Abstract
Recently developed molecular techniques have revolutionized the epidemiology of tuberculosis. Multiple studies have used these tools to examine the population structure of Mycobacterium tuberculosis isolates in different communities. The distributions of clusters of M. tuberculosis isolates in these settings may variously reflect social mixing patterns or the differential fitness of specific clones of the organism. We developed an individual-based microsimulation of tuberculosis transmission to explore social and demographic determinants of cluster distribution and to observe the effect of transmission dynamics on the empiric data from molecular epidemiologic studies. Our results demonstrate that multiple host-related factors contribute to wide variation in cluster distributions even when all strains of the organism are assumed to be equally transmissible. These host characteristics include interventions such as chemotherapy, vaccination and chemoprophylaxis, HIV prevalence, the age structure of the population, and the prevalence of latent tuberculosis infection. We consider the implications of these results for the interpretation of cluster studies of M. tuberculosis as well as the more general application of microsimulation models to infectious disease epidemiology.
Over the past 10 years, molecular tools have become available that have changed the way that epidemiologists study the transmission of infectious disease (1). In addition to their role in detecting unsuspected transmission links (2–4), molecular markers are increasingly being used to study transmission patterns within populations and to evaluate host- and strain-specific risk factors for disease spread (5, 6). Nowhere has this approach been used more rigorously than in the pioneering work on the molecular epidemiology of tuberculosis (TB). Since the development of standardized methods for DNA fingerprinting of Mycobacterium tuberculosis, molecular techniques have been used to estimate the fraction of cases attributable to recent transmission of M. tuberculosis (7–14), identify host-specific risk factors for disease spread (15–18), document exogenous reinfection (19–21), and study patterns of drug resistance (22–24). Investigators also have begun to use these methods to explore potential strain-specific differences in bacterial phenotypes such as tissue tropism, virulence, and transmissibility (25, 26).
This research has shown that the genetic diversity of M. tuberculosis isolates from different human communities can vary considerably (27, 28). Clusters of identical isolates are assumed to share DNA fingerprints as a result of the spread of the organism among the human hosts who harbor the isolates in the cluster. Patients with TB whose isolates cannot be grouped into clusters, i.e., those with unique DNA fingerprints, are assumed to have disease that results from the reactivation of latent infection acquired in the past. Variation in the distribution of clusters of M. tuberculosis isolates in different communities is thought to reflect different TB transmission dynamics and intensities in diverse parts of the world. A high proportion of clustered isolates in a community suggests ongoing TB transmission, whereas a predominance of unique cases implies that most TB cases are caused by reactivation of remote infection without further spread (29, 30).
The distribution of genotypes within bacterial populations also may reflect differences in the selective advantage of specific clones, especially those that are more transmissible or “fit” than their counterparts (31, 32). Clones that are especially transmissible would be expected to infect more human hosts per contact and therefore may belong to larger clusters than less-transmissible clones. Multiple molecular studies of Neisseria meningitides, for example, have used this approach to characterize pandemic strains that were shown to be genetically distinct from the strains isolated in sporadic cases of meningitis (33). Several recent epidemiologic studies of TB have focused on large clusters of cases caused by a single genotype or strain of M. tuberculosis (34–38). In some cases, these outbreaks were attributed to an increased capacity for transmission and/or replication by the specific strain (37). Other studies described highly mobile or complex social networks that may have facilitated the prolific spread of the organism (34–36, 38).
In practice, it can be difficult to distinguish between the effects of variability in strain behavior and the often complex transmission dynamics of TB in the analysis of differential cluster size. One way to address this problem is to consider the range of cluster sizes that might be expected in the absence of strain-specific variation in transmissibility, i.e., to establish an expectation of cluster distributions consistent with the null hypothesis that all strains are equally transmissible. Over the past decade, mathematical models of the transmission dynamics of TB have been developed that have helped elucidate the factors that impact TB epidemics (39–44). Several recent studies have begun to use some of the concepts developed in these models to examine the relationship between model parameters such as the basic reproductive rate and cluster size (45, 46). Although this research has important implications for interpreting the results of molecular epidemiology, few studies have explicitly examined the impact of the social and demographic variables that affect TB dynamics on the population structure of mycobacterial isolates. This report describes a stochastic model of TB transmission that generates cluster distributions as well as estimates of the annual incidences of TB infection and disease for a range of different input variables. Because few of the parameters relevant to TB transmission have been well characterized, the purpose of this study is not to predict distributions of cluster size in specific contexts. It is rather to explore the impact of different TB transmission dynamics on the population structure of isolates of M. tuberculosis and, more generally, to provide a link between the disciplines of infectious disease dynamics and molecular epidemiology.
Methods
Simulation Model.
A microsimulation model of TB transmission was used to generate dynamic transmission chains of TB. The model assumptions, parameters, and variables are described in detail in the text and Tables 2–5, which are published as supporting information on the PNAS web site, www.pnas.org. Briefly, the model specifies a distribution of discrete individuals, each of which is characterized by a vector of variables that determine the risks of TB infection, clinical disease, and transmitting infection once infected. Individuals are assigned to a series of social and physical spaces such as households, neighborhoods, and multineighborhood communities. The model also specifies the stochastic processes by which latent disease reactivates, infection is transmitted within and between social spaces, infection progresses to primary TB, vaccination or previous infection confers immunity, and individuals recover from infection. Because this model is meant to recapitulate the transmission dynamic described by state-compartmental models, parameter values were chosen to correspond to mean values used in previously published deterministic models of TB transmission. Individuals to whom disease is transmitted during the simulation acquire a variable reflecting the specific strain that is the source of their infection; thus chains of disease transmission can be identified as “clusters” of cases sharing a specific strain identifier. The model is run over a series of time steps, during which these stochastic processes may occur. To simulate the data collection of an epidemiologic study, the simulation was run for 208 1-week time steps for a total of 4 years. We assumed that molecular markers did not mutate during that period, and therefore all cases that shared a molecular marker were considered to belong to a single cluster. Output of the model includes standard measures of the incidence of infection and disease, the prevalence of infectious disease over time, and a count of cluster sizes.
TB Control Strategies.
To assess and compare the impact of specific interventions against TB on the distribution of cluster sizes, we considered the effects of several different TB control strategies on clustering patterns. These included the use of bacillus Calmette–Guérin vaccination, case finding and treatment, and the use of chemoprophylaxis in the exposed. Because it is widely held that effective control measures will diminish clustering, our aim was to explore the possibility that different control measures would differentially impact cluster distributions. We evaluated a range of coverage levels for each of these strategies, assuming that these levels were attained at the beginning of the simulated epidemic and maintained until the end of the simulation period.
HIV Prevalence.
We considered the possibility that HIV epidemics may have an impact on the clustering of TB cases. Although TB incidence clearly rises in areas with high HIV prevalence, this rise results from an increase in the rates of both primary and reactivation disease in the HIV-infected. Furthermore, because TB disease in the HIV-infected is often extrapulmonary, the overall effect of HIV on transmission and hence the size of clusters can be difficult to predict. In this analysis, we evaluated cluster distributions for logarithmic increases in HIV prevalence, assuming that other potential determinants of cluster distribution were held constant. In this way, we studied how much variability in cluster size across different communities might occur as the result of variations in levels of HIV prevalence.
Prevalence of Latent TB.
To study the link between transmission dynamics and cluster size, we examined and compared cluster distributions in areas with differing burdens of TB as reflected by the prevalence of latent infection at the beginning of the simulation run. Because previous investigators have noted more isolate diversity than expected in some high burden settings, this analysis was designed to explore the possibility that cluster sizes may be constrained by the presence of individuals who are partially immune to reinfection in areas of high prevalence of latent infection. We simulated TB epidemics for five different levels of latent TB prevalence holding other variables and parameters constant and assuming, unrealistically, that transmission parameters were identical in these five settings.
Demographics.
We evaluated model output for three different population age structures. Because TB behaves differently in different age groups, we hypothesized that cluster distributions may vary among populations with varying age composition. We chose age distributions consistent with those found in most-developed, moderately developed, and least-developed countries to simulate a wide range of demographic conditions.
Results
We examined the consistency of the basic assumptions of the model with current estimates of disease incidence by generating country-specific transmission scenarios and comparing the incidence of active disease to consensus estimates of disease burden in these settings. When transmission parameters were allowed to vary within reasonable bounds, each of these models generated an incidence approximately consistent with that reported from the specific setting modeled. Table 1 presents the modeled incidence of infection and disease, summary statistics of the cluster distributions, and the consensus estimates of incidence for nine of these settings. In these modeled settings, the proportion of unique isolates is not correlated strongly with either the incidence of clinical TB or the annual risk of infection.
Table 1.
Model-based output statistics for five settings from a microsimulation of tuberculosis transmission
| Output statistics | High burden
|
Moderate burden
|
Low burden
|
||
|---|---|---|---|---|---|
| Sudan | NY prison | Algeria | U.S. prison | Netherlands | |
| Tuberculosis incidence* | 190 | 581 | 32 | 82 | 14 |
| Consensus incidence estimates | 200 | NA† | 44 | NA† | 10 |
| ARI‡ | 0.025 | 0.046 | 0.003 | 0.005 | 0.001 |
| Maximum cluster size | 87 | 19 | 9 | 17 | 15 |
| Mean cluster size | 10.2 | 3.2 | 1.7 | 2.9 | 1.7 |
| Proportion unique isolates | 0.181 | 0.253 | 0.432 | 0.289 | 0.490 |
Consensus incidence estimates are shown for comparison with estimates obtained from the model.
Incidence per 100,000.
No data available.
Annual risk of infection.
Interventions.
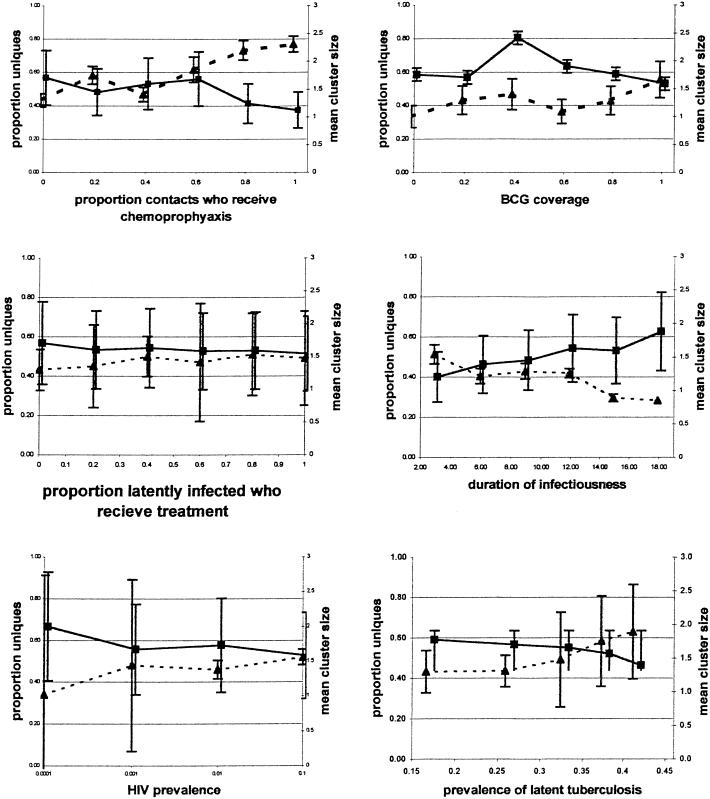
When other variables are held constant, modeled interventions that effectively reduce the transmission of M. tuberculosis such as a transmission-blocking vaccine, chemoprophylaxis of case contacts, and chemotherapy all cause a decrease in mean cluster size and a concomitant increase in the proportion of unique isolates (Fig. 1). Conversely, interventions that may reduce the incidence of reactivation but do not block transmission directly such as screening and treatment of latent infection or a vaccine that prevents endogenous reactivation in previously infected individuals reduce the number of clusters without impacting cluster size. Although both types of intervention ultimately reduce the number of cases of disease that results from ongoing transmission, they have very different effects on the distribution of cluster size and the proportions of unique and clustered isolates. These results suggest that monitoring the change in cluster sizes over time may not provide an adequate index of the impact of control programs on TB incidence.
Figure 1.
Effect of various factors on mean cluster size (bold line) and the proportion of uniques isolates (hatched line) of M. tuberculosis during modeled epidemics. BCG, bacillus Calmette–Guérin.
HIV Prevalence.
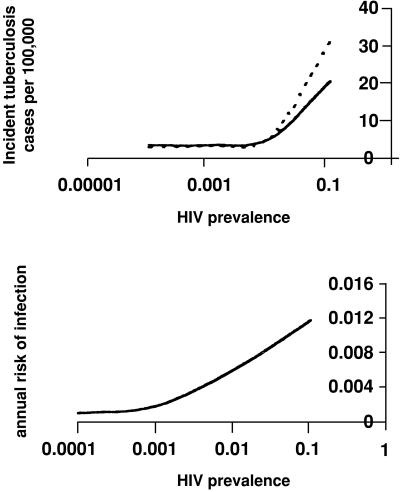
Fig. 1 shows the impact of HIV prevalence on model output. Not surprisingly, the model predicts that an increase in HIV prevalence will lead directly to an increased incidence of both TB infection and disease, the extent of which will depend on relative rates of reactivation and primary disease in the HIV-infected and the infectiousness of HIV-infected TB cases. More surprisingly, the model predicts that mean cluster size will fall as HIV prevalence increases despite the increase in incidence. This reduction in cluster size reflects a rise in the incidence of extrapulmonary disease and an early mortality and reduced duration of infectiousness in those coinfected with HIV and TB. The overall incidence of recently transmitted disease rises as HIV prevalence increases even though there are fewer secondary cases per infectious source. Because this effect is caused by a “tradeoff” between increased incidence of disease and reduced infectiousness, a realistic assessment of the magnitude of these effects must await more precise estimation of these parameters. Fig. 2 A and B show the relationships between rising HIV prevalence and the incidence of reactivation disease and recently transmitted disease for several possible rates of HIV-associated primary and reactivation disease. An important corollary of this observation is that factors associated with HIV may seem to be causally linked to small cluster size through confounding.
Figure 2.
The effect of increasing HIV prevalence on the incidence of cases caused by reactivation (dotted line) and primary disease (solid line) (A) and on the annual risk of infection (B). As HIV prevalence increases, the number of cases of reactivation TB increases precipitously, whereas the numbers of cases of infection and primary disease rise less steeply. Thus there are fewer cases of infection and primary disease for each “source” case.
Prevalence of Latent TB Infection.
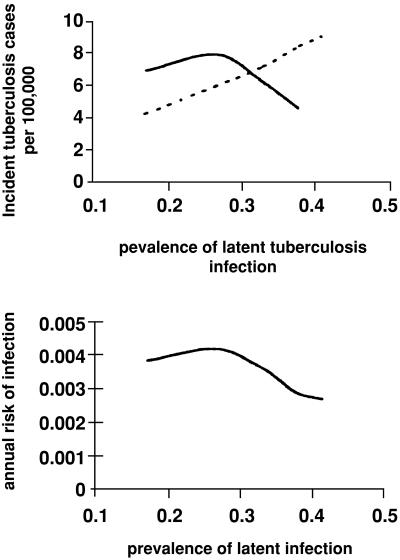
An increase in the prevalence of latent TB infection in this model leads to an increase in the incidence of active disease, with a rise in cases caused by reactivation and exogenous reinfection and a decline in those caused by primary disease (Fig. 1). Because the model assumes that previous infection with TB affords some degree of immunity to subsequent infection, an increase in the prevalence of latent infection translates into a corresponding decrease in the number of susceptibles available to be infected or reinfected. The effect of this “herd immunity” is a reduction in the size of clusters and in the proportion of clustered cases even though overall incidence has not been curtailed. Fig. 3 A and B illustrates the association between the prevalence of latent infection and the ratio of infectious cases to the annual risk of infection and indicates that an increase in the incidence of active disease may occur at the same time as a decline in TB transmission. Because the protective efficacy of previous TB infection for subsequent reinfection is not known, the quantitative impact of latent infection on cluster size cannot be determined. Nonetheless, this analysis does suggest that cluster size may be constrained in high burden areas even if the immunity conferred by previous disease is small.
Figure 3.
The effect of increasing the prevalence of latent TB on the incidence of cases caused by reactivation (dotted line) and primary disease (solid line) (A) and on the annual risk of infection (B). As prevalence increases, more source cases arise through reactivation. The falling number of susceptibles, however, means that transmission declines and the number of cases of infection and primary disease falls accordingly.
Demographics.
The model predicts that the age structure of a population can have a marked impact both on TB dynamics and cluster distribution (Table 4, which is published as supporting information on the PNAS web site). When the age structure of a population is skewed toward the young, the incidence of extrapulmonary disease increases, and transmission of the disease is reduced. Conversely, when older people make up a greater proportion of the population, the mean number of secondary infections produced by a single case will rise, because these patients are more likely to have pulmonary disease. Thus, the effect of raising the average age of a population in the absence of other changes is to increase mean cluster sizes and the proportion of clustered cases.
Despite these trends in mean cluster size with variation in specific determinants, the model output gave a wide range of cluster size both within and between simulations. The Monte Carlo-based standard deviations of cluster size are correspondingly large, reflecting the fact that this model can produce highly variable estimates across simulations for a single set of parameters. This variation largely stemmed from the underlying social structure incorporated into the model and the random distribution of an individual social-mixing factor that determined the number of contacts per case. When this mixing factor was fixed, the variance in cluster size was reduced substantially.
Discussion
This model identifies a number of factors that may have an effect on the cluster size and distribution of M. tuberculosis isolates in a specific community even under the “null” hypothesis that these isolates are identical phenotypically. These factors include interventions such as chemotherapy, vaccination, and chemoprophylaxis, HIV prevalence, the age structure of the population, and the prevalence of latent TB infection. Not surprisingly, these factors are those that have been shown to impact the disease dynamics in numerous deterministic models of TB transmission (39–44). Effecting a change in such parameters as the duration of infectiousness, the transmission probability, and the rates of progression to primary and reactivation disease alters the basic reproductive rate (R0) as well as the dynamic behavior and consequently the distribution of clusters of TB. Similarly, the finding that the expected number of new infections produced by an infectious case is reduced when some members of the population are immune to the disease is one of the most basic tenets of infectious disease dynamics (47). The observation that cluster size will be reduced in partially immune populations is entirely consistent with the well known nonlinear dynamics of communicable diseases. Despite the considerable recent focus on the dynamics of TB transmission (39–44, 46), few studies have explicitly extended these principles to understanding and interpreting data from molecular epidemiologic investigations of TB. Here, the use of an individual-based approach to modeling TB transmission allows us to generate the cluster distributions that comprise the empirical data from such studies and to observe the impact of transmission dynamics on the structure of this data directly. The wide range of cluster sizes found within simulations reflects variable levels of risk among different individuals as well as the social structure and mixing included in the model. Much recent analytical work has explored the impact of heterogeneous mixing and infectivity on estimates of R0 for infectious diseases (46, 48, 49); this work explores the implications of such heterogeneity for the molecular epidemiology of TB.
Although the cluster distributions generated by this model are not calibrated to represent realistic data sets, our results do have several implications for the practical interpretation of data from empirical studies. These are that (i) areas with high prevalence of latent TB may have a relatively high strain diversity because of the effect of partial herd immunity among infected hosts, (ii) the success of control measures may not always be reflected by a decrease in the mean cluster size in a community, and (iii) strain-specific characteristics associated with host factors may seem to be associated with trends in cluster size because of confounding of these characteristics by the host risk factor.
Although most systematic surveys of M. tuberculosis isolates have been conducted in low-prevalence communities, several recent studies have reported an unexpectedly high degree of genetic diversity in high-incidence communities in Africa. Warren et al. (50) hypothesized that epidemic areas would have relatively few circulating strains but instead found many unique isolates and small clusters among M. tuberculosis isolates sampled from two high-incidence suburban communities in Capetown, South Africa. Yang et al. (51) reported a similarly high level of strain diversity from Tanzania with 101 different strains noted among 134 isolates sampled. These results contrast markedly with the findings from low-incidence areas that have reported higher levels of clustering than anticipated. Our model predicts that, for a given basic reproductive rate, M. tuberculosis isolates from high-prevalence areas will be more diverse than isolates from low-prevalence areas. Because it is not clear how much the basic reproductive rate for TB varies between communities, this observation cannot be used to provide a quantitative expectation of how mean cluster sizes will vary in populations with differing prevalences of latent infection. However, it is consistent with the apparently paradoxical results observed in high- and low-incidence areas.
The second prediction of this model, that successful control measures will not always affect a reduction in mean cluster size, also may help explain the frequently reported finding of large clusters of cases in areas with low TB incidence. Numerous recent studies have documented the unsuspected spread of M. tuberculosis strains through diverse communities in low-incidence areas in the U.S. and have called attention to the inadequacy of standard contact-tracing techniques in identifying potential transmission events (34–38). Many of these communities focus on targeted testing and treatment of latent TB infection as a means of TB control (52). The impact of this strategy can be difficult to assess, because it is not clear how many of the potential cases averted would have gone on to transmit TB. Nonetheless, it is clear that reducing the number of newly reactivated cases will decrease incidence but may have little impact on the size of those clusters that do arise, especially if people in these clusters are not among those “targeted” for testing.
Point iii concerns epidemiologic inference as it applies to risk factors for clustering. In addition to identifying isolates that may be especially transmissible, some molecular studies have found that certain strains such as those that are isoniazid-resistant are less frequently clustered than others (53, 54). This finding has led to the inference that drug-resistant strains of M. tuberculosis may be less fit than their drug-sensitive counterparts and less likely to contribute to epidemic disease (55). The model presented here implicates a number of host and community-level factors as predictors of mean cluster size. In particular, determinants of reduced host capacity to spread infection such as HIV infection and young age directly reduce cluster size in this model. Many other host factors that were not addressed in this model may be associated with reduced capacity for transmission. Most notably, social-mixing patterns may be highly predictive of an individual's adherence to anti-TB therapy. If unmeasured confounders are risk factors both for the acquisition of drug resistance in TB and a reduced capacity to spread disease, drug-resistant strains will seem to be less frequently clustered even when they are no less transmissible than sensitive strains.
This study provides a first step in constructing individual-based models that link infectious disease dynamics and molecular epidemiologic approaches to studying strain diversity of pathogens. Such models can be refined and extended to incorporate recent innovative research on the social networks that facilitate TB transmission as well as a more specific approach to the molecular markers currently in use. Although the development of such models will require more computing capacity than current personal computers can provide, we believe that the elaboration of these methods will produce a powerful tool with which to study the epidemiology of infectious diseases.
Supplementary Material
Acknowledgments
I am grateful to Sid Atwood, Sam Bozeman, Barry Bloom, Marc Lipsitch, and James Robins for valuable help and advice with this paper. I was supported by National Institutes of Health Grant AI-01430-01.
Abbreviation
- TB
tuberculosis
References
- 1.Tibayrenc M. Annu Rev Genet. 1999;33:449–477. doi: 10.1146/annurev.genet.33.1.449. [DOI] [PubMed] [Google Scholar]
- 2.van Belkum A. Microb Drug Resist. 2000;6:173–188. doi: 10.1089/mdr.2000.6.173. [DOI] [PubMed] [Google Scholar]
- 3.Olsen S J, Hansen G R, Bartlett L, Fitzgerald C, Sonder A, Manjrekar R, Riggs T, Kim J, Flahart R, Pezzino G, Swerdlow D L. J Infect Dis. 2001;183:164–167. doi: 10.1086/317657. [DOI] [PubMed] [Google Scholar]
- 4.Knirsch C A, Jakob K, Schoonmaker D, Kiehlbauch J A, Wong S J, Della-Latta P, Whittier S, Layton M, Scully B. Am J Med. 2000;108:290–295. doi: 10.1016/s0002-9343(99)00459-3. [DOI] [PubMed] [Google Scholar]
- 5.Diaz R S, De Oliveira C F, Pardini R, Operskalski E, Mayer A J, Busch M P. AIDS Res Hum Retroviruses. 1999;15:1151–1156. doi: 10.1089/088922299310241. [DOI] [PubMed] [Google Scholar]
- 6.Wallinga J, Edmunds W J, Kretzschmar M. Trends Microbiol. 1999;7:372–377. doi: 10.1016/s0966-842x(99)01546-2. [DOI] [PubMed] [Google Scholar]
- 7.Alland D, Kalkut G E, Moss A R, McAdam R A, Hahn J A, Bosworth W, Drucker E, Bloom B R. N Engl J Med. 1994;330:1710–1716. doi: 10.1056/NEJM199406163302403. [DOI] [PubMed] [Google Scholar]
- 8.Small P M, Hopewell P C, Singh S P, Paz A, Parsonnet J, Ruston D C, Schecter G F, Daley C L, Schoolnik G. N Engl J Med. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- 9.van Deutekom H, Gerritsen J J, van Soolingen D, van Ameijden E J, van Embden J D, Coutinho R A. Clin Infect Dis. 1997;25:1071–1077. doi: 10.1086/516072. [DOI] [PubMed] [Google Scholar]
- 10.Shafer R W, Small P M, Larkin C, Singh S P, Kelly P, Sierra M F, Schoolnik G, Chirgwin K D. J Infect Dis. 1995;171:170–176. doi: 10.1093/infdis/171.1.170. [DOI] [PubMed] [Google Scholar]
- 11.Borgdorff M W, Nagelkerke N, van Soolingen D, de Haas P E W, Veen J, van Embden J D A. Am J Epidemiol. 1998;147:187–199. doi: 10.1093/oxfordjournals.aje.a009433. [DOI] [PubMed] [Google Scholar]
- 12.Hermans P W, Messadi F, Guebrexabher H, van Soolingen D, de Haas P E, Heersma H, de Neeling H, Ayoub A, Portaels F, Frommel D, et al. J Infect Dis. 1995;171:1504–1513. doi: 10.1093/infdis/171.6.1504. [DOI] [PubMed] [Google Scholar]
- 13.Barnes P F, Yang Z, Preston-Martin S, Pogoda J M, Jones B E, Otaya M, Eisenach K D, Knowles L, Harvey S, Cave M D. J Am Med Assoc. 1996;275:305–307. [Google Scholar]
- 14.van Soolingen D, Qian L, de Haas P E, Douglas J T, Traore H, Portaels F, Qing H Z, Enkhsaikan D, Nymadawa P, van Embden J D. J Clin Microbiol. 1995;33:3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coronado V G, Beck-Sague C M, Hutton M D, Davis B J, Nicholas P, Villareal C, Woodley C L, Kilburn J O, Crawford J T, Frieden T R, et al. J Infect Dis. 1993;168:1052–1055. doi: 10.1093/infdis/168.4.1052. [DOI] [PubMed] [Google Scholar]
- 16.Bishai W R, Graham N M, Harrington S, Pope D S, Hooper N, Astemborski J, Sheely L, Vlahov D, Glass G E, Chaisson R E. J Am Med Assoc. 1998;280:1679–1684. doi: 10.1001/jama.280.19.1679. [DOI] [PubMed] [Google Scholar]
- 17.Bifani P J, Mathema B, Liu Z, Moghazeh S L, Shopsin B, Tempalski B, Driscol J, Frothingham R, Musser J M, Alcabes P, Kreiswirth B N. J Am Med Assoc. 1999;282:2321–2327. doi: 10.1001/jama.282.24.2321. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Garcia M, Palacios-Martinez M, Ponce-de-Leon A, Jimenez-Corona M E, Jimenez-Corona A, Balandrano-Campos S, Olivera-Diaz H, Valdespino-Gomez J L, Small P M. Int J Tuberc Lung Dis. 2000;20:12–17. [PubMed] [Google Scholar]
- 19.Small P M, Schafer R W, Hopewell P C. N Engl J Med. 1993;328:1137–1144. doi: 10.1056/NEJM199304223281601. [DOI] [PubMed] [Google Scholar]
- 20.van Rie A, Waren R, Richardson M, Viater T C, Gia R P, Enarson D A, Beyers N, van Helden P D. N Engl J Med. 1999;341:353–360. doi: 10.1056/NEJM199910143411602. [DOI] [PubMed] [Google Scholar]
- 21.Schafer R W, Singh S P, Larkin C, Small P M. Tuber Lung Dis. 1995;76:575–577. doi: 10.1016/0962-8479(95)90537-5. [DOI] [PubMed] [Google Scholar]
- 22.Edlin B R, Tokars J I, Grieco M H, Crawford J T, Williams J, Sordillo E M, Ong K R, Kilburn J O, Dooley S W, Castro K G, et al. N Engl J Med. 1992;326:1514–1521. doi: 10.1056/NEJM199206043262302. [DOI] [PubMed] [Google Scholar]
- 23.Horn D L, Hewlett D, Jr, Haas W H, Butler W R, Alfalla C, Tan E, Levine A, Nayak A, Opal S M. Ann Intern Med. 1994;121:115–116. doi: 10.7326/0003-4819-121-2-199407150-00007. [DOI] [PubMed] [Google Scholar]
- 24.Davies G R, Pillay M, Sturm A W, Wilkinson D. Int J Tuberc Lung Dis. 1999;3:799–804. [PubMed] [Google Scholar]
- 25.Rhee J T, Piatek A S, Small P M, Harris L M, Chaparro S V, Kramer F R, Alland D. J Clin Microbiol. 1999;35:1764–1770. doi: 10.1128/jcm.37.6.1764-1770.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sreevatsan S, Pan X, Stockbauer K, Connell N D, Kreisworth B N, Whittam T S, Musser J M. Proc Natl Acad Sci USA. 1997;94:9869–9867. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato-Maeda M, Bifani P J, Kreiswirth B N, Small P M. J Clin Invest. 2001;10:533–537. doi: 10.1172/JCI11426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richner S, Meiring J, Kirby R. Electrophoresis. 1999;20:1800–1806. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1800::AID-ELPS1800>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 29.van Helden P D. Novartis Found Symp. 1998;217:178–194. doi: 10.1002/0470846526.ch13. [DOI] [PubMed] [Google Scholar]
- 30.Kato-Maeda M, Small P. West J Med. 2000;172:256–259. doi: 10.1136/ewjm.172.4.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levins B R, Lipsitch M, Bonhoeffer S. Science. 1999;283:806–809. doi: 10.1126/science.283.5403.806. [DOI] [PubMed] [Google Scholar]
- 32.Spratt B G, Maiden M C. Philos Trans R Soc London B. 1999;354:701–710. doi: 10.1098/rstb.1999.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caugant DA. APMIS. 1998;106:505–525. [PubMed] [Google Scholar]
- 34.Klovdahl A S, Graviss E A, Yaganehdoost A, Ross M W, Wanger A, Adams G J, Musser J M. Soc Sci Med. 2001;52:681–694. doi: 10.1016/s0277-9536(00)00170-2. [DOI] [PubMed] [Google Scholar]
- 35.Yaganehdoost A, Graviss E A, Ross M W, Adams G J, Ramaswamy S, Wanger A, Frothingham R, Soini H, Musser J M. J Infect Dis. 1999;180:1245–1251. doi: 10.1086/314991. [DOI] [PubMed] [Google Scholar]
- 36.Chin D P, Crane C M, Diul M Y, Sun S J, Agraz R, Taylor S, Desmond E, Wise F. J Am Med Assoc. 2000;283:2968–2974. doi: 10.1001/jama.283.22.2968. [DOI] [PubMed] [Google Scholar]
- 37.Yang Z, Barnes P F, Chaves F, Eisenach K D, Weis S E, Bates J H, Cave M D. J Clin Microbiol. 1998;36:1003–1007. doi: 10.1128/jcm.36.4.1003-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Classen C N, Warren R, Richardson M, Hauman J H, Gie R P, Ellis J H, van Helden P D, Beyers N. Thorax. 1999;54:136–140. doi: 10.1136/thx.54.2.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blower S M, McLean A R, Porco T C, Small P M, Hopewell P C, Sanchez M A, Moss A R. Nat Med. 1995;1:815–821. doi: 10.1038/nm0895-815. [DOI] [PubMed] [Google Scholar]
- 40.Blower S M, Gerberding J L. J Mol Med. 1998;76:624–636. doi: 10.1007/s001090050260. [DOI] [PubMed] [Google Scholar]
- 41.Vynnycky E, Fine P E. Epidemiol Infect. 1997;119:183–201. doi: 10.1017/s0950268897007917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dye C, Garnett G P, Sleeman K, Williams B G. Lancet. 1998;352:1886–1891. doi: 10.1016/s0140-6736(98)03199-7. [DOI] [PubMed] [Google Scholar]
- 43.Feng Z, Castillo-Chavez C, Capurro A F. Theor Popul Biol. 2000;57:235–247. doi: 10.1006/tpbi.2000.1451. [DOI] [PubMed] [Google Scholar]
- 44.Murray C J, Salamon J A. Proc Natl Acad Sci USA. 1998;95:13881–13886. doi: 10.1073/pnas.95.23.13881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borgdorff M W, Nagelkerke N, van Soolingen D, de Haas P E, Veen J, van Embden J D. Am J Epidemiol. 1998;147:187–195. doi: 10.1093/oxfordjournals.aje.a009433. [DOI] [PubMed] [Google Scholar]
- 46.Aparicio J P, Capurro A F, Castillo-Chavez C. J Theor Biol. 2000;206:327–341. doi: 10.1006/jtbi.2000.2129. [DOI] [PubMed] [Google Scholar]
- 47.Fine P E. Epidemiol Rev. 1993;15:265–302. doi: 10.1093/oxfordjournals.epirev.a036121. [DOI] [PubMed] [Google Scholar]
- 48.Hyman J M, Li J. Math Biosci. 2000;167:65–86. doi: 10.1016/s0025-5564(00)00025-0. [DOI] [PubMed] [Google Scholar]
- 49.Keeling M J, Grenfell B T. J Theor Biol. 2000;203:51–61. doi: 10.1006/jtbi.1999.1064. [DOI] [PubMed] [Google Scholar]
- 50.Warren R, Richardson M, van der Spuy G, Victor T, Sampson S, Beyers N, van Helden P. Electrophoresis. 1999;20:1807–1812. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1807::AID-ELPS1807>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 51.Yang Z H, Mtoni I, Chonde M, Mwasekaga M, Fuursted K, Askgard D S, Bennedsen J, de Haas P E, van Soolingen D, van Embden J D, et al. J Clin Microbiol. 1995;33:1064–1069. doi: 10.1128/jcm.33.5.1064-1069.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohn D L. Clin Infect Dis. 2000;31:120–124. doi: 10.1086/313891. [DOI] [PubMed] [Google Scholar]
- 53.Garcia-Garcia M L, Jimenez-Corona M E, Ponce-de-Leon A, Jimenez-Corona A, Palacios-Martinez M, Balandrano-Campos S, Ferreyra-Reyes L, Juarez-Sandino L, Sifuentes-Osornio J, Olivera-Diaz H, Valdespino-Gomez J L, Small P M. Int J Tuberc Lung Dis. 2000;4:S168–S170. [PubMed] [Google Scholar]
- 54.van Soolingen D, Borgdorff M W, de Haas P E, Sebek M M, Veen J, Dessens M, Kremer K, van Embden J D. J Infect Dis. 1999;180:726–736. doi: 10.1086/314930. [DOI] [PubMed] [Google Scholar]
- 55.Dye C, Williams B G. Proc Natl Acad Sci USA. 2000;97:8180–8185. doi: 10.1073/pnas.140102797. . (First Published June 20, 2000; 10.1073/pnas.140102797) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.