Abstract
Certain amino acid residues in a protein, when mutated, change the protein's function. We present an improved method of finding these specificity-determining positions that uses all the protein sequence data available for a family of homologous proteins. We study in detail two families of eukaryotic transcription factors, basic leucine zippers and nuclear receptors, because of the large amount of sequences and experimental data available. These protein families also have a clear definition of functional specificity: DNA-binding specificity. We compare our results to three other methods, including the evolutionary trace algorithm and a method that depends on orthology relationships. All of the predictions are compared to the available mutational and crystallographic data. We find that our method provides superior predictions of the known specificity-determining residues and also predicts residue positions within these families that deserve further study for their roles in functional specificity.
INTRODUCTION
Not all residue positions in a protein are equally important for the protein's function. When some residues are mutated, the protein can no longer carry out its function. Other residues, termed specificity-determining positions, when mutated can cause the protein to carry out a modified function. For example, if a certain residue in C/EBP is mutated from asparagine to arginine, the mutant will specifically bind a different DNA site (1). Finding these specificity-determining positions is our primary interest here.
Experimental studies can tell us a great deal about which positions are specificity-determining. For example, one could try the other 19 amino acids at every position in the DNA-binding region of C/EBP and measure their DNA binding to a wide range of DNA sequences. The problem is that even in this straightforward case where function is clearly defined (specificity for a particular DNA sequence), these experiments are expensive and time consuming.
There is a way in which protein sequence analysis can help. Instead of exhaustively testing all possible mutant proteins for potential functions, one can consider the extensive experimentation that has taken place within the living cells. An enormous amount of protein sequence data is now available for a wide variety of organisms. We would like to use these data to predict which positions are specificity-determining.
Others have used some of the available sequence data in the past. These methods depend on a certain feature of specificity-determining positions across proteins within the same family of homologous sequences. Because mutations at specificity-determining positions change the function of the protein, they are generally conserved between proteins with the same function, but tend to be distinct for proteins with different functions.
Three previous methods typify the techniques that have been used to consider the problem. First, some methods, such as that of Tian et al. (2), attempt to find discriminating, as opposed to specificity-determining residues. That is, they search for a pattern of highly conserved residues that are unique to proteins of a given function, yet also conserved by homologous proteins. While these positions may correspond to specificity-determining positions in some cases, the goal of these methods is different and only treats function in a binary way: proteins either have the correct function or they do not.
Second, the evolutionary trace (3) method looks for specificity-determining residues by using a gene tree to organize protein sequences. Beginning at the root, it then proceeds through different levels of the tree, looking at the conservation within each subtree. Proteins within a subtree are assumed to have the same function so conservation of residues within all subtrees may imply that they are important or specificity-determining. This method has been very successful in finding important residues and protein surfaces. The predictions, however, do not specify which residues are specificity-determining and which is important (e.g. for folding or stability) but not specificity-determining. In one paper (4), the authors do partition residues into important and specificity-determining groups. No general method for doing the partitioning, however, is described in that work; a certain number of residues closest to the root are described as important, while the next set of residues further from the root are taken to be specificity-determining.
Third, Mirny and Gelfand (5) presented, and others have further used (6–8), a different, statistically based method that searches specifically for specificity-determining residues. By considering the mutual information between the overall distribution and the different orthologous groupings, each of which is assumed to have a distinct function, they are able to find specificity-determining positions in several bacterial transcription factors. But this method has certain difficulties, especially when large eukaryotic families are considered. First, in many large protein families, particularly in eukaryotes, the orthology relationships are especially hard to define. There are often a large number of homologous sequences in each organism, making it hard to determine which protein in an organism is orthologous to a protein in another organism. Second, paralogs sometimes have the same general function, such as DNA-binding specificity, though they differ in other ways such as time of expression, location of expression or inclusion of other protein domains. Third, because the relationships are hard to define, the original method is limited to sequences where orthology relationships are well understood. This limitation removes useful sequence information from the dataset. For these reasons, we developed an alternative method.
Ideally, we would organize all proteins into groups based on experimental functional information. All of the methods presented in some way try to organize the proteins by their functions, using orthology or gene tree position to approximate a functional grouping. But since our experimental knowledge of the detailed function of these proteins is limited to only a small subset of known proteins, we would like to use a method that can group proteins into functional groups based on the information we do have, protein sequences.
In previous work, we developed a successful procedure for functionally grouping all known protein sequences in a protein family. The groups matched well with the experimentally known functions of several eukaryotic transcription factor families. Building off the statistical method of Mirny and Gelfand (5), we will use the larger functional groupings that our method provides, allowing us to consider all the available sequence information.
Because of the experimental difficulty in finding specificity-determining positions and in order to give the best test of the available methods, we decided to study in detail two well characterized families with a large amount of experimental information, basic leucine zippers and nuclear receptors. Many mutational studies have been carried out on these families, and several crystal structures for each family are known. In addition, the nuclear receptor family has been previously considered by evolutionary trace (3,4), allowing a direct comparison with our technique. Finally, we found that our method correctly predicts the specificity-determining positions for these two families. It outperforms the other available methods and allows us to make new predictions of other specificity-determining residues in these families.
MATERIALS AND METHODS
The methods used in this paper come chiefly from two sources which describes the respective algorithms more in detail (5,9). These methods are described briefly below. We also discuss the other methods used in this work.
Mutual information analysis
The maximum likelihood estimator (MLE) method, model 1 in Mirny and Gelfand's paper (5), was used for all of the different methods described below, except evolutionary trace, which was calculated elsewhere (4). The MLE method ranks each residue position by how much it shows the conservation pattern of a specificity-determining residue. It asks the question: Is the residue conserved within each subgroup but different between different subgroups? It does so by calculating, at a particular position in the multiple sequence alignment, the mutual information of the amino acid composition in each subgroup at that position with the overall amino acid composition at that position. The mutual information is then summed over all the subgroups.
Because mutual information can be biased for various reasons, the statistical significance is determined by comparing the mutual information to that found when the residues at a given position are shuffled vertically. Since proteins in each group are more similar to each other than the other groups, the shuffled mutual information will generally be lower than the calculated mutual information. To correct this, the expected mutual information is calculated based on fitting the shuffled scores to the actual mutual information by minimizing the sum of the squares of the Z-scores. These Z-scores are then used to measure the significance of each position as specificity-determining.
Only positions with <30% gaps in the alignment were used to ignore less important positions. For the main analysis and Figures 1 and 2, we shuffled the columns 105 times. For Figures 3 and 4, the columns were shuffled 104 times. Only minor differences are observed between the Z-scores produced by shuffling 104 or 105 times.
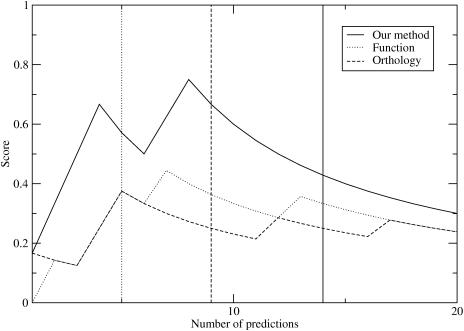
Figure 1.
Plot of score for basic leucine zipper DNA base specificity-determining residues versus the number of predictions considered. The vertical lines represent the number of the last prediction with a Z-score >3.0.
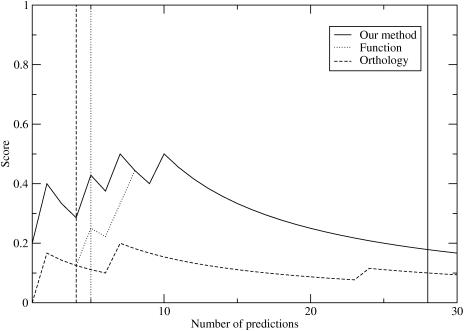
Figure 2.
Plot of score for nuclear receptor DNA base specificity-determining residues versus the number of predictions considered. The vertical lines represent the number of the last prediction with a Z-score >3.0.
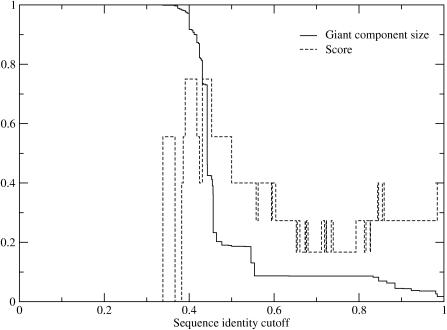
Figure 3.
Plot of giant component size and score for basic leucine zipper DNA base specificity-determining residues predicted using the top eight predictions for all possible different sequence identity cutoffs.
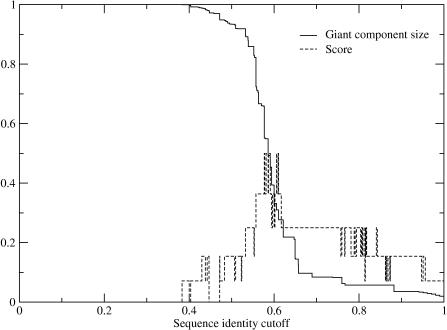
Figure 4.
Plot of giant component size and score for nuclear receptor DNA base specificity-determining residues predicted using the top ten predictions for all possible different sequence identity cutoffs.
Three methods of grouping proteins
We use the MLE method described above on three different groups of protein sequences for each family. In the ideal case, we would group all known sequences into groups based on their known function. For example, all basic leucine zippers that bind to TGACGTCA would be in one group while those binding to TGACTCA and [G/C]TCAY would be in different groups.
The problem is that the DNA-binding specificity is not known experimentally for the vast majority of known protein sequences. There are at least three ways of grouping the proteins given many sequences and a limited amount of experimental data. The most obvious method, which we call the ‘functional grouping method,’ groups the proteins based on their known binding specificities and discards the remaining protein sequences. Proteins are put in the same group if they share the same function (DNA-binding specificity), but different groups if they have different functions. While this may be an excellent way of grouping proteins, discarding the vast majority of sequences removes much of the information that could be used to determine whether a position is specificity-determining or not.
The other two methods described uses the sequences themselves to approximate which proteins share the same function. The ‘orthology method’ groups proteins by inferred orthology relationships based on their amino acid sequences and the organism in which it is found. Orthologs are expected to share the same function. Therefore, calculating the MLE method using this orthologous grouping might also find specificity-determining residues. In fact, this is the technique that Mirny and Gelfand (5) introduced in their paper. Because of the strict requirements of defining orthologs and the challenges of doing so in eukaryotes, the number of sequences that are used by this method is also much smaller than the total number of sequences. Again, much useful information is lost.
As a third alternative, our method uses the entire dataset of known sequences. The proteins are grouped by sequence similarity based on the giant component (described below and in our previous work). Because we do not discard any of the known homologous sequences, we do not lose the information that the other methods do.
Clustering based on giant component
We grouped the protein subsequences using single-linkage clustering by global sequence identity (9,10). Because different families have different levels of sequence identity between homologous sequences, we developed a method of estimating the sequence identity cutoff that would best group proteins of the same function together. The cutoff was chosen by considering the size of the largest cluster (termed the giant component in graph theory). As one scans across sequence identity cutoffs, the giant component transitions from containing all proteins under consideration (if 0% sequence identity is required) to only a single protein (if 100% sequence identity is required). In previous work (9), we found that the midpoint of the transition, when the transition is sharp, provides an excellent estimate of the best sequence identity cutoff. Likewise Figures 3 and 4 shows that, the most correct specificity-determining residues are predicted at the transition in the giant component for both families. For our analysis we will use the sequence identity cutoff at the midpoint of the giant component transition (where 50% of the sequences are in the giant component).
Bernoulli estimator
Following the method used by Kalinina et al. (7), we calculate the number of positions that are significant using a Bernoulli estimator. The estimator calculates the most likely number of significant Z-scores from a distribution of Z-scores, assuming a Gaussian distribution.
For basic leucine zippers our method and the functional grouping method predicted eight positions while the orthology method predicted four positions. For the nuclear receptor family, the methods predicted 11, 9 and 46 positions, respectively. In order to compare the results meaningfully, we used a uniform number of predictions for each method within a family, set at eight for basic leucine zippers and ten for nuclear receptors.
Score
To compare the different methods, we primarily use a simple scoring metric defined as:
where TP is the number of true positives (DNA specificity-determining residues predicted), FN is the number of false negatives (DNA specificity-determining residues not predicted) and FP is the number of false positives (residues not known to be specificity-determining predicted). Only residues that have been shown experimentally to change DNA base specificity are considered true specificity-determining residues; all other residues are considered as false positives, if predicted by a method. Because of the less important and less clear role in binding specificity, we do not consider as specificity-determining positions where mutations have been shown only to increase or decrease specificity, to change half-site spacing specificity or to affect orientational specificity. Since only a few positions have been exhaustively mutated, and these only for a single protein, there are likely to be other residue positions that are specificity-determining. The scores presented should therefore be considered a lower bound. This overly strict definition of specificity-determining residues allows us to compare the methods despite limited mutational data.
Because of the differences in the methods, particularly the much larger Z-scores for our method, we compared the methods in two different ways. First, we considered the top eight or ten predictions based on the results of the Bernoulli estimators. Second, we considered all residues above a certain level of significance, 3.0, to understand how statistical significance affects the methods' abilities to predict true specificity-determining residues. Tables showing all positions with Z-scores >3.0 are given in Supplementary Tables 1 and 2.
In Supplementary Figures 3 and 4, we also present the results of the Matthew's correlation coefficient (11). The coefficient ranges from −1 to 1, with 1 signifying the best predictive power. The Matthew's coefficient provides very similar results to the simple score described above. The coefficient is defined as follows:
Sequence selection
Following our previous work, we used protein family profiles and patterns from PROSITE (12) to select proteins from the NCBI non-redundant protein database, April 24, 2004 edition (13). The subsequence of each protein selected by the profile or pattern for the conserved domain was then used. Using only subsequences instead of whole protein sequences reduces the noise in the alignment and focuses the analysis in the most highly conserved part of the protein where the specificity-determining positions are most likely to occur.
For basic leucine zippers, the pfscan program (14) uses the PROSITE profile (PS50217) to select the basic region, hinge and leucine zipper region. The nuclear receptor pattern of PROSITE (PS00031) selects the N-terminal finger of the region; we extended this pattern to include the next 75 residues because experiments have shown that this region is also conserved and important for the proper functioning of the protein (15). 1255 basic leucine zippers and 1379 nuclear receptors were found. In our method we use these full sequence sets.
The functional grouping method limits the dataset to those proteins with known binding specificities, reducing the dataset to 139 basic leucine zippers and 209 nuclear receptors. The groupings were determined by an extensive literature search, described more in detail in our previous work (9). The subsequences were also taken from the non-redundant protein database dataset so that no small differences in protein sequence would affect the results. Because our functional data is for proteins listed in SWISSPROT (16), the subsequences were chosen by comparing the SWISSPROT subsequence to the full dataset using a BLAST search (blastall version 2.2.9) (17). The correct protein was then chosen by searching the highest hits for the protein that contained the correct protein name and organism. One basic leucine zipper was not found in the non-redundant dataset (TGA2_ARATH) and so was left out of this dataset.
Finally, for the orthology method, we used the KOG database of eukaryotic orthologous groups (18) to organize the proteins into orthologous groups. The same PROSITE search methods were used to find the proteins that contained basic leucine zipper or nuclear receptor motifs. This resulted in 101 basic leucine zippers and 140 nuclear receptors.
We primarily use the KOG database of orthologous groups because it is a consistent method of finding orthologs based on the success of the COG database (18,19). Because of the complexity of orthologous relationships in eukaryotes, we also present for a comparison an alternative method of grouping sequences by orthology. The results of this method are presented in Supplementary Figures 1 and 2 and Supplementary Tables 1 and 2. In the alternative method we used the PROSITE websites (12) (April 4, 2005 edition) to organize the proteins into orthologous groups based on their protein names. Proteins were grouped together if they had the same SWISSPROT (16) protein name but come from different organisms. Two proteins were not found in the non-redundant dataset for basic leucine zippers (FCR3_CANAL and TGA2_ARATH) and four for nuclear receptors (ERR3_PONPY, ERR3_RAT, THA_NECMA and THB_CAIMO) and so were left out of the dataset.
Numbering
There are different possible ways of numbering the proteins. For the basic leucine zippers, an alternative method is to count back from the first leucine of the leucine zipper. For the nuclear receptors, a standard numbering system has been developed for the DNA-binding domain. These numbering systems are presented in the supplementary material, along with the full protein numbering of a representative protein used in the text: yeast GCN4 for basic leucine zippers and rat glucocorticoid receptor for nuclear receptors.
Alignments
The alignments used for calculating mutual information were created with CLUSTALW (20), using the default parameters. The alignments used for our method are also shown in the Supplementary Materials.
RESULTS
We present the results of our method for two different protein families: basic leucine zippers and nuclear receptors. In order to test the validity of the predictions of our method, we compare the results to the large amount of experimental information, both mutational and structural, that is available for the DNA-binding domains for these proteins. While not every position in the proteins has been studied extensively, there is a large amount of information available. The known mutational data also allows us to compare the different methods with a simple scoring metric.
As described in Materials and Methods, there are several possible ways of grouping proteins that could be used with Mirny and Gelfand's mutual information method. Our method uses sequence information to group all known proteins. For comparison, we also present the results of three other methods, each of which limits the size of the dataset because of the information it needs to group the proteins. The functional grouping method groups proteins with known DNA-binding specificity. The orthology method limits the dataset to those proteins for which an orthology relationship can be determined and groups orthologs in the same group and paralogs in different groups. Finally, the evolutionary trace method uses a different method of finding specificity-determining residues but is presented for the nuclear receptor family for comparison.
Basic leucine zippers
The basic leucine zipper domain is an extended helix. The N-terminal end binds the DNA and is basic. The C-terminal end is used for dimerization and is amphipathic (21). Because a commonly studied basic leucine zipper is the yeast protein GCN4, we will use its full protein numbering to identify the residues. A different numbering scheme is shown in the Supplementary Materials.
Based on the Bernoulli estimator of Kalinina et al. (7) (Materials and Methods), we will focus particularly on the top eight predicted residues. Figure 5 shows the residues predicted by our method, all of which are found in the N-terminal DNA-binding region.
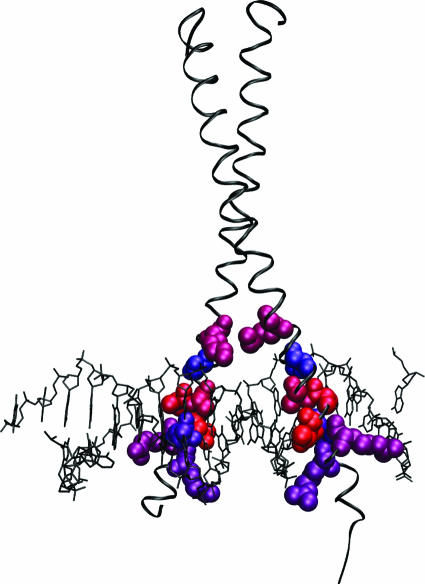
Figure 5.
Crystal structure of GCN4 bound to DNA (pdb:1YSA) (62). The eight predicted residues for the basic leucine zipper family are shown in VDW representation. The top ranked residue is colored red and the eighth is colored blue, with the colors shifted stepwise from red to blue for residues two through seven. The protein is shown in ribbon representation (colored black) and the DNA in lines representation (black). Figure made using VMD (63).
Agreement with mutational studies
Position 236 scores most highly with our method and is also found experimentally to be very important for determining specificity. A mutation at this position converts the DNA half-site binding specificities from C/EBP to TAF-1 (1) or from GCN4 to C/EBP (22). A double mutant, at this position and at position 238, converts GCN4 specificity to that of TAF-1 (1). In addition, an alanine scanning experiment found that this positions also plays a role in CREB determination of the correct spacer length between DNA half-sites (23). The experimental data at position 236 matches its selection by our algorithm as the most statistically significant position.
The second ranked position, 238, also has experimental data supporting its role in determining specificity. While no role in distinguishing between two plant basic leucine zipper sites was found (24), as mentioned above a double mutation involving this position converts GCN4 to TAF-1 DNA-binding specificity (1). Also, a mutation not found in known bzip proteins at this position broadens the specificity of GCN4 at the ±3 position of the binding site (25).
The third ranked position is 239. A mutation here has been found to change the specificity at the ±2 position of the binding site for GCN4 (25). A second mutation at position 242, the eighth ranked position, enhances this change in specificity. A triple mutant of positions 236, 238 and 239 has been shown to convert GCN4 specificity to that of C/EBP (1). A systematic mutational study of this position in GCN4 (26) has also shown that several mutants bind specifically to different half-site sequences. Also, another study showed that a mutation of this position in C/EBPβ affects the strength of DNA target specificity (27).
Position 246 is the fourth ranked position. Johnson did not find a role for this residue in the specificity difference between GCN4 and C/EBP when he considered a triple mutant (22). However, a mutation at this position, in combination with a different mutation at position 242, does change the specificity of GCN4 at the ±2 position of the binding site (25). Both mutations are necessary for this specificity change.
The fifth and sixth ranked positions, numbers 234 and 232, have not been studied extensively for a role in determining specificity. To our knowledge, the only mutational studies at these positions were carried out in an alanine scanning experiment (23). A replacement with alanine at position 234 in the CREB basic leucine zipper decreases specificity for the correct spacer length between DNA half-sites. Only two mutations had this behavior. The other residue was position 236, which is very important for determining DNA-binding specificity (see above). Therefore, we expect that residue 234 is also important for specificity.
Montclare et al. (23) also replaced the hydrophobic CREB residue (a leucine) at position 232 with alanine and found no major effect. For most basic leucine zippers, this residue is basic. Mutations to a basic residue at this position may have an important effect, especially since this residue is found to contact phosphate groups (28). More experimental study is needed to determine the role of this protein in DNA binding.
A particularly interesting specificity-determining residue is the seventh ranked residue, position 235. This position is highly conserved in natural bzip proteins, but two groups (29,30) found that mutating this residue changes the specificity of the protein. When GCN4 is bound to DNA with a two base pair spacer between the half-sites, an asparigine to tryptophan mutation alters specificity at the ±3 position of the binding site (29). When GCN4 is bound to DNA with a one base pair spacer, the same mutation alters specificity at the ±4 position of the binding site and discriminates much more strongly against a binding site mutation (30).
A small group of basic leucine zippers typified by the protein gadd153/Chop10 also do not have the conserved asparagine at this position; the proteins have a conserved glycine. A gadd153/Chop10-C/EBP dimer binds a DNA sequence unique to basic leucine zippers (31) and also inhibit certain other basic leucine zippers by forming an inactive heterodimer (32). Like the mutational data, this points to an important functional specificity-determining role for this residue.
Finally, position 242 is the last predicted specificity-determining residue. This position broadens the specificity of GCN4 when the native serine is mutated to cysteine, phenylalanine, histidine or tryptophan (33). As noted above, position 242 is also a part of double mutants with residue 239 and residue 246 that were found to cause half-site specificity changes (25).
The predicted residues, except for the relatively unstudied position 232, have all experimental data supporting their role as specificity-determining residues.
Agreement with structural studies
We also considered the crystal structure (28) of the commonly mutated bzip protein, GCN4 (Figure 5). The first three predicted positions, 236, 238 and 239, directly contact the DNA bases, as well as the seventh ranked position, 235. The fourth ranked position, 246, contacts a base through a water mediated contact. Finally, the remaining three significant positions (232, 234 and 242) contact the phosphate backbone.
Other methods
We next predicted specificity-determining residues using groups based on, (i) the limited set of known protein function (here DNA-binding specificity), and, (ii) known orthology relationships, closely following Mirny and Gelfand (5) (Materials and Methods). Table 1 shows the predictions of these methods more in detail, with residue positions shown to modify half-site base specificity underlined. Also shown are the Z-scores, which correspond to the predicted significance of the prediction.
Table 1.
Basic leucine zipper results
| Rank | Clustering | Function | Orthology |
|---|---|---|---|
| 1 | 236 (10.4) | 245 (6.35) | 236 (6.79) |
| 2 | 238 (9.54) | 238 (5.04) | 232 (6.50) |
| 3 | 239 (8.40) | 232 (3.94) | 234 (6.42) |
| 4 | 246 (7.59) | 236 (3.64) | 238 (6.32) |
| 5 | 234 (7.09) | 246 (3.23) | 242 (5.37) |
| 6 | 232 (6.68) | 234 (2.94) | 228 (4.43) |
| 7 | 235 (6.28) | 239 (2.92) | 241 (4.07) |
| 8 | 242 (6.24) | 247 (2.90) | 233 (3.56) |
Our method is labeled as ‘Clustering’, the functional grouping method is labeled as ‘Function,’ and the orthology based method is labeled as ‘Orthology.’ Residues that are experimentally known to play a role in DNA half-site specificity are underlined. Z-scores for the different methods are represented in parenthesis.
The functional grouping method ranks position 245 first, while the orthology method ranks position 241 in the seventh. Haas et al. (34) have shown that a double mutation of these two position in the basic leucine zipper VBP broadens the number of different sequences that are bound, but does not change the high affinity binding site consensus. Therefore, these positions appear not to be as important as several other positions, underlined in Table 1. With its final significant prediction, the functional method did predict one additional position that the other methods did not rank highly position 247, which affects spacer preference for GCN4 (25). This position does not change the actual DNA base specificity and so it also appears to play a less important role than some of the other residues, such as 235, which was not predicted by either the orthology or functional grouping methods.
The orthology method also predicts one residue, position 228, that enhances the 236–239 double mutation described above. However, it makes only an insignificant change in specificity as a single mutation (22). An alternative method of grouping proteins by orthology based on the protein names (Materials and Methods) was also used (Supplementary Figure 1 and Supplementary Table 1) and it was likewise unable to predict many of the half-site base specificity-determining residues.
Score
In order to have a consistent method of comparing our results, we selected the residues that have been shown through mutational studies to be able to change the DNA half-site bases specificity. We will use this overly strict definition of specificity-determining residues so that we can be confident that these positions should be predicted by any method. These are positions 235, 236, 238, 239, 242 and 246, which are described above.
Based on the selection of these residue positions as specificity-determining, we define a simple scoring metric, the number of true positives over the number of true positives, false positives and false negatives (Materials and Methods). The results are shown in Figure 1.
First, we look at the top eight predictions for our method and the other methods described above. Mirny and Gelfand's orthology method (5) predicts one-half of the specificity-determining positions in the top eight, while the functional grouping method predicts two-third in its top eight predictions. On the other hand, our method predicts all six.
Second, we considered only those predictions that are statistically significant by a cutoff in Z-score instead of using the Bernoulli estimator. For comparison purposes, we consider those predictions that have a Z-score >3.0 to be statistically significant. The vertical lines in Figure 1 show where the different methods reach the 3.0 cutoff. Our method is able to find all the specificity-determining residues, while both the orthology and functional grouping methods finds only one-half of these residues. It should also be noted that our method gives the most statistically significant predictions, probably because it includes a much larger number of sequences. It appears that the Bernoulli estimator provides a better estimate of the number of residues that are specificity-determining than a fixed Z-score cutoff.
Nuclear receptors
The nuclear receptor family conserves a DNA-binding structure with two zinc fingers (15). The N-terminal finger and the linker between these fingers are primarily responsible for DNA binding. The C-terminal end does contact the DNA in certain proteins and can extend the recognized DNA motif (35). This finger is also believed to play a role in dimerization. Nuclear receptors can dimerize in different orientations, termed head-to-head or head-to-tail, and different spacer lengths, with different dimerization partners. The different dimerization choices lead to the recognition of different DNA sequences. For this reason, the dimerization interface also plays an important role in nuclear receptor DNA-binding specificity. Rat glucocorticoid receptor is a commonly studied protein of this family, and we will use its full protein numbering to identify residue positions (Figure 6). Because the Bernoulli estimator did not give the same number of predictions for the different methods, we considered the top ten predictions for each method (Materials and Methods).
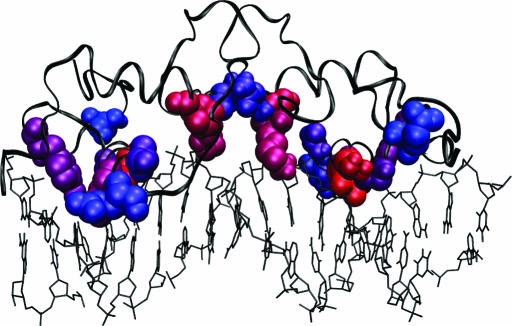
Figure 6.
Crystal structure of GCR bound to DNA (pdb:1R4R) (49). The ten predicted residues for the nuclear receptor family are shown in VDW representation. The top ranked residue is colored red and the tenth is colored blue, with the colors shifted stepwise from red to blue for residues two through nine. The protein is shown in ribbon representation (colored black) and the DNA in lines representation (black). Figure made using VMD (63).
Agreement with mutational studies
Nuclear receptors bind only to two main core six base sequences: AGGTCA, typified by the estrogen receptor, and AGAACA, typified by the glucocorticoid receptor (15). For many years, three residue positions (458, 459 and 462) have been known to determine this specificity (36,37). Our algorithm (Table 2) correctly chooses these residues, ranking them first (462), second (459) and seventh (458). In addition, a recent study (38) has determined that two more residues (465 and 469) are important for the distinct specificities of the vitamin D receptor (GGTCA) and the glucocorticoid receptor. These positions were also chosen by our method and ranked fifth and tenth.
Table 2.
Nuclear receptor results
| Rank | Clustering | Function | Orthology | ET |
|---|---|---|---|---|
| 1 | 462 (15.9) | 513 (4.89) | 465 (4.89) | 458 |
| 2 | 459 (13.4) | 459 (4.33) | 506 (3.16) | 459 |
| 3 | 493 (13.3) | 467 (4.23) | 468 (3.06) | 462? |
| 4 | 490 (11.7) | 493 (3.75) | 449 (3.02) | 465 |
| 5 | 465 (10.3) | 465 (3.11) | 442 (2.88) | 490 |
| 6 | 452 (9.86) | 450 (2.96) | 456 (2.84) | 493 |
| 7 | 458 (8.68) | 458 (2.73) | 459 (2.60) | 511 |
| 8 | 467 (8.31) | 462 (2.61) | 472 (2.54) | 513 |
| 9 | 491 (8.16) | 507 (1.75) | 441 (2.42) | |
| 10 | 469 (7.97) | 469 (1.65) | 450 (2.11) |
Our method is labeled as ‘Clustering’, the functional grouping method is labeled as ‘Function,’ and the orthology based method is labeled as ‘Orthology.’ ‘ET’ stands for evolutionary trace; these are the predictions presented unranked by Lichtarge et al. (4) and are listed sequentially here. The question mark for evolutionary trace predicted positions 462 signified that this position was not originally found by their algorithm but argued for because of improper pruning. Residues that are experimentally known to play a role in DNA half-site specificity are underlined.
Position 490, ranked fourth by our method, along with the unselected position 488, has also been shown to play a role in an important function of certain nuclear receptors, interference with the NF-κB pathway by interaction with RelA. Mutation of this position abolishes the ability of GCR to inhibit this pathway (39). While not a part of nuclear receptor DNA binding, this pathway is of great importance to the cell (40).
Position 493 has been studied structurally in detail by others (41,42). A mutation of the rat glucocorticoid receptor proline at this position to the arginine seen in other proteins causes the protein to become transcriptionally active even when not specifically bound to DNA. It has been proposed that this mutation causes a structural change that mimics the effect of binding to the specific DNA target. Mutations at this position affect the activation of these receptors, vital for the function of many of these proteins.
The ninth ranked position, 491, has not been shown to play a role in specificity through mutagenesis studies. The role of this position can be better understood by considering the known crystal structures.
Agreement with structural studies
Interestingly, position 491 is part of the dimer interface in all nuclear receptor dimer-DNA crystal structures we considered (43–51). It is in contact in the structures that form homodimers and heterodimers as well as head-to-head and head-to-tail structures. This position may play an important role in DNA specificity by modifying the way in which the proteins dimerize. In support of this, one mutational study (52) showed that changing this residue position in human estrogen receptor α from serine to glutamic acid does interfere with dimerization in the absence of estrogen. Otherwise, the position has not been well studied experimentally, particularly for its role in specificity of dimerization.
It is also interesting to note that none of the methods, including evolutionary trace, predict there to be specificity-determining residues in the D-box (37), involved in dimerization, or the A-box, involved in binding to the DNA minor groove in certain proteins (35).
Other methods
First, we consider the ability of the functional grouping and orthology methods to predict the known specificity-determining residues (Table 2). Again, our method has the highest statistical significance of its predictions. The functional grouping method is able to predict all of the known DNA base specificity-determining positions in its first ten predictions, though it ranks position 462 less highly. It also highly ranks two positions, 507 and 513, in the T-box (35). This box forms a third helix in the nuclear receptor RXRβ that is part of that protein's dimerization interface. In addition, both this method and the orthology method predict another residue position, 450, that Moraitis et al. (53) have shown to be one of the four positions involved in the comparative dimerization of RevErbAα and RORα.
However, the orthologous group method of Mirny and Gelfand (5) misses positions 458, 462 and 469. It also predicts one position in the T-box, 506, as significant. The fourth ranked position, 449, has been found to change the affinity of DNA binding for androgen receptors (54) but not for glucocorticoid receptors (55), and also affects androgen receptor binding to SNURF (56). The sixth highest ranked position, 456, has been shown (53) not to play a role in the different dimerizations of RevErbAα and RORα described above and no other molecular functional information is known. No functional information is available for several predicted positions, numbers 441, 442 and 472. In addition, a mutation in position 429 has only been reported in androgen receptors, and it has a very similar transactivation response to the wild-type (57). Again, the alternative method of grouping proteins by orthology based on the protein names (Materials and Methods) was also used (Supplementary Figure 2 and Supplementary Table 2). It was likewise unable to predict many of the half-site base specificity-determining residues.
Second, we are also able to consider our results against those of the evolutionary trace algorithm which has studied this family of proteins (4). Their method made six predictions (Table 2). They argue that misclassification of the v-erbA protein prevents them from predicting a seventh position, 462. Several of the predicted positions are not known to play a role in DNA base specificity. Two of these, 511 and 513, are in the T-box. The other, 489, may play a role in MAPK function (58), but no molecular evidence of its role is available, to our knowledge.
The score for the evolutionary trace method, when position 462 is included, is 0.44. If position 462 is not included, the score is 0.33. The score of our method when ten positions are predicted is 0.5 (Figure 2).
Score
For the nuclear receptor family, we repeat the same scoring technique that was used in the basic leucine zipper case. For this family, positions 458, 459, 462, 465 and 469 were classified as specificity-determining residues by our strict definition. Again, our method matches or outperforms the other methods, using both the top 10 rankings and the number of positions with Z-score >3.0 (Figure 2). As in the basic leucine zipper case, the Bernoulli estimate for our method and the functional grouping method (Materials and Methods) provides a better choice of the number of positions to consider.
DISCUSSION
We briefly describe some of the results of the method on basic leucine zippers and nuclear receptors more in detail. First we will compare our method to the results of the two other grouping methods, then to the evolutionary trace method for the nuclear receptor family. Our method outperforms each of the other available methods. We believe that this is primarily due to the fact that it is able to use approximately an order of magnitude more sequences than the other methods. Finally, we will present conclusions and future directions for this work.
Experimental evidence for predictions
All of the methods compared in this paper are able to find some of the residues that affect the specificity of the protein. When we compare the methods (below), our method, which uses all available sequences for a given protein family, is able to predict the largest number of specificity-determining residues. It also predicts the most important specificity-determining residues highest.
Correct overall predictions
We count only positions that cause a change in specific DNA half-site specificity as specificity-determining for the purposes of comparing the methods (underlined in Tables 1 and 2). Using this definition for the basic leucine zipper family, the clustering method is able to predict the largest number of these DNA specificity-determining positions, followed by the functional grouping and finally the orthology method of Mirny and Gelfand (5). According to our knowledge our method is able to predict all the mutations that affect specificity for bases in the DNA half-site. However, the functional grouping method misses one-third of the positions, and the orthology grouping method to one-half.
For the nuclear receptor family, we classified the DNA specificity-determining positions to be the five positions that have been shown to affect core half-site DNA sequences (36,37,59). Both our method and the functional grouping methods predict all five positions in this case. The orthology based grouping method, on the other hand, predicts only two-fifth, while the ET method classifies three-fifth or four-fifth while making a smaller total number of predictions.
Predicting the correct specificity-determining positions is most important in order to determine the best positions to study experimentally. Given that our method was best able to find the correct specificity-determining residues, it would also be helpful to rank these residues properly. That is, we would like the residues that are most important for determining specificity to be ranked highest.
Correct ranking
For the basic leucine zippers, the position with the most experimental data pointing to its being important for specificity is ranked first by our method and the orthology method. The functional grouping method lists this position fourth. Position 245 has been shown not to play a major role in specificity in VBP and has not been shown to have an important role in specificity for other proteins, but is ranked first by the functional grouping method.
In the nuclear receptor family results, our method and the functional grouping method both predict all five specificity-determining residues, and give very similar rankings. Our method does give a better ranking (first instead of eighth) of the very important position 462, one of the first three positions found experimentally. For both families, our method is better able to rank the correct specificity-determining residues.
Comparison to ET
ET is a very useful method for finding important residues in a family of proteins. It is also able to predict specificity-determining residues; although, a rigorous way of deciding which positions are important for other reasons such as folding and stability, and which are specificity-determining still needs to be developed. The original method requires a careful pruning of the sequences (60). For example, an online ET site (61) is unable to predict any positions when the full set of sequences used in this study was submitted. A recent paper (60) has presented a method that removes the requirement of pruning the sequence database, but it does not describe a way of finding specificity-determining residues. In the future, however, it may serve as a complimentary way of finding specificity-determining residues because it searches for these positions in a different way.
For the nuclear receptor family, the original evolutionary trace algorithm (3,4) was able to predict most of the known DNA specificity-determining residues. It originally misses one (position 462) because of incorrect sequence removal, as they discussed (4). This missed position is one of the most important positions (ranked first by our method). The method also misses position 469, a DNA specificity-determining position, and position 491, which may play an important role in dimerization. While potentially useful, the evolutionary trace method does not predict all of the known specificity-determining residues, nor does it provide a ranking of the positions it does predict. However, our method was successful in both of these tests.
New predictions
Given the verification of the results for our method, the most exciting next step is to be able to make new predictions. While we tested our method here on two very well studied families of proteins in order to provide the best benchmarks, we still are able to make a few predictions of residues that may play an important role in determining functional specificity of these families. For the basic leucine zippers, position 232 has not been well studied experimentally to our knowledge, but does contact the phosphate backbone and may play an important role in positioning the protein. For the nuclear receptor family, the prediction of position 491 matches its occurance in dimer interfaces, but, to our knowledge, an in-depth study of mutational effects on dimerization has not been carried out.
CONCLUSIONS
We have presented a simple, consistent method to find specificity-determining residues from a family of related protein sequences. The predicted residue positions closely match the experimentally known specificity-determining positions. New predictions are made for other residue positions that may also play a role in functional specificity.
One of the primary benefits of our method is that it is able to use all the available sequences from a particular protein family. Other methods generally need to remove many of the known sequences so that the algorithm works correctly, especially for large eukaryotic families. Typical ET, e.g. depends on a complicated or manual pruning method (3,60). Methods that depend on orthology can only use sequences where an orthology relationship can be determined. Likewise, the functional grouping method we present for comparison depends on our limited experimental knowledge of many proteins' functions. Since information is contained in every known protein sequence, methods that prune their dataset lose some of this information. Our method's superior results are likely due at least in part to the fact that it uses all of the available sequences. We expect that our method will be successful wherever there is a wealth of sequence information, and look forward to the results of studying other protein families.
Future directions
The method provides several directions for further study for these and other protein families. First of all, the results predict residues that can be tested for their roles in determining the specific function of the proteins in a given family. Predicted positions may also be used as a starting point for protein design on a family that has yet to be studied experimentally. In the future, we plan to provide a web server that will present the results of this method for these and many other protein families.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We would like to thank Leonid Mirny and Grigory Kolesov for helpful conversations. This work was supported by the National Science Foundation and the National Institutes of Health. J.E.D. is a recipient of a National Science Foundation Graduate Research Fellowship. Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Suckow M., von Wilcken-Bergmann B., Muller-Hill B. Identification of three residues in the basic regions of the bZIP proteins GCN4, C/EBP and TAF-1 that are involved in specific DNA binding. EMBO J. 1993;12:1193–1200. doi: 10.1002/j.1460-2075.1993.tb05760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tian W., Arakaki A.K., Skolnick J. EFICAz: a comprehensive approach for accurate genome-scale enzyme function inference. Nucleic Acids Res. 2004;32:6226–6239. doi: 10.1093/nar/gkh956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lichtarge O., Bourne H.R., Cohen F.E. An evolutionary trace method defines binding surfaces common to protein families. J. Mol. Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 4.Lichtarge O., Yamamoto K.R., Cohen F.E. Identification of functional surfaces of the zinc binding domains of intracellular receptors. J. Mol. Biol. 1997;274:325–337. doi: 10.1006/jmbi.1997.1395. [DOI] [PubMed] [Google Scholar]
- 5.Mirny L.A., Gelfand M.S. Using orthologous and paralogous proteins to identify specificity-determining residues in bacterial transcription factors. J. Mol. Biol. 2002;321:7–20. doi: 10.1016/s0022-2836(02)00587-9. [DOI] [PubMed] [Google Scholar]
- 6.Li L., Shakhnovich E.I., Mirny L.A. Amino acids determining enzyme-substrate specificity in prokaryotic and eukaryotic protein kinases. Proc. Natl Acad. Sci. USA. 2003;100:4463–4468. doi: 10.1073/pnas.0737647100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalinina O.V., Mironov A.A., Gelfand M.S., Rakhmaninova A.B. Automated selection of positions determining functional specificity of proteins by comparative analysis of orthologous groups in protein families. Protein Sci. 2004;13:443–456. doi: 10.1110/ps.03191704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalinina O.V., Novichkov P.S., Mironov A.A., Gelfand M.S., Rakhmaninova A.B. SDPpred: a tool for prediction of amino acid residues that determine differences in functional specificity of homologous proteins. Nucleic Acids Res. 2004;32:W424–W428. doi: 10.1093/nar/gkh391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donald J.E., Shakhnovich E.I. Determining functional specificity from protein sequences. Bioinformatics. 2005;21:2629–2635. doi: 10.1093/bioinformatics/bti396. [DOI] [PubMed] [Google Scholar]
- 10.Tian W., Skolnick J. How well is enzyme function conserved as a function of pairwise sequence identity? J. Mol. Biol. 2003;333:863–882. doi: 10.1016/j.jmb.2003.08.057. [DOI] [PubMed] [Google Scholar]
- 11.Matthews B.W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- 12.Hulo N., Sigrist C.J., Le Saux V., Langendijk-Genevaux P.S., Bordoli L., Gattiker A., De Castro E., Bucher P., Bairoch A. Recent improvements to the PROSITE database. Nucleic Acids Res. 2004;32:D134–D137. doi: 10.1093/nar/gkh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank: update. Nucleic Acids Res. 2004;32:D23–D26. doi: 10.1093/nar/gkh045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bucher P., Karplus K., Moeri N., Hofmann K. A flexible motif search technique based on generalized profiles. Comput. Chem. 1996;20:3–23. doi: 10.1016/s0097-8485(96)80003-9. [DOI] [PubMed] [Google Scholar]
- 15.Gronemeyer H., Laudet V. Transcription factors 3: nuclear receptors. Protein Profile. 1995;2:1173–1308. [PubMed] [Google Scholar]
- 16.Apweiler R., Bairoch A., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 18.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tatusov R.L., Koonin E.V., Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 20.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurst H.C. Transcription factors 1: bZIP proteins. Protein Profile. 1995;2:101–168. [PubMed] [Google Scholar]
- 22.Johnson P.F. Identification of C/EBP basic region residues involved in DNA sequence recognition and half-site spacing preference. Mol. Cell. Biol. 1993;13:6919–6930. doi: 10.1128/mcb.13.11.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montclare J.K., Sloan L.S., Schepartz A. Electrostatic control of half-site spacing preferences by the cyclic AMP response element-binding protein CREB. Nucleic Acids Res. 2001;29:3311–3319. doi: 10.1093/nar/29.16.3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niu X., Renshaw-Gegg L., Miller L., Guiltinan M.J. Bipartite determinants of DNA-binding specificity of plant basic leucine zipper proteins. Plant Mol. Biol. 1999;41:1–13. doi: 10.1023/a:1006206011502. [DOI] [PubMed] [Google Scholar]
- 25.Kim J., Tzamarias D., Ellenberger T., Harrison S.C., Struhl K. Adaptability at the protein–DNA interface is an important aspect of sequence recognition by bZIP proteins. Proc. Natl Acad. Sci. USA. 1993;90:4513–4517. doi: 10.1073/pnas.90.10.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suckow M., von Wilcken-Bergmann B., Muller-Hill B. The DNA binding specificity of the basic region of the yeast transcriptional activator GCN4 can be changed by substitution of a single amino acid. Nucleic Acids Res. 1993;21:2081–2086. doi: 10.1093/nar/21.9.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falvey E., Marcacci L., Schibler U. DNA-binding specificity of PAR and C/EBP leucine zipper proteins: a single amino acid substitution in the C/EBP DNA-binding domain confers PAR-like specificity to C/EBP. Biol. Chem. 1996;377:797–809. [PubMed] [Google Scholar]
- 28.Keller W., Konig P., Richmond T.J. Crystal structure of a bZIP/DNA complex at 2.2 A: determinants of DNA specific recognition. J. Mol. Biol. 1995;254:657–667. doi: 10.1006/jmbi.1995.0645. [DOI] [PubMed] [Google Scholar]
- 29.Suckow M., Schwamborn K., Kisters-Woike B., von Wilcken-Bergmann B., Muller-Hill B. Replacement of invariant bZip residues within the basic region of the yeast transcriptional activator GCN4 can change its DNA binding specificity. Nucleic Acids Res. 1994;22:4395–4404. doi: 10.1093/nar/22.21.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tzamarias D., Pu W.T., Struhl K. Mutations in the bZIP domain of yeast GCN4 that alter DNA-binding specificity. Proc. Natl Acad. Sci. USA. 1992;89:2007–2011. doi: 10.1073/pnas.89.6.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ubeda M., Wang X.Z., Zinszner H., Wu I., Habener J.F., Ron D. Stress-induced binding of the transcriptional factor CHOP to a novel DNA control element. Mol. Cell. Biol. 1996;16:1479–1489. doi: 10.1128/mcb.16.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen B.P., Wolfgang C.D., Hai T. Analysis of ATF3, a transcription factor induced by physiological stresses and modulated by gadd153/Chop10. Mol. Cell. Biol. 1996;16:1157–1168. doi: 10.1128/mcb.16.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suckow M., Madan A., Kisters-Woike B., von Wilcken-Bergmann B., Muller-Hill B. Creating new DNA binding specificities in the yeast transcriptional activator GCN4 by combining selected amino acid substitutions. Nucleic Acids Res. 1994;22:2198–2208. doi: 10.1093/nar/22.12.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haas N.B., Cantwell C.A., Johnson P.F., Burch J.B. DNA-binding specificity of the PAR basic leucine zipper protein VBP partially overlaps those of the C/EBP and CREB/ATF families and is influenced by domains that flank the core basic region. Mol. Cell. Biol. 1995;15:1923–1932. doi: 10.1128/mcb.15.4.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson T.E., Paulsen R.E., Padgett K.A., Milbrandt J. Participation of non-zinc finger residues in DNA binding by two nuclear orphan receptors. Science. 1992;256:107–110. doi: 10.1126/science.1314418. [DOI] [PubMed] [Google Scholar]
- 36.Mader S., Kumar V., de Verneuil H., Chambon P. Three amino acids of the oestrogen receptor are essential to its ability to distinguish an oestrogen from a glucocorticoid-responsive element. Nature. 1989;338:271–274. doi: 10.1038/338271a0. [DOI] [PubMed] [Google Scholar]
- 37.Umesono K., Evans R.M. Determinants of target gene specificity for steroid/thyroid hormone receptors. Cell. 1989;57:1139–1146. doi: 10.1016/0092-8674(89)90051-2. [DOI] [PubMed] [Google Scholar]
- 38.Hsieh J.C., Whitfield G.K., Jurutka P.W., Haussler C.A., Thatcher M.L., Thompson P.D., Dang H.T., Galligan M.A., Oza A.K., Haussler M.R. Two basic amino acids C-terminal of the proximal box specify functional binding of the vitamin D receptor to its rat osteocalcin deoxyribonucleic acid-responsive element. Endocrinology. 2003;144:5065–5080. doi: 10.1210/en.2003-0635. [DOI] [PubMed] [Google Scholar]
- 39.Liden J., Delaunay F., Rafter I., Gustafsson J., Okret S. A new function for the C-terminal zinc finger of the glucocorticoid receptor. Repression of RelA transactivation. J. Biol. Chem. 1997;272:21467–21472. doi: 10.1074/jbc.272.34.21467. [DOI] [PubMed] [Google Scholar]
- 40.De Bosscher K., Vanden Berghe W., Haegeman G. The interplay between the glucocorticoid receptor and nuclear factor-kappaB or activator protein-1: molecular mechanisms for gene repression. Endocr. Rev. 2003;24:488–522. doi: 10.1210/er.2002-0006. [DOI] [PubMed] [Google Scholar]
- 41.Lefstin J.A., Thomas J.R., Yamamoto K.R. Influence of a steroid receptor DNA-binding domain on transcriptional regulatory functions. Genes. Dev. 1994;8:2842–2856. doi: 10.1101/gad.8.23.2842. [DOI] [PubMed] [Google Scholar]
- 42.Stockner T., Sterk H., Kaptein R., Bonvin A.M. Molecular dynamics studies of a molecular switch in the glucocorticoid receptor. J. Mol. Biol. 2003;328:325–334. doi: 10.1016/s0022-2836(03)00316-4. [DOI] [PubMed] [Google Scholar]
- 43.Zhao Q., Chasse S.A., Devarakonda S., Sierk M.L., Ahvazi B., Rastinejad F. Structural basis of RXR–DNA interactions. J. Mol. Biol. 2000;296:509–520. doi: 10.1006/jmbi.1999.3457. [DOI] [PubMed] [Google Scholar]
- 44.Zhao Q., Khorasanizadeh S., Miyoshi Y., Lazar M.A., Rastinejad F. Structural elements of an orphan nuclear receptor-DNA complex. Mol. Cell. 1998;1:849–861. doi: 10.1016/s1097-2765(00)80084-2. [DOI] [PubMed] [Google Scholar]
- 45.Rastinejad F., Perlmann T., Evans R.M., Sigler P.B. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 46.Rastinejad F., Wagner T., Zhao Q., Khorasanizadeh S. Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1. EMBO J. 2000;19:1045–1054. doi: 10.1093/emboj/19.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sierk M.L., Zhao Q., Rastinejad F. DNA deformability as a recognition feature in the reverb response element. Biochemistry. 2001;40:12833–12843. doi: 10.1021/bi011086r. [DOI] [PubMed] [Google Scholar]
- 48.Devarakonda S., Harp J.M., Kim Y., Ozyhar A., Rastinejad F. Structure of the heterodimeric ecdysone receptor DNA-binding complex. EMBO J. 2003;22:5827–5840. doi: 10.1093/emboj/cdg569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luisi B.F., Xu W.X., Otwinowski Z., Freedman L.P., Yamamoto K.R., Sigler P.B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 50.Schwabe J.W., Chapman L., Finch J.T., Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 51.Shaffer P.L., Gewirth D.T. Structural basis of VDR–DNA interactions on direct repeat response elements. EMBO J. 2002;21:2242–2252. doi: 10.1093/emboj/21.9.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen D., Pace P.E., Coombes R.C., Ali S. Phosphorylation of human estrogen receptor alpha by protein kinase A regulates dimerization. Mol. Cell. Biol. 1999;19:1002–1015. doi: 10.1128/mcb.19.2.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moraitis A.N., Giguere V. Transition from monomeric to homodimeric DNA binding by nuclear receptors: identification of RevErbAalpha determinants required for RORalpha homodimer complex formation. Mol. Endocrinol. 1999;13:431–439. doi: 10.1210/mend.13.3.0250. [DOI] [PubMed] [Google Scholar]
- 54.Aarnisalo P., Santti H., Poukka H., Palvimo J.J., Janne O.A. Transcription activating and repressing functions of the androgen receptor are differentially influenced by mutations in the deoxyribonucleic acid-binding domain. Endocrinology. 1999;140:3097–3105. doi: 10.1210/endo.140.7.6792. [DOI] [PubMed] [Google Scholar]
- 55.Chen F., Watson C.S., Gametchu B. Multiple glucocorticoid receptor transcripts in membrane glucocorticoid receptor-enriched S-49 mouse lymphoma cells. J. Cell. Biochem. 1999;74:418–429. [PubMed] [Google Scholar]
- 56.Poukka H., Aarnisalo P., Santti H., Janne O.A., Palvimo J.J. Coregulator small nuclear RING finger protein (SNURF) enhances Sp1- and steroid receptor-mediated transcription by different mechanisms. J Biol. Chem. 2000;275:571–579. doi: 10.1074/jbc.275.1.571. [DOI] [PubMed] [Google Scholar]
- 57.Shi X.B., Ma A.H., Xia L., Kung H.J., de Vere White R.W. Functional analysis of 44 mutant androgen receptors from human prostate cancer. Cancer Res. 2002;62:1496–1502. [PubMed] [Google Scholar]
- 58.Yeh S., Hu Y.C., Wang P.H., Xie C., Xu Q., Tsai M.Y., Dong Z., Wang R.S., Lee T.H., Chang C. Abnormal mammary gland development and growth retardation in female mice and MCF7 breast cancer cells lacking androgen receptor. J. Exp. Med. 2003;198:1899–1908. doi: 10.1084/jem.20031233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsieh J.C., Jurutka P.W., Nakajima S., Galligan M.A., Haussler C.A., Shimizu Y., Shimizu N., Whitfield G.K., Haussler M.R. Phosphorylation of the human vitamin D receptor by protein kinase C. Biochemical and functional evaluation of the serine 51 recognition site. J. Biol. Chem. 1993;268:15118–15126. [PubMed] [Google Scholar]
- 60.Mihalek I., Res I., Lichtarge O. A family of evolution-entropy hybrid methods for ranking protein residues by importance. J. Mol. Biol. 2004;336:1265–1282. doi: 10.1016/j.jmb.2003.12.078. [DOI] [PubMed] [Google Scholar]
- 61.Innis C.A., Shi J., Blundell T.L. Evolutionary trace analysis of TGF-beta and related growth factors: implications for site-directed mutagenesis. Protein Eng. 2000;13:839–847. doi: 10.1093/protein/13.12.839. [DOI] [PubMed] [Google Scholar]
- 62.Ellenberger T.E., Brandl C.J., Struhl K., Harrison S.C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 63.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.