Abstract
Missense mutations in the gene encoding the microtubule-associated protein TAU (current and approved symbol is MAPT) cause autosomal dominant forms of frontotemporal dementia. Multiple models of frontotemporal dementia based on transgenic expression of human TAU in experimental model organisms, including Drosophila, have been described. These models replicate key features of the human disease but do not faithfully recreate the genetic context of the human disorder. Here we use CRISPR-Cas-mediated gene editing to model frontotemporal dementia caused by the TAU P301L mutation by creating the orthologous mutation, P251L, in the endogenous Drosophila tau gene. Flies heterozygous or homozygous for Tau P251L display age-dependent neurodegeneration, display metabolic defects, and accumulate DNA damage in affected neurons. To understand the molecular events promoting neuronal dysfunction and death in knock-in flies, we performed single-cell RNA sequencing on approximately 130,000 cells from brains of Tau P251L mutant and control flies. We found that expression of disease-associated mutant tau altered gene expression cell autonomously in all neuronal cell types identified. Gene expression was also altered in glial cells, suggestive of non-cell-autonomous regulation. Cell signaling pathways, including glial–neuronal signaling, were broadly dysregulated as were brain region and cell type–specific protein interaction networks and gene regulatory programs. In summary, we present here a genetic model of tauopathy that faithfully recapitulates the genetic context and phenotypic features of the human disease, and use the results of comprehensive single-cell sequencing analysis to outline pathways of neurotoxicity and highlight the potential role of non-cell-autonomous changes in glia.
The neuronal microtubule–associated protein TAU (current and approved symbol is MAPT) forms insoluble deposits termed neurofibrillary tangles and neuritic threads in neuronal soma and processes in a diverse group of age-dependent neurodegenerative diseases, including Alzheimer's disease and frontotemporal dementia. These disorders have collectively been termed “tauopathies” (Feany and Dickson 1996; Goedert 2004; Götz et al. 2019). Although wild-type TAU is deposited in Alzheimer's disease and other more common tauopathies, missense mutations in TAU occur in rarer familial forms of tauopathy, causing neurodegeneration and insoluble TAU deposition. Autosomal dominant disease-causing mutations occur throughout the TAU protein but are particularly frequent in exon 10, which contains one of four microtubule binding repeats (Ghetti et al. 2015). These repeats mediate microtubule (Lee et al. 1989; Butner and Kirschner 1991) and actin (Cabrales Fontela et al. 2017) binding and are important determinants of TAU aggregation (von Bergen et al. 2000). Experimental models of tauopathy have been created in diverse model organisms, from yeast to non-human primates, by expressing wild-type or frontotemporal dementia–associated mutant forms of human TAU in transgenic animals. Mutant forms of TAU are typically more toxic than wild-type TAU in transgenic model organisms. Work in these models has implicated a number of cellular pathways in mediating TAU neurotoxicity, including mitochondrial dysfunction (Rhein et al. 2009; DuBoff et al. 2012), oxidative stress (Dias-Santagata et al. 2007; Dumont et al. 2011), and aberrant cell-cycle reentry of postmitotic neurons (Andorfer et al. 2005; Khurana et al. 2006).
However, although transgenic models have been useful, they do not faithfully replicate the genetic underpinnings of the authentic human disorders and thus may not allow the identification and study of the full complement of important mediators of tauopathy pathogenesis. We have therefore used CRISPR-Cas9 gene editing to model familial frontotemporal dementia caused by missense mutations in TAU more precisely in Drosophila. Mutation of proline 301 to leucine in exon 10 is the most common mutation of TAU in frontotemporal dementia patients (Poorkaj et al. 2001) and has been frequently modeled in transgenic animals (Goedert and Jakes 2005). The overall structure and expression of TAU are conserved from mammals to Drosophila (Heidary and Fortini 2001), with proline 251 being orthologous to human proline 301. We have therefore replaced Drosophila Tau proline 251 with leucine (P251L) and phenotypically analyzed the resultant homozygous and heterozygous animals with age. We have additionally performed single-cell sequencing to identify cell populations, networks, and signaling systems altered by mutant tau expression.
Results
Phenotypic analysis of a Drosophila knock-in model of frontotemporal dementia
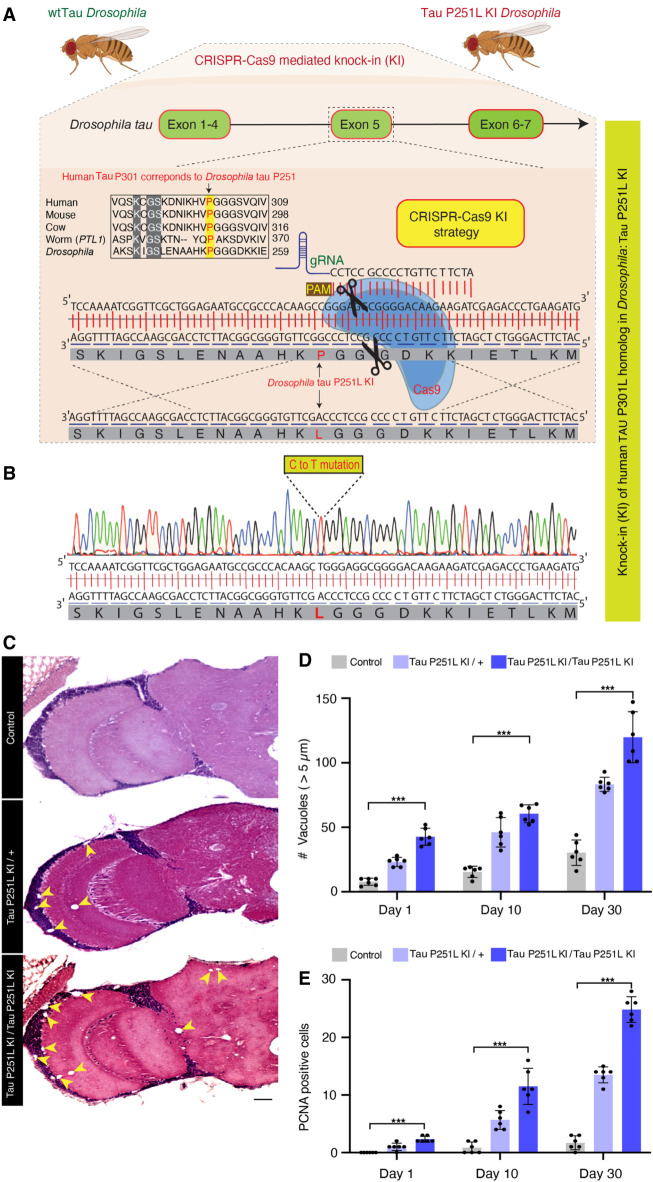
We used CRISPR-Cas9 gene editing to recapitulate the genetic basis of human frontotemporal dementia in the powerful genetic experimental organism Drosophila by modeling the disease-causing proline 301 to leucine in fly Tau. Protein sequence alignment shows that the microtubule-binding domains, including human TAU proline 301, are evolutionary conserved from Drosophila to humans (Supplemental Fig. S1). The homologous residue of the human TAU proline 301, Drosophila Tau proline 251, was mutated to leucine using a highly efficient guide RNA along with single-stranded oligodeoxynucleotides (Fig. 1A,B). Mutant Tau was expressed at equivalent levels to wild-type Tau (Supplemental Fig. S2A).
Figure 1.
CRISPR-Cas9-mediated knock-in model of frontotemporal dementia in Drosophila. (A) CRISPR-Cas9 gene editing strategy to knock-in the human TAU P301L homologous mutation in Drosophila, Tau P251L, located in exon 5 of Drosophila tau. (B) Successful mutation in homozygous Tau P251L knock-in flies. (C,D) Hematoxylin and eosin staining reveals evidence of neurodegeneration as seen by an increased number of brain vacuoles (arrowheads) with age in homozygous and heterozygous knock-in animals. (C) Scale bar represents 10 µm. (E) Neurodegeneration is accompanied by abnormal cell-cycle reentry as marked by proliferating cell nuclear antigen (PCNA) staining. Flies are 30 d old in C and the age indicated in the figure labels in D,E. (D,E) n = 6 per genotype and time point. Data are presented as mean ± SD. (***) P < 0.001, one-way ANOVA with Tukey post-hoc analysis.
The expression of frontotemporal dementia-linked forms of mutant TAU, including P301L, leads to age-dependent neuronal loss in patients and in transgenic models (Götz et al. 2001; Lewis et al. 2001; Yoshiyama et al. 2007; Ghetti et al. 2015). We thus examined the histology of brains of heterozygous (P251L/+) and homozygous (P251L) Tau knock-in animals with age. We found increased numbers of cortical and neuropil vacuoles in knock-in animals (Fig. 1C,D). Neurodegeneration in Drosophila is frequently accompanied by the formation of brain vacuoles (Heisenberg and Böhl 1979; Buchanan and Benzer 1993; Wittmann et al. 2001; Ordonez et al. 2018). Increasing numbers of vacuoles were observed with advancing age, as well as with two copies of the P251L compared with one copy (Fig. 1C,D). Inappropriate neuronal cycle reentry is a feature of human tauopathy (Husseman et al. 2000) and human TAU transgenic animals (Andorfer et al. 2005; Khurana et al. 2006). We stained control and Tau P251L knock-in brains with an antibody directed to proliferating cell nuclear antigen (PCNA) to assess cell-cycle activation (Khurana et al. 2006). We observed increasing cell-cycle reentry with age in Tau P251L knock-in brains, with more cell-cycle activation in homozygotes compared with heterozygotes (Fig. 1E; Supplemental Fig. S2B).
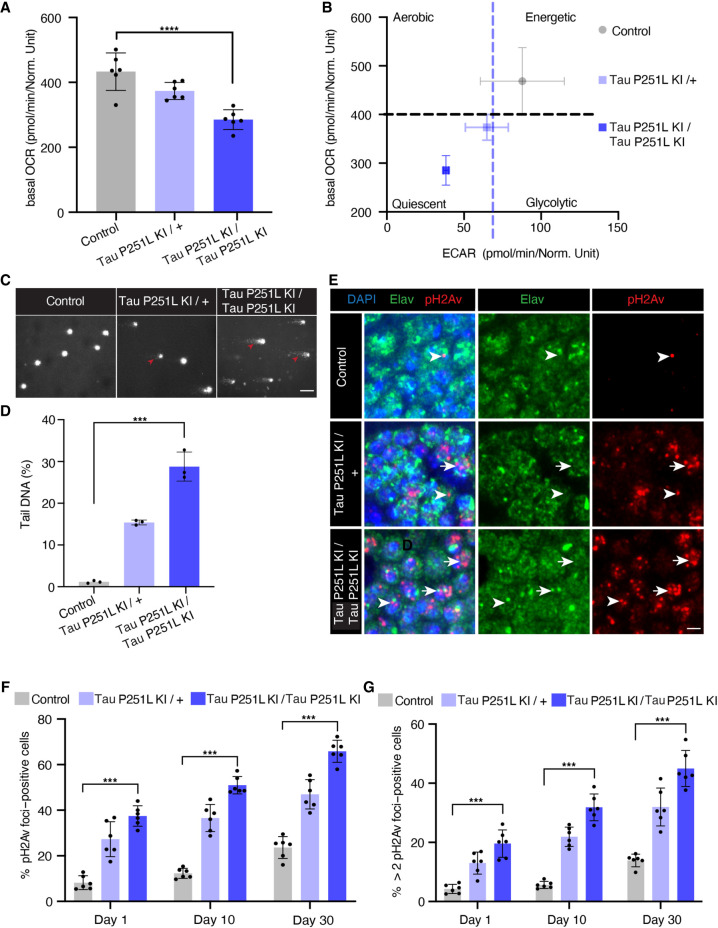
Metabolic alterations and mitochondrial dysfunction are pervasive features of neurodegenerative diseases, including tauopathies (DuBoff et al. 2013; Götz et al. 2019). We thus performed metabolic analysis on intact whole-fly brains using the Seahorse XFe96 analyzer (Neville et al. 2018). We observed a reduced basal oxygen consumption rate (OCR) and a shift in mitochondrial bioenergetics to quiescent metabolic state in Tau 251L knock-in animals, with homozygotes showing more impairment than heterozygotes (Fig. 2A,B).
Figure 2.
Mitochondrial dysfunction and DNA damage in Tau P251L knock-in brains. (A,B) Decreased oxygen consumption rate (OCR; A) and shift to a quiescent metabolic phenotype as indicated by plotting the OCR versus the extracellular acidification rate (ECAR; B) in homozygous and heterozygous Tau P251L knock-in brains compared with controls. n = 6 per genotype. (C,D) Elevated levels of DNA damage in Tau P251L knock-in brains as indicated by increased tail length (arrowheads) following electrophoresis of nuclei from dissociated brains in the comet assay. n = 3 per genotype. (E) Increase in the number of Kenyon cells neurons (identified by the neuronal marker elav) containing DNA double-strand breaks as marked by pH2Av foci (arrowheads; arrows indicate neuronal nuclei with more than two foci) in histological sections of mushroom bodies (Kenyon cells) from Tau P251L knock-in animals, as quantified in F,G. n = 6 per genotype and time point. Scale bars represent 5 µm. Flies are 10 d old in A–D, 30 d old in E, and the age indicated in the figure labels in F,G. Data are presented as mean ± SD. (***) P < 0.001, one-way ANOVA with Tukey post-hoc analysis.
Oxidative stress accompanying mitochondrial dysfunction results in damage to key cellular substrates, including DNA. DNA damage commonly occurs in age-related neurodegenerative diseases (Welch and Tsai 2022), including tauopathies (Khurana et al. 2012; Shanbhag et al. 2019; Thadathil et al. 2021). We took two approaches to examining DNA damage in Tau P251L knock-in animals. First, we used the comet assay, in which DNA single- or double-strand breaks are showed using single-cell gel electrophoresis (Khurana et al. 2012; Frost et al. 2014). We observed that nuclei from the brains of Tau P251L knock-in flies displayed almost twofold longer comet tails than those of controls (Fig. 2C, arrowheads, D).
As a second measure of DNA damage, we immunostained for the histone variant H2Av phosphorylated at serine 137 (pH2Av), a marker of DNA double-strand breaks (Madigan et al. 2002; Khurana et al. 2012; Frost et al. 2014). We found significantly increased numbers of double-strand breaks within neurons (Fig. 2E, arrows, arrowheads, F,G). DNA double-strand breaks were elevated with age and in homozygous compared with heterozygous Tau P251L knock-in flies (Fig. 2E–G). Increased DNA damage was assessed by counting both the numbers of Kenyon cell nuclei containing pH2Av foci and the numbers of Kenyon cell nuclei containing more than two foci (Fig. 2E, arrows, G), which correlate with increased numbers of DNA double-strand breaks (Hong and Choi 2013; Lapytsko et al. 2015).
Single-cell RNA sequencing reveals gene expression changes mediated by pathologic Tau
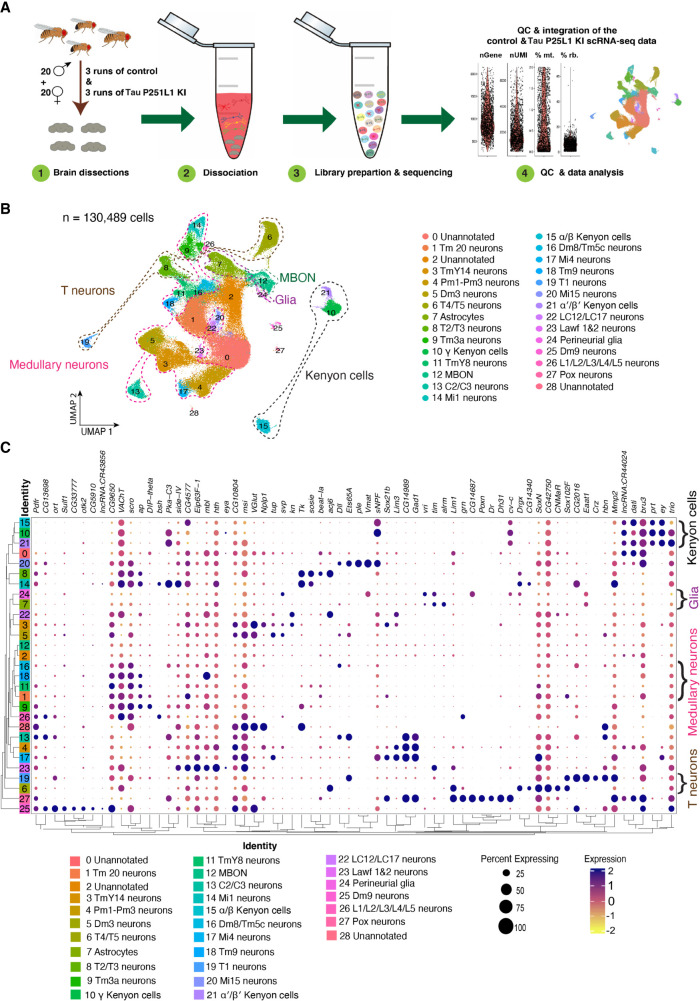
Our Tau P251L knock-in flies replicate important features of human tauopathies and transgenic models of the disorders. We therefore performed single-cell RNA sequencing (scRNA-seq) to investigate transcriptional programs and cellular pathways altered by expression of mutant Tau. Using an optimized brain dissociation method, 10x library preparation, sequencing, and a bioinformatics analysis pipeline, we implemented scRNA-seq on Tau P251L knock-in and control Drosophila brains at 10 d of age (Fig. 3A). The 10-d time point was chosen to identify early perturbations related to neuronal dysfunction and degeneration (Figs. 1, 2). After stringent quality control, 130,489 high-quality cells were retained in the final integrated data set, and 29 clusters of cells were identified. We annotated 26 clusters using a published fly cell atlas (Li et al. 2022). We used the most highly expressed marker genes within each cluster to identify clusters. For instance, we used dac, crb, and jdp to annotate Kenyon cells; Yp1, Yp2, and Yp3 for mushroom body output neurons (MBONs); Mtna, CG8369, and CG1552 for glia; CG34355, Gad1, and mamo for medullary neurons; and acj6, Li1, and sosie for T neurons (Supplemental Figs. S3–S5). The clustered dot plot illustrates enrichment of marker genes in annotated neuronal and glial clusters (Fig. 3C; Supplemental Fig. S4C). Based on prior published analyses (Croset et al. 2018; Davie et al. 2018; Li et al. 2022), we further outlined major groups of cells, including Kenyon cells, medullary neurons, MBONs, astrocytes, and perineurial glia (Fig. 3B; Supplemental Fig. S3; Supplemental Table S1). As previously observed (Davie et al. 2018), cholinergic neurons were the most common neuronal type defined by neurotransmitter phenotype, followed by GABAergic and glutamatergic neurons (Supplemental Fig. S3). Less abundant clusters of dopaminergic neurons were also identified (Supplemental Fig. S3). In summary, our scRNA sequencing in a precisely edited Drosophila tauopathy model, yielded 130,489 high-quality cells and identified cellular populations throughout diverse brain regions and cell types, including rarer cell populations such as astrocytes and perineurial glia.
Figure 3.
Single-cell RNA sequencing (scRNA-seq) of Tau P251L knock-in brains. (A) Schematic of the scRNA-seq analysis pipeline. Following dissection, brains were dissociated in the enzymatic solutions, and the single-cell suspension was encapsulated by the 10x Genomics Chromium platform. The 10x libraries were prepared and sequenced, and after quality control, data were analyzed. (B) UMAP representation of the six integrated scRNA-seq runs: three control and three Tau P251L knock-in. The integrated data set contains 130,489 cells, and 26 clusters out of 29 were annotated. (C) Percentage expression heatmap of the highly expressed marker genes within all clusters. Flies are 10 d old.
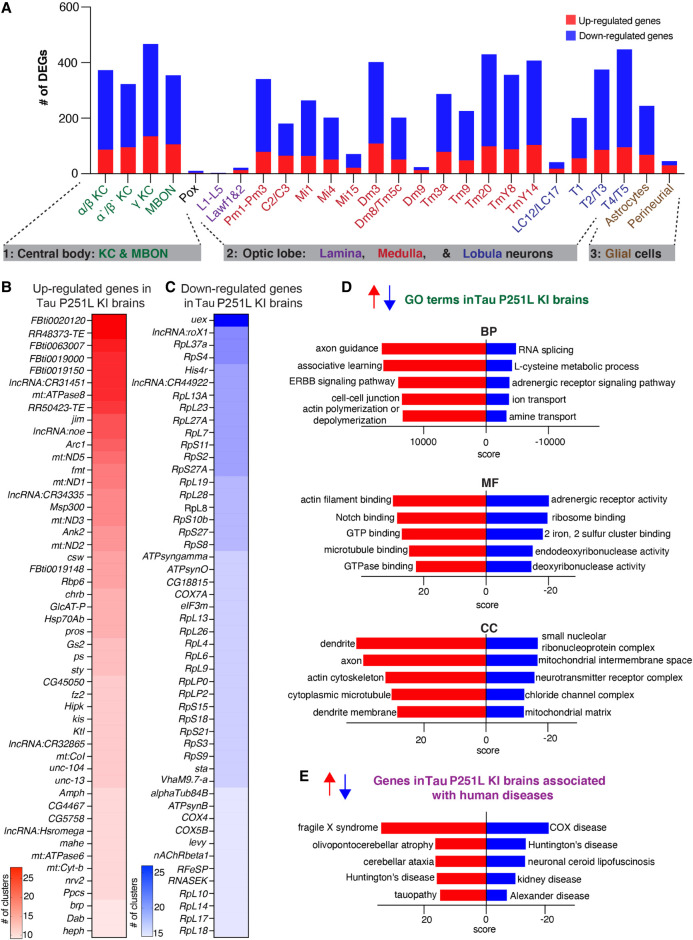
After sample integration, quality control, and cluster annotation, we performed differential gene expression (DEG) analysis to identify genes modulated by precise pathologic mutation modeling of tauopathy in the Drosophila brain. DEG analysis of all the 26 annotated clusters revealed that Tau P251L knock-in altered genes throughout the Drosophila brain, in both neurons and glia (Fig. 4A; Supplemental Table S2). We found that 472 genes were up-regulated across all clusters in Tau P251L knock-in brains, whereas 1145 genes were down-regulated (Supplemental Table S3). Transposable elements (FBti0020120 RR48373-transposable-element, FBti0063007, FBti0019000, FBti0019150, RR50423-transposable-element, FBti0019148) were frequently up-regulated in P251L knock-in brains (Fig. 4B), consistent with findings from Drosophila human TAU overexpression models and human Alzheimer's disease brain tissue (Guo et al. 2018; Sun et al. 2018). The set of commonly down-regulated genes was notable for multiple ribosomal protein genes (Fig. 4C), suggesting a translational defect in tauopathy. Multiple nuclear and mitochondrially encoded respiratory chain subunits and other mitochondrial proteins were notably present in the commonly up-regulated and down-regulated genes, as were genes encoding cytoskeletal and associated proteins (Arc1, Msp300, Ank2, unc-104, Amph, brp, alphaTub84B). Both categories of genes fit well with known mediators of tauopathy pathogenesis (DuBoff et al. 2013; Schulz et al. 2023).
Figure 4.
Differential gene expression and enrichment analysis of the scRNA-seq data set in Tau P251L knock-in brains compared with controls. (A) The number of differentially expressed genes (DEGs), both up-regulated and down-regulated genes, in all the annotated clusters of Tau P251L knock-in brains compared with controls. Results are displayed across three major anatomic and functional classes of cells: (1) central body containing three clusters of Kenyon cells (KCs), mushroom body output neurons (MBONs) and pox neurons; (2) optic lobe neurons containing lamina, medullary, and lobula neurons clusters; and (3) glia cells containing astrocytes and perineurial clusters. (B,C) Heatmaps of the top 50 up-regulated (B) and down-regulated (C) genes in all the clusters of Tau P251L knock-in brains compared with controls (Supplemental Table S3). (D) Gene Ontology (GO) enrichment analysis identified top up-regulated and down-regulated biological processes (BPs), molecular functions (MFs), and cellular components (CCs). (E) Analysis of human disease–associated genes revealed top up-regulated and down-regulated disease-associated gene sets. Score represents the combined score c = log(p) × z (Chen et al. 2013).
As expected, gene enrichment analyses (Fig. 4D; Supplemental Fig. S6) highlighted mitochondrial and cytoskeletal processes. In addition, diverse metabolic and neuronal function pathways, including associated learning, previously associated with Alzheimer's disease and related tauopathies emerged from Gene Ontology (GO) enrichment analyses. Enrichment analysis for human disease–associated genes revealed predominantly neurodegenerative disorders, including tauopathy (Fig. 4E).
Distinct and shared region- and cell-specific transcriptional programs in Tau P251L knock-in brains
Significant anatomic and cell type selectivity characterizes human neurodegenerative diseases, including tauopathies. We therefore analyzed gene expression changes separately in anatomically and functionally related groups of cells, including the central body (Kenyon cells, MBON, and Pox neurons), optic lobe (lamina, medulla, and lobula neurons), and glia (astrocytes and perineurial glia). Volcano plots in Supplemental Figure S7, A, C, and F, present up-regulated and down-regulated genes in each group of cells. Transposable element were the top up-regulated genes in each of the three groups. GO enrichment analyses (Supplemental Fig. S7B,D) identified distinct biological processes altered by mutant tau expression in the central body compared with the optic lobe. Both associative learning and cAMP metabolic process were specifically identified in the central body, correlating with the importance of Kenyon cells in learning and memory in flies and with the central role for cAMP underlying learning and memory (Feany and Quinn 1995; Guven-Ozkan and Davis 2014). Heterochromatin organization and DNA repair, both processes strongly implicated in tauopathy pathogenesis (Fig. 2; Khurana et al. 2012; Frost et al. 2014; Welch and Tsai 2022), emerged as enriched processes following analysis of the central body and optic lobe separately (Supplemental Fig. S7D). Direct comparison of differentially regulated genes in the central body compared with the optic lobe neurons revealed 239 commonly regulated genes and 562 distinct genes (Supplemental Fig. S7E). Consistent with analysis of the total transcriptome (Fig. 4), shared biological processes included down-regulation of mitochondrial genes and up-regulation of axon guidance–associated genes (Supplemental Fig. S7E; Supplemental Table S4).
Because tau is a predominantly neuronally expressed gene (Heidary and Fortini 2001; Goedert 2004; Götz et al. 2019), the observed changes in neuronal transcriptomes may reflect cell-autonomous effects of the frontotemporal dementia associated mutant TAU protein. Our single-cell approach also revealed significant changes in gene expression in glial cells in Tau P251L knock-in brains (Supplemental Fig. S7F,G). Expression of mutant tau may thus exert non-cell-autonomous control on glial transcriptional programs. Metabolic processes (Supplemental Fig. S7G) were down-regulated in glia in response to neuronal expression of mutant tau, consistent with the importance of glial metabolism in supporting a wide array of neuronal functions (Nedergaard and Verkhratsky 2012; Verkhratsky et al. 2012). The top two GO processes identified by analysis of up-regulated glial genes were associative learning and regulation of neuronal remodeling, suggesting that coordinate changes in neurons and glia may lead to impairment of critical neuronal functions when mutant tau is expressed by neurons.
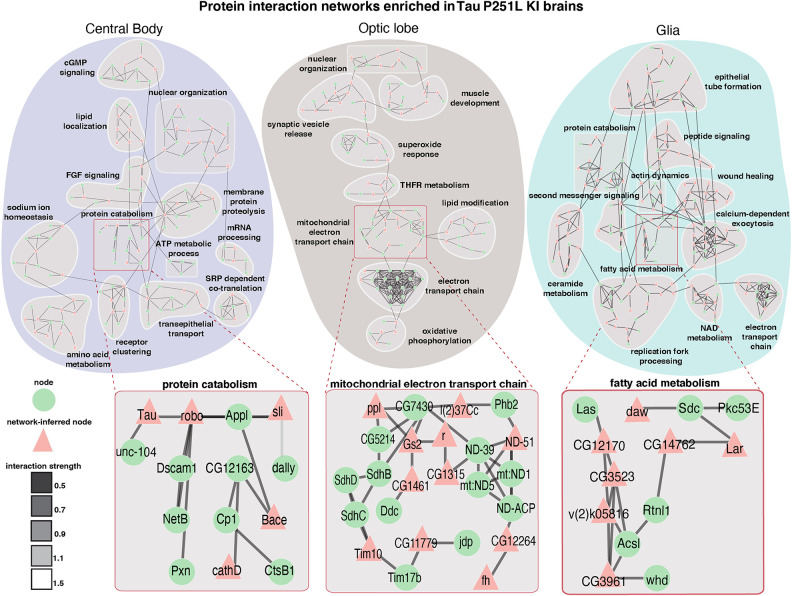
We next constructed protein interaction networks to explore further the biological pathways altered in Tau P251L knock-in brains compared with controls. We used the solution of the prize-collecting Steiner forest algorithm (Tuncbag et al. 2013) to map differentially expressed genes onto a network of physical protein interactions using Drosophila interactome data. Networks constructed from the central body, optic lobe, and glial cells were substantially distinct (Fig. 5), consistent with differential effects of mutant Tau on different anatomic regions and cell types. The electron transport chain was identified in subnetworks from both the optic lobe and glia, raising the possibility that mutant Tau can influence mitochondrial function in both a cell-autonomous and a non-cell-autonomous fashion (Figs. 2, 5). Regulation of nuclear function was commonly identified in both central body and optic lobe neurons, consistent with a strong influence of neuronally expressed Tau on chromatin structure mediated through the linker of nucleoskeleton and cytoskeleton (LINC) complex (Frost et al. 2014, 2016).
Figure 5.
Protein interaction networks enriched in the central body, optic lobe, and glia in Tau P251L knock-in brains compared with controls. Protein interaction networks are largely distinct among central body neurons, optic lobe neurons, and glia. Subnetworks including nodes enriched for protein catabolism (central body), electron transport chain (optic lobe), or fatty acid metabolism (glia) are highlighted. Interaction strength displayed in gray shows the stringency of the interaction: The lower the strength, the stronger the interaction.
Protein catabolism was a subnetwork in both central body and glial networks. Protein catabolism was connected to multiple other subnetworks in the central body network and contained multiple proteins previously implicated in Alzheimer's disease, including Appl (fly ortholog of APP), beta-site APP-cleaving enzyme (Bace; a fly homolog of BACE1), three members of the cathepsin family (CtsB1, cathD, CtsF/CG12163), and Tau itself identified as a computational network-inferred node. As expected from GO analysis (Supplemental Fig. S7G), multiple metabolic subnetworks were identified in the glial network, consistent with the role of glia in providing metabolic support to neurons (Nedergaard and Verkhratsky 2012; Verkhratsky et al. 2012). A subnetwork enriched for nodes associated with fatty acid metabolism was identified in the glial network (Fig. 5), correlating with the important role of glia in lipid metabolism and signaling in both flies and mammalian systems (Lee et al. 2021; Goodman and Bellen 2022). Detailed protein interaction networks identified in the central body, optic lobe, and glia are shown in Supplemental Figures S8 through S10.
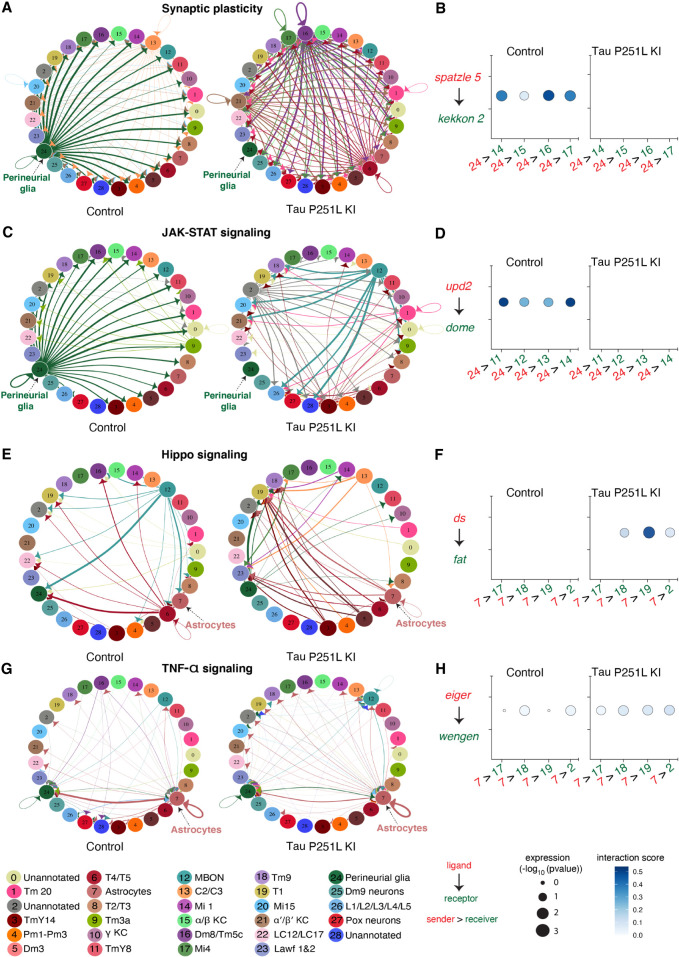
Cell–cell communication and pseudotime trajectory analyses highlight the role of glial cells in Tau P251L knock-in brains
Altered gene expression (Supplemental Fig. S7) and protein interaction networks (Fig. 5) in glia driven by neuronal-predominant expression of P251L mutant Tau suggest perturbed intercellular communication in P251L knock-in brains. We therefore calculated the interaction scores for 196 manually curated ligand–receptor pairs using the FlyPhoneDB quantification algorithm (Liu et al. 2022) in Tau P251L knock-in brains and controls. We found significant alterations predicted in major cellular signaling pathways (Fig. 6; Supplemental Fig. S11). Altered signaling is indicated in circle plots in Figure 6 (A,C,E,G) by nodes representing unique cell types and by edges representing a communication event. The thickness of an edge reflects the interaction strength of the communication event. Dot plots in Figure 6 (B,D,F,H) display the calculated score of selected ligand–receptor pairs from one cell type to another, with the shading of the dot indicating the interaction score and with the size of the dot indicating the the P-value. Many of predicated signaling changes support altered communication between glia and neurons. For instance, synaptic plasticity signaling, assessed by expression of the ligand spatzle and kekkon receptors, was mainly driven by perineurial glia in the control brain. However, perineurial glial cells in Tau P251L knock-in animals had reduced expression of the ligand spatzle 5, whereas recipient cells down-regulated kekkon receptors (Fig. 6B). Similarly, expression of the JAK-STAT ligand upd2 was significantly down-regulated in perineurial glia in Tau P251L knock-in brains compared with those of controls, whereas the receptor dome was reduced in expression in widespread target neuronal clusters (Fig. 6D). There was a predicted up-regulation of JAK-STAT signaling from MBONs to a restricted set of neuronal clusters in brains of flies expressing P251L mutant Tau (Fig. 6C). In contrast, predicted hippo signaling from MBONs to perineurial glial based on decreased levels of the ds ligand and receptor fat was decreased in Tau P251L knock-in brains compared with controls (Fig. 6E).
Figure 6.
Cell–cell communication analysis predicts altered signaling in Tau P251L knock-in brains compared with controls. (A) Altered ligand and receptor expression predicts regulation of synaptic plasticity signaling mainly via perineurial glial cells in control brains. (B) Signaling from perineurial glia is significantly reduced in Tau P251L knock-in brains, as predicted by levels of spaetzle ligand and kekkon receptor. (C,D) JAK-STAT signaling, as predicted by expression of the upd2 ligand and dome receptor, mediated by perineurial glia in control brains (C), is substantially reduced in brains from Tau P251L knock-in animals (D). (E,F) Hippo signaling, indicated by expression of ds ligand and fat receptor, is up-regulated in astrocytes of flies expressing P251L mutant Tau compared with controls. (G,H) Predicted TNF-α signaling from ligand eiger to receptor wengen is increased in astrocytes of Tau P251L knock-in flies. In panels B,D,F,H, the interactions from and to the specified cell types are indicated on the x-axis, the size of the circle indicates the P-value, and the intensity of the blue color illustrates the interaction score as defined in the figure label below the panels.
Astrocytic signaling also showed predicted changes in Tau P251L knock-in brains compared with controls. JAK-STAT signaling perineurial glia to astrocytes was reduced in mutant Tau-expressing brains (Fig. 6C), whereas hippo signaling from astrocytes to multiple neuronal subtypes was increased in Tau P251L knock-in brains (Fig. 6E,F). TNF-α signaling from astrocytes was also increased in flies expressing mutant Tau, as suggested by increased levels of the ligand eiger and receptor wengen (Fig. 6G,H). Altered astrocyte integrin, hedgehog, and insulin signaling was also suggested by changes in expression of ligand and cognate receptor pairs (Supplemental Fig. S11A,D,E).
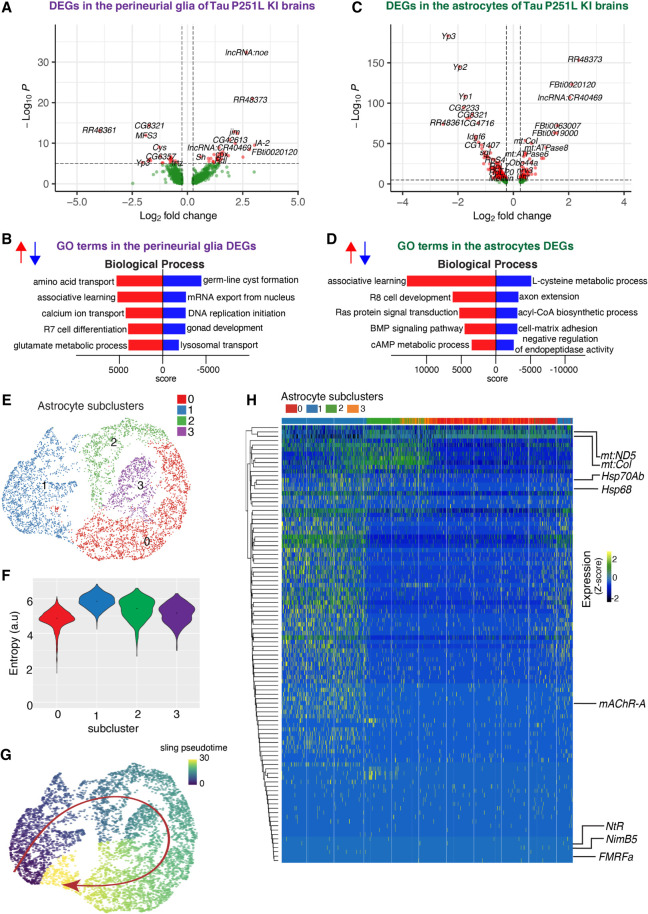
Given the altered gene expression (Fig. 4; Supplemental Fig. S7), protein interaction networks (Fig. 5), and predicted signaling (Fig. 6) in glia, we next examined gene expression profiles in these nonneuronal cells in more detail (Fig. 7). Transposable elements were significantly up-regulated in both types of glia (Fig. 7A,C; Supplemental Table S5), although one transposable element was highly down-regulated in both glia subsets (RR48361). GO enrichment analysis highlighted different metabolic pathways in the two cell types. Amino acid and glutamate metabolism pathways were enriched in perineurial glia, whereas L-cysteine, acyl-CoA, and cAMP metabolic pathways were enriched in astrocytes (Fig 7B,D).
Figure 7.
Gene expression and trajectory analysis in glia. (A) Differentially regulated genes, both up-regulated and down-regulated, in perineurial glia of Tau P251L knock-in brains compared with controls. (B) GO analysis shows biological processes associated with the up-regulated and down-regulated genes in perineurial glia from Tau P251L knock-in brains compared with controls. (C) Differentially regulated genes, both up-regulated and down-regulated, in astrocytes of Tau P251L knock-in brains compared with controls. (D) GO analysis shows biological process associated with up-regulated and down-regulated genes in astrocytes of Tau P251L knock-in brains. All dots on the volcano plots are significant at FDR < 0.05 and log2FC > 0.25 for up-regulated and log2FC< −0.25 for down-regulated genes. Score represents the combined score c = log(p) × z (Chen et al. 2013). Astrocytes from both control and Tau P251L knock-in brains were further subclustered into four groups. (E,F) Entropy analysis to define the root for trajectory analysis revealed cluster 1 to have the highest entropy. (G) Slingshot trajectory analysis on astrocyte clusters identified a single lineage passing sequentially from clusters 1 to 2, 3, and 0. (H) Differential gene expression between astrocyte subclusters adjacent in pseudotime were used to cluster genes along the pseudotime trajectory. Each row in the heat map represents a gene. The columns are astrocyte subclusters arranged according to pseudotime from left to right. Examples of differentially regulated genes from enriched GO biological processes are shown on the calculated trajectory.
Because we observed significant alterations in glial signaling in Tau P251L knock-in brains (Fig. 6; Supplemental Fig. S11), we investigated glial gene trajectories in our scRNA-seq, focusing on astrocytes because we obtained a large number (nearly 5800) of these cells (Supplemental Table S1). We first subclustered astrocytes into four groups (Fig. 7E). We then calculated the entropy of these clusters (Guo et al. 2017) and used cluster 1, which showed the highest entropy (Fig. 7F), as the root for trajectory analysis (Street et al. 2018). A single lineage starting from cluster 1 and progressing sequentially from cluster 2 through cluster 3 and finally to cluster 0 emerged (Fig. 7G). We then clustered differentially expressed genes along the calculated trajectory as presented in the heat map, in which pseudotime is represented in columns from left to right (Fig. 7H). Our pseudotemporal analysis suggests different stages of astrocytic response to tauopathy.
GO analysis across pseudotime revealed multiple genes involved in signaling pathways (FMRFa, NimB5), particularly in cholinergic signaling (nicotinic acetylcholine receptor subunit NtR, mAChR-A, ChAT) early in the glial trajectory. Cellular stress response emerged later in the trajectory with up-regulation of heat shock proteins (Hsp68, Hsp70Ab), whereas altered mitochondrial gene expression (mt:ND5, mt:Col) characterized astrocytes late in the calculated trajectory. These findings suggest that altered astrocyte signaling (Fig. 6; Supplemental Fig. S11) may emerge early in tauopathy pathogenesis and drive subsequent cell-autonomous and non-cell-autonomous stress responses and cytotoxicity. A complete list of all differentially expressed glial genes, genes associated with GO biological processes, and trajectory-associated genes is provided in Supplemental Table S5.
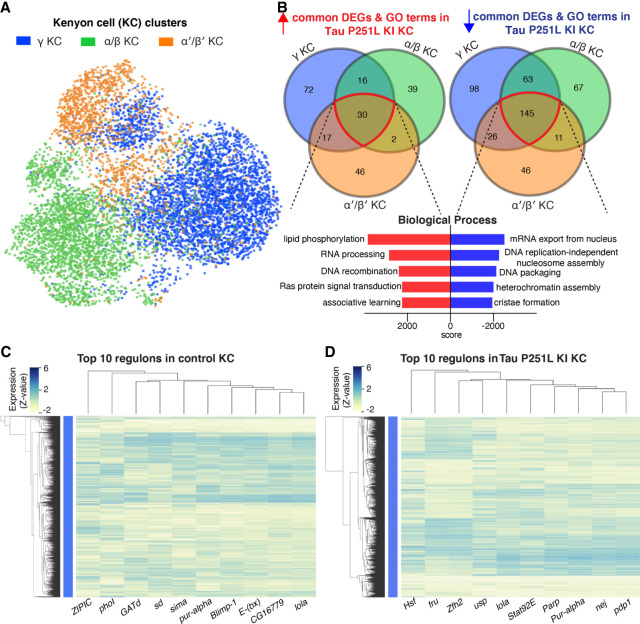
Gene regulatory networks in control and Tau P251L knock-in Kenyon cells
Kenyon cells are a major defined neuronal component of the central body of the Drosophila brain (Fig. 4). Together with their output neurons (MBONs), Kenyon cells play a central role in learning and memory in the Drosophila brain (Heisenberg 2003; Modi et al. 2020); memory loss is a key feature of human tauopathies (Grossman et al. 2023). Kenyon cells are cholinergic (Barnstedt et al. 2016), a neuronal type that is selectively vulnerable in previously described fly tauopathy models (Wittmann et al. 2001), and a pathway altered early in our trajectory analysis (Fig. 7). Our cell–cell communication analyses suggested altered signaling in Kenyon cells, or their output neurons, via multiple signaling pathways (Fig. 6; Supplemental Fig. S11). We therefore focused next on gene expression in Kenyon cells. We identified three Kenyon cells clusters: gamma-Kenyon cells, alpha/beta-Kenyon cells, and alpha′/beta′-Kenyon cells (Fig. 8A). Transposable elements were up-regulated in all Kenyon cell clusters in Tau P251L knock-in brains (Supplemental Fig. S12A,C,E), as observed in other neuronal and glial clusters (Fig. 7; Supplemental Fig. S7). Analysis of biological pathways associated with common up-regulated and down-regulated genes in all three Kenyon cell clusters identified key biological processes previously linked to tauopathy pathogenesis (Frost et al. 2015; Götz et al. 2019), including control of DNA and RNA structure and metabolism (Fig. 8B), as well as many pathways without prior links to tauopathy. A complete list of differentially expressed genes and associated biological processes is given in Supplemental Table S6.
Figure 8.
Gene expression and regulatory networks in Kenyon cells (KCs). (A,B) Three KC clusters—gamma-KC, alpha/beta-KC, and alpha′/beta′-KC—and biological process based on the common up-regulated and down-regulated genes in KC clusters in Tau P251L knock-in brains. Score represents the combined score c = log(p) × z (Chen et al. 2013). Control and Tau P251L knock-in Kenyon cells were clustered separately using SCENIC gene regulatory network analysis to identify regulons. (C,D) The top 10 regulons identified by SCENIC gene regulatory network analysis in control (C) and Tau P251L knock-in (D) KCs are presented in the heatmaps. Each row represents a KC; each column is a regulon.
Given the multiple lines of evidence connecting tauopathy pathogenesis to Kenyon cell function, we next determined the gene regulatory networks controlling disease-associated changes in gene expression in Kenyon cells. We implemented the single-cell regulatory network inference and clustering (SCENIC) (Aibar et al. 2017) workflow on gene expression data from control and Tau P251L knock-in Kenyon cells. The top 10 regulons identified in control cells compared with tauopathy model Kenyon cells are show in columns in the heat maps in Figure 8, C (control Kenyon cells) and D (Tau P251L knock-in Kenyon cells). Regulons were largely distinct in the two genotypes (Fig. 8C,D; Supplemental Table S7). The shared transcription factors among the top 10 regulons were lola and pur-alpha. Even for the shared regulons, the gene expression patterns per cell clustered and coexpressed with different transcription factors and are different among Kenyon cells of control versus Tau P251L knock-in animals. The distinct gene regulatory networks illustrated in the heatmap are concordant with altered gene expression (Fig. 8B) and cell–cell communication (Fig. 6) between control and Tau P251L knock-in Kenyon cells. The increase in HSF, Stat92E, and Parp expression (Supplemental Fig. S13) and regulons (Fig. 8D) in brains of tauopathy model flies are consistent with elevated cellular stress, DNA damage, and cell death in aging neurons exposed to mutant Tau P251L (Figs. 1, 2).
Discussion
Here we present a new model of tauopathy in the experimentally facile model organism Drosophila based on precise gene editing of the endogenous tau gene to introduce a mutation orthologous to human proline 301 to leucine (P301L), the most common TAU mutation in frontotemporal dementia patients (Poorkaj et al. 2001). We observe age-dependent neurodegeneration in our knock-in animals (Fig. 1C,D). Homozygous knock-in flies display early and greater total levels of degeneration compared with heterozygous animals. These findings are compatible with a toxic gain-of-function mechanism, as generally posited in familial frontotemporal tauopathies (Goedert et al. 2012; Frost et al. 2015; Bardai et al. 2018b; Götz et al. 2019). However, given the important role of microtubules in neurodevelopment, a loss-of-function component contribution cannot be excluded, even given the lack of clear neurodegeneration in Tau knockout mice (Harada et al. 1994; Dawson et al. 2001; Morris et al. 2013) and flies (Burnouf et al. 2016). As expected given that levels of mutant Tau are controlled by the endogenous tau promotor in our model compared with the strong exogenous promotor systems used in prior transgenic models, neurodegeneration in knock-in animals is observed at older ages and is milder (Wittmann et al. 2001; Bardai et al. 2018b; Law et al. 2022). However, we do observe key biochemical and cellular pathologies previously described in transgenic Drosophila tauopathy models, including metabolic dysfunction (Fig. 2A,B), elevated levels of DNA damage (Fig. 2C–G), and abnormal cell-cycle activation (Fig. 1E; Khurana et al. 2006; 2012; DuBoff et al. 2012; Bardai et al. 2018a). Importantly, these pathways are also perturbed in mouse tauopathy models and tauopathy patients (Herrup and Arendt 2002; Andorfer et al. 2005; Khurana et al. 2012; DuBoff et al. 2013; Frost et al. 2015; Götz et al. 2019; Welch and Tsai 2022).
The similarities of our knock-in model to human tauopathies and prior overexpression tauopathy models, recapitulated in a more faithful genetic knock-in context, motivated us to perform a comprehensive transcriptional analysis in our Tau P251L knock-in brains using scRNA-seq. We recovered a large number (130,489) of high-quality cells, which allowed us to identify the majority of previously annotated neuronal and glial groups from prior single-cell sequencing analyses in the adult fly brain (Davie et al. 2018; Li et al. 2022). Comparing gene expression profiles between control and Tau P251L knock-in animals revealed pervasive dysregulation of genes in neuronal (Figs. 4,8) and glial (Fig. 7) subtypes and throughout different anatomic regions (Fig. 4, Supplemental Fig. S6). These findings are consistent with prior single-cell sequencing studies in flies overexpressing mutant human TAU (Praschberger et al. 2023; Wu et al. 2023). We observed regulation of both common and distinct biological pathways when comparing differentially expressed genes across cell subtypes. Transposable elements were notably up-regulated in the complete gene expression set, as well as in specific anatomic regions and neuronal subtypes. These findings correlate with a previously described functional role for transposable element mobilization in Drosophila models of tauopathy and in tauopathy patients (Guo et al. 2018; Sun et al. 2018). Mitochondrial function has been strongly linked to neurotoxicity in tauopathies (DuBoff et al. 2013; Frost et al. 2015; Götz et al. 2019) and is a feature of our current model (Fig. 2). We accordingly observed altered expression of mitochondrial genes and biological processes in the complete expression data set (Fig. 4), as well as in separate analyses of the central body, optic lobe (Supplemental Fig. S7), and Kenyon cells (Fig. 8; Supplemental Fig. S12). More importantly, we observed significant alterations in multiple metabolic, cellular communication, and biological pathways not previously implicated in tauopathy pathogenesis (Figs. 4–6), which can now be assessed in tauopathy models and patients for mechanistic relevance and ultimately therapeutic targeting.
Cell type selectivity is a fundamental, and poorly understood, feature of human neurodegenerative diseases, including tauopathies. Our protein interaction networks highlighted regionally specific biology with predominantly distinct nodes appearing in the central body compared with the optic lobe (Fig. 5). Comparative analysis of genes differentially expressed in the central body compared with the optic lobe is consistent with substantial regional differences in the response to mutant tau expression with substantially greater numbers of unique compared with common genes up-regulated in the central body versus the optic lobe (Supplemental Fig. S7E). Even within subgroups of Kenyon cells, there are equivalent numbers or more uniquely up- or down-regulated genes compared with commonly regulated genes (Fig. 8B). Our data set thus highlights a substantial set of genes that may contribute to selective neuronal susceptibility in neurodegeneration, including many differentially regulated genes and processes not previously linked to TAU pathobiology.
Although TAU is a predominantly neuronal protein (Heidary and Fortini 2001; Goedert 2004; Götz et al. 2019), we observed significant alteration of glial gene expression in Tau P251L knock-in brains compared with controls (Figs. 4, 7), suggestive of non-cell-autonomous control of glia cell function by neuronally expressed Tau. GO (Fig. 7A,B) and protein interaction network (Fig. 5) analyses highlighted a number of metabolic processes altered in glia by expression of toxic Tau in neurons, including glutamate, lipid, and amino acid metabolism (Figs. 5, 7). Glial uptake and detoxification of neurotransmitters and their metabolites, as well as toxic lipid species, maintain neuronal function and viability. Lipid metabolism is further central to energy production by glial cells, which support highly energy consuming neurons with active synaptic transmission (Smolič et al. 2022; Jiwaji and Hardingham 2023). In addition to glial processes previously implicating in controlling neuronal health, our transcriptional analysis revealed new metabolic and signaling pathways in glia regulated by the expression of mutant Tau (Fig. 7A–C), which can now be explored as non-cell-autonomous mechanisms regulating neuronal function and viability in tauopathy.
An effect of mutant tau expression enriched in neurons on glial gene expression implies signaling, and possibly perturbed signaling, between the two cell types. Examination of the expression of 196 ligand–receptor pairs (Liu et al. 2022) indeed supported broad alterations in glial–neuronal communication in Tau P251L knock-in flies (Fig. 6; Supplemental Fig. S11), with mutant tau expression perturbing synaptic plasticity, JAK-STAT, hippo, TNF-α, integrin, and EGFR signaling between perineurial cells, astrocytes, and multiple neuronal subtypes. Although prior studies have implicated glial signaling, for example, the JAK-STAT pathway (Colodner and Feany 2010), in non-cell-autonomous control of neurotoxicity, the pervasive nature of the altered signaling suggested by our single-cell transcriptional analyses is unexpected and provides multiple targets for functional testing. Our findings further suggest that a systematic and broad perturbation of intercellular signaling is present in tauopathy, which may require manipulation of multiple pathways to correct and system-level analysis to monitor.
Trajectory analysis has been widely used to order temporal events along developmental pathways but has less often been applied to neurodegenerative disease progression (Fitz et al. 2021; Karademir et al. 2022; Wang et al. 2022; Dai et al. 2023). Given the evidence for altered glial–neuronal communication in our Tau knock-in model, we assessed possible trajectories in the four distinct subgroups of astrocytic glial cells that we defined. Using the astrocyte cluster with the highest entropy as the root (Guo et al. 2017), we identified a single astrocyte trajectory (Fig. 7G). DEG and GO analyses across the trajectory revealed altered expression of neurotransmitter and cell signaling genes first, followed by altered cell stress responses, and finally mitochondrial changes (Fig. 7H; Supplemental Table S5). A number of genes involved in cholinergic signaling were changed early in the glial trajectory. We have previously shown that cholinergic terminals are preferentially vulnerable and degenerate early in a tauopathy model based on transgenic human TAU expression in flies (Wittmann et al. 2001). Our trajectory analysis may thus help identify early events in glial-mediated neurodegeneration, including pathways not previously associated with tauopathy (Supplemental Table S5). Glial pathways contributing to neurodegeneration are increasingly recognized as attractive and understudied avenues for therapeutic intervention (Jiwaji and Hardingham 2023). Identifying and intervening in early glial–neuronal signaling events may prevent later, and possibly irreversible, neuronal damage.
Reversing pathological neuronal cell-autonomous programs may provide an alternative or additional method of preventing neuronal dysfunction and death in tauopathies. We focused on Kenyon cells as a group of neurons involved in the behaviorally relevant process of memory and composed of cholinergic neurons, a vulnerable cell type in Drosophila (Wittmann et al. 2001) and human (Whitehouse et al. 1981; Ishida et al. 2015) tauopathies, to define transcriptional programs driving neurodegeneration in response to mutant Tau expression. As expected by the multiple neuropathological and cell biological abnormalities present in our knock-in model flies (Figs. 1, 2), we observed substantially distinct regulons in Tau P251L knock-in Kenyon cells compared with controls (Fig. 8C,D). We identified regulons involved in stress responses (Hsf, Stat92E), including the DNA damage response (Parp), as would be expected from the presence of elevated DNA damage in Kenyon cells in our knock-in flies (Fig. 2E–G). We recovered nej, the fly ortholog of vertebrate CREB binding protein (also known as CBP), as a top regulon induced in knock-in flies. Increasing levels of nej/CBP are beneficial in fly (Cutler et al. 2015) and vertebrate (Caccamo et al. 2010) models relevant to Alzheimer's disease, suggesting that up-regulation of nej may represent a protective response in Kenyon cells. We also identified multiple regulons not previously associated with neurodegenerative tauopathies (Fig. 8C,D). Therapeutic manipulation of these programs or key transcriptionally regulated mediators will be attractive candidates for evaluation in patient tissue, patient-derived cellular models, and vertebrate models of tauopathy.
The mechanisms transducing the effects of mutant Tau on gene expression are likely multiple and, as yet, only partially characterized. We have previously defined a cascade in which cytosolic Tau binds and stabilizes F-actin (Fulga et al. 2007), leading to signal transduction through the LINC complex, nuclear lamin disruption (Frost et al. 2016), and consequent chromatin relaxation (Frost et al. 2014), promoting aberrant transposable element activation and neurodegeneration (Sun et al. 2018). Other cytosolic targets of Tau may promote transcriptional regulation through parallel mechanisms. For example, Tau-mediated actin hyperstabilization promotes mitochondrial dysfunction and excess production of oxidative free radicals by interfering with mitochondrial dynamics (DuBoff et al. 2012). Oxidative stress may directly contribute to elevated DNA damage in tauopathy (DuBoff et al. 2013; Frost et al. 2016; Bardai et al. 2018b; Götz et al. 2019). However, although TAU is best known as a cytosolic protein involved in regulation of the cytoskeleton, a number of studies have shown that TAU can also be detected in the nucleus (Loomis et al. 1990; Thurston et al. 1996; Cross et al. 2000), where the protein binds DNA (Hua et al. 2003; Sjöberg et al. 2006; Wei et al. 2008; Bukar Maina et al. 2016). Thus, TAU may play a direct role in instructing the nuclear transcriptional programs we have defined (Fig. 8C,D).
In summary, here we develop a genetically precise model of frontotemporal dementia caused by the most common TAU mutation found in patients and present a comprehensive picture of gene expression changes and derived protein interaction, cell signaling, and transcriptional networks. We recapitulate neurodegeneration, metabolic dysfunction, and DNA damage, common features of human tauopathies (Goedert 2004; Götz et al. 2019; Welch and Tsai 2022), and confirm that cellular pathways perturbed in overexpression tauopathy models are also dysregulated in the more faithful genetic knock-in context. More importantly, our work suggests previously unsuspected, pervasive alterations in glial–neuronal signaling in tauopathy pathogenesis, implicates many new genes and pathways, and provides a genetic model system in which to test the new hypotheses our data suggest.
Methods
Genetics and CRISPR-Cas9 editing
The Drosophila tau gene is located on the third chromosome. The guide RNAs targeting the tau gene to mutate proline 251 to leucine were identified using Harvard Medical School's DRSC/TRiP “find CRISPRs” tool. The gRNA 5′-CCGGGAGGCGGGGACAAGAAGAT-3′ was cloned into the pCDF3.1 plasmid and injected into the embryos of the TH_attP40 nos-Cas9 strain along with a single-stranded oligo nucleotide donor. The single-stranded oligo nucleotide donor was 150 bp in length and contained a C-to-T transition that resulted in alteration of the codon CCG (proline) to CTG (leucine). Embryos were injected (BestGene) and founder flies obtained. Founder flies were then balanced to obtain homozygous knock-in animals. The mutation was confirmed by PCR. The genotype of knock-in animals in most experiments (Figs. 1, 2C–E, 4–8) was elav-GAL4/+; tau-P251L knock-in (homozygous or heterozygous for tau-P251L knock-in as specified in figures and legends). In these experiments control animals were elav-GAL4/+. In Figure 2, A and B, the genotype of knock-in flies was w1118; tau-P251L knock-in/tau-P251L knock-in (homozygous) or w1118; tau-P251L knock-in/ + (heterozygous) as specified in the figure. In Figure 2, A and B, the genotype of control flies was w1118. The elav-GAL4 line was obtained from the Bloomington Drosophila Stock Center. Patrik Verstreken kindly provided tau knockout flies. All crosses and aging were performed at 25°C.
Assessment of neurodegeneration and metabolism
For sectioning, adult flies were fixed in formalin at 1, 10, and 30 d of age and embedded in paraffin. Vacuole, PCNA, and pH2Av levels were examined using previously described methodology (Fulga et al. 2007; Frost et al. 2014) with additional details provided in the Supplemental Methods. Primary antibodies used include pH2Av (Rockland 600-401-914, 1:100), elav (DSHB 9F8A9, 1:5), GAPDH (Thermo Fisher Scientific MA5-15738, 1:1000), and PCNA (DAKO MO879, 1:500). A polyclonal antibody to Drosophila Tau was prepared in rabbits immunized with full-length recombinant Tau protein (Thermo Fisher Scientific) and was used at 1:5,000,000 for western blotting. For all histological analyses, at least six brains were analyzed per genotype and time point. The comet assay and assessment of bioenergetics were performed as previously described (Frost et al. 2014; Sarkar et al. 2020) with additional details provided in the Supplemental Methods. The sample size (n), mean, and SEM are given in the figure legends. All statistical analyses were performed using GraphPad Prism 5.0. For comparisons across more than two groups, one-way ANOVA with Tukey post-hoc analysis was used. For comparison of two groups, Student's t-tests were performed.
scRNA-seq and downstream analyses
A standard sample preparation (Li et al. 2017; Davie et al. 2018), raw data processing (Satija et al. 2015), and downstream analyses such as cell cluster annotation (Hu et al. 2021) GO analysis (Kuleshov et al. 2016), protein–protein interaction network analysis (Tuncbag et al. 2016), cell–cell communication analysis (Liu et al. 2022), trajectory analysis (Street et al. 2018), and gene regulatory network analysis(Van de Sande et al. 2020) were performed as previously described. Detailed methods are presented in the Supplemental Methods.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE223345. R code (R Core Team 2016; Butler et al. 2018) that was used to perform Seurat-based integration, trajectory, cell–cell interaction, and PPI network analyses is available at GitHub (https://github.com/bwh-bioinformatics-hub/Single-cell-RNA-seq-of-the-CRISPR-engineered-endogenous-tauopathy-model) and as Supplemental Code.
Supplementary Material
Acknowledgments
We thank Tingting Zhao for help with bioinformatics analyses and Yi Zhong for excellent technical assistance. Fly stocks obtained from the Bloomington Drosophila Stock Center (National Institutes of Health [NIH] P40OD018537) were used in this study. We thank Dr. Patrik Verstreken for providing the Drosophila tau knockout line. Monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biology, Iowa City. This research was funded by NIH (National Institute on Aging) R01AG057331 and AG076214 and Aligning Science Across Parkinson's (grant number ASAP-000301) through the Michael J. Fox Foundation for Parkinson's Research (MJFF). For the purpose of open access, the author has applied a CC BY public copyright license to all Author Accepted Manuscripts arising from this submission.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.278576.123.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine J-C, Geurts P, Aerts J, et al. 2017. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 14: 1083–1086. 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andorfer C, Acker CM, Kress Y, Hof PR, Duff K, Davies P. 2005. Cell-cycle reentry and cell death in transgenic mice expressing nonmutant human τ isoforms. J Neurosci 25: 5446–5454. 10.1523/JNEUROSCI.4637-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardai FH, Ordonez DG, Bailey RM, Hamm M, Lewis J, Feany MB. 2018a. Lrrk promotes τ neurotoxicity through dysregulation of actin and mitochondrial dynamics. PLoS Biol 16: e2006265. 10.1371/journal.pbio.2006265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardai FH, Wang L, Mutreja Y, Yenjerla M, Gamblin TC, Feany MB. 2018b. A conserved cytoskeletal signaling cascade mediates neurotoxicity of FTDP-17 τ mutations in vivo. J Neurosci 38: 108–119. 10.1523/JNEUROSCI.1550-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnstedt O, Owald D, Felsenberg J, Brain R, Moszynski J-P, Talbot CB, Perrat PN, Waddell S. 2016. Memory-relevant mushroom body output synapses are cholinergic. Neuron 89: 1237–1247. 10.1016/j.neuron.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan RL, Benzer S. 1993. Defective glia in the Drosophila brain degeneration mutant drop-dead. Neuron 10: 839–850. 10.1016/0896-6273(93)90200-B [DOI] [PubMed] [Google Scholar]
- Bukar Maina M, Al-Hilaly YK, Serpell LC. 2016. Nuclear τ and its potential role in Alzheimer's disease. Biomolecules 6: 9. 10.3390/biom6010009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnouf S, Grönke S, Augustin H, Dols J, Gorsky MK, Werner J, Kerr F, Alic N, Martinez P, Partridge L. 2016. Deletion of endogenous τ proteins is not detrimental in Drosophila. Sci Rep 6: 23102. 10.1038/srep23102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butner KA, Kirschner MW. 1991. τ protein binds to microtubules through a flexible array of distributed weak sites. J Cell Biol 115: 717–730. 10.1083/jcb.115.3.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrales Fontela Y, Kadavath H, Biernat J, Riedel D, Mandelkow E, Zweckstetter M. 2017. Multivalent cross-linking of actin filaments and microtubules through the microtubule-associated protein τ. Nat Commun 8: 1981. 10.1038/s41467-017-02230-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccamo A, Maldonado MA, Bokov AF, Majumder S, Oddo S. 2010. CBP gene transfer increases BDNF levels and ameliorates learning and memory deficits in a mouse model of Alzheimer's disease. Proc Natl Acad Sci 107: 22687–22692. 10.1073/pnas.1012851108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colodner KJ, Feany MB. 2010. Glial fibrillary tangles and JAK/STAT-mediated glial and neuronal cell death in a Drosophila model of glial tauopathy. J Neurosci 30: 16102–16113. 10.1523/JNEUROSCI.2491-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset V, Treiber CD, Waddell S. 2018. Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. eLife 7: e34550. 10.7554/eLife.34550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross DC, Muñoz JP, Hernández P, Maccioni RB. 2000. Nuclear and cytoplasmic τ proteins from human nonneuronal cells share common structural and functional features with brain τ. J Cell Biochem 78: 305–317. [DOI] [PubMed] [Google Scholar]
- Cutler T, Sarkar A, Moran M, Steffensmeier A, Puli OR, Mancini G, Tare M, Gogia N, Singh A. 2015. Drosophila eye model to study neuroprotective role of CREB binding protein (CBP) in Alzheimer's disease. PLoS One 10: e0137691. 10.1371/journal.pone.0137691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DL, Li M, Lee EB. 2023. Human Alzheimer's disease reactive astrocytes exhibit a loss of homeostastic gene expression. Acta Neuropathol Commun 11: 127. 10.1186/s40478-023-01624-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie K, Janssens J, Koldere D, De Waegeneer M, Pech U, Kreft Ł, Aibar S, Makhzami S, Christiaens V, Bravo González-Blas C, et al. 2018. A single-cell transcriptome atlas of the aging Drosophila brain. Cell 174: 982–998.e20. 10.1016/j.cell.2018.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson HN, Ferreira A, Eyster MV, Ghoshal N, Binder LI, Vitek MP. 2001. Inhibition of neuronal maturation in primary hippocampal neurons from τ deficient mice. J Cell Sci 114: 1179–1187. 10.1242/jcs.114.6.1179 [DOI] [PubMed] [Google Scholar]
- Dias-Santagata D, Fulga TA, Duttaroy A, Feany MB. 2007. Oxidative stress mediates τ-induced neurodegeneration in Drosophila. J Clin Invest 117: 236–245. 10.1172/JCI28769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBoff B, Götz J, Feany MB. 2012. τ promotes neurodegeneration via DRP1 mislocalization in vivo. Neuron 75: 618–632. 10.1016/j.neuron.2012.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBoff B, Feany M, Götz J. 2013. Why size matters: balancing mitochondrial dynamics in Alzheimer's disease. Trends Neurosci 36: 325–335. 10.1016/j.tins.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Dumont M, Stack C, Elipenahli C, Jainuddin S, Gerges M, Starkova NN, Yang L, Starkov AA, Beal F. 2011. Behavioral deficit, oxidative stress, and mitochondrial dysfunction precede τ pathology in P301S transgenic mice. FASEB J 25: 4063–4072. 10.1096/fj.11-186650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feany MB, Dickson DW. 1996. Neurodegenerative disorders with extensive τ pathology: a comparative study and review. Ann Neurol 40: 139–148. 10.1002/ana.410400204 [DOI] [PubMed] [Google Scholar]
- Feany MB, Quinn WG. 1995. A neuropeptide gene defined by the Drosophila memory mutant amnesiac. Science 268: 869–873. 10.1126/science.7754370 [DOI] [PubMed] [Google Scholar]
- Fitz NF, Nam KN, Wolfe CM, Letronne F, Playso BE, Iordanova BE, Kozai TDY, Biedrzycki RJ, Kagan VE, Tyurina YY, et al. 2021. Phospholipids of APOE lipoproteins activate microglia in an isoform-specific manner in preclinical models of Alzheimer's disease. Nat Commun 12: 3416. 10.1038/s41467-021-23762-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Hemberg M, Lewis J, Feany MB. 2014. τ promotes neurodegeneration through global chromatin relaxation. Nat Neurosci 17: 357–366. 10.1038/nn.3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Götz J, Feany MB. 2015. Connecting the dots between τ dysfunction and neurodegeneration. Trends Cell Biol 25: 46–53. 10.1016/j.tcb.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Bardai FH, Feany MB. 2016. Lamin dysfunction mediates neurodegeneration in tauopathies. Curr Biol 26: 129–136. 10.1016/j.cub.2015.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulga TA, Elson-Schwab I, Khurana V, Steinhilb ML, Spires TL, Hyman BT, Feany MB. 2007. Abnormal bundling and accumulation of F-actin mediates τ-induced neuronal degeneration in vivo. Nat Cell Biol 9: 139–148. 10.1038/ncb1528 [DOI] [PubMed] [Google Scholar]
- Ghetti B, Oblak AL, Boeve BF, Johnson KA, Dickerson BC, Goedert M. 2015. Invited review: frontotemporal dementia caused by microtubule-associated protein τ gene (MAPT) mutations: a chameleon for neuropathology and neuroimaging: MAPT mutations and FTD. Neuropathol Appl Neurobiol 41: 24–46. 10.1111/nan.12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M. 2004. τ protein and neurodegeneration. Semin Cell Dev Biol 15: 45–49. 10.1016/j.semcdb.2003.12.015 [DOI] [PubMed] [Google Scholar]
- Goedert M, Jakes R. 2005. Mutations causing neurodegenerative tauopathies. Biochim Biophys Acta 1739: 240–250. 10.1016/j.bbadis.2004.08.007 [DOI] [PubMed] [Google Scholar]
- Goedert M, Ghetti B, Spillantini MG. 2012. Frontotemporal dementia: implications for understanding Alzheimer disease. Cold Spring Harb Perspect Med 2: a006254. 10.1101/cshperspect.a006254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman LD, Bellen HJ. 2022. Recent insights into the role of glia and oxidative stress in Alzheimer's disease gained from Drosophila. Curr Opin Neurobiol 72: 32–38. 10.1016/j.conb.2021.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz J, Chen F, Barmettler R, Nitsch RM. 2001. τ filament formation in transgenic mice expressing P301L τ. J Biol Chem 276: 529–534. 10.1074/jbc.M006531200 [DOI] [PubMed] [Google Scholar]
- Götz J, Halliday G, Nisbet RM. 2019. Molecular pathogenesis of the tauopathies. Annu Rev Pathol 14: 239–261. 10.1146/annurev-pathmechdis-012418-012936 [DOI] [PubMed] [Google Scholar]
- Grossman M, Seeley WW, Boxer AL, Hillis AE, Knopman DS, Ljubenov PA, Miller B, Piguet O, Rademakers R, Whitwell JL, et al. 2023. Frontotemporal lobar degeneration. Nat Rev Dis Primers 9: 40. 10.1038/s41572-023-00447-0 [DOI] [PubMed] [Google Scholar]
- Guo M, Bao EL, Wagner M, Whitsett JA, Xu Y. 2017. SLICE: determining cell differentiation and lineage based on single cell entropy. Nucleic Acids Res 45: e54. 10.1093/nar/gkw1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Jeong H-H, Hsieh Y-C, Klein H-U, Bennett DA, De Jager PL, Liu Z, Shulman JM. 2018. τ activates transposable elements in Alzheimer's disease. Cell Rep 23: 2874–2880. 10.1016/j.celrep.2018.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven-Ozkan T, Davis RL. 2014. Functional neuroanatomy of Drosophila olfactory memory formation. Learn Mem 21: 519–526. 10.1101/lm.034363.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Oguchi K, Okabe S, Kuno J, Terada S, Ohshima T, Sato-Yoshitake R, Takei Y, Noda T, Hirokawa N. 1994. Altered microtubule organization in small-calibre axons of mice lacking τ protein. Nature 369: 488–491. 10.1038/369488a0 [DOI] [PubMed] [Google Scholar]
- Heidary G, Fortini ME. 2001. Identification and characterization of the Drosophila τ homolog. Mech Dev 108: 171–178. 10.1016/S0925-4773(01)00487-7 [DOI] [PubMed] [Google Scholar]
- Heisenberg M. 2003. Mushroom body memoir: from maps to models. Nat Rev Neurosci 4: 266–275. 10.1038/nrn1074 [DOI] [PubMed] [Google Scholar]
- Heisenberg M, Böhl K. 1979. Isolation of anatomical brain mutants of Drosophila by histological means. Z Naturforsch C 34: 143–147. 10.1515/znc-1979-1-228 [DOI] [Google Scholar]
- Herrup K, Arendt T. 2002. Re-expression of cell cycle proteins induces neuronal cell death during Alzheimer's disease. J Alzheimers Dis 4: 243–247. 10.3233/JAD-2002-4315 [DOI] [PubMed] [Google Scholar]
- Hong S-T, Choi K-W. 2013. TCTP directly regulates ATM activity to control genome stability and organ development in Drosophila melanogaster. Nat Commun 4: 2986. 10.1038/ncomms3986 [DOI] [PubMed] [Google Scholar]
- Hu Y, Tattikota SG, Liu Y, Comjean A, Gao Y, Forman C, Kim G, Rodiger J, Papatheodorou I, Dos Santos G, et al. 2021. DRscDB: a single-cell RNA-seq resource for data mining and data comparison across species. Comput Struct Biotechnol J 19: 2018–2026. 10.1016/j.csbj.2021.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Q, He RQ, Haque N, Qu MH, del Carmen Alonso A, Grundke-Iqbal I, Iqbal K. 2003. Microtubule associated protein τ binds to double-stranded but not single-stranded DNA. Cell Mol Life Sci 60: 413–421. 10.1007/s000180300034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husseman JW, Nochlin D, Vincent I. 2000. Mitotic activation: a convergent mechanism for a cohort of neurodegenerative diseases. Neurobiol Aging 21: 815–828. 10.1016/S0197-4580(00)00221-9 [DOI] [PubMed] [Google Scholar]
- Ishida C, Kobayashi K, Kitamura T, Ujike H, Iwasa K, Yamada M. 2015. Frontotemporal dementia with parkinsonism linked to chromosome 17 with the MAPT R406W mutation presenting with a broad distribution of abundant senile plaques. Neuropathology 35: 75–82. 10.1111/neup.12154 [DOI] [PubMed] [Google Scholar]
- Jiwaji Z, Hardingham GE. 2023. The consequences of neurodegenerative disease on neuron-astrocyte metabolic and redox interactions. Neurobiol Dis 185: 106255. 10.1016/j.nbd.2023.106255 [DOI] [PubMed] [Google Scholar]
- Karademir D, Todorova V, Ebner LJA, Samardzija M, Grimm C. 2022. Single-cell RNA sequencing of the retina in a model of retinitis pigmentosa reveals early responses to degeneration in rods and cones. BMC Biol 20: 86. 10.1186/s12915-022-01280-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana V, Lu Y, Steinhilb ML, Oldham S, Shulman JM, Feany MB. 2006. TOR-mediated cell-cycle activation causes neurodegeneration in a Drosophila tauopathy model. Curr Biol 16: 230–241. 10.1016/j.cub.2005.12.042 [DOI] [PubMed] [Google Scholar]
- Khurana V, Merlo P, DuBoff B, Fulga TA, Sharp KA, Campbell SD, Götz J, Feany MB. 2012. A neuroprotective role for the DNA damage checkpoint in tauopathy. Aging Cell 11: 360–362. 10.1111/j.1474-9726.2011.00778.x [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44(W1): W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapytsko A, Kollarovic G, Ivanova L, Studencka M, Schaber J. 2015. FoCo: a simple and robust quantification algorithm of nuclear foci. BMC Bioinformatics 16: 392. 10.1186/s12859-015-0816-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law AD, Cassar M, Long DM, Chow ES, Giebultowicz JM, Venkataramanan A, Strauss R, Kretzschmar D. 2022. FTD-associated mutations in τ result in a combination of dominant and recessive phenotypes. Neurobiol Dis 170: 105770. 10.1016/j.nbd.2022.105770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Neve RL, Kosik KS. 1989. The microtubule binding domain of τ protein. Neuron 2: 1615–1624. 10.1016/0896-6273(89)90050-0 [DOI] [PubMed] [Google Scholar]
- Lee JA, Hall B, Allsop J, Alqarni R, Allen SP. 2021. Lipid metabolism in astrocytic structure and function. Semin Cell Dev Biol 112: 123–136. 10.1016/j.semcdb.2020.07.017 [DOI] [PubMed] [Google Scholar]
- Lewis J, Dickson DW, Lin WL, Chisholm L, Corral A, Jones G, Yen SH, Sahara N, Skipper L, Yager D, et al. 2001. Enhanced neurofibrillary degeneration in transgenic mice expressing mutant τ and APP. Science 293: 1487–1491. 10.1126/science.1058189 [DOI] [PubMed] [Google Scholar]
- Li H, Horns F, Wu B, Xie Q, Li J, Li T, Luginbuhl DJ, Quake SR, Luo L. 2017. Classifying Drosophila olfactory projection neuron subtypes by single-cell RNA sequencing. Cell 171: 1206–1220.e22. 10.1016/j.cell.2017.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Janssens J, De Waegeneer M, Kolluru SS, Davie K, Gardeux V, Saelens W, David FPA, Brbić M, Spanier K, et al. 2022. Fly Cell Atlas: a single-nucleus transcriptomic atlas of the adult fruit fly. Science 375: eabk2432. 10.1126/science.abk2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li JSS, Rodiger J, Comjean A, Attrill H, Antonazzo G, Brown NH, Hu Y, Perrimon N. 2022. FlyPhoneDB: an integrated web-based resource for cell-cell communication prediction in Drosophila. Genetics 220: iyab235. 10.1093/genetics/iyab235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis PA, Howard TH, Castleberry RP, Binder LI. 1990. Identification of nuclear τ isoforms in human neuroblastoma cells. Proc Natl Acad Sci 87: 8422–8426. 10.1073/pnas.87.21.8422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan JP, Chotkowski HL, Glaser RL. 2002. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res 30: 3698–3705. 10.1093/nar/gkf496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi MN, Shuai Y, Turner GC. 2020. The Drosophila mushroom body: from architecture to algorithm in a learning circuit. Annu Rev Neurosci 43: 465–484. 10.1146/annurev-neuro-080317-0621333 [DOI] [PubMed] [Google Scholar]
- Morris M, Hamto P, Adame A, Devidze N, Masliah E, Mucke L. 2013. Age-appropriate cognition and subtle dopamine-independent motor deficits in aged τ knockout mice. Neurobiol Aging 34: 1523–1529. 10.1016/j.neurobiolaging.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard M, Verkhratsky A. 2012. Artifact versus reality: how astrocytes contribute to synaptic events. Glia 60: 1013–1023. 10.1002/glia.22288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville KE, Bosse TL, Klekos M, Mills JF, Weicksel SE, Waters JS, Tipping M. 2018. A novel ex vivo method for measuring whole brain metabolism in model systems. J Neurosci Methods 296: 32–43. 10.1016/j.jneumeth.2017.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez DG, Lee MK, Feany MB. 2018. α-Synuclein induces mitochondrial dysfunction through spectrin and the actin cytoskeleton. Neuron 97: 108–124.e6. 10.1016/j.neuron.2017.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poorkaj P, Grossman M, Steinbart E, Payami H, Sadovnick A, Nochlin D, Tabira T, Trojanowski JQ, Borson S, Galasko D, et al. 2001. Frequency of τ gene mutations in familial and sporadic cases of non-Alzheimer dementia. Arch Neurol 58: 383–387. 10.1001/archneur.58.3.383 [DOI] [PubMed] [Google Scholar]
- Praschberger R, Kuenen S, Schoovaerts N, Kaempf N, Singh J, Janssens J, Swerts J, Nachman E, Calatayud C, Aerts S, et al. 2023. Neuronal identity defines α-synuclein and τ toxicity. Neuron 111: 1577–1590.e11. 10.1016/j.neuron.2023.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rhein V, Song X, Wiesner A, Ittner LM, Baysang G, Meier F, Ozmen L, Bluethmann H, Dröse S, Brandt U, et al. 2009. Amyloid-β and τ synergistically impair the oxidative phosphorylation system in triple transgenic Alzheimer's disease mice. Proc Natl Acad Sci 106: 20057–20062. 10.1073/pnas.0905529106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Murphy MA, Dammer EB, Olsen AL, Rangaraju S, Fraenkel E, Feany MB. 2020. Comparative proteomic analysis highlights metabolic dysfunction in α-synucleinopathy. NPJ Parkinsons Dis 6: 40. 10.1038/s41531-020-00143-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. 2015. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33: 495–502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz L, Ramirez P, Lemieux A, Gonzalez E, Thomson T, Frost B. 2023. τ-induced elevation of the activity-regulated cytoskeleton associated protein Arc1 causally mediates neurodegeneration in the adult Drosophila brain. Neuroscience 518: 101–111. 10.1016/j.neuroscience.2022.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Evans MD, Mao W, Nana AL, Seeley WW, Adame A, Rissman RA, Masliah E, Mucke L. 2019. Early neuronal accumulation of DNA double strand breaks in Alzheimer's disease. Acta Neuropathol Commun 7: 77. 10.1186/s40478-019-0723-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöberg MK, Shestakova E, Mansuroglu Z, Maccioni RB, Bonnefoy E. 2006. τ protein binds to pericentromeric DNA: a putative role for nuclear τ in nucleolar organization. J Cell Sci 119: 2025–2034. 10.1242/jcs.02907 [DOI] [PubMed] [Google Scholar]
- Smolič T, Zorec R, Vardjan N. 2022. Pathophysiology of lipid droplets in neuroglia. Antioxidants (Basel) 11: 22. 10.3390/antiox11010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, Dudoit S. 2018. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19: 477. 10.1186/s12864-018-4772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Samimi H, Gamez M, Zare H, Frost B. 2018. Pathogenic τ-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat Neurosci 21: 1038–1048. 10.1038/s41593-018-0194-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thadathil N, Delotterie DF, Xiao J, Hori R, McDonald MP, Khan MM. 2021. DNA double-strand break accumulation in Alzheimer's disease: evidence from experimental models and postmortem human brains. Mol Neurobiol 58: 118–131. 10.1007/s12035-020-02109-8 [DOI] [PubMed] [Google Scholar]
- Thurston VC, Zinkowski RP, Binder LI. 1996. τ as a nucleolar protein in human nonneural cells in vitro and in vivo. Chromosoma 105: 20–30. 10.1007/BF02510035 [DOI] [PubMed] [Google Scholar]
- Tuncbag N, Braunstein A, Pagnani A, Huang S-SC, Chayes J, Borgs C, Zecchina R, Fraenkel E. 2013. Simultaneous reconstruction of multiple signaling pathways via the prize-collecting Steiner forest problem. J Comput Biol 20: 124–136. 10.1089/cmb.2012.0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuncbag N, Gosline SJ, Kedaigle A, Soltis AR, Gitter A, Fraenkel E. 2016. Network-based interpretation of diverse high-throughput datasets through the omics integrator software package. PLoS Comput Biol 12: e1004879. 10.1371/journal.pcbi.1004879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Sande B, Flerin C, Davie K, De Waegeneer M, Hulselmans G, Aibar S, Seurinck R, Saelens W, Cannoodt R, Rouchon Q, et al. 2020. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc 15: 2247–2276. 10.1038/s41596-020-0336-2 [DOI] [PubMed] [Google Scholar]
- Verkhratsky A, Sofroniew MV, Messing A, deLanerolle NC, Rempe D, Rodríguez JJ, Nedergaard M. 2012. Neurological diseases as primary gliopathies: a reassessment of neurocentrism. ASN Neuro 4: AN20120010. 10.1042/AN20120010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bergen M, Friedhoff P, Biernat J, Heberle J, Mandelkow EM, Mandelkow E. 2000. Assembly of τ protein into Alzheimer paired helical filaments depends on a local sequence motif (306VQIVYK311) forming β structure. Proc Natl Acad Sci 97: 5129–5134. 10.1073/pnas.97.10.5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sudan R, Peng V, Zhou Y, Du S, Yuede CM, Lei T, Hou J, Cai Z, Cella M, et al. 2022. TREM2 drives microglia response to amyloid-β via SYK-dependent and -independent pathways. Cell 185: 4153–4169.e19. 10.1016/j.cell.2022.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Qu M-H, Wang X-S, Chen L, Wang D-L, Liu Y, Hua Q, He R-Q. 2008. Binding to the minor groove of the double-strand, τ protein prevents DNA from damage by peroxidation. PLoS One 3: e2600. 10.1371/journal.pone.0002600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch G, Tsai L-H. 2022. Mechanisms of DNA damage-mediated neurotoxicity in neurodegenerative disease. EMBO Rep 23: e54217. 10.15252/embr.202154217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse PJ, Price DL, Clark AW, Coyle JT, DeLong MR. 1981. Alzheimer disease: evidence for selective loss of cholinergic neurons in the nucleus basalis. Ann Neurol 10: 122–126. 10.1002/ana.410100203 [DOI] [PubMed] [Google Scholar]
- Wittmann CW, Wszolek MF, Shulman JM, Salvaterra PM, Lewis J, Hutton M, Feany MB. 2001. Tauopathy in Drosophila: neurodegeneration without neurofibrillary tangles. Science 293: 711–714. 10.1126/science.1062382 [DOI] [PubMed] [Google Scholar]
- Wu T, Deger JM, Ye H, Guo C, Dhindsa J, Pekarek BT, Al-Ouran R, Liu Z, Al-Ramahi I, Botas J, et al. 2023. τ polarizes an aging transcriptional signature to excitatory neurons and glia. eLife 12: e85251. 10.7554/eLife.85251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama Y, Higuchi M, Zhang B, Huang S-M, Iwata N, Saido TC, Maeda J, Suhara T, Trojanowski JQ, Lee VM-Y. 2007. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 53: 337–351. 10.1016/j.neuron.2007.01.010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.