Abstract
Cyclin-dependent kinase inhibitor 1A (Cip1/Waf1/CDKN1A/p21) is a well-established protein, primarily recognised for its pivotal role in the cell cycle, where it induces cell cycle arrest by inhibiting the activity of cyclin-dependent kinases (CDKs). Over the years, extensive research has shed light on various additional mechanisms involving CDKN1A/p21, implicating it in processes such as apoptosis, DNA damage response (DDR), and the regulation of stem cell fate. Interestingly, p21 can function either as an oncogene or as a tumour suppressor in these contexts. Complicating matters further, the expression of CDKN1A/p21 is elevated in certain tumour types while downregulated in others. In this comprehensive review, we provide an overview of the multifaceted functions of CDKN1A/p21, present clinical data pertaining to cancer patients, and delve into potential strategies for targeting CDKN1A/p21 as a therapeutic approach to cancer. Manipulating CDKN1A/p21 shows great promise for therapy given its involvement in multiple cancer hallmarks, such as sustained cell proliferation, the renewal of cancer stem cells (CSCs), epithelial–mesenchymal transition (EMT), cell migration, and resistance to chemotherapy. Given the dual role of CDKN1A/p21 in these processes, a more in-depth understanding of its specific mechanisms of action and its regulatory network is imperative to establishing successful therapeutic interventions.
Keywords: p21, CDKN1A, cyclin-dependent kinase inhibitor 1A, breast cancer, cell cycle, cancer stem cells
1. Introduction
1.1. Medical Need for Targeted Therapeutics and Biomarkers for the Management of Late-Stage Metastatic Breast Cancer
Breast cancer is the most prevalent cancer type, accounting for over 2 million cases in 2020 [1]. The accumulated knowledge in recent decades has resulted in advancements in treatment strategies, including radiotherapy, surgery, chemotherapy, and immunotherapy [2,3]. Nevertheless, the effectiveness of these treatment options significantly varies between late-stage (TNM stage IV) and early-stage (TNM stage I and II), with a 5-year survival rate around 30% for advanced stages, compared to 90–100% for tumours detected earlier [4,5]. Unfortunately, 20–30% of patients initially diagnosed at early stages eventually succumb to recurrent metastatic tumours, which exhibit heightened aggressiveness and drug resistance compared to primary tumours [6,7]. Therefore, there is a need for novel small molecules and innovative technologies/strategies aimed at specific factors driving the metastatic process.
In addition to the improvement of therapy options for metastasis, the identification of predictive biomarkers for patient stratification, and the early detection of metastatic spread would not only alleviate the burden on healthcare systems but also significantly enhance the quality of life for women suffering with metastatic breast cancer. In this review, we will focus on CDKN1A/p21, a protein central to key processes during metastatic spread, including cancer stem cell survival, epithelial–mesenchymal transition (EMT), and apoptosis, and discuss the described seemingly opposing roles [8] and potential uses as a predictive biomarker in metastatic breast cancer.
1.2. Cyclin-Dependent Kinase Inhibitor 1A
The cyclin-dependent kinase inhibitor 1A (CDKN1A, p21, CIP1, or WAF1) gene encodes for a potent cyclin-dependent kinase (CDK) inhibitor, the activity of which has been well documented regarding control of the mammalian cell cycle. Belonging to a larger family of cyclin-dependent kinase inhibitors, CIP/KIP proteins, CDKN1A/p21 can inhibit CDK activity both in vitro and in vivo via direct binding multiple CDKs and cyclin proteins [9,10]. Even though members of this family share a cyclin–CDK domain within the N-terminal, which aids in the inhibition of the cyclin–CDK complexes, they do not share additional sequence similarity, suggesting that they have unique functions and/or roles in cellular processes independent of the inhibition of CDKs and their role in cell cycle regulation.
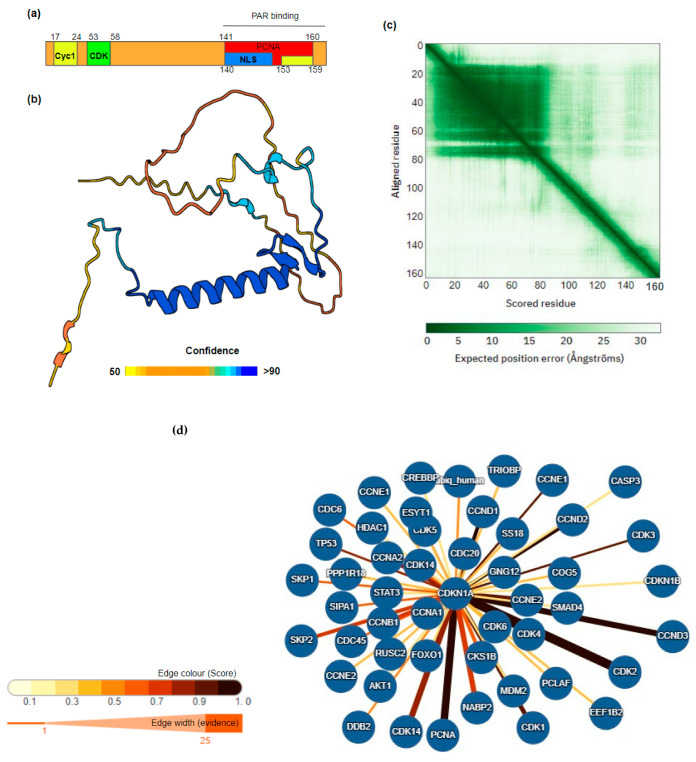
CDKN1A/p21 is a 164 amino acid protein, with a cyclin-dependent kinase inhibitor domain containing the cyclin- and CDK-binding motifs within the N-terminal and a nuclear localisation signal and binding sites for the replication protein, PCNA, and poly-ADP-ribose (PAR) within the C-terminal (Figure 1a–c). In addition, in terms of its 3D structure and organisation CDKN1A/p21 contains a disordered domain at the N-terminal and a longer C-terminal disordered domain (Figure 1b). Intrinsically disordered proteins (IDPs) and proteins containing large stretches of disordered regions play important roles in cell signalling, response to stimulus, and gene expression reprogramming as the disordered nature of this part of the polypeptide chain allows for the flexibility of binding, meaning that the protein can interact with multiple interaction partners depending upon the stimulus or pathway activated. The presence of these disordered regions in CDKN1A/p21 suggests that CDKN1A/p21 functions and interactions may be more complex than the canonical role in cell cycle regulation.
Although expressed in all mammalian tissues, p21 shows elevated expression signatures in the brain, connective tissue, and female reproductive tissues at the RNA and protein levels, respectively, clustering most significantly in smooth muscle tissue and extracellular matrix organisation [11]. At the cellular level, CDKN1A/p21 is located primarily within the nucleoplasm and nuclear bodies in normal cells, with the exception of monocytes [12,13], although cytoplasmic localisation has been observed in certain conditions and cancers, the significance of which will be later discussed.
Given the key role played by CDKN1A/p21 in the cell cycle, tight regulation of CDKN1A/p21 activity is essential. Regulation occurs via direct expression modulation but also via extensive post-translational modification. PTMs include phosphorylation, acetylation, methylation, and ubiquitination. These modifications result in a wide range of changes, including movement in terms of cellular localisation, increased protein stabilisation or degradation, and altering protein–protein interaction partners (Table 1). Indeed, the anti-cancer properties of berberine have been shown to be partly due to the increased nuclear localisation and stability of CDKN1A/p21 [14]. Human interacting proteins of CDKN1A include proteins involved in the cell cycle, DNA damage, and apoptosis (Figure 1d).
Figure 1.
Structure and interactors of CDKN1A/p21. (a) Domain structure CDKN1A/p21; binding sites for cyclins (Cyc), CDK, PCNA, and poly-ADP-ribose (PAR) are indicated. (b) Alpha fold prediction of p21/CDKN1A 3D structure, confidence score scale bar is shown below, the amino and larger C-terminal disordered domains are clearly visible. (c) Predicted aligned error (PAE) for each residue [11]. (d) Annotated human protein interactors of CDKN1A/p21 using IntAct molecular interaction database. Score and evidence weights are shown by colour width of interaction edges [15] as shown in the legend. Full interaction data are detailed in Supplementary Table S1.
Table 1.
Post-translational protein modifications of CDKN1A.
| Post-Translational Modification | Site | Effector | Observed Effect | Reference |
|---|---|---|---|---|
| Phosphorylation | Thr-55 | MELK (mouse) | Induce interaction CDK2/CDK4 | [16] |
| Thr-57 | MAPK1, MAPK14 and MAPK8 | Cytoplasmic localisation and degradation | [17,18] | |
| Thr-80 | LKB1 | Degradation | [19] | |
| Thr-145 | AKT1, CHEK1, DAPK1, PIM1, PIM2, PKA | Impairs binding to PCNA Enhances protein stability Alters protein localisation |
[20,21] | |
| Ser-31 | Unknown | Unknown/identified using mass spectrometry | [22] | |
| Ser-98 | ASK1 | Unknown | [23] | |
| Ser-114 | GSK3-beta | Enhances ubiquitination | [24] | |
| Ser-123 | Unknown | Protein stabilisation | [25] | |
| Ser-130 | CDK6, MAPK, MAPK14, MAPK8 | Impairs stability | [26,27] | |
| Ser 137 | Unknown | [28] | ||
| Ser-153 | DYRK1B, PKCA | Cytoplasmic localisation | [29] | |
| Ser-160 | PKC | Modulates binding to PCNA | [30] | |
| Ser-146 | AKT, CHEK1, DAPK1, LATS2, NUAK1, PKC, PRKCD, STK38 | Impairs binding to PCNA Protein stabilisation |
[31,32] | |
| Tyr-151 | Unknown | Unknown | [33] | |
| Ubiquitination | Lys-16 Lys-75 Lys-141 Lys-154 Lys-161 Lys-163 |
Breast cancer cell growth-induced | Protein degradation | [34] |
| Acetylation | Lys-141 Lys-154 Lys-161 Lys-163 |
HDAC1, TSA induced, cell growth and carcinogenesis inhibited | Protein stabilisation enhanced following acetylation | [35] |
| Methylation | Arg-156 | PRMT6 | Induction of cytoplasmic localisation | [36] |
2. Role of CDKN1A/p21 in Cancer
2.1. Clinical Significance of CDKN1A/p21 Expression Levels
2.1.1. CDKN1A/p21 in Cancer Progression
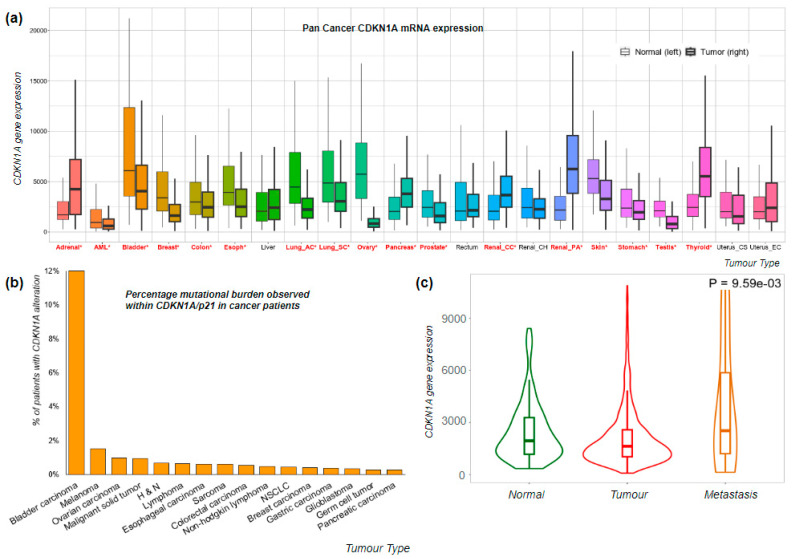
The expression levels of p21 vary when comparing tumour versus normal tissue, showing differences both in magnitude and direction across various tumour types. Using TMPlot, a tool that integrates expression data from repositories like NCBI-GEO, The Cancer Genome Atlas (TCGA), Therapeutically Applicable Research to Generate Effective Treatments (TARGET), and Genotype-Tissue Expression (GTEX), we can observe distinct patterns. In certain cancer types, such as adrenal, pancreatic, and renal, there is a significant (p < 0.01) increase in CDKN1A/p21 expression in the tumour tissue compared to normal tissue, while in others, such as AML, colon, and lung, the opposite trend is observed (Figure 2a).
The role of CDKN1A/p21 in various cancer types has been extensively studied using cell lines, primary cultures, and patient samples (Table 2). Hypermethylation of the CDKN1A/p21 promoter [37,38,39,40,41] and the binding of other regulatory proteins such as SOX2 and LMNB2 [42,43] lead to increased cancer cell proliferation by silencing its expression. Conversely, stabilisation of CDKN1A/p21 at both the mRNA [44] and protein levels [45] promotes cell cycle arrest in cancer cells. However, CDKN1A/p21 has also been documented to promote cancer progression by stabilising cyclin D1–CDK4 complexes [46] and inhibiting apoptosis [47].
Table 2.
Described actions of CDKN1A/p21 in different cancer studies.
| Cancer Type | Mechanism | Description | Ref |
|---|---|---|---|
| Colorectal | LMNB2 binding to the P3- and P6-binding regions of the CDKN1A promoter | Regulation of CDKN1A promoter by LMNB2 results in silencing of p21 expression and promotes cell proliferation in CRC tissues. | [42] |
| Lung | Suppression of CDKN1A via promoter methylation | Transcriptional inactivation of CDKN1A in lung cancer cell lines and MPM. | [37] |
| Prostate | Methylation within the CDKN1Apromoter, at the 5′ end of a CpG island and STAT1-binding site | Inactivation of CDKN1A expression by methylation promotes cell proliferation and DNA replication in metastatic prostate cancer. | [38] |
| Breast | Aberrant hypermethylation of CDKN1A promoter | Increased risk of breast cancer in hypermethylated CDKN1A promoter reduces CDKN1A mRNA expression in all age groups. Inhibition of G1 arrest. | [39] |
| Lymphoblastic leukaemia | Hypermethylation of CpG islands within the CDKN1A promoter regions led to a decrease in CDKN1A expression | Loss of CDKN1A is associated with disease progression and poorer DFS. Selective growth advantage and accelerated onset of other pathways. | [40] |
| Oligodendrogliomas | In the nucleus, p21 promotes ODG by stabilising cyclin D1–cdk4. Cytosolic p21 binds procaspase 3, desensitising apoptotic stimuli | PDGF signalling produces p21 accumulation in ODG, which associates with cyclinD-cdk4 complexes and increases proliferation. Binding to cytosolic components reduced apoptosis. | [46] |
| Renal | CPEB4 binds to the CDKN1A transcripts and stabilises its mRNA, increasing CDKN1A expression | Increasing CDKN1A expression modulates RCC cell proliferation by inhibiting cell cycle progression due to G1 cell cycle arrest. | [44] |
| Lymphomas | p21 provides survival to TNF- or Fas-triggered apoptosis by binding and inhibiting caspase-3 | Absence of p21 increases lifespan due to reduced incidence of thymic lymphomas. p21-proficient lymphomas have faster growth due to lower apoptotic rate. | [47] |
| Hepatocellular | p21 is stabilised by the repression of ubiquitin-proteasome proteolysis due to CMTM6 binding | Extension of CDKN1A half-life by CMTM6 suppresses HCC cell proliferation, sensitising patients to treatment and increasing survival rates. | [45] |
| Endometrial | p21/CDKN1A gene transcription is repressed via SOX2 binding to the CDKN1A promoter | Inhibition of CDKN1A by SOX2 overexpression correlates with poor prognosis in endometrial cancer due to cell cycle progression. | [43] |
| Pancreatic | DNMT1 silencing results in the upregulation of p21 expression | Upregulation of p21 decreases cell proliferation in pancreatic cancer cell lines. | [41] |
In mouse models, the deletion of CDKN1A/p21 does not affect carcinogenesis until around 16 months, where spontaneous tumours are observed [48,49]. Other phenotypes observed in CDKN1A/p21 knockout mice include morphological abnormalities during development, such as enlarged spleens, hearts, and smaller testes [50].
In terms of the mutational burden, CDKN1A mutations vary both in terms of the type of mutation and cancer. Overall, CDKN1A is altered in 0.96% of solid tumour malignancies [51] (Figure 2b). Among these, bladder carcinoma stands out with the highest mutation burden within CDKN1A/p21, affecting 11.99% of patients.
The cytoplasmic localisation of CDKN1A/p21 is predominantly associated with cancer, where it serves to promote tumorigenesis and inhibit apoptosis [52,53,54]. Notably, in breast cancer cell lines, the CDKN1A/p21 phosphorylation induced by the Akt pathway, especially in HER-2/neu-overexpressing cells, leads to cytoplasmic localisation. This event is central to cancer cell survival and resistance to apoptosis [52]. Moreover, the cytoplasmic translocation of CDKN1A/p21 in SUM159 breast cancer cells has been found to enhance chemoresistance [54].
In breast cancer patients, we observe a reduction in CDKN1A/p21 expression when compared to normal samples, although there is some variation among patients (Figure 2c). In addition, there is an increase in CDKN1A/p21 expression in metastatic patients compared to normal samples, although this observation is constrained by the limited number of metastatic cases sampled.
Figure 2.
Expression of CDKN1A/p21 in pan and breast cancer datasets. (a) CDKN1A gene expression levels in pan cancer dataset [55], including 15,648 normal, 40,442 tumour, and 848 metastatic samples; significant differences according to Mann–Whitney U tests are marked in red with an asterisk. (b) CDKN1A/p21 alteration frequency detected including all alterations, mutations, amplification, and loss. (c) CDKN1A gene expression analysis, RNA-Seq; normal, n = 403, tumour, n = 1097, and metastatic, n = 7 (TCGA Project “Breast Invasive Carcinoma” analysed using TNMplot), Kruskal–Wallis test T vs. N 1.9 × 10−51, N vs. M 8.86 × 10−3.
2.1.2. Expression of CDKN1A/p21 Shows Predictive Promise in Breast Cancer
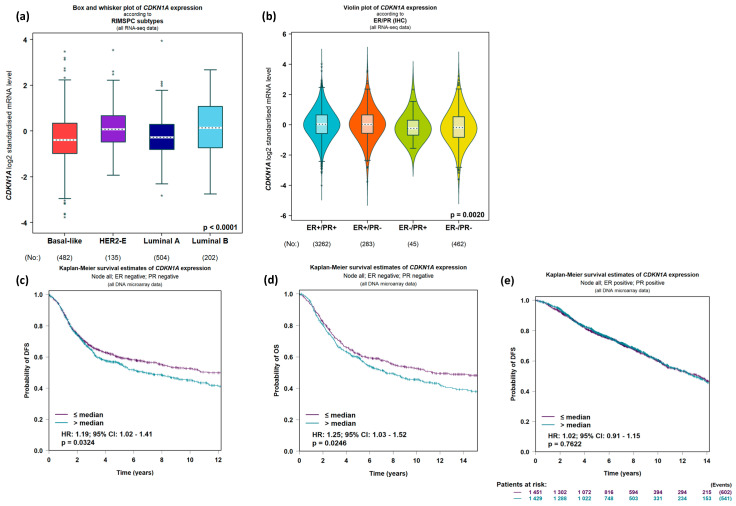
Patient data analysis reveals that CDKN1A/p21 shows a notable preference for Luminal A type (Figure 3a) and HR (Hormone Receptor) subtypes following cancer patient stratification, which could hold significant implications for treatment options (Figure 3b). HR-negative patients, in particular, exhibit a markedly poorer overall prognosis in comparison to HR-positive breast cancer patient datasets. Within this subset of HR-negative patients, stratification based on p21/CDKN1A expression levels emerges as a potent predictor of not only diminished overall survival but also an elevated risk of recurrence and metastasis (Figure 3c,d). Conversely, such trends are not observed within the HR+ patient datasets (Figure 3e).
Figure 3.
Expression levels and predictive power of CDKN1A/p21 in breast cancer patient datasets. Expression levels of CDKN1A log2 standardised mRNA grouping patients based on RIMSPC subtypes (a) or ER/PR expression levels detected using immunohistochemistry (IHC) (b); the number of patients in each dataset is indicated below each graph. (c) Disease-free survival (DFS) and overall survival (OS) analysis (d) of ER/PR-negative breast cancer patients stratifying patients based on high versus low expression levels of CDKN1A: significant p values based on CDKN1A of 0.0324 and 0.0246, respectively. (e) Disease-free survival (DFS) of ER/PR positive breast cancer patient dataset stratifying patients based on high versus low expression levels of CDKN1A. Data was analysed using GenExMiner 5.0.
2.1.3. Chemical Modulation of CDKN1A/p21 Expression and Action
One chemical modulator of CDKN1A/p21 is UC2288, which shares structural similarities with Sorafenib, a kinase inhibitor used in renal cancer therapy. Sorafenib, independent of its other targets, has been shown to reduce the protein levels of CDKN1A/p21 [56], which was also observed for UC2288 both at the mRNA and protein levels. By inhibiting cytosolic CDKN1A/p21, UC2288 impacts its anti-apoptotic function, resulting in decreased viability across various cancer cell lines [57]. Other small molecules, such as butyrolactone [58] and LLW10 [59], have also been shown to decrease CDKN1A/p21 protein levels, increasing degradation. However, their limited stability and lack of specificity make their use in clinical practice unlikely [60].
Histone deacetylase inhibitors (HDACis) represent promising agents in cancer therapy, inducing cell cycle arrest and apoptosis in diverse cell types [61,62]. Part of the mechanism of action of HDACis involves the upregulation of CDKN1A/p21 expression [63,64,65,66,67,68]. In one study, HDAC 2 knockdown using siRNA in HeLa cells resulted in increased CDKN1A/p21 expression at both the mRNA and protein levels, independently of p53 protein expression, leading to increased apoptosis [65]. Additionally, Trichostatin A (TSA), a well-known HDACi, induced the upregulation of apoptosis and growth arrest in the G0/G1 phase of human lymphatic endothelial cells (LEC), in a CDKN1A/p21-dependent and p53-independent manner [66]. In this case, the mRNA level of CDKN1A/p21 increased while the protein level remained constant; however, an accumulation of the protein within the nucleus instead of the cytoplasm was observed.
These observations highlight the dual role of CDKN1A/p21 in tumour growth. UC2288 and HDACi both enhance apoptosis, showing anti-tumour potential. TSA achieves this by increasing the expression levels of CDKN1A/p21, whereas UC2288 exerts its effects by inhibiting the CDKN1A/p21 function at the protein and cell localisation level. A more comprehensive understanding of their mechanisms of action may provide fresh insights into the role of CDKN1A/p21 in cancer progression.
2.2. Role of CDKN1A/p21 in Molecular Processes Important to Cancer Progression
2.2.1. Cell Cycle
The relationship between cancer and cell cycle control is tightly interconnected, as any dysregulation in this process can lead to uncontrolled cell proliferation and tumorigenesis [69]. Targeting specific molecules involved in cell cycle regulation holds great promise in cancer therapy [70].
In this context, the role of the CDKN1A/p21 protein in cell cycle regulation has been well established for many years, yet it appears more complex than initially thought. Firstly, CDKN1A/p21 directly binds to cyclin-dependent kinases (CDKs), hindering the progression of the replication machinery [71,72]. This cell cycle arrest via CDK binding predominantly occurs in the G1 phase and partially in the G2 phase, where higher levels of CDKN1A/p21 correlate with the duration of these phases [73].
Secondly, in normal cells, CDKN1A/p21 forms a complex with CDK and proliferating cell nuclear antigen (PCNA). Even in the absence of CDKs, CDKN1A/p21 can inhibit PCNA-dependent activation of DNA replication [74]. CDKN1A/p21 directly binds to PCNA vis a PIP-Box (PCNA-interacting protein) region near the C-terminal, competing for the same site of PCNA with elements of DNA replication [75]. This interaction during the S-phase may block DNA replication after initiation, providing an alternative safety mechanism if the blockade in the G1 phase proves ineffective. However, PCNA is integral to the DNA damage response (DDR) [76], and thus complete inhibition of PCNA is not beneficial to the cell. Consequently, the interaction of CDKN1A/p21 with PCNA is more intricate. In fact, endogenous levels of CDKN1A/p21 are insufficient to block DNA replication in the S-phase [77].
CDKN1A/p21 can also indirectly influence the cell cycle via its involvement in gene regulation. The overexpression of CDKN1A/p21 in human cells leads to the downregulation of specific genes associated with mitosis and the upregulation of genes linked to senescence, including those encoding extracellular matrix (ECM) proteins [78]. This is achieved via a functional interplay of CDKN1A/p21 with transcription factors. For instance, c-Myc, a known transcription factor promoting mitosis, can directly bind to the carboxyl-terminal region of CDKN1A/p21, the same binding site shared with PCNA. This results in the release of CDKN1A/p21’s inhibitory effect on PCNA activity. Simultaneously, CDKN1A/p21, via binding to c-Myc, represses its transcriptional activity. The balance between these two activities can determine whether the cell progresses through the cell cycle [79].
The DREAM (dimerisation partner, RB-like, E2F, and multi-vulval class B) complex is a protein complex acting as a transcriptional repressor for a significant group of cell cycle genes, via a p53-dependent pathway. In the absence of CDKN1A/p21, the cyclin/CDK complexes hinder the formation of the complete DREAM complex, thereby preventing cell cycle repression. In the presence of CDKN1A/p21, cyclin/CDK complexes are inhibited, permitting the formation of a complete DREAM complex [80].
The retinoblastoma protein (RB) is a tumour suppressor that regulates the cell cycle by forming complexes with the E2F family of transcription factors. CDKN1A/p21 prevents the phosphorylation of RB, resulting in complex repression [81]. Furthermore, p53 binds to the promoters of genes related to cell cycle regulation. Immunoprecipitation assays have shown that the CDKN1A/p21-p53 interaction is essential for p53 to bind to the promoters of PUMA (BBC3), MDM2, and GADD45A [82]. This presents another way in which CDKN1A/p21 indirectly influences the cell cycle via p53-dependent gene regulation.
Moreover, research has shown that CDKN1A/p21 can act as a transcription factor itself. Chromatin immunoprecipitation assays revealed that CDKN1A/p21 binds via its N-terminal region to the transcriptional start site of a set of cell cycle genes, repressing mRNA expression [83]. Additionally, CDKN1A/p21 can directly bind to the enhancer of the SOX2 gene, regulating its activity [84]. These multifaceted roles that CDKN1A/p21 play in cell cycle control within the nucleus make it an intriguing target for cancer therapy; however, the various mechanisms of its action and regulation require further global gene expression, chromatin binding, and pathway analysis to facilitate more sophisticated drug development approaches.
2.2.2. CDKN1A/p21 Is a Key Player in the DNA Damage Response
The DNA damage response (DDR) is a crucial cellular process responsible for safeguarding genomic stability, and defects in this process are a hallmark of tumorigenesis [85]. Given its paramount significance to cell health, many proteins involved in the DDR are targets of drug discovery projects for the treatment of cancer, particularly via the exploitation of deficits in DDR within cancerous cells and inducing synthetic lethality [86].
CDKN1A/p21 has been proposed as a regulator of the cellular response to DNA damage. Firstly, by directly inhibiting cell cycle progression, it affects the cell’s ability to repair the DNA before replication resumes. DNA damage being triggered by external sources prompts an increase in CDKN1A/p21 expression, primarily via a p53-dependent pathway [73,87]. Secondly, CDKN1A/p21 also affects the repair process via binding to PCNA [88,89], which in turns affects the interaction of PCNA with other proteins involved in the DNA repair processes [90]. The role of CDKN1A/p21 in the DNA damage response is dual. In general, CDKN1A/p21 inhibits the response mechanisms by binding to PCNA and preventing binding to regulatory proteins important for the completion of repair, but on the contrary, the presence of CDKN1A/p21 seems to promote repair [91] or regulate the repair process, for example, in the case of BER [92].
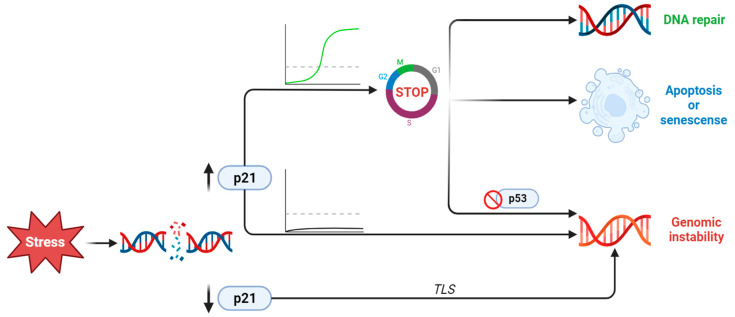
Cell cycle arrest is critical during the DNA damage response (DDR) [93], therefore making CDKN1A/p21, a cell cycle inhibitor, pivotal in this process. A study employing time-lapse imaging and single-cell tracking demonstrated that following damage, CDKN1A/p21 levels must rise above a threshold before the cell enters the S phase, pausing the cell cycle [94]. An interesting exception is Translesion DNA Synthesis (TLS), where pol η is incorporated and binds to proliferating cell nuclear antigen (PCNA). In the absence of replication stress, p21 binds to PCNA, preventing it from interacting with pol η, inhibiting TLS [95]. Furthermore, after DNA damage caused by UV, methylmethane sulfonate, or hydroxyurea, CDKN1A/p21 levels decrease, and TLS takes place [96]. At the single-cell level, it has been observed that after the initial increase via the p53 pathway, CDKN1A/p21 is actively degraded during the S phase to prevent genomic instability, as high levels of CDKN1A/p21 inhibit DNA repair [97]. When CDKN1A/p21 levels remain high, the cell may enter a state of senescence or apoptosis. Notably, Panagiotis Galanos et al. demonstrated that during sustained expression of CDKN1A/p21 in the absence of p53, a subpopulation arises that re-enters the cell cycle, a phenomenon known as re-replication stress, which can lead to genomic instability [98]. In conclusion, the precise regulation of p21 levels is a crucial aspect of the DNA damage response (DDR) (Figure 4).
Figure 4.
Summary of CDKN1A/p21 actions following DNA damage exposure. The protein levels of CDKN1A/p21 after damage may increase or decrease, leading to different outcomes. The decrease or increase under a certain threshold keeps the cell in the cell cycle without any repair of the damage and thus leads to genomic instability. Increases over the threshold may also lead to different outcomes. Initially, the cell arrests the cell cycle in order to repair the damage. If subsequently CDKN1A/p21 levels drop, repair continues. If the levels of CDKN1A/p21 remain high, then the cell becomes senescent, permanently exiting the cell cycle, or enters apoptosis.
Members of the poly-ADP-ribose polymerase (PARP) family, such as PARP1-3, play a vital role in DDR, from the recognition of DNA lesions to chromatin remodelling and repair [99]. PARP1’s involvement in Base Excision Repair (BER) has been extensively studied [100]. In human fibroblasts, the loss of CDKN1A/p21 results in increased chromatin binding of PARP1, leading to an impaired DDR after exposure to DNA-damaging agents. CDKN1A/p21 directly binds to the N-terminal region of PARP1, which encompasses the DNA binding and automodification domains [101]. CDKN1A/p21 also positively regulates the binding of PARP1 to other BER intermediates, as demonstrated in vitro utilising photoreactive BER intermediates [102]. Additionally, PARP1 interacts with p53 [103,104]. p53 is PARylated within the C-terminal domain, which facilitates p53 recruitment to DNA damage sites, as observed in time-lapse experiments measuring the exogenous p53 in p53 null Saos2 cells [105,106]. p53 serves as an upstream regulator of CDKN1A/p21, and its regulation by PARP1 may also influence CDKN1A/p21 expression. Indeed, DDX11-AS1, a long non-coding RNA found to be upregulated in liver cancer patients, affects the interaction between p53 and PARP1, resulting in decreased CDKN1A/p21 expression at both the transcriptional and protein levels. This interaction was demonstrated using a human hepatocellular carcinoma (HCC) cell line [107]. Sustained expression of CDKN1A/p21 in human fibroblasts after DDR activates the MAPK14 and TGFb signalling pathways, which induces the production of reactive oxygen species (ROS). Additionally, ROS-induced DNA damage activates the DDR, creating a positive feedback loop which keeps the cells in growth arrest until the damage is repaired or the cell exits the cell cycle or undergoes apoptosis [108].
2.2.3. CDKN1A/p21 Activation or Repression during EMT, Invasion, and Metastatic Transformation
Epithelial–mesenchymal transition (EMT) is a reversible process in which cells gradually transform from an epithelial-like phenotype to more a mesenchymal one. EMT is important during embryogenesis, tissue regeneration, and cancer. During EMT, a number of transcriptional factors, notably Snail1, TWIST (Twist Family BHLH Transcription Factor 1), and ZEB1/2, are activated, driving expression changes, which leads to alterations in the composition and structure of the cytoskeleton and cell adhesion molecules, a loss of cell junctions, and an increased invasive capability. During cancer progression, as the tumour grows, cells transition into mesenchymal phenotypes, which increases the metastatic ability, cancer stem cell (CSC) properties, and immuno- and drug-resistance [109,110,111,112].
It has been demonstrated in normal breast cell lines [113] and transgenic mice [114] that silencing of CDKN1A/p21 leads to the increased expression of EMT markers including TWIST and Snail1, decreased E-cadherin, and the induction of mesenchymal markers, suggesting that CDKN1A/p21 is a negative regulator of EMT, and may play an important role in the maintenance of the epithelial phenotype. CDKN1A/p21 downregulation is suggested to be necessary for Plasmacytoma Variant Translocation 1 (PVT1) lncRNA-induced EMT in triple-negative breast cancer cells [115] and in pancreatic cancer cells [116]. MiR-149-3p, a microRNA highly expressed in ovarian cancer patients, promotes EMT, in part by downregulating CDKN1A/p21 expression at both the RNA and protein levels [117]. In addition, loss of CDKN1A/p21 is associated with CMS4 colorectal cancer, an aggressive form of colorectal cancer characterised by the presence of mesenchymal-type cells [118].
The exact mechanism of CDKN1A/p21 regulation in EMT is not clear; however, there is emerging evidence to suggest an interplay between CDKN1A/p21 and important EMT transcriptional factors. CDKN1A/p21 forms a complex with ZEB1, inhibiting its action to repress miR-183-96-182, a miRNA that promotes EMT [119]. In another study, using lung carcinoma cells, it was shown that silencing of Snail1 leads to the upregulation of CDKN1A/p21 and cell cycle arrest. The cells also exhibited a more epithelial phenotype, decreased EMT marker expression, and reduced migratory capacity [120]. In contrast, during renal fibrosis, Snail1-induced EMT in animal models is accompanied by an upregulation of the p53–p21 axis and cell cycle arrest [121], suggesting that depending on the biological context, the role of CDKN1A/p21 may be different.
In triple-negative breast cancer, upregulation of the pro-inflammatory cytokine TNFα has been shown to be associated a more aggressive cancer type, via the activation of genes involved in invasion. An example is the metalloproteinase 9 (MMP9), which degrades ECM proteins in the local tumour environment via its proteolytic actions. Similar to MMP9, CDKN1A/p21 expression is upregulated in triple-negative breast cancer following TNFα exposure, which is not observed in normal cells. Importantly, MMP9 upregulation and subsequent increased invasiveness is directly dependent on the expression of CDKN1A/p21, indicating that CDKN1A/p21 may play a direct role in controlling the expression of key genes involved in EMT, invasion, and metastasis [122]. Angiogenesis is an important hallmark of cancer progression and consequently an interesting target for therapy [123]. However, the complexity and variety of mechanisms and pathways, even in the context of the same tumour, results in poor efficacy for anti-angiogenic treatments and acquired tumour resistance [124,125]. Concerning the role of CDKN1A/p21 in this process, studies suggest that CDKN1A/p21 in general promotes angiogenesis. The repression of CDKN1A/p21 in cancer cells decreases their angiogenic capacity in vitro by upregulating the angiogenic factor thioredoxin. Mechanistically, they demonstrated that CDKN1A/p21 binding to the promoter of thioredoxin-binding protein 2 (TBP2) (an inhibitor of thioredoxin) results in reduced TBP2 expression and thus increased thioredoxin [126]. More recently, it was shown that the long non-coding RNA MCM3A-AS1 carried in extracellular vesicles of cervical cancer cells promotes angiogenesis by upregulating CDKN1A/p21. The upregulation of CDKN1A/p21 is due to the binding of MCM3A-AS1 to microRNA-93, which when unbound, inhibits the expression of CDKN1A/p21 [127]. On the contrary, CDKN1A/p21 has been shown to also be able to repress the expression of the angiogenic factor VEGF in primary mouse embryonic fibroblasts (MEFs). Upon sustained hypoxia or DNA damage, CDKN1A/p21 is required for the p53-mediated downregulation of VEGF expression. In addition, CDKN1A/p21-null MEFs present increased angiogenic capacity compared with normal MEFs in xenotransplanted tumours [128]. Interestingly, using an in vivo model of retinal angiogenesis, it was shown that CDKN1A/p21 is spatially expressed in the tip endothelial cells (ECs) of the angiogenic vessels, accompanied by high VEGF stimuli, low Notch signalling, and high ERK signalling. This upregulation of CDKN1A/p21 results in a proportional increase in arrested ECs during highly mitogenic stimuli and consequently impaired angiogenesis. It appears that CDKN1A/p21 may serve as a defensive mechanism reducing EC proliferation and angiogenesis in the event that the ERK signalling pathway, which is known to induce proliferation, is activated [129]. Overall, the data suggest that throughout the process of metastatic progression, invasion, EMT, secondary site colonisation, and angiogenesis, CDKN1A/p21 may play an opposing role, depending on the cellular system, stimulus, and time of activation.
2.2.4. Stem Cells
Cancer stem cells (CSCs) are a small population of cells residing initially within the primary tumour. CSCs are self-renewing (pluripotent) and are considered to be the initiators of tumorigenesis, metastasis, and recurrence [130]. CSCs are often resistant to traditional radio- and chemotherapy treatments [131]; given this and their key role in metastasis, especially for breast cancer, targeting breast cancer stem cells (BCSCs) is a promising target for novel drug discovery and biomarker projects [132,133].
Stem cell renewal and differentiation rely on the coordination between cell cycle progression and exit [134], and it is not surprising that CDKN1A/p21, as a regulator of those functions, plays an important role in the determination of stem cell fate, especially in cancer stem cells [135]. CDKN1A/p21 is also involved in the DNA damage response in adult stem cells, implying that it plays a role in the survival of cancer stem cells after chemotherapy and therefore resistance [136]. Activation of the p53–p21 pathway, for example, after oxidative stress, induces senescence in breast cancer stem cells [137]. Similarly, depletion of the stem cell marker Musashi-1 (MSI1) leads to the upregulation of CDKN1A/p21 and increased apoptosis in breast cancer cells [138]. Additionally, silencing of CDKN1A/p21 in mice and in vitro results in the upregulation of stem cell gene signatures [114].
On the contrary, irradiation of hematopoietic stem cells (HSCs) leads to an upregulation of CDKN1A/p21, independently of p53, protects the cells from apoptosis, and induces self-renewal [139]. In this case, it appears cells adopt a different mechanism in response to DNA damage, protecting them from apoptosis, dependent on the inhibition of p53. To further support this, mesenchymal stem cells (MSCs) deficient in CDKN1A/p21 become more tumorigenic if p53 is also depleted [140].
On the other hand, in basal conditions, CDKN1A/p21 may protect adult stem cells from proliferative exhaustion [141], not only by affecting entry into the cell cycle, which could lead to asymmetrical divisions and the loss of a self-renewal capacity [142], but also by directly downregulating pluripotency factors such as SOX2 in neural stem cells (NSCs) [84]. In the case of embryonic stem cells (ESCs), suppression of CDKN1A/p21 by TET1 is necessary for the maintenance of a proliferative state [143].
It is clear that depending on the cellular system and the developmental stage, CDKN1A/p21 may have contradicting roles in the determination of stem cell fate. In breast cancer, in which we often observe a downregulation of CDKN1A/p21, this may serve as a predictive marker for elevated stem-like properties in tumours, and thus increased probability of metastasis and recurrence.
2.2.5. Apoptosis and Senescence
Dysregulation of apoptotic pathways is known to be a hallmark of cancer, where tumour cells can evade apoptosis via various mechanisms [144,145]. In addition, dysregulation in apoptotic pathways can lead to tumours that do not respond to chemotherapy, resulting in higher rates of recurrence, due to the development of a resistant population [146]. Especially for breast cancer, chemoresistance is a growing concern [147,148].
While nuclear CDKN1A/p21 mainly regulates the cell cycle and DNA damage response, cytoplasmic CDKN1A/p21 can inhibit apoptosis by directly interacting with several apoptotic proteins [149,150]. It has been proposed that CDKN1A/p21 interacts with pro-caspase-3 [151] or apoptosis signal-regulating kinase1 (ASK1) [152], inhibiting their activity. CDKN1A/p21 protects HCT116 cells from IR-induced apoptosis, a function that is diminished in cells mutated within the CDK binding site of CDKN1A/p21 [153]. CDKs play an active role in apoptotic pathways [154,155]; thus, CDKN1A/p21 in the nucleus can inhibit the initiation of apoptotic pathways independently of the effects of the cytoplasmic protein. It has been shown that the p53/p21 complex can directly inhibit anti-apoptotic protein Bcl-2 and induce apoptosis in lung [156] and prostate cancer cells [157].
CDKN1A/p21 is a known partner of many transcription factors, some of which are mediators of apoptotic signalling, for example, c-Myc. CDKN1A/p21 can repress the activity of c-Myc, which in turn can repress the expression of CDKN1A/p21 by binding to its promoter [158]. C-Myc can activate apoptotic pathways via p53-dependent and p53-independent mechanisms [159] and therefore this balance may determine the decision between cell cycle arrest or apoptosis.
All of the above suggests that the anti-apoptotic effect of CDKN1A/p21 is not as simple as previously thought, and it is not facilitated only by cytoplasmic CDKN1A/p21. Cytoplasmic CDKN1A/p21 may also induce apoptosis in certain cellular types. When the cell is faced with stress or DNA damage, CDKN1A/p21 is upregulated. The cell then either transiently arrests the cycle to allow the process of DNA repair to take place or permanently arrests and undergoes apoptosis. A lot of different mechanisms come into play considering arrest and repair or apoptosis, but the levels of CDKN1A/p21 and its cellular localisation seem to be two important factors. Senescence is a complex process with implications in development, ageing, and cancer [160,161,162,163]. Initially recognised for its anti-tumoural attributes due to the permanent cessation of the cell cycle, recent findings have suggested a potential oncogenic role: senescence may facilitate cellular migration and metastasis [164,165]. Notably, senescence induced by anti-cancer treatments emerges as a pivotal factor influencing recurrence and metastatic events [166,167,168], and key drivers of senescence are targets for cancer therapy drug development pipelines.
Considering senescence, the contradictory role of CDKN1A/p21 in cancer progression is important, as its upregulation induces a senescence-like phenotype in diverse cell lines [169,170,171]. The dual nature of senescence likely contributes to the perplexing role of CDKN1A/p21 in cancer biology. Pharmacological senescence was induced in non-small cell lung cancer cells using doxorubicin, and the expression level of CDKN1A/p21 was measured at the single-cell level. Intriguingly, cells exhibiting an early surge in CDKN1A/p21 levels (at 5.7 h post-treatment) entered a proliferative state, whereas those presenting a delayed increase (at 36 h) ended up in a senescent state. Notably, all senescent cells maintained elevated CDKN1A/p21 levels, whereas their proliferative counterparts reverted to lower levels [172]. In addition, it was demonstrated that cytoplasmic p21 may play a key role in CDK4/6 inhibitor-induced senescence in breast cancer cells [173]. These findings provoke intriguing ideas regarding the temporal dynamics of CDKN1A/p21 upregulation and its role in induced pharmacological senescence.
3. Discussion and Future Directions
In this review, we have discussed the implications of CDKN1A/p21 in cancer and present its different cell functions, which can both promote and suppress tumour growth. The dual role of CDKN1A/p21 is reflected in its expression levels in different cancer types compared with normal tissue (Figure 2a). Another interesting issue is the variability in the expression levels of CDKN1A/p21 among patients, for example, in the TCGA breast invasive carcinoma dataset (Figure 2c). This difference among patients could be used as a predictive tool for both outcomes and disease prognosis. Nevertheless, its binary role in cell functions, such as proliferation, apoptosis, stem cell regulation, and DNA damage response, make it clear that a simple intervention targeting CDKN1A/p21 in general may not be successful.
It is well established that CDKN1A/p21 interacts with the replication machinery by binding to CDKs and stopping the cell cycle. Apart from this, it is also an important regulatory factor in the control of gene expression via its interaction with transcriptional factors, such as C-Myc, the DREAM complex, the RB–E2F complex [79,80,81], and p53 [82]. There is strong evidence that CDKN1A/p21 is also a transcriptional factor itself [83,84], binding directly to chromatin and regulating gene expression. Another major function of CDKN1A/p21 is the regulation of DNA damage response (DDR), mainly by binding to PCNA and preventing interaction with DDR components [73]. Despite its inhibitory role, CDKN1A/p21 levels initially increase, in order for the cell cycle arrest and DDR to begin [77]. After the initial increase, the protein is either actively degraded [88], resulting in the initiation of the DNA repair process, or the cell becomes apoptotic or senescent. Despite the fact that cytoplasmic CDKN1A/p21 is mostly known to inhibit apoptosis by binding to pro-caspase 3 and ASK1 [151,152], nuclear CDKN1A/p21 induces apoptosis via complexing with p53 [156]. Nuclear CDKN1A/p21 also binds to CDKs, deactivating their pro-apoptotic function [153,154,155]. Therefore, the decision between apoptosis and survival is dependent on the levels of CDKN1A/p21, the specific timing of the cell cycle, and its cellular localisation. Interestingly, the levels of CDKN1A/p21 after DNA damage do not always increase as discussed previously regarding Translesion DNA Synthesis (TLS) [78,79]. TLS is a well-conserved process by which the cell can bypass DNA damage and continue the cell cycle, mediated by specialised polymerases; this process can result in tumour resistance to DNA-damaging reagents during cancer treatment [174,175,176].
It becomes clear that the regulation of the CDKN1A/p21 levels and its localisation in the cell have to be precisely regulated to prevent genomic instability (Figure 4). CDKN1A/p21 plays a regulatory role in almost all parts of the DDR and consequently determines the cell fate following damage. Also, CDKN1A/p21’s role in TLS and apoptosis suggests that it may be involved in the mechanism of drug resistance to chemotherapeutic reagents. Considering that a change in the levels and cellular localisation of CDKN1A/p21 can shift the balance in favour of cell survival against apoptosis, measurement of CDKN1A/p21 could be considered as a possible predictive marker for identifying patients with an increased risk of resistant disease.
Acquired resistance to chemotherapy is not considered a characteristic of all cancer cells, and presents more commonly within the cancer stem cell (CSC) niche. Drug resistance depends on the cell’s ability to tolerate stress and continue the cell cycle. Thus, it is not surprising that the function of CDKN1A/p21 is important for the conservation of stem cell properties in tumours [135]. In general, the loss of CDKN1A/p21 leads to the upregulation of stem cell properties [114,137,138]. A combined treatment of DNA damaging drugs and reagents aimed at the upregulation of CDKN1A/p21 may be beneficial for patients, especially in cancers where recurrence is frequent, such as breast cancer. Another interesting role of CDKN1A/p21 in cancer is regarding EMT. There is evidence that CDKN1A/p21 negatively regulates EMT [113,114], most probably by interacting with EMT transcriptional factors such as ZEB1 [119] and Snail1 [120]. The exact mechanisms via which it interacts with these and other transcriptional factors still needs to be better understood.
CDKN1A/p21 was originally described as an inhibitor of cell cycle binding to CDKs. Over the years, a lot of other functions, from DNA repair to apoptosis and senescence, were discovered to be part of its repertoire. It would be more suitable to say that CDKN1A/p21 is a regulator of cell fate. This regulation takes place both at the protein and gene expression levels, with the latter still not being well understood. CDKN1A/p21 plays a role in multiple processes important for cancer progression (cell cycle control, DNA damage, apoptosis, senescence, stem cell fate, EMT); however, its action can be both pro- and anti-tumorigenic. It varies depending on the cellular localisation, temporal regulation of its expression, total protein levels, as well as the intrinsic characteristics and gene expression signatures of the specific tissue and cell type. Given its central position in the regulation of cell fate, it makes CDKN1A/p21 a very promising target for cancer treatment. Therefore, CDKN1A/p21 is definitely a part of the problem, but it could also be a part of the solution, especially for breast cancer, where acquired drug resistance and metastasis are responsible for the majority of deaths. However, as discussed, given the dual role of CDKN1A/p21, a complete inhibition or silencing is unlikely to be beneficial. Therefore, a clearer understanding of the specific mechanisms, pathways, and regulators of its function is required so that it may be possible to develop novel specific therapeutic strategies.
Acknowledgments
The authors would like to acknowledge the support of the Basic Science department at the Universitat Internacional de Catalunya (UIC), Barcelona.
Abbreviations
| AKT | AKT serine/threonine kinase 1 |
| AML | Acute Myeloid Leukemia |
| ASK1 | Apoptosis signal-regulating kinase 1 |
| BER | Base Excision Repair |
| BCSC | Breast cancer stem cell |
| CDK | Cyclin-dependent kinase |
| CDKN1A | Cyclin-dependent kinase inhibitory protein 1A |
| CHEK1 | Serine/threonine-protein kinase Chk1 |
| CIP | CDK-interacting protein |
| CMTM6 | CKLF-Like MARVEL Transmembrane Domain-Containing Protein 6 |
| CMS4 | Consensus molecular subtype 4 |
| CPEB4 | Cytoplasmic polyadenylation element-binding protein 4 |
| CRC | Colorectal cancer |
| CSC | Cancer stem cell |
| DAPK1 | Death-associated protein kinase 1 |
| DFS | Disease-free survival |
| DNMT1 | DNA Methyltransferase 1 |
| DDR | DNA damage repair |
| DREAM | Dimerisation partner, RB-like, E2F, and multi-vulval class B |
| DYRK1B | Dual-specificity tyrosine-phosphorylation-regulated kinase 1B |
| EMT | Epithelial–mesenchymal transition |
| GSK | Glycogen synthase kinase |
| HCC | Hepatocellular carcinoma |
| HDAC | Histone Deacetylases |
| H and N | Head and Neck |
| HR | Homologous Recombination |
| KIP | Kinase inhibitor protein |
| LATS2 | Serine/threonine-protein kinase LATS2 |
| LEC | Lymphatic endothelial cells |
| LKB1 | Serine/threonine-protein kinase STK11 |
| LMNB2 | Lamin B2 |
| MAPK | MAP kinase-activated protein kinase |
| MELK | Maternal embryonic leucine zipper kinase |
| MPM | Malignant pleural mesotheliomas |
| MMR | Mismatch repair pathway |
| NER | Nucleotide excision repair |
| NHEJ | Non-homologous end joining |
| NSCLC | Non-small-cell lung carcinoma |
| NUAK1 | NUAK family SNF1-like kinase 1 |
| NUDT5 | Nudix hydrolase 5 |
| ODG | Oligodendrogliomas |
| PARP1 | Poly-ADP-ribose polymerase |
| PCNA | Proliferating cell nuclear antigen |
| PDGF | Platelet-Derived Growth Factor |
| PIM1 | Serine/threonine-protein kinase pim1 |
| PKA/B | Protein kinase B (Akt) |
| PRKCD | Protein kinase C delta type |
| PRMT1 | Protein arginine N-methyltransferase 1 |
| PVT1 | Plasmacytoma Variant Translocation 1 |
| RB | Retinoblastoma |
| RCC | Renal cell carcinoma |
| SOX2 | Sex-determining region Y-box 2 |
| STK38 | Serine/threonine-protein kinase 38 |
| STAT1 | Signal transducer and activator of transcription 1 |
| TLS | Translesion DNA synthesis |
| TSA | Trichostatin A |
| ZEB1 | Zinc finger E-box-binding homeobox 1 |
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms242417488/s1.
Author Contributions
Conceptualisation, E.M. and R.H.G.W.; formal analysis, E.M., R.H.G.W., M.G.E. and C.M.M.; writing—original draft preparation, E.M., R.H.G.W. and M.G.E.; writing—review and editing, R.H.G.W.; project administration, R.H.G.W.; funding acquisition, R.H.G.W. All authors have read and agreed to the published version of the manuscript.
Informed Consent Statement
Not applicable.
Data Availability Statement
All references associated with the public databases analysed in this review are listed in the reference section.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Ministerio de Ciencia e Innovación, grant number RYC2021-030923-I.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Waks A.G., Winer E.P. Breast Cancer Treatment: A Review. JAMA. 2019;321:288–300. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- 3.Agostinetto E., Gligorov J., Piccart M. Systemic therapy for early-stage breast cancer: Learning from the past to build the future. Nat. Rev. Clin. Oncol. 2022;19:763–774. doi: 10.1038/s41571-022-00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giaquinto A.N., Sung H., Miller K.D., Kramer J.L., Newman L.A., Minihan A., Jemal A., Siegel R.L. Breast Cancer Statistics, 2022. CA A Cancer J. Clin. 2022;72:524–541. doi: 10.3322/caac.21754. [DOI] [PubMed] [Google Scholar]
- 5.Nardin S., Mora E., Varughese F.M., D’Avanzo F., Vachanaram A.R., Rossi V., Saggia C., Rubinelli S., Gennari A. Breast Cancer Survivorship, Quality of Life, and Late Toxicities. Front. Oncol. 2020;10:864. doi: 10.3389/fonc.2020.00864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riggio A.I., Varley K.E., Welm A.L. The lingering mysteries of metastatic recurrence in breast cancer. Br. J. Cancer. 2021;124:13–26. doi: 10.1038/s41416-020-01161-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courtney D., Davey M.G., Moloney B.M., Barry M.K., Sweeney K., McLaughlin R.P., Malone C.M., Lowery A.J., Kerin M.J. Breast cancer recurrence: Factors impacting occurrence and survival. Ir. J. Med. Sci. 2022;191:2501–2510. doi: 10.1007/s11845-022-02926-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zohny S.F., Al-Malki A.L., Zamzami M.A., Choudhry H. p21Waf1/Cip1: Its paradoxical effect in the regulation of breast cancer. Breast Cancer. 2019;26:131–137. doi: 10.1007/s12282-018-0913-1. [DOI] [PubMed] [Google Scholar]
- 9.Besson A., Dowdy S.F., Roberts J.M. CDK Inhibitors: Cell Cycle Regulators and Beyond. Dev. Cell. 2008;14:159–169. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 10.Ullah Z., Lee C.Y., DePamphilis M.L. Cip/Kip cyclin-dependent protein kinase inhibitors and the road to polyploidy. Cell Div. 2009;4:10. doi: 10.1186/1747-1028-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uhlén M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F., et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 12.Asada M., Yamada T., Ichijo H., Delia D., Miyazono K., Fukumuro K., Mizutani S. Apoptosis inhibitory activity of cytoplasmic p21 Cip1/WAF1 in monocytic differentiation. EMBO J. 1999;18:1223–1234. doi: 10.1093/emboj/18.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asada M., Ohmi K., Delia D., Enosawa S., Suzuki S., Yuo A., Suzuki H., Mizutani S. Brap2 Functions as a Cytoplasmic Retention Protein for p21 during Monocyte Differentiation. Mol. Cell. Biol. 2004;24:8236–8243. doi: 10.1128/MCB.24.18.8236-8243.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tak J., Sabarwal A., Shyanti R.K., Singh R.P. Berberine enhances posttranslational protein stability of p21/cip1 in breast cancer cells via down-regulation of Akt. Mol. Cell. Biochem. 2019;458:49–59. doi: 10.1007/s11010-019-03529-4. [DOI] [PubMed] [Google Scholar]
- 15.del Toro N., Shrivastava A., Ragueneau E., Meldal B., Combe C., Barrera E., Perfetto L., How K., Ratan P., Shirodkar G., et al. The IntAct database: Efficient access to fine-grained molecular interaction data. Nucleic Acids Res. 2022;50:D648–D653. doi: 10.1093/nar/gkab1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seong H.-A., Ha H. Thr55 phosphorylation of p21 by MPK38/MELK ameliorates defects in glucose, lipid, and energy metabolism in diet-induced obese mice. Cell Death Dis. 2019;10:380. doi: 10.1038/s41419-019-1616-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang C.Y., Lee C., Kwon K.-S. Extracellular Signal-Regulated Kinase 2-Dependent Phosphorylation Induces Cytoplasmic Localization and Degradation of p21Cip1. Mol. Cell. Biol. 2009;29:3379–3389. doi: 10.1128/MCB.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foertsch F., Teichmann N., Kob R., Hentschel J., Laubscher U., Melle C. S100A11 is involved in the regulation of the stability of cell cycle regulator p21CIP1/WAF1 in human keratinocyte HaCaT cells. FEBS J. 2013;280:3840–3853. doi: 10.1111/febs.12378. [DOI] [PubMed] [Google Scholar]
- 19.Esteve-Puig R., Gil R., González-Sánchez E., Bech-Serra J.J., Grueso J., Hernández-Losa J., Moliné T., Canals F., Ferrer B., Cortés J., et al. A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21WAF1/CIP1) Degradation. PLOS Genet. 2014;10:e1004721. doi: 10.1371/journal.pgen.1004721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., Dowbenko D., Lasky L.A. AKT/PKB Phosphorylation of p21Cip/WAF1 Enhances Protein Stability of p21Cip/WAF1 and Promotes Cell Survival. J. Biol. Chem. 2002;277:11352–11361. doi: 10.1074/jbc.M109062200. [DOI] [PubMed] [Google Scholar]
- 21.Rössig L., Jadidi A.S., Urbich C., Badorff C., Zeiher A.M., Dimmeler S. Akt-Dependent Phosphorylation of p21Cip1 Regulates PCNA Binding and Proliferation of Endothelial Cells. Mol. Cell. Biol. 2001;21:5644–5657. doi: 10.1128/MCB.21.16.5644-5657.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stuart S.A., Houel S., Lee T., Wang N., Old W.M., Ahn N.G. A Phosphoproteomic Comparison of B-RAFV600E and MKK1/2 Inhibitors in Melanoma Cells. Mol. Cell. Proteom. 2015;14:1599–1615. doi: 10.1074/mcp.M114.047233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colleoni B., Paternot S., Pita J.M., Bisteau X., Coulonval K., Davis R.J., Raspé E., Roger P.P. JNKs function as CDK4-activating kinases by phosphorylating CDK4 and p21. Oncogene. 2017;36:4349–4361. doi: 10.1038/onc.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J.Y., Yu S.J., Park Y.G., Kim J., Sohn J. Glycogen Synthase Kinase 3β Phosphorylates p21WAF1/CIP1 for Proteasomal Degradation after UV Irradiation. Mol. Cell. Biol. 2007;27:3187–3198. doi: 10.1128/MCB.01461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X., Zhang J., Zhang M., Liu S., Yan W., Jung J., Chen X. Serine 123 Phosphorylation Modulates p21 Protein Stability and Activity by Suppressing Ubiquitin-independent Proteasomal Degradation. J. Biol. Chem. 2012;287:34410–34418. doi: 10.1074/jbc.M112.384990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi T., Zhai B., Yu Y., Kiyotsugu Y., Raschle T., Etzkorn M., Seo H.-C., Nagiec M., Luna R.E., Reinherz E.L., et al. Quantitative phosphoproteomic analysis reveals system-wide signaling pathways downstream of SDF-1/CXCR4 in breast cancer stem cells. Proc. Natl. Acad. Sci. USA. 2014;111:E2182–E2190. doi: 10.1073/pnas.1404943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Järviluoma A., Child E.S., Sarek G., Sirimongkolkasem P., Peters G., Ojala P.M., Mann D.J. Phosphorylation of the Cyclin-Dependent Kinase Inhibitor p21Cip1 on Serine 130 Is Essential for Viral Cyclin-Mediated Bypass of a p21Cip1-Imposed G1 Arrest. Mol. Cell. Biol. 2006;26:2430–2440. doi: 10.1128/MCB.26.6.2430-2440.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu H., Nie L., Maki C.G. Cdk2-dependent Inhibition of p21 Stability via a C-terminal Cyclin-binding Motif. J. Biol. Chem. 2005;280:29282–29288. doi: 10.1074/jbc.M407352200. [DOI] [PubMed] [Google Scholar]
- 29.Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V., Skrzypek E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2014;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodríguez-Vilarrupla A., Jaumot M., Abella N., Canela N., Brun S., Díaz C., Estanyol J.M., Bachs O., Agell N. Binding of Calmodulin to the Carboxy-Terminal Region of p21 Induces Nuclear Accumulation via Inhibition of Protein Kinase C-Mediated Phosphorylation of Ser153. Mol. Cell. Biol. 2005;25:7364–7374. doi: 10.1128/MCB.25.16.7364-7374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott M.T., Morrice N., Ball K.L. Reversible Phosphorylation at the C-Terminal Regulatory Domain of p21 Waf1/Cip1 Modulates Proliferating Cell Nuclear Antigen Binding. J. Biol. Chem. 2000;275:11529–11537. doi: 10.1074/jbc.275.15.11529. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z., Zhang Y., Gu J.J., Davitt C., Reeves R., Magnuson N.S. Pim-2 phosphorylation of p21Cip1/WAF1 enhances its stability and inhibits cell proliferation in HCT116 cells. Int. J. Biochem. Cell Biol. 2010;42:1030–1038. doi: 10.1016/j.biocel.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mertins P., Yang F., Liu T., Mani D.R., Petyuk V.A., Gillette M.A., Clauser K.R., Qiao J.W., Gritsenko M.A., Moore R.J., et al. Ischemia in Tumors Induces Early and Sustained Phosphorylation Changes in Stress Kinase Pathways but Does Not Affect Global Protein Levels. Mol. Cell. Proteomics. 2014;13:1690–1704. doi: 10.1074/mcp.M113.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu D., Richardson D.R. Iron chelation and regulation of the cell cycle: 2 mechanisms of posttranscriptional regulation of the universal cyclin-dependent kinase inhibitor p21CIP1/WAF1 by iron depletion. Blood. 2007;110:752–761. doi: 10.1182/blood-2007-03-076737. [DOI] [PubMed] [Google Scholar]
- 35.Li Z., Zhang Q., Mao J.-H., Weise A., Mrasek K., Fan X., Zhang X., Liehr T., Lu K.H., Balmain A., et al. An HDAC1-binding domain within FATS bridges p21 turnover to radiation-induced tumorigenesis. Oncogene. 2010;29:2659–2671. doi: 10.1038/onc.2010.19. [DOI] [PubMed] [Google Scholar]
- 36.Nakakido M., Deng Z., Suzuki T., Dohmae N., Nakamura Y., Hamamoto R. PRMT6 Increases Cytoplasmic Localization of p21CDKN1A in Cancer Cells through Arginine Methylation and Makes More Resistant to Cytotoxic Agents. Oncotarget. 2015;6:30957–30967. doi: 10.18632/oncotarget.5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teramen H., Tsukuda K., Tanaka N., Ueno T., Kubo T., Ando M., Soh J., Asano H., Pass H.I., Toyooka S., et al. Aberrant Methylation of p21 Gene in Lung Cancer and Malignant Pleural Mesothelioma. Acta Medica Okayama. 2011;65:179–184. doi: 10.18926/AMO/46629. [DOI] [PubMed] [Google Scholar]
- 38.Bott S.R.J., Arya M., Kirby R.S., Williamson M. p21WAF1/CIP1 gene is inactivated in metastatic prostatic cancer cell lines by promoter methylation. Prostate Cancer Prostatic Dis. 2005;8:321–326. doi: 10.1038/sj.pcan.4500822. [DOI] [PubMed] [Google Scholar]
- 39.Askari M., Sobti R.C., Nikbakht M., Sharma S.C. Aberrant promoter hypermethylation of p21 (WAF1/CIP1) gene and its impact on expression and role of polymorphism in the risk of breast cancer. Mol. Cell. Biochem. 2013;382:19–26. doi: 10.1007/s11010-013-1696-5. [DOI] [PubMed] [Google Scholar]
- 40.Roman-Gomez J., Castillejo J.A., Jimenez A., Gonzalez M.G., Moreno F., del Carmen Rodriguez M., Barrios M., Maldonado J., Torres A. 5 CpG Island Hypermethylation Is Associated with Transcriptional Silencing of the p21 CIP1/WAF1/SDI1 Gene and Confers Poor Prognosis in Acute Lymphoblastic Leukemia. 2002. [(accessed on 1 January 2023)]. Available online: http://ashpublications.org/blood/article-pdf/99/7/2291/1682050/2291.pdf. [DOI] [PubMed]
- 41.Xu M., Gao J., Du Y.Q., Gao D.-J., Zhang Y.Q., Li Z.S., Zhang Y.L., Li Y., Gong Y.F., Xu P. Reduction of pancreatic cancer cell viability and induction of apoptosis mediated by siRNA targeting DNMT1 through suppression of total DNA methyltransferase activity. Mol. Med. Rep. 2010;3:699–704. doi: 10.3892/mmr_00000320. [DOI] [PubMed] [Google Scholar]
- 42.Dong C.-H., Jiang T., Yin H., Song H., Zhang Y., Geng H., Shi P.-C., Xu Y.-X., Gao H., Liu L.-Y., et al. LMNB2 promotes the progression of colorectal cancer by silencing p21 expression. Cell Death Dis. 2021;12:331. doi: 10.1038/s41419-021-03602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamawaki K., Ishiguro T., Mori Y., Yoshihara K., Suda K., Tamura R., Yamaguchi M., Sekine M., Kashima K., Higuchi M., et al. Sox2-dependent inhibition of p21 is associated with poor prognosis of endometrial cancer. Cancer Sci. 2017;108:632–640. doi: 10.1111/cas.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di J., Wang H., Zhao Z., Zhao G., Qin X., Han Z., Liu Y. CPEB4 Inhibit Cell Proliferation via Upregulating p21 mRNA Stability in Renal Cell Carcinoma. Front. Cell Dev. Biol. 2021;9:687253. doi: 10.3389/fcell.2021.687253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Y., Zhu Y., Yang J., Pan Q., Zhao J., Song M., Yang C., Han Y., Tang Y., Wang Q., et al. CMTM6 inhibits tumor growth and reverses chemoresistance by preventing ubiquitination of p21 in hepatocellular carcinoma. Cell Death Dis. 2022;13:251. doi: 10.1038/s41419-022-04676-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y., Yeh N., Zhu X.-H., Leversha M., Cordon-Cardo C., Ghossein R., Singh B., Holland E., Koff A. Somatic cell type specific gene transfer reveals a tumor-promoting function for p21Waf1/Cip1. EMBO J. 2007;26:4683–4693. doi: 10.1038/sj.emboj.7601886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De la Cueva E., García-Cao I., Herranz M., López P., García-Palencia P., Flores J.M., Serrano M., Fernández-Piqueras J., Martín-Caballero J. Tumorigenic activity of p21Waf1/Cip1 in thymic lymphoma. Oncogene. 2006;25:4128–4132. doi: 10.1038/sj.onc.1209432. [DOI] [PubMed] [Google Scholar]
- 48.Benson E.K., Zhao B., Sassoon D.A., Lee S.W., Aaronson S.A. Effects of p21 deletion in mouse models of premature aging. Cell Cycle. 2009;8:2002–2004. doi: 10.4161/cc.8.13.8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng C., Zhang P., Harper J.W., Elledge S.J., Leder P., McLean M. Mice Lacking p21 c~P7/wAF7 Undergo Normal Development, but Are Defective in Gl Checkpoint Control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-X. [DOI] [PubMed] [Google Scholar]
- 50.Groza T., Gomez F.L., Mashhadi H.H., Muñoz-Fuentes V., Gunes O., Wilson R., Cacheiro P., Frost A., Keskivali-Bond P., Vardal B., et al. The International Mouse Phenotyping Consortium: Comprehensive knockout phenotyping underpinning the study of human disease. Nucleic Acids Res. 2023;51:D1038–D1045. doi: 10.1093/nar/gkac972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.The AACR Project GENIE Consortium AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017;7:818–831. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou B.P., Liao Y., Xia W., Spohn B., Lee M.-H., Hung M.-C. Cytoplasmic Localization of p21 Cip1/WAF1 by Akt-Induced Phosphorylation in HER-2/Neu-Overexpressing Cells. 2001. [(accessed on 1 January 2023)]. Available online: http://cellbio.nature.com. [DOI] [PubMed]
- 53.Zhou Y., Li G., Ji Y., Liu C., Zhu J., Lu Y. Cytoplasmic p21 induced by p65 prevents doxorubicin-induced cell death in pancreatic carcinoma cell line. J. Biomed. Sci. 2012;19:15. doi: 10.1186/1423-0127-19-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vincent A.J., Ren S., Harris L.G., Devine D.J., Samant R.S., Fodstad O., Shevde L.A. Cytoplasmic translocation of p21 mediates NUPR1-induced chemoresistance: NUPR1 and p21 in chemoresistance. FEBS Lett. 2012;586:3429–3434. doi: 10.1016/j.febslet.2012.07.063. [DOI] [PubMed] [Google Scholar]
- 55.Bartha Á., Győrffy B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021;22:2622. doi: 10.3390/ijms22052622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Inoue H., Hwang S.H., Wecksler A.T., Hammock B.D., Weiss R.H. Sorafenib attenuates p21 in kidney cancer cells and augments cell death in combination with DNA-damaging chemotherapy. Cancer Biol. Ther. 2011;12:827–836. doi: 10.4161/cbt.12.9.17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wettersten H.I., Hwang S.H., Li C., Shiu E.Y., Wecksler A.T., Hammock B.D., Weiss R.H. A novel p21 attenuator which is structurally related to sorafenib. Cancer Biol. Ther. 2013;14:278–285. doi: 10.4161/cbt.23374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sax J.K., Dash B.C., Hong R., Dicker D.T., El-Deiry W.S. The Cyclin-Dependent Kinase Inhibitor Butyrolactone Is a Potent Inhibitor of p21WAF1/CIP1 Expression. Cell Cycle. 2002;1:90–96. doi: 10.4161/cc.1.1.105. [DOI] [PubMed] [Google Scholar]
- 59.Park S.-H., Wang X., Liu R., Lam K.S., Weiss R.H. High throughput screening of a small molecule one-bead-one-compound combinatorial library to identify attenuators of p21 as chemotherapy sensitizers. Cancer Biol. Ther. 2008;7:2015–2022. doi: 10.4161/cbt.7.12.7069. [DOI] [PubMed] [Google Scholar]
- 60.Liu R., I Wettersten H., Park S.-H., Weiss R.H., Supuran C.T., Winum J.-Y., Shull A.Y., Farrell C.L., Wang R., Ma X., et al. Small-molecule inhibitors of p21 as novel therapeutics for chemotherapy-resistant kidney cancer. Futur. Med. Chem. 2013;5:991–994. doi: 10.4155/fmc.13.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y., Seto E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb. Perspect. Med. 2016;6:a026831. doi: 10.1101/cshperspect.a026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li G., Tian Y., Zhu W.-G. The Roles of Histone Deacetylases and Their Inhibitors in Cancer Therapy. Front. Cell Dev. Biol. 2020;8:576946. doi: 10.3389/fcell.2020.576946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gui C.-Y., Ngo L., Xu W.S., Richon V.M., Marks P.A. Histone Deacetylase (HDAC) Inhibitor Activation of p21 WAF1 Involves Changes in Promoter-Associated Proteins, Including HDAC1. 2004. [(accessed on 1 February 2023)]. Available online: https://www.pnas.org. [DOI] [PMC free article] [PubMed]
- 64.Bellucci L., Dalvai M., Kocanova S., Moutahir F., Bystricky K. Activation of p21 by HDAC Inhibitors Requires Acetylation of H2A.Z. PLoS ONE. 2013;8:e54102. doi: 10.1371/journal.pone.0054102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang B.H., Laban M., Leung C.H., Lee L., Lee C.K., Salto-Tellez M., Raju G.C., Hooi S.C. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1/WAF1 expression, independent of histone deacetylase 1. Cell Death Differ. 2005;12:395–404. doi: 10.1038/sj.cdd.4401567. [DOI] [PubMed] [Google Scholar]
- 66.Hrgovic I., Doll M., Kleemann J., Wang X.-F., Zoeller N., Pinter A., Kippenberger S., Kaufmann R., Meissner M. The histone deacetylase inhibitor trichostatin a decreases lymphangiogenesis by inducing apoptosis and cell cycle arrest via p21-dependent pathways. BMC Cancer. 2016;16:763. doi: 10.1186/s12885-016-2807-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kakiuchi A., Kakuki T., Ohwada K., Kurose M., Kondoh A., Obata K., Nomura K., Miyata R., Kaneko Y., Konno T., et al. HDAC inhibitors suppress the proliferation, migration and invasiveness of human head and neck squamous cell carcinoma cells via p63-mediated tight junction molecules and p21-mediated growth arrest. Oncol. Rep. 2021;45:46. doi: 10.3892/or.2021.7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kavoosi F., Sanaei M., Safari M. Effect of 5′-fluoro-2′-deoxycytidine and sodium butyrate on the gene expression of the intrinsic apoptotic pathway, p21, p27, and p53 genes expression, cell viability, and apoptosis in human hepatocellular carcinoma cell lines. Adv. Biomed. Res. 2023;12:24. doi: 10.4103/abr.abr_211_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matthews H.K., Bertoli C., de Bruin R.A.M. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2022;23:74–88. doi: 10.1038/s41580-021-00404-3. [DOI] [PubMed] [Google Scholar]
- 70.Suski J.M., Braun M., Strmiska V., Sicinski P. Targeting cell-cycle machinery in cancer. Cancer Cell. 2021;39:759–778. doi: 10.1016/j.ccell.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiong Y., Hannon G.J., Zhang H., Casso D., Kobayashi R., Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 72.Harper J.W., Adami G.R., Wei N., Keyomarsi K., Elledge S.J. The p21 Cdk-Interacting Protein Cipl Is a Potent Inhibitor of Gl Cyclin-Dependent Kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-G. [DOI] [PubMed] [Google Scholar]
- 73.Barr A.R., Cooper S., Heldt F.S., Butera F., Stoy H., Mansfeld J., Novák B., Bakal C. DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression. Nat. Commun. 2017;8:14728. doi: 10.1038/ncomms14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waga S., Hannon G.J., Beach D., Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 75.Bruning J.B., Shamoo Y. Structural and Thermodynamic Analysis of Human PCNA with Peptides Derived from DNA Polymerase-δ p66 Subunit and Flap Endonuclease-1. Structure. 2004;12:2209–2219. doi: 10.1016/j.str.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 76.Boehm E.M., Gildenberg M.S., Washington M.T. The Many Roles of PCNA in Eukaryotic DNA Replication. Enzymes. 2016;39:231–254. doi: 10.1016/bs.enz.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mansilla S.F., De La Vega M.B., Calzetta N.L., Siri S.O., Gottifredi V. CDK-Independent and PCNA-Dependent Functions of p21 in DNA Replication. Genes. 2020;11:593. doi: 10.3390/genes11060593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chang B.D., Watanabe K., Broude E.V., Fang J., Poole J.C., Kalinichenko T.V., Roninson I.B. Effects of p21 Waf1/Cip1/Sdi1 on Cellular Gene Expression: Implications for Carcinogenesis, Senescence, and Age-Related Diseases. 2000. [(accessed on 1 January 2023)]. Available online: www.pnas.org. [DOI] [PMC free article] [PubMed]
- 79.Kitaura H., Shinshi M., Uchikoshi Y., Ono T., Iguchi-Ariga S.M., Ariga H. Reciprocal Regulation via Protein-Protein Interaction between c-Myc and p21 cip1/waf1/sdi1 in DNA Replication and Transcription. J. Biol. Chem. 2000;275:10477–10483. doi: 10.1074/jbc.275.14.10477. [DOI] [PubMed] [Google Scholar]
- 80.Engeland K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 2018;25:114–132. doi: 10.1038/cdd.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Engeland K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ. 2022;29:946–960. doi: 10.1038/s41418-022-00988-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim U., Kim K.S., Park J.-K., Um H.-D. Involvement of the p53/p21 complex in p53-dependent gene expression. Biochem. Biophys. Res. Commun. 2022;621:151–156. doi: 10.1016/j.bbrc.2022.07.022. [DOI] [PubMed] [Google Scholar]
- 83.Ferrándiz N., Caraballo J.M., García-Gutierrez L., Devgan V., Rodriguez-Paredes M., Lafita M.C., Bretones G., Quintanilla A., Muñoz-Alonso M.J., Blanco R., et al. p21 as a Transcriptional Co-Repressor of S-Phase and Mitotic Control Genes. PLoS ONE. 2012;7:e37759. doi: 10.1371/journal.pone.0037759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marqués-Torrejón M., Porlan E., Banito A., Gómez-Ibarlucea E., Lopez-Contreras A.J., Fernández-Capetillo O., Vidal A., Gil J., Torres J., Fariñas I. Cyclin-Dependent Kinase Inhibitor p21 Controls Adult Neural Stem Cell Expansion by Regulating Sox2 Gene Expression. Cell Stem Cell. 2013;12:88–100. doi: 10.1016/j.stem.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hopkins J.L., Lan L., Zou L. DNA repair defects in cancer and therapeutic opportunities. Minerva Anestesiol. 2022;36:278–293. doi: 10.1101/gad.349431.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Groelly F.J., Fawkes M., Dagg R.A., Blackford A.N., Tarsounas M. Targeting DNA damage response pathways in cancer. Nat. Rev. Cancer. 2023;23:78–94. doi: 10.1038/s41568-022-00535-5. [DOI] [PubMed] [Google Scholar]
- 87.He G., Siddik Z.H., Huang Z., Wang R., Koomen J., Kobayashi R., Khokhar A.R., Kuang J. Induction of p21 by p53 following DNA damage inhibits both Cdk4 and Cdk2 activities. Oncogene. 2005;24:2929–2943. doi: 10.1038/sj.onc.1208474. [DOI] [PubMed] [Google Scholar]
- 88.Cazzalini O., Scovassi A.I., Savio M., Stivala L.A., Prosperi E. Multiple roles of the cell cycle inhibitor p21CDKN1A in the DNA damage response. Mutat. Res. Mol. Mech. Mutagen. 2010;704:12–20. doi: 10.1016/j.mrrev.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 89.Mailand N., Gibbs-Seymour I., Bekker-Jensen S. Regulation of PCNA–protein interactions for genome stability. Nat. Rev. Mol. Cell Biol. 2013;14:269–282. doi: 10.1038/nrm3562. [DOI] [PubMed] [Google Scholar]
- 90.Ticli G., Cazzalini O., Stivala L.A., Prosperi E. Revisiting the Function of p21CDKN1A in DNA Repair: The Influence of Protein Interactions and Stability. Int. J. Mol. Sci. 2022;23:7058. doi: 10.3390/ijms23137058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mostofa A., Punganuru S.R., Madala H.R., Srivenugopal K.S. S-phase Specific Downregulation of Human O6-Methylguanine DNA Methyltransferase (MGMT) and its Serendipitous Interactions with PCNA and p21cip1 Proteins in Glioma Cells. Neoplasia. 2018;20:305–323. doi: 10.1016/j.neo.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sukhanova M.V., Khodyreva S.N., Lebedeva N.A., Prasad R., Wilson S.H., Lavrik O.I. Human base excision repair enzymes apurinic/apyrimidinic endonuclease1 (APE1), DNA polymerase β and poly(ADP-ribose) polymerase 1: Interplay between strand-displacement DNA synthesis and proofreading exonuclease activity. Nucleic Acids Res. 2005;33:1222–1229. doi: 10.1093/nar/gki266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Campos A., Clemente-Blanco A. Cell Cycle and DNA Repair Regulation in the Damage Response: Protein Phosphatases Take over the Reins. Int. J. Mol. Sci. 2020;21:446. doi: 10.3390/ijms21020446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nathans J.F., Cornwell J.A., Afifi M.M., Paul D., Cappell S.D. Cell cycle inertia underlies a bifurcation in cell fates after DNA damage. Sci. Adv. 2021;7:eabe3882. doi: 10.1126/sciadv.abe3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Prives C., Gottifredi V. The p21 and PCNA partnership: A new twist for an old plot. Cell Cycle. 2008;7:3840–3846. doi: 10.4161/cc.7.24.7243. [DOI] [PubMed] [Google Scholar]
- 96.Soria G., Gottifredi V. PCNA-coupled p21 degradation after DNA damage: The exception that confirms the rule? DNA Repair. 2010;9:358–364. doi: 10.1016/j.dnarep.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sheng C., Mendler I.-H., Rieke S., Snyder P., Jentsch M., Friedrich D., Drossel B., Loewer A. PCNA-Mediated Degradation of p21 Coordinates the DNA Damage Response and Cell Cycle Regulation in Individual Cells. Cell Rep. 2019;27:48–58.e7. doi: 10.1016/j.celrep.2019.03.031. [DOI] [PubMed] [Google Scholar]
- 98.Galanos P., Vougas K., Walter D., Polyzos A., Maya-Mendoza A., Haagensen E.J., Kokkalis A., Roumelioti F.-M., Gagos S., Tzetis M., et al. Chronic p53-independent p21 expression causes genomic instability by deregulating replication licensing. Nature. 2016;18:777–789. doi: 10.1038/ncb3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sousa F.G., Matuo R., Soares D.G., Escargueil A.E., Henriques J.A., Larsen A.K., Saffi J. PARPs and the DNA damage response. Carcinogenesis. 2012;33:1433–1440. doi: 10.1093/carcin/bgs132. [DOI] [PubMed] [Google Scholar]
- 100.Chaudhuri A.R., Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017;18:610–621. doi: 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cazzalini O., Donà F., Savio M., Tillhon M., Maccario C., Perucca P., Stivala L.A., Scovassi A.I., Prosperi E. p21CDKN1A participates in base excision repair by regulating the activity of poly(ADP-ribose) polymerase-1. DNA Repair. 2010;9:627–635. doi: 10.1016/j.dnarep.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 102.Dutto I., Sukhanova M., Tillhon M., Cazzalini O., Stivala L.A., Scovassi A.I., Lavrik O., Prosperi E. p21CDKN1A Regulates the Binding of Poly(ADP-Ribose) Polymerase-1 to DNA Repair Intermediates. PLoS ONE. 2016;11:e0146031. doi: 10.1371/journal.pone.0146031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vaziri H., West M.D., Allsopp R.C., Davison T.S., Wu Y.-S., Arrowsmith C.H., Poirier G.G., Benchimol S. ATM-dependent telomere loss in aging human diploid fibroblasts and DNA damage lead to the post-translational activation of p53 protein involving poly(ADP-ribose) polymerase. EMBO J. 1997;16:6018–6033. doi: 10.1093/emboj/16.19.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wojciechowski J., Schmid G., Węsierska-Gądek J. Central and carboxy-terminal regions of human p53 protein are essential for interaction and complex formation with PARP-1. J. Cell. Biochem. 2003;89:220–232. doi: 10.1002/jcb.10521. [DOI] [PubMed] [Google Scholar]
- 105.Fischbach A., Krüger A., Hampp S., Assmann G., Rank L., Hufnagel M., Stöckl M.T., Fischer J.M., Veith S., Rossatti P., et al. The C-terminal domain of p53 orchestrates the interplay between non-covalent and covalent poly(ADP-ribosyl)ation of p53 by PARP1. Nucleic Acids Res. 2018;46:804–822. doi: 10.1093/nar/gkx1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Y.-H., Ho T.L.F., Hariharan A., Goh H.C., Wong Y.L., Verkaik N.S., Lee M.Y., Tam W.L., van Gent D.C., Venkitaraman A.R., et al. Rapid recruitment of p53 to DNA damage sites directs DNA repair choice and integrity. Proc. Natl. Acad. Sci. USA. 2022;119:e2113233119. doi: 10.1073/pnas.2113233119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu M., Zhao X., Zhao S., Yang Z., Yuan W., Han H., Zhang B., Zhou L., Zheng S., Li M.D. Landscape analysis of lncRNAs shows that DDX11-AS1 promotes cell-cycle progression in liver cancer through the PARP1/p53 axis. Cancer Lett. 2021;520:282–294. doi: 10.1016/j.canlet.2021.08.001. [DOI] [PubMed] [Google Scholar]
- 108.Passos J.F., Nelson G., Wang C., Richter T., Simillion C., Proctor C.J., Miwa S., Olijslagers S., Hallinan J., Wipat A., et al. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol. Syst. Biol. 2010;6:347. doi: 10.1038/msb.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ribatti D., Tamma R., Annese T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Transl. Oncol. 2020;13:100773. doi: 10.1016/j.tranon.2020.100773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dongre A., Weinberg R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 111.Huang Z., Zhang Z., Zhou C., Liu L., Huang C. Epithelial–mesenchymal transition: The history, regulatory mechanism, and cancer therapeutic opportunities. Medcomm. 2022;3:e144. doi: 10.1002/mco2.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bracken C.P., Goodall G.J. The many regulators of epithelial−mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2022;23:89–90. doi: 10.1038/s41580-021-00442-x. [DOI] [PubMed] [Google Scholar]
- 113.Zhang Y., Yan W., Jung Y.S., Chen X. PUMA Cooperates with p21 to Regulate Mammary Epithelial Morphogenesis and Epithelial-To-Mesenchymal Transition. PLoS ONE. 2013;8:e66464. doi: 10.1371/journal.pone.0066464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu M., Casimiro M.C., Wang C., Shirley L.A., Jiao X., Katiyar S., Ju X., Li Z., Yu Z., Zhou J., et al. p21 CIP1 attenuates Ras- and c-Myc-dependent breast tumor epithelial mesenchymal transition and cancer stem cell-like gene expression in vivo. Proc. Natl. Acad. Sci. USA. 2009;106:19035–19039. doi: 10.1073/pnas.0910009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang L., Wang R., Ye Z., Wang Y., Li X., Chen W., Zhang M., Cai C. PVT1 affects EMT and cell proliferation and migration via regulating p21 in triple-negative breast cancer cells cultured with mature adipogenic medium. Acta Biochim. Biophys. Sin. 2018;50:1211–1218. doi: 10.1093/abbs/gmy129. [DOI] [PubMed] [Google Scholar]
- 116.Wu B.-Q., Jiang Y., Zhu F., Sun D.-L., He X.-Z. Long Noncoding RNA PVT1 Promotes EMT and Cell Proliferation and Migration through Downregulating p21 in Pancreatic Cancer Cells. Technol. Cancer Res. Treat. 2017;16:819–827. doi: 10.1177/1533034617700559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang J., Liu L. MiR-149-3p promotes the cisplatin resistance and EMT in ovarian cancer through downregulating TIMP2 and CDKN1A. J. Ovarian Res. 2021;14:165. doi: 10.1186/s13048-021-00919-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bueno-Fortes S., Muenzner J.K., Berral-Gonzalez A., Hampel C., Lindner P., Berninger A., Huebner K., Kunze P., Bäuerle T., Erlenbach-Wuensch K., et al. A Gene Signature Derived from the Loss of CDKN1A (p21) Is Associated with CMS4 Colorectal Cancer. Cancers. 2022;14:136. doi: 10.3390/cancers14010136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li X.L., Hara T., Choi Y., Subramanian M., Francis P., Bilke S., Walker R.L., Pineda M., Zhu Y., Yang Y., et al. A p21-ZEB1 Complex Inhibits Epithelial-Mesenchymal Transition through the MicroRNA 183-96-182 Cluster. Mol. Cell. Biol. 2014;34:533–550. doi: 10.1128/MCB.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang X., Han M., Han H., Wang B., Li S., Zhang Z., Zhao W. Silencing Snail suppresses tumor cell proliferation and invasion by reversing epithelial-to-mesenchymal transition and arresting G2/M phase in non-small cell lung cancer. Int. J. Oncol. 2017;50:1251–1260. doi: 10.3892/ijo.2017.3888. [DOI] [PubMed] [Google Scholar]
- 121.Qi R., Wang J., Jiang Y., Qiu Y., Xu M., Rong R., Zhu T. Snai1-induced partial epithelial–mesenchymal transition orchestrates p53–p21-mediated G2/M arrest in the progression of renal fibrosis via NF-κB-mediated inflammation. Cell Death Dis. 2021;12:44. doi: 10.1038/s41419-020-03322-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zaremba-Czogalla M., Hryniewicz-Jankowska A., Tabola R., Nienartowicz M., Stach K., Wierzbicki J., Cirocchi R., Ziolkowski P., Tabaczar S., Augoff K. A novel regulatory function of CDKN1A/p21 in TNFα-induced matrix metalloproteinase 9-dependent migration and invasion of triple-negative breast cancer cells. Cell. Signal. 2018;47:27–36. doi: 10.1016/j.cellsig.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 123.Weis S.M., Cheresh D.A. Tumor angiogenesis: Molecular pathways and therapeutic targets. Nat. Med. 2011;17:1359–1370. doi: 10.1038/nm.2537. [DOI] [PubMed] [Google Scholar]
- 124.Liu Z.-L., Chen H.-H., Zheng L.-L., Sun L.-P., Shi L. Angiogenic signaling pathways and anti-angiogenic therapy for cancer. Signal Transduct. Target. Ther. 2023;8:198. doi: 10.1038/s41392-023-01460-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lugano R., Ramachandran M., Dimberg A. Tumor angiogenesis: Causes, consequences, challenges and opportunities. Cell. Mol. Life Sci. 2020;77:1745–1770. doi: 10.1007/s00018-019-03351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kuljaca S., Liu T., Dwarte T., Kavallaris M., Haber M., Norris M.D., Martin-Caballero J., Marshall G.M. The cyclin-dependent kinase inhibitor, p21WAF1, promotes angiogenesis by repressing gene transcription of thioredoxin-binding protein 2 in cancer cells. Carcinogenesis. 2009;30:1865–1871. doi: 10.1093/carcin/bgp225. [DOI] [PubMed] [Google Scholar]
- 127.Mo Y., Liang Z., Lan L., Xiong X., Zhang C., Liu W., Huang H., Fan J., Yang L. Extracellular vesicles derived from cervical cancer cells carrying MCM3AP-AS1 promote angiogenesis and tumor growth in cervical cancer via the miR-93/p21 axis. Exp. Cell Res. 2023;428:113621. doi: 10.1016/j.yexcr.2023.113621. [DOI] [PubMed] [Google Scholar]
- 128.Ghahremani M.F., Goossens S., Nittner D., Bisteau X., Bartunkova S., Zwolinska A., Hulpiau P., Haigh K., Haenebalcke L., Drogat B., et al. p53 promotes VEGF expression and angiogenesis in the absence of an intact p21-Rb pathway. Cell Death Differ. 2013;20:888–897. doi: 10.1038/cdd.2013.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pontes-Quero S., Fernández-Chacón M., Luo W., Lunella F.F., Casquero-Garcia V., Garcia-Gonzalez I., Hermoso A., Rocha S.F., Bansal M., Benedito R. High mitogenic stimulation arrests angiogenesis. Nat. Commun. 2019;10:2016. doi: 10.1038/s41467-019-09875-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Batlle E., Clevers H. Cancer stem cells revisited. Nat. Med. 2017;23:1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 131.Kharkar P.S. Cancer Stem Cell (CSC) Inhibitors in Oncology—A Promise for a Better Therapeutic Outcome: State of the Art and Future Perspectives. J. Med. Chem. 2020;63:15279–15307. doi: 10.1021/acs.jmedchem.0c01336. [DOI] [PubMed] [Google Scholar]
- 132.Pickup K.E., Pardow F., Carbonell-Caballero J., Lioutas A., Villanueva-Cañas J.L., Wright R.H.G., Beato M. Expression of Oncogenic Drivers in 3D Cell Culture Depends on Nuclear ATP Synthesis by NUDT5. Cancers. 2019;11:1337. doi: 10.3390/cancers11091337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang X., Powell K., Li L. Breast Cancer Stem Cells: Biomarkers, Identification and Isolation Methods, Regulating Mechanisms, Cellular Origin, and Beyond. Cancers. 2020;12:3765. doi: 10.3390/cancers12123765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ruijtenberg S., van den Heuvel S. Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle. 2016;15:196–212. doi: 10.1080/15384101.2015.1120925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xiao B.-D., Zhao Y.-J., Jia X.-Y., Wu J., Wang Y.-G., Huang F. Multifaceted p21 in carcinogenesis, stemness of tumor and tumor therapy. World J. Stem Cells. 2020;12:481–487. doi: 10.4252/wjsc.v12.i6.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sperka T., Wang J., Rudolph K.L. DNA damage checkpoints in stem cells, ageing and cancer. Nat. Rev. Mol. Cell Biol. 2012;13:579–590. doi: 10.1038/nrm3420. [DOI] [PubMed] [Google Scholar]
- 137.Zhong G., Qin S., Townsend D., Schulte B.A., Tew K.D., Wang G.Y. Oxidative stress induces senescence in breast cancer stem cells. Biochem. Biophys. Res. Commun. 2019;514:1204–1209. doi: 10.1016/j.bbrc.2019.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Troschel F.M., Palenta H., Borrmann K., Heshe K., Hua S.H., Yip G.W., Kiesel L., Eich H.T., Götte M., Greve B. Knockdown of the prognostic cancer stem cell marker Musashi-1 decreases radio-resistance while enhancing apoptosis in hormone receptor-positive breast cancer cells via p21WAF1/CIP1. J. Cancer Res. Clin. Oncol. 2021;147:3299–3312. doi: 10.1007/s00432-021-03743-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Insinga A., Cicalese A., Faretta M., Gallo B., Albano L., Ronzoni S., Furia L., Viale A., Pelicci P.G. DNA damage in stem cells activates p21, inhibits p53, and induces symmetric self-renewing divisions. Proc. Natl. Acad. Sci. USA. 2013;110:3931–3936. doi: 10.1073/pnas.1213394110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rodriguez R., Rubio R., Masip M., Catalina P., Nieto A., de la Cueva T., Arriero M., Martin N.S., de la Cueva E., Balomenos D., et al. Loss of p53 Induces Tumorigenesis in p21-Deficient Mesenchymal Stem Cells. Neoplasia. 2009;11:397–409. doi: 10.1593/neo.81620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ju Z., Choudhury A.R., Rudolph K.L. A Dual Role of p21 in Stem Cell Aging. Ann. N. Y. Acad. Sci. 2007;1100:333–344. doi: 10.1196/annals.1395.036. [DOI] [PubMed] [Google Scholar]
- 142.Zhang Z., Oh M., Sasaki J.-I., Nör J.E. Inverse and reciprocal regulation of p53/p21 and Bmi-1 modulates vasculogenic differentiation of dental pulp stem cells. Cell Death Dis. 2021;12:644. doi: 10.1038/s41419-021-03925-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chrysanthou S., Flores J.C., Dawlaty M.M. Tet1 Suppresses p21 to Ensure Proper Cell Cycle Progression in Embryonic Stem Cells. Cells. 2022;11:1366. doi: 10.3390/cells11081366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Letai A. Apoptosis and Cancer. Annu. Rev. Cancer Biol. 2017;1:275–294. doi: 10.1146/annurev-cancerbio-050216-121933. [DOI] [Google Scholar]
- 145.Carneiro B.A., El-Deiry W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020;17:395–417. doi: 10.1038/s41571-020-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Neophytou C.M., Trougakos I.P., Erin N., Papageorgis P. Apoptosis Deregulation and the Development of Cancer Multi-Drug Resistance. Cancers. 2021;13:4363. doi: 10.3390/cancers13174363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Prihantono, Faruk M. Breast cancer resistance to chemotherapy: When should we suspect it and how can we prevent it? Ann. Med. Surg. 2021;70:102793. doi: 10.1016/j.amsu.2021.102793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cao J., Zhang M., Wang B., Zhang L., Zhou F., Fang M. Chemoresistance and Metastasis in Breast Cancer Molecular Mechanisms and Novel Clinical Strategies. Front. Oncol. 2021;11:658552. doi: 10.3389/fonc.2021.658552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gartel A.L. The conflicting roles of the cdk inhibitor p21(CIP1/WAF1) in apoptosis. Leuk. Res. 2005;29:1237–1238. doi: 10.1016/j.leukres.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 150.Jänicke R.U., Sohn D., Essmann F., Schulze-Osthoff K. The Multiple Battles Fought by Anti-Apoptotic p21. Cell Cycle. 2007;6:407–413. doi: 10.4161/cc.6.4.3855. [DOI] [PubMed] [Google Scholar]
- 151.Suzuki A., Kawano H., Hayashida M., Hayasaki Y., Tsutomi Y., Akahane K. Procaspase 3/p21 Complex Formation to Resist Fas-Mediated Cell Death Is Initiated as a Result of the Phosphorylation of p21 by Protein Kinase A. [(accessed on 1 January 2023)]. Available online: www.nature.com/cdd. [DOI] [PubMed]
- 152.Shile H., Shu L., Dilling M.B., Easton J., Harwood F.C., Ichijo H., Houghton P.J. Sustained Activation of the JNK Cascade and Rapamycin-Induced Apoptosis Are Suppressed by p53/p21 Cip1. Mol. Cell. 2003;11:1491–1501. doi: 10.1016/s1097-2765(03)00180-1. [DOI] [PubMed] [Google Scholar]
- 153.Sohn D., Essmann F., Schulze-Osthoff K., Jänicke R.U. p21 Blocks Irradiation-Induced Apoptosis Downstream of Mitochondria by Inhibition of Cyclin-Dependent Kinase–Mediated Caspase-9 Activation. Cancer Res. 2006;66:11254–11262. doi: 10.1158/0008-5472.CAN-06-1569. [DOI] [PubMed] [Google Scholar]
- 154.Megyesi J., Tarcsafalvi A., Seng N., Hodeify R., Price P. Cdk2 phosphorylation of Bcl-xL after stress converts it to a pro-apoptotic protein mimicking Bax/Bak. Cell Death Discov. 2016;2:15066. doi: 10.1038/cddiscovery.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bhattacharya S., Ray R.M., Johnson L.R. Cyclin-dependent kinases regulate apoptosis of intestinal epithelial cells. Apoptosis. 2014;19:451–466. doi: 10.1007/s10495-013-0942-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kim E.M., Jung C.-H., Kim J., Hwang S.-G., Park J.K., Um H.-D. The p53/p21 Complex Regulates Cancer Cell Invasion and Apoptosis by Targeting Bcl-2 Family Proteins. Cancer Res. 2017;77:3092–3100. doi: 10.1158/0008-5472.CAN-16-2098. [DOI] [PubMed] [Google Scholar]
- 157.Choi K.-W., Suh H., Oh H.L., Ryou C., Lee C.-H. p21CIP1 Induces Apoptosis via Binding to BCL2 in LNCaP Prostate Cancer Cells Treated with MCS-C3, A Novel Carbocyclic Analog of Pyrrolopyrimidine. Anticancer Res. 2016;36:213–220. [PubMed] [Google Scholar]
- 158.Wu S., Cetinkaya C., Munoz-Alonso M.J., von der Lehr N., Bahram F., Beuger V., Eilers M., Leon J., Larsson L.-G. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene. 2003;22:351–360. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- 159.Hoffman B., A Liebermann D. Apoptotic signaling by c-MYC. Oncogene. 2008;27:6462–6472. doi: 10.1038/onc.2008.312. [DOI] [PubMed] [Google Scholar]
- 160.Wyld L., Bellantuono I., Tchkonia T., Morgan J., Turner O., Foss F., George J., Danson S., Kirkland J.L. Senescence and Cancer: A Review of Clinical Implications of Senescence and Senotherapies. Cancers. 2020;12:2134. doi: 10.3390/cancers12082134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Huang W., Hickson L.J., Eirin A., Kirkland J.L., Lerman L.O. Cellular senescence: The good, the bad and the unknown. Nat. Rev. Nephrol. 2022;18:611–627. doi: 10.1038/s41581-022-00601-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mylonas A., O’loghlen A. Cellular Senescence and Ageing: Mechanisms and Interventions. Front. Aging. 2022;3:866718. doi: 10.3389/fragi.2022.866718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Faggioli F., Velarde M.C., Wiley C.D. Cellular Senescence, a Novel Area of Investigation for Metastatic Diseases. Cells. 2023;12:860. doi: 10.3390/cells12060860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kumari R., Jat P. Mechanisms of Cellular Senescence: Cell Cycle Arrest and Senescence Associated Secretory Phenotype. Front. Cell Dev. Biol. 2021;9:645593. doi: 10.3389/fcell.2021.645593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kawaguchi K., Komoda K., Mikawa R., Asai A., Sugimoto M. Cellular senescence promotes cancer metastasis by enhancing soluble E-cadherin production. iScience. 2021;24:103022. doi: 10.1016/j.isci.2021.103022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Saleh T., Gewirtz D.A. Considering therapy-induced senescence as a mechanism of tumour dormancy contributing to disease recurrence. Br. J. Cancer. 2022;126:1363–1365. doi: 10.1038/s41416-022-01787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Perkins D.W., Haider S., Robertson D., Buus R., O’leary L., Isacke C.M. Therapy-induced senescence in normal tissue promotes breast cancer metastasis. Oncogene. 2022;41:4361–4370. doi: 10.1101/2020.10.17.343590. [DOI] [Google Scholar]
- 168.Gallanis G.T., Sharif G.M., Schmidt M.O., Friedland B.N., Battina R., Rahhal R., Davis J.E., Khan I.S., Wellstein A., Riegel A.T. Stromal Senescence following Treatment with the CDK4/6 Inhibitor Palbociclib Alters the Lung Metastatic Niche and Increases Metastasis of Drug-Resistant Mammary Cancer Cells. Cancers. 2023;15:1908. doi: 10.3390/cancers15061908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chen X., Zhang W., Gao Y.F., Qin X.S., Zhai Z.H. Senescence-Like Changes Induced by Expression of p21 Waf1/Cip1 in NIH3T3 Cell Line. 2002. [(accessed on 1 January 2023)]. Available online: http://www.cell-research.com. [DOI] [PubMed]
- 170.Englund D.A., Jolliffe A., Aversa Z., Zhang X., Sturmlechner I., Sakamoto A.E., Zeidler J.D., Warner G.M., McNinch C., White T.A., et al. p21 induces a senescence program and skeletal muscle dysfunction. Mol. Metab. 2023;67:101652. doi: 10.1016/j.molmet.2022.101652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shtutman M., Chang B.D., Schools G.P., Broude E.V. Cellular model of p21-induced senescence. Methods Mol. Biol. 2017;1534:31–39. doi: 10.1007/978-1-4939-6670-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hsu C.-H., Altschuler S.J., Wu L.F. Patterns of Early p21 Dynamics Determine Proliferation-Senescence Cell Fate after Chemotherapy. Cell. 2019;178:361–373.e12. doi: 10.1016/j.cell.2019.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kartika I.D., Kotani H., Iida Y., Koyanagi A., Tanino R., Harada M. Protective role of cytoplasmic p21Cip1/Waf1 in apoptosis of CDK4/6 inhibitor-induced senescence in breast cancer cells. Cancer Med. 2021;10:8988–8999. doi: 10.1002/cam4.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Anand J., Chiou L., Sciandra C., Zhang X., Hong J., Wu D., Zhou P., Vaziri C. Roles of trans-lesion synthesis (TLS) DNA polymerases in tumorigenesis and cancer therapy. NAR Cancer. 2023;5:zcad005. doi: 10.1093/narcan/zcad005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Stanzione M., Zhong J., Wong E., LaSalle T.J., Wise J.F., Simoneau A., Myers D.T., Phat S., Sade-Feldman M., Lawrence M.S., et al. Translesion DNA synthesis mediates acquired resistance to olaparib plus temozolomide in small cell lung cancer. Sci. Adv. 2022;8:eabn1229. doi: 10.1126/sciadv.abn1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zafar M.K., Eoff R.L. Translesion DNA Synthesis in Cancer: Molecular Mechanisms and Therapeutic Opportunities. Chem. Res. Toxicol. 2017;30:1942–1955. doi: 10.1021/acs.chemrestox.7b00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All references associated with the public databases analysed in this review are listed in the reference section.