Abstract
Signals generated by T cell receptor (TCR) and CD28 engagement are required for optimal T cell activation, but how these signals integrate within the cell is still largely unknown. We have used near genome-scale expression profiling to monitor T cell signal transduction pathways triggered via TCR and/or costimulatory receptors. Ligation of CD28 alone induced a set of short-lived early response transcripts in both Jurkat T cells and primary CD4 T cells, thus providing evidence that CD28 engagement can affect gene regulation independently of TCR engagement. Simultaneous signaling through both the TCR and CD28 resulted in altered expression of several thousand genes following several distinct temporal patterns. Most of these gene regulations were induced by TCR signaling alone and were augmented to varying degrees by CD28 costimulation. CD28 and ICOS costimulation had nearly identical effects on gene regulation, but a few transcripts (e.g., IL2, IL9) were significantly more affected by CD28. Therefore, the distinctive functional outcomes of costimulation via CD28 and ICOS are accompanied by relatively few distinct differences in gene expression. Cytotoxic T lymphocyte antigen 4 (CTLA-4) engagement selectively blocked augmentation of gene regulations by CD28-mediated costimulation, but did not ablate gene regulation induced by TCR triggering alone.
Antigen engagement of the T cell receptor (TCR) initiates a genetic program that results in anergy (1), apoptosis (2), clonal expansion, and/or differentiation into TH1, TH2 (3), or regulatory T cells (4). Interaction with antigen-presenting cells at the immunological synapse via costimulatory molecules provides an additional signal(s) that determines T cell fate after antigen recognition. CD28 is a costimulatory receptor whose ligation is required to rescue T cells from anergy or apoptosis and promote T cell clonal expansion (5). CTLA-4 (CD152) shares structural homology with CD28 and binds to the same B7 family ligands but functions as an immune attenuator (6). ICOS, a third member of the CD28 family, binds a different ligand and has a distinct in vivo function (7). Many of the proximal events following TCR engagement have been elucidated (5, 8), but how these events are integrated with costimulatory signaling is poorly understood. The technique of gene expression profiling by DNA microarray hybridization represents an alternative approach for studying the circuitry of signaling pathways. This technique allows systematic measurement of the expression of many thousands of discrete sequences in a single assay (9). Thus, one signaling pathway can be compared with another by measuring differences in both the amplitude and absolute number of gene regulations triggered (10). Additionally, insights into how signaling pathways integrate can be gained by measurement of gene expression changes induced by simultaneous triggering of two pathways. Here, we have used ink-jet DNA microarrays (11) to study TCR, CD28, ICOS, and CTLA-4 signaling pathways.
Materials and Methods
Cell Culture and Treatments.
PBLs were isolated by Percoll (Amersham Pharmacia Biotech) gradient centrifugation from leukopacks following apheresis and elutriation. CD4 T cells were purified by negative selection using magnetic beads (Dynal, Lake Success, NY) as described (12) and were routinely >98% CD3+, >98% CD28+, and <3% CD8+. The preparation of beads containing equal mixtures of CD3 and MHC I (CD3/MHC I), CD28 (CD3/28), and ICOS (CD3/ICOS) beads has been described (13, 14). The beads used to analyze the effects of CTLA-4 engagement were prepared as described (15). CD86 Ig (a generous gift of Beatriz Carreno, Genetics Institute, Cambridge, MA) and anti-CD28 (9.3) coated beads were made as per the manufacturer's instructions (Dynal). To stimulate cells, beads with were mixed with CD4 T cells at a 3:1 ratio and cultured in RPMI plus 10% FBS in a 5% CO2 atmosphere at 37°C at 1 × 106 per ml for the indicated time. TCR stimulation of Jurkat cells was performed by culturing on tissue culture vessels coated with optimal stimulating concentrations of purified anti-human Vβ8 TCR mAb (clone JR2, PharMingen). CD28 stimulation was performed by addition of anti-CD28 mAb to a concentration of 1 μg/ml (anti-CD28 mAbs 9.3 and 2E12, from Robert Mittler (Emory University, Atlanta), were used interchangeably in these experiments). Anti- CD2 mAb, clone RPA-2.10; anti-CXCR4 mAb, clone 12G5; and anti-CD11a mAb, clone HI111, were from PharMingen.
RNA Isolation, cRNA Amplification, Labeling, and Hybridization to hu25K Microarrays.
Following stimulation, cells were harvested and total cellular RNA was isolated using either TRIzol (Life Technologies, Rockville, MD) or RNeasy methods (Qiagen, Valencia, CA) including DNase treatment. Methods for mRNA amplification using in vitro transcription (IVT), cRNA labeling with Cy3 and Cy5, and hybridization have been described (11). cRNAs from stimulated and unstimulated cells were compared by competitive hybridization on hu25K ink-jet oligonucleotide microarrays. Sequences represented on the microarrays are listed in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. Hybridizations were performed in duplicate with fluor reversal. Slides were scanned using a confocal laser scanner from Agilent Technologies (Palo Alto, CA). Fluorescent intensities on images were quantified, corrected for background, and normalized (16). Transcript regulations are presented as stimulated/unstimulated ratios, which by convention are depicted as Cy5/Cy3 or Red/Green pseudocolor ratios (16). Hierarchical clustering using the agglomerative nesting technique was performed as described (17, 18).
Results and Discussion
CD28 Stimulation Alone Can Affect the Transcript Abundance of a Set of Genes.
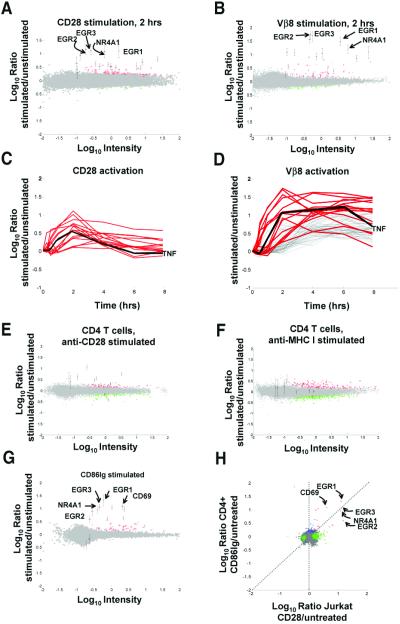
We first focused on determining whether expression profiling could demonstrate differences in CD28 and TCR signaling. CD28 triggering in the absence of TCR ligation induces the transient phosphorylation of several T cell signaling molecules and regulation of CD25 (19), but it is generally thought that CD28 only functions through its interactions with the TCR-generated signals. Initially, we focused our comparison of TCR and CD28 signals on Jurkat T cells. To compare CD28 and TCR signals, Jurkat T cells were treated with anti-TCR or anti-CD28 mAbs for 2 h, and RNA was harvested and amplified into cRNA by in vitro transcription (11). Competitive hybridization was performed using cRNA from untreated and treated Jurkat cells on a high-density DNA microarray by using a single 60-mer oligonucleotide to represent ≈25,000 different mRNA and EST sequences. A global view of gene regulations in all experiments presented in this manuscript is shown in Fig. 4, which is published as supporting information on the PNAS web site; the complete data set for all experiments is presented in Table 1. A comparison of transcripts regulated significantly (P < 0.01) by stimulation with CD28 and TCR is shown in Fig. 1 A and B. We reproducibly found that stimulation by anti-CD28 mAb led to regulation of a small group of genes that includes members of the Egr family of transcriptional activators (EGR1, EGR2, EGR3, EGR4). To ensure against erroneous identification of genes as regulated due to spurious fluctuations in hybridization signals, we combined array data from multiple independent experiments to select a consensus set of genes regulated by CD28 with defined statistical parameters (see Table 2, which is published as supporting information on the PNAS web site). Moreover, these genes were not induced by stimulation with other T cell specific mAbs, including anti-CD2, anti-CD11a, and anti-CXCR4 (see Fig. 5, which is published as supporting information on the PNAS web site). Additionally, we verified the up-regulation of two CD28-induced genes, EGR1 and CD69, by independent techniques (see Fig. 6, which is published as supporting information on the PNAS web site).
Figure 1.
CD28 stimulation induces gene regulations in T cells. (A) CD28 stimulation induces gene regulations in Jurkat T cells. Jurkat T cells were left untreated or were stimulated with soluble anti-CD28 mAb for 2 h. Gene regulations induced by treatment were detected by competitive hybridization to ink-jet DNA microarrays. Plotted is the average from duplicate microarrays of the Cy5/Cy3 hybridization ratio for each oligonucleotide versus the hybridization intensity for that oligonucleotide. In this representation, the y axis is an estimation of the ratio of expression for a given transcript in stimulated/unstimulated cells, whereas the scale on the x axis is proportional to transcript abundance. Red and green pseudocolors indicate Cy5 and Cy3 labeling, respectively. Genes expressed significantly (P < 0.01) more in treated cells (up-regulated genes) are colored red; significantly down regulated genes are colored green; genes whose expression was not significantly altered during stimulation are colored gray. Consensus Jurkat CD28-induced genes (see Table 2) are indicated with error bars (±SD). Selected CD28-induced genes are flagged: EGR1, X52541; EGR2, J04076; EGR3, X63741; NR4A1, D49728. (B) TCR stimulation alone induces gene regulations in Jurkat T cells. Jurkat T cells were left untreated or were stimulated with plate-bound anti-Vβ8 mAb for 2 h. Detection of gene regulations by DNA microarray analysis was performed and depicted as in A. Consensus Jurkat CD28-induced genes are indicated as in A. (C) CD28-induced genes have immediate-early kinetics. Jurkat T cells were left untreated or were stimulated with soluble anti-CD28 mAb for 0, 0.25, 0.5, 12, 2, 4, and 8 h. Detection of gene regulations by DNA microarray analysis was performed as in A. Red curves depict the kinetics of regulation for consensus Jurkat CD28-induced genes. No additional genes were regulated >2-fold, P < 0.01 for at least two time points. Genes not significantly regulated are not depicted. Black line, TNF, tumor necrosis factor, X01394. (D) CD28-induced genes are a subset of TCR-induced genes. Jurkat T cells were left untreated or were stimulated with plate-bound anti-Vβ8 mAb as in C. Red curves depict the kinetics of regulation for consensus Jurkat CD28-induced genes in Vβ8-stimulated cells. Gray curves indicate hybridization ratios for additional Vβ8-induced genes showing 2-fold regulation; P < 0.01 for at least two time points. Genes not significantly regulated are not depicted. (E) Anti-CD28 mAb induces gene expression changes in CD4 T cells. CD4 T cells were stimulated with anti-CD28 beads for 2 h. Consensus CD4 T cell CD86Ig-induced genes (see Table 2) are denoted with error bars. (F) CD28-induced genes are not induced by MHCI engagement in CD4 T cells. CD4 T cells were stimulated with anti-MHC beads for 2 h. Consensus CD4 CD28-induced genes are denoted with error bars. (G) CD28 stimulation by a natural ligand induces gene regulations in CD4 T cells. CD4 T cells were left untreated or were stimulated with CD86Ig beads for 2 h. Detection of gene regulations by DNA microarray analysis was performed and depicted as in A. Consensus CD4 T cell CD28-induced genes (see Table 2) are indicated with error bars (±SD). Selected CD28-induced genes are flagged as in A, with the addition of CD69, NM_001781. (H) Similar genes are induced by CD28 stimulation in Jurkat and CD4 T cells. Shown is a comparison (correlation) of genes significantly regulated (P < 0.01, log10 intensity >−1) in anti-CD28-treated Jurkat T cells (A, x axis) versus CD86Ig-treated CD4 T cells (G, y axis). Red, genes significantly regulated under both conditions; green, genes significantly regulated in X dimension; blue, genes significantly regulated in Y dimension; brown, genes showing opposite regulation in the two conditions; gray, genes not significantly regulated in either dimension. Selected genes significantly induced under both conditions are flagged.
TCR stimulation resulted in the up-regulation of a larger set of genes that included those regulated by CD28 (Fig. 1B). Next, we compared the kinetics of regulation of this set of genes following stimulation by either CD28 or TCR (red-shaded curves, Fig. 1 C and D). Most genes up-regulated by CD28 ligation reached maximal levels after 2 h of stimulation and returned to near resting levels after 8 h. In contrast, these same genes treated by TCR signaling remained up-regulated throughout 8 h, suggesting differences in the way TCR and CD28 signaling up-regulate this gene set. It was also important to determine whether engagement of CD28 alone would trigger these same gene regulations in nontransformed T cells. We therefore tested whether stimulation of CD28 by beads coated with anti-CD28 mAb (CD28 beads), or CD86Ig (CD86 beads), a natural ligand of CD28, could induce the same gene regulations in primary CD4 T cells. As shown in Fig. 1E, many of the same transcripts induced in Jurkat cells with soluble anti-CD28 mAb were also induced in CD4 T cells by CD28 beads but not anti-MHCI mAb-coated beads (Fig. 1 E and F), indicating that these genes are being induced as a direct result of CD28 ligation rather than a general nonspecific effect elicited by binding of mAb-coated beads. CD86Ig beads induced more pronounced gene regulations with CD4 T cells than CD28 mAb beads (Fig. 1G, consensus genes listed in Table 2). There was a correlation between the genes induced in primary and Jurkat T cells, indicating that this signaling pathway is conserved in both cell types (Fig. 1H). Although we recognize that these genes may be induced by many stimuli besides CD28, for the purposes of this manuscript we refer to these genes as “CD28-induced genes.” Taken together, the results in Fig. 1 indicate that CD28 ligation alone can alter the gene regulation of a subset of TCR-induced genes independent of TCR signaling.
The CD28-induced genes are noteworthy in that nearly all of them (7/9 or ≈78% of the known genes in the CD86Ig-induced gene set, Table 2) contain three or more copies of the sequence motif, AUUUA, in their 3′ untranslated regions (3′ UTR). In comparison, this motif was found in three or more copies in only 16/50 or 32% of randomly selected genes represented on the chip. Thus, transcripts containing multiple AUUUA motifs are ≈2.4-fold enriched amongst the CD28-induced transcripts. This motif is commonly found in the 3′ UTR of cytokines and other inflammatory mediators and may regulate mRNA stability of these transcripts (20). Lindsten et al. (21) demonstrated that CD28 costimulation altered mRNA stability for select cytokine genes having unstable transcripts characterized by AUUUA motifs in their 3′ UTR regions. The enrichment for transcripts containing multiple copies of this motif amongst the CD28-induced genes may indicate that signals initiated by CD28 triggering regulate mRNA stability of transcripts containing multiple AUUUA motifs. This signaling may contribute to desensitization of CD28 signaling following ligand engagement in the absence of TCR triggering (22). We speculate that signaling via engagement of CD28 alone may regulate costimulatory signals delivered through this receptor, particularly at sites of nonspecific inflammation where CD28 ligands, CD80 and CD86, are up-regulated on antigen-presenting cells.
Costimulation Through CD28 and ICOS Results in a Primarily Quantitative Effects on TCR-Induced Genes.
Simultaneous engagement of both TCR and CD28 is required for an optimal immune response. In vitro, this is demonstrated by the observation that stimulation of CD4 T cells by beads coated with anti-CD3 mAb (to trigger TCR signals) or anti-CD28 mAb alone results in minimal T cell proliferation. However, the combination of both mAbs on the same bead supports long-term T cell growth (data not shown). To better understand gene expression changes induced by simultaneous triggering of TCR and CD28 (CD28 costimulation) when both are necessary for T cell proliferation, we compared expression profiles of CD4 T cells stimulated with mAb-coated beads. CD4 T cells were left untreated or were stimulated with beads coated with an equal mixtures of anti-CD3 and anti-MHC I mAbs (CD3/MHC I beads); anti-CD3 and CD28 mAbs (CD3/CD28 beads); or anti-CD3 and ICOS (CD3/ICOS beads).
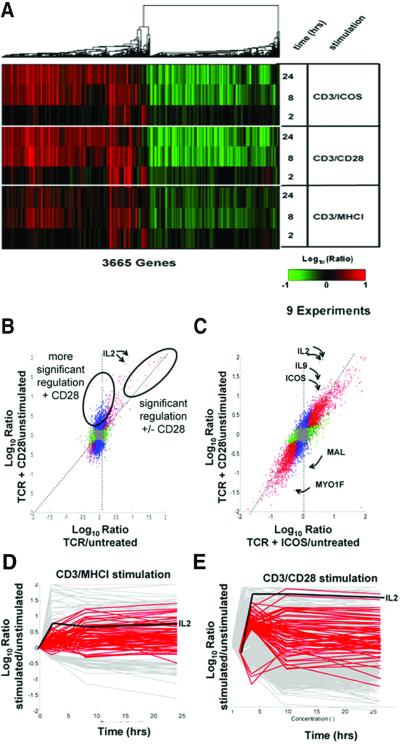
As shown in Fig. 2A, there were several obvious differences between overall gene expression profiles of stimulated Jurkat and primary CD4 T cells. Far more genes were induced by stimulation of primary CD4 T cells than Jurkat cells (≈3,500 genes vs. ≈120 genes, respectively; see Fig. 4). Furthermore, the ability of CD28 costimulation to alter gene expression was much more evident in primary CD4 cells than in Jurkat cells (Fig. 4). Stimulation of Jurkat T cells with an anti-TCR mAb plus a pharmacologic activator of T cell activation (PMA) resulted in more gene regulations, but the ability of CD28 to modulate these responses remained muted in comparison to what was observed in primary T cells (not shown). Finally, CD4 T cells treated with either CD3/MHCI or CD3/28 beads displayed approximately the same number of genes down-regulated as were induced. Stimulation of Jurkat T cells did not lead to as many down-regulated genes, suggesting that a mechanism(s) leading to these gene repressions is disabled in this transformed cell line. The approximately equivalent number of up- and down-regulated transcripts in CD4 T cells may suggest a dilution of transcripts present in resting cells with newly synthesized transcripts from activated cells. Alternatively, it may indicate a global homeostatic mechanism to regulate the total number of transcripts in primary T cells (23). Significantly, a much higher proportion of “down-regulated” than up-regulated transcripts are so poorly characterized that they lack gene names (i.e., they are ESTs): ≈41% of down-regulated transcripts (≈1,620 total) lacked gene names, whereas only ≈21% of up-regulated transcripts (≈1,870 total) lacked gene names. Thus, much less was known about genes “down-regulated” during T cell activation than about genes activated.
Figure 2.
CD28 costimulation primarily induces quantitative rather than qualitative changes in transcript levels. (A) Overall similarity of gene regulations induced by stimulation of TCR alone, TCR plus CD28, and TCR plus ICOS. Human peripheral blood CD4 T cells were cultured for the indicated times with CD3/MHCI, CD3/CD28, or CD3/ICOS beads. Shown is a display of regulations from 3665 genes significantly regulated (P < 0.01, >2-fold regulation, log10 intensity >−1 in at least two experiments) over a total of nine experiments. The dendrogram depicts the similarity of co-regulation of the indicated genes, with the length of the “branches” proportional to the degree of similarity of co-regulation. This experiment was repeated twice with equivalent results. (B) CD28 costimulation induces changes in levels of most but not all transcripts induced by TCR stimulation. CD4 T cells were left untreated or were stimulated with CD3/MHCI or CD3/CD28 beads for 2 h as in Fig. 2A. Shown are comparisons (correlations) of genes significantly regulated (P < 0.01, intensity >−1) in CD3/MHCI stimulated T cells (x axis) versus CD3/CD28 stimulated T cells (y axis). The plot is colored as in Fig. 1F. The dotted diagonal line indicates the expected positions of gene regulated equally by both stimulation conditions and the vertical dotted line, genes regulated only with CD28 costimulation. Ovals identify genes significantly regulated with or without CD28 costimulation or genes showing more significant regulation with CD28 costimulation. Genes flagged red, blue, and brown showed similar patterns of regulation in an independent experiment; those flagged green were not reproducible. The positions of S82692 and NM_000586, and IL2, are indicated. (C) CD28 and ICOS costimulation induces similar regulation in all but a few transcripts. CD4 T cells were left untreated or were stimulated with CD3/CD28 or CD3/ICOS beads for 8 h as in Fig. 2A. Shown are comparisons (correlations) of genes significantly regulated (P < 0.01, log10 intensity >−1) in CD3/ICOS -treated T cells (x axis) versus CD3/CD28-treated T cells (y axis). The positions of selected off diagonal genes regulated much more highly following CD28 costimulation are indicated by arrows:IL2; NM_000590, IL9, interleukin 9; AB023135; ICOS, inducible co-stimulator; NM_002371, MAL, mal T cell differentiation protein; X98411, MYO1F, myosin 1F. (D and E) Genes showing increased regulation with CD28 at 2 h show slower kinetics of activation in cells stimulated with TCR alone. CD4 T cells were left untreated or were stimulated with CD3/MHCI or CD3/CD28 beads for 2, 8, or 24 h as in Fig. 3A. Shown is a representation of the kinetics of gene regulation by CD3/MHCI beads (D) and CD3/CD28 beads (E). Gray curves indicate hybridization ratios for genes showing 2-fold regulation; P < 0.01 for three time points. Red curves depict genes showing higher regulation following CD28 costimulation (>5-fold regulation, P < 1 × 10−4, approximately depicted by the labeled oval in Fig. 2B). Genes not significantly regulated are not depicted. The regulation of IL2 is shown for reference.
Hundreds of gene regulations were triggered in CD4 T cells by CD3/MHCI and CD3/CD28 beads (Fig. 2A, Table 1). Overall, the gene expression changes triggered by these stimuli were very similar, suggesting that most genes regulated by CD3/CD28 costimulation were also regulated by CD3/MHCI stimulation, albeit to a lesser degree. This conclusion was supported by examination of regulations of selected individual genes (see Fig. 7, which is published as supporting information on the PNAS web site). T cell cytokines (IL2, IL4, IFNG, CSF2, TNF), T cell activation genes (CD69, IL2RA, CTLA4, ICOS), and protein biosynthetic genes (tRNA synthases, WARS, NARS, AARS, IARS, MARS) were up-regulated by stimulation with both CD3/MHCI beads and CD3/CD28 beads, with CD3/CD28 beads generally eliciting greater responses. Genes repressed during T cell activation were also regulated by both stimuli; this is demonstrated by regulations of certain tumor suppressor genes (BIN1, TOB1, TSC22, KLF2). The repression of these genes during T cell activation supports an emerging model that maintenance of a G0 cell cycle phenotype is an active process that must be disabled before a cell proceeds into the cell cycle (24).
A key feature of expression profiling is that patterns of expression shared by many genes can be identified and analyzed using statistical techniques. The similarity between gene expression changes induced by CD3/MHCI and CD3/CD28 bead stimulation suggested that the effects of CD28 costimulation on gene regulation were primarily quantitative rather than qualitative. To confirm this, we applied statistical comparisons to gene regulations induced by CD3/MHCI and CD3/CD28 beads. We compared gene regulations induced by CD3/MHCI and CD3/CD28 beads after 2 h of stimulation (both compared with untreated cells) by using correlation plots (Fig. 2B). This allowed identification of groups of genes differentially affected by CD28 costimulation.
Many genes were significantly regulated following both CD3/MHCI and CD3/CD28 beads stimulation (Fig. 2B, 282 red-shaded genes, listed in Table 3, which is published as supporting information on the PNAS web site). Because these genes were significantly regulated following stimulation with and without CD28 costimulation, they tend to lie on the diagonal of the comparisons between these two stimuli. A few genes were oppositely regulated with and without CD28 costimulation (13 brown-shaded genes). Some of these genes showed clearly reciprocal patterns of regulations throughout 24 h stimulation with CD3/MHCI and CD3/CD28 beads (see Fig. 8, which is published as supporting information on the PNAS web site); these patterns were repeated in an independent experiment (not shown). One of these genes, ID3, encodes a helix–loop–helix repressor protein with a role in T cell differentiation (25), and is down-regulated following CD3/CD28 bead stimulation, but transiently up-regulated during CD3/MHCI bead stimulation. This pattern of gene regulation suggests that reduction of ID3 protein levels may be important for the effects of CD28 costimulation on T cell activation. Some genes were modestly regulated by stimulation with CD3/MHCI beads but not CD3/CD28 beads (136 green-shaded genes). Because these genes did not show the same pattern of regulation in another identical experiment (not shown), we did not pursue them. By far the largest group of regulated genes was significantly regulated by CD3/CD28 but not CD3/MHCI (1,486 blue-shaded genes). Stimulation by CD3/MHCI and CD3/CD28 beads after 2 and 24 h stimulation gave similar results (data not shown). Thus, most gene regulations were quantitatively affected by CD28 costimulation.
If there were strictly qualitatively different regulations induced by CD28 costimulation (i.e., only induced with CD28 costimulation), then these should tend to lie on a vertical line parallel to the CD3/CD28 stimulation axis. However, Fig. 2B shows that the vast majority of genes lie intermediate between the vertical dimension and the diagonal occupied by genes significantly regulated by both stimuli. The lack of clear separation between the red- and blue-shaded genes suggested that these were not truly separate groups of genes but were arbitrarily grouped by the P value chosen. Supporting this view, we found that the number of blue-shaded genes decreased and the number of red-shaded genes correspondingly increased when the P value of detection was increased (data not shown). Thus, the increased significance of induction of these genes by CD28 costimulation reflects a limitation of the threshold of detection in the absence of costimulation rather than uniquely expressed genes.
It was important to determine whether gene expression changes elicited by CD28 costimulation were unique or whether they could also be induced by another costimulatory pathway(s). We therefore compared gene regulations induced by CD3/MHCI beads with those induced by CD3/ICOS beads. More gene regulations were triggered by CD3/ICOS beads than with CD3/MHCI beads, although there were fewer differences than with CD28 costimulation (data not shown). This finding suggests that ICOS costimulation induces quantitatively less regulation of many genes than CD28 costimulation. A comparison of gene expression triggered by CD28 and ICOS costimulation is shown in Fig. 2C. This comparison was made after 8 h of stimulation to allow for significant up-regulation of ICOS from the low levels found in resting T cells (14, 26). Gene regulations induced by ICOS and CD28 costimulation were very similar, although several genes highlighted in the figure lie off the diagonal line (IL-2, ICOS, IL-9, MAL, and MYO1F). This suggests that CD28 costimulation regulates these genes in a fundamentally different manner than ICOS costimulation.
Genes regulated more after CD28 costimulation may be regulated to different extents or with different kinetics. To distinguish these possibilities, we examined the kinetics of regulation of the most significantly differentially regulated genes after 2 h of stimulation (blue-shaded genes showing >5-fold regulation, P < 1e-4, Fig. 2B). Genes more significantly regulated by CD3/CD28 beads showed more rapid kinetics of induction or repression than with CD3/MHCI beads (Fig. 2 D and E). Many of these genes were regulated, but more slowly, following stimulation with CD3/MHCI beads. Therefore, many of the genes scored as qualitative changes at early time points are scored as quantitative changes at later time points. Similar results were seen with genes induced more strongly by CD3/CD28 beads for 8 h (not shown). Thus, CD28 costimulation alters the kinetics of expression of many genes induced by TCR triggering. The mechanism of regulation of these genes by CD28 costimulation probably does not involve the same enhancement of mRNA stability seen with cytokines (21), because this group of genes is not enriched for the presence of multiple AUUUA motifs in their 3′ UTRs: only 10/24 or ≈42% of known genes in this group contain three or more AUUUA motifs in their 3′ UTRs. The genes which do not contain multiple AUUUA motifs in their 3′ UTRs include several well characterized genes (e.g., POU2AF1, MAP2K3, SLAM). It is more likely that regulation of these genes is achieved by another mechanism, such as CD28-mediated induction of a new transcription factor(s) (27).
Although much evidence suggests that signaling through the CD28 receptor is distinct from signaling through the TCR (28), it remains unclear whether this difference is quantitative, qualitative, or both. Recently, Lanzavecchia and colleagues (29) suggested that CD28 costimulation amplifies or tunes CD3 signaling arguing for a mainly quantitative contribution of CD28 signaling. Our data provide clear evidence for both quantitative and qualitative differences in TCR and CD28 signaling. The data in Fig. 2 support the “TCR tuning model” by showing that the effect of costimulation on most gene regulations was primarily quantitative. However, several observations also support a “distinct signal model.” Some genes (see Table 3) were oppositely regulated in the presence or absence of CD28 costimulation (ID3,X69111 and other brown-shaded genes; Fig. 2B, Fig. 8). We observed similar behavior of ID3 in another independent experiment (data not shown). Moreover, CD28 costimulation did not simply modulate the levels of TCR responses, but rather altered the tempo of the gene regulation for a subset of genes (Fig. 2 D and E). Lastly, the comparison between ICOS and CD28 revealed several genes (i.e., IL-2, etc.) that were much more regulated by CD28 than ICOS costimulation. Thus, all genes are not “tuned” equally by different pathways of costimulation. Although our data provide evidence for both quantitative and qualitative mechanisms of costimulation, the statistical comparisons we present suggest that quantitative mechanisms predominate.
CTLA-4 Engagement Blocks the Effects of CD28 Costimulation.
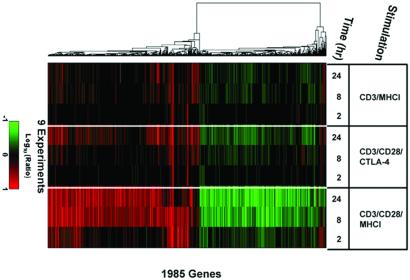
CTLA-4 engagement may function to prevent entry into the cell cycle by disrupting T cell signaling pathways (6). Previous studies suggested that CTLA-4 engagement antagonizes TCR-mediated signal transduction (30, 31) but these studies did not determine whether CTLA-4 engagement could block costimulatory pathways. We hypothesized that expression profiling would permit determination of whether CTLA-4 engagement ablated TCR signals, the augmentation of gene regulation by costimulation, or both. To distinguish between these alternatives, we compared expression profiles of cells stimulated with CD3/MHCI and CD3/CD28 or CD3/CD28/CTLA-4 beads (Fig. 3). Inclusion of anti-CTLA-4 mAb on CD3/CD28 beads inhibits CD3/CD28 bead-mediated proliferation and cytokine production (15). This analysis revealed that CTLA-4 ligation completely blocked the CD28 enhancement of gene regulations at 2 and 8 h after stimulation. In contrast, gene regulations induced by CD3/MHCI beads were unaltered by CTLA-4 engagement (Fig. 3). Correlation plots of these data also illustrate the lack of quantitative enhancement of CD3-regulated genes by CD28 costimulation at 2 and 8 h after stimulation (see Fig. 9, which is published as supporting information on the PNAS web site). Thus, CTLA-4 engagement affects costimulation rather than ablating TCR signals.
Figure 3.
CTLA-4 engagement blocks CD28-induced changes in gene regulation. CD4 T cells were stimulated with CD3, CD3/CD28/CTLA-4, or CD3/CD28/MHC I beads for 2, 8, or 24 h. To see the effects of CTLA-4 engagement, 10-fold less anti-CD3 mAb was used to coat beads used these experiments. Detection of gene regulations by DNA microarray analysis was performed and depicted as in Fig. 2. Shown are regulations of 1,985 genes significantly regulated (P < 0.01, log10 intensity >−1, >2-fold regulation in two or more experiments). This experiment was repeated twice with equivalent results.
After 24 h stimulation with CD3/CD28/CTLA-4 beads, we observed some CD28-enhanced gene regulation (“breakthrough genes,” Figs. 3 and 9), indicating that CTLA-4 is more effective at blocking the rapid and intermediate responses to CD28 costimulation. This finding is in agreement with previous reports that indicate CTLA-4 ligation affects early events in T cell activation (32). The breakthrough genes are regulated with more rapid kinetics in CD3/CD28-stimulated cells, but they are primarily not immediate-early genes (not shown). Thus, the breakthrough genes are regulated after CTLA-4 engagement even though the normal cascade of immediate-early gene regulations does not occur. Thus, CTLA-4 engagement alters, rather than blocks, CD28 costimulation.
Conclusion
Rather than measuring proximal signaling events (i.e., protein phosphorylation, etc.), this study measures signal transduction via a distal outcome, transcript regulation. This approach has provided several unique insights into T cell activation. By analyzing many thousand genes simultaneously, we demonstrated a previously unknown effect of triggering CD28 alone, the up-regulation of immediate-early genes (e.g., the Egr family). This gene family comprises pleiotropic mediators of growth and development in many systems, including T cell development (33). Our data suggest a role for these genes as mediators of CD28 signaling and possible utility as reporters for further elucidation of the CD28 signaling pathway(s). Moreover, our data have enabled a unique quantitative view of the global effect of CD28, ICOS, and CTLA-4 engagement on TCR signaling. Our data showed striking similarity in gene expression changes induced by CD28 and ICOS pathways suggesting that their unique functional properties may result from differential expression of only a few genes, such as IL2, IL9, etc. Likewise, although costimulation has been reported to induce both quantitative and qualitative differences in signaling, our data demonstrate that quantitative differences predominate on the mRNA level. It will be important to determine whether these conclusions are also valid at the protein level, because changes in mRNA expression sometimes do not result in proportional changes in protein levels. Finally, we demonstrated that CTLA-4 affects the costimulatory enhancement of TCR signaling rather than TCR signaling per se. The ability of CTLA-4 ligation to ablate CD28-augmented effects on gene expression as early as 2 h after stimulation is consistent with models that depict CTLA-4 ligation blocking early events of T cell proliferation (15, 32).
Supplementary Material
Acknowledgments
We thank Craig Thompson, Hugh Rosen, Richard Carroll, Daniel Shoemaker, Stephen Friend, Jeff Ledbetter, Michael Carlton, and Gary Koretzky for helpful comments on the manuscript; Dr. Beatriz Carreno (Genetics Institute) for the CTLA-4 mAb and CD86 Ig; Dr. Katsunari Tezuka (Japan Tobacco, Inc.) for the ICOS mAb; and the Rosetta North Creek group for performing the hybridizations.
Abbreviations
- CTLA-4
cytotoxic T lymphocyte antigen 4
- TCR
T cell receptor
References
- 1.Mueller D L, Jenkins M K, Schwartz R H. Annu Rev Immunol. 1989;7:445–480. doi: 10.1146/annurev.iy.07.040189.002305. [DOI] [PubMed] [Google Scholar]
- 2.Matzinger P. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- 3.Glimcher L H, Murphy K M. Genes Dev. 2000;14:1693–1711. [PubMed] [Google Scholar]
- 4.Salomon B, Bluestone J A. Annu Rev Immunol. 2001;19:225–252. doi: 10.1146/annurev.immunol.19.1.225. [DOI] [PubMed] [Google Scholar]
- 5.Rudd C E. Immunity. 1996;4:527–534. doi: 10.1016/s1074-7613(00)80479-3. [DOI] [PubMed] [Google Scholar]
- 6.Chambers C A, Kuhns M S, Egen J G, Allison J P. Annu Rev Immunol. 2001;19:565–594. doi: 10.1146/annurev.immunol.19.1.565. [DOI] [PubMed] [Google Scholar]
- 7.Linsley P S. Nat Immunol. 2001;2:139–140. doi: 10.1038/84233. [DOI] [PubMed] [Google Scholar]
- 8.Clements J L, Boerth N J, Lee J R, Koretzky G A. Annu Rev Immunol. 1999;17:89–108. doi: 10.1146/annurev.immunol.17.1.89. [DOI] [PubMed] [Google Scholar]
- 9.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 10.Roberts C J, Nelson B, Marton M J, Stoughton R, Meyer M R, Bennett H A, He Y D, Dai H, Walker W L, Hughes T R, et al. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 11.Hughes T R, Mao M, Jones A R, Burchard J, Marton M J, Shannon K W, Lefkowitz S M, Ziman M, Schelter J M, Meyer M R, et al. Nat Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 12.June C H, Ledbetter J A, Gillespie M M, Lindsten T, Thompson C B. Mol Cell Biol. 1987;7:4472–4481. doi: 10.1128/mcb.7.12.4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riley J L, Carroll R G, Levine B L, Bernstein W, St. Louis D C, Weislow O S, June C H. J Immunol. 1997;158:5545–5553. [PubMed] [Google Scholar]
- 14.Riley J L, Blair P J, Musser J T, Abe R, Tezuka K, Tsuji T, June C H. J Immunol. 2001;166:4943–4948. doi: 10.4049/jimmunol.166.8.4943. [DOI] [PubMed] [Google Scholar]
- 15.Blair P J, Riley J L, Levine B L, Lee K P, Craighead N, Francomano T, Perfetto S J, Gray G S, Carreno B M, June C H. J Immunol. 1998;160:12–15. [PubMed] [Google Scholar]
- 16.Hughes T R, Marton M J, Jones A R, Roberts C J, Stoughton R, Armour C D, Bennett H A, Coffey E, Dai H, He Y D, et al. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 17.Hartigan J A. Clustering Algorithms. New York: Wiley; 1975. [Google Scholar]
- 18.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ledbetter J A, Imboden J B, Schieven G L, Grosmaire L S, Rabinovitch P S, Lindsten T, Thompson C B, June C H. Blood. 1990;75:1531–1539. [PubMed] [Google Scholar]
- 20.Wilusz C J, Wormington M, Peltz S W. Nat Rev Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 21.Lindstein T, June C H, Ledbetter J A, Stella G, Thompson C B. Science. 1989;244:339–343. doi: 10.1126/science.2540528. [DOI] [PubMed] [Google Scholar]
- 22.Linsley P S, Bradshaw J, Urnes M, Grosmaire L, Ledbetter J A. J Immunol. 1993;150:3161–3169. [PubMed] [Google Scholar]
- 23.Teague T K, Hildeman D, Kedl R M, Mitchell T, Rees W, Schaefer B C, Bender J, Kappler J, Marrack P. Proc Natl Acad Sci USA. 1999;96:12691–12696. doi: 10.1073/pnas.96.22.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Santo J P. Nat Immunol. 2001;2:667–668. doi: 10.1038/90598. [DOI] [PubMed] [Google Scholar]
- 25.Rivera R R, Johns C P, Quan J, Johnson R S, Murre C. Immunity. 2000;12:17–26. doi: 10.1016/s1074-7613(00)80155-7. [DOI] [PubMed] [Google Scholar]
- 26.Hutloff A, Dittrich A M, Beier K C, Eljaschewitsch B, Kraft R, Anagnostopoulos I, Kroczek R A. Nature (London) 1999;397:263–266. doi: 10.1038/16717. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro V S, Truitt K E, Imboden J B, Weiss A. Mol Cell Biol. 1997;17:4051–4058. doi: 10.1128/mcb.17.7.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.June C H, Ledbetter J A, Linsley P S, Thompson C B. Immunol Today. 1990;11:211–216. doi: 10.1016/0167-5699(90)90085-n. [DOI] [PubMed] [Google Scholar]
- 29.Viola A, Lanzavecchia A. Science. 1996;273:104–106. doi: 10.1126/science.273.5271.104. [DOI] [PubMed] [Google Scholar]
- 30.Calvo C R, Amsen D, Kruisbeek A M. J Exp Med. 1997;186:1645–1653. doi: 10.1084/jem.186.10.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee K M, Chuang E, Griffin M, Khattri R, Hong D K, Zhang W, Straus D, Samelson L E, Thompson C B, Bluestone J A. Science. 1998;282:2263–2266. doi: 10.1126/science.282.5397.2263. [DOI] [PubMed] [Google Scholar]
- 32.Brunner M C, Chambers C A, Chan F K, Hanke J, Winoto A, Allison J P. J Immunol. 1999;162:5813–5820. [PubMed] [Google Scholar]
- 33.Kaye J. Immunol Res. 2000;21:71–81. doi: 10.1385/IR:21:2-3:71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.