Abstract
The task of transcribing nuclear genes is shared between three RNA polymerases in eukaryotes: RNA polymerase (pol) I synthesises the large rRNA, pol II synthesises mRNA and pol III synthesises tRNA and 5S rRNA. Although pol II has received most attention, pol I and pol III are together responsible for the bulk of transcriptional activity. This survey will summarise what is known about the process of transcription by pol I and pol III, how it happens and the proteins involved. Attention will be drawn to the similarities between the three nuclear RNA polymerase systems and also to their differences.
INTRODUCTION
The eukaryotic RNA polymerases I and III (pol I and pol III) transcribe only a limited set of genes. Pol I synthesises rRNA and pol III makes 5S rRNA, tRNA, 7SL RNA, U6 snRNA and a few other small stable RNAs, many involved in RNA processing. This contrasts with the huge variety of pol II-transcribed genes that encode proteins. The activities of pol I and pol III dominate cellular transcription, combining to exceed 80% of total RNA synthesis in growing cells! Therefore, tight regulation must be paramount if one considers the energy load on the cell. This review presents an overview of the factors and mechanisms of transcription by pol I and pol III, affectionately known as the ‘Oddpols’. Although the transcription factors are different, there are fundamental similarities in the mechanisms used by the two enzymes. In each case, a single factor binds at a similar position upstream of the transcription start site; it repetitively recruits the appropriate polymerase by protein–protein interaction, positioning it precisely to initiate transcription at a unique site.
For more comprehensive treatments, the reader is referred to two recent monographs by the authors covering transcription by pol I (1) and pol III (2).
LOCALISATION
It has long been known that pol I transcription is localised to discrete sites called nucleoli; these can be likened to ribosome factories, in which rRNA is synthesised by pol I in the fibrillar centres and then processed and assembled into ribosomes in the surrounding granular regions (3). This has always been regarded as a peculiarity of the pol I system, but a recent study has shown that pol II and pol III both also carry out transcription at discrete locations (4). Confocal and electron microscopy of HeLa cells revealed that pol III transcription occurs at ~2000 sites within the nucleoplasm (4). Each site has a radius of ~20 nm and contains, on average, five molecules of active pol III (4). Pol II is not found at these sites, but functions at its own spatially separate locations, of which there may be ~8000 per HeLa cell (4).
Although transcripts made by pol I and pol II are thought to be processed close to the site of their synthesis, at least part of the tRNA processing pathway may occur in the nucleolus (5). Thus, fluorescent in situ hybridisation reveals unprocessed pre-tRNAs within the nucleoli of Saccharomyces cerevisiae (5). Furthermore, the RNA subunit of RNase P, a pol III product which helps cleave pre-tRNA, can be found in both the nucleolus and the nucleoplasm in yeast and mammals (5,6). Processing of tRNA within the same compartment as ribosome synthesis may provide opportunies for coordinating production of distinct components of the translational apparatus.
SHARED POLYMERASE SUBUNITS
The eukaryotic nuclear RNA polymerases are complex enzymes, made up of 12 or more subunits (7). Five of these are gene products shared by all three enzymes (ABC10α, ABC10β, ABC14.5, ABC23 and ABC27 in S.cerevisiae). In addition, pol I and pol III share two subunits (AC19 and AC40 in S.cerevisiae) (8) that are not found in pol II, although the B12.5 and B44 pol II subunits are functionally equivalent, respectively (9–11). The presence of common subunits would seem to offer the opportunity for coordinate regulation. Indeed, the AC40 homologue is believed to be a target for regulation of rRNA synthesis in one instance (12), but the details of the mechanism have proven elusive. However, although this subunit is modified in both pol I and pol III, 5S rRNA expression is primarily controlled in this case through TFIIIA, not pol III (13).
PROMOTERS
Pol I promoters
Ribosomal RNA promoters have been extensively reviewed (14–17), so we will present only an outline here. Whereas the transcription machinery of pol II and pol III is often compatible with genes from widely different species, pol I transcription exhibits stringent (18), though not absolute (19), species specificity. This is housed in the interaction of the transcription factors with the promoter and, to a lesser extent, in the protein–protein interactions between the factors (20–24). There is very little sequence similarity between rRNA promoters from different species, but the general layout of functional promoter elements is highly conserved from yeast to humans (14–17). It even extends as far as African trypanosomes, in which both rRNA and two protein genes are expressed by pol I. This is the only known case in which pol I transcribes genes other than rRNA (reviewed in 25), though I-PpoI, an endonuclease encoded in a rRNA group I intron from Physarum polycephalum, is also expressed by pol I, but not from a dedicated promoter (26).
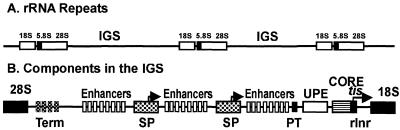
Multiple copies of rRNA genes are found as repeated clusters, usually arranged head-to-tail. Almost the entire promoter is housed in the intergenic spacer (IGS) between the transcribed units (Fig. 1). The ~50 bp region upstream of the initiation site (+1) is the core promoter and is necessary and sufficient for initiation of basal transcription in most species (14–17). The core promoter contains the only conserved rRNA promoter sequence element, the ribosomal initiator or rInr (27). The rInr is an AT-rich, TATA-like sequence surrounding +1. This element alone can very inefficiently mediate correct initiation and can be photocrosslinked to one of the factor subunits in the amoeba Acanthamoeba castellanii, suggesting that it functions similarly to pol II Inrs (28). Though its sequence resembles the TATA box, it is not an interaction site for TATA-binding protein (TBP). The remainder of the core promoter serves to bind the core factor which recruits pol I.
Figure 1.
Generic organisation of pol I transcription units. (A) The rRNA coding units are separated by intergenic spacers (IGS). (B) The IGS contains a series of terminators (term), enhancers, a spacer promoter (SP), a proximal terminator (PT), the upstream promoter element (UPE) and the promoter core, which includes the rInr. The sites of transcription initiation are indicated by tis and/or the bent arrows.
In most species, additional elements and factors help to assemble or stabilise the complex formed on the core promoter. One key player is the upstream promoter element (UPE) that extends 150–200 bp upstream of +1 (14–17). The UPE significantly stimulates transcription driven by the core promoter, but in most instances is not absolutely required for transcription initiation (29,30). The UPE binds an assembly factor that mediates efficient recruitment of the core factor to the promoter.
Non-essential DNA separates the core promoter and the UPE. However, the spacing and helical relationship between the elements is critical. Promoters with half-helical turn insertions or deletions (±5 or ±15 bp) are more severely compromised than those that retain the normal helical alignment (19,31); this likely reflects the interaction between proteins bound at the two elements. Curiously, half-helical spacing alterations can change species specificity of the promoter (19).
There is a transcription terminator just upstream of the UPE, the proximal terminator (PT), that serves many functions, including protection of the promoter from wandering polymerases (32,33), remodelling of chromatin over the promoter (34) and, possibly, architectural folding of the rRNA repeats in the nucleolus (35,36). The PT stimulates transcription significantly. In S.cerevisiae, the PT is in the wrong orientation to mediate termination. Therefore, it cannot protect the promoter, but it still binds the termination factor and stimulates transcription.
The remainder of the IGS contains elements that enhance transcription (37–39). Usually enhancers are repeated in the IGS, but only a single enhancer copy is found in S.cerevisiae (37,40,41). In many metazoans, the IGS also contains additional functional promoters called spacer promoters or SPs (42–47). SP transcripts, which have no known function, terminate at the PT (42). Phylogenetic comparisons and experimental data suggest that enhancers evolved from the spacer promoters by repetitive duplication and truncation (48).
Pol III promoters
The most striking and unusual feature of the promoters used by pol III is that the majority require sequence elements downstream of +1, within the transcribed region. These internal control regions are generally discontinuous structures composed of essential blocks separated by non-essential regions. A classic example is provided by the Xenopus laevis somatic 5S rRNA gene, which requires three internal elements for efficient transcription: an A block located between +50 and +64, an intermediate element at +67 to +72 and a C block from +80 to +97 (49) (Fig. 2). The promoter is relatively intolerant of changes in the spacing between individual elements (49). This type of internal control region is also found in the 5S rRNA genes of many lower organisms, including Drosophila melanogaster (50) and S.cerevisiae (51). It is unique to 5S rRNA genes and is known as the type I promoter.
Figure 2.
Organisation of the three general types of promoter used by pol III. The site of transcription initiation is indicated by +1 and the site of termination is indicated by Tn. Also shown are the positions of various promoter elements, including the intermediate element (IE), proximal sequence element (PSE) and distal sequence element (DSE).
The most common promoter arrangement used by pol III is found in the tRNA genes, the adenovirus VA genes and many middle repetitive gene families. It is called a type II promoter and consists of two highly conserved sequence blocks, A and B, within the transcribed region (Fig. 2). The A blocks of type I and II promoters are homologous and sometimes interchangeable (52), but in the latter case they are much closer to +1. For example, a Xenopus tRNALeu gene has its A block between +11 and +21, ~40 bp further upstream than the A block of the Xenopus 5S rRNA genes (53). The location of the B block is extremely variable, partly because some tRNA genes have short introns within their coding regions. Interblock separations of ~30–60 bp are optimal for transcription, although a distance of 365 bp can be tolerated (54,55). This flexibility is remarkable, because the A and B blocks are bound simultaneously by the single factor TFIIIC (56).
A minority of vertebrate pol III templates lack any requirement for intragenic promoter elements; these are referred to as type III promoters. For example, human and mouse U6 snRNA promoters have been identified that retain full activity following deletion of all sequences downstream of +1 (57–59). A similar situation is found for human 7SK and MRP/7-2 RNA genes (60,61). Although this arrangement is radically different from most pol III templates, it is, of course, the norm for genes transcribed by pol I and pol II, as well as bacterial genes. It is peculiar that an extragenic promoter organisation appears to have evolved relatively recently within the pol III system. In yeast, U6 genes have functional A and B blocks, albeit in unusual positions (62). A U6 gene with an entirely internal promoter has even been found in humans (63).
The best characterised type III promoter belongs to a human U6 gene (Fig. 2). The sequences required for efficient expression are a TATA box, between –30 and –25, a proximal sequence element (PSE) between –66 and –47 and a distal sequence element (DSE) between –244 and –214 (57–59,64–66). The U6 PSE and DSE are homologous and interchangeable with elements found at comparable positions in the U2 snRNA gene transcribed by pol II (59,64–66). However, a TATA box is not found in the U2 promoter; this is a curious anomaly, since TATA sequences are a classic feature of class II rather than class III genes. Even more paradoxical is the observation that inserting a TATA box can convert U2 into a pol III promoter, whereas crippling its TATA box allows U6 to be transcribed by pol II (59,67). Clearly, the U snRNA genes are a law unto themselves.
Several of the class III genes do not fall neatly into any of the three promoter types illustrated in Figure 2. For example, the EBER2 gene of Epstein–Barr virus has A and B blocks that are typical of type II promoters and are essential for transcription (68). However, deletion of sequences upstream of –46 reduces expression in transfected cells to 7% of the wild-type level (68). Upstream binding sites for Sp1 and ATF are thought to be responsible for this effect (68). The EBER2 promoter also has a TATA box between –28 and –23 that increases its activity 5-fold (68). Other examples of pol III promoters that rely on both internal and upstream sequences for efficient expression include silkworm tRNAAla genes (69), the Xenopus tRNASec gene (70), the rat vault RNA gene (71) and the human 7SL gene (72).
TRANSCRIPTION FACTORS AND ASSEMBLY OF THE INITIATION COMPLEX
Transcription complex assembly on pol I promoters
Compared to pol II, pol I requires fewer transcription factors for faithful initiation. In most systems only pol I (possibly with one or two tightly associated factors) plus a single core promoter-binding factor are required and sufficient. Additional factors augment transcription in vitro and may be necessary in vivo to yield the high levels of rRNA synthesis needed for cellular growth. Functionally related factors are grouped in Table 1. Note, however, that this grouping does not reflect true genetic homology, as the sequences of these factors from distantly related species show no similarities.
Table 1. Transcription factors associated with rRNA transcription.
| Core-binding factors |
| TIF-IB (mouse, A.castellanii) |
| SL1 (human, rat) |
| Rib1 (Xenopus) |
| CF (yeast) |
| Termination factors |
| TTF-I (mouse, human) |
| Reb1p (S.cerevisiae) |
| UPE-binding factors |
| UBF (human, mouse, rat, Xenopus) |
| UAF (S.cerevisiae) |
| Enhancer-binding factors |
| UBF (human, mouse, rat, Xenopus) |
| Reb1p (S.cerevisiae) |
| EBF (A.castellanii) |
| E1BF (rat) |
Formation of the committed complex. Ribosomal RNA genes are primed for transcription by binding a single factor to the promoter (30,73–76). This is mediated in most organisms by the core promoter, but the UPE, enhancer elements and possibly the proximal terminator facilitate this step. In some species, the UPE and its binding factor are nearly mandatory for assembling a ‘committed’ complex that is resistant to challenge by a second template and persists through multiple rounds of transcription (77–80). Indeed, the yeast UPE and its binding factor alone commit the template without the core-binding factor (81). About half the gene copies garner the appropriate factor(s) for activation (82).
Core promoter-binding factors. The core factor is the fundamental initiation factor for rRNA transcription that is necessary and sufficient to recruit pol I. In all species studied except S.cerevisiae, this factor consists of TBP plus three or four additional subunits known as pol I-specific TBP-associated factors, TAFIs (83–89). In S.cerevisiae, TBP interacts functionally but not stably with the core factor (90,91). The presence of TBP puts these factors into the same class as TFIID and TFIIIB in the pol II and pol III systems, respectively. These factors recruit RNA polymerase and remain bound to the promoter. Recruitment occurs directly in the cases of pol I and pol III and indirectly via additional factors in the case of pol II. Thus, these TBP-containing factors are functionally related.
The core factors from human, mouse and yeast contain three TAFIs, of roughly 50, 70 and 100 kDa (88–91). While the human and mouse TAFIs of similar size show strong sequence similarities, the yeast TAFI sequences bear no similarity to those of mammals. In contrast, A.castellanii TIF-IB consists of four TAFIs plus TBP (87) and these have sizes quite disparate from the above (145, 99, 96 and 91 kDa). Nevertheless, the A.castellanii TAFI96 sequence is related to the 56 kDa yeast TAFI (Rrn7p) (C.Radebaugh and M.Paule, unpublished results). All three mammalian TAFIs interact with each other and with TBP. Once assembly of TAFIs onto TBP has started, pol II-specific TAFIIs are excluded, preventing assembly of a mix of pol I and pol II TAFs (89). Yeast TBP can substitute for human TBP in vivo (92), suggesting that the non-conserved N-terminal domain of TBP is not required by this factor. This contrasts with a significant role of the N-terminal 96 amino acids of TBP in transcription of U6 genes by pol III (93). The yeast TAFIs, (Rrn6p, Rrn7p and Rrn11p) also mutually interact, but only one subunit, Rrn6p, associates significantly with TBP (94), probably explaining the facile loss of TBP from yeast core factor during purification (94).
Whereas transcription by pol II and pol III is inhibited strongly by oligonucleotides containing TATA box sequences, pol I is more resistant (86). This suggests that the DNA-binding domain of TBP is not used for rRNA promoter binding. Indeed, human TAFI48 has been shown to block the DNA-binding site of TBP (95). Whereas TFIID and TFIIIB induce severe bends in promoter DNA, Acanthamoeba TIF-IB only bends DNA by ~45° (96,97). In yeast, TBP is not required at all for basal pol I transcription in vitro (73). Thus the use of TBP in the pol I system is fundamentally different from the way it is used by pol II and pol III.
Extensive structural studies of the unusually stable committed complex of A.castellanii have shown that TIF-IB binds mainly the minor groove, contacting four successive minor grooves as the factor and DNA wrap around each other in a high pitched, right-handed superhelix (98). The most energetically significant interactions are with an AT-rich element with a minor groove compressed to half the normal width. Point mutations that alter this width result in significant loss of binding (G.Geiss, P.Kownin, M.Marilley and M.Paule, unpublished results), suggesting that recognition is primarily via backbone interactions rather than by hydrogen bonding to the bases. In addition, TAFI96 interacts with the conserved rInr; this interaction alone is able to support specific initiation, albeit very weakly (28). This may provide insight into the role of the yeast TAFI96 homologue Rrn7p.
UPE-binding factors. The UPE binds a transcription factor that aids core factor assembly onto the promoter. The need for the UPE and its cognate factor depends on the species; we see a spectrum of requirement for the committed complex, with A.castellanii at one end, yeast, rat and mouse in the middle and humans and Xenopus at the other. Human and Xenopus demonstrate an extreme reliance on UBF (79,80), but the others do not (29,30,98–100). In most cases, core factor alone can mediate basal transcription (see, however, Accessory Factors below).
The vertebrate UPE-binding factor is a dimer of a 90–100 kDa polypeptide called upstream binding factor (UBF) (101–105). UBF has an N-terminal dimerisation domain (106–108), followed by four to six HMG box DNA-binding domains, depending upon species, and a C-terminal acidic serine-rich tail (102,105). NMR solution structures of HMG boxes from other factors have shown that they bind to the minor groove of DNA and bend it, with the protein lying on the outside of the bend (109). Based on extensive footprinting studies (110), Moss proposed that a UBF dimer, with its 8–12 HMG boxes, could wrap 135–160 bp of DNA into a left-handed loop, a structure dubbed the ‘enhancesome’. Various studies (111,112) have confirmed this structure. Moss also proposed that the core and UPE promoter elements could be brought together on the surface of two adjacent enhancesomes. This might aid binding of core factor by allowing simultaneous interaction with both promoter elements (113). The requirement for correct helical alignment of the two elements supports this notion (31,114). UBF also makes direct contact with the core factor, aiding the latter’s recruitment to the promoter (79,101,115,116). In humans, contact is between TAFI48 and the C-terminal tail of UBF (95) and requires UBF phosphorylation (117,118). UBF also binds the multicopy ribosomal enhancers, suggesting that a series of enhancesomes could compress the DNA of the IGS.
In S.cerevisiae, the UPE is bound by upstream activating factor (UAF), which is unrelated to UBF. It contains six subunits (Rrn5p, 41.8 kDa; Rrn9p, 42.8 kDa; Rrn10p, 16.5 kDa; histone H3; p30, an unidentified 30 kDa protein; histone H4, in sub-stoichiometric amounts) (119,120). The presence of the two core histones implies that it may form a looped DNA similar to the enhancesome, but this has not yet been tested. Just as UBF is stimulatory but dispensable in vitro, UAF genes can be knocked out without loss of yeast viability; UAF– cells grow very slowly, a phenotype which is suppressed when rRNA is transcribed from a plasmid with a pol II promoter. Curiously, UAF– yeast transcribe their chromosomal rDNA using pol II instead of pol I, allowing more rapid growth (121,122). This trait is epigenetic. Similar polymerase switching has been observed when yeast rRNA genes are put under stress (123), and occurs in other organisms (124–126). It has been suggested that such switching plays a role in cellular ageing (127,128).
Role of enhancers. Ribosomal RNA enhancers stimulate transcription, but their precise mechanism is uncertain. Vertebrate repetitive enhancers increase formation of the committed complex and not later events (38,129–132); they can be cut away from the promoter following committed complex formation and stimulation continues unabated (131). Furthermore, the Xenopus enhancer can stimulate transcription when not truly in cis to the promoter. Dunaway and Dröge (133) concatenated enhancer- and promoter-containing DNA minicircles and injected the assembly into Xenopus oocytes; concatenation was rapidly lost following injection, but enhanced transcription persisted for at least 1 day. It has been suggested that enhancers act as loading agents for the promoter by supplying some limiting component. The best candidate in vertebrates is UBF, which binds the repetitive enhancers (47). Consistent with this notion is the kinetically rapid exchange of enhancer-bound and free UBF, which would maintain an elevated UBF concentration localised near the promoter (134). Also supporting this mechanism is the observation that when not in close proximity to the promoter, or when in high concentration in trans, enhancers inhibit transcription by competing for UBF (129,131). However, in vitro studies of the Xenopus enhancer have shown that they operate by a mechanism independent of UBF delivery; transcription can be enhanced by forms of UBF that are able to bind enhancers but are incapable of functioning at the promoter, such as UBF from other species or UBF2, a natually truncated form missing part of HMG box 2 (116). The authors suggested that enhancer-bound UBF recruits core factor by protein–protein interaction. It has also been suggested that formation of enhancesomes by UBF could compress the distance between the termination site of one repeat and the promoter of the next, thus stimulating the passage of pol I between repeats. However, this mechanism requires that enhancers operate in cis and are continuously present, neither of which is necessary in oocytes (133).
Spacer promoters also stimulate transcription when there are enhancers situated between them and the PT (135–137). Transcription from the spacer promoter is not required for this stimulation (138).
The single copy S.cerevisiae enhancer lies in its IGS, ~100 bp downstream of the transcription unit and overlapping the terminator element (40,41). It can act bi-directionally, in either orientation and at great distances from the gene promoter (139,140). It is argued that the yeast enhancer modulates the frequency of reinitiation, rather than altering the efficiency of gene activation as in vertebrates (139), though in vitro it stimulates promoter complex formation and not pol I loading (38). It binds the termination factor Reb1p (141) and binds Abf1p weakly (142). Mutation of the Abf1p site strongly inhibits rRNA transcription of single copy genes on a plasmid, but has little effect on genes integrated into the URA3 locus (143). In contrast, when tandem copies of rRNA test genes were inserted into the rDNA repeat, deletion of the Reb1p site led to a reduction in transcription. When both the ‘enhancer’ Reb1p site and the Reb1p site in the proximal terminator were deleted, there was a 90% drop in rRNA expression (144). It has been proposed that the single copy yeast enhancer, in concert with the PT, functions by looping the IGS to bring together the termination region of one gene copy with the promoter of another (140,144).
Recruitment of pol I. Recruitment of pol I is mediated by protein–protein interaction with core factor. The polymerase can contact any DNA sequence and, in the absence of the core factor, pol I initiates only randomly. When core factor is present, pol I binds specifically to the promoter and non-specific initiation is suppressed. Provided core factor is bound to its site, the DNA footprinted by pol I can be changed to any random sequence without affecting recruitment, but sequence alterations that disrupt core factor binding eliminate pol I recruitment (145). Site-specific DNA–protein photocrosslinking suggests that the A.castellanii TAFI96 is in a position to make contact with pol I (28). Addition of polymerase increases the proximity of this TAFI to the DNA, consistent with an interaction between polymerase and this TAFI (28).
Transcription complex assembly on pol III promoters
Type I and II promoters. The A and B block sequences found in most pol III promoters are recognised by a multisubunit complex called TFIIIC. This is one of the largest and most complex transcription factors known. In S.cerevisiae, TFIIIC consists of two globular domains, each of ~10 nm in diameter and ~300 kDa (56). It is composed of six subunits (Table 2), none of which seems able to bind specifically to DNA on its own. Photocrosslinking experiments have shown that the various subunits of TFIIIC extend across the entire length of a tRNATyr gene (146,147). Both the A and B blocks are contacted, although the latter is the major determinant of binding affinity (148). It is remarkable that these two promoter elements are recognised simultaneously by a single factor, since their separations can vary substantially among different genes. Furthermore, the relative helical orientation of the A and B blocks is not important for transcription efficiency (54). Electron microscopic analysis suggests that a linker region between the two domains of TFIIIC can stretch, giving the protein the appearance of a dumb-bell when bound to large tRNA genes (56). However, on promoters with very long interblock separations the ability to stretch is exceeded and the intervening DNA is looped out (56). This capacity of TFIIIC to accommodate such a diversity of promoter sizes constitutes an extraordinary feat of protein flexibility.
Table 2. TFIIIC subunits in S.cerevisiae.
| Subunit | Gene | Reference | Remarks |
|---|---|---|---|
| τ138 | TFC3 | (346) | Two regions of weak HMG homology |
| τ131 | TFC4 | (347) | Multiple TPR repeats; HLH homology |
| τ95 | TFC1 | (348) | HTH homology |
| τ91 | TFC6 | (349) | No significant homologies |
| τ60 | TFC8 | (351) | No significant homologies |
| τ55 | TFC7 | (350) | Chimeric protein generated by chromosomal rearrangement |
Human TFIIIC seems rather different from the yeast factor. It can be resolved by ion exchange chromatography into two components, called TFIIIC1 and TFIIIC2 (149–151). Whereas both components are required for expression of 5S rRNA, VAI and tRNA genes, U6 and 7SK transcription requires TFIIIC1 but not TFIIIC2 (151–153). The initial recognition of type II promoters is achieved by TFIIIC2, which binds to the B block and then serves to recruit TFIIIC1 and TFIIIB (154). The function of TFIIIC1 is unclear, although it enhances and extends the footprint produced by TFIIIC2 (149–151). Sedimentation analy-sis suggested that TFIIIC1 has a mass of up to 200 kDa (149), assuming it is globular, but little further progress has been made towards its characterisation. Human TFIIIC2 has been purified and shown to consist of five polypeptides, of 220, 110, 102, 90 and 63 kDa, with a cumulative mass approaching 600 kDa (150,155,156). TFIIIC2 can also be isolated in a transcriptionally inactive form that lacks the hTFIIIC110 subunit but still binds DNA (156). Cloning of cDNAs has been reported for all of the subunits (152,156–158,158a). Three of them, hTFIIIC220, hTFIIIC110 and hTFIIIC90, display no significant homology to S.cerevisiae TFIIIC. This is very surprising, because the A and B blocks are well conserved between mammals and yeast and crosslinking shows that hTFIIIC220 is responsible for contacting these promoter elements in humans (149). hTFIIIC63 shows only 22% homology to the τ95 subunit of yeast TFIIIC (158). However, hTFIIIC102 is 31% identical to τ131 and these subunits both contain 11 copies of a tetratricopeptide repeat (158).
Productive recruitment of TFIIIC to 5S rRNA promoters requires the presence of the gene-specific factor TFIIIA. Xenopus laevis TFIIIA was the first eukaryotic transcription factor to be purified to homogeneity (159) and the first to have its cDNA cloned (160). It was also the founding member of the zinc finger family, since most of its 344 residues are taken up by nine tandem, zinc-dependent DNA-binding domains (161). NMR and crystal structures have been published recently, showing three and six TFIIIA fingers, respectively, bound to fragments of a 5S rRNA gene (162,163). The C block is recognised by the N-terminal three fingers (fingers 1–3), which contribute ~95% of the total binding energy of full-length TFIIIA; these fingers wrap smoothly around the major groove in a manner that is typical of this class of DNA-binding domain (162–164). Fingers 7–9 are thought to contact the A block in a similar fashion but with lower affinity (164). The middle three fingers adopt a completely different configuration in order to span the interblock DNA, which is twice as long as the regions bound by fingers 1–3 or 7–9. They run along one side of the duplex in an open, extended structure (163). Of these, only finger 5 makes base contacts in the major groove, at the intermediate element, whereas fingers 4 and 6 straddle the neighbouring minor grooves and function primarily as spacer elements (163).
TFIIIA also has nine zinc fingers in S.cerevisiae (165). However, the spacings are very different from X.laevis TFIIIA and overall the two proteins are only ~20% identical, with most of the shared residues occurring in the finger motifs. Even amongst frog species there is considerable divergence, with TFIIIA from X.laevis showing only 84% identity to the Xenopus borealis protein and 63% identity to the Rana catesbeiana protein. This rapid evolution of TFIIIA, which is dedicated to the metabolism of 5S rRNA (166), is consistent with the lack of conservation of the pol I factors that are responsible for producing the other rRNA species.
TFIIIA serves as an adaptor, providing a platform that allows TFIIIC to be recruited onto 5S rRNA genes for which it has little affinity. Photocrosslinking experiments have shown that the locations of the various TFIIIC subunits, relative to each other and to the initiation site, are very similar on a yeast 5S rRNA gene to those seen on a tRNA gene (147). This is remarkable, given the dissimilarity in promoter structures and the additional presence of TFIIIA at the 5S rRNA gene. It is also notable that although tRNA and 5S rRNA genes both employ an A block, it is bound by TFIIIC in the former case but by TFIIIA in the latter.
Regardless of the promoter type, the primary function of TFIIIC is to recruit TFIIIB. This process has been characterised extensively in yeast, where S.cerevisiae TFIIIB has been reconstituted entirely from recombinant components (167–169). It is a complex of three polypeptides, one of which is TBP (170). The largest of the TBP-associated factors is a 90 kDa polypeptide called B′′, which displays little homology to other known proteins except for a putative SANT domain (167–169,171). This subunit is unusually resistant to truncation and will continue to support U6 transcription after all but 176 of its 594 residues have been deleted (172). The other component of S.cerevisiae TFIIIB is a 70 kDa subunit that displays 23% identity and 44% similarity to TFIIB in its N-terminal 320 residues (173–175). Because of this homology, it is often referred to as TFIIB-related factor or BRF, although it has also been called TDS4, PCF4 and TFIIIB70. Given their similarity, it was expected that BRF and TFIIB might interact with TBP in a similar manner. Indeed, a weak interaction is observed between TBP and the TFIIB-homologous region of BRF (176,177). However, the principal TBP-binding domain of BRF maps outside the region that is conserved with TFIIB (176–181). Extensive mutagenesis has shown that BRF contains two separate TBP-binding domains, which interact with opposite faces of the TBP–DNA complex (177,179,180). A remarkable feature of BRF is that it can be split down the middle to give two separate halves that continue to function when recombined (177). This property has allowed dissection of the roles of the separated domains (177).
As with TFIIIC and pol III, TFIIIB is less well characterised in metazoa. The human factor has been shown to contain TBP (182–185) and a homologue of BRF (186,187). The N-terminal domain of human BRF is 24% identical to human TFIIB and 41% identical to the corresponding region of S.cerevisiae BRF, but the C-terminal half is much less conserved (187). A BRF homologue has also been identified in Caenorhabditis elegans (188). Apart from TBP and BRF, human TFIIIB contains one or more additional, unidentified components (182,184,187,189,190). It seems highly probable that a B′′ equivalent will be involved, since B′′ from S.cerevisiae has been shown to support U6 transcription when reconstituted with human fractions (190). However, the TFIIIB requirements for type III promoters are distinct from those for types I and II (182,187,189,190).
Since it contains TBP, TFIIIB can bind independently to a TATA box (191). The affinity of binding is increased by a cooperative interaction between TBP and BRF, particularly at suboptimal TATA sites (192). However, most type I and II promoters lack a TATA sequence and so cannot be recognised directly by TFIIIB (170,185,193,194). In these cases, TFIIIB is recruited by protein–protein contacts with DNA-bound TFIIIC (193,195). Saccharomyces cerevisiae BRF has been shown to interact with τ131, the subunit of TFIIIC located furthest upstream along the promoter (176,196). Certain mutations in τ131 can stimulate transcription complex assembly, which has led to the suggestion that the TFIIIB-binding site may be masked prior to conformational rearrangements (196–198). Changes in the efficiency of photocrosslinking provide evidence that τ131 becomes reoriented during TFIIIB assembly (170). Once recruited, yeast TFIIIB occupies a region of ~40 bp immediately upstream of the transcription start site (147,177,193,199,200). From there it is able to bring pol III to the promoter and position it over the initiation region (199). All three subunits of TFIIIB are required for pol III recruitment, but direct interactions have only been identified in the case of BRF (176,201,202).
Several factors contribute to the selection of the transcription start site. The principal determinant is TFIIIC, which dictates the general location where TFIIIB is positioned. However, the interface between these factors is quite flexible and allows TBP to scan the region around –30 for an optimal site for TFIIIB (203). Initiation then occurs ~30 bp downstream of this site, but pol III has certain sequence preferences and will hunt within this small window for the optimal initiating nucleotide (203).
Once assembled, transcription complexes on class III genes display considerable stability towards high salt concentrations that would preclude their formation entirely (193,199,204,205). For example, ~45% of VAI transcriptional activity remains after a 6.5 min exposure of the preformed complex to 1 M KCl (205). Complexes formed on yeast tRNA and 5S rRNA genes retain full activity after exposure to 500 mM NaCl or 100 µg/ml heparin (193,199). In yeast, the interaction between TFIIIB and DNA is the most resistant to salt, whereas TFIIIC and TFIIIA are dissociated more readily (193,199). Once recruited to a promoter via TFIIIC, yeast TFIIIB will remain stably bound even in 1 M KCl (199). This is remarkable, given that TFIIIB alone is incapable of recognising a TATA-less class III gene. It seems that interaction with TFIIIC unmasks a latent DNA-binding capacity that locks TFIIIB onto a promoter. Indeed, a proteolytic fragment containing the C-terminus of BRF can bind DNA in a sequence-independent fashion, but this cryptic function is not detected with the isolated full-length polypeptide (206). Interactions between the TFIIIB subunits may constrain the DNA in such a way that it is unable to slide free; this might allow avid binding in the absence of sequence-specific recognition.
This unusual property of yeast TFIIIB was exploited to show that this factor is sufficient on its own to recruit pol III and direct multiple rounds of accurately initiated transcription (199). TFIIIC and TFIIIA were stripped from fully assembled transcription complexes by exposure to heparin, leaving TFIIIB alone on tRNA and 5S rRNA promoters; the efficiency of transcription was not compromised by this treatment (199). On this basis, TFIIIA and TFIIIC can be regarded as assembly factors that are dispensable for transcript initiation. It seems likely that this conclusion will also apply to metazoan systems, but this has not been demonstrated. Silkworm TFIIIB remains bound to promoters after the other factors have been stripped (K.Sprague, personal communication). However, with frog and human factors the dissociation pathway is the reverse of the assembly pathway; polymerase is lost before TFIIIB as the salt concentration is elevated (204,205,207). Thus, the exceptional salt stability of promoter-bound TFIIIB that is seen in simpler organisms appears not to be a feature of vertebrates.
The similarity between the committed complex of the rRNA genes and the TFIIIB complex of the pol III system is striking. In each case, the TBP-containing factor is positioned just upstream of +1. These complexes are sufficient to recruit RNA polymerases, positioning them over the initiation site. The complexes persist through multiple rounds of initiation. In some situations, the assembly factors can be dispensed with, allowing transcription in the presence of TIF-IB or TFIIIB alone.
Type III promoters. The type III promoters associated with vertebrate 7SK and U6 snRNA genes have distinct factor requirements from most pol III templates. They utilise TFIIIC1, but not TFIIIC2 (151,153). Furthermore, the TFIIIB employed by type III promoters is separable chromatographically from the form used by types I and II, probably representing a subcomplex (182,187,189,190). One study concluded on the basis of immunodepletion that BRF is not required for transcription of a human U6 gene (187). However, BRF can be immunoprecipitated in association with PTF, a factor employed by U6 genes (208).
The PSE of type III promoters is recognised by a factor called PTF or SNAPc. It is a complex of five subunits, all of which have been cloned (153,208–213). The largest subunit, SNAP190, contains a Myb DNA-binding domain and can be crosslinked to the PSE (153,213). The U1 and U2 snRNA genes that are transcribed by pol II also contain PSE promoter sequences and can be activated by PTF/SNAPc (208–212). Since pol III transcription of U6 and 7SK genes and pol II transcription of U1 and U2 is inhibited by immunodepletion with antisera against PTF/SNAPc and expression in the depleted extracts is restored by adding purified PTF/SNAPc (208–212), it seems that there is a single PSE-binding protein that is shared by the two polymerases.
The distance separating the PSE and the TATA box is constrained very precisely in type III promoters (214,215). This suggests that PTF/SNAPc interacts with TATA-bound TBP or TFIIIB. Indeed, subunits of PTF/SNAPc have been shown to bind directly to TBP (208–212). Both PSE and TATA binding are relatively slow and assembly of the initiation complex takes longer for U6 genes than it does for tRNA or 5S rRNA genes (216,217). SNAP190 contains a C-terminal extension that inhibits PSE recognition, perhaps by masking the DNA-binding domain (218). This self-repression can be relieved by Oct-1, which binds to the upstream DSE, makes direct contact with SNAP190 and thereby stimulates PSE occupancy (217,219,220). Association of PTF/SNAPc with the PSE of the human U6 gene takes over 1 h in the absence of Oct-1, but is complete in 15–30 min in its presence (217). In the case of the 7SK gene, Oct-1 produces a 10- to 20-fold increase in PSE occupancy and a similar increase in transcription (219,221).
Template competition experiments have shown that the PSE is essential for stable complex formation on the human U6 promoter (222). Mutation of the octamer sequence within the DSE reduced the stability of the complex (222). Although the TATA box helped stabilise the complex, it was not essential for resistance to template competition (222). Thus, both kinetic and template commitment experiments indicate that PSE occupancy is of primary importance in assembling an initiation complex on a type III promoter. PTF/SNAPc and TBP enhance each others recruitment to the U6 promoter, an effect that is dependent on the N-terminal 96 residues of TBP (93). Occupation of the U6 promoter, therefore, appears to be achieved by a series of cooperative interactions between Oct-1, PTF/SNAPc and TBP. It remains to be determined how TFIIIC1 is recruited, but this may be a late step in complex assembly on type III promoters.
TRANSCRIPT INITIATION AND ELONGATION
During initiation, the double-stranded DNA must be melted to allow access to the template strand. In pol II transcription, this process requires ATP βγ bond hydrolysis to energise DNA helicase activity of TFIIH. However, βγ bond hydrolysis is not necessary for transcription by pol I or pol III (205,223). Analyses using KMnO4 and diethylpyrocarbonate to detect single-stranded regions in the DNA show that a melted bubble forms spontaneously upon binding of pol I or pol III, in a temperature-dependent manner. For both enzymes, the melted bubble initiates upstream of +1 and propagates downstream. Pol I melts ~9–10 bp before initiation; following initiation and promoter clearance, the melted transcription bubble expands to ~19 bp. The length of the double-stranded DNA–RNA hybrid at steady-state is estimated to be 9 bp (224).
Pol III also melts the DNA helix around the initiation site (199,225). This process requires the active participation of TFIIIB, as certain mutations in BRF or B′′ allow normal polymerase recruitment and positioning but prevent the formation of a strand-separated open promoter complex (226). Indeed, B′′ can be dispensed with if a short segment of DNA is unpaired just upstream of +1 to form a pre-opened transcription bubble (227). Although promoter escape can limit the rate of pol II transcription, nearly all pol III molecules dissociate from promoter-bound TFIIIB without significant pausing or arrest (228). As pol III progresses into the gene, the bubble of melted DNA moves with it (225). At 20°C, yeast pol III elongates RNA at an average rate of ~20 nt/s, which is similar to the chain elongation rate measured for pol II in vivo (229). However, elongation does not proceed at a uniform rate; for example, at 20°C it takes pol III 3.0 s to traverse from nt 17 to nt 46 of the SUP4 tRNATyr gene and 4.1 s to travel the next 9 nt (229). This uneven progress results from pausing at internal sites, and the rate of extension at individual nucleotides can vary by 31-fold (229). Recognition of pause sites involves the C11 subunit of yeast pol III (230). This subunit bears significant homology to the pol II-specific elongation factor TFIIS (230). Whereas pol I and pol II employ dedicated elongation factors, pol III does not; this may not be necessary because of the TFIIS-like function of the C11 subunit (230) and because the class III genes are extremely short.
A large transcription complex within the transcribed region of a gene might be expected to block progression of polymerase or to be displaced by it. However, assembled transcription complexes are not removed from internal control regions by multiple passages of pol III (207,231). Furthermore, removing yeast TFIIIC from the SUP4 tRNATyr gene made no significant difference to the rate of elongation (229,232). During transcription in the normal direction, the presence of TFIIIC delays pol III for just 0.2 s at a single site upstream of the B block (229). Since the time required for promoter clearance limits initiation rates to <0.5/s, a downstream delay of 0.2 s will make no difference to the overall level of transcription (232). However, if pol III transcribes in the antisense direction and encounters TFIIIC from downstream, it pauses for around 9 s before continuing through the B block (232). In contrast, TFIIIB prevents the passage of pol III approaching from downstream for >1 h (232). A full X.laevis 5S RNA gene transcription complex has also been found not to impede transcription of either DNA strand by SP6 RNA polymerase and the complex remains stably bound following multiple polymerase transits (233). In contrast, a TFIIIA–5S rRNA gene complex in the absence of TFIIIB and TFIIIC is dissociated by passage of either SP6 RNA polymerase or pol III (233,234). Similarly, TFIIIC alone is rapidly displaced from DNA by pol III (232). These results suggest that the multiple contacts made by a complete transcription complex may be essential for continued integrity. An obvious possibility is that pol III transiently displaces a given factor from its binding site as it transcribes through the gene, but the factor remains associated due to protein–protein contacts with other factors bound to DNA sites not in the process of being transcribed. The interaction of TFIIIB with DNA upstream of the start site may be particularly important in preventing TFIIIC and TFIIIA from being displaced from the template as pol III transcribes through the internal promoter.
TRANSCRIPTION TERMINATION
Pol I transcription stops at a series of gene terminators found within the first 1000 bp downstream of the 28S rRNA coding sequence (235–238). Whereas pol III terminators are intrinsically recognised by the polymerase, pol I terminator function (both PT and the gene terminators) is mediated by the DNA-binding protein TTF-I (in mouse) or Reb1p (in yeast) (36,141,239–243). Termination requires binding of the terminator protein to specific DNA sequences downstream of the transcription unit. These factors bind in a specific orientation and termination is orientation dependent (235,236,240,244). In mouse, termination is pol I specific (245), but in yeast both pol I and pol III can be terminated in vitro by Reb1p in the correct sequence context (36,242); in contrast, pol II is only paused, requiring an additional factor for template and product release (246). It is striking that the sequence context for pol I and pol III termination is remarkably similar (246).
TTF-I and Reb1p have in their C-terminal halves direct repeats of a motif related to the DNA-binding domain of the protooncogene Myb (141,247–249). Deletion studies of the mouse factor suggest that a small sequence just upstream of these DNA-binding repeats (248) recruits a transcript releasing factor (250). A homologous element was not identified in Reb1p. However, the releasing factor from mouse will catalyse RNA release from pol I paused by the yeast factor (251).
In the mouse system, termination occurs when pol I encounters DNA-bound TTF-I (250). TTF-I then recruits the releasing factor, which catalyses 3′-end formation. Following release, there is some 3′ trimming of the transcript by an exonuclease to produce the mature 3′-end. Finally, polymerase dissociates from the template. In yeast, no releasing factor is needed and dissociation of the transcript depends upon instability of the nascent RNA–DNA hybrid in the active site of the polymerase (242). Instability is increased by base pair mismatching in the hybrid after the polymerase ‘retreats’ from the pause site.
The termination factors also bind to the PT. The PT stimulates transcription, and several mechanisms have been suggested to contribute to this effect. (i) The termination factors are capable of oligomerisation in vitro and, in the context of the repeated rRNA genes, could possibly link the end of one repeat to the beginning of the next, so that the intergenic spacer DNA is looped out (35,36,144). (ii) The PT serves a role in chromatin remodelling when bound by TTF-I, repositioning a nucleosome over the core promoter (252). (iii) The PT serves to protect the core and UPE from having their transcription factors stripped by a juggernaut pol I that initiated on a spacer promoter (32,33). However, the yeast PT is in the opposite orientation and non-functional in transcription termination.
The gene terminators are also DNA replication terminators in mouse (253). A DNA replication origin is found in each IGS, downstream of the gene terminator. Bi-directional replication starts here and proceeds through the downstream transcription unit. The replication fork moving in the opposite direction would encounter RNA polymerases transcribing the upstream repeat. However, the TTF-I-bound gene terminator acts as a replication fork barrier, stopping progression of the replication fork into the upstream transcription unit. The barrier function is strictly orientation dependent, but has opposite polarity to transcription termination.
In striking contrast to pol I and pol II, pol III can recognise termination sites accurately and efficiently in the apparent absence of other factors (254). Simple clusters of four or more T residues can serve as terminator signals in most cases (53,255,256). It has been proposed that the La autoantigen is involved in termination by pol III (257–259). HeLa extracts that have been immunodepleted of La synthesise very few pol III transcripts and these are truncated at their 3′-termini by between one and five U residues (257,258). Addition of purified La to immobilised DNA templates increases the release of RNA and the overall level of transcription (259). These observations led to the suggestion that La is involved in the production and release of full-length pol III transcripts in mammals (257–259). However, deletion of the gene encoding La in S.cerevisiae has no effect on pol III activity (260). Attempts to demonstrate a role for La in pol III transcription in Xenopus have also proved unsuccessful (261,262).
REINITIATION
Reinitiation on rRNA genes is extremely efficient, producing a high density of pol I molecules on the active repeats. This is at least in part because the core factor left on the promoter is fully functional, allowing for repetitive polymerase recruitment. In A.castellanii, the next polymerase can join the promoter once the transcribing polymerase has translocated beyond about +17 (224). It has been suggested that rRNA genes are arranged in bundles in the nucleolus such that all the terminators and promoters are pulled together in a central core with the transcribed portions and IGSs looped out (37,144). Such an arrangement would result in a very high concentration of polymerase in the vicinity of the promoter, thereby abetting frequent reinitiation.
After the initial round of transcription, a stable complex on a yeast tRNA gene can direct subsequent cycles 5- to 10-fold more rapidly than the first (263). Thus, during multiple round transcription, synthesis of each tRNA molecule takes ~35 s, whereas initiation of the first transcript can take ~5 min (at 22°C) (263). This is because pol III is recycled without being released from the template; as a consequence, the slow initial step of polymerase recruitment is avoided (263). Template commitment assays demonstrated that pol III remains associated with the gene on which it first initiated and is not released into a free pool (263). TFIIIA, TFIIIB and TFIIIC all bend DNA (96), and this may facilitate internal recycling by bringing the two ends of a class III gene into close proximity. Human pol III is also retained in the original transcription complex on VA and tRNA genes without dissociating after each round of synthesis (207). Evidence has been presented that limiting amounts of human pol III can be recycled to preinitiation complexes on the VAI promoter at least 5-fold more efficiently in the presence of La than in its absence (259,264). It remains to be determined whether La allows mammalian systems to utilise a facilitated recycling pathway that is similar to the one described in yeast.
ACCESSORY FACTORS
Pol I
In addition to the core- and UPE-binding factors, several additional pol I transcription factors have been described. An example is TIF-IA, which has only been identified in mouse (265–267). This is the factor regulated in response to growth conditions (266,268) and also in response to hormones, where it was dubbed TFIC (269,270). TIF-IA activity co-fractionates with a 75 kDa monomer, but extremely low yields have obstructed cloning (271). This factor is required for promoter clearance, but not for recruitment of polymerase to the promoter (271). TIF-IA is normally found tightly associated with a small fraction of the pol I in the cell (271). Several groups concluded that TIF-IA is utilised stoichiometrically rather than catalytically, suggesting that it is inactivated at each round of initiation (266,270). The nature of this inactivation has not been elucidated and it is difficult to distinguish experimentally from dilution into the reaction medium in vitro (270).
In yeast, an additional factor necessary for specific initiation has been identified genetically (272). The 72.4 kDa product of the RRN3 gene is required for viability, is purified as a monomer and associates with pol I (272), but at what step in initiation it functions is not known. Like TIF-IA, Rrn3p or its association with dimorphic forms of pol I may be regulated in response to entry into stationary phase (273,274). Since yeast Rrn3p and mouse TIF-IA exhibit similar functions (272,273), cloning may reveal that they are homologues.
TIF-IC (not to be confused with TFIC) has only been identified in mouse (275,276). Its composition is unknown, but it associates with pol I and is absolutely required for promoter clearance. TIF-IC also suppresses non-specific initiation and stimulates the elongation rate by depressing pausing (276), activities similar to the pol II-specific factor TFIIF. However, it is distinct from TFIIF in its gel filtration and ion exchange chromatographic behaviour. Both TIF-IC and TIF-IA may join the initiation complex in association with pol I or after pol I recruitment, with TIF-IA joining after TIF-IC. Unlike Rrn3p, these factors are known to have a role in transcription initiation at a step after polymerase promoter binding (100,277).
PAF53 was purified and cloned from mouse as part of a group of three polypeptides (53, 51 and 49 kDa) associated with a small fraction of highly purified pol I (278–280). PAF53 has significant sequence similarity to the S.cerevisiae pol I A49 subunit (280,281). A49 deletion yeast strains are viable, but grow slowly, for reasons that appear complex (282). Anti-PAF53 antibodies inhibit specific rRNA transcription, but not non-specific polymerisation activity in vitro, suggesting that PAF53 functions during initiation. Pull-down and far-western blot experiments show a weak interaction between UBF and PAF53, perhaps allowing UBF to participate in enzyme recruitment. Furthermore, PAF53 accumulates in nucleoli of actively transcribing cells, but is deficient in nucleoli of serum-starved cells, implicating it in regulation (280). However, another group found that the level of PAF53 was comparable in exponentially growing and growth-arrested cells, and concluded that it is the mouse A49 homologue (281).
TIF-IE stabilises kinetically the binding of TIF-IB to the core promoter (87). It has only been identified in A.castellanii, but unpublished data from a number of other systems suggest that similar activities are widespread. In the absence of TIF-IE, TIF-IB cannot form a complex on the promoter that is resistant to competition by another promoter-containing DNA. The apparent equilibrium binding constant of 50 pM is not significantly altered by the presence of TIF-IE, rather, the on–off rates are affected proportionately. TIF-IE has a native molecular mass of ~70 kDa by rate zonal sedimentation, but its subunit composition is not known. TIF-IE co-purifies with both pol I and TIF-IB, suggesting that it may interact with both. Functionally, it appears similar to yeast UAF and possibly to vertebrate UBF, but this is yet to be proven.
Rat E1BF/Ku, a clamp protein that tethers the catalytic subunit of DNA-dependent protein kinase to DNA, was reported to stimulate pol I initiation, but not elongation (283–288). Ku knockout mice exhibit an inability to repair DNA double-strand breaks and cannot rearrange DNA, thus they are immunodeficient (scid) mice (289). However, these knockouts develop normally except for slightly diminished size (289), so an absolute requirement for Ku in mammalian rRNA transcription has been excluded genetically.
Pol III
In addition to the basal factors described above, several additional factors have been proposed to influence pol III transcription. An S.cerevisiae protein called TFIIIE can stimulate transcription under certain conditions (169,290). Neither the molecular nature nor the mode of action have been characterised for TFIIIE, although suggestions include a role in facilitating TFIIIB recruitment, conformational rearrangements of TFIIIB or transcription complex stabilisation (169).
TFIIA has been proposed to stimulate human pol III transcription by interacting with TBP (291,292). However, deletion of the yeast TFIIA genes makes no difference to the level of pol III activity in vivo and TFIIA is not stimulatory with yeast cell extracts (293). Furthermore, BRF and B′′ prevent TFIIA from gaining access to TBP in the TFIIIB complex (168).
The human pol III transcription apparatus was reported to co-purify with an uncharacterised activity called TDF (294), as well as the pol II cofactor PC4 and DNA topoisomerase (topo) I (295). Both PC4 and topo I were found to enhance the footprints produced by TFIIIC in downstream promoter regions (295). In addition, they stimulated multiple round, but not single round transcription from preformed complexes, suggesting a role in reinitiation (295). Mutation of the catalytic site of topo I made no difference to these activities (295). No evidence has yet been presented that these factors contribute to pol III transcription in vivo.
A novel regulator called Staf has been isolated by screening a Xenopus oocyte expression library with an activator element from 200 bp upstream of the X.laevis tRNASec gene (296). This is an unusual tRNA gene that combines an internal B block with upstream elements that are similar to those of U6 genes (70). Staf can bind to the DSEs of U6, U1, U2 and 7SK genes and in each case substitution of the recognition motif can diminish transcription in vivo (297). Furthermore, recombinant Staf stimulates tRNASec expression when injected into oocytes (296). Staf is a 65 kDa polypeptide with a central DNA-binding domain containing seven tandem zinc fingers (296). It is likely to function cooperatively with Oct-1, since their respective binding sites are found close together in ~70% of all DSE elements and can activate synergistically if appropriately spaced (297).
Because of the high basal activity of most pol III promoters, transcriptional repression is likely to be important in controlling expression levels under some circumstances. Indeed, several previously known pol II regulators can also repress class III genes both in vitro and in vivo. The first example of this was provided by Dr1, a 19 kDa nuclear phosphoprotein that was isolated from HeLa cells as an inhibitor of pol II transcription (298). Saccharomyces cerevisiae contains a Dr1 homolog that is 37% identical to its human counterpart and is essential for viability (299). It binds directly to TBP and blocks its association with TFIIA and TFIIB, thereby inhibiting pol II transcription (298). In the TFIIIB complex, BRF contacts TBP at sites that overlap with both the TFIIA and TFIIB interaction surfaces (176–181). Accordingly, Dr1 can disrupt the association between TBP and BRF, thereby inactivating TFIIIB (300). Overexpression of Dr1 will repress transcription in vitro of a range of human pol III templates and will also inhibit tRNA synthesis in living yeast (299,300). In contrast, pol I transcription is not affected by Dr1 in either of these situations (299,300). This establishes the specificity of the effect and suggests that Dr1 might function to alter the balance of nuclear RNA polymerase activity, suppressing pol II and pol III under conditions where pol I transcription is limiting.
Overexpression of Dr1 slows cell growth in yeast (299). Two other unrelated growth suppressors that can also inhibit pol III transcription are RB and p53, but these are restricted to metazoa. Both RB and p53 can inhibit the expression of class III genes when transfected into cells or added to in vitro transcription reactions (301–305). This is not an artifact of overexpression, since the specific inactivation of endogenous RB or p53 using gene knockout technology results in elevated synthesis of pol III transcripts in living murine fibroblasts (302,305). Immunoprecipitation and co-fractionation have shown a physical association between endogenous cellular TFIIIB and both RB and p53 (303–305). Furthermore, TFIIIB activity is elevated specifically in fibroblasts derived from either RB or p53 knockout mice (304,305). Very similar results have been reported for the RB-related proteins p107 and p130 (306). However, it has yet to be determined why binding of these factors results in a loss of TFIIIB activity.
RB has also been shown to repress pol I transcription, in this case by targeting UBF (307,308). Immunoprecipitations with cellular extracts demonstrated interaction between endogenous RB and UBF (307). The amount of RB that co-precipitates with UBF increases as U937 cells differentiate, in parallel with a decrease in pol I transcription (307). Immunofluorescence has also shown that RB accumulates in the nucleolus as the differentiating cells down-regulate rRNA synthesis (307). Titration of RB can abolish the footprint produced by UBF on a mouse rDNA promoter, thereby providing a mechanism of action (308). Pol I transcription in vivo can be repressed by p53, although this effect may be indirect (309). It has been suggested that inhibiting rRNA and tRNA production may contribute to the growth suppression function of tumour suppressors (310,311).
CHROMATIN
Nucleosomes are associated with the coding regions of many genes transcribed by pol II and pol III (312–317). In contrast, eukaryotic rRNA genes lack nucleosomes on the transcribed repeats (318–319). Analysis of the DNA length within active rRNA genes in Miller spreads revealed a compaction ratio close to 1, whereas it would be near 6 if the genes were completely nucleosomal (reviewed in 318). These electron microscopic studies did not address the transcriptionally inactive copies, so their chromatin state has been controversial, largely owing to the difficulty of studying chromatin structure of multicopy genes in heterogeneous states of packaging. Sogo and co-workers (82,320) demonstrated two classes of rRNA genes using psoralen, a reagent that photocrosslinks one DNA strand to the other wherever it is not protected by protein; non-nucleosomal DNA crosslinks more extensively than nucleosomal and has diminished mobility in polyacrylamide gels. Their studies show that both the promoter and the transcribed portion of the active genes are stripped of nucleosomes even though both daughter rRNA genes are completely packaged immediately following DNA replication (321). This stripping does not occur in yeast mutants lacking active pol I, suggesting that the polymerase plays a critical role in nucleosome removal (320). Because the presence of inactive, nucleosomal copies of the rRNA genes interferes with analysis, it is not known whether the active copies are free of histones as well as of nucleosomes.
Simple pol III templates have provided extremely valuable systems for studying the organisation of chromatin and its effects on transcriptional activity, as has been reviewed extensively by Wolffe (322). A good example was provided by the use of photocrosslinking to probe the internal architecture of a nucleosome positioned on a Xenopus 5S rRNA gene (323). The same gene was used to demonstrate that histone acetylation can facilitate the access of transcription factors to chromatinised promoter sequences (324). For detailed reviews of the role of chromatin in pol III transcription, the reader is referred to White (2) and Wolffe (322).
The susceptibility of class III genes to nucleosomal repression is extremely template dependent. Most tRNA genes are remarkably resistant to chromatin-mediated repression, whereas middle repetitive genes such as B2 and Alu are highly susceptible. The clearest example of the differential effects of histones upon pol III transcription in vivo has been provided by studies in Xenopus. Injection of developing frog embryos with mRNA encoding histone H1 results in substantial repression of the oocyte 5S rRNA genes (325). Conversely, depletion of endogenous histone H1 using ribozymes causes increased expression of these genes (325,326). Neither of these manipulations has any significant effect on the transcription of tRNA and somatic 5S rRNA genes by pol III or of U1 and U2 snRNA genes by pol II (325,326).
tRNA genes are also highly resistant to repression by histones in other systems. Removal of H1 from murine chromatin makes little or no difference to the number of tRNA genes that are accessible to transcription factors (327). Synthesis of tRNA is not affected by a nucleosome deficiency in S.cerevisiae that can activate several class II genes (328). The SUP4 tRNATyr gene could even remain active in yeast cells when fused to nucleosome positioning signals that are capable of suppressing both pol II transcription and the initiation of DNA replication (329). When substitutions were introduced into the B block, these positioning signals were able to incorporate the start site and A block of the mutated tRNA gene into the centre of a nucleosome, although the wild-type SUP4 template could override the signals and stay free of nucleosomes (329). A yeast tRNAGlu gene also remained active after prolonged incubation in a Xenopus egg extract that assembled nucleosomes with physiological spacing (330).
The U6 gene of S.cerevisiae can also compete successfully with nucleosomes in vivo, as shown by the fact that its expression is unchanged following inactivation of the histone H4 gene (331). However, transcription of U6 promoter mutants was significantly enhanced by deletion of histone H4 (331). Thus, whereas the wild-type U6 gene competes efficiently with nucleosomes, mutant forms with weakened promoters are subject to histone-dependent repression. In vitro studies have shown that TFIIIC can protect this template against chromatin-mediated repression (330). The U6 promoter of S.cerevisiae has a functional B block, but this is only required for transcription in the presence of histones (330). Interaction with the B block allows TFIIIC to displace nucleosomes from the template, whereas TFIIIB does not have this effect (330). These results suggest that TFIIIC performs an additional role besides serving as an assembly factor for recruiting TFIIIB, namely to weaken the interaction of nucleosomes with the transcribed region of at least some class III genes. This is consistent with reports that human TFIIIC2 has histone acetyltransferase activity (158a,332). In yeast, the U6 gene is flanked on both sides by a series of positioned nucleosomes; this organisation disappeared upon inactivation of the B block with a 2 bp deletion (331). These results provided the first evidence that a class III gene can organise the chromatin around it. However, this ability was lost when the U6 gene was transferred from its chromosomal location to a centromeric vector (331).
The 500 000 Alu genes in the haploid human genome constitute 5% of the total chromosomal DNA. This number of templates could potentially provide an enormous sink for transcription factors, which might prove highly detrimental due to competition with essential class III genes. It may therefore be important for a cell to mask the majority of copies. Most of the functional Alu promoters are repressed in chromatin isolated from HeLa cells (333). Indeed, chromatin appears to silence ~99% of potentially active Alu genes, whereas the majority of tRNA and 5S rRNA genes are accessible to transcription factors in the same HeLa cell chromatin preparations (333). Reconstitution experiments using purified nucleosomes have shown that Alu elements position histone octamers over the start site and A block region, which results in repression (334). Intranuclear footprinting has shown that a substantial proportion of Alu elements position nucleosomes in this way in native chromatin (317). Silencing of Alu elements by nucleosomal core particles is so efficient that inclusion of H1 has little additional effect (333). Indeed, removal of H1 from HeLa cell chromatin raises Alu expression by only ~2-fold (333). In contrast, pol III transcription of the B2 repetitive gene family increases by ~20-fold when H1 is depleted from the chromatin of 3T3 cells (327,333). Thus, although chromatin-mediated repression may be common to middle repetitive class III genes, the molecular mechanism can vary between different families.
The Alu consensus sequence contains an unusually high CpG density, 9-fold above the average for the human genome (334). Indeed, Alu CpGs account for about one-third of the potential methylation sites in human DNA. The maintenance of this high CpG content may indicate that evolution exerts pressure to silence Alu transcription. Most Alu genes are highly methylated in DNA from a range of somatic human cell types (335,336). Treatment of HeLa cells with 5-azacytidine results in hypomethylation of several consensus Alu CpGs and a concomitant 5- to 8-fold increase in the abundance of Alu transcripts (337). Template methylation can repress pol III transcription of Alu, tRNA and VA genes in vitro and of VAI in transfected cells (336,338,339). It can also inhibit tRNA genes in injected Xenopus oocytes, although a 5S rRNA gene was unaffected in the same assay (340). When Xenopus oocytic 5S rRNA genes were purified from blood cells, in which they are not expressed, every CpG in the repeat unit was found to be heavily methylated (341,342). When this heavily methylated DNA was injected into Xenopus oocytes, it was transcribed as efficiently as unmethylated cloned 5S rRNA genes (343). Thus, methylation appears to affect only specific categories of class III genes. It may contribute to the suppression of inappropriate pol III transcription in higher organisms, such as that of middle repetitive genes.
Repression of methylated tRNA or Alu genes in vitro can be specifically relieved by the presence of methylated competitor DNA (339), which suggests that the inhibition is due to proteins that bind to methylated DNA and can be removed by the competitor. Complete methylation of an X.borealis 5S rRNA gene had no qualitative or quantitative influence upon its interaction with core or linker histones (344). The affinity of histone octamers for an Alu gene was also unaffected by methylation (334). However, CpG methylation increased the efficiency with which a histone tetramer could obstruct access to an Alu promoter and thereby repress transcription (334). Thus, histone tetramers reduce the expression of an unmethylated Alu gene by 2.5-fold but inhibit the same template by over 50-fold if it is methylated (334). When this gene was not reconstituted with histones, methylation decreased its transcription in a nuclear extract by ~2-fold (334). Factors such as MeCP1 and MeCP2, which bind specifically to methylated DNA in a sequence-independent fashion (345), might potentially play a role in class III gene regulation.
PERSPECTIVE
It will be appreciated from this survey that the pol I and pol III machineries each display many unique features and peculiarities. Nevertheless, it is also clear that the two systems have many properties in common. It is likely that the shared aspects are often the most evolutionarily ancient and the most fundamental to the underlying mechanics of the transcription process. Considerable insight can therefore be gained by continuing to compare and contrast the structure and function of the apparatuses used by the three eukaryotic nuclear RNA polymerases.
REFERENCES
- 1.Paule M.R. (1998) Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Springer-Verlag, New York, NY.
- 2.White R.J. (1998) RNA Polymerase III Transcription. Springer-Verlag, New York, NY.
- 3.Shaw P.J. and Jordan,E.G. (1995) Annu. Rev. Cell Dev. Biol., 11, 93–121. [DOI] [PubMed] [Google Scholar]
- 4.Pombo A., Jackson,D.A., Hollinshead,M., Wang,Z., Roeder,R.G. and Cook,P.R. (1999) EMBO J., 18, 2241–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertrand E., Houser-Scott,F., Kendall,A., Singer,R.H. and Engelke,D.R. (1998) Genes Dev., 12, 2463–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobson M.R., Cao,L.-G., Taneja,K., Singer,R.H., Wang,Y.-I. and Pederson,T. (1997) J. Cell Sci., 110, 829–837. [DOI] [PubMed] [Google Scholar]
- 7.Sentenac A., Riva,M., Thuriaux,P., Buhler,J.-M., Treich,I., Carles,C., Werner,M., Ruet,A., Huet,J., Mann,C. et al. (1992) In McKnight,S.L. and Yamamoto,K.R. (eds), Transcriptional Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 27–54.
- 8.Mann C., Buhler,J.-M., Treich,I. and Sentenac,A. (1987) Cell, 48, 627–637. [DOI] [PubMed] [Google Scholar]
- 9.Woychik N.A., McKune,K., Lane,W.S. and Young,R.A. (1993) Gene Expr., 3, 77–82. [PMC free article] [PubMed] [Google Scholar]
- 10.Kolodziej P.A. and Young,R.A. (1991) Mol. Cell. Biol., 11, 4669–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martindale D.W. (1990) Nucleic Acids Res., 18, 2953–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paule M.R. (1998) In Paule,M.R. (ed.), Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Springer-Verlag, New York, NY, pp. 195–200.
- 13.Matthews J.L., Zwick,M.G. and Paule,M.R. (1995) Mol. Cell. Biol., 15, 3327–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reeder R.H. (1984) Cell, 38, 349–351. [DOI] [PubMed] [Google Scholar]
- 15.Moss T. and Stefanovsky,V.Y. (1994) Prog. Nucleic Acids Res. Mol. Biol., 50, 25–66. [DOI] [PubMed] [Google Scholar]
- 16.Paule M.R. (1998) In Paule,M.R. (ed.), Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Springer-Verlag, New York, NY, pp. 39–50.
- 17.Planta R.J. (1998) In Paule,M.R. (ed.), Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Springer-Verlag, New York, NY, pp. 51–58.
- 18.Grummt I., Roth,E. and Paule,M.R. (1982) Nature, 296, 173–174. [DOI] [PubMed] [Google Scholar]
- 19.Pape L.K., Windle,J.J. and Sollner-Webb,B. (1990) Genes Dev., 4, 52–62. [DOI] [PubMed] [Google Scholar]
- 20.Muramatsu M. (1998) In Paule,M.R. (ed.), Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Springer-Verlag, New York, NY, pp. 295–308.
- 21.Safrany G., Tanaka,N., Kishimoto,T., Ishikawa,Y., Kato,H., Kominami,R. and Muramatsu,M. (1989) Mol. Cell. Biol., 9, 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishikawa Y., Safrany,G., Hisatake,K., Tanaka,N., Maeda,Y., Kato,H., Kominami,R. and Muramatsu,M. (1991) J. Mol. Biol., 218, 55–67. [DOI] [PubMed] [Google Scholar]
- 23.Bell S.P., Pikaard,C.S., Reeder,R.H. and Tjian,R. (1989) Cell, 59, 489–497. [DOI] [PubMed] [Google Scholar]
- 24.Rudloff U., Eberhard,D., Tora,L., Stunnenberg,H. and Grummt,I. (1994) EMBO J., 13, 2611–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee M.G. and Van der Ploeg,L.H. (1997) Annu. Rev. Microbiol., 51, 463–489. [DOI] [PubMed] [Google Scholar]
- 26.Lin J. and Vogt,V.M. (1998) Mol. Cell. Biol., 18, 5809–5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perna P.J., Harris,G.H., Iida,C.T., Kownin,P., Bugren,S. and Paule,M.R. (1992) Gene Expr., 2, 71–78. [PMC free article] [PubMed] [Google Scholar]
- 28.Radebaugh C.A., Gong,X., Bartholomew,B. and Paule,M.R. (1997) J. Biol. Chem., 272, 3141–3144. [DOI] [PubMed] [Google Scholar]
- 29.Henderson S.L. and Sollner-Webb,B. (1990) Mol. Cell. Biol., 10, 4970–4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith S.D., O’Mahony,D.J., Kinsella,B.T. and Rothblum,L.I. (1993) Gene Expr., 3, 229–236. [PMC free article] [PubMed] [Google Scholar]
- 31.Xie W. and Rothblum,L.I. (1992) Mol. Cell. Biol., 12, 1266–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bateman E. and Paule,M.R. (1988) Cell, 54, 985–992. [DOI] [PubMed] [Google Scholar]
- 33.Henderson S.L., Ryan,K. and Sollner-Webb,B. (1989) Genes Dev., 3, 212–223. [DOI] [PubMed] [Google Scholar]
- 34.Längst G., Becker,P.B. and Grummt,I. (1998) EMBO J., 17, 3135–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sander E.E. and Grummt,I. (1997) Nucleic Acids Res., 25, 1142–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mason S.W., Sander,E.E., Evers,R. and Grummt,I. (1998) In Paule,M.R. (ed.), Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Springer-Verlag, New York, NY, pp. 179–194.
- 37.Warner J.R., Ikonomova,R., Johnson,S.P., Nierras,C.R. and Wang,K. (1998) In Paule,M.R. (ed.), Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Springer-Verlag, New York, NY, pp. 241–252.
- 38.Schultz M.C., Choe,S.Y. and Reeder,R.H. (1993) Mol. Cell. Biol., 13, 2644–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paule M.R., Bateman,E., Hoffman,L., Iida,C., Imboden,M., Kubaska,W., Kownin,P., Li,H., Lofquist,A., Risi,P. et al. (1991) Mol. Cell. Biochem., 104, 119–126. [DOI] [PubMed] [Google Scholar]
- 40.Elion E.A. and Warner,J.R. (1984) Cell, 39, 663–673. [DOI] [PubMed] [Google Scholar]
- 41.Elion E.A. and Warner,J.R. (1986) Mol. Cell. Biol., 6, 2089–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgan G.T., Reeder,R.H. and Bakken,A.H. (1983) Proc. Natl Acad. Sci. USA, 80, 6490–6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.La Volpe A., Taggart,M., McStay,B. and Bird,A. (1983) Nucleic Acids Res., 11, 5361–5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgan G.T., Roan,J.G., Bakken,A.H. and Reeder,R.H. (1984) Nucleic Acids Res., 12, 6043–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moss T. and Birnstiel,M.L. (1979) Nucleic Acids Res., 6, 3733–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cassidy B.G., Yang-Yen,H.-F. and Rothblum,L.I. (1987) Mol. Cell. Biol., 7, 2388–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tower J., Henderson,S.L., Dougherty,K.M., Wejksnora,P.J. and Sollner-Webb,B. (1989) Mol. Cell. Biol., 9, 1513–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pikaard C.S. (1994) Proc. Natl Acad. Sci. USA, 91, 464–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pieler T., Hamm,J. and Roeder,R.G. (1987) Cell, 48, 91–100. [DOI] [PubMed] [Google Scholar]
- 50.Sharp S.J. and Garcia,A.D. (1988) Mol. Cell. Biol., 8, 1266–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee Y., Erkine,A.M., Van Ryk,D.I. and Nazar,R.N. (1995) Nucleic Acids Res., 23, 634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ciliberto G., Raugei,S., Constanzo,F., Dente,L. and Cortese,R. (1983) Cell, 32, 725–733. [DOI] [PubMed] [Google Scholar]
- 53.Galli G., Hofstetter,H. and Birnstiel,M.L. (1981) Nature, 294, 626–631. [DOI] [PubMed] [Google Scholar]
- 54.Baker R.E., Camier,S., Sentenac,A. and Hall,B.D. (1987) Proc. Natl Acad. Sci. USA, 84, 8768–8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fabrizio P., Coppo,A., Fruscoloni,P., Benedetti,P., Di Segni,G. and Tocchini-Valentini,G.P. (1987) Proc. Natl Acad. Sci. USA, 84, 8763–8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schultz P., Marzouki,N., Marck,C., Ruet,A., Oudet,P. and Sentenac,A. (1989) EMBO J., 8, 3815–3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Das G., Henning,D., Wright,D. and Reddy,R. (1988) EMBO J., 7, 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kunkel G.R. and Pederson,T. (1989) Nucleic Acids Res., 17, 7371–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lobo S.M. and Hernandez,N. (1989) Genes Dev., 58, 55–67. [Google Scholar]
- 60.Murphy S., Di Liegro,C. and Melli,M. (1987) Cell, 51, 81–87. [DOI] [PubMed] [Google Scholar]
- 61.Yuan Y. and Reddy,R. (1991) Biochim. Biophys. Acta, 1089, 33–39. [DOI] [PubMed] [Google Scholar]
- 62.Brow D.A. and Guthrie,C. (1990) Genes Dev., 4, 1345–1356. [DOI] [PubMed] [Google Scholar]
- 63.Tichelaar J.W., Knerer,B., Vrabel,A. and Wieben,E.D. (1994) Mol. Cell. Biol., 14, 5450–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bark C., Weller,P., Zabielski,J., Janson,L. and Pettersson,U. (1987) Nature, 328, 356–359. [DOI] [PubMed] [Google Scholar]
- 65.Carbon P., Murgo,S., Ebel,J.-P., Krol,A., Tebb,G. and Mattaj,I.W. (1987) Cell, 51, 71–79. [DOI] [PubMed] [Google Scholar]
- 66.Kunkel G.R. and Pederson,T. (1988) Genes Dev., 2, 196–204. [DOI] [PubMed] [Google Scholar]
- 67.Mattaj I.W., Dathan,N.A., Parry,H.D., Carbon,P. and Krol,A. (1988) Cell, 55, 435–442. [DOI] [PubMed] [Google Scholar]
- 68.Howe J.G. and Shu,M.-D. (1989) Cell, 57, 825–834. [DOI] [PubMed] [Google Scholar]
- 69.Sprague K.U., Larson,D. and Morton,D. (1980) Cell, 22, 171–178. [DOI] [PubMed] [Google Scholar]
- 70.Carbon P. and Krol,A. (1991) EMBO J., 10, 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vilalta A., Kickhoefer,V.A., Rome,L.H. and Johnson,D.L. (1994) J. Biol. Chem., 269, 29752–29759. [PubMed] [Google Scholar]
- 72.Ullu E. and Weiner,A.M. (1985) Nature, 318, 371–374. [DOI] [PubMed] [Google Scholar]
- 73.Keener J., Josaitis,C.A., Dodd,J.A. and Nomura,M. (1998) J. Biol. Chem., 273, 33795–33802. [DOI] [PubMed] [Google Scholar]
- 74.Wandelt C. and Grummt,I. (1983) Nucleic Acids Res., 11, 3795–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cizewski V. and Sollner-Webb,B. (1983) Nucleic Acids Res., 11, 7043–7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Iida C.T., Kownin,P. and Paule,M.R. (1985) Proc. Natl Acad. Sci. USA, 82, 1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haltiner-Jones M., Learned,R.M. and Tjian,R. (1988) Proc. Natl Acad. Sci. USA, 85, 669–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Learned R.M., Learned,T.K., Haltiner,M.M. and Tjian,R.T. (1986) Cell, 45, 847–857. [DOI] [PubMed] [Google Scholar]
- 79.Bell S.P., Learned,R.M., Jantzen,H.-M. and Tjian,R. (1988) Science, 241, 1192–1197. [DOI] [PubMed] [Google Scholar]
- 80.McStay B., Hu,C.H., Pikaard,C.S. and Reeder,R.H. (1991) EMBO J., 10, 2297–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Keys D.A., Lee,B.S., Dodd,J.A., Nguyen,T.T., Vu,L., Fantino,E., Burson,L.M., Nogi,Y. and Nomura,M. (1996) Genes Dev., 10, 887–903. [DOI] [PubMed] [Google Scholar]
- 82.Conconi A., Widmer,R.M., Koller,T. and Sogo,J.M. (1989) Cell, 57, 753–761. [DOI] [PubMed] [Google Scholar]
- 83.Comai L., Tanese,N. and Tjian,R. (1992) Cell, 68, 965–976. [DOI] [PubMed] [Google Scholar]
- 84.Cormack B.P. and Struhl,K. (1992) Cell, 69, 685–696. [DOI] [PubMed] [Google Scholar]
- 85.Schultz M.C., Reeder,R.H. and Hahn,S. (1992) Cell, 69, 697–702. [DOI] [PubMed] [Google Scholar]
- 86.Radebaugh C.A., Matthews,J.L., Geiss,G.K., Liu,F., Wong,J.-M., Bateman,E., Camier,S., Sentenac,A. and Paule,M.R. (1994) Mol. Cell. Biol., 14, 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Radebaugh C.A., Kubaska,W.M., Hoffman,L.H., Stiffler,K. and Paule,M.R. (1998) J. Biol. Chem., 273, 27708–27715. [DOI] [PubMed] [Google Scholar]
- 88.Heix J., Zomerdijk,J.C.B.M., Ravanpay,A., Tjian,R. and Grummt,I. (1997) Proc. Natl Acad. Sci. USA, 94, 1733–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Comai L., Zomerdijk,J.C.B.M., Beckmann,H., Zhou,S., Admon,A. and Tjian,R (1994) Science 266, 1966–1972. [DOI] [PubMed] [Google Scholar]
- 90.Keys D.A., Vu,L., Steffan,J.S., Dodd,J.A., Yamamoto,R.T., Nogi,Y. and Nomura,M. (1994) Genes Dev., 8, 2349–2362. [DOI] [PubMed] [Google Scholar]
- 91.Lin C.W., Moorefield,B., Pavel,J.P., Aprikian,P., Mitomo,K. and Reeder,R.H. (1996) Mol. Cell. Biol., 16, 6436–6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rudloff U., Stunnenberg,H.G., Keaveney,M. and Grummt,I. (1994) J. Mol. Biol., 243, 840–845. [DOI] [PubMed] [Google Scholar]
- 93.Mittal V. and Hernandez,N. (1997) Science, 275, 1136–1140. [DOI] [PubMed] [Google Scholar]
- 94.Steffan J.S., Keys,D.A., Dodd,J.A. and Nomura,M. (1996) Genes Dev., 10, 2551–2563. [DOI] [PubMed] [Google Scholar]
- 95.Beckmann H., Chen,J.L., O’Brien,T. and Tjian,R. (1995) Science, 270, 1506–1509. [DOI] [PubMed] [Google Scholar]
- 96.Braun B.R., Kassavetis,G.A. and Geiduschek,E.P. (1992) J. Biol. Chem., 267, 22562–22569. [PubMed] [Google Scholar]
- 97.Gong X., Radebaugh,C.A., Geiss,G.K., Simon,M.N. and Paule,M.R. (1995) Mol. Cell. Biol., 15, 4956–4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Geiss G.K., Radebaugh,C.A. and Paule,M.R. (1997) J. Biol. Chem., 272, 29243–29254. [DOI] [PubMed] [Google Scholar]
- 99.Bateman E., Iida,C.T., Kownin,P. and Paule,M.R. (1985) Proc. Natl Acad. Sci. USA, 82, 8004–8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schnapp A. and Grummt,I. (1991) J. Biol. Chem., 266, 24588–24595. [PubMed] [Google Scholar]
- 101.Pikaard C.S., McStay,B., Schultz,M.C., Bell,S.P. and Reeder,R.H. (1989) Genes Dev., 3, 1779–1788. [DOI] [PubMed] [Google Scholar]
- 102.Bachvarov D. and Moss,T. (1991) Nucleic Acids Res., 19, 2331–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.O’Mahony D.J. and Rothblum,L.I. (1991) Proc. Natl Acad. Sci. USA, 88, 3180–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dunaway M. (1989) Genes Dev., 3, 1768–1778. [DOI] [PubMed] [Google Scholar]
- 105.Jantzen H.-M., Admon,A., Bell,S.P. and Tjian,R. (1990) Nature, 344, 830–836. [DOI] [PubMed] [Google Scholar]
- 106.McStay B., Frazier,M.W. and Reeder,R.H. (1991) Genes Dev., 5, 1957–1968. [DOI] [PubMed] [Google Scholar]
- 107.O’Mahony D.J., Smith,S.D., Xie,W. and Rothblum,L.I. (1992) Nucleic Acids Res., 20, 1301–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jantzen H.-M., Chow,A.M., King,D.S. and Tjian,R. (1992) Genes Dev., 6, 1950–1963. [DOI] [PubMed] [Google Scholar]
- 109.Love J.J., Li,X., Case,D.A., Giese,K., Grosschedl,R. and Wright,P.E. (1995) Nature, 376, 791–795. [DOI] [PubMed] [Google Scholar]
- 110.Leblanc B., Read,C. and Moss,T. (1993) EMBO J., 12, 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bazett-Jones D.P., Leblanc,B., Herfort,M. and Moss,T. (1994) Science, 264, 1134–1137. [DOI] [PubMed] [Google Scholar]
- 112.Putnam C.D., Copenhaver,G.P., Denton,M.L. and Pikaard,C.S. (1994) Mol. Cell. Biol., 14, 6476–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moss T., Stefanovsky,V.Y. and Pelletier,G. (1998) In Paule,M.R. (ed.), Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Springer-Verlag, New York, NY, pp. 75–94.
- 114.Windle J.J. and Sollner-Webb,B. (1986) Mol. Cell. Biol., 6, 4585–4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bell S.P., Jantzen,H.-M. and Tjian,R. (1990) Genes Dev., 4, 943–954. [DOI] [PubMed] [Google Scholar]
- 116.McStay B., Sullivan,G.J. and Cairns,C. (1997) EMBO J., 16, 396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Voit R., Kuhn,A., Sander,E.E. and Grummt,I. (1995) Nucleic Acids Res., 23, 2593–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tuan J.C., Zhai,W.G. and Comai,L. (1999) Mol. Cell. Biol., 19, 2872–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Keys D.A., Lee,B.S., Dodd,J.A., Nguyen,T.T., Vu,L., Fantino,E., Burson,L.M., Nogi,Y. and Nomura,M. (1996) Genes Dev., 10, 887–903. [DOI] [PubMed] [Google Scholar]
- 120.Keener J., Dodd,J.A., Lalo,D. and Nomura,M. (1997) Proc. Natl Acad. Sci. USA, 94, 13458–13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vu L., Siddiqi,I., Lee,B.S., Josaitis,C.A. and Nomura,M. (1999) Proc. Natl Acad. Sci. USA, 96, 4390–4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Oakes M., Siddiqi,I., Vu,L., Aris,J. and Nomura,M. (2000) Mol. Cell. Biol., 19, 8559–8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Conrad-Webb H. and Butow,R.A. (1995) Mol. Cell. Biol., 15, 2420–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Smale S.T. and Tjian,R. (1985) Mol. Cell. Biol., 5, 352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lopata M.A., Cleveland,D.W. and Sollner-Webb,B. (1986) Proc. Natl Acad. Sci. USA, 83, 6677–6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Doelling J.H. and Pikaard,C.S. (1996) Nucleic Acids Res., 24, 4725–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sinclair D., Mills,K. and Guarente,L. (1998) Annu. Rev. Microbiol., 52, 533–560. [DOI] [PubMed] [Google Scholar]
- 128.Sinclair D.A. and Guarente,L. (1997) Cell, 91, 1033–1042. [DOI] [PubMed] [Google Scholar]
- 129.Reeder R.H., Roan,J.G. and Dunaway,M. (1983) Cell, 35, 449–456. [DOI] [PubMed] [Google Scholar]
- 130.Pikaard C.S. and Reeder,R.H. (1988) Mol. Cell. Biol., 8, 4282–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pape L.K., Windle,J.J., Mougey,E.B. and Sollner-Webb,B. (1989) Mol. Cell. Biol., 9, 5093–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Osheim Y.N., Mougey,E.B., Windle,J., Anderson,M., O’Reilly,M., Miller,O.L.,Jr, Beyer,A. and Sollner-Webb,B. (1996) J. Cell Biol., 133, 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dunaway M. and Dröge,P. (1989) Nature, 341, 657–659. [DOI] [PubMed] [Google Scholar]
- 134.Putnam C.D. and Pikaard,C.S. (1992) Mol. Cell. Biol., 12, 4970–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Grimaldi G., Fiorentini,P. and Di Nocera,P.P. (1990) Mol. Cell. Biol., 10, 4667–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.De Winter R.F.J. and Moss,T. (1986) Cell, 44, 313–318. [DOI] [PubMed] [Google Scholar]
- 137.Paalman M.H., Henderson,S.L. and Sollner-Webb,B. (1995) Mol. Cell. Biol., 15, 4648–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Labhart P. and Reeder,R.H. (1989) Proc. Natl Acad. Sci. USA, 86, 3155–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Banditt M., Koller,T. and Sogo,J.M. (1999) Mol. Cell. Biol., 19, 4953–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Johnson S.P. and Warner,J.R. (1989) Mol. Cell. Biol., 9, 4986–4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ju Q., Morrow,B.E. and Warner,J.R. (1990) Mol. Cell. Biol., 10, 5226–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kang J.J., Yokoi,T.J. and Holland,M.J. (1995) J. Biol. Chem., 270, 28723–28732. [DOI] [PubMed] [Google Scholar]
- 143.Morrow B.E., Johnson,S.P. and Warner,J.R. (1993) Mol. Cell. Biol., 13, 1283–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kulkens T., Van der Sande,C.A.F.M., Dekker,A.F., van Heerikhuizen,H. and Planta,R.J. (1992) EMBO J., 11, 4665–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kownin P., Bateman,E. and Paule,M.R. (1987) Cell, 50, 693–699. [DOI] [PubMed] [Google Scholar]
- 146.Bartholomew B., Kassavetis,G.A., Braun,B.R. and Geiduschek,E.P. (1990) EMBO J., 9, 2197–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Braun B.R., Bartholomew,B., Kassavetis,G.A. and Geiduschek,E.P. (1992) J. Mol. Biol., 228, 1063–1077. [DOI] [PubMed] [Google Scholar]
- 148.Baker R.E., Gabrielsen,O. and Hall,B.D. (1986) J. Biol. Chem., 261, 5275–5282. [PubMed] [Google Scholar]
- 149.Yoshinaga S.K., Boulanger,P.A. and Berk,A.J. (1987) Proc. Natl Acad. Sci. USA, 84, 3585–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang Z. and Roeder,R.G. (1996) Mol. Cell. Biol., 16, 6841–6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Oettel S., Hartel,F., Kober,I., Iben,S. and Seifart,K.H. (1997) Nucleic Acids Res., 25, 2440–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lagna G., Kovelman,R., Sukegawa,J. and Roeder,R.G. (1994) Mol. Cell. Biol., 14, 3053–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yoon J.-B., Murphy,S., Bai,L., Wang,Z. and Roeder,R.G. (1995) Mol. Cell. Biol., 15, 2019–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dean N. and Berk,A.J. (1988) Mol. Cell. Biol., 8, 3017–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yoshinaga S.K., L’Etoile,N.D. and Berk,A.J. (1989) J. Biol. Chem., 264, 10726–10731. [PubMed] [Google Scholar]
- 156.Sinn E., Wang,Z., Kovelman,R. and Roeder,R.G. (1995) Genes Dev., 9, 675–685. [DOI] [PubMed] [Google Scholar]
- 157.L’Etoile N.D., Fahnestock,M.L., Shen,Y. and Aebersold,R. (1994) Proc. Natl Acad. Sci. USA, 91, 1652–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hsieh Y.-J., Wang,Z., Kovelman,R. and Roeder,R.G. (1999) Mol. Cell. Biol., 19, 4944–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158a.Hsieh Y.-J., Kundu,T.K., Wang,Z., Kovelman,R. and Roeder,R.G. (1999) Mol. Cell. Biol., 19, 7697–7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Engelke D.R., Ng,S.-Y., Shastry,B.S. and Roeder,R.G. (1980) Cell, 19, 717–728. [DOI] [PubMed] [Google Scholar]
- 160.Ginsberg A.M., King,B.O. and Roeder,R.G. (1984) Cell, 39, 479–489. [DOI] [PubMed] [Google Scholar]
- 161.Miller J., McLachlan,A.D. and Klug,A. (1985) EMBO J., 4, 1609–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Foster M.P., Wuttke,D.S., Radhakrishnan,I., Case,D.A., Gottesfeld,J.M. and Wright,P.E. (1997) Nature Struct. Biol., 4, 605–608. [DOI] [PubMed] [Google Scholar]
- 163.Nolte R.T., Conlin,R.M., Harrison,S.C. and Brown,R.S. (1998) Proc. Natl Acad. Sci. USA, 95, 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Clemens K.R., Liao,X., Wolf,V., Wright,P.E. and Gottesfeld,J.M. (1992) Proc. Natl Acad. Sci. USA, 89, 10822–10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Archambault J., Milne,C.A., Schappert,K.T., Baum,B., Friesen,J.D. and Segall,J. (1992) J. Biol. Chem., 267, 3282–3288. [PubMed] [Google Scholar]
- 166.Camier S., Dechampesme,A.-M. and Sentenac,A. (1995) Proc. Natl Acad. Sci. USA, 92, 9338–9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kassavetis G.A., Nguyen,S.T., Kobayashi,R., Kumar,A., Geiduschek,E.P. and Pisano,M. (1995) Proc. Natl Acad. Sci. USA, 92, 9786–9790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Roberts S., Miller,S.J., Lane,W.S., Lee,S. and Hahn,S. (1996) J. Biol. Chem., 271, 14903–14909. [DOI] [PubMed] [Google Scholar]
- 169.Ruth J., Conesa,C., Dieci,G., Lefebvre,O., Dusterhoft,A., Ottonello,S. and Sentenac,A. (1996) EMBO J., 15, 1941–1949. [PMC free article] [PubMed] [Google Scholar]
- 170.Kassavetis G.A., Joazeiro,C.A.P., Pisano,M., Geiduschek,E.P., Colbert,T., Hahn,S. and Blanco,J.A. (1992) Cell, 71, 1055–1064. [DOI] [PubMed] [Google Scholar]
- 171.Aasland R., Stewart,A.F. and Gibson,T. (1996) Trends Biochem. Sci., 21, 87–88. [PubMed] [Google Scholar]
- 172.Kumar A., Kassavetis,G.A., Geiduschek,E.P., Hambalko,M. and Brent,C.J. (1997) Mol. Cell. Biol., 17, 1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Buratowski S. and Zhou,H. (1992) Cell, 71, 221–230. [DOI] [PubMed] [Google Scholar]
- 174.Colbert T. and Hahn,S. (1992) Genes Dev., 6, 1940–1949. [DOI] [PubMed] [Google Scholar]
- 175.Lopez-de-Leon A., Librizzi,M., Tuglia,K. and Willis,I. (1992) Cell, 71, 211–220. [DOI] [PubMed] [Google Scholar]
- 176.Khoo B., Brophy,B. and Jackson,S.P. (1994) Genes Dev., 8, 2879–2890. [DOI] [PubMed] [Google Scholar]
- 177.Kassavetis G.A., Kumar,A., Ramirez,E. and Geiduschek,E.P. (1998) Mol. Cell. Biol., 18, 5587–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kassavetis G.A., Bardeleben,C., Kumar,A., Ramirez,E. and Geiduschek,E.P. (1997) Mol. Cell. Biol., 17, 5299–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Colbert T., Lee,S., Schimmack,G. and Hahn,S. (1998) Mol. Cell. Biol., 18, 1682–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shen Y., Kassavetis,G.A., Bryant,G.O. and Berk,A.J. (1998) Mol. Cell. Biol., 18, 1692–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Andrau J.-C., Sentenac,A. and Werner,M. (1999) J. Mol. Biol., 288, 511–520. [DOI] [PubMed] [Google Scholar]
- 182.Lobo S.M., Tanaka,M., Sullivan,M.L. and Hernandez,N. (1992) Cell, 71, 1029–1040. [DOI] [PubMed] [Google Scholar]
- 183.Simmen K.A., Bernues,J., Lewis,J.D. and Mattaj,I.W. (1992) Nucleic Acids Res., 20, 5889–5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Taggart A.K.P., Fisher,T.S. and Pugh,B.F. (1992) Cell, 71, 1015–1028. [DOI] [PubMed] [Google Scholar]
- 185.White R.J. and Jackson,S.P. (1992) Cell, 71, 1041–1053. [DOI] [PubMed] [Google Scholar]
- 186.Wang Z. and Roeder,R.G. (1995) Proc. Natl Acad. Sci. USA, 92, 7026–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mital R., Kobayashi,R. and Hernandez,N. (1996) Mol. Cell. Biol., 16, 7031–7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Larminie C.G.C. and White,R.J. (1998) DNA Sequence, 9, 49–58. [DOI] [PubMed] [Google Scholar]
- 189.Teichmann M. and Seifart,K.H. (1995) EMBO J., 14, 5974–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Teichmann M., Dieci,G., Huet,J., Ruth,J., Sentenac,A. and Seifart,K.H. (1997) EMBO J., 16, 4708–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Joazeiro C.A.P., Kassavetis,G.A. and Geiduschek,E.P. (1994) Mol. Cell. Biol., 14, 2798–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Librizzi M.D., Brenowitz,M. and Willis,I.M. (1998) J. Biol. Chem., 273, 4563–4568. [DOI] [PubMed] [Google Scholar]
- 193.Kassavetis G.A., Riggs,D.L., Negri,R., Nguyen,L.H. and Geiduschek,E.P. (1989) Mol. Cell. Biol., 9, 2551–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Huet J. and Sentenac,A. (1993) Nucleic Acids Res., 20, 6451–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lassar A.B., Martin,P.L. and Roeder,R.G. (1983) Science, 222, 740–748. [DOI] [PubMed] [Google Scholar]
- 196.Chaussivert N., Conesa,C., Shaaban,S. and Sentenac,A. (1995) J. Biol. Chem., 270, 15353–15358. [DOI] [PubMed] [Google Scholar]
- 197.Rameau G., Puglia,K., Crowe,A., Sethy,I. and Willis,I. (1994) Mol. Cell. Biol., 14, 822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Moir R.D., Sethy-Coraci,I., Puglia,K., Librizzi,M.D. and Willis,I.M. (1997) Mol. Cell. Biol., 17, 7119–7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kassavetis G.A., Braun,B.R., Nguyen,L.H. and Geiduschek,E.P. (1990) Cell, 60, 235–245. [DOI] [PubMed] [Google Scholar]
- 200.Persinger J., Sengupta,S.M. and Bartholomew,B. (1999) Mol. Cell. Biol., 19, 5218–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bartholomew B., Durkovich,D., Kassavetis,G.A. and Geiduschek,E.P. (1993) Mol. Cell. Biol., 13, 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Werner M., Chaussivert,N., Willis,I.M. and Sentenac,A. (1993) J. Biol. Chem., 268, 20721–20724. [PubMed] [Google Scholar]
- 203.Joazeiro C.A.P., Kassavetis,G.A. and Geiduschek,E.P. (1996) Genes Dev., 10, 725–739. [DOI] [PubMed] [Google Scholar]
- 204.Setzer D.R. and Brown,D.D. (1985) J. Biol. Chem., 260, 2483–2492. [PubMed] [Google Scholar]
- 205.Carey M.F., Gerrard,S.P. and Cozzarelli,N.R. (1986) J. Biol. Chem., 261, 4309–4317. [PubMed] [Google Scholar]
- 206.Huet J., Conesa,C., Carles,C. and Sentenac,A. (1997) J. Biol. Chem., 272, 18341–18349. [DOI] [PubMed] [Google Scholar]
- 207.Jahn D., Wingender,E. and Seifart,K.H. (1987) J. Mol. Biol., 193, 303–313. [DOI] [PubMed] [Google Scholar]
- 208.Bai L., Wang,Z., Yoon,J.-B. and Roeder,R.G. (1996) Mol. Cell. Biol., 16, 5419–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Henry R.W., Sadowski,C.L., Kobayashi,R. and Hernandez,N. (1995) Nature, 374, 653–656. [DOI] [PubMed] [Google Scholar]
- 210.Henry R.W., Ma,B., Sadowski,C.L., Kobayashi,R. and Hernandez,N. (1996) EMBO J., 15, 7129–7136. [PMC free article] [PubMed] [Google Scholar]
- 211.Sadowski C.L., Henry,R.W., Kobayashi,R. and Hernandez,N. (1996) Proc. Natl Acad. Sci. USA, 93, 4289–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Yoon J.-B. and Roeder,R.G. (1996) Mol. Cell. Biol., 16, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wong M.W., Henry,R.W., Ma,B., Kobayashi,R., Klages,N., Matthias,P., Strubin,M. and Hernandez,N. (1998) Mol. Cell. Biol., 18, 368–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lescure A., Carbon,P. and Krol,A. (1991) Nucleic Acids Res., 19, 435–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Goomer R.S. and Kunkel,G.R. (1992) Nucleic Acids Res., 20, 4903–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Wanandi I., Waldschmidt,R. and Seifart,K.H. (1993) J. Biol. Chem., 268, 6629–6640. [PubMed] [Google Scholar]
- 217.Mittal V., Cleary,M.A., Herr,W. and Hernandez,N. (1996) Mol. Cell. Biol., 16, 1955–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Mittal V., Ma,B. and Hernandez,N. (1999) Genes Dev., 13, 1807–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Murphy S., Yoon,J.B., Gerster,T. and Roeder,R.G. (1992) Mol. Cell. Biol., 12, 3247–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ford E., Strubin,M. and Hernandez,N. (1998) Genes Dev., 12, 3528–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Murphy S., Pierani,A., Scheidereit,C., Melli,M. and Roeder,R.G. (1989) Cell, 59, 1071–1080. [DOI] [PubMed] [Google Scholar]
- 222.Kunkel G.R. and Danzeiser,D.A. (1992) J. Biol. Chem., 267, 14250–14258. [PubMed] [Google Scholar]
- 223.Lofquist A.K., Li,H., Imboden,M.A. and Paule,M.R. (1993) Nucleic Acids Res., 21, 3233–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kahl B.F., Li,H. and Paule,M.R. (2000) J. Mol. Biol., submitted for publication. [Google Scholar]
- 225.Kassavetis G.A., Blanco,J.A., Johnson,T.E. and Geiduschek,E.P. (1992) J. Mol. Biol., 226, 47–58. [DOI] [PubMed] [Google Scholar]
- 226.Kassavetis G.A., Kumar,A., Letts,G.A. and Geiduschek,E.P. (1998) Proc. Natl Acad. Sci. USA, 95, 9196–9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kassavetis G.A., Letts,G.A. and Geiduschek,E.P. (1999) EMBO J., 18, 5042–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bhargava P. and Kassavetis,G.A. (1999) J. Biol. Chem., 274, 26550–26556. [DOI] [PubMed] [Google Scholar]
- 229.Matsuzaki H., Kassavetis,G.A. and Geiduschek,E.P. (1994) J. Mol. Biol., 235, 1173–1192. [DOI] [PubMed] [Google Scholar]
- 230.Chedin S., Riva,M., Schultz,P., Sentenac,A. and Carles,C. (1998) Genes Dev., 12, 3857–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Bogenhagen D.F., Wormington,W.M. and Brown,D.D. (1982) Cell, 28, 413–421. [DOI] [PubMed] [Google Scholar]
- 232.Bardeleben C., Kassavetis,G.A. and Geiduschek,E.P. (1994) J. Mol. Biol., 235, 1193–1205. [DOI] [PubMed] [Google Scholar]
- 233.Wolffe A.P., Jordan,E. and Brown,D.D. (1986) Cell, 44, 381–389. [DOI] [PubMed] [Google Scholar]
- 234.Campbell F.E. and Setzer,D.R. (1991) Mol. Cell. Biol., 11, 3978–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Grummt I., Maier,U., Ohrlein,A., Hassouna,N. and Bachellerie,J.-P. (1985) Cell, 43, 801–810. [DOI] [PubMed] [Google Scholar]
- 236.Labhart P. and Reeder,R.H. (1987) Mol. Cell. Biol., 7, 1900–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Pfleiderer C., Smid,A., Bartsch,I. and Grummt,I. (1990) Nucleic Acids Res., 18, 4727–4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Labhart P. and Reeder,R.H. (1986) Cell, 45, 431–443. [DOI] [PubMed] [Google Scholar]
- 239.Reeder R.H. and Lang,W.H. (1998) In Paule,M.R. (ed.), Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Springer-Verlag, New York, NY, pp. 173–178.
- 240.Grummt I., Rosenbauer,H., Niedermeyer,I., Maier,U. and Ohrlein,A. (1986) Cell, 45, 837–846. [DOI] [PubMed] [Google Scholar]
- 241.McStay B. and Reeder,R.H. (1990) Mol. Cell. Biol., 10, 2793–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Lang W.H., Morrow,B.E., Ju,Q., Warner,J.R. and Reeder,R.H. (1994) Cell, 79, 527–534. [DOI] [PubMed] [Google Scholar]
- 243.Reeder R.H. and Lang,W.H. (1997) Trends Biochem. Sci., 22, 473–477. [DOI] [PubMed] [Google Scholar]
- 244.Lang W.H. and Reeder,R.H. (1993) Mol. Cell. Biol., 13, 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Kuhn A., Bartsch,I. and Grummt,I. (1990) Nature, 344, 559–562. [DOI] [PubMed] [Google Scholar]
- 246.Lang W.H., Platt,T. and Reeder,R.H. (1998) Proc. Natl Acad. Sci. USA, 95, 4900–4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Morrow B.E., Ju,Q. and Warner,J.R. (1993) Mol. Cell. Biol., 13, 1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Evers R., Smid,A., Rudloff,U., Lottspeich,F. and Grummt,I. (1995) EMBO J., 14, 1248–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Evers R. and Grummt,I. (1995) Proc. Natl Acad. Sci. USA, 92, 5827–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Mason S.W., Sander,E.E. and Grummt,I. (1997) EMBO J., 16, 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Mason S.W., Wallisch,M. and Grummt,I. (1997) J. Mol. Biol., 268, 229–234. [DOI] [PubMed] [Google Scholar]
- 252.Langst G., Blank,T.A., Becker,P.B. and Grummt,I. (1997) EMBO J., 16, 760–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Gerber J.K., Gögel,E., Berger,C., Wallisch,M., Müller,F., Grummt,I. and Grummt,F. (1997) Cell, 90, 559–567. [DOI] [PubMed] [Google Scholar]
- 254.Cozzarelli N.R., Gerrard,S.P., Schlissel,M., Brown,D.D. and Bogenhagen,D.F. (1983) Cell, 34, 829–835. [DOI] [PubMed] [Google Scholar]
- 255.Bogenhagen D.F. and Brown,D.D. (1981) Cell, 24, 261–270. [DOI] [PubMed] [Google Scholar]
- 256.Allison D.S. and Hall,B.D. (1985) EMBO J., 4, 2657–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Gottlieb E. and Steitz,J.A. (1989) EMBO J., 8, 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Gottlieb E. and Steitz,J.A. (1989) EMBO J., 8, 841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Maraia R.J., Kenan,D.J. and Keene,J.D. (1994) Mol. Cell. Biol., 14, 2147–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Yoo C.J. and Wolin,S.L. (1997) Cell, 89, 393–402. [DOI] [PubMed] [Google Scholar]
- 261.Campbell F.E. and Setzer,D.R. (1992) Mol. Cell. Biol., 12, 2260–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Lin-Marq N. and Clarkson,S.G. (1998) EMBO J., 17, 2033–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Dieci G. and Sentenac,A. (1996) Cell, 84, 245–252. [DOI] [PubMed] [Google Scholar]
- 264.Maraia R.J. (1996) Proc. Natl Acad. Sci. USA, 93, 3383–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Buttgereit D., Pflugfelder,G. and Grummt,I. (1985) Nucleic Acids Res., 13, 8165–8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Brun R.P., Ryan,K. and Sollner-Webb,B. (1994) Mol. Cell. Biol., 14, 5010–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Mahajan P.B. and Thompson,E.A.,Jr (1990) J. Biol. Chem., 265, 16225–16233. [PubMed] [Google Scholar]
- 268.Schnapp A., Pfleiderer,C., Rosenbauer,H. and Grummt,I. (1990) EMBO J., 9, 2857–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Mahajan P.B., Gokal,P.K. and Thompson,E.A.,Jr (1990) J. Biol. Chem., 265, 16244–16247. [PubMed] [Google Scholar]
- 270.Gokal P.K., Mahajan,P.B. and Thompson,E.A.,Jr (1990) J. Biol. Chem., 265, 16234–16243. [PubMed] [Google Scholar]
- 271.Schnapp A., Schnapp,G., Erny,B. and Grummt,I. (1993) Mol. Cell. Biol., 13, 6723–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Yamamoto R.T., Nogi,Y., Dodd,J.A. and Nomura,M. (1996) EMBO J., 15, 3964–3973. [PMC free article] [PubMed] [Google Scholar]
- 273.Milkereit P. and Tschochner,H. (1998) EMBO J., 17, 3692–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Paule M.R., Iida,C.T., Perna,P.J., Harris,G.H., Knoll,D.A. and D’Alessio,J.M. (1984) Nucleic Acids Res., 12, 8161–8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Schnapp A., Rosenbauer,H. and Grummt,I. (1991) Mol. Cell. Biochem., 104, 137–147. [DOI] [PubMed] [Google Scholar]
- 276.Schnapp G., Schnapp,A., Rosenbauer,H. and Grummt,I. (1994) EMBO J., 13, 4028–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Grummt I. (1998) In Paule,M.R. (ed.), Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Springer-Verlag, New York, NY, pp. 135–154.
- 278.Muramatsu M., Hanada,K., Yamamoto,K. and Song,C.Z. (1998) In Paule,M.R. (ed.), Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Springer-Verlag, New York, NY, pp. 95–106.
- 279.Song C.Z., Hanada,K., Yano,K., Maeda,Y., Yamamoto,K. and Muramatsu,M. (1994) J. Biol. Chem., 269, 26976–26981. [PubMed] [Google Scholar]
- 280.Hanada K., Song,C.Z., Yamamoto,K., Yano,K., Maeda,Y., Yamaguchi,K. and Muramatsu,M. (1996) EMBO J., 15, 2217–2226. [PMC free article] [PubMed] [Google Scholar]
- 281.Seither P., Zatsepina,O., Hoffmann,M. and Grummt,I. (1997) Chromosoma, 106, 216–225. [DOI] [PubMed] [Google Scholar]
- 282.Gadal O., Mariotte-Labarre,S., Chedin,S., Quemeneur,E., Carles,C., Sentenac,A. and Thuriaux,P. (1997) Mol. Cell. Biol., 17, 1787–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Zhang J. and Jacob,S.T. (1990) Mol. Cell. Biol., 10, 5177–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Ghosh A.K., Hoff,C.M. and Jacob,S.T. (1993) Gene, 125, 217–222. [DOI] [PubMed] [Google Scholar]
- 285.Hoff C.M. and Jacob,S.T. (1993) Biochem. Biophys. Res. Commun., 190, 747–753. [DOI] [PubMed] [Google Scholar]
- 286.Hoff C.M., Ghosh,A.K., Prabhakar,B.S. and Jacob,S.T. (1994) Proc. Natl Acad. Sci. USA, 91, 762–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Niu H. and Jacob,S.T. (1994) Proc. Natl Acad. Sci. USA, 91, 9101–9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Ghosh A.K. and Jacob,S.T. (1998) In Paule,M.R. (ed.), Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Springer-Verlag, New York, NY, pp. 229–240.
- 289.Pla M. and Mahouy,G. (1991) Nouv. Rev. Fr. Hematol., 33, 489–491. [PubMed] [Google Scholar]
- 290.Dieci G., Duimio,L., Coda-Zabetta,F., Sprague,K.U. and Ottonello,S. (1993) J. Biol. Chem., 268, 11199–11207. [PubMed] [Google Scholar]
- 291.Waldschmidt R. and Seifart,K.H. (1992) J. Biol. Chem., 267, 16359–16364. [PubMed] [Google Scholar]
- 292.Meibner W., Holland,R., Waldschmidt,R. and Seifart,K.H. (1993) Nucleic Acids Res., 21, 1013–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Kang J.J., Auble,D.T., Ranish,J.A. and Hahn,S. (1995) Mol. Cell. Biol., 15, 1234–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Wang Z., Luo,T. and Roeder,R.G. (1997) Genes Dev., 11, 2371–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Wang Z. and Roeder,R.G. (1998) Mol. Cell, 1, 749–757. [DOI] [PubMed] [Google Scholar]
- 296.Schuster C., Myslinski,E., Krol,A. and Carbon,A. (1995) EMBO J., 14, 3777–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Schaub M., Myslinski,E., Schuster,C., Krol,A. and Carbon,P. (1997) EMBO J., 16, 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Inostroza J.A., Mermelstein,F.H., Ha,I., Lane,W.S. and Reinberg,D. (1992) Cell, 70, 477–489. [DOI] [PubMed] [Google Scholar]
- 299.Kim S., Na,J.G., Hampsey,M. and Reinberg,D. (1997) Proc. Natl Acad. Sci. USA, 94, 820–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.White R.J., Khoo,B.C.-E., Inostroza,J.A., Reinberg,D. and Jackson,S.P. (1994) Science, 266, 448–450. [DOI] [PubMed] [Google Scholar]
- 301.Chesnokov I., Chu,W.-M., Botchan,M.R. and Schmid,C.W. (1996) Mol. Cell. Biol., 16, 7084–7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.White R.J., Trouche,D., Martin,K., Jackson,S.P. and Kouzarides,T. (1996) Nature, 382, 88–90. [DOI] [PubMed] [Google Scholar]
- 303.Chu W.-M., Wang,Z., Roeder,R.G. and Schmid,C.W. (1997) J. Biol. Chem., 272, 14755–14761. [DOI] [PubMed] [Google Scholar]
- 304.Larminie C.G.C., Cairns,C.A., Mital,R., Martin,K., Kouzarides,T., Jackson,S.P. and White,R.J. (1997) EMBO J., 16, 2061–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Cairns C.A. and White,R.J. (1998) EMBO J., 17, 3112–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Sutcliffe J.E., Cairns,C.A., McLees,A., Allison,S.J., Tosh,K. and White,R.J. (1999) Mol. Cell. Biol., 19, 4255–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Cavanaugh A.H., Hempel,W.M., Taylor,L.J., Rogalsky,V., Todorov,G. and Rothblum,L.I. (1995) Nature, 374, 177–180. [DOI] [PubMed] [Google Scholar]
- 308.Voit R., Schafer,K. and Grummt,I. (1997) Mol. Cell. Biol., 17, 4230–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Budde A. and Grummt,I. (1999) Oncogene, 19, 1119–1124. [DOI] [PubMed] [Google Scholar]
- 310.Nasmyth K. (1996) Nature, 382, 28–29. [DOI] [PubMed] [Google Scholar]
- 311.White R.J. (1997) Trends Biochem. Sci., 22, 77–80. [DOI] [PubMed] [Google Scholar]
- 312.Foe V.E., Wilkinson,L.E. and Laird,C.D. (1976) Cell, 9, 131–146. [DOI] [PubMed] [Google Scholar]
- 313.Scheer U. (1978) Cell, 13, 535–549. [DOI] [PubMed] [Google Scholar]
- 314.Gottesfeld J.M. and Bloomer,L.S. (1980) Cell, 21, 751–760. [DOI] [PubMed] [Google Scholar]
- 315.Louis C., Schedl,P., Samal,B. and Worcel,A. (1980) Cell, 22, 387–392. [DOI] [PubMed] [Google Scholar]
- 316.Chipev C.C. and Wolffe,A.P. (1992) Mol. Cell. Biol., 12, 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Englander E.W. and Howard,B.H. (1995) J. Biol. Chem., 270, 10091–10096. [DOI] [PubMed] [Google Scholar]
- 318.Lucchini R. and Sogo,J.M. (1998) In Paule,M.R. (ed.), Transcription of Eukaryotic Ribosomal RNA Genes by RNA Polymerase I. Springer-Verlag, New York, NY, pp. 253–274.
- 319.Lucchini R., Pauli,U., Braun,R., Koller,T. and Sogo,J.M. (1987) J. Mol. Biol., 196, 829–843. [DOI] [PubMed] [Google Scholar]
- 320.Dammann R., Lucchini,R., Koller,T. and Sogo,J.M. (1995) Mol. Cell. Biol., 15, 5294–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Lucchini R. and Sogo,J.M. (1995) Nature, 374, 276–280. [DOI] [PubMed] [Google Scholar]
- 322.Wolffe A. (1998) Chromatin Structure and Function. Academic Press, London, UK.
- 323.Pruss D., Bartholomew,B., Persinger,J., Hayes,J., Arents,G., Moudrianakis,E.N. and Wolffe,A.P. (1996) Science, 274, 614–617. [DOI] [PubMed] [Google Scholar]
- 324.Lee D.Y., Hayes,J.J., Pruss,D. and Wolffe,A.P. (1993) Cell, 72, 73–84. [DOI] [PubMed] [Google Scholar]
- 325.Bouvet P., Dimitrov,S. and Wolffe,A.P. (1994) Genes Dev., 8, 1147–1159. [DOI] [PubMed] [Google Scholar]
- 326.Kandolf H. (1994) Proc. Natl Acad. Sci. USA, 91, 7257–7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Carey M.F. and Singh,K. (1988) Proc. Natl Acad. Sci. USA, 85, 7059–7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Han M. and Grunstein,M. (1988) Cell, 55, 1137–1145. [DOI] [PubMed] [Google Scholar]
- 329.Morse R.H., Roth,S.Y. and Simpson,R.T. (1992) Mol. Cell. Biol., 12, 4015–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Burnol A.-F., Margottin,F., Huet,J., Almouzni,G., Prioleau,M.-N., Mechali,M. and Sentenac,A. (1993) Nature, 362, 475–477. [DOI] [PubMed] [Google Scholar]
- 331.Marsolier M.-C., Tanaka,S., Livingstone-Zatchej,M., Grunstein,M., Thoma,F. and Sentenac,A. (1995) Genes Dev., 9, 410–422. [DOI] [PubMed] [Google Scholar]
- 332.Kundu T.K., Wang,Z. and Roeder,R.G. (1999) Mol. Cell. Biol., 19, 1605–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Russanova V.R., Driscoll,C.T. and Howard,B.H. (1995) Mol. Cell. Biol., 15, 4282–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Englander E.W., Wolffe,A.P. and Howard,B.H. (1993) J. Biol. Chem., 268, 19565–19573. [PubMed] [Google Scholar]
- 335.Schmid C.W. (1991) Nucleic Acids Res., 19, 5613–5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Kochanek S., Renz,D. and Doerfler,W. (1993) EMBO J., 12, 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Liu W.-M., Maraia,R.J., Rubin,C.M. and Schmid,C.W. (1994) Nucleic Acids Res., 22, 1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Juttermann R., Hosokawa,K., Kochanek,S. and Doerfler,W. (1991) J. Virol., 65, 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Liu W.-M. and Schmid,C.W. (1993) Nucleic Acids Res., 21, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Besser D., Gotz,F., Schulze,F.K., Wagner,H., Kroger,H. and Simon,D. (1990) FEBS Lett., 269, 358–362. [DOI] [PubMed] [Google Scholar]
- 341.Fedoroff N.V. and Brown,D.D. (1978) Cell, 13, 701–716. [DOI] [PubMed] [Google Scholar]
- 342.Miller J.R., Cartwright,E.M., Brownlee,G.G., Fedoroff,N.V. and Brown,D.D. (1978) Cell, 13, 717–725. [DOI] [PubMed] [Google Scholar]
- 343.Brown D.D. and Gurdon,J. (1977) Proc. Natl Acad. Sci. USA, 74, 2064–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Nightingale K. and Wolffe,A.P. (1995) J. Biol. Chem., 270, 4197–4200. [DOI] [PubMed] [Google Scholar]
- 345.Kass S.U., Pruss,D. and Wolffe,A.P. (1997) Trends Genet., 13, 444–449. [DOI] [PubMed] [Google Scholar]
- 346.Lefebvre O., Carles,C., Conesa,C., Swanson,R.N., Bouet,F., Riva,M. and Sentenac,A. (1992) Proc. Natl Acad. Sci. USA, 89, 10512–10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Marck C., Lefebvre,O., Carles,C., Riva,M., Chaussivert,N., Ruet,A. and Sentenac,A. (1993) Proc. Natl Acad. Sci. USA, 90, 4027–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Swanson R.N., Conesa,C., Lefebvre,O., Carles,C., Ruet,A., Quemeneur,E., Gagon,J. and Sentenac,A. (1991) Proc. Natl Acad. Sci. USA, 88, 4887–4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Arrebola R., Manaud,N., Rozenfeld,S., Marsolier,M.-C., Lefebvre,O., Carles,C., Thuriaux,P., Conesa,C. and Sentenac,A. (1998) Mol. Cell. Biol., 18, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Manaud N., Arrebola,R., Buffin-Meyer,B., Lefebvre,O., Voss,H., Riva,M., Conesa,C. and Sentenac,A. (1998) Mol. Cell. Biol., 18, 3191–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Deprez E., Arrebola,R., Conesa,C. and Sentenac,A. (1999) Mol. Cell. Biol., 19, 8042–8051. [DOI] [PMC free article] [PubMed] [Google Scholar]