Abstract
Liquid-liquid phase separation of various transcription factors into biomolecular condensates plays an essential role in gene regulation. Here, using cellular models and in vitro studies, we show the spatiotemporal formation and material properties of p53 condensates that might dictate its function. In particular, p53 forms liquid-like condensates in the nucleus of cells, which can bind to DNA and perform transcriptional activity. However, cancer-associated mutations promote misfolding and partially rigidify the p53 condensates with impaired DNA binding ability. Irrespective of wild-type and mutant forms, the partitioning of p53 into cytoplasm leads to the condensate formation, which subsequently undergoes rapid solidification. In vitro studies show that abundant nuclear components such as RNA and nonspecific DNA promote multicomponent phase separation of the p53 core domain and maintain their liquid-like property, whereas specific DNA promotes its dissolution into tetrameric functional p53. This work provides mechanistic insights into how the life cycle and DNA binding properties of p53 might be regulated by phase separation.
p53 phase separation is associated with its life cycle and function.
INTRODUCTION
The cell is organized into subcellular compartments for specific biological reactions, which are essential for the cell’s functions and survival (1, 2). While several of these compartments are membrane bound, multiple membraneless organelles exist, including nucleoli, promyelocytic leukemia protein (PML) bodies, and centrioles (3–6). Studies over the last decade have revealed that liquid-liquid phase separation (LLPS) leading to biomolecular condensate formation is a critical phenomenon involved in the formation, function, and regulation of these membraneless organelles in the cell (5–13). Some well-known examples of biomolecular condensates are nuclear speckles, stress granules, and cell signaling clusters, which play essential roles in cellular activities, including transcriptional regulation, cellular stress response, and signal transduction (4, 14–19). Proteins containing intrinsically disordered regions (IDRs) or prion-like domains, in the presence or absence of other biomolecules such as DNA/RNA, are known to undergo phase separation and form biomolecular condensates (12, 20–28). Apart from having physiological functions, LLPS is also known to be associated with disease pathology spanning from neurodegenerative diseases to cancer (5, 29–31).
Previously, it has been shown that many transcription factors undergo LLPS, where a phase-separated condensate state is linked to their transcriptional activities (4, 32, 33). p53 is a transcription factor that regulates many biological processes, including cell cycle, DNA repair, and apoptosis (34–36). Moreover, the transcriptional regulator p53 plays an essential role in tumor suppression (37–41). This is further supported by the fact that most cancers are often characterized by p53 mutations wherein p53 frequently accumulates in the nucleus as well as in the cytoplasm (42–46). Recent studies suggested that p53 misfolding, aggregation, and amyloid formation, which are further promoted by cancer-associated mutations, are shown to be associated with p53 loss of native function and gain of oncogenic functions (45, 47–52). Further, it was shown that p53 phase separation into biomolecular and/or mesoscopic condensates could act as a potential precursor for p53 amyloid formation (53–55). Moreover, partitioning of p53 into cellular condensates such as Cajal and PML bodies may also regulate the posttranslational modifications and function of p53 (56, 57).
p53 is a nuclear phosphoprotein, and its nuclear localization is essential for growth-suppressing activity in the late G1 stage (58, 59) of the cell cycle. In normal cells, p53 levels are tightly regulated as a result of a short half-life (15 to 30 min) (60), and it is undetectable by immunocytochemistry (61). Upon stress signals such as DNA damage, p53 gets stabilized and its local concentration increases in the nucleus for its tumor-suppressive functions (62). We hypothesized that p53, upon its stabilization and/or high expression, may undergo LLPS and form biomolecular condensates in the nucleus, which might act as the local reservoir of the functional protein for immediate functions such as DNA binding. The stimulated emission depletion (STED) microscopy of two cancer cells (MDA-MB-231 and MCF7 cells) with known nuclear p53 stabilization and exogenous expression of p53 in HeLa and SaOS2 cells (p53 null) showed p53 condensate formation in the nucleus. Our cellular model suggests that initial p53 condensate formation predominantly localizes in the nucleus in a liquid-like state; however, with time, cytoplasmic p53 condensates appear, which transition into a solid-like state. The spatiotemporal formation, modulation (under cisplatin treatment), and material properties of the p53 condensates may modulate p53 function and cell fate. The oncogenic mutants p53 R175H and p53 R248Q show an enhanced transition to a dynamically arrested state both in the nucleus and in the cytoplasm, which inhibits their binding to p53-specific DNA. Consistent with cellular data, the p53 core domain (DNA binding domain, p53C) also undergoes LLPS in vitro and shows solidification with time, which is further promoted by cancer-associated p53 mutations. On the basis of the observation that nuclear p53 condensates maintain their liquid-like state over time, whereas cytoplasmic condensates undergo transition to an arrested state, we hypothesized that the nucleo-cytoplasmic microenvironment (particularly DNA/RNA) might modulate the formation and material properties of p53 condensates. Both RNA and nontarget DNA (NTR-DNA) promote the p53C phase separation and maintain liquid-like properties. In contrast, the target DNA (TR-DNA) promotes the soluble state of p53, which might be a functional tetrameric form. Overall, our study provides mechanistic insights into the physical state of the p53 condensates in the cell, its correlation to p53 transcriptional function, and cell fate.
RESULTS
p53 forms liquid condensates in cancer cells
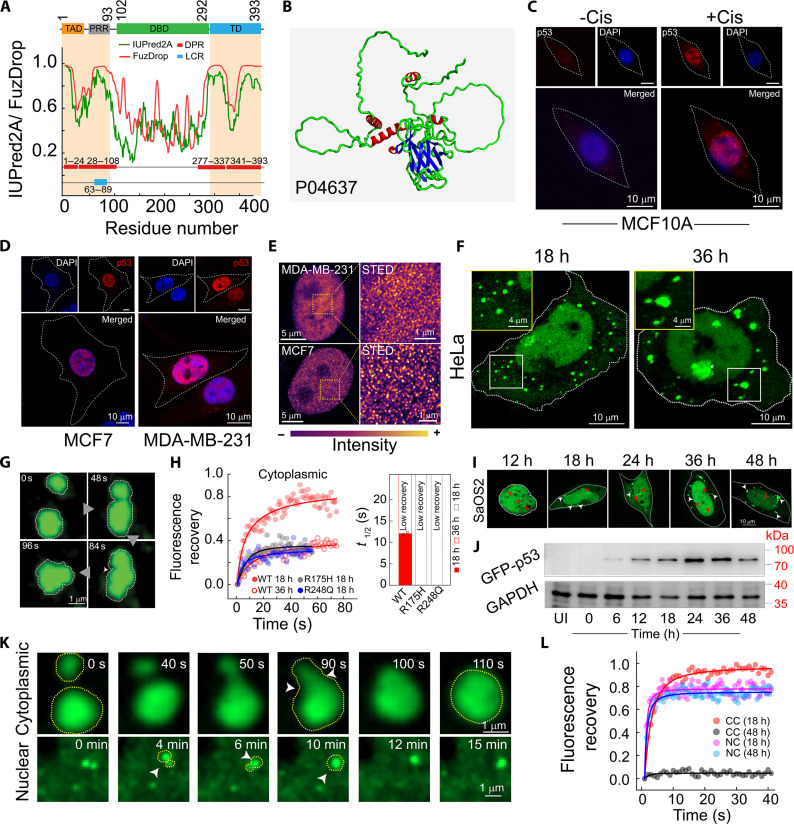
p53 is a 393–amino acid tumor suppressor protein and transcriptional factor consisting of four major domains: transactivation domain (TAD), proline-rich region (PRR), DNA binding domain (DBD), and tetramerization domain (TD) (63) (Fig. 1A). Bioinformatics analysis suggests that p53 not only contains a low-complexity region (63 to 89 residues) but also has multiple disordered regions (residues 30 to 98, 283 to 328, and 343 to 393) (Fig. 1A). The droplet forming propensity of p53 predicted by FuzDrop (64) revealed four major droplet-promoting regions at the N terminus (residues 1 to 24 and 28 to 108) and C terminus (amino acids 227 to 337 and 341 to 393) (Fig. 1A). The secondary structure prediction by Alphafold (65) and recent experimental studies (66) revealed that the N and C termini of the protein mainly comprises a random coil structure, whereas the core DNA binding region consists of α helix and β sheet conformations (Fig. 1B).
Fig. 1. p53 condensate formation in HeLa and SaOS2 cells.
(A) Domain organization of p53 showing TAD (1 to 61), PRR (64 to 93), DBD (102 to 292), and TD (325 to 393) (top), low-complexity regions (blue) using SMART, disorder tendency (green) using IUPRED2A, residue-based droplet promoting probabilities (red), and droplet-promoting regions (DPR) (red box) using FuzDrop (bottom). (B) Structure of full-length p53 protein as predicted by Alphafold from 229 structures in PDB (UniProt ID P04637). (C) Confocal images of MCF10A cells showing p53 localization with (right) and without (left) cisplatin treatment. (D) The confocal microscopy showing the p53 (red) localization in MCF7 and MDA-MB-231. (E) STED microscopy of MDA-MB-231 and MCF7 cells showing p53 condensate state in the nucleus. Pseudocolor (LUT, mpl-plasma) has been used for representative purposes. (F) HeLa cells overexpressing WT GFP-p53 at 18 and 36 hours show cytoplasmic condensates. h, hours. (G) Time-lapse microscopic images showing fusion of WT GFP-p53 condensates in the cytoplasm of HeLa cells. (H) FRAP curves of cytoplasmic condensates in HeLa cells at 18 and 36 hours showing fluorescence recovery with (right) their corresponding t1/2. Note that the t1/2 for condensates with low recovery was not estimated. (I) Time-dependent expression of WT GFP-p53 in SaOS2 cells showing nuclear (red arrowheads) and cytoplasmic condensates (white arrowheads). (J) Western blot images showing the expression of GFP-p53 with time in SaOS2 cells. (K) Time-dependent confocal images of cytoplasmic and nuclear condensates of WT GFP-p53 showing the fusion event of two condensates. (L) FRAP curves of nuclear (NC) and cytoplasmic condensate (CC) at 18 and 48 hours. All the experiments [(C) to (L)] were repeated two times with similar observations.
In healthy cells, the basal level of p53 remains low due to its rapid turnover (67). In the presence of stress (such as hypoxia or DNA damage), p53 undergoes posttranslational modifications (68, 69) and is stabilized (70), which is eventually transported to the nucleus for the functions. When epithelial cells MCF10A are treated with cisplatin [DNA damage agent (71)], we observed nuclear p53 stabilization in contrast to untreated cells (Fig. 1C). This is also consistent with p53 status in two cancer cells (MDA-MB-231 and MCF7) (Fig. 1D), where nuclear stabilization of functional p53 is known (47). To further examine the nuclear p53 state, we performed super-resolution imaging studies using STED microscopy (72, 73) of MDA-MB-231 and MCF7 cells (Fig. 1E). The STED microscopy revealed the presence of dense nuclear p53 condensate-like state (Fig. 1E). This indicates that p53 might naturally exist in the phase-separated state when stabilized in cells. These phase-separated p53 in the nucleus are unlikely to be amyloidogenic in nature as nuclear p53 in these cells were previously shown to be OC (amyloid-specific antibody) negative (47). Further, to understand the mechanism of p53 phase separation, we exogenously expressed green fluorescent protein (GFP)–p53 wild-type (WT) in HeLa and p53-deficient SaSO2 cells. p53 is generally maintained at a low basal level with a rapid turnover (67) [undetectable under immunofluorescence study (61, 74, 75)], but under stress (such as DNA damage), the half-life of p53 increases by 10- to 12-fold (76, 77) and accumulates in large amounts when the protein is needed to function for DNA repair and/or apoptosis. Therefore, our exogenous expression model can recapitulate the scenario when p53 accumulates in the nucleus under stress conditions.
The exogenous expression of GFP-p53 in HeLa cells mainly showed nuclear localization (at ~12 hours) (fig. S1B); however, at later time points (~18 hours onwards), the cells showed cytoplasmic condensates (Fig. 1F; fig. S1, A and B; and movie S1). At this stage, we only observed distinct p53 condensate formation in the cytoplasm. However, intense p53 signals with few p53 condensates were visible in the nucleus. HeLa cells expressing only GFP did not show any appearance of condensates but rather showed a diffuse pan-cellular signal, confirming that liquid condensate formation is a property of p53 and not influenced by the GFP tag (fig. S1C).
Because cytoplasmic condensates were distinct, further characterization of these condensates showed that at the early time points (18 hours), they were highly dynamic and spherical in nature (diameter ~1 μm) and underwent fusion events to form larger condensates (diameter ~3 μm) (Fig. 1G and movie S2), suggesting their liquid-like behavior. This is further supported by their high fluorescence recovery after photobleaching (FRAP) (~80% recovery) with a short t1/2 (~12 s) (Fig. 1H and fig. S1D). At a later time point (36 hours), p53 condensates in the cytoplasm not only lose their spherical shape (fig. S1E) but also show a substantial decrease in FRAP recovery (~30%), suggesting their transition toward a solid-like state. Condensates formed by two cancer-associated hotspot p53 mutants (R175H and R248Q) exhibited an arrested state (very low FRAP recovery) at the early time point (18 hours) (Fig. 1H and fig. S1, D and F) compared to WT p53. To examine the protein expression level, Western blot analysis was done, which showed a gradual increase in expression of GFP-p53 (up to 36 hours) that was sustained until 48 hours (fig. S1, G and H). The data indicate that p53 is retained in the nucleus upon exogenous expression, but with time, p53 partitions into the cytoplasm, where it undergoes liquid-to-solid state transition (fig. S1, A and B).
To nullify the effect of endogenous p53 and to establish functional consequences due to p53 condensate formation, we examined p53 LLPS in p53 null SaOS2 cells (78, 79). Similar to HeLa cells, p53 was mainly localized in the nucleus with the appearance of a few p53 condensates at an early time point (12 hours). With time, when protein load increases (Fig. 1J and fig. S2B), the distinct cytoplasmic condensates of p53 started to appear after 18 hours of transfection (Fig. 1I). The SaOS2 cells expressing only GFP did not show any condensate formation (fig. S2A), suggesting that GFP does not influence the p53 condensate formation similar to HeLa cells. The p53 condensates formed at the nucleus and cytoplasm were liquid-like immediately after their formation as confirmed by their fusion events (Fig. 1K and movie S3) as well as high FRAP recovery (~80 to 100%) (Fig. 1L). At the late time point (48 hours), the cytoplasmic condensates underwent liquid-to-solid-like transition with no FRAP recovery (less than 10%) in contrast to nuclear condensates (~80% FRAP recovery) (Fig. 1L and fig. S2, C and D). This suggests that once p53 is expelled from the nucleus, the condensates undergo a liquid-to-solid state transition in the cytoplasmic environment. This is further consistent with morphological analysis, where the cytoplasmic condensates showed an increase in size and a decrease in circularity with time (fig. S2E).
Spatiotemporal partitioning of p53 condensates in cells
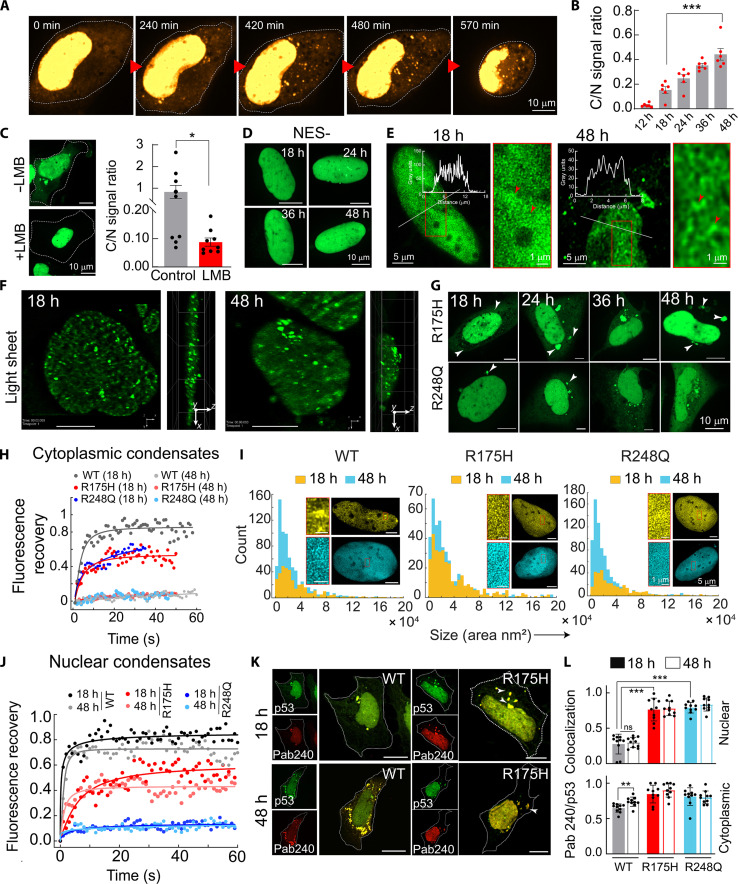
To understand nucleo-cytoplasmic partitioning and condensate formation of p53 with time, we performed live-cell imaging experiments with SaOS2 cells. Initially, the p53 signal was mostly localized in the nucleus, but with time, p53 appeared in the cytoplasm and formed cytoplasmic condensates (Fig. 2A and movie S4). With time, the increased cytoplasmic/nuclear (C/N) ratio of the GFP-p53 signal indicates the nucleo-cytoplasmic shuttling of p53 (Fig. 2B), possibly through the nuclear pore complex. This is further confirmed as treatment with Leptomycin B (LMB, inhibitor of nuclear export (80)) decreased the C/N ratio (Fig. 2C). Moreover, the expression of the GFP-p53 construct without nuclear exclusion signal sequence (GFP-p53 NES−) in SaOS2 cells showed only nuclear localization of p53 (from 18 to 48 hours) (Fig. 2D and fig. S2F), further supporting the idea that the cytoplasmic p53 condensates are formed after expulsion of p53 from the nucleus. These cytoplasmic condensates did not exhibit any colocalization with Nile red [specific for intracellular lipid (81)], Lysotracker red (for lysosomes), and Mitotracker red (for mitochondria), suggesting their membrane-less state (fig. S3, A to C). Moreover, they are not associated with the aggresome formation in the cytoplasm (fig. S3D).
Fig. 2. Spatiotemporal formation of p53 condensates in p53-null SaOS2 cells.
(A) Time-lapse confocal microscopy images showing spatiotemporal formation of p53 condensates. (B) Cytoplasmic/nuclear (C/N) ratio of WT GFP-p53 fluorescence signal in SaOS2 cells with time (mean ± SEM for n = 6 transfected cells). (C) Confocal microscopy images showing the effect of Leptomycin B (LMB) treatment (left) and corresponding C/N ratio calculation (right) of SaOS2 cells (mean ± SEM for n = 9 transfected cells). (D) The time-lapse confocal microscopy showing nuclear localization of the p53 with p53 NES− in SaOS2 cells. (E) STED microscopy of nuclear condensates of WT GFP-p53 at 18 and 48 hours. The line profile represents the intensity of GFP-p53 across the nucleus. (F) Lattice light-sheet imaging of nuclear condensates of SaOS2 transfected with WT GFP-p53 at 18 and 48 hours. Scale bars, 10 μm. (G) p53 condensates of R175H and R248Q in SaOS2 cells monitored over time using confocal microscopy. (H) FRAP recovery profiles of WT, R175H, and R248Q cytoplasmic condensates at 18 and 48 hours. (I) Size (area) and number distribution of nuclear condensates of WT, R175H, and R248Q at 18 and 48 hours, calculated from STED microscopy. Scale bars, 5 μm. (J) FRAP recovery profile of WT, R175H, and R248Q nuclear condensates at 18 and 48 hours. (K) Immunofluorescence study of WT GFP-p53 condensates with misfolded p53 specific antibody (Pab240). Scale bars, 10 μm. (L) Quantification of Pab240 colocalization with GFP-p53 nuclear and cytoplasmic condensates at 18 and 48 hours (mean with SEM, for N = 2). For (B), (C), and (L), the statistical significance was calculated using a two-tailed t test [***P < 0.001, **P < 0.002, *P < 0.033, and P > 0.012 (ns), with 95% confidence interval]. The experiments (A), (C) to (H), (J), and (K) were performed two independent times.
To further characterize the p53 condensates in the nucleus and cytoplasm, STED microscopy of GFP-p53 in SaOS2 cells was performed. The data showed that the nucleus of the cells is rather crowded with p53 condensates, which sometimes clump together to form local regions of high density (Fig. 2E). Further analysis also revealed that the majority of the nuclear p53 (>70%) exists as condensates (fig. S2G), where the morphology appeared to be that of interconnected microphase-separated condensates (percolation clusters) as reported previously (82–84). The STED microscopy of cytoplasmic condensates suggests that both WT and mutant p53 exist in the form of small condensates (fig. S4). Further, to investigate the dynamic and reversible nature of these nuclear condensates, we treated the cells with sorbitol and 1,6-hexanediol. Our data suggest that 1,6-hexanediol and sorbitol treatment does not alter the major characteristics of nuclear condensates. We observe subtle changes in the size and number of p53 condensates inside the nucleus with both treatments. For example, in the presence of 1,6-hexanediol, there are a higher number of smaller p53 condensates, while sorbitol-treated cells showed more clustering of nuclear condensates (fig. S5). In this context, although hexanediol has been shown to dissolve the condensates (85), many recent studies, however, suggest that the dissolution due to hexanediol depends on the nature of intermolecular interactions. For example, condensates formed mostly by hydrophobic interactions will dissolve in the presence of hexanediol (85). Further, one needs to consider the effect of 1,6-hexanediol/sorbitol on cells, and cells, in turn, modulate the condensate behavior. To further support STED microscopy analysis and the nature of the relatively small-sized condensates, we also performed lattice light-sheet microscopy of SaOS2 cells expressing WT GFP-p53 in live cells. The data showed largely dynamic p53 nuclear condensates throughout the nucleus (Fig. 2F and movie S5).
We further studied the effect of two hotspot p53 mutants associated with cancer (R175H and R248Q) and characterized their condensates in both cytoplasm and nucleus in SaOS2 cells. The data showed that both mutants formed slightly larger cytoplasmic condensates (18 hours) as compared to WT protein (~1 μm), but their decrease in circularity with time was similar to WT protein (Fig. 2G and fig. S2, H and I). FRAP analysis of the cytoplasmic condensates showed (in contrast to WT p53 with ~80% recovery) only ~50% recovery for both the mutants, even at an early time point (18 hours), which is further reduced to almost no recovery at 48 hours (Fig. 2H and fig. S2J). The data suggest a faster liquid-to-solid state transition of the mutant p53 condensates than WT p53 in the cytoplasm. We did not find any major morphological differences between the cytoplasmic condensates of WT and mutants by STED microscopy (fig. S6). Further, the data revealed that both mutants formed a dense population of condensates in the nucleus at 18 hours, having an apparent higher density of accumulation at certain local regions (fig. S6) compared to the WT protein. At a later time point (48 hours), larger-sized (in terms of area parameter) and a higher population of nuclear condensates were seen for both mutants, as observed for WT protein (Fig. 2I). When FRAP analysis of nuclear condensates was performed, both mutants showed substantially reduced recovery (~40% for R175H and ~10% for R248Q), at both time points (18 and 48 hours). This indicates a solid-like nature of nuclear condensates of mutants even at an early time point (18 hours) (Fig. 2J and fig. S2K). Note that p53 R175H condensates showed a relatively higher FRAP recovery compared to R248Q.
Thus, the data suggest that while p53 cytoplasmic condensates transitioned from a liquid-to-solid state, which is further promoted by both cancer-associated mutants, the nuclear WT p53 condensates, however, remained in a liquid-like state in SaOS2 cells. The rigidification of nuclear condensates of p53 mutants and cytoplasmic condensates of both WT and mutants in SaOS2 cells could be due to stronger assembly of p53 caused by misfolding and/or aggregation. The immunofluorescence study using the Pab240 antibody (specific to misfolded p53 (86)) showed that mutant p53 condensates in the nucleus are composed of misfolded proteins compared to that of WT protein. In addition, most of the p53 in cytoplasmic condensates (both WT and mutants) were misfolded, where the mutants showed a higher colocalization of p53 with Pab240 compared to WT (Fig. 2, K and L, and fig. S7A). However, the amyloid-specific OC antibody staining showed no colocalization with GFP-p53 WT and mutants GFP-p53 R175H and GFP-p53 R248Q (fig. S7B). This suggests that p53 remains in a misfolded conformation in the cytoplasm without attaining amyloid conformation. Further, we also performed the A11 staining, which is known as an amyloid oligomer–specific antibody (87). Our data suggest the absence of A11 staining, and therefore, no colocalization was observed in both nucleus and cytoplasm at the 24- and 48-hour time points (fig. S8). This suggests that in our cellular conditions, no amyloid oligomers were formed. In this context, our previous studies showed that p53 aggregates in cells do not have an amyloid state in contrast to cancer tissues, suggesting that extracellular matrix component [e.g., chondroitin sulfate A (CSA)] plays a role in amyloid formation (47).
The functional consequence of p53 phase separation in cells
To investigate the consequences of p53 nuclear condensate formation on DNA binding and transcriptional activity, we monitored the expression of p53 downstream gene p21 by Western blot analysis. The data suggest a time-dependent increase in p21 expression up to 24 hours and then plateaued (Fig. 3, A and B), indicating the presence of functional p53 in the nucleus. This is further apparent as the Annexin-PI assay followed by FACS (fluorescence-activated cell sorting) analysis showed a gradual increase in the apoptotic population of cells with time (Fig. 3C and fig. S9), indicating p53-mediated apoptosis. This increase in p21 expression and apoptosis might be due to the higher expression and condensate formation by p53 with time (Fig. 1). We further examined p53 binding to its direct target, p21, by enzyme-linked immunosorbent assay (ELISA) and chromatin immunoprecipitation (ChIP) assay at 18 hours of transfection for both WT and mutants. While WT p53 showed higher p53-DNA binding, both cancer-associated mutants (R175H and R248Q) showed much less DNA binding activity (Fig. 3, D and E). This is consistent with the fact that mutant p53 condensates are composed of misfolded p53, which promotes condensate solidification even at 18 hours (Fig. 2, J and K), where p53 is most likely nonfunctional. To further examine the effect of p53 condensates on DNA damage, SaOS2 cells were transfected with WT GFP-p53 and treated with cisplatin. After 18 hours, STED imaging indicated a higher number of p53 condensates in the nucleus for cisplatin-treated cells than untreated cells (Fig. 3, F to H, and fig. S10, A to C). Increased p53 condensate formation also correlated with increased p53 stabilization (Fig. 3, I and J) and p21 mRNA levels (Fig. 3K) in cells upon treatment with cisplatin compared to untreated cells. The data suggest that the p53 condensates might be functional in terms of DNA binding activity, which might be further enhanced when DNA damage occurs for tumor-suppressive functions.
Fig. 3. Stabilization and activity of p53 condensates.
(A and B) Western blot showing time-dependent expression of p21 (p53 target) and the corresponding fold change of p21. (C) Time-dependent apoptotic cell death of SaOS2 cells using FACS analysis. (D) p53 DNA binding capacity of p21-specific DNA sequence by ELISA at 18 hours for WT and p53 mutants. (E) ChIP assay of p53 DNA binding to p21-response element at 18 hours. For ELISA and ChIP assays, one-way analysis of variance (ANOVA) (***P < 0.001, **P < 0.01, *P < 0.05). (F) SaOS2 cells transfected with WT GFP-p53 after treatment with cisplatin showing p53 condensates at 18 hours (left). Scale bar, 10 μm. Super-resolution STED imaging showing the p53 clusters in the nucleus after treatment at 18 hours (right). (G) Size/number distribution of nuclear condensates for cisplatin-treated and untreated cells. (H) Condensates count in uniform area of (5 μm by 5 μm, N = 2) in the nucleus of cisplatin-treated SaOS2 cells. (I and J) Western blot of p53 expression with and without treatment of cisplatin (10 μM, 9 hours). (K) The p21 mRNA expression of SaOS2 cells at 18 hours in the presence and absence of cisplatin treatment {(H) and (K), two-tailed t test [***P < 0.001, **P < 0.002, *P < 0.033, and P > 0.012 (ns)]}, [(A) to (F) and (I) to (K)] N = 2. (L) Immunofluorescence of SaOS2 cells showing no colocalization with WT p53 and MDM2. In a few cells, cytoplasmic condensates of large size partially colocalizes with MDM2. (M) SaOS2 cells transfected with WT GFP-p53 after treatment with Nutlin showing p53 cytoplasmic condensates at 18 hours (left). Scale bar, 10 μm. STED imaging showing the presence of p53 clusters in the nucleus after treatment at 18 hours (right). (N) Estimation of condensate count in the nucleus of Nutlin-treated (Nut) and untreated (WT) cells. (O) FRAP recovery profiles of cytoplasmic condensates with Nutlin-treated cells.
Further, it is known that the expression, function, and regulation of p53 and MDM2 are interdependent via a feedback mechanism (88). MDM2 controls p53 activities, including the inhibition of transactivation, nuclear export, and degradation by the proteasome. To further examine the role of MDM2 in p53 phase separation, SaOS2 cells were transfected with p53-GFP WT and immunostained for MDM2 antibody 24 hours after transfection. We did not find any colocalization of p53 condensate and MDM2 in the nucleus, suggesting that the nuclear condensates are possibly protected against MDM2-mediated degradation and are functional in nature. Larger cytoplasmic p53 condensates were partially colocalized with MDM2, implying that these condensates could be sites of MDM2-mediated regulation and/or degradation of p53 (88, 89) (Fig. 3L and fig. S10D).
Treatment with Nutlin 3a (inhibitor of MDM2) resulted in the deposition of large condensates in the cell cytoplasm, indicating either a higher release of p53 from the nucleus or stabilization of p53 in the cytoplasm. Super-resolution imaging revealed dense p53 clusters in the nucleus (Fig. 3M and fig. S10E). Further, STED imaging indicated a higher number of p53 condensates in the nucleus for Nutlin-treated cells as compared to untreated cells (Fig. 3N). FRAP studies indicated a liquid-like nature of the early cytoplasmic condensates while it showed an arrested nature at a later time point (Fig. 3O).
p53 DNA binding domain (p53 core, p53C) undergoes LLPS and liquid-to-solid state transition in vitro
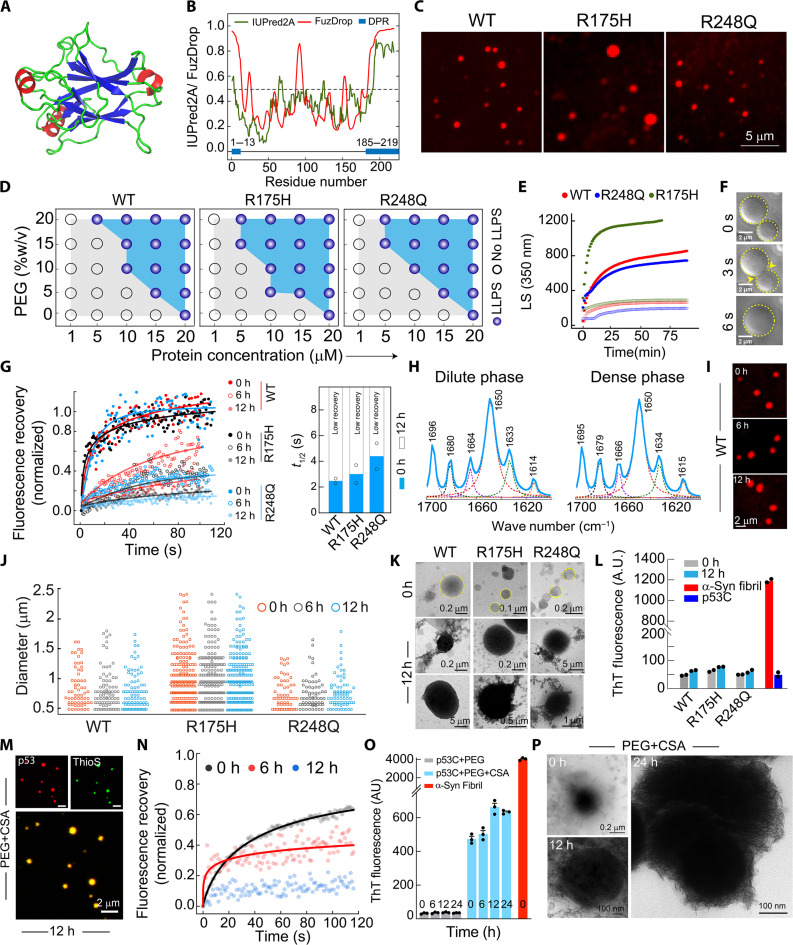
The p53 core domain (p53C) represents the DNA binding domain of full-length p53, which is often used for in vitro studies due to the unstable nature of full-length p53 protein (90–92). The previously reported crystal structure of p53C (93) shows the organization of its secondary structure into α-helical, β sheet, and random coil conformations (Fig. 4A). Further, this domain also contains IDRs [predicted by IUPRED 2A (94, 95)] and two droplet-promoting regions [DPR, predicted by FuzDrop (64)], which are present at the N terminus and C terminus of the p53C (Fig. 4B and fig. S11, A and B). We also included two mutants, R175H and R248Q, to study the effect of cancer-related hotspot mutations. For in vitro studies, WT p53C and mutants were recombinantly expressed and purified (fig. S11, C and D), and NHS-Rhodamine–labeled p53C (10:90 labeled:unlabeled protein) was incubated in the presence of varying percentages of PEG-8000 (polyethylene glycol, molecular weight 8000, as molecular crowder) at pH 7.4 in 50 mM phosphate buffer. When examined under microscopy, we observed that both WT and mutant (R175H and R248Q) p53C readily phase separates to form condensates at as low as 10 μM concentration in the presence of 10% PEG-8000 (Fig. 4C and fig. S11E) with no significant difference in their phase regime (Fig. 4D; fig. S11, F to H; and movie S6). The mutant R175H showed a slightly higher propensity for phase separation than WT and R248Q. The condensate formation was further examined by static light scattering (LS) experiments (at 350 nm), which showed that all proteins spontaneously phase separate within a few minutes in the presence of 10% PEG-8000. Consistent with the microscopic phase regime, R175H also showed higher scattering than WT and R248Q in the presence of PEG, suggesting its higher propensity for LLPS (Fig. 4E). The liquid-like property of phase-separated condensates by these proteins was confirmed by the fusion (for WT condensates) (Fig. 4F) and high FRAP recovery (~100%) with shorter t1/2 (~<5 s), immediately after their formation (0 hours) (Fig. 4G and fig. S12A). These condensates, however, showed reduced FRAP recovery with time, suggesting compromised dynamicity of the molecules inside the condensates, which is further promoted by both cancer-associated mutants (Fig. 4G). Further, the Fourier transform infrared (FTIR) spectroscopy study of the dense and dilute fraction of WT p53C (separated by centrifugation) LLPS (at 0 hours) showed no difference in their secondary structure after phase separation (Fig. 4H). This indicates that condensate formation by p53 might preserve the functionality of the protein.
Fig. 4. Liquid-liquid phase separation of p53C condensates in vitro and their transition from liquid to arrested state.
(A) The PDB structure of p53C (PDB ID: 2OCJ) shows random coil and β sheet conformation. (B) WT p53C shows disorder tendency (green), residue-based droplet-promoting probabilities (red), and droplet-promoting regions (DPR) (blue box). (C) Microscopy images showing the condensate formation of WT, R175H, and R248Q p53C (N = 3). (D) Phase regimes (schematic) of WT, R175H, and R248Q. Opaque and open circles represent LLPS and no LLPS, respectively. (E) Static light scattering (LS at 350 nm) showing condensate formation by WT p53C, R175H, and R248Q in the presence (filled circles) and absence (open circles) of 10% PEG-8000. (F) Time-lapse DIC images showing the fusion event of two WT p53 condensates with time. Scale bars, 2 μm. (G) FRAP experiment at different time intervals of WT p53C, R175H, and R248Q variants with t1/2 calculation (right). (H) FTIR spectroscopy of dense and dilute phase of WT p53 after centrifugation. (I) Confocal microscopy images WT p53C condensates at different time intervals. (J) The size distribution of WT, R175H, and R248Q condensates with time. (K) TEM micrographs WT p53C, R175H, and R248Q condensates at 0 and 12 hours. (L) The ThT binding p53C condensates at 12 hours. α-Synuclein fibrils were used as a positive control. N = 3 for (C), (D), and (I), and N = 2 for (E) to (H), (K), and (L). (M) ThioS staining of WT p53C condensates formed in the presence and absence of CSA at indicated time points (n = 3 independent experiments). (N) The FRAP of WT p53C condensates in the presence of CSA. (O) The ThT binding for p53C condensates in the presence of CSA. α-Synuclein fibrils were used as a positive control (N = 3). (P) TEM micrographs of condensates in the presence of CSA.
Further, the time-dependent study of condensate formation/growth showed no change in the size and number of condensates over time (up to 12 hours) (Fig. 4, I and J, and fig. S12B). However, with an increase in protein concentration, although the condensate size did not alter, the number of condensates increased, as shown for WT p53C (fig. S12, C and D). Further, the morphology of the condensates was characterized using transmission electron microscopy (TEM), which showed circular morphology at 0 hours, but with time, dark-round shape condensates along with the appearance of amorphous aggregates originating from condensates were observed (Fig. 4K). However, the lack of thioflavin T (ThT) (96) and Congo red (CR) (97) dye (specific dye for amyloid aggregates) binding suggests that these aggregates might not be amyloid in nature (Fig. 4L and fig. S12E). The data, therefore, suggest that p53C undergoes LLPS and liquid-to-solid state transition with time, which is further promoted by both the cancer-associated p53 mutations. It is important to note that our cellular study and in vitro studies did not show any indication of amyloid formation through LLPS, suggesting that p53 amyloid formation might require different conditions, such as the presence of extracellular components like glycosaminoglycans (GAGs) (47). To investigate the role of GAGs in p53C LLPS and liquid-to-solid transition, we performed LLPS studies of p53C in the presence of CSA. A rapid phase separation of p53C in the presence of CSA, where more condensates have a larger size, were observed in contrast to p53C (Fig. 4M and fig. S12, F and G). The FRAP data suggest that in contrast to p53C alone condensate, the condensates formed in the presence of CSA showed reduced fluorescence recovery immediately after their formation as well as rapid solidification with time (Fig. 4N and fig. S12H). Further, thioflavin S (ThioS) readily gets copartitioned with condensates formed in the presence of CSA, which was not seen in WT condensates formed by p53C alone (Fig. 4M and fig. S12F). The data indicate that p53C condensate formation in the presence of CSA might transform into an amyloid state with time, which is further confirmed by the high ThT binding of the condensates (Fig. 4O). The amyloid formation through LLPS by p53C was further confirmed by TEM study where the morphology of the condensates in the presence of CSA shows condensates containing amyloid-like fibrils at 12 hours (Fig. 4P).
DNA/RNA modulates the formation and fate of multicomponent p53C phase separation
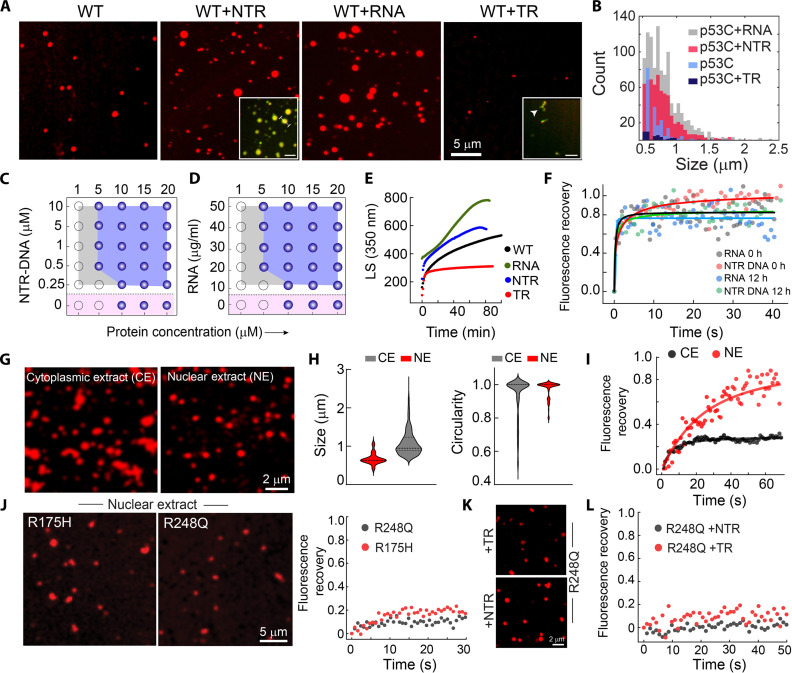
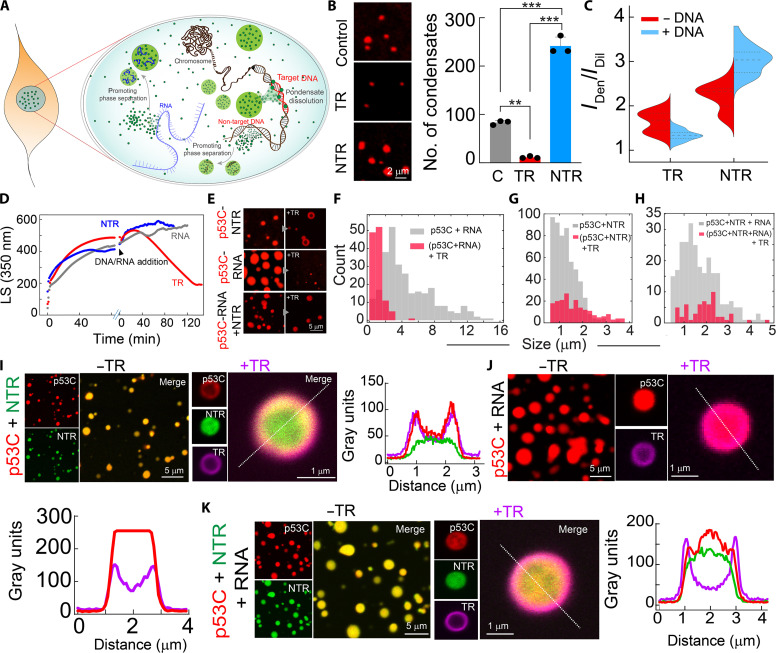
Cellular studies have shown that the microenvironment of the nucleus and cytoplasm might dictate the formation, fate, and material properties of p53 condensates, as nuclear p53 condensates maintain liquid-like properties and cytoplasmic condensates solidify with time (Figs. 1 and 2). Because p53 is a transcription factor that requires extensive interactions with nucleic acids in the cellular milieu, we studied the effect of nucleic acids on phase separation of p53C. To examine this, p53C LLPS experiment was performed (with 10% PEG-8000) in the presence and absence of RNA, TR-DNA (corresponding to the consensus sequence of the p53 response element for the p21 gene), and NTR-DNA (98) (table S2). Compared to condensates formed in the absence of any nucleic acid (p53C alone in the presence of 10% PEG-8000), the presence of both NTR-DNA and RNA increases the size and number of the p53 condensates (Fig. 5, A and B). In contrast, in the presence of TR-DNA, p53C did not show a significant number of condensates; rather, a few smaller condensates were observed (Fig. 5, A and B). The dual fluorescence labeling study suggests that WT p53C condensates in the presence of NTR-DNA resulted in multicomponent phase separation (yellow condensate in the inset), while in the presence of TR-DNA, few residual DNA-protein conjugates were observed (fig. S13, A and B), similar to the previous study (99). The facilitated p53C LLPS in the presence of RNA and NTR-DNA was further evident from the phase regime of p53C, which showed a decrease in the p53C concentration required for phase separation (Fig. 5, C and D, and fig. S13, C and D). Consistent with this, the LS experiment also showed enhanced LLPS kinetics of WT p53C in the presence of RNA and NTR-DNA but reduced LLPS kinetics in the presence of TR-DNA (Fig. 5E) compared to the p53C alone. The NTR-DNA and RNA not only promote p53 phase separation but also help to maintain its liquid-like state as FRAP analysis showed >80% recovery at both 0 and 12 hours (Fig. 5F and fig. S13E), in contrast to p53C alone where liquid-to-solid state transition was observed (Fig. 4G).
Fig. 5. DNA and RNA modulate p53C phase separation.
(A) Confocal images of WT p53C condensates formed in the presence and absence of target DNA (TR-DNA), nontarget DNA (NTR-DNA), and RNA. The inset for NTR-DNA and TR-DNA represents multicomponent condensate formation. (B) Size (Feret) distributions of p53C condensates formed in the presence and absence of TR-DNA, NTR-DNA, and RNA. (C and D) Phase regime of WT p53C with varying concentrations of NTR-DNA and RNA. Opaque and open circles represent LLPS and no LLPS, respectively. Pink shaded region showing p53 condensates in the absence of any nucleic acid. (E) Static light scattering (LS at 350 nm) showing the effect of RNA, NTR-DNA, and TR-DNA for p53C condensate formation. (F) FRAP profile of p53C condensates in the presence of NTR-DNA and RNA showing a complete recovery at the early (0 hours) and late (12 hours) time points, suggesting maintenance of the liquid-like state. (G) The confocal image of NHS-Rhodamine–labeled p53C condensates formed in the cellular fraction of the nuclear extract (NE) and cytoplasmic extract (CE). Scale bar, 2 μm. (H) The size and circularity distribution of p53C condensates formed in the presence of CE and NE showing larger size in CE with lesser circularity in comparison to condensate formed in the NE. (I) FRAP of p53C condensates formed in NE and CE. The experiments (A), (C), (D), and (G) were repeated three independent times and (E), (F), and (I) were repeated two independent times. (J) The confocal image of p53C mutant (R175H and R248Q) condensates formed in NE (left). FRAP profile of R175H and R248Q condensates formed in NE (right). (K) Representative confocal images of preformed R248Q condensates after adding TR-DNA and NTR-DNA. (L) FRAP of preformed R248Q condensates after adding TR-DNA and NTR-DNA, showing negligible fluorescence recovery.
Further, to directly probe the p53C condensates’ property in nuclear versus cytoplasmic microenvironment, we examined p53C condensate formation in nuclear extract (NE) and cytoplasmic extract (CE) of SaOS2 cells (100) (fig. S14, A and B). The data showed condensate formation of p53C in both extracts, but with a lower concentration regime at 0 hours (fig. S14C). The size of the p53C condensates in CE was higher, with lower circularity than the condensates formed in NE (Fig. 5, G and H). This indicates the solidification of the condensates formed in CE. The FRAP recovery of p53C condensates in NE was higher than the condensates formed in the CE (Fig. 5I and fig. S14D), suggesting that nuclear condensates remain in a relatively more liquid-like state. In contrast, the cytoplasmic microenvironment promotes the solidification of p53 condensates, consistent with in cellulo data. Further, when cancer-associated hotspot mutants (R175H and R248Q) were allowed to form condensates in NE, the FRAP study showed almost no recovery immediately after their formation (Fig. 5J). This study directly demonstrated that p53 mutations promote solidification even in the nuclear microenvironment, suggesting that their lack of DNA binding ability leads to loss of function. It is further evident that R248Q p53C condensates do not get dissolved after the addition of TR-DNA, and also for NTR-DNA, suggesting irreversible condensates with rapid solidification for mutant in contrast to WT protein. This indicates that WT p53C binding to DNA inhibits the formation of condensates, while the lack of DNA binding of mutant p53C with TR-DNA leads to phase separation and subsequent solidification (Fig. 5, K and L).
Effect of TR-DNA on preformed p53C multicomponent condensates
On the basis of our data, we hypothesize that the abundance of nonspecific DNA and RNA in the nucleus might promote and stabilize the p53 liquid condensate (demixed state). In contrast, cues such as exposure to specific DNA might promote their dissolution (mixed state) (Fig. 6A). This demixing/mixing might be part of the p53 life cycle and function. When preformed WT p53C condensates were added on NTR-DNA–coated coverslips, significantly more p53C condensates were seen. In contrast, the disappearance of the p53C condensates was seen with TR-DNA–coated coverslips (Fig. 6B and fig. S15A). This is also consistent with the p53 partitioning analysis in a single p53 condensate, where a significant reduction of p53 intensity in the dense phase was observed when TR-DNA was added to preformed p53C condensate. In contrast, the dense phase signal increased with the addition of NTR-DNA (Fig. 6C and fig. S15, B and C). The demixing and mixing due to NTR-DNA versus TR-DNA, respectively, were further confirmed by LS experiments, where, after the formation of p53C condensates when RNA, TR-DNA, and NTR-DNA were added, the intensity values reduced significantly in the presence of TR-DNA, but slightly increased with the addition of NTR-DNA/RNA (Fig. 6D).
Fig. 6. Effect of DNA/RNA on preformed p53C condensates.
(A) Schematic showing the possible mode of operation of p53 in the nuclear microenvironment. (B) Left: Representative images of preformed p53 condensates after the addition onto the uncoated and TR- and NTR-DNA-coated coverslips. Right: The calculated number of condensates with various nucleic acid–coated surfaces and uncoated coverslips were taken as control (C). The statistical significance was determined by one-way ANOVA using Newman-Keuls (SNK) post hoc analysis (***P < 0.001, **P < 0.01). (C) Split-violin plot represents the change in the partitioning of p53C after adding TR-DNA and NTR-DNA. (D) Static light scattering (LS at 350 nm) showing the kinetics of p53C in the presence of 10% (w/v) PEG-8000 and the effect after the addition of TR-DNA, NTR-DNA, and RNA. The data showing a decrease in light scattering value in the presence of TR-DNA, suggesting the dissolution of preformed p53C condensates. (E) The confocal microscopy images showing the effect of TR-DNA on preformed p53C condensates formed in the presence of NTR-DNA (p53C-NTR DNA), RNA (P53C-RNA), and RNA + NTR-DNA (p53-RNA-NTR-DNA) condensates. (F to H) Size distribution plot showing the reduction in the size/numbers of condensates after the addition of TR-DNA to (F) p53C-RNA condensates, (G) p53C-NTR-DNA condensates, and (H) p53C-RNA-NTR-DNA condensates. (I to K) The confocal microscopy images show few residual condensates observed after condensate dissolution due to the addition of TR-DNA. The left panel represents Rhod-p53 condensates formed in the presence of (I) Atto 488–labeled NTR DNA, (J) RNA, and (K) RNA and Atto 488–labeled NTR-DNA. The middle panel represents residual p53C condensates that remained after the addition of Atto 647N–labeled TR-DNA. The right panel represents a line profile across the condensates to show the spatial distribution of protein and DNA. The experiments (B), (D), (E), and (I) were repeated two independent times.
We further examined the effect of TR-DNA on multicomponent p53 condensates formed in the presence of NTR-DNA (p53C-NTR-DNA) and RNA (p53C-RNA), which is more relevant in the nuclear microenvironment. Adding TR-DNA to preformed p53C-RNA condensates led to the dissolution of p53C-RNA condensates. However, few residual condensates of smaller sizes were seen (Fig. 6, E and F), possibly due to their attachment to the surface (99). Similarly, the addition of TR-DNA to preformed p53C-NTR-DNA and p53C-NTR-DNA+RNA multicomponent condensates resulted in the dissolution of most of the preformed p53 condensates as evident from the reduced number and size of the condensates (Fig. 6, E, G, and H). The residual p53C-NTR-DNA condensates present after the addition of TR-DNA resulted in two distinct morphologies; one was spherical, and the other one was hollow ring–like condensates (Fig. 6E).
To further characterize the spatial distribution of protein and nucleic acids in the multicomponent system, we performed the fluorescence study using Atto 647N–labeled TR-DNA, Atto 488–labeled NTR-DNA, and NHS-Rhodamine–labeled p53C. For residual NTR-p53C condensates (which are large enough to resolve spatially), we observed that TR-DNA and p53C both partitioned at the periphery of the condensate, while NTR-DNA is mainly localized at the center of the condensate (Fig. 6I and fig. S15D). Similar results were also obtained when TR-DNA was added to p53C-RNA and NTR-DNA-RNA-p53C multicomponent condensates, resulting in a few residual condensates, where TR-DNA remained at the periphery and p53C localized at the center of the condensates (Fig. 6, J and K, and fig. S15, D and E). These data suggest that TR DNA might interact with the condensate periphery and drive their dissolution, which could also result in the formation of hollow condensate (101).
Mixing/demixing modulates the temporal storage and DNA binding for p53 functions
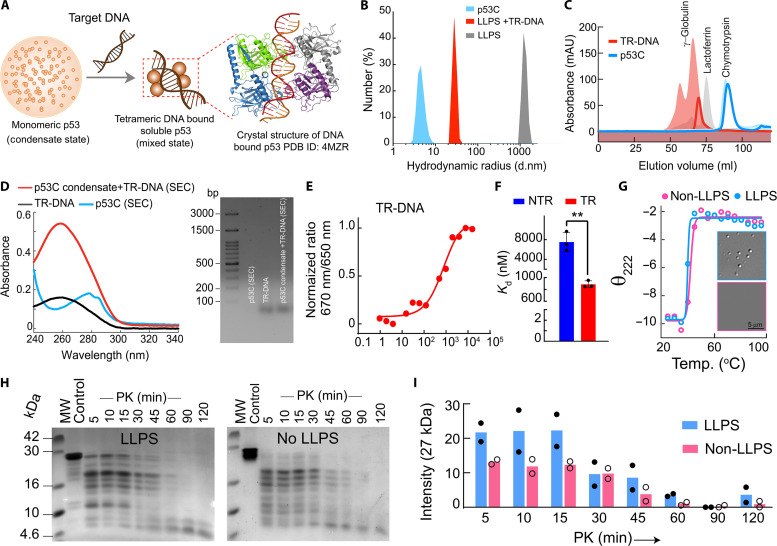
Previously, it was known that p53, upon activation, goes to the nucleus and, once bound to the DNA, stabilizes as a tetrameric structure, which is known to be transcriptionally active. Similar to full-length p53, it was also shown that the p53 core domain/DNA binding domain, in the presence of p53-specific DNA, forms a tetrameric structure, as confirmed by the crystal structure (102). Because our data show that in the presence of specific DNA, the p53 condensates get dissolved (mixed phase), we hypothesized that upon stress signal when p53 concentration gets high with an increased half-life in the nucleus, p53 phase separation might serve as a temporal depot for the functional p53, which could still maintain its structural integrity (Fig. 4H) of monomeric p53 and bound by weak interactions forming condensates. However, upon binding to the specific DNA, it forms a functional tetrameric structure, which is known to be the transcriptionally active state (Fig. 7A). This way, p53 in condensates remains as a ready pool of the protein, and in demand, the protein is released out of the condensates and binds to TR-DNA where it functions as a transcription factor to regulate its downstream genes.
Fig. 7. Specific DNA modulates the p53C condensates for functionality.
(A) The proposed model of p53 regulation through p53 phase separation. Crystal structure showing the core domain bound with DNA consensus element of p53 in tetrameric form. (B) Dynamic light scattering profile of p53C monomer, p53C condensates alone, and p53C condensates dissolved in the presence of the specific DNA. (C) SEC measurement of p53C monomer and p53C condensate in the presence of target DNA showing a peak approximately at 100 kDa, confirming tetrameric state. γ-Globulin (150 kDa), lactoferrin (78 kDa), and chymotrypsin (25 kDa) are used as markers in SEC. (D) The SEC eluted the oligomeric fraction of p53C condensates in the presence of TR-DNA, showing the presence of DNA bound with protein using UV spectroscopy (left) and agarose gel electrophoresis (right). (E) Spectral shift measurement of p53C in the presence of TR-DNA showing binding affinity of p53C with nucleic acids. (F) Bar graph representing the binding affinity (Kd) of p53C with TR-DNA and NTR-DNA showing a significantly higher binding affinity for TR-DNA. Error bars represent mean ± SEM for N = 3 independent experiments. The statistical significance was estimated with an unpaired t test with 95% confidence interval (**P < 0.002). (G) The change in molar ellipticity at θ222 with an increase in temperature shows no major changes in temperature denaturing profiles between the LLPS and non-LLPS state of p53C. Representative microscopic images of p53C in LLPS and non-LLPS states for the temperature-dependent CD experiment are shown. (H) Proteinase K (PK) digestion was followed by SDS-PAGE and showed the higher PK resistivity of the p53 LLPS state. (I) SDS-PAGE quantification of PK digestion assay of LLPS and non-LLPS p53C showing faster PK digestion with time of p53C in the non-LLPS state compared to the LLPS state. The experiments (B) to (H) were repeated two independent times.
To examine the possibility that it forms tetrameric structure after the dissolution of p53 condensates in the presence of TR-DNA, we performed dynamic light scattering (DLS) and size exclusion chromatography (SEC) with p53 monomer and p53 condensate in the presence of TR-DNA. As expected, the p53C monomer exhibited a hydrodynamic radius (RH) 5 to 8 nm, whereas p53C condensates dissolved in the presence of TR-DNA showed an RH of ~30 nm, suggesting multimeric p53C. The p53C condensates were also used as a control, showing a large scattering peak at ~>1000 nm (Fig. 7B). When we performed SEC, we observed that p53C-TR-DNA eluted at a volume corresponding to the molecular weight of tetrameric p53C (peak between 150 and 78 kDa). In contrast, the p53C alone was eluted at volumes corresponding to monomeric p53 (Fig. 7C). This suggests that in the presence of TR-DNA, it is likely that p53 forms a tetramer, which could be DNA bound. The protein eluted at ~73 ml was found to contain DNA as suggested by ultraviolet (UV) spectroscopy (high-intensity absorbance at ~260 nm) and DNA band in agarose gel electrophoresis (Fig. 7D).
Further, the binding strength of nucleic acids with p53 might dictate whether p53 will remain in a tetrameric mixed or demixed state in the presence of nucleic acid. Our binding study using spectral shift assay (103) suggests strong binding of p53 with TR-DNA over NTR-DNA (Fig. 7, E and F, and fig. S16A). Therefore, stronger binding to TR-DNA dissolved the preformed condensate by forming a tetrameric DNA bound structure by specific interaction between TR-DNA and p53C. In contrast, NTR DNA might provide a template for p53 condensate formation through weak interactions.
We further hypothesized that in contrast to the monomeric/mixed state, the condensed form of p53 might have the advantage of gaining temporal stability and might work as a depot of p53 supply as required. Apart from instability and low half-life, p53 is also known to be degraded by the proteasomal system in the nucleus (67). We examined the thermal stability assay using differential scanning calorimetry (DSC) and CD spectroscopy to address whether p53C condensate formation has any stability advantage over soluble states. Estimation of melting temperature by DSC and ϴ222 by CD did not show any difference between p53C LLPS and non-LLPS state (Fig. 7G and fig. S16B). However, the p53C condensate state showed higher proteinase K (PK) resistivity, as shown in Fig. 7 (H and I), than non-LLPS p53. These data suggest that p53 condensate not only has an advantage as a ready pool for immediate action (p53 DNA binding for apoptosis, cell cycle arrest when needed) but also provides protease resistivity, countering their cellular instability.
DISCUSSION
LLPS of biomolecules (such as proteins and nucleic acids) has been implicated in driving the formation of membraneless compartments in cells, commonly designated as biomolecular condensates (5, 6, 9–12). These liquid-like condensates quickly respond to immediate environmental changes and thus perform various cellular functions (17, 104–109). Recent data suggests that the cell nucleus is not homogeneous but consists of a heterogeneous mixture of subnuclear membraneless compartments capable of performing different biological processes (33, 110, 111). These membraneless organizations include the Cajal body, nuclear speckles, nucleoli, etc. (5, 6, 16, 57). In this context, many transcription factors are known to be heterogeneously localized in the nucleus, which are indeed in a biomolecular condensate state (15, 112–115). Further, the nuclear condensates consisting of transcription factors and coactivators are shown to regulate the transcription process of key genes associated with cellular identity and functionality (15, 33, 115–118).
p53 is a tumor suppressor protein and transcription factor, generally maintaining a low-level concentration in normal cells (69, 119). However, upon stress signals such as DNA damage, p53 gets phosphorylated and translocated to the nucleus, where it is concentrated and binds to DNA. This event is responsible for many activities regulating tumor suppressor functions such as DNA repair, cell cycle arrest, or apoptosis (70, 120). Mutation of p53 not only results in the loss of tumor-suppressive function but also results in their gain of oncogenic function by inactivating WT p53 and other tumor suppressors, known as the “dominant negative effect” (45). Recently, it was suggested that p53 aggregation and amyloid formation may also be linked with tumorigenic potential in cells (48, 121). Previous studies have also indicated that p53 aggregation might be associated with p53 LLPS, where liquid-to-solid-like transition leads to amyloid fibril formation (53–55), similar to many other neurodegenerative disorders (5, 29, 31, 122). Further, p53 condensate formation is also shown to be modulated by posttranslational modifications (123). It was shown that phase separation was linked with p53 oscillation, and Ostwald ripening helps p53 condensates to repair double-stranded DNA break. Oscillation occurs to maintain cellular integrity or cellular functions, including transcription factor functions or apoptosis (124). p53 condensates, thus, can have widespread cellular functions. However, the mechanism and role of various nuclear constituents (such as RNA and DNA) on spatiotemporal regulation of p53 phase separation in cells are unknown. Further, it is unclear whether p53 condensates are functional and how their material properties might be linked with cancer pathogenesis. In this context, several studies have shown the formation of biomolecular condensates by cancer-associated proteins such as SPOP (125), BRD4 (116), and fusion oncoproteins (126, 127).
In our current work, we have investigated the condensate formation ability of p53 in mammalian cells, followed by a molecular-level investigation using an in vitro approach. We showed that once p53 gets stabilized in the cancer cells or is overexpressed in cells, p53 is heterogeneously concentrated in the nucleus (Fig. 1D), as shown for other transcription factors (4, 15, 33, 115). After overexpression in cells, initially, p53 was shown to be localized inside the nucleus as liquid-like condensates, but with time, it partially translocated to the cytoplasm and underwent LLPS, which eventually transitioned into a solid-like state. In contrast, the nuclear p53 condensates remained liquid-like (high FRAP recovery and condensate fusion) with time (Figs. 1K and 2J). The nuclear to cytoplasmic partitioning and subsequent solidification are also shown for other transcription factors, such as ARF (128). These data indicate that liquid-like p53 condensates in the nucleus might be functional (transcriptionally active). In contrast, cytoplasmic condensates, which become a solid-like state over time, could be degraded for overall cellular fitness. p53 mutations led to a solid-state transition of nuclear condensates and accelerated the cytoplasm’s liquid-to-solid state transition, suggesting a nonfunctional state of mutants. This is further supported by staining for misfolded p53, where mutant p53 condensates showed a higher degree of misfolding in the nucleus and cytoplasm than WT protein (Fig. 2, K and L). Therefore, mutations promote p53 misfolding in the condensate state, and their solid-state transition could be associated with p53 loss of function, which is shown by the loss of DNA binding (Fig. 3, D and E). In this context, it was shown previously that transcription factors also undergo a liquid-to-solid state transition, where this transition results in the loss of transcriptional activity (129).
It has been shown that transcription factors bind to various nuclear components, including nuclear RNA and/or DNA, which promote and modulate the condensate state and their transcriptional activity (4, 25, 33, 99, 130–133). Further, nuclear RNAs are also known to participate in multicomponent phase separation with various proteins for membraneless organelle formation in the nucleus, such as nucleoli and nuclear bodies, including Cajal bodies (16, 33, 134, 135). To understand the role of different nucleic acids in the formation of p53 condensate and its material properties, p53C LLPS was studied in vitro. Our data showed that p53C readily undergoes LLPS upon incubation (Fig. 4C), and with time, it transforms into a solid-like state (Fig. 4G). The LLPS and subsequent solidification of p53C condensates are further fastened by cancer-associated hotspot mutations. Both RNA and nonspecific DNA enhanced the p53C phase separation and helped maintain its liquid-like nature (Fig. 5, A to F). RNA and nonspecific DNA might template or promote the multivalent protein-protein and protein-DNA/RNA interaction for p53C phase separation (23). Further, it was shown previously that a low RNA (ribosomal RNA from Escherichia coli and Bacillus subtilis)–to–protein ratio (~1:50) promotes aggregation of p53C, while a higher ratio (>1:8) inhibits this process. Our findings resonate with these observations, as we noted that RNA facilitates the assembly of p53C (136) and, therefore, LLPS. These data are consistent with our in cellulo data of p53 condensates. The presence of specific DNA not only inhibits the p53 demixing, but also dissolves the preformed p53 condensates (formed either by p53C alone or in the presence of DNA and/or RNA) (Fig. 6). Previous studies also suggest that nucleic acids modulate the formation of multicomponent LLPS and material property of the condensates (23, 113, 137). Our data thus suggest that upon entry into the nucleus, p53 readily undergoes LLPS into a microphase-separated condensate state (83, 138) in the presence of nucleic acid, which might act as a ready pool of functional p53. This process might help p53 stay in the vicinity of specific DNA (upon exposure), which may get dissolved into a soluble state upon binding to DNA and forming DNA-bound tetrameric structure (transcriptionally active form) for performing the tumor suppressor/transcriptional activity. This dissolution upon contact with specific DNA could also be linked with the wetting phenomenon of p53 condensates on DNA (99) (Fig. 6A). This is consistent with the previous study that the binding of p53C to its consensus DNA sequence enhances the conformational stability of p53, while nonspecific DNA does not influence this stability (139).
Further, the exposure of p53 target genes might also direct the movement of p53 condensates, where p53 becomes a mixed state for attaining higher thermodynamic stability. In this direction, it was shown that droplets containing urease move toward the gradient of urea, resulting in a mixed state (140). Further, once the p53 load increases in the nucleus, some nuclear p53 population is transported into the cytoplasm through the nuclear pore complex, where it undergoes LLPS and subsequent liquid-to-solid transition with time. The MDM2 colocalization with cytoplasmic p53 condensates suggests that some of these cytoplasmic condensates might eventually be degraded by proteasomal degradation machinery or could also attain an amyloid state upon exposure to GAGs and other amyloid-promoting factors.
Further, the p53 phase separation not only enhances the protein’s stability against its degradation but also increases the local concentration of p53, which may increase the availability of the protein to function as a transcription factor. The LLPS of p53 and its misregulated transition to a solid-like state followed by misfolding might result in the loss of tumor-suppressive and gain of oncogenic function, which may be linked to cancer.
MATERIALS AND METHODS
Chemicals and reagents
The chemicals and reagents used for experimental purposes were mainly procured from Sigma-Aldrich (St. Louis, MO, USA) or Merck (Darmstadt, Germany) unless mentioned otherwise. The water was double distilled and deionized by a Milli-Q water purification system (Millipore Corp., USA).
In silico analysis of p53 protein
Full-length p53 and the p53C sequence were used for in silico analysis. The disordered regions of the primary structure of the protein were identified using the Online tool IUPred2A (94, 95) and the droplet-promoting region was predicted using Fuzpred (64). SMART (141) (Simple Modular Architecture Research Tool) was used to identify low-complexity regions (LCRs) along the protein sequence. Output data were plotted using OriginPro 2021 (Origin Lab, USA) software. The secondary structure of p53 and p53C was obtained from Alphafold (65) and Protein Data Bank (93) (PDB ID: 6OCJ). The output PDB structures were represented using PyMOL (142).
Site-directed mutagenesis
Plasmids for expression of GFP-p53 WT, GFP-p53 NES−, and p53C WT [p53C WT (residues 94 to 312)] were obtained from Addgene (Addgene ID nos. 12091, 12092, and 24866, respectively). The mammalian and bacterial expression constructs for p53 mutants were constructed via polymerase chain reaction (PCR)–mediated site-directed mutagenesis. The plasmid containing GFP-p53 WT was used as the template for two mammalian expression constructs of p53 mutants GFP-p53 R175H and GFP-p53 R248Q. For bacterial expression constructs containing mutant p53C, namely, p53C R175H and p53C R248Q, p53C WT (residues 94 to 312) was used as the template. Primers were designed against the template by altering the codon at the mutation site such that the mutation site lies at the primer’s center (table S1). Pfu DNA Polymerase (SRL Ltd., India) was used for PCR. After 20 thermal cycles, the PCR product was selected via Dpn1 (Invitrogen, USA) digestion. The digested PCR product transformed XL10 (Gold) competent cells, and colonies containing the construct were selected using relevant antibiotic resistance (kanamycin for GFP-p53 variants and ampicillin for p53C variants) in the Luria-Agar medium. The colonies were thereby grown in Luria broth (LB) media. Plasmids were isolated by a mini-prep kit (Favorgen Biotech Corp., Taiwan) and quantified using a NanoDrop UV photometer (Implen, USA). The newly generated plasmids were verified by sequencing analysis.
Cell lines and transfection studies
MDA-MB-231 (human breast epithelial adenocarcinoma) cells, MCF7 (human breast epithelial adenocarcinoma) cells, HeLa cells (human cervical epithelial adenocarcinoma), MCF10A cells (human mammary gland epithelial cells), and SaOS2 (human bone epithelial osteosarcoma) cells were obtained from the National Centre for Cell Science (NCCS, Pune, India). MCF10A, MDA-MB-231, and MCF7 cells were seeded on coverslips, allowed to grow until 80% confluency, and fixed for immunostaining. For transfection studies, cells were seeded on 35-mm confocal dishes and allowed to grow to 70% (HeLa) and 90% (SaOS2) confluency. Hereafter, the cells were transfected with 1 μg of DNA using Lipofectamine and P3000 following the manufacturer’s protocol (L3000-008, Invitrogen, USA). HeLa cells were cultured in DMEM containing 10% fetal bovine serum (FBS, Gibco, USA), while SaOS2 cells were cultured in McCoy’s 5A Medium (catalog no. M4892, Sigma-Aldrich) with 15% FBS. The cells were maintained at 37°C with 5% CO2. For microscopic studies, cisplatin (catalog no. P4394, Sigma-Aldrich) and LMB (catalog no. L2913, Sigma-Aldrich, USA; 20 ng/ml) were added after 12 hours after transfection at concentrations of 10 μM and 20 ng/ml, respectively.
Immunostaining of cells
MCF10A, MCF7, and MDA-MB-231 cells were first seeded on 12-mm coverslips. At 80% confluency, cells were fixed with 4% paraformaldehyde (PFA) solution for 20 min at 37°C. Fixed cells were hereby permeabilized using 0.2% Triton X-100 in phosphate-buffered saline (PBS) for 15 min, and nonspecific antigen sites were blocked with 2% bovine serum albumin (BSA) for another 1 hour. Cells were then stained with mouse monoclonal anti-human p53 (DO-1) (catalog no. sc-126, Santa Cruz Biotechnology, USA) antibody (1:200) overnight at 4°C. The coverslips were then washed with PBST (0.1% Tween 20 in PBS). The coverslips were incubated with Alexa Fluor 555 (catalog no. A32727, Invitrogen, USA)– or Alexa Fluor 488 (catalog no. A11001, Invitrogen, USA)–conjugated goat anti-mouse secondary antibody diluted in PBST (1:500) for 2 hours at 37°C for MDA-MB-231 and MCF10A cells and (1:250) for 3 hours at 37°C for MCF7 cells. Cells were washed with PBST and 4′,6-diamidino-2-phenylindole (DAPI) (1 μg/ml) was added for 2 min for nuclear stain. Cells were then washed with PBS and mounted on a glass slide with mounting medium containing 1% (w/v) 1,4-diazabicyclo [2.2.2] octane and 90% (v/v) glycerol in PBS. Images were acquired using a laser scanning confocal microscope (Zeiss LSM 780, Carl Zeiss, Germany) with iPlan-apochromat 100×/1.4 NA (numerical aperture) objective.
For SaOS2, cells were seeded on 12-mm coverslips and transfected with GFP-p53 WT, R175H, and R248Q and fixed at early (18 hours) and late (48 hours) time points with ice-cold 4% PFA in PBS for 5 min at 37°C for immunostaining of misfolded p53 and amyloid fibril. Using a similar protocol as mentioned before, cells were stained with mouse monoclonal anti-human Pab240 antibody (1:500) (catalog no. sc-99, Santa Cruz Biotechnology, USA) followed by Alexa Fluor 555–conjugated anti-mouse secondary antibody (1:500) and OC antibody (1:500) (catalog no. AB2286, Sigma-Aldrich) followed by Alexa Fluor 555–conjugated anti-rabbit secondary antibody (catalog no. A32732, Invitrogen) (1:1000). Images were acquired using a Spinning Disk confocal microscope (Zeiss Axio-Observer Z1 with Nipkow spinning disc), using iPlan-apochromat 63×/1.4 NA objective. For A11 staining, SaOS2 cells were seeded as mentioned previously and transfected with GFP-p53 WT and fixed at 24 and 48 hours. The staining was done using anti-rabbit A11 antibody (1:500) (catalog no. AB9234, Sigma-Aldrich) followed by Alexa Fluor 555 (catalog no. A32732, Invitrogen)–conjugated anti-rabbit secondary antibody (1:1000). For MDM2 staining, cells were stained with anti-human MDM2 antibody (1:200) (catalog no. 86934, Cell Signaling Technology) followed by Alexa Fluor 555 (catalog no. A32732, Invitrogen)–conjugated anti-rabbit secondary antibody (1:500). Images were acquired using an Olympus FV3000 laser scanning confocal microscope with 100× objective. Further, SaOS2 cells were transfected with GFP-p53 WT, as described previously. After transfection for 18 and 48 hours, cells were stained with MitoTracker Red (catalog no. M7513, Thermo Fisher Scientific, USA) and LysoTracker Red DND-99 (catalog no. L7528, Thermo Fisher Scientific) as per the manufacturer’s protocol. For the Nile red staining, Nile red (10 μg/ml) was added to cells at 18 and 48 hours after transfection and incubated for 3 min. The cells were washed using PBS after incubation and Opti-MEM (catalog no. 11058021, Gibco) was added before capturing the images. For the ProteoStat binding assay, 0.5 μl of ProteoStat solution (5 μM) was diluted in 1 ml of assay buffer provided in the kit (Enzo Life Sciences, USA). The solution was added to the fixed cells (at 18 and 48 hours after transfection) and incubated for 30 min at room temperature as per the manufacturer’s protocol. The cells were washed using PBS and Opti-MEM was added before imaging. All the images were acquired using a Zeiss laser scanning confocal microscope (LSM 780, Carl Zeiss, Germany) with iPlan-apochromat 100×/1.4 oil objective. The images were processed using FIJI software.
Time-lapse confocal microscopy imaging
Live cell imaging was done for time-dependent studies after transfection of GFP-p53 with HeLa and SaOS2 cells. The time-dependent cellular imaging was done at 12, 18, 24, 36, and 48 hours (512 × 512, 8 bit) and images were captured using a Spinning Disk confocal microscope (Zeiss Axio-Observer Z1 with Nipkow spinning disc) using 63×/1.4 NA (oil) or 100×/1.4 NA (oil) objectives. Opti-MEM (catalog no. 11058021, Gibco) was used as media for all live cell imaging and incubation was maintained at 5% CO2 and 37°C. For nuclear stain, Hoechst dye (catalog no. 33342, Enzo, USA) was used. For in vitro characterization of p53C LLPS, NHS-Rhodamine–labeled p53C was used for imaging at 37°C. The excitation source of 488-nm solid-state laser was used for GFP-p53 and Atto 488–conjugated DNA and 561-nm solid-state laser was used for NHS-Rhodamine–labeled p53C. Images and movies were processed using Zen Blue software (Zen Lite 2012).
Western blotting
At various time points after transfection, HeLa and SaOS2 cells were suspended in 1× Laemmli sample buffer (62.5 mM tris, pH 6.8, 2% SDS, and 10% glycerol) for protein extraction. The suspension was centrifuged at 20,817g at 20°C for 1 hour, and the supernatant was collected for protein estimation using a BCA kit (Pierce BCA Protein Assay Kit, catalog no. 23227, Thermo Fisher Scientific, USA). Further, 40 μg of protein was loaded for separation by SDS-PAGE electrophoresis and blotted onto a 0.22-μm polyvinylidene difluoride (PVDF) membrane (catalog no. BSP0161, Pall Life Sciences). After transfer, the PVDF membrane was blocked with 5% BSA in TBST for 1 hour at room temperature. The blot was probed with the respective primary antibodies made in 2% BSA in TBST with 1:2000 dilution and incubated overnight at 4°C. The PVDF membrane was washed with TBST and the blot was incubated with the horseradish peroxidase (HRP)–conjugated secondary antibody (1:2000 dilution prepared in 2% BSA in TBST) and incubated at room temperature for 1 hour. The PVDF membrane was then washed with TBST and signals were detected using a Clarity Western ECL detection kit (catalog no. 1705060, Bio-Rad) according to the manufacturer’s instructions. The images were captured using the Image Quant LAS 500 (GE Life Sciences, USA). The captured images were quantified using the Bio-Rad Image lab software. The antibodies used for Western blotting experiments are anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (catalog no. sc-365062, Santa Cruz Biotechnology, USA), anti-p53 (catalog no. MA5-16387, Invitrogen, USA), anti-p21 (catalog no. 2947, Cell Signaling Technology, USA), and anti-histone H3 (D1H2) (catalog no. 4499, Cell Signaling Technology). The secondary antibodies used were goat anti-mouse immunoglobulin G (IgG), H & L chain-specific peroxidase conjugate (catalog no. 401253, Merck, USA) and goat anti-rabbit IgG, H & L chain-specific peroxidase conjugate (catalog no. 401353, Merck, USA).
FRAP experiments
FRAP studies were done using a laser scanning confocal microscope (Zeiss LSM 780 Axio-Observer Z1 microscope). HeLa cells and SaOS2 were seeded on 30-mm glass-bottom confocal dishes (Genetix, India) and transfected with plasmids containing mammalian expression vectors with GFP-p53 WT, R175H, and R248Q, separately. At various time points, the dish was mounted on the microscope with an incubation temperature 37°C, 5% CO2 and under optimal humidity conditions during imaging. At each time point (18 and 48 hours), photobleaching of nuclear and cytoplasmic condensates was done using a 488-nm laser (100% laser power). Along with the region of interest (ROI) for photobleaching, two more ROIs were selected within and outside condensates, respectively, for bleaching correction. Time-lapse images were taken with laser excitation at 488 nm and emission was recorded at 534 nm. Recovery was monitored until there was no signal strength change with time, indicating the limit of recovery of the bleached region. For FRAP studies of nuclear condensates, due to small size and high dynamicity, translational correction of the images was done post-acquisition using the FIJI linear stack alignment tool (143). For this, new ROIs were selected in the aligned stacked image, and the intensity of the whole stack for each ROI was recorded and processed to calculate fluorescence recovery in a similar method as stated before.
For FRAP studies, labeled p53C protein (10% v/v, NHS-Rhodamine labeled to unlabeled protein) was used. Protein labeling was done as per previously published protocols (47). At different time intervals, photobleaching of the droplets was done using a 561-nm DPSS 561-10 laser (100% laser power). For every bleaching, 2 unbleached ROIs of the same diameter inside and outside of the droplets were taken as controls for bleaching correction. After bleaching, the recovery was monitored until a plateau in the signal was reached. For the FRAP, iPlan-apochromat 100×/1.40 (oil) objective with GaAsP detector was used for cytoplasmic, nuclear, and in vitro p53C condensates. The intensity profiles were recorded using Zen Pro (Zeiss, Germany) software with 8-bit depth. A similar protocol was used for the FRAP study of condensate formed in the presence of CSA. For both in cellulo and in vitro FRAP studies, the data were normalized after bleach correction and fitted in Origin Pro 2021 (Origin Lab, USA) software using exponential growth function for calculation of half-time (t1/2) as per the protocol mentioned in Ray et al. (144)
STED imaging of p53 condensates
Super-resolution studies of p53 nuclear and cytoplasmic condensates were done using STED microscopy (72, 73) (Leica Microsystems, Germany). For cells, MDA-MB-231 and MCF7 cells were seeded, fixed on coverslips, and immunostained as described in the previous section. After staining, the cells were mounted on glass slides with Mowiol mounting medium [10% (w/v) Mowiol 4-88, 25% (v/v) glycerol in 100 mM tris-HCl (pH 8.5)]. For GFP-p53 variants, SaOS2 cells were seeded and transfected with GFP-p53 WT, R175H, and R148Q plasmids and fixed at early (18 hours) and late (48 hours) time points with ice-cold 4% PFA in PBS for 5 min. For stressor treatment, 10 μM cisplatin (catalog no. P4394, Sigma-Aldrich) was added at an early time point (18 hours) and fixed after 3 hours. For 1,6-hexanediol and sorbitol treatment, SaOS2 cells were seeded and transfected with GFP-p53 WT plasmid. After 24 hours, cells were treated with 10% 1,6-hexanediol (catalog no. 240117, Sigma-Aldrich) or 0.5 M sorbitol (catalog no. S1876, Sigma-Aldrich) for 30 min followed by fixing using ice-cold 4% PFA in PBS for 5 min. For all the cells, coverslips containing cells were mounted on a glass slide with Mowiol mounting medium. Images were captured with a STED microscope (Leica Microsystems, Germany), and after acquisition, images were deconvoluted using the Classical Maximum Likelihood Estimation (CMLE) algorithm (145) in Huygens Professional Suite. Rendering of microscopic images for the visualization of condensates was carried out using IMARIS 8.3.1 (Bitplane, Switzerland). For quantification of the percentage of p53 condensate occupancy with the total pool of p53 in the nucleus, the thresholding tool (146, 147) of FIJI software was used. The whole area of the nucleus was first selected before thresholding to limit the area of estimation. The intensity thresholding tool of p53 was used to estimate the total p53 in the nuclear region, and the local thresholding tool (148, 149) was used to identify the portion of nuclear condensates that had higher local intensity relative to its surroundings due to clustering. The percentage of nuclear occupancy was calculated as % = (area of the region above local threshold/area of the region above threshold) × 100. Trainable Weka Segmentation tool (150) of FIJI was used for estimating the area of the nuclear condensates. Each image was trained using 50 regions each for background (region 1) and 50 for condensates (region 2). The segmentation classifier was then run and the resultant segmented image was obtained, defining the clusters and background. Thresholding was done on this segmented image to obtain the area, and the Feret diameter was calculated for each condensate. For the high irregularity of the shape of the nuclear clusters, the area was taken as a measure of the size of the condensates. Considering the lowest possible resolution of images obtained by STED to be 50 nm, areas above 2500 nm2 were counted for estimation. The area distribution plots for each pair for each variant (GFP-p53 WT, R175H, and R248Q) were pairwise normalized before plotting to account for the different sizes of the nucleus. The data were plotted using MATLAB (MathWorks) 2019a software.
Lattice light-sheet microscopy of p53 condensates
Lattice light-sheet microscopy (LLSM) was done to examine nuclear condensates in live SaOS2 cells. For this, the cells were seeded on 5-mm coverslips and allowed to grow to ~80% confluency. Hereafter, cells were transfected with WT GFP-p53 plasmid and imaged at 18 and 48 hours after transfection using a 3i Lattice light-sheet microscope (3i, Colorado, USA). Volumetric image stacks were generated using a square lattice in dithered mode. A square lattice was made for the 488-nm channel with 51 beams spaced at 0.99-μm intervals between the beams, and the pattern was cropped with a cropping factor of 0.150. The deskewed step size was 0.163 μm after scanning the sample stage in 0.3-μm steps. The excitation laser power was adjusted between 0.14 and 0.43 mW at the image plane to obtain a good signal-to-noise ratio. In the emission path, a custom-made 488-640T/560 R dichroic beam splitter was used. A quad-notch filter (Semrock FF01-446/523/600/677) was added to the transmitted path to collect the emitted signal. The final fluorescence signal was imaged on a Hamamatsu ORCA-Fusion Bt sCMOS camera. Each image plane was captured with a 10-ms exposure time. The frame size of the image in X, Y, and Z directions was between 20 and 50 μm, 20 μm, and 140 to 200 planes (23 to 30 μm), respectively, to ensure the entire nuclear region was imaged while avoiding any strong fluorescence signal near the edges of the frame. Hence, a volumetric image stack of 50 × 20 × 25 μm was captured within ~ 2.5 s (100 planes for 1.2 s).
After acquisition, the images were cropped from the raw datasets. Because the images were captured at an angle of approximately 32° using LLSM, the images were deskewed and corrected for coverslip orientation in the Slidebook software. Photobleach correction was applied when necessary. The images were further deconvoluted using the Richardson Lucy Constraint Iterative (CI) algorithm with 10 iterations using the theoretical point spread function and a Gaussian noise smoothening radius of 0.3. Final images were processed in FIJI for representative purposes.
Apoptosis studies using flow cytometry
For studying the time-dependent (6, 12, 18, 24, 36, and 48 hours) apoptotic cell death upon WT p53 transfection cells, SaOS2 (7 × 105 cells per well) were seeded in six-well plates (Corning, USA). After 24 hours, cells were transfected with GFP-p53 WT plasmid in a time-dependent manner, i.e., 48, 36, 24, 18, 12, and 6 hours before the acquisition using flow cytometry. The cells were trypsinized and collected by centrifugation at 664g for 4 min at 25°C. The cell pellets were washed with ice-cold PBS and centrifuged at 664g. The cell pellets were suspended in 100 μl of 1× binding buffer followed by doubling staining with V450 annexin V and PI (catalog no. 560506, BD Horizon) and incubated for 15 to 20 min at room temperature. The samples were diluted to 400 μl with the 1× binding buffer and quantified by flow cytometry (FACS Aria, BD Biosciences). The gating was performed using untransfected cells and ~10,000 GFP-p53 WT positive cells were counted for each sample. The values are normalized using untransfected control cells. The recorded data were plotted using FlowJo v10 software (Tree Star Inc.).
ChIP-qPCR assay
ChIP-qPCR assay was performed to determine the functional status of the p53 protein. For this, SaOS2 cells were transfected with WT p53 and mutant p53 (R175H and R248Q) as per the described protocol (151). The cells were collected at 18 hours and chromatin was cross-linked using 1% formaldehyde for 30 min and added directly to the media. Further, the cross-linking was quenched using 125 mM glycine for 5 min. To attain higher degree of resolution during detection, the samples were sonicated to obtain DNA fragments that are ~200 to 1000 bp in size. Sonication was done at 30% amplitude for a total of three cycles with 1 min on and 1 min off on ice. The samples were centrifuged at 10,000g at 4°C for 10 min to remove insoluble material. The cells were incubated overnight using 5 μg (25 μl) of p53 antibody (DO-1) as described earlier (151). The chromatin-bound DNA was eluted with 50 μl of elution buffer using the Magna ChIP G Tissue Kit (Millipore, USA) as per the manufacturer’s instructions. A QIAquick PCR purification kit (QIAGEN, Valencia, USA) was used for the purification of all DNA samples (IPs and inputs) in accordance with the manufacturer’s protocol. Further, qPCR was performed with SYBR Green reaction mixture (20 μl) using an Agilent qPCR machine. Both ChIP and qPCR experiments were performed in duplicate. Calculations for enrichment/input values were made using Eq. 1
| (1) |
where E represents the specific primer efficiency value and % Enrichment/Input = E. The sequence of primers for the p53 response element for p21 was as follows:
Forward primer: 5′-GTGGCTCTGATTGGCTTTCTG-3′
Reverse primer: 5′-CTGAAAACAGGCAGCCCAAG-3′
p53 DNA binding assay in cells
An ELISA for p53 was performed to determine the ability of p53 to bind to the DNA element. For this, SaOS2 cells were transfected with GFP-p53 WT, R175H, and R248Q. The cells were collected at 18 hours after transfection. The cells were harvested and lysed using radioimmunoprecipitation assay (RIPA) buffer supplemented with a Roche protease inhibitor cocktail. The lysate was centrifuged at 4°C to remove the cell debris. The supernatant obtained was added onto the 96-well plate immobilized with double-stranded DNA sequence containing p53 response element using the p53 Transcription Factor Assay kit (catalog no. 600020, Cayman Chemical, Michigan) as per the manufacturer’s protocol. p53 present in the cell lysate binds specifically to the p53 response element and is detected by adding a p53 antibody (DO-1). Anti-mouse secondary antibody conjugated to HRP was added to provide a sensitive colorimetric readout at 450 nm. Nutlin-3–stimulated MCF7 nuclear extract was used as a positive control for the assay.
RNA isolation and qRT-PCR
SaOS2 cells were seeded on 6-well plates and transfected with GFP-p53 WT. For stressor treatment, 10 μM cisplatin was added at 9 hours after transfection. The cells were trypsinized for both treated and untreated cases and pelleted down at 18 hours. The RNA isolation was carried out using TRizol reagent (catalog no. 15596018, Ambion by Life Technologies, USA) according to the manufacturer’s protocol. Briefly, the harvested cell pellets were lysed in 1 ml of TRIzol reagent, followed by the addition of 350 μl of chloroform. The tubes were vortexed vigorously and kept in ice for 5 min. The tubes containing lysed samples were centrifuged at 12,000g for 15 min at 4°C. The upper aqueous phase containing RNA was carefully transferred into new tubes, and half a volume of isopropyl alcohol was used for the precipitation of RNA. The samples were mixed and incubated for 30 min at room temperature and centrifuged at 12,000g for 10 min at 4°C. The white gel-like RNA pellet was collected and washed with 1 ml of ethanol (70%) and centrifuged at 7500g for 5 min at 4°C. The supernatant was discarded and the RNA pellet was allowed to air dry and resuspended in nuclease-free water. The samples were digested with DNase I to remove the DNA contamination from the isolated RNA sample as per the manufacturer’s instruction (Turbo DNA-free kit, catalog no. AM-1907, Thermo Fisher Scientific, USA). The concentration of the purified RNA was measured using a nanodrop spectrophotometer (Implen, USA). Hereafter, cDNA was synthesized from total RNA using the RevertAid First Strand cDNA Synthesis Kit (catalog no. K1622, Molecular Biology, Thermo Fisher Scientific, USA) as per the manufacturer’s instruction. Oligo (dT) primers were used for the reverse transcriptase reaction. Real-time PCR (qRT-PCR) was further carried out on a CFX96 Touch Real-Time PCR detection system (Bio-Rad, USA) using the SYBR Green method. Maxima SYBR Green/ROX qPCR Master Mix (2×) (catalog no. K0221, Thermo Fisher Scientific, USA) was used according to the manufacturer’s protocol with the predesigned primers for the required genes (table S3). Two independent experiments in duplicates were performed for each reaction.
p53C protein expression and purification
Plasmid (pet15b vector carrying 6×His-tagged 94 to 312 amino acid gene sequence) for p53C (WT, R175H, and R248Q) was transformed into BL21(DE3) competent cells using the standard protocol. Transformed cells were selected based on ampicillin resistance. A single colony was inoculated in 100 ml of LB and grown to an OD600 (optical density at 600 nm) of 0.5 as a starter culture. Hereafter, 20 ml of starter culture was added to 1 liter of LB with antibiotic and allowed to grow until an OD600 of 0.7 is reached. The protein expression of WT, R175H, and R248Q was induced by 1, 0.25, and 1 mM isopropyl β-d-1-thiogalactopyranoside, respectively, for 12 hours at 18°C. The cells were then harvested and resuspended in lysis buffer (50 mM sodium phosphate, 300 mM sodium chloride) and 1× protease inhibitor cocktail (catalog no. 05056489001, Roche cOmplete, Mini, EDTA-free, Roche Diagnostics, Germany) was added to prevent proteolysis. The cell suspension was then sonicated using a probe sonicator (Sonics & Materials Inc., pulse of 2 s on, 2 s off; 40% amplitude per cycle, for five cycles). The soluble fraction was collected and passed through the column of the Qiagen Nickel NTA (catalog no. 1018244, Qiagen, Germany). The column was washed with lysis buffer containing 50 mM imidazole (4 column volumes) and protein fractions were eluted against gradient concentration of imidazole from 100 to 500 mM. Each gradient was run on SDS-PAGE to identify the fraction containing the maximum yield of the protein. Hereafter, the protein was passed through SEC (Hi Load 16/60, Superdex 200 TM 10/300, catalog no. 17-1069-01, Cytiva Life sciences) column preequilibrated with 2 column volumes of 50 mM sodium phosphate buffer (PB) (pH 7.4, 0.01% sodium azide) for further purification. The eluent protein was then further used for labeling and experimental purposes.
The protein concentration was measured using UV spectroscopy following Beer-Lambert’s law. The absorbance was recorded at 280 nm and concentration was estimated considering the molar absorptivity constant as 17,420 M−1 cm−1.
For experimental studies involving labeled p53C, the protein was labeled with NHS-Rhodamine (catalog no. 46406, Thermo Fisher Scientific, USA), using the manufacturer’s instructions in 50 mM PB. A fivefold molar excess of NHS-Rhodamine was added to p53C and incubated at 4°C for 3 hours with constant stirring. Hereafter, the excess dye in the labeled protein mixture was then dialyzed out using a 12.4-kDa cutoff membrane (catalog no. D0655, Sigma-Aldrich, USA) in 50 mM PB. The efficiency of labeling was verified by calculating the degree of labeling before proceeding with the experiment as per the manufacturer’s protocol. The concentration of the labeled protein was determined using spectroscopic methods as per instructions.
In vitro p53C phase separation studies
SEC-purified WT, R175H, and R248Q p53C were studied for phase separation at varied concentrations in the presence of various concentrations of molecular crowder PEG-8000 (0, 5, 10, 15, and 20%) in 50 mM PB, pH 7.4, to construct the phase regime. For microscopy-based studies, 10% (v/v) NHS-Rhodamine–labeled protein was used. Different concentrations of protein in the presence and absence of PEG-8000 were prepared in 50 mM PB (pH 7.4). Glass coverslips (no. 1) and glass slides were thoroughly washed with 1% Hellmanex solution and dried, and 10 μl of the reaction mixture was spotted on the coverslips. The coverslips were mounted on the glass slides and were sealed properly using transparent nail paint and incubated at 37°C in a hydrated chamber for time-dependent studies. Hereafter, at regular time intervals, the samples were visualized for condensate formation under the Zeiss Spinning Disk confocal microscope using 63×/1.40 oil objective. For p53C studies along with nonlabeled DNA, Atto labeled target (TR) and nontarget (NTR) DNA (Sigma-Aldrich, USA) was used. Labeled DNA [10% (v/v)] was used in all the experiments. The imaging of LLPS of p53C samples in the presence of unlabeled DNA and RNA was captured using Zeiss laser scanning confocal microscope under 63×/1.40 oil or 100×/1.40 oil objective. Quantification of size and number of condensates was done using the Thresholding tool (148) of FIJI and plotted using KaleidaGraph (v 4.03) and GraphPad Prism 8 software.
Light Scattering(LS) studies
LS studies were performed for 10 μM p53C in 50 mM PB (pH 7.4) in the presence of PEG-8000 (10% w/v) as crowder at 37°C. The experiments were performed in continuous mode, with a 1-min time interval using JASCO FP8500 (USA) spectrofluorimeter, with an excitation and emission wavelength of 350 nm and 5 nm of slit width. One hundred twenty microliters of p53C WT, R175H, and R248Q samples were loaded into a quartz cuvette and scattering was recorded immediately. Each experiment was performed twice. For studying the effect of DNA on the LLPS of p53C, 10 μM TR-DNA, 10 μM NTR-DNA, and ~20 nM (30 μg/ml) RNA were added to 10 μM p53C in 50 mM PB, pH 7.4, in the presence of 10% (w/v) PEG-8000 and scattering profile was recorded. Similarly, to study the effect of DNA and RNA on preformed p53C condensates, first, scattering reactions were set up for WT p53C in the presence of 10% (w/v) PEG-8000. Hereafter, immediately on attaining saturation in the scattering profile, TR-DNA, NTR-DNA, and RNA were added to a final concentration of 10 μM, 10 μM, and 30 μg/ml, respectively, and the scattering profile was further recorded after addition in the same settings. The data were plotted using Origin Pro 8 software (Origin Lab, USA).
FTIR spectroscopy
FTIR spectroscopic studies were done to elucidate the secondary structure of the WT p53C in the dilute and dense phase after phase separation. Initially, 500 μl of 20 μM protein was taken and that was allowed to phase separate in 50 mM PB in the presence of 10% PEG-8000 for 1 hour. The formation of condensates was verified after observation under the microscope. Hereafter, the protein sample was centrifuged at high speed (20,000g) for separation of the dilute phase and dense phase (152). Five microliters of dilute phase (supernatant) and dense phase (pellet) were spotted on separate thin KBr pellets and were subjected to dry under an infrared (IR) lamp. Then, the spectrum was obtained in the amide I stretching frequency in a range of 1800 to 1500 cm−1, with a resolution of 4 cm−1, by using a Bruker VERTEX 80 spectrometer (Bruker, Leipzig, Germany) attached to a DTGS detector. The recorded spectrum was baseline corrected and then deconvoluted using the Fourier Self Deconvolution (FSD) method at the frequency range of 1700 to 1600 cm−1. The Lorentzian curve fitting method was used to fit the spectrum using OPUS-65 software (Bruker, Germany) according to the manufacturer’s instructions. Curve fitting and the area under the curve were determined and the frequencies were noted. Three independent experiments were performed for each sample. The data were plotted using KaleidaGraph (v 4.03) software.
Transmission electron microscopy
WT R175H and R248Q p53C samples (10 μM) in 50 mM PB (pH 7.4, in the presence of 10% PEG-8000) were set up for the formation of condensates on coverslips mounted on glass slides. After the formation of condensates (confirmed via observation under the microscope), the samples from the coverslip were transferred to a copper formvar EM grid (Electron Microscopy Sciences, USA) and incubated for 5 min. The grids were subjected to staining with uranyl formate (1% w/v) for 5 min. The excess liquid on the grid was dried using filter paper and the sample was set to air dry. Before imaging, the samples were further dried using an IR lamp. TEM sample preparation and image acquisition were done for condensates formed at 0 hours (immediately after formation) and 12 hours. A similar protocol was used for TEM imaging of condensate formed in the presence of CSA at different time points. Image acquisition was done using a 200-kV transmission electron microscope (JEOL JEM 2100F, Japan) with 10,000× magnification. The images were recorded digitally using the Gatan microscopy suite (Gatan, USA).
ThT and CR binding assay
Two hundred microliters of 10 μM p53C was set to phase separate in the presence and absence of equimolar concentration of CSA in 50 mM PB (pH 7.4) in 10% PEG-8000. ThT (1 mM) was prepared in 20 mM tris-HCl buffer, pH 8.0, containing 0.01% sodium azide. Then, 2 μl of ThT was added to 200 μl of protein sample. Fluorescence was measured with a cuvette (Hellma; volume, 500 μl; path length, 10 mm) using a Spectrofluorometer (JASCO FB 8500, USA) with excitation wavelength at 450 nm and emission wavelength was recorded from 460 to 500 nm. A slit width of 5 nm was used for both emission and excitation. For both time points, i.e., at 0 and 12 hours, the fluorescence obtained at 480 nm was plotted as a function of incubation time. α-Synuclein fibril was taken as positive control and freshly purified p53C was taken as negative control.
For the CR binding assay, 100 μM CR was dissolved in 50 mM PB containing 10% ethanol. p53C condensates were formed in the presence of 10% PEG-8000. CR solution (5 μl) was mixed with 95 μl of LLPS reaction mixture at 0 and 12 hours. For CR binding, absorbance was measured from 300 to 700 nm. Five microliters of CR with 95 μl of 50 mM PB was taken as control. The data were plotted using GraphPad Prism 8 software.
ThioS staining
For the ThioS staining assay, 0.0625% (w/v) ThioS was prepared in 50 mM sodium phosphate buffer (pH 7.4). p53C protein sample (10 μM) in the presence of 10% (w/v) PEG-8000 and equimolar concentration of CSA was prepared in 50 mM PB, pH 7.4. ThioS (2 μl) was added to 20 μl of the LLPS reaction mixtures. The reaction mixture was spotted on the slides and sandwiched using 12-mm glass coverslips. The coverslips were sealed using nail paint available commercially. The confocal microscopy imaging was done at different time points (0, 6, and 12 hours) using an Olympus FV3000 laser scanning confocal microscope (100×/1.4 NA oil immersion objective). The images were analyzed using ImageJ (NIH, Bethesda, USA) software.
Effect of DNA on p53C condensate formation
Microscopy-based studies were conducted to understand the effect of DNA on p53C condensate formation. For this, 10 μl of 100 μM TR-DNA and NTR-DNA was separately spin coated at the center of a rectangular cover glass and 12-mm glass coverslips, using photoresist spinner (PRS-6K, Ducom Ltd.) at 956g for 20 s. The cover glass that was not coated with DNA was taken as a control. After drying the sample for 10 min, the coverslip was mounted on a Zeiss laser scanning confocal microscope [LSM 780 Zeiss Axio-Observer Z1 microscope (inverted)] using 100×/1.4 NA oil immersion objective. In addition, 10 μM WT p53C was incubated with 10% PEG-8000 in 50 mM PB, pH 7.4, to form condensates, which was confirmed using a microscope. Freshly preformed condensates (10 μl) were spotted at the center of the coating on the rectangular coverslip mounted on the microscope. The 12-mm coated coverslips with the same DNA on the corresponding rectangular coverslip were mounted over the sample to avoid evaporation. Preformed condensates were also spotted on uncoated cover glass and covered with circular uncoated coverslips, taken as control. For condensates spotted on each case (TR-DNA–coated, NTR-DNA–coated, and uncoated surfaces), images were acquired using a laser scanning confocal microscope. For each of these conditions, the total number of condensates was calculated using the thresholding tool of FIJI and plotted in GraphPad Prism 8 software.
In another experimental study, we studied the effect of DNA on the extent of partitioning of p53. Here, rubber gaskets (Silicone isolators, catalog no. 666505, Grace Bio-Labs, USA) were mounted over rectangular glass coverslips to create a chamber for holding the LLPS solution. Sixty microliters of 20 μM protein and PEG-8000 (10% w/v) solution was added to two chambers of the gasket and the solution was allowed to phase separate. Once condensates were formed, as observed under the microscope, images were acquired using a laser scanning confocal microscope and the coordinates of the specific positions were marked. Hereafter, TR-DNA and NTR-DNA were added separately to each of the chambers to an equimolar concentration. Images were acquired for the same positions after the addition of DNA, using the same laser settings for the same set of positions. This enabled comparison at a single droplet stage to elucidate the effect of DNA. The extent of partitioning was calculated as the ratio of the intensity of the dense phase to the intensity of the liquid phase using FIJI software. Average fluorescence intensity from ROI of identical size, inside and outside the condensate, was measured, and their ratio was calculated as a measure of the extent of partitioning. The images were acquired in a laser scanning confocal microscope using 100× oil objectives.
Dynamic light scattering
Dynamic light scattering (DLS) study was performed to determine the particle size for p53C and p53C condensate in the presence of an equimolar concentration of TR-DNA. To do that, 100 μl of 20 μM p53C protein sample (without PEG-8000) in 50 mM PB, pH 7.4, was incubated at 37°C. The formation of p53C condensate was confirmed using a fluorescence microscope (DMi8 microscope, Leica Microsystems, Germany). After condensate formation, 20 μM TR-DNA was added to the 100 μl of condensate solution and incubated for 2 hours at 37°C. p53C condensate dissolution was confirmed using a fluorescence microscope (DMi8 microscope, Leica Microsystems, Germany). One hundred microliters of these dissolved condensate solutions were then loaded into the ZEN0040 cuvette and DLS measurements were recorded. The DLS laser (405 nm) was set to 100% power and all size analysis measurements were performed at 25°C. The autocorrelation functions from individual samples were used to obtain the size (hydrodynamic radius) distribution profile. Freshly prepared 20 μM p53C monomer and condensates were used as controls. All the DLS measurements in this study were carried out using the MALVERN polyanalytical Zetasizer Ultra instrument. The data were plotted using the Origin Pro 8 (Origin Lab, USA).
Size exclusion chromatography
To determine the tetrameric state of p53C condensate in the presence of TR-DNA, size exclusion chromatography (SEC) was performed. Six hundred microliters of 50 μM p53C protein sample (without PEG-8000) in 50 mM PB, pH 7.4, was incubated at 37°C and condensate formation was confirmed using a fluorescence microscope (DMi8 microscope, Leica Microsystems, Germany). After condensate formation, the solution was mixed with 50 μM TR-DNA. The mixture was incubated for 2 hours at 37°C. p53C condensate dissolution was confirmed using a fluorescence microscope (DMi8 microscope, Leica Microsystems, Germany). Five hundred microliters of this dissolved condensate sample was injected into the SEC column. Freshly prepared 50 μM p53C monomer was used as a control. γ-Globulin (150 kDa), lactoferrin (78 kDa), and chymotrypsin (25 kDa) were used as markers. All the elution profiles were plotted using the GraphPad Prism 8 software. The SEC eluted oligomeric fraction of p53C condensates in the presence of TR-DNA was collected and analyzed for the presence of DNA using UV spectroscopy and agarose gel electrophoresis.
Determination of binding affinity of p53 with the TR-DNA, NTR-DNA, and RNA
p53C was labeled using the RED-NHS second-generation protein labeling kit (catalog no. MO-L011, NanoTemper Technologies GmbH) (103). Briefly, threefold molar excess dye was added to 10 μM p53C and incubated for 30 min at room temperature. The excess unbound dye was removed using the B-column of the labeling kit. The protein concentration was measured using a Nanodrop spectrophotometer (Implen, USA), and the final concentration was diluted to 1 μM in 50 mM PB (pH 7.4). Stock solution of 250 μM TR-DNA and 250 μM NTR-DNA was prepared. For determination of binding affinity, serial dilution of ligands (TR-DNA and NTR-DNA) was prepared to a final volume of 10 μl (15,625 to 0.95 nM for TR-DNA and 50,000 to 39 nM for NTR-DNA in 50 mM PB, pH 7.4). Ten microliters of 200 nM labeled p53, which is the target of the ligand, was added to each of these dilutions to a final protein concentration of 100 nM, mixed and incubated for 10 min in the dark. The samples were loaded into capillaries (Monolith Premium Capillaries, catalog no. MO-K025) and inserted into the Monolith X device (NanoTemper, Germany). The spectral shift measurements at 650 and 670 nm were recorded. The experiment was performed three times. The data were normalized and fitted, and Kd (binding affinity) was determined using the binding saturation Eq. (2)
| (2) |
where y is the normalized ratio of 670 nm/650 nm, x is ligand concentration, maximum specific binding is denoted by Bmax, the equilibrium dissociation constant is represented by Kd, the slope of nonlinear regression is represented by NS, and background is measured binding with no added ligand. The data were plotted using the GraphPad Prism 8 software.
PK digestion assay
p53C protein sample (50 μM, without PEG-8000, to avoid the effect of PEG for PK digestion) in 50 mM PB, pH 7.4, was incubated for 12 hours at 37°C to form condensates without any crowder, as confirmed under a fluorescence microscope (DMi8 microscope, Leica Microsystems, Germany). In addition, 10 μM p53C was incubated at identical conditions, as no-LLPS control. Hereafter, just before the digestion, 50 μM LLPS sample was quickly diluted to 10 μM in buffer. Both samples (LLPS and no LLPS) (10 μM) were separately mixed with PK (10 μg/ml; catalog no. 49936, SRL Pvt. Ltd. India) from a stock of 200 μg/ml. The samples were hereby incubated at 37°C and the fixed volumes (50 μl) from the reaction mixture were aliquoted at different time intervals (5, 10, 15, 30, 45, 60, 90, and 120 min). The protein samples (LLPS and no LLPS) just before the addition of PK were taken as samples of 0 min digestion. The reactions at different time points were immediately stopped upon the addition of SDS-PAGE sample buffer followed by heating for 10 min at 95°C. All the samples were analyzed using 15% Tricine SDS-PAGE (Bio-Rad, USA) and visualized using Coomassie Brilliant Blue stain.
DSC analysis
p53C LLPS and no-LLPS samples (10 μM) were prepared the same way as for PK digestion. DSC measurements were run on the MicroCal PEAQ-DSC system (Malvern, Worcestershire, UK). The DSC cells were washed with 14% (v/v) Decon 90 solution, followed by a thorough wash with Milli-Q filtered water. First, water measurement was done to verify the performance of the instrument and the cleanliness of the DSC cell. Once repeatable baselines were achieved with water, 50 mM PB, pH 7.4, scan was performed three times to establish the thermal history of the instrument. Then, three sample scans were examined with the buffer wash in between the sample runs, where freshly prepared samples were loaded for every cycle of heating. For the measurement, 250 μl of 10 μM samples was scanned in the range of 20° to 95°C at a scan rate of 60°C/h with feedback set to “high” for stability profiling of the protein. Data were analyzed using the MicroCal PEAQ-DSC Analysis software (Malvern, Worcestershire, UK). The DSC thermograms of protein samples were corrected for the instrument baseline by subtraction of the corresponding buffer scan. The baseline-subtracted data were fitted to a two-state fitting model to obtain apparent Tm values. The resulting baseline-corrected DSC traces of the p53 protein in the LLPS condition and non-LLPS condition were analyzed for Tm, and the acquired spectra were plotted in Origin Pro 8 (Origin Lab, USA). The experiment was repeated twice with independent sample preparations.
Separation of nuclear and cytoplasmic fraction of SaOS2 cells
Nuclear and cytoplasmic fractions of SaOS2 cells were extracted using a protocol by Senichkin et al. (153, 154). Briefly, SaOS2 cells were grown in 60-mm cell culture dishes up to 90% confluency. Hereafter, the cells were washed with PBS, trypsinized, and harvested after centrifugation. The pellet-containing cells were washed with PBS once and then resuspended in 200 μl of hypotonic solution [20 mM tris-HCl (pH 7.4), 10 mM KCl, 2 mM MgCl2, 1 mM EGTA, 0.5 mM dithiothreitol (DTT), and 0.5 mM phenylmethylsulfonyl fluoride (PMSF)] and incubated for 5 min at 4°C. Membrane lysis was done with 0.1% NP-40 for 3 min at 4°C. The solution was then centrifuged at 1000g for 5 min at 4°C. The supernatant was collected as the cytoplasmic fraction. The pellet containing the nuclear fraction was washed with isotonic buffer [20 mM tris-HCl (pH 7.4), 150 mM KCl, 2 mM MgCl2, 1 mM EGTA, 0.5 mM DTT, and 0.5 mM PMSF] containing 0.3% NP-40 in the isotonic buffer for 5 min at 4°C. The solution was centrifuged and the pellet containing nuclei was dissolved in RIPA buffer [25 mM tris-HCl (pH 7.4), 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40, and cOmplete Protease Inhibitor Cocktail (Roche Diagnostics)] and incubated for 20 min at 4°C. The solution was again centrifuged at 2000g for 3 min at 4°C and the supernatant was collected as the nuclear fraction. The separated cytoplasmic fraction was centrifuged at 15,000g to remove the residual debris and the supernatant was collected. After extraction, both the NE and the CE were dialyzed using the 1-kDa cutoff membrane (Spectra/Por Dialysis membrane, catalog no. 132638, Spectrum Laboratories Inc., USA) in PBS for 2 hours. The efficient separation of nuclear and cytoplasmic fractions of SaOS2 cells was further verified by Western blotting of the two fractions, following the previously mentioned protocol. An equal protein amount (30 μg), estimated by the Bradford test, was loaded for SDS-PAGE. Anti-GAPDH antibody (1:2000) was used as a cytoplasmic marker and anti-Histone H3 antibody (1:1000, catalog no. 4499, Cell Signaling Technology) was used as a nuclear marker. The images were captured using the Image Quant LAS 500 (GE Life Sciences, USA).
Acknowledgments
We thank the IIT Bombay central facilities, namely, the microscopy and FACS facility at BSBE, and the TEM and FTIR facilities at SAIF. We also thank R. Padinhateeri, BSBE, IIT Bombay, for valuable suggestions. We likewise thank N. Singh, RCB Faridabad; S. Panigrahi, P. Shinde, and V. Soman from the microscopy facility at IIT Bombay; V. Vittal, Technical Assistant for the microscopy facility at IISER Pune; S. Menon, Senior Field Application Scientist; R. Dadhich, Application Scientist at Nanotemper Technologies; and D. Chatterjee for valuable inputs in experiments.
Funding: We thank DST-SERB (file no. CRG/2019/001133) and TATA Innovation (BT/HRD/35/01/03/2020) for financial support. S.S. acknowledges the DBT/Wellcome Trust India Alliance Fellowship (IA/E/17/1/503663) for financial support. D.D. acknowledges MoE, Government of India, for the GATE fellowship.
Author contributions: Conceptualization: S.K.M. Methodology: S.K.M. and D.D. Validation: S.K.M., D.D., A.N., A.S., A.P., K.P., S.Mas., L.G., S.Man., A.B., S.S., M.P., J.D., A.S.S., R.S., S.P., S.Muk., and N.G. Formal analysis: D.D., A.N., A.S., A.P., K.P., S.Mas., L.G., S.S., J.D., and R.S. Investigation: D.D., A.N., A.S., A.P., K.P., S.Mas., L.G., S.Man., A.B., S.S., M.P., J.D., A.S.S., R.S., S.P., S.Muk., and N.G. Resources: S.K.M. and K.S. Data curation: D.D., A.N., A.S., A.P., K.P., S.Mas., L.G., S.Man., A.B., S.S., M.P., J.D., A.S.S., R.S., S.P., S.Muk., and N.G. Writing—original draft preparation: S.K.M., D.D., and A.N. Writing—review and editing preparation: S.K.M., D.D., A.N., A.S., K.P., A.P., and S.Muk. Visualization: D.D., A.N., A.S., A.P., K.P., S.Mas.., L.G., S.Man., A.B., S.S., M.P., J.D., A.S.S., P.K., R.S., and S.Muk. Supervision: S.K.M. Project administration: S.K.M. Funding acquisition: S.K.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The tools used for plotting and analysis have been mentioned in the main text and the Supplementary Materials.
Supplementary Materials
The PDF file includes:
Figs. S1 to S16
Tables S1 to S3
Legends for movies S1 to S6
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S6
REFERENCES AND NOTES
- 1.Friedman J. R., Nunnari J., Mitochondrial form and function. Nature 505, 335–343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luzio J. P., Pryor P. R., Bright N. A., Lysosomes: Fusion and function. Nat. Rev. Mol. Cell Biol. 8, 622–632 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Boisvert F.-M., van Koningsbruggen S., Navascués J., Lamond A. I., The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Banani S. F., Rice A. M., Peeples W. B., Lin Y., Jain S., Parker R., Rosen M. K., Compositional control of phase-separated cellular bodies. Cell 166, 651–663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shin Y., Brangwynne C. P., Liquid phase condensation in cell physiology and disease. Science 357, 1253 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodruff J. B., Hyman A. A., Boke E., Organization and function of non-dynamic biomolecular condensates. Trends Biochem. Sci. 43, 81–94 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Hyman A. A., Simons K., Beyond oil and water—Phase transitions in cells. Science 337, 1047–1049 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Hyman A. A., Weber C. A., Julicher F., Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Lyon A. S., Peeples W. B., Rosen M. K., A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol. 22, 215–235 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alberti S., Phase separation in biology. Curr. Biol. 27, R1097–R1102 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Boeynaems S., Alberti S., Fawzi N. L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L., Tompa P., Fuxreiter M., Protein phase separation: A new phase in cell biology. Trends Cell Biol. 28, 420–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alberti S., Gladfelter A., Mittag T., Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu H., Yu D., Hansen A. S., Ganguly S., Liu R., Heckert A., Darzacq X., Zhou Q., Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hnisz D., Shrinivas K., Young R. A., Chakraborty A. K., Sharp P. A., A phase separation model for transcriptional control. Cell 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fox A. H., Nakagawa S., Hirose T., Bond C. S., Paraspeckles: Where long noncoding RNA meets phase separation. Trends Biochem. Sci. 43, 124–135 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Riback J. A., Katanski C. D., Kear-Scott J. L., Pilipenko E. V., Rojek A. E., Sosnick T. R., Drummond D. A., Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168, 1028–1040.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cramer P., Organization and regulation of gene transcription. Nature 573, 45–54 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Kribelbauer J. F., Rastogi C., Bussemaker H. J., Mann R. S., Low-affinity binding sites and the transcription factor specificity paradox in eukaryotes. Annu. Rev. Cell Dev. Biol. 35, 357–379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calabretta S., Richard S., Emerging roles of disordered sequences in RNA-binding proteins. Trends Biochem. Sci. 40, 662–672 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Martin E. W., Holehouse A. S., Peran I., Farag M., Incicco J. J., Bremer A., Grace C. R., Soranno A., Pappu R. V., Mittag T., Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pak C. W., Kosno M., Holehouse A. S., Padrick S. B., Mittal A., Ali R., Yunus A. A., Liu D. R., Pappu R. V., Rosen M. K., Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell 63, 72–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maharana S., Wang J., Papadopoulos D. K., Richter D., Pozniakovsky A., Poser I., Bickle M., Rizk S., Guillen-Boixet J., Franzmann T. M., Jahnel M., Marrone L., Chang Y.-T., Sterneckert J., Tomancak P., Hyman A. A., Alberti S., RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bremer A., Farag M., Borcherds W. M., Peren I., Martin E. W., Pappu R. V., Mittag T., Deciphering how naturally occurring sequence features impact the phase behaviours of disordered prion-like domains. Nat. Chem. 14, 196–207 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Y., Protter D. S. W., Rosen M. K., Parker R., Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., Choi J.-M., Holehouse A. S., Lee H. O., Zhang X., Jahnel M., Maharana S., Lemaitre R., Pozniakovsky A., Drechsel D., Poser I., Pappu R. V., Alberti S., Hyman A. A., A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saha S., Weber C. A., Nousch M., Adame-Arana O., Hoege C., Heim M. Y., Osborne-Nishimura E., Mahamid J., Jahnel M., Jawerth L., Pozniakovski A., Eckmann C. R., Jülicher F., Hyman A. A., Polar positioning of phase-separated liquid compartments in cells regulated by an mRNA competition mechanism. Cell 166, 1572–1584.e16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brangwynne C. P., Tompa P., Pappu R. V., Polymer physics of intracellular phase transitions. Nat. Phys. 11, 899–904 (2015). [Google Scholar]
- 29.Alberti S., Hyman A. A., Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 22, 196–213 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Jiang S., Fagman J. B., Chen C., Alberti S., Liu B., Protein phase separation and its role in tumorigenesis. eLife 9, e60264 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alberti S., Dormann D., Liquid-liquid phase separation in disease. Annu. Rev. Genet. 53, 171–194 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Wei M. T., Chang Y. C., Shimobayashi S. F., Shin Y., Strom A. R., Brangwynne C. P., Nucleated transcriptional condensates amplify gene expression. Nat. Cell Biol. 22, 1187–1196 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Bhat P., Honson D., Guttman M., Nuclear compartmentalization as a mechanism of quantitative control of gene expression. Nat. Rev. Mol. Cell Biol. 22, 653–670 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Levine A. J., p53, the cellular gatekeeper for growth and division. Cell 88, 323–331 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Fuster J. J., Sanz-Gonzalez S. M., Moll U. M., Andres V., Classic and novel roles of p53: Prospects for anticancer therapy. Trends Mol. Med. 13, 192–199 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Zilfou J. T., Lowe S. W., Tumor suppressive functions of p53. Cold Spring Harb. Perspect. Biol. 1, a001883 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine A. J., Momand J., Finlay C. A., The p53 tumour suppressor gene. Nature 351, 453–456 (1991). [DOI] [PubMed] [Google Scholar]
- 38.Harms K., Nozell S., Chen X., The common and distinct target genes of the p53 family transcription factors. Cell. Mol. Life Sci. 61, 822–842 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vousden K. H., Lu X., Live or let die: The cell’s response to p53. Nat. Rev. Cancer 2, 594–604 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Kruiswijk F., Labuschagne C. F., Vousden K. H., p53 in survival, death and metabolic health: A lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 16, 393–405 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Bieging K. T., Mello S. S., Attardi L. D., Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 14, 359–370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moll U. M., LaQuaglia M., Benard J., Riou G., Wild-type p53 protein undergoes cytoplasmic sequestration in undifferentiated neuroblastomas but not in differentiated tumors. Proc. Natl. Acad. Sci. U.S.A. 92, 4407–4411 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moll U. M., Ostermeyer A. G., Haladay R., Winkfield B., Frazier M., Zanbetti G., Cytoplasmic sequestration of wild-type p53 protein impairs the G1 checkpoint after DNA damage. Mol. Cell. Biol. 16, 1126–1137 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moll U. M., Riou G., Levine A. J., Two distinct mechanisms alter p53 in breast cancer: Mutation and nuclear exclusion. Proc. Natl. Acad. Sci. U.S.A. 89, 7262–7266 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu J., Reumers J., Couceiro J. R., Smet F. D., Gallardo R., Rudyak S., Cornelis A., Rozenski J., Zwolinska A., Marine J.-C., Lambrechts D., Suh Y.-A., Rousseau F., Schymkowitz J., Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat. Chem. Biol. 7, 285–295 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Smet F. D., Rubio M. S., Hompes D., Naus E., Baets G. D., Langenberg T., Hipp M. S., Houben B., Claes F., Charbonneau S., Blanco J. D., Plaisance S., Ramkissoon S., Ramkissoon L., Simons C., Brandt P. V. D., Weijenberg M., England M. V., Lambrechts S., Amant F., Hoore A. D., Ligon K. L., Sagaert X., Schymkowitz J., Rousseau F., Nuclear inclusion bodies of mutant and wild-type p53 in cancer: A hallmark of p53 inactivation and proteostasis remodelling by p53 aggregation. J. Pathol. 242, 24–38 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Ghosh S., Salot S., Sengupta S., Navalkar A., Ghosh D., Jacob R., Das S., Kumar R., Jha N. N., Sahay S., Mehra S., Mohite G. M., Ghosh S. K., Komrabail N., Krishnamoorthy G., Chaudhari P., Maji S. K., p53 amyloid formation leading to its loss of function: Implications in cancer pathogenesis. Cell Death Differ. 24, 1784–1798 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Navalkar A., Pandey S., Singh N., Patel K., Datta D., Mohanty B., Jadhav S., Chaudhari P., Maji S. K., Direct evidence of cellular transformation by prion-like p53 amyloid infection. J. Cell Sci. 134, jcs258316 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Navalkar A., Ghosh S., Pandey S., Paul A., Datta D., Maji S. K., Prion-like p53 amyloids in cancer. Biochemistry 59, 146–155 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Joerger A. C., Fersht A. R., The p53 pathway: Origins, inactivation in cancer, and emerging therapeutic approaches. Annu. Rev. Biochem. 85, 375–404 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Silva J. L., Gallo C. V. D. M., Costa D. C. F., Rangel L. P., Prion-like aggregation of mutant p53 in cancer. Trends Biochem. Sci. 39, 260–267 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Costa D. C. F., Oliveira G. A. P. D., Cino E. A., Soares I. N., Rangel L. P., Silva J. L., Aggregation and prion-like properties of misfolded tumor suppressors: Is cancer a prion disease? Cold Spring Harb. Perspect. Biol. 8, a023614 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Safari M. S., Wang Z., Tailor K., Kolomeisky A. B., Conrad J. C., Vekilov P. G., Anomalous dense liquid condensates host the nucleation of tumor suppressor p53 fibrils. Iscience 12, 342–355 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang D. S., Saeedi A., Davtyan A., Fathi M., Sherman M. B., Safari M. S., Klindziuk A., Barton M. C., Varadarajan N., Kolomeisky A. B., Vekilov P. G., Mesoscopic protein-rich clusters host the nucleation of mutant p53 amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 118, e2015618118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petronilho E. C., Pedrote M. M., Marques M. A., Passos Y. M., Mota M. F., Jakobus B., Sousa G. D. S. D., Costa F. P. D., Felix A. L., Ferretti G. D. S., Almeida F. P., Cordeiro Y., Vieira T. C. R. G., de Oliveira G. A. P., Siva J. L., Phase separation of p53 precedes aggregation and is affected by oncogenic mutations and ligands. Chem. Sci. 12, 7334–7349 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo A., Salomoni P., Luo J., Shih A., Zhong S., Gu W., Paolo P. P., The function of PML in p53-dependent apoptosis. Nat. Cell Biol. 2, 730–736 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Cioce M., Lamond A. I., CAJAL BODIES: A long history of discovery. Annu. Rev. Cell Dev. Biol. 21, 105–131 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Cannon J. V., Lane D. P., Protein synthesis required to anchor a mutant p53 protein which is temperature-sensitive for nuclear transport. Nature 349, 802–806 (1991). [DOI] [PubMed] [Google Scholar]
- 59.Shaulsky G., Goldfinger N., Peled A., Rotter V., Involvement of wild-type p53 protein in the cell cycle requires nuclear localization. Cell Growth Differ. 2, 661–667 (1991). [PubMed] [Google Scholar]
- 60.Oren M., Maltzman W., Levine A. J., Post-translational regulation of the 54K cellular tumor antigen in normal and transformed cells. Mol. Cell. Biol. 1, 101–110 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Slade N., Moll U. M., Mutational analysis of p53 in human tumors: Immunocytochemistry. Methods Mol. Biol. 234, 231–243 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Meek D. W., The p53 response to DNA damage. DNA Repair 3, 1049–1056 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Joerger A. C., Fersht A. R., Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 77, 557–582 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Miskei M., Horvath A., Vendruscolo M., Fuxreiter M., Sequence-based prediction of fuzzy protein interactions. J. Mol. Biol. 432, 2289–2303 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., Hassabis D., Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Solares M. J., Jonaid G. M., Luqiu W. Y., Berry S., Khadela J., Liang Y., Evans M. C., Pridham K. J., Dearnaley W. J., Sheng Z., Kelly D. F., High-resolution imaging of human cancer proteins using microprocessor materials. Chembiochem 23, e202200310 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Maki C. G., Huibregtse J. M., Howley P. M., In vivo ubiquitination and proteasome-mediated degradation of p53(1). Cancer Res. 56, 2649–2654 (1996). [PubMed] [Google Scholar]
- 68.Moll U. M., Petrenko O., The MDM2-p53 interaction. Mol. Cancer Res. 1, 1001–1008 (2003). [PubMed] [Google Scholar]
- 69.Kubbutat M. H., Jones S. N., Vousden K. H., Regulation of p53 stability by Mdm2. Nature 387, 299–303 (1997). [DOI] [PubMed] [Google Scholar]
- 70.Vogelstein B., Lane D., Levine A. J., Surfing the p53 network. Nature 408, 307–310 (2000). [DOI] [PubMed] [Google Scholar]
- 71.Hu J., Lieb J. D., Sancar A., Adar S., Cisplatin DNA damage and repair maps of the human genome at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 113, 11507–11512 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blom H., Widengren J., Stimulated emission depletion microscopy. Chem. Rev. 117, 7377–7427 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Hell S. W., Wichmann J., Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19, 780–782 (1994). [DOI] [PubMed] [Google Scholar]
- 74.Marine J. C., Lozano G., Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 17, 93–102 (2010). [DOI] [PubMed] [Google Scholar]
- 75.Francoz S., Froment P., Bogaerts S., De Clercq S., Maetens M., Doumont G., Bellefroid E., Marine J.-C., Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc. Natl. Acad. Sci. U.S.A. 103, 3232–3237 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baresova P., Musilova J., Pitha P. M., Lubyova B., p53 tumor suppressor protein stability and transcriptional activity are targeted by Kaposi’s sarcoma-associated herpesvirus-encoded viral interferon regulatory factor 3. Mol. Cell. Biol. 34, 386–399 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maltzman W., Czyzyk L., UV irradiation stimulates levels of p53 cellular tumor antigen in nontransformed mouse cells. Mol. Cell. Biol. 4, 1689–1694 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ottaviano L., Sehaefer K.-L., Gajewski M., Huckenbeck W., Baldus S., Rogel U., Mackintosh C., Alava E. D., Myklebost O., Kresse S. H., Meza-Zepeda L. A., Serra M., Cleton-Jansen A.-M., Hogendoorn P. C. W., Buerger H., Aigner T., Gabbert H. E., Poremba C., Molecular characterization of commonly used cell lines for bone tumor research: A trans-European EuroBoNet effort. Genes Chromosomes Cancer 49, 40–51 (2010). [DOI] [PubMed] [Google Scholar]
- 79.Chen X., Ko L. J., Jayaraman L., Prives C., p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 10, 2438–2451 (1996). [DOI] [PubMed] [Google Scholar]
- 80.Kudo N., Matsumori N., Taoka H., Fujiwara D., Schreiner E. P., Wolff B., Woshida M., Horinouchi S., Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. U.S.A. 96, 9112–9117 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Greenspan P., Mayer E. P., Fowler S. D., Nile red: A selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 100, 965–973 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choi J. M., Holehouse A. S., Pappu R. V., Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys. 49, 107–133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kar M., Dar F., Welsh T. J., Vogel L. T., Kuhnemuth R., Majumdar A., Krainer G., Franzmann T. M., Alberti S., Seidel C. A., Knowles T. P. J., Hyman A. A., Pappu R. V., Phase-separating RNA-binding proteins form heterogeneous distributions of clusters in subsaturated solutions. Proc. Natl. Acad. Sci. U.S.A. 119, e2202222119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Choi J.-M., Hyman A. A., Pappu R. V., Generalized models for bond percolation transitions of associative polymers. Phys. Rev. E 102, 042403 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kroschwald S., Maharana S., Simon A., Hexanediol: A chemical probe to investigate the material properties of membrane-less compartments. Matters 3, e201702000010 (2017). [Google Scholar]
- 86.Stephen C. W., Lane D. P., Mutant conformation of p53: Precise epitope mapping using a filamentous phage epitope library. J. Mol. Biol. 225, 577–583 (1992). [DOI] [PubMed] [Google Scholar]
- 87.Kayed R., Head E., Thompson J. L., Mclntire T. M., Milton S. C., Cotman C. W., Glabe C. G., Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489 (2003). [DOI] [PubMed] [Google Scholar]
- 88.Chene P., Inhibiting the p53–MDM2 interaction: An important target for cancer therapy. Nat. Rev. Cancer 3, 102–109 (2003). [DOI] [PubMed] [Google Scholar]
- 89.Shirangi T. R., Zaika A., Moll U. M., Nuclear degradation of p53 occurs during down-regulation of the p53 response after DNA damage. FASEB J. 16, 420–422 (2002). [DOI] [PubMed] [Google Scholar]
- 90.Brandt T., Kaar J. L., Fersht A. R., Veprintsev D. B., Stability of p53 homologs. PLOS ONE 7, e47889 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bullock A. N., Henckel J., DeDecker B. S., Johnson C. M., Nikolova P. V., Proctor M. R., Lane D. P., Fersht A. R., Thermodynamic stability of wild-type and mutant p53 core domain. Proc. Natl. Acad. Sci. U.S.A. 94, 14338–14342 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ang H. C., Joerger A. C., Mayer S., Fersht A. R., Effects of common cancer mutations on stability and DNA binding of full-length p53 compared with isolated core domains. J. Biol. Chem. 281, 21934–21941 (2006). [DOI] [PubMed] [Google Scholar]
- 93.Wang Y., Rosengarth A., Luecke H., Structure of the human p53 core domain in the absence of DNA. Acta Crystallogr. D Biol. Crystallogr. 63, 276–281 (2007). [DOI] [PubMed] [Google Scholar]
- 94.Meszaros B., Erdos G., Dosztanyi Z., IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 46, W329–W337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Erdős G., Dosztányi Z., Analyzing protein disorder with IUPred2A. Curr. Protoc. Bioinformatics 70, e99 (2020). [DOI] [PubMed] [Google Scholar]
- 96.LeVine H. III, Quantification of β-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 309, 274–284 (1999). [DOI] [PubMed] [Google Scholar]
- 97.Klunk W. E., Jacob R. F., Mason R. P., Quantifying amyloid by congo red spectral shift assay. Methods Enzymol. 309, 285–305 (1999). [DOI] [PubMed] [Google Scholar]
- 98.Kamagata K., Kanbayashi S., Honda M., Itoh Y., Takahashi H., Kameda T., Nagatsugi F., Takahashi S., Liquid-like droplet formation by tumor suppressor p53 induced by multivalent electrostatic interactions between two disordered domains. Sci. Rep. 10, 580 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Morin J. A., Wittmann S., Choubey S., Klosin A., Golfier S., Hyman A. A., Julicher F., Grill S. W., Sequence-dependent surface condensation of a pioneer transcription factor on DNA. Nat. Phys. 18, 271–276 (2022). [Google Scholar]
- 100.Poudyal M., Patel K., Gadhe L., Sawner A. S., Kadu P., Datta D., Mukherjee S., Ray S., Navalkar A., Maiti S., Chatterjee D., Devi J., Bera R., Gahlot N., Joseph J., Padinhateeri R., Maji S. K., Intermolecular interactions underlie protein/peptide phase separation irrespective of sequence and structure at crowded milieu. Nat. Commun. 14, 6199 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alshareedah I., Moosa M. M., Raju M., Potoyan D. A., Banerjee P. R., Phase transition of RNA-protein complexes into ordered hollow condensates. Proc. Natl. Acad. Sci. U.S.A. 117, 15650–15658 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen Y., Dey R., Chen L., Crystal structure of the p53 core domain bound to a full consensus site as a self-assembled tetramer. Structure 18, 246–256 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Langer A., Ludecke A., Bartoschik T., Cehlar O., Duhr S., Baaske P., Streicher W., A new spectral shift-based method to characterize molecular interactions. Assay Drug Dev. Technol. 20, 83–94 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Franzmann T. M., Jahnel M., Pozniakovsky A., Mahamid J., Holehouse A. S., Nüske E., Richter D., Baumeister W., Grill S. W., Pappu R. V., Hyman A. A., Alberti S., Phase separation of a yeast prion protein promotes cellular fitness. Science 359, eaao5654 (2018). [DOI] [PubMed] [Google Scholar]
- 105.Hubstenberger A., Courel M., Bénard M., Souquere S., Ernoult-Lange M., Chouaib R., Yi Z., Morlot J.-B., Munier A., Fradet M., Daunesse M., Bertrand E., Pierron G., Mozziconacci J., Kress M., Weil D., P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell 68, 144–157.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 106.Sheu-Gruttadauria J., MacRae I. J., Phase transitions in the assembly and function of human miRISC. Cell 173, 946–957.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brangwynne C. P., Eckmann C. R., Courson D. S., Rybarska A., Hoege C., Gharakhani J., Julicher F., Hyman A. A., Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009). [DOI] [PubMed] [Google Scholar]
- 108.Mitrea D. M., Kriwacki R. W., Phase separation in biology; functional organization of a higher order. Cell Commun. Signal 14, 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Franzmann T. M., Alberti S., Protein phase separation as a stress survival strategy. Cold Spring Harb. Perspect. Biol. 11, a034058 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Strom A. R., Emelyanov A. V., Mir M., Fyodorov D. V., Darzacq X., Karpen G. H., Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sabari B. R., Dall’Agnese A., Young R. A., Biomolecular condensates in the nucleus. Trends Biochem. Sci. 45, 961–977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Boija A., Klein I. A., Sabari B. R., Agnese A. D., Coffey E. L., Zanudio A. V., Li C. H., Shrinivas K., Manteiga J. C., Hannett N. M., Abraham B. J., Afeyan L. K., Guo Y. E., Rimel J. K., Fant C. B., Schuijers J., Lee T. I., Taatjes D. J., Young R. A., Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gibson B. A., Doolittle L. K., Schneider M. W., Jensen L. E., Gamarra N., Henry L., Gerlich D. W., Redding S., Rosen M. K., Organization of chromatin by intrinsic and regulated phase separation. Cell 179, 470–484.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lu Y., Wu T., Gutman O., Lu H., Zhou Q., Henis Y. I., Luo K., Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nat. Cell Biol. 22, 453–464 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kimura H., Sato Y., Imaging transcription elongation dynamics by new technologies unveils the organization of initiation and elongation in transcription factories. Curr. Opin. Cell Biol. 74, 71–79 (2022). [DOI] [PubMed] [Google Scholar]
- 116.Sabari B. R., Dallagnese A., Boija A., Klein I. A., Coffey E. L., Shrinivas K., Abraham B. J., Hannett N. M., Zamudio A. V., Manteiga J. C., Li C. H., Guo Y. E., Day D. S., Schuijers J., Vasile E., Malik S., Hnisz D., Lee T. I., Cisse I. I., Roeder R. G., Sharp P. A., Chakraborty A. K., Young R. A., Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cho W. K., Spille J. H., Hecht M., Lee C., Li C., Grube V., Cisse I. I., Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Donovan B. T., Larson D. R., Regulating gene expression through control of transcription factor multivalent interactions. Mol. Cell 82, 1974–1975 (2022). [DOI] [PubMed] [Google Scholar]
- 119.Oren M., Regulation of the p53 tumor suppressor protein. J. Biol. Chem. 274, 36031–36034 (1999). [DOI] [PubMed] [Google Scholar]
- 120.Harris S. L., Levine A. J., The p53 pathway: Positive and negative feedback loops. Oncogene 24, 2899–2908 (2005). [DOI] [PubMed] [Google Scholar]
- 121.Navalkar A., Paul A., Sakunthala A., Pandey S., Dey A. K., Saha S., Sahoo S., Jolly M. K., Maiti T. K., Maji S. K., Oncogenic gain of function due to p53 amyloids occurs through aberrant alteration of cell cycle and proliferation. J. Cell Sci. 135, jcs259500 (2022). [DOI] [PubMed] [Google Scholar]
- 122.Alberti S., Hyman A. A., Are aberrant phase transitions a driver of cellular aging? Bioessays 38, 959–968 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dai Z., Li G., Chen Q., Yang X., Ser392 phosphorylation modulated a switch between p53 and transcriptional condensates. Biochim. Biophys. Acta Gene Regul. Mech. 1865, 194827 (2022). [DOI] [PubMed] [Google Scholar]
- 124.Heltberg M. S., Lucchetti A., Hsieh F. S., Nguyen D. P. M., Chen S. H., Jensen M. H., Enhanced DNA repair through droplet formation and p53 oscillations. Cell 185, 4394–4408.e10 (2022). [DOI] [PubMed] [Google Scholar]
- 125.Bouchard J. J., Otero J. H., Scott D. C., Szulc E., Martin E. W., Sabri N., Granata D., Marzahn M. R., Lindorff-Larsen K., Salvatella X., Schulman B. A., Mittag T., Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol. Cell 72, 19–36.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chandra B., Michmerhuizen N. L., Shirnekhi H. K., Tripathi S., Pioso B. J., Baggett D. W., Mitrea D. M., Iacobucci I., White M. R., Chen J., Park C.-G., Wu H., Pounds S., Medyukhina A., Khairy K., Gao Q., Qu C., Abdelhamed S., Gorman S. D., Bawa S., Maslanka C., Kinger S., Dogra P., Ferrolino M. C., Giacomo D. D., Mecucci C., Klco J. M., Mullighan C. G., Kriwacki R. W., Phase separation mediates NUP98 fusion oncoprotein leukemic transformation. Cancer Discov. 12, 1152–1169 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zuo L., Zhang G., Massett M., Cheng J., Guo Z., Wang L., Gao Y., Li R., Huang X., Li P., Qi Z., Loci-specific phase separation of FET fusion oncoproteins promotes gene transcription. Nat. Commun. 12, 1491 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Powers S. K., Holehouse A. S., Korasick D. A., Schreiber K. H., Clark N. M., Jing H., Emenecker R., Han S., Tycksen E., Hwang I., Sozzani R., Jez J. M., Pappu R. V., Strader L. C., Nucleo-cytoplasmic partitioning of ARF proteins controls auxin responses in Arabidopsis thaliana. Mol. Cell 76, 177–190.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Patel A., Lee O. H., Zawerth L., Maharana S., Jahnel M., Hein M. K., Stoynov S., Mahamid J., Saha S., Franzmann T. M., Pozniakovski A., Poser I., Maghelli N., Royer L. A., Weigert M., Myers E. W., Grill S., Drechsel D., Hyman A. A., Alberti S., A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 130.Xiao R., Chen J. Y., Liang Z., Luo D., Chen G., Lu Z. J., Chen Y., Zhou B., Li H., Du X., Yang Y., San M., Wei X., Liu W., Lécuyer E., Graveley B. R., Yeo G. W., Burge C. B., Zhang M. Q., Zhou Y., Fu X.-D., Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell 178, 107–121.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oksuz O., Henninger J. E., Warneford-Thomson R., Zheng M. M., Erb H., Vancura A., Overholt K. J., Hawken S. W., Banani S. F., Lauman R., Reich L. N., Robertson A. L., Hannett N. M., Lee T. I., Zon L. I., Bonasio R., Young R. A., Transcription factors interact with RNA to regulate genes. Mol. Cell 83, 2449–2463.e13 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Riback J. A., Zhu L., Ferrolino M. C., Tolbert N., Mitrea D. M., Sanders D. W., Wei M. T., Kriwacki R. W., Brangwynne C. P., Composition dependent phase separation underlies directional flux through the nucleolus. Nature 581, 209–214 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Elbaum-Garfinkle S., Kim Y., Szczepaniak K., Chen C. C., Eckmann C. R., Yong S. M., Branngwynne C. P., The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U.S.A. 112, 7189–7194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Weber S. C., Brangwynne C. P., Getting RNA and protein in phase. Cell 149, 1188–1191 (2012). [DOI] [PubMed] [Google Scholar]
- 135.Caudron-Herger M., Rippe K., Nuclear architecture by RNA. Curr. Opin. Genet. Dev. 22, 179–187 (2012). [DOI] [PubMed] [Google Scholar]
- 136.Kovachev P. S., Banerjee D., Rangel L. P., Eriksson J., Pedrote M. M., Martins-Dinis M. M., Edwards K., Cordeiro Y., Silva J. L., Sanyal S., Distinct modulatory role of RNA in the aggregation of the tumor suppressor protein p53 core domain. J. Biol. Chem. 92, 9345–9357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang H., Elbaum-Garfinkle S., Langdon E. M., Taylor N., Occhipinti P., Bridges A. A., Brangwynne C. P., Gladfelter A. S., RNA controls polyQ protein phase transitions. Mol. Cell 60, 220–230 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Adame-Arana O., Bajpai G., Lorber D., Volk T., Safran S., Regulation of chromatin microphase separation by binding of protein complexes. eLife 12, e82983 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ishimaru D., Bom A. P. A., Lima L. M., Quesado P. A., Oyama M. F., Gallo C. V. D. M., Cordeiro Y., Silva J. L., Cognate DNA stabilizes the tumor suppressor p53 and prevents misfolding and aggregation. Biochemistry 48, 6126–6135 (2009). [DOI] [PubMed] [Google Scholar]
- 140.Jambon-Puillet E., Testa A., Lorenz C., Style R. W., Rebane A. A., Dufresne E. R., Phase-separated droplets swim to their dissolution. Nat. Commun. 15, 3919 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Letunic I., Bork P., 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.DeLano W. L., PyMOL: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 40, 82–92 (2002). [Google Scholar]
- 143.Lowe D. G., Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 60, 91–110 (2004). [Google Scholar]
- 144.Ray S., Singh N., Kumar R., Patel K., Pandey S., Datta D., Mahato J., Panigrahi R., Navalkar A., Mehra S., Gadhe L., Chatterjee D., Sawner A. S., Maiti S., Bhatia S., Gerez J. A., Chowdhury A., Kumar A., Padinhateeri R., Riek R., Krishnamoorthy G., Maji S. K., α-Synuclein aggregation nucleates through liquid–liquid phase separation. Nat. Chem. 12, 705–716 (2020). [DOI] [PubMed] [Google Scholar]
- 145.Schoonderwoert V., Dijkstra R., Luckinavicius G., Kobler O., van der Voort H., Huygens STED deconvolution increases signal-to-noise and image resolution towards 22 nm. Micros. Today 21, 38–44 (2013). [Google Scholar]
- 146.Yen J.-C., Chang F.-J., Chang S., A new criterion for automatic multilevel thresholding. IEEE Trans. Image Process. 4, 370–378 (1995). [DOI] [PubMed] [Google Scholar]
- 147.Sezgin M., Sankur B., Survey over image thresholding techniques and quantitative performance evaluation. J. Electron. Imaging 13, 146–168 (2004). [Google Scholar]
- 148.Sauvola J., Pietikainen M., Adaptive document image binarization. Pattern Recognit. 33, 225–236 (2000). [Google Scholar]
- 149.P. Neerad, M. Sumit, S. Ashish, J. Madhuri, Adaptive local thresholding for detection of nuclei in diversity stained cytology images, in 2011 International Conference on Communications and Signal Processing (IEEE, 2011), pp. 218–220. [Google Scholar]
- 150.Arganda-Carreras I., Kaynig V., Rueden C., Eliceiri K. W., Schindelin J., Cardona A., Seung H. S., Trainable Weka Segmentation: A machine learning tool for microscopy pixel classification. Bioinformatics 33, 2424–2426 (2017). [DOI] [PubMed] [Google Scholar]
- 151.Sengupta S., Singh N., Paul A., Datta D., Chatterjee D., Mukherjee S., Gadhe L., Devi J., Mahesh Y., Jolly M. K., Maji S. K., p53 amyloid pathology is correlated with higher cancer grade irrespective of the mutant or wild-type form. J. Cell Sci. 136, jcs261017 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Milkovic N. M., Mittag T., Determination of protein phase diagrams by centrifugation. Methods Mol. Biol. 2141, 685–702 (2020). [DOI] [PubMed] [Google Scholar]
- 153.Senichkin V. V., Prokhorova E. A., Zhivotovsky B., Kopeina G. S., Simple and efficient protocol for subcellular fractionation of normal and apoptotic cells. Cells 10, 852 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Freibaum B. D., Messing J., Yang P., Kim H. J., Taylor J. P., High-fidelity reconstitution of stress granules and nucleoli in mammalian cellular lysate. J. Cell Biol. 220, e202009079 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S16
Tables S1 to S3
Legends for movies S1 to S6
Movies S1 to S6