Abstract
Although preB cell-receptor (pre-BCR) formation and cell-surface expression is essential for B cell development, pre-BCR generation of signal transduction remains elusive. Here, we report that recombinant pre-BCRs and the surrogate light chain bind specifically to the bone marrow stromal cell galectin-1 (GAL1), an S-type lectin. The surrogate light chain/GAL1 association is a direct protein–protein interaction (Ka = 2 × 106 M−1), and the NH2 extra loop of λ-like is the major binding element. Pre-BCR binding to stromal cells depends upon GAL1 anchoring to glycosylated counter-receptors, and these complexes completely relocalize to form a synapse at the contact zone between preB and stromal cells. This immune developmental synapse is accompanied by the initiation of intracellular tyrosine kinase activity and signal transduction from the pre-BCR.
B lymphocytes mature from hematopoietic stem cells through a series of developmental stages that are characterized by cell-surface markers and sequential DNA rearrangements of Ig gene segments (1). Although bone marrow (BM) stromal cells are an essential support for normal precursor B cell development, factors driving the earliest stages of B lymphopoiesis remain poorly understood. The α4 integrin-mediated interactions between B cell progenitors and BM stromal cells are crucial for normal development, and secretion of soluble factors by stromal cells (IL7, SCF, Flt3L, SDF-1) regulates precursor B cells growth, maturation, and survival (2).
To progress through the different B cell differentiation stages, B cells must pass several checkpoints. Igμ chain synthesized by developing preBII cells must associate with surrogate light chain (SLC) made of λ-like (λ5 in mouse) and VpreB (3, 4) and with the CD79a/CD79b transducing complex to form a functional pre- BCR (5, 6).
In mice lacking the transmembrane portion of Igμ chain (μmT), B cell development is blocked at the transition between preBI to large preBII cells (7). VH to D-JH rearrangements can occur, but in the absence of membrane-bound pre-BCRs, cells do not proliferate, and Igμ chains are not allelically excluded (7, 8). Surprisingly, in mice lacking the λ5 or the two VpreB genes, Igμ chains are allelically excluded (4, 9), and B cell development is not totally blocked. However in these mice, preBII cells do not proliferate (9, 10). These data suggest that the allelic exclusion process totally depends upon Igμ membrane deposition but does not involve the SLC, and that pre-BCR mediates preBII cell expansion. Indeed, Igμ chains that fail to pair with SLC lose the ability to be expressed at the cell surface and are counterselected (11). Thus, the SLC seems to be essential for preBII cells to enter into cell cycle and for the selection of an early Igμ repertoire. These functions necessitate the generation of pre-BCR intracellular signaling involving tyrosine activation motifs (ITAMs) of both the CD79a and CD79b chains (12). Recently, it has been shown that a fraction of the pre-BCR is constitutively associated to raft structures and that pre-BCR engagement enhances this association, leading to calcium flux and changes in protein tyrosine phosphorylation (13). However, the question of the generation of pre-BCR signaling remains completely unanswered. Whether pre-BCR cell-surface expression is sufficient to generate a constitutive signal, or whether the pre-BCR is triggered by its interaction with an environmental ligand, remain to be determined. The only reported observation concerns the binding of soluble pre-BCR to mouse ST2 stromal cells (14).
In this paper, we report the identification of a pre-BCR ligand, which turns out to be the galectin-1 (GAL1), expressed by stromal cells. GAL1 fixation on preB cells leads to formation of a pre-BCR/GAL1 lattice, polarized at the contact zone between preB and stromal cells, which results in pre-BCR triggering.
Materials and Methods
Cell Lines.
Human and mouse cell lines used in this study are reported in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. The murine MS5.1 stromal cell line subclone was selected on the basis of its high binding property to recombinant SLC. Cell lines and normal bone marrow stromal cells were cultured as described (15, 16).
Antibodies and Reagents.
Anti-human VpreB 4G7 mAb and anti-HEL HH5 mAb were generated as described (17); anti-Flag M2 mAb, HRP-conjugated goat anti-mouse IgG antibody, HRP-conjugated protein A, thiodigalactoside, lactose, maltose, digitonin, saponin, and sodium orthovanadate were obtained from Sigma Aldrich; anti-human IgM biotin-coupled antibody was obtained from PharMingen; anti-human CD79a mAb, FITC-labeled goat anti-mouse IgM+IgG antibody, FITC-labeled donkey anti-rabbit IgG antibody, Texas red-labeled goat anti-mouse IgG antibody, Texas red-labeled goat anti-rabbit antibody, FITC-labeled streptavidin, and rhodamine-labeled streptavidin were obtained from Beckman-Coulter-Immunotech; anti-phosphotyrosine 4G10 (anti-P-Tyr) mAb was a gift of H. T. He (Marseille, France); rabbit anti-human GAL1 antiserum was a generous gift of R. Caron-Joubert (Paris) (18); rabbit anti-human VpreB and anti-NH2-terminal λ-like peptide were generated as described (19).
Production and Purification of Recombinant Proteins.
The scSLC (λ-like linked to VpreB by a linker peptide), the two Fab-like (VH-CH1μ and scSLC) using VH-CH1μ from Nalm6 or 1E8 preB/B cell lines, and a conventional Fab (VH-CH1μ 1E8 and Igκ) recombinant protein expressing an M2 COOH-terminal flag were produced in insect cells (15). For production in Escherichia coli; scSLC*, λ-like, and VpreB coding segments were amplified from the pGm16-scΨL transfer plasmid (15, 20) and cloned into the pQE-60 expression vector (Qiagen). His-tags are present at the COOH-terminal for scSLC* and λ-like and at the NH2-terminal for VpreB. Proteins expressed in BL21-RP strain (Stratagene) were purified from inclusion bodies on Talon Superflow Metal Affinity Resin (CLONTECH) according to manufacturer's instructions. Refolding of the scSLC*, λ-like, and VpreB purified proteins was performed at room temperature by decreasing the urea concentration. For SLC* refolding, λ-like and VpreB proteins were mixed with a 1/1 molar ratio. Soluble refolded proteins were purified on a Superdex 200 size-exclusion column (AKTA system, Pharmacia Biosensor).
Flow Cytometry.
Adherent cells were detached with nonenzymatic dissociation solution (Sigma). Cells (2–5 × 105) were stained for 30 min at 4°C in ice-cold PBSBN (PBS buffer supplemented with 0.2% BSA/0.05% NaN3), using the different recombinant proteins (5–20 μg/ml). Binding was revealed by incubation with anti-VpreB 4G7 (2.5 μg/ml) or M2 (5 μg/ml) mAbs for 30 min followed by fluorochrome-conjugated secondary Abs. Analyses were performed by using the FACSCalibur apparatus (Becton Dickinson).
Before incubation with recombinant scSLC* (20 μg/ml) or anti-GAL1 antibodies, HeLa cells were incubated for 15 min with recombinant GAL1 (3.5 μg/ml) in PBSBN supplemented with 0.5 M lactose or maltose. For inhibition experiments, MS5.1 cells were cultivated for 2 h in the presence of 0.5 M lactose or maltose before flow cytometry analysis.
BIAcore Analysis.
Surface plasmon resonance measurements were performed on a BIAcore apparatus (Pharmacia Biosensor) using the BIAlogue Kinetics Evaluation program (BIAEVALUATION V.3.1, Pharmacia Biosensor). Recombinant GAL1 protein was immobilized on a Sensor Chip B1 as described (15).
Confocal Microscopy.
Stromal cells seeded at 30,000 cells per cm2 on glass coverslips (Marienfeld, Lauda-Königshofen, Germany) were allowed to grow for 2 days. Lymphoid cells (75,000 per well) were cultured on stromal cells for 2 h at 37°C. After fixation, cells were stained with the appropriate antibodies and examined with an LSM-510 Carl Zeiss confocal microscope. Adherent stromal cells were preincubated for 20 min with medium supplemented with 0.1 M lactose, maltose, or thiodigalactoside and lymphoid cells were cultured on stromal cells with medium supplemented with the appropriate sugars. Slides were imaged by laser-scanning microscopy by using the 16× objective. Slides were first scanned by differential interferential contrast (DIC) imaging (21), and 300–1,500 lymphoid cells in close contact with stromal cells were examined further for their surface fluorescence distribution. SDs were calculated from different independent experiments. Nalm6 preB cells were incubated with recombinant GAL1 (3.5 μg/ml) for 30 min at 37°C, washed, and allowed to grow for 90 min at 37°C before deposition on polyl-lysine-treated coverslips and stained.
Phosphotyrosine intracellular staining was performed according to Muller et al. (22) using the 4G10 anti-P-Tyr mAb and the anti-human IgM biotin-coupled antibody.
Tridimensional (3D) Iso-Surface Construction.
For 3D fluorescent iso-surface constructions, cells were imaged by the LSM-510 Zeiss microscope with a 43× objective. For individual cells, 40–50 confocal slices of 0.14-μm thickness were acquired. Z-stack slices then were analyzed by using IMARIS3 software (Bitplane, Zurich) on a Silicon Graphics Octane 2 SGI computer.
Biochemical Analysis.
MS5.1 (3 × 108) cells were lysed with 25 ml of IGEPAL lysis buffer [1% (vol/vol) IGEPAL CA-630/50 mM Na2HPO4, pH 7.5/NaCl 300 mM/PMSF 1 mM/protease inhibitor mixture; Sigma)]. Two lysate fractions were separately incubated for 2 h at 4°C scSCL* with 1 ml of nickel Sepharose loaded with 4 mg of His-tag scSCL or with nickel Sepharose beads alone. Proteins were eluted by 6 M urea (3 × 1 ml) to dissociate bounded material without removing the scSLC* from the beads, followed by 6 M urea/200 mM imidazole (3 × 1 ml) buffer. Urea-eluted fractions were analyzed on an SDS/7.5–17.5% gradient PAGE, and fractions containing specifically captured proteins were concentrated and separated on a preparative SDS/17.5% PAGE. The corresponding fractions from unloaded beads were similarly treated and used as negative controls.
MS5.1 cells were seeded at 60,000 cells per cm2 in Petri dishes and cultured for 1 day at 37°C. Nalm6 or Laz221 cells (20 × 106 cells) were cocultured with MS5.1 for 5 min, 30 min, or 2 h. Cells (20 × 106 per ml) were lysed in ice-cold DLB (50 mM Tris, pH 7.5/150 mM NaCl/1% digitonin/4 mM Na3VO4/protease inhibitors), and proteins were immunoprecipitated by using anti-human CD79a mAb and protein G-Sepharose beads for 4 h at 4°C. Samples were separated on an SDS/7.5–17.5% gradient PAGE. For Laz221, cocultures with MS5.1 were performed in the presence of scSLC* (80 μg/ml) or lactose (0.1 M) for 30 min.
Western blotting were performed as described (15) by using anti-VpreB, anti-CD79a, or anti-P-Tyr mAbs.
Mass Spectrometry Analysis.
SDS/PAGE-resolved protein bands were excised from the gel and “in-gel” tryptic digestion was performed for 4 h at 37°C with 0.5 μg of trypsin (Promega sequencing grade). Matrix-assisted laser desorption ionization mass spectra of trypsin peptide digestions were obtained by using an Autoflex mass spectrometer (Bruker Daltonik). Monoisotopic peptide masses were assigned and used for database searching by using the program MS-FIT (http://prospector.ucsf.edu/). Tandem mass spectrometry experiments were carried out on a Q-TOF hybrid mass spectrometer (Micromass, Manchester, England) to sequence isolated tryptic peptides. MS/MS sequence data were used for database searching with the programs MS-PATTERN (http://prospector.ucsf.edu/) or PEPTIDESEARCH (www.mann.embl-heidelberg.de/).
Results
Binding of Recombinant Pre-BCRs to Adherent Stromal Cells.
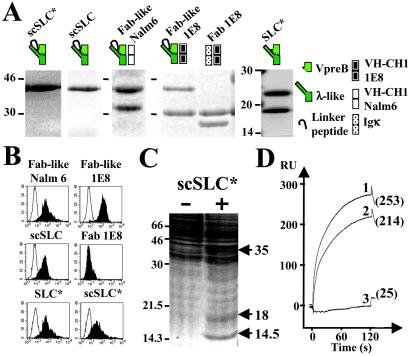
Recombinant Fab-like, Fab 1E8, and SLC proteins have been produced in various expression systems and their purity has been verified by SDS/PAGE analysis (Fig. 1A). Flow cytometry analysis shows that Fab-like and SLC proteins bind to the bone marrow-derived murine MS5.1 cell line (Fig. 1B), to a series of adherent cell lines, and to normal bone marrow-derived stromal cells from human fetuses and adult mice (Table 1 and Fig. 6, which are published as supporting information on the PNAS web site). The regular Fab 1E8, composed of the 1E8 VH-CH1μ chain and a κ chain, did not bind to stromal cells, suggesting that the SLC is the critical component in pre-BCR/stromal cell interactions. Indeed, identical cell-binding patterns were observed for the two scSLC, the native SLC* and the Fab-like recombinant proteins (Fig. 1B).
Figure 1.
Binding of soluble pre-BCRs or SLC components to MS5.1 cell line and biochemical identification of GAL1 as the SLC ligand. (A) Purified recombinant proteins were separated on SDS/15% polyacrylamide gels under reducing conditions and visualized by Coomassie blue staining. scSLC* (λ-like is fused to VpreB by a linker peptide) and SLC* (λ-like is reassociated with VpreB) were produced in E. coli and recombinant scSLC, Fab-like Nalm6 (scSLC+Nalm6 VH-CH1μ), Fab-like 1E8 (scSLC + 1E8 VH-CH1μ), and Fab 1E8 (κ+1E8 VH-CH1μ) by using baculoviruses. Schematic representations of the recombinant proteins are shown on the top of the figure. (B) Binding of the two Fab-like, Fab 1E8 and the different SLC recombinant proteins to MS5.1 cell line, were analyzed by flow cytometry. For each analysis, 10 μg/ml of recombinant proteins were used, and bindings were revealed by incubation with the 4G7 (2.5 μg/ml) anti-VpreB mAb (γ1, κ) for E. coli-derived proteins or with the M2 (5 μg/ml) anti-Flag mAb (γ1, κ) for baculovirus-derived proteins (black). For negative controls, the recombinant proteins are revealed with the HH5 (5 μg/ml) anti-HEL mAb (γ1, κ) (white). (C) Preparative MS5.1 cell lysate was incubated with nickel-Sepharose scSLC-loaded (+) and unloaded (−) beads (1.5 × 108 MS5.1 cells were used for each experimental condition). Material eluted in 6 M urea was separated on a preparative SDS/17.5% PAGE and revealed by Coomassie staining. Molecular weights of standard proteins are indicated in kDa. Arrows indicate differentially captured proteins. (D) BIAcore analysis of scSLC* (1), λ-like (2), and VpreB (3) binding to immobilized GAL1. The three analytes (40 μl at 10 μg/ml) were injected at a flow rate of 20 μl/min in HBS buffer on a dextran layer containing 500 resonance units (RU) of GAL1. The resulting sensorgrams are superimposed and are representative of two independent experiments. RU values at 125 s after the injection start are indicated.
Thus, soluble pre-BCRs or the SLC alone are able to interact with adherent cells from different species and can be derived from various tissues, including those supporting B cell differentiation.
Identification of a Stromal Cell-Derived SLC-Binding Protein.
To identify stromal cell-derived SLC-binding proteins, a large-scale preparative MS5.1 cell lysate (3 × 108 cells) was incubated with 4 mg of His-tag scSCL* loaded onto nickel-Sepharose beads. Urea-eluted fractions were analyzed on an SDS/7.5–17.5% gradient PAGE (data not shown), and fractions containing specifically captured proteins were concentrated and separated on an SDS/17.5% preparative PAGE (Fig. 1C). Unloaded nickel-Sepharose beads served as negative controls. Differentially detected proteins were analyzed by mass spectrometry. Proteins at 35 kDa and 18 kDa were characterized as the scSLC* and an scSLC*-derived product, respectively. The 14.5-kDa protein was identified as the GAL1 by peptide mass fingerprint (Swissprot, P16045) and tandem mass spectrometry by sequencing four internal tryptic peptides. GAL1 belongs to a large family of calcium-independent S-type lectins with affinity for β-galactosides.
Evidence for a Direct and Efficient SLC and GAL1 Binding.
BIAcore analysis demonstrate that recombinant Fab-like 1E8 binds to the immobilized human recombinant GAL1, in contrast to the Fab control (Figs. 7 and 8, which are published as supporting information on the PNAS web site). Moreover, the nonglycosylated scSLC* interacts efficiently with GAL1 (Fig. 1D), indicating that SLC–GAL1 interaction is a direct protein–protein binding. Apparent affinity constants (Ka values) were roughly similar whenever kinetics (Ka: 3.12 × 106 M−1) or equilibrium (Ka: 2.34 × 106 M−1) measurements were performed (Fig. 9, which is published as supporting information on the PNAS web site). We observed that VpreB fails to bind to immobilized GAL1 protein, whereas λ-like interacts (Fig. 1D) with a Ka of 1.29 × 106 M−1, a value very close to that of the complete scSLC* (Fig. 9). Preincubation of scSLC* with antiserum against the NH2-terminal peptide of λ-like blocks the binding of scSLC* to the immobilized GAL1 (Fig. 9), suggesting that the λ-like extra loop (20) is involved in the binding. Finally, we observed that the molar ratio of scSLC*/GAL1 never exceeded 0.5 (data not shown), suggesting that only one SLC molecule binds to one GAL1 homodimer.
These data indicate that the GAL1 protein interacts directly with the SLC and that the NH2-terminal extra loop peptide of λ-like is the major component of the binding. The fact that a GAL1 homodimer associates to a single SCL seems to indicate that the SLC-binding site is shared by two GAL1 polypeptides.
Direct Binding of SLC to Stromal Cell-Anchored GAL1.
As GAL1 is captured at the cell surface by glycosylated receptors (23, 24), we examined whether stromal cell membrane-anchored GAL1 is the target of SLC binding.
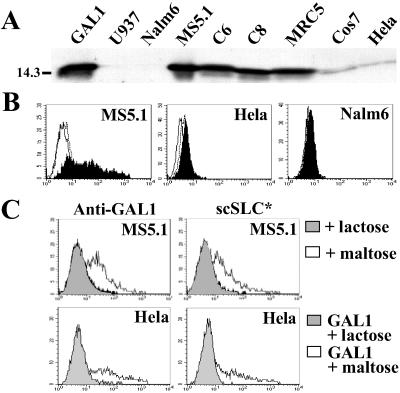
GAL1 is detected by Western blotting from total cell lysates of human, murine, and simian-adherent cell lines but not from lymphoid and myeloid cells (Fig. 2A). GAL1 is expressed at the cell surface of the MS5.1 cell line but is not detected on HeLa or Nalm6 cells (Fig. 2B). These data are in agreement with Fab-like or SLC-binding patterns (Table 1 and Fig. 6) and suggest that cell-surface GAL1 is indeed the SLC target. To confirm this, the MS5.1 surface-anchored GAL1 was removed by cultivating the cells in the presence of lactose, a known GAL1 ligand, or maltose as a control. Labeling with rabbit anti-GAL1 antibodies or with scSLC* (Fig. 2C Upper) shows that anti-GAL1 and SLC binding are totally inhibited by lactose (gray) and not by maltose (white) treatment.
Figure 2.
Expression of GAL1 by different cell lines and binding inhibition of SLC to stromal cells by lactose treatment. (A) Western blot analysis of GAL1 expression of total cell lysate from different cell lines by using rabbit anti-GAL1 antiserum. Molecular mass of standard proteins is in kDa. Cell lines: U937 (human myeloid), Nalm6 (human preB), MS5.1 (murine BM stromal), C6 and C8 (human BM stromal), MRC5 (human embryonic fibroblast), HeLa (human epithelium), and Cos7 (simian epithelium). As positive control, 0.1 μg of recombinant GAL1 was loaded. (B) Flow cytometry analysis of GAL1 expression on MS5.1, HeLa, and Nalm6 cells, using anti-GAL1 antiserum and FITC-labeled donkey anti-rabbit IgG antibodies (black). For negative controls, the anti-GAL1 antiserum was omitted (white) and an irrelevant rabbit anti-serum was used (dashed). (C) Flow cytometry analysis of MS5.1 stromal cells (Upper) cultured for 2 h in presence of 0.5 M lactose, a GAL1-specific sugar (gray), and 0.5 M maltose (white) by using anti-GAL1 antiserum or scSLC* (20 μg/ml). HeLa cells (Lower) were incubated for 15 min with recombinant GAL1 (3.5 μg/ml) in the presence of 0.5 M lactose (gray) or maltose (white) before staining with anti-GAL1 antibodies or scSLC* (20 μg/ml). Histograms corresponding to MS5.1 or GAL1-loaded HeLa cells in the absence of sugar treatments are identical to those with maltose treatment (not shown).
To demonstrate that SCL stromal cell-surface labeling is caused by a direct interaction with GAL1, we have treated HeLa cells with recombinant GAL1 (3.5 μg/ml) and analyzed scSLC* fixation. First, we controlled the GAL1 interaction to HeLa cells by binding to glycosylated counter-receptors (Fig. 2C Lower). Then, we observed that scSLC* binds to GAL1-loaded HeLa cells in a lactose-sensitive way (Fig. 2C Lower).
Lactose or maltose treatments have not effect on direct scSLC*–GAL1 interactions (Fig. 9), indicating that the inhibition of scSLC* binding to stromal cells is not caused by a direct competition of SLC and lactose for GAL1. This result also indicates that SLC- and sugar-binding sites are independent.
Altogether, these data confirm that GAL1 is anchored at the cell surface of stromal cells by glycosylated receptors, and that SLC binding to stromal cell surface is caused by the presence of membrane-associated GAL1 proteins.
Relocalization of Pre-BCR and GAL1 Proteins at the Contact Zone Between PreB and Stromal Cells.
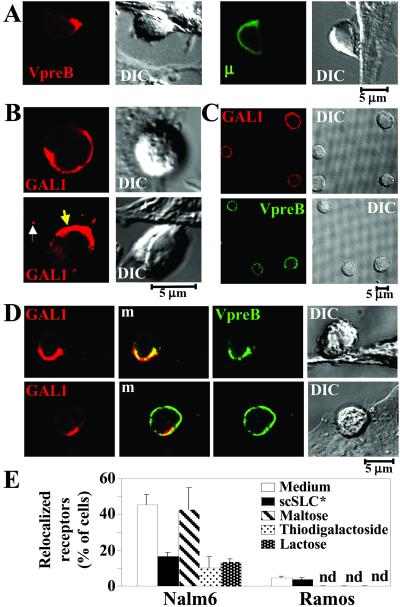
To analyze SLC–GAL1 interactions in the context of cell-surface environment, the behavior of GAL1 and pre-BCR were followed by confocal microscopy after preB/stromal cell cocultures. DIC imaging shows that the preB cells have established a close interaction with stromal cells (Fig. 3A and Fig. 10, which is published as supporting information on the PNAS web site). They present morphological modifications consecutive to the formation of a large contact area between the two cells and show a clear and remarkable translocation of the pre-BCR at the contact zone. This is the case for Nalm6 preB cells cultivated on human C8 or mouse MS5.1 stromal cells and for the murine 70Z3 preB on MS5.1 cells (Fig. 3A and Fig. 10). Pre-BCRs translocate completely to the junction to form one or few patches that do not cover the entire contact area. By contrast, for Ramos mature B cells, no modification of the BCR cell-surface distribution was detected (Fig. 3A).
Figure 3.
Interaction between preB and stromal cells induces the relocalization of pre-BCR and GAL1 at the contact zone between the two cells and depends on GAL1 binding to glycosylated receptors. PreB and stromal cell conjugates were analyzed by confocal microscopy with a Carl Zeiss confocal microscope. (A) DIC images show fixed lymphoid/MS5.1 stromal cell conjugates after a 2-h incubation. Immunofluorescent stainings of the same conjugates done with the 4G7 anti-VpreB mAb (Nalm6, Left) and the anti-human IgM mAb (Ramos, Right) are shown. (B) Anti-GAL1 staining of Nalm6 and MS5.1 cells after 2-h coculture. White and yellow arrows show stromal cell-surface GAL1 containing dots and GAL1 polarization toward the contact zone between preB and stromal cells, respectively. (C) Anti-GAL1 (Upper) and 4G7 anti-VpreB (Lower) staining of Nalm6 cells incubated 30 min with recombinant GAL1 in the absence of stromal cells. (D) Double staining of Nalm6 (Upper) and Ramos (Lower) cells cultivated on MS5.1 using anti-GAL1 antiserum, 4G7 anti-VpreB (Nalm6), and anti-human IgM (Ramos) mAbs. m, merge view. (E) Specific inhibition of pre-BCR relocalization with soluble recombinant scSLC* and GAL1 carbohydrate ligands. Nalm6 and Ramos were cocultured with MS5.1 cells for 2 h under various conditions, and the percentage of relocalized pre-BCR or BCR cells was analyzed by confocal microscopy after staining with the anti-human IgM mAb. Recombinant scSLC* (40 μg/ml) was added at the beginning of the coculture. MS5.1 cells were incubated for 20 min with 0.1 M of the different carbohydrates before the addition of Nalm6 cells. Data are representative of four independent experiments. For Nalm6, a total of 1,500, 1,200, 330, 350, and 383 cells were examined in medium without scSLC*, with scSLC*, with maltose, with thiodigalactoside, and with lactose, respectively. For Ramos, a total of 1,050 and 780 cells were examined in medium without scSLC* and with scSLC*, respectively.
Labeling of stromal cells with anti-GAL1 antiserum shows a patchy distribution with relatively rare small isolated dots disseminated all over the cell surface (data not shown). This peculiar staining is also visible when these cells are cultivated in the presence of the Nalm6 preB cell line (Fig. 3B, white arrow). In addition, a remarkable accumulation of GAL1 at the surface of preB cells is observed: few preB cells present a homogeneous cell-surface distribution (Fig. 3B Upper), whereas for the majority, surface-anchored GAL1 is polarized at the contact zone between preB and stromal cells (Fig. 3B Lower, yellow arrow). Thus, GAL1 produced by stromal cells is able to bind to preB cells and also to relocalize at the contact zone between stromal and preB cells.
When preB cells are decorated by recombinant GAL1 in the absence of stromal cells, polarized cell-surface GAL1 distribution is never observed, and the pre-BCR is not relocalized (Fig. 3C). By contrast, when preB cells are cultured on stromal cells, double labeling with anti-GAL1 and anti-VpreB reagents shows that GAL1 and pre-BCR colocalize and are polarized at the contact area between the two cells, with a molecular surface organization presenting the characteristic of a synapse (Fig. 3D Upper). Although a faint accumulation of GAL1 is observed at the contact zone between some mature B cells and stromal cells, no modification of BCR cell-surface distribution occurs (Fig. 3D Lower).
To confirm that SLC–GAL1 interactions are directly implicated in pre-BCR relocalization, we analyzed the influence of soluble SLC and sugar treatments on this process. Nalm6 preB cells and Ramos B cells were cocultured with MS5.1 cells for 2 h in various conditions, and the frequency of pre-BCR or BCR relocalization was determined by confocal microscopy. According to Fig. 3 A and D, a low frequency (3%) of BCR relocalization was noted for Ramos cells, which are not modified by the presence of soluble scSLC* (Fig. 3E). By contrast, for preB/stromal cocultures in medium alone, the frequency of relocalized cells reaches 45%. scSLC* addition had no detectable effect on Nalm6 adhesion to stromal cells (data not shown) but inhibited by 65% the relocalization process. Coculture treatments with β-galactoside sugars such as thiogalactoside and lactose result in 76 and 69% inhibition of the pre-BCR relocalization process, respectively, whereas maltose addition has no effect (Fig. 3E).
These results clearly demonstrate that the relocalization process is driven by direct binding of the pre-BCR to the stromal GAL1 ligand and is dependent on GAL1 binding to glycosylated receptors. The fact that stromal cells are required for pre-BCR and GAL1 relocalization points to a probable implication of stromal glycosylated counter-receptors in synapse formation.
The PreB/Stromal Cell Synapse Is a Signaling Platform.
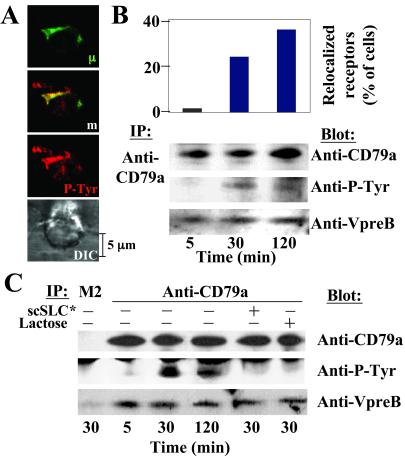
To demonstrate that transduction signals are generated upon synapse formation, we looked for tyrosine kinase activity in the vicinity of relocalized pre-BCR. The synapse formation results in the concentration of phosphotyrosine-containing proteins, which colocalize with the pre-BCR in Nalm6 (Fig. 4A) and Laz221 (data not shown) preB cell lines. These results indicate that most of the phosphorylation-based signal events are generated within the pre-BCR-containing complexes.
Figure 4.
Evidence for a functional PreB/stromal cell synapse. (A) Double staining of Nalm6 preB cells cultivated on MS5.1 using anti-P-Tyr and the anti-human IgM mAbs. m, merge view. (B Upper) Diagram representation of the frequency of Nalm6-relocalized cells for 5-, 30-, and 120-min coculture on MS5.1. For each point, 300–500 stromal-adhering preB cells were examined, and the results are expressed as percent of total lymphoid cells. (Lower) CD79a proteins were immunoprecipitated from Nalm6 lysates after coculture with MS5.1. Immunoprecipitates were blotted with anti-CD79a, anti-P-Tyr, and anti-VpreB mAbs after separation on an SDS/7.5–15% PAGE under reducing conditions. (C) CD79a proteins were immunoprecipitated from Laz221 lysates and analyzed as in B. In addition, CD79a proteins were immunoprecipitated after 30 min of coculture in the presence of scSLC* (80 μg/ml) or lactose (0.1 M). Control includes immunoprecipitation with an isotype-matched (M2) mAb.
To demonstrate the direct involvement of the pre-BCR in synaptic signal transduction, Nalm6 and Laz221 were cocultured on MS5.1 cells for different incubation times (5, 30, and 120 min), and the percentage of relocalized cells and the phosphorylation status of the CD79a transducing molecule were determined (Fig. 4 B and C). Pre-BCR-associated molecules are coimmunoprecipitated with CD79a, because VpreB was revealed by Western blotting. Among immunoprecipitated molecules, one at the expected size of CD79a (approximately 40 kDa) presents a modification of its phosphorylation status during coculture. Comigration with CD79a is confirmed by Western blotting. For Nalm6 and Laz221, a fraction of CD79a is phosphorylated at 30 min and, to a lesser extent, at 120 min of coculture. CD79a phosphorylation correlates with the dynamic of the relocalization process (Fig. 4 B and C). For Laz221, the inhibition of CD79a phosphorylation with scSLC* or lactose treatments during coculture revealed that pre-BCR-signaling specifically results from pre-BCR–GAL1 interactions (Fig. 4C).
These data suggest that pre-BCR recruitment into the preB/stromal cell synapse is responsible for the initiation of pre-BCR signal transduction.
Discussion
In this article, we identified GAL1 produced by stromal cells as a pre-BCR ligand. Galectins are a large family of calcium-independent S-type lectins, widely conserved in animals, plants, and microorganisms and with specificity for galactose derivatives. Fourteen galectins have been identified in mammals (25, 26). GAL1 consists of a single carbohydrate recognition domain (CRD) with a short NH2 sequence and occurs naturally as noncovalently bonded dimers (27). High GAL1 expression is observed in sensory, motor, and olfactory neurons and in hematopoietic and lymphoid organs; GAL1 is also expressed by stromal cells (28). The implication of GAL1 in various biological processes, including immune-cell homeostasis, has been extensively reported (for a review, see ref. 29). GAL1 is an apoptotic factor for thymocytes and mature T cells (30); it is involved in cell-cycle regulation and in various cell-adhesion processes. Cellular responses depend on the signals delivered by the different counter-receptors (31).
We have shown that the SLC binds to GAL1 by direct protein–protein interactions and that the λ-like chain is the major element of the binding (Fig. 1D). The VpreB partner is neither directly involved in GAL1 binding nor is it necessary for a conformational imprinting of the SLC-binding site, because SLC and λ-like bind to GAL1 with close affinities (Ka values of 3.12 × 106 M−1 and 1.29 × 106 M−1, respectively). These affinities are on the same order of magnitude as those measured for GAL1 cell-surface counter-receptors (32). The SLC λ-like NH2-terminal extra loop seems to be the major GAL1-binding element, which points to a clear biological role for this SLC region. The direct SLC binding to GAL1, with a binding site independent of the CRD carbohydrate pocket, is an interesting observation because, most frequently, galectins bind to their targets in a sugar-dependent manner. However, direct protein–protein interactions have been reported for other galectin family members (33).
Stromal cell-secreted GAL1 is able to bind to the preB cell surface (Fig. 3B), and we have demonstrated that pre-BCRs are GAL1 targets. However, when preB cells are in contact with stromal cells, as the pre-BCR cell-surface distribution is always included in a large GAL1 area (Figs. 3D and 5A), other GAL1 counter-receptors expressed by preB cells (cis-counter-receptors) must exist. Accordingly, it was shown that GAL1 binds to CD45, CD43, and CD7 on T cells (34) and to CD45 on mature B cells (35). Although CD45 could be a good candidate for a cis-counter-receptor, it is dispensable for the formation of relocalized SLC/GAL1 complexes, because this phenomenon was observed for Nalm6 cells that do not express CD45 (ref. 36; Fig. 11, which is published as supporting information on the PNAS web site). On stromal cells, glycosylated counter-receptors (transcounter-receptor) have not been identified so far. One possible candidate could be the 135-kDa protein precipitated from ST2 murine stromal cells by a recombinant pre-BCR (14).
Figure 5.
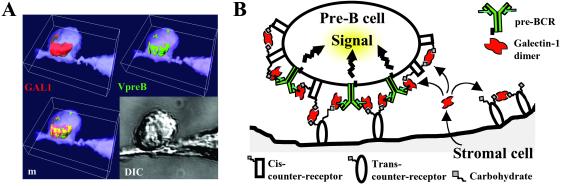
Model visualizing the molecular organization of the preB/stromal cell synapse. (A) 3D isosurface construction of GAL1 and pre-BCR cell surface distribution after preB and stromal cell synapse formation. Isosurfaces have been reconstructed from experimental data presented in Fig. 3D. This representation shows that pre-BCRs are included in the GAL1 relocalization zone. (B) GAL1 secreted by stromal cells is captured by pre-BCR and cis and trans counter-receptors. The model proposes that GAL1 is a supramolecular organizer of a 3D lattice that clusters together counter-receptors and pre-BCRs. The preB/stromal cell synapse formation results in pre-BCR relocalization and in the initiation of pre-BCR signaling.
We observed that SLC/GAL1-binding results in pre-BCR clustering into a GAL1/counter-receptor complex lattice, polarized at the preB/stromal cell-synaptic junction (Figs. 3D and 5A). Taking into account the predicted stoichiometry, one SLC for one GAL1 dimer, direct pre-BCR crosslinking by GAL1 seems unlikely. Thus pre-BCR aggregation by GAL1 should necessitate the participation of counter-receptors. The binding affinity between SLC and GAL1 allows us to predict a high avidity of the divalent pre-BCR to counter-receptor-anchored GAL1 in this context. As preB cell decoration by GAL1 does not induce pre-BCR clustering and polarization in the absence of stromal cells (Fig. 3C), GAL1 anchoring to stromal transcounter-receptors is essential to drive the pre-BCR relocalization process. In addition, we showed that pre-BCR translocation to the preB/stromal cells synapse initiates intracellular tyrosine kinase activity and signal transduction from the pre-BCR (Fig. 4), indicating that such a phenomenon is associated with important biological functions. From these observations, we present a structural and functional model for the preB/stromal cell synapse in which pre-BCR/GAL1 crosslinked complexes could contain glycosylated counter-receptors from both preB and stromal cells and could form a signaling-competent platform (Fig. 5B).
We hypothesize that pre-BCR/GAL1 clustering and triggering could have physiological consequences for the progression of precursor cells through the B cell-differentiation pathway. According to the lack of preBII population in λ5 or VpreB-mutant mice (9, 10), preB/stromal cells synapse formation could be essential for the cycling initiation of large preBII cells and for assuring the transition between large and small preBII cells. Although GAL1 is implicated in many different biological aspects (30, 31), inactivation of the GAL1 gene results in a mild phenotype. These mutant mice exhibit normal viability, fertility (37), and normal bone marrow B cell development (data not shown), thus pointing to a probable functional complementation by other galectin family members during development.
The existence of a stromal pre-BCR ligand has been the object of controversy for a long time. Transgenes encoding truncated Igμ chains unable to bind to the SLC allow complete B cell differentiation (38, 39). However, in these cases, self-aggregation of the truncated receptors could have mimicked pre-BCR triggering. Various anti-SLC mAbs do not interfere on early B cell development in a fetal liver organ culture system (40), but as these mAbs are unable to block pre-BCR binding to murine ST2 stromal cells (14), they could not interfere with SLC–GAL1 interactions. Finally, precursor B cells have been shown to differentiate into immature B cells in vitro in the absence of stromal cells (41). However, cells do not proliferate more than once or twice, pointing to the probable implication of pre-BCR/GAL1 lattice formation to preB cell proliferation commitment. In contrast, a report using the BLIN2 preB cell line has emphasized the cooperative signaling between pre-BCR and the BM microenvironment for B cell survival and proliferation (2). In this case, the fact that pre-BCR crosslinking without stromal cells is not sufficient to induce proliferation suggests a potential role of GAL1 counter-receptors to obtain an efficient proliferative signal.
Supplementary Material
Acknowledgments
We thank M. Fallet for excellent assistance with confocal microscopy. We are indebted to Dr. Joubert-Caron for providing us with anti-GAL1 antiserum and recombinant GAL1 protein and to Dr. D. Baty for the Igκ baculovirus. We gratefully acknowledge Dr. J. Garin for advice in mass spectrometry analysis. We thank Dr. R. Guinamard and Dr. A. M. Lellouch for critical reading of the manuscript. This work was supported by Centre National de la Recherche Scientifique and Institut National de la Santé et de la Recherche Médicale (INSERM) and by a training grant from the INSERM Provence-Alpes-Côte d'Azur region (to B.R.).
Abbreviations
- BM
bone marrow
- SLC
surrogate light chain
- GAL1
galectin-1
- pre-BCR
pre-B cell receptor
- 3D
tridimensional
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ghia P, ten Boekel E, Sanz E, de la Hera A, Rolink A, Melchers F. J Exp Med. 1996;184:2217–2229. doi: 10.1084/jem.184.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertrand F E, Eckfeldt C E, Fink J R, Lysholm A S, Pribyl J A, Shah N, LeBien T W. Immunol Rev. 2000;175:175–186. [PubMed] [Google Scholar]
- 3.Kline G H, Hartwell L, Beck-Engeser G B, Keyna U, Zaharevitz S, Klinman N R, Jack H M. J Immunol. 1998;161:1608–1618. [PubMed] [Google Scholar]
- 4.ten Boekel E, Melchers F, Rolink A G. Immunity. 1998;8:199–207. doi: 10.1016/s1074-7613(00)80472-0. [DOI] [PubMed] [Google Scholar]
- 5.Karasuyama H, Kudo A, Melchers F. J Exp Med. 1990;172:969–972. doi: 10.1084/jem.172.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bossy D, Milili M, Zucman J, Thomas G, Fougereau M, Schiff C. Int Immunol. 1991;3:1081–1090. doi: 10.1093/intimm/3.11.1081. [DOI] [PubMed] [Google Scholar]
- 7.Kitamura D, Roes J, Kuhn R, Rajewsky K. Nature (London) 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 8.Kitamura D, Rajewsky K. Nature (London) 1992;356:154–156. doi: 10.1038/356154a0. [DOI] [PubMed] [Google Scholar]
- 9.Mundt C, Licence S, Shimizu T, Melchers F, Martensson I L. J Exp Med. 2001;193:435–446. doi: 10.1084/jem.193.4.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rolink A, Karasuyama H, Grawunder U, Haasner D, Kudo A, Melchers F. Eur J Immunol. 1993;23:1284–1288. doi: 10.1002/eji.1830230614. [DOI] [PubMed] [Google Scholar]
- 11.ten Boekel E, Melchers F, Rolink A G. Immunity. 1997;7:357–368. doi: 10.1016/s1074-7613(00)80357-x. [DOI] [PubMed] [Google Scholar]
- 12.Papavasiliou F, Jankovic M, Suh H, Nussenzweig M C. J Exp Med. 1995;182:1389–1394. doi: 10.1084/jem.182.5.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo B, Kato R M, Garcia-Lloret M, Wahl M I, Rawlings D J. Immunity. 2000;13:243–253. doi: 10.1016/s1074-7613(00)00024-8. [DOI] [PubMed] [Google Scholar]
- 14.Bradl H, Jack H M. J Immunol. 2001;167:6403–6411. doi: 10.4049/jimmunol.167.11.6403. [DOI] [PubMed] [Google Scholar]
- 15.Gauthier L, Lemmers B, Guelpa-Fonlupt V, Fougereau M, Schiff C. J Immunol. 1999;162:41–50. [PubMed] [Google Scholar]
- 16.Gauthier L, Fougereau M, Tonnelle C. Exp Hematol. 1998;26:534–540. [PubMed] [Google Scholar]
- 17.Lemmers B, Gauthier L, Guelpa-Fonlupt V, Fougereau M, Schiff C. Blood. 1999;93:4336–4346. [PubMed] [Google Scholar]
- 18.Fouillit M, Levi-Strauss M, Giudicelli V, Lutomski D, Bladier D, Caron M, Joubert-Caron R. J Chromatogr. 1998;706:B167–B171. doi: 10.1016/s0378-4347(97)00336-8. [DOI] [PubMed] [Google Scholar]
- 19.Bossy D, Salamero J, Olive D, Fougereau M, Schiff C. Int Immunol. 1993;5:467–478. doi: 10.1093/intimm/5.5.467. [DOI] [PubMed] [Google Scholar]
- 20.Guelpa-Fonlupt V, Bossy D, Alzari P, Fumoux F, Fougereau M, Schiff C. Mol Immunol. 1994;31:1099–1108. doi: 10.1016/0161-5890(94)90105-8. [DOI] [PubMed] [Google Scholar]
- 21.Allen R D, David G B, Nomarski G. Z Wiss Mikrosk Tech. 1969;69:193–221. [PubMed] [Google Scholar]
- 22.Muller S, Demotz S, Bulliard C, Valitutti S. Immunology. 1999;97:287–293. doi: 10.1046/j.1365-2567.1999.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barondes S H, Castronovo V, Cooper D N, Cummings R D, Drickamer K, Feizi T, Gitt M A, Hirabayashi J, Hughes C, Kasai K, et al. Cell. 1994;76:597–598. doi: 10.1016/0092-8674(94)90498-7. [DOI] [PubMed] [Google Scholar]
- 24.Perillo N L, Marcus M E, Baum L G. J Mol Med. 1998;76:402–412. doi: 10.1007/s001090050232. [DOI] [PubMed] [Google Scholar]
- 25.Hughes R C. Biochem Soc Trans. 1997;25:1194–1198. doi: 10.1042/bst0251194. [DOI] [PubMed] [Google Scholar]
- 26.Dunphy J L, Barcham G J, Bischof R J, Young A R, Nash A, Meeusen E N. J Biol Chem. 2002;277:14916–14924. doi: 10.1074/jbc.M200214200. [DOI] [PubMed] [Google Scholar]
- 27.Bourne Y, Bolgiano B, Liao D I, Strecker G, Cantau P, Herzberg O, Feizi T, Cambillau C. Nat Struct Biol. 1994;1:863–870. doi: 10.1038/nsb1294-863. [DOI] [PubMed] [Google Scholar]
- 28.Chiariotti L, Salvatore P, Benvenuto G, Bruni C B. Biochimie. 1999;81:381–388. doi: 10.1016/s0300-9084(99)80085-6. [DOI] [PubMed] [Google Scholar]
- 29.Rabinovich G A, Baum L G, Tinari N, Paganelli R, Natoli C, Liu F T, Iacobelli S. Trends Immunol. 2002;23:313–320. doi: 10.1016/s1471-4906(02)02232-9. [DOI] [PubMed] [Google Scholar]
- 30.Perillo N L, Uittenbogaart C H, Nguyen J T, Baum L G. J Exp Med. 1997;185:1851–1858. doi: 10.1084/jem.185.10.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes R C. Biochimie. 2001;83:667–676. doi: 10.1016/s0300-9084(01)01289-5. [DOI] [PubMed] [Google Scholar]
- 32.Ramkumar R, Podder S K. J Mol Recognit. 2000;13:299–309. doi: 10.1002/1099-1352(200009/10)13:5<299::AID-JMR504>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 33.Yang R Y, Hsu D K, Liu F T. Proc Natl Acad Sci USA. 1996;93:6737–6742. doi: 10.1073/pnas.93.13.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pace K E, Lee C, Stewart P L, Baum L G. J Immunol. 1999;163:3801–3811. [PubMed] [Google Scholar]
- 35.Fouillit M, Joubert-Caron R, Poirier F, Bourin P, Monostori E, Levi-Strauss M, Raphael M, Bladier D, Caron M. Glycobiology. 2000;10:413–419. doi: 10.1093/glycob/10.4.413. [DOI] [PubMed] [Google Scholar]
- 36.Salamero J, Fougereau M, Seckinger P. Eur J Immunol. 1995;25:2757–2764. doi: 10.1002/eji.1830251007. [DOI] [PubMed] [Google Scholar]
- 37.Poirier F, Robertson E J. Development (Cambridge, UK) 1993;119:1229–1236. doi: 10.1242/dev.119.4.1229. [DOI] [PubMed] [Google Scholar]
- 38.Corcos D, Dunda O, Butor C, Cesbron J Y, Lores P, Bucchini D, Jami J. Curr Biol. 1995;5:1140–1148. doi: 10.1016/s0960-9822(95)00230-2. [DOI] [PubMed] [Google Scholar]
- 39.Shaffer A L, Schlissel M S. J Immunol. 1997;159:1265–1275. [PubMed] [Google Scholar]
- 40.Ceredig R, Rolink A G, Melchers F, Andersson J. Eur J Immunol. 2000;30:759–767. doi: 10.1002/1521-4141(200003)30:3<759::AID-IMMU759>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 41.Rolink A G, Winkler T, Melchers F, Andersson J. J Exp Med. 2000;191:23–32. doi: 10.1084/jem.191.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.