Abstract
Tetracycline-inducible HEK293S stable cell lines have been prepared that express high levels (up to 10 mg/liter) of WT opsin and its mutants only in response to the addition of tetracycline and sodium butyrate. The cell lines were prepared by stable transfection of HEK293S-TetR cells with expression plasmids that contained the opsin gene downstream of a cytomegalovirus promoter containing tetO sequences as well as the neomycin resistance gene under control of the weak H2Ld promoter. The inducible system is particularly suited for overcoming problems with toxicity either due to the addition of toxic compounds, for example, tunicamycin, to the growth medium or due to the expressed protein products. By optimization of cell growth conditions in a bioreactor, WT opsin, a constitutively active opsin mutant, E113Q/E134Q/M257Y, presumed to be toxic to the cells, and nonglycosylated WT opsin obtained by growth in the presence of tunicamycin have been prepared in amounts of several milligrams per liter of culture medium.
Keywords: G protein-coupled receptors‖HEK293S cells‖cytomegalovirus promoter‖ glycosylation‖sodium butyrate
Certain studies on rhodopsin and its mutants require relatively large amounts of purified proteins. To meet these needs, stable mammalian cell lines for high-level expression of the opsin gene and its mutants were developed (1, 2). The application of NMR spectroscopy to structure–function studies of rhodopsin thus became possible (3–6). Recently, evidence has been accumulating that certain gene products either fail to be expressed in adequate amounts in the constitutive expression systems above or that viable stable cell lines cannot be generated for certain mutant opsin genes. For example, stable cell lines carrying the rhodopsin kinase gene expressed the kinase at levels much lower than those predicted from the transient transfection experiments (7). Further, we failed to isolate a viable stable cell line for expression of the strongly constitutively active opsin mutant, E113Q/E134Q/M257Y. It seems likely that the proteins produced by unregulated expression of the genes above are cytotoxic and, indeed, in certain cases may be lethal. To overcome these difficulties, we wanted to develop a strategy frequently used in bacterial expression systems (8, 9) in which membrane proteins toxic to the bacterial cells are expressed by induction of the desired gene only after achieving growth of cells to near-maximum density. Previously, this strategy enabled large-scale production in Escherichia coli of numerous toxic bacterio-opsin mutants (10). Therefore, our present aim was to develop an inducible expression system in stable mammalian cell lines.
A tetracycline (tet)-regulated gene expression system in mammalian cells was first described by Gossen and Bujard (11). By fusing the bacterial tet repressor (TetR) with the activating domain of the viral protein 16 (VP-16), a tetracycline-controlled transactivator was generated that stimulated transcription of a desired gene from a minimal cytomegalovirus (CMV) promoter containing tet operator (tetO) sequences. This inducible system has been used extensively for regulated gene expression in diverse systems (for a recent review, see ref. 12). More recently, Yao et al. (13) devised an alternative tetracycline-inducible expression system that is based on repression by TetR. The system uses a full-length CMV promoter that contains two tetO sequences in tandem. In the present work, we chose the repressor-based induction system because of our previous successful use of the full-length CMV promoter in high-level expression of the bovine opsin gene in HEK293S cells (1). We now have integrated the tetracycline-inducible system above into our previously developed methodology for constructing stable HEK293S cell lines (14). The cell lines thus constructed require the combined addition of tetracycline and sodium butyrate to growth-saturated cultures for gene expression at maximum levels. We describe application of the inducible system to the large-scale production of two desired opsin mutants where, in each case, it was necessary to overcome toxicity during expression. First, we have expressed WT opsin in its nonglycosylated form for its potential use in crystallization work. This has been achieved by overcoming toxicity due to the drug tunicamycin added to prevent glycosylation. Second, a stable cell line has been prepared that enables the preparation in milligram amounts of a strongly constitutively active opsin mutant, E113Q/E134Q/M257Y, presumed to be toxic to the cells.†
Materials and Methods
Materials.
Frozen bovine retinae were from W. L. Lawson (Lincoln, NE). The silica column (10 μm, Econosphere, 250 mm × 22 mm i.d.) used for purification of 11-cis-retinal was from Alltech Associates. Sepharose-4B was from Amersham Pharmacia. The detergents n-dodecyl-β-D-maltoside (DM) and n-nonyl-β-D-glucoside (NG) were purchased from Anatrace (Maumee, OH). The anti-rhodopsin antibody, rho-1D4 (15), was prepared by the Cell Culture Center (Minneapolis) from a cell line provided by R. S. Molday (University of British Columbia, Vancouver). The epitope for rho-1D4, the nonapeptide corresponding to the C-terminal sequence of rhodopsin, was prepared by the Massachusetts Institute of Technology Biopolymer Laboratory. FBS, tetracycline, all-trans-retinal, and dextran sulfate (average Mr 5,000) were purchased from Sigma, and sodium butyrate was from J. T. Baker (Mallinchrodt Baker, Phillipsburg, NJ). Primatone RL-UF was a gift from Quest International (Hoffman Estates, IL), calcium-free DMEM was from Atlanta Biologicals (Norcross, GA), and tunicamycin was from Calbiochem. Blasticidin and plasmids pCEP4 and pCDNA6-TR were from Invitrogen. Geneticin was from GIBCO/BRL. DNA oligonucleotides were purchased from Genosys (The Woodlands, TX). Restriction and other DNA modification enzymes were from New England Biolabs and Roche Molecular Biochemicals. The enhanced chemiluminesence detection kit was from Amersham Pharmacia.
Buffers used were as follows: buffer A (137 mM NaCl/2.7 mM KCl/1.5 mM K2HPO4, pH 7.2); buffer B (buffer A + 1% wt/vol DM); buffer C (buffer A + 0.1% wt/vol DM); buffer D {10 mM 1,3-bis[tris(hydroxymethyl)-methylamino] propane, pH 6.0 + 0.1% DM}; buffer E (buffer D + 100 μM epitope nonapeptide); buffer F (buffer C + 100 μM epitope nonapeptide); buffer G (buffer A + 2% wt/vol NG); buffer H (buffer A + 0.5% wt/vol NG); buffer I {10 mM 1,3-bis[tris(hydroxymethyl)-methylamino] propane, pH 6.0 + 0.5% NG}; buffer J (buffer I + 100 μM epitope nonapeptide); and buffer K (buffer H + 100 μM epitope nonapeptide).
Methods.
Rod outer segments from bovine retinae were prepared as described (16). Purification of the expressed rhodopsin and its mutants was by immunoaffinity chromatography with rho-1D4-Sepharose (17). 11-cis-Retinal was prepared by illumination (>435 nm) of all-trans-retinal in ethanol (18) by using light from a slide projector equipped with a 300-W bulb. The isomers formed were separated by using isocratic HPLC with the silica column described above. The solvent mixture used was hexane containing ethyl acetate (1.3%) and isopropyl alcohol (0.1%) (vol/vol) in hexane.
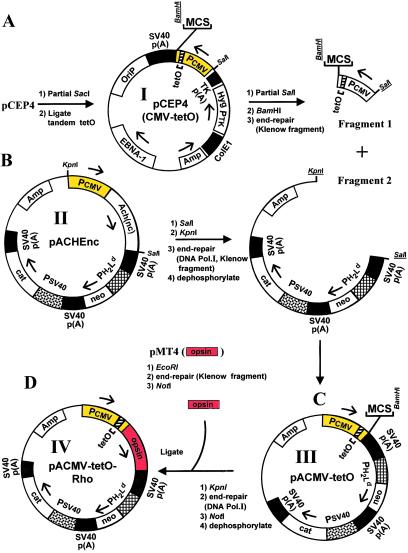
Construction of the Tetracycline-Regulated Expression Plasmid, pACMV-tetO-Rho (Fig. 1D IV).
Figure 1.
Preparation of the vector pACMV-tetO-Rho (IV) for tetracycline-inducible opsin expression. The detailed procedure is in Materials and Methods. In brief, the construction involved four main steps, A–D. In A, the tetO operator sequences were inserted into the plasmid pCEP4(I) and the fragment 1 containing the CMV(tetO) promoter was isolated. In B, fragment 2 was prepared from the plasmid pACHEnc(II) by removal of CMV + Ach(nc). In C, fragments 1 and 2 were ligated to give the plasmid pACMV-tetO (IV). In D, the opsin gene isolated from the plasmid pMT4 was inserted into the plasmid pACMV-tetO(III) to give the vector pACMV-tetO-Rho. The multiple cloning site (MCS) located downstream of the CMV-tetO promoter in pACMV-tetO contains [(5′-3′) KpnI, PvuII, NheI, HindIII, NheI, NotI, XhoI, SfiI, BamHI].
The total steps in the construction are illustrated in Fig. 1. An oligonucleotide cassette containing two tetO sequences in tandem was inserted, as described by Yao et al. (13), into the SacI site at the 3′ end of the CMV promoter of plasmid pCEP4 (Invitrogen) to give pCEP4-(CMV-tetO) (Fig. 1A I). The DNA fragment corresponding to the CMV-tetO promoter was excised from this plasmid by partial digestion with SalI followed by complete digestion with BamHI. This DNA, designated fragment 1 (Fig. 1A), was then end-repaired with E. coli DNA polymerase I Klenow fragment and purified by using agarose gel electrophoresis. Fragment 2 (Fig. 1B) was prepared from the plasmid, pACHEnc (Fig. 1B II) (19) by excising a DNA fragment containing both the entire CMV promoter and the acetylcholine esterase, Ach(nc), gene by digestion with KpnI and SalI (Fig. 1B). Fragment 2 was then end-repaired with both DNA polymerase (Pol-I) and Klenow fragment, treated with alkaline phosphatase, and purified by agarose gel electrophoresis. Blunt-end ligation of fragments 1 and 2 gave the plasmid pACMV-tetO (Fig. 1C III). The opsin gene was prepared from the plasmid pMT4 (17). The latter was digested with EcoRI, the resulting linear DNA end-repaired by using the DNA polymerase I Klenow fragment, and then digested with NotI to liberate the opsin gene, which was purified by agarose gel electrophoresis. For preparation of the plasmid, pACMV-tetO-Rho (Fig. 1D IV), the plasmid pACMV-tetO (Fig. 1C III) was digested with KpnI and the resulting linear DNA end-repaired by using DNA Pol-I, then digested with NotI, treated with alkaline phosphatase, and purified by agarose gel electrophoresis. Subsequent ligation with the opsin gene gave IV, pACMV-tetO-Rho (Fig. 1D).
Construction of Stable HEK293S Cell Lines for Tetracycline-Induced Opsin Gene Expression.
HEK293S cells maintained as described (1) were transfected with the plasmid pCDNA6-TR (Invitrogen) carrying a gene encoding the selectable marker, blasticidin, and a gene coding for the tet operon repressor protein (TetR). The transfection method used calcium phosphate precipitation as described (1), except that the proportion of CO2 in the incubator was reduced to 1.5%. This reduction further increased the transfection efficiency. Stably transfected cell lines resistant to blasticidin (5 μg/ml) were selected. The pool of the resulting colonies was then expanded under blasticidin selection and transfected with pACMV-tetO-Rho (Fig. 1). Cell lines stably transfected with this plasmid were selected by using Geneticin (2 mg/ml), and individual colonies appearing after 14 days were isolated and expanded as described (1). Cell lines were expanded in triplicate to 107 cells in 10-cm-diameter culture dishes. Cells from one dish were stored in liquid nitrogen. Another dish was supplemented with growth medium alone, whereas the third dish was supplemented with growth medium containing both tetracycline (2 μg/ml) and sodium butyrate (5 mM). When necessary, tunicamycin was added to the growth medium 3–4 h before induction of the opsin gene. The cells were incubated for a further 48 h. For harvesting, the cells were removed from the dish, pelleted by using a clinical centrifuge, and washed with 10 ml of buffer A. The cells were repelleted by using a clinical centrifuge and resuspended in 500 μl of buffer A. 11-cis-Retinal (5 μM) was then added to the whole-cell suspension, which was mixed end-over-end by using a nutator in the dark at 4°C for 2 h. The cells were solubilized by addition of DM to the final concentration in buffer B and incubation for 1 h. The clear supernatant was collected by centrifugation for 30 min at 50 K (TL-100 rotor) and analyzed by UV-visible absorbance spectroscopy before and after photobleaching (1 min, >495 nm light from a fiber optic).
Growth of HEK293S Cell Line Suspension Cultures in a Bioreactor.
Preparation of the medium.
The growth vessels (2.4- and 14-liter capacity) of a Celligen Plus bioreactor were equipped with pitch blade impellers and primed for use as directed by the manufacturer (New Brunswick Scientific). The growth medium used was a calcium-free, high-glucose (4.5 g/liter) DMEM custom formulation prepared as a powder by Atlanta Biologicals. The dissolved medium was supplemented with sodium bicarbonate (3.7 g/liter) and 0.3g/liter of Primatone RL-UF. After filtration through a 0.2-μm membrane the medium was supplemented further with the following sterile components: heat-treated FBS (10% vol/vol), penicillin G (100 units/ml), streptomycin (100 μg/ml), glutamine (292 μg/ml), dextran sulfate (300 μg/ml), and pluronic F-68 (0.1% wt/vol).
Growth of 1.1-liter cultures in a 2.4-liter vessel.
The growth parameters were set to 37°C, pH 7, and 50% dissolved oxygen, the latter two parameters being achieved and maintained by interactive control delivery of a four-gas mixture (air, N2, CO2, and O2) split between a direct sparge (up to 21 ml/min) and an overlay sparge (up to 185 ml/min). The medium was inoculated by using cells obtained by trypsinization of six confluent 15-cm-diameter dishes that had been fed the day before inoculation. Cells were stirred by using the pitch black impeller at the stirrer speed of 30–35 rpm. The cell density at inoculation was 3–5 × 105 cells per milliliter. On day 5 after inoculation, supplements, 10 ml of 20% (wt/vol) glucose and 30 ml of 10% (wt/vol) Primatone RL-UF, were added, and expression of the opsin gene was induced on the sixth day by addition of both tetracycline (2 μg/ml) and sodium butyrate (5 mM) to the growth medium. Samples (10 ml) were removed every 24 h for determining the viable count and for analysis of protein. For determining the viable cell count, 1 ml of pelleted cells was washed with 1 ml of buffer A, repelleted, and then incubated for 1 min with 100 μl of trypsin (0.05%). The trypsinized cells were diluted to 1 ml by using the complete DMEM, diluted further if necessary, mixed with trypan blue, and counted by using a hemocytometer. Cells were harvested on day 7 or 8.
Growth of 5.5-liter cultures in a 14-liter vessel.
The same growth medium, growth parameters, supplementation, and gene induction strategy as described above were used with the following changes. The inoculation cell density (3–5 × 105 cells per ml) was achieved by scaling up the number of inoculum dishes to 30. For agitation the stirrer speed was set at 30 rpm and aeration was by direct sparge only starting at a flow rate of 21 ml/min on day 1 with daily increments to 374 ml/min toward the end of the incubation period.
Immunoaffinity Purification.
Nonglycosylated rhodopsin.
Growth of the WT opsin-producing cell line was performed by using a 5.5-liter culture volume. Cells were grown as described above and treated with tunicamycin (2 μg/ml) on day 6, 4 h before induction of opsin expression. The cells were harvested 1 day later, and a portion (one-seventh) was suspended in buffer A (40 ml final volume) containing PMSF (0.1 mM) and treated with 11-cis-retinal (15 μM) for 15 h at 4°C to constitute the pigment (20). Pigment constitution and all subsequent operations were performed in the dark or under dim red light. Cells were solubilized in buffer G [by addition of the detergent NG from a 10% (wt/vol) stock to the concentration as in buffer G] followed by end-over-end mixing for 1.5 h at 4°C. The solubilized cell extract was clarified by centrifugation (Ti-45 rotor, 35,000 rpm for 40 min), and the total amount of rhodopsin was determined by UV-visible difference spectroscopy analysis with a small portion of the solubilized material. A 50% slurry (10 ml) of Sepharose-rho-1D4 beads (capacity, 0.7 mg of rhodopsin per ml of settled beads) was added, followed by incubation for a further 1.5 h. The beads were packed into a column (1.6 cm i.d. × 8.5 cm) and washed sequentially with 200 ml of buffer H and 50 ml of buffer I. Elution of rhodopsin (5-ml fractions) was performed at 22°C by application of buffer J followed by buffer K.
The rhodopsin mutant, E113Q/E134Q/M257Y.
Cell growth was performed on a 1.1-liter scale as described above, and cells were harvested 1 day after induction of opsin expression. A one-third portion of the harvested cells was suspended in 25 ml of buffer A containing PMSF (0.1 mM) and treated with 11-cis-retinal (40 μM) for 15 h in the dark. Cells were solubilized in buffer B (by addition of DM from a 20% stock to the concentration as in buffer B) for 1 h. The solubilized cell extract was clarified by centrifugation (Ti-45 rotor, 35 K, 40 min) and to this centrifugate was added 4 ml of Sepharose-rho-1D4 beads. After 3–5 h, the beads were packed into a column (1.0 cm i.d. × 7.0 cm) and the column washed with 500 ml of buffer C followed by 50 ml of buffer D. Elution of the pigment was with buffer E followed by buffer F; 2-ml fractions were collected.
Results
Tetracycline and Sodium Butyrate-Dependent Expression of WT Opsin in HEK293S Cell Lines.
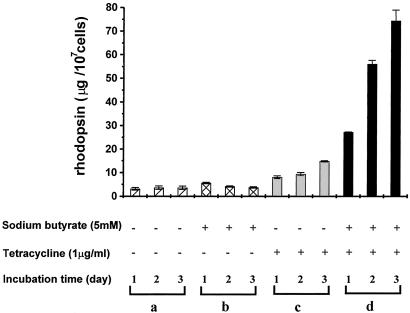
Regulatable expression of WT opsin in one stable HEK293S cell line (Materials and Methods) was probed as shown in Fig. 2. Cells were grown to near confluence in multiple 10-cm-diameter dishes. They were then supplied with fresh medium supplemented with tetracycline or sodium butyrate or both. Cells were harvested at days 1, 2, and 3, treated with 11-cis-retinal (5 μM), and after solubilization of the cells in buffer B, the yield of constituted rhodopsin was quantitated by UV-visible difference spectrum analysis (1). As analyzed in Fig. 2, uninduced cells seemed to make insignificant (<5 μg/dish) amounts of rhodopsin as did those treated only with sodium butyrate. However, the more sensitive immunodetection method (Fig. 3B) suggests that opsin gene expression in the absence of induction is even lower than that indicated in Fig. 2. Cells supplemented with tetracycline alone showed significant production of rhodopsin in a time-dependent manner. However, the presence of both tetracycline and sodium butyrate resulted in the highest level of opsin expression, the yield being up to ≈75 μg/dish after 72 h.
Figure 2.
Expression of the opsin gene under different conditions after growth of a stable cell line. Cells were grown in dishes to near confluence and then incubated for the time period shown after addition of fresh growth medium supplemented as indicated (a–d). Each bar shows the average amount of opsin produced by cells from duplicate culture dishes as measured by constituted rhodopsin (UV-visible difference spectroscopy; see Materials and Methods).
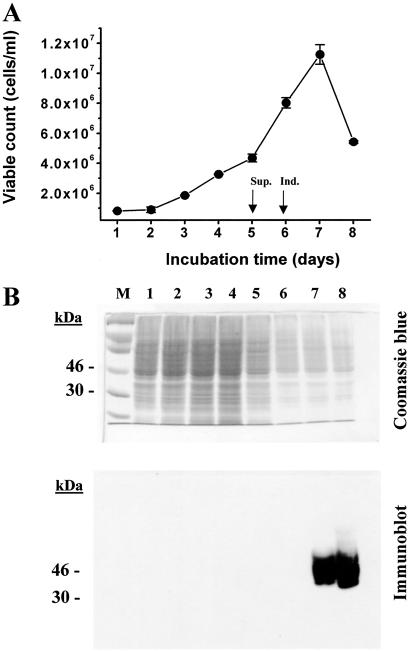
Figure 3.
Growth of a cell line inducible for WT opsin expression in 1.1-liter suspension culture by using a bioreactor. The cell line was grown in the bioreactor as described in Materials and Methods. (A) Viable count calculated by counting cells samples removed from the bioreactor at 24-h intervals. (Error bars represent average of two counts from same time point; Materials and Methods.) The culture was supplemented with the growth medium and opsin expression was induced as indicated by arrows. (B) Samples collected throughout growth were solubilized and proteins were separated by SDS/PAGE (10%). Whole-cell proteins were detected by Coomassie blue staining. For immunodetection, the proteins were transferred from gels to nitrocellulose by electroblotting and opsin was visualized by immunodetection with anti-rhodopsin monoclonal antibody rho-1D4. Sup., supplementation; Ind., induction.
Inducible High-Level Expression of WT Opsin in a Bioreactor.
An HEK293S cell line carrying an inducible opsin gene was grown in a bioreactor by using optimal growth conditions established in Materials and Methods. Primatone RL-UF is a source of short polypeptides and amino acids and thus enriches the medium (21). Dextran sulfate (22) was added, but calcium chloride was omitted to reduce the tendency of the cells to aggregate in suspension culture. In this experiment, 1.1 liter of the medium was inoculated with cells from six confluent 15-cm dishes. An initial lag phase of 24 h was usually observed (Fig. 3A), followed by cell growth with a doubling time of ≈24–30 h. The growth medium was supplemented (Materials and Methods) on day 5, and opsin gene expression was induced on day 6 by addition of tetracycline and sodium butyrate to the medium. The cells reached a maximum density of 107 cells per milliliter on day 7 (Fig. 3A). (In separate experiments cell densities of >5 × 106 were obtained consistently.) Opsin expression was monitored in cell samples taken at 24-h intervals. Detergent-solubilized cell lysates were investigated by SDS/PAGE (10%), and the separated total proteins were visualized by Coomassie blue, whereas rhodopsin was selectively visualized by immunoblotting with rho-1D4 antibody (Fig. 3B). No opsin was detected by immunoblotting in the 6 days before induction (Fig. 3B, lanes 1–6), even when overloading of the protein occurred (Fig. 3B, Coomassie blue). Opsin production was clearly evident in the samples taken 24 and 48 h after induction (days 7 and 8). The final level of opsin at the point of harvest on day 8 was 9 mg/liter.
Expression of Nonglycosylated Opsin in the Presence of Tunicamycin.
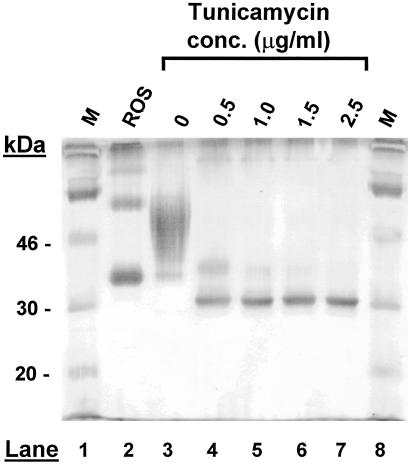
Rhodopsin purified from bovine rod outer segments migrates at an apparent Mr of ≈35 kDa (Fig. 4, lane 2), whereas rhodopsin purified from tetracycline and sodium butyrate induced HEK293S cells migrates predominantly as a smear of 40–60 kDa (Fig. 4, lane 3). Induction of the opsin gene expression and inhibition of opsin glycosylation was studied in the presence of tunicamycin. As seen in lanes 4–7 (Fig. 4) tunicamycin inhibited glycosylation in a concentration-dependent (0.5–2.5 μg/ml) manner. By using tunicamycin at a level of 2 μg/ml, opsin synthesis was ≈50% of that from non-tunicamycin-treated cells. However, the opsin made in the presence of tunicamycin was fully constituted to rhodopsin with 11-cis-retinal, as shown (23) with λmax at 500 nm and A280/500 ratio of 1.6 (data not shown).
Figure 4.
Effect of tunicamycin concentration on extent of opsin N-glycosylation. The tetracycline-inducible cell line producing WT opsin was grown to near confluence in 10-cm-diameter cell culture dishes. Spent medium was removed and replaced with fresh medium containing different concentrations (0–2.5 μg/ml) of tunicamycin. Three hours later, expression of opsin was induced by further supplementation of the growth medium with tetracycline and sodium butyrate. Cells were harvested 48 h later and treated with 11-cis-retinal to constitute the rhodopsin pigment. Rhodopsin was purified and examined (0.5 μg) by SDS/PAGE with silver stain for detection. Rhodopsin purified from bovine rod outer segments was loaded in lane 2, whereas molecular weight protein standards (M) were in lanes 1 and 8.
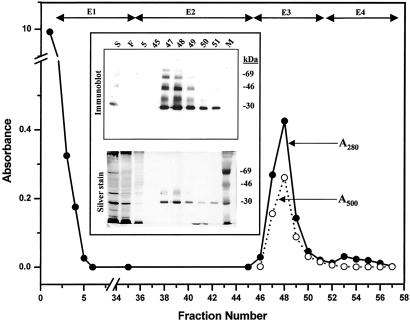
For preparation of milligram levels of nonglycosylated rhodopsin, the inducible opsin-producing HEK293S cell line was grown in 5.5-liter suspension exactly as described (Materials and Methods). On day 6 after inoculation, the cells were treated with tunicamycin (2 μg/ml), and then 4 h later with tetracycline and sodium butyrate. The cells were incubated for another 24 h before being harvested. The growth profile of the suspension culture and SDS/PAGE of samples removed daily from the bioreactor were similar to those shown in Fig. 3, except that a decline in cell viability occurred on day 7. The maximum cell density reached in this experiment was ≈5 × 106 cells per milliliter. The cells were harvested on day 7 when the total amount of rhodopsin produced was 3 mg/liter as determined by UV-visible difference spectrum with an aliquot of 11-cis-retinal-treated cells. SDS/PAGE analysis showed that opsin was synthesized only after induction of the opsin gene (data not shown). Purification of nonglycosylated rhodopsin is shown in Fig. 5. The elution profile of nonglycosylated rhodopsin from the column is identical with that of fully glycosylated rhodopsin (1) with the majority eluting in the third column volume (fraction 48). The total amount of rhodopsin recovered from the rho-1D4-Sepharose column was 3 mg (from 3.3 mg applied with a recovery of >90%). Examination by SDS/PAGE shows the greater part of rhodopsin thus produced migrated as a relatively sharp band (Fig. 5 Inset, silver stain) with an apparent Mr of ≈30 kDa. Some glycosylated rhodopsin was detected by overdeveloped immunoblot (Fig. 5 Inset). The slower migrating species with mobilities around 42 and 65 kDa presumably correspond to dimer and trimer forms, respectively, of the opsin resulting from slow denaturation.
Figure 5.
Purification of nonglycosylated rhodopsin from a portion of the cells grown in a 5.5-liter bioreactor by rho-1D4 immunoaffinity chromatography. Elution buffers were: E1, buffer H; E2, buffer I; E3, buffer J; E4, buffer K. Absorption at 280 nm (A280) (filled circles) and at 500 nm (A500) (open circles) was recorded as indicated. (Inset) SDS/PAGE (10% gel) examination of selected fractions eluted from the column as visualized by silver stain and immunoblot after electroblotting to nitrocellulose. M, molecular weight standards; S, solubilized cell extract; F, flowthrough containing proteins that do not bind to rho-1D4-Sepharose.
High-Level Inducible Expression of a Constitutively Active Opsin Mutant.
Several attempts by using the previous methodology (1) to construct a stable HEK293S cell line that would express the opsin triple mutant, E113Q/E134Q/M257Y, were unsuccessful. This failure suggests strongly that even low-level expression of this protein is cytotoxic. This mutant opsin gene was cloned into the plasmid used for constructing inducible HEK293S cell lines and used to transfect HEK293S-TetR cells. Isolated Geneticin-resistant colonies were expanded and stable cell lines exhibiting inducible opsin expression were identified by rho-1D4 immunodetection after transferring soluble cell extracts to nitrocellulose. One stable cell line displaying inducible expression of this mutant was chosen for further study. Monolayer growth of cells in 10-cm-diameter culture dishes followed by induction of opsin expression by treatment with tetracycline and sodium butyrate for 24 h showed that this mutant protein was produced in the range of 30 μg of rhodopsin per 107 cells (data not shown). This expression level is similar to that observed for the WT protein in the same incubation period. Incubation in the presence of tetracycline and sodium butyrate for this cell line was extremely toxic and most cells detached from the culture dish within 24 h after induction of expression.
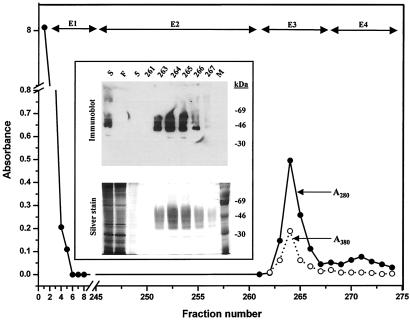
The cell line expressing this mutant opsin gene was grown in suspension culture (1.1 liter) as described in Materials and Methods. Induction of expression was restricted to 24 h. The purification of this mutant is shown in Fig. 6. The total amount of rhodopsin purified was 1.1 mg. This mutant rhodopsin combines with 11-cis-retinal to form a pigment with a λmax of 380 nm. The glycosylation pattern (Fig. 6 Inset) is identical with WT rhodopsin produced by HEK293S cells (1) as evidenced by the migration pattern on SDS/PAGE.
Figure 6.
Purification by rho-1D4 immunoaffinity chromatography of rhodopsin mutant, E113Q/E134Q/M257Y, from a portion of the cells grown in a 1.1-liter bioreactor culture. Elution buffers were: E1, buffer C; E2, buffer D; E3, buffer E; E4, buffer F. Absorption at 280 nm (A280) (filled circles) and 380 nm (A380) (open circles) was recorded as indicated. (Inset) SDS/PAGE (10% gel) examination of selected fractions eluted from the column as visualized by silver stain and immunoblot after electroblotting to nitrocellulose. M, molecular weight standards; S, solubilized cell extract; F, flowthrough containing proteins that do not bind to rho-1D4-Sepharose.
Discussion
Specific impetus for integration of tetracycline-inducible expression into our previously designed stable HEK293S mammalian cell lines came from our inability to express adequately certain opsin mutants that currently are under focus in our structure–function studies on rhodopsin. Conditional gene expression is an established method for high-level production of recombinant proteins in bacteria (8, 9), and it was necessary, for example, for the high-level expression of bacteriorhodopsin mutant genes in E. coli (10). The aim of the present work was to use this principle for expression of proteins in mammalian cell lines, and the examples now provided demonstrate preparative scale expression of opsins that could not be produced by previously developed constitutive expression methods.
Inducible expression was found to require both tetracycline and sodium butyrate; the two components acting synergistically to give high-level expression of the opsin gene (Fig. 2). In the absence of tetracycline, the addition of butyrate was not sufficient for opsin gene expression, thus demonstrating that regulation by tetracycline is dominant. Sodium butyrate is an inhibitor of histone deacetylase and likely acts by disrupting the chromatin complexes that regulate gene expression. Enhancement of gene expression by sodium butyrate has been reported (24), and we have also shown that in stable cell lines expression of the opsin gene under control of the WT CMV promoter is enhanced 2- to 3-fold by its addition (2).
The main focus in the present work has been on the inducible expression of two proteins, the WT opsin in its nonglycosylated form and a strongly constitutively active opsin mutant. In each case, toxicity to the growing cells was concluded to be the problem. In the first case, toxicity was due to the presence of tunicamycin; and in the second case, toxicity was presumably derived from the accumulation of the constitutively active mutant, even though the specific effect of the protein on HEK293 cells is not understood. Thus, the first application described is production of nonglycosylated rhodopsin (23), which is a potentially important candidate for crystallization attempts. Rhodopsin obtained from stable 293S cell lines (1), including the present tetracycline-inducible system, contains complex N-glycans as is evident by the diffuse band seen on SDS/PAGE (Fig. 4, lane 3). (See also accompanying paper, ref. 25.) N-glycosylation can be prevented by addition of the antibiotic tunicamycin to the growth medium (it could also be prevented by mutating the consensus sites for N-glycosylation) (23). By delaying addition of tunicamycin to cultures until near the end of the growth phase, problems associated with toxicity of this compound are overcome. The second example chosen, the constitutively active mutant, E113Q/E134Q/M257Y, forms a topic of our current study of the mechanism of receptor activation.
The techniques now developed should be useful for relatively large-scale production of membrane proteins, including other G protein-coupled receptors, that may impart cytotoxicity when constitutively expressed.
Acknowledgments
We are grateful to Professor U. L. RajBhandary of our Biology Department for valuable discussions. The enthusiastic assistance of Ms. Judy Carlin in the preparation of the manuscript is acknowledged. This work was supported by National Institutes of Health Grant GM28289.
Abbreviations
- DM
n-dodecyl-β-d-maltoside
- NG
n-nonyl-β-d-glucoside
- CMV
cytomegalovirus
Footnotes
This is paper 52 in the series “Structure–Function in Rhodopsin.” The preceding paper is ref. 14.
References
- 1.Reeves P J, Thurmond R L, Khorana H G. Proc Natl Acad Sci USA. 1996;93:11487–11492. doi: 10.1073/pnas.93.21.11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reeves P J, Klein-Seetharaman J, Getmanova E V, Eilers M, Loewen M C, Smith S O, Khorana H G. Biochem Soc Trans. 1999;27:950–955. doi: 10.1042/bst0270950. [DOI] [PubMed] [Google Scholar]
- 3.Eilers M, Reeves P J, Ying W, Khorana H G, Smith S O. Proc Natl Acad Sci USA. 1999;96:487–492. doi: 10.1073/pnas.96.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein-Seetharaman J, Getmanova E V, Loewen M C, Reeves P J, Khorana H G. Proc Natl Acad Sci USA. 1999;96:13744–13749. doi: 10.1073/pnas.96.24.13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein-Seetharaman J, Reeves P J, Loewen M C, Getmanova E V, Chung J, Schwalbe H, Wright P E, Khorana H G. Proc Natl Acad Sci USA. 2002;99:3452–3457. doi: 10.1073/pnas.052713999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loewen M C, Klein-Seetharaman J, Getmanova E V, Reeves P J, Schwalbe H, Khorana H G. Proc Natl Acad Sci USA. 2001;98:4888–4892. doi: 10.1073/pnas.051633098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruel C, Cha K, Reeves P J, Getmanova E, Khorana H G. Proc Natl Acad Sci USA. 2000;97:3004–3009. doi: 10.1073/pnas.97.7.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mieschendahl M, Petri T, Hanggi U. Bio/Technology. 1986;4:802–808. [Google Scholar]
- 9.Remaut E, Tsao H, Fiers W. Gene. 1983;22:103–113. doi: 10.1016/0378-1119(83)90069-0. [DOI] [PubMed] [Google Scholar]
- 10.Karnik S S, Nassal M, Doi T, Jay E, Sgaramella V, Khorana H G. J Biol Chem. 1987;262:9255–9263. [PubMed] [Google Scholar]
- 11.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fussenegger M. Biotechnol Prog. 2001;17:1–51. doi: 10.1021/bp000129c. [DOI] [PubMed] [Google Scholar]
- 13.Yao F, Svensjo T, Winkler T, Lu M, Eriksson C, Eriksson E. Hum Gene Ther. 1998;9:1939–1950. doi: 10.1089/hum.1998.9.13-1939. [DOI] [PubMed] [Google Scholar]
- 14.Niu L, Kim J-M, Khorana H G. Proc Natl Acad Sci USA. 2002;99:13409–13412. doi: 10.1073/pnas.212518899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molday R S, MacKenzie D. Biochemistry. 1983;22:653–660. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- 16.Papermaster D S. Methods Enzymol. 1982;81:240–246. doi: 10.1016/s0076-6879(82)81037-9. [DOI] [PubMed] [Google Scholar]
- 17.Oprian D D, Molday R S, Kaufman R J, Khorana H G. Proc Natl Acad Sci USA. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knowles A, Priestly A. Vision Res. 1978;18:115–116. doi: 10.1016/0042-6989(78)90086-x. [DOI] [PubMed] [Google Scholar]
- 19.Velan B, Kronman C, Ordentlich A, Flashner Y, Leitner M, Cohen S, Shafferman A. Biochem J. 1993;296:649–656. doi: 10.1042/bj2960649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reeves P J, Hwa J, Khorana H G. Proc Natl Acad Sci USA. 1999;96:1927–1931. doi: 10.1073/pnas.96.5.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlaeger E-J. J Immunol Methods. 1996;194:191–199. doi: 10.1016/0022-1759(96)00080-4. [DOI] [PubMed] [Google Scholar]
- 22.Dee K U, Shuler M L, Wood H A. Biotechnol Bioeng. 1997;54:191–205. doi: 10.1002/(SICI)1097-0290(19970505)54:3<191::AID-BIT1>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 23.Kaushal S, Ridge K D, Khorana H G. Proc Natl Acad Sci USA. 1994;91:4024–4028. doi: 10.1073/pnas.91.9.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmero D P, Degraaf M E, Marotti K R, Rehberg E, Post L E. J Biotechnol. 1991;19:35–47. doi: 10.1016/0168-1656(91)90073-5. [DOI] [PubMed] [Google Scholar]
- 25.Reeves P J, Callewaert N, Contreras R, Khorana H G. Proc Natl Acad Sci USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]