Abstract
Purpose
There are no effective treatment strategies for children with highest-risk posterior fossa ependymoma (PFA). Chromosome 1q gains (1q+) are present in approximately 25% of newly diagnosed PFA tumors and this number doubles at recurrence. Seventy percent of children with chromosome 1q+ PFA will die because of the tumor, highlighting the urgent need to develop new therapeutic strategies for this population.
Experimental Design
In this study we utilize 1q+ PFA in vitro and in vivo models to test the efficacy of combination radiation and chemotherapy in a preclinical setting.
Results
5FU enhances radiotherapy in 1q+ PFA cell lines. Specifically, 5FU increases p53 activity mediated by the extra copy of UCK2 located on chromosome 1q in 1q+ PFA. Experimental down regulation of UCK2 resulted in decreased 5FU sensitivity in 1q+ PFA cells. In in vitro studies, combination of 5FU, tretinoin (ATRA) and radiation provided the greatest reduction in cellular proliferation and greatest increase in markers of apoptosis in 1q+ PFA cell lines compared to other treatment arms. Similarly in vivo experiments demonstrated significant enhancement of survival in mice treated with combination radiation and 5FU and ATRA.
Conclusion
These results are the first to identify a chromosome 1q+ specific therapy approach in 1q+ PFA. Existing phase 1 studies have already established single-agent pediatric safety and dosages of 5FU and ATRA, allowing for expedited clinical application as phase 2 trials for children with high-risk PFA.
Introduction
Ependymoma (EPN) is an aggressive pediatric brain tumor with poor survival and significant morbidity, particularly in the context of multiple recurrences1. Most pediatric EPN arise in the posterior fossa (PF) and are predominantly subgroup A EPN (PFA). Over 70% of children with PFA tumors will relapse, and once relapsed, effective therapies are lacking2,3. Specific negative prognostic risk factors include PFA and gain of chromosome 1q (1q+) based on multiple national and international study cohorts. Children’s Oncology Group (COG) ANCS0121 trial demonstrated a 5-year progression-free survival rate for posterior fossa EPN patients with 1q+ of 47.4% and 1q wild type (WT) of 82.8% (p=0.189)2. In a more recent multi-center study of matched primary and recurrent pediatric EPN tumors, 25% had gain of 1q+ however this number doubled at recurrence with 50% of tumors at first recurrence having 1q+4.
The role of chemotherapy in treatment of EPN remains uncertain. Several ongoing clinical trials in US and Europe are evaluating standard chemotherapy agents, agents that are known to cross the blood-brain barrier, as opposed to drugs that have biologic rationale specific to EPN. Although the COG ACNS0831 trial, which randomized posterior fossa EPN patients to a multiagent chemotherapy regimen or observation following radiation, has completed accrual, data analysis is ongoing. In the meantime, several groups have conducted high-throughput drug screens in models of pediatric EPN, including PFA cell lines with 1q+, to identify rational chemotherapeutic agents to combine with or follow standard radiation therapy5,6. These preclinical screening studies identified purine antimetabolite 5-fluorouracil (5FU) as an EPN selective therapy. 5FU is a well-established chemotherapeutic agent that rapidly enters cells and is converted to several active metabolites causing downstream DNA and RNA damage. 5FU is used for standard treatment of cancers, such as colorectal adenocarcinoma, hepatoblastoma, and nasopharyngeal carcinoma. Of specific relevance to PFA, a recent report found that gain of 1q+ was associated with significantly higher sensitivity to 5FU compared to wildtype 1q tumor cells of multiple tumor types7.
A pharmacokinetic study demonstrated optimal drug concentrations in brains of murine models of ependymoma with IV bolus administration of 5FU, as opposed to continuous infusion methods prevalent among cancer regimens8. Preclinical pharmacokinetic-pharmacodynamic analysis from this study confirmed the dosage selected for IV bolus 5FU achieved target exposure for antitumor effect8. A St Jude phase I study was subsequently designed to establish the safety profile of single-agent weekly IV bolus 5FU in a cohort of recurrent EPN9. Twenty-three relapsed pediatric EPN patients (16 posterior fossa; 6 supratentorial; 1 spinal tumor location) were enrolled. Of the enrolled PF patients, five (31%) patients experienced partial responses (duration 6–54 weeks, median 12 weeks); and patients who demonstrated objective tumor responses were treated at dosage 500 mg/m2/dose or higher9. While this study was encouraging, there was not a clear mechanism for 5FU sensitivity in pediatric EPN patients and further clinical trials were not explored.
Considering the known radiosensitization properties of 5FU in multiple tumor types, the efficacy of combining 5FU during radiation in in vitro and in vivo preclinical models of 1q+ PFA was tested. Additionally, we identify the mechanism by which 5FU sensitivity is enhanced in 1q+ PFA cells. Further, we identify retinoid tretinoin (ATRA), that was previously identified as a PFA-selective FDA-approved oncology drug5, as a rational and effective maintenance strategy for combination with 5FU and chemotherapy, that in vivo showed significant improvement of long-term survival of mice with 1q+ PFA. This manuscript provides a biologically tested therapy approach in both in vitro and in vivo preclinical models of highest risk pediatric EPN subtype, that combines already FDA approved agents with known safety profiles in children, making it readily translatable to large scale clinical trials.
Materials and Methods
Cell Lines
EPN cell lines, MAF-811 and MAF-928, were established from recurrent pediatric PFA that have been well characterized10. Both cell lines harbor highest risk phenotype of chromosome 1q gain (1q+) and loss of chromosome 6q (6q−). These cell lines are established from flank tumors propagated in NSG mice and viably cryopreserved in Bambankers (BB001, GC Lymphotec, Tokyo, Japan). Cell lines are routinely screened for mycoplasma, and short tandem repeat sequencing to ensure cell line identities. For all in vitro experiments, cells are cultured as monolayers in Opti-MEM with 15% FBS and 1% pen/strep (O15) (Gibco). HEK 293 FT cells (RRID:CVCL_6911, ATCC) were cultured in DMEM supplemented with 10% FBS and 1% pen/strep. All in vitro experiments were performed using cells below passage 5 following thawing.
Chemotherapy agents
Fluorouracil (5FU) (Sigma PHR1227) and tretinoin (ATRA) (Sigma PHR-1187) were dissolved in DMSO and stored in small aliquots at −80°C. For in vitro experiments, 5FU and ATRA stocks were further diluted in serum free Opti-MEM. For in vivo experiments, 5FU stocks were diluted in sterile PBS and ATRA was made fresh daily by resuspending the powder in sterile corn oil (Sigma C8267). All work involving chemotherapy agents are performed under institutional approved safety protocol (IBC-00001304).
IncuCyte Growth Assays
MAF-811 and MAF-928 cells were seeded at 4,000 cells per well in six 96-well plates (Corning, Corning, NY) in O15. Cells were cultured at 37°C and 5% CO2 and monitored using an Incucyte Zoom (RRID: SCR_019874, Essen BioScience, Ann Arbor, MI). After 24hours, plates were treated with radiation on a cesium source irradiator. Each dose was delivered in 1.25Gy fractions, rotating the plates at each fraction until the final radiation dose was achieved. Immediately following radiation CellEvent Caspase-3/7 Green Detection Reagent (Life Technologies, Grand Island, NY) and 5FU curve starting at 10μM was added to the plates. For combination experiments using ATRA, radiation and 5FU treatments were repeated as described and cells incubated for 48hrs before 10μM ATRA was added. Images were captured at 4-hour intervals from four separate regions per well using a 10x objective over 10 days. Each experiment was done in triplicate and accumulation of caspase 3/7 over time was normalized to confluence of cells.
Cell Viability Assays
Following the 10 days of incubation on the IncuCyte, cell viability was measured using Cell Titer Glo (Promega) following manufacturer’s protocol. The optical density of each well was measured using Cell Titer Glo protocol template on GloMax Explorer (RRID:SCR_015575, Promega). The proportion of cells per treatment group was normalized to control wells of DMSO control-treated cells.
Transcriptional Analyses
Gene expression of MDM4 and UCK2 was obtained from existing RNAseq transcriptomic data published as part of a recent study (GSE226961) in accordance with local and federal human research protection guidelines and institutional review board regulations (COMIRB 95-500). These data were supplemented with new RNAseq data from 16 “normal” brain samples (GSE244124) obtained predominantly from autopsy material that is collected after parent approval and deidentified in accordance with HIPPA regulations. RNA was isolated from autopsy samples and sequenced using the Illumina Novaseq6000. Sequencing data was aligned using STAR (RRID: SCR_004463) and quantification of RNA expression as fragments per kilobase per million and derived by Cufflinks (RRID: SCR_014597) as described previously11.
Quantitative Real-Time PCR
Total RNA was isolated from cells using DNA/RNA All Prep Kit (Qiagen Cat. 80204) analyzed for gene expression using qRT-PCR performed on a StepOne Plus Real Time PCR System with Taqman Gene Assay Reagents (Applied Biosystems), according to manufacturer’s protocols. cDNA was generated from total RNA using High-Capacity cDNA Kit (Applied Biosystems). Resulting cDNA was loaded at 50ng per well for qRT-PCR (four replicates), along with Taqman gene-specific probe for UCK2 (Hs00989900_m1) and CDKN1A (p21) (Hs00355782_m1). Relative gene expression was calculated using ΔΔCt method with 18S (Hs03003631_g1) as endogenous control.
Proteome Profiler
To investigate the mechanism of cell death from 5FU and radiation treated cells, we used the Human Apoptosis Array Kit (RRID: AB_3086729, R&D ARY009). The apoptosis array is a nitrocellulose membrane that detects 35 apoptosis related proteins including positive controls. To generate lysates, MAF-811 and MAF-928 cells were plated 106 cells in 10 cm dishes. Cells were irradiated 24 hours later, and 5FU was added immediately after radiation. Cells were harvested per manufacturer’s protocol 72 hours after treatment with radiation and 5FU. Briefly, cells were washed in PBS then homogenized in lysis buffer using a rotorstator. Cell lysates were gently rocked for 30 min at 4°C and centrifuged at 14,000×g for 5 min (4°C). The supernatants removed and stored at −80°C until use. A total of 250 μg of protein, as determined by BCA assay, was used for each array and developed using chemiluminescent reagent provided. Positive signals on developed film were analyzed using a transmission mode scanner and collected at pixel density using Image J software (RRID:SCR_003070, https://imagej.nih.gov/ij).
shRNA Transfection
Genetic knockdown of UCK2 (TRC number TRCN0000037849 and TRCN00000380355) was performed using a Sigma Aldrich pLKO system. ShRNA plasmids were packaged into virus by transfection of HEK 293FT cells with TransIT-LT1 Reagent (Mirus) and pMD2G (RRID:Addgene_12259) and psPax (RRID:Addgene_12260) packaging plasmids. Viral supernatant was harvested and centrifuged to remove debris. Polybrene was added to virus and to EPN cell lines prior to viral transduction. Virus was removed after 24 hours, and transduced cells were positively selected with puromycin. Knockdown validation was performed using qRT-PCR.
Animal models
We have previously published the characteristics of MAF-928_XC patient-derived orthotopic xenograft models (PDX)12. Briefly, 250,000 cells from viably frozen stocks of disaggregated flank tumor cells were implanted into the 4th ventricle (lat: 0.000, ant: −6.000, ventr: −4.3000; with respect to the bregma), under stereotactic control, of 6–8week old NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (RRID:IMSR_JAX:005557, bred in house) mice. Sequential T2-weighted magnetic resonance imaging (T22-MRI, field strength 9.4 Tesla) to confirm tumor engraftment were performed between 6-8 weeks post-surgery with a 90-100% take rate for this PDX model. All mouse work was approved by CU Anschutz Institutional Animal Care and Use Committee (IACUC 00784). All mice were housed in CU Anschutz Vivarium. Mice were maintained at five per cage under humidity and temperature-controlled conditions with a light/dark cycle set at 12-hours. Mice were given plastic huts, cotton pads and autoclaved cardboard toilet paper rolls to provide enrichment. Handling was performed with universal sterile precautions and experienced personnel perform all procedures. There were two full-time veterinarian technicians and a staff of animal caretakers. Animals were monitored for discomfort daily by laboratory animal technicians. Due to severe stress that can be created by tumor development, mice were sacrificed as soon as signs are evident in consultation with our veterinarians.
Preclinical in vivo experiments
Mouse numbers for each group were determined a priori using 25% increase in survival with a power of 90% and an alpha of 0.05. Six mice were needed per treatment arm to achieve significance with three experimental replicates. Following confirmation of tumor formation by T2w-MRI scan, cages were arranged in numerical order and mice numbered 1-36. Treatment assignment was given based on order in which drugs were administered (Vehicle Control (n=5), 5FU only (n=4), 5FU+ATRA (n=5), 10Gy+Vehicle (n=12),10Gy+5FU (n=12) and 10Gy+5FU+ATRA (n=12). For chemo only groups in the initial experiment, cage floods resulted in death unrelated to tumor. With the numbers that survived, chemotherapy showed little effect. This prompted elimination of chemotherapy only groups in repeat experiments to prevent unnecessary harm. MRI analysts, who provided read-outs on tumor volumes, were blinded to treatment group assignment. High-resolution T2w-MRI scans were performed on a Bruker 9.4 Tesla BioSpec scanner and all analysis was carried out using ParaVision NEO v.2.2 as previously described12.
Irradiation was performed under IACUC protocol 00784. Using the X-RAD SmART image-guided irradiator (RRID:SCR_021996, Precision X-Ray Inc., North Branford CT) at 225kVp with a 0.3mm copper filter, mice received 10Gy in five daily fractions at a dose rate of 5.9Gy/min. Under isoflurane anesthesia, each mouse was positioned in prone orientation and aligned to isocenter in two orthogonal planes by fluoroscopy12. Half the dose was delivered from each side in opposing lateral beams. Monte-Carlo simulation in SmART-ATP (SmART Scientific Solutions B.V., Maastricht, Netherlands) showed 95% coverage of the target volume (4th ventricle) receiving the prescribed dose.
5FU dose of 75mg/kg was previously established in preclinical study for ependymoma8. However, this dose proved to be too toxic in our 1q+ PFA PDX models when combined with radiation and we reduced the dose moving forward (Figure S1). 5FU (30mg/kg) or equivalent PBS volume was administered once weekly via intraperitoneal injections. Mice receiving radiation in combination with 5FU were given one dose a day prior to radiation and one dose on the final day of irradiation. This was to simulate a human trial in which weekly 5FU injections are given during 52 weeks of fraction radiation. Mice were given a two-week recovery period following radiation before weekly cycles of 5FU or PBS were continued. Cycles were comprised of weekly 5FU for six weeks followed by a 2-week break and repeated until mice reached endpoint. ATRA (40mg/kg) or equivalent volume of corn oil was administered through oral gavage five days per week. ATRA treatments began after the two-week recovery from radiation and were given in six-week cycles with two-week break to coincide with 5FU schedule until mice reached study endpoint. Three MRI scans were performed following radiation during rest periods to monitor tumor growth. Mice were provided moist chow or nutrigel (S5769, Bio-Serv, Flemington, NJ) during chemotherapy cycles to support weight loss due to treatment. Mice were monitored every day and weights documented twice weekly.
Survival was the primary outcome measurement for preclinical studies. Chemotherapy toxicities (for example, weight loss) and tumor volume were secondary outcome measurements. Endpoint criteria for this study was approved through IACUC 00784. When mice were moribund and/or had lost more than 15% of their initial body weight, they were euthanized. Mice were euthanized by CO2 asphyxiation followed by neck dislocation by lab personnel. Following neck dislocation, brains were removed and preserved in 10% Buffer Formalin for 24 hours before being transferred to 70% ethanol. Fixed tumor was processed for immunohistochemistry analysis as previously described12.
Statistical Analysis
Survival analysis was performed using Kaplan-Meier analysis. All data was pooled from replicate experiments and reported as the mean with standard deviation for error bars. Quantification of tumor burden was achieved by hands-free drawing of the region of interest (ROI on axial and sagittal T2w-MRI scans, by two analyst.) The total of ROI areas on all affected slices was multiplied by slice thickness to achieve 3D assessment of tumor burden, using Bruker ParaVison NEO v.2.2. software. Statistical significance was defined as p<0.05 using student t-test (RRID:SCR_002798, GraphPadv10).
Data availability
The normal brain gene expression data generated in this study are publicly available in Gene Expression Omnibus (GEO, RRID: SCR_005012) at GSE 244124 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi=GSE244124). All other data generated in this study is available upon request from the corresponding author.
Results
5FU sensitivity is enhanced with radiation in in vitro models of 1q+ PFA
Our original work evaluating an FDA approved drug screen panel identified 5FU analog, floxuridine, as the top PFA selective inhibitor5. However, fluorouracil (5FU), also a top PFA selective compound in this study, was chosen as the agent to move forward based on existing pharmacokinetic and safety studies available in children8,9. Two pediatric PFA cell lines harboring 1q+/6q−, MAF-811 and MAF-928, were seeded in 96-well plates and treated with a dose curve of fractioned radiation (Figure 1A). Immediately following radiation treatment, a range of 5FU doses for dose curve modelling was added to each plate. We measured cell proliferation by phase confluency (Figure 1B) and caspase 3/7 cleavage (Figure S2A) every 4 hours using the incucyte live cell imaging system. MAF-928 was much more sensitive to both 5FU and radiation alone with IC-50s of 2.5μM and 2.5Gy respectively. MAF-811, which has more differentiated expression profile10, was sensitive to higher doses of 5FU and similar radiation doses with IC-50s of 5μM and 2.5Gy respectively. These IC-50 doses are in range of those found in previous published studies that identified 5FU as a candidate for ependymoma treatment and correlate with in situ pharmacokinetic-pharmacodynamic simulation model for 5FU dosing in pediatrics6,8. When 5FU was combined with radiation, we found significant growth impairment compared to single treatment alone (Figure 1C and Figure S2B). This corresponded with greater loss of cell viability after 10 days, particularly in MAF-928 cells in which radiation alone had little effect on cell viability (Figure 1D). We did not see a significant difference in cleaved caspase 3/7 over time (Figure S2A).
Figure 1. In vitro evaluation of combination 5FU and radiation in 1q+ PFA cell lines.
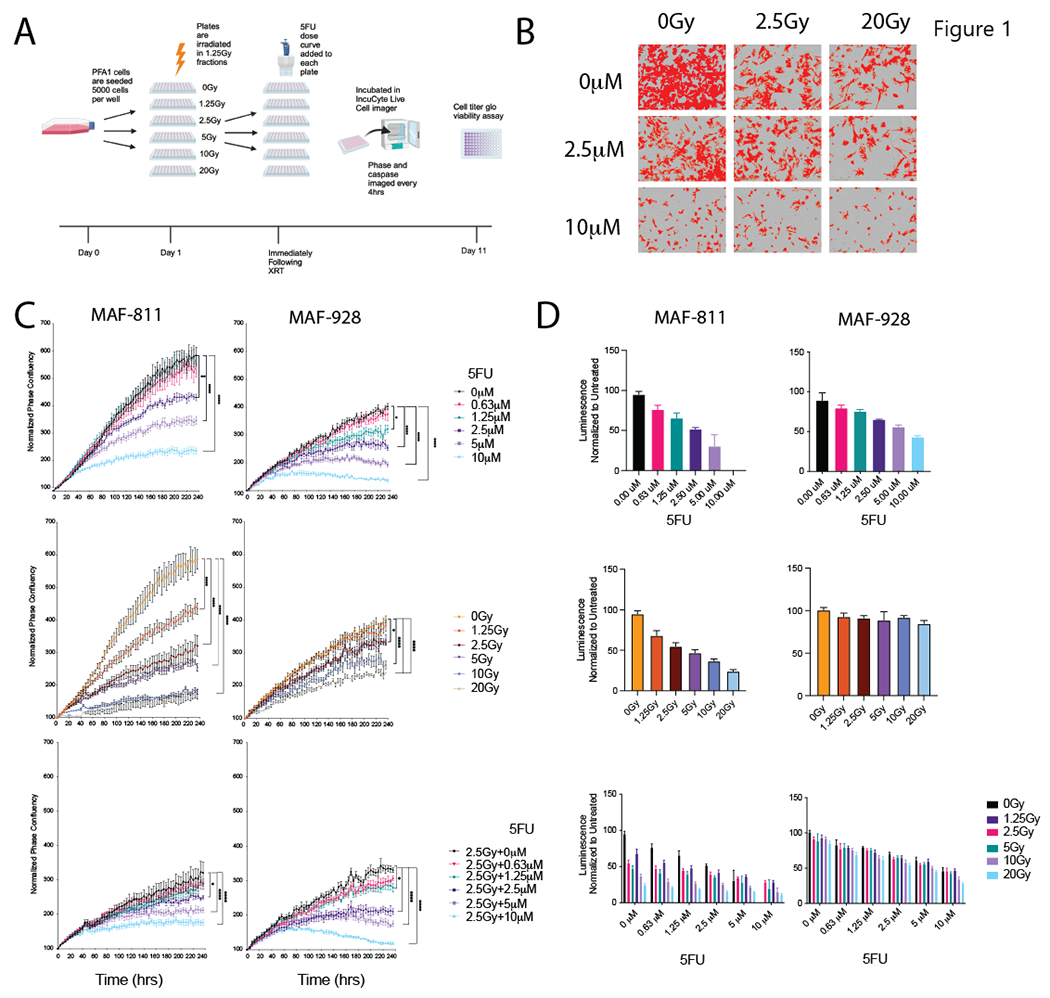
A. Schematic of timing and order of treatments for in vitro studies. Graphic made using BioRender. B. Representative phase contrast images from IncuCyte live-cell imager with confluence mask overlay (red). C. Proliferation curves following treatment with 5FU alone (top), radiation alone (middle) and combination 2.5Gy and 5FU (bottom) Data represented as normalized phase confluency. * Denotes p-value<0.05, **** denotes p-value<0.0001. D. Viability assays measured by Cell Titer Glo after 10 days of treatment with 5FU alone (top), radiation alone (middle) and combination (bottom). Data is represented as normalized luminescence to the untreated controls.
5FU treatment upregulates p53 signaling
Gain of 1q in pediatric PFA results in trisomy chromosome 1q. Girish et al. recently reported that trisomy 1q mimics p53 deactivating mutations in part due to the third copy of MDM4, known to be negative regulator of p537. PFA do not harbor p53 mutations, nor have any other known mutational drivers been identified in PFA. We investigated the expression of MDM4 across our PFA tumor bank to determine whether 1q+ PFA tumors had increased expression. Both 1q WT and 1q+ PFA tumors have higher MDM4 gene expression compared to normal pediatric brain (Figure 2A). Consistent with Girish et al.’s findings, PFA tumors with gain of chromosome 1q have significant higher gene expression of MDM4 compared to WT PFA tumors (FPKM mean difference 4.885, 95%CI 1.644 to 8.127, p-value=0.0018) (Figure 2A). Interestingly, CDKN1A (p21) gene expression was significantly increased in 5FU-treated cell lines in a dose response manner as measured by qRT-PCR (Figure 2B). As CDKN1A is upregulated by p53 activity, this would suggest that 5FU might restore p53 activity in 1q+ PFA cell lines.
Figure 2. Mechanism for 5FU sensitivity in 1q+ PFA.
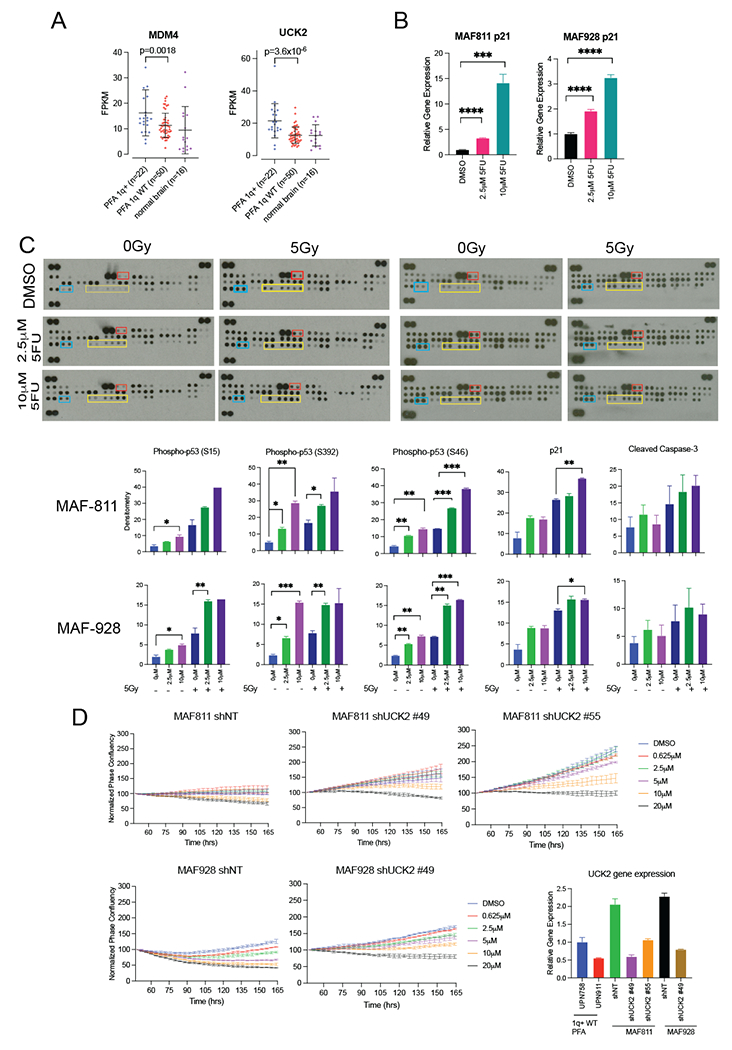
A. Gene expression levels of MDM4 and UCK2 in 1q+ PFA, 1q wild type (WT) PFA and normal brain. P-value<0.0001. B. CDKN1A (p21) relative gene expression following treatment with 5-FU in MAF-811 and MAF-928. C. Representative apoptosis proteomic dot blots for combination 5-FU and radiation treatment in MAF-811 and MAF-928. Yellow boxes outline the dots corresponding to p-p53 proteins. The blue boxes outline p21 protein and red boxes outline cleaved caspase-3 protein. Bar graphs show densitometry values of each protein normalized to the positive controls for the dot blot D. 5-FU proliferation curves of Non-Targeting shRNA transfected cells (shNT) and shRNA targeting UCK2 (shUCK2). UCK2 reduction was measured by qRT-PCR and normalized to 1q WT PFA tumors, UPN 758 and UPN 911.
We therefore sought to measure p53 protein activity and other apoptosis pathway effectors using an apoptosis proteomic array in both MAF-811 and MAF-928 cell lines following 5FU treatment (Figure 2C and S3). In both cell lines, there was an increase in phosphorylated p53 on all three serine sites in response to 5FU treatment (Figure 2C). Consistent with qRT-PCR data (Figure 2B) we also observed an increase of p21 protein in 5FU treated cells (Figure 2C). We found p53 protein phosphorylation was further increased with radiation treatment (Figure 2C). While we did not see a significant change in real-time imaging of cleaved caspase 3/7, we observed higher levels cleaved caspase-3 in the proteomic array analysis with radiation treatment alone and with combination 5FU and radiation (Figure 2C). We performed MDM4 qRT-PCR on both cell lines following 5FU treatment and found MDM4 increased with 5FU (Figure S4). These data suggest that 5FU treatment, especially in combination with radiation, appears to overcome trisomy MDM4-mediated suppression of p53 activity but this effect is independent of MDM4 gene expression. While increased phosphorylation of p53 does not mean all p53 functions are restored, our findings do provide a partial mechanism for the significant decrease in proliferation and viability in combined 5FU and radiation treated 1q+ PFA cell lines.
UCK2 is required for 5FU sensitivity in 1q+ PFA cell lines
Another therapeutically relevant trisomy gene in 1q+ tumors identified by Girish et al. is Uridine-cytidine kinase 2 (UCK2)7. UCK2 is a rate limiting protein that facilitates incorporation of uracil during DNA replication, and in 1q+ tumors were shown to confer sensitivity to anticancer nucleotide analogs that include 5FU7. We therefore hypothesized reduction of UCK2 would decrease 1q+ PFA cell lines’ sensitivity to 5FU. Consistent with findings of Girish et al., 1q+ PFA tumors have significantly higher UCK2 gene expression compared to wild type 1q PFA tumors (FPKM mean difference 8.905, 95% CI 5.245 to 12.56, p-value <0.0001) (Figure 2A). We used qRT-PCR to compare UCK2 gene expression following stable introduction of shRNA targeting UCK2 and compared these levels to two 1q WT PFA tumors (UPN 758 and UPN 911). Interestingly, while MAF-811 and MAF-928 are trisomy for UCK2, we found relative gene expression by qRT-PCR was 2-fold higher than both 1q WT PFA tumors (Figure 2D). Both shRNA constructs were able to reduce UCK2 gene expression to levels of 1q WT PFA tumors (Figure 2D). Following selection of shUCK2 and shNT MAF-811 and MAF-928 with puromycin, we performed proliferation assays with the same dose curve of 5FU as previous experiments. In cells containing non-targeting control, we saw effects on growth with 5μM and higher of 5FU in MAF-811 and 2.5μM and higher of 5FU in MAF-928 (Figure 2D). Knockdown of UCK2 gene expression reversed 5FU sensitivity with growth suppression starting to be seen only with the highest 5FU dose of 10μM in both MAF-811 and MAF-928 (Figure 2D). This would suggest that pediatric patients with 1q+ PFA would be more sensitive to 5FU treatments given trisomy UCK2 levels.
Radiation improves 5FU efficacy in vivo
Standard of care for pediatric EPN is maximal surgery followed by radiation treatment. We therefore evaluated the efficacy of 5FU treatment alone and in combination with fractionated radiation in vivo using our novel 1q+ PFA patient derived xenograft models12. 5FU was given twice during fractionated radiation to the posterior fossa in tumor-bearing mice (Figure 3A). 5FU alone was not effective and experiments without radiation were humanely discontinued (Figure 3B). Overall survival in mice treated with 10Gy radiation combined with 5FU was prolonged compared to radiation alone though this data did not reach significance (Figure 3B). Tumor volume from MRI scans also showed a trend in response to 10Gy+5FU compared to radiation alone, more numbers would be needed to achieve significance. Immunohistochemistry analysis of tumor removed from radiation alone mice showed occurrence of metastatic tumor lesions in the olfactory bulb and lateral ventricles (Figure S5).
Figure 3. In Vivo efficacy of combination radiation and 5FU treatment.
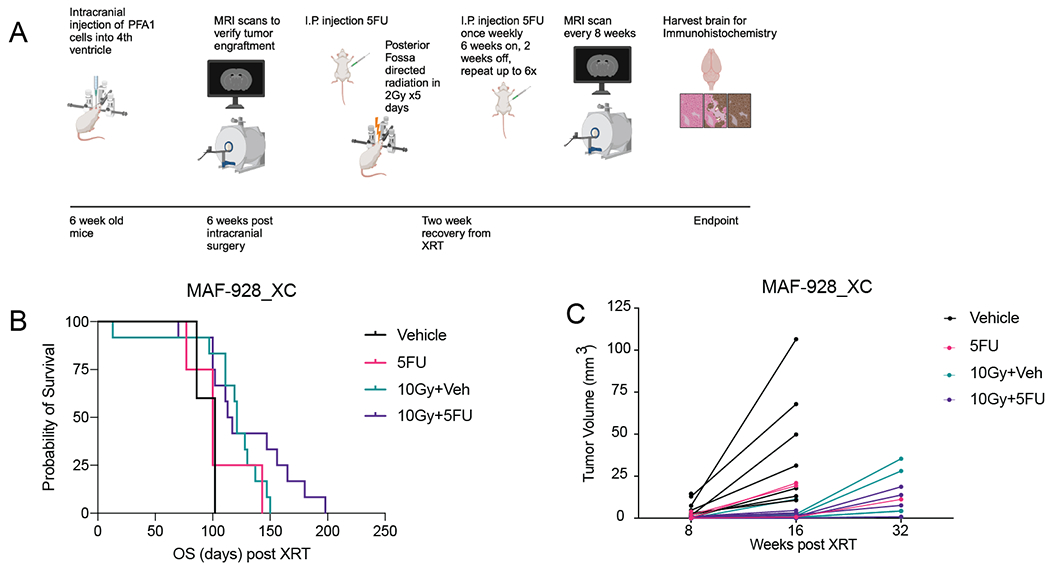
A. Treatment strategy for in vivo experiments. Graphic created using BioRender. B. Survival for 1q+ PFA mice treated with 5-FU, radiation, or combination radiation and 5-FU. C. Tumor volume as measured from MRI scans that were available. D. Representative immunohistochemistry of Ki67 and Caspase 3 stains. E. Quantification of Ki67 and Caspase 3 counts from immunohistochemistry analysis that were available.
Experiments in our second patient derived xenograft model were significantly hindered by the COVID19 pandemic. MAF-811_XC mice were only given one six-week cycle of 5FU maintenance therapy, and this was not sufficient to prolong survival in combination with radiation (Figure S6).
ATRA further enhances 5FU and radiation in vitro
While we observed a significant decrease in tumor growth in vitro with 5FU in combination with radiation, we wanted to evaluate whether efficacy could be improved by adding a second chemotherapy agent. Tretinoin (ATRA) was identified as a top PFA specific inhibitor from our original FDA-drug screen5(Figure S7A). ATRA is a standard component of certain pediatric leukemia therapy regimens. To simulate using ATRA as a maintenance chemotherapy, we treated MAF-811 and MAF-928 with 5FU and radiation as before and 48 hours later treated with a single dose of ATRA (Figure 4A). ATRA alone reduced cell proliferation and viability in both MAF-811 and MAF-928 (Figure 4B). Combining 5FU and ATRA resulted in similar loss of cell proliferation as 5FU alone though still significant compared to ATRA alone (Figure 4B top panel). We also see a similar pattern when ATRA was added following just radiation (Figure 4B middle panel) However, when ATRA was added following 2.5Gy radiation and 5FU treatment, we observed a greater loss of proliferation for all doses 5FU (Figure 4B bottom panel). For MAF-811, we saw decrease cell viability after 10 days of cells treated with combination treatment, however viability was relatively similar for MAF-928 (Figure S7B). This is likely due to the number of cells left after 10 days in MAF-928 combination treated wells (Figure 4B). As we - observed in previous experiments, there was not a significant difference in real-time imaging of cleaved caspase 3/7 (Figure S7C).
Figure 4. ATRA treatment enhances combination radiation and 5FU treatment in vitro.
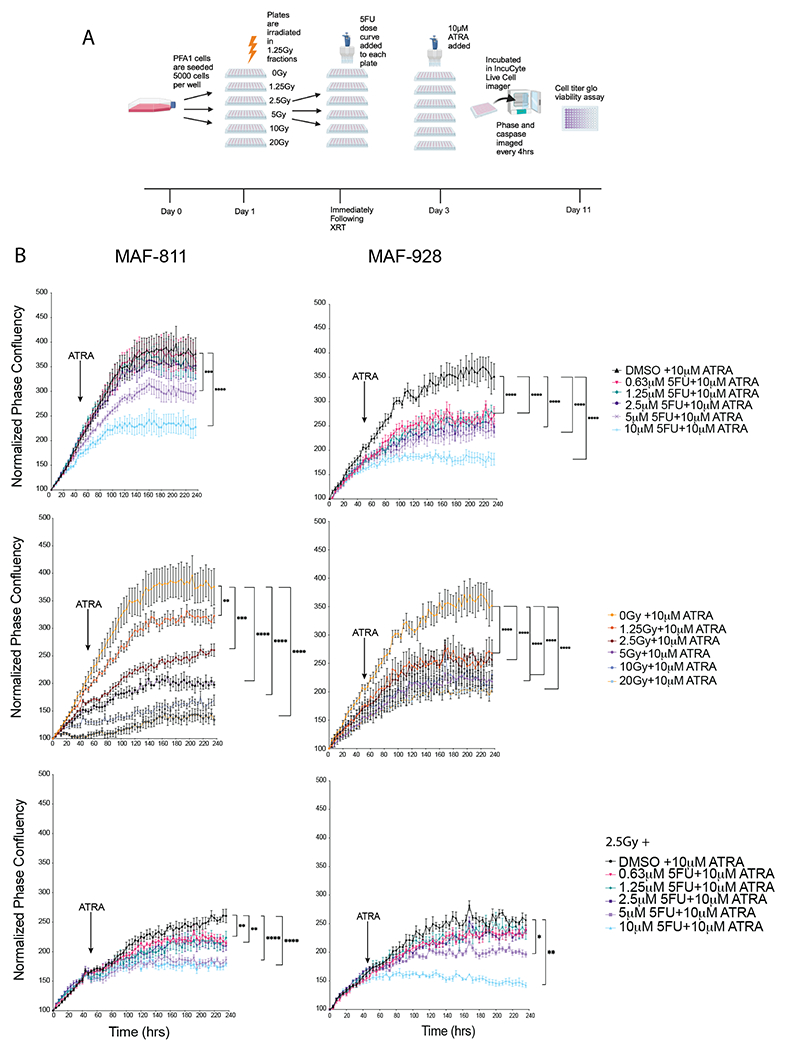
A. Timeline and order of in vitro experiments. Graphic created in BioRender. B. Representative phase confluency images from IncuCyte live-cell imager with confluency mask (red). C. Proliferation growth curves of combination radiation, 5FU and ATRA treated cells. * Denotes p-value<0.05. D. Cell viability measured using Cell Titer Glo 10 days following combination treatments.
Combination radiation, 5FU and ATRA significant prolongs survival in vivo
We next evaluated the efficacy of adding an ATRA maintenance chemotherapy regime to our in vivo model. Following recovery from radiation treatment, mice were given both weekly 5FU and daily ATRA treatment for 6 weeks with a 2-week break (Figure 5A). For ATRA alone, we did see a significant survival advantage over vehicle alone (Figure 5B). However, there did not seem to be a benefit to adding 5FU with ATRA without radiation. When combination 5FU and ATRA was given following radiation treatment, there was a significant increase in mouse survival (95% CI 1.868 to 14.87, p-value=0.0017). Additionally, a decrease in tumor growth was evident with combination treatment and was nearly significant at 32 weeks following radiation (Average tumor volume 10Gy+Veh= 14.55, Average tumor volume 10Gy+5FU+ATRA= 4.8, 95%CI −22.06 to 2.556, p-value 0.055) (Figure 5C).
Figure 5. Pre-clinical evaluation combination chemotherapy and radiation in 1q+ PFA in vivo model.
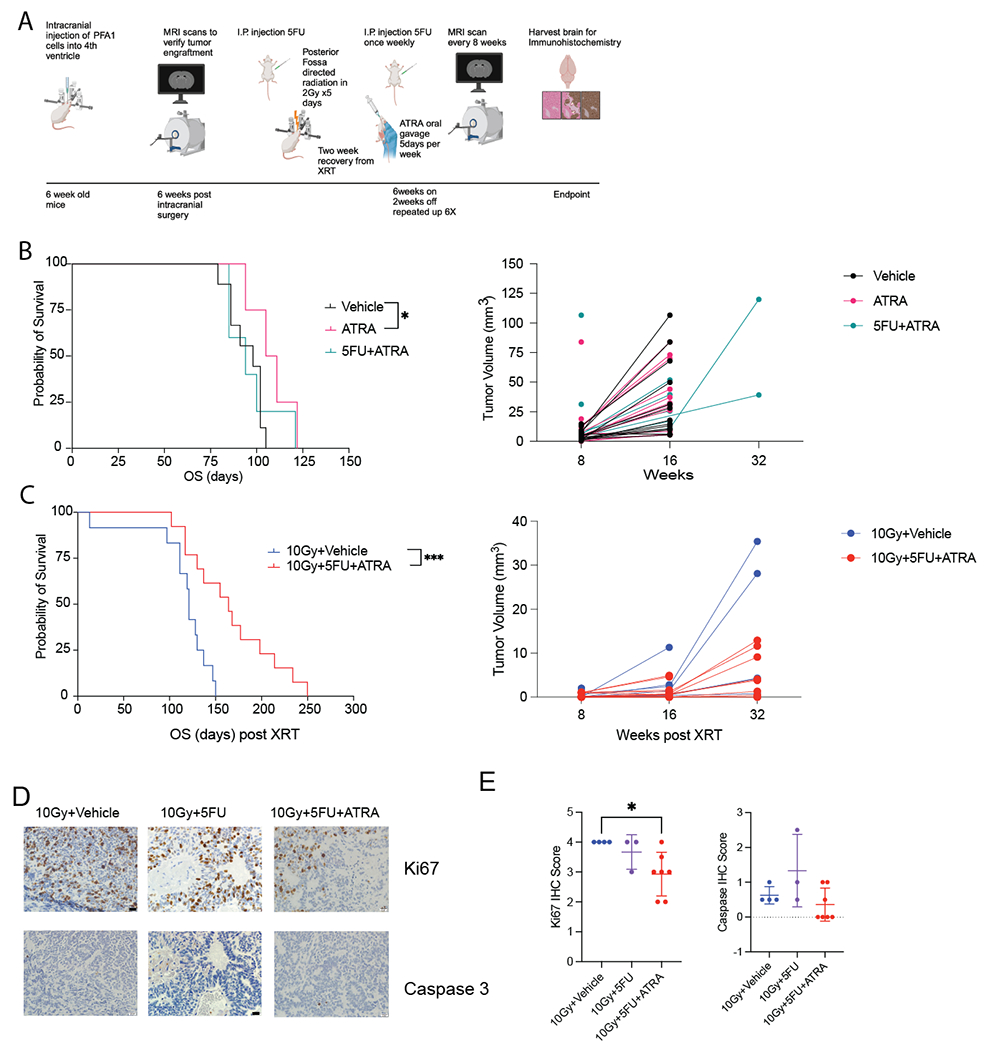
A. Timeline and order of in vivo experiments. Graphic made in BioRender. B. Kaplan Meier survival analysis of mice treated with 5FU+ATRA and in combination with radiation. ** denotes p-value <0.01. C. Tumor volume measured by MRI scans. D. Representative immunohistochemistry of Ki67 and caspase 3 stains. E. Quantification of Ki67 and caspase 3 counts from immunohistochemistry analysis. * Denotes p-value<0.05.
Immunohistochemistry analysis of tumor removed from mice postmortem shows significant tumor burden in radiation alone mice (Figure 5D). Ki67 counts for combination chemotherapy and radiation were significantly lower than with radiation alone (Figure 5D, p-value<0.05). However, there was not a significant difference in caspase counts between treatment conditions (Figure 5D).
We also performed these experiments with MAF-811 xenograft models (Figure S8). Again, these mice were only given a single 6-week cycle of chemotherapy. We did however observe significance in overall survival in combination 5FU+ATRA (Figure S8A). MAF-811 is a much slower growing xenograft compared to MAF-928 which is likely the reason we observed a response with just chemotherapy alone. However, when combined with radiation, we did not see a survival advantage (Figure S8B).
Discussion
Chromosome 1q gain (1q+) PFA have poor outcomes, and most patients experience recurrence or death within 2 years. The incidence of 1q+ at primary diagnosis in PFA is approximately 25% and this number doubles at first recurrence4. Time to disease recurrence and death is considerably shorter for children with 1q+ PFA than children with 1q WT PFA4. There is an urgent need for therapy designed specifically for this high-risk pediatric population with 1q+ PFA both at diagnosis and recurrence.
Treatment advances in posterior fossa EPN have stalled due to lack of relevant pre-clinical models. We were first to publish in vitro and in vivo models of PFA with both models harboring high-risk phenotype of 1q+10,12. In this study we utilized these models to test the efficacy of 1q+ specific chemotherapy strategy that provides rationale for clinical testing. 5FU and ATRA were identified as top PFA cell line specific inhibitors from an FDA drug screen5. Here we show that combining 5FU with radiation leads to significant loss in cell proliferation in vitro, which was further enhanced when ATRA was added as a maintenance chemotherapy approach. A limitation of the in vitro data is that true synergy was unable to be measured with the analyses performed. However, in vivo studies of combination radiation and 5FU and ATRA led to significant survival advantage compared to just radiation alone. This also corresponded to less tumor burden and decreased Ki67 counts on postmortem histology slides.
While our 1q+ PFA xenograft models are important for preclinical evaluation of any treatment strategy there are still limitations to in vivo studies. Timing of treatment initiation in vivo was shown to impact efficacy, and we have found that, like the human patient disease, no therapy was effective when there was significant tumor burden. The early deaths in the 5FU+radiation arm was not seen in replicate cohort when we started treatment when tumor burden was small on MRI scans. Additionally, the COVID pandemic severely impacted in vivo experiments done in MAF-811 with lockdown delaying initiation of treatment and staff illness preventing continuation of chemotherapy cycles.
5FU is a highly potent and widely available chemotherapy agent. Its use in pediatric EPN was first proposed in 2011 by Atkinson et al. in which a high throughput in vitro and in vivo drug screen was performed to identify possible inhibitors6. 5FU was the top inhibitor identified from their screen, which prompted a Phase 1 study evaluating safety and dosage strategy for children with relapsed EPN9. 5FU and other 5FU analogues were also identified in our drug screen as a top PFA-selective inhibitors5. More recently, a plausible mechanism for 5FU sensitivity was identified. Girish et al., found the gene UCK2, which is located on the long arm of chromosome 1q, is amplified in cancers that have a trisomy 1q phenotype7. UCK2 is a DNA synthesis rate limiting protein that facilitates incorporation of uracil into newly synthesized DNA. Loss of one copy of UCK2 in trisomy 1q cell lines reversed sensitivity of the trisomy cells to 3-Deazauridine, an UCK2-mediated nucleotide analog7. We found 1q+ PFA also had elevated levels of UCK2 compared to 1q WT PFA tumors. We found reducing expression of UCK2 in our 1q+ PFA cells to levels seen in 1q WT PFA was sufficient to reduce sensitivity of the cells to 5FU treatment, providing a mechanism for 5FU as a selective inhibitor in 1q+ PFA. A limitation of our in vitro and in vivo models is both 1q gain and 6q loss high risk chromosomal abnormalities are present. We are therefore unable to determine the extent 6q loss has on the 5FU effect that was observed. However, the loss of sensitivity to 5FU with UCK2 knock down would suggest that loss of 6q does not affect response to 5FU. There are no known models of posterior fossa ependymoma with chromosome 1q gain and wild type chromosome 6q to validate findings or to determine whether ATRA sensitivity correlates with the loss of 6q.
There are no known mutational drivers in PFA. In a recent report, chromosome 1q+, behaves like a p53 mutation through trisomy expression of MDM4, a negative regulator of p53 downstream activation7. We have shown 1q+ PFA tumors have significantly higher MDM4 gene expression compared to 1q WT PFA and normal brain. Interestingly, 5FU treatment, especially when combined with radiation, induced activation of p53 signaling pathway. This finding suggests that 5FU treatment may abrogate the effect of trisomy MDM4 in 1q+ PFA tumors.
We, along with other researchers have reported PFA tumors are a mixture of molecular subpopulations: including undifferentiated, stem-like cells (UEC), mesenchymal cells (MEC), and ependymally-differentiated ciliated (CEC) and transportive EPN cells (TEC)13,14. We hypothesized identifying an inhibitor that would target differentiated UEC toward less aggressive CEC and TEC cells would enhance effect of 5FU and radiation on PFA cell lines. From our FDA drug screen, ATRA was identified as the second most potent PFA-selective FDA approved oncology drug5. Retinoids are potent differentiation agents and it is therefore plausible that ATRA is working by differentiating UEC cells toward low-risk ependymally-differentiated CEC and TEC subpopulations. Further studies need to be completed to determine the mechanism by which ATRA is working in PFA.
Both 5FU and ATRA are agents that are commonly used in pediatric oncology, making them readily translatable into combination clinical trials for high-risk EPN. Based on this work, this multimodal therapy is most promising for the highest risk EPN patients, specifically those children with PFA tumors that harbor 1q gain. Multiple consortia in the US and Europe are planning to develop clinical trials with this combination therapy, as there is an urgent need to improve clinical outcomes for this patient population.
Supplementary Material
Translational Relevance.
Gain of chromosome 1q (1q+) is fatal in most pediatric posterior fossa group A ependymoma (PFA) patients and the incidence of 1q+ is 25% in newly diagnosed PFA increases to 50% at recurrence. Standard therapy of complete resection and radiation have very limited benefit to children with 1q+ PFA which has not seen improvement in trials of additional chemotherapy. In this study, we provide preclinical efficacy in vitro and in vivo models of 1q+ PFA. We identify overexpression of 1q gene UCK2 as a mechanism of sensitivity to 5FU in 1q+ PFA. Further we show preclinical efficacy by combination of 5FU with 1q+ PFA selective retinoid tretinoin (ATRA) and radiation in in vitro and in vivo models of 1q+ PFA. Existing pediatric phase 1 trials have established dosing for 5FU and ATRA agents allowing for rapid translation to phase 1 and 2 clinical trials for children with 1q+ PFA.
Acknowledgements
The genomics core is funded through University of Colorado Cancer Center (P30CA046934). All MRI scans were acquired at the Animal Imaging Shared Resources and by P30CA046934 and the NIH high-end shared instrumentation grant (S10OD023485). Animal studies were funded by The Tanner Seebaum Foundation and Department of Defense (CA190494).
This study was supported by NIH/NCATS Colorado CTSA Grant Number UM1 TR004399. Contents are the authors’ sole responsibility and do not necessarily represent official NIH views.
The authors would like to thank The Morgan Adams Foundation for their continued support.
The authors would like to thank the veterinarians, animal care team and vet technicians at University of Colorado Anschutz vivarium.
Funding sources
All funding sources for this study are listed in the “acknowledgments” section of the manuscript.
Footnotes
Institutional affiliations
All affiliations are listed on the title page of the manuscript.
Competing financial interests
We, the authors and our immediate family members, have no financial interests to declare.
Advisory/management and consulting positions
We, the authors and our immediate family members, have no positions to declare and are not members of the journal’s advisory board.
Patents
We, the authors and our immediate family members, have no related patents to declare.
References
- 1.Marinoff AE, Ma C, Guo D, Snuderl M, Wright KD, Manley PE, Al-Sayegh H, Sinai CE, Ullrich NJ, Marcus K, et al. (2017). Rethinking childhood ependymoma: a retrospective, multi-center analysis reveals poor long-term overall survival. J Neurooncol 135, 201–211. 10.1007/s11060-017-2568-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merchant TE, Bendel AE, Sabin ND, Burger PC, Shaw DW, Chang E, Wu S, Zhou T, Eisenstat DD, Foreman NK, et al. (2019). Conformal Radiation Therapy for Pediatric Ependymoma, Chemotherapy for Incompletely Resected Ependymoma, and Observation for Completely Resected, Supratentorial Ependymoma. J Clin Oncol 37, 974–983. 10.1200/JCO.18.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ridley L, Rahman R, Brundler MA, Ellison D, Lowe J, Robson K, Prebble E, Luckett I, Gilbertson RJ, Parkes S, et al. (2008). Multifactorial analysis of predictors of outcome in pediatric intracranial ependymoma. Neuro Oncol 10, 675–689. 10.1215/15228517-2008-036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donson AM, Bertrand KC, Riemondy KA, Gao D, Zhuang Y, Sanford B, Norris GA, Chapman RJ, Fu R, Willard N, et al. (2023). Significant increase of high-risk chromosome 1q gain and 6q loss at recurrence in posterior fossa group A ependymoma: a multicenter study. Neuro Oncol. 10.1093/neuonc/noad096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donson AM, Amani V, Warner EA, Griesinger AM, Witt DA, Levy JMM, Hoffman LM, Hankinson TC, Handler MH, Vibhakar R, et al. (2018). Identification of FDA-Approved Oncology Drugs with Selective Potency in High-Risk Childhood Ependymoma. Mol Cancer Ther 17, 1984–1994. 10.1158/1535-7163.MCT-17-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atkinson JM, Shelat AA, Carcaboso AM, Kranenburg TA, Arnold LA, Boulos N, Wright K, Johnson RA, Poppleton H, Mohankumar KM, et al. (2011). An integrated in vitro and in vivo high-throughput screen identifies treatment leads for ependymoma. Cancer Cell 20, 384–399. 10.1016/j.ccr.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girish V, Lakhani AA, Thompson SL, Scaduto CM, Brown LM, Hagenson RA, Sausville EL, Mendelson BE, Kandikuppa PK, Lukow DA, et al. (2023). Oncogene-like addiction to aneuploidy in human cancers. Science 381, eadg4521. 10.1126/science.adg4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daryani VM, Patel YT, Tagen M, Turner DC, Carcaboso AM, Atkinson JM, Gajjar A, Gilbertson RJ, Wright KD, and Stewart CF (2016). Translational Pharmacokinetic-Pharmacodynamic Modeling and Simulation: Optimizing 5-Fluorouracil Dosing in Children With Pediatric Ependymoma. CPT Pharmacometrics Syst Pharmacol 5, 211–221. 10.1002/psp4.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright KD, Daryani VM, Turner DC, Onar-Thomas A, Boulos N, Orr BA, Gilbertson RJ, Stewart CF, and Gajjar A (2015). Phase I study of 5-fluorouracil in children and young adults with recurrent ependymoma. Neuro Oncol 17, 1620–1627. 10.1093/neuonc/nov181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amani V, Donson AM, Lummus SC, Prince EW, Griesinger AM, Witt DA, Hankinson TC, Handler MH, Dorris K, Vibhakar R, et al. (2017). Characterization of 2 Novel Ependymoma Cell Lines With Chromosome 1q Gain Derived From Posterior Fossa Tumors of Childhood. J Neuropathol Exp Neurol 76, 595–604. 10.1093/jnen/nlx040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu R, Norris GA, Willard N, Griesinger AM, Riemondy KA, Amani V, Grimaldo E, Harris F, Hankinson TC, Mitra S, et al. (2023). Spatial transcriptomic analysis delineates epithelial and mesenchymal subpopulations and transition stages in childhood ependymoma. Neuro Oncol 25, 786–798. 10.1093/neuonc/noac219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierce AM, Witt DA, Donson AM, Gilani A, Sanford B, Sill M, Van Court B, Oweida A, Prince EW, Steiner J, et al. (2019). Establishment of patient-derived orthotopic xenograft model of 1q+ posterior fossa group A ependymoma. Neuro Oncol 21, 1540–1551. 10.1093/neuonc/noz116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillen AE, Riemondy KA, Amani V, Griesinger AM, Gilani A, Venkataraman S, Madhavan K, Prince E, Sanford B, Hankinson TC, et al. (2020). Single-Cell RNA Sequencing of Childhood Ependymoma Reveals Neoplastic Cell Subpopulations That Impact Molecular Classification and Etiology. Cell Rep 32, 108023. 10.1016/j.celrep.2020.108023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gojo J, Englinger B, Jiang L, Hubner JM, Shaw ML, Hack OA, Madlener S, Kirchhofer D, Liu I, Pyrdol J, et al. (2020). Single-Cell RNA-Seq Reveals Cellular Hierarchies and Impaired Developmental Trajectories in Pediatric Ependymoma. Cancer Cell 38, 44–59 e49. 10.1016/j.ccell.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The normal brain gene expression data generated in this study are publicly available in Gene Expression Omnibus (GEO, RRID: SCR_005012) at GSE 244124 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi=GSE244124). All other data generated in this study is available upon request from the corresponding author.


