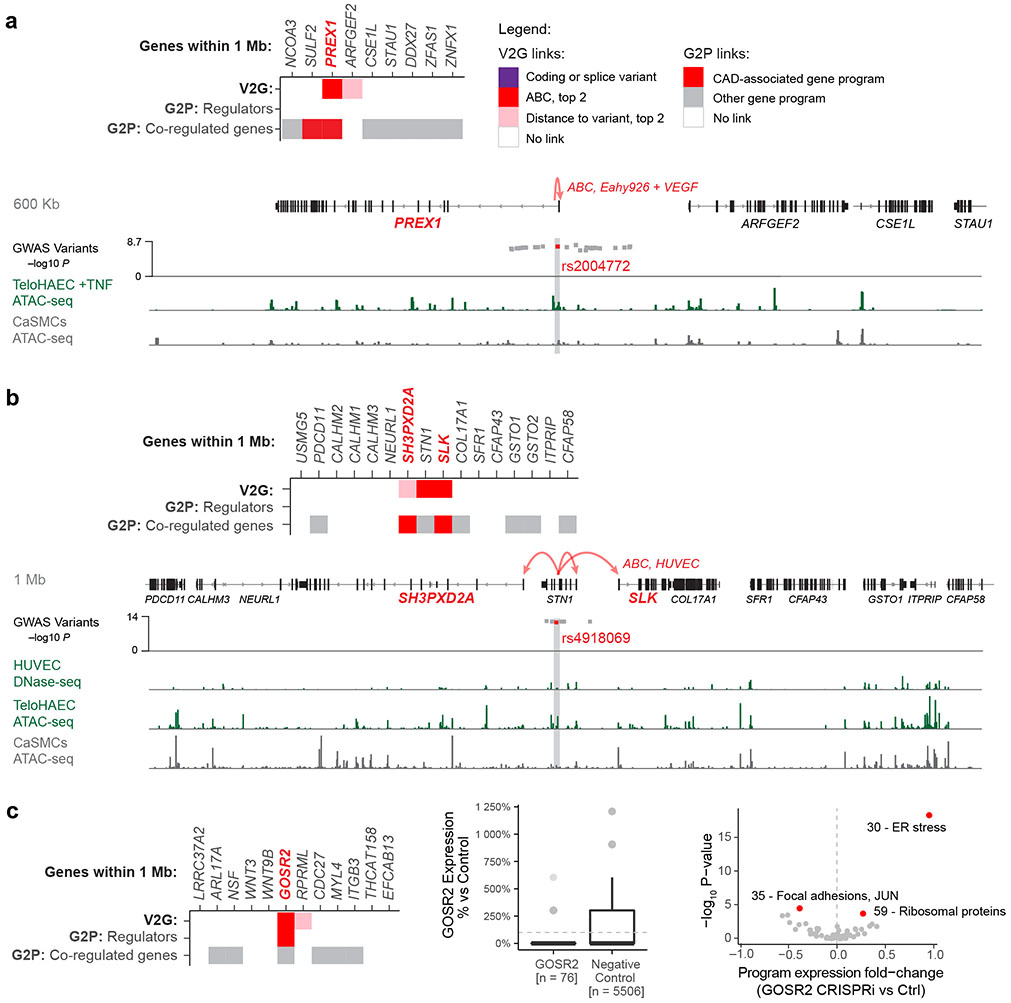
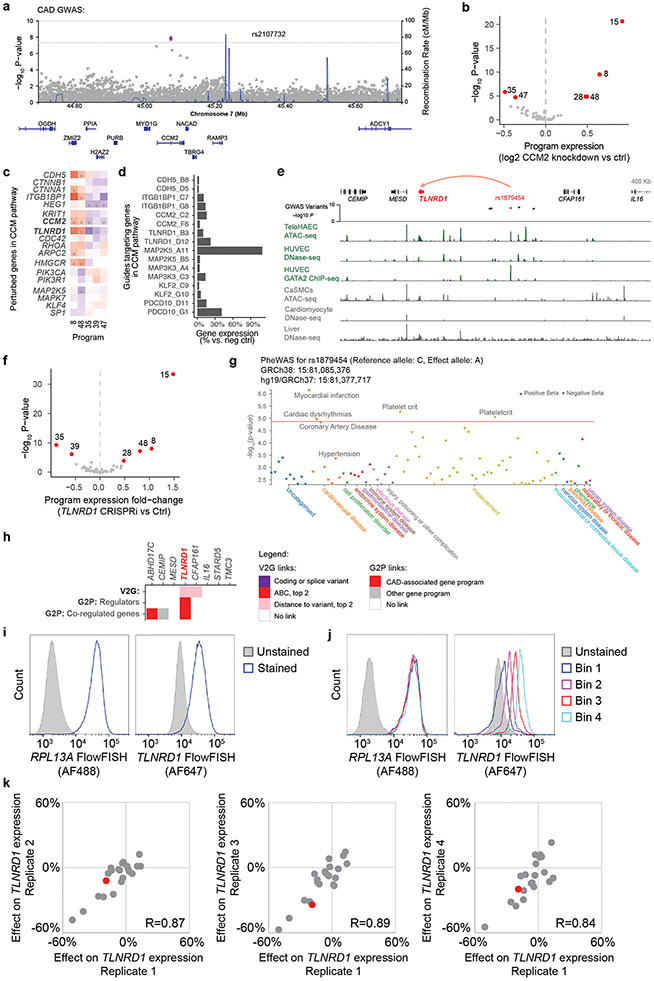
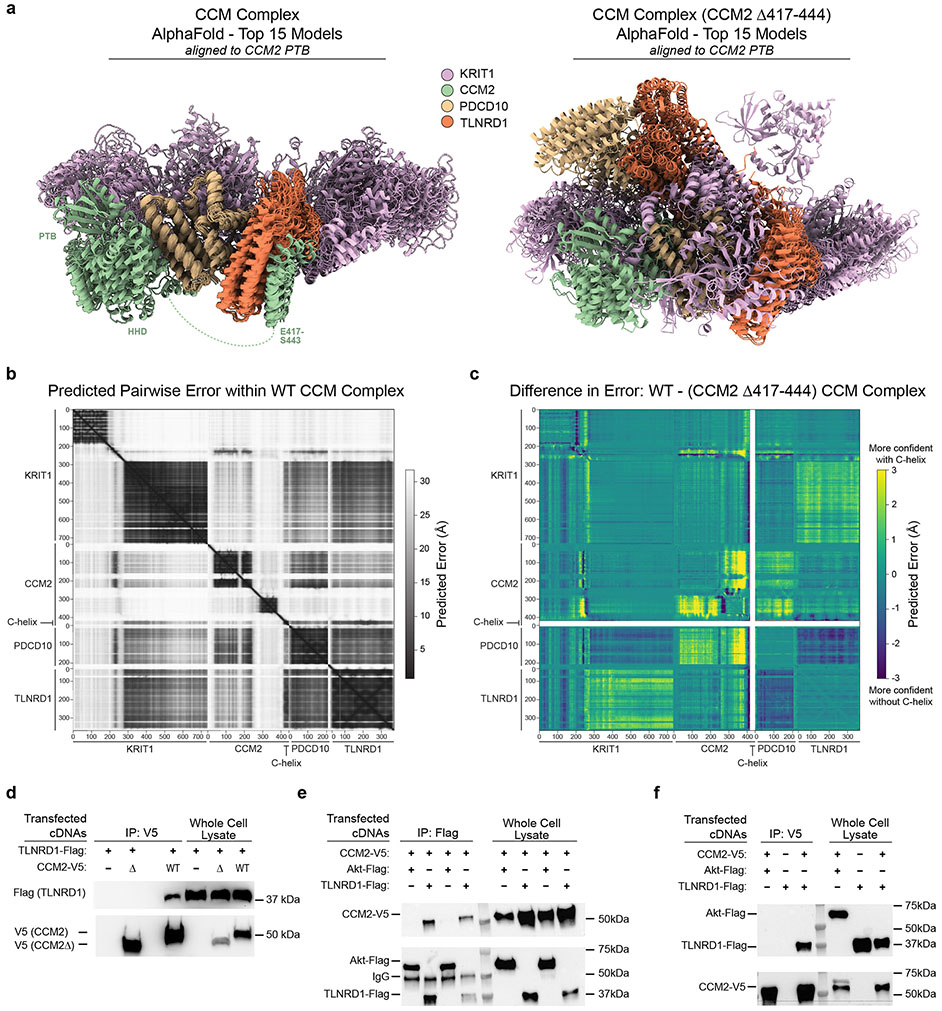
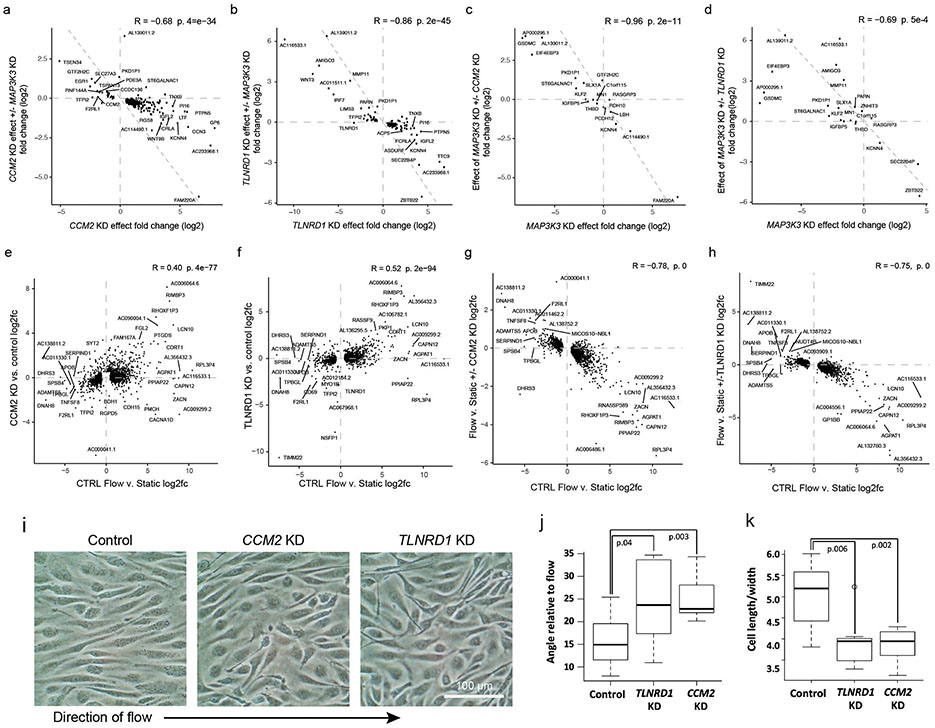
Abstract
Linking variants from genome-wide association studies (GWAS) to underlying mechanisms of disease remains a challenge1,4,6. For some diseases, a successful strategy has been to look for cases where multiple GWAS loci contain genes that act in the same biological pathway1-6. However, our knowledge of which genes act in which pathways is incomplete, particularly for cell-type specific pathways or understudied genes. Here we introduce a new method to connect GWAS variants to functions, which links variants to genes using epigenomic data, links genes to pathways de novo using Perturb-seq, and integrates these data to identify convergence of GWAS loci onto pathways. We apply this approach to study the role of endothelial cells in genetic risk for coronary artery disease (CAD), and discover that 43 CAD GWAS signals converge on the cerebral cavernous malformations (CCM) signaling pathway. Two regulators of this pathway, CCM2 and TLNRD1, are each linked to a CAD risk variant, regulate other CAD risk genes, and affect atheroprotective processes in endothelial cells. These results suggest a model where CAD risk is driven in part by the convergence of causal genes onto a particular transcriptional pathway in endothelial cells, highlight shared genes between common and rare vascular diseases (CAD and CCM), and identify TLNRD1 as a new, previously uncharacterized member of the CCM signaling pathway. This approach will be widely useful for linking variants to functions for other common polygenic diseases.
Introduction
Genetic variants that influence complex traits are thought to regulate genes that work together in biological pathways. Identifying convergence on particular pathways can help in discovering genes and cellular functions that causally influence disease risk1-6. However, it is often challenging to identify such convergence: complex traits involve contributions from multiple cell types; most risk variants are noncoding and can regulate multiple nearby genes; and it remains unclear which genes work together in which pathways in which cell types7-9.
GWAS for coronary artery disease have discovered over 300 independent signals10-12(Supplementary Table 1). 75% of these signals are not associated with circulating lipids (Supplementary Table 1), indicating the presence of undiscovered disease mechanisms that may function through cells in the coronary artery where atherosclerosis that causes CAD develops. Endothelial cells (ECs) are one of the most important of these arterial cells, controlling cholesterol uptake and efflux, smooth muscle cell responses, blood clotting and inflammatory immune cell recruitment13,14, and are highly enriched for CAD heritability15. At a few individual CAD GWAS loci, noncoding risk variants have been shown to regulate the expression of key EC genes such as endothelial nitric oxide synthase (NOS3), endothelin 1 (EDN1), and others16. It remains unclear, however, which other genes in CAD GWAS loci might work together in which EC pathways to modulate disease risk.
To address these challenges, we have developed a new approach that systematically and unbiasedly links GWAS variants to genes and identifies their convergence onto specific disease-associated transcriptional programs. The 5 steps of this Variant-to-Gene-to-Program (V2G2P) approach (Fig. 1a, Supplementary Note 1), and their application to EC functions in CAD, are summarized below:
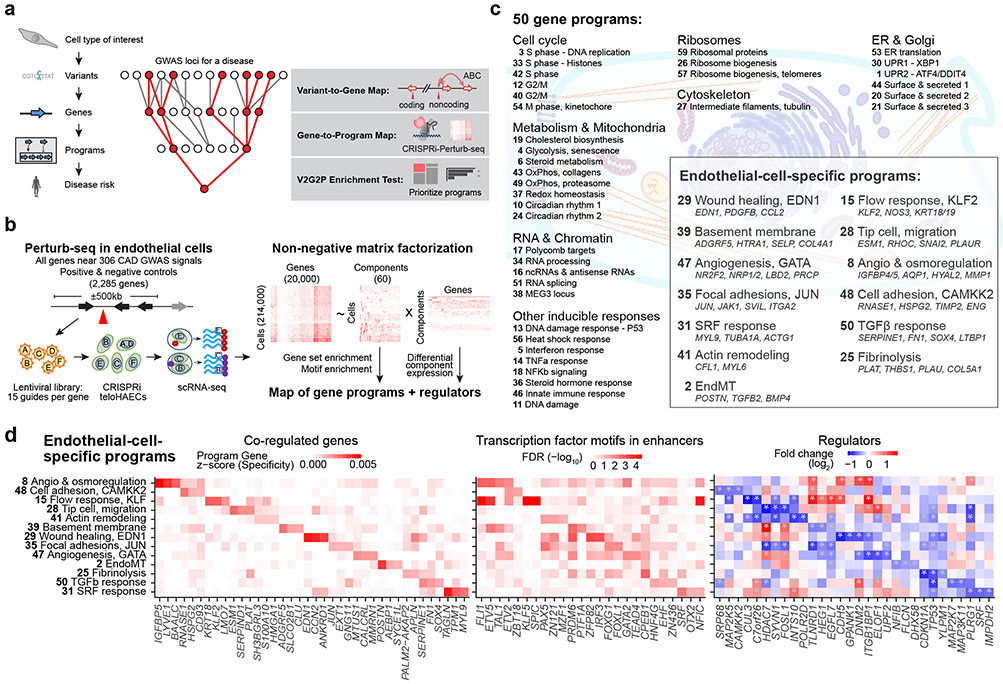
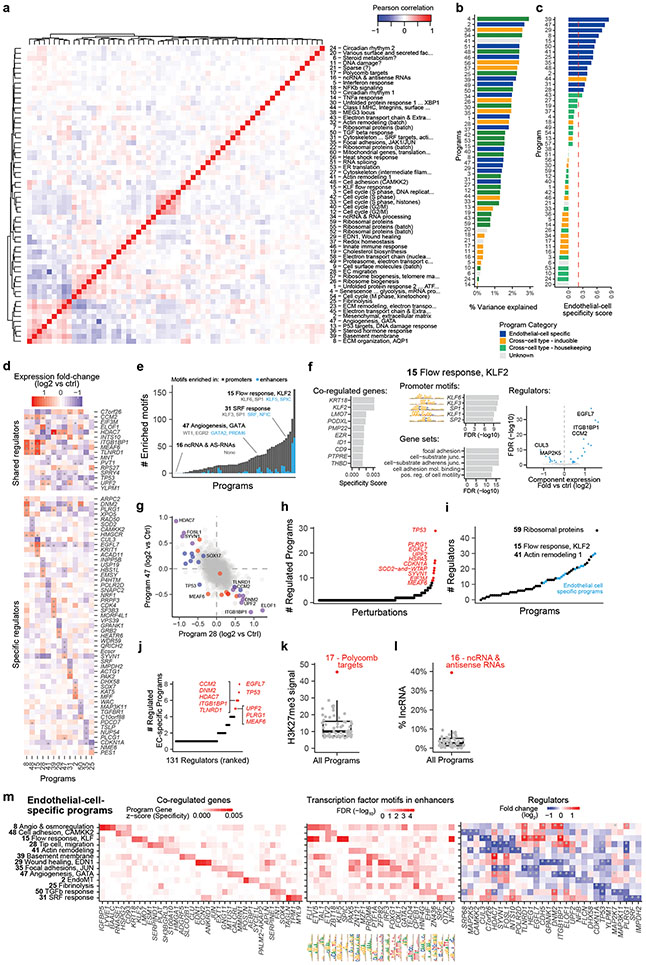
Fig. 1. Building a map of gene programs in endothelial cells using Perturb-seq.
a. Overview of the Variant-to-Gene-to-Program (V2G2P) approach. Red nodes outline an example of the predicted convergence of multiple risk variants onto a small number of cell type-specific programs related to disease.
b. Diagram of the approach to create a map of gene programs and regulators using Perturb-seq (Gene to Program, G2P).
c. 50 gene programs were identified de novo from Perturb-seq and cNMF. For the 13 EC-specific programs, selected co-regulated genes in the program are shown in italics.
d. EC-Specific Programs. Heatmaps show, for each program, the top 3 program co-regulated genes (left, ranked by specificity to the program, see Methods); the top 3 transcription factor motifs in enhancers (middle, ranked by enrichment FDR); and top 3 regulators (right, ranked by fold-change in component expression upon perturbation; see also Extended Data Fig. 3e).
Identify a cell type and cellular model relevant to disease genetics, through enrichment of disease risk variants in enhancers in that cell type. Here, we focused on human arterial ECs, using telomerase-immortalized human aortic ECs (teloHAEC) as a model.
Build a map of variant-to-gene (V2G) links in that cell type, to link disease-associated variants to potential target genes. Here, we consider evidence from variants in EC enhancers, as well as coding regions and splice sites.
Build a map of gene-to-program (G2P) links in that cell type, by using Perturb-seq17-20 to systematically knock down all possible candidate disease genes and identify sets of genes that act together in biological pathways. Here, we knock down all expressed genes within ±500kb of 306 CAD GWAS signals, read out the effects of each perturbation with single cell RNA-seq, and use unsupervised machine learning to define gene “programs,” unbiased by prior knowledge of gene sets or pathways.
Identify “disease-associated programs”, by developing a statistical test to determine whether the genes with links to risk variants are enriched in (that is, converge on) particular programs. Here, we find that many CAD GWAS loci converge on 5 gene programs identified de novo with Perturb-seq, which appear to correspond to branches of the cerebral cavernous malformations (CCM) signal transduction pathway.
Study the genes in disease-associated programs. Here, we nominate 41 genes likely to influence CAD risk through effects in ECs, and dissect two in detail: showing that knockdown of TLNRD1 or CCM2 mimics the effects of atheroprotective laminar blood flow, and that the poorly-characterized gene, TLNRD1, is a novel regulator in the CCM pathway.
In summary, the V2G2P approach defines cellular programs de novo using Perturb-seq, intersects these programs with enhancer-to-gene maps from the same cell type, and provides an interpretable, systematic, and unbiased framework for tracing the path from variant to gene to disease program simultaneously for all GWAS loci for a given disease and cell type.
A variant-to-gene map in ECs
To implement this V2G2P approach, we collected GWAS signals for coronary artery disease10,12, and defined a set of “nearby genes” for each GWAS signal to include the 2 closest genes on either side, plus all genes within +/−500 kb. We focused on the 228 “non-lipid” GWAS signals that were not associated with circulating lipid levels (see Methods: “Defining variants in CAD GWAS signals”), because lipid-associated signals likely act in hepatocytes or other non-endothelial cell types. This yielded 1,942 total candidate genes, with a median of 8 nearby genes per GWAS signal (Supplementary Table 1).
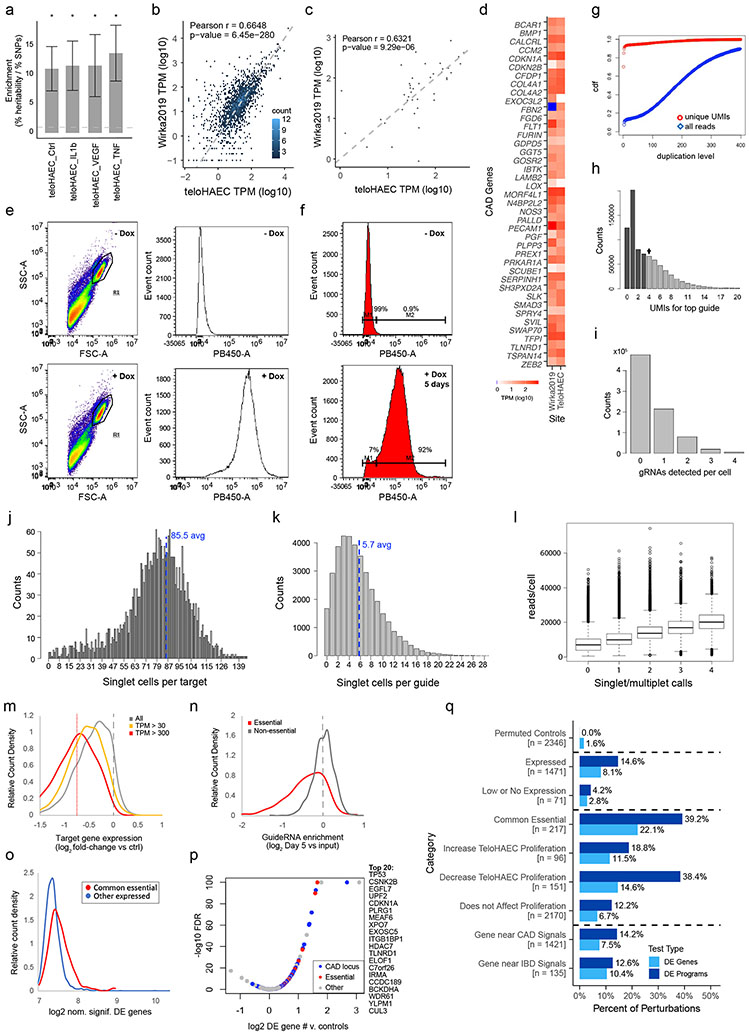
We selected telomerase-immortalized primary human aortic ECs (teloHAEC) as a well-established arterial endothelial cell model21, and collected bulk RNA-seq, ATAC-seq, and H3K27ac ChIP-seq data in resting and several stimulated conditions (+IL1β, TNFα, VEGFA) to identify expressed genes and candidate enhancers (Supplementary Table 2). Variants in teloHAEC enhancers were 11-to-13-fold enriched for CAD heritability by stratified linkage disequilibrium score regression (S-LDSC, Extended Data Fig. 1a), and the genes near CAD GWAS loci that were expressed in teloHAEC were also expressed in primary coronary artery ECs in vivo (Extended Data Fig. 1b-d), supporting the choice of this cellular model.
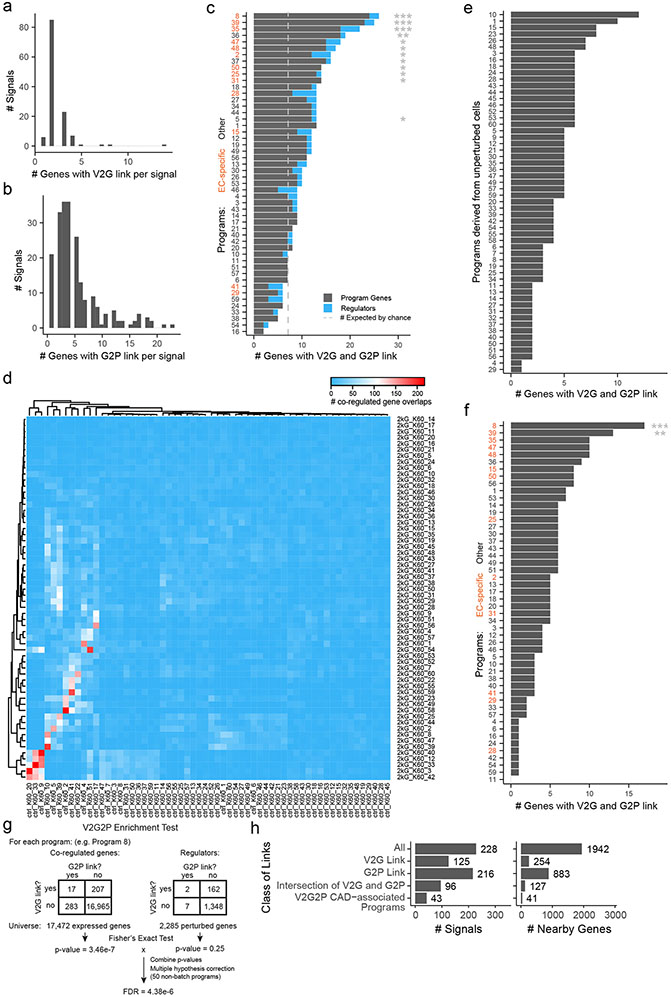
To link risk variants to genes (V2G), we identified genes predicted to be regulated by EC enhancers containing CAD variants using the Activity-by-Contact model (ABC9,22). We also considered genes containing coding variants (see Methods: Linking variants to genes). We identified 254 of 1,942 nearby genes with a link to a CAD risk variant (“genes with a V2G link”, or simply “V2G genes”), at 125 of 228 non-lipid GWAS signals (range: 1–5 genes per signal, Supplementary Tables 3 & 26).
A gene-to-program map in ECs
To link genes to programs (G2P), we applied CRISPR interference (CRISPRi)-Perturb-seq to identify de novo sets of genes that act in the same transcriptional pathways. While Perturb-seq is emerging as a powerful tool to study gene pathways17-20,23,24, new design and analysis approaches are needed to discover pathways enriched for genetic risk for common diseases. Accordingly, we developed an approach in which we systematically knocked down all expressed genes near all CAD GWAS signals, applied an unsupervised matrix factorization approach to identify sets of co-regulated genes, and linked upstream perturbed genes with the downstream genes they regulate to define “gene programs” in a systematic fashion, unbiased by previous knowledge of annotated pathways or gene sets (Fig. 1b).
We engineered teloHAEC to express dCas9-KRAB (CRISPRi, Extended Data Fig. 1e,f) and transduced these cells with a guide library targeting all 1,661 expressed genes nearby CAD GWAS signals and 624 control genes (15 guides/promoter), plus 1000 control guides, for a total of 37,637 guides (Supplementary Tables 4, 5 & 6). After 5 days of doxycycline induction of CRISPRi, we collected 20 lanes of 10x 3’ single-cell RNA-seq (see Methods). In total, we obtained data for 214,449 cells expressing a single guide at an average depth of 929,000 total transcript UMIs per targeted promoter (Extended Data Figs. 1 g-l & 2 a-b, Supplementary Table 7). We found that target genes were effectively knocked down, that knockdown of common essential genes decreased fitness, and that 10.7% of perturbations of expressed targets significantly impacted the transcriptome (Extended Data Fig. 1m-q; Supplementary Tables 8, 9 & 10).
We applied an unsupervised approach to this Perturb-seq data to discover gene programs, independent of previous knowledge of annotated pathways or gene sets (Fig. 1b, right). First, we used consensus non-negative matrix factorization (cNMF)25 to model the gene-by-cell matrix as a linear combination of latent components representing co-regulated gene sets that covary in the population of cells (see Methods, Extended Data Figs. 2c-g, Supplementary Table 11). From each cNMF component, we defined a “program”: a set of genes comprised of both “co-regulated genes” (the 300 marker genes whose expression is most specific to that component) and “regulators” (the 0 to 35 genes whose perturbations significantly affected the expression of each component, relative to negative control guideRNAs, FDR < 0.05, Extended Data Fig. 2h). This analysis established a gene-to-program map that included 18,606 links from 7,692 unique genes to 50 programs (Fig. 1c, Extended Data Fig. 3a-c; Supplementary Tables 12, 13 & 14).
After defining these 50 programs using an unsupervised approach, we annotated each program based on their regulators and co-regulated genes; including manual curation, analysis of transcription factor (TF) motifs in their promoters and predicted enhancers, and gene set enrichment (see Methods: Definition and annotation of gene expression programs). We identified programs representing an array of cellular functions: from ubiquitously expressed (“housekeeping”) processes, to a wide variety of inducible responses (e.g., unfolded protein response (UPR), DNA damage, heat shock, and inflammation) despite the absence of stimuli for these responses in our culture system (Fig. 1c). We annotated 13 programs as “EC-specific” because they included genes that were on average more highly expressed in ECs than in other cell types (Fig. 1c, Extended Data Fig. 3c, Supplementary Table 13, see Methods: Defining EC-specific programs). These EC-specific programs included distinct combinations of genes enriched for roles in angiogenesis, extracellular matrix remodeling, barrier function, and the endothelial-to-mesenchymal transition (endoMT), and the promoters of their co-regulated genes were enriched for different transcription factor motifs (Fig. 1d, Extended Data Fig. 3d-f). Analysis of regulators (perturbations) identified cases where programs were coordinately or oppositely regulated by the same perturbations (e.g. Extended Data Fig. 3g), and identified 10 genes that were regulators of 5 or more EC-specific programs, including genes known to have important functions in ECs such as EGFL7 and ITGB1BP1/ICAP126,27 (Extended Data Fig. 3h-l).
Taken together, this gene-to-program map represents a wide range of cellular pathways, links upstream regulators to coherent sets of downstream genes, and provides a resource for understanding the functions and potential disease-relevance of genes in ECs.
CAD GWAS signals converge on 5 programs
We next applied a simple statistical test (“V2G2P enrichment”) to determine, in an unbiased fashion, whether GWAS variants for a trait would converge onto particular gene programs. Specifically, we tested whether genes for each program (Genes with a G2P link, Fig. 2a) were more highly enriched in genes likely to be affected by CAD risk variants (Genes with a V2G link, Fig. 2a) than expected by chance (see Methods: Identifying CAD-associated programs via variant-to-gene-to-program analysis).
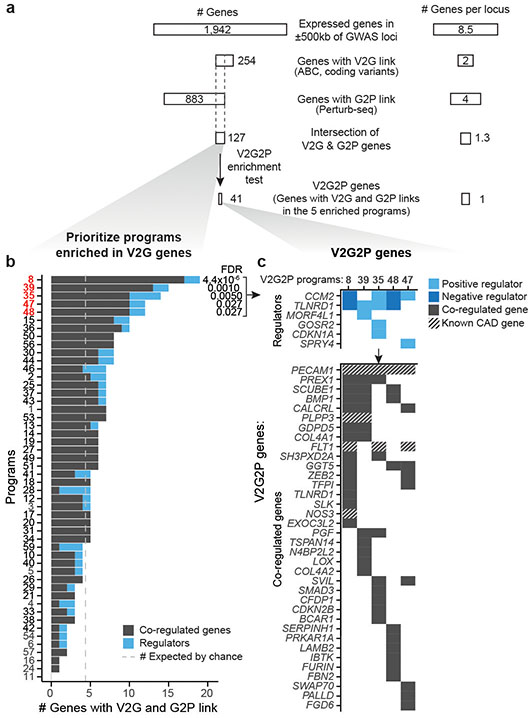
Fig. 2. CAD genes converge on 5 programs in endothelial cells.
a. Path to the convergence of 5 V2G2P programs and 41 V2G2P genes for coronary artery disease.
b. Identification of V2G2P programs for CAD. The 50 programs are ordered (y-axis) by the number of program genes linked to CAD variants (x-axis). We define the 5 programs with FDR < 0.05 as V2G2P programs. Gray dashed line: the number of genes linked to CAD variants that would be expected by chance.
c. Relationships among the 41 V2G2P genes for CAD and the 5 V2G2P programs. Top: 6 V2G2P genes were regulators of one or more V2G2P programs (FDR < 0.05). Light blue boxes indicate positive regulators (genes where loss-of-function leads to a decrease in program expression); dark blue indicates negative regulators (genes where loss-of-function leads to an increase in program expression). Bottom: 36 V2G2P genes for CAD were co-regulated genes in one or more V2G2P program. Cross hatching indicates members of the 8 gold standard EC CAD genes, previously known to affect CAD risk through effects in ECs (Supplementary Table 16).
We identified significant V2G2P enrichment for 5 programs, each including 12 to 18 genes linked to CAD variants (versus 4.5 expected by chance; 2.6- to 4-fold enrichment, FDR < 0.05, Fig. 2b, Supplementary Table 13). Together, these 5 programs included 41 unique V2G2P genes (genes linked to CAD variants and part of at least one of the 5 V2G2P programs), including genes near 43 of 228 non-lipid GWAS signals (Fig. 2c, Supplementary Tables 1 & 15).
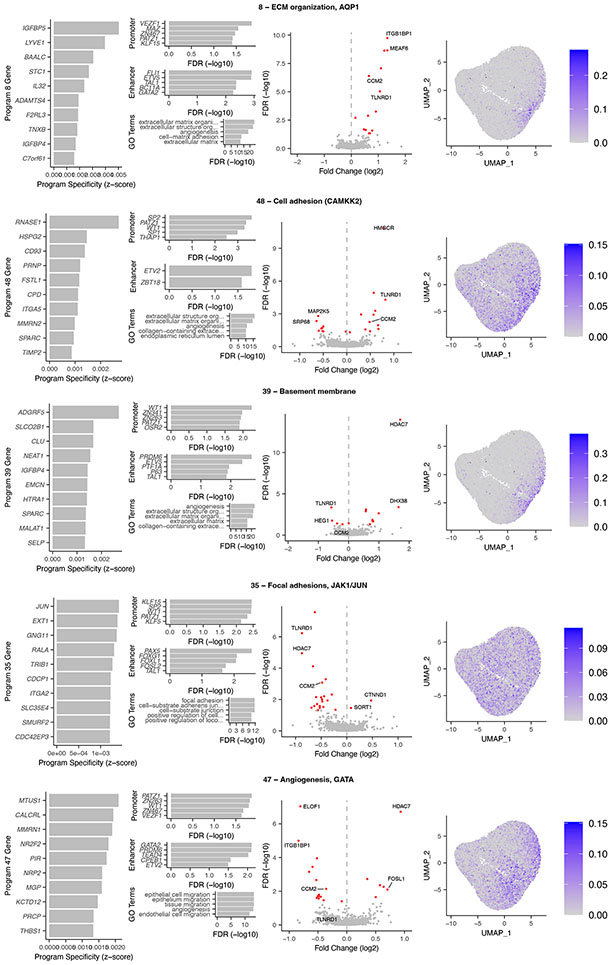
The 5 V2G2P programs corresponded to distinct sets of genes related to extracellular matrix (ECM) organization, cell migration, and angiogenesis (Fig. 2b, Extended Data Fig. 4). Program 8 included genes involved in negative regulation of angiogenesis (IGFBP4 and IGFBP5) and osmotic balance (SLC12A2 and AQP1). Program 48 included genes involved in cell adhesion and migration such as FSLT1 and TIMP2, and was regulated by MEK5/MAP2K5, ERK5/MAPK7, and calcium/calmodulin- dependent (CAMKK2) signaling. Program 39 expressed genes involved in the basement membrane (COL4A1/2) and platelet recruitment (VWF, SELP). Program 35 expressed genes involved in focal adhesions (ITGA2) and the JAK/STAT signaling pathway. Program 47 expressed genes involved in angiogenesis including NR2F2 and NRP1/2, including two genes specifically associated with a stalk cell phenotype (VWF, EHD4).
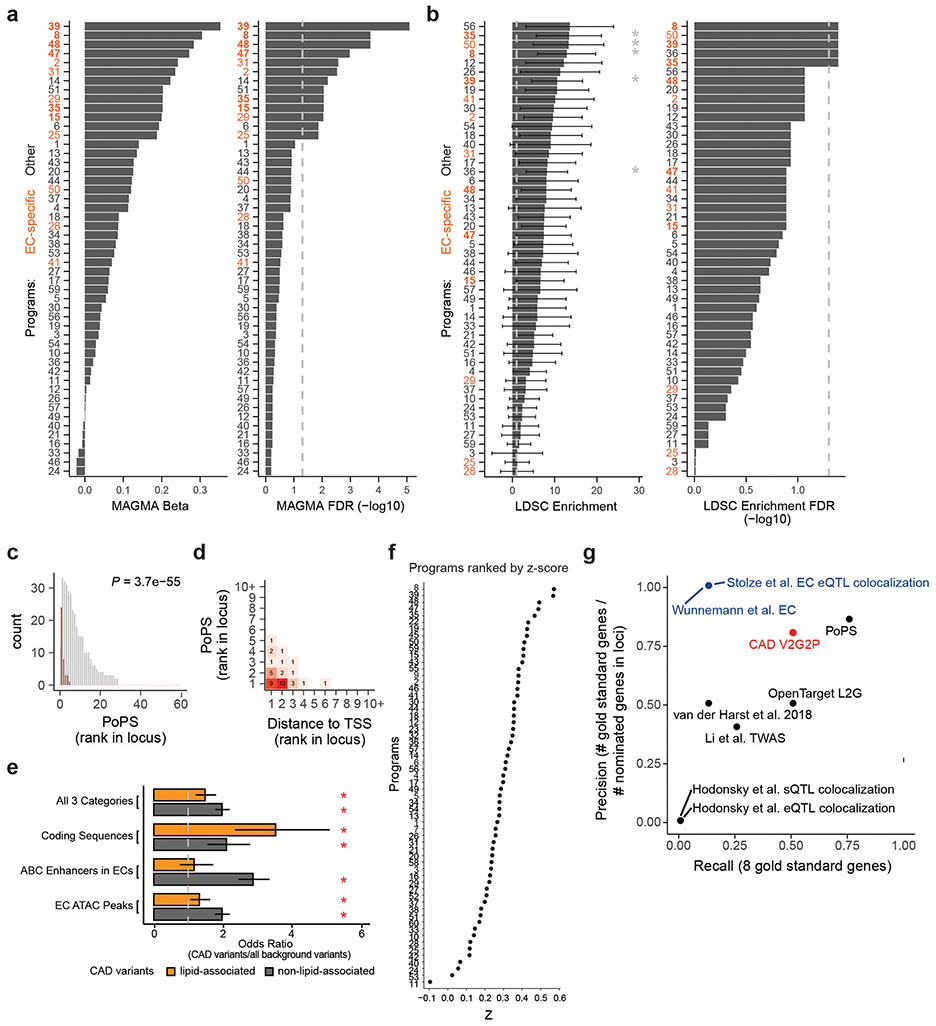
Several independent lines of evidence supported the associations of these 5 programs and 41 genes with CAD. (i) All 5 V2G2P programs were EC-specific programs that included at least 1 of the 8 gold standard genes whose variant-to-gene-to-disease effects in ECs have previously been characterized (“known endothelial cell CAD genes” in Fig. 2c, Supplementary Tables 15 & 16). Program 8 included four such genes: NOS3, PLPP3, FLT1, and PECAM1. (ii) All 5 V2G2P programs were significantly enriched for CAD heritability by MAGMA2, and two were significantly enriched for CAD heritability by S-LDSC28 (FDR < 0.05, see Methods; Extended Data Fig. 5a,b). (iii) The 41 V2G2P genes were highly ranked by an independent gene prioritization method, PoPS3, compared to other nearby genes at the same GWAS signals (rank-sum test P = 2.5 x 10−53, Extended Data Fig. 5c,d, Supplementary Table 15). (iv) 9 of the 41 V2G2P genes have previously been found to affect atherosclerosis and/or vascular barrier integrity via studies in mouse models, in a way that is consistent with their acting in ECs (Supplementary Table 15).
In summary, the V2G2P approach identifies the convergence of CAD GWAS signals onto 5 EC-specific gene programs that are enriched in CAD heritability and include 41 unique genes linked to CAD risk variants.
Benchmarking, and methods comparisons
We compared our V2G2P-prioritized genes to those from seven previous studies that used a variety of approaches to prioritize genes in CAD GWAS loci (Supplementary Note 2, Supplementary Table 17). We found that 31 out of the 41 V2G2P genes were not prioritized in the two EC-specific studies, and 17 were not prioritized by any of these seven studies (Supplementary Note 2, Supplementary Table 17). These 17 novel genes included the two strongest regulators of the 5 V2G2P programs, TLNRD1 and CCM2, which we will explore in detail below.
We also benchmarked the ability of V2G2P to identify the 8 gold standard EC CAD genes (Supplementary Note 2). Compared to other studies that nominated CAD genes in an EC-specific fashion (based on eQTL colocalization29 or variant-targeted CRISPR screens30), V2G2P achieved much higher recall for the gold standard genes (50% vs 12.5% for the others), while also achieving high precision (80%). V2G2P also performed well compared to studies that nominated CAD genes without specificity for ECs, and compared to methods to prioritize gene sets/programs from GWAS data (Supplementary Note 2, Extended Data Fig. 5g).
We found that both variant-to-gene data from ABC and gene-to-program data from Perturb-seq were essential for the ability of V2G2P analysis to identify disease-associated genes and programs (Fig. 2a, Supplementary Note 3, Extended Data Figs. 5, 6 & 7), consistent with recent observations that combining locus-specific variant-to-gene links with genome-wide enrichments for gene pathways can improve the specificity of disease gene identification3,31,32. In particular, at most GWAS signals, neither V2G nor G2P information alone was sufficient to identify likely disease genes: 119 GWAS signals had 2 or more genes with a V2G link (up to 5), and 195 GWAS signals had 2 or more genes with a G2P link (up to 25), including links to all 50 programs. These observations are consistent with the expectation that noncoding variants often regulate multiple nearby genes9,33, and that, by chance, a given GWAS signal might have several nearby genes involved in various cellular pathways. Combining these two layers of information in the V2G2P enrichment test provided far more specificity: for the 43 signals with V2G2P links to these programs, only 6 had more than 1 linked gene (up to 2, Extended Data Fig. 6h). We performed other internal benchmarking studies to confirm the value of each component of the V2G2P approach (including cell type-specific versus cell-type-nonspecific ABC data, and Perturb-seq versus scRNAseq without perturbations, Supplementary Note 3).
In summary, we show that V2G2P identifies known CAD genes more accurately than other cell-type specific gene prioritization studies, identifies 17 new genes not nominated by any prior study of CAD loci, and requires both cell-type specific variant-to-gene data and systematic Perturb-seq data for its ability to identify disease-relevant programs and genes.
Linking the CCM pathway to CAD risk
A key feature of the V2G2P approach is that it provides mechanistic hypotheses linking variants to genes to pathways at all prioritized loci, and thereby can accelerate further functional studies to understand the molecular mechanisms that drive effects on disease risk. We used this information to propose potential mechanisms for previously uncharacterized GWAS loci (see Supplementary Note 4), and explored in detail 2 specific genes—CCM2 and TLNRD1—that were the strongest regulators of the 5 CAD-associated programs (Fig. 2c). These investigations revealed that both CCM2 (a known member of the cerebral cavernous malformations (CCM) complex) and TLNRD1 (a previously poorly studied gene with no known function in ECs) act together in the CCM signaling pathway to regulate many other CAD genes.
We first examined CCM2, which was prioritized in our V2G2P analysis because it harbors a missense coding variant associated with a decreased risk of CAD10,12 (rs2107732, V74I; odds ratio: 0.92, P = 1.53 x 10−8), and because its knockdown in Perturb-seq significantly regulated 4 of the 5 CAD-associated programs (Fig. 2c, Extended Data Fig. 8a-b).
CCM2 encodes one of three known components of the cerebral cavernous malformations (CCM) complex. Rare loss-of-function mutations in CCM complex proteins are known to cause rare monogenic vascular malformations via effects on microvascular ECs including activation of MEKK3/MEK5/ERK5 signaling to KLF2/434,35. However, no mechanistic link between CCM signaling and genetic risk for CAD has been previously described.
Notably, examination of the Perturb-seq data revealed that CCM2 and other known members of the CCM signaling pathway regulate the CAD-associated programs in a consistent pattern (Fig. 3a, b). Knockdown of CCM2, another member of the CCM complex (KRIT1), and other genes in the pathway that act upstream of the CCM complex (CDH5, ITGB1BP1, CTNNA1, HEG1) showed directionally concordant effects on the V2G2P programs (upregulating programs 8 & 48 and downregulating programs 35, 39 & 47, Fig. 3a, b, Extended Data Fig. 8c). Knockdown of downstream genes known to be repressed by the CCM complex — including MEK5 (MAP2K5), ERK5 (MAPK7), and KLF4 — affected the expression of the 5 CAD-associated programs in the opposite direction (Fig. 3a, b).
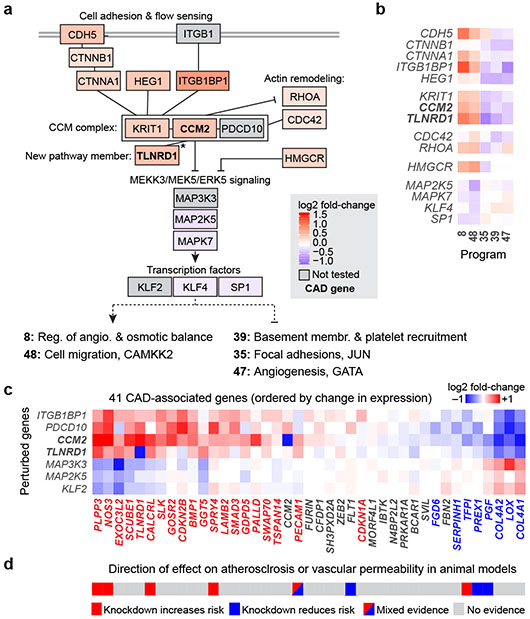
Fig. 3. Regulatory connections among CAD genes in the CCM pathway.
a. Genes that are members of the CCM complex and pathways regulate V2G2P programs for CAD. Color scale: average log2 fold-change of effect of the perturbed gene on the 5 programs, with red shading indicating knockdown leads to increased expression of Programs 8 and 48 and reduced expression of Programs 35, 39, and 47. Solid black lines indicate previously known physical or functional interactions (see Methods). *: TLNRD1 is newly linked to the CCM complex via our analysis (see next section). Gray boxes indicate functionally related genes that were not tested in the Perturb-seq experiment. Bold text: V2G2P genes for CAD.
b. Effects of genes in panel (a) on the 5 V2G2P programs. Color scale: log2 fold-change on program expression.
c. Effects of perturbing CCM pathway members on expression of the 41 V2G2P genes for CAD. Color scale: log2 fold-change on gene expression in individual knockdown experiments assayed by bulk RNA-seq (average for two guides to each target). Bold row names: V2G2P genes. Colored text in columns: Genes significantly regulated by one or more CCM pathway perturbation (FDR < 0.05), red: upregulated by upstream signaling gene perturbations or downregulated by downstream gene perturbations, blue: vice versa.
d. Likely direction of effect of V2G2P genes on atherosclerosis or vascular barrier dysfunction based on prior genetic studies in mouse models (see Supplementary Table 15 for citations).
To validate observations from the Perturb-seq screen and further characterize the role of the CCM signaling pathway on gene expression, we individually knocked down CCM2 and 5 other genes in the CCM pathway (ITGB1BP1, CCM2, PDCD10, MAP3K3, MAP2K5, and KLF2) and measured effects using bulk RNA-seq. 28 of the 41 V2G2P genes were significantly differentially expressed upon CCM pathway perturbation (FDR < 0.05, Fig. 3c, Extended Data Fig. 8d, Supplementary Tables 15, 18).
Interestingly, the directionality of the effects on downstream CAD V2G2P genes indicated that inhibition of CCM signaling likely has a protective effect on CAD — opposite of its direction of effect on risk for monogenic CCM disease36. In particular, 8 of the V2G2P genes that are regulated by the CCM pathway have previously been studied in mice, and show effects on atherosclerosis and or vascular permeability in ways that are consistent with functions in ECs (Supplementary Table 15). The direction of effect on disease phenotypes and response to CCM2 knockdown were similar (Fig. 3d): of the 5 genes previously shown to maintain vascular barrier function or be protective for atherosclerosis, 4 (NOS3, PLPP3, CALCRL, and SPRY4) were up-regulated in response to CCM2 knockdown, whereas both of the genes previously shown to promote atherosclerosis or vascular dysfunction (PGF and PREX1) were down-regulated. One additional gene (PECAM1) has been observed to have mixed directions of effect on disease depending on the genetic model (Fig. 3d, Supplementary Table 15). Thus, down-regulation of the CCM complex leads to changes in gene expression that may be protective for CAD.
Together, these data show that many of the V2G2P CAD genes can be placed in a transcriptional pathway downstream of CCM2, implicating the CCM complex in genetic risk for CAD beyond its known role in rare monogenic CCM disease.
Variant to gene to programs for TLNRD1
We next examined TLNRD1, the V2G2P gene with the strongest combined effect on the 5 V2G2P programs (Fig. 4a, Fig. 2c, Extended Data Fig. 8e-h). TLNRD1 (talin rod domain containing 1) is a poorly studied gene that has previously been found to interact with F-actin37 and to affect cell migration in a cancer cell line38, but has not been linked to CAD or any function in ECs. Surprisingly, the transcriptional effect of knocking down TLNRD1 was almost identical to that of knocking down CCM2 (Fig. 4b). TLNRD1 regulated the 5 CAD V2G2P programs in the same direction as CCM2 and other upstream CCM signaling components (Fig. 3a,b), and had a similar effect on the expression of the 41 V2G2P genes (Fig. 3c). These observations suggested that TLNRD1 could be a novel regulator in the CCM signaling pathway and that its down-regulation should be protective for CAD.
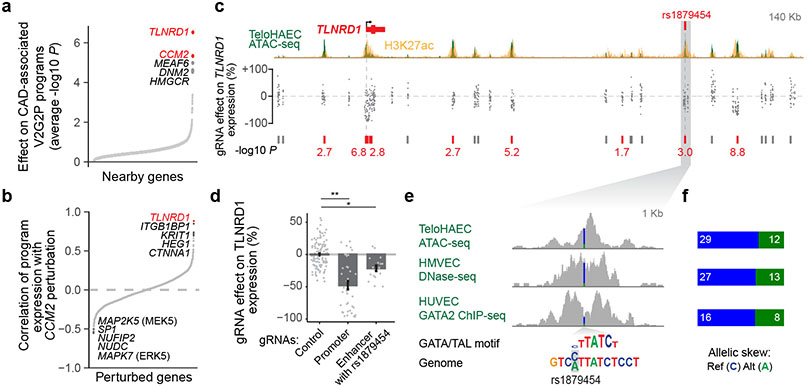
Fig. 4. Linking CAD risk variants at 15q25.1 to TLNRD1.
a. 1,503 perturbed nearby genes to CAD GWAS loci, ordered by effect on the 5 V2G2P programs for CAD (average −log10 p-value, two-sided statistical test from MAST39). Labels: top 5 genes. Red: V2G2P genes.
b. 2,284 perturbed genes ordered by their similarity with CCM2 perturbation (correlation in log2 effects on Program expression). Labels: as in (a).
c. CRISPRi-FlowFISH targeting chromatin accessible elements around TLNRD1. Each point represents the average effect on TLNRD1 gene expression of a single gRNA across 4 replicate FlowFISH experiments. Bars: elements in which CRISPRi leads to either no significant change (gray) or a significant decrease (red) in expression. Red numbers: −log10 FDR (Heteroscedastic two-sided t-test).
d. FlowFISH quantitation for guides targeting the indicated elements. Bar and whiskers: mean ± SEM. Dots: average effects, across 4 replicates, of individual gRNAs (117 negative controls (Control), 37 targeting the promoter of TLNRD1, and 17 targeting the enhancer containing rs1879454). *:FDR 2e-7. **:FDR 0.001.
e. Zoom-in on the enhancer containing rs1879454. Colored bar in signal tracks indicates read coverage of the reference (C, blue) and alternate (A, green) alleles. Bottom shows the position-weight matrix for a composite GATA/TAL motif and the genome sequence with reference and alternate alleles highlighted in gray.
f. Quantitation for allele-specific counts at rs1879454, from (e). Reads were re-aligned to both reference and alternate alleles to avoid bias toward the reference allele (see Methods).
We experimentally validated the predicted variant-to-gene link for TLNRD1 by testing whether the protective CAD allele would down-regulate TLNRD1 expression in ECs. TLNRD1 is located in the 15q25.1 CAD risk locus (lead variant P=2.63 x 10−10; rank: 159 of 241), where our V2G2P analysis identified a noncoding variant in a predicted EC-specific enhancer (Extended Data Fig. 8e, rs1879454; hg19 chr15:81377717: C (major, risk allele) → A (minor, protective allele); MAF = 0.16). We used CRISPRi-FlowFISH22 to perturb this and other enhancers near TLNRD1, and found that the chromatin accessible element containing rs1879454 indeed regulated TLNRD1 expression (estimated −21% effect, FDR corrected two-sided Student’s t-test, P=0.001, Fig. 4c,d, Extended Data Fig. 8i-k). rs1879454 also appeared to affect the regulatory activity of this element. The protective A allele was predicted to disrupt a GATA motif, and, in cells heterozygous for this variant, the A allele was associated with a 2-fold decrease in allele-specific GATA2 ChIP-seq signal in human umbilical vein ECs (HUVEC, binomial P = 0.0758), a 2.4-fold decrease in allele-specific ATAC-seq signal in teloHAEC (P = 0.0058) and a 1.9-fold decrease in allele-specific DNAse-seq signal in human microvascular ECs (HMVEC, P = 0.0192) (Figs. 4e,f).
These data show that the effects of TLNRD1 knockdown are very similar to those of CCM2 knockdown, and link a protective noncoding CAD allele to decreased TLNRD1 expression in ECs, suggesting that these genes function together and that a decrease of either one may protect against CAD.
TLNRD1 interacts with the CCM complex
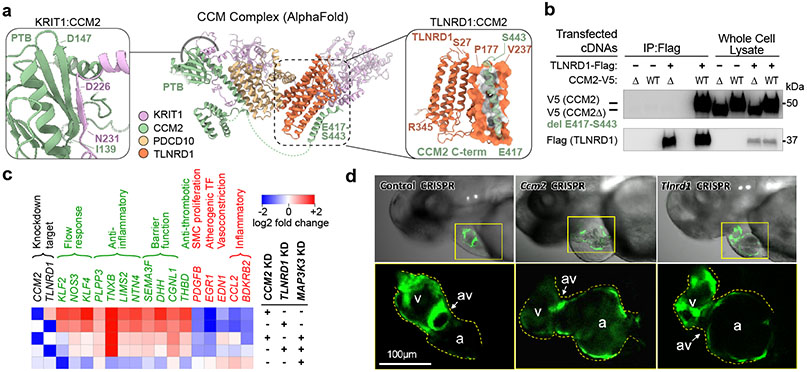
Given the strong similarity in the transcriptional effects of TLNRD1 and CCM2, we sought to gain further insight into the molecular role of this previously poorly characterized gene in the CCM signaling pathway.
We first considered whether the two proteins might physically interact. CCM2 is known to physically interact with other proteins in the CCM complex and downstream pathways36, and a recent genome-wide yeast-2-hybrid screen provided preliminary evidence of a direct interaction between CCM2 and TLNRD140. We used AlphaFold2.3 Multimer to model potential interactions between the three core CCM proteins and TLNRD1, and found that TLNRD1 was predicted to directly bind the C-terminal helix of CCM2 (C-helix, residues 417-443, Fig. 5a, right inset), as part of a consistent high confidence arrangement that also recapitulated the known CCM2/KRIT1 binding site in the PDB domain of CCM241 (Fig. 5a, left inset), as well as published interactions with PDCD1042 (Extended Data Fig. 9a-c). The predicted CCM2/TLNRD1 interaction depends on the C-helix of CCM2, which binds the TLNRD1 nine-helix bundle (Fig. 5a). We tested the TLNRD1-CCM2 interaction in human cells, and found that TLNRD1 immunoprecipitated with CCM2 pulldown, and vice versa, and that this interaction was lost upon deletion of the C-helix of CCM2 (Fig. 5b & Extended Data Fig. 9d-f, Supplementary Fig. 1).
Fig. 5. TLNRD1 interacts with CCM2 and phenocopies CCM2 in ECs and zebrafish.
a. AlphaFold2.3 Multimer model for TLNRD1, CCM2, PDCD10 and KRIT1. Right: predicted interaction between TLNRD1 (residues P177-V237) and the C-terminal helix of CCM2 (residues E417-S443). Left: Recapitulation of the known CCM2/KRIT1 binding site in the PTB (phosphotyrosine binding) domain of CCM2 with KRIT1 residues D225-N33141. Dashed green lines: flexible loops. Amino acid positions: boundaries of predicted alpha-helix and beta-sheet features.
b. FLAG-tagged TLNRD1 and/or V5-tagged CCM2 full length (“WT”) or C-terminal truncation (“Δ”) were expressed in HEK293T cells, as indicated. Extracts were co-immunoprecipitated with mouse anti-FLAG and blotted with rabbit anti-V5 (top) or anti-TLNRD1 (bottom). For gel source data, see Supplementary Figure 1a. Similar results were seen in 2 separate experiments.
c. Heatmap of top genes regulated by both TLNRD1 and CCM2 that affect CAD-relevant EC functions (see Methods), in cells with the indicated knockdowns. Black text: the TLNRD1 and CCM2 knockdown targets. Green text: likely atheroprotective genes. Red text: likely atherogenic genes.
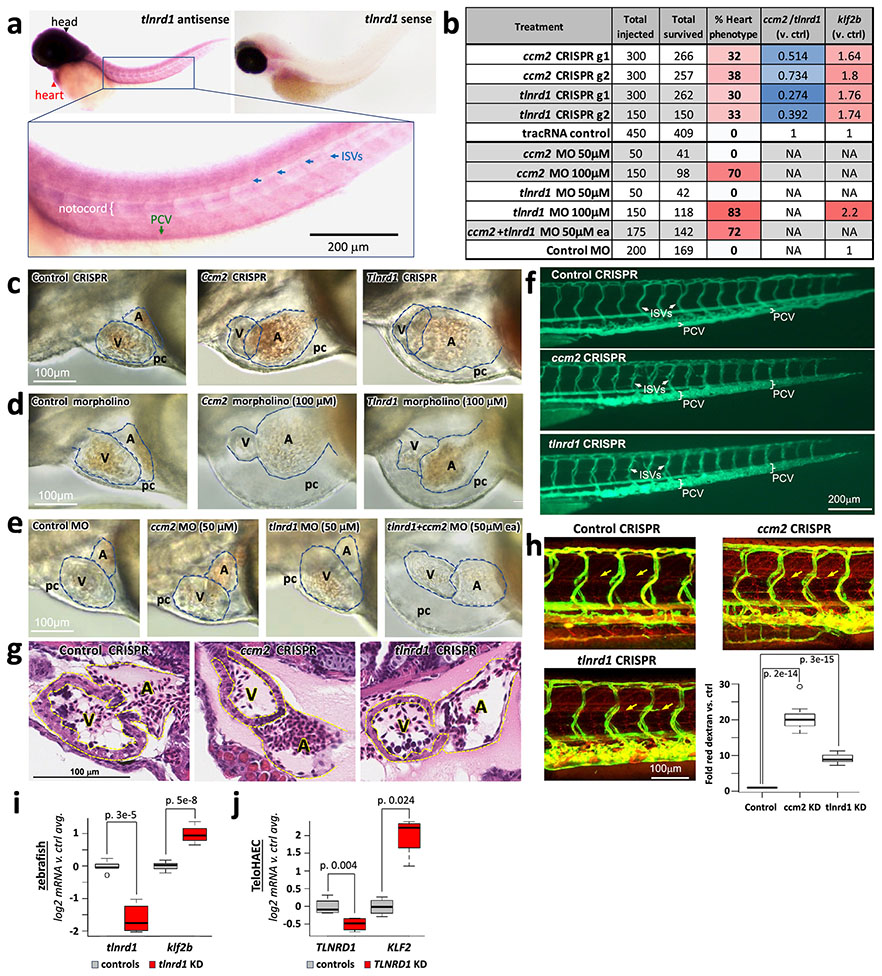
d. ccm2 and tlnrd1 knockdowns induce atrial enlargement and atrioventricular valve (AV) dilation in zebrafish embryos. Top: Representative merged light microscopic and fluorescent (cardiac myosin light chain 2/cmlc2-GFP in cardiomyocytes) confocal microscopic images of 50 hour post-fertilization zebrafish embryos (anterior to the left). Bottom: 3x zoomed-in fluorescent-only image of the heart (yellow boxes, above). N=5 embryos were analyzed per group. a: atrium, v: ventricle, av: atrioventricular valve.
A key molecular function of the CCM complex is to repress downstream signaling through MAP3K3/MEKK334,36,43 (Fig. 3a). To test if the transcriptional effects of TLNRD1 knockdown were also related to MAP3K3 signaling, we knocked down these genes alone or in combination. Individual knockdown of TLNRD1 or CCM2 upregulated KLF2, KLF4, NOS3 and other likely atheroprotective genes and downregulated likely atherogenic genes, MAP3K3 knockdown had the opposite effect, and double knockdown of MAP3K3 and TLNRD1 or MAP3K3 and CCM2 partially rescued the transcriptional effect of each individual knockdown (Fig. 5c; Extended Data Fig. 10a-d, Supplementary Table 19).
To determine if the relationship between TLNRD1 and CCM2 might extend beyond human ECs in vitro, we tested tlnrd1 function in zebrafish—a model system in which ccm2 has been shown to have characteristic effects in heart and vascular development34,44,45. We targeted tlnrd1 or ccm2 with CRISPR and found highly similar effects on cardiac and vascular development, including atrial chamber enlargement, pericardial edema, atrioventricular valve defects, and thin ventricular walls (Fig. 5d, Extended Data Fig. 11b,c,g, Supplementary Table 20). Both tlnrd1 and ccm2 CRISPR embryos also had vascular defects, including posterior cardinal vein (PCV) dilation and increased vascular permeability to red dextran particles (Extended Data Fig. 11f,h). Tlnrd1 was expressed in the heart and vasculature (Extended Data Fig. 11a), and tlnrd1 knockdown led to increased klf2b expression, similar to the effect of human TLNRD1 knockdown on KLF2 expression in teloHAECs (Extended Data Fig. 11i,j). Finally, whereas a 100 μM dose of tlnrd1 or ccm2 morpholino had similar effects as CRISPR perturbations, and a 50 μM dose of either morpholino had no effect, 50 μM of both morpholinos had similar effects as 100 μM of either morpholino alone (Extended Data Fig. 11b,d,e), consistent with both proteins functioning in the same pathway.
Together, these data indicate that TLNRD1 is a previously unrecognized, evolutionarily conserved member of the CCM signaling pathway, and provide an example of molecular convergence in which V2G2P analysis identifies two novel CAD genes that not only regulate the same transcriptional pathway but also physically interact.
CAD-relevant phenotypes of CCM2 & TLNRD1
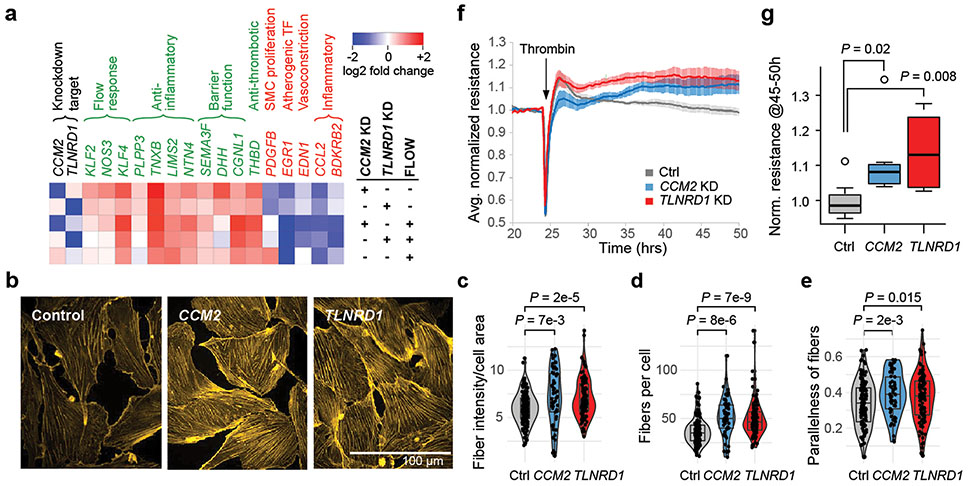
We further experimentally characterized how TLNRD1 and CCM2 might affect EC phenotypes relevant to CAD. Because atherosclerosis predominantly develops in regions of disrupted or turbulent blood flow13, we tested how the effects of TLNRD1 or CCM2 knockdowns compared to the effects of laminar flow. First, we noted that all of the genes most strongly upregulated by both TLNRD1 and CCM2 knockdowns, and whose effects on relevant EC functions have previously been assessed, were likely atheroprotective (including NOS3, damaging mutations in which have recently been shown to be a non-lipid driver of CAD 46), while downregulated genes were likely atherogenic (Fig. 6a, top 2 rows, Supplementary Table 29), suggesting molecular mechanisms by which reduction of TLNRD1 and CCM2 could decrease CAD risk. Strikingly, the transcriptional effects of TLNRD1 or CCM2 knockdown in static culture were similar to the effects of laminar shear stress (“flow”, 12 dynes/cm2) on control cells (Pearson R = 0.40, P = 1.5e-54 for CCM2; R = 0.52, P = 1.6e-94 for TLNRD1; Fig. 6a, Extended Data Fig. 10e,f, Supplementary Table 19).
Figure 6. CCM2 and TLNRD1 knockdowns mimic the atheroprotective effects of laminar flow in ECs.
a. Heatmap of genes strongly regulated by both TLNRD1 and CCM2 that affect CAD-relevant endothelial cell functions (as per Fig. 5c), in CRISPRi TeloHAEC with the indicated treatments vs. control cells in static culture.
b. Representative maximum projection images of phalloidin-stained CRISPRi teloHAEC with control, TLNRD1 or CCM2 guides.
c. Quantitation of actin fiber (phalloidin stain) intensity per cell area (see Methods). N: Control=145, CCM2=47, TLNRD1=117. Boxplot: center line, median; box limits, upper and lower quartiles. Significance was assessed by two-sided T-test.
d. As in (c), but showing the number of actin fibers per cell.
e. As in (c), but showing parallelness of actin fibers. A score of 0 indicates randomly oriented fibers, and a score of 1 indicates all fibers in a cell are parallel to each other.
f. Trans-endothelial electrical resistance (TEER) measurements for CRISPRi teloHAEC with the indicated guides (2 guides per target), each normalized to average resistance over the 4 hours before thrombin was added to disrupt cell junctions. N=8 (control), 7 (CCM2 KD) and 6 (TLNRD1 KD). Ranges: SEM.
g. Boxplot of normalized TEER signal, from (f), averaged for hours 45 to 50 (20-25 hrs post-thrombin). Quantitation as in (c). In addition, boxplot whiskers=1.5x interquartile range and points=outliers. Note that the CCM2 KD effect we see differs from prior studies of human dermal microvascular ECs48,49, which showed decreased resistance with CCM2 perturbation. This could indicate a difference between ECs from arteries (where atherosclerosis develops) vs. capillaries.
Consistent with the similar transcriptional effects of flow and either TLNRD1 or CCM2 knockdown, we found that CCM2 or TLNRD1 knockdown in static culture increased the number and parallelness of actin stress fibers (Fig. 6b-e), a characteristic of flow response in unperturbed ECs13, consistent with prior studies of CCM2 knockdown in HUVEC47. On the other hand, TLNRD1 or CCM2 knockdown cells showed reduced alignment to flow, relative to control cells (Extended Data Fig. 10i-k), and a weaker transcriptional response to flow, perhaps because the flow-response program was already partly active (Extended Data Fig. 10g,h). Endothelial dysfunction that is thought to contribute to CAD is also characterized by decreased barrier function (a leaky endothelium that allows inflammatory cells into the arterial wall13). We found that CRISPRi knockdown of either TLNRD1 or CCM2 in teloHAEC reproducibly increased EC barrier function, as measured by trans-endothelial electrical resistance (Fig. 6f,g).
Together, these observations demonstrate that TLNRD1 and CCM2 similarly regulate arterial EC phenotypes relevant to CAD. They indicate that down-regulation of TLNRD1 or CCM2 by common variants may be atheroprotective by conferring a “flow-like” response and improving barrier function in ECs not exposed to laminar flow, which are most prone to atherogenesis13.
V2G2P generalizes to other traits
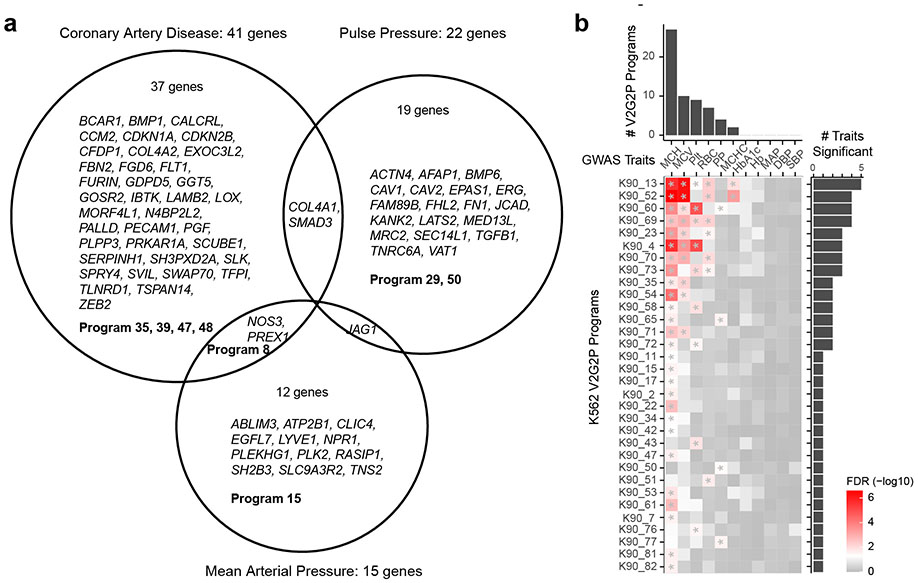
To test whether the V2G2P approach would generalize to other complex traits, we applied our ABC maps and Perturb-seq data from ECs to study two other vascular traits: mean arterial blood pressure (MAP) and pulse pressure (PP) (see Methods). The V2G2P enrichment test identified programs significantly associated with each of these two traits, which were distinct from those we identified for CAD (Extended Data Fig. 12a, Supplementary Table 21). For example, for pulse pressure, the V2G2P test identified significant enrichment for Program 50 (TGFβ response, FDR = 0.0046) and Program 29 (EDN1, wound healing, FDR = 0.0316), and identified genes known to regulate vascular tone and stiffness such as FHL2, SMAD3, and TGFB1 (Extended Data Fig. 12a). These observations confirm that V2G2P does not simply identify generic EC programs, but rather identifies different cell-type specific programs relevant to each vascular trait.
To test whether our approach is generalizable to other cell cell types, we applied V2G2P to study 7 traits related to red blood cells using ABC maps9 and a recent genome-scale Perturb-seq dataset from K562 erythroleukemia cells19 (Extended Data Fig. 12b, Supplementary Table 22). We again found that different traits showed significant enrichment for different, relevant programs. For example, genes linked to variants associated with mean corpuscular hemoglobin were most significantly enriched in K562 Program 13, which included many hemoglobin genes and known regulators (including GFI1B and CBFA2T3), while variants associated with platelet count showed most significant enrichment in K562 Program 4, whose program genes showed high promoter enrichment of motifs for the known megakaryocyte regulators SP1/3, and included genes known to be involved in megakaryocyte differentiation and platelet count such as VASP and TPM4 (Extended Data Fig. 12b, Supplementary Table 22).
In summary, we find that variants associated with different traits map onto different programs corresponding to relevant biological pathways, even within a single cell type, and that the V2G2P approach can be successfully applied to a second cellular model. Thus, the V2G2P analysis pipeline is likely to be generally applicable to provide insights for complex traits and cell types beyond CAD.
Discussion
GWAS have identified hundreds of loci for many common complex diseases such as CAD. An emerging paradigm for understanding their function is that risk variants should map onto genes that act together in biological pathways1-6. Yet, such pathway-level convergence has been difficult to identify for many diseases, because existing approaches can be limited to studying one gene or pathway at a time, underpowered, and/or biased toward rediscovering known genes and pathways.
Our study introduces a novel method to address this challenge, in which we build unbiased maps of genome function using epigenomic data and Perturb-seq, and then combine these maps to identify convergence of risk variants onto pathways. The method is systematic in that it measures the full transcriptomic effects of all genes in all relevant GWAS loci, facilitating the discovery of novel disease-associated pathways and new functions for uncharacterized genes (Supplementary Note 5). Applying this method to ECs revealed that 43 of 306 CAD GWAS signals indeed converge onto 5 transcriptional programs, all related to CCM signaling. We find that two newly prioritized CAD genes, CCM2 and TLNRD1, strongly regulate these programs (which include dozens of other CAD genes), show highly similar transcriptional and cellular phenotypes, and physically interact with one another. This strong signature of molecular convergence identifies the poorly-characterized gene TLNRD1 as a new member of the CCM signaling pathway, implicates the CCM pathway in genetic risk for CAD, and demonstrates that Perturb-seq indeed identifies transcriptional programs that help to interpret disease risk variants.
Our data suggest a model where protective alleles affecting CCM2 and TLNRD1 down-regulate activity of the CCM complex in ECs, increasing the expression of atheroprotective genes (including two V2G2P-prioritized gold standard genes, NOS3 and PLPP350,51) and downregulating atherogenic genes (Fig. 3, Fig. 5c, Fig. 6a, Supplementary Table 15). Importantly, downregulation of CCM2 or TLNRD1 in cells in static culture mimics the effect of laminar flow, promoting atheroprotective gene regulation, actin stress fiber formation and endothelial barrier function (Fig. 6). Given that atherosclerosis in vivo predominantly develops in areas of disrupted or turbulent blood flow13, these data suggest that reduction of CCM2 or TLNRD1 function might confer a resistant phenotype on ECs precisely in those regions of disrupted flow that are most prone to atherogenesis.
The convergence of 43 CAD risk loci onto CCM signaling related to flow responses in ECs (a number of loci comparable to the 78 CAD loci that are associated by GWAS with circulating lipid levels) suggests that this is a key mechanism controlling risk for CAD in the human population. We anticipate that future application of the V2G2P approach in other atherosclerosis-relevant cell types, including smooth muscle cells and monocytes, will be a powerful approach to provide a more comprehensive analysis of genetic risk for CAD.
Interestingly, the novel link we find between CCM signaling and common, polygenic coronary artery disease is in the opposite direction of its known role in rare, monogenic CCM disease: whereas complete loss of function of CCM complex proteins causes cavernous malformations in the brain and spinal cord, our results indicate that common variant alleles that quantitatively down-regulate CCM complex function in arterial ECs reduce CAD risk. Further studies in vivo will be required to understand how CCM signaling could have such different effects in these two diseases. Additionally, our finding that TLNRD1 is a novel regulator of CCM signaling suggests that future studies could also evaluate mutations in TLNRD1 as an alternative possible cause of disease in the 47% of sporadic CCM cases with multiple lesions that lack pathogenic mutations in the three previously known members of the CCM complex52.
In summary, our approach establishes a new, generalizable path to systematically link risk variants to disease genes and to convergent transcriptional programs, providing a rich foundation for further studies to dissect novel disease mechanisms. By applying Perturb-seq and the V2G2P approach across many cell types and states relevant to various complex diseases, it should be possible to nominate causal disease genes for a large fraction of GWAS loci and map how they converge onto particular cellular pathways. Such a project is becoming increasingly feasible, and would provide a foundation for systematic efforts to leverage human genetic data to discover disease mechanisms.
Methods
Cell culture & creation of CRISPRi TeloHAEC
Telomerase-immortalized human aortic endothelial cells (TeloHAEC) were purchased from ATCC (CRL-4052), and grown in Lifeline VEGF endothelial cell media (LL-0005) with 1x Penn/Strep. Cells were plated at a density of 0.5-1.0 x 106 cells per 10 cm plate and split before reaching 4 x 106/plate (3 to 4 days). To create the TeloHAEC CRISPRi line, cells were transduced with lentiviral vectors containing 1) dox-inducible (tetracycline operator controlled) dCas9-KRAB-BFP (CRISPRi machinery, which targets epigenetic repressors to efficiently silence enhancers or promoters53-55, Addgene #85449) and 2) rtTA (tetracycline activator) with a hygromycin marker (Addgene #66810). After hygromycin selection (250 μg/ml for 4 days), cells were treated with 1 μg/ml doxycycline (dox, a stable tetracycline analogue) for 3 days before FACS sorting for the top 15% of BFP positive cells, and after a period in culture without dox, treated again with dox and re-sorted (Extended Data Fig. 1e). Diagnostic FACS performed immediately before the Perturb-seq screen showed no leaky BFP expression in the absence of dox, and 93% BFP positive cells in the presence of dox (Extended Data Fig. 1f). CRISPRi TeloHAEC were passaged for routine maintenance in the absence of dox. Eahy926 cells (a HUVEC + A549 hybrid line) and HEK293T cells were purchased from ATCC (CRL-2922 and CRL-3216, respectively), and grown in DMEM + 10% FBS. To study responses to CAD-associated cytokines, cells were untreated (control), or treated with 10 ng/ml recombinant human IL-1β (Millipore IL038), 10 ng/ml recombinant TNFα (Millipore GF023), or with normal media lacking VEGF (for TeloHAEC) or supplemented with VEGF (1x concentration from LifeLine VEGF media, for Eahy926), for 24 hours. All cell lines were mycoplasma-free. In addition to authentication by the provider, we further authenticated each line by analysis of microscopic morphology (e.g. TeloHAEC displayed the characteristic EC cobblestone morphology and showed localization of VE-Cadherin to endothelial cell junctions), functionality (high transfectability and protein expression for HEK293T), mapping of ATAC-seq, ChIP-seq and RNAseq reads to the human genome, and RNAseq profiles and responses (e.g., for Eahy926 & TeloHAEC, expression of EC-specific genes and observation of previously-observed responses to stimuli, such as IL-1β).
Bulk RNA-seq
Total RNA was harvested from TeloHAEC (parental or CRISPRi lines) by Qiagen RNeasy kit (74016, Qiagen), DNAse treated (TURBO DNAse, InVitrogen AM2238, 15’ 37°C), and purified on MyOne Silane beads. For flow response and MAP3K3 knockdown studies, DNAse treatment was performed on the spin column between two buffer RW1 washes, for 20 mins at room temperature with Purelink DNAse (InVitrogen 12185010), 10 μl in 80 μl of 1x buffer. mRNA was purified from 400ng to 1 μg of total RNA using the NEBNext Poly(A) mRNA Magnetic Isolation module (NEB), processed for RNA-seq library generation using the NEBNext Ultra II RNA Library Kit for Illumina (NEB), and sequenced to a depth of 10 to 30 million reads/library. Reads were mapped to the human hg19 genome build, and counts per gene tables assembled as per 9,22, or, for flow response and MAP3K3 knockdown studies, using kallisto 56. Differential expression calls were made using Limma VOOM57 (for parental TeloHAEC & Eahy926) or edgeR58 (for all other libraries). Bulk RNA-seq data is available from the Gene Expression Omnibus (GEO). For cytokine treatment of parental lines and single guide knockdowns use accession GSE210522. For flow response and MAP3K3 double knockdown studies use accession GSE232400.
ATAC-seq, H3K27ac ChIP-seq & identification of TeloHAEC enhancers
For ATAC-seq, one well of a 12-well plate (~200,000 cells) was directly lysed using a custom TN5 buffer (33 mM Tris Acetate pH 7.8, 66 mM Potassium Acetate, 10 mM Magnesium Acetate, 16% dimethylformamide & 0.1% NP40). 47.5 μl of lysed cells was added to 2.5 μl Tn5 tagmentation enzyme (Illumina) & incubated at 37°C for 1 hr, and the reaction stopped by addition of 20 μl buffer RLT (Qiagen). Products were purified by addition of 1.8 volumes Ampure XP beads (Beckman-Coulter) & magnetic separation of beads, followed by two 80% ethanol washes, brief drying of pellets & resuspension in 23 μl water. Barcoded ATAC-seq libraries were then generated as described in 9,22, and sequenced to a depth of 10-20 million reads per library. Chromatin immunoprecipitation for histone H3 lysine 27 acetylation (H3K27ac) was performed as described in 9,22, using anti H3K27ac antibody (#39685, Active Motif) at 1:200 dilution. ChIP-seq libraries were prepared using the KAPA Hyper Prep Kit (KAPA Biosystems). ATAC-seq libraries were prepared in biological triplicate, and ChIP-seq libraries in biological duplicate. For both types of libraries, reads were mapped to the human genome (hg19 build) using Bowtie2, and peaks identified using MACS2, essentially as per 9,22. Raw and processed data are available on GEO: GSE210489 (ATAC-seq) and GSE210491 (ChIP-seq). Enhancers and their predicted target genes were identified by applying the Activity-by-Contact (ABC) model to these data, using ATAC-seq and H3K27ac ChIP-seq as the measures of enhancer Activity, and using a cross-cell type average of Hi-C maps as the measure of 3D enhancer-promoter contact frequency (https://github.com/broadinstitute/ABC-Enhancer-Gene-Prediction 7,8). We used an ABC fractional score threshold of ≥0.0159.
Selection of genes for the Perturb-seq library
We constructed a library of promoter-targeted CRISPRi guides to all potential causal CAD genes (Fig. 1b). First, we identified all coding genes within a 1 megabase window surrounding the lead SNPs from CAD loci identified in either or both of van der Harst et al.10 and Aragam et al.12 that were expressed in TeloHAEC (1+ TPM, from bulk RNA-seq). If fewer than 2 expressed genes were found within 500kb up- or downstream of the lead SNP, the window was expanded to include the closest 2 genes to each side (for a total of 1661 genes). Non-coding genes were generally excluded, unless there was strong evidence for regulatory functions, particularly in ECs. Selected genes with TPM <1 were included, particularly if they were known to be important for CAD in tissues where they were more highly expressed (e.g. PCSK9), or were regulated by IL1-beta in bulk RNA-seq data in TeloHAEC (FDR<0.05, fold change >1.3). As negative controls, we included guides targeting 48 coding genes expressed in other cell types but not detectably expressed in ECs, and the 132 expressed coding genes within 1 Mb of 16 randomly-selected lead SNPs associated with Inflammatory bowel disease, Crohn’s disease or Ulcerative colitis 59, and which did not overlap with CAD loci. As positive controls, and to aid in connecting candidate CAD genes to known pathways in ECs, we targeted the promoters of an additional 284 genes with known roles in a wide range of CAD-relevant EC functions such as barrier formation, TGF-beta signaling and inflammation, as well as major classes of expressed transcription factors and common essential genes. We also targeted an additional 160 promoters of expressed genes predicted to be regulated by EC enhancers containing fine-mapped variants associated with other disease phenotypes expected to be modulated by ECs (migraine, blood clotting in leg, systolic blood pressure, diastolic blood pressure & mean arterial blood pressure, from UKBB, see Supplementary Table 23). This gave a total of 2285 genes, some of which were members of more than one category.
Guide library production and validation
sgRNA guides were designed to target promoters of the chosen CAD and control genes (15 guides spanning from -150 to +100 relative to the Transcription Start Site (TSS)), using our established pipeline (9,22, https://github.com/EngreitzLab/CRISPRDesigner). We included 400 non-targeting guides (that do not have close matches to any region in the human genome) and 600 safe targeting guides (targeting non-genic regions lacking enhancer marks) 53. Because TeloHAEC are puromycin resistant, we adapted the CROP-opti vector (20, Addgene, #106280) for Blasticidin resistance (“CROP-opti Blast”), by digesting the vector with BsiWI and MluI, PCR-amplifying the Blasticidin resistance gene from lenti-dCas-VP64_Blast (Addgene, #61425) with added homology arms, and performing Gibson Assembly (Gibson Master mix, New England Biolabs). To create “CROP-opti-BC-Blast”, we added HyPR-Seq barcodes between the WPRE element and the U6 promoter of CROP-opti-Blast, as described in 60. A pool of oligos encoding the guide sequences, plus extensions with homology to the U6 promoter and downstream scaffold (TATCTTGTGGAAAGGACGAAACACCG & GTTTAAGAGCTATGCTGGAAACAGCATAG) was synthesized by Agilent Technologies, and cloned into Crop-Opti-BC-Blast by Gibson assembly and bacterial electroporation as described53, at an average coverage of 202 transformants per guide. Note that, since the vector was prepared from a single clone, diversity of the HyPR-seq barcodes (which were not required for Perturb-seq) was not preserved. The library was sequenced and shown to include all 37,637 designed guides with relatively equal coverage of each (the difference in count frequency between the top and bottom 10th percentiles of guides was 2.8). A lentiviral library was produced using a standard 3-plasmid protocol53, at a scale to yield 10 ml of virus, stored in aliquots at -80°C, with each aliquot thawed only once.
Perturb-seq: Experimental procedure
To transduce this library into CRISPRi TeloHAEC, cells were resuspended in media containing 10 μg/ml polybrene at a density of 1e6 cells per ml, mixed with virus and plated 4 ml per well to 6-well plates, centrifuged at 2000 rpm for 2 hrs at 30°C, and incubated at 37°C for 2 hrs before addition of another 4 ml media without polybrene. The next day, cells were harvested and plated to 15 cm plates and treated with 15 μg/ml blasticidin for 4 days. The effective viral titre was determined using this same protocol, and a volume of virus was chosen that gave a final measured 15.7% infection rate (such that most successfully transduced cells have only 1 guideRNA). For the Perturb-seq study, 127.5 million CRISPRi TeloHAEC were transduced and selected for blasticidin resistance, for a coverage of approximately 360 cells per guide (as back-calculated from yield at the first post-blasticidin split, using the 36.7 hr doubling time observed in routine culture) to 461 cells per guide (as estimated from initial number of cells and infection rate). After blasticidin selection, cells were treated with 2 μg/ml dox for 5 days (plating 18e6 cells at each split, to maintain complexity of the library). We reasoned that, since atherosclerotic plaques develop slowly, the longer-term transcriptional effects of causal CAD gene disruption would provide the greatest insights into disease mechanisms. Thus, while we have found that knock down of guide-targeted genes is near maximal after 2 days of doxycycline treatment (inducing the CRISPRi machinery), we treated guide-containing cells with 2 μg/ml doxycycline for 5 days, to measure the longer-term consequences of each perturbation. We also used this same 5-day dox treatment protocol for downstream validation studies (e.g. bulk RNAseq of single guideRNA clones).
The presence of guideRNAs in cells allows multiplets (droplets containing 2 or more cells) to be unambiguously identified, as droplets containing more than one guide. This allowed us to load ~10-fold more cells per 10X Genomics lane than the maximum number recommended in the manufacturer’s protocol. Briefly, cells were harvested, resuspended in PBS with 1% BSA, counted, and loaded at 150,000 cells per lane on a 10X Genomics Chromium Controller using a 3’ scRNA-seq V3 kit (20 lanes, for a total of 3 million cells). Cells were isolated in two batches, with 6 lanes for the first batch, and 14 lanes, across 2 cassettes, for the 2nd batch, 6 hours later. scRNA-seq libraries were generated using the 10X Genomics protocol, and given lane-specific indexes. From the initial amplified cDNA, we used a two stage PCR protocol to generate “dialout” libraries, for each lane. Because the CROP-seq vector expresses a Pol II polyadenylated transcript that ends just downstream of the guide sequence, the dialout libraries identify the guideRNA sequences associated with each droplet20. PCR1 oligos for the guide dialout PCR were: CTACACGACGCTCTTCCGATCT & GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTGTGGAAAGGACGAAACACC, and PCR2 oligos were AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTC & CAAGCAGAAGACGGCATACGAGAT-8bp index sequence-GTGACTGGAGTTCAG.
Assignment of guideRNAs to cells
To get complete information about guide assignments, dialout libraries were sequenced to approximately 40-fold saturation. Guides were identified from read 1 sequences, using Bowtie2 to align dialout reads to a “genome” composed of all 37,637 guide sequences, requiring no-mismatches. Aligning read 1 and read 2 sequences linked gRNA sequences with cell barcodes (CBCs, unique to each bead/droplet) and unique molecular identifiers (UMIs). To avoid low-frequency PCR chimeras, we required that each CBC-UMI-guide combination be duplicated at least 4 times (Extended Data Fig. 1g). We then identified the guides associated with each CBC, and the number of different UMIs for each CBC-guide combination. We selected 4 UMIs for any single guide as the threshold to call a cell as containing a guide (Extended Data Fig. 1h). We defined singlets (one cell & one guide per CBC) as having ≥4 UMIs for the most frequent guide and ≥4x less than this for the 2nd most frequent guide (choosing these thresholds to give a good balance between power to detect transcriptional effects and accuracy in measuring the magnitude of these effects, as described under Selection of Singlet Thresholds, below). Doublets and higher multimers, were cells with ≥4 UMIs for the top guide, and one or more additional guides with more than 1/4 this number of UMIs. The counts of identified singlets, doublets and higher multiplets are shown in Extended Data Fig. 1i.
scRNA-seq data pre-processing and subsetting to singlets
scRNA-seq libraries were sequenced on two Illumina NovaSeq S4 flowcells, yielding 20,245,734,673 total reads, across all 20 libraries. The FASTQ files were processed on the 10X Cloud to run CellRanger count with the hg38 reference genome. We used the “filtered” features (i.e., cell barcodes corresponding to droplets that contain a cell), and combined the outputs from all twenty 10X lanes into a single genes x cell matrix. This analysis identified 822,156 cell-containing droplets (see Supplementary Table 7 for other CellRanger output statistics). To measure the effects of individual guides on individual cells, we selected only those CBCs identified in the dialout analysis as corresponding to singlet cells. This identified 214,449 singlets (droplets containing one cell and one guide), defined as 4+ unique molecular identifiers (UMIs) for the top guide and ≥4-fold fewer UMIs for any other guide (Extended Data Fig. 1i). This gave an average of 5.7 cells per guide and 85.5 cells per target promoter. Average sequencing depth was 10,870 transcriptome-mapped UMIs per singlet cell, and 929,000 transcript UMIs, across all 15 guides, for each target promoter. Raw and processed data, as well as supplemental files for downstream analyses, are available from GEO: GSE210681.
Estimation of fitness effects
To estimate the fitness effects of guides, we compared the relative frequency of all 15 guides to a given target in the original library to the frequency of the same guides in singlet cells, and estimated significance by Benjamini-Hochberg adjusted binomial tests. Essential genes were defined as those that scored as fitness reducing in 5 of 7 tested lines in 61.
Differential gene expression (DE) analysis & knockdown efficacy
To measure the differential effects of guides to specific target promoters on individual genes, we used edgeR58, with settings for scRNA-seq from 62, comparing all singlet cells with guides to each target to all singlet cells with any of the 1,000 non-targeting and safe targeting guides. Genes with fewer than 10 UMI counts across all singlet cells were excluded from the analysis. To control for possible batch effects, we included the 10X lane number as a covariate. For average knockdown efficacy for each perturbation (across all 15 guides), we used the log2 fold change and p-values reported by edgeR. To measure the knockdown efficacy of individual guides, we performed binomial tests on: the number of transcripts for the guide’s target in singlet cells with that guide (hits), all transcripts in singlets with that guide (tests) versus a background frequency of (transcripts to the target in other singlet cells)/(all transcripts in other singlet cells). Note that with an average of 5.7 cells per guide, assigning significance for knockdown effects of individual guides was only possible for genes with high expression in unperturbed cells (e.g., TPM>100). To identify perturbations with a significant effect on the transcriptome, we used the edgeR results for the 48 negative control promoters (for genes not detectably expressed in TeloHAEC) to estimate the number of DE genes found by chance, at thresholds of nominal p-value < 0.01 and fold change > 1.15. Perturbations with a significant effect on the transcriptome (across all 15 guides to each target) were defined as having more DE genes, by these same thresholds, than the 48 non-expressed controls (using binomial tests with a background rate equal to the average DE gene count for controls over all genes tested, and multiple hypothesis correction by the Benjamini Hochberg method).
Selection of singlet thresholds
Expression of the CROP-seq guide mRNA in TeloHAEC is lower than in some other cell lines, such as K562 & HEK293T12 resulting in the absence of a clear gap between noise (low UMI CBC-guide combinations that are likely PCR chimeras) and higher UMI-count true guide reads (Extended Data Fig. 1h). We hypothesized that reducing stringency for singlet calls could potentially reduce power to detect perturbation effects on transcription (due to increased noise from mis-calling some true doublets as singlets), or could increase power (by increasing the total number of called singlets analyzed). To test which of these was true, we measured the correlation between differential expression calls for cells with guides to a given target in the full Perturb-seq library versus a smaller pilot library tested in resting TeloHAEC, reasoning that parameters that improved the correlation between these separate studies would also increase the power of the full scale library to detect real transcriptional effects. Information about guides, as well as raw and processed data for the “200 gene” pilot library can be found on GEO, with accession number GSE212396. For the pilot library, we chose singlets with the very stringent threshold of 6 UMIs for the top guide and more than 5-fold less for the next most frequent guide (“6&<5x”). For the full Perturb-seq dataset we chose 4 UMIs for the top guide and equal to or more than 4-fold less than the next most frequent guide (“4&<=4x”, our final applied standard, yielding 214,449 singlets), or the relaxed thresholds “3&<=3x” (284,466 singlets) and “2&<=2x” (389,792 singlets). We identified 37 gene targets that were shared between libraries, and which also showed an FDR<0.1 effect on the transcriptome in the full Perturb-seq 4&<=4x dataset (measured as described above). We then ran EdgeR58,62 for differential expression testing (cells with guides to each of these 37 targets versus cells with control guides), for each library and singlet definition (pilot 6&<5x, or full library 4&<=4x, 3&<=3x, and 2&<=2x). Then, for all genes called as differentially expressed in either the pilot library or the full library (raw p-value < 0.01), we measured the correlation in log2 fold changes between the pilot & full scale data, repeating this analysis for each singlet definition.
Lastly, we measured the difference in correlation coefficients (R) between the relaxed threshold comparisons (pilot v. full library 3&<=3x, and pilot v. full library 2&<=2x) and the base comparison (pilot vs. full library 4&<=4x). We found that the median correlation between pilot & full-scale studies significantly improved with the relaxed singlet thresholds (Supplementary Fig. 2a, with significance assessed by two-sided t-test). This indicates that the increased number of called singlets with the relaxed thresholds increased the power to detect real transcriptional effects, despite an expected increase in doublets mis-assigned as singlets. Plotting change in R for each target for the 2&<=2x singlet definition ((R for pilot v. full library 2&<=2x) - (R for pilot v. full library 4&<=4x), y-axis) against the R value for the base correlation (between the pilot and the 4&<=4x full library singlet definition, x-axis), we found that in all 13 cases where R started high (>0.15, likely real correlations between strong transcriptional effects), R increased (Supplementary Fig. 2b). R also increased in all but one case where it started out negative (correcting anti-correlations likely driven by noise). Weak positive base correlations were adjusted up or down, potentially improving true correlations and correcting spurious ones. As such, relaxed singlet thresholds might improve power to detect reproducible transcriptional changes more than is indicated by simple mean differences in R values. On the other hand, we found that lower stringencies reduced the apparent knock down effect on these target genes, themselves (Supplementary Fig. 2c, median log2 fold changes: -0.53 for 4&<=4x, -0.41 for 3&<=3x and -0.42 for 2&<=2x), likely due to the fact that a mis-called singlet that was actually 2 cells with different guides would show half-magnitude transcriptional effects of each guide. Reduced singlet thresholds also decreased median log2-fold changes for target genes across all targets in the full-scale library (Supplementary Fig. 2d, -0.368 for the 4&<=4x singlet definition, and -0.327 for the 2&<=2x singlet definition). Based on these observations, we chose the thresholds of 4 UMIs for the top guide and <=¼ this for the next (4&<=4x), to provide a good balance between overall power and accurate detection of the magnitude of effects.
Data processing prior to defining gene programs
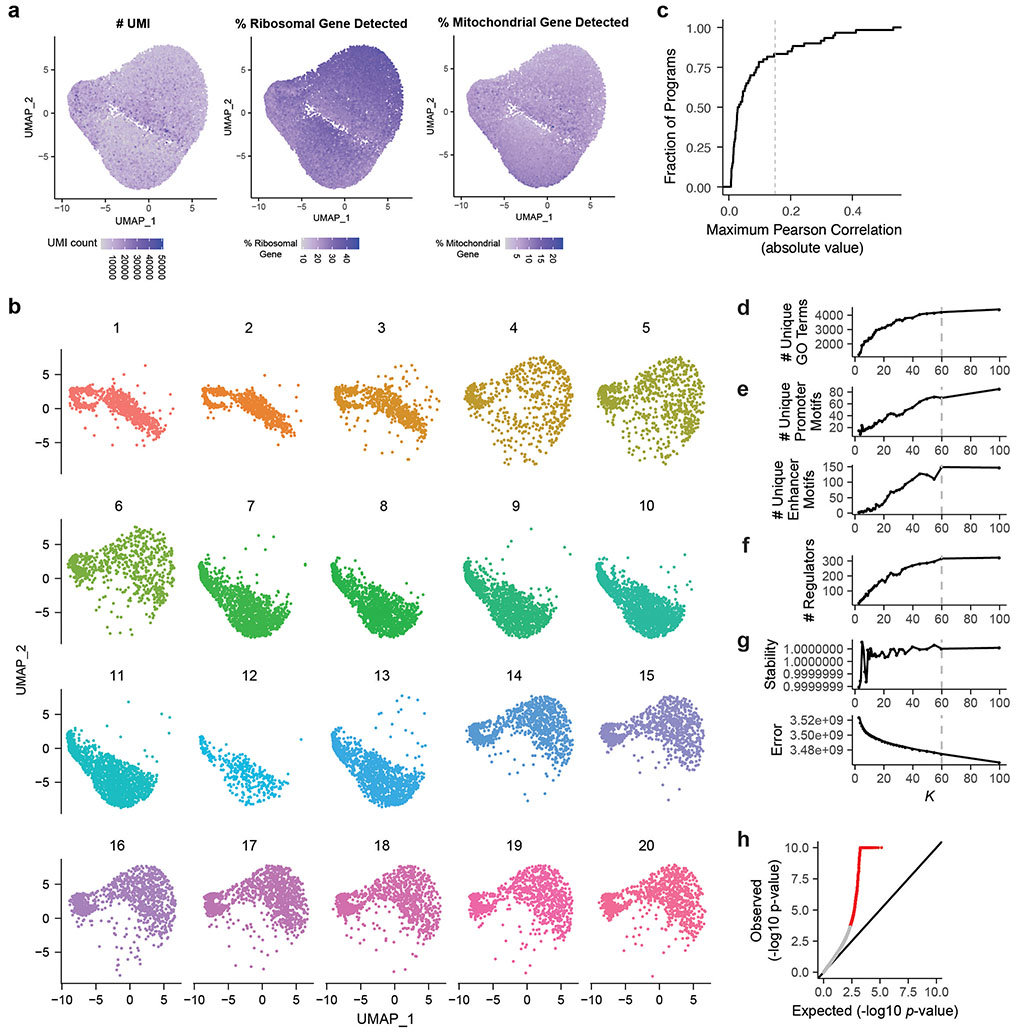
To remove noncoding RNA from the analysis, we removed genes with names starting with “LINC” and gene names with patterns starting with two letters and six digits. We retained cells with a minimum of 200 unique detected genes and a minimum of 200 UMIs. We retained genes detected in a minimum of 10 cells.
Consensus non-negative matrix factorization (cNMF)
To identify sets of genes that are co-expressed across single cells in a dataset, we used non-negative matrix factorization (NMF). NMF decomposes an input cell x gene UMI count matrix (X) into a cell x component matrix (W) and a component x gene matrix (H), such that X = W • H + E, where E is the error term. The cell x component matrix W represents the contribution of each component to the cell’s transcriptional profile, and the component x gene matrix H encodes information about gene expression programs. The number of components (K) is a hyperparameter defined prior to performing matrix factorization (see below). To account for the fact that the NMF algorithm is a stochastic algorithm that depends on the initial seed, we used the consensus NMF (cNMF) method developed by Kotliar et al25. The cNMF method, after normalizing each gene’s expression to unit standard deviation, factorizes the normalized matrix multiple times (here, 100 repeats); clusters the components from the repeat runs based on their pairwise Euclidean distances; removes the components that show low similarity to any other component (here, threshold on Euclidean distance = 0.2); defines “consensus components” as the median of each of the component clusters; and recomputes the cell x component matrix W using these consensus components. As one technical note about applying the cNMF pipeline as described by Kotliar et al.25, we found that including all genes, as opposed to the 2000 most variable genes, was important for finding certain programs observed only infrequently in the dataset (data not shown). This is because genes whose expression changes in only a small fraction of cells (e.g. cells with a particular perturbation) would not end up being included in the 2000 most variable genes.
Choosing the number of components for cNMF analysis
To choose the free parameter K (number of components, Extended Data Fig. 2d-e), we defined a set of benchmarking statistics and compared the results of cNMF run for K = [3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 17, 19, 21, 23, 25, 27, 29, 30, 35, 40, 45, 50, 55, 60, 100]. We ultimately chose K=60 for all downstream analyses.
We examined the following benchmarking statistics:
(i) Number of unique GO terms enriched in program co-regulated genes (Extended Data Fig. 2d, see below)
(ii) Number of unique enriched TF motifs in the promoters or enhancers of program co-regulated genes (Extended Data Fig. 2e, see below)
(iii) Number of perturbations significantly regulating any component (Extended Data Fig. 2f).
(iv) Error of cNMF (difference between the original normalized data and reconstructed data, calculated by taking the sum of squares of the element-wise difference of the data, Extended Data Fig. 2g)
(v) Stability of cNMF (a measure of consistency of the components output from repeated runs, represented by the silhouette score25, Extended Data Fig. 2g)
We chose K = 60 for further analysis, as the number of components that gave a low cNMF error value while near-maximizing each other metric.
Excluding components associated with batch effects
We examined whether some components identified by cNMF were likely to represent batch effects. To do so, we calculated the Pearson correlation between each of the 20 batches (i.e., 10X lanes) and the expression of each component across all cells. Based on the distribution of batch x program Pearson correlation (Supplementary Tables 11 & 13), we assigned 10 components with Pearson correlation > 0.15 as likely representing batch effects (Extended Data Fig. 2c). We chose the threshold of 0.15 because: 1) most components showed very low correlation with batch, 2) above this threshold sample correlation with batch increases greatly (indicating particularly strong association with batch for these 10 components), 3) 8 of 10 of these components were associated with mitochondrial or ribosomal genes - sets of genes commonly observed as batch effects in 10x scRNAseq, 4) these components showed abs(R)>.15 across multiple 10X lanes (average of 9), and 5) importantly, none of these components showed enrichment by the V2G2P test (described below), and so would not have been identified as significant for CAD risk. We used the remaining 50 components for further analysis. This approach (including batch as a covariate in the differential expression test) has theoretical advantages, in particular reducing bias when groups (here, perturbed genes) are not distributed evenly across batches63.
Defining co-regulated genes for each program
We defined ‘co-regulated genes’ for each cNMF component as the 300 marker genes with the highest z-score regression coefficient as defined by cNMF25. Essentially, cNMF uses a linear regression model to identify coefficients indicating the number of standard deviations each gene’s expression would change with the increased usage of a given component. A component’s marker genes, then, are those with the highest “marker gene regression coefficients” (or “specificity scores”) for that component, and we selected the top 300 of these marker genes as the set of “co-regulated genes” for each gene expression “program” (as defined below).
Defining regulators for each program
We tested whether gRNAs targeting a given gene led to a significant change in expression of each component from the cNMF model. We used the Model-based Analysis of Single Cell Transcriptomics package (MAST)39 to compare the expression of each component in cells carrying gRNAs targeting a given gene vs. cells carrying control gRNAs (1,000 safe-targeting and negative control guides), including 10X lane as a covariate to account for batch effects. We removed the guides present in fewer than 3 singlet cells and the perturbations with fewer than 2 guides. We used the Benjamini-Hochberg method to account for multiple hypothesis testing on the MAST p-values (Extended Data Fig. 2h), and assigned ‘regulators’ of a program as those genes whose perturbation affected component expression with FDR < 0.05 accounting for 140,760 total tests (60 programs x 2,346 perturbations, which includes the 2,285 targeted TSSes, as well as targeted enhancers that were not further analyzed in this study, Supplementary Table 6).
To confirm that these FDRs were well-calibrated, we also conducted a simulation-based test. For each perturbed gene, we sampled from the control cells (all singlet cells with non-targeting or safe-targeting guides) the same number of cells, and compared these sampled cells to the rest of the control cells using the same MAST39 procedure. We identified 0 significant regulators in this approach (Extended Data Fig. 1q), indicating that our FDR < 0.05 threshold is a conservative estimate. We also performed the same procedure to estimate the background rate for perturbations called as having a significant effect on the transcriptome using EdgeR58,64,65.
Definition and annotation of gene expression programs
We defined a gene expression program as the set of genes comprised of both the 300 “co-regulated genes” and the significant “regulators” for each cNMF component. We annotated programs based on features of their co-regulated genes and regulators, including: by manual curation of genes with known biological functions, by enrichment of transcription factor (TF) motifs in the promoters and predicted enhancers of co-regulated genes, and by GO term enrichment (see below). The manual labels we assigned to each program (e.g., “Program 8 – Angiogenesis and osmoregulation” in Fig. 1c) represent our attempt to annotate the program based on functions of known genes, but we note each program includes many genes and regulators that have not been studied in combination before and may represent new, specific gene-pathway relationships that we do not currently have the vocabulary to describe.
Identifying motifs enriched in promoters and enhancers
To identify transcription factors that might regulate program co-regulated genes, we calculated enrichment of human transcription factor motifs in the sequences of the promoter and enhancers of the top 300 genes ranked by component specificity score (Extended Data Fig. 2e, Supplementary Table 24).
We obtained promoter sequences by taking 500 bp surrounding the TSS as previously annotated22 and enhancer regions from the Activity by Contact model at an ABC score threshold of 0.015 in the TeloHAEC control condition. For a gene that had multiple enhancers, we counted motif instances across all of its enhancers. To match motifs to sequences, we used HOCOMOCO v11 human full scan motifs (https://hocomoco11.autosome.ru/downloads_v11), and Find Individual Motif Occurrences (FIMO) (https://meme-suite.org/meme/meme_5.3.2/tools/fimo), with the default settings, and p-value thresholds of 10−6 for enhancers or 10−4 for promoters.
For a given motif and a given program, we counted the number of occurrences of a motif in the promoter sequences of either (i) the top 300 program co-regulated genes, or (ii) all expressed genes in teloHAEC, and compared these two vectors of motif counts using a two-sided t-test. We computed enrichment by dividing the program gene’s average motif match count by the rest of the expressed gene’s average motif match count. We tested all pairs of matched motifs (570 for promoter and 590 for enhancer) x 60 programs, and used the Benjamini-Hochberg method to account for multiple hypothesis testing on the t-test p-values.
Determining enrichment of annotated gene sets in components
To determine if the gene expression programs align with annotated and publicly available pathways, we tested whether the co-regulated genes in each component were enriched in gene sets from the Molecular Signatures Database (MSigDB). To do so, we used the clusterProfiler R package66 and MSigDB gene sets67 (here, the gene sets labeled as “all” for all gene sets and “c5” for GO terms only). We filtered the MSigDB gene sets to only those with more than 3 genes and less than 800 genes. We annotated each program with the gene sets that showed significant enrichment among the program genes (FDR < 0.05, Supplementary Table 25), and compared the number of gene sets showing significant enrichment as a function of the number of programs K (Extended Data Fig. 2d).
Defining endothelial-cell-specific programs
To annotate programs as “endothelial-cell-specific”, we analyzed the degree to which program co-regulated genes were expressed in endothelial cells versus other cell types. We took gene expression transcript per million (TPM) data across all available cell types from FANTOM5 and calculated the expression z-score of each gene across all cell types. To give each gene an endothelial-cell specificity score, we calculated the average of all z-scores for a gene across endothelial cell samples. We defined endothelial-cell specificity scores for each program as the average of the 300 co-regulated genes’ specificity scores, and selected 0.19 (90% percentile) as the threshold to call programs as “endothelial-cell-specific” (Extended Data Fig. 3c, Supplementary Table 13).
Variance explained by all cNMF components
To quantify the fraction of variance explained by all 60 programs jointly, we compared the residual variance in the dataset after subtracting the consensus matrix factorization to the total variance in the dataset: V = 1 - Var(X - WH)/Var(X), where X is the (cell x gene) normalized data matrix input to cNMF, W is the (cell x program) usage matrix, H is the (program x gene) spectra or weight matrix, and matrix variance is defined by summing the column- or gene-level variances: Var(X) = ΣjVar(Xj). Note that cNMF normalizes the input data so each Var(Xj) = 1.
Variance explained by individual gene programs
To rank gene programs by variance explained, we devised a method to quantify variance explained by NMF or cNMF components separately. For the k’th program Hk, we consider the effective matrix decomposition given only this program; the effective usage matrix Bk in this case is given simply by orthogonal projection or ordinary least squares: Bk = XH’k/∥Hk∥2, where the prime indicates transposition. We then define the variance explained in terms of the residual fraction as above: Vk = 1 - Var(X - BkHk)/Var(X). Our method may be generalized to any set of programs, but with more than one program the effective usage matrix must be obtained by nonnegative least squares (a single iteration of NMF).
Defining variants in CAD GWAS signals for variant-to-gene analysis
CAD lead GWAS variants were derived from both Aragam et al.12 and Harst et al.10. We excluded lead variants from Harst et al. if the variants were in strong LD (r2 ≥ 0.7) with an Aragam et al.12 lead variant or were ≤5Kb away from an Aragam et al. lead variant. An LD-expansion was performed to include variants that are both within a 1 Mb window of, and are in strong LD (r2 ≥ 0.9) with the any of these lead GWAS variants in 1000 Genome European ancestry (plink --ld-window-kb 1000 --ld-window 99999 --ld-window-r2 0.9). For each lead variant, we also included variants prioritized through functionally informed fine-mapping (PIP ≥ 0.1) in either study12,10. We defined a “GWAS Signal” as this collection of variants around, and including, each lead variant.
Identifying CAD variants associated with lipid levels
We classified CAD GWAS signals as “lipid” or “non-lipid” based on their association with lipid levels in other GWAS studies, because the CAD GWAS signals also associated with lipids are presumed to act through non-endothelial cells such as hepatocytes. For lead signals included in Aragam et al.12, we defined a CAD GWAS signal to be associated with lipids if the lead variant was linked to “LDL-direct”, “Triglycerides”, “Cholesterol”, “HDL-cholesterol”, “Apolipoprotein A”, “Apolipoprotein B”, “HDLC” or “LDLC” in the phenome-wide association scan (PheWas) conducted by Aragam et al.12 For GWAS signals exclusively nominated by Harst et al10, we used a different procedure in which we considered a signal to be associated with lipids if its lead variant was associated (P < 5 × 10−8) with HDLC, LDLC, TG, ApoA, or ApoB based on GWAS from the UK Biobank (Hilary Finucane and Jacob Ulirsch: https://www.finucanelab.org/data). We refer to the remaining GWAS signals not associated with lipid levels as “non-lipid CAD GWAS signals”, and focused on this subset of signals as cases where CAD variants might plausibly act in endothelial cells.
Linking variants to genes
We used a combination of variant-to-gene methods to identify a list of genes linked to CAD variants that could plausibly act in endothelial cells. At each CAD GWAS signal, we considered as candidate genes at least two genes upstream or downstream of the lead GWAS SNP, and all the genes within +/− 500Kb of the lead variant to be potentially regulated by the GWAS signal. We focused our analysis on protein-coding genes and excluded long noncoding RNAs (“^LINC”), gene isoforms (“-AS”), microRNAs (“^MIR”), small nuclear RNAs (“RNU”), and genes of uncertain functions ("^LOC"). To link CAD variants to genes, we intersected the variants with ABC enhancers9 in endothelial cells to identify the top two genes most likely to be regulated by each variant (highest 2 ABC fractional scores over 0.015). Specifically, we used ABC data, for enhancers and predicted target genes, from TeloHAEC and Eahy926 (control, or treated with IL1β, TNFα or VEGF, this study), and from prior ABC analysis of HUVEC (‘endothelial_cell_of_umbilical_vein_Roadmap’, ‘endothelial_cell_of_umbilical_vein_VEGF_stim_12_hours-Zhang2013’, and ‘endothelial_cell_of_umbilical_vein_VEGF_stim_4_hours-Zhang2013’ datasets from 9). To account for cell state-specific regulation that was not predicted by ABC, we also intersected candidate CAD variants at each signal with ATAC peaks and considered the 2 genes closest to variant-containing peaks as plausibly regulated. We also linked variants to genes if the variant was in a coding sequence or within 10 bp of a splice site annotated in the RefGene database (downloaded from UCSC Genome Browser on 24 June 2017)68. We confirmed that these candidate CAD variants were significantly enriched for matching any or all of these criteria (Extended Data Fig. 5e). We identified 254 candidate CAD genes, defined as “genes with V2G (variant-to-gene) links”, at 125 of 228 non-lipid CAD GWAS signals (Supplementary Table 1).
Transcription profile comparisons between teloHAEC and human right coronary artery endothelial cell (RCAEC)
To confirm the validity of teloHAEC as a relevant model for endothelial cells in human coronary artery (where atherosclerosis that leads to CAD develops), we compared single cell RNA-seq gene expression from control guide carrying teloHAEC from our Perturb-seq screen to scRNAseq data from explanted human right coronary artery endothelial cells (RCAECs)69. We compared the gene expression at two levels: for all perturbed genes (2,285 genes) and for the 41 CAD associated genes. Among the perturbed genes in teloHAEC, 2,107 genes are expressed at TPM > 1 in healthy or disease RCAECs. We observed high correlation of gene expression in transcripts per million (TPM) between teloHAECs and RCAECs (Pearson correlation = 0.66, p-value = 6.45 x 10−280, Extended Data Fig. 1b). We observed similar correlations of gene expression for the 41 CAD associated genes (Pearson correlation = 0.63, p-value = 9.29 x 10−6, Extended Data Fig. 1c). Furthermore, 40 out of 41 CAD associated genes are expressed at >1 TPM in RCAECs (Extended Data Fig. 1d).
Identifying CAD-associated programs via variant-to-gene-to-program analysis
We developed an approach to identify gene programs likely to affect CAD risk through functions in endothelial cells. To do so, we tested whether the 254 genes with V2G links (between CAD variants and enhancers/coding regions in endothelial cells) were enriched in each Perturb-seq program. Specifically, we performed a one-tailed Fisher exact test separately for co-regulated genes and for regulators. For co-regulated genes, we constructed a contingency table for whether a gene is a co-regulated gene (out of 17,472 expressed genes) and whether a gene has a V2G link. For regulators, we constructed a contingency table for whether a gene is a regulator (out of all perturbed genes) and whether a gene has a V2G link. We then multiplied the p-values from co-regulated gene and regulator Fisher exact tests together to get a final program enrichment p-value. We use Benjamini-Hochberg method for multiple hypothesis correction across all 50 non-batch programs. For an example of this analysis, see Extended Data Fig. 6g. 5 programs showed significant enrichment by this method (FDR < 0.05: Programs 8, 35, 39, 47, 48), referred to as “V2G2P programs for CAD”.
Defining CAD-associated V2G2P genes
We defined “V2G2P genes for CAD” as those 41 genes that were both (i) a gene with a V2G link to a CAD variant and (ii) a member of one of the 5 CAD-associated programs (as a regulator and/or co-expressed gene). The 41 genes were linked to 43 GWAS signals due to cases where independent GWAS signals are linked to the same gene.
Identifying enriched programs via MAGMA
We tested whether the co-regulated genes in each program were significantly enriched near variants associated with CAD using MAGMA. To do so, we took the CAD summary statistics from Aragam et al.12 (https://data.mendeley.com/public-files/datasets/2zdd47c94h/files/5b4eb0d7-96e8-4c7e-b109-046107ebd480/file_downloaded), and used the MAGMA --annotate function to summarize CAD association p-values for variants within a 50 kb window of all human genes, using the 1000 genomes European reference data for base allele frequencies (https://ctg.cncr.nl/software/MAGMA/ref_data/g1000_eur.zip). We then ran MAGMA to test for enrichment of CAD heritability within 50 kb of the top 300 program genes, and corrected for multiple testing (60 components) using the Benjamini-Hochberg method.
Identifying programs and cell types enriched for CAD heritability via stratified LD score regression
We used S-LDSC to estimate the enrichment of CAD heritability linked to program genes and to enhancers in TeloHAEC. While the original implementations of S-LDSC linked variants to genes based on genomic distance28,70, we additionally required that variants either overlap exonic regions of the gene or overlap nearby candidate enhancers in endothelial cells (as in 32,71). In particular, for co-regulated genes in each program, we derived an annotation for S-LDSC by including exonic regions (exons from transcripts with Ensembl_canonical, appris_principal, appris_candidate, or appris_candidate_longest tags, as indicated in the GENCODE v38lift37 annotations) as well as endothelial cis-regulatory elements derived from snATAC-seq72, from which we merged the 9 adult and 8 fetal sets of endothelial peaks into a single annotation, and for each geneset included all peaks within 50 kb of the gene starts and ends. For all peaks, we first converted coordinates from the GRCh38 to the GRCh37 reference assembly using UCSC LiftOver, discarding peaks that could not be converted. To estimate the enrichment of CAD heritability in TeloHAEC enhancers, we required the variants to overlap enhancers predicted by ABC from ATAC-seq and H3K27ac ChIP-seq data in TeloHAEC under control conditions or treated with IL1β, TNFα or VEGF (ABC score > 0.015). For each set of variants (programs or TeloHAEC enhancers) we ran S-LDSC using 1000G EUR Phase3 genotype data to estimate LD scores, baseline v2.2 annotations as recommended by the LDSC developers73, and HapMap 3 SNPs excluding the MHC region as regression SNPs. We ranked programs by their enrichments and reported the p-values of these enrichments (Extended Data Fig. 5b). Full S-LDSC results for TeloHAEC enhancers can be found in Supplementary Table 27.
Polygenic Priority Score (PoPS)
PoPS is a method to nominate likely causal genes in a GWAS locus, which prioritizes genes based on their being members of many gene sets enriched for heritability genome-wide3. We applied PoPS to summary statistics from Aragam et al.12 using the predefined set of gene sets as previously described3 (Extended Data Fig. 5c,d). For each GWAS signal, we calculated the PoPS rank among “nearby genes” (2 to either side of the lead SNP, and all within +/−500kb). Previously we have shown that genes with the highest PoP score in the locus are strongly enriched for likely causal genes, as identified by fine-mapped coding variants3, and that this enrichment increases when further focusing on genes that are both the closest gene and have the highest PoP score. In this analysis, we did not use any features from Perturb-seq and, as such, this method represents an entirely independent method that validates the high likelihood of causality of the set of CAD-associated V2G2P genes.
Defining gene expression programs for cells carrying control guides
To examine the gene programs in normal, unperturbed teloHAECs, we used the same analysis pipeline on the subset of cells carrying control guides (5,506 cells). We used cNMF to discover K=60 components, and defined 60 “control programs” based solely on the 300 co-regulated genes defining each component (because control guides did not target any genes, so there was no regulator information). Of the 60 programs, 4 programs correlated with batch (Programs 2, 17, 22, 41). We compared the program co-regulated genes between control cells and full library programs (Extended Data Fig. 6d). Control program 10 highly overlapped with full library programs 8 and 39. The four control programs that correlated with batch also had high overlap in co-regulated genes with the full library’s batch programs. We then utilized the V2G2P approach to prioritize these programs, and found that none of the control programs was enriched for genes with V2G links (Extended Data Fig. 6e).
Identifying genes in the CCM pathway
Fig. 3 shows a curated set of genes previously reported to interact physically or functionally with the CCM complex and/or downstream ERK5/MEK5 signaling47,48,74-78, plus one additional gene (TLNRD1) that we identify here as a member of the CCM pathway. These genes were manually selected through an iterative process involving examining genes known to interact with the CCM complex and that were found to regulate the enriched programs in Perturb-seq.
Allelic imbalance analysis for a variant linked to TLNRD1
We calculated allelic imbalance in ATAC-seq and ChIP-seq signal for the rs1879454 variant, accounting for any mapping bias toward the reference allele following methods previously described79. Specifically, we created two reference genome FASTA files that harbored the reference or alternate alleles at rs1879454; aligned ATAC-seq data to both genome files; selected reads that overlapped the variant coordinate; and used PySuspenders79 and PySAM (https://github.com/pysam-developers/pysam) to assign and count reads that uniquely aligned to one or the other allele. We applied this procedure to ATAC-seq data from TeloHAEC and the ENCODE datasets ENCSR000EVW (GATA2 ChIP-seq on HUVEC) and ENCSR000EOB (DNase-seq and DGF on HMVEC-dLy-Neo).
CRISPRi-FlowFISH for TLNRD1
We used CRISPRi-FlowFISH to test the effects of 61 candidate enhancers on TLNRD1 expression in teloHAEC, including the enhancer containing rs1879454. We designed gRNAs tiling across all accessible regions (here, defined as the union of the peaks in the chromatin accessibility dataset called by MACS2 with a lenient P-value cut-off of 0.1, and 150-bp regions on either side of the MACS2 summit) in the range chr15:81,267,614-81,427,246 in ATAC-seq data from TeloHAEC. We excluded gRNAs with low specificity scores or low-complexity sequences as previously described 22. We infected teloHAECs with the gRNA lentiviral library with 15μg/mL blasticidin selection for 3 days, and activated CRISPRi with 2μg/mL doxycycline incubation for 5 days. We performed FlowFISH using ThermoFisher PrimeFlow (ThermoFisher 88-18005-210) as previously described 22, using ThermoFisher probesets VA1-3010837-PF for TLNRD1 and VA4-13187-PF for RPL13A. We observed an approximately 2.6-fold signal for TLNRD1 in cells with all probes applied (“stained”) versus cells without target gene probes applied (“unstained”) (Extended Data Fig. 8j). We analyzed these data as previously described 22. In brief, we counted gRNAs in each bin using Bowtie to map reads to a custom index, normalized gRNA counts in each bin by library size, then used a maximum-likelihood estimation approach to compute the effect size for each gRNA. We used the limited-memory Broyden–Fletcher–Goldfarb–Shanno algorithm (implemented in the R stats4 package) to estimate the most likely log-normal distribution that would have produced the observed guide counts, and the effect size for each gRNA is the mean of its log-normal fit divided by the average of the means from all negative-control gRNAs. As previously described, we scaled the effect size of each gRNA in a screen linearly, so that the strongest 20-guide window at the TSS of the target gene has an 85% effect, in order to account for non-specific probe binding in the RNA FISH assay (this is based on our observation that promoter CRISPRi typically shows 80–90% knockdown by qPCR). We averaged the effect sizes of each gRNA across replicates and computed the effect size of an element as the average of all gRNAs targeting that element. We assessed significance using a two-sided t-test comparing the mean effect size of all gRNAs in a candidate element to all negative-control guides. We computed the false-discovery rate (FDR) for elements using the Benjamini–Hochberg procedure and used an FDR threshold of 0.05 to call significant regulatory effects.
Generation of single-guide CRISPRi TeloHAEC derivatives
Paired oligos for individual guides (newly-designed, as described for the Perturb-seq library, or with the best KD efficacy in Perturb-seq) were annealed and cloned into the BsmBI site of a CROP-Opti-Blast vector (plasmid available upon request), which were then used to generate lentivirus (as per 53). CRISPRi TeloHAEC were infected with each virus, in separate wells, and selected for blasticidin (15 μg/ml 4 days), before 5 day dox induction and analysis by bulk RNA-seq, fluorescence imaging or physiological assays. Guides (TargetGene_CloneIndex: ForwardSequence) were: CCM2_C2: GGCAAGAAGGTGAGCGTGCG, CCM2_F6: GAGCCGCTACATGCTCGACCC, CDH5_B8: GCCAGCTGGAAAACCTGAAG, CDH5_D5: GTTGGACTGCCTGTCCGTCCA, ITGB1BP1_C7: GAAGGCCGCGGCACTCCCACG, ITGB1BP1_G8: GAAGTCCGCAACCCGGGGAT, KLF2_C9: GGACCCGGGGAGAAAGGACG, KLF2_G10: GCCGCGGTATATAAGCCGGC, MAP2K5_A11: GCCGAGGCCGCGCGGACTGG, MAP2K5_B5: GTCTGCCCCACCCGGAGACAC, MAP3K3_A4: GTTCCTGAGGTGGAGAACGG, MAP3K3_C3: GCCAATAACAAGAAGGAAGT, MEF2A_C10: GCGGCGCGAAGCGCTGGTGG, MEF2A_H10: GACTGAATTATCCTCTCGGT, Negative_control_B6: GCAACGGTGTACCGCGGATC, Negative_control_D2: GTGGTTCACAACCGGACCCA, Negative_control_D8: GGTGGTTCGGTTTGCGTGGCC, Negative_control_F4: GCTGGGCGGACGTTGGGATA, NFAT5_D4: GGCCTCGCTTCCTGCCGGCG, NFAT5_D7: GGTCCCCGTCCCGCCGGGGG, PDCD10_D11: GACCGAGCAGAAGAGGTCTA, PDCD10_G1: GCCGCTTTACGCCACTCGCGT, TLNRD1_B3: GTGGCTGCGCCGCCGCCCGCA, TLNRD1_D12: GCCTCCGGCAGCCCCTGCGGG.
Ribonucleoprotein-based CRISPR/Cas9 genome editing
For some experiments, we used Synthego’s ribonucleoprotein (RNP) technology as an orthologous method to knock down target genes, as previously described 80. Briefly, TeloHAEC were nucleofected with Synthego’s Gene Knockout Kit v2 for non-targeting negative control, CCM2, TLNRD1 or MAP3K3 using the Lonza 4D-Nucleofector system. For each nucleofection reaction, we used 150,000 cells with 20 pmol of Cas9 and 50 pmol of sgRNA. The cells were then nucleofected (program CA-210) using SG cell line nucleofection solution (Lonza; V4XC-3024). The nucleofected cells were seeded in TeloHAEC culture medium, and harvested 48 hrs later for RNA extraction for qRT-PCR analysis and/or RNAseq to measure gene knockdown efficiency and perturbation effects. For MAP3K3 knockdown in single-guide CRISPRi lines, cells were treated with 2 μg/ml doxycycline for 72 hours before nucleofection, and for the 48 hours afterwards.
Identification of disease-relevant genes regulated by both TLNRD1 and CCM2
For the heatmaps in Figs. 5c & 6a, we identified genes that were most-strongly regulated by both TLNRD1 and CCM2 CRISPRi knockdowns from bulk RNAseq data as follows. 1) We included genes that were members of the 8 gold standard, known EC-acting CAD genes, that were significantly regulated by both TLNRD1 and CCM2 knockdowns (both p.<.05). 2) We included genes that were in the top 40 TLNRD1 and CCM2 up- or down-regulated genes (by highest p.value across both knockdowns), had an average fold change in TLNRD1 and CCM2 knockdowns of >2 (either positive or negative), and had prior evidence for functions predicted to increase or decrease CAD-relevant endothelial cell phenotypes (Supplementary Table 29).
Computational prediction of the TLNRD1 and CCM protein structure
AlphaFold2.3 Multimer v3 81 was run using sequences for KRIT1 (UniProt O00522), CCM2 (Uniprot Q9BSQ5, with and without deletion of residues 417-444), PDCD10 (Uniprot Q9BUL8), and TLNRD1 (Uniprot Q9H1K6). Models were visualized using UCSF ChimeraX v1.61. Predicted Alignment Error (PAE) was extracted using AlphaPickle 82 and plotted using combinations of AlphaPickle, Matplotlib v3.7.0, and Seaborn.
Co-immunoprecipitation of CCM2 and TLNRD1
HEK293 cells were transfected with V5-tagged CCM2 full length, V5-tagged CCM2 C-terminal truncation, Flag-tagged TLNRD1 and/or Flag-tagged Akt1, as indicated in Fig. 5b and Extended Data Fig. 9d-f, using FuGENE (E2311, Promega) or PEI MAX (Polysciences). Two days after the transfection, cell lysates were extracted with IP lysis buffer (87787, Thermo Scientific) supplemented with 1x Halt Protease Inhibitor Cocktail (1862209, Thermo Scientific). Protein concentration was determined using the Pierce BCA Assay (ThermoFisher), and equal mass of protein used for each sample. Immunoprecipitation was carried out using magnetic beads (88805, Thermo Scientific) conjugated with 5 μg of either rabbit anti-V5 (13202, Cell Signaling Technology) or mouse anti-Flag (F1804, Millipore Sigma) antibody. Cell lysates were incubated with the antibody-conjugated beads for 20 to 30 mins at room temperature. Beads were then washed three times with IP lysis buffer (1861603, Thermo Scientific), and precipitants were eluted using 2xLDS sample buffer (NP0007, Thermo Fisher Scientific). Precipitants and input lysates were separated by 10% SDS-PAGE and transblotted to nitrocellulose. For the anti-FLAG IP, blots were immunoblotted with 1:1000 rabbit anti-V5 (13202, Cell Signaling Technology), followed by 1: 5000 anti-rabbit HRP secondary (7074, Cell Signaling), then stripped (21059, Thermo Scientific) and re-probed with 1:1000 rabbit anti-TLNRD1 (HPA071766, Sigma). For the anti-V5 IP, blots were immunoblotted with 1:1000 primary mouse anti-Flag (F1804, Millipore Sigma) and 1:5000 secondary anti-mouse HRP (7076, Cell Signaling), and stripped and reprobed with 1:1000 mouse anti-V5 (ab27671, Abcam). The Akt1-FLAG vector is Addgene #9021. CCM2-V5 (ccsbBroad304_04281) and TLNRD1-V5 (ccsbBroad304_03872) vectors were obtained from the Broad Institute Gene Perturbation Platform83. For TLNRD1-FLAG, cDNA sequences were amplified from the TLNRD1-V5 vector using primers that incorporated an in-frame FLAG tag, and cloned into the pcDNA3.1 backbone. The CCM2 C-terminal truncation, was created in the CCM2-V5 vector by site-directed mutagenesis using the QuickChange II Site-Directed Mutagenesis kit (Agilent 200523-5), and the oligos F-CACCCTCAGAGGGGTCAGCATGCCCAAC and R-GTTGGGCATGCTGACCCCTCTGAGGGTG, which resulted in a deletion of amino acids 419-442 at the C-terminus of CCM2, in frame with the V5 tag.
Trans-endothelial electrical resistance (TEER) measurements
For TEER measurements, we used the ECIS Z-Theta instrument from Applied BioPhysics in the 96-well plate system (Applied BioPhysics; 96W10idf). CRISPRi TeloHAEC expressing individual guides to TLNRD1, CCM2, or non-targeting guides (2 guides each) were treated for 5 days with 2 μg/ml doxycycline. A gold electrode-containing 96-well ECIS plate was incubated at 37°C and 5% CO2 with culture media for 30 min to equilibrate before coating with 2.5 mg/mL fibronectin in 0.1 M bicarbonate buffer at pH 8.0. Then, the coated wells were inoculated with 45,000 cells in 100 mL media. An additional 100 mL of media was added to each well before initiating the measurements at 4000-Hz AC. At 25 hours, after the cells formed a confluent layer, the culture media was replaced with 200 mL of fresh culture media with 1 U/mL thrombin to disrupt cell-cell junctions, and measurements continued until 50 hrs to observe cell junction recovery after thrombin treatment.
Measurement of endothelial cell responses to laminar flow
200,000 CRISPRi TeloHAEC cells with individual control, CCM2 or TLNRD1 guides were seeded on flow chamber slides (80176, Ibidi) that had been pre-coated with 0.2% gelatin. After 24 hours, cells were cultured under laminar flow (12 dynes/cm2) for 48 hours (10902, Ibidi pump system). Static culture controls were seeded at the same density. Cells were treated with 2 μg/ml doxycycline for 2 days prior to seeding, and throughout, for a total of 5 days. RNA was harvested with 300 μl of Trizol and extracted with 60 μl of chloroform. After addition of 1 volume 70% ethanol, RNA was loaded onto a Qiagen RNeasy spin column, washed with 350 μl of buffer RW1 and treated for 20 mins at room temperature with 10 μl Purelink DNAse (InVitrogen 12185010) in 80 μl of 1x buffer. Subsequent RNA purification steps were as per the Qiagen RNeasy protocol.
Fluorescence imaging and quantitation of TeloHAEC
For quantitation of actin fiber characteristics, CRISPRi teloHAEC expressing individual guideRNAs (targeting CCM2, TLNRD1, or negative control) were treated with 2 μg/ml doxycycline for 5 days. Cells were fixed in situ with by addition of paraformaldehyde to 3.2% for 30 mins at 37°C, washed with PBS, permeabilized by addition of PBS with 0.1% triton X100 for 15 mins at room temperature, washed with PBS and stained with PerkinElmer Cell Painting dyes (Phenovue Fluor 568 - Phalloidin, Phenovue Fluor 488 - Concanavalin A, Phenovue Hoechst 33342 Nuclear Stain & Phenovue 512 Nucleic Acid Stain) according to the manufacturer’s instructions. Cells were imaged in four channels as described in 84 on a Perkin Elmer Opera Phenix Imaging System-106513, confocal 63x magnification with 1x binning. The stacks of images for the Phalloidin and Hoechst channels were converted to single images using maximum projection, output ranges standardized, and images exported. Cell boundaries were drawn by hand on a Phalloidin/Hoechst composite image in FIJI and saved as regions of interest (ROI). Phalloidin channel images were loaded into FIJI, converted to 16-bit grayscale, and cell areas and dimensions for each ROI were extracted using the Measure function (reporting Area and Fit Ellipse). Actin fibers were detected and quantified using the LPX FIJI plugin as described in 85, with lineExtract parameters: giwsiter = 5, mdnmsLen = 8, pickup = above (10.0), shaveLen = 3, delLen = 5, and line properties for each ROI measured using LineFeature. Parallelness (a_normAvgRad) ranges from 0 (for randomly-oriented fibers) to 1 (all fibers parallel).
Zebrafish husbandry and transgenic lines
Adult wild type AB, transgenic Tg(flk1:EGFP) (that express EGFP at the surface of blood vessels) and transgenic Tg(cmlc2:EGFP) (that express EGFP in heart muscle) zebrafish lines were maintained at 28.5 °C in circulating system water on a 14-h light/10-h dark cycle under standard conditions. Male and female embryos and larvae (≤ 5dpf) were kept in the dark in an incubator at 28.5 °C for subsequent experiments. At the end point, embryos were euthanized by tricaine overdose (MS-222; Western Chemical Inc.) followed by freezing (for RNA isolation), PFA-fixing (for histological analysis) or bleach treatment. All animal experiments were performed in accordance with relevant guidelines and regulations and with approval from the Mayo Clinic Institutional Animal Care and Use Committee.
tlnrd1 and ccm2 CRISPR knockdown in zebrafish
crRNAs for both ccm2 and tlnrd1 were designed using the Alt-R Predesigned Cas9 crRNA Selection Tool using the Integrated DNA Technologies (IDT) database. All the crRNAs were selected based on published criteria 86. For ccm2, guides were designed to target two distinct exons shared by all transcripts (AA: TTGAACGGAGACACGATACC, AF: ATGGAGCCACAACACCCACC). For tlnrd1, guides either targeted the 5’ untranslated region (UTR, AN.1: GGAAACACAAGGGACGTCTC, AF: GCTGAAAGTTACACCCAACG) or the single tlnrd1 exon (AN.2: CTGCCGCTAAGGATGTTGGT, DG: CAAGAGCAAAATGCAGCTGG). For ccm2 and tlnrd1, RNPs were prepared as described; briefly, the crRNA (bearing the guide sequence) was annealed with an equal molar amount of tracrRNA (bearing the gRNA scaffold, IDT, #1072532) in duplex buffer (IDT, #11010301), to form gRNA, by heating at 95 °C for 5 min and subsequently cooling on ice. Guide RNA was assembled with an equal molar amount of Alt-R S.p. Cas9 Nuclease V3 (IDT, #1081058) to form the RNP complex (28.5 μM final concentration), by incubation at 37 °C for 5 min followed by storage at − 20 °C, following the published protocol 86,87. RNP complexes prepared from the tracrRNA/scaffold only were used as a negative control. 3 nl of each RNP complex (28.5 μM final concentration) was injected into the yolk of one-to-two cell stage embryos (wildtype, Tg;Fli:EGFP (for the permeability analysis) or Tg;cmlc2:EGFP (for visualization of the atrioventricular valve, AV)).
tlnrd1 and ccm2 morpholino knockdown in zebrafish
Morpholinos (MOs) to knock down tlnrd1 and ccm2 were designed and injected using standard protocols88. The ccm2 morpholino has been validated to cause cardiovascular phenotypes at the 100 μM dose78. A custom morpholino for Tlnrd1 (TTCCCCGAGCCACTACTAGCCATAG) was designed to target the translation start site and ordered from Gene Tools, LLC. The control oligo is a single sequence, CCTCTTACCTCAGTTACAATTTATA, that is a validated negative control88. Wildtype zebrafish embryos were injected with 3 nl of diluted morpholinos at multiple concentrations (50 μM, 100 μM, 200 μM, 300 μM, of control, tlnrd1 or ccm2 morpholinos) at the one cell stage, using a pico-injector (Harvard Apparatus). For coinjection, tlnrd1 and ccm2 MOs were mixed to give 50 μM of each, and 3nl of the mixture was injected.
Zebrafish imaging and phenotyping
Embryos were observed for mortality and visible phenotypes at 2 days post-fertilization (dpf) and 3 dpf using a light microscope. Images were captured at 2 and 3 dpf on an EVOS microscope (Life technology) and Zeiss Axio-observer Z1. 3 dpf embryos (knock down or control) were scored as having a heart phenotype if they displayed visible atrial chamber enlargement, moderate to severe pericardial edema and slow blood flow in the tail veins. Note that, normal zebrafish undergo cardiac looping between approximately 2dpf and 3dpf (wherein the atrium and ventricle change from a linear posterior-to-anterior arrangement to a right-to-left asymmetric arrangement). Most of the ccm2 or tlnrd1 knockdown embryos that scored positive by the criteria above also showed a looping defect, maintaining the posterior-to-anterior arrangement of atrium and ventricle at 3dpf. However, because looping is a time dependent phenomenon that normally occurs near the 3dpf time when we examined the embryos for heart phenotypes, we did not include this as a scoring criterion. For the additional phenotypic analyses described below (confocal imaging, H&E staining, tail vein morphology, blood flow & vascular permeability), we selected ccm2 or tlnrd1 knockdown embryos that scored as positive for heart phenotype at 2dpf. High resolution images for the vascular permeability and cardiac chamber analyses, were acquired using a confocal microscope LSM 800 (Zeiss).
Histological staining of zebrafish embryos for atrial/ventricular thickness
H&E staining was performed by the Mayo Clinic Comprehensive Cancer Center Histology core lab. Jacksonville, FL. Briefly, zebrafish 3dpf larvae were fixed in 4 % paraformaldehyde overnight at 4 °C. To obtain paraffin sections, fixed larvae were dehydrated stepwise in ethanol/1x PBS dilutions (5, 25, 50, 75 and 100% ethanol). Transverse sections at a thickness of 5 μm using a microtom (MICROME) were produced from the anterior beginning of the otic vesicle and included posterior structures until the cloacal vent. The sectioned region therefore spanned from the glomerulus up to the cloaca and included the complete pronephros. Sections were stained with Gills 1, eosin Y and Harris hematoxylin (Richard Allan Scientific) according to the manufacturer protocol.
FITC-Dextran 2000 kDa & Texas Red-Dextran 70 kDa injections, & imaging for tail vein morphology and vascular permeability
Microangiography was performed as described 89,90. Briefly, at 3-days post-fertilization (3-dpf), Crispr/Cas9-injected embryos were anesthetized in 0.015% tricaine methanesulfonate (Western Chemical, Inc) and microangiography was performed by inserting a glass microneedle (World precision Instruments, Sarasota, FL) through the pericardium directly into the ventricle. For assessment of vascular morphology, 2000 kDa FITC dextran (Sigma, FD2000S-100MG) was diluted to 2 mg/ml in Zebrafish embryo medium 91, and a total of 4.5 nL was injected. For measurement of vascular permeability, Texas Red-dextran with a molecular weight of 70 kDa was solubilized in embryo medium at a 2 mg/mL concentration and a total of 4.5 nL was injected. Images were acquired after 30 minutes, using a Zeiss LSM 880 confocal microscope, and standard FITC and dsRed filter sets, and 10X objective, at room temperature. For quantitation of permeability, the Raw “.czi” images were preprocessed using the Zeiss software (ZEN2) to generate a maximum intensity projection image. The maximum intensity projection images of controls as well as Crispr mutants were then processed using the MATLAB programming platform, as described in our recent publication 90. Movies for the blood flow in the heart and tail veins were taken by capturing 60 second bright field-time-lapse images at 60 frames per second, using an EVOS microscope at 20x magnification, as described previously 92.
qRT-PCR assays in zebrafish
klf2b, ccm2 & tlnrd1 expression was measured by qRT-PCR on RNA isolated from 100 μM tlnrd1 morpholino embryos or CRISPR tlnrd1, ccm2 or control embryos at 3 dpf, using primers for klf2b, F: GAAGAGACACCTGTGAGGGC & R: GGACACCGATTCGTAGGACC, for ccm2, F: GGCGGATCAGATGAGGGAAC & R: CAGACAGCAATACGGACCGA, and for tlnrd1, F: ACACGCGAGAGTACCTGTTG & R: TCATCCCGCGACAAATCCAA.
In situ hybridization for tlnrd1 expression in zebrafish.
In situ hybridization was performed using previously validated methods93. Briefly, a 437 bp fragment of tlnrd1 was amplified from genomic DNA using the PCR primers, F: CATTAACGGAATGGCAGGCG and R: TGCCCGGATAAAGGCAAAGT, subcloned and verified by sequencing. Antisense in situ hybridization probes were generated using an M13 reverse primer with SpeI-linearized plasmid, while sense (negative control) probes were generated using an M13 forward primer with NotI-linearized plasmid. In situ hybridization of embryos was conducted at 24 and 72 hrs post-fertilization using these anti-sense or sense (control) probes against tlrnd1.
Applying the Variant-to-Gene-to-Program Approach to additional GWAS traits and cell types.
We tested whether the V2G2P method was generally applicable to other traits beyond CAD in endothelial cells, and to other cell types.
We first examined whether the same Perturb-seq dataset in endothelial cells could be applied to interpret variants for other vascular traits related to endothelial cell functions, beyond CAD. We applied V2G2P to 2 additional GWAS traits (Pulse Pressure (PP) and Mean Arterial Pressure (MAP), from the UK Biobank 94, with finemapping information from Hilary Finucane and Jacob Ulirsch: https://www.finucanelab.org/data). We performed V2G analysis by mapping variants associated with these traits onto the same endothelial cell enhancer map we used for CAD, and identified genes linked to PP or MAP variants in endothelial cells. We then performed V2G2P analysis, by testing for enrichment of the PP or MAP V2G gene sets in the 50 endothelial cell programs we identified from Perturb-seq. Note, that we performed the V2G2P enrichment test using only the 300 co-regulated genes in each program, because not all the genes at GWAS loci for these blood pressure traits were targeted for perturbation in our endothelial cell Perturb-seq screen.
We next examined whether the entire analysis framework could be applied to another cell type: K562 erythroid cells, which are a relevant model for red blood cell and platelet traits. Here, we examined 7 GWAS traits for red blood cell and platelet measures: Mean Corpuscular Hemoglobin (MCH), Mean Corpuscular Volume (MCV), Platelet Count (Plt), Red Blood Cell count (RBC), Mean Corpuscular Hemoglobin Concentration (MCHC), Hemoglobin A1c (HbA1c) and Hemoglobin (Hb), along with 4 traits for which K562 cells are not likely to be an appropriate model: pulse pressure (PP), mean arterial pressure (MAP), systolic blood pressure (SBP) & diastolic blood pressure (DBP), from the UK biobank 94, with finemapping by Hilary Finucane and Jacob Ulirsch: https://www.finucanelab.org/data.
We constructed V2G maps for each trait using ABC data in K562 cells (K562-Roadmap95), to identify variant-containing enhancers, and identified the set of V2G genes for each trait (genes with links to variants associated, by GWAS, with each trait). We, then, constructed a gene-to-program map by applying cNMF to the genome-scale Perturb-seq data previously collected in K562 cells 19. We tested K values over a broad range, and selected K=90 as the number of components that minimized cNMF error and maximized other ranking metrics (see “Choosing the number of components for cNMF analysis” above). Finally, we performed the V2G2P enrichment test (considering both the 300 co-regulated genes for each program and the regulators of each program, identified as the perturbations significantly affecting expression of each program, from Perturb-seq). Of the 90 programs, we found, 32 programs were prioritized for at least one of 6 GWAS traits (Extended Data Fig. 12b, Supplementary Table 22).
Curating previously-identified CAD prioritization gene sets.
To assess the ability of V2G2P to prioritize disease-associated genes, we surveyed several CAD studies that used more than just GWAS and genomic positioning data to prioritize CAD loci and genes. Below is a summary of how each study created their gene set and how we accessed this data.
Aragam et al.12, polygenic prioritization score (PoPS):
Computed PoPS score for all protein-coding genes within 500 kb of all GWAS signals and prioritized the gene with the highest PoPS score in each locus, resulting in 221 genes. Obtained from their Supplementary Table 25.
Hodonsky et al.96, eQTL and sQTL colocalization:
Bulk RNA-seq was collected from human coronary artery tissue samples from explanted transplant tissue, or collected from rejected transplant donors (138 individuals, from left anterior descending coronary artery, right coronary artery, and left circumflex artery). eQTL colocalization was performed to find eQTL-associated genes (eGenes), or to find splice QTLs (sQTLs). The eQTL list was from Supplementary Table 12 column “vdh_CAD_PPH4” with posterior probability > 0.8 (Methods: "PPH4 >0.8 to support evidence of a shared causal variant"). The slice variant list was from Supplementary Table 22 and subset for variants with posterior probability > 0.8 (column “vdh_CAD_PPH4”). We then identified sGenes linked to the colocalized sQTLs by finding matching genes in Supplementary Table 20 (columns “gene_id” and “spliceid”).
Li et al.97, transcriptome-wide association study (TWAS):
Associated genotype and expression data across 15 tissues (7 from STARNET and 8 from GTEx). We used Supplementary Table 4 for significant TWAS genes (114 genes).
OpenTarget L2G98 :
Used a supervised machine-learning model to learn the weights of multiple evidence sources (distance, molecular QTL colocalization, chromatin interaction, and variant pathogenicity) based on a gold standard of previously identified causal genes. The authors applied this model to the van der Harst coronary artery disease GWAS dataset10. Prioritized genes had an L2G model score > 0.5 (table downloaded from https://genetics.opentargets.org/Study/GCST005194/associations).
Stolze et al.29, endothelial cell-specific eQTL colocalization:
Human aortic endothelial cells (HAECs) were isolated from deceased heart donor aortic trimmings and cultured +/− IL-1beta (53 individuals, bulk RNA-seq), as well as 157 EC donors’ cultured ECs +/− oxPL treatment (microarray). They performed eQTL mapping using Matrix eQTL and used the R package "coloc" for colocalization. We obtained their data from Table S5.
van der Harst and Verweij10:
Prioritized variants using Probabilistic Annotation Integrator based on several features such as LD information, p-value distribution, coding genes, and H3K4me1 sites. Data were obtained from Table 2 and Online Table XX.
Wunnemann et al.30, endothelial cell CRISPR screen for 6 phenotypes:
The authors used a CRISPR screening approach to identify CAD risk variant-containing regulatory elements in 83 CAD GWAS loci that altered FACS-sortable signals for any of 6 pre-selected phenotypes in endothelial cells (E-selectin, ICAM1, VCAM1, nitric oxide, reactive oxygen species, and intracellular calcium). The identified 26 loci where perturbation of a variant-containing element affected one or more of these phenotypes (prioritizing a single gene in 21 of these loci). Data was obtained from their Fig. 3a and Supplementary Table 4.
Extended Data
Extended Data Fig. 1. Establishing the TeloHAEC CRISPRi model and Perturb-seq details.
a. Enrichment of CAD heritability in TeloHAEC enhancers, from Stratified Linkage Disequilibrium Score Regression analysis (S-LDSC, see Methods), where enrichment is the percentage of heritability explained by variants in enhancers (%heritability), divided by the percentage of variants in enhancers (%SNPs). Enhancers in TeloHAEC (treated under the indicated conditions) were identified from ATAC-seq and H3K27ac ChIP-seq data (n=6 for control ATAC, 3 for IL-1β, TNFα or VEGF ATAC, 4 for control ChIP, and 2 for IL-1β, TNFα or VEGF ChIP) by the Activity-by-Contact model. Error bars: standard error around the enrichment estimate, calculated by S-LDSC using jackknife (which resamples the data used for calculating heritability enrichment). P.values were calculated using the S-LDSC method28, and FDR by the Benjamini-Hochberg method. *: FDR<0.05, with specific FDR values of: Ctrl; 0.037, IL-1β; 0.015, TNFα; 0.020 and VEGF; 0.041. Full S-LDSC results can be found in Supplementary Table 27.
b. Scatter density plot of human right coronary artery endothelial cell single cell RNA-seq pseudobulk gene expression (from 69) versus teloHAEC pseudobulk gene expression, for genes perturbed in this study. Among the perturbed genes in teloHAEC, 2,107 genes are expressed at TPM > 1 in healthy or diseased RCAECs. R and p.values from two sided Pearson correlation test.
c. Scatter plot of the 41 V2G2P genes, comparing single cell RNA-seq pseudobulk expression (in TPM) in human right coronary artery endothelial cells to TeloHAEC. R and p.values from two sided Pearson correlation test.
d. Heatmap of gene expression (log10 TPM) of the 41 V2G2P genes in diseased right coronary artery ECs and in teloHAEC. 40 out of 41 CAD associated genes are expressed at >1 TPM in RCAECs. FBN2 is lowly expressed in the human right coronary artery endothelial cells.
e. FACS showing dox inducibility of KRAB-dCas9-IRES-BFP in TeloHAEC, after sorting but before the screen. Left panels: gating for viable individual cells. Right panels: Counts of gated cells by fluorescence intensity in the BFP/PB450 channel.
f. BFP channel counts of cells grown in parallel and concurrently with cells for the Perturb-seq screen. After expansion to 120M cells, transduction, selection and 5-day doxycycline treatment, 92% of cells remain BFP positive.
g. Cumulative distribution fraction for duplication levels of unique CBC-UMI-Guide combinations in deeply-sequenced dialout libraries (“unique UMIs”, red) or all guide reads (blue) versus duplication level. Requiring 4 duplicates (dotted line) eliminates 90% of CBC-UMI-guide combinations (likely PCR chimeras), while retaining >85% of total guide reads.
h. UMIs for top guide per CBC. Arrow: the chosen 4 UMI threshold.
i. Counts of singlets (1 gRNA, black bar), doublets (2) and higher multimers, as well as cells with no guide called (0), at the chosen thresholds of 4 UMIs for the top guide and 4 or more fold fewer for the next most frequent guide.
j. Histogram of counts of singlet cells per target. Dotted line: average.
k. Histogram of counts of singlet cells per guide. Dotted line: average.
l. Read UMI counts for all transcripts per cell by singlet/multiplet status. Median UMIs per singlet cell was 9,997, and average was 10,870. The median for cells with no guide called was 7,125, indicating that low guide UMI count is associated with low overall UMI count. Median UMIs for doublets was 13,723, 37.3% more than singlets. Assuming that droplets with two cells will have double the number of reads, this suggests 37% of doublets are due to two cells (9.3% of cells with guides) while the remainder (15.7% of cells with guides) are due to two guides in one cell, very close to the expectation from the infection MOI of 15%. n=352686, 214449, 79744, 19195 and 5345 cells with 0, 1, 2, 3, or 4 guides, respectively. Boxplot center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers.
m. Distribution of knockdown efficiency across target genes (log2 expression in cells containing guideRNAs targeting the gene versus in cells containing negative control guideRNAs). Gray line: all targeted genes. Yellow and red lines: Genes expressed at >30 and >300 TPM, respectively. Red dotted vertical line: 40% knockdown (average for 300+ TPM target genes).
n. Distribution of fitness effects across all guideRNAs (log2 ratio of guide frequency in singlet cells from the Perturb-seq experiment after 5 days of CRISPRi induction compared to guide frequency in the original guideRNA library). Guides targeting common essential genes (red) were depleted more frequently than guideRNAs targeting other genes.
o. Number of nominally significant differentially expressed (DE) genes per perturbed target (genes with raw p <0.01, and fold change >1.15 from EdgeR DE analysis). Perturbations that affected the transcriptome were those that significantly increased the number of nominally significant DE genes relative to the 48 targeted negative control genes (not expressed in TeloHAEC). Dotted line: 95th percentile number of DE genes for negative controls. 245 perturbations had a significant effect on the transcriptome, FDR <0.05 (10.7% of all targets that were not negative controls: including 31.9% of common essential genes (red, as per panel n), and 9.0% of other genes (blue)).
p. Volcano plot showing log2 (# DE genes for target)/(avg. # DE genes for non-expressed controls) versus −log10 FDR (capped at 100). Right: Symbols for target genes with the strongest effects.
q. Percent of perturbations that have a significant transcriptional effect in Perturb-seq, as defined by either (i) “DE Genes”: perturbations with significant effect on the transcriptome, as compared to 48 non-expressed negative control promoters, by binomial test (see Methods) or (ii) “DE Programs”: perturbations that lead to significant changes in program expression by MAST with 10X lane correction (FDR < 0.05).
Permuted Controls: Simulated negative controls, where statistical tests were performed on randomly drawn cells that carry negative control or safe-targeting guides.
Expressed: Genes with >1 transcripts per million (TPM) in teloHAEC control bulk RNA-seq.
Low or No Expression: Genes with less than or equal to 1 TPM.
Common Essential: Common essential genes from DepMap147.
TeloHAEC Proliferation: Fitness effects observed in the Perturb-seq experiment, by comparing guide frequencies (see Methods). Increase: >15% increase in guide frequency (FDR < 0.05), Decrease: >15% decrease in guide frequency (FDR < 0.05).
Gene near CAD GWAS signals: Expressed genes nearby any CAD GWAS signal (2 closest on each side, and all within +/−500kb).
Gene near IBD signals: Expressed genes nearby 10 selected IBD GWAS signals (closest 2 genes & all within +/−500kb), with no genes overlapping those for CAD signals.
Extended Data Fig. 2. QC metrics for single cells, and selection of number of components for cNMF.
a. UMAPs showing number of UMIs per cell (left), percent ribosomal genes detected per cell (middle), percent mitochondrial genes detected per cell (right).
b. UMAPs showing cells from each of the twenty 10X lanes. The differences in clustering along the UMAP_2 axis indicates a technical batch effect between 10X lanes.
c. Cumulative distribution function (CDF) plot of the maximum absolute value of Pearson correlation between cNMF component expression in cells and batch. Dotted line: the R>=0.15 threshold used to call programs associated with batch.
d. Gene set enrichment analysis for GO terms among co-regulated genes, as a function of the number of components in the cNMF model (K). y-axis: The number of unique GO terms enriched across all programs for a given K.
e. Number of unique motifs enriched among the promoters (top) or enhancers (bottom) of co-regulated genes across all components, as a function of K.
f. Number of unique perturbations that have significant effect (FDR < 0.05) on one or more programs, as a function of K.
g. Model-based evaluation of the choice of K. Stability of the components over 100 NMF runs (top) and element-wise square of error (bottom, see Methods).
h. Quantile-quantile plot for effects of perturbations on program expression. X-axis: Expected uniform distribution. Y-axis: −log10 p-value computed from MAST package39. Red: p-value < 0.05.
Extended Data Fig. 3. Catalog of gene programs.
a. Correlation heatmap of cNMF components. Color: Pearson’s correlation of log2 fold-change in component expression across all perturbed genes.
b. 50 programs ordered by variance explained (see Methods).
c. 50 programs ordered by endothelial-cell specificity score — that is, the degree to which the co-regulated genes in the program are specifically expressed in endothelial cells versus in other cell types from FANTOM5 CAGE data (see Methods). Red line: z-score corresponding to top 10% of genes most specifically expressed in endothelial cells.
d. Effects of selected regulators on the 13 endothelial-cell-specific programs. Heatmap: log2 fold-change in component expression in perturbation vs control. Top: 16 regulators shared between multiple endothelial cell-specific programs. Bottom: the 4 significant regulators (experiment-wide FDR < 0.05) per program with the most specific effects on that program relative to other endothelial-cell-specific programs.
e. Programs ordered by number of enriched transcription factor motifs (See Methods). Gray: promoters. Blue: enhancers. Some programs only have enrichment for motifs in promoters. Some programs showed enrichment of distinct motifs in enhancers versus promoters, such as Program 47 (Angiogenesis, GATA2), with promoter enrichment in WT1 and EGR2 motifs, and enhancer enrichment in GATA2 and PRDM6 motifs. Among the programs with few or no enriched transcription factor motifs, we identified other likely proximal regulatory mechanisms: Program 17 expressed genes whose promoters were marked by H3K27me3 in endothelial cells (see also panel k), and the most significant regulator of this program was SUZ12, a component of the complex (PRC2) that writes this histone modification; and Program 16 pointed to a potential RNA surveillance program, since 40% of its program genes were noncoding RNAs (panel l), and its regulators included a component of the RNA exosome (EXOSC5) and the chromatin remodeler INO80E, which has previously been shown to regulate a subset of noncoding transcripts in yeast148 (see also Supplementary Table 12).
f. Annotations for an example program: 15. Left: Top 10 program co-regulated genes. Middle, top: Motifs enriched in promoters of the 300 program co-regulated genes. Middle, bottom: Gene Ontology terms enriched in the 300 program co-regulated genes. Right: Volcano plot of the effects of regulators on cNMF component 15 genes. Program 15 (Flow response, KLF2) appeared to correspond to a canonical endothelial cell response to laminar shear stress defined by the known flow-responsive transcription factor KLF2: the program was highly enriched for KLF motifs in promoters; included known flow-responsive genes such as KRT18/19, NOS3, and KLF2 itself; and was significantly reduced by perturbations to MAP2K5 (MEK5), a kinase known to activate the signaling pathway upstream of KLF235,149.
g. Log2 fold change in expression of programs 28 versus 47 for each perturbed gene relative to controls. Program 28 (Tip cell, migration) includes co-regulated genes that mark tip cell specification during sprouting angiogenesis (ESM1, RHOC, PLAUR), and Program 47 (Angiogenesis, GATA) includes co-regulated genes that are enriched in GATA2 & TAL1 motifs and that include NRP2, a co-receptor for VEGF-A, previously shown to act downstream of GATA2150). Blue, red, and purple mark genes that are regulators of Program 28, Program 47, or both programs, respectively. Note that regulators that affect both programs do so in opposite directions.
h. Perturbations ordered by the number of regulated programs. Red: top 10 perturbed genes.
i. Programs ordered by the number of regulators. Blue: endothelial-cell-specific programs. The top 3 programs, by number of regulators, are labeled.
j. 131 perturbed genes that are regulators of at least one endothelial-cell-specific program, ordered by the number of such programs that they regulate. Top 10 regulators are labeled, and included genes known to have important functions in ECs such as EGFL7 and ITGB1BP1/ICAP126,27.
k. Average H3K27me3 ChIP-seq signal in co-regulated gene promoters. The top program is Program 17 (Polycomb targets). See legend to (e) for more details. N=50 programs. Boxplot center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, all data points.
l. Percent of noncoding RNA genes in program co-regulated genes. The top program is Program 16 (ncRNA & antisense RNAs). See legend to (e) for more details. N=50 programs. Boxplot center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, all data points.
Extended Data Fig. 4. Annotations for CAD-associated programs: 8, 35, 39, 47, 48.
Left panels. Top 10 program co-regulated genes. Program Specificity z-scores are the cNMF marker gene coefficients, indicating how specific this gene is to this program, relative to other programs (see Methods).
Middle left panels. Top: Top 5 motifs enriched in the promoters or enhancers of the program co-regulated genes. Bottom: Top 5 GO terms enriched in program co-regulated genes.
Middle right panels. Regulators of the program. Volcano plot shows effects of all perturbed genes on program expression. Red: FDR < 0.05. Labeled: top 2 significant regulators in each direction, plus CCM2 and TLNRD1.
Right panels. UMAP of program expression in a subset of cells (24,000, randomly selected).
Extended Data Fig. 5. Prioritization of CAD-associated programs and candidate CAD genes.
a. Using MAGMA to prioritize gene programs enriched for CAD heritability (linking variants to program genes and 50kb of flanking sequence, see Methods). Barplots show beta regression coefficient (left) and −log10 FDR (Benjamini-Hochberg adjusted enrichment p-value, right). Programs are ordered separately by beta or FDR value. Dotted line: FDR = 0.05.
b. Using S-LDSC to prioritize gene programs enriched for CAD heritability (linking variants in endothelial cell chromatin accessible regions to genes within 50 Kb, see Methods). Barplots show enrichment (left) and −log10 FDR (Benjamini-Hochberg adjusted enrichment p-value, right). N = 300 (co-regulated program genes ranked by z-score coefficient, for each program). Error bars: standard error around the enrichment estimate, calculated by S-LDSC using jackknife (which resamples the data used for calculating heritability enrichment). P.values were calculated using the S-LDSC method28, and FDR by the Benjamini-Hochberg method. *: FDR<0.05. Dotted lines: 1 fold enrichment (left), or FDR 0.05 (right).
c. CAD-associated V2G2P genes are ranked highly by an independent gene prioritization method, the Polygenic priority score (PoPS). For each of the 43 CAD GWAS signals including a CAD-associated V2G2P gene, we ranked nearby genes based on their PoPS scores. Red: 39 CAD-associated V2G2P genes (2 genes, EXOC3L2 and PECAM1, were not assigned scores by PoPS). Gray: all other nearby genes. p-value: two-sided Mann-Whitney U-test.
d. Contingency table of PoPS and distance-to-TSS ranks for the 39 CAD-associated V2G2P genes. (2 CAD-associated V2G2P genes were not assigned scores by PoPS).
e. Odds ratios of variants in lipid-associated (N=1,181) or non-lipid-associated (N=3,313) CAD GWAS signals in (i) ATAC peaks in endothelial cells (N=373,630 unique non-overlapping non-promoter features from 11 epigenomic datasets in ECs, see Methods), (ii) ABC enhancers in endothelial cells (N=47,112 unique non-overlapping non-promoter features from 11 epigenomic datasets in ECs), (iii) coding sequences (N=189,232 unique non-overlapping non-promoter features), or (iv) all three categories combined (N=519,046 unique non-overlapping non-promoter features), compared to background variants (all SNPs from 1000 Genomes, excluding lipid-associated or non-lipid associated CAD GWAS variants, N=9,955,2088 or N=9,953,076, respectively, see Methods). Odds ratios were calculated as ((CAD variants within the indicated genomic features)/(all background variants within these features))/((CAD variants outside of these features)/(all background variants outside of these features), and significance assessed by application of a two-sided Fisher’s exact test to the contingency table of this data, with columns=CAD variants v. background variants and rows=inside features v. outside features. Error bars: 95% confidence interval. *: FDR < 0.05. Specific FDR values, from top to bottom, were 1.1e-4, 3.3e-33, 1.5e-8, 3.2e-6, 0.39, 6.0e-32, 0.011, 7.5e-31. Dotted line: odds ratio of 1.
f. sc-linker prioritization for 60 EC Perturb-seq gene programs, ranked by z-score. The ranking of programs was similar to V2G2P analysis, but none of the programs reached significance.
g. Precision/Recall (PR) plot for V2G2P and seven prior approaches to prioritize CAD locus genes. Recall: the fraction of the 8 “gold standard” genes (with strong prior evidence for endothelial cell-specific roles in CAD) detected by each method. Precision: [number of “gold standard” genes called] / [number of genes called within these gold standard loci]. Red: V2G2P. Blue: Other studies that prioritized CAD GWAS genes in endothelial cells.
Extended Data Fig. 6. Details for V2G2P analysis.
a. Number of genes with V2G links, per non-lipid CAD GWAS signal.
b. Number of genes with G2P links, per non-lipid CAD GWAS signal.
c. The cell-type specificity of V2G links appeared to be important for identifying endothelial-cell-specific programs. Here, we repeated the V2G2P analysis (as outlined in Fig 2), but linked variants to genes using cell-type-agnostic criteria (including ABC scores from any cell type and not just endothelial cells). The 50 programs are ordered (y-axis) by the number of program genes linked to CAD variants (x-axis). Gray dashed line: the number of genes linked to CAD variants that would be expected by chance. Orange labels: endothelial-cell-specific programs. Two significant non-endothelial-cell-specific programs were identified. Fisher exact test with FDR correction, as outlined in panel g. (*FDR <0.05, **FDR <0.005, ***FDR <5e-4)
d. Number of overlapping co-regulated genes between control programs (ctrl, x-axis) and full library (2kG, y-axis) programs.
e. Using the full Perturb-seq dataset appeared to be important for identifying the 5 CAD-associated programs. Programs discovered through cNMF analysis of only the “unperturbed” cells carrying negative control guideRNAs. In this version of the analysis, none of the programs are enriched for genes with V2G link. Fisher exact test with FDR correction, see procedure in panel g. (All programs have FDR > 0.05).
f. Enrichment of genes with V2G links, from the full library but only using co-regulated genes (not regulators,***: FDR < 0.0005, **: FDR < 0.005)
g. Steps of V2G2P enrichment test. Numbers shown as examples are from program 8.
h. V2G2P analysis prioritizes a small subset of genes and GWAS signals compared to either V2G or G2P information alone. Barplots: Counts for signals (left) or nearby genes (right), total (“All”) or those that have: a V2G link, a G2P link, both a V2G link and G2P link to any program, or both a V2G link and a G2P link to a significantly enriched V2G2P program.
Extended Data Fig. 7. Variant-to-gene to program links refine causal gene predictions.
a. V2G2P evidence at the 20p13.1 CAD GWAS locus. Top: Heatmap lists genes within 1 Mb of the CAD GWAS signal in genomic order, and shows variant-to-gene (V2G) and gene-to-pathway (G2P) evidence, with the prioritized CAD-associated V2G2P gene(s) labeled in red bold font. Legend details: “ABC, top 2”: A noncoding variant overlaps a chromatin accessible peak in endothelial cells, and the ABC score is at least the second highest of all genes near the GWAS signal. “Distance to variant, top 2”: A noncoding variant overlaps a chromatin accessible peak in endothelial cells, and the gene is one of the two closest genes to the variant. Bottom: Zoom-in on genes near the CAD GWAS signal, where rs2004772 is predicted by ABC to regulate PREX1 in the Eahy926 endothelial cell line treated with VEGF. Red dot: Prioritized variant in predicted enhancer. Gray dots: Other variants within R2 < 0.9 of the lead variant in the locus. Signal tracks below show ATAC-seq or DNase-seq for endothelial and coronary artery smooth muscle cells (CaSMCs, another cell type relevant to CAD).
b. As per a, showing V2G2P evidence at the 10p24.33 CAD GWAS signal, where three genes had V2G links (to an enhancer containing rs4918069) and 2 had gene to CAD-associated program links. HUVEC: human umbilical vein endothelial cells.
c. V2G2P evidence at the 17q21.3 CAD GWAS locus, where we have previously linked rs17608766 to GOSR2129. Heatmap, as in panel (a). Middle: Box plot of GOSR2 reads per cell, normalized to control cell average. n: number of cells. Dotted line: control average (100%). Boxplot center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers. Counts for outliers, from top to bottom: GOSR2; 1 & 17, Control; 4 & 15. Right: Volcano plot shows effect of GOSR2 knockdown in Perturb-seq on the expression of the 50 non-batch gene programs. Red: FDR < 0.05, from two-sided statistical tests on program expression between perturbed vs. control cells by the MAST package39.
Extended Data Fig. 8. Regulatory connections amongst perturbed genes and the CCM pathway, and variant-to-gene links to TLNRD1.
a. Locus zoom plot (http://locuszoom.org/) for CAD GWAS in a 1-Mb region around CCM2. P.values are from the joint association analysis in Aragam et al. 12.
b. Volcano plot showing effect of CCM2 knockdown in Perturb-seq on the expression of the 50 programs. Red: FDR < 0.05. Significance was assessed by two-sided statistical test on program expression between perturbed vs. control cells by the MAST package39.
c. Effects of selected perturbed genes on CAD-associated programs (Same as Fig. 3b, with significant effects marked with a * (FDR < 0.05)). Color scale: log2 fold-change on program expression in Perturb-seq. Bold text: CAD-associated V2G2P genes.
d. Knockdown efficiency for genes in the CCM pathway, in bulk RNA-seq (Fig. 3c). x-axis: Gene expression for each target gene in cells receiving target guides, vs. cells with control guides. y-axis: guide IDs for CCM pathway genes.
e. 15q25.1 CAD risk locus, where rs1879454 is predicted to regulate TLNRD1 (red arc). GWAS variants: −log10 GWAS P-value12 for variants with LD R2 > 0.9 with the lead SNP. Green signal tracks: Epigenomic data from ECs. Gray signal tracks: data from other cell types. HUVEC: human umbilical vein ECs. CaSMCs: coronary artery smooth muscle cells.
f. Volcano plot showing the effect of TLNRD1 knockdown in Perturb-seq on the expression of the 50 programs. Red: FDR < 0.05. Statistical test as per (b).
g. Phenome-wide association study (PheWAS) for rs1879454, from Open Targets98,151. P.values are the best GWAS p.values for association of this variant with each trait across all GWAS sources used by Open Targets (February 25, 2022 release), including UKBB. Orange line: p.value required for significance across all traits. No lipid measure met the p.value threshold for inclusion in the plot, of 0.005. Note: The p.value for CAD is higher than that observed in Aragam et al.12 because Open Targets does not currently contain summary statistics for the latest CAD GWAS. There were no measures of circulating lipids or blood pressure associated with this GWAS signal in a PheWAS analysis in Aragam et al.12.
h. Variant-to-gene-to-program evidence at the 15q25.1 CAD GWAS locus. Heatmap shows variant-to-gene (V2G) and gene-to-pathway (G2P) evidence for all genes within 1 Mb of the CAD GWAS signal, in genomic order, with the CAD-associated V2G2P gene labeled in red bold font. Legend details: “ABC, top 2”: A noncoding variant overlaps a predicted enhancer linked to this gene in endothelial cells, and ABC score is at least the second highest of all genes in the locus. “Distance to variant, top 2”: A noncoding variant overlaps a chromatin accessible peak near this gene in endothelial cells, and the gene is one of the two closest genes to the peak. “CAD-associated gene programs” are the 5 V2G2P programs: 8, 35, 39, 47, 48.
i. Histograms of FlowFISH signal (arbitrary units of fluorescence) for RPL13A (left) and TLNRD1 (right) in unstained versus stained teloHAEC expressing the gRNA pool against promoter and potential enhancers. The complete FlowFISH data can be found in Supplementary Table 28.
j. As in (d), but showing the RPL13A (left) and TLNRD1 (right) signal after sorting of cells into 4 bins based on expression of TLNRD1. Results are typical of cells across the 4 independent samples.
k. Scatter plots showing the strong correlation between effects on TLNRD1 gene expression for all E-G pairs measured in each of 4 independent CRISPRi-FlowFISH screens. The enhancer containing rs1879454 is colored in red, all others in gray. R: Pearson correlation coefficient.
Extended Data Fig. 9. TLNRD1 interaction with the CCM complex depends on the CCM2 C-terminal helix.
a. The 15 top-ranking AlphaFold2 models of the CCM complex are shown, as predicted with and without the presence of CCM2 residues 417-444, and aligned to the CCM2 PTB domain (residues 55-237). The complex is consistently predicted in a high-confidence arrangement (left panel), with most variability in positions of the HHD domain of CCM2 and flexible regions of Krit1. Interactions are predicted between all members of the complex, including published interactions between CCM2-PDCD10. In contrast, multiple conformations are predicted in the absence of the CCM2 C-terminal helix (right panel).
b. Predicted Alignment Error (PAE, Å) of all pairwise residue combinations in the WT CCM complex (extracted and plotted using AlphaPickle 82); lower error indicates higher confidence.
c. Differences in PAE (Å) between the full CCM complex and the CCM complex lacking the CCM2 C-terminal helix; larger numbers represent higher confidence in the presence of the helix. The thick white lines correspond to deleted helix residues, which are omitted from comparison. Predictions within individual domains and proteins are largely unaffected, but within CCM2, the HHD and subsequent loops are predicted with reduced confidence upon helix deletion (yellow). Between domains and proteins, the largest differences are reduced confidence interactions between CCM2 and PDCD10 and between Krit1 and TLNRD1. Summing over the entire matrices, the increase in predicted error with deletion of the helix is 0.7Å (22.9 vs 23.6 Å, p=10−56, two-tailed t-test).
d. FLAG-tagged TLNRD1 and/or V5-tagged CCM2 full length (“WT”) or C-terminal truncation (“Δ”) were expressed in HEK293T cells, as indicated. Extracts were co-immunoprecipitated with rabbit anti-V5 and blotted with mouse anti-Flag (top) or mouse anti-V5 (bottom). For gel source data, see Supplementary Figure 1b. Similar results were seen in 2 separate experiments.
e. HEK293 cells were transfected with V5-tagged CCM2 and either Flag-tagged TLNRD1 or Flag-tagged Akt (negative control). Cell lysates were either immunoprecipitated with mouse anti-Flag antibody-bound beads (IP Flag), or loaded directly on the gel (Input). The membranes were first probed with rabbit anti-V5 to detect CCM2 in the Flag precipitant and confirm the transfection of CCM2-V5. The membranes were then re-blotted with rabbit anti-TLNRD1 to evaluate the efficiency of Flag immunoprecipitation and validate the transfection of Akt-Flag and TLNRD1-Flag. Each pair of lanes came from independent biological replicates. For gel source data, see Supplementary Figure 1c. Similar results were seen in 2 separate experiments.
f. HEK293 cells were transfected with V5-tagged CCM2 and Flag-tagged TLNRD1, or, as negative controls, either with CCM2-V5 and Akt-Flag or only TLNRD1-Flag. Cell lysates were either immunoprecipitated with anti-V5 beads (IP V5), or loaded directly on the gel (Input). The membranes were first blotted for Flag to detect TLNRD1 in the V5 precipitant and validate the transfection of Akt-Flag and TLNRD1-Flag. The membranes were re-blotted for V5 to evaluate the efficiency of V5 immunoprecipitation and confirm the transfection of CCM2-V5. For gel source data, see Supplementary Figure 1d. Similar results were seen in 2 separate experiments.
Extended Data Fig. 10. Effects of CCM2 and TLNRD1 knockdown relative to MAP3K3 knockdown and laminar flow.
a-d. CRISPRi TeloHAEC with control non-targeting guides or with guides to CCM2 or TLNRD1 (2 guides apiece) were nucleofected with Cas9 particles containing control non-targeting guides or with 3 guides targeting exon 3 of MAP3K3 (Synthego). Cells were treated with 2μg/ml doxycycline for a total of 5 days (3 prior to nucleofection & 2 afterwards), to induce the CRISPRi machinery. RNA was harvested 48 hours later (after phase contrast imaging), and RNA-seq libraries sequenced to a depth of 10-12 million reads. The CCM2 guides reduced target gene expression, on average, by 3.4-3.6 fold (p.<2e-9), while TLNRD1 guides reduced target gene expression by 9.4-9.9 fold (p. <2e-43), consistent with the effects of these guides in our other bulk RNAseq data (Fig. 3c). MAP3K3 transcript levels were not significantly reduced, but genome-mappable reads for the targeted exon (#3) were greatly reduced, and most of the remaining reads showed multiple mismatches, indicating efficient introduction of Cas9-targeted deletions. N=2 per condition (from one experiment, 1 RNAseq library for each of 2 CRISPRi guides per target - CCM2, TLNRD1 or non-targeting controls). Correlation coefficient (R), and p.values given in each panel are from a two-sided Pearson correlation test.
a. The difference between the effect of CCM2 knockdown in cells with MAP3K3 knockdown and the effect of CCM2 knockdown in control cells ([CCM2kd_with_MAP3K3kd/Control_with_MAP3K3kd] / [CCM2kd/Control], Y-axis) was plotted against the effect of CCM2 knockdown in control cells ([CCM2kd/Control], X-axis, plotting all genes regulated at p. <5e-4 in either contrast). Labeled genes are the top 5 up- or down-regulated genes by log2 fold change, on each axis. Diagonal line: slope -1 reference.
b. As in (a), but for the difference between the effect of TLNRD1 knockdown in cells with MAP3K3 knockdown and the effect of TLNRD1 knockdown in control cells (Y-axis), versus the effect of TLNRD1 knockdown in control cells (X axis). The negative correlations in (a) & (b) indicate that MAP3K3 perturbation partially reverses the transcriptomic effects of CCM2 or TLNRD1 knock down, consistent with a role of MEKK3/MAP3K3 signaling in regulating transcription downstream of both CCM2 & TLNRD1.
c. The difference between the effect of MAP3K3 knockdown in cells with CCM2 knockdown and the effect of MAP3K3 knockdown in control cells (Y-axis) versus the effect of MAP3K3 knockdown alone (X axis, plotting all genes regulated p. < 5e-4 in either contrast). Diagonal line: slope -1 reference.
d. As in (c), but for the difference between the effect of MAP3K3 knockdown in TLNRD1 knockdown cells and the effect of MAP3K3 knockdown in control cells (Y-axis) versus the effect of MAP3K3 knockdown in control cells (X axis). The negative correlations in both (c) & (d) indicate that perturbation of CCM2 or TLNRD1 partially reverses the transcriptional effects of MAP3K3 knockdown, consistent with the expectation that decreased expression of upstream inhibitors can compensate for decreased expression of MEKK3.
e-h. CRISPRi TeloHAEC with control non-targeting guides or with guides to CCM2 or TLNRD1 (2 guides apiece) were grown in static culture or subjected to flow in an Ibidi flow chamber for 48 hours. In each case, cells were treated with 2μg/ml doxycycline to induce the CRISPRi machinery for 5 days (3 days prior & 2 days after introduction of laminar flow). After phase contrast imaging, RNAseq libraries were prepared and sequenced to a depth of 10–12 million reads. N=2 per condition (one experiment, with 1 RNAseq library for each of 2 CRISPRi guides per target - CCM2, TLNRD1 or non-targeting controls). R and p.values for panels (e-h) as per (a-d).
e. The effects of CCM2 knockdown in static culture (Y-axis) compared to the effect of flow in control cells (X-axis, showing all genes regulated at p. < 5e-4 in either contrast). Labeled genes are the top 5 up- or down-regulated genes by log2 fold change, on each axis. Diagonal line: slope = +1 reference.
f. As in (e), but for the effects of TLNRD1 knockdown in static culture. The positive correlations indicate that TLNRD1 or CCM2 knockdown in static culture is similar to the effect of flow, consistent with the observation that TLNRD1 or CCM2 knockdown increases the number and parallelness of actin stress fibers (Fig. 6b-e), a characteristic of flow response in unperturbed ECs 13.
g. The difference between the effect of flow in cells with CCM2 knockdown and the effect of flow in control cells ([Flow_CCM2kd/Static_CCM2kd] / [Flow_Ctrl/Static_Ctrl], Y-axis) versus the effect of flow in control cells ([Flow_Ctrl/Static_Ctrl], X-axis). Diagonal line: slope = -1 reference.
h. As in (g), but showing the difference between the effect of flow in cells with TLNRD1 knock down and the effect of flow in control cells (Y-axis) versus the effect of flow in control cells (X-axis). The negative correlations in (g & h) indicate that CCM2 or TLNRD1 knockdown cells have a weaker transcriptional response to flow than control cells. The negative correlations are also consistent with the observations in (e & f) that CCM2 or TLNRD1 knockdown in static culture are similar to the effects of flow, such that lesser fold-changes in gene expression would be required for CCM2 or TLNRD1 knockdown cells to achieve the full normal transcriptional response to flow.
i. Representative images of CRISPRi teloHAEC with control non-targeting guides or with guides to CCM2 or TLNRD1, that were subjected to flow in an Ibidi flow chamber for 48 hours. Cells were imaged by phase contrast microscopy using a 20x objective. N=2 per condition from one (one experiment, with 2 CRIPSRi guides to CCM2, TLNRD1 or controls), and with 4 images per guide.
j. The normal alignment to flow in control teloHAEC (measured as the angle, relative to flow, of the long axis of each cell) is significantly abrogated in both CCM2 & TLNRD1 KD cells (increased average angle relative to flow). Average values for all cells in each of 4 images for each of 2 guides per target were calculated (35 to 103 cells per image). Significance was assessed by two sided T-test on these average values. N=8. Boxplot center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers. Note that alignment to flow is not completely blocked in CCM2 or TLNRD1 KD cells, since the average angle relative to flow does not reach the 45% value expected if orientation were entirely random.
k. As per (j), but measuring the ratio of the long vs. short axis lengths for each cell (“length/width”) from the fit ellipse function in FiJi.
Extended Data Fig. 11. Additional analysis of ccm2 and tlnrd1 CRISPR and morpholino knockdown zebrafish embryos.
Zebrafish is a model system which has been extensively used to study CCM gene functions, and where ccm2 has been shown to have characteristic effects in heart and vascular development34,35,44,45,78,152.
(a) In-situ analysis of tlnrd1 mRNA expression. tlnrd1 mRNA, detected by the anti-sense in situ probe, is expressed in the head (black arrowhead) and heart (red arrowhead, top left), and in the notochord (white bracket), posterior cardinal vein (PCV, green arrow) and intersegmental vessels (ISVs, blue arrows, bottom panel). This staining pattern is not seen with the negative control sense probe (top right). N=10 from one experiment. This staining pattern is consistent with TLNRD1 being most highly expressed in human endothelial cells (Tabula Sapiens atlas of gene expression, https://tabula-sapiens-portal.ds.czbiohub.org/).
(b) Quantitation of ccm2 & tlnrd1 CRISPR/morpholino heart phenotypes, knockdown efficacy & effects on klf2b expression. The table summarizes the number of embryos injected with each guide, or with tracRNA control (for CRISPR experiments), or with each experimental or control morpholinos at the indicated concentration(s), the number that survived, and the percent that showed a heart phenotype characterized by enlarged atrium, pericardial edema and slow blood flow in tail veins. “ccm2/tlnrd1 (v. ctrl)” summarizes qRT-PCR analysis of knockdown efficacy (ccm2 or tlnrd1 levels in embryos with CRISPR guides to each of these target genes, versus control embryos). “klf2b (v. ctrl)” summarizes qRT-PCR quantification of klf2b in CRISPR or morpholino knockdown animals versus controls. “NA”: Effects on ccm2 or tlnrd1 were not measured for the morpholino studies, because morpholinos generally function by inhibiting translation; and klf2b levels were not tested for the indicated morpholino treatments. See also Supplementary Table 20.
(c) Light microscopic images of CRISPR embryos. Representative images of Zebrafish 3dpf embryos injected with control, ccm2 or tlnrd1 gRNA and Cas9 protein. A: atrium. V: ventricle. pc: pericardial space. For N and experimental replicates see panel (b) and Supplementary Table 20.
(d) Similar heart phenotypes in ccm2 & tlnrd1 morpholino embryos. Knockdown of either tlnrd1 or ccm2 with 100 μM anti-tlnrd1 or anti-ccm2 morpholino caused similar heart defects as seen by CRISPR knockdown, with no heart defects seen using 100 μM control morpholino. For N and experimental replicates see panel (b) and Supplementary Table 20.
(e) Synergistic effect of ccm2 & tlnrd1 morpholinos. As in (d), but showing the synergistic phenotype of 50 μM tlnrd1 & 50 μM ccm2 morpholinos, which, individually, showed no phenotype, but together showed the heart phenotype in 72% of embryos. For N and experimental replicates see panel (b) and Supplementary Table 20.
(f) Vascular phenotype in ccm2 & tlnrd1 morpholino embryos. Representative microangiogram images showing FITC dextran green (2000 kDa) injected in the vasculature in control, ccm2 and tlnrd1 gRNA injected 3 dpf larvae. Brackets mark the thickness of the posterior cardinal vein (PCV). Arrows indicate the intersegmental vessels (ISVs). Experiments were repeated 3 times. N=6 for control, and 5 for ccm2 or tlnrd1 gRNAs, from one experiment.
(g) Ventricle wall thinning in ccm2 & tlnrd1 morpholino embryos. Hematoxylin & eosin (H&E) stained sections of 3dpf embryos. A: atrium. V: ventricle. The space between dotted lines in the ventricle indicates ventricular wall thinning in ccm2 or tlnrd1 CRISPR embryos. Cells within each chamber are blood cells. N=3 for each treatment, from one experiment.
(h) More permeable vasculature in ccm2 & tlnrd1 morpholino embryos. Representative images from vascular permeability analysis in control, ccm2 and tlnrd1 gRNA injected zebrafish at 3 dpf. Red color indicates texas red dextran 70 KD, which was injected into the vasculature before imaging. Green: Green fluorescence protein expression in the vasculature (Tg:Fli GFP). Both ccm2 and tlnrd1 gRNA injected embryos displayed higher levels of red dye in the interspace between the vessels (arrows). Bottom right: Quantitation of permeability (ratio of red dextran in interspace vs. controls. n=10 for control, 13 for ccm2 & 13 for tlnrd1). Significance was assessed by two-sided T-test. Boxplot center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers. For the complete data, see Supplementary Table 31.
(i) tlnrd1 knockdown upregulates klf2b in zebrafish. qRT-PCR for knockdown of tlnrd1 & induction of klf2b in zebrafish embryos treated with CRISPR guides to tlnrd1, or with control tracrRNA. Signal was normalized to Actin, and then to the average for controls. n=9 for klf2b (6 for guide AF, 3 for guide AN.2). n=5 for tlnrd1 (2 for guide AF, 3 for guide AN.2). Quantitation and boxplot features in in (h). For the complete data, see Supplementary Table 30.
(j) TLNRD1 knockdown upregulates KLF2 in TeloHAEC. qRT-PCR for knockdown of TLNRD1 & induction of KLF2 in TeloHAEC with Cas9-guide nucleofection knock down of TLNRD1 (or non-targeting guides, “Control”). Signal was normalized to GAPDH, and then to the average for controls. n=4 separate samples. Quantitation and boxplot features as in (h). For the complete data, see Supplementary Table 30.
Extended Data Fig. 12. Application of V2G2P to other traits and other cell models.
a. Venn diagram of V2G2P genes for coronary artery disease (CAD), pulse pressure (PP), and mean arterial pressure (MAP) GWAS traits in teloHAEC (using the same ABC-maps and Perturb-seq data, but disease variants for each trait). For MAP, we prioritized program 8 (ECM organization, AQP1, FDR = 0.0135) and program 15 (KLF2, flow response, FDR = 0.0289). For PP we prioritized program 50 (TGFβ response, FDR = 0.0046) and program 29 (EDN1, wound healing, FDR = 0.0316). Several genes in the PP programs are known to regulate vascular tone and stiffness, including FHL2, SMAD3, and TGFB1153-155.
b. K562 V2G2P programs for mean corpuscular hemoglobin (MCH), mean corpuscular volume (MCV), platelet count (Plt), red blood cell count (RBC), pulse pressure (PP), mean corpuscular hemoglobin concentration (MCHC), average blood glucose level (HbA1c), hemoglobin count (Hb), mean arterial pressure (MAP), diastolic blood pressure (DBP), systolic blood pressure (SBP). Overall, 32 programs were prioritized for 6 GWAS traits, ranging from 27 programs associated with MCH to 2 programs for MCHC. In general, traits that were not relevant to K562 erythroleukemia cells had no K562 programs significantly associated with them (e.g. MAP, DBP & SBP). Programs associated with each trait contained genes related to that trait. For instance, the most significantly-enriched mean corpuscular hemoglobin program was K562 Program 13, which included many hemoglobin genes as well as the known regulators GFI1B156 and CBFA2T3157, while variants associated with platelet count showed most significant enrichment in K562 Program 4, which included genes known to be involved in megakaryocyte differentiation and platelet count such as VASP158 and TPM4159, and which showed high enrichment of motifs for the known megakaryocyte regulators SP1/3160.
Supplementary Material
Acknowledgements
This work was supported by the Variant-to-Function Initiative at the Broad Institute (to R.M.G. and J.M.E.); NHLBI R01HL159176 and R01HL164811 (to J.M.E. and R.M.G.); the NHGRI Impact of Genomic Variation on Function Consortium (UM1HG011972 to J.M.E.); a NHGRI Genomic Innovator Award (R35HG011324 to J.M.E.); Gordon and Betty Moore and the BASE Research Initiative at the Lucile Packard Children’s Hospital at Stanford University (J.M.E.); a NIH Pathway to Independence Award (K99HG009917 and R00HG009917 to J.M.E.); the Novo Nordisk Foundation (NNF21SA0072102); the Harvard Society of Fellows (J.M.E.); an NIH New Innovator Award (DP2HL152423 to R.M.G); NHLBI U01HL166060 (R.M.G); a Khoury Innovation Award and Braunwald Scholar Award (R.M.G); the Broad Institute (E.S.L.); NIH HL70567 (D.M.); Florida Department of Health Cancer Research Chair's Fund Grant 3J-02 (to D.M.); and K08DK129824 (to M.S.T). We thank members of the Engreitz and Gupta research groups for discussions and technical assistance, Joyce Bischoff and Sana Nasim (Harvard Medical School), for assistance with the TEER assay, Hyung-Jin Yoo for assistance with the Ibidi shear stress assay, Kwangwoon Lee, Minja Velimirovic, and Trevor van Eeuwen for assistance with computation and visualization of modeling data, and the Harvard O2 cluster for computational resources.
Footnotes
Data Analysis: Data was processed using all packages required for running cNMF v1.2 25. We additionally used R-3.6.1 and R-3.6.3 99, edgeR 3.28.0 58,62,64,65, seqLogo 1.52.0, MAST 1.12.0 39, clusterProfiler 3.14.0 66,100, org.Hs.eg.db 3.10.0, Seurat 3.0.2 101, SeuratObject 4.0.0 101-103, stats4 v3.6.1 and SingleCellExperiment 1.8.0 104 for downstream analysis in R. The supporting packages in R used for data processing and figure generation were ggplot2 3.3.5 105, ggpubr 0.4.0, ggrepel 0.9.1, gplots 3.1.1, gridExtra 2.3, scales 1.1.1, cowplot 1.1.1, dplyr 1.0.7, tidyr 1.1.3, textshape 1.7.1, reshape2 1.4.4, stringi 1.7.5 106, conflicted 1.0.4, data.table 1.14.0, purrr 0.3.4, readxl 1.3.1, writexl 1.5.0, ramify 0.3.3, optparse 1.6.6, and all dependencies. Data was further processed using pysuspenders 0.2.6 107, pysam 0.19.1 108, python 2.7.15, and LDSC v1.0.1 109. Other software packages used were kallisto 0.48.0 56, limma 3.42.2 57, Bowtie v 1.3.0 and Bowtie2 v2.4.2 110, Macs2 2.2.7 111, CellRanger 7.0.0 112, FIMO (https://meme-suite.org/meme/meme_5.3.2/tools/fimo), plink v1.90b6.21 113, MAGMA 2, S-LDSC 28,70, PoPS 3 , FloJo v10.8.1, AlphaFold2.3 Multimer v3 81, UCSF ChimeraX v1.61, AlphaPickle 82, Matplotlib v3.7.0, Seaborn (https://seaborn.pydata.org/) and MATLAB R2018a (MathWorks). Phalloidin analysis was done using Fiji/ImageJ 2.9.0/1.53t with the LPX plugin. CRISPRi primer design used CRISPRDesigner, https://github.com/EngreitzLab/CRISPRDesigner.
Data Collection: The Opera Phenix imager (used for phalloidin stain analysis of TeloHAEC) was run using Harmony 4.9.2137.273, Acapella 5.0.1.124082 & Oda 4.9.2137.273. The Zeiss confocal microscope LSM 880 (used for zebrafish imaging) was run using ZEN 2.3 SP1 software. The Sony MA900FP cell sorter (used for FlowFISH) was run using “Cell Sorter Software” v3.2. Other instruments were used with the manufacturer-supplied intrinsic software, including EVOS microscopes, Applied Biosystems QuantStudio 5 (for qRT-PCR) and the ECIS Z-Theta instrument (Applied BioPhysics).
Competing interests
J.M.E. is a shareholder of Illumina, Inc. and 10X Genomics; has received materials from 10X Genomics unrelated to this work, is an equity holder in and consultant for Martingale Labs, Inc., and has received guest speaker honoraria from GSK plc. M.S.T. holds equity and has received consulting fees from ROME Therapeutics, which is not related to this work. G.R.S., R.M.G., J.M.E, H.K and X.R.M. are inventors on a provisional patent related to this work. All other authors declare no competing interests.
Additional Information
Supplementary Information is available for this paper.
Data Availability
Raw and processed data for Perturb-seq, ATAC-seq, H3K27ac ChIP-seq, and RNA-seq in TeloHAEC were deposited in NCBI’s Gene Expression Omnibus under accession number GSE210523. This superseries is composed of subseries: GSE210489 (ATAC-seq), GSE210491 (ChIP-seq), GSE210522 (bulk RNAseq of cytokine-treated parental lines & single guide CRISPRi knockdowns), GSE232400 (bulk RNAseq of cells under flow, and MAP3K3 double knockdowns), GSE212396 (pilot scRNA-seq studies) and GSE210681 (comprehensive Perturb-seq).
Other datasets used in these studies: CAD lead GWAS variants were derived from both Aragam et al.12 and Harst et al.10, PheWas data was from Aragam et al.12, GWAS summary statistics for other traits, and finemapping analysis, were from Hilary Finucane and Jacob Ulirsch’s analysis of UKBB data (https://www.finucanelab.org/data), coding and splice site annotations were from the RefGene database (from the UCSC Genome Browser dated 24 June 2017) 68, 1000 Genome European ancestry LD data was accessed using “plink --ld-window-kb 1000 --ld-window 99999 --ld-window-r2 0.9”, TF binding site information was from HOCOMOCO v11 human full scan motifs (https://hocomoco11.autosome.ru/downloads_v11), gene sets were from the Molecular Signatures Database (MSigDB) 67. We also used scRNAseq data from explanted human right coronary artery endothelial cells 69, endothelial cis-regulatory elements derived from snATAC-seq 72, ENCODE datasets ENCSR000EVW (GATA2 ChIP-seq on HUVEC) and ENCSR000EOB (DNase-seq and DGF on HMVEC-dLy-Neo), protein structure models for KRIT1 (UniProt O00522), CCM2 (Uniprot Q9BSQ5, with and without deletion of residues 417-444), PDCD10 (Uniprot Q9BUL8), and TLNRD1 (Uniprot Q9H1K6). Previous prioritization calls for genes in CAD GWAS loci were from these studies: Aragam et al.12, Hodonsky et al.96, Li et al.97, OpenTarget L2G 98, Stolze et al.29, van der Harst and Verweij 10 and Wunnemann et al.30 (with details in the Methods).
Code Availability
The Variant-to-Gene-to-Program (V2G2P) approach snakemake pipeline is available at: https://github.com/EngreitzLab/V2G (V2G)
https://github.com/EngreitzLab/cNMF_pipeline/ (G2P and V2G2P enrichment).
References
- 1.Uffelmann E. et al. Genome-wide association studies. Nature Reviews Methods Primers 1, 1–21 (2021). [Google Scholar]
- 2.de Leeuw CA, Mooij JM, Heskes T & Posthuma D MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weeks EM et al. Leveraging polygenic enrichments of gene features to predict genes underlying complex traits and diseases. medRxiv (2020) doi: 10.1101/2020.09.08.20190561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visscher PM et al. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet 101, 5–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pers TH et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun 6, 5890 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claussnitzer M. et al. A brief history of human disease genetics. Nature 577, 179–189 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westra H-J & Franke L From genome to function by studying eQTLs. Biochim. Biophys. Acta 1842, 1896–1902 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Maurano MT et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nasser J. et al. Genome-wide enhancer maps link risk variants to disease genes. Nature 593, 238–243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Harst P, van der Harst P & Verweij N Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circulation Research 122, 433–443 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tcheandjieu C. et al. Large-scale genome-wide association study of coronary artery disease in genetically diverse populations. Nat. Med 28, 1679–1692 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aragam KG et al. Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. Nat. Genet 54, 1803–1815 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gimbrone MA Jr & García-Cardeña G Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ. Res 118, 620–636 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta RM et al. A Genetic Variant Associated with Five Vascular Diseases Is a Distal Regulator of Endothelin-1 Gene Expression. Cell 170, 522–533.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner AW et al. Single-nucleus chromatin accessibility profiling highlights regulatory mechanisms of coronary artery disease risk. Nat. Genet 54, 804–816 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pepin ME & Gupta R The Role of Endothelial Cells in Atherosclerosis: Insights from Genetic Association Studies. Am. J. Pathol (2023) doi: 10.1016/j.ajpath.2023.09.012. [DOI] [PubMed] [Google Scholar]
- 17.Dixit A. et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1866.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adamson B. et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell 167, 1867–1882.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Replogle JM et al. Mapping information-rich genotype-phenotype landscapes with genome-scale Perturb-seq. Cell 185, 2559–75.e38 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Datlinger P. et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouïs D, Hospers GA, Meijer C, Molema G & Mulder NH Endothelium in vitro: a review of human vascular endothelial cell lines for blood vessel-related research. Angiogenesis 4, 91–102 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Fulco CP, Nasser J, Jones TR & Munson G Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nature Genetics 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norman TM et al. Exploring genetic interaction manifolds constructed from rich single-cell phenotypes. Science 365, 786–793 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris JA et al. Discovery of target genes and pathways at GWAS loci by pooled single-cell CRISPR screens. Science 380, eadh7699 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotliar D. et al. Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-Seq. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nichol D & Stuhlmann H EGFL7: a unique angiogenic signaling factor in vascular development and disease. Blood 119, 1345–1352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brütsch R. et al. Integrin cytoplasmic domain-associated protein-1 attenuates sprouting angiogenesis. Circ. Res 107, 592–601 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stolze LK et al. Systems Genetics in Human Endothelial Cells Identifies Non-coding Variants Modifying Enhancers, Expression, and Complex Disease Traits. Am. J. Hum. Genet 106, 748–763 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wünnemann F. et al. Multimodal CRISPR perturbations of GWAS loci associated with coronary artery disease in vascular endothelial cells. PLoS Genet. 19, e1010680 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stacey D. et al. ProGeM: a framework for the prioritization of candidate causal genes at molecular quantitative trait loci. Nucleic Acids Res. 47, e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jagadeesh KA et al. Identifying disease-critical cell types and cellular processes across the human body by integration of single-cell profiles and human genetics. bioRxiv (2021) doi: 10.1101/2021.03.19.436212. [DOI] [Google Scholar]
- 33.GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snellings DA et al. Cerebral Cavernous Malformation: From Mechanism to Therapy. Circ. Res 129, 195–215 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Z. et al. The cerebral cavernous malformation pathway controls cardiac development via regulation of endocardial MEKK3 signaling and KLF expression. Dev. Cell 32, 168–180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riolo G, Ricci C & Battistini S Molecular Genetic Features of Cerebral Cavernous Malformations (CCM) Patients: An Overall View from Genes to Endothelial Cells. Cells 10, 704 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gingras AR et al. Central Region of Talin Has a Unique Fold That Binds Vinculin and Actin. Journal of Biological Chemistry 285, 29577–29587 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cowell AR et al. Talin rod domain–containing protein 1 (TLNRD1) is a novel actin-bundling protein which promotes filopodia formation. J. Cell Biol 220, e202005214 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finak G. et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luck K. et al. A reference map of the human binary protein interactome. Nature 580, 402–408 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fisher OS et al. Structural basis for the disruption of the cerebral cavernous malformations 2 (CCM2) interaction with Krev interaction trapped 1 (KRIT1) by disease-associated mutations. J. Biol. Chem 290, 2842–2853 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Draheim KM et al. CCM2-CCM3 interaction stabilizes their protein expression and permits endothelial network formation. J. Cell Biol 208, 987–1001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Z. et al. Cerebral cavernous malformations arise from endothelial gain of MEKK3-KLF2/4 signalling. Nature 532, 122–126 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Renz M. et al. Regulation of β1 integrin-Klf2-mediated angiogenesis by CCM proteins. Dev. Cell 32, 181–190 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Donat S. et al. Heg1 and Ccm1/2 proteins control endocardial mechanosensitivity during zebrafish valvulogenesis. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khera AV et al. Gene Sequencing Identifies Perturbation in Nitric Oxide Signaling as a Nonlipid Molecular Subtype of Coronary Artery Disease. Circ Genom Precis Med 15, e003598 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macek Jilkova Z. et al. CCM proteins control endothelial β1 integrin dependent response to shear stress. Biol. Open 3, 1228–1235 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitehead KJ et al. The cerebral cavernous malformation signaling pathway promotes vascular integrity via Rho GTPases. Nat. Med 15, 177–184 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng X. et al. CCM3 signaling through sterile 20-like kinases plays an essential role during zebrafish cardiovascular development and cerebral cavernous malformations. J. Clin. Invest 120, 2795–2804 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knowles JW et al. Enhanced atherosclerosis and kidney dysfunction in eNOS−/−Apoe−/− mice are ameliorated by enalapril treatment. J. Clin. Invest 105, 451–458 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mueller PA et al. Coronary Artery Disease Risk-Associated Plpp3 Gene and Its Product Lipid Phosphate Phosphatase 3 Regulate Experimental Atherosclerosis. Arterioscler. Thromb. Vasc. Biol 39, 2261–2272 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Denier C. et al. Genotype-phenotype correlations in cerebral cavernous malformations patients. Ann. Neurol 60, 550–556 (2006). [DOI] [PubMed] [Google Scholar]
Additional References
- 53.Fulco CP, Munschauer M, Anyoha R & Munson G Systematic mapping of functional enhancer–promoter connections with CRISPR interference. Science 354, 769–773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thakore PI et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12, 1143–1149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilbert LA et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bray NL, Pimentel H, Melsted P & Pachter L Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Law CW, Chen Y, Shi W & Smyth GK voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y, Lun ATL & Smyth GK From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 5, 1438 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang H. et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547, 173–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marshall JL et al. HyPR-seq: Single-cell quantification of chosen RNAs via hybridization and sequencing of DNA probes. Proc. Natl. Acad. Sci. U. S. A 117, 33404–33413 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hart T & Moffat J BAGEL: a computational framework for identifying essential genes from pooled library screens. BMC Bioinformatics 17, 164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soneson C & Robinson MD Bias, robustness and scalability in single-cell differential expression analysis. Nat. Methods 15, 255–261 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Nygaard V, Rødland EA & Hovig E Methods that remove batch effects while retaining group differences may lead to exaggerated confidence in downstream analyses. Biostatistics 17, 29–39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCarthy DJ, Chen Y & Smyth GK Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu T. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karolchik D, Hinrichs AS & James Kent W The UCSC Genome Browser. Current Protocols in Human Genetics 71, 18.6.1–18.6.33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wirka RC et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat. Med 25, 1280–1289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Finucane HK et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet 50, 621–629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dey KK et al. Contribution of enhancer-driven and master-regulator genes to autoimmune disease revealed using functionally informed SNP-to-gene linking strategies. bioRxiv 2020.09.02.279059 (2021) doi: 10.1101/2020.09.02.279059. [DOI] [Google Scholar]
- 72.Zhang K. et al. A single-cell atlas of chromatin accessibility in the human genome. Cell 184, 5985–6001.e19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hujoel MLA, Gazal S, Hormozdiari F, van de Geijn B & Price AL Disease Heritability Enrichment of Regulatory Elements Is Concentrated in Elements with Ancient Sequence Age and Conserved Function across Species. Am. J. Hum. Genet 104, 611–624 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Padarti A & Zhang J Recent advances in cerebral cavernous malformation research. Vessel Plus 2, 21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wei S. et al. Cerebral Cavernous Malformation Proteins in Barrier Maintenance and Regulation. Int. J. Mol. Sci 21, 275 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fischer A, Zalvide J, Faurobert E, Albiges-Rizo C & Tournier-Lasserve E Cerebral cavernous malformations: from CCM genes to endothelial cell homeostasis. Trends Mol. Med 19, 302–308 (2013). [DOI] [PubMed] [Google Scholar]
- 77.Cullere X, Plovie E, Bennett PM, MacRae CA & Mayadas TN The cerebral cavernous malformation proteins CCM2L and CCM2 prevent the activation of the MAP kinase MEKK3. Proc. Natl. Acad. Sci. U. S. A 112, 14284–14289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kleaveland B. et al. Regulation of cardiovascular development and integrity by the heart of glass-cerebral cavernous malformation protein pathway. Nat. Med 15, 169–176 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Engreitz JM et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Atri DS et al. CRISPR-Cas9 genome editing of primary human vascular cells in vitro. Curr Protoc 1, e291 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jumper J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.mattarnoldbio. mattarnoldbio/alphapickle: Release v1.4.0. (2021). doi: 10.5281/zenodo.5708709. [DOI] [Google Scholar]
- 83.Yang X. et al. A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 8, 659–661 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bray M-A et al. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat. Protoc 11, 1757–1774 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Higaki T. Quantitative evaluation of cytoskeletal organizations by microscopic image analysis. Plant Morphology 29, 15–21 (2017). [Google Scholar]
- 86.Kroll F. et al. A simple and effective F0 knockout method for rapid screening of behaviour and other complex phenotypes. Elife 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu F, Leach LL & Gross JM A CRISPR-Cas9-mediated F0 screen to identify pro-regenerative genes in the zebrafish retinal pigment epithelium. Sci. Rep 13, 3142 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moulton JD & Yan Y-L Using Morpholinos to control gene expression. Curr. Protoc. Mol. Biol Chapter 26, Unit 26.8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hoeppner LH et al. Revealing the role of phospholipase Cβ3 in the regulation of VEGF-induced vascular permeability. Blood 120, 2167–2173 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Y. et al. Dissecting VEGF-induced acute versus chronic vascular hyperpermeability: Essential roles of dimethylarginine dimethylaminohydrolase-1. iScience 24, 103189 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zebrafish embryo medium. Cold Spring Harb. Protoc 2011, db.rec12478 (2011). [Google Scholar]
- 92.Machikhin AS, Volkov MV, Burlakov AB, Khokhlov DD & Potemkin AV Blood Vessel Imaging at Pre-Larval Stages of Zebrafish Embryonic Development. Diagnostics (Basel) 10, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thisse C & Thisse B High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc 3, 59–69 (2008). [DOI] [PubMed] [Google Scholar]
- 94.Sudlow C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hodonsky CJ et al. Integrative multi-ancestry genetic analysis of gene regulation in coronary arteries prioritizes disease risk loci. medRxiv (2023) doi: 10.1101/2023.02.09.23285622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li L. et al. Transcriptome-wide association study of coronary artery disease identifies novel susceptibility genes. Basic Res. Cardiol 117, 6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mountjoy E. et al. An open approach to systematically prioritize causal variants and genes at all published human GWAS trait-associated loci. Nat. Genet 53, 1527–1533 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.R Core Team: R Foundation for Statistical Computing, Vienna, Austria. R A Language and Environment for Statistical Computing. https://www.R-project.org/ (2022). [Google Scholar]
- 100.Yu G, Wang L-G, Han Y & He Q-Y clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stuart T. et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Macosko EZ et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Amezquita RA et al. Orchestrating single-cell analysis with Bioconductor. Nat. Methods 17, 137–145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wickham H. ggplot2. (Springer-Verlag, 2016). [Google Scholar]
- 106.Gagolewski M. stringi: Fast and Portable Character String Processing in R. J. Stat. Softw 103, 1–59 (2022). [Google Scholar]
- 107.Holt J, Huang S, McMillan L & Wang W Read Annotation Pipeline for High-Throughput Sequencing Data. in Proceedings of the International Conference on Bioinformatics, Computational Biology and Biomedical Informatics 605–612 (Association for Computing Machinery, 2013). [Google Scholar]
- 108.Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Langmead B, Wilks C, Antonescu V & Charles R Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics 35, 421–432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gaspar JM Improved peak-calling with MACS2. bioRxiv 496521 (2018) doi: 10.1101/496521. [DOI] [Google Scholar]
- 112.Zheng GXY et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Turner AW et al. Author Correction: Single-nucleus chromatin accessibility profiling highlights regulatory mechanisms of coronary artery disease risk. Nat. Genet 54, 1259 (2022). [DOI] [PubMed] [Google Scholar]
- 115.Emdin CA et al. Phenotypic Consequences of a Genetic Predisposition to Enhanced Nitric Oxide Signaling. Circulation 137, 222–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jones PD et al. JCAD, a Gene at the 10p11 Coronary Artery Disease Locus, Regulates Hippo Signaling in Endothelial Cells. Arterioscler. Thromb. Vasc. Biol 38, 1711–1722 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Klarin D. et al. Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat. Genet 49, 1392–1397 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Touat-Hamici Z. et al. Role of lipid phosphate phosphatase 3 in human aortic endothelial cell function. Cardiovasc. Res 112, 702–713 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lalonde S. et al. Integrative analysis of vascular endothelial cell genomic features identifies AIDA as a coronary artery disease candidate gene. Genome Biol. 20, 133 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Medfai H. et al. Human peroxidasin 1 promotes angiogenesis through ERK1/2, Akt, and FAK pathways. Cardiovasc. Res 115, 463–475 (2019). [DOI] [PubMed] [Google Scholar]
- 121.Damoulakis G. et al. P-Rex1 directly activates RhoG to regulate GPCR-driven Rac signalling and actin polarity in neutrophils. J. Cell Sci 127, 2589–2600 (2014). [DOI] [PubMed] [Google Scholar]
- 122.Naikawadi RP et al. A critical role for phosphatidylinositol (3,4,5)-trisphosphate-dependent Rac exchanger 1 in endothelial junction disruption and vascular hyperpermeability. Circ. Res 111, 1517–1527 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Seals DF et al. The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer Cell 7, 155–165 (2005). [DOI] [PubMed] [Google Scholar]
- 124.Wagner S, Flood TA, O’Reilly P, Hume K & Sabourin LA Association of the Ste20-like kinase (SLK) with the microtubule. Role in Rac1-mediated regulation of actin dynamics during cell adhesion and spreading. J. Biol. Chem 277, 37685–37692 (2002). [DOI] [PubMed] [Google Scholar]
- 125.Lahm H. et al. Congenital heart disease risk loci identified by genome-wide association study in European patients. J. Clin. Invest 131, e141837 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wild PS et al. Large-scale genome-wide analysis identifies genetic variants associated with cardiac structure and function. J. Clin. Invest 127, 1798–1812 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pirruccello JP et al. Deep learning of left atrial structure and function provides link to atrial fibrillation risk. bioRxiv (2021) doi: 10.1101/2021.08.02.21261481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pirruccello JP et al. Genetic analysis of right heart structure and function in 40,000 people. Nat. Genet 54, 792–803 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yu M. et al. Computational estimates of annular diameter reveal genetic determinants of mitral valve function and disease. JCI Insight 7, e146580 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pan CS et al. Adrenomedullin ameliorates the development of atherosclerosis in apoE−/− mice. Peptides 31, 1150–1158 (2010). [DOI] [PubMed] [Google Scholar]
- 131.Nakayama A. et al. Disturbed flow-induced Gs-mediated signaling protects against endothelial inflammation and atherosclerosis. JCI Insight 5, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Iring A. et al. Shear stress-induced endothelial adrenomedullin signaling regulates vascular tone and blood pressure. J. Clin. Invest 129, 2775–2791 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Barkefors I. et al. Exocyst complex component 3-like 2 (EXOC3L2) associates with the exocyst complex and mediates directional migration of endothelial cells. J. Biol. Chem 286, 24189–24199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Netherton SJ, Sutton JA, Wilson LS, Carter RL & Maurice DH Both protein kinase A and exchange protein activated by cAMP coordinate adhesion of human vascular endothelial cells. Circ. Res 101, 768–776 (2007). [DOI] [PubMed] [Google Scholar]
- 135.Sun W. et al. SCUBE1 Controls BMPR2-Relevant Pulmonary Endothelial Function: Implications for Diagnostic Marker Development in Pulmonary Arterial Hypertension. JACC Basic Transl Sci 5, 1073–1092 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lin Y-C et al. Endothelial SCUBE2 Interacts With VEGFR2 and Regulates VEGF-Induced Angiogenesis. Arterioscler. Thromb. Vasc. Biol 37, 144–155 (2017). [DOI] [PubMed] [Google Scholar]
- 137.Wu M-Y et al. Inhibition of the plasma SCUBE1, a novel platelet adhesive protein, protects mice against thrombosis. Arterioscler. Thromb. Vasc. Biol 34, 1390–1398 (2014). [DOI] [PubMed] [Google Scholar]
- 138.Tsumura Y, Toshima J, Leeksma OC, Ohashi K & Mizuno K Sprouty-4 negatively regulates cell spreading by inhibiting the kinase activity of testicular protein kinase. Biochem. J 387, 627–637 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Taniguchi K. et al. Suppression of Sproutys has a therapeutic effect for a mouse model of ischemia by enhancing angiogenesis. PLoS One 4, e5467 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Taniguchi K. et al. Sprouty4 deficiency potentiates Ras-independent angiogenic signals and tumor growth. Cancer Sci. 100, 1648–1654 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Edelstein LC et al. Human genome-wide association and mouse knockout approaches identify platelet supervillin as an inhibitor of thrombus formation under shear stress. Circulation 125, 2762–2771 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zoldhelyi P, Chen ZQ, Shelat HS, McNatt JM & Willerson JT Local gene transfer of tissue factor pathway inhibitor regulates intimal hyperplasia in atherosclerotic arteries. Proc. Natl. Acad. Sci. U. S. A 98, 4078–4083 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang J. et al. Endothelial cell-anchored tissue factor pathway inhibitor regulates tumor metastasis to the lung in mice. Mol. Carcinog 55, 882–896 (2016). [DOI] [PubMed] [Google Scholar]
- 144.White TA et al. Endothelial-derived tissue factor pathway inhibitor regulates arterial thrombosis but is not required for development or hemostasis. Blood 116, 1787–1794 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Westrick RJ et al. Deficiency of tissue factor pathway inhibitor promotes atherosclerosis and thrombosis in mice. Circulation 103, 3044–3046 (2001). [DOI] [PubMed] [Google Scholar]
- 146.Chen D. et al. Expression of human tissue factor pathway inhibitor on vascular smooth muscle cells inhibits secretion of macrophage migration inhibitory factor and attenuates atherosclerosis in ApoE−/− mice. Circulation 131, 1350–1360 (2015). [DOI] [PubMed] [Google Scholar]
- 147.Tsherniak A. et al. Defining a Cancer Dependency Map. Cell 170, 564–576.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Alcid EA & Tsukiyama T ATP-dependent chromatin remodeling shapes the long noncoding RNA landscape. Genes Dev. 28, 2348–2360 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.SenBanerjee S. et al. KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J. Exp. Med 199, 1305–1315 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Coma S. et al. GATA2 and Lmo2 control angiogenesis and lymphangiogenesis via direct transcriptional regulation of neuropilin-2. Angiogenesis 16, 939–952 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ghoussaini M. et al. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 49, D1311–D1320 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hogan BM, Bussmann J, Wolburg H & Schulte-Merker S ccm1 cell autonomously regulates endothelial cellular morphogenesis and vascular tubulogenesis in zebrafish. Hum. Mol. Genet 17, 2424–2432 (2008). [DOI] [PubMed] [Google Scholar]
- 153.Neuman NA et al. The Four-and-a-half LIM Domain Protein 2 Regulates Vascular Smooth Muscle Phenotype and Vascular Tone*. J. Biol. Chem 284, 13202–13212 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang W. et al. Essential Role of Smad3 in Angiotensin II–Induced Vascular Fibrosis. Circ. Res 98, 1032–1039 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tsai S. et al. TGF-β through Smad3 signaling stimulates vascular smooth muscle cell proliferation and neointimal formation. American Journal of Physiology-Heart and Circulatory Physiology (2009) doi: 10.1152/ajpheart.91478.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Crispino JD & Weiss MJ Erythro-megakaryocytic transcription factors associated with hereditary anemia. Blood 123, 3080–3088 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gruber TA & Downing JR The biology of pediatric acute megakaryoblastic leukemia. Blood 126, 943–949 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hauser W. et al. Megakaryocyte hyperplasia and enhanced agonist-induced platelet activation in vasodilator-stimulated phosphoprotein knockout mice. Proc. Natl. Acad. Sci. U. S. A 96, 8120–8125 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pleines I. et al. Mutations in tropomyosin 4 underlie a rare form of human macrothrombocytopenia. J. Clin. Invest 127, 814–829 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Meinders M. et al. Sp1/Sp3 transcription factors regulate hallmarks of megakaryocyte maturation and platelet formation and function. Blood 125, 1957–1967 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data for Perturb-seq, ATAC-seq, H3K27ac ChIP-seq, and RNA-seq in TeloHAEC were deposited in NCBI’s Gene Expression Omnibus under accession number GSE210523. This superseries is composed of subseries: GSE210489 (ATAC-seq), GSE210491 (ChIP-seq), GSE210522 (bulk RNAseq of cytokine-treated parental lines & single guide CRISPRi knockdowns), GSE232400 (bulk RNAseq of cells under flow, and MAP3K3 double knockdowns), GSE212396 (pilot scRNA-seq studies) and GSE210681 (comprehensive Perturb-seq).
Other datasets used in these studies: CAD lead GWAS variants were derived from both Aragam et al.12 and Harst et al.10, PheWas data was from Aragam et al.12, GWAS summary statistics for other traits, and finemapping analysis, were from Hilary Finucane and Jacob Ulirsch’s analysis of UKBB data (https://www.finucanelab.org/data), coding and splice site annotations were from the RefGene database (from the UCSC Genome Browser dated 24 June 2017) 68, 1000 Genome European ancestry LD data was accessed using “plink --ld-window-kb 1000 --ld-window 99999 --ld-window-r2 0.9”, TF binding site information was from HOCOMOCO v11 human full scan motifs (https://hocomoco11.autosome.ru/downloads_v11), gene sets were from the Molecular Signatures Database (MSigDB) 67. We also used scRNAseq data from explanted human right coronary artery endothelial cells 69, endothelial cis-regulatory elements derived from snATAC-seq 72, ENCODE datasets ENCSR000EVW (GATA2 ChIP-seq on HUVEC) and ENCSR000EOB (DNase-seq and DGF on HMVEC-dLy-Neo), protein structure models for KRIT1 (UniProt O00522), CCM2 (Uniprot Q9BSQ5, with and without deletion of residues 417-444), PDCD10 (Uniprot Q9BUL8), and TLNRD1 (Uniprot Q9H1K6). Previous prioritization calls for genes in CAD GWAS loci were from these studies: Aragam et al.12, Hodonsky et al.96, Li et al.97, OpenTarget L2G 98, Stolze et al.29, van der Harst and Verweij 10 and Wunnemann et al.30 (with details in the Methods).
The Variant-to-Gene-to-Program (V2G2P) approach snakemake pipeline is available at: https://github.com/EngreitzLab/V2G (V2G)
https://github.com/EngreitzLab/cNMF_pipeline/ (G2P and V2G2P enrichment).