Abstract
α-RIMs and Munc13s are active zone proteins that control priming of synaptic vesicles to a readily releasable state, and interact with each other via their N-terminal sequences. The α-RIM N-terminal sequence also binds to Rab3s (small synaptic vesicle GTPases), an interaction that regulates presynaptic plasticity. We now demonstrate that α-RIMs contain adjacent but separate Munc13- and Rab3-binding sites, allowing formation of a tripartite Rab3/RIM/Munc13 complex. Munc13 binding is mediated by the α-RIM zinc-finger domain. Elucidation of the three-dimensional structure of this domain by NMR spectroscopy facilitated the design of a mutation that abolishes α-RIM/Munc13 binding. Selective disruption of this interaction in the calyx of Held synapse decreased the size of the readily releasable vesicle pool. Our data suggest that the ternary Rab3/RIM/Munc13 interaction approximates synaptic vesicles to the priming machinery, providing a substrate for presynaptic plasticity. The modular architecture of α-RIMs, with nested binding sites for Rab3 and other targets, may be a general feature of Rab effectors that share homology with the α-RIM N-terminal sequence.
Keywords: Munc13/unc13, neurotransmitter release, protein NMR, Rab proteins, RIM
Introduction
The release of neurotransmitters is controlled by a complex protein machinery (Lin and Scheller, 2000; Rizo and Südhof, 2002). This machinery contains components that are conserved in most types of intracellular membrane fusion, including, among others, SNARE and Rab proteins. SNARE proteins form tight ‘core complexes' that bring the synaptic vesicle and plasma membranes together and may induce membrane fusion (Hanson et al, 1997). Formation of the core complex by the neuronal SNAREs syntaxin, SNAP-25, and synaptobrevin/VAMP is thought to be tightly regulated, and involves a transition between the so-called ‘closed' and ‘open' conformations of syntaxin (see Dulubova et al, 1999). Rabs are small GTPases that cycle between active, GTP-bound and inactive, GDP-bound states (Zerial and McBride, 2001). Rab3s, the most abundant neuronal Rab proteins, are attached to synaptic vesicles in their GTP-bound state and serve as key regulators of neurotransmitter release (Schlüter et al, 2004), although it is unknown how this role is connected with SNARE function.
The release machinery also contains specialized proteins that control the exquisite temporal and spatial regulation that characterizes synaptic exocytosis. These include Munc13s and α-RIMs, large multidomain proteins (see Figure 1A) localized at active zones. Munc13s comprise three isoforms in mammals (Munc13-1, -2, and -3) (Brose et al, 1995) that are homologues of Caenorhabditis elegans unc-13 (Maruyama and Brenner, 1991). Disruption of Munc13/unc-13 genes in different species abolishes evoked synaptic transmission (Aravamudan et al, 1999; Augustin et al, 1999; Richmond et al, 1999; Varoqueaux et al, 2002). This severe phenotype is thought to reflect a key role for Munc13/unc13 in priming synaptic vesicles to a release-ready state. In addition, Munc13s have an essential role in short-term synaptic plasticity (Rhee et al, 2002). α-RIMs are Rab3 effectors with two isoforms in mammals (RIM1α and RIM2α) (Wang et al, 1997, 2000; Wang and Südhof, 2003) that are homologues of C. elegans unc-10 (Koushika et al, 2001). Like Munc13s, α-RIMs are required for normal synaptic vesicle priming and are also involved in short- and long-term synaptic plasticity (Koushika et al, 2001; Castillo et al, 2002; Schoch et al, 2002; Calakos et al, 2004). The critical importance of these multiple functions is emphasized by the severe alterations in memory and learning exhibited by RIM1α knockout mice (Powell et al, 2004).
Figure 1.
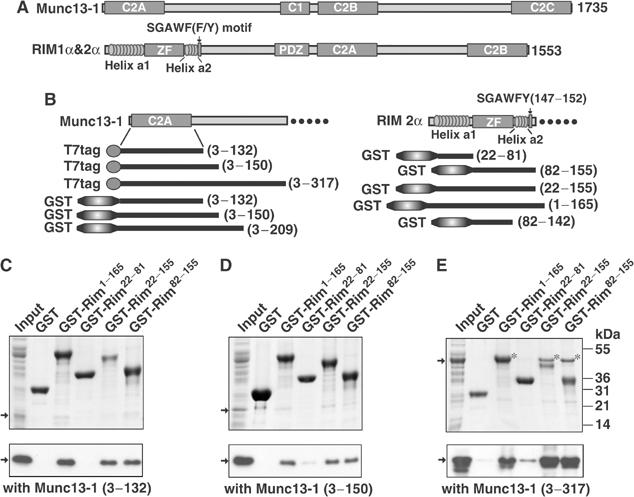
Identification of the Munc13-1/RIM2α binding regions. (A) Domain structures of Munc13-1 and α-RIMs. The α-RIM sequences that share homology with rabphilin are indicated. (B) Recombinant fragments of rat Munc13-1 and rat RIM2α used in the study. (C–E) Pulldowns of T7-tagged N-terminal fragments of Munc13-1 with GST-fusion proteins of different N-terminal sequences of RIM2α. Input and bound proteins were analyzed by Coomassie blue staining (upper panels) and immunoblotting with a T7-tag antibody visualized by ECL (lower panels). The positions of molecular mass standards are shown on the right. Arrows point out the positions of the Munc13-1 fragments. The asterisks indicate the Munc13-13−317 bands. Lower molecular weight bands observed by ECL in (E) are degradation products.
The implication of both Munc13 and α-RIMs in synaptic vesicle priming, and the finding that overexpression of a constitutively open syntaxin mutant in C. elegans partially rescues the phenotypes caused by deletion of unc-13 or unc-10 (Koushika et al, 2001; Richmond et al, 2001), indicate that the functions of Munc13s and RIMs are closely related. These observations, together with the report of a Munc13-1/syntaxin interaction (Betz et al, 1997), led to a model which predicts that Munc13-1 acts in vesicle priming by promoting SNARE complex formation, and that α-RIM assists in this function. Although this model remains to be proven, the observation of a direct interaction between the N-terminal sequences of α-RIMs and Munc13s (Betz et al, 2001) further suggests a coupling between α-RIM and Munc13 function. The importance of this interaction is highlighted by the fact that deletion of RIM1α in mice causes an ∼60% reduction in the Munc13-1 level (Schoch et al, 2002). Furthermore, overexpression of the N-terminal, RIM-binding sequence of Munc13-1 impairs neurotransmitter release in cultured hippocampal neurons (Betz et al, 2001).
The α-RIM N-terminal sequence also binds Rab3s (Wang et al, 1997, 2000), and is homologous to that of rabphilin and other Rab effectors. Both Rab3A and RIM1α are essential for mossy-fiber LTP, suggesting that the RIM1α/Rab3A interaction mediates this form of synaptic plasticity (Castillo et al, 1997, 2002). The crystal structure of a rabphilin N-terminal fragment complexed to Rab3A revealed a zinc-finger (ZF) domain that is preceded by a long α-helix (helix a1) and followed by a short α-helix (helix a2) (Ostermeier and Brunger, 1999). Helix a1 and a SGAWFF motif at the end of helix a2 are responsible for binding Rab3A. The α-RIM N-terminal region is expected to have the same structural features (Figure 1A), but some studies indicated that Rab3A binding to RIM1α is primarily mediated by helix a1 (Wang et al, 2001; Fukuda, 2004), while others suggested that helix a1 is not required for binding (Betz et al, 2001). In addition, Rab3A and Munc13-1 binding to RIM1α was found to be mutually exclusive (Betz et al, 2001).
The studies summarized above suggest that the α-RIM N-terminal sequence is a ‘hotspot' for regulation of presynaptic activity that may link Rab and SNARE function via Munc13-1, and may couple synaptic vesicle priming with presynaptic plasticity. However, the relation between Rab3A and Munc13-1 binding to α-RIMs is still unclear, in part due to a lack of structural data on their interactions that contrasts with the wealth of genetic and electrophysiological data available. To shed light on how the functions of α-RIMs, Munc13s and Rab3s are coupled, we have dissected the mechanisms of Rab3A- and Munc13-1 binding to α-RIMs using a panel of biochemical and biophysical techniques. We find that Rab3A and Munc13-1 bind to different regions of the N-terminal sequence of α-RIMs, allowing the formation of a tripartite Rab3A/α-RIM/Munc13-1 complex. We have determined the three-dimensional (3D) structure of the RIM2α ZF domain by NMR spectroscopy and designed a mutation that selectively disrupts Munc13-1 binding. Electrophysiological analyses of the effects of wild-type (WT) and mutant ZF domains on neurotransmitter release at the calyx of Held synapse show that the interaction of α-RIMs with Munc13s is a critical determinant of the number of primed, readily releasable vesicles. Our results suggest a model whereby the Munc13/α-RIM/Rab3 complex tethers docked vesicles in the active zone, facilitating priming and making them substrates for short- and long-term synaptic plasticity.
Results
Identification of the minimal binding regions of RIM2α and Munc13-1
RIM1α and RIM2α are highly homologous proteins with redundant functions (Wang et al, 2000; S Schoch, P Kaeser, and TC Südhof, unpublished data). All experiments described below were performed with RIM2α fragments, since they were readily expressed in bacteria in soluble and stable form. To define the minimal binding regions of RIM2α and Munc13-1, we performed pulldown assays with different GST-RIM2α N-terminal fragments and T7-tagged Munc13-1 N-terminal fragments (Figure 1B–E). Our results showed that a minimal RIM2α fragment containing the ZF domain, helix a2, and the SGAWFY motif (residues 82–155) is sufficient for tight binding to Munc13-1. In contrast, a RIM2α fragment spanning helix a1 (residues 22–81) exhibits no significant binding. Immunodetection revealed weak binding of RIM2α to Munc13-1 fragments containing the C2A domain alone (residues 1–132), or with a short C-terminal extension (residues 1–150), but only an Munc13-1 fragment including the C2A domain and a long C-terminal extension (residues 3–317) bound strongly to Rim2α, as detected by Coomassie blue staining (Figure 1C–E). These data suggest that the minimal RIM-binding region of Munc13-1 that was previously identified by yeast-two hybrid assays (residues 1–150; Betz et al, 2001) needs a C-terminal extension for tight binding to RIM2α.
To further delineate the RIM-binding region of Munc13-1, we acquired two-dimensional (2D) 1H–15N heteronuclear single-quantum correlation (HSQC) spectra using the 15N-labeled Rim2α ZF domain (residues 82–142) and a series of Munc13-1 fragments. The 1H–15N HSQC spectrum of RIM2α82−142 was well dispersed (Figure 2A), showing that the ZF domain is autonomously folded. Addition of a four-fold excess of unlabeled Munc13-1 C2A domain (residues 3–132) induced only small changes in the RIM282−142 1H–15N HSQC spectrum (Figure 2B), suggesting that these two fragments interact very weakly. However, massive changes were observed upon addition of the longer Munc13-13−150 fragment (Figure 2C, red contours), and Munc13-13−209 induced further crosspeak shifts (Figure 2C, blue contours). No additional changes were caused by Munc13-13−228 and Munc13-13−252 (data not shown). A control Munc13-1105−228 fragment lacking most of the C2A domain did not bind to the Rim2α ZF domain (Figure 2D). These data confirmed that the Munc13 C2A domain is necessary but not sufficient for tight binding to Rim2α, and that Munc13-1 sequences C-terminal to its C2A domain (up to residue 209) are required to strengthen the interaction.
Figure 2.
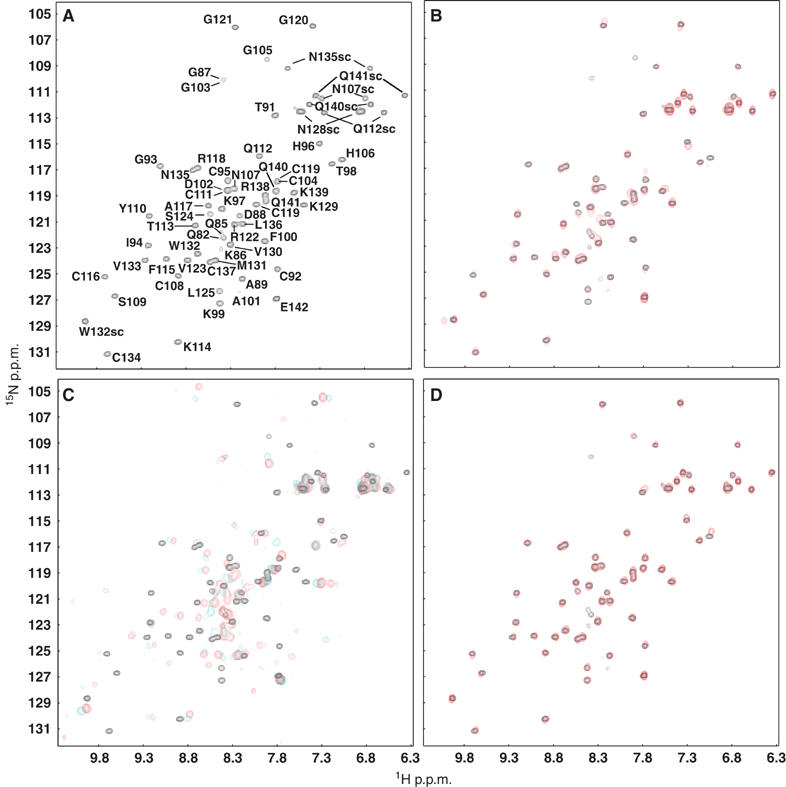
NMR analysis of the interactions between the RIM2α ZF domain and different Munc13-1 N-terminal fragments. (A) 1H–15N HSQC spectrum of 15N-labeled RIM2α82−142 (20 μM). Crosspeak assignments are indicated. (B–D) Superpositions of 1H–15N HSQC spectra of 15N-labeled RIM2α82−142 (20 μM) in the absence (black contours) or presence (red or cyan contours) of the following unlabeled Munc13-1 fragments: (B) 80 μM Munc13-13−132 (red); (C) 60 μM Munc13-13−150 (red) or 70 μM Munc13-13−209 (cyan); (D) 65 μM Munc13-1105−228 (red).
To assess the energetic contributions of different sequences to the RIM2α/Munc13-1 interaction, we employed isothermal titration calorimetry (ITC). These experiments showed that both Munc13-13−150 and Munc13-13−209 form 1:1 complexes with the RIM2α ZF domain, but with different apparent affinities (kD 351 and 70 nM, respectively; Table I). The kD observed for binding of the RIM2α ZF domain (residues 82–142) to the Munc13-13−209 fragment was comparable to those measured for the longer RIM2α fragments (Table I). Altogether, our data establish that: (i) the ZF domain of Rim2α alone is responsible for the interaction with Munc13-1; (ii) high-affinity binding to RIM2α requires the C2A domain and sequences within residues 132–209 of Munc13-1; and (iii) this interaction has a nanomolar apparent affinity.
Table 1.
Dissociation constants (kD) for the RIM2α/Munc13-1 and RIM2α/Rab3A binary interactions determined by ITC
| RIM2α fragment | Binding partner | kD (μM)a |
|---|---|---|
| RIM2α82–142 | Munc13-13–209 | 0.070±0.015 (n=2) |
| RIM2α82–142 | Munc13-13–150 | 0.351±0.0244 |
| RIM2α22–155 | Munc13-13–209 | 0.057±0.0068 |
| RIM2α1–165 | Munc13-13–209 | 0.064±0.0103 |
| RIM2α82–142 (K97E/K99E) | Munc13-13–209 | b |
| RIM2α22–155 | Rab3A15–217(Q81L) | 1.09±0.145 (n=4) |
| RIM2α22–155 | Rab3A15–217(T36N) | b |
| RIM2α22–81 | Rab3A15–217(Q81L) | 1.51±0.05 |
| RIM2α1–165 |
Rab3A15–217(Q81L) |
0.552±0.047 (n=2) |
| aThe errors indicated correspond to standard deviations for experiments performed multiple times (number indicated in parenthesis) or the errors yielded by the curve-fitting algorithm for single experiments. | ||
| bNo detectable binding. | ||
A tripartite complex between RIM2α, Munc13-1, and Rab3A
Since the ZF domain of rabphilin does not contact Rab3A in the rabphilin/Rab3A complex (Ostermeier and Brunger, 1999), our finding that Munc13-1 binds to the RIM2α ZF domain suggested that Munc13-1 and Rab3A binding to RIM2α might be compatible. To test this possibility, we performed gel filtration experiments using four protein fragments: Munc13-13−209, T7-tagged RIM2α22−155, Rab3A15−217(Q81L), and Rab3A15−217(T36N). Rab3A15−217(Q81L) is a Rab3A fragment that locks the enzyme in its ‘active' form and is sufficient for effector binding (Ostermeier and Brunger, 1999), while the T36N mutation locks Rab3A in its inactive conformation, which does not interact with effectors. The isolated Munc13-13−209 fragment forms a homodimer that dissociates upon interaction with RIM2α, yielding a heterodimer (Lu et al, in preparation). Thus, the Munc133−209 fragment migrated as a homodimer by gel filtration column and RIM2α22−155 eluted as a monomer, while an equimolar mixture of Munc133−209 and RIM2α22−155 co-eluted as a 1:1 heterodimer (Figure 3A).
Figure 3.
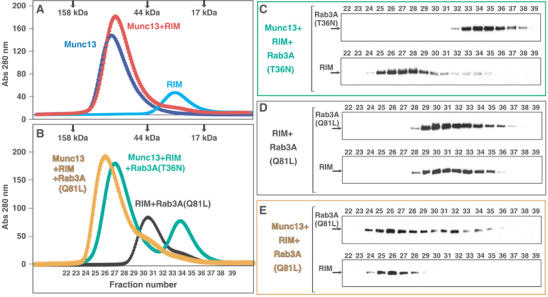
A tripartite Munc13-1/RIM2α/Rab3A complex revealed by gel filtration. (A, B) Superdex S200 (10/300GL) elution profiles of Munc13-13−209, T7-tagged RIM22−155, Rab3A15−217(Q81L), and Rab3A15−217(T36N) applied individually or in different combinations (see color-coded labels). Positions of molecular mass markers are shown on the top. (C–E) Immunoblotting analysis of the elution fractions indicated in (B). Aliquots of each fraction were separated by SDS–PAGE, followed by Western blotting with Rab3A (top panels) or T7-tag antibodies (bottom panels).
To resolve potential ambiguities, additional gel filtration experiments were analyzed by SDS–PAGE, followed by Western blotting with Rab3A and T7-tag antibodies. As expected, a mixture of the Munc133−209, RIM2α22−155, and the inactive Rab3A(T36N) mutant eluted in two separate peaks corresponding to the Munc133−209/RIM2α22−155 heterodimer and the Rab3A15−217 monomer (Figure 3B and C). RIM2α22−155 and the constitutively active Rab3A15−217(Q81L) mutant coeluted in a major peak, confirming the formation of a complex between them (Figure 3B and D). Importantly, injection of a mixture of Munc13-13−209, RIM2α22−155, and the Rab3A15−217(Q81L) mutant resulted in a single major peak at ca. 76 kDa (Figure 3B). The coelution of RIM2α22−155 and Rab3A15−217(Q81L) under this peak, at a substantially higher molecular weight than the RIM2α22−155/Rab3A15−217(Q81L) complex (compare Figure 3D and E), unambiguously demonstrates the formation of a tripartite Munc13-1/RIM2α/Rab3A complex.
NMR analysis of the interactions of RIM2α with Munc13-1 and Rab3A
To explore the structural basis of the RIM2α/Munc13-1 and RIM2α/Rab3A interactions, we prepared several 15N-labeled RIM2α fragments (see Figure 1B) and examined their 1H–15N HSQC spectra before and after addition of unlabeled Munc13-13−209 and Rab3A15−217(Q81L) (Figure 4 and data not shown). The model of Figure 5 can help to visualize the basis of the NMR spectral changes discussed below.
Figure 4.
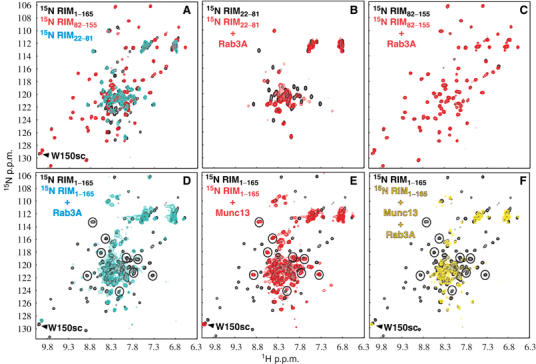
NMR analysis of the interactions of RIM2α with Rab3A and Munc13-1. (A–F) Superpositions of 1H–15N HSQC spectra of samples containing the following fragments: (A) 15N-labeled RIM2α1−165 (black), RIM2α22−81 (cyan) and RIM2α82−155 (red); (B) 15N-labeled RIM2α22−81 in the absence (black) or presence (red) of unlabeled Rab3A15−217(Q81L); (C) 15N-labeled RIM2α82−155 in the absence (black) or presence (red) of unlabeled Rab3A15−217(Q81L); (D) 15N-labeled RIM2α1−165 in the absence (black) or presence (cyan) of unlabeled Rab3A15−217(Q81L); (E) 15N-labeled RIM2α1−165 in the absence (black) or presence (red) of unlabeled Munc13-13−209; (F) 15N-labeled RIM2α1−165 in the absence (black) or presence (yellow) of unlabeled Munc13-13−209 and Rab3A15−217(Q81L). Protein concentrations ranged from 30 to 60 μM. In (D–F), the well-resolved cross-peaks of helix a1 involved in Rab3A binding are circled.
Figure 5.
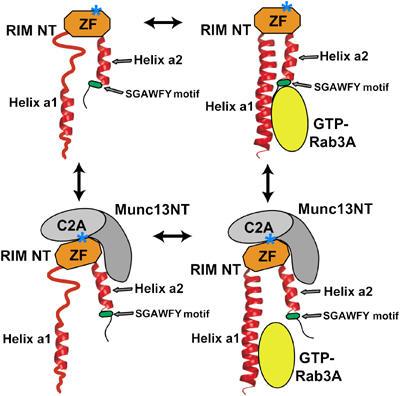
Models of the interactions of RIM2α with Munc13-1 and Rab3A. RIM NT: α-RIM N-terminal sequence; Munc13 NT: Munc13-1 N-terminal sequence; ZF: α-RIM ZF domain. The positions of the two α-helices (a1 and 2) and the SGAWFY motif are modeled according to the crystal structure of the rabphilin/Rab3A complex (Ostermeier and Brunger, 1999). Note that the helices may not be completely structured in the absence of Rab3A. A star indicates the approximate location of the mutation that disrupts Munc13-1 binding.
RIM2α82−155 contains the ZF domain, helix a2, and the SGAWFY motif. Comparison of the 1H–15N HSQC spectra of RIM2α82−155 (Figure 4A, red contours) and RIM2α82−142 (Figure 2A) allowed us to unambiguously identify the crosspeak from the W150 side chain of the SGAWFY motif. The 1H–15N HSQC spectrum of RIM2α22−81 (corresponding to helix a1) exhibits only a few well-dispersed crosspeaks (Figure 4A, cyan contours), indicating that this fragment is only partially structured. Most crosspeaks from the 1H–15N HSQC spectrum of RIM2α1−165 (Figure 4A, black contours) superimpose well with crosspeaks of either RIM2α22−81 or RIM2α82−155, showing that the sequence corresponding to helix a1 is flexibly attached to the ZF domain and remains only partially structured. The additional black crosspeaks correspond to residues 1–21 and 156–165 of RIM2α1−165. This analysis allowed us to assign distinct sequences of RIM2α1−165 to different subsets of 1H–15N HSQC crosspeaks that can be used to probe binding to the various sequences.
1H–15N HSQC spectra of RIM2α22−81 and RIM2α82−155 before and after addition of Rab3A15−217(Q81L) (Figure 4B and C) revealed substantial shifts and broadening only for the former fragment. Thus, helix a1 of RIM2α is sufficient for Rab3A binding, whereas the SGAWFY motif is not. Addition of Rab3A15−217(Q81L) caused widespread changes in the 1H–15N HSQC spectrum of RIM2α1−165 (Figure 4D). These changes included not only the perturbations in selected crosspeaks of helix a1 (circled), but also broadening of the W150 side chain NH. Thus, the SGAWFY motif participates in binding to Rab3A in the context of this longer RIM2α fragment. In addition, Rab3A15−217(Q81L) also induced broadening and slight shifts in crosspeaks from the ZF domain of RIM2α1−165 (Figure 4D). These changes are not unexpected, since at least parts of helix a1 and the SGAWFY motif should become more structured upon binding to Rab3A, which may induce small conformational changes in the ZF domain. In contrast, addition of Munc13-13−209 to 15N-labeled RIM2α1−165 broadened the HSQC crosspeaks from the ZF domain, but those from helix a1 were largely unaffected (Figure 4E). This observation confirms that the ZF domain is responsible for Munc13-1 binding, and shows that helix a1 is flexibly linked to the ZF domain/Munc13-13−209 complex. Munc13-13−209 binding did not broaden the crosspeak from the W150 side chain, showing that W150 from the SGAWFY motif is not involved in Munc13-1 binding.
Simultaneous addition of Rab3A15−217(Q81L) and Munc13-13−209 to 15N-labeled RIM2α1−165 broadened the same crosspeaks of the ZF domain and helix a1 that were affected by separate addition of each protein (Figure 4F). Compared to the perturbations induced by Rab3A15−217(Q81L) alone (Figure 4D), addition of both Rab3A15−217(Q81L) and Munc13-13−209 induced further broadening of the crosspeaks from the ZF domain (Figure 4F). These results confirm the formation of a tripartite Munc13-1/RIM2α/Rab3A complex. However, the W150 side chain crosspeak was not perturbed upon simultaneous addition of Rab3A15−217(Q81L) and Munc13-13−209. Hence, the interaction of Rab3A with the SGAWFY motif of RIM2α1−165 is released upon Munc13-13−209 binding, while the interaction of Rab3A with helix a1 persists.
Overall, our NMR results lead to a model whereby the primary binding sites for Rab3A and Munc13-1 on RIM2α are helix a1 and the ZF domain, respectively, allowing the formation of a ternary complex (Figure 5). This conclusion agrees with a surface plasmon resonance (SPR) analysis of Rab3A/RIM1α binding (Wang et al, 2001). The SGAWFY motif of RIM2α appears to contribute to Rab3A binding, but this interaction does not persist upon Munc13-1 binding (Figure 5). This observation explains why Munc13-1 competed with Rab3A binding to RIM1α in previous GST-pulldown experiments, which were performed with a RIM1α fragment lacking helix a1 (Betz et al, 2001). To further test our model, we measured the affinity of Rab3A15−217(Q81L) for different RIM2α fragments using ITC (Table I). These experiments strongly supported the conclusion that helix a1 is primarily responsible for RIM2α binding to Rab3A. The kD values measured for the RIM2α22−81 (helix a1) and RIM2α22−155 fragments were 1.51 and 1.09 μM, respectively, showing that inclusion of the SGAWFY motif in the latter fragment does not dramatically increase the affinity. The kD measured for binding of Rab3A15−217(Q81L) to RIM2α1−165 was 0.55 μM (Table I), indicating that residues 1–21 and 156–165 of RIM2α make an additional, small contribution to Rab3A binding. Overall, these affinities are comparable to those measured by SPR for RIM1α/Rab3A binding (Wang et al, 2001).
3D structure of the RIM2α ZF domain
The RIM2α ZF domain directly binds Munc13-1, a property that is not shared by rabphilin. The RIM and rabphilin ZF domains have the same distribution of zinc-binding cysteine residues, but otherwise exhibit low sequence similarity (Figure 6A). To investigate potential structural distinctions between these ZF domains that may underlie their different properties, we determined the structure in solution of the RIM2α ZF domain using NMR spectroscopy. Final structures of the domain were calculated with a set of 961 experimental restraints (structural statistics are shown in Supplementary Table 1). A ribbon diagram of a representative structure is shown in Figure 6B, and a backbone superposition of the 20 lowest energy structures is shown in Supplementary Figure 1. The structure is formed mostly by loops that surround two zinc ions, but also includes a short, two-stranded β-sheet in the center, and a β-hairpin that protrudes on one side of the domain. A short α-helix that corresponds to the beginning of helix a2 is observed at the C-terminus. A trace superposition with the rabphilin ZF domain (Figure 6C) shows that the two domains indeed share a similar architecture, with an r.m.s. deviation of 2.8 Å for 51 equivalent Cα carbons. However, there are marked differences in the N-terminal sequences that precede strand b1. Thus, whereas the position of loop 1, which contains two zinc ligands, is similar in both domains, the locations of loop 2 and the sequence connecting loops 1 and 2 are strikingly divergent. These results suggest that the eight zinc-binding cysteines and a few additional residues that are conserved in the rabphilin and RIM2α ZF domains are sufficient to dictate a similar overall structure, although there are structural differences that could have critical functional consequences.
Figure 6.
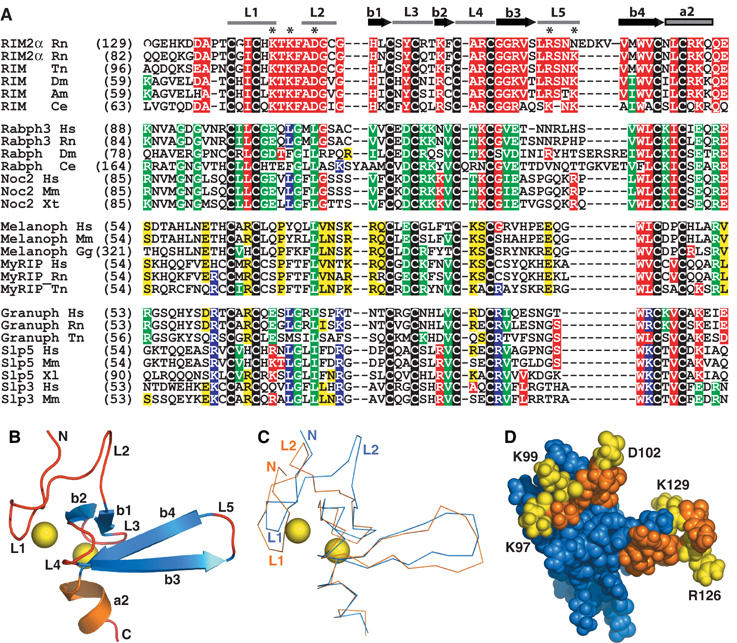
The RIM2α ZF domain contains exposed, selectively conserved sequences that may be involved in protein–protein interactions. (A) Sequence alignment of the ZF domains from α-RIMs, rabphilin, and related Rab effectors. The sequences were classified into four groups based on the sequence conservation of their entire N-terminal fragments and a phylogenetic tree created using VectorNTI (InforMax). The eight cysteines conserved in all proteins are shown in white on a black background. Identical or highly homologous (K=R, E=D, L=I=V) residues that are selectively conserved in one group (appear in >80% of group sequences), but may partially propagate into another group, are shown on different backgrounds: RIMs—red, rabphilins/NOCs—green, melanophilin/MyRIP—yellow, and granuphilins/Slp5/Slp3—blue. Hs, Homo sapiens; Rn, Rattus norvegicus; Tn, Tetraodon nigroviridis; Dm, Drosophila melanogaster; Am, Apis mellifera; Ce, Caenorhabditis elegans; Mm, Mus musculus; Xt, Xenopus tropicalis; Xl, Xenopus laevis; Gg, Gallus gallus. (B) Ribbon diagram of the RIM2α ZF domain. Loops are labeled L1–L5, β-strands are labeled b1–b4, and the C-terminal helix is labeled a2. The zinc ions are represented by yellow spheres. (C) Trace superposition of the ZF domains from rabphilin (blue) and RIM2α (orange). The N-terminus and the two N-terminal loops, which are most divergent in the two structures, are labeled. (D) Space-filling model of the RIM2α ZF domain. The atoms from the loop1–loop2 and b3–loop5 sequences that are selectively conserved in α-RIMs are colored in yellow and orange. The residues selected for mutagenesis (in yellow) are labeled in (D) and are indicated by a ‘*' in (A).
A double-point mutation that disrupts RIM2α/Munc13-1 binding
Since Munc13-1 and Rab3A interact with different regions of RIM2α, we sought to design point mutations that specifically abolish Munc13-1 binding without affecting Rab3A binding. Munc133−209 binding induces changes in almost all 1H–15N HSQC crosspeaks of the RIM2α ZF domain (Figure 2C), which hindered mapping of the Munc13-binding site. We hypothesized that Munc13-1 binding might be mediated by exposed sequences that are highly conserved in α-RIMs, but not in rabphilin. Sequence alignments revealed two such regions that correspond to loop1/loop2 and strand b3/loop 5 of the RIM2α ZF domain (Figure 6A and D). We thus prepared four 15N-labeled single mutants of the RIM2α ZF domain, where charged, exposed residues in these two sequences were replaced by residues of the opposite charge (referred to as K97E, D102K, R126E, and K129D mutants), and a double mutant (K97E/K99E). These mutations did not perturb the overall structure of the domain (Supplementary Figure 2).
Binding of Munc133−209 to WT and mutant 15N-labeled ZF domains was analyzed by gel filtration and a one-dimensional (1D) 15N-edited 1H-NMR method similar to our previously described 13C-edited technique (Araç et al, 2003). In this method, binding of an unlabeled protein to an 15N-labeled protein causes overall decreases in the resonance intensities of the 1D 15N-edited 1H-NMR spectrum of the labeled protein due to general broadening upon complex formation. This method can be performed at concentrations lower than conventional 2D 1H–15N HSQC spectra, and allows detection of interactions that may be too weak to be observed by gel filtration. The gel filtration and NMR experiments showed that none of the single mutations in the RIM2α ZF domain disrupt Munc13-13−209 binding (Figure 7 and Supplementary Figure 3). However, the double K97E/K99E mutation abolishes binding, as assessed by both gel filtration and NMR (Figure 7C and D). These results were corroborated by ITC experiments, which yielded no detectable Munc13-13−209 binding to the K97E/K99E mutant ZF domain (Table I). Moreover, no significant binding of Munc13-13−209 to RIM2α22−155(K97E/K99E) was observed by 1H–15N HSQC experiments (not shown). Note that the K97E/K99E mutation is distant from the Rab3A-binding site (see Figure 5) and should not affect Rab3A binding, as confirmed with 1H–15N HSQC experiments (not shown).
Figure 7.
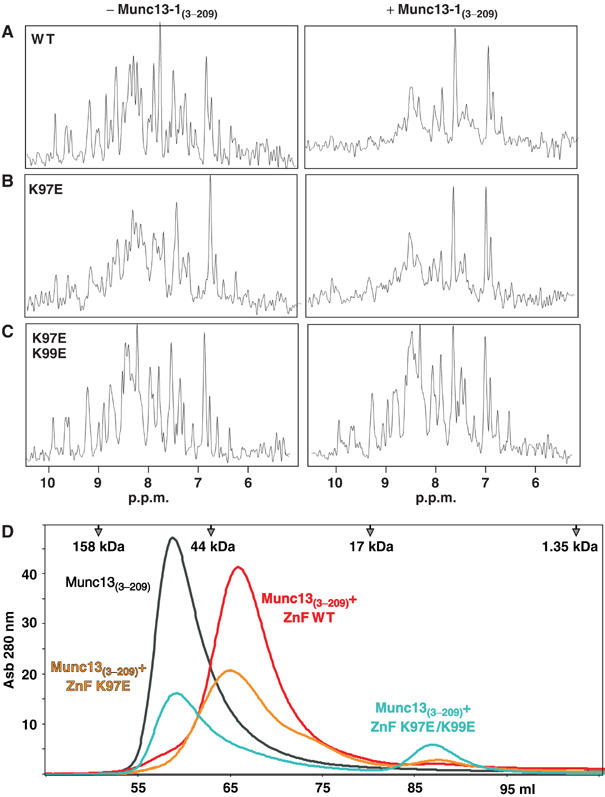
The K97E/K99E double-point mutation in the RIM2α ZF domain abolishes Munc13-1 binding. (A–C) Amide region of 1D 15N-edited 1H-NMR spectra of 1H–15N-labeled WT (A), K97E (B), and K97E/K99E (C) RIM2α82−142 (7.5 μM) in the absence or presence of the unlabeled Munc13-13−209 fragment (20 μM). (D) Superdex S75 (16/60) elution profiles of the Munc13-13−209 fragment alone (in black) or together with an equimolar amount of WT (red), K97E (yellow), or K97E/K99E (blue) RIM2α82−142.
Interference with the α-RIM/Munc13-1 interaction impairs neurotransmitter release
The availability of a mutation that specifically disrupts binding of RIM2α to Munc13-1 provided an ideal control to study the role of this interaction in neurotransmitter release. For this purpose, we compared the physiological effects induced by presynaptic application of recombinant WT and K97E/K99E mutant RIM2α ZF domains using simultaneous pre- and postsynaptic whole-cell recordings at the calyx of Held synapse. In these experiments, the cells were stimulated with a strong, presynaptic voltage-clamp depolarization that is expected to deplete a fast and a slow subpool of the readily releasable vesicle pool (RRP) (Sakaba and Neher, 2001; Woelfel et al, 2003), allowing us to quantitatively probe the size of each subpool.
Figure 8A and C show example recordings of presynaptic Ca2+ currents and excitatory postsynaptic currents (EPSCs) observed in calyces perfused with the WT (Figure 8A) or K97E/K99E mutant ZF domain (Figure 8C). Traces in response to stimuli applied early (∼1 min; black traces) and late (∼8–10 min; red traces) after the start of presynaptic whole-cell recording are shown. Plots of the time dependence of the EPSC amplitudes normalized to that of the first response are shown in Figure 8E (bottom panel). Perfusing the WT ZF domain led to a marked decrease in EPSC amplitude which, after ∼8–10 min, had reached an average fraction of 0.35±0.08 of the first EPSC amplitude. The mutant ZF domain led to a substantially smaller reduction of the EPSC amplitudes (to 0.81±0.04 of control) that parallels the reduction caused by perfusion of buffer alone and can be attributed to rundown of the release machinery during the course of the experiment. The accelerated decrease of EPSC amplitudes caused by the WT ZF domain, together with the lack of an effect upon application of the K97E/K99E ZF domain mutant, suggests that the WT ZF domain affects transmitter release by interfering with the endogenous α-RIM/Munc13-1 interaction.
Figure 8.
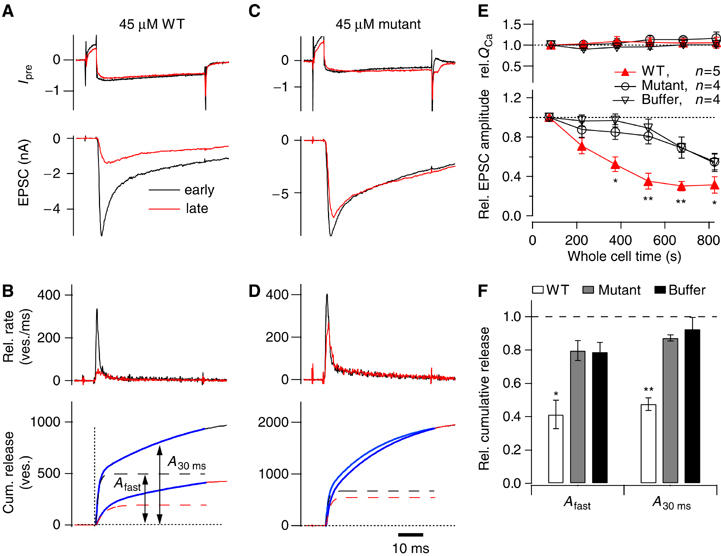
Disrupting the α-Rim–Munc13-1 interaction reduces the size of the RRP at the calyx of Held. (A–D) Paired pre- and postsynaptic whole-cell patch-clamp recordings were made at the calyx of Held synapse, with 45 μM WT (A, B) or K97E/K99E mutant (C, D) RIM2α ZF domain included in the presynaptic patch pipette solution. (A, C) Presynaptic Ca2+ current (upper panels) and EPSCs (bottom panels) in response to a 50-ms depolarization of the presynaptic terminal (to 0 mV, preceded by a step to +60 ms for 6 ms). In (A–D), responses obtained early (1–2 min) and late (8–10 min) after obtaining the presynaptic whole-cell recording condition are represented by black and red traces, respectively. (B, D) Transmitter release rates obtained by EPSC deconvolution (upper panel) and integrated release rate traces (lower panel) for the responses shown in (A, C), respectively. The estimated fast component of release (Afast) and the total release after 30 ms (A30 ms) are indicated in (B, lower panel). (E) Plot of normalized presynaptic Ca2+-current integral (upper panel) and normalized EPSC amplitude as a function of the presynaptic whole-cell recording time, for three experimental conditions. Symbols indicate the statistical significance of WT ZF domain with respect to the control data groups (*P<0.05; **P<0.01). (F) Cumulative release at late recording times relative to the control values obtained early during recording time in each experiment. The amount of release estimated from the fast component (Afast), or of the total release reached after 30 ms (A30ms), was analyzed (see (B), lower panel), for the three experimental conditions. Note that inclusion of the WT ZF domain leads to a significant (*P<0.05) reduction of both the fast component (left) and the total transmitter release reached after 30 ms.
To investigate the effects of the WT and mutant ZF domains on transmitter release in more detail, we deconvolved the evoked EPSCs with the estimated miniature EPSC (mEPSC) waveform and derived the time course and amount of quantal release (Figure 8B and D, top panels) (see Sakaba and Neher 2001). In addition, we integrated the release rate traces to estimate the cumulative release time course (Figure 8B and D, bottom panels). The time course of the release rate could not be fitted by a single exponential, and required double-exponential fits, indicating the expected release of a rapidly (τfast∼1 ms) and a more slowly releasable (τslow∼20 ms) subpool of RRP vesicles. We operationally define the amplitude of the fast release component (Afast) as the size of a fast-releasing subpool (FRP), and the total cumulative release after 30 ms (A30ms) as the size of the readily releasable pool (RRP). We found that infusing the WT ZF domain decreased the amplitude of both the FRP and the total RRP, while the mutant ZF domain had no significant effect (Figure 8F). Finally, we also checked for effects on the recovery of the RRP when a second 50 ms depolarization is applied at an interstimulus interval of 500 ms, but neither the WT nor the mutant ZF domain caused a significant change in the fraction of the FRP or RRP recovered after 500 ms (not shown). Thus, our data indicate that the WT ZF domain reduces the size of the RRP, but the recovery from pool depletion was unchanged.
Discussion
Munc13s and α-RIMs are multidomain active zone proteins that act in a ‘priming' reaction that prepares synaptic vesicles for neurotransmitter release and are also involved in short- and long-term synaptic plasticity. However, it is unclear how α-RIMs and Munc13s perform these multiple functions, and an immense gap separates the extensive genetic and physiological data available on α-RIMs and Munc13s from a molecular understanding of their roles. This gap is in part due to a lack of structural information about how α-RIMs and Munc13s interact with each other and with other synaptic proteins. Our data now provide a detailed analysis of Rab3 and Munc13 binding to α-RIMs.
Architecture of the Rab/α-RIM/Munc13 complex
Previous studies of the α-RIM/Rab3A and α-RIM/Munc13-1 interactions yielded contradictory results, perhaps because one of the assays used is sensitive to the choice of detergent (Betz et al, 2001; Wang et al, 2001). To avoid such ambiguities, we analyzed the basis for the RIM2α/Munc13-1 and RIM2α/Rab3A interactions in the absence of detergents using a range of biophysical techniques. Our data show that the long α-helix preceding the ZF domain of RIM2α is primarily responsible for Rab3A binding, in agreement with previous SPR experiments with RIM1α (Wang et al, 2001), while the SGAWFY motif makes only a small contribution to the interaction. We also find that RIM2α binds to Munc13-1 through the ZF domain, and that tight binding requires the Munc13-1 C2A domain and a sequence of approximately 80 residues C-terminal to this domain. Importantly, our data reveal that, although Munc13-1 binding to RIM2α alters slightly the RIM2α/Rab3A binding mode by releasing the SGAWFY motif, the Munc13-1/RIM2α and Rab3A/RIM2α interactions are compatible and lead to formation of a tripartite Munc13-1/RIM2α/Rab3A complex. These results show that the N-terminal sequence of α-RIMs has a modular design that places Rab3A and Munc13-1 into close proximity, but not into competition. Furthermore, elucidation of the 3D structure of the RIM2α ZF domain, together with sequence comparisons, led to the design of a double-point mutation that specifically disrupts Munc13-1 binding and that provides a powerful tool to study the importance of the α-RIM/Munc13-1 interaction for neurotransmitter release.
Functional implications
The notion that the α-RIM/Munc13-1 interaction plays a role in vesicle priming was supported by the precise correlation between the decrease in priming and the reduction in Munc13-1 levels observed in RIM1α-deficient mice (Schoch et al, 2002; Calakos et al, 2004), as well as by the decrease in the RRP caused by overexpression of a 451-residue N-terminal fragment of Munc13-1 (Betz et al, 2001). However, these experiments did not rule out a role for other interactions involving this sequence. Our electrophysiological measurements in the calyx of Held synapse show that application of a short RIM2α fragment spanning the ZF domain (which is responsible for Munc13-1 binding) severely decreases the RRP of vesicles, while the mutant RIM2α ZF domain (which is unable to bind to Munc13-1) does not. However, the recovery of vesicles into the RRP was unchanged. These results imply that the α-Rim/Munc13-1 interaction determines the total amount of primed and readily releasable vesicles, maybe by providing a site for vesicle priming, but does not affect the rate of replenishment of new readily releasable vesicles after depletion. In addition, our data show that the α-Rim/Munc13-1 interaction is equally important for both a fast-releasing and a more slowly releasable subpool of readily releasable vesicles, which can be distinguished at the calyx of Held synapse (Sakaba and Neher, 2001).
Since both Rab3A and RIM1α are required for mossy fiber LTP (Castillo et al, 1997, 2002) and the α-RIM N-terminal sequence is involved in Rab3A and Munc13-1 binding, it is likely that this sequence provides a link between presynaptic plasticity and synaptic vesicle priming. Based on GST-pulldown assays, it was originally thought that Rab3s and Munc13s compete for α-RIM binding (Betz et al, 2001), suggesting that α-RIMs may be present in two alternative states that mediate LTP (through Rab3A) and vesicle priming (through Munc13-1). Our present results indicate that this view needs to be revised: Munc13-1 and α-RIMs are likely to form a constitutive complex, considering their colocalization and the high affinity of their interaction; this complex is likely to be the substrate for Rab3 binding, which may be regulated during mossy fiber LTP. This notion is more satisfying from a physiological point of view since vesicle priming, if anything, should be enhanced during LTP. Thus, a direct connection between LTP and vesicle priming seems more plausible than a competition between the two processes.
No neurotransmitter release is observed in the absence of Munc13s (Richmond et al, 1999; Varoqueaux et al, 2002), whereas release is impaired but not abolished upon deletion of RIMs or Rab3s (Koushika et al, 2001; Schlüter et al, 2004). Moreover, Munc13s were suggested to interact with SNAREs and Munc18-1, which are central components of the fusion machinery (reviewed in Rizo and Südhof, 2002). While the physiological relevance of these interactions needs to be validated, a possible interpretation of the available data is that Munc13s mediate synaptic vesicle priming via direct or indirect interactions with SNAREs or Munc18-1, serving as the common final target that integrates signals activated by diverse presynaptic receptors involved in plasticity. In this model, α-RIMs indirectly participate in vesicle priming by facilitating the recruitment of Munc13-1 to the synapse. Our data showing that Munc13-1 and Rab3A binding to RIM2α are compatible with each other suggest that formation of a tripartite Munc13/α-RIM/Rab3 complex may move synaptic vesicles into close proximity to Munc13-1, perhaps facilitating priming during mossy fiber LTP. Detailed characterization of additional interactions involving Munc13s and α-RIMs will be required to test this and other potential models of their function.
Implications for Rab effector signaling in exocytosis
α-RIMs belong to a class of putative Rab effectors that share a similar N-terminal Rab-interacting sequence and also include Rabphilin, Noc2, Slp1, Slp2, Slp3, Slp5, granuphilin, melanophilin, and MyRIP (Fukuda, 2005). These proteins bind either Rab3 proteins, Rab27 proteins, or both (Fukuda, 2003, 2005), and contain the α-helical sequences and SGAWF(Y/F) motif that are involved in Rab3 binding to α-RIMs, rabphilin, and Noc2 (see Figure 4; Joberty et al, 1999; Ostermeier and Brunger, 1999; Haynes et al, 2001; Wang et al, 2001). Most of these proteins have a ZF domain (aligned in Figure 6A) and share the same modular design observed for α-RIMs and rabphilins in their N-terminal sequences. The finding that this architecture provides a basis for simultaneous binding of two different proteins, Rab3A and Munc13-1, to RIM2α suggest that the ZF domains of these other Rab effectors could generally constitute independent binding sites that bring another class of proteins (effectors or regulators) to the Rab/Rab effector pairs. This idea is supported by the high level of sequence conservation observed in several regions of the ZF domains within a given subclass of Rab effectors (Figure 6A). Thus, the mode of the RIM2α interactions uncovered here may be paradigmatic for the entire group of Rab effectors containing a homologous N-terminal sequence.
Materials and methods
Protein expression and purification
DNA constructs for protein expression are described in Supplementary data. Unlabeled and isotopically labeled proteins were expressed in bacteria as GST fusions as described (Dulubova et al, 1999). The fusion proteins were affinity-purified on glutathione-Sepharose (Pharmacia), cleaved from the GST moiety with thrombin (Sigma) and further purified by size-exclusion or ion-exchange chromatography. Gel-filtration binding experiments were performed on Superdex S75 or S200 columns (Pharmacia) in 50 mM Tris–HCl buffer containing 150 mM NaCl, 5 mM MgCl2, and 2 mM DTT at pH 7.3. Equal amount of fractions were separated by SDS–PAGE in duplicates, transferred to nitrocellulose and probed with either anti-T7·Tag (Novagen) or anti-Rab3A (Cl 42.2; Matteoli et al, 1991) monoclonal antibodies. Immunoblots were developed using enhanced chemiluminescence (ECL) detection (Pharmacia).
GST-pulldown assays
T7-tagged Munc13-1 fragments were expressed in bacteria (Escherichia coli BL21(DE3)), the soluble fractions of the bacterial lysates in binding buffer (PBS containing 1 mM EGTA, 1 mM EDTA, 1 mM AEBSF, 2 mM DTT, and Sigma protease inhibitors cocktail) were precleared by incubation with glutathione-agarose (4 h at 4°C), and Triton X-100 was added (0.5% final concentration). Bacterially expressed GST-fusion proteins of different RIM2α fragments or GST alone were affinity purified on glutathione-Sepharose beads (see above). Beads containing about ∼100 μg of protein were combined with 1.8 ml of the bacterial extracts containing the T7-tagged Munc13-1 fragments. After overnight incubation at 4°C, the beads were washed with binding buffer containing 1% Triton X-100 (4 × 1 ml), and the same buffer containing an additional 0.4 M NaCl (4 × 1 ml). Aliquots of the beads (ca. 2.5 μg of fusion protein) and 5 μl of the corresponding bacterial extract were analyzed by SDS–PAGE, followed by Coomassie blue staining and immunoblotting.
NMR spectroscopy
NMR data were acquired at 25°C on Varian INOVA500 or INOVA600 spectrometers using H2O/D2O 95:5 (v/v) as the solvent. 1H–15N HSQC spectra were obtained at 25–100 μM protein concentration in 50 mM Tris–HCl (pH 7.3) containing 150 mM NaCl and 2 mM DTT or 1 mM TCEP; 5 mM MgCl2 was included for samples containing Rab3A. For 1D 15N-edited 1H-NMR spectra, we acquired the first increment of a 1H–15N HSQC spectrum using 7.5 μM 15N-labeled WT or mutant ZF domain with or without addition of 20 μM unlabeled Munc13-1 (3–209) fragment. Elucidation of the structure of the RIM2α ZF domain was performed with 1 mM uniformly 15N- or 15N,13C-labeled samples of RIM2α82−142 in buffer containing 20 mM Pipes, pH 6.9, 150 mM NaCl, and 1 mM TCEP, using standard triple resonance techniques (see the legend to Supplementary Figure 1). The structure has been deposited in the PDB (accession code 2A20).
Isothermal titration calorimetry (ITC)
ITC experiments were performed using a VP-ITC system (MicroCal) at 20°C in a buffer composed of 50 mM Tris, 150 mM NaCl, 5 mM MgCl2, and 1 mM TCEP at pH 7.3, which was optimal for the solubility of all protein fragments analyzed. Typically, 150–200 μM protein solution was injected 30 times in 8 μl aliquots into the 1.4 ml sample cell containing another protein at 14–18 μM concentration. Data were fitted using a nonlinear least-squares routine using a single-site binding model with Origin for ITC v.5.0 (Microcal), varying the stoichiometry (n), the enthalpy of the reaction (ΔH) and the association constant (Ka).
Electrophysiology
Paired pre- and postsynaptic whole-cell recordings at the calyx of Held synapse were made as described (Felmy et al, 2003), using 200 μm brainstem slices of 8–10-days-old Wistar rats. Both cells were voltage-clamped at −80 mV, and series resistance (Rs) compensation of up to 85% was employed. The extracellular solution was a bicarbonate buffered Ringer solution with 2 mM CaCl2 and 1 mM MgCl2, to which 0.5 μM tetrodotoxin (TTX), 10 mM tetraethylammonium-chloride (TEA), 50 μM D-2-amino-5-phosphonovaleric acid, 100 μM cyclothiazide (CTZ), and 1 mM kynurenic acid (KYN) were added. CTZ and KYN were employed to minimize desensitization, and saturation of postsynaptic glutamate receptors of the AMPA subtype (Neher and Sakaba, 2001). The intracellular solution for the postsynaptic recording was a Cs-gluconate-containing solution with 5 mM EGTA (see Felmy et al, 2003). The presynaptic pipette solution contained (in mM): 135 CsCl, 20 TEA-Cl, 10 Hepes, 5 NaCl, 1 MgCl2, 2 DTT, 4 Mg-ATP, 0.3 Na-GTP, 0.2 EGTA, and 45 μM WT, or mutant RIM2α ZF domain (residues 82–142). ZF domain-containing solutions were prepared by first dialyzing the protein solution against the buffer in the absence of ATP and GTP, yielding final protein concentrations of ca. 200 μM. Stock solutions were diluted to 45 μM, and ATP and GTP were added to yield the indicated final concentrations. In control experiments, the same intracellular solution was used without added ZF domain (Figure 8E and F, ‘buffer').
To derive the transmitter release rates (Figure 8B and D), EPSCs were deconvolved with an idealized mEPSC waveform approximated by a double-exponentially decaying function (Neher and Sakaba, 2001). The deconvolution assumes that EPSCs arise from linearly summed mEPSCs with amplitudes of 15 pA (for the presence of CTZ and KYN), and an additional current component generated by glutamate spillover, that was subtracted. The release rate traces resulting from deconvolution were integrated, and fitted with double-exponential functions. The size of an FRP of vesicles was estimated from the amplitude value of the fast component (Afast; Figure 8B). In addition, the total number of vesicles released after 30 ms was analyzed (A30ms).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary data
Supplementary Table 1
Acknowledgments
JL was a fellow from the American Heart Association. This work was supported by a grant from the Welch Foundation and NIH grant NS40944 to JR.
References
- Araç D, Murphy T, Rizo J (2003) Facile detection of protein–protein interactions by one-dimensional NMR spectroscopy. Biochemistry 42: 2774–2780 [DOI] [PubMed] [Google Scholar]
- Aravamudan B, Fergestad T, Davis WS, Rodesch CK, Broadie K (1999) Drosophila UNC-13 is essential for synaptic transmission. Nat Neurosci 2: 965–971 [DOI] [PubMed] [Google Scholar]
- Augustin I, Rosenmund C, Südhof TC, Brose N (1999) Munc13-1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature 400: 457–461 [DOI] [PubMed] [Google Scholar]
- Betz A, Okamoto M, Benseler F, Brose N (1997) Direct interaction of the rat unc-13 homologue Munc13-1 with the N terminus of syntaxin. J Biol Chem 272: 2520–2526 [DOI] [PubMed] [Google Scholar]
- Betz A, Thakur P, Junge HJ, Ashery U, Rhee JS, Scheuss V, Rosenmund C, Rettig J, Brose N (2001) Functional interaction of the active zone proteins Munc13-1 and RIM1 in synaptic vesicle priming. Neuron 30: 183–196 [DOI] [PubMed] [Google Scholar]
- Brose N, Hofmann K, Hata Y, Südhof TC (1995) Mammalian homologues of Caenorhabditis elegans unc-13 gene define novel family of C2-domain proteins. J Biol Chem 270: 25273–25280 [DOI] [PubMed] [Google Scholar]
- Calakos N, Schoch S, Südhof TC, Malenka RC (2004) Multiple roles for the active zone protein RIM1alpha in late stages of neurotransmitter release. Neuron 42: 889–896 [DOI] [PubMed] [Google Scholar]
- Castillo PE, Janz R, Südhof TC, Tzounopoulos T, Malenka RC, Nicoll RA (1997) Rab3A is essential for mossy fibre long-term potentiation in the hippocampus. Nature 388: 590–593 [DOI] [PubMed] [Google Scholar]
- Castillo PE, Schoch S, Schmitz F, Südhof TC, Malenka RC (2002) RIM1alpha is required for presynaptic long-term potentiation. Nature 415: 327–330 [DOI] [PubMed] [Google Scholar]
- Dulubova I, Sugita S, Hill S, Hosaka M, Fernandez I, Südhof TC, Rizo J (1999) A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J 18: 4372–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felmy F, Neher E, Schneggenburger R (2003) Probing the intracellular calcium sensitivity of transmitter release during synaptic facilitation. Neuron 37: 801–811 [DOI] [PubMed] [Google Scholar]
- Fukuda M (2003) Distinct Rab binding specificity of Rim1, Rim2, rabphilin, and Noc2. Identification of a critical determinant of Rab3A/Rab27A recognition by Rim2. J Biol Chem 278: 15373–15380 [DOI] [PubMed] [Google Scholar]
- Fukuda M (2004) Alternative splicing in the first alpha-helical region of the Rab-binding domain of Rim regulates Rab3A binding activity: is Rim a Rab3 effector protein during evolution? Genes Cells 9: 831–842 [DOI] [PubMed] [Google Scholar]
- Fukuda M (2005) Versatile role of Rab27 in membrane trafficking: focus on the Rab27 effector families. J Biochem (Tokyo) 137: 9–16 [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE (1997) Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell 90: 523–535 [DOI] [PubMed] [Google Scholar]
- Haynes LP, Evans GJ, Morgan A, Burgoyne RD (2001) A direct inhibitory role for the Rab3-specific effector, Noc2, in Ca2+-regulated exocytosis in neuroendocrine cells. J Biol Chem 276: 9726–9732 [DOI] [PubMed] [Google Scholar]
- Joberty G, Stabila PF, Coppola T, Macara IG, Regazzi R (1999) High affinity Rab3 binding is dispensable for Rabphilin-dependent potentiation of stimulated secretion. J Cell Sci 112 (Part 20): 3579–3587 [DOI] [PubMed] [Google Scholar]
- Koushika SP, Richmond JE, Hadwiger G, Weimer RM, Jorgensen EM, Nonet ML (2001) A post-docking role for active zone protein Rim. Nat Neurosci 4: 997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RC, Scheller RH (2000) Mechanisms of synaptic vesicle exocytosis. Annu Rev Cell Dev Biol 16: 19–49 [DOI] [PubMed] [Google Scholar]
- Maruyama IN, Brenner S (1991) A phorbol ester/diacylglycerol-binding protein encoded by the unc-13 gene of Caenorhabditis elegans. Proc Natl Acad Sci USA 88: 5729–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matteoli M, Takei K, Cameron R, Hurlbut P, Johnston PA, Südhof TC, Jahn R, De Camilli P (1991) Association of Rab3A with synaptic vesicles at late stages of the secretory pathway. J Cell Biol 115: 625–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher E, Sakaba T (2001) Combining deconvolution and noise analysis for the estimation of transmitter release rates at the calyx of held. J Neurosci 21: 444–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier C, Brunger AT (1999) Structural basis of Rab effector specificity: crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell 96: 363–374 [DOI] [PubMed] [Google Scholar]
- Powell CM, Schoch S, Monteggia L, Barrot M, Matos MF, Feldmann N, Südhof TC, Nestler EJ (2004) The presynaptic active zone protein RIM1alpha is critical for normal learning and memory. Neuron 42: 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond JE, Davis WS, Jorgensen EM (1999) UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat Neurosci 2: 959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond JE, Weimer RM, Jorgensen EM (2001) An open form of syntaxin bypasses the requirement for UNC-13 in vesicle priming. Nature 412: 338–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Südhof TC (2002) SNAREs and munc18 in synaptic vesicle fusion. Nat Rev Neurosci 3: 641–653 [DOI] [PubMed] [Google Scholar]
- Rhee JS, Betz A, Pyott S, Reim K, Varoqueaux F, Augustin I, Hesse D, Südhof TC, Takahashi M, Rosenmund C, Brose N (2002) Beta phorbol ester- and diacylglycerol-induced augmentation of transmitter release is mediated by Munc13s and not by PKCs. Cell 108: 121–133 [DOI] [PubMed] [Google Scholar]
- Sakaba T, Neher E (2001) Calmodulin mediates rapid recruitment of fast-releasing synaptic vesicles at a calyx-type synapse. Neuron 32: 1119–1131 [DOI] [PubMed] [Google Scholar]
- Schlüter OM, Schmitz F, Jahn R, Rosenmund C, Südhof TC (2004) A complete genetic analysis of neuronal Rab3 function. J Neurosci 24: 6629–6637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch S, Castillo PE, Jo T, Mukherjee K, Geppert M, Wang Y, Schmitz F, Malenka RC, Südhof TC (2002) RIM1alpha forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature 415: 321–326 [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Sigler A, Rhee JS, Brose N, Enk C, Reim K, Rosenmund C (2002) Total arrest of spontaneous and evoked synaptic transmission but normal synaptogenesis in the absence of Munc13-mediated vesicle priming. Proc Natl Acad Sci USA 99: 9037–9042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hu B, Zimmermann B, Kilimann MW (2001) Rim1 and rabphilin-3 bind Rab3-GTP by composite determinants partially related through N-terminal alpha-helix motifs. J Biol Chem 276: 32480–32488 [DOI] [PubMed] [Google Scholar]
- Wang Y, Okamoto M, Schmitz F, Hofmann K, Südhof TC (1997) Rim is a putative Rab3 effector in regulating synaptic-vesicle fusion. Nature 388: 593–598 [DOI] [PubMed] [Google Scholar]
- Wang Y, Sugita S, Südhof TC (2000) The RIM/NIM family of neuronal C2 domain proteins. Interactions with Rab3 and a new class of Src homology 3 domain proteins. J Biol Chem 275: 20033–20044 [DOI] [PubMed] [Google Scholar]
- Wang Y, Südhof TC (2003) Genomic definition of RIM proteins: evolutionary amplification of a family of synaptic regulatory proteins. Genomics 81: 126–137 [DOI] [PubMed] [Google Scholar]
- Woelfel M, Neher E, Schneggenburger R (2003) Presynaptic Ca2+ elevations by Ca2+ uncaging produce two kinetically distinct components of transmitter release at the calyx of Held. Soc Neurosci Abstr 686: 10 [Google Scholar]
- Zerial M, McBride H (2001) Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2: 107–117 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary data
Supplementary Table 1


