Abstract
Skin cancer is a growing global concern, with cases steadily rising. Typically, malignant moles are identified through visual inspection, using dermatoscopy and patient history. Active thermography has emerged as an effective method to distinguish between malignant and benign lesions. Our previous research showed that spatio-temporal features can be extracted from suspicious lesions to accurately determine malignancy, which was applied in a distance-based classifier.
In this study, we build on that foundation by introducing a set of novel spatial and temporal features that enhance classification accuracy and can be integrated into any machine learning approach. These features were implemented in a support-vector machine classifier to detect malignancy. Notably, our method addresses a common limitation in existing approaches—manual lesion selection—by automating the process using a U-Net convolutional neural network.
We validated our system by comparing U-Net's performance with expert dermatologist segmentations, achieving a 17% improvement in the Jaccard index over a semi-automatic algorithm. The detection algorithm relies on accurate lesion segmentation, and its performance was evaluated across four segmentation techniques. At an 85% sensitivity threshold, expert segmentation provided the highest specificity at 87.62%, while non-expert and U-Net segmentations achieved comparable results of 69.63% and 68.80%, respectively. Semi-automatic segmentation lagged behind at 64.45%. This automated detection system performs comparably to high-accuracy methods while offering a more standardized and efficient solution. The proposed automatic system achieves 3% higher accuracy compared to the ResNet152V2 network when processing low-quality images obtained in a clinical setting.
Keywords: Skin cancer screening, Dynamic thermal imaging, Image segmentation, Convolutional neural networks, Machine learning
Graphical abstract
Highlights
-
•
Design and evaluation of the first automatic algorithm to detect skin cancer using active thermography.
-
•
Base scheme to attack skin cancer detection problem with active thermography and machine learning.
-
•
Design and evaluation of U-Net network for segmenting lesions in low-quality images acquired in a clinical setting.
-
•
Comparison of U-Net segmentation quality with expert manual, non-expert, and semi-automatic segmentation methods.
-
•
Comparison of thermography-based detection with convolutional networks processing dermoscopic images.
1. Introduction
Skin cancer is considered the most common type of cancer, and its early detection is quite relevant, since it has a direct impact on the decrease in mortality rates [1]. Moreover, early detection provides the additional benefit of requiring less aggressive and lower-cost therapies [2]. It is estimated that globally in 2018, 1.28 million new cases of skin cancer were diagnosed, with 1 million corresponding to non-melanoma skin cancer and 280,000 to Malignant Melanoma (MM), which are associated with 65,000 and 60,000 deaths, respectively [3].
Improving the detection rate along with the use of drugs such as viniferin [4] and simvastatin [5] is important to reduce the mortality rate from this disease.
Currently, skin cancer is diagnosed by anatomical pathology analysis of the biopsy of each suspicious lesion, normally indicated by a dermatologist. Typically, a suspicious lesion is analyzed by a dermatologist by performing visual inspection based on Asymmetry, Border, Color, Diameter and Evolution over time using the ABCDE rule [6]. In addition, the specialist considered the patient's medical history and other factors to determine if the lesion was probably cancerous and referred the patient for biopsy.
Biopsy is a surgical intervention; therefore, it is an invasive and expensive procedure. Normally, there is a waiting list for patients requiring biopsy because of the limited availability of both specialized professionals and resources. Moreover, several patients are referred for biopsy solely because the practitioner may want to avoid false-negative identification of some lesions [7]. Several technological approaches have been developed to optimize resources and minimize the number of patients for whom biopsies can be avoided. These technologies include dermoscopy [8], developments based on thermography [9], [10], [11], [12], [13], [14], [15], visible image analysis systems [16], [17], [18], [19], [20], [21], ultrasound-based tools [22], [23], [24], and multispectral digital skin lesion analysis [25].
Dermoscopy is commonly performed by dermatologists. This technique enables better visualization of the spatial patterns and structures of lesions. When compared with naked-eye evaluation, it increases the sensitivity from 71% to 90% and the specificity from 81% to 90%. Moreover, dermoscopy decreases the biopsy ratio from 15:1 (1 of 15 exams corresponds to MM, while the others are benign lesions) to 5:1 [3]. A key drawback is that specialists require years of training to achieve this performance. Motivated by the importance of improving diagnosis, several tools have been developed based on visible image analysis and infrared thermography (IRT).
Technologies that analyze visible images have been of great interest to the scientific community in recent years. Various systems have been developed to assist in the diagnosis of skin cancer using machine learning techniques, such as support vector machines, artificial neural networks, and K-nearest neighbors [26]. Currently, deep learning techniques are the most commonly used because of the successful processing of visible-range images. There are different deep learning approaches that involve pre-processing stages, such as artifact removal, contrast enhancement, and the use of different color spaces [27]. Deep learning approaches have proven to be effective in two main areas: lesion segmentation and classification of skin cancer lesions [28], [29], [30], [27]. First, the results reported in the literature demonstrate that deep learning can be effectively used to detect the contour of lesions versus the surrounding skin. The similarity reaches a Jaccard index of up to 0.9 [31], which is relevant in the proposed work, as explained in Section 2.4.
Second, outstanding results have been obtained by classifying skin lesions, particularly when using strategies that involve pre-trained networks, exceeding 90% accuracy [18], [19], [32], [33], [27], [34], [35]. However, most of these studies focused on the problem of detecting melanoma lesions, unlike this study, which sought to detect any type of malignant lesions.
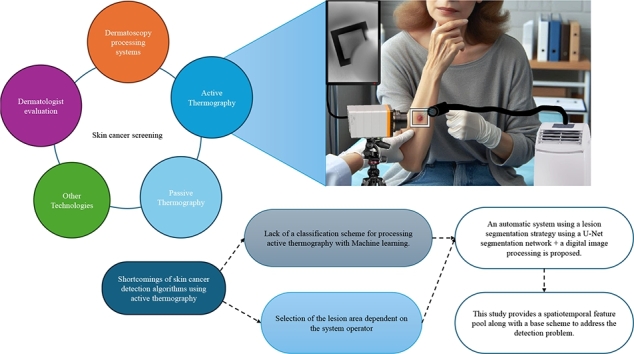
Another approach explored for skin cancer detection is the use of IRT, which can be either passive or active [36]. The main difference between these techniques is that active thermography requires a thermal stimulus that produces a thermal contrast between the area of interest and background. Passive thermography is applied to problems where the area of interest is naturally at a higher or lower temperature with respect to the background [37]. Passive IRT has not been successful in detecting skin cancer lesions [38], and no developments related to skin cancer have been observed using this technology until a few years ago. Through digital image processing techniques and statistical analysis, it was determined that passive thermography images can distinguish cancerous from healthy skin areas [39], [40]. For example, Magalhaes et al. [15] proposed a deep learning configuration to process passive IRT. This network could differentiate melanoma from nevi and melanoma from non-melanoma skin cancer lesions with precisions of 96.65% and 92.59%, respectively. However, the performance was inferior when working on the problem of distinguishing benign from malignant lesions, with a sensitivity of 62.81% and specificity of 57.58%. In contrast, active IRT has been successful in the detection of skin cancer [9], [10], [11], [12], [13], [14], with sensitivity and specificity of up to 99% in sensitivity and specificity [13]. Another advantage of active IRT is that it provides useful information for lesion modeling, which provides more insight into lesion morphology. Verstockt et al. [41] proposed lesion modeling using the finite element method (FEM), technique that has been successful in modeling different structures present in the human body [42], [43], [44]. Skin cancer detection can be considered solved based on the results reported by Godoy et al. [13]. However, this method and other methods based on active thermography require expert knowledge of the selection of lesion and non-lesion areas, as these systems are sensitive to the operator. Moreover, machine-learning techniques have not been well explored in the detection of skin cancer using active thermography, probably owing to the key aspects of data pre-processing for successful detection.
In this study, an automatic image-based skin cancer detection scheme is proposed for a real clinical environment, where lower-quality images, partially blurred areas owing to patient movement, poor illumination, and similar factors are common. This scheme is intended to boost development using infrared (IR) technology, as IR technology development currently provides low-cost technology suitable for clinical use. This scheme provides a key pre-processing stage for the extraction of discriminative features, along with the proposal of a pool of features that can be used with any machine learning technique, such as a support vector machine (SVM) classifier. Another novel point addressed in this work is the use of a convolutional neural networks (CNN) U-Net to segment the lesion area, which was trained to work with low-quality imagery. This was performed to assess the performance of the algorithm in the worst-case scenario.
2. Materials and methods
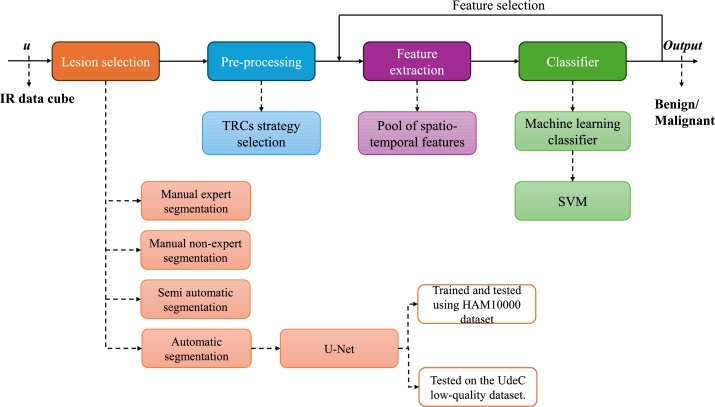
The proposed detection scheme addresses two problems in the area of study: the systems are not automatic, so the results vary depending on who operates them, and there is a lack of exploration of machine-learning techniques. In this study, an automatic system for skin cancer detection is proposed, which is a base scheme for exploring machine learning techniques. A diagram of the methodology used is presented in Fig. 1, which is composed of four blocks: Lesion selection, Pre-processing, Feature extraction, and Classifier. Lesion selection addresses the automatic detection of the lesion area using a U-Net segmentation CNN, which was trained and tested with the high-quality dermoscopic dataset HAM10000 and was then tested with the low-quality visible image set UdeC, which is the target dataset. The segmentation quality was measured with respect to expert manual segmentation, non-expert manual segmentation, and the semi-automatic segmentation method proposed by Diaz et al. [45]. The next block corresponds to Pre-processing, which corresponds to the selection considerations of features to be processed, based on the work proposed by Godoy et al. [12]. Feature extraction corresponds to the computation of the parameters that are then processed by the classifier. In this study, a large set of features is proposed, which are described in Section 2.3. Finally, there is a classifier in which any machine-learning technique can be used. In this study, an SVM classifier was used because in a previous study [46], the classifier that reported the best performance. In this scheme, a feedback loop was observed because, in this study, we searched for the best combination of up to five features.
Figure 1.
Diagram of the methodology used in the study.
2.1. Datasets
The development of this work considered the use of two datasets: the HAM10000 [47] set, which was used for training and validating the CNN. The HAM10000 dataset contains 10,015 dermoscopic images, of which 3,327 were selected as belonging to common skin cancer categories, namely, lesions declared as MM, Basal Cell Carcinoma (BCC), and benign. Second, a dataset called the UdeC set was used for the testing. This dataset is private and was acquired by one of the authors of his doctoral research with the support of the University of New Mexico Dermatology Clinic staff. The data were captured with informed consent, which did not allow public release. As explained below, the UdeC set contained lower-quality, poorly illuminated imagery acquired in clinical trials conducted by our team. The UdeC set contained 144 image cubes of size , where K is the number of images contained in the cube, M is the number of rows, and N is the number of columns in each picture. of K images are IR images corresponding to the thermal recovery of the skin after cold excitation. The remaining image corresponds to a visible image that was captured using a smartphone, unlike the HAM10000 set. The spatial dimensions of the data were variable because the size of the lesions was variable. A lesion typically involves an area of 5x5 cms., which corresponds to images of 100x100 pixels, i.e., . The IR data were captured for 1.5 minutes at a rate of 60 frames per second, so each cube contained IR images. This set was composed of MM, BCC, Squamous Cell Carcinoma (SCC) and benign lesions. Visible images of the set were used to evaluate the performance of the segmentation algorithm by contrasting the segmentation with manually generated masks. Table 1 shows the distribution of the datasets according to lesion type.
Table 1.
Distribution of lesions captures from the HAM10000 and UdeC datasets according to type of lesion: Malignant Melanoma (MM), Basal Cell Carcinoma (BCC), Squamous Cell Carcinoma (SCC) and benign (BN).
| Dataset | MM | BCC | SCC | BN | Total |
|---|---|---|---|---|---|
| HAM10000 | 1113 | 514 | 0 | 1700 | 3327 |
| UdeC | 7 | 41 | 9 | 87 | 144 |
It is important to note that for all images in both sets, there is a mask that defines the lesion area. For the HAM10000 set, the mask was defined as the pigmented area of the skin, i.e., the area whose color stands out from the healthy skin. For the UdeC set, the lesion area may coincide with the pigmented area; however, the expert who made the masks considered the nature of the lesions; thus, sometimes he defined that the suspicious parts of the lesion encompassed more or less of the area than solely the pigmented area.
2.2. Skin-cancer detection algorithms
It has been shown that in the detection of skin cancer using active thermography, the comparison between a descriptive thermoregulation curve (TRC) from a healthy skin area with the descriptive TRC contained in the lesion area is key to obtaining good classification results. The algorithm proposed by Godoy et al. [12] provides a solid foundation for feature extraction and subsequent classification, with the key stage being the selection of TRCs to obtain features that facilitate discrimination between benign and malignant lesions. For this reason, it is proposed to use the pre-processing stage of this algorithm as a basis for detection (Section 2.2.1), incorporating new techniques for feature extraction and classification described in Section 2.2.2.
2.2.1. Euclidean distance detection algorithm
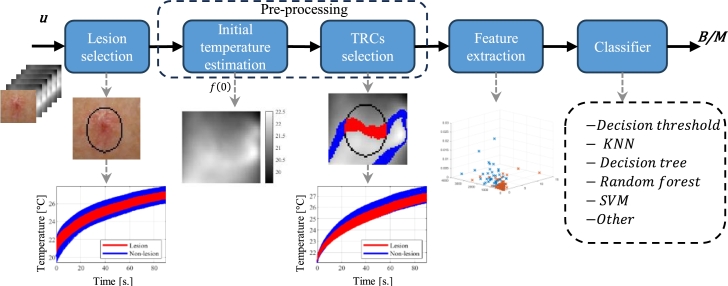
The Euclidean distance detection algorithm (ED) proposed by Godoy et al. [12] comprises the following five stages, as shown in Fig. 2.
-
1.
Lesion selection. A mask was generated over the visible image, which defined the lesion area. The original algorithm proposes creating a mask that manually encloses the pigmented area to generate the set L which contains the list of pixel locations of the lesion area (mask). With the remaining pixel locations of the visible image, another set, named N is created. It contains a list of pixel locations that are not within the lesion, but the surrounding skin.
-
2.Initial temperature estimation. The next stage corresponds to the selection of the TRCs, which depends on the initial temperature of each TRC. To select curves whose initial temperature is less affected by non-uniformities in the cooling process, the TRC of the -th pixel is modeled as a double exponential function by
whose parameters are computed using a non-linear least-squares fitting approach. With this model, the initial temperature is estimated by setting , i.e., .(1) -
3.
TRCs selection. The reference temperature is calculated as , where E is the expectation operator, and is the vector function of the initial temperature of the TRCs of the modeled lesion area within set L. Simply put, pixels within set L are selected, and the average initial temperature is calculated to determine .
The selection of points for processing by the algorithm considers a margin of error of with respect to . Thus, the set of points of interest for the algorithm is defined as
Experimentally was defined as a good value. Using the set S of all the location within a p% of , the sets of TRCs with similar initial temperature from the lesion and non-lesion areas are defined as and , respectively.(2) -
4.
Feature extraction. From the representative TRCs, a combination of features was extracted (feature vector). The detection algorithm proposed by Godoy et al. [12] uses only Euclidean distance as a feature to feed the next step. The formula used to compute this feature is given in Eq. (3).
-
5.
Classifier. In the final step, the feature vector is processed by a classifier that defines whether the lesion is suspicious. The original detection algorithm used a simple threshold to discriminate between malignant and benign lesions. This threshold was defined by the training set.
Figure 2.
Base detection scheme for skin cancer using active thermography and machine learning.
2.2.2. Proposed detection algorithm
The proposed detection algorithm considers the following 3 modifications to the algorithm described above:
-
1.
Lesion selection. The main drawback of the base algorithm is that when a non-expert selects the lesion the performance drops from 86% to 75%, as shown in Section 3.2. As such, the first proposed modification to use the CNN U-Net described in Section 2.4 to automatically select the lesion by the segmentation of the pigmented area.
-
2.
Feature extraction. As already mentioned, the base algorithm uses only one feature that accounts for the difference between the thermal responses. In Section 2.3, 32 feature extraction techniques are described, of which 31 were proposed in this study. Using the complete detection scheme, the feature vector that provides the highest accuracy was selected, considering a maximum of five features out of the 32 available.
-
3.
Classifier. Based on prior work, the SVM classifier with a radial basis function (RBF) kernel provides the best performance compared to techniques such as XGBoost and Random Forest for skin cancer detection using active thermography [46]. SVM works by finding a hyperplane that best separates the data points of different classes in a way that maximizes the margin, i.e., the distance between the hyperplane and the closest data points from both classes. The closest data points are called support vectors. The hyperplane's position and orientation are determined by the data distribution, and the SVM aims to find the hyperplane that minimizes the classification error and generalizes well to new, unseen data [48]. SVM classifiers are known for their ability to handle complex decision boundaries, their resistance to overfitting, and their effectiveness in high-dimensional spaces. Owing to the property of projecting the features onto a kernel such as RBF, it is possible to map the characteristics into a higher-dimensional space where the data might become separable or enhance their separability. As such, whereas the base algorithm only uses thresholding of one feature, here the best combination of up to five features selected by an SVM classifier was used.
2.3. Descriptive feature extracted from TRCs
Initially, Euclidean distance was a good discriminating parameter between benign and malignant cases of TRCs, as demonstrated by Godoy et al. [12]. However, when considering new data acquired after the publication of the article, this performance decreased from an accuracy of 89% to 60%. Therefore, new features are proposed in this study to improve the differentiation between malignant and benign cases in a more general manner.
From the representative TRC of the lesion () area and non-lesion area (), the following features were extracted:
-
1.Euclidean distance (d). This feature is calculated as the norm of the difference , normalized by the number of points, i.e., as shown in the following equation:
(3) The idea behind this feature is that if the Euclidean distance is small, the curves are similar, and it is very likely that the lesion is benign (i.e., the thermal recovery of the lesion behaves like normal tissue). Conversely, if the Euclidean distance is large, the curves show differences in thermal recovery, and the lesion is likely to be malignant because the lesion thermal recovery does not behave like a normal tissue [12].
-
2.Energy difference (). Let , i.e., the unbiased TRC of the area. The energy difference is calculated as follows:
(4) This feature is closely related to d. However, because of the triangular inequality , thus quantifies smaller differences than d.
-
3.
Rise time difference (). To calculate this characteristic, unbiased characteristic TRCs of each area were used. Over and the time at which 63.2% of the maximum amplitude is reached is calculated, which is referred to as and , respectively. The rise time difference was then calculated as .
-
4.Area difference (). Let and be the unbiased characteristic TRCs of lesion and non-lesion areas, respectively. The normalized area of a is calculated as:
(5) Then, was calculated as .
-
5.
Statistical similitude features. Here, six base features were defined to measure the similarity of a set of TRCs with a normalized modeled TRC using the dual exponential model shown in Eq. (1). It is assumed here that the modeled TRC has K sample points, and thus, . With this, it inner product is computed by , where and T is the column-vector transpose. was obtained by computing the five model parameters using a non-linear least-squares fitting approach. These parameters were averaged to obtain the descriptive TRC for each class (cancerous and non-cancerous TRCs). Then is forced to have a unit norm, i.e., .
Let and be a model TRC and an arbitrary TRC, respectively, both modeled and normalized. (Here, it was assumed that .) The characteristics of projection, correlation, and Euclidean distance are described below.-
a.Projection. The projection of onto is calculated as . This operation is performed for a set of at least 10 TRCs, and the mean projection, , and the standard deviation of the projection, , are then calculated over this set of projections.
-
b.Correlation. The correlation between and was calculated using the following expression:
where is the covariance between and , and and are the standard deviations (in the time samples) of and , respectively. This operation was performed for a set of at least 10 TRCs, and then the (the mean correlation) and (the standard deviation of the correlation) were calculated.(6) -
c.Distance. This parameter was calculated according to Eq. (3) but using as a reference and a family of at least 10 TRCs, . The mean value and standard deviation were calculated for this set of distance values.Considering a model of malignant and benign TRC, and , respectively, for each data cube, the projection characteristics and , correlation and , and distance and were calculated for the sets of TRCs and . In this manner, 24 features were extracted and grouped into four categories based on the origin of the model curve and the analysis group. For example, the malignant model over a non-lesion area allows the extraction , , , , and features.
-
a.
-
6.Difference in temperature at 20 seconds (). Let and be the characteristic TRCs of lesion and non-lesion areas, respectively. Each TRC is normalized and modeled as a simple exponential function , which is adjusted the least-squares method. The benign, , and malignant, , models are evaluated at to obtain the temperature value for each case. The difference in temperature is defined by
Experimentally, was chosen because after this time, the slope of change between malignant and benign TRCs starts to look alike. Thus, the difference in the time evolution of the thermal recovery was more easily appreciated at the chosen time.(7) -
7.
Principal component analysis (PCA) of the difference of the exponential model parameters. By using the double-exponential model shown in Eq. (1), the TRCs representative of the lesion and non-lesion areas were modeled to obtain sets of parameters and , respectively. The difference between these parameters is calculated as , generating a vector of five components.
Considering the training set, the dimension reduction technique PCA was applied to the five difference parameters Δθ. The covariance matrix was analyzed to reduce the dimensions of these five characteristics to three principal components: , and .
2.4. Skin lesion segmentation algorithm
As mentioned previously, the main drawback of the ED algorithm [12] is that it requires good manual selection of the lesion to perform well. The algorithm breaks down by wrongfully selecting the lesion tissue. As such, this work focuses on automatically identifying the lesions with respect to the surrounding skin to eliminate this weakness of the ED algorithm.
This is not a trivial problem to address because the segmentation algorithm to do so should work with low resolution visible-range images acquired in a clinical environment. Namely, the patient may move, and the angle between the lesion and the camera may not be the best. Moreover, the main problem with the proposed application is that the images are not acquired with controlled light, as they are commonly imaged in the literature.
In recent years, several algorithms have been reported in which a U-Net convolutional network or its variations are used to perform this task [49], [50], [31]. U-Net was specially designed to solve biomedical image-segmentation problems [51]. The network merges a convolutional network architecture with a deconvolutional architecture to generate a semantic segmentation. As described by Ashraf et al. [31]: It is an fully connected network that adds network layers to a traditional contracting network, replacing pooling operators with up-sampling operators to improve the resolution of the predicted image. A subsequent convolution layer assembles a more specific output for localization and backward propagation by concatenating high resolution features from up-sampled output and contraction path. The network can spread context information to higher resolution layers with up-sampling components, resulting in a u-shaped architecture. The architecture utilized in this study was a U-Net model with modifications proposed by Bisla et al. [52]. The main difference from the original U-Net is the consideration of up-sample stages and the use of dilatation kernels of size and . This allowed us to obtain an output with the same spatial resolution, without being clipped. To reiterate, these algorithms work well, but are trained and tested over high-quality imagery, which may not always be available in a clinical trial. Here, the U-Net is pushed to the limit so that it can operate under these extreme circumstances.
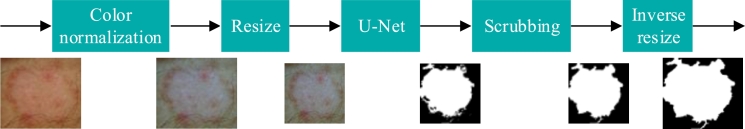
Fig. 3 shows the proposed segmentation scheme, which comprises the following five stages:
-
1.
Color normalization. Standardize the color conditions of images, which can be affected by illumination conditions and the use of different setups to acquire images. This study considers the use of the algorithm Shades of Gray [53], which has been shown to be effective in obtaining greater performance in the analysis of dermoscopic images [54].
-
2.
Resize. The size of the image was adjusted to dimensions 380×380, to meet the input conditions considered by the implemented U-Net network. As previously mentioned, images may have been acquired with different setups; therefore, image size standardization is necessary.
-
3.
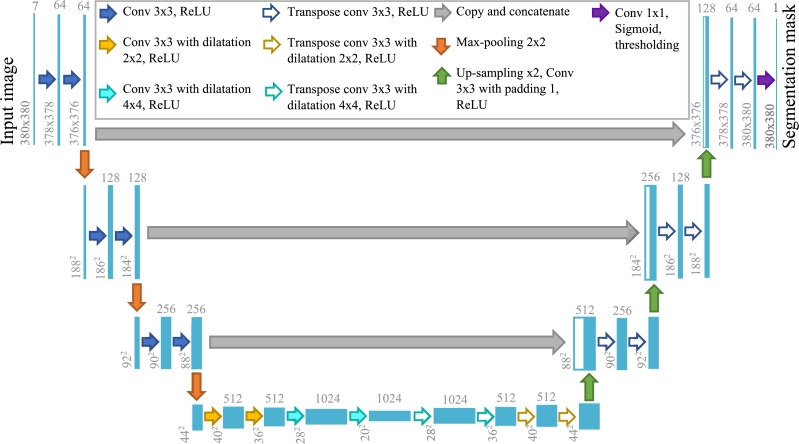
U-Net. Segmentation using a variant of the U-Net network proposed by Bisla et al. [52]. This network requires as input an RGB image concatenated with the transformation of the same image to HSV and L*a*b format, generating a cube of size . This size was considered in the design of the network because it is a representative size of the images of the UdeC set, which is the objective set to be segmented. This model has nine blocks and 21 convolutional layers as illustrated in Fig. 4.
-
4.
Scrubbing. Morphological dilation and erosion filters were applied to smooth the segmented area and fill the holes. An analysis by connected components was also carried out; in the event that there was more than one segmentation, the segment with the largest area was selected.
-
5.
Inverse resize. The output mask is adjusted to the original dimensions of the input image.
Figure 3.
Proposed segmentation scheme.
Figure 4.
Scheme of U-Net architecture proposed by Bisla et al. [52].
The segmentation algorithm was trained using the HAM10000 dataset [47], which was split by allocating 70% to train and to validate the algorithm, and the remaining 30% was used for testing. This proportion was selected for being a typical division ratio used in machine learning, which according to Nguyen et al. [55] reports better performance. As a final test, the algorithm was evaluated over the visible images of the UdeC dataset.
The training parameters of the U-Net network were summarized in Table 2. To obtain an algorithm that is robust to variations in lighting conditions and the use of different cameras, the following data augmentation techniques were used in the training stage: vertical and horizontal flip with a 50% probability of being flipped, adjustments shuffled brightness, shuffled saturation, and shuffled contrast, and blur was added using a Gaussian kernel filter of size and a variable standard deviation randomly selected between 0.1 and 20.
Table 2.
Detail of U-Net training parameters.
| Parameter | Value |
|---|---|
| Learning rate | 0.0001 |
| Optimizer | Adam |
| Function loss | BCE |
| Data augmentation | Vertical and horizontal flip, brightness, saturation, contrast and blurring adjustments |
| Maximum training iterations | 100 |
| Best performace iteration | 33 |
| Training/validation samples (HAM10000 dataset) | 2340 |
| Testing samples 1 (HAM10000 dataset) | 987 |
| Testing samples 2 (UdeC dataset) | 144 |
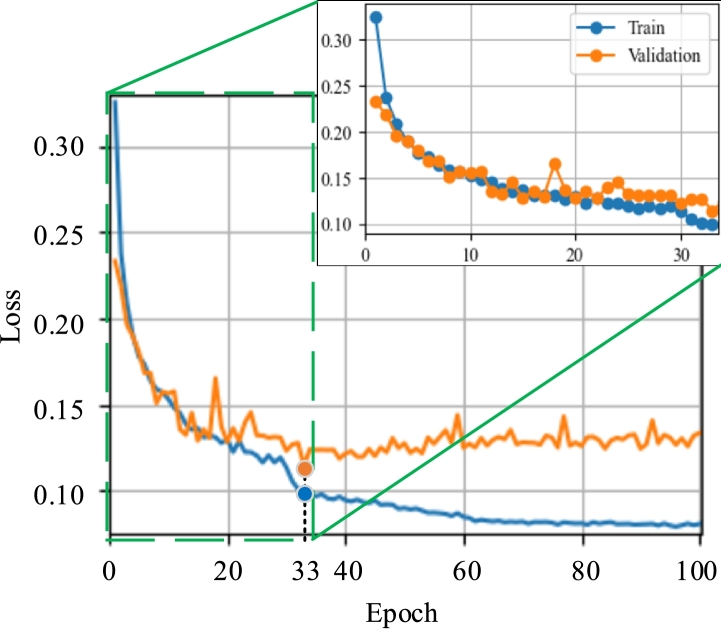
In the training process, different learning rates were evaluated, whose value varied by modifying r between 3 and 5 in steps of 1. The best performance was achieved by training the network with a learning rate of 0.0001, Adam's optimizing function [56], and the BCE loss function with sigmoid [57], adjusting the network for 33 epochs. The training and validation curves obtained over 100 epochs are shown in Fig. 5. Because 33 the result did not improve from epoch 33, it was decided not to continue with the training. It can be seen that after epoch, the loss in the training set was reduced. However, the validation loss increased, indicating that the network did not learn, thus overfitting the training set.
Figure 5.
Training and validation curves of U-Net network.
3. Results and discussion
3.1. Segmentation performance
As mentioned above, the segmentation algorithm was evaluated on two datasets: with the data partition of the HAM10000 set intended for testing and with the UdeC set. The well-known metrics of accuracy [58], sensitivity (also known as true positive rate (TPR)) [59], specificity (also known as true negative rate (TNR)) [59], precision (also known as positive predictive value (PPV)) [59] and Jaccard index [60] were used as performance measures.
Table 3 shows the performance of the algorithm on the HAM10000 and UdeC sets, reporting the aforementioned metrics. A considerable reduction in performance was observed when evaluating the segmentation of images of the UdeC set, decreasing the Jaccard index by 0.19 points and the precision by 15%. This is because the images of the UdeC set were captured under different lighting conditions, with a low-cost visible camera that, unlike a dermatoscope, is not made to provide the highest quality of clinical conditions.
Table 3.
Performance of the segmentation algorithm in terms of accuracy, sensitivity (TPR), specificity (TNR), precision (PPV) and Jaccard index.
| Dataset | Acc. | TPR | TNR | PPV | Jaccard |
|---|---|---|---|---|---|
| HAM10000 | 0.93 | 0.85 | 0.96 | 0.89 | 0.79 |
| UdeC | 0.98 | 0.82 | 0.99 | 0.74 | 0.60 |
Moreover, the HAM10000 set contained segmentations faithfully based on discriminating the pigmented area from the background, while the masks of the UdeC set were created considering expert knowledge, adjusting the masks to the criteria of a dermatologist, who determined that part of the lesion was malignant or benign. Because the segmentation algorithm was trained with the HAM10000 set, its performance was expected to be lower when evaluating masks created with expert criteria. To measure the impact of the proposed segmentation algorithm, using the UdeC set, the segmentation performance was contrasted with manual masks (non-experts) adjusted solely to the pigmented areas and masks created with the semi-automatic segmentation algorithm proposed by Díaz et al. [45], hereinafter referred to as the Díaz algorithm.
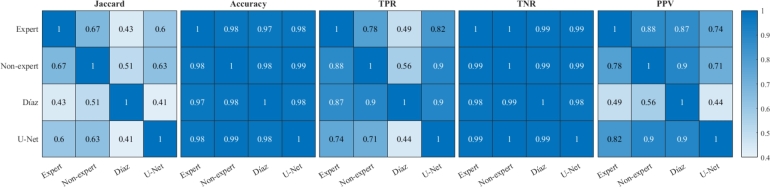
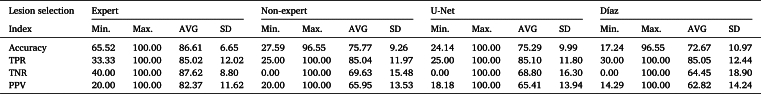
Fig. 6 shows the cross performance indices resulting from different segmentation techniques. In contrast to expert segmentation, it is observed that the highest Jaccard index is obtained by non-expert segmentation, with a value of 0.67, followed by U-Net segmentation with 0.60, with the worst performance obtained with the Díaz algorithm. Similarly, the best accuracy performance was obtained by non-expert segmentation with 98.26%, followed by the U-Net and Díaz segmentation algorithms with 98.00% and 97.37%, respectively. Regarding TPR, the best performance was obtained by U-Net segmentation with 82.02%, followed by inexperienced segmentation and Díaz segmentation with 77.71% and 48.58%, respectively. Whereas in specificity the worst result was obtained by the U-Net segmentation with 98.92%. This last result is consistent with the precision, in which the worst result was obtained by the U-Net segmentation. Thus, despite the fact that the U-Net segmentation presents a high specificity, the difference that exists with the other techniques generates a lower precision of approximately 15%, since most of the images present a much larger zone of no lesion than of injury, which means that generating a mask slightly larger than the reference produces a large drop in precision performance.
Figure 6.
Comparison of segmentation performance indices using different benchmarks. The rows represent the benchmarks and the columns are the evaluated masks.
As expected, the indices indicated that the U-Net segmentation algorithm is similar to non-expert segmentation, with the advantage of being a approach with standardized criteria, unlike manual segmentation, which depends on the criteria of each user, whose decision can be affected by conditions such as stress and fatigue, among others.
In relation to the multiple image processing stages that include the proposed segmentation scheme, the best result is obtained considering all stages, with a Jaccard index value of 0.5963. By deleting the scrubbing step, the index is reduced to 0.5894, while removing the color normalization and scrubbing steps, the index is reduced to 0.5721. The segmentation results are reflected in the detection performance, such that the complete scheme provides an Area Under Curve (AUC) of 0.792, whereas deleting the scrubbing step reduces the AUC to 0.778, and removing the color normalization and the scrubbing steps reduces the AUC to 0.759.
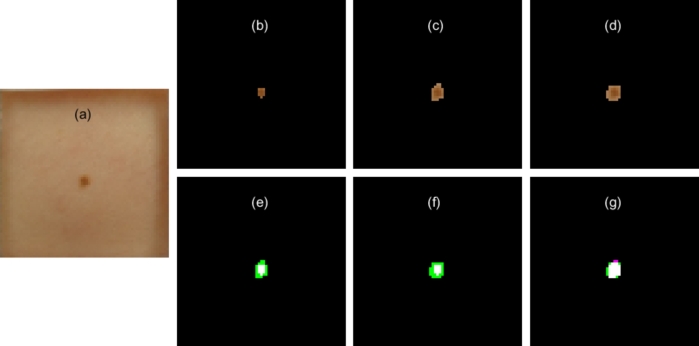
Fig. 7(a) shows an image of a lesion of the UdeC set, together with manual segmentation of the lesion using expert knowledge (Fig. 7(b)), manual non-expert segmentation (Fig. 7(c)), and the proposed automatic segmentation (Fig. 7(d)). It was observed that all the segmentation algorithms selected the lesion. It should be noted that the expert segmentation selects the inner area of the lesion, whereas the other segmentation output contains the lesion with a border of non-pigmented skin. However, the lesion area was small with respect to the total area of the image, which implies that small differences in the selection of the lesion negatively affect the TPR, PPV and Jaccard indices. This situation is characteristic in the UdeC set, where the lesion area covers an average of of the image. While with the HAM10000 set it addresses . Despite the fact that to the human eye the segmentation results are quite similar, the performance index used is highly reduced if the lesion is composed by few pixels. Considering an extreme case where lesion is composed by 5 pixels, not detecting one implies reducing the Jaccard index from 1 to 0.8. It could be worse if the segmentation algorithm generates an over or under segmentation with a 1-pixel border. Therefore, the fewer the pixels the lesion has, the worse the index when the algorithm mis-segments one pixel. This problem will always be present when the suspicious lesion is imaged with low quality sensors.
Figure 7.
Segmentation contrast. (a) Image with lesion to be segmented, (b) segmentation with expert knowledge, (c) manual non-expert segmentation, (d) automatic segmentation using U-Net network, (e) contrast of expert segmentation with non-expert segmentation, (f) expert segmentation contrast with automatic segmentation, (g) non-expert segmentation contrast with automatic segmentation.
3.2. Detection performance
The detection algorithms described in Section 2.2 were evaluated using the four lesion segmentation techniques previously described: (i) Expert, (ii) Non-expert, (iii) Díaz, and (iv) U-Net. Both expert and non-expert segmentations are manually performed, whereas Díaz is semi-automatic in the sense that it requires interaction with the user. In contrast, the proposed approach (U-Net) is fully automatic in the sense that it does not require any input from the user.
The proposed detection algorithm considers a novel feature-extraction stage that feeds an SVM classifier. From the set of features described in Section 2.3, a combination of up to five characteristics that provided the greatest accuracy was sought. When working with expert segmentation, the best feature combinations were . Whereas, with the other types of segmentations, the following feature vectors were selected: .
The performance and variability of the ED algorithm and proposed detection algorithm based on the new features were evaluated using the bootstrap method. This technique entails drawing multiple samples with replacements from the original dataset, creating a new dataset of the same size. The modified dataset was then divided by allocating 80% of the data for training and 20% for testing. The entire procedure was repeated 2,000 times such that the mean results were presented, as well as the minimum and maximum results for each approach.
Table 4 lists the performance of the ED detection algorithm on the active thermography dataset. Considering a sensitivity of 85% as a reference point, the best result was achieved with expert segmentation, followed by the U-Net, non-expert, and Díaz segmentation algorithms, with an average specificity of 42.72%, 32.39%, 28.90%, and 17.03%, respectively. Similarly, Table 5 shows the performance of the detection algorithm using a SVM detection algorithm; it is observed that the best result is obtained with the expert segmentation, followed by the non-expert, U-Net, and Díaz segmentation algorithms, with average specificities of 87.62%, 69.63%, 68.50%, and 64.45%, respectively.
Table 4.
Detection performance using Euclidean classifier and different lesion area selection techniques.
Table 5.
Detection performance using SVM classifier and different lesion area selection techniques.
Both detection algorithms exhibited their best performance using expert segmentation as input, followed by detection when non-expert and U-Net segmentation algorithms were used. The worst result was obtained using Díaz segmentation as input. Analyzing these results, it is evident that the selection of TRCs is crucial for the performance of skin cancer detection algorithms that process active thermography. These results indicate that the detection algorithm used, which is based on active thermography analysis, requires expert selection of the lesion area to obtain the best performance. Thus, expert knowledge is relevant for determining the key pigmented area to determine whether a lesion is benign or malignant.
By linking the performance of the detection algorithm using different lesion segmentation techniques with the performance in segmentation quality, it is observed that these are highly correlated. The proposed detection algorithm, when working with U-Net segmentation, exhibited superior performance compared to the Díaz segmentation algorithm, and similar performance to that of non-expert segmentation. Thus, it was demonstrated that accurate selection of the lesion area is relevant for successful detection. Thus, it is important to automate these types of algorithms so that they are not susceptible to the operator. Or, in contrast, utilize algorithms that does not use the time difference of the TRCs (or forms of it) as their discriminant factor.
Regarding the proposed modifications to feature extraction techniques and the detection algorithm, it is highlighted that the combination of the proposed features along with the SVM classification technique enables an increase in accuracy of 26.58%, 24.14%, 21.63%, and 28.16% when selecting the lesion area with expert, non-expert, U-Net, and Díaz segmentations, respectively. Thus, these modifications make the proposed detection algorithm less sensitive to segmentation differences than the ED algorithm. However, its performance suffers if the input segmentation is incorrect.
The presented results seem to be lacking in impact when compared to others that process dermatoscopic images, which achieved an accuracy higher than 95%, as shown in Table 6. However, they mainly addressed the classification MM vs. non-MM or MM vs. benign. It is important to mention that the proposed approach tackles the problem of detecting benign lesions vs. malignant lesions, i.e., all types of cancer are malignant cases for our approach. In this scenario, the detection of skin cancer is not trivial owing to the small variability between the two classes.
Table 6.
Comparison of related articles for detecting skin cancer using different types of imaging, differentiating the classification problems, where MM: Malignant melanoma; Malignant includes MM, Basal-cell carcinomas and Squamous-cell carcinomas.
| Reference | Methodology | Classification problem | Dataset | Technology | Performance |
|---|---|---|---|---|---|
| [61] | Nasnet Mobile with transfer learning | MM vs. non-MM | HAM10000 | Dermoscopy | 97.90% accuracy |
| [62] | MobileNetV2-based transfer learning | MM vs. benign | ISIC2020 | Dermoscopy | 98.20% accuracy |
| [63] | CNN DenseNet-121 with multi-layer perceptron (MLP) | MM vs. non-MM | ISIC 2016, ISIC 2017, PH2 y HAM10000 | Dermoscopy | Accuracy of 98.33%, 80.47%, 81.16% and 81.00% on PH2, ISIC 2016, ISIC 2017 and HAM10000 datasets. |
| Own implementation | ResNet152V2-based transfer learning | MM vs. benign | HAM10000 | Dermoscopy | 90.63% accuracy |
| Own implementation | ResNet152V2-based transfer learning | MM vs. benign | UdeC | Low-quality visible images | 72.49% accuracy |
| [15] | Deep learning | MM vs.benign | Passive IRT | 96.91% accuracy | |
| [15] | Deep learning | Malignant vs.benign | Passive IRT | 57.58% accuracy | |
| Proposed method using automatic U-Net segmentation | Machine learning | Malignant vs.benign | UdeC | Active IRT | 75.29% accuracy |
| Proposed method using manual expert segmentation | Machine learning | Malignant vs.benign | UdeC | Active IRT | 86.61% accuracy |
This was reflected in the work of Magalhaes et al. [15], who obtained 62.81% sensitivity and 57.58% specificity. Their study is one of the most recent approaches in the literature that uses thermography. The performance reported in this manuscript surpasses that of Magalhaes, achieving an 85% sensitivity and 69% specificity when working with automatic segmentation and an SVM classifier, respectively.
By implementing our own classifier based on a ResNet152V2 network, which was trained using the HAM10000 dataset in conjunction with the U-Net network, the ResNet152V2 achieved an accuracy of 90.63% on the HAM10000 test set and 72.49% on the UdeC visible image test set. This result demonstrates the great potential of transfer learning techniques. However, their performance declines by almost 20% when processing poorer quality data, obtained in a clinical environment with limited resources. This result is concordant with the study by Akilandasowmya et al. [35], which showed high variability in the performance of systems processing transfer learning-based dermoscopy images by varying the training set sizes.
4. Conclusions
This study presents two improvements to a previously published detection algorithm, resulting in the first automatic skin cancer detection algorithm that processes active thermography. The first improvement involved automating the lesion area selection stage using U-Net CNN. The second improvement involves a new selection of features to be classified, which are processed using an SVM-based algorithm.
Regarding the lesion segmentation algorithm, training was performed using a set of dermoscopic images with the aim of segmenting lower-quality images, which were acquired with standard smartphone cameras under varying degrading conditions. The algorithm presented a Jaccard index of 0.79 using dermoscopic images, a value similar to that reported by other authors. As expected, when evaluating a set generated with a smartphone camera under varying acquisition conditions, the performance dropped to 0.60. The performance on the target image set is considered satisfactory because it achieved 0.07 Jaccard points lower than that obtained using non-expert segmentation, which is based on pigmentation. It is consistent with the fact that the training dataset (HAM10000) considers pigmented pixels as lesions. This drop was consistent with the degraded resolution, lower illumination, and blurred areas of the test dataset. Even though this drop is significant, the segmentation result that can be performed without the training of a dermatologist.
The performances of both detection algorithms were evaluated using expert, non-expert, and Díaz (semi-automatic algorithm) segmentation. Considering a sensitivity of 85%, both detection algorithms showed their best performance with expert segmentation and the worst with Díaz segmentation, whereas U-Net segmentation provided a detection performance similar to that of non-expert segmentation.
The use of machine learning in classification increased the detection accuracy, reaching values of 86.61%, 75.77%, 75.29%, and 72.67% when using expert, non-expert, U-Net, and Díaz lesion selection, respectively. All of these results outperformed the original detection algorithm, which achieved an accuracy of 60.03%.
When comparing the performance of the automatic detection system with that of the ResNet152V2 network on the UdeC visible image set, the thermography-based approach demonstrated a 3% increase in the accuracy. However, when using expert segmentation, the thermography system outperformed it by nearly 14 percentage points in accuracy. These results indicate that the proposed automatic detection system has significant potential for further improvement.
This leads to the conclusion that the main problem with the proposed automatic algorithm is that the automatic segmentation fits the pigmented area of the skin very well, which is correct according to the HAM10000 set. However, there are differences with the expert criterion, which considers knowledge of the nature of the lesion, such that sometimes the lesion area does not necessarily match only the pigmented area, meaning that the actual lesion may be larger or smaller than the pigmented area. Thus, the pigmented area is only a projection of the lesion over the skin surface, and its malignancy may spread below the surface in a random fashion.
The difference between selecting only the pigmented area with respect to the expert criterion implies a low accuracy of the detection algorithm of approximately 11%, owing to the manner in which the classifiers were designed. This implies the detection of more benign and malignant cases, that is, when using the automatic segmentation, more biopsies would be requested than when using the segmentation performed by an expert. To improve this aspect, it is necessary to teach the segmentation network about segmentations performed using expert criteria, which is the main limitation of the proposed algorithm. As there is no set of hundreds of lesions labeled with expert criteria, it is not yet possible to teach this knowledge to the network, but we are currently working on it.
We consider that skin cancer detection using active thermography is still relatively unexplored, possibly due to the challenges involved in addressing the detection problem with this technology, such as the high cost that this technology implied some years ago, and the knowledge required to successfully process active thermography data. Regarding processing, we consider the judicious selection of TRCs crucial for more successful discrimination between benign and malignant cases. Therefore, this study proposes a generic framework for skin cancer detection using active thermography. Owing to the high cost of IR technology, in the last decade, low-cost microbolometer technology has emerged, which may have lower image quality, but makes it feasible to transfer the technology developed for skin cancer detection.
It is expected that by using more robust detection algorithms, such as deep-learning algorithms over dynamic infrared imagery, their performance will be less affected by the lesion area selection stage. This is an ongoing research by our group, and we expect to report a novel detection algorithm that uses only deep-learning techniques within a few months. The purpose of this novel algorithm is to work with lower-quality IR images acquired using low-cost equipment.
Ethics declarations
In this study, two datasets captured from humans were used. The first dataset corresponds to the public dataset HAM10000, which is protected by a Creative Commons Attribution-NonCommercial International Public License 4.0.
The other dataset, called UdeC dataset was prepared by S. E. Godoy in his doctoral research with the support of the University of New Mexico Dermatology Clinic staff. This dataset was captured by the project “Functional infrared imaging for melanoma in patients”, which was conducted in accordance with the Declaration of Helsinki, and approved by the Human Research Review Committee of the University of New Mexico (protocol code 10-304 approved on 15-Oct-2010 and renewed annually until the year 2013).
CRediT authorship contribution statement
Ricardo F. Soto: Writing – original draft, Validation, Software, Methodology, Investigation, Formal analysis, Conceptualization. Sebastián E. Godoy: Writing – review & editing, Supervision, Resources, Conceptualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This research was funded by the National Agency of Research and Development (ANID) of Chile under the doctoral scholarship PCHA/Doctorado Nacional Folio 2019-21191485.
Data availability
The dataset HAM10000 is public and available on the Harvard Dataverse website. The other dataset used in this study is of a private nature, due to the confidentiality restrictions imposed by the ethics committee.
References
- 1.Wild C., Weiderpass E., Stewart B.W. IARC Press; 2020. World Cancer Report: Cancer Research for Cancer Prevention. [PubMed] [Google Scholar]
- 2.Sabatini-Ugarte N., Molgó M., Vial G. Melanoma en Chile ¿Cuál es nuestra realidad? Rev. Med. Clin. Las Condes. 2018;29(4):468–476. doi: 10.1016/j.rmclc.2018.06.006. [DOI] [Google Scholar]
- 3.Dorrell D.N., Strowd L.C. Skin cancer detection technology. Dermatol. Clin. 2019;37(4):527–536. doi: 10.1016/j.det.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 4.Fuloria S., Sekar M., Khattulanuar F.S., Gan S.H., Rani N.N.I.M., Ravi S., Subramaniyan V., Jeyabalan S., Begum M.Y., Chidambaram K., Sathasivam K.V., Safi S.Z., Wu Y.S., Nordin R., Maziz M.N.H., Kumarasamy V., Lum P.T., Fuloria N.K. Chemistry, biosynthesis and pharmacology of viniferin: potential resveratrol-derived molecules for new drug discovery, development and therapy. Molecules. 2022;27(16) doi: 10.3390/molecules27165072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malviya R., Raj S., Fuloria S., Subramaniyan V., Sathasivam K., Kumari U., Unnikrishnan Meenakshi D., Porwal O., Hari Kumar D., Singh A., Chakravarthi S., Kumar Fuloria N. Evaluation of antitumor efficacy of chitosan-tamarind gum polysaccharide polyelectrolyte complex stabilized nanoparticles of simvastatin. Int. J. Nanomed. 2021;16:2533–2553. doi: 10.2147/IJN.S300991. https://doi.org/10.2147/IJN. S300991 pMID: 33824590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rigel D.S., Friedman R.J., Kopf A.W., Polsky D. Abcde—an evolving concept in the early detection of melanoma. Arch. Dermatol. 2005;141(8):1032–1034. doi: 10.1001/archderm.141.8.1032. [DOI] [PubMed] [Google Scholar]
- 7.Bibbins-Domingo K., Grossman D.C., Curry S.J., Davidson K.W., Ebell M., Epling J.W., García F.A., Gillman M.W., Kemper A.R., Krist A.H., et al. Screening for skin cancer: US preventive services task force recommendation statement. JAMA. 2016;316(4):429–435. doi: 10.1001/jama.2023.4342. [DOI] [PubMed] [Google Scholar]
- 8.Yélamos O., Braun R.P., Liopyris K., Wolner Z.J., Kerl K., Gerami P., Marghoob A.A. Usefulness of dermoscopy to improve the clinical and histopathologic diagnosis of skin cancers. J. Am. Acad. Dermatol. 2019;80(2):365–377. doi: 10.1016/j.jaad.2018.07.072. [DOI] [PubMed] [Google Scholar]
- 9.Buzug T.M., Schumann S., Pfaffmann L., Reinhold U., Ruhlmann J. 2006 International Conference of the IEEE Engineering in Medicine and Biology. Society, IEEE; 2006. Functional infrared imaging for skin-cancer screening; pp. 2766–2769. [DOI] [PubMed] [Google Scholar]
- 10.Çetingül M.P., Herman C. Quantification of the thermal signature of a melanoma lesion. Comput. Therm. Sci. 2011;50(4):421–431. doi: 10.1016/j.ijthermalsci.2010.10.019. [DOI] [Google Scholar]
- 11.Di Carlo A., Elia F., Desiderio F., Catricalà C., Solivetti F.M., Laino L. Can video thermography improve differential diagnosis and therapy between basal cell carcinoma and actinic keratosis? Dermatol. Ther. 2014;27(5):290–297. doi: 10.1111/dth.12141. [DOI] [PubMed] [Google Scholar]
- 12.Godoy S.E., Ramirez D.A., Myers S.A., von Winckel G., Krishna S., Berwick M., Padilla R.S., Sen P., Krishna S. Dynamic infrared imaging for skin cancer screening. Infrared Phys. Technol. 2015;70:147–152. doi: 10.1016/j.infrared.2014.09.017. [DOI] [Google Scholar]
- 13.Godoy S.E., Hayat M.M., Ramirez D.A., Myers S.A., Padilla R.S., Krishna S. Detection theory for accurate and non-invasive skin cancer diagnosis using dynamic thermal imaging. Biomed. Opt. Express. 2017;8(4):2301–2323. doi: 10.1364/BOE.8.002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magalhaes C., Vardasca R., Mendes J. Recent use of medical infrared thermography in skin neoplasms. Skin Res. Technol. 2018;24(4):587–591. doi: 10.1111/srt.12469. [DOI] [PubMed] [Google Scholar]
- 15.Magalhaes C., Tavares J.M.R., Mendes J., Vardasca R. Comparison of machine learning strategies for infrared thermography of skin cancer. Biomed. Signal Process. Control. 2021;69 doi: 10.1016/j.bspc.2021.102872. [DOI] [Google Scholar]
- 16.Nasr-Esfahani E., Samavi S., Karimi N., Soroushmehr S.M.R., Jafari M.H., Ward K., Najarian K. 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology. Society (EMBC), IEEE; 2016. Melanoma detection by analysis of clinical images using convolutional neural network; pp. 1373–1376. [DOI] [PubMed] [Google Scholar]
- 17.Kawahara J., BenTaieb A., Hamarneh G. 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI) IEEE; 2016. Deep features to classify skin lesions; pp. 1397–1400. [DOI] [Google Scholar]
- 18.Dorj U.-O., Lee K.-K., Choi J.-Y., Lee M. The skin cancer classification using deep convolutional neural network. Multimed. Tools Appl. 2018;77(8):9909–9924. doi: 10.1007/s11042-018-5714-1. [DOI] [Google Scholar]
- 19.Rajasekhar K., Babu T.R. Skin lesion classification using convolution neural networks. Indian J. Publ. Health Res. Dev. 2019;10(12) doi: 10.37506/v10/i12/2019/ijphrd/192205. [DOI] [Google Scholar]
- 20.Rezvantalab A., Safigholi H., Karimijeshni S. Dermatologist level dermoscopy skin cancer classification using different deep learning convolutional neural networks algorithms. 2018. arXiv:1810.10348 arXiv preprint.
- 21.Acosta M.F.J., Tovar L.Y.C., Garcia-Zapirain M.B., Percybrooks W.S. Melanoma diagnosis using deep learning techniques on dermatoscopic images. BMC Med. Imaging. 2021;21(1):1–11. doi: 10.1186/s12880-020-00534-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wortsman X., Wortsman J. Clinical usefulness of variable-frequency ultrasound in localized lesions of the skin. J. Am. Acad. Dermatol. 2010;62(2):247–256. doi: 10.1016/j.jaad.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 23.Meyer N., Lauwers-Cances V., Lourari S., Laurent J., Konstantinou M.-P., Lagarde J.-M., Krief B., Batatia H., Lamant L., Paul C. High-frequency ultrasonography but not 930-nm optical coherence tomography reliably evaluates melanoma thickness in vivo: a prospective validation study. Br. J. Dermatol. 2014;171(4):799–805. doi: 10.1111/bjd.13129. [DOI] [PubMed] [Google Scholar]
- 24.Kozárová A., Kozar M., Tonhajzerová I., Pappova T., Minariková E. The value of high-frequency 20 MHz ultrasonography for preoperative measurement of malignant melanoma thickness. Acta Dermatovenerol. Croat. 2018;26(1):15–20. [PubMed] [Google Scholar]
- 25.Rigel D.S., Roy M., Yoo J., Cockerell C.J., Robinson J.K., White R. Impact of guidance from a computer-aided multispectral digital skin lesion analysis device on decision to biopsy lesions clinically suggestive of melanoma. Arch. Dermatol. 2012;148(4):541–543. doi: 10.1001/archdermatol.2011.3388. [DOI] [PubMed] [Google Scholar]
- 26.Hameed N., Ruskin A., Hassan K.A., Hossain M.A. 2016 10th International Conference on Software, Knowledge, Information Management & Applications (SKIMA) IEEE; 2016. A comprehensive survey on image-based computer aided diagnosis systems for skin cancer; pp. 205–214. [DOI] [Google Scholar]
- 27.Zafar M., Sharif M.I., Sharif M.I., Kadry S., Bukhari S.A.C., Rauf H.T. Skin lesion analysis and cancer detection based on machine/deep learning techniques: a comprehensive survey. Life. 2023;13(1) doi: 10.3390/life13010146. https://www.mdpi.com/2075-1729/13/1/146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adegun A., Viriri S. Deep learning techniques for skin lesion analysis and melanoma cancer detection: a survey of state-of-the-art. Artif. Intell. Rev. 2021;54(2):811–841. doi: 10.1007/s10462-020-09865-y. [DOI] [Google Scholar]
- 29.Dildar M., Akram S., Irfan M., Khan H.U., Ramzan M., Mahmood A.R., Alsaiari S.A., Saeed A.H.M., Alraddadi M.O., Mahnashi M.H. Skin cancer detection: a review using deep learning techniques. Int. J. Environ. Res. Public Health. 2021;18(10):5479. doi: 10.3390/ijerph18105479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alzubaidi L., Zhang J., Humaidi A.J., Al-Dujaili A., Duan Y., Al-Shamma O., Santamaría J., Fadhel M.A., Al-Amidie M., Farhan L. Review of deep learning: concepts, cnn architectures, challenges, applications, future directions. J. Big Data. 2021;8(1):1–74. doi: 10.1186/s40537-021-00444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashraf H., Waris A., Ghafoor M.F., Gilani S.O., Niazi I.K. Melanoma segmentation using deep learning with test-time augmentations and conditional random fields. Sci. Rep. 2022;12(1):1–16. doi: 10.1038/s41598-022-07885-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar M., Alshehri M., AlGhamdi R., Sharma P., Deep V. A de-ann inspired skin cancer detection approach using fuzzy c-means clustering. Mob. Netw. Appl. 2020;25:1319–1329. doi: 10.1007/s11036-020-01550-2. [DOI] [Google Scholar]
- 33.Damarla A., Doraikannan S. Optimized one-shot neural architecture search for skin cancer classification. J. Electron. Imaging. 2022;31(6) doi: 10.1117/1.JEI.31.6.063053. [DOI] [Google Scholar]
- 34.Singh S., Pilania U., Kumar M., Awasthi S.P. 2023 3rd International Conference on Intelligent Technologies (CONIT) IEEE; 2023. Image processing based skin cancer recognition using machine learning; pp. 1–7. [DOI] [Google Scholar]
- 35.Akilandasowmya G., Nirmaladevi G., Suganthi S., Aishwariya A. Skin cancer diagnosis: leveraging deep hidden features and ensemble classifiers for early detection and classification. Biomed. Signal Process. Control. 2024;88 doi: 10.1016/j.bspc.2023.105306. [DOI] [Google Scholar]
- 36.Lahiri B., Bagavathiappan S., Jayakumar T., Philip J. Medical applications of infrared thermography: a review. Infrared Phys. Technol. 2012;55(4):221–235. doi: 10.1016/j.infrared.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verstockt J., Verspeek S., Thiessen F., Tjalma W.A., Brochez L., Steenackers G. Skin cancer detection using infrared thermography: measurement setup, procedure and equipment. Sensors. 2022;22(9) doi: 10.3390/s22093327. https://www.mdpi.com/1424-8220/22/9/3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cristofolini M., Perani B., Piscioli F., Recchia G., Zumiani G. Uselessnes of thermography for diagnosis and follow-up of cutaneous malignant melanoma. Tumori J. 1981;67(2):141–143. doi: 10.1177/030089168106700211. [DOI] [PubMed] [Google Scholar]
- 39.Shaikh S., Akhter N., Manza R. Application of image processing techniques for characterization of skin cancer lesions using thermal images. Indian J. Sci. Technol. 2016;9(15):1–7. doi: 10.17485/ijst/2016/v9i15/89794. [DOI] [Google Scholar]
- 40.Benjumea E., Morales Y., Torres C., Vilardy J. vol. 1221. IOP Publishing; 2019. Characterization of Thermographic Images of Skin Cancer Lesions Using Digital Image Processing; p. 012076. (Journal of Physics: Conference Series). [DOI] [Google Scholar]
- 41.Verstockt J., Somers R., Thiessen F., Hoorens I., Brochez L., Steenackers G. Finite element skin models as additional data for dynamic infrared thermography on skin lesions. Quant. InfraRed Thermogr. J. 2023;0(0):1–20. doi: 10.1080/17686733.2023.2256998. [DOI] [Google Scholar]
- 42.Güvercin Y., Yaylacı M., Dizdar A., Kanat A., Yaylacı E.U., Ay S., Abdioğlu A.A., Şen A. Biomechanical analysis of odontoid and transverse atlantal ligament in humans with ponticulus posticus variation under different loading conditions: finite element study. Injury. 2022;53(12):3879–3886. doi: 10.1016/j.injury.2022.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Zagane M.E.S., Abdelmadjid M., Yaylacı M., Abderahmen S., Yaylacı E.U. Finite element analysis of the femur fracture for a different total hip prosthesis (Charnley, Osteal, and Thompson) Struct. Eng. Mech. Int. J. 2023;88(6):583–588. doi: 10.12989/sem.2023.88.6.583. [DOI] [Google Scholar]
- 44.Kurt A., Yaylacı M., Dizdar A., Naralan M.E., Yaylacı E.U., Öztürk Ş., Çakır B. Evaluation of the effect on the permanent tooth germ and the adjacent teeth by finite element impact analysis in the traumatized primary tooth. Int. J. Paediatr. Dent. 2024;34(6):822–831. doi: 10.1111/ipd.13183. [DOI] [PubMed] [Google Scholar]
- 45.Díaz S., Krohmer T., Moreira Á., Godoy S.E., Figueroa M. An instrument for accurate and non-invasive screening of skin cancer based on multimodal imaging. IEEE Access. 2019;7:176646–176657. doi: 10.1109/ACCESS.2019.2956898. [DOI] [Google Scholar]
- 46.Soto R.F., Godoy S.E. 2023 IEEE Latin American Conference on Computational Intelligence (LA-CCI) IEEE; 2023. A novel feature extraction approach for skin cancer screening using active thermography; pp. 1–6. [DOI] [Google Scholar]
- 47.Tschandl P., Rosendahl C., Kittler H. The ham10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data. 2018;5(1):1–9. doi: 10.1038/sdata.2018.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cortes C., Vapnik V. Support vector machine. Mach. Learn. 1995;20(3):273–297. doi: 10.1007/BF00994018. [DOI] [Google Scholar]
- 49.Seeja R., Suresh A. Deep learning based skin lesion segmentation and classification of melanoma using support vector machine (svm) Asian Pac. J. Cancer Prev. 2019;20(5):1555. doi: 10.31557/APJCP.2019.20.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nathan S., Kansal P. Lesion net–skin lesion segmentation using coordinate convolution and deep residual units. 2020. arXiv:2012.14249 arXiv preprint.
- 51.Ronneberger O., Fischer P., Brox T. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [DOI] [Google Scholar]
- 52.Bisla D., Choromanska A., Stein J.A., Polsky D., Berman R. Skin lesion segmentation and classification with deep learning system. 2019. arXiv:1902.06061 arXiv preprint. pp. 1–6.
- 53.Finlayson G.D., Trezzi E. vol. 2004. Society for Imaging Science and Technology; 2004. Shades of Gray and Colour Constancy; pp. 37–41.https://ueaeprints.uea.ac.uk/id/eprint/23682 (Color and Imaging Conference). [Google Scholar]
- 54.Barata C., Celebi M.E., Marques J.S. Improving dermoscopy image classification using color constancy. IEEE J. Biomed. Health Inform. 2015;19(3):1146–1152. doi: 10.1109/JBHI.2014.2336473. [DOI] [PubMed] [Google Scholar]
- 55.Nguyen Q.H., Ly H.-B., Ho L.S., Al-Ansari N., Le H.V., Tran V.Q., Prakash I., Pham B.T. Influence of data splitting on performance of machine learning models in prediction of shear strength of soil. Math. Probl. Eng. 2021;2021 doi: 10.1155/2021/4832864. [DOI] [Google Scholar]
- 56.Kingma D.P., Ba J. Adam: a method for stochastic optimization. 2014. arXiv:1412.6980 arXiv preprint.
- 57.Jadon S. 2020 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB) IEEE; 2020. A survey of loss functions for semantic segmentation; pp. 1–7. [DOI] [Google Scholar]
- 58.Olson D.L., Delen D. Advanced Data Mining Techniques. Springer; 2008. Performance evaluation for predictive modeling; pp. 137–147. [DOI] [Google Scholar]
- 59.Fawcett T. An introduction to roc analysis. Pattern Recognit. Lett. 2006;27(8):861–874. doi: 10.1016/j.patrec.2005.10.010. [DOI] [Google Scholar]
- 60.Hancock J.M. Dictionary of Bioinformatics and Computational Biology. 2004. Jaccard distance (Jaccard index, Jaccard similarity coefficient) [DOI] [Google Scholar]
- 61.Çakmak M., Tenekecı M.E. 2021 29th Signal Processing and Communications Applications Conference (SIU) IEEE; 2021. Melanoma detection from dermoscopy images using nasnet mobile with transfer learning; pp. 1–4. [DOI] [Google Scholar]
- 62.Rashid J., Ishfaq M., Ali G., Saeed M.R., Hussain M., Alkhalifah T., Alturise F., Samand N. Skin cancer disease detection using transfer learning technique. Appl. Sci. 2022;12(11):5714. doi: 10.3390/app12115714. [DOI] [Google Scholar]
- 63.Gajera H.K., Nayak D.R., Zaveri M.A. A comprehensive analysis of dermoscopy images for melanoma detection via deep cnn features. Biomed. Signal Process. Control. 2023;79 doi: 10.1016/j.bspc.2022.104186. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset HAM10000 is public and available on the Harvard Dataverse website. The other dataset used in this study is of a private nature, due to the confidentiality restrictions imposed by the ethics committee.