Abstract
The appearance of modular proteins is a widespread phenomenon during the evolution of proteins. The combinatorial arrangement of different functional and/or structural domains within a single polypeptide chain yields a wide variety of activities and regulatory properties to the modular proteins. In this review, we will discuss proteins, that in addition to their catalytic, transport, structure, localization or adaptor functions, also have segments resembling the helix-loop-helix EF-hand motifs found in Ca2+-binding proteins, such as calmodulin (CaM). These segments are denoted CaM-like domains (CaM-LDs) and play a regulatory role, making these CaM-like proteins sensitive to Ca2+ transients within the cell, and hence are able to transduce the Ca2+ signal leading to specific cellular responses. Importantly, this arrangement allows to this group of proteins direct regulation independent of other Ca2+-sensitive sensor/transducer proteins, such as CaM. In addition, this review also covers CaM-binding proteins, in which their CaM-binding site (CBS), in the absence of CaM, is proposed to interact with other segments of the same protein denoted CaM-like binding site (CLBS). CLBS are important regulatory motifs, acting either by keeping these CaM-binding proteins inactive in the absence of CaM, enhancing the stability of protein complexes and/or facilitating their dimerization via CBS/CLBS interaction. The existence of proteins containing CaM-LDs or CLBSs substantially adds to the enormous versatility and complexity of Ca2+/CaM signaling.
Keywords: α-Actinin, Calcineurin, Calpain, Epidermal growth factor receptor, Glycerol-3-phosphate dehydrogenase, NADPH oxidases, Na+/H+ exchanger, Plasma membrane Ca2+-ATPase, Protein kinases
Introduction
The calcium ion plays a fundamental signaling role in all eukaryotic organisms. The transduction of the Ca2+ signal is mostly achieved by a great variety of Ca2+-binding proteins containing helix–loop–helix EF-hand motifs. The name of this motif is derived from the Ca2+-binding sites of parvalbumin, where it was first identified [1]. It is formed by a loop of 12 amino acids rich in acidic residues, which coordinates Ca2+, and links the two α-helical segments E and F in a perpendicular way. The phylogeny of these proteins shows an evolutionary history, in which gene duplication played a fundamental role. By this mechanism, a predicted ancient gene encoding a single EF-hand-containing protein can be seen as a precursor yielding a plethora of different proteins working as Ca2+-binding sensors and buffers [2–4]. Nevertheless, this did not occur in all cases, because events of convergent evolution took place as well [3, 4]. The best-studied EF-hand containing protein is calmodulin (CaM). The structure of CaM consists in two globular lobes located at the N- and C-termini of the protein each containing two EF-hand Ca2+-binding sites and a flexible linker connecting both globular lobes. This structure permits the interaction of CaM with its targets adopting different conformations, and to work as a linker between different proteins and/or segments of the same polypeptide chain. This contributes to the dimerization of identical or different proteins, and to attain structural conformers of the target protein modulating their functionality (reviewed in [5]). CaM is universally expressed in all eukaryotic cells and interacts with several hundred enzymes, channels, and other proteins without catalytic or transport activity including transcription factors, adaptors, signaling and structural proteins, regulating a myriad of cellular functions (reviewed in [5–10]). Although CaM mainly works in a Ca2+-dependent manner, it also regulates the function of many proteins in a Ca2+-free form (apo-CaM) [11, 12]. CaM has an identical sequence in all vertebrates but is coded from two or three independent genes, depending on the species [13, 14]. Plants, however, express several CaM isoforms coded from different genes (for example seven in Arabidopsis) (reviewed in [15, 16]). In some cases, CaM may work as an integral component of the target proteins, rather than just interacting with them when the concentration of Ca2+ rises. This consists in the integration of CaM as a constitutive subunit of the functional target protein even when the Ca2+ concentration is low. Well-known examples of such proteins are phosphorylase b kinase (reviewed in [17]), the inducible nitric oxide synthase isoform (reviewed in [18]), and it has been proposed to be the case as well for some ion channels (reviewed in [19]).
Another level of complexity of Ca2+ sensors/target signaling consists of the presence of EF-hand modules in proteins that have an enzymatic, transport, adaptor or another functional domain. This enabled the proteins to be directly regulated by Ca2+ and thereby transmit the signal downstream without the need of a distinct Ca2+ sensor protein, such as CaM or other proteins containing EF-hand motifs. Nevertheless, there are examples, in which in addition to the direct binding of Ca2+ to the EF-hands of the CaM-like domain (CaM-LD) containing proteins, Ca2+/CaM binds to these proteins and plays a regulatory role (see “Protein kinases (CDPKs)”). This adds complexity to the functionality of this group of proteins.
In the first section of this review, we will discuss a series of these CaM-LD containing proteins on a structural and functional level. This will include some enzymes (kinases, proteases, dehydrogenases and oxidases), and proteins with transport, hormonal, adaptor or structural functions. In the second section, we will discuss CaM-binding proteins that in addition to the CBS also contain a CLBS as defined by Jarrett and Madhavan [20]. There is no sequence homology of the CBSs or CLBSs in different proteins, except that they are rich in basic and acidic residues, respectively. These CLBSs lack EF-hand motifs, and are proposed to interact electrostatically with the CBSs when the latter are free of CaM. Therefore, they could provide a regulatory decoy system by binding to the CBS and preventing the action of Ca2+/CaM leading to autoinhibition. We will discuss the presence and function of CBS/CLBS motifs in some enzymes, transport proteins, and growth factor receptors. To highlight the communality of these regulatory domains in this diverse group of proteins, Fig. 1 depicts the organization of the major domains of some enzymes classified as containing either CaM-LD or CLBS/CBS domains discussed in this review. This arrangement allows either the direct and/or indirect (CaM-mediated) regulation of these enzymes by Ca2+ with great precision. Interestingly, the presence of an autoinhibitory domain that may have an independent identity, or to coincide with theirs CLBS or CBS appears to be common features of these enzymes to avoid unwanted activation in the absence of Ca2+ and/or the Ca2+ sensor CaM.
Fig. 1.
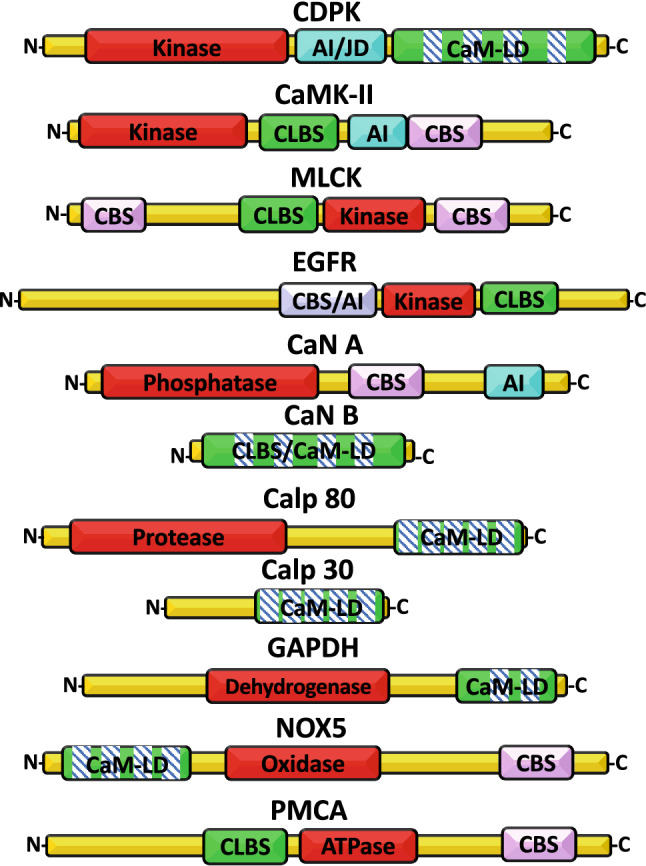
Domain organization of some calmodulin-like domain (CaM-LD)- and calmodulin-like binding site (CLBS)-containing proteins with enzymatic activity. The figure depicts the linear organization of the relevant domains of some enzymes with either a CaM-LD or CLBS. The domains shown are: catalytic site (red); CaM-LD or CLBS (green and EF-hands, boxes with bars, where applicable); autoinhibitory (AI)/junction (JD) domains (magenta); and calmodulin-binding site (CBS) (pink). CDPK CaM-like domain protein kinase, CaMK-II CaM-dependent protein kinase-II, MLCK myosin light-chain kinase, EGFR epidermal growth factor receptor, CaN A calcineurin A, CaN B calcineurin B, Calp 80 calpain 80 kDa subunit, Calp 30 calpain 30 kDa subunit, GAPDH glycerol-3-phosphate dehydrogenase, NOX5 NADPH oxidase 5, PMCA plasma membrane Ca2+-ATPase. The length of the proteins and different domains is not drawn to scale. The CBS and AI domains of the EGFR overlap [183]
Proteins with EF-hand calmodulin-like domains (CaM-LDs)
The occurrence of proteins containing a segment with an enzymatic (e.g., kinase or protease), or non-enzymatic domain (e.g., with transport or adaptor cytoskeletal function) and a segment resembling the helix-loop-helix EF-hand motifs found in Ca2+-binding proteins, such as CaM, could be the result of the fusion of two genes with completely different functions and evolutionary origin. This has been proposed to be the case for calpains, a group of Ca2+-dependent cysteine endo-peptidases [21], as well as plant and protist calmodulin-like domain protein kinases [22, 23]. It is likely that this is also the origin of other proteins containing canonical EF-hand CaM-LDs, which are the focus of this section. Table 1 presents the amino acid sequence of the Ca2+-binding sites of selected proteins containing EF-hands.
Table 1.
Sequences of Ca2+-binding sites from selected EF-hand-containing proteins
Acidic amino acid residues are marked (red)
CDPK CaM-like domain protein kinase, CPK Ca2+-dependent protein kinase
aUndefined isoenzyme
bPotential Ca2+-binding sites are underlined
cBinds 11–14 calcium ions per subunit, the indicated sequences correspond to Ca2+-binding sites in EGF-like domains 2 and 3
CaM-LD proteins with enzymatic activity
Among the proteins with calmodulin-like domains and enzymatic activity most thoroughly studied are enzymes implicated in phosphorylation processes such as certain protein kinases in plants and protists denoted CDPKs, and proteases of the calpain family in vertebrates. In addition, enzymes involved in redox processes such as some dehydrogenases and oxidases also have calmodulin-like domains.
Protein kinases (CDPKs)
In many higher plants, algae and protists, a multi-gene family of Ca2+-dependent serine/threonine protein kinases denoted CDPKs presenting many isoforms was identified. These proteins contain as major structural features a variable N-terminal region, a catalytic domain similar to the one in calmodulin-dependent kinases (CaMKs), a regulatory junction domain connected to an adjoined C-terminal regulatory domain similar to CaM named the CaM-like domain (CaM-LD) containing four EF-hand Ca2+ binding sites, although some isoforms with fewer sites have been identified (Figs. 1, 2a) [24]. The phylogeny of this extensive protein kinase family resulting from the fusion of an ancestral kinase gene and a CaM gene has been reviewed [23]. CDPKs have a high affinity for Ca2+ and do not require an exogenous Ca2+-binding protein for their activity, as they are directly activated by this cation due to their in-built CaM-like domain (reviewed in [25, 26]). Through their enzymatic activity, these kinases transmit the Ca2+ signal generated by hormones, light and other effectors to diverse substrates, thereby regulating distinct physiological processes.
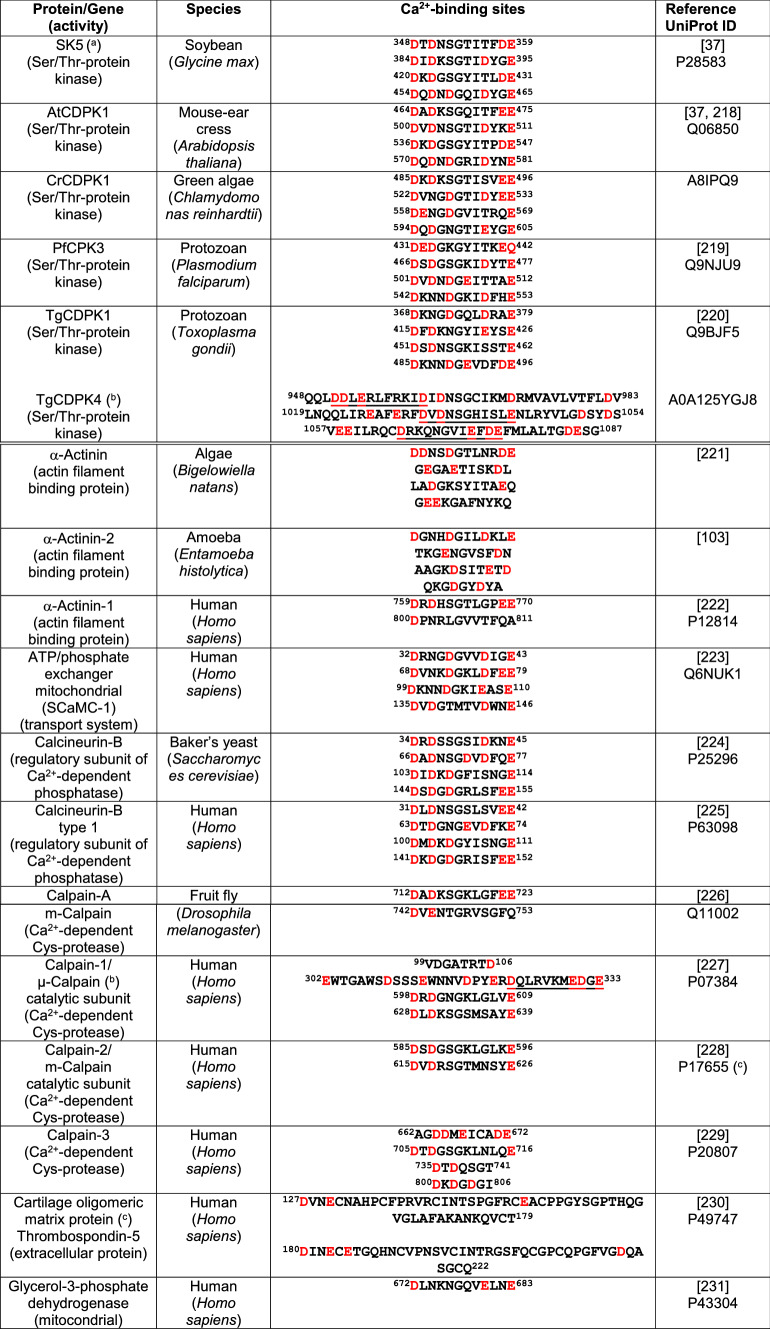
Fig. 2.
Structure and Ca2+-mediated activation of a calmodulin-like domain protein kinase. a The figure depicts the X-ray crystallographic structure of Ca2+-dependent protein kinase 1 (AtCDPK1) from Arabidopsis thaliana at 2 Å resolution (Q06850 CDPK1_ARATH based on template 4m97A, residue range 142–588) taken from UniProt SWISS-MODEL database. The crystallographic structure in the left panel is shown using a rainbow color code going from the N-terminal (blue) to the C-terminal (red). The Ca2+-binding sites in the four EF-hands are highlighted in the right panel in different colors indicating the amino acid residues involved. b The figure depicts a model for the Ca2+-dependent activation of a CaM-like domain protein kinase (CDPK). In the absence of Ca2+, a non-physiological condition, when the four EF-hands of the CDPK are free of Ca2+ (apo-CDPK), a sector of the junction domain (JD) blocks the catalytic site of the protein kinase (PK) rendering the enzyme inactive (top panel). In basal conditions, when the cytosolic Ca2+ concentration is very low (< 50–100 nM) the high-affinity Ca2+-binding sites located in the distal EF-hands 3 and 4 are occupied and both of them interact with the JD (marked in green), the catalytic site is partially released and the enzyme becomes partially active (central panel). The interrogation mark indicates a discrepancy in the literature as to whether binding of Ca2+ to EF-hands 3 and 4 is sufficient to at least partially release the auto-inhibition (see references [27–31]). When the cytosolic Ca2+ concentration increases at saturating concentrations (> 0.5–1 µM), the two low-affinity Ca2+-binding sites located in EF-hands 1 and 2 are also occupied by Ca2+, and all EF-hands interact with the JD (marked in green) which is totally released from the catalytic site rendering the enzyme fully active (bottom panel). Ca2+ ions are represented by gray spheres. CaM-LD calmodulin-like domain.
The junction between the kinase domain and the CaM-LD appears to exert an auto-inhibitory role acting as a pseudosubstrate-like structure blocking the catalytic site [27–29]. It was demonstrated that when the CaM-LD interacts with the junction this auto-inhibition is partially released. Nevertheless, for the total release of autoinhibition, not only Ca2+ binding to the high-affinity sites in the C-lobe but also Ca2+ occupancy of the lower affinity sites in the N-lobe is required [30]. Binding of Ca2+ to the C-lobe appears to increase the affinity of this cation for the N-lobe, facilitating activation of the kinase in a cooperative manner (reviewed in [31]). Figure 2b shows a putative model depicting the Ca2+-mediated activation of a CDPK. Nevertheless, structural analysis of the CDPK of Arabidopsis thaliana, AtCPK1, showed that this junction almost exclusively interacts with the high Ca2+ affinity C-lobe of the CaM-LD to release autoinhibition. This suggests activation of the kinase even at low basal Ca2+ concentrations [32]. Although this mechanism does not appear to be sufficient to attain full activation of the enzyme, mutations at the CaM-LD and the linker connecting this domain with the kinase domain block its enzymatic activity, and the activity of a mutant lacking the CaM-LD can be reconstituted upon addition of either this isolated domain or CaM [33–35]. CDPKs and CDPK-related kinases from plants and protists are able to undergo autophosphorylation, and a common locus of the autophosphorylation sites in the N-terminal variable domain has been identified [36].
The first Ca2+-dependent kinase (CDPK) was discovered in 1991 in soybean (Glycine max), later denoted CDPKα [37], followed by the identification of isoforms CDPKβ and CDPKγ with different kinetic properties and Ca2+ affinities indicating distinct functional roles [38]. Further, CDPKs were later on found in many other higher plants, green algae and protists. Table 1 shows that the EF-hands in these proteins are highly conserved, especially the Ca2+ coordinating acidic residues D and E at position 1 and 12 (highlighted in red), and Table 2 shows similarities in the sequence of the autoinhibitory/junction domain. Interestingly, a phenylalanine (underlined in bold) in higher plant CDPKs appears to have an important functional role [32], similarly to a highly conserved segment in protozoan CDPKs (underlined in bold). Figure 2a depicts the crystallographic structure of A. thaliana CDPK1 highlighting the four EF-hands Ca2+ binding sites in the C-terminal CaM-LD.
Table 2.
Sequence of selected CDPK autoinhibitory/junction domains
| CDPK | Species | Autoinhibitory/junction domain | Identity (%) UniProt ID |
|---|---|---|---|
| SK5 | Soybean (Glycine max) | 298APDKPLDSAVLSRLKQFSAMNKLKKMALRVI328 |
– |
| OsCPK11 | Rice (Oryza sativa)_ | 377APDKPLDSAVLSRLKQFSAMNKLKKMALRVI407 |
100 |
| NtCDPK3 | Tobacco (Nicotiana tabacum) | 378APDKPLDSAVLSRMKQFSAMNKLKKMALRVI408 |
96.8 |
| AtCPK11 | Mouse-ear cress (Arabidopsis thaliana) | 290APDKPLDPAVLSRLKQFSQMNKIKKMALRVI320 |
90.3 |
| PfCPK1a | Protozoan (Plasmodium falciparum) | 344LSNMRKFEGSQKLAQAAILFI364 |
– |
| TgCDPK1b | Protozoan (Toxoplasma gondii) |
320VPSLDNAILNIRQFQGTQKLAQAALLYMGSKLTSQDETKELTAIFHKMDK369 420GYIEYSEFVTVAMDRKTLLSRERLERAFRMFD451 |
81.8 |
In higher plant CDPKs the residues in bold indicate changes with respect to soybean CDPK, and a conserved functionally important phenylalanine residue is underlined in bold [32]. In P. falciparum and T. gondii the underlined segment is highly conserved and may represent a functionally important segment
aSmall segment of the autoinhibitory/junction domain [32]
bTwo helical segments of the autoinhibitory/junction domain [232]
Some CDPKs bind and are regulated by CaM. It was shown that A. thaliana CPK28 binds Ca2+/CaM at its junctional domain with high affinity inhibiting its activity. However, when CPK28 is autophosphorylated it primes the enzyme for Ca2+ binding at low concentration, and this reduces the inhibitory effect of Ca2+/CaM [39]. Of interest, the activation loop of some CDPKs contains acidic amino acid residues, which prevent Ser/Thr phosphorylation [40]. Therefore, it could be speculated that this acidic site may work as well as a CLBS (see “Calmodulin-binding proteins with non-EF-hand CaM-like binding sites (CLBSs)”) possibly interacting with the CBS of these enzymes. The junction domain of soybean CDPKα induces a significant structural change in the C-terminal domain of the isolated CaM-LD in the presence of Ca2+ without forming a stable complex as determined by NMR spectroscopy [41]. Moreover, the N- and C-lobes of the CaM-LD adopt a side-by-side position, unlike CaM, and similar to calcineurin B [42]. Using FRET, it was demonstrated that the CaM-LD is partially unfolded in the absence of Ca2+ and folds upon Ca2+ binding [43] (see Fig. 2b).
Autophosphorylation is a Ca2+-dependent process and considered to be a requirement for plant CDPK activation, as demonstrated in groundnut (Arachis hypogaea) CDPK [44]. This CDPK, an example of a group of atypical structurally similar CDPKs, containing a bipartite NLS at their auto-inhibitory junction domain and non-canonical EF-hands, binds Ca2+ with very low affinity. The presence of a bipartite NLS is shared by many other CDPKs including ginger (Zingiber oficinale) ZoCDPK1 and Arabidopsis AtCPK30 among others [45]. As expected, this kinase is localized in the nucleus and its junction domain in its activated form has been shown to interact with importins, which are localized at the nuclear pores participating in the translocation process [46]. As autophosphorylation at a tyrosine residue has been reported recently, it was suggested that CDPKs could work as dual-specific protein kinases [47]. In some instances, however, autophosphorylation has been shown to be Ca2+-independent and to induce inhibition of the enzyme [48]. The functionality of these kinases appears to be diverse as they act on many different targets. Table 3 collects some functional roles of selected examples of these kinases in different plants and other organisms. Relevant in this context is the fundamental role of CDPKs in defense response to pathogens and herbivores attack (reviewed in [49]).
Table 3.
Biological functions and some properties of selected CDPKs
| CDPK isoform | Species | Biological functions and properties | References |
|---|---|---|---|
| MsCPK3 | Alfalfa (Medicago sativa) | Sensitive to CaM antagonists. Responsive to auxin, role in embryogenesis and thermal stress response. Phosphorylates a CDK inhibitor enhancing its inhibitory activity and blocking the cell cycle | [233–235] |
| VfCDPKa | Bean (Vicia faba) |
Phosphorylates and inhibits the K+ rectifying channel KAT1 in guard cells; leads to closure of stomas This CDPK works upstream of a 48 kDa Ca2+-independent kinase responsive to abscisic acid involved in stomatal closure |
[236] [237] |
|
DcCPK1 CRK |
Carrot (Daucus carota) |
Sensitive to CaM antagonists. Activated by acidic phospholipids CDPK-related kinase. Degenerated EF-hands. N-terminal extension with myristoylation consensus sequence |
[240] |
| CaCPK1, CaCPK2 | Chickpea (Cicer arietinum) | Sensitive to CaM antagonists | [241] |
|
ZmCDPKa ZmCPK11 (ZmCPKp54) CRPK1, CRPK2 |
Corn (Zea mays) |
Phosphorylates the C-terminal region of plasma membrane H+-ATPase in vitro, no effect on ATPase activity. Phosphorylates ADF/cofilin; leads to actin cytoskeleton disassembly Involved in drought response Involved in wound response. Activated by acidic phospholipids; present in seeds, plant seedling and other organs Root-specific isoforms |
[40] [246] |
| CsCDPK5 | Cucumber (Cucumis sativus) |
Upregulated by phytohormones (benzyladenine, abscisic acid, gibberellic acid and indole acetic acid) in a light-dependent and independent manner N-Myristoylation motif absent |
[247] |
| ZoCDPK1 | Ginger (Zingiber officinale) | Involved in response to multiple stresses including high salt and drought. Contains a bipartite NLS at its junction domain | [45] |
| AhCPK2 | Groundnut (Arachis hypogaea) | Translocates to the nucleus in response to hypertonic stress. Non-functional second EF-hand. Very low Ca2+ affinity (Kd = 392 µM). Bipartite NLS at the junction domain | [46] |
| IiCPK2 | Indigowood (Isatis indigotica) | Involved in stress response to cold and high salinity | [248] |
| LjCDPKa | Birds’ foot trefoil (Lotus japonicas) | Phosphorylates aquaporin LIMP-2 (nodulin-26 orthologue) in N2-fixing root nodules | [249] |
|
AtCDPK AK1-6Ha CPK9 |
Mouse-ear cress (Arabidopsis thaliana) |
Express many isoforms. The ~ 72-kDa isoform has a N-terminal extension and is synergistically activated by Ca2+ and phospholipids Involved in drought response Phosphorylates and activates NADPH oxidase when ectopically expressed in tomato protoplasts enhancing ROS production. Phospho-NADPH oxidase dephosphorylated by protein phosphatase 2A. Involved in parasite defense mechanisms Transferred DNA (T-DNA) of the mutagenic tumor-inducing bacteria Agrobacterium tumefaciens is inserted in the gene inactivating this isoform |
[40] [252] [253] |
|
PiCDPK1 PiCDPK2 |
Petunia (Petunia inflata) | Controls the growth and polarity of the pollen tube | [254] |
| PrCDPKa | Poppy (Papaver rhoeas) | Phosphorylates a pollen 26-kDa protein to prevent self-pollination | [255] |
| StCPK1 | Potato (Solanum tuberosum) | CDPK-related kinase with canonical EF-hands | [256] |
| CmCPK1 | Pumpkin (Cucurbita maxima) | N-terminal proteolytically cleavaged form enters the phloem and phosphorylates sap proteins | [257] |
|
OsCDPKa SPKa |
Rice (Oryza sativa) |
Involved in drought response Phosphorylates sucrose synthase; regulates starch storage in grain |
[40] |
| SbCDPKa | Sorghum (Sorghum bicolor) | Involved in drought response | [40] |
|
CDPKα CDPKβ CDPKγ GmCDPKa |
Soybean (Glycine max) |
CDPKα higher Ca2+ affinity (K0.5 = 0.06 µM) than CDPKβ (K0.5 = 0.4 µM) and CDPKγ (K0.5 = 1 µM) Phosphorylates the symbiosome membrane protein nodulin-26 in N2-fixing root nodules regulating its voltage-dependent channel activity |
[260] |
| SoCDPKa | Spinach (Spinacia oleracea) |
Phosphorylates leaf nitrate reductase; decreases nitrate assimilation via 14-3-3 protein binding Phosphorylates and inhibits leaf sucrose-phosphate synthase arresting sucrose biosynthesis |
[261] [262] |
| FaCDPK1 | Strawberry (Fragaria ananassa) | Fruit development, transcript increases at low temperature. Three EF-hands only | [263] |
| BvCDPKa | Sugar beet (Beta vulgaris) | Phosphorylates and inhibits plasma membrane H+-ATPase in root cells | [264] |
| NtCDPK1 | Tobacco (Nicotiana tabacum) | Responsive to phytohormones and tissue wounding | [265] |
|
LeCPK1 LeCPK2 LeCRK1 |
Tomato (Solanum lycopersicum) |
LeCPK1 phosphorylates the C-terminus of plasma membrane H+-ATPase. LeCPK2 responsive to phytohormones and mechanical injuries. Myristoylation of LeCPK1 N-terminus required for plasma membrane localization; Ca2+-binding sites of low (Kd = 55 µM) and high (Kd = 0.6 µM) affinity LeCPK2 very high affinity for Ca2+ (K0.5 = 45–49 nM) Degenerated CaM-LD unable to bind Ca2+. Contains a CaM-binding site as assayed in vitro Involved in ripening |
[269] |
| TaCDPK1 | Wheat (Triticum aestivum) | Expression upregulated by sucrose | [270] |
| PtCDPK/WbCDPK | Winged bean (Psophocarpus tetragonolobus) |
Sensitive to CaM antagonists The 70-kDa isoform is unable to undergo autophosphorylation Dephosphorylated by serine-phosphatase WbPP |
[272] |
| EtCDPKa | Protozoan (Eimeria tenella) |
Involved in the invasion of host cells by the sporozoite Located in the apical filament-like structure of the sporozoite |
[60] |
|
PfCDPK1 PfCDPK3 PfCDPK4 |
Protozoan (Plasmodium falciparum) |
Phosphorylates the protease PfSERA5. Implicated in invasion and the egress of merozoites from the erythrocyte Implicated in gametogenesis of the parasite Sensitive to CaM antagonists |
[219] [275] |
| PbCDPK4 | Protozoan (Plasmodium berghei) | Translates the Ca2+ signal generated by xanthurenic acid in gametocytes inducing its differentiation to mature gametes | [276] |
|
TgCDPK1 TgCDPK4 |
Protozoan (Toxoplasma gondii) |
Involved in motility, host cell attachment and the invasive capacity of the parasite. Expressed in tachyzoites Phosphorylates aldolase-1 forming part of the glideosome apparatus which is required for parasite invasiveness |
[107] [277] |
ADF actin-depolymerizing factor, CaM calmodulin, CDK cyclin-dependent kinase, CDPK CaM-like domain protein kinase, CPK Ca2+-dependent protein kinase, NLS nuclear localization sequence, ROS reactive oxygen species
aIsoform not described
CaM-like domain protein kinases similar to the above-described plant enzymes have been identified in green algae [50, 51] and protists (see Table 3). Protist CDPKs exhibit a more elaborated mechanism of activation. Binding of Ca2+ to the CaM-LD releases the auto-inhibition, as in the case of plant CDPKs, but the junction/CaM-LD complex further interacts with a segment of the kinase domain and a ‘latch’ with the variable N-terminal domain (not shown in Fig. 2b). This contributes to the allosteric activation of the enzyme. Nevertheless, it is likely that plant CDPKs may have a similar activation mechanism (reviewed in [52]).
In the protozoan Plasmodium falciparum, the causing agent of malaria, up to seven CDPKs are expressed, what aroused interest as potential therapeutic targets due to their important role in transducing essential Ca2+-mediated signaling as well as the fact that mammals do not express similar proteins (reviewed in [53]). The structural and functional features of the Ca2+-binding sites at the N- and C-lobes of the CaM-like domain of CDPK3 in this parasite has been studied in detail using recombinant proteins of each lobe [54]. It was found that the C-lobe was constitutively occupied by Ca2+, as it could not be prepared in its Ca2+-free form, and was found in a dimeric form. This is in contrast to the N-lobe, which did not form dimers and changed its structure upon binding of a single Ca2+ ion. This may indicate if confirmed with the full-length protein, that the C-lobe is Ca2+ saturated even at basal intracellular concentration and that activation occurs due to structural changes induced by Ca2+ binding to the N-lobe as suggested by the authors of this report. Using the CDPK4 isoform of this parasite, a model of Ca2+-mediated activation of this kinase was proposed, in which the junctional domain occupies its catalytic domain, acting as a pseudo-substrate when the CaM-LD is in its Ca2+-free form, as in other CDPKs [55] (see Fig. 2b). Upon Ca2+ binding, the junctional domain is detached exposing this site and allowing its auto-phosphorylation at Thr234, which activates the enzyme.
The causative agent of toxoplasmosis, the pathogenic intracellular protozoan Toxoplasma gondii, expresses Ca2+-dependent/phospholipid-independent CDPKs. These kinases could be valid targets to prevent invasion of the parasite transmitted by cats gravely affecting the fetus in pregnant women. In this context, imidazopyridine, an inhibitor of PKG used against this and other coccidial parasites, has been shown to specifically inhibit as well CDPK1 in T. gondii with very high (sub-nanomolar) affinity [56]. The so-called “bumped” kinase inhibitors are those that compete for ATP. Thereby they inhibit protozoan kinases but not mammalian kinases because the latter usually contain a bulky gatekeeper residue in the catalytic site that prevents entry of the inhibitor (reviewed in [57]). These CDPK inhibitors have been found useful for the treatment of protozoal diseases. However, resistance to this class of inhibitors has been found when MAPK1, but not CDPK1, in T. gondii is mutated [58]. Based on these and other results it has been suggested to develop CDPK-based vaccines against T. gondii (reviewed in [59]). Parasitic protozoans of the genus Eimeria, also expressing CDPKs, are the agents causing coccidiosis [60].
The calpain protein family
Calpains form a family of Ca2+-dependent cysteine neutral proteases with at least fourteen isoforms in human coded from different genes. These proteases play a variety of important roles, as they perform restricted controlled proteolysis on target substrates modulating in this fashion their functionality (reviewed in [61–63]). They also participate in the onset of different pathologies including cancer (reviewed in [64]). The major functional classification system of calpains is based on the affinity of these enzymes for Ca2+. This includes μ-calpain (calpain-1) with high affinity for Ca2+ (3–50 μM), and m-calpain (calpain-2) with a very low affinity for this cation (0.2–1 mM). A third type is calpain-3, which is also dependent on Na+ and has some peculiar properties, such as the propensity to become reactivated by intermolecular complementation of the fragments after auto-proteolysis. Ca2+ binding induces the autoproteolytic activation of calpains. Interestingly, calpain-5 was shown to be able to undergo Ca2+-dependent processing in a human neuroblastoma cell line even though it does not contain a CaM-LD [65].
Calpains are heterodimers constituted of 80 kDa and 30 kDa subunits. Five EF-hands domains are found in both subunits at their C-terminal region, of which only three or four in the large subunit and some in the smaller subunit are able to bind Ca2+ (Fig. 1). The catalytic domain of calpain is divided in two sub-domains, IIa and IIb, separated by cleft allowing access of the substrate to the catalytic center (Fig. 3a) [66, 67]. The activity of a recombinant μ-calpain lacking the CaM-LD (domain IV) was shown to be independent of Ca2+, and a chimeric μ/m-calpain in which the native CaM-LD (domain IV) [68] of μ-calpain was replaced by the one of m-calpain presents the highest affinity for Ca2+ [69]. The physiological concentration of cytosolic Ca2+ appears to be too low for full activation of either μ-calpain or m-calpain in living cells. Several mechanisms have been suggested to solve this dilemma, from increasing Ca2+ availability by co-localization with Ca2+-channels, and/or to increase the effective Ca2+ affinity in vivo by binding to membranes and subsequent activation by acidic phospholipids or regulation by a phosphorylation/dephosphorylation mechanism, and the existence of a hypothetical Ca2+ activator similar to CaM (reviewed in [70]). However, some of these concepts have been challenged, and alternative views have been suggested, and it has been proposed that the relatively low Ca2+ affinity to fulfill in vivo working conditions may represent an evolutionary safety device to prevent unwanted proteolysis (reviewed in [71]). The differential Ca2+ affinity of μ-calpain and m-calpain is not due to differences in the sequence of the EF-hand Ca2+-binding sites which are almost 100% conserved, but it has been ascribed to differences in the N-terminal and the C-terminal EF-hand-containing domain IV of the 80 kDa catalytic subunit, contributing 20–25% and 65%, respectively, to the difference in Kd for Ca2+ between the two calpain types as determined by mutagenesis studies [72].
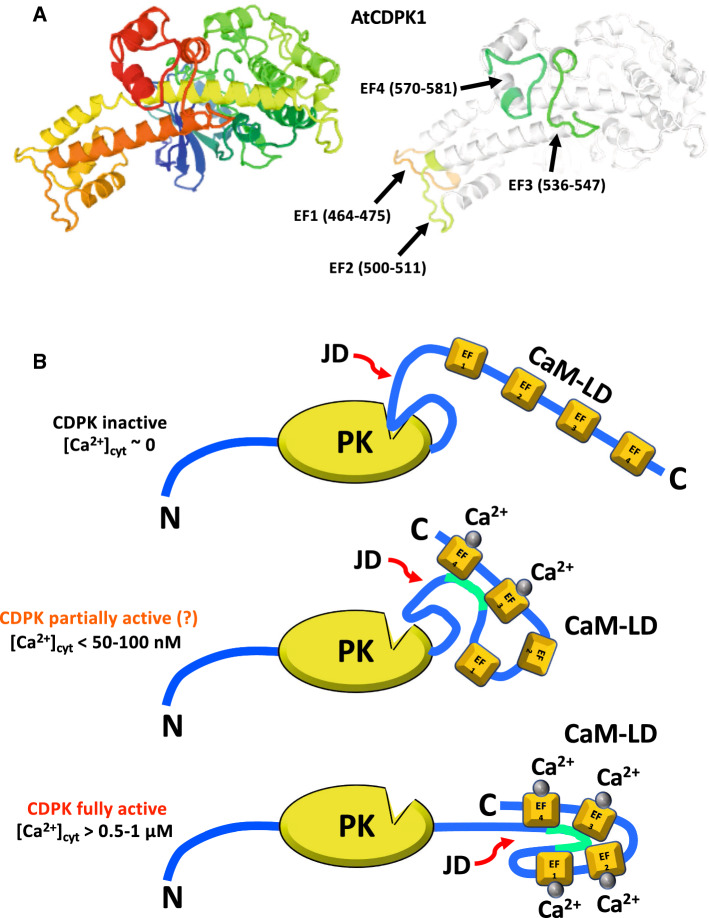
Fig. 3.
Structure and function of the Ca2+ binding sites of m-calpain, α-actinin, ATP/phosphate exchanger and calcineurin. a X-ray crystallographic structure of the rat m-calpain/calpain-2 dimer with Ca2+ bound in complex with the physiological inhibitor calpastatin at 2.4 Å resolution. The structure was obtained from PDB: ID 3BOW. The EF-hand with Ca2+-bound (gray spheres), the loop forming the acidic electrostatic switch (AES) and domains IIa, IIb, III and IV in the large catalytic subunit (pink), and domain VI in the small regulatory subunit (blue) are indicated [66]. The red arrow shows the cleft between the two catalytic sub-domains IIa and IIb [66, 67]. A segment (B-peptide, residues 595–629) of calpastatin (brown) blocking the catalytic site in domains IIa and IIb is also shown [68, 212]. b X-ray crystallographic structure of a human muscle α-actinin-2 (α-Act-2) dimer (residues 16–874) at 3.5 Å resolution is shown in pink and blue colors. The location of EF-hands 1 and 2 and the actin-binding domain (ABD) are indicated (left). Cryo-electron microscope derived structure of tandem calponin-homology domains (residues 1–109) of human skeletal muscle α-actinin-3 (α-Act-3) bound to a F-actin filament formed by G-actin subunits (residues 1–374) at 15 Å resolution. Each calponin-1 homology domain (CH) and actin subunit is represented in different colors (right). The structures were obtained from PDB ID: 4D1E and 3LUE, respectively [104, 213, 214]. c The CaM-like binding domain (CaM-BD), also denoted Ca2+ sensor, of human mitochondrial ATP/phosphate exchanger-1 (SCaMC-1) located at its N-terminus is shown in a rainbow color code (residues 22–176). The four EF-hands bound to Ca2+ (gray sphere) and diethylene glycol molecule are indicated. The structure was obtained from PDB ID 4N5X [121]. d The figure depicts the calcineurin A (CaN-A, pink)/calcineurin B (CaN-B, blue) heterodimer in which the four EF-hands of the regulatory CaN-B subunit are saturated with Ca2+ (cyan spheres), and the calmodulin-binding site (CBS) of CaN-A (residues 391–414, dashed box) is already separated from the CaN-B binding region (residues 348–370) considered to be a calmodulin-like binding site (CLBS). Binding of the Ca2+/calmodulin complex (Ca2+/CaM, rainbow color coded) to the CBS allows the C-terminal auto-inhibitory site (AIS) (residues 457–482) to be separated from the Zn/Fe-containing (brown spheres) catalytic site (residues 71–342) releasing the auto-inhibition and inducing full activation of the enzyme. The structures were obtained from PDB ID: 1AUI (human CaN-A/CaN-B heterodimer at 2.1 Å resolution) [215] and 1CLL (human CaM at 1.7 Å resolution) [216]. See text and Ref. [135] for more details on the activation mechanism
Due to its high affinity for Ca2+, µ-calpain can be isolated by Ca2+-dependent affinity chromatography using an immobilized peptide containing the CBS of the plasma membrane Ca2+-ATPase, indicative of the interaction between the CaM-LD of calpain with the CBS in analogy to CaM. However, the CaM-LD of the irreversibly inactivated protease only mildly activated the hydrolytic activity of the ATPase, in contrast to the strong effect exerted by CaM [73]. As deduced from the crystallographic structure of m-calpain in its Ca2+-free form, the CaM-LD is separated by a β-sandwich domain III also denoted C2-like domain, which binds phospholipids, participating together with potential non-EF-hand Ca2+-binding sites in a cooperative way during Ca2+-induced enzyme activation [68]. This structure also shows that in the absence of Ca2+ key amino acids in the catalytic site are separated by more than 10 Å, rendering the enzyme inactive, as more proximity is required to become active upon Ca2+ binding [67, 74]. Interestingly, a tissue-specific calpain homologue identified in Drosophila has an insert in the CaM-LD containing a hydrophobic segment, that likely participates in membrane attachment [75].
In addition to the CaM-LD found in the large subunit, the 30 kDa subunit has two domains denoted V and VI, and the CaM-LD is located in domain VI [68] (Fig. 1), possibly contributing to Ca2+-induced activation of the complex. The CaM-LDs in both m-calpain subunits interact with CaM antagonists, in addition to Ca2+ [76]. Interestingly, the isolated CaM-LDs of the large subunits of μ-calpain and m-calpain and their small subunits all appear to dimerize in the absence of Ca2+, recovering their monomeric status upon addition of this cation [77]. This reversible association suggests the involvement of this region in the formation of active heterodimers and/or the formation of enzymatic oligomers. The crystallographic structure of the full-length Ca2+-free m-calpain heterodimer reveals how Ca2+ binding to a highly negatively charged loop in domain III, denoted electrostatic switch, may induce the necessary molecular reorganization to create a functional catalytic center located between domains IIa and IIb [68] (Fig. 3a). In addition, Ca2+ could as well facilitate the binding of the enzyme to membranes, where the concentration of this cation is high, which may explain the low requirement for Ca2+ of this calpain isoform [66].
Calpains have various modulatory activities. An example of modulatory calpain action is the removing of the CaM-binding region of the plasma membrane Ca2+-ATPase inducing in this manner its CaM-independent activity [78, 79]. Very strikingly, both μ-calpain and m-calpain appear to have some preference to proteolyze CaM-binding proteins within regions enriched in proline, aspartic/glutamic acids, and serine/threonine residues, which are denoted PEST sequences (reviewed in [80]). The voracity of calpain for CaM-binding proteins is intriguing, and it could be speculated that the CBS of the target protein is the region recognized by the CaM-LD of the protease to recruit its targets acting as a surrogate of CaM. In this context, it has been shown that the PEST domain of IκBα, an inhibitor of the transcription factor NFκB, binds to the CaM-LD of µ-calpain previous to its degradation by the protease. As a consequence, this allows the transfer of sequestered cytosolic NFκB to the nucleus and subsequent activation of gene transcription [81]. Nevertheless, the proteolytic capacity of calpain toward the CaM-binding proteins caldesmon and calponin does not depend on the CBS of those proteins, as deletion of these domains does not affect their cleavage by the protease [82].
Calpains are regulated by a physiological inhibitory protein denoted calpastatin, which requires Ca2+ for its function and is found in several isoforms, ranging in size from ~ 19 to 85 kDa (reviewed in [61, 62]). Both, the CaM-LD of the 80 kDa subunit of μ-calpain and the one of the 30 kDa subunit, participate in Ca2+-dependent calpastatin binding with very high affinity via one of their N-terminal inhibitory repetitive acidic regions [83–85] (Fig. 3a). However, it is not yet clear how binding of the different inhibitory domains of calpastatin to the CaM-LD of calpain participate in the inhibition of its enzymatic activity [86]. The penta-peptide LSEAL, with high homology to conserved regions of calpastatin, binds to the CaM-LD of m-calpain and μ-calpain mimicking the inhibition of calpastatin on the catalytic activity [87].
Redox enzymes
GPDH is a tetrameric enzyme that catalyzes the formation of glycerol-3-phosphate from dihydroxyacetone-phosphate, which is important for shuttling glycerol-3-phosphate across the inner mitochondrial membrane. This enzyme also has nitrosylase activity, and is strongly activated by Ca2+ enhancing the affinity of the enzyme for its substrate. It contains two EF-hand motifs (Fig. 1). However, one of them does not participate in Ca2+ binding. The presence of a lysine in this modified EF-hand prevents the binding of this cation. Moreover, this enzyme is not inhibited by CaM antagonists [88].
NADPH oxidases are membrane-bound enzymes involved in the generation of anion superoxide (O2−) both in animal and plant cells. They are mainly involved in host defense, as for example inducing the so-called oxygen burst by leukocytes and macrophages. In addition, they participate in signaling processes, as exerted by their product and other ROS species (reviewed in [89, 90]). Human NOX5 contains a CaM-LD located at its N-terminus with three canonical EF-hands Ca2+-binding sites and an additional non-canonical EF-hand. The EF-hands 1 and 2 pair binds Ca2+ with low affinity, while the EF-hands 3 and 4 pair binds this cation with high affinity. The binding of Ca2+ to the CaM-LD activates the enzyme by interaction of this domain with the catalytic site [91]. In addition, the C-terminus of NOX5 contains a CaM-binding site. Ca2+-dependent CaM binding to this site increases the Ca2+ sensitivity of the N-terminus CaM-LD [92] (Fig. 1). Furthermore, in bovine aorta endothelial cells CaMK-II phosphorylates NOX5 at different Ser/Thr residues activating the enzyme, where phosphorylation of Ser475 appears to play a particular important role in the activation process, as determined by mutagenesis studies [93]. In plants, NADPH oxidases are also known as respiratory burst oxidase homologues (RBOHs). In Arabidopsis six RBOHs were initially identified containing two EF-hand Ca2+-binding sites located at the N-terminal region [94, 95]. Ca2+ binding to the EF-hands of these oxidases induces activation of the enzyme synergistically with a concomitant phosphorylation process [95, 96]. In rice RBOH two additional EF-hand-like motifs were identified. This enzyme is able to form dimers, where the two canonical EF-hands are swapped adopting the dimer a head-to-tail conformation, that together with the EF-hand-like motifs has a three-dimensional structure similar to calcineurin B and recoverin [97].
The cytoskeleton protein α-actinin
The group of CaM-LD-containing proteins is not restricted to the group of enzymes, but is extended to proteins with other functional roles, such as proteins with mechanical/structural functions of the cytoskeleton. This underscores the functional versatility of CaM-LD proteins. In this section we will discuss the case of the cytoskeleton protein α-actinin. α-Actinin is a stick shape protein that crosslinks F-actin filaments forming bundles of stress fibers connected to focal adhesions. This multi-domain protein belongs to the spectrin family and is structurally organized with an N-terminal actin-binding domain (ABD), formed by two sequential calponin-homology domains, followed by a linker that connects four sequential spectrin repeat segments and a CaM-LD at its C-terminus (reviewed in [98, 99]). In the case of α-spectrin, of the two EF-hand motifs located in the C-terminus only the EF-hand closer to the spectrin repeat segments binds Ca2+ [100]. α-Actinin makes anti-parallel homo-dimers through the formation of a ternary complex between the N-terminal actin-binding domain plus the adjacent linker region of one monomer, and the C-terminal CaM-LD of the apposed monomer [101] (Fig. 3b). The CaM-LD has four EF-hand motifs that bind Ca2+ only in the non-muscle isoforms (1 and 4) while the muscle isoforms (2 and 3) lack the capacity to bind Ca2+ due to mutations in key amino acids involved in the coordination of this cation [102]. Structural data of the CaM-LD of α-actinin-2 from Entamoeba histolytica in its native conformation obtained by NMR in the absence of Ca2+ shows that the linker region between the two lobes harboring the EF-hand motifs is flexible, as in the case of CaM [103]. In human α-actinin-2 similar NMR structural studies show that in the absence of Ca2+ the linker region is indeed flexible, but upon Ca2+ binding a stiffness of the linker region occurs limiting the rotation of the N- and C-terminal lobes leading to the loss of the capacity to crosslink actin through the juxtaposed actin-binding domain [104]. Modeling studies suggest that binding of the CaM-LD to the linker region regulates the distance between F-actin and the actin-binding site of α-actinin [105]. The gene coding for the α-actinin-binding protein UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (GNE) is mutated in some congenital myopathies (reviewed in [106]). This enzyme binds with high affinity (Kd ≈ 9 nM) to α-actinin-2 and with tenfold lower affinity to α-actinin-1. The CaM-LD of α-actinin-2 participates in the interaction with GNE. Most relevant in connection with myopathies is the finding that the myopathic M743T mutation in GNE leads to a tenfold decrease of its affinity for α-actinin-2 [107]. The α-actinin-binding protein palladin is part of the dense region of stress fibers and microfilaments where it plays an important role in the regulation of the actin cytoskeleton (reviewed in [108]). The interaction site of palladin has been mapped to the CaM-LD of α-actinin [109]. In addition, it was shown that the interaction between palladin and α-actinin is of functional importance as it plays a role in directing α-actinin to specific subcellular foci.
In non-muscle cells α-actinin bridges the actin filaments in the cytoskeleton but loses the capacity to bind actin when the concentration of Ca2+ exceed values above 0.1 μM [99]. Based on structural data it has been proposed that PIP2 binds to the linker between the actin-binding domain and the first spectrin-like repeat of α-actinin, and therefore it may regulate the interaction of its CaM-LD to this linker region [110]. Cryo-electron microscopy reveals that in smooth muscle α-actinin the CaM-LD of one monomer is oriented at approximately an angle of 90° to the axis between the two calponin-homology domains of the appose monomer laying just between both of them [111]. By inference, the authors of this report proposed that binding of Ca2+ to the CaM-LD of non-muscle α-actinin facilitates its interaction with the linker between the two calponin-homology domains separating both of them, and therefore explaining the loss of capacity to interact with actin in the presence of high calcium. Vinculin forms part of focal adhesions, and it has been shown that a segment of this adaptor protein, comprised between amino acids 1–258, interacts with the CaM-LD of smooth muscle α-actinin [112]. NMR structural analysis of the CaM-LD of the major non-muscle α-actinin-1 indicates that a single Ca2+ ion binds to the N-lobe of this domain [104]. Based on this structure, these authors proposed a model for the regulation of the interaction between α-actinin and actin confirming the above-proposed view that Ca2+ works negatively on this interaction by stabilizing the actin binding sites. α-Actinin also participates in linking transmembrane receptors, channels and other signaling proteins to the cytoskeleton, regulating in this manner a variety of signaling pathways (reviewed in [99, 113]). α-Actinin-4 contains a domain that interacts with nuclear receptors, such as estrogen receptor-α (ERα). This potentiates its transcriptional activity. However, when the CaM-LD of α-actinin, which is required to interact with histone deacetylase-4, is lacking its co-activator activity is lost [114]. The CaM-LD of α-actinin also interacts with the long C-terminal tail of adenosine A2A receptor (A2AR) in the absence of Ca2+. This fixes the receptor to the actin cytoskeleton, as α-actinin acts as a bridging molecule, and participates in receptor internalization upon ligand binding. When the cytosolic Ca2+ concentration rises, Ca2+/CaM binds to the site where the CaM-LD of α-actinin binds to the receptor because of its higher affinity. This reduces ligand-dependent A2AR internalization and its detachment from the cytoskeleton [115].
The α-actinin isoforms expressed in striated muscle are localized at the Z-disk and analogous dense bodies. They crosslink the actin filaments of adjacent sarcomeres to hold them together forming a stabilizing lattice in the contractile apparatus (reviewed in [98, 99]). The C-lobe of the CaM-LD of muscle α-actinin interacts with a region close to the N-terminus of titin, a gargantuan filamentous ~ 3 MDa protein expanding the sarcomere from the Z-disk to the M-line. This interaction induces a structural change of the titin/α-actinin-binding region into a helical conformation, participating in this manner in the assembly of the Z-disk as shown by NMR analysis using the CaM-LD sequence of α-actinin and a Z-repeat of titin [116]. The interaction of the CaM-LD with the Z-repeat of titin is very weak and changes depending on the angle in which the force is applied using optical tweezers at a single molecular level. Therefore, a model was proposed in which multiple cooperative interactions are required to attain stable titin anchoring to the Z-disk while individual components are dynamically exchanged [117]. The three-dimensional structure of muscle α-actinin has shown that upon Ca2+ binding the C-terminal CaM-LD gets close to the linker region between the two calponin-homology domains forming the actin-binding region. This suggests a Ca2+-dependent interaction of α-actinin with the actin filaments [118] as later also shown for non-muscle α-actinin [111], described above.
Other CaM-LD proteins
The mitochondrial inner membrane has an ATP transport system unrelated to the ATP/ADP exchanger, and is hence insensitive to the ATP/ADP translocase inhibitor atractyloside [119]. This transporter was identified as an ATP-Mg/phosphate exchanger, with at least eleven isoforms in mammals that are denominated SCaMCs. It has highly conserved orthologues in lower eukaryotes as for example in yeast (denoted Sal1p). It is positively regulated by micromolar levels of Ca2+ and contain six transmembrane segments and an N-terminal extension corresponding to a CaM-LD with four EF-hands (Fig. 3c), although some isoforms lack one or several of these EF-hand motifs. The CaM-LD of SCaMC faces the space between the inner and outer mitochondrial membrane and therefore senses the Ca2+ concentration that is at equilibrium with the cytosol (reviewed in [119]). The N-terminal extension of SCaMC1 (NTD) comprising the first 193 amino acids has been crystallized in its Ca2+-bound form showing that the EF-hands are sequestered by an endogenous helical segment in a compact structure [120]. NMR studies indicate that the apo-NTD could interact with the transmembrane domain [121]. The carrier works as an electroneutral exchanger between ATP-Mg and divalent phosphate. Although it is able to work in both directions, the net influx and efflux of ATP into the mitochondria depends on the pH in the medium, as the uptake of phosphate into the mitochondria is driven by the pH gradient (alkaline inside), and the chemical potential of phosphate secondarily drives ATP-Mg uptake in exchange for phosphate exit (reviewed in [119]). At least three ATP/phosphate exchangers exist in Arabidopsis. Interestingly, in vitro analysis shows that the three isoforms prefer ATP-Ca, over of ATP-Mg, exchanged by phosphate [122]. From mutagenesis experiments, it was concluded that the Ca2+ effect on ATP transport is not primarily dependent on the CaM-LD of these transporters.
COMP, also named thrombospondin-5, is a pentameric Ca2+-binding glycoprotein of the cartilage extracellular matrix that interacts with multiple other proteins of the extracellular matrix, including growth factors, integrins and extracellular proteases among others (reviewed in [123, 124]). COMP forms a lattice to present the growth factors to the cells, participating in this manner in chondrogenesis (reviewed in [125]). Its structure comprises an N-terminal domain followed by four EGF-like repeats, eight CaM-like repeats and a globular C-terminal region. The five thrombospondin family members contain similar Ca2+-binding repeats, although the general structure of these proteins and their oligomerization status differ (reviewed in [123, 124]). COMP/thrombospondin-5 is particularly abundant surrounding the chondrocytes. Interestingly, mutations or single amino acid deletions affecting their CaM-like domains most likely in their Ca2+-binding capacity are responsible for the occurrence of pseudoachondroplasia and multiple epiphyseal dysplasias in humans. These are autosomal dominant congenital conditions characterized by short limbs and hands, ligament laxity and scoliosis among other skeletal anomalies leading to the onset of osteoarthrosis [126–128].
Genes coding for atypical short class XIV myosins (type A and B) were found in three species of Plasmodium. These proteins contain a conserved catalytic head that binds actin and hydrolyzes ATP, and a neck region, however lacking the characteristic tail region of other myosins. These class XIV myosins are expressed during the motile stages of the parasite implying a functional role during the invasion process. The light-chain of myosin-B contains a CaM-LD at its C-terminus that has similarity to other EF-hand proteins such as CaM. However, this protein does not bind Ca2+, as inferred from the lack of change in the circular dichroism spectrum upon addition of this cation. The CaM-LD of myosin-light chain-B interacts with the neck region of myosin-B. Blast analysis of the genome of T. gondii shows that the sequence of this CaM-LD is conserved in a related myosin-B light-chain [129].
Clonorchiasis is a liver infestation by the planaria worm Clonorchis sinensis that is transmitted by contaminated food. Tegumental proteins in the outer surface membrane of this parasite are vital for its physiology and therefore interesting targets for vaccine development. A tegumental-allergen-like protein (CsTAL3) has been identified in this worm containing a CaM-LD at its N-terminus and a dynein light chain-like domain at its C-terminus. The CaM-LD has two Ca2+-binding motifs, however, only the closest to the N-terminus binds Ca2+. Proteins with similar CaM-LDs were identified in Schistosoma mansoni, another pathogenic worm. CsTAL3 is antigenic and implicated in the host immune response mediated by IgA but not IgG. The crystal structure of this protein has been described [130].
FSH is a heterodimeric hormone synthesized in the anterior pituitary gland that regulates growth, development, maturation and reproductive processes in both males and females in concert with other hormones. Using a battery of synthetic peptides corresponding to the sequence of the β-subunit of human FSH to study receptor interaction, it was uncovered that a peptide, corresponding to amino acids 1–15 of the hormone was able to bind Ca2+ and presents similarities in affinity to the third EF-hand of CaM [131]. The authors of this report suggested that this Ca2+-binding site may participate in the interaction of the hormone with its receptor. Whether the binding site of this peptide in the receptor is a non-canonical CaM-LD, although likely, remains an open issue. Another intriguing issue not yet clarified, is whether there is any physiological relevance to the fact that this peptide was able to form transmembrane Ca2+ channels in liposomes.
Calmodulin-binding proteins with non-EF-hand calmodulin-like binding sites (CLBSs)
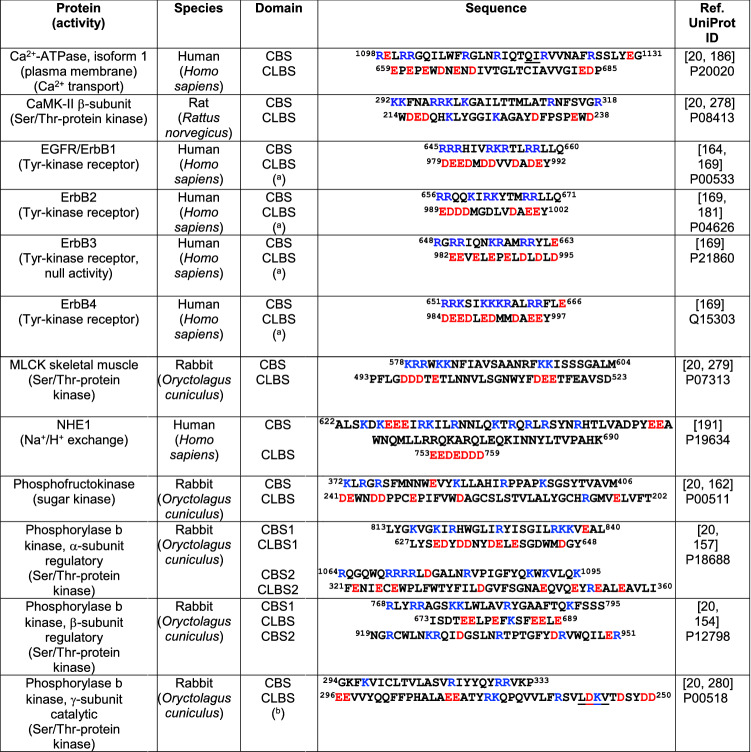
In this section, we will discuss different types of proteins, unrelated to the phylogenetic origin of those discussed in the previous section, in which gene fusion events appear to be the origin. It refers to the existence of regions in CaM-binding proteins that are proposed to interact with their CBS in the absence of CaM. These regions, lacking EF-hand motifs but having binding properties similar to CaM, are generally anionic and are denoted CaM-like binding sites (CLBSs), in contrast to the CBSs that are rich in basic residues. The CLBSs are predicted to bind the potent CaM antagonist melittin, which is present in bee venom. Several CaM-binding enzymes, including MLCK, phosphofructokinase, phosphorylase b kinase, plasma membrane Ca2+-ATPase and CaMK-II have been proposed to comply with these criteria as their putative CLBSs were identified [20]. Nevertheless, experimental evidence for this assertion is lacking for some of these proteins, including CaMK-II and MLCK. In this section we also include calcineurin, where the CBS and CLBS are located in distinct subunits. As discussed by Jarrett and Madhavan [20], the functional role of the CLBS is to exert an auto-inhibitory function by blocking the CBS. In addition, a flip–flop model for CaM-binding enzyme activation was proposed, where shuttling between the inactive form of the enzyme (CBS occupied by the CLBS) and the active conformation (CBS occupied by Ca2+/CaM), was envisioned. This also explains why the proteolytic removal of the CBS of some enzymes induces their activation in the absence of CaM, as for example in the PMCA when proteolyzed by trypsin [132], or in a more physiological setting by calpain [78, 79, 133]. In what follow we will discuss some of those “old” CLBS containing proteins as well as some newly discovered and analyzed examples. Table 4 shows collected data on the amino acid sequences of the CBS and CLBS of selected proteins containing non-EF-hand CLBSs.
Table 4.
The CBS and CLBS from selected non-EF-hand proteins
Basic amino acids (blue) and acidic amino acids (red) are indicated. CBS calmodulin-binding site, CaMK-II calmodulin-dependent protein kinase, CLBS calmodulin-like binding site, EGFR epidermal growth factor receptor, ErbB1/2/3/4 erythroblastic leukemia viral oncogene homologues 1, 2, 3 and 4, MLCK myosin light chain kinase
aSequence position of the mature receptor lacking the signal peptide
bThe CaM-BD sequence in UniProt P00518 starts at position 302; and the CaM-LD sequence lacks of the amino acid residues LDKV (underlined)
The protein phosphatase calcineurin
The CaM-dependent serine/threonine phosphatase calcineurin (also denoted protein phosphatase 2B) is representative of heterodimeric proteins with a CBS and CLBS located in distinct subunits (Fig. 3d). Melittin interacts with calcineurin, indicative that this CaM-binding peptide binds to CLBS that interacts with the CBS as described above and in Fig. 3d [20]. This phosphatase regulates multiple cellular functions, best studied is the dephosphorylation of the transcription factor NFAT inducing its nuclear translocation to initiate its transcriptional program (reviewed in [134]). This enzyme is also target of immunosuppressive drugs used in the clinic, such as cyclosporin A and FK506 (tacrolimus) (reviewed in [135, 136]). Calcineurin is formed by a 60 kDa catalytic subunit (CaN-A) that binds calmodulin, and a 19 kDa regulatory subunit denoted CaN-B with four EF-hands Ca2+-binding sites (see Figs. 1, 3d). CaN-B is myristoylated at its N-terminus (reviewed in [137]) and has structural similarity to CaM [138]. At low Ca2+ concentration the binding of Ca2+ to the two high-affinity sites of CaN-B maintains the CBS of CaN-A bound to a CLBS located in the same subunit, allowing auto-inhibition. At high Ca2+ concentration the low-affinity Ca2+ binding sites at CaN-B are also occupied and the CBS dissociates from this site. Thereafter, Ca2+/CaM binds to the CBS in CaN-A displacing an auto-inhibitory domain located at the C-terminus of its catalytic site yielding the fully active enzyme (reviewed in [135, 136, 139]). Apparently, a disordered fragment of the auto-inhibitory domain blocks access to the catalytic site playing a central role in the auto-inhibition mechanism [140]. Recent structural studies on the interaction of CaM with a peptide corresponding to CaN-A further refined this model for the activation of calcineurin by Ca2+/CaM and its regulatory subunit [141]. However, the CaN-Aβ1 isoform shows a distinct non-canonical regulation by Ca2+ and CaM, as it lacks an auto-inhibitory domain but a four amino acid motif (462LAVP465) competitively inhibits the enzyme conferring basal and Ca2+-dependent activity decreasing its need of CaM [142]. Using luminescent lanthanides, as surrogate cations for Ca2+, it was demonstrated that the highest affinity for Tb3+ is located in the C-terminal region of CaN-B [143]. Using 113Cd NMR and Ca2+ flow-dialysis the existence of a single EF-hand with high affinity for Ca2+ (close to 10 nM) and three others of lower affinity (close to 10 µM) were identified [144]. Mutational inactivation of CaN-B impairing Ca2+-binding to EF-hands 1, 2 and 3, but not EF-hand 4, has shown that these sites are required for the catalytic activity of CaN-A in the absence of CaM, with an essential role of EF-hand 2. Moreover, eliminating the Ca2+-binding capacity of EF-hand 2 on CaN-B decreases the affinity of CaM for CaN-A at low Ca2+ concentration [145].
Kinases
CaMK-II is a ubiquitous and multifunctional CaM-dependent kinase, activated by autophosphorylation in a Ca2+/CaM-dependent way that also contains an acidic CLBS (Fig. 1). CaMK-II mediates Ca2+ signals by phosphorylating numerous targets important for signaling processes leading to synaptic plasticity, ion homeostasis, transcription and immunoregulation among many other processes (reviewed in [146, 147]). Due to its multimeric structure and autoregulation, it is able to switch into a form, which is no more dependent of initial Ca2+/CaM as autophosphorylation increases the affinity to CaM by a factor of 1000, slowing the release of Ca2+/CaM, and therefore, it is able to participate in learning and memory function [148]. CaMK-II exists in four isoenzymes and 30 splice forms, which determine its specific localization, activation mechanism and target interaction/phosphorylation. This kinase forms a dodecameric complex, and each monomer has a N-terminal kinase domain followed by a regulatory segment, a short linker region and a hub domain. Jarrett and Madhavan [20] compared a number of CaM-binding proteins and found that CaMK-II and skeletal and smooth muscle MLCK (described below) among others bind to melittin, and as melittin is a well-known target of CaM they proposed the existence of a putative CLBS. In the two kinases, the CLBS with the main characteristic of having minimally two regions containing acidic residues adjacent to hydrophobics and aromatics among the hydrophobics one could be identified. The authors [20], predicted CLBS sequences based on their proposed potential to interact with the CBS. They found a striking alignment of the CBS (with positive charges) and CLBS (with negative charges) for both CaMK-II and MLCK (Table 4), suggesting that these sites could interact electrostatically inhibiting the enzymatic activity in the absence of Ca2+/CaM. Nevertheless, the dodecameric structure of the auto-inhibited human CaMK-II holoenzyme contradicts this model, as the autoinhibitory domains of the complex are docked to the central hub, in which the CaM-binding sites are located. Ca2+/CaM-binding displaces the regulatory segment allowing auto-phosphorylation and activation of the enzyme [149]. As the CaM-binding site in the inactive form is inaccessible according to this structure and as mentioned previously is contiguous to a distinct autoinhibitory domain (Fig. 4a, b), it is therefore questionable whether the CLBS interacts with the CBS in CaMK-II to induce autoinhibition.
Fig. 4.
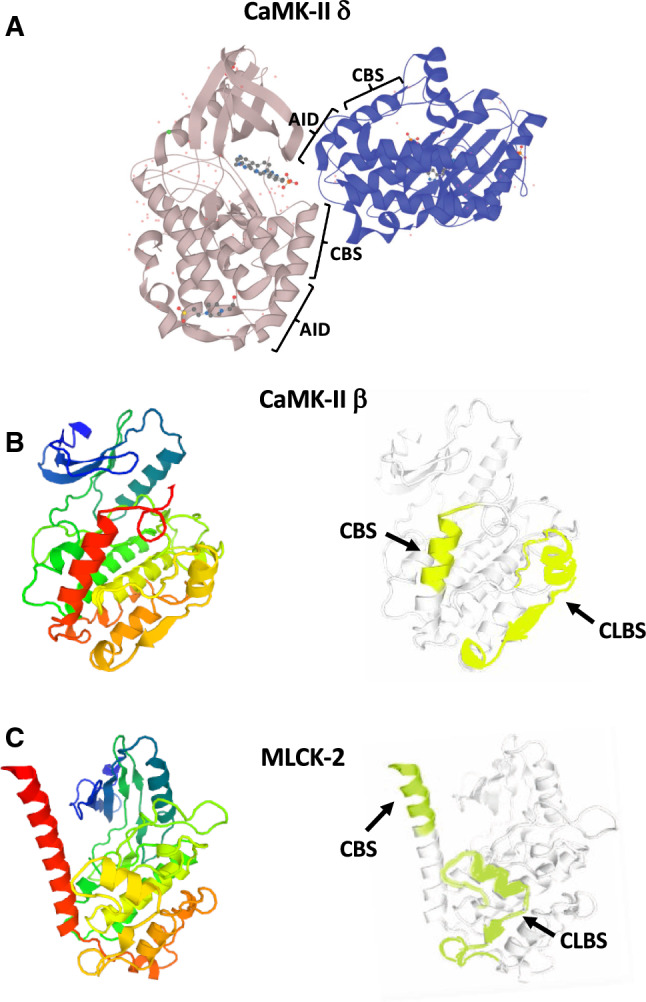
Structure and CBS/CLBS location in CaMK-II and MLCK. The figure depicts two chains of the autoinhibited dodecameric human CaMK-II δ isoform showing the contiguous autoinhibitory domain (AID) and CBS in both chains (a); the X-ray crystallographic structure and the location of the calmodulin-binding domain (CBS) and calmodulin-like binding site (CLBS) of the β-subunit of calmodulin-dependent protein kinase-II (CaMK-II) from rat (b); and myosin light-chain kinase-2 (MLCK-2) from rabbit skeletal/cardiac muscle showing the CBS and CLBS (c). In panels b and c left the structures are shown using a rainbow color code going from the N-terminal (blue) to the C-terminal (red), and in panels b and c right highlighting in color the CBS and CLBS. In panel a two chains of CaMK-II are shown in different colors. Crystallographic 3D models for rat and human CaMK-II at 2.3 Å resolution (P08413 KCC2B_RAT based on template 2vn9A, residue range 10–309, and Q13557 KCC2D_HUMAN based on template 2vn9, residue range 11–309), and rabbit MLCK-2 at 2.2 Å resolution (P07313 MYLK2_RABIT based on template 2x0gA, residue range 290–596) were taken and modified from UniProt SWISS-MODEL database
MLCK is another Ca2+/CaM-dependent kinase containing a potential CLBS only predicted because of its interaction with melittin, and as for CaMK-II it was proposed to play an inhibitory role [20]. Figure 1 shows the domains in MLCK and Fig. 4c the location of the major CBS and CLBS of MLCK-2. This kinase is most known as an essential part of the regulation of smooth muscle contraction through phosphorylation of regulatory myosin II light chains. However, the smooth muscle MLCK is also found in many other cells involved in cell motility processes driven by myosin II. There exists also a skeletal and a heart muscle-specific MLCK form, expressed from different genes. Next to the catalytic domain on both sides, Ca2+/CaM-binding domains are found and a myosin binding domain is placed at the C-terminus. In smooth muscle MLCK, one CBS is located in the N-terminus and another major CBS in the C-terminus distal of the catalytic site [150]. An actin-binding domain exists in the N-terminal region but only in the smooth muscle form, which keeps the enzyme in place. For becoming active MLCK has to be phosphorylated by the c-Src kinase following Ca2+/CaM binding. This leads to autophosphorylation on 19 serine and threonine residues resulting in actin–myosin cross-bridging needed for smooth muscle contraction (reviewed in [151, 152]). In addition to c-Src, the tyrosine kinase c-Abl phosphorylates non-muscle MLCK inducing its activation [153]. As in the case CaMK-II the autoinhibitory site has been localized close to the CBS, and experimental evidence for CBS/CLBS interaction is lacking. However, it could be speculated that the CLBS in these kinases may bind to a CBS during structural transition states in activation/inactivation of the same polypeptide or other subunits.
Phosphorylase b kinase is a multimeric Ca2+/CaM-regulated enzyme that changes the poorly active glycogen phosphorylase b into a more active conformation (form-a) that catalyzes the release of glucose-1-phosphate from glycogen to fuel sugar metabolism. The three subunits (α, β, γ) of phosphorylase b kinase constitutively bind CaM (denoted integral δ subunit) [154–157]. These subunits also have acidic regions that could represent CLBSs [20]. The CBS located in the catalytic γ subunit was found to interact with the regulatory α subunit that has an inhibitory function [158]. This suggests that this interaction may play an auto-inhibitory role when CaM (δ subunit) is free of Ca2+. Nevertheless, a demonstration of direct CBS/CLBS interaction in the whole enzyme is lacking.
Phosphofructokinase is a key rate-limiting allosteric enzyme in glycolysis that catalyzes the phosphorylation of fructose-6-phosphate resulting in fructose-1,6-bisphosphate. In mammals, it forms homo-tetramers, and the formation of active tetramers is negatively regulated by Ca2+/CaM [159, 160]. Phosphofructokinase, together with hexokinase, pyruvate kinase and lactic dehydrogenase, is overexpressed in tumor cells and responsible for the high rate of aerobic glycolysis also called the Warburg effect [161]. The identified CBS was found to be located in the region where two dimers interact to form tetramers. The binding of Ca2+/CaM stabilizes the inactive dimer, allowing in this manner an inactive-active equilibrium following the dimer-tetramer status of the enzyme [162, 163]. Phosphofructokinase also has a CLBS, and the CBS in one subunit of the enzyme was proposed to interact with the CLBS in another subunit contributing to maintaining the stability of the tetramer [20].
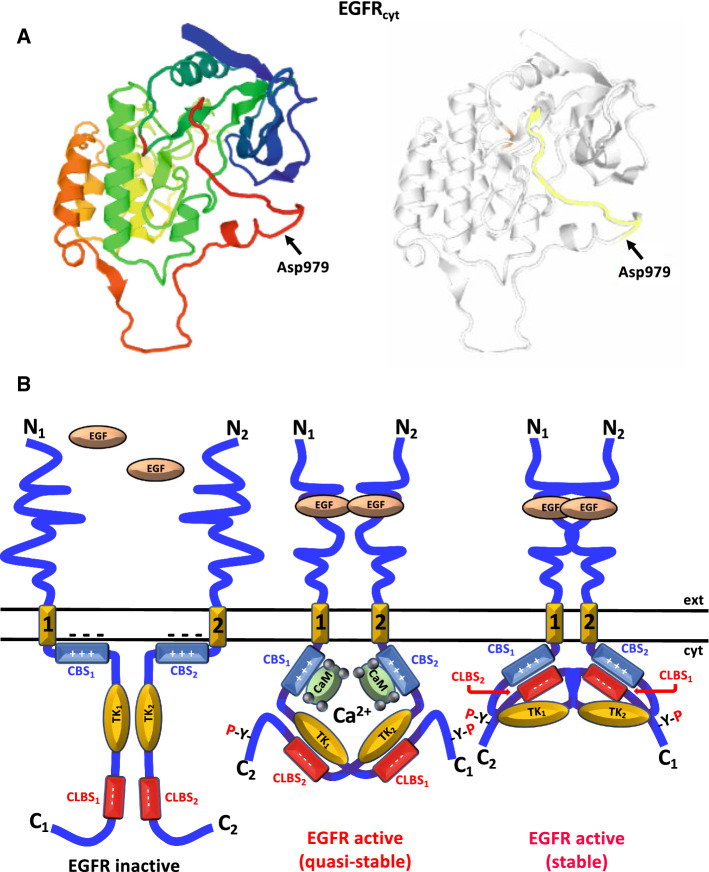
We have demonstrated that the EGFR is a Ca2+-dependent CaM-binding protein containing a CBS located at the cytosolic juxtamembrane region [164–166]. We also demonstrated that Ca2+/CaM appears to participate in the ligand-dependent activation of the receptor in living cells [167], and that phospho-Tyr-CaM is a positive regulator of the EGFR when activated by its ligand [168]. We also noticed the existence of a potential CLBS in the receptor due to its acidic nature (979DEEDMDDVVDADEY992) and relative sequence similarity to a segment of CaM located at its C-terminus (118DEEVDEMIREADI130), suggesting that it may interact with the CBS of the receptor [169] (Table 4). Figure 5a shows the location of the CLBS in the cytosolic region of the receptor (EGFRcyt). Modeling studies further suggested that indeed the electrostatic interaction between CBS and CLBS in the EGFR is sterically possible and that it could facilitate the stabilization of its dimers upon ligand binding [170–172] (reviewed in [173]). Figure 5b depicts a model where hetero-CBS/CLBS interaction stabilizes the ligand-bound EGFR dimer. The CLBS is located distal to the tyrosine kinase domain and within the Ca2+ internalization (CAIN) domain which is implicated in Ca2+ mobilization and receptor internalization (reviewed in [174]). However, direct experimental evidence of interaction between the CBS and the CLBS of the EGFR is lacking.
Fig. 5.
Structure and functional role of CBS/CLBS interaction in the EGFR. a The figure depicts the X-ray crystallographic structure of the cytosolic region of the human epidermal growth factor receptor (EGFRcyt) at 3.2 Å resolution (P00533 EGFR_HUMAN based on template 3rcdA, residue range 702–1015 corresponding to residues 678–991 in the mature receptor lacking the 24 amino acids of the signal peptide) taken and modified from UniProt MODBASE database using a rainbow color code (left panel) going from the N-terminal (blue) to the C-terminal (red), and the location of the CaM-like domain (CLBS) highlights in color starting at Asp979 (right panel). b At resting basal conditions (left panel), the monomeric ligand-free epidermal growth factor receptor (EGFR) has the positively charged juxtamembrane calmodulin-binding domain (CBS) (blue segment) electrostatically bound to the negatively charged inner leaflet of the plasma membrane (minus symbols), which is rich in acidic phosphoinositides, maintaining the receptor in an auto-inhibited state [183]. Upon binding of the ligand epidermal growth factor (EGF) (central panel), the receptor initiates its dimerization and the Ca2+/CaM complex binds to the CBS helping to its detachment from the inner leaflet of the plasma membrane [183], and therefore contributing to the ligand-induced EGFR activation by trans-phosphorylation of C-terminal tyrosine residues (-Y-P). In this model, we propose that the EGFR dimer is active but in a quasi-stable conformation. Subsequently, the active EGFR dimer releases the Ca2+/CaM complex and adopts a more stable conformation (right panel) by the electrostatic interaction between of positively charged CBS (blue segment) of one EGFR monomer (labeled 1) with the negatively charged CaM-like domain (CLBS) (red segment) of the apposed EGFR monomer (labeled 2). For clarity, the N- and C-termini, transmembrane region (yellow segment), CBS, CLBS, and tyrosine kinase domain (TK) of each EGFR monomer are labeled with numbers 1 and 2 to document CBS/CLBS hetero-interaction of apposed monomers. Ca2+ ions are represented by gray spheres. ext extracellular medium, cyt cytosol.
Adapted from Ref. [173]
A series of 89 human glial tumors, including grade IV glioblastomas, grade III anaplastic astrocytomas and grade II astrocytomas, were analyzed for mutations affecting the coding regions of the CBS and CLBS of the EGFR and other ErbB family members [175, 176]. No point mutations were detected affecting the analyzed domains in any of the ErbB family members in these studies, but an in-frame tandem duplication of exons 18–25/18–26 of the EGFR was found, that comprise the tyrosine kinase domain and the whole or part of the CAIN domain, where the CLBS is located, resulting in 190/185 kDa mutant receptors with two CLBSs [175, 176], as was previously reported in different glioma cell lines [177, 178]. The CLBS of the EGFR includes Tyr992 at its C-terminal end, which upon phosphorylation recruits and activates PLCγ [179], leading to Ca2+ release from the ER. Deletion of the CAIN domain, and hence the lack of the CLBS, or mutation of its acidic residues or Tyr992, increases the transforming activity of the EGFR and of the N-terminal truncated c-erbB homologue encoded by the avian erythroleukemia virus, suggesting that this region plays a role in decreasing their transformation potential [180].
ErbB2 was also demonstrated to be a Ca2+/CaM-binding protein, with a CBS located in the cytosolic juxtamembrane region [181], as in the case of the EGFR (Table 4). More recently, White et al. identified two CBS in ErbB2 [182]. Similar juxtamembrane CBSs where identified in ErbB4 and ErbB3, although the latter receptor has lower homology to the one identified in EGFR [169]. Consistent with this, the CBS of ErbB3 has a lower affinity (Ka = 3 μM) for Ca2+/CaM, as compared to the other three receptors, with Ka values of 10 nM in EGFR, 0.2 μM in ErbB4, and 0.6 μM in ErbB2 [183]. Of notice, the lower affinity of ErbB3 for Ca2+/CaM correlates with the absence of intrinsic tyrosine kinase activity of this receptor, which only forms active heterodimers with other family members such as ErbB2, which lacks of ligand-binding capacity [184]. This underscores the importance of CaM in ErbB receptors activation. Likewise, CLBSs were also identified in ErbB2, ErbB3 and ErbB4, similar to the one in EGFR (Table 4), and once again the one in ErbB3 has the lowest similarity to EGFR, as compare to the other two receptors [169].
Transport systems
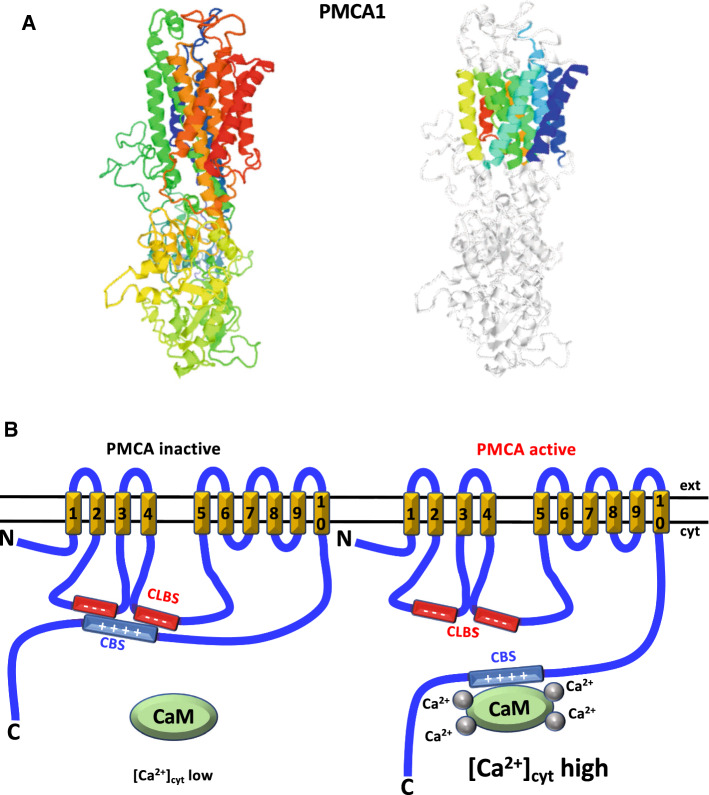
The CaM-dependent plasma membrane Ca2+-ATPase (PMCA) is one of the major Ca2+-transport systems maintaining cytosolic Ca2+ at low concentration by extruding this cation to the extracellular fluid. It has ten transmembrane segments and a bulky cytosolic region where the catalytic site resides formed by two major and two minor loops and a C-terminal tail (Figs. 1, 6a, b). Two acidic regions in the first and second intracellular loops denoted the CLBS have an important function in regulating the enzyme, as it induces auto-inhibition in the absence of Ca2+/CaM likely by blocking CaM binding to the CBS of the pump [185, 186] (Fig. 6b). Removal of the CaM-BD by proteolytic digestion with trypsin [187–189], or the endogenous Ca2+-dependent protease calpain [78, 79, 133], prevents its interaction with the auto-inhibitory CLBS and results in a CaM-independent constitutively active Ca2+-ATPase (reviewed in [80, 190]). This underscores the interaction of the CBS with the autoinhibitory region of the enzyme.
Fig. 6.
Structure and functional role of CBS/CLBS interaction in the plasma membrane Ca2+-ATPase. a The figure depicts the X-ray crystallographic structure of human plasma membrane Ca2+-ATPase isoform 1 (PMCA1) at 3 Å resolution (P20020 AT2B1_HUMAN based on template 2c9mA, residue range 52–1063) taken from UniProt SWISS-MODEL database. The crystallographic structure in the left panel is shown using a rainbow color code going from the N-terminal (blue) to the C-terminal (red), and in the right panel highlights in different colors the ten transmembrane segments. b At low cytosolic Ca2+ concentration the positively charged CaM-binding site (CBS) (blue segment), located at the C-terminal tail of the enzyme, is free of calmodulin (CaM) and interacts with two acidic regions representing a bi-partite calmodulin-like binding site (CLBS) (red segments), respectively located in the first and second intracellular bulky loops of the enzyme. The first loop goes between the second and third transmembrane region, and the second loop goes between the fourth and fifth transmembrane region (yellow segments). The CBS has two interaction sites separated by a 38 amino acids segment (not shown). The ten transmembrane regions are numbered, and the N- and C-termini of the enzyme indicated. The interaction of the CBS with the CLBS maintains the enzyme in an auto-inhibited state (left panel). When the cytosolic Ca2+ concentration increases the Ca2+/CaM complex is formed, binding with high affinity to the CBS and detaching itself from the CLBS, which renders the enzyme active (right panel). Ca2+ ions are represented by gray spheres. ext extracellular medium, cyt cytosol, PMCA plasma membrane Ca2+-ATPase.
Adapted from Ref. [217]
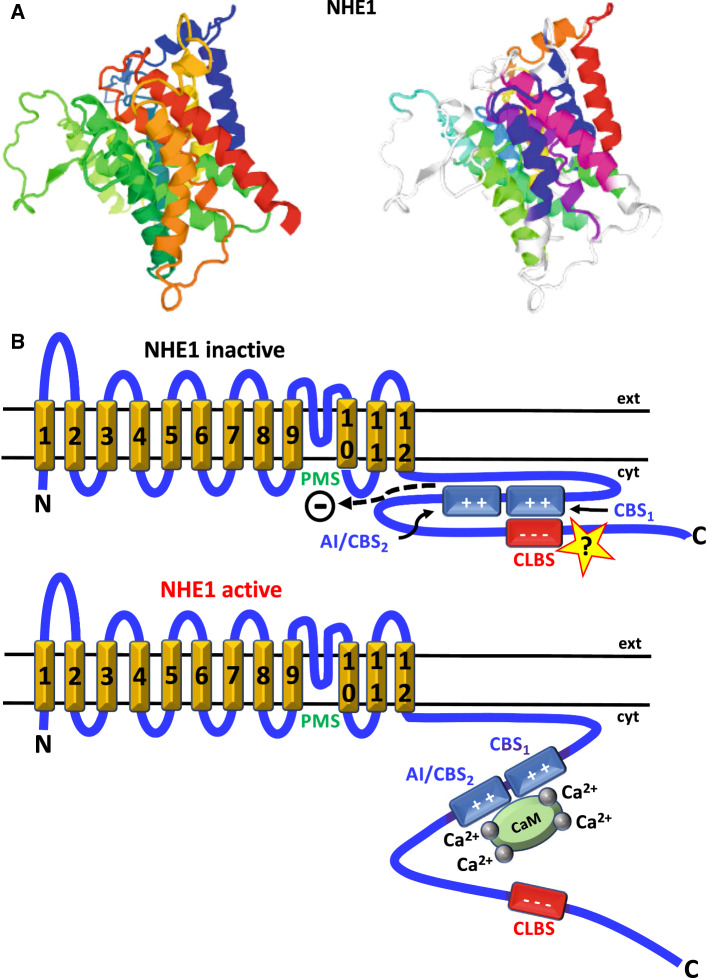
The formation of a complex between CaM and a regulatory region in NHE1, an important pH, salt concentration and volume regulator, has been shown to occur by structural analysis, facilitating our understanding of the role of Ca2+/CaM in its activation [191] (Fig. 7a, b). This report shows that the C-terminal tail of this transporter contains two nearby CBSs located at amino acid residues 629–652 (proximal) and 658–671 (distal), respectively. It has been proposed that in resting cells, when the cytosolic concentration of Ca2+ is very low, the distal CBS interacts with the so-called proton modifier site (PMS), located in the C-terminal end of an intramembrane loop, inducing auto-inhibition [191, 192]. Moreover, it has been hypothesized that the proximal CBS also interacts electrostatically with a highly acidic cluster (amino acid residues 753–759) distal of the CBSs [191, 193]. This region may have the functional characteristics of a non-EF-hand CLBS described above and has been proposed to be involved in maintaining an adequate conformation of the cytosolic domain, promoting CaM binding and activation of the exchanger [191–193]. When the cytosolic Ca2+ concentration increases, Ca2+/CaM binds with the N-lobe to distal CBS and with the C-lobe to proximal CBS [191] and this releases the autoinhibitory/distal CBS from the PMS leading to activation of the exchanger, at the same time that the CLBS is detached from the CBSs. Figure 7b depicts a model illustrating this concept.
Fig. 7.
Structure and functional role of CBS/CLBS interaction in the Na+/H+-exchanger. a The figure depicts the X-ray crystallographic structure of rat Na+/H+-exchanger 1 (NHE1) at 3.5 Å resolution (P26431 SL9A1_RAT based on template 1zcda, residue range 109–510) taken from UniProt MODBASE database using a rainbow color code (left panel) going from the N-terminal (blue) to the C-terminal (red), and the location of eleven transmembrane and one intramembrane segment highlighted in different colors (right panel). b In resting conditions, the Na+/H+-exchanger 1 (NHE1) remains auto-inhibited (top panel) as the distal autoinhibitory/calmodulin-binding site (AI/CBS2) has autoinhibitory function interacting with the proton modifier site (PMS) (dashed arrow), located in an intramembrane loop region. It has been hypothesized that CBS1 (proximal CBS) interacts electrostatically with the distal-located negatively charged CaM-like site (CLBS) (marked with a question mark). When the cytosolic Ca2+ concentration increases the Ca2+/CaM complex is formed, binding to both the proximal and distal CBSs (CBS1 and CBS2, respectively) the autoinhibition is released by detachment of AI/CBS2 from the regulatory PMS, rendering the NHE1 active (bottom panel). Ca2+ ions are represented by gray spheres. ext extracellular medium, cyt cytosol. The twelve transmembrane segments (yellow) are numbered, and the N- and C-termini of the exchanger are indicated.
The potential role of decoy proteins on calmodulin-binding proteins
In addition to the occurrence of CLBS in CaM-binding proteins, in which CBS/CLBS interaction is expected [20], as we have discussed in “Calmodulin-binding proteins with non-EF-hand CaM-like binding sites (CLBSs)”, an intriguing emerging possibility is the binding of the CLBS of CaM-binding proteins to decoy proteins behaving as CaM but exerting a different functional role. This appears to be the case for the apoptosis-inducing protein Bak, as this endoplasmic reticulum (ER)-associated protein is able to interact with both the kinase domain and the CBS of DAPK1, facilitating the entry of ER-Ca2+ into the mitochondria [194]. This suggests the existence of at least one non-canonical CLBS in Bak. The search for interaction of CaM-binding proteins with proteins having CLBS, and the study of their functional roles, would be of interest to investigate. We suggest that a potential role of this type of CBS/CLBS interaction could be to regulate the action of CaM on the target proteins. In addition, myristoylated Ca2+-binding proteins that participate in diverse physiological functions exhibit great sequence similarity to CaN-B [195–197]. Like the CaM-like proteins, many CaN-B-like proteins are expressed in a great variety of plants (reviewed in [198]). However, the function of these proteins appears to be unrelated to CaN-A regulation, participating instead in membrane trafficking, microtubule transport activity or interacting with the platelet fibrinogen receptor. Nevertheless, the possibility that myristoylated Ca2+-binding proteins could act as decoys to block the action of CaN-B on CaN-A is a possibility that warrant further investigation.
Perspectives
A large number of ion channels bind and are regulated by CaM, either in its Ca2+-bound and/or Ca2+-free form. This includes the IP3 and ryanodine receptors (reviewed in [199, 200]), ORAI channels (reviewed in [201]), TRP channels (reviewed in [202, 203]), ligand-gated Ca2+ channels (reviewed in [204]), voltage-dependent Ca2+ channels (reviewed in [205]), voltage-dependent Na+ channels (reviewed in [206, 207]), voltage-dependent K+ channels [208, 209], Ca2+-activated K+ channels (reviewed in [210]), and Ca2+-activated Cl− channels (reviewed in [211]). Although in certain cases CaM is constitutively bound to some ion channels and can be considered an intrinsic subunit of the channel [19], it is important to consider whether the CBS of these channels, when free of CaM, could interact with other regions of the same or different subunit of the channel. If so, these regions could be considered CLBSs. However, to our knowledge no information about the existence of these potential CLBSs within ion channels is available. This could be an interesting topic of future research, as its study may underscore the functionality of these potential regulatory regions. As mentioned in the different sections, the interaction between the CBS and its CLBS has only been established with certainty in a few non-EF-hand proteins discussed in this review. Therefore, effort for further investigations in this area of research should be of interest, as this could give additional information on the functional roles of these domains.
Acknowledgements
Original work in the authors laboratories were funded by the Secretaría de Estado de Investigación, Desarrollo e Innovación Grant SAF2014-52048-R, and the Consejería de Educación, Juventud y Deportes—Comunidad de Madrid Grant B2017/BMD-36 involving contributions from the European Funds for Regional Development (EFRD) and the Social European Fund (SEF) (to AV); the Danish Research Council Grant DFF-4004-00560, the AP Møller Foundation, Dagmar Marshalls Foundation, Einar Willumsen Foundation, Aase and Ejnar Danielsen Foundation, Wedell Wedellsborg Foundation, Frænkels Foundation and the Danish Heart Foundation Grant 13-04-R94-A4547-2280 (to MWB).
Abbreviations
- ADF
Actin-depolymerizing factor
- Bak
Bcl2-antagonist/killer
- CAIN
Calcium and internalization
- CaM
Calmodulin
- CaM-BD
CaM-binding domain
- CaMK-II
CaM-dependent protein kinase-II
- CaM-LD
CaM-like domain
- CaN-A
Calcineurin A
- CaN-B
Calcineurin B
- CBS
Calmodulin binding site
- CDK
Cyclin-dependent kinase
- CDPK/CPK
CaM-like domain protein kinase/Ca2+-dependent protein kinase
- c-erbB
Cellular erythroblastic leukemia viral oncogene homologue
- CLBS
Calmodulin-like binding site
- COMP
Cartilage oligomeric matrix protein
- DAPK1
Death-associated protein kinase 1
- ER
Endoplasmic reticulum
- ErbB1/2/3/4
Erythroblastic leukemia viral oncogene homologues 1/2/3/4
- EGF
Epidermal growth factor
- EGFR
EGF receptor
- EGFRcyt
EGFR cytosolic region
- FRET
Fluorescence resonance energy transfer
- FSH
Follicle-stimulating hormone
- GPDH
Glycerol-3-phosphate dehydrogenase
- IgA/IgG
Immunoglobulins A/G
- IP3
Inositol-1,4,5-trisphosphate
- MAPK1
Mitogen-activated protein kinase 1
- MLCK
Myosin light-chain kinase
- NHE1
Na+/H+ exchanger 1
- NLS
Nuclear localization sequence
- NMR
Nuclear magnetic resonance
- NOX5
NADPH oxidase 5
- ROS
Reactive oxygen species
- LIMP-2
Lotus intrinsic membrane protein 2
- PIP2
Phosphatidylinositol 4,5-bisphosphate
- PKG
cGMP-dependent protein kinase
- PLCγ
Phospholipase Cγ
- PMCA
Plasma membrane Ca2+-ATPase
- SERA5
Serine-repeat antigen 5
- TRP
Transient receptor potential
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Antonio Villalobo, Email: antonio.villalobo@iib.uam.es, Email: antonio.villalobo@idipaz.es.
Martin W. Berchtold, Email: mabe@bio.ku.dk
References
- 1.Kretsinger RH. Hypothesis: calcium modulated proteins contain EF hands. In: Carafoli E, Clementi F, Drabikowski W, Margreth A, editors. Calcium transport in contraction and secretion. Amsterdam: Elsevier; 1975. pp. 469–478. [Google Scholar]
- 2.Berchtold MW. Evolution of EF-hand calcium-modulated proteins. V. The genes encoding EF-hand proteins are not clustered in mammalian genomes. J Mol Evol. 1993;36:489–496. doi: 10.1007/BF02406724. [DOI] [PubMed] [Google Scholar]
- 3.Kawasaki H, Nakayama S, Kretsinger RH. Classification and evolution of EF-hand proteins. Biometals. 1998;11:277–295. doi: 10.1023/A:1009282307967. [DOI] [PubMed] [Google Scholar]
- 4.Kawasaki H, Kretsinger RH. Structural and functional diversity of EF-hand proteins: evolutionary perspectives. Protein Sci. 2017;26:1898–1920. doi: 10.1002/pro.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villalobo A, Ishida H, Vogel HJ, Berchtold MW. Calmodulin as a protein linker and a regulator of adaptor/scaffold proteins. Biochim Biophys Acta. 2018;1865:507–521. doi: 10.1016/j.bbamcr.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Klee CB, Vanaman TC. Calmodulin. Adv Protein Chem. 1982;35:213–321. doi: 10.1016/S0065-3233(08)60470-2. [DOI] [PubMed] [Google Scholar]
- 7.Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/S0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 8.Benaim G, Villalobo A. Phosphorylation of calmodulin. Functional implications. Eur J Biochem. 2002;269:3619–3631. doi: 10.1046/j.1432-1033.2002.03038.x. [DOI] [PubMed] [Google Scholar]
- 9.Berchtold MW, Villalobo A. The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochim Biophys Acta. 2014;1843:398–435. doi: 10.1016/j.bbamcr.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 10.Villarroel A, Taglialatela M, Bernardo-Seisdedos G, Alaimo A, Agirre J, Alberdi A, Gomis-Perez C, Soldovieri MV, Ambrosino P, Malo C, Areso P. The ever changing moods of calmodulin: how structural plasticity entails transductional adaptability. J Mol Biol. 2014;426:2717–2735. doi: 10.1016/j.jmb.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 11.Jurado LA, Chockalingam PS, Jarrett HW. Apocalmodulin. Physiol Rev. 1999;79:661–682. doi: 10.1152/physrev.1999.79.3.661. [DOI] [PubMed] [Google Scholar]
- 12.Bähler M, Rhoads A. Calmodulin signaling via the IQ motif. FEBS Lett. 2002;513:107–113. doi: 10.1016/S0014-5793(01)03239-2. [DOI] [PubMed] [Google Scholar]
- 13.Berchtold MW, Egli R, Rhyner JA, Hameister H, Strehler EE. Localization of the human bona fide calmodulin genes CALM1, CALM2, and CALM3 to chromosomes 14q24-q31, 2p21.1-p21.3, and 19q13.2-q13.3. Genomics. 1993;16:461–465. doi: 10.1006/geno.1993.1211. [DOI] [PubMed] [Google Scholar]
- 14.Putkey JA, Ts’ui KF, Tanaka T, Lagace L, Stein JP, Lai EC, Means AR. Chicken calmodulin genes. A species comparison of cDNA sequences and isolation of a genomic clone. J Biol Chem. 1983;258:11864–11870. [PubMed] [Google Scholar]
- 15.Perochon A, Aldon D, Galaud JP, Ranty B. Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie. 2011;93:2048–2053. doi: 10.1016/j.biochi.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 16.Zeng H, Xu L, Singh A, Wang H, Du L, Poovaiah BW. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front Plant Sci. 2015;6:600. doi: 10.3389/fpls.2015.00600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brushia RJ, Walsh DA. Phosphorylase kinase: the complexity of its regulation is reflected in the complexity of its structure. Front Biosci. 1999;4:D618–D641. doi: 10.2741/Brushia. [DOI] [PubMed] [Google Scholar]
- 18.Marletta MA. Nitric oxide synthase: aspects concerning structure and catalysis. Cell. 1994;78:927–930. doi: 10.1016/0092-8674(94)90268-2. [DOI] [PubMed] [Google Scholar]
- 19.Saimi Y, Kung C. Calmodulin as an ion channel subunit. Annu Rev Physiol. 2002;64:289–311. doi: 10.1146/annurev.physiol.64.100301.111649. [DOI] [PubMed] [Google Scholar]
- 20.Jarrett HW, Madhavan R. Calmodulin-binding proteins also have a calmodulin-like binding site within their structure. The flip–flop model. J Biol Chem. 1991;266:362–371. [PubMed] [Google Scholar]
- 21.Ohno S, Emori Y, Imajoh S, Kawasaki H, Kisaragi M, Suzuki K. Evolutionary origin of a calcium-dependent protease by fusion of genes for a thiol protease and a calcium-binding protein? Nature. 1984;312:566–570. doi: 10.1038/312566a0. [DOI] [PubMed] [Google Scholar]
- 22.Zhang XS, Choi JH. Molecular evolution of calmodulin-like domain protein kinases (CDPKs) in plants and protists. J Mol Evol. 2001;53:214–224. doi: 10.1007/s002390010211. [DOI] [PubMed] [Google Scholar]
- 23.Chen F, Zhang L, Cheng ZM. The calmodulin fused kinase novel gene family is the major system in plants converting Ca2+ signals to protein phosphorylation responses. Sci Rep. 2017;7:4127. doi: 10.1038/s41598-017-03367-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breviario D, Morello L, Giani S. Molecular cloning of two novel rice cDNA sequences encoding putative calcium-dependent protein kinases. Plant Mol Biol. 1995;27:953–967. doi: 10.1007/BF00037023. [DOI] [PubMed] [Google Scholar]
- 25.Roberts DM. Calcium-modulated proteins: targets of intracellular calcium signals in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:375–414. doi: 10.1146/annurev.pp.43.060192.002111. [DOI] [Google Scholar]
- 26.Roberts DM. Protein kinases with calmodulin-like domains: novel targets of calcium signals in plants. Curr Opin Cell Biol. 1993;5:242–246. doi: 10.1016/0955-0674(93)90110-C. [DOI] [PubMed] [Google Scholar]
- 27.Harper JF, Huang JF, Lloyd SJ. Genetic identification of an autoinhibitor in CDPK, a protein kinase with a calmodulin-like domain. Biochemistry. 1994;33:7267–7277. doi: 10.1021/bi00189a031. [DOI] [PubMed] [Google Scholar]
- 28.Harmon AC, Yoo BC, McCaffery C. Pseudosubstrate inhibition of CDPK, a protein kinase with a calmodulin-like domain. Biochemistry. 1994;33:7278–7287. doi: 10.1021/bi00189a032. [DOI] [PubMed] [Google Scholar]
- 29.Weljie AM, Clarke TE, Juffer AH, Harmon AC, Vogel HJ. Comparative modeling studies of the calmodulin-like domain of calcium-dependent protein kinase from soybean. Proteins. 2000;39:343–357. doi: 10.1002/(SICI)1097-0134(20000601)39:4<343::AID-PROT70>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 30.Christodoulou J, Malmendal A, Harper JF, Chazin WJ. Evidence for differing roles for each lobe of the calmodulin-like domain in a calcium-dependent protein kinase. J Biol Chem. 2004;279:29092–29100. doi: 10.1074/jbc.M401297200. [DOI] [PubMed] [Google Scholar]
- 31.Liese A, Romeis T. Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK) Biochim Biophys Acta. 2013;1833:1582–1589. doi: 10.1016/j.bbamcr.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 32.Chandran V, Stollar EJ, Lindorff-Larsen K, Harper JF, Chazin WJ, Dobson CM, Luisi BF, Christodoulou J. Structure of the regulatory apparatus of a calcium-dependent protein kinase (CDPK): a novel mode of calmodulin-target recognition. J Mol Biol. 2006;357:400–410. doi: 10.1016/j.jmb.2005.11.093. [DOI] [PubMed] [Google Scholar]
- 33.Yoo BC, Harmon AC. Intramolecular binding contributes to the activation of CDPK, a protein kinase with a calmodulin-like domain. Biochemistry. 1996;35:12029–12037. doi: 10.1021/bi9606612. [DOI] [PubMed] [Google Scholar]
- 34.Huang JF, Teyton L, Harper JF. Activation of a Ca2+-dependent protein kinase involves intramolecular binding of a calmodulin-like regulatory domain. Biochemistry. 1996;35:13222–13230. doi: 10.1021/bi960498a. [DOI] [PubMed] [Google Scholar]
- 35.Vitart V, Christodoulou J, Huang JF, Chazin WJ, Harper JF. Intramolecular activation of a Ca2+-dependent protein kinase is disrupted by insertions in the tether that connects the calmodulin-like domain to the kinase (correction in Biochemistry 39: 12102, 2000) Biochemistry. 2000;39:4004–4011. doi: 10.1021/bi992373m. [DOI] [PubMed] [Google Scholar]
- 36.Hegeman AD, Rodriguez M, Han BW, Uno Y, Phillips GN, Jr, Hrabak EM, Cushman JC, Harper JF, Harmon AC, Sussman MR. A phyloproteomic characterization of in vitro autophosphorylation in calcium-dependent protein kinases. Proteomics. 2006;6:3649–3664. doi: 10.1002/pmic.200500926. [DOI] [PubMed] [Google Scholar]
- 37.Harper JF, Sussman MR, Schaller GE, Putnam-Evans C, Charbonneau H, Harmon AC. A calcium-dependent protein kinase with a regulatory domain similar to calmodulin. Science. 1991;252:951–954. doi: 10.1126/science.1852075. [DOI] [PubMed] [Google Scholar]
- 38.Lee JY, Yoo BC, Harmon AC. Kinetic and calcium-binding properties of three calcium-dependent protein kinase isoenzymes from soybean. Biochemistry. 1998;37:6801–6809. doi: 10.1021/bi980062q. [DOI] [PubMed] [Google Scholar]
- 39.Bender KW, Blackburn RK, Monaghan J, Derbyshire P, Menke FL, Zipfel C, Goshe MB, Zielinski RE, Huber SC. Autophosphorylation-based calcium (Ca2+) sensitivity priming and Ca2+/calmodulin inhibition of Arabidopsis thaliana Ca2+-dependent protein kinase 28 (CPK28) J Biol Chem. 2017;292:3988–4002. doi: 10.1074/jbc.M116.763243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mittal S, Mallikarjuna MG, Rao AR, Jain PA, Dash PK, Thirunavukkarasu N. Comparative analysis of CDPK family in maize, arabidopsis, rice, and sorghum revealed potential targets for drought tolerance improvement. Front Chem. 2017;5:115. doi: 10.3389/fchem.2017.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weljie AM, Gagne SM, Vogel HJ. Solution structure and backbone dynamics of the N-terminal region of the calcium regulatory domain from soybean calcium-dependent protein kinase α. Biochemistry. 2004;43:15131–15140. doi: 10.1021/bi048751r. [DOI] [PubMed] [Google Scholar]
- 42.Weljie AM, Vogel HJ. Unexpected structure of the Ca2+-regulatory region from soybean calcium-dependent protein kinase-α. J Biol Chem. 2004;279:35494–35502. doi: 10.1074/jbc.M311520200. [DOI] [PubMed] [Google Scholar]
- 43.Weljie AM, Robertson KM, Vogel HJ. Conformational changes in the Ca2+-regulatory region from soybean calcium-dependent protein kinase-α: fluorescence resonance energy transfer studies. J Biol Chem. 2003;278:43764–43769. doi: 10.1074/jbc.M306799200. [DOI] [PubMed] [Google Scholar]
- 44.Chaudhuri S, Seal A, Gupta MD. Autophosphorylation-dependent activation of a calcium-dependent protein kinase from groundnut. Plant Physiol. 1999;120:859–866. doi: 10.1104/pp.120.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vivek PJ, Tuteja N, Soniya EV. CDPK1 from ginger promotes salinity and drought stress tolerance without yield penalty by improving growth and photosynthesis in Nicotiana tabacum. PLoS One. 2013;8:e76392. doi: 10.1371/journal.pone.0076392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raichaudhuri A, Bhattacharyya R, Chaudhuri S, Chakrabarti P, Dasgupta M. Domain analysis of a groundnut calcium-dependent protein kinase: nuclear localization sequence in the junction domain is coupled with nonconsensus calcium binding domains. J Biol Chem. 2006;281:10399–10409. doi: 10.1074/jbc.M511001200. [DOI] [PubMed] [Google Scholar]
- 47.Oh MH, Wu X, Kim HS, Harper JF, Zielinski RE, Clouse SD, Huber SC. CDPKs are dual-specificity protein kinases and tyrosine autophosphorylation attenuates kinase activity. FEBS Lett. 2012;586:4070–4075. doi: 10.1016/j.febslet.2012.09.040. [DOI] [PubMed] [Google Scholar]
- 48.Saha P, Singh M. Characterization of a winged bean (Psophocarpus tetragonolobus) protein kinase with calmodulin-like domain: regulation by autophosphorylation. Biochem J. 1995;305(Pt 1):205–210. doi: 10.1042/bj3050205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romeis T, Herde M. From local to global: CDPKs in systemic defense signaling upon microbial and herbivore attack. Curr Opin Plant Biol. 2014;20:1–10. doi: 10.1016/j.pbi.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 50.Guo YL, Roux SJ. Partial purification and characterization of a Ca2+-dependent protein kinase from the green alga, Dunaliella salina. Plant Physiol. 1990;94:143–150. doi: 10.1104/pp.94.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siderius M, Henskens H, Porto-leBlanche A, van Himbergen J, Musgrave A, Haring M. Characterisation and cloning of a calmodulin-like domain protein kinase from Chlamydomonas moewusii (Gerloff) Planta. 1997;202:76–84. doi: 10.1007/s004250050105. [DOI] [PubMed] [Google Scholar]
- 52.Bender KW, Zielinski RE, Huber SC. Revisiting paradigms of Ca2+signaling protein kinase regulation in plants. Biochem J. 2018;475:207–223. doi: 10.1042/BCJ20170022. [DOI] [PubMed] [Google Scholar]
- 53.Kadian K, Gupta Y, Kempaiah P, Gupta N, Sharma A, Rawat M. Calcium dependent protein kinases (CDPKs): key to malaria eradication. Curr Top Med Chem. 2017;17:2215–2220. doi: 10.2174/1568026617666170130112714. [DOI] [PubMed] [Google Scholar]
- 54.Andresen C, Niklasson M, Cassman Eklof S, Wallner B, Lundstrom P. Biophysical characterization of the calmodulin-like domain of Plasmodium falciparum calcium dependent protein kinase 3. PLoS One. 2017;12:e0181721. doi: 10.1371/journal.pone.0181721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ranjan R, Ahmed A, Gourinath S, Sharma P. Dissection of mechanisms involved in the regulation of Plasmodium falciparum calcium-dependent protein kinase 4. J Biol Chem. 2009;284:15267–15276. doi: 10.1074/jbc.M900656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Donald RG, Zhong T, Wiersma H, Nare B, Yao D, Lee A, Allocco J, Liberator PA. Anticoccidial kinase inhibitors: identification of protein kinase targets secondary to cGMP-dependent protein kinase. Mol Biochem Parasitol. 2006;149:86–98. doi: 10.1016/j.molbiopara.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 57.Van Voorhis WC, Doggett JS, Parsons M, Hulverson MA, Choi R, Arnold SLM, Riggs MW, Hemphill A, Howe DK, Mealey RH, Lau AOT, Merritt EA, Maly DJ, Fan E, Ojo KK. Extended-spectrum antiprotozoal bumped kinase inhibitors: a review. Exp Parasitol. 2017;180:71–83. doi: 10.1016/j.exppara.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sugi T, Kobayashi K, Takemae H, Gong H, Ishiwa A, Murakoshi F, Recuenco FC, Iwanaga T, Horimoto T, Akashi H, Kato K. Identification of mutations in TgMAPK1 of Toxoplasma gondii conferring resistance to 1NM-PP1. Int J Parasitol Drugs Drug Resist. 2013;3:93–101. doi: 10.1016/j.ijpddr.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Foroutan M, Ghaffarifar F. Calcium-dependent protein kinases are potential targets for Toxoplasma gondii vaccine. Clin Exp Vaccine Res. 2018;7:24–36. doi: 10.7774/cevr.2018.7.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dunn PP, Bumstead JM, Tomley FM. Sequence, expression and localization of calmodulin-domain protein kinases in Eimeria tenella and Eimeria maxima. Parasitology. 1996;113(Pt 5):439–448. doi: 10.1017/S0031182000081506. [DOI] [PubMed] [Google Scholar]
- 61.Murachi T. Calcium-dependent proteinases and specific inhibitors: calpain and calpastatin. Biochem Soc Symp. 1984;49:149–167. [PubMed] [Google Scholar]
- 62.Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 63.Ono Y, Ojima K, Shinkai-Ouchi F, Hata S, Sorimachi H. An eccentric calpain, CAPN3/p94/calpain-3. Biochimie. 2016;122:169–187. doi: 10.1016/j.biochi.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 64.Storr SJ, Carragher NO, Frame MC, Parr T, Martin SG. The calpain system and cancer. Nat Rev Cancer. 2011;11:364–374. doi: 10.1038/nrc3050. [DOI] [PubMed] [Google Scholar]
- 65.Waghray A, Wang DS, McKinsey D, Hayes RL, Wang KK. Molecular cloning and characterization of rat and human calpain-5. Biochem Biophys Res Commun. 2004;324:46–51. doi: 10.1016/j.bbrc.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 66.Strobl S, Fernandez-Catalan C, Braun M, Huber R, Masumoto H, Nakagawa K, Irie A, Sorimachi H, Bourenkow G, Bartunik H, Suzuki K, Bode W. The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium. Proc Natl Acad Sci USA. 2000;97:588–592. doi: 10.1073/pnas.97.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hosfield CM, Elce JS, Davies PL, Jia Z. Crystal structure of calpain reveals the structural basis for Ca2+-dependent protease activity and a novel mode of enzyme activation. EMBO J. 1999;18:6880–6889. doi: 10.1093/emboj/18.24.6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suzuki K, Hata S, Kawabata Y, Sorimachi H. Structure, activation, and biology of calpain. Diabetes. 2004;53(Suppl 1):S12–S18. doi: 10.2337/diabetes.53.2007.S12. [DOI] [PubMed] [Google Scholar]
- 69.Vilei EM, Calderara S, Anagli J, Berardi S, Hitomi K, Maki M, Carafoli E. Functional properties of recombinant calpain I and of mutants lacking domains III and IV of the catalytic subunit. J Biol Chem. 1997;272:25802–25808. doi: 10.1074/jbc.272.41.25802. [DOI] [PubMed] [Google Scholar]
- 70.Goll DE, Thompson VF, Taylor RG, Zalewska T. Is calpain activity regulated by membranes and autolysis or by calcium and calpastatin? BioEssays. 1992;14:549–556. doi: 10.1002/bies.950140810. [DOI] [PubMed] [Google Scholar]
- 71.Friedrich P. The intriguing Ca2+ requirement of calpain activation. Biochem Biophys Res Commun. 2004;323:1131–1133. doi: 10.1016/j.bbrc.2004.08.194. [DOI] [PubMed] [Google Scholar]
- 72.Dutt P, Spriggs CN, Davies PL, Jia Z, Elce JS. Origins of the difference in Ca2+ requirement for activation of μ- and m-calpain. Biochem J. 2002;367:263–269. doi: 10.1042/bj20020485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Molinari M, Maki M, Carafoli E. Purification of μ-calpain by a novel affinity chromatography approach. New insights into the mechanism of the interaction of the protease with targets. J Biol Chem. 1995;270:14576–14581. doi: 10.1074/jbc.270.24.14576. [DOI] [PubMed] [Google Scholar]
- 74.Reverter D, Strobl S, Fernandez-Catalan C, Sorimachi H, Suzuki K, Bode W. Structural basis for possible calcium-induced activation mechanisms of calpains. Biol Chem. 2001;382:753–766. doi: 10.1515/bchm.2001.382.5.753. [DOI] [PubMed] [Google Scholar]
- 75.Theopold U, Pinter M, Daffre S, Tryselius Y, Friedrich P, Nassel DR, Hultmark D. CalpA, a Drosophila calpain homolog specifically expressed in a small set of nerve, midgut, and blood cells. Mol Cell Biol. 1995;15:824–834. doi: 10.1128/MCB.15.2.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hong H, Johnson P, El-Saleh SC. Effects of calcium and calmodulin antagonists on calpain II subunit conformations. Int J Biol Macromol. 1990;12:269–272. doi: 10.1016/0141-8130(90)90008-X. [DOI] [PubMed] [Google Scholar]
- 77.Suo S, Koike H, Sorimachi H, Ishiura S, Suzuki K. Association and dissociation of the calcium-binding domains of calpain by Ca2+ Biochem Biophys Res Commun. 1999;257:63–66. doi: 10.1006/bbrc.1999.0407. [DOI] [PubMed] [Google Scholar]
- 78.Wang KK, Villalobo A, Roufogalis BD. Activation of the Ca2+-ATPase of human erythrocyte membrane by an endogenous Ca2+-dependent neutral protease. Arch Biochem Biophys. 1988;260:696–704. doi: 10.1016/0003-9861(88)90498-5. [DOI] [PubMed] [Google Scholar]
- 79.Wang KK, Roufogalis BD, Villalobo A. Further characterization of calpain-mediated proteolysis of the human erythrocyte plasma membrane Ca2+-ATPase. Arch Biochem Biophys. 1988;267:317–327. doi: 10.1016/0003-9861(88)90037-9. [DOI] [PubMed] [Google Scholar]
- 80.Wang KK, Villalobo A, Roufogalis BD. Calmodulin-binding proteins as calpain substrates. Biochem J. 1989;262:693–706. doi: 10.1042/bj2620693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shumway SD, Maki M, Miyamoto S. The PEST domain of IκBα is necessary and sufficient for in vitro degradation by μ-calpain. J Biol Chem. 1999;274:30874–30881. doi: 10.1074/jbc.274.43.30874. [DOI] [PubMed] [Google Scholar]
- 82.Croall DE, Chacko S, Wang Z. Cleavage of caldesmon and calponin by calpain: substrate recognition is not dependent on calmodulin binding domains. Biochim Biophys Acta. 1996;1298:276–284. doi: 10.1016/S0167-4838(96)00138-0. [DOI] [PubMed] [Google Scholar]
- 83.Yang HQ, Ma H, Takano E, Hatanaka M, Maki M. Analysis of calcium-dependent interaction between amino-terminal conserved region of calpastatin functional domain and calmodulin-like domain of μ-calpain large subunit. J Biol Chem. 1994;269:18977–18984. [PubMed] [Google Scholar]
- 84.Takano E, Ma H, Yang HQ, Maki M, Hatanaka M. Preference of calcium-dependent interactions between calmodulin-like domains of calpain and calpastatin subdomains. FEBS Lett. 1995;362:93–97. doi: 10.1016/0014-5793(95)00219-Y. [DOI] [PubMed] [Google Scholar]
- 85.Ma H, Yang HQ, Takano E, Hatanaka M, Maki M. Amino-terminal conserved region in proteinase inhibitor domain of calpastatin potentiates its calpain inhibitory activity by interacting with calmodulin-like domain of the proteinase. J Biol Chem. 1994;269:24430–24436. [PubMed] [Google Scholar]
- 86.Ma H, Yang HQ, Takano E, Lee WJ, Hatanaka M, Maki M. Requirement of different subdomains of calpastatin for calpain inhibition and for binding to calmodulin-like domains. J Biochem. 1993;113:591–599. doi: 10.1093/oxfordjournals.jbchem.a124088. [DOI] [PubMed] [Google Scholar]
- 87.Deshmukh L, Wu L, Guttmann RP, Vinogradova O. NMR structural characterization of the penta-peptide calpain inhibitor. FEBS Lett. 2009;583:135–140. doi: 10.1016/j.febslet.2008.11.037. [DOI] [PubMed] [Google Scholar]
- 88.MacDonald MJ, Brown LJ. Calcium activation of mitochondrial glycerol phosphate dehydrogenase restudied. Arch Biochem Biophys. 1996;326:79–84. doi: 10.1006/abbi.1996.0049. [DOI] [PubMed] [Google Scholar]
- 89.Brandes RP, Weissmann N, Schroder K. Nox family NADPH oxidases: molecular mechanisms of activation. Free Radic Biol Med. 2014;76:208–226. doi: 10.1016/j.freeradbiomed.2014.07.046. [DOI] [PubMed] [Google Scholar]
- 90.Marino D, Dunand C, Puppo A, Pauly N. A burst of plant NADPH oxidases. Trends Plant Sci. 2012;17:9–15. doi: 10.1016/j.tplants.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 91.Banfi B, Tirone F, Durussel I, Knisz J, Moskwa P, Molnar GZ, Krause KH, Cox JA. Mechanism of Ca2+ activation of the NADPH oxidase 5 (NOX5) J Biol Chem. 2004;279:18583–18591. doi: 10.1074/jbc.M310268200. [DOI] [PubMed] [Google Scholar]
- 92.Tirone F, Cox JA. NADPH oxidase 5 (NOX5) interacts with and is regulated by calmodulin. FEBS Lett. 2007;581:1202–1208. doi: 10.1016/j.febslet.2007.02.047. [DOI] [PubMed] [Google Scholar]
- 93.Pandey D, Gratton JP, Rafikov R, Black SM, Fulton DJ. Calcium/calmodulin-dependent kinase II mediates the phosphorylation and activation of NADPH oxidase 5. Mol Pharmacol. 2011;80:407–415. doi: 10.1124/mol.110.070193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Torres MA, Onouchi H, Hamada S, Machida C, Hammond-Kosack KE, Jones JD. Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox) Plant J. 1998;14:365–370. doi: 10.1046/j.1365-313X.1998.00136.x. [DOI] [PubMed] [Google Scholar]
- 95.Ogasawara Y, Kaya H, Hiraoka G, Yumoto F, Kimura S, Kadota Y, Hishinuma H, Senzaki E, Yamagoe S, Nagata K, Nara M, Suzuki K, Tanokura M, Kuchitsu K. Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J Biol Chem. 2008;283:8885–8892. doi: 10.1074/jbc.M708106200. [DOI] [PubMed] [Google Scholar]
- 96.Takahashi S, Kimura S, Kaya H, Iizuka A, Wong HL, Shimamoto K, Kuchitsu K. Reactive oxygen species production and activation mechanism of the rice NADPH oxidase OsRbohB. J Biochem. 2012;152:37–43. doi: 10.1093/jb/mvs044. [DOI] [PubMed] [Google Scholar]
- 97.Oda T, Hashimoto H, Kuwabara N, Akashi S, Hayashi K, Kojima C, Wong HL, Kawasaki T, Shimamoto K, Sato M, Shimizu T. Structure of the N-terminal regulatory domain of a plant NADPH oxidase and its functional implications. J Biol Chem. 2010;285:1435–1445. doi: 10.1074/jbc.M109.058909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Broderick MJ, Winder SJ. Spectrin, α-actinin, and dystrophin. Adv Protein Chem. 2005;70:203–246. doi: 10.1016/S0065-3233(05)70007-3. [DOI] [PubMed] [Google Scholar]
- 99.Sjöblom B, Salmazo A, Djinovic-Carugo K. α-actinin structure and regulation. Cell Mol Life Sci. 2008;65:2688–2701. doi: 10.1007/s00018-008-8080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Travé G, Pastore A, Hyvonen M, Saraste M. The C-terminal domain of α-spectrin is structurally related to calmodulin. Eur J Biochem. 1995;227:35–42. doi: 10.1111/j.1432-1033.1995.tb20357.x. [DOI] [PubMed] [Google Scholar]
- 101.Travers T, Shao H, Wells A, Camacho CJ. Modeling the assembly of the multiple domains of α-actinin-4 and its role in actin cross-linking. Biophys J. 2013;104:705–715. doi: 10.1016/j.bpj.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Beggs AH, Byers TJ, Knoll JH, Boyce FM, Bruns GA, Kunkel LM. Cloning and characterization of two human skeletal muscle α-actinin genes located on chromosomes 1 and 11. J Biol Chem. 1992;267:9281–9288. [PubMed] [Google Scholar]
- 103.Karlsson G, Persson C, Mayzel M, Hedenstrom M, Backman L. Solution structure of the calmodulin-like C-terminal domain of Entamoeba α-actinin2. Proteins. 2016;84:461–466. doi: 10.1002/prot.24992. [DOI] [PubMed] [Google Scholar]
- 104.Drmota Prebil S, Slapsak U, Pavsic M, Ilc G, Puz V, de Almeida Ribeiro E, Anrather D, Hartl M, Backman L, Plavec J, Lenarcic B, Djinovic-Carugo K. Structure and calcium-binding studies of calmodulin-like domain of human non-muscle α-actinin-1. Sci Rep. 2016;6:27383. doi: 10.1038/srep27383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shams H, Golji J, Garakani K, Mofrad MR. Dynamic regulation of α-actinin’s calponin homology domains on F-actin. Biophys J. 2016;110:1444–1455. doi: 10.1016/j.bpj.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huizing M, Carrillo-Carrasco N, Malicdan MC, Noguchi S, Gahl WA, Mitrani-Rosenbaum S, Argov Z, Nishino I. GNE myopathy: new name and new mutation nomenclature. Neuromuscul Disord. 2014;24:387–389. doi: 10.1016/j.nmd.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Harazi A, Becker-Cohen M, Zer H, Moshel O, Hinderlich S, Mitrani-Rosenbaum S. The interaction of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (GNE) and α-actinin 2 is altered in GNE myopathy M743T mutant. Mol Neurobiol. 2017;54:2928–2938. doi: 10.1007/s12035-016-9862-x. [DOI] [PubMed] [Google Scholar]
- 108.Goicoechea SM, Arneman D, Otey CA. The role of palladin in actin organization and cell motility. Eur J Cell Biol. 2008;87:517–525. doi: 10.1016/j.ejcb.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rönty M, Taivainen A, Moza M, Otey CA, Carpen O. Molecular analysis of the interaction between palladin and α-actinin. FEBS Lett. 2004;566:30–34. doi: 10.1016/j.febslet.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 110.Franzot G, Sjoblom B, Gautel M, Djinovic Carugo K. The crystal structure of the actin binding domain from α-actinin in its closed conformation: structural insight into phospholipid regulation of α-actinin. J Mol Biol. 2005;348:151–165. doi: 10.1016/j.jmb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 111.Liu J, Taylor DW, Taylor KA. A 3-D reconstruction of smooth muscle α-actinin by CryoEm reveals two different conformations at the actin-binding region. J Mol Biol. 2004;338:115–125. doi: 10.1016/j.jmb.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 112.Kelly DF, Taylor DW, Bakolitsa C, Bobkov AA, Bankston L, Liddington RC, Taylor KA. Structure of the α-actinin-vinculin head domain complex determined by cryo-electron microscopy. J Mol Biol. 2006;357:562–573. doi: 10.1016/j.jmb.2005.12.076. [DOI] [PubMed] [Google Scholar]
- 113.Pavalko FM, Otey CA, Simon KO, Burridge K. α-actinin: a direct link between actin and integrins. Biochem Soc Trans. 1991;19:1065–1069. doi: 10.1042/bst0191065. [DOI] [PubMed] [Google Scholar]
- 114.Khurana S, Chakraborty S, Cheng X, Su YT, Kao HY. The actin-binding protein, actinin α 4 (ACTN4), is a nuclear receptor coactivator that promotes proliferation of MCF-7 breast cancer cells. J Biol Chem. 2011;286:1850–1859. doi: 10.1074/jbc.M110.162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Piirainen H, Taura J, Kursula P, Ciruela F, Jaakola VP. Calcium modulates calmodulin/α-actinin 1 interaction with and agonist-dependent internalization of the adenosine A2A receptor. Biochim Biophys Acta. 2017;1864:674–686. doi: 10.1016/j.bbamcr.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 116.Atkinson RA, Joseph C, Dal Piaz F, Birolo L, Stier G, Pucci P, Pastore A. Binding of α-actinin to titin: implications for Z-disk assembly. Biochemistry. 2000;39:5255–5264. doi: 10.1021/bi991891u. [DOI] [PubMed] [Google Scholar]
- 117.Grison M, Merkel U, Kostan J, Djinovic-Carugo K, Rief M. α-Actinin/titin interaction: a dynamic and mechanically stable cluster of bonds in the muscle Z-disk. Proc Natl Acad Sci USA. 2017;114:1015–1020. doi: 10.1073/pnas.1612681114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tang J, Taylor DW, Taylor KA. The three-dimensional structure of α-actinin obtained by cryoelectron microscopy suggests a model for Ca2+-dependent actin binding. J Mol Biol. 2001;310:845–858. doi: 10.1006/jmbi.2001.4789. [DOI] [PubMed] [Google Scholar]
- 119.Satrústegui J, Pardo B, Del Arco A. Mitochondrial transporters as novel targets for intracellular calcium signaling. Physiol Rev. 2007;87:29–67. doi: 10.1152/physrev.00005.2006. [DOI] [PubMed] [Google Scholar]
- 120.Yang Q, Bruschweiler S, Chou JJ. Purification, crystallization and preliminary X-ray diffraction of the N-terminal calmodulin-like domain of the human mitochondrial ATP-Mg/Pi carrier SCaMC1. Acta Crystallogr Sect F Struct Biol Commun. 2014;70:68–71. doi: 10.1107/S2053230X1303241X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang Q, Bruschweiler S, Chou JJ. A self-sequestered calmodulin-like Ca2+ sensor of mitochondrial SCaMC carrier and its implication to Ca2+-dependent ATP-Mg/P(i) transport. Structure. 2014;22:209–217. doi: 10.1016/j.str.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lorenz A, Lorenz M, Vothknecht UC, Niopek-Witz S, Neuhaus HE, Haferkamp I. In vitro analyses of mitochondrial ATP/phosphate carriers from Arabidopsis thaliana revealed unexpected Ca2+-effects. BMC Plant Biol. 2015;15:238. doi: 10.1186/s12870-015-0616-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bornstein P, Armstrong LC, Hankenson KD, Kyriakides TR, Yang Z. Thrombospondin 2, a matricellular protein with diverse functions. Matrix Biol. 2000;19:557–568. doi: 10.1016/S0945-053X(00)00104-9. [DOI] [PubMed] [Google Scholar]
- 124.Chen H, Herndon ME, Lawler J. The cell biology of thrombospondin-1. Matrix Biol. 2000;19:597–614. doi: 10.1016/S0945-053X(00)00107-4. [DOI] [PubMed] [Google Scholar]
- 125.Acharya C, Yik JH, Kishore A, Van Dinh V, Di Cesare PE, Haudenschild DR. Cartilage oligomeric matrix protein and its binding partners in the cartilage extracellular matrix: interaction, regulation and role in chondrogenesis. Matrix Biol. 2014;37:102–111. doi: 10.1016/j.matbio.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 126.Briggs MD, Hoffman SM, King LM, Olsen AS, Mohrenweiser H, Leroy JG, Mortier GR, Rimoin DL, Lachman RS, Gaines ES, et al. Pseudoachondroplasia and multiple epiphyseal dysplasia due to mutations in the cartilage oligomeric matrix protein gene. Nat Genet. 1995;10:330–336. doi: 10.1038/ng0795-330. [DOI] [PubMed] [Google Scholar]
- 127.Cohn DH, Briggs MD, King LM, Rimoin DL, Wilcox WR, Lachman RS, Knowlton RG. Mutations in the cartilage oligomeric matrix protein (COMP) gene in pseudoachondroplasia and multiple epiphyseal dysplasia. Ann N Y Acad Sci. 1996;785:188–194. doi: 10.1111/j.1749-6632.1996.tb56258.x. [DOI] [PubMed] [Google Scholar]
- 128.Susic S, McGrory J, Ahier J, Cole WG. Multiple epiphyseal dysplasia and pseudoachondroplasia due to novel mutations in the calmodulin-like repeats of cartilage oligomeric matrix protein. Clin Genet. 1997;51:219–224. doi: 10.1111/j.1399-0004.1997.tb02458.x. [DOI] [PubMed] [Google Scholar]
- 129.Yusuf NA, Green JL, Wall RJ, Knuepfer E, Moon RW, Schulte-Huxel C, Stanway RR, Martin SR, Howell SA, Douse CH, Cota E, Tate EW, Tewari R, Holder AA. The plasmodium class XIV myosin, MyoB, has a distinct subcellular location in invasive and motile stages of the malaria parasite and an unusual light chain. J Biol Chem. 2015;290:12147–12164. doi: 10.1074/jbc.M115.637694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jo CH, Son J, Kim S, Oda T, Kim J, Lee MR, Sato M, Kim HT, Unzai S, Park SY, Hwang KY. Structural insights into a 20.8-kDa tegumental-allergen-like (TAL) protein from Clonorchis sinensis. Sci Rep. 2017;7:1764. doi: 10.1038/s41598-017-02044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Santa-Coloma TA, Grasso P, Reichert LE., Jr Synthetic human follicle-stimulating hormone-β-(1-15) peptide-amide binds Ca2+ and possesses sequence similarity to calcium binding sites of calmodulin. Endocrinology. 1992;130:1103–1107. doi: 10.1210/endo.130.3.1537277. [DOI] [PubMed] [Google Scholar]
- 132.Benaim G, Zurini M, Carafoli E. Different conformational states of the purified Ca2+-ATPase of the erythrocyte plasma membrane revealed by controlled trypsin proteolysis. J Biol Chem. 1984;259:8471–8477. [PubMed] [Google Scholar]
- 133.Wang KK, Roufogalis BD, Villalobo A. Calpain I activates Ca2+ transport by the reconstituted erythrocyte Ca2+ pump. J Membr Biol. 1989;112:233–245. doi: 10.1007/BF01870954. [DOI] [PubMed] [Google Scholar]
- 134.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 135.Li H, Rao A, Hogan PG. Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 2011;21:91–103. doi: 10.1016/j.tcb.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Klee CB, Ren H, Wang X. Regulation of the calmodulin stimulated protein phosphatase, calcineurin. J Biol Chem. 1998;273:13367–13370. doi: 10.1074/jbc.273.22.13367. [DOI] [PubMed] [Google Scholar]
- 137.Rusnak F, Mertz P. Calcineurin: form and function. Physiol Rev. 2000;80:1483–1521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]
- 138.Anglister J, Grzesiek S, Wang AC, Ren H, Klee CB, Bax A. 1H, 13C, 15N nuclear magnetic resonance backbone assignments and secondary structure of human calcineurin B. Biochemistry. 1994;33:3540–3547. doi: 10.1021/bi00178a010. [DOI] [PubMed] [Google Scholar]
- 139.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 140.Ye Q, Feng Y, Yin Y, Faucher F, Currie MA, Rahman MN, Jin J, Li S, Wei Q, Jia Z. Structural basis of calcineurin activation by calmodulin. Cell Signal. 2013;25:2661–2667. doi: 10.1016/j.cellsig.2013.08.033. [DOI] [PubMed] [Google Scholar]
- 141.Chyan CL, Irene D, Lin SM. The recognition of calmodulin to the target sequence of calcineurin-A novel binding mode. Molecules. 2017;22:1584. doi: 10.3390/molecules22101584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bond R, Ly N, Cyert MS. The unique C terminus of the calcineurin isoform CNA1 confers non-canonical regulation of enzyme activity by Ca2+and calmodulin. J Biol Chem. 2017;292:16709–16721. doi: 10.1074/jbc.M117.795146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Burroughs SE, Horrocks WD, Jr, Ren H, Klee CB. Characterization of the lanthanide ion-binding properties of calcineurin-B using laser-induced luminescence spectroscopy. Biochemistry. 1994;33:10428–10436. doi: 10.1021/bi00200a026. [DOI] [PubMed] [Google Scholar]
- 144.Kakalis LT, Kennedy M, Sikkink R, Rusnak F, Armitage IM. Characterization of the calcium-binding sites of calcineurin B. FEBS Lett. 1995;362:55–58. doi: 10.1016/0014-5793(95)00207-P. [DOI] [PubMed] [Google Scholar]
- 145.Feng B, Stemmer PM. Ca2+ binding site 2 in calcineurin-B modulates calmodulin-dependent calcineurin phosphatase activity. Biochemistry. 2001;40:8808–8814. doi: 10.1021/bi0025161. [DOI] [PubMed] [Google Scholar]
- 146.Hudmon A, Schulman H. Structure-function of the multifunctional Ca2+/calmodulin-dependent protein kinase II. Biochem J. 2002;364:593–611. doi: 10.1042/bj20020228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wayman GA, Lee YS, Tokumitsu H, Silva AJ, Soderling TR. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008;59:914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Meyer T, Hanson PI, Stryer L, Schulman H. Calmodulin trapping by calcium-calmodulin-dependent protein kinase. Science. 1992;256:1199–1202. doi: 10.1126/science.256.5060.1199. [DOI] [PubMed] [Google Scholar]
- 149.Chao LH, Stratton MM, Lee IH, Rosenberg OS, Levitz J, Mandell DJ, Kortemme T, Groves JT, Schulman H, Kuriyan J. A mechanism for tunable autoinhibition in the structure of a human Ca2+/calmodulin-dependent kinase II holoenzyme. Cell. 2011;146:732–745. doi: 10.1016/j.cell.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ye L-H, Kishi H, Nakamura A, Okagaki T, Tanaka T, Oiwa K, Kohama K. Myosin light-chain kinase of smooth muscle stimulates myosin ATPase activity without phosphorylating myosin light chain. Proc Natl Acad Sci USA. 1999;96:6666–6671. doi: 10.1073/pnas.96.12.6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Khapchaev AY, Shirinsky VP. Myosin light chain kinase MYLK1: anatomy, interactions, functions, and regulation. Biochemistry (Mosc) 2016;81:1676–1697. doi: 10.1134/S000629791613006X. [DOI] [PubMed] [Google Scholar]
- 152.Stull JT, Lin PJ, Krueger JK, Trewhella J, Zhi G. Myosin light chain kinase: functional domains and structural motifs. Acta Physiol Scand. 1998;164:471–482. doi: 10.1111/j.1365-201X.1998.tb10699.x. [DOI] [PubMed] [Google Scholar]
- 153.Dudek SM, Chiang ET, Camp SM, Guo Y, Zhao J, Brown ME, Singleton PA, Wang L, Desai A, Arce FT, Lal R, Van Eyk JE, Imam SZ, Garcia JG. Abl tyrosine kinase phosphorylates nonmuscle myosin light chain kinase to regulate endothelial barrier function. Mol Biol Cell. 2010;21:4042–4056. doi: 10.1091/mbc.e09-10-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kilimann MW, Zander NF, Kuhn CC, Crabb JW, Meyer HE, Heilmeyer LM., Jr The α and β subunits of phosphorylase kinase are homologous: cDNA cloning and primary structure of the β subunit. Proc Natl Acad Sci USA. 1988;85:9381–9385. doi: 10.1073/pnas.85.24.9381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dasgupta M, Honeycutt T, Blumenthal DK. The γ-subunit of skeletal muscle phosphorylase kinase contains two noncontiguous domains that act in concert to bind calmodulin. J Biol Chem. 1989;264:17156–17163. [PubMed] [Google Scholar]
- 156.Heilmeyer LM, Jr, Gerschinski AM, Meyer HE, Jennissen HP. Interaction sites on phosphorylase kinase for calmodulin. Mol Cell Biochem. 1993;127–128:19–30. doi: 10.1007/BF01076754. [DOI] [PubMed] [Google Scholar]
- 157.Zander NF, Meyer HE, Hoffmann-Posorske E, Crabb JW, Heilmeyer LM, Jr, Kilimann MW. cDNA cloning and complete primary structure of skeletal muscle phosphorylase kinase (α subunit) Proc Natl Acad Sci USA. 1988;85:2929–2933. doi: 10.1073/pnas.85.9.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rice NA, Nadeau OW, Yang Q, Carlson GM. The calmodulin-binding domain of the catalytic γ subunit of phosphorylase kinase interacts with its inhibitory α subunit: evidence for a Ca2+ sensitive network of quaternary interactions. J Biol Chem. 2002;277:14681–14687. doi: 10.1074/jbc.M201229200. [DOI] [PubMed] [Google Scholar]
- 159.Mayr GW, Heilmeyer LM., Jr Phosphofructokinase is a calmodulin binding protein. FEBS Lett. 1983;159:51–57. doi: 10.1016/0014-5793(83)80415-3. [DOI] [PubMed] [Google Scholar]
- 160.Martin SR, Biekofsky RR, Skinner MA, Guerrini R, Salvadori S, Feeney J, Bayley PM. Interaction of calmodulin with the phosphofructokinase target sequence. FEBS Lett. 2004;577:284–288. doi: 10.1016/j.febslet.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 161.Li XB, Gu JD, Zhou QH. Review of aerobic glycolysis and its key enzymes—new targets for lung cancer therapy. Thorac Cancer. 2015;6:17–24. doi: 10.1111/1759-7714.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Buschmeier B, Meyer HE, Mayr GW. Characterization of the calmodulin-binding sites of muscle phosphofructokinase and comparison with known calmodulin-binding domains. J Biol Chem. 1987;262:9454–9462. [PubMed] [Google Scholar]
- 163.Mayr GW. Interaction of calmodulin with muscle phosphofructokinase. Interplay with metabolic effectors of the enzyme under physiological conditions. Eur J Biochem. 1984;143:521–529. doi: 10.1111/j.1432-1033.1984.tb08401.x. [DOI] [PubMed] [Google Scholar]
- 164.Martin-Nieto J, Villalobo A. The human epidermal growth factor receptor contains a juxtamembrane calmodulin-binding site. Biochemistry. 1998;37:227–236. doi: 10.1021/bi971765v. [DOI] [PubMed] [Google Scholar]
- 165.Li H, Villalobo A. Evidence for the direct interaction between calmodulin and the human epidermal growth factor receptor. Biochem J. 2002;362:499–505. doi: 10.1042/bj3620499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li H, Ruano MJ, Villalobo A. Endogenous calmodulin interacts with the epidermal growth factor receptor in living cells. FEBS Lett. 2004;559:175–180. doi: 10.1016/S0014-5793(04)00067-5. [DOI] [PubMed] [Google Scholar]
- 167.Li H, Panina S, Kaur A, Ruano MJ, Sanchez-Gonzalez P, la Cour JM, Stephan A, Olesen UH, Berchtold MW, Villalobo A. Regulation of the ligand-dependent activation of the epidermal growth factor receptor by calmodulin. J Biol Chem. 2012;287:3273–3281. doi: 10.1074/jbc.M111.317529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Stateva SR, Salas V, Benguria A, Cossio I, Anguita E, Martin-Nieto J, Benaim G, Villalobo A. The activating role of phospho-(Tyr)-calmodulin on the epidermal growth factor receptor. Biochem J. 2015;472:195–204. doi: 10.1042/BJ20150851. [DOI] [PubMed] [Google Scholar]
- 169.Martin-Nieto J, Cusidó-Hita DM, Li H, Benguría A, Villalobo A. Regulation of ErbB receptors by calmodulin. Recent Res Dev Biochem. 2002;3:41–58. [Google Scholar]
- 170.Aifa S, Aydin J, Nordvall G, Lundstrom I, Svensson SP, Hermanson O. A basic peptide within the juxtamembrane region is required for EGF receptor dimerization. Exp Cell Res. 2005;302:108–114. doi: 10.1016/j.yexcr.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 171.Aifa S, Frikha F, Miled N, Johansen K, Lundstrom I, Svensson SP. Phosphorylation of Thr654 but not Thr669 within the juxtamembrane domain of the EGF receptor inhibits calmodulin binding. Biochem Biophys Res Commun. 2006;347:381–387. doi: 10.1016/j.bbrc.2006.05.200. [DOI] [PubMed] [Google Scholar]
- 172.Aifa S, Miled N, Frikha F, Aniba MR, Svensson SP, Rebai A. Electrostatic interactions of peptides flanking the tyrosine kinase domain in the epidermal growth factor receptor provides a model for intracellular dimerization and autophosphorylation. Proteins. 2006;62:1036–1043. doi: 10.1002/prot.20780. [DOI] [PubMed] [Google Scholar]
- 173.Sánchez-González P, Jellali K, Villalobo A. Calmodulin-mediated regulation of the epidermal growth factor receptor. FEBS J. 2010;277:327–342. doi: 10.1111/j.1742-4658.2009.07469.x. [DOI] [PubMed] [Google Scholar]
- 174.Gill GN. Regulation of EGF receptor expression and function. Mol Reprod Dev. 1990;27:46–53. doi: 10.1002/mrd.1080270110. [DOI] [PubMed] [Google Scholar]
- 175.Arjona D, Bello MJ, Alonso ME, Gonzalez-Gomez P, Lomas J, Aminoso C, Lopez-Marin I, Isla A, De Campos JM, Vaquero J, Gutierrez M, Villalobo A, Rey JA. Molecular analysis of the erbB gene family calmodulin-binding and calmodulin-like domains in astrocytic gliomas. Int J Oncol. 2004;25:1489–1494. [PubMed] [Google Scholar]
- 176.Arjona D, Bello MJ, Alonso ME, Aminoso C, Isla A, De Campos JM, Sarasa JL, Gutierrez M, Villalobo A, Rey JA. Molecular analysis of the EGFR gene in astrocytic gliomas: mRNA expression, quantitative-PCR analysis of non-homogeneous gene amplification and DNA sequence alterations. Neuropathol Appl Neurobiol. 2005;31:384–394. doi: 10.1111/j.1365-2990.2005.00653.x. [DOI] [PubMed] [Google Scholar]
- 177.Fenstermaker RA, Ciesielski MJ, Castiglia GJ. Tandem duplication of the epidermal growth factor receptor tyrosine kinase and calcium internalization domains in A-172 glioma cells. Oncogene. 1998;16:3435–3443. doi: 10.1038/sj.onc.1202156. [DOI] [PubMed] [Google Scholar]
- 178.Ciesielski MJ, Fenstermaker RA. Oncogenic epidermal growth factor receptor mutants with tandem duplication: gene structure and effects on receptor function. Oncogene. 2000;19:810–820. doi: 10.1038/sj.onc.1203409. [DOI] [PubMed] [Google Scholar]
- 179.Rotin D, Margolis B, Mohammadi M, Daly RJ, Daum G, Li N, Fischer EH, Burgess WH, Ullrich A, Schlessinger J. SH2 domains prevent tyrosine dephosphorylation of the EGF receptor: identification of Tyr992 as the high-affinity binding site for SH2 domains of phospholipase C γ. EMBO J. 1992;11:559–567. doi: 10.1002/j.1460-2075.1992.tb05087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Chang CM, Shu HK, Ravi L, Pelley RJ, Shu H, Kung HJ. A minor tyrosine phosphorylation site located within the CAIN domain plays a critical role in regulating tissue-specific transformation by erbB kinase. J Virol. 1995;69:1172–1180. doi: 10.1128/jvi.69.2.1172-1180.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Li H, Sanchez-Torres J, Del Carpio A, Salas V, Villalobo A. The ErbB2/Neu/HER2 receptor is a new calmodulin-binding protein. Biochem J. 2004;381:257–266. doi: 10.1042/BJ20040515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.White CD, Li Z, Sacks DB. Calmodulin binds HER2 and modulates HER2 signaling. Biochim Biophys Acta. 2011;1813:1074–1082. doi: 10.1016/j.bbamcr.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.McLaughlin S, Smith SO, Hayman MJ, Murray D. An electrostatic engine model for autoinhibition and activation of the epidermal growth factor receptor (EGFR/ErbB) family. J Gen Physiol. 2005;126:41–53. doi: 10.1085/jgp.200509274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Citri A, Skaria KB, Yarden Y. The deaf and the dumb: the biology of ErbB-2 and ErbB-3. Exp Cell Res. 2003;284:54–65. doi: 10.1016/S0014-4827(02)00101-5. [DOI] [PubMed] [Google Scholar]
- 185.Brandt P, Zurini M, Neve RL, Rhoads RE, Vanaman TC. A C-terminal, calmodulin-like regulatory domain from the plasma membrane Ca2+-pumping ATPase. Proc Natl Acad Sci USA. 1988;85:2914–2918. doi: 10.1073/pnas.85.9.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Verma AK, Filoteo AG, Stanford DR, Wieben ED, Penniston JT, Strehler EE, Fischer R, Heim R, Vogel G, Mathews S, et al. Complete primary structure of a human plasma membrane Ca2+ pump. J Biol Chem. 1988;263:14152–14159. [PubMed] [Google Scholar]
- 187.Caroni P, Zurini M, Clark A, Carafoli E. Further characterization and reconstitution of the purified Ca2+-pumping ATPase of heart sarcolemma. J Biol Chem. 1983;258:7305–7310. [PubMed] [Google Scholar]
- 188.Zurini M, Krebs J, Penniston JT, Carafoli E. Controlled proteolysis of the purified Ca2+-ATPase of the erythrocyte membrane. A correlation between the structure and the function of the enzyme. J Biol Chem. 1984;259:618–627. [PubMed] [Google Scholar]
- 189.Benaim G, Clark A, Carafoli E. ATPase activity and Ca2+ transport by reconstituted tryptic fragments of the Ca2+ pump of the erythrocyte plasma membrane. Cell Calcium. 1986;7:175–186. doi: 10.1016/0143-4160(86)90021-7. [DOI] [PubMed] [Google Scholar]
- 190.Wang KK, Villalobo A, Roufogalis BD. The plasma membrane calcium pump: a multiregulated transporter. Trends Cell Biol. 1992;2:46–52. doi: 10.1016/0962-8924(92)90162-G. [DOI] [PubMed] [Google Scholar]
- 191.Köster S, Pavkov-Keller T, Kuhlbrandt W, Yildiz O. Structure of human Na+/H+ exchanger NHE1 regulatory region in complex with calmodulin and Ca2+ J Biol Chem. 2011;286:40954–40961. doi: 10.1074/jbc.M111.286906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wakabayashi S, Bertrand B, Ikeda T, Pouyssegur J, Shigekawa M. Mutation of calmodulin-binding site renders the Na+/H+ exchanger (NHE1) highly H+-sensitive and Ca2+ regulation-defective. J Biol Chem. 1994;269:13710–13715. [PubMed] [Google Scholar]
- 193.Li X, Ding J, Liu Y, Brix BJ, Fliegel L. Functional analysis of acidic amino acids in the cytosolic tail of the Na+/H+ exchanger. Biochemistry. 2004;43:16477–16486. doi: 10.1021/bi048538v. [DOI] [PubMed] [Google Scholar]
- 194.Mebratu YA, Leyva-Baca I, Wathelet MG, Lacey N, Chand HS, Choi AMK, Tesfaigzi Y. Bik reduces hyperplastic cells by increasing Bak and activating DAPk1 to juxtapose ER and mitochondria. Nat Commun. 2017;8:803. doi: 10.1038/s41467-017-00975-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Barroso MR, Bernd KK, DeWitt ND, Chang A, Mills K, Sztul ES. A novel Ca2+-binding protein, p22, is required for constitutive membrane traffic. J Biol Chem. 1996;271:10183–10187. doi: 10.1074/jbc.271.17.10183. [DOI] [PubMed] [Google Scholar]
- 196.Naik UP, Patel PM, Parise LV. Identification of a novel calcium-binding protein that interacts with the integrin αIIb cytoplasmic domain. J Biol Chem. 1997;272:4651–4654. doi: 10.1074/jbc.272.8.4651. [DOI] [PubMed] [Google Scholar]
- 197.Timm S, Titus B, Bernd K, Barroso M. The EF-hand Ca2+-binding protein p22 associates with microtubules in an N-myristoylation-dependent manner. Mol Biol Cell. 1999;10:3473–3488. doi: 10.1091/mbc.10.10.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.DeFalco TA, Bender KW, Snedden WA. Breaking the code: Ca2+ sensors in plant signalling. Biochem J. 2009;425:27–40. doi: 10.1042/BJ20091147. [DOI] [PubMed] [Google Scholar]
- 199.Patel S, Joseph SK, Thomas AP. Molecular properties of inositol 1,4,5-trisphosphate receptors. Cell Calcium. 1999;25:247–264. doi: 10.1054/ceca.1999.0021. [DOI] [PubMed] [Google Scholar]
- 200.Tang W, Sencer S, Hamilton SL. Calmodulin modulation of proteins involved in excitation–contraction coupling. Front Biosci. 2002;7:d1583–d1589. doi: 10.2741/tang. [DOI] [PubMed] [Google Scholar]
- 201.Zhu J, Feng Q, Stathopulos PB. The STIM-Orai pathway: STIM-Orai structures: isolated and in complex. Adv Exp Med Biol. 2017;993:15–38. doi: 10.1007/978-3-319-57732-6_2. [DOI] [PubMed] [Google Scholar]
- 202.Thiel G, Rubil S, Lesch A, Guethlein LA, Rossler OG. Transient receptor potential TRPM3 channels: pharmacology, signaling, and biological functions. Pharmacol Res. 2017;124:92–99. doi: 10.1016/j.phrs.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 203.Zhu MX. Multiple roles of calmodulin and other Ca2+-binding proteins in the functional regulation of TRP channels. Pflugers Arch. 2005;451:105–115. doi: 10.1007/s00424-005-1427-1. [DOI] [PubMed] [Google Scholar]
- 204.Pankratov Y, Lalo U. Calcium permeability of ligand-gated Ca2+ channels. Eur J Pharmacol. 2014;739:60–73. doi: 10.1016/j.ejphar.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 205.Neely A, Hidalgo P. Structure-function of proteins interacting with the α1 pore-forming subunit of high-voltage-activated calcium channels. Front Physiol. 2014;5:209. doi: 10.3389/fphys.2014.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Abriel H, Kass RS. Regulation of the voltage-gated cardiac sodium channel Nav1.5 by interacting proteins. Trends Cardiovasc Med. 2005;15:35–40. doi: 10.1016/j.tcm.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 207.Marionneau C, Abriel H. Regulation of the cardiac Na+ channel NaV1.5 by post-translational modifications. J Mol Cell Cardiol. 2015;82:36–47. doi: 10.1016/j.yjmcc.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 208.Marques-Carvalho MJ, Oppermann J, Munoz E, Fernandes AS, Gabant G, Cadene M, Heinemann SH, Schonherr R, Morais-Cabral JH. Molecular insights into the mechanism of calmodulin inhibition of the EAG1 potassium channel. Structure. 2016;24:1742–1754. doi: 10.1016/j.str.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Sachyani D, Dvir M, Strulovich R, Tria G, Tobelaim W, Peretz A, Pongs O, Svergun D, Attali B, Hirsch JA. Structural basis of a Kv7.1 potassium channel gating module: studies of the intracellular C-terminal domain in complex with calmodulin. Structure. 2014;22:1582–1594. doi: 10.1016/j.str.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 210.Kohler R. Single-nucleotide polymorphisms in vascular Ca2+-activated K+-channel genes and cardiovascular disease. Pflugers Arch. 2010;460:343–351. doi: 10.1007/s00424-009-0768-6. [DOI] [PubMed] [Google Scholar]
- 211.Yang T, Colecraft HM. Calmodulin regulation of TMEM16A and 16B Ca2+-activated chloride channels. Channels (Austin) 2016;10:38–44. doi: 10.1080/19336950.2015.1058455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hanna RA, Campbell RL, Davies PL. Calcium-bound structure of calpain and its mechanism of inhibition by calpastatin. Nature. 2008;456:409–412. doi: 10.1038/nature07451. [DOI] [PubMed] [Google Scholar]
- 213.de Ribeiro EA, Pinotsis N, Ghisleni A, Salmazo A, Konarev PV, Kostan J, Sjoblom B, Schreiner C, Polyansky AA, Gkougkoulia EA, Holt MR, Aachmann FL, Zagrovic B, Bordignon E, Pirker KF, Svergun DI, Gautel M, Djinovic-Carugo K. The structure and regulation of human muscle α-actinin. Cell. 2014;159:1447–1460. doi: 10.1016/j.cell.2014.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Galkin VE, Orlova A, Salmazo A, Djinovic-Carugo K, Egelman EH. Opening of tandem calponin homology domains regulates their affinity for F-actin. Nat Struct Mol Biol. 2010;17:614–616. doi: 10.1038/nsmb.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kissinger CR, Parge HE, Knighton DR, Lewis CT, Pelletier LA, Tempczyk A, Kalish VJ, Tucker KD, Showalter RE, Moomaw EW, et al. Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature. 1995;378:641–644. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
- 216.Chattopadhyaya R, Meador WE, Means AR, Quiocho FA. Calmodulin structure refined at 1.7 A resolution. J Mol Biol. 1992;228:1177–1192. doi: 10.1016/0022-2836(92)90324-D. [DOI] [PubMed] [Google Scholar]
- 217.Di Leva F, Domi T, Fedrizzi L, Lim D, Carafoli E. The plasma membrane Ca2+ ATPase of animal cells: structure, function and regulation. Arch Biochem Biophys. 2008;476:65–74. doi: 10.1016/j.abb.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 218.Harper JF, Binder BM, Sussman MR. Calcium and lipid regulation of an Arabidopsis protein kinase expressed in Escherichia coli. Biochemistry. 1993;32:3282–3290. doi: 10.1021/bi00064a010. [DOI] [PubMed] [Google Scholar]
- 219.Li JL, Baker DA, Cox LS. Sexual stage-specific expression of a third calcium-dependent protein kinase from Plasmodium falciparum. Biochim Biophys Acta. 2000;1491:341–349. doi: 10.1016/S0167-4781(00)00032-4. [DOI] [PubMed] [Google Scholar]
- 220.Kieschnick H, Wakefield T, Narducci CA, Beckers C. Toxoplasma gondii attachment to host cells is regulated by a calmodulin-like domain protein kinase. J Biol Chem. 2001;276:12369–12377. doi: 10.1074/jbc.M011045200. [DOI] [PubMed] [Google Scholar]
- 221.Backman L. α-actinin of the chlorarchiniophyte Bigelowiella natans. PeerJ. 2018;6:e4288. doi: 10.7717/peerj.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Millake DB, Blanchard AD, Patel B, Critchley DR. The cDNA sequence of a human placental α-actinin. Nucleic Acids Res. 1989;17:6725. doi: 10.1093/nar/17.16.6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.del Arco A, Satrustegui J. Identification of a novel human subfamily of mitochondrial carriers with calcium-binding domains. J Biol Chem. 2004;279:24701–24713. doi: 10.1074/jbc.M401417200. [DOI] [PubMed] [Google Scholar]
- 224.Kuno T, Tanaka H, Mukai H, Chang CD, Hiraga K, Miyakawa T, Tanaka C. cDNA cloning of a calcineurin B homolog in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1991;180:1159–1163. doi: 10.1016/S0006-291X(05)81188-X. [DOI] [PubMed] [Google Scholar]
- 225.Guerini D, Krinks MH, Sikela JM, Hahn WE, Klee CB. Isolation and sequence of a cDNA clone for human calcineurin B, the Ca2+-binding subunit of the Ca2+/calmodulin-stimulated protein phosphatase. DNA. 1989;8:675–682. doi: 10.1089/dna.1.1989.8.675. [DOI] [PubMed] [Google Scholar]
- 226.Emori Y, Saigo K. Calpain localization changes in coordination with actin-related cytoskeletal changes during early embryonic development of Drosophila. J Biol Chem. 1994;269:25137–25142. [PubMed] [Google Scholar]
- 227.Aoki K, Imajoh S, Ohno S, Emori Y, Koike M, Kosaki G, Suzuki K. Complete amino acid sequence of the large subunit of the low-Ca2 + -requiring form of human Ca2 + -activated neutral protease (μCANP) deduced from its cDNA sequence. FEBS Lett. 1986;205:313–317. doi: 10.1016/0014-5793(86)80919-X. [DOI] [PubMed] [Google Scholar]
- 228.Imajoh S, Aoki K, Ohno S, Emori Y, Kawasaki H, Sugihara H, Suzuki K. Molecular cloning of the cDNA for the large subunit of the high-Ca2+-requiring form of human Ca2+-activated neutral protease. Biochemistry. 1988;27:8122–8128. doi: 10.1021/bi00421a022. [DOI] [PubMed] [Google Scholar]
- 229.Richard I, Broux O, Allamand V, Fougerousse F, Chiannilkulchai N, Bourg N, Brenguier L, Devaud C, Pasturaud P, Roudaut C, et al. Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell. 1995;81:27–40. doi: 10.1016/0092-8674(95)90368-2. [DOI] [PubMed] [Google Scholar]
- 230.Newton G, Weremowicz S, Morton CC, Copeland NG, Gilbert DJ, Jenkins NA, Lawler J. Characterization of human and mouse cartilage oligomeric matrix protein. Genomics. 1994;24:435–439. doi: 10.1006/geno.1994.1649. [DOI] [PubMed] [Google Scholar]
- 231.Lehn DA, Brown LJ, Simonson GD, Moran SM, MacDonald MJ. The sequence of a human mitochondrial glycerol-3-phosphate dehydrogenase-encoding cDNA. Gene. 1994;150:417–418. doi: 10.1016/0378-1119(94)90469-3. [DOI] [PubMed] [Google Scholar]
- 232.Wernimont AK, Artz JD, Finerty P, Jr, Lin YH, Amani M, Allali-Hassani A, Senisterra G, Vedadi M, Tempel W, Mackenzie F, Chau I, Lourido S, Sibley LD, Hui R. Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium. Nat Struct Mol Biol. 2010;17:596–601. doi: 10.1038/nsmb.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Davletova S, Meszaros T, Miskolczi P, Oberschall A, Torok K, Magyar Z, Dudits D, Deak M. Auxin and heat shock activation of a novel member of the calmodulin like domain protein kinase gene family in cultured alfalfa cells. J Exp Bot. 2001;52:215–221. doi: 10.1093/jexbot/52.355.215. [DOI] [PubMed] [Google Scholar]
- 234.Pettko-Szandtner A, Meszaros T, Horvath GV, Bako L, Csordas-Toth E, Blastyak A, Zhiponova M, Miskolczi P, Dudits D. Activation of an alfalfa cyclin-dependent kinase inhibitor by calmodulin-like domain protein kinase. Plant J. 2006;46:111–123. doi: 10.1111/j.1365-313X.2006.02677.x. [DOI] [PubMed] [Google Scholar]
- 235.Dudits D, Abraham E, Miskolczi P, Ayaydin F, Bilgin M, Horvath GV. Cell-cycle control as a target for calcium, hormonal and developmental signals: the role of phosphorylation in the retinoblastoma-centred pathway. Ann Bot. 2011;107:1193–1202. doi: 10.1093/aob/mcr038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Li J, Lee YR, Assmann SM. Guard cells possess a calcium-dependent protein kinase that phosphorylates the KAT1 potassium channel. Plant Physiol. 1998;116:785–795. doi: 10.1104/pp.116.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Mori IC, Muto S. Abscisic acid activates a 48-kDa protein kinase in guard cell protoplasts. Plant Physiol. 1997;113:833–839. doi: 10.1104/pp.113.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Farmer PK, Choi JH. Calcium and phospholipid activation of a recombinant calcium-dependent protein kinase (DcCPK1) from carrot (Daucus carota L.) Biochim Biophys Acta. 1999;1434:6–17. doi: 10.1016/S0167-4838(99)00166-1. [DOI] [PubMed] [Google Scholar]
- 239.Suen KL, Choi JH. Isolation and sequence analysis of a cDNA clone for a carrot calcium-dependent protein kinase: homology to calcium/calmodulin-dependent protein kinases and to calmodulin. Plant Mol Biol. 1991;17:581–590. doi: 10.1007/BF00037045. [DOI] [PubMed] [Google Scholar]
- 240.Lindzen E, Choi JH. A carrot cDNA encoding an atypical protein kinase homologous to plant calcium-dependent protein kinases. Plant Mol Biol. 1995;28:785–797. doi: 10.1007/BF00042065. [DOI] [PubMed] [Google Scholar]
- 241.Prakash SRS, Jayabaskaran C. Heterologous expression and biochemical characterization of two calcium-dependent protein kinase isoforms CaCPK1 and CaCPK2 from chickpea. J Plant Physiol. 2006;163:1083–1093. doi: 10.1016/j.jplph.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 242.Camoni L, Fullone MR, Marra M, Aducci P. The plasma membrane H+-ATPase from maize roots is phosphorylated in the C-terminal domain by a calcium-dependent protein kinase. Physiol Plant. 1998;104:549–555. doi: 10.1034/j.1399-3054.1998.1040405.x. [DOI] [Google Scholar]
- 243.Allwood EG, Smertenko AP, Hussey PJ. Phosphorylation of plant actin-depolymerising factor by calmodulin-like domain protein kinase. FEBS Lett. 2001;499:97–100. doi: 10.1016/S0014-5793(01)02528-5. [DOI] [PubMed] [Google Scholar]
- 244.Szczegielniak J, Liwosz A, Jurkowski I, Loog M, Dobrowolska G, Ek P, Harmon AC, Muszynska G. Calcium-dependent protein kinase from maize seedlings activated by phospholipids. Eur J Biochem. 2000;267:3818–3827. doi: 10.1046/j.1432-1327.2000.01420.x. [DOI] [PubMed] [Google Scholar]
- 245.Szczegielniak J, Klimecka M, Liwosz A, Ciesielski A, Kaczanowski S, Dobrowolska G, Harmon AC, Muszynska G. A wound-responsive and phospholipid-regulated maize calcium-dependent protein kinase. Plant Physiol. 2005;139:1970–1983. doi: 10.1104/pp.105.066472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Takezawa D, Patil S, Bhatia A, Poovaiah BW. Calcium-dependent protein kinase genes in corn roots. J Plant Physiol. 1996;149:329–335. doi: 10.1016/S0176-1617(96)80130-1. [DOI] [PubMed] [Google Scholar]
- 247.Kumar KG, Ullanat R, Jayabaskaran C. Molecular cloning, characterization, tissue-specific and phytohormone-induced expression of calcium-dependent protein kinase gene in cucumber (Cucumis sativus L.) J Plant Physiol. 2004;161:1061–1071. doi: 10.1016/j.jplph.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 248.Lu B, Ding R, Zhang L, Yu X, Huang B, Chen W. Molecular cloning and characterization of a novel calcium-dependent protein kinase gene IiCPK2 responsive to polyploidy from tetraploid Isatis indigotica. J Biochem Mol Biol. 2006;39:607–617. doi: 10.5483/bmbrep.2006.39.5.607. [DOI] [PubMed] [Google Scholar]
- 249.Guenther JF, Roberts DM. Water-selective and multifunctional aquaporins from Lotus japonicus nodules. Planta. 2000;210:741–748. doi: 10.1007/s004250050675. [DOI] [PubMed] [Google Scholar]
- 250.Binder BM, Harper JF, Sussman MR. Characterization of an Arabidopsis calmodulin-like domain protein kinase purified from Escherichia coli using an affinity sandwich technique. Biochemistry. 1994;33:2033–2041. doi: 10.1021/bi00174a008. [DOI] [PubMed] [Google Scholar]
- 251.Hrabak EM, Dickmann LJ, Satterlee JS, Sussman MR. Characterization of eight new members of the calmodulin-like domain protein kinase gene family from Arabidopsis thaliana. Plant Mol Biol. 1996;31:405–412. doi: 10.1007/BF00021802. [DOI] [PubMed] [Google Scholar]
- 252.Xing T, Wang XJ, Malik K, Miki BL. Ectopic expression of an Arabidopsis calmodulin-like domain protein kinase-enhanced NADPH oxidase activity and oxidative burst in tomato protoplasts. Mol Plant Microbe Interact. 2001;14:1261–1264. doi: 10.1094/MPMI.2001.14.10.1261. [DOI] [PubMed] [Google Scholar]
- 253.Krysan PJ, Young JC, Tax F, Sussman MR. Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc Natl Acad Sci USA. 1996;93:8145–8150. doi: 10.1073/pnas.93.15.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Yoon GM, Dowd PE, Gilroy S, McCubbin AG. Calcium-dependent protein kinase isoforms in Petunia have distinct functions in pollen tube growth, including regulating polarity. Plant Cell. 2006;18:867–878. doi: 10.1105/tpc.105.037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Rudd JJ, Franklin F, Lord JM, Franklin-Tong VE. Increased phosphorylation of a 26-kD pollen protein is induced by the self-incompatibility response in Papaver rhoeas. Plant Cell. 1996;8:713–724. doi: 10.2307/3870346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Lakatos L, Hutvagner G, Banfalvi Z. Potato protein kinase StCPK1: a putative evolutionary link between CDPKs and CRKs. Biochim Biophys Acta. 1998;1442:101–108. doi: 10.1016/S0167-4781(98)00148-1. [DOI] [PubMed] [Google Scholar]
- 257.Yoo BC, Lee JY, Lucas WJ. Analysis of the complexity of protein kinases within the phloem sieve tube system. Characterization of Cucurbita maxima calmodulin-like domain protein kinase 1. J Biol Chem. 2002;277:15325–15332. doi: 10.1074/jbc.M200382200. [DOI] [PubMed] [Google Scholar]
- 258.Asano T, Kunieda N, Omura Y, Ibe H, Kawasaki T, Takano M, Sato M, Furuhashi H, Mujin T, Takaiwa F, Wu Cy CY, Tada Y, Satozawa T, Sakamoto M, Shimada H. Rice SPK, a calmodulin-like domain protein kinase, is required for storage product accumulation during seed development: phosphorylation of sucrose synthase is a possible factor. Plant Cell. 2002;14:619–628. doi: 10.1105/tpc.010454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Shimada H, Koishihara H, Saito Y, Arashima Y, Furukawa T, Hayashi H. A rice antisense SPK transformant that lacks the accumulation of seed storage substances shows no correlation between sucrose concentration in phloem sap and demand for carbon sources in the sink organs. Plant Cell Physiol. 2004;45:1105–1109. doi: 10.1093/pcp/pch122. [DOI] [PubMed] [Google Scholar]
- 260.Lee JW, Zhang Y, Weaver CD, Shomer NH, Louis CF, Roberts DM. Phosphorylation of nodulin 26 on serine 262 affects its voltage-sensitive channel activity in planar lipid bilayers. J Biol Chem. 1995;270:27051–27057. doi: 10.1074/jbc.270.45.27051. [DOI] [PubMed] [Google Scholar]
- 261.Douglas P, Moorhead G, Hong Y, Morrice N, MacKintosh C. Purification of a nitrate reductase kinase from Spinacia oleracea leaves, and its identification as a calmodulin-domain protein kinase. Planta. 1998;206:435–442. doi: 10.1007/s004250050419. [DOI] [PubMed] [Google Scholar]
- 262.Huang JZ, Huber SC. Phosphorylation of synthetic peptides by a CDPK and plant SNF1-related protein kinase. Influence of proline and basic amino acid residues at selected positions. Plant Cell Physiol. 2001;42:1079–1087. doi: 10.1093/pcp/pce137. [DOI] [PubMed] [Google Scholar]
- 263.Llop-Tous I, Dominguez-Puigjaner E, Vendrell M. Characterization of a strawberry cDNA clone homologous to calcium-dependent protein kinases that is expressed during fruit ripening and affected by low temperature. J Exp Bot. 2002;53:2283–2285. doi: 10.1093/jxb/erf103. [DOI] [PubMed] [Google Scholar]
- 264.Lino B, Baizabal-Aguirre VM, Gonzalez de la Vara LE. The plasma-membrane H+-ATPase from beet root is inhibited by a calcium-dependent phosphorylation. Planta. 1998;204:352–359. doi: 10.1007/s004250050266. [DOI] [PubMed] [Google Scholar]
- 265.Yoon GM, Cho HS, Ha HJ, Liu JR, Lee HS. Characterization of NtCDPK1, a calcium-dependent protein kinase gene in Nicotiana tabacum, and the activity of its encoded protein. Plant Mol Biol. 1999;39:991–1001. doi: 10.1023/A:1006170512542. [DOI] [PubMed] [Google Scholar]
- 266.Rutschmann F, Stalder U, Piotrowski M, Oecking C, Schaller A. LeCPK1, a calcium-dependent protein kinase from tomato. Plasma membrane targeting and biochemical characterization. Plant Physiol. 2002;129:156–168. doi: 10.1104/pp.000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Chang W, Fu G, Chen X, Zhu J, Zhang Z. Biochemical characterization of a calcium-sensitive protein kinase LeCPK2 from tomato. Indian J Biochem Biophys. 2011;48:148–153. [PubMed] [Google Scholar]
- 268.Chang WJ, Su HS, Li WJ, Zhang ZL. Expression profiling of a novel calcium-dependent protein kinase gene, LeCPK2, from tomato (Solanum lycopersicum) under heat and pathogen-related hormones. Biosci Biotechnol Biochem. 2009;73:2427–2431. doi: 10.1271/bbb.90385. [DOI] [PubMed] [Google Scholar]
- 269.Leclercq J, Ranty B, Sanchez-Ballesta MT, Li Z, Jones B, Jauneau A, Pech JC, Latche A, Ranjeva R, Bouzayen M. Molecular and biochemical characterization of LeCRK1, a ripening-associated tomato CDPK-related kinase. J Exp Bot. 2005;56:25–35. doi: 10.1093/jxb/eri003. [DOI] [PubMed] [Google Scholar]
- 270.Martinez-Noël G, Nagaraj VJ, Calo G, Wiemken A, Pontis HG. Sucrose regulated expression of a Ca2+-dependent protein kinase (TaCDPK1) gene in excised leaves of wheat. Plant Physiol Biochem. 2007;45:410–419. doi: 10.1016/j.plaphy.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 271.Ganguly S, Singh M. Characterization of a second calcium-dependent protein kinase from winged bean. Phytochemistry. 1998;48:61–70. doi: 10.1016/S0031-9422(97)01096-0. [DOI] [PubMed] [Google Scholar]
- 272.Ganguly S, Singh M. Purification and characterization of a protein phosphatase from winged bean. Phytochemistry. 1999;52:239–246. doi: 10.1016/S0031-9422(98)00473-7. [DOI] [PubMed] [Google Scholar]
- 273.Iyer GR, Singh S, Kaur I, Agarwal S, Siddiqui MA, Bansal A, Kumar G, Saini E, Paul G, Mohmmed A, Chitnis CE, Malhotra P. Calcium-dependent phosphorylation of Plasmodium falciparum serine repeat antigen 5 triggers merozoite egress. J Biol Chem. 2018;293:9736–9746. doi: 10.1074/jbc.RA117.001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Bansal A, Singh S, More KR, Hans D, Nangalia K, Yogavel M, Sharma A, Chitnis CE. Characterization of Plasmodium falciparum calcium-dependent protein kinase 1 (PfCDPK1) and its role in microneme secretion during erythrocyte invasion. J Biol Chem. 2013;288:1590–1602. doi: 10.1074/jbc.M112.411934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Cavagnino A, Rossi F, Rizzi M. The potent antiplasmodial calmodulin-antagonist trifluoperazine inhibits plasmodium falciparum calcium-dependent protein kinase 4. Protein Pept Lett. 2011;18:1273–1279. doi: 10.2174/092986611797642742. [DOI] [PubMed] [Google Scholar]
- 276.Billker O, Dechamps S, Tewari R, Wenig G, Franke-Fayard B, Brinkmann V. Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite. Cell. 2004;117:503–514. doi: 10.1016/S0092-8674(04)00449-0. [DOI] [PubMed] [Google Scholar]
- 277.Sugi T, Kato K, Kobayashi K, Pandey K, Takemae H, Kurokawa H, Tohya Y, Akashi H. Molecular analyses of Toxoplasma gondii calmodulin-like domain protein kinase isoform 3. Parasitol Int. 2009;58:416–423. doi: 10.1016/j.parint.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 278.Bennett MK, Kennedy MB. Deduced primary structure of the β subunit of brain type II Ca2+/calmodulin-dependent protein kinase determined by molecular cloning. Proc Natl Acad Sci USA. 1987;84:1794–1798. doi: 10.1073/pnas.84.7.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Takio K, Blumenthal DK, Walsh KA, Titani K, Krebs EG. Amino acid sequence of rabbit skeletal muscle myosin light chain kinase. Biochemistry. 1986;25:8049–8057. doi: 10.1021/bi00372a038. [DOI] [PubMed] [Google Scholar]
- 280.Reimann EM, Titani K, Ericsson LH, Wade RD, Fischer EH, Walsh KA. Homology of the γ subunit of phosphorylase b kinase with cAMP-dependent protein kinase. Biochemistry. 1984;23:4185–4192. doi: 10.1021/bi00313a027. [DOI] [PubMed] [Google Scholar]