Abstract
Hematopoietic stem cells (HSCs) are primarily dormant in a cell-cycle quiescence state to preserve their self-renewal capacity and long-term maintenance, which is essential for the homeostasis of hematopoietic system. Dysregulation of quiescence causes HSC dysfunction and may result in aberrant hematopoiesis (e.g., myelodysplastic syndrome and bone marrow failure syndromes) and leukemia transformation. Accumulating evidence indicates that both intrinsic molecular networks and extrinsic signals regulate HSC quiescence, including cell-cycle regulators, transcription factors, epigenetic factors, and niche factors. Further, the transition between quiescence and activation of HSCs is a continuous developmental path driven by cell metabolism (e.g., protein synthesis, glycolysis, oxidative phosphorylation, and autophagy). Elucidating the complex regulatory networks of HSC quiescence will expand the knowledge of HSC hemostasis and benefit for clinical HSC use. Here, we review the current understanding and progression on the molecular and metabolic regulation of HSC quiescence, providing a more complete picture regarding the mechanisms of HSC quiescence maintenance.
Keywords: Mitochondrial metabolism, Autophagy, Akt-mTOR, p38MAPK, Cell niche, Extracellular matrix
Introduction
Hematopoietic stem cells (HSCs) are a population of blood stem cells that primarily originate from endothelial cells during the embryonic period [1, 2]. Postnatal HSCs predominantly reside in the bone marrow (BM) and are heterogeneous, containing two well-recognized subpopulations: long-term HSCs (LT-HSCs) and short-term HSCs (ST-HSCs). LT-HSCs are largely cell-cycle-quiescent (G0 phase) with low biosynthetic activity, while ST-HSCs are broadly considered as activated stem cells. LT-HSCs and ST-HSCs can both respond to stimulation and stress, undergoing proliferation and differentiation into hematopoietic stem/progenitor cells (HSPCs), which further differentiate and produce all lineages of blood cells [3, 4]. A fraction of activated HSCs regenerates fully potent, non-committed HSCs via asymmetric cell division, a process called self-renewal [5, 6]. Although ST-HSCs respond faster to activation, LT-HSCs exhibit the highest self-renewal capacity and are responsible for life-long hematopoiesis [7, 8]. The long-term maintenance of HSCs requires a quiescent state against multiple cytotoxic and exhausted stresses, such as DNA damage and reactive oxygen stress (ROS) [5, 9, 10]. Dysregulated transitions between quiescence and activation impair HSC self-renewal and result in HSC dysfunction and eventual hematopoietic disorders [10, 11]. For instance, myeloproliferative neoplasm (MPN) has been reported to be associated with the increased proliferation and cell cycle entry of HSCs, leading to splenomegaly, leukocytosis, and myeloid hypercellularity [12]. Growth suppression of HSCs accelerates their exhaustion, which is a part of the cause of bone marrow failure diseases, such as Fanconi anemia (FA) [13, 14]. Thus, quiescence maintenance is pivotal for HSCs to preserve their function.
Emerging evidence indicates that the quiescence state of HSCs is regulated by a tightly coordinated molecular network, including both intracellular and extracellular molecules. It has been revealed that cell cycle regulators, transcription factors, epigenetic molecules, niche factors, and even physical factors govern HSC quiescence [6, 15]. Moreover, accumulating evidence suggests that intracellular metabolism drives HSC transitions between quiescence and activation [16–18]. This review aims to summarize and discuss the complicated regulatory networks of HSC quiescence, providing deep insight into HSC hemostasis and hematopoiesis.
Assessment of HSC quiescence
Quiescent HSCs are dormant cells in the cell cycle G0 phase. Based on the fundamental and associated characteristics of quiescent HSCs, several methods have been developed to assess the quiescence of HSCs. In general, these methods determine the cell cycle state based on nucleic acid content, proliferative markers, cell-surface markers, and other indirect markers (Table 1).
Cells in G0 and G1 stages have 2 N DNA content, while cells in S and G2–M stage have 2–4 N and 4 N DNA content, respectively. Moreover, G0-stage cells have lower RNA content than cells in the G1 stage. Thus, the cell cycle phase of HSCs can be detected using DNA-binding dyes (e.g., propidium iodide, DRAQ-5, DAPI, 7-AAD, and Hoechst 33,342) with RNA-binding dyes (Pyronin Y and SYTO) [5, 19, 20]. It has been demonstrated that most LT-HSCs are in the G0 phase (~ 70%) and G1 phase (~ 20%) by Hoechst 33,342 and Pyronin Y staining [20, 21]. Goodell et al. reported a small subset of Hoechst 33,342-stained BM cells that were separated from other cells and named them Hoechst-stained side population cells. These cells have cell surface markers of HSCs and the highest reconstitution capacity; moreover, only 1–3% of these HSCs are in the S–G2–M phase, indicating that the Hoechst-stained side population can be used for the enrichment or purification of quiescent HSCs [19]. Moreover, label incorporation-based assays can precisely monitor proliferative HSCs, as the nucleotide analogs, EdU and BrdU can be incorporated and detected in the newly synthesized DNA in HSCs [22, 23]. For instance, through the combination of DNA content staining and BrdU incorporation assays, the cell cycle stage of HSCs can be detected, G0–G1 stage (2 N DNA content without BrdU incorporation) and S stage (2–4 N DNA content with BrdU incorporation) [24, 25].
Several intracellular proteins have been revealed to exhibit cell-cycle stage dependence, with Ki-67 expression in G1 and S–G2–M phase, proliferating cell nuclear antigen (PCNA) expression in S phase, minichromosome maintenance-2 (MCM-2) expression in S phase, and phosphohistone H3 expression in M phase [26, 27]. Ki-67 and DNA staining can clearly distinguish HSCs in G0, G1, and S–G2–M phases [28, 29]. Furthermore, Ki-67RFP knock-in mice have been constructed and provide an effective method to trace the division history of HSCs in vivo [30].
Detection of DNA and intracellular protein in HSCs requires a process of cell fixation, which results in cell death and hinders the further use of HSCs. Quiescent HSCs reportedly have unique cell surface markers. CD41− cells in LT-HSCs (Lin−c-Kit+Sca-1+CD48−CD150+) are nearly all quiescent (~ 96%); thus, CD41 is an effective candidate to mark quiescent HSCs [31]. Moreover, CD229 and CD244 can sub-fractionate LT-HSCs, as CD229−/lowCD244− cells in LT-HSCs are quiescent with almost undetectable Ki-67 expression [32]. CD63 and CD82 are also quiescent markers of HSCs, which sustain transforming growth factor β (TGFβ) signaling activity [33, 34]. Interestingly, a recent study reported that CD93 designates HSCs that are at the edge of quiescence and prime to activation [35].
Several new methods have been developed for assessing the cell cycle of HSCs using multiple indirect markers. Similar to BrdU incorporation, the histone H2B-GFP protein can be incorporated into nucleosomes; thus, H2B-GFP transgenic mice provide a model for analyzing HSC quiescence in vivo [36, 37]. Furthermore, it has been revealed that mitochondrial membrane potential (MMP)-low HSCs are quiescent (~ 85% in G0 phase), while MMP-high HSCs are activated (~ 40% in G0 phase), indicating that MMP represents a potential marker for distinguishing between quiescent and activated HSCs [37]. Similarly, quiescent HSCs exhibit relatively low ROS levels than activated HSCs [38]. Recently, the advanced single-cell-RNA-sequence (scRNA-seq) method has been applied to distinguish HSCs in different cell cycle phases based on functional annotations [39, 40].
Table 1.
Current methods to determine HSC quiescence
| Label | Labeling | Reference |
|---|---|---|
| Content of DNA and RNA | Hoechst + Pyronin Y | [20, 21] |
| DAPI/7AAD + BrdU | [23] | |
| DAPI/7AAD + EdU | [24, 25] | |
| Proliferative markers and content of DNA | Ki-67 + DAPI/Hoechst | [28, 29] |
| Cell surface markers | CD41 | [31] |
| CD229 + CD244 | [32] | |
| CD63 | [33] | |
| CD82 | [34] | |
| Content of nucleosomes | H2B-GFP | [36, 37] |
| Mitochondrial membrane potential | Tetramethylrhodamine ethyl ester | [37] |
| Reactive oxygen stress | DCFH-DA | [38] |
| Transcriptional dynamics | Single-cell-RNA-sequence | [39, 40] |
In brief, the above-mentioned methods provide a variety of strategies for assessing HSC quiescence; however, more methods for labeling quiescent HSCs are required, especially for unique cell-surface markers.
Intracellular molecular networks of HSC quiescence
In addition to metabolism-regulating HSC quiescence that is process-driven, many intracellular factors have been demonstrated to play a vital role in the cell cycle of HSCs. These factors form molecular networks that govern HSC quiescence and self-renewal; chiefly, these molecular networks include cell cycle regulators, transcription factors, kinase factors, and epigenetic factors.
Cell cycle regulators
The cell cycle process is directly governed by several master regulators, cyclins, cyclin-dependent kinases (CDKs), and cyclin-dependent kinase inhibitors (CKIs) [6]. Cyclins are synthesized during the cell cycle at specific times, bind to the CDKs and form CDK–cyclin complexes that drive a series of events in order, such as DNA synthesis and mitosis [41]. The activity of CDK–cyclin complexes is tightly regulated by CKIs, which interact with CDKs and can impede cell cycle progression [41, 42]. The well-recognized regulators involved in the cell cycle include cyclin classes (cyclin A, cyclin B, cyclin C, and cyclin D), CDK classes (CDK1, CDK2, CDK4, CDK6), and CKI classes (p21, p27, and p57) [41, 42]. Abundant evidence has demonstrated that the above-mentioned regulators play direct roles in HSC maintenance via regulation of cell division or quiescence. Loss of cyclin A inhibits proliferation of HSCs and results in an acute reduction of the HSC pool, suggesting that HSCs rely on cyclin A to sustain cell division [43]. Moreover, deletion of cyclin D1/2/3 leads to decreased HSPC numbers and severe anemia [44]. Cyclin D binds and activates CDK4 and CDK6, and overexpression of the cyclin D1–CDK4 complex promotes HSC exit from the G0 phase (quiescence) and shortens the G1 phase. This results in increased reconstitution capacity of HSCs, suggesting a direct role of cyclin A in HSC quiescence [45]. Consistently, deficiency of CDK4 and CDK6 impedes the quiescence exit of HSCs, resulting in severe anemia in mice [46]. Notably, ST-HSCs exhibit higher CDK6 protein levels than LT-HSCs, and enforcing CDK6 expression in LT-HSCs promotes quiescence exist without impairing self-renewal [8]. It seems that enforcing the expression of CDK4 or CDK6 is an effective strategy to promote HSC proliferation and subsequent hematopoiesis without the expense of impaired HSC maintenance.
The members of the CKI family, including p21, p27, and p57, primarily interact with and inhibit CDK2 and CDK4. It has been demonstrated that CKIs have predominant roles in quiescence maintenance in HSCs, especially p21 and p57. Deficiency of p21 or p57 promotes quiescence HSC entry into the cell cycle and results in HSC exhaustion [47, 48]. Interestingly, loss of p18 remarkably rescues loss of the HSC pool in p21-deficient mice, suggesting diverse functions of CKIs [49]. Loss of p27 in mice has no obvious effect on cell cycling of HSCs, but it has been revealed that p27 cooperates with p57 to maintain quiescence of HSCs by regulating the localization of heat shock cognate protein 70 (Hsc70)–cyclin D complex [50]. In brief, the above-mentioned evidence reveals that cyclin–CDK complexes are essential for HSC proliferation, and HSCs rely on CKIs to restrict activity of cyclin–CDK complexes to preserve quiescence. In addition to the above-mentioned cell cycle regulators, G(0)/G(1) switch gene 2 (G0S2) has been shown to be enriched in LT-HSCs and to preserve HSC quiescence by sequestering nucleolin in the cytosol [51].
Transcription factors
HSC quiescence is intrinsically governed by the expression of a series of genes associated with metabolism and the cell cycle. Transcription factors are a type of proteins that directly bind DNA, promoting gene expression [52]. Several transcription factors have been revealed to play pivotal roles in quiescence maintenance in HSCs by regulating gene expression [6, 53]. Tumor suppressor p53 regulates cell proliferation and apoptosis via its multiple transcriptional targets [54]. Loss of p53 substantially promotes HSC proliferation, shown by 30% to 60% BrdU+ HSCs, suggesting that p53 is essential for quiescence maintenance in HSCs [55]. Mechanistically, p53 preserves quiescence of HSCs primarily by transcriptionally promoting expression of growth factor independent-1 (Gfi-1) and Necdin, which are inhibitors of HSC proliferation [55–58]. The Myc oncogenes (c-Myc, N-Myc, and L-Myc) transcriptionally regulate multiple genes required for cell survival and proliferation [59]. Double-deficiency of c-Myc and N-Myc inhibits proliferation of myelomonocytic populations but promotes cell cycling of HSCs and results in depleted HSC pool [60]. In contrast, deletion of c-Myb transcription factor substantially reduces proliferation but promotes differentiation of HSCs [61]. Similarly, it has been revealed that several transcription factors or repressors are essential for quiescence maintenance in HSCs, including HLF, Pbx1, Evi-1, Nurr1, Nrf2, C/EBPα, PU.1, YY1, Gfi-1, Fhl2, and Tcf15 [29, 40, 56, 62–70]. Deletion of several transcription factors, such as MEF (ELF4) and Id1, preserves quiescence in HSCs [71, 72]. Moreover, several transcriptional families have been demonstrated to govern the quiescence of HSCs. Hox genes encode a family of transcriptional regulators, and deletion of Hoxb3 and Hoxb4 impairs the proliferation of HSCs, while overexpression of Hoxb4 or Hoxb7 substantially enhances the expansion capacity of HSCs [73–76]. Importantly, Hoxb5 marks long-term HSCs, and these quiescent HSCs are almost attached to VE-cadherin-positive cells, suggesting a vital role of Hoxb5 in quiescence maintenance in HSCs [77]. Notably, GATA transcriptional family member Gata2 inhibits proliferation and increases quiescence in HSCs by repressing gene expression of CCND1, CDK4, and CDK6, while Gata3 is essential for HSC entry into the cell cycle [78, 79]. Forkhead box (Fox) genes encode several transcription factors that are categorized into subclasses FoxA to FoxS; it has been demonstrated that FoxM1 is essential for quiescence maintenance in HSCs by inducing expression of Nurr1, which preserves quiescence of HSCs via inducing expression of p21 and p27 [65, 80, 81].
Deletion of FoxO1, FoxO3, and FoxO4 increases cell cycling and apoptosis in quiescent HSCs via alteration of the expression of their target genes, including cyclin G2, cyclin D, p21, and p27 [82, 83]. In addition to the above-mentioned mechanisms, several transcription factors regulate quiescence via cell metabolism. Also, increased ROS has been shown to contribute to quiescence loss in FoxO1/3/4-deficient HSCs [82, 83]. Moreover, FoxO3 is reported to direct autophagy in HSCs by maintaining the expression of autophagy-related genes, suggesting that autophagy substantially mediates the effect of FoxO transcription factors on HSC quiescence [84]. Similarly, Runx1 transcriptionally induces the expression of multiple ribosome protein genes and controls ribosome biogenesis, suggesting that metabolism is one of the main processes involved in the regulation of HSC quiescence by transcription factors [85]. In summary, multiple transcription factors play a vital role in quiescence maintenance in HSC primarily by regulating the expression of genes related to cell metabolism and cell cycling, and this regulatory network requires further investigation.
Epigenetic factors
Emerging evidence for cell-intrinsic regulators of HSC quiescence includes epigenetic factors, such as DNA methyltransferases, RNA methylases and demethylases, histone modification regulators, and non-coding RNA. Gene expression is tightly associated with DNA methylation that is catalyzed by DNA methyltransferases; it has been revealed that loss of Dnmt3a promotes HSC self-renewal at the expense of differentiation, as well as reducing the percentage of Ki-67+ HSCs [86]. N6-methyladenosine (m6A) is the most prevalent type of RNA modification and is involved in splicing regulation, RNA stability, translocation, and translation [87]. Recent evidence suggests that m6A plays a crucial role in quiescence maintenance in HSCs; for instance, deletion of the m6A methyltransferase Mettl3 decreases quiescence in HSCs primarily via modulating Myc mRNA [88]. DNA and histones (H2A, H2B, H3, and H4) form condensed chromatin and the structure of chromatin controls gene expression [89]. It has been reported that HSC quiescence is governed by 3D genome reorganization, and CCCTC-binding factor (CTCF) is a key factor mediating chromatin interactions [90]. More precisely, histone modifications tightly regulate chromatin structure and gene expression, including acetylation, methylation, phosphorylation, and others [91]. Deficiency of SET domain containing 2 (Setd2), a histone methyltransferase, promotes dormant HSC entry into the cell cycle, suggesting a critical role of histone methylation in HSC quiescence maintenance [92]. Similarly, loss of histone methyltransferase mixed-lineage leukemia (MLL) or absent, small, or homeotic 1-like (Ash1l) reduces quiescence in HSCs by decreasing gene expression of CKI [93, 94]. Nuclear receptor corepressor 1 (NCoR1) binds its partner histone deacetylase 3 (HDAC3) and modulates H3 lysine 27 acetylation (H3K27ac), while deletion of NCoR1 substantially promotes quiescent HSC proliferation by regulating the expression of myeloid-differentiation genes [95]. The latest evidence has revealed that deletion of histone acetyltransferase MOF, a regulator of H4 lysine 16 acetylation (H4K16ac), promotes CD93 expression in HSCs and results in HSCs having quiescent transcriptional features but elevated proliferative capacity, suggesting that H4K16ac sustains a balance of HSCs between quiescence and activation [35, 96].
Non-coding RNAs
In addition to the above-mentioned epigenetic factors, multiple pieces of evidence has revealed that non-coding RNAs also regulate quiescence in HSCs. An imprinted gene at the H19-Igf2 locus is essential for quiescence maintenance in HSCs by transcribing microRNA-675 (miR-675) [97]. Similarly, the imprinted Dlk1-Glt2 locus preserves quiescence state of HSCs by producing a miRNA cluster that targets the PI3K-Akt pathway [98]. MicroRNAs are ubiquitously expressed in cells and function as post-transcriptional regulators of gene expression by targeting transcripts [99]. miR-675 targets insulin-like growth factor 1 receptor (Igfr1) and restricts Igf2-Igfr1, preserving quiescence in HSCs [97, 100]. Moreover, inhibited miR-126 expression increases HSCs proliferation without exhaustion by targeting multiple transcripts associated with the PI3K-Akt pathway [101]. Loss of miR-21 impairs quiescence maintenance in HSCs by targeting programmed cell death 4 (PDCD4) [102]. Interestingly, miR-29a has been reported to be essential for quiescence maintenance partially by regulating Dnmt3a [103]. In summary, epigenetic factors can be considered one hub of quiescence regulatory networks within HSCs, primarily by governing gene expression.
Intracellular signaling pathways
In addition to the above-mentioned intracellular regulators of HSC quiescence, several factors control cell cycling in HSCs via unique pathways. For instance, ubiquitination is a critical process for inducing protein degradation [104]. It has been demonstrated that COP9 signalosome subunit 5 (CSN5), F-box and WD-40 domain protein 7 (Fbxw7), and Huwe1 control HSC quiescence by mediating ubiquitin-dependent degradation of proteins associated with cell cycle regulation [105–108]. S-phase kinase-associated protein-2 (Skp2) forms an Skp2 complex with its partners that induces ubiquitination and degradation of cell cycle inhibitors; however, loss of Skp2 triggers quiescent HSC entry into the cell cycle by promoting gene expression of cyclin D1 [109]. Moreover, multiple kinase-related pathways play a vital role in sensing intracellular and extracellular signaling and regulating cellular behaviors, such as proliferation, differentiation, migration, and apoptosis. Among these kinases, several kinase-related signaling transduction pathways have been demonstrated to govern HSC quiescence. The Janus kinases (JAKs) are critical factors responding to cytokines, while deletion of JAK1 increases HSC quiescence and impedes proliferation, thus impairing HSC self-renewal [110]. The ataxia telangiectasia mutated (ATM) kinase is a key factor responding to replication stress and DNA damage; it has been demonstrated that ATM is essential for quiescence maintenance in HSCs by restricting oxidative stress [111, 112]. Loss of Ptpn21, a protein tyrosine phosphatase, results in reduced quiescence in HSCs partially by decreasing HSC stiffness and increasing HSC motility, suggesting that cellular deformability is a critical regulator of HSC quiescence [113].
Moreover, a wide variety of kinases-related pathways synergistically maintain the balance of HSCs between quiescence and activation, to preserve HSC homeostasis and hematopoiesis. The phosphoinositide 3-kinase (PI3K)-protein kinase B (Akt)-mammalian target of rapamycin (mTOR) pathway is a universal enzyme-related signaling pathway, and mTOR functions as a key effector that regulates protein synthesis, mitochondrial metabolism, and autophagy via multiple targets, such as ribosome S6 kinase (S6K1), eukaryotic translation initiation factor 4E-binding protein (4E-BP1), YY1, Atg1, and Atg3 [114, 115]. It has been revealed that activation of Akt or mTOR substantially induces quiescent HSCs to exit dormancy, while inhibition of Akt causes HSCs to persist in G0 phase, a process largely dependent on mitochondrial metabolism [116–119]. PTEN restricts activity of Akt-mTOR signaling by inhibiting phosphorylation of PI3K, and loss of PTEN promotes quiescent HSC proliferation [120–122]. AMP-activated protein kinase (AMPK) inhibits mTOR activity by phosphorylation of tuberous sclerosis complex (TSC); moreover, AMPK promotes FOX transcription factors [123]. Deficiency of AMPK and its upstream-regulator Lkb1 increases the division of quiescent HSC and results in a depleted HSC pool [124]. The mitogen-activated protein kinase (MAPK) pathway regulates multiple cellular processes and senses stress, primarily including three kinases, MAPK kinase kinase, MAPK kinase, and MAPK [125]. It has been revealed that p38MAPK is essential for quiescent HSC entry into the cell cycle in response to stress primarily by promoting purine metabolism [126]. Similarly, deletion of another MAPK family member, extracellular signal-regulated kinase 1/2 (ERK1/2), inhibits HSC proliferation [127]. Interestingly, ERK reportedly mediates activated HSCs returning to quiescence by phosphorylation of its upstream-regulator MEK and limiting the activation of Akt-mTOR pathway, which further regulates mitochondrial metabolism and protein synthesis [128]. Collectively, cell cycle regulators, transcription factors, epigenetic factors, non-coding RNAs, and kinase-related factors all have been revealed as critical regulators of HSC quiescence. Notably, these factors are interrelated. Cell cycle regulators directly drive cell cycle progression; moreover, metabolism, which we will discuss in the next section, is also a hub of HSC quiescence regulation. Transcription factors, epigenetic factors, and non-coding RNAs govern HSC quiescence mainly via indirect pathways that affect the expression of their targets. For instance, transcription factor Gata2 regulates the expression of cell cycle regulators CCND1, CDK4, and CDK6 to preserve HSC quiescence [78, 79]. In brief, the above-mentioned intracellular factors synergistically form an intrinsic regulator network that governs HSC equilibrium between quiescence and activation; however, this network has not been entirely elucidated and requires further exploration.
Intracellular molecular networks of HSC quiescence, especially for metabolism
It has been revealed that the transition of HSCs between quiescence and activation is a continuous developmental path associated with altered cellular metabolism rather than a stepwise progression [129]. Emerging evidence indicates that intracellular metabolism plays a pivotal role in quiescence maintenance in HSCs, especially energy metabolism, protein synthesis, and autophagy [6, 130].
Glycolysis and mitochondrial metabolism
Adenosine triphosphate (ATP) is the basic substance that provides energy for cellular processes. There are two metabolic pathways to synthesize ATP in cells, glycolysis, and oxidative phosphorylation (OXPHOS) [131]. Glycolysis is involved in the breakdown of glucose to pyruvate via sequential enzyme-catalyzed reactions, which occurs in the cytosol in an oxygen-independent manner [131–133]. In mitochondria, pyruvate or other oxidative fuels can be further oxidized to generate acetyl-coenzyme A (acetyl-CoA), which leads to ATP production via the tricarboxylic acid (TCA) cycle and the electron transport chain (ETC). The OXPHOS process requires oxygen and produces more ATP than glycolysis with higher efficiency but also generates ROS that may damage cells [132, 134, 135].
HSCs primarily localize in a hypoxic BM environment, and quiescent HSCs primarily utilize anaerobic glycolysis to synthesize ATP [136–138]. It has been revealed that quiescent LT-HSCs have higher glycolytic capacity than other cells in the BM [139, 140]. Pyruvate dehydrogenase (PDH) is a key enzyme that converts pyruvate to acetyl-CoA; HSCs exhibit activated PDH kinases (PDKs) that inhibit PDH by phosphorylation to maintain glycolysis [141, 142]. Deletion of Pdk2 and Pdk4 in mice suppresses glycolysis but promotes OXPHOS in HSCs, leading to impaired quiescence maintenance and exhaustion of HSCs and demonstrating that HSCs rely on glycolysis to sustain quiescence [142]. Hypoxia-inducible factor 1α (HIF-1α) is a transcription factor that is activated by hypoxia and promotes the expression of glycolytic genes, such as PDKs and lactate dehydrogenase (LDA) [143]. It has been revealed that the location of HSCs in the BM is uneven but that they prefer hypoxic regions; accordingly, LT-HSCs exhibit high expression of HIF-1α [140, 144]. HIF-1α and its up-regulator, Meis1, are both enriched in LT-HSCs and preserve HSC glycolysis [145]. Knock-out of HIF-1α in mice results in loss of quiescence in HSCs (G0 phase ~ 83% to ~ 65%) and impairs HSC maintenance [146]. The unique energy metabolism in HSCs is tightly associated with cell cycle quiescence in HSCs, suggesting that glycolysis preserves HSC quiescence [140, 145].
Glycolysis meets the demand of ATP synthesis and maintains HSC quiescence; upon activation, HSCs switch their metabolism to utilize mitochondrial OXPHOS to synthesize more ATP, meeting the robust energy demand [147]. Previous studies have revealed reduced mitochondria levels in HSCs compared to other hematopoietic cells by MitoTracker Green staining; however, dye efflux affects the accuracy of MitoTracker Green staining. Thus, the results of recent studies using mtDNA quantification, an artificial mitochondrial reporter, and enumeration of mitochondrial nucleoids indicate higher mitochondrial mass in HSCs than in lineage-committed progenitors and mature cells [16, 148–150]. The ETC generates the mitochondrial membrane potential (MMP) across the mitochondrial membrane, which reflects the polarization of mitochondria and promotes dye intake; thus, researchers proposed that HSCs have highest the MMP but relatively low mitochondrial volumes and amount than that in multipotent progenitors [151]. Despite the above-mentioned controversy, it has been demonstrated that low mitochondrial activity or MMP marks quiescent HSCs and these MMPlow HSCs exhibit higher potency than MMPhigh HSCs, suggesting that mitochondrial activity governs HSC quiescence and self-renewal [37, 149]. Furthermore, it has been demonstrated that increased mitochondrial Ca2+ flux drives elevated MMP levels and subsequent HSC division [152]. Indeed, deficiency of mitochondrial carrier homologue 2 induces increased mitochondrial size and OXPHOS, subsequently promoting HSCs proliferation [153]. Mitochondrial metabolism triggers HSC entry into the cell cycle primarily through generating ROS, which has been demonstrated to induce damage and cell-cycling in quiescent HSCs [16, 116, 154, 155]. Like MMP, ROS also distinguishes quiescent from activated HSCs, and these ROSlow HSCs are more quiescent and have higher self-renewal capacity than that in ROShigh HSCs [38]. Moreover, metabolites derived from glycolysis and mitochondrial metabolism affect epigenetic modifications, such as histone acetylation and DNA methylation [136, 147]. For instance, acetyl-CoA is primarily generated by glycolysis-derived pyruvate and functions as a substrate of histone acetyltransferases (HATs). It has been demonstrated that acetylation of H4K16 and H3K27 plays a pivotal role in HSC transition between quiescence and activation [35, 147, 156, 157]. Further establishment of how metabolism and epigenetic modifications synergistically control HSC quiescence will greatly improve our understanding of this process.
Quiescent HSCs rely on glycolysis to generate ATP and require restricted mitochondrial metabolism to preserve quiescence; consistently, inhibition of mitochondrial activity using trifluoromethoxyphenylhydrazone to uncouple the ETC induces quiescence in HSCs and benefits HSC maintenance [149]. More directly, deletion of Rieske iron sulfur protein (RISP), a subunit of mitochondrial complex III, results in aberrant HSC entry into the cell cycle and eventual HSC exhaustion [148]. Similarly, deficiency of PTPMT1, a PTEN-like mitochondrial phosphatase localized at the mitochondrial membrane, impairs mitochondrial aerobic metabolism and considerably reduces quiescence in HSCs by ~ 80% to ~ 50%, leading to HSC exhaustion and BM failure in mice [158]. In summary, HSCs require balanced mitochondrial activity to preserve quiescence, and both excessively activated and inhibited mitochondrial metabolism impair quiescence maintenance in HSCs.
Protein synthesis at ribosomes and the endoplasmic reticulum
Like energy production, protein synthesis is one of the most fundamental forms of metabolism in cells. Emerging evidence has revealed that HSC quiescence is tightly regulated by protein synthesis that primarily occurs at the ribosome and endoplasmic reticulum (ER). Ribosomes consist of ribosomal RNA and ribosomal proteins (RPs) and are responsible for mRNA translation into proteins [159]. Mutations in several ribosomal proteins lead to hematopoietic diseases, such as Diamond Blackfan anemia [159–161]. Using an O-propargyl-puromycin (OP-Puro) incorporation assay, it has been found that HSCs exhibit the lowest rate of protein synthesis compared to other BM cells, while inhibition of ribosome function by haploinsufficiency of large ribosomal subunit protein 24 (Rpl24) results in fewer HSCs being in S/G2/M phases, suggesting that the reduced ribosome function inhibits proliferation of HSCs [162]. Unexpectedly, haploinsufficiency of small ribosomal subunit protein 14 (Rps14) remarkably reduces the quiescence in HSCs (~ 70% to ~ 42%) [163]. Moreover, deletion of Notchless impairs maturation of the ribosomal pre-60S subunit, resulting in quiescent HSC entry into the cell cycle, suggesting that ribosome functions are necessary for quiescence maintenance in HSCs [164]. Nevertheless, existing evidence suggests that increased ribosome biogenesis may also impair quiescence maintenance in HSCs. For instance, deficiency of phosphatase and tension homolog (PTEN) increases protein synthesis and promotes HSC cell cycling, while blocking ribosome biogenesis largely restores the function of PTEN-deficient HSCs [162]. It seems that both increased and inhibited ribosome biogenesis impairs quiescence maintenance in HSCs; however, more direct and convincing evidence is required to confirm these findings.
Abundant ribosomes are found attached to the ER, comprising 60% of the cell membrane and playing critical roles in protein synthesis, folding, and modification, as well as calcium storage and lipid metabolism [165, 166]. The ER senses the quantity and quality of synthesized proteins and assists protein folding into native structures, while the accumulation of misfolded/unfolded proteins induces ER stress [167]. Emerging evidence has revealed the critical roles of the ER in quiescence maintenance in HSCs. Knock-out of glucose-regulated protein 94 (Grp94), an ER chaperone regulating protein folding, assembly, and secretion, substantially reduces HSC quiescence [168]. The Sel1L/Hrd1/ER-associated degradation (ERAD) complex localizes to the ER membrane, and assists in recognizing and subsequent degradation of misfolded proteins, thus reducing ER stress. Increased ER protein folding or reduced ER stress promotes the self-renewal and reconstituted capacity of HSCs [169, 170]. It has been revealed that quiescent HSCs exhibit relatively higher expression of ERAD genes. Deletion of Sel1L remarkably induces proliferation of quiescent HSCs via mammalian target of rapamycin (mTOR) pathway and niche interaction, suggesting that ER stress drives HSCs to exist quiescence [171, 172]. In summary, ribosome biogenesis and ER stress are key regulators of HSC quiescence.
Autophagy and lysosomal metabolism
Like materials synthesis, degradation of pre-existing materials is also vital for cell renovation [173]. There are two primary cellular degradation systems, the proteasome, and the lysosome; the former exhibits high selectivity recognizing ubiquitinated substrates, while the latter has broader substrate selection [173]. Autophagy is a pathway by which cytoplasmic materials are delivered to lysosomes for degradation; furthermore, the degraded products can be reutilized [174]. Autophagy is governed by a series of autophagy-related genes (Atg) and is divided into three categories: macroautophagy, microautophagy, and chaperone-mediated autophagy [173, 174]. It has been demonstrated that autophagy is essential for cellular longevity and plays a critical role in hematopoiesis [175]. Importantly, loss of autophagy substantially promotes quiescent HSC entry into the cell cycle, resulting in excessive HSC activation and subsequent exhaustion through deletion of FIP200, Atg5, Atg7, or Atg12 [175–179]. It has been revealed that loss of autophagy largely increased mitochondrial metabolism and ROS level in HSCs; the activated metabolism leads to stepwise epigenetic remodeling (DNA methylation) and impairs HSC self-renewal [175–177, 179]. It has been revealed that mitophagy, a process of quality control in mitochondria, is essential for the self-renewal of Tie2+ HSCs [180]. Loss of O-linked N-acetylglucosamine transferase impairs quiescence maintenance of HSCs due to inhibited mitophagy [181]. HSCs require autophagy to clear excess active, healthy mitochondria; thus, restricted mitochondrial metabolism maintains quiescence [179]. Chaperone-mediated autophagy (CMA) has been consistently reported to be upregulated in activated HSCs, promoting activated HSC self-renewal and preserving HSC pool [182]. Moreover, HSC aging is associated with decreased autophagy, while activation of autophagy enhances HSC function, including quiescence reentry [179, 182]. Lysosomes are major mediators of endocytic and autophagic pathways [183]. A recent study has revealed that inhibition of lysosomal activity by concanamycin A (ConA) decreases MMP and promotes quiescence in HSCs, suggesting that HSC quiescence requires restrained lysosomal activity [37].
In addition to the above-mentioned metabolic regulators of HSC quiescence, the metabolism of fatty acids, amino acids, and nucleic acids has been reported to affect HSC proliferation [126, 184, 185]. For instance, it has been demonstrated that hematopoietic stem and progenitor cells have a higher level of fatty acid oxidation (FAO) compared with differentiated hematopoietic cells. Inhibition of the PML-PPAR-δ-FAO pathway impairs asymmetric division of HSCs and results in their exhaustion [186]. In summary, emerging evidence suggests that unique metabolic status can be used as a hallmark of quiescent HSCs. Glycolysis, OXPHOS, and protein synthesis directly drive the cell cycle progression of HSCs, suggesting that metabolism is a determinant of HSC quiescence, similar to cell cycle regulators. Certainly, metabolism is also interrelated and hierarchical; for instance, mitophagy maintains HSC quiescence largely via the regulation of mitochondrial metabolism [180, 181]. However, many questions regarding HSC quiescence and activation remain unanswered. What is the integrated change of cell metabolism during HSC transition between quiescence and activation? How do energy production, protein synthesis, autophagy, and fatty acid metabolism synergistically preserve HSC quiescence? Thus, it is necessary to further explore these areas to provide a more integrated landscape of metabolism governing HSC quiescence.
Extracellular regulatory networks regulating HSC quiescence
Adult HSCs primarily reside in the BM throughout life, and this niche is extremely complicated and provides a supportive microenvironment for HSCs [9, 187]. It is well-recognized that the maintenance of stem cells relies on the surrounding microenvironment defined as the stem cell niche [188, 189]. Dormant HSCs rapidly lose quiescence and are activated upon exiting the BM niche, suggesting a critical role for the microenvironment in governing HSC quiescence [190]. Current evidence indicates that multiple niche factors regulate HSC quiescence, such as extracellular matrix (ECM), niche-secreted factors, and cell–cell interactions [6, 190].
Cell-ECM interactions
The ECM is a three-dimensional macromolecular structure that provides a basic scaffold for cells and is composed of up to 300 components, primarily including collagens, laminin, fibronectin, and proteoglycans [191, 192]. The composition and structure of the ECM exhibit tissue specificity; moreover, it has been demonstrated that the ECM plays critical roles in cellular behaviors, such as proliferation, differentiation, and migration, primarily through its physical properties and chemical factors [191, 192]. It has been revealed that physical properties, such as stiffness and elasticity, regulate HSC function in vivo; for instance, quiescent HSCs require a stiffer endosteal niche while softer vascular niches promote HSC activation [193–196]. The increased spatial confinement of 3D scaffold benefits preserving quiescence in HSCs in vivo, suggesting that biophysical cues of ECM play critical roles in quiescence maintenance in HSC [197]. In addition to physical properties, components of the ECM can directly interact with adhesion receptors to regulate HSC quiescence, and disruption of ECM remodeling by deletion of metalloproteinase-1 (TIMP-1) impairs quiescence maintenance and self-renewal of HSCs [196, 198–200]. For instance, the ECM protein Matrilin-4 is highly expressed in LT-HSCs and represses proliferation under stress, while the ECM protein Del-1 promotes LT-HSC proliferation [201, 202]. Integrins, cadherins, CD44, and selectins are the main adhesion receptors of HSCs; it has been demonstrated that disruption of ECM-adhesion receptor interactions impairs quiescence maintenance in HSCs [6, 196]. CD44 interacts with hyaluronic acid and drives HSC homing to the BM, sustaining their quiescence state [203]. The periostin-integrin-αv interaction preserves the quiescence state in HSCs by inhibiting the FAK-PI3K-Akt pathway [204]. Consistently, inhibition of N-cadherin impairs the self-renewal of HSCs, while overexpression of N-cadherin substantially increases quiescence in HSCs [205]. In contrast, loss of E-selectin markedly increases quiescence in HSCs, suggesting the diverse functions of adhesion receptors in regulating HSC quiescence [206]. Besides the above-mentioned physical and chemical properties, the ECM may also regulate HSC cell cycling by retaining niche-secreted factors, such as TGF-β [6]. In summary, the present evidence suggests the critical roles of the ECM in HSC quiescence; considering the complicated composition and structure of the ECM, the exact function of the ECM requires further investigation.
Niche cells and niche-secreted factors
At present, the most well-recognized extracellular regulatory factors of HSC quiescence are niche-secreted [6]. Adult HSCs localize in the BM niche, where factors produced by other cells (e.g., osteoblasts, endothelial cells, adipocytes, and megakaryocytes) and niche cell-HSC interactions, as well as factors secreted by distal tissues, substantially regulate HSC quiescence [6, 190]. It has been demonstrated that niche-produced factors, such as CXC motif chemokine ligand 4 (CXCL4), CXCL12, TGF-β, thrombopoietin (THPO), osteopontin (OPN), angiopoietin-1 (Ang-1), and prostaglandin E2 (PGE2), are essential for quiescence maintenance in HSCs (Table 2). Chemokines are transmembrane cytokines that interact with chemokine receptors on cells to regulate multiple cellular behaviors; several CXCL genes are relatively highly expressed in quiescence human HSCs compared to activated HSCs [207]. Deficiency of CXCL12 secreted by leptin receptor positive (LepR+) stromal cells compromises quiescence maintenance in HSCs; moreover, inhibition of CXCR4 (the CXCL12 receptor) induces excessive proliferation of HSCs [208, 209]. Deletion of CXCL4 or its receptor CXCR2 both promotes HSC proliferation and impairs HSC self-renewal, while injection of CXCL4 increases HSC quiescence [207, 210]. Megakaryocytes are one of the main producers of CXCL4 and have been demonstrated to maintain HSC quiescence by CXCL4 secretion [210]. Although other research reported that deletion of megakaryocytes results in quiescent HSC entry in cell cycle, they propose different mechanisms in which megakaryocytes secrete TGF-β and maintain HSC quiescence via the TGF-β-Smad pathway [211]. In addition to megakaryocytes, Schwann cells in the BM also sustain HSC quiescence by producing TGF-β, which further regulates the expression of genes involved in cell cycling via the TGF-β-Smad pathway [212]. THPO is also a critical cytokine for quiescence maintenance in HSCs, and HSCs are closely associated with THPO-secreted osteoblasts where THPO interacts with its receptor MPL to activate β-integrin and CKIs signaling [213, 214]. Moreover, osteoblasts produce OPN and angiopoietin-1, which both sustain a quiescence state in HSCs [215, 216]. Among the multiple BM stromal cells, macrophages have been demonstrated to produce cyclooxygenase, a precursor of PGE2, which preserves HSC quiescence by inhibiting Akt signaling and repressing ROS production in HSCs [217].
Table 2.
Niche-secreted factors regulating HSC quiescence
| Factor | Source | Effect on HSC quiescence | Reference |
|---|---|---|---|
| CXCL4 | Megakaryocytes and plateles | Preserving quiescence | [207, 210] |
| CXCL12 | LepR+ stromal cells | Preserving quiescence | [208, 209] |
| TGF-β | Megakaryocytes and Schwann cells | Preserving quiescence | [211, 212] |
| Thrombopoietin | Osteoblasts | Preserving quiescence | [213, 214] |
| Osteopontin | Osteoblasts | Preserving quiescence | [215] |
| Angiopoietin-1 | Osteoblasts | Preserving quiescence | [216] |
| Prostaglandin E2 | Macrophages | Preserving quiescence | [217] |
| Histamine | Myeloid cells | Preserving quiescence | [219] |
| Adiponectin | Adipocytes | Inducing quiescence exit | [225] |
| Leptin | Adipocytes | Inducing quiescence exit | [227] |
| IGF1 | Hepatocytes and osteoblasts | Inducing quiescence exit | [228] |
Recently, emerging evidence has uncovered several unreported niche factors that participate in preserving HSC quiescence. Vitamin A and its metabolite, retinoic acid have been found to sustain HSC quiescence by restricting protein synthesis and ROS production [129]. Angiogenin (ANG), a niche-produced ribonuclease, represses HSC proliferation primarily by inducing the generation of tiRNA and restricting protein synthesis [218]. Niche histamine, produced by a subpopulation of myeloid cells, preserves quiescence and self-renewal in myeloid-biased HSCs by activation the H2 receptor [219]. Extracellular vesicles (EVs) are membrane bound and transfer bioactive molecules that regulate the cell cycle of HSPCs. Mesenchymal stromal cells release EVs and the MyD88 adapter protein in EVs reduces quiescence of HSPCs [220]. Recently, it has been reported that EVs produced by osteoblastic cells promote proliferation of hematopoietic progenitor cells via processed tiRNA in EVs, suggesting the critical role of EVs and tiRNA in HSPC quiescence [221]. Notably, several immune factors have been to impact HSC quiescence, including interleukin-1 (IL-1), IL-3, and interferon gamma (INFγ) [222–224]. HSC hemostasis requires an equilibrium between quiescence and activation and several niche factors have been reported to induce cell cycling of HSCs. For instance, adipocyte-secreted adiponectin induces quiescence exit of HSCs by activating mTOR signaling [225]. Niche fibroblast growth factor 2 (FGF2) promotes the proliferation of HSCs predominantly by activation of the c-kit receptor and phosphorylation of STAT5 [226]. Leptin, primarily secreted by adipose tissue, promotes the proliferation of HSCs by regulating niche cells to secrete other niche factors, such as CXCL12 [227]. IGF1 is mainly produced by the liver and osteoblasts, which has been demonstrated to selectively promote the proliferation of a subset of HSCs by activating Akt signaling and restoring mitochondrial metabolism [228].
In addition to the above-mentioned factors secreted by niche cells, physical interaction between HSCs and niche cells may also regulate HSC cell cycling. It has been reported that multiple stromal cells physically interact with HSCs, including osteoblasts, BM mesenchymal stem cells (BMSCs), megakaryocytes, and macrophages [34, 212, 229]. The direct interaction between macrophages and HSCs promotes stabilization of CD82 on HSCs, and CD82 preserves HSC quiescence, implying a critical role of physical interaction in HSC homeostasis [34]. Collectively, ECM, niche factors, and niche cells all have critical roles in preserving HSC quiescence. Notably, these extracellular regulatory networks regulate HSC quiescence via intracellular signaling transduction, indicating a signaling axis from extracellular to intracellular. Nevertheless, how these extracellular signaling pathways synergistically function and whether certain pathways are more critical to HSC quiescence remains poorly understood. Moreover, identifying novel niche factors and developing strategies for utilizing cytokines to preserve quiescence and self-renewal in HSCs will benefit the clinical application of HSCs.
Conclusion and future perspectives
In this review, we summarized the definition, assessment methods, and regulatory networks of HSC quiescence.
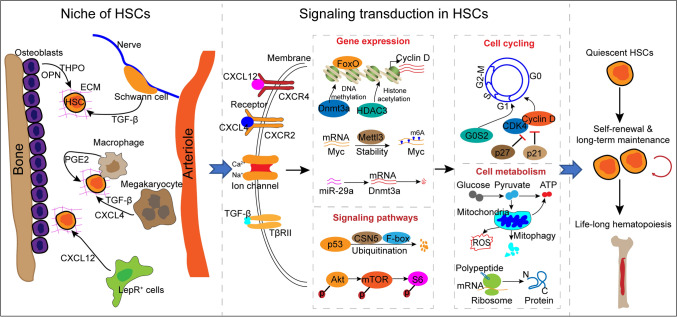
Quiescent HSCs are dormant cells in the G0 phase, which can be precisely detected using multiple direct and indirect markers. HSC quiescence is regulated by multiple factors, and intrinsically, cell cycle-associated factors directly govern HSC quiescence. Moreover, emerging evidence indicates that HSC transition between quiescence and activation is driven by cell metabolism, including mitochondrial metabolism, protein synthesis, and autophagy. The expression of cell cycle-associated factors and mitochondrial metabolism are governed by multiple intracellular molecules, such as transcription factors, kinase-related factors, and epigenetic factors that synergistically comprise a regulatory network of HSC quiescence. In addition to intracellular factors, the ECM, niche cells, and niche factors have been reported to affect HSC proliferation, depending on receptors on the HSCs and downstream signal transduction. Overall, cell cycle regulators and cell metabolism can be considered the hubs of the quiescence regulatory network of HSCs, and other intracellular and extracellular factors sustain HSC quiescence primarily through the basic cell cycling and metabolism pathways. Thus, the above-mentioned factors form a multilevel regulatory network of HSC quiescence (Fig. 1).
Fig. 1.
Overview of the regulatory network of HSC quiescence. HSC quiescence is regulated by multiple factors, and intrinsically, cell cycle-associated factors and cell metabolism directly govern HSC quiescence. Niche cells, niche-secreted factors, kinases, transcription factors, and epigenetic factors comprise a complicated network. The network transduces multiple signaling and primarily affects cell cycling and cell metabolism, eventually preserving HSC quiescence to sustain long-term maintenance of HSCs and life-long hematopoiesis
Quiescence maintenance is a pivotal property for HSC self-renewal, and dysregulated quiescence impairs HSC homeostasis and subsequent hematopoiesis. Specifically, several hematopoietic diseases are tightly associated with abnormal cell cycling in HSCs, such as myeloproliferative disease and Fanconi anemia [12–14]. Moreover, HSCs lose quiescence and self-renewal capacity upon exiting their niche, which impede HSC culture in vitro and further clinical utilization. Thus, several questions and directions should be considered in future research. First, the metabolic and molecular regulatory network of HSC quiescence still needs to be completely illustrated. Second, the exact effects and underlying mechanisms of dysregulated HSC quiescence on hematopoiesis require further clarification. Third, attention should be paid to developing efficient strategies to preserve HSC quiescence and potential in vivo and in vitro, such as by leveraging cytokines.
Acknowledgements
This work was supported by grants from the National Key R&D Program of China (2017YFA0106700) and the National Science Foundation of China (82170115).
Author contributions
ZC and QG wrote the manuscript. GBS and YH designed and revised the manuscript. All authors have approved this version of the article.
Funding
National Key R&D Program of China, 2017YFA0106700, Yu Hou, National Science Foundation of China, 82170115, Yu Hou
Availability of data and material
Not applicable.
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Yu Hou: Lead contact.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhe Chen and Qian Guo contributed equally.
Contributor Information
Guanbin Song, Email: song@cqu.edu.cn.
Yu Hou, Email: houyuxn@vip.126.com.
References
- 1.McGrath KE, Frame JM, Fromm GJ, et al. A transient definitive erythroid lineage with unique regulation of the β-globin locus in the mammalian embryo. Blood. 2011;117:4600–4608. doi: 10.1182/blood-2010-12-325357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dharampuriya PR, Scapin G, Wong C, John Wagner K, Cillis JL, Shah DI. Tracking the origin, development, and differentiation of hematopoietic stem cells. Curr Opin Cell Biol. 2017;49:108–115. doi: 10.1016/j.ceb.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bigas A, Waskow C. Blood stem cells: from beginning to end. Development. 2016;143:3429–3433. doi: 10.1242/dev.142828. [DOI] [PubMed] [Google Scholar]
- 4.Dzierzak E, Bigas A. Blood development: hematopoietic stem cell dependence and independence. Cell Stem Cell. 2018;22:639–651. doi: 10.1016/j.stem.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura-Ishizu A, Takizawa H, Suda T. The analysis, roles and regulation of quiescence in hematopoietic stem cells. Development. 2014;141:4656–4666. doi: 10.1242/dev.106575. [DOI] [PubMed] [Google Scholar]
- 6.O'Reilly E, Zeinabad HA, Szegezdi E. Hematopoietic versus leukemic stem cell quiescence: Challenges and therapeutic opportunities. Blood Rev. 2021 doi: 10.1016/j.blre.2021.100850. [DOI] [PubMed] [Google Scholar]
- 7.Wilson A, Laurenti E, Oser G, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 8.Laurenti E, Frelin C, Xie S, et al. CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell Stem Cell. 2015;16:302–313. doi: 10.1016/j.stem.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calvi LM, Link DC. The hematopoietic stem cell niche in homeostasis and disease. Blood. 2015;126:2443–2451. doi: 10.1182/blood-2015-07-533588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamashita M, Dellorusso PV, Olson OC, Passegué E. Dysregulated haematopoietic stem cell behaviour in myeloid leukaemogenesis. Nat Rev Cancer. 2020;20:365–382. doi: 10.1038/s41568-020-0260-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh S, Jakubison B, Keller JR. Protection of hematopoietic stem cells from stress-induced exhaustion and aging. Curr Opin Hematol. 2020;27:225–231. doi: 10.1097/MOH.0000000000000586. [DOI] [PubMed] [Google Scholar]
- 12.Chu SH, Heiser D, Li L, et al. FLT3-ITD knockin impairs hematopoietic stem cell quiescence/homeostasis, leading to myeloproliferative neoplasm. Cell Stem Cell. 2012;11:346–358. doi: 10.1016/j.stem.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Kozono DE, O'Connor KW, et al. TGF-β inhibition rescues hematopoietic stem cell defects and bone marrow failure in fanconi anemia. Cell Stem Cell. 2016;18:668–681. doi: 10.1016/j.stem.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ceccaldi R, Parmar K, Mouly E, et al. Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 2012;11:36–49. doi: 10.1016/j.stem.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Göttgens B. Regulatory network control of blood stem cells. Blood. 2015;125:2614–2620. doi: 10.1182/blood-2014-08-570226. [DOI] [PubMed] [Google Scholar]
- 16.Hu M, Wang J. Mitochondrial metabolism and the maintenance of hematopoietic stem cell quiescence. Curr Opin Hematol. 2019;26:228–234. doi: 10.1097/MOH.0000000000000507. [DOI] [PubMed] [Google Scholar]
- 17.Ito K, Ito K. Hematopoietic stem cell fate through metabolic control. Exp Hematol. 2018;64:1–11. doi: 10.1016/j.exphem.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z, Jiang H. A chromatin perspective on metabolic and genotoxic impacts on hematopoietic stem and progenitor cells. Cell Mol Life Sci. 2020;77:4031–4047. doi: 10.1007/s00018-020-03522-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gothot A, Pyatt R, McMahel J, Rice S, Srour EF. Functional heterogeneity of human CD34(+) cells isolated in subcompartments of the G0 /G1 phase of the cell cycle. Blood. 1997;90:4384–4393. doi: 10.1182/blood.V90.11.4384. [DOI] [PubMed] [Google Scholar]
- 21.Passegué E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med. 2005;202:1599–1611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheshier SH, Morrison SJ, Liao X, Weissman IL. In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc Natl Acad Sci USA. 1999;96:3120–3125. doi: 10.1073/pnas.96.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkinson AC, Ishida R, Kikuchi M, et al. Long-term ex vivo haematopoietic-stem-cell expansion allows nonconditioned transplantation. Nature. 2019;571:117–121. doi: 10.1038/s41586-019-1244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z, Huo D, Li L, et al. Nuclear DEK preserves hematopoietic stem cells potential via NCoR1/HDAC3-Akt1/2-mTOR axis. J Exp Med. 2021;218:e20201974. doi: 10.1084/jem.20201974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheng Y, Ma R, Yu C, et al. Role of c-Myc haploinsufficiency in the maintenance of HSCs in mice. Blood. 2021;137:610–623. doi: 10.1182/blood.2019004688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rumman M, Dhawan J, Kassem M. Concise review: quiescence in adult stem cells: biological significance and relevance to tissue regeneration. Stem Cells. 2015;33:2903–2912. doi: 10.1002/stem.2056. [DOI] [PubMed] [Google Scholar]
- 27.Gerdes J, Li L, Schlueter C, et al. Immunobiochemical and molecular biologic characterization of the cell proliferation-associated nuclear antigen that is defined by monoclonal antibody Ki-67. Am J Pathol. 1991;138(4):867–873. [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Diao D, Shi Z, et al. SIRT6 Controls Hematopoietic stem cell homeostasis through epigenetic regulation of wnt signaling. Cell Stem Cell. 2016;18:495–507. doi: 10.1016/j.stem.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 29.Lu Z, Hong CC, Kong G, et al. Polycomb group protein YY1 is an essential regulator of hematopoietic stem cell quiescence. Cell Rep. 2018;22:1545–1559. doi: 10.1016/j.celrep.2018.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grinenko T, Eugster A, Thielecke L, et al. Hematopoietic stem cells can differentiate into restricted myeloid progenitors before cell division in mice. Nat Commun. 2018;9:1898. doi: 10.1038/s41467-018-04188-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 32.Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu M, Lu Y, Wang S, et al. CD63 acts as a functional marker in maintaining hematopoietic stem cell quiescence through supporting TGFβ signaling in mice. Cell Death Differ. 2021 doi: 10.1038/s41418-021-00848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hur J, Choi JI, Lee H, et al. CD82/KAI1 maintains the dormancy of long-term hematopoietic stem cells through interaction with DARC-expressing macrophages. Cell Stem Cell. 2016;18:508–521. doi: 10.1016/j.stem.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 35.Pessoa Rodrigues C, Akhtar A. Differential H4K16ac levels ensure a balance between quiescence and activation in hematopoietic stem cells. Sci Adv. 2021;7:abi5987. doi: 10.1126/sciadv.abi5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foudi A, Hochedlinger K, Van Buren D, et al. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol. 2009;27:84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang R, Arif T, Kalmykova S, et al. Restraining lysosomal activity preserves hematopoietic stem cell quiescence and potency. Cell Stem Cell. 2020;26:359–376. doi: 10.1016/j.stem.2020.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jang YY, Sharkis SJ. A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood. 2007;110:3056–3063. doi: 10.1182/blood-2007-05-087759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kowalczyk MS, Tirosh I, Heckl D, et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25:1860–1872. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez-Fraticelli AE, Weinreb C, Wang SW, et al. Single-cell lineage tracing unveils a role for TCF15 in haematopoiesis. Nature. 2020;583:585–589. doi: 10.1038/s41586-020-2503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 42.Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 43.Kalaszczynska I, Geng Y, Iino T, et al. Cyclin A is redundant in fibroblasts but essential in hematopoietic and embryonic stem cells. Cell. 2009;138:352–365. doi: 10.1016/j.cell.2009.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozar K, Ciemerych MA, Rebel VI, et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 2004;118:477–491. doi: 10.1016/j.cell.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 45.Mende N, Kuchen EE, Lesche M, et al. CCND1-CDK4-mediated cell cycle progression provides a competitive advantage for human hematopoietic stem cells in vivo. J Exp Med. 2015;212:1171–1183. doi: 10.1084/jem.20150308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malumbres M, Sotillo R, Santamaría D, et al. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell. 2004;118:493–504. doi: 10.1016/j.cell.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Cheng T, Rodrigues N, Shen H, et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- 48.Matsumoto A, Takeishi S, Kanie T, et al. p57 is required for quiescence and maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2011;9:262–271. doi: 10.1016/j.stem.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 49.Yu H, Yuan Y, Shen H, Cheng T. Hematopoietic stem cell exhaustion impacted by p18 INK4C and p21 Cip1/Waf1 in opposite manners. Blood. 2006;107:1200–1206. doi: 10.1182/blood-2005-02-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zou P, Yoshihara H, Hosokawa K, et al. p57(Kip2) and p27(Kip1) cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell Stem Cell. 2011;9:247–261. doi: 10.1016/j.stem.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 51.Yamada T, Park CS, Burns A, Nakada D, Lacorazza HD. The cytosolic protein G0S2 maintains quiescence in hematopoietic stem cells. PLoS One. 2012;7:e38280. doi: 10.1371/journal.pone.0038280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haberle V, Stark A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Biol. 2018;19:621–637. doi: 10.1038/s41580-018-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hao S, Chen C, Cheng T. Cell cycle regulation of hematopoietic stem or progenitor cells. Int J Hematol. 2016;103:487–497. doi: 10.1007/s12185-016-1984-4. [DOI] [PubMed] [Google Scholar]
- 54.Parrales A, Iwakuma T. Targeting oncogenic mutant p53 for cancer therapy. Front Oncol. 2015;5:288. doi: 10.3389/fonc.2015.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y, Elf SE, Miyata Y, et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009;4:37–48. doi: 10.1016/j.stem.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hock H, Hamblen MJ, Rooke HM, et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431:1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 57.Hu B, Wang S, Zhang Y, Feghali CA, Dingman JR, Wright TM. A nuclear target for interleukin-1alpha: interaction with the growth suppressor necdin modulates proliferation and collagen expression. Proc Natl Acad Sci USA. 2003;100:10008–10013. doi: 10.1073/pnas.1737765100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Asai T, Liu Y, Di Giandomenico S, et al. Necdin, a p53 target gene, regulates the quiescence and response to genotoxic stress of hematopoietic stem/progenitor cells. Blood. 2012;120:1601–1612. doi: 10.1182/blood-2011-11-393983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. 2018;3:5. doi: 10.1038/s41392-018-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laurenti E, Varnum-Finney B, Wilson A, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3:611–624. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lieu YK, Reddy EP. Conditional c-myb knockout in adult hematopoietic stem cells leads to loss of self-renewal due to impaired proliferation and accelerated differentiation. Proc Natl Acad Sci USA. 2009;106:21689–21694. doi: 10.1073/pnas.0907623106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Komorowska K, Doyle A, Wahlestedt M, et al. Hepatic leukemia factor maintains quiescence of hematopoietic stem cells and protects the stem cell pool during regeneration. Cell Rep. 2017;21:3514–3523. doi: 10.1016/j.celrep.2017.11.084. [DOI] [PubMed] [Google Scholar]
- 63.Ficara F, Murphy MJ, Lin M, Cleary ML. Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell. 2008;2:484–496. doi: 10.1016/j.stem.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goyama S, Yamamoto G, Shimabe M, et al. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell. 2008;3:207–220. doi: 10.1016/j.stem.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 65.Sirin O, Lukov GL, Mao R, Conneely OM, Goodell MA. The orphan nuclear receptor Nurr1 restricts the proliferation of haematopoietic stem cells. Nat Cell Biol. 2010;12:1213–1219. doi: 10.1038/ncb2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsai JJ, Dudakov JA, Takahashi K, et al. Nrf2 regulates haematopoietic stem cell function. Nat Cell Biol. 2013;15:309–316. doi: 10.1038/ncb2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ye M, Zhang H, Amabile G, et al. C/EBPa controls acquisition and maintenance of adult haematopoietic stem cell quiescence. Nat Cell Biol. 2013;15:385–394. doi: 10.1038/ncb2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Staber PB, Zhang P, Ye M, et al. Sustained PU.1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Mol Cell. 2013;49:934–946. doi: 10.1016/j.molcel.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chavez JS, Rabe JL, Loeffler D, et al. PU.1 enforces quiescence and limits hematopoietic stem cell expansion during inflammatory stress. J Exp Med. 2021;218:e20201169. doi: 10.1084/jem.20201169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hou Y, Wang X, Li L, et al. FHL2 regulates hematopoietic stem cell functions under stress conditions. Leukemia. 2015;29:615–624. doi: 10.1038/leu.2014.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lacorazza HD, Yamada T, Liu Y, et al. The transcription factor MEF/ELF4 regulates the quiescence of primitive hematopoietic cells. Cancer Cell. 2006;9:175–187. doi: 10.1016/j.ccr.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 72.Singh SK, Singh S, Gadomski S, et al. Id1 ablation protects hematopoietic stem cells from stress-induced exhaustion and aging. Cell Stem Cell. 2018;23:252–265.e8. doi: 10.1016/j.stem.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu M, Zhan J, Zhang H. HOX family transcription factors: Related signaling pathways and post-translational modifications in cancer. Cell Signal. 2020;66:109469. doi: 10.1016/j.cellsig.2019.109469. [DOI] [PubMed] [Google Scholar]
- 74.Björnsson JM, Larsson N, Brun AC, et al. Reduced proliferative capacity of hematopoietic stem cells deficient in Hoxb3 and Hoxb4. Mol Cell Biol. 2003;23:3872–3883. doi: 10.1128/MCB.23.11.3872-3883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Antonchuk J, Sauvageau G, Humphries RK. HOXB4 overexpression mediates very rapid stem cell regeneration and competitive hematopoietic repopulation. Exp Hematol. 2001;29:1125–1134. doi: 10.1016/s0301-472x(01)00681-6. [DOI] [PubMed] [Google Scholar]
- 76.Carè A, Valtieri M, Mattia G, et al. Enforced expression of HOXB7 promotes hematopoietic stem cell proliferation and myeloid-restricted progenitor differentiation. Oncogene. 1999;18:1993–2001. doi: 10.1038/sj.onc.1202498. [DOI] [PubMed] [Google Scholar]
- 77.Chen JY, Miyanishi M, Wang SK, et al. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature. 2016;530:223–227. doi: 10.1038/nature16943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tipping AJ, Pina C, Castor A, et al. High GATA-2 expression inhibits human hematopoietic stem and progenitor cell function by effects on cell cycle. Blood. 2009;113:2661–2672. doi: 10.1182/blood-2008-06-161117. [DOI] [PubMed] [Google Scholar]
- 79.Ku CJ, Hosoya T, Maillard I, Engel JD. GATA-3 regulates hematopoietic stem cell maintenance and cell-cycle entry. Blood. 2012;119:2242–2251. doi: 10.1182/blood-2011-07-366070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Golson ML, Kaestner KH. Fox transcription factors: from development to disease. Development. 2016;143:4558–4570. doi: 10.1242/dev.112672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hou Y, Li W, Sheng Y, et al. The transcription factor Foxm1 is essential for the quiescence and maintenance of hematopoietic stem cells. Nat Immunol. 2015;16:810–818. doi: 10.1038/ni.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tothova Z, Kollipara R, Huntly BJ, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 83.Miyamoto K, Araki KY, Naka K, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 84.Warr MR, Binnewies M, Flach J, et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494:323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cai X, Gao L, Teng L, et al. Runx1 deficiency decreases ribosome biogenesis and confers stress resistance to hematopoietic stem and progenitor cells. Cell Stem Cell. 2015;17:165–177. doi: 10.1016/j.stem.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Challen GA, Sun D, Jeong M, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2011;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qin Y, Li L, Luo E, et al. Role of m6A RNA methylation in cardiovascular disease (review) Int J Mol Med. 2020;46:1958–1972. doi: 10.3892/ijmm.2020.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cheng Y, Luo H, Izzo F, et al. m6A RNA methylation maintains hematopoietic stem cell identity and symmetric commitment. Cell Rep. 2019;28:1703–1716. doi: 10.1016/j.celrep.2019.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takayama N, Murison A, Takayanagi SI, et al. The transition from quiescent to activated states in human hematopoietic stem cells is governed by dynamic 3D genome reorganization. Cell Stem Cell. 2021;28:488–501. doi: 10.1016/j.stem.2020.11.001. [DOI] [PubMed] [Google Scholar]
- 91.Beerman I, Rossi DJ. Epigenetic control of stem cell potential during homeostasis, aging, and disease. Cell Stem Cell. 2015;16:613–625. doi: 10.1016/j.stem.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou Y, Yan X, Feng X, et al. Setd2 regulates quiescence and differentiation of adult hematopoietic stem cells by restricting RNA polymerase II elongation. Haematologica. 2018;103:1110–1123. doi: 10.3324/haematol.2018.187708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McMahon KA, Hiew SY, Hadjur S, et al. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 2007;1:338–345. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 94.Jones M, Chase J, Brinkmeier M, et al. Ash1l controls quiescence and self-renewal potential in hematopoietic stem cells. J Clin Invest. 2015;125:2007–2020. doi: 10.1172/JCI78124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wan X, Liu L, Zhou P, et al. The nuclear receptor corepressor NCoR1 regulates hematopoiesis and leukemogenesis in vivo. Blood Adv. 2019;3:644–657. doi: 10.1182/bloodadvances.2018022756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Valerio DG, Xu H, Eisold ME, Woolthuis CM, Pandita TK, Armstrong SA. Histone acetyltransferase activity of MOF is required for adult but not early fetal hematopoiesis in mice. Blood. 2017;129:48–59. doi: 10.1182/blood-2016-05-714568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Venkatraman A, He XC, Thorvaldsen JL, et al. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature. 2013;500:345–349. doi: 10.1038/nature12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qian P, He XC, Paulson A, et al. The Dlk1-Gtl2 locus preserves LT-HSC function by inhibiting the PI3K-mTOR pathway to restrict mitochondrial metabolism. Cell Stem Cell. 2016;18:214–228. doi: 10.1016/j.stem.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shi Y, Wang Q, Song R, Kong Y, Zhang Z. Non-coding RNAs in depression: Promising diagnostic and therapeutic biomarkers. EBio Med. 2021;71:103569. doi: 10.1016/j.ebiom.2021.103569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Keniry A, Oxley D, Monnier P, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lechman ER, Gentner B, van Galen P, et al. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell. 2012;11:799–811. doi: 10.1016/j.stem.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hu M, Lu Y, Zeng H, et al. MicroRNA-21 maintains hematopoietic stem cell homeostasis through sustaining the NF-κB signaling pathway in mice. Haematologica. 2021;106:412–423. doi: 10.3324/haematol.2019.236927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hu W, Dooley J, Chung SS, et al. miR-29a maintains mouse hematopoietic stem cell self-renewal by regulating Dnmt3a. Blood. 2015;125:2206–2216. doi: 10.1182/blood-2014-06-585273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pohl C, Dikic I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science. 2019;366:818–822. doi: 10.1126/science.aax3769. [DOI] [PubMed] [Google Scholar]
- 105.Sinha S, Dwivedi TR, Yengkhom R, et al. Asrij/OCIAD1 suppresses CSN5-mediated p53 degradation and maintains mouse hematopoietic stem cell quiescence. Blood. 2019;133:2385–2400. doi: 10.1182/blood.2019000530. [DOI] [PubMed] [Google Scholar]
- 106.Iriuchishima H, Takubo K, Matsuoka S, et al. Ex vivo maintenance of hematopoietic stem cells by quiescence induction through Fbxw7α overexpression. Blood. 2011;117:2373–2377. doi: 10.1182/blood-2010-07-294801. [DOI] [PubMed] [Google Scholar]
- 107.Thompson BJ, Jankovic V, Gao J, et al. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J Exp Med. 2008;205:1395–1408. doi: 10.1084/jem.20080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.King B, Boccalatte F, Moran-Crusio K, et al. The ubiquitin ligase Huwe1 regulates the maintenance and lymphoid commitment of hematopoietic stem cells. Nat Immunol. 2016;17:1312–1321. doi: 10.1038/ni.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang J, Han F, Wu J, et al. The role of Skp2 in hematopoietic stem cell quiescence, pool size, and self-renewal. Blood. 2011;118:5429–5438. doi: 10.1182/blood-2010-10-312785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kleppe M, Spitzer MH, Li S, et al. Jak1 integrates cytokine sensing to regulate hematopoietic stem cell function and stress hematopoiesis. Cell Stem Cell. 2017;21:489–501. doi: 10.1016/j.stem.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Maryanovich M, Oberkovitz G, Niv H, et al. The ATM-BID pathway regulates quiescence and survival of haematopoietic stem cells. Nat Cell Biol. 2012;14:535–541. doi: 10.1038/ncb2468. [DOI] [PubMed] [Google Scholar]
- 112.Fortin J, Bassi C, Ramachandran P, et al. Concerted roles of PTEN and ATM in controlling hematopoietic stem cell fitness and dormancy. J Clin Invest. 2021;131:e131698. doi: 10.1172/JCI131698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ni F, Yu WM, Wang X, et al. Ptpn21 controls hematopoietic stem cell homeostasis and biomechanics. Cell Stem Cell. 2019;24:608–620. doi: 10.1016/j.stem.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vadlakonda L, Dash A, Pasupuleti M, Anil Kumar K, Reddanna P. The paradox of Akt-mTOR interactions. Front Oncol. 2013;3:165. doi: 10.3389/fonc.2013.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Manning BD, Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen C, Liu Y, Liu R, et al. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med. 2008;205:2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kharas MG, Okabe R, Ganis JJ, et al. Constitutively active AKT depletes hematopoietic stem cells and induces leukemia in mice. Blood. 2010;115:1406–1415. doi: 10.1182/blood-2009-06-229443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Juntilla MM, Patil VD, Calamito M, Joshi RP, Birnbaum MJ, Koretzky GA. AKT1 and AKT2 maintain hematopoietic stem cell function by regulating reactive oxygen species. Blood. 2010;115:4030–4038. doi: 10.1182/blood-2009-09-241000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hemmati S, Sinclair T, Tong M, et al. PI3 kinase alpha and delta promote hematopoietic stem cell activation. JCI Insight. 2019;5:e125832. doi: 10.1172/jci.insight.125832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yilmaz OH, Valdez R, Theisen BK, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 121.Zhang J, Grindley JC, Yin T, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- 122.Kim I, Kim YJ, Métais JY, Dunbar CE, Larochelle A. Transient silencing of PTEN in human CD34(+) cells enhances their proliferative potential and ability to engraft immunodeficient mice. Exp Hematol. 2012;40:84–91. doi: 10.1016/j.exphem.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Carling D. AMPK signalling in health and disease. Curr Opin Cell Biol. 2017;45:31–37. doi: 10.1016/j.ceb.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 124.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468:653–658. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Drosten M, Barbacid M. Targeting the MAPK pathway in KRAS-driven tumors. Cancer Cell. 2020;37:543–550. doi: 10.1016/j.ccell.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 126.Karigane D, Kobayashi H, Morikawa T, et al. p38α activates purine metabolism to initiate hematopoietic stem/progenitor cell cycling in response to stress. Cell Stem Cell. 2016;19:192–204. doi: 10.1016/j.stem.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 127.Staser K, Park SJ, Rhodes SD, et al. Normal hematopoiesis and neurofibromin-deficient myeloproliferative disease require Erk. J Clin Invest. 2013;123:329–334. doi: 10.1172/JCI66167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Baumgartner C, Toifl S, Farlik M, et al. An ERK-dependent feedback mechanism prevents hematopoietic stem cell exhaustion. Cell Stem Cell. 2018;22:879–892. doi: 10.1016/j.stem.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cabezas-Wallscheid N, Buettner F, Sommerkamp P, et al. Vitamin A-retinoic acid signaling regulates hematopoietic stem cell dormancy. Cell. 2017;169:807–823. doi: 10.1016/j.cell.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 130.Nakamura-Ishizu A, Ito K, Suda T. Hematopoietic stem cell metabolism during development and aging. Dev Cell. 2020;54:239–255. doi: 10.1016/j.devcel.2020.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rabinowitz JD, Enerbäck S. Lactate: the ugly duckling of energy metabolism. Nat Metab. 2020;2:566–571. doi: 10.1038/s42255-020-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yellen G. Fueling thought: management of glycolysis and oxidative phosphorylation in neuronal metabolism. J Cell Biol. 2018;217:2235–2246. doi: 10.1083/jcb.201803152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Moreno-Sánchez R, Rodríguez-Enríquez S, Marín-Hernández A, Saavedra E. Energy metabolism in tumor cells. FEBS J. 2007;274:1393–1418. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- 134.du Plessis SS, Agarwal A, Mohanty G, van der Linde M. Oxidative phosphorylation versus glycolysis: what fuel do spermatozoa use? Asian J Androl. 2015;17:230–235. doi: 10.4103/1008-682X.135123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kobayashi CI, Suda T. Regulation of reactive oxygen species in stem cells and cancer stem cells. J Cell Physiol. 2012;227(2):421–430. doi: 10.1002/jcp.22764. [DOI] [PubMed] [Google Scholar]
- 136.Chandel NS, Jasper H, Ho TT, Passegué E. Metabolic regulation of stem cell function in tissue homeostasis and organismal ageing. Nat Cell Biol. 2016;18:823–832. doi: 10.1038/ncb3385. [DOI] [PubMed] [Google Scholar]
- 137.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol. 2014;15:243–256. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Snoeck HW. Mitochondrial regulation of hematopoietic stem cells. Curr Opin Cell Biol. 2017;49:91–98. doi: 10.1016/j.ceb.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Klimmeck D, Hansson J, Raffel S, Vakhrushev SY, Trumpp A, Krijgsveld J. Proteomic cornerstones of hematopoietic stem cell differentiation: distinct signatures of multipotent progenitors and myeloid committed cells. Mol Cell Proteom. 2012;11:286–302. doi: 10.1074/mcp.M111.016790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang CC, Sadek HA. Hypoxia and metabolic properties of hematopoietic stem cells. Antioxid Redox Signal. 2014;20:1891–1901. doi: 10.1089/ars.2012.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Halvarsson C, Eliasson P, Jönsson JI. Pyruvate dehydrogenase kinase 1 is essential for transplantable mouse bone marrow hematopoietic stem cell and progenitor function. PLoS One. 2017;12:e0171714. doi: 10.1371/journal.pone.0171714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Takubo K, Nagamatsu G, Kobayashi CI, et al. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell. 2013;12:49–61. doi: 10.1016/j.stem.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rankin EB, Giaccia AJ. Hypoxic control of metastasis. Science. 2016;352:175–180. doi: 10.1126/science.aaf4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chow DC, Wenning LA, Miller WM, Papoutsakis ET. Modeling pO(2) distributions in the bone marrow hematopoietic compartment. I Krogh’s model Biophys J. 2001;81:675–684. doi: 10.1016/S0006-3495(01)75732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Simsek T, Kocabas F, Zheng J, et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7:380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Takubo K, Goda N, Yamada W, et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7:391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 147.Verovskaya EV, Dellorusso PV, Passegué E. Losing sense of self and surroundings: hematopoietic stem cell aging and leukemic transformation. Trends Mol Med. 2019;25:494–515. doi: 10.1016/j.molmed.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ansó E, Weinberg SE, Diebold LP, et al. The mitochondrial respiratory chain is essential for haematopoietic stem cell function. Nat Cell Biol. 2017;19:614–625. doi: 10.1038/ncb3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vannini N, Girotra M, Naveiras O, et al. Specification of haematopoietic stem cell fate via modulation of mitochondrial activity. Nat Commun. 2016;7:13125. doi: 10.1038/ncomms13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.De Almeida MJ, Luchsinger LL, Corrigan DJ, Williams LJ, Snoeck HW. Dye-independent methods reveal elevated mitochondrial mass in hematopoietic stem cells. Cell Stem Cell. 2017;21:725–729. doi: 10.1016/j.stem.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bonora M, Ito K, Morganti C, Pinton P, Ito K. Membrane-potential compensation reveals mitochondrial volume expansion during HSC commitment. Exp Hematol. 2018;68:30–37. doi: 10.1016/j.exphem.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Umemoto T, Hashimoto M, Matsumura T, Nakamura-Ishizu A, Suda T. Ca2+-mitochondria axis drives cell division in hematopoietic stem cells. J Exp Med. 2018;215:2097–2113. doi: 10.1084/jem.20180421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Maryanovich M, Zaltsman Y, Ruggiero A, et al. An MTCH2 pathway repressing mitochondria metabolism regulates haematopoietic stem cell fate. Nat Commun. 2015;6:7901. doi: 10.1038/ncomms8901. [DOI] [PubMed] [Google Scholar]
- 154.Mantel CR, O'Leary HA, Chitteti BR, et al. Enhancing hematopoietic stem cell transplantation efficacy by mitigating oxygen shock. Cell. 2015;161:1553–1565. doi: 10.1016/j.cell.2015.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rimmelé P, Liang R, Bigarella CL, et al. Mitochondrial metabolism in hematopoietic stem cells requires functional FOXO3. EMBO Rep. 2015;16:1164–1176. doi: 10.15252/embr.201439704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Katsumoto T, Yoshida N, Kitabayashi I. Roles of the histone acetyltransferase monocytic leukemia zinc finger protein in normal and malignant hematopoiesis. Cancer Sci. 2008;99:1523–1527. doi: 10.1111/j.1349-7006.2008.00865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cabal-Hierro L, van Galen P, Prado MA, et al. Chromatin accessibility promotes hematopoietic and leukemia stem cell activity. Nat Commun. 2020;11:1406. doi: 10.1038/s41467-020-15221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yu WM, Liu X, Shen J, et al. Metabolic regulation by the mitochondrial phosphatase PTPMT1 is required for hematopoietic stem cell differentiation. Cell Stem Cell. 2013;12:62–74. doi: 10.1016/j.stem.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pelletier J, Thomas G, Volarević S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer. 2018;18:51–63. doi: 10.1038/nrc.2017.104. [DOI] [PubMed] [Google Scholar]
- 160.Cai P, Mao X, Zhao J, Luo L. Ribosome biogenesis protein Urb2 regulates hematopoietic stem cells development via P53 pathway in zebrafish. Biochem Biophys Res Commun. 2018;497:776–782. doi: 10.1016/j.bbrc.2018.02.153. [DOI] [PubMed] [Google Scholar]
- 161.Le Goff S, Boussaid I, Floquet C, et al. p53 activation during ribosome biogenesis regulates normal erythroid differentiation. Blood. 2021;137:89–102. doi: 10.1182/blood.2019003439. [DOI] [PubMed] [Google Scholar]
- 162.Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schneider RK, Schenone M, Ferreira MV, et al. Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9. Nat Med. 2016;22:288–297. doi: 10.1038/nm.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Le Bouteiller M, Souilhol C, Beck-Cormier S, et al. Notchless-dependent ribosome synthesis is required for the maintenance of adult hematopoietic stem cells. J Exp Med. 2013;210:2351–2369. doi: 10.1084/jem.20122019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Schwarz DS, Blower MD. The endoplasmic reticulum: structure, function and response to cellular signaling. Cell Mol Life Sci. 2016;73:79–94. doi: 10.1007/s00018-015-2052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Guzel E, Arlier S, Guzeloglu-Kayisli O, et al. Endoplasmic reticulum stress and homeostasis in reproductive physiology and pathology. Int J Mol Sci. 2017;18:792. doi: 10.3390/ijms18040792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529:326–335. doi: 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- 168.Luo B, Lam BS, Lee SH, et al. The endoplasmic reticulum chaperone protein GRP94 is required for maintaining hematopoietic stem cell interactions with the adult bone marrow niche. PLoS One. 2011;6:e20364. doi: 10.1371/journal.pone.0020364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.van Galen P, Kreso A, Mbong N, et al. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature. 2014;510:268–272. doi: 10.1038/nature13228. [DOI] [PubMed] [Google Scholar]
- 170.Miharada K, Sigurdsson V, Karlsson S. Dppa5 improves hematopoietic stem cell activity by reducing endoplasmic reticulum stress. Cell Rep. 2014;7:1381–1392. doi: 10.1016/j.celrep.2014.04.056. [DOI] [PubMed] [Google Scholar]
- 171.Liu L, Inoki A, Fan K, et al. ER-associated degradation preserves hematopoietic stem cell quiescence and self-renewal by restricting mTOR activity. Blood. 2020;136:2975–2986. doi: 10.1182/blood.2020007975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Xu L, Liu X, Peng F, et al. Protein quality control through endoplasmic reticulum-associated degradation maintains haematopoietic stem cell identity and niche interactions. Nat Cell Biol. 2020;22:1162–1169. doi: 10.1038/s41556-020-00581-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 174.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liu F, Lee JY, Wei H, et al. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood. 2010;116:4806–4814. doi: 10.1182/blood-2010-06-288589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mortensen M, Soilleux EJ, Djordjevic G, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hashimoto M, Umemoto T, Nakamura-Ishizu A, et al. Autophagy is dispensable for the maintenance of hematopoietic stem cells in neonates. Blood Adv. 2021;5:1594–1604. doi: 10.1182/bloodadvances.2020002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Jung HE, Shim YR, Oh JE, Oh DS, Lee HK. The autophagy protein Atg5 plays a crucial role in the maintenance and reconstitution ability of hematopoietic stem cells. Immune Netw. 2019;19:e12. doi: 10.4110/in.2019.19.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ho TT, Warr MR, Adelman ER, et al. Autophagy maintains the metabolism and function of young and old stem cells. Nature. 2017;543:205–210. doi: 10.1038/nature21388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ito K, Turcotte R, Cui J, et al. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science. 2016;354:1156–1160. doi: 10.1126/science.aaf5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Murakami K, Kurotaki D, Kawase W, et al. OGT regulates hematopoietic stem cell maintenance via PINK1-dependent mitophagy. Cell Rep. 2021;34:108579. doi: 10.1016/j.celrep.2020.108579. [DOI] [PubMed] [Google Scholar]
- 182.Dong S, Wang Q, Kao YR, et al. Chaperone-mediated autophagy sustains haematopoietic stem-cell function. Nature. 2021;591:117–123. doi: 10.1038/s41586-020-03129-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Saftig P, Haas A. Turn up the lysosome. Nat Cell Biol. 2016;18:1025–1027. doi: 10.1038/ncb3409. [DOI] [PubMed] [Google Scholar]
- 184.Taya Y, Ota Y, Wilkinson AC, et al. Depleting dietary valine permits nonmyeloablative mouse hematopoietic stem cell transplantation. Science. 2016;354:1152–1155. doi: 10.1126/science.aag3145. [DOI] [PubMed] [Google Scholar]
- 185.Liu X, Zhang F, Zhang Y, et al. PPM1K regulates hematopoiesis and leukemogenesis through CDC20-mediated ubiquitination of MEIS1 and p21. Cell Rep. 2018;23:1461–1475. doi: 10.1016/j.celrep.2018.03.140. [DOI] [PubMed] [Google Scholar]
- 186.Ito K, Carracedo A, Weiss D, et al. A PML–PPAR-δ pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18:1350–1358. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yu VW, Scadden DT. Hematopoietic stem cell and its bone marrow niche. Curr Top Dev Biol. 2016;118:21–44. doi: 10.1016/bs.ctdb.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells. 1978;4:7–25. [PubMed] [Google Scholar]
- 189.Crane GM, Jeffery E, Morrison SJ. Adult haematopoietic stem cell niches. Nat Rev Immunol. 2017;17:573–590. doi: 10.1038/nri.2017.53. [DOI] [PubMed] [Google Scholar]
- 190.Gao X, Xu C, Asada N, Frenette PS. The hematopoietic stem cell niche: from embryo to adult. Development. 2018;145:dev139691. doi: 10.1242/dev.139691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, Werb Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat Commun. 2020;11:5120. doi: 10.1038/s41467-020-18794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Choi JS, Harley BA. The combined influence of substrate elasticity and ligand density on the viability and biophysical properties of hematopoietic stem and progenitor cells. Biomaterials. 2012;33:4460–4468. doi: 10.1016/j.biomaterials.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 194.Li D, Chiu G, Lipe B, et al. Decellularized Wharton jelly matrix: a biomimetic scaffold for ex vivo hematopoietic stem cell culture. Blood Adv. 2019;3:1011–1026. doi: 10.1182/bloodadvances.2018019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Choi JS, Harley BA. Marrow-inspired matrix cues rapidly affect early fate decisions of hematopoietic stem and progenitor cells. Sci Adv. 2017;3:e1600455. doi: 10.1126/sciadv.1600455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Saw S, Weiss A, Khokha R, Waterhouse PD. Metalloproteases: on the watch in the hematopoietic niche. Trends Immunol. 2019;40:1053–1070. doi: 10.1016/j.it.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 197.Gvaramia D, Müller E, Müller K, et al. Combined influence of biophysical and biochemical cues on maintenance and proliferation of hematopoietic stem cells. Biomaterials. 2017;138:108–117. doi: 10.1016/j.biomaterials.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 198.Ikeda K, Ueda T, Yamasaki N, et al. Maintenance of the functional integrity of mouse hematopoiesis by EED and promotion of leukemogenesis by EED haploinsufficiency. Sci Rep. 2016;6:29454. doi: 10.1038/srep29454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bruns I, Czibere A, Fischer JC, et al. The hematopoietic stem cell in chronic phase CML is characterized by a transcriptional profile resembling normal myeloid progenitor cells and reflecting loss of quiescence. Leukemia. 2009;23:892–899. doi: 10.1038/leu.2008.392. [DOI] [PubMed] [Google Scholar]
- 200.Rossi L, Ergen AV, Goodell MA. TIMP-1 deficiency subverts cell-cycle dynamics in murine long-term HSCs. Blood. 2011;117:6479–6488. doi: 10.1182/blood-2009-10-248955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Uckelmann H, Blaszkiewicz S, Nicolae C, et al. Extracellular matrix protein Matrilin-4 regulates stress-induced HSC proliferation via CXCR4. J Exp Med. 2016;213:1961–1971. doi: 10.1084/jem.20151713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mitroulis I, Chen LS, Singh RP, et al. Secreted protein Del-1 regulates myelopoiesis in the hematopoietic stem cell niche. J Clin Invest. 2017;127:3624–3639. doi: 10.1172/JCI92571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Avigdor A, Goichberg P, Shivtiel S, et al. CD44 and hyaluronic acid cooperate with SDF-1 in the trafficking of human CD34+ stem/progenitor cells to bone marrow. Blood. 2004;103:2981–2989. doi: 10.1182/blood-2003-10-3611. [DOI] [PubMed] [Google Scholar]
- 204.Khurana S, Schouteden S, Manesia JK, et al. Outside-in integrin signalling regulates haematopoietic stem cell function via Periostin-Itgav axis. Nat Commun. 2016;7:13500. doi: 10.1038/ncomms13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Arai F, Hosokawa K, Toyama H, Matsumoto Y, Suda T. Role of N-cadherin in the regulation of hematopoietic stem cells in the bone marrow niche. Ann N Y Acad Sci. 2012;1266:72–77. doi: 10.1111/j.1749-6632.2012.06576.x. [DOI] [PubMed] [Google Scholar]
- 206.Winkler IG, Barbier V, Nowlan B, et al. Vascular niche E-selectin regulates hematopoietic stem cell dormancy, self renewal and chemoresistance. Nat Med. 2012;18:1651–1657. doi: 10.1038/nm.2969. [DOI] [PubMed] [Google Scholar]
- 207.Sinclair A, Park L, Shah M, et al. CXCR2 and CXCL4 regulate survival and self-renewal of hematopoietic stem/progenitor cells. Blood. 2016;128:371–383. doi: 10.1182/blood-2015-08-661785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Asada N, Kunisaki Y, Pierce H, et al. Differential cytokine contributions of perivascular haematopoietic stem cell niches. Nat Cell Biol. 2017;19:214–223. doi: 10.1038/ncb3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Nie Y, Han YC, Zou YR. CXCR4 is required for the quiescence of primitive hematopoietic cells. J Exp Med. 2008;205:777–783. doi: 10.1084/jem.20072513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bruns I, Lucas D, Pinho S, et al. Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nat Med. 2014;20:1315–1320. doi: 10.1038/nm.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zhao M, Perry JM, Marshall H, et al. Megakaryocytes maintain homeostatic quiescence and promote post-injury regeneration of hematopoietic stem cells. Nat Med. 2014;20:1321–1326. doi: 10.1038/nm.3706. [DOI] [PubMed] [Google Scholar]
- 212.Yamazaki S, Ema H, Karlsson G, et al. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell. 2011;147:1146–1158. doi: 10.1016/j.cell.2011.09.053. [DOI] [PubMed] [Google Scholar]
- 213.Qian H, Buza-Vidas N, Hyland CD, et al. Critical role of thrombopoietin in maintaining adult quiescent hematopoietic stem cells. Cell Stem Cell. 2007;1:671–684. doi: 10.1016/j.stem.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 214.Yoshihara H, Arai F, Hosokawa K, et al. Thrombopoietin/MPL signaling regulates hematopoietic stem cell quiescence and interaction with the osteoblastic niche. Cell Stem Cell. 2007;1:685–697. doi: 10.1016/j.stem.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 215.Nilsson SK, Johnston HM, Whitty GA, et al. Osteopontin, a key component of the hematopoietic stem cell niche and regulator of primitive hematopoietic progenitor cells. Blood. 2005;106:1232–1239. doi: 10.1182/blood-2004-11-4422. [DOI] [PubMed] [Google Scholar]
- 216.Arai F, Hirao A, Ohmura M, et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 217.Ludin A, Itkin T, Gur-Cohen S, et al. Monocytes-macrophages that express α-smooth muscle actin preserve primitive hematopoietic cells in the bone marrow. Nat Immunol. 2012;13:1072–1082. doi: 10.1038/ni.2408. [DOI] [PubMed] [Google Scholar]
- 218.Goncalves KA, Silberstein L, Li S, et al. Angiogenin promotes hematopoietic regeneration by dichotomously regulating quiescence of stem and progenitor cells. Cell. 2016;166:894–906. doi: 10.1016/j.cell.2016.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Chen X, Deng H, Churchill MJ, et al. Bone marrow myeloid cells regulate myeloid-biased hematopoietic stem cells via a histamine-dependent feedback loop. Cell Stem Cell. 2017;21:747–760. doi: 10.1016/j.stem.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Goloviznina NA, Verghese SC, Yoon YM, Taratula O, Marks DL, Kurre P. Mesenchymal stromal cell-derived extracellular vesicles promote myeloid-biased multipotent hematopoietic progenitor expansion via Toll-like receptor engagement. J Biol Chem. 2017;292:3541. doi: 10.1074/jbc.A116.745653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kfoury YS, Ji F, Mazzola M, et al. tiRNA signaling via stress-regulated vesicle transfer in the hematopoietic niche. Cell Stem Cell. 2021;28:2090–2103.e9. doi: 10.1016/j.stem.2021.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Nitsche A, Junghahn I, Thulke S, et al. Interleukin-3 promotes proliferation and differentiation of human hematopoietic stem cells but reduces their repopulation potential in NOD/SCID mice. Stem Cells. 2003;21:236–244. doi: 10.1634/stemcells.21-2-236. [DOI] [PubMed] [Google Scholar]
- 223.Ye H, Qian L, Zhu S, et al. IL-1Ra protects hematopoietic cells from chemotoxicity through p53-induced quiescence. FASEB J. 2019;33:12135–12145. doi: 10.1096/fj.201900788RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Florez MA, Matatall KA, Jeong Y, et al. Interferon gamma mediates hematopoietic stem cell activation and niche relocalization through BST2. Cell Rep. 2020;33:108530. doi: 10.1016/j.celrep.2020.108530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Masamoto Y, Arai S, Sato T, et al. Adiponectin enhances quiescence exit of murine hematopoietic stem cells and hematopoietic recovery through mTORC1 potentiation. Stem Cells. 2017;35:1835–1848. doi: 10.1002/stem.2640. [DOI] [PubMed] [Google Scholar]
- 226.Itkin T, Ludin A, Gradus B, et al. FGF-2 expands murine hematopoietic stem and progenitor cells via proliferation of stromal cells, c-Kit activation, and CXCL12 down-regulation. Blood. 2012;120:1843–1855. doi: 10.1182/blood-2011-11-394692. [DOI] [PubMed] [Google Scholar]
- 227.Frodermann V, Rohde D, Courties G, et al. Exercise reduces inflammatory cell production and cardiovascular inflammation via instruction of hematopoietic progenitor cells. Nat Med. 2019;25:1761–1771. doi: 10.1038/s41591-019-0633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Young K, Eudy E, Bell R, et al. Decline in IGF1 in the bone marrow microenvironment initiates hematopoietic stem cell aging. Cell Stem Cell. 2021;28:1473–1482.e7. doi: 10.1016/j.stem.2021.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Méndez-Ferrer S, Michurina TV, Ferraro F, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.