Abstract
HIV-1 entry into cells is mediated by the envelope glycoprotein receptor-binding (gp120) and membrane fusion-promoting (gp41) subunits. The gp41 heptad repeat 1 (HR1) domain is the molecular target of the fusion-inhibitor drug enfuvirtide (T20). The HR1 sequence is highly conserved and therefore considered an attractive target for vaccine development, but it is unknown whether antibodies can access HR1. Herein, we use gp41-based peptides to select a human antibody, 5H/I1-BMV-D5 (D5), that binds to HR1 and inhibits the assembly of fusion intermediates in vitro. D5 inhibits the replication of diverse HIV-1 clinical isolates and therefore represents a previously unknown example of a crossneutralizing IgG selected by binding to designed antigens. NMR studies and functional analyses map the D5-binding site to a previously identified hydrophobic pocket situated in the HR1 groove. This hydrophobic pocket was proposed as a drug target and subsequently identified as a common binding site for peptide and peptidomimetic fusion inhibitors. The finding that the D5 fusion-inhibitory antibody shares the same binding site suggests that the hydrophobic pocket is a “hot spot” for fusion inhibition and an ideal target on which to focus a vaccine-elicited antibody response. Our data provide a structural framework for the design of new immunogens and therapeutic antibodies with crossneutralizing potential.
Keywords: envelope, fusion, prehairpin, vaccine
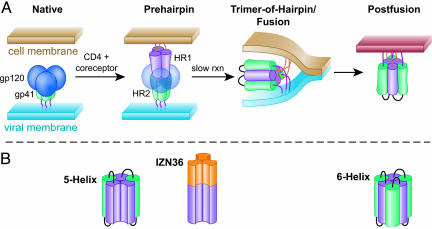
HIV and other enveloped viruses enter host cells by promoting fusion of the viral membrane with a host cell plasma or endosomal membrane, thus delivering the viral core into the host cell cytoplasm. For HIV, the viral envelope glycoprotein that drives membrane fusion is composed of a trimer of gp120:gp41 heterodimers. Binding of gp120 to CD4 and a coreceptor initiates a series of conformational changes in gp41, exposing the prehairpin intermediate and culminating in formation of a six-helical bundle, or trimer-of-hairpins, conformation (1) (Fig. 1A). The six-helical bundle is formed from three gp41 protomers, each of which contributes one heptad repeat (HR) 1 (HR1) segment and one HR2 segment. Three HR1 segments form a three-stranded coiled-coil, and three HR2 segments pack around the HR1 core in an antiparallel orientation (2-4). Formation of the six-helical bundle is critical for driving membrane fusion (5).
Fig. 1.
Working model of HIV entry pathway and gp41 conformational intermediates. (A) Schematic diagram of gp41 function during HIV entry, adapted from Chan and Kim (1). The gp41 HR1 and HR2 regions are depicted in magenta and green, respectively. (B) Schematic diagrams of the synthetic gp41 HR1 mimetics IZN36, 5H, and six-helix. The HR1 and HR2 regions are colored as in A. Synthetic linker sequences in 5H and six-helix are shown in black, and the synthetic IZ leucine zipper is shown in orange.
Extensive amino acid sequence variability and a continuously shifting antigenic surface of the HIV-1 glycoprotein (6-8) present formidable challenges to the development of effective humoral immunity. However, the sequences of the HR1 and HR2 regions are significantly less variable, probably because they are a critical driver of viral membrane fusion and are exposed only transiently during this process. Despite their transient exposure, HR1 and HR2 can be accessed by small proteins and peptides that bind the prehairpin intermediate and disrupt fusion by preventing subsequent folding into a six-helical bundle. Examples of such inhibitors include cyclic d-peptides that bind to HR1 (9) and synthetic peptides [e.g., IQN17 (10)] and designed proteins [e.g., five-helix, hereafter referred to as 5H (11)] that bind to HR2. Importantly, the prehairpin intermediate is a clinically validated target for the antiretroviral drug enfuvirtide (T20, DP-178) (12), a 36-aa synthetic peptide that binds to HR1 and inhibits HIV entry.
To test the possibility that antibodies directed to the prehairpin intermediate could also block HIV infection, we have used synthetic peptides and proteins as immunogens to select human monoclonal antibodies specifically targeted to HR1 in vitro. Here, we provide proof of concept that a human monoclonal antibody directed at these conserved structures can block HIV-1 infection and characterize this antibody-antigen interaction to define an epitope in HIV-1 gp41 with the potential for eliciting broadly neutralizing antibodies.
Materials and Methods
Peptides and Proteins. T20, C34, the severe acute respiratory syndrome (SARS) coronavirus (CoV) N3/N6 control peptide (13), IZN36, IQN17, IZN17, and the Ala-scan mutants were produced by standard Fmoc solid-phase methods (14). 5H was expressed and purified by a published procedure (11) modified as described in Supporting Text, which is published as supporting information on the PNAS web site.
In Vitro Isolation of 5H/I1-BMV-D5 (D5) Single-Chain Variable Region Fragment (scFv). The selection strategy designed to isolate cross-specific scFvs from large naive scFv libraries (Fig. 6, which is published as supporting information on the PNAS web site) was based upon methods described (15). Phage supernatants were screened by bacteriophage ELISA as described (16, 17), where the biotinylated forms of 5H and IZN36 were immobilized onto 96-well ABGene, Surrey, U.K., streptavidin plates. For viral neutralization assays, immobilized metal ion affinity chromatography-purified soluble scFv fragments were prepared by using standard methods (18).
Antiviral Assays. IMAC-purified scFvs were tested in the HIV reporter particle (HIVRP) assay essentially as described (19). Measurement of HIV infection of p4-2/R5 cells by using a chemiluminescent β-galactosidase substrate was done as described (20). BaL and HXB2 were purchased from Advanced Biotechnologies (Columbia, MD); 89.6 was grown in peripheral blood mononuclear cells, and vesicular stomatitis virus-G-pseudotyped HIV was made by transfection as described (21). The luciferase-based pseudotyped viral neutralization assay was done as described (7). In brief, envelope genes were amplified by PCR, cloned into an expression vector, and cotransfected with a proviral plasmid to generate pseudotyped luciferase-encoding viruses. Viruses were used to infect U87/CD4/CXCR4/CCR5 cells in the presence of varying amounts of inhibitors. Luciferase production was measured 72 h after infection and IC50s calculated as described (7).
AlphaScreen-Based Peptide/D5 Interaction Assays. An AlphaScreen detection kit (PerkinElmer) was used to measure binding. Biotinylated peptides (5H, IZN36, IZN17, or IQN17) were bound to streptavidin-conjugated donor beads, and D5 IgG was bound to Protein A-conjugated acceptor beads. Beads were mixed in the presence or absence of competitors, incubated overnight at room temperature, and analyzed on a Fusion α-FP HT instrument (Perkin-Elmer), as suggested by the manufacturer. Six-helical bundle formation was measured by using the peptide C34-HA (22). Serial dilutions of inhibitors (D5-IgG1, C34, C34AAA, and 2F5) were preincubated with biotinylated 5H (final concentration, 10 nM) for 40 min at room temperature, then C34-HA was added to a final concentration of 3.3 nM along with AlphaScreen beads for detection of HA-tagged proteins (Amersham Pharmacia) and read on the Fusion instrument.
Results
In Vitro Selection of a Human HIV-Neutralizing Antibody. We selected human-derived scFvs from phage display libraries by binding to IZN36 and 5H, antigens designed to mimic HR1 as it may exist in the prehairpin intermediate (Fig. 1). IZN36 is a homotrimeric peptide in which 36 amino acids of HR1 are fused to a stable coiled-coil peptide (IZ) to yield a soluble discrete trimeric form of the HR1 three-stranded coiled-coil in the absence of HR2 (10). In 5H, the three-stranded HR1 core is associated with two bound HR2 peptides, presenting a single binding site for HR2 (11).
As a source of antibodies, we used large diverse well characterized libraries of bacteriophage bearing scFvs derived from normal human B cells (15). From a starting population of ≈1011 independent scFv-displaying bacteriophage, a total of 481 target-specific scFvs were obtained after two rounds of sequential selection for binding to biotinylated forms of 5H and IZN36 (schematic shown in Fig. 6). Nucleotide sequencing identified 100 unique sequences within this population of 481 scFvs.
Using the HIVRP assay (19), we screened purified scFvs produced from 5H/IZN36-binding bacteriophage and identified an scFv that blocks HIV entry. The HIVRP assay relies on incorporation of β-lactamase into infectious HIV particles so that fusion of the viral and cellular membranes delivers β-lactamase into the target cell, where it is detected by using a cell-permeant fluorescent β-lactamase substrate. This assay is particularly well suited to screening scFvs, which are inherently less durable than IgGs, because it requires only a 3- to 4-h 37°C incubation of viral particles with cells to allow viral entry.
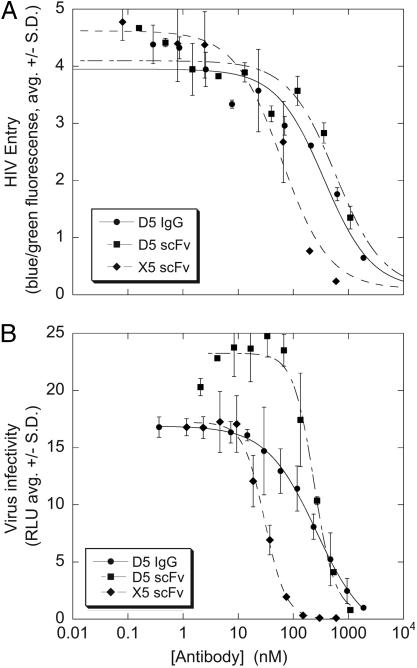
One scFv, designated 5H/I1-BMV-D5 (hereafter referred to as D5), specifically inhibited the HIVRP assay in a dose-dependent manner (Fig. 2A). The HIVRP assay was inhibited by the anti-gp120 scFv X5 (Fig. 2A) but not by the fluorescein-specific scFv COLIN (data not shown), thus confirming the specificity. The D5 scFv also blocked HIV infection in a single-cycle infectivity assay (20) (Fig. 2B), indicating that D5 can inhibit HIV entry in multiple assay formats.
Fig. 2.
Antiviral activity of D5. (A) D5 scFv and IgG1 inhibit HIV entry in the HIVRP assay. The blue/green fluorescence ratio is proportional to virus entry. X5 scFv is shown as a control. (B) D5 scFv and IgG1 inhibit HIV infectivity in a single-cycle infection assay. X5 scFv is shown as a control.
Previous reports found that the HIV-neutralizing activity of the X5 scFv was dramatically reduced upon conversion to an IgG, presumably because the larger IgG could not gain access to its binding site (23). Unlike X5, the human IgG1 form of D5 retained antiviral activity against HIV in both the HIVRP (Fig. 2A) and a single-cycle infectivity (Fig. 2B) assay with potency similar to the scFv (for infectivity assay, scFv IC50 = 240 nM, IgG1 IC50 = 260 nM). However, in the infectivity assay, the inhibition curves by the monovalent scFv reproducibly display a Hill slope of ≈2, whereas the bivalent IgG1 reproducibly displays a Hill slope of ≈1 (note differences in curve shape in Fig. 2). The reason for this difference is unknown but may be related to differences in binding valence or molecular size.
Broader testing of additional HIV isolates in the same single-cycle infectivity assay showed that D5-IgG1 also neutralized the laboratory isolates NL4-3 and MN-1 along with the primary isolates BaL (R5) and 89.6 (X4/R5) (Table 1). These findings distinguish the neutralization activity of D5-IgG1 from that of X5-IgG1, which showed poor activity against isolates other than HXB2 (23). Importantly, D5-IgG1 did not block infectivity of HIV pseudotyped with the vesicular stomatitis virus-G protein (Table 1), supporting the conclusion that the antiviral target of D5-IgG1 is the HIV envelope glycoprotein.
Table 1. Antiviral activity of D5.
| Viral envelope | D5 lgG1 lC50, μg/ml | D5 lgG1 lC50, nM |
|---|---|---|
| Hxb2 | 46.5 | 310 (n = 6) |
| BaL | 14 | 93 (n = 4) |
| 89.6 | 262 | 1750 (n = 2) |
| MN-1 | 59 | 393 (n = 4) |
| NL4-3 | 34 | 226 (n = 1) |
| VSVG | Not active | Not active |
Experiments were performed as described in the legend to Fig. 4. IC50s represent the average of the indicated number of determinations (n = x).
D5 Binds in a Highly Conserved gp41 HR1 Hydrophobic Pocket Critical for Six-Helical Bundle Formation. Initial mapping by AlphaScreen and surface plasmon resonance-binding assays identified the gp41 target of D5-IgG1 as the C-terminal half of the HR1 segment. As expected, D5-IgG1 bound to biotinylated forms of 5H and IZN36, the peptides used to select this antibody (data not shown). D5-IgG1 also bound peptides IZN17 and IQN17, which contain only the C-terminal 17 amino acids of the HR1 segment fused to different trimerization domains (Table 3, which is published as supporting information on the PNAS web site) but did not bind to peptides representing the SARS-CoV S protein HR1 segment (13) (data not shown). Surface plasmon resonance analysis showed that D5-IgG1 binds 5H, IZN36, and IZN17 with approximately the same affinity (Kd = 0.26, 0.17, and 0.1 nM, respectively). Collectively, these findings indicate the D5 epitope resides in the C-terminal (N17) half of HR1.
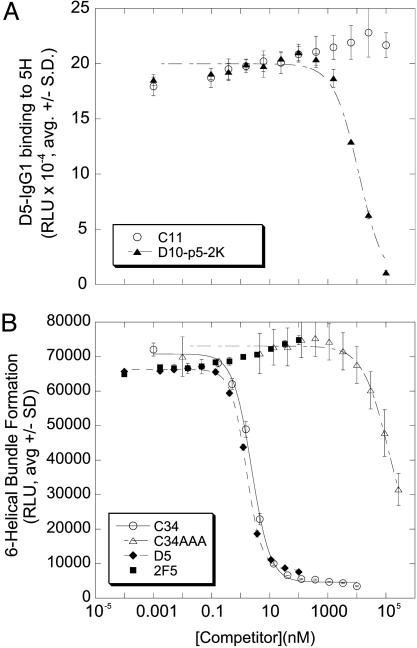
In the trimer-of-hairpins (postfusion) gp41 structure (3), the HR1 N17 region includes a deep cavity, known as the hydrophobic pocket, which provides an important set of contacts for the cognate HR2 and was proposed to be a potential drug target for HIV-1 fusion inhibitors (9, 24). To test whether this hydrophobic pocket is involved in D5 binding, we used a specific pocket-binding cyclic d-peptide, D10-p5-2K (9), as a probe. D10-p5-2K blocked the binding of D5-IgG1 to 5H, whereas a nonbinding linear control peptide (C11) did not block D5-IgG1 binding to 5H (Fig. 3A). This finding indicates that the D5 antibody epitope overlaps the HR1 hydrophobic pocket.
Fig. 3.
D5 binds the hydrophobic pocket and inhibits gp41 six-helical bundle formation in vitro. (A) The pocket-binding cyclic d-peptide D10-p5-2K (closed squares) or the control peptide C11 (closed triangles) were tested as competitors in a biotin-5H/D5-IgG1-binding assay. (B) D5-IgG1, 2F5, C34, and C34AAA were tested as competitors in a biotin-5H/C34-HA-binding assay.
Based on the binding data, we hypothesized that D5 inhibits HIV entry by binding to gp41 and preventing six-helical bundle formation in a manner analogous to T20. To test this hypothesis, we devised a homogeneous binding assay that measures binding of an epitope-tagged HR2 peptide (C34-HA) to a biotinylated form of 5H. Inhibitors of six-helical bundle formation should block binding of C34-HA to 5H, and indeed untagged C34 blocked binding in a dose-dependent manner. Neither a mutant C34 peptide (C34AAA) in which three critical amino acids (W628, W631, and I635) (24) were changed to alanine nor a SARS-CoV-derived HR2 peptide with similar size and isoelectric point were effective inhibitors, confirming the specificity of competition (Fig. 3B and data not shown). D5-IgG1 blocked six-helix bundle assembly with IC50 ≈1 nM, but the human IgG1 2F5, which binds to an epitope on gp41 not present in either 5H or C34-HA, did not inhibit at 100-fold higher concentrations (Fig. 3B). These results support a model in which D5-IgG1 inhibits HIV entry by preventing six-helical bundle formation in a manner analogous to HR2 peptides such as C34 and T20.
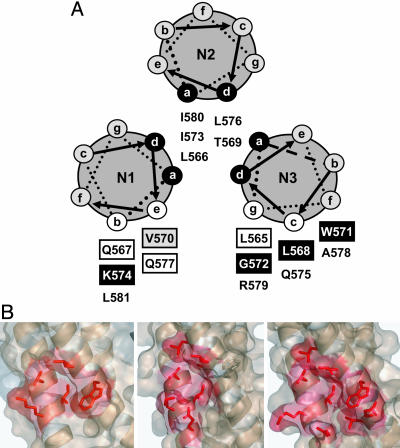
To identify the amino acids of the hydrophobic pocket that form the D5 epitope, a series of IZN17 mutant peptides was tested for the ability to block D5-IgG1 binding to biotinylated IQN17. Each amino acid in IZN17 except the heptad repeat a and d positions, which form the trimerization interface (Fig. 4A), was mutated to alanine (Table 3). CD studies showed that all mutant peptides were fully helical and had Tm >90°C (data not shown) and Tm similar to IZN17 when tested in 2 M guanidine hydrochloride (Table 4, which is published as supporting information on the PNAS web site). Thus, any reduction in inhibitory potency resulting from alanine substitution could be safely interpreted as a loss of antibody binding.
Fig. 4.
Mapping the D5 epitope. (A) Results of IZN17 alanine-scanning mutagenesis are summarized in a coiled-coil helical wheel representation of IZN17. Residues forming the hydrophobic pocket (3) are boxed; shading indicates the effect of alanine substitution on D5-IgG1 binding: white, mutant ≈ WT; gray, mutant < WT; black, mutant ≪ WT (Table 4). (B) Comparison of binding surfaces on 5H occupied by D5 (Left), GCN4-gp41/C7Mn34Mn42 [Center; Protein Data Bank (PDB) ID code 1FAV] (33), and IQN17/D10-p1 (Right, PDB ID code 1CZQ) (9). The D5 surface was modeled based on binding and NMR data, with side chains positioned as in the uncomplexed 5H. NMR showed that the L568, W571, and G572 residues contacted by D5 are located on N3. We have modeled the K574 contact residue on N1, although NMR could not distinguish whether this K574 is on N1 or N3.
When tested as competitors of IQN17/D5-IgG1 binding, peptides with alanine substitutions at positions L568, W571, and K574 were completely ineffective (IC50 >1,000 nM), indicating that these three residues are critical components of the D5 epitope (Fig. 4A, Table 4). The V570A mutant competed for D5 binding less effectively than wild-type IZN17, suggesting that this residue may represent an additional, minor contact point for D5-IgG1. As expected, the critical residues for D5 binding are all located in the hydrophobic pocket (Fig. 4A) (3). Accordingly, mutant G572D, which positions an Asp residue in the bottom of the hydrophobic pocket, was also inactive.
We used NMR spectroscopy to determine whether all of the contact residues map to one or more N helices. Having assigned the relevant resonances of isolated 5H (M.M., F. Talamo, L. Orsatti, and G. B., unpublished work), we monitored those residues whose chemical shift changed upon addition of D5-IgG1 (Figs. 7-9, which are published as supporting information on the PNAS web site). The magnitude of the observed changes was small but sufficient to allow unambiguous definition of an asymmetric interaction surface composed of residues I573 and V570 on helix N1, and residues L568, G572, and W571 on helix N3. A smaller chemical-shift change was observed for N3-L565, likely due to propagation of the perturbation rather than to a direct interaction based on L565A-binding data (Table 4). Because our analysis was limited to chemical shifts of amide, aromatic, and methyl groups, which could be unequivocally assigned, the contribution of K574 to binding could not be confirmed by NMR. Nevertheless, NMR defined the D5 interaction surface as a conformational epitope overlapping the hydrophobic pocket (Fig. 4B).
The Amino Acids Critical for D5 Binding Are Highly Conserved Among HIV Isolates. Using the Los Alamos National Laboratory HIV sequence database (25), we extracted all sequences spanning the N17 region but removed any sequences with errors, insertions, or deletions, leaving 5,326 sequences in total. Only 4.9% (259 of 5,326) of these sequences varied at one or more of the four residues (L568, V570, W571, or K574) implicated in D5 binding. Most (216 of 259 sequences, 83%) of these variant sequences had the dominant substitution K574R, 98% of which were from group O isolates (Table 5, which is published as supporting information on the PNAS web site). These observations show that the D5 epitope is highly conserved in HIV-1 and suggest that the D5 antibody may have a broad neutralization profile across HIV-1 group M viruses.
The gp41 HR1 Hydrophobic Pocket Is the Target of D5 Antiviral Activity. To confirm that D5-IgG1 blocks HIV infection by binding to the gp41 HR1 region, we changed each D5-binding residue to alanine in an infectious proviral clone. Although L568 and K574 are nearly invariant, viruses with an alanine substitution at either residue were viable, albeit with reduced specific infectivity in a single-cycle assay (data not shown). Viruses with substitution of W571 were essentially noninfectious.
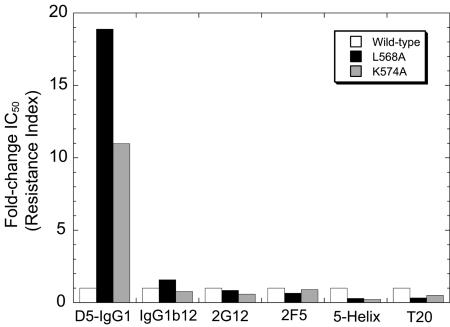
Viruses containing substitutions at either L568 or K574 were resistant to D5-IgG1: L568A and K574A conferred ≈19- and ≈11-fold resistance, respectively (Fig. 5). However, these mutants were not globally resistant to entry inhibitors, because they were as sensitive as wild-type virus to neutralizing antibodies IgG1b12, 2G12, and 2F5, and both mutants were somewhat sensitized to two inhibitors of six-helical bundle formation, 5H and T20. Because T20 potency is sensitive to fusion kinetics (26), these results rule out the trivial possibility that the L568A and K574A mutations simply confer D5-IgG1 resistance by accelerating fusion kinetics. Rather, these findings constitute additional evidence that the gp41 HR1 hydrophobic pocket is the antiviral target of D5-IgG1.
Fig. 5.
Amino acid changes in gp41 HR1 confer resistance to D5-IgG1. Amino acid residues L568 and K574 in gp41 were changed to alanine in the proviral clone R8.HXB2 and virus stocks were produced by transfection. The antiviral potencies of the indicated entry inhibitors against wild-type and mutant viruses were assessed by using a single-cycle infection assay. Fold-change IC50 refers to the IC50 of the test virus divided by the IC50 of the wild-type virus; fold change > 1 indicates resistance, fold change < 1 indicates hypersensitivity.
D5-IgG1 Neutralizes Diverse HIV Isolates. The high sequence conservation of the D5 epitope suggests that D5 might be able to neutralize a broad array of HIV isolates. When tested in a commercial assay for neutralizing antibodies (ViroLogic, South San Francisco, CA), D5-IgG1 blocked infection of diverse HIV isolates but did not neutralize HIV pseudotyped with the amphotrophic murine leukemia virus envelope (Table 2). In this analysis, D5-IgG1 neutralized 9 of 19 viruses tested. We note that the BaL envelope used in this assay was insensitive to D5 inhibition, whereas the BaL stock used in our single-cycle infectivity assay (Table 1) was sensitive; the reason for this discrepancy is unknown but may be related to different origins of the BaL stocks or different assay protocols. Sensitive viruses included examples from subtypes B, C, and F, as well as CRFs AE and BF. D5-IgG potency against various HIV isolates varied considerably (Tables 1 and 2). In principle, potency differences could arise from differences in the D5 epitope sequences. However, that cannot be the only explanation for differential sensitivity, as the HR1 sequences of MN (D5-IgG1 IC50 = 393 nM) and 89.6 (D5-IgG1 IC50 = 1,750 nM) are identical (Table 1; sequence data not shown). This is likely an example of indirect resistance, which can occur for a variety of reasons such as differences in coreceptor affinity and fusion kinetics (26). In comparison, the well characterized broadly neutralizing antibodies IgG1b12 and 2F5 neutralized 11/19 and 14/19 isolates, respectively. IgG1b12 and 2F5 are much more potent than D5-IgG1 (Table 2). However, D5-IgG1 neutralized isolates that were not neutralized by IgG1b12 (e.g., isolate 93BR029) or 2F5 (e.g., isolate 21068) at the highest concentrations tested. Overall, the range of isolates neutralized by D5-IgG1, albeit with lower potency, exceeds that of type-specific antibodies and approaches the range of isolates inhibited by the “broadly neutralizing” antibodies 2F5 and IgG1b12.
Table 2. D5 neutralizes diverse HIV isolates.
| Viral envelope
|
IC50, nM
|
|||||
|---|---|---|---|---|---|---|
| Subtype | C34 | T20 | IgG1b12 | 2F5 | D5-IgG1 | |
| 1168 | B | 17 | 54 | >667 | 80 | >2,333 |
| 21068 | C | 2.5 | 43 | 18 | >667 | 766 |
| 92RW008 | A | 3.3 | 14 | 57 | 39 | >2,333 |
| 92UG005 | D | 10 | 11 | >667 | 42 | >2,333 |
| 92UG031 | A | 11 | 38 | >667 | 26 | >2,333 |
| 93BR029 | F | 5 | 20 | >667 | 9.8 | 546 |
| 94KE105 | AC | 3 | 17 | >667 | >667 | >2,333 |
| 97ZA012 | C | 3.1 | 33 | >667 | >667 | 1175 |
| 98CN009 | C | 0.6 | 7.4 | 6.6 | >667 | 963 |
| 98INO22 | C | 3.4 | 13 | 6.0 | >667 | >2,333 |
| BAL | B | 2.0 | 7.6 | 0.4 | 17 | >2,333 |
| CMU02 | AE | 40 | 27 | 54 | 5.0 | 1886 |
| HXB2r | B | 0.45 | 3.0 | 0.04 | 0.5 | 1588 |
| JRFL | B | 36 | 25 | 0.2 | 40 | >2,333 |
| SF162 | B | 14 | 43 | 0.09 | 8.9 | >2,333 |
| VLGCBF2 | BF | 1.5 | 1.5 | >667 | 96 | 918 |
| VLGCJ1 | J | 6.7 | 2.3 | >667 | 33 | >2,333 |
| JRCSF | B | 6.5 | 10 | 1.6 | 23 | 1,385 |
| NL43 | B | 1.6 | 66 | 0.4 | 10.0 | 922 |
| aMLV | N/A | >8,828 | >5,618 | >667 | >667 | >2,333 |
Five entry inhibitors (C34, T20, IgG1b12, 2F5, and D5-IgG1) were titrated in the ViroLogic PhenoSense Entry assay by using HIV test viruses pseudotyped with the indicated envelopes. IC50s are shown; IC50 > x indicates that there was <50% inhibition at x nM, the highest concentration tested.
N/A, not applicable.
Discussion
The suggestion that gp41 may undergo a major conformational change in a manner analogous to the influenza fusion protein (2, 27) and the first x-ray structures of gp41 (3, 4) together suggested a model for gp41 function (1). This model in turn spawned the idea that gp41 conformational intermediates might serve as targets for HIV-neutralizing antibodies (9). These intermediates have nevertheless proved very difficult targets: despite reports of polyclonal neutralizing antibodies possibly directed at such structures (28, 29), until now there have been no examples of a defined antibody that unambiguously blocks normal HIV infection by binding to the gp41 prehairpin intermediate. Some have speculated that the HR1 coiled-coil region may not be accessible to proteins as large as an IgG (30), and some data may bolster that view (31).
We have now shown that an epitope overlapping the HR1 hydrophobic pocket can be accessed by a human IgG1 molecule, and that D5-IgG1 binding to that epitope can block HIV infection. We have reached this conclusion by a series of experiments that include: (i) Observation of direct binding of D5 to the C-terminal 17 amino acids of HR1; (ii) inhibition of D5 binding by peptides that occupy the hydrophobic pocket; (iii) disruption of D5-IgG1 binding to N17 by mutation of pocket-forming residues; (iv) observation of D5-IgG1 binding to the hydrophobic pocket of 5H by 2D NMR; and (v) demonstration that mutation of key pocket residues of gp41 on HIV virions confers resistance to neutralization by D5.
The gp41 hydrophobic pocket was previously proposed as an important target to inhibit viral fusion (24), a concept subsequently confirmed by several studies. In one study, cyclic d-peptides identified by mirror-image phage display blocked HIV entry and were shown by x-ray crystallography to bind the hydrophobic pocket (9). The most potent peptide in this series, D10-p5-2K, competes with D5 mAb for binding to 5H (Fig. 2B), and the surface of IQN17 in contact with another member of this peptide series (D10-p1) is shown in Fig. 4B Right (9). In a second study, constrained peptides of similar size, but featuring the natural pocket-binding sequence of HR2, were also shown to inhibit HIV-1 entry, and an x-ray crystal costructure confirmed that the peptides bound to the hydrophobic pocket (32). In a third study, inhibitors were selected from a biased combinatorial library of nonnatural binding elements fused to a peptide corresponding to HR2 amino acids 636-653, residues immediately adjacent to the pocket-binding residues (33). The best binders were able to inhibit cell fusion mediated by HIV envelope glycoproteins, and the x-ray structure of one compound (C7Mn34Mn42) showed that it bound to the same surface on the HR1 trimer (Fig. 4B Center) (34).
The D5-binding surface on gp41 HR1 as defined in the present study is depicted in Fig. 4B Left. Comparison with the surface bound by the above inhibitors shows a striking conservation of the core-binding residues. The difference observed in the shape of the three complexes is consistent with the notion that protein-binding sites can be highly adaptive, the specific shape and size of the contact surface being defined by the ligand (35). Although we cannot formally extend this comparison to small-molecule inhibitors, for lack of structural data, several reports have appeared of compounds putatively targeting the hydrophobic pocket and displaying fusion-inhibitory activity.
We conclude that the hydrophobic pocket in the inner coil of gp41, which was already recognized as a common binding solution for peptide (9) and peptidomimetic (32-34) fusion inhibitors, also represents the binding site for a neutralizing antibody. A small complementary set of contact residues that contributes the majority of the binding energy within a larger protein-protein interface is increasingly being recognized in a variety of protein-protein interactions and has been termed a “hot spot” (36). The privileged nature of the gp41 HR1 hydrophobic pocket qualifies it as a “hot spot” for fusion inhibition.
D5-IgG1 inhibits infectivity of a diverse range of HIV isolates (Table 2), including viruses from different subtypes and viruses with reduced sensitivity to other entry inhibitors such as T20 and the broadly neutralizing antibodies IgG1b12 and 2F5. However, D5 IgG is much less potent than the latter antibodies. Indeed, because of the lower potency, we may have underestimated the breadth of isolates that are sensitive to it, because there is a limit to the concentration of antibody achievable in the antiviral assays. In this study, the peptide entry inhibitor T20 neutralized all of the 19 viruses tested. Because D5 IgG inhibits viral fusion by a T20-like mechanism and targets a conserved epitope, it seems reasonable to expect a wider neutralization profile than the current data suggest. To address this question, as well as to answer questions important for vaccine development, we are attempting to select more potent neutralizing antibodies directed at this epitope using modern in vitro antibody evolution methods (37). Given the extremely conserved nature of the D5 epitope, an extraordinarily potent variant of D5, if identified, may represent an attractive therapeutic agent for the treatment of HIV infection.
By a variety of criteria, D5 is different from all other broadly neutralizing anti-HIV monoclonal antibodies reported to date, most of which were derived from HIV-infected subjects (reviewed in ref. 29). D5 was derived from B cells of HIV-naïve subjects and has not been subject to extensive somatic hypermutation, with only seven non-complementarity-determining regions (CDR) amino acid changes from germline sequences (four changes in VH and three in VL; data not shown). Unlike b12, 2F5, and 4E10, D5 does not have an atypically long heavy chain CDR3 region (10 amino acids; data not shown). Unlike 2G12, D5 does not require a “domain-swapped” structure for neutralization (data not shown). Unlike X5, D5 retains antiviral activity against primary HIV isolates when converted to an IgG1 format. Finally, and most importantly, D5 was selected by binding to IZN36 and 5H, synthetic antigens with well defined structures.
The ability of IZN36 and 5H to select a neutralizing antibody immediately suggests they might serve as vaccine candidates. In experimental animals including rhesus macaques, both structures have repeatedly elicited high-titer nonneutralizing antibodies (data not shown). The reason these antibodies fail to neutralize HIV is unknown; perhaps the polyclonal antibodies are present in insufficient quantity, have affinities that are too low, or are directed at irrelevant epitopes. In this regard, we have found other human and mouse antibodies that bind to 5H and/or IZN36 but do not block HIV infection; four such mouse antibodies all bind to physiologically irrelevant structures not present in native gp41, such as the ends of the synthetic antigens (unpublished work). D5's ability to inhibit HIV infection probably derives from its ability to bind a physiologically relevant epitope with high affinity.
Therefore, although IZN36 and 5H do contain the D5-IgG1 epitope as present in the authentic gp41 structure on virions, these structures are still not ideal immunogens. With D5-IgG as a yardstick, we can now evaluate the epitope structures of new candidate immunogens that might elicit a more powerful neutralizing antibody response. The identification of a new neutralizing epitope on gp41, the accessibility of that epitope to an IgG molecule, and the ability to mimic that epitope on designed antigens all provide new hope for HIV vaccine designs aimed at eliciting neutralizing antibodies.
Supplementary Material
Acknowledgments
We thank Chris Petropoulos, Yolanda Lie, and Terri Wrin of ViroLogic (South San Francisco, CA) for entry assays; Hermann Katinger of the Institute for Applied Microbiology (Vienna) for 2G12 and 2F5; Dennis Burton (The Scripps Research Institute, La Jolla, CA) for IgG1b12; Christopher Aiken (Vanderbilt University, Nashville, TN) for R8; Ned Landau of The Salk Institute (La Jolla, CA) for P4-2/R5 cells; Prof. R. Boelens (Utrecht University, Utrecht, The Netherlands) and N. van Nuland (Utrecht University, Utrecht, The Netherlands) for time on the Utrecht University (Utrecht, The Netherlands) Large Scale Facility 900-MHz NMR; and the AIDS Research and Reference Reagent Program for SupT1 cells.
Abbreviations: HRn, heptad repeat n; 5H, five-helix; scFv, single-chain variable region fragment; HIVRP, HIV reporter particle; D5, 5H/I1-BMV-D5; T20, enfuvirtide.
References
- 1.Chan, D. C. & Kim, P. S. (1998) Cell 93, 681-684. [DOI] [PubMed] [Google Scholar]
- 2.Lu, M., Blacklow, S. C. & Kim, P. S. (1995) Nat. Struct. Biol 2, 1075-1082. [DOI] [PubMed] [Google Scholar]
- 3.Chan, D. C., Fass, D., Berger, J. M. & Kim, P. S. (1997) Cell 89, 263-273. [DOI] [PubMed] [Google Scholar]
- 4.Weissenhorn, W., Dessen, A., Harrison, S. C., Skehel, J. J. & Wiley, D. C. (1997) Nature 387, 426-430. [DOI] [PubMed] [Google Scholar]
- 5.Melikyan, G. B., Markosyan, R. M., Hemmati, H., Delmedico, M. K., Lambert, D. M. & Cohen, F. S. (2000) J. Cell Biol. 151, 413-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwong, P. D., Wyatt, R., Robinson, J., Sweet, R. W., Sodroski, J. & Hendrickson, W. A. (1998) Nature 393, 648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richman, D. D., Wrin, T., Little, S. J. & Petropoulos, C. J. (2003) Proc. Natl. Acad. Sci. USA 100, 4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei, X., Decker, J. M., Wang, S., Hui, H., Kappes, J. C., Wu, X., Salazar-Gonzalez, J. F., Salazar, M. G., Kilby, J. M., Saag, M. S., et al. (2003) Nature 422, 307-312. [DOI] [PubMed] [Google Scholar]
- 9.Eckert, D. M., Malashkevich, V. N., Hong, L. H., Carr, P. A. & Kim, P. S. (1999) Cell 99, 103-115. [DOI] [PubMed] [Google Scholar]
- 10.Eckert, D. M. & Kim, P. S. (2001) Proc. Natl. Acad. Sci. USA 98, 11187-11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Root, M. J., Kay, M. S. & Kim, P. S. (2001) Science 291, 884-888. [DOI] [PubMed] [Google Scholar]
- 12.Kilby, J. M., Lalezari, J. P., Eron, J. J., Carlson, M., Cohen, C., Arduino, R. C., Goodgame, J. C., Gallant, J. E., Volberding, P., Murphy, R. L., et al. (2002) AIDS Res. Hum. Retroviruses 18, 685-693. [DOI] [PubMed] [Google Scholar]
- 13.Ingallinella, P., Bianchi, E., Finotto, M., Cantoni, G., Eckert, D. M., Supekar, V. M., Bruckmann, C., Carfi, A. & Pessi, A. (2004) Proc. Natl. Acad. Sci. USA 101, 8709-8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atherton, E. & Sheppard, R. C. (1989) Solid-Phase Peptide Synthesis, A Practical Approach (IRL, Oxford).
- 15.Vaughan, T. J., Williams, A. J., Pritchard, K., Osbourn, J. K., Pope, A. R., Earnshaw, J. C., McCafferty, J., Hodits, R. A., Wilton, J. & Johnson, K. S. (1996) Nat. Biotechnol. 14, 309-314. [DOI] [PubMed] [Google Scholar]
- 16.McCafferty, J., Griffiths, A. D., Winter, G. & Chiswell, D. J. (1990) Nature 348, 552-554. [DOI] [PubMed] [Google Scholar]
- 17.Lou, J., Marzari, R., Verzillo, V., Ferrero, F., Pak, D., Sheng, M., Yang, C., Sblattero, D. & Bradbury, A. (2001) J. Immunol. Methods 253, 233-242. [DOI] [PubMed] [Google Scholar]
- 18.Osbourn, J. K., Field, A., Wilton, J., Derbyshire, E., Earnshaw, J. C., Jones, P. T., Allen, D. & McCafferty, J. (1996) Immunotechnology 2, 181-196. [DOI] [PubMed] [Google Scholar]
- 19.Tobiume, M., Lineberger, J. E., Lundquist, C. A., Miller, M. D. & Aiken, C. (2003) J. Virol. 77, 10645-10650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joyce, J. G., Hurni, W. M., Bogusky, M. J., Garsky, V. M., Liang, X., Citron, M. P., Danzeisen, R. C., Miller, M. D., Shiver, J. W. & Keller, P. M. (2002) J. Biol. Chem. 277, 45811-45820. [DOI] [PubMed] [Google Scholar]
- 21.Wyma, D. J., Jiang, J., Shi, J., Zhou, J., Lineberger, J. E., Miller, M. D. & Aiken, C. (2004) J. Virol. 78, 3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furuta, R. A., Wild, C. T., Weng, Y. & Weiss, C. D. (1998) Nat. Struct. Biol. 5, 276-279. [DOI] [PubMed] [Google Scholar]
- 23.Labrijn, A. F., Poignard, P., Raja, A., Zwick, M. B., Delgado, K., Franti, M., Binley, J., Vivona, V., Grundner, C., Huang, C. C., et al. (2003) J. Virol. 77, 10557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan, D. C., Chutkowski, C. T. & Kim, P. S. (1998) Proc. Natl. Acad. Sci. USA 95, 15613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuiken, C. L., Foley, B., Freed, E., Hahn, B., Korber, B., Marx, P. A., McCutchan, F., Mellors, J. W. & Wolinsky, S., eds. (2002) HIV Sequence Compendium (Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM), LA-UR 03-3564.
- 26.Reeves, J. D., Gallo, S. A., Ahmad, N., Miamidian, J. L., Harvey, P. E., Sharron, M., Pohlmann, S., Sfakianos, J. N., Derdeyn, C. A., Blumenthal, R., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carr, C. M. & Kim, P. S. (1993) Cell 73, 823-832. [DOI] [PubMed] [Google Scholar]
- 28.Louis, J. M., Nesheiwat, I., Chang, L., Clore, G. M. & Bewley, C. A. (2003) J. Biol. Chem. 278, 20278-20285. [DOI] [PubMed] [Google Scholar]
- 29.Golding, H., Zaitseva, M., de Rosny, E., King, L. R., Manischewitz, J., Sidorov, I., Gorny, M. K., Zolla-Pazner, S., Dimitrov, D. S. & Weiss, C. D. (2002) J. Virol. 76, 6780-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burton, D. R., Desrosiers, R. C., Doms, R. W., Koff, W. C., Kwong, P. D., Moore, J. P., Nabel, G. J., Sodroski, J., Wilson, I. A. & Wyatt, R. T. (2004) Nat. Immunol 5, 233-236. [DOI] [PubMed] [Google Scholar]
- 31.Hamburger, A. E., Kim, S., Welch, B. D. & Kay, M. S. (2005) J. Biol. Chem. 280, 12567-12572. [DOI] [PubMed] [Google Scholar]
- 32.Sia, S. K., Carr, P. A., Cochran, A. G., Malashkevich, V. N. & Kim, P. S. (2002) Proc. Natl. Acad. Sci. USA 99, 14664-14669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrer, M., Kapoor, T. M., Strassmaier, T., Weissenhorn, W., Skehel, J. J., Oprian, D., Schreiber, S. L., Wiley, D. C. & Harrison, S. C. (1999) Nat. Struct. Biol. 6, 953-960. [DOI] [PubMed] [Google Scholar]
- 34.Zhou, G., Ferrer, M., Chopra, R., Kapoor, T. M., Strassmaier, T., Weissenhorn, W., Skehel, J. J., Oprian, D., Schreiber, S. L., Harrison, S. C., et al. (2000) Bioorg Med. Chem. 8, 2219-2227. [DOI] [PubMed] [Google Scholar]
- 35.Ma, B., Shatsky, M., Wolfson, H. J. & Nussinov, R. (2002) Protein Sci. 11, 184-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeLano, W. L. (2002) Curr. Opin. Struct. Biol. 12, 14-20. [DOI] [PubMed] [Google Scholar]
- 37.Jermutus, L., Honegger, A., Schwesinger, F., Hanes, J. & Pluckthun, A. (2001) Proc. Natl. Acad. Sci. USA 98, 75-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.