Abstract
The human immunodeficiency virus type 1 (HIV-1) envelope glycoproteins (Envs) function as a trimer, mediating virus entry by promoting the fusion of the viral and target cell membranes. HIV-1 Env trimers induce membrane fusion through a pH-independent pathway driven by the interaction between an Env trimer and its cellular receptors, CD4 and CCR5/CXCR4. We studied viruses with mixed heterotrimers of wild-type and dominant-negative Envs to determine the number (T) of Env trimers required for HIV-1 entry. To our surprise, we found that a single Env trimer is capable of supporting HIV-1 entry; i.e., T = 1. A similar approach was applied to investigate the entry stoichiometry of envelope glycoproteins from amphotropic murine leukemia virus (A-MLV), avian sarcoma/leukosis virus type A (ASLV-A), and influenza A virus. When pseudotyped on HIV-1 virions, the A-MLV and ASLV-A Envs also exhibit a T = 1 entry stoichiometry. In contrast, eight to nine influenza A virus hemagglutinin trimers function cooperatively to achieve membrane fusion and virus entry, using a pH-dependent pathway. The different entry requirements for cooperativity among Env trimers for retroviruses and influenza A virus may influence viral strategies for replication and evasion of the immune system.
The human immunodeficiency virus type 1 (HIV-1) establishes persistent infections in humans and eventually causes AIDS in most infected individuals. The entry of HIV-1 into host cells is mediated by the viral envelope glycoproteins (Envs), which function as a trimer consisting of three gp120 exterior and three gp41 transmembrane Env proteins (31, 123). The gp120 subunits interact noncovalently with the ectodomain of the gp41 glycoprotein, which is anchored in the viral membrane by a transmembrane region. The sequential binding of gp120 to the CD4 receptor and one of the coreceptors, CCR5 or CXCR4, is thought to promote major structural changes in the Env complex. These changes eventually lead to the formation of a six-helix bundle by the gp41 ectodomain, a process that is proposed to drive the fusion of the viral and target cell membranes (1, 5, 12, 17, 19, 21, 25, 33, 55, 84, 112, 119, 120). Membrane fusion is generally believed to result in the creation of a pore connecting the interior of the virion with the target cell cytoplasm, facilitating the entry of the viral core into the host cell cytoplasm.
A number of enveloped viruses utilize trimeric helical bundles to achieve viral-cell membrane fusion and entry into the host cell (9, 24, 64, 118, 119). The conformational changes in Envs required for membrane fusion are triggered by distinct mechanisms in different viruses (30). The pH-dependent viruses are endocytosed after receptor binding, and the low pH within endosomal compartments induces major Env conformational changes leading to virus entry (9, 24, 64, 118, 119). Many viruses, including orthomyxoviruses (such as influenza viruses), togaviruses, rhabdoviruses, and filoviruses, enter cells via pH-dependent membrane fusion (22, 104, 106). Viruses that employ pH-independent entry include most retroviruses (including HIV-1 and murine leukemia viruses), paramyxoviruses, and coronaviruses (13, 22, 78, 86). For at least some of these viruses, receptor binding can trigger the Env conformational changes required for membrane fusion and entry (18). One retrovirus, avian sarcoma/leukosis virus type A (ASLV-A), requires both receptor binding and a decrease in pH to achieve entry into the host cell (67, 79). Helical bundles are also employed by higher eukaryotes to fuse membranes in such processes as directional trafficking among cell organelles, endocytosis and exocytosis, and fertilization of eggs (2, 109).
Significant insights into the mechanisms of protein-mediated membrane fusion have been derived from the study of the influenza virus hemagglutinin (HA) (104). Even in this relatively well-characterized system, the precise number of HA molecules required for membrane fusion is uncertain. Based on observations of cell-cell fusion mediated by HA, it has been proposed that two to three trimers engaged with the cellular receptors form a prefusion structure, and six to eight trimers are needed to create the stable fusion pore (3, 4, 6, 20, 72). Experiments using in vitro reconstituted liposomes with the HA proteins have produced conflicting results with respect to the number of trimers required for membrane fusion (14, 32, 45, 101). Detailed quantitative studies on live influenza viruses have been frustrated by the difficulty of labeling the virions for convenient and accurate monitoring of the progress of the fusion reactions (91).
Conventional approaches such as quantitative biochemical measurement and morphological observation using electron microscopy have not been successful in studying the stoichiometry of HIV-1 entry due to several well-known properties of HIV-1 Envs and virions. These include the replication defectiveness of the vast majority (greater than 99%) of HIV-1 virions (8, 61), the small number of intact Env trimers per virion (15, 47, 61, 128), the spontaneous and ligand-induced dissociation (“shedding”) of gp120 from the Env complexes (77, 99), and the potential heterogeneity among HIV-1 Env complexes (8, 49, 90). Although the study of cell-cell fusion mediated by HIV-1 Envs might be more convenient, there are indications that quantitative differences exist between the number of functional Env proteins required for cell-cell fusion and that needed for virus infection. For example, HIV-1 Env mutants with alterations in membrane-fusing capacity often demonstrate dramatic reductions in the ability to form syncytia, with little or only modest decreases in virus replication (11, 126). Furthermore, cell-cell fusion and virus entry mediated by HIV-1 Envs exhibit different sensitivities to neutralizing antibodies and other inhibitors (44, 89, 120). As a result of these difficulties, the number of HIV-1 Env trimers required to support virus entry is unknown.
Recently, we investigated the stoichiometry of antibody-mediated neutralization by studying heterotrimers composed of wild-type and neutralization-escape mutant Envs of HIV-1 in 15 combinations of nine different antibodies and Envs from four different HIV-1 strains. The data suggested that the target of anti-HIV-1 neutralizing antibodies is composed of a single Env trimer (125). This result was independent of HIV-1 strain variation, applying to primary isolates and T-cell-adapted viruses, CCR5- and CXCR4-using strains, and neutralization-resistant and neutralization-sensitive viruses. Although the target size of neutralizing antibodies cannot be formally equated with the functional unit of HIV-1 Env trimers operative during the normal entry process, it should reflect the size of the latter, suggesting that only a small number of HIV-1 Env trimers is required for virus entry.
The heterogeneity and defectiveness of HIV-1 preparations necessitate the use of approaches that assess the functional subset of virion Env complexes to achieve insight into stoichiometric parameters. Here we use previously described dominant-negative HIV-1 Env mutants to form heterotrimers with the wild-type Env proteins and to titrate downward the functional Env trimers in HIV-1 stocks. We compare the empirically observed infectivity of these viruses with the expected frequency of functional trimers predicted by models that assume different requirements for cooperativity among individual Env trimers. Thus, the study of each dominant-negative mutant yields an independent determination of the number of functional Env trimers that are required for virus entry. We also used a similar approach to investigate the stoichiometry of virus entry mediated by the envelope glycoproteins of amphotropic murine leukemia virus (A-MLV), ASLV-A, and influenza A virus.
MATERIALS AND METHODS
Plasmids expressing Env proteins.
The wild-type and mutant HIV-1 Env proteins were expressed from the pSVIIIenv vector (111). The Env mutants were created by the PCR-based QuikChange protocol (Stratagene). The presence of the desired mutations and the absence of unintended changes were confirmed by sequencing the entire env reading frames. The names of the mutants designate the wild-type amino acid residue in single-letter code, the residue number, and the substituted amino acid. The residue numbering is based upon that of the prototypic HIV-1HXBc2 gp160, according to current convention (58).
The A-MLV (strain 4070A) gp90 envelope glycoprotein was expressed under the control of the cytomegalovirus (CMV) immediate-early promoter in the pVPack-ampho vector (Stratagene). Site-directed mutagenesis was used to generate the cleavage-defective mutant, using the QuikChange protocol.
The wild-type ASLV-A Env protein was expressed from the pCB6/WTA vector (a gift from S. Delos and J. White, University of Virginia) (43). The cleavage-defective mutant was generated using the standard QuikChange protocol.
The wild-type HA and the HAfd mutant of influenza A virus (strain FPV/Rostock/34) were expressed under the control of the CMV immediate-early promoter in the pCMVHA and pCMVHAfd vectors, respectively (a gift from P. Cannon, University of Southern California) (63, 115).
All plasmids expressing wild-type and mutant Envs used in one experimental session were prepared using the QIAFilter Kit (QIAGEN) and then were quantified, stored, and used as a set.
Reporter virus stocks and single-round infection assays of HIV-1.
Recombinant HIV-1 encoding firefly luciferase was produced as previously described (50). Briefly, 293T cells in 100-mm tissue culture dishes were cotransfected by the Lipofectamine reagent with 2 μg of pSVIIIenv plasmid expressing the HIV-1 Env variants, 2 μg of the pCMVΔP1ΔenvpA plasmid, and 6 μg of the pHIV-1Luc plasmid. The pCMVΔP1ΔenvpA plasmid encodes the Gag/Pol and Tat proteins of HIV-1. The pHIV-1Luc plasmid encodes an HIV-1 RNA genome that is defective in all HIV-1 genes except tat and contains a firefly luciferase reporter gene. The virus stocks, which are capable of a single round of infection, were harvested 2 days later, aliquoted, and stored at −80°C. In some experiments, viruses containing different ratios of wild-type and mutant envelope glycoproteins were produced. The fM value describes the frequency of the mutant Env monomers in the total population of Env monomers. For each Env mutant, viral stocks associated with one series of different fM values were produced and tested as a set. To produce virus stocks with different fM values, 293T cells were transfected as described above except that the pSVIIIenv plasmids expressing the wild-type and mutant Envs were mixed at a given ratio, keeping the total amount of plasmid DNA at 2 μg. To minimize the potential for pipetting errors in creating viruses with very low or high fM values, the pSVIIIenv plasmids were first diluted so that all pipetting could be performed in the 4- to 20-μl range. Recombinant viral production was measured with a standard reverse transcriptase assay, and all viral stocks had reverse transcriptase levels within 20% of each other (data not shown).
The infectivity of recombinant viruses was measured by incubating the viruses with Cf2Th-CD4/CCR5 cells. Target cells (6 × 103 cells per well) were seeded into 96-well tissue culture Isoplates (EG&G Wallac) and cultured for 16 to 24 h. After medium was thoroughly removed from the target cells, 100 μl of virus suspension in fresh culture medium was added to each well. After 48 h, viral infectivity was quantified by measuring luciferase activity using a luciferase detection kit (Pharmingen) and an automated luminometer (Microlumat Plus; EG&G Berthod). For each sample of virus, multiple (four to eight) wells of target cells were infected in parallel, and the mean value of luciferase activity was obtained.
In pilot experiments using wild-type HIV-1 viral stocks in which infectivity was used as a readout, we determined that normalization of the amount of virus by reverse transcriptase assays resulted in more variation than using the same volume of virus stocks produced in parallel (data not shown). The main source of variation in assays normalized by reverse transcriptase activities was from the variability of the reverse transcriptase measurements, and this experimental variability is also likely to apply to p24 measurements of viral stocks. Therefore, luciferase signals generated from the same volumes of viral stocks were used for comparison of the infectivity of different viral stocks in all experiments. The infectivities of recombinant viruses with different ratios (fM values) of the dominant-negative mutant and the wild-type Envs were determined. After several preliminary experiments that generated similar results, one experiment involving eight parallel measurements of the infectivity of each viral sample was conducted. The infectivity of the viral stocks was expressed as a percentage of the infectivity of the wild-type viruses to generate a value for relative infectivity [RI(%)]. The relationship among RI(%), fM (the frequency of mutant Env in the pool of total envelope glycoproteins, where 0 ≤ fM ≤ 1), and T (the number of Env trimers in the infectious unit) was then analyzed using a nonlinear regression model by resolving the following formula: RI (%) = (1 − fM)3T × 100% + e, where e is the residual error. The derivation of this formula is described in Results. Parametrical estimations of T and its 95% confidence interval (CI) were produced by a nonlinear regression analysis using the PROC GNL procedure, SAS package (SAS/STAT. 1999. User's Guide, version 8, vol. 2. SAS Institute Inc., Cary, N.C.).
Incorporation of HIV Env proteins in virions.
To evaluate the efficiency with which HIV-1 mutant Env proteins were incorporated into virions, virus-like particles were produced and radiolabeled for 48 h in 293T cells after cotransfecting 4 μg of the pSVIIIenv plasmids expressing the wild-type HIV-1 Env or the V513E or R508S/R511S mutant Envs with 4 μg of the pCMVΔP1ΔenvpA plasmid and 1 μg each of the pc-Tat and pc-Rev plasmids. The viral particles were pelleted by ultracentrifugation in an SW41 rotor at 28,000 rpm for 2 h and lysed in the lysis buffer (0.5% NP-40, 150 mM NaCl, and 10 mM Tris-HCl, pH 7.5, with a cocktail of protease inhibitors). The virion-associated Envs were then immunoprecipitated by pooled sera from HIV-1-infected individuals. The cell-associated Env proteins were immunoprecipitated from the cell lysates in parallel. After being resolved by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the levels of Env proteins associated with the cells and the virions were quantified using a PhosphorImager.
Reporter virus stocks and single-round infection assay of Env proteins from A-MLV, ASLV-A, and influenza A virus.
For pseudotyping A-MLV Env onto HIV-1 virions, 3 μg of the pVPack-ampho vector were cotransfected with 1.5 μg of the pCMVΔP1ΔenvpA plasmid, 4.5 μg of the pHIV-1Luc vector, and 1 μg of a pc-Rev plasmid. The pc-Rev plasmid expresses the HIV-1 Rev protein and is included to facilitate the transport of the HIV-1 transcripts from the nucleus into the cytoplasm. The virus infectivities were assessed on Cf2Th target cells in the presence of 1 μM polybrene.
For pseudotyping ASLV-A Env onto HIV-1 virions, 3 μg of the pCB6/WTA vector was cotransfected with 1.5 μg of the pCMVΔP1ΔenvpA plasmid, 4.5 μg of the pHIV-1Luc vector, and 1 μg of a pc-Rev plasmid. To create target cells, 293T cells were transiently transfected by Lipofectamine reagent with 10 μg of the pCB7/PG950 vector to express the membrane-anchored form of Tva, which serves as a receptor for ASLV-A (a gift from S. Delos and J. White, University of Virginia) (43). The virus infectivities were assessed on the target cells in the presence of 1 μM polybrene, as described above. To study entry into receptor-negative 293T cells mediated through the soluble Tva protein, viral stocks were incubated on ice for 30 min in 100 μl of growth medium containing 1 μM polybrene with 10 nM soluble Tva (a gift from James Cunningham, Harvard Medical School) (107). The resulting mixtures were then incubated with 293T cells in 96-well plates for 2 days at 37°C.
To pseudotype the influenza A virus HA proteins onto HIV-1 virions, a total of 3 μg of either the pCMVHA or the pCMVHAfd expression plasmid or various ratios of the two plasmids were cotransfected with 1.5 μg of the pCMVΔP1ΔenvpA plasmid, 4.5 μg of the pHIV-1Luc vector, and 1 μg of a pc-Rev plasmid expressing the HIV-1 Rev protein. The infectivity of the resulting reporter viruses was detected in the single-round entry assay using Cf2Th target cells.
Viral stocks with different fM values were generated by cotransfecting the plasmids expressing the wild-type Envs with their corresponding cleavage-defective mutants, keeping the total amounts of Env-expressing plasmids at 3 μg per transfection. All viral stocks with a series of fM values were studied as a set in a single experimental session. The infectivity measurement and data analysis were conducted as described above for HIV-1 viruses.
Predictions of infectivity of viruses with high fM values based on the Poisson distribution.
In some experiments, HIV-1 viral stocks were produced in cells expressing a high proportion of the dominant-negative Env (high fM values). In these cases, the expected frequency of virions that carry a given number of wild-type homotrimers was calculated according to the Poisson formula: P(x) = e−λ λx/x!, where P(x) is the frequency of virions carrying x number of wild-type homotrimers, e is the base of the system of natural logarithms, and λ is the average number of wild-type homotrimers per virion in the whole viral stock. When fM is 0.9, the frequency of wild-type homotrimers in the total population is 0.1% and the value of λ is either 0.01 or 0.072, assuming that each virus carries either 10 or 72 Env trimers, respectively. These values represent the observed average and theoretical maximum number, respectively, of Env spikes on HIV-1 virions (15, 40, 61, 127, 128). We first calculated the values for P(0) and P(1) using the above formula; then P(≥2) was calculated by the following formula: 1 − (P(0) + P(1)).
RESULTS
Building theoretical models of Env trimer function during the entry of HIV-1.
HIV-1 gp160 trimerizes soon after its synthesis in the endoplasmic reticulum (28). Subsequently, proteolytic cleavage produces the mature, functional trimer of gp120 and gp41 subunits (110). When two different species of viral glycoproteins are coexpressed in a cell, heterotrimers can form. Heterotrimer formation has been documented for HIV-1 Envs with different coreceptor preferences and with different sensitivities to neutralizing antibodies (96, 100). In fact, even the divergent HIV-1 and HIV-2 Envs are able to assemble into heterotrimers (23). Heterotrimers between two species of closely related Env proteins can form with an efficiency close to that predicted by random mixing of the two species of monomeric subunits; random mixing of influenza A virus HA variants has been documented by biochemical measurements (7, 105).
To investigate the number of Env trimers involved in HIV-1 entry, we determined the efficiency of infection of recombinant viruses containing different ratios of wild-type HIV Envs and dominant-negative mutants. By using amounts of plasmid DNA in the linear range of the relationship between transfected DNA and expressed protein, the ratios between the two species of Env monomers were experimentally controlled. Assuming that monomers from the two Env species mix randomly, the frequency of different trimers in the viral population can be calculated for mutant homotrimers as fM3, for heterotrimers with two mutant subunits as 3fM2(1 − fM), for heterotrimers with one mutant subunit as 3fM(1 − fM)2, and for wild-type homotrimers as (1 − fM)3, where fM, ranging from 0 to 1, is the frequency of the mutant monomers in the total population of Env monomers.
Recombinant HIV-1 viruses containing HIV-1 envelope glycoproteins or pseudotyped with different Envs were employed in this study. HIV-1 exhibits a very high level of defectiveness, a substantial fraction of which has been attributed to defects in env and pol gene products (54, 70, 81, 88). Infectivity-to-particle ratios for HIV-1 are typically less than 10−4. We define an infectious unit as a collection of the minimum number of Env trimers that have the potential to mediate infection. Because of the extremely high level of defectiveness of virions in HIV-1 stocks, the probability that any single virion will comprise more than one infectious unit is negligible; therefore, the infectious units within a viral stock can be considered to function independently. The infectivity of a viral stock containing wild-type and mutant Envs, relative to that of a virus with only wild-type Envs, can then be predicted based upon two parameters: (i) the frequency (F) of Env trimers that are potentially functional in the population and (ii) the number (T) of trimers required to achieve infection (i.e., the number of trimers in the infectious unit). The relative infectivity as a percentage of that of the wild-type viruses can be calculated as follows: RI(%) = FT × 100%.
This model is built on the assumption that each Env trimer forms an infectious unit only with its immediately neighboring trimers and makes no assumptions about the size or internal configuration of the functional unit. F can be deduced from the expected frequencies of the homo- and heterotrimers and knowledge of the relative ability of these forms to function in mediating virus entry. In our experiments, we employ Env mutants that have been demonstrated to be strongly dominant negative. The extremely low ability of the mutant homotrimers to support HIV-1 infection can be experimentally documented. Heterotrimers containing both wild-type and mutant Env subunits are also expected to be nonfunctional due to the very nature of the dominant-negative mutant. Thus, for an effective dominant-negative mutant, only the wild-type homotrimers in a viral stock are potentially functional, i.e., F = (1 − fM)3. Consequently, the relative infectivity of a viral stock containing wild-type and dominant-negative mutant Envs is predicted by the following formula: RI(%) = (1 − fM)3T × 100%. Empirical control of the fM values of viral stocks and precise measurement of the infectivity or RI(%) of the resulting viruses should therefore permit estimation of the T value.
Individual HIV-1 Env trimers independently mediate viral infection.
Although many defective HIV-1 Env mutants exist, only a few mutants act in a dominant-negative fashion to disrupt HIV-1 entry. For this study, we used two Env mutants from HIV-1YU2 reported to have potent dominant-negative activity. The first mutant, R508S/R511S, is totally defective for proteolytic cleavage of the gp160 Env precursor due to changes in two amino acid residues in the furin recognition sequence (37, 46, 68). This HIV-1 Env mutant is transported efficiently to the cell surface, and its antigenicity has been shown to resemble closely that of the wild-type Env (103). Because oligomerization of HIV-1 Envs precedes proteolytic maturation, heterotrimers composed of the R508S/R511S mutant and wild-type Envs should form efficiently (26, 28, 110, 121). Furthermore, an Env trimer with one or more uncleaved subunits would be expected to be completely nonfunctional because the exposure of fusion peptides capable of freely inserting into the target membrane is a prerequisite for Env function, according to our current understanding of the membrane fusion process (13, 123). We constructed the R508S/R511S mutant of the HIV-1YU2 gp160 envelope glycoprotein. The resulting mutant exhibited at least a 7-log reduction in supporting HIV-1 infection compared with the wild-type HIV-1YU2 envelope glycoprotein and functioned dominant-negatively when coexpressed with the wild-type HIV-1YU2 gp160 (data not shown).
The second mutant, V513E, contains a charged amino acid residue in the hydrophobic fusion peptide at the amino terminus of gp41. This mutant, originally created in the HIV-1HXBc2 gp160 envelope glycoprotein, exhibited an exceptionally strong dominant-negative phenotype in viral replication and cell-cell fusion assays (35). We constructed the analogous mutant in the HIV-1YU2 gp160 envelope glycoprotein using site-directed mutagenesis. As expected, this mutant exhibited at least a 5-log reduction in the ability to mediate HIV-1 infection, compared with the wild-type envelope glycoprotein, and was confirmed to be strongly dominant negative in this context (data not shown).
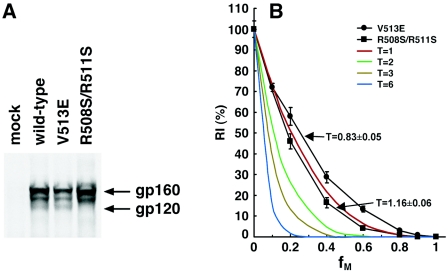
The wild-type and V513E and R508S/R511S mutant Envs were transiently expressed in 293T cells and immunoprecipitated by pooled sera from HIV-1-infected individuals. The V513E mutant was expressed and processed similarly to the wild-type envelope glycoproteins; moreover, the association of gp120 and gp41 subunits was also comparable to that of the wild-type Envs, as indicated by the retention of cleaved gp120 in the cells (Fig. 1A). The R508S/R511S mutant was totally devoid of processed gp120 glycoprotein, confirming its cleavage-defective phenotype (Fig. 1A).
FIG. 1.
Stoichiometry of HIV-1 entry. (A) 293T cells were transfected with 9 μg of plasmids expressing the wild-type and mutant Envs and 1 μg of pc-Tat1. The lysates of [35S]Met/Cys-labeled cells were precipitated with 3 μl of pooled sera from HIV-1-infected individuals following a standard protocol (124). The precipitated proteins were analyzed by 8% SDS-PAGE under reducing conditions. The gp160 precursor and the mature gp120 glycoproteins are indicated. The band migrating between gp160 and gp120 represents an under-glycosylated gp160 precursor. (B) HIV-1 luciferase reporter viruses with various ratios (fM) of the wild-type HIV-1YU2 gp160 and a dominant-negative mutant (either V513E or R508S/R511S) were incubated with Cf2Th-CD4/CCR5 cells. The luciferase activity in the target cells was measured, and the RI(%) value for each virus stock at a given fM value was calculated by normalizing to that of the wild-type viruses assayed in the same experiment. The means and ranges of variation observed in eight parallel infections are shown (black). Also shown are the theoretical curves expected if the value of T in the infectious unit is 1, 2, 3, or 6. Parametrical estimations of the T value were generated using a nonlinear regression between fM and RI(%) values by resolving the general relationship formula, RI(%) = (1 − fM)3T × 100% + e, using the GNL procedure of the SAS package.
To evaluate whether these mutant Env proteins could be efficiently incorporated into virions, radiolabeled virus-like particles were produced in 293T cells and concentrated by ultracentrifugation. The virion-associated Envs were then immunoprecipitated by pooled sera from HIV-1-infected individuals. The cell-associated Env proteins were immunoprecipitated from the cell lysates in parallel, and the levels of Env proteins associated with the cells or the virions were quantified. The ratio of cell to virion Env proteins was 7.00, 7.10, and 8.65 for the wild-type, V513E, and R508S/R511S Envs, respectively. Thus, compared with the wild-type HIV-1 Env, the two mutant Envs were incorporated into viruses efficiently.
A precise assay measuring a single round of HIV-1 infection was used to determine the infectivity of recombinant viruses containing wild-type or mutant Envs or mixtures of the two glycoproteins. Stocks of HIV-1 reporter viruses were generated by cotransfecting the Env-expressing plasmid(s) with the pCMVΔP1ΔenvpA packaging plasmid and the pHIV-1luc luciferase vector, as previously described (50). The recombinant HIV-1 viruses, which encode firefly luciferase, were incubated with Cf2Th-CD4/CCR5 cells that express high levels of the receptors utilized by HIV-1YU2. Two days later, the luciferase activity in the target cells was measured, providing an indication of the efficiency of infection. By controlling the ratio between the plasmid DNAs in the transfections, the relative expression levels of the wild-type and mutant Envs in the virus-producing cells were modulated. Specific measures were employed to minimize experimental variation (see Materials and Methods). For each of the dominant-negative mutants, the infectivity of virus stocks with different ratios (fM values) of wild-type and mutant Envs was measured and then normalized to that of viruses with only wild-type gp160 to generate an RI(%) value for that given viral stock. The means and ranges of variation of RI(%) as a function of fM for viruses containing each of the two dominant-negative mutants are shown in Fig. 1B. Also shown are the theoretical curves expected if the value of T is 1, 2, 3, or 6, according to the models described above.
The relationship between RI(%) and fM observed for viruses with the R508S/R511S and V513E Env mutants was close to that expected if T equals 1. A nonlinear regression was performed to summarize the relationship between the T, fM, and RI(%) values by resolving the formula established above, RI(%) = (1 − fM)3T × 100% + e, in which e is the statistical residue for modeling, using the GNL procedure of the SAS package (SAS/STAT, User's Guide, version 8, vol. 2). A parametrical estimation of T for viruses with the mixed wild-type and R508S/R511S Env was 1.16, with a 95% CI of 1.10 to 1.23. For viruses with the mixed wild-type and V513E Envs, T was estimated to be 0.83, with a 95% CI of 0.78 to 0.88. Similar results were also produced using the wild-type and cleavage-defective Envs of another virus strain, HIV-1HXBc2 (data not shown).
HIV-1 virions with a single functional Env trimer are infectious.
When mixtures of wild-type and dominant-negative mutant Envs are formed at high fM values, the expected frequency of wild-type homotrimers in the total population of Env spikes is very low. For example, when fM is 0.9, the percentage of wild-type homotrimers in the Env pool is (1 − 0.9)3 × 100%, or 0.1%. Like other rare events, the allocation of wild-type Env homotrimers to individual HIV-1 virions in such a viral stock should follow a Poisson distribution (94). Assuming that each virion in a typical HIV-1 stock carries 10 Env spikes, a value derived by empirical observations (15, 61, 127, 128), the expected frequencies of viruses carrying 0, 1, and 2 or more wild-type homotrimers were calculated according to the Poisson distribution. Early electron microscopic observations of the structure of the viral matrix in immature HIV-1 virions suggested that immature HIV-1 virions might have as many as 72 Env spikes (40). This level of Env content is consistent with observations of virions of several simian immunodeficiency virus strains, a lentivirus closely related to HIV-1 (127, 128), but is significantly higher than the estimation derived from recent biochemical and structural studies of mature HIV-1 virions (15, 61, 127, 128). To cover the entire theoretical range of possibilities, we estimated the frequencies of the viruses with a single wild-type homotrimer in viral stocks with an fM of 0.9, assuming that each virion carries either 10 or 72 Env spikes. When fM is 0.9 and virions carry an average of 10 trimers, viruses with a single wild-type Env homotrimer constitute the dominant population (>99%) among viruses with at least one wild-type homotrimer. Such dominance is still predicted even with the unlikely and conservative assumption that all HIV-1 virions carry 72 spikes; in this scenario, 96.4% of all viruses with at least one wild-type homotrimer contain only a single wild-type homotrimer. If HIV-1 virions carrying a single wild-type Env homotrimer are infectious, the infectivity of a viral stock with an fM value of 0.9 should approach 0.1% of that of the wild-type viruses. In the event that virions bearing a single wild-type homotrimer are not infectious, the infectivity of a viral stock with an fM of 0.9 should be less than or equal to 0.1% × (100% − 99%), or 0.001% of that of the wild-type virus, assuming 10 spikes per virion. If we assume 72 spikes per virion, the 96.4% of virions bearing a single wild-type homotrimer should carry 93.1% of the total wild-type homotrimers in the viral stock with an fM of 0.9; in this case, the expected infectivity of a viral stock with an fM of 0.9 could not exceed 0.1% × (100% − 93.1%), or 0.0069% of that of the wild-type virus. Thus, under these conditions, at least 15- to 100-fold differences in infectivity can be expected depending on the ability of HIV-1 particles with a single wild-type Env homotrimer to infect cells.
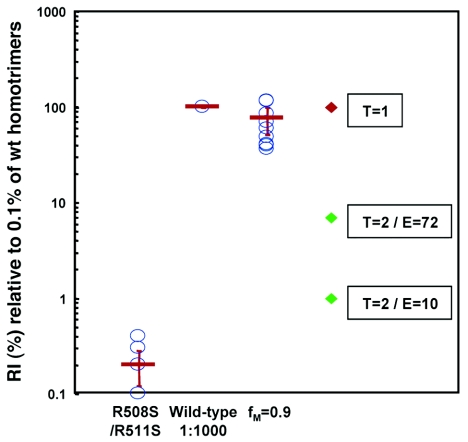
Viral stocks produced with mixed wild-type and dominant-negative mutant Envs with an fM of 0.9 are expected to contain a large proportion (99.9%) of nonfunctional Env proteins. Because the R508S/R511S mutant used in this study is still able to bind the CD4 and CCR5 receptors, these nonfunctional Envs can compete with the wild-type homotrimers for receptors on the target cells (103). This competition could complicate a comparison of the infectivity of wild-type viruses and viruses with mixtures of wild-type and R508S/R511S glycoproteins, particularly at high fM values. By using the Cf2Th-CD4/CCR5 target cells, which express very high levels of CD4 and CCR5 proteins, the functional consequences of receptor competition should be minimized. Nonetheless, to ensure that this potential factor was compensated for, the infectivity of the viruses with mixtures of wild-type and dominant-negative Envs was compared with the infectivity of mixtures of the wild-type viruses and the R508S/R511S viruses at a 1:1,000 ratio. This mixture of two different viral stocks should contain similar overall levels of functional as well as nonfunctional Envs as the viral stock with mixed wild-type and R508S/R511S Envs with an fM of 0.9. In 11 independent assays using four sets of independently prepared virus stocks, the infectivity of the virus stock with an fM of 0.9 was on average 75.6% (range, 36.1% to 149.6%; 95% CI, 51.1 to 100%) of that of the wild-type viruses diluted 1:1,000 with viruses bearing only the R508S/R511S Env (Fig. 2). The maximum potential values of infectivity predicted if two trimers on a virion were required for infection, assuming an average of either 10 or 72 spikes per virion, are also shown in Fig. 2. Models in which more than two Env trimers are required for viral infection predict even lower infectivities for the viral stocks with fM values of 0.9 than the model in which T is 2 (data not shown). Thus, the infectivity of the virus stocks with fM values of 0.9 was 11- to 75-fold greater than the maximal value possible if viruses with a single wild-type Env homotrimer were not infectious. We conclude that HIV-1 virions with one wild-type Env homotrimer can infect cells.
FIG. 2.
Relative infectivity of viruses with a high percentage of a dominant-negative Env mutant. The infectivity of viruses with only the R508S/R511S Env was very close to the background of the assays. Recombinant viruses with wild-type HIV-1YU2 Env were mixed at a 1:1,000 ratio with viruses bearing only the R508S/R511S mutant glycoproteins, and the luciferase activity of these viruses was assigned a relative infectivity of 100%. The infectivity of viruses with wild-type and R508S/R511S heterotrimers at an fM value of 0.9 was determined in parallel and was normalized to that of the above mixture of two virus stocks. The mean values for RI(%) observed in 11 independent measurements using four independently generated viral stocks with an fM value of 0.9 are shown as individual blue circles. A 1:1,000 dilution of the wild-type viral stock was included in each of the 11 individual experiments and served as the basis for normalization. Some of the 11 values observed for the R508S/R511S mutant were very close and thus appear as overlapping circles. The average of the 11 mean values and its 95% CI are also shown (red bars). The relative infectivity predicted by the Poisson distribution for a T value of 1 is 100%, as indicated. The relative infectivities predicted by the Poisson distribution for a T value of 2 are indicated, assuming that the number of Env trimers (E) on an average HIV-1 virion is either 10 or 72.
HIV-1 entry stoichiometry is independent of the gp41 cytoplasmic tail or the matrix protein.
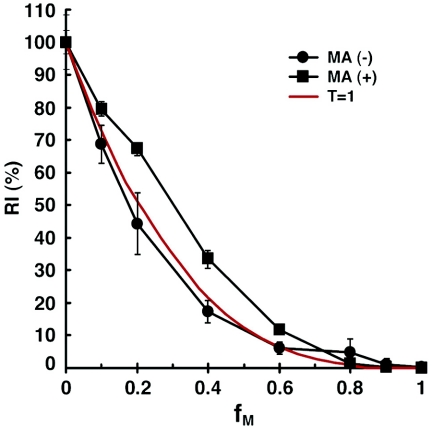
The cytoplasmic tail of HIV-1 gp41 is unusually long among retroviral transmembrane glycoproteins (59). This long cytoplasmic tail is accommodated by the viral matrix (MA) protein, which is associated with the inner surface of the viral membrane (36, 65). We investigated whether the gp41 cytoplasmic tail is important for allowing one Env trimer to achieve virus entry. Plasmids expressing cytoplasmic tail-deleted versions of the HIV-1YU2 wild-type and R508S/R511S mutant Envs were created by replacing the codon for tyrosine 712 with a stop codon. Recombinant reporter viruses with different ratios (fM values) of the cytoplasmic tail-deleted versions of the wild-type and the R508S/R511S mutant Envs were studied as described above. The relationship between fM and RI(%) of these viral stocks was close to the theoretical curve expected if the T value is 1 (Fig. 3). To examine the potential influence of the MA protein on HIV-1 entry stoichiometry, the MA coding sequence was deleted in the pCMVΔP1ΔenvpA packaging plasmid, as previously described (92). Because the MA-deleted Gag protein does not support efficient encapsidation of HIV-1 Envs with long cytoplasmic tails (36), the cytoplasmic tail-deleted versions of the wild-type and R508S/R511S mutant Envs were used to produce reporter viruses with the MA deletion. Viruses with deletions of the gp41 cytoplasmic tail and the MA protein were able to infect cells with an efficiency approximately 12-fold lower than that of the wild-type virus with only the gp41 cytoplasmic tail deletion (data not shown). Even with this reduced infectivity, the relationship between fM and RI(%) remained consistent with the theoretical curve when the T value is 1 (Fig. 3). Thus, the ability of HIV-1 to achieve entry using a single Env trimer is not dependent upon the matrix protein or the gp41 cytoplasmic tail.
FIG. 3.
Relative infectivities of HIV-1 variants lacking the gp41 cytoplasmic tail and/or the matrix protein. Recombinant HIV-1 expressing luciferase and containing either wild-type matrix [MA(+)] sequences or a matrix-deleted [MA(−)] mutant Gag protein was produced. The viruses containing versions of the wild-type and R508S/R511S HIV-l YU2 Envs with the cytoplasmic tail deleted were studied at the indicated fM values in the presence or absence of the MA proteins. The RI(%) values of the viruses were determined, relative to that of the corresponding MA(+) or MA(−) viruses containing cytoplasmic tail-deleted Envs with wild-type HIV-1YU2 ectodomains. The means and ranges of variation observed from eight parallel wells of infection are shown. The experiments were repeated at least once with consistent results. The theoretical curve predicted by the model in which T is 1 is depicted in red.
The entry stoichiometry of A-MLV Env.
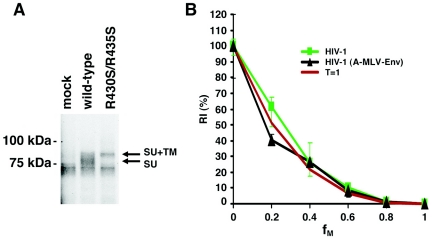
Our observation that a T = 1 stoichiometry of entry is associated with HIV-1 envelope glycoproteins with deleted cytoplasmic tails prompted an examination of the entry requirements associated with other viral envelope glycoproteins, all of which have shorter cytoplasmic segments than the HIV-1 gp41 glycoprotein. A-MLV enters target cells using a pH-independent pathway, similar to HIV-1. The main receptor for A-MLV Env is Pit2 which, like the HIV-1 coreceptors CXCR4 and CCR5, spans the membrane multiple times (16, 114). The A-MLV Envs, gp70 and p15E, are derived by proteolytic cleavage of the gp90 precursor (52, 83). As for HIV-1, this processing event is essential for infection by MLV. By analogy to the R508S/R511S cleavage-defective mutant of HIV-1, we made a mutant gp90 of A-MLV (strain 4070A) by replacing two arginine residues with serines in the cleavage recognition sequence. The resulting mutant, R430S/R435S, has the sequence … QLEQS430 TKYKS435 EPV…. The R430S/R435S mutant was expressed efficiently in transfected cells and was not processed into the mature gp70 glycoprotein, as determined by immunoprecipitation using a monoclonal antibody against gp70 (83A25; a gift from L. Evans of NIAID/NIH) (Fig. 4A).
FIG. 4.
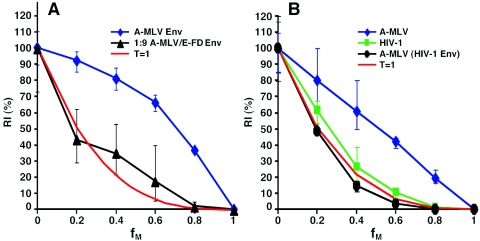
The entry stoichiometry of A-MLV Env. (A) The wild-type and R430S/R435S mutant A-MLV Envs were transiently expressed in 293T cells, which were radiolabeled with [35S]methionine/cysteine. The Envs in cell lysates were precipitated with 50 μl of culture medium containing the 83A25 anti-gp70 monoclonal antibody (a gift from L. Evans of NIAID/NIH). The mature SU glycoprotein and the gp90 precursor (SU+TM) are indicated. (B) The infectivities of reporter viruses containing different ratios (fM values) of the wild-type and R430S/R435S mutant A-MLV glycoproteins pseudotyped on HIV-1 virions are measured in Cf2Th cells, relative to that of a virus with the wild-type glycoproteins [HIV-1(A-MLV-Env)]. The RI(%) curve for HIV-1 with different ratios of wild-type and R508S/R511S Envs is shown for comparison (HIV-1). The means and ranges of variation from six parallel wells of infection are shown. The theoretical curve predicted by the model in which T is 1 is also shown in red.
To pseudotype the wild-type and R430S/R435S mutant A-MLV Envs onto HIV-1 virions, 3 μg of the pVPack-ampho vector was cotransfected with 1.5 μg of the pCMVΔP1ΔenvpA plasmid, 4.5 μg of the pHIV-1Luc vector, and 1 μg of a pc-Rev plasmid. When pseudotyped on HIV-1 reporter viruses, the R430S/R435S mutant exhibited at least a 3-log reduction in the ability to support virus infection, compared with the wild-type A-MLV Env (data not shown). A set of reporter viruses with different fM values was generated by cotransfecting different amounts of the wild-type and R430S/R435S mutant Envs with the above plasmids encoding the HIV-1 packaging system and vector. The RI(%) of each viral stock was determined as above for HIV-1 envelope glycoproteins. The relationship between the RI(%) and fM values for these hybrid viruses with A-MLV Env proteins was consistent with that expected when the T value is 1 and was indistinguishable from that of authentic HIV-1 virions (Fig. 4B). Thus, one Env trimer of A-MLV is capable of supporting virus entry.
The effect of high Env density and functional redundancy on the measurement of entry stoichiometry.
We also investigated the entry stoichiometry of authentic A-MLV virions using a similar approach. Reporter A-MLV viruses with different fM values were made by cotransfecting 293T cells with a total of 2 μg of plasmids expressing the wild-type and R430S/R435S Envs, 4 μg of the pVPack-GP packaging plasmid, and 4 μg of the pFB-luc vector plasmid (Stratagene). The pVPack-GP plasmid expresses the gag and pol gene products of Friend murine leukemia virus, and the pFB-luc plasmid contains a Friend murine leukemia virus vector encoding a firefly luciferase reporter gene. The relationship between fM and RI(%) was studied as described above. The infectivity of these viruses was measured on Cf2Th target cells. Interestingly, the RI(%) values were higher than those predicted by all of the theoretical models (Fig. 5A). Such a surprising result could not be explained by trivial reasons such as differences in the expression levels of the wild-type and R430S/R435S mutant Envs or the low efficiency of mixing between these two Env variants, because the same Env-expressing plasmids were used in the experiment above that yielded a T value of 1 (Fig. 4B).
FIG. 5.
The effect of Env density on the measurement of entry stoichiometry. (A) A-MLV reporter viruses with different fM values were created and their infectivity was measured in Cf2Th target cells in the presence of 1 μM polybrene (Sigma). The RI(%) curve is shown for viruses with different ratios of the wild-type and R430S/R435S mutant A-MLV Envs (A-MLV). The RI(%) curve is also shown for viruses made in cells transfected with 2.7 μg of a plasmid expressing a cleavage-defective mutant of E-MLV Env (E-FD Env) and 0.3 μg total plasmids expressing the wild-type and R430S/R435S mutant A-MLV Envs in different ratios (1:9 for A-MLV/E-FD Env). The theoretical curve predicted by a model in which T is 1 is shown in red. (B) The RI(%) curves for authentic HIV-1 and A-MLV viruses are shown in green and blue, respectively. The curve for A-MLV virions pseudotyped with HIV-1YU2 Env is in black. The means and ranges of variation observed from four parallel wells of infection are shown. The experiments were repeated at least once with consistent results.
One possible explanation for this phenotype is that the infectious units do not function independently in viral stocks of A-MLV, violating one of the model-building assumptions. If individual A-MLV virions carry multiple infectious units because of high Env content, reduction of the Env concentration on A-MLV virions should result in an alteration of the observed relationship between RI(%) and fM. To reduce the Env density on A-MLV virions, reporter viruses were generated by cotransfection of the packaging and vector plasmids with a total of 0.3 μg of the plasmids expressing the wild-type and R430S/R435S A-MLV Envs and 2.7 μg of a plasmid expressing a cleavage-defective mutant gp90 of the ecotropic MLV (E-MLV). The E-MLV Env does not bind the Pit2 receptor (117). The reporter virus stock produced by cotransfecting 2.7 μg of the E-MLV cleavage-defective mutant Env plasmid and 0.3 μg of the wild-type A-MLV Env plasmid had a similar level of infectivity to that produced by cotransfecting 2.7 μg of the pcDNA3.1(Zeo/−) vector DNA and 0.3 μg of the wild-type A-MLV Env plasmid. Conversely, the reporter virus stock produced by cotransfecting 2.7 μg of the A-MLV cleavage-defective mutant Env plasmid and 0.3 μg of the wild-type E-MLV Env plasmid exhibited a similar level of infectivity to that produced by cotransfecting 2.7 μg of the pcDNA3.1(Zeo/−) vector DNA and 0.3 μg of the wild-type E-MLV Env plasmid (data not shown). Thus, the E-MLV and A-MLV Envs apparently do not form heterotrimers when coexpressed. Because both the A-MLV and E-MLV Envs can be efficiently encapsidated on MLV virions, the mutant E-MLV Env trimers can theoretically compete with wild-type A-MLV Env trimers for incorporation into virions, resulting in a net reduction of the A-MLV Env trimer density on the reporter viruses. Viruses with different fM values were generated by cotransfecting 2.7 μg of the plasmid expressing the cleavage-defective E-MLV Env and 0.3 μg total of the plasmids expressing the wild-type and R430S/R435S mutant A-MLV Envs, varying the ratio of the A-MLV Env-expressing plasmids. In this context, the relationship between RI(%) and fM values for MLV virions was close to that expected if T is 1 (Fig. 5A).
Another plausible explanation of the observed RI(%)/fM relationship for authentic MLV virions is that some unique properties of MLV virions might influence our measurement of the entry stoichiometry of these viruses. Thus, we studied the entry stoichiometry of A-MLV in additional contexts in which the wild-type and R508S/R511S mutant HIV-1YU2 Envs were pseudotyped on A-MLV virions. To make reporter viruses with HIV-1 Envs and MLV virion components, reporter viruses were generated by cotransfecting 3 μg of the pVPack packaging plasmid, 3 μg of the pFB-luc genome vector, 3 μg of pSVIIIenv vector, and 1 μg of the pc-Tat plasmid. The pc-Tat construct expresses the HIV-1 Tat protein, which stimulates the expression of the HIV-1 Env protein from the pSVIIIenv vector. The infectivity of the resulting viruses was measured on Cf2Th-CCR5/CD4 cells. The RI(%)/fM curve of these chimeric viruses was consistent with that of the model in which the T value is 1 (Fig. 5B). These results indicate that the A-MLV Gag-Pol proteins can support virus entry with a T = 1 stoichiometry.
Taken together, these results indicate that A-MLV Envs can support virus entry using a T = 1 stoichiometry. The capability of individual Env trimers to support entry means that the large numbers of Env spikes on normal A-MLV virions probably function redundantly during the normal virus entry process.
The entry stoichiometry of ASLV-A Env.
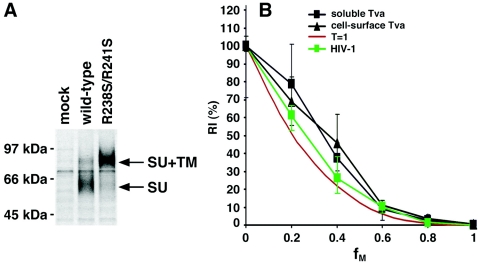
ASLV-A, unlike most retroviruses, uses a two-stage process to enter cells (29, 67, 79). In the first stage, ASLV-A Env binds in a pH-independent fashion to its sole receptor, Tva. Tva is a type I transmembrane protein with a single membrane-spanning domain (43, 67, 95). After viral attachment and endocytosis, the acidic pH environment in the endosome triggers the second stage, in which viral and cellular membranes are fused by Env. The ASLV-A Env is expressed as a gp85 precursor that is subsequently cleaved into SU and TM (75). The wild-type sequence for the cleavage site is…TGIR238RKR241STS… . To create a cleavage-defective ASLV-A Env mutant analogous to the R504S/R511S mutant of HIV-1 gp160, the two highlighted arginine residues were replaced with serines in the R238S/R241S construct. After transient transfection, the radiolabeled wild-type and R238S/R241S mutant ASLV-A Envs were immunoprecipitated from the cell lysates using a monoclonal antibody (8C5; a gift from C. Ochsenbauer-Jambor, University of Alabama at Birmingham). As expected, the R238S/R241S mutant was well expressed (Fig. 6A, SU+TM precursor) but was not cleaved to produce the mature SU product (Fig. 6A).
FIG. 6.
The entry stoichiometry of ASLV-A Env. (A) The wild-type and R238S/R241S mutant ASLV-A gp85s were transiently expressed in 293T cells. Radiolabeled ASLV-A Envs were immunoprecipitated from cell lysates using an anti-SU monoclonal clonal antibody and resolved by 10% SDS-PAGE under reducing conditions following previously reported procedures (85). The mature SU glycoprotein and the gp85 precursor (SU+TM) are indicated. (B) The infectivities of reporter viruses containing different ratios (fM values) of the wild-type and R238S/R241S mutant ASLV-A glycoproteins pseudotyped on HIV-1 virions were measured in Tva-expressing 293T cells, relative to that of a virus with the wild-type ASLV-A glycoproteins (cell surface Tva). The same viruses were used to infect 293T cells after incubation with 10 mM soluble Tva (soluble Tva). The means and ranges of variation from six parallel wells of infection are shown. The RI(%) curve for HIV-1 with different ratios of wild-type and R508S/R511S Envs is shown in green for comparison (HIV-1). The theoretical curve predicted by the model in which T is 1 is also shown in red.
To pseudotype the wild-type and R238S/R241S mutant ASLV-A Envs onto HIV-1 virions, 3 μg of the pCB6/WTA or pCB6/R238S/R241S plasmid was cotransfected with 1.5 μg of the pCMVΔP1ΔenvpA plasmid, 4.5 μg of the pHIV-1Luc vector, and 1 μg of a pc-Rev plasmid. The infectivity of these viruses was measured on 293T target cells that transiently expressed the Tva receptor after transfection. When pseudotyped onto HIV-1 virions, the R238S/R241S mutant exhibited an approximately 2-log reduction in the ability to support virus entry, compared with the wild-type ASLV-A Env (data not shown). A set of reporter viruses with different fM values was generated by cotransfecting different amounts of plasmids expressing the wild-type and R238S/R241S mutant Envs, and the RI(%) of each viral stock was determined as described above for HIV-1 envelope glycoproteins. The relationship between RI(%) and fM values for the chimeric viruses with ASLV-A Env proteins was close to that expected for an entry model in which T equals 1 (Fig. 6B). However, all of the viruses containing the R238S/R241S mutant Env glycoproteins demonstrated infectivities higher than those predicted by the model in which T equals 1. This likely resulted from the slight level of function (0.9% of the wild-type ASLV-A Env) retained by the R238S/R241S mutant. In comparison, the R508S/R511S mutant of the HIV-1YU2 gp160 glycoprotein exhibited approximately 7 logs less function than the wild-type HIV-1 Env glycoproteins.
The ectodomain of Tva is sufficient to support ASLV-A infection when either expressed on the cell surface as a fusion with other membrane proteins or provided as a soluble protein (48, 107, 108). This created an opportunity to determine whether a fixed position of the Tva receptor in the target cell membrane is important for a T = 1 entry stoichiometry. We conducted the measurement of the entry of HIV-1(ASLV-A Env) viruses into Tva-negative target cells in the presence of soluble Tva protein. The soluble Tva protein (sTva) contains the first 56 amino acids of the Tva ectodomain and was expressed in and purified from Escherichia coli (107). The viral stocks were first incubated with 10 nM sTva on ice for 30 min before being applied to 293T cells for infectivity measurements. The RI(%) curve produced with sTva was indistinguishable from that obtained when the Tva receptor was expressed on the cell surface (Fig. 6B). Thus, the association of the Tva receptor with the target membrane is not required for a T = 1 entry stoichiometry in this experimental setting, suggesting that a specific distance or position of the cellular receptor is not a determining factor of the T = 1 entry stoichiometry of ASLV-A.
Multiple HA trimers of influenza A virus are required for virus entry.
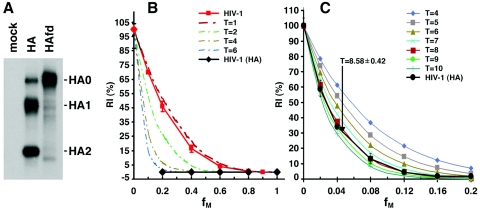
Influenza A viruses, which utilize a pH-dependent entry pathway, contain eight independent ribonucleoprotein complexes, thus complicating the development of a reporter virus through reverse genetics that can be conveniently adopted in our experimental system (82). Fortunately, influenza A virus HA can be readily pseudotyped on retroviral virions to allow precise measurement of HA-mediated virus entry (97). The proteolytic cleavage of the HA0 precursor into HA1 and HA2 subunits is essential for the entry of all influenza viruses. For most influenza A viruses, the freshly produced viral stocks are not infectious, and trypsin treatment is used to cleave and activate the HA proteins for viral infection in tissue-cultured cells (91). Interestingly, the HA proteins from avian influenza A viruses are naturally cleaved when produced in cultured cells; these viral stocks are readily infectious without trypsin treatment and thus are uniquely suited to our study design (115). The FPV/Rostock/34 strain of influenza A virus encodes an HA0 precursor with the sequence…PEPSKKREKR*LFG… where cleavage occurs after the indicated arginine residue (indicated by italics and an asterisk). Insertion of a glycine after this arginine residue completely inhibits HA0 cleavage, resulting in the HAfd mutant (115). HAfd is expressed at a level similar to the wild-type HA and is totally defective for function (115). The wild-type HA and HAfd were transiently expressed in 293T cells, radiolabeled, and precipitated by a polyclonal anti-HA serum. Our results verified that HAfd was expressed at a level similar to that of wild-type HA and was defective for proteolytic cleavage (Fig. 7A). HIV-1(HA) reporter viruses were created by cotransfecting the HA and HAfd expression plasmids with the pHIV-1Luc vector, the pCMVΔP1ΔenvpA packaging plasmid, and a plasmid expressing the HIV-1 Rev protein. The HIV-1(HA) reporter viruses were incubated with Cf2Th canine thymocytes, and luciferase activity was measured in the target cells. The reporter viruses with wild-type HA infected the Cf2Th cells efficiently, whereas the viruses with HAfd produced luciferase signals at least 10,000-fold lower and not significantly above those observed in the uninfected control cells (data not shown). Viruses with a range of fM values were produced by cotransfecting different ratios of the plasmids expressing HA and HAfd. The RI(%) values of these reporter viruses were determined, as described above. The relationship between RI(%) and fM for viruses with the influenza A virus HA proteins was clearly distinct from the curve observed for viruses with the HIV-1 Envs and from that predicted by the model in which T is 1 (Fig. 7B). The data points associated with the HIV-1(HA) viruses suggested a T value greater than 6. To estimate this T value more accurately, a set of reporter viruses with fM values between 0 and 0.2 was generated and studied. The observed relationship between RI(%) and fM for these viruses was close to the theoretical curves predicted by models in which the T value is 8 or 9 (Fig. 7C). A nonlinear regression analysis yielded a parametric estimation of T equals 8.58 with a 95% CI of 8.16 to 9.00. These results indicate that approximately eight to nine HA trimers are required to mediate virus entry in these circumstances. Because this was studied in the context of chimeric viruses containing influenza A virus HA and nonenvelope virion components of HIV-1, we can conclude that the HA glycoproteins alone dictate an entry stoichiometry different from that of authentic HIV-1.
FIG. 7.
The stoichiometry of virus entry mediated by the HA proteins of influenza A virus. (A) The wild-type HA and HAfd mutant of influenza A virus (FPV/Rostock/34) were transiently expressed in 293T cells and immunoprecipitated by 2 μl of a rabbit anti-HA serum (a gift from R. Wagner and H. Klenk, Marburgh University, Germany). The glycoproteins were resolved by 14% SDS-PAGE under reducing conditions. The HA0 precursor as well as mature HA1 and HA2 glycoproteins are indicated. (B) The infectivities of reporter viruses containing different ratios (fM values) of the HA and HAfd glycoproteins in Cf2Th cells, relative to that of a virus with wild-type HA proteins, are shown [HIV-1(HA)]. The RI(%) curve for HIV-1 with different ratios of wild-type and R508S/R511S Envs is shown in orange for comparison. The means and ranges of variation from eight parallel wells of infection are shown. The theoretical curves predicted by models in which the T values are 1, 2, 4, and 6 are shown. (C) HIV-1(HA) reporter viruses were generated and studied at very low fM values between 0 and 0.2. The data from six parallel wells of infection are presented and were used to parametrically estimate the T value using a nonlinear regression. The theoretical curves predicted by models in which T has a value from 4 to 10 are shown. The estimation of T is 8.58 with a 95% CI of 8.16 to 9.0.
DISCUSSION
We examined the stoichiometry of HIV-1 entry. Our data indicate that individual HIV-1 Env trimers can function independently to achieve HIV-1 infection without apparent cooperativity; i.e., that T is 1. This result is surprising because conventional models, largely derived from the well-characterized influenza virus system, require cooperativity among multiple envelope spikes for membrane fusion (3, 4, 6, 20, 32, 45, 53, 74, 91). Our conclusion is supported by the results from the study of dominant-negative mutants of Env proteins from two different HIV-1 strains. These mutants are very different in nature: one disrupts the proteolytic cleavage of the HIV-1 Env precursor, and the second has a dysfunctional fusion peptide. The ability of our experimental system to document distinct stoichiometric requirements for virus entry was demonstrated by our results with HIV-1 virions bearing HIV-1 Env proteins (T value of 1) or the influenza A virus HA proteins (T value of 8 or 9). Moreover, we demonstrated that HIV-1 virions carrying a single wild-type homotrimer are infectious, providing qualitative support for our conclusion. Importantly, our conclusion that the value of T is 1 for HIV-1 is supported by results using an independent approach suggesting that the target of anti-HIV-1 neutralizing antibodies is composed of a single Env trimer (123). The target size for neutralizing antibodies is expected to be close or equivalent to the size of an independent infectious unit that was measured in this study. Collectively, our results indicate that for HIV-1 glycoproteins, a single Env trimer is sufficient for membrane fusion and virus entry.
Our conclusion that the value of T is 1 for HIV-1 derives from fitting the experimental measurements to discrete models built on theoretical grounds. We realize that the strength of our conclusions is dependent on the validity of the assumptions used for model building. (i) The wild-type and dominant-negative Envs are expressed in the same cells after cotransfection, the relative levels of the paired Env proteins are proportional to the ratios of the transfected plasmids, and the monomers of the paired Env proteins form trimers by random association. (ii) Each HIV-1 virion particle contains no more than one infectious unit; in other words, the infectious units in HIV-1 viral stocks function independently. (iii) All heterotrimers containing dominant-negative mutant subunits are totally nonfunctional.
The first two assumptions, i.e., random mixing and independence of infectious units, were utilized to build theoretical models in this study and in our study of the neutralization stoichiometry of HIV-1 (123). Because both approaches utilized recombinant HIV-1 produced in the identical system, any violation of these assumptions must affect the construction of theoretical models in both approaches and must be compatible with the data and deductions derived from both approaches. Each of our assumptions is discussed in turn.
(i) Random mixing of two species of Env monomers.
Random mixing of two species of Env monomers is assumed in all of the experiments reported herein. If some of the wild-type and dominant-negative Envs are not expressed in the same cells after cotransfection or if the relative levels of the paired Env proteins are not proportional to the ratios of the transfected plasmids, the functional consequences to model fitting would be equivalent to that of nonrandomness in mixing of the two species of Env monomers. In our system for production of recombinant viruses, only cells that have been transfected with the pCMVΔP1ΔenvpA and pHIV-1Luc plasmids can produce viruses that deliver the luciferase gene and are consequently detected. In a system in which greater than 90% of the cells are successfully transfected, the frequency of cells that take up these two plasmids but only one of the two Env-expressing plasmids is expected to be very low. The relative expression of the wild-type and mutant Env proteins is likewise expected to parallel the levels of the Env-expressing plasmids, as the plasmids are identical except for the introduced mutations, and the steady-state levels of the individually expressed Env proteins are comparable. The largely defective nature of HIV-1 virions precludes any attempt to rule out nonrandom mixing of wild-type and mutant Env proteins by biochemical approaches, which have been successfully applied to influenza A HA protein variants (103). However, we can estimate the degree to which nonrandom mixing of HIV-1 Env glycoproteins needs to have occurred to produce the data presented in Fig. 2 if, in fact, T is not equal to 1. By calculating the potential number of the wild-type homotrimers on average virions in viral stocks with an fM of 0.9 according to the Poisson distribution, the levels of nonrandom mixing between two species of Env monomers had to reach 15.7% and 40.4% in order to produce the observed RI(%) values for models in which the value of T is 2 or 3, respectively. Attempts to retrofit the models in which T is 2 or 3 with these levels of nonrandom mixing to the data in Fig. 1B resulted in significant deviation of the observed results from either model. Further, we applied a similar retrofitting analysis to our results from the antibody neutralization stoichiometry experiments; this analysis showed that 40.4% nonrandom mixing did not conform to the observed results, and 15.7% nonrandom mixing was compatible with only a minority of the data. Thus, systematic nonrandom mixing of two different species of Env monomers when the T value is greater than 1 would not explain the majority of the data generated in the two experimental approaches employed to estimate T.
(ii) Independence of infectious units in HIV-1 viral stocks.
It is well established that in HIV-1 stocks, the majority of virions (>99.9%) are not infectious (8, 61). Pseudotyping HIV-1 particles with heterologous viral Env proteins, such as the vesicular stomatitis virus G glycoprotein, indicates that at least 2 logs of this defectiveness can be attributed to the HIV-1 Env glycoproteins (X. Yang, unpublished observations). Thus, if envelope glycoproteins are packed on virions randomly, the Poisson distribution dictates that HIV-1 particles with more than one infectious unit must be very rare. The presence of redundant infectious units on virions would result in an underestimation of the actual T value, in both this study and in the analysis of T using antibody neutralization (123). This point is vividly illustrated by our results with authentic A-MLV viruses and pseudotyped viruses presented in Fig. 4B and 5A and B. By contrast, in our study of neutralizing antibody stoichiometry (123), the presence of multiple infectious units on HIV-1 virions would result in an overestimation of N, the number of antibody molecules required to inactivate the HIV-1 Env trimer. As the data in that study suggest that N equals 1, which cannot be an overestimate (i.e., N cannot be 0), serious deviations from independently functioning infectious units cannot be occurring in the recombinant virions produced in this system.
(iii) Lack of function of heterotrimers containing dominant-negative mutant subunit(s).
Our study of the stoichiometry of antibody-mediated neutralization, which also yielded a T value close to 1 (123), did not involve the use of mutants that are defective in entry function. Therefore, the assumption that Env heterotrimers containing one or more dominant-negative mutants are nonfunctional pertains only to this study. Violation of this assumption could result in an underestimation of T. However, by using two types of dominant-negative HIV-1 Env mutants that are structurally distinct and thus should exert their dominant-negative effects through different mechanisms, we increase our level of confidence in making this assumption. Moreover, analogous mutants of A-MLV and ASLV Envs produced interpretable data, also supporting the model in which the T value is 1. Finally, our current conceptions of HIV-1 Env structure and function are incompatible with the possibility that heterotrimers containing one or more uncleaved gp160 precursor molecules are functional (123).
Most of the experiments in this study require no assumptions about the number of Env spikes on HIV-1 virions. Indeed, when the amounts of transfected Env-expressing plasmids were titrated downwards (1, 2, and 3 μg total), experiments similar to those shown in Fig. 1B yielded identical results (data not shown). Thus, although decreasing the level of Env proteins on the HIV-1 virions decreases the infectivity of the viruses (38), this does not alter the T = 1 entry stoichiometry. Only in the analysis presented in Fig. 2 are assumptions made about the number of Env trimers on HIV-1 virions. Here we employ numbers derived from both biochemical measurements (14, 59) and morphological studies (125, 126), which suggest that HIV-1 virions carry an average of 10 Env trimers per particle. These virions were derived from both lymphocyte cell lines chronically infected by HIV-1 (14, 59, 126) and from transiently transfected 293T cells (125) and thus are relevant to virions produced in our experimental system. We also considered the possibility that HIV-1 virions contain as many as 72 Env trimers, but this did not alter the validity of the conclusions of the experiment shown in Fig. 2.
In summary, even an unlikely, systematic violation of the assumptions that we used to construct the candidate models would not alter the validity of our conclusion that a small number of Env trimers can mediate retroviral infection. Even allowing a significant degree of deviation from these assumptions, the observed data in toto would accommodate a T value that is 1 or, at most, approaches 2 for the retroviral envelope glycoproteins. The relatively low spike density on some retroviruses and the immobility of Env spikes on retroviral virions due to the interaction with the matrix are compatible with low but not high T values for entry. The use of a single HIV-1 Env trimer to achieve virus entry is consistent with previous estimates of four to six CCR5 receptors cooperating to support virus entry, considering that these receptors can dimerize and that three CCR5-binding moieties exist in one Env trimer (60, 73, 116). Using pseudotyped HIV-1 viruses, we found that eight to nine HA trimers of influenza A virus are required to achieve virus entry. This result is consistent with estimates of the number of HA trimers required for membrane fusion and pore formation derived from studies of cells and liposomes (3, 4, 6, 20, 32, 45, 72, 74).
Distinct patterns of transmission, strategies for spread within the host, and requirements for evasion of the host humoral immune response may have dictated the optimal value of T associated with different enveloped viruses. Viruses like influenza A virus cause short-lived infections that are controlled by strain-specific neutralizing antibodies elicited by the HA glycoproteins (42). Under these circumstances, a larger T value may be acceptable. For a virus like HIV-1 that needs to persist for many years in the presence of the host humoral immune system, the utilization of a single Env trimer to achieve entry is a major advantage (10, 62, 76). This property permits HIV-1 virions to carry few Env trimers while maintaining infectivity, thus minimizing Env exposure to the humoral immune system of the host. Furthermore, when the T value is 1, many of the Env spikes on a virion are dispensable for infectivity, allowing a high level of defectiveness. Functionally defective HIV-1 Env proteins, including shed gp120 and the gp41 remnants of subunit dissociation, have been suggested to act as decoy antigens that promote nonneutralizing antibody responses (57, 69, 87). Low requirements for viral glycoproteins on the retrovirus surface allow the envelope to be densely populated with cellular adhesion molecules, which can facilitate virus-cell attachment in a process that is invisible to the host immune system (34, 71, 93, 98). Finally, for a virus in which T is 1, essentially every functional Env trimer must be incapacitated by bound antibodies for neutralization of the virus to be achieved, maximizing the chances of viral escape from antibody responses. Persistence in vivo is common in retroviruses, and the shared T = 1 entry stoichiometry among HIV-1, A-MLV, and ASLV-A attest to the biological importance of using only one or very few trimers for infection by these viruses.
Differences in the T values associated with virus entry may require distinct structural features in the viruses and/or receptors. Although influenza A viruses and HIV-1 are of similar size, the influenza A virion carries about 200 to 300 HA trimers, in striking contrast to the 7 to 14 Env spikes per virion for HIV-1 (15, 41, 61, 113, 122, 127, 128). The high density of HA spikes on influenza virions may be essential for the cooperativity that is needed for successful membrane fusion (56, 80). During the pH-dependent entry process utilized by influenza A virus, the simultaneous exposure of all HA trimers to acidic pH in the endosome facilitates the coordination of conformational changes among the Env spikes (4, 18, 66, 102, 109). By contrast, for the hybrid ASLV-A and pH-independent viruses such as HIV-1 and A-MLV, conformational changes in the Env complex are set in motion by receptor engagement (86). For these viruses, factors such as the density, affinity, mobility, and configuration of the Env spikes and receptors could limit the coordinated activation of multiple trimers. The ability of a single Env trimer to mediate entry obviates the need to coordinate temporally and spatially the binding of receptors and activation of many trimers. A low T value may thus be preferentially associated with the entry of viruses that depend on receptor engagement to trigger fusion-related conformational changes. Of note, it has been reported that multiple trimers of the paramyxovirus F fusion glycoprotein are required to mediate cell-to-cell fusion, although paramyxoviruses enter cells in a pH-independent manner (27). Therefore, in the future, it would be of interest to test how many paramyxovirus F fusion glycoprotein trimers are needed to mediate the virus entry process.
The membrane fusion process is not only used for viral entry but is also employed for important cellular functions such as subcellular organelle trafficking, exo- and endocytosis, and fertilization of eggs (51, 109). Current models of membrane fusion propose that the cooperative function of several membrane-fusing complexes is necessary (4, 18). Our conclusion that the value of T is 1 for the entry of three retroviruses raises the question of how a single Env trimer could mediate membrane fusion and allow access of the viral capsid into the host cell cytoplasm. The different stoichiometries of virus entry mediated by different Env proteins that are pseudotyped on HIV-1 virions clearly demonstrate that the viral glycoproteins specify the entry stoichiometry. Moreover, the unusually long cytoplasmic tail of the HIV-1 transmembrane glycoprotein and its interaction with the viral matrix protein do not influence the T = 1 entry stoichiometry. Both HIV-1 and A-MLV virions could support viral entry with a T value of 1 under specific experimental conditions, despite the structural differences between the virions of these two viruses (39). Therefore, the ectodomains and membrane-spanning anchors of the HIV-1 and A-MLV Env glycoproteins and/or their cellular receptors determine the ability of these viruses to use low numbers of spikes to achieve infection.
Noteworthily, the three viruses that exhibit a T = 1 entry stoichiometry utilize protein receptors (86). As discussed above, receptor binding results in conformational changes in the retroviral Env trimer that drive the membrane fusion process. This situation differs markedly from that of influenza A virus, in which the binding of the HA1 glycoprotein to cell surface sialic acid serves mostly to attach the virus and allow targeting to the endosome (18, 86, 104). The receptors utilized by the three retroviruses that exhibit a T = 1 entry stoichiometry are structurally quite distinct. Although the receptors for HIV-1 and A-MLV, CCR5/CXCR4 and Pit2, respectively, have multiple membrane-spanning regions, the ASLV-A receptor, Tva, has only a single transmembrane domain (86). Thus, the T = 1 entry stoichiometry does not rely on a specific type of protein receptor. Furthermore, entry mediated by the ASLV-A Env glycoproteins still exhibits a T = 1 stoichiometry when supported by soluble Tva, suggesting that a specific spatial position and distance of the receptor/Env complex relative to the target membrane prior to the actual fusion reaction are not necessary for this stoichiometry. The main determinant for the T = 1 entry stoichiometry may be the structural and functional properties of the Env ectodomains of these viruses. Future studies will be needed to understand the mechanism whereby a single Env trimer promotes membrane fusion and virus infection.
Acknowledgments
We thank Y. McLaughlin and S. Farnum for manuscript preparation; and R. Wagner, H. Klenk, C. Jambor, and L. Evans for antibodies; J. Cunningham for soluble Tva protein; and P. Cannon, S. Delos, and J. White for plasmid constructs.
This work was supported by grants from NIH (AI24755, AI31783, AI46725, and a Center for AIDS Research award), the International AIDS Vaccine Initiative, the Bristol-Myers Squibb Foundation, the William A. Haseltine Foundation for the Arts and Sciences, and a gift from William F. McCarty-Cooper.
REFERENCES
- 1.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 2.Bentz, J. 2000. Membrane fusion mediated by coiled coils: a hypothesis. Biophys. J. 78:886-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentz, J. 2000. Minimal aggregate size and minimal fusion unit for the first fusion pore of influenza hemagglutinin-mediated membrane fusion. Biophys. J. 78:227-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentz, J., and A. Mittal. 2003. Architecture of the influenza hemagglutinin membrane fusion site. Biochim. Biophys. Acta 1614:24-35. [DOI] [PubMed] [Google Scholar]
- 5.Bleul, C. C., M. Farzan, H. Choe, C. Parolin, I. Clark-Lewis, J. Sodroski, and T. A. Springer. 1996. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature 382:829-833. [DOI] [PubMed] [Google Scholar]
- 6.Blumenthal, R., D. P. Sarkar, S. Durell, D. E. Howard, and S. J. Morris. 1996. Dilation of the influenza hemagglutinin fusion pore revealed by the kinetics of individual cell-cell fusion events. J. Cell Biol. 135:63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulay, F., R. W. Doms, R. G. Webster, and A. Helenius. 1988. Posttranslational oligomerization and cooperative acid activation of fixed influenza hemagglutinin trimers. J. Cell Biol. 106:629-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourinbaiar, A. S. 1994. The ratio of defective HIV-1 particles to replication-competent infectious virions. Acta Virol. 38:59-61. [PubMed] [Google Scholar]
- 9.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 10.Bukrinskaya, A. G. 2004. HIV-1 assembly and maturation. Arch. Virol. 149:1067-1082. [DOI] [PubMed] [Google Scholar]
- 11.Cao, J., B. Vasir, and J. G. Sodroski. 1994. Changes in the cytopathic effects of human immunodeficiency virus type 1 associated with a single amino acid alteration in the ectodomain of the gp41 transmembrane glycoprotein. J. Virol. 68:4662-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 13.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 14.Chernomordik, L. V., V. A. Frolov, E. Leikina, P. Bronk, and J. Zimmerberg. 1998. The pathway of membrane fusion catalyzed by influenza hemagglutinin: restriction of lipids, hemifusion, and lipidic fusion pore formation. J. Cell Biol. 140:1369-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chertova, E., J. W. Bess Jr., Jr., B. J. Crise, I. R. Sowder, T. M. Schaden, J. M. Hilburn, J. A. Hoxie, R. E. Benveniste, J. D. Lifson, L. E. Henderson, and L. O. Arthur. 2002. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 76:5315-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chien, M. L., J. L. Foster, J. L. Douglas, and J. V. Garcia. 1997. The amphotropic murine leukemia virus receptor gene encodes a 71-kilodalton protein that is induced by phosphate depletion. J. Virol. 71:4564-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 18.Colman, P. M., and M. C. Lawrence. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 4:309-319. [DOI] [PubMed] [Google Scholar]
- 19.Dalgleish, A. G., P. C. Beverley, P. R. Clapham, D. H. Crawford, M. F. Greaves, and R. A. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763-767. [DOI] [PubMed] [Google Scholar]
- 20.Danieli, T., S. L. Pelletier, Y. I. Henis, and J. M. White. 1996. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J. Cell Biol. 133:559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 22.Dimitrov, D. S. 2004. Virus entry: molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2:109-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doms, R. W., P. L. Earl, S. Chakrabarti, and B. Moss. 1990. Human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus env proteins possess a functionally conserved assembly domain. J. Virol. 64:3537-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doms, R. W., and A. Helenius. 1986. Quaternary structure of influenza virus hemagglutinin after acid treatment. J. Virol. 60:833-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doranz, B. J., J. Rucker, Y. Yi, R. J. Smyth, M. Samson, S. C. Peiper, M. Parmentier, R. G. Collman, and R. W. Doms. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149-1158. [DOI] [PubMed] [Google Scholar]
- 26.Dubay, J. W., S. R. Dubay, H. J. Shin, and E. Hunter. 1995. Analysis of the cleavage site of the human immunodeficiency virus type 1 glycoprotein: requirement of precursor cleavage for glycoprotein incorporation. J. Virol. 69:4675-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutch, R. E., S. B. Joshi, and R. A. Lamb. 1998. Membrane fusion promoted by increasing surface densities of the paramyxovirus F and HN proteins: comparison of fusion reactions mediated by simian virus 5 F, human parainfluenza virus type 3 F, and influenza virus HA. J. Virol. 72:7745-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Earl, P. L., B. Moss, and R. W. Doms. 1991. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J. Virol. 65:2047-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Earp, L. J., S. E. Delos, R. C. Netter, P. Bates, and J. M. White. 2003. The avian retrovirus avian sarcoma/leukosis virus subtype A reaches the lipid mixing stage of fusion at neutral pH. J. Virol. 77:3058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Earp, L. J., S. E. Delos, H. E. Park, and J. M. White. 2005. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 285:25-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 32.Ellens, H., J. Bentz, D. Mason, F. Zhang, and J. M. White. 1990. Fusion of influenza hemagglutinin-expressing fibroblasts with glycophorin-bearing liposomes: role of hemagglutinin surface density. Biochemistry 29:9697-9707. [DOI] [PubMed] [Google Scholar]
- 33.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 34.Fortin, J. F., R. Cantin, G. Lamontagne, and M. Tremblay. 1997. Host-derived ICAM-1 glycoproteins incorporated on human immunodeficiency virus type 1 are biologically active and enhance viral infectivity. J. Virol. 71:3588-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freed, E. O., E. L. Delwart, G. L. Buchschacher, Jr., and A. T. Panganiban. 1992. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc. Natl. Acad. Sci. USA 89:70-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freed, E. O., and M. A. Martin. 1995. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J. Virol. 69:1984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freed, E. O., D. J. Myers, and R. Risser. 1989. Mutational analysis of the cleavage sequence of the human immunodeficiency virus type 1 envelope glycoprotein precursor gp160. J. Virol. 63:4670-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gabuzda, D. H., A. Lever, E. Terwilliger, and J. Sodroski. 1992. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 66:3306-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gelderblom, H. R. 1991. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS 5:617-637. [PubMed] [Google Scholar]
- 40.Gelderblom, H. R., E. H. Hausmann, M. Ozel, G. Pauli, and M. A. Koch. 1987. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology 156:171-176. [DOI] [PubMed] [Google Scholar]
- 41.Gentile, M., T. Adrian, A. Scheidler, M. Ewald, F. Dianzani, G. Pauli, and H. R. Gelderblom. 1994. Determination of the size of HIV using adenovirus type 2 as an internal length marker. J. Virol. Methods 48:43-52. [DOI] [PubMed] [Google Scholar]
- 42.Gerhard, W. 2001. The role of the antibody response in influenza virus infection. Curr. Top. Microbiol. Immunol. 260:171-190. [DOI] [PubMed] [Google Scholar]
- 43.Gilbert, J. M., L. D. Hernandez, J. W. Balliet, P. Bates, and J. M. White. 1995. Receptor-induced conformational changes in the subgroup A avian leukosis and sarcoma virus envelope glycoprotein. J. Virol. 69:7410-7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graham, B. S., J. M. Rowland, A. Modliszewski, and D. C. Montefiori. 1990. Antifusion activity in sera from persons infected with human immunodeficiency virus type 1. J. Clin. Microbiol. 28:2608-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gunther-Ausborn, S., P. Schoen, I. Bartoldus, J. Wilschut, and T. Stegmann. 2000. Role of hemagglutinin surface density in the initial stages of influenza virus fusion: lack of evidence for cooperativity. J. Virol. 74:2714-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo, H. G., F. M. Veronese, E. Tschachler, R. Pal, V. S. Kalyanaraman, R. C. Gallo, and M. S. Reitz, Jr. 1990. Characterization of an HIV-1 point mutant blocked in envelope glycoprotein cleavage. Virology 174:217-224. [DOI] [PubMed] [Google Scholar]
- 47.Hart, T. K., A. M. Klinkner, J. Ventre, and P. J. Bugelski. 1993. Morphometric analysis of envelope glycoprotein gp120 distribution on HIV-1 virions. J. Histochem. Cytochem. 41:265-271. [DOI] [PubMed] [Google Scholar]
- 48.Hernandez, L. D., R. J. Peters, S. E. Delos, J. A. Young, D. A. Agard, and J. M. White. 1997. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J. Cell Biol. 139:1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herrera, C., C. Spenlehauer, M. S. Fung, D. R. Burton, S. Beddows, and J. P. Moore. 2003. Nonneutralizing antibodies to the CD4-binding site on the gp120 subunit of human immunodeficiency virus type 1 do not interfere with the activity of a neutralizing antibody against the same site. J. Virol. 77:1084-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hofmann, W., D. Schubert, J. LaBonte, L. Munson, S. Gibson, J. Scammell, P. Ferrigno, and J. Sodroski. 1999. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 73:10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jahn, R., T. Lang, and T. C. Sudhof. 2003. Membrane fusion. Cell 112:519-533. [DOI] [PubMed] [Google Scholar]
- 52.Kamps, C. A., Y. C. Lin, and P. K. Wong. 1991. Oligomerization and transport of the envelope protein of Moloney murine leukemia virus-TB and of ts1, a neurovirulent temperature-sensitive mutant of MoMuLV-TB. Virology 184:687-694. [DOI] [PubMed] [Google Scholar]
- 53.Kaplan, D., J. Zimmerberg, A. Puri, D. P. Sarkar, and R. Blumenthal. 1991. Single cell fusion events induced by influenza hemagglutinin: studies with rapid-flow, quantitative fluorescence microscopy. Exp. Cell Res. 195:137-144. [DOI] [PubMed] [Google Scholar]
- 54.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klatzmann, D., E. Champagne, S. Chamaret, J. Gruest, D. Guetard, T. Hercend, J. C. Gluckman, and L. Montagnier. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767-768. [DOI] [PubMed] [Google Scholar]
- 56.Kohama, M. T., J. M. Cardenas, and J. T. Seto. 1981. Immunoelectron microscopic study of the detection of the glycoproteins of influenza and Sendai viruses in infected cells by the immunoperoxidase method. J. Virol. Methods 3:293-301. [DOI] [PubMed] [Google Scholar]
- 57.Kohler, H., J. Goudsmit, and P. Nara. 1992. Clonal dominance: cause for a limited and failing immune response to HIV-1 infection and vaccination. J. Acquir. Immune Defic. Syndr. 5:1158-1168. [PubMed] [Google Scholar]
- 58.Korber, B. F., F. Kuiken, C. Pillai, S. Sodroksi, J. 1998. Numbering positions in HIV relative to HXBc2, p. II-A-54-II-A-69. In B. K. Korber, C. Foley, F. Hahn, B. McCutchan, F. Mellor, and J. Sodroski (ed.), Human retroviruses and AIDS. Los Alamos National Laboratories, Los Alamos, N. Mex.
- 59.Korber, B. T., S. Osmanov, J. Esparza, and G. Myers. 1994. The World Health Organization Global Programme on AIDS proposal for standardization of HIV sequence nomenclature. W.H.O. Network for HIV Isolation and Characterization. AIDS Res. Hum. Retrovir. 10:1355-1358. [DOI] [PubMed] [Google Scholar]
- 60.Kuhmann, S. E., E. J. Platt, S. L. Kozak, and D. Kabat. 2000. Cooperativity of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J. Virol. 74:7005-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Layne, S. P., M. J. Merges, M. Dembo, J. L. Spouge, S. R. Conley, J. P. Moore, J. L. Raina, H. Renz, H. R. Gelderblom, and P. L. Nara. 1992. Factors underlying spontaneous inactivation and susceptibility to neutralization of human immunodeficiency virus. Virology 189:695-714. [DOI] [PubMed] [Google Scholar]
- 62.Levy, J. A. 1988. The human immunodeficiency virus and its pathogenesis. Infect. Dis. Clin. N. Am. 2:285-297. [PubMed] [Google Scholar]
- 63.Lin, A. H., and P. M. Cannon. 2002. Use of pseudotyped retroviral vectors to analyze the receptor-binding pocket of hemagglutinin from a pathogenic avian influenza A virus (H7 subtype). Virus Res. 83:43-56. [DOI] [PubMed] [Google Scholar]
- 64.Maerz, A. L., R. J. Center, B. E. Kemp, B. Kobe, and P. Poumbourios. 2000. Functional implications of the human T-lymphotropic virus type 1 transmembrane glycoprotein helical hairpin structure. J. Virol. 74:6614-6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mammano, F., F. Salvatori, S. Indraccolo, A. De Rossi, L. Chieco-Bianchi, and H. G. Gottlinger. 1997. Truncation of the human immunodeficiency virus type 1 envelope glycoprotein allows efficient pseudotyping of Moloney murine leukemia virus particles and gene transfer into CD4+ cells. J. Virol. 71:3341-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Markovic, I., E. Leikina, M. Zhukovsky, J. Zimmerberg, and L. V. Chernomordik. 2001. Synchronized activation and refolding of influenza hemagglutinin in multimeric fusion machines. J. Cell Biol. 155:833-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsuyama, S., S. E. Delos, and J. M. White. 2004. Sequential roles of receptor binding and low pH in forming prehairpin and hairpin conformations of a retroviral envelope glycoprotein. J. Virol. 78:8201-8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCune, J. M., L. B. Rabin, M. B. Feinberg, M. Lieberman, J. C. Kosek, G. R. Reyes, and I. L. Weissman. 1988. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell 53:55-67. [DOI] [PubMed] [Google Scholar]
- 69.McDougall, B., M. H. Nymark, G. Landucci, D. Forthal, and W. E. Robinson, Jr. 1997. Predominance of detrimental humoral immune responses to HIV-1 in AIDS patients with CD4 lymphocyte counts less than 400/mm3. Scand. J. Immunol. 45:103-111. [DOI] [PubMed] [Google Scholar]
- 70.McKeating, J. A., A. McKnight, and J. P. Moore. 1991. Differential loss of envelope glycoprotein gp120 from virions of human immunodeficiency virus type 1 isolates: effects on infectivity and neutralization. J. Virol. 65:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meerloo, T., M. A. Sheikh, A. C. Bloem, A. de Ronde, M. Schutten, C. A. van Els, P. J. Roholl, P. Joling, J. Goudsmit, and H. J. Schuurman. 1993. Host cell membrane proteins on human immunodeficiency virus type 1 after in vitro infection of H9 cells and blood mononuclear cells. An immuno-electron microscopic study. J. Gen. Virol. 74:129-135. [DOI] [PubMed] [Google Scholar]
- 72.Melikyan, G. B., W. D. Niles, and F. S. Cohen. 1995. The fusion kinetics of influenza hemagglutinin expressing cells to planar bilayer membranes is affected by HA density and host cell surface. J. Gen. Physiol. 106:783-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mellado, M., J. M. Rodriguez-Frade, S. Manes, and A. C. Martinez. 2001. Chemokine signaling and functional responses: the role of receptor dimerization and TK pathway activation. Annu. Rev. Immunol. 19:397-421. [DOI] [PubMed] [Google Scholar]
- 74.Mittal, A., T. Shangguan, and J. Bentz. 2002. Measuring pKa of activation and pKi of inactivation for influenza hemagglutinin from kinetics of membrane fusion of virions and of HA expressing cells. Biophys. J. 83:2652-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moelling, K., and M. Hayami. 1977. Analysis of precursors to the envelope glycoproteins of avian RNA tumor viruses in chicken and quail cells. J. Virol. 22:598-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moore, J., and A. Trkola. 1997. HIV type 1 coreceptors, neutralization serotypes, and vaccine development. AIDS Res. Hum. Retrovir. 13:733-736. [DOI] [PubMed] [Google Scholar]
- 77.Moore, J. P., J. A. McKeating, R. A. Weiss, and Q. J. Sattentau. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139-1142. [DOI] [PubMed] [Google Scholar]
- 78.Morrison, T. G. 2003. Structure and function of a paramyxovirus fusion protein. Biochim. Biophys. Acta 1614:73-84. [DOI] [PubMed] [Google Scholar]
- 79.Mothes, W., A. L. Boerger, S. Narayan, J. M. Cunningham, and J. A. Young. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103:679-689. [DOI] [PubMed] [Google Scholar]
- 80.Murti, K. G., and R. G. Webster. 1986. Distribution of hemagglutinin and neuraminidase on influenza virions as revealed by immunoelectron microscopy. Virology 149:36-43. [DOI] [PubMed] [Google Scholar]
- 81.Nara, P. L., and P. J. Fischinger. 1988. Quantitative infectivity assay for HIV-1 and -2. Nature 332:469-470. [DOI] [PubMed] [Google Scholar]
- 82.Neumann, G., and Y. Kawaoka. 2004. Reverse genetics systems for the generation of segmented negative-sense RNA viruses entirely from cloned cDNA. Curr. Top. Microbiol. Immunol. 283:43-60. [DOI] [PubMed] [Google Scholar]
- 83.Ng, V. L., T. G. Wood, and R. B. Arlinghaus. 1982. Processing of the env gene products of Moloney murine leukaemia virus. J. Gen. Virol. 59:329-343. [DOI] [PubMed] [Google Scholar]
- 84.Oberlin, E., A. Amara, F. Bachelerie, C. Bessia, J. L. Virelizier, F. Arenzana-Seisdedos, O. Schwartz, J. M. Heard, I. Clark-Lewis, D. F. Legler, M. Loetscher, M. Baggiolini, and B. Moser. 1996. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature 382:833-835. [DOI] [PubMed] [Google Scholar]
- 85.Ochsenbauer-Jambor, C., S. E. Delos, M. A. Accavitti, J. M. White, and E. Hunter. 2002. Novel monoclonal antibody directed at the receptor binding site on the avian sarcoma and leukosis virus Env complex. J. Virol. 76:7518-7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Overbaugh, J., A. D. Miller, and M. V. Eiden. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol. Mol. Biol. Rev. 65:371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Parren, P. W., M. C. Gauduin, R. A. Koup, P. Poignard, P. Fisicaro, D. R. Burton, and Q. J. Sattentau. 1997. Relevance of the antibody response against human immunodeficiency virus type 1 envelope to vaccine design. Immunol. Lett. 57:105-112. [DOI] [PubMed] [Google Scholar]
- 88.Piatak, M., Jr., M. S. Saag, L. C. Yang, S. J. Clark, J. C. Kappes, K. C. Luk, B. H. Hahn, G. M. Shaw, and J. D. Lifson. 1993. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science 259:1749-1754. [DOI] [PubMed] [Google Scholar]
- 89.Pleskoff, O., M. Seman, and M. Alizon. 1995. Amphotericin B derivative blocks human immunodeficiency virus type 1 entry after CD4 binding: effect on virus-cell fusion but not on cell-cell fusion. J. Virol. 69:570-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Poignard, P., M. Moulard, E. Golez, V. Vivona, M. Franti, S. Venturini, M. Wang, P. W. Parren, and D. R. Burton. 2003. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J. Virol. 77:353-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ramalho-Santos, J., and M. C. de Lima. 1998. The influenza virus hemagglutinin: a model protein in the study of membrane fusion. Biochim. Biophys. Acta 1376:147-154. [DOI] [PubMed] [Google Scholar]
- 92.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Gottlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rizzuto, C. D., and J. G. Sodroski. 1997. Contribution of virion ICAM-1 to human immunodeficiency virus infectivity and sensitivity to neutralization. J. Virol. 71:4847-4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rohlf, F. J., and R. R. Sokal. 1994. Biometry: the principles and practice of statistics in biological research. W. H. Freeman, New York, N.Y.
- 95.Rong, L., and P. Bates. 1995. Analysis of the subgroup A avian sarcoma and leukosis virus receptor: the 40-residue, cysteine-rich, low-density lipoprotein receptor repeat motif of Tva is sufficient to mediate viral entry. J. Virol. 69:4847-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Salzwedel, K., and E. A. Berger. 2000. Cooperative subunit interactions within the oligomeric envelope glycoprotein of HIV-1: functional complementation of specific defects in gp120 and gp41. Proc. Natl. Acad. Sci. USA 97:12794-12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sandrin, V., B. Boson, P. Salmon, W. Gay, D. Negre, R. Le Grand, D. Trono, and F. L. Cosset. 2002. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood 100:823-832. [DOI] [PubMed] [Google Scholar]
- 98.Saphire, A. C., M. D. Bobardt, and P. A. Gallay. 1999. Host cyclophilin A mediates HIV-1 attachment to target cells via heparans. EMBO J. 18:6771-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schneider, J., O. Kaaden, T. D. Copeland, S. Oroszlan, and G. Hunsmann. 1986. Shedding and interspecies type sero-reactivity of the envelope glycopolypeptide gp120 of the human immunodeficiency virus. J. Gen. Virol. 67:2533-2538. [DOI] [PubMed] [Google Scholar]
- 100.Schonning, K., O. Lund, O. S. Lund, and J. E. Hansen. 1999. Stoichiometry of monoclonal antibody neutralization of T-cell line-adapted human immunodeficiency virus type 1. J. Virol. 73:8364-8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shangguan, T., D. Alford, and J. Bentz. 1996. Influenza-virus-liposome lipid mixing is leaky and largely insensitive to the material properties of the target membrane. Biochemistry 35:4956-4965. [DOI] [PubMed] [Google Scholar]
- 102.Shangguan, T., D. P. Siegel, J. D. Lear, P. H. Axelsen, D. Alford, and J. Bentz. 1998. Morphological changes and fusogenic activity of influenza virus hemagglutinin. Biophys. J. 74:54-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Si, Z., N. Phan, E. Kiprilov, and J. Sodroski. 2003. Effects of HIV type 1 envelope glycoprotein proteolytic processing on antigenicity. AIDS Res. Hum. Retrovir. 19:217-226. [DOI] [PubMed] [Google Scholar]
- 104.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 105.Sklyanskaya, E. I., M. Shie, Y. S. Komarov, S. S. Yamnikova, and N. V. Kaverin. 1988. Formation of mixed hemagglutinin trimers in the course of double infection with influenza viruses belonging to different subtypes. Virus Res. 10:153-165. [DOI] [PubMed] [Google Scholar]
- 106.Smith, A. E., and A. Helenius. 2004. How viruses enter animal cells. Science 304:237-242. [DOI] [PubMed] [Google Scholar]
- 107.Smith, J. G., W. Mothes, S. C. Blacklow, and J. M. Cunningham. 2004. The mature avian leukosis virus subgroup A envelope glycoprotein is metastable, and refolding induced by the synergistic effects of receptor binding and low pH is coupled to infection. J. Virol. 78:1403-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Snitkovsky, S., and J. A. Young. 1998. Cell-specific viral targeting mediated by a soluble retroviral receptor-ligand fusion protein. Proc. Natl. Acad. Sci. USA 95:7063-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stegmann, T. 2000. Membrane fusion mechanisms: the influenza hemagglutinin paradigm and its implications for intracellular fusion. Traffic 1:598-604. [DOI] [PubMed] [Google Scholar]
- 110.Stein, B. S., and E. G. Engleman. 1990. Intracellular processing of the gp160 HIV-1 envelope precursor. Endoproteolytic cleavage occurs in a cis or medial compartment of the Golgi complex. J. Biol. Chem. 265:2640-2649. [PubMed] [Google Scholar]
- 111.Sullivan, N., Y. Sun, J. Li, W. Hofmann, and J. Sodroski. 1995. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J. Virol. 69:4413-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tan, K., J. Liu, J. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 94:12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Taylor, H. P., S. J. Armstrong, and N. J. Dimmock. 1987. Quantitative relationships between an influenza virus and neutralizing antibody. Virology 159:288-298. [DOI] [PubMed] [Google Scholar]
- 114.van Zeijl, M., S. V. Johann, E. Closs, J. Cunningham, R. Eddy, T. B. Shows, and B. O'Hara. 1994. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc. Natl. Acad. Sci. USA 91:1168-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vey, M., M. Orlich, S. Adler, H. D. Klenk, R. Rott, and W. Garten. 1992. Hemagglutinin activation of pathogenic avian influenza viruses of serotype H7 requires the protease recognition motif R-X-K/R-R. Virology 188:408-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vila-Coro, A. J., M. Mellado, A. Martin de Ana, P. Lucas, G. del Real, A. C. Martinez, and J. M. Rodriguez-Frade. 2000. HIV-1 infection through the CCR5 receptor is blocked by receptor dimerization. Proc. Natl. Acad. Sci. USA 97:3388-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang, H., M. P. Kavanaugh, R. A. North, and D. Kabat. 1991. Cell-surface receptor for ecotropic murine retroviruses is a basic amino-acid transporter. Nature 352:729-731. [DOI] [PubMed] [Google Scholar]
- 118.Weissenhorn, W., L. J. Calder, S. A. Wharton, J. J. Skehel, and D. C. Wiley. 1998. The central structural feature of the membrane fusion protein subunit from the Ebola virus glycoprotein is a long triple-stranded coiled coil. Proc. Natl. Acad. Sci. USA 95:6032-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 120.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Willey, R. L., J. S. Bonifacino, B. J. Potts, M. A. Martin, and R. D. Klausner. 1988. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc. Natl. Acad. Sci. USA 85:9580-9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wrigley, N. G. 1979. Electron microscopy of influenza virus. Br. Med. Bull. 35:35-38. [DOI] [PubMed] [Google Scholar]
- 123.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 124.Yang, X., L. Florin, M. Farzan, P. Kolchinsky, P. D. Kwong, J. Sodroski, and R. Wyatt. 2000. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J. Virol. 74:4746-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang, X., S. Kurteva, S. Lee, and J. Sodroski. 2005. Stoichiometry of antibody neutralization of human immunodeficiency virus type 1. J. Virol. 79:3500-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang, X., E. Mahony, G. H. Holm, A. Kassa, and J. Sodroski. 2003. Role of the gp120 inner domain beta-sandwich in the interaction between the human immunodeficiency virus envelope glycoprotein subunits. Virology 313:117-125. [DOI] [PubMed] [Google Scholar]
- 127.Yuste, E., J. D. Reeves, R. W. Doms, and R. C. Desrosiers. 2004. Modulation of Env content in virions of simian immunodeficiency virus: correlation with cell surface expression and virion infectivity. J. Virol. 78:6775-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhu, P., E. Chertova, J. Bess, Jr., J. D. Lifson, L. O. Arthur, J. Liu, K. A. Taylor, and K. H. Roux. 2003. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc. Natl. Acad. Sci. USA 100:15812-15817. [DOI] [PMC free article] [PubMed] [Google Scholar]