Abstract
Autism is a heterogeneous neurodevelopmental condition, and functional magnetic resonance imaging–based studies have helped advance our understanding of its effects on brain network activity. We review how predictive modeling, using measures of functional connectivity and symptoms, has helped reveal key insights into this condition. We discuss how different prediction frameworks can further our understanding of the brain-based features that underlie complex autism symptomatology and consider how predictive models may be used in clinical settings. Throughout, we highlight aspects of study interpretation, such as data decay and sampling biases, that require consideration within the context of this condition. We close by suggesting exciting future directions for predictive modeling in autism.
Autism spectrum disorder (hereafter, autism) is a neurodevelopmental condition characterized by difficulties with social communication and interaction as well as restricted and repetitive behaviors (1) and atypical responses to sensory information. There are limited empirically validated treatments for autistic features, especially with respect to medical interventions. Methods that improve our understanding of the brain-based characteristics underlying this condition could ultimately guide clinical research and practice by identifying targets for individualized interventions.
Functional magnetic resonance imaging (fMRI) connectivity analyses (2) have yielded tools that localize brain circuits supporting specific behaviors. These approaches can be used to infer brain-behavior relationships at the individual level that are validated through predictive models. Prediction-based approaches offer a statistically rigorous framework (by using separate data for model training and testing) to study individual differences (3–5), particularly in neurodevelopmental conditions (6). Here, we assert that models have two broad areas of utility in autism: 1) to deepen our understanding of how functional connections coalesce to give rise to the complex autism symptomatology (hereafter, biological insight) and 2) to potentially assist in diagnosis, prognostication, intervention planning, and monitoring of intervention response (hereafter, clinical utility) (7) (Figure 1A).
Figure 1.
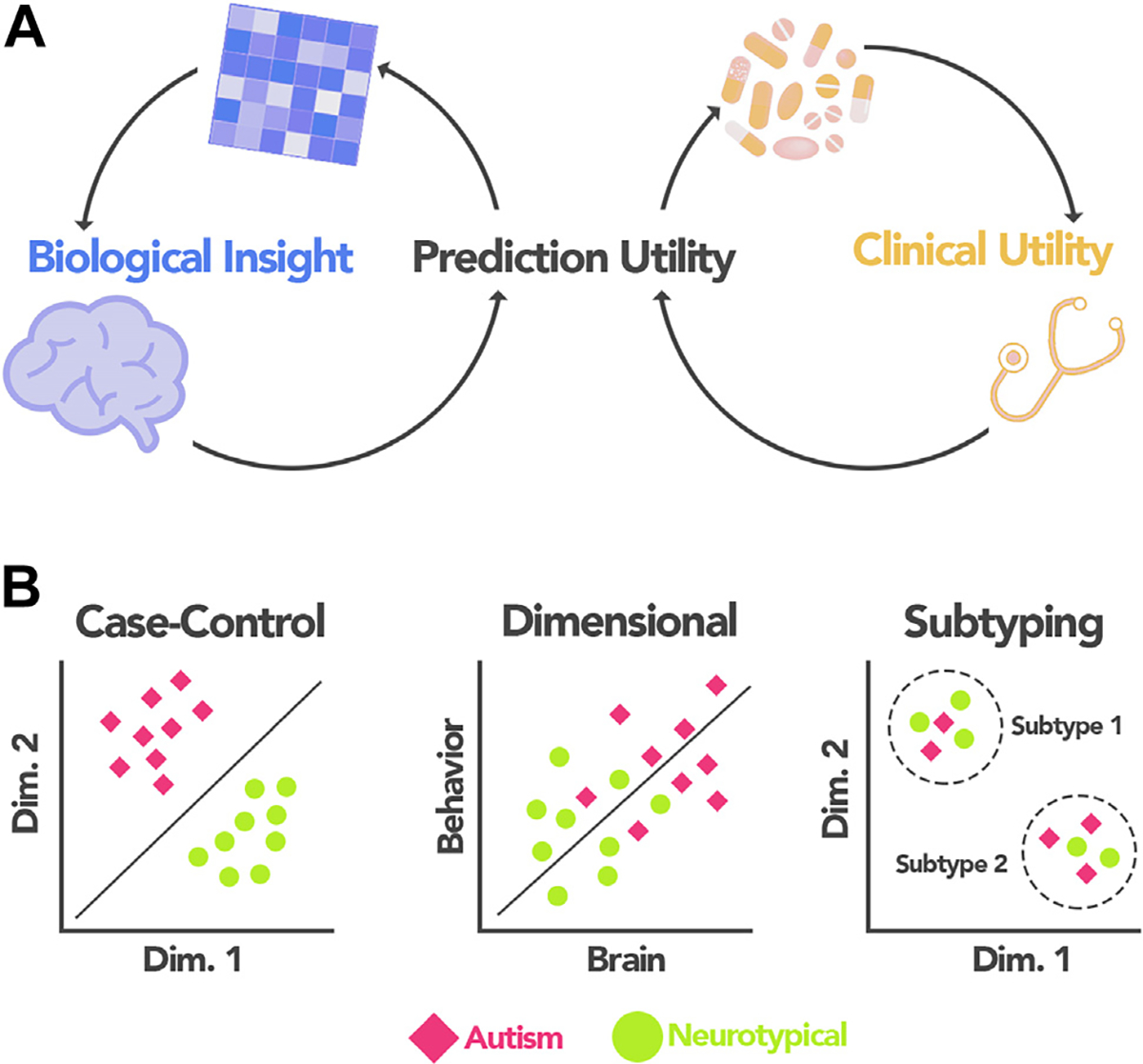
Predictive modeling applications in autism. (A) Prediction-based approaches can serve two needs in autism research: they can help to disentangle the complex brain-based features giving rise to autism symptomatology (biological insight) or be used to potentially inform decisions related to providing care for individuals with autism (clinical utility). Because brain-based insights and clinically useful models are interdependent, their discussion is interwoven throughout the manuscript. (B) Three frameworks for prediction-based modeling using functional connectivity data that we discuss in this review: case-control classification, dimensional prediction, and subtyping. Dim., dimension.
With these two areas in mind, we review the autism predictive modeling literature, focusing on studies using MRI functional connectivity data. Consistent with the lifelong nature of autism, we consider studies across a wide range of participant ages (6 months to 65 years). After detailing autism-specific study design considerations, we discuss three predictive modeling frameworks: case-control classification, dimensional prediction, and subtyping applications (Figure 1B). In each section, we emphasize brain-based insights and identify areas in which we expect predictive models to yield clinical utility. Because brain-based insights underlie clinically useful models (and vice versa), we weave their discussion together throughout the text to stress their interdependence. The goal of this review is to highlight key papers of interest and discuss conceptual considerations that can make autism predictive models more useful (8). Our goal is not to perform an exhaustive, systematic review of machine learning approaches/algorithms in autism prediction studies; the reader is referred to (9–11) for comprehensive reviews summarizing recent progress.
AUTISM-SPECIFIC CONSIDERATIONS FOR PREDICTIVE MODELING
Predictive Models Offer Biological Insight and Potential Clinical Utility
For the purposes of this review, predictive modeling encompasses approaches using statistics to relate MRI functional connectivity measures to phenotypic measures (diagnostic status/symptoms) (4) (see the Supplement for background about predictive modeling/machine learning). These methods separate a dataset into training and testing samples, then apply cross-validation or use external data to test the model. Here, we place emphasis on the functional features (connections and networks) selected through predictive modeling and the potential biological insights/clinical relevance they offer. For example, consider a model implicating connections in frontoparietal areas as important for social attention. Such a model yields biological insight into a complex phenotype by localizing circuits. The model may show clinical utility in the future by predicting which individuals are most likely to respond to behavioral interventions.
Balancing Large Sample Sizes, Concerns About Data Decay, and Site Effects
Predictive modeling studies in participants with autism (12) and neurotypical participants (13) have demonstrated that large samples are needed to obtain reproducible results. In the autism field, using large datasets generally means using data from the Autism Brain Imaging Data Exchange (ABIDE) (14,15) and/or the European Autism Interventions Multicenter Study for Developing New Medications (EU-AIMS) (16). A concern with these samples is data decay (17) and is related to sensitivity and specificity (concepts of relevance for case-control classification studies; sensitivity is an algorithm’s ability to correctly classify individuals with autism who actually have the condition; specificity is an algorithm’s ability to correctly classify neurotypical individuals who do not have the condition). Data decay means that over time, the capacity of a sample to reveal new, statistically significant relationships (such as sensitivity/specificity) decreases as the number of statistical tests performed in the sample increases (17). Concerns about data decay are not unique to autism research; a similar issue has been noted for those using the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset (18).
In addition, ABIDE and EU-AIMS comprise data from multiple sites. Care must be taken to ensure that site effects are not confounding results (4); ComBat is one method investigators have used to minimize site effects (19,20). To help further mitigate concerns about data decay and site effects, other samples could be used to further validate predictive models (21), as has been done with other phenotypes (22). Using multiple datasets to ensure that results hold across samples is one way to increase generalizability of results (23). We describe these issues to increase awareness; we strongly advocate for openly sharing datasets.
Confounds, Tolerability of the Scanning Environment, and Consequences for Predictive Modeling
Confounds, or variables that relate to both the independent and dependent variables in a model, can drive spurious statistical relationships and lead to false conclusions. In-scanner head motion is a notorious confound in measures of functional connectivity (24) and is a concern in the autism field (25). Performing global signal regression decreases motion artifact (26) and strengthens brain-behavior relationships in those with autism (27) and in neurotypical individuals (28). Implementing global signal regression is not without controversy (29); see (30) for a full discussion, including how global signal regression can alter functional correlation structure and affect between-group comparisons.
Barring a consensus approach to remove the effects of head motion, individuals with high-motion data are often excluded in model building (27,31). This practice influences the participants included in predictive modeling studies. Relatedly, individuals with autism who tolerate fMRI scans and produce low-motion data tend to have fewer language/cognitive difficulties and higher IQs. These facts must be kept in mind when considering the feasibility of using predictive modeling in clinical settings. To diversify individuals with autism that meet data quality criteria, the length of the imaging protocol is often minimized by shortening functional scans (<5 minutes) and eliminating task scans. The trade-off is the limited scope of the data obtained. Fewer, shorter scans result in less reliable functional connectomes (32), and resting-state data tends to generate poorer prediction performance (in neurotypical individuals) (31). Exact solutions to confounds depend on analysis goals, but we point the reader to (33) for an examination of confounds in the UK Biobank (and ways to address them). To increase reliability of functional connectomes, we recommend collecting more scanning data (both task and rest data) and/or using methods to increase the quality of scanning data [Framewise Integrated Real-time MRI Monitoring (34), mock scan protocols (35), Inscapes (36)].
Comorbidities and Phenotypic Overlap
Individuals with autism have high rates of co-occurring conditions, including attention-deficit/hyperactivity disorder (ADHD) (37), anxiety disorders (38), and intellectual disabilities (39). Comorbidities can pose challenges for researchers, including how to covary for different diagnoses. Linking analytic approaches (i.e., dimensional and subtyping approaches) is one solution. For example, individuals with and without anxiety symptoms could be grouped into a priori subtypes [as in (40)], and separate dimensional models could be generated to predict autism symptoms in each group. Approaches allowing participants to express subtype characteristics to varying degrees (multidimensional subtyping) (41) have also shown success in parsing heterogeneity.
Sex Imbalance in Autism and Sex-Specific Effects in Predictive Modeling
Sex effects play a role in predictive modeling in autism. There is an estimated 3:1 male:female imbalance in diagnoses (42), and there are sex differences in the neurobiology of autism (43). Females and males tend to exhibit different symptoms, often leading to missed diagnoses in females (44). Further, models predicting fluid intelligence in neurotypical individuals show higher accuracies when generated separately for each sex, and the functional features underlying the models are sex specific (31). Sex specificity aligns with the high degree of accuracy with which sex can be predicted using connectivity data in neurotypical individuals (45). Therefore, investigators should include equal numbers of males and females in analyses (when feasible) and/or build sex-specific models.
We next highlight how all of these factors can impact the biological and clinical utility of predictive modeling in autism as we review three different approaches: case-control classification, dimensional phenotype prediction, and subtype-specific prediction.
CASE-CONTROL CLASSIFICATION: THE CASE FOR FOCUSING ON DIAGNOSIS
Case-control classification studies (12,46–68) constitute most of the prediction literature in autism (Table 1). A strength of these studies is their unambiguous nature: participants are either correctly classified or not. Another strength is the large number compared with dimensional and subtyping prediction studies, allowing for broad trends to be observed. Below, we highlight the biological and clinical utility of a few of these studies through a developmental lens (69), spanning infancy into older adulthood (65+ years).
Table 1.
Representative Autism Case-Control Classification Studies
| Study | Data Source | n | Age, Years, Mean ± SD or Range | Male, % | Algorithm | Features | Validation | Accuracy (Sensitivity, Specificity) | Notes |
|---|---|---|---|---|---|---|---|---|---|
| (68) | Lab-specific | 40 autism 40 NT |
8–42 | 100% | – | FC from 7266 × 7266 ROIs | Internal (leave-one-out CV); external validation in independent sample | 79% (83%, 75%) | External validation results: 71% accuracy, 75% sensitivity, and 69% specificity |
| (50) | Lab-specific | 13 autism 14 NT |
Autism: 21.4 ± 3.9 NT: 22.6 ± 4.2 |
100% | LRC | FC from 102 × 102 ROIs (AAL atlas) (139); seed-based connectivity | Internal (leave-one-out CV) | 77.8% (76.9%, 78.6%) | Whole-brain classification results reported; seed-based accuracies ranged from 70% to 96% |
| (53) | Lab-specific | 29 autism 29 NT |
– | 83% | LRC | FC from 106 × 106 ROIs (AAL atlas) (139) | Internal (leave-one-out CV) | (82.8%, 82.8%) | – |
| (52) | Lab-specific | 20 autism 20 NT |
Autism: 9.96 ± 1.59 NT: 9.95 ± 1.60 |
80% | LRC | Network FC maps (10 components) | Internal (leave-one-out CV); external validation in independent sample | 78% (75%, 80%) | Best performing network (salience network) is shown External validation results: 83% accuracy, 67% sensitivity, 100% specificity |
| (46) | ABIDE | 126 autism 126 NT |
6–36 | 85% | RF | FC from 220 × 220 ROIs (Power atlas) (140) | Bootstrapping CV (one third left out); external validation set | 91% (89%, 93%) | Various SVM pipelines also tested, with the highest accuracy obtained = 66% |
| (51) | Lab-specific | 59 autism 59 NT |
Autism: 17.66 ± 2.72 NT: 18.3 ± 3.05 |
100% | Various tested | Various tested | Internal (leave-one-out, stratified 3-fold, stratified 10-fold CV) | 76.7% (70%, 83.3%) | Pipeline with the highest accuracy and positive predictive values is shown Classifier: SVM; features: FC from 162 × 162 ROIs (Destrieux atlas); validation: leave-one-out CV |
| (49) | ABIDE | 312 autism 328 NT |
6–19 | 84% | NN | FC from 90 × 90 ROIs (AAL atlas) (139) | Internal (leave-one-out, various k-fold CV strategies tested) | 89.4% (92%, 87%) | Showing results for leave-one-out |
| (47) | ABIDE | 112 autism 128 NT |
12–18 | 85% | SVM | FC from 142 × 142 ROIs (Dosenbach atlas) (141); multiple frequency bands used | Internal (leave-one-out, 10-fold CV); leave-one-site-out CV | 79.2% (77.8%, 80.5%) | Showing results for leave-one-out |
| (54) | Lab-specific, ABIDE | Lab-specific: 74 autism, 107 NT ABIDE: 44 autism, 44 NT |
Lab-specific: ~30 ± 8 per site ABIDE: site-specific |
82% | LRC | FC from 140 × 140 ROIs (extended BrainVisa Sulci atlas) (142) | Internal (leave-one-out CV); external validation | 85% (80%, 89%) | Results are reported for leave-one-out; accuracy on external data = 75% |
| (48) | IBIS | 11 autism 48 HR |
2 | 69% | SVM | FC from 230 × 230 ROIs (including nodes from Power atlas) (140) | Internal (leave-one-out CV) | 96.6% (81.8%, 100%) | – |
| (55) | ABIDE | 55 autism 55 NT |
8–19 | 76% | NN | FC from 116 × 116 ROIs (AAL atlas) (139) | Internal (5-fold nested CV) | 86.36% | – |
| (57) | ABIDE | 126 autism 126 NT |
7–36 | 81% | RF, CRF | FC from 220 × 220 ROIs (Power atlas) (140) | External validation sample | 66.7% | Highest accuracy using CRF in validation dataset shown here; highest accuracy using RF in validation data = 71% |
| (12) | ABIDE | 403 autism 468 NT |
Site-specific | Site-specific | SVM | Various tested | Internal (10-fold CV); leave-one-site-out CV | 66.8% | Highest accuracy obtained in leave-one-site-out analyses using a dictionary learning-based atlas (143) |
| (56) | ABIDE | 505 autism 530 NT |
Site-specific | Site-specific | NN | FC from 200 × 200 ROIs (Craddock atlas) (144) | Internal (5-fold, 10-fold CV); leave-one-site-out CV | 70 (74%, 63%) | Showing results for 10-fold CV |
| (58) | ABIDE | 816 autism + NT | 5–65 | – | SVM | GT properties (AAL atlas) (139) | Internal (10-fold CV) | 95% (97%, 91%) | Highest accuracy obtained across various pipelines, age groups (obtained in 30+ year olds) using sparse inverse covariance to estimate connectivity GT properties included various measures of integration, segregation, and centrality |
| (59) | ABIDE | 505 autism 530 NT |
Site-specific | Site-specific | NN | FC from 200 × 200 ROIs (Craddock atlas) (144) | Internal (5-fold, 10-fold CV) | 70.3% (68.3%, 72.2%) | Showing results for 10-fold CV |
| (60) | ABIDE | 408 autism 401 NT |
Autism: 16.5 ± 6.7 NT: 16.8 ± 7.8 |
84% | NN | Various tested | Internal (various k-fold CV strategies tested); leave-one-site-out CV | 73.2% (74.5%, 71.7%) | Showing results for 10-fold CV using a combination of features from AAL + HO + Craddock atlases, along with demographic data |
| (61) | ABIDE | 506 autism 548 NT |
16.86 ± 7.55 | 85% | SVM | FC from 200 × 200 ROIs (Craddock atlas) (144) | Internal (10-fold nested CV) | 72.2% (68.6%, 75.4%) | – |
| (62) | ABIDE | 505 autism 530 NT |
Site-specific | Site-specific | NN | FC from 392 × 392 ROIs (Craddock atlas) (144) | Internal (10-fold CV); leave-one-site-out CV | 70.2% (77.5%, 61.8%) | Showing results for 10-fold CV |
| (63) | ABIDE | 505 autism 530 NT |
– | – | NN | FC from 116 × 116 ROIs (AAL atlas) (139); voxel-wise × ROI FC | Internal (test set validation) | 74% | – |
| (64) | ABIDE | 45 autism 47 NT |
7–15 | 78% | SVM | FC from 116 × 116 ROIs (AAL atlas) (139); various dFC measures | Internal (6-fold nested CV) | 83% (82%, 84%) | Showing results from a combination of FC and dFC measures that resulted in best performance |
| (65) | ABIDE | 505 autism 530 NT |
– | – | NN | FC from 200 × 200 ROIs (Craddock atlas) (144) | Internal (10-fold nested CV); leave-one-site-out CV | 76.4% (77.8%, 75%) | Showing results from 10-fold CV |
| (66) | ABIDE | 403 autism 468 NT |
– | – | NN | FC from 264 × 264 ROIs (Power atlas) (140) | Internal (10-fold CV) | 79.2% | – |
| (67) | ABIDE | 306 autism 350 NT |
6–18 | Varied by analysis | RF | FC from 237 × 237 ROIs [Gordon (145) + HO subcortical (146) + cerebellar (147) atlases] | Bootstrapping CV (one third left out) | 62.5% (60%, 65%) | Main sample size is shown here; different subsamples were tested consisting of n = 200 with autism and n = 200 NT participants Results from subsample including males and females with no ADOS score cutoffs |
Table adapted with permission from (9). Note that studies are arranged in chronological order such that more recent studies are at the bottom of the table.
AAL, automated anatomical labeling; ABIDE, Autism Brain Imaging Data Exchange; ADOS, Autism Diagnostic Observation Schedule; CRF, conditional random forest; CV, cross-validation; dFC, dynamic functional connectivity; FC, functional connectivity; GT, graph theory; HO, Harvard-Oxford atlas; HR, high risk; IBIS, Infant Brain Imaging Study; LRC, logistic regression classifier; NN, neural network; NT, neurotypical; RF, random forest; ROI, region of interest; SVM, support vector machine.
Brain-Based Features Implicated in Autism Classification Differ Across the Life Span
Autism is a lifelong condition, with symptoms changing across an individual’s lifetime (70). The developmental changes are reflected in the neurobiological correlates differentiating individuals with autism from neurotypical participants. For instance, using a Gaussian kernel support vector machine and resting-state data from ABIDE, Kazeminejad and Sotero (58) have shown that the functional features most discriminative of autism status in 5- to 15-year-olds (connections involving the parietal and ventrolateral prefrontal cortices) differ from those most discriminative in 15- to 30-year-olds (with more connections involving the dorsolateral prefrontal cortex and temporal cortex). Across studies, the general theme of developmental effects holds: the functional network organization discriminating autism cases from neurotypical participants seems to be different at different stages of the lifespan (58,71,72). Differences across the lifespan also hold in case-control studies using T1-weighted structural MRI data (73) and align with the dynamic nature of brain maturation (74,75).
From this evidence, we draw two conclusions. First, predictive models that do not generalize across different age groups should not be viewed as model failures (4). Age-specific models for autism case-control classification might be necessary to maximize model utility. This observation is in line with longitudinal work conducted in children (11–18 years old), suggesting that functional networks change at different rates among those with autism and those without (76). Second, given the growing evidence, we can make some overarching observations and devise new hypotheses for testing. For instance, maturational trajectories of cortical areas tend to follow a hierarchical sensory-association axis (74). Unimodal sensory areas mature during childhood, and heteromodal association areas mature later in adolescence and young adulthood. Disruptions have been observed in this hierarchy in autism (77,78), making it intriguing to consider this axis in the context of classification. Perhaps developmental deviations along this functional axis could be used to more accurately delineate between individuals with and without autism? In the future, researchers could investigate this hypothesis in large datasets while keeping in mind autism-specific predictive modeling issues (particularly data decay; most recent case-control studies have been conducted in ABIDE) (Table 1).
Clinical Utility: Toward Early Diagnosis
A major push of clinically relevant research is to identify individuals with autism using objective biological markers at early stages of development (Figure 2), when support services can be most effective (79) [see (80) for a review of imaging markers of autism in infants]. Accurate prediction of case-control status using functional connectivity data has been demonstrated in individuals under 5 years of age (81). In a study of even younger ages, Emerson et al. (48) used functional connectivity data from 6-month-old infants imaged while sleeping and showed that a support vector machine could be used to predict autism status at 24 months of age (Figure 3A). The network models driving correct classification were complex (Figure 3B), comprising short- and long-range connections distributed across the brain, with many of these clustered in the parietal cortex. The neuroanatomical complexity of successful models is a theme we will note throughout this review.
Figure 2.
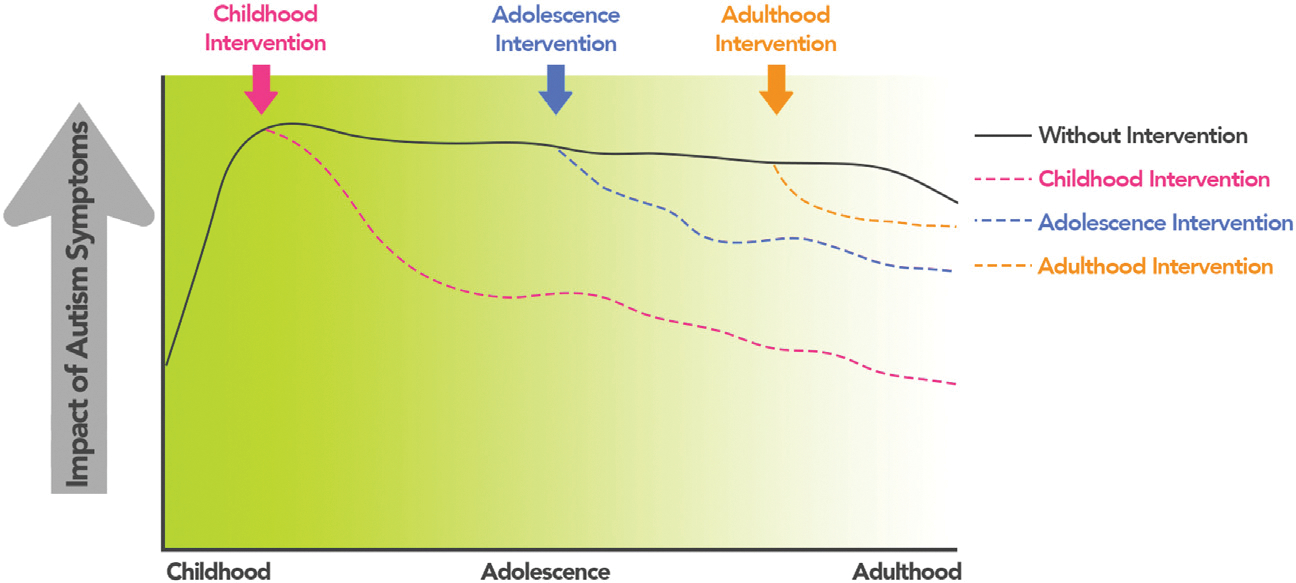
Windows of intervention in autism. The schematic illustrates the clinical utility of correctly identifying a hypothetical individual with autism and then acting on that information to provide appropriate support services. The dark line indicates the individual with autism and the impact of their symptoms (broadly conceived, on the y-axis) over time if no support services are accessed. If autism is diagnosed early (in childhood and adolescence), resources can be allocated to the individual and their caregivers (pink and blue dotted lines, respectively). If correct diagnosis and interventions are delayed, resources can still be leveraged later in life, although they might be less efficacious. The green shading indicates the utility of correct diagnosis and allocation of resources; the darker the green color, the more responsive individuals might be to support services. We stress that this is a hypothetical example; symptoms might not increase from childhood to adolescence, and individuals with late diagnoses might not necessarily have more significant symptoms overall. Indeed, trajectories of symptoms vary across individuals and can vary at different points in the lifespan.
Figure 3.
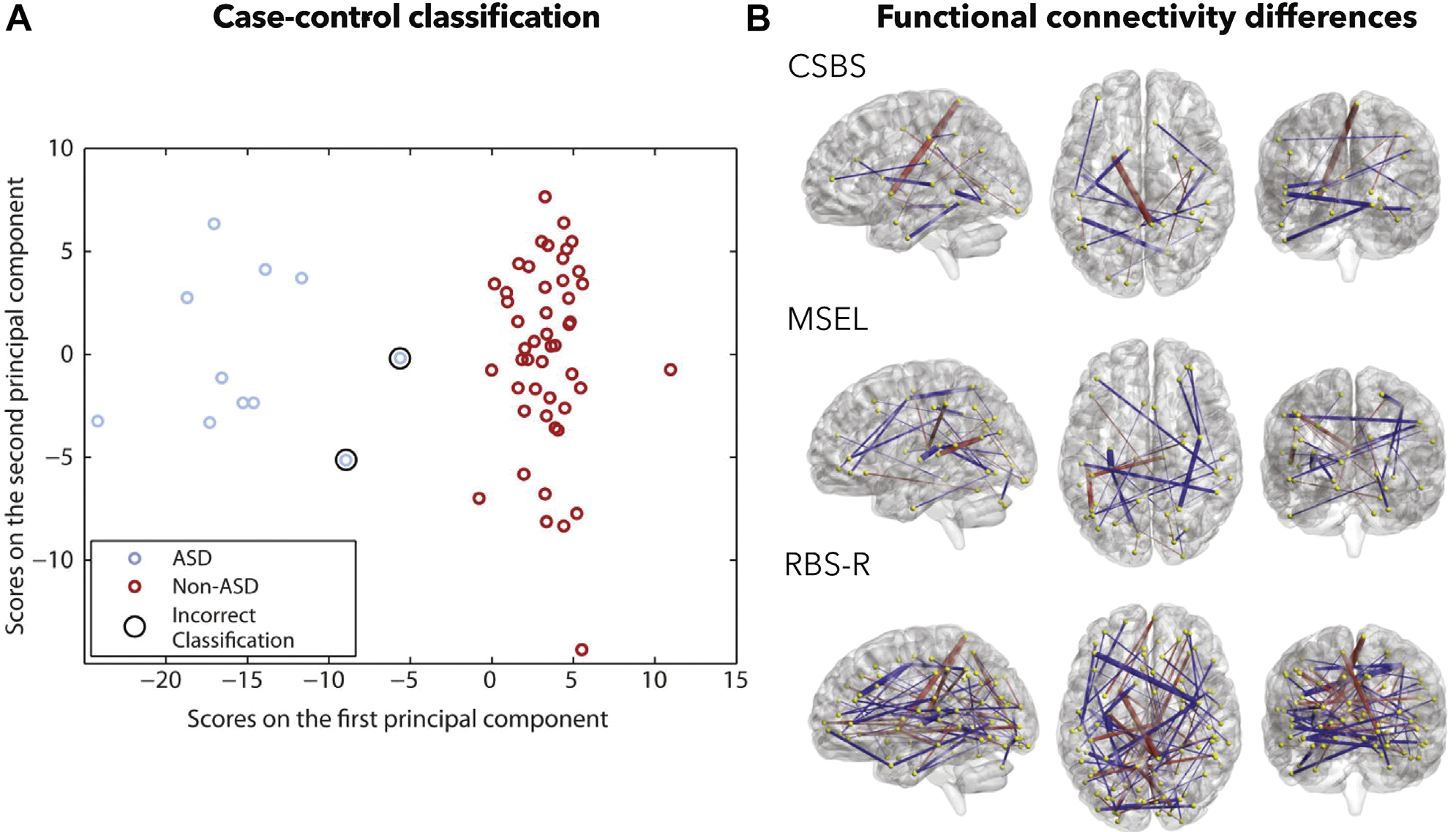
Case-control prediction is possible using measures of infant brain functional connectivity. (A) Classifying 24-month-olds using 6-month-old imaging data. Classification accuracy was 96.6%. (B) Post hoc visualization of functional connections and their relationship to different phenotypic scales. A red line indicates a connection that shows more negative connectivity in the autism group, whereas a blue line indicates more positive connectivity. ASD, autism spectrum disorder; CSBS, Communication and Symbolic Behavior Scales; MSEL, Mullen Scales of Early Learning; RBS-R, Repetitive Behaviors Scale-Revised. Adapted with permission from (48).
Evidence that autism diagnoses can be predicted at young ages is promising and sets the stage for imaging even earlier in life. Findings from genetic studies suggest that changes in transcriptional pathways specific to autism may be evident during gestation (82). Given the advent of fetal imaging (83), future predictive models may be generated to gauge autism likelihood prenatally, thus enabling support services to be made available at birth (see the Supplement for a discussion of the ethics of such a scenario and the ethics of predictive modeling in general).
DIMENSIONAL PREDICTION: ACCOUNTING FOR COMPLEX SYMPTOMATOLOGY
Symptoms in a number of psychiatric conditions (84), including autism, exist on a continuum, and the line between what constitutes adaptive versus divergent behavior is often unclear. From a biological perspective, dimensional approaches can be used to characterize function in specific behavioral domains and identify underlying patterns of brain connectivity. The implicated functional circuits can then be monitored clinically following interventions (85). Despite the advantages, there are only a handful of dimensional prediction studies (27,54,78,86–89) (Table 2). Below, we highlight work of interest in two areas: prediction of symptoms and prediction of cognitive phenotypes important for adaptive function.
Table 2.
Dimensional Studies Using Functional Connectivity Data
| Study | Data Source | n | Age, Years, Mean ± SD or Range | Male, % | Algorithm/Approach | Features | Validation | Symptoms/Phenotypes Predicted | Highlights |
|---|---|---|---|---|---|---|---|---|---|
| (27) | ABIDE, ADHD-200 | ABIDE: 122 autism, 230 NTa ADHD-200: 77 ADHD, 35 NT |
6–24 | 71%b | CPM (linear regression) | FC from 268 × 268 ROIs (Shen atlas) (148) | Internal (split half, leave-one-out CV); leave-one-site-out CV; external validation in independent sample | SRS, ADOS, ADHD-RS | SRS, ADOS predictive network models were largely distinct; SRS model generalized to predict inattention in a separate ADHD sample |
| (54) | Lab-specific | 58 autismc | Not specified for dim. analyses | Not specified for dim. analyses | Linear regression | 16 FC | Internal (leave-one-out CV) | ADOS, ADI-R | Functional connections identified as correctly classifying autism vs. NT status generalized to predict the communication domain of ADOS |
| (78) | ABIDE | 103 autism | 20.8 ± 8.1 | 100% | SVR | First score of principal connectivity gradient, stepwise connectivity maps | Internal (5-fold CV); leave-one-site-out CV | ADOS | Prediction of ADOS total and social cognition; DMN and primary visual areas highly represented in predictive models |
| (86) | ABIDE, LEAP | 232 autismd | 14.8 ± 6.5 | 74% | CCA | FC from 415 × 415 ROIs [Schaefer atlas (149) 1 subcortical atlas] | Leave-one-site-out CV | Various autism, social abilities scales | Multiple canonical variates predictive of left-out-site; connections within and between somatomotor, DMN, attention, visual networks implicated in models |
| (87) | Lab-specific | 31 autism | 17.9 ± 3.4 | 100% | Ridge regression | FC from DMN, SN, FPN networks (Power atlas) (140) | Internal (leave-one-out CV) | SRS, ABAS | Prediction of change in SRS and ABAS scores and prediction of time 2 SRS scores ~ 3 years after scanning |
| (88) | ABIDE | 85 autism 191 NT |
8–13 | 66% | CPM (linear regression) | FC from 268 × 268 ROIs (Shen atlas) (148) | Internal (leave-one-out, split-half CV); site 1 to site 2 prediction | BRIEF | Complex, brain-wide model with many edges in somatomotor, visual, and cerebellar areas; also edges in DMN and temporal lobe |
| (89) | ABIDE | 82 autism | 7–12 | 100% | Lasso regression | FC from 227 × 227 ROIs (Power atlas) (140) | Internal (leave-one-out CV) | ADOS | Separate predictive brain networks predicting communication and social interaction phenotypes can be merged to predict social affect scores |
ABAS, Adaptive Behavior Assessment System; ABIDE, Autism Brain Imaging Data Exchange; ADHD, attention-deficit/hyperactivity disorder; ADHD-RS, ADHD Rating Scale; ADI-R, Autism Diagnostic Interview–Revised; ADOS, Autism Diagnostic Observation Schedule; BRIEF, Behavior Rating Inventory of Executive Function; CCA, canonical correlation analysis; CPM, connectome-based predictive modeling; CV, cross-validation; dFC, dynamic functional connectivity; dim., dimension; DMN, default mode network; FC, functional connectivity; FPN, frontoparietal network; LEAP, European Autism Interventions Multicenter Study for Developing New Medications Longitudinal European Autism Project; NT, neurotypical; ROI, region of interest; SN, salience network; SRS, Social Responsiveness Scale; SVR, support vector regression.
Here, we report sample size used for most of the SRS analyses (SRS total scores as well as the following subscales: communication, motivation, and mannerisms). The sample size was n = 180 NT and 80 autism for predicting SRS cognition and awareness subscales and n = 79 autism and n = 58 autism for predicting ADOS modules 3 and 4, respectively.
Across SRS analyses, ~70% of the sample was male; in ADOS analyses, ~85% of the sample was male. Sex was not reported in the ADHD-200 sample.
We report the sample size used in the ADOS dimensional prediction analyses. A total of 27 participants with autism were used in the ADI-R dimensional prediction analyses.
A total of 125 individuals with autism and 78 control subjects from the LEAP cohort were used in Short Sensory Profile subscales analyses; demographics were roughly similar to the rest of the sample.
Predicting Autism Symptoms
One of the first works demonstrating dimensional symptom prediction was conducted by Plitt et al. (87). In a sample of adolescents and young adults, the authors used resting-state connectivity data from a priori networks (default mode [DMN], salience, and frontoparietal) to predict (using ridge regression) changes in social behavior 3 years later. This early report was cause for excitement, in that a prediction approach could be used to interrogate the functional connections associated with a complex symptom.
Work since has used larger samples from ABIDE to search for brain-wide correlates of symptoms. For example, using resting-state data and connectome-based predictive modeling, Lake et al. (27) generated network models predictive of Social Responsiveness Scale (SRS) scores, as well as separate models predictive of Autism Diagnostic Observation Schedule scores (Figure 4A, B). While the two models shared some common regions (cerebellum and subcortical areas, regions increasingly recognized as important in cognitive and social processes) (90), they were largely distinct. The fact that different functional circuits were detected is encouraging: despite both instruments measuring social ability, SRS and Autism Diagnostic Observation Schedule scores are only somewhat correlated (27), arguing that potentially subtle relationships between brain and phenotype are detectable using predictive methods.
Figure 4.
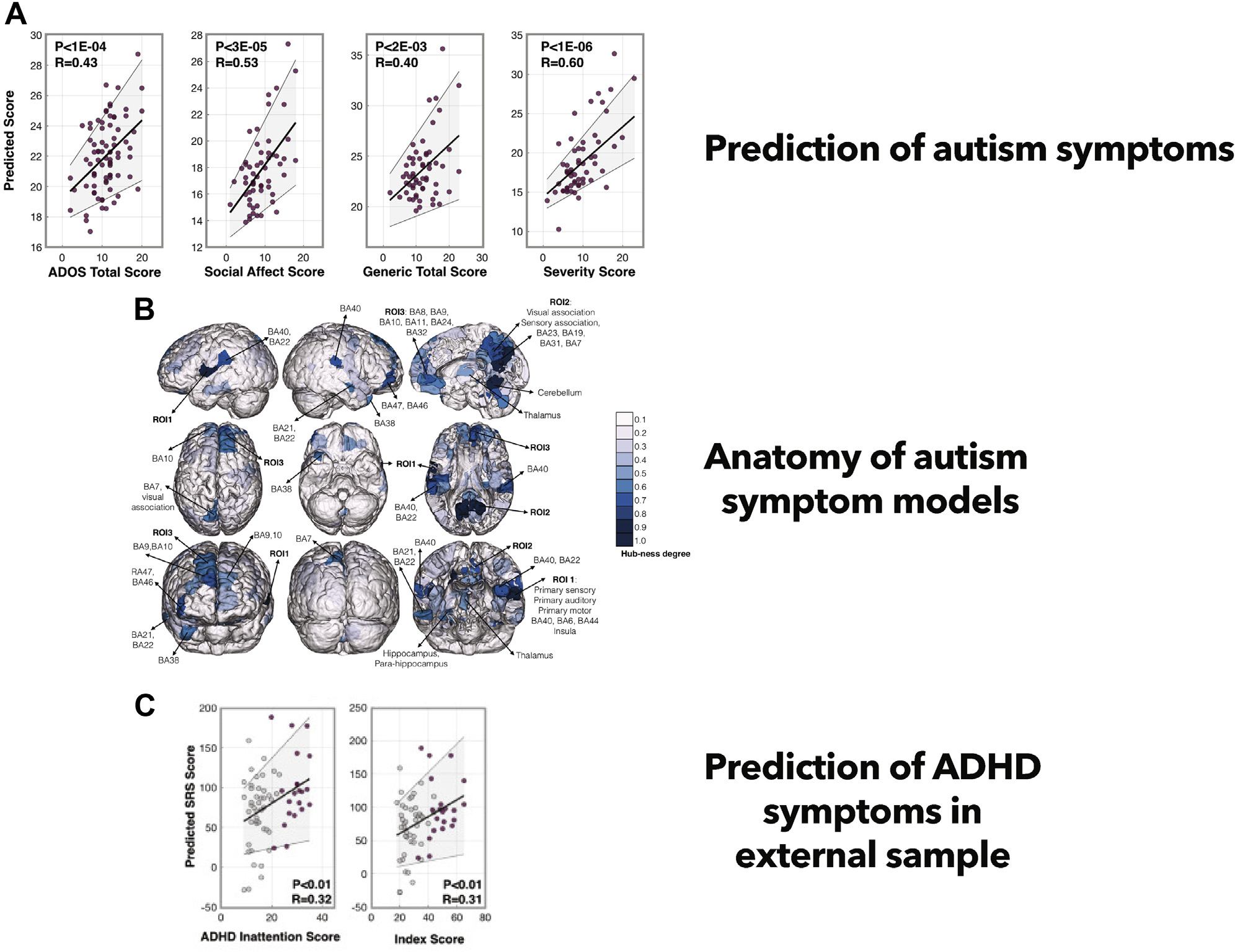
Dimensional prediction of autism symptoms. (A) Models predictive of autism symptoms are built on training data and then validated on left-out testing data within the same dataset. Predicted symptom scores from this process are shown on the y-axis; observed symptom scores are shown on the x-axis. (B) Post hoc visualization of predictive functional features (data are summarized at the node level and are shaded according to degree). (C) Application of the predictive model derived from autism symptoms to an external dataset to predict attention-deficit/hyperactivity disorder (ADHD) symptoms in young children. ADOS, Autism Diagnostic Observation Schedule; BA, Brodmann area; ROI, region of interest [as defined in (33)]; SRS, Social Responsiveness Scale. Adapted with permission from (27).
Once built, models can be applied to different datasets to test generalizability and to determine if different populations or phenotypes share neurobiological correlates. For example, Lake et al. (27) applied a connectome-based predictive modeling network built to predict SRS scores in individuals with autism (generated in ABIDE) to an independent sample of children with ADHD (ADHD-200) and found that the model predicted symptoms of inattention (Figure 4C). This is of note because of the high co-occurrence of autism and ADHD (91) and because of the brain regions (cerebellum, subcortical areas, and DMN) present within the model, which have been implicated as important for mediating aspects of internal and external attention (92). The DMN has also been found to play a significant role in theory of mind and making social inferences, processes commonly atypical in autism [reviewed in (93,94)]. Through a combination of two network models (89)—one for predicting communication, the other for predicting social interaction ability—the DMN also emerged as key for predicting social affect in autism. These results suggest that despite the complexity of symptoms in autism, it is possible to home in on neurobiological commonalities across studies.
Predicting Phenotypes Relevant for Adaptive Functioning
An avenue of clinical interest is generating dimensional predictive models for adaptive functioning. To this end, Rohr et al. (88), using resting-state data from ABIDE and connectome-based predictive modeling, generated network models predictive of a component of adaptive functioning—the ability to resist inappropriate behavioral impulses. Their behavioral inhibition model consisted of distributed, whole-brain functional features, mostly within and between default mode, somatomotor, visual, and cerebellar areas, consistent with other work (92). These findings point to the feasibility of identifying relevant markers that can be tracked to measure improvement after behavioral interventions.
The notion of monitoring adaptive function in response to interventions goes hand in hand with predicting individual outcomes in the future (6,95). The work of Plitt et al. (87) suggests that this is possible for individuals with autism, in that changes in overall adaptive function could (remarkably) be predicted 3 years after imaging. Normative modeling approaches (96) have proven useful in disentangling heterogeneity in autism brain-behavior relationships using structural (97,98) and functional (99) MRI data; future work could apply these models to generate longitudinal phenotypic predictions. Future studies could also take a multidimensional approach to predict combinations of different phenotypes (86), as well as incorporating measures of functional connectivity dynamics (100).
SUBTYPING: SIMPLIFYING COMPLEXITY BY FINDING COMMONALITIES
There has been interest in identifying autism subtypes [reviewed in (101)]. This work aims to identify homogeneous clusters to interrogate the biological basis of each subgroup, offering more specific information for potential interventions. The existence of distinct clusters in autism is supported by results in multiple modalities, including structural MRI (102,103), electroencephalography (104), eye-tracking (105,106), and symptom-level measures (70,107).
Initial Efforts at Subtyping Connectomes
Subtyping methods based on clustering functional connectomes suggest at least two or three autism subtypes (41,108–112) (Table 3). Consistent with the distributed brain features identified by dimensional models, subtyping methods indicate that there is no focal brain area differentiating subtypes; brain-based features distinguishing subgroups are complex and spatially distributed. However, the DMN and the frontoparietal network (implicated in dimensional models) (27,78,89) seem to be most consistently involved in discriminating subtypes (101). To date, the majority of studies have been conducted in ABIDE and tend to be male focused. Future work should assess the reliability/generalizability of subtypes in different datasets, include more female participants, and use a combination of rest and task data (31,113). While most studies have focused on identifying nonoverlapping subtypes, sophisticated analytic approaches allowing participants to express different subtypes to varying degrees—dimensional subtyping—are beginning to be reported (41) and are reason for enthusiasm.
Table 3.
Subtyping Studies Using FC Data
| Study | Data Source | n | Age, Years, Mean ± SD or Range | Male, % | Subtyping Approach | Features | Validationa | No. of Subtypes | Highlights |
|---|---|---|---|---|---|---|---|---|---|
| (41) | ABIDE, GENDAAR | 306 autismb | 15 ± 8 | 77% | LFAc | FC from 418 × 418 ROIs [Schaefer atlas (149) + subcortical atlas] | Multiple clinical and demographic measures | 3 | Subtypes had dissociable whole-brain hypo/hyper-FC and shared atypical FC in DMN. Individuals expressed multiple subtypes to different degrees. Subtype 1 = decreased FC (DAN, SM, SN, VN) and increased FC (including in DMN) in autism and with symptom severity. Subtype 2 = opposite patterns of FC (compared with subtype 1) with comorbid symptoms. Subtype 3 = complex mixture of increased and decreased FC; preferentially expressed in older male individuals. |
| (108) | POND | 175 autism 93 ADHD 55 OCD 84 NT |
12 ± ~4 diagnostic group | 73% | k-means clustering | FC from 76 × 76 ROIs (Desikan-Killiany-Tourville atlas) (150) | Diagnostic and behavioral measures | 2 | NT participants and participants with autism split across two subtypes; ~80% of individuals with ADHD in subtype 1, ~80% of individuals with OCD in subtype 2; participants’ distance ratio between two subtypes was significantly correlated with general adaptive functioning, social deficits, and inattention symptoms. |
| (109) | ABIDE | 145 autism 121 NT |
7–39 | 100% | k-means clustering | FC from 160 × 160 ROIs (Dosenbach atlas) (141) | SRS, IQ, ADOS | 2 | Subtype 1 (59% autism, 45% NT), subtype 2 (41% autism, 55% NT). Subtype 2 had decreased FC between networks and increased FC within networks relative to subtype 1. Subtypes did not differ in behavior, demographics, or IQ. PLS brain-behavior analyses showed FC correlations with a combination of symptom scores unique to each subtype. |
| (110) | Lab-specific | 57 autism | 9–18 | 82% | k-means clustering | FC from occipital cortex to frontal pole cortex | IQ, ADI-R, ADOS, comorbidities, medication use, age, sex | 2 | Post hoc clustering of FC in ROIs identified by group mean. Subtypes had opposite FC patterns and did not differ in clinical and demographic metrics. |
| (111) | ABIDE, ADHD-200 | 369 autism 284 ADHD 652 NTd |
7–21 | 100% | LFAc | FC from 21 × 21 ROIs (in DMN, SN, DAN) | Diagnostic labels, symptom questionnaires (unspecified) | 3 | Subtype 1 = increased DMN-DAN, medium DMN–SN, decreased intra-DMN and intra-DAN FC, and positive association with ADHD; subtype 2 = decreased DMN-DAN and DMN–SN, positive association with autism diagnosis and total IQ; subtype 3 = decreased DMN-DAN with no behavioral associations. |
| (112) | ABIDE | 210 autism | Most sites ~12 ± 2 | 85% | Hierarchical clustering | Relationship between FC from 200 × 200 ROIs (Craddock atlas) (144) with clinical symptoms | Multiclass SVM of brain-clinical symptom relationships | 3 | Subtype 1 = increased within-network connectivity, high IQ and RRB scores; subtype 2 = decreased connectivity including in DMN and cerebellar regions, increased ADI-R and SRS scores; subtype 3 = hypoconnectivity between subcortical and DMN nodes, hyperconnectivity involving the DMN, low IQ; greater social motivation difficulties and verbal difficulties (relative to subtype 1). |
Table adapted with permission from (101).
ABIDE, Autism Brain Imaging Data Exchange; ADHD, attention-deficit/hyperactivity disorder; ADI-R, Autism Diagnostic Interview–Revised; ADOS, Autism Diagnostic Observation Schedule; DAN, dorsal attention network; DMN, default mode network; FC, functional connectivity; GENDAAR, Gender Exploration of Neurogenetics and Development to Advance Autism Research; LFA, latent factor analysis; NT, neurotypical; OCD, obsessive-compulsive disorder; PLS, partial least squares; POND, Province of Ontario Neurodevelopment Disorders dataset; ROI, region of interest; RRB, restricted repetitive behavior; SM, somatomotor network; SN, salience network; SRS, Social Responsiveness Scale; SVM, support vector machine; VN, visual network.
Reporting validation based on domains distinct from the features originally used to identify subtypes.
NT sample (n = 348) from ABIDE II 1 GENDAAR was used to generate FC z scores in individuals with autism.
LFA used a Bayesian model based on latent Dirichlet allocation.
303 NT from ADHD-200, 349 NT from ABIDE I.
After connectome-based subtypes have been identified, they are typically validated by determining if some other measure, usually symptom information, differs between subgroups (114). For example, Easson et al. (109) applied k-means clustering to resting-state functional connectivity matrices from ABIDE and observed two distinct subtypes (Figure 5). The subtypes were composed of a mixture of individuals with autism and those without. Both subtypes showed wide-scale differences in connectivity. The hallmark feature of the first subtype was stronger connectivity between the DMN and cingulo-opercular, somatomotor, and visual networks. The second subtype exhibited stronger within-network connectivity. Further, each subtype showed differences in brain-behavior relationships. That is, unique connectivity signatures in each subtype differentially predicted SRS and Autism Diagnostic Observation Schedule scores.
Figure 5.
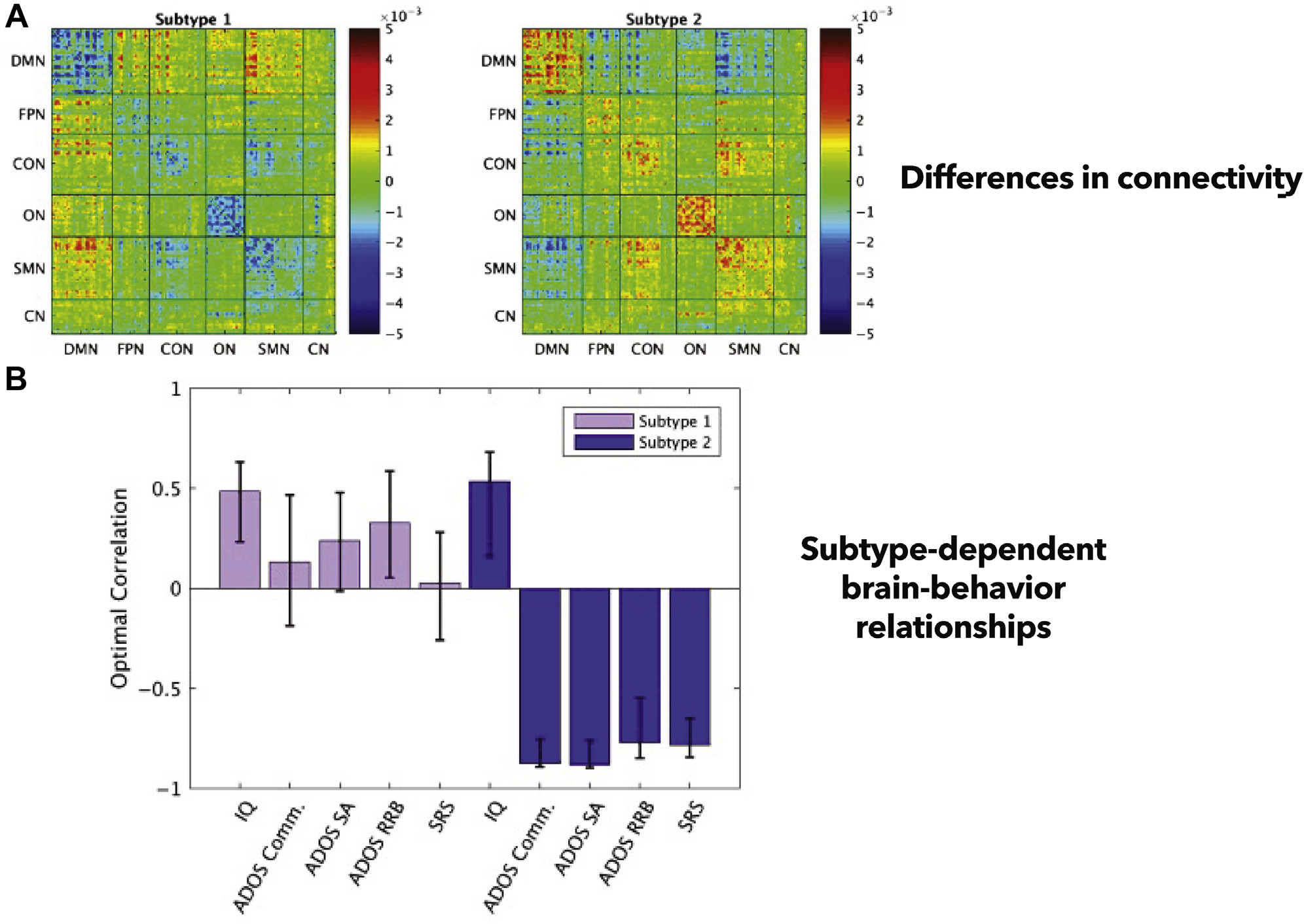
Subtyping connectomes in autism. (A) Easson et al. (109) identified two subtypes. Each is composed of individuals with and without autism. These subtypes exhibit differences in functional connectivity patterns; an average matrix for each subtype is shown. (B) A multivariate brain-behavior analysis (partial least squares regression) reveals that subtypes exhibit unique brain-behavior relationships among a set of key behavioral measures in autism. ADOS, Autism Diagnostic Observation Schedule; CN, cerebellar network; Comm., communication; CON, cingulo-opercular network; DMN, default mode network; FPN, frontoparietal network; ON, occipital network; RRB, restricted repetitive behaviors; SA, social affect; SMN, sensorimotor network; SRS, Social Responsiveness Scale. Adapted with permission from (109).
Toward Subtyping of Brain-Behavior Predictive Models
The fact that the subtypes identified by Easson et al. exhibited distinct brain-behavior relationships hints at the possibility of subtyping brain-behavioral predictive models. Crucially, these are subtypes based not on brain or phenotype alone, but on the relationship between them (112), setting them apart from work assuming that a single brain-phenotype predictive model is adequate across a sample (115). The groupings revealed by model-based subtyping may help to uncover clusters of individuals crossing diagnostic and demographic boundaries. In addition to data-driven approaches, hypothesis-driven model-based subtypes might also prove useful, whether based on symptom profiles (116,117) or other variables less expensive to measure, such as biological sex. Overall, the brain-based features derived through model-based subtyping will help yield insight into the biological underpinnings of autism (112,116). The phenotypic and demographic features differing across subtypes may help triage individuals for better care management.
LIMITATIONS
Concern has been expressed about the reliability of functional connections (118). There is work suggesting that with enough data per participant (>15 minutes/scan, allowing more reliable estimates of connections) (32), connectomes between individuals with autism and neurotypical individuals become quite similar (119). Most of the studies reported here include only a 5-minute scan. More work could be conducted to determine how increasing the amount of data affects predictive models, in terms of both accuracy and reliability (120). Aside from reliability, the precise biological nature of a functional connection remains elusive, a concern that must be acknowledged in predictive modeling studies.
An issue with case-control studies is the grouping of individuals into a single category. Individuals with autism have unique symptom profiles and complex neurobiological correlates of symptoms. Categorical diagnoses render it difficult to determine how specific aspects of a phenotype are supported by underlying brain circuits (84). Further, predicting a diagnosis is insufficient clinically; more individual-level information is needed to optimize care.
Concerns have been raised about dimensional studies in psychiatry (121,122). For example, severe communication difficulties in a person with autism might be the result of a different neurobiological process than the process supporting communication capacities in a neurotypical individual; it might be incorrect to assume that all individuals can be situated on a single dimension for a given phenotype (121). Certain dimensional indices (SRS) rely on parent/self-report measures; such measures may be weakly related to the symptom or behavioral constructs of interest (123). It is possible that a dimensional approach cannot be used to model all brain-phenotype relationships (124), and computational constraints might limit the practicality of dimensional methods due to the curse of dimensionality (122).
Subtypes in some psychiatric conditions have proved difficult to replicate across datasets (125,126), and a recent study reported an inability to define reliable subgroups in autism (127). It will be crucial to continue to test reproducibility and generalizability of autism subtypes. Additionally, interpretation of subtypes may be complicated by unmeasured, sample-dependent covariates. Collecting precise and inclusive demographic/clinical data can be used to correct for confounds (128), although hidden confounds may persist (129).
FUTURE DIRECTIONS AND CONCLUSIONS
We have reviewed how predictive modeling frameworks can offer insight into neurobiological correlates of autism, as well as potential clinical utility. Presently, case-control classification studies comprise most of the literature, allowing developmental trends to be observed. Due to the heterogeneity of individuals with autism, more dimensional and subtyping prediction studies are needed. All three prediction frameworks can be affected by the autism-specific modeling considerations discussed here. Classification approaches may one day enable early diagnoses (perhaps even in utero) using objective, biological data. Meanwhile, dimensional and subtyping studies may both deepen our understanding of the brain-based features behind autism and discover means of improving management through imaging-based prognostication and monitoring of intervention response.
Consistent with the complexity of autism symptoms, brain-based predictive models are complex and reveal large-scale networks supporting specific behaviors. To aid interpretation and translation, continuing to collect large datasets is essential (21). Ideally, the datasets will be broad (large numbers of diverse individuals with and without autism) (71) and deep (comprising many data modalities) (130). An example of a biological insight gained by a broad and deep approach is determining if specific genetic signatures underlie different connectivity phenotypes (131), and elegant work linking genes to complex brain activity patterns to behavioral phenotypes in autism is beginning to appear (40). A deep, multimodal focus might offer a marker common to fMRI and functional near-infrared spectroscopy (132) or electroencephalography (133), offering complementary information that can be used clinically (and is less expensive and better tolerated by some than fMRI).
Dense scanning approaches—imaging the same participants many times—have proven useful in neurotypical adults (134). Combined with innovative task paradigms, such as movie watching (135), dense scanning could provide large amounts of individual-level data during naturalistic social settings. Such an approach could help autism researchers better parse participant-specific trajectories (95). Ideally, dense scanning initiatives would comprise many individuals to maximize the detection of individual differences (see the Supplement for more about dense scanning in autism).
We do not suggest that the path forward will be easy. While expectations have been high, fMRI has largely failed to benefit individuals with autism to date. Aside from the difficulty in producing reliable fMRI results (136), there are numerous points at which findings can fail to translate (137). Research and clinical priorities do not always align (138), so it will be essential to maintain open channels between researchers, clinicians, individuals with autism, and their caregivers. Going forward, we envision predictive modeling approaches continuing to aid the quest to understand the complex neurobiology of autism.
Supplementary Material
ACKNOWLEDGMENTS AND DISCLOSURES
This work was supported by the National Institute of Mental Health (Grant Nos. P50MH115716 [to KC], R21 MH122202 [to JCM], and U19 MH108206 [to JCM]), the National Institutes of Health (NIH)/National Institute of General Medical Sciences Medical Scientist Training Program training grant (Grant No. T32GM007205 [to CH and ASG]), the NIH (Grant No. TR001864 [to ASG]), and the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant (Grant No. 101025785 [to DLF]).
We thank Dr. Andres Martin and Dr. Jennifer B. Dwyer for helpful discussions related to this manuscript.
Footnotes
The authors report no biomedical financial interests or potential conflicts of interest.
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.biopsych.2022.04.008.
Contributor Information
Corey Horien, Interdepartmental Neuroscience Program, MD-PhD Program, Yale School of Medicine.
Dorothea L. Floris, Methods of Plasticity Research, Department of Psychology, University of Zürich, Zurich, Switzerland Donders Center for Brain, Cognition and Behavior, Radboud University Nijmegen, Nijmegen, The Netherlands.
Abigail S. Greene, Interdepartmental Neuroscience Program, MD-PhD Program, Yale School of Medicine
Stephanie Noble, Department of Radiology and Biomedical Imaging, Yale School of Medicine.
Max Rolison, Yale Child Study Center, New Haven, Connecticut.
Link Tejavibulya, Interdepartmental Neuroscience Program, Yale School of Medicine.
David O’Connor, Department of Biomedical Engineering, Yale University.
James C. McPartland, Department of Psychology, Yale University Yale Child Study Center, New Haven, Connecticut.
Dustin Scheinost, Interdepartmental Neuroscience Program, Yale School of Medicine; Department of Radiology and Biomedical Imaging, Yale School of Medicine; Department of Biomedical Engineering, Yale University; Department of Statistics and Data Science, Yale University; Yale Child Study Center, New Haven, Connecticut.
Katarzyna Chawarska, Department of Pediatrics, Yale School of Medicine; Department of Statistics and Data Science, Yale University; Yale Child Study Center, New Haven, Connecticut.
Evelyn M.R. Lake, Department of Radiology and Biomedical Imaging, Yale School of Medicine
R. Todd Constable, Interdepartmental Neuroscience Program, Yale School of Medicine; Department of Neurosurgery, Yale School of Medicine; Department of Radiology and Biomedical Imaging, Yale School of Medicine; Department of Biomedical Engineering, Yale University.
REFERENCES
- 1.McPartland JC, Reichow B, Volkmar FR (2012): Sensitivity and specificity of proposed DSM-5 diagnostic criteria for autism spectrum disorder. J Am Acad Child Adolesc Psychiatry 51:368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswal B, Yetkin FZ, Haughton VM, Hyde JS (1995): Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med 34:537–541. [DOI] [PubMed] [Google Scholar]
- 3.Gabrieli JDE, Ghosh SS, Whitfield-Gabrieli S (2015): Prediction as a humanitarian and pragmatic contribution from human cognitive neuroscience. Neuron 85:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheinost D, Noble S, Horien C, Greene AS, Lake EM, Salehi M, et al. (2019): Ten simple rules for predictive modeling of individual differences in neuroimaging. Neuroimage 193:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yarkoni T, Westfall J (2017): Choosing prediction over explanation in psychology: Lessons from machine learning. Perspect Psychol Sci 12:1100–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberg MD, Casey BJ, Holmes AJ (2018): Prediction complements explanation in understanding the developing brain. Nat Commun 9:589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McPartland JC (2017): Developing clinically practicable biomarkers for autism spectrum disorder. J Autism Dev Disord 47:2935–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finn ES, Rosenberg MD (2021): Beyond fingerprinting: Choosing predictive connectomes over reliable connectomes. Neuroimage 239:118254. [DOI] [PubMed] [Google Scholar]
- 9.Liu M, Li B, Hu D (2021): Autism spectrum disorder studies using fMRI data and machine learning: A review. Front Neurosci 15:697870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uddin LQ, Dajani DR, Voorhies W, Bednarz H, Kana RK (2017): Progress and roadblocks in the search for brain-based biomarkers of autism and attention-deficit/hyperactivity disorder. Transl Psychiatry 7:e1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfers T, Floris DL, Dinga R, van Rooij D, Isakoglou C, Kia SM, et al. (2019): From pattern classification to stratification: Towards conceptualizing the heterogeneity of autism spectrum disorder. Neurosci Biobehav Rev 104:240–254. [DOI] [PubMed] [Google Scholar]
- 12.Abraham A, Milham MP, Di Martino A, Craddock RC, Samaras D, Thirion B, Varoquaux G (2017): Deriving reproducible biomarkers from multi-site resting-state data: An autism-based example. Neuroimage 147:736–745. [DOI] [PubMed] [Google Scholar]
- 13.Marek S, Tervo-Clemmens B, Calabro FJ, Montez DF, Kay BP, Hatoum AS, et al. (2022): Reproducible brain-wide association studies require thousands of individuals [published correction appears in Nature 2022; 605(7911):E11]. Nature 603(7902):654–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Martino A, O’Connor D, Chen B, Alaerts K, Anderson JS, Assaf M, et al. (2017): Enhancing studies of the connectome in autism using the Autism Brain Imaging Data Exchange II. Sci Data 4:170010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, et al. (2014): The Autism Brain Imaging Data Exchange: Towards a largescale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry 19:659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loth E, Charman T, Mason L, Tillmann J, Jones EJH, Wooldridge C, et al. (2017): The EU-AIMS Longitudinal European Autism Project (LEAP): Design and methodologies to identify and validate stratification biomarkers for autism spectrum disorders. Mol Autism 8:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson WH, Wright J, Bissett PG, Poldrack RA (2020): Dataset decay and the problem of sequential analyses on open datasets. Elife 9:e53498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith SM, Nichols TE (2018): Statistical challenges in “big data” human neuroimaging. Neuron 97:263–268. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita A, Yahata N, Itahashi T, Lisi G, Yamada T, Ichikawa N, et al. (2019): Harmonization of resting-state functional MRI data across multiple imaging sites via the separation of site differences into sampling bias and measurement bias. PLoS Biol 17:e3000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu M, Linn KA, Cook PA, Phillips ML, McInnis M, Fava M, et al. (2018): Statistical harmonization corrects site effects in functional connectivity measurements from multi-site fMRI data. Hum Brain Mapp 39:4213–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horien C, Noble S, Greene AS, Lee K, Barron DS, Gao S, et al. (2021): A hitchhiker’s guide to working with large, open-source neuroimaging datasets. Nat Hum Behav 5:185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg MD, Scheinost D, Greene AS, Avery EW, Kwon YH, Finn ES, et al. (2020): Functional connectivity predicts changes in attention observed across minutes, days, and months. Proc Natl Acad Sci U S A 117:3797–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yarkoni T (2020): The generalizability crisis. Behav Brain Sci 45:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Power JD, Schlaggar BL, Petersen SE (2015): Recent progress and outstanding issues in motion correction in resting state fMRI. Neuroimage 105:536–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yerys BE, Jankowski KF, Shook D, Rosenberger LR, Barnes KA, Berl MM, et al. (2009): The fMRI success rate of children and adolescents: Typical development, epilepsy, attention deficit/hyperactivity disorder, and autism spectrum disorders. Hum Brain Mapp 30:3426–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Power JD, Plitt M, Laumann TO, Martin A (2017): Sources and implications of whole-brain fMRI signals in humans. Neuroimage 146:609–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lake EMR, Finn ES, Noble SM, Vanderwal T, Shen X, Rosenberg MD, et al. (2019): The functional brain organization of an individual allows prediction of measures of social abilities transdiagnostically in autism and attention-deficit/hyperactivity disorder. Biol Psychiatry 86:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Kong R, Liégeois R, Orban C, Tan Y, Sun N, et al. (2019): Global signal regression strengthens association between resting-state functional connectivity and behavior. Neuroimage 196:126–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gotts SJ, Saad ZS, Jo HJ, Wallace GL, Cox RW, Martin A (2013): The perils of global signal regression for group comparisons: A case study of autism spectrum disorders. Front Hum Neurosci 7:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy K, Fox MD (2017): Towards a consensus regarding global signal regression for resting state functional connectivity MRI. Neuroimage 154:169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greene AS, Gao S, Scheinost D, Constable RT (2018): Task-induced brain state manipulation improves prediction of individual traits. Nat Commun 9:2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Birn RM, Molloy EK, Patriat R, Parker T, Meier TB, Kirk GR, et al. (2013): The effect of scan length on the reliability of resting-state fMRI connectivity estimates. Neuroimage 83:550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alfaro-Almagro F, McCarthy P, Afyouni S, Andersson JLR, Bastiani M, Miller KL, et al. (2021): Confound modelling in UK Biobank brain imaging. Neuroimage 224:117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dosenbach NUF, Koller JM, Earl EA, Miranda-Dominguez O, Klein RL, Van AN, et al. (2017): Real-time motion analytics during brain MRI improve data quality and reduce costs. Neuroimage 161:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horien C, Fontenelle S 4th, Joseph K, Powell N, Nutor C, Fortes D, et al. (2020): Low-motion fMRI data can be obtained in pediatric participants undergoing a 60-minute scan protocol. Sci Rep 10:21855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vanderwal T, Kelly C, Eilbott J, Mayes LC, Castellanos FX (2015): Inscapes: A movie paradigm to improve compliance in functional magnetic resonance imaging. Neuroimage 122:222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antshel KM, Zhang-James Y, Faraone SV (2013): The comorbidity of ADHD and autism spectrum disorder. Expert Rev Neurother 13:1117–1128. [DOI] [PubMed] [Google Scholar]
- 38.Zaboski BA, Storch EA (2018): Comorbid autism spectrum disorder and anxiety disorders: A brief review. Future Neurol 13:31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matson JL, Shoemaker M (2009): Intellectual disability and its relationship to autism spectrum disorders. Res Dev Disabil 30:1107–1114. [DOI] [PubMed] [Google Scholar]
- 40.Lombardo MV, Pramparo T, Gazestani V, Warrier V, Bethlehem RAI, Carter Barnes C, et al. (2018): Large-scale associations between the leukocyte transcriptome and BOLD responses to speech differ in autism early language outcome subtypes. Nat Neurosci 21:1680–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang S, Sun N, Floris DL, Zhang X, Di Martino A, Yeo BTT (2020): Reconciling dimensional and categorical models of autism heterogeneity: A brain connectomics and behavioral study [published correction appears in Biol Psychiatry 2021; 90:275]. Biol Psychiatry 87:1071–1082. [DOI] [PubMed] [Google Scholar]
- 42.Loomes R, Hull L, Mandy WPL (2017): What is the male-to-female ratio in autism spectrum disorder? A systematic review and meta-analysis. J Am Acad Child Adolesc Psychiatry 56:466–474. [DOI] [PubMed] [Google Scholar]
- 43.Lai MC, Lombardo MV, Auyeung B, Chakrabarti B, Baron-Cohen S (2015): Sex/gender differences and autism: Setting the scene for future research. J Am Acad Child Adolesc Psychiatry 54:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ratto AB, Kenworthy L, Yerys BE, Bascom J, Wieckowski AT, White SW, et al. (2018): What about the girls? Sex-based differences in autistic traits and adaptive skills. J Autism Dev Disord 48:1698–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weis S, Patil KR, Hoffstaedter F, Nostro A, Yeo BTT, Eickhoff SB (2020): Sex classification by resting state brain connectivity. Cereb Cortex 30:824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen CP, Keown CL, Jahedi A, Nair A, Pflieger ME, Bailey BA, Müller RA (2015): Diagnostic classification of intrinsic functional connectivity highlights somatosensory, default mode, and visual regions in autism. Neuroimage Clin 8:238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen H, Duan X, Liu F, Lu F, Ma X, Zhang Y, et al. (2016): Multivariate classification of autism spectrum disorder using frequency-specific resting-state functional connectivity—A multi-center study. Prog Neuropsychopharmacol Biol Psychiatry 64:1–9. [DOI] [PubMed] [Google Scholar]
- 48.Emerson RW, Adams C, Nishino T, Hazlett HC, Wolff JJ, Zwaigenbaum L, et al. (2017): Functional neuroimaging of high-risk 6-month-old infants predicts a diagnosis of autism at 24 months of age. Sci Transl Med 9:eaag2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iidaka T (2015): Resting state functional magnetic resonance imaging and neural network classified autism and control. Cortex 63:55–67. [DOI] [PubMed] [Google Scholar]
- 50.Murdaugh DL, Shinkareva SV, Deshpande HR, Wang J, Pennick MR, Kana RK (2012): Differential deactivation during mentalizing and classification of autism based on default mode network connectivity. PLoS One 7:e50064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plitt M, Barnes KA, Martin A (2014): Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. Neuroimage Clin 7:359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uddin LQ, Supekar K, Lynch CJ, Khouzam A, Phillips J, Feinstein C, et al. (2013): Salience network-based classification and prediction of symptom severity in children with autism. JAMA Psychiatry 70:869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H, Chen C, Fushing H (2012): Extracting multiscale pattern information of fMRI based functional brain connectivity with application on classification of autism spectrum disorders. PLoS One 7: e45502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yahata N, Morimoto J, Hashimoto R, Lisi G, Shibata K, Kawakubo Y, et al. (2016): A small number of abnormal brain connections predicts adult autism spectrum disorder. Nat Commun 7:11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo X, Dominick KC, Minai AA, Li H, Erickson CA, Lu LJ (2017): Diagnosing autism spectrum disorder from brain resting-state functional connectivity patterns using a deep neural network with a novel feature selection method. Front Neurosci 11:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heinsfeld AS, Franco AR, Craddock RC, Buchweitz A, Meneguzzi F (2017): Identification of autism spectrum disorder using deep learning and the ABIDE dataset. Neuroimage Clin 17:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jahedi A, Nasamran CA, Faires B, Fan J, Müller RA (2017): Distributed intrinsic functional connectivity patterns predict diagnostic status in large autism cohort. Brain Connect 7:515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kazeminejad A, Sotero RC (2018): Topological properties of resting-state fMRI functional networks improve machine learning-based autism classification. Front Neurosci 12:1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eslami T, Mirjalili V, Fong A, Laird AR, Saeed F (2019): ASD-DiagNet: A hybrid learning approach for detection of autism spectrum disorder using fMRI data. Front Neuroinform 13:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Niu K, Guo J, Pan Y, Gao X, Peng X, Li N, Li H (2020): Multichannel deep attention neural networks for the classification of autism spectrum disorder using neuroimaging and personal characteristic data. Complexity 2020:1–9. [Google Scholar]
- 61.Liu Y, Xu L, Li J, Yu J, Yu X (2020): Attentional connectivity-based prediction of autism using heterogeneous rs-fMRI data from CC200 atlas. Exp Neurobiol 29:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sherkatghanad Z, Akhondzadeh M, Salari S, Zomorodi-Moghadam M, Abdar M, Acharya UR, et al. (2020): Automated detection of autism spectrum disorder using a convolutional neural network. Front Neurosci 13:1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang M, Kumar P, Chen H, Shrivastava A (2020): Deep multimodal learning for the diagnosis of autism spectrum disorder. J Imaging 6:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao F, Chen Z, Rekik I, Lee SW, Shen D (2020): Diagnosis of autism spectrum disorder using central-moment features from low- and high-order dynamic resting-state functional connectivity networks. Front Neurosci 14:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang ZA, Zhu Z, Yau CH, Tan KC (2021): Identifying autism spectrum disorder from resting-state fMRI using deep belief network. IEEE Trans Neural Netw Learn Syst 32:2847–2861. [DOI] [PubMed] [Google Scholar]
- 66.Yin W, Mostafa S, Wu FX (2021): Diagnosis of autism spectrum disorder based on functional brain networks with deep learning. J Comput Biol 28:146–165. [DOI] [PubMed] [Google Scholar]
- 67.Reiter MA, Jahedi A, Jac Fredo AR, Fishman I, Bailey B, Müller RA (2021): Performance of machine learning classification models of autism using resting-state fMRI is contingent on sample heterogeneity. Neural Comput Appl 33:3299–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson JS, Nielsen JA, Froehlich AL, DuBray MB, Druzgal TJ, Cariello AN, et al. (2011): Functional connectivity magnetic resonance imaging classification of autism. Brain 134:3742–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Di Martino A, Fair DA, Kelly C, Satterthwaite TD, Castellanos FX, Thomason ME, et al. (2014): Unraveling the miswired connectome: A developmental perspective. Neuron 83:1335–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fountain C, Winter AS, Bearman PS (2012): Six developmental trajectories characterize children with autism. Pediatrics 129:e1112–e1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Benkarim O, Paquola C, Park BY, Kebets V, Hong SJ, de Wael RV, et al. (2021): The cost of untracked diversity in brain-imaging prediction. bioRxiv. 10.1101/2021.2006.2016.448764. [DOI] [Google Scholar]
- 72.Lanka P, Rangaprakash D, Dretsch MN, Katz JS, Denney TS Jr, Deshpande G (2020): Supervised machine learning for diagnostic classification from large-scale neuroimaging datasets. Brain Imaging Behav 14:2378–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ferrari E, Bosco P, Calderoni S, Oliva P, Palumbo L, Spera G, et al. (2020): Dealing with confounders and outliers in classification medical studies: The Autism Spectrum Disorders case study. Artif Intell Med 108:101926. [DOI] [PubMed] [Google Scholar]
- 74.Sydnor VJ, Larsen B, Bassett DS, Alexander-Bloch A, Fair DA, Liston C, et al. (2021): Neurodevelopment of the association cortices: Patterns, mechanisms, and implications for psychopathology. Neuron 109:2820–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Casey BJ, Tottenham N, Liston C, Durston S (2005): Imaging the developing brain: What have we learned about cognitive development? Trends Cogn Sci 9:104–110. [DOI] [PubMed] [Google Scholar]
- 76.Lawrence KE, Hernandez LM, Bookheimer SY, Dapretto M (2019): Atypical longitudinal development of functional connectivity in adolescents with autism spectrum disorder. Autism Res 12:53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Benkarim O, Paquola C, Park BY, Hong SJ, Royer J, Vos de Wael R, et al. (2021): Connectivity alterations in autism reflect functional idiosyncrasy. Commun Biol 4:1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hong SJ, Vos de Wael R, Bethlehem RAI, Lariviere S, Paquola C, Valk SL, et al. (2019): Atypical functional connectome hierarchy in autism. Nat Commun 10:1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Whitehouse AJO, Varcin KJ, Pillar S, Billingham W, Alvares GA, Barbaro J, et al. (2021): Effect of preemptive intervention on developmental outcomes among infants showing early signs of autism: A randomized clinical trial of outcomes to diagnosis. JAMA Pediatr 175: e213298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Molnar-Szakacs I, Kupis L, Uddin LQ (2021): Neuroimaging markers of risk and pathways to resilience in autism spectrum disorder. Biol Psychiatry Cogn Neurosci Neuroimaging 6:200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dinstein I, Pierce K, Eyler L, Solso S, Malach R, Behrmann M, Courchesne E (2011): Disrupted neural synchronization in toddlers with autism. Neuron 70:1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Uddin M, Tammimies K, Pellecchia G, Alipanahi B, Hu P, Wang Z, et al. (2014): Brain-expressed exons under purifying selection are enriched for de novo mutations in autism spectrum disorder. Nat Genet 46:742–747. [DOI] [PubMed] [Google Scholar]
- 83.van den Heuvel MI, Thomason ME (2016): Functional connectivity of the human brain in utero. Trends Cogn Sci 20:931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, et al. (2010): Research domain criteria (RDoC): Toward a new classification framework for research on mental disorders. Am J Psychiatry 167:748–751. [DOI] [PubMed] [Google Scholar]
- 85.McPartland JC (2016): Considerations in biomarker development for neurodevelopmental disorders. Curr Opin Neurol 29:118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ilioska I, Oldehinkel M, Llera A, Chopra S, Looden T, Chauvin R, et al. (2022): Connectome-wide mega-analysis reveals robust patterns of atypical functional connectivity in autism. medRxiv. 10.1101/2022.2001.2009.22268936. [DOI] [PubMed] [Google Scholar]
- 87.Plitt M, Barnes KA, Wallace GL, Kenworthy L, Martin A (2015): Resting-state functional connectivity predicts longitudinal change in autistic traits and adaptive functioning in autism. Proc Natl Acad Sci U S A 112:E6699–E6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rohr CS, Kamal S, Bray S (2020): Building functional connectivity neuromarkers of behavioral self-regulation across children with and without autism spectrum disorder. Dev Cogn Neurosci 41:100747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xiao J, Chen H, Shan X, He C, Li Y, Guo X, et al. (2021): Linked social-communication dimensions and connectivity in functional brain networks in autism spectrum disorder. Cereb Cortex 31:3899–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Buckner RL (2013): The cerebellum and cognitive function: 25 years of insight from anatomy and neuroimaging. Neuron 80:807–815. [DOI] [PubMed] [Google Scholar]
- 91.Reiersen AM, Constantino JN, Volk HE, Todd RD (2007): Autistic traits in a population-based ADHD twin sample. J Child Psychol Psychiatry 48:464–472. [DOI] [PubMed] [Google Scholar]
- 92.Rosenberg MD, Finn ES, Scheinost D, Papademetris X, Shen X, Constable RT, Chun MM (2016): A neuromarker of sustained attention from whole-brain functional connectivity. Nat Neurosci 19:165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Buckner RL, DiNicola LM (2019): The brain’s default network: Updated anatomy, physiology and evolving insights. Nat Rev Neurosci 20:593–608. [DOI] [PubMed] [Google Scholar]
- 94.Padmanabhan A, Lynch CJ, Schaer M, Menon V (2017): The default mode network in autism. Biol Psychiatry Cogn Neurosci Neuroimaging 2:476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yip SW, Konova AB (2022): Densely sampled neuroimaging for maximizing clinical insight in psychiatric and addiction disorders. Neuropsychopharmacology 47:395–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marquand AF, Rezek I, Buitelaar J, Beckmann CF (2016): Understanding heterogeneity in clinical cohorts using normative models: Beyond case-control studies. Biol Psychiatry 80:552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bethlehem RAI, Seidlitz J, Romero-Garcia R, Trakoshis S, Dumas G, Lombardo MV (2020): A normative modelling approach reveals age-atypical cortical thickness in a subgroup of males with autism spectrum disorder. Commun Biol 3:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shan X, Uddin LQ, Xiao J, He C, Ling Z, Li L, et al. (2022): Mapping the heterogeneous brain structural phenotype of autism spectrum disorder using the normative model. Biol Psychiatry 91:967–976. [DOI] [PubMed] [Google Scholar]
- 99.Looden T, Floris DL, Llera A, Chauvin RJ, Charman T, Banaschewski T, et al. (2022): Patterns of connectome variability in autism across five functional activation tasks. Findings from the LEAP project. bioRxiv. 10.1101/2022.2002.2022.481408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xie Y, Xu Z, Xia M, Liu J, Shou X, Cui Z, et al. (2022): Alterations in connectome dynamics in autism spectrum disorder: A harmonized mega- and meta-analysis study using the autism brain imaging data exchange dataset. Biol Psychiatry 91:945–955. [DOI] [PubMed] [Google Scholar]
- 101.Hong SJ, Vogelstein JT, Gozzi A, Bernhardt BC, Yeo BTT, Milham MP, Di Martino A (2020): Toward neurosubtypes in autism. Biol Psychiatry 88:111–128. [DOI] [PubMed] [Google Scholar]
- 102.ChenH UddinLQ, GuoX WangJ, WangR WangX, et al. (2019):Parsing brain structural heterogeneity in males with autism spectrum disorder reveals distinct clinical subtypes. Hum Brain Mapp 40:628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hong SJ, Valk SL, Di Martino A, Milham MP, Bernhardt BC (2018): Multidimensional neuroanatomical subtyping of autism spectrum disorder. Cereb Cortex 28:3578–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Duffy FH, Als H (2019): Autism, spectrum or clusters? An EEG coherence study. BMC Neurol 19:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pierce K, Conant D, Hazin R, Stoner R, Desmond J (2011): Preference for geometric patterns early in life as a risk factor for autism. Arch Gen Psychiatry 68:101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pierce K, Marinero S, Hazin R, McKenna B, Barnes CC, Malige A (2016): Eye tracking reveals abnormal visual preference for geometric images as an early biomarker of an autism spectrum disorder subtype associated with increased symptom severity. Biol Psychiatry 79:657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Feczko E, Balba NM, Miranda-Dominguez O, Cordova M, Karalunas SL, Irwin L, et al. (2018): Subtyping cognitive profiles in autism spectrum disorder using a functional random forest algorithm. Neuroimage 172:674–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Choi EJ, Vandewouw MM, Taylor MJ, Arnold PD, Brian J, Crosbie J, et al. (2020): Beyond diagnosis: Cross-diagnostic features in canonical resting-state networks in children with neurodevelopmental disorders. Neuroimage Clin 28:102476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Easson AK, Fatima Z, McIntosh AR (2019): Functional connectivity-based subtypes of individuals with and without autism spectrum disorder. Netw Neurosci 3:344–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jao Keehn RJ, Nair S, Pueschel EB, Linke AC, Fishman I, Müller RA (2019): Atypical local and distal patterns of occipito-frontal functional connectivity are related to symptom severity in autism. Cereb Cortex 29:3319–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kernbach JM, Satterthwaite TD, Bassett DS, Smallwood J, Margulies D, Krall S, et al. (2018): Shared endo-phenotypes of default mode dsfunction in attention deficit/hyperactivity disorder and autism spectrum disorder. Transl Psychiatry 8:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Reardon AM, Li K, Langley J, Hu XP (2022): Subtyping autism spectrum disorder via joint modeling of clinical and connectomic profiles. Brain Connect 12:193–205. [DOI] [PubMed] [Google Scholar]
- 113.Finn ES, Scheinost D, Finn DM, Shen X, Papademetris X, Constable RT (2017): Can brain state be manipulated to emphasize individual differences in functional connectivity? Neuroimage 160:140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Agelink van Rentergem JA, Deserno MK, Geurts HM (2021): Validation strategies for subtypes in psychiatry: A systematic review of research on autism spectrum disorder. Clin Psychol Rev 87:102033. [DOI] [PubMed] [Google Scholar]
- 115.Beaty RE, Kenett YN, Christensen AP, Rosenberg MD, Benedek M, Chen Q, et al. (2018): Robust prediction of individual creative ability from brain functional connectivity. Proc Natl Acad Sci U S A 115:1087–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lombardo MV, Eyler L, Moore A, Datko M, Carter Barnes C, Cha D, et al. (2019): Default mode-visual network hypoconnectivity in an autism subtype with pronounced social visual engagement difficulties. Elife 8:e47427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lombardo MV, Pierce K, Eyler LT, Carter Barnes C, AhrensBarbeau C, Solso S, et al. (2015): Different functional neural substrates for good and poor language outcome in autism. Neuron 86:567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Noble S, Scheinost D, Constable RT (2019): A decade of test-retest reliability of functional connectivity: A systematic review and meta-analysis. Neuroimage 203:116157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Byrge L, Kennedy DP (2020): Accurate prediction of individual subject identity and task, but not autism diagnosis, from functional connectomes. Hum Brain Mapp 41:2249–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tian Y, Zalesky A (2021): Machine learning prediction of cognition from functional connectivity: Are feature weights reliable? Neuroimage 245:118648. [DOI] [PubMed] [Google Scholar]
- 121.Ross CA, Margolis RL (2019): Research domain criteria: Strengths, weaknesses, and potential alternatives for future psychiatric research. Mol Neuropsychiatry 5:218–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Feczko E, Miranda-Dominguez O, Marr M, Graham AM, Nigg JT, Fair DA (2019): The heterogeneity problem: Approaches to identify psychiatric subtypes. Trends Cogn Sci 23:584–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dang J, King KM, Inzlicht M (2020): Why are self-report and behavioral measures weakly correlated? Trends Cogn Sci 24:267–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shen X, Finn ES, Scheinost D, Rosenberg MD, Chun MM, Papademetris X, Constable RT (2017): Using connectome-based predictive modeling to predict individual behavior from brain connectivity. Nat Protoc 12:506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dinga R, Schmaal L, Penninx BWJH, van Tol MJ, Veltman DJ, van Velzen L, et al. (2019): Evaluating the evidence for biotypes of depression: Methodological replication and extension of Drysdale et al. (2017). Neuroimage Clin 22:101796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Esterman M, Stumps A, Jagger-Rickels A, Rothlein D, DeGutis J, Fortenbaugh F, et al. (2020): Evaluating the evidence for a neuroimaging subtype of posttraumatic stress disorder. Sci Transl Med 12: eaaz9343. [DOI] [PubMed] [Google Scholar]
- 127.Dajani DR, Burrows CA, Nebel MB, Mostofsky SH, Gates KM, Uddin LQ (2019): Parsing heterogeneity in autism spectrum disorder and attention-deficit/hyperactivity disorder with individual connectome mapping. Brain Connect 9:673–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Linn KA, Gaonkar B, Doshi J, Davatzikos C, Shinohara RT (2016): Addressing confounding in predictive models with an application to neuroimaging. Int J Biostat 12:31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rao A, Monteiro JM, Mourao-Miranda J, Alzheimer’s Disease Initiative (2017): Predictive modelling using neuroimaging data in the presence of confounds. Neuroimage 150:23–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lombardo MV, Lai MC, Baron-Cohen S (2019): Big data approaches to decomposing heterogeneity across the autism spectrum. Mol Psychiatry 24:1435–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Richiardi J, Altmann A, Milazzo AC, Chang C, Chakravarty MM, Banaschewski T, et al. (2015): Brain networks. Correlated gene expression supports synchronous activity in brain networks. Science 348:1241–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mazzoni A, Grove R, Eapen V, Lenroot RK, Bruggemann J (2019): The promise of functional near-infrared spectroscopy in autism research: What do we know and where do we go? Soc Neurosci 14:505–518. [DOI] [PubMed] [Google Scholar]
- 133.Mash LE, Keehn B, Linke AC, Liu TT, Helm JL, Haist F, et al. (2020): Atypical relationships between spontaneous EEG and fMRI activity in autism. Brain Connect 10:18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gordon EM, Laumann TO, Gilmore AW, Newbold DJ, Greene DJ, Berg JJ, et al. (2017): Precision functional mapping of individual human brains. Neuron 95:791–807.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vanderwal T, Eilbott J, Castellanos FX (2019): Movies in the magnet: Naturalistic paradigms in developmental functional neuroimaging. Dev Cogn Neurosci 36:100600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Milham MP, Vogelstein J, Xu T (2021): Removing the reliability bottleneck in functional magnetic resonance imaging research to achieve clinical utility. JAMA Psychiatr 78:587–588. [DOI] [PubMed] [Google Scholar]
- 137.Chekroud AM, Koutsouleris N (2018): The perilous path from publication to practice. Mol Psychiatry 23:24–25. [DOI] [PubMed] [Google Scholar]
- 138.Chekroud AM (2017): Bigger data, harder questions–Opportunities throughout mental health care. JAMA Psychiatry 74:1183–1184. [DOI] [PubMed] [Google Scholar]
- 139.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, et al. (2002): Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15:273–289. [DOI] [PubMed] [Google Scholar]
- 140.Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, et al. (2011): Functional network organization of the human brain. Neuron 72:665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dosenbach NUF, Nardos B, Cohen AL, Fair DA, Power JD, Church JA, et al. (2010): Prediction of individual brain maturity using fMRI [published correction appears in Science 2010; 330:756]. Science 329:1358–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Perrot M, Rivière D, Mangin JF (2011): Cortical sulci recognition and spatial normalization. Med Image Anal 15:529–550. [DOI] [PubMed] [Google Scholar]
- 143.Abraham A, Dohmatob E, Thirion B, Samaras D, Varoquaux G (2013): Extracting brain regions from rest fMRI with total-variation constrained dictionary learning. Med Image Comput Comput Assist Interv 16:607–615. [DOI] [PubMed] [Google Scholar]
- 144.Craddock RC, James GA, Holtzheimer PE 3rd, Hu XP, Mayberg HS (2012): A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum Brain Mapp 33:1914–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, Petersen SE (2016): Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb Cortex 26:288–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. (2006): An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31:968–980. [DOI] [PubMed] [Google Scholar]
- 147.Diedrichsen J, Balsters JH, Flavell J, Cussans E, Ramnani N (2009): A probabilistic MR atlas of the human cerebellum. Neuroimage 46:39–46. [DOI] [PubMed] [Google Scholar]
- 148.Shen X, Tokoglu F, Papademetris X, Constable RT (2013): Groupwise whole-brain parcellation from resting-state fMRI data for network node identification. Neuroimage 82:403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Schaefer A, Kong R, Gordon EM, Laumann TO, Zuo XN, Holmes AJ, et al. (2018): Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb Cortex 28:3095–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Klein A, Tourville J (2012): 101 labeled brain images and a consistent human cortical labeling protocol. Front Neurosci 6:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


