Abstract
The gut microbiota plays a key role in host health and disease, particularly through their interactions with the immune system. Intestinal homeostasis is dependent on the symbiotic relationships between the host and the diverse gut microbiota, which is influenced by the highly co-evolved immune–microbiota interactions. The first step of the interaction between the host and the gut microbiota is the sensing of the gut microbes by the host immune system. In this review, we describe the cells of the host immune system and the proteins that sense the components and metabolites of the gut microbes. We further highlight the essential roles of pattern recognition receptors (PRRs), the G protein-coupled receptors (GPCRs), aryl hydrocarbon receptor (AHR) and the nuclear receptors expressed in the intestinal epithelial cells (IECs) and the intestine-resident immune cells. We also discuss the mechanisms by which the disruption of microbial sensing because of genetic or environmental factors causes human diseases such as the inflammatory bowel disease (IBD).
Keywords: mucosal immunology, pattern recognition receptors, protein-coupled receptors, intestinal epithelial cells, immune cells, gut microbiota, enteric viruses, inflammatory bowel disease
Introduction
The human gut lumen contains trillions of microorganisms, including bacteria, viruses, fungi, and archaea. These microorganisms are collectively termed as the “microbiota” and harbor 100-fold more genes than the humans (Sommer and Bäckhed, 2013). The human microbiota is estimated to consist of nearly 4 × 1013 microbial cells, which is approximately equivalent to the total number of cells in the human body (Elson and Alexander, 2015; Sender et al., 2016a, 2016b). The Human Microbiome Project (HMP) was established in 2008 and aimed to (i) characterize the microbial communities found in different parts of the human body; (ii) identify the correlation between changes in the microbiome and the human health; and (iii) demonstrate opportunities to improve human health by manipulating the human microbiome (Group et al., 2009). The HMP initiative and other “omics” studies including metabolomics, transcriptomics, and proteomics, have significantly expanded our understanding of the diversity and complexity of the human microbiota (Palm et al., 2015). In the last 2 decades, lots of studies have shown that the microbiota-host crosstalk plays an important role in maintaining the intestinal homeostasis for both the host and the microbiota, and also plays a critical role in the systemic health, physiology, and development of the host. Current progress in the field has shown that the gut microbiota significantly impacts human health and disease. The host immune system recognizes antigens derived from the structural components of the microbiota and their metabolites and mount an optimal immune response to maintain the physiological functions of the host (Franzosa et al., 2015; Rooks and Garrett, 2016).
The three distinct inter-linked layers of the mammalian gastrointestinal tract, namely, (i) the luminal mucus layer; (ii) the 10-μm monolayer of intestinal epithelial cells; and (iii) the internal layer of the mucosal immune system called as the lamina propria, maintained a delicate balance between the immune system and the tissue homeostasis in the gut lumen (Mowat and Agace, 2014). The immune cells in the gut barrier such as the macrophages, dendritic cells (DCs), innate lymphoid cells (ILCs), T cells and B cells, and the non-immune intestinal epithelial cells (IECs) play a significant role in the gut–microbiota interactions. The pattern recognition receptors (PRRs) play a crucial role in the innate immune system and constitute a large collection of proteins that play a critical role in maintaining the immune-microbiota homeostasis by rapidly recognizing signaling molecules derived from the non-commensal microbes and the injured host intestinal tissue components (Medzhitov and Janeway, 2000; Burgueno and Abreu, 2020). For example, mutations in NOD2, a well-characterized PRR, is significantly associated with the inflammatory bowel disease (IBD) (Hugot et al., 2001; Ogura et al., 2001; Khor et al., 2011). The host cells also express receptors, which sense and respond to metabolites derived either from the anaerobic fermentation of undigested dietary components or from compounds that are de novo synthesized by the microbes or the host cells (Eshleman and Alenghat, 2021). The immune system-microbiota homeostasis is maintained by the activity of several signaling pathways. However, genetic mutations in the host genome, infections, antibiotics, and/or diet can disrupt the delicate host-microbiota balance causing dysbiosis, which contributes significantly to the pathogenesis of infections, autoimmune diseases, inflammatory diseases, obesity, cancer, and other diseases (Gopalakrishnan et al., 2018). In this review, we summarize the mechanisms by which the host intestinal cells sense the microbes and gut metabolites derived from the microbe to maintain gut homeostasis. We also describe the mechanisms by which disruption of the host-microbiota homeostasis in the gut adversely affects human health and disease.
Sensing of microbes and metabolites by receptors in intestine
Skin and mucosa act as physical barriers and prevent the entry of pathogens such as bacteria, viruses, and fungi into the host. However, when these pathogens breach the first line of defense, the innate immune system immediately responds and produces antimicrobial products and recruits a wide variety of immune cells to eliminate them (Liu and Cao, 2016). Microbial ligands (e.g., nucleic acids and the bacterial cell wall components) are detected by the germline encoded PRRs to initiate downstream immune responses against the invading microbes. The microbial metabolites are recognized by the G protein-coupled receptors (GPCRs), aryl hydrocarbon receptors (AHRs), nuclear receptors, and other receptors, which modulate innate immune signaling pathways, impact epithelium integrity, and importantly, regulate hormonal signals release. Gut hormones have a wide range of targets in the whole body and play systemic physiological roles especially in the control of metabolism. Except for products of food digestion (e.g., glucose, amino acids, and fatty acids), microbial products [such as short-chain fatty acids (SCFAs), lipopolysaccharide (LPS) and secondary bile acids] also act as stimuli for local enteroendocrine cells (EECs) to generate hormonal signals that reflect dietary intake, microbial composition and epithelial integrity (Gribble and Reimann, 2019).
Microbial pattern recognition by the intestinal PRRs
The microbial pattern recognition model was first proposed by Charles Janeway Jr. It has since been established through the discovery and characterization of numerous PRRs (Janeway and Medzhitov, 2002). PRRs are classified into five families based on their protein domain homology—(i) Toll-like receptors (TLRs), (ii) NOD-like receptors (NLRs), (iii) retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), (iv) C-type lectin receptors (CLRs), and (v) cytosolic DNA sensors including absence in melanoma 2 (AIM2)-like receptors (ALRs) and cyclic GMP-AMP synthase (cGAS). PRRs recognize evolutionarily conserved pathogen-associated molecular patterns (PAMPs) such as LPS, flagellin, bacterial DNA, and bacterial RNA. PRRs also sense danger-associated molecular patterns (DAMPs) according to the “danger theory” proposed by Matzinger (2002). DAMPs include molecules such as ATP, high mobility group box 1 protein (HMGB1), and uric acid, which are released from the injured cells. They subsequently bind to the PRRs and activate specific downstream signaling pathways. The activation of PRR signaling by PAMPs or DAMPs plays a significant role in pathogen clearance and tissue homeostasis. In this section, we review the intestinal PRRs, including (i) specific cell membrane-expressed TLRs, which respond to the extracellular microbes; (ii) TLRs in the endosomes or lysosomes, which regulate inflammatory cytokine production; and (iii) cytoplasm-expressed NLRs or RLRs, which respond to microbes or components, enter the nucleus to induce transcription of genes encoding cytokines such as interleukin (IL)-1β and IL-18, and trigger inflammatory cell death termed as “pyroptosis” (Fig. 1).
Figure 1.
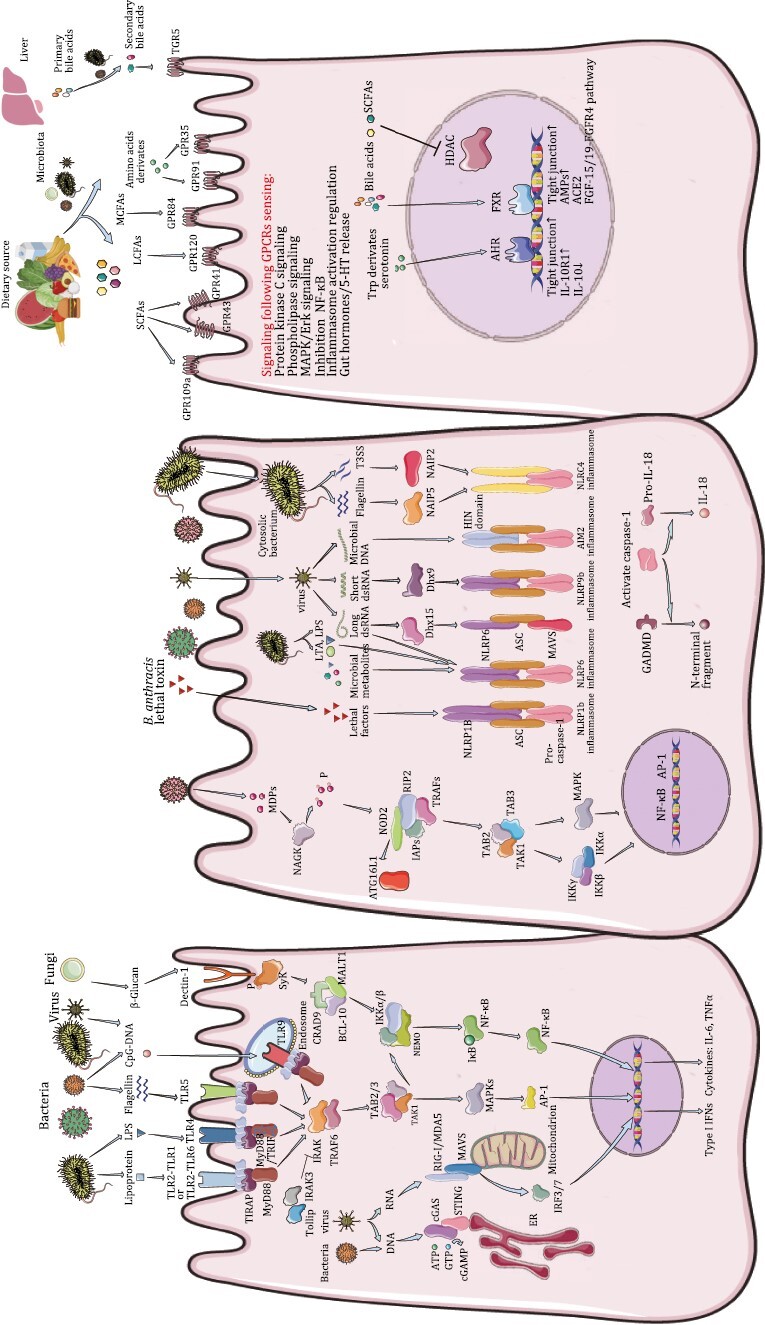
Microbial signal–host sensor interactions in IECs and associated signaling events. (Left) Microorganism-associated molecular patterns (MAMPs) from the bacteria, fungi and viruses are sensed by extensive types of pattern recognition receptors (PRRs), which are broadly expressed in/on intestinal epithelial cells (IECs). For toll-like receptors (TLRs), TLR4 senses bacterial lipopolysaccharide (LPS), TLR2-TLR1 or TLR2-TLR6 dimers sense bacterial lipoprotein, TLR5 senses bacterial flagellin and TLR9, which mainly localized on endosome membrane, is triggered by CpG DNA from both bacteria and viruses. Once binding ligands, TLR homologous or heterodimers are formed and MyD88-dependent and/or MyD88-independent signaling pathways are activated by different adaptor proteins (AP) such as MyD88, Toll/IL-1 receptor (TIR) domain-containing adaptor protein (TIRAP), TIR domain-containing adaptor molecule (TRAM) and/or TIR domain-containing adaptor inducing IFNβ (TRIF). However, TLR2 and TLR4 need TIRAP to recruit MyD88. TLR4 can initiate downstream signals using both MyD88 and TRIF as signaling adaptors, but it also needs TRAM to recruit TRIF and TIRAP to recruit MyD88. MyD88 attracts IL-1 receptor-associated kinases (IRAKs) to TLRs through the interaction of two molecular death domains and IRAK-1 is activated by phosphorylation, which then interacts with TNF-receptor-associated factor (TRAF), a ubiquitin ligase. Typically, TRAF6 forms complexes with TGF-beta activated kinase (TAK) and TAK1-associated binding protein 2 (TAB), and TAK activates downstream mitogen activated protein (MAP) kinase [Jun N-terminal kinases (JNK), p38 MAPK] and inhibitor of nuclear factor kappa-B kinase (IKK), leading to activation of nuclear factor kappa-B (NF-κB) and activator protein 1 (AP-1), and therefore regulating the expression of pro-inflammatory cytokines and other immune-related genes. TLR4 and TLR3 could induce TRIF-dependent IFN-I pathway, and activate similar downstream signals through TRAF3 or receptor-interacting serine/threonine-protein kinase 1 (RIPK1)/TRAF6. Both Tollip and IRAK-3 interact with IRAK-1 and negatively regulate TLR-mediated signal transduction pathways. Viruses are sensed by retinoic acid-inducible gene I (RIG-I), which detect 5ʹ-ppp RNA and short-chain dsRNA, and melanoma differentiation-associated gene 5 (MDA5), which detect long-chain dsRNA respectively, and lead to type I IFN production in a mitochondrial antiviral signaling protein (MAVS)- and stimulator of interferon response cGAMP interactor (STING)-dependent manner. DNA viruses activate the cyclic GMP-AMP synthase (cGAS)-STING pathway. In the presence of DNA, cGAS enzymatically converts ATP and GTP into the cytosolic dinucleotide, cGAMP, which then binds to STING and activates IFN signaling. Dectin-1, officially named as C-type lectin domain-containing 7A (CLEC7A), a member of the C-type lectin family, senses β-glucans from fungi and promotes spleen tyrosine kinase (SyK) phosphorylation and subsequent signaling activation. (Mid) NLRs are a subset of cytoplasmic PRRs. On sensing their respective peptidoglycan ligands [d-glutamyl-meso-diaminopimelic acid (DAP) or phosphorylated muramyl dipeptide (MDP)], nucleotide-binding oligomerization domain-containing 1 (NOD1) and NOD2 undergo auto-oligomerization and induce NF-κB signaling by activating receptor-interacting serine/threonine kinase 2 (RIP2). Or NODs can interact with autophagy related 16 like 1 (ATG16L1) to induce autophagy. ATG16L1 is also a negative regulator of NOD signaling. NOD-like receptors (NLRs) such as NLRP1b, NLRP6 and NLRP9b oligomerize upon activation to form inflammasome complexes, which serve as platforms for the recruitment, cleavage, and activation of the inflammatory Caspase-1 (CASP-1). Subsequently, activated CASP-1 can process pro-IL-1β, pro-IL-18 or gasdermin D (GSDMD) into active forms, which in turn stimulate cytokine secretion or pyroptosis. Furthermore, NLRP6 can sense and bind the long viral dsRNA directly or through the RNA helicase DEAH-box helicase 15 (DHX15), which forms liquid–liquid phase separation (LLPS) to integrate inflammasome and and ISG. Microbial metabolites such as SCFAs could regulate NLRP6 inflammasome function. NLRP9b senses and binds the short viral dsRNA through DHX9 and activates the inflammasome. Flagellin and PrgJ, a conserved type III secretion system (TTSS) rod component from bacteria, bind to their corresponding adaptors, NAIP5 and NAIP2, respectively. Subsequently, they interact with NLRC4 to initiate formation of the NLRC4 inflammasome. (Right) Microbiota-processed metabolites could be sensed mainly by G Protein-Coupled Receptors (GPCRs) on cell membrane or by aryl hydrocarbon receptor (AHR) and nuclear receptors in cell nucleus and induce downstream signaling pathways. GPR41/43/109a are part of typical receptors for short-chain fatty acids (SCFAs, <6C), GPR120 could sense long -chain fatty acids (LCFAs, >12C) and GPR84 is stimulated by medium-chain fatty acids (MCFAs, 6C~12C). SCFAs could directly inhibit the activity of Histone deacetylase (HDAC). Amino acids derivates such as tryptophan and tyrosine catabolism products could activate GPR35, and succinate binds GPR91. Tryptophan derivatives, for example, indole-3-aldehyde could also stimulate nuclear AHR. Primary bile acids are produced in liver and some of them would be metabolized into secondary bile acids by microbiota when come to distal intestine. Nuclear receptor farnesoid X-activated receptor (FXR) can bind bile acids and regulate bile acid metabolism. G protein-coupled bile acid receptor 1 (GP-BAR1, also named TGR5) is a membrane GPCR and mainly binds to secondary bile acids.
TLRs
TLRs are type I transmembrane receptors that are characterized by an extracellular leucine-rich repeat (LRR) domain and an intracellular Toll/IL-1 receptor (TIR) domain (Rock et al., 1998). In the PRRs, LRR domain is responsible for ligand recognition and the TIR domain is required for signal transduction. Ten functional TLRs (TLR1–10) have been reported in humans and 12 TLRs (TLR1–9 and TLR11–13) have been reported in mice (Chuang and Ulevitch, 2001; Temperley et al., 2008; Mathur et al., 2016; Li and Wu, 2021). TLRs play a key role in shaping the intestinal microbiota and maintaining gut homeostasis. Myeloid differentiation factor 88 (MyD88) is an adaptor protein (AP) that is used by most TLRs and MyD88-deficent mice demonstrate defective T cell function and aggravated colitis (Takeuchi and Akira, 2010; Pandey et al., 2014; Wang et al., 2015b). TLR3 mediates transcriptional induction of type I interferons (IFNs), pro-inflammatory cytokines, and chemokines through the TIR domain-containing adaptor inducing IFNβ (TRIF) dependent pathway; TLR4 activates both MyD88- and TRIF-dependent pathways (Takeuchi and Akira, 2010; Pandey et al., 2014). MyD88-dependent microbial sensing is required to maintain the intestinal epithelial homeostasis and the production of antimicrobial peptides in the intestine (Rakoff-Nahoum et al., 2004; Vaishnava et al., 2008; Menendez et al., 2013; Bhinder et al., 2014).
TLRs are expressed in different cell types of the gut, including the IECs, immune cells, stromal cells, and the neuronal cells (Burgueño et al., 2016; Price et al., 2018; Burgueno and Abreu, 2020). The expression pattern of 5 TLRs (TLR2, TLR4, TLR5, TLR7, and TLR9) in the gut epithelium was established by experiments with the TLR-specific fluorescent reporter mice (Price et al., 2018). The expression levels of TLRs 6–9 and 11–13 were significantly low in both the small intestinal (SI) and the colonic IECs; the expression levels of TLRs 1, 2, 4, and 5 were moderate to low in the SI but significantly high in the colonic IECs; TLR3 expression levels were comparable in the SI and the colonic enterocytes (Price et al., 2018). IECs of the specific pathogen free (SPF) mice showed higher expression levels of most TLRs compared to those from the germ-free mice (Hörmann et al., 2014; Huhta et al., 2016; Fang et al., 2022). These data suggested that the expression levels of TLRs in the IECs were associated with the increased abundances of the commensal microbes.
TLR2 shows constitutive expression in the gastrointestinal epithelium and the enteric neurons, and mononuclear cells in the lamina propria even could release soluble forms of the TLR2 ectodomain, especially in patients with enteric inflammation (Aliprantis et al., 1999; Candia et al., 2012; Burgueño et al., 2016). The heterodimers of TLR1 or TLR6 with TLR2 (TLR1/TLR2 or TLR6/TLR2) on the plasma membrane recognized small-molecule agonists such as lipoteichoic acid (LTA) and lipopeptide (Cheng et al., 2015; Li and Wu, 2021). Pam3CSK4 is a synthetic tri-acylated lipopeptide that can activate TLR2. Dextran sulfate sodium (DSS)-induced colitis model mice pre-treated with Pam3CSK4 showed lower body weights and rectal bleeding (Horuluoglu et al., 2020). Pam3CSK4 stimulation of TLR2 effectively preserved the intestinal epithelial tight junction (TJ)-associated barrier assembly against stress-induced damage by inducing Phosphatidylinositol-3-kinase (PI3K)/Akt-mediated cell survival via MyD88 (Cario et al., 2007). TLR2-deficient mice showed severe chemotherapy-induced SI mucositis that was characterized by the accumulation of damaged DNA and infiltration of the CD11b+ myeloid cells in the proximal jejunum (Frank et al., 2015). Moreover, wild-type mice treated with antibiotics to deplete gut microbiota showed increased susceptibility to chemotoxicity, but the chemo-toxic effects were alleviated by pre-treatment with a TLR2 agonist (Frank et al., 2015). The abrogation of epithelial TLR2 signaling impaired mucosal repair because of reduced numbers of goblet cells (Podolsky et al., 2009; Walker et al., 2022). Diacyl phosphatidylethanolamine with two branched chains (a15:0-i15:0 PE) derived from Akkermansia muciniphila exhibited immunomodulatory selectivity and weak immunoregulatory effects through the non-canonical TLR2–TLR1 signaling pathway (Bae et al., 2022). Moreover, genetic ablation of TLR2 disrupted the structure and function of the enteric nervous system (ENS) in mice (Brun et al., 2013; Burgueño et al., 2016; Schill et al., 2022). The stimulation of colonic TLR2 signaling by gram-positive bacteria promoted colonic neurogenesis to maintain the nitrergic neurons in the longitudinal muscle myenteric plexus (Yarandi et al., 2020). These data demonstrated that sensing of microbial lipopeptides by TLR2 was critical for maintaining the integrity of the intestinal epithelium, ENS function, and immune tolerance to the bacteria.
TLR4 is a well-characterized PRR that is expressed on the cell membrane and the endosomes of the immune cells, enteric neurons, and the enterocytes (Neal et al., 2006). Activation of TLR4 on the cell surface by the extracellular LPS derived from the outer membrane of the Gram-negative bacteria mediates the downstream immune responses (Lu et al., 2022). For LPS cytosolic recognition, outer membrane vesicles produced by the Gram-negative bacteria could deliver LPS into the cytosol and trigger caspase 11-dependent effector responses (Shi et al., 2014; Vanaja et al., 2016). TLR4 expression is increased in the IECs during intestinal inflammation such as IBD; moreover, increased TLR4 signaling disables host protection against intestinal inflammation (Hausmann et al., 2002; Neal et al., 2006; Yang et al., 2017). A high-fat diet (HFD) causes gut microbiota dysfunction and exacerbates intestinal inflammation through the TLR4 signaling pathway (Kim et al., 2012). Conversely, in the murine acute colitis model, TLR4 deficiency caused severe mucosal damage, which was characterized by impaired epithelial proliferation and significant bacterial invasion (Fukata et al., 2005). Furthermore, TLR4 protected against intestinal inflammation by modulating the inhibitory immune responses associated with A. muciniphila (Liu et al., 2022b). TLR4 also regulated the gut microbiota by altering gastrointestinal motility to drive pathogen clearance, thereby regulating maintenance of the commensal microbiota, goblet cell differentiation, and production of the antimicrobial peptides (Vora et al., 2004; Sodhi et al., 2012). TLR4 demonstrates two-sided function during bowel inflammation which looks like to result from distinct TLR4 responses induced by different microbial species. TLR4 is expressed on the intestinal stem cells (ISCs) and induced stem cell apoptosis through innate immune signaling and therefore inhibits their proliferation (Neal et al., 2012; Afrazi et al., 2014; Hackam and Sodhi, 2022). However, hyperactivation of TLR4 triggered a neoplastic developmental program in the mouse models (Santaolalla et al., 2013). Furthermore, epithelial TLR4 deficiency increased the abundance of goblet cells and suppressed Notch signaling in the SI IECs (Sodhi et al., 2012; Hackam and Sodhi, 2022). LPS-triggered TLR4 signaling regulates the intestinal inflammatory responses and modulated the crypt stem cell functions, but the regulation differs under different circumstances. Similar to TLR2, TLR4 was also expressed in the gut neuronal cells and played a key role in the gut neuron cell signaling and development (Anitha et al., 2012; Kovler et al., 2021; Schill et al., 2022; Bódi et al., 2023).
TLR5 is localized on the cell surface and is highly expressed in the crypts of the small intestine and IECs of the colon; it is activated by bacterial flagellins (Gewirtz et al., 2001; Price et al., 2018). A screening study of flagellins in the human gut metagenomes that recognize and activate TLR5 resulted in the discovery of a class of silent flagellins, which were produced by the gut bacteria called Lachnospiraceae; silent flagellins induced weak TLR5 activation and innate immune tolerance (Clasen et al., 2023). X-linked inhibitor of apoptosis protein (XIAP)-mediated resilience of TLR5 signaling in the TLR5-expressing Paneth cells and the intestinal DCs enabled them to maintain tissue integrity and microbiota homeostasis during ileitis (Wahida et al., 2021). Null or epithelial-specific deletion of TLR5 impaired pathogen clearing and increased the susceptibility to microbial-induced colitis and metabolic syndrome (Vijay-Kumar et al., 2010; Letran et al., 2011; Chassaing et al., 2014). This suggested that dysfunction of the innate immune system promoted development of the metabolic syndrome. Mice with TLR5 deficiency showed increased body mass at 20 weeks of age. Magnetic resonance imaging (MRI) data demonstrated increased visceral and epididymal fat mass in the TLR5-deficient mice and correlated with increased serum triglycerides and high blood pressure. HFD-fed TLR5-deficient mice exhibited inflammatory infiltrates in the pancreatic islets and hepatic steatosis, but this phenotype was reversed by depleting the gut microbiota using broad-spectrum antibiotics. Moreover, germ-free mice transplanted with microbiota from the TLR5-deficient mice showed various symptoms of metabolic syndrome such as obesity, insulin resistance, hyperglycemia, and elevated levels of pro-inflammatory cytokines, which were characteristic features of the TLR5-deficient mice. These results suggested that the altered gut microbiota in the TLR5-deficient mice contributed to the development of the metabolic syndrome (Vijay-Kumar et al., 2010). In the small intestine, TLR5 expression was restricted to the Paneth cells and gradually decreased during the neonatal period (Price et al., 2018). TLR5 recognition of the flagellin from Roseburia intestinalis upregulated the TJ protein Occludin and restored the gut microbiota through elevated expression of IL-22 and REG3γ (Fulde et al., 2018; Seo et al., 2020). Moreover, TLR5 activation promoted early defense mechanisms against pathogen invasion of the host tissues by inducing the production of IL-17 and IL-22 on the mucosal surface (Frosali et al., 2015; Song et al., 2016). TLR5/NCLR-4-mediated synthesis of IL-22 and IL-18 during bacterial flagellin treatment was critical for the prevention and cure of rotavirus (RV) infection in mice (Zhang et al., 2014a). However, TLR5-deficient mice were resistant to Salmonella infection. The small intestine and colon tissues in the TLR5-deficient mice showed upregulated levels of the host defensive genes, antimicrobial peptides, and the serum and fecal Immunoglobulin A (IgA) (Uematsu et al., 2006; Vijay-Kumar et al., 2008a). TLR5 was also unexpectedly found to be co-expressed with the neurofilament-200 in the large-diameter A-fiber neurons of the dorsal root ganglion (DRG). Activation of TLR5 with its flagellin ligand induced TLR5-dependent blockade of the sodium currents, predominantly in the A-fiber neurons of the DRG and silenced the Aβ-fibers (Xu et al., 2015). Together, these studies demonstrated that TLR5 played a significant role in the intestinal pathogen defense and inflammation. TLR5 deficiency correlated with metabolic dysfunction. Further applications of the TLR5 ligand flagellins require in-depth investigations.
Other TLRs have also been reported to play significant roles in the host–microbiota interactions. Mice pre-treated with the TLR3 agonist poly(I:C) and the TLR7 agonist, imiquimod, were protected from DSS-induced colitis, whereas mice deficient in both TLR3 and TLR7 were more susceptible to DSS-induced colitis. A genome-wide association study (GWAS) showed that rs3775291 in TLR3 was significantly associated with susceptibility to both ulcerative colitis (UC) and Crohn’s disease (CD). This was consistent with the results from animal studies. Human subjects with combined TLR3 and TLR7 genetic variations showed severe UC, with a higher cumulative rate of hospitalization for patients with two combined genetic variants (rs3775291 in TLR3 and rs3853839 in TLR7) than those with or without a single variant (Yang et al., 2016). In the bat intestinal organoids, rapid and robust induction of the TLR3 response induced the bat cells to prevent virus propagation in the early phase of infection (Liu et al., 2022a). Several studies have reported that majority of the TLRs are localized to the basolateral membrane, whereas TLR3 and TLR9 are mostly located at the apical surface (Yu and Gao, 2015; Price et al., 2018). Viral infections from the basolateral side of the IECs elicit a stronger intrinsic immune response in comparison to the luminal apical infections; moreover, clathrin-sorting AP-1B mediated polarized sorting of the TLR3 towards the basolateral side of the IECs (Stanifer et al., 2020). In the colonocytes, basolateral TLR9 signaling induced IκBα degradation and activation of the nuclear factor kappa-B (NF-κB) pathway, whereas apical TLR9 stimulation invoked a unique response in which ubiquitinated IκB accumulated in the cytoplasm and prevented NF-κB activation (Lee et al., 2006). TLR9 also showed a protective function in the DSS colitis model; TLR9-deficient mice showed decreased expression levels of the intestinal repair genes such as hairy enhancer of split 1 and the vascular endothelial growth factor (Rose et al., 2012). TLR9 also induced intestinal inflammation against pathogens by sensing the bacterial and viral CpG DNA (Chen et al., 2020b; Zhong et al., 2022).
NLRs
NLRs are a group of cytosolic receptors that mediate caspase-1 activation, secretion of the pro-inflammatory cytokines such as IL-1β and IL-18, and inflammatory cell death in response to various cytoplasmic stimuli (de Zoete et al., 2014). NLRs consist of (i) a ligand-binding LRR domain similar to the TLRs, (ii) a central nucleotide-binding domain (NACHT) that is involved in oligomerization of the NLRs, (iii) a N-terminal interaction domain for signal transduction to activate downstream target proteins such as the caspases, and (iv) a caspase recruitment domain (CARD) or a pyrin domain (PYD) (Fritz et al., 2006; Kim et al., 2016; Eshleman and Alenghat, 2021). Recent studies have reported that NLRs play a critical role in maintaining intestinal homeostasis by responding to the intestinal microbes or their derivatives (de Zoete and Flavell, 2013; Guo et al., 2020).
Both nucleotide-binding oligomerization domain-containing 1 (NOD1) and NOD2 sense cytosolic bacterial peptidoglycan fragments. NOD1 senses d-glutamyl-meso-diaminopimelic acid (DAP)-containing peptidoglycan fragments, which are mainly found in the Gram-negative bacteria, whereas NOD2 responds to muramyl dipeptide (MDP), which is found in all the bacterial strains (Chamaillard et al., 2003; Girardin et al., 2003a, 2003b). The results of a forward genetic screening experiment to identify factors required for MDP detection showed that N-acetylglucosamine kinase (NAGK) was essential for the immunostimulatory activity of MDP and NAGK-phosphorylated MDP was an agonist for NOD2 (Stafford et al., 2022). NOD2 used the AP mitochondrial antiviral signaling protein (MAVS) after recognizing the viral single-stranded RNA (ssRNA) to activate IRF3 and induced IFN-β synthesis and secretion (Sabbah et al., 2009). Both NOD1 and NOD2 are expressed in the gut-resident DCs, macrophages, and the IECs; NOD1 is constitutively expressed in the IECs, whereas NOD2 expression is restricted to the Paneth cells in the small intestine (Lala et al., 2003; Eshleman and Alenghat, 2021). In mice, the recognition of Gram-negative bacteria by NOD1 in the epithelial cells was necessary and sufficient to induce genesis of the isolated lymphoid follicles (ILFs) (Bouskra et al., 2008). NOD1 ligands derived from the intestinal bacteria also acted as circulating signal molecules and directly modulated insulin trafficking in the pancreatic beta cells via sensing by NOD1 and its downstream adaptor, receptor-interacting-serine/threonine-protein kinase 2 (RIP2) (Zhang et al., 2019). NOD2 activation in the Paneth cells by the microbial peptidoglycans induced cytokine release, antimicrobial peptide production, autophagy, and epithelial cell regeneration (Nigro et al., 2014). NOD2-deficient mice inoculated with Helicobacter hepaticus induced ileal granulomatous inflammation, but this phenotype was reversed by the transgenic expression of α-defensins in the Paneth cells (Biswas et al., 2010). However, multiple studies reported contradictory results suggesting that NOD2 was not a direct regulator of the antimicrobial function of the Paneth cells. These studies demonstrated that the previously reported NOD2-dependent changes in the gut microbial composition could be overcome by environmental factors such as co-housing with the wild-type littermates (Petnicki-Ocwieja et al., 2009; Shanahan et al., 2014; Wilson et al., 2015b). NOD2 deficiency caused a pro-inflammatory environment because of dysbiosis and increased the risk of colitis and colitis-associated carcinogenesis, but this risk was reduced by transplanting normal microbiota (Couturier-Maillard et al., 2013). Moreover, the expression of NOD2 was dependent on the gut microbiota. In the germ-free mice, NOD2 expression was significantly reduced in the terminal ileum, but was re-induced by colonization with the commensal microbiota (Petnicki-Ocwieja et al., 2009). Furthermore, GWAS and functional screening studies showed that NOD2 polymorphisms were significantly associated with IBD susceptibility in humans (Hugot et al., 2001; Ogura et al., 2001; Tan et al., 2015; Ntunzwenimana et al., 2021). As a follow-up of these reports, several studies have investigated mechanisms underlying the induction of intestinal inflammation and dysbiosis by NOD2 in patients with CD. NOD2 promoted IgA transport through the human and mouse microfold cells (M cells) by downregulating the expression of two retrograde transport receptors, Dectin-1 and Siglec-5; moreover, secretory IgA (sIgA) transport in the NOD2-mutated CD patients significantly increased compared to the CD patients without NOD2 mutations or healthy subjects (Rochereau et al., 2021). NOD2 also induced microbiota dependent endoplasmic reticulum (ER)-stress-mediated regulation of mucus secretion in the goblet cells and the ER stress-mediated antimicrobial pathway in the macrophages (Ranjan et al., 2021; Naama et al., 2023). Conversely, NOD2 induced the early IL-33-dependent expansion of group 2 ILCs during CD pathogenesis (De Salvo et al., 2021). In patients with NOD2-driven CD, gp130 blockade rescued the activated inflammatory program in the NOD2-deficient cells and complemented anti-tumor necrosis factor (TNF) therapy (Nayar et al., 2021). This demonstrated that NOD2 was significantly associated with IBD by sensing bacterial MDP and modulated the antimicrobial cell function. NOD2 also played a key role in the metabolic syndrome. NOD2 in the IECs was necessary for the production of insulin-like growth factor-1 (IGF-1) mediated by a strain of Lactiplantibacillus plantarum (strain LpWJL) and also promoted postnatal growth in the malnourished animals (Schwarzer et al., 2023). This suggested that the bacteria cell wall components or purified NOD2 ligands (MDP and mifamurtide) may alleviate growth stunting.
NLR family, CARD domain-containing 4 (NLRC4) is an N-terminal CARD domain-containing NLR that discriminates commensal bacteria from the pathogens (Franchi et al., 2012; Nordlander et al., 2014). NLRC4 is highly expressed in the intestinal mononuclear phagocytes and the epithelial crypts. NLR family apoptosis inhibitory protein (NAIP)-NLRC4 inflammasomes coordinated the expulsion of IECs through the release of eicosanoids and IL-18 via activation of caspases-1 and -8 (Rauch et al., 2017). Pathogenic Gram-negative bacteria such as Salmonella and Pseudomonas promoted the release of NLRC4-dependent IL-1β by stimulating the intestinal mononuclear phagocytes. However, the release of IL-1β was not observed when the intestinal mononuclear phagocytes were stimulated by the commensal bacteria such as Bacteroides fragilis, Enterococcus faecalis and Lactobacillus (Franchi et al., 2012). NLRC4 responded to the invading pathogens by sensing flagellin and the bacterial Type III secretion systems (T3SS) needle protein PrgJ/CprI (Franchi et al., 2006; Miao et al., 2006, 2010; Molofsky et al., 2006; Kofoed and Vance, 2011; Zhao et al., 2011). In the cytosol, flagellin and PrgJ/CprI interacted with NLRC4 to initiate oligomerization by binding to the APs NAIP2 and NAIP5, respectively, and activated the inflammasomes (Conforti-Andreoni et al., 2011; Kofoed and Vance, 2011; Zhao et al., 2011). Flagellin also promoted immediate elimination of the RV-infected cells via induction of the NLRC4-dependent IL-18 (Zhang et al., 2014a, 2020). NLRC4 protected against DSS-induced experimental colitis and azoxymethane/dextran sodium sulfate (AOM/DSS)-induced tumorigenesis by regulating epithelial cell proliferation and apoptosis during injury (Hu et al., 2010; Nordlander et al., 2014). In mice challenged with Burkholderia thailandensis and Salmonella typhimurium, NLRC4 was activated by a specific constituent of the murine intestinal microbiota, Escherichia coli O21:H+; moreover, challenge induced translocation of E. coli O21:H+ to the white adipose tissue (WAT) sustained systemic insulin and IGF-1 signaling, thereby promoting disease tolerance and protection against wasting (Schieber et al., 2015). In summary, the sensing of flagellin and T3SS rod proteins from the pathogenic bacteria or the commensal bacteria by NLRC4 initiates immunogenic or immunotolerant responses.
NLR family, pyrin domain-containing 3 (NLRP3) is one of the best characterized NLRs to date. NLRP3 senses numerous ligands, including ATP, pore-forming toxins such as nigericin and viroporins, uric acid crystals, amyloid-β (Aβ) depositions, asbestos, silica particles, and nucleic acids (Yang et al., 2019; Coll et al., 2022; Harapas et al., 2022). Activated NLRP3 oligomerizes, binds and activates the ASC complex, and reactivates the effector complex composed of CARD and caspase-1. This NLRP3–ASC–pro-Caspase-1 complex is denoted as the NLRP3 inflammasome, which activates critical pro-inflammatory factors (Yang et al., 2019; Li and Wu, 2021). In select patients with active IBD, unfermented dietary β-fructan fibers such as oligofructose (FOS) ~8 sugars induced the pro-inflammatory cytokines by activating NLRP3- and TLR2-mediated signaling pathways (Singh et al., 2019b; Armstrong et al., 2023). In vivo mouse studies have demonstrated that NLRP3 is a key player in maintaining the mucosal barrier. For example, NLRP3-deficient mice were highly susceptible to DSS-induced colitis because of impaired epithelial integrity in the colon (Zaki et al., 2010; Hirota et al., 2011). Several studies demonstrated that hyperactivation of the NLRP3 inflammasome adversely impacted gut epithelial cell proliferation and intestine–blood barrier integrity (Zaki et al., 2010; Huang et al., 2019; Cui et al., 2020). However, lower levels of pro-inflammatory cytokines in the colon tissues of the NLRP3-deficient mice suggested that decreased NLRP3 inflammasome activation is required for amelioration of acute and chronic intestinal inflammation (Bauer et al., 2010; Wang et al., 2018b; Mehto et al., 2019; Sanchez-Lopez et al., 2019). These contrasting results may be due to differences in the experimental approaches between different studies. However, NLRP3 is closely associated with IBD. Several GWAS studies have identified single nucleotide polymorphisms (SNPs) in the NLRP3 (rs35829419, rs10733113, rs4925648, and rs10925019), which are associated with CD (Schoultz et al., 2009; Villani et al., 2009; Cummings et al., 2010; Roberts et al., 2010). Furthermore, mice carrying the Nlrp3R258W mutation maintained gut homeostasis in the cryopyrin-associated periodic syndrome (CAPS) model (Yao et al., 2017). CAPS disease-associated mutations are mostly located in the NACHT domain, but few mutations are located in the LRR domain of NLRP3 (Touitou et al., 2004; Booshehri and Hoffman, 2019; Zhou et al., 2021).
NLRP6 also plays a significant role in the intestine and is predominantly expressed in the enterocytes and the goblet cells (Chen et al., 2011; Elinav et al., 2011; Wlodarska et al., 2014). NLRP6 sensed viral RNA, microbiota-associated metabolites, bacteria cell wall component LTA, and LPS (Levy et al., 2015; Wang et al., 2015a; Hara et al., 2018; Leng et al., 2020; Shen et al., 2021). NLRP6 can also form an inflammasome (Li et al., 2022a). Microbiota-related metabolites such as taurine, histamine, and spermine stimulated the formation of an ASC-dependent NLRP6 inflammasome, which induced synthesis and secretion of the downstream pro-inflammatory cytokines (Levy et al., 2015). Stimulation of the NLRP6 inflammasome by the SCFAs protected against intestinal epithelial barrier impairment and abrogated high-fructose diet-induced hippocampal neuroinflammation and neuronal loss (Li et al., 2019a). The activation of NLRP6 resulted in the formation of specks in the cells (Ghimire et al., 2018; Hara et al., 2018). Interaction with the NLRP6 agonists and double-stranded RNA (dsRNA) caused NLRP6 to undergo liquid–liquid phase separation (LLPS), an essential step for recruiting other components of the inflammasome (Shen et al., 2021). The intrinsically disordered poly-lysine sequence (K350-354) of NLRP6 was required for the formation of the NLRP6 puncta upon interaction with the dsRNA (Shen et al., 2021). And further investigation is still needed to solve problems like how is dsRNA/NLRP6 LLPS physiologically or pathologically regulated in cells (Li et al., 2022a). NLRP6 plays a crucial role in regulating inflammation and host defense against microorganisms in the intestine. NLRP6 deficiency in the mouse colonic epithelial cells decreased the production of IL-18 and altered the composition of the fecal microbiota, with increased representation of Prevotellaceae and TM7. NLRP6-deficient mice showed spontaneous intestinal hyperplasia, inflammatory cell recruitment, and exacerbated DSS-induced colitis. Further antibiotic treatment and electron microscopy studies revealed the role of Prevotellaceae in the microbiota-associated colitis disease (Elinav et al., 2011). De-ubiquitination of the NLRP6 inflammasome by CYLD lysine 63 deubiquitinase (CYLD) decreased IL-18 levels in the colonic mucosa and regulated intestinal inflammation (Mukherjee et al., 2020). However, contradictory reports have shown that both wild-type and Nlrp6−/− littermate mice display comparable sensitivity to DSS-induced colitis, (Lemire et al., 2017; Mamantopoulos et al., 2017). This suggested that NLRP6 did not influence the composition of the commensal gut microbiota. The host NLRP6 also aggravated the gastrointestinal graft-versus-host disease (GVHD) independent of the gut microbial composition (Toubai et al., 2019). Furthermore, NLRP6-deficient goblet cells showed defective mucus secretion into the lumen of the large intestine because of aberrant autophagy and inflammasome activation (Wlodarska et al., 2014; Birchenough et al., 2016). However, another study showed that the basal inner mucus layer formation and function was independent of the inflammasome activity and was determined by the gut microbiota (Volk et al., 2019). NLRP6 plays a vital role in the defense mechanisms against viral infection in the intestine. Systemic challenge of NLRP6-deficient and control mice with the encephalomyocarditis virus (ECMV; transmitted through the oral-fecal route) showed similar mortality rates but the viral loads in the gastrointestinal tract of the NLRP6-deficient mice were higher compared to the control mice. Moreover, NLRP6-deficient mice showed higher mortality and viremia compared to the control mice when challenged by ECMV or murine norovirus (MNV) through oral administration. Mechanistically, NLRP6 binds to the viral RNA through DEAH-box helicase 15 [DHX15, an RNA helicase, and subsequently interacted with MAVS to induce the synthesis of type I/III IFNs and IFN-stimulated genes (ISGs) (Wang et al., 2015a)]. NLRP6 also modulated the death program in the intrinsic enteric-associated neurons, including the microbe-responsive subset of viscerofugal CART+ (cocaine- and amphetamine-regulated transcript) neurons, which are enriched in the ileum and the colon (Matheis et al., 2020; Muller et al., 2020). Microbiota depletion caused impaired glucose regulation because of NLRP6- and caspase 11-dependent loss of the CART+ neurons (Muller et al., 2020). In summary, NLRP6 is activated by the bacteria and the viruses. It functions in the enterocytes, goblet cells, and the ENS by regulating pathogen defense, intestine inflammation, and neuronal death.
Like NLRP6, NLRP9b is also specifically expressed in the IECs and is associated with protection against rotavirus infection. Rotavirus is another type of enteric virus with dsRNA that directly infects the IECs and causes severe diarrheal illness. In contrast to NLRP6, NLRP9b sensed the short double-stranded RNA generated during replication of the rotavirus dsRNA outside the viroplasm through DHX9 and activated the inflammasome, which in turn induced IL-18 secretion and pyroptosis to restrict rotavirus replication (Zhu et al., 2017b). NLRP9b and NLRP6 recognize different viral RNA patterns. Therefore, the intestinal regions in which NLRP9b is expressed are different from those expressing NLRP6. Therefore, these two IEC-specific NLRs cooperatively protected the host against enteric viral infections (Li and Zhu, 2020).
NLRP10 is a member of the NLR family that lacks pathogen sensing ability because it does not contain a putative LRR domain (Wang et al., 2004). NLRP10 function was elusive for more than a decade. NLRP10 was considered as an anti-inflammatory NLR that inhibited ASC-caspase 1 activation (Imamura et al., 2010). A recent study showed that NLRP10 monitored mitochondrial integrity in an mtDNA-independent manner and assembled the inflammasome in the differentiated human keratinocytes (Próchnicki et al., 2023). A recent study showed that NLRP10 played a protective role in intestinal inflammation (Zheng et al., 2023). In the intestine, NLRP10 was expressed in the distal colonic IECs. NLRP10 mRNA levels were higher in the GF mice than in the SPF mice. This suggested that the microbiome regulated NLRP10 expression. Furthermore, NLRP10 forms an ASC-dependent inflammasome. Whole-body or conditional IEC depletion of NLRP10 reduced caspase-1 activation in the IECs and increased the susceptibility to DSS-induced colitis. In conclusion, NLRP10 is a potential target for treating autoinflammatory diseases. However, the regulation of NLRP10 expression by the microbiota requires further investigation.
NLRP12 is expressed in the immune cells. However, the ligands of NLRP12 are unknown. NLRP6 and NLRP12 demonstrate dual functions. They can function both as an inflammasome and as a negative regulator of immune signaling (Tuladhar and Kanneganti, 2020). NLRP12-deficient mice are hyper-resistant to infection by S. typhimurium (Zaki et al., 2014). The expression of NLRP12 was lower in patients with UC compared to the other patient cohorts. Furthermore, mice with NLRP12 deficiency showed enhanced DSS-induced colitis and AOM/DSS-induced colon cancer due to dysbiosis and increased NF-κB signaling (Zaki et al., 2011; Allen et al., 2012; Chen et al., 2017). NLRP12 also played a role in HFD-induced obesity. HFD-fed Nlrp12−/− mice showed increased weight, adipose deposition, and blood glucose levels compared to the wild-type mice; dysbiosis in the HFD-fed Nlrp12−/− was marked by increased obesity-associated Erysipelotrichaceae and reduced Lachnospiraceae family and enzymes required for SCFA biosynthesis (Truax et al., 2018).
NLRC3 is a member of the NLR family that is significantly downregulated in the colorectal cancer tissues (Liu et al., 2015b). NLRC3 is an inhibitory nucleic acid sensor that is activated by the TLRs that attenuates type I IFN response by sequestering and inhibiting the stimulator of IFN genes (STING) protein (Schneider et al., 2012; Zhang et al., 2014b; Li et al., 2019c). NLRC3-deficient mice were highly susceptible to DSS-induced colitis and AOM/DSS-induced colorectal tumorigenesis. Mechanistically, NLRC3 inhibited activation of the mechanistic target of rapamycin kinase (mTOR) signaling pathway by blocking the activation of PI3K-dependent, which was triggered by engagement of the growth factor receptors or TLR4 (Karki et al., 2016).
RLRs
RIG-I, melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) are three major proteins belonging to the RIG-I-like receptor family. RIG-1 and MDA5 are PRRs with a RNA helicase domain and a CARD domain, whereas LGP2 contains only the RNA helicase domain and does not have a CARD domain (Moon and Stappenbeck, 2012; Brisse and Ly, 2019). RLRs are involved in viral infection. RIG-I binds to the 5ʹ-triphosphate RNA, whereas MDA5 recognizes long dsRNA structures. The recognition of viral RNA by the RLRs activated of the MAVS-dependent signaling pathway that results in the production of type I IFNs and ISGs (Li and Wu, 2021). LGP2 assisted MDA5–RNA interactions and enhanced MDA5-mediated antiviral signaling. LGP2 increased the initial rate of MDA5–RNA interactions and regulated MDA5 filament assembly, including the formation of numerous shorter MDA5 filaments with equivalent or higher activity than the longer filaments with only MDA5 (Bruns et al., 2014).
Both RIG-I and MDA5 are involved in the defense mechanisms against rotavirus infections in the intestine (Broquet et al., 2011). RIG-I and MDA5 are both upregulated in the IECs during rotavirus infection. Silencing of RIG-I, MDA5 or MAVS significantly decreased IFN-β production and increased rotavirus titers in the infected IECs. Rotavirus-infected mice with MAVS deficiency showed lower IFN-β levels, increased rotavirus titers in the IECs, and increased viral shedding in the feces. These results collectively demonstrated that the RIG-I/MDA5/MAVS signaling pathway played a significant role in protecting against viral infection in the intestine (Broquet et al., 2011). RIG-I in the antigen presenting cells (APCs) recognized the commensal viruses and maintained intestinal intraepithelial lymphocytes (IELs) through a type I IFN-independent, but MAVS/IRF1/IL-15 axis-dependent manner (Liu et al., 2019). Enteric bacteria and fungi are mostly linked to the IBD phenotypes. However, a recent study showed that perturbations in the intestinal virome or altered ability of RIG-I and MDA5 to sense the virome contributed to the induction of IBD (Adiliaghdam et al., 2022). IECs with the loss-of-function MDA5 mutations (rs35744605, E627X) were associated with detrimental consequences for patients with IBD when exposed to viromes (Adiliaghdam et al., 2022). MAVS was downstream of RLRs and played a pivotal role in monitoring the commensal bacteria and preventing DSS-induced colitis by inducing multiple cytokines and antimicrobial peptides, including IFN-β and REG3γ (Li et al., 2011a). During acute intestinal tissue injury in mice, activation of RIG-I/MAVS or STING pathways induced IFN-I signaling and maintained integrity of the gut epithelial barrier and reduced the severity of GVHD (Fischer et al., 2017). Therefore, functional role of the RLR family members includes sensing the viral RNA, regulating the protective response against enteric virus infections, and maintaining intestinal homeostasis by sensing the commensal viruses.
Other PRRs
Several other PRRs are also involved in modulating the host–microbiota interactions in the intestine. AIM2 is a cytosolic DNA sensor belonging to the ALR family. AIM2-deficient mice are highly susceptible to DSS-induced colitis and have a high colonic burden of E. coli, a commensal bacterium. The colonization of germ-free mice with microbiota from the AIM2-deficient mice caused higher susceptibility to colitis compared to the colonization of germ-free mice with microbiota from the wild-type mice. AIM2-deficient mice showed reduced production of IL-1β and IL-18, but infusion of IL-18 alleviated the higher colonic burden of E. coli and susceptibility to colitis (Hu et al., 2015). AIM2 sensed radiation-induced DNA damage in the nucleus and induced activation of the inflammasome and triggered cell death. This suggested that AIM2 was a promising therapeutic target for reducing the damage caused by exposure to ionizing radiations to tissues with actively proliferating cells such as the bone marrow and the gastrointestinal tract (Hu et al., 2016). Chemotherapeutic agent irinotecan (CPT-11) induced massive release of double-strand DNA from the intestine through exosome secretion. This dsDNA entered the cytosol of innate immune cells and activated the AIM2 inflammasome, which subsequently induced the synthesis and secretion of IL-1β and IL-18, both of which caused intestinal mucositis and late-onset diarrhea. The abrogation of AIM2 signaling, either in the AIM2-deficient mice or by a pharmacological inhibitor such as thalidomide, significantly reduced the incidence of drug-induced diarrhea without affecting the anti-cancer efficacy of CPT-11 (Lian et al., 2017). The expression of AIM2 is reduced in several cancer types. AIM2 suppressed colon tumorigenesis by interacting with the DNA-dependent protein kinase (Man et al., 2015; Wilson et al., 2015a). This function of AIM2 was independent of the inflammasome activation and IL-1β release. In the AOM/DSS and APCMin models of CRC, Aim2-deficient mice showed higher tumor load than the control mice (Man et al., 2015; Wilson et al., 2015a). Therefore, AIM2 is a DNA sensor that responds to intestine damage and participates in colitis, pathogen invasion, and bowel cancer.
The cGAS-cGAMP-STING pathway is another cytosolic DNA-sensing pathway. In the presence of DNA, cGAS uses GTP and ATP to synthesize cyclic-di-GMP/AMP (cGAMP), which activated IFN signaling by binding with high affinity to STING (Brubaker et al., 2015). Alterations in the cGAS-STING DNA-sensing pathway adversely affected intestinal homeostasis but the underlying mechanisms are not clear. K63-linked ubiquitination of STING in the intestinal myeloid cells is elicited by the bacterial products including cGMP and the corresponding positive feedback loop drives intestinal inflammation (Shmuel-Galia et al., 2021). However, cGAS/STING-dependent IFN-β response triggered intestinal regeneration and recovery from radiation injury in the animal models (Leibowitz et al., 2021). In the colon cancer models under Lactobacillus rhamnosus GG therapy, the cGAS/STING-dependent type I IFN pathway improved the response to immune checkpoint blockade therapy (Si et al., 2022).
Dectin-1 is a member belonging to the CLR family that is expressed in the human IECs and human IEC lines, HT29 and SW480. Dectin-1 recognizes and responds to the fungal cell wall component β-glucan, and promotes chemokine secretion through phosphorylation of its downstream mediator Syk (Chan et al., 2009). Furthermore, β-glucan-induced chemokine secretion was not affected by the inhibition of v-raf-1 murine leukemia viral oncogene homolog 1 (RAF-1), which is involved in an alternate Dectin-1 responsive pathway. Therefore, it is critical to determine if Dectin-1 pathway in the IECs involves other downstream mediators such as CARD9/Bcl10/MALT1 (Cohen-Kedar et al., 2014). Mice lacking Dectin-1 exhibited increased susceptibility to chemically-induced colitis because of altered responses to indigenous fungi; Dectin-1 gene polymorphism (CLEC7A) in humans is significantly associated with a severe form of UC (Iliev et al., 2012). Conversely, inhibition of Dectin-1 signaling ameliorated colitis by inducing Lactobacillus-mediated expansion of Treg cells in the intestine (Tang et al., 2015b). A recent study showed that deletion of both the Dectin-1 and Dectin-2 receptors significantly modified the bacterial microbiota but did not affect the fungal microbiota; moreover, these changes induced protective effects during colitis through members of the Lachnospiraceae family (Wang et al., 2022a). These conflicting results may be caused by the complex and variable microbiota populations and the gut environment. Specific intracellular adhesion molecule-3 grabbing non-integrin homolog-related 3 (SIGNR3), a mouse homolog of human DC-SIGN, belongs to the CLR family and recognizes fungi in the intestine (Eriksson et al., 2013). SIGNR3-deficient mice exhibited increased weight loss because of severe colitis symptoms compared to the wild-type littermates in a DSS-induced colitis model; SIGNR3-deficient mice showed increased inflammation in the colon with higher levels of TNF-α. Binding of the surface layer protein A derived from Lactobacillus to SIGNR3 exerted regulatory signals that mitigated colitis (Lightfoot et al., 2015).
Metabolite-sensing receptors in the intestine
GPCRs
Intestinal GPCRs sense metabolites including SCFAs, medium-chain fatty acids (MCFAs), long-chain fatty acids (LCFAs), and others. SCFAs such as acetate, propionate, and butyrate are produced by the microbial fermentation of the dietary fiber and are the main energy sources for the colonocytes (Kaiko et al., 2016). SCFAs exert diverse effects on the functions of the mucosal immune cells and the IECs (Willemsen et al., 2003; Kaiko et al., 2016; Park et al., 2016; Zhao et al., 2018; Lavelle and Sokol, 2020). GPR43, GPR41, and GPR109a are the three known receptors for the SCFAs and are expressed on the IECs and the immune cells. SCFAs significantly increased the expression levels of GPR43 and GPR41 in the adipose tissue and reduced their expression in the colon (Lu et al., 2016). GPR43-deficient mice showed exacerbated or unresolved inflammation in the colitis models, thereby showing that stimulation of GPR43 by the SCFAs was necessary for the normal resolution of inflammatory responses under specific conditions (Maslowski et al., 2009). Furthermore, SCFA induced RegIIIγ and β-defensins in the intestinal epithelial enteroids from the wild-type mice but this was suppressed in the GPR43−/− mice. This demonstrated that SCFAs promoted the synthesis of antimicrobial peptides in the IECs in a GPR43-dependent manner (Zhao et al., 2018). GPR109a, encoded by the Niacr1 gene, is a receptor for butyrate and niacin, which are produced by the gut microbiota. GPR109a signaling promotes anti-inflammatory activity by inducing the differentiation of regulatory T cells (Tregs) and IL-10 production. Moreover, activation of GPR109a signaling by butyrate promoted IL-18 secretion in the colonic epithelium. Niacr1-deficient mice were highly susceptible to the development of colitis and colorectal tumorigenesis (Thangaraju et al., 2009; Singh et al., 2014). GPR109a also protected against colitis and facilitated dietary fiber-induced gut homeostasis by regulating the inflammasomes (Macia et al., 2015). GPR84 sensed MCFAs, which were derived from milk, especially coconut milk. GPR40 was also a sensor of MCFAs. GPR40 and GPR120 are involved in a spectrum of metabolic diseases and are sensors of LCFAs such as omega-3 fatty acids, which are derived mainly from fish (Tan et al., 2017; Mao et al., 2023). GPR35 can sense and bind products from tryptophan (Trp) and tyrosine catabolism. GPR91 binds succinate, an intermediate of the citric acid cycle. GPR119 is activated by three N-acylethanolamines (oleoylethanolamine, palmitoleoylethanolamine, and linoleylethanolamine) and 2-oleoylglycerol. Furthermore, linoleylethanolamine and 2-oleoylglycerol served as physiologically relevant endogenous GPR119 agonists that mediated receptor activation upon nutrient uptake (Syed et al., 2012). Lipid sensing in the enteroendocrine cells of the distal intestine by the fat receptor GPR119 regulates gut hormone levels, food intake, and body weight (Higuchi et al., 2020). Therefore, GPR119 plays a key role in metabolic functions. The G protein-coupled bile acid receptor (GP-BAR1, also named as TGR5) recognized secondary bile acids and was highly expressed in the ileum and the colon (Li and Chiang, 2014). Morphology of the colonic mucous cells and molecular architecture of the epithelial TJs were altered in the TGR5−/− mice, thereby causing increased intestinal permeability and susceptibility to develop severe colitis in response to DSS (Cipriani et al., 2011). GPCRs consist of multiple orphan receptors for several ligands and have diverse functions. Therefore, further in-depth investigations are necessary in this area of research. Cohen and colleagues used bioinformatics and synthetic biology to mine the human microbiota and identify GPCR-interacting N-acyl amides. They reported that GPCRs targeted by the human microbial N-acyl amides were localized to the gastrointestinal tract and the associated immune cells (Cohen et al., 2015, 2017). In the mouse models, GPCRs were associated with diverse mucosal functions, including metabolism (GPR119, GPR120), immune cell differentiation (S1PR4, PTGIR, and PTGER4), immune cell trafficking (S1PR4, G2A) and tissue repair (PTGIR) (Cohen et al., 2017). Vmn2r26 is an olfactory receptor (a type of chemosensory GPCR) that is highly expressed on the Tuft-2 cells. It stimulated prostaglandin D2 (PGD2) production upon recognizing the Shigella metabolite N-undecanoylglycine (N-C11-G) and enhanced mucus secretion by the goblet cells in response to bacterial infections (Xiong et al., 2022). GPCRs also play a role in human UC. Inflamed UC regions are distinguished by the MAS related GPR family member X2 (MRGPRX2)-mediated activation of mast cells and decreased activation is observed in a UC-protective genetic variant of MRGPRX2 (Asn62Ser) (Chen et al., 2021b). Although several studies have characterized a whole plethora of GPCRs, the precise roles of each of these receptors are still evolving, especially in the immune responses (Tan et al., 2017).
AHRs
AHRs are ligand-inducible transcription factors that are expressed in the immune cells, epithelial cells, and few types of tumor cells. AHRs maintain homeostasis at the mucosal surfaces by recognizing xenobiotics, natural compounds such as Trp metabolites, and dietary components such as serotonin, melatonin, and vitamin B3 (Schiering et al., 2017). AhR expression was suppressed upon depletion of intestinal microbiota in the animal models and the intestinal tissue of patients with IBD, thereby suggesting a relationship between AhR expression and the gut microbiota (Monteleone et al., 2011; Wang et al., 2018a). In mice, deficiency of AhR in the IECs exacerbated inflammation in the DSS colitis model, but deletion of AhR in the T cells attenuated colitis and was manifested by the infiltration of Th17 cells into the lamina propria (Chinen et al., 2015). A recent study showed that mice with AHR repressor deficiency in the IELs were susceptible to infection with Clostridium difficile and DSS-induced colitis because excessive AHR signaling lead to oxidative stress and ferroptosis of the IELs and suppression of the intestinal immune responses (Panda et al., 2023). Highly adaptive lactobacilli produce indole-3-aldehyde, a Trp-indole derivative. Indole-3-aldehyde acted as an AHR ligand and contributed to AHR-dependent IL-22 transcription (Zelante et al., 2013; Agus et al., 2018). IL-22-dependent mucosal response resisted colonization of the fungus Candida albicans, protected the mucosa from inflammation, and was required for the survival of mixed microbial communities. Therefore, the microbiota–AHR axis represented an important strategy developed through co-evolutionary commensalism for fine-tuning host mucosal reactivity in accordance with Trp catabolism (Zelante et al., 2013; Stockinger et al., 2021). AHR signaling was also linked to intestinal inflammation through the downstream NF-κB-C/EBPβ signaling axis in the T cells and the DCs (Sanmarco et al., 2022). AHR signaling protected the stem cell niche and maintained the integrity of the intestinal barrier (Metidji et al., 2018). Besides, AHR played a key role in the regulation of IL-10R1 expression in the colon (Lanis et al., 2017). Dietary and microbiota-derived oxazole suppressed the production of IL-10 by the IECs and inflammation by activating AhR in the intestinal epithelium (Iyer et al., 2018). AhR expression in the gut also regulated the function and maintenance of the ILCs. Genetic or pharmacological activation of AhR suppressed ILC2 function but enhanced ILC3 maintenance to protect the host from Citrobacter rodentium infection (Li et al., 2018). Furthermore, AHR signaling in the enteric neurons acted as a regulatory node to maintain gut homeostasis and health; neuron-specific deletion of AhR reduced peristaltic activity of the colon; expression of AhR in the enteric neurons of mice partially restored intestinal motility when treated with antibiotics (Obata et al., 2020). In summary, AhR was expressed widely in the gut and sensed several metabolites, especially derivatives of Trp to regulate the function of T cells, IECs, and ILCs, and maintain gut homeostasis.
Nuclear receptors
Nuclear receptors are a group of ligand-activated transcription factors that play important roles in embryogenesis, development, and metabolism (Li and Chiang, 2014). Farnesoid X receptor (FXR), Pregnane X receptor (PXR), and Vitamin D receptor (VDR) are three nuclear receptors that directly bind to the bile acids, which are small molecules with a steroidal structure and are synthesized from cholesterol by the liver hepatocytes. Primary bile acids are secreted into the small intestine, reabsorbed in the terminal ileum, transported back to the liver via portal circulation, and further metabolized by the gut microbiota to generate secondary bile acids. Bile acids facilitate intestinal digestion and absorption of dietary fat, steroids, drugs, and lipophilic vitamins. FXR and PXR are highly expressed in the tissues exposed to the bile acids such as the liver and the intestine, whereas VDR is widely expressed in most tissues (Li and Chiang, 2014). Both primary and secondary bile acids activated FXR signaling, which regulates bile acid synthesis, metabolism, and intake by the host. Mice lacking FXR showed increased levels of bacteria in the ileum and compromised integrity of the epithelial barrier (Inagaki et al., 2006). FXR activation inhibited inflammation, promoted the synthesis of cathelicidin, an antimicrobial peptide (D’Aldebert et al., 2009), and protected the integrity of the intestinal barrier in IBD (Gadaleta et al., 2011; Eshleman and Alenghat, 2021; Chen et al., 2022). The extracts of Citrus aurantium L. exerted protective effects by modulating the FXR/fibroblast growth factor 15 (FGF15) pathway and the FXR-targeted proteins and restored the composition of the intestinal microbiota, reshaped the barrier integrity, and maintained homeostasis of the bile acids (Liu et al., 2020). Ursodeoxycholic acid (UDCA) is an off-patent drug that reduced ACE2 levels by inhibiting FXR signaling in the lungs, cholangiocytes, and intestinal organoids of humans, mice, and hamsters, thereby suggesting that FXR inhibition may protect against SARS-CoV-2 infection (Brevini et al., 2023a). In the pluripotent stem cell (PSC)-derived hepatocyte like cells (HLCs), which demonstrated a liver-intestine hybrid state, combined FXR expression plus agonist exposure enhanced the expression of hepatocyte-associated genes and increased the ability of bile canalicular secretion as well as the lipid droplet formation (Nell et al., 2022). These data demonstrate that FXR signaling played a significant role in the metabolic, gastrointestinal, and liver diseases, but the exact details require further investigation (Sun et al., 2021). Several studies have shown that vitamin D-VDR signaling promotes innate immune responses (Ismailova and White, 2022). The downstream VDR target genes were upregulated in the colon tissues of the HFD-fed mice; moreover, combinatorial treatment of the intestinal HT29 epithelial cells with lipids and bile acids, and modulation of the vitamin D targeting pathways protected against colitis and colitis-associated cancer risk (O’Mahony et al., 2023).
These data showed that PRRs and other metabolite receptors played pivotal roles in maintaining gut homeostasis by sensing the intestinal microbes and the dietary and microbiota-derived metabolites.
In summary, the microbial sensing receptors influence intestinal homeostasis through several mechanisms. Firstly, the PRRs sense foreign pathogens and activate downstream signaling mechanisms to promote synthesis of the pro-inflammatory cytokines, which eventually protect against pathogen evasion and maintain the gut homeostasis. Secondly, the highly mutualistic relationship between the host and the gut microbiota is partly mediated through PRR signaling. Thirdly, microbiota-derived metabolites regulated the differentiation and functions of host intestinal cells through metabolite sensor signaling pathways. Although significant data has been generated to understand the interactions between the sensors and the microbiota, several questions remain that require further investigations.
Intestinal cells in the host–microbiota interactions
Host immune cells in the intestine maintain intestinal homeostasis by sensing the microbial signals. The intestinal immune cells exert an “inside-out” control to determine the microbiota localization and composition of the microbial community. At the same time, microbiota exerts an “outside-in” effect on the intestinal immune cell activity and development. In this section, we will discuss interactions between the gut microbiota and the immune cells that govern innate or adaptive immune responses (Fig. 2).
Figure 2.
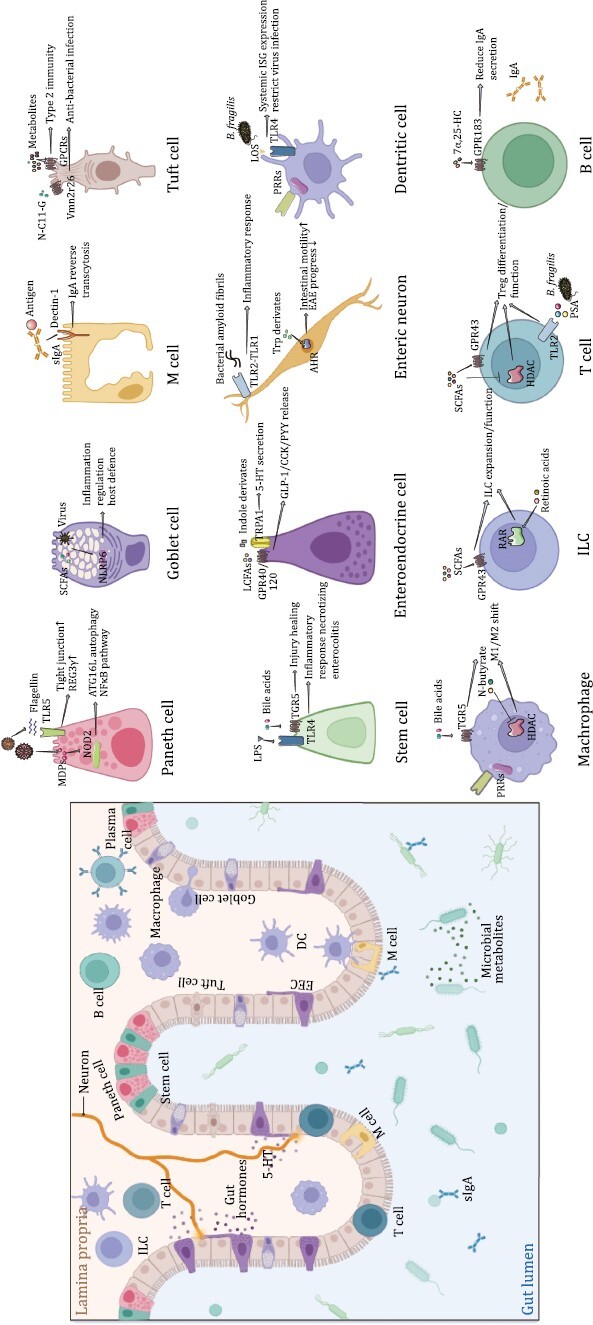
Ligands and functions of microbial sensors in different cell types in the intestine. The diagram on the left depicts the distribution of different intestinal epithelial cells (IECs) and immune cell types in the intestine, and the diagram on the right elaborates several typical signaling pathways through which specific cell types from the intestine take function. Paneth cell expresses NOD2 and TLR5 respectively to respond to bacterial MDP and flagellin. Genetically, Nod2 is highly associated with inflammatory bowel disease (IBD). NLRP6 is important for goblet cell to prime inflammatory responses, defend virus infection and mucus secretion. Dectin-1 is required for the reverse endocytosis of the secretory IgA (sIgA)–antigen complex in M cells. Tuft cells would express lots of GPCRs to recognize different metabolites, such as Tuft-2 cells express orphan receptor Vmn2r26 to recognize metabolite N-undecanoylglycine (N-C11-G). LPS–TLR4 interaction help intestinal stem cells to induce inflammatory responses and bile acids–TGR5 interaction is important for stem cell proliferation to proceed wound healing. Enteroendocrine cell (EEC) could also be considered as nervous cell and is important in producing gut hormones. TRPA1 and GPR40/120 expressed in EECs could be activated by corresponding ligands and induce 5-Hydroxytryptamine (5-HT) or glucagon-like peptide-1 (GLP-1)/cholecystokinin (CCK)/peptide tyrosine tyrosine (PYY) release. Recognition of microbial signals occurring on enteric neurons is also important for host health. Bacteria can release amyloid protein to stimulate TLR2–TLR1 dimers on neurons and induce neuroinflammation. Meanwhile, tryptophan derivatives released by bacteria can also activate AHR in the nucleus of neurons and increase intestinal motility. As classic antigen presenting cells, myeloid cells including dendritic cells (DCs) and macrophages play an important role in host immune response and tolerance formation. They both express a lot of PRRs for microbial recognition. Symbiotic microorganisms such as Bacteroides fragilis regulate natural resistance to viral infection by inducing IFN-I via intestinal DCs through its outer membrane component lipooligosaccharide (LOS) associated polysaccharide A (PSA). TGR5 signaling and HDAC activation regulated by microbial metabolites could modulate macrophage M1/M2 polarization shift. ILC3s expressed GPR43 in colon to sense SCFAs, and take retinoic acid (RA)-RAR signaling in SI to promote cell expansion. Lymphocytes have always been the important members of the intestinal immune system. For Treg cells, SCFAs signal through GPR43 or HDAC are both important to maintain Treg population and function. Furthermore, PSA signals from B. fragilis directly activate TLR2 on Treg cells to promote induced Treg (iTreg) cell differentiation. With the participation of MyD88-dependent intestinal flora recognition, IECs absorb dietary cholesterol to produce 7α,25-dihydroxycholesterol (7α,25-HC). 7α,25-HC can be then sensed by plasma B cells via chemotactic receptor GPR183 and finally inhibits antigen-specific IgA secretion.
Classical and non-classical innate immune cells
Normal functioning of the innate immune system is required to maintain healthy gut microbiota. Dysbiosis causes inflammation because innate immune cells are aberrantly activated. The innate immune cells are the first defense barrier against the foreign agents. Therefore, they maintain gut homeostasis by expressing significantly higher levels of PRRs and other microbial sensors and act as effective sensing and signaling units.
IECs
IECs function as a physical barrier between the host’s internal milieu and the gut luminal environment. Therefore, the barrier function is impaired because of defects in the TJ components such as ZO-1, Claudin-4, and Occludin between the IECs. This causes increased exposure to the gut microbiota and uptake of the luminal antigens, and leads to aberrant activation of the mucosal immune system (Cario et al., 2007; Hartmann et al., 2012). Urolithin A is a microbial metabolite derived from the polyphenols that are naturally found in fruits such as berries and pomegranate. Urolithin A and its potent synthetic analogue, UAS03, protect against colon diseases by enhancing barrier functions and reducing inflammation through upregulation of epithelial TJ proteins via activation of AhR-nuclear factor erythroid 2-related factor 2 (NRF2)-dependent pathways (Singh et al., 2019a). Furthermore, the physical barrier is reinforced by various biochemical adaptations. For example, IECs secrete a broad range of antimicrobial peptides, including defensins, cathelicidins, and calprotectins (Ganz, 2003; Salzman et al., 2003). These diverse group of peptides confer broad-spectrum antimicrobial properties by forming pores in the bacterial cell wall and obstruct the entry of commensal and pathogenic bacteria into the underlying lamina propria (Artis, 2008). PRR signaling represents one of the mechanisms through which IECs respond to the microbes in the lumen. Furthermore, IECs utilize various mechanisms to regulate PRR signaling and optimize immune responses. For example, Single Ig IL-1 Related Receptor (SIGIRR), a negative regulator of IL-1 and TLR signaling, is responsible for the hypo-responsiveness of the IECs and promotes resistance of the commensal microbiota against the bacterial pathogens (Sham et al., 2013). Therefore, IECs acquire TLR tolerance immediately after birth by exposure to exogenous endotoxins to facilitate microbial colonization through posttranscriptional down-regulation of the interleukin 1 receptor-associated kinase 1, which is essential for the in vitro epithelial TLR4 signaling (Lotz et al., 2006).
IECs play an integral role in the discrimination between pathogenic and commensal bacteria and the subsequent regulation of immune responses in the intestinal microenvironment. IECs include a wide variety of cells such as the enterocytes, stem cells, EECs, Paneth cells, goblet cells, M cells, and Tuft cells, which express a wide range of PRRs that are activated upon recognizing the microbial components and maintain intestinal homeostasis by activating immune responses (Pott and Hornef, 2012). The niche of ISCs are at the forefront of host-microbe activity and are critical for regulating nutrient absorption, endocrine signaling, energy homeostasis, immune responses, and systemic health (Peck et al., 2017). TGR5 senses bile acids and promotes renewal of ISCs to drive regeneration in response to injury (Sorrentino et al., 2020). Paneth cells express elevated levels of NOD2, which are activated by the microbial peptidoglycans. These induce pro-inflammatory cytokines, autophagy, and epithelial regeneration, which in turn regulate the composition of the gut microbiota (Couturier-Maillard et al., 2013; Nigro et al., 2014; Ramanan et al., 2014). NLRP6-dependent mechanisms contribute to the secretion of mucus by the goblet cells. NLRP6 deficiency significantly reduced mucus secretion in the large intestinal lumen because of defective autophagy in the goblet cells. This caused dysbiosis and hyper-susceptibility to enteric infections (Elinav et al., 2011; Wlodarska et al., 2014; Levy et al., 2015; Birchenough et al., 2016). A subset of Trp-derived indole derivatives produced by Edwardsiella tarda directly stimulate epithelial sensory EECs through the human and mouse receptor transient receptor potential ankyrin A1 (TRPA1); moreover, intestinal 5-Hydroxytryptamine (5-HT) secretion by the activated Trpa1+ EECs regulated the enteric and vagal neuronal pathways (Ye et al., 2021). Tuft cells orchestrated antiparasitic immunity in the gut by acting as sentinels of type 2 immunity against the intestinal parasites (Howitt et al., 2016). Tuft cells triggered type 2 immunity by expressing a myriad of GPCRs, which sensed microbes and metabolites such as succinate that are derived from the host, microbes, and diet (Nadjsombati et al., 2018; O’Leary et al., 2019; Banerjee et al., 2020). CD45+ Tuft-2 cells quickly expand in response to the bacterial infections by sensing the bacterial metabolite N-undecanoylglycine through the vomeronasal receptor Vmn2r26 (Xiong et al., 2022). ALK2, a bone morphogenetic protein (BMP) receptor, played a key role in the negative feedback loop that regulated immune type 2-driven tuft cell hyperplasia (Lindholm et al., 2022). M cells are highly specialized epithelial cells of the gut-associated lymphoid tissue (GALT). They take up intestinal microbial antigens, transport them from the gut lumen to the underlying immune system, and stimulate an efficient mucosal immune response. M cells sense and bind to the secretory IgA-antigen complex through Dectin-1. Then, the bound secretory IgA-antigen complex is transferred and internalized via DC-SIGN by the DCs, which induce the mucosal immune responses (Rochereau et al., 2013). NOD2 modulated SIgA transport through the human and mouse M cells by downregulating the expression of two retrograde transport receptors, Dectin-1 and Siglec-5 (Rochereau et al., 2021). Transmembrane mucins were expressed on the apical surface of the enterocytes and participated in the terminal digestion of polysaccharides and peptides as well as absorption of various nutrients. The dysfunction or damage of enterocytes cause intestinal diseases. In vivo infection with E. coli induced the formation of phosphorylated H2AX foci in the mouse colon enterocytes and contributed to the development of sporadic colorectal cancer (Cuevas-Ramos et al., 2010). Thus, IECs serve not only as a mucosal barrier but also discriminate between the pathogens and the commensals.
Myeloid cells
Myeloid cells located in the lamina propria play a significant role in the defense against pathogenic antigens and perform a regulatory role in the immune homeostasis. The commensal-bearing DCs induce protective secretory IgAs, which are distributed throughout all the mucosal surfaces through recirculation of the activated B and T cells (Macpherson and Uhr, 2004). Gut macrophages develop a unique phenotype called as “inflammation anergy”, which is characterized by the induction of a non-inflammatory profile when the intestinal macrophages encounter microbial stimuli under homeostatic conditions (Smythies et al., 2005). Macrophages and DCs are critical APCs that require PRR signaling for maturation and efficient antigen presentation and T cell activation. Therefore, they play a crucial role in the crosstalk between innate and adaptive immune responses (Geremia et al., 2014; Spindler et al., 2022).
Microbiota composition affects the development and function of the myeloid cells. For example, in vivo aging analysis in mice showed that neutrophil y was driven by the microbiota through TLR- and MyD88-dependent signaling pathways. Furthermore, depletion of the microbiota reduced the number of circulating aged neutrophils and improved the pathogenesis and inflammation-related organ damage in the mouse models of endotoxin-induced septic shock and sickle cell disease (Zhang et al., 2015). Bacterial strains such as E. faecalis, Streptococcus gallolyticus, and E. coli induced differentiation and function of the colon-resident macrophages (Wang and Huycke, 2007; Raisch et al., 2015). Fusobacterium nucleatum induced immunosuppression by triggering M2 macrophage polarization through TLR4 and IL-6/STAT3/c-MYC signaling, thereby enhancing colorectal tumor growth (Chen et al., 2018). Intestinal muscularis macrophages responded to luminal infection by upregulating a neuroprotective program through β2-adrenergic receptor signaling, and limited enteric-associated neuronal damage through the arginase 1-polyamine axis (Matheis et al., 2020). Gut microbiota-derived metabolites also orchestrated functions of the resident macrophages (Levy et al., 2016). Stimulation of GPR109a signaling by niacin or butyrate promoted anti-inflammatory properties of the colon-resident macrophages, which then induced differentiation of the Treg cells and the IL-10-producing T cells (Singh et al., 2014). N-butyrate regulated selective functions of the lamina propria macrophages and maintained intestinal homeostasis by directly inhibiting the histone deacetylases (Chang et al., 2014; Schulthess et al., 2019). Tryptamine and indole-3-acetate reduced fatty acid- and LPS-stimulated production of pro-inflammatory cytokines by the macrophages and inhibited the migration of macrophages towards the chemokine in an AhR-dependent manner (Krishnan et al., 2018). TGR5 is a G protein-couple receptor for secondary bile acids expressed on the circulating monocytes and the colon-resident macrophages. It promoted an IL-10- and age-dependent shift towards the alternative M1/M2 macrophage phenotypes (Biagioli et al., 2017; Becker et al., 2019). Putrescine is a polyamine that functions as a substrate for symbiotic metabolism. It increased the abundance of anti-inflammatory macrophages in the colon and ameliorated the symptoms of DSS-induced colitis in mice (Nakamura et al., 2021). Furthermore, germ-free animals showed extensive defects in the development of intestinal innate immune cells. This indicated that microbiota was essential for the normal development and functioning of the host innate immune system. The number of intestinal macrophages was normal in the germ-free mice, but the functions of peritoneal macrophages including chemotaxis, phagocytosis and antimicrobial activity were aberrant (Morland and Midtvedt, 1984; Williams et al., 2006). The number of DCs was also reduced in the intestines of the germ-free mice, but mono-colonization with E. coli was sufficient to recruit the DCs into the intestinal tissues of the germ-free mice (Williams et al., 2006). Intestinal DCs established oral tolerance against antigens derived from the food, commensal microbiota, and metabolites while initiating immune responses against the mucosal pathogens in the intestine (Sun et al., 2020). The activation of PRRs such as TLRs promoted the maturation of DCs with upregulation of the costimulatory molecules and secretion of the pro-inflammatory cytokines (Iwasaki and Medzhitov, 2010; Schenten and Medzhitov, 2011). And these days, several studies proved that intestinal microbiome could restrict virus infection through activating PRRs in gut DCs followed by downstream type I IFN release which could induce systemic ISGs expression and natural virus resistance (Stefan et al., 2020; Winkler et al., 2020). DCs also used the lysosomal histidine/peptide solute carrier transporter SLC15A4 to couple phagocytosis (the necessary first step to sense foreign antigens) with NLRC4 inflammasome assembly and activity (López-Haber et al., 2022).
ILCs
ILCs constitute a newly identified lymphoid lineage that protects the intestine from pathogen invasion by contributing to development of the gut lymphoid tissue and cooperating with the IECs and other innate and adaptive immune cells to repair gut injury (Bostick and Zhou, 2016). ILCs are classified into three main groups based on the expression of specific transcriptional factors: (i) T-bet+ ILC1s (ILC1s and natural killer (NK) cells); (ii) GATA binding protein 3 (GATA3+) ILC2s; and (iii) retinoic acid receptor-related orphan receptor-γt (RORγt+) ILC3s (ILC3s and lymphoid tissue-inducer (LTi) cells) (Spits and Di Santo, 2011; Walker et al., 2013). ILCs are mainly located at the epithelial surfaces and sense nutrients, environmental xenobiotics, and microbiota through the AhR, Retinoic acid receptors (RARs), retinoid X receptors (RXRs), TLRs, and other cell surface receptors (Kiss et al., 2011; Sawa et al., 2011; Spencer et al., 2014; van de Pavert et al., 2014). The genomic landscape of the intestinal ILCs has been characterized through integrated genome-wide RNA-seq, ChIP-seq, ATAC-seq, and single-cell transcriptome analyses (Gury-BenAri et al., 2016). The immune function of the ILC3s is most well-studied among the ILCs in the intestine. ILC3s are comprised of a heterogeneous population of cells that protect against bacterial and fungal invasion, and regulate the commensal microbiota populations and lymphoid tissue development. Activated ILC3s produce IL-17A, IL-22, TNF, and GM-CSF (Takatori et al., 2009; Mortha et al., 2014). GPR43 expressed on the colon-resident ILC3s senses SCFAs and promotes expansion and function of the ILC3s; however, in the small intestine, retinoic acid signaling promotes expansion and function of the ILC3s (Chun et al., 2019). Furthermore, microbiota-derived SCFAs regulated the optimal numbers of ILCs and promoted IL-22 production in the ILCs through GPCRs and AhRs by inhibiting the histone deacetylase (Yang et al., 2020; Sepahi et al., 2021). ILC3s also produce soluble lymphotoxin-α, which regulate T cell-dependent IgA production in the lamina propria (Kruglov et al., 2013). ILC populations and functions play an important role in maintaining intestinal homeostasis and during inflammatory conditions such as IBD (Saez et al., 2021; Coman et al., 2022).
Adaptive immune cells
RAG1-deficient mice lacking adaptive immunity showed alterations in their microbiota populations (Kato et al., 2014). Furthermore, germ-free mice showed extensive defects in the development of GALT (Round and Mazmanian, 2009). This suggested a critical link between adaptive immunity and the gut microbiota. Although the role of T cells or B cells in microbial sensing is not discussed generally, several molecules belonging to the microbial sensing pathways are expressed in the adaptive immune cells and serve important functions (Kabelitz, 2007). Extensive studies have shown that the antigen-specific adaptive immune responses influence the mutualistic relationship between host and gut microbiota.
T cells
The complicated environmental factors and the unique structure of the mammalian intestine influence the distribution of a wide range of T lymphocyte subsets in the IECs and their scattered distribution in the lamina propria. These intestine-resident T cells show distinct effector functions and play a pivotal role in maintaining the gut homeostasis. The innate antigen presenting cells play a significant role in the function, differentiation, and proliferation of the intestinal T cells. For example, microbiota-induced intestinal RORγt+ Tregs depend on the DCs and the major histocompatibility complex (MHC) class II to differentiate into a specific subset of Th17 cells (Ohnmacht et al., 2015). TLRs are primary sensors of the microbiota and are expressed on both the innate and the adaptive immune systems. T cell activation and functions (Kabelitz, 2007) are regulated by TLR signaling and TLR components such as TLR2 (Liu et al., 2006; Sutmuller et al., 2006; Wang et al., 2006; Chen et al., 2009), TLR3 (Wesch et al., 2006), TLR4 (Caramalho et al., 2003), TLR8 (Peng et al., 2005), MyD88 (Gelman et al., 2006; Fukata et al., 2008; Geuking et al., 2011), TRIF (Geuking et al., 2011), and other PRRs regulating TLR signaling (Negishi et al., 2012).
Treg cells play an important role in maintaining immune tolerance to the gut microbiota and the dietary antigens (Round and Mazmanian, 2010; Geuking et al., 2011; Bilate and Lafaille, 2012). Both thymus-derived Tregs (tTregs) and peripherally differentiated Tregs (pTregs) are found in the intestine (Ohnmacht et al., 2015; Sefik et al., 2015). Germ-free mice lack pTregs. This suggested that the pTregs were induced and maintained by the microbiota. Furthermore, in-depth investigation of the fate of immature T cells expressing the transgenic T cell receptor (TCR) showed that the differentiation of Treg cells did not occur in the thymus but occurred in the colon after exposure to the microbiota (Lathrop et al., 2011). Tregs in the intestine use the TLRs to directly sense the microbes. Polysaccharide A derived from a commensal bacterium B. fragilis signaled directly through TLR2 on the Foxp3+ Treg cells and promotes iTreg cell differentiation (Caesar et al., 2015; Cao et al., 2015) and suppressed immunity by inhibiting the Th17 cell responses (Round et al., 2011).
T cell-specific MyD88 signaling is required for the induction of both Th1 and Th17 responses after overcoming Treg suppression through IL-1 signaling (Schenten et al., 2014). Treg-specific Myd88 deletion mice showed deficiency in the intestinal Treg cells, increased numbers of TH17 cells, and enhanced IL-17-dependent inflammation in experimental colitis (Wang et al., 2015b). This demonstrated that TLR-MyD88 signaling in the Treg cells promoted mucosal tolerance by inducing the anti-commensal IgA-responses (Wang et al., 2015b). Experiments with the Clostridiales species-associated food allergy model showed that the commensal microbiota activated the MyD88–RORγt pathway in the nascent Treg cells to mediate the effects of bacteriotherapy against food allergy and this mechanisms included restoring the RORγt+ Treg population and re-establishing tolerance; however, dysbiosis impaired this Treg cell response (Abdel-Gadir et al., 2019). These data demonstrated that MyD88 was indispensable for mucosal T cell induction and activities.
Other PRR signaling pathways also exhibit regulatory functions in the gut T cells. For example, transfer of the CD4+CD45RBhiNlrp12−/− T cells into the immunodeficient mice exacerbated severe colitis and showed that NLRP12 was an intrinsic negative regulator of T cell activation (Lukens et al., 2015). NLRC3 attenuated IFN-γ and TNF expression in the CD4+ T cells and reduced Th1 and Th17 cell proliferation in a T cell-intrinsic manner (Uchimura et al., 2018). The cytosolic DNA sensor AIM2 also showed an intrinsic role in the T cells for promoting the stability of Treg cells during inflammation. AIM2 was highly expressed by both the human and mouse Treg cells. AIM2 altered metabolism and enhanced the stability of Treg cells by reducing AKT-mTOR signaling (Chou et al., 2021). These data demonstrated a novel role for the PRRs in the T cell functions.
Th17 cell differentiation was reduced and Treg cell differentiation was increased by the microbiota-derived metabolites, including bile acids. Furthermore, SCFAs enhanced the differentiation of Treg cells in the intestinal lamina propria (Furusawa et al., 2013; Hang et al., 2019). SCFAs influenced the activity and differentiation of the peripheral T cells, especially Tregs, by inhibiting HDACs. In mice, the production of butyrate by the commensal microorganisms during starch fermentation facilitated extrathymic generation of the Treg cells. Propionate was another SCFA of microbial origin that regulated de novo generation of peripheral Tregs through HDAC inhibition. However, acetate, another SCFA generated by the microbiota, lacked HDAC-inhibitory activity. SCFAs transmitted signals through the GPCRs. For example, SCFAs regulated the size and function of the colon-resident Tregs and protected against colitis in a GPR43-dependent manner (Smith et al., 2013). These results demonstrated that the bacterial metabolites mediated the communication between the commensal microbiota and the immune system and influenced the fate of gut-resident T cells (Arpaia et al., 2013). However, several studies suggested that SCFAs did not induce the RORγt+ Treg cells effectively (Sefik et al., 2015; Abdel-Gadir et al., 2019). Trp derivate indole-3-aldehyde generated by the Lactobacillus reuteri D8 stimulated the LPLs through AhR to secrete IL-22 and induced recovery of the damaged intestinal mucosa by accelerating the proliferation of IECs through increased STAT3 phosphorylation (Hou et al., 2018). Fatty acid sensor GPR120 regulated IL-10 expression in the intestinal CD4+ T cells and suppressed DSS-induced colitis development in mice (Yang et al., 2022). In the CD patients, treatment with oral vitamin D reduced VDR expression in the CD4+ T cells (Bendix et al., 2015). However, in mice, VDR binding to NLRP3 restricted IL-4 expression and prevented biased Th2 polarization (Bendix et al., 2015; Huang et al., 2018). Bacterial metabolites in the gut–liver axis and the tissue stromal factors regulated liver immunity by driving the myeloid instruction of CD8+ T cells with immunomodulatory ability (Pallett et al., 2023). These data demonstrated that the microbiota-derived metabolites significantly modulated immune function by directly regulating signaling in the T cells. Conversely, acute SFB colonization stimulated Th17 cells, which then initiated a dynamic IL-17A-CXCR2-neutrophil axis that limited microbiota colonization (Flannigan et al., 2017).
B cells
Intestinal B cell activation and differentiation is highly dependent on the gut microbiota. In turn, B cells regulated the gut microbiota and maintained intestinal homeostasis, through the production of immunoglobulins (Igs). IgA is the most abundant antibody isotype that is secreted into the gut lumen. It binds to the gut microbiota and prevents their direct interaction with the host. The function of IgA in shaping the microbiota was first reported in mice that are deficient in the activation-induced cytidine deaminase (AID), which is critical for antibody isotype switching. Mice lacking AID showed hyperplasia of the ILFs and a 100-fold expansion of the anaerobic commensal bacteria in the intestine (Fagarasan et al., 2002). SIgA binds or “coats” microbes and prevents direct interaction between the microbes or other antigens and the host via immune exclusion (Yang and Palm, 2020). The coating of commensal microbes such as Lactobacillus, Akkermansia and some colitogenic bacteria with sIgA was higher in the healthy humans and mice compared to those with malnutrition or colitis (Palm et al., 2014; Kau et al., 2015; Huus et al., 2020). Furthermore, MyD88-mediated signaling was required for the development of intestinal IgA+ B cells. Loss of MyD88-mediated signaling diminished targeting of the gut microbiota by high-affinity IgA and resulted in a failure to regulate the growth and homeostasis of the bacterial communities (Kubinak et al., 2015). TLR5-deficient mice showed reduced levels of specific anti-flagellin IgA and contributed to the overexpression of flagellar genes by the commensal microbiota (Cullender et al., 2013). A subset of TLR5+CD11chiCD11bhi DCs in the lamina propria induced the differentiation of naive B cells into IgA-producing plasma cells in the intestine (Uematsu et al., 2008). The absorption of dietary cholesterol and the recognition of commensal bacteria by the duodenal IECs stimulated the production of oxysterols such as 7α,25-dihydroxycholesterol, which are evolutionarily conserved lipids that suppress IgA secretion by the plasma cells through the GPR183 receptor. This suggested that B cells were regulated by the dietary metabolites (Ceglia et al., 2023). IgA production by the B cells was critical for maintaining a diverse and healthy composition of the gut microbiota. Diverse and selective IgA secretion in the gut reduced microbiota complexity in the T cell-deficient mice caused gut mucosal dysbiosis (Kawamoto et al., 2014). Murein lipoprotein is a highly conserved outer membrane protein of the Gram-negative bacteria and is a major antigen that induces TLR4-dependent IgG production in both mice and humans (Zeng et al., 2016). Multiple studies have demonstrated that the presence of IL-10–producing regulatory B cells in humans prevents chronic colitis. Resident enteric bacteria stimulate the intestinal regulatory B cells through the TLR2 signaling pathways to produce IL-10, which in turn suppresses colonic T cell activation and maintains mucosal homeostasis (Mishima et al., 2019). In conclusion, B cells regulate homeostasis of both the immune responses and the gut microbiota by secreting sIgA and through other cellular sensors.
In summary, the gut microbiota sensing pathways have direct consequences on the regulation of barrier-forming epithelial cells, APCs, cytokine-secreting T cells including T helper cells, and the IgA-producing B cells in the gut. The signals from the local environment in the intestinal tract act on the innate immune cells and control the differentiation, migration, and maintenance of the adaptive immune cells. These cells maintain immune homeostasis by suppressing the host immune responses against harmless antigens and enforcing the integrity and functioning of the gut mucosal barrier.
Microbial sensing in health and disease
Symbiotic relationship between the gut microbiota and the host has co-evolved over millions of years and is mutually beneficial. This mutualistic relationship is critical for intestinal homeostasis and overall human health. Therefore, dysbiosis caused by decreased or increased intestinal microbial signaling plays a central role in several human diseases, including infections, metabolic syndrome, cancer, neurological disorders, and autoimmune disorders (Tlaskalova-Hogenova et al., 2011). Dysbiosis causes failure of immunological tolerance, persistent inflammation, and immune regulation of remote organs, all of which contributes to the initiation and progression of these diseases (Tables 1 and 2).
Table 1.
Sensors of microbial components in the intestine.
IEC, intestine epithelial cell; PC, Paneth cell; GC, Goblet cell; CSC, crypt stem cell; M, M cell; MC, myeloid cell; Mф, macrophage; neu, neutrophil; ENS, enteric nervous system; DC, dentritic cell; TC, T cell; BC, B cell; RBC, red blood cell; CAPS: cryopyrin-associated periodic fever syndromes; SBC, small bowel cancer; GVHD, graft-versus-host disease; UC, ulcerative colitis; CRC, colorectal cancer; IBD, inflammatory bowel disease.
Table 2.
Sensors of microbial metabolites in the intestine.
IEC, intestine epithelial cell; MC, myeloid cell; ENS, enteric nervous system; ILC, innate lymphoid cell; TC, T cell; BC, B cell; RBC, red blood cell; GVHD, graft-versus-host disease; UC, ulcerative colitis; CRC, colorectal cancer; LCA, lithocholic acid; DCA, deoxycholic acid; CDCA, chenodeoxycholic acid; CA, cholic acid.
Inflammatory bowel disease
IBD represents a group of chronic inflammatory disorders that affect the colon, small intestine, and the extra-intestinal organs. UC and CD are the two most prevalent forms of IBD. There is increasing evidence that the gut microbiota plays a central role in the pathogenesis of IBD (Baumgart and Carding, 2007; Lloyd-Price et al., 2019). GWAS identified several IBD-related susceptibility genes that showed involvement of the gut microbiota in the pathogenesis of IBD. A meta-analysis of GWAS for the two most common forms of IBD, namely, CD and UC, identified 71 new associations and 163 IBD-related loci (Jostins et al., 2012). Furthermore, other large genome sequencing studies reported several IBD susceptibility loci in genes encoding PRRs and other factors across various human populations (Liu et al., 2015a; Somineni et al., 2021). NOD2 gene locus is highly associated with IBD risk (Hampe et al., 2001; Hugot et al., 2001; Ogura et al., 2001). NOD2 is an intracellular receptor for MDP, which is found in all the bacteria. CD patients with NOD2 mutations showed significantly lower levels of antimicrobial peptides such as α-defensin (Wehkamp et al., 2004). ATG16L1, an abbreviation of autophagy related 16 like 1, is another well-established susceptibility gene for UC. The NOD1/NOD2/ATG16L pathway was critical for the pathogenesis of CD because both NOD2 and NOD1 recruited ATG16L to the site of bacterial entry and triggered autophagy (Cooney et al., 2010). Interaction between a specific enteric viral infection and a specific mutation in the ATG16L1 locus determines susceptibility to IBD (Cadwell et al., 2010). Higher proportions of abnormal Paneth cells were associated with shorter time to recurrence of CD after surgery; moreover, the proportion of abnormal cells was associated with the cumulative numbers of NOD2 and ATG16L1 risk alleles (VanDussen et al., 2014). dl-Endopeptidase, a Firmicutes peptidoglycan remodeling enzyme, increased NOD2 levels in the gut and impacted colitis outcomes (Gao et al., 2022b). This study showed that depletion of dl-endopeptidase contributed to CD pathogenesis through NOD2 signaling. Supplementation of dl-endopeptidases or dl-endopeptidase-producing bacteria strains and a synthetic lipophilic analog of MDP protected against colitis in mice by restoring the NOD2 pathway. This provided a therapeutically modifiable target for the CD patients.
Changes in the fungal microbiota, an integral part of the complex microbial community, are also linked to IBD. Deep sequencing of the rRNA genes showed that the abundance of specific Candida taxa was increased in the IBD samples (Chehoud et al., 2015). The abundance and preferential localization of Debaryomyces hansenii in the incompletely healed intestinal wounds of mice and the inflamed mucosal tissues of human subjects with CD impaired mucosal healing through the myeloid cell-specific type 1 IFN–CCL5 axis (Jain et al., 2021). Candida albicans-secreted peptide toxin candidalysin significantly influenced inflammation and altered gut immunity in patients with IBD; moreover, C. albicans strains with a high immune cell damaging capacity induced intestinal inflammation through IL-1β-dependent mechanisms in the UC models (Li et al., 2022b). Intestinal fungi also promoted intestinal host-microbiota homeostasis. Mucosal fungi promoted host-protective immunity and epithelial barrier function through type 17 immunity (Leonardi et al., 2022). Therefore, host–fungal interactions are novel diagnostic and therapeutic targets for IBD.
Type I diabetes
Autoimmune diseases are complex diseases caused by a combination of genetic, environmental, and life-style factors. Recent studies have demonstrated that the dysregulation of the composition and function of the gut microbiota are involved in autoimmune disorders such as type I diabetes (T1D) (Miyauchi et al., 2023). MyD88 deficiency leads to changes in the composition of the gut microbiota. Germ-free non-obese diabetic (NOD) mice with MyD88 deficiency developed severe diabetes, but colonization of these mice with defined gut microbiota attenuated diabetes (Wen et al., 2008). Non-synonymous mutants of human RIG-I (S183I) and MDA5 (E627*, I923V, and A946T) are implicated in the resistance against type I diabetes (Smyth et al., 2006; Shigemoto et al., 2009). AIM2−/− mice treated with broad-spectrum antibiotics are protected from streptozotocin-induced T1D and lower pancreatic pro-inflammatory response, thereby implicating the role of gut microbiota in T1D (Leite et al., 2020). In obese subjects, intestinal microbial DNA-containing vesicles pass through the gut barrier and are transferred into islet β cells, resulting in enhanced inflammation and impaired insulin secretion through activation of the cGAS/STING signaling pathway (Gao et al., 2022a). Moreover, diabetic incidence is higher in the NOD mice under germ-free conditions than those under specific pathogen-free conditions. This finding was inconsistent with the higher prevalence of T1D is in countries with better hygiene (Bach, 2002; Vatanen et al., 2018; Kim et al., 2020).
Metabolic syndrome
There is extensive evidence that the gut microbiota is important for the host in harvesting energy from the diet and increasing fat storage (Turnbaugh et al., 2006; Samuel et al., 2008). Germ-free mice ingested 29% more calories but showed 40% lower body fat than the conventional mice (Turnbaugh et al., 2006). The weight gain of the germ-free mice was lower and were protected against development of insulin resistance (Piya et al., 2013). In mice, administration of a specific protein isolated from the outer membrane of A. muciniphila interacted with TLR2 and prevented the development of obesity and associated complications (Plovier et al., 2017). A proof-of-concept study (Clinical trial No. NCT02637115) showed that supplementation with A. muciniphila improved several metabolic parameters associated with the obesity-related disorders but the overall structure of the gut microbiome was unaffected (Depommier et al., 2019). The gut microbiota contributed to metabolic disease through chronic low-grade inflammation (Gregor and Hotamisligil, 2011). For example, chronically increased plasma LPS concentration triggered inflammation-related obesity and metabolic disease in mice (Cani et al., 2007). The binding of LPS to TLR4 triggered NF-κB- and mitogen activated protein kinase (MAPK)-dependent secretion of pro-inflammatory cytokines (Neal et al., 2006). TLR4-deficient mice were protected against HFD-induced insulin resistance (Shi et al., 2006b). Furthermore, ob/ob mice with CD14 deficiency were unable to induce LPS signaling and were resistant to weight gain and insulin hypersensitivity (Cani et al., 2008). The gut microbiota also affects host immunity via other microbial-derived metabolites in addition to LPS. Indole is a microbial-derived metabolite that is produced by the bacterial tryptophanase, which is present in many commensal bacteria (DeMoss and Moser, 1969). Indole is metabolized into 3-indoxyl sulfate in the liver after absorption into blood from the intestine. Indoxyl sulfate is related to obesity because it promoted synthesis and secretion of the pro-inflammatory cytokine IL-6 through AhR (Ramadoss et al., 2005; Russell et al., 2013). The dysfunction of fatty acid and bile acid sensors also caused metabolic syndrome. The activation of GPR120, a lipid sensor, mediated the secretion of glucagon-like peptide-1 (GLP-1). GPR120 deficiency or mutations (p.R270H in the European populations) decreased insulin sensitivity and caused glucose intolerance and fatty liver, thereby increasing the risk of obesity in the mice models and humans (Hirasawa et al., 2005; Ichimura et al., 2012; Paschoal et al., 2020). TGR5 signaling pathway was triggered by the bile acids and played a significant role in regulating intestinal GLP-1 secretion (Thomas et al., 2009). Cholic acid-7-sulfate is an endogenous bile acid and a TGR5 agonist that enhanced glucose tolerance in the insulin-resistant mice by increasing GLP-1 secretion (Chaudhari et al., 2021). However, activation of another bile acid receptor, FXR in the enteroendocrine L cells decreased GLP-1 secretion by inhibiting glycolysis. Inhibition of FXR signaling in the intestine improved metabolic parameters in the mouse models of obesity (Jiang et al., 2015; Trabelsi et al., 2015). In the T2D mouse model, metformin acted partially through the B. fragilis—glycoursodeoxycholic acid (GUDCA)-intestinal FXR axis to improve metabolic dysfunction (Sun et al., 2018a). Conversely, a gut-restricted FXR agonist fexaramine robustly induced enteric fibroblast growth factor 15 (FGF15) and induced alterations in the bile acid composition and metabolic improvements without activating the FXR-target genes in the liver (Fang et al., 2015). This suggested that tissue restricted FXR activation was potentially a novel approach for treating obesity and metabolic syndrome.
Neurological disorders
The gut microbiota also plays a significant role in the bidirectional communication along the “gut–brain axis”, which regulates the central nervous system development, function, and pathogenesis (Socała et al., 2021). Emerging evidence suggests that the NLRs and the TLRs in the gut–brain axis play a critical role in the pathogenetic mechanisms underlying neurodevelopmental and neurodegenerative disorders (Keogh et al., 2021). IEC-specific Nod1 knockout mice showed stress-induced anxiety-like behavior and cognitive impairment by affecting 5-HT biosynthesis and signaling (Pusceddu et al., 2019). Microbiota-derived LPS promoted survival of enteric neurons and gastrointestinal motility in mice by activating TLR4 and NF-κB (Anitha et al., 2012). SNPs including TLR1 rs4833095, TLR2 rs3804099 and TLR4 rs1927914 were associated with increased risk of Parkinson’s disease (PD) (Zhao et al., 2015; Gorecki et al., 2020). Precursors of neurotransmitters like serotonin, GABA, dopamine, and norepinephrine are generated by the gut microbiota. Serotonin, dopamine, and norepinephrine act on the ENS through specific receptors in the brain, whereas transporter for GABA is expressed in the blood–brain barrier (Lyte, 2011; Reardon, 2014; Dinan et al., 2015; Hubbard et al., 2015). Amyloid protein in the gut bacteria demonstrate molecular mimicry and can elicit cross-seeded misfolding, inflammation and cellular cytotoxicity that initiates or influences the pathogenesis underlying PD, Alzheimer’s disease (AD), and other neurological disorders (Friedland, 2015). Bacterial amyloid proteins induce molecular mimicry pathways via TLR1/2 signaling, which enhanced inflammatory responses to brain amyloid proteins such as α-synuclein (Oppong et al., 2013; Chen et al., 2016; Sampson et al., 2020). The upregulation of TLR2 signaling in the microglia was associated with neuroinflammation in PD (Beraud and Maguire-Zeiss, 2012). Metabolites of dietary Trp produced by the commensal flora activated AHR and suppressed CNS inflammation due to pathological activities of the microglia and the astrocytes in the experimental autoimmune encephalomyelitis (EAE) mouse model of multiple sclerosis (Rothhammer et al., 2018). In the intestinal neural circuits, AHR is a microbiota-related biosensor that impacts intestinal motility (Obata et al., 2020).
Cancer
Colorectal cancer (CRC) is a disease of the ISCs that is characterized by dysregulation of multiple signaling pathways and components in the intestine. Inflammation associated with IBD, heritable genetic defects, diets with low fiber content and high red meat content, and imbalance in the gut microbiota are known risk factors of CRC (Janney et al., 2020). The expression of TLR2 is significantly higher in the tumor tissues compared with the adjacent normal intestinal tissues of the CRC patients. TLR2 knockdown significantly inhibited proliferation of the CRC cells but exacerbated colitis symptoms in the mouse colitis-associated cancer model (CAC) (Meng et al., 2020). Epigenetic modifying drugs are used to treat several cancers. A recent study showed potential immunoregulatory role of these drugs in response to viral stimulation, especially changes in the TLR3 responses (Hennessy et al., 2019). Peptostreptococcus anaerobius activated TLR2 and TLR4 on the colon cells and increased the intracellular levels of reactive oxygen species (ROS), which promoted cholesterol synthesis and cancer cell proliferation. Peptostreptococcus anaerobius levels were higher in the human CRC tissues and adenomas compared to the non-tumor tissues (Tsoi et al., 2017). Pro-inflammatory effects induced by the activation of TLRs promoted malignant progression of colon cancer (Yesudhas et al., 2014; Gao et al., 2018; Moradi-Marjaneh et al., 2018). Furthermore, infections promoted cancer progression by enhancing tumor invasiveness through the TLR9-mediated matrix metalloproteinases (MMPs) (Merrell et al., 2006). However, IRAK-M, a negative regulator of TLR signaling, was expressed in the CRC cells through combined TLR and Wnt activation and promoted CRC cell proliferation by stabilizing the STAT3 protein (Kesselring et al., 2016). These conflicting data may be due to the negative feedback of the pro-inflammatory signaling pathways related to the TLRs. Furthermore, TLR5-dependent commensal microbiota drives the malignant progression of tumors by increasing IL-6 levels and depletion of the commensal bacteria abrogates TLR5-dependent tumor progression. However, TLR5 agonists acted as organ-specific immunoadjuvants and enabled efficient anti-tumor vaccination by stimulating the NK-DCs-CD8+ T cell axis (Brackett et al., 2016). NOD1 is another PRR that is highly expressed in the human CRC tissues and the human and murine CRC cell lines and augments CRC cell metastasis through the p38 MAPK pathway (Jiang et al., 2020). AIM2 gene mutations were frequent in patients with intestinal cancers (Schulmann et al., 2005). AIM2 suppressed uncontrolled expansion of ISCs by dysregulating the Wnt signaling pathway. In the CAC model, AIM2-deficient mice developed significantly higher number of colon tumors that were mediated by DNA-PK-Akt activation but were independent of inflammasome activation (Man et al., 2015; Wilson et al., 2015a). Epithelial metabolite receptors, such as GPR120, VDR and AHR, are essential for maintaining the mucosal barrier integrity, restoring the gut barrier homeostasis, and preventing CRC development (Metidji et al., 2018; Rubbino et al., 2022; Zhang et al., 2022). Several VDR gene polymorphisms are significantly associated with colorectal cancer development and progression (Latacz et al., 2021; Messaritakis et al., 2022).
Outlook
In the recent decades, several studies have investigated the gut microbial sensing and shown that co-evolution of the gut microbiota and the immune system plays a pivotal role in human health and disease. However, the relationship between the gut microbiota, microbiota-derived metabolites, and the host immune system is far from being understood fully because of the extensive size of the gut microbiota and the limitations of current technology. The mechanistic details of many microbial sensors have been discovered. However, studies have also reported controversial results about the roles of the microbial sensors in the intestine. For example, TLR4 signaling reinforced by LPS from the Eubacterium rectale-deficit microbiota promoted intestinal inflammation. However, TLR4 ameliorated colitis by upregulating Treg cells by interacting with A. muciniphila (Liu et al., 2022b; Lu et al., 2022). These bidirectional roles of TLR4 in the intestinal inflammation may be due to the structural differences in the microbial components such as LPS. In 2022, a study reported that NOD2 recognized phosphorylated MDP instead of unmodified MDP (Stafford et al., 2022). Therefore, precise elucidation of the structure and functions of the microbial ligands is necessary to determine the exact microbe-host signaling connections. Recent studies have investigated the mechanisms by which the commensal microorganisms modulate the innate immune sensors using high throughput bacterial screening and the mechanisms by which complex dietary or microbe-derived metabolites modulate the host receptors (Cohen et al., 2017; Spindler et al., 2022). Furthermore, in-depth studies are necessary to determine the exact role of several orphan ligands or receptors including NLR family members such as NLRP12 and other orphan GPCRs. Thus, the “one microbe, one response” approach needs to be replaced by systematic studies. Moreover, development of advanced computational tools and other integrative technologies are needed for unraveling the mechanisms underlying the host-microbiota relationship, especially in relation to human health and disease.
Acknowledgements
We would like to thank W. Tao, H. Ma, and other members of Zhu laboratory and Z. Wei in Richard Flavell’s lab for helpful discussions. This work was supported by grants from the National Key R&D Program of China (Grant No. 2018YFA0508000; S.Z.), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB29030101; S.Z.), the CAS Project for Young Scientists in Basic Research (Grant No. YSBR-074; S.Z.); and the National Natural Science Foundation of China (Grant No. 82061148013, S.Z.).
Abbreviations
- Aβ
amyloid-β
- AD
Alzheimer’s disease
- AHR
aryl hydrocarbon receptor
- AID
activation-induced cytidine deaminase
- AIM2
absence in melanoma 2
- ALR
AIM2-like receptor
- AOM
azoxymethane
- AP
adaptor protein
- APC
antigen presenting cell
- ATG16L1
autophagy related 16 like 1
- BMP
bone morphogenetic protein
- CAC
colitis-associated cancer model
- CAPS
cryopyrin-associated periodic syndrome
- CARD
caspase recruitment domain
- CART
cocaine- and amphetamine-regulated transcript
- CD
Crohn’s disease
- cGAMP
cyclic-di-GMP/AMP
- cGAS
cyclic GMP-AMP synthase
- CLR
C-type lectin receptor
- CRC
colorectal cancer
- CYLD
CYLD lysine 63 deubiquitinase
- DAMPs
danger-associated molecular patterns
- DAP
d-glutamyl-meso-diaminopimelic acid
- DC
dendritic cell
- DHX15
DEAH-box helicase 15
- DRG
dorsal root ganglion
- DSS
dextran sulfate sodium
- EAE
experimental autoimmune encephalomyelitis
- ECMV
encephalomyocarditis virus
- EEC
enteroendocrine cell
- ENS
enteric nervous system
- ER
endoplasmic reticulum
- FGF15
fibroblast growth factor 15
- FOS
oligofructose
- FXR
Farnesoid X receptor
- GALT
gut-associated lymphoid tissue
- GP-BAR1
G protein-coupled bile acid receptor, also named as TGR5
- GPCR
G protein-coupled receptor
- GUDCA
glycoursodeoxycholic acid
- GVHD
graft-versus-host disease
- GWAS
genome-wide association study
- 5-HT
5-Hydroxytryptamine
- HFD
high-fat diet
- HLCs
hepatocyte like cells
- HMGB1
high mobility group box 1 protein
- HMP
Human Microbiome Project
- IBD
inflammatory bowel disease
- IEC
intestinal epithelial cell
- IEL
intraepithelial lymphocyte
- IFN
interferon
- IgA
Immunoglobulin A
- IGF-1
insulin-like growth factor-1
- IL
interleukin
- ILCs
innate lymphoid cells
- ILF
isolated lymphoid follicle
- ISG
IFN-stimulated gene
- LCFA
long-chain fatty acid
- LGP2
laboratory of genetics and physiology 2
- LLPS
liquid–liquid phase separation
- LPS
lipopolysaccharide
- LRR
leucine-rich repeat
- LTA
lipoteichoic acid
- LTi
lymphoid tissue-inducer
- M cells
microfold cells
- MAPK
mitogen activated protein kinase
- MAVS
mitochondrial antiviral signaling protein
- MCFAs
medium-chain fatty acids
- MDA5
melanoma differentiation-associated gene 5
- MDP
muramyl dipeptide
- MHC
major histocompatibility complex
- MMP
matrix metalloproteinase
- MNV
murine norovirus
- MRGPRX2
MAS related GPR family member X2
- MRI
magnetic resonance imaging
- mTOR
mechanistic target of rapamycin kinase
- MyD88
myeloid differentiation factor 88
- NAGK
N-acetylglucosamine kinase
- NAIP
NLR family apoptosis inhibitory protein
- NACHT
NAIP, CIITA, HET-E and TP1
- N-C11-G
N-undecanoylglycine
- NF-κB
nuclear factor kappa-B
- NK
natural killer
- NLR
NOD-like receptor
- NLRC4
NLR family, CARD domain-containing 4
- NLRP3
NLR family, pyrin domain-containing 3
- NOD model
non-obese diabetic model
- NOD1
nucleotide-binding oligomerization domain-containing 1
- NRF2
nuclear factor erythroid 2-related factor 2
- PAMPs
pathogen-associated molecular patterns
- PD
Parkinson’s disease
- PI3K
phosphatidylinositol-3-kinase
- PRRs
pattern recognition receptors
- PSC
pluripotent stem cell
- PXR
Pregnane X receptor
- PYD
pyrin domain
- RAF-1
v-raf-1 murine leukemia viral oncogene homolog 1
- RAR
retinoic acid receptor
- RIG-I
retinoic acid-inducible gene I
- RIP2
receptor-interacting-serine/threonine-protein kinase 2
- RLR
RIG-I-like receptor
- RORγt
retinoic acid receptor-related orphan receptor-γt
- ROS
reactive oxygen species
- RV
rotavirus
- RXR
retinoid X receptor
- SCFAs
short-chain fatty acids
- sIgA
secretory IgA
- SIGNR3
specific intracellular adhesion molecule-3 grabbing non-integrin homolog-related 3
- SNP
single nucleotide polymorphism
- SPF
specific pathogen free
- STING
sequestering and inhibiting the stimulator of IFN genes
- T1D
type I diabetes
- T3SS
Type III secretion systems
- TIR
Toll/IL-1 receptor
- TJ
tight junction
- TLR
Toll-like receptor
- Treg
regulatory T cell
- TRIF
TIR domain-containing adaptor inducing IFNβ
- Trp
tryptophan
- Trpa1
transient receptor potential ankyrin A1
- UC
ulcerative colitis
- UDCA
ursodeoxycholic acid
- VDR
Vitamin D receptor
- WAT
white adipose tissue
- XIAP
X-linked inhibitor of apoptosis protein
Contributor Information
Tingting Wan, Division of Life Sciences and Medicine, The CAS Key Laboratory of Innate Immunity and Chronic Disease, Institute of Immunology, School of Basic Medical Sciences, University of Science and Technology of China, Hefei 230027, China.
Yalong Wang, Division of Life Sciences and Medicine, The CAS Key Laboratory of Innate Immunity and Chronic Disease, Institute of Immunology, School of Basic Medical Sciences, University of Science and Technology of China, Hefei 230027, China.
Kaixin He, Division of Life Sciences and Medicine, The CAS Key Laboratory of Innate Immunity and Chronic Disease, Institute of Immunology, School of Basic Medical Sciences, University of Science and Technology of China, Hefei 230027, China.
Shu Zhu, Division of Life Sciences and Medicine, The CAS Key Laboratory of Innate Immunity and Chronic Disease, Institute of Immunology, School of Basic Medical Sciences, University of Science and Technology of China, Hefei 230027, China; Department of Digestive Disease, Division of Life Sciences and Medicine, The First Affiliated Hospital of USTC, University of Science and Technology of China, Hefei 230001, China; Institute of Health and Medicine, Hefei Comprehensive National Science Center, Hefei 230601, China.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- Abdel-Gadir A, Stephen-Victor E, Gerber GKet al. Microbiota therapy acts via a regulatory T cell MyD88/RORγt pathway to suppress food allergy. Nat Med 2019;25:1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adiliaghdam F, Amatullah H, Digumarthi Set al. Human enteric viruses autonomously shape inflammatory bowel disease phenotype through divergent innate immunomodulation. Sci Immunol 2022;7:eabn6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afrazi A, Branca MF, Sodhi CPet al. Toll-like receptor 4-mediated endoplasmic reticulum stress in intestinal crypts induces necrotizing enterocolitis. J Biol Chem 2014;289:9584–9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshan FU, Masood A, Nissar Bet al. Promoter hypermethylation regulates vitamin D receptor (VDR) expression in colorectal cancer—a study from Kashmir valley. Cancer Genet 2021;252–253:96–106. [DOI] [PubMed] [Google Scholar]
- Agus A, Planchais J, Sokol H.. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 2018;23:716–724. [DOI] [PubMed] [Google Scholar]
- Aksentijevich I, Nowak M, Mallah Met al. De novo CIAS1 mutations, cytokine activation, and evidence for genetic heterogeneity in patients with neonatal-onset multisystem inflammatory disease (NOMID): a new member of the expanding family of pyrin-associated autoinflammatory diseases. Arthritis Rheum 2002;46:3340–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albaugh VL, Banan B, Antoun Jet al. Role of bile acids and GLP-1 in mediating the metabolic improvements of bariatric surgery. Gastroenterology 2019;156:1041–1051.e1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemi F, Poole DP, Chiu Jet al. The receptor TGR5 mediates the prokinetic actions of intestinal bile acids and is required for normal defecation in mice. Gastroenterology 2013;144:145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliprantis AO, Yang RB, Mark MRet al. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science 1999;285:736–739. [DOI] [PubMed] [Google Scholar]
- Allam R, Maillard MH, Tardivel Aet al. Epithelial NAIPs protect against colonic tumorigenesis. J Exp Med 2015;212:369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen IC, TeKippe EM, Woodford RMet al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med 2010;207:1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen IC, Wilson JE, Schneider Met al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-kappaB signaling. Immunity 2012;36:742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand PK, Malireddi RK, Lukens JRet al. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature 2012;488:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anitha M, Vijay-Kumar M, Sitaraman SVet al. Gut microbial products regulate murine gastrointestinal motility via Toll-like receptor 4 signaling. Gastroenterology 2012;143:1006–1016.e1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong HK, Bording-Jorgensen M, Santer DMet al. Unfermented β-fructan fibers fuel inflammation in select inflammatory bowel disease patients. Gastroenterology 2023;164:228–240. [DOI] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan Xet al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013;504:451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol 2008;8:411–420. [DOI] [PubMed] [Google Scholar]
- Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med 2002;347:911–920. [DOI] [PubMed] [Google Scholar]
- Bae M, Cassilly CD, Liu Xet al. Akkermansia muciniphila phospholipid induces homeostatic immune responses. Nature 2022;608:168–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Herring CA, Chen Bet al. Succinate produced by intestinal microbes promotes specification of tuft cells to suppress ileal inflammation. Gastroenterology 2020;159:2101–2115.e2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C, Duewell P, Mayer Cet al. Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut 2010;59:1192–1199. [DOI] [PubMed] [Google Scholar]
- Baumgart DC, Carding SR.. Inflammatory bowel disease: cause and immunobiology. Lancet 2007;369:1627–1640. [DOI] [PubMed] [Google Scholar]
- Becker L, Spear ET, Sinha SRet al. Age-related changes in gut microbiota alter phenotype of Muscularis macrophages and disrupt gastrointestinal motility. Cell Mol Gastroenterol Hepatol 2019;7:243–245.e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilmann-Lehtonen I, Hagström J, Mustonen Het al. High tissue TLR5 expression predicts better outcomes in colorectal cancer patients. Oncology (Huntingt) 2021;99:589–600. [DOI] [PubMed] [Google Scholar]
- Bendix M, Dige A, Deleuran Bet al. Flow cytometry detection of vitamin D receptor changes during vitamin D treatment in Crohn’s disease. Clin Exp Immunol 2015;181:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beraud D, Maguire-Zeiss KA.. Misfolded alpha-synuclein and Toll-like receptors: therapeutic targets for Parkinson’s disease. Parkinsonism Relat Disord 2012;18:S17–S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhinder G, Stahl M, Sham HPet al. Intestinal epithelium-specific MyD88 signaling impacts host susceptibility to infectious colitis by promoting protective goblet cell and antimicrobial responses. Infect Immun 2014;82:3753–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagioli M, Carino A, Cipriani Set al. The bile acid receptor GPBAR1 regulates the M1/M2 phenotype of intestinal macrophages and activation of GPBAR1 rescues mice from murine colitis. J Immunol 2017;199:718–733. [DOI] [PubMed] [Google Scholar]
- Bilate AM, Lafaille JJ.. Induced CD4+Foxp3+ regulatory T cells in immune tolerance. Annu Rev Immunol 2012;30:733–758. [DOI] [PubMed] [Google Scholar]
- Birchenough GM, Nyström EE, Johansson MEet al. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 2016;352:1535–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas A, Liu YJ, Hao Let al. Induction and rescue of Nod2-dependent Th1-driven granulomatous inflammation of the ileum. Proc Natl Acad Sci USA 2010;107:14739–14744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bódi N, Egyed-Kolumbán A, Onhausz Bet al. Intestinal region-dependent alterations of toll-like receptor 4 expression in myenteric neurons of Type 1 diabetic rats. Biomedicines 2023;11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booshehri LM, Hoffman HM.. CAPS and NLRP3. J Clin Immunol 2019;39:277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick JW, Zhou L.. Innate lymphoid cells in intestinal immunity and inflammation. Cell Mol Life Sci 2016;73:237–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouskra D, Brezillon C, Berard Met al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 2008;456:507–510. [DOI] [PubMed] [Google Scholar]
- Brackett CM, Kojouharov B, Veith Jet al. Toll-like receptor-5 agonist, entolimod, suppresses metastasis and induces immunity by stimulating an NK-dendritic-CD8+ T-cell axis. Proc Natl Acad Sci USA 2016;113:E874–E883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brevini T, Maes M, Webb GJet al. FXR inhibition may protect from SARS-CoV-2 infection by reducing ACE2. Nature 2023a;615(7950):134–142. 10.1038/s41586-022-05594-0Epub 2022 Dec 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brevini T, Maes M, Webb GJet al. UK-PBC Consortium. FXR inhibition may protect from SARS-CoV-2 infection by reducing ACE2. Nature 2023b;615:134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisse M, Ly H.. Comparative structure and function analysis of the RIG-I-like receptors: RIG-I and MDA5. Front Immunol 2019;10:1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broquet AH, Hirata Y, McAllister CSet al. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J Immunol 2011;186:1618–1626. [DOI] [PubMed] [Google Scholar]
- Brubaker SW, Bonham KS, Zanoni Iet al. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol 2015;33:257–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun P, Giron MC, Qesari Met al. Toll-like receptor 2 regulates intestinal inflammation by controlling integrity of the enteric nervous system. Gastroenterology 2013;145:1323–1333. [DOI] [PubMed] [Google Scholar]
- Bruns AM, Leser GP, Lamb RAet al. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5–RNA interaction and filament assembly. Mol Cell 2014;55:771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgueno JF, Abreu MT.. Epithelial toll-like receptors and their role in gut homeostasis and disease. Nat Rev Gastroenterol Hepatol 2020;17:263–278. [DOI] [PubMed] [Google Scholar]
- Burgueño JF, Barba A, Eyre Eet al. TLR2 and TLR9 modulate enteric nervous system inflammatory responses to lipopolysaccharide. J Neuroinflammation 2016;13:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgueño JF, Fritsch J, González EEet al. Epithelial TLR4 signaling activates DUOX2 to induce microbiota-driven tumorigenesis. Gastroenterology 2021;160:797–808.e796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Maloney NSet al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell 2010;141:1135–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar R, Tremaroli V, Kovatcheva-Datchary Pet al. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab 2015;22:658–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candia E, Díaz-Jiménez D, Langjahr Pet al. Increased production of soluble TLR2 by lamina propria mononuclear cells from ulcerative colitis patients. Immunobiology 2012;217:634–642. [DOI] [PubMed] [Google Scholar]
- Cani PD, Amar J, Iglesias MAet al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007;56:1761–1772. [DOI] [PubMed] [Google Scholar]
- Cani PD, Bibiloni R, Knauf Cet al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008;57:1470–1481. [DOI] [PubMed] [Google Scholar]
- Cao AT, Yao S, Gong Bet al. Interleukin (IL)-21 promotes intestinal IgA response to microbiota. Mucosal Immunol 2015;8:1072–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramalho I, Lopes-Carvalho T, Ostler Det al. Regulatory T cells selectively express toll-like receptors and are activated by lipopolysaccharide. J Exp Med 2003;197:403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cario E, Podolsky DK.. Differential alteration in intestinal epithelial cell expression of toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect Immun 2000;68:7010–7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cario E, Gerken G, Podolsky DK.. Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function. Gastroenterology 2007;132:1359–1374. [DOI] [PubMed] [Google Scholar]
- Carvalho FA, Koren O, Goodrich JKet al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe 2012a;12:139–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho FA, Nalbantoglu I, Aitken JDet al. Cytosolic flagellin receptor NLRC4 protects mice against mucosal and systemic challenges. Mucosal Immunol 2012b;5:288–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho FA, Nalbantoglu I, Ortega-Fernandez Set al. Interleukin-1β (IL-1β) promotes susceptibility of Toll-like receptor 5 (TLR5) deficient mice to colitis. Gut 2012c;61:373–384. [DOI] [PubMed] [Google Scholar]
- Castro FA, Försti A, Buch Set al. TLR-3 polymorphism is an independent prognostic marker for stage II colorectal cancer. Eur J Cancer 2011;47:1203–1210. [DOI] [PubMed] [Google Scholar]
- Ceglia S, Berthelette A, Howley Ket al. An epithelial cell-derived metabolite tunes immunoglobulin A secretion by gut-resident plasma cells. Nat Immunol 2023;24(3):531–544. 10.1038/s41590-022-01413-w Epub 2023 Jan 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamaillard M, Hashimoto M, Horie Yet al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol 2003;4:702–707. [DOI] [PubMed] [Google Scholar]
- Chan GC, Chan WK, Sze DM.. The effects of beta-glucan on human immune and cancer cells. J Hematol Oncol 2009;2:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PV, Hao L, Offermanns Set al. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA 2014;111:2247–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaniotou Z, Giannogonas P, Theoharis Set al. Corticotropin-releasing factor regulates TLR4 expression in the colon and protects mice from colitis. Gastroenterology 2010;139:2083–2092. [DOI] [PubMed] [Google Scholar]
- Chassaing B, Ley RE, Gewirtz AT.. Intestinal epithelial cell toll-like receptor 5 regulates the intestinal microbiota to prevent low-grade inflammation and metabolic syndrome in mice. Gastroenterology 2014;147:1363–77.e1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee I, Zhang Y, Zhang Jet al. Overexpression of Vitamin D receptor in intestinal epithelia protects against colitis via upregulating tight junction protein Claudin 15. J Crohns Colitis 2021;15:1720–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari SN, Harris DA, Aliakbarian Het al. Bariatric surgery reveals a gut-restricted TGR5 agonist with anti-diabetic effects. Nat Chem Biol 2021;17:20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehoud C, Albenberg LG, Judge Cet al. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis 2015;21:1948–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Davidson TS, Huter ENet al. Engagement of TLR2 does not reverse the suppressor function of mouse regulatory T cells, but promotes their survival. J Immunol 2009;183:4458–4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GY, Liu M, Wang Fet al. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J Immunol 2011;186:7187–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SG, Stribinskis V, Rane MJet al. Exposure to the functional bacterial amyloid protein Curli enhances alpha-synuclein aggregation in aged Fischer 344 rats and Caenorhabditis elegans. Sci Rep 2016;6:34477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wilson JE, Koenigsknecht MJet al. NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat Immunol 2017;18:541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Li Q, Wu Jet al. Fusobacterium nucleatum promotes M2 polarization of macrophages in the microenvironment of colorectal tumours via a TLR4-dependent mechanism. Cancer Immunol Immunother 2018;67:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Jin D, Huang Set al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett 2020a;469:456–467. [DOI] [PubMed] [Google Scholar]
- Chen X, Yang X, de Anda Jet al. Clostridioides difficile toxin A remodels membranes and mediates DNA entry into cells to activate toll-like receptor 9 signaling. Gastroenterology 2020b;159:2181–2192.e2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Fang S, Wei Het al. Prevotella copri increases fat accumulation in pigs fed with formula diets. Microbiome 2021a;9:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Chuang LS, Giri Met al. Inflamed ulcerative colitis regions associated With MRGPRX2-mediated mast cell degranulation and cell activation modules, defining a new therapeutic target. Gastroenterology 2021b;160:1709–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Jiao T, Liu Wet al. Hepatic cytochrome P450 8B1 and cholic acid potentiate intestinal epithelial injury in colitis by suppressing intestinal stem cell renewal. Cell Stem Cell 2022;29:1366–1381.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K, Gao M, Godfroy JIet al. Specific activation of the TLR1–TLR2 heterodimer by small-molecule agonists. Sci Adv 2015;1(3):e1400139. 10.1126/sciadv.1400139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinen I, Nakahama T, Kimura Aet al. The aryl hydrocarbon receptor/microRNA-212/132 axis in T cells regulates IL-10 production to maintain intestinal homeostasis. Int Immunol 2015;27:405–415. [DOI] [PubMed] [Google Scholar]
- Chou WC, Guo Z, Guo Het al. AIM2 in regulatory T cells restrains autoimmune diseases. Nature 2021;591:300–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Khosravi A, Kusumawardhani IPet al. Gene–microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 2016;352:1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang T, Ulevitch RJ.. Identification of hTLR10: a novel human Toll-like receptor preferentially expressed in immune cells. Biochim Biophys Acta 2001;1518:157–161. [DOI] [PubMed] [Google Scholar]
- Chun E, Lavoie S, Fonseca-Pereira Det al. Metabolite-sensing receptor Ffar2 regulates colonic Group 3 innate lymphoid cells and gut immunity. Immunity 2019;51:871–884.e876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriani S, Mencarelli A, Chini MGet al. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS One 2011;6:e25637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasen SJ, Bell MEW, Borbón Aet al. Silent recognition of flagellins from human gut commensal bacteria by Toll-like receptor 5. Sci Immunol 2023;8:eabq7001. [DOI] [PubMed] [Google Scholar]
- Cleynen I, González JR, Figueroa Cet al. Genetic factors conferring an increased susceptibility to develop Crohn’s disease also influence disease phenotype: results from the IBDchip European Project. Gut 2013;62:1556–1565. [DOI] [PubMed] [Google Scholar]
- Cohen LJ, Kang HS, Chu Jet al. Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist. Proc Natl Acad Sci USA 2015;112:E4825–E4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen LJ, Esterhazy D, Kim SHet al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 2017;549:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Kedar S, Baram L, Elad Het al. Human intestinal epithelial cells respond to beta-glucans via Dectin-1 and Syk. Eur J Immunol 2014;44:3729–3740. [DOI] [PubMed] [Google Scholar]
- Coll RC, Schroder K, Pelegrín P.. NLRP3 and pyroptosis blockers for treating inflammatory diseases. Trends Pharmacol Sci 2022;43:653–668. [DOI] [PubMed] [Google Scholar]
- Coman D, Coales I, Roberts LBet al. Helper-like Type-1 innate lymphoid cells in inflammatory bowel disease. Front Immunol 2022;13:903688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforti-Andreoni C, Ricciardi-Castagnoli P, Mortellaro A.. The inflammasomes in health and disease: from genetics to molecular mechanisms of autoinflammation and beyond. Cell Mol Immunol 2011;8:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney R, Baker J, Brain Oet al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med 2010;16:90–97. [DOI] [PubMed] [Google Scholar]
- Couturier-Maillard A, Secher T, Rehman Aet al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest 2013;123:700–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Ramos G, Petit CR, Marcq Iet al. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci USA 2010;107:11537–11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Wang C, Bai Wet al. CD1d1 intrinsic signaling in macrophages controls NLRP3 inflammasome expression during inflammation. Sci Adv 2020;6(43):eaaz7290. 10.1126/sciadv.aaz7290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullender TC, Chassaing B, Janzon Aet al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe 2013;14:571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JR, Cooney RM, Clarke Get al. The genetics of NOD-like receptors in Crohn’s disease. Tissue Antigens 2010;76:48–56. [DOI] [PubMed] [Google Scholar]
- D’Aldebert E, Biyeyeme Bi Mve MJ, Mergey Met al. Bile salts control the antimicrobial peptide cathelicidin through nuclear receptors in the human biliary epithelium. Gastroenterology 2009;136:1435–1443. [DOI] [PubMed] [Google Scholar]
- Dang EV, McDonald JG, Russell DWet al. Oxysterol restraint of cholesterol synthesis prevents AIM2 inflammasome activation. Cell 2017;171:1057–1071.e1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMoss RD, Moser K.. Tryptophanase in diverse bacterial species. J Bacteriol 1969;98:167–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depommier C, Everard A, Druart Cet al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med 2019;25:1096–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Salvo C, Buela KA, Creyns Bet al. NOD2 drives early IL-33-dependent expansion of group 2 innate lymphoid cells during Crohn’s disease-like ileitis. J Clin Invest 2021;131(5):e140624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vadder F, Kovatcheva-Datchary P, Goncalves Det al. Microbiota-generated metabolites promote metabolic benefits via gut–brain neural circuits. Cell 2014;156:84–96. [DOI] [PubMed] [Google Scholar]
- de Zoete MR, Flavell RA.. Interactions between nod-like receptors and intestinal bacteria. Front Immunol 2013;4:462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zoete MR, Palm NW, Zhu Set al. Inflammasomes. Cold Spring Harb Perspect Biol 2014;6:a016287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Díaz CJ, Ronnekleiv-Kelly SM, Nukaya Met al. The Aryl Hydrocarbon Receptor is a repressor of inflammation-associated colorectal tumorigenesis in mouse. Ann Surg 2016;264:429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinan TG, Stilling RM, Stanton Cet al. Collective unconscious: how gut microbes shape human behavior. J Psychiatr Res 2015;63:1–9. [DOI] [PubMed] [Google Scholar]
- Ding L, Sousa KM, Jin Let al. Vertical sleeve gastrectomy activates GPBAR-1/TGR5 to sustain weight loss, improve fatty liver, and remit insulin resistance in mice. Hepatology 2016;64:760–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docampo MD, da Silva MB, Lazrak Aet al. Alloreactive T cells deficient of the short-chain fatty acid receptor GPR109A induce less graft-versus-host disease. Blood 2022;139:2392–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Chen Y, Shi Yet al. 1,25-Dihydroxyvitamin D protects intestinal epithelial barrier by regulating the Myosin Light Chain Kinase Signaling Pathway. Inflamm Bowel Dis 2015;21:2495–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan JL, He HQ, Yu Yet al. E3 ligase c-Cbl regulates intestinal inflammation through suppressing fungi-induced noncanonical NF-κB activation. Sci Adv 2021;7(19):eabe5171. 10.1126/sciadv.abe5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau ALet al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 2011;145:745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson CO, Alexander KL.. Host–microbiota interactions in the intestine. Dig Dis 2015;33:131–136. [DOI] [PubMed] [Google Scholar]
- Eriksson M, Johannssen T, von Smolinski Det al. The C-Type lectin receptor SIGNR3 binds to fungi present in commensal microbiota and influences immune regulation in experimental colitis. Front Immunol 2013;4:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshleman EM, Alenghat T.. Epithelial sensing of microbiota-derived signals. Genes Immun 2021;22:237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasan S, Muramatsu M, Suzuki Ket al. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science 2002;298:1424–1427. [DOI] [PubMed] [Google Scholar]
- Fang S, Suh JM, Reilly SMet al. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat Med 2015;21:159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Yan C, Zhao Qet al. The association between gut microbiota, toll-like receptors, and colorectal cancer. Clin Med Insights Oncol 2022;16:11795549221130549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann J, Prieur AM, Quartier Pet al. Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am J Hum Genet 2002;71:198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Barral A, Costales-Carrera A, Buira SPet al. Vitamin D differentially regulates colon stem cells in patient-derived normal and tumor organoids. FEBS J 2020;287:53–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JC, Bscheider M, Eisenkolb Get al. RIG-I/MAVS and STING signaling promote gut integrity during irradiation- and immune-mediated tissue injury. Sci Transl Med 2017;9(386):eaag2513. 10.1126/scitranslmed.aag2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannigan KL, Ngo VL, Geem Det al. IL-17A-mediated neutrophil recruitment limits expansion of segmented filamentous bacteria. Mucosal Immunol 2017;10:673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Amer A, Body-Malapel Met al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol 2006;7:576–582. [DOI] [PubMed] [Google Scholar]
- Franchi L, Kamada N, Nakamura Yet al. NLRC4-driven production of IL-1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol 2012;13:449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Hennenberg EM, Eyking Aet al. TLR signaling modulates side effects of anticancer therapy in the small intestine. J Immunol 2015;194:1983–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Hsu T, Sirota-Madi Aet al. Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nat Rev Microbiol 2015;13:360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland RP. Mechanisms of molecular mimicry involving the microbiota in neurodegeneration. J Alzheimers Dis 2015;45:349–362. [DOI] [PubMed] [Google Scholar]
- Fritz JH, Ferrero RL, Philpott DJet al. Nod-like proteins in immunity, inflammation and disease. Nat Immunol 2006;7:1250–1257. [DOI] [PubMed] [Google Scholar]
- Frosali S, Pagliari D, Gambassi Get al. How the intricate interaction among toll-like receptors, microbiota, and intestinal immunity can influence gastrointestinal pathology. J Immunol Res 2015;2015:489821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu T, Coulter S, Yoshihara Eet al. FXR regulates intestinal cancer stem cell proliferation. Cell 2019;176:1098–1112.e1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara H, Docampo MD, Riwes Met al. Microbial metabolite sensor GPR43 controls severity of experimental GVHD. Nat Commun 2018;9:3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata M, Michelsen KS, Eri Ret al. Toll-like receptor-4 is required for intestinal response to epithelial injury and limiting bacterial translocation in a murine model of acute colitis. Am J Physiol Gastrointest Liver Physiol 2005;288:G1055–G1065. [DOI] [PubMed] [Google Scholar]
- Fukata M, Breglio K, Chen Aet al. The myeloid differentiation factor 88 (MyD88) is required for CD4+ T cell effector function in a murine model of inflammatory bowel disease. J Immunol 2008;180:1886–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulde M, Sommer F, Chassaing Bet al. Neonatal selection by Toll-like receptor 5 influences long-term gut microbiota composition. Nature 2018;560:489–493. [DOI] [PubMed] [Google Scholar]
- Furman D, Chang J, Lartigue Let al. Expression of specific inflammasome gene modules stratifies older individuals into two extreme clinical and immunological states. Nat Med 2017;23:174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda Set al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013;504:446–450. [DOI] [PubMed] [Google Scholar]
- Gadaleta RM, van Erpecum KJ, Oldenburg Bet al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 2011;60:463–472. [DOI] [PubMed] [Google Scholar]
- Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol 2003;3:710–720. [DOI] [PubMed] [Google Scholar]
- Gao C, Qiao T, Zhang Bet al. TLR9 signaling activation at different stages in colorectal cancer and NF-kappaB expression. Onco Targets Ther 2018;11:5963–5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Luo Z, Ji Yet al. Accumulation of microbial DNAs promotes to islet inflammation and β cell abnormalities in obesity in mice. Nat Commun 2022a;13:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Zhao X, Hu Set al. Gut microbial dl-endopeptidase alleviates Crohn’s disease via the NOD2 pathway. Cell Host Microbe 2022b;30:1435–1449.e9. [DOI] [PubMed] [Google Scholar]
- Gao H, Zhou H, Zhang Zet al. Vitamin D3 alleviates inflammation in ulcerative colitis by activating the VDR-NLRP6 signaling pathway. Front Immunol 2023;14:1135930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman AE, LaRosa DF, Zhang Jet al. The adaptor molecule MyD88 activates PI-3 kinase signaling in CD4+ T cells and enables CpG oligodeoxynucleotide-mediated costimulation. Immunity 2006;25:783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geremia A, Biancheri P, Allan Pet al. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun Rev 2014;13:3–10. [DOI] [PubMed] [Google Scholar]
- Geuking MB, Cahenzli J, Lawson MAet al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 2011;34:794–806. [DOI] [PubMed] [Google Scholar]
- Gewirtz AT, Navas TA, Lyons Set al. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol 2001;167:1882–1885. [DOI] [PubMed] [Google Scholar]
- Ghimire L, Paudel S, Jin Let al. NLRP6 negatively regulates pulmonary host defense in Gram-positive bacterial infection through modulating neutrophil recruitment and function. PLoS Pathog 2018;14:e1007308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles DA, Moreno-Fernandez ME, Stankiewicz TEet al. Thermoneutral housing exacerbates nonalcoholic fatty liver disease in mice and allows for sex-independent disease modeling. Nat Med 2017;23:829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Carneiro LAet al. Nod1 detects a unique muropeptide from Gram-negative bacterial peptidoglycan. Science 2003a;300:1584–1587. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Viala Jet al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem 2003b;278:8869–8872. [DOI] [PubMed] [Google Scholar]
- Goettel JA, Gandhi R, Kenison JEet al. AHR activation is protective against colitis driven by T Cells in humanized mice. Cell Rep 2016;17:1318–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan MA, Liu W, Shi Yet al. Transgenic expression of Vitamin D receptor in gut epithelial cells ameliorates spontaneous colitis caused by Interleukin-10 deficiency. Dig Dis Sci 2015;60:1941–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V, Helmink BA, Spencer CNet al. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell 2018;33:570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorecki AM, Bakeberg MC, Theunissen Fet al. Single nucleotide polymorphisms associated with gut homeostasis influence risk and age-at-onset of Parkinson’s Disease. Front Aging Neurosci 2020;12:603849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor MF, Hotamisligil GS.. Inflammatory mechanisms in obesity. Annu Rev Immunol 2011;29:415–445. [DOI] [PubMed] [Google Scholar]
- Gribble FM, Reimann F.. Function and mechanisms of enteroendocrine cells and gut hormones in metabolism. Nat Rev Endocrinol 2019;15:226–237. [DOI] [PubMed] [Google Scholar]
- Gronke K, Hernández PP, Zimmermann Jet al. Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature 2019;566:249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Group NHW, Peterson J, Garges Set al. The NIH Human Microbiome Project. Genome Res 2009;19:2317–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Sun Y, Liu Wet al. Small molecule-driven mitophagy-mediated NLRP3 inflammasome inhibition is responsible for the prevention of colitis-associated cancer. Autophagy 2014;10:972–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Gibson SA, Ting JPY.. Gut microbiota, NLR proteins, and intestinal homeostasis. J Exp Med 2020;217(10):e20181832. 10.1084/jem.20181832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gury-BenAri M, Thaiss CA, Serafini Net al. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 2016;166:1231–1246.e1213. [DOI] [PubMed] [Google Scholar]
- Hackam DJ, Sodhi CP.. Bench to bedside—new insights into the pathogenesis of necrotizing enterocolitis. Nat Rev Gastroenterol Hepatol 2022;19:468–479. [DOI] [PubMed] [Google Scholar]
- Hampe J, Cuthbert A, Croucher PJet al. Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet 2001;357:1925–1928. [DOI] [PubMed] [Google Scholar]
- Hang S, Paik D, Yao Let al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature 2019;576:143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H, Seregin SS, Yang Det al. The NLRP6 inflammasome recognizes lipoteichoic acid and regulates gram-positive pathogen infection. Cell 2018;175:1651–1664.e1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harapas CR, Idiiatullina E, Al-Azab Met al. Organellar homeostasis and innate immune sensing. Nat Rev Immunol 2022;22:535–549. [DOI] [PubMed] [Google Scholar]
- Hartmann P, Haimerl M, Mazagova Met al. Toll-like receptor 2-mediated intestinal injury and enteric tumor necrosis factor receptor I contribute to liver fibrosis in mice. Gastroenterology 2012;143:1330–1340.e1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Yang K, Hashimoto Met al. Differential release and distribution of Nod1 and Nod2 immunostimulatory molecules among bacterial species and environments. J Biol Chem 2006;281:29054–29063. [DOI] [PubMed] [Google Scholar]
- Hausmann M, Kiessling S, Mestermann Set al. Toll-like receptors 2 and 4 are up-regulated during intestinal inflammation. Gastroenterology 2002;122:1987–2000. [DOI] [PubMed] [Google Scholar]
- He L, Liu T, Shi Yet al. Gut epithelial Vitamin D receptor regulates microbiota-dependent mucosal inflammation by suppressing intestinal epithelial cell apoptosis. Endocrinology 2018;159:967–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Ma Y, Yang Set al. Gut microbiota-derived ursodeoxycholic acid from neonatal dairy calves improves intestinal homeostasis and colitis to attenuate extended-spectrum β-lactamase-producing enteroaggregative Escherichia coli infection. Microbiome 2022;10:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimesaat MM, Nogai A, Bereswill Set al. MyD88/TLR9 mediated immunopathology and gut microbiota dynamics in a novel murine model of intestinal graft-versus-host disease. Gut 2010;59:1079–1087. [DOI] [PubMed] [Google Scholar]
- Heinsbroek SE, Oei A, Roelofs JJet al. Genetic deletion of dectin-1 does not affect the course of murine experimental colitis. BMC Gastroenterol 2012;12:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Mejia J, Elinav E, Jin Cet al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 2012;482:179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy C, O’Connell S, Egan LJet al. Inhibition of anti-viral responses in intestinal epithelial cells by epigenetic modifying drugs is mediated by a reduction in viral pattern recognition receptor expression and activity. Immunopharmacol Immunotoxicol 2019;41:527–537. [DOI] [PubMed] [Google Scholar]
- Higuchi S, Ahmad TR, Argueta DAet al. Bile acid composition regulates GPR119-dependent intestinal lipid sensing and food intake regulation in mice. Gut 2020;69:1620–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa A, Tsumaya K, Awaji Tet al. Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nat Med 2005;11:90–94. [DOI] [PubMed] [Google Scholar]
- Hirota SA, Ng J, Lueng Aet al. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm Bowel Dis 2011;17:1359–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman HM, Mueller JL, Broide DHet al. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat Genet 2001;29:301–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörmann N, Brandão I, Jäckel Set al. Gut microbial colonization orchestrates TLR2 expression, signaling and epithelial proliferation in the small intestinal mucosa. PLoS One 2014;9:e113080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horuluoglu BH, Kayraklioglu N, Tross Det al. PAM3 protects against DSS-induced colitis by altering the M2:M1 ratio. Sci Rep 2020;10:6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Q, Ye L, Liu Het al. Lactobacillus accelerates ISCs regeneration to protect the integrity of intestinal mucosa through activation of STAT3 signaling pathway induced by LPLs secretion of IL-22. Cell Death Differ 2018;25:1657–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt MR, Lavoie S, Michaud Met al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 2016;351:1329–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Elinav E, Huber Set al. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Proc Natl Acad Sci USA 2010;107:21635–21640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Peng L, Kwak YTet al. The DNA sensor AIM2 maintains intestinal homeostasis via regulation of epithelial antimicrobial host defense. Cell Rep 2015;13:1922–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Jin C, Li HBet al. The DNA-sensing AIM2 inflammasome controls radiation-induced cell death and tissue injury. Science 2016;354:765–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Fang Y, Chen Xet al. cGAS restricts colon cancer development by protecting intestinal barrier integrity. Proc Natl Acad Sci USA 2021;118(23):e2105747118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Fang M, Jostins Let al. International Inflammatory Bowel Disease Genetics Consortium. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 2017;547:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Hong JY, Wu YJet al. Vitamin D receptor interacts with NLRP3 to restrict the allergic response. Clin Exp Immunol 2018;194:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Feng Z, Jiang Yet al. VSIG4 mediates transcriptional inhibition of Nlrp3 and Il-1β in macrophages. Sci Adv 2019;5:eaau7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TD, Murray IA, Perdew GH.. Indole and tryptophan metabolism: endogenous and dietary routes to Ah receptor activation. Drug Metab Dispos 2015;43:1522–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali Het al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001;411:599–603. [DOI] [PubMed] [Google Scholar]
- Huhta H, Helminen O, Kauppila JHet al. The expression of Toll-like receptors in normal human and murine gastrointestinal organs and the effect of microbiome and cancer. J Histochem Cytochem 2016;64:470–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo X, Li D, Wu Fet al. Cultivated human intestinal fungus Candida metapsilosis M2006B attenuates colitis by secreting acyclic sesquiterpenoids as FXR agonists. Gut 2022;71:2205–2217. [DOI] [PubMed] [Google Scholar]
- Huus KE, Bauer KC, Brown EMet al. Commensal bacteria modulate immunoglobulin a binding in response to host nutrition. Cell Host Microbe 2020;27:909–921.e905. [DOI] [PubMed] [Google Scholar]
- Ichimura A, Hirasawa A, Poulain-Godefroy Oet al. Dysfunction of lipid sensor GPR120 leads to obesity in both mouse and human. Nature 2012;483:350–354. [DOI] [PubMed] [Google Scholar]
- Iliev ID, Funari VA, Taylor KDet al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012;336:1314–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura R, Wang Y, Kinoshita Tet al. Anti-inflammatory activity of PYNOD and its mechanism in humans and mice. J Immunol 2010;184:5874–5884. [DOI] [PubMed] [Google Scholar]
- Inagaki T, Moschetta A, Lee YKet al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci USA 2006;103:3920–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismailova A, White JH.. Vitamin D, infections and immunity. Rev Endocr Metab Disord 2022;23:265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R.. Regulation of adaptive immunity by the innate immune system. Science 2010;327:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer SS, Gensollen T, Gandhi Aet al. Dietary and microbial Oxazoles induce intestinal inflammation by modulating aryl hydrocarbon receptor responses. Cell 2018;173:1123–1134.e1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain U, Ver Heul AM, Xiong Set al. Debaryomyces is enriched in Crohn’s disease intestinal tissue and impairs healing in mice. Science 2021;371:1154–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA Jr., Medzhitov R.. Innate immune recognition. Annu Rev Immunol 2002;20:197–216. [DOI] [PubMed] [Google Scholar]
- Janney A, Powrie F, Mann EH.. Host-microbiota maladaptation in colorectal cancer. Nature 2020;585:509–517. [DOI] [PubMed] [Google Scholar]
- Jia DJ, Wang QW, Hu YYet al. Lactobacillus johnsonii alleviates colitis by TLR1/2-STAT3 mediated CD206(+) macrophages(IL-10) activation. Gut Microbes 2022;14:2145843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Xie C, Lv Yet al. Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction. Nat Commun 2015;6:10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HY, Najmeh S, Martel Get al. Activation of the pattern recognition receptor NOD1 augments colon cancer metastasis. Protein Cell 2020;11:187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RKet al. International IBD Genetics Consortium (IIBDGC). Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabelitz D. Expression and function of Toll-like receptors in T lymphocytes. Curr Opin Immunol 2007;19:39–45. [DOI] [PubMed] [Google Scholar]
- Kaiko GE, Ryu SH, Koues OIet al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell 2016;165:1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantari P, DeOliveira RB, Chan Jet al. Dual engagement of the NLRP3 and AIM2 inflammasomes by plasmodium-derived hemozoin and DNA during malaria. Cell Rep 2014;6:196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki R, Man SM, Malireddi RKSet al. NLRC3 is an inhibitory sensor of PI3K-mTOR pathways in cancer. Nature 2016;540:583–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki R, Malireddi RKS, Zhu Qet al. NLRC3 regulates cellular proliferation and apoptosis to attenuate the development of colorectal cancer. Cell Cycle 2017;16:1243–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi I, Morita R, Schichita Tet al. Smad2 and Smad3 inversely regulate TGF-β autoinduction in Clostridium butyricum-activated dendritic cells. Immunity 2015;43:65–79. [DOI] [PubMed] [Google Scholar]
- Kataoka K, Muta T, Yamazaki Set al. Activation of macrophages by linear (1right-arrow3)-beta-d-glucans. Implications for the recognition of fungi by innate immunity. J Biol Chem 2002;277:36825–36831. [DOI] [PubMed] [Google Scholar]
- Kato LM, Kawamoto S, Maruya Met al. The role of the adaptive immune system in regulation of gut microbiota. Immunol Rev 2014;260:67–75. [DOI] [PubMed] [Google Scholar]
- Kau AL, Planer JD, Liu Jet al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med 2015;7:276ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto S, Maruya M, Kato LMet al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 2014;41:152–165. [DOI] [PubMed] [Google Scholar]
- Kawashima T, Kosaka A, Yan Het al. Double-stranded RNA of intestinal commensal but not pathogenic bacteria triggers production of protective interferon-β. Immunity 2013;38:1187–1197. [DOI] [PubMed] [Google Scholar]
- Kaye DM, Shihata WA, Jama HAet al. Deficiency of prebiotic fiber and insufficient signaling through gut metabolite-sensing receptors leads to cardiovascular disease. Circulation 2020;141:1393–1403. [DOI] [PubMed] [Google Scholar]
- Keogh CE, Rude KM, Gareau MG.. Role of pattern recognition receptors and the microbiota in neurological disorders. J Physiol 2021;599:1379–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesselring R, Glaesner J, Hiergeist Aet al. IRAK-M expression in tumor cells supports colorectal cancer progression through reduction of antimicrobial defense and stabilization of STAT3. Cancer Cell 2016;29:684–696. [DOI] [PubMed] [Google Scholar]
- Khor B, Gardet A, Xavier RJ.. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011;474:307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KA, Gu W, Lee IAet al. High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PLoS One 2012;7:e47713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Kang SG, Park JHet al. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 2013;145:396–406.e1. [DOI] [PubMed] [Google Scholar]
- Kim YK, Shin JS, Nahm MH.. NOD-Like receptors in infection, immunity, and diseases. Yonsei Med J 2016;57:5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Lee JC, Im SHet al. Amelioration of autoimmune diabetes of NOD mice by immunomodulating probiotics. Front Immunol 2020;11:1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura I, Miyamoto J, Ohue-Kitano Ret al. Maternal gut microbiota in pregnancy influences offspring metabolic phenotype in mice. Science 2020;367(6481):eaaw8429. 10.1126/science.aaw8429 [DOI] [PubMed] [Google Scholar]
- Kiss EA, Vonarbourg C, Kopfmann Set al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 2011;334:1561–1565. [DOI] [PubMed] [Google Scholar]
- Kofoed EM, Vance RE.. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 2011;477:592–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo JE, Shin SW, Um SHet al. X-shaped DNA potentiates therapeutic efficacy in colitis-associated colon cancer through dual activation of TLR9 and inflammasomes. Mol Cancer 2015;14:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovler ML, Gonzalez Salazar AJ, Fulton WBet al. Toll-like receptor 4-mediated enteric glia loss is critical for the development of necrotizing enterocolitis. Sci Transl Med 2021;13:eabg3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S, Ding Y, Saedi Net al. Gut microbiota-derived tryptophan metabolites modulate inflammatory response in hepatocytes and macrophages. Cell Rep 2018;23:1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglov AA, Grivennikov SI, Kuprash DVet al. Nonredundant function of soluble LTalpha3 produced by innate lymphoid cells in intestinal homeostasis. Science 2013;342:1243–1246. [DOI] [PubMed] [Google Scholar]
- Kubinak JL, Petersen C, Stephens WZet al. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe 2015;17:153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunac N, Degoricija M, Viculin Jet al. Activation of cGAS-STING Pathway Is associated with MSI-H Stage IV colorectal cancer. Cancers (Basel) 2022;15(1):221. 10.3390/cancers15010221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo WT, Lee TC, Yang HYet al. LPS receptor subunits have antagonistic roles in epithelial apoptosis and colonic carcinogenesis. Cell Death Differ 2015;22:1590–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lala S, Ogura Y, Osborne Cet al. Crohn’s disease and the NOD2 gene: a role for paneth cells. Gastroenterology 2003;125:47–57. [DOI] [PubMed] [Google Scholar]
- Lamas B, Richard ML, Leducq Vet al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 2016;22:598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanis JM, Alexeev EE, Curtis VFet al. Tryptophan metabolite activation of the aryl hydrocarbon receptor regulates IL-10 receptor expression on intestinal epithelia. Mucosal Immunol 2017;10:1133–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latacz M, Rozmus D, Fiedorowicz Eet al. Vitamin D Receptor (VDR) gene polymorphism in patients diagnosed with colorectal cancer. Nutrients 2021;13:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop SK, Bloom SM, Rao SMet al. Peripheral education of the immune system by colonic commensal microbiota. Nature 2011;478:250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle A, Sokol H.. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 2020;17:223–237. [DOI] [PubMed] [Google Scholar]
- Lax S, Schauer G, Prein Ket al. Expression of the nuclear bile acid receptor/farnesoid X receptor is reduced in human colon carcinoma compared to non-neoplastic mucosa independent from site and may be associated with adverse prognosis. Int J Cancer 2012;130:2232–2239. [DOI] [PubMed] [Google Scholar]
- Layunta E, Latorre E, Forcén Ret al. NOD1 downregulates intestinal serotonin transporter and interacts with other pattern recognition receptors. J Cell Physiol 2018;233:4183–4193. [DOI] [PubMed] [Google Scholar]
- Lee J, Mo JH, Katakura Ket al. Maintenance of colonic homeostasis by distinctive apical TLR9 signalling in intestinal epithelial cells. Nat Cell Biol 2006;8:1327–1336. [DOI] [PubMed] [Google Scholar]
- Lee GS, Subramanian N, Kim AIet al. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature 2012;492:123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz BJ, Zhao G, Wei Let al. Interferon b drives intestinal regeneration after radiation. Sci Adv 2021;7:eabi5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite JA, Pessenda G, Guerra-Gomes ICet al. The DNA sensor AIM2 protects against streptozotocin-induced Type 1 diabetes by regulating intestinal homeostasis via the IL-18 pathway. Cells 2020;9:959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire P, Robertson SJ, Maughan Het al. The NLR protein NLRP6 does not impact gut microbiota composition. Cell Rep 2017;21:3653–3661. [DOI] [PubMed] [Google Scholar]
- Leng F, Yin H, Qin Set al. NLRP6 self-assembles into a linear molecular platform following LPS binding and ATP stimulation. Sci Rep 2020;10:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi I, Gao IH, Lin WYet al. Mucosal fungi promote gut barrier function and social behavior via Type 17 immunity. Cell 2022;185:831–846.e814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letran SE, Lee SJ, Atif SMet al. TLR5-deficient mice lack basal inflammatory and metabolic defects but exhibit impaired CD4 T cell responses to a flagellated pathogen. J Immunol 2011;186:5406–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Thaiss CA, Zeevi Det al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 2015;163:1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Thaiss CA, Elinav E.. Metabolites: messengers between the microbiota and the immune system. Genes Dev 2016;30:1589–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Chiang JY.. Bile acid signaling in metabolic disease and drug therapy. Pharmacol Rev 2014;66:948–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Wu M.. Pattern recognition receptors in health and diseases. Signal Transduct Target Ther 2021;6:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhu S.. NLRP6 inflammasome. Mol Aspects Med 2020;76:100859. [DOI] [PubMed] [Google Scholar]
- Li XD, Chiu YH, Ismail ASet al. Mitochondrial antiviral signaling protein (MAVS) monitors commensal bacteria and induces an immune response that prevents experimental colitis. Proc Natl Acad Sci USA 2011a;108:17390–17395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Innocentin S, Withers DRet al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 2011b;147:629–640. [DOI] [PubMed] [Google Scholar]
- Li XX, Sun GP, Meng Jet al. Role of toll-like receptor 4 in colorectal carcinogenesis: a meta-analysis. PLoS One 2014;9:e93904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Bostick JW, Ye Jet al. Aryl hydrocarbon receptor signaling cell intrinsically inhibits intestinal Group 2 innate lymphoid cell function. Immunity 2018;49:915–928.e915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JM, Yu R, Zhang LPet al. Dietary fructose-induced gut dysbiosis promotes mouse hippocampal neuroinflammation: a benefit of short-chain fatty acids. Microbiome 2019a;7:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhou R, Wang Het al. Gut microbiota-stimulated cathepsin K secretion mediates TLR4-dependent M2 macrophage polarization and promotes tumor metastasis in colorectal cancer. Cell Death Differ 2019b;26:2447–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Deng M, Petrucelli ASet al. Viral DNA binding to NLRC3, an inhibitory nucleic acid sensor, unleashes STING, a cyclic dinucleotide receptor that activates Type I interferon. Immunity 2019c;50:591–599.e596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Song A, Yu H.. Interaction between toll-like receptor 4 (TLR4) gene and alcohol drinking on Parkinson’s disease risk in Chinese Han population. J Clin Neurosci 2019d;62:128–132. [DOI] [PubMed] [Google Scholar]
- Li R, Zan Y, Sui Ket al. The latest breakthrough on NLRP6 inflammasome. Precis Clin Med 2022a;5:pbac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XV, Leonardi I, Putzel GGet al. Immune regulation by fungal strain diversity in inflammatory bowel disease. Nature 2022b;603:672–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Q, Xu J, Yan Set al. Chemotherapy-induced intestinal inflammatory responses are mediated by exosome secretion of double-strand DNA via AIM2 inflammasome activation. Cell Res 2017;27:784–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot YL, Selle K, Yang Tet al. SIGNR3-dependent immune regulation by Lactobacillus acidophilus surface layer protein A in colitis. EMBO J 2015;34:881–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JD, Devlin JC, Yeung Fet al. Rewilding Nod2 and Atg16l1 mutant mice uncovers genetic and environmental contributions to microbial responses and immune cell composition. Cell Host Microbe 2020;27:830–840.e834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm HT, Parmar N, Drurey Cet al. BMP signaling in the intestinal epithelium drives a critical feedback loop to restrain IL-13-driven tuft cell hyperplasia. Sci Immunol 2022;7:eabl6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cao X.. Cellular and molecular regulation of innate inflammatory responses. Cell Mol Immunol 2016;13:711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Komai-Koma M, Xu Det al. Toll-like receptor 2 signaling modulates the functions of CD4+ CD25+ regulatory T cells. Proc Natl Acad Sci USA 2006;103:7048–7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, van Sommeren S, Huang Het al. International Multiple Sclerosis Genetics Consortium. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015a;47:979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Truax AD, Chen Let al. Expression profile of innate immune receptors, NLRs and AIM2, in human colorectal cancer: correlation with cancer stages and inflammasome components. Oncotarget 2015b;6:33456–33469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Gong T, Tao Wet al. Commensal viruses maintain intestinal intraepithelial lymphocytes via noncanonical RIG-I signaling. Nat Immunol 2019;20:1681–1691. [DOI] [PubMed] [Google Scholar]
- Liu L, Liu Z, Li Het al. Naturally occurring TPE-CA maintains gut microbiota and bile acids homeostasis via FXR signaling modulation of the liver–gut axis. Front Pharmacol 2020;11:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li C, Wan Zet al. Analogous comparison unravels heightened antiviral defense and boosted viral infection upon immunosuppression in bat organoids. Signal Transduct Target Ther 2022a;7:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yang M, Tang Let al. TLR4 regulates RORγt(+) regulatory T-cell responses and susceptibility to colon inflammation through interaction with Akkermansia muciniphila. Microbiome 2022b;10:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Price J, Arze C, Ananthakrishnan ANet al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodes MJ, Cong Y, Elson COet al. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest 2004;113:1296–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Haber C, Netting DJ, Hutchins Zet al. The phagosomal solute transporter SLC15A4 promotes inflammasome activity via mTORC1 signaling and autophagy restraint in dendritic cells. EMBO J 2022;41:e111161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotz M, Gütle D, Walther Set al. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J Exp Med 2006;203:973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Fan C, Li Pet al. Short chain fatty acids prevent high-fat-diet-induced obesity in mice by regulating G protein-coupled receptors and gut microbiota. Sci Rep 2016;6:37589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Shang M, Zhang YGet al. Lactic acid bacteria isolated From Korean Kimchi activate the Vitamin D receptor-autophagy signaling pathways. Inflamm Bowel Dis 2020;26:1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Zhang YG, Xia Yet al. Paneth cell alertness to pathogens maintained by Vitamin D receptors. Gastroenterology 2021;160:1269–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Xu X, Fu Det al. Butyrate-producing Eubacterium rectale suppresses lymphomagenesis by alleviating the TNF-induced TLR4/MyD88/NF-κB axis. Cell Host Microbe 2022;30:1139–1150.e1137. [DOI] [PubMed] [Google Scholar]
- Lukasova M, Malaval C, Gille Aet al. Nicotinic acid inhibits progression of atherosclerosis in mice through its receptor GPR109A expressed by immune cells. J Clin Invest 2011;121:1163–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens JR, Gurung P, Shaw PJet al. The NLRP12 sensor negatively regulates autoinflammatory disease by modulating Interleukin-4 production in T cells. Immunity 2015;42:654–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q, Zeng L, Tang Cet al. TLR9 induces colitis-associated colorectal carcinogenesis by regulating NF-κB expression levels. Oncol Lett 2020;20:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyte M. Probiotics function mechanistically as delivery vehicles for neuroactive compounds: microbial endocrinology in the design and use of probiotics. Bioessays 2011;33:574–581. [DOI] [PubMed] [Google Scholar]
- Macia L, Tan J, Vieira ATet al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun 2015;6:6734. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Uhr T.. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 2004;303:1662–1665. [DOI] [PubMed] [Google Scholar]
- Magalhaes JG, Fritz JH, Le Bourhis Let al. Nod2-dependent Th2 polarization of antigen-specific immunity. J Immunol 2008;181:7925–7935. [DOI] [PubMed] [Google Scholar]
- Maisonneuve C, Tsang DKL, Foerster EGet al. Nod1 promotes colorectal carcinogenesis by regulating the immunosuppressive functions of tumor-infiltrating myeloid cells. Cell Rep 2021;34:108677. [DOI] [PubMed] [Google Scholar]
- Mamantopoulos M, Ronchi F, Van Hauwermeiren Fet al. Nlrp6- and ASC-dependent inflammasomes do not shape the commensal gut microbiota composition. Immunity 2017;47:339–348.e4. [DOI] [PubMed] [Google Scholar]
- Man SM, Zhu Q, Zhu Let al. Critical role for the DNA sensor AIM2 in stem cell proliferation and cancer. Cell 2015;162:45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Xiao P, Tao XNet al. Unsaturated bond recognition leads to biased signal in a fatty acid receptor. Science 2023:380(6640):eadd6220. 10.1126/science.add6220 Epub 2023 Apr 7. [DOI] [PubMed] [Google Scholar]
- Maran RR, Thomas A, Roth Met al. Farnesoid X receptor deficiency in mice leads to increased intestinal epithelial cell proliferation and tumor development. J Pharmacol Exp Ther 2009;328:469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariño E, Richards JL, McLeod KHet al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol 2017;18:552–562. [DOI] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng Aet al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009;461:1282–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheis F, Muller PA, Graves CLet al. Adrenergic signaling in Muscularis macrophages limits infection-induced neuronal loss. Cell 2020;180:64–78.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur R, Zeng W, Hayden MSet al. Mice lacking TLR11 exhibit variable Salmonella typhi susceptibility. Cell 2016;164:829–830. [DOI] [PubMed] [Google Scholar]
- Matzinger P. The danger model: a renewed sense of self. Science 2002;296:301–305. [DOI] [PubMed] [Google Scholar]
- McAlpine W, Sun L, Wang KWet al. Excessive endosomal TLR signaling causes inflammatory disease in mice with defective SMCR8–WDR41–C9ORF72 complex function. Proc Natl Acad Sci USA 2018;115:E11523–E11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckel K, Li YC, Lim Jet al. Serum 25-hydroxyvitamin D concentration is inversely associated with mucosal inflammation in patients with ulcerative colitis. Am J Clin Nutr 2016;104:113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Janeway C Jr. Innate immune recognition: mechanisms and pathways. Immunol Rev 2000;173:89–97. [DOI] [PubMed] [Google Scholar]
- Mehto S, Jena KK, Nath Pet al. The Crohn’s Disease Risk Factor IRGM limits NLRP3 inflammasome activation by impeding its assembly and by mediating its selective autophagy. Mol Cell 2019;73:429–445.e427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez A, Willing BP, Montero Met al. Bacterial stimulation of the TLR-MyD88 pathway modulates the homeostatic expression of ileal Paneth cell alpha-defensins. J Innate Immun 2013;5:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng S, Li Y, Zang Xet al. Effect of TLR2 on the proliferation of inflammation-related colorectal cancer and sporadic colorectal cancer. Cancer Cell Int 2020;20:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell MA, Ilvesaro JM, Lehtonen Net al. Toll-like receptor 9 agonists promote cellular invasion by increasing matrix metalloproteinase activity. Mol Cancer Res 2006;4:437–447. [DOI] [PubMed] [Google Scholar]
- Messaritakis I, Koulouridi A, Sfakianaki Met al. The role of Vitamin D receptor gene polymorphisms in colorectal cancer risk. Cancers (Basel) 2020;12(6):1379. 10.3390/cancers12061379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaritakis I, Koulouridi A, Boukla Eet al. Investigation of microbial translocation, TLR and VDR gene polymorphisms, and recurrence risk in Stage III colorectal cancer patients. Cancers (Basel) 2022;14(18):4407. 10.3390/cancers14184407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metidji A, Omenetti S, Crotta Set al. The environmental sensor AHR protects from inflammatory damage by maintaining intestinal stem cell homeostasis and barrier integrity. Immunity 2018;49:353–362.e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miani M, Le Naour J, Waeckel-Enée Eet al. Gut microbiota-stimulated innate lymphoid cells support β-defensin 14 expression in pancreatic endocrine cells, preventing autoimmune diabetes. Cell Metab 2018;28:557–572.e556. [DOI] [PubMed] [Google Scholar]
- Miao EA, Alpuche-Aranda CM, Dors Met al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol 2006;7:569–575. [DOI] [PubMed] [Google Scholar]
- Miao EA, Mao DP, Yudkovsky Net al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci USA 2010;107:3076–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y, Oka A, Liu Bet al. Microbiota maintain colonic homeostasis by activating TLR2/MyD88/PI3K signaling in IL-10-producing regulatory B cells. J Clin Invest 2019;129:3702–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi E, Shimokawa C, Steimle Aet al. The impact of the gut microbiome on extra-intestinal autoimmune diseases. Nat Rev Immunol 2023;23:9–23. [DOI] [PubMed] [Google Scholar]
- Mobraten K, Haug TM, Kleiveland CRet al. Omega-3 and omega-6 PUFAs induce the same GPR120-mediated signalling events, but with different kinetics and intensity in Caco-2 cells. Lipids Health Dis 2013;12:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modica S, Murzilli S, Salvatore Let al. Nuclear bile acid receptor FXR protects against intestinal tumorigenesis. Cancer Res 2008;68:9589–9594. [DOI] [PubMed] [Google Scholar]
- Modica S, Gofflot F, Murzilli Set al. The intestinal nuclear receptor signature with epithelial localization patterns and expression modulation in tumors. Gastroenterology 2010;138:636–48, 648.e631–612. [DOI] [PubMed] [Google Scholar]
- Molofsky AB, Byrne BG, Whitfield NNet al. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med 2006;203:1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone I, Rizzo A, Sarra Met al. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology 2011;141:237–48, 248.e231. [DOI] [PubMed] [Google Scholar]
- Moon C, Stappenbeck TS.. Viral interactions with the host and microbiota in the intestine. Curr Opin Immunol 2012;24:405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradi-Marjaneh R, Hassanian SM, Fiuji Het al. Toll like receptor signaling pathway as a potential therapeutic target in colorectal cancer. J Cell Physiol 2018;233:5613–5622. [DOI] [PubMed] [Google Scholar]
- Mori Y, Yin J, Rashid Aet al. Instabilotyping: comprehensive identification of frameshift mutations caused by coding region microsatellite instability. Cancer Res 2001;61:6046–6049. [PubMed] [Google Scholar]
- Morland B, Midtvedt T.. Phagocytosis, peritoneal influx, and enzyme activities in peritoneal macrophages from germfree, conventional, and ex-germfree mice. Infect Immun 1984;44:750–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortha A, Chudnovskiy A, Hashimoto Det al. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science 2014;343:1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat AM, Agace WW.. Regional specialization within the intestinal immune system. Nat Rev Immunol 2014;14:667–685. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Kumar R, Tsakem Lenou Eet al. Deubiquitination of NLRP6 inflammasome by Cyld critically regulates intestinal inflammation. Nat Immunol 2020;21:626–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Matheis F, Schneeberger Met al. Microbiota-modulated CART(+) enteric neurons autonomously regulate blood glucose. Science 2020;370:314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münzker J, Haase N, Till Aet al. Functional changes of the gastric bypass microbiota reactivate thermogenic adipose tissue and systemic glucose control via intestinal FXR-TGR5 crosstalk in diet-induced obesity. Microbiome 2022;10:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naama M, Telpaz S, Awad Aet al. Autophagy controls mucus secretion from intestinal goblet cells by alleviating ER stress. Cell Host Microbe 2023;31:433–446.e434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadjsombati MS, McGinty JW, Lyons-Cohen MRet al. Detection of succinate by intestinal tuft cells triggers a Type 2 innate immune circuit. Immunity 2018;49:33–41.e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Kurihara S, Takahashi Det al. Symbiotic polyamine metabolism regulates epithelial proliferation and macrophage differentiation in the colon. Nat Commun 2021;12:2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natividad JM, Agus A, Planchais Jet al. Impaired aryl hydrocarbon receptor ligand production by the gut microbiota is a key factor in metabolic syndrome. Cell Metab 2018;28:737–749.e734. [DOI] [PubMed] [Google Scholar]
- Nayar S, Morrison JK, Giri Met al. A myeloid-stromal niche and gp130 rescue in NOD2-driven Crohn’s disease. Nature 2021;593:275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal MD, Leaphart C, Levy Ret al. Enterocyte TLR4 mediates phagocytosis and translocation of bacteria across the intestinal barrier. J Immunol 2006;176:3070–3079. [DOI] [PubMed] [Google Scholar]
- Neal MD, Sodhi CP, Jia Het al. Toll-like receptor 4 is expressed on intestinal stem cells and regulates their proliferation and apoptosis via the p53 up-regulated modulator of apoptosis. J Biol Chem 2012;287:37296–37308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi H, Yanai H, Nakajima Aet al. Cross-interference of RLR and TLR signaling pathways modulates antibacterial T cell responses. Nat Immunol 2012;13:659–666. [DOI] [PubMed] [Google Scholar]
- Nell P, Kattler K, Feuerborn Det al. Identification of an FXR-modulated liver-intestine hybrid state in iPSC-derived hepatocyte-like cells. J Hepatol 2022;77:1386–1398. [DOI] [PubMed] [Google Scholar]
- Nigro G, Rossi R, Commere PHet al. The cytosolic bacterial peptidoglycan sensor Nod2 affords stem cell protection and links microbes to gut epithelial regeneration. Cell Host Microbe 2014;15:792–798. [DOI] [PubMed] [Google Scholar]
- Nordlander S, Pott J, Maloy KJ.. NLRC4 expression in intestinal epithelial cells mediates protection against an enteric pathogen. Mucosal Immunol 2014;7:775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntunzwenimana JC, Boucher G, Paquette Jet al. iGenoMed Consortium. Functional screen of inflammatory bowel disease genes reveals key epithelial functions. Genome Med 2021;13:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary CE, Schneider C, Locksley RM.. Tuft cells-systemically dispersed sensory epithelia integrating immune and neural circuitry. Annu Rev Immunol 2019;37:47–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Mahony C, Clooney A, Clarke SFet al. Dietary-induced bacterial metabolites reduce inflammation and inflammation-associated cancer via Vitamin D Pathway. Int J Mol Sci 2023;24(3):1864. 10.3390/ijms24031864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata Y, Castaño A, Boeing Set al. Neuronal programming by microbiota regulates intestinal physiology. Nature 2020;578:284–289. [DOI] [PubMed] [Google Scholar]
- Obermeier F, Dunger N, Strauch UGet al. CpG motifs of bacterial DNA essentially contribute to the perpetuation of chronic intestinal inflammation. Gastroenterology 2005;129:913–927. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara Net al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 2001;411:603–606. [DOI] [PubMed] [Google Scholar]
- Oh DY, Talukdar S, Bae EJet al. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 2010;142:687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnmacht C, Park JH, Cording Set al. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science 2015;349:989–993. [DOI] [PubMed] [Google Scholar]
- Ooi JH, Li Y, Rogers CJet al. Vitamin D regulates the gut microbiome and protects mice from dextran sodium sulfate-induced colitis. J Nutr 2013;143:1679–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppong GO, Rapsinski GJ, Newman TNet al. Epithelial cells augment barrier function via activation of the Toll-like receptor 2/phosphatidylinositol 3-kinase pathway upon recognition of Salmonella enterica serovar Typhimurium curli fibrils in the gut. Infect Immun 2013;81:478–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallett LJ, Swadling L, Diniz Met al. Tissue CD14(+)CD8(+) T cells reprogrammed by myeloid cells and modulated by LPS. Nature 2023;614:334–342. [DOI] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Cullen TWet al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014;158:1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Flavell RA.. Immune–microbiota interactions in health and disease. Clin Immunol 2015;159:122–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda SK, Peng V, Sudan Ret al. Repression of the aryl-hydrocarbon receptor prevents oxidative stress and ferroptosis of intestinal intraepithelial lymphocytes. Immunity 2023;56(4):797–812.e4. 10.1016/j.immuni.2023.01.023 Epub 2023 Feb 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Kawai T, Akira S.. Microbial sensing by Toll-like receptors and intracellular nucleic acid sensors. Cold Spring Harb Perspect Biol 2014;7:a016246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Kotani T, Konno Tet al. Promotion of intestinal epithelial cell turnover by commensal bacteria: role of short-chain fatty acids. PLoS One 2016;11:e0156334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschoal VA, Walenta E, Talukdar Set al. Positive reinforcing mechanisms between GPR120 and PPARγ modulate insulin sensitivity. Cell Metab 2020;31:1173–1188.e1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak P, Xie C, Nichols RGet al. Intestine farnesoid X receptor agonist and the gut microbiota activate G-protein bile acid receptor-1 signaling to improve metabolism. Hepatology 2018;68:1574–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Pavert SA, Ferreira M, Domingues RGet al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature 2014;508:123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck BCE, Shanahan MT, Singh APet al. Gut microbial influences on the mammalian intestinal stem cell Niche. Stem Cells Int 2017;2017:5604727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G, Guo Z, Kiniwa Yet al. Toll-like receptor 8-mediated reversal of CD4+ regulatory T cell function. Science 2005;309:1380–1384. [DOI] [PubMed] [Google Scholar]
- Perez-Pardo P, Dodiya HB, Engen PAet al. Role of TLR4 in the gut–brain axis in Parkinson’s disease: a translational study from men to mice. Gut 2019;68:829–843. [DOI] [PubMed] [Google Scholar]
- Petnicki-Ocwieja T, Hrncir T, Liu YJet al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci USA 2009;106:15813–15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi Y, Wu Y, Zhang Xet al. Gut microbiota-derived ursodeoxycholic acid alleviates low birth weight-induced colonic inflammation by enhancing M2 macrophage polarization. Microbiome 2023;11:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik M, Joossens S, Van Steen Ket al. Toll-like receptor-1, -2, and -6 polymorphisms influence disease extension in inflammatory bowel diseases. Inflamm Bowel Dis 2006;12:1–8. [DOI] [PubMed] [Google Scholar]
- Piya MK, McTernan PG, Kumar S.. Adipokine inflammation and insulin resistance: the role of glucose, lipids and endotoxin. J Endocrinol 2013;216:T1–T15. [DOI] [PubMed] [Google Scholar]
- Plovier H, Everard A, Druart Cet al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 2017;23:107–113. [DOI] [PubMed] [Google Scholar]
- Podolsky DK, Gerken G, Eyking Aet al. Colitis-associated variant of TLR2 causes impaired mucosal repair because of TFF3 deficiency. Gastroenterology 2009;137:209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole DP, Godfrey C, Cattaruzza Fet al. Expression and function of the bile acid receptor GpBAR1 (TGR5) in the murine enteric nervous system. Neurogastroenterol Motil 2010;22:814–25, e227–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott J, Hornef M.. Innate immune signalling at the intestinal epithelium in homeostasis and disease. EMBO Rep 2012;13:684–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AE, Shamardani K, Lugo KAet al. A map of toll-like receptor expression in the intestinal epithelium reveals distinct spatial, cell type-specific, and temporal patterns. Immunity 2018;49:560–575.e566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Próchnicki T, Vasconcelos MB, Robinson KSet al. Mitochondrial damage activates the NLRP10 inflammasome. Nat Immunol 2023;24:595–603. [DOI] [PubMed] [Google Scholar]
- Pusceddu MM, Barboza M, Keogh CEet al. Nod-like receptors are critical for gut–brain axis signalling in mice. J Physiol 2019;597:5777–5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao S, Lv C, Tao Yet al. Arctigenin disrupts NLRP3 inflammasome assembly in colonic macrophages via downregulating fatty acid oxidation to prevent colitis-associated cancer. Cancer Lett 2020;491:162–179. [DOI] [PubMed] [Google Scholar]
- Qiu J, Heller JJ, Guo Xet al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 2012;36:92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Guo X, Chen ZMet al. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity 2013;39:386–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachmilewitz D, Katakura K, Karmeli Fet al. Toll-like receptor 9 signaling mediates the anti-inflammatory effects of probiotics in murine experimental colitis. Gastroenterology 2004;126:520–528. [DOI] [PubMed] [Google Scholar]
- Raisch J, Rolhion N, Dubois Aet al. Intracellular colon cancer-associated Escherichia coli promote protumoral activities of human macrophages by inducing sustained COX-2 expression. Lab Invest 2015;95:296–307. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh Fet al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004;118:229–241. [DOI] [PubMed] [Google Scholar]
- Ramadoss P, Marcus C, Perdew GH.. Role of the aryl hydrocarbon receptor in drug metabolism. Expert Opin Drug Metab Toxicol 2005;1:9–21. [DOI] [PubMed] [Google Scholar]
- Ramanan D, Tang MS, Bowcutt Ret al. Bacterial sensor Nod2 prevents inflammation of the small intestine by restricting the expansion of the commensal Bacteroides vulgatus. Immunity 2014;41:311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan K, Hedl M, Sinha Set al. Ubiquitination of ATF6 by disease-associated RNF186 promotes the innate receptor-induced unfolded protein response. J Clin Invest 2021;131(17):e145472. 10.1172/JCI145472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratsimandresy RA, Indramohan M, Dorfleutner Aet al. The AIM2 inflammasome is a central regulator of intestinal homeostasis through the IL-18/IL-22/STAT3 pathway. Cell Mol Immunol 2017;14:127–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch I, Deets KA, Ji DXet al. NAIP-NLRC4 inflammasomes coordinate intestinal epithelial cell expulsion with Eicosanoid and IL-18 release via activation of Caspase-1 and -8. Immunity 2017;46:649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon S. Gut–brain link grabs neuroscientists. Nature 2014;515:175–177. [DOI] [PubMed] [Google Scholar]
- Reiser ML, Mosaheb MM, Lisk Cet al. The TLR2 binding Neisserial Porin PorB enhances antigen presenting cell trafficking and cross-presentation. Sci Rep 2017;7:736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RL, Topless RK, Phipps-Green AJet al. Evidence of interaction of CARD8 rs2043211 with NALP3 rs35829419 in Crohn’s disease. Genes Immun 2010;11:351–356. [DOI] [PubMed] [Google Scholar]
- Rochereau N, Drocourt D, Perouzel Eet al. Dectin-1 is essential for reverse transcytosis of glycosylated SIgA-antigen complexes by intestinal M cells. PLoS Biol 2013;11:e1001658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochereau N, Roblin X, Michaud Eet al. NOD2 deficiency increases retrograde transport of secretory IgA complexes in Crohn’s disease. Nat Commun 2021;12:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock FL, Hardiman G, Timans JCet al. A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci USA 1998;95:588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooks MG, Garrett WS.. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 2016;16:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose WA 2nd, Sakamoto K, Leifer CA.. TLR9 is important for protection against intestinal damage and for intestinal repair. Sci Rep 2012;2:574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer V, Borucki DM, Tjon ECet al. Microglial control of astrocytes in response to microbial metabolites. Nature 2018;557:724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK.. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009;9:313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK.. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA 2010;107:12204–12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Lee SM, Li Jet al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 2011;332:974–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbino F, Garlatti V, Garzarelli Vet al. GPR120 prevents colorectal adenocarcinoma progression by sustaining the mucosal barrier integrity. Sci Rep 2022;12:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz PA, Morón B, Becker HMet al. Titanium dioxide nanoparticles exacerbate DSS-induced colitis: role of the NLRP3 inflammasome. Gut 2017;66:1216–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell WR, Duncan SH, Scobbie Let al. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol Nutr Food Res 2013;57:523–535. [DOI] [PubMed] [Google Scholar]
- Ryan KK, Tremaroli V, Clemmensen Cet al. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature 2014;509:183–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah A, Chang TH, Harnack Ret al. Activation of innate immune antiviral responses by Nod2. Nat Immunol 2009;10:1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Gomez-Bris R, Herrero-Fernandez Bet al. Innate lymphoid cells in intestinal homeostasis and inflammatory bowel disease. Int J Mol Sci 2021;22(14):7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainathan SK, Bishnupuri KS, Aden Ket al. Toll-like receptor-7 ligand Imiquimod induces type I interferon and antimicrobial peptides to ameliorate dextran sodium sulfate-induced acute colitis. Inflamm Bowel Dis 2012;18:955–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman NH, Ghosh D, Huttner KMet al. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 2003;422:522–526. [DOI] [PubMed] [Google Scholar]
- Sampson TR, Challis C, Jain Net al. A gut bacterial amyloid promotes α-synuclein aggregation and motor impairment in mice. Elife 2020;9:e53111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel BS, Shaito A, Motoike Tet al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci USA 2008;105:16767–16772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Lopez E, Zhong Z, Stubelius Aet al. Choline uptake and metabolism modulate macrophage IL-1β and IL-18 production. Cell Metab 2019;29:1350–1362.e1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmarco LM, Chao CC, Wang YCet al. Identification of environmental factors that promote intestinal inflammation. Nature 2022;611:801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaolalla R, Sussman DA, Ruiz JRet al. TLR4 activates the β-catenin pathway to cause intestinal neoplasia. PLoS One 2013;8:e63298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S, Lochner M, Satoh-Takayama Net al. RORγt+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol 2011;12:320–326. [DOI] [PubMed] [Google Scholar]
- Scheeren FA, Kuo AH, van Weele LJet al. A cell-intrinsic role for TLR2-MYD88 in intestinal and breast epithelia and oncogenesis. Nat Cell Biol 2014;16:1238–1248. [DOI] [PubMed] [Google Scholar]
- Scheithauer TPM, Herrema H, Yu Het al. Gut-derived bacterial flagellin induces beta-cell inflammation and dysfunction. Gut Microbes 2022;14:2111951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenten D, Medzhitov R.. The control of adaptive immune responses by the innate immune system. Adv Immunol 2011;109:87–124. [DOI] [PubMed] [Google Scholar]
- Schenten D, Nish SA, Yu Set al. Signaling through the adaptor molecule MyD88 in CD4+ T cells is required to overcome suppression by regulatory T cells. Immunity 2014;40:78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieber AM, Lee YM, Chang MWet al. Disease tolerance mediated by microbiome E. coli involves inflammasome and IGF-1 signaling. Science 2015;350:558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiering C, Wincent E, Metidji Aet al. Feedback control of AHR signalling regulates intestinal immunity. Nature 2017;542:242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schill EM, Floyd AN, Newberry RD.. Neonatal development of intestinal neuroimmune interactions. Trends Neurosci 2022;45:928–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Zimmermann AG, Roberts RAet al. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-κB. Nat Immunol 2012;13:823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoultz I, Verma D, Halfvarsson Jet al. Combined polymorphisms in genes encoding the inflammasome components NALP3 and CARD8 confer susceptibility to Crohn’s disease in Swedish men. Am J Gastroenterol 2009;104:1180–1188. [DOI] [PubMed] [Google Scholar]
- Schulmann K, Brasch FE, Kunstmann Eet al. German HNPCC Consortium. HNPCC-associated small bowel cancer: clinical and molecular characteristics. Gastroenterology 2005;128:590–599. [DOI] [PubMed] [Google Scholar]
- Schulthess J, Pandey S, Capitani Met al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 2019;50:432–445.e437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer M, Gautam UK, Makki Ket al. Microbe-mediated intestinal NOD2 stimulation improves linear growth of undernourished infant mice. Science 2023;379:826–833. [DOI] [PubMed] [Google Scholar]
- Sefik E, Geva-Zatorsky N, Oh Set al. Individual intestinal symbionts induce a distinct population of RORγ⁺ regulatory T cells. Science 2015;349:993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Pruijssers AJ, Dermody TSet al. The early interferon response to rotavirus is regulated by PKR and depends on MAVS/IPS-1, RIG-I, MDA-5, and IRF3. J Virol 2011;85:3717–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R, Fuchs S, Milo R.. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 2016a;164:337–340. [DOI] [PubMed] [Google Scholar]
- Sender R, Fuchs S, Milo R.. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 2016b;14:e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo B, Jeon K, Moon Set al. Roseburia spp. abundance associates with alcohol consumption in humans and its administration ameliorates alcoholic fatty liver in mice. Cell Host Microbe 2020;27:25–40.e26. [DOI] [PubMed] [Google Scholar]
- Sepahi A, Liu Q, Friesen Let al. Dietary fiber metabolites regulate innate lymphoid cell responses. Mucosal Immunol 2021;14:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham HP, Yu EY, Gulen MFet al. SIGIRR, a negative regulator of TLR/IL-1R signalling promotes microbiota dependent resistance to colonization by enteric bacterial pathogens. PLoS Pathog 2013;9:e1003539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan MT, Carroll IM, Grossniklaus Eet al. Mouse Paneth cell antimicrobial function is independent of Nod2. Gut 2014;63:903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X, Sun S, Zhou Yet al. Bacteroides fragilis restricts colitis-associated cancer via negative regulation of the NLRP3 axis. Cancer Lett 2021;523:170–181. [DOI] [PubMed] [Google Scholar]
- Sharifinejad N, Mozhgani SH, Bakhtiyari Met al. Association of LRRK2 rs11564258 single nucleotide polymorphisms with type and extent of gastrointestinal mycobiome in ulcerative colitis: a case-control study. Gut Pathog 2021;13:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Li R, Negro Ret al. Phase separation drives RNA virus-induced activation of the NLRP6 inflammasome. Cell 2021;184:5759–5774.e5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D, Das J, Das G.. Inflammatory bowel disease requires the interplay between innate and adaptive immune signals. Cell Res 2006a;16:70–74. [DOI] [PubMed] [Google Scholar]
- Shi H, Kokoeva MV, Inouye Ket al. TLR4 links innate immunity and fatty acid-induced insulin resistance. J Clin Invest 2006b;116:3015–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Yet al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 2014;514:187–192. [DOI] [PubMed] [Google Scholar]
- Shigemoto T, Kageyama M, Hirai Ret al. Identification of loss of function mutations in human genes encoding RIG-I and MDA5: implications for resistance to type I diabetes. J Biol Chem 2009;284:13348–13354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmuel-Galia L, Humphries F, Lei Xet al. Dysbiosis exacerbates colitis by promoting ubiquitination and accumulation of the innate immune adaptor STING in myeloid cells. Immunity 2021;54:1137–1153.e1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si W, Liang H, Bugno Jet al. Lactobacillus rhamnosus GG induces cGAS/STING-dependent type I interferon and improves response to immune checkpoint blockade. Gut 2022;71:521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira TN, Gomes MT, Oliveira LSet al. NLRP12 negatively regulates proinflammatory cytokine production and host defense against Brucella abortus. Eur J Immunol 2017;47:51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinal CJ, Tohkin M, Miyata Met al. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 2000;102:731–744. [DOI] [PubMed] [Google Scholar]
- Singh N, Gurav A, Sivaprakasam Set al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014;40:128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Chassaing B, Zhang Let al. Microbiota-dependent hepatic lipogenesis mediated by Stearoyl CoA Desaturase 1 (SCD1) promotes metabolic syndrome in TLR5-deficient mice. Cell Metab 2015;22:983–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Chandrashekharappa S, Bodduluri SRet al. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat Commun 2019a;10:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Yeoh BS, Walker REet al. Microbiota fermentation–NLRP3 axis shapes the impact of dietary fibres on intestinal inflammation. Gut 2019b;68:1801–1812. [DOI] [PubMed] [Google Scholar]
- Sinha SR, Haileselassie Y, Nguyen LPet al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe 2020;27:659–670.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov Net al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013;341:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DJ, Cooper JD, Bailey Ret al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet 2006;38:617–619. [DOI] [PubMed] [Google Scholar]
- Smythies LE, Sellers M, Clements RHet al. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest 2005;115:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socała K, Doboszewska U, Szopa Aet al. The role of microbiota–gut–brain axis in neuropsychiatric and neurological disorders. Pharmacol Res 2021;172:105840. [DOI] [PubMed] [Google Scholar]
- Sodhi CP, Neal MD, Siggers Ret al. Intestinal epithelial Toll-like receptor 4 regulates goblet cell development and is required for necrotizing enterocolitis in mice. Gastroenterology 2012;143:708–718.e701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somineni HK, Nagpal S, Venkateswaran Set al. Whole-genome sequencing of African Americans implicates differential genetic architecture in inflammatory bowel disease. Am J Hum Genet 2021;108:431–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, Bäckhed F.. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol 2013;11:227–238. [DOI] [PubMed] [Google Scholar]
- Song X, He X, Li Xet al. The roles and functional mechanisms of interleukin-17 family cytokines in mucosal immunity. Cell Mol Immunol 2016;13:418–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhao W, Zhang Xet al. Mutant RIG-I enhances cancer-related inflammation through activation of circRIG-I signaling. Nat Commun 2022;13:7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino G, Perino A, Yildiz Eet al. Bile acids signal via TGR5 to activate intestinal stem cells and epithelial regeneration. Gastroenterology 2020;159:956–968.e958. [DOI] [PubMed] [Google Scholar]
- Spencer SP, Wilhelm C, Yang Qet al. Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science 2014;343:432–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindler MP, Siu S, Mogno Iet al. Human gut microbiota stimulate defined innate immune responses that vary from phylum to strain. Cell Host Microbe 2022;30:1481–1498.e1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H, Di Santo JP.. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat Immunol 2011;12:21–27. [DOI] [PubMed] [Google Scholar]
- Stafford CA, Gassauer AM, de Oliveira Mann CCet al. Phosphorylation of muramyl peptides by NAGK is required for NOD2 activation. Nature 2022;609:590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanifer ML, Mukenhirn M, Muenchau Set al. Asymmetric distribution of TLR3 leads to a polarized immune response in human intestinal epithelial cells. Nat Microbiol 2020;5:181–191. [DOI] [PubMed] [Google Scholar]
- Stefan KL, Kim MV, Iwasaki Aet al. Commensal microbiota modulation of natural resistance to virus infection. Cell 2020;183:1312–1324.e1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner A, Reygaerts T, Pontillo Aet al. Recessive NLRC4-autoinflammatory disease reveals an ulcerative colitis locus. J Clin Immunol 2022;42:325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger B, Shah K, Wincent E.. AHR in the intestinal microenvironment: safeguarding barrier function. Nat Rev Gastroenterol Hepatol 2021;18:559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Xie C, Wang Get al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat Med 2018a;24:1919–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Wu W, Chen Let al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat Commun 2018b;9:3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Nguyen A, Gommerman JL.. Dendritic cell subsets in intestinal immunity and inflammation. J Immunol 2020;204:1075–1083. [DOI] [PubMed] [Google Scholar]
- Sun L, Cai J, Gonzalez FJ.. The role of farnesoid X receptor in metabolic diseases, and gastrointestinal and liver cancer. Nat Rev Gastroenterol Hepatol 2021;18:335–347. [DOI] [PubMed] [Google Scholar]
- Sutmuller RP, den Brok MH, Kramer Met al. Toll-like receptor 2 controls expansion and function of regulatory T cells. J Clin Invest 2006;116:485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed SK, Bui HH, Beavers LSet al. Regulation of GPR119 receptor activity with endocannabinoid-like lipids. Am J Physiol Endocrinol Metab 2012;303:E1469–E1478. [DOI] [PubMed] [Google Scholar]
- Takatori H, Kanno Y, Watford WTet al. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J Exp Med 2009;206:35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S.. Pattern recognition receptors and inflammation. Cell 2010;140:805–820. [DOI] [PubMed] [Google Scholar]
- Tan G, Li RH, Li Cet al. Down-regulation of human enteric antimicrobial peptides by NOD2 during differentiation of the paneth cell lineage. Sci Rep 2015;5:8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, McKenzie C, Vuillermin PJet al. Dietary fiber and bacterial SCFA enhance oral tolerance and protect against food allergy through diverse cellular pathways. Cell Rep 2016;15:2809–2824. [DOI] [PubMed] [Google Scholar]
- Tan JK, McKenzie C, Mariño Eet al. Metabolite-sensing G protein-coupled receptors-facilitators of diet-related immune regulation. Annu Rev Immunol 2017;35:371–402. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Katsuma S, Adachi Tet al. Free fatty acids induce cholecystokinin secretion through GPR120. Naunyn Schmiedebergs Arch Pharmacol 2008;377:523–527. [DOI] [PubMed] [Google Scholar]
- Tang Y, Chen Y, Jiang Het al. G-protein-coupled receptor for short-chain fatty acids suppresses colon cancer. Int J Cancer 2011;128:847–856. [DOI] [PubMed] [Google Scholar]
- Tang C, Ahmed K, Gille Aet al. Loss of FFA2 and FFA3 increases insulin secretion and improves glucose tolerance in type 2 diabetes. Nat Med 2015a;21:173–177. [DOI] [PubMed] [Google Scholar]
- Tang C, Kamiya T, Liu Yet al. Inhibition of Dectin-1 signaling ameliorates colitis by inducing lactobacillus-mediated regulatory T cell expansion in the intestine. Cell Host Microbe 2015b;18:183–197. [DOI] [PubMed] [Google Scholar]
- Temperley ND, Berlin S, Paton IRet al. Evolution of the chicken Toll-like receptor gene family: a story of gene gain and gene loss. BMC Genom 2008;9:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangaraju M, Cresci GA, Liu Ket al. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res 2009;69:2826–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Gioiello A, Noriega Let al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab 2009;10:167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn AN, McKenzie CI, Shen Set al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun 2015;6:7320. [DOI] [PubMed] [Google Scholar]
- Tlaskalova-Hogenova H, Stepankova R, Kozakova Het al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol Immunol 2011;8:110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubai T, Fujiwara H, Rossi Cet al. Host NLRP6 exacerbates graft-versus-host disease independent of gut microbial composition. Nat Microbiol 2019;4:800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touitou I, Lesage S, McDermott Met al. Infevers: an evolving mutation database for auto-inflammatory syndromes. Hum Mutat 2004;24:194–198. [DOI] [PubMed] [Google Scholar]
- Trabelsi MS, Daoudi M, Prawitt Jet al. Farnesoid X receptor inhibits glucagon-like peptide-1 production by enteroendocrine L cells. Nat Commun 2015;6:7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Yadava Ket al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 2014;20:159–166. [DOI] [PubMed] [Google Scholar]
- Truax AD, Chen L, Tam JWet al. The inhibitory innate immune sensor NLRP12 maintains a threshold against obesity by regulating gut microbiota homeostasis. Cell Host Microbe 2018;24:364–378.e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi H, Chu ESH, Zhang Xet al. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology 2017;152:1419–1433.e1415. [DOI] [PubMed] [Google Scholar]
- Tuladhar S, Kanneganti TD.. NLRP12 in innate immunity and inflammation. Mol Aspects Med 2020;76:100887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MAet al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006;444:1027–1031. [DOI] [PubMed] [Google Scholar]
- Uchimura T, Oyama Y, Deng Met al. The innate immune sensor NLRC3 acts as a rheostat that fine-tunes T Cell responses in infection and autoimmunity. Immunity 2018;49:1049–1061.e1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu S, Jang MH, Chevrier Net al. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nat Immunol 2006;7:868–874. [DOI] [PubMed] [Google Scholar]
- Uematsu S, Fujimoto K, Jang MHet al. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat Immunol 2008;9:769–776. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Behrendt CL, Ismail ASet al. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA 2008;105:20858–20863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanaja SK, Russo AJ, Behl Bet al. Bacterial outer membrane vesicles mediate cytosolic localization of LPS and Caspase-11 activation. Cell 2016;165:1106–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandanmagsar B, Youm YH, Ravussin Aet al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med 2011;17:179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDussen KL, Liu TC, Li Det al. Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn’s disease. Gastroenterology 2014;146:200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhove W, Peeters PM, Staelens Det al. Strong upregulation of AIM2 and IFI16 inflammasomes in the mucosa of patients with active inflammatory bowel disease. Inflamm Bowel Dis 2015;21:2673–2682. [DOI] [PubMed] [Google Scholar]
- Vatanen T, Franzosa EA, Schwager Ret al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 2018;562:589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Sanders CJ, Taylor RTet al. Deletion of TLR5 results in spontaneous colitis in mice. J Clin Invest 2007;117:3909–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Kumar Aet al. Toll-like receptor 5-deficient mice have dysregulated intestinal gene expression and nonspecific resistance to Salmonella-induced typhoid-like disease. Infect Immun 2008a;76:1276–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Sanders CJet al. Flagellin treatment protects against chemicals, bacteria, viruses, and radiation. J Immunol 2008b;180:8280–8285. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FAet al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 2010;328:228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani AC, Lemire M, Fortin Get al. Common variants in the NLRP3 region contribute to Crohn’s disease susceptibility. Nat Genet 2009;41:71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladimer GI, Weng D, Paquette SWet al. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity 2012;37:96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk JK, Nyström EEL, van der Post Set al. The Nlrp6 inflammasome is not required for baseline colonic inner mucus layer formation or function. J Exp Med 2019;216:2602–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora P, Youdim A, Thomas LSet al. Beta-defensin-2 expression is regulated by TLR signaling in intestinal epithelial cells. J Immunol 2004;173:5398–5405. [DOI] [PubMed] [Google Scholar]
- Wada K, Tanaka H, Maeda Ket al. Vitamin D receptor expression is associated with colon cancer in ulcerative colitis. Oncol Rep 2009;22:1021–1025. [DOI] [PubMed] [Google Scholar]
- Wada T, Sunaga H, Miyata Ket al. Aryl hydrocarbon receptor plays protective roles against High Fat Diet (HFD)-induced hepatic steatosis and the subsequent lipotoxicity via direct transcriptional regulation of Socs3 Gene Expression*. J Biol Chem 2016;291:7004–7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahida A, Müller M, Hiergeist Aet al. XIAP restrains TNF-driven intestinal inflammation and dysbiosis by promoting innate immune responses of Paneth and dendritic cells. Sci Immunol 2021;6:eabf7235. [DOI] [PubMed] [Google Scholar]
- Walker JA, Barlow JL, McKenzie AN.. Innate lymphoid cells—how did we miss them? Nat Rev Immunol 2013;13:75–87. [DOI] [PubMed] [Google Scholar]
- Walker NM, Liu J, Young SMet al. Goblet cell hyperplasia is not epithelial-autonomous in the Cftr knockout intestine. Am J Physiol Gastrointest Liver Physiol 2022;322:G282–G293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Huycke MM.. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology 2007;132:551–561. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hasegawa M, Imamura Ret al. PYNOD, a novel Apaf-1/CED4-like protein is an inhibitor of ASC and caspase-1. Int Immunol 2004;16:777–786. [DOI] [PubMed] [Google Scholar]
- Wang Q, McLoughlin RM, Cobb BAet al. A bacterial carbohydrate links innate and adaptive responses through Toll-like receptor 2. J Exp Med 2006;203:2853–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang HX, Sun YPet al. Rig-I−/− mice develop colitis associated with downregulation of G alpha i2. Cell Res 2007;17:858–868. [DOI] [PubMed] [Google Scholar]
- Wang P, Zhu S, Yang Let al. Nlrp6 regulates intestinal antiviral innate immunity. Science 2015a;350:826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Charbonnier LM, Noval Rivas Met al. MyD88 adaptor-dependent microbial sensing by regulatory T cells promotes mucosal tolerance and enforces commensalism. Immunity 2015b;43:289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang P, Tian Het al. Aryl hydrocarbon receptor/IL-22/Stat3 signaling pathway is involved in the modulation of intestinal mucosa antimicrobial molecules by commensal microbiota in mice. Innate Immun 2018a;24:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Lin Y, Yuan Xet al. REV-ERBα integrates colon clock with experimental colitis through regulation of NF-κB/NLRP3 axis. Nat Commun 2018b;9:4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Spatz M, Da Costa Get al. Deletion of both Dectin-1 and Dectin-2 affects the bacterial but not fungal gut microbiota and susceptibility to colitis in mice. Microbiome 2022a;10:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wei B, Wang Det al. DNA damage repair promotion in colonic epithelial cells by andrographolide downregulated cGAS‒STING pathway activation and contributed to the relief of CPT-11-induced intestinal mucositis. Acta Pharm Sin B 2022b;12:262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Kitani A, Murray PJet al. Nucleotide binding oligomerization domain 2 deficiency leads to dysregulated TLR2 signaling and induction of antigen-specific colitis. Immunity 2006;25:473–485. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Motooka D, Yamasaki S.. The kinetics of signaling through the common FcRγ chain determine cytokine profiles in dendritic cells. Sci Signal 2023;16:eabn9909. [DOI] [PubMed] [Google Scholar]
- Wehkamp J, Harder J, Weichenthal Met al. NOD2 (CARD15) mutations in Crohn’s disease are associated with diminished mucosal alpha-defensin expression. Gut 2004;53:1658–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Ye J, Pei Yet al. Extracellular vesicles from colorectal cancer cells promote metastasis via the NOD1 signalling pathway. J Extracell Vesicles 2022;11:e12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PYet al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 2008;455:1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesch D, Beetz S, Oberg HHet al. Direct costimulatory effect of TLR3 ligand poly(I:C) on human gamma delta T lymphocytes. J Immunol 2006;176:1348–1354. [DOI] [PubMed] [Google Scholar]
- Willemsen LE, Koetsier MA, van Deventer SJet al. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E(1) and E(2) production by intestinal myofibroblasts. Gut 2003;52:1442–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AM, Probert CS, Stepankova Ret al. Effects of microflora on the neonatal development of gut mucosal T cells and myeloid cells in the mouse. Immunology 2006;119:470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Petrucelli AS, Chen Let al. Inflammasome-independent role of AIM2 in suppressing colon tumorigenesis via DNA-PK and Akt. Nat Med 2015a;21:906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SS, Tocchi A, Holly MKet al. A small intestinal organoid model of non-invasive enteric pathogen–epithelial cell interactions. Mucosal Immunol 2015b;8:352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler ES, Shrihari S, Hykes BL Jr.et al. The intestinal microbiome restricts alphavirus infection and dissemination through a bile acid-Type I IFN signaling axis. Cell 2020;182:901–918.e918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Thaiss CA, Nowarski Ret al. NLRP6 inflammasome orchestrates the colonic host–microbial interface by regulating goblet cell mucus secretion. Cell 2014;156:1045–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Zhang YG, Lu Ret al. Intestinal epithelial Vitamin D receptor deletion leads to defective autophagy in colitis. Gut 2015;64:1082–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang W, Shi R, Zhang Det al. Dietary fats suppress the peritoneal seeding of colorectal cancer cells through the TLR4/Cxcl10 axis in adipose tissue macrophages. Signal Transduct Target Ther 2020;5:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Li XX, Chung HKet al. RNA-binding protein HuR regulates paneth cell function by altering membrane localization of TLR2 via post-transcriptional control of CNPY3. Gastroenterology 2019;157:731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Zhou X, Fang Met al. DHX15 is required to control RNA virus-induced intestinal inflammation. Cell Rep 2021;35:109205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Zhu X, Geng Jet al. Intestinal Tuft-2 cells exert antimicrobial immunity via sensing bacterial metabolite N-undecanoylglycine. Immunity 2022;55:686–700.e687. [DOI] [PubMed] [Google Scholar]
- Xu ZZ, Kim YH, Bang Set al. Inhibition of mechanical allodynia in neuropathic pain by TLR5-mediated A-fiber blockade. Nat Med 2015;21:1326–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P, Ellinghaus D, Rémy Get al. International IBD Genetics Consortium. Genetic factors interact with tobacco smoke to modify risk for inflammatory bowel disease in humans and mice. Gastroenterology 2017;153:550–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Jiang W, Spinetti Tet al. Omega-3 fatty acids prevent inflammation and metabolic disorder through inhibition of NLRP3 inflammasome activation. Immunity 2013;38:1154–1163. [DOI] [PubMed] [Google Scholar]
- Yan X, Zhang H, Fan Qet al. Dectin-2 deficiency modulates Th1 differentiation and improves wound healing after myocardial infarction. Circ Res 2017;120:1116–1129. [DOI] [PubMed] [Google Scholar]
- Yang Y, Palm NW.. Immunoglobulin A and the microbiome. Curr Opin Microbiol 2020;56:89–96. [DOI] [PubMed] [Google Scholar]
- Yang JY, Kim MS, Kim Eet al. Enteric viruses ameliorate gut inflammation via Toll-like receptor 3 and Toll-like receptor 7-mediated interferon-beta production. Immunity 2016;44:889–900. [DOI] [PubMed] [Google Scholar]
- Yang WH, Heithoff DM, Aziz PVet al. Recurrent infection progressively disables host protection against intestinal inflammation. Science 2017;358(6370):eaao5610. 10.1126/science.aao5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang H, Kouadir Met al. Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death Dis 2019;10:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Yu T, Huang Xet al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat Commun 2020;11:4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Long D, Hu Let al. AIM2 deficiency in B cells ameliorates systemic lupus erythematosus by regulating Blimp-1-Bcl-6 axis-mediated B-cell differentiation. Signal Transduct Target Ther 2021;6:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Liu H, Xu Let al. GPR120 inhibits colitis through regulation of CD4(+) T Cell Interleukin 10 production. Gastroenterology 2022;162:150–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Zhang C, Xing Yet al. Remodelling of the gut microbiota by hyperactive NLRP3 induces regulatory T cells to maintain homeostasis. Nat Commun 2017;8:1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarandi SS, Kulkarni S, Saha Met al. Intestinal bacteria maintain adult enteric nervous system and nitrergic neurons via Toll-like receptor 2-induced neurogenesis in mice. Gastroenterology 2020;159:200–213.e208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Bae M, Cassilly CDet al. Enteroendocrine cells sense bacterial tryptophan catabolites to activate enteric and vagal neuronal pathways. Cell Host Microbe 2021;29:179–196.e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesudhas D, Gosu V, Anwar MAet al. Multiple roles of toll-like receptor 4 in colorectal cancer. Front Immunol 2014;5:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yore MM, Syed I, Moraes-Vieira PMet al. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell 2014;159:318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Gao N.. Compartmentalizing intestinal epithelial cell toll-like receptors for immune surveillance. Cell Mol Life Sci 2015;72:3343–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Li S, Guo Jet al. Farnesoid X receptor antagonizes Wnt/β-catenin signaling in colorectal tumorigenesis. Cell Death Dis 2020;11:640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss MM, Rapin A, Lebon Let al. The intestinal microbiota contributes to the ability of helminths to modulate allergic inflammation. Immunity 2015;43:998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki MH, Boyd KL, Vogel Pet al. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 2010;32:379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki MH, Vogel P, Malireddi RKet al. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell 2011;20:649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki MH, Man SM, Vogel Pet al. Salmonella exploits NLRP12-dependent innate immune signaling to suppress host defenses during infection. Proc Natl Acad Sci USA 2014;111:385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T, Iannitti RG, Cunha Cet al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013;39:372–385. [DOI] [PubMed] [Google Scholar]
- Zeng MY, Cisalpino D, Varadarajan Set al. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity 2016;44:647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Chassaing B, Shi Zet al. Viral infection. Prevention and cure of rotavirus infection via TLR5/NLRC4-mediated production of IL-22 and IL-18. Science 2014a;346:861–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Mo J, Swanson KVet al. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity 2014b;40:329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Chen G, Manwani Det al. Neutrophil ageing is regulated by the microbiome. Nature 2015;525:528–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Pan Y, Zeng Bet al. Intestinal lysozyme liberates Nod1 ligands from microbes to direct insulin trafficking in pancreatic beta cells. Cell Res 2019;29:516–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zou J, Shi Zet al. IL-22-induced cell extrusion and IL-18-induced cell death prevent and cure rotavirus infection. Sci Immunol 2020;5(52):eabd2876. 10.1126/sciimmunol.abd2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Garrett S, Carroll REet al. Vitamin D receptor upregulates tight junction protein claudin-5 against colitis-associated tumorigenesis. Mucosal Immunol 2022;15:683–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Shi Jet al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 2011;477:596–600. [DOI] [PubMed] [Google Scholar]
- Zhao J, Han X, Xue Let al. Association of TLR4 gene polymorphisms with sporadic Parkinson’s disease in a Han Chinese population. Neurol Sci 2015;36:1659–1665. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chen F, Wu Wet al. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol 2018;11:752–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Chen T, Jiang Ret al. Hyocholic acid species improve glucose homeostasis through a distinct TGR5 and FXR signaling mechanism. Cell Metab 2021;33:791–803.e797. [DOI] [PubMed] [Google Scholar]
- Zheng D, Mohapatra G, Kern Let al. Epithelial Nlrp10 inflammasome mediates protection against intestinal autoinflammation. Nat Immunol 2023;24:585–594. [DOI] [PubMed] [Google Scholar]
- Zhong X, Du G, Wang Xet al. Nanovaccines mediated subcutis-to-intestine cascade for improved protection against intestinal infections. Small 2022;18:e2105530. [DOI] [PubMed] [Google Scholar]
- Zhou X, Cao L, Jiang Cet al. PPARα–UGT axis activation represses intestinal FXR-FGF15 feedback signalling and exacerbates experimental colitis. Nat Commun 2014;5:4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Liu T, Huang Bet al. Excessive deubiquitination of NLRP3-R779C variant contributes to very-early-onset inflammatory bowel disease development. J Allergy Clin Immunol 2021;147:267–279. [DOI] [PubMed] [Google Scholar]
- Zhu H, Xu WY, Hu Zet al. RNA virus receptor Rig-I monitors gut microbiota and inhibits colitis-associated colorectal cancer. J Exp Clin Cancer Res 2017a;36:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Ding S, Wang Pet al. Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature 2017b;546:667–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Wang Z, Cai Jet al. VDR signaling via the enzyme NAT2 inhibits colorectal cancer progression. Front Pharmacol 2021;12:727704. [DOI] [PMC free article] [PubMed] [Google Scholar]


