Abstract
Cancer cells show increased genome rearrangements, although it is unclear what defects cause these rearrangements. Previous studies have implicated the Saccharomyces cerevisiae replication checkpoint in the suppression of spontaneous genome rearrangements. In the present study, low doses of methyl methane sulfonate that activate the intra-S checkpoint but not the G1 or G2 DNA damage checkpoints were found to cause increased accumulation of genome rearrangements in both wild-type strains and to an even greater extent in strains containing mutations causing defects in the intra-S checkpoint. The rearrangements were primarily translocations or events resulting in deletion of a portion of a chromosome arm along with the addition of a new telomere. Combinations of mutations causing individual defects in the RAD24 or SGS1 branches of the intra-S checkpoint or the replication checkpoint showed synergistic interactions with regard to the spontaneous genome instability rate. PDS1 and the RAD50–MRE11–XRS2 complex were found to be important members of all the S-phase checkpoints in suppressing genome instability, whereas RAD53 only seemed to play a role in the intra-S checkpoints. Combinations of mutations that seem to result in inactivation of the S-phase checkpoints and critical effectors resulted in as much as 12,000–14,000-fold increases in the genome instability rate. These data support the view that spontaneous genome rearrangements result from DNA replication errors and indicate that there is a high degree of redundancy among the checkpoints that act in S phase to suppress such genome instability.
Saccharomyces cerevisiae and other organisms contain a number of checkpoints that respond to DNA damage and aberrant DNA structures that occur when DNA replication is blocked. The DNA damage checkpoints in S. cerevisiae result in cell-cycle arrest in either G1 or G2 in response to DNA damage during these phases of the cell cycle (1). The DNA damage checkpoint also results in slowing of DNA replication and cell-cycle progression when DNA damage occurs during S phase (2, 3); this latter checkpoint response often is called the intra-S checkpoint. A second checkpoint, sometimes called the replication checkpoint, also functions in S phase to cause cell-cycle arrest and suppression of late replication origins in response to blocked DNA replication (4, 5). Checkpoints are critical for preventing DNA damage-induced genome instability, because they cause cell-cycle delay or arrest in response to DNA damage to allow repair to occur (1, 3, 6, 7). Activation of checkpoints by DNA damage causes activation of signal-transduction pathways resulting in phosphorylation of a number of proteins including recombination and repair proteins as well as increased transcription of a number of genes (8–21). As a result, checkpoints directly target a number of DNA metabolic processes in addition to delaying or arresting cell-cycle progression. Checkpoints also act to prevent spontaneous genome instability (22). The importance of checkpoints in human disease is evidenced by the observations that ataxia telangiectasia, Nijmegen breakage syndrome, Bloom syndrome, and inherited breast cancer susceptibility syndromes (BRCA) 1 and 2 as well as p53 defects have been linked to defects in DNA damage responses and/or DNA repair (23–28).
The G1 and G2 DNA damage checkpoints involve several groups of proteins that function in conjunction with a central signal-transduction cascade. These proteins include the “RFC-like” complex RAD24–RFC2-5 and the “PCNA-like” complex RAD17–MEC3–DDC1 complex, which act as DNA damage sensors (12, 29–35). RAD9 also functions in the G1 and G2 DNA damage checkpoints, in which it acts as a scaffold that recruits RAD53, resulting in the activation of RAD53 (12). The DNA damage checkpoint also acts in S phase and in this context is often called the intra-S checkpoint (2, 3). The intra-S checkpoint also requires RAD9 and the RAD24–RFC2-5 and RAD17–MEC3–DDC1 complexes. SGS1, the yeast homologue of the BLM (Bloom syndrome; ref. 36) and WRN (Werner syndrome; ref. 37) proteins, functions in an intra-S checkpoint pathway that is parallel to the RAD24-dependent branch, but SGS1 does not function in the G1 or G2 DNA damage checkpoints (38). A second type of checkpoint, sometimes called the replication checkpoint, also functions in S phase to cause cell-cycle arrest and suppress late replication origins in response to blocked DNA replication (4, 5). The replication checkpoint is independent of RAD9, RAD17, RAD24, and MEC3 but requires RFC5, DPB11, DRC1, POL2, and possibly other proteins to sense replication blocks (39–44). The intra-S and replication checkpoints may not be entirely separate, because mutations in RFC5, DPB11, DRC1, and POL2 also cause defects in the intra-S checkpoint (39, 42–44) in addition to causing defects in the replication checkpoint. However, it is not clear yet in which branch of the intra-S checkpoint RFC5, DPB11, DRC1, and POL2 function. An sgs1 mutation causes a small defect in the replication checkpoint, although it is not known whether SGS1 functions along with RFC5, DPB11, DRC1, and POL2 or whether it functions in a minor, independent pathway (38).
The intra-S and replication checkpoints activate a phosphorylation-mediated signal-transduction cascade (1, 3, 7). The central protein kinase in this cascade is MEC1, an ataxia telangiectasia-mutated (ATM) homologue (45, 46). MEC1 and its interacting factor, DDC2, apparently interact with damaged DNA independently of the different proposed damage sensors RAD24–RFC2-5, SGS1, and RFC1-5 (47, 48). This observation suggests that the MEC1–DDC2 complex, the damage sensors, and possibly other proteins assemble on the DNA at sites of damage (48, 49). TEL1, a second ATM homologue, is redundant with MEC1 in some way; tel1 mutations do not cause sensitivity to DNA-damaging agents but enhance the sensitivity of mec1 mutants to DNA damage, and overexpression of TEL1 suppresses the sensitivity of mec1 mutants to DNA damage (50, 51). Checkpoint activation causes MEC1-dependent phosphorylation of a number of proteins including RAD9 (12, 13), RAD53 (9, 10), RPA (11), RAD55 (16), and CHK1 (14). DUN1, which controls a DNA damage-regulated transcription response, is targeted by MEC1 through RAD53 (8), whereas PDS1, an inhibitor of anaphase, is targeted by MEC1 directly and through CHK1 (15, 17, 18). MRE11–RAD50–XRS2 is phosphorylated on the MRE11 and XRS2 subunits primarily by TEL1 in response to checkpoint activation (19–21, 27). Interestingly, there seems to be differential regulation of different checkpoint effectors depending on how the checkpoints are activated.
Two important effectors of the damage, intra-S and replication checkpoints, are PDS1 and the MRE11–RAD50–XRS2 complex. PDS1 is stabilized as a result of phosphorylation by CHK1 in a MEC1-dependent reaction in response to DNA damage checkpoint activation (14). Similarly, it is stabilized as a result of phosphorylation through a MEC1-dependent CHK1-independent S-phase response, resulting in maintenance of sister-chromatid cohesion during S phase (15, 18). Thus, chk1 mutations cause DNA damage checkpoint defects but not replication checkpoint defects (14). MRE11–RAD50–XRS2 initially was implicated in the intra-S checkpoint (26) and also plays a role in adaptation to DNA damage-induced cell-cycle arrest (52). MRE11–RAD50–XRS2 is required for full activity of intra-S checkpoint responses involving MEC1-dependent phosphorylation of RAD53 and TEL1-dependent phosphorylation of MRE11 and XRS2 (19–21) but does not seem to be required for the replication checkpoint (52). In mec1 mutants, MRE11–RAD50–XRS2 is required for a TEL1-dependent checkpoint that is activated by both intra-S damage and replication blocks (19, 22). Thus, MRE11–RAD50–XRS2 seems to function both as part of the damage sensor system and as a downstream effector.
In previous studies, we observed that defects in the replication checkpoint, but not the intra-S or G1 and G2 damage checkpoints, caused increased levels of spontaneous genome rearrangements (22). This result suggested that DNA replication errors can lead to genome rearrangements, and the replication checkpoint acts to suppress these rearrangements. A mec1 tel1 double mutant had a much higher rate of accumulating genome rearrangements than was caused by other mutations causing checkpoint defects. One interpretation of these results is that there might be a high degree of redundancy among the checkpoints that function in S phase. In the present study, we present evidence that different S-phase checkpoint systems interact in suppressing spontaneous genome instability.
Materials and Methods
General Genetic Methods.
Media for propagation of strains and for determining gross chromosomal rearrangement (GCR) rates were as described (22, 53). All S. cerevisiae strains were propagated at 30°C except for rfc5-1, dpb11-1, or pds1Δ mutants, which were grown at 23°C. All strains were made by standard PCR-based gene-disruption methods, and correct gene disruptions were verified by PCR as described (22). The sequences of primers used to generate disruption cassettes and confirm disruption of indicated genes are available on request. All strains were derived from the S288c parental strain RDKY3023 [MATa, ura3-52, leu2Δ1, trp1Δ63, his3Δ200, lys2ΔBgl, hom3-10, ade2Δ1, ade8] and in addition contained the hxt13∷URA3 insertion used in the GCR assay (22, 53). Relevant genotypes of these strains are: RDKY3615 wild type; RDKY3633 mre11∷HIS3; RDKY3715 mad3∷TRP1; RDKY3717 bub3∷TRP1; RDKY3719 rad9∷HIS3; RDKY3721 rad17∷HIS3; RDKY3723 rad24∷HIS3; RDKY3725 mec3∷HIS3; RDKY3727 rfc5-1; RDKY3729 pds1∷TRP1; RDKY3731 tel1∷HIS3; RDKY3735 sml1∷KAN, mec1∷HIS3; RDKY3743 sml1∷KAN, mec1∷HIS3, tel1∷LEU2; RDKY3745 chk1∷HIS3; RDKY3749 sml1∷KAN, rad53∷HIS3; RDKY3751 sml1∷KAN, rad53∷HIS3, chk1∷TRP1; RDKY3753 sml1∷KAN, mec1∷HIS3, rad53∷TRP1; RDKY3759 mre11∷HIS3, tel1∷TRP1; RDKY3761 sml1∷KAN, mec1∷HIS3, mre11∷TRP1; RDKY3767 sml1∷KAN, mec1∷HIS3, tel1∷TRP1; RDKY3773 tel1∷TRP1, mec3∷HIS3; RDKY3775 tel1∷HIS3, rfc5-1; RDKY3813 sgs1∷HIS3; RDKY3821 pds1∷TRP1, tel1∷HIS3; RDKY3823 pds1∷TRP1, sml1∷KAN, mec1∷HIS3; RDKY4343 pif1-m2; RDKY4347 est2∷TRP1; RDKY4403 pif1-m2, rfc5-1; RDKY4405 pif1-m2, mec3∷HIS3; RDKY4407 pif1-m2, sml1∷KAN, mec1∷HIS3; RDKY4413 est2∷HIS3, rfc5-1; RDKY4415 sml1∷KAN, mec1∷HIS3, est2∷TRP1; RDKY4500 sml1∷KAN, mec1∷HIS3, rfc5-1; RDKY4505 tel1∷TRP1, rad9∷HIS3; RDKY4507 tel1∷TRP1, rad17∷HIS3; RDKY4509 tel1∷TRP1, rad24∷HIS3; RDKY4511 est2∷TRP1, rad9∷HIS3; RDKY4513 est2∷TRP1, rad17∷HIS3; RDKY4515 est2∷TRP1, rad24∷HIS3; RDKY4517 est2∷TRP1, mec3∷HIS3; RDKY4521 rfc5-1, rad9∷HIS3; RDKY4523 rfc5-1, rad17∷HIS3; RDKY4525 rfc5-1, mec3∷HIS3; RDKY4527 sml1∷KAN, mec1∷TRP1, rad9∷HIS3; RDKY4529 sml1∷KAN, mec1∷TRP1, rad17∷HIS3; RDKY4531 sml1∷KAN, mec1∷TRP1, mec3∷HIS3; RDKY4533 pif1-m2, rad9∷HIS3; RDKY4535 pif1-m2, rad17∷HIS3; RDKY4538 dpb11-1; RDKY4543 sml1∷KAN, mec1∷TRP1, rad24∷HIS3; RDKY4545 rfc5-1, rad24∷HIS3; RDKY4561 rfc5-1, sgs1∷HIS3; RDKY4562 rad9∷HIS3, sgs1∷TRP1; RDKY4563 rad24∷HIS3, sgs1∷TRP1; RDKY4564 rfc5-1, rad9∷HIS3, sgs1∷KAN; RDKY4565 sml1∷KAN, mec1∷HIS3, sgs1∷TRP1; RDKY4566 tel1∷HIS3, sgs1∷TRP1; RDKY4567 pif1-m2, sgs1∷HIS3; RDKY4569 pds1∷TRP1, sgs1∷HIS3; RDKY4570 pds1∷TRP1, dun1∷HIS3; RDKY4571 pds1∷TRP1, sml1∷KAN, rad53∷HIS3; RDKY4572 pds1∷TRP1, chk1∷HIS3; RDKY4573 pds1∷TRP1, rad24∷HIS3; RDKY4583 est2∷TRP1, sgs1∷HIS3; RDKY4584 mad2∷HIS3; RDKY4585 mad2∷HIS3, pds1∷TRP1; RDKY4586 sml1∷KAN, rad53∷HIS3, rfc5-1; RDKY4587 sml1∷KAN, rad53∷TRP1, rad24∷HIS3; RDKY4588 sml1∷KAN, rad53∷HIS3, sgs1∷TRP1; RDKY4589 mre11∷HIS3, rfc5-1; RDKY4590 mre11∷TRP1, rad24∷HIS3; RDKY4591 mre11∷KAN, sgs1∷HIS3; RDKY4592 mre11∷HIS3, pds1∷TRP1; RDKY4609 rfc5-1, rad24∷HIS3, sgs1∷KAN; RDKY4611 pif1-m2, rad24∷HIS3; RDKY4612 rad9∷HIS3, rad24∷TRP1; RDKY4613 sae2∷TRP1; RDKY4617 pds1∷HIS3, rfc5-1.
Characterization of GCR Rates and Breakpoints.
All GCR rates were determined independently by fluctuation analysis two or more times by using either 5 or 11 cultures, and the average value is reported (22, 53). The sequences of independent rearrangement breakpoints were determined and classified as described (22, 53).
Induction of GCRs by Treatment with Methyl Methanesulfonate (MMS).
Log-phase S. cerevisiae cells (2–4 × 107 cells per ml) were washed with water two times, suspended in an equal volume of water, and incubated with the indicated concentrations of MMS (Sigma) for 2 h at 30°C except for strains containing the rfc5-1, dpb11-1, or pds1 mutations, which were incubated at 23°C. The MMS concentration used resulted in 10–20% survival of the strain analyzed as determined by plating cells immediately after the 2-h MMS treatment. The treated cells were washed with water two times, resuspended in 10 volumes of yeast extract/peptone/dextrose (YPD), and incubated at 30 or 23°C, as appropriate, overnight until the culture reached saturation. The cells then were plated onto YPD plates and FC plates, which contain both 5-fluoroorotic acid (United States Biochemical) and canavanine (Sigma). After 2–4 days of incubation at 30 or 23°C, the frequency of cells resistant to both drugs was determined. Three to five independent cultures of each strain were treated with MMS in each experiment, and each experiment was performed at least twice. The average fold increase in the frequency of GCRs relative to no MMS treatment is reported.
Results
MMS Treatment Increases Genome Instability.
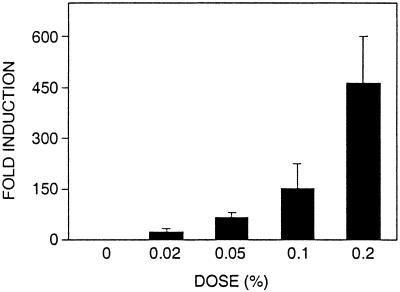
Mutations that inactivate the DNA damage checkpoints only cause small increases in the rate of accumulating spontaneous GCRs (22). To investigate whether the DNA damage checkpoints might play a role in suppressing GCRs induced by DNA-damaging agents, the effect of treatment with MMS on the induction of GCRs was tested. Treatment with MMS increased the GCR frequency in a dose-dependent fashion (Fig. 1). Low concentrations of MMS (0.02–0.1%), which result in slowing of S phase but do not induce G1 or G2 arrest (38, 54), caused induction of GCRs; this suggests that the intra-S checkpoint functions in the suppressions of GCRs. The majority of GCRs induced by MMS treatment were terminal deletions of the left arm of chromosome V associated with de novo telomere addition (75%) called “telomere additions.” Also observed were three translocations with microhomology at the rearrangement breakpoints (25%). In the absence of MMS treatment, five telomere additions (83%) and one translocation with a nonhomology breakpoint were observed (17%). These rearrangement sequences are available on request.
Figure 1.
Induction of GCRs by treatment of a wild-type S. cerevisiae strain with MMS. The strain RDKY3615 was treated with the indicated MMS concentration for 2 h, and the resulting GCR frequency was determined as described under Materials and Methods. The results reported are the average fold induction of the GCR frequency relative to the GCR frequency of untreated wild-type cells. The error bars indicate the SD of the results from 3–6 independent experiments.
The effect of different checkpoint defects on MMS induction of GCRs then was analyzed (Table 1). The concentration of MMS was varied to yield 10–20% survival to allow detection of induction of GCRs. MMS (0.07%) treatment of strains containing rad9, rad17, rad24, or mec3 mutations that cause defects in the DNA damage and intra-S checkpoints and MMS (0.05%) treatment of strains containing rfc5-1 or dpb11-1 mutations that cause defects in the replication and intra-S checkpoints resulted in a significant increased induction of GCRs. MMS (0.07%) treatment of an sgs1 strain resulted in significantly increased induction of GCRs. Mutation of MEC1 and MRE11, which function as a signal modifier in the intra-S checkpoint, resulted in increased induction of GCRs by MMS (0.02%). Mutations of the TEL1 gene and mutation of the mitotic checkpoint genes BUB3 and MAD3 (data not shown) had no effect on the induction of GCRs by MMS. Finally, mutation of PDS1 resulted in a modest increase in the induction of GCRs by MMS. Overall, these results are consistent with the view that the intra-S checkpoint plays an important role in suppressing MMS-induced GCRs.
Table 1.
Effect of checkpoint defects on the induction of GCRs by MMS
| Relevant genotype | Strain | GCR frequency × 1010 | Fold induction by MMS (+MMS/−MMS) | |
|---|---|---|---|---|
| No treatment | MMS treatment | |||
| 0.02% MMS | ||||
| Wild type | 3615 | 8.5 (1) ± 2.0 | 260 (30) ± 120 | 30 |
| mec1Δsml1Δ | 3735 | 400 (47) ± 220 | 2,300 (270) ± 890 | 6 |
| mre11Δ | 3633 | 10,000 (1,176) ± 2,500 | 19,000 (2,235) ± 2,500 | 2 |
| 0.05% MMS | ||||
| Wild type | 3615 | 8.5 (1) ± 2.0 | 650 (76) ± 200 | 76 |
| rfc5-1 | 3727 | 620 (73) ± 290 | 9,000 (1,059) ± 2,800 | 15 |
| dpb11-1 | 4538 | 550 (65) ± 320 | 8,600 (1,012) ± 1,300 | 16 |
| 0.07% MMS | ||||
| Wild type | 3615 | 8.5 (1) ± 2.0 | 830 (98) ± 380 | 98 |
| rad9Δ | 3719 | 29 (7) ± 9.3 | 10,000 (1,176) ± 4,800 | 168 |
| rad17Δ | 3721 | 37 (4) ± 20 | 8,000 (941) ± 3,400 | 235 |
| rad24Δ | 3723 | 21 (3) ± 20 | 5,900 (694) ± 2,500 | 231 |
| mec3Δ | 3725 | 43 (9) ± 24 | 8,400 (988) ± 3,500 | 109 |
| sgs1Δ | 3813 | 12 (2) ± 8.5 | 5,000 (588) ± 1,300 | 294 |
| tel1Δ | 3731 | 5.1 (0.6) ± 3.7 | 530 (62) ± 50 | 103 |
| 0.1% MMS | ||||
| Wild type | 3615 | 8.5 (1) ± 2.0 | 1,300 (153) ± 290 | 151 |
| pds1Δ | 3729 | 1,300 (153) ± 200 | 3,200 (374) ± 800 | 2 |
( ) indicates fold induction of the GCR frequency relative to wild type without MMS treatment. The MMS treatment used resulted in 10–20% survival of the strains. Note that the wild-type GCR frequency is 8.5 × 10−10.
Redundancy Among S-Phase Checkpoint Sensor Functions.
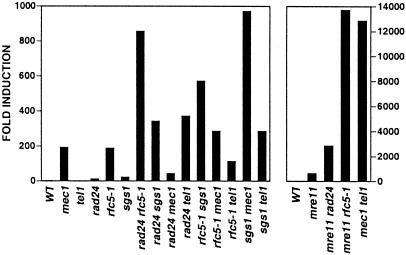
In contrast to our previous analysis of the suppression of spontaneous GCRs (22), the MMS induction of GCRs by doses that activate the intra-S checkpoint but not the G1 or G2 damage checkpoints suggests that the intra-S checkpoint can suppress spontaneous GCRs. Accordingly, the role of the intra-S checkpoint in suppression of spontaneous GCRs was investigated by examining the effect of combining mutations in multiple checkpoint genes (Tables 2 and 3 and Fig. 2). Mutations in RAD9, RAD17, RAD24, MEC3, and SGS1 had relatively small effects on the GCR rate compared with mutations in RFC5 and DPB11. Combining mutations from the rad9, rad17, rad24, and mec3 group with an sgs1 mutation resulted in a synergistic increase in the GCR rate. Similarly, combining the rfc5-1 or dpb11-1 mutations with either mutation from the rad9, rad17, rad24, and mec3 group or an sgs1 mutation resulted in a synergistic increase in the GCR rate. Significant interaction between a rad9 mutation and rad17 or rad24 mutations was not observed. The rfc5-1 rad9 sgs1 and rfc5-1 rad24 sgs1 triple mutant strains showed increased GCR rates of up to almost 4,000-fold (Table 2), although this was lower than that of a mec1 tel1 double mutant (ref. 22; Table 4 and Fig. 2). The majority of the GCRs seen in the rfc5-1, rad9, mec3, and sgs1 single mutants were telomere additions (Table 3). The rfc5-1 rad9, rad9 sgs1, rfc5-1 sgs1, and rad9 rfc5-1 sgs1 strains showed significant increases in the rate of accumulating telomere additions, although the rfc5-1 sgs1 strain also showed an ≈20-fold increase in the rate of accumulating translocations. These results are consistent with previous observations that checkpoint defects primarily lead to rate increase of accumulating telomere additions (22). Overall, these results support the view that SGS1 and RAD24 define partially redundant branches of the intra-S checkpoint and that there is redundancy among intra-S and replication checkpoints in suppression of genome rearrangements.
Table 2.
Genetic interactions between mutations affecting damage-sensing functions: GCR rate analysis
| Relevant genotype | Wild type
|
rfc5-1
|
sgs1Δ
|
|||
|---|---|---|---|---|---|---|
| Strain | GCR rate × 1010 | Strain | GCR rate × 1010 | Strain | GCR rate × 1010 | |
| Wild type | 3615 | 3.5 (1)* | 3727 | 660 (189)* | 3813 | 77 (22)† |
| rad9Δ | 3719 | 20 (6)* | 4521 | 3,200 (914) | 4562 | 1,300 (371) |
| rad17Δ | 3721 | 30 (9)* | 4523 | 3,300 (943) | ND | |
| rad24Δ | 3723 | 40 (11)* | 4545 | 3,000 (857) | 4563 | 1,200 (343) |
| mec3Δ | 3725 | 190 (54)* | 4525 | 1,800 (514) | ND | |
| sgs1Δ | 3813 | 77 (22)† | 4561 | 2,000 (571) | NA | |
( ) indicates the GCR rate relative to the wild-type GCR rate. The GCR rate of RDKY4564 (rfc5-1 rad9Δ sgs1Δ) was 1.3 × 10−6 (3,714). The GCR rate of RDKY4612 (rad9Δ rad24Δ) was 1.6 × 10−9 (5). The GCR rate of RDKY4609 (rfc5-1 rad24Δ sgs1Δ) was 3.3 × 10−7 (943). Note that the wild type GCR rate is 3.5 × 10−10. ND, not determined; NA, not applicable.
Data from Myung et al. (22).
Data from Myung et al. (64).
Table 3.
Genetic interactions between mutations affecting damage-sensing functions: Rearrangement breakpoint analysis
| Relevant genotype | Strain | Telomere addition | Translocation, non,micro |
|---|---|---|---|
| Wild type* | 3615 | 5 | 1,0 |
| rfc5-1* | 3727 | 10 | 0,0 |
| sgs1Δ† | 3813 | 10 | 2,4‡ |
| rad9Δ | 3719 | 8 | 0,1‡ |
| mec3Δ* | 3725 | 8 | 0,2‡ |
| rfc5-1 rad9Δ | 4521 | 12 | 0,0 |
| rad9Δ sgs1Δ | 4562 | 10 | 0,0 |
| rfc5-1 sgs1Δ | 4561 | 6 | 0,3‡ |
| rad9Δ rfc5-1 sgs1Δ | 4564 | 9 | 0,1 |
The numbers reported are the actual numbers of each type of event seen. The designation n,n indicates the number of nonhomology breakpoints observed followed by the number of microhomology breakpoints observed.
Data from Myung et al. (22).
Data from Myung et al. (64).
These breakpoints had more homology (homeology) than most microhomology breakpoints.
Figure 2.
Combination of mutations in different checkpoint genes cause synergistic increases in the GCR rate. The GCR rates of the indicated mutant strains were determined as described under Materials and Methods. The values for fold induction were calculated by dividing the GCR rate of the indicated mutant strain by the GCR rate of the wild-type (WT) strain. The GCR rates are from Tables 2, 4, and 8.
Table 4.
Genetic interaction between mutations affecting damage-sensing functions and signal-transduction cascade components: GCR rate analysis
| Relevant genotype | Wild type
|
mec1Δ sml1Δ
|
tel1Δ
|
|||
|---|---|---|---|---|---|---|
| Strain | GCR rate × 1010 | Strain | GCR rate × 1010 | Strain | GCR rate × 1010 | |
| Wild type | 3615 | 3.5 (1)* | 3735 | 680 (194)* | 3731 | 2.0 (0.6)* |
| rad9Δ | 3719 | 20 (6)* | 4527 | 270 (77) | 4505 | 230 (66) |
| rad17Δ | 3721 | 30 (9)* | 4529 | 330 (94) | 4507 | 430 (123) |
| rad24Δ | 3723 | 40 (11)* | 4543 | 150 (43) | 4509 | 1,300 (371) |
| mec3Δ | 3725 | 190 (54)* | 4531 | 210 (60) | 3773 | 4,200 (1,200)* |
| rfc5-1 | 3727 | 660 (189)* | 4500 | 1,000 (286)* | 3775 | 400 (114)* |
| sgs1Δ | 3813 | 77 (22)† | 4565 | 3,400 (971) | 4566 | 1,000 (286) |
| mec1Δ sml1Δ | 3735 | 680 (194)* | NA | 3743 | 45,000 (12,857)* | |
| rad53Δ sml1Δ | 3749 | 95 (27)* | 3753 | 1,000 (286)* | 3767 | 750 (214)* |
The MEC1 and TEL1 Proteins Play Redundant Roles in the Suppression of GCRs.
MEC1 seems to be more important than TEL1 in suppressing GCRs in that only mec1 mutations and not tel1 mutations cause checkpoint defects and increased GCR rates (22, 50, 51, 55, 56). However, MEC1 and TEL1 clearly are redundant, because mec1 and tel1 mutations show synergistic interactions when combined (refs. 10, 22, 50, and 57; Table 4 and Fig. 2). A tel1 mutation showed a synergistic effect on the GCR rate when combined with all mutations effecting the intra-S checkpoint including rad9, rad17, rad24, mec3, sgs1, and rad53 but did not significantly change the GCR rate caused by the rfc5-1 mutation. Consistent with previous results, combining a tel1 mutation with rfc5-1, sgs1, rad9, and mec3 mutations significantly increased the proportion of GCRs that were translocations (Table 5). In addition, combining a tel1 mutation with rad9 and mec3 mutations also significantly increased the rate of accumulating telomere additions, consistent with the view that a tel1 mutation increases the extent of the checkpoint defect caused by rad9 and mec3. Overall, these results suggest that the intra-S checkpoint functions are redundant with a TEL1-dependent function or pathway.
Table 5.
Genetic interaction between mutations affecting damage-sensing functions and signal-transduction cascade components: Rearrangement breakpoint analysis
| Relevant genotype | Strain | Telomere addition | Translocation, non,micro |
|---|---|---|---|
| Wild type* | 3615 | 5 | 1,0 |
| mec1Δ sml1Δ* | 3735 | 9 | 0,0 |
| tel1Δ* | 3731 | 0 | 0,6 |
| tel1Δ rfc5-1* | 3775 | 2 | 4,4 |
| tel1Δ sgs1Δ | 4566 | 0 | 4,6 |
| tel1Δ rad9Δ | 4505 | 7 | 1,4 |
| tel1Δ mec3Δ* | 3773 | 4 | 1,4 |
| mec1Δ sml1Δ rad9Δ | 4527 | 9 | 0,2 |
The numbers reported are the actual numbers of each type of event seen. The designation n,n indicates the number of nonhomology breakpoints observed followed by the number of microhomology breakpoints observed.
Data from Myung et al. (22).
Quite different results were obtained when double-mutant analysis was performed with a mec1 mutation (note that all mec1 mutants also contain a sml1 mutation). Combining a mec1 mutation with either the rfc5-1 or rad53 mutations resulted in a GCR rate that was similar to that caused by the mec1 mutation. These results suggest that RFC5 and RAD53 primarily function in a MEC1-dependent pathway. Combining a mec1 mutation with a sgs1 mutation resulted in a synergistic interaction and a GCR rate that was significantly higher than that observed for the sgs1 tel1 double mutant. Thus, SGS1 seems to be redundant with both TEL1- and MEC1-dependent functions or pathways. Combining rad9, rad17, rad24, or mec3 mutations with a mec1 mutation resulted in a 2–4-fold decrease in the GCR rate compared with a mec1 mutant; however, these GCR rates were significantly higher than those caused by rad9, rad17, rad24, and mec3 single mutations. These results suggest that some component of mec1-induced genome instability such as the de novo telomere-addition reaction depends on RAD9, RAD17, RAD24, and MEC3. This reduction of the GCR rate caused by a mec1 mutation is similar to that observed when mutations inactivating telomerase function are combined with a mec1 mutation (ref. 58; Table 6), and interestingly, mutations in at least two of these genes, RAD17 and MEC3, are known to effect telomere length (59, 60). Consistent with this, analysis of the rearrangement breakpoints (Table 5) from a rad9 mec1 sml1 mutant revealed a higher proportion of translocations than seen in a mec1 sml1 strain (all telomere additions). The rate of accumulating telomere additions in the rad9 mec1 mutant was 3-fold lower than in a mec1 mutant, and the rate of accumulating translocations was 22-fold higher in the rad9 mec1 mutant compared with the rad9 mutant. This suggests that the rad9 mutation selectively reduced the telomere addition class of rearrangements in a mec1 mutant background. Alternatively, RAD9, RAD17, and RAD24 have been suggested to control the degradation of damaged DNA, which may play a role in the production of GCRs in a mec1 mutant (29).
Table 6.
Genetic interaction between mutations affecting damage-sensing and telomere-maintenance functions
| Relevant genotype | Wild type
|
est2Δ
|
pif1-m2
|
|||
|---|---|---|---|---|---|---|
| Strain | GCR rate × 1010 | Strain | GCR rate × 1010 | Strain | GCR rate × 1010 | |
| Wild type | 3615 | 3.5 (1)* | 4347 | 1.2 (0.3)† | 4343 | 830 (237)† |
| rad9Δ | 3719 | 20 (6)* | 4511 | 4.5 (1.3) | 4533 | 1,310 (374) |
| rad17Δ | 3721 | 30 (9)* | 4513 | 6.0 (1.7) | 4535 | 1,150 (329) |
| rad24Δ | 3723 | 40 (11)* | 4515 | 8.5 (2.4) | 4611 | 1,760 (503) |
| mec3Δ | 3725 | 190 (54)* | 4517 | 50 (14) | 4405 | 3,100 (886) |
| rfc5-1 | 3727 | 660 (189)* | 4413 | 440 (125)† | 4403 | 13,300 (3,800)† |
| sgs1Δ | 3813 | 77 (22) | 4583 | 7.3 (2) | 4567 | 2,900 (829) |
| mec1Δ sml1Δ | 3735 | 680 (194)* | 4415 | 280 (80)* | 4407 | 18,000 (5,200) |
Interaction Between Mutations Effecting Telomere Maintenance and Checkpoints.
Proteins participating in the maintenance of telomeres play important roles both in suppressing GCRs and generating the telomere-addition class of GCRs (61). The effect of combining rad9, rad17, rad24, mec3, and sgs1 mutations with either a mutation that eliminates the catalytic activity of telomerase (est2; ref. 62) or increases the de novo telomere-addition reaction (pif1-m2; refs. 61 and 63) was tested. Combining rad9, rad17, rad24, mec3, and sgs1 mutations with an est2 mutation resulted in significant 3–4-fold reductions in the GCR rate (Table 6), consistent with the observation that the majority of GCRs caused by rad9, mec3, or sgs1 mutations were telomere additions (Table 3). Combining an est2 mutation with either a rfc5-1 or mec1 mutation, which causes much larger increases in the GCR rate, resulted in at most a 2-fold reduction in GCR rate; a previous study showed that mutations in genes required for telomere maintenance in rfc5-1 or mec1 mutants resulted in a shift from telomere additions to translocations and chromosome fusions (22). Combining the pif1-m2 mutation with rad9, rad17, and rad24 mutations resulted in small, ≈2-fold increases in the GCR rate above that caused by the pif1-m2 mutation alone (Table 6). These results are consistent with the observation that rad9, rad17, and rad24 mutations cause very small defects in the suppression of genome instability (22). Consistent with the observation that sgs1 and mec3 mutations cause higher increases in the GCR rate (22, 64), a larger, greater than additive on the order of a 3–4-fold increase in GCR rate was observed when the pif1-m2 mutation was combined with the sgs1 and mec3 mutations. These synergistic interactions were less than those observed when a pif1-m2 mutation was combined with mutations that cause significantly larger checkpoint defects such as mec1 or rfc5-1 mutations (Table 6). Overall, these results are consistent with the idea that the RAD9, RAD17, RAD24, MEC3, and SGS1 checkpoint functions act to suppress GCRs, and the GCR precursors involved are subject to telomere-addition reactions.
Analysis of the Downstream Effectors PDS1 and RAD53.
PDS1 was identified as an inhibitor of the initiation of anaphase (65). Most relevant to the studies presented here, PDS1 seems to maintain sister-chromatid cohesion in a MEC1-dependent, CHK1-independent manner during S phase and also functions in the DNA damage checkpoints in a MEC1- and CHK1-dependent manner (14). Recently, we showed that a pds1 mutation caused a significant increase in the rate of accumulating GCRs (22). To understand the role of PDS1 in suppressing GCRs better, the effect of a pds1 mutation in combination with other checkpoint mutations was analyzed (Table 7). When a pds1 mutation was combined with sgs1, rad53, or chk1 mutations, the resulting GCR rate was not significantly different from the GCR rate caused by the pds1 mutation. The GCR rates of the rfc5-1 pds1 and rad24 pds1 double mutants were 2-fold higher than the GCR rate of the pds1 single mutant, which is a small but significant difference. Combining the pds1 mutation with mec1, tel1, or dun1 mutations resulted in more striking increases in the GCR rate, with the most dramatic increase being observed with the tel1 pds1 double mutant. Mutations in BUB3 and MAD3 (22) and MAD2 had no effect on the GCR rate, and the GCR rate of a mad2 pds1 double mutant was the same as that of a pds1 single mutant. These results suggest that the role of PDS1 in suppressing GCRs is independent of the microtubule (spindle) checkpoint (66, 67) and more likely is related to its role in maintaining sister-chromatid cohesion during S phase (15, 18).
Table 7.
Effect of pds1Δ and rad53Δ mutations on GCR rates
| Relevant genotype | Wild type
|
pds1Δ
|
rad53Δ sml1Δ
|
|||
|---|---|---|---|---|---|---|
| Strain | GCR rate × 1010 | Strain | GCR rate × 1010 | Strain | GCR rate × 1010 | |
| Wild type | 3615 | 3.5 (1)* | 3729 | 670 (190)* | 3749 | 95 (27)* |
| mad2Δ | 4584 | 5.6 (1.6) | 4585 | 660 (189) | ND | |
| rad24Δ | 3723 | 40 (11)* | 4573 | 1,300 (371)‡ | 4587 | 140 (40) |
| rfc5-1 | 3727 | 660 (189)* | 4617 | 1,400 (400)‡ | 4586 | 500 (143) |
| sgs1Δ | 3813 | 77 (22)† | 4569 | 390 (111)§ | 4588 | 460 (131) |
| mec1Δ sml1Δ | 3735 | 680 (194)* | 3823 | 3,200 (914)* | 3753 | 1,000 (286)* |
| tel1Δ | 3731 | 2.0 (0.6)* | 3821 | 14,000 (4,000) | 3767 | 750 (214)* |
| dun1Δ | 3739 | 730 (208)* | 4570 | 2,000 (571)‡ | ND | |
| rad53Δ sml1Δ | 3749 | 95 (27)* | 4571 | 490 (140)§ | NA | |
| chk1Δ | 3745 | 130 (37)* | 4572 | 830 (237)§ | 3751 | 230 (64)* |
| mre11Δ | 3633 | 2,200 (629)* | 4592 | 18,000 (5,143) | ND | |
( ) indicates the GCR rate relative to the wild-type GCR rate. ND, not determined; NA, not applicable.
Data from Myung et al. (22).
Data from Myung et al. (64).
Significant difference compared to the GCR rate of the pds1 strain at the 95% confidence interval (Student's t test).
No significant difference compared to the GCR rate of the pds1 strain at the 95% confidence interval (Student's t test). Note that the wild-type GCR rate is 3.5 × 10−10.
Our previous studies showed that a rad53 mutation caused only a small increase in the GCR rate (22). The observation that rad53 and tel1 mutations showed a synergistic interaction (ref. 22; Table 7) suggested that RAD53 might function in one of the redundant checkpoint pathways. The effect of a rad53 mutation in combination with rfc5-1, rad24, or sgs1 mutations was analyzed (note that all rad53 mutants also contain a sml1 mutation; Table 7). The rad53 rfc5-1 double mutant had the same GCR rate as the rfc5-1 single mutant, and the rad53 rad24 double mutant had essentially the same GCR rate as the rad53 single mutant. In contrast, combining the rad53 mutation with the sgs1 mutation resulted in a highly synergistic effect on the GCR rate. The GCR rate of the sgs1 rad53 mutant was similar to that of the rad53 tel1 mutant. These results suggest that RAD53 functions in suppression of GCRs by a pathway that is redundant with the SGS1 pathway that acts through TEL1, and this pathway probably is the RAD24-dependent pathway.
Analysis of the Role of MRE11–RAD50–XRS2 in Suppression of Genome Instability.
The MRE11–RAD50–XRS2 complex is known to function in homologous recombination, nonhomologous end joining, and telomere maintenance (19, 20, 68, 69). Mutations inactivating the MRE11–RAD50–XRS2 complex result in high GCR rates (22, 53). The MRE11–RAD50–XRS2 complex has been found to be required for full activity of the intra-S checkpoint involving both MEC1- and TEL1-dependent responses and to act in both intra-S and replication checkpoint responses when MEC1 function is absent (19, 20). To understand the role of the MRE11–RAD50–XRS2 complex better, the effect of a mre11 mutation in combination with other mutations in checkpoint genes was analyzed (Table 8 and Fig. 2). Combining a mre11 mutation with a sgs1 mutation resulted in the same GCR rate as caused by the mre11 mutation, whereas combining a mre11 mutation with a rad24, pds1, or mec1 mutation resulted in a synergistic increase in the GCR rate. However, these synergistic increases in GCR rate were not as high as the ≈13,000-fold increase in GCR rate observed in a mec1 tel1 double mutant (Table 4 and Fig. 2), which presumably completely lacks DNA damage and S-phase checkpoint responses as well as possibly lacking full downstream effector activity caused by loss of basal levels of phosphorylation by these two kinases (10, 22, 50). The rfc5-1 mre11 double mutant and rad24 sgs1 mre11 triple mutant strains showed synergistic increases in the GCR rate that were as high or slightly higher than that seen in the mec1 tel1 double mutant. Because a mre11 mutant is not defective completely in either checkpoint response (19, 20) or suppression of GCRs (22, 53), these results are most consistent with the view that the MRE11–RAD50–XRS2 complex is critical for both the activity of the S-phase checkpoints and downstream repair events that suppress genome instability.
Table 8.
Effect of a mre11Δ mutation on GCR rates
| Relevant genotype | Wild type
|
mre11Δ
|
||
|---|---|---|---|---|
| Strain | GCR rate × 1010 | Strain | GCR rate × 1010 | |
| Wild type | 3615 | 3.5 (1)* | 3633 | 2,200 (629)* |
| rfc5-1 | 3727 | 660 (189)* | 4589 | 48,000 (13,714) |
| rad24Δ | 3723 | 40 (11)* | 4590 | 10,000 (2,857) |
| sgs1Δ | 3813 | 77 (22)† | 4591 | 2,700 (743) |
| pds1Δ | 3729 | 670 (190)* | 4592 | 18,000 (5,143) |
| mec1Δ sml1Δ | 3735 | 680 (194)* | 3761 | 29,000 (8,286)* |
| tel1Δ | 3731 | 2.0 (0.6)* | 3759 | 2,200 (629)* |
Discussion
In previous studies, we found that defects in the replication checkpoint caused increased GCR rates, whereas defects in G1 and G2 DNA damage checkpoint and spindle checkpoint did not cause increased GCR rates (22). The results presented here indicate that the intra-S DNA damage checkpoint functions to suppress GCRs induced by low doses of MMS. The intra-S checkpoint also functions to suppress spontaneous GCRs; however, because the RAD24 and SGS1 branches of the intra-S checkpoint are redundant (38), defects in only one branch cause little if any increase in GCR rate. The intra-S checkpoint also seems to be redundant with the replication checkpoint such that defects in both checkpoints result in much higher spontaneous GCR rates than individual defects. The major class of rearrangement observed in intra-S checkpoint-defective mutants was deletion of part of a chromosome arm combined with de novo addition of a telomere, similar to that seen for defects in the replication checkpoint (22). Both checkpoints seem to interact with MEC1- and TEL1-dependent signal-transduction pathways as well as effectors such as RAD53, DUN1, PDS1, and MRE11–RAD50–XRS2 albeit to different extents. MRE11–RAD50–XRS2 plays a particularly important role in suppressing GCRs, apparently being required for both the full activity of the S-phase checkpoints and downstream repair events (19, 20, 22, 26, 53). Overall, our analysis indicates there is extensive redundancy among S-phase checkpoints that function to suppress spontaneous genome instability, and the complete loss of these functions results in very large increases (12,000–14,000-fold) in the GCR rate.
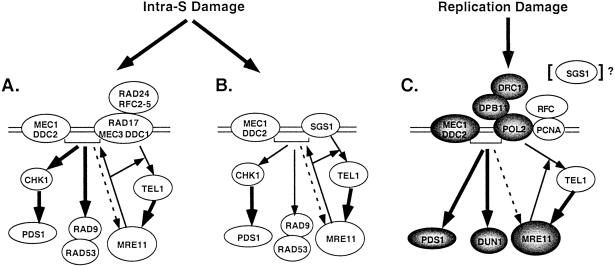
The data presented here along with the results of other studies are consistent with the following model of S-phase checkpoint function in the suppression of spontaneous GCRs (refs. 1, 19–22, and 61; Fig. 3). Three different sensor proteins or groups of sensor proteins can assemble independently at the site of DNA damage that occurs during DNA replication: (i) SGS1, (ii) RAD24–RFC2-5 and RAD17–MEC3–DDC1, and (iii) RFC1-5, DPB11, POL2, and probably others. These likely independently interact with the MEC1–DDC2 complex, which can bind to DNA damage by itself (48, 49), and TEL1; whether TEL1 is in a complex with other proteins and can bind to DNA damage is not known. These complexes interact with other components (i.e., signal) that are critical to suppression of GCRs (e.g., RAD53, PDS1, DUN1, and MRE11–RAD50–XRS2; refs. 1 and 3). With regard to the intra-S checkpoint branches, the RAD24 branch seems to act predominantly, but not completely, through MEC1, and the SGS1 branch seems to act predominantly, but not completely, through TEL1. This view is consistent with the following observations: (i) compared with a tel1 mutation, a mec1 mutation shows a larger synergistic interaction with a sgs1 mutation, (ii) a tel1 mutation shows synergistic interactions with mutations in RAD9, RAD17, RAD24, MEC3, and RAD53, whereas a mec1 mutation does not show a synergistic interaction with rad9, rad17, rad24, mec3, or rad53 mutations, and (iii) the GCR rate of the sgs1 rad53 and tel1 rad53 double mutants are essentially the same and significantly higher than that of the respective single mutants. The replication checkpoint involving RFC1-5, DPB11, POL2, and others seems to be the most important checkpoint for suppressing spontaneous GCRs, because mutations inactivating this checkpoint cause much larger increases in the GCR rate than mutations inactivating the intra-S checkpoint (22). The replication checkpoint predominantly acts through MEC1, although clearly TEL1 plays a redundant role (20, 22, 39–44). However, all the S-phase checkpoints cooperate in suppressing spontaneous genome instability as evidenced by the synergistic interactions observed between rad24, sgs1, and rfc5-1 mutations. Inactivation of all the checkpoints in a mec1 tel1 double mutant results in an extremely large increase in the GCR rate (22), although this probably is caused by the combined loss of the checkpoints and the function of critical effectors that are phosphorylated by these kinases in normally growing cells (10, 22, 50).
Figure 3.
Model for the action of S-phase checkpoints in the suppression of spontaneous genome instability. Three different checkpoints that respond to DNA damage (errors) that occur during DNA replication are illustrated. (A) The RAD24 intra-S checkpoint branch in which the MEC1–DDC2 complex and RAD17–MEC3–DDC1 complex (loaded by RAD24–RFC2-5) assemble onto DNA damage and then interact with RAD9–RAD53 and PDS1 through CHK1. Interaction with the RAD50–MRE11–XRS2 (MRE11) complex occurs through TEL1 in a MEC1-independent manner. The MRE11 complex appears to be required for the full activity of MEC1- and TEL1-dependent responses. (B) The SGS1 intra-S checkpoint branch, which is similar to that in A except that SGS1 substitutes for the RAD17–MEC3–DDC1 and RAD24–RFC2-5 complexes. (C) The replication checkpoint in which the MEC1–DDC2 complex and the POL2–DPB11–DRC1 complex and presumably PCNA, loaded by RFC, assemble onto replication damage and interact with PDS1 and DUN1. Interaction with the MRE11 complex occurs through TEL1 in a MEC1-independent manner. The MRE11 complex appears to be required for the full activity of TEL1-dependent but not MEC1-dependent responses. SGS1 also may play a minor role in this checkpoint. Within each illustrated checkpoint branch, the thick arrows indicate the major signaling pathways consistent with genetic interaction analysis, and the thin arrows indicate the less important signaling pathway. The dashed arrow indicates that MEC1 may only weakly interact with the MRE11 complex. The arrows from the MRE11 complex to the central checkpoint proteins indicate the cases in which MRE11 complex activity is required for full activity of the checkpoint. The shading indicates those genes in which single mutations cause major increases in genome instability, presumably through inactivation of the replication checkpoint.
Each of the checkpoints seems to interact with a unique set of effectors that plays roles in suppressing GCRs. DUN1 was suggested previously to act downstream of MEC1 in the replication checkpoint (8, 22, 70). Our previous study of RAD53 suggested it only played a minor role in suppression of GCRs (22). The results presented here on the analysis of double-mutant combinations of rad53 with tel1, sgs1, rad24, and other mutations indicate that RAD53 functions in the RAD24, MEC1 intra-S checkpoint branch in suppression of GCRs. These results are consistent with previous studies of the role of RAD53 in the intra-S DNA damage response but are different from previous results on the role of RAD53 in response to high concentrations of hydroxyurea (i.e., replication checkpoint; refs. 4, 5, and 70–72). PDS1 has two roles in these checkpoint responses: it is regulated by MEC1 to maintain sister-chromatid cohesion during S phase, and it prevents the onset of anaphase in response to the damage checkpoints and the spindle checkpoint (17, 18, 66, 67). Because pds1 mutations but not bub3, mad2, or mad3 mutations cause high rates of accumulation of GCRs (this study and ref. 22), it seems likely that the critical role of PDS1 in maintaining genome instability is its role in maintaining sister-chromatid cohesion (18) and not its role in preventing anaphase (66). The observation that a pds1 mutation shows additive or greater effects on the GCR rate when combined with rad24, rfc5-1, dun1, mec1, and tel1 mutations indicates that the S-phase checkpoints regulate other responses that are important for suppressing GCRs besides PDS1. The MRE11–RAD50–XRS2 complex is a critical player in suppressing GCRs (22, 53). It is required for full activity of MEC1- and TEL1-dependent phosphorylation in response to activation of the intra-S checkpoint and full activity of TEL1-dependent, MEC1-independent phosphorylation in response to activation of the replication checkpoint, and it is phosphorylated also by TEL1 (19, 20, 69, 73, 74). The observation that mutations that inactivate the MRE11–RAD50–XRS2 complex show synergistic interactions with rfc5-1, rad24, and mec1 mutations is consistent with the idea that the MRE11–RAD50–XRS2 complex is required for both the full activity of these checkpoints and downstream repair events (19, 20, 22, 26). That mre11 mutation shows a larger interaction with rfc5-1 as compared with rad24 may reflect that the rfc5-1 mutation causes defects in both the intra-S and replication checkpoints. That mre11 tel1 and sgs1 mre11 double mutants have the same GCR rate as a mre11 single mutant and the mre11 pds1 and pds1 tel1 double mutants have essentially the same GCR rate is consistent with the idea that the SGS1 pathway is predominantly TEL1-dependent, and MRE11–RAD50–XRS2 is a major target of TEL1.
Genome instability is characteristic of cancer cells (75–80). In previous studies, we have shown that the types of GCRs seen in the S. cerevisiae mutants studied here and elsewhere are similar to those seen in cancer cells (22, 53, 61, 64). In many cases, defects in the mouse and human genes that are homologs of the S. cerevisiae genes identified are associated with genome instability and/or cancer susceptibility (77, 79–87). In addition, the proteins encoded by the cancer susceptibility genes BRCA1 and BRCA2 either interact directly with proteins that function in the genome instability suppression pathways described or are phosphorylated by proteins that function in these pathways (24, 88, 89). The studies presented here have extended the results of previous studies by identifying additional genes that function in suppression of genome instability as well as by further identifying the extensive level of redundancy among the pathways that regulate genome stability. This high level of redundancy and the extremely large increases in genome instability that result when multiple pathways are inactivated indicate that there are high levels of spontaneous DNA damage in normal cells that can result in genome instability. Our results further suggest that if cancer cells acquire defects that result in increased genome instability, then there may be a large number of genes in which such defects can be found.
Acknowledgments
We thank Ellen Kats, Vincent Pennaneach, Chris Putnam, and Nobu Sasaki for comments on the manuscript and J. Weger and J. Green for DNA sequencing. This work was supported by National Institutes of Health Grant GM26017 (to R.D.K.) and a fellowship from the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation (to K.M.).
Abbreviations
- GCR
gross chromosomal rearrangement
- MMS
methyl methanesulfonate
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 2, 2000.
Contribued by Richard D. Kolodner, December 26, 2001
References
- 1.Michelson R J, Weinert T. BioEssays. 2000;22:966–969. doi: 10.1002/1521-1878(200011)22:11<966::AID-BIES2>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 2.Paulovich A G, Margulies R U, Garvik B M, Hartwell L H. Genetics. 1997;145:45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowndes N, Murguia J. Curr Opin Genet Dev. 2000;10:17–25. doi: 10.1016/s0959-437x(99)00050-7. [DOI] [PubMed] [Google Scholar]
- 4.Santocanale C, Diffley J F X. Nature (London) 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 5.Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Nature (London) 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 6.Hartwell L. Cell. 1992;71:543–546. doi: 10.1016/0092-8674(92)90586-2. [DOI] [PubMed] [Google Scholar]
- 7.Zhou B B, Elledge S J. Nature (London) 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Z, Elledge S J. Cell. 1993;75:1119–1127. doi: 10.1016/0092-8674(93)90321-g. [DOI] [PubMed] [Google Scholar]
- 9.Sun Z, Fay D F, Marini F, Foiani M, Stern D F. Genes Dev. 1996;10:295–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez Y, Desany B A, Jones W J, Liu Q, Wang B, Elledge S J. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 11.Brush G S, Morrow D M, Hieter P, Kelly T J. Proc Natl Acad Sci USA. 1996;93:15075–15080. doi: 10.1073/pnas.93.26.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emili A. Mol Cell. 1998;2:183–189. doi: 10.1016/s1097-2765(00)80128-8. [DOI] [PubMed] [Google Scholar]
- 13.Vialard J E, Gilbert C S, Green C M, Lowndes N F. EMBO J. 1998;17:5679–5688. doi: 10.1093/emboj/17.19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, Elledge S. Science. 1999;286:1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- 15.Clarke D J, Segal M, Mondesert G, Reed S I. Curr Biol. 1999;9:365–368. doi: 10.1016/s0960-9822(99)80163-8. [DOI] [PubMed] [Google Scholar]
- 16.Bashkirov V I, King J S, Bashkirova E V, Schmuckli-Maurer J, Heyer W-D. Mol Cell Biol. 2000;20:4393–4404. doi: 10.1128/mcb.20.12.4393-4404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Liu D, Wang Y, Qin J, Elledge S J. Genes Dev. 2001;15:1361–1372. doi: 10.1101/gad.893201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke D J, Segal M, Jensen S, Reed S I. Nat Cell Biol. 2001;3:619–627. doi: 10.1038/35083009. [DOI] [PubMed] [Google Scholar]
- 19.Usui T, Ogawa H, Petrini J H J. Mol Cell. 2001;7:1255–1266. doi: 10.1016/s1097-2765(01)00270-2. [DOI] [PubMed] [Google Scholar]
- 20.D'Amours D, Jackson S P. Genes Dev. 2001;15:2238–2249. doi: 10.1101/gad.208701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grenon M, Gilbert C, Lowndes N F. Nat Cell Biol. 2001;3:844–847. doi: 10.1038/ncb0901-844. [DOI] [PubMed] [Google Scholar]
- 22.Myung K, Datta A, Kolodner R D. Cell. 2001;104:397–408. doi: 10.1016/s0092-8674(01)00227-6. [DOI] [PubMed] [Google Scholar]
- 23.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 24.Zhong Q, Chen C F, Li S, Chen Y, Wang C C, Xiao J, Chen P L, Sharp Z D, Lee W H. Science. 1999;285:747–750. doi: 10.1126/science.285.5428.747. [DOI] [PubMed] [Google Scholar]
- 25.Cortez D, Wang Y, Qin J, Elledge S J. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 26.Petrini J H. Curr Opin Cell Biol. 2000;12:293–296. doi: 10.1016/s0955-0674(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 27.Khanna K K, Jackson S P. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 28.Moynahan M E, Pierce A J, Jasin M. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 29.Lydall D, Weinert T. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- 30.Weinert T A, Hartwell L H. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 31.Weinert T A, Hartwell L H. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naiki T, Shimomura T, Kondo T, Matsumoto K, Sugimoto K. Mol Cell Biol. 2000;20:5888–5896. doi: 10.1128/mcb.20.16.5888-5896.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimomura T, Ando S, Matsumoto K, Sugimoto K. Mol Cell Biol. 1998;18:5485–5491. doi: 10.1128/mcb.18.9.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kondo T, Matsumoto K, Sugimoto K. Mol Cell Biol. 1999;19:1136–1143. doi: 10.1128/mcb.19.2.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H-S, Brill S J. Mol Cell Biol. 2001;21:3725–3737. doi: 10.1128/MCB.21.11.3725-3737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.German J, Ellis N A. In: The Genetic Basis of Human Cancer. Vogelstein B, Kinzler K M, editors. New York: McGraw–Hill; 1998. [Google Scholar]
- 37.Moser M J, Shima J, Monnat J R J. Hum Mutat. 1999;13:271–279. doi: 10.1002/(SICI)1098-1004(1999)13:4<271::AID-HUMU2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 38.Frei C, Gasser S M. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 39.Araki H, Leem S H, Phongdara A, Sugino A. Proc Natl Acad Sci USA. 1995;92:11791–11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navas T A, Zhou Z, Elledge S J. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 41.Longhese M P, Fraschini R, Plevani P, Lucchini G. Mol Cell Biol. 1996;16:3235–3244. doi: 10.1128/mcb.16.7.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugimoto K, Shimomura T, Hashimoto K, Araki H, Sugino A, Matsumoto K. Proc Natl Acad Sci USA. 1996;93:7048–7052. doi: 10.1073/pnas.93.14.7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugimoto K, Ando S, Shimomura T, Matsumoto K. Mol Cell Biol. 1997;17:5905–5914. doi: 10.1128/mcb.17.10.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H, Elledge S J. Proc Natl Acad Sci USA. 1999;96:3824–3829. doi: 10.1073/pnas.96.7.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foiani M, Pellicioli A, Lepes M, Lucca C, Ferrari M, Liberi G, Muzi Falconi M, Plevani P. Mutat Res. 2000;451:187–196. doi: 10.1016/s0027-5107(00)00049-x. [DOI] [PubMed] [Google Scholar]
- 46.Elledge S J. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 47.Paciotti V, Clerici M, Lucchini G, Longhese M P. Genes Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- 48.Melo J A, Cohen J, Toczyski D P. Genes Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kondo T, Wakayama T, Naiki T, Matsumoto K, Sugimoto K. Science. 2001;294:867–870. doi: 10.1126/science.1063827. [DOI] [PubMed] [Google Scholar]
- 50.Morrow D M, Tagle D A, Shiloh Y, Collins F S, Hieter P. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 51.Greenwell P W, Kronmal S L, Porter S E, Gassenhuber J, Obermaier B, Petes T D. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- 52.Lee S E, Moore J K, Holmes A, Umezu K, Kolodner R D, Haber J E. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 53.Chen C, Kolodner R D. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- 54.Sidorova J M, Breeden L L. Genes Dev. 1997;11:3032–3045. doi: 10.1101/gad.11.22.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kato R, Ogawa H. Nucleic Acids Res. 1994;22:3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weinert T A, Kiser G L, Hartwell L H. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 57.Ritchie K B, Mallory J C, Petes T D. Mol Cell Biol. 1999;19:6065–6075. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Craven R J, Petes T D. Mol Cell Biol. 2000;20:2378–2384. doi: 10.1128/mcb.20.7.2378-2384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Corda Y, Schramke V, Longhese M P, Smokvina T, Paciotti V, Brevet V, Gilson E, Geli V. Nat Genet. 1999;21:204–208. doi: 10.1038/5991. [DOI] [PubMed] [Google Scholar]
- 60.Longhese M P, Paciotti V, Neecke H, Lucchini G. Genetics. 2000;155:1577–1591. doi: 10.1093/genetics/155.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Myung K, Chen C, Kolodner R D. Nature (London) 2001;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- 62.Lundblad V. Mutat Res. 2000;451:227–240. doi: 10.1016/s0027-5107(00)00052-x. [DOI] [PubMed] [Google Scholar]
- 63.Zhou J-Q, Monson E K, Teng S-C, Schulz V P, Zakian V A. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 64.Myung K, Datta A, Chen C, Kolodner R D. Nat Genet. 2001;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- 65.Yamamoto A, Guacci V, Koshland D. J Cell Biol. 1996;133:85–97. doi: 10.1083/jcb.133.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Amon A. Curr Opin Genet Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]
- 67.Taylor S S. Curr Biol. 1999;9:562–564. doi: 10.1016/s0960-9822(99)80355-8. [DOI] [PubMed] [Google Scholar]
- 68.Haber J E. Cell. 1998;95:583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- 69.Ritchie K B, Petes T D. Genetics. 2000;155:475–479. doi: 10.1093/genetics/155.1.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gardner R, Punam C W, Weinert T. EMBO J. 1999;18:3173–3185. doi: 10.1093/emboj/18.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon C S, Foiani M. Nature (London) 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 72.Tercero J A, Diffley J F X. Nature (London) 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 73.Boulton S J, Jackson S P. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mallory J C, Petes T D. Proc Natl Acad Sci USA. 2000;97:13749–13754. doi: 10.1073/pnas.250475697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kolodner R. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 76.Jiricny J. EMBO J. 1998;17:6427–6436. doi: 10.1093/emboj/17.22.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kinzler K W, Vogelstein B. Science. 1998;280:1036–1037. doi: 10.1126/science.280.5366.1036. [DOI] [PubMed] [Google Scholar]
- 78.Lengauer C, Kinzler K W, Voelstein B. Nature (London) 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 79.Coleman W B, Tsongalis G J. Anticancer Res. 1999;19:4645–4664. [PubMed] [Google Scholar]
- 80.Vessey C J, Norbury C J, Hickson I D. Prog Nucleic Acid Res Mol Biol. 1999;63:189–221. doi: 10.1016/s0079-6603(08)60723-0. [DOI] [PubMed] [Google Scholar]
- 81.Shiloh Y. Anuu Rev Genet. 1997;31:635–662. doi: 10.1146/annurev.genet.31.1.635. [DOI] [PubMed] [Google Scholar]
- 82.Varon R, Vissinga C, Platzer M, Cerosaletti K M, Chrzanowska K H, Saar K, Beckmann G, Seemanova E, Cooper P R, Nowak N J, et al. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 83.Carney J P, Maser R S, Olivares H, Davis E M, Le Buau M, Yates III J R, Hays L, Morgan W F, Petrini J H. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 84.Stewart G S, Maser R S, Stankovic T, Bressan D A, Kaplan M I, Jaspers N G J, Raams A, Byrd P J, Petrini J H J, Taylor A M R. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 85.Lavin M F, Khanna K K. Int J Radiat Biol. 1999;75:1201–1214. doi: 10.1080/095530099139359. [DOI] [PubMed] [Google Scholar]
- 86.Bell D W, Varley J M, Szydlo T E, Kang D H, Wahrer D C, Shannon K E, Lubratovich M, Verselis S J, Isselbacher K J, Fraumeni J F, et al. Science. 1999;286:2528–2531. doi: 10.1126/science.286.5449.2528. [DOI] [PubMed] [Google Scholar]
- 87.Brown E J, Baltimore D. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 88.Sharan S K, Bradley A. J Mammary Gland Biol Neoplasia. 1998;3:413–421. doi: 10.1023/a:1018788132560. [DOI] [PubMed] [Google Scholar]
- 89.Scully R. Breast Cancer Res. 2000;2:324–330. doi: 10.1186/bcr76. [DOI] [PMC free article] [PubMed] [Google Scholar]