Abstract
Maize (Zea mays subsp mays) was domesticated from teosinte (Z. mays subsp parviglumis) through a single domestication event in southern Mexico between 6000 and 9000 years ago. This domestication event resulted in the original maize landrace varieties, which were spread throughout the Americas by Native Americans and adapted to a wide range of environmental conditions. Starting with landraces, 20th century plant breeders selected inbred lines of maize for use in hybrid maize production. Both domestication and crop improvement involved selection of specific alleles at genes controlling key morphological and agronomic traits, resulting in reduced genetic diversity relative to unselected genes. Here, we sequenced 1095 maize genes from a sample of 14 inbred lines and chose 35 genes with zero sequence diversity as potential targets of selection. These 35 genes were then sequenced in a sample of diverse maize landraces and teosintes and tested for selection. Using two statistical tests, we identified eight candidate genes. Extended gene sequencing of these eight candidate loci confirmed that six were selected throughout the gene, and the remaining two exhibited evidence of selection in the 3′ portion of each gene. The selected genes have functions consistent with agronomic selection for nutritional quality, maturity, and productivity. Our large-scale screen for artificial selection allows identification of genes of potential agronomic importance even when gene function and the phenotype of interest are unknown.
INTRODUCTION
One prominent goal of plant molecular biology is to identify and characterize the genes responsible for phenotypic variation. Historically, quantitative trait locus (QTL) mapping has been used to localize genomic regions contributing to phenotypic variation, but this approach has rarely led to candidate gene isolation. In crop systems, for example, only a limited number of genes has been isolated, cloned, and characterized based on information from a QTL analysis (e.g., fw2.2, Frary et al., 2000; Hd1, Yano et al., 2000; tga1, Wang et al., 2005). Another approach to defining the genes that control phenotypic variation is association analysis (Thornsberry et al., 2001). This approach is especially promising when prior information identifies potential candidate genes thought to contribute to the trait of interest. In the absence of candidate genes, a full-genome scan is necessary, and the potential for false positives (Type I error) in crop plants with their large genome size is extremely high.
Both QTL and association approaches rely on segregating phenotypic and molecular genetic variation. In crop systems, however, some genes underlying agronomic traits are expected to be bereft of segregating genetic variants because artificial selection has substantially decreased genetic diversity. One can think of selection occurring in two stages: domestication and crop improvement. Domestication resulted in the original landrace varieties, which were adapted to a wide range of environmental conditions. These landraces provided the genetic material for modern plant breeders to select improved varieties and inbred lines by enhancing traits controlling agricultural productivity and performance, such as yield and resistance to biotic and abiotic stresses. Consequently, crop varieties experienced strong selection at genes controlling traits of agronomic importance during their domestication and improvement by plant breeding (Wang et al., 1999; Whitt et al., 2002; Palaisa et al., 2003; Gallavotti et al., 2004; Wang et al., 2005). One result from selection during domestication and improvement is that QTL and association methods may miss the most interesting class of genes (i.e., those genes that lack genetic diversity because of a history of selection on their key role in controlling desirable agronomic traits).
How then, can one identify this selected class of genes that contributes to agronomic traits? In this and a previous article (Wright et al., 2005), we have been using population genetics approaches to identify selected genes in maize (Zea mays subsp mays). Maize was domesticated from teosinte (Z. mays subsp parviglumis) through a single domestication event in southern Mexico between 6000 and 9000 years ago (Piperno and Flannery, 2001; Matsuoka et al., 2002). Maize and teosinte differ in many aspects of plant morphology and productivity. Despite its selection history, most maize genes retain high levels of nucleotide diversity (Tenaillon et al., 2001). Maize is an outcrossing species and exhibits a high level of recombination (Fu et al., 2002), and the historical population size was large (Vigouroux et al., 2002a), all factors that contribute to a rapid decay of linkage disequilibrium (LD) among current maize inbred lines (Remington et al., 2001; Tenaillon et al., 2001). Therefore, maize is a model crop for performing association analysis to identify genes controlling agronomic traits (Thornsberry et al., 2001; Rafalski and Morgante, 2004). Several studies in maize have also shown that there is a large contrast in nucleotide diversity between genes with a history of selection and neutral genes (Wang et al., 1999; Whitt et al., 2002; Gallavotti et al., 2004; Tenaillon et al., 2004).
We recently began to address the genome-wide effects of artificial selection in maize by an analysis of single nucleotide polymorphisms (SNPs) in 774 genes in 16 teosinte accessions and 14 maize inbred lines (Wright et al., 2005). In that study, we concluded that 2 to 4% or ∼1200 maize genes show evidence of selection. Because the sequencing involved a single amplification product from each gene, the power to detect selected genes may have been limited. All genes significant for selection retained minimal, if any polymorphism, among the maize inbred lines. This result suggests a simplified method to conduct large-scale screens for selected genes in maize, which is sequencing a short region of each gene in inbred lines and only sequencing in teosinte those genes with very low inbred polymorphism. Another major question unaddressed by Wright et al. (2005) is the extent to which selection on any particular gene occurred at domestication rather than during subsequent crop improvement. This question is of both academic interest in understanding genetic consequences of selection and of practical importance to plant breeders concerned with diversifying germplasm used in maize breeding programs. For example, improvement genes, while lacking variation in elite inbreds, could be studied via QTL methods in crosses with landraces, and the germplasm diversity for these genes may be enhanced using exotic maize resources. For domestication genes, the plant breeder would have to extend crosses to at least the teosintes to introduce novel alleles.
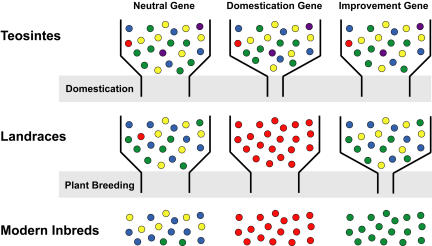
In this study, we conduct a large-scale screen to discover genes responsible for maize domestication and improvement. Based on previous studies (Wang et al., 1999; Whitt et al., 2002; Palaisa et al., 2003; Gallavotti et al., 2004; Tenaillon et al., 2004), it is clear that the domestication event resulted in a loss of genetic diversity between teosinte and maize landraces and that modern plant breeding reduced the genetic diversity in maize inbreds relative to maize landraces (Figure 1). Moreover, genetic diversity in neutral (unselected) genes is expected to be reduced only by bottleneck effects, thereby retaining more diversity than selected genes (Figure 1). This reasoning leads to the prediction that genes strongly impacted by domestication or improvement are enriched in the subset of genes that exhibit low nucleotide diversity in modern improved varieties (Wright et al., 2005). Hence, we sequenced a large set of randomly chosen genes in diverse maize inbreds and identified genes with zero nucleotide diversity as potential targets of selection. These candidate genes were sequenced in additional diverse sets of maize landraces and teosintes. Finally, statistical analyses contrasting DNA sequence diversity among maize inbreds, landraces, and teosintes were performed to document selection and to determine whether selection occurred primarily during domestication or crop improvement. For the selection candidates with the strongest evidence of selection, we performed extended sequencing throughout the available gene sequence to confirm selection and to define the regions of the genes under selection. The selected genes have functions consistent with agronomic selection for nutritional quality, maturity, and productivity.
Figure 1.
Effect of Domestication and Plant Breeding on Genetic Diversity of Maize Genes.
The colored circles represent different alleles. The shaded areas indicate bottleneck effects placed on all genes by the processes of domestication and improvement (plant breeding). Our model assumes that there will be three types of genes: neutral genes that show reduction of diversity by the general bottleneck effects, domestication genes in which diversity is greatly reduced by selection between the teosintes and landraces, and improvement genes in which diversity is greatly reduced by selection between the landraces and inbreds.
RESULTS
Genetic Diversity in Maize Inbreds, Landraces, and Teosintes
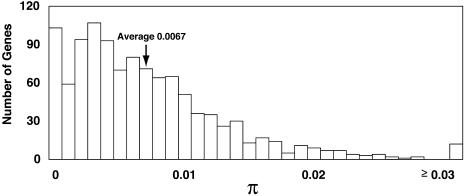
To efficiently identify potential selected genes, a panel of inbred lines was established to represent as much of the genetic diversity of modern maize inbreds as possible. Simple sequence repeat (SSR) polymorphism data were used to choose 14 maize inbreds lines with maximum allelic diversity (Liu et al., 2003), with the constraint that final composition would include seven temperate inbreds and seven tropical inbreds (Table 1). This constraint was added to allow us to contrast genetic diversity in the temperate and tropical maize germplasm breeding pools. Sequence alignments for a single PCR product were obtained for 1095 randomly selected maize genes. An average of ∼13 inbred sequences per gene using 14 diverse inbreds and an average length of alignment of 280 bp without gaps were obtained (see Supplemental Table 1 online; summarized in Table 2). Among these 1095 loci, we identified 6169 SNPs, an average of 5.6 SNPs per alignment. This corresponds to one SNP approximately every 50 bp per alignment length or every 150 bp between any randomly chosen pair of maize inbreds. We found 2848 SNPs shared in temperate and tropical inbreds, 1742 SNPs specifically within temperate inbreds and 1579 SNPs within the tropical inbreds. The number of segregating sites (S) was not significantly different between temperate and tropical inbreds (Mann-Whitney U test, P > 0.12). There also was not a significant difference between temperate and tropical inbreds in the nucleotide diversity measure (π; Student's t test, P > 0.22), which is average proportion of pairwise nucleotide differences per nucleotide site (Tajima, 1983). The average π in all maize inbreds was 0.0067 (Figure 2), confirming the high level of sequence diversity previously reported for maize inbred lines (Tenaillon et al., 2001; Wright et al., 2005).
Table 1.
Plant Materials Used in This Study
| Type | Inbred/Race (Accession or Type) | Origin |
|---|---|---|
| Inbred line | B73 (temperate) | Iowa |
| Z. mays subsp mays | Hp301 (temperate) | Indiana |
| Il14H (temperate) | Illinois | |
| Ky21 (temperate) | Kentucky | |
| M37W (temperate) | South Africa | |
| Mo17 (temperate) | Missouri | |
| Oh43 (temperate) | Ohio | |
| CML69 (tropical) | Mexico | |
| CML247 (tropical) | Mexico | |
| CML322 (tropical) | Mexico | |
| CML333 (tropical) | Mexico | |
| Ki3 (tropical) | Thailand | |
| Ki11 (tropical) | Thailand | |
| NC350 (tropical) | North Carolina | |
| Landrace | Assinboine (PI213793) | Northern U.S. |
| Z. mays subsp mays | Bolita (OAX 68) | Southern Mexico |
| Cateto Sulino (URG 11) | Uruguay | |
| Chalqueno (MEX 48) | Central Mexico | |
| Chapalote (SIN 2) | Western Mexico | |
| Conico (PUE 32) | Central Mexico | |
| Costeno (VEN 453) | Venezuela | |
| Cristalino Norteno (CHI 349) | Chile | |
| Dzit Bacal (GUA 131) | Guatemala | |
| Gordo (CHH 160) | Northern Mexico | |
| Guirua (MAG 450) | Colombia | |
| Nal-tel (YUC 7) | Southern Mexico | |
| Pissccotunto (APC 13) | Peru | |
| Sabanero (SAN 329) | Colombia | |
| Serrano (GUA 14) | Guatemala | |
| Zapalote Chico (OAX 70) | Southern Mexico | |
| Teosinte | Balsas (Beadle and Kato Site 4) | Mexico |
| Z. mays subsp parviglumis | Balsas (CIMMYT 8783) | Mexico |
| Balsas (CIMMYT 11355) | Mexico | |
| Balsas (INIFAP JSG 374) | Mexico | |
| Balsas (INIFAP JSG 378) | Mexico | |
| Balsas (INIFAP JSG y LOS 109) | Mexico | |
| Balsas (INIFAP JSG y LOS 119) | Mexico | |
| Balsas (INIFAP JSG y LOS 161) | Mexico | |
| Balsas (INIFAP JSP y Lo5130) | Mexico | |
| Balsas (Kato Site 4) | Mexico | |
| Balsas (USDA PI566686) | Mexico | |
| Balsas (Wilkes Site 6) | Mexico | |
| Jalisco (Benz 967) | Mexico | |
| Jalisco (INIFAP JSG y MAS 264) | Mexico | |
| Jalisco (INIFAP JSG y MAS 401) | Mexico | |
| Oaxaca (INIFAP JSG 197) | Mexico | |
| T. dactyloides | WW-2120 | Oklahoma |
| MIA 34597 | Colombia |
Table 2.
Sequence Statistics for 1095 Genes in Diverse Maize Inbreds
| Type | N | L | Total L | S | Total S | π |
|---|---|---|---|---|---|---|
| All maize inbreds | 13.1 | 280.3 | 306,895 | 5.6 | 6,169 | 0.0067 |
| Temperate inbreds | 6.7 | 292.2 | 310,306 | 4.3 | 4,590 | 0.0065 |
| Tropical inbreds | 6.6 | 290.6 | 308,649 | 4.2 | 4,427 | 0.0061 |
N, average number of sequences in the final alignment; L, average length of the core alignments in which all sequences contain bases, excluding gaps; S, average number of segregating sites (SNPs) per alignment; total L and total S, sums for all 1095 alignments; π, average proportion of pairwise differences among all shared positions for all sequences in an alignment.
Figure 2.
Distribution of Genetic Diversity π for 1095 Maize Genes.
The genes were sequenced in the panel of 14 diverse maize inbreds (Table 1).
From the 1095 genes, we selected 35 genes with alignments >200 bp and zero nucleotide diversity, either from SNPs or from insertion/deletion polymorphisms that are also very common among maize inbreds (Bhattramakki et al., 2002). Although the lack of nucleotide diversity at these genes could reflect a history of selection during domestication or improvement, the low diversity in maize could also reflect low diversity in teosinte and/or the demographic effects of domestication, plant breeding, and/or chance events (e.g., genetic drift). In order to distinguish between selection and other effects and also to determine if selection occurred primarily during domestication or crop improvement, we sequenced the same region of these 35 genes in 16 diverse teosinte accessions and 16 diverse maize landraces (Tables 1 and 3; sequence alignments are presented in Supplemental Table 2 online). There were significant differences for number of haplotypes (h) among the three populations: inbreds < landraces < teosintes (Kruskal-Wallis H test, P < 0.001; multiple comparison Scheffé's F test, all P < 0.001) (Table 4). The Kruskal-Wallis H test for differences in π among the populations was also significant (P < 0.001). Although π in inbreds and landraces was significantly lower than π in teosintes (Scheffé's F test, P < 0.001), π in inbreds versus landraces was not significantly different (Scheffé's F test, P > 0.05). For comparison, we also sequenced the same three populations for four documented neutral (unselected) genes: adh1, bz2, fus6, and glb1 (Eyre-Walker et al., 1998; Hilton and Gaut, 1998; Tenaillon et al., 2001). For these neutral genes, h and π were not significantly different among the three populations (Kruskal-Wallis H test, P > 0.40 and P > 0.66, respectively). In addition, use of the Tajima's D test (Tajima, 1989) supported neutrality for adh1, bz2, fus6, and glb1in all three populations (P > 0.10).
Table 3.
Sequence Diversity in Maize Inbreds, Landraces, and Teosintes at 35 Genes That Exhibited Zero Sequence Diversity in Maize Inbreds and Four Neutral Genes
| Inbreds
|
Landraces
|
Teosintes
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | N | L | S | h | π | N | L | S | h | π | N | L | S | h | π |
| AY108876 | 14 | 390 | 0 | 1 | 0.00000 | 15 | 482 | 1 | 2 | 0.00028 | 15 | 445 | 9 | 8 | 0.00428 |
| AY106123 | 14 | 327 | 0 | 1 | 0.00000 | 14 | 373 | 5 | 3 | 0.00250 | 15 | 374 | 5 | 4 | 0.00514 |
| AY106190 | 14 | 326 | 0 | 1 | 0.00000 | 14 | 365 | 1 | 2 | 0.00072 | 16 | 330 | 2 | 3 | 0.00197 |
| AY108504 | 14 | 321 | 0 | 1 | 0.00000 | 15 | 336 | 1 | 2 | 0.00040 | 15 | 336 | 3 | 3 | 0.00119 |
| AY108957 | 14 | 315 | 0 | 1 | 0.00000 | 16 | 329 | 0 | 1 | 0.00000 | 14 | 309 | 1 | 2 | 0.00046 |
| AY108255 | 14 | 300 | 0 | 1 | 0.00000 | 15 | 319 | 0 | 1 | 0.00000 | 14 | 318 | 8 | 5 | 0.00473 |
| AY107195 | 14 | 288 | 0 | 1 | 0.00000 | 14 | 274 | 2 | 2 | 0.00104 | 15 | 302 | 10 | 7 | 0.00826 |
| AY105850 | 13 | 271 | 0 | 1 | 0.00000 | 16 | 311 | 3 | 2 | 0.00314 | 16 | 311 | 5 | 6 | 0.00271 |
| AY110109 | 14 | 271 | 0 | 1 | 0.00000 | 16 | 280 | 6 | 4 | 0.00307 | 16 | 273 | 10 | 8 | 0.01029 |
| AY106826 | 14 | 265 | 0 | 1 | 0.00000 | 15 | 277 | 2 | 2 | 0.00096 | 16 | 277 | 4 | 4 | 0.00253 |
| AY109101 | 14 | 260 | 0 | 1 | 0.00000 | 16 | 285 | 2 | 3 | 0.00088 | 13 | 288 | 3 | 4 | 0.00285 |
| AY105752 | 14 | 246 | 0 | 1 | 0.00000 | 16 | 293 | 2 | 4 | 0.00191 | 14 | 309 | 2 | 3 | 0.00217 |
| AY105060 | 14 | 248 | 0 | 1 | 0.00000 | 15 | 297 | 2 | 3 | 0.00090 | 15 | 285 | 21 | 8 | 0.02185 |
| AY108178 | 14 | 239 | 0 | 1 | 0.00000 | 15 | 244 | 6 | 2 | 0.00609 | 14 | 230 | 12 | 8 | 0.01529 |
| AY109011 | 14 | 237 | 0 | 1 | 0.00000 | 16 | 275 | 1 | 2 | 0.00045 | 15 | 273 | 6 | 6 | 0.00433 |
| AY107827 | 13 | 229 | 0 | 1 | 0.00000 | 13 | 240 | 3 | 2 | 0.00192 | 14 | 261 | 5 | 4 | 0.00564 |
| AY107821 | 14 | 228 | 0 | 1 | 0.00000 | 16 | 253 | 3 | 3 | 0.00148 | 14 | 258 | 3 | 3 | 0.00554 |
| AY107949 | 13 | 229 | 0 | 1 | 0.00000 | 16 | 223 | 0 | 1 | 0.00000 | 16 | 231 | 2 | 3 | 0.00155 |
| AY106125 | 14 | 217 | 0 | 1 | 0.00000 | 14 | 248 | 1 | 1 | 0.00058 | 15 | 242 | 9 | 7 | 0.00543 |
| AY107673 | 12 | 220 | 0 | 1 | 0.00000 | 9 | 312 | 2 | 4 | 0.00258 | 12 | 292 | 2 | 2 | 0.00280 |
| AY108759 | 14 | 214 | 0 | 1 | 0.00000 | 14 | 224 | 1 | 2 | 0.00235 | 16 | 219 | 5 | 4 | 0.00285 |
| AY108231 | 14 | 213 | 0 | 1 | 0.00000 | 14 | 239 | 0 | 1 | 0.00000 | 16 | 238 | 4 | 5 | 0.00326 |
| AY106616 | 14 | 209 | 0 | 1 | 0.00000 | 16 | 197 | 8 | 4 | 0.01193 | 16 | 226 | 6 | 5 | 0.00830 |
| AY106734 | 14 | 193 | 0 | 1 | 0.00000 | 15 | 194 | 0 | 1 | 0.00000 | 15 | 242 | 1 | 2 | 0.00055 |
| AY107952 | 13 | 531 | 0 | 1 | 0.00000 | 12 | 476 | 1 | 2 | 0.00064 | 12 | 541 | 12 | 6 | 0.00370 |
| AY108388 | 14 | 441 | 0 | 1 | 0.00000 | 13 | 479 | 13 | 3 | 0.00418 | 16 | 476 | 22 | 6 | 0.00840 |
| AY108552 | 13 | 313 | 0 | 1 | 0.00000 | 14 | 295 | 0 | 1 | 0.00000 | 14 | 338 | 2 | 3 | 0.00085 |
| AI737881 | 14 | 294 | 0 | 1 | 0.00000 | 16 | 323 | 5 | 4 | 0.00421 | 14 | 306 | 12 | 11 | 0.00991 |
| AY108194 | 13 | 253 | 0 | 1 | 0.00000 | 10 | 274 | 1 | 2 | 0.00073 | 16 | 280 | 4 | 4 | 0.00179 |
| AY106702 | 14 | 247 | 0 | 1 | 0.00000 | 15 | 258 | 2 | 3 | 0.00103 | 14 | 257 | 3 | 4 | 0.00167 |
| AY106371 | 14 | 235 | 0 | 1 | 0.00000 | 12 | 282 | 0 | 1 | 0.00000 | 15 | 284 | 6 | 6 | 0.00604 |
| AY106889 | 14 | 240 | 0 | 1 | 0.00000 | 8 | 141 | 0 | 1 | 0.00000 | 15 | 266 | 3 | 4 | 0.00150 |
| AY107535 | 14 | 237 | 0 | 1 | 0.00000 | 14 | 265 | 0 | 1 | 0.00000 | 16 | 264 | 3 | 4 | 0.00218 |
| AY107529 | 14 | 235 | 0 | 1 | 0.00000 | 16 | 255 | 4 | 3 | 0.00275 | 14 | 255 | 6 | 5 | 0.00612 |
| AY107818 | 14 | 203 | 0 | 1 | 0.00000 | 15 | 258 | 3 | 3 | 0.00199 | 15 | 253 | 4 | 4 | 0.00256 |
| adh1 | 14 | 1217 | 42 | 6 | 0.01237 | 16 | 1309 | 58 | 10 | 0.01480 | 7 | 1217 | 55 | 5 | 0.01808 |
| bz2 | 14 | 524 | 11 | 5 | 0.00910 | 16 | 594 | 17 | 8 | 0.00885 | 9 | 522 | 15 | 7 | 0.00702 |
| fus6 | 14 | 238 | 10 | 4 | 0.01057 | 15 | 234 | 8 | 5 | 0.01213 | 10 | 227 | 10 | 6 | 0.01429 |
| glb1 | 14 | 958 | 52 | 11 | 0.01747 | 14 | 957 | 86 | 14 | 0.02169 | 12 | 939 | 99 | 12 | 0.02433 |
Gene, the GenBank accession number of the original unigene sequence; N, number of sampled sequences; L, length of the core alignments in which all sequences contain bases, excluding gaps; S, total number of segregating sites; h, number of unique sequences (haplotypes); π, average proportion of pairwise differences per base pair.
Table 4.
Sequence Statistics for 35 Genes with Zero Inbred Diversity
| Type | N | L | h | π |
|---|---|---|---|---|
| Inbreds | 13.8 | 271.0 | 1.0 | 0.0000 |
| Landraces | 14.3 | 290.7 | 2.2 | 0.0017 |
| Teosintes | 14.8 | 296.8 | 4.9 | 0.0048 |
N, average number of sequences in the final alignment; L, average length of the core alignments in which all sequences contain bases, excluding gaps; h, average number of unique sequences (haplotypes) in the alignments; π, average proportion of pairwise differences per base pair among all shared positions for all sequences in an alignment.
Statistical Tests for Selection
Two separate tests of selection were conducted for each of the 35 candidate genes: Hudson-Kreitman-Aguadé (HKA) tests (Hudson et al., 1987) and coalescent simulations of domestication (CS) tests (Tenaillon et al., 2004). The HKA test requires an outgroup sequence to compare rates of divergence between species to levels of polymorphism within species. The test also requires reference loci that are not believed to have been affected by selection. The ratio of divergence to polymorphism is then compared between a putatively selected gene and reference gene(s). If the putatively selected gene has a significantly higher ratio of divergence to polymorphism, it is reasonable to postulate that polymorphism in the putative selected gene has been diminished by selection. Simulation studies of the genetic effects of artificial selection have demonstrated that the HKA is an appropriate and powerful test to find genes affected by artificial selection (Innan and Kim, 2004).
To perform the HKA test, we obtained an orthologous sequence of Tripsacum dactyloides for 31 of 35 candidate genes. We used a multiple locus implementation of the HKA test (see Methods) to compare each of our candidate genes to the four neutral genes adh1, bz2, fus6, and glb1 at three population levels: teosintes, landraces, and inbreds. Given the results of these HKA comparisons, we identified a gene as a candidate domestication gene if it was significant for selection both in inbreds and landraces but not significant in teosintes; this pattern indicates that selection occurred primarily between teosintes and landraces (Figure 1). Similarly, we identified a candidate improvement gene if significant for selection in inbreds but not significant in landraces and teosintes, indicating that diversity was reduced by modern plant breeding (Figure 1). Using this approach, we detected six candidate genes for domestication and six for improvement by the HKA analysis (Table 5, Figure 3). For example, HKA tests on gene AY108876 were significant for both the inbred (P = 0.010) and landrace sample (P = 0.014) (Table 5), suggesting that polymorphism in this gene is aberrantly low in both the landrace and inbred samples. Given our model in Figure 1, we therefore conclude that this gene was selected during domestication, prior to improvement. Similarly, HKA tests on gene AY110109 were significant only for the inbreds (P = 0.046), leading to the conclusion that this gene was selected during improvement.
Table 5.
Results of the Tests of Selection and Homology Searches
| P Values in HKA
|
Candidate Status by HKA
|
P Values in CS
|
Candidate Status by CS
|
|||||
|---|---|---|---|---|---|---|---|---|
| Gene | I | L | T | I versus T | L versus T | Homology Search | ||
| AY108876 | 0.010* | 0.014* | 0.225 | Domestication | 0.0152* | 0.0145* | Domestication | Amino acid transporter |
| AY106123 | 0.155 | 0.022* | 0.014* | – | 0.0730 | 0.6184 | – | Putative poly(A) binding protein binding protein |
| AY106190 | 0.047* | 0.047* | 0.046* | – | 0.1579 | 0.5561 | – | Hypothetical protein |
| AY108504 | 0.030* | 0.013* | 0.036* | – | 0.1219 | 0.4699 | – | |
| AY108957 | 0.057 | 0.023* | 0.028* | – | 0.3032 | 0.1827 | – | |
| AY108255 | 0.323 | 0.158 | 0.848 | – | 0.0294* | 0.0038** | Domestication | Transcriptional factor |
| AY107195 | 0.067 | 0.383 | 0.858 | Improvement | 0.0175* | 0.1471 | Improvement | Auxin response factor |
| 0.047* | 0.250 | 0.833 | ||||||
| AY105850 | NA | NA | NA | – | 0.0393* | 0.5508 | Improvement | |
| AY110109 | 0.046* | 0.644 | 0.822 | Improvement | 0.0211* | 0.7939 | Improvement | GTP binding protein |
| AY106826 | 0.102 | 0.308 | 0.541 | – | 0.0898 | 0.6554 | – | |
| AY109101 | 0.082 | 0.217 | 0.251 | – | 0.0847 | 0.6766 | – | Ser/Thr protein kinase |
| AY105752 | 0.040* | 0.116 | 0.109 | Improvement | 0.1915 | 0.7282 | – | Putative casein kinase |
| AY105060 | 0.016* | 0.034* | 0.863 | Domestication | 0.0065** | 0.0008*** | Domestication | |
| AY108178 | 0.007** | 0.053 | 0.217 | Improvement | 0.0059** | 0.2970 | Improvement | Circadian clock |
| 0.010* | 0.039* | 0.208 | Domestication | |||||
| AY109011 | 0.028* | 0.028* | 0.234 | Domestication | 0.0403* | 0.1042 | Improvement | NLI interacting factor |
| AY107827 | 0.085 | 0.691 | 0.536 | – | 0.0731 | 1.0000 | – | |
| AY107821 | 0.030* | 0.105 | 0.083 | Improvement | 0.1350 | 0.7468 | – | Universal stress protein |
| AY107949 | 0.033* | 0.011* | 0.037* | – | 0.1713 | 0.0878 | – | |
| AY106125 | 0.175 | 0.305 | 0.864 | – | 0.0160* | 0.0435* | Domestication | |
| AY107673 | 0.027* | 0.027* | 0.018* | – | 0.2279 | 0.7457 | – | |
| 0.023* | 0.029* | 0.020* | ||||||
| AY108759 | 0.053 | 0.062 | 0.292 | – | 0.0662 | 0.2380 | – | |
| AY108231 | 0.530 | 0.308 | 0.879 | – | 0.0588 | 0.0140* | – | |
| AY106616 | 0.013* | 0.291 | 0.101 | Improvement | 0.0499* | 0.5148 | Improvement | Ankyrin repeat-like protein |
| AY106734 | NA | NA | NA | – | 0.3544 | 0.2543 | – | |
| AY107952 | 0.008** | 0.011* | 0.363 | Domestication | 0.0161* | 0.0460* | Domestication | Putative fruit protein, oxidoreductase |
| AY108388 | NA | NA | NA | – | 0.0047** | 0.6850 | Improvement | Cys synthase |
| AY108552 | 0.029* | 0.008** | 0.026* | – | 0.1654 | 0.0938 | – | Arm repeat-containing protein |
| AI737881 | 0.052 | 0.279 | 0.781 | – | 0.0034** | 0.0992 | Improvement | Putative alcohol oxidase |
| AY108194 | 0.148 | 0.183 | 0.483 | – | 0.1020 | 0.4527 | – | |
| 0.164 | 0.264 | 0.589 | ||||||
| AY106702 | 0.019* | 0.025* | 0.041* | – | 0.0808 | 0.6868 | – | Putative suppressor of actin 1, inositol 5-phosphatase 3-like |
| AY106371 | 0.023* | 0.010* | 0.196 | Domestication | 0.0375* | 0.0052** | Domestication | Putative methyl binding domain protein |
| AY106889 | 0.025* | 0.019* | 0.031* | – | 0.0991 | 0.1456 | – | |
| AY107535 | 0.082 | 0.026* | 0.245 | – | 0.0938 | 0.0284* | – | Putative integral membrane protein |
| AY107529 | NA | NA | NA | – | 0.0479* | 0.8760 | Improvement | Putative Ser carboxypeptidase |
| AY107818 | 0.010* | 0.017* | 0.011* | – | 0.1105 | 0.7098 | – | Putative polygalacturonase |
Gene, the GenBank accession of the original unigene sequence. HKA tests: P value of the candidate gene by multiple locus HKA against the four neutral genes by the maximum cell value test. I, inbreds; L, landraces; T, teosintes. For AY107195, AY108178, AY107673, and AY108194, two distinct T. dactyloides haplotypes were used. NA, T. dactyloides sequence was not obtained. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
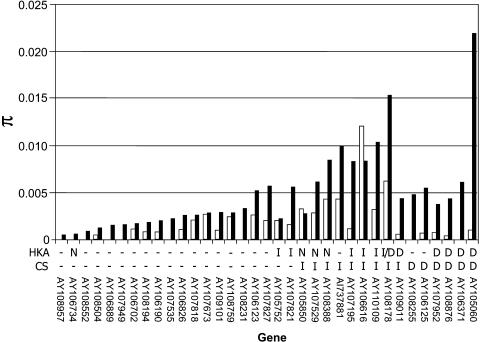
Figure 3.
The Genetic Diversity π in Landraces and Teosintes for Each of 35 Genes with Zero Diversity in Inbred Lines.
The black bars indicate genetic diversity in the teosintes, and the white bars indicate genetic diversity in the landraces. Where no white bars are present, the genetic diversity in the landraces was zero. Inbreds are not shown as all are π = zero. The two rows under the figure indicate the test results for selection by HKA and CS analyses. The “-” indicates no significance; n = not tested in the HKA test due to inability to amplify a T. dactyloides orthologous sequence; I, improvement candidate; D, domestication candidate. The GenBank accession of the original Maize Mapping Project/DuPont unigene sequence is indicated under the bar.
The HKA test is valid, but the distribution of test statistics can be influenced by demographic history (Wright and Gaut, 2005). By contrast, CS analyses incorporate summary statistics, including information about recombination, to estimate the duration and severity of the bottleneck, based on data from the four reference genes. The CS then tests whether the loss of diversity in inbreds versus teosintes and landraces versus teosintes at a candidate locus is too great to be explained by demographic effects alone. By the CS analyses, a gene is considered a domestication gene if the gene was significant for selection in both landraces versus teosintes and inbreds versus teosintes, and a gene is inferred to be an improvement gene if inbreds versus teosintes, but not landraces versus teosintes, were significant for selection. Because the CS analysis directly tests for a major consequence of artificial selection, the specific loss of diversity at target genes, in addition to expected diversity loss from bottleneck effects on all genes, the CS analysis complements the HKA test as a second independent and powerful test for selected genes.
Note that for identification of an improvement gene, we did not apply the CS test directly between landraces to inbreds. An implicit assumption of the CS test is that the ancestral population follows a neutral equilibrium model. Although the teosinte population demonstrates some deviation from neutrality (Wright et al., 2005), the equilibrium neutral assumption is reasonable for any single gene in teosinte. By contrast, we cannot reasonably assume that the landraces fit a neutral equilibrium model because they have recently experienced a population bottleneck.
We identified six candidate genes for domestication and nine candidates for improvement by the CS analysis (Table 5, Figure 3). There were four domestication and four improvement genes identified in common between the HKA and CS analyses (Table 5, Figure 3). Because these eight genes were identified by two distinct tests for selection, one that relies on divergence information and the other that corrects for demographic history, they are our strongest candidates for selected genes.
Extended Sequencing of Candidate-Selected Genes
One limitation to our approach for identifying selected genes is that the short length of the alignment restricts the power of the approach. Longer sequences would increase the power to identify selection. Although the short alignments are expected to result in more false negatives rather than false positives, it is also important to characterize the false positive rate. We conducted extended sequencing in the maize inbreds and the teosintes for the entire available sequence of the eight genes identified by both HKA and CS analyses (Table 6, Figure 4; sequence alignments are presented in Supplemental Table 3 online).
Table 6.
Sequence Diversity in Maize Inbreds and Teosintes at Eight Candidate Genes and Results of the Tests of Selection
| Inbreds
|
Teosintes
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | N | L | S | h | πtotal | πsilent | P Value in HKAtotal | P Value in HKAsilent | N | L | S | h | πtotal | πsilent | P Value in HKAtotal | P Value in HKAsilent | Candidate Status | Homology Search |
| AY108876 | 14 | 1055 | 1 | 2 | 0.00025 | 0.00050 | <0.0068** | <0.0120* | 16 | 1026 | 13 | 8 | 0.00245 | 0.00403 | <0.0433* | <0.1849 | Selected | Amino acid transporter |
| AY107195 | 14 | 3119 | 1 | 2 | 0.00005 | 0.00007 | <0.0058** | <0.0087** | 11 | 3097 | 81 | 11 | 0.00820 | 0.01171 | <0.5889 | <0.6761 | Selected | Auxin response factor |
| AY110109 | 14 | 1466 | 1 | 2 | 0.00036 | 0.00000 | <0.0051** | <0.0054** | 14 | 1355 | 43 | 11 | 0.00697 | 0.00999 | <0.3613 | <0.5321 | Selected | GTP binding protein |
| AY105060 | 14 | 1090 | 0 | 1 | 0.00000 | 0.00000 | <0.0041** | <0.0053** | 15 | 1112 | 59 | 13 | 0.01566 | 0.01890 | <0.7005 | <0.7631 | Selected | |
| AY108178 | 14 | 1259 | 0 | 1 | 0.00000 | 0.00000 | <0.0054** | <0.0082** | 13 | 1224 | 54 | 10 | 0.01174 | 0.01458 | <0.3233 | <0.4719 | Selected | Circadian clock |
| AY106616 | 14 | 2745 | 84 | 9 | 0.01132 | 0.01888 | <0.4395 | <0.2205 | 7 | 2619 | 97 | 7 | 0.01509 | 0.02446 | <0.6859 | <0.7214 | Selected | Ankyrin repeat-like protein |
| AY107952 | 14 | 2469 | 23 | 5 | 0.00333 | 0.00446 | <0.1193 | <0.0927 | 14 | 2599 | 38 | 12 | 0.00390 | 0.00502 | <0.1453 | <0.1678 | Selected | Putative fruit protein, oxidoreductase |
| AY106371 | 14 | 1574 | 4 | 5 | 0.00091 | 0.00000 | <0.0094** | <0.0061** | 15 | 1615 | 65 | 12 | 0.01078 | 0.01197 | <0.4603 | <0.4047 | Selected | Putative methyl binding domain protein |
Gene, the GenBank accession of the original unigene sequence; N, number of sampled sequences; L, length of the core alignments in which all sequences contain bases, excluding gaps; S, total number of segregating sites; h, number of unique sequences (haplotypes); πtotal and πsilent, average proportion of pairwise differences per base pair at all sites and silent sites, respectively. HKA tests: P value of the candidate genes by multiple locus HKA against the four neutral genes by the maximum cell value test. HKAtotal and HKAsilent: HKA test at all sites and silent sites, respectively. *, P < 0.05; **, P < 0.01. Candidate status: genes shown in bold are selected throughout the gene length, and genes not in bold are only selected at the 3′ region.
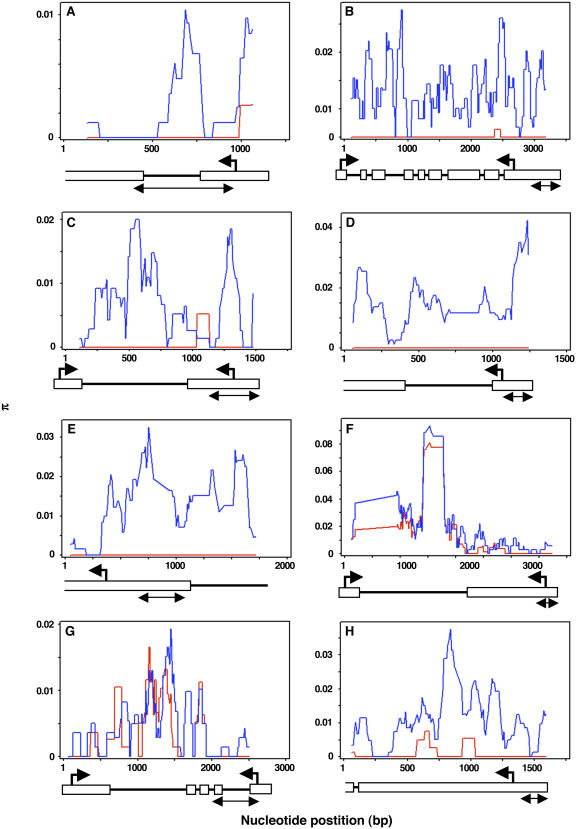
Figure 4.
Sliding-Window Analysis of the Genetic Diversity π in Maize Inbreds and Teosintes.
For sliding-window analysis, π was calculated for segments of 100 bp at 10-bp intervals. Horizontal and vertical axes on the graphs indicate DNA sequence position and genetic diversity π, respectively. Red lines indicate genetic diversity in the inbreds, whereas blue lines indicate genetic diversity in the teosintes. For gene structure under the sliding-window graphs, white bars indicate the predicted exons, and black lines indicate introns or genomic regions. Left arrows and right arrows indicate the positions for predicted start codons and stop codons, respectively. The lines with two arrows under the gene structure indicate the sequencing regions in our initial screening. (A) AY108876; (B) AY107195; (C) AY110109; (D) AY105060; (E) AY108178; (F) AY106616; (G) AY107952; (H) AY106371.
Five candidate genes, AY107195, AY110109, AY105060, AY108178, and AY106371, exhibited low nucleotide diversity in maize inbreds and high (normal) levels of nucleotide diversity in teosintes throughout the entire length of the gene (Table 6, Figure 4). The HKA tests at all sites (HKAtotal) verified selection at these five candidates (P < 0.01), whereas HKAtotal in teosintes was not significant. The relative ratios of π at all nucleotide sites (πtotal) in inbreds versus teosintes ranged from zero to 0.08, indicating that the inbreds have lost >92% of genetic variation in the teosinte sample. Thus, these five genes exhibit evidence of selection throughout their length, and extended sequencing unambiguously demonstrates that these are selected genes.
The evidence for selection on the remaining three genes is less striking. For AY108876, HKAtotal was significant in both teosintes and inbreds, but the HKA test at silent sites (HKAsilent; synonymous and noncoding positions) was significant for selection only in the inbreds (Table 6). One interpretation of these results is that this gene experienced selection in teosinte (as evidenced by low diversity in teosinte) and has undergone additional artificial selection (as evidenced by loss of silent site polymorphism from teosinte to maize).
For the remaining genes, AY106616 and AY107952, neither HKAtotal nor HKAsilent were significant for selection of the entire gene. However, the entire second exon and 3′ untranslated region at AY106616 was significant for selection (Figure 4F; Pinbreds < 0.0218 and Pteosintes < 0.4169). Because the region of reduced polymorphism in this gene extends for more than 1 kb, the distribution of polymorphism for this gene is also strongly consistent with selection. For AY107952, two small exons exhibit marked decreases in polymorphism between maize and teosinte (Figure 4). For only this single gene out of eight is the decreased polymorphism limited to the initially sequenced region. In summary, for seven of our eight candidate genes, the extended sequencing clearly supports selection over genetic drift for the cause of the low inbred diversity, affirming the validity of our approach to identifying selected genes.
DISCUSSION
Our overarching goal in this article was to identify selected genes in maize because they represent candidates to contribute to agronomic traits. These genes will permit us to reconstruct the selection history of maize and provide novel candidate genes for maize improvement. To identify these genes, we first determined that the average genetic diversity π for 1095 maize genes was 0.0067. This result confirms that the average maize gene retains high levels of sequence polymorphism. Our sample of inbred maize contains more nucleotide diversity than species-wide samples of sorghum (Sorghum bicolor), soybean (Glycine max), Arabidopsis thaliana, or Arabidopsis lyrata (Arabidopsis Genome Initiative, 2000; Wright et al., 2003; Zhu et al., 2003; Hamblin et al., 2004; Ramos-Onsins et al., 2004; Wright and Gaut, 2005). In addition to providing the basis for identifying candidates for selection, our sequence data identifying 6169 SNPs in 1095 genes also provide the groundwork for the development of a public SNP mapping resource for maize. As all genes selected for sequencing were from the Maize Mapping Project/Dupont unigene set (see Methods), the physical and genetic location of the majority of these genes is known by association to anchored BACs of the maize fingerprint contig map (www.genome.arizona.edu/fpc/maize).
In choosing the panel of inbreds for sequencing, we intentionally established a comparison of genetic diversity between temperate and tropical maize lines. Our results indicate that the amount of genetic diversity contained within the two germplasm pools is similar. Using SSR markers, Liu et al. (2003) reported that tropical inbreds contained greater genetic diversity than temperate inbreds. This discrepancy may be more apparent than real because of the different sampling strategies in the two studies. First, our temperate sample uniquely contains both popcorn (Hp301) and sweet maize (Il14H) inbreds, expected to extend the diversity within the temperate sample. Second, while we sampled a larger number of loci than Liu et al. (2003), our shallow sampling of only seven lines each from temperate and tropical germplasm pools is primarily detecting SNP that occur at moderate to high frequency in maize. If tropical maize contains more rare SNP variants than temperate inbred lines, these rare SNPs would go largely undetected by our sampling strategy. However, our data clearly indicate that introducing tropical maize inbred lines into U.S. maize breeding programs will not greatly increase the genetic diversity of most maize genes over using diverse temperate inbred lines.
In a previous publication, we demonstrated that maize genes containing SSR sequences that were invariant among maize inbred lines, but polymorphic among teosintes, were enriched for selected genes (Vigouroux et al., 2002b). Although successful in finding candidates, SSR screening for selected genes has many limitations: only ∼10% of EST contigs for maize contain an SSR sequence (M.D. McMullen, unpublished data), and the high mutation rate of SSR may allow some recovery of diversity since domestication (Vigouroux et al., 2002a). For example, the promoter region of a maize domestication gene tb1 is a well-characterized region of reduced nucleotide variation, but an SSR in this region exhibits high diversity (Tenaillon et al., 2002). In this study, we have generalized the approach of random searches of low diversity genes to those identified by DNA sequence data from a large set of randomly chosen genes. This approach can be used to test any maize gene for a role in domestication and crop improvement.
In theory, diversity screens for identifying selected genes can be applied to any animal or plant domesticate, but the power of the approach depends critically on relative levels and patterns of diversity in neutral genes, selected genes, and genes in the wild taxon. If neutral genes retain very little diversity after domestication, it is difficult to discriminate neutral from selected genes without sequence fragments that are much longer than the 500-bp fragments used in this study. Sorghum may be one crop for which it will be difficult to identify selected genes without long sequence fragments because diversity levels in the domesticate are very low for all loci sampled to date (Hamblin et al., 2004). By contrast, maize has relatively high levels of polymorphism among plants (Wright and Gaut, 2005) so that screens for selection can be efficient with short sequences. Another important issue that will vary from taxon to taxon is the probability that selected genes were targets of selection or were hitchhiking with another target of selection. The distinction between target and hitchhiking depends critically on LD. In maize, LD generally decays very rapidly (Remington et al., 2001; Tenaillon et al., 2001), thus improving the possibility that an identified gene was a target rather than hitchhiked. Generally, self-pollinating species like soybean (Zhu et al., 2003) are expected to have high levels of LD, but it is important to note that this is not always true (Morrell et al., 2005). Thus, even self-pollinating taxa may be suitable species for diversity screens.
The HKA and CS analyses test selection on two separate selection criteria: the HKA test compares genetic diversity of the candidate gene with neutral genes and explicitly incorporates divergence information (Hudson et al., 1987), whereas the CS test examines reduction of diversity within a gene compared with its estimated demographic history (Tenaillon et al., 2004). Therefore, the two tests are complementary rather than redundant, and our strongest selection candidates are the four domestication and four improvement genes identified by both criteria (Table 5, Figure 3). The CS tests assigned three additional genes as improvement genes that could not be tested by HKA due to our inability to obtain an orthologous T. dactyloides sequence (Table 5, Figure 3).
We recognize at least four limitations in our analysis. First, the fact that average length of sequence alignment is short leads to a conservative test for selection. From Figure 3 it is clear that the candidate genes with no inbred diversity that failed tests of selection were of low diversity in the teosintes. There must be a high number of segregating sites in the teosintes before significant loss of diversity can be detected. Longer sequences would provide more power to test for selection in genes with lower genetic diversity levels in all three populations. However, our extended sequencing results clearly indicate that the short alignment length does not result in an appreciable false positive rate. For the eight genes identified by both the initial HKA and CS tests, the extended sequencing demonstrated that six genes were significant for selection throughout their sequence, and one additional gene was selected for an extended region. For only one of the eight genes, we could not distinguish between selection and genetic drift as the cause of the initial identification of selection based on a short alignment. Second, despite the rapid decay of LD in maize (Remington et al., 2001; Tenaillon et al., 2001), the genes we have identified may only be hitchhiking with neighboring selected genes. The selective sweep at tb1 extended only 60 to 90 kb upstream of the gene and does not contain other genes (Clark et al., 2004). However, for the Y1 locus, a gene under recent and strong selection in maize breeding programs, the effects of selection were evident up to 600 kb downstream of the target gene in the yellow endosperm subset of maize lines (Palaisa et al., 2004). The extent of the low diversity region around selected genes needs to be determined for many additional genes before definitive statements of selection can be made, but it is reasonable to expect that hitchhiked regions may be larger for improvement genes than domestication genes due to a more recent history of selection. However, it is interesting to note that for the six genes with evidence of selection throughout their sequence, three are domestication candidates (AY108876, AY105060, and AY106371) and three are improvement candidates (AY107195, AY110109, and AY108178). Third, if a causal site under selection within a gene, such as a single amino acid difference, is at moderate allele frequency in the population and at low LD with other polymorphisms within that gene before selection, this selected gene will remain polymorphic at the linked sites after selection and evade detection by either the HKA or CS analysis (Innan and Kim, 2004). Fourth, we acknowledge multiple test issues. However, the use of a standard correction such as the Bonferroni with genomic scans, such as this study, would result in an unacceptably high false negative rate, eliminating many biologically significant genes from further study (Storey and Tibshirani, 2003). Our extended sequencing empirically demonstrates that requiring candidates to be significant by both HKA and CS analyses provides a stringent selection criterion that resulted in a very low false positive rate.
A BLAST analysis was used to identify the protein function of the selected genes and to provide clues as to the traits under selection (Table 5). For the domestication genes, AY107952 has homology to fruit protein of kiwifruit (Ledger and Gardner, 1994). AY106371 encodes a putative methyl binding domain protein in maize. DNA methylation is a common factor in epigenetic gene regulation in plants (Martienssen, 1998; Bender, 2004) and may affect expression of an undetermined target gene. AY108876 has significant homology to an amino acid transporter, suggesting a role in amino acid synthesis or metabolism. Other genes for amino acid synthesis have also been identified as candidates for selection in maize (Wright et al., 2005), suggesting that nutritional quality has been a major target of human selection. This result complements the finding that many genes in the starch synthesis pathways in maize also show evidence of human selection (Whitt et al., 2002). AY105060 has no homology to known genes and proteins. One of the advantages of our scan is that genes of unknown function like AY105060 can only be identified as selection candidates by an unbiased approach such as used in this article. Of the improvement genes, AY107195 encodes an auxin response transcription factor (Ulmasov et al., 1997) and may have been selected for altered growth response. A second auxin response transcription factor, AY104948, was identified as a selected gene by Wright et al. (2005). Taken together, the data suggest that auxin-regulated growth responses may have been a major target of artificial selection for enhanced maize productivity. AY106616 encodes ankyrin repeat-like protein, which mediates protein–protein interactions. AY110109 encodes a GTP binding protein; this class of proteins is involved in signal transduction, cell differentiation, or membrane vesicle transport (Kang et al., 1995). AY108178 encodes an F-box protein with homology to circadian clock genes, ZTL and FKF1, controlling flowering time in Arabidopsis (Nelson et al., 2000; Somers et al., 2000) and may have been selected for an aspect of maize maturity. Another F-box protein, AY104147, was also identified as selected by Wright et al. (2005). From conducting multiple genomic searches for selection, key gene families and biological processes essential to maize improvement are becoming apparent.
Early studies on potential domestication genes in maize focused on the obvious morphological and developmental differences between maize and teosinte (Dorweiler et al., 1993; Hanson et al., 1996; Doebley et al., 1997). Recent studies have indicated that a number of the genes for enzymes in the starch synthesis pathway also exhibit signs of selection, demonstrating that selection has affected specific biochemical pathways (Whitt et al., 2002). However, performing individual tests for candidate genes postulated in controlling adaptive processes is laborious and time consuming. Taking an unbiased approach to the identification of selected genes is a more efficient path to understanding the genetic consequences of domestication and crop improvement. The results of this study suggest that genes controlling a wide range of traits may have been targets of selection (Table 5), and that our approach can also identify unknown function genes as important to domestication or improvement demonstrates an important advantage of our approach over a priori selection of candidates to test.
The productivity of crop species advances by the selection of favorable alleles at genes controlling the traits targeted by the plant breeder. However, as we have demonstrated, the genes that have undergone the most stringent selection (and thereby the greatest reduction in diversity) have little remaining genetic variation and cannot easily be further improved by standard plant breeding or even identified by QTL analysis because all inbred lines will have similar alleles. Indeed, where classical QTL mapping experiments between inbreds may fail to identify these genes as important genetic factors for agronomic traits, our strategy of genome screening for selected genes proved successful for detecting novel candidate domestication and improvement genes, providing new target genes for crop improvement. There are two approaches to reintroduce variation at these genes into maize breeding programs. The alleles from teosintes for domestication genes and from landraces for improvement genes could be introduced into maize breeding programs. It is specifically these genes that need to be added to maize breeding from exotic sources to broaden the genetic base of maize breeding efforts. Alternatively, transgenic alteration of the expression patterns of selected genes can be tested for desired effects on the relevant agronomic traits.
METHODS
Plant Materials
We used three diverse sets of maize (Zea mays) materials for DNA sequence analysis: inbred lines, landraces, and teosinte accessions (Table 1). The 14 maize inbreds, composed of seven temperate and seven tropical inbreds, were chosen based on SSR polymorphism data to maximize allelic diversity (Liu et al., 2003). The 16 maize landraces represent all areas in which maize was grown at the time of the discovery of the New World (Tenaillon et al., 2001). The 16 teosinte accessions were chosen based on geographic criteria to represent all areas where Z. mays subsp parviglumis is found. Each teosinte accession was self-pollinated twice or three times to derive partially inbred material. Two accessions of the wild relative, Tripsacum dactyloides (gammagrass), were used as an outgroup species for statistical analyses. The DNA was extracted with standard protocols (Saghai-Maroof et al., 1984) with minor modification.
Sequence Analysis
All genes for DNA sequencing were selected from the Maize Mapping Project/Dupont unigene set (http://www.agron.missouri.edu/files_dl/MMP/Cornsensus). The PCR primers were designed using the Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) to amplify 300 to 500 bp surrounding the 3′ untranslated region. The PCR was performed using Red Taq (Sigma-Aldrich) in a DNA Engine Tetrad thermocycler (MJ Research) with touchdown PCR (one cycle of 45 s at 95°C, 45 s at 65°C, and 55 s at 72°C; 1°C decrement in annealing temperature per cycle until annealing temperature is 55°C; then 25 cycles of 45 s at 95°C, 45 s at 55°C, and 1 min at 72°C). Following PCR amplification, unincorporated primers and deoxynucleotide triphosphates were removed by ethanol precipitation prior to sequencing. The PCR products were sequenced with each forward and reverse primer using BigDye terminator version 3.1 or dRhodamine terminator cycle sequencing kits (Applied Biosystems) and analyzed on ABI 3100 or 3700 sequencers (Applied Biosystems). The PCR products from T. dactyloides accession WW-2120 were cloned into pGEM-T vector (Promega), and eight clones were sequenced in both directions to eliminate Taq polymerase errors.
For extended sequencing of the eight selected genes, several sets of PCR primers were designed using the Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) to amplify 500- to 1000-bp products. The genomic PCR was performed using PCR Master Mix (Promega) or TaKaRa LA Taq with GC buffer (TaKaRa Bio) with a DNA Engine Tetrad thermocycler (one cycle of 2 min at 95°C; 35 cycles of 1 min at 95°C, 1 min at 50 to 65°C [dependent on primers] and 30 s to 3 min [dependent on PCR product] at 72°C; final extension of 3 to 5 min at 72°C). Following PCR amplification, unincorporated primers and deoxynucleotide triphosphates were removed by exonuclease I and shrimp alkaline phosphatase (United States Biochemical) and ethanol precipitation prior to sequencing. The PCR products were sequenced using BigDye terminator version 3.1 and analyzed on an ABI 3100 sequencer. The PCR products from two T. dactyloides accessions, WW-2120 and MIA 34597, were cloned into pGEM-T vector, and several clones were sequenced to eliminate Taq polymerase errors.
Base calling, quality assessment, and trimming of trace files were conducted with PHRED (Ewing and Green, 1998; Ewing et al., 1998), and sequence assembly was performed by PHRAP. The multiple sequences for each gene were aligned with ClustalW (Thompson et al., 1994) and edited manually.
Statistical Analyses
For each locus, two individuals, B73 and Mo17, were sequenced twice. The inclusion of replicates permitted assessment of empirical error rates. With an SNP identification criterion based on a PHRAP quality score >30, we found 45 mismatched sites between either B73 or Mo17 sequences. In total, 628,071 replicate nucleotide sites were compared, yielding an empirical error rate of ∼1 error in 7290 bp. However, the errors were nonrandomly clustered among loci. The 45 errors were found in only 11 loci, and five loci accounted for 90% of the errors. This distribution suggests that some of the errors may be due to sample switching. Such switching is not problematic for identifying SNPs and measuring diversity but could cause problems for association analyses. After removal of errors apparently due to sample switching, the sequencing error rate is essentially zero.
For population genetic analyses, sequences were removed if they had an average PHRAP quality score <30 or were <80% of the average length of the sequences in the alignment. Only loci with sequences from four or more individuals were analyzed for SNP identification and population genetic analysis. Only SNP variants with a PHRAP quality score >30 were used for analysis. For population genetic analyses of the candidate genes, we determined the number of polymorphic sites (S), the number of unique sequences (haplotypes; h), and the average proportion of pairwise nucleotide differences per nucleotide site (π ; Tajima, 1983) for each gene in DnaSP version 4.00 (Rozas et al., 2003). DnaSP was also used to determine Tajima's D (Tajima, 1989) for the four neutral genes. For sequence data comparisons, Mann-Whitney U, Student's t, Kruskal-Wallis H, and Scheffé's F tests were performed using Statcel (Yanai, 1998).
The HKA tests (Hudson et al., 1987) were conducted with adh1, bz2, fus6, and glb1 (Eyre-Walker et al., 1998; Hilton and Gaut, 1998; Tenaillon et al., 2001) as neutral control genes and with sequence from T. dactyloides representing the outgroup using HKA software (http://lifesci.rutgers.edu/∼heylab/HeylabSoftware.htm#HKA). The HKA test of selection is similar to a χ2 test and asks whether the relative levels of intraspecific polymorphism and interspecific divergence for a locus are consistent across loci. The table for observation values was arranged for an interest gene and four neutral genes, including one for variation in a species (maize inbreds, landraces, or teosintes), one for variation in T. dactyloides, and one for the divergence between the species. Like a χ2 test, two tables for expectations under a null model and standardized discrepancies between observations and expectations were generated. Rejection of the null hypothesis requires that the sum of standardized discrepancies between observations and expectations be greater than expected by chance (Hudson et al., 1987). If selection has occurred at the interest gene in a species, only one cell in the standardized discrepancy table may show a large value, and the overall test may not detect the discrepancy. The HKA software analyzes the single cell departure from the null hypothesis with multiple loci. In this outlier test, the test statistic is the maximum standardized discrepancy (MSD) observed for any polymorphism value. If the species has experienced a selective sweep at the interest locus, the standardized discrepancy for that observation will be high. This test statistic was then compared with a neutral distribution of the MSD that was generated by independent HKA coalescent simulations (Hudson et al., 1987). For each simulation, the MSD observed for any locus, for polymorphism in either species, is the MSD measure for that simulation and is recorded. If the observed MSD value is >95% of the entire frequency distribution of simulated MSD values, then the outlier test is significant (Wang and Hey, 1996). If this test was not possible due to low deviation for the interest gene, the overall P value based on simulated MSD distribution was used. Any gene in which the HKA tests indicated significance in teosintes was considered a low diversity gene for the teosintes and excluded from consideration for selection in maize.
Coalescent Simulations
CS tests were used to model the impact of a bottleneck on sequence diversity, using protocols detailed previously (Tenaillon et al., 2004). The method first estimates the demographic history of reference genes. Two demographic scenarios were estimated separately: a domestication bottleneck based on comparisons between teosinte and landrace data and an improvement bottleneck based on comparisons between teosinte and inbred data. For both scenarios, the maximum likelihood of the ratio k of bottleneck duration and strength was estimated with data from four reference genes (adh1, bz2, fus6, and glb1; Eyre-Walker et al., 1998; Hilton and Gaut, 1998; Tenaillon et al., 2001). For the landraces, the estimated neutral multilocus parameter was k = 4.65, and for the inbreds, the estimated parameter was k = 1.25. We also estimated the maximum likelihood of domestication and improvement bottlenecks for each candidate gene. The CS statistic is a likelihood ratio of the best fitting demographic scenario for the candidate gene against the multilocus reference estimate, and it tests whether the candidate gene has a history concordant with the reference loci at P = 0.05. For all likelihood estimation and all tests, initial simulation parameters and multilocus likelihoods were calculated as described previously (Tenaillon et al., 2004) except that (1) only Hudson's estimator (Hudson, 2001) was used as the recombination-population parameter in simulations and (2) k was explored over a grid of 32 values, with a fixed bottleneck duration of 1000 generations. Fixing the duration of the bottleneck does not affect estimation of k (Tenaillon et al., 2004). For all simulations, the goodness-of-fit statistic was the number of segregating sites in maize.
Homology Search
We utilized both nucleotide–nucleotide (BLASTN) and translated query versus protein database (BLASTX) BLAST routines (http://www.ncbi.nlm.nih.gov/BLAST) to identify protein functions for candidate genes. All reported homologies exceed a 1e−20 threshold for at least one of the two BLAST routines. The gene prediction was performed using the automated annotation system RiceGAAS (Sakata et al., 2002; http://ricegaas.dna.affrc.go.jp/).
Accession Numbers
All sequence data from this article were deposited in GenBank (BV106362 to BV123527), and all alignments are available from www.panzea.org. The 35 initial sequence alignments and the eight extended sequencing alignments are also available in Supplemental Tables 2 and 3 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Sequence Diversity in Temperate and Tropical Maize Inbreds for 1095 Maize Unigenes.
Supplemental Table 2. Sequence Alignments in Maize Inbreds, Landraces, Teosintes, and Tripsacum at 35 Genes That Exhibited Zero Sequence Diversity in Maize Inbreds.
Supplemental Table 3. Extended Sequence Alignments in Maize Inbreds, Teosintes, and Tripsacum at Eight Candidate Genes.
Supplementary Material
Acknowledgments
This research was supported by National Science Foundation Plant Genome Awards DBI0096033, DBI9872655, and DBI0321467, by research funds provided by the USDA Agricultural Research Service (M.D.M.), and by the Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad in 2004 (M.Y.). We thank Kate Houchins, Linda Schultz, and Ngozi Duru for technical assistance. We thank Ed Buckler and Jack Liu for assistance in choosing the 14 diverse maize inbred lines. Names of products are necessary to report factually on available data; however, neither the USDA nor any other participating institution guarantees or warrants the standard of the product, and the use of the name does not imply approval of the product to the exclusion of others that may also be suitable.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Michael D. McMullen (mcmullenm@missouri.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.037242.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Bender, J. (2004). DNA methylation and epigenetics. Annu. Rev. Plant Biol. 55, 41–68. [DOI] [PubMed] [Google Scholar]
- Bhattramakki, D., Dolan, M., Hanafey, M., Wineland, R., Vaske, D., Register III, J.C., Tingey, S.V., and Rafalski, A. (2002). Insertion-deletion polymorphisms in 3′ regions of maize genes occur frequently and can be used as highly informative genetic markers. Plant Mol. Biol. 48, 539–547. [DOI] [PubMed] [Google Scholar]
- Clark, R.M., Linton, E., Messing, J., and Doebley, J.F. (2004). Pattern of diversity in the genomic region near the maize domestication gene tb1. Proc. Natl. Acad. Sci. USA 101, 700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488. [DOI] [PubMed] [Google Scholar]
- Dorweiler, J., Stec, A., Kermicle, J., and Doebley, J. (1993). Teosinte glume architecture 1: A genetic locus controlling a key step in maize evolution. Science 262, 233–235. [DOI] [PubMed] [Google Scholar]
- Ewing, B., and Green, P. (1998). Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8, 186–194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. (1998). Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8, 175–185. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker, A., Gaut, R.L., Hilton, H., Feldman, D.L., and Gaut, B.S. (1998). Investigation of the bottleneck leading to the domestication of maize. Proc. Natl. Acad. Sci. USA 95, 4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary, A., Nesbitt, T.C., Frary, A., Grandillo, S., van der Knaap, E., Cong, B., Liu, J., Meller, J., Elber, R., Alpert, K.B., and Tanksley, S.D. (2000). fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 289, 85–88. [DOI] [PubMed] [Google Scholar]
- Fu, H., Zheng, Z., and Dooner, H.K. (2002). Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proc. Natl. Acad. Sci. USA 99, 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti, A., Zhao, Q., Kyozuka, J., Meeley, R.B., Ritter, M.K., Doebley, J.F., Enrico Pè, M., and Schmidt, R.J. (2004). The role of barren stalk1 in the architecture of maize. Nature 432, 630–635. [DOI] [PubMed] [Google Scholar]
- Hamblin, M.T., Mitchell, S.E., White, G.M., Gallego, J., Kukatla, R., Wing, R.A., Paterson, A.H., and Kresovich, S. (2004). Comparative population genetics of the panicoid grasses: Sequence polymorphism, linkage disequilibrium and selection in a diverse sample of Sorghum bicolor. Genetics 167, 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, M.A., Gaut, B.S., Stec, A.O., Fuerstenberg, S.I., Goodman, M.M., Coe, E.H., and Doebley, J.F. (1996). Evolution of anthocyanin biosynthesis in maize kernels: The role of regulatory and enzymatic loci. Genetics 143, 1395–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton, H., and Gaut, B.S. (1998). Speciation and domestication in maize and its wild relatives: Evidence from the Globulin-1 gene. Genetics 150, 863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R.R. (2001). Two-locus sampling distributions and their application. Genetics 159, 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R.R., Kreitman, M., and Aguadé, M. (1987). A test of neutral molecular evolution based on nucleotide data. Genetics 116, 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan, H., and Kim, Y. (2004). Pattern of polymorphism after strong artificial selection in a domestication event. Proc. Natl. Acad. Sci. USA 101, 10667–10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, K.K., Sano, H., and Kameya, T. (1995). Characterization of cDNAs encoding small GTP-binding proteins from maize. Plant Physiol. 107, 275–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledger, S.E., and Gardner, R.C. (1994). Cloning and characterization of five cDNA for genes differentially expressed during fruit development of kiwifruit (Actinidia deliciosa var. deliciosa). Plant Mol. Biol. 25, 877–886. [DOI] [PubMed] [Google Scholar]
- Liu, K., Goodman, M., Muse, S., Smith, J.S., Buckler, E., and Doebley, J. (2003). Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165, 2117–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen, R. (1998). Transposons, DNA methylation and gene control. Trends Genet. 14, 263–264. [DOI] [PubMed] [Google Scholar]
- Matsuoka, Y., Vigouroux, Y., Goodman, M.M., Jesus Sanchez, G., Buckler, E., and Doebley, J. (2002). A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. USA 99, 6080–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell, P.L., Toleno, D.M., Lundy, K.E., and Clegg, M.T. (2005). Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp. spontaneum) despite high rates of self-fertilization. Proc. Natl. Acad. Sci. USA 102, 2442–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.C., Lasswell, J., Rogg, L.E., Cohen, M.A., and Bartel, B. (2000). FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101, 331–340. [DOI] [PubMed] [Google Scholar]
- Palaisa, K., Morgante, M., Tingey, S., and Rafalski, A. (2004). Long-range patterns of diversity and linkage disequilibrium surrounding the maize Y1 gene are indicative of an asymmetric selective sweep. Proc. Natl. Acad. Sci. USA 101, 9885–9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaisa, K.A., Morgante, M., Williams, M., and Rafalski, A. (2003). Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci. Plant Cell 15, 1795–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, D.R., and Flannery, K.V. (2001). The earliest archaeological maize (Zea mays L.) from highland Mexico: New accelerator mass spectrometry dates and their implications. Proc. Natl. Acad. Sci. USA 98, 2101–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski, A., and Morgante, M. (2004). Corn and humans: Recombination and linkage disequilibrium in two genomes of similar size. Trends Genet. 20, 103–111. [DOI] [PubMed] [Google Scholar]
- Ramos-Onsins, S.E., Stranger, B.E., Mitchell-Olds, T., and Aguadé, M. (2004). Multilocus analysis of variation and speciation in the closely related species Arabidopsis halleri and A. lyrata. Genetics 166, 373–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington, D.L., Thornsberry, J.M., Matsuoka, Y., Wilson, L.M., Whitt, S.R., Doebley, J., Kresovich, S., Goodman, M.M., and Bucker IV, E.S. (2001). Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 98, 11479–11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., Sánchez-DelBarrio, J.C., Messeguer, X., and Rozas, R. (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof, M.A., Soliman, K.M., Jorgensen, R.A., and Allard, R.W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 81, 8014–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata, K., Nagamura, Y., Numa, H., Antonio, B.A., Nagasaki, H., Idonuma, A., Watanabe, W., Shimizu, Y., Horiuchi, I., Matsumoto, T., Sasaki, T., and Higo, K. (2002). RiceGAAS: An automated annotation system and database for rice genome sequence. Nucleic Acids Res. 30, 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers, D.E., Schultz, T.F., Milnamow, M., and Kay, S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101, 319–329. [DOI] [PubMed] [Google Scholar]
- Storey, J.D., and Tibshirani, R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F. (1983). Evolutionary relationship of DNA sequences in finite populations. Genetics 105, 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, M.I., Sawkins, M.C., Anderson, L.K., Stack, S.M., Doebley, J., and Gaut, B.S. (2002). Patterns of diversity and recombination along chromosome 1 of maize (Zea mays L. ssp. mays). Genetics 162, 1401–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, M.I., Sawkins, M.C., Long, A.D., Gaut, R.L., Doebley, J.F., and Gaut, B.S. (2001). Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays L. ssp. mays). Proc. Natl. Acad. Sci. USA 98, 9161–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, M.I., U'Ren, J., Tenaillon, O., and Gaut, B.S. (2004). Selection versus demography: A multilocus investigation of the domestication process in maize. Mol. Biol. Evol. 21, 1214–1225. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry, J.M., Goodman, M.M., Doebley, J., Kresovich, S., Nielsen, D., and Buckler IV, E.S. (2001). Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28, 286–289. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- Vigouroux, Y., Jaqueth, J.S., Matsuoka, Y., Smith, O.S., Beavis, W.D., Smith, J.S.C., and Doebley, J. (2002. a). Rate and pattern of mutation at microsatellite loci in maize. Mol. Biol. Evol. 19, 1251–1260. [DOI] [PubMed] [Google Scholar]
- Vigouroux, Y., McMullen, M., Hittinger, C.T., Houchins, K., Schulz, L., Kresovich, S., Matsuoka, Y., and Doebley, J. (2002. b). Identifying genes of agronomic importance in maize by screening microsatellites for evidence of selection during domestication. Proc. Natl. Acad. Sci. USA 99, 9650–9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Nussbaum-Wagler, T., Li, B., Zhao, Q., Vigouroux, Y., Faller, M., Bomblies, K., Lukens, L., and Doebley, J.F. (2005). The origin of naked grains of maize. Nature 436, 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R.-L., and Hey, J. (1996). The speciation history of Drosophila pseudoobscura and close relatives: Inferences from DNA sequence variation at the period locus. Genetics 144, 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R.-L., Stec, A., Hey, J., Lukens, L., and Doebley, J. (1999). The limits of selection during maize domestication. Nature 398, 236–239. [DOI] [PubMed] [Google Scholar]
- Whitt, S.R., Wilson, L.M., Tenaillon, M.I., Gaut, B.S., and Buckler IV, E.S. (2002). Genetic diversity and selection in the maize starch pathway. Proc. Natl. Acad. Sci. USA 99, 12959–12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S.I., and Gaut, B.S. (2005). Molecular population genetics and the search for adaptive evolution in plants. Mol. Biol. Evol. 22, 506–519. [DOI] [PubMed] [Google Scholar]
- Wright, S.I., Lauga, B., and Charlesworth, D. (2003). Subdivision and haplotype structure in natural populations of Arabidopsis lyrata. Mol. Ecol. 12, 1247–1263. [DOI] [PubMed] [Google Scholar]
- Wright, S.I., Vroh Bi, I., Schroeder, S.G., Yamasaki, M., Doebley, J.F., McMullen, M.D., and Gaut, B.S. (2005). The effects of artificial selection on the maize genome. Science 308, 1310–1314. [DOI] [PubMed] [Google Scholar]
- Yanai, H. (1998). Statcel: The Useful Addin Forms on Excel. (Saitama, Japan: OMS Publishing Company).
- Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., Baba, T., Yamamoto, K., Umehara, Y., Nagamura, Y., and Sasaki, T. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y.L., Song, Q.J., Hyten, D.L., Van Tassell, C.P., Matukumalli, L.K., Grimm, D.R., Hyatt, S.M., Fickus, E.W., Young, N.D., and Cregan, P.B. (2003). Single-nucleotide polymorphisms in soybean. Genetics 163, 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.