Abstract
During lignin biosynthesis in angiosperms, coniferyl and sinapyl aldehydes are believed to be converted into their corresponding alcohols by cinnamyl alcohol dehydrogenase (CAD) and by sinapyl alcohol dehydrogenase (SAD), respectively. This work clearly shows that CAD-C and CAD-D act as the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis thaliana by supplying both coniferyl and sinapyl alcohols. An Arabidopsis CAD double mutant (cad-c cad-d) resulted in a phenotype with a limp floral stem at maturity as well as modifications in the pattern of lignin staining. Lignin content of the mutant stem was reduced by 40%, with a 94% reduction, relative to the wild type, in conventional β-O-4–linked guaiacyl and syringyl units and incorportion of coniferyl and sinapyl aldehydes. Fourier transform infrared spectroscopy demonstrated that both xylem vessels and fibers were affected. GeneChip data and real-time PCR analysis revealed that transcription of CAD homologs and other genes mainly involved in cell wall integrity were also altered in the double mutant. In addition, molecular complementation of the double mutant by tissue-specific expression of CAD derived from various species suggests different abilities of these genes/proteins to produce syringyl-lignin moieties but does not indicate a requirement for any specific SAD gene.
INTRODUCTION
Lignin is a complex polymer of high carbon content distinct from carbohydrates that impregnates the plant cell wall. This phenolic polymer has no extended sequences of repeating units and is characterized by a set of variable cross-linkages. Angiosperm lignins are composed of three main units named p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units. These components originate from the polymerization of the three monolignols, the p-coumaryl, coniferyl, and sinapyl alcohols, respectively. The monolignols are synthesized from Phe through successive deamination, reduction, hydroxylation, and methylation steps (Boerjan et al., 2003). The proportions of these three units in the cell wall vary according to plant species and tissue type (Campbell and Sederoff, 1996).
Cinnamyl alcohol dehydrogenase (CAD) is a specialized enzyme involved in the reduction of cinnamaldehydes into cinnamyl alcohols, the last step of monolignol biosynthesis before oxidative polymerization in the cell wall. CAD genes and their proteins in different taxa display distinct features depending on their phylogenetic origin. Duplication of CAD genes has only been observed in angiosperms (Knight et al., 1992; Brill et al., 1999). In gymnosperms, CAD genes are reported to be monogenic despite the fact that different alleles could be detected in some species (Galliano et al., 1993b; MacKay et al., 1995). In addition, conifer CAD proteins are believed to be highly specific for the reduction of coniferaldehyde (Kutsuki et al., 1982; Galliano et al., 1993a), whereas angiosperm CAD proteins have been shown to have a significant affinity for both coniferaldehyde and sinapaldehyde (Mansell et al., 1974; Grima-Pettenati et al., 1994; Hawkins and Boudet, 1994; Brill et al., 1999). These characteristics, combined with the fact that angiosperm lignin displays mainly both G and S units, whereas lignin from most gymnosperms contains predominantly G units, has led some authors to hypothesize that specialized CAD proteins could be specifically involved in either sinapyl or coniferyl alcohol biosynthesis (Hawkins and Boudet, 1994). This hypothesis has been supported by the identification of a novel enzyme in aspen (Populus tremuloides), sinapyl alcohol dehydrogenase (SAD), which was shown to be specifically involved in the reduction of sinapaldehyde in vitro (Li et al., 2001). These authors suggested that all the previously characterized CAD proteins were involved only in the reduction of coniferaldehyde. Together with aldehyde-5-hydroxylase and aldehyde 5-O-methyl transferase, SAD is proposed to be the third enzyme involved in sinapyl alcohol biosynthesis. The biological role of SAD could be of practical importance because it may regulate the channeling of S units during lignification. Indeed, modulation of the S:G ratio in transgenic lines could potentially lead to advantageous alterations in wood properties, such as reduced resistance to kraft delignification (reviewed in Baucher et al., 1998, 2003; Mellerowicz et al., 2001).
Approaches using mutants and transgenic plants have previously been shown to be of considerable use for studying the impact of metabolic disruption upon secondary cell wall structure. For instance, the cell wall properties of the naturally CAD-deficient mutants brown midrib1 (bm1) of maize (Zea mays) and cad-n1 of loblolly pine (Pinus taeda) have been intensively studied (Lapierre et al., 2000; Marita et al., 2003). bm1 showed a 20% decrease in lignin content with no alteration in the S:G ratio, although lignin-associated aldehydes were detected (Halpin et al., 1998). The loblolly pine CAD− mutant was shown to have a very slight reduction in lignin content (<5%) but to also contain an unusually high content of lignin-associated coniferaldehyde and dihydroconiferyl alcohols. The production of downregulated CAD plants via antisense strategies has been performed in various species, including alfalfa (Medicago sativa), poplar (Populus spp), tobacco (Nicotiana tabacum), and, most recently, fescue (Festuca arundinacea) (Halpin et al., 1994; Baucher et al., 1996, 1999; Chen et al., 2003). Lignin analysis of these transgenic plants led to controversial interpretations of the potential role of CAD (Li et al., 2001; Anterola and Lewis, 2002). Indeed, lignin content was found to be impacted only modestly, if at all. Moreover, even though the S:G ratio was found reduced in some studies, there was a general lack of consistency (Halpin et al., 1994; Higuchi et al., 1994; Baucher et al., 1996; Stewart et al., 1997; Chen et al., 2003), a result that could be related to incomplete disruption of CAD expression. In addition, the potential for functional redundancy, as for example produced by duplicate CAD genes, could lead to further ineffectiveness of antisense strategies. Transcriptomic tools such as large-scale microarrays in lignin-altered plants could help confirm or reject such hypotheses by revealing the extent of transcriptional modification in these transgenic plants.
A better understanding of the complexity of gene families involved in lignification has been provided by the complete genome sequencing and annotation of rice (Oryza sativa) and Arabidopsis thaliana (Sasaki and Sederoff, 2003). For example, Tobias and Chowk (2005) have recently shown that there are 12 CAD genes in rice, consistent with duplication of many of the genes coding for enzymes involved in the phenylpropanoid pathway of Arabidopsis, although the precise role of these duplicates in lignin metabolism remains to be elucidated (Costa et al., 2003; Goujon et al., 2003; Raes et al., 2003). Previously, we surveyed the complete CAD gene family in Arabidopsis (Sibout et al., 2003), which led to the identification of nine CAD-like proteins distributed into four different classes based on their amino acid similarity. CAD-C (At3g19450) and CAD-D (At4g34230) belong to one class that is highly similar to other well-characterized angiosperm CAD proteins (eucalyptus [Eucalyptus gunnii] CAD2, alfalfa CAD2, aspen CAD, and tobacco CAD14) (reviewed in Baucher et al., 2003). The second class contains CAD-A (At4g34970), CAD-B1 (At4g37980), and CAD-B2 (At4g37990), which are most closely related to the poplar SAD identified by Li et al. (2001). The third class contains CAD1 (At4g34930), CAD-E (At2g21730), and CAD-F (At2g21890), which are most closely related to the alfalfa CAD2 identified by Brill et al. (1999). The fourth class contains CAD-G (At1g72680), which does not have well-identified homologs. Importantly, two mutants (cad-c and cad-d) were also characterized with decreased CAD activities, although only cad-d produced a modest reduction in lignin content along with a substantial decrease in conventional S lignin (Sibout et al., 2003).
In this study, we show that the double cad-c cad-d mutant in Arabidopsis produces the strongest phenotype achieved to date in any CAD-disrupted dicotyledonous angiosperm. This mutant possesses a highly condensed lignin enriched in aldehydes. Thioacidolysis results also show that conventional H, G, and S subunits are drastically altered. Using Affymetrix Arabidopsis 24K GeneChip and real-time PCR, we showed that this double mutation alters the expression of several genes, including CAD genes. Moreover, Fourier transform infrared (FTIR) spectroscopy analysis clearly showed that the metabolic impact of CAD mutations affects both fiber and xylem cell wall composition. Finally, the complementation of the double mutant by various CAD genes from different species suggests different abilities of these genes/proteins to produce syringyl-lignin moieties but does not indicate the requirement for any specific SAD gene.
RESULTS
Phenotype of the cad-c cad-d Double Mutant in Arabidopsis
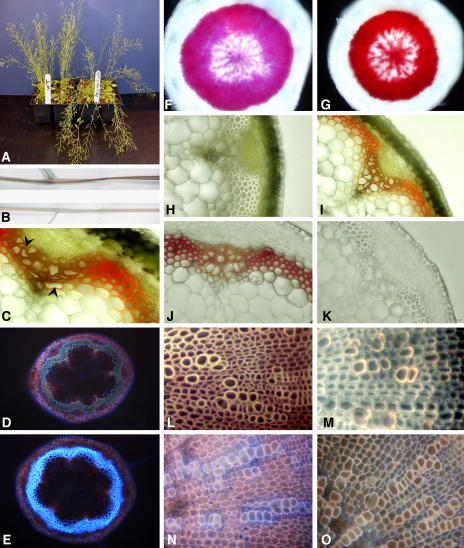
Double mutant lines were identified by PCR from F2 crosses of cad-d × cad-c. Protein gel blot analysis using an antiserum directed against tobacco CAD, which was used previously to detect both CAD in single mutants (Sibout et al., 2003), failed to detect both the CAD-D and CAD-C proteins in stems of the double mutant (Figure 1). Visible phenotypes of the double mutant included a slight delay in growth, compared with the wild type, that resulted in a delay in bolting. During the first week after bolting, the length and appearance of the stems were similar to the wild type (data not shown). However, by the third week, a reduction in stem elongation became evident, resulting in shorter stems at maturation. In addition, the mutants underwent precocious senescence, the severity of which was highly dependent on growth conditions. Moreover, mature stems of the mutant had reduced rigidity resulting in a bending of the floral stems, a phenotype that was enhanced during growth under continuous light and under dry growth conditions (Figure 2A). After senescence, dry stems became completely flat and twisted, with a small and irregular circumference (Figure 2B). An intense purple color was also visible throughout the green epidermal layer, particularly toward the bottom of the mature stem. Transverse sections of stems revealed that this coloration was localized to lignified xylem bundles and interfascicular fibers, along with a frequent occurrence of collapsed xylem cells (Figure 2C).
Figure 1.
Protein Blot Analysis Assayed by Immunoblotting Using Anti-Tobacco CAD Antibodies.
Crude extracts from wild-type and cad-c cad-d stems were analyzed. The apparent molecular masses corresponding to Arabidopsis CAD-D and CAD-C (44/42 kD) are indicated.
Figure 2.
Phenotype of cad-c cad-d and Histochemical Analysis of Lignified Tissues.
(A) Stems of the cad-c cad-d mutant tend to lay down at a mature stage.
(B) Dried cad-c cad-d stems are flat and twisted and show a reddish-brown color (top half) compared with the yellow color and round shape of the wild-type stems (bottom half).
(C) to (J) Light microscopy of cross sections of stems or hypocotyls.
(C) Transverse section of mature cad-c cad-d stem (arrowheads show xylem vessels).
(D) and (E) transverse sections under UV light fluorescence of the base of a stem of cad-c cad-d (D) and the wild type (E).
(F) and (G) Variations in phloroglucinol staining in the vascular tissues of the wild type (F) and mutant (G) hypocotyls.
(H) to (K) Color of lignified tissues in the wild-type stem ([H] and [J]) and the cad-c cad-d stem ([I] and [K]) before ([H] and [I]) and after ([J] and [K]) Mäule staining.
(L) and (M) UV light microscopy of hypocotyls from the wild type (L) and cad-c cad-d (M) previously treated with the Mäule staining procedure.
(N) and (O) Fiber and xylem elements of the wild type (N) and cad-c cad-d (O) under UV previously treated with a solution of Safranin O/Alcian Blue.
Because of an apparent association of these phenotypes with lignin formation and deposition, we performed a series of histological analyses using cross sections of various lignified tissues. UV light microscopy demonstrated that the typical blue autofluorescence produced by lignin within both the fibers and xylem of the wild-type stem was drastically reduced in the double mutant (Figures 2D and 2E). Staining of lignin by phloroglucinol-HCl within tissues of the double mutant resulted in a more intense coloration within mature hypocotyls (Figures 2F and 2G) as well as in stems (data not shown). Of all the methods typically used to detect lignin in angiosperms, the Mäule protocol is the most noteworthy due to its ability to differentiate S units from G units (Lin and Dence, 1992). Surprisingly, Mäule staining of stem sections from the cad-c cad-d double mutant produced no coloration in either the fibers or xylem (Figures 2H to 2K) or in transverse sections of mature hypocotyls (Figures 2L and 2M), suggesting that the syringyl moiety content was below the detection limits of this stain. Safranin/Alcian Blue staining, which allows polysaccharide elements to be distinguished from phenolic compounds, showed that polysaccharides within mature hypocotyls were only detectable in the cell walls surrounding xylem vessels but rarely in fiber cell walls, confirming the highly lignified nature of the fiber secondary wall in wild-type Arabidopsis (Figure 2N). By contrast, the fiber cell walls of the double mutant stained more intensely (Figure 2O), consistent with a significantly lower phenolic content.
All these observations suggested that the lignin of the double mutant is reduced within various organs and tissues and that the S unit content is reduced below that detectable by Mäule staining. It is also evident that the polysaccharides of the cell wall fibers in the double mutant were more accessible to Safranin/Alcian Blue staining analysis as compared with the wild type, which may be due to lower lignin content. These features prompted us to perform additional analyses to better define the lignin characteristics of the double mutant.
Cell Wall Modifications of the cad-c cad-d Mutant
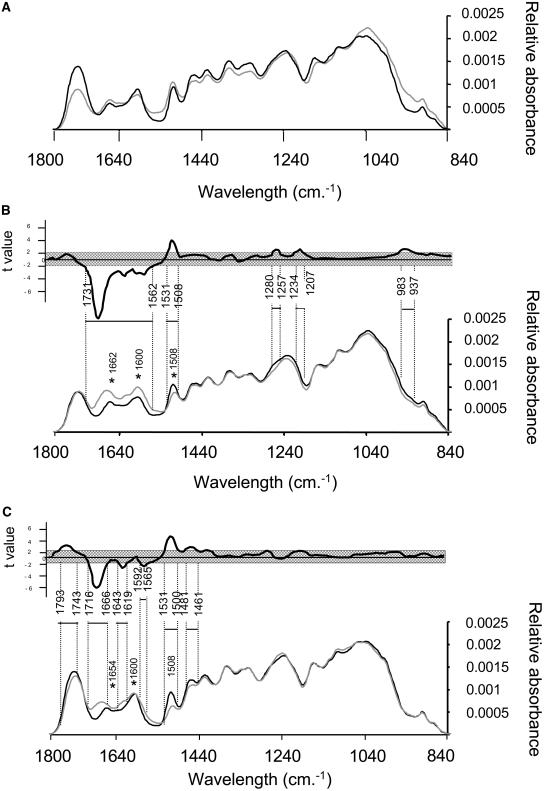
The previous histological analysis showed that interfascicular fiber elements and xylem bundles can be distinguished easily within transverse sections, despite the small size of Arabidopsis stems. These tissues are of interest because lignin composition is substantially different: xylem cell walls are mainly lignified with G units, whereas fiber lignin is composed of both G and S units. We performed localized FTIR analysis on these specialized tissues and analyzed spectra modifications within the range of 1790 to 839 cm−1 in both wild-type and double mutant plants.
First, we compared the absorbance of xylem and fiber tissues from wild-type plants. Although the spectra from the xylem and fibers in wild-type plants were similar, the magnitude of absorbance was often different at specific wavelengths (Figure 3A). For example, the difference in peak height at 1743 cm−1 may be due to the relative concentration of hemicellulose polymer in the fibers as compared with the xylem (Figure 3A). Owen and Thomas (1989) suggested that strong absorption at that wavelength is likely produced by the stretching of the free carbonyl group of the hemicellulose polymer. Similarly, variation in absorbance in the range of 1190 to 850 cm−1 is characteristic of changes in polysaccharide structure or content such as cellulose (Mouille et al., 2003).
Figure 3.
FTIR Analysis.
(A) FTIR spectra in the fiber (black line) and xylem (gray line) areas of wild-type stem sections.
(B) FTIR spectra of the wild type (black line) and cad-c cad-d (gray line) collected in the xylem area.
(C) FTIR spectra of the wild type (black line) and cad-c cad-d (gray line) collected in the fiber area. Student's t test values corresponding to the differences between the mutant and wild-type spectra are plotted against the wavelengths in the top part of (B) and (C). Nonsignificant values (P = 0.05) are shadowed. The wavelength ranges of areas significantly different between mutant and wild-type spectra are shown. Asterisks refer to discussed peaks (see text).
Second, we compared xylem and fiber tissue absorbance in the mutant and in the wild type. A t test (P = 0.05) indicated that the absorbance spectra from 1712 to 1500 cm−1 were significantly different in both the xylem and the fibers of the mutant (Figures 3B and 3C) relative to the wild type. These modifications are consistent with an alteration of lignin structure and content as described by Faix (1992). On the one hand, the peak at 1508 cm−1 was decreased in both the fibers and the xylem of the double mutant, consistent with a reduction in lignin content. On the other hand, amplitudes of the peak at 1662 cm−1 in the xylem (Figure 3B) and 1654 cm−1 in the fibers (Figure 3C) were considerably amplified in the mutant and slightly shifted to 1673 and 1681 cm−1, respectively, suggesting an enrichment in conjugated aldehydes or ketones (Buta and Galetti, 1989; Faix, 1992). Absorbance at 1600 cm−1 was found increased in the xylem of the double mutant but not in fibers (Figures 3B and 3C). Numerous studies (Faix, 1992; Stewart et al., 1997; Zhong et al., 2000) have shown that vibrations of G condensed units (with C–C linkage) result in greater absorption than etherified G units at 1600 cm−1. Furthermore, the absorption maxima at 1600 cm−1 have also been attributed to aromatic skeletal vibrations and the C=O stretch present in aldehydes such as vanillin (Sarkanen et al., 1967). Analysis of cad-c cad-d xylem also showed slight modifications (not visible in fibers) in the spectral regions of 1265 to 1214 cm−1 and 964 cm−1 (Figures 3B and 3C), previously assigned to C–C, C–O, and C=O stretching and –CH=CH– out-of-plane deformation (Faix, 1992). By contrast, absorbance at 1461 cm−1 was significantly decreased in the fibers but not in the xylem (Figure 3C). This modification, assigned by Faix (1992) to C–H deformation and asymmetry in –CH3 and –CH2–, has been observed previously in milled lignin of CAD-deficient tobacco plants (Stewart et al., 1997). In summary, FTIR analysis showed significant but different repercussions of CAD mutations within the xylem and fiber cell walls of the double mutant. Furthermore, these results suggest that the double mutant is characterized by higher amounts of aldehydes and reduced lignin content with a specific G condensed structure, especially in the xylem of the mutant.
In a previous study, cad-c was not found to have a reduced Klason lignin content, whereas cad-d produced a very modest reduction in lignin content compared with wild-type plants (Sibout et al., 2003). By contrast, the double mutant showed a drastic reduction (40%) in Klason lignin content (Table 1). We also quantified the susceptibility of the double mutant to cellulase using a method adapted from Rexen (1977). We observed a 57% dry mass loss in the double mutant after cellulase treatment but only a 29% dry mass loss in the wild type. The H, G, and S lignin-derived units released by thioacidolysis were also analyzed by gas chromatography–mass spectrometry (GC-MS). When calculated on the basis of the Klason lignin content, the yield of lignin-derived monomers was drastically reduced in the mutant (Table 1). This result clearly shows a severe reduction in conventional units specifically involved in β-O-4 linkage and suggests that the lignin is considerably enriched in condensed bonds that resist the thioacidolysis procedure. Besides the conventional H, G, and S thioacidolysis monomers, we found relatively large amounts of indene derivatives (Table 1). These lignin-derived monomers specifically originate from β-O-4–linked sinapaldehyde and coniferaldehyde units, as shown in recent articles (Kim et al., 2002; Lapierre et al., 2004). In addition, the dithioketal derivatives of coniferaldehyde, vanillin, and syringaldehyde were also found to be released in relatively larger amounts from the thioacidolysis of the extract-free double mutant sample (data not shown), suggesting that these aldehydes were incorporated in the mutant lignin by ether bonds at C4OH. The reduced thioacidolysis S:G ratio of the conventional S and G units (0.38 in the wild type versus 0.08 in the mutant, Table 1) suggests that CAD mutations more specifically affect the formation of conventional S units than that of G units. In addition to the determination of the lignin-derived monomers, the amount of ferulic and sinapic acids released by thioacidolysis was found to be higher in the mutant (Table 1).
Table 1.
Lignin in Wild-Type and Double Mutant Extract-Free Floral Stems and Amount of Thioacidolysis-Released Ferulic and Sinapic Acids
| Thioacidolysis Monomers from β-O-4–Linked H, G, and S Conventional Lignin Unitsb
|
Thioacidolysis Indene Monomers from C6C3 Aldehydes Linked at Cβ by β-O-4 Bondsb
|
Thioacidolysis-Released Ferulic (Fe) and Sinapic (Si) Acidsc
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Line | Acid-Insoluble Klason Lignina | H | G | S | S:G | Coniferaldehyde | Sinapaldehyde | Fe | Si |
| Wild type | 19.5 ± 0.65 | 7.8 ± 0.2 | 1049 ± 29 | 404 ± 15 | 0.38 | 0.6 ± 0.03 | 0.8 ± 0.01 | 0.85 ± 0.1 | 0.20 ± 0.01 |
| cad-c cad-d | 11.9 ± 0.5 | Trace | 84 ± 1 | 7.1 ± 0.6 | 0.08 | 32.7 ± 0.57 | 21.8 ± 0.13 | 2.0 ± 0.4 | 0.67 ± 0.13 |
Klason lignin content (percentage of weight of the extract-free stem).
Yield in main lignin-derived thioacidolysis monomers (expressed in micromoles per gram of Klason lignin) recovered from conventional H, G, or S β-O-4–linked lignin units or from coniferaldehyde or sinapaldehyde linked at Cβ by β-O-4 bonds. The data correspond to the means and standard errors from duplicate or triplicate analyses.
Calculated from ion chromatograms reconstructed at mass-to-charge ratios of 338 and 368 (molecular ions of their trimethylsilylated derivatives).
In addition to the stems, we analyzed dried extracts from hypocotyls of the wild type and cad-c cad-d, in which we induced secondary growth as described by Chaffey et al. (2002). Table 2 shows the percentage of the three monomers (H+G+S) in the mutant released by thioacidolysis. We observed that the amount of H units was substantially higher in the hypocotyls than in the stems of wild-type plants (2.5% versus <0.5%, respectively). In the mutant, the recovery of H units was also higher in the hypocotyls than in the stems (5% in hypocotyls versus traces in stem).
Table 2.
Thioacidolysis of cad-c cad-d and Wild-Type Hypocotyls
| Lines | S+G+H (μmol·g−1)a | S:G | H (%) | G (%) | S (%) |
|---|---|---|---|---|---|
| Wild type | 258.8 ± 8.5 | 0.31 | 2.4 ± 0.1 | 74.6 ± 0.4 | 23 ± 0.5 |
| cad-c cad-d | 29.8 ± 2.2 | 0.07 | 5.1 ± 0.1 | 90 ± 0.86 | 6.5 ± 0.1 |
Yield in main lignin-derived thioacidolysis monomers (expressed in micromoles per gram of dry milled stem).
Large-Scale Transcript Profiling and Analysis of Transcript Levels of CAD Genes in cad-c cad-d Stems
We performed expression profiling experiments using the Affymetrix ATH1-24K GeneChip (representing ∼24,000 genes). Our first goal was to use the technology as a tool to explore gene expression profiles in the double mutant transcriptome. In particular, we investigated in detail the expression profile of CAD homologs as well as other genes upstream in the phenylpropanoid pathway. Four biological repetitions of RNA extracted from the bases of stems (which share differentiated interfascicular fibers and xylem) of wild-type and mutant plants were used for the microarray analysis. The complete array data for each sample is presented in Supplemental Tables 1 to 8 online. A statistical treatment of eight ATH1 GeneChips (unpaired Student's t test, P = 0.05) using Affymetrix Microarray Suite (MAS 5.0) software detected 5789 genes in the double mutant that had significantly different expression compared with the wild type (see Supplemental Table 9 online). Two screen parameters were chosen to avoid little or unreliable evidence of differential expression: probe set variations between mutant and wild-type hybridization were tagged “significant” when the change in raw signal was higher than 200 units with a minimum twofold change. A preliminary analysis was done to validate the stringency of our screen parameters and to reveal the reproducibility between ATH1 GeneChip hybridization experiments. Therefore, scatterplots of the average signal intensity of each probe set from two control ATH1 GeneChips were compared with those of two other control ATH1 GeneChips. Different combinations were done with all control or all cad-c cad-d ATH1 DNA chips (see Supplemental Figure 1 online). After application of our screen parameters to the 5789 genes with altered expression by computing all ATH1 GeneChip data (four wild-type versus four mutant ATH1 GeneChips), we found 546 genes with altered expression in the double mutant. Means of signals with standard errors, fold changes, and the Student's t test P value of each probe set are presented in Supplemental Table 10 online.
A few small differences in signal were observed for the genes involved in monolignol biosynthesis, from Phe to the cinnamaldehydes (Table 3). For example, signals corresponding to Phenylalanine ammonia-lyase 3 (PAL3; At5g04230) and 4-Coumarate-CoA ligase 3 (4CL3; At1g65060) suggest that these genes are overexpressed 2-fold (Student's t test, P = 0.0087) and 2.6-fold (Student's t test, P = 0.0092), respectively, relative to the wild type. Furthermore, cinnamyl CoA reductase 1 (CCR1; At1g15950) and ferulate 5-hydroxylase (F5H; At4g36220) seem to be underexpressed 1.7-fold (Student's t test, P = 0.0004) and 1.8-fold (Student's t test, P = 0.014), respectively, relative to the wild type. However, low hybridization signal (<200) and/or weak change in expression (less than twofold) were not relevant to our screen parameters and were consequently not considered in the analysis. Contrastingly, transcripts of CAD-G were found to be 5.5 times more abundant in the double mutant. In addition, five laccase-like and two peroxidase-like genes were found to be downregulated in cad-c cad-d. Several other genes encoding proteins known to be involved in cell wall biosynthesis also had altered expression, including arabinogalactans, pectin esterases, proline-rich protein (PRP), and glycine-rich protein (GRP) (Table 3). It is noteworthy that among several glucosyl transferases with altered expression in the mutant (see Supplemental Table 10 online), the IRX3 (At5g17420) and IRX5 (At5g44030) genes that encode cellulose synthase subunits showed at least fourfold lower expression than the wild type (Table 3). Nevertheless, we did not observe an alteration in the transcript levels of the UDP glycosyl transferase genes (see Supplemental Table 2 online), which are specifically involved in the glycosylation of sinapyl and coniferyl alcohol or related acids (Lim et al., 2001). Interestingly, distinct increases in transcript abundance were observed for 10 genes encoding glutathione S-transferase (GST), some being increased >10-fold (At2g29480), 29-fold (At2g29490), and 40-fold (At1g17170) (see Supplemental Table 10 online). The 1-aminocyclopropane-1-carboxylic acid oxidase, which is involved in ethylene biosynthesis, exhibited a twofold increase in transcript abundance in the double mutant relative to the wild type. This enzyme is known to play a role in wood formation and also in stress response (for review, see Andersson-Gunnerås et al., 2003). The double mutant also showed alterations in the expression level of several transcription factors (WRKY, MYB, HD-ZIP, BZIP, and MADS box). We did not detect, however, any significant alteration in the expression of PAP1, SHATTERPROOF1 or 2, BREVIPEDICELLUS, or MYB61, all of which have been suggested to be involved in lignin biosynthesis (Borevitz et al., 2000; Liljegren et al., 2000; Mele et al., 2003; Newman et al., 2004).
Table 3.
Selected Gene Chip Hybridization Data
| Putative Gene Function | Accession Number | Mutant Callsa | Control Callsa | Student's t Test P Valueb | Foldc | Sigd |
|---|---|---|---|---|---|---|
| Monolignol biosynthesis | ||||||
| Phenylalanine ammonia-lyase 1 (PAL1) | At2g37040 | 13,419 ± 1,029 | 19,644 ± 3,727 | 1.81E-02 | −1.4 | No |
| Phenylalanine ammonia-lyase 2 (PAL2) | At3g53260 | 2,299 ± 268 | 3,086 ± 893 | 1.42E-01 | −1.3 | No |
| Phenylalanine ammonia-lyase 3 (PAL3) | At5g04230 | 221 ± 52 | 106e ± 30 | 8.73E-03 | +2.0 | No |
| Phenylalanine ammonia-lyase (PAL4) | At3g10340 | 13,336 ± 452 | 13,580 ± 2,444 | 8.51E-01 | −1.0 | No |
| Cinnamic acid 4-hydroxylase (C4H) | At2g30490 | 16,963 ± 317 | 19,500 ± 2,655 | 1.07E-01 | −1.1 (1.0) | No |
| 4-Coumarate-CoA ligase 1 (4CL1) | At1g51680 | 24,109 ± 5,232 | 26,798 ± 2,103 | 3.77E-01 | −1.1(−1.1) | No |
| 4-Coumarate-CoA ligase 2 (4CL2) | At3g21240 | 6,865 ± 344 | 7,446 ± 1,258 | 4.07E-01 | −1.1 | No |
| 4-Coumarate-CoA ligase 3 (4CL3) | At1g65060 | 294e ± 76 | 115e ± 56 | 9.27E-03 | +2.6 | No |
| 4-Coumarate-CoA ligase (4CL4) | At3g21230 | 4,035 ± 426 | 4,986 ± 460 | 2.30E-02 | −1.2 | No |
| Transferase family protein (CST) | At5g48930 | 13,683 ± 519 | 14,060 ± 2,217 | 7.52E-01 | −1.0 (−1.2) | No |
| Coumarate 3-hydroxylase (C3H) | At2g40890 | 6,591 ± 244 | 6,315 ± 1,472 | 7.24E-01 | −1.1 | No |
| Ferulate-5-hydroxylase (F5H) | At4g36220 | 2,746 ± 246 | 4,910 ± 1,252 | 1.47E-02 | −1.8 | No |
| O-methyltransferase (OMT1) | At5g54160 | 25,852 ± 4,798 | 30,285 ± 3,491 | 1.86E-01 | −1.2 (+1.3) | No |
| Caffeoyl-CoA 3-O-methyltransferase | At4g34050 | 31,965 ± 1,940 | 35,561 ± 3,779 | 1.41E-01 | −1.1 (−1.2) | No |
| Caffeoyl-CoA 3-O-methyltransferase | At1g24735 | 150e ± 41 | 195 ± 25 | 1.09E-01 | −1.3 | No |
| Caffeoyl-CoA 3-O-methyltransferase | At4g26220 | 1,287 ± 459 | 1,578 ± 430 | 3.92E-01 | −1.2 | No |
| Caffeoyl-CoA 3-O-methyltransferase | At1g67990 | 104e ± 55 | 78e ± 43 | 4.95E-01 | +1.3 | No |
| Caffeoyl-CoA 3-O-methyltransferase | At1g67980 | 131e ± 40 | 51e ± 37 | 2.55E-02 | +2.6 | No |
| Caffeoyl-CoA 3-O-methyltransferase | At3g61990 | 835 ± 101 | 881 ± 166 | 6.50E-01 | −1.1 | No |
| Caffeoyl-CoA 3-O-methyltransferase | At3g62000 | 230e ± 105 | 201e ± 48 | 6.23E-01 | +1.1 | No |
| Cinnamoyl-CoA reductase (CCR1) | At1g15950 | 7,982 ± 663 | 13,652 ± 1,476 | 4.20E-04 | −1.7 (−1.3) | No |
| Cinnamoyl-CoA reductase (CCR2) | At1g80820 | 119e ± 87 | 60e ± 62 | 3.10E-01 | +2.0 | No |
| Cinnamyl-alcohol dehydrogenase AtCAD1 | At4g39330 | 637 ± 52 | 362 ± 86 | 1.51E-03 | +1.8 (+2.0) | No |
| Cinnamyl-alcohol dehydrogenase AtCAD-A | At4g37970 | 641 ± 216 | 600 ± 82 | 7.38E-01 | +1.1 (+1.8) | No |
| Cinnamyl-alcohol dehydrogenase AtCAD-B1 | At4g37980 | 27e ± 26 | 12e ± 9 | 3.23E-01 | +2.2 (−1.2) | No |
| Cinnamyl-alcohol dehydrogenase AtCAD-B2 | At4g37990 | 62e ± 45 | 53e ± 46 | 7.81E-01 | +1.2 (−1.3) | No |
| Cinnamyl-alcohol dehydrogenase AtCAD-F | At2g21890 | 524 ± 78 | 417 ± 157 | 2.69E-01 | +1.3 | No |
| Cinnamyl-alcohol dehydrogenase AtCAD-C | At3g19450 | 94e ± 74 | 2,271 ± 846 | 2.16E-03 | −24.1 | Yes |
| Cinnamyl-alcohol dehydrogenase AtCAD-D | At4g34230 | 24e ± 31 | 15,517 ± 2,290 | 1.01E-05 | −651.3 | Yes |
| Cinnamyl-alcohol dehydrogenase AtCAD-G | At1g72680 | 1,607 ± 286 | 295 ± 57 | 1.06E-04 | +5.5 (+5.4) | Yes |
| Monolignol polymerization | ||||||
| Laccase putative/diphenol oxidase | At5g60020 | 763 ± 539 | 2,644 ± 940 | 1.33E-02 | −3.5 | Yes |
| Laccase putative/diphenol oxidase | At5g05390 | 548 ± 262 | 1,506 ± 662 | 3.60E-02 | −2.7 | Yes |
| Laccase putative/diphenol oxidase | At2g40370 | 159e ± 99 | 424 ± 152 | 2.64E-02 | −2.7 | Yes |
| Laccase putative/diphenol oxidase | At2g29130 | 976 ± 478 | 2,577 ± 863 | 1.75E-02 | −2.6 | Yes |
| Laccase putative/diphenol oxidase | At2g38080 | 6,763 ± 3,674 | 23,742 ± 5,828 | 2.63E-03 | −3.5 | Yes |
| Peroxidase | At5g42180 | 1,167 ± 755 | 4,960 ± 1,898 | 9.92E-03 | −4.2 | Yes |
| Peroxidase | At3g42570 | 124e ± 53 | 350 ± 68 | 1.89E-03 | −2.8 | Yes |
| Cell wall–related proteins | Yes | |||||
| Arabinogalactan-protein (AGP18) | At4g37450 | 97e ± 32 | 412 ± 117 | 2.08E-03 | −4.3 | Yes |
| Arabinogalactan-protein (AGP4) | At5g10430 | 354 ± 172 | 3,611 ± 1,316 | 2.69E-03 | −10.2 | Yes |
| Arabinogalactan-protein (AGP9) | At2g14890 | 2,222 ± 877 | 7,299 ± 1,472 | 1.03E-03 | −3.3 | Yes |
| Arabinogalactan-protein (AGP22) | At5g53250 | 177e ± 59 | 525 ± 161 | 6.55E-03 | −3.0 | Yes |
| Arabinogalactan-protein (FLA11) | At5g03170 | 7,086 ± 4,048 | 25,441 ± 6,368 | 2.81E-03 | −3.6 | Yes |
| Arabinogalactan-protein (FLA12) | At5g60490 | 2,521 ± 1,552 | 11,676 ± 3,781 | 4.19E-03 | −4.6 | Yes |
| Arabinogalactan-protein (FLA2) | At4g12730 | 267e ± 104 | 750 ± 209 | 6.15E-03 | −2.8 | Yes |
| Arabinogalactan-protein (FLA13) | At5g44130 | 547 ± 161 | 1,999 ± 335 | 2.32E-04 | −3.7 | Yes |
| Pectinacetylesterase putative | At4g19410 | 2,435 ± 255 | 683 ± 49 | 1.04E-05 | +3.6 | Yes |
| Pectinesterase family protein | At2g45220 | 105e ± 82 | 354 ± 154 | 2.86E-02 | −3.4 | Yes |
| Pectinesterase family protein | At2g43050 | 1,039 ± 282 | 2,337 ± 743 | 1.71E-02 | −2.2 | Yes |
| Proline-rich family protein (PRP4) | At4g38770 | 122e ± 117 | 419 ± 120 | 1.20E-02 | −3.4 | Yes |
| Glycine/proline-rich protein | At5g17650 | 433 ± 58 | 868 ± 175 | 3.32E-03 | −2.0 | Yes |
| Hydroxyproline-rich glycoprotein | At1g70985 | 178e ± 26 | 395 ± 108 | 7.97E-03 | −2.2 | Yes |
| Glycine-rich protein (GRP3S) | At2g05380 | 1,903 ± 493 | 7e ± 2 | 2.52E-04 | +269.3 | Yes |
| Cellulose synthase catalytic subunit (IRX3) | At5g17420 | 4,106 ± 2,124 | 17,145 ± 4,342 | 1.67E-03 | −4.1 | Yes |
| Cellulose synthase catalytic subunit (IRX5) | At5g44030 | 1,993 ± 1,090 | 8,611 ± 2,314 | 2.06E-03 | −4.3 | Yes |
Mean and standard deviation of four biological repetitions.
Student's t test P values represent the level of confidence as to whether a gene is altered in the mutant.
The minus sign indicates that the gene is underrepresented in the mutant. The plus sign indicates that the gene is overrepresented in the mutant. Ratios in parentheses were obtained from real-time PCR analysis performed on an independent experimental replication.
Sig, significant. Expression of a gene is significantly altered when the P value is <0.05, signal call is increased/decreased twofold at least, and magnitude between mutant call (Atcad cd) and control call (wild type) is >200.
At least two biological repetitions were tagged with an A for “absent call” (see Methods).
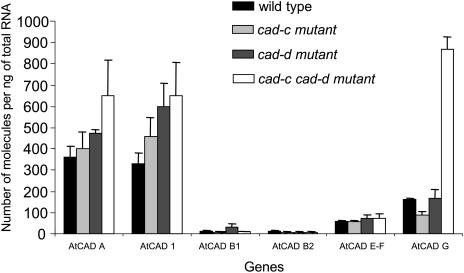
Absolute quantification of the transcript levels of the CAD homologs in cad-c, cad-d, and the double mutant was performed using real-time PCR in a duplicate experiment to confirm the data obtained with ATH1 GeneChip hybridization and to further investigate the molecular analysis of the cad mutants. Specific primers were designed for each member of the Arabidopsis CAD family, with the exception of the homologous CAD-E and -F genes, which are 98% identical at the nucleotide level. In this specific case, common primers were designed to amplify both genes. Transcripts of CAD-G and CAD1 were found to be significantly overrepresented in the double mutant 5.4-fold and 2-fold, respectively (Figure 4), which is consistent with the 5.5-fold and 1.8-fold differences in the microarray analysis (Table 3). CAD-A was also found overexpressed 1.8-fold in the double mutant in real-time PCR analysis, whereas it was not significantly modified in the microarray experimental data (Table 3). Quantification of CAD-E, -F, -B1, and -B2 did not allow detection of perturbation in transcription of these genes in either cad-c, cad-d, or the double mutant, a result also previously observed with the microarray data. CAD-B1 and ACAD-B2 were found poorly expressed in the stem base of Arabidopsis (Table 3; see Supplemental Tables 1 to 9 online). Remarkably, the slight upregulation of CAD-A and CAD1 within the double mutant was also detected in cad-d and cad-c, whereas the strong overexpression of CAD-G in the double mutant was not found in the single mutants.
Figure 4.
Absolute Quantification of Arabidopsis Wild-Type and cad Mutant Transcripts in the Bases of Stems.
For reverse transcription, total RNA from the stem bases of cad-c, cad-d, and cad-c cad-d was used. Bars represent the means and standard errors of three to five biological repetitions.
Finally, real-time PCR analysis of other genes encoding proteins involved in lignin biosynthesis, including caffeic acid O-methyl transferase 1, caffeoyl CoA O-methyl transferase 1, CCR1, hydroxycinnamoyl CoA:Shikimate/quinate hydroxycinnamoyl transferase, coumarate 3-hydroxylase (C3H), 4-coumarate-CoA ligase 1, and cinnamic acid 4-hydroxylase, did not reveal substantial alterations, which is also consistent with our microarray analysis (Table 3).
Complementation of cad-c cad-d with CAD Genes from Forest Trees
An obvious way to demonstrate that the physiological features observed in the double mutant were really due to the tagged mutations in both CAD genes was to rescue the double mutant with CAD-D and CAD-C expression. In addition, to broaden our study in the context of recently published data, we attempted to complement the double mutant with three CADs known to be phylogenetically distinct and supposedly displaying different substrate affinities. The first gene was CAD (Z19568) from Populus deltoides, which has previously been used in antisense strategies in poplar (Baucher et al., 1996). P. deltoides CAD is 97.5% identical at the amino acid level to P. tremuloides CAD, the isoform suggested to be involved in the production of guaiacyl units in aspen by Li et al. (2001). The second gene was CAD (AJ868574) from spruce (Picea abies), and the third, SAD1 (AY830131), was cloned in our laboratory from Populus tremula × tremuloides. P. tremula × tremuloides SAD1 is 99.2% identical at the amino acid level to P. tremuloides SAD, a protein found to have high affinity with sinapaldehyde in vitro (Li et al., 2001). Both a constitutive expression system (35S Cauliflower mosaic virus promoter) and a tissue-specific expression system using the Arabidopsis CAD-D promoter were used to perform complementation experiments. Because the results obtained using both approaches were similar, we present here only the lignin features of the complemented cad-c cad-d lines constructed using the Arabidopsis CAD-D promoter.
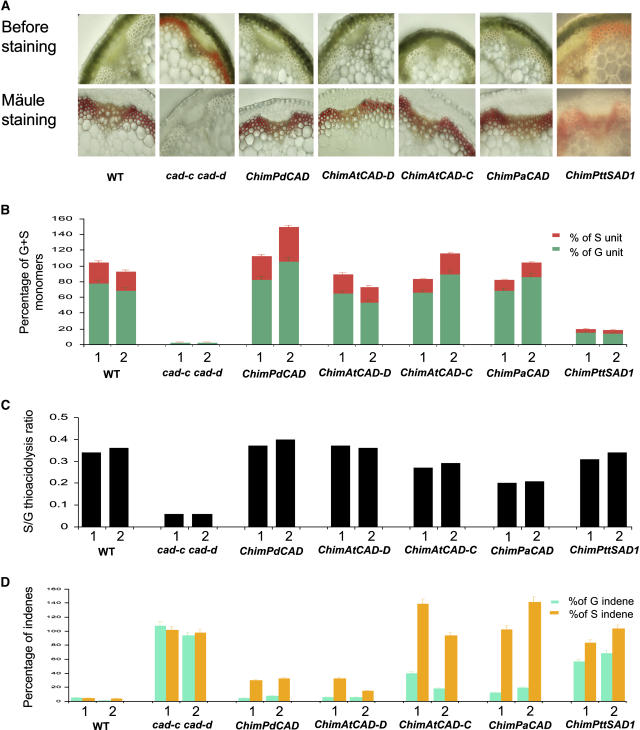
First, we selected 16 transgenic lines per type of molecular construct expressing Arabidopsis CAD-C, Arabidopsis CAD-D, P. deltoides CAD, or P. abies CAD and named these chimeric lines ChimAtCAD-C, ChimAtCAD-D, ChimPdCAD, and ChimPaCAD, respectively. Most of the 16 lines for each construct reacted to Mäule staining, allowing positive correlation with the disappearance of the natural typical reddish color observed in the double mutant (Figure 5A). Moreover, typical phenotypes observed in cad-c cad-d, such as bending of stems, were not present in those lines (data not shown). Among 25 ChimPttSAD1 lines transformed with the P. tremula × tremuloides SAD1 gene, only six reacted very weakly to Mäule staining. Moreover, all ChimPttSAD1 lines still showed a reddish-brown color in lignified tissues typical of the lignin-deficient mutant (Figure 5A).
Figure 5.
Characterizations of Complemented Lines with Different CAD cDNAs.
(A) Stem cross sections of wild-type, cad-c cad-d, and complemented lines before and after Mäule staining.
(B) Yield of thioacidolysis monomers from β-O-4–linked G and S lignin units released from dry milled stems. Two lines for each molecular construct are shown. Amounts of G and S units are represented by green and red bars, respectively. Results are expressed as a percentage of the average of the two control plants.
(C) S:G ratio found after thioacidolysis analysis of two lines per type of molecular construction.
(D) Thioacidolysis indene monomers from C6C3 aldehydes linked at Cβ by β-O-4 bonds released from dried stems. Two lines per each molecular construct were analyzed. Light green and orange histograms represent indene corresponding to coniferaldehydes and sinapaldehydes, respectively. Results are expressed as a percentage of the average of the two control plants. Absolute quantity of thioacidolysis products are shown in Supplemental Table 11 online.
Second, two lines from each construct showing the highest reactivity to Mäule staining were analyzed by GC-MS after thioacidolysis. GC-MS samples of thioacidolysis monomers (see Supplemental Table 11 online) were collected from whole dry mass; thus, monomer yields were expressed as a percentage of the control plants (Figure 5B). Figure 5C shows the S:G thioacidolysis ratio found in those lines, and Figure 5D shows the percentage of indene monomers from C6C3 aldehydes linked at Cβ by β-O-4 bonds released by thioacidolysis in complemented lines.
Except for the ChimPttSAD1 lines, most of the constructs were able to restore the G and S yield to levels similar to the wild type (Figure 5B). Nevertheless, the S:G ratios were not equally restored between constructs (Figure 5C). ChimAtCAD-D and ChimPdCAD lines showed a complete recovery of the proportion of S and G thioacidolysis main monomers. The S:G ratios were found similar to those of the wild-type plants (0.34 and 0.36 for each wild-type line). In addition, the unusually high level of indene monomers released by thioacidolysis of the double mutant lignin was reduced drastically in those lines, although still detectable. ChimAtCAD-C lines showed S:G ratios of 0.27 and 0.29, consistent with the S:G ratio observed in the Arabidopsis cad-d mutant (Sibout et al., 2003) deficient in CAD-D but not CAD-C transcripts. In agreement with this finding, indenes corresponding to coniferaldehydes were found significantly lower in ChimAtCAD-C than indenes corresponding to sinapaldehydes that were detected at a level comparable to that of the mutant. Similar results were observed with ChimPaCAD; however, the S:G ratio was further reduced (0.20 and 0.21 for each line). Total yield of conventional thioacidolysis monomers in the ChimPttSAD1 lines was found increased relative to the double mutant but still 80% lower than the wild type. Surprisingly, the S:G ratio was restored to levels close to that of the wild type (0.31 and 0.34), suggesting that these lines could produce S units (Figure 5C). Nevertheless, yield of syringyl units was still 82% lower than that of the wild type, and indene monomers were still present in those lines at a level comparable to that of the mutant. P. tremula × tremuloides SAD1 mRNAs were detected in those lines (data not shown), and a protein blot analysis using a specific antibody allowed the detection of P. tremula × tremuloides SAD1 (see Supplemental Figure 2 online).
Thioacidolysis analysis of four other ChimpttSAD1 lines found positive in the Mäule test revealed similar results (data not shown). Finally, results obtained with P. tremula × tremuloides SAD1 were later confirmed with transgenic lines expressing the P. tremuloides SAD gene kindly provided by V. Chiang and L. Li (data not shown).
DISCUSSION
According to the results of our previous study (Sibout et al., 2003), we hypothesized that CAD-C and -D could both be involved in lignification in Arabidopsis. Although both cad-c and -d mutants each had a significant decrease in CAD activity using coniferyl and sinapyl alcohol as substrates, only cad-d showed a significant decrease in lignin content. Expression patterns of both genes overlapped in lignified tissues, although CAD-C expression had a broader range. In fact, both genes are homologous and belong to the same phylogenetic clade (Sibout et al., 2003). To bypass a suspected synergistic effect of those genes on monolignol biosynthesis in the corresponding mutants, we crossed the lines to obtain a double cad-c cad-d mutant.
The cad-c cad-d Double Mutation Affects Plant Growth and Xylem/Fiber Cell Walls
The cad-c cad-d mutant has a delay in both vegetative growth and bolting, combined with reduced size. Similar growth characteristics have previously been observed in irx (irregular xylem) mutants (Turner and Somerville, 1997), such as irx3, which is mutated in a cellulose synthase gene, or irx4, which has incorrect splicing of CCR transcripts (Jones et al., 2001) and a strong decrease in lignin content. In addition to altered growth characteristics, irx mutants are distinguished by stems that tend to recline, a trait also observed with the cad-c cad-d double mutant. Moreover, xylem elements in the double mutant were found to be distorted and collapsed, whereas interfascicular fibers were not, features that were also observed in irx mutants. Consequently, it is likely that the collapse of the functioning xylem elements in the cad-c cad-d double mutant is also due to the negative pressure generated by water in this tissue, as was deduced for irx by Turner and Somerville (1997). This alteration in the conducting vessels could impact water and nutrient movements to the upper part of the stem and reduce the stem size of the double mutant. This trait might make the double mutant susceptible to environmental conditions. Another possible explanation for the delay in growth and shorter stems in this mutant could be the high content of accumulated soluble aldehydes that have been associated with the coloration of lignin. Such coloration has often been observed in transgenic or mutant plants with altered expression of caffeic acid O-methyl transferase, caffeoyl CoA O-methyl transferase, CCR, or CAD (reviewed in Baucher et al., 2003). Cinnamaldehydes have been shown to be growth inhibitors of bacterial and fungal strains (Rutten and Gocke, 1988; Utama et al., 2002), with recent studies reporting cytostatic properties of cinnamaldehyde derivatives by inducing mitotic arrest in mammalian cells (Jeong et al., 2003). Brown et al. (2001) have previously suggested some phenolics to be endogenous modulators of auxin transport. Similar impacts of aldehydes on growth of the double mutant should not be excluded.
Fibers also play an important role in the upright growth habit of Arabidopsis stems, as shown by studies of the ifl1 mutant, in which fibers are lacking (Zhong et al., 1997). The strong tendency of cad-c cad-d stems to collapse further suggested that physical properties of the fibers could be altered. Whereas most CAD genes and/or proteins have been detected in both the xylem and sclerenchyma cells of different species (Feuillet et al., 1995; Hawkins et al., 1997), Li et al. (2001) showed, using a stringent in situ hybridization technique, that P. tremuloides SAD expression is restricted to the fibers and colocalized with G-S units but not with the xylem elements that display only G units. Because we suspected cad-c cad-d to be deficient not only in G, but also in G-S units, we took advantage of FTIR spectroscopy to determine whether both the xylem and fibers were affected within the double mutant without compromising the integrity of these tissues. Xylem bundles can be easily distinguished from the interfascicular fibers within an Arabidopsis stem, facilitating the spectral properties of the tissues of the mutant to be compared with that of the wild type. Statistical analysis of FTIR spectra showed unambiguously that cell walls of both lignified tissues were impacted in the cad-c cad-d stem. It is important to note that the results of FTIR analysis were strongly consistent with those of the thioacidolysis analysis, allowing FTIR to be further developed to detect Arabidopsis mutants differentially altered in fibers and/or vessels. The impact of the CAD deficiency on both fiber and xylem tissues is in accordance with the expression patterns of Arabidopsis CAD-C and Arabidopsis CAD-D (Sibout et al., 2003). It is also interesting to note that the impact of the mutations is different between the two tissue types. This distinction is likely due to differences in cell wall composition between the fibers and xylem elements as suggested in Figure 3A. Limited knowledge exists on the chemical composition of the cell walls of Arabidopsis fiber and xylem tissues, and new techniques of dissections, such as laser capture coupled with chemical cell wall analysis, could help considerably to increase our understanding.
The inability of not only the fibers but also the xylem cell walls of the double mutant to be stained by the Mäule procedure as they are in the wild-type plants is astonishing. This result could be attributed to lignin units under detectable levels and particularly to the structure of the lignin polymer in the double mutant. Further experiments are currently underway to explain this phenomenon.
Alteration in the Transcript Levels of Cell Wall–Associated Genes
Although perturbations of lignin-related genes such as CCR and C3H could induce severe phenotypes (Jones et al., 2001; Franke et al., 2002), most CAD upregulated or downregulated single mutants or transgenics did not result in a strong phenotype and/or modifications in lignin content. Baucher et al. (2003) explained this phenomenon by the ability of CAD-modified plants to incorporate aldehyde precursors into lignin to compensate for the reduced availability of corresponding monolignols. Because the cad-c cad-d double mutant produced the strongest impact on lignin described to date in CAD downregulated plants, we were interested in determining whether other genes of the phenylpropanoid pathway were altered in this mutant and contributed to its phenotype. We took advantage of the Affymetrix Arabidopsis ATH1-24K GeneChip to analyze alterations of whole genome expression in the lignified tissues of the mutant compared with the wild type.
The consistent expression level of the majority of the genes encoding enzymes positioned upstream of the CAD step in both lines suggested that the double mutation does not induce substantial feedback on their transcription in the tissues harvested. This finding is in accordance with the unchanged expression of most transcription factors previously shown to promote lignification (Borevitz et al., 2000; Liljegren et al., 2000; Mele et al., 2003; Newman et al., 2004). By contrast, alterations in transcript abundance of other genes encoding enzymes downstream from the CAD step, such as laccases and peroxidases, suggest different regulation for those genes compared with genes involved in monolignol biosynthesis. For example, altered transcript levels of laccase and peroxidase genes could be related to a decrease in the amount of conventional monomer availability for oxidative polymerization. However, microarray analyses were only performed at one time point during the growth and development of the Arabidopsis stem. Therefore, it is possible that expression of these genes might be impacted in the double mutant at other developmental stages.
Alteration of transcription levels of other genes involved in cell wall biosynthesis, such as cellulose synthases, arabinogalactans, pectinacetylesterases, glycosyl hydrolases, and UDP-glucose transferases, suggests that the cell wall–related phenotype of the double mutant goes beyond modified lignin deposition, potentially affecting synthesis of cell wall polysaccharides. However, additional chemical analysis is needed to substantiate this possibility. The correlation of GRP transcript abundance with the decrease in PRP transcripts, as well as the decrease in IRX3 and IRX5 transcripts, which are specific to secondary cell wall biosynthesis, suggests a delay in secondary cell wall formation in this double mutant. Indeed, under normal conditions, GRP abundance in the primary wall is negatively correlated with lignification (Condit, 1993). It has been suggested that GRP may be part of a structural mechanism stabilizing conductive elements to prevent water flow between adjacent protoxylem (Ringli et al., 2001), and the increased level of GRP transcripts in the double mutant may thus reflect activation of a compensatory mechanism in response to a mechanical deficiency.
Interestingly, transcripts related to detoxification, such as GST proteins, are also overrepresented in the double mutant. GST catalyzes the conjugation of a variety of hydrophobic, electrophilic, and cytotoxic substrates that are often xenobiotics. Aldehyde derivatives that accumulate in the double mutant are also members of a group suggested to be GST substrates, based upon a common chemical signal named the Michael acceptor, consisting of a carbon–carbon double bond adjacent to an electron-withdrawing group (Marrs, 1996). GST-encoding genes can be induced by their own substrates (Marrs, 1996). One could therefore speculate that a detoxifying process is activated in stem tissues of the double mutant in response to aldehyde accumulation.
Neither microarray nor real-time PCR analyses revealed a significant decrease in transcript levels for any of the Arabidopsis CAD genes in the double mutant other than the expected CAD-C and CAD-D. These findings suggest that the other CAD members are not associated with the dramatic phenotype of cad-c cad-d. Notably, CAD-A and CAD1 transcripts were slightly more abundant in cad-c, cad-d, and cad-c cad-d mutants, and this increase in transcript level seems to be correlated with the severity of lignin phenotypes in these mutants (Sibout et al., 2003; this work). One could therefore speculate that a CAD compensatory mechanism may take place in the double mutant. This mechanism could be responsible to some extent for the biosynthesis of the conventional monolignols still weakly detectable in the double mutant during thioacidolysis despite the lack of significantly detectable in vitro CAD activity of CAD1, CAD-A, and CAD-G (Kim et al., 2004). However, β-glucuronidase promoter expression analysis for CAD-G confirmed a weak but significant expression pattern in lignified tissues of wild-type stems and also a strong expression pattern in fiber tissues of the double mutant stems (data not shown). Finally, a robust analysis of CAD gene transcription in wild-type and cad mutant plants was obtained by absolute quantification with real-time PCR. This method allowed detection of the magnitude of CAD gene expression in a lignified tissue—the base of the Arabidopsis stem. Thus, CAD-B1 and -B2 were shown to be very poorly expressed, which would support an insignificant role in stem lignification.
Precursors of Syringyl and Guaiacyl Units Are Synthesized by CAD-C and -D
Our results suggest that both CAD-C and -D are involved in the synthesis of syringyl and guaiacyl precursors in Arabidopsis for the following reasons. First, both of the single mutants were affected in sinapyl and coniferyl alcohol activities, with cad-d being more deficient in sinapyl alcohol activity than cad-c (Sibout et al., 2003). Second, the slight decrease in lignin-derived thioacidolysis monomers observed in the single mutant cad-d, combined with a dramatic decrease in the double mutant, highlights the importance of the redundant expression pattern observed previously for the two genes (Sibout et al., 2003), validating the synergistic roles for supplying G and S precursors. Third, the three types of lignin units (G, S, and H) are drastically affected in the double mutant, resulting in the production of a highly condensed lignin in which sinapaldehyde and coniferaldehyde are incorporated through a β-O-4 bonding pattern. Finally, additional confirmation of a complementary role for CAD-D and CAD-C in G- and S-unit biosynthesis was provided by the ability of each gene to complement the double mutant when expressed under the control of the CAD-D promoter.
That two genes should be involved in both coniferyl and sinapyl alcohol biosynthesis is intriguing. The possibility that they are the result of a gene duplication event (Tavares et al., 2000) that led to the evolution of additional functions is supported by expression analysis that showed that CAD-C has a broader pattern of expression when compared with CAD-D (Sibout et al., 2003). A range of aldehyde substrates are potentially available to CAD-C in the Arabidopsis leaf (for review, see Kirch et al., 2004). CAD-C may also be able to modulate the pools of ferulate and sinapate in Arabidopsis leaves because an aldehyde dehydrogenase REF1 has been discovered recently (Nair et al., 2004). The authors showed that cell wall–linked ferulate esters were decreased in the ref1 plants. The slight amount of ferulic and sinapic acids released by thioacidolysis of extract-free stems of the double mutant is consistent and suggests that aldehydes accumulated in the mutant because of the CAD deficiency could be oxidized by REF1 to produce their corresponding acids. The high raw signal detected for REF1 in our microarray analysis of Arabidopsis stems (see Supplemental Table 9 online) is consistent with this hypothesis.
cad-c cad-d as a Tool to Study CAD/SAD Genes from Woody Plants
The absence of S units in gymnosperm lignin has long been attributed to a lack of the enzymatic function (such as 5-hydroxylase or 5-O-methyl transferase activity) required for the transformation of coniferaldehyde into sinapyl alcohol. CAD proteins isolated from gymnosperms showed very low sinapyl alcohol activity in vitro, whereas most angiosperm CAD proteins displayed suitable activity for both sinapyl and coniferyl alcohols, consistent with the lignin structure and the taxonomy of these plants. The ability of the spruce CAD to produce S units in the double mutant was surprising at first. However, the low but bona fide sinapaldehyde affinity often detected in gymnosperm CAD proteins and the amounts of syringyl moieties found in some conifers support our results (Fullerton and Franisch, 1983; Lewis and Yamamoto, 1990). Our data clearly show that the absence of S units in some conifers (at least in spruce) is not due to the CAD gene. Therefore, introduction of angiosperm CAD genes into transgenic conifers to produce enriched S lignins for kraft pulping improvement may not be necessary, although it may be useful for increasing syringyl yield.
It is interesting to note that whereas the tissue-specific expression of Arabidopsis CAD-D restored the lignin phenotype of the mutant with a recovery of the S:G thioacidolysis ratio and decrease of aldehydes, CAD-C overexpression only partially restored the S:G ratio, suggesting slight differences in substrate affinity between these enzymes. In addition, restored lignin in ChimAtCAD-C lines is comparable to lignin of ChimPaCAD lines, suggesting similar protein properties between them, consistent with results obtained by Kim et al. (2004), who found that Arabidopsis CAD-C protein showed higher affinity to coniferaldehyde than to sinapaldehyde.
Recently, Lapierre et al. (2004) reported a detailed analysis of structural traits of lignins in CAD-deficient poplars. They showed that the lignin of their CAD-deficient poplar lines had fewer syringyl and β-O-4 units and displayed a slight but significant level of sinapaldehyde. The amount of coniferaldehyde units, however, was not enhanced. In our work, we show that the same gene complements cad-c cad-d, completely restoring the level of conventional G units in the lignin, but also that of the conventional S units. These observations suggest a bifunctional activity for this protein. These results, however, are not consistent with the role proposed by Li et al. (2001) for P. tremuloides CAD, which is homologous to P. deltoides CAD, which is supposed to be involved in coniferyl alcohol biosynthesis only. Our study also shows that the role of SAD genes in lignification requires further investigation. The ability of P. tremula × tremuloides SAD1 to restore the thioacidolysis S:G ratio in ChimPttSAD1 lines confirms the existence of an aldehyde reduction activity of the SAD protein in accordance with in vitro assays shown by Li et al. (2001) and by Bomati and Noel (2005). Nevertheless, the very low yield in syringyl units and the amount of sinapaldehyde remaining in ChimPttSAD1 lines compared with other restored lines (including ChimPaCAD) do not support an essential role for this gene in the incorporation of S units into the constitutive lignin polymer. Nevertheless, the similarity of SAD to defense-related proteins suggests that this gene might be involved in synthesis of syringyl units or monolignol-like compounds in very specific conditions in poplar (Bomati and Noel, 2005).
In conclusion, the unusual incorporation of sinapaldehydes and coniferaldehydes in the lignin of the double mutant and the significant sinapaldehyde amounts remaining in lines overexpressing spruce CAD supports an aldehyde pathway (Osakabe et al., 1999) rather than an alcohol pathway for the production of sinapyl alcohol in Arabidopsis, despite the fact that an alcohol pathway may also be present (Humphreys et al., 1999). Also, accumulation of ferulic and sinapic acids in the double mutant is consistent with our model (Figure 6) and is supported by the recent description of an aldehyde dehydrogenase (REF1; Nair et al., 2004). However, our work does not exclude involvement of at least a third enzyme displaying very weak CAD activity responsible for the low content of conventional units (mainly guaiacyl units) remaining within the lignin of the double mutant. Thus, our work shows that the last step of monolignol biosynthesis in Arabidopsis is under the control of a multigenic process. CAD-D and CAD-C, which display a synergistic role in reducing coniferaldehyde and sinapaldehyde into corresponding alcohols, are the two main protagonists of the last reduction step in monolignol biosynthesis in Arabidopsis stems.
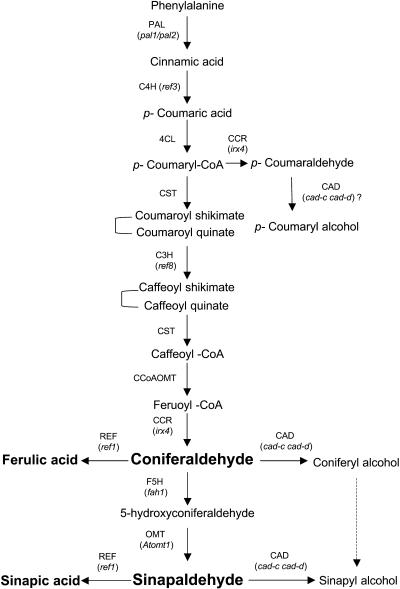
Figure 6.
Proposed Phenylpropanoid Pathway from Phe to Monolignols.
The enzymes are caffeoyl CoA O-methyltransferase (CCoAOMT), cinnamate 4-hydroxylase (C4H), CAD, hydroxycinnamoyl CoA reductase (CCR), C3H, 4-coumarate CoA ligase (4CL), ferulate 5-hydroxylase (F5H), hydroxycinnamoyl CoA:shikimate/quinate hydroxycinnamoyltransferase (CST), phenylammonia-lyase (PAL), caffeic acid/5-hydroxyferulic acid O-methyltransferase (COMT), and bifunctional hydroxycinnamaldehyde dehydrogenase (REF1). Mutants identified for corresponding enzymes are shown in bold between brackets. Compounds accumulating in the cad-c cad-d mutant are in italic.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana cv Wassilewskija wild type and the cad-c and cad-d mutants (Sibout et al., 2003) were used in this study. The cad-c cad-d double mutant lines were isolated from the F2 population of cad-d × cad-c crosses. The putative double mutant lines were identified and confirmed using genomic PCR analyses.
For RNA isolation and lignin and histological analyses of mature stems, each line was grown in the same growth chamber with random arrangement for 8 weeks at 23°C under 12 h of light per day and low moisture. Plants were grown as described by Chaffey et al. (2002) for inducing hypocotyl secondary growth.
Protein Gel Blot Analysis
Protein gel blot analysis was conducted as described by Sibout et al. (2003). Briefly, total protein extracts were obtained by homogenization of mature stems in 100 mM Tris-HCl, pH 7.5, containing 10% polyvinylpolypyrrolidone (Sigma-Aldrich, St. Louis, MO) and 10 μM β-mercaptoethanol. Protein samples (15 μg) were heated at 95°C for 5 min and loaded on a 15% acrylamide SDS-PAGE with a 12% resolving gel using a Bio-Rad Protean II apparatus (Hercules, CA). Proteins were transferred onto 0.45-μm nitrocellulose membrane (Amersham Biosciences, Piscataway, NJ) by electroblotting. The polyclonal antibodies directed against Nicotiana tabacum xylem CAD2 were used at a 1:800 dilution. Blots were developed using the Western Light Plus kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions.
Arabidopsis Transformation
Binary vectors were introduced into the Agrobacterium tumefaciens strain C58pMP90 (Koncz and Schell, 1986) by electroporation. Plants were transformed using the flower infiltration protocol (Bechtold and Pelletier, 1998). T1 transgenic plants were selected on medium containing hygromycin (50 mg/L) or kanamycin (100 mg/L).
Histology
Fresh hand-cut sections were subjected to histochemical analysis or immediately observed under a Zeiss Axioskop microscope (Jena, Germany). Wiesner staining was performed by incubating sections in 1% phloroglucinol in ethanol:water (7:3) with 30% HCl. Mäule staining was performed by first incubating sections in KMnO4. After 10 min, sections were washed and acidified with HCl for 1 min, washed again, and then incubated in NaHCO3. For detection of carbohydrates and phenolics in the hypocotyls, sections were incubated 15 min in a Safranin O/Alcian Blue 1:1 (v/v) solution and then washed before observation under UV light.
RNA Extraction and Real-Time PCR
Because cad-c cad-d is delayed in growth and bolting and to assure uniformity of samples, the first emerging stems (5 to 10 cm long) were removed by cutting at the base and not used for analysis. This removal of the first stem allowed us to standardize the growth of all secondary stems that emerged subsequently at the same time for all lines. The development of the first 20 cm of stem was identical between mutants and controls under our growth conditions. At this stage of growth, the base of the stems (first 5 cm) was harvested at random from individual plants, three of which were pooled to constitute one sample, thereby providing enough material for analysis. Cauline leaves and secondary buds were systematically removed from stem tissues. For each line (the wild type, cad- d, cad-c, and cad-c cad-d), five samples were harvested. Samples were immediately frozen in liquid nitrogen and stored at −80°C until extraction.
RNA from each sample was extracted using the RNeasy extraction kit (Qiagen, Valencia, CA) according to the manufacturer's recommendations, with an extra DNase I treatment. RNA samples were quantified using a Beckman spectrophotometer (Fullerton, CA). Reverse transcriptase (Superscript II; Invitrogen, Carlsbad, CA) reactions were performed using 1 μg of total RNA in a 20-μL volume according to the manufacturer's instructions. These reactions were done at the same time for all samples. The reverse-transcribed cDNA was then treated with RNase H and diluted fivefold before PCR analysis.
Real-time PCR analyses were performed using the Quantitec SYBR green system (Qiagen) and the Opticon PCR machine (MJ Research, Waltham, MA). Data were treated using the Opticon 2 software provided by MJ Research. Because Arabidopsis CAD genes belong to a small family, oligonucleotide primers were designed to take advantage of the unique sequences on the 3′ untranslated cDNA regions and had a Tm close to 70°C (see Supplemental Table 12 online). PCR amplification was done in two steps: DNA denaturation at 94°C for 10 s and elongation at 62°C for 2 min. Fluorescence was evaluated at the end of the 2-min elongation. PCR reactions were maintained for 45 cycles. Absolute quantity of transcripts was calculated using standard curves as described by Rutledge and Côté (2003).
Microarray Analysis
Samples were harvested and prepared as described for real-time PCR analysis, except that 10 bases of stems of individual plants were pooled to provide enough material for one sample. Four samples were harvested for each line (cad-c cad-d and control). Ten micrograms of total RNA from each biological sample were used for each ATH1 GeneChip hybridization (Arabidopsis Genome ATH1 Array). Complementary RNA was prepared as described in the Affymetrix ATH1 GeneChip Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA; available at http://genomequebec.mcgill.ca). Data generated from Affymetrix Microarray Suite software (MAS5.0) was downloaded in Excel format for each single ATH1 GeneChip (see Supplemental Tables 1 to 8 online) from the Affymetrix Web site to verify the level of confidence (P value) for the probe set of interest. Each signal value was assigned a detection call of P (present, indicating that the signal is of high quality), A (absent, indicating that the signal is generally too low and/or unreliable), or M (marginal, indicating that the hybridization signal is negligible [no transcript detected]). The unpaired Student's t test (P = 0.05) was performed on the data from the eight ATH1 GeneChip samples using MAS5.0 software to select a first set of genes potentially upregulated or downregulated in the mutant. False positives were eliminated by subtracting probe set signals that showed a difference lower than 200 and a fold change of less than two.
Transformation Vectors and Cloning Procedures
The binary plasmid pCAMBIA-1390-HygR (Cambia, Canberra, Australia) was modified and used for this study. A plant expression system was used to express specifically different CAD genes with the goal to complement the cad-c cad-d mutant. In each vector, the different CAD genes were under the control of the Arabidopsis CAD-D promoter and the nopaline synthase gene terminator. The CAD-D promoter, including the 5′ untranslated region, was amplified from the previous construction pCAD-D:β-glucuronidase (see Sibout et al., 2003) using primers EcoRIpcad3const 5′-GGAATTCGAAATTCTCCACTCGTAGCTCTTCGTTCTG-3′ and SpeIpcad3const 5′-GACTAGTTCTTTCTTCTTTCTTATCTTGATCTGCTGC-3′. The PCR product was cloned using the pGEM-T Easy vector cloning kit (Promega, Madison, WI). Selected clones were sequenced to verify the absence of sequence errors and digested to insert the amplicon between the EcoRI-SpeI sites of the binary vector pCAMBIA 1390. This new vector, named CHIMpCAMBIA 1390, was used for all subsequent constructions.
cDNAs corresponding to Populus tremula × tremuloides SAD1 (AY850131), Picea abies CAD (AJ868574), and Populus deltoides CAD (Z19568) were cloned from P. tremula × tremuloides, P. abies, and P. deltoides, respectively. P. abies CAD was a kind gift from M.H. Walter (Leibniz Institute of Plant Biochemistry, Halle, Germany), whereas cDNAs for Arabidopsis CAD-D (At4g34230) and Arabidopsis CAD-C (At3g19450) were cloned from stem RNA using RT-PCR (ecotype Wassilewskija). Besides the SpeI site inserted into both primers and used to amplify each cDNA, five adenosines were added between the SpeI site and the ATG start codon of each gene (only the translated region, not the 5′ and 3′ untranslated regions, was amplified for each gene). Resulting amplicons were cloned into the pGEM-T easy vector system, sequenced, digested with SpeI, and inserted into the binary vector CHIMpCAMBIA 1390. Clones with the appropriate sense of open reading frames were selected after sequencing.
FTIR Spectroscopy
Stems from 3-week-old plants were used for analysis. Three biological replicates of wild-type or mutant stems were embedded in 0.8% agarose and then sectioned with a vibratome to assure similar thickness of sections. Five different sections per replicate were subjected to FTIR analysis. An area of 50 μm × 50 μm in the xylem bundles or in the interfascicular fibers was selected for spectral collection. Spectra were collected using a ThermoNicolet Nexus spectrometer (Madison, WI) with a Continμum microscope accessory. Fifty interferograms were collected in transmission mode with 8 cm−1 resolution and coadded to improve the signal-to-noise ratio of the spectrum. Spectra were then baselined and normalized as described by Robin et al. (2003). Student's t test was applied as described by Mouille et al. (2003).
Lignin Analysis
Dried mature stems were collected after removal of the leaves and siliques. Extract-free samples were prepared using a Soxhlet apparatus by sequentially extracting the ground material with toluene:ethanol (2:1, v/v), ethanol, and water. The determination of lignin content was performed on the extract-free samples using the standard Klason procedure. The evaluation of lignin structure was performed on whole plant material or on extract-free material, using the thioacidolysis procedure. The lignin-derived monomers were identified by GC-MS as their trimethyl-silylated derivatives.
Cellulolysis
The susceptibility to cellulolysis was evaluated using a method adapted from Rexen (1977): 200 mg of the extract-free ground sample were placed in 30 mL of 0.05 M sodium acetate buffer, pH 4.5, containing 2 mg/mL of commercial cellulase (cellulase Onozuka-R10; Serva, Heidelberg, Germany). Cellulolysis was performed for 48 h at 37°C with magnetic stirring. After incubation, the reaction medium was filtered over a tared filtering crucible. The residue was then washed with water, oven-dried, and gravimetrically determined.
Sequence data from this article have been deposited previously in the EMBL/GenBank libraries under the following accession numbers: P. tremula × tremuloides SAD1 (AY850131), P. tremuloides SAD (AF273256), P. abies CAD (AJ868574), and P. deltoides CAD (Z19568). AGI locus identifiers are as follows: CAD-A (At4g34970), CAD-B1 (At4g37980), CAD-B2 (At4g37990), CAD-C (At3g19450), CAD-D (At4g34230), CAD-CAD1 (At4g34930), CAD-F (At2g21890), and CAD-G (At1g72680).
Supplementary Material
Acknowledgments
We thank Frédérick Legée and Laurent Cézart (both of Institut National de la Recherche Agronomique–Institut National d'Agronomie de Paris-Grignon) for Klason and thioacidolysis analyses, respectively. We are grateful to Robert Rutledge (Natural Resources Canada) for critical reading of the manuscript and technical advice on quantitative PCR and also to Anic Levasseur (Natural Resources Canada) for her help in Arabidopsis stem sections of transgenic lines. Harry Zuzan (McGill University and Genome Quebec Innovation Centre, Montreal, Canada) also deserves credit for his help with the microarray statistical analysis. We thank Denis Lachance (Natural Resources Canada), Christian Hardtke, and Tamara Western (both of McGill University) for helpful discussion as well as the knowledgeable reviewers for their constructive criticism and suggestions on an earlier version of this manuscript. We are grateful to C. Halpin (University of Dundee, UK) and L. Li (North Carolina State University, Raleigh, NC) for providing CAD and SAD antibodies, respectively. We also thank M.H. Walter and V. Chiang (North Carolina State University) for providing us P. abies CAD and P. tremuloides SAD cDNAs, respectively. Work in L.J.'s laboratory is partly supported by Genoplante Program 2001-009. Work in A.S.'s laboratory is supported by a Canadian Biotechnology Strategy grant and a Natural Sciences and Engineering Research Council of Canada Discovery grant.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Armand Séguin (armand.seguin@nrcan.gc.ca).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.030767.
References
- Andersson-Gunnerås, S., Hellgren, J.M., Björklund, S., Regan, S., Moritz, T., and Sundberg, B. (2003). Asymmetric expression of a poplar ACC oxidase controls ethylene production during gravitational induction of tension wood. Plant J. 34, 339–349. [DOI] [PubMed] [Google Scholar]
- Anterola, A.M., and Lewis, N.G. (2002). Trends in lignin modification: A comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry 61, 221–294. [DOI] [PubMed] [Google Scholar]
- Baucher, M., Bernard-Vailhé, M.A., Chabbert, B., Besle, J.-M., Opsomer, C., Van Montagu, M., and Botterman, J. (1999). Down-regulation of cinnamyl alcohol dehydrogenase in transgenic alfalfa (Medicago sativa L.) and the effect on lignin composition and digestibility. Plant Mol. Biol. 39, 437–447. [DOI] [PubMed] [Google Scholar]
- Baucher, M., Chabbert, B., Pilate, G., Van Doorsselaere, J., Tollier, M.-T., Petit-Conil, M., Cornu, D., Monties, B., Van Montagu, M., Inzé, D., Jouanin, L., and Boerjan, W. (1996). Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in poplar. Plant Physiol. 112, 1479–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucher, M., Halpin, C., Petit-Conil, M., and Boerjan, W. (2003). Lignin: Genetic engineering and impact on pulping. Crit. Rev. Biochem. Mol. Biol. 38, 305–350. [DOI] [PubMed] [Google Scholar]
- Baucher, M., Monties, B., Van Montagu, M., and Boerjan, W. (1998). Biosynthesis and genetic engineering of lignin. Crit. Rev. Plant Sci. 17, 125–197. [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. [DOI] [PubMed] [Google Scholar]
- Bomati, E.K., and Noel, J.P. (2005). Structural and kinetic basis for substrate selectivity in Populus tremuloides sinapyl alcohol dehydrogenase. Plant Cell 17, 1598–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz, J.O., Xia, Y., Blount, J., Dixon, R.A., and Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill, E.M., Abrahams, S., Hayes, C.M., Jenkins, C.L.D., and Watson, J.M. (1999). Molecular characterisation and expression of a wound-inducible cDNA encoding a novel cinnamyl-alcohol dehydrogenase enzyme in lucerne (Medicago sativa L.). Plant Mol. Biol. 41, 279–291. [DOI] [PubMed] [Google Scholar]
- Brown, D.E., Rashotte, A.M., Murphy, A.S., Normanly, J., Tague, B.W., Peer, W.A., Taiz, L., and Muday, G.K. (2001). Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 126, 524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buta, J.G., and Galetti, G.C. (1989). FT-IR investigation of lignin components in various agricultural lignocellulosic by-products. J. Sci. Food Agric. 49, 37–43. [Google Scholar]
- Campbell, M.M., and Sederoff, R.R. (1996). Variation in lignin content and composition. Mechanisms of control and implications for the genetic improvement of plants. Plant Physiol. 110, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffey, N., Cholewa, E., Regan, S., and Sundberg, B. (2002). Secondary xylem development in Arabidopsis: A model for wood formation. Physiol. Plant. 114, 594–600. [DOI] [PubMed] [Google Scholar]
- Chen, L., Auh, C.-K., Dowling, P., Bell, J., Chen, F., Hopkins, A., Dixon, R.A., and Wang, Z.-Y. (2003). Improved forage digestibility of tall fescue (Festuca arundinacea) by transgenic down-regulation of cinnamyl alcohol dehydrogenase. Plant Biotechnol. J. 1, 437–449. [DOI] [PubMed] [Google Scholar]
- Condit, C.M. (1993). Developmental expression and localization of petunia glycine-rich protein 1. Plant Cell 5, 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, M.A., Collins, R.E., Anterola, A.M., Cochrane, F.C., Davin, L.B., and Lewis, N.G. (2003). An in silico assessment of gene function and organization of the phenylpropanoid pathway metabolic networks in Arabidopsis thaliana and limitations thereof. Phytochemistry 64, 1097–1112. [DOI] [PubMed] [Google Scholar]
- Faix, O. (1992). Fourier transform infrared spectroscopy. In Methods in Lignin Chemistry, S.Y. Lin and C.W. Dence, eds (Berlin: Springer-Verlag), pp. 83–109.
- Feuillet, C., Lauvergeat, V., Deswarte, C., Pilate, G., Boudet, A., and Grima-Pettenati, J. (1995). Tissue- and cell-specific expression of a cinnamyl alcohol dehydrogenase promoter in transgenic poplar plants. Plant Mol. Biol. 27, 651–667. [DOI] [PubMed] [Google Scholar]
- Franke, R., Humphreys, J.M., Hemm, M.R., Denault, J.W., Ruegger, M.O., Cusumano, J.C., and Chapple, C. (2002). The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 30, 33–45. [DOI] [PubMed] [Google Scholar]
- Fullerton, T.J., and Franisch, R.A. (1983). Lignin analysis by pyrolysis-GC-MS: Characterisation of ethanol lignin pyrolysates and identification of syringyl units in Pinus radiata milled wood lignin. Holzforschung 37, 267–269. [Google Scholar]
- Galliano, H., Cabane, M., Eckerskorn, C., Lottspeich, F., Sandermann, H., Jr., and Ernst, D. (1993. b). Molecular cloning, sequence analysis and elicitor-/ozone-induced accumulation of cinnamyl alcohol dehydrogenase from Norway spruce (Picea abies L.). Plant Mol. Biol. 23, 145–156. [DOI] [PubMed] [Google Scholar]
- Galliano, H., Heller, W., and Sandermann, H., Jr. (1993. a). Ozone induction and purification of spruce cinnamyl alcohol dehydrogenase. Phytochemistry 32, 557–563. [Google Scholar]
- Goujon, T., Sibout, R., Eudes, A., MacKay, J., and Jouanin, L. (2003). Genes involved in the biosynthesis of lignin precursors in Arabidopsis thaliana. Plant Physiol. Biochem. 41, 677–687. [Google Scholar]
- Grima-Pettenati, J., Campargue, C., Boudet, A., and Boudet, A.M. (1994). Purification and characterization of cinnamyl alcohol dehydrogenase isoforms from Phaseolus vulgaris. Phytochemistry 37, 941–947. [DOI] [PubMed] [Google Scholar]
- Halpin, C., Holt, K., Chojecki, J., Oliver, D., Chabbert, B., Monties, B., Edwards, K., Barakate, A., and Foxon, G.A. (1998). Brown-midrib maize (bm1)—A mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J. 14, 545–553. [DOI] [PubMed] [Google Scholar]
- Halpin, C., Knight, M.E., Foxon, G.A., Campbell, M.M., Boudet, A.M., Boon, J.J., Chabbert, B., Tollier, M.-T., and Schuch, W. (1994). Manipulation of lignin quality by downregulation of cinnamyl alcohol dehydrogenase. Plant J. 6, 339–350. [Google Scholar]
- Hawkins, S., Samaj, J., Lauvergeat, V., Boudet, A., and Grima-Pettenati, J. (1997). Cinnamyl alcohol dehydrogenase: Identification of new sites of promoter activity in transgenic poplar. Plant Physiol. 113, 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins, S.W., and Boudet, A.M. (1994). Purification and characterization of cinnamyl alcohol dehydrogenase isoforms from the periderm of Eucalyptus gunnii Hook. Plant Physiol. 104, 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, T., Ito, T., Umezawa, T., Hibino, T., and Shibata, D. (1994). Red-brown color of lignified tissues of transgenic plants with antisense CAD gene: Wine-red lignin from coniferyl aldehyde. J. Biotechnol. 37, 151–158. [Google Scholar]
- Humphreys, J.M., Hemm, M.R., and Chapple, C. (1999). New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc. Natl. Acad. Sci. USA 96, 10045–10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, H.-W., Han, D.C., Son, K.-H., Han, M.Y., Lim, J.-S., Ha, J.-H., Lee, C.W., Kim, H.M., Kim, H.-C., and Kwon, B.-M. (2003). Antitumor effect of the cinnamaldehyde derivative CB403 through the arrest of cell cycle progression in the G2/M phase. Biochem. Pharmacol. 65, 1343–1350. [DOI] [PubMed] [Google Scholar]
- Jones, L., Ennos, A.R., and Turner, S.R. (2001). Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J. 26, 205–216. [DOI] [PubMed] [Google Scholar]
- Kim, H., Ralph, J., Lu, F., Pilate, G., Leplé, J.C., Pollet, B., and Lapierre, C. (2002). Identification of the structure and origin of thioacidolysis marker compounds for cinnamyl alcohol dehydrogenase deficiency in angiosperms. J. Biol. Chem. 49, 47412–47419. [DOI] [PubMed] [Google Scholar]
- Kim, S.-J., Kim, M.-R., Bedgar, D.L., Moinuddin, S.G.A., Cardenas, C.L., Davin, L.B., Kang, C., and Lewis, N.G. (2004). Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc. Natl. Acad. Sci. USA 101, 1455–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch, H.-H., Bartels, D., Wei, Y., Schnable, P.S., and Wood, A.J. (2004). The ALDH gene superfamily of Arabidopsis. Trends Plant Sci. 9, 371–377. [DOI] [PubMed] [Google Scholar]
- Knight, M.E., Halpin, C., and Schuch, W. (1992). Identification and characterisation of cDNA clones encoding cinnamyl dehydrogenase from tobacco. Plant Mol. Biol. 19, 793–801. [DOI] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204, 383–396. [Google Scholar]
- Kutsuki, H., Shimada, M., and Higuchi, T. (1982). Regulatory role of cinnamyl alcohol dehydrogenase in the formation of guaiacyl and syringyl lignins. Phytochemistry 21, 19–23. [Google Scholar]
- Lapierre, C., Pilate, G., Pollet, B., Mila, I., Leplé, J.-C., Jouanin, L., Kim, H., and Ralph, J. (2004). Signatures of cinnamyl alcohol dehydrogenase deficiency in poplar lignins. Phytochemistry 65, 313–321. [DOI] [PubMed] [Google Scholar]
- Lapierre, C., Pollet, B., MacKay, J.J., and Sederoff, R.R. (2000). Lignin structure in a mutant pine deficient in cinnamyl alcohol dehydrogenase. J. Agric. Food Chem. 48, 2326–2331. [DOI] [PubMed] [Google Scholar]
- Lewis, N.G., and Yamamoto, E. (1990). Lignin: Occurrence, biogenesis and biodegradation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41, 455–496. [DOI] [PubMed] [Google Scholar]
- Li, L., Cheng, X.F., Leshkevich, J., Umezawa, T., Harding, S.A., and Chiang, V.L. (2001). The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding sinapyl alcohol dehydrogenase. Plant Cell 13, 1567–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren, S.J., Ditta, G.S., Eshed, Y., Savidge, B., Bowman, J.L., and Yanofsky, M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. [DOI] [PubMed] [Google Scholar]
- Lim, E.-K., Li, Y., Parr, A., Jackson, R., Ashford, D.A., and Bowles, D.J. (2001). Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J. Biol. Chem. 276, 4344–4349. [DOI] [PubMed] [Google Scholar]
- Lin, S.Y., and Dence, C.W. (1992). Methods in Lignin Chemistry. (Berlin: Springer-Verlag).
- MacKay, J.J., Liu, W., Whetten, R., Sederoff, R.R., and O'Malley, D.M. (1995). Genetic analysis of cinnamyl alcohol dehydrogenase in loblolly pine: Single gene inheritance, molecular characterization and evolution. Mol. Gen. Genet. 247, 537–545. [DOI] [PubMed] [Google Scholar]
- Mansell, R.L., Gross, G.G., Stoeckigt, J., Franke, H., and Zenk, M.H. (1974). Purification and properties of cinnamyl alcohol dehydrogenase from higher plants involved in lignin biosynthesis. Phytochemistry 13, 2427–2436. [Google Scholar]
- Marita, J.M., Vermerris, W., Ralph, J., and Hatfield, R.D. (2003). Variations in the cell wall composition of maize brown midrib mutants. J. Agric. Food Chem. 51, 1313–1321. [DOI] [PubMed] [Google Scholar]
- Marrs, A.K. (1996). The functions and regulation of glutathione S-transferases in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 127–158. [DOI] [PubMed] [Google Scholar]
- Mele, G., Ori, N., Sato, Y., and Hake, S. (2003). The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 17, 2088–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerowicz, E.J., Baucher, M., Sundberg, B., and Boerjan, W. (2001). Unravelling cell wall formation in the woody dicot stem. Plant Mol. Biol. 47, 239–274. [PubMed] [Google Scholar]
- Mouille, G., Robin, S., Lecomte, M., Pagant, S., and Höfte, H. (2003). Classification and identification of Arabidopsis cell wall mutants using Fourier-Transform InfraRed (FT-IR) microspectroscopy. Plant J. 35, 393–404. [DOI] [PubMed] [Google Scholar]
- Nair, R.B., Bastress, K.L., Ruegger, M.O., Denault, J.W., and Chapple, C. (2004). The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 16, 544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, L.J., Perazza, D.E., Juda, L., and Campbell, M.M. (2004). Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J. 37, 239–250. [DOI] [PubMed] [Google Scholar]
- Osakabe, K., Tsao, C.C., Li, L., Popko, J.L., Umezawa, T., Carraway, D.T., Smeltzer, R.H., Joshi, C.P., and Chiang, V.L. (1999). Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc. Natl. Acad. Sci. USA 96, 8955–8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen, N.L., and Thomas, D.W. (1989). Infrared studies of “hard” and “soft” woods. Appl. Spectrosc. 43, 451–455. [Google Scholar]
- Raes, J., Rohde, A., Christensen, J.H., Van de Peer, Y., and Boerjan, W. (2003). Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133, 1051–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexen, B. (1977). Enzyme solubility: A method for evaluating the digestibility of alkali-treated straw. Anim. Feed Sci. Technol. 2, 205–218. [Google Scholar]
- Ringli, C., Keller, B., and Ryser, U. (2001). Glycine-rich proteins as structural components of plant cell walls. Cell. Mol. Life Sci. 58, 1430–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin, S., Lecomte, M., Höfte, H., and Mouille, G. (2003). A procedure for the clustering of cell wall mutants in the model plant Arabidopsis based on Fourier-transform infrared (FT-IR) spectroscopy. J. Appl. Stat. 30, 669–681. [Google Scholar]
- Rutledge, R.G., and Côté, C. (2003). Mathematics of quantitative kinetic PCR and the application of standard curves. Nucleic Acids Res. 31, e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutten, B., and Gocke, E. (1988). The “antimutagenic” effect of cinnamaldehyde is due to a transient growth inhibition. Mutat. Res. 201, 97–105. [DOI] [PubMed] [Google Scholar]
- Sarkanen, K.V., Chang, H.-M., and Ericsson, B. (1967). Species variation in lignins. TAPPI (Tech. Assoc. Pulp Pap. Ind.) 50, 572–575. [Google Scholar]
- Sasaki, T., and Sederoff, R.R. (2003). Genome studies and molecular genetics. The rice genome and comparative genomics of higher plants. Curr. Opin. Plant Biol. 6, 97–100. [Google Scholar]
- Sibout, R., Eudes, A., Pollet, B., Goujon, T., Mila, I., Granier, F., Séguin, A., Lapierre, C., and Jouanin, L. (2003). Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis. Isolation and characterization of the corresponding mutants. Plant Physiol. 132, 848–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, D., Yahiaoui, N., McDougall, G.J., Myton, K., Marque, C., Boudet, A.M., and Haigh, J. (1997). Fourier-transform infrared and Raman spectroscopic evidence for the incorporation of cinnamaldehydes into the lignin of transgenic tobacco (Nicotiana tabacum L.) plants with reduced expression of cinnamyl alcohol dehydrogenase. Planta 201, 311–318. [DOI] [PubMed] [Google Scholar]
- Tavares, R., Aubourg, S., Lecharny, A., and Kreis, M. (2000). Organization and structural evolution of four multigene families in Arabidopsis thaliana: AtLCAD, AtLGT, AtMYST and AtHD-GL2. Plant Mol. Biol. 42, 703–717. [DOI] [PubMed] [Google Scholar]
- Tobias, C.M., and Chowk, E.K. (2005). Structure of the cinnamyl-alcohol dehydrogenase gene family in rice and promoter activity of a member associated with lignification. Planta 220, 678–688. [DOI] [PubMed] [Google Scholar]
- Turner, S.R., and Somerville, C.R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9, 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utama, I.M.S., Wills, R.B.H., Ben-yehoshua, S., and Kuek, C. (2002). In vitro efficacy of plant volatiles for inhibiting the growth of fruit and vegetable decay microorganisms. J. Agric. Food Chem. 22, 6371–6377. [DOI] [PubMed] [Google Scholar]
- Zhong, R., Jennifer, J.J., and Ye, Z.-H. (1997). Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 9, 2159–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Morrison III, W.H., Himmelsbach, D.S., Poole II, F.L., and Ye, Z.-H. (2000). Essential role of caffeoyl coenzyme A O-methyltransferase in lignin biosynthesis in woody poplar plants. Plant Physiol. 124, 563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.