Evaluation of a large panel of human cancer cells revealed that gain-of-function effects of mutant TP53 are not required for tumor growth and therapy resistance, whereas loss of wild-type TP53 functions is essential for these phenotypes.
Abstract
Mutations in the tumor suppressor TP53 cause cancer and impart poor chemotherapeutic responses, reportedly through loss-of-function, dominant-negative effects and gain-of-function (GOF) activities. The relative contributions of these attributes is unknown. We found that removal of 12 different TP53 mutants with reported GOFs by CRISPR/Cas9 did not impact proliferation and response to chemotherapeutics of 15 human cancer cell lines and colon cancer–derived organoids in culture. Moreover, removal of mutant TP53/TRP53 did not impair growth or metastasis of human cancers in immune-deficient mice or growth of murine cancers in immune-competent mice. DepMap mining revealed that removal of 158 different TP53 mutants had no impact on the growth of 391 human cancer cell lines. In contrast, CRISPR-mediated restoration of wild-type TP53 extinguished the growth of human cancer cells in vitro. These findings demonstrate that LOF but not GOF effects of mutant TP53/TRP53 are critical to sustain expansion of many tumor types.
Significance:
This study provides evidence that removal of mutant TP53, thereby deleting its reported GOF activities, does not impact the survival, proliferation, metastasis, or chemotherapy responses of cancer cells. Thus, approaches that abrogate expression of mutant TP53 or target its reported GOF activities are unlikely to exert therapeutic impact in cancer.
See related commentary by Lane, p. 211 .
This article is featured in Selected Articles from This Issue, p. 201
INTRODUCTION
TP53/TRP53/p53 suppresses tumor development through oncogenic stress-induced transcriptional upregulation of genes involved in several cellular responses, including apoptotic cell death, cell-cycle arrest, cellular senescence, and DNA repair (1, 2). Mutations in the TP53 gene are found in human cancers of diverse cellular origin with an overall frequency of approximately 50%, often occurring alongside loss of the other TP53 allele (loss of heterozygosity; LOH; ref. 3). Although some of these mutations cause loss of the TP53 protein (modeled by Trp53 knockout mice), most TP53 mutations in human cancers result in substitution of single amino acids, usually in the DNA-binding domain. High levels of stabilized mutant TP53 proteins are a common feature of such malignant cells (4, 5). Mutant TP53 proteins have been proposed to drive malignant transformation and sustain tumor growth via three not mutually exclusive processes: (i) loss-of-function (LOF), that is, an inability of mutant TP53 to activate expression of the genes that are transcriptionally activated by wt TP53 to suppress tumorigenesis; (ii) dominant-negative effects (DNE), that is, mutant TP53 repressing wt TP53 function through engagement in mixed tetramers; and (iii) gain-of-function (GOF) activities (Fig. 1A; refs. 3, 4, 6, 7). The GOF activities are thought to be mediated through neomorphic interactions of mutant TP53 proteins with other transcriptional regulators, activating cellular responses that are not impacted by wt TP53 (8). The LOF and DNE of mutant TP53 are accepted to be critical for malignant transformation, with the importance of the DNE restricted to cells that coexpress wt TP53 and mutant TP53, most notably nascent neoplastic cells during early stages of tumorigenesis (9–11). Sustained LOF was proven to be critical for continued tumor growth using mouse models in which expression of wt TP53 could first be switched off to drive tumorigenesis but then restored in the malignant cells. This suppressed tumor expansion through induction of either apoptosis or cell senescence, depending on the tumor type (12–14). It is less clear how important the GOF activities of mutant TP53 proteins are for sustained tumor growth. This issue has been resolved for several oncoproteins. For example, genetic removal or pharmacologic inhibition of mutant KRAS diminishes the growth of cancers, validating this oncogenic protein as a therapeutic target (15). If mutant TP53 and its reported GOF activities were likewise essential for sustained tumor expansion, drugs that could abolish its expression (e.g., approaches based on PROTAC technology; ref. 16) or block its GOF activities would be predicted to have therapeutic impact. A reduction in tumor expansion, albeit minor, was reported in a study using knock-in mice expressing R248Q mutant TRP53, where its expression could be extinguished in lymphoma cells through CRE-mediated gene deletion (17). Resolving the issue whether mutant TP53 is essential for sustained survival and proliferation of malignant cells has important ramifications for the design of novel cancer therapies.
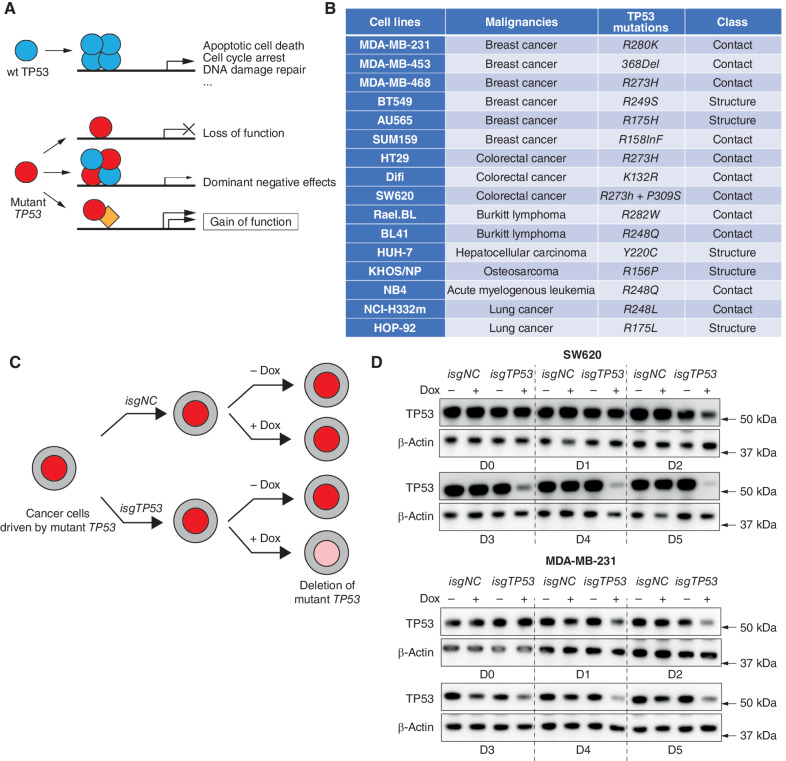
Figure 1.
Removal of mutant TP53 in human cancer cell lines using doxycycline-inducible CRISPR/Cas9 technology. A, Schematic of how wt TP53 functions as a tumor suppressor and mechanisms by which mutant TP53/TRP53 proteins are postulated to promote neoplastic transformation. B, Table listing the names and cellular origin of the 16 mutant TP53–expressing human cancer–derived cell lines examined, with their respective TP53 mutations indicated. C, Schematic to illustrate the strategy for removing mutant TP53 proteins using an inducible CRISPR/Cas9 platform. D, Western blotting to demonstrate the progressive removal of mutant TP53 protein in the human cancer cell lines SW620 and MDA-MB-231 after transduction with the vector containing the doxycycline-inducible sgRNAs targeting TP53 (isgTP53) and treatment with doxycycline. One control included was to not treat these cells with doxycycline. The same cancer cell lines transduced with a doxycycline-inducible nontargeting control sgRNA (isgNC) and treated, or not treated, with doxycycline were used as further controls. Probing for β-actin was used as a protein loading control. The Western blots shown are representative of 2 or 3 independent blots from independent experiments. Removal of the respective mutant TP53 proteins from the other human cancer–derived cell lines used in this study is documented in Supplementary Fig. S1.
RESULTS
Inducible Removal of Mutant TP53 Does not Reduce Proliferation or Survival of Diverse Human Cancer Cell Lines
Substantial resources are being invested into the development of drugs that abrogate mutant TP53 expression or its reported GOF activities as potential cancer therapeutics. However, the evidence that such drugs would be effective is conflicting (17, 18). This motivated us to undertake a wide-ranging and detailed examination of the requirement for sustained expression of mutant TP53 protein for the in vitro growth of a panel of fifteen human cancer cell lines of diverse origin (breast, colorectal, lung, and hepatocellular carcinomas, lymphoma, leukemia, and an osteosarcoma; Fig. 1B). To achieve this, we used an inducible CRISPR/Cas9 platform to genetically inactivate 12 distinct endogenous mutant TP53 genes that have been reported to engender GOF activities (19) in the panel of human cancer cell lines. These cancer cell lines were transduced with a vector for stable expression of Cas9 plus doxycycline-inducible vectors for the expression of a TP53-specific guide RNA (isgTP53) or a control guide RNA (isgNC; ref. 19). The transduced cancer cells were treated with doxycycline to induce sgRNA expression or were left untreated, and their survival and proliferation were monitored every two days from day 0 to day 12 (Fig. 1C). The progressive removal of mutant TP53 protein was verified by Western blotting and sequencing of the relevant TP53 exons (Fig. 1D; Supplementary Fig. S1A and S1B; the potential caveats with respect to any residual small amount of mutant TP53 protein found after 6 or more days of treatment of isgTP53-transduced cancer cells with doxycycline are addressed in later experiments). The inducible removal of mutant TP53 had no impact on the in vitro survival or proliferation of the cancer cell lines tested. Regardless of their TP53 state, all cancer cell lines proliferated vigorously during log expansion until reaching confluence, followed by a reduction in viable cells and an increase in dead cells (Fig. 2A and B; Supplementary Fig. S2A, Supplementary Tables S1 and S2). Flow cytometric analysis confirmed that the removal of mutant TP53 had no impact on cell cycling (Fig. 2C; Supplementary Fig. S2B). Because these experiments were conducted during and immediately after the removal of mutant TP53, it appears unlikely that acquisition of mutations that provide compensation for the loss of GOF activities of mutant TP53 would explain these findings.
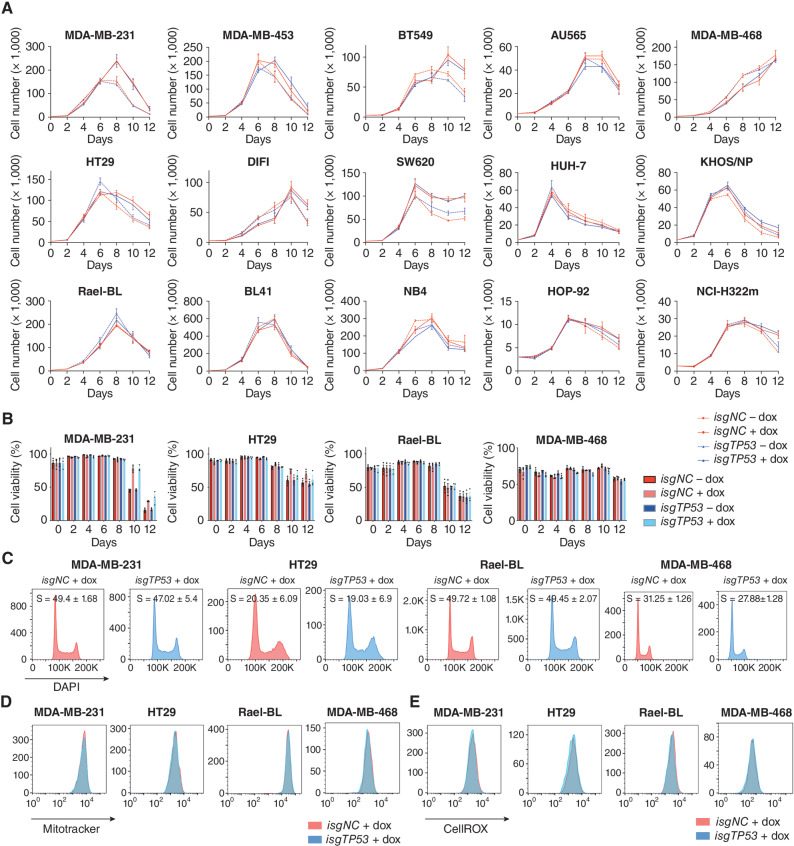
Figure 2.
Removal of mutant TP53 does not impact the proliferation, survival, mitochondrial content, and ROS levels in human cancer cell lines. A,In vitro growth of the indicated human cancer cell lines with or without doxycycline-mediated induction of a mutant TP53-specific inducible sgRNA (isgTP53) or an inducible control sgRNA (isgNC). B,In vitro survival of the cancer cells described in A. C, Cell-cycle analysis of the cancer cells described in A. D, Mitotracker staining of the cancer cells described in A. E, CellROX staining of the cancer cells described in A. The analyses described in C–E were conducted 2 days after the cancer cells had been treated with doxycycline for 5 days (see A). Data in A and B are presented as mean ± SEM of three independent experiments. There were no consistent significant differences between the mutant TP53–deleted cancer cells versus the control cancer cells in any of the experiments shown (see Supplementary Tables S1 and S2 for details of the statistical analyses).
Inducible Removal of Mutant TP53 Does Not Impact Mitochondrial Activity or ROS Levels in Human Cancer Cell Lines
Adaptation of cell metabolism (i.e., the Warburg effect; ref. 20) is a hallmark of cancer (21), and high intracellular levels of reactive oxygen species (ROS) are often associated with increased cellular metabolism (22, 23). Mutant TP53 has been reported to regulate metabolic changes and impact the levels of ROS in cancer cells (24). Mitotracker and CellROX staining revealed that the removal of mutant TP53 had no impact on mitochondrial content/activity or the intracellular levels of ROS in the cancer cell lines tested (Fig. 2D and E; Supplementary Fig. S2C and S2D). These data show that sustained expression of mutant TP53 is not required for the metabolic adaptation of these cancer cells.
Removal of Mutant TP53 Does Not Impact Responses of Human Cancer Cell Lines to Nutrient Deprivation or Cytotoxic Drugs
The ability of cancer cells to tolerate diverse stresses, such as deprivation of nutrients or growth factors, is critical for ensuring their survival and proliferation (25). The GOF effects of mutant TP53 have been reported to assist malignant cells in adapting to such stress (26). To determine the importance of the alleged GOF activities of mutant TP53 on cancer cells under stress, we cultured the control cancer cell lines and the mutant TP53–depleted derivatives in medium containing only 3% or 1% FCS, as a model of nutrient deprivation (27). Culturing in 1% FCS plus dox treatment had similar impact on cell expansion and induced similar extent of death in both the control and mutant TP53–depleted SW620 cells (∼50%), whereas no significant death was observed in the other cell lines tested (Fig. 3A and B; Supplementary Fig. S3A). The death seen in the SW620 cells is likely due to their sensitivity to doxycycline when cultured in low serum and was not associated with the removal of mutant TP53. Furthermore, we could not detect any impact of induced removal of mutant TP53 on cell growth, cell cycling, mitochondrial content/activity or ROS levels in any of the cancer cell lines tested in medium containing 1% FCS (Fig. 3A; Supplementary Fig. S3B–S3D) or 3% FCS (Supplementary Figs. S4A and S4B and S5A–S5C; Supplementary Tables S1 and S2).
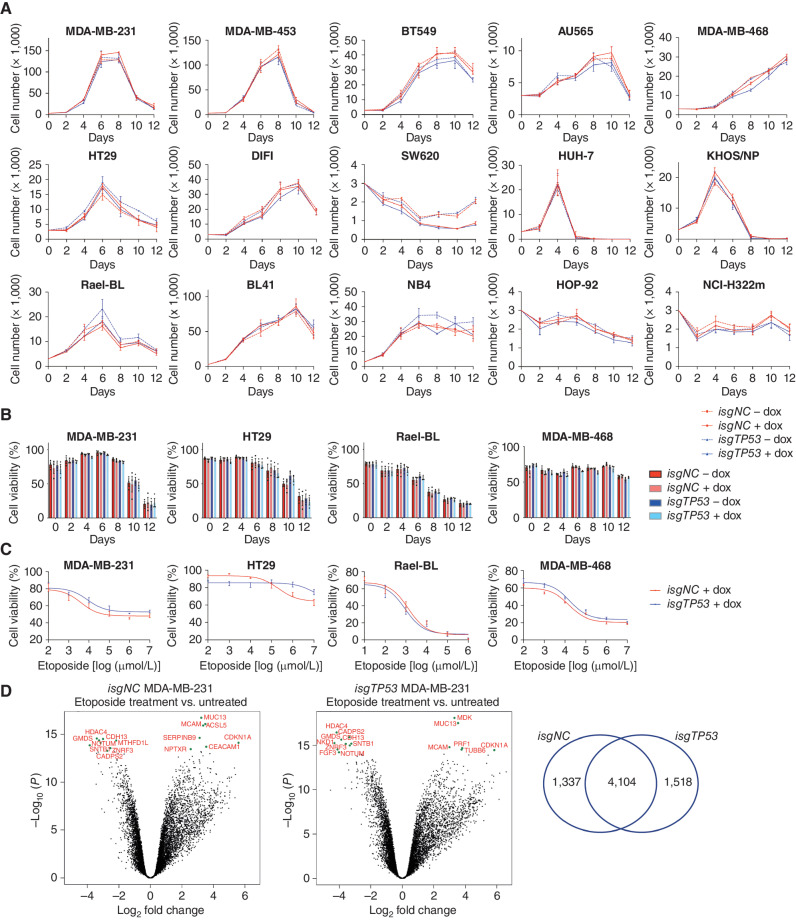
Figure 3.
Removal of mutant TP53 does not impair the ability of cancer cells to adapt to conditions of stress. A,In vitro growth of the indicated human cancer cell lines with or without doxycycline-mediated induction of an inducible mutant TP53–specific sgRNA (isgTP53) or an inducible control sgRNA (isgNC) grown in medium with 1% FCS. B,In vitro survival of the cancer cells described in A. C, The viability of the cancer cells described in A after treatment with the indicated concentrations of etoposide for 48 hours. D, Volcano plots for the control mutant TP53–expressing MDA-MB-231 cells treated with etoposide versus treatment with vehicle (left) and for the mutant TP53–deleted derivatives treated with etoposide treatment versus treatment with vehicle (right). The x-axis shows the log2 fold change in gene expression whereas the y-axis shows −log10 (P). Significantly differentially expressed (DE) genes are those above −log10 (0.05) = 1.3 on the y-axis. The top 15 significantly DE genes are indicated by green dots. The Venn diagram shows that nearly 60% of DE genes after treatment with etoposide are the same between the control and the mutant TP53–deleted MDA-MB-231 cancer cells. Data in A–C are presented as mean ± SEM of three independent experiments. There were no consistent significant differences between the TP53-deleted cancer cells versus the control cancer cells in any of the experiments shown (see Supplementary Tables S1 and S2 for details of the statistical analyses).
Mutant TP53 has been reported to render malignant cells resistant to diverse anticancer agents (28). We found that removal of mutant TP53 did not impact the killing of the cancer cell lines tested in response to treatment with etoposide, 5-fluorouracil (5-FU), taxol, or cisplatin (Fig. 3C; Supplementary Fig. S6A–S6D; Supplementary Table S3). RNA-sequencing analysis revealed a large overlap of the differentially expressed (DE) genes between the control MDA-MB-231 cells and their mutant TP53-deleted derivatives after treatment with etoposide (Fig. 3D). This indicates that these cancer cells deal with this cytotoxic drug by engaging the same signaling pathways. Some similarities in changes in gene expression were seen in previous reports but those studies did not compare side-by-side isogenic parental cells with their mutant TP53–deleted derivatives (29, 30). Collectively, our results demonstrate that sustained expression of mutant TP53 is not required for the ability of cancer cells to adapt to conditions of stress, including treatment with anticancer agents.
Clonal Populations of Mutant TP53–Deleted Human Cancer Cell Lines Behave Similarly to Their Control Mutant TP53–Expressing Counterparts
A potential caveat to the data presented above is that within the polyclonal populations of cancer cells that had been induced to express the mutant TP53 targeting sgRNA, a small amount of mutant TP53 protein remained, which was detectable by Western blotting and confirmed by TP53 exon sequencing (Fig. 1D; Supplementary Fig. S1A and S1B). To determine whether the residual, albeit markedly reduced, levels of mutant TP53 could have influenced the read-out of the experiments, single cell–derived clones (2–3 per cancer cell line) lacking detectable mutant TP53 protein were established from the MDA-MB-231 (breast cancer, R280K), HT29 (colorectal cancer, R273H), Rael-BL (Burkitt lymphoma, R282W), and MDA-MB-468 (breast cancer, R273H) human cancer cell lines. The complete absence of mutant TP53, verified by Western blotting, had no impact on the proliferation, survival, cycling, mitochondrial content/activity, ROS levels, and response of these cells to anticancer agents, even in medium containing only 3% or 1% FCS (Supplementary Figs. S7A–S7G, S8A–S8G, S9A–S9G, and S10A–S10G).
Competitive coculture of two isogenic cell populations where one carries a mutation is a sensitive assay to detect even subtle defects in cell proliferation and survival. When mutant TP53–deleted MDA-MB-231, HT29, AU565, and BT549 human cancer cells were each mixed at a 50/50 ratio with their mutant TP53–expressing controls, no competitive disadvantage was observed during 15 days of coculture (Supplementary Fig. S11A), even when the cells were continuously treated with etoposide (Supplementary Fig. S11B). This demonstrates that the removal of mutant TP53 does not cause a detectable competitive disadvantage in these human cancer cell lines.
shRNA-Mediated Reduction of Mutant TP53 Can Impair the Survival and Proliferation of Human Cancer Cell Lines Through Off-Target Toxicity
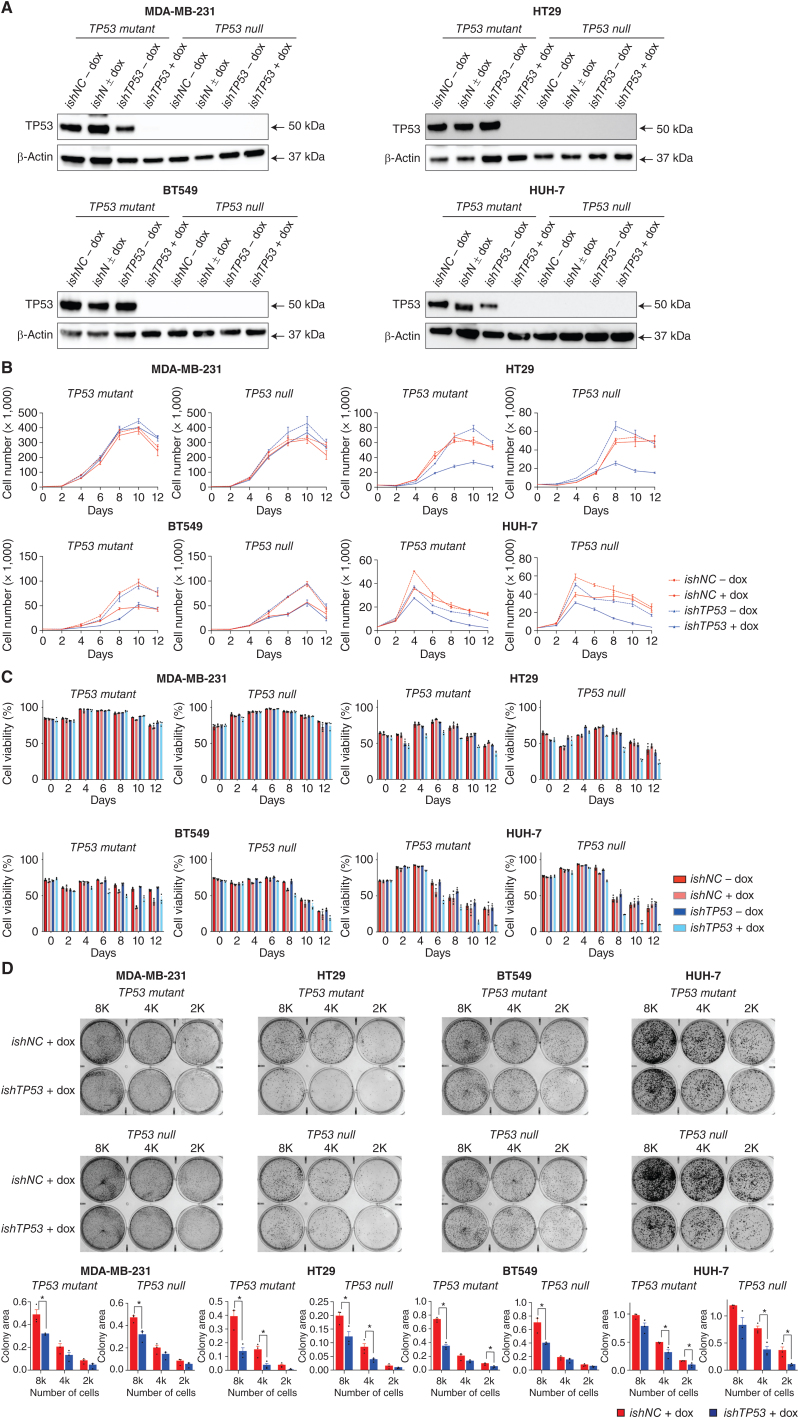
Our findings contrast with some previous studies that showed that reduction of mutant TP53 expression using RNA interference inhibited the in vitro growth and survival of certain human cancer cell lines (31, 32). To try to explain these discrepancies, the previously published TP53 targeting (ishTP53) and control (ishNC) shRNA sequences were cloned into a doxycycline-inducible vector and transduced into the same human cancer cell lines used in those studies: SKBR3 (breast cancer, R175H), HUH-7 (hepatocellular carcinoma, Y220C), T47D (breast cancer, L194F), MDA-MB-231 (breast cancer, R280K), SW620 (colon cancer, R273H + P309S), and HT29 (colon cancer, R273H). Despite efficient reduction in mutant TP53 levels (Supplementary Fig. S12A), only minor cell death and reduction in cell proliferation were observed in both the control shNC and shTP53 variants from day 0 to day 5 (Supplementary Fig. S12B and S12C). To clarify the minor killing induced by RNAi, ishTP53 and ishNC vectors were also transduced into derivatives of the MDA-MB-231, HT29, BT549, and HUH-7 cancer cell lines in which mutant TP53 had already been removed by CRISPR (Fig. 4A). Notably, significant killing and inhibition of cell growth were observed in the TP53-deleted HUH-7, HT29, and BT549 cells with doxycycline-induced expression of ishTP53 (Fig. 4B and C). Accordingly, the induction of ishTP53 decreased colony formation to a similar extent in the mutant TP53–expressing control cells and the mutant TP53–deleted CRISPR derivatives in each of these cancer cell lines (Fig. 4D). These findings demonstrate that the doxycycline-mediated induction of the shRNA targeting mutant TP53 can inhibit the survival and proliferation of certain cancer cells through nonspecific toxic effects, rather than by removing a critical function of mutant TP53.
Figure 4.
Doxycycline-mediated induction of an inducible shRNA targeting mutant TP53 inhibits proliferation and survival of human cancer cell lines through an off-target toxic effect. A, Western blot analysis showing the reduction of mutant TP53 protein in the indicated human cancer cell lines with or without doxycycline-mediated induction of an inducible TP53-specific shRNA (ishTP53) or an inducible control shRNA (ishNC). Probing for β-Actin was used as a protein loading control. The Western blots shown are representative of 2 or 3 independent blots from independent experiments. B,In vitro growth of the indicated human cancer cell lines with or without doxycycline-mediated induction of an inducible TP53-specific shRNA (ishTP53) or an inducible control shRNA (ishNC) for 12 days. C,In vitro survival of the cancer cells described in A and B. D, The cancer cell lines described in A and B were seeded at 8,000 (8K), 4,000 (4K), or 2,000 (2K) cells per well. The photographs show examples of colony formation assays. Colonies were stained with 1% crystal violet. Measurements of colony areas as determined by software ImageJ. Three independent assays were performed for each cancer cell line. Data in B–D are presented as mean ± SEM of three independent experiments.
Mutant TP53-Expressing Human Cancer Cell Lines Retain the Machinery to Respond to Wild-Type TP53
We next addressed whether the mutant TP53–expressing human cancer cell lines retained the ability to respond to wt TP53, provided mutant TP53 was removed to obviate its DNE. To examine this, wt TP53 was ectopically expressed in control mutant TP53–expressing MDA-MB-231 (R280K) and SW620 (R273H + P309S) human cancer cells and their mutant TP53–deleted derivatives (Supplementary Fig. S13A). In the mutant TP53–depleted derivatives, enforced expression of wt TP53 caused transcriptional activation of the TP53 target genes BBC3 (encoding proapoptotic PUMA) and CDKN1A (encoding the cyclin-dependent kinase inhibitor p21; Supplementary Fig. S13B) and increased their sensitivity to the MDM2 inhibitor nutlin-3a and etoposide (Supplementary Fig. S13C and S13D). As expected, this was not observed in the control MDA-MB-231 and SW620 cells, owing to the DNE exerted by endogenous mutant TP53 on the ectopically introduced wt TP53.
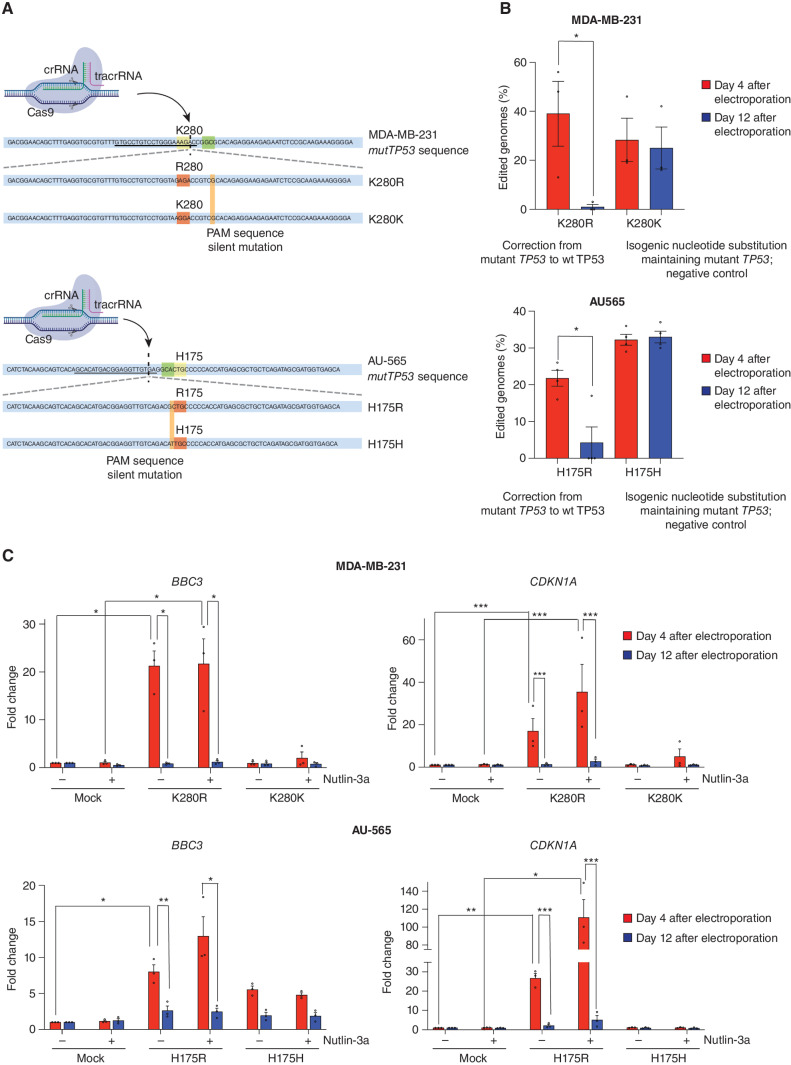
Next, we attempted CRISPR-mediated repair of mutant TP53 into wt TP53 gene sequences in the MDA-MB-231 (R280K), AU565 (R175H), SKBR3 (R175H), and HCC70 (R248Q) human cancer cell lines, relying on simultaneous electroporation of a Cas9 ribonucleoprotein (RNP) along with a ssDNA HDR donor (Fig. 5A; Supplementary Fig. S14A; ref. 33). Remarkably, 4 days after electroporation, 13%–56% of genomes in these cancer cells encoded wt TP53 but after 12 days, wt TP53 genomes were undetectable in all four cancer cell lines (Fig. 5B; Supplementary Fig. S14B) because of potent selection against cells with restored wt TP53 expression. The restored wt TP53 protein was functional, given that 4 days, but not 12 days, after RNP electroporation, the MDA-MB-231 and AU565 cells both displayed a significant increase in BBC3 and CDKN1A mRNA upon treatment with nutlin-3a (Fig. 5C). This was not seen in cancer cells electroporated with a control HDR donor that introduced a silent mutation avoiding correction of the mutant TP53 gene sequence (Fig. 5C). These findings demonstrate that these cancer cells retain the machinery to respond to wt TP53 by activating its cell death and cell-cycle arrest programs.
Figure 5.
Impact of correction of the mutant TP53 sequence to wt TP53 in the MDA-MB-231 and AU565 human cancer cell lines. A, Schematic of the components used for electroporation to correct the mutant TP53 gene sequence to wt TP53 in human cancer cells. The crRNA recognition site (underlined), PAM sequence (in green), and the cut site (dashed vertical line) for the MDA-MB-231 and AU565 mutTP53 target sequence are presented (K280 and H175 residues in yellow). Bottom, the ssDNA HDR donor sequences are presented: the modified codon (in red) together with the PAM sequence silent mutation (in orange) are shown. B, Percentages of genomes carrying the intended TP53 edit at the indicated time points. N = 3 independent experiments. Individual values of biological replicates their mean and SEM are reported. C, The mRNA abundance for the indicated genes at the indicated time points in the absence (-) or presence (+) of treatment with 5 μmol/L nutlin-3a for 12 hours. N = 3 independent experiments. The data are presented as the mean and SEM of three independent experiments.
Removal of Mutant TP53 Does Not Reduce Growth or Metastasis of Human or Mouse Cancer Cells In Vivo
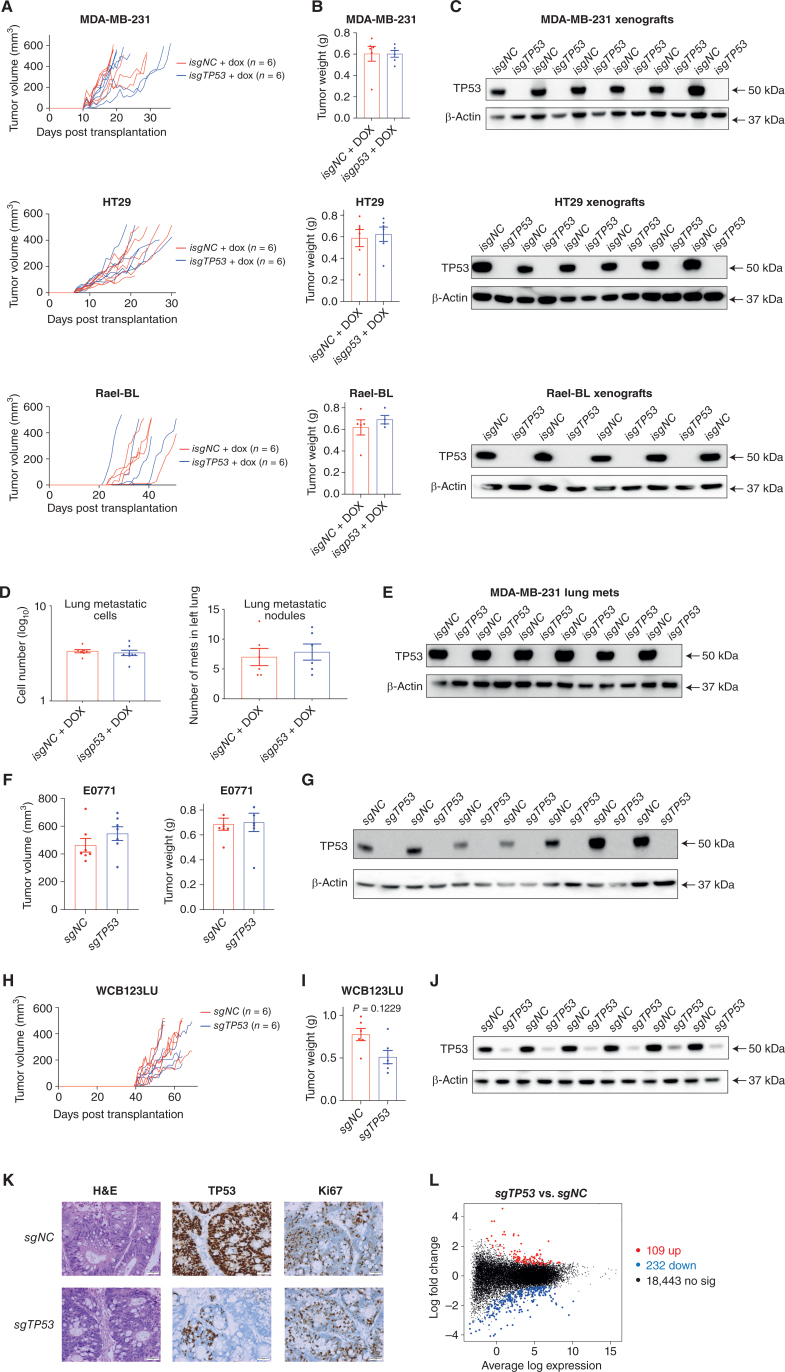
We next examined whether removal of mutant TP53 impacted the growth of cancer cells in vivo. MDA-MB-231, HT29, and Rael-BL–derived clones with complete loss of mutant TP53 were able to grow in immune-deficient NSG mice at rates comparable to their mutant TP53–expressing controls (Fig. 6A; Supplementary Fig. S15), reaching similar tumor weights at ethical endpoint (Fig. 6B). For MDA-MB-231 and HT29 cancer cells, even after transplantation with limited cell numbers (200K, 20K, or 2K), no impact of removal of mutant TP53 on tumor growth in vivo and tumor weight at ethical endpoint were observed (Supplementary Fig. S16A and S16B). Western blotting confirmed the presence or absence of mutant TP53 in the tumors derived from the control cancer cells or their mutant TP53–deleted derivatives, respectively (Fig. 6C; Supplementary Fig. S16C). Consistent with these findings from in vivo experiments, removal of mutant TP53 had no impact on colony formation of MDA-MB-231, MDA-MB-468, HT29, HUH-7, and AU565 cancer cells in vitro (Supplementary Fig. S17A and S17B).
Figure 6.
Removal of mutant TP53 does not impair tumor growth and metastasis in vivo. A, Growth of the human cancer cell lines MDA-MB-231, SW620, and Rael-BL, either mutant TP53-expressing control cells or the mutant TP53-deleted derivatives, in NSG mice (N = 6 mice per cell line) with tumor volumes presented in mm3. B, Weights of the tumors from A at the ethical endpoint. C, Western blot analysis of the tumors from A to verify the presence of mutant TP53 protein in the control cancer cells and to confirm its absence in the tumors arising from the mutant TP53–deleted cancer cells. Probing for β-Actin was used as a protein loading control. D, Numbers of metastatic cells and nodules in the left lungs of NSG mice that had been injected with MDA-MB-231 breast cancer cells, either mutant TP53-expressing control cells or the mutant TP53-deleted derivatives, into their mammary fat pads (N = 6 mice for each cancer cell line). The primary breast tumors were resected when they had reached 200 mm3 to enable analysis of metastasis thereafter. E, Western blot analysis of the metastases from D to verify the presence or absence of mutant TP53, respectively. Probing for β-Actin was used as a protein loading control. F, Volume and weight of the primary tumors in immune-competent mice transplanted with EO771 mouse breast cancer cells, either mutant Trp53–expressing control cells or their mutant Trp53–deleted derivatives. The breast cancer cells were transplanted into mammary fat pads of Cas9 transgenic mice (C57BL/6 background) to prevent immune rejection caused by Cas9 expression. G, Western blot analysis of the tumors from F to verify the presence of mutant TRP53 in the control cells or its absence in the mutant Trp53–deleted derivatives. Probing for β-Actin was used as a protein loading control. H, Growth of the human colon cancer–derived organoids WCB123LU, either the mutant TP53–expressing controls or the mutant TP53–deleted derivatives, in NSG mice (n = 6 mice per organoid line) with tumor volume presented in mm3. I, Weights of the tumors from H at the ethical endpoint. J, Western blot analysis of the tumors from H to verify the presence of mutant TP53 in the tumors derived from the control organoids or its absence in the tumors derived from the mutant TP53–deleted derivatives. K, Hematoxylin and eosin staining and IHC of the tumors from H to verify the presence of mutant TP53 in the control tumors or its absence in the mutant TP53–deleted derivatives. IHC analysis of Ki-67 in the tumors from H to reveal the expression of this marker of cell proliferation in tumors derived from the control mutant TP53–expressing colon cancer organoids or their mutant TP53–deleted derivatives. Magnification, 200×. L, Mean difference plot for the RNA-seq differential gene expression analysis comparing tumors in NSG mice that had been derived from mutant TP53–expressing control WCB123LU colon cancer organoids with tumors in NSG mice derived from the mutant TP53–deleted derivatives. The x-axis shows the average gene log expression, whereas the y-axis shows gene log2 fold change. Points colored red and blue indicate genes that are significantly upregulated or downregulated, respectively, in the tumors derived from the mutant TP53–deleted colon cancer organoids compared with the mutant TP53–expressing control tumors. Data in B, D, and I are presented as mean ± SEM of results from experiments conducted in triplicate.
To examine the impact of removal of mutant TP53 on cancer metastasis, MDA-MB-231 (R280K) and SUM159 (R158InF) human breast cancer cells were implanted into the mammary fat pads of NSG mice, and the primary tumors were resected when they had reached 200 mm3 to enable analysis of metastasis thereafter. No significant differences in the numbers of lung metastases were observed between the control cancer cells versus their mutant TP53–deleted derivatives (Fig. 6D; Supplementary Fig. S18A). Western blotting confirmed the presence of mutant TP53 protein in the lung metastases from the former and its absence in the latter (Fig. 6E; Supplementary Fig. S18B). Histologic staining demonstrated that the control SUM159 cells and their mutant TP53–deleted derivatives had similar ability to form metastases in the lung (Supplementary Fig. S18C). Accordingly, transwell assays revealed that control AU565 as well as BT549 cancer cells and their mutant TRP53–deleted derivatives had similar ability to migrate in vitro (Supplementary Fig. S19A and S19B).
Furthermore, for NSG mice transplanted with HT29 human colon cancer cells, both the mutant TP53–expressing control tumors and the mutant TP53–deleted counterparts responded to 5-FU in vivo in a similar manner, with their expansion reduced to a similar extent compared with the growth of these tumors in untreated host mice (Supplementary Fig. S20A). Immunostaining revealed similarly decreased numbers of Ki-67–positive (i.e., proliferating) control or mutant TP53–depleted HT29 colon cancer cells in the 5-FU–treated host mice compared with the untreated tumor-bearing mice (Supplementary Fig. S20B).
To allow studies of tumor growth in mice with a competent immune system, we examined the impact of removal of mutant TP53 in Eμ-Myc mouse lymphoma cells, where spontaneous mutation of Trp53 is found in approximately 25% of lymphomas (34). We used the MRE412 cell line derived from a lymphoma that had spontaneously acquired an R246Q-mutant TRP53 (equivalent to R249Q in humans) during its development. Removal of mutant TRP53 had no impact on the survival and proliferation of these lymphoma cells in vitro (Supplementary Fig. S21A and S21B) or their ability to grow in immune-competent mice (Supplementary Fig. S21C). Removal of mutant TRP53 was confirmed in the lymphomas grown in vivo by Western blotting (Supplementary Fig. S21D), and flow cytometric analysis of GFP and Ly5.2 expression further verified that they were derived from the transplanted Eμ-Myc lymphoma cells (Supplementary Fig. S22). Furthermore, we transplanted EO771 mouse breast cancer cells (S364R), both controls and their mutant Trp53–deleted derivatives, into the mammary fat pads of isogenic C57BL/6 recipient mice with a functional immune system. Removal of mutant TRP53 protein had no impact on the weight and volume of the primary tumors collected 11 days after transplantation (Fig. 6F) or the composition of immune cells in the microenvironment of these tumors (Supplementary Fig. S23). The loss of mutant TRP53 in the primary tumors derived from the sgTrp53-transduced derivative cancer cells was confirmed by Western blotting (Fig. 6G).
Removal of Mutant TP53 Does Not Impair the Growth of Human Colon Cancer–Derived Organoids in Culture or as Tumor Xenografts in NSG Mice
Organoids derived from human cancers and their transplantation into NSG mice are considered an ideal model for studying the growth of human cancer while enabling the genetic manipulation of these cells (35). We derived organoids, WCB123LU and WCB139T, from human colorectal cancers expressing R248W or R175H mutant TP53, respectively, and used CRISPR/Cas9 to generate derivatives lacking mutant TP53. The mutant TP53–deleted colon cancer organoids grew in culture (Supplementary Fig. S24) and formed tumors in NSG mice (Fig. 6H; Supplementary Fig. S25 and S26A) in a manner comparable with the control organoids, reaching similar tumor weights at ethical endpoint (Fig. 6I; Supplementary Fig. S26B). Removal of mutant TP53 was confirmed by Western blotting (Fig. 6J; Supplementary Fig. S26C) and IHC (Fig. 6K; Supplementary Fig. S26D). Staining for Ki-67 revealed that the in vivo proliferation rate was comparable between tumor cells derived from the control organoids and their mutant TP53–deleted derivatives (Fig. 6K; Supplementary Fig. S26D). RNA-sequencing analysis revealed extensive similarity in gene expression between tumors from the mutant TP53–deleted WCB123LU organoids compared with tumors derived from the control organoids, with only 109 genes upregulated and 232 downregulated (Fig. 6L). The removal of mutant TP53 in colon cancer–derived organoids did not increase their sensitivity to anticancer drugs in culture (Supplementary Fig. S27). Moreover, the control organoids and the mutant TP53–deleted derivatives displayed similar responses to the anticancer agent 5-FU in vivo, with their growth rate reduced to a comparable extent relative to the expansion of these tumors in untreated host mice (Supplementary Fig. S28A). Ki-67–positive cells were consistently reduced in the 5-FU–treated tumors compared with untreated tumors, with no differences detected between the control organoid versus the mutant TP53–deleted organoid-derived tumors (Supplementary Fig. S28B). Collectively, these results demonstrate that mutant TP53/TRP53 is not critical for the sustained in vivo growth of diverse human and mouse cancer cells, the metastasis of human breast cancer cells, or the response of human cancer cells to anticancer agents.
DepMap Database Mining Reveals that Mutant TP53 Is Not a Cancer Vulnerability
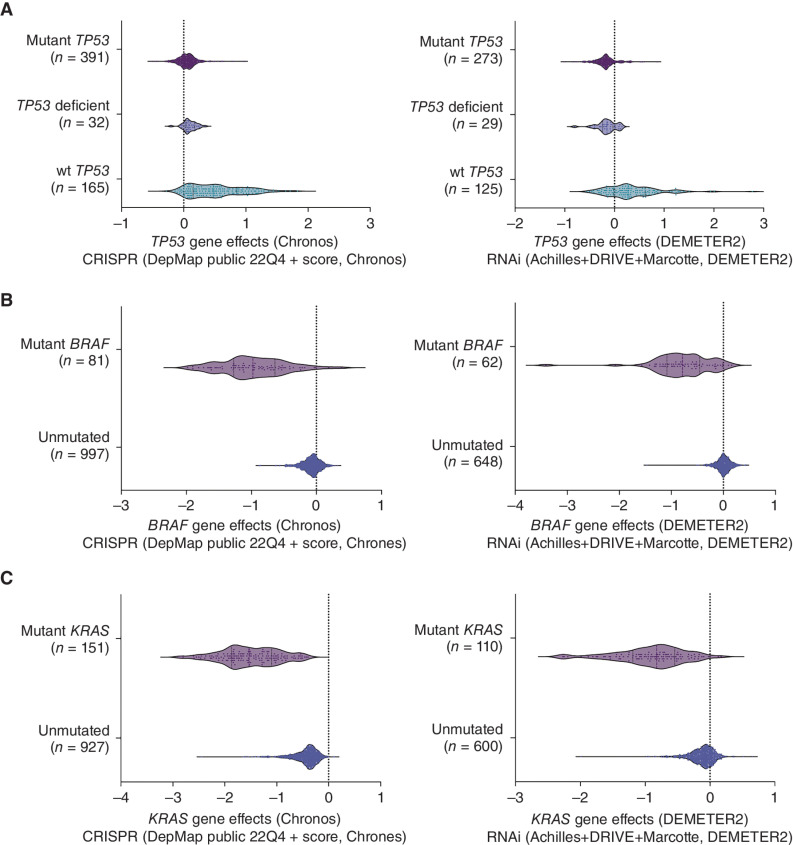
To extend beyond our own study, we mined the data within DepMap (Cancer Dependency Map) to examine the effect of deleting mutant TP53 by CRISPR on the growth and survival of 391 diverse types of human cancer cell lines, covering 158 mutant TP53 proteins. This showed unequivocally that removal of mutant TP53 did not affect the growth of any of these cancer cell lines, with them behaving in a manner comparable with the TP53-deficient cancer cell lines after transduction with a TP53 sgRNA (Fig. 7A). Interestingly, the studies reported in DepMap that used a TP53-specific RNAi showed that this treatment inhibited the growth of a few cancer cell lines, but tellingly, these growth-inhibitory effects were also seen in several TP53-deficient human cancer cell lines (Fig. 7A). This is consistent with our observation of off-target growth-inhibitory effects of TP53 targeting RNAi (Fig. 4). In contrast, and consistent with our findings (Fig. 5; Supplementary Fig. S12 and S13), targeting wt TP53 by either CRISPR or RNAi caused a growth advantage in many cancer cell lines expressing wt TP53 (Fig. 7A). As a positive control, we further mined the DepMap database to show that targeting mutant KRAS or mutant BRAF either by CRISPR or RNAi greatly impaired the growth of human cancer cell lines driven by these mutant proteins (Fig. 7B and C). This demonstrates that mutant KRAS and mutant BRAF proteins but not mutant TP53 proteins are cancer cell dependencies.
Figure 7.
Analysis of the DepMap database does not identify mutant TP53 as a cancer cell dependency. A, Analysis of the DepMap database shows that the deletion of mutant TP53 using CRISPR had no impact on the growth of 391 human cancer cell lines encompassing 158 different TP53 mutations. RNAi-mediated removal of mutant TP53 impaired the growth of not only a small number of cancer cell lines expressing mutant TP53 but also of some cancer cell lines that are TP53 deficient, demonstrating the off-target effects of RNAi. Removal of wt TP53 by either CRISPR or RNAi led to a growth advantage in many cancer cell lines expressing wt TP53. B, Mining of the DepMap database shows that the in vitro growth of human cancer cell lines expressing mutant BRAF is impaired when mutant BRAF is removed by using CRISPR/Cas9 or RNAi technology. C, Mining of the DepMap database shows that the in vitro growth of human cancer cell lines expressing mutant RAS is impaired when mutant RAS is removed by using CRISPR/Cas9 or RNAi technology.
DISCUSSION
In this study, we used a doxycycline-inducible lentiviral CRISPR platform to remove endogenous mutant TP53 in cancer cells. This approach enabled the continuous monitoring of the proliferation and survival of cancer cells during the process of removal of mutant TP53 in vitro. Importantly, this provided a full range of controls with which the potential cytotoxic effects of specific sgRNA vectors or doxycycline on cancer cells could be reliably observed, thereby excluding off-target effects to the greatest possible extent. Our findings show that in 16 human cancer cell lines and two mouse cancer cell lines of diverse cellular origin encompassing 15 mutant TP53/TRP53 proteins with proposed GOF activities, removal of mutant TP53 had no impact on cell survival, proliferation, migration, metabolism, or response to stress, including treatment with anticancer drugs. Our findings are supported by studies of human AML and mouse B lymphoma in which mutant TP53/TRP53 proteins exerted their tumorigenic effects through their DNE with no evidence for a critical role of alleged GOF effects (9, 11), and the theory that aneuploidy may explain the behaviour of certain cancer cells expressing mutant TP53 (36). However, our findings contradict some previous publications. For example, it was reported that mouse embryonic fibroblasts (MEF) homozygous for the Trp53 R172H mutation proliferated more quickly in culture and underwent transformation more readily than Trp53−/− MEFs (37). The differences between these findings compared with our results may be explained by the fact that we used isogenic cancer cell lines to directly compare the TP53/TRP53 mutant versus TP53/TRP53–deficient state, whereas the previous study compared cells that were not isogenic, which obviates accurate comparison (37). Moreover, some studies reported that RNA interference–mediated removal of mutant TP53 inhibited the growth of a small number of human cancer cell lines in vitro (31, 32), supporting an essential role for its GOF activities in sustaining tumor expansion. Importantly, we found that doxycycline-induced expression of the same shTP53 caused death and inhibited proliferation of the same human cancer cell lines not only in the mutant TP53–expressing parental cells but also in their CRISPR-generated mutant TP53–deficient derivatives. TRP53 RNAi-mediated inhibition of in vitro growth of human cancer cell lines that lack TRP53 was also evident in the DepMap data. These findings demonstrate that the inhibition of cancer cell proliferation and survival observed in previous studies may be due to off-target toxic effects of RNA interference. Off-target toxic effects of RNAi technologies have been demonstrated (38). For example, several genes that were identified as cancer dependencies in screens using RNAi (e.g., HDAC6, MAPK14, CASP3, and PAK4) could not be validated when these genes were deleted using CRISPR/Cas9 or when the encoded proteins were targeted with specific inhibitors. In conclusion, identification of mutant TP53 as a cancer dependency by RNAi is not reliable.
The notion that pharmacologic removal of mutant TP53 can impair cancer growth, which was based on studies using compounds that allegedly do this indirectly, has also been challenged (17). For example, one HDAC inhibitor, SAHA, was reported to inhibit tumor growth by causing specific degradation of mutant TP53 protein (17), but another HDAC inhibitor, FK228, was shown to kill tumor cells in a manner independent of the removal of mutant TP53 (18). Notably, CRISPR/Cas9-mediated removal of HDAC6 did not inhibit the proliferation of these tumor cells (38). These observations challenge the notion that mutant TP53 is the critical target of HDAC inhibition and that removal of mutant TP53 would inhibit tumor growth.
Our unprecedented expansive xenograft studies of human and mouse cancer cell lines and patient colon cancer–derived organoids in NSG mice (the latter widely regarded as the gold standard in vivo model for studying human tumor growth; ref. 39) revealed that removal of mutant TP53 did not impair tumor expansion or metastasis. The tumors formed from the mutant TP53–deleted human colon cancer organoids exhibited the same architecture and rate of cell proliferation as tumors derived from the control mutant TP53–expressing colon cancer organoids. RNA-seq analysis confirmed that there were only very few differences in gene expression between the control versus the mutant TP53–deleted tumors growing in NSG mice. Thus, we found no evidence that mutant TP53, and hence its reported GOF activities, are critical for sustained tumor growth in vivo. Our findings are consistent with observations that tumors from TRP53-deficient mice, the state resulting from genetic loss of mutant TP53/TRP53, are highly aggressive (40, 41) and relatively insensitive to diverse anticancer agents (42, 43).
In conclusion, our studies overall using 16 cancer cell lines representing 7 different types of human cancers, mouse lymphoma, and breast cancer cells and human colon cancer–derived organoids, spanning 15 different TP53/TRP53 mutants, extended by mining the DepMap database encompassing 391 cancer cell lines with 158 different mutations in TP53, reveal that the GOF activities of mutant TP53 are not universally required for the sustained survival and proliferation of malignant cells in vitro or tumor growth in vivo. It remains possible that GOF effects of certain mutant TP53/TRP53 proteins, alongside their DN effects, may play critical roles during early stages of tumor development when nascent neoplastic cells express both mutant TP53 and wild-type TP53. Moreover, it cannot be excluded that GOF activities of certain TP53 mutant proteins, not tested here and not present in the DepMap database, may be critical for sustained tumor expansion. However, our findings suggest that drugs capable of abrogating expression of mutant TP53 proteins or blocking their GOF activities would be unlikely to afford substantial general therapeutic benefit in mutant TP53–expressing cancers. Rather, our finding that CRISPR-mediated correction of mutant TP53 into wt TP53 could extinguish the proliferation of cancer cells in vitro suggests that drugs that can restore wt TP53 activity to mutant TP53 proteins, such as PC14586, a structural corrector specifically targeting Y220C mutant TP53, should be effective.
METHODS
Cancer-Derived Cell Lines
The Burkitt lymphoma–derived cell lines, BL41 and Rael-BL, were cultured in RPMI1640 medium (Thermo Fisher Scientific) supplemented with 10% FCS (Sigma), 2 mmol/L l-glutamine (Thermo Fisher Scientific), 1 mmol/L sodium pyruvate (Thermo Fisher Scientific), 50 μmol/L α-thioglycerol (Sigma), 100 U/mL penicillin (Gibco), and 100 U/mL streptomycin (Gibco). SW620, HT29, HUH-7, MDA-MB-231, and MDA-MB-453 cells were cultured in DMEM with 10% FCS, 100 U/mL penicillin, and 100 U/mL streptomycin. HOP-92, NCI-H322m, Difi, HCC70, SUM159, AU565, SKBR3, BT549, and NB4 cells were cultured in RPMI1640 medium with 10% FCS, 100 U/mL penicillin, and 100 U/mL streptomycin. KHOS/NP cells were cultured in MEM with 10% FCS, 100 U/mL penicillin, and 100 U/mL streptomycin. Eμ-Myc lymphoma cells were isolated from the lymphoid organs of sick Eμ-Myc transgenic mice and cultured in high-glucose DMEM (Thermo Fisher Scientific) supplemented with 10% FCS, 50 μmol/L β-mercaptoethanol (Sigma), 100 μmol/L asparagine (Sigma), 100 U/mL penicillin, and 100 U/mL streptomycin. EO771 mouse breast cancer cells were cultured in RPMI1640 medium supplemented with 10% FCS, 2 mmol/L l-glutamine, 25 mmol/L HEPES, nonessential amino acids, 1 mmol/L sodium pyruvate, 100 U/mL penicillin, and 100 U/mL streptomycin. The authenticity of all cell lines used was routinely verified by genomic analysis. All cell lines were also routinely tested to confirm the absence of Mycoplasma infection (Lonza).
Human Colon Cancer–Derived Organoids
Human colon cancer–derived organoids were cultured as previously described (44). In brief, the organoids were maintained in the basal medium [DMEM/F12 supplemented with penicillin/streptomycin, Glutamax, HEPES, Nicatinamide, N2, and B27 (all from Thermo Fisher Scientific) with Matrigel (BD Biosciences) adding growth factors (N-Acetyl-l-cysteine, hbFGF, and EGF)]. The medium was changed twice a week, and organoids were passaged 1:3 every ten days. This study was conducted in accordance with the Declaration of Helsinki, the NHMRC Statement on Ethical Conduct in Human Research, and Institutional Human Research Ethics approval (HREC 2016.249). Patients gave written informed consent.
Virus Packaging and Infection of Cancer Cell Lines and Colon Cancer–Derived Organoids
The construction of the inducible sgRNA and shRNA expression vectors and virus production were performed as previously described (19). A negative control sgRNA (isgNC) which targets mouse Bim exon 2 (5′-GACAATTGCAGCCTGCTGAG) and a sgRNA targeting human TP53 exon 5 (5′- GAGCGCTGCTCAGATAGCGA) were used. The sgRNA targeting mouse Trp53 exon 4 (5′-GGCAACTATGGCTTCCACCT) was used in mouse lymphoma cells, whereas the sgRNA targeting human BIM exon 3 (5′- GACAATTGCAGCCTGCGGAG) was used as a negative control in mouse lymphoma cells. The shRNA targeting human TP53 (5′-GACTCCAGTGGTAATCTAC) was used, whereas the shRNA targeting rat CD8 (5′-AGCAAGCTGAACGATATA) was used as a negative control.
For adherent human cancer cell lines, 5 × 105 cells were plated into 6-well plates along with 3 mL viral supernatant and cultured overnight. For suspension cancer cell lines, 1 × 105 cells were suspended in 5 mL viral supernatant and centrifuged at 2,200 rpm for 2 hours at 32°C. GFP (indication of sgRNA) and mCherry (marker of Cas9 expression) double positive cells were sorted for subsequent experiments.
The same sgRNA sequences were also cloned into a constitutive expression vector, LCV2, which contains the sequences for Cas9, to delete mutant TP53 in human colon cancer–derived organoids. A total of 1 × 105 single-cell suspensions of organoids were mixed with 2 mL viral supernatant and centrifuged at 2,200 rpm for 2 hours at 32°C. After 72 hours, cells were selected with 5 μg/mL puromycin (Sigma) to enrich for those containing the vector.
RNA-Seq Analysis
Total RNA was isolated from cancer cells in vitro or tumors ex vivo by using TRIzol reagent according to the manufacturer's protocol (Thermo Fisher Scientific). The mRNA libraries were generated by using the Truseq RNA Sample Prep Kit (Illumina) and sequenced on a NextSeq 500 instrument using High Output kit (150 cycles; Illumina). For both data sets, sequencing reads were aligned to the human reference genome, hg38, using Rsubread package v2.2.6 (45). Over 97% of reads mapped to the reference genome for each sample in the breast cancer cell data and more than 94% for the colon cancer cell data samples. Successfully mapped reads were summarized into gene-level counts using featureCounts (46). Genes were identified using GENCODE annotation v37 (47). For all samples, 74%–82% read pairs were assigned to a gene. Genes labeled as “to be experimentally confirmed (TEC)” as well as long noncoding RNAs were excluded. Further data preprocessing and differential expression analyses were performed separately for the two datasets. Differential gene expression analyses were undertaken using the limma v3.46.0 (48) and edgeR v3.32.1 (49) software packages. Lowly expressed genes were filtered out using the filterByExpr function in edgeR with a minimum count of 30 for the in vitro breast cancer cell line data and a minimum count of 10 for the in vivo colon cancer data. This resulted in 14,793 genes and 18,784 genes being retained for downstream analysis in the breast and colon cancer datasets, respectively. Compositional differences between the libraries were normalized using the trimmed mean of M-values (TMM) method (50). Read counts were converted to log2 counts per million (logCPM), and differential expression between groups of samples was assessed using linear models and robust empirical Bayes moderated t-statistics with a trended prior variance relative to a fold-change threshold of 1.2 (limma-trend pipeline with TREAT; refs. 51, 52). False discovery rate was controlled below 5% using the Benjamini and Hochberg method. Volcano plots and mean-difference plots were generated using the volcano plot and plotMD functions in limma. Pathway analyses were performed on differentially expressed genes to test for over-representation of biological pathways as defined by Gene Ontology and KEGG pathways using limma's goana and kegga functions, respectively. Gene-set enrichment analyses were performed using ROAST (53), and barcode plots were generated using the barcode plot function in limma.
HDR-Mediated Correction of Mutant TP53 in Human Cancer Cell Lines
A total of 2 × 105 MDA-MB-231 cancer cells and 3 × 105 AU-565, HCC-70 or SK-BR-3 cancer cells were electroporated as previously described (33): 120 pmol of Cas9, 1.5 μL Alt-R CRISPR-Cas9 crRNA (100 μmol/L, IDT), 1.5 μL Alt-R CRISPR-Cas9 tracrRNA (100 μmol/L, IDT), 1.2 μL Ultramer DNA Oligonucleotide (100 μmol/L, IDT), and 1.2 μL of Alt-R Cas9 Electroporation enhancer (100 μmol/L, IDT), in SE (MDA-MB-231) or SF (AU-565, HCC-70, SK-BR-3) Cell Line Nucleofector Solution (Lonza). After electroporation, cells were treated with the DNA-PKcs inhibitor NU7441 (1 μmol/L, Selleck Chemicals) for 48 hours. Genomic DNA was isolated 4 and 12 days after electroporation using NucleoSpin Tissue columns (Macherey-Nagel) and subjected to Sanger sequencing and ICE analysis (bioRxiv 2019.08.10.251082).
Total RNA was isolated using NucleoSpin RNA Plus kit (Macherey-Nagel) and reverse transcribed using RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific) with random hexamer primers. BBC3 and CDKN1A mRNA levels were determined via quantitative PCR using qPCRBIO Probe Mix (PCR Biosystems) and the following probe sets: BBC3 probe (PCR amplicon within exon 1; Hs.PT.58.38345739.g), CDKN1A (PCR amplicon spanning exon 4 and 5; Hs.PT.58.40874346.g), and ACTB (PCR amplicon spanning exons 1 and 2; Hs.PT.39a.22214847). All probes were conjugated with a 6-FAM/ZEN/IBFQ dye/quencher mode. qPCR assays were performed on a CFX Touch Real-Time PCR Detection System (Bio-Rad), and Ct values were extracted using a Bio-Rad CFX Manager software. Expression values were normalized to the values for β-actin mRNA, and the relative quantifications are presented as linearized Ct values (2−ΔΔCt), normalized to the wild-type untreated reference values.
Mice and Tumor Transplants
All experiments with mice were approved and carried out in accordance with the guidelines of both the Melbourne Directorate Animal Ethics Committee and The Walter and Eliza Hall Institute of Medical Research Ethics Committee. Six- to 7-week-old Nod/scid/common γ chain–deficient (NSG) female mice were obtained from The Walter and Eliza Hall Institute mouse breeding facility. For the human cancer cell lines MDA-MB-231, HT29, and Rael-BL as well as the human colon cancer–derived organoids, the mutant TP53–expressing control cells and their mutant TP53–deleted derivatives were transplanted subcutaneously into the flanks of NSG mice (2 × 106 tumor cells per injection for MDA-MB-231 and HT29 cell lines, 4 × 106 tumor cells per injection for Rael-BL cell line, and 1 × 106 cells per injection for the human colon cancer–derived organoids). Tumor size was measured with calipers until mice had reached the mandated animal ethics endpoint (i.e., when the first tumor on one flank had reached a size of 500 mm3). To examine the growth of tumor xenografts initiated with limiting cell numbers, 200K, 20K, and 2K MDA-MB-231 cells and HT29 cells were transplanted subcutaneously into the flanks of NSG mice. Tumor size was monitored as described above.
To examine metastasis, MDA-MB-231 and SUM159 human breast cancer cells (0.5 × 106) were injected into mammary fat pads of NSG mice, and primary tumors were surgically removed when tumors had reached a volume of 200 mm3 to ensure prolonged survival of the mice. The lungs of tumor-burdened mice were excised, and single-cell suspensions were generated. GFP/mCherry double positive cells were sorted for further analysis.
For in vivo treatment with the anticancer agent 5-FU, NSG mice were transplanted with HT29 human colon cancer cells or WCB123LU human colon cancer–derived organoids (2 × 106 tumor cells per injection). When the tumor size reached 100 mm3, 75 mg/kg 5-FU was injected intraperitoneally (i.p.) into tumor-bearing NSG mice on day 0 and day 3. The tumor size was measured as described above for 7 days after treatment with 5-FU.
MRE412 Eu-Myc mouse lymphoma cells (C57BL/6J-Ly5.2 background), the mutant Trp53–expressing control cells or their mutant Trp53–deleted derivatives, were injected intravenously (i.v.) into 7- to 9-week-old immune-competent C57BL/6J-Ly5.1 male mice that had been purchased from The Walter and Eliza Hall Institute mouse breeding facility. Mice were sacrificed when deemed having reached ethical endpoint by an animal research technician who was blinded to the genotype of the transplanted lymphoma cells.
EO771 mutant Trp53 mouse breast cancer cells and their mutant Trp53–deleted derivatives were injected into mammary fat pads of 8-week-old Cas9 transgenic immune-competent C57BL/6J mice (54) to avoid immune rejection caused by Cas9 expressed in the EO771 cells. Primary tumors were excised 11 days after transplantation by surgery. IHC was used to detect the immune cells in the tumor microenvironment.
Data Mining and Analysis of the DepMap Database
The DepMap portal is designed to empower discoveries related to cancer cell vulnerabilities. The cancer cell lines listed in the database were grouped by their mutations in the TP53, KRAS, or BRAF genes, respectively. The corresponding gene-dependence scores of the effects of TP53, KRAS, or BRAF gene deletion or silencing using either CRISPR or RNAi, respectively, in the indicated cancer cell lines were derived from the datasets DepMap public 22Q4 (CRISPR) and Achilles+DRIVE+Marcotte (RNAi), which are listed in the Supplementary Tables S4–S9. Scatter plots were generated by using GraphPad Prism 9.
Statistical Analysis
GraphPad Prism was used for statistical analysis. Error bars indicate the SEM of three independent experiments. Two-tailed t tests were used to compare two data sets. Definition of P values in the figures and Supplementary Data figures is as follows: P > 0.05 (not significant; n.s.), P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***).
Data Availability Statement
Two RNA-seq datasets are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181011 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181016, respectively. Other data and reagents are available on request from A. Strasser and G.L. Kelly.
Supplementary Material
Description of Supplementary Materials and Methods
Supplementary Table S1 showing statistical analysis of differences in the numbers of human cancer cells in vitro with or without removal of mutant TP53
Supplementary Table S2 showing statistical analysis of differences in the survival of human cancer cells in vitro with or without removal of mutant TP53
Supplementary Table S3 showing the IC50 values of human cancer cell lines with or without removal of mutant TP53 treated with anti-cancer drugs
Supplementary Table S4 showing the impact of removal of mutant TP53 in human cancer cell lines in vitro by CRISPR/Cas9 by mining the DepMap database
Supplementary Table S5 showing the impact of removal of mutant TP53 in human cancer cell lines in vitro by RNAi by mining the DepMap database
Supplementary Table S6 showing the impact of removal of mutant BRAF in human cancer cell lines in vitro by CRISPR/Cas9 by mining the DepMap database
Supplementary Table S7 showing the impact of removal of mutant BRAF in human cancer cell lines in vitro by RNAi by mining the DepMap database
Supplementary Table S8 showing the impact of removal of mutant KRAS in human cancer cell lines in vitro by CRISPR/Cas9 by mining the DepMap database
Supplementary Table S9 showing the impact of removal of mutant KRAS in human cancer cell lines in vitro by RNAi by mining the DepMap database
Supplementary Figure S1 confirming the removal of mutant TP53 in diverse human cancer cell lines
Supplementary Figure S2 showing that removal of mutant TP53 has no impact on survival, proliferation, mitochondrial content and ROS levels in diverse human cancer cell lines
Supplementary Figure S3 showing that removal of mutant TP53 does not impair the ability of diverse human cancer cell lines to adapt to conditions of stress
Supplementary Figure S4 showing that removal of mutant TP53 has no impact on survival and proliferation of diverse human cancer cell lines when cultured in 3% FCS
Supplementary Figure S5 showing that removal of mutant TP53 has no impact on cycling, mitochondrial content or ROS levels in diverse human cancer cell lines when cultured in 3% FCS
Supplementary Figure S6 showing that removal of mutant TP53 does not impact the chemotherapy responses of diverse human cancer cell lines in culture
Supplementary Figure S7 showing that complete removal of mutant TP53 in MDA-MB-231 cells does not impair survival and proliferation in culture
Supplementary Figure S8 showing that complete removal of mutant TP53 in HT29 cells does not impair survival and proliferation in culture
Supplementary Figure S9 showing that complete removal of mutant TP53 in Rael-BL cells does not impair survival and proliferation in culture
Supplementary Figure S10 showing that complete removal of mutant TP53 in MDA-MB-468 cells does not impair survival and proliferation in culture
Supplementary Figure S11 showing that removal of mutant TP53 does not cause a competitive growth disadvantage in diverse human cancer cell lines in culture
Supplementary Figure S12 showing that removal of mutant TP53 by RNAi has minor impact on the survival and proliferation of certain human cancer cell lines in culture
Supplementary Figure S13 showing that mutant TP53 deficient human cancer cell lines retain the machinery to respond to the activation of wt TP53
Supplementary Figure S14 showing that CRISPR-HDR mediated conversion of mutant TP53 into wt TP53 causes a growth disadvantage in human cancer cell lines
Supplementary Figure S15 showing that removal of mutant TP53 does not impair in vivo tumor growth of certain human cancer cell lines
Supplementary Figure S16 showing that removal of mutant TP53 does not impair in vivo tumor growth of human cancer lines even when limited numbers of cancer cells are transplanted
Supplementary Figure S17 showing that removal of mutant TP53 does not impact colony formation of human cancer cell lines in vitro
Supplementary Figure S18 showing that removal of mutant TP53 in SUM159 human breast cancer cells does not impair metastasis in vivo
Supplementary Figure S19 showing that removal of mutant TP53 does not impair the migration of human cancer cell lines in vitro
Supplementary Figure S20 showing that removal of mutant TP53 does not impair the chemotherapy responses of the human colon cancer cell line HT29 in vivo
Supplementary Figure S21 showing that removal of mutant TRP53 does not impact the growth and survival of Eu-Myc mouse lymphoma cells in vitro and in vivo
Supplementary Figure S22 showing flow cytometric analysis of the transplanted Eu-Myc mouse lymphoma cells expanding in recipient mice
Supplementary Figure S23 showing that removal of mutant TRP53 in the E0771 mouse breast cancer cell line has no impact on the tumor microenvironment in vivo
Supplementary Figure S24 showing that removal of mutant TP53 does not impair the in vitro growth of human colon cancer derived organoids
Supplementary Figure S25 showing that removal of mutant TP53 has no impact on in vivo tumor growth upon transplantation of human colon cancer derived organoids into NSG mice
Supplementary Figure S26 showing that removal of mutant TP53 does not impact in vivo tumor growth upon transplantation of WCB139T human colon cancer derived organoids into NSG mice
Supplementary Figure S27 showing that removal of mutant TP53 does not impact the in vitro response of human colon cancer derived organoids to cisplatin
Supplementary Figure S28 showing that removal of mutant TP53 has no impact on the response of tumors derived from human colon cancer derived organoids to 5-FU in vivo
Acknowledgments
The authors thank G. Siciliano, D. Fayle, L. Spencer, and E. Loza for mouse husbandry, S. Monard and his team at the WEHI Flow Cytometry Unit for their assistance and Drs F. De Souza-e-Melo, F. De Sauvage (both from Genentech, South San Francisco, CA) for advice on colon cancer derived organoids, and Dr. K. Rajewsky (Max Delbrueck Centre for Genetics, Berlin, Germany) for Cas9 transgenic mice. This work was supported by funding from the Victorian Cancer Agency Fellowship (MCRF 17028, awarded to G.L. Kelly), Cancer Council Victoria, grants-in-aid #1086157 and #1147328 (awarded to G.L. Kelly); the National Health and Medical Research Council, Project Grant #1086291 and Ideas grants #2002618 and #2001201 (awarded to G.L. Kelly), program grant #101671 (awarded to A. Strasser and J.E. Visvader), fellowship #1020363 (awarded to A. Strasser), fellowship #1102742 (awarded to J.E. Visvader), fellowship #1136119 (awarded to O.M. Sieber); the Leukaemia Foundation Australia grant (awarded to G.L. Kelly and A. Strasser), the Leukaemia and Lymphoma Society grant #7001–13 (awarded to A. Strasser); the Giovanni Armenise-Harvard Foundation (CDA 2017; awarded to L.L. Fava); AIRC under MFAG 2019 - ID. 23560 project (awarded to L.L. Fava); the estate of Anthony (Toni) Redstone OAM, The Craig Perkins Cancer Research Foundation, The Harry Secomb Trust and the Dyson Bequest; and operational infrastructure grants through the Australian Government NHMRCS IRIISS and the Victorian State Government Operational Infrastructure Support.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
D.S. Simpson reports other support from The Walter and Eliza Hall Institute of Medical Research during the conduct of the study, personal fees and non-financial support from The Walter and Eliza Hall Institute of Medical Research, and personal fees and non-financial support from The University of Melbourne outside the submitted work. E. Lieschke reports grants from HSANZ/Leukemia Foundation of Australia during the conduct of the study. O.M. Sieber reports grants from National Health and Medical Research Council during the conduct of the study. L.L. Fava reports grants from Fondazione AIRC per la Ricerca sul Cancro and grants from Giovanni Armenise Harvard Foundation during the conduct of the study. G.L. Kelly reports grants from Servier, other support from Abbvie, and other support from Genentech outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
Z. Wang: Conceptualization, resources, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. M. Burigotto: Conceptualization, formal analysis, validation, investigation, writing–review and editing. S. Ghetti: Validation, investigation. F. Vaillant: Formal analysis, supervision, investigation. T. Tan: Formal analysis, investigation. B.D. Capaldo: Investigation, methodology. M. Palmieri: Investigation, methodology. Y. Hirokawa: Formal analysis, methodology. L. Tai: Resources, methodology. D.S. Simpson: Resources, methodology. C. Chang: Resources, methodology. A.S. Huang: Investigation, methodology. E. Lieschke: Resources, data curation, investigation. S.T. Diepstraten: Resources, supervision, investigation. D. Kaloni: Data curation, validation. C. Riffkin: Resources, validation. D.C. Huang: Resources, supervision. C.S. Li Wai Suen: Data curation, formal analysis, investigation. A.L. Garnham: Data curation, formal analysis, supervision. P. Gibbs: Resources, supervision. J.E. Visvader: Resources, supervision, writing–review and editing. O.M. Sieber: Resources, supervision. M.J. Herold: Conceptualization, resources, methodology, writing–review and editing. L.L. Fava: Conceptualization, resources, formal analysis, supervision, investigation. G.L. Kelly: Conceptualization, formal analysis, supervision, funding acquisition, validation, methodology, writing–original draft, writing–review and editing. A. Strasser: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Aylon Y, Oren M. Living with p53, dying of p53. Cell 2007;130:597–600. [DOI] [PubMed] [Google Scholar]
- 2. Janic A, Valente LJ, Wakefield MJ, Di Stefano L, Milla L, Wilcox S, et al. DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat Med 2018;24:947–53. [DOI] [PubMed] [Google Scholar]
- 3. Kastenhuber ER, Lowe SW. Putting p53 in context. Cell 2017;170:1062–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol 2007;8:275–83. [DOI] [PubMed] [Google Scholar]
- 5. Vousden KH, Prives C. P53 and prognosis: new insights and further complexity. Cell 2005;120:7–10. [DOI] [PubMed] [Google Scholar]
- 6. Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol 2010;2:a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer 2009;9:749–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev 2012;26:1268–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aubrey BJ, Janic A, Chen Y, Chang C, Lieschke EC, Diepstraten ST, et al. Mutant TRP53 exerts a target gene-selective dominant-negative effect to drive tumor development. Genes Dev 2018;32:1420–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell 2006;127:1323–34. [DOI] [PubMed] [Google Scholar]
- 11. Boettcher S, Miller PG, Sharma R, McConkey M, Leventhal M, Krivtsov AV, et al. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science 2019;365:599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shchors K, Persson AI, Rostker F, Tihan T, Lyubynska N, Li N, et al. Using a preclinical mouse model of high-grade astrocytoma to optimize p53 restoration therapy. Proc Natl Acad Sci U S A 2013;110:E1480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, et al. Restoration of p53 function leads to tumour regression in vivo. Nature 2007;445:661–5. [DOI] [PubMed] [Google Scholar]
- 14. Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 2007;445:656–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McCormick F. Targeting KRAS directly. Annu Rev Cancer Biol 2018;2:81–90. [Google Scholar]
- 16. Veggiani G, Gerpe MCR, Sidhu SS, Zhang W. Emerging drug development technologies targeting ubiquitination for cancer therapeutics. Pharmacol Ther 2019;199:139–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alexandrova EM, Yallowitz AR, Li D, Xu S, Schulz R, Proia DA, et al. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature 2015;523:352–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Panicker J, Li Z, McMahon C, Sizer C, Steadman K, Piekarz R, et al. Romidepsin (FK228/depsipeptide) controls growth and induces apoptosis in neuroblastoma tumor cells. Cell Cycle 2010;9:1830–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aubrey BJ, Kelly GL, Kueh AJ, Brennan MS, O'Connor L, Milla L, et al. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep 2015;10:1422–32. [DOI] [PubMed] [Google Scholar]
- 20. Liberti MV, Locasale JW. The warburg effect: how does it benefit cancer cells? Trends Biochem Sci 2016;41:211–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 22. Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X, et al. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun 2013;4:2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cordani M, Butera G, Pacchiana R, Masetto F, Mullappilly N, Riganti C, et al. Mutant p53-associated molecular mechanisms of ROS regulation in cancer ells. Biomolecules 2020;10:361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eriksson M, Ambroise G, Ouchida AT, Lima Queiroz A, Smith D, Gimenez-Cassina A, et al. Effect of mutant p53 proteins on glycolysis and mitochondrial metabolism. Mol Cell Biol 2017;37:e00328–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fulda S, Gorman AM, Hori O, Samali A. Cellular stress responses: cell survival and cell death. Int J Cell Biol 2010;2010:214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sicari D, Fantuz M, Bellazzo A, Valentino E, Apollonio M, Pontisso I, et al. Mutant p53 improves cancer cells’ resistance to endoplasmic reticulum stress by sustaining activation of the UPR regulator ATF6. Oncogene 2019;38:6184–95. [DOI] [PubMed] [Google Scholar]
- 27. Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab 2016;23:27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Do PM, Varanasi L, Fan S, Li C, Kubacka I, Newman V, et al. Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev 2012;26:830–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wei F, Hao P, Zhang X, Hu H, Jiang D, Yin A, et al. Etoposide-induced DNA damage affects multiple cellular pathways in addition to DNA damage response. Oncotarget 2018;9:24122–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vinall R, Chen Q, Talbott G, Ramsamooj R, Dang A, Tepper CG, et al. Use of RNA-seq and a transgenic mouse model to identify genes which may contribute to mutant p53-driven prostate cancer initiation. Biology 2022;11:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bossi G, Lapi E, Strano S, Rinaldo C, Blandino G, Sacchi A. Mutant p53 gain of function: reduction of tumor malignancy of human cancer cell lines through abrogation of mutant p53 expression. Oncogene 2006;25:304–9. [DOI] [PubMed] [Google Scholar]
- 32. Vikhanskaya F, Lee MK, Mazzoletti M, Broggini M, Sabapathy K. Cancer-derived p53 mutants suppress p53-target gene expression–potential mechanism for gain of function of mutant p53. Nucleic Acids Res 2007;35:2093–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ghetti S, Burigotto M, Mattivi A, Magnani G, Casini A, Bianchi A, et al. CRISPR/Cas9 ribonucleoprotein-mediated knockin generation in hTERT-RPE1 cells. STAR Protoc 2021;2:100407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 1985;318:533–8. [DOI] [PubMed] [Google Scholar]
- 35. de Sousa e Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, et al. A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature 2017;543:676–80. [DOI] [PubMed] [Google Scholar]
- 36. Redman-Rivera LN, Shaver TM, Jin H, Marshall CB, Schafer JM, Sheng Q, et al. Acquisition of aneuploidy drives mutant p53-associated gain-of-function phenotypes. Nat Commun 2021;12:5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 2004;119:861–72. [DOI] [PubMed] [Google Scholar]
- 38. Lin A, Giuliano CJ, Palladino A, John KM, Abramowicz C, Yuan ML, et al. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci Transl Med 2019;11:eaaw8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yan C, Brunson DC, Tang Q, Do D, Iftimia NA, Moore JC, et al. Visualizing engrafted human cancer and therapy responses in immunodeficient zebrafish. Cell 2019;177:1903–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA Jr, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 1992;356:215–21. [DOI] [PubMed] [Google Scholar]
- 41. Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol 1994;4:1–7. [DOI] [PubMed] [Google Scholar]
- 42. Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell 1993;74:957–67. [DOI] [PubMed] [Google Scholar]
- 43. Strasser A, Harris AW, Jacks T, Cory S. DNA-damage can induce apoptosis in proliferating lymphoid-cells via p53-independent mechanisms inhibitable by Bcl-2. Cell 1994;79:329–39. [DOI] [PubMed] [Google Scholar]
- 44. Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 2011;141:1762–72. [DOI] [PubMed] [Google Scholar]
- 45. Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 2019;47:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 47. Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 2019;47:D766–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010;11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann Appl Stat 2016;10:946–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McCarthy DJ, Smyth GK. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics 2009;25:765–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu D, Lim E, Vaillant F, Asselin-Labat ML, Visvader JE, Smyth GK. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 2010;26:2176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chu VT, Graf R, Wirtz T, Weber T, Favret J, Li X, et al. Efficient CRISPR-mediated mutagenesis in primary immune cells using CrispRGold and a C57BL/6 Cas9 transgenic mouse line. Proc Natl Acad Sci U S A 2016;113:12514–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Supplementary Materials and Methods
Supplementary Table S1 showing statistical analysis of differences in the numbers of human cancer cells in vitro with or without removal of mutant TP53
Supplementary Table S2 showing statistical analysis of differences in the survival of human cancer cells in vitro with or without removal of mutant TP53
Supplementary Table S3 showing the IC50 values of human cancer cell lines with or without removal of mutant TP53 treated with anti-cancer drugs
Supplementary Table S4 showing the impact of removal of mutant TP53 in human cancer cell lines in vitro by CRISPR/Cas9 by mining the DepMap database
Supplementary Table S5 showing the impact of removal of mutant TP53 in human cancer cell lines in vitro by RNAi by mining the DepMap database
Supplementary Table S6 showing the impact of removal of mutant BRAF in human cancer cell lines in vitro by CRISPR/Cas9 by mining the DepMap database
Supplementary Table S7 showing the impact of removal of mutant BRAF in human cancer cell lines in vitro by RNAi by mining the DepMap database
Supplementary Table S8 showing the impact of removal of mutant KRAS in human cancer cell lines in vitro by CRISPR/Cas9 by mining the DepMap database
Supplementary Table S9 showing the impact of removal of mutant KRAS in human cancer cell lines in vitro by RNAi by mining the DepMap database
Supplementary Figure S1 confirming the removal of mutant TP53 in diverse human cancer cell lines
Supplementary Figure S2 showing that removal of mutant TP53 has no impact on survival, proliferation, mitochondrial content and ROS levels in diverse human cancer cell lines
Supplementary Figure S3 showing that removal of mutant TP53 does not impair the ability of diverse human cancer cell lines to adapt to conditions of stress
Supplementary Figure S4 showing that removal of mutant TP53 has no impact on survival and proliferation of diverse human cancer cell lines when cultured in 3% FCS
Supplementary Figure S5 showing that removal of mutant TP53 has no impact on cycling, mitochondrial content or ROS levels in diverse human cancer cell lines when cultured in 3% FCS
Supplementary Figure S6 showing that removal of mutant TP53 does not impact the chemotherapy responses of diverse human cancer cell lines in culture
Supplementary Figure S7 showing that complete removal of mutant TP53 in MDA-MB-231 cells does not impair survival and proliferation in culture
Supplementary Figure S8 showing that complete removal of mutant TP53 in HT29 cells does not impair survival and proliferation in culture
Supplementary Figure S9 showing that complete removal of mutant TP53 in Rael-BL cells does not impair survival and proliferation in culture
Supplementary Figure S10 showing that complete removal of mutant TP53 in MDA-MB-468 cells does not impair survival and proliferation in culture
Supplementary Figure S11 showing that removal of mutant TP53 does not cause a competitive growth disadvantage in diverse human cancer cell lines in culture
Supplementary Figure S12 showing that removal of mutant TP53 by RNAi has minor impact on the survival and proliferation of certain human cancer cell lines in culture
Supplementary Figure S13 showing that mutant TP53 deficient human cancer cell lines retain the machinery to respond to the activation of wt TP53
Supplementary Figure S14 showing that CRISPR-HDR mediated conversion of mutant TP53 into wt TP53 causes a growth disadvantage in human cancer cell lines
Supplementary Figure S15 showing that removal of mutant TP53 does not impair in vivo tumor growth of certain human cancer cell lines
Supplementary Figure S16 showing that removal of mutant TP53 does not impair in vivo tumor growth of human cancer lines even when limited numbers of cancer cells are transplanted
Supplementary Figure S17 showing that removal of mutant TP53 does not impact colony formation of human cancer cell lines in vitro
Supplementary Figure S18 showing that removal of mutant TP53 in SUM159 human breast cancer cells does not impair metastasis in vivo
Supplementary Figure S19 showing that removal of mutant TP53 does not impair the migration of human cancer cell lines in vitro
Supplementary Figure S20 showing that removal of mutant TP53 does not impair the chemotherapy responses of the human colon cancer cell line HT29 in vivo
Supplementary Figure S21 showing that removal of mutant TRP53 does not impact the growth and survival of Eu-Myc mouse lymphoma cells in vitro and in vivo
Supplementary Figure S22 showing flow cytometric analysis of the transplanted Eu-Myc mouse lymphoma cells expanding in recipient mice
Supplementary Figure S23 showing that removal of mutant TRP53 in the E0771 mouse breast cancer cell line has no impact on the tumor microenvironment in vivo
Supplementary Figure S24 showing that removal of mutant TP53 does not impair the in vitro growth of human colon cancer derived organoids
Supplementary Figure S25 showing that removal of mutant TP53 has no impact on in vivo tumor growth upon transplantation of human colon cancer derived organoids into NSG mice
Supplementary Figure S26 showing that removal of mutant TP53 does not impact in vivo tumor growth upon transplantation of WCB139T human colon cancer derived organoids into NSG mice
Supplementary Figure S27 showing that removal of mutant TP53 does not impact the in vitro response of human colon cancer derived organoids to cisplatin
Supplementary Figure S28 showing that removal of mutant TP53 has no impact on the response of tumors derived from human colon cancer derived organoids to 5-FU in vivo
Data Availability Statement
Two RNA-seq datasets are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181011 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181016, respectively. Other data and reagents are available on request from A. Strasser and G.L. Kelly.