Characterization of 80 tumor-associated missense variants of AXIN1 reveals a subset of 18 mutations that disrupt its β-catenin regulatory function, whereas the majority are passenger mutations.
Abstract
AXIN1 is a major component of the β-catenin destruction complex and is frequently mutated in various cancer types, particularly liver cancers. Truncating AXIN1 mutations are recognized to encode a defective protein that leads to β-catenin stabilization, but the functional consequences of missense mutations are not well characterized. Here, we first identified the GSK3β, β-catenin, and RGS/APC interaction domains of AXIN1 that are the most critical for proper β-catenin regulation. Analysis of 80 tumor-associated variants in these domains identified 18 that significantly affected β-catenin signaling. Coimmunoprecipitation experiments revealed that most of them lost binding to the binding partner corresponding to the mutated domain. A comprehensive protein structure analysis predicted the consequences of these mutations, which largely overlapped with the observed effects on β-catenin signaling in functional experiments. The structure analysis also predicted that loss-of-function mutations within the RGS/APC interaction domain either directly affected the interface for APC binding or were located within the hydrophobic core and destabilized the entire structure. In addition, truncated AXIN1 length inversely correlated with the β-catenin regulatory function, with longer proteins retaining more functionality. These analyses suggest that all AXIN1-truncating mutations at least partially affect β-catenin regulation, whereas this is only the case for a subset of missense mutations. Consistently, most colorectal and liver cancers carrying missense variants acquire mutations in other β-catenin regulatory genes such as APC and CTNNB1. These results will aid the functional annotation of AXIN1 mutations identified in large-scale sequencing efforts or in individual patients.
Significance:
Characterization of 80 tumor-associated missense variants of AXIN1 reveals a subset of 18 mutations that disrupt its β-catenin regulatory function, whereas the majority are passenger mutations.
Introduction
The β-catenin signaling pathway is one of the most commonly deregulated pathways among cancers (1). In normal cells, β-catenin signaling is maintained at low levels by a destruction complex consisting of the APC tumor suppressor, scaffold proteins AXIN1, AXIN2, and the kinases GSK3β and CK1α. When cells are exposed to Wnt ligands, activity of this destruction complex is temporarily inhibited, leading to the nuclear translocation of β-catenin. In the nucleus, it associates with members of the TCF/LEF family of transcription factors, thereby regulating the expression of specific Wnt/β-catenin target genes.
In many tumor types this pathway is constitutively activated through mutational (in)activation of one of the core elements of the destruction complex. For example, in about 70%–80% of colorectal cancers loss-of-function mutations are observed in the APC gene (2). Oncogenic β-catenin (CTNNB1) mutations that make the protein more resistant to proteolytic breakdown, are more commonly observed in desmoid-type fibromatosis, and hepatocellular and endometrial carcinomas (3–5). Large-scale sequencing has also identified many AXIN1 variants in several tumor types. They are especially common in liver cancers, but also observed frequently in skin and uterus cancers, and cancers of the gastrointestinal tract (https://www.cbioportal.org/; refs. 6, 7). The mutational spectrum consists mostly of missense variants and mutations leading to AXIN1 truncation. The latter are regarded to result in at least a partial loss of β-catenin signaling regulation (8), which is currently the best-studied and accepted function of AXIN1. Anecdotally, this has also been reported for some missense variants (8–10), but for the majority of tumor-associated variants, this is still unknown. However, from a tumor perspective, it is important to identify whether these AXIN1 variants represent driver mutations or can be considered passenger mutations. This remains one of the main challenges in cancer genomics, that is, to make sense of the large amount of gene alterations observed in cancers (11).
Here, we first determined which functional domains of the AXIN1 protein are more likely to affect β-catenin signaling when lost, and then determined whether specific amino acid alterations may represent driver mutations that increase β-catenin signaling. Our results show that most AXIN1 variants have no clear effect on β-catenin signaling, but a selected set of mutations clearly leads to aberrant activation. In addition, we establish at endogenous levels an inverse correlation between AXIN1 truncation length and level of β-catenin regulation. Our results are important to interpret the functional consequences of AXIN1 mutations observed in cancers or the germline of patients.
Materials and Methods
Cell culture
HEK293T (RRID:CVCL_0063), JHH7 (RRID:CVCL_2805), and SNU449 AXIN1-repaired cells (clone 20, RRID:CVCL_0454) were cultured in DMEM (Gibco) supplemented with 10% (v/v) FBS (Gibco). SNU449 AXIN1-repaired cells were generated using CRISPR-Cas9 genome editing, as previously described (12). Cells were cultured in a humidified incubator maintained at 37°C with 5% CO2. All cell lines tested negative for Mycoplasma based on the real-time PCR method at Eurofins GATC-Biotech. Identity of all cell lines and clones thereof was confirmed by the Erasmus Molecular Diagnostics Department using Powerplex-16 STR genotyping (Promega).
For the preparation of conditioned medium, L-Control [L-M(TK-), RRID:CVCL_4536] and L-Wnt3A (RRID:CVCL_0635) cells were cultured in complete DMEM medium, followed by collection and filtration of medium according to the standard procedures.
Plasmids used in the present study
Wild-type (WT) FLAG-AXIN1 (cat. #109370, RRID:Addgene_109370) and HA GSK3β wt pcDNA3 (cat. #14753, RRID:Addgene_14753) were purchased from Addgene. EGFP-APC plasmid was constructed previously (13). The EGFP-β-catenin plasmid was generated by inserting the mouse Ctnnb1 ORF into the EGFP-C1 vector. On the basis of the WT FLAG–AXIN1 vector, all variants were generated using Q5 site-directed mutagenesis (NEB). The RGS domain (P81_R212) was inserted in front of the AXIN1 ORF, using the Gibson assembly method. Primer sequences for generating deletions, cancer-related variants and additional RGS domain are depicted in Supplementary Table S1A–S1C, respectively.
Genome editing with CRISPR/Cas9 system
CRISPR/Cas9 genome editing was performed as described previously (13, 14). Briefly, single-guide RNAs (sgRNA) were designed using the following CRISPR design tool (http://crispor.tefor.net/, RRID:SCR_015935) and cloned into pSpCas9(BB)-2A-GFP (PX458, cat. #48138, RRID:Addgene_48138). AXIN1 exon5 knockout cells were generated by seeding them into 10-cm round culture dishes. At approximately 60%–80% confluency, they were transfected with 3 μg of each exon5 flanking gRNA PX458 plasmid using Lipofectamine 2000 (cat. #10696153, Thermo Fisher Scientific). To obtain HEK293T cells expressing various truncated versions of AXIN1, cells were grown in 6-well plates and transfected with 1 μg PX458 plasmid. Coincidentally, this experiment also yielded one clone with a homozygous G425_G508del deletion encompassing the β-catenin–binding domain. To produce HEK293T cells expressing either the R395P or R395H variants endogenously, we cultured the cells in 6-well plates and performed transfections with 2.5 μg of the PX458 plasmid (which shares the same gRNA sequence as generated for the R395-truncating clones) and 1.25 μg of ssODN template. The ssODN template was sourced through the IDT-company Alt-R HDR Design Tool and Templates program.
After 48 hours, GFP-positive cells were sorted out and plated into 96-well plates by a FACS FACSAria II cell sorter (BD Biosciences). Two weeks later, genomic DNA was extracted from expanded single-cell clones by QuickExtract DNA Extraction Solution (cat. #QE09050, Lucigen). To verify that the correct AXIN1 alteration was obtained, a PCR fragment encompassing the sgRNA site was generated, followed by Sanger sequencing (Macrogen). We also sequenced AXIN1 cDNA to ensure that all generated variants are correctly expressed.
The AXIN1 D94_Q108 deletion present in JHH7 cells was repaired using CRISPR/Cas9 technology. To achieve this, a 2.1 kb genomic PCR fragment from WT DNA was cloned into the pGEM-T Easy Vector (cat. #A1360, Promega, RRID:Addgene_86229), serving as a homology-directed repair (HDR) template. In addition, a sgRNA was cloned into pSpCas9(BB)-2A-Puro (PX459, cat. #48139, RRID:Addgene_48139). Next, JHH7 cells were seeded in 3 wells of a 6-well plate at 40% confluency. Once the cells reached 80% confluency, we transfected them with 1 μg of the PX459 plasmid and 5 μg of the HDR plasmid using a 3:1 ratio of Lipofectamine 2000 reagent. After 6 hours, the cells were trypsinized and transferred equally into 9 dishes (x200 Petri dishes 100×20 mm style Falcon, cat. #353003, Fisher Scientific) at cell densities of 1/7, 2/7, and 4/7, respectively. Puromycin (working concentration 1 μg/mL) was added to the cells 48 hours later to select 2 days for transfected cells. Cells were constantly cultured containing 1:10 diluted L-Wnt3A and R-spondin conditioned medium to maintain high levels of β-catenin signaling. After 14 days, DNA from clones grown successfully was isolated using the QuickExtract DNA Extraction Solution (Epicentre). All related primers and ssODN sequences can be found in Supplementary Table S1D–S1H.
Coimmunoprecipitation
HEK293T cells in a 6-well plate were transiently transfected with 200 ng of FLAG-AXIN1 variants or empty plasmid control and equal amounts of HA-GSK3β, GFP-APC or GFP-β-catenin. After 48 hours, cells were washed with cold PBS once, and then 500 μL of cold lysis buffer (30 mmol/L Tris-HCl, pH 7.4; 150 mmol/L NaCl; 1% Triton-100; 5 mmol/L EDTA; 5 mmol/L NaF) containing Halt Protease and Phosphatase Inhibitor Cocktail (100×, cat. #78442, Thermo Fisher Scientific) was added to each well for 15 minutes on ice. Cells were collected by scraping, transferred into low-adhesion tubes, and lysate was cleared at 4°C by centrifugation at 11,000 × g for 15 minutes. From the cleared lysate, 10% was taken as input control, to which the same volume of 2×Laemmli/0.1 mol/L dithiothreitol (DTT) was directly added, followed by heating for 7 minutes at 95°C. To the remainder of the supernatant, we added 50 μL prewashed ANTI-FLAG M2 Affinity Gel (cat. #A2220, Sigma-Aldrich, RRID:AB_10063035), followed by incubation at 4°C for 2 hours. Next, FLAG-beads were centrifuged and washed with lysis buffer for 3 times. Finally, the pellet was dissolved in 75 μL 2×Laemmli sample buffer with 0.1 mol/L DTT and heated.
Western blotting and antibodies
Following a PBS wash, cells were lysed in 2×Laemmli Sample Buffer (4% SDS, 20% glycerol, 0.004% bromophenol blue, 0.15 mol/L Tris-HCl, pH 6.8) with 0.1 mol/L DTT and heated for 7 minutes at 95°C. Proteins samples were run using 10% SDS-PAGE. Proteins were then transferred onto Immobilon-PVDF membranes (Millipore) and blocked with Odyssey blocking buffer (cat. #927–70001, Licor-Biosciences). The membranes were scanned on the Odyssey Infrared Imaging System (Licor-Biosciences). Secondary antibodies were incubated with IRDye 680 Goat anti-Mouse (1:10,000, cat. #926–68070, Licor-Biosciences, RRID:AB_10956588) or IRDye 800 Goat anti-Rabbit (1:10,000, cat. #926–32211, Licor-Biosciences, RRID:AB_621843). The intensity of protein bands was analyzed by software Image Studio Lite version 5.2 (Licor-Biosciences, RRID:SCR_013715).
The primary antibodies used in this study are as follows: anti-FLAG (1:1,000, cat. # F1804, Sigma-Aldrich, RRID:AB_262044), anti-GSK3β (D5C5Z) XP Rabbit mAb (1:1,000, cat. #12456S, Cell Signaling Technology, RRID:AB_2636978), anti-GFP (D5.1) rabbit mAb (1:1,000, cat. #2956S, Cell Signaling Technology, RRID:AB_1196615), anti–α-tubulin (11H10) rabbit mAb (1:1,000, cat. #2125S, Cell Signaling Technology, RRID:AB_2619646), anti–β-actin (1:1,000, cat. #sc-47778, Santa Cruz Biotechnology, RRID:AB_626632), anti–β-catenin (1:1,000. cat. #610154, BD Biosciences, RRID:AB_397555), anti–β-catenin (D10A8) XP(R) rabbit mAb (1:1,000, cat. #8480S, Cell Signaling Technology, RRID:AB_11127855), and AXIN1 (1:1,000, Cell Signaling Technology, cat. #2087S (RRID:AB_2274550), and cat. #3323S; RRID:AB_2258885). Anti-Tankyrase1/2 (1:500, cat. #sc-365897, Santa Cruz Biotechnology, RRID:AB_10844977).
For AXIN1 Western blot analysis, we used an enhanced chemiluminescence (ECL)-based detection method. Membranes for ECL detection were blocked and incubated using Immobilon Block-CH reagent (cat. #WBAVDCH01, Millipore). The secondary antibodies were goat anti-rabbit immunoglobulins/HRP (horseradish peroxidase; 1:10,000, cat. #P044801–2, Agilent Technologies, RRID:AB_2617138) or goat anti-mouse immunoglobulins/HRP (1:10,000, cat. #P026002–2, Agilent Technologies, RRID:AB_2636929). All original Western blot images can be found in Supplementary Data S1.
Immunofluorescence analysis
Cells were grown on cover slides (cat. #0111580–18 mm, Marienfeld-superior) prewashed in 75% and 100% ethanol. HEK293T cells were seeded at a concentration of 30,000 cells/well in a 12-well plate containing the prewashed glass slides. On the second day, 50 ng of AXIN1 plasmid was transfected using Lipofectamine 2000. After 48 hours, the cell medium was removed, and cells were washed with cold PBS twice. Then, cells were fixed with 4% paraformaldehyde (PFA) in 50 mmol/L sodium phosphate, pH 7.4, buffered at room temperature for 10 minutes. After washing with PBS twice, cells were quenched in 50 mmol/L NH4Cl for 10 minutes at room temperature, washed with PBS twice, and incubated with blocking and permeabilization buffer (2% BSA and 0.1% Saponin in PBS) for 1 hour at room temperature. Cells were then stained with anti-FLAG antibody (1:500, cat. #F1804, Sigma-Aldrich, RRID:AB_262044) overnight at 4°C. After washing 2 times with PBST (0.05% Tween in PBS) and once with PBS, cells were incubated for 1 hour at room temperature with goat anti-mouse Alexa488 secondary antibody (1:1,000, cat. #A21203, Invitrogen, RRID:AB_11071160) protected from light. All antibody incubation steps were performed in the blocking/permeabilization buffer described above. Following 2 times washing with PBST and once with PBS, cover glasses were moved from the 12-well plate to the microscope slide, embedded in Vectashield/DAPI (cat. #H-1200, Vector Laboratories), and sealed with colorless nail polish. Images were captured by an SP5 604 confocal laser scanning microscope using Leica Application Suite X (LAS X) software with constant parameter settings.
Β-Catenin reporter assays
The β-catenin reporter assays were performed basically as previously described (12). To determine the effects of AXIN1-variant plasmids on β-catenin signaling, HEK293T cells were seeded in 24-well plates, and when reaching approximately 50%–60% confluency, were cotransfected with 100-ng Wnt responsive element (WRE) vector, 100-ng FLAG–AXIN1 variants and 10 ng CMV-Renilla using Lipofectamine 2000 Transfection Reagent at a ratio of 3:1 for 48 hours. After 24 hours, L-Wnt3A (1/10) conditioned medium was added into an empty-vector transfected well, serving as a positive control for β-catenin signaling induction. For CRISPR/Cas9-generated AXIN1-variant cell clones, 100-ng WRE plasmid or its mutant responsive element (MRE) control vector were cotransfected with 10-ng CMV-Renilla. The effect of AXIN2 siRNA-mediated knockdown in these clones was determined using ON-TARGET plus SMART pool AXIN2 (cat. #L-008809–00–0005) and the nontargeting pool (cat. #D-001810–10–05), both from Dharmacon. For this experiment, 0.25 μL (20 μmol/L) siRNA was cotransfected with 100-ng WRE and 10-ng CMV-Renilla.
We measured luciferase activities and normalized the data for the transfection efficiency using the Dual Luciferase Reporter Assay system (cat. #E1910, Promega). Luciferase activities were measured in a LumiStar Optima luminescence counter (BMG LabTech, Offenburg, Germany). The β-catenin reporter activities are shown as WRE/CMV-Renilla or WRE/MRE ratios. Each experiment was performed in triplicate and repeated at least twice.
The β-catenin reporter system represents one of the most sensitive and well-accepted systems to demonstrate alterations in nuclear β-catenin signaling. Nevertheless, to ensure that the observed effects on β-catenin reporter activity for selected variants really reflect alterations in nuclear translocation, we performed an immunofluorescence assay for β-catenin on AXIN1-variant transfected HEK293T cells. These results are depicted in Supplementary Fig. S1, and are in line with the reporter assay. Expression of more defective mutants such as R395P and V478G leads to similar levels of nuclear β-catenin staining as exposure to Wnt3A conditioned medium.
qRT-PCR
Total RNA was isolated using the NucleoSpin RNA II kit (Macherey-Nagel), and then the RNA was reverse transcribed with the Primescript RT Reagent Kit (Perfect Real-Time, cat. #RR036A, Takara) according to the manufacturer's instructions. Quantitative PCR was performed in the StepOne Real-Time PCR System (Applied Biosystems). Analyses were performed using the StepOne version 2.0 software (Applied Biosystems) and normalized with the human housekeeping gene GAPDH. All experiments were performed in triplicate. Primer sequences are provided in Supplementary Table S1I.
Mice and hydrodynamic tail injection
For the hydrodynamic tail vein injection experiments, N-terminal MYC-tagged AXIN1 variants were inserted in between the attL1/L2 sites of the pEN_TmiRc3 entry vector (RRID:Addgene_25748) using Gibson cloning. Next, using the Gateway LR Clonase II Enzyme mix (cat. #11791020, Invitrogen) the AXIN1 variants were inserted in the pET3-EF1a-GW vector. Hydrodynamic tail vein injection was performed basically as previously described (15, 16). Briefly, 20 μg of either pT3-EF1α-AXIN1-WT, C121F, L202P, R395P or V478G construct was mixed with 20 μg pT3-EF1α-c-Met and 1.6-μg pCMV/SB in 2-mL saline. This solution was injected in the lateral tail vein of 6–8 week WT FVB/N mice purchased from The Jackson Laboratory (RRID:MGI:3528175). Mice were monitored via abdominal palpation every three days, and were euthanized when they developed a palpable abdominal mass based on the Institutional Animal Care & Use Committee protocol. Mice were housed and fed in accordance with protocols approved by the Committee for Animal Research at the University of Hawaii Cancer Center.
IHC
Immunostaining was performed on 4-μm-thick whole-slide sections from formalin-fixed, paraffin-embedded embedded tissue blocks by using the Ventana Benchmark Discovery (Ventana Medical Systems Inc.). The following antibodies were used: anti–β-catenin (1:200, BD Biosciences, clone 14, RRID:AB_397555); anti–Myc-tag (1:400, cat. #2276, Cell Signaling Technology, RRID:AB_331783); anti–Ki-67 (2 μg/mL, Ventana, clone 30–9, RRID:AB_2631262). In brief, heat-induced antigen retrieval with CC1 (cat. #950–500, Ventana) was performed at 97°C for 64 minutes (β-catenin/Myc-tag) or 40 minutes (Ki-67). Next, samples were incubated with antibodies at 37°C for 60 minutes (β-catenin/Myc-tag) or 28 minutes (Ki-67), followed by an amplification step (cat. #760–080, Ventana). Β-catenin and Myc-tag incubations were detected with unconjugated rabbit anti-mouse (1:500; Abcam, cat. #AB133469, RRID:AB_2910607) for 32 minutes at 37°C followed by omnimap anti-rabbit HRP for 20 minutes at 37°C, followed by visualization with DAB (cat. #760–159, Ventana). For the Ki-67 antibody incubations, the unconjugated rabbit anti-mouse was omitted. Counterstain was done by hematoxylin II (cat. #790–2208, Ventana) for 20 minutes and a Bluing reagent (cat. #3760–2037, Ventana) for 4 minutes.
Database analysis
The data reported in Supplementary Fig. S2 were obtained from the cBioPortal website by retrieving all AXIN1 mutations from the curated set of nonredundant studies. To obtain the cancer-related AXIN1 mutations depicted in Supplementary Table S1B, we analyzed the COSMIC and cBioPortal websites, filtering them on amino acids D65_S228, V383_M418, and P434_P503. We also combined it with the evolutionary conservation score obtained using the ConSurf tool (https://consurf.tau.ac.il/), which is shown in Supplementary Fig. S3. Data were updated until January 2022. Except when specifically stated, variants were not selected on the basis of the tumor type. For the cBioPortal analyses on co-occurrence with other β-catenin–activating gene mutations, we retrieved mutations in Wnt/β-catenin–related genes specifically for colorectal and liver cancers carrying AXIN1 variants. As in most cases information about zygosity of the mutations was not available, this feature was not taken along in the analysis.
Structure analysis
The interfaces of AXIN1 complexes with β-catenin, APC, and GSK3β were analyzed using experimentally determined structures. To analyze the β-catenin-AXIN1 complex, 1qz7.pdb was obtained from the PDB–REDO databank (17). In this crystal structure human β-catenin is bound to Xenopus Axin (18). We refer to the amino acids in Axin according to the corresponding human AXIN1 residues (register shift of 2 residues). For the AXIN1–APC interface, 1emu.pdb was obtained from the PDB–REDO databank (this structure has a 37-residue register shift in the amino acid residue numbering; ref. 19). For the AXIN1–GSK3β complex, 1o9u.pdb was obtained from the RCSB Protein Data Bank (20). The predicted structure of full-length human AXIN1 was obtained from the AlphaFold Protein Structure Database (21). Mutations were mapped onto the structures and categorized as located at the interface, surface exposed but not at interface, or present within the hydrophobic core. The effect of the mutation was predicted taking into account the known amino acid side chain properties (shape, size, charge, hydrophobicity, and hydrogen bond formation capacity). Figures were created using the PyMOL (RRID:SCR_000305) Molecular Graphics System, Version 2.0 Schrödinger, LLC.
Statistical analysis
All results are presented as the mean ± SD. Statistical analyses were carried out using GraphPad Prism version 8.0.2 software (GraphPad Software Inc.; RRID:SCR_002798). Comparisons between groups were performed with the Mann–Whitney test. Differences were considered significant at a P value of less than 0.05 (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
Data availability
Tumor associated mutations analyzed in this study were obtained from the cBioPortal (https://www.cbioportal.org/) and COSMIC (https://cancer.sanger.ac.uk/cosmic) databases. The predicted structure of full-length human AXIN1 was obtained from the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/entry/O15169). The PDB–REDO databank (https://pdb-redo.eu/) was used to obtain the optimized crystal structures for the β-catenin–AXIN1 complex (1qz7.pdb) and AXIN1–APC interface (1emu.pdb). The RCSB Protein Data Bank (https://www.rcsb.org/) was used to obtain AXIN1–GSK3β complex (1o9u.pdb). The ConSurf tool (https://consurf.tau.ac.il/) was used to obtain the AXIN1 evolutionary conservation score. Other data generated in this study are available upon request from the corresponding author.
Results
Analysis of the AXIN1 mutational landscape
Analyzing AXIN1 mutations in cBioPortal showed that tumor-associated truncating and missense variants appear evenly distributed along the AXIN1 coding region (Supplementary Fig. S2). Germline AXIN1 amino acid variants identified using the gnomAD database are observed only at low frequencies, and accordingly they are rarely observed in tumor samples (Supplementary Table S2). Mutations are especially common in liver cancers (8%), with a predominance of truncating mutations (Supplementary Fig. S4). Other tumor types with AXIN1 mutation frequencies above 2% are those of the skin, uterus, and cancers of the gastrointestinal tract.
Deletion of the GSK3β and β-catenin–binding domains increases β-catenin signaling
To test the functionality of a large number of AXIN1 variants and deletions, we made use of transient transfection of an AXIN1 expression vector, which leads to a more than 3,000-fold higher AXIN1 expression on RNA level (Supplementary Fig. S5). Assuming that this also translates into a similar overexpression on protein level, it means that transfected AXIN1 protein is in high excess of endogenously expressed AXIN1/AXIN2. Thus, basically all β-catenin breakdown complexes will have incorporated the transfected variant, and when (partially) defective this will lead to increased β-catenin signaling.
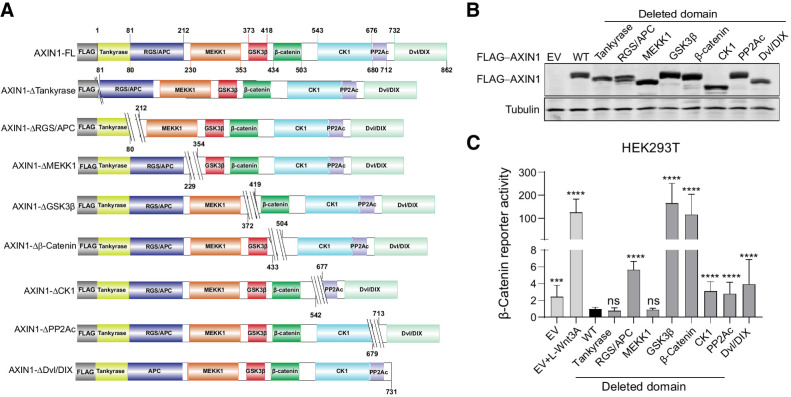
To explore which functional domains of AXIN1 affect β-catenin signaling when lost, we constructed a series of deletion mutants (Fig. 1A and B; refs. 22–26). When cotransfected with a β-catenin reporter construct into HEK293T cells, the constructs that lack either the GSK3β or β-catenin–binding domain strongly increase signaling (Fig. 1C). Weaker but significant increases in signaling are observed when the APC, CK1, PP2Ac, and Dvl/DIX–binding domains are deleted.
Figure 1.
Deletion of the GSK3β and β-catenin–binding domains strongly increases β-catenin signaling. A, Schematic diagram depicting the FLAG-tagged AXIN1 mutant expression vectors that were generated. Amino acid numbers are based on the long 862 amino acid isoform of AXIN1. For clarity, we refer to each domain by one reported binding partner, but especially for the MEKK1, CK1, and PP2A domains; additional binding proteins have been reported. B, Immunoblot for FLAG-tagged AXIN1 to demonstrate expression of the expected variants after HEK293T transfection. Tubulin was used as loading control. C, A β-catenin reporter assay was conducted in HEK293T cells expressing WT AXIN1 and variants lacking the depicted domains. The β-catenin reporter activities are shown as WRE/CMV-Renilla ratios (in triplicate, three independent experiments), in which the value obtained for wild-type AXIN1 was arbitrarily set to 1. As a positive control for increased β-catenin signaling, Wnt3A conditioned medium was added to empty vector (EV)-transfected cells (EV+Wnt3A). Data are shown as mean ± SD. Statistical significance was analyzed using a Mann–Whitney test. ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant.
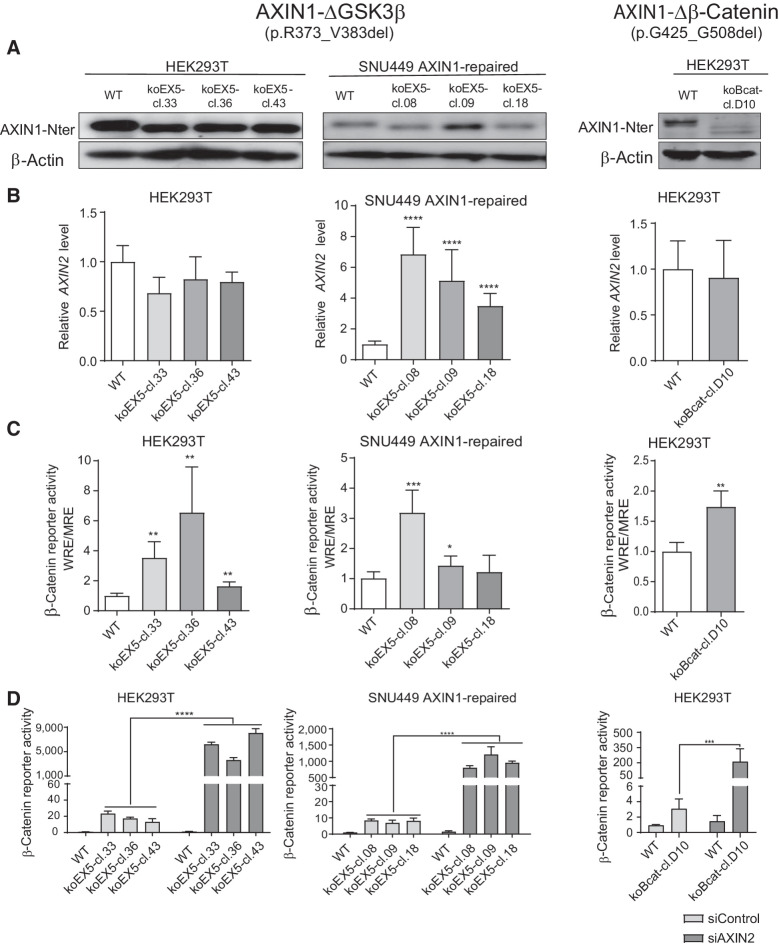
To determine whether loss of the GSK3β domain also affects β-catenin signaling at endogenous levels, we deleted exon5 (R373_M418del) in HEK293T cells and the hepatocellular carcinoma SNU449 cell line in which we previously restored WT AXIN1 expression (12). For each cell line, three independent clones were identified that correctly express the shortened AXIN1 protein (Fig. 2A). We also identified one HEK293T clone carrying a homozygous G425_G508del deletion encompassing the β-catenin–binding domain.
Figure 2.
Analysis of β-catenin signaling in cell clones with endogenous knockout of the GSK3β and β-catenin domains. HEK293T and SNU449 AXIN1-repaired cells (12) were used to knockout the GSK3β-binding domain encoded by exon 5. We also identified one HEK293T clone with an endogenous knockout of the β-catenin–binding region. A, Immunoblot using an N-terminal AXIN1 antibody (cat. #3323, Cell Signaling Technology) to show expression of the shortened AXIN1 proteins. β-Actin served as a loading control. B, qRT-PCR assay to demonstrate the mRNA expression level of AXIN2 (in triplicate, n = 2 independent experiments). Expression levels were depicted relative to the housekeeping gene GAPDH. The value for the WT control was arbitrarily set to 1. C, A β-catenin luciferase reporter assay was performed to determine the β-catenin signaling activity in all knockout clones. Values are depicted as WRE/MRE ratios (in triplicate, n = 3 independent experiments). The value for the WT control was arbitrarily set to 1. D, siRNA-mediated knockdown of AXIN2 was performed in all knockout clones, followed by a β-catenin reporter assay. Values are depicted relative to the WRE/CMV-Renilla ratios obtained for the siControl-WT (five replicates, two independent experiments), which was arbitrarily set to 1. Data are shown as mean ± SD. Statistical significance for all experiments was analyzed using a Mann–Whitney test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
In the ΔGSK3β SNU449 clones, AXIN2 RNA levels, a well-established β-catenin target gene, were increased about 10-fold, whereas a β-catenin reporter was significantly increased in 2 of 3 clones (Fig. 2B and C). Reporter activity was also enhanced in all ΔGSK3β HEK293T clones and the Δβ-catenin HEK293T clone, whereas AXIN2 RNA levels did not significantly change. The latter may result from low responsiveness of AXIN2 in HEK293T cells (Supplementary Fig. S6A and S6B).
AXIN2 can partially compensate for the functional impairment caused by AXIN1 mutation (12, 20, 27, 28), thereby obscuring the effects of AXIN1 mutations. SMARTpool siRNA-mediated knockdown of AXIN2 resulted in a 100–500-fold increase of β-catenin reporter activity in all ΔGSK3β and Δβ-catenin clones, while signaling was barely affected in WT control cells (Fig. 2D), thus confirming that AXIN1 proteins missing these functional domains are strongly impaired in their β-catenin regulatory function.
Most missense variants in the GSK3β-binding domain retain functionality
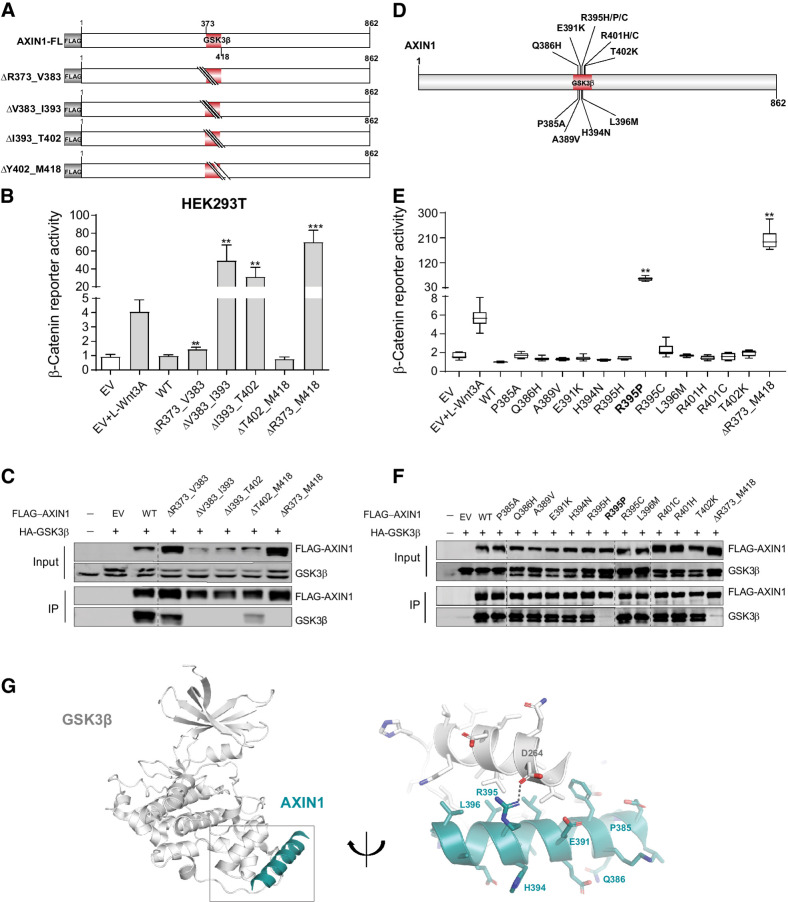
To identify tumor-associated missense mutations within the GSK3β domain that affect β-catenin signaling, we first constructed four sequential 10–17 amino acid deletions (Fig. 3A and B), of which, only the V383_I393del and I393_T402del constructs are clearly defective. Immunoprecipitation (IP) experiments showed that constructs that increased β-catenin signaling, also lost binding to endogenous and HA-tagged GSK3β (Fig. 3C).
Figure 3.
Analysis of missense variants in the GSK3β-binding domain. Tumor-associated missense variants within the GSK3β-binding domain were tested for their effect on β-catenin signaling regulation. A, Schematic diagram indicating the sequential amino acid deletions within the GSK3β domain that were generated in the AXIN1 expression plasmid. B, A β-catenin reporter assay was performed to show the defect in β-catenin regulation associated with the variants shown in A. C, Immunoprecipitation experiment following cotransfection with HA-tagged GSK3β to identify which deletion variants shown in A affect GSK3β binding. AXIN1 and GSK3β were detected using anti-FLAG and anti-GSK3β antibodies, respectively. Transfection with empty vector (EV) and nontransfected cells were used as negative controls. D, Schematic diagram depicting the selected tumor-associated missense variants within the V383-T402 domain. These 12 variants were the only ones reported in the cBioPortal and COSMIC databases until January 2021. E, A β-catenin reporter assay to determine the defect in β-catenin regulation for the missense variants is shown in D. F, Immunoprecipitation experiment following cotransfection with HA-tagged GSK3β to identify which missense variants shown in D affect GSK3β binding. Image is a composite of two original blots (see Supplementary Data S1) in which the AXIN1 variants have been arranged in numerical order. G, Cartoon representation of GSK3β (gray) with bound Axin1 α-helix (deep teal). Right, details of the interaction interface with amino acid sidechains in stick representation with nitrogen atoms (blue) and oxygen (red). Dashed line, hydrogen bond between Aspartate D264 in GSK3β and Arginine R395 in Axin1. AXIN1 residues mutated in this study are labeled. All β-catenin reporter activities are depicted as WRE/CMV-Renilla ratios (in triplicate, two independent experiments), in which the value obtained for the empty vector was arbitrarily set to 1. Data are shown as mean ± SD. Statistical significance for all experiments was analyzed using a Mann–Whitney test. **, P < 0.01; ***, P < 0.001.
Next, 12 tumor-associated variants within the V383_T402 domain were investigated (Fig. 3D). All, except the Q386H variant, are alterations of evolutionarily conserved residues (Supplementary Fig. S3). The L396M variant was previously suggested to increase signaling (9). In addition, we included 6 variants at conserved residues directly following the GSK3β domain. The β-catenin reporter and IP experiments indicated that 17 of 18 variants behaved as silent mutations (Fig. 3E and F; Supplementary Fig. S7A and S7B). Only the R395P variant lost GSK3β binding and was defective in regulating β-catenin signaling.
To confirm this at endogenouse levels, we generated R395P-knockin HEK293T clones and one clone homozygously expressing R395H AXIN1 (Supplementary Fig. S8A and S8B). R395P clones show a similar increase in β-catenin signaling as clones expressing short truncated AXIN1 (Supplementary Fig. S8C), whereas the R395H clone yields lower values. AXIN2 knockdown strongly increased signaling in the truncating and R395P clones, but barely affected signaling in WT and R395H expressing cells. Combined, these results indicate that at endogenous levels the R395P variant carries no dominant-negative activity, as it would otherwise have interfered with endogenous AXIN2 and other breakdown complex components, but instead behaves as a loss-of-function mutant.
AlphaFold predicts an α-helical structure for residues P385_E421, which is partially visible in the crystal structure of an AXIN1–GSK3β complex (residues E384_V399), in which it packs against GSK3β via a predominantly hydrophobic interface (Fig. 3G; Supplementary Fig. S9; refs. 21, 29). Only the R395P mutation is predicted to have a significant effect on the stability of the interface by losing a salt bridge and because prolines cannot be accommodated in the middle of a regular α-helix. All other mutations are expected to have no or only a minor effect (Supplementary Table S3).
Taken together, these results indicate that proper GSK3β binding to AXIN1 is important for its function, but tumor-associated variants within this domain rarely impair functionality. The functional consequences can be largely inferred through careful analysis of available protein structures.
Missense variants in the β-catenin–binding domain mostly retain functionality
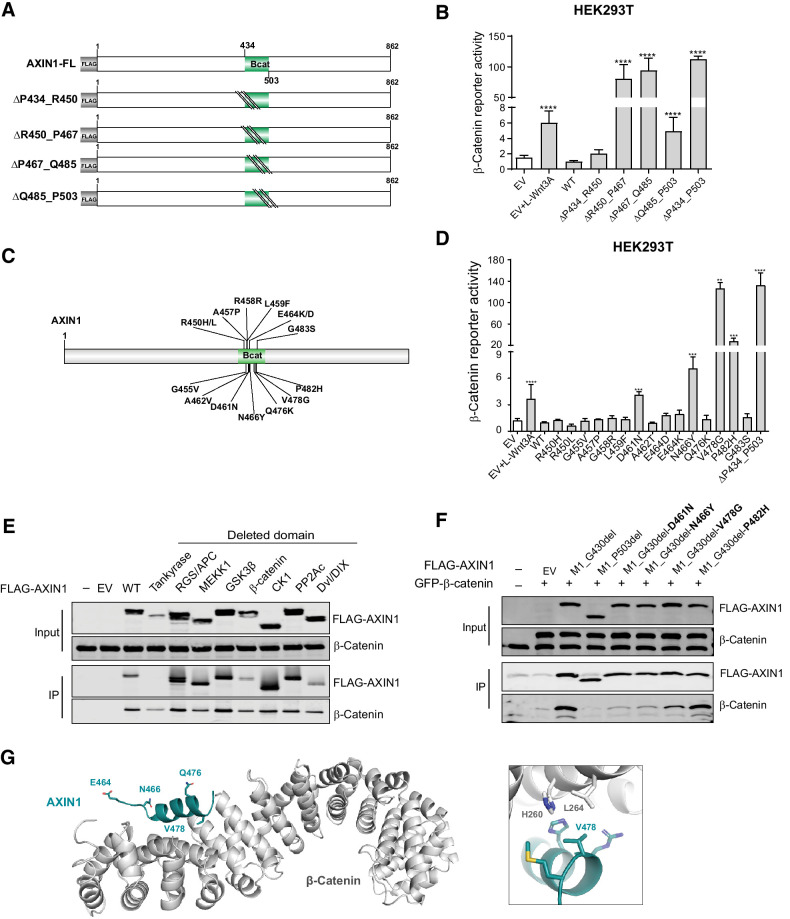
Four sequential deletion, constructs for the β-catenin domain were generated, of which, the R450_P467del, P467_Q485del, and Q485_P503del all led to enhanced signaling (Fig. 4A and B). These three constructs encompass the conserved R460_P494 region (Supplementary Fig. S3), which is known to directly associate with β-catenin (18). Fifteen tumor-associated variants within or directly preceding this domain were tested. Especially the V478G variant strongly increased signaling, whereas the D461N, N466Y, and P482H variants showed intermediate defects (Fig. 4C and D). The remaining 11 variants behaved similarly to WT.
Figure 4.
Missense variants in the β-catenin–binding domain mostly retain functionality. Tumor-associated missense variants within the β-catenin–binding domain were tested for their effect on β-catenin signaling regulation. A, Schematic diagram of the indicated sequential amino acid deletions that were generated in the AXIN1 expression plasmid. B, A β-catenin reporter assay was performed to show the defect in β-catenin regulation associated with the variants shown in A. C, Schematic diagram depicting the 15 selected tumor-associated missense variants within the R450_P503 domain. D, A β-catenin reporter assay to determine the defect in β-catenin regulation for the missense variants shown in C. Reporter activities are depicted as WRE/CMV-Renilla ratios (mean ± SD; three independent experiments), in which the value obtained for the empty vector (EV) was arbitrarily set to 1. E, Immunoprecipitation experiment with FLAG-tagged AXIN1 variants to identify which domain deletions affect endogenous β-catenin binding. F, Immunoprecipitation experiment with depicted FLAG-tagged AXIN1 missense variants to determine β-catenin–binding capacity. Deletion of the N-terminal half of AXIN1 up to the β-catenin domain (M1_G430del) was used to allow a better evaluation. A larger deletion (M1_P503del) also removing the β-catenin domain was used as negative control. Cotransfected GFP–β-catenin and endogenous β-catenin were detected using a β-catenin antibody. In all IP experiments, transfection with EV and nontransfected cells was used as negative controls. Data are shown as mean ± SD. G, Cartoon representation of β-catenin (gray) with bound AXIN1 α-helix (deep teal). Side chains of amino acid residues that are mutated in this study are indicated in stick representation, with nitrogen atoms (blue) and oxygen (red). Right, close-up of the region around AXIN1 residue Valine 478. Statistical significance for all experiments was analyzed using a Mann–Whitney test. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
IP experiments using AXIN1 variants with loss of functional domains, unexpectedly showed that AXIN1 lacking the entire β-catenin–binding domain still coprecipitated β-catenin (Fig. 4E). This can be explained by coprecipitated GSK3β and APC protein, which can independently bind β-catenin. Following removal of the N-terminal portion (M1_G430del), the D461N, N466Y, and V478G variants showed reduced β-catenin binding, whereas the P482H variant retained normal binding affinity (Fig. 4F).
Five of 15 mutations can be mapped onto the crystal structure between human β-catenin and Xenopus Axin (18). Only V478 is located directly at the interface where it forms a hydrophobic surface patch with surrounding residues on β-catenin (Fig. 4G; ref. 30). The V478G variant creates a hole in this hydrophobic interface, leading to its destabilization. The E464D/K and Q476K variants are located outside this interface and not expected to significantly influence complex formation. N466 forms an interhelical hydrogen bond with an adjacent residue and its mutation to tyrosine may affect the stability of the AXIN1 helix, indirectly influencing complex formation. A more detailed analysis of the protein structure is provided in Supplementary Table S4.
Taken together, most variants within the β-catenin domain retain normal function, but a selected number of variants can aberrantly increase β-catenin signaling. For the five variants that are visible in the crystal structure, the functional consequences are in line with those predicted from the protein structure.
Analysis of variants in the RGS/APC-binding domain
Deletion of the entire RGS/APC-binding domain (P81_R212del) significantly increased β-catenin signaling (Fig. 1C), which contrasts with another report claiming no effect upon losing the entire RGS domain (10). However, their deletion construct also removed amino acids essential for proper binding to tankyrases that are responsible for PARsylation and subsequent degradation of AXIN1 (25). By doing so, it may lead to a more stable AXIN1 protein, which is expected to retain more overall β-catenin regulatory activity. To test this speculation, we generated two RGS/APC deletion constructs also encompassing the C-terminal part of the tankyrase domain (D65_S228del and D65_R212del), which led to increased AXIN1 stability that is no longer regulated by tankyrase inhibition (Supplementary Fig. S10A and S10B). Furthermore, it restores β-catenin regulation to WT levels, whereas a more accurate removal increases β-catenin signaling (Supplementary Fig. S10C). Finally, we tested their ability to form β-catenin degradasomes, visualized as cytoplasmic puncta in cells (19). WT AXIN1 readily forms these large aggregates, which was also observed for both variants lacking part of the tankyrase domain (Supplementary Fig. S10D). In contrast, the P81_R212del variant fails to form these puncta and instead yields a diffuse staining pattern. Taken together, the accurate removal of the RGS/APC domain is required to observe the consequences following deletion of this domain.
To confirm that a deletion within the RGS/APC domain also affects β-catenin signaling at endogenous levels, we made use of the JHH7 hepatocellular carcinoma cell line carrying a homozygous D94_Q108del AXIN1 deletion. An expression construct carrying this deletion demonstrated a clear increase in β-catenin reporter activity and loss of APC binding (Supplementary Fig. S11A and S11B). Using gene editing, we generated three JHH7 clones with homozygous restoration of WT AXIN1 expression, which all showed significantly reduced expression of AXIN2 and decreased β-catenin reporter activity (Supplementary Fig. S11C–S11E), thus confirming that losing part of the RGS/APC domain also affects AXIN1 activity at endogenous levels.
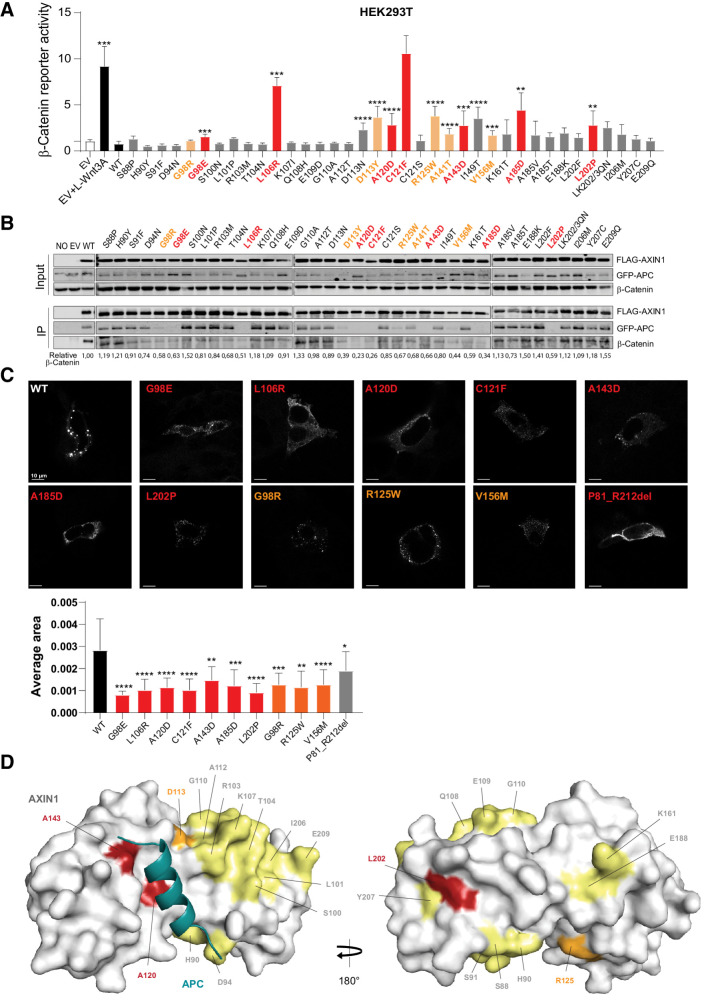
Next, we selected 37 tumor-associated variants along the RGS/APC domain (P81_R212), mainly based on a high conservation level and homology with AXIN2 (Supplementary Table S5). In addition, we analyzed 10 variants observed in the C-terminal part of the tankyrase domain, of which, only the P72T variant resulted in a statistically significant albeit modest effect on β-catenin signaling (Supplementary Fig. S12). Among the RGS/APC domain variants, we identified 13 variants that significantly increased β-catenin reporter activity 2–10-fold (Fig. 5A). Next, we tested their ability to co-precipitate GFP–APC and/or β-catenin (Fig. 5B). Of the 13 variants, 7 showed a near-complete loss of APC binding accompanied by a strongly reduced binding to β-catenin (Fig. 5A, red bars). Four variants showed a partially reduced APC and β-catenin binding (orange bars), whereas the remaining two variants appeared to associate correctly. We also identified one variant (G98R) with partially reduced binding, but no significant increase in β-catenin reporter activity. Taken together, we observe a good correlation between increased β-catenin reporter activity and loss of APC/β-catenin binding, although some exceptions are observed. Ten variants with complete or partial loss of APC binding were tested for their ability to efficiently polymerize into puncta. Compared with WT AXIN1, all variants lost the ability to form large AXIN1 aggregations (Fig. 5C).
Figure 5.
Analysis of missense variants within the RGS/APC domain. Tumor-associated missense variants in the RGS/APC domain were analyzed for their defect in β-catenin regulation, APC and β-catenin binding, and capacity to induce intracellular puncta formation. A, A β-catenin reporter assay was performed to determine the defect in β-catenin regulation for 37 RGS/APC domain variants. Red-marked bars represent variants that show a near-complete loss of GFP-APC and/or β-catenin binding (see B), whereas orange bars represent variants with partial loss. All β-catenin reporter activities are depicted as WRE/CMV-Renilla ratios (in triplicate, three independent experiments), in which the value obtained for the empty vector (EV) was arbitrarily set to 1. B, Immunoprecipitation assay to determine the binding capacity of missense variants to cotransfected GFP-APC or endogenous β-catenin. C, Immunofluorescence analysis of transfected AXIN1 missense variants to determine their puncta formation capacity. Average dot size was determined using ImageJ software on at least 6 independent cells. Data are shown as mean ± SD. D, Surface representation of AXIN1 (gray) with bound APC helix (deep teal). Amino acid residues mutated in this study that are located on the surface of AXIN1 are labeled and colored according to extent of loss of APC binding (B), with yellow indicating no loss, orange indicating partial loss, and red indicating near-complete loss of APC binding. Statistical significance for all experiments was analyzed using a Mann–Whitney test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
These data suggest that loss of APC binding may be responsible for the increased β-catenin signaling. To investigate this further, we added an extra RGS domain (Supplementary Fig. S13A) at the front of all 7 AXIN1 variants with a near-complete loss of APC binding (Fig. 5A, red bars). In all cases, APC and β-catenin binding were restored (Supplementary Fig. S13B and S13C). The additional RGS domain at the N-terminus of ΔRGS–AXIN1 was also able to restore β-catenin reporter activity to WT levels, indicating that an RGS domain at this position is functional (Supplementary Fig. S13D). This was also observed for the variants with a moderate increase in β-catenin reporter activity (A120D, A143D, and L202P), with the exception of G98E that shows an unexplained increase in signaling. The three variants that more strongly impacted on β-catenin signaling, that is, L106R, C121F, and A185D, all showed a significant reduction in β-catenin reporter activity, but were not fully restored to WT levels. Thus, restoring APC binding could rescue the defect of most “weaker” variants, and partially rescued variants that strongly impair β-catenin regulation.
An available crystal structure shows how the third AXIN-binding repeat of human APC forms an α-helix that binds in a surface groove of AXIN1 (Fig. 5D; ref. 31). At large, three types of consequences are observed for the missense variants affecting β-catenin signaling; (i) those directly located at the interface (Fig. 5D; Supplementary Fig. S14A and S14B); (ii) located on the surface within helices that stabilize the interaction with APC (Fig. 5D); and (iii) located within the hydrophobic core of the AXIN1 RGS domain (Supplementary Fig. S14B). A120D and A143D belong to the first category, where the alteration to aspartate introduces a large polar sidechain with a negative charge, severely reducing the strength of the interaction. D113N/D113Y and R125W are examples of surface-located variants that likely destabilize helices indirectly involved in forming the binding interface with APC. In two variants, surface-exposed Leucine residues residing within an α-helix are mutated to Proline (L101P and L202P), which will distort the geometry of the α-helix. Indeed, we found increased signaling for L202P; however, this was not observed for L101P. Last, 8 mutations (G98E, G98R, L106R, C121F, A141T, I149T, V156M, and A185D) that occur in the hydrophobic core of the AXIN1 domain, are predicted to render the RGS domain unstable. Their new sidechains sterically will not fit within the core, and/or there are no polar neighboring residues available that would satisfy the charges and hydrogen bond capacity of the new polar side chains.
Most other mutations are predicted not to affect the interaction between AXIN1 and APC. An extensive description and discussion on the expected structural consequences is provided in Supplementary Table S6. Overall, most of the observed consequences on β-catenin signaling are in accordance with the structural analysis.
Testing the defective nature of selected variants through hydrodynamic transfection
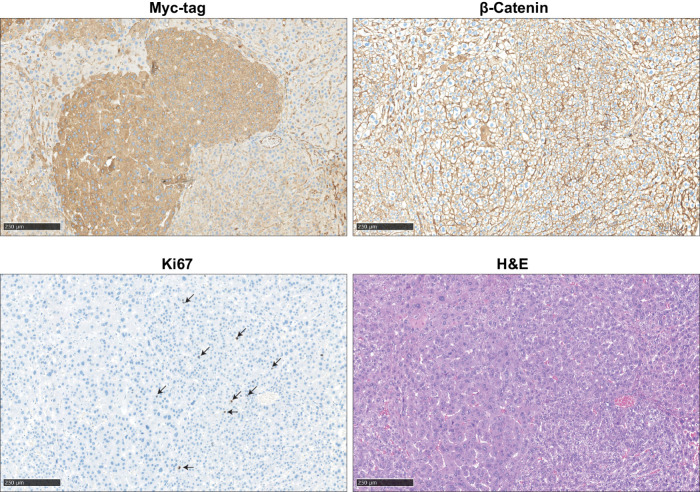
To determine the defective nature of four selected variants in vivo (C121F, L202P, R395P, and V478G), we hydrodynamically co-injected Myc-tagged AXIN1 variants with c-Met in mice (15, 32, 33). In a direct side-by-side β-catenin reporter assay, the R395P and V478G variants show a much bigger defect in β-catenin regulation compared with both RGS/APC domain variants (Supplementary Fig. S15), which is also reflected in their potential to induce liver lesions. Five out of 6 mice injected with the R395P GSK3β domain mutant developed multiple lesions within 10 weeks after injection, whereas this was the case for 1 of 4 V478G β-catenin domain mutant injected mice (Supplementary Fig. S16A–S16C). In stark contrast, none of the WT or C121F/L202P-injected animals showed signs of tumor formation when sacrificed 5 months following injection. Lesions show an identical histopathology as previously reported for AXIN1-deficient mouse tumors, with mostly slightly increased hepatocyte size and mild to moderate nuclear atypia (Fig. 6; ref. 34). A low number of Ki67-positive proliferating cells is observed. IHC for β-catenin reveals only membranous staining patterns, which is in accordance with previous AXIN1-deficient liver tumors (34, 35). As outlined by various reports, absence of nuclear identifiable β-catenin does not exclude that a low level of biologically relevant signaling is active (36), and is also observed in tumors carrying oncogenic β-catenin mutations within the armadillo repeats and a subset of S45–β-catenin mutant cancers (13, 37). It merely represents a technical shortcoming of β-catenin IHC to detect low levels of nuclear β-catenin above background levels. This analysis reveals that the most defective R395P and V478G AXIN1 variants are more likely to induce liver tumor formation when hydrodynamically overexpressed than weaker RGS/APC domain variants.
Figure 6.
Immunohistochemical analysis of induced liver tumors in mice. An example is shown of two adjacent AXIN1–R395P-induced liver lesions. IHC was performed to reveal expression of transfected Myc-tagged AXIN1, endogenous location of β-catenin, and Ki67 to identify proliferating cells. Hematoxylin and eosin (H&E) staining revealed mostly slight increases in hepatocyte size and mild-to-moderate nuclear atypia. Scale bar, 250 μm.
An inverse correlation between AXIN1 truncation length and β-catenin regulatory function
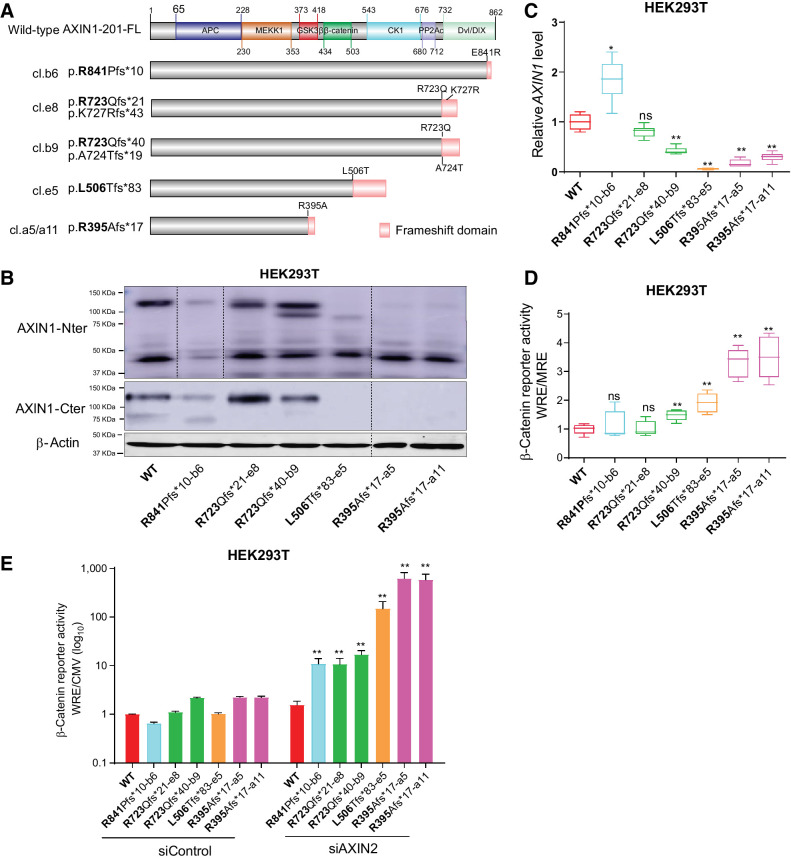
cBioPortal regards all truncating AXIN1 mutations as likely tumor-driving mutations. To confirm whether this also holds true for C-terminal truncations retaining most functional domains, we generated various HEK293T clones with AXIN1 truncations (Fig. 7A). The most C-terminal R841Pfs*10 truncation removes the fifth and last β-strand of the DIX/DVL domain, most likely resulting in improper formation of this structure (38). Other truncations sequentially remove entire functional domains. Immunoblot analysis confirmed expression of the pR723Qfs variants at comparable levels with WT (Fig. 7B). The observed reduction in protein levels in the R841-truncating clone is consistent with a report stating that SUMOylation of the most C-terminal AXIN1 residues protects it from degradation (39). Reduced protein levels are also observed for the R395Afs*17 and L506Tfs*83 clones. Interestingly, in these latter clones, AXIN1 RNA levels are also significantly reduced (Fig. 7C).
Figure 7.
Characterization of AXIN1-truncating mutations in regulating β-catenin signaling. Analysis of clones carrying various endogenous AXIN1 truncations generated in HEK293T cells using CRISPR/Cas9-mediated gene editing. A, Schematic diagram showing the AXIN1 truncation variants generated in HEK293T cells using gene editing. The red domain at the end of truncated variants represents amino acid stretches resulting from the introduced frameshift mutation. B, Immunoblot analysis of generated clones using AXIN1 antibodies with N- and more C-terminal epitopes (cat. # 3323S and cat. #2087S, Cell Signaling Technology). Images were generated from one blot for each antibody, with irrelevant lanes removed. Original blots can be seen in Supplementary Data S1. C, qRT-PCR assay to determine AXIN1 mRNA expression levels (in triplicate, two independent experiments). Expression levels are depicted relative to the housekeeping gene GAPDH. The value for the WT control was arbitrarily set to 1. D, A β-catenin reporter assay was performed to determine β-catenin signaling levels in AXIN1-truncated clones. Values are depicted as WRE/MRE ratios (mean ± SD; in triplicate; three independent experiments). The value for the WT control was arbitrarily set to 1. E, Following siRNA-mediated knockdown of AXIN2, a β-catenin reporter assay was performed in all AXIN1-truncated clones. Values are depicted relative to the WRE/CMV-Renilla ratios obtained for the siControl-WT, which was arbitrarily set to 1. Note the logarithmic scale. Data are shown as mean ± SD. Statistical significance for all experiments was analyzed using a Mann–Whitney test. *, P < 0.05; **, P < 0.01; ns, nonsignificant.
A β-catenin reporter assay showed no change in the R841 and the first R723 clone, and a weak but significant 1.5-fold increase in the second R723 clone (Fig. 7D). The L506 truncation, which is expected to retain GSK3β and β-catenin binding, shows a significant 2-fold increase, whereas both R395-truncating clones show a 3-fold increase. We hypothesized that these relatively weak alterations in β-catenin signaling may result from functional compensation by AXIN2, and indeed, robust increases were observed following siRNA mediated knockdown of AXIN2 (Fig. 7E). WT cells do not show a clear increase in β-catenin reporter activity, whereas an inverse correlation is observed in the AXIN1 truncation clones relative to AXIN1 protein length.
Taken together, all tested AXIN1 truncations show a reduced capability to regulate β-catenin signaling. Loss of functionality appears to be inversely correlated with the length of the truncated AXIN1 protein, with longer proteins retaining more functionality. For the shorter AXIN1 variants, this is the result of a combination of loss of functional domains and potentially reduced mRNA stability.
AXIN1 missense mutations often co-occur with other β-catenin–activating gene mutations
Thus, far our data suggest that most AXIN1-truncating mutations can be regarded as (partial) loss of function, whereas most missense mutations behave as WT. Colorectal and liver cancers are characterized by a high proportion of mutations that aberrantly increase β-catenin signaling (2, 37). On the basis of these assumptions, one can hypothesize that colorectal and liver cancers with AXIN1 missense mutations will frequently be accompanied by other gene mutations that result in β-catenin activation. In such scenarios, the latter mutation is likely the driver mutation for tumor growth, whereas the AXIN1 missense mutation is probably a passenger mutation. Similarly, in cancers with AXIN1- truncating mutations, these mutations alone may be sufficient to sustain tumor growth without the need for other mutations in β-catenin–related genes.
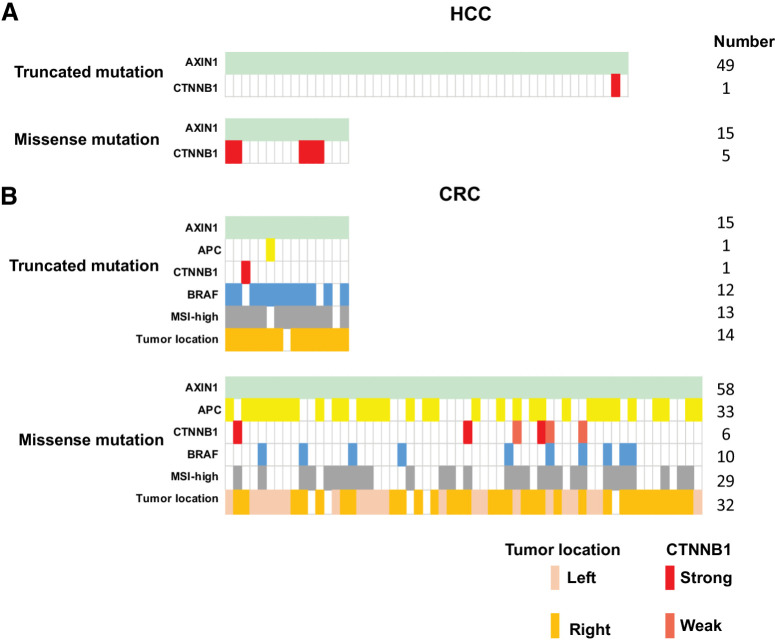
To explore this hypothesis, we selected all liver and colorectal cancers carrying AXIN1 mutations from cBioPortal (Supplementary Table S7). Among 15 liver cancers with missense AXIN1 variants, we identified 5 cancers (5/15, 33.3%) with highly active β-catenin mutations (Fig. 8A; ref. 37). Only one such mutation was observed among 49 liver cancers with truncating AXIN1 mutations, whereas two other tumors carried a truncating AXIN2 mutation that may affect β-catenin signaling (3/49, 6.1%). Using a Fisher's exact test this difference is significant (P = 0.0139), supporting our assumption. The 5 missense AXIN1 variants observed in tumors carrying β-catenin mutations have not been investigated in our study, but they are located in AXIN1 domains less likely to increase signaling. Among the remaining 10 tumors three carry variants that are partially defective (G98E, A120D, and C121F), which may support tumor growth by weakly increasing β-catenin signaling.
Figure 8.
Co-occurrence of mutations in β-catenin–related genes observed in AXIN1-mutant HCC and colorectal cancer tumors. From the cBioPortal database, we obtained information from AXIN1-mutant HCC and colorectal cancer (CRC) tumors about APC, CTNNB1, and BRAF mutation status, in addition to colorectal cancer tumor location and MSI-status. A, Identification of oncogenic β-catenin mutations in HCC tumors carrying either truncating (n = 49) or missense (n = 15) AXIN1 mutations. B, Identification of oncogenic β-catenin or inactivating APC mutations in colorectal cancer tumors carrying either truncating (n = 15) or missense (n = 58) AXIN1 mutations. The BRAF mutation (light blue) is the classical BRAF-V600E variant. Gray, microsatellite instability-high colorectal cancer tumors carrying a mismatch repair defect. Colorectal cancer tumor location is defined as being located on left- or right-sided of the splenic flexure. Identity of the specific β-catenin and APC mutations can be found in Supplementary Table S7.
The same analysis performed on colorectal cancers is more challenging to interpret correctly due to the existence of a mismatch-repair defective subgroup that leads to a high mutation frequency in many genes. Nevertheless, the analysis seems to support our assumption (Fig. 8B). Colorectal cancers with truncating AXIN1 mutations are highly enriched for BRAF-V600E mutation, a mismatch-repair defect, and right-sided tumor location (12/15, 13/15, and 14/15, respectively). Accordingly, most AXIN1-truncating mutations are frameshift mutations that arise at G or C mononucleotide repeats, prone for mutation in a mismatch-repair deficient background (2). Only 2 tumors carried either an additional truncating APC or oncogenic CTNNB1 mutation (2/15, 13.3%), two genes classically linked to activated β-catenin signaling. The observed mutations are regarded as rather weak activators of β-catenin signaling, that is APC-T1556Nfs*3 and β-catenin S45F (2, 37), suggesting that they cooperate with the truncating AXIN1 mutations to elevate β-catenin signaling to sufficiently high levels to support tumor growth.
Colorectal cancers with missense AXIN1 mutations do not show a clear association with BRAF-V600E (10/58, 17.2%) or tumor location (right-sided 32/58, 55.1%), but are somewhat enriched for a mismatch-repair defect (29/58, 50%). Importantly, 38 of 58 tumors (65.5%) carry either inactivating APC (33/58, 56.9%) or oncogenic β-catenin mutations (6/58, 10,3%) mutations (2, 13, 37), which is significantly higher compared with tumors with truncating AXIN1 mutations (P = 0.0004, Fisher's exact). However, interpretation is complicated by additional mutations in genes such as AXIN2, ZNRF3, and RNF43 in the truncated AXIN1 samples (Supplementary Table S7), generally believed to result in a modest β-catenin activation. When considering these genes in the analysis, it is found that 12/15 AXIN1-truncating colorectal cancers and 49/58 AXIN1-missense colorectal cancers carry potential additional β-catenin–activating mutations, which is not significantly different.
Thus, on the basis of the co-occurrence of mutations with the more classical tumor-related genes APC and CTNNB1, it reinforces our observations that truncating mutations probably support tumor formation, whereas AXIN1 missense mutations mostly represent passenger mutations, with the exception of specific defective variants as identified here.
Discussion
In this era of large-scale sequencing, many genetic variants are being detected in an increasing number of tumor samples. This has, however, also created a new challenge in that many variants are detected for which the clinical relevance is largely unknown (11). Which ones can be regarded as passenger mutations and which ones are likely to support tumor growth? Such knowledge is important to better understand the mechanisms underlying tumorigenesis, and is or will become important for deciding which treatment may benefit the patient. However, currently most detected gene alterations are so-called variants with unknown significance, meaning that their functional consequences are uncertain (40). This is also the case for the great majority of AXIN1 missense variants that are observed at comparable frequencies as truncating mutations. Before our analysis, a large proportion could theoretically represent driver mutations. Here, we investigated a total of 80 tumor-associated missense variants in three structural relevant domains of AXIN1. Our results indicate that most variants can be regarded as silent mutations, but in each domain-specific variants can be identified that are impaired in correctly regulating β-catenin signaling. In total, we identify 18 variants impaired in β-catenin regulation, observed in 22 individual cancers originating mainly from the liver, skin, pancreas or gastrointestinal tract. In addition, we establish at endogenous levels an inverse correlation between AXIN1 truncation length and remaining β-catenin regulatory function.
Deleting the GSK3β and β-catenin–binding domains from AXIN1 had the biggest impact on β-catenin signaling, followed by an accurate removal of the RGS/APC domain. In all three domains, we identified tumor-associated variants that were (partially) defective in β-catenin regulation. The observed defects overlapped in most cases with predicted consequences of our protein structure analysis. Unfortunately, such structural information is only available for the RGS/APC and DIX/DVL domain and short stretches of helical structures associating with GSK3β or β-catenin. The remainder of AXIN1 is apparently intrinsically disordered (Supplementary Fig. S9; refs. 41, 42), meaning that for the greater part of AXIN1 such an analysis is not possible. These disordered domains show a lower level of evolutionary conservation (Supplementary Fig. S3), and thus may show greater tolerance to amino acid variation, meaning that we expect only a small proportion of reported variations in these regions to be pathogenic. Likewise, mutations that reduce binding to proteolytic regulators of AXIN1, that is, tankyrases and SIAH1/2 (25, 43), are not expected to contribute to tumorigenesis, as they would theoretically lead to a more stable AXIN1 variant that in fact will be more effective in β-catenin regulation. Although our results indicate that most missense variants are silent, reliable conclusions can only be drawn using functional analyses such as performed here. In a Supplementary Discussion, we provide a more thorough description of the structural and functional consequences of our findings. Below we will focus on implications for AXIN1-driven tumorigenesis.
Our database analysis of colorectal and liver tumors (Fig. 8), supported our notion that most missense variants are silent ones, as they mostly co-occur with other gene mutations expected to increase β-catenin signaling. This latter mutation is then more likely to drive tumor formation. One limitation of this cBioPortal analysis is that no reliable information can be retrieved as to whether these mutations are heterozygous or homozygous. Except for oncogenic β-catenin mutations, all other mutated genes linked to increased β-catenin signaling are tumor-suppressor genes, meaning that both copies must acquire inactivating mutations. Thus, until we obtain a reliable zygosity status of gene mutations such as for APC, this analysis will not be entirely conclusive.
The AXIN1 gene must also obtain mutations on both copies to impair its function. We consider this also to be the case for the identified defective missense mutants. Strongly defective missense variants such as R395P, lead to a loss of function similar to early truncations, whereas selected missense variants in, for example, the RGS/APC domain, result in a partial loss of function, possibly equivalent to the long truncating AXIN1 variants we also examined. When missense mutations are observed in a tumor, it must first be determined whether it affects β-catenin regulation and to which extent. It must then be established whether this mutation is homozygous, or whether there is another second hit on the remaining gene copy. Only then one can conclude that AXIN1 function is sufficiently impaired to potentially contribute to tumorigenesis.
With respect to truncated AXIN1 proteins, loss of functionality appears to be inversely correlated to the length of the truncated AXIN1 protein. Longer truncating proteins retained β-catenin regulation that was almost indistinguishable from WT cells, and their partially defective nature only became apparent after simultaneous AXIN2 knockdown. Shorter variants, that is, before amino acid 506, led to significant increases and were more strongly activated by simultaneous AXIN2 knockdown. These latter truncations do not only lose more functional domains, but were also associated with reduced mRNA levels, possibly resulting from nonsense mediated decay (44). The observed increases may not seem strong, but as we and others have shown extensively in the past, high signaling levels are often not required in various tumor types or are even disadvantageous, whereas they nevertheless contribute to tumor growth (2). Moreover, being mutant for AXIN1 will make those tumors more liable to aberrant β-catenin activation, as they become strongly dependent on AXIN2 to counterbalance signaling activity.
The importance of truncating AXIN1 mutations is best established for hepatocellular cancers. Up to 10% carry AXIN1 mutations, mostly truncating ones (4, 45–47). Their relevance for colorectal cancer has remained more uncertain (8), as they mostly occur in tumors with a mismatch-repair defect that accumulate many mutations anyway, including genes irrelevant for tumorigenesis. Our database analysis suggests, however, that they appear positively selected in colorectal cancers, as AXIN1 truncations rarely co-occur with APC or CTNNB1 mutations, two genes classically linked to activated β-catenin signaling. Instead, they associate with truncating mutations in genes such as AXIN2, RNF43 or ZNRF3 (Supplementary Table S7), all mutations that are believed to lead to a weak activation of β-catenin signaling. Previously, we have shown that right-sided colorectal tumors select for mutations that result in more modest “just-right” levels of β-catenin signaling compared with their distal counterparts (2). All truncated AXIN1 colorectal cancers were right-sided cancers, and thus these mutations, possibly in synergy with mutations in other genes leading to weak β-catenin activation, may suffice to bring β-catenin signaling to the optimal level for this side of the colon. This could represent an alternative route compared with the more classical APC truncations or oncogenic β-catenin mutations. Further support for a role of AXIN1 truncations in colorectal cancer is provided by the observation that intestinal organoids grown to select for increased β-catenin signaling in a genome-wide CRISPR screen, repeatedly acquired sgRNAs targeting the AXIN1 gene (48).
Taken together, we show that most of AXIN1 missense variants behave as passenger mutations, but importantly, some variants can clearly impact AXIN1 function and should be regarded as driver mutations. Our results will aid the functional annotation of AXIN1 mutations identified in large-scale sequencing efforts or those from individual patients. They will also be relevant to correctly stratify samples when comparing AXIN1 mutant versus WT samples in various bioinformatics approaches. Last, we establish an inverse correlation of AXIN1 truncation and remaining residual function at endogenous levels.
Supplementary Material
Immunofluorescence analysis of β-catenin following Wnt3A addition or transfection with various AXIN1 variants
Analysis of the AXIN1 mutational landscape
Analysis of evolutionary conserved AXIN1 residues using the ConSurf tool.
The AXIN1 mutation landscape for various cancer types
Comparison of overexpressed and endogenous levels of AXIN1 mRNA.
response of β-catenin signaling in HEK293T cells following Wnt3A or GSK3β inhibitor addition
Additional analysis of missense variants near the GSK3β domain
β-catenin reporter analysis of AXIN1 R395P and R395H HEK293T knockin clones
AlphaFold structure prediction for human AXIN1
An accurate removal of the RGS/APC binding domain is required to correctly determine the functional consequences of losing this domain
Deleting part of the RGS/APC domain at endogenous levels results in increased β-catenin signaling
Analysis of additional tumor-associated AXIN1 variants located at conserved residues within the C-terminal part of the Tankyrase domain for their ability to impact β-catenin signaling.
Addition of an extra RGS/APC domain restores APC and β-catenin binding to selected RGS/APC domain variants, and (partially) restores β-catenin regulation
Cartoon representation of AXIN1 with bound APC
Side-by-side comparison of selected AXIN1 variants in different domains
Strong AXIN1 missense variants induce liver tumor formation in mice
Primer sequences
AXIN1 gnomAD analysis
Overview of the interactions at the interface between AXIN1 and GSK3β for the mutated amino acids and their predicted effects
Overview of the interactions at the interface between AXIN1 and β-catenin for the mutated amino acids and their predicted effects.
Reported missense mutations in D65_S228 domain of AXIN1
Overview of the interactions at the interface between AXIN1 and APC for the mutated amino acids and their predicted effects
Supplementary Table NO. 7
supplementary discussion
Original Western blot images
Acknowledgments
This research was financially supported by a China Scholarship Council PhD fellowship (file no. 201808530490 to R. Zhang) and (file no. 201909370083 to S. Li). J.H.G. Lebbink is supported by the gravitation program CancerGenomiCs.nl from the Netherlands Organisation for Scientific Research (NWO), part of the Oncode Institute, which is partly financed by the Dutch Cancer Society.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
S. Li reports personal fees from China Scholarship Council during the conduct of the study. M.P. Peppelenbosch reports grants, personal fees, and nonfinancial support from Pfizer, European Commission, Minzu NWU, and NWO/KWF, as well as personal fees from Narodowe Centrum Nauki, FWO, and ZONMW outside the submitted work. J.H.G. Lebbink reports grants from Oncode during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
R. Zhang: Data curation, software, formal analysis, supervision, investigation, writing–original draft, project administration, writing–review and editing. S. Li: Data curation, software, formal analysis, investigation, writing–original draft, writing–review and editing. K. Schippers: Data curation. Y. Li: Data curation, visualization. B. Eimers: Data curation, supervision. M. Lavrijsen: Data curation, visualization. L. Wang: Data curation. G. Cui: Data curation. X. Chen: Data curation, supervision. M.P. Peppelenbosch: Data curation, software, supervision. J.H.G. Lebbink: Data curation, software, supervision, project administration, writing–review and editing. R. Smits: Conceptualization, data curation, software, supervision, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene 2017;36:1461–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Albuquerque C, Bakker ER, van Veelen W, Smits R. Colorectal cancers choosing sides. Biochim Biophys Acta 2011;1816:219–31. [DOI] [PubMed] [Google Scholar]
- 3. Timbergen MJM, Smits R, Grünhagen DJ, Verhoef C, Sleijfer S, Wiemer EAC, et al. Activated signaling pathways and targeted therapies in desmoid-type fibromatosis: a literature review. Front Oncol 2019;9:397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 2015;149:1226–39. [DOI] [PubMed] [Google Scholar]
- 5. Ledinek Ž, Sobočan M, Knez J. The role of CTNNB1 in endometrial cancer. Dis Markers 2022;2022:1442441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 2015;43:D805–D11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mazzoni SM, Fearon ER. AXIN1 and AXIN2 variants in gastrointestinal cancers. Cancer Lett 2014;355:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Webster MT, Rozycka M, Sara E, Davis E, Smalley M, Young N, et al. Sequence variants of the axin gene in breast, colon, and other cancers: an analysis of mutations that interfere with GSK3 binding. Genes Chromosomes Cancer 2000;28:443–53. [PubMed] [Google Scholar]
- 10. Anvarian Z, Nojima H, van Kappel EC, Madl T, Spit M, Viertler M, et al. Axin cancer mutants form nanoaggregates to rewire the Wnt signaling network. Nat Struct Mol Biol 2016;23:324–32. [DOI] [PubMed] [Google Scholar]
- 11. Martínez-Jiménez F, Muiños F, Sentís I, Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer 2020;20:555–72. [DOI] [PubMed] [Google Scholar]
- 12. Wang W, Liu P, Lavrijsen M, Li S, Zhang R, Li S, et al. Evaluation of AXIN1 and AXIN2 as targets of tankyrase inhibition in hepatocellular carcinoma cell lines. Sci Rep 2021;11:7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu P, Liang B, Liu M, Lebbink JHG, Li S, Qian M, et al. Oncogenic mutations in armadillo repeats 5 and 6 of β-catenin reduce binding to APC, increasing signaling and transcription of target genes. Gastroenterology 2020;158:1029–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang W, Li S, Liu P, Sideras K, van de Werken HJG, van der Heide M, et al. Oncogenic STRAP supports hepatocellular carcinoma growth by enhancing wnt/beta-catenin signaling. Mol Cancer Res 2019;17:521–31. [DOI] [PubMed] [Google Scholar]
- 15. Qiao Y, Xu M, Tao J, Che L, Cigliano A, Monga SP, et al. Oncogenic potential of N-terminal deletion and S45Y mutant β-catenin in promoting hepatocellular carcinoma development in mice. BMC Cancer 2018;18:1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Adebayo Michael AO, Ko S, Tao J, Moghe A, Yang H, Xu M, et al. Inhibiting glutamine-dependent mTORC1 activation ameliorates liver cancers driven by β-catenin mutations. Cell Metab 2019;29:1135–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Beusekom B, Touw WG, Tatineni M, Somani S, Rajagopal G, Luo J, et al. Homology-based hydrogen bond information improves crystallographic structures in the PDB. Protein Sci 2018;27:798–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xing Y, Clements WK, Kimelman D, Xu W. Crystal structure of a beta-catenin/axin complex suggests a mechanism for the beta-catenin destruction complex. Genes Dev 2003;17:2753–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martino-Echarri E, Brocardo MG, Mills KM, Henderson BR. Tankyrase inhibitors stimulate the ability of tankyrases to bind axin and drive assembly of β-catenin degradation-competent axin puncta. PLoS ONE 2016;11:e0150484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bernkopf DB, Brückner M, Hadjihannas MV, Behrens J. An aggregon in conductin/axin2 regulates Wnt/β-catenin signaling and holds potential for cancer therapy. Nat Commun 2019;10:4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021;596:583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jin LH, Shao QJ, Luo W, Ye ZY, Li Q, Lin SC, et al. Detection of point mutations of the Axin1 gene in colorectal cancers. Int J Cancer 2003;107:696–9. [DOI] [PubMed] [Google Scholar]
- 23. Choi EJ, Kim S, Jho EH, Song KJ, Kee SH. Axin expression enhances herpes simplex virus type 1 replication by inhibiting virus-mediated cell death in L929 cells. J Gen Virol 2013;94:1636–46. [DOI] [PubMed] [Google Scholar]
- 24. Neo SY, Zhang Y, Yaw LP, Li P, Lin SC. Axin-induced apoptosis depends on the extent of its JNK activation and its ability to down-regulate beta-catenin levels. Biochem Biophys Res Commun 2000;272:144–50. [DOI] [PubMed] [Google Scholar]
- 25. Morrone S, Cheng Z, Moon RT, Cong F, Xu W. Crystal structure of a tankyrase-axin complex and its implications for axin turnover and tankyrase substrate recruitment. Proc Natl Acad Sci USA 2012;109:1500–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang X, Farrell AS, Daniel CJ, Arnold H, Scanlan C, Laraway BJ, et al. Mechanistic insight into Myc stabilization in breast cancer involving aberrant axin1 expression. Proc Natl Acad Sci USA 2012;109:2790–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Picco G, Petti C, Centonze A, Torchiaro E, Crisafulli G, Novara L, et al. Loss of AXIN1 drives acquired resistance to WNT pathway blockade in colorectal cancer cells carrying RSPO3 fusions. EMBO Mol Med 2017;9:293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu J, Li P, Wang L, Li M, Ge Z, Noordam L, et al. Modelling liver tumor organoids and cancer-associated fibroblasts interaction reveals the robust effects of stromal niche in cancer nurturing and treatment resistance. J Hepatol 2020;73:S638–S9. [Google Scholar]
- 29. Dajani R, Fraser E, Roe SM, Yeo M, Good VM, Thompson V, et al. Structural basis for recruitment of glycogen synthase kinase 3beta to the axin-APC scaffold complex. Embo J 2003;22:494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. von Kries JP, Winbeck G, Asbrand C, Schwarz-Romond T, Sochnikova N, Dell'Oro A, et al. Hot spots in β-catenin for interactions with LEF-1, conductin, and APC. Nat Struct Biol 2000;7:800–7. [DOI] [PubMed] [Google Scholar]
- 31. Spink KE, Polakis P, Weis WI. Structural basis of the axin-adenomatous polyposis coli interaction. EMBO J 2000;19:2270–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen X, Calvisi DF. Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am J Pathol 2014;184:912–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tao J, Xu E, Zhao Y, Singh S, Li X, Couchy G, et al. Modeling a human hepatocellular carcinoma subset in mice through coexpression of met and point-mutant β-catenin. Hepatology 2016;64:1587–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qiao Y, Wang J, Karagoz E, Liang B, Song X, Shang R, et al. Axis inhibition protein 1 (Axin1) deletion-induced hepatocarcinogenesis requires intact β-catenin but not notch cascade in mice. Hepatology 2019;70:2003–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abitbol S, Dahmani R, Coulouarn C, Ragazzon B, Mlecnik B, Senni N, et al. AXIN deficiency in human and mouse hepatocytes induces hepatocellular carcinoma in the absence of β-catenin activation. J Hepatol 2018;68:1203–13. [DOI] [PubMed] [Google Scholar]
- 36. Fodde R, Tomlinson I. Nuclear beta-catenin expression and wnt signalling: in defence of the dogma. J Pathol 2010;221:239–41. [DOI] [PubMed] [Google Scholar]
- 37. Rebouissou S, Franconi A, Calderaro J, Letouzé E, Imbeaud S, Pilati C, et al. Genotype-phenotype correlation of CTNNB1 mutations reveals different ß-catenin activity associated with liver tumor progression. Hepatology 2016;64:2047–61. [DOI] [PubMed] [Google Scholar]
- 38. Schwarz-Romond T, Fiedler M, Shibata N, Butler PJ, Kikuchi A, Higuchi Y, et al. The DIX domain of Dishevelled confers wnt signaling by dynamic polymerization. Nat Struct Mol Biol 2007;14:484–92. [DOI] [PubMed] [Google Scholar]
- 39. Kim MJ, Chia IV, Costantini F. SUMOylation target sites at the C terminus protect axin from ubiquitination and confer protein stability. FASEB J 2008;22:3785–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weile J, Roth FP. Multiplexed assays of variant effects contribute to a growing genotype-phenotype atlas. Hum Genet 2018;137:665–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Noutsou M, Duarte AM, Anvarian Z, Didenko T, Minde DP, Kuper I, et al. Critical scaffolding regions of the tumor suppressor axin1 are natively unfolded. J Mol Biol 2011;405:773–86. [DOI] [PubMed] [Google Scholar]
- 42. Xue B, Romero PR, Noutsou M, Maurice MM, Rüdiger SG, William AM Jr, et al. Stochastic machines as a colocalization mechanism for scaffold protein function. FEBS Lett 2013;587:1587–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ji L, Jiang B, Jiang X, Charlat O, Chen A, Mickanin C, et al. The SIAH E3 ubiquitin ligases promote wnt/β-catenin signaling through mediating wnt-induced axin degradation. Genes Dev 2017;31:904–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Behm-Ansmant I, Kashima I, Rehwinkel J, Saulière J, Wittkopp N, Izaurralde E, et al. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett 2007;581:2845–53. [DOI] [PubMed] [Google Scholar]
- 45. Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dahmani R, Just P-A, Perret C. The wnt/β-catenin pathway as a therapeutic target in human hepatocellular carcinoma. Clin Res Hepatol Gastroenterol 2011;35:709–13. [DOI] [PubMed] [Google Scholar]
- 47. Wang W, Pan Q, Fuhler GM, Smits R, Peppelenbosch MP. Action and function of wnt/β-catenin signaling in the progression from chronic hepatitis C to hepatocellular carcinoma. J Gastroenterol 2017;52:419–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ringel T, Frey N, Ringnalda F, Janjuha S, Cherkaoui S, Butz S, et al. Genome-scale CRISPR screening in human intestinal organoids identifies drivers of TGFβ resistance. Cell Stem Cell 2020;26:431–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunofluorescence analysis of β-catenin following Wnt3A addition or transfection with various AXIN1 variants
Analysis of the AXIN1 mutational landscape
Analysis of evolutionary conserved AXIN1 residues using the ConSurf tool.
The AXIN1 mutation landscape for various cancer types
Comparison of overexpressed and endogenous levels of AXIN1 mRNA.
response of β-catenin signaling in HEK293T cells following Wnt3A or GSK3β inhibitor addition
Additional analysis of missense variants near the GSK3β domain
β-catenin reporter analysis of AXIN1 R395P and R395H HEK293T knockin clones
AlphaFold structure prediction for human AXIN1
An accurate removal of the RGS/APC binding domain is required to correctly determine the functional consequences of losing this domain
Deleting part of the RGS/APC domain at endogenous levels results in increased β-catenin signaling
Analysis of additional tumor-associated AXIN1 variants located at conserved residues within the C-terminal part of the Tankyrase domain for their ability to impact β-catenin signaling.
Addition of an extra RGS/APC domain restores APC and β-catenin binding to selected RGS/APC domain variants, and (partially) restores β-catenin regulation
Cartoon representation of AXIN1 with bound APC
Side-by-side comparison of selected AXIN1 variants in different domains
Strong AXIN1 missense variants induce liver tumor formation in mice
Primer sequences
AXIN1 gnomAD analysis
Overview of the interactions at the interface between AXIN1 and GSK3β for the mutated amino acids and their predicted effects
Overview of the interactions at the interface between AXIN1 and β-catenin for the mutated amino acids and their predicted effects.
Reported missense mutations in D65_S228 domain of AXIN1
Overview of the interactions at the interface between AXIN1 and APC for the mutated amino acids and their predicted effects
Supplementary Table NO. 7
supplementary discussion
Original Western blot images
Data Availability Statement
Tumor associated mutations analyzed in this study were obtained from the cBioPortal (https://www.cbioportal.org/) and COSMIC (https://cancer.sanger.ac.uk/cosmic) databases. The predicted structure of full-length human AXIN1 was obtained from the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/entry/O15169). The PDB–REDO databank (https://pdb-redo.eu/) was used to obtain the optimized crystal structures for the β-catenin–AXIN1 complex (1qz7.pdb) and AXIN1–APC interface (1emu.pdb). The RCSB Protein Data Bank (https://www.rcsb.org/) was used to obtain AXIN1–GSK3β complex (1o9u.pdb). The ConSurf tool (https://consurf.tau.ac.il/) was used to obtain the AXIN1 evolutionary conservation score. Other data generated in this study are available upon request from the corresponding author.