Summary
Degenerative bone disorders have a significant impact on global health, and regeneration of articular cartilage remains a challenge. Existing cell therapies using mesenchymal stromal cells (MSCs) have shown limited efficacy, highlighting the necessity for alternative stem cell sources. Here, we have identified and characterized MSX1+ mesenchymal progenitor cells in the developing limb bud with remarkable osteochondral-regenerative and microenvironment-adaptive capabilities. Single-cell sequencing further revealed the presence of two major cell compositions within the MSX1+ cells, where a distinct PDGFRAlow subset retained the strongest osteochondral competency and could efficiently regenerate articular cartilage in vivo. Furthermore, a strategy was developed to generate MSX1+PDGFRAlow limb mesenchyme-like (LML) cells from human pluripotent stem cells that closely resembled their mouse counterparts, which were bipotential in vitro and could directly regenerate damaged cartilage in a mouse injury model. Together, our results indicated that MSX1+PDGFRAlow LML cells might be a prominent stem cell source for human cartilage regeneration.
Keywords: MSX1, PDGFRA, limb bud-like cells, cartilage, regeneration, hPSC, differentiation, in vivo
Highlights
-
•
MSX1+ cells in the developing limb bud retain strong osteochondral potential
-
•
MSX1+PDGFRAlow cells were further identified as the key osteochondral progenitors
-
•
MSX1+PDGFRAlow cells were successfully derived from human pluripotent stem cells
-
•
MSX1+PDGFRAlow cells enabled the efficient regeneration of articular cartilage in vivo
In this article, Liao et al. have identified and characterized a group of MSX1+PDGFRAlow mesenchymal progenitors from the developing mouse limb buds that exhibited strong osteochondral-regenerative and microenvironment-adaptive potential. A stepwise induction method was then established to derive MSX1+PDGFRAlow limb mesenchyme-like cells directly from hPSCs for efficient repair of articular cartilage in a joint injury mouse model.
Introduction
Musculoskeletal conditions affect approximately 1.71 billion individuals and are the leading contributor to pain and disability worldwide (Cieza et al., 2021). With the aging population, the incidence of musculoskeletal conditions has continued to rise, and joint disorders such as osteoarthritis are most prevalent in people aged 65 years and older (Hunter et al., 2020; Long et al., 2020). Articular cartilage in the joint has severely limited capability to self-repair because of its distinct anatomy, with chondrocytes encased in an extracellular matrix composed of their secretions, such as collagens, proteoglycans, and other non-collagenous proteins, as well as the absence of a regional blood supply or neural innervations (Kwon et al., 2019). Thus, the majority of current clinical interventions focus on symptom relief and disease management (Arden et al., 2021; Katz et al., 2021) and progression, and patients are often left with no alternative options except surgery.
Cell-based therapies have been recognized as a prominent approach to restoring damaged articular cartilage. Currently, autologous chondrocyte implantation (ACI) is the only US Food and Drug Administration (FDA)-approved cellular treatment (Makris et al., 2015). However, its practicality is limited by several risk factors, such as donor site morbidity due to chondrocyte harvesting, periosteal hypertrophy, surgery-related complications, and age-related concerns (Harris et al., 2010; Madeira et al., 2015). Meanwhile, other stem cells, including mesenchymal stromal cells (MSCs) from multiple sources (Le et al., 2020; Liu et al., 2022; Madeira et al., 2015; McGonagle et al., 2017), skeletal stem cells (SSCs) (Murphy et al., 2020; Ono et al., 2019; Serowoky et al., 2020), and neural crest stem cells (NCSCs) (Achilleos and Trainor, 2012; Dash and Trainor, 2020; Liu and Cheung, 2016), were also being explored pre-clinically and clinically to examine their potential use in cartilage regeneration. Among these cells, MSCs were one of the most intensively studied stem cells in cartilage regeneration, but the results were often less encouraging, with a high incidence of fibrosis tissue formation reported in a clinical trial using endogenous bone marrow-derived MSCs (BM-MSCs) (Steadman et al., 2003) and abnormal cartilage repair observed in 76% patients when using exogenous ones (Koh et al., 2014). One of the key issues for the use of MSCs for cartilage regeneration is its lack of developmental relevance to the joint chondrocytes. Even for BM-MSCs, joint cartilage was already formed when MSCs were harvested, indicating that MSCs and cartilage chondrocytes were at distinct developmental stages, despite MSCs’ capability to differentiate into chondrocytes in vitro. Thus, there is growing enthusiasm to explore and test other more developmentally relevant stem cell sources for cartilage regeneration.
During development, mammalian synovial joint cartilages were formed during the endochondral ossification process of condensed mesenchymal cells in the osteochondral primordium through interzone formation and joint cavitation (Chijimatsu and Saito, 2019). The cell origin of joint cartilage was typically derived from three lineages: neural crest, sclerotome, and lateral plate mesoderm (LPM) (Humphreys et al., 2022). Among them, mesenchymal progenitor cells from LPM give rise to all the appendicular skeleton, such as the radius, humerus, femur, and tibia (Prummel et al., 2020). In mice, the formation of limb buds marks the initial development of the appendicular skeleton, where an interplay of Wnt, Sonic hedgehog (SHH), FGF, and retinoic acid (RA) signaling pathways determines the further patterning and formation of limb bones and cartilage (McQueen and Towers, 2020; Royle et al., 2021). Interestingly, it was found that the transcription factors Msx1 and Msx2 played critical roles in early limb bud development (Bensoussan-Trigano et al., 2011; Lallemand et al., 2005), and Msx1+ mesenchymal progenitors were the key mediator for digit tip regeneration (Lehoczky et al., 2011). Recent publication of single-cell analysis on developing limb bud also indicated Msx1, as well as Lhx2 and Lhx9, marked the naive progenitor population (Markman et al., 2023), suggesting that MSX1+ cells might be a potential cell source for osteochondral regeneration. However, the regenerative capabilities of these cells were not systematically investigated yet and neither was the strategy to derive them from human pluripotent stem cells (hPSCs) established.
In this study, we used a Msx1P2A-tdTomato knock-in reporter mouse model established previously (Hu et al., 2022) to investigate the regenerative potential of limb-bud-derived MSX1+ mesenchymal cells across multiple developmental stages and discovered that MSX1+ progenitors from E10.5 possessed remarkable osteochondral-regenerative and microenvironment-adaptive capabilities. Single-cell RNA sequencing (scRNA-seq) with fate mapping further revealed that two major cell compositions were present in these cells, where a distinct PDGFRAlow subset retained the strongest osteochondral competency and could efficiently regenerate articular cartilage in vivo. Furthermore, we developed a strategy to generate MSX1+PDGFRAlow limb mesenchyme-like (LML) cells from hPSCs that closely resemble their mouse counterparts. These cells were bipotential in vitro and able to directly regenerate damaged cartilage in a mouse injury model. Together, our findings indicated that MSX1+PDGFRAlow LML cells might be a prominent stem cell source for human cartilage regeneration.
Results
Primary MSX1+ mesenchymal progenitors from limb buds exhibited strong osteochondral-regenerative capabilities
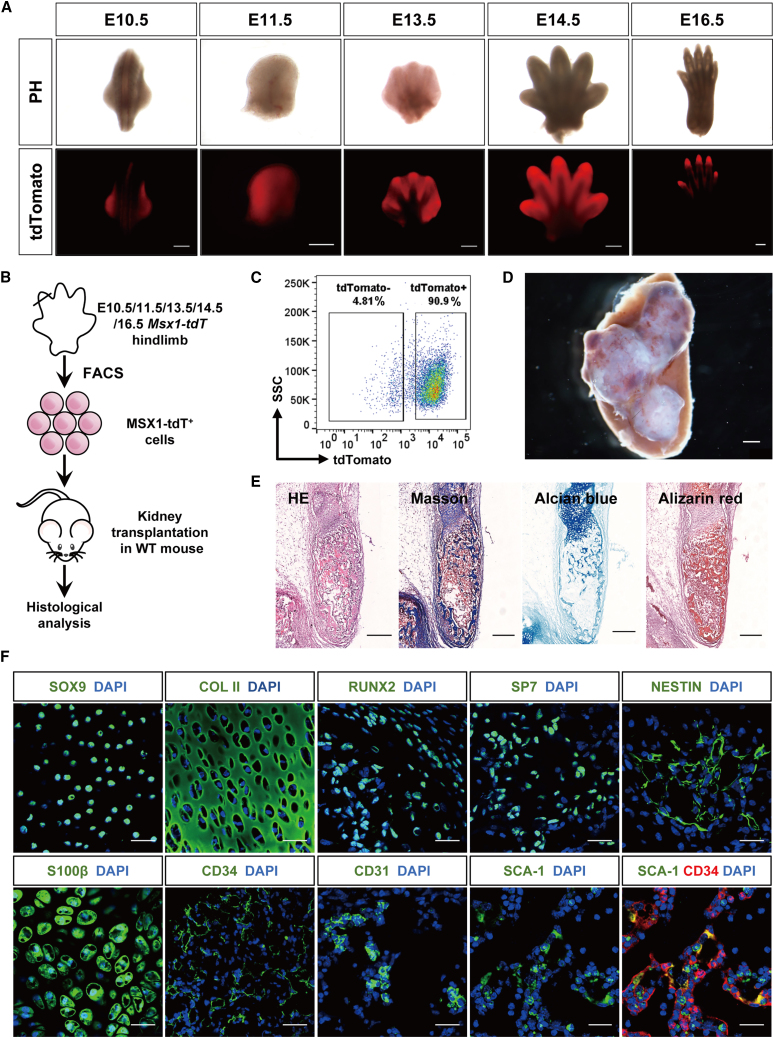
Previously, we constructed a Msx1P2A-tdTomato knockin mouse in which Msx1 expression was tracked through P2A-mediated tdTomato expression (Hu et al., 2022). The tdTomato expression was observed throughout early limb development from embryonic day 10.5 (E10.5) to E16.5 (hindlimb), where it was initially expressed broadly across the limb bud primordium and gradually concentrated on interdigital and distal digit tip regions (Figures 1A and S1A). To evaluate the developmental potential of these cells, MSX1+ cells were sorted by flow cytometry from mouse hindlimb at various stages (E10.5, E11.5, E13.5, E14.5, and E16.5) and transplanted into renal capsules of the recipient mice (Figures 1B, 1C, and S1B). Three weeks post-transplantation, large areas with bone-like structures were formed in the transplanted regions (Figures 1D, S1B, and S1C), especially for E10.5 MSX1+ cells, which yielded the largest grafts and positive staining for Masson, Alcian blue, and Alizarin red (Figure 1E), suggesting them at a more progenitor state. Immunostaining in the graft sections further revealed the expression of various markers, including SOX9 and collagen II (COL II) for chondrocyte and cartilage (Bi et al., 1999; Lefebvre et al., 1997), RUNX2 and SP7 for osteoblasts (Franceschi and Xiao, 2003; Hojo et al., 2016), NESTIN and S100β for stromal (Bernal and Arranz, 2018) and neural cells (Carr and Johnston, 2017), and vascular endothelial cell makers CD31 and CD34, as well as hematopoietic stem cell (HSC) and stromal marker SCA-1 (Tokoyoda et al., 2010; Wilson and Trumpp, 2006) (Figure 1F). Co-localization of SCA-1 and CD34 indicated the association between potential HSCs and endothelial cells (Figure 1F).
Figure 1.
Limb bud MSX1+ mesenchymal progenitors exhibited strong bone regeneration capability
(A) Expression pattern of MSX1 (tdTomato) in primary hindlimbs of different developmental stages. Scale bars: 500 μm.
(B) Schematic illustration of limb bud MSX1+ (tdTomato+) mesenchymal progenitor cells transplantation in vivo. MSX1+ cells isolated from the mouse primary hindlimbs (E10.5, E11.5, E13.5, E14.5, and E16.5 hindlimbs) were first dissociated into single cells and embedded in collagen I for incubation overnight at 37°C. These cells were then transplanted into the kidney capsule of recipient C57BL6 WT mice. Samples were harvested for analysis at three weeks post-transplantation.
(C) Isolation of E10.5 MSX1+ limb bud mesenchymal progenitors by flow cytometry.
(D) Representative image of bone-like tissues formed by transplanted MSX1+ cells under the kidney capsule. Samples were harvested at three weeks post-transplantation. Scale bar: 1 mm.
(E) H&E, Masson, Alcian blue, and alizarin red staining confirmed the presence of collagen, proteoglycan, and calcium salts in the bone-like tissues in MSX1+ cell transplants. Scale bars: 250 μm.
(F) Immunostaining of osteochondral markers in the regenerated bone-like tissues. Chondrocytes: SOX9 and COL II; osteoblasts: RUNX2 and SP7; neural lineage: NESTIN and S100β; vascular endothelial cells (VECs): CD31 and CD34; hematopoietic and stromal cells: SCA-1. Scale bars: 25 μm (SOX9, COL II, RUNX2, and SP7) and 30 μm (NESTIN, S100β, CD31, CD34, and SCA-1).
To investigate whether the cells in the grafts were derived from the transplanted ones, sorted MSX1+ cells were transplanted in a ZsGreen reporter mouse, in which all the host cells were labeled as ZsGreen+ (Figure S1D). Indeed, most regions of the MSX1+ graft were ZsGreen− (Figure S1E). Immunostaining of bone, cartilage, neural, and vascular markers further confirmed that most bone and cartilage signals were from MSX1+ cells, while ∼25%–35% of neural and vascular ones were from the host (Figures S1F and S1G). Moreover, quantitative analysis of the grafts from different MSX1+ cell transplants revealed that MSX1+ cells from E10.5 limb buds had the highest regenerative capabilities (Figure S2). Together, these data indicated that MSX1+ mesenchymal progenitor cells from the developing limb bud possessed strong osteochondral induction potential and could regenerate bone/cartilage-like tissues efficiently upon transplantation.
Limb-bud-derived MSX1+ mesenchymal progenitors could directly repair defects in the joint cartilage
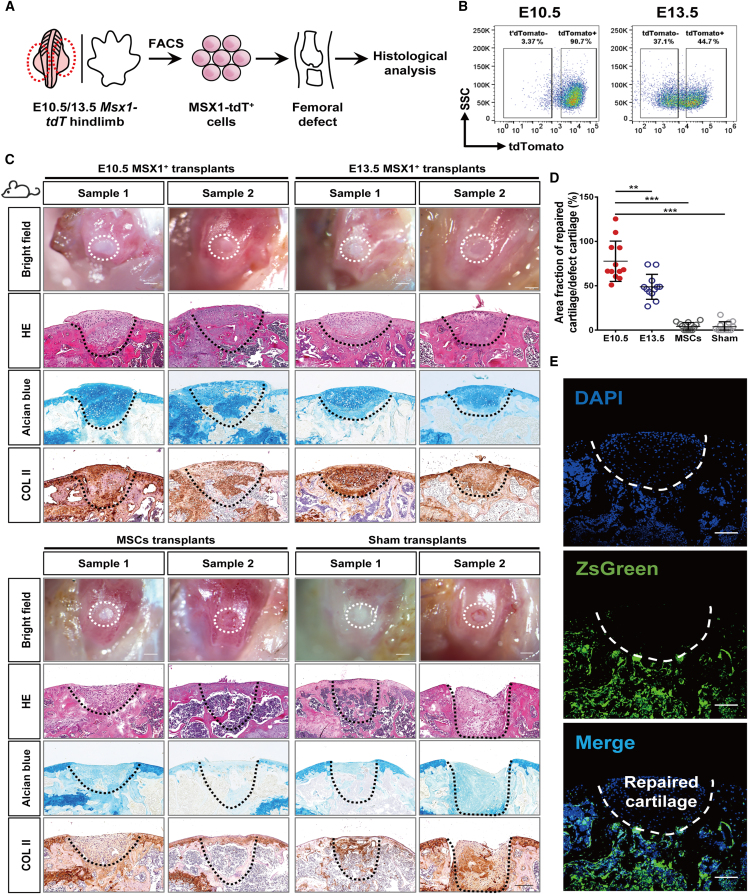
To investigate if the limb-bud-derived mesenchymal progenitors were able to respond to microenvironmental cues and directly regenerate cartilage without prior induction for chondrocyte lineage commitment, MSX1+ cells from E10.5 and 13.5 limb buds were sorted using fluorescence-activated cell sorting (FACS) and transplanted in a joint injury mouse model with 0.8 mm diameter critical sized articular cartilage damage (Figures 2A and 2B) (Fitzgerald et al., 2008). In comparison, besides sham control, MSCs isolated from compact bones were also included (Zhu et al., 2010). The expression of MSC markers as well as their osteochondral differentiation capacities were confirmed before use (Figure S3). Interestingly, three weeks post-transplantation, both E10.5 and E13.5 cells showed remarkable regenerative capabilities to repair the defect sites and form hyaline cartilage. However, the thickness of the cartilage layer was greater in the E10.5 group (Figures 2C and 2D). Histological analysis with Alcian blue staining and COL II immunostaining further confirmed the cartilage formation (Figure 2C). In contrast, the MSC and sham groups exhibited incomplete repair and formed fibrotic tissues but no cartilage (Figure 2C). To verify that the regenerated cartilage was derived directly from the transplanted MSX1+ cells but not any host cells recruited to the damaged site, the experiments were repeated in the H11-ZsGreen transgenic mice, in which host cells would be visualized as ZsGreen+. Indeed, the regenerated hyaline cartilages were completely derived from the transplanted MSX1+ cells but not the host (Figure 2E).
Figure 2.
Microenvironmental adaption of MSX1+ mesenchymal progenitors enabled efficient articular cartilage regeneration
(A) Schematic illustration of limb-bud-derived MSX1+ (tdTomato+) mesenchymal progenitor cells transplantation in the articular cartilage injury model. MSX1+ cells isolated from E10.5 and E13.5 hindlimbs were embedded in collagen I for incubation overnight before being transplanted into the femoral defect sites of recipient mice. Samples were harvested for analysis at three weeks post-transplantation. Transplantation of MSCs was used for comparison. Animals transplanted with collagen matrix only were used as the sham control.
(B) Isolation of MSX1+ mesenchymal progenitors by flow cytometry.
(C) MSX1+ mesenchymal progenitors could efficiently regenerate articular cartilage at the injury site. Representative pictures of two samples with bright-field (BF; white dashed line) and H&E (black dashed line) tissue morphology are shown. Alcian blue and COL II staining were used to detect cartilage formation. Scale bars: 1 mm (BF) and 200 μm (H&E, Alcian blue, and COL II).
(D) Quantitative analysis of cartilage repair efficiency in defect sites. The results confirmed that E10.5 MSX1+ cells had the most efficient regenerative potential. Error bars represent data from twelve sections of six mice from three independent experiments (mean ± SD). Statistics: one-way ANOVA followed by Tamhane’s T2 post hoc multiple comparisons using SPSS version 22.0. ∗∗p < 0.01 and ∗∗∗p<0.001.
(E) Regenerated articular cartilage was derived from transplanted MSX1+ cells. Cells were transplanted in ZsGreen-expressing hosts to verify the cell origin of regenerated cartilage tissues. Immunofluorescence images showing repaired cartilage tissues were ZsGreen negative. Scale bars: 100 μm.
Together, these data suggested that the primary limb-bud-derived MSX1+ mesenchymal progenitors were highly adaptive to the local microenvironment, and upon transplantation in the joint region, could directly regenerate hyaline cartilage without recruitment of host cells.
scRNA-seq analysis revealed the osteochondral potential of limb-bud-derived MSX1+PDGFRAlow progenitors
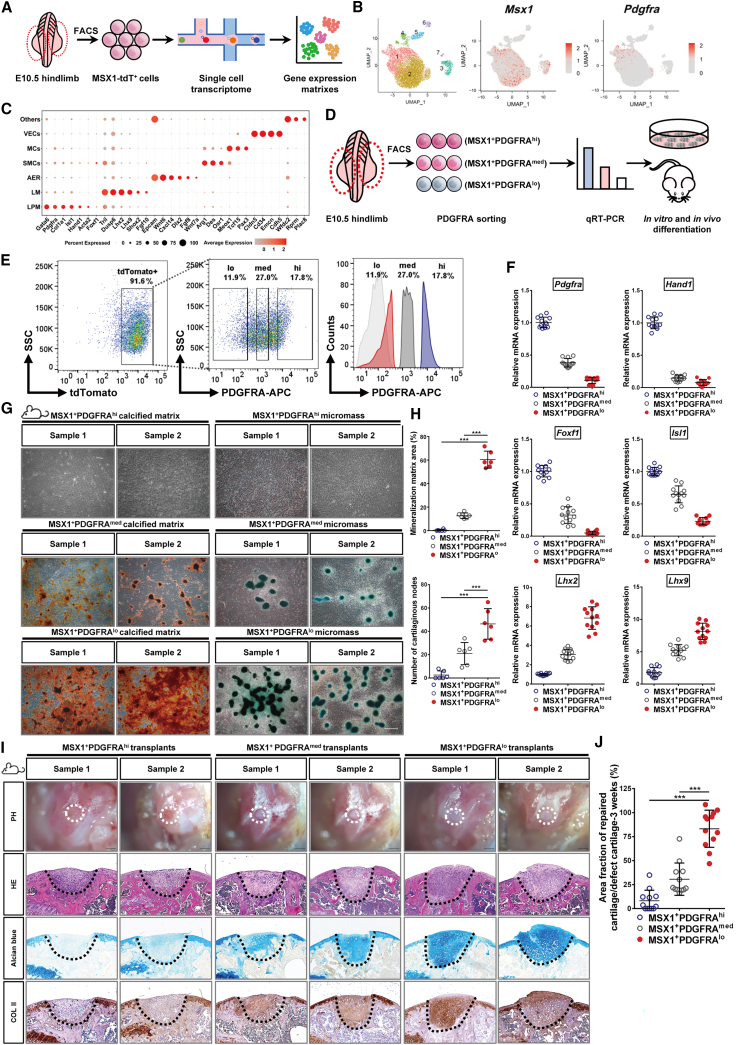
To investigate what cell populations within the developing limb buds might retain the osteochondral differentiation capabilities, single-cell transcriptome analysis was performed using E10.5 mouse limb buds (Figure 3A). Cluster analysis with UMAP (uniform manifold approximation and projection) identified seven cell subgroups, each with unique gene expression features (Figures 3B and 3C). Among them, Msx1 expression was detected in clusters 1, 2, and 3 (Figure 3B). Clusters 1 and 2 were further defined as LPM and limb mesenchyme (LM), respectively, on the basis of their marker expression (Gata6, Pdgfra, and Hand1 in cluster 1; Lhx2, Lhx9, and Dusp6 in cluster 2) (Figure 3C). Cluster 3 was defined as the apical ectodermal ridge (AER), as it expressed Epcam, Wnt6, and Fgf8 (Figure 3C). Pseudotime analysis of mesenchymal cells, identified through Prrx2 and Twist-1 expression (Figure S4A), revealed a differentiation path from LPM to LM (Figures S4B–D), suggesting that LPM cells were at a more progenitor state in development. As LPM was positive for Pdgfra expression, which was also a surface marker (Figure 3C), we separated the limb-bud-derived MSX1+ cells into three groups, PDGFRAhigh, PDGFRAmedium, and PDGFRAlow (Figure 3D) and using FACS to enrich each cell population (Figure 3E). qRT-PCR analysis further confirmed correct cell sorting and demonstrated that MSX1+PDGFRAhigh cells were highly enriched with LPM markers such as Foxf1, Hand1, and Isl1, while MSX1+PDGFRAlow cells were enriched with LM ones such as Lhx2 and Lhx9 (Figure 3F).
Figure 3.
A distinct PDGFRAlow subset retained the osteochondral competency of MSX1+ mesenchymal progenitors
(A) Schematic diagram showing scRNA-seq analysis of E10.5 limb bud MSX1+ (tdTomato+) mesenchymal progenitor cells.
(B) Seven cell clusters were identified from scRNA-seq analysis (UMAP plots). Expression of Msx1 and Pdgfra are shown.
(C) Specific marker expression of different cell clusters. Cluster 1: lateral plate mesoderm (LPM); cluster 2: limb mesenchyme (LM); cluster 3: apical ectodermal ridge (AER).
(D) Schematic diagram of MSX1+ cell sorting and characterization strategy.
(E) Isolation of MSX1+ subpopulations by cell sorting. Left: flow cytometry analysis of MSX1+ cells from E10.5 embryos. Middle and right: gating strategy to isolate PDGFRA-expressing cells.
(F) qRT-PCR analysis of marker gene expressions for LPM (Foxf1, Hand1, Pdgfra, and Isl1) and LM (Lhx2 and Lhx9) in sorted cells. Error bars represent data from four independent experiments with triplicates.
(G) Confirmation of the osteochondral competency of MSX1+PDGFRAlow subpopulation in vitro. Differentiated cells from the sorted subsets were stained with alizarin red for osteogenic (left) and Alcian blue for chondrogenic lineages (right). Scale bars: 100 μm.
(H) Quantitative analysis of the differentiated mineralized matrix and cartilaginous nodes by sorted cells. MSX1+PDGFRAlow cells formed the largest area of the mineralized matrix and the highest number of cartilage nodes. Error bars represent data from six samples of three independent experiments (mean ± SD). Statistics: one-way ANOVA followed by Tamhane’s T2 (mineralized matrix) and Tukey (cartilaginous nodes) post hoc multiple comparisons using SPSS version 22.0. ∗∗∗p < 0.001.
(I) MSX1+PDGFRAlow cells could efficiently regenerate articular cartilage in vivo. Representative images of 3 week samples are shown. Alcian blue and COL II staining (black dashed line) were used to detect cartilage formation. Scale bars: 1 mm (BF) and 200 μm (H&E, Alcian blue, and COL II).
(J) Quantitative analysis of cartilage repair in the defect sites at 3 weeks. Error bars represent data from twelve sections of six mice in three independent experiments (mean ± SD). Statistics: one-way ANOVA followed by Tukey post hoc multiple comparisons using SPSS version 22.0. ∗∗∗p < 0.001.
To evaluate the osteochondral competence of the sorted cells, in vitro differentiation assays for osteogenesis and chondrogenesis were carried out using sorted PDGFRAhigh, PDGFRAmedium, and PDGFRAlow MSX1+ cells. The results demonstrated that the MSX1+PDGFRAlow cells retained the highest potential for both types of differentiation, as evidenced by the formation of more osteoblastic and cartilaginous nodules compared with the other two cells (Figures 3G and 3H). Renal capsule transplantation experiments further confirmed these findings, where MSX1+PDGFRAlow cells gave rise to the largest bone-like tissues, while tissues formed by MSX1+PDGFRAhigh cells were the smallest (Figure S4E). It was further supported by quantitative measurement of the regenerated bone-like tissues (Figure S4F). To evaluate if the MSX1+PDGFRAlow cells were also microenvironment adaptive, sorted cells were transplanted directly in the joint detect sites in the injury mouse model. Indeed, the cells exhibited remarkable cartilage regenerative potential, even without prior induction of chondrocyte lineage commitment (Figures 3I and 3J), and the regenerated cartilage was still maintained after 8 weeks (Figures S4G and S4H). Thus, these results indicated that among the primary cells derived from the developing limb buds, MSX1+PDGFRAlow cells were the key progenitors that retained high osteochondral potential and could adapt to the local microenvironment and regenerate hyaline cartilage efficiently.
Stepwise induction of MSX1+PDGFRAlow LML cells from hPSCs
To test if the developmental trajectory and cell compositions were conserved between human and mouse limb buds, we reanalyzed scRNA-seq data from human embryos at 5 weeks post-conception (WPC) (He et al., 2021). Hindlimb cells were identified through specific markers expressed at this stage, including Pdgfra and Sox9 (Xi et al., 2020), as well as Tbx4 (Ranganath et al., 2020) (Figure S5A). LPM and LM cells within the hindlimb populations and AER cells were further identified through their marker expression (LPM: PDGFRA+COL1A2+COL3A1+TSHZ2+; LM: LHX2+HOXC10+HMMR+MKI67+; AER: EPCAM+FGF8+DLX5+KRT8+) (Figure S5B). Pseudotime trajectory analysis indeed confirmed that similar developmental paths from LPM to LM were conserved in human cells as well (Figures S5C and S5D). Previously, it was reported that AER played a crucial role in the induction of LM in mouse limbs (Street et al., 2018). Therefore, we analyzed the ligand-receptor signals between the LM and AER cells. CellChat analysis (Jin et al., 2021) revealed that Wnts and FGFs were the main stimulating signals from AER to LM cells (Figure S5E), suggesting that the derivation of human LML cells from hPSCs might also require such signals.
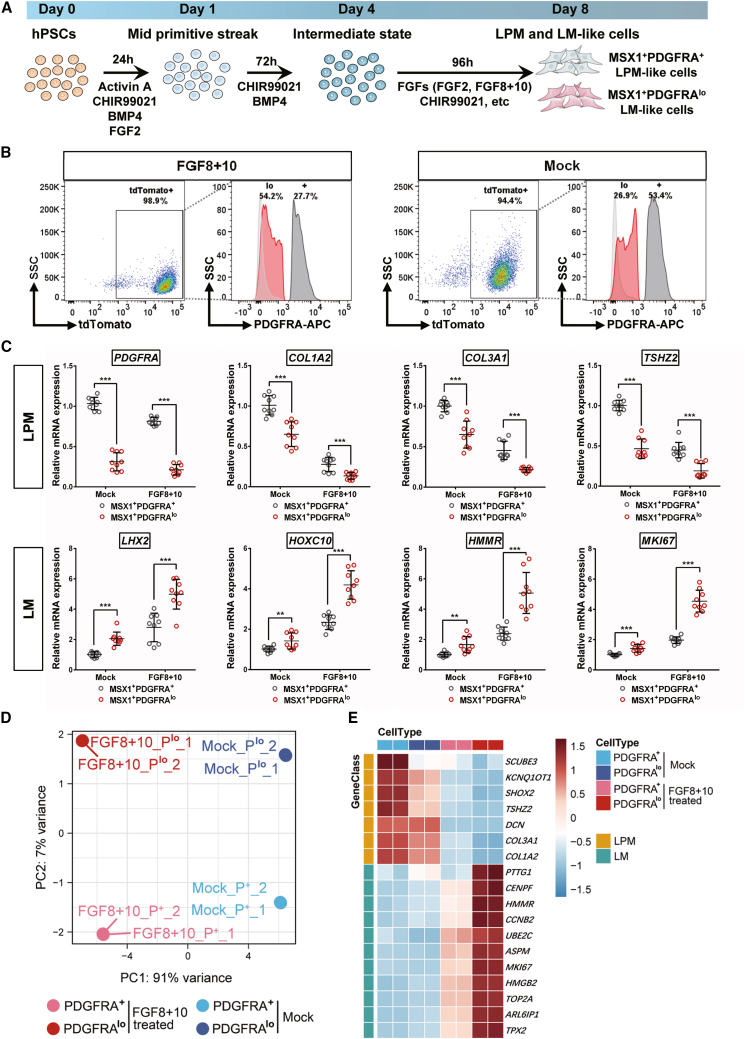
To mimic human limb bud development in vitro and derive LML cells, a MSX1:P2A-tdTomato knockin hPSC cell line was first established by using CRISPR-Cas9 (Figure S6A) and confirmed by Sanger sequencing (Figure S6B). Immunostaining of OCT4 and SOX2 further confirmed the self-renewal state of the knock-in cell line (Figure S6C). To induce LML cells, hPSCs were first differentiated toward mid-primitive streak (MPS) by a cocktail of activin A, CHIR99021 (CHIR), FGF2, and BMP4 and then further induced to an intermediated state by CHIR and BMP4 only for three days before final differentiation using either CHIR99021 to activate Wnt signaling or FGFs (FGF2 or FGF8+10) to stimulate FGF signaling (Figure 4A). As limb bud development is a three-dimensional (3D) process characterized by distinct patterning along the proximal-distal (P-D), anterior-posterior (A-P), and dorsal-ventral (D-V) axes (Huangfu et al., 2008), we compared the effects of both traditional 2D culture and 3D culture using spheroid formation. After 8 days of differentiation, cells in all the treatment groups displayed various degrees of MSX1/tdTomato expression (Figure S6D). qRT-PCR analysis of marker genes for LPM (, PDGFRA, COL1A2, and TSHZ2), LM (LHX2 and HOXC9), and general mesenchymal makers such as HAND2 and PRRX1, as well as the hindlimb-specific marker TBX4, all confirmed that 3D spheroid culture exhibited stronger gene expression induction and the FGF8+10 group was the highest (Figure S6E). The presence of MSX1+PDGFRA+ and MSX1+PDGFRAlow cells was also evaluated by flow cytometry, where 3D-cultured FGF8+10 treated cells, together with mock control, were chosen for further characterization because of robust differentiation of both MSX1+PDGFRA+ and MSX1+PDGFRAlow cells (Figures 4B and S6F).
Figure 4.
Induction of MSX1+PDGFRAlow limb mesenchyme-like cells from human pluripotent stem cells
(A) Schematic illustration of the strategy for stepwise induction of MSX1+ cells from human pluripotent stem cells (hPSCs).
(B) FACS analysis confirmed the induction of MSX1+PDGFRA+ LPM-like and MSX1+PDGFRAlow LM-like cells from 3D-cultured MSX1P2A-tdTomato hPSCs with FGF8+10 and mock treatments.
(C) qRT-PCR analysis confirmed the stronger induction of LM markers in the FGF8+10 treated cells. LPM makers: PDGFRA, COL1A2, COL3A1, and TSHZ2. LM markers: LHX2, HOXC10, HMMR, and MKI67. Error bars represent data from three independent experiments with triplicates. Statistics: independent-sample t test using SPSS version 22.0. ∗∗p < 0.01 and ∗∗∗p < 0.001.
(D) Principal component (PC) analysis of bulk RNA-seq data from hPSC-derived MSX1+PDGFRAlow and MSX1+PDGFRA+ cells. FGF8+10-treated cells were compared with mock control.
(E) Heatmaps of marker gene expression in FGF8+10 treated cells compared with mock control. The range of transcriptional expression is illustrated by a color change as depicted on the extreme right of the figure (dark red correlates to high expression, whereas light blue correlates to low expression).
MSX1+PDGFRA+ and MSX1+ PDGFRA low cells were separated by FACS and the expression of LPM and LM markers were analyzed using qRT-PCR (Figure 4C). For all the LM markers (LHX2, HOXC10, HMMR, and MKI67), MSX1+PDGFRAlow cells from the FGF8+10 treated group exhibited stronger induction during differentiation than mock control (Figure 4C). Transcriptome analysis by RNA-seq comparing FGF8+10 and mock-derived cells further indicated that the cells were at distinct states and FGF8+10 treatment promoted the cells to differentiate toward a LML profile (Figures 4D and 4E). Together, our results indicated that by using 3D spheroid culture with FGF8+10 cytokine stimulation, hPSCs could be induced step by step to acquire LML state, and MSX1+PDGFRAlow LML cells could be readily derived from such differentiation strategy.
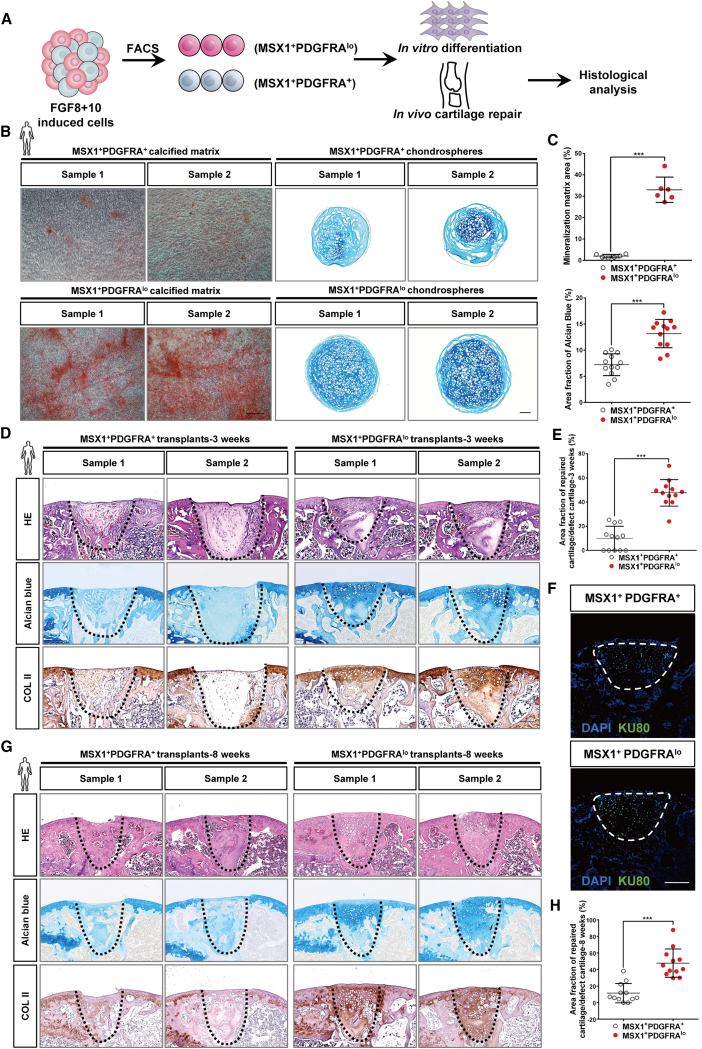
Direct regeneration of joint hyaline cartilage by hPSC-derived MSX1+PDGFRAlow LML cells
To evaluate the developmental potential and cartilage repair abilities of differentiated LPM-like and LML cells, we conducted both in vitro and in vivo differentiation assays using hPSC-derived MSX1+PDGFRA+ and MSX1+PDGFRAlow cells from the 3D-FGF8+10 group (Figure 5A). Indeed, compared with MSX1+PDGFRA+ cells, MSX1+PDGFRAlow LML cells exhibited stronger differentiation capabilities in both osteogenic and chondrogenic differentiation in vitro (Figure 5B), which were also supported by the quantitative measurement of the regenerated calcified matrix and chondrospheres (Figure 5C). We then transplanted both cells into a joint cartilage defect model and found that MSX1+PDGFRAlow cells were also highly microenvironment adaptive, similar to their mouse counterparts, and could directly regenerate articular hyaline cartilage in vivo (Figure 5D). Quantification of the cartilage also confirmed the enhanced regeneration competency of MSX1+PDGFRAlow cells (Figure 5E). Immunostaining of human nuclear protein KU80 indicated that the repairment was not due to the recruitment of the host cells but by the transplanted cells themselves (Figure 5F). In addition, the regenerated cartilage was still maintained after 8 weeks post-transplantation (Figures 5G and 5H).
Figure 5.
MSX1+PDGFRAlow LM-like cells exhibited strong osteochondral competence
(A) Schematic illustration for the strategy to characterize the osteochondral competence of hPSC-derived LM-like cells.
(B) Confirmation of osteogenic and chondrogenic capability of LPM- and LM-like cells in vitro. Stronger staining of alizarin red and Alcian blue was seen in LM-like cells (MSX1+PDGFRAlow) in comparison with LPM-like ones (MSX1+PDGFRA+), suggesting the enhanced osteochondral potential of the LM-like cells. Scale bars: 100 μm.
(C) Quantitative analysis of the alizarin red (top) and Alcian blue (bottom) staining. Msx1+PDGFRAlow cells had better potential to form more mineralized matrix and larger chondrospheres. Top: error bars represent data from six samples in three independent experiments (mean ± SD). Bottom: error bars represent data from twelve chondrospheres from three independent experiments (mean ± SD). Statistics: independent-sample t test using SPSS version 22.0. ∗∗∗p < 0.001.
(D) Efficient regeneration of articular cartilage by LM-like cells in vivo. Representative images are shown. Alcian blue and COL II staining (black dashed line) were used to detect cartilage formation. Scale bars: 250 μm.
(E) Quantitative analysis of the regenerated cartilage. Alcian blue staining was performed in 3 week samples. Error bars represent data from twelve samples of three independent experiments (mean ± SD). Statistics: independent-sample t test using SPSS version 22.0. ∗∗∗p < 0.001.
(F) Confirmation of human origin in the transplanted cells. Representative images of KU80 immunostaining (white dashed line) in the articular cartilage defect sites are shown. Scale bars: 250 μm.
(G) Repaired articular cartilages were maintained after 8 weeks. Alcian blue and COL II staining (black dashed line) were used to detect cartilage formation. Scale bars: 250 μm.
(H) Quantitative analysis of the regenerated cartilage. Error bars represent data from twelve samples of three independent experiments (mean ± SD). Statistics: independent-sample t test using SPSS version 22.0. ∗∗∗p < 0.001.
Thus, these results indicated that the hPSC-derived MSX1+PDGFRAlow LML cells retained osteochondral biopotency and were highly microenvironment adaptive that could regenerate joint articular cartilage without prior induction of chondrocyte fate commitment.
Discussion
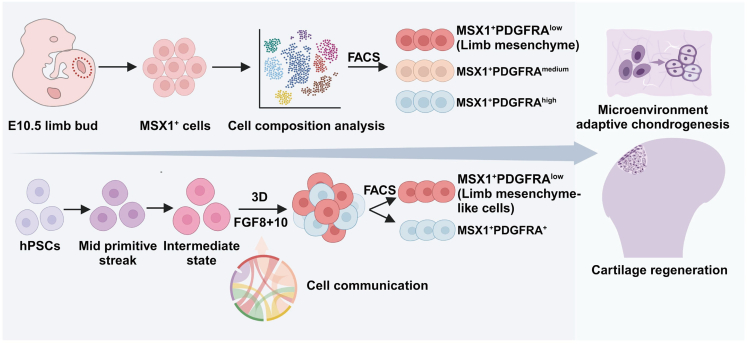
Stem cell-based therapy holds great promise for cartilage regeneration, but identifying suitable seed cells remains a challenge. In the present study, we investigated the osteochondral potential of MSX1+ mesenchymal progenitors isolated from the developing mouse limb buds. Kidney capsule transplantation and animal injury model repairment assays confirmed that these cells retained remarkable osteochondral differentiation capabilities and could adapt to the joint microenvironment and regenerate hyaline cartilage without prior lineage induction. scRNA-seq and pseudotime analysis revealed the developmental trajectory of LPM to LM, where Pdgfra expression was a marker to separate the two populations. Subsequent characterization of LPM (MSX1+PDGFRAhigh) and LM (MSX1+PDGFRAlow) cells further discovered that LM cells retained the strongest osteochondral potential, suggesting them as a promising cell source for cartilage repair. To derive LML cells from hPSCs, we developed a protocol with 3D culture and a combination of FGF8 and FGF10 to promote lineage commitment toward LM. Indeed, hPSC-derived MSX1+PDGFRAlow LML cells exhibited robust osteochondral competency in vitro and could also adapt to the joint microenvironment and directly regenerate damaged cartilage in vivo. Therefore, our work highlighted the potential of MSX1+PDGFRAlow LML cells as a promising cell source for hyaline cartilage regeneration (Figure 6).
Figure 6.
Illustrative model for limb mesenchyme-like cells as a promising stem cell source for cartilage regeneration
An important MSX1+PDGFRAlow cell subpopulation was identified from mouse E10.5 hindlimb; these cells were remarkably osteochondral competent, were microenvironment adaptive, and could directly regenerate articular cartilage. A stepwise protocol was then developed to derive such cells from hPSCs, which is a promising approach for human cartilage regeneration.
Although MSCs have been widely studied for cartilage repair, their heterogeneity and limited expansion pose challenges to clinical translation. In contrast, hPSCs offer a potentially unlimited source of cells with the ability to differentiate into various lineages. Recent studies have shown that allogeneic primate iPSC-derived organoids elicited a minimal immune reaction in repairing articular cartilage defects (Abe et al., 2023), suggesting that hPSC-derived cells might be a viable option for cartilage repair. To identify the proper cell source, our initial research focused on early limb buds, which possessed the potential to develop into a complete limb and could regenerate osteoblasts, chondroblasts, and neural and endothelial cells. In addition, mouse limb bud cells demonstrated superior cartilage repair effects compared with compact bone-derived MSCs. More importantly, we were able to establish a differentiation strategy to derive MSX1+PDGFRAlow LML cells from hPSCs and provide evidence that these cells were like their mouse counterparts in retaining the osteochondral potential.
Finally, it is worth noting that our differentiation strategy would also need further improvement, as the differentiation efficiency is limited currently. In addition to that, although hPSC-derived MSX1+PDGFRAlow cells exhibited upregulated expression of LM enriched signature genes when compared with MSX1+PDGFRA+ counterparts, they did have differences from mouse primary limb mesenchymal cells. One such difference was that for mouse primary MSX1+PDGFRAlow cells, their signature gene expressions were much stronger (usually 30- to 60-fold higher than MSX1+PDGRFRAhigh cells) and thus generally exhibited stronger osteochondral potential. This suggested that the differentiation protocol of hPSC to LM required further optimization. For example, AER cells are known to play critical roles in LM development by providing many inductive signals. Other signals secreted by AER besides FGF8+10 might have synergistic effects on LM derivation from hPSCs. Therefore, further investigations were warranted by combining those signals to derive more matured LML cells. Although our work was ongoing, it was reported that Prrx1+ limb bud-like mesenchymal cells were derived from hPSCs as well (Yamada et al., 2021), which exhibited the potential to form hyaline cartilaginous-like tissues in vitro and in vivo. However, these cells were induced by activating WNT but inhibiting BMP, TGF-β, and HH signaling, while in our case the cells were responsive to FGF8+10 induction, suggesting that the MSX1+PDFGRAlow cells reported here might be more physiologically relevant. Meanwhile, both studies did support that LML cells derived from hPSCs could serve as a promising stem cell source for cartilage regeneration in humans.
Experimental procedures
Resource availability
Lead contact
Zhonghan Li (Zhonghan.Li@scu.edu.cn).
Materials availability
The materials included in this study are available upon reasonable request to the lead contact.
Data and code availability
The datasets generated in the present study are available from the lead contact upon reasonable request. The accession number for the scRNA-seq data reported in this paper is GEO: GSE232586.
Mouse strains and animal care
All animal experiments were approved by the Institutional Animal Care and Use Committee at the College of Life Sciences, Sichuan University. All animals were maintained under standardized conditions with the temperature and light controlled (25°C, 12 h light/dark cycle), in individually ventilated cages, and had free access to food and water. Knockin C57BL6-Msx1P2A-tdTomato mice and H11-ZsGreen mice were custom generated by Biocytogen, Inc. (Beijing, China). Mouse offspring from these strains were routinely genotyped using standard PCR protocols. C57BL6 wild-type (WT) mice and NOD-SCID mice were purchased from GemPharmatech Co., Ltd. (Chengdu, China). C57BL6 WT mice were used as recipients for renal subcapsular and articular cartilage transplantation of mouse MSX1+ cells, while NOD-SCID mice were used as recipients for articular cartilage transplantation of differentiated human LPM- and LML cells.
Single-cell preparation and scRNA sequencing of E10.5 MSX1+ cells
About 20 hindlimb buds were dissected from E10.5 Msx1P2A-tdTomato mice. These limb buds were dissociated into single cells first and resuspended in PBS with 1% BSA. The sorted MSX1+ cells were both counted and adjusted to the concentration of about 1 × 106/mL. Then the suspension was centrifuged at 550 × g for 5 min at 4°C and repeated twice. Cells were counted and cell viability was confirmed by Countess II Automated Cell Counter (catalog #AMQAX1000; Thermo Fisher Scientific). Samples were then used for scRNA-seq with the 10X Genomics system (library preparation and sequencing were performed by Berry Genomics Inc., Beijing, China).
Other experimental procedures can be found in supplemental information.
Acknowledgments
We would like to thank the Core Facilities in the College of Life Sciences and College of Polymer Sciences & Engineering for their technical assistance. This work was supported by the National Key Research and Development Program of China (grants 2022YFA1104401 and 2021YFA1100601), the National Natural Science Foundation of China (grants 32071455 and 32271295), SCU grant 020SCUNL109, the Research and Development Program of West China Hospital of Stomatology Sichuan University (grant RD-03-202106), and the Fundamental Research Funds for the Central Universities (grant SCU2019D013).
Author contributions
Z.L., Q.Z., and Y.L. conceived the study. Y.L., J.X. K.X., and L.Y. performed most of the wet experiments. F.K., H.C., and H.C. performed the bioinformatic analysis. Y.W. and G.Y. provided experimental assistance. Z.L., Q.Z., H.C., W.G., and Y.Y. oversaw the collection of results and data interpretation. Z.L., Q.Z., and Y.L. wrote the manuscript. All authors have seen and approved the final version of the paper.
Declaration of interests
The authors declare no competing interests.
Published: February 29, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2024.02.001.
Contributor Information
Qing Zhu, Email: zhuqing@scu.edu.cn.
Zhonghan Li, Email: zhonghan.li@scu.edu.cn.
Supplemental information
References
- Abe K., Yamashita A., Morioka M., Horike N., Takei Y., Koyamatsu S., Okita K., Matsuda S., Tsumaki N. Engraftment of allogeneic iPS cell-derived cartilage organoid in a primate model of articular cartilage defect. Nat. Commun. 2023;14:804. doi: 10.1038/s41467-023-36408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achilleos A., Trainor P.A. Neural crest stem cells: discovery, properties, and potential for therapy. Cell Res. 2012;22:288–304. doi: 10.1038/cr.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arden N.K., Perry T.A., Bannuru R.R., Bruyère O., Cooper C., Haugen I.K., Hochberg M.C., McAlindon T.E., Mobasheri A., Reginster J.-Y. Non-surgical management of knee osteoarthritis: comparison of ESCEO and OARSI 2019 guidelines. Nat. Rev. Rheumatol. 2021;17:59–66. doi: 10.1038/s41584-020-00523-9. [DOI] [PubMed] [Google Scholar]
- Bensoussan-Trigano V., Lallemand Y., Saint Cloment C., Robert B. Msx1 and Msx2 in limb mesenchyme modulate digit number and identity. Dev. Dynam. 2011;240:1190–1202. doi: 10.1002/dvdy.22619. [DOI] [PubMed] [Google Scholar]
- Bernal A., Arranz L. Nestin-expressing progenitor cells: function, identity and therapeutic implications. Cell. Mol. Life Sci. 2018;75:2177–2195. doi: 10.1007/s00018-018-2794-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W., Deng J.M., Zhang Z., Behringer R.R., de Crombrugghe B. Sox9 is required for cartilage formation. Nat. Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- Carr M.J., Johnston A.P. Schwann cells as drivers of tissue repair and regeneration. Curr. Opin. Neurobiol. 2017;47:52–57. doi: 10.1016/j.conb.2017.09.003. [DOI] [PubMed] [Google Scholar]
- Chijimatsu R., Saito T. Mechanisms of synovial joint and articular cartilage development. Cell. Mol. Life Sci. 2019;76:3939–3952. doi: 10.1007/s00018-019-03191-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieza A., Causey K., Kamenov K., Hanson S.W., Chatterji S., Vos T. Global estimates of the need for rehabilitation based on the Global Burden of Disease study 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2021;396:2006–2017. doi: 10.1016/s0140-6736(20)32340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S., Trainor P.A. The development, patterning and evolution of neural crest cell differentiation into cartilage and bone. Bone. 2020;137 doi: 10.1016/j.bone.2020.115409. [DOI] [PubMed] [Google Scholar]
- Fitzgerald J., Rich C., Burkhardt D., Allen J., Herzka A.S., Little C.B. Evidence for articular cartilage regeneration in MRL/MpJ mice. Osteoarthritis Cartilage. 2008;16:1319–1326. doi: 10.1016/j.joca.2008.03.014. [DOI] [PubMed] [Google Scholar]
- Franceschi R.T., Xiao G. Regulation of the osteoblast-specific transcription factor, Runx2: responsiveness to multiple signal transduction pathways. J. Cell. Biochem. 2003;88:446–454. doi: 10.1002/jcb.10369. [DOI] [PubMed] [Google Scholar]
- Harris J.D., Siston R.A., Pan X., Flanigan D.C. Autologous chondrocyte implantation: a systematic review. J. Bone Joint Surg. Am. 2010;92:2220–2233. doi: 10.2106/jbjs.J.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Yan J., Wang J., Zhao L., Xin Q., Zeng Y., Sun Y., Zhang H., Bai Z., Li Z., et al. Dissecting human embryonic skeletal stem cell ontogeny by single-cell transcriptomic and functional analyses. Cell Res. 2021;31:742–757. doi: 10.1038/s41422-021-00467-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojo H., Ohba S., He X., Lai L.P., McMahon A.P. Sp7/Osterix Is Restricted to Bone-Forming Vertebrates where It Acts as a Dlx Co-factor in Osteoblast Specification. Dev. Cell. 2016;37:238–253. doi: 10.1016/j.devcel.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Duan Y., Wang K., Fu H., Liao Y., Wang T., Zhang Z., Kang F., Zhang B., Zhang H., et al. Dental niche cells directly contribute to tooth reconstitution and morphogenesis. Cell Rep. 2022;41 doi: 10.1016/j.celrep.2022.111737. [DOI] [PubMed] [Google Scholar]
- Huangfu D., Osafune K., Maehr R., Guo W., Eijkelenboom A., Chen S., Muhlestein W., Melton D.A. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Humphreys P.A., Mancini F.E., Ferreira M.J.S., Woods S., Ogene L., Kimber S.J. Developmental principles informing human pluripotent stem cell differentiation to cartilage and bone. Semin. Cell Dev. Biol. 2022;127:17–36. doi: 10.1016/j.semcdb.2021.11.024. [DOI] [PubMed] [Google Scholar]
- Hunter D.J., March L., Chew M. Osteoarthritis in 2020 and beyond: a Lancet Commission. Lancet. 2020;396:1711–1712. doi: 10.1016/s0140-6736(20)32230-3. [DOI] [PubMed] [Google Scholar]
- Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.H., Myung P., Plikus M.V., Nie Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz J.N., Arant K.R., Loeser R.F. Diagnosis and Treatment of Hip and Knee Osteoarthritis: A Review. JAMA. 2021;325:568–578. doi: 10.1001/jama.2020.22171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh Y.G., Choi Y.J., Kwon O.R., Kim Y.S. Second-Look Arthroscopic Evaluation of Cartilage Lesions After Mesenchymal Stem Cell Implantation in Osteoarthritic Knees. Am. J. Sports Med. 2014;42:1628–1637. doi: 10.1177/0363546514529641. [DOI] [PubMed] [Google Scholar]
- Kwon H., Brown W.E., Lee C.A., Wang D., Paschos N., Hu J.C., Athanasiou K.A. Surgical and tissue engineering strategies for articular cartilage and meniscus repair. Nat. Rev. Rheumatol. 2019;15:550–570. doi: 10.1038/s41584-019-0255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand Y., Nicola M.-A., Ramos C., Bach A., Cloment C.S., Robert B. Analysis of Msx1; Msx2 double mutants reveals multiple roles for Msx genes in limb development. Development. 2005;132:3003–3014. doi: 10.1242/dev.01877. [DOI] [PubMed] [Google Scholar]
- Le H., Xu W., Zhuang X., Chang F., Wang Y., Ding J. Mesenchymal stem cells for cartilage regeneration. J. Tissue Eng. 2020;11 doi: 10.1177/2041731420943839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V., Huang W., Harley V.R., Goodfellow P.N., de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol. Cell Biol. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehoczky J.A., Robert B., Tabin C.J. Mouse digit tip regeneration is mediated by fate-restricted progenitor cells. Proc. Natl. Acad. Sci. USA. 2011;108:20609–20614. doi: 10.1073/pnas.1118017108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.A., Cheung M. Neural crest stem cells and their potential therapeutic applications. Dev. Biol. 2016;419:199–216. doi: 10.1016/j.ydbio.2016.09.006. [DOI] [PubMed] [Google Scholar]
- Liu T.P., Ha P., Xiao C.Y., Kim S.Y., Jensen A.R., Easley J., Yao Q., Zhang X. Updates on mesenchymal stem cell therapies for articular cartilage regeneration in large animal models. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.982199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H., Zeng X., Liu Q., Wang H., Vos T., Hou Y., Lin C., Qiu Y., Wang K., Xing D., et al. Burden of osteoarthritis in China, 1990–2017: findings from the Global Burden of Disease Study 2017. Lancet. Rheumatol. 2020;2:e164–e172. doi: 10.1016/S2665-9913(19)30145-6. [DOI] [PubMed] [Google Scholar]
- Madeira C., Santhagunam A., Salgueiro J.B., Cabral J.M.S. Advanced cell therapies for articular cartilage regeneration. Trends Biotechnol. 2015;33:35–42. doi: 10.1016/j.tibtech.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Makris E.A., Gomoll A.H., Malizos K.N., Hu J.C., Athanasiou K.A. Repair and tissue engineering techniques for articular cartilage. Nat. Rev. Rheumatol. 2015;11:21–34. doi: 10.1038/nrrheum.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markman S., Zada M., David E., Giladi A., Amit I., Zelzer E. A single-cell census of mouse limb development identifies complex spatiotemporal dynamics of skeleton formation. Dev. Cell. 2023;58:565–581.e4. doi: 10.1016/j.devcel.2023.02.013. [DOI] [PubMed] [Google Scholar]
- McGonagle D., Baboolal T.G., Jones E. Native joint-resident mesenchymal stem cells for cartilage repair in osteoarthritis. Nat. Rev. Rheumatol. 2017;13:719–730. doi: 10.1038/nrrheum.2017.182. [DOI] [PubMed] [Google Scholar]
- McQueen C., Towers M. Establishing the pattern of the vertebrate limb. Development. 2020;147:dev177956. doi: 10.1242/dev.177956. [DOI] [PubMed] [Google Scholar]
- Murphy M.P., Koepke L.S., Lopez M.T., Tong X., Ambrosi T.H., Gulati G.S., Marecic O., Wang Y., Ransom R.C., Hoover M.Y., et al. Articular cartilage regeneration by activated skeletal stem cells. Nat. Med. 2020;26:1583–1592. doi: 10.1038/s41591-020-1013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono N., Balani D.H., Kronenberg H.M. In: Current Topics in Developmental Biology. Olsen B.R., editor. Academic Press; 2019. Chapter One - Stem and progenitor cells in skeletal development; pp. 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prummel K.D., Nieuwenhuize S., Mosimann C. The lateral plate mesoderm. Development. 2020;147:dev175059. doi: 10.1242/dev.175059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganath P., Perala S., Nair L., Pamu P.K., Shankar A., Murugan S., Dalal A. A newly recognized multiple malformation syndrome with caudal regression associated with a biallelic c.402G>A variant in TBX4. Eur. J. Hum. Genet. 2020;28:669–673. doi: 10.1038/s41431-020-0572-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle S.R., Tabin C.J., Young J.J. Limb positioning and initiation: An evolutionary context of pattern and formation. Dev. Dynam. 2021;250:1264–1279. doi: 10.1002/dvdy.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serowoky M.A., Arata C.E., Crump J.G., Mariani F.V. Skeletal stem cells: insights into maintaining and regenerating the skeleton. Development. 2020;147 doi: 10.1242/dev.179325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steadman J.R., Briggs K.K., Rodrigo J.J., Kocher M.S., Gill T.J., Rodkey W.G. Outcomes of microfracture for traumatic chondral defects of the knee: Average 11-year follow-up. Arthroscopy. 2003;19:477–484. doi: 10.1053/jars.2003.50112. [DOI] [PubMed] [Google Scholar]
- Street K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokoyoda K., Hauser A.E., Nakayama T., Radbruch A. Organization of immunological memory by bone marrow stroma. Nat. Rev. Immunol. 2010;10:193–200. doi: 10.1038/nri2727. [DOI] [PubMed] [Google Scholar]
- Wilson A., Trumpp A. Bone-marrow haematopoietic-stem-cell niches. Nat. Rev. Immunol. 2006;6:93–106. doi: 10.1038/nri1779. [DOI] [PubMed] [Google Scholar]
- Xi H., Langerman J., Sabri S., Chien P., Young C.S., Younesi S., Hicks M., Gonzalez K., Fujiwara W., Marzi J., et al. A Human Skeletal Muscle Atlas Identifies the Trajectories of Stem and Progenitor Cells across Development and from Human Pluripotent Stem Cells. Cell Stem Cell. 2020;27:158–176.e10. doi: 10.1016/j.stem.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada D., Nakamura M., Takao T., Takihira S., Yoshida A., Kawai S., Miura A., Ming L., Yoshitomi H., Gozu M., et al. Induction and expansion of human PRRX1+ limb-bud-like mesenchymal cells from pluripotent stem cells. Nat. Biomed. Eng. 2021;5:926–940. doi: 10.1038/s41551-021-00778-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Guo Z.K., Jiang X.X., Li H., Wang X.Y., Yao H.Y., Zhang Y., Mao N. A protocol for isolation and culture of mesenchymal stem cells from mouse compact bone. Nat. Protoc. 2010;5:550–560. doi: 10.1038/nprot.2009.238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated in the present study are available from the lead contact upon reasonable request. The accession number for the scRNA-seq data reported in this paper is GEO: GSE232586.
Mouse strains and animal care
All animal experiments were approved by the Institutional Animal Care and Use Committee at the College of Life Sciences, Sichuan University. All animals were maintained under standardized conditions with the temperature and light controlled (25°C, 12 h light/dark cycle), in individually ventilated cages, and had free access to food and water. Knockin C57BL6-Msx1P2A-tdTomato mice and H11-ZsGreen mice were custom generated by Biocytogen, Inc. (Beijing, China). Mouse offspring from these strains were routinely genotyped using standard PCR protocols. C57BL6 wild-type (WT) mice and NOD-SCID mice were purchased from GemPharmatech Co., Ltd. (Chengdu, China). C57BL6 WT mice were used as recipients for renal subcapsular and articular cartilage transplantation of mouse MSX1+ cells, while NOD-SCID mice were used as recipients for articular cartilage transplantation of differentiated human LPM- and LML cells.
Single-cell preparation and scRNA sequencing of E10.5 MSX1+ cells
About 20 hindlimb buds were dissected from E10.5 Msx1P2A-tdTomato mice. These limb buds were dissociated into single cells first and resuspended in PBS with 1% BSA. The sorted MSX1+ cells were both counted and adjusted to the concentration of about 1 × 106/mL. Then the suspension was centrifuged at 550 × g for 5 min at 4°C and repeated twice. Cells were counted and cell viability was confirmed by Countess II Automated Cell Counter (catalog #AMQAX1000; Thermo Fisher Scientific). Samples were then used for scRNA-seq with the 10X Genomics system (library preparation and sequencing were performed by Berry Genomics Inc., Beijing, China).
Other experimental procedures can be found in supplemental information.