SUMMARY
The “innate-like” T cell compartment, known as Tinn, represents a diverse group of T cells that straddle the boundary between innate and adaptive immunity. We explore the transcriptional landscape of Tinn compared to conventional T cells (Tconv) in the human thymus and blood using single-cell RNA sequencing (scRNA-seq) and flow cytometry. In human blood, the majority of Tinn cells share an effector program driven by specific transcription factors, distinct from those governing Tconv cells. Conversely, only a fraction of thymic Tinn cells displays an effector phenotype, while others share transcriptional features with developing Tconv cells, indicating potential divergent developmental pathways. Unlike the mouse, human Tinn cells do not differentiate into multiple effector subsets but develop a mixed type 1/type 17 effector potential. Cross-species analysis uncovers species-specific distinctions, including the absence of type 2 Tinn cells in humans, which implies distinct immune regulatory mechanisms across species.
In brief
Our study presents a comprehensive atlas of human innate T (Tinn) cells in the thymus and blood, highlighting their blended type 1 and type 17 transcriptional profiles. Loh et al. use single-cell RNA sequencing and flow cytometry to reveal species-specific distinctions and potential divergent developmental pathways of Tinn compared to conventional T cells.
Graphical Abstract
INTRODUCTION
Conventional CD4+ and CD8+ T cells (Tconv) are essential for adaptive immunity, recognizing peptide antigens presented by HLA class I or II proteins via their T cell antigen receptors (TCRs). Upon antigenic stimulation, these T cells undergo transcriptional and epigenetic changes, secreting pro-inflammatory cytokines, acquiring cytotoxic abilities to clear pathogens, or forming memory T cells for rapid response upon reencounter. Thus, Tconv cells within the circulation are heterogeneous and are commonly classified into naive (Tn), central memory (Tcm), effector memory (Tem), and terminally differentiated effector memory (Temra) subsets.1–3
Innate-like T cells (Tinn), such as invariant natural killer T (iNKT) cells, mucosal-associated invariant T (MAIT) cells, and certain γδ T cells, maintain consistent TCRs among individuals and function without prior pathogen exposure. They account for 10%–20% of human T cells.4,5 Tinn cells, originating from thymic progenitors, do not recognize peptides presented by HLA class I or II. Instead, iNKT cells use semi-invariant TCRs to recognize lipid antigens presented by CD1D.6 MAIT cells, also characterized by semi-invariant TCRs,7 recognize riboflavin precursor-derived metabolites presented by MR1.8,9 Some γδ T cells recognize self- and foreign phosphoantigens in conjunction with the transmembrane butyrophilin-family receptors BTN2A1-BTN3A1-BTN3A2 complex.10–12 The antigens recognized by other human γδ T cell populations remain unclear.13 Thus, Tinn cells enhance immune detection of diverse threats.14–16 Furthermore, the secretion of cytokines at steady state by mouse iNKT cells influences surrounding cells and Tconv development,17–19 suggesting that they may also function as gatekeepers, ensuring proper T cell development and maturation.
The conservation of Tinn cells throughout mammalian evolution indicates their crucial and non-redundant role in the immune system, which may be attributed to their rapid activation kinetics and their ability for TCR-independent activation.20–22 In mice, the rapid effector capacity of Tinn cells stems from unique transcriptional programs formed during their development in the thymus.23,24 Analogous to CD4 Tconv cells, which can be polarized by cytokines into T helper (Th) phenotypes such as Th1, Th2, and Th17 that secrete interferon-γ (IFN-γ), interleukin-4 (IL-4), and IL-17, respectively, mouse Tinn cells diverge into distinct, terminally differentiated subsets that can be readily identified based on the expression of specific transcription factors (TFs) such as PLZF, GATA3, T-bet, and RORγt.17
In this study, using single-cell genomics and flow cytometry, we assessed the range of phenotypic states Tconv and Tinn cells can adopt in vivo in the human thymus and blood. We uncovered that unlike adult blood Tinn cells most postnatal thymic Tinn cells resemble naive Tconv cells, with few displaying an “effector” state. A distinct transcriptional program is shared among human Tinn cells, and major TFs driving this program are also expressed in mice. However, unlike the mouse, human Tinn cells do not form distinct subsets but develop a mixed type 1/type 17 effector program. We further highlight differences in CD1D and SLAMF6 expression in the thymus between the two species, which could potentially impact the maturation process of iNKT cells in humans. This comprehensive transcriptomic dataset of human Tinn cells provides a valuable resource to explore their fundamental properties and may serve as a guide for their strategic application in immunotherapeutic approaches.
RESULTS
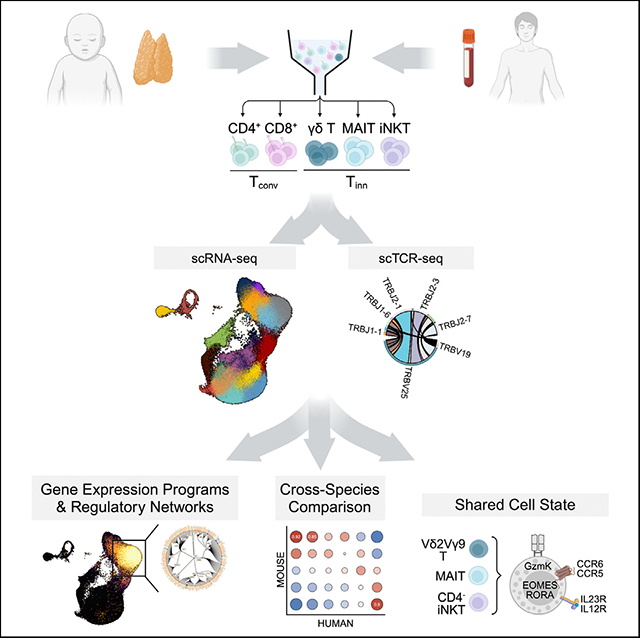
Single-cell RNA-sequencing analysis of T cell maturation and post-maturation stages in humans
We explored the transcriptional profiles of Tinn and Tconv cells through single-cell RNA sequencing (scRNA-seq) of antibody DNA-barcoded (“hashtags”) tetramer-sorted iNKT (PBS57-CD1d tetramer+, TRAV10+), MAIT (5-OP-RU-MR1 tetramer+, TRAV1–2+), and total γδ T cells, along with single-positive (SP) CD4 and CD8 Tconv cells from pediatric thymus and adult blood samples (Figure S1). A subset of samples was also subjected to VDJ sequencing (Figure 1A and Table S1). We analyzed 78,355 cells (37,369 pediatric thymus and 40,986 adult blood) after quality control, integrating them into a reference dataset (Figures 1B, 1C, S2A, and S2B). Using unsupervised clustering, we identified 18 distinct and stable clusters (c), primarily separating into thymus (c0–c9) or blood-associated regions (c12–c17, Figures 1C and S2C), with transitioning clusters (c10 and c11) showing cells from both tissues (Figures 1D and 1E), representing naive T cells prepared to leave the thymus or just entering the blood, in agreement with their over-representation of an “egress” gene signature25 (Figure 1F and Table S3).
Figure 1. Integrative view on Tinn and Tconv development and peripheral function.
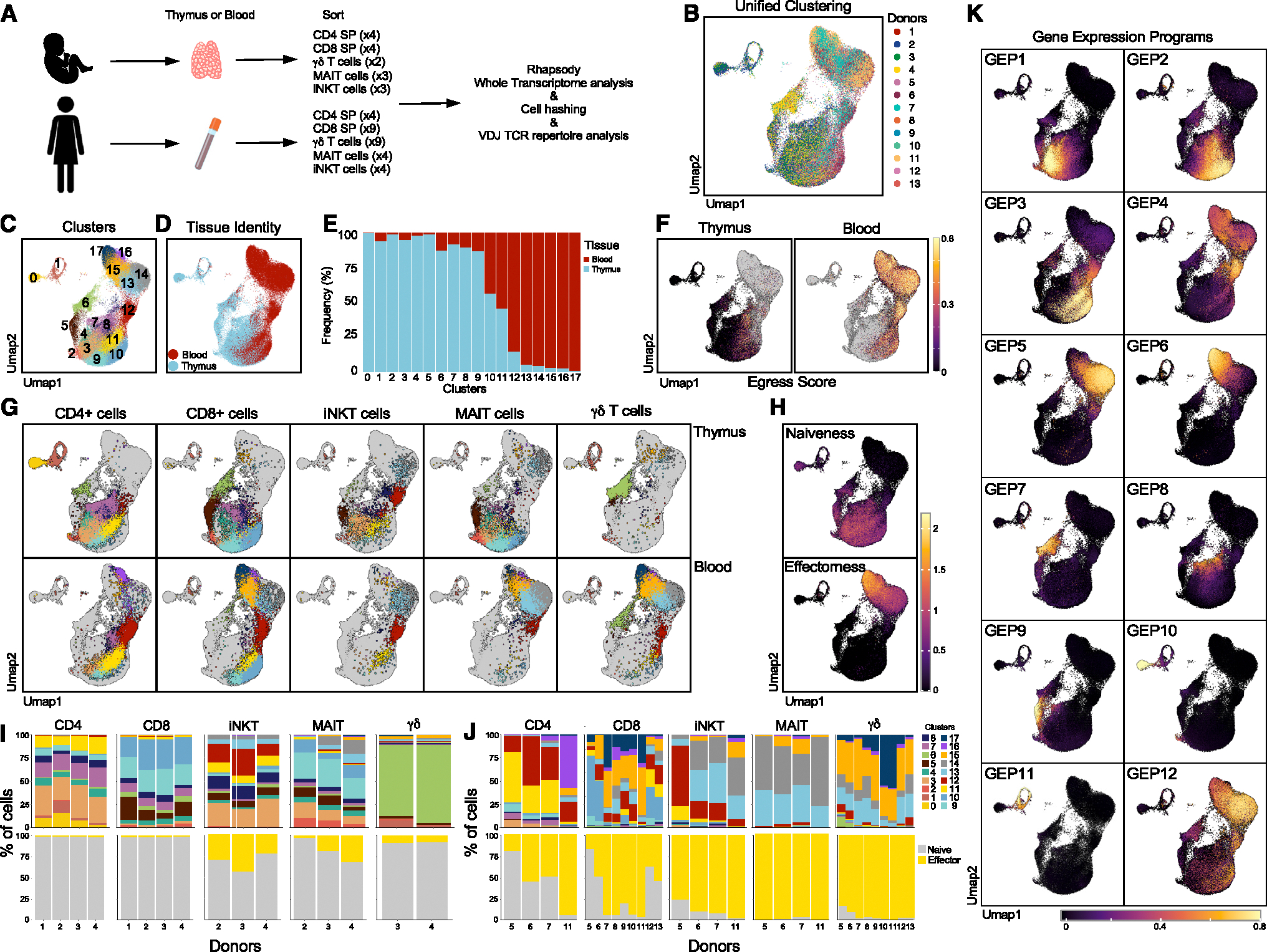
(A) Experimental setup specifying donor type (postnatal/adult), tissue, and sorted cell types.
(B) Harmony batch-corrected and integrated dataset across donors, tissues, and cell types.
(C–E) (C) Stable Louvain-derived cell clusters distributed across (D) both blood and thymus-derived cells and (E) their respective frequencies in these clusters.
(F) “Egress” score on thymus- and blood-derived cells.
(G) Cells color coded by cluster (as in C) visualized by their hashtag-sorted cell type (columns) and the tissue they originated from (rows).
(H–J) (H) Projection of naive and effector scores and the proportion of (I) thymic and (J) blood cell types per donor, classified on the basis of these scores (bottom row); top row shows analogous proportions by cell cluster (as in C).
(K) Gene-expression programs (GEPs) in thymus and blood identified using cNMF, with color scale representing GEP usage. Sample numbers for all panels as depicted in (A). Score defining genes as described in text.
Cluster identities and transcriptional states were determined using reference genes (Figures S3A and S3B; Table S2), with cell types validated by neighbor voting26 with the human thymus atlas27 (Figure S4). Early T cell development included immature single positive (ISP) cells (c0 and c1), double-positive (DP) thymocytes (c2), and early SP stages characterized by CCR9 expression (c3 for CD4 SP and c9 for CD8 SP). Later stages expressed CCR7 (c11 for CD4 SP, c10 for CD8 SP; Figure S5A). Differentially expressed gene (DEG) analysis comparing CD4 SP cells (c3 and c11) to CD8 SP cells (c9 and c10) further confirms these annotations (Figure S5B). Specialized lineages were also detected, including CD8αα cells (GNG4 and NUCB2), thymic γδ T cells (TRDC, TRGC2), and regulatory T cells (Tregs) with high expression of FOXP3 (c5, c6, and c7, respectively). Other signaling states included “agonist” cells with high levels of TF transcripts associated with TCR signaling (c4; NR4A1, EGR1, EGR3, and NFKBID), cells with high expression of type I IFN signaling genes (IFI6, MX1, and IFI44L in c8), and AP-1 TFs (JUN, FOS, JUNB in c12). We also found cells (c13 through c17) expressing effector-encoding genes (GZMK, GZMH, GZMB, PRF1, and CCL5), suggesting involvement in effector functions of these cells (Figures S3A, S3B, and S5A). Altogether, we observed cells with distinct transcriptional profiles, representing unique cell types (CD8αα, Tregs) and stages of T cell development and maturation.
Identification of the gene-expression programs that characterize T cell populations in thymus and blood
Deciphering scRNA-seq data can be challenging due to the complexity of gene-expression patterns reflecting cell identity and activity. We used consensus non-negative matrix factorization (cNMF)28 to reduce dimensionality, identifying 12 gene-expression programs (GEPs; Figure 1K and Table S4). Each cell type’s contribution to these GEPs was assessed, revealing some GEPs shared across cell types, with others unique to specific clusters (Figures 1G and 1K). GEPs 7–11 were linked to thymic γδ T cells, Tregs, thymic CD8αα T cells, quiescent ISP, and proliferating ISP, respectively (Figure 1K). GEP 12 was excluded due to batch effects (Figure S6). GEPs 1 and 2, marked by CCR9 and CCR7, were active in early and late thymic T cells, respectively. GEP3 was expressed by naive T cells. GEPs 4, 5, and 6, associated with “effectorness,” were active in clusters 12, 13–14, and 15–17, respectively (Figures 1G–1K). Leveraging insights from these GEPs, we conducted an in-depth analysis of thymic and blood T cell populations, providing an integrated understanding of T cell differentiation and function.
Gene-expression similarities between αβ thymocyte lineages
We separated each sample by cell type and tissue, using the identifying tag (Figure 1G). CD4+ thymic cells were found in clusters 0–4, 7, 8, and 11, while CD8+ thymic cells were in clusters 2 and 5–10. Cell proportions in each cluster were consistent across samples, with about 1% (1.1% ± 0.4%) of the cells showing an effector signature (Figures 1G–1I and Table S3). Thymic iNKT cells were distributed similarly to conventional CD4+ SP, and thymic MAIT cells shared clusters with CD8+ SP (Figure 1G). Thymic γδ T cells were distinct, mainly in cluster 6 and GEP7 (Figures 1G and 1K).
Global gene-expression analysis between thymocyte lineages revealed three distinct patterns. First, effector stages of iNKT, MAIT, and γδ thymocytes (clusters 12–17) exhibited high gene-expression similarity. Second, non-effector γδ thymocytes showed no expression similarity to other lineages. Third, non-effector iNKT and MAIT thymocytes (clusters 0–11) resembled CD4+ and CD8+ thymocytes, respectively (Figure 2A). This suggests that specific subpopulations of iNKT thymocytes engage transcriptional programs akin to those of conventional CD4+ thymocytes, with a parallel observation between MAIT and CD8+ thymocytes. Pseudo-bulk DEG analysis between non-effector iNKT/CD4+ and non-effector MAIT/CD8+ further demonstrated that non-effector iNKT and CD4+ thymocytes share 43 commonly upregulated and 38 commonly downregulated genes (Figure 2B). Key genes such as GATA3, ZBTB7B, and CD40LG were upregulated in non-effector iNKT thymocytes, indicating CD4 lineage differentiation, while CD8A, CD8B, and RUNX3 were elevated in non-effector MAIT thymocytes. Using DEG analysis between CD4+/MAIT and CD8+/iNKT cells as a negative control revealed only EPHB6, ZFAS1, and EEF1A1 as differentially expressed (Figure 2B). Comparing conventional CD4+/CD8+ thymocytes with non-effector iNKT/MAIT cells highlighted 11 DEGs (Figure 2C), including ZBTB16 and SLAM family receptors SLAMF5 (CD84) and SLAMF6, underscoring specific pathways crucial for effector differentiation, similar to observations in mice.23
Figure 2. Gene-expression patterns in thymocyte lineages.
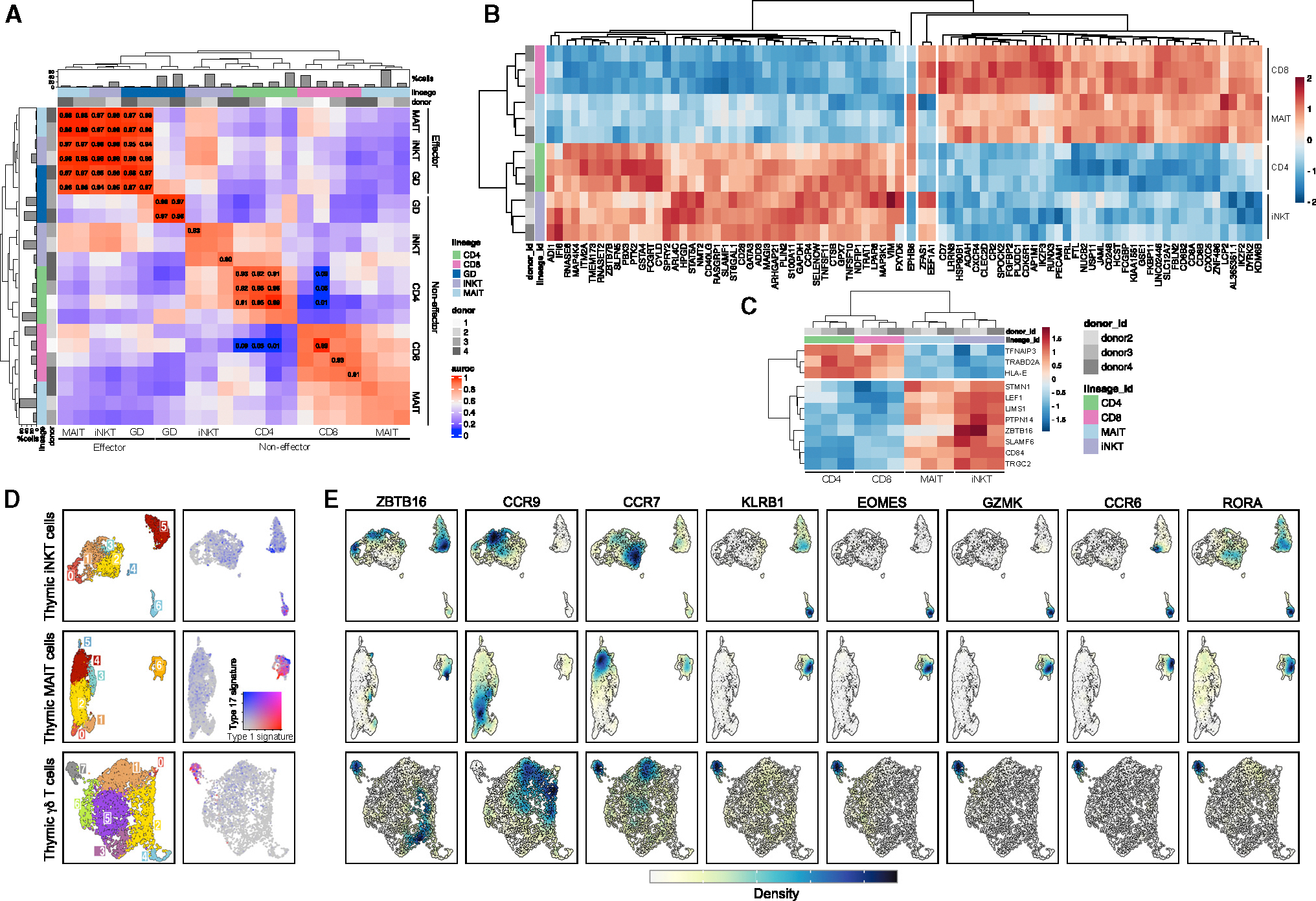
(A) Heatmap showing MetaNeighbor’s AUROC scores between thymocytes split by donor, lineage, and non-effector (c0–11) versus effector (c12–17) clusters. Barplots indicate thymocyte proportions per lineage.
(B) Pseudo-bulk differential expression analysis between CD4+/iNKT and CD8+/MAIT thymocytes in naive clusters (3, 9, 10, 11). As a negative control, the only three genes that were differentially expressed between CD4+/MAIT and CD8+/iNKT thymocytes are displayed in the center of the heatmap.
(C) Pseudo-bulk differential expression analysis between CD4+/CD8+ and iNKT/MAIT thymocytes in naive clusters (3, 9, 10, 11). For both (B) and (C), heatmap displays the expression level of genes (represented with color scale as a Z score of the average normalized expression) that are significantly differentially expressed (padj < 0.01).
(D) Clustering of hashtag-separated thymic iNKT cells (top), MAIT cells (middle), and γδ T cells (bottom). Right panel shows the score of type I and type III effector gene signature for the corresponding thymic lineage.
(E) Kernel density estimates of the normalized expression level of genes of interest. The expression level distribution varies between genes and lineage. The range of kernel density estimate values also varies between each panel (from 0 to 0.04 for the smallest range and 0 to 0.4 for the largest range). A unique color scale was represented to indicate the direction of the values.
Unbiased transcriptomic analysis of human Tinn differentiation
To further explore the transcriptional heterogeneity of human Tinn thymocytes, we re-analyzed iNKT and MAIT cell populations individually (Figures 2D–2E and S7). We identified seven clusters for both cell types (Figure 2D), with some variability in their proportion across donors (Figures S7B and S7F). Five major cell signatures were shared across Tconv, iNKT, and MAIT cells (Figures S3C and S3D). First, we observed a distinctive gene signature associated with CD8αα T cells (captured by GEP9; Figure S8; Tables S5 and S6). This signature was characterized by the heightened expression of NUCB2, MINDY2, and HIVEP3 (Figures S3C and S3D), and intriguingly, it was observed in both iNKT and MAIT cells (termed NKT_c0 and MAIT_c1; Figures S7C and S7G). This subset of thymic CD8A-expressing iNKT cells, which also exhibited some PLZF expression while lacking KLRB1 (coding for CD161), EOMES, and GZMK expression (Figure 2E), could be readily identified using flow cytometry (Figure S9). Second, we identified a shared pattern of expression for the CCR9 and CCR7 chemokine receptors across Tconv cells, iNKT cells, and MAIT cells (Figure 2E). Initially, iNKT and MAIT cells exhibited an upregulation of CCR9 in conjunction with TOX and SATB1 (Figures S3C and S3D), resembling the developmental program of early developing CD4 SP and CD8 SP cells, respectively. Subsequently, the elevated expression of CCR7 marked cells at a seemingly more advanced developmental stage (termed NKT_c2 and MAIT_c4). These sequential waves of chemokine receptor expression align with gene modules GEP1 and GEP2 (Figure S8 and Table S4), suggesting their sequential induction during the development of CD4, CD8, iNKT, and MAIT cells. These findings establish that the human thymus harbors iNKT and MAIT cells with a transcriptome resembling that of developing naive Tconv cells. The existence of such naive-like populations (CD161−EOMES−GZMK−) of iNKT and MAIT cells in the human thymus was confirmed by flow cytometry (Figure S9).
Third, we discovered iNKT and MAIT cells characterized by upregulation of genes associated with type I IFN signaling such as MX1 and IFI6, similar to CD4 and CD8 SP cells (NKT_c3 and MAIT_c5; Figures S3C and S3D). Fourth, TCR signaling/AP-1 signatures were found in iNKT cells, with AP-1 TFs FOS and JUN upregulated alongside ZBTB16 and KLRB1. These cells also expressed CD4 transcripts but not CD8A (NKT_c5; Figures 2E and S3C). The TCR signature was more pronounced in MAIT cells, where a small subset showed clear upregulation of genes involved in TCR signaling (NR4A1, NFKBID, REL; MAIT_c3; Figure S3D). Unlike Tconv, a proportion of both thymic iNKT and MAIT cells displayed an “effector” signature. iNKT cells in cluster 6 (NKT_c6) express classically iNKT-associated genes along with upregulation of effector genes usually associated with type 1 or type 17 immunity (EOMES/GZMK and RORA/CCR6, respectively; Figures 2E and S3C). Some of these cells expressed CD8A transcripts, suggesting that CD4+ and CD8+ iNKT cells might develop into transcriptionally distinct subsets, with CD8+ iNKT cells having a more effector-associated signature. We found the same type 1 (EOMES/GZMK) and type 17 (RORA/CCR6) immunity effector co-expression signatures in MAIT cells (MAIT_c6; Figures 2D and S3D), with upregulation of genes encoding for granzymes (GZMA and GZMK) and chemokines (CCL5), as well as genes previously associated with MAIT cells29,30 (KLRB1, SLC4A10, IL23R; Figure 2E).
Analysis of the thymic γδ T cells identified eight distinct clusters (Figures 2D, S3E, and S7I–S7L; Table S7), confirming previous findings on pediatric γδ thymocytes25 with immature populations (GD_c0, 1, 2), TCR activation profiles (GD_c3), type I IFN response (GD_c6), and effector γδ T cells with egress and mixed type 1/type 17 effector potential (GD_c7). We also observed cells with a cycling gene signature (GD_c4), which was notably absent in iNKT and MAIT cells. Overall, we found a shared utilization of a mixed type 1/type 17 effector programs by iNKT, MAIT, and γδ T cells, with 28.7% ± 22.3% of thymic Tinn cells displaying this effector signature (Figures 1G and 1I, in clusters 12–17).
Lastly, because effector Tinn cells in the thymus might represent cells that initially acquired an effector signature in the blood and subsequently recirculated back to the thymus, we compared expression profiles between thymic and blood effector iNKT and MAIT cells. We found distinctive tissue-specific gene-expression profiles for both cell types (Figure S5C), indicating that thymic effector Tinn cells are unlikely to be derived from recirculating blood cells.
Effect of clonal selection on iNKT, MAIT, and γδ T cells’ effector states
To investigate whether Tinn cells with an effector transcriptome had distinct TCR repertoires compared to naive Tinn cells, we conducted paired VDJ sequencing (Figure 3).
Figure 3. Innate T cell TCR diversity during development.
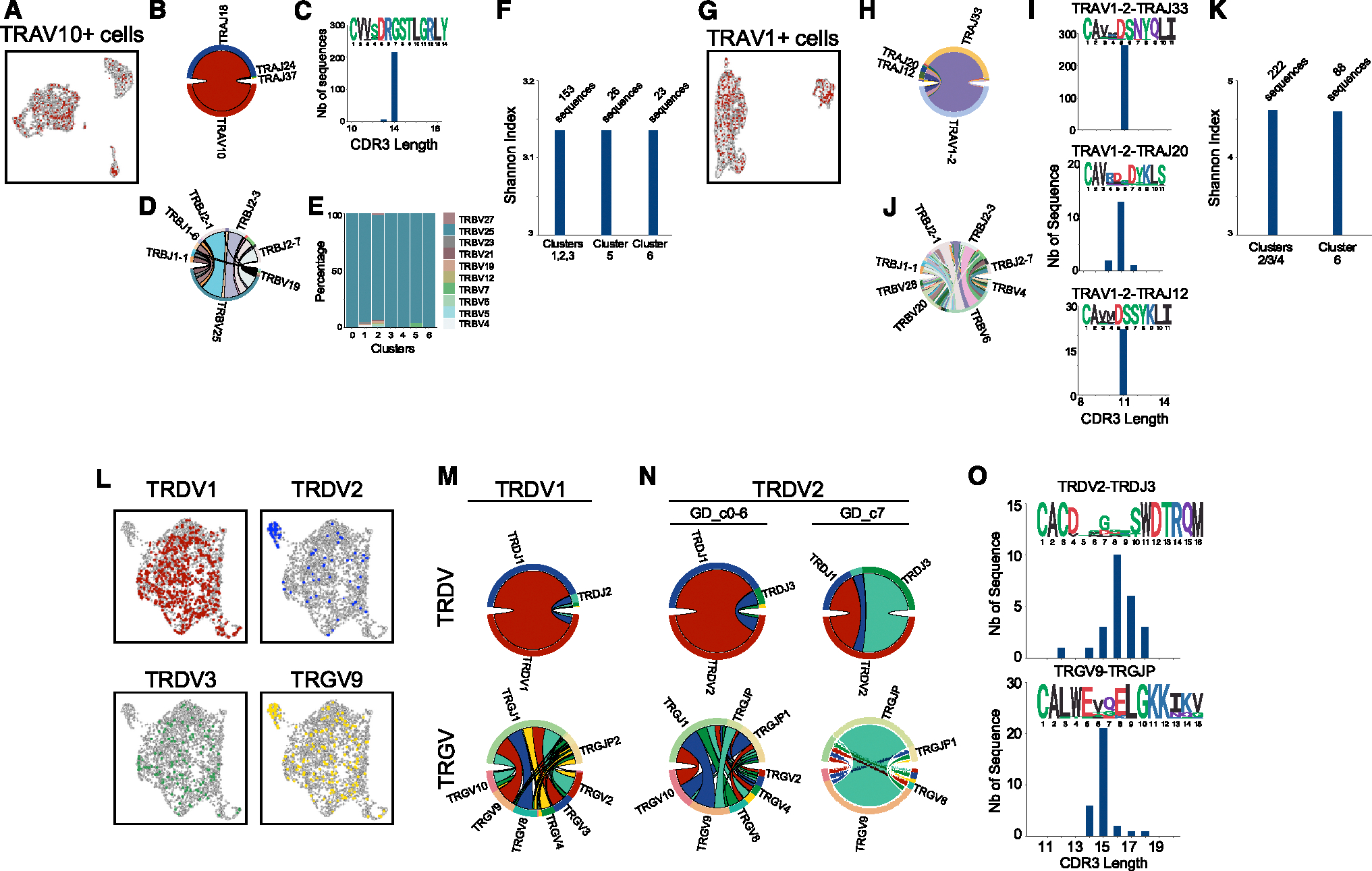
Cells with VDJ sequencing and their cell-type-specific characteristic chain arrangement for thymic iNKT cells (A), MAIT cells (G), and γδ T cells (L). For each cell type, the respective proportions of gene segment usage in each chain (B and D; H and J; M and N) are shown together with their CDR3 length and sequence logo (C, I, O) and their cluster-specific usage (E, with clusters as in Figure 2A). Shannon index as an estimation of TCR diversity in the naive-like and effector-like iNKT (F) and MAIT (K) cells, based on clusters in Figure 2D. n = 1 human thymus sample for all panels.
For iNKT cells, most VDJ sequenced cells used the TRAV10 gene segment (encoding for the Vα24 chain) rearranged with TRAJ18 (Figures 3A and 3B), resulting in a canonical CDR3α sequence of 14 amino acids (Figure 3C), crucial for antigen recognition.31 This invariant TCRα chain paired with diverse TCRβ rearrangements, mainly involving the TBV25 chain (Figure 3D), and was used evenly across all clusters (Figure 3E). The Shannon index, a measure of TCR diversity, revealed that no clonal selection had occurred or was associated with effector transcriptome development (Figure 3F), and no shared TCR clonotypes were identified between naive- and effector-like cells.
Thymic MAIT cells primarily used the TRAV1–2 gene segment (encoding for the Vα7.2 chain) with TRAJ33, TRAJ20, and TRAJ12 (Figures 3G and 3H), maintaining a conserved Y95 residue in the CDR3α loop (Figure 3I) essential for MAIT cell activation.32,33 These TCRα chains paired with diverse TCRβ chains (Figure 3J) dominated by TRBV6, TRBV20, and TRBV4 gene segments.34,35 Like iNKT cells, MAIT cells showed no shared TCR clonotypes and no clonal selection in effector transcriptome cells (MAIT_c6) compared to naive-like cells (MAIT_c2–4), based on the Shannon index (Figure 3K).
Effector γδ T cells (GD_c7) were enriched for cells expressing TRDV2 and TRGV9 gene segments, while TRDV1 and TRDV3 segments were excluded from this cluster (Figure 3L). Some TRDV2+ or TRGV9+ cells were also found in non-effector clusters, suggesting a potential role for these gene segments in the development of effector γδ T cells in the postnatal human thymus. Supporting this hypothesis, we observed that the rearrangements of both the Vδ2 chains and associated Vγ9 chains differed largely between cells in the effector versus non-effector clusters (Figure 3N). Specifically, the Vγ9 chains of effector cells were found to be preferentially rearranged with the TRGJP gene segment and enriched for the public CDR3 sequence typically found among Vδ2Vγ9 γδ T cells in adult blood36 (Figure 3O), whereas Vδ2+ cells in the non-effector clusters showed more diverse Vg gene usage and rearrangements (Figure 3N). In summary, the acquisition of effector programs in iNKT and MAIT cells is not associated with changes in TCR diversity, whereas Vδ2 and Vγ9 chain rearrangements in γδ T cells suggest a predisposition toward the effector program.
Gene-expression programs that characterize T cell effector functions
To further characterize the functionality of Tinn and Tconv cells in the blood, we examined the distribution of these cell types across transcriptional clusters. Conventional CD4+ T cells were primarily located in cluster 11 (naive CD4 T cells) and cluster 7 (Tregs). CD4+ T cells also appeared in effector clusters, particularly clusters 12 and 16, with proportions varying among donors (18.4%–93.7%; Figures 1G and 1J). Blood CD8+ T cells were predominantly in cluster 10 (naive CD8 T cells), with varying proportions in effector clusters, reflecting differences in donor immunological history (16.5%–94%; Figure 1J). In contrast, the majority of blood Tinn cells (~94.3% ± 6.7%) were distributed across effector clusters 12–17, regardless of donor (Figure 1J).
We next investigated transcriptional states in blood T cell populations using cell hashtags to reanalyze blood iNKT cells, MAIT cells, γδ T cells, and Tconv CD4+ and CD8+ T cells individually (Figures 4A and 4B). Each cell type was found within previously identified GEPs (GEP3–6; Figure 1K; Tables S8, S9, S10, S11, and S12), albeit with varying proportions for each cell type (Figures 4C and 4D). To contextualize these GEPs, we computed overlap scores and statistically assessed their enrichment with literature-derived signatures.37–40 Subsequently, we scored the joint signature-GEP interactions in our dataset (Figure S10). GEP3 was found to be closely associated with signatures of naive T cell characteristics. In contrast, GEP4 displayed similarities with Tcm, Tem, or literature-derived signatures classified as a mix thereof (Tcm/Tem), while GEP6 exhibited characteristics akin to Temra. GEP5, on the other hand, shared elements with Tem cells and previously identified CD8 MAIT signatures (Figure S10A).
Figure 4. Gene-expression programs in circulating Tinn and Tconv.
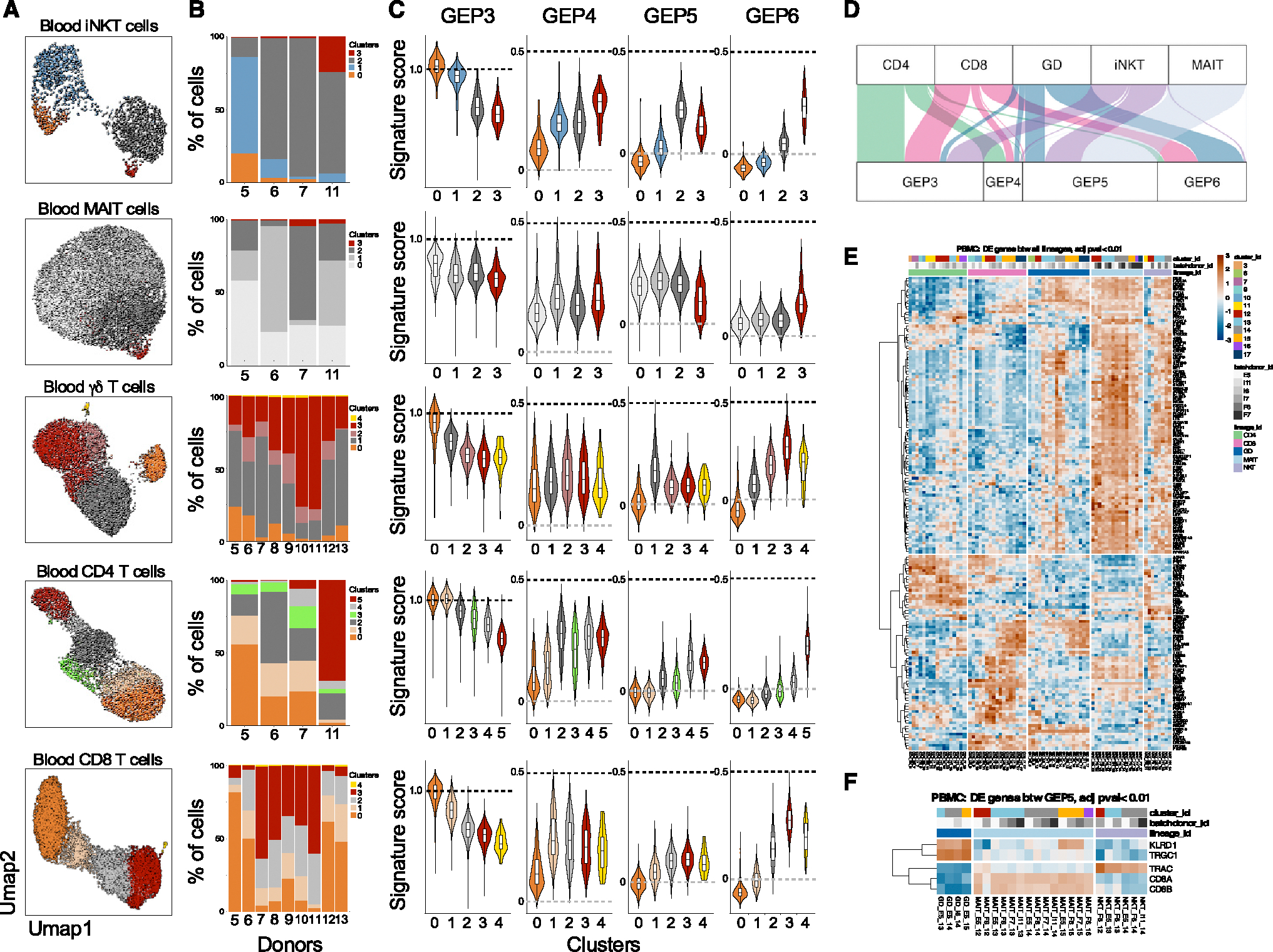
(A–C) (A) Clustering of hashtag-separated blood iNKT, MAIT, γδ T, CD4, and CD8 T cells, (B) the respective proportion of cells per cluster and donor, and (C) the effector GEP signature scores (as in Figure 1K) per cell type and cluster.
(D) Top GEP usage for each cell type, based on cNMF-derived usage matrix.
(E) Pseudo-bulk, pairwise differential gene expression between cell types.
(F) Cell-type-specific genes among Tinn cells using GEP5. For both (E) and (F), heatmaps depict the expression level of genes (represented with color scale as a Z score of the average normalized expression) that are significantly differentially expressed (padj < 0.01). n = 4, 4, 9, 4, and 9 for iNKT, MAIT, γδ, CD4, and CD8 cells, respectively.
Blood iNKT cells predominantly expressed GEP3 or GEP5, with some expressing GEP4 or GEP6 (Figure 4D). GEP4-expressing iNKT cells had CD4 transcripts, while GEP5 and GEP6 cells lost CD4 expression (Figures S11A and S11B). To validate this observation, we examined the cellular phenotype of blood iNKT cells. Blood CD4+ iNKT cells were mostly PLZF−CD161−EOMES−GZMK− but CCR7+, indicating a naive GEP3 program (Figure S11C). In contrast, CD8+ and double-negative iNKT cells were mostly PLZF+CD161+ and displayed an effector phenotype (EOMES+GZMK+CCR7−CD62L−; Figure S11). Interestingly, the distribution of these programs varied significantly among different donors (Figure 4B). These findings are in line with previous data indicating that CD4-negative iNKT cells become more prevalent in the blood with age, eventually becoming the dominant population in the adult blood iNKT cell compartment.41,42 This suggests that CD4-negative iNKT cells may originate from CD4+ iNKT cells that undergo a loss of CD4 expression as they transition toward a more effector-like state. Conversely, when examining MAIT cells in the blood, we observed that the majority of them exhibited the GEP5 program, with only a minor fraction utilizing the GEP6 program (Figure 4D). This GEP phenotype was further confirmed by flow cytometry, revealing that most MAIT cells were CD8+ and, interestingly, that all MAIT cells displayed a uniform effector state (characterized by PLZF+CD161+EOMES+GZMK+CCR7−CD62L−), regardless of CD8 expression (Figure S12). These findings indicate that MAIT cells in the bloodstream primarily exist in an exclusive transcriptional state.
Blood γδ T cells were stratified into five clusters: naive (c0, GEP3), cycling (c4, GEP11), and clusters c1–c3 categorized into GEP5 or GEP6 programs (Figures 4C and 4D). This division in GEP utilization closely mirrored the specific TCR usage among these cells. TRDV2/TRGV9-expressing cells were predominantly GEP5, while TRDV1+ or TRDV3+ cells were enriched in GEP6 (Figures S13A–S13C). Flow cytometry confirmed Vγ9+Vδ2+ T cells primarily expressed PLZF and GZMK, while Vδ2− T cells were increased in GZMB+ cells (Figures S13D and S13E). Thus, GEP5 is an effector gene module unique to Tinn cells, indicating common transcriptional states in human Tinn cells.
The distribution of CD4 and CD8 Tconv cells in the blood revealed two primary patterns. Tconv cells were found either within clusters containing naive cells (clusters 0 and 1) characterized by high expression of GEP3 or dispersed across clusters of cells displaying a gradient of the GEP6 program, with intermediary cells expressing GEP4 (Figure S14). The proportions of cells in these clusters exhibited variations among donors (Figure 4B). Together, these findings underscore the distinct associations between different T cell types and effector programs. In the blood, CD4− iNKT, MAIT, and Vγ9+Vδ2+ γδ T cells predominantly employ the GEP5 program, a program also shared by effector Tinn cells in the thymus (Figure S8). Conversely, conventional CD4+ and CD8+ T cells transition into effector cells along a gradient defined by the GEP6 program. Notably, this GEP6 program is also shared by Vδ3+ and Vδ1+ γδ T cells.
Next, we used pairwise DEG analyses between T cell lineages in human blood to identify lineage-specific genes. We uncovered a total of 167 genes that exhibited significant differential expression (padj < 0.01) in at least two of the comparisons we conducted (Figure 4E). These distinct patterns of DEGs provided insights into changes linked to the transition from a “naive” state to an “effector” state across cell types. Furthermore, we identified genes that were commonly expressed by two distinct cell types when compared to the others. Nevertheless, we did not readily discern any gene-expression patterns specific to a particular cell type. However, intriguingly, among these, a group of 104 genes distinguished γδ, MAIT, and iNKT cells from Tconv CD4 and CD8 T cells, with 63% overlapping with GEP5. Given that only Vγ9+/Vδ2+ T cells share the GEP5 program with iNKT and MAIT cells, while Vδ2− T cells exhibit greater similarity to Tconv cells as they share the GEP6 program (Figure S13C), we explored whether we could identify cell-type-specific gene signatures specifically among GEP5-expressing cells using the same analytical approach. Surprisingly, the results demonstrated that the only significant DEGs between iNKT, MAIT, and γδ T cells employing the GEP5 program (Figure 4F) were genes encoding the constant regions of the TCR genes (TRGC1, TRAC), the CD8 coreceptor (CD8A, CD8B), and the CD94 receptor (encoded by KLRD1). Thus, human blood Tinn cells, which encompass iNKT, MAIT, and Vγ9+/Vδ2+ cells, distinguish themselves from Tconv cells by employing a specific gene program, but there is minimal transcriptional difference among Tinn cells themselves.
The effector GEPs exhibit distinct migration, cytokine, chemokine, and integrin characteristics established by distinct gene-regulatory networks
The differentiation states of T cells are intricately linked to their phenotypic, functional, and migratory attributes, making their characterization clinically relevant. Each GEP aligns with distinct sets of chemokine and cytokine receptors as well as molecules related to cytotoxicity, NK receptors, and integrins (Figure 5A). For example, the GEP4 program, shared by Tcm/Tem (Figure S10) and some iNKT cells depending on the donor (Figures 4B–4D), shows high expression of chemokine receptors CXCR3 and CCR4, sphingosine-1-phosphate receptor 4, and oxysterol receptor GPR183, with the latter offering survival and migratory signals to thymocytes and CD4 T follicular helper cells.43 GEP4 also features high levels of IL2RA, IL6R, IL4R, and ITGB1, while conspicuously lacking cytotoxic molecules (Figure 5A). In contrast, the GEP5 program, predominant in Tinn cells across most donors (Figures 4B–4D), shows elevated expression of CCR1, CCR2, CCR5, and CCR6, as well as CXCR6 and cytokine receptors such as IL18R1, IL18RAP, IL12RB1, IL12RB2, IL23R, and IFNGR1 (Figure 5A). This pattern includes GZMA and GZMK but lacks GZMB and GZMH (Figures 5A and S5A) and features the NK receptor KLRB1. On the other hand, the GEP6 program, mainly associated with Tem/Temra cells (Figure S10) and Vδ1+ and Vδ3+ γδ T cells across the majority of donors (Figures 4B, 4C, and S13), shows increased CX3CR1 expression (Figure S14), correlating with effector state differentiation in CD4 and CD8 T cells.44 GEP6 includes IFNG, CCL4, CCL5, and KLRD1, several integrins (ITGAL, ITGB2, ITGAM), and cytotoxicity genes such as GZMB, GZMH, and granulysin (GNLY), with reduced GZMK compared to GEP5 (Figure 5A). These findings align with studies indicating that GZMK+ and GZMB+ cells delineate Tcm and Tem/Temra cell populations.45,46
Figure 5. Effector gene-expression programs in Tinn and Tconv.
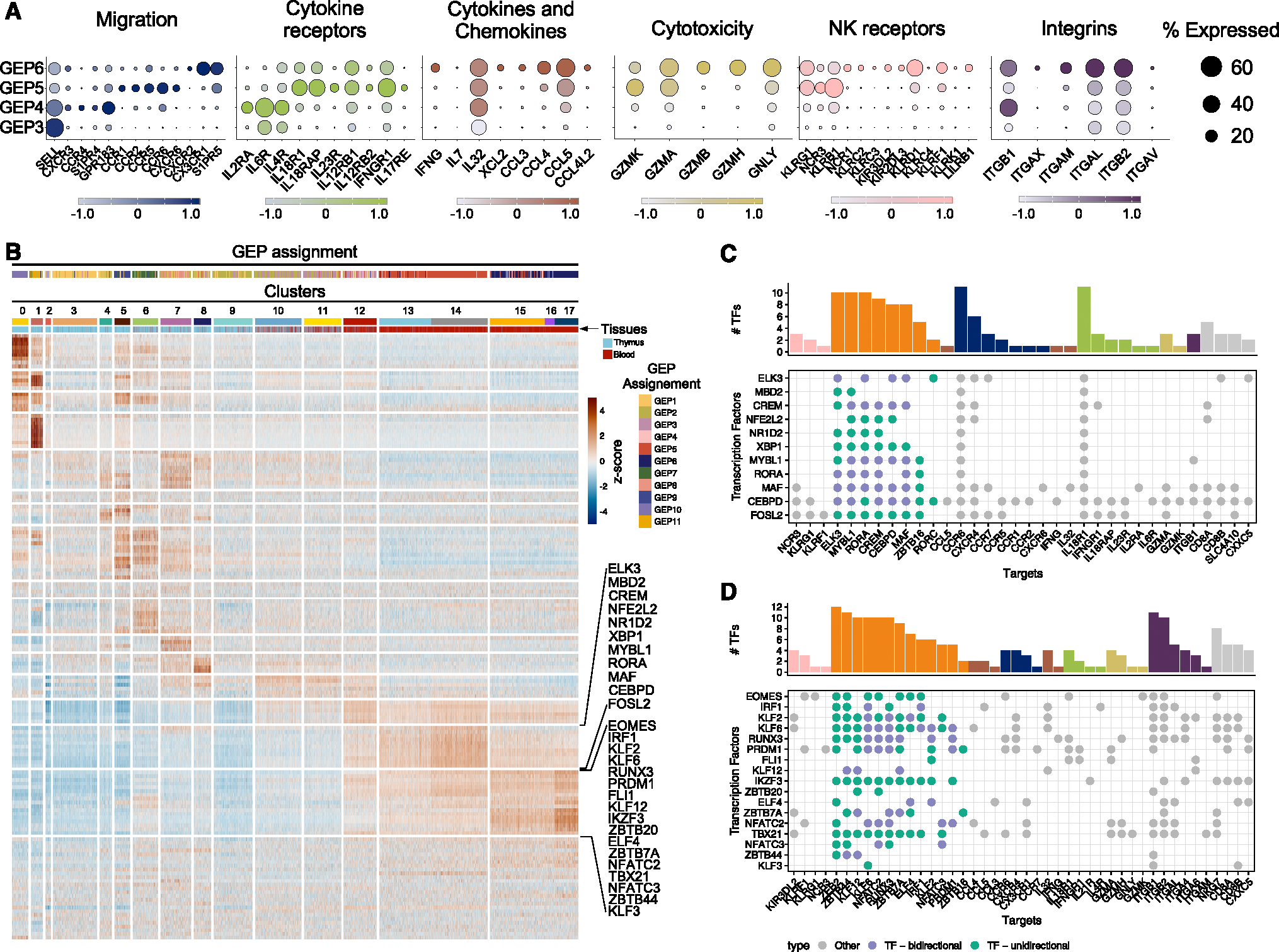
(A) Key genes categorized by function and depicted by their expression level (Z-score color scale) and percentage of expression in cells belonging to the indicated GEPs.
(B–D) (B) Single-cell regulatory network inference and clustering of TFs and enrichment score per cell (as row-scaled Z scores), ordered by cluster (as in Figure 1C), with tissue of origin and GEP assignment (based on cNMF usage) indicated by color bar. Two row clusters are marked, which are preferentially enriched in Tinn (upper bracket) and Tconv (lower bracket). (C and D) TFs with pronounced activity in (C) Tinn and (D) Tconv (corresponding to brackets in B) and their targets. Green dots indicate TFs (y axis) that have other TFs as their target (x axis), where purple labels TFs that can interact in either direction. The marginal bar chart shows the number of TFs per target, color coded by their functional categorization (as in A).
We then used a network inference approach to identify the activity of regulons—TFs and their targets—in single cells.47 We identified 149 of such regulons, with 11 regulons more active in Tinn compared to Tconv cells (Figure 5B). These regulons were governed by TFs such as ELK3, MBD2, CREM, NFE2L2, NR1D2, XBP1, MYBL1, RORA, MAF, CEBPD, and FOSL2. Curated analysis of their predicted target genes indicates that these TFs may play a central role in shaping the unique transcriptional profile observed in Tinn cells during steady-state conditions. This role encompasses the regulation of chemokine and cytokine receptors as well as other genes associated with Tinn cells, including ZBTB16 (encoding PLZF), the master regulator of the Tinn cell lineage (Figure 5C). A second group of regulons exhibited enriched activity within effector Tconv cells, although some shared activity with Tinn cells (including EOMES, RUNX3, PRDM1, and FLI1; Figure 5B). As Tconv cells differentiate into Tem/emra cells, there is an increased activity of regulons driven by TBX21, KLF, and NFAT family TFs (Figures 5B and 5D), in agreement with their functions in regulating the cytolytic activity of CD8 T cells.48–50 Taken together, we discovered novel candidate regulators of Tinn and Tconv effector programs, along with their predicted target genes, which warrant further experimental validation.
Cross-species analysis of thymic Tinn cell development
Only a minority of human Tinn thymocytes display an effector phenotype, contrasting with the predominant effector association of mouse Tinn thymocytes, which develop into distinct effector subsets.23,51,52 To explore transcriptional similarities between mouse and human Tinn cells in the thymus, we constructed a reference mouse Tinn dataset from nine studies.51–59 This dataset revealed 13 transcriptionally distinct clusters, with iNKT, MAIT, and γδ T cells coexisting in variable proportions (Figures 6A and S15; Table S13). Lineage-specific clusters included those unique to γδ T cells (c1 and c2, immature Cd24a+Gzma+ cells51,58), signaling cells (c3 and c4), cycling cells (c5), type 1 cells (c8 and c9), type 2 cells (c6 and c7), and type 17 cells (c10 and c11). Cluster 0, expressing markers such as Sell (encoding for Cd62l), Klf2, Ccr7, Foxo1, and S1pr1, likely represents Tinn cells positively selected on thymic epithelial cells (TECs), bypassing the “innate” pathway.23,60 We performed a cross-species comparison of cell identities by assessing the pairwise correspondence between murine Tinn signatures and human iNKT, MAIT, and γδ T cell clusters (Figure 6B). Human iNKT cells in cluster 0 (NKT_c0), exhibiting a CD8αα T cell gene signature, showed the strongest resemblance to signaling cells (Figure 6B), likely due to shared TCR activation genes. Conversely, cells with an effector profile (NKT_c5 and NKT_c6) showed the closest relationship to mouse type 1 and to a lower extent type 17 cells (Figure 6B). Importantly, we did not find human clusters corresponding uniquely to specific mouse subsets, confirming that human iNKT cells do not differentiate into distinct subsets but rather acquire a mixed type 1/type 17 transcriptome. We also did not detect any human iNKT cell clusters that matched with the mouse type 2 subset with a high degree of confidence (area under the receiver operator characteristic curve [AUROC] > 0.65; Figure 6B), suggesting that type 2 iNKT cells are likely absent in the human thymus. Corroborating this finding, we did not detect any expression of IL-4- or IL-13-encoding transcripts in human thymic iNKT cells, which are typically associated with mouse type 2 thymic iNKT cells. Similar patterns were observed for MAIT and γδ T cells in the human thymus, with effector cells resembling more closely mouse type 1 and type 17 effector cells (Figure 6B). This indicates that human Tinn cells follow a distinctive path with mixed effector potential, unlike the mouse model with multiple effector subsets.
Figure 6. Cross-species comparison of mouse and human Tinn development.
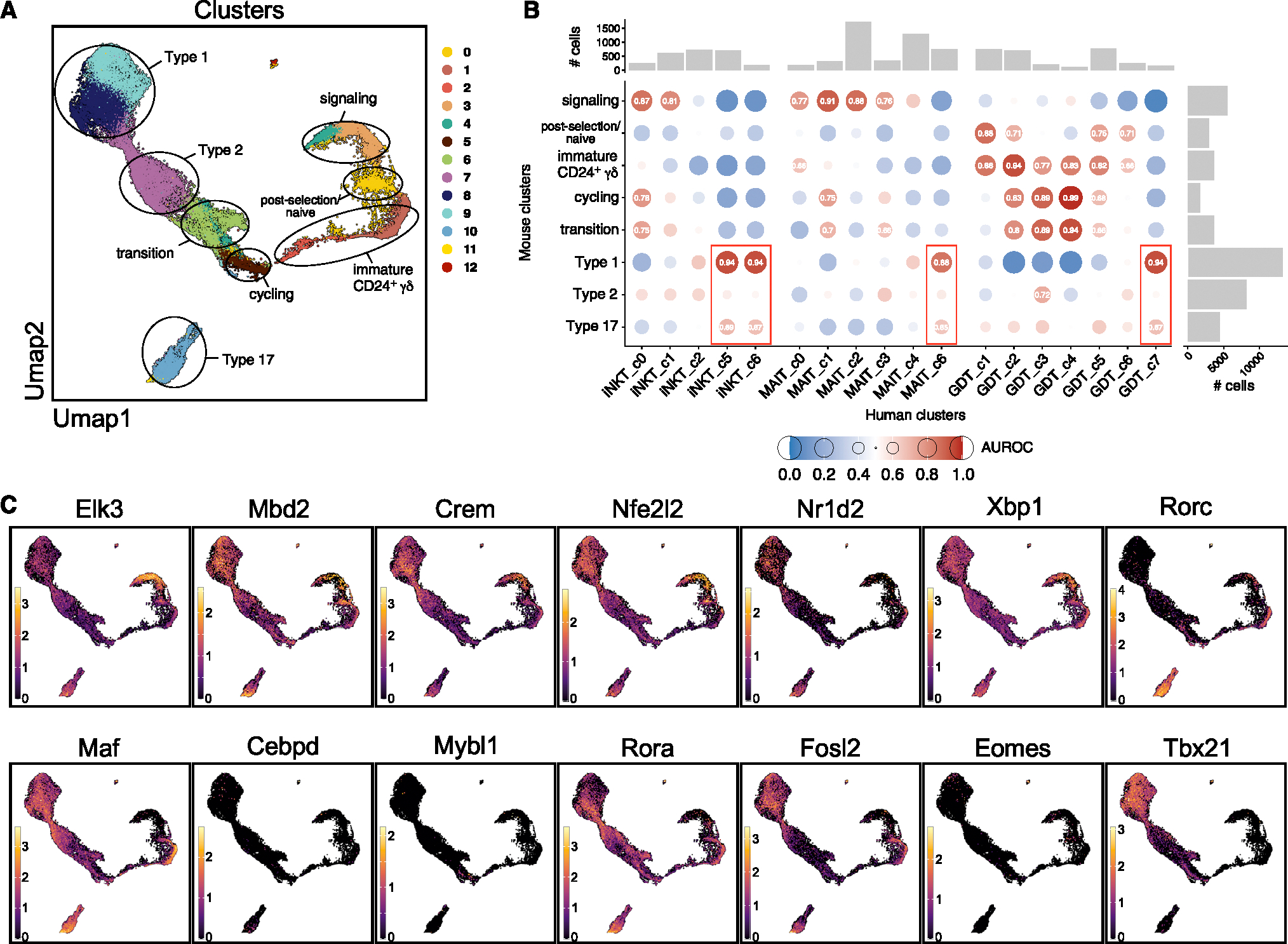
(A) Mouse Tinn reference atlas with seven characteristic cell states highlighted, which are found across lineages (as in Figure S15).
(B) MetaNeighbor analyses showing pairwise correspondence (AUROC scores) between murine Tinn (as in A) and human iNKT, MAIT, and γδ T cell clusters (as in Figure 2D). Marginal bar charts indicate number of cells in the corresponding clusters.
(C) Expression of human regulon-driving TFs (as in Figure 5) together with murine TFs of importance in Tinn development (Rorc, Tbx21) projected on mouse Tinn reference atlas (as in A).
We next assessed whether the TFs driving human Tinn cell regulons (Figure 5A) were also expressed in mouse Tinn cells (Figure 6C). Most TFs were indeed expressed in mouse Tinn cells, although their expression varied across clusters. However, exceptions included CEBPD, EOMES, and MYBL1, which were highly expressed in human Tinn cells (Figure S5A) but barely detectable in mouse Tinn cells (Figure 6C). Conversely, mouse type 1 Tinn cells exhibited high T-bet levels (encoded by Tbx21, Figure 6C), while human Tinn cells had low T-bet expression (Figures S5A, S11B, and S12B). These findings highlight some species-specific differences in TF expression that could play a role in modulating Tinn cell development and functions.
CD1D, SLAMF6, and SLAMF1 expression in the mouse and human thymus
The existence of Tinn cells in the human thymus with a transcriptome similar to developing Tconv cells raises questions about their origin. In mice, a subset of MAIT cells is positively selected by radiation-resistant TECs, which do not lead to a memory or effector phenotype acquisition.52,59 This is because TECs lack SLAM receptors, crucial for Tinn commitment.61 Although this is more common among MAIT cells, some thymic mouse iNKT cells also exhibit a similar transcriptome.23
We hypothesized that naive Tinn cells in humans might result from a similar TEC-mediated selection process. Given the challenge of detecting surface MR1 expression at steady state, we investigated CD1D protein expression in the thymus instead. Mouse TECs have been reported to express CD1d on their surface.62 scRNA-seq of the mouse thymus27 confirmed Cd1d1 expression across various cell types, including thymocytes and cortical and medullary TECs (Figures 7A and 7B). Flow-cytometry analyses corroborated these findings (Figures 7C and 7D).
Figure 7. CD1D, SLAMF1, and SLAMF6 gene and protein expression in mouse and human thymus.
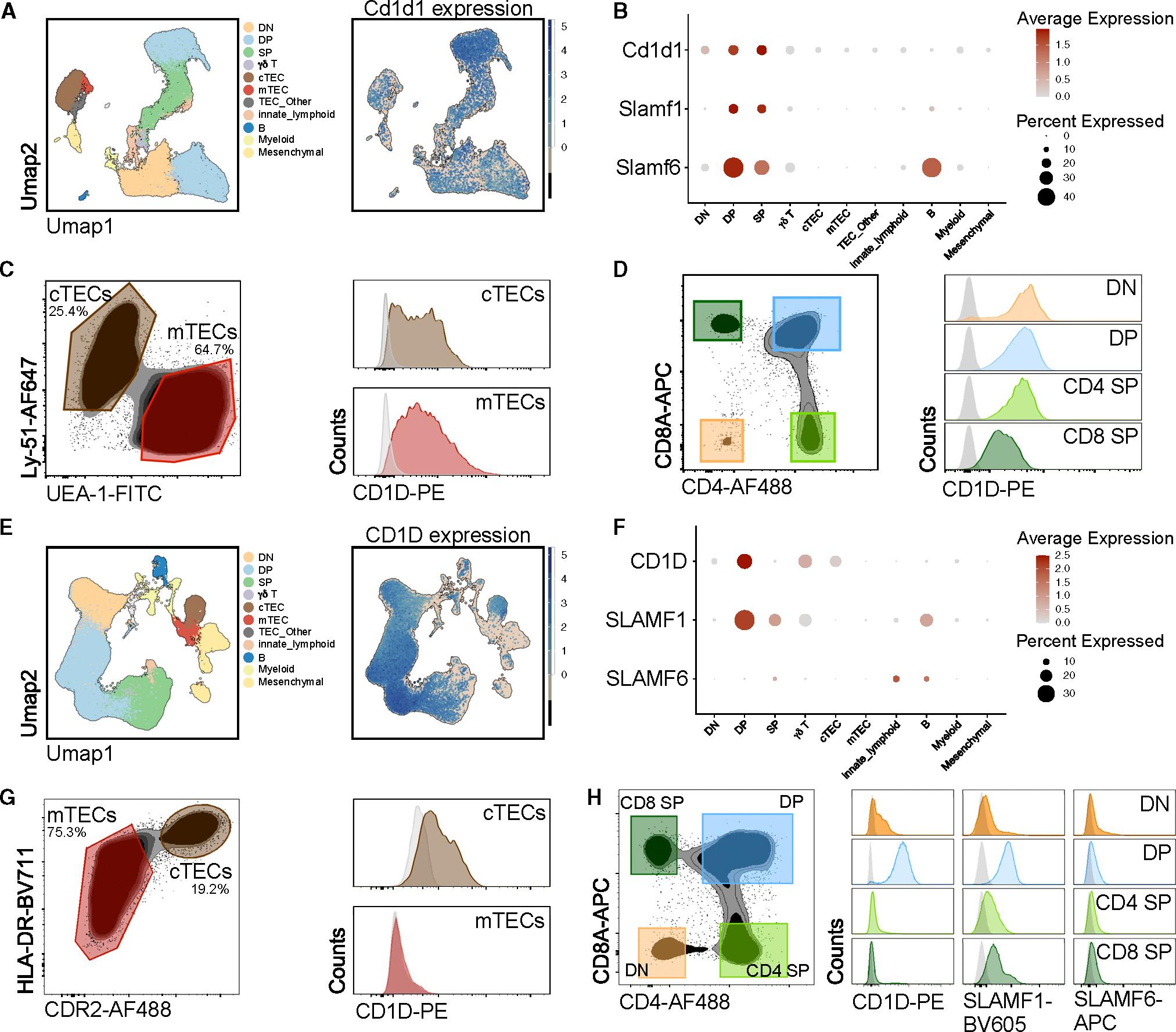
(A) Clustering of thymic cell populations and (E) their expression of Cd1d1 (mouse)/CD1D (human) derived from the mouse and human thymus cell atlas, respectively.27 (B and F) Normalized expression of Cd1d1/CD1D and Slam/SLAM transcripts across thymic cell populations. Flow cytometry of (C and G) mouse and human TECs and (D and H) thymocyte subsets.
In contrast, human thymus scRNA-seq data27 showed a more limited pattern of CD1D and SLAMF6 expression (Figures 7E and 7F). Human DP thymocytes express CD1D transcripts and surface CD1D molecules, but this expression is lost in mature SP thymocytes. Flow cytometry confirmed that, as in mice,61 SLAMF1 is expressed by human DP and SP thymocytes, but SLAMF6 is not detectable on the surface of human thymocytes (Figure 7H). Additionally, human cortical TECs (cTECs) express CD1D transcripts and have surface CD1D protein, whereas medullary TECs (mTECs) do not (Figures 7G and 7H). This interspecies difference in CD1D and SLAM family members’ expression might affect iNKT cell development, given the crucial role of mTECs in murine iNKT cell development.19,63
DISCUSSION
In this study, we employed multi-modal single-cell transcriptomics to explore the diverse phenotypic states of Tinn cells within the human thymus and blood. By comparing these states to those of Tconv cells, we provided insights into human T cell biology and a comprehensive resource for further studies of health and disease. Our work emphasizes Tinn cells as promising candidates for immunotherapies.64–66
Our study demonstrated that the majority of Tinn cells in adult human blood exhibit a distinct transcriptional program shared by most iNKT, MAIT, and Vδ2Vγ9+ T cells under steady-state conditions. This program implies a blended type 1/type 17 transcriptional pattern, driven by specific TFs that enable the expression of distinct chemokine and cytokine receptors, NK receptors, and cytotoxic molecules. This equips Tinn cells to swiftly respond to cytokines such as IL-12, IL-18, and IL-23 independently of TCR signaling.22,67 Notably, human Tinn cells constitutively express GZMK but lack GZMB while also expressing cathepsins necessary for activating granzymes.68 This suggests that Tinn cells are prepared to release active GZMK upon stimulation,69 which can induce pro-inflammatory cytokines45,70 and activate complement,71 implying a role in immune regulation and inflammatory responses. In contrast, mouse Tinn cells do not express GZMK transcripts but possess pre-formed cytokine-encoding transcripts, allowing for immediate responses.72,73 Hence, despite their evolutionary conservation, Tinn cells may have evolved species-specific mechanisms to provide early signaling and amplification of the adaptive immune response.
We identified TFs and their predicted target genes with increased transcriptional activity in human Tinn cells compared to naive and effector Tconv cells. Many of these TFs are associated with IFN-γ, cytotoxicity,48–50,74 and IL-17 production,75–77 consistent with the type 1/type 17 transcriptional program observed in Tinn cells. In mice, the Th1/Th17 paradigm identifies IL-12 and IL-23 as cytokines that induce IFN-γ and IL-17 production, respectively. By contrast, activating human MAIT cells through their TCR or with IL-12 and IL-18, and stimulating Tinn cells with IL-23, results in IFN-γ production.67,78 Yet, only a subset of these cells produces IL-17 in the same conditions, a phenomenon thought to be influenced by epigenetic modifications at the IL-17 gene loci.78 Underlining the importance of understanding the regulation of IL-17 production in Tinn cells, IL-17 production in human MAIT cells is increased in diseases such as severe asthma, community-acquired pneumonia in children,79,80 and colorectal cancer patients.81 Interestingly, NR1D family TFs, associated with Th17 cell regulation,77,82 drive regulons in human Tinn cells. These factors are regulated by the circadian clock, suggesting that circadian rhythms might affect IL-17 production in Tinn cells, a hypothesis that warrants further investigation.
While many TFs essential for the human Tinn program are also expressed in mouse Tinn cells, there are notable exceptions such as CEBPD, EOMES, and MYBL1, which are highly expressed in human Tinn cells but barely detectable in mouse Tinn. CEBPD regulates CCR6 in human MAIT cells83 and may play a crucial role in the human Tinn program. MYBL1 is expressed in human Tinn cells,84 but its function remains to be defined. EOMES, essential for mouse iNKT cell development, shows low expression under steady-state conditions in mice85,86 but is highly expressed in human Tinn cells. By contrast, T-bet is highly expressed in type 1 mouse Tinn cells and is essential for their development and functions.86,87 However, human effector Tinn cells, which are most similar to mouse type 1 Tinn cells, express relatively low levels of T-bet. Instead, T-bet’s expression and activity were correlated with the acquisition of the GEP6 program by Tconv cells in humans. These findings suggest the possibility of species-specific transcriptional regulation of Tinn cells, which could be relevant for their future therapeutic applications. Curiously, high-confidence regulons such as PLZF and Rorγt were not identified in the gene-regulatory network of human Tinn cells, possibly due to the relatively low gene detection in this context.
In the postnatal thymus, iNKT and MAIT cells display a transcriptional profile similar to that of developing conventional CD4+ and CD8+ T cells. Unlike conventional thymocytes selected by cTECs, murine iNKT and MAIT cells undergo selection by DP thymocytes via SLAM family receptors, leading to PLZF expression and effector differentiation.61,88,89 A minor fraction of murine MAIT cells with a naive-like phenotype is TEC selected.52 The corresponding expression patterns of CD1d on cTECs and DP thymocytes in both murine and human thymus suggests a similar selection of human iNKT thymocytes by cTECs or DP thymocytes. The expression of PLZF in non-effector iNKT cells, as observed through both scRNA-seq and flow cytometry, supports the hypothesis of DP selection. However, the initial selection process may not provide all necessary signals for complete maturation, as cells just experiencing positive selection are more akin to CD4+ and CD8+ Tconv cells. Additional signals may be required for human iNKT and MAIT cells to acquire effector functionalities. This hypothesis aligns with the prevalence of naive iNKT and MAIT cells in human cord blood and the gradual increase in effector Tinn cells with age.41,42,90 Unlike murine Tinn cells, which use SLAMF6 and SLAMF1 to induce PLZF expression and effector maturation,61 human thymocytes do not express SLAMF6, possibly affecting Tinn cell maturation. Additionally, human mTECs lack CD1D expression, which could also contribute to the variations in iNKT cell maturation observed in cross-species analyses.19,63
Our study highlights a distinct path taken by Tinn cells with an effector program in the human postnatal thymus, characterized by a mixed type 1/type 17 effector potential, contrasting with mice where Tinn cells split into multiple effector subsets. We did not observe specific clusters of proliferative human iNKT and MAIT thymocytes. In mice, proliferative thymic iNKT and MAIT cell clusters are identifiable52,54,55 and reflect the proliferative burst following positive selection,91 crucial for establishing a substantial Tinn cell pool. While Tinn cells constitute 1%–2% of thymocytes in mice, their proportion is much lower in pediatric humans. Moreover, our analysis did not reveal any type 2 Tinn cells in humans, unlike in mice, where thymus-resident iNKT2 cells significantly impact the thymic environment through IL-4 production.17,18,63,92,93 The scarcity of type 2 Tinn cells in the human thymus suggests that these phenomena may be species specific or regulated by different cell types in humans.
Taken together, our findings hold significance in elucidating the diverse functional attributes of human Tinn cells and their potential applications in immunotherapeutic contexts.
Limitations of the study
Our study presents a comprehensive atlas of human Tinn cells in the thymus and blood, revealing a blended type 1 and type 17 transcriptional profile. However, there are several limitations to consider. We noted variations in effector cell percentages among donors, which may be influenced by factors such as age, sex, or immunological history. Additionally, our analysis focused on steady-state transcriptional profiles, leaving unclear how these profiles might change in various disease states. Comparing pediatric thymic T cells with adult blood samples introduces age-related differences that could impact transcriptional profiles. Ideally, matched samples would offer a more accurate comparison, but the limited availability of iNKT and MAIT cells in neonates presents challenges in obtaining sufficient cell quantities. Future research should explore thymus-derived fetal Tinn cells to investigate whether they exhibit specific type 1 and type 17 profiles akin to those described in mice. Recent studies on fetal γδ T cells suggest that fetal Tinn cells might also develop unique transcriptional subsets.25 Given that iNKT and MAIT cells appear from gestational weeks 18–2394,95 and fetal hematopoietic stem and progenitor cells are predisposed to innate-like lymphocytes,96 examining these cells could yield insights into their development and function. Furthermore, the potential for distinct waves of Tinn cells arising from fetal versus adult hematopoietic stem cells in mice97 highlights the need for further investigation into their transcriptional differences.
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Laurent Gapin (laurent.gapin@cuanschutz.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Data that support the findings of this study, including the raw data and Seurat object, were deposited in NCBI GEO with the accession code GEO: GSE249684. The data from this study is also displayed as a ShinyCell application at https://xspeciestcells.cshl.edu/.
The code used for all analyses presented in this study is publicly available on GitHub: https://github.com/meyer-lab-cshl/xspeciestcells. The code for the browser application can be found at https://github.com/meyer-lab-cshl/xspeciestcells-shiny.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
STAR★METHODS
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Mice
C57BL/6 mice were purchased from Jackson Laboratories. The Cd1d1d2−/− mice backcrossed to the C57BL/6 background were previously described.99 All mice used were between 6 and 15 weeks and age-matched for each experiment. Mice were raised in a specific pathogen-free environment at the Office of Laboratory Animal Research at the University of Colorado Anschutz Medical Campus or the Animal Core Facility at Cold Spring Harbor Laboratory. Animal procedures were approved by the UCD (00065) Institutional Animal Care and Use Committees and the Cold Spring Harbor Laboratory IACUC (23–1); all procedures were carried out in accordance with the approved guidelines.
Human
Thymus tissues were obtained from anonymous human donors who were undergoing medically necessary surgery where removal of a portion of the thymus was required toa facilitate exposure of the operative field. No tissues were obtained specifically for the purposes of this study. This use of discarded tissue was approved by the Institutional Review Board (IRB) of the University of Colorado Anschutz Medical Campus (IRB-17–2159). Additional samples were collected under the Mount Sinai Biorepository with a Waiver of Consent and under the Northwell Health Biospecimen Repository (IRB 20–0150) with patient consent; the use of these human tissues was reviewed by Cold Spring Harbor Institutional Review Board. Pediatric thymus samples for scRNAseq came from individuals between 10 and 20 weeks old (Table S1), and samples used for flow cytometry experiments came from individuals between 4 days and 5 months old. Plateletpheresis leukoreduction filters (LRS chambers) were purchased from Vitalant Blood Center (Denver, CO, USA). Additional PBMCs were collected under COMIRB #17–2159 at the University of Colorado Clinical and Translation Research Centers (CTRC) which is a part of the Colorado Clinical and Translation Sciences Institute (CCTSI), with all donors having provided written informed consent. Overview of sample metadata is provided in Table S1.
METHOD DETAILS
Murine thymus samples
To isolate thymocytes, thymus tissue was immersed in RPMI 1640 media (Corning, #10–040-CV) and gently pressed through a 40mm cell strainer using the plunger of a 1 mL syringe. For TEC isolation, the thymus tissue was cut into small fragments and submerged in RPMI 1640 media without phenol red (Gibco, #11835030), supplemented with 20mM HEPES, 1.3 U/mL Liberase TH, and 100 U/mL DNase I. These tissue fragments were incubated for 5 min on ice followed by an additional 20 min at 37°C. After digestion, the solution was repeatedly mixed with a micropipette to ensure complete tissue disintegration. To stop the digestion process, cells were suspended in HBSS, 4% heat-inactivated FBS (HI-FBS, FBS preheated for 20 min at 56°C), 20mM HEPES, and 10U/mL DNase I. To remove immune cells, the cell suspension was incubated with rat anti-mouse CD90.2 (clone 53–2.1), anti-mouse CD45 (clone 30-F11), and anti-mouse CD45-BV605 (clone 30-F11) antibodies for 30 min at 4°C. Subsequently, the cell suspension was placed on panning plates coated with goat anti-rat IgG for 20 min at room temperature. Unattached cells were then transferred to new panning plates for a second depletion round. The remaining cell suspension, following this depletion process, was prepared for flow cytometry analysis.
Human thymus samples – Thymocytes
To extract thymocytes for both scRNAseq and flow cytometry, the thymus tissue was placed in complete RPMI 1640 media (Gibco, #22400–071), with 10% HI-FBS, 1% non-essential amino acids, 1% sodium pyruvate, 1X GlutaMAX, 1% Penicillin/Streptomycin, and 1X 2-Mercaptoethanol (Sigma-Aldrich #21985–023). The tissue was cut into small pieces, and gently pressed with the back of a 10 mL syringe to release thymocytes. The resulting suspension was passed through a 70 μm filter. Thymocytes were isolated using a Ficoll-Paque density gradient provided by Cytiva. Tetramer staining for MAIT and iNKT cells, and surface staining for CD4+, CD8+ and γδ T cells was performed on freshly isolated thymocytes.
Human thymus samples – thymic epithelial cells
To enrich TECs for flow cytometry, thymus tissue was cut into small pieces and placed in RPMI 1640 media without phenol red (Gibco, #11835030), 5% HI-FBS, 1% Penicillin/Streptomycin, 10mM HEPES, and 0.55mM 2-Mercaptoethanol (Gibco, #21985023). The thymus tissue in this media was stirred on a magnetic plate for 40 min. The supernatant was removed and replaced with fresh media every 10 min to remove released thymocytes. The remaining tissue chunks were placed in a digestion buffer consisting of RPMI 1640 media without phenol red (Gibco, #11835030), 2% HI-FBS, 20mM HEPES, 80 U/mL DNase I, 1.6 U/mL Dispase I, and 0.3 mg/mL Collagenase IV, for digestion at 37°C with gentle shaking. This digestion process was conducted in two sessions of 25 min each, in between which the supernatant was extracted and replaced with fresh digestion buffer. At the end of the digestion, the tissue chunks had nearly entirely disintegrated, and the digestion was halted by resuspending cells in the same buffer used for thymocyte release (RPMI 1640 media without phenol red, 5% HI-FBS, 1% Penicillin/Streptomycin, 10mM HEPES, 0.55mM 2-Mercaptoethanol). The combined supernatants were further incubated in TrypLE Express Enzyme, 1mM MgCl2, 2mM CaCl2, 100U/mL DNase I for 5 min at 37°C to obtain a single-cell suspension. The digestion was stopped by resuspending cells in the previously described thymocyte release buffer. To remove immune cells and erythrocytes, cells were incubated with mouse anti-human CD3 (clone UCHT1), anti-human CD4 (clone RPA-T4), anti-human CD8ɑ (clone RPA-T8), anti-human CD45 (clone HI30) and anti-human CD235a (clone HI264) antibodies in HBSS, 4%HI-FBS and 20U/mL DNase I, for 30 min at 4°C. Cells were then placed on panning plates coated with goat anti-mouse IgG for 20 min at room temperature, and the unadhered cells were transferred to new panning plates for a second round of depletion. The remaining cells following depletion were then stained for flow cytometry.
Human peripheral blood samples
De-identified peripheral blood samples from the Human Immune Tissue Network Biobank (COMIRB # 17–2159) were collected using sodium heparin tubes. De-identified peripheral blood samples from the Vitalant Blood Center were acquired from plateletpheresis leukoreduction filter chambers (LRS). Peripheral blood mononuclear cells (PBMCs) were isolated from these samples using a Ficoll-Paque density gradient provided by Cytiva. PBMCs were cryopreserved in FBS with 10% DMSO and stored in liquid nitrogen.
Magnetic-bead enrichment of iNKT and MAIT cells
To enrich for thymic MAIT and thymic/blood iNKT cells, up to 2 × 109 cells were incubated with MR1–5-OP-RU-PE or CD1d-PBS57-PE tetramers respectively in MACS buffer (1X PBS, 0.5% BSA, 2mM EDTA), for 25 min at room temperature. Cells were washed twice and incubated with anti-PE microbeads, followed by separation using an autoMACS Pro Separator (Miltenyi) according to manufacturer’s instructions. PE-microbead-labelled cells in the enriched fraction were stained with the specified panel of antibodies listed below.
Fluorescence-activated cell sorting
To sort CD4+ T, CD8+ T, γδ T, or peripheral blood MAIT cells, 2 × 106 cells unenriched cell suspensions were used. To sort iNKT cells (thymocytes or PBMCs) or thymic MAIT cells, the staining and sorting was performed on magnetic bead-enriched cell suspensions from 0.5 to 10 × 108 cells (above). All single cell suspensions were stained with efluor780 viability dye for 10 min at room temperature and washed once prior to cell surface staining. The cell suspensions were then stained in MACS buffer (1X PBS, 0.5% BSA, 2mM EDTA) at room temperature for 20 min, both with cell surface markers and a unique oligonucleotide-tagged antibody sample tag (Human Single Cell Sample Multiplexing Kit, BD Biosciences) to allow separation in downstream scRNAseq analyses. The following cell surface markers were included in the staining: CD3-AF488 (clone OKT3), CD14-eFluor450 (clone 61D3), CD19-eFluor450 (clone H1B19), Vα7.2-BV785 (clone 3C10), Vα24-PerCP-Cy5.5 (clone C15), CD4-redFluor710 (clone OKT4), CD8α-PE-Cy7 (clone SK1), TCRγδ-BV650 (clone 11F2), FcγR block. Following cell surface staining, cells were washed twice and resuspended in MACS buffer prior to cell sorting on the Aria 3 (BD Biosciences). Purified cell populations were sorted into MACS buffer. Validation of the cell sorting panel was performed on the Cytek Aurora flow cytometry system using SpectroFlo software (v3.0). Overall, from infant thymus and PBMC donors up to 5 populations were sorted after doublet, viability, B cell (CD19+CD3−) and monocyte (CD14+CD3−) discrimination: 1. MAIT cells (MR1–5-OP-RU-Tet+Vα7.2+CD3+), 2. iNKT cells (CD1d-PBS57-Tet+Vα24+CD3+), 3. γδ T cells (CD3+TCRγδ+), 4. CD4+ T cells (CD4+CD8α−CD3+) and CD8+ T cells (CD8α+CD4−CD3+). Cell subsets sorted for the different donors are listed in Table S1 and the gating strategy is shown in Figure S16.
Flow cytometry — Thymic and peripheral blood T cells
To confirm gene expression from scRNAseq analysis, MAIT and iNKT cells were enriched from the human thymus as described above, as were iNKT cells from human blood. Peripheral blood MAIT and γδ T cells were stained directly without enrichment. Single cell suspensions were stained as above with efluor780 viability dye prior to incubation at 37°C for 10 min with CCR7-APC-Fire810 (clone G043H7) and FcγR block. A combination of the following cell surface markers were subsequently added and cells were stained at room temperature for 15 min: CD3-BUV496 (clone UCHT1), CD14-PE-Cy5 (clone 61D3), CD19-PE Cy5 (clone H1B19), Vα7.2-BV785 (clone 3C10), Vα24-PerCP-Cy5.5 (clone C15), CD4-BV570 (clone RPA-T4), CD8α-BUV395 (clone RPA-T8), TCRγδ-BV650 (clone 11F2), Vδ1-PerCP-Vio700 (clone REA173), Vδ2-FITC (clone 123R3), Vγ9 Vγ9-PE (clone B3), CD161-BUV805 (clone HP-3G10), CD62L-BV650 (clone DREG-56). Cells were then washed twice with MACS buffer, and intracellular staining was performed with BD Transcription Factor Buffer Set according to the manufacturer’s specification. The following antibodies were used to stain for intracellular proteins: PLZF-PE-CF594 (clone R17–809), Eomes-BUV737 (clone X4–83), T-bet-BV605 (clone 4B10), GZMK-eFluor660 (clone G3H69), GZMB-AF700 (clone GB11). Phenotypic analyses were performed on the Cytek Aurora flow cytometry system using SpectroFlo software (v3.0). Data were analyzed using FlowJo software (v10.7.1).
Flow cytometry -— CD1d, SLAMF1, SLAMF6
For murine experiments, thymocytes were resuspended in PBS, 5% FBS, 4mM EDTA and stained for 30 min at 4°C with: Fc blocker anti-CD16/32 (clone 93), CD4-AF488 (clone GK1.5), CD8α-APC (clone 53–6.7), CD1d-PE (clone 1B1). For murine thymus samples which were depleted of immune cells, the single cell suspension was resuspended in HBSS, 4% HI-FBS, 20mM HEPES, 10U/mL DNase I, 2.5mM EDTA, and stained for 30 min at 4°C with: Fc blocker anti-CD16/32 (clone 93), EpCAM-BV421 (clone G8.8), CD45-BV605 (clone 30-F11), UEA1-FITC, Ly-51-AF647 (clone 6C3), and CD1d-PE (clone 1B1). For flow cytometry experiments on human samples, thymocytes were resuspended in PBS, 2% FBS, and stained for 30 min at 4°C with: TruStain FcX, CD45-BV421 (clone HI30), CD4-AF488 (clone OKT4), CD8α-APC (clone RPA-T8), CD1d-PE (clone 51.1). For human samples which were depleted of immune cells and erythrocytes, cells were resuspended in PBS, 2% FBS, and stained for 30 min at 4°C with: TruStain FcX, CD45-AF647 (clone QA17A19), EPCAM-BV421 (clone 9C4), CDR2-AF488 (purified CDR2 antibody kindly provided by Dr. Sheena Pinto, conjugated with the AF488 antibody labeling kit from ThermoFisher Scientific), HLADR-BV711 (clone L243), CD1d-PE (clone 51.1). In all experiments, to measure viability cells were stained with the live/dead Fixable Near-IR dead cell stain kit, simultaneously with cell surface markers. Flow cytometry was performed on a BD LSR Fortessa Cell Analyzer (BD Biosciences).
Single-cell RNA sequencing
Prior to cDNA library preparation for the whole transcriptome (WTA) and VDJ libraries, all cell subsets from the different donors were pooled, with up to 12 unique sample tags combined per library. Single cell WTA and VDJ sequencing libraries were prepared using the BD Rhapsody Single-Cell Analysis System (BD Biosciences) according to the manufacturer’s specifications. Libraries were quantified and pooled according to equivalent molar concentrations and sequenced on the NovaSeq sequencing platform at the University of Colorado Genomics Core with the following read lengths: read 1–150 cycles; read 2–150 cycles; and i7 index - 8 cycles.
Single-cell RNA-seq data analysis
The quality of sequencing reads was evaluated using FastQC and MultiQC. Sequencing reads (FASTQ) were mapped and sample tags were deconvoluted with The BD Rhapsody WTA Analysis Pipeline on the GRCh38 genome sequence. This pipeline produced a gene expression matrix for each sample, which records the number of UMIs for each gene associated with each cell barcode. Aggregated data were then imported into the R environment (version ≥4.0.3) and analyzed with Seurat (v4.3.0). Low-quality cells were filtered using the cutoffs nFeature_RNA ≥ 500 & nFeature_RNA <3000. Cells with a high mitochondrial content were removed using a batch-dependent threshold with the isOutlier function from the scater package (v1.26.1).100 Genes expressed in less than 20 cells were ignored. This resulted in 78,607 cells with 17,204 genes for downstream analyses. The NormalizeData function of Seurat was performed using default parameters to remove the differences in sequencing depth across cells. Dimensionality reduction was performed prior to integration for visualization purposes (Figure S2A), by selecting 2000 highly variable genes for principal component analysis (PCA) and uniform manifold approximation and projection (UMAP). To integrate the data and remove batch-effects from the PCA subspaces based on the correct cell alignment, we used Harmony101 following PCA to project cells into a shared embedding in which cells group by cell type rather than dataset-specific conditions. We then applied the RunUMAP function on 20 dimensions of the harmony embedding to obtain bidimensional coordinates for each cell. We determined the k-nearest neighbors of each cell using the FindNeighbors function and used this knn graph to cluster cells using the Louvain algorithm from FindClusters based on the same harmony dimensions as the RunUMAP function (20 dimensions, resolution 1.2). For analyses performed on individual lineages (CD4+ T, CD8+ T, iNKT, MAIT or γδ T cells) in Figures 2 and 4, the dataset was split up based on cell hashing tags. Each lineage from each tissue was re-analyzed individually using the same steps to obtain UMAPs and clusters in Figures 2 and 4. Plots displaying cells on UMAPs were generated using the SCpubR package (v2.0.2).102
LISI metric and analysis of cluster stability
The local inverse Simpson’s index (LISI) was used to assess the degree of mixing during batch correction and dataset integration in scRNAseq analysis.101 This approach helps evaluate the effectiveness of data integration methods by quantifying how well datasets are merged without introducing artificial batch effects. To assess the integration process, we employed the “integration LISI” (iLISI) score. iLISI measures the effective number of datasets within a neighborhood and provides an indication of how effectively the individual datasets were harmoniously integrated into a unified whole during the process. In addition, we used the “cell-type LISI” (cLISI) score to evaluate the accuracy of cell-type assignments in the integrated dataset. cLISI is a modified version of the LISI score, but instead of assessing dataset labels, it focuses on the accuracy of cell type assignments within the integrated data. As the specific identities of individual cells were not known beforehand, we assigned mock cell identities based on anticipated gene expression patterns. These mock identities were determined using prior knowledge of gene expression markers associated with distinct cell types. For instance, we identified DN thymocytes as cells expressing PTCRA > 1; B cells as cells expressing CD19 > 1 and IGKC > 1; Tregs as cells expressing FOXP3 > 1; MAIT cells as cells expressing SLC4A10 > 1 and FOXP3 < 1; CD4+ T cells as cells expressing CD4 > 1, CD8A < 1, SLC4A10 < 1, FOXP3 < 1, and CCR7 > 1; DP thymocytes as cells expressing RAG1 > 1 and CD1C > 1; and CD8αα thymocytes as cells expressing CD8A > 1 and GNG4 > 1. These mock identities were used as initial cell type assignments and served as the basis for assessing the success of integration, as indicated by increased iLISI scores and the maintenance of a cLISI score of 1 (Figure S2B). Only cells with assigned mock identities were included in the cLISI analysis. To evaluate the stability of clusters, we conducted a bootstrapping procedure in which cells from each predefined cluster were repeatedly sampled and then subjected to re-clustering. Cluster stability was assessed by examining co-assignment probabilities (CP), where higher CP values indicated greater cluster stability (Figure S2C). In essence, a high CP suggests that the cells within a cluster consistently grouped together across multiple iterations, reinforcing the reliability and robustness of that cluster’s identity.
TCR analysis
V(D)J single cell sequencing data were mapped and quantified using the BD Rhapsody WTA Analysis Pipeline and the GRCh38 genome sequence. To connect the VDJ data with transcripts data for each cell, we established links based on cell indexes extracted from the consensus annotation files (VDJ_percell.csv) and MolsPerCell.csv files from each demultiplexed sample. Only TCR paired sequences were retained for subsequent analyses. TCR data from each VDJ-sequenced sample were combined together and added to the metadata of the Seurat object. Clonotypes were defined based on unique TCR VJ usage and complementary-determining region (CDR3) motifs. Basic TCR statistics, such as the distribution of length and counts were computed using the tidyverse package (v1.3.2). The assessment of clonotype diversity was conducted using the mean value of the Shannon index, computed through the diversity function of the vegan R package (v2.6–4) after 100 iterations. Prior to the diversity calculation, the data was subjected to rarefaction to match the lowest sequence count found within the studied groups. Chord diagrams were generated using the circlize package (v0.4.15)103 and CDR3 motif logos using the ggseqlogo package (v0.1).104 The stacked letters’ cumulative height at each position signifies the degree of sequence conservation, portraying the relative abundance of amino acids, which is further depicted by the varying heights of individual letters within the stack.
Identification of differentially expressed genes between clusters
We identified cluster-enriched genes by using the FindAllMarkers function in Seurat with test.use = wilcox. This function identified differentially expressed genes for each cluster by comparing the gene expression for cells belonging to a cluster versus cells belonging to all other clusters. Only those genes that passed an adjusted p value (Benjamini-Hochberg) cutoff of 0.05, log fold change >0.4 and min.pct = 0.3 were included in Figure S3.
Characterizing the replicability of cell types defined by scRNAseq between studies and between species
We assessed the consistency of cell clusters in our integrated thymic data by comparing them with the human thymus atlas from the Park et al. dataset.27 To do this, we focused exclusively on thymocytes, totaling 37,369 cells in our dataset. We also acquired the annotated AnnData object from the Park et al. dataset, which specifically contained T cells. To enable a meaningful comparison, we combined the two raw count matrices, concentrating on the top 2000 highly variable genes shared across both datasets. This resulted in a matrix containing 3,106 genes and 114,363 cells. To evaluate the consistency of cell types between these datasets, we employed the pyMN package to perform unsupervised MetaNeighbor analysis.26 MetaNeighbor assesses the similarity of cell types by constructing a network of cells based on the correlation of their gene expression profiles. It then predicts cell type labels, hiding them from one dataset while using the other. The result is expressed as a mean area under the receiver operator characteristic (AUROC) score, which measures the probability of correctly identifying a cell’s type based on its gene expression profile. We used the ggplot2 package to visualize the AUROC scores obtained from pyMN, comparing our integrated clusters with the thymocyte clusters defined in the Park et al. dataset. For assessing gene expression similarity between thymic lineages in Figure 2A, we considered donors as distinct datasets (study_id parameter); and cell lineages (based on cell hashing) as the cell groups of interest to compare (cell_type parameter), more particularly splitting thymocytes in each lineage into non-effector (clusters 0–11) versus effector states (clusters 12–17). MetaNeighbor was run on the top 2000 highly variable genes chosen on our integrated dataset. For assessing the replicability of cell clusters across species in Figure 6, we utilized the reference scRNAseq murine Tinn dataset (see below) and our human thymic iNKT, MAIT, and γδT individual Seurat objects from Figure 2. To ensure an appropriate comparison, we obtained orthologous genes between mouse and human with the biomaRt package.105,106 We filtered the murine count matrix to retain only genes with known 1:1 orthologs in humans. Then, we performed unsupervised MetaNeighbor analysis with pyMN on the combined set of highly variable genes from both human and mouse datasets. Finally, we used ggplot2 to create visualizations of the AUROC scores returned by pyMN, including clusters that contained at least 1% of the cells in each species to ensure greater confidence in assessing the replicability of clusters across species.
Identification of gene expression programs
The count matrix was used for conducting consensus non-negative matrix factorization (cNMF).28 This process enabled us to infer both identity and activity programs, along with their respective contributions in each cell. The usage of each program for each cell was added to the metadata of the Seurat object and displayed as a feature plot (Figure 1K). To determine the genes associated with each program, we plotted the gene ranks (ranging from most associated to least associated) against the gene_spectra_score output from the cNMF analysis. The plotted gene ranks were fitted to a sigmoid curve and the slope at the first elbow point was calculated as the minimum threshold for genes to be retained in a given GEP. The same slope was applied to every GEP to prevent bias in ranked gene selection, as the gene rankings between GEPs are not comparable and are relative to each GEP (as depicted in Figure S17). Cells from blood samples were assigned to the GEP with the highest usage (as provided by cNMF), to display an alluvial plot with ggalluvial in Figure 4D (v0.12.5).107
Scoring of gene signatures
Gene signatures were scored on our Seurat object, or on other dataset’s Seurat or AnnData objects using either the function AddModuleScore in Seurat, or scanpy.tl.score_genes in scanpy. In both cases, the score is computed as the average expression of all genes contained in the gene list, and subtracting the average expression of 100 control genes (randomly chosen to match the expression bins of the gene list). Gene signatures used throughout this manuscript and their source can be found in Table S2.
Gene regulatory network inference
To deduce gene regulatory networks, we employed pySCENIC from a pre-built singularity container, aertslab/pyscenic:0.12.1, a tool utilizing cis-regulatory motif analysis to identify potential transcription factors (TFs) that might govern a cluster of co-expressed genes within individual cells.47 pySCENIC was run using the –mask-dropouts flag and a normalized enrichment score threshold of 2 to help mitigate the effects of the varying degrees of sparsity across the datasets we generated. The initial step involved generating modules composed of transcription factors and co-expressed genes using GRNboost2.108 These modules were pruned to remove indirect targets that lacked significant enrichment for the corresponding TF motif within ±10 kb from the transcription starting site of the putative target (cisTarget). This process yielded a collection of transcription factor regulons. Considering the inherent stochasticity in gene regulatory network inference using GRNBoost2, each run of pySCENIC may yield different quantities of regulons, along with distinct target genes associated with each TF. To mitigate this variability, we performed 100 pySCENIC runs and retained regulons present in 100% of the runs. We also removed regulons that did not have at least 5 target genes defining the regulon activity. Due to the high degree of noise in target genes, we retained target genes that appeared within a regulon in at least 95% of the runs. Furthermore, each target gene also had to overlap with the union of all possible retained ranked gene expression targets across all GEPs generated from cNMF. To identify regulons that were specific to the underlying biology of our cell types and GEPs, we calculated the AUC scores using the R package AUCell, located in the pySCENIC container, for each regulon based on the pruned target gene list. A regulon was deemed specific to a defined cell population if at least 20% of the cells within the annotated population scored in the 90th percentile of the overall AUC score for all cells.
Comparison of gene expression programs with gene signatures from the literature
To compare the genes characterizing each GEP with known peripheral blood T cell states defined in the literature (Figure S10), we obtained gene signatures identified from (1) differential expression (DE) analysis from bulk RNAseq between sorted naive, Tcm, Tem CD4+ and CD8+ T cell populations by Rose et al.38; (2) DE genes between cell clusters defined from scRNAseq of naive and memory CD4+ T cells isolated from PBMCs by Cano-Gamez et al.37; (3) DE genes between cell clusters defined from scRNAseq of blood immune cells by Terekhova et al.40 (4) DE genes between cell clusters defined from scRNAseq of T cells across nine human tissues by Poon et al.39 In the Rose dataset, we kept genes that defined their Figures 2E and 2H (adjusted p-value ≤%0.05). In the Cano-Gamez and Poon datasets, we kept DE genes with a minimum log fold-change of 0.25 (adjusted p-value threshold ≤%0.05 or 0.01, respectively). In the Terekhova dataset, we used the top 100 differentially expressed genes shared in their Table S5. We computed a weighted Jaccard Index (JI) between the gene lists derived from our GEPs and those from the Rose, Cano-Gamez, Terekhova and Poon datasets (see quantification and statistical analysis). For the co-expression analysis of GEPs and gene lists from other datasets, we scored the gene lists on the entire integrated dataset. This was done using functions like Seurat’s AddModuleScore with the blend = TRUE parameter. Additionally, GEP4, GEP5, and GEP6 were scored on the Poon et al. dataset using scanpy’s tl.score_genes function, and their scores in specific cell clusters of interest were displayed in Figure S10B.
Pseudo-bulk differential expression analysis
To investigate for differentially expressed genes between αβ thymocytes lineages in Figure 2, we grouped cells by donor and lineage, restricting our analysis to only donors 2–4 where all αβ lineages were sorted, and also restricting ourselves to thymocytes found in clusters 3, 9, 10 and 11 in the integrated dataset (where CD4+/iNKT and CD8+/MAIT cells showed high gene expression similarity). We then used DESeq2 (v1.40.2)109 to perform pseudo-bulk DE analysis between lineages using a Wald test. We restricted our analysis to genes which were significantly upregulated (padj < 0.01) in both CD4+ and iNKT cells compared to CD8+ and MAIT cells, and likewise for other comparisons shown in Figure 2. To investigate for cell lineage-specific gene signatures in PBMCs in Figure 4, we grouped cells by batch, cluster and lineage, restricting our analysis to only batches E, F and I where at least 3 or more cell lineages were sorted and sequenced within the same batch. We then used DESeq2 (v1.34.0)109 to perform pseudo-bulk DE analysis with a likelihood-ratio test (LRT), where the full model included batch + cluster + lineage, and the reduced model included batch + cluster, in order to detect genes whose expression can be explained by lineage, and not by batch or cell state (i.e., cluster). We used the LRT test by computing pairwise comparisons, contrasting all lineages against each other (CD4vsCD8, CD4vsiNKT, etc.), for each comparison keeping DE genes with an adjusted p-value of 0.01. Then, to extract lineage-specific genes, for each lineage we kept genes that were commonly upregulated in at least 3 or more contrasts. We normalized the raw counts with the rlog function from DESeq2 and batch-corrected them with removeBatchEffect from the limma package (v3.56.2),110 before displaying the final list of DE genes on a heatmap with the pheatmap package (v1.0.12).
Creation of a reference scRNAseq mouse Tinn dataset
ScRNAseq data from mouse thymic iNKT, MAIT and γδ T cells51–59 were downloaded from Gene Expression Omnibus or the European Bioinformatics Institute (see key resources table). Data were analyzed using the Seurat package. Analyzed cells were selected to express between 800 and 4200 genes per cell, with less than 5% of mitochondrial reads. Datasets were merged and integrated using the FastMNN algorithm,111 using 5000 variable features, k = 20 and auto.merge = TRUE. Cell clustering was carried out with a resolution parameter set at 0.5, and potential doublets were detected using scDblFinder112 and subsequently eliminated. To discern differential gene expression between clusters, the FindAllMarkers function was employed, utilizing the MAST algorithm. The analysis considered latent features, specifically the number of genes per cell, and the sample identity, with a log2 fold change threshold of 0.3.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Human PBS57 CD1d tetramer PE | NIH tetramer core facility | N/A |
| Human 5-OP-RU MR1 tetramer PE | NIH tetramer core facility | N/A |
| Anti-human CCR7 - APC-Fire810 (clone G043H7) | Biolegend | Cat#353263; RRID: AB_2894483 |
| Anti-human CD1d - PE (clone 51.1) | Biolegend | Cat#350305; RRID: AB_10642028 |
| Anti-human CD3 purified (clone UCHT1) | Biolegend | Cat#300402; RRID: AB_314056 |
| Anti-human CD3 - AF488 (clone OKT3) | Biolegend | Cat#317310; RRID: AB_571877 |
| Anti-human CD3 - BUV496 (clone UCHT1) | BD Biosciences | Cat#612941; RRID: AB_2916883 |
| Anti-human CD4 purified (clone RPA-T4) | Biolegend | Cat#300570; RRID: AB_2810427 |
| Anti-human CD4 - AF488 (clone OKT4) | Thermo Fisher Scientific | Cat#53-0048-42; RRID: AB_10735503 |
| Anti-human CD4 - redFluor710 (clone OKT4) | Tonbo biosciences | Cat#80-0048; RRID: AB_2621976 |
| Anti-human CD4 - BV570 (clone RPA-T4) | Biolegend | Cat# 300533; RRID: AB_10896788 |
| Anti-human CD8ɑ purified (clone RPA-T8) | Biolegend | Cat#301002; RRID: AB_314120 |
| Anti-human CD8ɑ - APC (clone RPA-T8) | Thermo Fisher Scientific | Cat#17-0088-42; RRID: AB_10669564 |
| Anti-human CD8ɑ - BUV395 (clone RPA-T8) | BD Biosciences | Cat#563795; RRID: AB_2722501 |
| Anti-human CD8ɑ - PE-Cy7 (clone SK1) | Tonbo biosciences | Cat#60-0087; RRID: AB_3106994 |
| Anti-human CD14 - eFluor450 (clone 61D3) | Thermo Fisher Scientific | Cat#48-0149-42; RRID: AB_1272050 |
| Anti-human CD14 - PE-Cy5 (clone 61D3) | Thermo Fisher Scientific | Cat#15-0149-42; RRID: AB_2573058 |
| Anti-human CD19 - eFluor450 (clone H1B19) | Thermo Fisher Scientific | Cat#48-0199-42; RRID: AB_1272053 |
| Anti-human CD19 - PE-Cy5 (clone H1B19) | Thermo Fisher Scientific | Cat#15-0199-42; RRID: AB_10853658 |
| Anti-human CD45 purified (clone HI30) | Biolegend | Cat#304002; RRID: AB_314390 |
| Anti-human CD45 - AF647 (clone QA17A19) | Biolegend | Cat#393406; RRID: AB_2750083 |
| Anti-human CD45 - BV421 (clone HI30) | Biolegend | Cat#304032; RRID: AB_2561357 |
| Anti-human CD62L - BV650 (clone DREG-56) | Biolegend | Cat#304832; RRID: AB_2563821 |
| Anti-human CD161 - BUV805 (clone HP-3G10) | BD Biosciences | Cat#749221; RRID: AB_2873599 |
| Anti-human CD235a (clone HI264) | Biolegend | Cat#349102; RRID: AB_10612565 |
| Anti-human Eomes - BUV737 (clone X4-83) | BD Horizon | Cat# 567170; RRID: AB_2916487 |
| Anti-human EPCAM - BV421 (clone 9C4) | Biolegend | Cat#324220; RRID: AB_2563847 |
| Anti-human GZMB - AF700 (clone GB11) | BD Biosciences | Cat#560213; RRID: AB_1645453 |
| Anti-human GZMK - eFluor660 (clone G3H69) | Thermo Fisher Scientific | Cat#50-8897-42; RRID: AB_2574299 |
| Anti-human HLADR - BV711 (clone L243) | Biolegend | Cat#307643; RRID: AB_11218794 |
| Anti-human PLZF - PE-CF594 (clone R17-809) | BD Horizon | Cat#565738; RRID: AB_2739339 |
| Anti-human T-bet - BV605 (clone 4B10) | Biolegend | Cat#644817; RRID: AB_11219388 |
| Anti-human TCR γδ - BV650 (clone 11F2) | BD Biosciences | Cat#569510; RRID: AB_3106997 |
| Anti-human Vɑ7.2 - BV785 (clone 3C10) | Biolegend | Cat#351721; RRID: AB_2566041 |
| Anti-human Vɑ24 - PerCP-Cy5.5 (clone C15) | Biolegend | Cat#360004; RRID: AB_2562495 |
| Anti-human Vδ1 - PerCP-Vio700 (clone REA173) | Miltenyi Biotec | Cat#130-120-441; RRID: AB_2784469 |
| Anti-human Vδ2 - FITC (clone 123R3) | Miltenyi Biotec | Cat#130-125-095; RRID: AB_2819745 |
| Anti-human Vγ9 - PE (clone B3) | BD Biosciences | Cat#555733; RRID: AB_396076 |
| CDR2 purified98 | Dr. Sheena Pinto | N/A |
| Anti-mouse CD1d - PE (clone 1B1) | Biolegend | Cat#123509; RRID: AB_1236547 |
| Anti-mouse CD4 - AF488 (clone GK1.5) | Biolegend | Cat#100423; RRID: AB_389302 |
| Anti-mouse CD8ɑ - APC (clone 53-6.7) | Biolegend | Cat#100711; RRID: AB_312750 |
| Anti-mouse CD45 (clone 30-F11) | Thermo Fisher Scientific | Cat#14-0451-85; RRID: AB_467252 |
| Anti-mouse CD45 - BV605 (clone 30-F11) | Biolegend | Cat#103139; RRID: AB_2562341 |
| Anti-mouse CD90.2 purified (clone 53-2.1) | Biolegend | Cat#140302; RRID: AB_10641692 |
| Anti-mouse EpCAM - BV421 (clone G8.8) | Biolegend | Cat#118225; RRID: AB_2563983 |
| Goat anti-mouse IgG | Vector Laboratories | Cat#AI-9200-1.5; RRID: AB_3107016 |
| Anti-mouse Ly51 - AF647 (clone 6C3) | Biolegend | Cat#108312; RRID: AB_2099613 |
| Goat anti-rat IgG | Vector Laboratories | Cat#BA-9400-1.5; RRID: AB_3107017 |
| Human Fc receptor blocking FcγR block | Miltenyi Biotec | Cat# 130-059-901; RRID: AB_2892112 |
| Human Fc receptor blocking TruStain FcX | Biolegend | Cat#422302; RRID: AB_2818986 |
| Mouse Fc receptor blocking anti-CD16/32 (clone 93) | Thermo Fisher Scientific | Cat#14-0161-85; RRID: AB_467134 |
| UEA1 - FITC | Vector Laboratories | Cat#FL-1061; RRID: AB_2336767 |
| Human Single Cell Sample Multiplexing Kit | BD Biosciences | Cat# 633781; RRID: AB_2870299 |
|
| ||
| Biological samples | ||
|
| ||
| Healthy human whole blood (Human Immune Tissue Network Biobank); COMIRB protocol #17-2159 | Colorado Anschutz Medical Campus Clinical and Translational Sciences Institute, Aurora (USA) | N/A |
| Healthy human blood (from plateletpheresis leukoreduction filter chambers); Vitalant | Vitalant Blood Donation Center, Denver (USA) | N/A |
| Human pediatric thymic tissue; IRB protocol #20-0150 | Children's Hospital Colorado, Aurora (USA) | N/A |
| Human pediatric thymic tissue; IRB protocol #IRB-23-4 | Mount Sinai, New York City (USA) | N/A |
| Human pediatric thymic tissue; protocol #NHBR2101 | Northwell Health Biospecimen Repository, Lake Success (USA) | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Bovine Serum Albumin (BSA) | ThermoFisher Scientific | Cat#J10857-22 |
| Collagenase type IV | StemCell Technologies | Cat#07427 |
| Dispase I | Sigma-Aldrich | Cat#04942086001 |
| DMSO | Sigma-Aldrich | Cat#D8418-100ML |
| DNase I | Sigma-Aldrich | Cat#11284932001 |
| Fixable Viability Dye efluor780 | ThermoFisher Scientific | Cat#65-0865-14 |
| Fetal Bovine Serum (FBS) | Corning | Cat#35-010-CV |
| GlutaMAX | Gibco | Cat#35050-061 |
| Liberase TH | Sigma-Aldrich | Cat#5401135001 |
| 2-Mercaptoethanol | Sigma-Aldrich | Cat#21985-023 |
| 2-Mercaptoethanol | Gibco | Cat#21985023 |
| Penicillin/streptomycin | Gibco | Cat#15140-122 |
| Non-essential amino acids | Sigma-Aldrich | Cat#11140-050 |
| Sodium pyruvate | Sigma-Aldrich | Cat#11360-070 |
| Ficoll-paque Premium | Cytiva | Cat#17544203 |
|
| ||
| Critical commercial assays | ||
|
| ||
| AF488 antibody labeling kit | ThermoFisher Scientific | Cat#A20181 |
| Anti-PE microbeads | Miltenyi | Cat#130-048-801 |
| BD Transcription Factor Buffer Set | BD Biosciences | Cat#562574 |
| Live/dead Fixable Near-IR dead cell stain kit | ThermoFisher Scientific | Cat#L10119 |
| BD Rhapsody Cartridge Kit | BD Biosciences | Cat#633773 |
| BD Rhapsody Enhanced Cartridge Kit | BD Biosciences | Cat#664887 |
| BD Rhapsody Cartridge Reagent Kit | BD Biosciences | Cat#633773 |
| BD Rhapsody cDNA Kit | BD Biosciences | Cat#633773 |
| BD Rhapsody TCR/BCR Amplification Kit | BD Biosciences | Cat#665345 |
| BD Rhapsody WTA Amplification Kit | BD Biosciences | Cat#633801 |
|
| ||
| Deposited data | ||
|
| ||
| Raw data (fastq files) Preprocessed Seurat object | This study | GEO: GSE249684 |
| Web browser exploration tool (ShinyCell) of human and murine integrated datasets | This study | https://xspeciestcells.cshl.edu/ |
| Code | This study |
https://doi.org/10.5281/zenodo.13737916. https://doi.org/10.5281/zenodo.13251142 |
|
| ||
| Experimental models: organisms/strains | ||
|
| ||
| C57BL/6J | Jackson Laboratories | Strain #000664; RRID:IMSR_JAX:000664 |
| C57BL/6J CD1d1d2-/- | Bred in house | Chen et al.99 |
|
| ||
| Software and algorithms | ||
|
| ||
| Flow cytometry software FlowJo (v10.7.1) | BD Biosciences | RRID: SCR_008520 |
| Flow cytometry software SpectroFlo (v3.0) | Cytek Biosciences | RRID: SCR_019826 |
| FastQC (v0.11.9) | https://github.com/s-andrews/FastQC | RRID: SCR_014583 |
| MultiQC (v1.12) | https://github.com/MultiQC/MultiQC | RRID: SCR_014982 |
| cNMF (v1.5) | https://github.com/dylkot/cNMF | RRID: SCR_025495 |
| Python (version ≥3.8) | https://www.python.org/ | RRID: SCR_008394 |
| R (version ≥4.0.3) | https://www.r-project.org/ | RRID: SCR_001905 |
|
| ||
| Other | ||
|
| ||
| Public dataset - peripheral blood CD4+ T cell gene signatures (Cano-Gamez et al.) | Cano-Gamez et al.37 | Supplementary Data 6 |
| Public dataset - peripheral blood T cell gene signatures (Rose et al.) | Rose et al.38 | Supplementary Data 2, Supplementary Data 3 |
| Public dataset - peripheral blood T cell gene signatures (Terekhova et al.) | Terekhova et al.40 | Table S5 |
| Public dataset - T cell gene signatures across human tissues (Poon et al.) | Poon et al.39 | Table S6 |
| Public dataset - scRNAseq of human thymus (Park et al.) | Park et al.27 | https://datasets.cellxgene.cziscience.com/de8665a2-0476-4865-b4af-c7b8d3b1b87f.h5ad |
| Public dataset - scRNAseq of murine iNKT thymocytes (Baranek et al.) | Baranek et al.54 | https://datasets.cellxgene.cziscience.com/de8665a2-0476-4865-b4af-c7b8d3b1b87f.h5ad |
| Public dataset - scRNAseq of murine iNKT thymocytes (Krovi et al.) | Krovi et al.55 | GEO: GSE152786 |
| Public dataset - scRNAseq of murine iNKT thymocytes (Maas-Bauer et al.) | Maas-Bauer et al.56 | GEO: GSE172169 |
| Public dataset - scRNAseq of murine iNKT thymocytes (Wang et al.) | Wang et al.57 | GEO: GSE130184 |
| Public dataset - scRNAseq of murine iNKT, MAIT, γδ thymocytes (Lee et al.) | Lee et al.51 | SRA: PRJNA549112 |
| Public dataset - scRNAseq of murine MAIT thymocytes (Chandra et al.) | Chandra et al.59 | GEO: GSE189484 |
| Public dataset - scRNAseq of murine MAIT thymocytes (Koay et al.) | Koay et al.53 | GEO: GSE137350 |
| Public dataset - scRNAseq of murine MAIT thymocytes (Legoux et al.) | Legoux et al.52 | EMBL-EBI: E-MTAB-7704 |
| Public dataset - scRNAseq of murine γδ thymocytes (Li et al.) | Li et al.58 | GEO: GSE179422 |
| HBSS | Gibco | Cat#14175079 |
| HEPES (1M) | Gibco | Cat#15630080 |
| RPMI 1640 | Corning | Cat#10-040-CV |
| RPMI 1640 with 25mM HEPES | Gibco | Cat#22400071 |
| RPMI 1640 without phenol red | Gibco | Cat#11835030 |
| TrypLE Express Enzyme | Gibco | Cat#12604013 |
QUANTIFICATION AND STATISTICAL ANALYSIS
Pseudo-bulk differential expression analysis in thymic samples
Differentially expressed genes in Figures 2B and 2C between αβ thymocyte lineages were identified using a Wald test from the DE-Seq2 package (v1.40.2),109 with a padj < 0.01 threshold. This analysis was restricted only to thymocytes from three sequencing batches (B, C, D; see Table S1) where all αβ lineages were sorted; and to thymocytes in clusters 3, 9, 10 and 11 where naive-like CD4+/iNKT and CD8+/MAIT had shown transcriptional similarity (based on Figures 1G, 1I, and 2A).
Pseudo-bulk differential expression analysis in PBMC samples
Differential expression analysis in Figure 4 was performed with an LRT analysis, to take into account the heterogeneity of batches and cell states among peripheral blood T cells (see method details). This analysis was restricted only to PBMCs from three sequencing batches (E, F, I; see Table S1), where at least three distinct T cell lineages were sorted and sequenced in the same batch. Only DE genes with padj < 0.01 were kept.
Comparison of GEPs with gene signatures from the literature
In Figure S10, we computed the Jaccard Index (JI) between the gene lists derived from our GEPs and those from the Rose, Cano-Gamez, Terekhova and Poon datasets. Since the gene lists varied in length, we weighted the JI to make it comparable across pairwise comparisons. This was achieved by dividing the JI by the maximal theoretical JI for each pairwise comparison, which is the ratio of the length of the smaller list to the length of the larger list. This “weighted Jaccard Index” is shown on the x axis of Figure S10A. To assess the significance of the observed JI, we performed a permutation analysis. We generated 1000 random gene lists A′ and B′, matching in length and expression pattern to the original lists A and B. We computed the weighted JI between these random lists and defined an empirical p-value by counting how many of these weighted JIs were greater than the observed weighted JI divided by the number of permutations. To account for multiple comparisons, we applied a Bonferroni correction to the empirical p-values. Adjusted p-values below 0.01 are shown as text in the figure.
Supplementary Material
Highlights.
Comprehensive atlas of human Tinn cells in thymus and blood
Most postnatal thymic Tinn cells resemble naive Tconv cells
Adult blood Tinn cells display a mixed type 1/type 17 effector program
Human Tinn cells do not form distinct subsets like mouse Tinn cells
ACKNOWLEDGMENTS
We thank members of our laboratories for thoughtful discussions and critical comments on the manuscript; Drs. Jennifer Matsuda, James Scott-Browne, Ross Kedl, Jesse Gillis, Douglas Fearon, and Leslie Berg for critical comments and support; the Flow Core and the University of Colorado flow cytometry shared resource facility for assistance with cell sorting; Dr. Roberta Pelanda for the development of the Human Immune Tissue Network Biobank, the Clinical Research Support Team (CReST), and the Clinical and Translational Research Center (supported by NIH/NCATS Colorado CTSA grant UL1 TR002535) at the University of Colorado Anschutz Medical Campus; the Genomics Core at the University of Colorado Anschutz Medical Campus for sequencing and the National Institutes of Health core facility for CD1d and MR1-tetramers; Sarah Chapin for help in setting up the Shiny app; Eun Seo Park and Jong Kyoung Kim for sharing their mouse γδ T cells Seurat object; Daniel P. Caron and Donna L. Farber for sharing their across-tissues T cell AnnData object; and Ania Lorenc and Gosia Trynka for sharing their Seurat object of human peripheral blood T cells. This work was supported by National Institutes of Health grants R21AI163454 and R01AI130198 (to L.G.), R01AI158410 (to P.J.N.), and R01AI167862–01 (to H.M.). S.C. is supported through the Florence Gould and Annette Kade Fellowships with the Cold Spring Harbor Laboratory of Biological Sciences. This research was further supported by the Simons Center for Quantitative Biology at Cold Spring Harbor Laboratory, the Cold Spring Harbor Laboratory and Northwell Health Affiliation, and the Pershing Square Foundation. It was performed with assistance from the Cancer Center Pilot Awards Program and the CSHL Shared Resources, including the CSHL Flow Cytometry Shared Resource, which are supported by the Cancer Center support grant 5P30CA045508 as well as assistance from the NIH grant S10OD028632–01 at Cold Spring Harbor Laboratory. Computational infrastructure was supported by the Alpine HPC system, which is jointly funded by the University of Colorado Boulder, the University of Colorado Anschutz Medical Campus, Colorado State University, and the National Science Foundation (award 2201538). Additionally, this study was partly supported by the NIH P30CA046934 Bioinformatics and Biostatistics Shared Resource core at the University of Colorado Anschutz Medical Campus.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
DECLARATION OF GENERATIVE AI AND AI-ASSISTED TECHNOLOGIES IN THE WRITING PROCESS
During the preparation of this work the authors used ChatGPT in order to improve language and readability. After using this tool/service, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114705.
REFERENCES
- 1.Sallusto F, Lenig D, Förster R, Lipp M, and Lanzavecchia A (1999). Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401, 708–712. 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 2.Kaech SM, and Cui W (2012). Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761. 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jameson SC, and Masopust D (2018). Understanding Subset Diversity in T Cell Memory. Immunity 48, 214–226. 10.1016/j.immuni.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, and Moody DB (2015). The burgeoning family of unconventional T cells. Nat. Immunol. 16, 1114–1123. 10.1038/ni.3298. [DOI] [PubMed] [Google Scholar]
- 5.Mayassi T, Barreiro LB, Rossjohn J, and Jabri B (2021). A multilayered immune system through the lens of unconventional T cells. Nature 595, 501–510. 10.1038/s41586-021-03578-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuda JL, Mallevaey T, Scott-Browne J, and Gapin L (2008). CD1d-restricted iNKT cells, the ‘Swiss-Army knife’ of the immune system. Curr. Opin. Immunol. 20, 358–368. 10.1016/j.coi.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Legoux F, Salou M, and Lantz O (2017). Unconventional or Preset alphabeta T Cells: Evolutionarily Conserved Tissue-Resident T Cells Recognizing Nonpeptidic Ligands. Annu. Rev. Cell Dev. Biol. 33, 511–535. 10.1146/annurev-cellbio-100616-060725. [DOI] [PubMed] [Google Scholar]
- 8.Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, et al. (2012). MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723. 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- 9.Corbett AJ, Eckle SBG, Birkinshaw RW, Liu L, Patel O, Mahony J, Chen Z, Reantragoon R, Meehan B, Cao H, et al. (2014). T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509, 361–365. 10.1038/nature13160. [DOI] [PubMed] [Google Scholar]
- 10.Harly C, Guillaume Y, Nedellec S, Peigné CM, Mönkkönen H, Mönkkönen J, Li J, Kuball J, Adams EJ, Netzer S, et al. (2012). Key implication of CD277/butyrophilin-3 (BTN3A) in cellular stress sensing by a major human gammadelta T-cell subset. Blood 120, 2269–2279. 10.1182/blood-2012-05-430470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rigau M, Ostrouska S, Fulford TS, Johnson DN, Woods K, Ruan Z, McWilliam HEG, Hudson C, Tutuka C, Wheatley AK, et al. (2020). Butyrophilin 2A1 is essential for phosphoantigen reactivity by gammadelta T cells. Science 367, eaay5516. 10.1126/science.aay5516. [DOI] [PubMed] [Google Scholar]
- 12.Karunakaran MM, Willcox CR, Salim M, Paletta D, Fichtner AS, Noll A, Starick L, Nohren A, Begley CR, Berwick KA, et al. (2020). Butyrophilin-2A1 Directly Binds Germline-Encoded Regions of the Vgamma9Vdelta2 TCR and Is Essential for Phosphoantigen Sensing. Immunity 52, 487–498.e486. 10.1016/j.immuni.2020.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deseke M, and Prinz I (2020). Ligand recognition by the gammadelta TCR and discrimination between homeostasis and stress conditions. Cell. Mol. Immunol. 17, 914–924. 10.1038/s41423-020-0503-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godfrey DI, Koay HF, McCluskey J, and Gherardin NA (2019). The biology and functional importance of MAIT cells. Nat. Immunol. 20, 1110–1128. 10.1038/s41590-019-0444-8. [DOI] [PubMed] [Google Scholar]
- 15.Hayday AC (2019). gammadelta T Cell Update: Adaptate Orchestrators of Immune Surveillance. J. Immunol. 203, 311–320. 10.4049/jimmunol.1800934. [DOI] [PubMed] [Google Scholar]
- 16.Chandra S, and Kronenberg M (2015). Activation and Function of iNKT and MAIT Cells. Adv. Immunol. 127, 145–201. 10.1016/bs.ai.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, and Hogquist KA (2013). Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 14, 1146–1154. 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breed ER, Voboril M, Ashby KM, Martinez RJ, Qian L, Wang H, Salgado OC, O’Connor CH, and Hogquist KA (2022). Type 2 cytokines in the thymus activate Sirpalpha(+) dendritic cells to promote clonal deletion. Nat. Immunol. 23, 1042–1051. 10.1038/s41590-022-01218-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui G, Shimba A, Jin J, Ogawa T, Muramoto Y, Miyachi H, Abe S, Asahi T, Tani-Ichi S, Dijkstra JM, et al. (2022). A circulating subset of iNKT cells mediates antitumor and antiviral immunity. Sci. Immunol. 7, eabj8760. 10.1126/sciimmunol.abj8760. [DOI] [PubMed] [Google Scholar]
- 20.Harly C, Robert J, Legoux F, and Lantz O (2022). gammadelta T, NKT, and MAIT Cells During Evolution: Redundancy or Specialized Functions? J. Immunol. 209, 217–225. 10.4049/jimmunol.2200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leite-De-Moraes MC, Hameg A, Arnould A, Machavoine F, Koezuka Y, Schneider E, Herbelin A, and Dy M (1999). A distinct IL-18-induced pathway to fully activate NK T lymphocytes independently from TCR engagement. J. Immunol. 163, 5871–5876. [PubMed] [Google Scholar]
- 22.Ussher JE, Bilton M, Attwod E, Shadwell J, Richardson R, de Lara C, Mettke E, Kurioka A, Hansen TH, Klenerman P, and Willberg CB (2014). CD161++ CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur. J. Immunol. 44, 195–203. 10.1002/eji.201343509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krovi SH, Loh L, Spengler A, Brunetti T, and Gapin L (2022). Current insights in mouse iNKT and MAIT cell development using single cell transcriptomics data. Semin. Immunol. 60, 101658. 10.1016/j.smim.2022.101658. [DOI] [PubMed] [Google Scholar]
- 24.Baranek T, de Amat Herbozo C, Mallevaey T, and Paget C (2022). Deconstructing iNKT cell development at single-cell resolution. Trends Immunol. 43, 503–512. 10.1016/j.it.2022.04.012. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez Sanchez G, Papadopoulou M, Azouz A, Tafesse Y, Mishra A, Chan JKY, Fan Y, Verdebout I, Porco S, Libert F, et al. (2022). Identification of distinct functional thymic programming of fetal and pediatric human gammadelta thymocytes via single-cell analysis. Nat. Com mun. 13, 5842. 10.1038/s41467-022-33488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crow M, Paul A, Ballouz S, Huang ZJ, and Gillis J (2018). Characterizing the replicability of cell types defined by single cell RNA-sequencing data using MetaNeighbor. Nat. Commun. 9, 884. 10.1038/s41467-018-03282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park JE, Botting RA, Dominguez Conde C, Popescu DM, Lavaert M, Kunz DJ, Goh I, Stephenson E, Ragazzini R, Tuck E, et al. (2020). A cell atlas of human thymic development defines T cell repertoire formation. Science 367, eaay3224. 10.1126/science.aay3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotliar D, Veres A, Nagy MA, Tabrizi S, Hodis E, Melton DA, and Sabeti PC (2019). Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-Seq. Elife 8, e43803. 10.7554/eLife.43803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D, Milder M, Le Bourhis L, Soudais C, Treiner E, and Lantz O (2011). Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood 117, 1250–1259. 10.1182/blood-2010-08-303339. [DOI] [PubMed] [Google Scholar]
- 30.Park D, Kim HG, Kim M, Park T, Ha HH, Lee DH, Park KS, Park SJ, Lim HJ, and Lee CH (2019). Differences in the molecular signatures of mucosal-associated invariant T cells and conventional T cells. Sci. Rep. 9, 7094. 10.1038/s41598-019-43578-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott-Browne JP, Matsuda JL, Mallevaey T, White J, Borg NA, McCluskey J, Rossjohn J, Kappler J, Marrack P, and Gapin L (2007). Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat. Immunol. 8, 1105–1113. 10.1038/ni1510. [DOI] [PubMed] [Google Scholar]
- 32.Reantragoon R, Kjer-Nielsen L, Patel O, Chen Z, Illing PT, Bhati M, Kostenko L, Bharadwaj M, Meehan B, Hansen TH, et al. (2012). Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J. Exp. Med. 209, 761–774. 10.1084/jem.20112095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young MH, U’Ren L, Huang S, Mallevaey T, Scott-Browne J, Crawford F, Lantz O, Hansen TH, Kappler J, Marrack P, and Gapin L (2013). MAIT cell recognition of MR1 on bacterially infected and uninfected cells. PLoS One 8, e53789. 10.1371/journal.pone.0053789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reantragoon R, Corbett AJ, Sakala IG, Gherardin NA, Furness JB, Chen Z, Eckle SBG, Uldrich AP, Birkinshaw RW, Patel O, et al. (2013). Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J. Exp. Med. 210, 2305–2320. 10.1084/jem.20130958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tilloy F, Treiner E, Park SH, Garcia C, Lemonnier F, de la Salle H, Bendelac A, Bonneville M, and Lantz O (1999). An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J. Exp. Med. 189, 1907–1921. 10.1084/jem.189.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davey MS, Willcox CR, Hunter S, Kasatskaya SA, Remmerswaal EBM, Salim M, Mohammed F, Bemelman FJ, Chudakov DM, Oo YH, and Willcox BE (2018). The human Vdelta2(+) T-cell compartment comprises distinct innate-like Vgamma9(+) and adaptive Vgamma9(−) subsets. Nat. Commun. 9, 1760. 10.1038/s41467-018-04076-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cano-Gamez E, Soskic B, Roumeliotis TI, So E, Smyth DJ, Baldrighi M, Willé D, Nakic N, Esparza-Gordillo J, Larminie CGC, et al. (2020). Single-cell transcriptomics identifies an effectorness gradient shaping the response of CD4(+) T cells to cytokines. Nat. Commun. 11, 1801. 10.1038/s41467-020-15543-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rose JR, Akdogan-Ozdilek B, Rahmberg AR, Powell MD, Hicks SL, Scharer CD, and Boss JM (2023). Distinct transcriptomic and epigenomic modalities underpin human memory T cell subsets and their activation potential. Commun. Biol. 6, 363. 10.1038/s42003-023-04747-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poon MML, Caron DP, Wang Z, Wells SB, Chen D, Meng W, Szabo PA, Lam N, Kubota M, Matsumoto R, et al. (2023). Tissue adaptation and clonal segregation of human memory T cells in barrier sites. Nat. Immunol. 24, 309–319. 10.1038/s41590-022-01395-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terekhova M, Swain A, Bohacova P, Aladyeva E, Arthur L, Laha A, Mogilenko DA, Burdess S, Sukhov V, Kleverov D, et al. (2023). Single-cell atlas of healthy human blood unveils age-related loss of NKG2C(+)GZMB(−)CD8(+) memory T cells and accumulation of type 2 memory T cells. Immunity 56, 2836–2854.e9. 10.1016/j.immuni.2023.10.013. [DOI] [PubMed] [Google Scholar]
- 41.Berzins SP, Cochrane AD, Pellicci DG, Smyth MJ, and Godfrey DI (2005). Limited correlation between human thymus and blood NKT cell content revealed by an ontogeny study of paired tissue samples. Eur. J. Immunol. 35, 1399–1407. 10.1002/eji.200425958. [DOI] [PubMed] [Google Scholar]
- 42.Sandberg JK, Stoddart CA, Brilot F, Jordan KA, and Nixon DF (2004). Development of innate CD4+ alpha-chain variable gene segment 24 (Valpha24) natural killer T cells in the early human fetal thymus is regulated by IL-7. Proc. Natl. Acad. Sci. USA 101, 7058–7063. 10.1073/pnas.0305986101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, Lu E, Yi T, and Cyster JG (2016). EBI2 augments Tfh cell fate by promoting interaction with IL-2-quenching dendritic cells. Nature 533, 110–114. 10.1038/nature17947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zwijnenburg AJ, Pokharel J, Varnaite R, Zheng W, Hoffer E, Shryki I, Comet NR, Ehrstrom M, Gredmark-Russ S, Eidsmo L, and Gerlach C (2023). Graded expression of the chemokine receptor CX3CR1 marks differentiation states of human and murine T cells and enables cross-species interpretation. Immunity 56, 1955–1974.e1910. 10.1016/j.immuni.2023.06.025. [DOI] [PubMed] [Google Scholar]
- 45.Jonsson AH, Zhang F, Dunlap G, Gomez-Rivas E, Watts GFM, Faust HJ, Rupani KV, Mears JR, Meednu N, Wang R, et al. (2022). Granzyme K(+) CD8 T cells form a core population in inflamed human tissue. Sci. Transl. Med. 14, eabo0686. 10.1126/sci-translmed.abo0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duquette D, Harmon C, Zaborowski A, Michelet X, O’Farrelly C, Winter D, Koay HF, and Lynch L (2023). Human Granzyme K Is a Feature of Innate T Cells in Blood, Tissues, and Tumors, Responding to Cytokines Rather than TCR Stimulation. J. Immunol. 211, 633–647. 10.4049/jimmunol.2300083. [DOI] [PubMed] [Google Scholar]
- 47.Aibar S, Gonzá lez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, et al. (2017). SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086. 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, et al. (2005). Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6, 1236–1244. 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 49.Klein-Hessling S, Muhammad K, Klein M, Pusch T, Rudolf R, Flöter J, Qureischi M, Beilhack A, Vaeth M, Kummerow C, et al. (2017). NFATc1 controls the cytotoxicity of CD8(+) T cells. Nat. Commun. 8, 511. 10.1038/s41467-017-00612-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nah J, and Seong RH (2022). Kruppel-like factor 4 regulates the cytolytic effector function of exhausted CD8 T cells. Sci. Adv. 8, eadc9346. 10.1126/sciadv.adc9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee M, Lee E, Han SK, Choi YH, Kwon DI, Choi H, Lee K, Park ES, Rha MS, Joo DJ, et al. (2020). Single-cell RNA sequencing identifies shared differentiation paths of mouse thymic innate T cells. Nat. Commun. 11, 4367. 10.1038/s41467-020-18155-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Legoux F, Gilet J, Procopio E, Echasserieau K, Bernardeau K, and Lantz O (2019). Molecular mechanisms of lineage decisions in metabolite-specific T cells. Nat. Immunol. 20, 1244–1255. 10.1038/s41590-019-0465-3. [DOI] [PubMed] [Google Scholar]
- 53.Koay HF, Su S, Amann-Zalcenstein D, Daley SR, Comerford I, Miosge L, Whyte CE, Konstantinov IE, d’Udekem Y, Baldwin T, et al. (2019). A divergent transcriptional landscape underpins the development and functional branching of MAIT cells. Sci. Immunol. 4, eaay6039. 10.1126/sciimmunol.aay6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baranek T, Lebrigand K, de Amat Herbozo C, Gonzalez L, Bogard G, Dietrich C, Magnone V, Boisseau C, Jouan Y, Trottein F, et al. (2020). High Dimensional Single-Cell Analysis Reveals iNKT Cell Developmental Trajectories and Effector Fate Decision. Cell Rep. 32, 108116. 10.1016/j.celrep.2020.108116. [DOI] [PubMed] [Google Scholar]
- 55.Harsha Krovi S, Zhang J, Michaels-Foster MJ, Brunetti T, Loh L, Scott-Browne J, and Gapin L (2020). Thymic iNKT single cell analyses unmask the common developmental program of mouse innate T cells. Nat. Commun. 11, 6238. 10.1038/s41467-020-20073-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maas-Bauer K, Lohmeyer JK, Hirai T, Ramos TL, Fazal FM, Litzenburger UM, Yost KE, Ribado JV, Kambham N, Wenokur AS, et al. (2021). Invariant natural killer T-cell subsets have diverse graft-versus-host-disease-preventing and antitumor effects. Blood 138, 858–870. 10.1182/blood.2021010887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J, Adrianto I, Subedi K, Liu T, Wu X, Yi Q, Loveless I, Yin C, Datta I, Sant’Angelo DB, et al. (2023). Integrative scATAC-seq and scRNA-seq analyses map thymic iNKT cell development and identify Cbfbeta for its commitment. Cell Discov. 9, 61. 10.1038/s41421-023-00547-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Z, Yang Q, Tang X, Chen Y, Wang S, Qi X, Zhang Y, Liu Z, Luo J, Liu H, et al. (2022). Single-cell RNA-seq and chromatin accessibility profiling decipher the heterogeneity of mouse gammadelta T cells. Sci. Bull. 67, 408–426. 10.1016/j.scib.2021.11.013. [DOI] [PubMed] [Google Scholar]
- 59.Chandra S, Ascui G, Riffelmacher T, Chawla A, Ramírez-Suástegui C, Castelan VC, Seumois G, Simon H, Murray MP, Seo GY, et al. (2023). Transcriptomes and metabolism define mouse and human MAIT cell populations. Sci. Immunol. 8, eabn8531. 10.1126/sciimmunol.abn8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salou M, Legoux F, and Lantz O (2021). MAIT cell development in mice and humans. Mol. Immunol. 130, 31–36. 10.1016/j.molimm.2020.12.003. [DOI] [PubMed] [Google Scholar]
- 61.Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, Mamchak AA, Terhorst C, and Bendelac A (2007). Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell line-age development. Immunity 27, 751–762. 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Forestier C, Park SH, Wei D, Benlagha K, Teyton L, and Bendelac A (2003). T cell development in mice expressing CD1d directed by a classical MHC class II promoter. J. Immunol. 171, 4096–4104. 10.4049/jimmunol.171.8.4096. [DOI] [PubMed] [Google Scholar]
- 63.White AJ, Jenkinson WE, Cowan JE, Parnell SM, Bacon A, Jones ND, Jenkinson EJ, and Anderson G (2014). An essential role for medullary thymic epithelial cells during the intrathymic development of invariant NKT cells. J. Immunol. 192, 2659–2666. 10.4049/jimmunol.1303057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Delfanti G, Cortesi F, Perini A, Antonini G, Azzimonti L, de Lalla C, Garavaglia C, Squadrito ML, Fedeli M, Consonni M, et al. (2022). TCR-engineered iNKT cells induce robust antitumor response by dual targeting cancer and suppressive myeloid cells. Sci. Immunol. 7, eabn6563. 10.1126/sciimmunol.abn6563. [DOI] [PubMed] [Google Scholar]
- 65.Dogan M, Karhan E, Kozhaya L, Placek L, Chen X, Yigit M, and Unutmaz D (2022). Engineering Human MAIT Cells with Chimeric Antigen Receptors for Cancer Immunotherapy. J. Immunol. 209, 1523–1531. 10.4049/jimmunol.2100856. [DOI] [PubMed] [Google Scholar]
- 66.Lee D, Dunn ZS, Guo W, Rosenthal CJ, Penn NE, Yu Y, Zhou K, Li Z, Ma F, Li M, et al. (2023). Unlocking the potential of allogeneic Vdelta2 T cells for ovarian cancer therapy through CD16 biomarker selection and CAR/IL-15 engineering. Nat. Commun. 14, 6942. 10.1038/s41467-023-42619-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Philippot Q, Ogishi M, Bohlen J, Puchan J, Arias AA, Nguyen T, Martin-Fernandez M, Conil C, Rinchai D, Momenilandi M, et al. (2023). Human IL-23 is essential for IFN-gamma-dependent immunity to mycobacteria. Sci. Immunol. 8, eabq5204. 10.1126/sciimmunol.abq5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.D’Angelo ME, Bird PI, Peters C, Reinheckel T, Trapani JA, and Sutton VR (2010). Cathepsin H is an additional convertase of pro-granzyme B. J. Biol. Chem. 285, 20514–20519. 10.1074/jbc.M109.094573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kurioka A, Ussher JE, Cosgrove C, Clough C, Fergusson JR, Smith K, Kang YH, Walker LJ, Hansen TH, Willberg CB, and Klenerman P (2015). MAIT cells are licensed through granzyme exchange to kill bacterially sensitized targets. Mucosal Immunol. 8, 429–440. 10.1038/mi.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaiserman D, Zhao P, Rowe CL, Leong A, Barlow N, Joeckel LT, Hitchen C, Stewart SE, Hollenberg MD, Bunnett N, et al. (2022). Granzyme K initiates IL-6 and IL-8 release from epithelial cells by activating protease-activated receptor 2. PLoS One 17, e0270584. 10.1371/journal.pone.0270584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jonsson A, Donado C, Theisen E, Jones D, Nathan A, Zhang F, Raychaudhuri S, and Brenner M (2023). Accelerating Medicines Partnership (AMP) RA/SLE. Granzyme K Elicits a New Pathway for Complement Activation in RA Synovium [abstract]. Arthritis Rheumatol. 75, (Suppl 9). [Google Scholar]
- 72.Matsuda JL, Gapin L, Baron JL, Sidobre S, Stetson DB, Mohrs M, Locksley RM, and Kronenberg M (2003). Mouse V alpha 14i natural killer T cells are resistant to cytokine polarization in vivo. Proc. Natl. Acad. Sci. USA 100, 8395–8400. 10.1073/pnas.1332805100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Govindarajan S, Gaublomme D, Van der Cruyssen R, Verheugen E, Van Gassen S, Saeys Y, Tavernier S, Iwawaki T, Bloch Y, Savvides SN, et al. (2018). Stabilization of cytokine mRNAs in iNKT cells requires the serine-threonine kinase IRE1alpha. Nat. Commun. 9, 5340. 10.1038/s41467-018-07758-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Istaces N, Splittgerber M, Lima Silva V, Nguyen M, Thomas S, Le A, Achouri Y, Calonne E, Defrance M, Fuks F, et al. (2019). EOMES interacts with RUNX3 and BRG1 to promote innate memory cell formation through epigenetic reprogramming. Nat. Commun. 10, 3306. 10.1038/s41467-019-11233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu JS, Hamada M, Ohtsuka S, Yoh K, Takahashi S, and Miaw SC (2017). Differentiation of IL-17-Producing Invariant Natural Killer T Cells Requires Expression of the Transcription Factor c-Maf. Front. Immunol. 8, 1399. 10.3389/fimmu.2017.01399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkhurst CN, Muratet M, et al. (2012). A validated regulatory network for Th17 cell specification. Cell 151, 289–303. 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chang C, Loo CS, Zhao X, Solt LA, Liang Y, Bapat SP, Cho H, Kamenecka TM, Leblanc M, Atkins AR, et al. (2019). The nuclear receptor REV-ERBalpha modulates Th17 cell-mediated autoimmune disease. Proc. Natl. Acad. Sci. USA 116, 18528–18536. 10.1073/pnas.1907563116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garner LC, Amini A, FitzPatrick MEB, Lett MJ, Hess GF, Filipowicz Sinnreich M, Provine NM, and Klenerman P (2023). Single-cell analysis of human MAIT cell transcriptional, functional and clonal diversity. Nat. Immunol. 24, 1565–1578. 10.1038/s41590-023-01575-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lezmi G, Abou-Taam R, Garcelon N, Dietrich C, Machavoine F, Delacourt C, Adel-Patient K, and Leite-de-Moraes M (2019). Evidence for a MAIT-17-high phenotype in children with severe asthma. J. Allergy Clin. Immunol. 144, 1714–1716.e6. 10.1016/j.jaci.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 80.Lu B, Liu M, Wang J, Fan H, Yang D, Zhang L, Gu X, Nie J, Chen Z, Corbett AJ, et al. (2020). IL-17 production by tissue-resident MAIT cells is locally induced in children with pneumonia. Mucosal Immunol. 13, 824–835. 10.1038/s41385-020-0273-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Borras DM, Verbandt S, Ausserhofer M, Sturm G, Lim J, Verge GA, Vanmeerbeek I, Laureano RS, Govaerts J, Sprooten J, et al. (2023). Single cell dynamics of tumor specificity vs bystander activity in CD8(+) T cells define the diverse immune landscapes in colorectal cancer. Cell Discov 9, 114. 10.1038/s41421-023-00605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu X, Rollins D, Ruhn KA, Stubblefield JJ, Green CB, Kashiwada M, Rothman PB, Takahashi JS, and Hooper LV (2013). TH17 cell differentiation is regulated by the circadian clock. Science 342, 727–730. 10.1126/science.1243884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee CH, Zhang HH, Singh SP, Koo L, Kabat J, Tsang H, Singh TP, and Farber JM (2018). C/EBPdelta drives interactions between human MAIT cells and endothelial cells that are important for extravasation. Elife 7, e32532. 10.7554/eLife.32532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gutierrez-Arcelus M, Teslovich N, Mola AR, Polidoro RB, Nathan A, Kim H, Hannes S, Slowikowski K, Watts GFM, Korsunsky I, et al. (2019). Lymphocyte innateness defined by transcriptional states reflects a balance between proliferation and effector functions. Nat. Commun. 10, 687. 10.1038/s41467-019-08604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shimizu K, Sato Y, Kawamura M, Nakazato H, Watanabe T, Ohara O, and Fujii SI (2019). Eomes transcription factor is required for the development and differentiation of invariant NKT cells. Commun. Biol. 2, 150. 10.1038/s42003-019-0389-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, Gapin L, and Glimcher LH (2004). T-bet Regulates the Terminal Maturation and Homeostasis of NK and Vα14i NKT Cells. Immunity 20, 477–494. 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 87.Matsuda JL, Zhang Q, Ndonye R, Richardson SK, Howell AR, and Gapin L (2006). T-bet concomitantly controls migration, survival, and effector functions during the development of Valpha14i NKT cells. Blood 107, 2797–2805. 10.1182/blood-2005-08-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bendelac A (1995). Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J. Exp. Med. 182, 2091–2096. 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Coles MC, and Raulet DH (2000). NK1.1+ T cells in the liver arise in the thymus and are selected by interactions with class I molecules on CD4+CD8+ cells. J. Immunol. 164, 2412–2418. 10.4049/jimmunol.164.5.2412. [DOI] [PubMed] [Google Scholar]
- 90.Ben Youssef G, Tourret M, Salou M, Ghazarian L, Houdouin V, Mondot S, Mburu Y, Lambert M, Azarnoush S, Diana JS, et al. (2018). Ontogeny of human mucosal-associated invariant T cells and related T cell subsets. J. Exp. Med. 215, 459–479. 10.1084/jem.20171739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Benlagha K, Kyin T, Beavis A, Teyton L, and Bendelac A (2002). A thymic precursor to the NK T cell lineage. Science 296, 553–555. 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 92.White AJ, Baik S, Parnell SM, Holland AM, Brombacher F, Jenkinson WE, and Anderson G (2017). A type 2 cytokine axis for thymus emigration. J. Exp. Med. 214, 2205–2216. 10.1084/jem.20170271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weinreich MA, Odumade OA, Jameson SC, and Hogquist KA (2010). T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat. Immunol. 11, 709–716. 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Loh L, Ivarsson MA, Michaëlsson J, Sandberg JK, and Nixon DF (2014). Invariant natural killer T cells developing in the human fetus accumulate and mature in the small intestine. Mucosal Immunol. 7, 1233–1243. 10.1038/mi.2014.13. [DOI] [PubMed] [Google Scholar]
- 95.Leeansyah E, Loh L, Nixon DF, and Sandberg JK (2014). Acquisition of innate-like microbial reactivity in mucosal tissues during human fetal MAIT-cell development. Nat. Commun. 5, 3143. 10.1038/ncomms4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yuan J, Nguyen CK, Liu X, Kanellopoulou C, and Muljo SA (2012). Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science 335, 1195–1200. 10.1126/science.1216557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rudd BD (2020). Neonatal T Cells: A Reinterpretation. Annu. Rev. Immunol. 38, 229–247. 10.1146/annurev-immunol-091319-083608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rouse RV, Bolin LM, Bender JR, and Kyewski BA (1988). Monoclonal antibodies reactive with subsets of mouse and human thymic epithelial cells. J. Histochem. Cytochem. 36, 1511–1517. 10.1177/36.12.2461413. [DOI] [PubMed] [Google Scholar]
- 99.Chen YH, Chiu NM, Mandal M, Wang N, and Wang CR (1997). Impaired NK1+ T cell development and early IL-4 production in CD1-deficient mice. Immunity 6, 459–467. 10.1016/s1074-7613(00)80289-7. [DOI] [PubMed] [Google Scholar]
- 100.McCarthy DJ, Campbell KR, Lun ATL, and Wills QF (2017). Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186. 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR, and Raychaudhuri S (2019). Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296. 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blanco-Carmona E (2022). Generating publication ready visualizations for Single Cell transcriptomics using SCpubr. bioRxiv. 10.1101/2022.02.28.482303. [DOI] [Google Scholar]
- 103.Gu Z, Gu L, Eils R, Schlesner M, and Brors B (2014). circlize Implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812. 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 104.Wagih O (2017). ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics 33, 3645–3647. 10.1093/bioinformatics/btx469. [DOI] [PubMed] [Google Scholar]
- 105.Durinck S, Spellman PT, Birney E, and Huber W (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191. 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, and Huber W (2005). BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440. 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 107.Brunson JC (2020). ggalluvial: Layered Grammar for Alluvial Plots. J. Open Source Softw. 5, 2017. 10.21105/joss.02017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Moerman T, Aibar Santos S, Bravo Gonzá lez-Blas C, Simm J, Moreau Y, Aerts J, and Aerts S (2019). GRNBoost2 and Arboreto: efficient and scalable inference of gene regulatory networks. Bioinformatics 35, 2159–2161. 10.1093/bioinformatics/bty916. [DOI] [PubMed] [Google Scholar]
- 109.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Haghverdi L, Lun ATL, Morgan MD, and Marioni JC (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421–427. 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Germain PL, Lun A, Garcia Meixide C, Macnair W, and Robinson MD (2021). Doublet identification in single-cell sequencing data using scDblFinder. F1000Res. 10, 979. 10.12688/f1000research.73600.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that support the findings of this study, including the raw data and Seurat object, were deposited in NCBI GEO with the accession code GEO: GSE249684. The data from this study is also displayed as a ShinyCell application at https://xspeciestcells.cshl.edu/.
The code used for all analyses presented in this study is publicly available on GitHub: https://github.com/meyer-lab-cshl/xspeciestcells. The code for the browser application can be found at https://github.com/meyer-lab-cshl/xspeciestcells-shiny.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


