, machine learning, and morphological binning. Biochim. Biophys. Acta 1853, 348–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhu XL Yang WC, and Yang GF (2014). Comparative kinetics of Qi site inhibitors of cytochrome bc1 complex: Picomolar antimycin and micromolar cyazofamid. Chem. Biol. Drug Des 83, 71–80. [DOI] [PubMed] [Google Scholar]
- Loomis WF, and Lipmann F. (1948). Reversible inhibition of the coupling between phosphorylation and oxidation. J. Biol. Chem 173, 807. [PubMed] [Google Scholar]
- Marroquin LD, Hynes J, Dykens JA, Jamieson JD, and Will Y. (2007). Circumventing the Crabtree effect: Replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicol. Sci 97, 539–547. [DOI] [PubMed] [Google Scholar]
- Mehta R, Chan K, Lee O, Tafazoli S, and O’Brien PJ (2008) Drug-associated mitochondrial toxicity. In Drug-induced Mitochondrial Dysfunction, 1st ed. (Dykens JA and Will Y, Eds.). John Wiley & Sons, Inc., Hoboken, NJ. [Google Scholar]
- Metcalf RL, and Horowitz AR (2014) Insect Control, 2. Individual Insecticides. Ullmann’s Encyclopedia of Industrial Chemistry. Wiley-VCH Verlag GmbH & Co., Weinheim. [Google Scholar]
- Meyer JN, and Chan SSL (2017). Sources, mechanisms, and consequences of chemical-induced mitochondrial toxicity. Toxicology 391, 2–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JN, Hartman JH, and Mello DF (2018). Mitochondrial toxicity. Toxicol. Sci 162, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, and Suomalainen A. (2012). Mitochondria: In sickness and in health. Cell 148, 1145–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessayre D, Fromenty B, Berson A, Robin MA, Lettéron P, Moreau R, and Mansouri A. (2012). Central role of mitochondria in drug-induced liver injury. Drug Metab. Rev 44, 34–87. [DOI] [PubMed] [Google Scholar]
- Rodriguez RJ, and Acosta D. (1996). Inhibition of mitochondrial function in isolated rate liver mitochondria by azole antifungals. J. Biochem. Toxicol 11, 127–131. [DOI] [PubMed] [Google Scholar]
- Rolo AP, and Palmeira CM (2006). Diabetes and mitochondrial function: Role of hyperglycemia and oxidative stress. Toxicol. Appl. Pharmacol 212, 167–178. [DOI] [PubMed] [Google Scholar]
- Sakamuru S, Li X, Attene-Ramos MS, Huang R, Lu J, Shou L, Shen M, Tice RR, Austin CP, and Xia M. (2012). Application of a homogenous membrane potential assay to assess mitochondrial function. Physiol. Genomics 44, 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders JE, Beeson CC, and Schnellmann RG (2013). Characterization of functionally distinct mitochondrial subpopulations. J. Bioenerg. Biomembr 45, 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierotzki H. (2015) Respiration inhibitors: Complex III. In Fungicide Resistance in Plant Pathogens (Ishii H. and Hollomon D, Eds.). Springer, Tokyo. [Google Scholar]
- Stammler G, Wolf A, Glaettli A, and Klappach K. (2015) Respiration inhibitors: Complex II. Fungicide Resistance in Plant Pathogens (Ishii H. and Hollomon D, Eds.). Springer, Tokyo. [Google Scholar]
- Stockdale M, and Selwyn MJ (1971). Effects of ring substituents on the activity of phenols as inhibitors and uncouplers of mitochondrial respiration. Eur. J. Biochem 21, 565–574. [DOI] [PubMed] [Google Scholar]
- Wallace DC (2005). A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu. Rev. Genet 39, 359–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace KB (2018). Historical Perspective of mitochondria in the toxicological sciences. Toxicol. Sci 162, 12–14. [DOI] [PubMed] [Google Scholar]
- Wallace KB, and Starkov AA (2000). Mitochondrial targets of drug toxicity. Annu. Rev. Pharmacol. Toxicol 40, 353–388. [DOI] [PubMed] [Google Scholar]
- Wills LP (2017). The use of high-throughput screening techniques to evaluate mitochondrial toxicity. Toxicology 391, 34–41. [DOI] [PubMed] [Google Scholar]
- Wills LP, Beeson GC, Hoover DB, Schnellmann RG, and Beeson CC (2015). Assessment of ToxCast Phase II for mitochondrial liabilities using a high-throughput respirometric assay. Toxicol. Sci 146, 226–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills LP, Beeson GC, Trager RE, Lindsey CC, Peterson YK, Beeson CC, and Schnellmann RG (2013). High-throughput respirometric assay identifies predictive toxicophore of mitochondrial injury. Toxicol. Appl. Pharmacol 272, 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.