Abstract
Reduced nicotinamide adenine dinucleotide phosphate (NADPH) is a crucial reducing cofactor for reductive biosynthesis and protection from oxidative stress. To fulfill their heightened anabolic and reductive power demands, cancer cells must boost their NADPH production. Progrowth and mitogenic protein kinases promote the activity of cytosolic NAD kinase (NADK), which produces NADP+, a limiting NADPH precursor. However, the molecular architecture and mechanistic regulation of human NADK remain undescribed. Here, we report the cryo–electron microscopy structure of human NADK, both in its apo-form and in complex with its substrate NAD+ (nicotinamide adenine dinucleotide), revealing a tetrameric organization with distinct structural features. We discover that the amino (N)- and carboxyl (C)-terminal tails of NADK have opposing effects on its enzymatic activity and cellular NADP(H) levels. Specifically, the C-terminal region is critical for NADK activity, whereas the N-terminal region exhibits an inhibitory role. This study highlights molecular insights into the regulation of a vital enzyme governing NADP(H) production.
The cryo-EM structures of human NADK in its free form and bound to NAD+ uncover key insights into its regulatory mechanisms.
INTRODUCTION
Reduced nicotinamide adenine dinucleotide phosphate (NADPH) serves as a vital reducing cofactor for anabolic biosynthetic pathways, including lipid, nucleotide, and amino acid synthesis (1, 2). In addition, NADPH fuels antioxidant systems, protecting cells from the reactive oxygen species (3). Cancer cells enhance NADPH production to meet their increased anabolic and reductive power demands (4, 5). Cells maintain NADPH pools in different subcellular compartments, such as the cytosol and mitochondria. NADPH levels are regulated by NAD kinases (NADKs), specifically cytosolic NADK and mitochondrial NADK2, which catalyze the synthesis of NADP+, the limiting precursor for NADPH production, within their respective locations (6–9). However, until recently, we had a limited understanding of the regulation of NADPH biosynthesis.
At the molecular level, several protein kinases, including Akt, protein kinase C (PKC), and extracellular signal–regulated kinase (ERK), have been shown to phosphorylate NADK at conserved serine residues within the N-terminal regulatory domain (10–13). These phosphorylation events stimulate NADK activity, increasing the cellular NADPH pool to support cancer cell growth. Emerging evidence suggests that NADK is a promising therapeutic target in cancer (14). Activating mutations of NADK have been observed in pancreatic cancer (15), while suppression of its activity hinders tumor growth in various cancers, including pancreatic cancer. Furthermore, NADK activity has been implicated in promoting breast cancer metastasis (16) and lymph node metastasis of lung cancer (17), highlighting the crucial role of NADK-dependent NADPH synthesis in cancer growth and spread. To elucidate the impact of cancer mutations and enable precise therapeutic targeting, detailed structural information on human NADK (hsNADK) is necessary.
In this study, we report the cryo–electron microscopy (cryo-EM) structure of hsNADK, both in its free form and bound to its substrate nicotinamide adenine dinucleotide (NAD+). In contrast to human mitochondrial NADK2 (18, 19), hsNADK, akin to its counterparts in most species, including bacteria and yeast, exists as a tetramer. Animal cytosolic NADKs, including hsNADK, have unique extensions that may regulate the conserved catalytic core (7, 20). Through biochemical and metabolic characterization of various deletion and point mutants, we highlight the significance of critical structural elements, including the N- and C-terminal disordered extensions, in hsNADK cellular function.
RESULTS
The cryo-EM structure of full-length hsNADK reveals tetrameric organization primed for catalysis
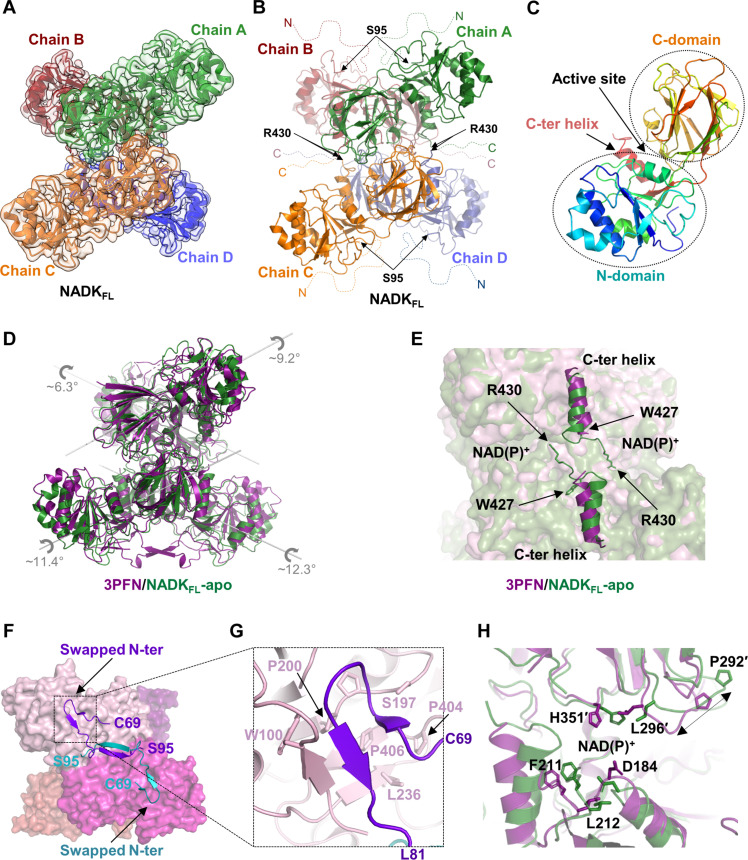
We solved the cryo-EM structure of full-length human NADK (NADKFL) at an overall resolution of 3.2 Å with D2 symmetry (Fig. 1, A and B, and Table 1). NADKFL with a C-terminal FLAG tag was purified from human embryonic kidney (HEK) 293 cells and was frozen on the grid before imaging with an electron microscope. The cryo-EM structure revealed a well-organized tetramer, similar to its bacterial and yeast counterparts but distinct from the dimeric human mitochondrial NADK2. Each NADKFL monomer comprises an N-terminal domain (residues 96 to 237) and a C-terminal domain (residues 238 to 430), both of which are well conserved (Fig. 1C and figs. S1, A and B, and S2A). The C-terminal domain includes a conserved helix at its distal end (residues 413 to 424), positioned between the two domains, with its C-terminal end oriented toward the active site. The active site is built similarly to other NADKs and located at the interface between the N-terminal domain from one monomer and the C-terminal domain from a second monomer (21, 22) (Fig. 1C).
Fig. 1. Cryo-EM structure of NADKFL revealing a tetrameric architecture.
(A) Cryo-EM structure of hsNADKFL purified from mammalian cells, revealing a tetrameric organization. Tetrameric NADK is shown embedded within its electron density, with each monomer in a distinct color that are labeled as chains A to D. (B) The organization of the tetrameric hsNADKFL is depicted with each monomer shown in a distinct color as in (A), and their disordered tails are indicated by dashed lines. The boundaries of the catalytic domain visible in the cryo-EM structure are indicated by arrows showing the first (S95) and last (R430) residues, modeled in the density for the N-tail and C-tail, respectively. (C) Domain organization of hsNADKFL, showing the N-terminal domain, the C-terminal domain, the C-terminal helix (residues 413 to 424), and the position of the active site. The monomer is colored in rainbow from its N-terminal (blue) to its C-terminal (red). (D) Comparison of the apo hsNADKFL cryo-EM structure in green and the crystal structure of a truncated construct of hsNADK (PDB: 3PFN) in violet. The rotation axis for each monomer is shown. (E) Comparison of the orientation of the C-terminal helix (residues 413 to 424) in the crystal structure of hsNADK (3PFN) (violet) and the hsNADKFL cryo-EM structure (green). (F) Illustration of the domain swapping for the N-tail (residues 69 to 94) shown in violet and blue in the crystal structure 3PFN. NADK monomers are shown in different shades of pink. (G) Zoom of the contacts between the catalytic domain (pink ribbon) and the swapped N-tail (in violet ribbon). (H) Close-up of the active site of NADK from the 3PFN and cryo-EM structures, showing the indicated structural rearrangements. The black double arrow highlights the translation of P292. (A) to (H) were drawn in PyMOL.
Table 1. Cryo-EM data collection and refinement statistics.
| hsNADKFL (EMD-45831) (PDB: 9CR3) | NADKesv (EMD-45832) (PDB: 9CR4) | NADKesv-NAD (EMD-45856) (PDB: 9CRA) | |
|---|---|---|---|
| Data collection and processing | |||
| Microscope | Titan Krios | ||
| Nominal magnification | 105,000 | 105,000 | 165,000 |
| Voltage (kV) | 300 | ||
| Electron exposure (e−/Å−2) | 50 | ||
| Defocus range (μm) | −0.9 to −2.2 | ||
| Pixel size (Å) | 0.83 | 0.834 | 0.738 |
| Symmetry imposed | D2 | ||
| Initial particle images (no.) | 1,820,219 | 4,587,353 | 1,716,164 |
| Final particle images (no.) | 77,606 | 316,306 | 251,813 |
| Map resolution (Å) | 3.18 | 2.81 | 2.34 |
| FSC threshold | 0.143 | ||
| Map pixel size (Å) | 0.83 | 1.043 | 1.046 |
| Refinement | |||
| Initial model used | ab initio | ||
| Model composition | |||
| Nonhydrogen atoms | 9,299 | 9,099 | 9,842 |
| Number of protein residues | 1,200 | 1,179 | 1,247 |
| Number of ligands | 0 | 0 | 4 |
| RMSDs | |||
| Bond lengths (Å) | 0.003 | 0.003 | 0.003 |
| Bond angles (°) | 0.620 | 0.560 | 0.606 |
| Validation | |||
| MolProbity score | 1.94 | 1.89 | 2.01 |
| Clashscore | 10.49 | 7.48 | 10.51 |
| Poor rotamers (%) | 0 | 0 | 0 |
| Ramachandran plot | |||
| Favored (%) | 94.0 | 92.3 | 92.4 |
| Allowed (%) | 6.0 | 7.7 | 7.6 |
| Disallowed (%) | 0 | 0 | 0 |
The cryo-EM structure resembles the crystal structure of a truncated human NADK form (residues 68 to 426), which lacks most of the N- and C-terminal regions [deposited in the Protein Data Bank (PDB), 3PFN]. Both structures aligned well, particularly in the catalytic core, with root mean square deviation (RMSD) reaching 2.1 Å for the whole tetramer and ranging from 1.0 to 1.5 Å at the monomer level (Fig. 1D and fig. S2B). While the lack of extra density prevented us from modeling part of the highly flexible N- and C-terminal extensions (residues 1 to 94 and 431 to 446), the cryo-EM structure allowed us to visualize four additional conserved residues at the end of the C-terminal domain (amino acids W427 to R430) (Fig. 1E). This C-terminal tail appears to point to the active site of another monomer in the cryo-EM structure but is absent in the crystal structure 3PFN (Fig. 1E). On the contrary, a segment of the N-terminal extension (residues 72 to 94), which was not visible in our cryo-EM structure, is clearly resolved in the crystal structure 3PFN, where it appears to swap between two monomers (Fig. 1F). This region, composed of two small β strands (residues 72 to 74 and 77 to 80), folds back against the N-terminal domain of another monomer and aligns with a short extended region (residues 97 to 101) to form a small β sheet (Fig. 1F). Upon folding, residues F74, G75, A78, and V80 from the N-terminal tail make hydrophobic contacts with residues 197 to 200, L236, and P404 to P406 from the catalytic domain of a second monomer (Fig. 1G). These contacts, near two hinge pivots (V201 and S407) in the catalytic domain, likely stabilize the N-terminal domains in a resting conformation (fig. S2C) in the crystalline state because of domain swapping (fig. S2D). Hence, upon folding against the catalytic domain, the N-tail could influence the catalytic core’s flexibility and activity.
Other differences between these two structures arise from two hinge motions: one involving the N-terminal domain and the other involving the C-terminal helix (residues 413 to 424), with rotations relative to the C-terminal domain by ~10° and ~20°, respectively (Fig. 1, D and E, and figs. S2, B and D). Their respective pivots (V201 and S407) are very close in space (~4.5 Å), suggesting that their movements are coordinated, possibly through tight contacts between the C-terminal helix and a short segment (residues 209 to 214) in the N-terminal domain (fig. S2C). Noteworthily, within this segment, two highly conserved residues, F211 and L212, position their backbone nitrogens toward the catalytic aspartate (D184), which is oriented inward in the cryo-EM structure as it is in the liganded forms of bacterial (21) and yeast (22) NADKs (fig. S2C). In contrast, D184 points outward to the solvent in the 3PFN crystal structure (Fig. 1H and fig. S2C). Last, a small seven-residue loop (residues 290 to 297) shifts (by up to ~5.7 Å at proline P292) outward the ligand binding pocket in the cryo-EM structure (Fig. 1H). Together, all these changes seem to prime the active site for catalysis in the cryo-EM structure, while the crystal structure cannot readily accommodate NAD+ or NADP+.
The cryo-EM structure of engineered hsNADK in apo- and NAD+-bound forms illuminates the catalytic core
To better visualize the catalytic core, we aimed to solve the cryo-EM structure of hsNADK bound to its substrate NAD+. Before obtaining the cryo-EM structure, we faced numerous challenges in freezing hsNADK on grids, as the proteins often formed aggregates (Fig. 2A). This aggregation was likely due to oligomerization facilitated by disulfide bridges involving surface cysteine residues.
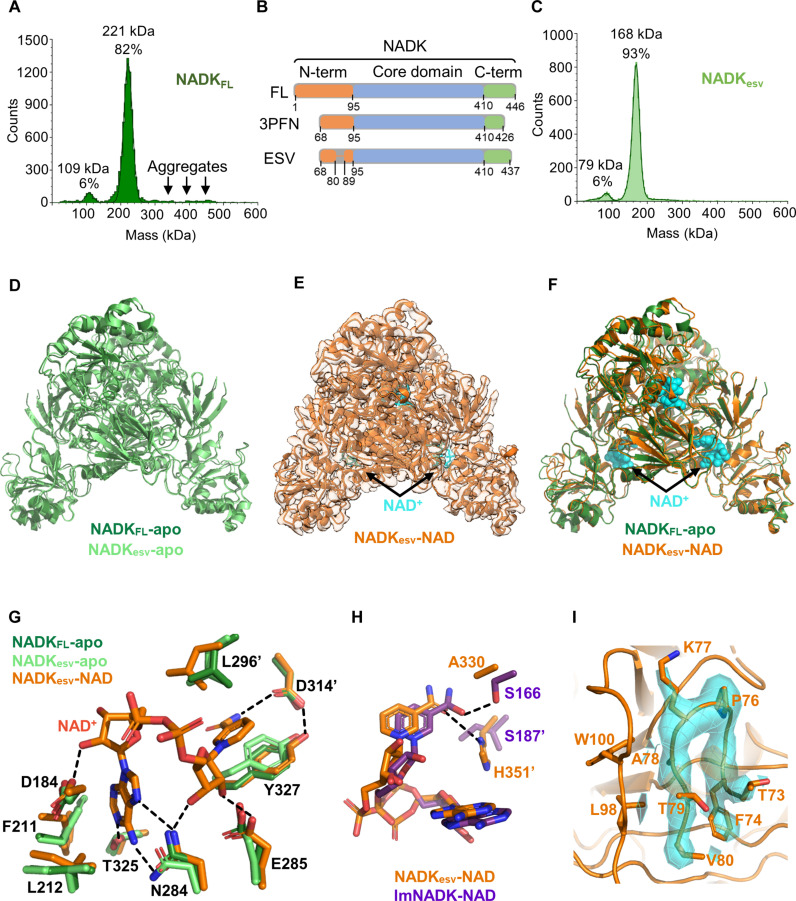
Fig. 2. Cryo-EM structures of the engineered stable variant, hsNADKesv, in apo- and NAD+-bound forms.
(A) Mass photometry of hsNADKFL purified from HEK-293 cells, showing a tetrameric NADK (82%) with some high-molecular-weight aggregates. (B) Schematic of the various constructs of hsNADK (WT; PDB: 3PFN; hsNADKesv). The catalytic domain is shown in blue, the N-tail in orange, and the C-tail in green (including the very C-terminal helix). (C) Mass photometry of hsNADKesv, showing a predominant tetramer (93%). (D) Superimposition of the cryo-EM structures of the apo-forms of hsNADKesv (light green ribbons) and hsNADKFL (dark green ribbon), indicating subtle differences. (E) The cryo-EM structure of the NAD+-bound form of hsNADKesv in orange ribbons and sticks is displayed embedded in its electron density. The NAD+ molecule is shown in cyan. (F) Superimposition of the cryo-EM structures of the apo-form of hsNADKFL (dark green ribbon) and the NAD+-bound form of hsNADKesv (orange ribbons). The NAD+ molecule is shown as cyan spheres. (G) Interactions between hsNADK and the bound NAD+ molecule. Hydrogen bonds are shown as dashed lines. (H) Comparison of the recognition of the amide group of the nicotinamide moiety in hsNADKesv (orange ribbons and sticks) and in the bacterial NADK from Listeria monocytogenes (PDB: 2I29) (violet ribbons and sticks). (I) Electron density of the N-tail: The electron density for the visible part of the N-tail (residues 73 to 80) in the NAD+-bound NADKesv structure is shown as an orange surface and mesh. The backbone and side chains are depicted as sticks, while the rest of the structure is shown as orange ribbons. (D), (F), and (G) were drawn using PyMOL.
To enhance the structural integrity of hsNADK for cryo-EM analysis, we developed a construct that removes the exposed cysteines (except for C424, which points to the active site) and omits the first 67 residues and the last 9 residues from NADKFL. We also introduced eight amino acid deletions (residues 81 to 88) and two substitutions (H89E and I90T) within the N-terminal tail to prevent any domain swapping (shown in Fig. 1F), which may stabilize hsNADK in its inactive form. We named this construct “NADKesv,” which stands for engineered stable version (Fig. 2B). NADKesv displayed reduced thermostability compared to the full-length enzyme in its apo-form but higher thermostability in the presence of NADP+ (fig. S3A). It also displayed reduced activity compared to the full-length enzyme (fig. S3B), possibly because of stronger inhibition by its product NADP+. However, it showed a similar binding affinity to NAD+, which was measured via isothermal titration calorimetry (ITC) (fig. S3, C and D). Nonetheless, it achieved a higher degree of homogeneity, as judged by mass photometry (Fig. 2C), and was successfully applied to cryo-EM grids, resulting in fewer high-molecular-mass assemblies.
We first solved the apo-form of this engineered hsNADK by cryo-EM at a 2.8-Å resolution. This structure was highly similar to the full-length wild type (WT) with an RMSD of 0.6 to 0.7 Å at the monomer level and 0.9 Å at the tetramer level (Fig. 2D and Table 1).
Subsequently, we resolved a high-resolution cryo-EM structure of the NADKesv in a complex with NAD+ at a 2.34-Å resolution (Fig. 2E and Table 1). The NAD+ molecule is clearly visible in the electron density map (fig. S3E). This structure closely resembles the apo-form of the same construct (RMSD of 0.7 to 0.8 Å at the monomer level and 0.9 Å at the tetramer level), showing only subtle changes in the orientation of the N-terminal domain, along with minor rearrangements within the active site (Fig. 2, F and G). In agreement with the absence of substantial structural reorganization besides the hinge motion, the proteolysis profiles of hsNADKs were nearly identical in the presence or absence of NAD+ (fig. S4, A and B).
Analysis of the active site of hsNADK revealed similar features conserved in most known NADK structures. In both cryo-EM apo-forms of NADKFL and NADKesv, the active site appears ready to accommodate its substrate NAD+ (Fig. 2G). Notably, in the C-terminal domain, the side chain of tyrosine Y327 is held in place by a neighboring aspartate D314′ (denotes a residue from another monomer), positioning it to readily accommodate a nicotinamide moiety upon a slight rotation of its phenol ring (Fig. 2G). D314′ is also poised to form a hydrogen bond with the amide group of the nicotinamide moiety, whereas asparagine N284 and glutamate E285 are positioned to interact with the adenine and ribose moieties of NAD+, respectively (Fig. 2G). Threonine T325 is also aligned to form a hydrogen bond with the N1 nitrogen atom of adenine, similar to other NADKs. In the N-terminal domain, the hydrophobic side chains of F211 and L212 are also arranged to accommodate the adenine moiety (Fig. 2G). Their backbone nitrogen atoms hold the catalytic aspartate D184 in place, ready to catalyze the phosphotransfer.
The interactions between the NAD+ molecule and the NADKesv protein closely resemble those observed in other tetrameric NADKs. These interactions are characterized by conserved hydrophobic contacts and hydrogen bond networks that recognize the adenosine and ribose-nicotinamide parts of NAD+, while there are no strong interactions with the diphosphate part of the NAD+ molecule (Fig. 2G). The only notable difference in hsNADK compared to all other NADKs of known structures is the absence of a highly conserved serine in the active site. This is replaced by alanine A330, which cannot form a hydrogen bond with the amide group of the nicotinamide moiety (Fig. 2H). The cryo-EM structure suggests that the recognition of this amide group may instead involve the nearby side chain of H351′ (Fig. 2H), which adopts a distinct orientation in the apo-form compared to the NAD+-bound form (fig. S3, F and G). Both residues A330 and H351, along with the other residues that point into the active site (noted above), are strictly conserved within the cytoplasmic NADKs from animals.
In the NADK-NAD+ complex, we also observed an additional density in the N-terminal tail, spanning residues F74 to V80, which shows similar hydrophobic contacts as in the 3PFN structure (Fig. 2I). Albeit the weak density, these residues appear connected to S95, indicating a lack of domain swapping, because of the deletion of residues 81 to 88. Comparison of domain-swapped apo-form and the NADK-NAD+ complex further highlights structural rearrangements at the juncture between the folded catalytic domain and the N- and C-terminal tails, characteristic of animal cytosolic NADKs. These changes are likely linked to specific functional regulations.
The N-terminal tail acts as an inhibitory regulator in hsNADK
The cryo-EM structure suggests substantial flexibility in the N-terminal region of hsNADK. Specifically, the segment encompassing the first 68 amino acids appears unfolded in NADKFL (Fig. 3A). This observation aligns with AlphaFold (23) predictions and is consistent with the presence of many polar residues in this region.
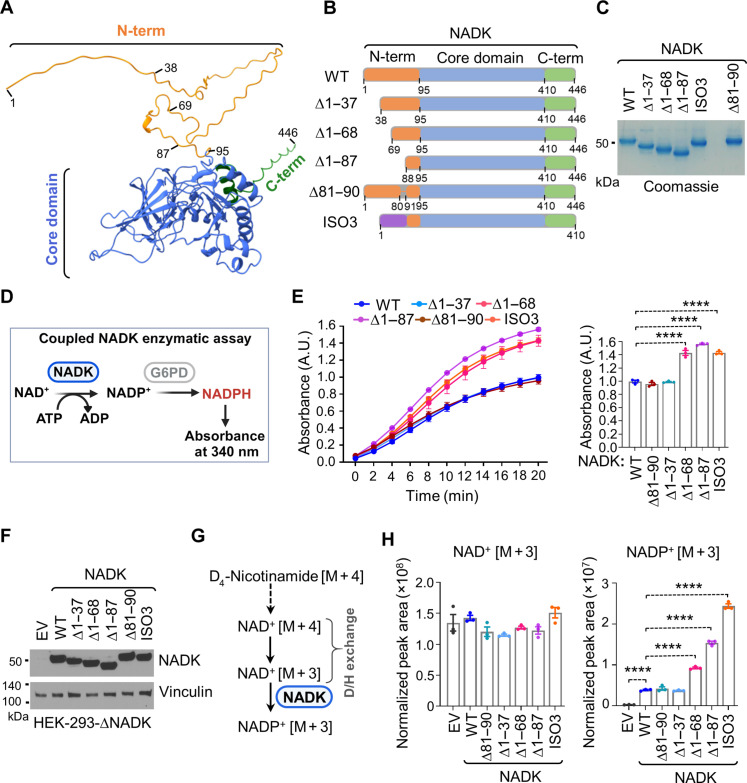
Fig. 3. Disordered N-terminal domain inhibits NADK activity and cellular NADP+ biosynthesis.
(A) Ribbon structure of NADK showing the disordered N terminus (1 to 95) in orange, the kinase domain (96 to 429) in blue, and the C-terminal (410 to 446) region in green. PyMOL was used to depict the various regions using the AlphaFold model of human NADK. (B) Schematics of NADK (full length) or various deletion (Δ) variants, including ∆1 to 37, ∆1 to 68, ∆1 to 87, ∆81 to 90, and ISO3. The N-terminal domain is depicted in orange, the kinase domain in blue, and the C terminus in green. (C) Coomassie blue staining of the indicated purified NADK variants from HEK-293 cells. (D) Schematics of the coupled NADK enzymatic assay. The absorbance of NADPH is detected at A340. (E) In vitro enzymatic activity assay for the indicated immunopurified NADK variants (WT, ∆1 to 37, ∆1 to 68, ∆1 to 87, ∆81 to 90, and ISO3). Bar graphs show A340 values after 20 min of the reaction. The absorbance values are indicated as the means ± SEM of biological triplicates. The data are representative of three independent experiments. ****P < 0.001 was calculated with the one-way ANOVA. A.U., arbitrary units. (F) Immunoblotting from NADK-deficient HEK-293 cells expressing either an empty vector (EV) or the indicated NADK variants (WT, ∆1 to 37, ∆1 to 68, ∆1 to 87, ∆81 to 90, and ISO3). (G) Schematics of labeling of NAD+ and NADP+ from 2D4-nicotinamide. (H) Normalized peak areas for newly synthesized NAD+ [M + 3] and NADP+ [M + 3] from NADK-deficient HEK-293 cells expressing either EV or the indicated NADK variants (WT, ∆1 to 37, ∆1 to 68, ∆1 to 87, ∆81 to 90, and ISO3) after 2 hours of labeling with 2,4,5,6-2D-nicotinamide. Data are shown as the means ± SEM of biological triplicates. The data are representative of at least two independent experiments. ****P < 0.001 was calculated with the one-way ANOVA.
Noteworthily, the N-terminal domain of hsNADK undergoes numerous posttranslational modifications (PTMs), including phosphorylation at S44, S46, S48, S50, and S64, among others (www.phosphositeplus.org) (24). Several protein kinases target this region to stimulate NADK activity and NADP(H) generation. Notably, Akt predominantly phosphorylates S44, followed by S46 (10), and PKC targets both S46 and S64 (11, 12), while ERK1/2 has been recently reported to target S48 and S50 (13). Analyses of PTMs on hsNADK purified from HEK-293 cells cultured in 10% serum confirmed that the N-terminal domain of NADK is extensively modified, including phosphorylation at S15, S46, S48, and S64, as well as arginine methylation (e.g., R39, R41, and R45) and lysine methylation and/or acetylation (K12, K57, and K77) (table S1). Notably, the proteolysis profiles of modified hsNADK from HEK-293 cells and those from bacteria (lacking PTMs) were quite similar, with only trypsin, which primarily targets basic residues, showing distinct differences between the two forms (fig. S4, A and B). Deletion of this modified N-terminal domain Δ1 to 87 showed enhanced stability of the catalytic core (fig. S4, A and B). Thermal shift assays on samples purified from HEK-293 or Escherichia coli cells (fig. S5) revealed slight changes in thermostability, with a 2.8°C reduction observed in the absence of PTMs. These findings suggest that the N-terminal tail of hsNADK integrates multiple signals that can modulate its thermostability.
To assess the impact of different regions of the N-terminal extension on NADK activity, we generated several deletion (Δ) variants, including Δ1 to 37, Δ1 to 68, Δ1 to 87, and Δ81 to 90, a small loop within the N-terminal extension (Fig. 3B). After expressing and purifying these variants from HEK-293 cells, we evaluated their in vitro activities alongside a hyperactive naturally occurring variant of NADK [isoform 3 (ISO3)], which lacks most of this N-terminal region (10) (Fig. 3, B to D).
Deletion of the 1 to 37 region (Δ1 to 37) or the 81 to 90 loop (Δ81 to 90) had no effect on NADK activity. On the contrary, deletions of the 1 to 68 (Δ1 to 68) and 1 to 87 (Δ1 to 87) regions, which encompass the heavily posttranslationally modified area, resulted in substantially increased activity, comparable to ISO3 (Fig. 3E), confirming an inhibitory role for the N-terminal tail.
Next, we examined the impact of these variants on NADP+ generation in cells. We expressed these variants in NADK-deficient HEK-293 cells and performed a 2-hour pulse labeling with D4-nicotinamide to assess the newly synthesized NAD+ (M + 3) and NADP+ (M + 3) (10, 25) (Fig. 3, F and G, and fig. S6A). Consistent with the in vitro data, Δ1 to 37 and Δ81 to 90 showed no appreciable differences from WT in NADP+ synthesis. In contrast, Δ1 to 68, Δ1 to 87, and ISO3 exhibited increased levels of newly synthesized NADP+ (Fig. 3H) as well as total NADP+ and NADPH pools, without significantly affecting NAD+ abundance (fig. S6B). These results indicate a critical inhibitory role for the N-terminal tail for NADP(H) generation.
The C-terminal tail is crucial for the activity of hsNADK
The cryo-EM map of NADKFL purified from mammalian cells allowed us to visualize the C-terminal domain (W427 to R430) (Fig. 1E). This C-terminal tail appears to be embedded in the active site, located between the N- and C-terminal domains (Fig. 4A). Tryptophan W427 within the C-tail is buried at the interface of two C-terminal domains in the three cryo-EM structures (fig. S7A). This feature is also observed in crystal structures of NADKs (22, 26) from Gram-negative bacteria (PDB: 2AN1; PDB: 7QVS; PDB: 4HAO) and the yeast mitochondrial enzyme POS5 (PDB: 3AFO). Notably, the side chain of W427 folds back against the very C-terminal helix (Fig. 4B and fig. S7A) and makes contact with L421 and F418 from the very same monomer (fig. S7B). In addition, it interacts with hydrophobic side chains F355′ and I358′ from a second monomer (fig. S7B). This configuration is further stabilized by a hydrogen bond network involving asparagine N428 and polar residues, such as T309′, T310′, and S352′, from a second monomer (fig. S7C).
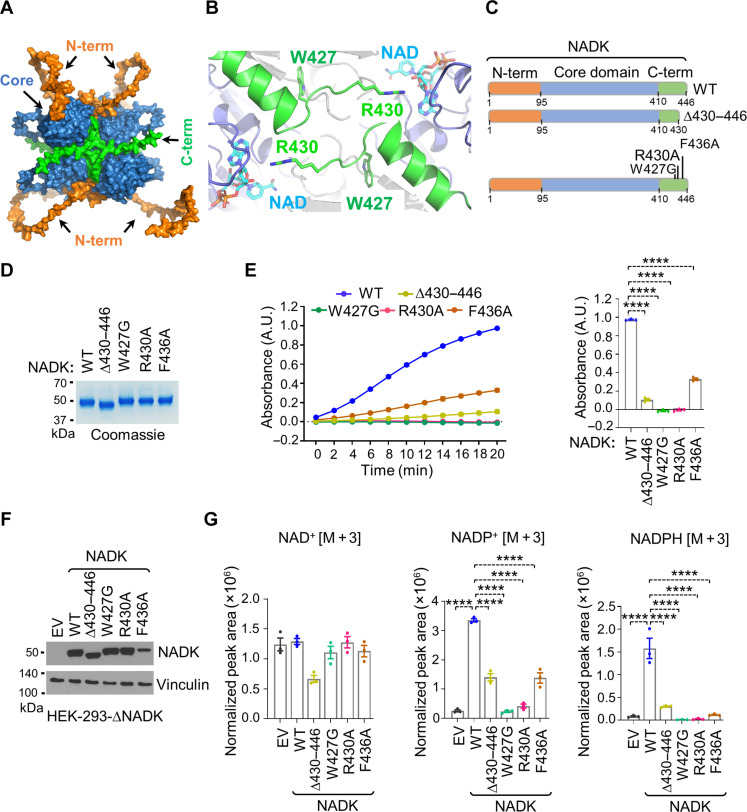
Fig. 4. The C terminus is essential for maintaining NADK activity.
(A) AlphaFold model of WT hsNADK as a tetramer. The N-tail, catalytic core, and C-tail of hsNADK are shown in orange, blue, and green, respectively. (B) Zoom in the cryo-EM structure of hsNADKesv. The very C-terminal helix (residues 413 to 424) of the catalytic core and the start of the C-tail (residues 427 to 430) are shown as green ribbons. The NAD+ binding site is highlighted with the NAD+ molecules drawn as sticks. The side chains of W427 and R430 are also shown as sticks. Figure drawn using PyMOL as (A). (C) Schematics of NADK (full length) and different C-terminal variants, including ∆430 to 446, W427G, R430A, and F436A, are color coded as follows: N-terminal domain (orange), the kinase domain (blue), and C terminus (green). (D) Coomassie blue staining of the indicated purified NADK C-terminal variants from HEK-293 cells. (E) In vitro enzymatic activity assay for the indicated immunopurified NADK variants (WT, ∆430 to 446, W427G, R430A, and F436A). Bar graphs show A340 values after 20 min of the reaction. The absorbance values are shown as the means ± SEM of biological triplicates. The data are representative of three independent experiments. (F) Immunoblotting from NADK-deficient HEK-293 cells expressing either EV or the indicated NADK variants (WT, ∆430 to 446, W427G, R430A, and F436A). (G) Normalized peak areas for newly synthesized NAD+ [M + 3], NADP+ [M + 3], and NADPH [M + 3], from NADK-deficient HEK-293 cells expressing either EV or the indicated NADK NADP+ [M + 3], after 2 hours of labeling with 2,4,5,6-2D-nicotinamide. Data are shown as the means ± SEM of biological triplicates. The data are representative of at least two independent experiments. ****P < 0.001 was calculated with the one-way ANOVA.
The orientation of the remaining C-terminal tail varies across NADK structures, as it points into the active site only in hsNADK and mitochondrial yeast POS5 (PDB: 3AFO), while it extends outward in bacterial counterparts (fig. S7D). Notably, in the NAD+-bound form, the side chain of R430 points toward the adenosine ribose of NAD+, positioning it well to assist in the entry of the transferred phosphate (Fig. 4B and fig. S7E). The R430A mutation or deletion of the C-tail (Δ431 to 446) did not lead to pronounced changes to the proteolytic profile or thermostability compared to the WT construct, suggesting that it adopts a similar overall fold (figs. S4 and S5).
We also observed a weak but continuous electron density in the full-length apo-form between the N- and C-terminal domains, which likely represents the highly flexible C-terminal tail, although it could not be accurately modeled because of insufficient electron density. This elongated conformation agrees with the model predicted by AlphaFold (Fig. 4A). Accordingly, we slightly reoriented this segment to better fit the extra density to the AlphaFold model (fig. S7E).
While the C-terminal end (residues 431 to 446) was barely visible in our cryo-EM structures, it contains two conserved positions, glutamine Q433 and (to a lesser extent) F436, whose side chains point toward the NAD(P)+ binding groove in our AlphaFold/NADKFL hybrid model (fig. S7F).
To test the function of the C-terminal domain, we generated point mutations in the conserved residues (W427G, R430A, and F436A) and truncated the last 16 amino acids from the C-tail (Δ430 to 446) (Fig. 4C). We expressed and purified these variants from HEK-293 cells (Fig. 4D) and tested their activity in vitro (Fig. 4E). All mutants exhibited a substantial reduction in NADK activity, with W427G and R430A showing almost no activity (Fig. 4E). Consistently expressing these C-terminal variants in NADK-deficient HEK-293 cells, alongside WT, resulted in a marked reduction in NADP+ biosynthesis and NADPH generation (Fig. 4G and fig. S6). Together, these findings highlight the critical role of the C-terminal domain in NADK activity, likely through its influence on the active site.
DISCUSSION
Here, we report the cryo-EM structures of functional human NADK variants, both a full-length variant and engineered stable variant, both in apo-form and in complex with its substrate NAD+, highlighting structural and functional features typical of most tetrameric NADKs. We also reveal unique aspects, such as the presence of functionally important extensions at the N and C termini [see the schematic model of hsNADK regulation (Fig. 5)].
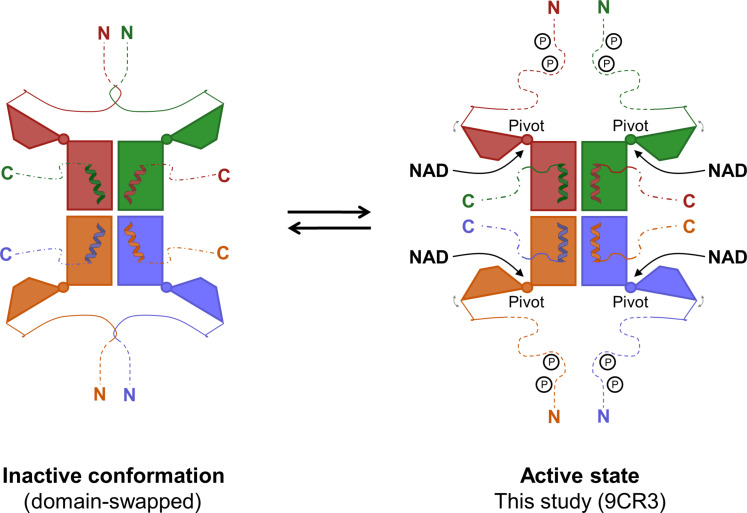
Fig. 5. Schematic model illustrating the regulation of hsNADK by its terminal regions.
In hsNADK, intertwined pairs of N-terminal tails lock the catalytic domains in an inactive conformation, preventing the proper orientation of the C-terminal helix and the folding of the C-terminal tail over the active site. PTMs, such as phosphorylation, may trigger the separation of the N-tails, enabling the hinge motion of the N-terminal domain to accommodate NAD. Concurrently, the C-terminal helix readjusts to anchor the C-tail, forming a fully functional active site.
Our findings that partial and complete deletions of the N-terminal disordered region (Δ1 to 68, Δ1 to 87, and ISO3) significantly enhance NADK’s enzymatic activity in vitro, as well as NADP+ biosynthesis and NADP(H) pools in cells, support a model in which the N-terminal domain of NADK exerts an autoinhibitory effect on its function (Fig. 5) (10). Notably, the N-terminal tail of human NADK undergoes extensive modifications, including multiple phosphorylation events mediated by various protein kinases such as Akt, PKC, and ERK (10–13). These modifications would fine tune NADK activity, thereby calibrating cellular signaling with the metabolic state. Although we were unable to fully resolve this N-terminal tail, future studies focused on identifying NADK binding partners that could stabilize this region are expected to illuminate the detailed mechanisms by which phosphorylation events enhance NADK activity.
By contrast, the C-terminal tail has an opposing effect to that of the N-terminal domain and is essential for NADK enzyme function and NADP(H) production. Our high-resolution structures reveal that part of the C-terminal tail is integrated into the catalytic core, with R430 directly oriented toward the active site. This alignment provides insight into how mutations within the C-terminal domain can abrogate NADK function. It is likely that the C-terminal tail’s proximity to the active site affects either NAD+ binding or the phosphotransfer process.
Comparative analysis of NADK structures in inactive (3PFN) and active (NADKFL) states suggests a model in which the N-terminal tails intertwine to lock the catalytic domains in an inactive conformation, preventing the proper orientation of the C-terminal helix and the folding of the C-terminal tail over the active site. PTMs, such as phosphorylation, may release this autoinhibition by separating the N-tails, thereby enabling hinge motions of the N-terminal domain to accommodate NAD+, while the C-terminal helix and C-tail realign to form a fully functional active site. In addition, other PTMs, including acetylation and methylation within the N-terminal domain (table S1), likely modulate NADK’s conformational state and activity, providing a versatile regulatory mechanism responsive to the cellular state.
In line with the key role of the flexible NADK tails in regulating its enzyme activity, numerous mutations are observed in both the N- and C-terminal regions in cancer patients, as well as within the catalytic core (27, 28). However, further studies, including mutational analysis and investigations into their effects on tumorigenesis, are necessary to determine which mutations functionally alter NADK activity.
Last, a structural comparison of various NADKs to date reveals an original phylogenetic evolution, as the cytosolic hsNADK shares more similarities with bacterial enzymes, particularly those from Gram-negative pathogens, than the human mitochondrial variant (fig. S1).
In summary, the high-resolution structure of the NADK-NAD+ complex paves the way for in silico screening of chemical libraries and structure-based drug design to identify NADK inhibitors as potential drug candidates for cancer therapy.
MATERIALS AND METHODS
Reagents and antibodies
Adenosine triphosphate (ATP) salt (Sigma-Aldrich, A2383), anti-FLAG M2 Affinity Gel (Sigma-Aldrich, A2220), 3× FLAG peptide bicinchoninic acid kit (Thermo Fisher Scientific, 23225), Bradford assay reagent (Bio-Rad, 5000006), β-nicotinamide adenine dinucleotide hydrate (NAD+; Sigma-Aldrich, N7004), d-glucose 6-phosphate disodium salt hydrate (G6P; Sigma-Aldrich, G7250), glucose-6-phosphate dehydrogenase from baker’s yeast (G6PD; Sigma-Aldrich, G6378), KOD Xtreme Hot Start DNA Polymerase (Millipore, 71975-3), protease inhibitor cocktail (Sigma-Aldrich, P8340), Hepes (Thermo Fisher Scientific, 15630080), Hepes (Sigma-Aldrich, H0887), PEI MAX Linear Polyethylenimine Hydrochloride (Polysciences, 24765-1), InstantBlue Coomassie Protein Stain (ISB1L) (abcam, ab119211), and SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, 34580) are commercially available.
Commercially available antibodies, such as anti-FLAG antibody (Cell Signaling Technology, cat. no. 14793; RRID: AB_2572291; 1:1000 for Western blotting), vinculin (Cell Signaling Technology, cat. no. 13901; RRID: AB_2728768; 1:1000 for Western blotting), and horseradish peroxidase–conjugated secondary anti-rabbit (Cell Signaling Technology, cat. no. 7074; RRID: AB_2099233; 1:5000), were used.
cDNA constructs for mammalian expression
NADKFL isoform 1 (RC200544) and NADK ISO3 (RC231219) with a C-terminal FLAG tag were purchased from Origene and described previously (10). NADK and deletion variants were subcloned into the pcDNA FRT/TO vector with a C-terminal FLAG tag. Point mutations and several deletion variants (Δ81 to 90) were generated by site-detected mutagenesis using KOD Xtreme Hot Start DNA Polymerase, followed by DPN1 digestion of the parental DNA. All the different mutants of hsNADK used in this study were confirmed using the Sanger sequencing methods at the McDermott Center Sequencing Core at UT Southwestern Medical Center.
Engineering a stable variant of hsNADK
The NADKesv amino acid sequence was derived from the full-length cytosolic human NADKFL. (EC 2.7.1.23; Uniprot ID: O95544). The changes were as follows: (i) deletions of residues 1 to 67 and 81 to 88 and the polyglutamate chain 436 to 444, (ii) the substitutions of H89E, I90T, and Q96V residues, and (iii) S/T substitution of surface-exposed cysteines, including C69S, C79T, C162T, and C402T.
Bacterial expression and purification of full-length and engineered hsNADKs
The synthetic genes coding for hsNADK and NADKesv were cloned in the plasmid pET22b to produce a recombinant C-terminal His-tagged protein. The resulting plasmid was transformed in BL21(DE3) (C2527 NEB), and bacteria were grown under aerobic conditions in LB at 37°C with the appropriate antibiotics [ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml)]. The expression of the His-tagged NADKesv was induced during the exponential phase (OD600 = 0.6 to 0.8) by adding 0.7 mM isopropyl-β-d-thiogalactopyranoside for 5 hours at 37°C. Afterward, bacterial cells were harvested by centrifugation at 6000g for 15 min at 8°C and suspended in lysis buffer [50 mM tris (pH 7.8), 50 mM NaCl, and 2 mM dithiothreitol (DTT)] with lysozyme (10 μg/ml) and a cocktail of protease inhibitors (cOmplete, EDTA-free Protease Inhibitor cocktail, Roche Life Science, Germany), and the lysate was lastly frozen at −40°C for 72 hours. Once thawed, the lysate was sonicated four times (6 min; 2-s pulses; amplitude, 55; Sonicator BIOBLOCK Scientific Vibra Cell 72405) and clarified by centrifugation (18,000g, 20 min, 8°C). The supernatant was collected, passed through a 0.45-μm filter, and placed on the cOmplete His-Tag Purification Column (Roche Life Science, Germany) previously equilibrated with 50 mM tris (pH 7.8), 50 mM NaCl, and 2 mM DTT. The His-tagged protein was eluted with a linear gradient of imidazole [12 to 300 mM imidazole in 50 mM tris (pH 7.8), 50 mM NaCl, and 2 mM DTT]. The protein was subjected to size exclusion chromatography with a Superdex 200 column (GE Healthcare) equilibrated with 50 mM tris (pH 7.8), 50 mM NaCl, and 2 mM DTT, and the presence and purity of NADKesv were confirmed by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) (Bolt 4 to 12%, bis-tris, Invitrogen, US).
Cell culture
The HEK-293 cells [a gift from C. Mackintosh (University of Dundee)] were grown in Dulbecco’s modified Eagle’s medium (Corning, 10-017-CV) supplemented with 10% fetal bovine serum (R&D Systems, S11550). The cells used for the experiments were tested regularly for mycoplasma.
Cell line–based purification of NADK for in vitro activity assays
Briefly, HEK-293 cells were seeded in 15-cm dishes for transfection. The next day, cells were transfected with 20 μg of plasmids using the polyethylenimine transfection reagent according to the manufacturer’s instructions. Thirty-six hours after transfection, cells were washed with ice-cold phosphate-buffered saline (PBS) and were lysed with 1% Triton lysis buffer [1% Triton X-100, 40 mM Hepes (pH 7.4), 120 mM NaCl, 10 mM sodium pyrophosphate, 10 mM glycerol 2-phosphate, 1 mM EDTA, and protease inhibitor cocktail (Sigma-Aldrich, P8340)]. Cell lysates were centrifuged at 21,000g for 15 min at 4°C. Next, cell lysates were further incubated with 30 μl of a 50% slurry of anti-FLAG M2 Affinity Gel for 4 hours at 4°C and washed thrice with lysis buffer and once with equilibration buffer (10 mM Hepes, 50 mM NaCl, and protease inhibitors). NADK-FLAG was eluted in 50 μl of elution buffer [3× FLAG peptide (0.2 mg/ml), 10 mM Hepes, 50 mM NaCl, and protease inhibitors] for 1.5 hours at 4°C. The eluted product was passed through 0.45-μm Costar Spin-X Centrifuge Tube Filters, and protein concentrations were determined with a Bradford assay (Bio-Rad, 500-0006). For further confirmation, proteins were visualized with Coomassie staining.
Protein purification for cryo-EM
Full-length NADK purified from mammalian cells
NADKFL isoform 1 (RC200544) with a C-terminal FLAG tag was obtained from Origene. FLAG-tagged NADK was transiently transfected in HEK-293 cells. Cells were lysed with 1% Triton lysis buffer [1% Triton X-100, 40 mM Hepes (pH 7.4), 120 mM NaCl, 10 mM sodium pyrophosphate, 10 mM glycerol 2-phosphate, and 1 mM EDTA] supplemented with 0.1% β-mercaptoethanol and protease inhibitor cocktail before cell lysis (9, 29). Eighty milligrams of protein was incubated with 1 ml of anti-FLAG M2 Affinity Gel for 4 hours, followed by three washes with 1% Triton lysis buffer containing 0.5 M NaCl, two washes with equilibration buffer (10 mM Hepes, 50 mM NaCl, and protease inhibitors), and subsequent elution with FLAG peptide [3× FLAG peptide (0.2 mg/ml), 10 mM Hepes, 50 mM NaCl, and protease inhibitors] (10). The eluate was passed through 0.45-μm Costar Spin-X Centrifuge Tube Filters before concentrating the samples using Amicon Ultra columns (Millipore Sigma, UFC501024). Samples were prepared at a concentration of 5 mg/ml in 10 mM Hepes, with 0.01% NP-40 and 1 mM DTT, and further diluted for cryo-EM analysis.
Cryo-EM sample preparation and data acquisition
The protein samples were prepared and used as follows: For sample 1 (NADKFL), a stock (5 mg/ml) was diluted to 0.2 mg/ml in 10 mM Hepes, 0.01% NP-40, and 1 mM DTT. For sample 2 (stable NADKesv for the apo-structure), a stock (11 mg/ml) was used at 0.3 mg/ml in 50 mM tris (pH 7.8), 50 mM NaCl, and 2 mM DTT. For sample 3 (NADKesv-NAD+ complex), it was prepared at 0.55 mg/ml in 50 mM tris (pH 7.8), 50 mM NaCl, 2 mM DTT, and 5 mM NAD+.
The protein samples were then applied to Quantifoil 300 mesh R1.2/1.3 grids (Quantifoil, samples 1 and 2) or C-flat 300 mesh R1.2/1.3 grids (EMS, sample 3) pretreated using a Pelco EasiGlow instrument (Ted Pella). The grid was flash frozen into liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific), with the following settings: blot time, 3 s; wait time, 3 s; relative humidity, 95%; 4°C.
Grid screening was performed on a Talos Arctica or Talos Glacios microscope (Thermo Fisher Scientific) at the UTSW Cryo-Electron Microscopy Facility. The best grid (from sample 1 or 2) was used for a 24-hour data collection on a Titan Krios microscope (Thermo Fisher Scientific) at the UTSW Cryo-Electron Microscopy Facility, equipped with the postcolumn BioQuantum Imaging Filter (Gatan) and a K3 direct electron detector (Gatan). Sample 3 was collected similarly with the Selectris X Imaging Filter (Thermo Fisher Scientific) and a Falcon 4i direct electron detector (Thermo Fisher Scientific).
All using SerialEM (30), cryo-EM data of samples 1 and 2 were collected in super-resolution mode with a 20-eV energy filter slit, while sample 3 was collected with a 10-eV energy filter slit. For the full-length NADK, 6013 movies were acquired at a pixel size of 0.415 Å. For NADKesv-Apo, 6732 movies were acquired at a pixel size of 0.417 Å. For the NADKesv-NAD+ complex, 9105 movies were acquired in EER format at a pixel size of 0.738 Å. The accumulated total dose was 50 e−/Å2 for each movie stack. The defocus range of the images was set to be −0.9 to −2.2 μm.
Cryo-EM data processing for full-length NADKFL
Cryo-EM data were first processed in Relion (31–33) to obtain an initial map and used as the template for particle picking, and then CryoSPARC was used (34). Raw movies were motion corrected, dose weighted, and then averaged into single images with a binning factor of 2 using patch motion correction, resulting in a pixel size of 0.83 Å per pixel. Contrast transfer function (CTF) parameters were estimated using patch CTF estimation. A total of 5779 micrographs were selected for further processing after manual curation. A total of 1,820,219 particles were picked and extracted and went through two rounds of two-dimensional (2D) classification, from which 305,975 particles were selected. These particles were subjected to one round of 3D heterogeneous refinement, nonuniform refinement, and global CTF refinement. It was then followed by another round of 3D heterogeneous refinement, and 77,606 particles remained. A final nonuniform refinement generated a reconstruction with an overall resolution of 3.18 Å according to the gold-standard Fourier shell correlation (FSC) using the 0.143 criterion (35). Local resolution was estimated using CryoSPARC. The map was further processed with DeepEMhancer (35) for model building and refinement (Table 1 and fig. S9).
Cryo-EM data processing for NADKesv (apo)
Cryo-EM data were processed using CryoSPARC (34). Raw movies were motion corrected, dose weighted, and then averaged into single images with a binning factor of 2 using patch motion correction, resulting in a pixel size of 0.834 Å per pixel. CTF parameters were estimated using patch CTF estimation. A total of 5901 micrographs were selected for further processing after manual curation. A total of 4,587,353 particles were extracted and went through two rounds of 2D classification, from which 543,841 particles were selected. These particles were subjected to one round of 3D heterogeneous refinement, and 316,309 remained. It is then followed by global CTF refinement, homogeneous refinement, and nonuniform refinement, resulting in a reconstruction with an overall resolution of 2.81 Å, according to the gold-standard FSC using the 0.143 criterion (36). Local resolution was estimated using Relion (31–33). The map was further processed with DeepEMhancer (35) for model building and refinement (Table 1 and fig. S10).
Cryo-EM data processing for the NADKesv-NAD+ complex
Cryo-EM data were processed using CryoSPARC (34). Raw movies were motion corrected, dose weighted, and then averaged into single images. CTF parameters were estimated using patch CTF estimation. A total of 7007 micrographs were selected for further processing after manual curation. A structure obtained from CryoSPARC-Live (34) was used as the template for particle picking. A total of 1,716,164 particles were extracted and went through two rounds of 2D classification, from which 451,850 particles were selected. These particles were subjected to one round of 3D heterogeneous refinement, and 251,813 remained. It is then followed by homogeneous refinement, nonuniform refinement, global CTF refinement, and local CTF refinement, resulting in a reconstruction with an overall resolution of 2.34 Å, according to the gold-standard FSC using the 0.143 criterion (36). Local resolution was estimated using CryoSPARC (34). The map was further processed with DeepEMhancer (35) for model building and refinement (Table 1 and fig. S11).
In vitro enzymatic activity assay on bacterially expressed hsNADK
The in vitro enzymatic activities of NADKFL (E. coli) and NADKesv (E. coli) were assessed using a glucose-6-phosphate dehydrogenase–coupled assay (10). Final concentrations were 5 mM MgATP, 5 mM glucose-6-phosphate, 0.1 to 2 mM NAD+ (12 different concentrations), G6PD (1 U/ml), and 25 nM NADK in 50 mM bis-tris (pH 7.0), 100 mM NaCl, and 1 mM MgCl2 with a final volume of 100 μl. Experiments were carried out at 30°C in UV-Star 96-half-well plates (Greiner) in a CLARIOstar plate reader (BMG Labtech), measuring absorbance at 340 nm (A340) every 32 s for 30 min after enzyme addition. A340 was converted into NADH (reduced form of NAD+) concentration using an absorption coefficient of 6.22 mM−1·cm−1. The steady-state rate constant, kss, was determined at each NAD+ concentration by linear regression of the initial reaction phase (first 10 min). Catalytic rate constants, kcat, and KM values were obtained by fitting the dependence of kss on NAD+ concentration using a hyperbolic function in GraFit software (Erithacus Software Ltd., version 7.0.3).
In vitro enzymatic activity assay for hsNADK expressed in mammalian cells
The in vitro enzymatic activity of hsNADK was assayed using the glucose-6-phosphate dehydrogenase coupling assay, as previously described (10). Briefly, for each reaction, approximately 0.8 μg of purified NADK or NADK variants was subjected to an enzymatic assay that couples its generation of NADP+ to G6PD-mediated production of NADPH, which is then measured as a change in A340 over time.
Experiments were carried out at 37°C in a SpectraMax iD3 Microplate reader (Molecular Devices) with a final volume of 100 μl. The assay was performed in 100 μl of reaction in a 96-well plate containing 10 mM ATP, 10 mM glucose-6-phosphate, 0.5 U G6PD, 10 mM MgCl2, 100 mM tris-HCl (pH 8.0), and 2 mM NAD+. Measurements of A340 were made every 2 min for 20 min at 37°C. A control reaction containing all the components except the enzyme (no NADK) was included. The presented data show normalized absorbance (A), with the control (no enzyme) subtracted.
PTM analysis by mass spectrometry
HEK-293 cells [10 dishes (15 cm each)] were transiently transfected with NADK-FLAG (20 μg per 15-cm dish) and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Forty-eight hours after transfection, cells were lysed with 1% Triton X-100 in lysis buffer, and NADK-FLAG was immunoprecipitated using anti-FLAG M2 Affinity Gel (Sigma-Aldrich, A2220) for 3.5 hours. The beads were washed three times with lysis buffer, followed by two washes with elution buffer (10 mM Hepes, 50 mM NaCl, protease inhibitors, and 1 μM microcystin). NADK-FLAG was then eluted by incubating the beads in elution buffer containing 3× FLAG peptide (0.2 mg/ml) for 1.5 hours at 4°C. The eluate was subjected to SDS-PAGE run and Coomassie blue staining (Abcam, ab119211). The NADK band was excised, and samples were then processed for PTM analysis as previously described (18).
Thermal shift assay for hsNADK
Thermal denaturation was performed in duplicate by increasing the temperature from 25 to 93°C in a quantitative reverse transcriptase polymerase chain reaction device (Mx3005P, Stratagene) using a SYAL emission filter at a gain of 8. The final concentrations were 2 μM NADK, 5× Sypro Orange (Sigma-Aldrich), and 10 mM ligand in 50 mM bis-tris (pH 7.0), 100 mM NaCl, and 1 mM MgCl2 and with a final volume of 20 μl. Melting temperature values (Tm) were determined using the wTSA-CRAFT web server (https://bioserv.cbs.cnrs.fr/TSA_CRAFT/) (37).
Limited proteolysis
Ten microliters of protein (0.2 mg/ml; without or with 5 mM NAD+) was incubated with 10 μl of different proteases (0.01 mg/ml; α-chymotrypsin, endoproteinase Glu-C, elastase, trypsin, thermolysin, bromelain, pepsin, clostripain, actinase-E, and subtilisin from Hampton Research) at 37°C during 1 hour. Laemmli buffer was then added to the reaction, and the mix was heated for 5 min at 95°C and deposed on SDS-PAGE (Bolt 4 to 12%, bis-tris, Invitrogen, USA).
ITC on bacterially expressed hsNADK
The enzymes were purified in a buffer containing 50 mM Hepes (pH 7.8), 50 mM NaCl, and 5 mM DTT, and ligand solutions were prepared in the same buffer and an equimolar concentration of MgCl2. Experiments were carried out at 25°C in a MicroCal PEAQ-ITC device (Malvern Panalytical). The enzyme was used at a final concentration of 300 μM in the ITC cell and the ligands at 5 mM in the syringe. Titrations consisted of 30 injections of 1 μl of ligand every 150 s with a stir speed of 750 rpm. Kd values were determined using MicroCal PEAQ-ITC analysis software version 1.41.
Mass photometry
Mass photometry was performed as previously described (18). Engineered NADK (NADKesv) purified from E. coli (stock of 11 mg/ml) was diluted in PBS to make 1 μM. Before mass photometry measurements, it was further diluted 1:10 in PBS (final concentration of 0.1 μM). For each measurement, 2 μl of protein was used, and 60-s movies were recorded. Data were analyzed using DiscoverMP (Refeyn).
Western blotting
Western blotting was performed as previously described (10, 29, 38). Cells were lysed in ice-cold 1% Triton cell lysis buffer. After determining the protein concentration using a Bio-Rad Protein Assay kit (Bio-Rad, 5000006), 15 μg of protein was separated by SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad, 1620112). The membranes were blocked with 5% nonfat milk for 1 hour at room temperature and incubated overnight with primary antibodies, followed by incubation with horseradish peroxidase–conjugated secondary antibodies. Last, a chemiluminescence detection reagent was used to visualize the proteins.
Isotope tracing for de novo NADP+ generation
HEK-293 NADK knockout cells (400 × 103 cells per well) were grown in six-well plates overnight followed by transfection with 3 μg of various NADK variants using transfection reagent according to the manufacturer’s instructions. Twenty-four hours after transfection, cells were subjected to isotope tracing in nicotinamide-free media supplemented with 10% dialyzed serum and [2,4,5,6-2H] nicotinamide (0.2 mg/ml) (Cambridge Isotope Laboratories, DLM-6883-PK) for 2 hours. Metabolites were extracted in 200 μl of ice-cold extraction buffer (methanol:acetonitrile:water in a 40:40:20 ratio). The tubes were incubated on ice for 15 min and centrifuged at 20,000g for 10 min at 4°C. Supernatants were collected and subjected to liquid chromatography–mass spectrometry. NADP+ and NADPH were chromatographically resolved as described previously (18, 39). Tracing 2H4-nicotinamide into these metabolites was achieved with a Thermo Lumos 1M tribrid mass spectrometer operated in a selected ion monitoring (SIM) scan mode, switching between positive and negative polarities. The mass spectrometer collected data with a resolving power of 500,000 full width at half maximum to resolve the deuterium-labeled metabolite from its naturally abundant 13C4 isotope. All analytes in our SIM scans were isolated with a 13-Da window with no offset. This wide isolation window ensured that we could capture all predicted isotopologs and any naturally occurring isotopologs in our labeling strategy. The AGC target for these scans was set to 100%, and the maximum injection time was set to Auto to maximize method efficiency. We used the following mass/charge ratio values for our inclusion lists: 664.1164 (NAD+, positive mode), 744.0827 (NADP+, positive mode), 664.11675 (NADH, negative mode), and 744.0838 (NADPH, negative mode). SIM scans were not scheduled and cycled throughout the method. Source conditions were the same as described in our previously reported paper (18). Data were analyzed using the Thermo Fisher Scientific TraceFinder 5.1 software package. Theoretical mass/charge ratio values were calculated out to M + 10 for 2H isotopologs by using the exact mass shift of a hydrogen atom to a deuterium atom (1.0063) and the data processing criteria described previously (40). For normalization of each sample, the insoluble pellet from the metabolite extractions was solubilized with 8 M urea in 10 mM tris (pH 8), and the protein was quantified with a bicinchoninic acid assay kit. The data are presented as normalized peak area; each data point is the means ± SEM of three biological replicates. Experiments are representative of at least two independent experiments.
Phylogenetic analysis of NADKs
The multiple sequence alignments of various NADKs shown in fig. S1 were generated using Espript (https://espript.ibcp.fr/ESPript/ESPript/) (41). The phylogenic tree of various NADKs was built using the software Ibis2Analyzer (http://ibis2analyzer.lcqb.upmc.fr/) (42).
Statistical analysis
GraphPad Prism 10 is used for all the statistical analysis. For comparative analysis, two-tailed Student’s t tests were used for two conditions, and analyses of variance (ANOVAs) were used for experiments with more than two conditions to determine significance. Data are from at least three independent experiments and represented as the means ± SEM of individual data points.
Acknowledgments
We thank the Structural Biology Laboratory and the Cryo-Electron Microscopy Facility at UT Southwestern Medical Center, partially supported by grant RP220582 from the Cancer Prevention & Research Institute of Texas (CPRIT), for assistance with cryo-EM studies. We also thank the CRI Metabolomics Core, supported by the CPRIT Core Facilities Award RP240494, for help with liquid chromatography–mass spectrometry–based studies and the Proteomics Core at UT Southwestern Medical Center for assistance with proteomics. Special thanks to C. Brautigam and S.-C. Tso for assistance with mass photometry. Some data in this study were acquired with a mass photometer, supported by award S10OD030312-01 from the National Institutes of Health. We thank A. Devisch for assistance with hsNADK purification and S. Trapani for providing in-house software to compute and draw rotation axes in PyMOL.
Funding: This research was supported by the grant from the NIH [R01GM143236 (to G.H.)] and Welch Foundation awards [I-2067-20210327 and I-2067-20240404 (to G.H.)]; G.H. is a recipient of a Pew-Stewart Scholar, CPRIT Scholar (CPRIT; RR190087), and ACS Scholar awards (RSG-22-177-01-TBE). This work was also supported by the CNRS and the INSERM and by grants from the ANR (ANR-17-CE18-0011 and ANR-20-CE18-0025) and the charity FRM (EQU202303016265).
Author contributions: Conceptualization: G.H. and G.L. Methodology: P.P.P., Y.L., C.M., M.H.S., D.T., H.R., A.L., L.G.Z., J.S.P., T.P.M., G.H., and G.L. Software: Y.L. Validation: P.P.P., Y.L., C.M., D.T., H.T., T.P.M., C.L., and G.H. Formal analysis: P.P.P., Y.L., H.T., T.P.M., D.T., C.L., and G.H. Investigation: P.P.P., Y.L., C.M., D.K., T.D., M.G., T.P.M., C.L., G.H., and G.L. Resources: H.T., M.G., C.L., G.H., and G.L. Data curation: P.P.P., Y.L., M.G., T.P.M., G.H., and G.L. Writing—original draft: P.P.P., Y.L., C.M., C.L., G.H., and G.L. Writing—review and editing: D.K., C.L., G.H., and G.L. Visualization: P.P.P., Y.L., C.M., H.T., D.T., C.L., G.H., and G.L. Supervision: G.H. and G.L. Project administration: G.H. and G.L. Funding acquisition: G.H., G.L., C.L., and T.P.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The cryo-EM maps have been deposited to the Electron Microscopy Data Bank with accession codes EMD-45856, EMD-45832, and EMD-45831. Atomic coordinates have been deposited to the PDB with accession codes 9CRA, 9CR4, and 9CR3.
Supplementary Materials
This PDF file includes:
Table S1
Figs. S1 to S11
REFERENCES AND NOTES
- 1.Pollak N., Dölle C., Ziegler M., The power to reduce: Pyridine nucleotides – Small molecules with a multitude of functions. Biochem. J. 402, 205–218 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ju H. Q., Lin J. F., Tian T., Xie D., Xu R. H., NADPH homeostasis in cancer: Functions, mechanisms and therapeutic implications. Signal Transduct. Target. Ther. 5, 231 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandel N. S., NADPH—The forgotten reducing equivalent. Cold Spring Harb. Perspect. Biol. 13, a040550 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoxhaj G., Manning B. D., The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 20, 74–88 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pramono A. A., Rather G. M., Herman H., Lestari K., Bertino J. R., NAD- and NADPH-contributing enzymes as therapeutic targets in cancer: An overview. Biomolecules 10, 358 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohashi K., Kawai S., Murata K., Identification and characterization of a human mitochondrial NAD kinase. Nat. Commun. 3, 1248 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lerner F., Niere M., Ludwig A., Ziegler M., Structural and functional characterization of human NAD kinase. Biochem. Biophys. Res. Commun. 288, 69–74 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Pollak N., Niere M., Ziegler M., NAD kinase levels control the NADPH concentration in human cells. J. Biol. Chem. 282, 33562–33571 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Tran D. H., Kesavan R., Rion H., Soflaee M. H., Solmonson A., Bezwada D., Vu H. S., Cai F., Phillips J. A. III, DeBerardinis R. J., Hoxhaj G., Mitochondrial NADP+ is essential for proline biosynthesis during cell growth. Nat. Metab. 3, 571–585 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoxhaj G., Ben-Sahra I., Lockwood S. E., Timson R. C., Byles V., Henning G. T., Gao P., Selfors L. M., Asara J. M., Manning B. D., Direct stimulation of NADP+ synthesis through Akt-mediated phosphorylation of NAD kinase. Science 363, 1088–1092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schild T., McReynolds M. R., Shea C., Low V., Schaffer B. E., Asara J. M., Piskounova E., Dephoure N., Rabinowitz J. D., Gomes A. P., Blenis J., NADK is activated by oncogenic signaling to sustain pancreatic ductal adenocarcinoma. Cell Rep. 35, 109238 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rabani R., Cossette C., Graham F., Powell W. S., Protein kinase C activates NAD kinase in human neutrophils. Free Radic. Biol. Med. 161, 50–59 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Liang Z., Liu T., Li Q., Zhang G., Zhang B., Du X., Liu J., Chen Z., Ding H., Hu G., Lin H., Zhu F., Luo C., Deciphering the functional landscape of phosphosites with deep neural network. Cell Rep. 42, 113048 (2023). [DOI] [PubMed] [Google Scholar]
- 14.Tedeschi P. M., Bansal N., Kerrigan J. E., Abali E. E., Scotto K. W., Bertino J. R., NAD+ kinase as a therapeutic target in cancer. Clin. Cancer Res. 22, 5189–5195 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Tsang Y. H., Dogruluk T., Tedeschi P. M., Wardwell-Ozgo J., Lu H., Espitia M., Nair N., Minelli R., Chong Z., Chen F., Chang Q. E., Dennison J. B., Dogruluk A., Li M., Ying H., Bertino J. R., Gingras M. C., Ittmann M., Kerrigan J., Chen K., Creighton C. J., Eterovic K., Mills G. B., Scott K. L., Functional annotation of rare gene aberration drivers of pancreatic cancer. Nat. Commun. 7, 10500 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ilter D., Drapela S., Schild T., Ward N. P., Adhikari E., Low V., Asara J., Oskarsson T., Lau E. K., DeNicola G. M., McReynolds M. R., Gomes A. P., NADK-mediated de novo NADP(H) synthesis is a metabolic adaptation essential for breast cancer metastasis. Redox Biol. 61, 102627 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng Z., Gao J., Chen T., Zhang Z., Li M., Fan Q., Liu G., Li X., Li Z., Zhong C., Yao F., Sun L., Deng Y., Li M., Nicotinamide adenine dinucleotide kinase promotes lymph node metastasis of NSCLC via activating ID1 expression through BMP pathway. Int. J. Biol. Sci. 19, 3184–3199 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mary C., Soflaee M. H., Kesavan R., Gelin M., Brown H., Zacharias G., Mathews T. P., Lemoff A., Lionne C., Labesse G., Hoxhaj G., Crystal structure of human NADK2 reveals a dimeric organization and active site occlusion by lysine acetylation. Mol. Cell 82, 3299–3311.e8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du J., Estrella M., Solorio-Kirpichyan K., Jeffrey P. D., Korennykh A., Structure of human NADK2 reveals atypical assembly and regulation of NAD kinases from animal mitochondria. Proc. Natl. Acad. Sci. U.S.A. 119, e2200923119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love N. R., Pollak N., Dolle C., Niere M., Chen Y., Oliveri P., Amaya E., Patel S., Ziegler M., NAD kinase controls animal NADP biosynthesis and is modulated via evolutionarily divergent calmodulin-dependent mechanisms. Proc. Natl. Acad. Sci. U.S.A. 112, 1386–1391 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poncet-Montange G., Assairi L., Arold S., Pochet S., Labesse G., NAD kinases use substrate-assisted catalysis for specific recognition of NAD. J. Biol. Chem. 282, 33925–33934 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Ando T., Ohashi K., Ochiai A., Mikami B., Kawai S., Murata K., Structural determinants of discrimination of NAD+ from NADH in yeast mitochondrial NADH kinase Pos5. J. Biol. Chem. 286, 29984–29992 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., Hassabis D., Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., Skrzypek E., PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L., Su X., Quinn W. J. III, Hui S., Krukenberg K., Frederick D. W., Redpath P., Zhan L., Chellappa K., White E., Migaud M., Mitchison T. J., Baur J. A., Rabinowitz J. D., Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab. 27, 1067–1080.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahimova R., Nogaret P., Huteau V., Gelin M., Clement D. A., Labesse G., Pochet S., Blanc-Potard A. B., Lionne C., Structure-based design, synthesis and biological evaluation of a NAD(+) analogue targeting Pseudomonas aeruginosa NAD kinase. FEBS J. 290, 482–501 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., Schultz N., The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N., Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran D. H., Kim D., Kesavan R., Brown H., Dey T., Soflaee M. H., Vu H. S., Tasdogan A., Guo J., Bezwada D., Al Saad H., Cai F., Solmonson A., Rion H., Chabatya R., Merchant S., Manales N. J., Tcheuyap V. T., Mulkey M., Mathews T. P., Brugarolas J., Morrison S. J., Zhu H., DeBerardinis R. J., Hoxhaj G., De novo and salvage purine synthesis pathways across tissues and tumors. Cell 187, 3602–3618.e20 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandez-Leiro R., Scheres S. H. W., A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D Struct. Biol. 73, 496–502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheres S. H. W., Amyloid structure determination in RELION-3.1. Acta Crystallogr. D Struct. Biol. 76, 94–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Sanchez-Garcia R., Gomez-Blanco J., Cuervo A., Carazo J. M., Sorzano C. O. S., Vargas J., DeepEMhancer: A deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henderson R., Sali A., Baker M. L., Carragher B., Devkota B., Downing K. H., Egelman E. H., Feng Z., Frank J., Grigorieff N., Jiang W., Ludtke S. J., Medalia O., Penczek P. A., Rosenthal P. B., Rossmann M. G., Schmid M. F., Schroder G. F., Steven A. C., Stokes D. L., Westbrook J. D., Wriggers W., Yang H., Young J., Berman H. M., Chiu W., Kleywegt G. J., Lawson C. L., Outcome of the first electron microscopy validation task force meeting. Structure 20, 205–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reys V., Kowalewski J., Gelin M., Lionne C., wTSA-CRAFT: An open-access web server for rapid analysis of thermal shift assay experiments. Bioinform. Adv. 3, vbad136 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soflaee M. H., Kesavan R., Sahu U., Tasdogan A., Villa E., Djabari Z., Cai F., Tran D. H., Vu H. S., Ali E. S., Rion H., O'Hara B. P., Kelekar S., Hallett J. H., Martin M., Mathews T. P., Gao P., Asara J. M., Manning B. D., Ben-Sahra I., Hoxhaj G., Purine nucleotide depletion prompts cell migration by stimulating the serine synthesis pathway. Nat. Commun. 13, 2698 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aurora A. B., Khivansara V., Leach A., Gill J. G., Martin-Sandoval M., Yang C., Kasitinon S. Y., Bezwada D., Tasdogan A., Gu W., Mathews T. P., Zhao Z., DeBerardinis R. J., Morrison S. J., Loss of glucose 6-phosphate dehydrogenase function increases oxidative stress and glutaminolysis in metastasizing melanoma cells. Proc. Natl. Acad. Sci. U.S.A. 119, 2698 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faubert B., Tasdogan A., Morrison S. J., Mathews T. P., DeBerardinis R. J., Stable isotope tracing to assess tumor metabolism in vivo. Nat. Protoc. 16, 5123–5145 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oteri F., Sarti E., Nadalin F., Carbone A., iBIS2Analyzer: A web server for a phylogeny-driven coevolution analysis of protein families. Nucleic Acids Res. 50, W412–W419 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Figs. S1 to S11