Abstract
Metabolic syndrome (MetS) is a complex hereditary condition comprising various metabolic traits as risk factors. Although the genetics of individual MetS components have been investigated actively through large-scale genome-wide association studies, the conjoint genetic architecture has not been fully elucidated. Here, we performed the largest multivariate genome-wide association study of MetS in Europe (nobserved = 4,947,860) by leveraging genetic correlation between MetS components. We identified 1,307 genetic loci associated with MetS that were enriched primarily in brain tissues. Using transcriptomic data, we identified 11 genes associated strongly with MetS. Our phenome-wide association and Mendelian randomization analyses highlighted associations of MetS with diverse diseases beyond cardiometabolic diseases. Polygenic risk score analysis demonstrated better discrimination of MetS and predictive power in European and East Asian populations. Altogether, our findings will guide future studies aimed at elucidating the genetic architecture of MetS.
Subject terms: Genome-wide association studies, Metabolic disorders, Population genetics
Large-scale multivariate analyses across populations of European ancestry identify risk loci for the metabolic syndrome, improving polygenic prediction models and highlighting associations with diverse traits beyond cardiometabolic diseases.
Main
MetS is a cluster of interrelated risk factors that predispose individuals to cardiovascular disease (CVD) and type 2 diabetes (T2D). These factors, known as MetS components, include central obesity, dyslipidemia, hypertension (HTN) and impaired glucose tolerance1. They often run within families, suggesting a shared genetic basis. MetS components are moderately heritable, with heritability estimates ranging from 0.26 for T2D2 to 0.61 for HTN3, with MetS having a heritability of 0.10–0.30 (ref. 4). Recent genome-wide association studies (GWASs) revealed numerous genetic loci associated with individual MetS components. For example, a GWAS conducted by the Global Lipids Genetics Consortium has identified 380 and 388 genetic variants associated with high-density lipoprotein cholesterol (HDL) and triglycerides (TG), respectively, in populations of European ancestry5. However, the genetic basis of the shared risk across these components remains unclear.
The co-occurrence of unhealthy metabolic traits has prompted ongoing endeavors to unveil their common genetic underpinnings. For instance, five categories of T2D mechanistic pathways were identified using T2D-associated variants through clustering analyses, and the genes and traits associated with each cluster were investigated6,7. Similarly, colocalization analyses were conducted between cardiometabolic traits and quantitative trait loci (QTL) to identify shared genes across various traits8. However, these clustering analyses were constrained to T2D-associated variants, and the mere overlap between colocalized genes may lack substantial evidence of a shared genetic basis across all components of MetS.
Most previous MetS GWAS have focused on a binary definition of MetS. Kraja et al.9 expanded on this by conducting a GWAS for MetS and pairwise combinations of its components and identified 29 common variant associations; however, these studies lacked robust evidence for a consistent association across all MetS components. Lind10 examined 291,107 individuals from the UK Biobank (UKB) and identified 93 independent loci associated with MetS. Despite the different definitions of MetS, both studies categorized MetS based on the number of MetS criteria met. Such an approach may potentially introduce variability due to the different combinations of criteria met, thus limiting the representativeness of MetS and leading to an incomplete understanding of the genetic architecture of MetS11.
Multivariate GWAS, such as genomic structural equation modeling (Genomic SEM)12 could be a more suitable approach for studying the broad genetic liability across several phenotypes. Genomic SEM incorporates a genetic covariance structure to model the shared genetic architecture across indicators with several advantages, such as accommodating varying and unknown sample overlaps, flexibility in constructing factor models, and comparing the fit indices across different models. Additionally, it has been used widely to distinguish the shared genetic architecture of several diseases, ranging from psychiatric disorders13,14 to immune diseases15.
In this study, we investigated the common genetic liability for MetS in a European population by constructing a comprehensive factor model using large-scale GWASs with an observed sample size of 4,947,860 individuals. We examined the genetic relationships between key indicators, including body mass index (BMI), waist circumference (WC), T2D, fasting glucose (FG), HTN, HDL and TG, using Genomic SEM. Extensive functional annotation, enrichment analyses and gene prioritization were performed to determine the underlying biological mechanisms. We evaluated the performance of the MetS polygenic risk score (PRS) in European and East Asian populations. Because MetS is highly interrelated with various disorders16, we examined its association with health outcomes using a phenome-wide association study with PRS (PRS-PheWAS). We further explored the causal relationship between significantly associated health outcomes using two-sample Mendelian randomization (TSMR). Collectively, our results provided new insights into the complex genetic structure of MetS (Supplementary Fig. 1).
Results
Factor analysis of MetS components
We collected summary statistics from previous GWASs conducted in European populations for seven MetS components: BMI, WC, T2D, FG, HTN, HDL, and TG (Supplementary Table 1). Quality control was applied to the GWAS summary statistics, where single-nucleotide polymorphisms (SNPs) were filtered based on six criteria (Supplementary Table 2; Methods). We conducted a meta-analysis on T2D and HTN to create a GWAS with a larger sample size and greater statistical power. Minimal evidence of sample overlap was observed between the GWAS summary statistics included in the meta-analysis, as indicated by the bivariate linkage disequilibrium score (LDSC) regression17,18 intercept (Supplementary Table 3).
The effective sample sizes (neff) of the seven MetS components ranged from 151,188 to 1,253,277. Although our findings were based on GWAS summary statistics for UKB individuals, we prepared an additional set of GWAS summary statistics (referred to as the UKB-excluded cohort) to avoid sample overlap when conducting the following post-GWAS analyses: PRS analyses, PRS-PheWAS and TSMR. The univariate LDSC regression intercept of each GWAS summary statistic suggested polygenicity rather than bias from cryptic confounders such as population stratification (Supplementary Table 4).
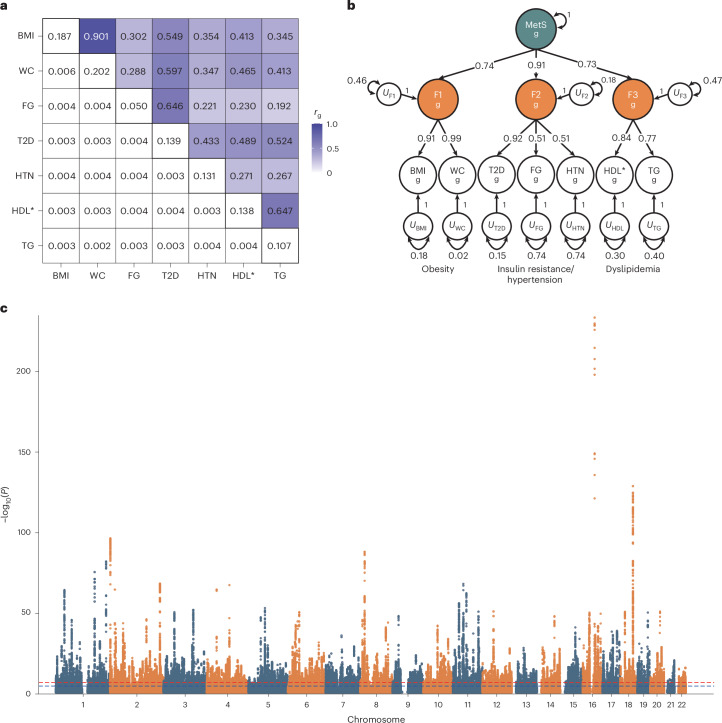
The genetic correlations among all pairs of MetS components were computed using LDSC regression (Fig. 1a). These correlations were then used for the Kaiser–Meyer–Olkin (KMO)19 test and parallel analysis. The KMO test estimated a sampling adequacy of 0.73, suggesting that the data were suitable for subsequent factor analysis. Parallel analysis suggested that the extraction of the three factors was optimal (Supplementary Table 5 and Supplementary Fig. 2). We then conducted exploratory factor analysis (EFA) and concluded that the three-factor solution was optimal, accounting for 70.2% of the variance in the seven MetS components (Supplementary Table 6).
Fig. 1. Genetic correlations, multivariate genetic factor model and multivariate GWAS of MetS.
a, SNP-based heritability and pairwise genetic correlations for the seven MetS components were estimated using LD score regression. The off-diagonal upper, lower and diagonal triangles represent the pairwise genetic correlation, the standard error of the genetic correlation and SNP-based heritability, respectively. b, Path diagram with standardized factor loadings in the hierarchical model estimated using Genomic SEM. The U represents the residual variance that is not explained by the latent factor. The subscript g indicates that the model was built based on genetic covariances between the MetS components. c, Manhattan plot of Genomic SEM-based GWAS associations. The x axis represents the chromosomal position, and the y axis represents the uncorrected −log10(P) from two-sided z tests for SNP associations with MetS. *Reverse-coded.
Before conducting confirmatory factor analysis (CFA) based on the EFA results using Genomic SEM12, we examined whether a single latent construct could represent MetS by constructing a common factor model. However, the model fit indices were insufficient, suggesting that MetS is a complex trait that cannot be described adequately by a common factor (Supplementary Table 7).
Subsequently, we constructed a correlated three-factor model based on the EFA results, and it exhibited a good model fit (χ2(11) = 185.98; Akaike information criterion (AIC) = 219.98; comparative fit index (CFI) = 0.981; standardized root mean square residual (SRMR) = 0.043) (Supplementary Table 8). The latent factors were clustered with MetS components having similar characteristics and labeled as follows: F1, obesity; F2, insulin resistance/hypertension and F3, dyslipidemia. Since a moderate genetic correlation was observed between the three latent factors and the goal of this study was to define and identify shared genetic effects for MetS among its components, we further constructed a hierarchical factor model (χ2(11) = 185.98; AIC = 219.98; CFI = 0.981; SRMR = 0.043) (Fig. 1b and Supplementary Table 9). An identical model could be constructed using UKB-excluded cohorts, which showed a good fit (χ2(11) = 107.09; AIC = 141.09; CFI = 0.996; SRMR = 0.036) (Supplementary Tables 10 and 11).
Association analyses using multivariate GWAS for MetS
Genomic SEM12 was used to conduct a multivariate GWAS to analyze the association between SNPs and MetS (Fig. 1c and Supplementary Fig. 3). The neff of the MetS factor was estimated to be 1,384,348 and the SNP-based heritability (h2) was estimated to be 0.11. We identified 1,650 lead SNPs for MetS across 939 genetic loci using the FUMA standard clumping method20. We further conducted a conditional and joint analysis (COJO)21 on 1,650 MetS lead SNPs to report statistically significant and independent SNPs, of which 1,307 COJO MetS SNPs were identified (Supplementary Table 12). Among them, 26 were nonsynonymous, 44 had a RegulomeDB score of 1 (indicating a high likelihood of having a regulatory function) and 19 were located in the active transcription start site (TSS) with the highest accessibility. Moreover, 414 genes were mapped using three gene-mapping strategies, including positional mapping, expression quantitative trait loci mapping (eQTL) mapping and chromatin interaction mapping, in the FUMA and MAGMA gene-based analyses (Supplementary Note, Supplementary Tables 13–15 and Supplementary Figs. 4 and 5).
We then assessed the independence of the COJO MetS SNPs, with a window size of 500 kb and r2 threshold of <0.01, from previously reported signals. Among the 1,307 COJO MetS SNPs, 82 (6.3%) were independent of the GWAS signals of the seven MetS components included in the Genomic SEM, 848 (64.9%) were independent of previous MetS GWAS (Lind10 and van Walree et al.22), and 159 (12.2%) were previously unreported in the NHGRI-EBI GWAS Catalog23 using FUMA (Table 1, Supplementary Tables 11 and 16 and Supplementary Fig. 6). The GWAS effect of COJO MetS SNPs exhibited a high correlation with GWAS results conducted using UKB-excluded cohorts (Pearson’s correlation (ρ) of 0.95, 95% confidence interval (CI) = 0.95–0.96; Supplementary Fig. 7). We identified a substantial number of COJO SNPs (718, 496 and 608) associated with obesity (F1), insulin resistance/hypertension (F2) and dyslipidemia (F3), respectively.
Table 1.
Summary of multivariate GWAS for MetS factor model
| Factor | neff | h2 (s.e.) | Mean χ2 | λGC | LDSC intercept (s.e.) | Attenuation ratio (s.e.) | n COJO SNP | Independent from QSNP | Independent from corresponding MetS components | Independent from previous studiesa | Unreported in GWAS catalog |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MetS | 1,384,348 | 0.1109 (0.003) | 4.2501 | 2.8971 | 1.1208 (0.0259) | 0.0372 (0.008) | 1,307 | 811 | 82 | 848 | 159 |
| F1 | 679,472 | 0.1721 (0.0048) | 3.515 | 2.5641 | 1.1097 (0.0191) | 0.0436 (0.0076) | 718 | 677 | 14 | – | 40 |
| F2 | 728,556 | 0.1136 (0.0039) | 2.7305 | 1.9923 | 1.0529 (0.0208) | 0.0306 (0.012) | 496 | 346 | 57 | – | 57 |
| F3 | 1,086,560 | 0.0993 (0.0069) | 3.0111 | 1.6831 | 0.8766 (0.0234) | <0 | 608 | 329 | 0 | – | 85 |
We then conducted a heterogeneity test of QSNP (that is, SNPs having heterogeneous effects on one or more indicators rather than pleiotropic effects via latent factors) for MetS using Genomic SEM and identified 1,203 QSNP lead SNPs using an identical clumping approach (Supplementary Note and Supplementary Fig. 8). Among the 1,307 COJO MetS SNPs, 62.1% (n COJO SNPs = 811) were independent of QSNP and 20.9% (n COJO SNPs = 273) were genome-wide significant (GWS) in the heterogeneity test of QSNP. When we compared the direction of the effects of the COJO MetS SNPs directly with the corresponding SNPs in the GWAS of MetS components, we observed that 59% of the COJO MetS SNPs (n = 772) had a perfect match in terms of effect direction. Additionally, 37.9% of the COJO MetS SNPs (n = 495) showed consistency in the effect direction with five or six MetS components. The Cohen’s kappa (κ) test indicated agreement in the effect direction between MetS and each of the seven MetS components, with κ values ranging from 0.47 to 0.96 (Supplementary Table 17). These findings suggest that the identified COJO MetS SNPs exhibit consistent and horizontal pleiotropic effects across MetS components via shared genetic liability (that is, MetS factor).
Genetic correlation analysis of MetS with external traits
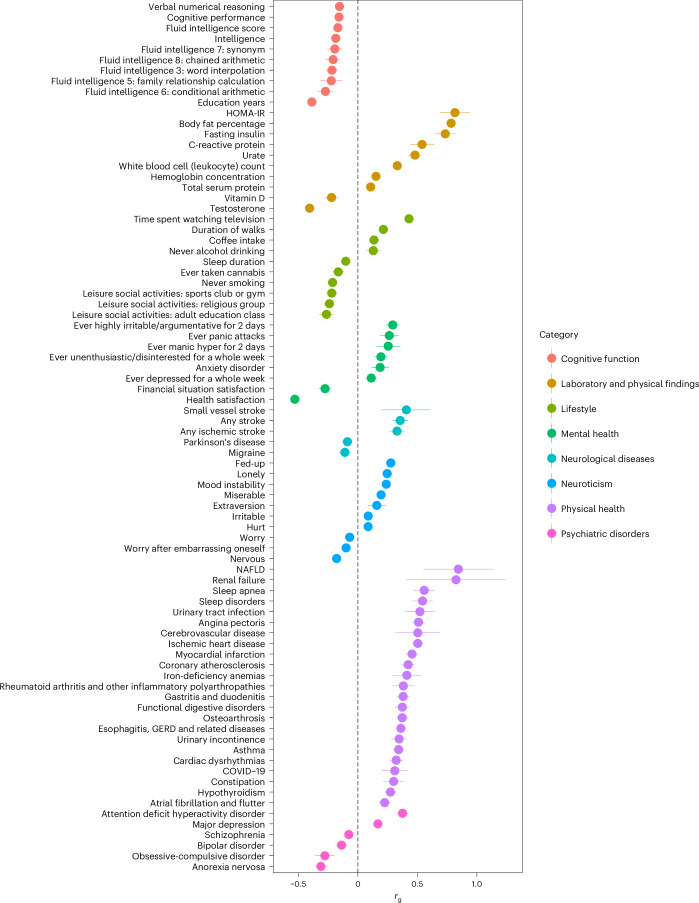
We estimated the genetic correlation (rg) between MetS and 119 external traits categorized into eight groups using LDSC regression (Fig. 2 and Supplementary Table 18). Among the 119 examined traits, 82 exhibited significant associations after Bonferroni correction. Given that the link between MetS and CVD is acknowledged widely24, our findings reveal a moderate genetic overlap with angina pectoris (rg = 0.51; 95% CI = 0.46–0.56) and ischemic heart disease (rg = 0.50; 95% CI = 0.46–0.55).
Fig. 2. Genetic correlation between MetS and external traits.
Among the 119 external traits, 82 Bonferroni significant rg values are illustrated (Supplementary Tables 19 and 18 report all rg values with 119 traits and corresponding sample size, respectively). Error bars represent 95% CIs for rg, calculated as rg ± 1.96 × s.e. The black dashed line indicates an rg of 0. HOMA-IR, homeostatic model assessment of insulin resistance.
We also observed a substantial positive genetic correlation between MetS and nonalcoholic fatty liver disease (NAFLD) (rg = 0.84, 95% CI = 0.55–1.14) and a negative correlation with health satisfaction (rg = −0.53; 95% CI = −0.57 to −0.49) (Supplementary Table 19). Given that MetS is genetically correlated with various cognitive functions and psychiatric disorders, such as education years (rg = −0.39; 95% CI = −0.41 to −0.36) and attention deficit hyperactivity disorder (rg = 0.38, 95% CI = 0.33–0.42), we additionally investigated its correlation with magnetic resonance imaging phenotypes in 101 brain regions. We observed a significant negative correlation with gray matter volume (rg = −0.24; 95% CI = −0.31 to −0.17) after applying Bonferroni correction (P < 4.95 × 10−4) (Supplementary Table 20).
Heritability and enrichment in functional categories
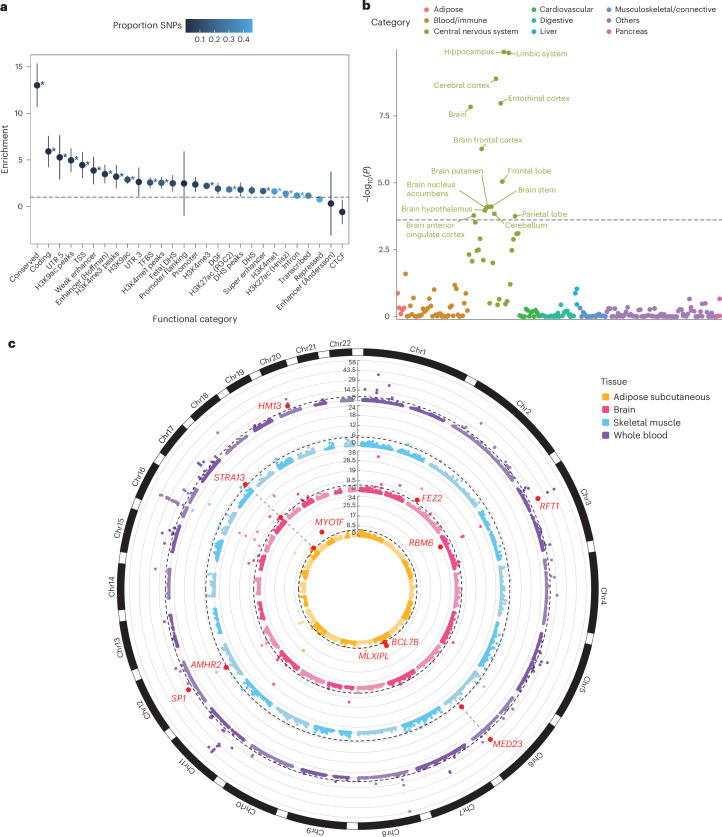
To determine whether specific functional categories of SNPs were enriched for heritability, we performed a partitioned SNP heritability analysis using stratified LDSC (S-LDSC)25 regression on the 53 annotations. We observed significant enrichment of SNP heritability in 35 of these annotations even after Bonferroni correction (P < 9.43 × 10−4). The highest levels of enrichment were observed in conserved genomic regions (enrichment = 13.0; s.e. = 1.2), coding regions (enrichment = 5.91; s.e. = 0.86), and the 5′ untranslated regions (UTRs) of genes (enrichment = 5.28; s.e. = 1.22) (Fig. 3a and Supplementary Table 21).
Fig. 3. Post-GWAS analyses of MetS GWAS.
a, Heritability enrichment for functional SNP categories. The x axis represents functional categories, and the y axis represents enrichment. Error bars represent the 95% CIs, calculated as enrichment ± 1.96 × s.e. Asterisks represent significant enrichment after Bonferroni correction (uncorrected P from two-sided enrichment test <9.43 × 10−4), and exact P is available in Supplementary Table 21. b, Tissue-specific enrichment analysis using LDSC-SEG. The x axis represents tissue categories, and the y axis represents the uncorrected −log10(P) from one-sided z test for enrichment in specific tissues. The black dashed line represents the Bonferroni significance threshold of P = 1.02 × 10-4. c, Circos plot62 for gene prioritization using SMR in four tissues. The y axis represents the uncorrected −log10(P) from two-sided χ²-test with 1 degree of freedom for the gene association. Red dots with annotations show replicated genes. The dashed black line represents the Bonferroni significance threshold for the corresponding tissue. Chr, chromosome; CTCF, CCCTC-binding factor; DGF, digital genomic footprint; DHS, DNase I hypersensitivity site; TFBS, transcription factor binding site; TSS, transcription start site; UTR, untranslated region.
LDSC regression for specifically expressed genes (LDSC-SEG)26 was used to investigate whether MetS genetic signals were enriched in specific cells or tissue types based on gene expression or activating histone marks. We observed significant enrichment in various brain tissues, including the hippocampus (P = 1.30 × 10−10), entorhinal cortex (P = 1.08 × 10−08) and frontal cortex (P = 5.46 × 10−7), using gene expression data sourced from a previous study27 (Fig. 3b). This enrichment in brain tissues was consistent with the significant genetic correlation observed between brain volume and MetS and was further validated using multitissue chromatin-based annotation data and MAGMA gene property analyses (Supplementary Note and Supplementary Tables 22–26). Furthermore, both F1 and F2 were enriched in brain tissues, whereas F3 was enriched in adipose and liver tissues (Supplementary Tables 27 and 28).
MAGMA gene set analysis identified 25 gene sets that were associated significantly with MetS, including a range of functions related to neuronal, molecular and lipid metabolism (Supplementary Table 29).
Prioritization of MetS genes
We used summary data-based Mendelian randomization (SMR) using gene expression data from the following tissues to prioritize MetS genes: brain cortex (BrainMeta v.2 (ref. 28), n = 2,865), whole blood (eQTLGen29, n = 31,684), subcutaneous adipose (GTEx30, n = 581), skeletal muscle (GTEx, n = 706), liver (GTEx, n = 208), aorta (GTEx, n = 387) and coronary artery (GTEx, n = 213) (Supplementary Table 30). The Heterogeneity in Dependent Instruments (HEIDI) test was simultaneously conducted to distinguish pleiotropic gene expression from MetS associations due to linkage. Significant associations with MetS were identified in various tissues, including the brain cortex, whole blood, subcutaneous adipose, skeletal muscle, liver, aorta and coronary artery, with 43, 136, 14, 16, 5, 14 and 5 genes, respectively, meeting the criteria after Bonferroni correction (P < 0.05/n genes tested) and passing the HEIDI test (P > 0.05) (Fig. 3c, Table 2 and Supplementary Table 31). Among these, 11 genes (AMHR2, BCL7B, FEZ2, HM13, MED23, MLXIPL, MYO1F, RBM6, RFT1, SP1 and STRA13) were replicated in an independent cohort across the corresponding tissues, except for the liver, aorta and coronary artery because of the lack of available independent data (Supplementary Table 32). Nonetheless, among the 11 replicated genes, significant associations were observed for STRA13 in the aorta and coronary artery tissues, and MYO1F in coronary artery tissue. Additionally, HM13, AMHR2, RFT1 and SP1 were identified through positional, eQTL and chromatin interaction mapping using FUMA, and demonstrated significant gene associations with MAGMA.
Table 2.
MetS genes prioritized from the SMR analysis
| Gene | Tissue | Chr | Top SNP | PeQTL | PGWAS | Beta | s.e. | PSMR | PHEIDI |
|---|---|---|---|---|---|---|---|---|---|
| RBM6 | Adipose subcutaneous | 3 | rs9814664 | 1.27 × 10−116 | 1.28 × 10−47 | 0.034 | 0.003 | 1.54 × 10−34 | – |
| MLXIPL | Adipose subcutaneous | 7 | rs17145813 | 5.19 × 10−12 | 1.03 × 10−34 | 0.111 | 0.018 | 1.78 × 10−9 | 6.78 × 10−2 |
| BCL7B | Adipose subcutaneous | 7 | rs11972595 | 1.54 × 10−14 | 4.55 × 10−9 | −0.054 | 0.012 | 3.15 × 10−6 | 5.69 × 10−1 |
| MYO1F | Adipose subcutaneous | 19 | rs4804311 | 2.59 × 10−20 | 8.05 × 10−30 | 0.095 | 0.013 | 7.98 × 10−13 | 1.30 × 10−1 |
| STRA13 (CENPX) | Adipose subcutaneous | 17 | rs4995642 | 2.78 × 10−47 | 1.61 × 10−7 | −0.016 | 0.003 | 8.41 × 10−7 | 1.32 × 10−1 |
| STRA13 (CENPX) | Brain cortex | 17 | rs4995642 | 2.17 × 10−192 | 1.61 × 10−7 | −0.007 | 0.001 | 2.48 × 10−7 | 6.13 × 10−2 |
| FEZ2 | Brain cortex | 2 | rs10172196 | 2.72 × 10−88 | 1.05 × 10−9 | 0.013 | 0.002 | 5.42 × 10−9 | 1.00 × 10−1 |
| MED23 | Skeletal muscle | 6 | rs2608954 | 7.35 × 10−16 | 1.44 × 10−16 | 0.048 | 0.008 | 7.89 × 10−9 | 3.10 × 10−1 |
| AMHR2 | Skeletal muscle | 12 | rs2272002 | 3.15 × 10−11 | 6.98 × 10−12 | −0.046 | 0.010 | 1.84 × 10−6 | 1.92 × 10−1 |
| STRA13 (CENPX) | Skeletal muscle | 17 | rs4995642 | 2.99 × 10−45 | 1.61 × 10−7 | −0.016 | 0.003 | 9.00 × 10−7 | 1.03 × 10−1 |
| HM13 | Whole blood | 20 | rs6120704 | 0 | 1.41 × 10−11 | 0.021 | 0.003 | 2.14 × 10−11 | 7.32 × 10−2 |
| MED23 | Whole blood | 6 | rs2245133 | 3.75 × 10−181 | 2.04 × 10−19 | 0.051 | 0.006 | 8.14 × 10−18 | 6.95 × 10−2 |
| RFT1 | Whole blood | 3 | rs2336725 | 6.08 × 10−263 | 7.37 × 10−21 | −0.038 | 0.004 | 1.52 × 10−19 | – |
| SP1 | Whole blood | 12 | rs10876447 | 1.66 × 10−139 | 9.12 × 10−14 | −0.032 | 0.004 | 8.95 × 10−13 | 9.21 × 10−1 |
PeQTL is the uncorrected P value from two-sided z test for the top associated cis-eQTL in the eQTL, PGWAS is the uncorrected P value from two-sided z test for the top associated cis-eQTL in the GWAS, Beta is the effect estimate from SMR, s.e. is the corresponding standard error, PSMR is the uncorrected P value from two-sided χ2-test with 1 d.f. for SMR and PHEIDI is the uncorrected P value from two-sided HEIDI test.
Chr, chromosome.
PRS analysis
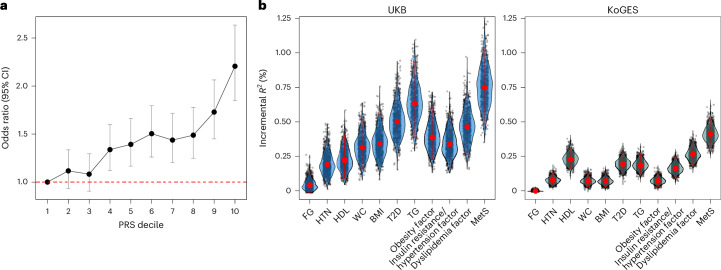
We computed the genome-wide PRS for MetS using MetS GWAS conducted with UKB-excluded cohorts and PRS-CS31. The MetS PRS was used to investigate its relationship with dichotomized MetS in the UKB cohort of 11,139 individuals (ncase = 4,641; ncontrol = 6,498) and the CVD incidence rate from UKB32 (Supplementary Note).
First, we investigated the odds ratio (OR) of MetS risk within the UKB cohort for each PRS decile (Fig. 4a and Supplementary Table 33). OR increased as the PRS decile increased, with the top decile showing a 2.21-fold increased risk (95% CI = 1.85–2.63) compared with the first decile.
Fig. 4. MetS PRS analysis in European and East Asian populations.
a, Line plot representing the change in OR across deciles of the MetS PRS in 11,139 UKB individuals; error bars indicate 95% CIs, calculated as OR ± 1.96 × s.e. The red dashed line represents an OR of 1. b, Violin plot illustrating the incremental proportion of variance explained (ΔR2) by the PRS of seven MetS components and four latent factors for predicting MetS in UKB and KoGES cohorts. Each dot represents ΔR2 from a single iteration of bootstrapping. Red dots represent ΔR2 obtained with all individuals and error bars indicate 95% CIs estimated using the percentile bootstrapping method with 1,000 iterations.
We then investigated how the PRS of latent factors (that is, obesity, insulin resistance/hypertension, dyslipidemia and MetS) and individual MetS components contributed to the prediction of dichotomized MetS in the UKB. To this end, we compared the incremental variance explained (ΔR2) between the baseline and PRS models (Fig. 4b and Supplementary Table 34). In all PRS models, the estimates were statistically significant after Bonferroni correction (P < 0.05/11). The MetS PRS explained the largest variance for predicting MetS (ΔR2 = 0.75%; 95% CI = 0.49–1.04%), followed by the TG PRS (ΔR2 = 0.63%; 95% CI = 0.39–0.93%) and the T2D PRS (ΔR2 = 0.5%; 95% CI = 0.3–0.75%).
We further investigated the relationship between MetS PRS and the incidence rate of CVD in the UKB. The MetS PRS showed distinct differences in CVD incidence rates across stratified genetic risk groups, and a notable association between the MetS PRS and CVD incidence rate (hazard ratio = 1.11; 95% CI = 1.10–1.13) was identified through multivariable Cox regression analysis (Supplementary Fig. 9 and Supplementary Table 35).
MetS GWAS and PRS transferability in East Asians
Using PLINK v.2.0 (ref. 33), we performed a GWAS for dichotomized MetS based on the AHA/NHLBI criteria, including 62,314 independent Korean individuals from the Korean Genome and Epidemiology Study (KoGES) cohort34 (Supplementary Fig. 10). We identified 19 lead SNPs across six genomic risk loci using the standard clumping method within FUMA20, of which 10 SNPs were significant even after conditional analysis using COJO (Supplementary Table 36). We computed the genetic correlation between European and East Asian MetS GWAS using Popcorn35 and observed a substantial genetic correlation of rg = 0.70 (s.e. = 0.18) (Supplementary Table 37).
To evaluate the consistency and transferability of European COJO MetS SNPs in the East Asian population, we investigated GWAS effect consistency and locus transferability. We observed a moderate ρ of 0.34 (P = 3.16 × 10−34; Supplementary Fig. 11) and a high power-adjusted transferability (PAT)36 of 1.66.
We further evaluated the performance of PRS computed using the European GWAS in KoGES (Fig. 4b and Supplementary Table 38). Similar to the UKB target, the MetS PRS demonstrated the largest ΔR2 in KoGES (UKB ΔR2 = 0.75%; 95% CI = 0.49–1.04%; KoGES ΔR2 = 0.41%; 95% CI = 0.31–0.54%), and a similar ΔR2 pattern was evident in PRS computed using PRSice-2 (ref. 37) (Supplementary Note and Supplementary Table 39). These findings suggest the potential transferability of European MetS GWAS findings to diverse populations.
PRS-PheWAS
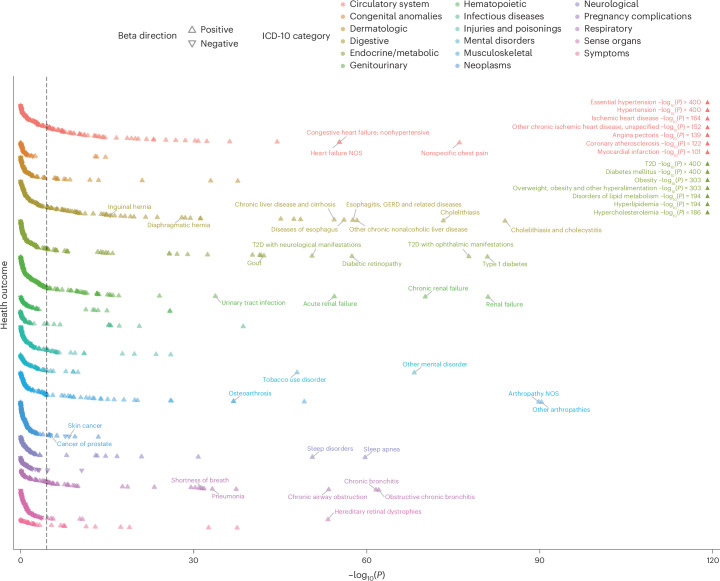
To investigate the health outcomes associated with MetS, we conducted a PRS-PheWAS on UKB individuals with a sample size ranging from 196,837 to 294,024. We applied logistic regression to analyze 1,621 distinct health outcomes after adjusting for sex, age and ten principal components (PCs) of genetic ancestry. After Bonferroni correction (P < 0.05/1,621), 350 medical outcomes were associated significantly with MetS, and the OR ranged from 0.84 to 1.83 per s.d. increase in the MetS PRS (Fig. 5 and Supplementary Table 40). Additionally, we conducted PRS-PheWAS for F1, F2 and F3, and 205, 213 and 294 health outcomes exhibited significant associations, respectively (Supplementary Table 41). Moreover, 43 health outcomes showed significant associations solely with the MetS PRS, including paroxysmal tachycardia (OR = 1.09; 95% CI = 1.06–1.13) and delirium (OR = 1.10; 95% CI = 1.06–1.14) (Supplementary Fig. 12).
Fig. 5. Phenome-wide association study using the MetS PRS.
Scatterplot illustrating the association between the genome-wide MetS PRS and 1,621 health outcomes in the UKB. The x axis represents uncorrected −log10(P) from the two-sided z test for beta (that is, the log of the OR), and the y axis represents health outcomes. Health outcomes are color-coded by category; triangles indicate the direction of beta. Associations with high significance are labeled independently in the upper-right corner of the plot. The black dashed line represents the Bonferroni-corrected significance threshold of P < 3.08 × 10−5. NOS, nitric oxide synthase; GERD, gastroesophageal reflux disease.
These associations extended beyond the expected links with health outcomes closely related to MetS components, such as hypertension, obesity, diabetes, hyperlipidemia and hypercholesterolemia. Thus, the MetS PRS exhibited significant associations with health outcomes across a wide range of bodily systems, including the circulatory (for example, chest pain and heart failure), digestive (for example, cholelithiasis, liver disease and gastroesophageal reflux disease), renal (for example, renal failure and urinary tract infection), and respiratory (for example, pneumonia) systems, and mental disorders (for example, tobacco use and anxiety disorders).
Two-sample Mendelian randomization
We conducted TSMR to investigate the potential causal association between MetS and health outcomes that showed a significant association in the PRS-PheWAS analysis. Among the 350 significant health outcomes, we collated UKB GWAS summary statistics for 325 medical outcomes. After harmonization, 262 instrumental variables (IVs) were extracted from the MetS GWAS. Four causal estimation methods—inverse-variance weighted (IVW)38, weighted median (WM)39, MR-Egger40 and MR pleiotropy residual sum and outlier (MR-PRESSO)41—were used to estimate causal effects. We selected the most robust causal effect estimate based on its significance after Bonferroni correction (P < 0.05/325) and results from heterogeneity and pleiotropy tests following the approach of Kim et al.42.
MetS was associated significant and causally with 29 health outcomes spanning various bodily systems (Table 3), such as ischemic heart disease (OR = 2.32; 95% CI = 2.00–2.68), other mental disorders (OR = 1.34; 95% CI = 1.18–1.52), spondylosis without myelopathy (OR = 1.61; 95% CI = 1.28–2.03), cholelithiasis and cholecystitis (OR = 1.52; 95% CI = 1.29–1.80) and renal failure (OR = 1.47; 95% CI = 1.21–1.77) (Supplementary Tables 42 and 43). We observed robustness in the identified causal associations through constrained maximum likelihood and model averaging-based MR (cML-MA)43, and the bidirectional TSMR suggested insufficient evidence for the potential reverse causation (Supplementary Tables 44–46 and Supplementary Figs. 13 and 14). We further validated these results by leveraging GWASs from an independent cohort, the Michigan Genomics Initiative (MGI)44 freeze 3 (we note that one health outcome was unavailable in the MGI). Among the 28 health outcomes, 13 remained significant in the MGI after Bonferroni correction (P < 0.05/28) (Supplementary Tables 47 and 48).
Table 3.
Results of TSMR analysis
| ICD-10 category | Outcome | Method | OR (95% CI) | P value | Heterogeneity P value | MR-Egger intercept P value | MR-PRESSO global test P value |
|---|---|---|---|---|---|---|---|
| Circulatory | Peripheral angiopathy in diseases classified elsewhere | Inverse variance weighted | 5.58 (2.42, 12.87) | 5.43 × 10−5 | 0.056 | 0.44 | – |
| Circulatory | Other disorders of arteries and arterioles | Inverse variance weighted | 2.68 (1.75, 4.11) | 6.34 × 10−6 | 0.343 | 0.63 | – |
| Circulatory | Coronary atherosclerosis | MR-PRESSO | 2.49 (2.10, 2.95) | 1.54 × 10−21 | 2.95 × 10−37 | 0.31 | <0.001 |
| Circulatory | Angina pectoris | MR-PRESSO | 2.46 (2.05, 2.95) | 3.92 × 10−19 | 6.34 × 10−24 | 0.22 | <0.001 |
| Circulatory | Other chronic ischemic heart disease, unspecified | MR-PRESSO | 2.39 (2.00, 2.85) | 1.34 × 10−18 | 1.20 × 10−16 | 0.50 | <0.001 |
| Circulatory | Unstable angina (intermediate coronary syndrome) | MR-PRESSO | 2.34 (1.82, 3.02) | 2.48 × 10−10 | 1.74 × 10−7 | 0.77 | <0.001 |
| Circulatory | Peripheral vascular disease, unspecified | Inverse variance weighted | 2.33 (1.68, 3.23) | 4.08 × 10−7 | 0.058 | 0.84 | – |
| Circulatory | Ischemic heart disease | MR-PRESSO | 2.32 (2.00, 2.68) | 8.93 × 10−24 | 3.05 × 10−45 | 0.19 | <0.001 |
| Circulatory | Other forms of chronic heart disease | Inverse variance weighted | 2.29 (1.58, 3.30) | 1.00 × 10−5 | 0.393 | 0.69 | – |
| Circulatory | Myocardial infarction | MR-PRESSO | 2.25 (1.84, 2.75) | 7.13 × 10−14 | 4.47 × 10−23 | 0.28 | <0.001 |
| Circulatory | Heart failure NOS | MR-PRESSO | 1.79 (1.36, 2.35) | 3.88 × 10−5 | 2.42 × 10−4 | 0.58 | <0.001 |
| Circulatory | Congestive heart failure; nonhypertensive | MR-PRESSO | 1.75 (1.37, 2.22) | 7.93 × 10−6 | 2.62 × 10−4 | 0.56 | <0.001 |
| Circulatory | Peripheral vascular disease | Inverse variance weighted | 1.74 (1.34, 2.27) | 3.86 × 10−5 | 0.057 | 0.25 | – |
| Circulatory | Circulatory disease NEC | MR-PRESSO | 1.36 (1.18, 1.56) | 2.28 × 10−5 | 1.87 × 10−4 | 0.02 | <0.001 |
| Circulatory | Other disorders of the circulatory system | MR-PRESSO | 1.36 (1.18, 1.56) | 2.19 × 10−5 | 2.40 × 10−4 | 0.02 | <0.001 |
| Circulatory | Nonspecific chest pain | MR-PRESSO | 1.34 (1.20, 1.51) | 1.11 × 10−6 | 3.17 × 10−12 | 0.01 | <0.001 |
| Digestive | Other chronic nonalcoholic liver disease | MR-PRESSO | 3.3 (2.16, 5.04) | 9.31 × 10−8 | 2.60 × 10−9 | 0.14 | <0.001 |
| Digestive | Chronic liver disease and cirrhosis | MR-PRESSO | 2.12 (1.53, 2.95) | 1.13 × 10−5 | 1.83 × 10−7 | 0.04 | <0.001 |
| Digestive | Cholelithiasis and cholecystitis | MR-PRESSO | 1.52 (1.29, 1.80) | 1.50 × 10−6 | 6.59 × 10−28 | 0.12 | <0.001 |
| Digestive | Cholelithiasis | MR-PRESSO | 1.45 (1.21, 1.73) | 6.72 × 10−5 | 7.70 × 10−23 | 0.07 | <0.001 |
| Endocrine/metabolic | Gout | MR-PRESSO | 2.07 (1.48, 2.90) | 3.38 × 10−5 | 2.28 × 10−8 | 0.14 | <0.001 |
| Genitourinary | Renal failure | Inverse variance weighted | 1.47 (1.21, 1.77) | 7.05 × 10−5 | 0.291 | 0.58 | – |
| Mental disorders | Tobacco use disorder | MR-PRESSO | 1.35 (1.16, 1.56) | 1.28 × 10−4 | 1.35 × 10−19 | 0.03 | <0.001 |
| Mental disorders | Other mental disorder | MR-PRESSO | 1.34 (1.18, 1.52) | 1.20 × 10−5 | 7.68 × 10−14 | 0.30 | <0.001 |
| Musculoskeletal | Spondylosis without myelopathy | Inverse variance weighted | 1.61 (1.28, 2.03) | 4.23 × 10−5 | 0.119 | 0.99 | – |
| Musculoskeletal | Unspecified monoarthritis | MR-PRESSO | 1.59 (1.34, 1.90) | 3.65 × 10−7 | 7.14 × 10−22 | 0.01 | <0.001 |
| Musculoskeletal | Arthropathy NOS | MR-PRESSO | 1.46 (1.29, 1.66) | 1.62 × 10−8 | 8.40 × 10−32 | 0.02 | <0.001 |
| Musculoskeletal | Other arthropathies | MR-PRESSO | 1.4 (1.24, 1.59) | 2.97 × 10−7 | 5.23 × 10−34 | 0.04 | <0.001 |
| Respiratory | Asthma | MR-PRESSO | 1.35 (1.18, 1.55) | 2.39 × 10−5 | 1.58 × 10−30 | 0.77 | <0.001 |
P value is uncorrected P value from two-sided z test for inverse variance weighted and two-sided t-test for MR-PRESSO, Heterogeneity P value is uncorrected P value from one-sided χ2-test with number of IV − 1 as degrees of freedom, MR-Egger intercept P value is uncorrected P value from two-sided t-test with number of IV − 1 as d.f., and MR-PRESSO global test P value is uncorrected P value from two-sided global test.
NOS, nitric oxide synthase.
Discussion
In this study, we used Genomic SEM to investigate genetic MetS factors using seven MetS components: BMI, WC, T2D, FG, HTN, HDL and TG. We developed a hierarchical model comprising three first-order factors (obesity, insulin resistance/hypertension and dyslipidemia), which effectively described the shared genetic liability of MetS. We identified the genomic loci associated with MetS and 11 potential therapeutic target genes. We demonstrated the performance of the MetS PRS in individuals of European ancestry and its potential utility in individuals of East Asian ancestry. Additionally, we examined the association between MetS and various health outcomes.
Genomic SEM identified genomic loci associated with MetS, of which 1,307 were significant COJO SNPs even after conditional analysis. Furthermore, 6.3% (n = 82) of the COJO MetS SNPs were independent of the genomic loci of the MetS components, and only 21% (n = 273) were GWS in the QSNP heterogeneity test. A previous study by Van Walree et al.22 used Genomic SEM to construct a common MetS factor based on genetic clustering across five main criteria. Although they identified numerous MetS lead SNPs (n = 318), their study has several limitations, including a lack of validity assessment through a heterogeneity test, limited exploration of potential relationships with other health outcomes and a lack of validation in independent cohorts, especially in populations of different ancestry. Therefore, our findings are notable, as we identified additional MetS SNPs compared with the largest MetS GWAS conducted to date and rigorously assessed their validity.
MetS has strong connections with brain functions45, and its association with various psychiatric disorders has been consistently suggested46,47. For example, individuals with schizophrenia (SCZ) or bipolar disorder (BD) exhibit an increased prevalence of MetS (SCZ: 37–63%; BD: 30–49%) and a higher relative risk than the general population (SCZ: two- to threefold; BD: 1.5- to twofold)48. Additionally, MetS is associated with Alzheimer disease, with a more than threefold increased risk49. In this study, we observed a significant association between MetS and psychiatric disorders through genetic correlation analysis and enrichment of MetS genetic signals in the hippocampus and brain tissues. As psychiatric disorders have a heritable component, our results provide genetic evidence that MetS may be associated with brain function and, consequently, various mental disorders.
We prioritized 11 potential genes (AMHR2, BCL7B, FEZ2, HM13, MED23, MLXIPL, MYO1F, RBM6, RFT1, SP1 and STRA13) associated strongly with MetS through SMR analyses across MetS-relevant tissues. Additionally, whole blood tissue from the eQTLGen cohort was included because of its drug delivery suitability, biomarker measurement convenience and large sample size enhancing statistical power50–52. Using resources such as GeneCards53, the Open Targets Platform54 and the International Mouse Phenotyping Consortium (IMPC)55, most of these genes were linked to MetS components (Supplementary Note). STRA13 regulates adipogenesis and lipogenesis56 and SP1 is crucial for the transcription of genes associated with hyperinsulinemia, T2D and MetS in response to insulin levels57. Both BCL7B and MLXIPL are associated with MetS and inflammation, whereas MLXIPL is associated specifically with lipid metabolism58. Furthermore, Med23 knockout mice had higher HDL and lean body mass, whereas Hm13 and Rbm6 deletions led to lower fasting glucose levels, which is concordant with the direction of the genetic effect from SMR analyses. The prioritized MetS genes demonstrated substantial evidence of their relevance to metabolic traits, supporting their use as potential targets for therapeutic interventions in MetS.
The utility of the MetS PRS in the European population was examined by stratifying the MetS PRS into deciles, where individuals with MetS PRS in the highest decile had a 2.21-fold greater risk of developing MetS. Furthermore, the MetS PRS exhibited slightly improved discrimination in identifying individuals at an elevated risk of developing CVD compared with the PRS of its components. These findings emphasize the utility of the MetS PRS in identifying individuals at high CVD risk and in implementing proactive lifestyle adjustments and clinical interventions.
Building on these findings, we further investigated the performance of the MetS PRS computed from a European GWAS for predicting dichotomized MetS in identical and different target populations. The MetS PRS demonstrated superior predictive power for dichotomized MetS in both cohorts compared with the PRSs of its components, which is consistent with MetS exhibiting the highest PAT. In contrast, FG accounted for the least MetS variance in both cohorts. This may be attributed to the fact that the performance of PRS depends on the GWAS sample size59, and the sample size of the FG GWAS was comparatively smaller than that of the other components. These findings highlight a promising scope for wider application of the MetS PRS across different populations, yet they stress the need for GWAS with larger sample sizes.
As mentioned previously, MetS is associated with various health outcomes16. We investigated the associations across various health outcomes in different bodily systems using the PRS-PheWAS, and 350 health outcomes showed significant associations. As an extension of the association analysis, we investigated the causal relationship between MetS and a broad range of health outcomes using the TSMR. Most health outcomes with a causal relationship to MetS are related to the circulatory system. However, the association between digestive, respiratory, and genitourinary system disorders and mental disorders was notable.
Our study had several limitations. First, we focused on individuals of European ancestry for the main discovery of the MetS genetic architecture. Genomic SEM is based on LDSC, which uses ancestry-dependent linkage disequilibrium scores that restrict the analysis to a homogenous population. However, the potential heterogeneity in SNP effects on MetS components among various ancestral backgrounds suggests that genetic signals for MetS may vary across populations60,61. These findings highlight the importance of conducting analyses that include several ancestral groups. Second, the genetic factor model of MetS described in this study may not be the true model. Genomic SEM can deduce an optimal factor model only from the genetic covariance of indicators included in the analysis. Hence, various post hoc analyses are essential to elucidate the usefulness of the suggested latent factors in biological mechanisms. Furthermore, considering that the definition of MetS undergoes periodic updates, encompassing a broader spectrum of metabolic traits or diseases, such as insulin resistance and NAFLD, will provide more comprehensive insights into the genetic underpinnings of MetS. Third, apart from MetS GWAS signals, we observed a substantial number of SNPs that showed heterogeneous effects (that is, QSNP) on the MetS components included in the analysis. Further investigation of QSNP may potentially identify phenotype-specific genetic signals. Fourth, because of the limited availability of GWAS for MetS components in East Asian populations, the genetic model of MetS, which was identified specifically in individuals of European ancestry, could not be replicated. Therefore, we highlight the need for an increase in the sample size across diverse populations.
In conclusion, our results reveal the complex genetic architecture of MetS and its relationship with various health outcomes.
Methods
Ethics statement
We received approval to use data from the UKB and KoGES under application number 33002 and NBK-2022-063, respectively. Our study adhered to all the conditions and access procedures established by the corresponding biobanks.
Selection of GWAS on MetS components and meta-analysis
Seven MetS components (BMI, WC, T2D, FG, HTN, HDL and TG) were selected to define MetS1 (Supplementary Note). We gathered the largest European GWAS summary statistics for these components from previous studies or public repositories, including the Genetic Investigation of Anthropometric Traits, Meta-Analyses of Glucose and Insulin-related Traits Consortium, Global Lipids Genetics Consortium, Million Veterans Program (MVP), FinnGen Release 7 and UKB. Additionally, we prepared a UKB-excluded cohort, in which UKB individuals were absent from the GWAS summary statistics. The purpose of the UKB-excluded cohort was to avoid sample overlap with the target cohort (that is, UKB) when conducting post-GWAS analyses such as PRS analyses, PRS-PheWAS and TSMR. A detailed summary of the individual GWAS summary statistics used in this study is provided in Supplementary Table 1.
We conducted a fixed-effects meta-analysis of T2D and HTN using METAL63 to increase the sample size of the corresponding GWAS. FinnGen and UKB were included in the meta-analysis of HTN whereas FinnGen, MVP and Vujkovic et al.64 were included in the meta-analysis of T2D. For the UKB-excluded cohort, we performed a T2D-specific meta-analysis using FinnGen and MVP. The intercept of the bivariate LDSC regression indicated no sample overlap between cohorts (Supplementary Table 3).
Quality control of GWAS
Quality control (QC) of the GWAS summary statistics was conducted using EasyQC65 v.23.8. The European 1000 Genomes Phase 3 (1kGP3) was used as a reference for QC. The SNPs were filtered based on the following criteria: (1) SNPs with alleles other than A, T, C and G, or mismatched with the reference; (2) monomorphic SNPs; (3) SNPs with a minor allele frequency (MAF) below 0.5%; (4) duplicated or multiallelic SNPs; (5) low-quality SNPs (INFO < 0.9 if imputation quality is provided); and (6) SNPs absent in the reference (Supplementary Table 2). All the GWAS summary statistics were aligned to the genome build GRCh37. For the FinnGen data, we converted them from GRCh38 to GRCh37 using the UCSC liftOver66.
KMO test
The KMO19 test was used to assess the suitability of the data for factor analysis. The KMO test computes measures of sampling adequacy for each and all overall indicators, and a value above 0.5 indicates data adequacy for factor analysis. We used the R package psych v.2.1.9 to conduct the KMO test.
Parallel analysis
Parallel analysis67 determined the number of factors retained in the following EFA. The eigenvalues from the genetic correlation matrix were compared with those from a Monte-Carlo-simulated genetic correlation matrix by introducing random noise from the sampling covariance matrix. We used the ldsc function in Genomic SEM12 v.0.0.5 to compute the genetic correlation matrix and the corresponding sampling covariance matrices for the seven MetS components. The suggested number of factors was determined by selecting the last factor with an eigenvalue greater than that of the simulated genetic correlation matrix. The scree plot for the parallel analysis is illustrated in Supplementary Figs. 2 and 15.
Exploratory factor analysis
EFA was performed using the factanal function in R package stats v.4.0.5. We fitted the single-factor model to the three-factor model, as suggested by the parallel analysis. Promax rotation accounted for the correlation between the latent factors, and we retained a factor that explained more than 15% of the variance68.
Confirmatory factor analysis
A CFA was performed using Genomic SEM12 v.0.0.5. The result from EFA was leveraged to construct the genetic factor model, where the indicators with factor loadings ≥0.3 were retained for the corresponding latent factor. In addition, the factor models in the CFA were specified using unit variance identification (that is, fixing the variance of the latent factor to one) for a better interpretation of the model fit result. CFA parameters were estimated using the diagonally weighted least-squares fit method12.
Model fit depends on the closeness of the model-implied genetic covariance matrix to the observed genetic covariance matrix. We assessed the model fit using four indices conventionally used in SEM: χ2-statistic, CFI, AIC and SRMR. The χ2-statistic indicates the exact fit of the SEM model. The CFI indicates whether the proposed model fits better than one that assumes that all phenotypes are heritable and genetically uncorrelated. The AIC is a relative fit index used to compare models regardless of their nested states. SRMR represents an approximate model fit, calculated as the difference in SRMR between the model-implied and observed genetic covariance matrices. We defined a good model fit based on CFI and SRMR, where CFI ≥ 0.95 and SRMR < 0.08, as proposed by Hu and Bentler69.
MetS multivariate GWAS with Genomic SEM
A multivariate GWAS was conducted to estimate the association of SNPs with MetS factors from a hierarchical factor model using Genomic SEM. The factor construct was specified by unit loading identification (that is, one of the factor loadings of indicators on the latent factor was fixed at 1), where the factor loadings of BMI and the F1 factor (that is, obesity factor) were fixed for the F1 factor and MetS factor, respectively. The identical parameter estimation fit function (that is, diagonally weighted least-squares) was used for the SNP association test. Additional QC steps for individual-level GWAS were performed by filtering SNPs with MAF > 1% and SNPs commonly observed in all individual-level GWAS and the European 1kGP3 reference panel. In total, 2,265,555 SNPs were included in the multivariate GWAS for MetS. The neff for the latent factor was estimated by following Mallard et al.14, where neff for the latent factor is calculated as using the SNPs with MAF between 10 and 40%.
FUMA20 v.1.4.1 was used to identify independent lead SNPs, perform functional annotation and conduct functional gene mapping. The lead SNPs were identified through a two-step clumping procedure: the independent significant SNPs were first defined by clumping the SNPs with a P < 0.05 and r2 ≥ 0.6; the lead SNPs were defined by applying a second round of clumping with a P < 5 × 10−8 and r2 ≥ 0.1. The identified lead SNPs were subjected to GCTA-COJO21 v.1.94.1 and determined whether they were conditionally and jointly associated SNPs through a stepwise model selection procedure with default parameters (that is, P < 5 × 10−8, window of 10 Mb, and collinearity <0.9). To identify MetS COJO SNPs independent of other GWAS, we compared the lead SNPs with their independent significant SNPs within the 500-kb window of the MetS COJO SNP. A MetS COJO SNP was classified as ‘independent’ if any lead SNPs and their independent significant SNPs with P < 5 × 10−8 and r2 > 0.01 were not identified. A MetS COJO SNP was classified as ‘previously reported’ if the SNP was mapped from FUMA using NHGRI-EBI GWAS catalog23 (last updated on 2 August 2023).
We conducted a heterogeneity test using genome-wide QSNP statistics to evaluate the potential SNP effect heterogeneity across the indicators. The null hypothesis of the heterogeneity test posits that the effect of an SNP operates through a latent factor (that is, the common pathway model). The deviation from this null hypothesis indicates that the model with direct SNP effects on the indicators (that is, the independent pathway model) provides a better explanation. QSNP statistics were derived from the χ2 difference test comparing the common and independent pathway models.
SNP-based heritability and genetic correlation
LDSC17,18 v.1.0.1 was used to estimate genomic inflation, SNP-based heritability, and bivariate genetic correlation. For all estimations, precalculated LD scores from the European 1kGP3 and approximately 1.3 million high-quality SNPs were defined by the HapMap3 Consortium70. Genomic inflation was determined based on the LDSC intercept and the attenuation ratio. The LDSC intercept was close to 1 and the attenuation ratio was close to 0, indicating that the observed inflation in the association test statistics was due to polygenicity rather than bias from population stratification.
Genetic correlations were computed between the MetS components used in the Genomic SEM and between MetS factors and external traits (Supplementary Table 18). The SNPs in the corresponding GWAS summary statistics were filtered using the following thresholds: MAF > 0.01 and INFO > 0.9 (if available). For dichotomous traits, we used the effective sample size rather than the total sample size (that is, the sum of ncase and ncontrol).
Partitioned heritability
We partitioned SNP heritability using S-LDSC25. The SNPs were categorized into 53 annotations as defined by Finucane et al.25. Heritability enrichment in each category was computed as the proportion of SNP heritability in that category to the proportion of total SNPs annotated in the corresponding category. We used the precomputed LD scores of the European 1kGP3 reference panel and annotations available online. The Bonferroni-corrected significance threshold for 53 annotations was indicated by P < 0.05/53 = 9.43 × 10−4.
Cell- and tissue-type-specific analysis
LDSC-SEG26 was used to analyze cell- and tissue-specific enrichment by testing the heritability enrichment of genes with high cell- or tissue-specific expression. The human leukocyte antigen region was excluded from all analyses because of its complex LD patterns26. Precomputed annotation files (LD scores) from Finucane et al.26 were utilized, covering 205 tissues and cell types from Franke laboratory27, 489 tissue-specific chromatin-based annotations from Roadmap Epigenomics71 and the ENCODE project72, and three mouse brain cell types from Cahoy et al.73.
MAGMA gene-based, gene set and gene property analyses
MAGMA74 v.1.08, implemented in FUMA, was used for gene-based gene sets and gene property analyses75. In the gene-based analysis, SNPs were linked to protein-coding genes by aggregating the P values of SNPs using the SNP-wise mean model. The European 1kGP3 reference panel was used to account for LD between SNPs. This mapping process encompassed 17,674 protein-coding genes from Ensembl Build 85, with Bonferroni-corrected significance set at P < 2.83 × 10−6.
Gene set analysis was performed to identify the genes (grouped by specific biological pathways, cellular components or molecular functions) associated with the phenotype through competitive analysis. In total, 15,481 gene sets (5,497 curated genes and 9,984 gene ontology terms) obtained from MsigDB v.7 (ref. 76) were tested, with the Bonferroni-corrected significance set at P < 3.23 × 10−6.
Gene property analysis was performed to test the relationship between tissue-specific gene expression profiles and the phenotype-gene association P value obtained from the gene-based analysis. We evaluated 54 specific tissues from GTEx30 v.8 and 11 brain developmental stages from BrainSpan77. Gene expression values in both sets were log2 transformed average reads per kilobase million per tissue type (with reads per kilobase million exceeding 50 replaced with 50). Bonferroni-corrected significance thresholds were set at P < 9.26 × 10−4 for GTEx tissues and at P < 0.005 for BrainSpan developmental stages.
Gene prioritization
To prioritize genes potentially mediating the effects of SNPs on the target phenotype, we performed SMR78 analysis using SMR v.1.03. We used publicly available cis-eQTL data from BrainMeta v.2 (ref. 28) (n = 2,865) for the brain, eQTLGen29 (n = 31,684) for whole blood and GTEx v.8 (ref. 30) for subcutaneous adipose tissue (n = 581), skeletal muscle (n = 706), liver (n = 208), aorta (n = 387) and coronary arteries (n = 213) as our discovery set. The results were then validated in brain cortical tissues (cortex (n = 205), frontal cortex (n = 175), anterior cingulate cortex (n = 147)) and whole blood (n = 670) using GTEx v.8. Brain cortical tissues in the GTEx were considered specifically because the genotype and RNA sequencing data of BrainMeta v.2 were obtained from brain cortical tissue. The results for subcutaneous adipose and skeletal muscles were validated using METSIM79 (n = 434) and FUSION80 (n = 301), respectively (an overview of the eQTL data used can be obtained in Supplementary Table 30).
We randomly selected 10,000 individuals of European ancestry from the UKB to compute the LD between SNPs. The significance of the SMR results was determined using the Bonferroni correction for the number of genes tested in each cohort. The resulting genes were subjected to the HEIDI78 test to distinguish the associations between pleiotropy and linkage. The null hypothesis of the HEIDI test was that there is a single causal SNP associated with gene expression and phenotype; hence, P > 0.05, suggesting that a pleiotropic effect due to linkage is unlikely.
PRS analysis
We computed the PRS for all seven MetS components (BMI, WC, T2D, FG, HTN, HDL and TG) included in the Genomic SEM and latent factors using PRS continuous shrinkage (PRS-CS)31, while GWAS summary statistics from the UKB-excluded cohort were used. The PRS-CS uses a Bayesian approach that infers the posterior mean effect size of the SNP, where continuous shrinkage is applied before the SNP effect estimates from the GWAS. We used the global shrinkage parameter (Φ) of 0.01, the neff for the input of GWAS sample size if the GWAS was of dichotomous phenotype, the LD reference panel from European 1kGP3, and the rest of the parameters set to default values (that is, Bayesian gamma-gamma prior of 1 and 0.5; Monte-Carlo iterations of 1,000; burn-in iterations of 500, and Markov chain thinning factor of 5). We then computed the PRS for individuals in the UKB using PLINK v.2.0 (ref. 33) by summing the variants weighted by the inferred posterior mean effect size for the effect allele.
The computed PRS of the seven MetS components and latent factors predicted the MetS status in UKB individuals (Supplementary Note describes the phenotyping of MetS in the UKB cohort). We constructed baseline and PRS models, in which the percentage of incremental pseudo-R2 was computed. The baseline model was a logistic regression model with baseline covariates of sex, age and the ten genetic PCs. The PRS model was identical to the baseline model, except for PRS as an additional covariate. The adjusted McFadden’s pseudo-R2 was computed for each model to penalize McFadden’s pseudo-R2 as more covariates were added to the model. The 95% CI for incremental pseudo-R2 was estimated using the percentile bootstrapping method with 1,000 iterations.
MetS GWAS in the East Asian population
We conducted a GWAS on MetS in Korean individuals using the KoGES. The KoGES comprises three population-based studies. We gathered 62,314 individuals from the Ansan and Ansung (Ansan/Ansung) and Health Examinee studies that had both genotype and phenotype data after QC. Among the 62,314 individuals, 10,684 were MetS cases and 51,630 were MetS controls (Supplementary Note describes KoGES, quality control and MetS phenotyping). The GWAS was performed using PLINK v.2.0 (ref. 33), and logistic regression with the firth-fallback option was used. The covariates included sex, age and the presence of ten genetic PCs.
Crosspopulation genetic correlation
The crosspopulation genetic correlation between the European and East Asian MetS GWAS was estimated using Popcorn software v.1.0 (ref. 35). We included SNPs with MAF > 0.01 and precomputed crosspopulation scores for European and East Asian 1kGP3 populations. We discuss the genetic effect correlation and correlation coefficients of the per-allele SNP effect sizes.
Power-adjusted transferability
We computed PAT to evaluate the reproducibility of MetS GWAS loci from European individuals in East Asians, regardless of effect size and considering differences in power or LD patterns. PAT was calculated by dividing the observed number of transferable loci by the expected number of transferable loci. The observed number of transferable loci is obtained by the following steps: (1) generation of credible sets from European MetS GWAS that consist of lead SNPs (available in East Asian MetS GWAS) and corresponding proxy SNPs (r2 ≥ 0.8) within a 50-kb window with P < 100 × PleadSNP; (2) defining each credible set as being transferable if at least one SNP is associated with P < 0.05 in East Asian MetS GWAS. The expected number of loci was obtained by adding the computed power (alpha = 0.05) for each lead SNP from the European MetS GWAS.
Phenome-wide association study with PRS
We performed PheWAS using PRS, computed using GWAS from the UKB-excluded cohort, and determined health outcomes from the UKB cohort. We fitted a logistic regression to 1,621 disease phenotypes, adjusted for sex, age and ten genetic PCs. The number of samples for each disease phenotype varied between 196,837 and 294,024, respectively.
Two-sample Mendelian randomization
We conducted TSMR to investigate the causal relationship between MetS and the health outcomes identified in the PRS-PheWAS. MetS was considered an exposure, and its GWAS summary statistics were sourced from the UKB-excluded cohort to avoid sample overlap with the outcome GWAS. From the 350 significant Bonferroni associations from PRS-PheWAS, we extracted GWAS summary statistics for 325 medical outcomes from three sources (UKB SAIGE81, PanUKB and UKB Neale Laboratory) after excluding outcomes related to or overlapping with MetS (Supplementary Table 49). The TSMR results from the UKB were further validated using data from the MGI44 Freeze 3. TSMR analysis was conducted using the R package TwoSampleMR v.0.5.6.
The SNPs in the exposure and outcome GWAS were filtered with an MAF below 0.5%. IVs from the exposure GWAS were selected using the default clumping options in TwoSampleMR (that is, P < 5 × 10−8, r2 = 0.001, clumping distance = 10 Mb, and the European 1kGP3 reference panel). They harmonized with the outcome GWAS, with the effect alleles aligned between the exposure and outcome GWAS. We further excluded any IV palindromic or absent cases from the GWAS outcomes. A total of 262 IVs were extracted from MetS, and the number of IVs utilized for TSMR ranged from 176 to 253, depending on the outcome data sources and estimation methods (Supplementary Table 50).
Bidirectional TSMR (that is, considering health outcome as exposure and MetS as outcome) was performed for 22 of 29 health outcomes that showed a significant causal association with MetS (note that seven health outcomes were excluded as IV was absent after filtering by GWS threshold and palindromic SNP). The IVs for bidirectional TSMR were extracted using an approach identical to that described above. The number of extracted IVs ranged from 1 to 14, depending on the health outcomes (Supplementary Table 51).
Five methods were used for each exposure and outcome pair to obtain causal estimates, depending on the number of IVs available. The Wald ratio82 method was used when a single IV was available, and the IVW38 method was used when more than two IVs were available. The WM39, MR-Egger regression40 and MR-PRESSO41 methods were used when more than three IVs were available where the MR-PRESSO method was not performed if either the MR-PRESSO global test was insignificant or the number of IVs retained after outlier removal was insufficient.
The Wald ratio method estimates the causal effect by dividing the regression of the outcome coefficient by exposure. The IVW method assumes that all IVs are valid and estimates the causal effect through a weighted regression of the coefficient from the outcome and exposure while constraining the intercept to zero. The WM method estimates the effect by varying the weights of the IVs, which provides a more consistent estimate even if 50% of the IVs are valid. The MR-Egger regression method detects and accounts for the potential horizontal pleiotropy. The MR-PRESSO method estimates the causal effects by correcting horizontal pleiotropy and removing pleiotropic outliers. We examined the heterogeneity of the IVW model using Cochran’s Q-test83. The presence of horizontal pleiotropy was determined based on the significance of either the MR-Egger intercept test or the MR-PRESSO global test. We distinguished the best causal effect estimation method by following the decision tree from Kim et al.42, which is based on heterogeneity and horizontal pleiotropy. When a single IV was available, neither heterogeneity nor pleiotropy tests could be performed. Hence, the Wald ratio method was selected as the best causal effect estimation method.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41588-024-01933-1.
Supplementary information
Supplementary Notes 1–11 and Figs. 1–15.
Supplementary Tables 1–51.
Acknowledgements
We acknowledge the participants and investigators of UKB and KoGES cohorts. This research was conducted using the UKB Resource under Application Number 33002. The UKB (https://www.ukbiobank.ac.uk/about-biobank-uk) was approved by the National Research Ethics Committee (REC reference 11/NW/0382). This study was supported by the National Research Foundation of Korea Grant funded by the Ministry of Science and Information and Communication Technologies, South Korea (grant no. NRF-2022R1A2C2009998 to H.-H.W. and NRF-2021R1A2C4001779 and RS-2024-00335261 to W.M.). The funders had no role in study design, data collection, analysis, decision to publish or preparation of the manuscript. This study was conducted with bioresources from the National Biobank of Korea, the Korea Disease Control and Prevention Agency, Republic of Korea (NBK-2022-063).
Author contributions
The paper was written by S.P., W.M. and H.-H.W. with input from A.C.F., P.N., P.T.E., A.T., W.-Y.P. and T.Y.Y. S.P. conducted all analyses and visualized results with the following additional inputs: S.K., B.K. and D.S.K. provided critical statistical insights, S.K., Y.A. and H.K. curated the GWAS summary statistics for genetic correlation analysis, I.S. and B.K. prepared the UKB and KoGES genotype data for polygenic risk analysis, C.C. and M.S. curated phenotypic data for PRS-PheWAS analysis, J.K. provided statistical insights in cross-ancestry genetic correlation and power-adjusted transferability analyses, S.-H.J. curated phenotypic data and provided statistical insight in survival analysis, and S.L. visualized the result. S.H. and H.J. managed and resolved critical issues for all software used in analyses. The study was supervised by W.M. and H.-H.W. All authors agreed to the final version of the manuscript.
Peer review
Peer review information
Nature Genetics thanks Xueling Sim and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
Summary statistics of the multivariate and KoGES MetS GWAS are available through GWAS Catalog (https://www.ebi.ac.uk/gwas/) under accession IDs GCST90444487–GCST90444489. The PRS weights developed for MetS are available through PGS Catalog (https://www.pgscatalog.org/) with publication ID PGP000664 and score ID PGS004928. The individual-level phenotypic and genetic data for UKB (https://www.ukbiobank.ac.uk/) and KoGES (https://is.kdca.go.kr/) can be accessed through the application. The reference panel for 1kGp3 can be obtained from the following URL: https://mathgen.stats.ox.ac.uk/impute/1000GP_Phase3.html. The NHGRI-EBI GWAS Catalog is available at the following URL: https://www.ebi.ac.uk/gwas/docs/file-downloads. The GWAS summary statistics from the following consortia and biobanks are publicly available at the corresponding URL: GIANT (https://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files), MAGIC (https://magicinvestigators.org/downloads/), GLGC (http://www.lipidgenetics.org/#data-downloads-title), FinnGen (https://www.finngen.fi/en/access_results), UKB from Neale laboratory (http://www.nealelab.is/uk-biobank), UKB from Lee lab (https://www.leelabsg.org/resources) and PanUKB (https://pan.ukbb.broadinstitute.org/downloads). The GWAS summary statistics of the MVP cohort were obtained using an approved dbGaP application (phs001672.v9.p1). The GWAS summary statistics from the MGI cohort are available upon request from https://precisionhealth.umich.edu/our-research/documents-for-researchers. The eQTL summary statistics are available from GTEx (https://gtexportal.org/home/downloads/adult-gtex#qtl), eQTLGen (https://www.eqtlgen.org/cis-eqtls.html), BrainMeta v.2 (https://yanglab.westlake.edu.cn/software/smr/#DataResource), METSIM (https://mohlke.web.unc.edu/data/1702-2/) and FUSION (https://www.ebi.ac.uk/birney-srv/FUSION/).
Code availability
The script used to conduct the parallel analysis was written by Javier de la Fuente and is available via GitHub (https://github.com/AnnaFurtjes/genomicPCA/blob/main/data); also available on GitHub are the Genomic SEM software (https://github.com/GenomicSEM/GenomicSEM), the LDSC software including S-LDSC and SEG-LDSC (https://github.com/bulik/ldsc) and the Popcorn software (https://github.com/brielin/Popcorn). LDScore is available at https://alkesgroup.broadinstitute.org/LDSCORE and FUMA at https://fuma.ctglab.nl. The analysis codes are deposited with the repository name Park-Nature-Genetics-2024 via Zenodo at 10.5281/zenodo.13137680 (ref. 84).
Competing interests
A.C.F. is cofounder of Goodpath. P.T.E. reports personal fees from Bayer AG, Novartis and MyoKardia. P.N. reports personal fees from Allelica, Apple, AstraZeneca, Blackstone Life Sciences, Creative Education Concepts, CRISPR Therapeutics, Eli Lilly & Co., Foresite Labs, Genentech/Roche, GV, HeartFlow, Magnet Biomedicine, Merck and Novartis; scientific advisory board membership at Esperion Therapeutics, Preciseli and TenSixteen Bio; scientific cofounder status at TenSixteen Bio; equity in MyOme, Preciseli and TenSixteen Bio; and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. W.-Y.P. was employed by the commercial company GENINUS. A.T. declares he is a cofounder and equity shareholder of GeneXwell Inc. A.T. is an advisor to InsideTracker. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Woojae Myung, Hong-Hee Won.
Contributor Information
Woojae Myung, Email: wmyung@snu.ac.kr.
Hong-Hee Won, Email: wonhh@skku.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41588-024-01933-1.
References
- 1.Alberti, K. G. et al. Harmonizing the metabolic syndrome: a joint interim statement of the international diabetes federation task force on epidemiology and prevention; National Heart, Lung, And Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation120, 1640–1645 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Poulsen, P., Ohm Kyvik, K., Vaag, A. & Beck-Nielsen, H. Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance—a population-based twin study. Diabetologia42, 139–145 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Kupper, N. et al. Heritability of daytime ambulatory blood pressure in an extended twin design. Hypertension45, 80–85 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Musani, S. K. et al. Heritability of the severity of the metabolic syndrome in whites and Blacks in 3 large cohorts. Circ. Cardiovasc. Genet.10, e001621 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham, S. E. et al. The power of genetic diversity in genome-wide association studies of lipids. Nature600, 675–679 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Udler, M. S. et al. Type 2 diabetes genetic loci informed by multi-trait associations point to disease mechanisms and subtypes: a soft clustering analysis. PLoS Med.15, e1002654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimas, A. S. et al. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes63, 2158–2171 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gloudemans, M. J. et al. Integration of genetic colocalizations with physiological and pharmacological perturbations identifies cardiometabolic disease genes. Genome Med.14, 31 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kraja, A. T. et al. A bivariate genome-wide approach to metabolic syndrome: STAMPEED consortium. Diabetes60, 1329–1339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lind, L. Genome-wide association study of the metabolic syndrome in UK Biobank. Metab. Syndr. Relat. Disord.17, 505–511 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Lotta, L. A. et al. Definitions of metabolic health and risk of future type 2 diabetes in BMI categories: a systematic review and network meta-analysis. Diabetes Care38, 2177–2187 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grotzinger, A. D. et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat. Hum. Behav.3, 513–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grotzinger, A. D. et al. Genetic architecture of 11 major psychiatric disorders at biobehavioral, functional genomic and molecular genetic levels of analysis. Nat. Genet.54, 548–559 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallard, T. T. et al. Multivariate GWAS of psychiatric disorders and their cardinal symptoms reveal two dimensions of cross-cutting genetic liabilities. Cell Genomics2, 100140 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demela, P., Pirastu, N. & Soskic, B. Cross-disorder genetic analysis of immune diseases reveals distinct gene associations that converge on common pathways. Nat. Commun.14, 2743 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendrick, D. L. et al. Metabolic syndrome and associated diseases: from the bench to the clinic. Toxicol. Sci.162, 36–42 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet.47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet.47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser, H. F. & Rice, J. Little Jiffy, Mark Iv. Educ. Psychol. Meas.34, 111–117 (1974). [Google Scholar]
- 20.Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun.8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet.44, 369–375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Walree, E. S. et al. Disentangling genetic risks for metabolic syndrome. Diabetes71, 2447–2457 (2022). [DOI] [PubMed] [Google Scholar]
- 23.MacArthur, J. et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res.45, D896–D901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grundy, S. M. Obesity, metabolic syndrome, and cardiovascular disease. J. Clin. Endocrinol. Metab.89, 2595–2600 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet.47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finucane, H. K. et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet.50, 621–629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fehrmann, R. S. N. et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat. Genet.47, 115–125 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Qi, T. et al. Genetic control of RNA splicing and its distinct role in complex trait variation. Nat. Genet.54, 1355–1363 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Võsa, U. et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat. Genet.53, 1300–1310 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aguet, F. et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ge, T., Chen, C. Y., Ni, Y., Feng, Y. A. & Smoller, J. W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun.10, 1776 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yun, J.-S. et al. Polygenic risk for type 2 diabetes, lifestyle, metabolic health, and cardiovascular disease: a prospective UK Biobank study. Cardiovasc. Diabetol.21, 131 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, Y., Han, B.-G. & KoGES Group. Cohort profile: the Korean Genome and Epidemiology Study (KoGES) consortium. Int. J. Epidemiol.46, e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown, B. C., Ye, C. J., Price, A. L. & Zaitlen, N. Transethnic genetic-correlation estimates from summary statistics. Am. J. Hum. Genet.99, 76–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang, Q. Q. et al. Transferability of genetic loci and polygenic scores for cardiometabolic traits in British Pakistani and Bangladeshi individuals. Nat. Commun.13, 4664 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi, S. W. & O’Reilly, P. F. PRSice-2: polygenic risk score software for biobank-scale data. GigaScience8, giz082 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol.37, 658–665 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol.40, 304–314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burgess, S. & Thompson, S. G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol.32, 377–389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verbanck, M., Chen, C. Y., Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet.50, 693–698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim, M. S. et al. Causal effect of adiposity on the risk of 19 gastrointestinal diseases: a Mendelian randomization study. Obes. (Silver Spring)31, 1436–1444 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue, H., Shen, X. & Pan, W. Constrained maximum likelihood-based Mendelian randomization robust to both correlated and uncorrelated pleiotropic effects. Am. J. Hum. Genet.108, 1251–1269 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zawistowski, M. et al. The Michigan Genomics Initiative: a biobank linking genotypes and electronic clinical records in Michigan Medicine patients. Cell Genom.3, 100257 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Rahilly, S. The metabolic syndrome: all in the mind? Diabet. Med.16, 355–357 (1999). [DOI] [PubMed] [Google Scholar]
- 46.Rojas, M. et al. Metabolic syndrome: is it time to add the central nervous system? Nutrients13, 2254 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waterson, M. J. & Horvath, T. L. Neuronal regulation of energy homeostasis: beyond the hypothalamus and feeding. Cell Metab.22, 962–970 (2015). [DOI] [PubMed] [Google Scholar]
- 48.De Hert, M., Schreurs, V., Vancampfort, D. & Van Winkel, R. Metabolic syndrome in people with schizophrenia: a review. World Psychiatry8, 15–22 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Razay, G., Vreugdenhil, A. & Wilcock, G. The metabolic syndrome and Alzheimer disease. Arch. Neurol.64, 93–96 (2007). [DOI] [PubMed] [Google Scholar]
- 50.van Rheenen, W. et al. Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat. Genet.53, 1636–1648 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Storm, C. S. et al. Finding genetically-supported drug targets for Parkinson’s disease using Mendelian randomization of the druggable genome. Nat. Commun.12, 7342 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, Z. et al. Genome-wide association analyses of physical activity and sedentary behavior provide insights into underlying mechanisms and roles in disease prevention. Nat. Genet.54, 1332–1344 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stelzer, G. et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics54, 1.30.1–1.30.33 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Ochoa, D. et al. The next-generation open targets platform: reimagined, redesigned, rebuilt. Nucleic Acids Res.51, D1353–D1359 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Groza, T. et al. The International Mouse Phenotyping Consortium: comprehensive knockout phenotyping underpinning the study of human disease. Nucleic Acids Res.51, D1038–D1045 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ow, J. R., Tan, Y. H., Jin, Y., Bahirvani, A. G. & Taneja, R. Stra13 and Sharp-1, the non-grouchy regulators of development and disease. Curr. Top. Dev. Biol.110, 317–338 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Solomon, S. S., Majumdar, G., Martinez-Hernandez, A. & Raghow, R. A critical role of Sp1 transcription factor in regulating gene expression in response to insulin and other hormones. Life Sci.83, 305–312 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Kraja, A. T. et al. Pleiotropic genes for metabolic syndrome and inflammation. Mol. Genet. Metab.112, 317–338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruan, Y. et al. Improving polygenic prediction in ancestrally diverse populations. Nat. Genet.54, 573–580 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wan, J. Y. et al. Genome-wide association analysis of metabolic syndrome quantitative traits in the GENNID multiethnic family study. Diabetol. Metab. Syndr.13, 59 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Willems, E. L., Wan, J. Y., Norden-Krichmar, T. M., Edwards, K. L. & Santorico, S. A. Transethnic meta-analysis of metabolic syndrome in a multiethnic study. Genet. Epidemiol.44, 16–25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin, L. et al. rMVP: A memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genomics Proteomics Bioinformatics19, 619–628 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vujkovic, M. et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat. Genet.52, 680–691 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winkler, T. W. et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc.9, 1192–1212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hinrichs, A. S. et al. The UCSC genome browser database: update 2006. Nucleic Acids Res.34, D590–D598 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Horn, J. L. A rationale and test for the number of factors in factor analysis. Psychometrika30, 179–185 (1965). [DOI] [PubMed] [Google Scholar]
- 68.Karlsson Linnér, R. et al. Multivariate analysis of 1.5 million people identifies genetic associations with traits related to self-regulation and addiction. Nat. Neurosci.24, 1367–1376 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu, L. T. & Bentler, P. M. Cutoff criteria for fit indexes in covariance structure analysis: conventional criteria versus new alternatives. Struct. Equ. Model.6, 1–55 (1999). [Google Scholar]
- 70.Altshuler, D. M. et al. Integrating common and rare genetic variation in diverse human populations. Nature467, 52–58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dunham, I. et al. An integrated encyclopedia of DNA elements in the human genome. Nature489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cahoy, J. D. et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci.28, 264–278 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol.11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watanabe, K., Umićević Mirkov, M., de Leeuw, C. A., van den Heuvel, M. P. & Posthuma, D. Genetic mapping of cell type specificity for complex traits. Nat. Commun.10, 3222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kang, H. J. et al. Spatio-temporal transcriptome of the human brain. Nature478, 483–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet.48, 481–487 (2016). [DOI] [PubMed] [Google Scholar]
- 79.Raulerson, C. K. et al. Adipose tissue gene expression associations reveal hundreds of candidate genes for cardiometabolic traits. Am. J. Hum. Genet.105, 773–787 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taylor, D. L. et al. Integrative analysis of gene expression, DNA methylation, physiological traits, and genetic variation in human skeletal muscle. Proc. Natl Acad. Sci. USA116, 10883–10888 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou, W. et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet.50, 1335–1341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wald, A. The fitting of straight lines if both variables are subject to error. Ann. Math. Stat.11, 284–300 (1940). [Google Scholar]
- 83.Burgess, S., Bowden, J., Fall, T., Ingelsson, E. & Thompson, S. G. Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology28, 30–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Park, S. sanghyeonp/Park-Nature-Genetics-2024: Park-Nature-Genetics v1.0.0. Zenodo10.5281/zenodo.13137680 (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Notes 1–11 and Figs. 1–15.
Supplementary Tables 1–51.
Data Availability Statement
Summary statistics of the multivariate and KoGES MetS GWAS are available through GWAS Catalog (https://www.ebi.ac.uk/gwas/) under accession IDs GCST90444487–GCST90444489. The PRS weights developed for MetS are available through PGS Catalog (https://www.pgscatalog.org/) with publication ID PGP000664 and score ID PGS004928. The individual-level phenotypic and genetic data for UKB (https://www.ukbiobank.ac.uk/) and KoGES (https://is.kdca.go.kr/) can be accessed through the application. The reference panel for 1kGp3 can be obtained from the following URL: https://mathgen.stats.ox.ac.uk/impute/1000GP_Phase3.html. The NHGRI-EBI GWAS Catalog is available at the following URL: https://www.ebi.ac.uk/gwas/docs/file-downloads. The GWAS summary statistics from the following consortia and biobanks are publicly available at the corresponding URL: GIANT (https://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files), MAGIC (https://magicinvestigators.org/downloads/), GLGC (http://www.lipidgenetics.org/#data-downloads-title), FinnGen (https://www.finngen.fi/en/access_results), UKB from Neale laboratory (http://www.nealelab.is/uk-biobank), UKB from Lee lab (https://www.leelabsg.org/resources) and PanUKB (https://pan.ukbb.broadinstitute.org/downloads). The GWAS summary statistics of the MVP cohort were obtained using an approved dbGaP application (phs001672.v9.p1). The GWAS summary statistics from the MGI cohort are available upon request from https://precisionhealth.umich.edu/our-research/documents-for-researchers. The eQTL summary statistics are available from GTEx (https://gtexportal.org/home/downloads/adult-gtex#qtl), eQTLGen (https://www.eqtlgen.org/cis-eqtls.html), BrainMeta v.2 (https://yanglab.westlake.edu.cn/software/smr/#DataResource), METSIM (https://mohlke.web.unc.edu/data/1702-2/) and FUSION (https://www.ebi.ac.uk/birney-srv/FUSION/).
The script used to conduct the parallel analysis was written by Javier de la Fuente and is available via GitHub (https://github.com/AnnaFurtjes/genomicPCA/blob/main/data); also available on GitHub are the Genomic SEM software (https://github.com/GenomicSEM/GenomicSEM), the LDSC software including S-LDSC and SEG-LDSC (https://github.com/bulik/ldsc) and the Popcorn software (https://github.com/brielin/Popcorn). LDScore is available at https://alkesgroup.broadinstitute.org/LDSCORE and FUMA at https://fuma.ctglab.nl. The analysis codes are deposited with the repository name Park-Nature-Genetics-2024 via Zenodo at 10.5281/zenodo.13137680 (ref. 84).