Abstract
Recruitment of non-canonical BCOR-PRC1.1 to non-methylated CpG islands via KDM2B plays a fundamental role in transcription control during developmental processes and cancer progression. However, the mechanism is still largely unknown on how this recruitment is regulated. Here, we unveiled the importance of the Poly-D/E regions within the linker of BCOR for its binding to KDM2B. Interestingly, we also demonstrated that these negatively charged Poly-D/E regions on BCOR play autoinhibitory roles in liquid-liquid phase separation (LLPS) of BCORANK-linker-PUFD/PCGF1RAWUL. Through neutralizing negative charges of these Poly-D/E regions, Ca2+ not only weakens the interaction between BCOR/PCGF1 and KDM2B, but also promotes co-condensation of the enzymatic core of BCOR-PRC1.1 with KDM2B into liquid-like droplet. Accordingly, we propose that Ca2+ could modulate the compartmentation and recruitment of the enzymatic core of BCOR-PRC1.1 on KDM2B target loci. Thus, our finding advances the mechanistic understanding on how the tethering of BCOR-PRC1.1 enzymatic core to KDM2B is regulated.
Subject terms: X-ray crystallography, Molecular biophysics
Mechanistic studies on KDM2B interaction with BCOR/PCGF1 sheds light on the role of calcium and liquid-liquid phase separation in KDM2B recruiting BCOR-PRC1.1 on chromatin.
Introduction
In eukaryotic cells, gene transcription or expression depends heavily on the conformation of chromatin, which is modulated by DNA methylation or post-translation modification of histone tail via chromatin modifying complexes1,2. Among the epigenetic regulators involving in histone modification, polycomb repressive complexes (PRCs) have gained much attention, playing fundamental roles in cellular processes, such as development, carcinogenesis and self-renewal of embryonic stem cells3–6. Based on enzymatic activity toward histone proteins, PRCs are mainly composed of two multi-protein complexes named PRC1 and PRC27,8. PRC1 catalyzes the mono-ubiquitination of K119 on histone H2A through the heterodimer of RING1 A/B and PCGF protein9. PRC2 is a methyltransferase that methylates K27 on histone H3, performed by its core subunits EZH1/EZH2, EED and SUZ1210,11.
In addition to possessing E3 ligase activity, RING1-PCGF heterodimer also forms scaffold for PRC1 assembly by recruiting accessory proteins via the C-terminal RAWUL domains12. Based on PCGF member (PCGF1-6), mammalian PRC1 complexes have been classified into six groups, PRC1.1-PRC1.613. Canonical PRC1 (cPRC1), containing RING1A/B, PCGF2 or 4, CBX protein and PHC protein14, is recruited to H3K27me3 enriched site of chromatin through the chromodomain of CBX protein15. However, in non-canonical PRC1 (ncPRC1), CBX protein is replaced by RYBP or its homolog YAF2, suggesting that ncPRC1 binds to chromatin independent of the histone mark H3K27me3 deposited by PRC215. In contrast to canonical PRC1, a variety of ncPRC1s are much more active on ubiquitylating histone H2A at Lys119 and repressing gene expression, therefore are fundamental in maintaining mESCs identity3,16–18.
PRC1.1 is a non-canonical PRC1 that has been well identified as a critical regulator in identity maintenance of ESCs, as well as in solid and hematologic cancers19–22. In addition to RING1 A/B protein and PCGF1, PRC1.1 also comprises non-PcG proteins, including KDM2B, BCOR or its homolog BCORL1, SKP1 and RYBP. Although BCOR and BCORL1 are homologous, these two proteins exhibit different expression profile. Different from BCORL1, BCOR is highly expressed in hESCs and many tissues22,23. In addition to acting as a tumor suppressor in normal cells, recent studies also showed that BCOR in PRC1.1 is essential for leukemogenesis and proliferation of prostate cancer24,25.
Via the CxxC domain of KDM2B that specifically binds to the non-methylated CpG dinucleotide, PRC1.1 can be recruited to CpG islands (CGIs)26–28. The importance of PRC1.1 regulating gene expression was evident by the fact that CGIs are frequently enriched in promoters of ~70% of mammalian genes, including housekeeping genes, as well as genes that are active in early developmental stages26,29,30. To recruit enzymatic core RING1-PCGF1, KDM2B must cooperate with BCOR or BCORL1. Mutations on BCOR/BCORL1 unlinking PRC1.1 core to KDM2B have been demonstrated contributing to the progression and poor prognosis of acute myeloid leukemia (AML)24,31. Structural studies on BCORL1-PRC1.1 revealed that the leucine rich repeats (LRRs) domain of KDM2B binds to the interface formed by heterodimer of C-terminal PUFD domain of BCORL1 and RAWUL domain of PCGF132. Conversely, biochemical data showed that heterodimer of BCORPUFD and PCGF1RAWUL is not sufficient to interact with KDM2B, while the linker region between ANK and PUFD domain of BCOR is indispensable for its binding to KDM2B22,32,33.
Interestingly, several studies had revealed that, almost all of CGI are highly occupied by KDM2B, but only a fraction of them are enriched for RING1B27,28,34. This implies that KDM2B recruiting enzymatic core of PRC1.1 to non-methylated CpG islands is tightly controlled. Compared to lowly expressed BCORL1, the importance of BCOR in PRC1.1 has been well established in cell fate decision and cancer development19,31,35. However, mechanism details are still largely unknown on how BCOR links the enzymatic core of PRC1.1 to KDM2B. In this study, we constructed a structure model of BCORANK-linker-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 using integrated methods, including X-ray crystallography, XL-MS and SAXS. Our structural and biochemical studies revealed that electrostatic interactions, contributed by Poly-D/E regions on linker of BCOR and positively charged residues on the F-box and LRRs of KDM2B, are essential for BCOR/PCGF1 interacting with KDM2B/SKP1. This interaction can be weakened by Ca2+ through neutralizing the Poly-D/E regions. Intriguingly, Ca2+ also induces the liquid-liquid phase separation of BCORANK-linker-PUFD/PCGF1RAWUL, leading to condensation of enzymatic core of BCOR-PRC1.1 together with KDM2B. Taken together, this study suggests that the tethering of BCOR-PRC1.1 enzymatic core to KDM2B may be modulated by Ca2+.
Results
Poly-D/E on the linker region of BCOR is critical for its association with KDM2B
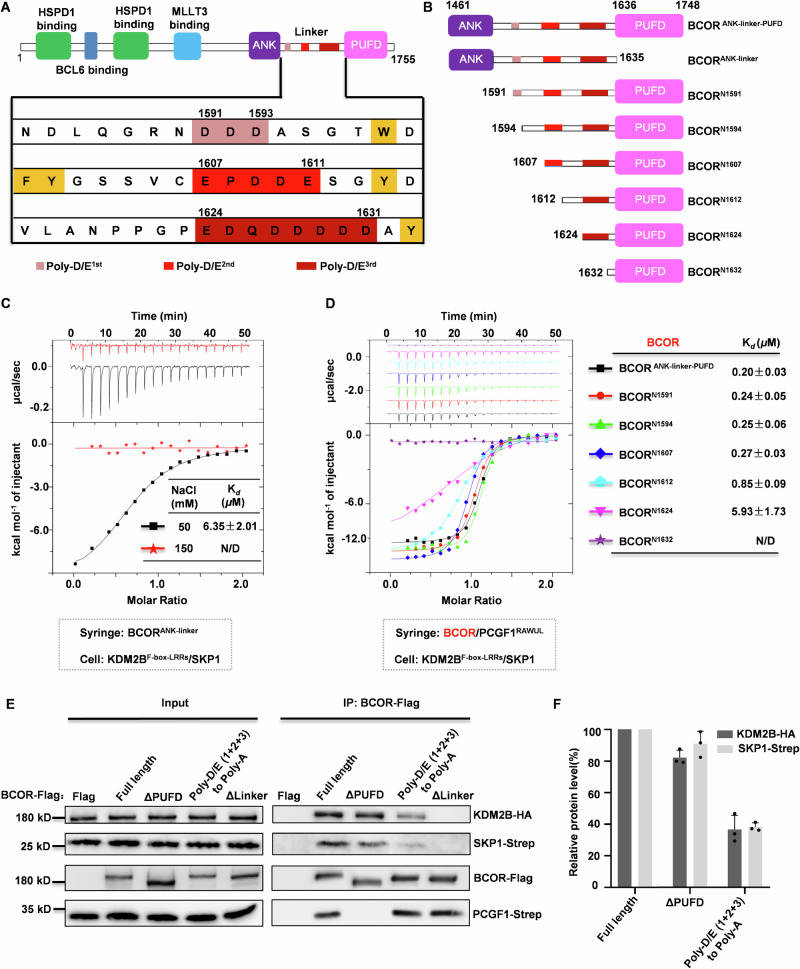
The mechanism for BCORL1 binding to KDM2B was reported that heavily relies on E1664 of BCORL1PUFD domain32. However, for the homologous BCOR, the mechanism on its binding to KDM2B remains unknown, due to the substitution of the critical residue E1664 by L1705 on the PUFD domain of BCOR. Previously, Co-IP experiments showed that the linker region connecting ANK and PUFD domain may be solely responsible for BCOR association with KDM2B22. However, more recently, Wang and colleagues failed to detect association between BCORlinker and KDM2B using biolayer interferometry (BLI)33. Although contradicting in the role of linker region in BCOR association with KDM2B, both groups agreed that ANK contributes moderately to BCOR binding to KDM2B. To confirm the contribution made by ANK domain and the linker region, we performed Isothermal Titration Calorimetry (ITC) assays to determine the interaction between BCORANK-linker and KDM2BF-box-LRRs/SKP1 dimer (KDM2BF-box-LRRs indicates a truncation of KDM2B containing C-terminal F-box domain and LRR repeats domain) (Fig. 1A–C). Interestingly, we failed to detect interaction between BCORANK-linker and KDM2B/SKP1 dimer with buffer containing 150 mM NaCl, whereas this binding was detected with a Kd value of 6.35 ± 2.01 μM when concentration of NaCl in buffer was decreased to 50 mM (Fig. 1C). Then, we tested whether the interaction between BCORANK-linker-PUFD/PCGF1RAWUL dimer and KDM2B/SKP1 dimer could be detected in buffer containing 150 mM NaCl. As shown in Fig. 1D, BCORANK-linker-PUFD/PCGF1RAWUL dimer exhibits strong interaction with KDM2B/SKP1 dimer with a Kd value of 0.20 ± 0.03 μM.
Fig. 1. Determination of the role of Poly-D/E regions on BCOR binding to KDM2B.
A Schematic representation of the domain architecture of BCOR. Three Poly-D/E regions on linker are highlighted with light red for Poly-D/E1st, red for Poly-D/E2nd, and dark red for Poly-D/E3rd. B Truncations of BCOR used in this study. C Binding affinity between BCORANK-linker and KDM2BF-box-LRRs/SKP1 was measured by ITC with buffer containing 50 mM or 150 mM NaCl. D Binding affinity measurement between dimer of PCGF1RAWUL/indicated BCOR truncation and the KDM2BF-box-LRRs/SKP1 dimer using ITC with buffer containing 150 mM NaCl. N/D: Cannot be determined. For (C, D), The Kd values are shown as mean ± SD for triplicate experiments. E The importance of Poly-D/E regions on the linker of BCOR binding to KDM2B/SKP1 is assessed using Co-IP assay. Expressing plasmids for BCOR (wild or mutant), PCGF1, KDM2B or SKP1, were co-transfected into HEK293T cells. Co-IP was performed with anti-Flag magnetic beads, after 48 h transfection. The western-blotting data is representative of three independent experiments. F Relative level of KDM2B-HA or PCGF1-strep protein co-purified with BCOR-Flag as shown in (E). Data are represented as mean ± SD for triplicate experiments.
BCORANK-linker binding to KDM2B can be disrupted by increasing salt concentration, which indicates that the interaction is mainly electrostatic. Upon a closer look at the linker sequence within BCOR, three regions enriched with negatively charged residues (Poly-D/E) were identified. Hereinafter, the three Poly-D/E regions are named Poly-D/E1st (residues 1591–1593), Poly-D/E2nd (residues 1607–1611) and Poly-D/E3rd (residues 1624–1631), respectively (Fig. 1A). To further explore the roles of these Poly-D/E regions in BCOR for binding to KDM2B, Co-IP experiments were performed with Flag-tagged BCOR (wild type or mutant), PCGF1, KDM2B and SKP1 over-expressed in HEK293T cells. As shown in Fig. 1E, F, BCOR binding to KDM2B or SKP1 was apparently impaired by mutating D/E residues on three Poly-D/E regions to Ala, implying the importance of the Poly-D/E regions for BCOR binding to KDM2B.
To explore which Poly-D/E region is involved in BCOR binding to KDM2B via electrostatic interactions, here, we constructed several truncations for the linker of BCOR (Fig. 1B), and ITC assays were used to measure the affinities of heterodimers of these truncations and PCGF1RAWUL binding to KDM2B/SKP1 with buffer containing 150 mM NaCl. The results showed that the binding affinity of BCORN1591 / PCGF1RAWUL to KDM2B/SKP1 was 0.24 ± 0.05 μM, which is comparable to that of BCORANK-linker-PUFD/PCGF1RAWUL dimer (Fig. 1D). This suggested that the ANK domain is not critical for BCOR binding to KDM2B. Then, we determined the affinity of BCORN1594 /PCGF1RAWUL binding to KDM2B/SKP1 to be 0.25 ± 0.06 μM, suggesting Poly-D/E1st (1591–1593) is not required for BCOR binding to KDM2B (Fig. 1D). The determined Kd values for BCORN1607/PCGF1RAWUL dimer, BCORN1612 /PCGF1RAWUL dimer and BCORN1624 /PCGF1RAWUL dimer to KDM2B/SKP1 are 0.27 ± 0.03 μM, 0.85 ± 0.09 μM and 5.93 ± 1.73 μM, respectively (Fig. 1D). When Poly-D/E3rd region was further truncated to just preceding the PUFD domain, the interaction between BCORPUFD/PCGF1RAWUL and KDM2B/SKP1 could not be detected (Fig. 1D). These results suggest that Poly-D/E2nd (1607–1611), Poly-D/E3rd (1624–1631), as well as residues between them, contribute to BCOR binding to KDM2B.
Crystal structure reveals that the conserved NPPGP motif in linker region is also involved in BCOR binding to KDM2B
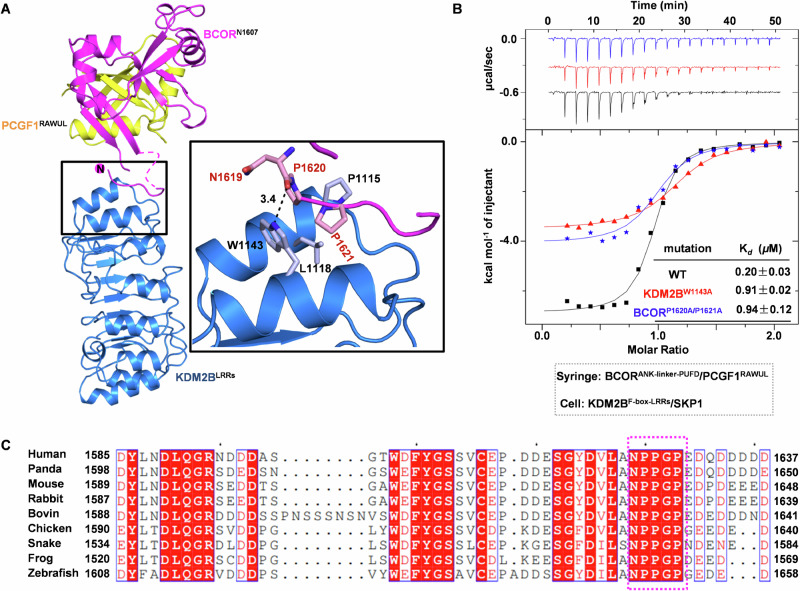
To explore the detailed mechanism on how BCOR binds to KDM2B, we performed structural studies on BCOR/PCGF1 in complex with KDM2B/SKP1 or LRR repeats alone of KDM2B (KDM2BLRRs). Eventually, we successfully obtained high quality crystals for BCORN1607/PCGF1RAWUL/KDM2BLRRs heterotrimer. The crystal structure was determined to resolution of 2.2 Å in space group P43212 with one heterotrimer in the asymmetric unit.
For the linker of BCORN1607, the electron density is only visible for residues from N1619 to D1625, with electron density missing for residues E1607–A1618 and Q1626-S1634. In the crystal structure, N1619–D1625 occupies a narrow cavity formed by the first and second LRR motif of KDM2BLRRs. This binding is stabilized by both hydrophobic and hydrophilic interactions. Key residues contributing to hydrophobic interactions include P1620 and P1621 from BCOR, and P1115/L1118/W1143 from KDM2B. Hydrophilic interactions include hydrogen bond formed by main chain carbonyl of N1619 from BCOR and side chain nitrogen of W1143 from KDM2B (Fig. 2A). These interactions observed in the crystal structure corroborate well with our affinity determination. Residues between Poly-D/E2nd (1607–1611) and Poly-D/E3rd (1624–1631) contribute to BCOR binding with KDM2B/SKP1, as BCORN1612/PCGF1RAWUL dimer exhibited stronger KDM2B/SKP1 binding capacity than that of BCORN1624/ PCGF1RAWUL (Fig. 1D). To further confirm that these interactions revealed in crystal structure contribute to BCOR binding to KDM2B, we constructed mutations on P1620 and P1621 of BCOR, and W1143 of KDM2B. The results clearly showed that binding affinity of BCORANK-linker-PUFD/PCGF1RAWUL dimer and KDM2B/SKP1 dimer was reduced by P1620A/P1621A double mutation of BCOR or W1143A mutation of KDM2B (Fig. 2B). Sequence alignment showed that, 1619NPPGP1623 motif on linker of BCOR is conserved among species from zebrafish to human (Fig. 2C). Taken together, these data revealed that the conserved NPPGP motif preceding the Poly-D/E3rd (1624–1631) also contributes to BCOR association with KDM2B.
Fig. 2. Crystal structure of BCORN1607/PCGF1RAWUL dimer in complex with KDM2BLRRs.
A Structure of BCORN1607/PCGF1RAWUL/KDM2BLRRs heterotrimer is presented as cartoon. BCORN1607, PCGF1RAWUL and KDM2BLRRs are colored as magenta, yellow and blue, respectively. Residues for BCORN1607 interacting with KDM2BLRRs are shown as stick. B Binding affinity between BCORANK-linker-PUFD (wild type or P1620A/P1621A mutant)/PCGF1RAWUL and KDM2BF-box-LRRs (wild type or W1143A mutant)/SKP1 was measured using ITC. The Kd values are shown as mean ± SD for triplicate experiments. C Sequence alignment of BCORlinker among species from zebrafish to human. NPPGP motif on linker of BCOR is boxed with magenta dash.
Structures of BCOR/PCGF1/KDM2B/SKP1 tetramer constructed by integrative approaches
The affinity measurement data presented above demonstrates that NPPGP motif, Poly-D/E2nd and Poly-D/E3rd together are involved in BCOR binding to KDM2B (Fig. 1D). However, in the crystal structure, interactions between these Poly-D/E regions and KDM2B were not observed (Fig. 2A). One possible explanation is that this interaction may be disturbed during the crystallization process. To better understand the mechanism on how BCOR can be recruited by KDM2B, structure models of BCORN1607 /PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 and BCORANK-linker-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 were constructed using integrative approaches, including Small Angle X-ray Scattering (SAXS), molecular docking and cross-linking mass spectrometry (XL-MS).
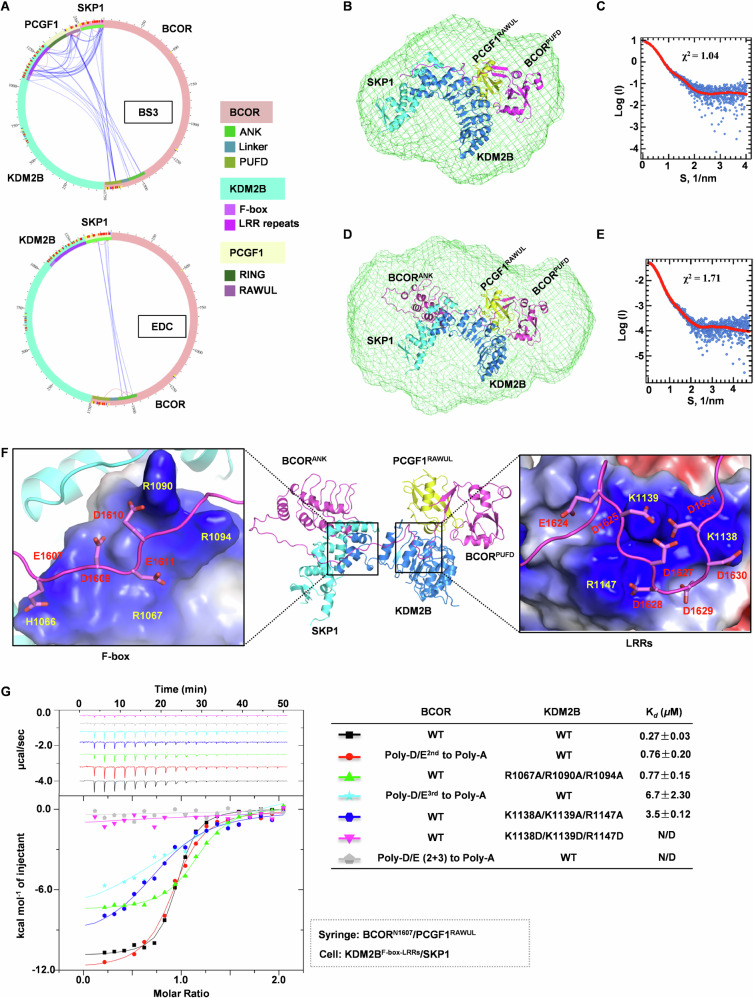
To obtain distance information between domains of BCORANK-linker-PUFD/ PCGF1RAWULdimer and KDM2BF-box-LRRs/SKP1 dimer in the tetramer, the complex was subjected to crosslinking, followed by mass spectrometry analysis. Crosslinking analysis showed that LRRs of KDM2B were extensively cross-linked with RAWUL domain of PCGF1 by crosslinker BS3 (Fig. 3A, Supplementary Table 1 and 2), and ANK domain of BCOR was significantly cross-linked with SKP1 by crosslinker EDC (Fig. 3A, Supplementary Table 3). These results suggest that, in the complex of BCORANK-linker-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1, BCORPUFD/PCGF1RAWUL and KDM2BLRRs are in close proximity, and ANK domain of BCOR is close to SKP1.
Fig. 3. Structural models of BCOR/PCGF1/KDM2B/SKP1 tetramer.
A A Circular plot illustrating the XL-MS analysis of the BCORANK-linker-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 complex cross-linked by crosslinker BS3 or EDC. B The structure model of BCORN1607/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 is overlaid with the ab initio structure envelope. The structure of BCORN1607/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 is modeled using FoXSDock and ModLoop. The ab initio envelope is determined by program GASBOR and averaged by DAMAVER. C Experimental SAXS profile overlaid with the theoretical profile calculated from modeled structure of BCORN1607/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1. D The structure model of BCORANK-linker-PUFD /PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 is overlaid with the ab initio structure. The structure of BCORANK-linker-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 is modeled using FoXSDock and ModLoop. The ab initio envelope is determined by program GASBOR and averaged by DAMAVER. E Experimental SAXS profile overlaid with the theoretical profile calculated from modeled structure of BCORANK-linker-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1. F Details for Poly-D/E2nd and Poly-D/E3rd on linker of BCOR interacting with positive patches on F-box and LRRs of KDM2B, respectively. BCOR, PCGF1 and SKP1 are shown cartoon, and colored as magenta, yellow and cyan, respectively; KDM2B is shown as blue cartoon or electrostatic surface. G Binding affinity between BCORN1607 (wild type or mutation on Poly-D/E)/PCGF1RAWUL and KDM2BF-box-LRRs (wild type or mutation on positive patch)/SKP1 was measured using ITC. N/D: Cannot be determined. The Kd values are shown as mean ± SD for triplicate experiments.
To construct the structure of BCORN1607/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1, we collected SAXS data for the complex (Supplementary Table 4, Supplementary Fig. 1A, B). BCORPUFD (1634–1746)/PCGF1RAWUL was docked onto KDM2BF-box-LRRs/SKP1 with SAXS profile using software FoXSDock, then section of N1619-D1625 from BCOR linker was generated based on crystal structure of BCORN1607 /PCGF1RAWUL/KDM2BLRRs heterotrimer, and finally residues E1607-A1618 and Q1626-S1633 from linker of BCOR were built using software ModLoop (Fig. 3B). The theoretical profile calculated from the derived model of BCORN1607 /PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 is in excellent agreement with the experimental SAXS profile, with a χ2 value of 1.04 (Fig. 3C). Additionally, the BCORN1607 /PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 model agrees well with the ab initio envelope (Fig. 3B).
To construct the structure of BCORANK-linker-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1, ANK domain of BCOR predicted by AlphaFold2 was docked onto the complex of BCOR1607/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1, together with SAXS profile (Supplementary table 4, Supplementary Fig. 1C, D) and distance constrains based on EDC crosslinking data using software FoXSDock. The linker of BCOR was further completed with software ModLoop (Fig. 3D). The theoretical profile calculated from the derived model of BCORANK-linker-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 is in agreement with the experimental SAXS profile, with a χ2 value of 1.73 (Fig. 3E). This model also fits well with the ab initio envelope (Fig. 3D).
Molecular mechanism of Poly-D/E regions of BCOR binding with KDM2B
In the modeled structure of BCORANK-linker-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 tetramer, we not only fit the NPPGP motif of BCOR as that in the crystal structure, but also obtain insights into the interaction of Poly-D/E regions on linker of BCOR. The Poly-D/E1st (1591–1593) does not interact with KDM2B/SKP1 dimer, which is consistent with our affinity measurement that the Poly-D/E1st does not contribute to the binding (Fig. 1D). On the other hand, the Poly-D/E2nd (1607–1611) can form salt bridges with H1066, R1067, R1090 and R1094 from the F-box of KDM2B (Fig. 3F). These interactions were then confirmed by mutagenesis analysis that, the affinity of BCORN1607/PCGF1RAWUL binding to KDM2BF-box-LRRs/SKP1 was reduced >2.5 fold, when D/E on Poly-D/E2nd (1607–1611) were mutated to Ala, or R1067/R1090/R1094 from the F-box of KDM2B were mutated to Ala (Fig. 3G). The Poly-D/E3rd (1624–1631) was predicted to form electrostatic interactions with K1138/K1139/R1147 from the second LRR of KDM2B (Fig. 3F). The binding site of Poly-D/E3rd (1624–1631) was further confirmed by our findings that, the affinity of BCORN1607 /PCGF1RAWUL binding to KDM2BF-box-LRRs/SKP1 was significantly reduced (>12 fold), when D/E on Poly-D/E3rd (1624–1631) were mutated to Ala, or K1138/K1139/R1147 from KDM2B were mutated to Ala, and was almost abolished by mutating K1138/K1139/R1147 from KDM2B to Asp (Fig. 3G). Moreover, by mutating D/E residues on both Poly-D/E2nd (1607–1611) and Poly–D/E3rd (1624–1631) to Ala, interactions between BCORN1607 /PCGF1RAWUL and KDM2BF-box-LRRs/SKP1 cannot be detected by ITC experiments (Fig. 3G). Our results further confirmed that electrostatic interactions mediated by Poly-D/E2nd (1607–1611) and Poly-D/E3rd (1624–1631) from linker of BCOR and positive patches on F-box and LRR repeats from KDM2B are indispensable for KDM2B recruiting BCOR-PRC1.1. Thus, within this BCORANK-linker-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 tetramer model, we observed extensive interactions contributed by Poly-D/E regions and NPPGP motif. The dependence of the Poly-D/E regions of linker for BCOR tethering enzymatic core with KDM2B is distinguished from the homologous BCORL1, in which PUFD domain alone is sufficient to mediate BCORL1-PRC1.1 assembly28,33.
The binding between KDM2B and BCOR/PCGF1 was weakened by magnesium or calcium ions
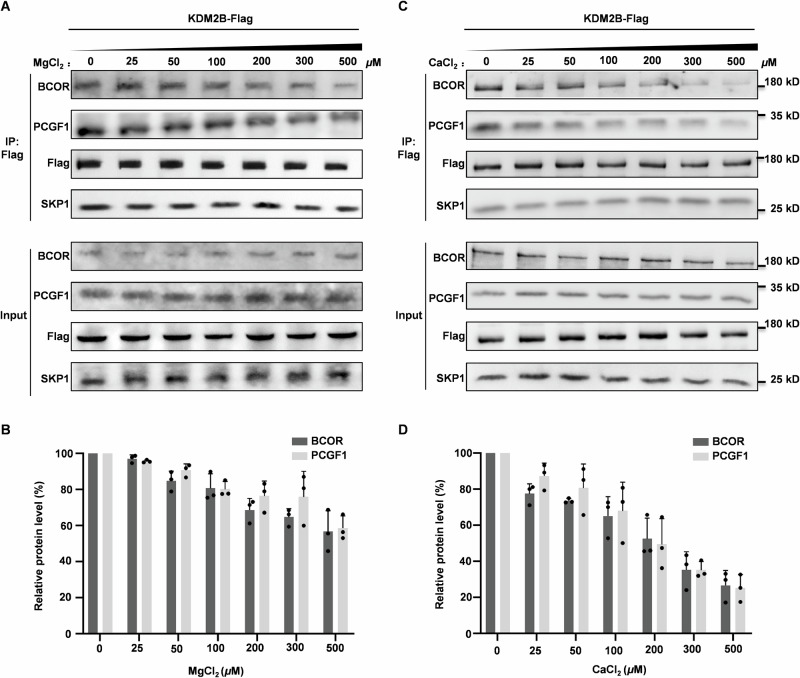
Above analysis clearly shows that electrostatic interactions attributed by Poly-D/E2nd (1607–1611) and Poly-D/E3rd (1624–1631) are indispensable for BCOR binding to KDM2B. Intriguingly, strong interaction between Poly-D/E3rd (1624–1631) and KDM2BLRRs was not observed in the crystal structure of BCORN1607/PCGF1RAWUL/KDM2BLRRs heterotrimer. We noticed that the crystallization buffer contains 100 mM magnesium acetate. We hypothesized that Mg2+ may disrupt BCOR association with KDM2B by neutralizing Poly-D/E2nd (1607–1611) and Poly-D/E3rd (1624–1631). To test this notion, we determined the binding between BCORN1607 /PCGF1RAWUL dimer and KDM2BF-box-LRRs/SKP1 dimer or KDM2BLRRs in the presence of magnesium chloride. Indeed, BCORN1607 /PCGF1RAWUL dimer binding to KDM2BF-box-LRRs/SKP1 dimer or KDM2BLRRs was impaired by magnesium chloride in a concentration dependent manner (Supplementary Fig. 2A, B). To further verify whether Mg2+ can impair BCOR binding to KDM2B under cellular condition, Co-IP experiments were performed to analyze the interaction of exogenous Flag-tagged KDM2B with endogenous BCOR-PRC1.1 subunits in the presence of magnesium ion. As shown in Fig. 4A, B, binding affinity of KDM2B and BCOR/PCGF1 was significantly reduced in the presence of Mg2+ in IP buffer with concentration >200 μM.
Fig. 4. Effect of magnesium ion or calcium ion on BCOR binding to KDM2B.
A Analyzing the effect of magnesium ion on over-expressed KDM2B-Flag binding to endogenous BCOR, PCGF1 and SKP1 using Co-IP assay. The western-blotting data is representative of three independent experiments. B Relative level of BCOR or PCGF1 protein co-purified with KDM2B-Flag as shown in (A). Data are represented as mean ± SD for triplicate experiments. C Analyzing the effect of calcium on over-expressed KDM2B-Flag binding to endogenous BCOR, PCGF1 and SKP1 using Co-IP assay. The western-blotting data is representative of three independent experiments. D Relative level of BCOR or PCGF1 protein co-purified with KDM2B-Flag as shown in (C). Data are represented as mean ± SD for triplicate experiments. Expressing plasmid for KDM2B-Flag was transfected into HEK293T cells. Co-IP was performed with anti-Flag magnetic beads, after 48 h transfection. MgCl2 or CaCl2 at indicated concentration is supplemented to IP buffer.
It has been well demonstrated that calcium (Ca2+) is one of the most important second messengers, in some instances specifically functioning in the nucleus, that regulate a variety of cellular functions such as senescence, cell death, proliferation36–38.It is interesting to test whether KDM2B recruiting BCOR can be regulated by Ca2+, when PRC1.1 complex serving as a transcription regulator. To this end, ITC and biolayer interferometry (BLI) were used to determine the interaction between BCORANK-linker-PUFD/PCGF1RAWULdimer and KDM2BF-box-LRRs/SKP1 dimer in the presence of Ca2+. ITC experiments showed the binding affinity of BCORANK-linker-PUFD/PCGF1RAWUL to KDM2BF-box-LRRs/SKP1 was significantly reduced with increased concentration of Ca2+, indicating that this interaction is inhibited by Ca2+ (Supplementary Fig. 2C). Meanwhile, BLI assay also showed that Ca2+ can inhibit this interaction in a concentration dependent manner (Supplementary Fig. 2D). IP assay also showed that the interaction was impaired for exogenous Flag-tagged KDM2B and endogenous BCOR-PRC1.1 subunits, in the presence of calcium ions with concentration >25 μM (Fig. 4C, D). As a highly localized messenger, the concentration of Ca2+ can reaches over 100 μM near the mouth of ion channels upon stimulation39. Thus, these results suggest that KDM2B association with the core of BCOR-PRC1.1 can be weakened by calcium at increased physiological concentration.
Calcium ion induces liquid-liquid phase separation of BCORANK-linker-PUFD/PCGF1RAWUL heterodimer
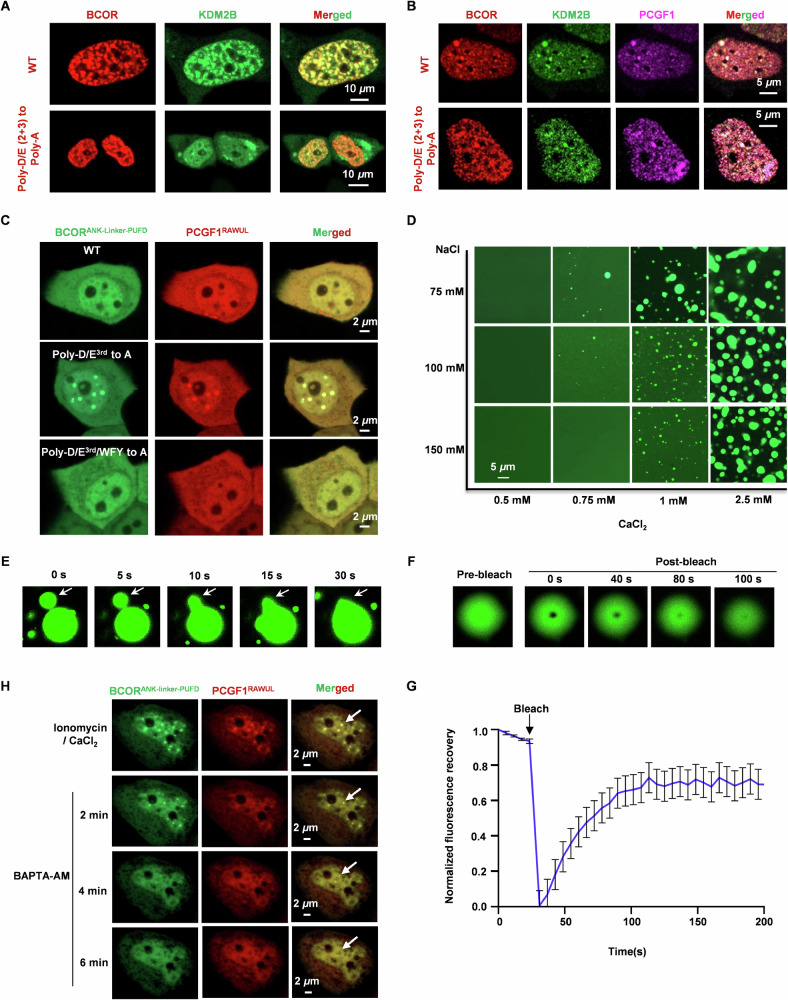
Our structural studies and affinity determination have clearly identified that Poly-D/E2nd (1607–1611) and Poly-D/E3rd (1624–1631) on BCOR are critical for BCOR/PCGF1 associating with KDM2B (Fig. 3F, G). As expected, confocal microscopy showed that co-localization was substantially reduced for over-expressed BCOR-mCherry and KDM2B-EGFP, upon mutating Poly-D/E2nd and Poly-D/E3rd to Poly-A, without over-expressed PCGF1 (Fig. 5A). Interestingly, with the presence of over-expressed PCGF1, robust co-localization into punctate structures can be observed for KDM2B, PCGF1 and mutated BCOR (Poly-D/E2nd and Poly-D/E3rd to Poly-A) (Fig. 5B). This led us to speculate that the co-localization of KDM2B with PCGF1 and mutated BCOR (Poly-D/E2nd and Poly-D/E3rd to Poly-A) may be maintained by liquid-liquid phase separation (LLPS) mechanism, which was mediated by multivalent weak interactions. To test this, we explored the LLPS property of Poly-D/E mutated BCORANK-linker-PUFD /PCGF1RAWUL dimer. As shown in Supplementary Fig. 3A, just mutating Poly-D/E3rd to Poly-A, BCORANK-linker-PUFD mutant /PCGF1RAWUL dimer, but not the monomeric BCORANK-linker-PUFD mutant, formed phase separation in protein concentration dependent manners (Supplementary Fig. 3B). Additionally, the mutated BCORANK-linker-PUFD/PCGF1RAWUL also formed condensates in live HeLa cells (Fig. 5C).
Fig. 5. Liquid-liquid phase separation of BCORANK-linker-PUFD/PCGF1RAWUL hetero-dimer.
A Co-localization analysis of BCOR-mCherry with KDM2B-EGFP in live HeLa cells. B Co-localization analysis of BCOR-mCherry, PCGF1-ECFP and KDM2B-EGFP in live HeLa cells. C Observation of condensates for BCORANK-linker-PUFD (wild type or mutant)/PCGF1RAWUL in HeLa cells. WFY indicates the 5 aromatic residues on linker of BCOR, including W1598, F1600, Y1601, Y1614 and Y1633. D Effect of CaCl2 and NaCl on the phase separation of BCORANK-linker-PUFD/PCGF1RAWUL hetero-dimer with concentration of 60 μM. E Representative fusion event of BCORANK-linker-PUFD/PCGF1RAWUL droplets. F In vitro FRAP analysis of the droplet of BCORANK-linker-PUFD/PCGF1RAWUL. G Normalized FRAP recovery curves for BCORANK-linker-PUFD-EGFP/PCGF1RAWUL droplet in (F). Data are mean ± SEM at each time point and combined data from three independent repeats. H Plasmids expressing BCORANK-Linker-PUFD or PCGF1RAWUL were co-transfected into HeLa cell. Before imaging for LLPS, cells were pretreated with 10 μM ionomycin and 5 mM CaCl2 for 2 h. To observe the effect of calcium-chelator BAPTA-AM on the ionomycin induced LLPS, culture containing 10 μM ionomycin and 5 mM CaCl2 was discarded, the cell was washed with PBS, then culture containing 10 μM BAPTA-AM was added to the cell.
Then, we tested whether Ca2+ could induce phase separation of BCORANK-linker-PUFD/PCGF1RAWUL by neutralizing negative charged Poly-D/E region. Indeed, BCORANK-linker-PUFD/PCGF1RAWUL formed phase separation in CaCl2 and NaCl concentration dependent manners (Fig. 5D,). To investigate the liquid-like nature of BCORANK-linker-PUFD/PCGF1RAWUL condensates, we performed fusion events observation and fluorescence recovery after photobleaching (FRAP) experiments. Rapid fusion of BCORANK-linker-PUFD/PCGF1RAWUL condensates can be observed under time lapse microscopy (Fig. 5E). In addition, rapid recovery of fluorescence in BCORANK-linker-PUFD/PCGF1RAWUL condensates was observed after photo-bleaching (Fig. 5F). Evidently, liquid-like property of BCORANK-linker-PUFD/PCGF1RAWUL condensate is induced by CaCl2. While Mg2+ is the most abundant bivalent cation in cell, we also performed LLPS experiments in the presence of MgCl2. The ability of MgCl2, at concentration as high as 25 mM, to induce LLPS of BCORANK-linker-PUFD/PCGF1RAWUL (at 100 μM), is comparable with that of CaCl2 at concentration of 1 mM (Supplementary Fig. 4).
It is interesting to uncover which sequence motif directly contributes to the condensation of BCORANK-linker-PUFD/PCGF1RAWUL, besides the Ca2+. π-interaction was suggested as one of the common drivers for LLPS formation40. There are five aromatic residues on the linker of BCOR, including W1598, F1600, Y1601, Y1614 and Y1633 (Fig. 1A), and named as “WFY”. After mutating these residues to Ala, CaCl2 failed to induce LLPS of BCORANK-linker-PUFD/PCGF1RAWUL (Supplementary Fig. 5A). Consistently, these aromatic residues were required for LLPS of BCORANK-linker-PUFD (Poly-D/E3rd to Poly-A)/PCGF1RAWUL, in solution or in cells (Fig. 5C and Supplementary Fig. 5B).
Given the fact that BCORANK-linker-PUFD/PCGF1RAWUL could not phase separate spontaneously in live Hela cells (Fig. 5C), physiologically relevant concentration of Mg2+ is insufficient to induce LLPS of BCORANK-linker-PUFD/PCGF1RAWUL. We then tested whether phase separation of BCORANK-linker-PUFD/PCGF1RAWUL in live cell could be induced by cellular calcium. As shown in Fig. 5H, after calcium influx is induced by 10 μM ionomycin, liquid-like droplet of BCORANK-linker-PUFD/PCGF1RAWUL can be observed in live HeLa cells. Moreover, the ionomycin induced LLPS can be reversed by treatment with 10 μM calcium-chelator BAPTA-AM (Fig. 5H).
Taken together, these results demonstrated that, via neutralizing the negatively charged Poly-D/E region on BCOR, Ca2+ or Mg2+ can induce the liquid-liquid phase separation of BCORANK-linker-PUFD/PCGF1RAWUL in solution. More importantly, elevated Ca2+ concentration is required to trigger the phase separation of BCORANK-linker-PUFD/PCGF1RAWUL under cellular condition.
Co-condensation of enzymatic core of BCOR–PRC1.1 with KDM2B via Ca2+ stimulated LLPS mechanism
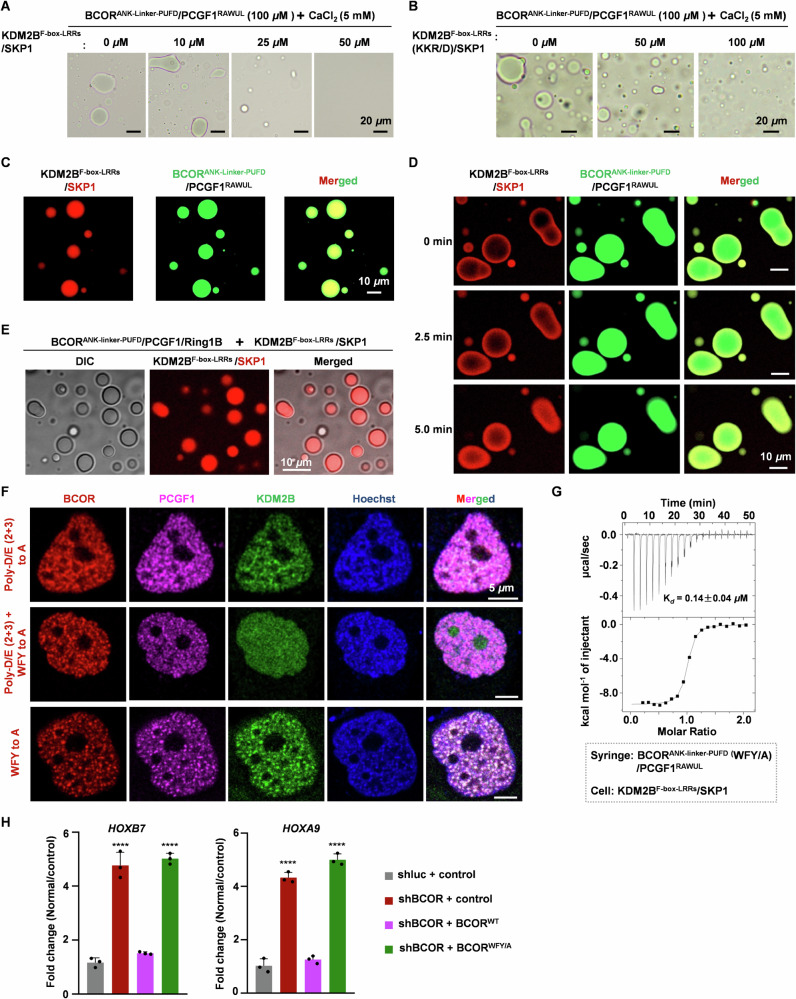
Given that KDM2B tightly associates with the linker on BCOR, here, we test whether KDM2BF-box-LRRs/SKP1 can influence Ca2+ induced LLPS of BCORANK-linker-PUFD/PCGF1RAWUL. We performed LLPS experiments in the presence of KDM2BF-box-LRRs/SKP1 dimer. Under microscopy observation, we found that CaCl2 induced LLPS of BCORANK-linker-PUFD/PCGF1RAWUL was significantly decreased, with increasing concentration of KDM2BF-box-LRRs/SKP1 dimer (Fig. 6A). After mutating K1138/K1139/R1147 on KDM2B to Asp, KDM2BF-box-LRRs (K1138D/K1139D/R1147D mutant)/SKP1 dimer showed reduced capacity to impair Ca2+ induced LLPS of BCORANK-linker-PUFD/PCGF1RAWUL (Fig. 6B), most likely resulting from its decreased binding capacity to BCORANK-linker-PUFD/PCGF1RAWUL. These suggest that, by binding to BCORANK-linker-PUFD/PCGF1RAWUL, KDM2BF-box-LRRs/SKP1 competitively inhibits Ca2+ induced LLPS of BCORANK-linker-PUFD/PCGF1RAWUL.
Fig. 6. KDM2B co-segregates together with BCOR /PCGF1 in Ca2+ induced droplets.
A, B Effects of KDM2BF-box-LRRs/SKP1(A) or KDM2BF-box-LRRs(KKR/D)/SKP1 (B) on condensates of BCORANK-linker-PUFD/PCGF1RAWUL. Before mixing with KDM2BF-box-LRRs/SKP1 or KDM2BF-box-LRRs(KKR/D)/SKP1, BCORANK-linker-PUFD/PCGF1RAWUL (100 μM) was mixed with 5 mM CaCl2 to stimulate droplets formation. C, D Co-localization analysis of KDM2BF-box-LRRs/SKP1 and BCORANK-linker-PUFD/PCGF1RAWUL in Ca2+ induced droplets. For (C), KDM2BF-box-LRRs/SKP1-mCherry or KDM2BF-box-LRRs(KKR/D)/SKP1-mCherry (10 μM) was mixed with GFP-BCORANK-linker-PUFD/PCGF1RAWUL (100 μM), then 5 mM CaCl2 was added to induce droplet formation. For (D), BCORANK-linker-PUFD/PCGF1RAWUL (100 μM) was incubated with 5 mM CaCl2 to stimulate droplets formation, then KDM2BF-box-LRRs/SKP1-mCherry or KDM2BF-box-LRRs(KKR/D)/SKP1-mCherry (10 μM) was added to the mixture immediately. KDM2BF-box-LRRs(KKR/D) indicates that residues K1138, K1139 and R1147 on KDM2BF-box-LRRs were mutated to Asp. E Co-localization analysis of KDM2BF-box-LRRs/SKP1-mCherry with CaCl2 induced BCORANK-linker-PUFD/PCGF1ΔN35/Ring1B droplets. KDM2BF-box-LRRs/SKP1-mCherry (6 μM) was mixed with BCORANK-linker-PUFD/PCGF1ΔN35/Ring1B (60 μM), then 5 mM CaCl2 was added to induce droplet formation. F Co-localization analysis of BCOR-mCherry mutant, PCGF1-ECFP and KDM2B-EGFP in live HeLa cells. DNA was stained by Hoechst 33258 (Blue). G Binding affinity between BCORANK-linker-PUFD (WFY to A)/PCGF1RAWUL and KDM2BF-box-LRRs/SKP1 was measured using ITC. The Kd values are shown as mean ± SD for triplicate experiments. H Effect of BCORWFY/A mutant on the transcription of HOXB7 and HOXA9 in HEK293T cells. Empty vector was used as a negative control. shluc was used as control shRNA. ∗∗∗∗p < 0.0001. Error bars represent standard deviation from 3 individual samples.
As shown in Fig. 6A, with KDM2BF-box-LRRs/SKP1 dimer at <25 μM, robust Ca2+ (5 mM) stimulated condensates of BCORANK-linker-PUFD/PCGF1RAWUL (100 μM) can be observed under microscopy observation. Then, we performed co-localization analysis for KDM2BF-box-LRRs/SKP1-mCherry (10 μM) and LLPS droplets of EGFP-BCORANK-linker-PUFD/PCGF1RAWUL (100 μM) induced by Ca2+. Surprisingly, we found that KDM2BF-box-LRRs/SKP1 can be partitioned together with BCORANK-linker-PUFD/PCGF1RAWUL, when KDM2BF-box-LRRs/SKP1-mCherry (10 μM) was mixed with EGFP-BCORANK-linker-PUFD/PCGF1RAWUL (100 μM) before treatment with 5 mM CaCl2 (Fig. 6C). When BCORANK-linker-PUFD/PCGF1RAWUL (100 μM) was pretreated with 5 mM CaCl2 before mixing with KDM2BF-box-LRRs/SKP1 (10 μM), KDM2BF-box-LRRs/SKP1 was enriched on the periphery of BCORANK-linker-PUFD/PCGF1RAWUL condensates (Fig. 6D). With time lapse microscopy, we observed that KDM2BF-box-LRRs/SKP1 diffused toward inner side of the condensates (Fig. 6D). These results suggest that Ca2+ induces co-segregation of BCORANK-linker-PUFD/PCGF1RAWUL with KDM2B/SKP1.
To further validate that Ca2+ can induce con-condensation of BCOR-PRC1.1 with KDM2B, we performed LLPS experiments for BCORANK-linker-PUFD/PCGF1ΔN35/Ring1B. As shown in Supplementary Fig. 6, BCORANK-linker-PUFD/PCGF1ΔN35/Ring1B formed LLPS dependent on the presence of CaCl2. Co-localization analysis also showed that KDM2BF-box-LRRs/SKP1 could localize to Ca2+ induced droplets of BCORANK-linker-PUFD/PCGF1ΔN35/Ring1B (Fig. 6E). We have demonstrated that mutating Poly-D/E to Poly-A mimics Ca2+ induced LLPS of BCORANK-linker-PUFD/PCGF1RAWUL. To further confirm that Ca2+ induced LLPS can modulate co-condensation of core subunits of BCOR-PRC1.1 with KDM2B in cell, we determined whether the co-localization of KDM2B together with PCGF1 and mutated BCOR (Poly-D/E2nd and Poly-D/E3rd to Poly-A) was dependent on LLPS of BCORANK-linker-PUFD/PCGF1RAWUL. To this end, aromatic residues on the linker of BCOR, which is critical for the LLPS of BCORANK-linker-PUFD/PCGF1RAWUL induced by Ca2+ or Poly-D/E mutation, were mutated to Ala. As shown in Fig. 6F, by mutating these residues, KDM2B could not condensate with PCGF1/ BCOR (Poly-D/E2nd and Poly-D/E3rd to Poly-A). Given the fact that aromatic residues on linker of BCOR were dispensable for the affinity of BCOR/PCGF1 binding to KDM2B (Fig. 6G, and Supplementary Fig. 7) and the core subunits of BCOR-PRC1.1 binding to chromatin (Supplementary Fig. 8), we concluded that co-condensation of BCOR (Poly-D/E2nd and Poly-D/E3rd to Poly-A) mutant, PCGF1 and KDM2B, was maintained through LLPS mechanism. Furthermore, heterochromatin puncta were overlapped well with condensates of mutated BCOR (Poly-D/E2nd and Poly-D/E3rd to Poly-A) together with PCGF1 and KDM2B (Fig. 6F). Interestingly, when mutating aromatic residues on linker of BCOR (BCORWFY/A) thereby disrupting Ca2+ induced LLPS of BCORANK-linker-PUFD/PCGF1RAWUL, the expression of two well-demonstrated PRC1.1 targets HOXB725 and HOXA941 was significantly increased (Fig. 6H, and Supplementary Fig. 9). This suggests that Ca2+ induced LLPS of BCORANK-linker-PUFD/PCGF1RAWUL is functionally relevant for PRC1.1. Taken together, these results indicate that Ca2+ may modulate condensation of BCOR-PRC1.1 on KDM2B target loci, through neutralizing the Poly-D/E region on linker of BCOR, which may promote gene silencing and compaction of chromatin on target loci.
Discussion
Despite the well-established fact that KDM2B recruiting PRC1.1 to CpG islands is essential for repressing expression of development regulator genes20,27,28, there are still fundamental gaps in understanding how to regulate PRC1.1 recruitment to KDM2B occupied CpG-rich promoters. In this study, we clarified the molecular mechanism on BCORANK-linker-PUFD/PCGF1RAWUL interaction with KDM2BF-box-LRRs/SKP1 through integrative structural approach. With this, we unveiled that the enzymatic core of BCOR-PRC1.1 can be loosened from KDM2B in the presence Ca2+. Interestingly, our results also showed that Ca2+ could induce condensation of BCOR-PRC1.1 together with KDM2B on heterochromatin puncta through liquid-liquid phase separation.
Previous studies have identified the importance of the linker region of BCOR located between ANK repeats and PUFD domain in the process of KDM2B recruiting BCOR-PRC1.1 to targeted loci22. The mechanistic details on the linker region of BCOR binding to KDM2B remain elusive, due to the lack of structural information. In this study, we demonstrated that Poly-D/E2nd (1607–1611) and Poly-D/E3rd (1624–1631) on the linker of BCOR form electrostatic interaction with positively charged patch on the F-box and LRRs of KDM2B. We proposed that BCOR interaction with KDM2B may be weakened via neutralizing the Poly-D/E regions by metal ion. As a universal second messenger, calcium (Ca2+) signaling is involved in various cellular processes, including fertilization, stem cell identity, muscle contraction, secretion, synaptic transmission, memory, gene transcription and cell death42–46. Calcium can regulate gene transcription, often through binding to calmodulin, and stimulates translocation of transcription factor from cytosol to nucleus or exportation of epigenetic regulator from the nucleus37,47,48. However, the direct role of calcium on epigenetic regulator has not been demonstrated in previous studies. In this study, we found that calcium can act on epigenetic regulator BCOR-PRC1.1 by loosening BCOR/PCGF1 heterodimer from KDM2B/SKP1 heterodimer. Our results provide the first example that calcium directly regulates activity of epigenetic regulator.
In resting state, the cellular level of free Ca2+ is usually maintained at low concentration of ~100 nM45,49. In response to stimuli, Ca2+ is released from extracellular or intracellular stores, and the local concentration can reach over 100 μM (at the mouth of channels)39. With Co-IP experiment, we identified that BCOR/PCGF1 interaction with KDM2B was largely weakened in the presence of 100 μM Ca2+. This may suggest that rapidly released Ca2+ may regulate the association of enzymatic core of BCOR-PRC1.1 with KDM2B.
In addition to calcium signaling, liquid-liquid phase separation has also been implicated in transcription control through regulating chromatin organization50–52. Polycomb proteins have been demonstrated to concentrate into membrane-less organelle called Polycomb body via phase separation mechanism53–55. Polycomb protein CBX2, a component of canonical PRC1, condensates CBX2-PRC1 together with nucleosome via LLPS driven by charged low-complexity disordered region (LCDR)56. Our work uncovers another phase separation driver of PRC1 complex. We find that BCOR/PCGF1 dimer, components of non-canonical BCOR-PRC1.1, forms phase separated condensates driven by Ca2+. CpG reader KDM2B can be concentrated into Ca2+ induced BCOR/PCGF1 condensates. Thus, our work suggests that repression effect of BCOR-PRC1.1 on transcription would be amplified by Ca2+ via LLPS mechanism.
Our ITC experiments and Co-IP assays clearly showed that BCOR/PCGF1 interaction with KDM2B could also be weakened by Mg2+. In additional, the LLPS of BCORANK-linker-PUFD/PCGF1RAWUL also can be induced by high concentration of Mg2+. Unlike calcium, free magnesium is abundant in cellular, with concentration in the range of 203.68 to 673.50 μM (in platelets)57. This suggests that Ca2+ may cooperate with Mg2+ to reduce the affinity of enzymatic core of BCOR-PRC1.1 with KDM2B, and to trigger the LLPS of BCORANK-linker-PUFD/PCGF1RAWUL.
BCORL1 is a homolog of BCOR. Previous study revealed that, PUFD domain of BCORL1 is sufficient to associate core subunits of PRC1.1 with KDM2B, and E1664 is indispensable for this activity of BCORL132. In BCOR, the Glu is substituted by L1705. The consequence of this substitution is that PUFD domain of BCOR is insufficient to associate core subunits of PRC1.1 with KDM2B. Whereas, the linker region is indispensable for BCOR to connect core subunits of PRC1.1 with KDM2B. Sequence alignment showed that, D/E is less enriched in linker of BCORL1 than that of BCOR (Supplementary Fig. 10). These analyses suggest that calcium regulation may be unique in the assembly of BCOR-PRC1.1 at CpG islands. Interestingly, sequence analysis shows that L1705 is only conserved in mammalian BCOR (Supplementary Fig. 10). In species from chicken to zebrafish, this L1705 is replaced by Glu, same as that in BCORL1, which may imply that the assembly of BCOR-PRC1.1 cannot be regulated by calcium in these species. We speculate that the inhibitory effect of calcium on KDM2B recruiting BCOR-PRC1.1 to CpG islands is the consequence of evolution.
Previous studies showed that components of PRC1.1 are frequently up-regulated in acute myeloid leukemia (AML) patients24. In line with this finding, Leukemogenesis was suppressed by down-regulating PRC1.1 components including BCOR, PCGF1 and Ring1B24,58. In addition to AML cells, BCOR-PRC1.1 mediated H2A monoubiquitylation is critical for prostate cancer by repressing expression of a subset of androgen receptor target genes25. These imply that disrupting KDM2B recruitment of enzymatic core of BCOR-PRC1.1 is a potential strategy to treat leukemia and prostate cancer.
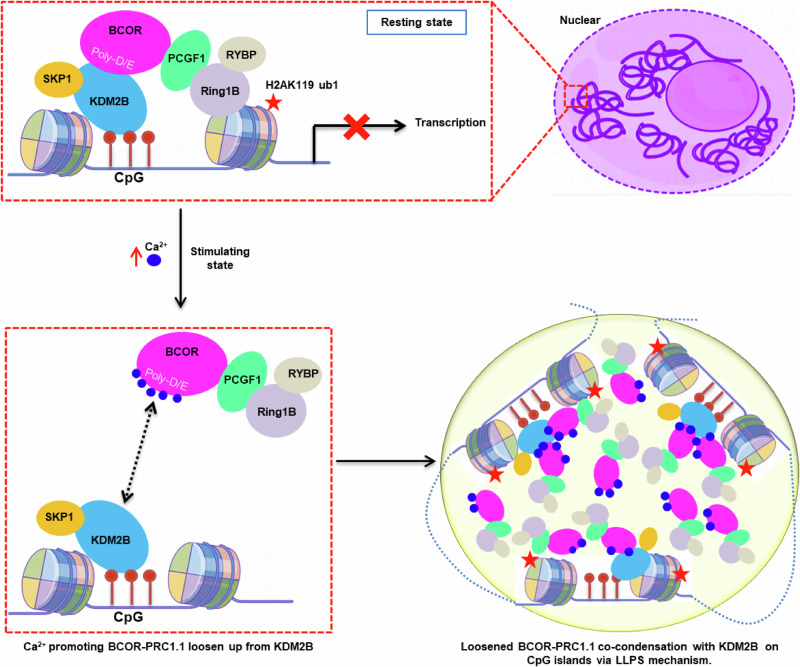
Taken together, the results presented here clearly showed that, by neutralizing Poly-D/E regions on BCOR, Ca2+ not only can reduce the affinity of KDM2B and enzymatic core of BCOR-PRC1.1, but also can condensate them into liquid-like droplet through liquid-liquid phase separation mechanism (Fig. 7). Via liquid-liquid phase separation, Ca2+ may effectively enhance the action of BCOR-PRC1.1 on target loci. Thus, our finding on Ca2+ induced liquid-liquid phase separation of BCOR/PCGF1, not only underpin the understanding how the behavior of BCOR-PRC1.1 is physiologically regulated, but also facilitate the development of potential strategy against cancers by modulating the action of BCOR-PRC1.1 on target loci.
Fig. 7. Proposed mechanisms on calcium ion modulating KDM2B recruitment enzymatic core of BCOR-PRC1.1 to CpG islands.
In resting state, BCOR-PRC1.1 was recruited to CpG islands through interaction with KDM2B, leading to gene silence. After stimulation, concentration of nuclear Ca2+ is elevated. The elevated Ca2+ weakens the association between BCOR/PCGF1 and KDM2B, through neutralizing highly acidic Poly-D/E regions on linker of BCOR. Ca2+ also induces phase separation of BCOR/PCGF1. The elevated Ca2+ further promotes co-condensation of enzymatic core of BCOR-PRC1.1 with KDM2B on target loci via LLPS mechanism.
Methods
Expression and purification
For bacterial expression, cDNA encoding truncations of BCOR, KDM2BF-box-LRRs (residues 1059–1336) and KDM2BLRRs (residues 1103–1336) were cloned into pET-28a with N-terminal hexa-histidine tag and TEV cleavage site; cDNA encoding PCGF1RAWUL (residues 150–255) and SKP1 were cloned into pET-21a without his-tag. BCORANK-linker (residues 1461-1635), BCOR/PCGF1RAWUL dimer, KDM2BF-box-LRRs/SKP1 dimer and KDM2BLRRs (residues 1103–1336) were expressed in BL21 (DE3) cells. Cells were grown in LB medium at 37 °C until OD600 of 0.6 ~ 0.8, then expression was induced with 0.2 mM isopropyl-b-D-thiogalactoside (IPTG) for overnight at 16 °C. Cells were collected and re-suspected with lysis buffer (50 mM Tris pH 8.0, 300 mM NaCl, 10 mM imidazole, 1 mM PMSF, 5 mM β-mercaptoethanol and 5% glycerol). Proteins were purified using Ni2+ affinity chromatography. Truncations of BCOR and KDM2B were digested with TEV to remove the N-terminal his-tag. The proteins were further purified by ion exchange chromatography, and followed by gel filtration equilibrated against buffer containing 20 mM Tris pH 7.5, 150 mM NaCl and 0.5 mM TCEP. The BCORANK-linker-PUFD (Poly-D/E3rd to Poly-A)/PCGF1RAWUL was purified by Ni2+ affinity chromatography and gel filtration with buffer containing 1 M NaCl.
Isothermal titration calorimetry (ITC)
ITC experiments were performed using a Microcal ITC200 instrument. Protein sample in syringe at concentration of 300 μM was titrated into protein sample in cell at concentration of 30 μM in 20 × 2 μL. All Protein samples were dissolved in 20 mM Tris pH7.5, 50 mM NaCl or 150 mM NaCl, 0.5 mM TCEP. Data was analyzed and the figure was prepared using Origin 7.
Co-immunoprecipitation (Co-IP)
HEK293T cells in 10 cm dish were cultured to 70%-80% density to conduct gene transfection. For the interaction identification via co-immunoprecipitation, the plasmid of BCOR-Flag (wild type or mutant) was co-transfected with PCGF1-Strep, KDM2B-HA and SKP1-Strep. Each plasmid of experiment group was transfected with 10 μg. Cells were collected after 48 h and lysed with lysis buffer (50 mM Tris pH7.4, 150 mM NaCl, 5% glycerol, 1% NP-40) with 1 mM protease inhibitor cocktail (Roche) and 1 mM PMSF at 4 °C for 30 min. Cell lysates were clarified by centrifugation at 4 °C for 30 min. Then the 20 μL anti-Flag magnetic beads, which was washed with IP buffer (50 mM Tris pH7.4, 150 mM NaCl, 5% glycerol) for 3 times, was added into the supernatant with 2 times the volume IP buffer and incubated at 4 °C overnight. The magnetic bead was washed with IP wash buffer (50 mM Tris pH 7.4, 150 mM NaCl, 5% glycerol, 0.1% NP-40) with 1 mM PMSF for 3 times. Finally, the magnetic bead was eluted by the IP wash buffer with 120 μL 3 x Flag peptide and 1 mM protease inhibitor cocktail (Roche) and 1 mM PMSF at 4 °C for 2 h. The eluent was then analyzed by western blot.
To verify the effect of magnesium or calcium ions on the binding of KDM2B to BCOR, the plasmid of KDM2B-Flag was transfected to HEK293F cells. The cells were counted every 24 h after transfection, and 15 × 106 cells per EP tube were collected after about 72 h of culture when the viability of the cells was about 80%. Cells were lysed with lysis buffer (50 mM Tris pH7.4, 150 mM NaCl, 5% glycerol, 1% NP-40) with 1 mM protease inhibitor cocktail (Roche) and 1 mM PMSF at 4 °C for 30 min. Cell lysates were clarified by centrifugation at 4 °C for 30 min. The supernatant was collected, divided into aliquots and added with different concentrations of calcium chloride. Then, Co-IP experiment was performed using anti-Flag magnetic beads with calcium chloride at indicated concentration.
Crystallization and structure determination
To obtain BCORN1607/PCGF1RAWUL/KDM2BLRRs heterotrimer, BCORN1607/PCGF1RAWUL was mixed with KDM2BLRRs heterotrimer at mole ratio 1:1, and purified with gel filtration. For crystallization screening, BCORN1607/PCGF1RAWUL/KDM2BLRRs was concentrated to 10 mg/mL and sitting drop vapor diffusion method was used by mixing protein with reservoir solution at volume ratio 1:1 at 20 °C. Crystals of BCORN1607/PCGF1RAWUL/KDM2BLRRs heterotrimer were grown with reservoir solution containing 0.1 M magnesium acetate, 0.1 M sodium citrate pH 6.1, 4% w/v PEG5000 MME.
Crystals were cryoprotected in mother liquid with addition of 30% glycerol, prior to diffraction. Diffraction data was collected at Beam Line 19U1 (BL19U1)59 of National center for Protein Science Shanghai and the Shanghai Synchrotron Radiation Facility. Diffraction data was processed with automatic data-processing system Aquarium, and scaled with Aimless60. Crystal structure was solved by molecule replacement with software Molrep61 using crystal structures of KDM2BLRRs (PDB code 5JH5) and BCORPUFD/PCGF1RAWUL heterodimer (PDB code 4HPL) as starting model. Coot was used to complete the model building. Refinement of structure was conducted using Refmac562. Data collection and refinement statistics are shown in Table 1.
Table 1.
Data collection and refinement statistics
| Data collection | |
|---|---|
| Space group | P43212 |
| Cell dimensions | |
| a, b, c (Å) | 67.29, 67.29, 252.04 |
| Resolution (Å) | 126.02-2.20 (2.27-2.20)a |
| Total observations | 421860 (37997) |
| Unique refection | 30637 (2605) |
| Rmerge | 0.114 (1.180) |
| Rpim | 0.045 (0.453) |
| I / σI | 12.0 (2.4) |
| Completeness (%) | 100.0 (99.9) |
| Redundancy | 13.8 (14.6) |
| Refinement | |
| Resolution (Å) | 65.01-2.20 |
| No. reflections | 29020 |
| Rwork / Rfree (%)b | 20.02/23.84 |
| No. atoms | |
| Protein | 3522 |
| Solvent | 70 |
| Average B-factors | |
| Protein | 67.76 |
| Solvent | 59.84 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 1.17 |
| Ramachandran (%) | |
| Most favored | 98.13 |
| Allowed | 1.87 |
| Outlier | 0.00 |
aThe values in parentheses refer to statistics in the highest bin.
bRfactor = ∑( | Fobs| − k|Fcalc | )/∑|Fobs |; Rfree is the R-value for a test set of reflections consisting of a random 5% of the diffraction data not used in refinement.
Cross-linking mass spectrometry (XL-MS)
BCORANK-linker-PUFD/PCGF1RAWUL/ KDM2BF-box-LRRs/SKP1 complex was purified with Superdex 200 (GE) equilibrated against crosslinking buffer (20 mM HEPES pH 7.5, 150 mM NaCl). For cross-linking with bis[sulfosuccinimidyl] suberate (BS3), BCORANK-linker-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 complex (1 mg/mL) was reacted with 20-50 folds molar excess of BS3 for 2 h at 4 °C, and reaction was quenched with Tris-base to final concentration of 50 mM and incubated for 15 min at room temperature. For cross-linking with 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC), BCORANK-linker-PUFD/PCGF1RAWUL/ KDM2BF-box-LRRs/SKP1 complex (1 mg/mL) was reacted with 10-30 folds molar excess of EDC for 1 h at room temperature. Cross-linking reaction was quenched with 50 mM Tris and 20 mM β-me, and incubated for 15 min at room temperature.
Crosslinked complexes were separated by SDS-PAGE, and stained by coomassie blue. Coomassie blue-stained gel bands were cut into small pieces and washed sequentially with water, 50 mM NH4HCO3 in 50% acetonitrile, and 100% acetonitrile. Disulfide bonds of the protein were reduced with 10 mM TCEP in 100 mM NH4HCO3 for 30 min at room temperature and followed by 55 mM iodoacetamide treatment in 100 mM NH4HCO3 for 30 min at room temperature in the dark. Then, the gel pieces were washed sequentially with 100 mM NH4HCO3 and 100% acetonitrile, and dried using a SpeedVac. Samples were then digested with trypsin (12.5 ng/μL) in 50 mM NH4HCO3 for 16 h at 37 °C, and the peptides were extracted twice with 50% acetonitrile/5% formic acid. The sample was evaporated to dryness, and resuspended with 0.1% formic acid, and desalted using MonoSpinTM C18 column, and then dried with SpeedVac.
The crosslinked peptides were then analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS) using an Easy-nLC 1200 nano HPLC, and the temperature of the analytical column was set to 55 °C during the experiment. Data-dependent tandem mass spectrometry (MS/MS) analysis was performed with a Q Exactive Orbitrap mass spectrometer. The crosslinked peptides were identified using pLink2 software (pFind Team, Beijing, China) as described previously63,64. Carbamidomethylation of cysteine was set as fixed modification, and oxidation of methionine was set as variable modification.
Small-Angel X-ray scattering (SAXS) data collection and processing
The small angle X-ray scattering data were obtained at the BL19U2 beamline of National center for Protein Science Shanghai (NCPSS) and Shanghai Synchrotron Radiation Facility (SSRF). SAXS data were collected from 100 μL of BCORANK-linker-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 hetero-tetramer or BCORN1607/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 hetero-tetramer loaded onto Superdex 200 Increase 10/300 GL column equilibrated with 20 mM Tris-HCl pH 7.5, 150 mM NaCl. Scattering data were averaged, scaled and background-corrected using software BioXTAS RAW. ATSAS program suite (version 3.1) was used for further process of SAXS data. GNOM software was used to calculate the pair distances distribution function P(γ) and maximum diameter (Dmax). Ab initio modeling was carried out using the software GASBOR. 40 independent GASBOR models of each complex were averaged using DAMAVER. SAXS parameters were shown in Supplementary table 4.
Model building for BCORN1607/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 hetero-tetramer and BCORANK-linker-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 hetero-tetramer
To build structure model of BCORN1607/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 hetero-tetramer, software FoXSDock65,66 was employed to dock BCORPUFD (1634–1746)/PCGF1RAWUL dimer (isolated from our crystal structure of BCORN1607/PCGF1RAWUL/KDM2BLRRs) onto KDM2BF-box-LRRs/SKP1 (PDB code 5JH5) with SAXS profile of the complex. Top ranked poses were collected for further analysis. Section of N1619-D1625 was generated from crystal structure of BCORN1607 /PCGF1RAWUL/KDM2BLRRs heterotrimer. Then, modeling of the remaining residues E1607-A1618 and Q1626-S1633 from BCOR linker was performed using online software ModLoop67.
For the modeling of BCORANK-linker-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 hetero-tetramer, ANK domain predicted by AlphaFold2 was docked on model structure of BCORN1607 /PCGF1RAWUL/KDM2BLRRs using FoXSDock, with the SAXS profile of the complex and distance constraints based on cross-linking data with EDC cross-linker. For the distance constraints, the distance between Cα atom of E1484 from BCOR-ANK domain (residues 1461–1606) and Cα atom of K121 from SKP1 was set to 0-20 Å, and the distance between Cα atom of D1555 from BCOR-ANK domain and Cα atom of K142 from SKP1 was set to 0-20 Å. The linker of BCOR was further completed with software ModLoop.
Biolayer Interferometry Assay (BLI)
The inhibitory potency of calcium ion against BCORANK-linker-PUFD/PCGF1RAWUL dimer interaction with KDM2BF-box-LRRs/SKP1 dimer was evaluated by the Octet R8 (Sartorius) using the bio-layer interferometry approach. Purified BCORANK-linker-PUFD/PCGF1RAWUL dimer was labeled with biotin and diluted with the buffer (20 mM HEPES pH8.0, 150 mM NaCl, 0.02% Tween 20) to 10 μg/mL and loaded onto a streptavidin biosensor (SAX). KDM2BF-box-LRRs/SKP1 dimer (333 nM) acts as the mobile phase, in the presence or absence of calcium ion.
DIC and fluorescent imaging for LLPS in solution
All the LLPS samples of BCORANK-linker-PUFD/PCGF1RAWUL and mutant BCORANK-linker-PUFD (Poly-D/E3rd to Poly-A)/PCGF1RAWUL were prepared in the buffer containing 20 mM Tris pH 7.5, 150 mM NaCl and 0.5 mM TCEP. For Differential interference contrast (DIC) imaging of LLPS, the samples were loaded onto a glass slide and images were acquired on a Leica DM4B microscope. For fluorescence imaging of BCORANK-linker-PUFD/PCGF1RAWUL, KDM2BF-box-LRRs/SKP1 and mutant KDM2BF-box-LRRs (K1137D/K1138D/R1147D)/SKP1, BCORANK-linker-PUFD was tagged with GFP at N-terminal, and SKP1 was tagged with mCherry at C-terminal. The samples were loaded onto a glass bottom culture dish. Imaging was performed with Zeiss LSM 800 Confocal Laser Scanning Microcopy.
Fluorescence recovery after photobleaching (FRAP) assay
The LLPS sample of BCORANK-linker-PUFD/PCGF1RAWUL (100 μM) induced by 5 mM CaCl2 was papered for FRAP assay. The FRAP assay was performed using the FRAP module of a Leica SP8 X STED Laser Scanning Confocal Microscopy with a 100 x objective (oil immersion). The fluorescent labeled assemblies were bleached using laser beam. Region of interest loop (ROI) was bleached with laser power of 30% and exposure time 30 ms and recovery was imaged at low laser intensity with power of 10%. After photobleaching, images were continuously captured (1 image/5 s). Data from three independent experiments were used for analysis.
Imaging LLPS in live cells
cDNA encoding BCORANK-linker-PUFD (wild type or mutant) and PCGF1RAWUL were cloned into vector pCDNA3.1 with EGFP-tag and mCherry-tag at the C-terminal, respectively. HeLa cells were seeded at 2 × 105 in a 6-well glass bottom culture plate, the plasmid pCDNA3.1-BCORANK-linker-PUFD (wild type or mutant)-EGFP (2 μg) and pCDNA3.1-PCGF1RAWUL-mCherry (2 μg) were co-transfected with PEI after 18 h. Cells were transfected for 36 h before proceeding with subsequent live imaging. Imaging was performed with Zeiss LSM 800 Confocal Laser Scanning Microcopy.
Co-localization assay for BCOR, PCGF1 and KDM2B
cDNA encoding BCOR, PCGF1 and KDM2B were cloned into vector pCDNA3.1 with C-terminal mCherry-tag, ECFP-tag and EGFP-tag, respectively. The plasmids were co-transfected into HeLa cell with Lipo2000. After transfected for 36 h, imaging was performed with confocal microscope (Leica SP8 X STED).
Construct shRNA targeting BCOR
To knockdown endogenous BCOR in HEK293T cells, shRNA (5´ GGCACTTGGTGATATAACTCTCGAGA GTTATATCACCAAGTGCC3´) targeting the 3’-UTR of BCOR was constructed to pLKO.1 vector. shluc (5´ CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG 3´) targeting luciferase was set as negative control. Constitutive expression of shRNA was performed by lentiviral infection of the pLKO.1 vector with puromycin (2.5 μg/ml) selection. Knockdown efficiency was validated by western blot (anti-BCOR, 1:500, Mouse, Sigma-SAB4200843) and RT-qPCR. The primers used in qPCR were: forward 5′ TATGCAGATTCCAGTCAGCTC 3′, and reverse 5′GGGCTGAATTTGCACATCTC 3′.
Subcellular fractionation isolation
Cell pellets were homogenized on ice in Buffer A (10 mM HEPES pH 8.0, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 0.2% NP-40 (v/v), 1 x protease inhibitor cocktail) and nuclei were collected by centrifugation (2500 g for 10 min at 4 °C) and washed with Buffer B (Buffer A without NP-40). The collected supernatant was used as the cytoplasmic fraction and the pellets were the total nuclear. Then, the nuclear pellets were lysed in Buffer C (3 mM EDTA, 2 mM EGTA, inhibitors) for 30 min at 4 °C, and the nuclear soluble fractions were separated from the tight chromatin pellets by centrifugation (14,000 g for 10 min at 4 °C). The tight chromatin pellets were lysed with 4% SDS as chromatin fraction and H3 immunoblotting served as chromatin fraction control.
Gene expression analysis
HEK293T cells were grown on six-well plates (300,000 cells/ well) and cultured to 70%–80% density to conduct gene transfection. The 1 μg plasmid of BCOR (WT or WFY/A mutant) was transfected into the HEK293T cells that endogenous BCOR was knockdown by shBCOR. The empty vector (control) was transfected into sh-luc and sh-BCOR cells. Cells were collected after 48 h and total RNA was extracted with Trizol and chloroform, and quantified by nanodrop. 2.5 μg of total RNA was reverse to generate cDNA. Resultant cDNA was diluted 20-fold for RT-qPCR reaction. Reactions were run on a CFX96 instrument using 2 x RealStar Fast SYBR qPCR Mix (GenStar).Statistical significance was analyzed according to unpaired student’s t-test.
Statistics and reproducibility
Isothermal titration calorimetry experiments and Gene expression analysis were repeated for three times. Co-immunoprecipitation and subcellular fractionation isolation experiments were repeated for two or three times. Statistical analyses were performed using GraphPad Prism 7 software.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank the staff from BL19U1 and BL19U2 beamlines at National Facility for Protein Science in Shanghai (NFPS) and Shanghai Synchrotron Radiation Facility, for assistance during data collection. The work was financially supported by grants from the National Key Research and Development Program (2022YFA1303101, 2017YFA0504104), Guangdong Provincial Key Laboratory of Biocomputing (2016B030301007), and Key Laboratory of Regenerative Biology, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences (KLRB202004)
Author contributions
J.X., J.L., and C.P. designed experiment. R.C., F.S., Y.Z., M.S., Y.D., Y.Y., and C.S. performed biochemical and cellular experiments. J.X., F.S., R.C., and J.L. performed structural studies. J.X., J.L., and R.C. wrote and edited the manuscript.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Christina Karlsson Rosenthal. A peer review file is available.
Data availability
Atomic coordinates and structure factors amplitudes have been deposited into Protein Data Bank with accession code (8HCU). Small-angle neutron scattering (SANS) datasets and models for BCORlinker(N1607)-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 hetero-tetramer and BCORANK-linker-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 hetero-tetramer were deposited to Small-Angle Scattering Biological Data Bank (SASBDB), with entry number SASDS67 and SASDS66, respectively. Replicated ITC data is available in Supplementary Data 1. Replicated WB data is available in Supplementary Data 2. Uncropped western blot images are available in Supplementary Data 3. The source data for the graphs presented in the figures of this paper are available in Supplementary Data 4.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Rui Chen, Feng Shen.
Contributor Information
Jinsong Liu, Email: liu_jinsong@gibh.ac.cn.
Jinxin Xu, Email: xu_jinxin@gibh.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06820-3.
References
- 1.Jaenisch, R. & Bird, A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet.33, 245–254 (2003). 10.1038/ng1089 [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides, T. Chromatin modifications and their function. Cell128, 693–705 (2007). 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- 3.Blackledge, N. P. & Klose, R. J. The molecular principles of gene regulation by polycomb repressive complexes. Nat. Rev. Mol. Cell Biol.22, 815–833 (2021). 10.1038/s41580-021-00398-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaenisch, R. & Young, R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell132, 567–582 (2008). 10.1016/j.cell.2008.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Margueron, R. & Reinberg, D. The polycomb complex PRC2 and its mark in life. Nature469, 343–349 (2011). 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sparmann, A. & van Lohuizen, M. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer6, 846–856 (2006). 10.1038/nrc1991 [DOI] [PubMed] [Google Scholar]
- 7.Schuettengruber, B., Bourbon, H. M., Di Croce, L. & Cavalli, G. Genome regulation by polycomb and trithorax: 70 years and counting. Cell171, 34–57 (2017). 10.1016/j.cell.2017.08.002 [DOI] [PubMed] [Google Scholar]
- 8.Simon, J. A. & Kingston, R. E. Occupying chromatin: polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell49, 808–824 (2013). 10.1016/j.molcel.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbour, H., Daou, S., Hendzel, M. & Affar, E. B. Polycomb group-mediated histone H2A monoubiquitination in epigenome regulation and nuclear processes. Nat. Commun.11, 5947 (2020). 10.1038/s41467-020-19722-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hojfeldt, J. W. et al. Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nat. Struct. Mol. Biol.25, 225–232 (2018). 10.1038/s41594-018-0036-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margueron, R. et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell32, 503–518 (2008). 10.1016/j.molcel.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chittock, E. C., Latwiel, S., Miller, T. C. & Muller, C. W. Molecular architecture of polycomb repressive complexes. Biochem. Soc. Trans.45, 193–205 (2017). 10.1042/BST20160173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao, Z. et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell45, 344–356 (2012). 10.1016/j.molcel.2012.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aranda, S., Mas, G. & Di Croce, L. Regulation of gene transcription by polycomb proteins. Sci. Adv.1, e1500737 (2015). 10.1126/sciadv.1500737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gil, J. & O’Loghlen, A. PRC1 complex diversity: where is it taking us? Trends Cell Biol.24, 632–641 (2014). 10.1016/j.tcb.2014.06.005 [DOI] [PubMed] [Google Scholar]
- 16.Fursova, N. A. et al. Synergy between variant PRC1 complexes defines polycomb-mediated gene repression. Mol. Cell74, 1020–1036 e8 (2019). 10.1016/j.molcel.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taherbhoy, A. M., Huang, O. W. & Cochran, A. G. BMI1-RING1B is an autoinhibited RING E3 ubiquitin ligase. Nat. Commun.6, 7621 (2015). 10.1038/ncomms8621 [DOI] [PubMed] [Google Scholar]
- 18.Zhu, Y. et al. Functional redundancy among polycomb complexes in maintaining the pluripotent state of embryonic stem cells. Stem. Cell Rep.17, 1198–1214 (2022). 10.1016/j.stemcr.2022.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aldera, A. P. & Govender, D. Gene of the month: BCOR. J. Clin. Pathol.73, 314–317 (2020). 10.1136/jclinpath-2020-206513 [DOI] [PubMed] [Google Scholar]
- 20.He, J. et al. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat. Cell Biol.15, 373–384 (2013). 10.1038/ncb2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isshiki, Y. et al. KDM2B in polycomb repressive complex 1.1 functions as a tumor suppressor in the initiation of T-cell leukemogenesis. Blood Adv.3, 2537–2549 (2019). 10.1182/bloodadvances.2018028522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, Z. et al. A non-canonical BCOR-PRC1.1 complex represses differentiation programs in human ESCs. Cell Stem. Cell22, 235–251 e9 (2018). 10.1016/j.stem.2017.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tiacci, E. et al. The corepressors BCOR and BCORL1: two novel players in acute myeloid leukemia. Haematologica97, 3–5 (2012). 10.3324/haematol.2011.057901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Boom, V. et al. Non-canonical PRC1.1 targets active genes independent of H3K27me3 and is essential for leukemogenesis. Cell Rep14, 332–346 (2016). 10.1016/j.celrep.2015.12.034 [DOI] [PubMed] [Google Scholar]
- 25.Lempiainen, J. K. et al. BCOR-coupled H2A monoubiquitination represses a subset of androgen receptor target genes regulating prostate cancer proliferation. Oncogene39, 2391–2407 (2020). 10.1038/s41388-020-1153-3 [DOI] [PubMed] [Google Scholar]
- 26.Deaton, A. M. & Bird, A. CpG islands and the regulation of transcription. Genes Dev.25, 1010–1022 (2011). 10.1101/gad.2037511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farcas, A. M. et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife1, e00205 (2012). 10.7554/eLife.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu, X., Johansen, J. V. & Helin, K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol. Cell49, 1134–1146 (2013). 10.1016/j.molcel.2013.01.016 [DOI] [PubMed] [Google Scholar]
- 29.Xie, W. et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell153, 1134–1148 (2013). 10.1016/j.cell.2013.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saxonov, S., Berg, P. & Brutlag, D. L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl Acad. Sci. USA103, 1412–1417 (2006). 10.1073/pnas.0510310103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaefer, E. J. et al. BCOR and BCORL1 mutations drive epigenetic reprogramming and oncogenic signaling by unlinking PRC1.1 from target genes. Blood Cancer Discov.3, 116–135 (2022). 10.1158/2643-3230.BCD-21-0115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong, S. J. et al. KDM2B recruitment of the polycomb group complex, PRC1.1, requires cooperation between PCGF1 and BCORL1. Structure24, 1795–1801 (2016). 10.1016/j.str.2016.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong, S. J. et al. Structure and role of BCOR PUFD in noncanonical PRC1 assembly and disease. Biochemistry59, 2718–2728 (2020). 10.1021/acs.biochem.0c00285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boulard, M., Edwards, J. R. & Bestor, T. H. FBXL10 protects polycomb-bound genes from hypermethylation. Nat. Genet.47, 479–485 (2015). 10.1038/ng.3272 [DOI] [PubMed] [Google Scholar]
- 35.Sportoletti, P., Sorcini, D. & Falini, B. BCOR gene alterations in hematologic diseases. Blood138, 2455–2468 (2021). 10.1182/blood.2021010958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Miranda, M. C. et al. Epidermal growth factor (EGF) triggers nuclear calcium signaling through the intranuclear phospholipase Cdelta-4 (PLCdelta4). J. Biol. Chem.294, 16650–16662 (2019). 10.1074/jbc.RA118.006961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alonso, M. T. & Garcia-Sancho, J. Nuclear Ca(2+) signalling. Cell Calcium49, 280–289 (2011). 10.1016/j.ceca.2010.11.004 [DOI] [PubMed] [Google Scholar]
- 38.Kunrath-Lima, M. et al. Phospholipase C delta 4 (PLCdelta4) is a nuclear protein involved in cell proliferation and senescence in mesenchymal stromal stem cells. Cell Signal49, 59–67 (2018). 10.1016/j.cellsig.2018.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chun, J.T., Santella, L. & Miller, J.J. Calcium | intracellular calcium waves. In Encyclopedia of Biological Chemistry III (ed. Jez, J.) 669–677 (Elsevier, 2021).
- 40.Vernon, R.M. et al. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife7, e31486 (2018). [DOI] [PMC free article] [PubMed]
- 41.Nakajima-Takagi, Y. et al. Polycomb repressive complex 1.1 coordinates homeostatic and emergency myelopoiesis. eLife12, e83004 (2023). [DOI] [PMC free article] [PubMed]
- 42.Sato, K., Fukami, Y. & Stith, B. J. Signal transduction pathways leading to Ca2+ release in a vertebrate model system: lessons from Xenopus eggs. Semin Cell Dev. Biol.17, 285–292 (2006). 10.1016/j.semcdb.2006.02.008 [DOI] [PubMed] [Google Scholar]
- 43.Monteith, G. R., Prevarskaya, N. & Roberts-Thomson, S. J. The calcium-cancer signalling nexus. Nat. Rev. Cancer17, 367–380 (2017). 10.1038/nrc.2017.18 [DOI] [PubMed] [Google Scholar]
- 44.So, C. L., Saunus, J. M., Roberts-Thomson, S. J. & Monteith, G. R. Calcium signalling and breast cancer. Semin. Cell Dev. Biol.94, 74–83 (2019). 10.1016/j.semcdb.2018.11.001 [DOI] [PubMed] [Google Scholar]
- 45.Izquierdo-Torres, E., Hernandez-Oliveras, A., Fuentes-Garcia, G. & Zarain-Herzberg, A. Calcium signaling and epigenetics: a key point to understand carcinogenesis. Cell Calcium91, 102285 (2020). 10.1016/j.ceca.2020.102285 [DOI] [PubMed] [Google Scholar]
- 46.Shi, X. et al. Ca2+ regulates T-cell receptor activation by modulating the charge property of lipids. Nature493, 111–115 (2013). 10.1038/nature11699 [DOI] [PubMed] [Google Scholar]
- 47.Berridge, M. J., Bootman, M. D. & Roderick, H. L. Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol.4, 517–529 (2003). 10.1038/nrm1155 [DOI] [PubMed] [Google Scholar]
- 48.Snoeck, H. W. Calcium regulation of stem cells. EMBO Rep.21, e50028 (2020). 10.15252/embr.202050028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clapham, D. E. Calcium signaling. Cell131, 1047–1058 (2007). 10.1016/j.cell.2007.11.028 [DOI] [PubMed] [Google Scholar]
- 50.Wang, L. et al. Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol. Cell76, 646–659 e6 (2019). 10.1016/j.molcel.2019.08.019 [DOI] [PubMed] [Google Scholar]
- 51.Sanulli, S. et al. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature575, 390–394 (2019). 10.1038/s41586-019-1669-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, H. et al. Liquid-liquid phase separation in biology: mechanisms, physiological functions and human diseases. Sci. China Life Sci.63, 953–985 (2020). 10.1007/s11427-020-1702-x [DOI] [PubMed] [Google Scholar]
- 53.Pirrotta, V. & Li, H. B. A view of nuclear polycomb bodies. Curr. Opin. Genet. Dev.22, 101–109 (2012). 10.1016/j.gde.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eeftens, J. M., Kapoor, M. & Brangwynne, C. P. Epigenetic memory as a time integral over prior history of polycomb phase separation. bioRvix10.1101/2020.08.19.254706 (2020).
- 55.Guo, Y., Zhao, S. & Wang, G. G. Polycomb gene silencing mechanisms: PRC2 chromatin targeting, H3K27me3 ‘readout’, and phase separation-based compaction. Trends Genet.37, 547–565 (2021). 10.1016/j.tig.2020.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plys, A. J. et al. Phase separation of polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev.33, 799–813 (2019). 10.1101/gad.326488.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fox, C. H. et al. A method for measuring intracellular free magnesium concentration in platelets using flow cytometry. Magnes Res.20, 200–207 (2007). [PubMed] [Google Scholar]
- 58.Maat, H. et al. The USP7-TRIM27 axis mediates non-canonical PRC1.1 function and is a druggable target in leukemia. iScience24, 102435 (2021). 10.1016/j.isci.2021.102435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, W.-Z. et al. The protein complex crystallography beamline (BL19U1) at the shanghai synchrotron radiation facility. Nuclear Sci. Tech.30, 170 (2019). 10.1007/s41365-019-0683-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Evans, P. R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta. Crystallogr. D Biol. Crystallogr.67, 282–292 (2011). 10.1107/S090744491003982X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lebedev, A. A., Vagin, A. A. & Murshudov, G. N. Model preparation in MOLREP and examples of model improvement using X-ray data. Acta. Crystallogr. D Biol. Crystallogr.64, 33–39 (2008). 10.1107/S0907444907049839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kovalevskiy, O., Nicholls, R. A., Long, F., Carlon, A. & Murshudov, G. N. Overview of refinement procedures within REFMAC5: utilizing data from different sources. Acta. Crystallogr. D Struct. Biol.74, 215–227 (2018). 10.1107/S2059798318000979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang, B. et al. Identification of cross-linked peptides from complex samples. Nat. Methods9, 904–906 (2012). 10.1038/nmeth.2099 [DOI] [PubMed] [Google Scholar]
- 64.Lu, S. et al. Mapping native disulfide bonds at a proteome scale. Nat. Methods12, 329–331 (2015). 10.1038/nmeth.3283 [DOI] [PubMed] [Google Scholar]
- 65.Schneidman-Duhovny, D., Hammel, M. & Sali, A. Macromolecular docking restrained by a small angle X-ray scattering profile. J. Struct. Biol.173, 461–471 (2011). 10.1016/j.jsb.2010.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schneidman-Duhovny, D., Hammel, M., Tainer, J. A. & Sali, A. FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res.44, W424–W429 (2016). 10.1093/nar/gkw389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fiser, A., Do, R. K. & Sali, A. Modeling of loops in protein structures. Protein Sci.9, 1753–1773 (2000). 10.1110/ps.9.9.1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Atomic coordinates and structure factors amplitudes have been deposited into Protein Data Bank with accession code (8HCU). Small-angle neutron scattering (SANS) datasets and models for BCORlinker(N1607)-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 hetero-tetramer and BCORANK-linker-PUFD/PCGF1RAWUL/KDM2BF-box-LRRs/SKP1 hetero-tetramer were deposited to Small-Angle Scattering Biological Data Bank (SASBDB), with entry number SASDS67 and SASDS66, respectively. Replicated ITC data is available in Supplementary Data 1. Replicated WB data is available in Supplementary Data 2. Uncropped western blot images are available in Supplementary Data 3. The source data for the graphs presented in the figures of this paper are available in Supplementary Data 4.