Abstract
In the era of renewed space exploration, comprehending the effects of the space environment on human health, particularly for deep space missions, is crucial. While extensive research exists on the impacts of spaceflight, there is a gap regarding female reproductive risks. We hypothesize that space stressors could have enduring effects on female health, potentially increasing risks for future pregnancies upon return to Earth, particularly related to small-for-gestational-age (SGA) fetuses. To address this, we identify a shared microRNA (miRNA) signature between SGA and the space environment, conserved across humans and mice. These miRNAs target genes and pathways relevant to diseases and development. Employing a machine learning approach, we identify potential FDA-approved drugs to mitigate these risks, including estrogen and progesterone receptor antagonists, vitamin D receptor antagonists, and DNA polymerase inhibitors. This study underscores potential pregnancy-related health risks for female astronauts and proposes pharmaceutical interventions to counteract the impact of space travel on female health.
Subject terms: Systems biology, Systems analysis, Reproductive disorders
A circulating miRNA signature linked to birth defect risk, (i.e. as small-for-gestational-age (SGA) fetuses), was identified for females exposed to the space environment. AI/ML tools were used to predict potential countermeasures to mitigate this risk. @AfshinBeheshti @PittTweet
Introduction
We are amidst a new space age marked by a surge in missions from governmental and commercial space agencies. These missions encompass deep space explorations, including Moon missions like NASA’s Artemis campaign, with ambitions to reach Mars ultimately1. The rapid expansion in space endeavors necessitates a profound understanding of how the inhospiSupplementary Table pace environment affects human health and development. In addition, it is crucial to develop pharmacological countermeasures to mitigate human physiological alterations.
The space environment significantly contrasts with our terrestrial habitat and introduces primary hazards that elevate health risks, with space ionizing radiation and microgravity being the most deleterious2. Previously, we elucidated spaceflight-associated health risks in human biology, including six key physiological aspects: oxidative stress, DNA damage, mitochondrial dysregulation, epigenetic and gene regulation alterations, telomere-length dynamics, and shifts in the microbiome2. Certain altered health risks may recede to preflight levels depending on the duration spent in space. However, evidence indicates that the health risks associated with extended deep space missions will exert a lasting impact on human well-being.
Most prior spaceflight health studies are biased toward male participants. Women comprised just 15% of NASA’s astronaut corps until a decade ago. Nowadays, the representation of women in astronaut training has increased to nearly 50%3. Emerging studies are beginning to shed light on sex-specific health risks associated with spaceflight, supplementing the traditional sex-independent spaceflight research4. Yet, our understanding of how space travel can affect women’s reproductive systems remains limited.
The typical physiological responses to spaceflight can potentially trigger early menopause, reduce fertility, and even contribute to the development of conditions like endometriosis3. Recent research also indicates that spaceflight-induced changes in estrogen and insulin levels, particularly in women, can adversely impact the female reproductive system5. Notably, the National Academies Decadal Survey on Biological and Physical Sciences Research in Space for 2023–2032 has highlighted the research gap concerning female health risks in space and underscores the need for further investigation. Specifically, it emphasizes the importance of comprehending the space environment’s effects on the female reproductive system6. Lastly, this research is timely since NASA’s Space Biology program launched the Rodent Research-20 mission in November of 2023 to the International Space Station to investigate ovarian function and whether space-flown female mice have temporary or permanent alterations to their reproductive capabilities.
A pivotal question surrounding the impact of space travel on the female reproductive system is whether prolonged exposure to the space environment might elevate the risk of birth defects or have any adverse impact on future pregnancies. One concern revolves around small-for-gestational-age (SGA) births, where infants have birth weights below the 10th percentile for their gestational age7. On average, 16% of infants are SGA, ranging from 7% in developed countries to over 40% in developing countries8. While nearly 90% of SGA infants exhibit catch-up growth within two years, the remaining 10% do not9. SGA infants need to be monitored carefully till adulthood9. These infants face various health risks, including hypertension, cardiovascular disease, and type 2 diabetes mellitus10,11, which can be exacerbated by rapid postnatal weight gain12. The risk of SGA births is influenced by various factors, including maternal lifestyle choices (e.g., smoking), placental insufficiency (e.g., in pre-eclampsia), gestational weight gain, twin pregnancies, paternal factors (e.g., SGA birth, height), and environmental elements (e.g., pollution)13. In this work, we hypothesize that the risk of SGA development increases after maternal exposure to space stressors, and we further hypothesize that microRNAs (miRNAs) provide a relevant biosignature for diagnostics and monitoring.
The rising interest in miRNAs as prognostic and diagnostic biomarkers stems from their non-invasive sample collection methods. miRNAs, small non-coding RNAs typically spanning 19-22 nucleotides, exert post-transcriptional and translational regulation over hundreds to thousands of mRNAs7. These versatile molecules govern diverse biological processes such as cell proliferation, differentiation14, inflammation15, mitochondrial function16, and apoptosis17. Furthermore, miRNAs display high conservation across species, including humans and mice18. Aberrant miRNA expression has been linked to complex pregnancies19,20, maternal smoking21, and fetal growth restriction22, indicating their potential role in placental-related diseases. Here, we seek to identify an SGA-associated miRNA signature, serving as a potential biomarker and target for future countermeasures. Given this signature, machine learning techniques were applied to identify FDA-approved drugs that could potentially be repurposed to address elevated miRNA levels.
Our study utilized a two-step approach to explore the miRNA signature associated with SGA and its potential links to spaceflight. Initially, we leveraged data from the ImmPort database (www.immport.org), a resource offered by the National Institute of Allergy and Infectious Diseases (NIAID) at the National Institutes of Health (NIH), to establish a miRNA signature for SGA. This database, focusing primarily on immunology data, includes diverse experimental results such as PCR, ELISA, and flow cytometry. We examined miRNA profiles in maternal plasma from SGA deliveries7.
Subsequently, we turned our attention to space biology data available through NASA’s Open Science Data Repository (OSDR) and GeneLab platforms (https://www.nasa.gov/osdr/). These resources offer a rich collection of omics datasets from space biology experiments, including miRNA-sequence data from simulated spaceflight conditions23. By comparing the SGA-associated miRNA signature from ImmPort with the spaceflight data from GeneLab/OSDR, particularly focusing on female C57BL/6 mice, we identified 13 shared miRNAs. This overlap suggests a possible impact of space conditions on similar biological pathways involved in SGA.
Results
miRNA response linked to both SGA and spaceflight
We started by analyzing datasets from NIAID’s ImmPort and NASA’s OSDR/GeneLab platforms to understand whether there are datasets that could help us elucidate a miRNA signature for SGA-associated risks during spaceflight. Our focus was to identify datasets that shed light on how clinically relevant miRNAs associated with SGA are affected in the circulation of females during spaceflight. We used the dataset SDY1871 from ImmPort, concentrating on miRNA expression in human maternal plasma obtained from healthy pregnancies and those with SGA births7.
We considered three distinct gestational time points: (time-point A), (time-point B), and (time-point C) weeks7. A gestation time-dependent analysis unveiled differences in miRNA expression profiles between SGA and control patient samples (Supplementary Fig. 1 and Fig. 1a, e). However, a time-independent analysis revealed a separation between miRNA samples from SGA samples vs. controls, both in terms of global miRNA profiles and significantly regulated miRNAs (with a p-value < 0.05; see Methods) (Fig. 1b, f and Supplementary Fig. 1). Hereafter, we will focus on a time-independent analysis to investigate the miRNAs present in circulation before and during gestation.
Fig. 1. The global response of miRNA expression comparing SGA patients to controls and spaceflight samples to controls and the core pathways being influenced.
Principal Component Analysis (PCA) plots demonstrate the clustering of the samples for each group. a Comparative miRNA expression analysis between Small for Gestational Age (SGA) patients (triangles) and control (healthy) patients (circles) at three distinct time-points: A ( weeks) (blue), B ( weeks) (yellow), and C ( weeks) (green). Individual timepoint PCA plots can be found in Supplementary Fig. 1. For all SGA analysis there were a total of N = 29 patients included in the study with N = 16 normal birth outcomes and N = 13 SGA births for each time point. b miRNA expression PCA plots comparing SGA and control patients, irrespective of time. c miRNA expression clustering of OSD-336 mouse plasma under various treatments: 0.5 Gy Galactic Cosmic Radiation (GCR, red), 1 Gy Solar Particle Event (SPE, yellow), 5 Gy Gamma radiation (blue), and Sham (black); in both microgravity (circles) and normal gravity (triangles). Individual radiation-type PCA plots can be found in Supplementary Fig. 2. d miRNA expression analysis of OSD-55 human peripheral blood leukocytes (PBLs) under normal Earth gravity (circles) and microgravity (triangles). e miRNA expression analysis comparing SGA patients and control patients at three different time-points: A, B, and C, focusing exclusively on significant genes (p < 0.05). f Time-independent analysis of significant miRNAs differences (p < 0.05) between SGA and control patients. g Significant miRNA expression analysis (p < 0.05) of OSD-336 mouse plasma under different simulated radiation and microgravity conditions. h Significant miRNA expression analysis (p < 0.05) of OSD-55 human PBLs under microgravity and normal gravity conditions. Gene set analysis of miRNAs on common (i) Hallmark pathways and (j) MitoPathways in SGA and simulated spaceflight pathways compared to control, using the Molecular Signatures Database (MSigDB) Hallmark pathways to assess miRNA expressions impact on these pathways and functions. SGA human miRNA regulation is compared to control both time-independently and at different timepoints (left). Space radiation miRNAs with and without simulated microgravity are compared to Sham (right). The x-axis represents a coefficient term indicating pathway inhibition (negative value) or activation (positive value). The point size indicates the degree of significance, denoted by False Discovery Rate (FDR). Only significant values (FDR < 0.25) are displayed. The full list of significantly regulated pathways can be found in Supplementary Figs. 3 and 4.
To investigate miRNA profile changes during spaceflight in females, we employed two datasets from NASA’s OSDR/GeneLab platform: OSD-55 and OSD-336. OSD-336 consists of miRNA sequencing from plasma samples from female C57BL/6 mice subjected to simulated spaceflight experiments24 (Fig. 1c, g and Supplementary Fig. 2). To simulate microgravity, the mice underwent hindlimb unloading (HU), a well-established murine model for microgravity that has previously yielded similar biological responses to actual spaceflight25. For simulating space radiation, the mice were exposed to two types of radiation: 1) an acute, simplified seven-ion Galactic Cosmic Radiation (GCR) beam26 at 0.5 Gy, equivalent to the estimated dose an astronaut would receive during a yearlong round-trip mission to Mars, and 2) a simulated Solar Particle Event (SPE) radiation at 1 Gy, consisting of protons with energies ranging from 50 MeV to 150 MeV. Another group of mice was exposed to 5 Gy of gamma radiation for comparison with terrestrial radiation. Although not visible, when all samples are considered (Fig. 1c), for significantly regulated genes, a distinction emerges between the GCR-irradiated samples and the sham (unirradiated control group) (Fig. 1g). When analyzing the individual irradiation groups, it becomes evident that, for significantly regulated circulating miRNAs (i.e., p < 0.05), the samples cluster separately between the GCR and sham groups (Supplementary Fig. 2d). In contrast, both SPE and, to a greater extent, gamma-irradiated samples do not clearly separate the sham and irradiated groups (Supplementary Fig. 2b, c, e, f). This suggests that miRNA expression is more likely to be affected in samples exposed to GCR irradiation.
Lastly, to assess the influence of microgravity on the miRNA signature, we conducted miRNA expression analysis on OSD-55, an in vitro experiment performed on human peripheral blood lymphocytes (PBLs) under simulated microgravity conditions using a rotating wall vessel (RWV) bioreactor27. While these miRNAs are not derived from plasma like the SGA data and OSD-336, their inclusion is merited since they also circulate in the blood. Clear differentiation is observed between the PBLs exposed to modeled microgravity (MMG) and the 1 g control group (Fig. 1d, h). As demonstrated in the original publication by Girardi et al., 42 miRNAs in MMG-incubated PBLs were dysregulated compared to 1g-incubated PBLs, with 25 up-regulated and 17 down-regulated. These miRNAs predominantly play roles in crucial biological processes, including cell proliferation, apoptosis, signal transduction, and immune/inflammatory response27.
To understand the global miRNA response in each condition, we analyzed the core pathways influenced by miRNAs in both the spaceflight and SGA experimental groups. We specifically employed the curated Molecular Signatures Database (MSigDB) Hallmark pathways28 genesets to assess how miRNA expression impacts essential pathways and functions. Our pathway analysis unveiled commonalities and distinctions in the miRNA effects between SGA and spaceflight conditions (Fig. 1i and Supplementary Figs. 3 and 5). Notably, miRNAs overexpressed in SGA patient plasma displayed a down-regulation of adipogenesis, an effect unrelated to the space environment. In contrast, microgravity and space radiation had distinct impacts, including an up-regulation of coagulation and a down-regulation of pancreas beta cells, neither of which were observed in SGA patients (Supplementary Figs. 3 and 5). For the shared pathways between SGA and spaceflight responses (Fig. 1i and Supplementary Fig. 5), several anticipated spaceflight-related pathways overlapped with SGA patients, such as the up-regulation of DNA repair pathways29, downregulation of cell cycle pathways30, and suppression of immune response-related factors31.
As we have previously published, mitochondrial dysregulation is a prominent biological consequence of spaceflight32. To ascertain whether the mitochondrial damage observed during spaceflight is a universally shared response between SGA and spaceflight, we focused on the global miRNA response and its specific impact on mitochondrial pathways. Our analysis employed the hand-curated MitoCarta pathway database33. We uncovered distinct pathways for SGA patients and those exclusive to the space environment (Fig. 1j and Supplementary Figs. 4 and 6). Notably, samples experiencing spaceflight conditions exhibited an up-regulation of complex V, coenzyme Q metabolism, and mtDNA maintenance (Supplementary Figs. 4 and 6). In contrast, SGA patients displayed relatively less mitochondrial pathway damage, alongside an upregulation of vitamin B1 metabolism and the TIM22 carrier pathways, which were not observed in the simulated spaceflight data. Key shared pathways between SGA and simulated spaceflight included increased oxidative phosphorylation (OXPHOS) complex I, lipid metabolism, mitochondrial ribosome functions, and mitochondrial translation (Fig. 1j and Supplementary Fig. 6). Complex I plays a pivotal role in the cellular energy production process, serving as the initial entry point for electrons in the respiratory complex chain34. Dysfunction in complex I has been associated with an elevated risk of many diseases34, including cardiac35, cognitive and central nervous system36, and developmental defects37. Dysfunction in complex I has also been associated with the release of reactive oxygen species (ROS) and increased lipid and fatty acid metabolism34,38,39, which we observe as common elements in the miRNA response in both SGA and spaceflight conditions (Fig. 1i, j and Supplementary Figs. 5 and 6). Within these shared mitochondrial and general pathways, the space environment exhibits a more pronounced miRNA dysregulation than SGA, involving upregulation or downregulation (Fig. 1i, j and Supplementary Figs. 5 and 6). This striking contrast underscores significant concerns about the potential repercussions of these dysregulations for female astronauts upon their return to Earth’s environment and their potential role in elevating the risk of SGA associated with spaceflight if these signals remain elevated for a prolonged period of time.
Identifying key miRNAs responsible for spaceflight-associated SGA risk
We aim to pinpoint miRNAs displaying consistent changes in both SGA and spaceflight to serve as potential biomarkers and molecular targets for pharmacological countermeasure development to mitigate the spaceflight-induced damage. The presence of shared miRNAs between space and SGA raises concerns that female astronauts may have a higher risk of potential pregnancy-related health risks after spaceflight missions if the observed miRNA dysregulation continues after returning to Earth.
To assess the correlation between the space environment and SGA, we compared miRNA expression in SGA patients and mice exposed to simulated space conditions. While there were 34 shared miRNAs involved in both SGA and space (Fig. 2a), we chose to focus exclusively on miRNAs exhibiting a consistent trend in both datasets: either up-regulated (depicted in red) or down-regulated (depicted in blue). Our analysis revealed 13 common miRNAs with a consistent trend (Fig. 2b), all up-regulated, except one miRNA, miR-146b-5p, displayed down-regulation. Furthermore, we note a significant (p < 0.05) positive correlation in miRNA expression when comparing SGA vs control samples and spaceflight vs terrestrial conditions for the 13 common miRNAs out of which two shared the same seed sequence AGCACCA (miR-29b-3p, miR-29c-3p) (Fig. 2c). The upregulation of these miRNAs has been linked to various diseases, including osteoarthritis (miR-24-3p40), atherosclerosis (miR-148a-3p41), cardiac diseases (miR-24-3p42, miR-29b-3p43), neonatal encephalopathy (let-7b-5p44) and also spontaneous abortion (miR-146b-5p45). Notably, a significant portion of these miRNAs are implicated in cancer. It is common in miRNA research that many of these miRNAs were initially discovered within the context of cancer46. Although individual miRNAs can play roles in various diseases, including cancer, our working hypothesis, supported by previous research, suggests that the collective impact of a group of miRNAs, known as a miRNA signature, is often associated with specific diseases47–49. In the case of these specific 13 miRNAs, we hypothesized that this miRNA signature could potentially contribute to birth defects and/or SGA if the mother has previously been exposed to the space environment50.
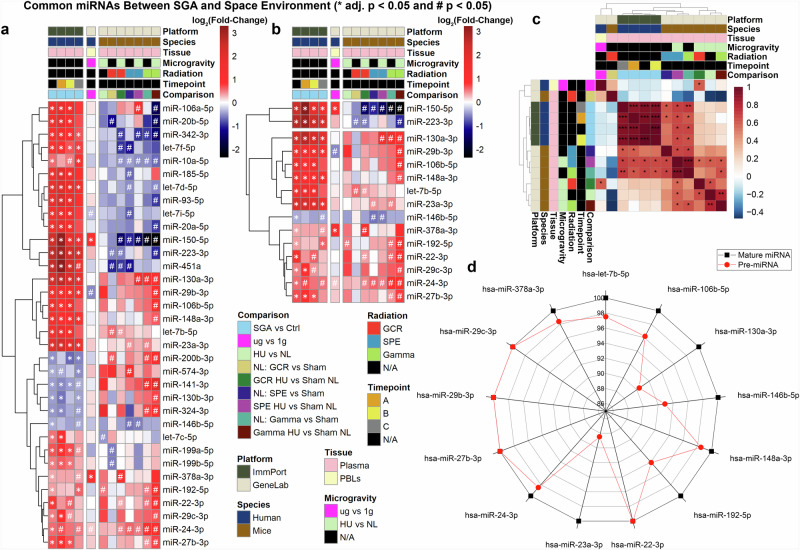
Fig. 2. Common miRNAs in SGA and space environment.
a Heatmap depicting miRNAs significantly regulated in SGA vs. controls (adj. p < 0.05) present in simulated spaceflight datasets. b Heatmap displaying miRNAs consistently regulated in the same direction between SGA vs controls and PBL samples or mouse plasma. Analysis focuses on 13 common miRNAs from the plasma comparison. For the heatmaps, log2(Fold-Change) is color-coded, with red shades indicating upregulated genes and blue shades indicating downregulated genes. Significance is denoted by * adj. p-value < 0.05 and # p-value < 0.05. c Correlation plot of log2(Fold-Change) values for the 13 miRNAs, indicating positive correlation in red shades and negative correlation in blue shades. d Radar plot illustrating homology of pre-miRNAs (red) and mature miRNAs (black) in humans and mice for the 13 common miRNAs in SGA and space.
Our research seeks to ascertain the potential risk of birth defects in female astronauts upon their return to Earth. These 13 common miRNAs were identified through a comparative analysis of miRNAs in both mice and humans. It is widely acknowledged in the literature that miRNAs exhibit a high degree of conservation across species, including mice and humans51. Our objective is to evaluate the extent to which these 13 miRNAs are conserved between humans and mice, providing evidence that our findings in murine models are likely to translate well to humans. To begin, we compared precursor-miRNAs (pre-miRNAs) between the two species. Pre-miRNAs represent the hairpin precursors of miRNAs52, which are cleaved to generate mature miRNAs. Although pre-miRNA sequences are not entirely conserved, mature miRNAs often retain most of their sequence and target conservation across different species51. Surprisingly, we discovered for the 13 miRNAs that pre-miRNA sequences were highly conserved, with a minimum of 88% homology between humans and mice (Fig. 2d). Notably, the 13 mature miRNAs exhibited 100% homology between the two species. This level of conservation between miRNAs in humans and mice suggests that they share conserved biological functions53. Moreover, it allows us to assess and determine similar behavior between the two models more confidently by comparing human miRNA data from SGA to murine miRNA spaceflight data.
The diseases, biological functions, genes, and pathways affected by the miRNA signature associated with spaceflight-induced SGA
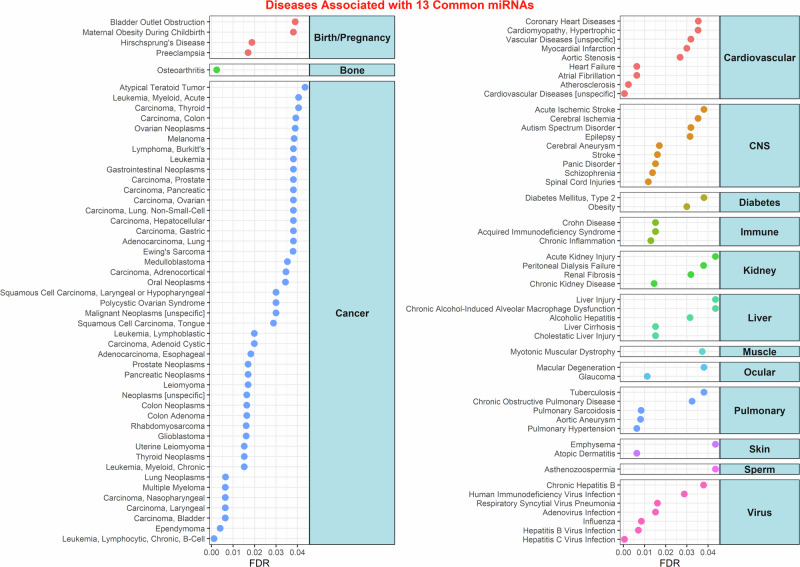
The 13 common miRNAs identified in our study target various genes that can contribute to the development of disease pathways (identified through miRNet predictions as described in the Methods) (Fig. 3). In addition to the well-known diseases associated with birth defects and pregnancies, such as bladder outlet obstruction and preeclampsia, several other pathways exhibit correlations with SGA. These pathways may lead to conditions such as bone diseases (osteoarthritis), cardiovascular diseases (atherosclerosis, heart failure), diabetes, liver injury, and pulmonary diseases in children born with SGA (Fig. 3). A significant portion of the diseases associated with these 13 miRNAs are related to cancer. Notably, Östling et al. identified similarities between the expression of placental miRNAs in SGA and cancer, particularly in terms of invasiveness and rapid growth50. Although the exact explanation for this correlation between cancer-related miRNAs and placental miRNAs in SGA remains elusive, it is worth noting that space radiation has been shown to impact the human body, often leading to cancer development2. It is important to acknowledge that the abundance of literature on these miRNAs in cancer may bias the pathway results, potentially leading to an overabundance of associations with cancer.
Fig. 3. Diseases linked to common SGA and space miRNAs.
Predicted diseases associated with the 13 miRNAs identified through miRNet. Curated into main groups, only diseases with FDR < 0.05 are displayed on the x-axis.
The 13 common miRNAs play a pivotal role in various developmental biological functions that can have long-lasting effects on children’s development. Jeong et al.54 demonstrated from miRNAs of SGA children compared to appropriate for gestational age children that 22 miRNAs were found differentially expressed in the two groups. It is interesting to note that two of the 13 miRNAs from our analysis, hsa-miR-29b-3p and hsa-miR-29c-3p, are upregulated in SGA children. In particular, the authors suggest that hsa-miR-29c-3p is responsible for the recovery in growth in SGA children.
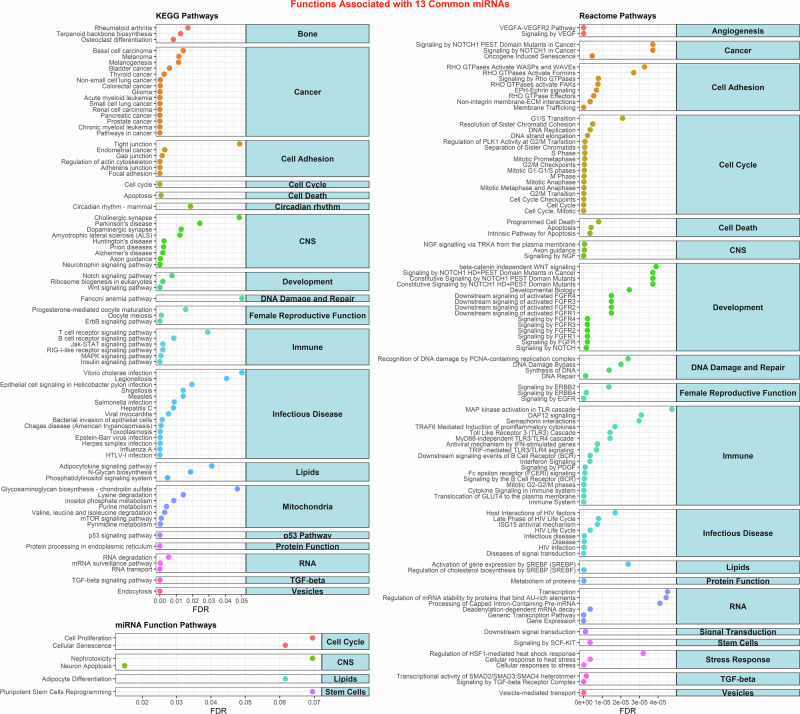
These 13 miRNAs target specific functions related to organs associated with defects in SGA infants (Fig. 4). These miRNAs influence bone regeneration and cytoskeleton remodeling processes, potentially leading to conditions like osteoarthritis. Moreover, they affect angiogenesis55, contributing to the development of cardiac diseases. There is a strong correlation between these miRNAs and processes involving cell cycle and apoptosis, which can result in SGA babies. Notably, these miRNAs also target DNA damage and repair, a well-documented effect of space radiation2. Furthermore, they influence genes related to immune system regulation and female reproductive functions, aligning with our earlier analysis (Fig. 1). As anticipated, this miRNA signature is involved with mitochondrial dysfunction through mTOR signaling, pyrimidine and purine metabolism, and other metabolic pathways. The impact on mitochondrial function can lead to impaired energy production, potentially resulting in deficiencies during pregnancy and birth that affect the child’s development56. To determine if these pathways and functions are impacted in females long-term after exposure to the space environment, we included RNA-seq analysis of blood samples from male and female C57BL/6J mice exposed to 50 cGy GCR simulated irradiation57. The blood samples were collected 14 days post-irradiation to assess any long-term biological impact. Although this data is not from human subjects, it demonstrates the long-term impact of space radiation exposure on specific miRNA-related pathways. Our results show that the majority of the pathways related to the 13 miRNAs (Fig. 4) are dysregulated in the female mice, with some pathways similarly affected in both sexes and a few unique to males when compared to the non-irradiated control mice (Supplementary Fig. 7 and Supplementary Data 1). Interestingly, pathways related to the immune system and mitochondria are suppressed long-term exclusively in female mice, raising further concerns about the health risks associated with these 13 miRNAs.
Fig. 4. Biological functions linked to common SGA and space miRNAs.
Predicted KEGG, Reactome, and miRNA functions associated with the 13 miRNAs identified through miRNet. Curated into main groups, only pathways with FDR < 0.05 are displayed on the x-axis.
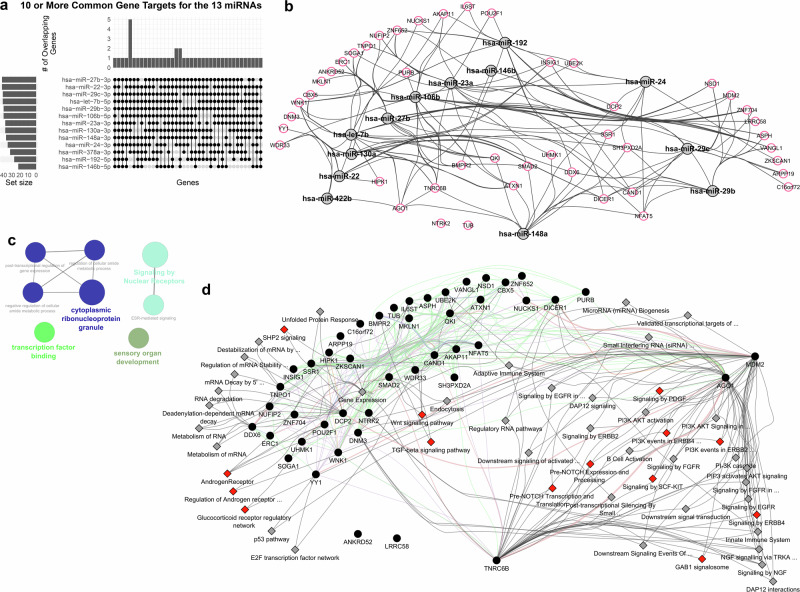
We identified key gene targets associated with the miRNA signature, to uncover central hubs in the genetic network affected by these miRNAs and provide insights into the biological alterations linked to the increased risk of SGA during spaceflight for females. Of note, when miRNAs target genes, they bind to the mRNA, effectively silencing them47. Through our established methods48, we identified 9,387 gene targets for all 13 miRNAs (Supplementary Data 2). We focused on gene targets shared by 10 or more of the 13 miRNAs (Fig. 5a), which we consider central hubs in the key pathways influenced by the miRNA signature. This approach led us to 45 genes, with NSD1 being the sole gene targeted by all 13 miRNAs (Supplementary Data 2). NSD1 has been associated with Sotos syndrome, a genetic disorder that can be prenatally diagnosed and is linked to abnormal growth in children58. Some miRNAs appear to have more gene targets than others, allowing the relative importance of the 13 miRNAs to be gauged (Fig. 5b and Supplementary Data 2). When utilizing a single miRNA-gene target database, some gene-miRNA interactions might not appear (see Fig. 5b as an example). In examining the overarching pathways affected by these 45 gene targets, we find connections to development and estrogen-related signaling (Fig. 5c). Unbiased analysis of these 45 gene targets using Ingenuity Pathway Analysis (IPA) tool identified that estrogen signaling receptor (ESR)-mediated signaling was the third most significant canonical pathway (p value = 6.78E-05) (Supplementary Fig. 8a). Four candidate genes identified to be participating were AGO1, POU2F1, TNRC6B, and YY1. All of these genes were found to be associated with reproductive system diseases including female genital tract adenocarcinoma, endometrioid endometrial adenocarcinoma and adenosquamous ovarian carcinoma (IPA, p value range 1.43E-3 to 2.6 E-2) (Supplementary Fig. 8b). Some of the key Subsequent analysis using upstream analysis tool of IPA predicted 10 genes in different cellular compartment to be regulated by ESR1 (Supplementary Fig. 8c). Additionally, the top 50 pathways linked to these gene targets encompass several pathways related to the regulation of female-specific functions and disorders (see red nodes in Fig. 5d). Assuming that the miRNAs inhibit these 45 genes, we can anticipate potential downregulation in many pathways. This includes the suppression of androgen signaling pathways, which are crucial for maintaining the female reproductive system59. Additionally, various immune-related pathways are targeted, covering both adaptive and innate immunity. PI3K-AKT pathways, vital for metabolism, proliferation, and cell survival, are also heavily influenced by these miRNAs60. These pathways are essential in the female reproductive system, especially in embryo development and female germ cells61. To further determine the long-term impact of these pathways after exposure to the space environment, we analyzed the mice blood data collected 14 days post-exposure to 50 cGy GCR irradiation (described above). The pathway analysis revealed that certain pathways were suppressed exclusively in female mice over the long term, including immune-related pathways associated with both the adaptive and innate immune systems and RNA metabolism (Supplementary Fig. 9). Interestingly, signal transduction pathways were upregulated only in male mice, indicating that these pathways might not be as critical in females for contributing to the long-term risk associated with SGA.
Fig. 5. Gene targets of the SGA-associated spaceflight miRNA signature.
a Upset plot depicting the 45 gene targets shared by 10 or more of the 13 miRNAs. b Network illustrating the connections between the 13 miRNAs (gray nodes) and the 45 gene targets (red highlighted nodes). The gene target interactions with the miRNAs were determined by CluePedia. c Network displaying the global pathways associated with the 45 genes, predicted by CluePedia and visualized in Cytoscape. d Top 50 pathways regulated by the 45 genes, determined by GeneMANIA, with black nodes representing genes and gray diamonds indicating pathways. The red diamond nodes indicate the pathways that are related to female-specific functions and disorders. The full list of gene targets for the miRNAs can be found in Supplementary Data 2.
Since there is a lack of data and experiments available for tissues from female reproductive organs from mice exposed to the space environment, our investigation extended to the assessment of the impact of spaceflight on 13 miRNAs and 45 genes across various tissues in mice sent to the International Space Station (ISS) (Supplementary Fig. 10). While we theorize that the alterations in these miRNAs are likely specific to tissues and organs related to SGA, there is a possibility of broader effects on other organs owing to the regulatory influence of these miRNAs in circulation. Examination of the 13 miRNAs in other tissues from mice subjected to simulated spaceflight experiments (Supplementary Fig. 10a) revealed a notable upregulation in key miRNAs, with the liver exhibiting the highest, followed by the heart, with the soleus muscle showing the least upregulated miRNAs (Supplementary Fig. 10c). It has previously been shown that issues with the liver, specifically nonalcoholic fatty liver disease (NAFLD), can lead to a higher risk of SGA and other preterm issues during pregnancy62. We have also previously shown that spaceflight will lead to lipid accumulation in the liver and also an increased risk of NAFLD63. From this data, we hypothesize that the identified circulating miRNAs may exert an impact on the liver, serving as an additional potential marker for the risk of spaceflight-associated SGA. For the top 45 gene targets of the miRNAs, we have further analyzed multiple tissues from mice flown for different durations to the ISS (Supplementary Fig. 10d). This analysis highlighted the potential for miRNA interaction in specific tissues, resulting in the inhibition of the top 45 genes. The kidney, spleen, thymus, and liver emerged as the most affected organs, exhibiting inhibition of these genes. While this does not conclusively confirm the upregulation of the miRNAs, it does propose a hypothesis that warrants further quantification of these miRNAs in the respective tissues from mice subjected to spaceflight. We also utilized the NASA Twin Study miRNA-sequence data47,64 to determine if these miRNAs are present in humans during spaceflight (Supplementary Fig. 10b). The study involved male twins, with one twin spending 340 days on the ISS while the other remained on Earth. Blood samples from both were analyzed using miRNA sequencing across different sorted cell populations, in addition to other assays. As this is the only available miRNA-seq data from humans in space, we aimed to see if these miRNAs are expressed during spaceflight regardless of sex. Our analysis showed that 7 out of 13 miRNAs were overexpressed during spaceflight in one of the cell types. Upon return to Earth the miRNAs start to decrease back to control levels, but fourf of the miRNAs still remain elevated above control levels after flight (Supplementary Fig. 10b). Although this data is from a male astronaut, it indicates that these miRNAs might be overexpressed during spaceflight in a sex-independent manner, potentially posing long-term risks if upregulated in females. This highlights the need for more sex-dependent studies on miRNA changes in astronauts and additional confirmation from further human studies. Overall, the gene targets of these miRNAs may pose a risk of increased birth defects if inhibited over time, as indicated by our findings related to spaceflight.
As the NASA Twin Study suggests, there is potential for these miRNAs to remain overexpressed for a period after returning to Earth. Although there is no evidence indicating whether these miRNAs will remain dysregulated in females or in a larger cohort of astronauts, potentially increasing the risk of SGA, we observed a long-lasting impact on the dysregulation of these 13 miRNAs due to factors such as the environment, diet, or exercise (Supplementary Table 1). A comprehensive literature search revealed that a high-fat diet causes long-lasting dysregulation in miRNAs similar to the signature observed with spaceflight and SGA (Supplementary Table 1). Additionally, certain pollutants and toxic environments, such as exposure to polychlorinated biphenyls65 or specific hydrocarbons66,67, can cause long-lasting miRNA upregulation. While this is not direct evidence that these miRNAs will persist long-term after astronauts return to Earth, it indicates that long-term dysfunction associated with these 13 miRNAs can significantly impact human health, influencing downstream pathways and genes.
Confirmation of key SGA-related molecular changes in astronauts
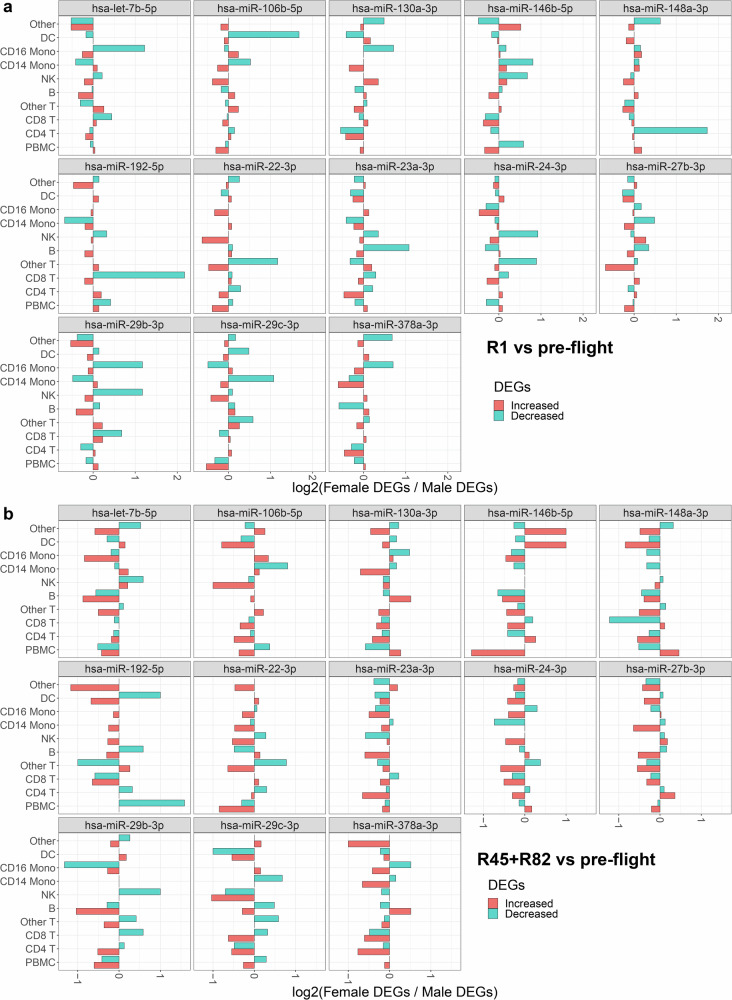
The changes with the miRNAs and their gene targets that we have reported so far are focused on the publicly available murine data. To provide relevance to human spaceflight, we draw upon data from the first civilian commercial SpaceX Inspiration4 (I4) mission, which spanned a brief 3 days of spaceflight1. This mission, featuring two male and two female astronauts, involved single-nuclei RNA sequencing (snRNA-seq) on the blood at four key time points: pre-flight, 1-day post-flight (R1), 45 days post-flight (R45), and 82 days post-flight (R82). Despite its brevity, it is crucial to note that the I4 mission occurred at a higher altitude (590 km above Earth’s elevation) than typical ISS experiments (420 km above elevation), resulting in reduced protection from Earth’s Van Allen magnetic field, so that astronauts are exposed to an accumulated radiation dose equivalent to nine months on the ISS. The recorded accumulated dose for I4 astronauts was 4.72 mSv or 0.472 cGy1, significantly lower than the 0.5 Gy Galactic Cosmic Radiation (GCR) estimated for a round trip to Mars. Nonetheless, changes associated with the top 45 miRNA gene targets were observed even at these lower doses (Figs. 6 and 7, and Supplementary Fig. 11).
Fig. 6. Sex difference comparison of gene expression changes in the top 45 gene targets of the 13 specific miRNAs from the Inspiration 4 (I4) astronaut data derived from scRNA-sequence analysis of whole blood.
Bar plots depict the ratio of increased (red) or decreased (cyan) gene expression values between female and male I4 astronauts. a The values obtained 1 day after return to Earth (R1) versus the average of the 4 pre-flight time points. b The average values for 45 days (R45) and 82 days (R82) after return to Earth compared to the pre-flight average across the 4 time points.
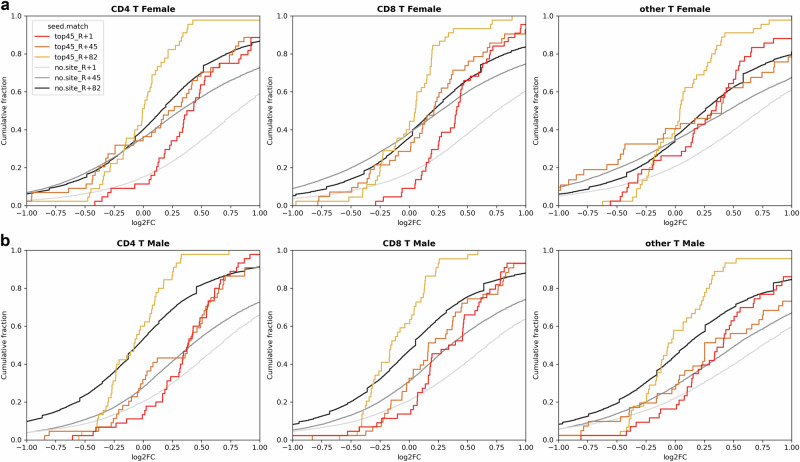
Fig. 7. Sex-specific cumulative plots illustrate the impact on the top 45 gene targets of 13 specific miRNAs in Inspiration 4 (I4) astronaut data, derived from scRNA-sequence analysis of whole blood.
These cumulative plots focus on T cells (i.e., CD4 and CD8 T cells and others) in the I4 astronaut scRNA-seq data, comparing 1 day after return to Earth (R1) (red line), 45 days after return to Earth (R45) (orange line), and 82 days after return to Earth (R82) (gold line) to pre-flight levels. The x-axis represents log2(fold-change) values for the comparisons, while the “no-site” line serves as a baseline for genes without targets to the 13 miRNAs. Various shades of gray in the no-site lines correspond to specific comparisons, as indicated in the figure legend. a Cumulative plots specifically for the female astronauts. b Cumulative plots specifically for the male astronauts.
In examining overall up- and down-regulation of the top 45 miRNA gene targets, a sex-dependent analysis revealed distinct cell types contributing to more inhibition in females (Fig. 6). At R1, T cell populations (CD8, CD4, or other T cells) or CD16 monocyte populations exhibited greater inhibition in females compared to males (Fig. 6a). As time progressed post-flight (45 and 82 days), the changes in females diminished, but certain miRNAs (e.g., miR-29b, miR-29c, and miR-22) still indicated a larger number of inhibited genes in females than males (Fig. 6b). Notably, a cell type-independent analysis of miRNA binding to the top 45 genes, assessed through cumulative plots (Supplementary Fig. 11 and Fig. 7), revealed that T cell populations (CD4, CD8, and other T cells) in females exhibited the most significant inhibition of top 45 gene targets, starting prominently at R1 and persisting up to the last measured time point (R82) (Fig. 7a). In contrast, males displayed a much lesser degree of inhibition at R1 and no deviation from baseline at R45 and R82 (Fig. 7b). This suggests potential impact and suppression of regulatory T cells due to the identified miRNAs associated with SGA during spaceflight. Intriguingly, prior studies have indicated that maternal T cell exhaustion and senescence, leading to SGA and other preterm complications during pregnancy68, may be increased. Specifically, Steinborn et al. demonstrated a significant reduction in fetal regulatory T cells (Tregs) from SGA infants68. CD4 Treg cells at the maternal-fetal interface are important for establishing tolerance in early pregnancy. A reduction in CD4 Tregs within the overall population suggests suppressed tolerance and elevated inflammation, which could contribute to numerous labor and birth issues, including potentially SGA69. Furthermore, our previous investigations have demonstrated the suppression of CD4 and CD8 T cells within 24 hours following exposure to 0.5 Gy GCR irradiation, both with and without simulated microgravity25. This provides additional evidence of the occurrence of T-cell population suppression and supports the heightened risk of SGA after spaceflight. While indirect, the suppression of gene targets for SGA-associated miRNAs in female astronauts necessitates further studies to conclusively assess the potentially heightened risk of SGA in female astronauts desiring future pregnancies after returning to Earth.
Lastly, we aimed to determine if the key pathways impacted by the 13 miRNAs and their gene targets are also dysregulated in astronauts, specifically the I4 astronauts. Utilizing the I4 snRNA-seq data, we performed pathway analysis and identified key pathways similarly regulated by the miRNAs (Figs. 4 and 5). Since the I4 crew included two males and two females, we were able to perform sex-dependent analysis and observe the key pathways regulated immediately upon return to Earth and the long-term post-flight effects (Supplementary Figs. 12–15 and Supplementary Data 3). The I4 astronaut data showed similar results to the simulated space radiation experiments performed on mice (Supplementary Figs. 7 and 9). Specifically, female astronauts exhibited similar suppression of the majority of pathways observed for the 13 miRNAs (Supplementary Fig. 12). A similar pattern was observed in male I4 astronauts (Supplementary Fig. 13). Although there were overlapping pathways between males and females, distinct pathway patterns specific to female astronauts were not present in males. Additionally, we observed similar suppression of pathways as in the mouse study (Supplementary Fig. 9) in both female and male astronauts for the 45 gene targets (Supplementary Figs. 14 and 15). Interestingly, the RNA metabolism pathway was suppressed across all cell types. Through this analysis, we provide indirect confirmation that miRNAs potentially influence key pathways long-term, which may increase health risks associated with SGA.
Identification of prospective countermeasures to target the miRNA signature associated with spaceflight-induced SGA
We have established that these miRNAs are associated with both spaceflight and SGA, making them potentially valuable biomarkers for monitoring the risk of SGA development in space. We set out to see whether any countermeasures currently exist that could target these miRNAs. It is important to note that future studies are needed to determine whether this miRNA signature is causal or merely associated with both spaceflight and SGA.
Clinically, women at risk of having SGA (either because they already had a baby with SGA, or they have risk factors such as hypertension, are obese, are smokers, have twins, etc.)70 are started on Aspirin 75 mg in the first trimester, as soon as an ultrasound scan confirms that there is a fetal heart70. If a baby needs to be delivered earlier than the due date for a clinical reason (including SGA), the mother is given antenatal intramuscular corticosteroids (dexamethasone or betamethasone), in order to boost the baby’s lungs, and minimize the risk of neonatal respiratory distress syndrome71. However, glucocorticoids administered prenatally may induce intrauterine growth restriction72. When an SGA baby is born, growth hormone may be administered later in infancy9. Folic acid, in many cases vitamin D is started pre-conception and continued during pregnancy73. Vitamin D has also been shown to be beneficial for fetal growth74. Other medications including progesterone and antioxidants, such as vitamins C and E, have been implicated to help in preventing placental insufficiency, which is the underlying factor in SGA75,76.
To identify any countermeasures that currently exist for targeting the SGA-spaceflight miRNA signature, we leveraged our well-established machine-learning tool, sChemNET77. Using sChemNET, we predicted potential FDA-approved small molecule drugs that could target these miRNAs and be further developed as countermeasures. The goal of this analysis was to find small molecule drugs that could potentially inhibit spaceflight-associated SGA miRNAs both during spaceflight and before pregnancy. Any potential toxicity concerns related to these treatments during pregnancy and potential placental transfer of the drugs to the fetus are alleviated, as the drugs will not be administered during pregnancy, given that currently, pregnant women do not participate in spaceflight.Theoretically, the associated miRNAs should be suppressed due to implemented countermeasures.
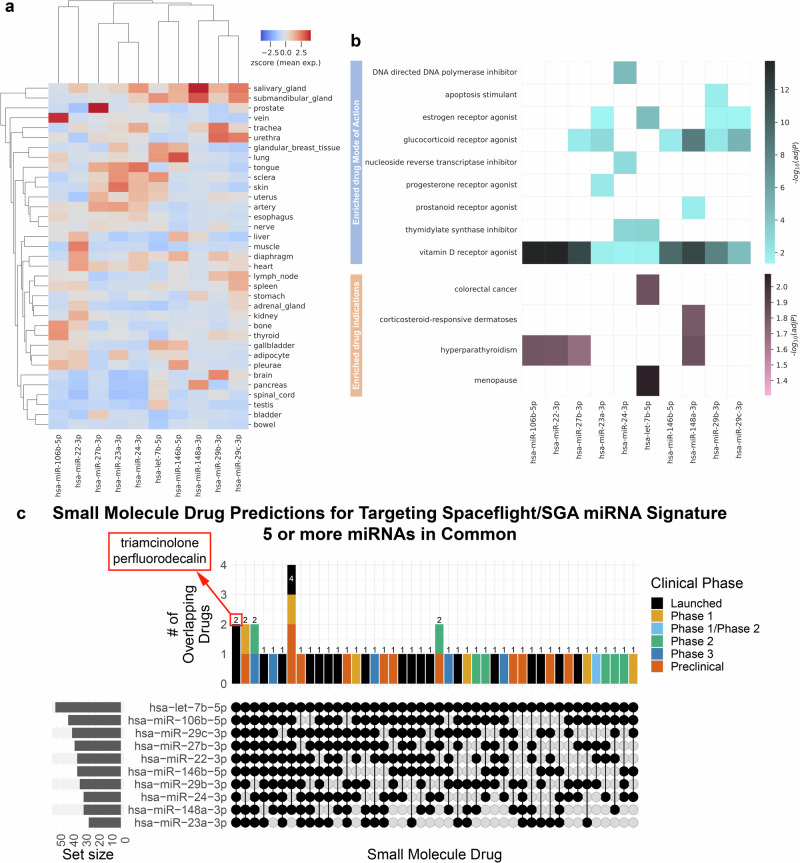
When developing countermeasures targeting miRNAs, it is crucial to establish the baseline levels of these molecules in healthy tissues. For the spaceflight-associated SGA miRNA signature, we have observed that the expression of miRNAs in healthy tissues is generally low (Fig. 8a). In the bladder and bowel, miRNA expression is minimal, and in the uterus, most miRNAs exhibit low expression, except for four highly expressed miRNAs (miR-27b-3p, miR-23a-3p, miR-24-3p, and let-7b-5p). Additionally, we assessed artery expression because we are investigating circulating miRNAs. Arterial miRNA expression is generally low, with three miRNAs showing higher expression that aligns with the uterus (miR-27b-3p, miR-23a-3p, miR-24-3p). Consequently, in healthy tissues, these miRNAs exhibit low expression levels (Fig. 8a), whereas in both SGA patients and spaceflight they are upregulated (Fig. 2).
Fig. 8. Basal miRNA expression in healthy tissues and predicted small molecule drugs.
a Heatmap presenting miRNA expression levels as z-scores across various healthy tissues, sourced from the miRNA Tissue Atlas. b Heatmap illustrating the mode of action and enriched drug indicators for predicted small molecule drugs targeting the miRNAs, as determined by sChemNET. Color intensity represents −log10(adj. p-value). c Upset plot revealing specific predicted small molecule drugs that target the miRNA signature associated with Small-for-Gestational-Age (SGA) in spaceflight. The drugs are in common with 5 or more of the 13 miRNAs. Notably, two small molecule drugs, triamcinolone and perfluorodecalin, are shared among all the identified miRNAs. All predicted small molecule drugs can be found in Supplementary Fig. 11 and Supplementary Data 3.
Using sChemNET, we successfully predicted small molecule drugs that target the miRNA signature. It’s important to highlight that our predictions do not discern whether the small molecule drugs will inhibit or promote the miRNAs, and instead these results serve as a starting point for future clinical studies. It is essential to recognize that this tool serves specifically to predict potential FDA-approved drugs, streamlining the validation and repurposing process as countermeasures targeting the miRNAs. In this study, we lean on prior research involving our predicted drugs to offer potential indicators of a positive response to these small molecule interventions. Further research is required to screen the effects of our predicted drugs.
We determined the drug indications and their modes of action, shedding light on the potential impact of these drugs (Fig. 8b). Our analysis revealed that these drugs include estrogen and progesterone receptor agonizts, as well as vitamin D receptor agonizts, each with distinct modes of action (Fig. 8b). Of particular importance is the DNA polymerase inhibitor, as space radiation has been shown to cause DNA damage, which aligns with our findings in the global miRNA response (Fig. 1). Furthermore, we observed an upregulation of the DNA repair system in SGA. One noteworthy target is the glucocorticoid receptor agonist, which is pivotal in regulating physiological processes, including glycemic levels. Dysregulation in this area could lead to conditions such as diabetes or even more severe cerebral diseases78. Estrogen and progesterone receptor agonizts are also targeted and are relevant to SGA, as they play crucial roles in sustaining pregnancies and ensuring placental health79. Vitamin D, targeted by all 13 miRNAs, is notable as it is commonly used as a supplement by both male and female astronauts. Vitamin D is integral to mitochondrial activity, which is known to be dysregulated in both SGA and space conditions80 (Fig. 1).
Many of the drugs predicted to target the miRNA signature are already in clinical use for treating specific conditions (Fig. 8b). These conditions include hyperparathyroidism and menopause (Fig. 8b). Hyperparathyroidism is a common disease in women. It can even develop during pregnancy81. Notably, it has been associated with potential risks during space travel82. Upon returning to Earth, women astronauts may face premature menopause, which can significantly hinder their ability to conceive83. Therefore, if these miRNAs can impact women’s fertility, they could also contribute to birth defects and placental disorders such as SGA. The drugs we identified have the potential to reduce these risks and serve as effective countermeasures.
We identified a total of 128 small molecule drugs at various stages of clinical development or approval that target the 13 miRNAs (Supplementary Data 4 and Supplementary Fig. 16). Although each predicted drug can target multiple miRNAs (Supplementary Fig. 16), our focus was on small molecule drugs that can target five or more miRNAs (Fig. 8c). This strategy aimed to address the majority of the miRNAs within the signature responsible for the potential increased risk of SGA during spaceflight. Upon further refinement, we found that only two small molecule drugs, triamcinolone, and perfluorodecalin, were both commercially available and FDA-approved for targeting all 13 miRNAs (Fig. 8c). Triamcinolone is a synthetic corticosteroid with applications in treating respiratory inflammation, asthma in both adults84 and children85, rheumatoid arthritis86, retinal deposits87, and other inflammatory conditions. In contrast, perfluorodecalin is a biologically and chemically inert biomaterial with versatility in various medical applications. It is radiopaque, hydrophobic, and possesses high gas solute capacity, particularly for oxygen88. It finds utility in bone regeneration88, lung distention in infants89, and, most notably, in delivering additional oxygen to specific organs through aerosol delivery90. Although perfluorodecalin is considered as part of per- and poly-fluoroalkyl substances, which are classified as pollutants91 and might be toxic92, perfluorodecalin has been declared safe for human health93. Overall, our machine learning prediction identified drugs that may prove useful for targeting miRNA that are associated with both SGA and spaceflight, but extensive preclinical and clinical testing would be needed to assess whether they are effective and safe in women and provide the functional impact on the miRNAs as predicted.
Discussion
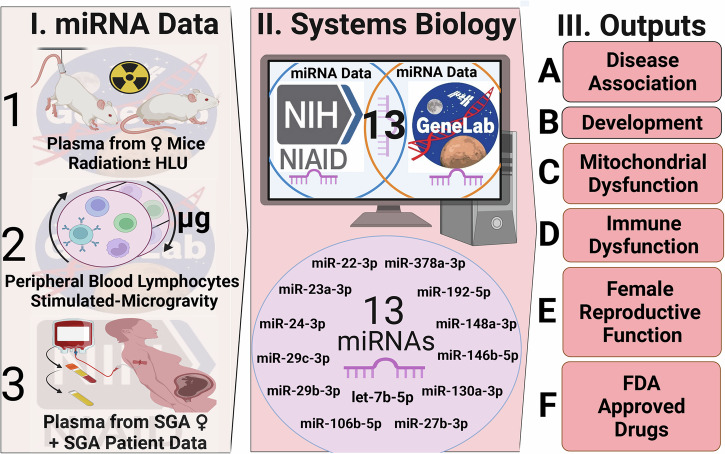
Spaceflight, influenced by factors like space radiation, microgravity, and isolation, presents unique health risks, especially in the realm of reproductive health for both women and men. These risks, often related to oxidative stress, sleep disruption, and microgravity94, can manifest in several sex-specific disorders. Our focus here was to investigate the likelihood of birth defects, specifically SGA fetuses, in female astronauts upon their return to Earth. Normal placenta formation early in the first trimester sets the stage for the rest of the pregnancy. It involves a delicate balance between maternal-fetal crosstalk via an interplay of finely orchestrated cellular and molecular pathways95. The resulting intrauterine environment from peri- and pre-conception onwards plays a vital role in fetal programming and the future health of the offspring, which is further modulated by genetic and epigenetic factors. Abnormalities in the controlled spiral artery invasion can lead to placental ischemic disorders and consequent adverse obstetrical outcomes96. These include SGA due to intrauterine growth restriction, with or without preeclampsia, placental abruption, and/or preterm labor. Utilizing human data from the ImmPort database and comparing it with miRNA data from mice exposed to simulated space environments from NASA’s GeneLab, we identified 13 shared miRNAs (Fig. 9).
Fig. 9. Overall summary of the findings from this study.
In panel (I) we define the experimental data utilized for this study with: (1) the mice under simulated space environment experiments, (2) the simulated microgravity experiments done in vitro, and (3) the plasma data obtained from females during pregnancy with and without SGA outcome. For panel II) we define the databases (i.e. ImmPort and GeneLab) that the data was obtained from and the 13 miRNAs identified to be key for potential SGA risk during spaceflight. In panel III) we defined the key endpoints of the study which are (A) disease association, (B) development, (C) mitochondrial dysfunction, (D) immune dysfunction, (E) female reproductive function, and (F) predicted FDA-approved drugs as potential countermeasures. This figure was created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
During pregnancy, miRNAs have been shown to play dual roles, contributing to both healthy stages of pregnancy and potential complications for the fetus. For instance, Kamity et al. discuss in a comprehensive review of miRNAs related to inflammation and immunity how certain miRNAs expressed in circulation prior to pregnancy can persist during pregnancy, potentially increasing the risk of complications for both the pregnancy and the fetus97. In contrast, a study by Cécilia Légaré et al. identified 191 miRNAs commonly expressed between pregnant and nonpregnant women98. Among these, only two of our 13 miRNAs show differential expression: hsa-miR-22-3p and hsa-miR-378a-3p, which are less abundant in pregnant women compared to nonpregnant women. Although this study does not specifically investigate the potential risk of SGA, it is noteworthy that these miRNAs we have identified as associated with SGA are less abundant during normal pregnancy. The 13 miRNAs we identified as elevated during spaceflight in nonpregnant female mice may potentially contribute to the risk of SGA upon women’s return to Earth, as there was no overlap or increase of these miRNAs observed in healthy pregnant women. When conducting a comprehensive literature review of the remaining 11 miRNAs, we discovered that these miRNAs are heavily involved in various pregnancy complications, including preeclampsia, gestational diabetes mellitus (GDM), preterm birth, and miscarriages. These complications are associated with specific pathways such as the TGF-β pathway, metabolic pathways, and immune-related pathways (see Supplementary Table 2 for a complete list and related references).
These miRNAs are implicated in a range of health conditions, from pregnancy issues to bone and liver diseases (Figs. 3–5). Notably, the miRNA signature that we obtained shows a significant overlap with pathways involved in DNA repair, mitochondrial function, and cardiovascular health (Figs. 3, 4). In addition, utilizing data from the I4 mission we start to show the impact these key miRNAs (via the miRNA gene targets) have on female human astronauts (Figs. 6, 7, and Supplementary Fig. 11). Our research highlights the significance of integrating diverse federal databases, particularly those from NASA and the NIH, in understanding health risks associated with spaceflight, especially for females. Our study exemplifies how space biology, with its unique environmental challenges, serves as a crucial model for Earth applications. By examining conditions in space, we gain valuable insights into physiological and molecular responses under extreme conditions, which can be instrumental in developing effective pharmacological solutions for similar health issues on Earth. This interdisciplinary approach advances our knowledge of spaceflight’s impact on human health and paves the way for innovative medical treatments and preventive strategies applicable in terrestrial settings.
Oxygen and miRNAs regulate the normal expression of the renin-angiotensin system (RAS) in the placenta, leading to its normal development, and also play a role in disease states involving dysregulation of angiogenesis in which the RAS is overexpressed55. The placental RAS has been shown to be downregulated by miRNAs that are suppressed during the physiologically normal ‘hypoxic’ phase of early placentation, thus allowing the placental RAS to stimulate placental growth and angiogenesis.
Oxidative stress is manifested at the maternal-fetal interface from early pregnancy, where it modulates placental development99. The potent antioxidant properties of estrogen94 play a significant role in reducing oxidative damage through ROS. Other antioxidants include vitamin C and vitamin E, which have been shown to be beneficial in improving placental health in maternal-fetal dyads100,101 (Supplementary Data 4). This aspect is particularly relevant given that spaceflight conditions tend to elevate ROS levels, leading to DNA damage and cellular aging (Fig. 1i and Supplementary Fig. 5). Oxidative stress in space can disrupt the balance of pro-oxidants and antioxidants, crucial for women’s fertility regulation94, potentially contributing to complications like miscarriages, preeclampsia, and fetal growth restriction102.
Both SGA and space conditions exhibit mitochondrial dysregulation (Fig. 1j and Supplementary Fig. 6), leading to a loss of biological properties and programmed cell death. This shared characteristic raises concerns about downstream disruptions in biological functions, including lipid metabolism—a key process regulated by mitochondria. Our identified SGA-spaceflight miRNA signature targets functions related to lipid metabolism and adipogenesis (Fig. 4), suggesting potential concerns for female astronauts if these miRNAs persist in circulation upon return to Earth.
Prior studies indicate that steroid hormones can enhance mitochondrial respiration by interacting with the inner mitochondrial membrane103. Our analysis suggests a potential class of drugs targeting the 13 miRNAs as hormone receptor antagonists (Fig. 8b), including estrogen, glucocorticoid, and progesterone. Moreover, the top gene targets for these miRNAs regulate estrogen signaling receptor (ESR)-mediated pathways (Fig. 5c). Estrogen, known for its protective role on mitochondria, influences various mitochondrial functions such as ATP production, mitochondrial membrane potential, and calcium handling104. Estrogen receptors (ERs) binding to mitochondrial DNA (mtDNA) regulate mitochondrial gene expression and contribute to mitochondrial biogenesis105. Estrogen’s role extends to antioxidation, reducing ROS production and regulating antioxidant genes94. Its interaction with mitochondria also provides cardio-protective functions106. Consequently, cardiovascular disease risk is lower in pre-menopausal women than in men, but it rises in postmenopausal women compared to age-matched men107. Finally, our published findings underscore the substantial impact of spaceflight on estrogen and insulin-resistance pathways5, offering additional confirmation of estrogen-signaling dysfunction occurring during spaceflight. The association of the SGA-spaceflight miRNA signature with estrogen signaling raises concerns about both mitochondrial dysfunction and fundamental female sex hormone signaling in future pregnancies.
The pragmatic challenges of menstruating in space often lead female astronauts to opt for medically induced amenorrhea, which will impact estrogen levels108. While this addresses logistical concerns, it inadvertently eliminates the protective effects of estrogen, potentially affecting bone health and elevating the risk of osteopenia in the already demanding conditions of space. Importantly, these disorders may be partially linked to estrogen’s antioxidant role94, known to influence conditions such as hypertension109, diabetes78, and cardiovascular diseases110. Women, leveraging their naturally higher estrogen levels, potentially enjoy added protection against specific health risks associated with spaceflight compared to men. However, inducing amenorrhea can exacerbate health risks by further diminishing estrogen levels, emphasizing the delicate balance between mitigating challenges, and maintaining hormonal safeguards in the unique environment of space. There is a critical need for comprehensive education and informed decision-making among female astronauts regarding menstrual suppression in space. Furthermore, a study on reproductive endocrine markers in female mice has provided insights into the estrous cycle and spaceflight, underscoring the necessity for further research in this area111. In the context of this research, the long-term impact of such disruption on SGA remains to be fully understood.
To mitigate the effects of spaceflight on future SGA risk, we explored potential countermeasures for nonpregnant women on future spaceflight missions. Using our machine learning framework, sChemNET77, we aim to provide a basis for future clinical studies. We identified two FDA-approved drugs targeting all 13 miRNAs in the signature: triamcinolone and perfluorodecalin (Fig. 8c). These drugs, known for their roles in modulating ROS levels and mitochondrial function, could be pivotal in developing strategies to reduce SGA risk among female astronauts. Triamcinolone, a versatile corticosteroid, is known for its application in preventing the rise of ROS during hypoxia by altering mitochondrial function112. Despite its effectiveness in reducing the inflammatory response, triamcinolone may induce immunosuppression and delayed wound healing113. However, its ability to prevent an increase in ROS by upregulating mitochondrial respiration makes it a potential candidate for mitigating the impact of spaceflight112, it does so by preventing a rise in TNF-β1. Although we are not recommending that this countermeasure should be given to pregnant women, Bérard et al.114 demonstrated that the administration of triamcinolone intranasally to pregnant women does not increase the risk of SGA, malformation or spontaneous abortion, but is associated with potential risk of respiratory system defects. Perfluorodecalin, a fluorocarbon renowned for its remarkable oxygen dissolving capacity, serves a crucial role as an oxygen carrier115,116. Its versatile applications span both topical and aerosol administration. Topically, it has demonstrated efficacy in enhancing skin wound healing caused by burns117 and treating ocular damage resulting from chemical burns116. In aerosol form, it has exhibited benefits in improving pulmonary functions among preterm births118, fostering lung distention in infants89, and ameliorating respiratory distress syndrome in preterm infants90. Additionally, its utility extends to diverse areas such as bone regeneration88 and addressing retinal dysfunction in the eyes119. Perfluorodecalin also possesses the intriguing capability to target mitochondria120, and its high solute capacity for oxygen suggests a potential role in mitigating ROS production. As stated in the results, it is important to note that both perfluorodecalin and triamcinolone are predictions made by our sChemNet ML model. In the original manuscript for sChemNet77, we experimentally validated the model’s predictions and provided functional validations for a few miRNAs as proof of principle. For instance, we demonstrated that a few predicted drugs for miR-451 can rescue oxidative stress in a zebrafish animal model, and predicted vitamin D targeting miR-181 can reduce cell proliferation in breast cancer cell lines. Although this provides some proof that our model can produce drug predictions with functional relevance to the miRNAs of interest, further functional experimental assays are necessary to validate their impact on the 13 miRNAs and their potential to mitigate the progression of SGA. This validation is crucial to assess their effectiveness as countermeasures for spaceflight.
Micronutrient availability has an important role in the development of the placenta121. Another potential pharmaceutical intervention we identified for targeting miRNAs is a vitamin D receptor agonist (Fig. 8b). Vitamin D plays a crucial role in lipid metabolism regulated by the mitochondria122. The weight loss experienced during spaceflight contributes to deficiencies in vitamin D, calcium, and vitamin K123, consequently leading to a decline in bone mineral density. Presently, vitamin D supplementation is the only regular supplement administered to astronauts124. Low maternal vitamin D levels are associated with incomplete human extravillous trophoblast (EVT) invasion of the decidua and maternal spiral arteries125. This leads to placental insufficiency, which is a hallmark of SGA, as well as other placental disorders such as preeclampsia. A recent systematic review indicates that early pregnancy vitamin D supplementation may have a role in reducing the risk of suboptimal placental formation, and hence reduce the risk of SGA126. Our approach has led us to pinpoint potential countermeasures that should be examined in future studies. These interventions hold the potential to reduce the risk of SGA among female astronauts.
Our primary methodology involved employing an in silico approach to identify a miRNA signature associated with SGA risk during spaceflight and assessing its downstream impact on health. Additionally, we utilized a machine learning technique to determine potential countermeasures for mitigating this identified risk. The inclusion of astronaut data served as an initial confirmation of the predicted biological changes in females during spaceflight. While this marks the beginning of experimental validation, we recognize the necessity for robust validation in space and simulated space flight conditions to instill complete confidence in the results. The predicted countermeasures show promise but warrant further experimental validation. Given the challenges of spaceflight-related experiments, leveraging resources such as ImmPort and GeneLab becomes essential for expediting the discovery of biological health risks. Timely dissemination of these findings within the scientific community is crucial, facilitating further experiments and validation for the development of SGA-related biomarkers and countermeasures in the future.
Our findings pave the way for future studies that can offer clinical therapeutics, extending the boundaries of science into space and accelerating our understanding of various diseases. This discussion underscores the critical need for further investigation into the female reproductive system in space. Female reproductive health is currently understudied but essential for understanding and mitigating health risks associated with space travel. In space research, it is crucial to address potential health risks before they manifest during long-term missions, as they can cause long-term health issues that will be difficult to treat at that point. Our work generates a specific hypothesis and identifies a potential gap in knowledge that needs to be further explored in future research. Although it is currently challenging to assess the potential SGA risk due to spaceflight, we have provided some evidence that this research avenue, which could impact the female reproductive system, should be investigated further. Prevention is key in space biology research, and identifying potential novel health risks for future investigation is essential. Finally, the use of integrating space biology databases with NIH databases has been instrumental in this research, offering a unique perspective on biological processes under extreme environmental stress.
Methods
ImmPort SGA dataset and data processing
We accessed the NIAID’s ImmPort platform127 to obtain the relevant dataset, SDY18717, focusing on miRNA expression in Small-for-Gestational-Age (SGA) cases. The original study7 involved whole blood collection from N = 29 women, including N = 16 with normal birth outcomes and N = 13 with SGA births. Samples were collected at three gestation time points: (time-point A), (time-point B), and (time-point C) weeks. Plasma purification was followed by Nanostring nCounter miRNA assays covering 800 miRNAs. Normalized miRNA count data, available on ImmPort, underwent analysis using the R package DESeq2 (ver.1.36.0)128 to identify differentially expressed miRNAs in SGA vs. Controls at each time point and across all samples (time-independent). Principal Component Analysis (PCA) plots, based on normalized miRNA counts, and heatmaps, using log2(fold-change) values from DESeq2 analysis, were generated with ggplot2 (ver. 3.4.3) and pheatmap (ver. 1.0.12) R packages, respectively.
GeneLab simulated spaceflight miRNA datasets and data processing
We accessed datasets OSD-55129 and OSD-33624 from NASA’s Open Science Data Repository (OSDR) and GeneLab for our analysis. OSD-336 involved a simulated space environment experiment on mice, with miRNA-seq data available on GeneLab. The method and data analysis for this experiment are briefly outlined below. OSD-55129 was a previous in vitro experiment simulating modeled microgravity (MMG) on human peripheral blood lymphocytes (PBLs) using a Rotating Wall Vessel (RWV) bioreactor. In the original publication27, PBLs from 12 healthy donors were split for MMG and 1 g control conditions. At 24 hours, RNA was isolated from 107 PBLs from each replicate, and miRNA profiling was performed using the Whole Human Genome Oligo Microarray (Agilent). The miRNA microarray data was deposited on the Gene Expression Omnibus (GEO) website (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE57400. Data processing involved “Analyze with GEOR” on GEO, applying log transformation and determining significant miRNAs by Benjamini and Hochberg (FDR) with a cutoff < 0.05. Principal Component Analysis (PCA) plots and heatmaps were generated using ggplot2 (ver. 3.4.3) and pheatmap (ver. 1.0.12) R packages, respectively.
Murine simulated space environment experiments
We utilized murine data from our prior experiments reported in Malkani et al.47 and Paul et al.25 and is available on NASA’s OSDR as dataset OSD-336 as mentioned above. Briefly, here we have also provided a description of the murine simulated space experiments. C57Bl/6 J wildtype female mice (15 weeks ±3 days old) were obtained from Jackson Laboratories and housed at Brookhaven National Laboratory (BNL, Upton, NY). After quarantine and acclimation to a 12:12 hour light:dark cycle with controlled temperature/humidity for a week, mice were cage acclimated (n = 10 mice per group; 2 mice per cage) 3 days before hindlimb unloading (HU). Food and water were given ad libitum, and standard bedding was changed once per week. The normally loaded (NL) mice, used in parallel experiments not reported here, underwent the same acclimation. HU was conducted for 14 days, with irradiation on day 13 (0.5 Gy GCR, 1 Gy SPE, 5 Gy Gamma, and 0 Gy Sham control). SPE simulated irradiation consisted of protons or different energies ranging from 50 MeV to 150 MeV. On day 13, mice were transported to NASA Space Radiation Laboratory (NSRL). They were individually placed in HU boxes and exposed to GCR or Sham control (no irradiation) in the plateau region of the Bragg curve at room temperature. Dosimetry was performed by NSRL physics staff. The radiation dose simulated the exposure an astronaut might receive during a Mars mission, modeled as a single 25-min exposure rather than the actual chronic exposures over 1.5 years. A 60×60 beam was utilized at the NSRL for irradiation. Sham controls underwent the same procedures without irradiation. Blood tissues were collected 24 hours post-irradiation and post-euthanasia. Plasma was separated, flash-frozen, and stored at −80 °C. A cellular fraction aliquot was stored for RNA analyses, while the remaining fraction underwent flow cytometric preparation. All organs (i.e., heart, liver, and soleus muscle) were flash frozen at dissection and stored at -80°C. Body weight tracking was performed on days −3, 0, 7, and 14.
All experiments were approved by Brookhaven National Laboratory’s (BNL) Institutional Animal Care and Use Committee (IACUC) (protocol number: 506) and all experiments were performed by trained personnel in AAALAC accredited animal facilities at BNL, while conforming to the U.S. National Institutes of Health Guide for the Care and Use of Laboratory Animals. All methods were carried out in accordance with the relevant guidelines and regulations and are reported in accordance with ARRIVE guidelines.
Simulated Galactic Cosmic Radiation exposure
The animals were irradiated at the NASA Space Radiation Laboratory (NSRL) at Brookhaven National Laboratory (BNL). Positioned in the plateau region of the Bragg curve, mice received irradiation at room temperature. NSRL physics staff conducted dosimetry using a 60 cm x 60 cm beam. All mice were exposed to 0.5 Gy of simplified simulated Galactic Cosmic Radiation (GCR). The irradiation utilized ions, energy, and doses determined by a NASA consensus formula for five ions: protons at 1000 MeV, 28Si at 600 MeV/n,4He at 250 MeV/n, 16O at 350 MeV/n, 56Fe at 600 MeV/n, and protons at 250 MeV in the following proportions – 1000 MeV protons at 34.8%, 250 MeV protons at 39.3%, 28Si at 1.1%, 4He at 18%, 16O at 5.8%, and 56Fe at 1%. This simplified mixture mirrors the ion proportions in space, making it relevant to exploratory class missions26. While low Linear Energy Transfer (LET) particles (Protons and Helium) dominate, high LET ions generally have a greater relative biological effect (RBE).
miRNA extraction from murine tissues
MiRNA extractions from plasma was carried out using the Qiagen miRNeasy serum/plasma kit (Cat# 217184). Quantitation of miRNA samples was done using a NanoDrop 2000 Spectrophotometer (ThermoFisher Scientific).
miRNA sequencing on murine samples
For miRNA library construction and sequencing, plasma-derived miRNAs from the aforementioned mouse experiments were isolated using the QIAgen miRNeasy kit (#217004). Total RNA quality and quantity were assessed with a Bioanalyzer 2100 (Agilent, CA, USA), ensuring a RIN number > 7. A TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, USA) protocol was followed, utilizing approximately 1 μg of total RNA to prepare a small RNA library. Single-end sequencing with 50 bp was conducted on an Illumina Hiseq 2500 at LC Sciences (Hangzhou, China), adhering to the vendor’s recommended protocol. Raw miRNA-sequence data is available on NASA Open Science Data Repository with the following identifiers: heart-related data: OSD-334, DOI: 10.26030/cg2g-as49, liver-related data: OSD-335, DOI: 10.26030/72ke-1k67, plasma-related data: OSD-336, DOI: 10.26030/qasa-rr29, and soleus muscle-related data: OSD-337, DOI: 10.26030/m73g-2477.
Analysis of miRNA sequencing from murine samples
Raw reads underwent preprocessing with ACGT101-miR software (LC Sciences, Houston, Texas, USA), eliminating adapter dimers, junk, low complexity, and common RNA families (rRNA, tRNA, snRNA, snoRNA), as well as repeats. Unique sequences, spanning 18 ~ 26 nucleotides, were aligned to miRBase 22.0 by BLAST search for identification of known miRNAs and novel 3p- and 5p-derived miRNAs. Allowances for length variations at both ends and one mismatch inside the sequence were applied. Sequences mapping to species-specific mature miRNAs were recognized as known miRNAs, while those on the opposite arm of annotated miRNA-containing arms were deemed novel 5p- or 3p-derived candidates. Unmapped sequences were subjected to BLAST against specific genomes, and hairpin RNA structures were predicted using RNAfold software based on predefined criteria. Known miRNAs were identified using the same criteria. Differential expression analysis of miRNAs, utilizing normalized deep-sequencing counts, employed Fisher exact test, Chi-squared 2×2 test, Chi-squared nXn test, Student t test, or ANOVA, with significance thresholds set at 0.01 and 0.05 for each test.
It is important to also consider the issue of hemolysis with our analysis. Although hemolysis is unavoidable for blood/plasma samples obtained from in vivo studies similar to those described in OSD-336, there are ways to account for this in the analysis when hemolysis is not measured. Numerous papers discuss the impact of hemolysis on circulating miRNAs, concluding that while hemolysis in in vivo samples cannot be entirely avoided, procedures can significantly reduce its effects130 Given the constraints of space biology experiments, the primary step to reduce hemolysis variations for miRNAs is by handling all samples identically. Both the controls and experimental samples were handled under the exact same conditions throughout the study. Thus, if miRNA levels had been altered due to hemolysis, all miRNA levels would have been altered similarly across our comparisons. For the miRNA-seq data, the variation across the biological replicates was minimal, and the miRNAs that showed high variation were either not significant or filtered out during the preprocessing step. This suggests that while hemolysis can alter miRNA levels130, the levels of the remaining miRNAs after data processing should have been altered identically in our controls and experimental conditions, reducing variability between results.
miRNA pathway analysis
To determine Hallmark28 and MitoPathway33 pathways being regulated by the miRNAs, we performed miRNA gene set analysis utilizing the RBiomirGS131 v0.2.12 R package from the processed miRNA analysis for all conditions in the plasma, PBLs, and SGA data. From the pathways we chose an FDR < 0.25 cutoff for significantly regulated pathways. We plotted the specific pathways as lollipop plots with R package ggplot2 (ver. 3.4.3).
Determining common miRNAs between SGA and spaceflight data
To identify common miRNAs between the SGA and spaceflight datasets, we overlaid significantly regulated miRNAs (adj. p-value < 0.05) from the SGA dataset (SY1871) with simulated space environment data (OSD-55 and OSD-336). We narrowed down the list to include only miRNAs that exhibited consistent regulation in the same direction between OSD-336 and SY1871. This refined process resulted in the identification of 13 common miRNAs, forming the miRNA signature associated with both SGA and spaceflight.
Conserved miRNA analysis between humans and mice
Selected precursor and mature miRNA sequences from human and mouse were extracted from miRBase v.22.1132. After BLASTN alignment, conservation between mouse and human sequences was determined as the percentage of aligned nucleotide identities in mature and pre-miRNA sequences.
Analysis of pathway, disease, and gene targets for miRNAs and network generation
To predict the functions and diseases associated with the 13 common miRNAs, we employed miRNet133 and visualized the results using ggplot2 in R (v3.4.3). For identifying gene targets of each miRNA, we utilized six different miRNA-gene target databases: miRmap134. miRwalk135, miRnet, miRDB136, miRTarBase137, and mirDIP138. To ensure robust predictions, we considered only the gene targets shared by three or more databases (Supplementary Table 2). Further refinement narrowed down the essential gene targets for the miRNA signature associated with SGA and spaceflight to those common across 10 or more of the 13 miRNAs, resulting in 45 genes. Upset plots were made with ComplexUpset (ver.1.3.3) R package. Using Cytoscape’s139 ClueGo/CluePedia plugin140, we created a network map illustrating the connectivity between the genes and miRNAs. To explore the global pathways regulated by the 45 genes, we employed CluePedia, presenting the key pathways as a connected network. Additionally, a more detailed pathway interaction analysis was conducted using the GeneMANIA plugin141 in Cytoscape. GeneMANIA settings included 0 related genes, 50 attributes with automated weighting, revealing the top 50 pathways associated with the 45 genes.
We also performed additional pathway analysis using the Ingenuity Pathway Analysis (IPA) database (QIAGEN Inc., Redwood City, CA, www.qiagen.com/ingenuity) which predicts biological networks enriched in the dataset (Supplementary Fig. 8). Significantly differentially regulated miRNAs and their shared targets were subjected to pathways analysis using IPA as reported by us previously142. The enriched canonical pathways and upstream regulators was obtained using the pathway and upstream regulator analysis (subcellular layout) functions in IPA. The bar plot (Fig. 8a) was created with ggplot2 (ver 3.3.5) and the network display (Fig. 8b, c) were used to visualize the canonical pathways and upstream regulators respectively.
Analysis on plasma RNA-seq data from mice exposed to GCR radiation
We analyzed publicly available RNA-seq data from experiments that were done on 24-week-old male and female C57BL/6 J mice that were irradiated with 50 cGy simplified GCR simulated beam as described above. This data is available on NASA’s OSDR with identifier OSD-719143 and details for the experiment are described in the original publication by Burke et al.57. Briefly we will provide some of the details from these experiments. Plasma from the mice were collected 14 days post irradiation and total RNA was extracted using Trizol (Thermo Fisher) and purified with the Quick-RNA Miniprep Kit (Zymo Research). Libraries were prepared using Illumina’s total RNA kit according to the manufacturer’s instructions. Integrity of RNA (500 ng) was assessed using Agilent’s TapeStation and eRIN values greater than 7 were processed. Ribosomal RNA was depleted, first- and second-strand cDNA were synthesized, and adapter index ligation and strand selection were generated (Illumina Stranded Total RNA Prep with Ribo-Zero Plus). Unique indices (IDT for Illumina) were used for sample multiplexing. Libraries were amplified with a Mastercycler Pro (Eppendorf) and purified using RNAClean XP Agencourt beads (Beckman Coulter). Sequencing was carried out on a NextSeq1000 (Illumina) with 30 million, 75-base pair paired-end reads per sample. Raw reads were assessed for quality and trimmed using FastQC. Passed reads were quasi-mapped to the Ensembl (GRCm39) using Salmon v1.9.0 and imported into R using tximport. Normalized gene expression analysis on male and female blood samples exposed to 50 cGy radiation were compared to controls using DESeq2. A p-value less than 0.05 (adjusted for multiple testing using the Benjamini–Hochberg method) were considered significant genes.
For pathway analysis on the data (seen in Supplementary Figs. 7 and 9 with data available in Supplementary Data 1), we utilized fast Gene Set Enrichment Analysis (fGSEA)144. Pathway analysis was done utilizing both MSigDB gene sets (specifically, Hallmark and Reactome)28,145 and custom-made mitochondria-specific Gene Set file from MitoPathway33. Using fGSEA, all samples were compared to controls and the ranked list of genes was defined by the t-score statistics. The statistical significance was determined by 1000 permutations of the genesets146. We utilized a statistical cutoff of FDR < 0.05 and lollipop plots were made using ggplot2 (ver 3.3.5).
Meta-analysis of GeneLab RNA-seq data from 817 samples across 27 datasets encompassing 10 different mouse tissues
FASTQ files were programmatically downloaded from GeneLab/OSDR (https://www.nasa.gov/osdr/) and processed using the MTD pipeline147. A meta-analysis of the GeneLab data was performed in two steps. Firstly, we performed batch correction using Combat-seq148 to standardize data across different datasets. Subsequently, DESeq2128 was used to perform differential expression analysis between the spaceflight and ground control mice while adjusting for age, sex, tissue, sacrifice site, and mission duration. Then, we visually confirmed that the miRNA targets (top 45 or all) across datasets. The following GeneLab datasets were used: OSD-98, 99, 100, 101, 102, 103, 104, 105, 137, 161, 162, 163, 168, 194, 238, 239, 240, 241, 242, 243, 244, 245, 246, 247, 248, 253, and 254.
Inspiration4 (I4) astronaut sample collection
Detailed methods for sample collection and data processing are described in Overbey et al.149. and Kim et al.150. Blood samples were collected before (Pre-launch: L-92, L-44, and L-3) and after (Return; R + 1, R + 45, and R + 82) the spaceflight. Chromium Next GEM Single Cell 5′ v2, 10x Genomics was used to generate single-cell multi-omics data from isolated PBMCs. Subpopulations were annotated based on Azimuth human PBMC reference.
In summary, blood samples were collected before (prelaunch; L-92 days, 44 days, and 3 days) and after (return; R + 1 day, R + 45 days, and R + 82 days) the spaceflight. Top45 or all miRNA target genes suggested in the paper has been used for identifying DEGs from i4 PBMCs and subpopulations. The Seurat R packagewas used to normalize RNA count data and calculate average expression of each gene. We used the average expression of the genes from four astronauts comparing post-flight (R + 1) vs pre-flight (L-92, L-44, and L-3) to identify DEGs with the Wilcoxon signed-rank test (adjusted p-value < 0.05). Average expression of the miRNA target genes (Top45 or all) was used to generate heatmaps plotted by the pheatmap R package. Sex-specific analysis was performed with fGSEA using MSigDB gene sets (specifically, Hallmark and Reactome) and mitspecific Gene Set file from MitoPathway (data available in Supplementary Data 3). Using fGSEA, all samples were compared to controls and the ranked list of genes was defined by the t-score statistics. We utilized a statistical cutoff of FDR < 0.001 and lollipop plots were made using ggplot2 (ver 3.3.5). The I4 data can be found on NASA’s Open Science Data Repository (OSDR) with the following identifiers: OSD-570 (10.26030/41s1-j243).
The procedure followed guidelines set by Health Insurance Portability and Accountability Act (HIPAA) and operated under Institutional Review Board (IRB) approved protocols and informed consent was obtained. Experiments were conducted in accordance with local regulations and with the approval of the IRB at the Weill Cornell Medicine (IRB #21-05023569). All ethical regulations relevant to human research participants were followed.
Cumulative plots for the miRNA targets, and statistical test
Cumulative plots for miRNA gene targets were constructed by retrieving information on 8mer sites matching the seed region of miRNAs from the TargetScan v8.0 database151. mRNAs were categorized based on miRNA seed sequences (8mer, or ‘no site’), and the cumulative plot was generated using mRNA log2 fold change values with the ecdfplot method from seaborn152. To assess the distribution differences in mRNA fold change values between each seed match and the ‘no site’ scenario, the Kolmogorov–Smirnov test was calculated using the kstest function from scipy in Python153.
NASA Twin Study miRNA-seq analysis
Specific details related to all the methods related to the NASA Twin Study miRNA-seq data can be found in the following references47,64. Briefly, we will highlight the key methods. Small RNA libraries were prepared from 50 ng total RNA using the NEBNext Multiplex Small RNA Library Prep Set for Illumina (NEB #E7560) per manufacturer’s recommendations with the following modifications: adapters and RT primer were diluted fourfold, 17 cycles of PCR were used for library amplification, and no size selection was performed. The i7 primers in the NEBNext Multiplex Oligos for Illumina Dual Index Primers (NEB# E7600, NEB#E7780) were used to supplement the index primers in NEB #E7560. The libraries were sequenced in an Illumina NextSeq instrument (1x50bp).
Standard libraries were preprocessed and quality controlled using miRTrace8. Subsequently, reads were mapped against MirGeneDB sequences9 using the miRDeep210 quantifier module. Expression values were normalized to reads per million (RPM) considering only miRNA counts. The normalized RPM value was utilized for all analysis. If the value for the miRNA was zero for all samples that miRNA was excluded from the analysis. To determine statistically significant miRNAs, ANOVA analysis with p-value < 0.2 was independently performed for each cell type. For flight only comparisons, the same statistical significance was applied for all time points excluding the ground samples and all ambient return samples. All statistics were run independently for each cell condition/type. To determine the overlap of the miRNA signature in this paper with the Twin Study miRNA-seq data, the proposed miRNAs were used as well as all mature components of the miRNAs included in the miRNA family. Specific miRNAs related to globin pathways were plotted as heatmaps on the expression values using R package pheatmap version 1.0.12.
The NASA Twin Study subjects signed informed consent according to the declaration of Helsinki and 14 CFR Part 1230 for collection and use of sample materials and data in research protocols at NASA and the collaborating institutions. Study protocols were approved by the NASA Flight Institutional Review Board (protocol number Pro1245) and all participating institutions. Also the data is hosted at the NASA Life Sciences Data Archive (LSDA). Informed consent was provided by all participants in the NASA Twin Study. All ethical regulations relevant to human research participants were followed.
Machine learning analysis for small molecule drug predictions for targeting miRNAs
We used the sChemNET machine learning framework77 to predict small molecules that might affect miRNAs or their gene targets. sChemNET is a deep learning approach that takes as input the chemical structure of the small molecule in the form of a binary fingerprint and predicts a score that a given small molecule might affect a given miRNA. We trained sChemNET on Homo sapiens small molecule-miRNA data from the SM2miR database, from which we used 1,102 associations between 131 small molecules and 126 miRNA targets. Unlabeled small molecules were obtained from the Drug Repurposing Hub (see details in Galeano et al.77).
We also performed an enrichment analysis of the drug’s mode of action and indications from the obtained drugs with sChemNET. To this end, we trained sChemNET for each miRNA using all the available labeled small molecules and a randomly selected set of 2400 unlabeled small molecules. Then, sChemNET was used to rank the remaining set of unlabeled small molecules based on the average prediction score of 20 random independent repetitions. Small molecules amongst the 98th percentile score were then kept as predictions for the miRNA. We then retrieved the mode of action and indication information of small molecules from the Drug Repurposing Hub database. The enrichment score was calculated based on p-values calculated using Fisher’s Exact Test and adjusted with Benjamin-Hochberg correction for multiple testing.
Statistics and reproducibility
The statistics, samples sizes, and number of replicates for the analysis has been discussed throughout each section of the methods. The study utilized publicly available datasets from the ImmPort and GeneLab/OSDR platforms. For all omics analysis we used a minimum statistically stringency of p-value < 0.05 and when FDR < 0.05. For the SGA dataset (N = 29), differential miRNA expression was assessed using DESeq2 with a significance threshold of adjusted p-value < 0.05. For the GeneLab datasets (OSD-336 and OSD-55130), significant miRNAs were determined using FDR correction < 0.05, and various statistical tests (Fisher’s exact test, Chi-squared, Student’s t-test, ANOVA) were applied for differential expression analysis. The murine simulated space environment experiments included N = 10 mice per group, with consistent handling to minimize variability and ensure reproducibility. For miRNA pathway analysis, an FDR < 0.25 cutoff was used for identifying significant pathways. Comparative miRNA analysis between SGA and spaceflight data identified 13 common miRNAs, with stringent criteria for inclusion (adj. p-value < 0.05). The NASA Twin Study miRNA-seq data used ANOVA with p-value < 0.2 for flight-only comparisons. Machine learning predictions for small molecule-miRNA interactions used a 98th percentile score and were validated with enrichment analysis, applying Benjamini–Hochberg correction. Overall, the study maintained stringent statistical controls and used robust methodologies to ensure accurate and reproducible results across all datasets.
Supplementary information
Description of Additional Supplementary File
Acknowledgements
Part of the work was performed under funding from NASA’s Biological and Physical Sciences Division to NASA Ames under the Task: ID016GLS, Contract Number: NNA14AB82C awarded to A.B. C.E.M. thanks the Scientific Computing Unit (SCU) at WCM, the WorldQuant and GI Research Foundation, NASA (NNX14AH50G, NNX17AB26G, NNH18ZTT001N-FG2, 80NSSC22K0254, 80NSSC23K0832, the National Institutes of Health (R01ES032638, U54AG089334), and the LLS (7029-23). J.K. was supported by Basic Science Research Program through the National Research Foundation of Korea(NRF), funded by the Ministry of Education(RS-2023-00241586). C.E.M and J.K. acknowledges Boryung for their financial support and research enhancement ground, provided through their Global Space Healthcare Initiative, Humans In Space, including mentorship and access to relevant expert networks. L.I.G. was supported by the US Army Medical Research Command (award W81XWH2110402).
Author contributions
Conceptualization: A.B.; Methodology: A.B.; Formal Analysis: A.B., J.K. on all I4 data and Supplementary Fig. 10d, M.F. on preprocessing for Supplementary Fig. 10d, F.E. on miRNA conservation analysis, J.M.C. on blood RNA-seq data for male and female mice experiments, K.S. with generation of Supplementary Fig. 8 and D.G. with the ML model and results; Investigation: A.B., J.K., F.E., and D.G.; Inspiration4 (I4) data and omics: C.E.M. and J.K.; Resources: A.B.; Original Draft: A.B. and G.C.; Review & Editing: G.C., J.K, F.E., R.T.S., D.F., M.F., L.M.S., A.M., J.G., C.E.M., D.G., M.B., L.I.G., S.V.C., J.M.C., A.M.P., J.C-A, K.S., C.K.S., and A.B.; Figures and Visualization: A.B., J.K. (for Figs. 6 and 7, and Supplementary Figs. 10d and 11), F.E. (for Fig. 2d), J.G. (for Fig. 9), and D.G. (for Fig. 8); Funding Acquisition: A.B. and C.E.M. (for I4 study); Supervision: A.B.
Peer review
Peer review information
Communications Biology thanks Sung Hye Kim and Vasso Terzidou for their contribution to the peer review of this work. Primary Handling Editors: Kaliya Georgieva. A peer review file is available.
Data availability
The datasets used are publicly available via the NASA’s OSDR and NIAID’s ImmPort (www.immport.org) with identifiers: for miRNA data: OSD-55 (10.26030/9thk-dv75, OSD-334 (10.26030/cg2g-as49), OSD-335 (10.26030/72ke-1k67), OSD-336 (10.26030/qasarr29), and OSD-337 (10.26030/m73g-2477) from OSDR and SDY1871 (10.21430/M3TU8P1DQ0) on ImmPort; for genes/RNA_seq data: OSD-98 (10.26030/n1jq-2364), OSD-99 (10.26030/1h3m-3q49), OSD-100 (10.26030/whek-4p98), OSD-101 (10.26030/sdmt-ae51), OSD- 102 (10.26030/yn9m-2d19), OSD-103 (10.26030/9vzk-b116), OSD-104 (10.26030/em9rw619), OSD-105 (10.26030/xgw6-6t64), OSD-137 (10.26030/9k6w-4c28), OSD- 161 (10.26030/js8g-yq79), OSD-162 (10.26030/pys4-6t29), OSD-163 (10.26030/q8vt-7p92), OSD-168 (10.26030/rwyp-9325), OSD-194 (10.26030/pev7-5695), OSD-238 (10.26030/pev7-5695), OSD-239 (10.26030/s7k9-7958), OSD-240 (10.26030/s7k9-7958), OSD-241 (10.26030/6eg2-wz66), OSD-242 (10.26030/fmkc-8h31), OSD-243 (10.26030/jngj-0y14), OSD-244 (10.26030/34je-wd95), OSD-245 (10.26030/34jewd95), OSD-246 (10.26030/m33d-yq24), OSD-247 (10.26030/g9z7-7b61), OSD-248 (10.26030/d56m-7e45), OSD-253 (10.26030/4mx6-5x80), OSD-254 (10.26030/dcq8-6c70), and OSD-719 (10.26030/r998-cw71) from OSDR. The I4 data can be found on OSDR at OSD-570 (10.26030/41s1-j243).
Competing interests
The authors declare no competing interests.
Ethics and inclusion statement
This manuscript has included authors from all backgrounds from the scientific international community and the results are held at the highest ethical standards.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Giada Corti, JangKeun Kim.
Change history
10/16/2024
A Correction to this paper has been published: 10.1038/s42003-024-07022-7
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06944-6.
References
- 1.Mason, C. E. et al. A second space age spanning omics, platforms and medicine across orbits. Nature632, 995–1008 (2024). [DOI] [PMC free article] [PubMed]
- 2.Afshinnekoo, E. et al. Fundamental biological features of spaceflight: advancing the field to enable deep-space exploration. Cell183, 1162–1184 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rose, B. I. Female astronauts: Impact of space radiation on menopause. Eur. J. Obstet. Gynecol. Reprod. Biol.271, 210–213 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Reyes, D. P., Masterova, K. S., Walton, M., Kerstman, E. L. & Antonsen, E. L. Assessment of sex-dependent medical outcomes during spaceflight. J. Women’s Health (Larchmt.)31, 1145–1155 (2022). [DOI] [PubMed] [Google Scholar]
- 5.Mathyk, B. A. et al. Spaceflight induces changes in gene expression profiles linked to insulin and estrogen. Commun. Biol.7, 1–17 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thriving in Space: Ensuring the Future of Biological and Physical Sciences Research: A Decadal Survey for 2023–2032. (National Academies Press, Washington, D.C., 2023). 10.17226/26750. [PubMed]
- 7.Kim, S. H. et al. Maternal plasma miRNAs as potential biomarkers for detecting risk of small-for-gestational-age births. eBioMedicine62, 103145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campisi, S. C., Carbone, S. E. & Zlotkin, S. Catch-up growth in full-term small for gestational age infants: a systematic review. Adv. Nutr.10, 104–111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hokken-Koelega, A. C. S. et al. International consensus guideline on small for gestational age: etiology and management from infancy to early adulthood. Endocr. Rev.44, 539–565 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whincup, P. H. et al. Birth weight and risk of type 2 diabetes: a systematic review. JAMA300, 2886–2897 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Cauzzo, C., Chiavaroli, V., Di Valerio, S. & Chiarelli, F. Birth size, growth trajectory and later cardio-metabolic risk. Front. Endocrinol.14, 1187261 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leunissen, R. W. J., Kerkhof, G. F., Stijnen, T. & Hokken-Koelega, A. Timing and tempo of first-year rapid growth in relation to cardiovascular and metabolic risk profile in early adulthood. JAMA301, 2234–2242 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Wołejszo, S. et al. Insights into prevention of health complications in small for gestational Age (SGA) births in relation to maternal characteristics: a narrative review. J. Clin. Med.12, 531 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang, H.-W. & Mendell, J. T. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer94, 776–780 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonkoly, E. & Pivarcsi, A. microRNAs in inflammation. Int. Rev. Immunol.28, 535–561 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Rosolen, D. et al. MiRNAs action and impact on mitochondria function, metabolic reprogramming and chemoresistance of cancer cells: a systematic review. Biomedicines11, 693 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jovanovic, M. & Hengartner, M. O. miRNAs and apoptosis: RNAs to die for. Oncogene25, 6176–6187 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Friedman, R. C., Farh, K. K.-H., Burge, C. B. & Bartel, D. P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res.19, 92–105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, L. et al. Differentially expressed microRNAs and affected biological pathways revealed by modulated modularity clustering (MMC) analysis of human preeclamptic and IUGR placentas. Placenta34, 599–605 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hromadnikova, I., Kotlabova, K., Hympanova, L. & Krofta, L. Cardiovascular and cerebrovascular disease associated microRNAs are dysregulated in placental tissues affected with gestational hypertension, preeclampsia and intrauterine growth restriction. PLoS ONE10, e0138383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maccani, M. A. et al. Maternal cigarette smoking during pregnancy is associated with downregulation of miR-16, miR-21, and miR-146a in the placenta. Epigenetics5, 583–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higashijima, A. et al. Characterization of placenta-specific microRNAs in fetal growth restriction pregnancy. Prenat. Diagn.33, 214–222 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Berrios, D. C., Galazka, J., Grigorev, K., Gebre, S. & Costes, S. V. NASA GeneLab: interfaces for the exploration of space omics data. Nucleic Acids Res.49, D1515–D1522 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beheshti, A. miRNA signature detection and countermeasures against HZE radiation exposure for tissue degeneration-Plasma. NASA Open Science Data Repository. 10.26030/qasa-rr29 (2020).
- 25.Paul, A. M. et al. Beyond low-earth orbit: characterizing immune and microrna differentials following simulated deep spaceflight conditions in mice. iScience23, 101747 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simonsen, L. C., Slaba, T. C., Guida, P. & Rusek, A. NASA’s first ground-based Galactic Cosmic Ray Simulator: enabling a new era in space radiobiology research. PLoS Biol.18, e3000669 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girardi, C. et al. Integration analysis of microRNA and mRNA expression profiles in human peripheral blood lymphocytes cultured in modeled microgravity. Biomed. Res. Int.2014, 296747 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst.1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stalman, S. E. et al. Genetic analyses in small-for-gestational-age newborns. J. Clin. Endocrinol. Metab.103, 917–925 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Liu, Z. et al. USP22 regulates the formation and function of placental vasculature during the development of fetal growth restriction. Placenta111, 19–25 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Lin, F. et al. The maternal–fetal interface in small-for-gestational-age pregnancies is associated with a reduced quantity of human decidual NK cells with weaker functional ability. Front. Cell Dev. Biol.8, 633 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.da Silveira, W. A. et al. Comprehensive multi-omics analysis reveals mitochondrial stress as a central biological hub for spaceflight impact. Cell183, 1185–1201.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rath, S. et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res.49, D1541–D1547 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mimaki, M., Wang, X., McKenzie, M., Thorburn, D. R. & Ryan, M. T. Understanding mitochondrial complex I assembly in health and disease. Biochim. Biophys. Acta1817, 851–862 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Zhao, Q., Sun, Q., Zhou, L., Liu, K. & Jiao, K. Complex regulation of mitochondrial function during cardiac development. J. Am. Heart Assoc.8, e012731 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergman, O. & Ben-Shachar, D. Mitochondrial oxidative phosphorylation system (OXPHOS) deficits in schizophrenia: possible interactions with cellular processes. Can. J. Psychiatry61, 457–469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valsecchi, F. et al. Complex I disorders: causes, mechanisms, and development of treatment strategies at the cellular level. Dev. Disabil. Res. Rev.16, 175–182 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Ghezzi, D. & Zeviani, M. Human diseases associated with defects in assembly of OXPHOS complexes. Essays Biochem.62, 271–286 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zorov, D. B., Juhaszova, M. & Sollott, S. J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev.94, 909–950 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu, J., Qian, X. & Ding, R. MiR-24-3p attenuates IL-1β-induced chondrocyte injury associated with osteoarthritis by targeting BCL2L12. J. Orthop. Surg. Res.16, 371 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, K., Huang, X.-T., Miao, Y.-P., Bai, X.-L. & Jin, F. MiR-148a-3p attenuates apoptosis and inflammation by targeting CNTN4 in atherosclerosis. Ann. Transl. Med.10, 1201 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, Y. et al. MicroRNA-24-3p alleviates cardiac fibrosis by suppressing cardiac fibroblasts mitophagy via downregulating PHB2. Pharmacol. Res.177, 106124 (2022). [DOI] [PubMed] [Google Scholar]
- 43.He, D. & Yan, L. MiR-29b-3p aggravates cardiac hypoxia/reoxygenation injury via targeting PTX3. Cytotechnology73, 91–100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ponnusamy, V. et al. Neuronal let-7b-5p acts through the Hippo-YAP pathway in neonatal encephalopathy. Commun. Biol.4, 1143 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding, J. et al. Extracellular vesicles derived from M1 macrophages deliver miR-146a-5p and miR-146b-5p to suppress trophoblast migration and invasion by targeting TRAF6 in recurrent spontaneous abortion. Theranostics11, 5813–5830 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakrabortty, A., Patton, D. J., Smith, B. F. & Agarwal, P. miRNAs: potential as biomarkers and therapeutic targets for cancer. Genes14, 1375 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malkani, S. et al. Circulating miRNA spaceflight signature reveals targets for countermeasure development. Cell Rep.33, 108448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McDonald, J. T. et al. Role of miR-2392 in driving SARS-CoV-2 infection. Cell Rep.37, 109839 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beheshti, A. et al. Identification of circulating serum multi-microRNA signatures in human DLBCL models. Sci. Rep.9, 17161 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Östling, H., Kruse, R., Helenius, G. & Lodefalk, M. Placental expression of microRNAs in infants born small for gestational age. Placenta81, 46–53 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Lee, C.-T., Risom, T. & Strauss, W. M. Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol.26, 209–218 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Zhang, Y. Encyclopedia of Systems Biology (eds. Dubitzky, W., Wolkenhauer, O., Cho, K.-H. & Yokota, H.) 1735–1736 (Springer, 2013).
- 53.Weber, M. J. New human and mouse microRNA genes found by homology search. FEBS J.272, 59–73 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Jeong, H. R. et al. Exosomal miRNA profile in small-for-gestational-age children: a potential biomarker for catch-up growth. Genes13, 938 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delforce, S. J., Lumbers, E. R. & Pringle, K. G. Regulation of the prorenin - angiotensin system by oxygen and miRNAs; parallels between placentation and tumour development? Placenta56, 27–33 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Bartho, L. A., Fisher, J. J., Walton, S. L., Perkins, A. V. & Cuffe, J. S. M. The effect of gestational age on mitochondrial properties of the mouse placenta. Reprod. Fertil.3, 19–29 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burke, M. et al. Sexual Dimorphism during Integrative Endocrine and Immune Responses to Ionizing Radiation in Mice. Sci. Rep.14, 7334 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waggoner, D. J. et al. NSD1 analysis for Sotos syndrome: insights and perspectives from the clinical laboratory. Genet. Med.7, 524–533 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Chang, C., Lee, S. O., Wang, R.-S., Yeh, S. & Chang, T.-M. Androgen receptor (AR) physiological roles in male and female reproductive systems: lessons learned from AR-knockout mice lacking AR in selective cells. Biol. Reprod.89, 21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hemmings, B. A. & Restuccia, D. F. PI3K-PKB/Akt pathway. Cold Spring Harb. Perspect. Biol.4, a011189 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kalous, J., Aleshkina, D. & Anger, M. A role of PI3K/Akt signaling in oocyte maturation and early embryo development. Cells12, 1830 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stokkeland, K., Ebrahim, F., Hultcrantz, R., Ekbom, A. & Stephansson, O. Mothers with alcoholic liver disease and the risk for preterm and small-for-gestational-age birth. Alcohol Alcohol.48, 166–171 (2013). [DOI] [PubMed] [Google Scholar]
- 63.Beheshti, A. et al. Multi-omics analysis of multiple missions to space reveal a theme of lipid dysregulation in mouse liver. Sci. Rep.9, 19195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garrett-Bakelman, F. E. et al. The NASA Twins Study: a multidimensional analysis of a year-long human spaceflight. Science364, eaau8650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petri, B. J. et al. Multiomics analysis of the impact of polychlorinated biphenyls on environmental liver disease in a mouse model. Environ. Toxicol. Pharmacol.94, 103928 (2022). [DOI] [PubMed] [Google Scholar]
- 66.Deng, Q. et al. Co-exposure to metals and polycyclic aromatic hydrocarbons, microRNA expression, and early health damage in coke oven workers. Environ. Int.122, 369–380 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Huang, S. et al. Polycyclic aromatic hydrocarbons-associated microRNAs and heart rate variability in coke oven workers. J. Occup. Environ. Med.58, e24–e31 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Steinborn, A. et al. Small for gestational age (SGA) neonates show reduced suppressive activity of their regulatory T cells. Clin. Immunol.134, 188–197 (2010). [DOI] [PubMed] [Google Scholar]
- 69.Gomez-Lopez, N. et al. Regulatory T cells play a role in a subset of idiopathic preterm labor/birth and adverse neonatal outcomes. Cell Rep.32, 107874 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bendix, I., Miller, S. L. & Winterhager, E. Editorial: causes and consequences of intrauterine growth restriction. Front Endocrinol. (Lausanne)11, 205 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roberts, D., Brown, J., Medley, N. & Dalziel, S. R. Antenatal corticosteroids for accelerating fetal lung maturation for women at risk of preterm birth. Cochrane Database Syst. Rev.3, CD004454 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arias, A. et al. Dexamethasone-induced intrauterine growth restriction modulates expression of placental vascular growth factors and fetal and placental growth. Mol. Hum. Reprod.27, gaab006 (2021). [DOI] [PubMed] [Google Scholar]
- 73.Liu, C., Liu, C., Wang, Q. & Zhang, Z. Supplementation of folic acid in pregnancy and the risk of preeclampsia and gestational hypertension: a meta-analysis. Arch. Gynecol. Obstet.298, 697–704 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roth, D. E. et al. Vitamin D supplementation in pregnancy and lactation and infant growth. N. Engl. J. Med.379, 535–546 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Joo, E. H. et al. Effect of endogenic and exogenic oxidative stress triggers on adverse pregnancy outcomes: preeclampsia, fetal growth restriction, gestational diabetes mellitus and preterm birth. Int. J. Mol. Sci.22, 10122 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alawadhi, M., Mouihate, A., Kilarkaje, N. & Al-Bader, M. Progesterone partially recovers placental glucose transporters in dexamethasone-induced intrauterine growth restriction. Reprod. Biomed. Online44, 595–607 (2022). [DOI] [PubMed] [Google Scholar]
- 77.Galeano, D. et al. sChemNET: a deep learning framework for predicting small molecules targeting microRNAs. Nat. Commun. (in press) (2024).
- 78.Haidara, M. A., Yassin, H. Z., Rateb, M., Ammar, H. & Zorkani, M. A. Role of oxidative stress in development of cardiovascular complications in diabetes mellitus. Curr. Vasc. Pharmacol.4, 215–227 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Chatuphonprasert, W., Jarukamjorn, K. & Ellinger, I. Physiology and pathophysiology of steroid biosynthesis, transport and metabolism in the human placenta. Front Pharm.9, 1027 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen, B., Chen, Y. & Xu, Y. Vitamin D deficiency in pregnant women: influenced by multiple risk factors and increase the risks of spontaneous abortion and small-for-gestational age. Medicine100, e27505 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Som, M. & Stroup, J. S. Primary hyperparathyroidism and pregnancy. Proc. (Bayl. Univ. Med Cent.)24, 220–223 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith, S. M. et al. Space flight calcium: implications for astronaut health, spacecraft operations, and earth. Nutrients4, 2047–2068 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mishra, B. & Luderer, U. Reproductive hazards of space travel in women and men. Nat. Rev. Endocrinol.15, 713–730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peters, M. C. et al. The impact of insulin resistance on loss of lung function and response to treatment in asthma. Am. J. Respir. Crit. Care Med.206, 1096–1106 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gaffin, J. M. et al. Determinants of lung function across childhood in the Severe Asthma Research Program (SARP) 3. J. Allergy Clin. Immunol.151, 138–146.e9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen, T.-Y. et al. Development of triamcinolone acetonide-hyaluronic acid conjugates with selective targeting and less osteoporosis effect for rheumatoid arthritis treatments. Int. J. Biol. Macromol.237, 124047 (2023). [DOI] [PubMed] [Google Scholar]
- 87.Freire, F. S., Lang, R., Abalem, M. F. & Johnson, M. W. Retinal deposits of triamcinolone-moxifloxacin after dropless cataract surgery. Retin. Cases Brief. Rep.17, 577 (2023). [DOI] [PubMed] [Google Scholar]
- 88.Tamimi, F. et al. Perfluorodecalin and bone regeneration. Eur. Cell Mater.25, 22–36 (2013). [DOI] [PubMed] [Google Scholar]
- 89.Walker, G. M. et al. Early perfluorodecalin lung distension in infants with congenital diaphragmatic hernia. J. Pediatr. Surg.38, 17–20 (2003). [DOI] [PubMed] [Google Scholar]
- 90.Aramendia, I. et al. Experimental and numerical modeling of aerosol delivery for preterm infants. Int J. Environ. Res. Public Health15, 423 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Solan, M. E., Koperski, C. P., Senthilkumar, S. & Lavado, R. Short-chain per- and polyfluoralkyl substances (PFAS) effects on oxidative stress biomarkers in human liver, kidney, muscle, and microglia cell lines. Environ. Res.223, 115424 (2023). [DOI] [PubMed] [Google Scholar]
- 92.Aimuzi, R. et al. Perfluoroalkyl and polyfluroalkyl substances and maternal thyroid hormones in early pregnancy. Environ. Pollut.264, 114557 (2020). [DOI] [PubMed] [Google Scholar]
- 93.Céline, C., Catherine, B., Romane, C. & Laurence, C. Per- and polyfluoroalkyls used as cosmetic ingredients - Qualitative study of 765 cosmetic products. Food Chem. Toxicol.187, 114625 (2024). [DOI] [PubMed] [Google Scholar]
- 94.Steller, J. G., Alberts, J. R. & Ronca, A. E. Oxidative stress as cause, consequence, or biomarker of altered female reproduction and development in the space environment. Int. J. Mol. Sci.19, 3729 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kroener, L., Wang, E. T. & Pisarska, M. D. Predisposing factors to abnormal first trimester placentation and the impact on fetal outcomes. Semin. Reprod. Med.34, 27–35 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Burton, G. J., Jauniaux, E. & Murray, A. J. Oxygen and placental development; parallels and differences with tumour biology. Placenta56, 14–18 (2017). [DOI] [PubMed] [Google Scholar]
- 97.Kamity, R., Sharma, S. & Hanna, N. MicroRNA-mediated control of inflammation and tolerance in pregnancy. Front. Immunol.10, 718 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Légaré, C. et al. Human plasma pregnancy-associated miRNAs and their temporal variation within the first trimester of pregnancy. Reprod. Biol. Endocrinol.20, 14 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schoots, M. H., Gordijn, S. J., Scherjon, S. A., van Goor, H. & Hillebrands, J.-L. Oxidative stress in placental pathology. Placenta69, 153–161 (2018). [DOI] [PubMed] [Google Scholar]
- 100.Chappell, L. C. et al. Vitamin C and E supplementation in women at risk of preeclampsia is associated with changes in indices of oxidative stress and placental function. Am. J. Obstet. Gynecol.187, 777–784 (2002). [DOI] [PubMed] [Google Scholar]
- 101.Jhamb, I. et al. Evaluation of vitamin E isoforms in placental tissue and their relationship with maternal dietary intake and plasma concentrations in mother–infant dyads. Antioxidants12, 1797 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pereira, A. C. & Martel, F. Oxidative stress in pregnancy and fertility pathologies. Cell Biol. Toxicol.30, 301–312 (2014). [DOI] [PubMed] [Google Scholar]
- 103.Gómez-Puyou, A., Peña-Días, A., Guzmán-García, J. & Laguna, J. Effect of triamcinolone and other steroids on the oxidative phosphorylation reaction. Biochem. Pharmacol.12, 331–340 (1963). [DOI] [PubMed] [Google Scholar]
- 104.Ventura-Clapier, R., Piquereau, J., Veksler, V. & Garnier, A. Estrogens, estrogen receptors effects on cardiac and skeletal muscle mitochondria. Front. Endocrinol. (Lausanne)10, 557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Klinge, C. M. Estrogenic control of mitochondrial function. Redox Biol.31, 101435 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Czubryt, M. P., Espira, L., Lamoureux, L. & Abrenica, B. The role of sex in cardiac function and diseaseThis paper is one of a selection of papers published in this Special Issue, entitled Young Investigator’s Forum. Can. J. Physiol. Pharmacol.84, 93–109 (2006). [DOI] [PubMed] [Google Scholar]
- 107.Appelman, Y., van Rijn, B. B., Ten Haaf, M. E., Boersma, E. & Peters, S. A. E. Sex differences in cardiovascular risk factors and disease prevention. Atherosclerosis241, 211–218 (2015). [DOI] [PubMed] [Google Scholar]
- 108.Jain, V. & Wotring, V. E. Medically induced amenorrhea in female astronauts. npj Microgravity2, 1–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lopez-Ruiz, A., Sartori-Valinotti, J., Yanes, L. L., Iliescu, R. & Reckelhoff, J. F. Sex differences in control of blood pressure: role of oxidative stress in hypertension in females. Am. J. Physiol. Heart Circ. Physiol.295, H466–H474 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Reckelhoff, J. F. Gender differences in the regulation of blood pressure. Hypertension37, 1199–1208 (2001). [DOI] [PubMed] [Google Scholar]
- 111.Hong, X. et al. Effects of spaceflight aboard the International Space Station on mouse estrous cycle and ovarian gene expression. npj Microgravity7, 1–8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Purandare, N. et al. Molecular mechanisms regulating lysophosphatidylcholine acyltransferase 1 (LPCAT1) in human pregnancy. Placenta106, 40–48 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Risberg, B. Adhesions: preventive strategies. Eur. J. Surg. Suppl.577, 32–39 (1997). [PubMed]
- 114.Bérard, A., Sheehy, O., Kurzinger, M.-L. & Juhaeri, J. Intranasal triamcinolone use during pregnancy and the risk of adverse pregnancy outcomes. J. Allergy Clin. Immunol.138, 97–104.e7 (2016). [DOI] [PubMed] [Google Scholar]
- 115.Castro, C. I. & Briceno, J. C. Perfluorocarbon-based oxygen carriers: review of products and trials. Artif. Organs34, 622–634 (2010). [DOI] [PubMed] [Google Scholar]
- 116.Li, S., Pang, K., Zhu, S., Pate, K. & Yin, J. Perfluorodecalin-based oxygenated emulsion as a topical treatment for chemical burn to the eye. Nat. Commun.13, 7371 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li, J. et al. A topical aqueous oxygen emulsion stimulates granulation tissue formation in a porcine second-degree burn wound. Burns41, 1049–1057 (2015). [DOI] [PubMed] [Google Scholar]
- 118.Murgia, X., Mielgo, V., Valls-i-Soler, A., Ruiz-del-Yerro, E. & Rey-Santano, C. Aerosolized perfluorocarbon improves gas exchange and pulmonary mechanics in preterm lambs with severe respiratory distress syndrome. Pediatr. Res.72, 393–399 (2012). [DOI] [PubMed] [Google Scholar]
- 119.Saatci, A. O. & Koçak, N. Retained submacular perfluorodecalin. Can. J. Ophthalmol.38, 293–296 (2003). [PubMed] [Google Scholar]
- 120.Ji, C. et al. Mitochondria-targeted and ultrasound-responsive nanoparticles for oxygen and nitric oxide codelivery to reverse immunosuppression and enhance sonodynamic therapy for immune activation. Theranostics11, 8587–8604 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sainty, R., Silver, M. J., Prentice, A. M. & Monk, D. The influence of early environment and micronutrient availability on developmental epigenetic programming: lessons from the placenta. Front. Cell Dev. Biol.11, 1212199 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Silvagno, F. & Pescarmona, G. Spotlight on vitamin D receptor, lipid metabolism and mitochondria: Some preliminary emerging issues. Mol. Cell. Endocrinol.450, 24–31 (2017). [DOI] [PubMed] [Google Scholar]
- 123.Iwamoto, J., Takeda, T. & Sato, Y. Interventions to prevent bone loss in astronauts during space flight. Keio J. Med.54, 55–59 (2005). [DOI] [PubMed] [Google Scholar]
- 124.Smith, S. M., Zwart, S. R., Block, G., Rice, B. L. & Davis-Street, J. E. The nutritional status of astronauts is altered after long-term space flight aboard the International Space Station. J. Nutr.135, 437–443 (2005). [DOI] [PubMed] [Google Scholar]
- 125.Chan, S. Y. et al. Vitamin D promotes human extravillous trophoblast invasion in vitro. Placenta36, 403–409 (2015). [DOI] [PubMed] [Google Scholar]
- 126.Dahma, G. et al. The effects of vitamin D supplementation before 20 weeks of gestation on preeclampsia: a systematic review. J. Pers. Med.13, 996 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bhattacharya, S. et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci. Data5, 180015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lanfranchi, G. et al. microRNA expression profiles in human peripheral blood lymphocytes cultured in modeled microgravity. NASA Open Science Data Repository. 10.26030/9thk-dv75 (2014).
- 130.Pizzamiglio, S. et al. A methodological procedure for evaluating the impact of hemolysis on circulating microRNAs. Oncol. Lett.13, 315–320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang, J. & Storey, K. B. RBiomirGS: an all-in-one miRNA gene set analysis solution featuring target mRNA mapping and expression profile integration. PeerJ6, e4262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kozomara, A., Birgaoanu, M. & Griffiths-Jones, S. miRBase: from microRNA sequences to function. Nucleic Acids Res.47, D155–D162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chang, L. & Xia, J. MicroRNA regulatory network analysis using miRNet 2.0. Methods Mol. Biol.2594, 185–204 (2023). [DOI] [PubMed] [Google Scholar]
- 134.Vejnar, C. E. & Zdobnov, E. M. MiRmap: comprehensive prediction of microRNA target repression strength. Nucleic Acids Res.40, 11673–11683 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sticht, C., Torre, C. D. L., Parveen, A. & Gretz, N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS ONE13, e0206239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen, Y. & Wang, X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res.48, D127–D131 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huang, H.-Y. et al. miRTarBase update 2022: an informative resource for experimentally validated miRNA–target interactions. Nucleic Acids Res.50, D222–D230 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tokar, T. et al. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res.46, D360–D370 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bindea, G., Galon, J. & Mlecnik, B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics29, 661–663 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Warde-Farley, D. et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res.38, W214–W220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Singh, K. et al. Genome-wide DNA hypermethylation opposes healing in patients with chronic wounds by impairing epithelial-mesenchymal transition. J. Clin. Invest.132, e157279 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Paul, A. Sexual dimorphism during integrative endocrine and immune responses to ionizing radiation in mice – blood data. NASA Open Sci. Data Repos.14, 7334 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Korotkevich, G. et al. Fast gene set enrichment analysis. 060012 Preprint at 10.1101/060012 (2021).
- 145.Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wu, F., Liu, Y.-Z. & Ling, B. MTD: a unique pipeline for host and meta-transcriptome joint and integrative analyses of RNA-seq data. Brief. Bioinform.23, bbac111 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang, Y., Parmigiani, G. & Johnson, W. E. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform.2, lqaa078 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Overbey, E. G. et al. The Space Omics and Medical Atlas (SOMA) and international astronaut biobank. Nature632, 1145–1154 (2024). [DOI] [PMC free article] [PubMed]
- 150.Kim, J. et al. Single-cell multi-ome and immune profiles of the Inspiration4 crew reveal conserved, cell-type, and sex-specific responses to spaceflight. Nat. Commun.15, 4954 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.McGeary, S. E. et al. The biochemical basis of microRNA targeting efficacy. Science366, eaav1741 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Waskom, M. L. Seaborn: Statistical Data Visualization. J. Open Source Softw.6, 3021 (2021). [Google Scholar]
- 153.Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary File
Data Availability Statement
The datasets used are publicly available via the NASA’s OSDR and NIAID’s ImmPort (www.immport.org) with identifiers: for miRNA data: OSD-55 (10.26030/9thk-dv75, OSD-334 (10.26030/cg2g-as49), OSD-335 (10.26030/72ke-1k67), OSD-336 (10.26030/qasarr29), and OSD-337 (10.26030/m73g-2477) from OSDR and SDY1871 (10.21430/M3TU8P1DQ0) on ImmPort; for genes/RNA_seq data: OSD-98 (10.26030/n1jq-2364), OSD-99 (10.26030/1h3m-3q49), OSD-100 (10.26030/whek-4p98), OSD-101 (10.26030/sdmt-ae51), OSD- 102 (10.26030/yn9m-2d19), OSD-103 (10.26030/9vzk-b116), OSD-104 (10.26030/em9rw619), OSD-105 (10.26030/xgw6-6t64), OSD-137 (10.26030/9k6w-4c28), OSD- 161 (10.26030/js8g-yq79), OSD-162 (10.26030/pys4-6t29), OSD-163 (10.26030/q8vt-7p92), OSD-168 (10.26030/rwyp-9325), OSD-194 (10.26030/pev7-5695), OSD-238 (10.26030/pev7-5695), OSD-239 (10.26030/s7k9-7958), OSD-240 (10.26030/s7k9-7958), OSD-241 (10.26030/6eg2-wz66), OSD-242 (10.26030/fmkc-8h31), OSD-243 (10.26030/jngj-0y14), OSD-244 (10.26030/34je-wd95), OSD-245 (10.26030/34jewd95), OSD-246 (10.26030/m33d-yq24), OSD-247 (10.26030/g9z7-7b61), OSD-248 (10.26030/d56m-7e45), OSD-253 (10.26030/4mx6-5x80), OSD-254 (10.26030/dcq8-6c70), and OSD-719 (10.26030/r998-cw71) from OSDR. The I4 data can be found on OSDR at OSD-570 (10.26030/41s1-j243).