Abstract
Using a genome-wide approach, we asked how many transporter genes contribute to symbiotic phosphate uptake and analyzed their evolutionary conservation. Considering the sequenced rice genome at hand, only the Oryza sativa phosphate transporter (OsPT) gene OsPT11 was specifically induced during the arbuscular mycorrhizal symbiosis. This induction was confined to the root system and was tightly correlated with the degree of root colonization by Glomus intraradices. OsPT11 activation was independent of the nutritional status of the plant and phosphate availability in the rhizosphere. Moreover, infection of roots with the fungal pathogens Rhizoctonia solani and Fusarium moniliforme did not activate OsPT11, corroborating the high signal specificity for OsPT11 activation in the arbuscular mycorrhizal symbiosis. OsPT11 expression complemented a defect in phosphate uptake in a yeast strain mutated in its high-affinity Pi transporter (pho84), thereby confirming its function. Recently, a phosphate transporter gene in potato was shown to be induced during arbuscular mycorrhizal symbiosis. Assessment of the phylogenetic relationship of the rice and potato protein revealed that the rice is nonorthologous to the potato protein. Further, there are no structural commonalities in the promoter regions. Thus, although cytological and physiological features of the arbuscular mycorrhizal symbiosis seem to be conserved, the molecular components may differ significantly between distantly related plant species.
Arbuscular mycorrhizal (AM) fungi associate intimately with the roots of more than 80% of terrestrial plants, growing inter- and intracellularly in the root cortex. It is well documented that AM fungi enhance nutrient availability to plants, in particular, inorganic (ortho)phosphate (Pi), by forming far-reaching extraradical mycelia which operate as functional extensions of the plant root system (1, 2). In the absence of the symbiosis, Pi is taken up directly by plant roots in the form of orthophosphate; however, its concentration rarely exceeds 10 μM in the soil fluid (3). Plants have acquired a number of different strategies to maximize Pi uptake under such Pi-limiting conditions. Similar to yeast, plants explore high- and low-affinity Pi-transporter systems. Although low-affinity Pi transporters are constitutively expressed and operate at Pi concentrations in the millimolar range, genes for high-affinity Pi transport are transcriptionally induced at low Pi availability and contribute to Pi uptake at limiting, micro-molar concentrations (3–5). An additional set of Pi transporters participates in the translocation of Pi throughout the plant. Furthermore, symbiosis-mediated Pi uptake probably involves a plant-encoded acquisition activity (6, 7). Two dicotyledonous high-affinity Pi transporter genes have been characterized for their possible involvement in the AM symbiosis. Messenger of the tomato LePT1 gene was moderately expressed in arbusculated cortex cells (7) but was also present in epidermis, root cap, root hairs, the vascular system, and in cells of the palisade parenchyma of Pi-deprived plants (8). Thus, it seems to participate rather generally in Pi uptake and translocation. In contrast, StPT3 mRNA was found to be absent from nonmycorrhizal roots but strongly induced during AM symbiosis (6). No data are available for Pi transporters from monocotyledonous plants.
Little is known about how many Pi transporters exist in plant genomes or how many of these play a role in symbiosis-mediated Pi uptake. The present availability of fully sequenced genomes allows identification of the Pi transporters determining the entire network of Pi-relocation activities. The genome of Arabidopsis thaliana contains nine putative high-affinity transporters (www.cbs.umn.edu/Arabidopsis). However, because Arabidopsis belongs to the small minority of plants that do not associate with AM fungi, only symbiosis-independent Pi uptake can be considered. In contrast, the genome of rice has recently been sequenced (9, 10), thus providing a unique opportunity to reveal the complete picture of Pi acquisition operating in the absence or presence of symbiotic uptake. We have identified the entire set of rice high-affinity Pi transporter genes in Oryza sativa cv. Nipponbare (9) and found that only one gene responded to mycorrhizal colonization with a strong and remarkably specific induction of expression. Interestingly, in contrast to the StPT3 protein, which clustered well with other solanaceous Pi transporters, the OsPT11 protein appeared divergent from StPT3 and from the other known plant Pi transporters. Moreover, the absence of commonalities within the promoter regions of StPT3 and OsPT11 suggests divergent evolution of their symbiotic functions. Thus, although the symbiosis is an ancient process assumed to have accompanied the origin of early land plants (1, 11, 12), its molecular components have undergone significant diversification during the subsequent coevolution of AM fungi and plant species.
Materials and Methods
Plant Growth Conditions.
Rice plants were grown in sterilized soil in phytochambers with a 12-h day/12-h night cycle at 28°C. Tissue samples were harvested at various developmental stages (Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org). For studies of the AM symbiosis, seeds were surface-sterilized and planted into sterilized sand; plants were watered every second day with half-strength Hoagland solution supplemented with KH2PO4 concentrations of 5–500 μM.
Determination of Phosphorus Content in Plant Tissue.
Ion content of air-dried root and leaf tissue was determined after tissue digestion at 65°C for 16 h with 2 ml concentrated HNO3 with a Varian Vista Simultaneous Axial ICP-AES (inductively coupled plasma atomic emission spectrometer, Varian). The obtained values were normalized to magnesium (Mg) as an internal standard.
Inoculation of Plants with Glomus intraradices.
G. intraradices was grown in aseptic conditions according to (13). For plant inoculation, equal volumes of medium either containing 1,000 spores or without fungal spores (mock) were added to the sand at 1.5-cm depth upon planting.
Rice Inoculation with Rhizoctonia solani and Fusarium moniliforme.
Sterilized rice seeds were pregerminated on potato dextrose agar (Difco) for 3 days and subsequently aseptically grown in M medium (pH 5.5 with KOH; ref. 13) without sucrose but supplemented with glucose (1 g/liter) at 24°C (12-h day/12-h night). Roots of 10-day-old rice seedlings were inoculated with pieces of solidified medium containing mycelium of R. solani AG 1-IA or F. moniliforme. The plants were inspected regularly for symptom development and intraradical fungal growth until the plants collapsed. At 8 days post inoculation (dpi), symptoms appeared on roots, stems, and leaves, and fungal hyphae had penetrated into the roots and grew within the cortex of the roots as well as on the root surface. After prolonged incubation, extensive fungal mycelium developed on the surface of roots and leaves of inoculated plants, and 15 to 18 dpi the plants collapsed. Because the plants were heavily infected but the root tissue was little disintegrated at 8 dpi, we collected tissue from this time point for RNA extraction.
Microscopic Examination of Mycorrhizae and Root Pathogens.
The degree of mycorrhizal colonization was determined as described in (14) by using the gridline-intersect procedure according to (15). To estimate the degree of root infection by R. solani and F. moniliforme, root tissue was fixed in 3% (vol/vol) glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) for 2 h at room temperature and washed in the same buffer. Root pieces were dehydrated in a series of ethanol washes and embedded in LR White resin (16). Sections of ≈1 μm were cut and stained with toluidine blue.
Computational Identification and Characterization of Rice Pi Transporters.
The amino acid sequence of the 13 rice Pi transporter proteins were determined as described in ref. 9. The programs CLUSTALW (17) and T-COFFEE (18) were used to generate multiple amino acid sequence alignments. The two alignments were trimmed at the N- and C-terminal ends to include only the well aligned block of amino acids (Fig. 8, which is published as supporting information on the PNAS web site). These trimmed alignments were compared for phylogenetic signals with the phylogenetic analysis package PAUP 4.0b8 (19) by using maximum parsimony (with random addition of sequences) and bootstrap analysis (2,000 pseudoreplicates). The T-COFFEE alignment resulted in a longer, most parsimonious tree but one with a higher consistency index and stronger bootstrap support. Therefore, this alignment was used to estimate the phylogenetic relationship of the Pi transporter proteins. DNA sequences were analyzed for possible motifs by internally developed TMRI programs and were also examined for redundancies by comparing to a TMRI internal database of rice repeats.
RNA Isolation and Analysis.
Total RNA was isolated from 1 g of fresh weight of plant tissue using the RNeasy Maxi kit (Qiagen, Chatsworth, CA). DNaseI-treated RNA (10 ng) was used with TaqMan One-Step RT-PCR reagents and quantitatively analyzed in an ABI 7700 (Applied Biosystems). Reactions were carried out as described by the manufacturers. Primers and 6-FAM 5′ end-labeled probes (Applied Biosystems and Genosys, The Woodlands, TX, respectively) were designed for the 3′UTR of all Pi transporter genes and the rice polyubiquitin 1 gene (RubQ1) mRNA as a control (20), employing the PRIMER EXPRESS software (Applied Biosystems; Table 1, which is published as supporting information on the PNAS web site). Expression levels were calculated by using standard curves with a correlation coefficient of ≥0.99. The amount of transcript for each gene was normalized after determining the constitutive level of the RubQ1 mRNA. We examined RubQ1 gene expression in a set of three independent experiments under a broad range of environmental stimuli and various developmental stages by using the Affymetrix rice GeneChip. The results of these experiments suggested constitutive RubQ1 expression (data not shown) and, therefore, suitability to serve as a standard for normalization.
Isolation of the OsPT11 Gene and Its cDNA.
Genomic DNA and total RNA were extracted by using the Qiagen DNeasy and RNeasy kits, respectively (Qiagen). The Superscript II kit (Invitrogen) was used for the reverse transcription reaction. PCR amplification of the OsPT11 cDNA and genomic DNA was carried out by using the reverse primer and sequence 5′-ATGGGCTTCTTCACCGTCGCCTAC-3′ located at the translation start of the gene. PCR amplification was performed with high-fidelity Taq polymerase (Invitrogen). The cDNA of OsPT11 then was inserted into the yeast expression vector p426Adh (ATCC).
Yeast Manipulations.
The deletion mutant pho84 lacking a functional high-affinity Pi transporter gene was obtained from the stock center (Saccharomyces Genome Deletion Project, Stanford; www-sequence.stanford.edu/group/yeast_deletion_project/). The yeast expression vector p426Adh or the vector containing the OsPT11 gene was transformed into the pho84 mutant after the protocol of ref. 21. Cells of both yeast strains were prepared for 32P uptake studies according to ref. 8. The radioactivity taken up by cells was measured by scintillation counting and normalized for the total amount of protein (22). The kinetics values were examined by SIGMAPLOT (Jandel, San Rafael, CA).
Results
Identification of Rice Genes Homologous to High-Affinity Pi Transporters.
A total of 13 sequences were identified as being related to high-affinity Pi transporters (9). The identified genes were named according to the Commission on Plant Gene Nomenclature (http://mbclserver.rutgers.edu/CPGN/Guide.html) as ORYsa;Pht1;1 through ORYsa;Pht1;13; for simplification, they will be called OsPT1 through OsPT13 in this article. The rice Pi transporters shared 44.5–74% sequence identity (SI) with AtPT1 and 38.1–87.4% with each other; the highest degree of SI was found between OsPT4 and OsPT5 (87.4%; Fig. 9A, which is published as supporting information on the PNAS web site). The predicted rice proteins are structurally homologous to the Pi transporter proteins from dicotyledonous plants, containing the conserved sequence motifs as phosphorylation sites for protein kinase C and casein kinase II and a site for potential N-glycosylation (Fig. 9B; ref. 23).
Interaction of Rice with G. intraradices.
To our knowledge, the interaction of rice with aseptically produced inoculum of AM fungi has never been cytologically or functionally documented. Therefore, we established a controlled and efficient procedure for the colonization of rice by G. intraradices. Eight weeks after inoculation, the fungus had reproducibly formed all typical symbiotic structures such as intercellular hyphae, arbuscules, and vesicles (data not shown), and colonization reached levels of 85% (±9), as revealed by quantitative microscopic inspection (14, 15). To verify whether or not the AM symbiosis had led to enhanced Pi uptake, the total phosphorus (P) contents of roots and shoots of mycorrhizal and control plants were compared. When grown under Pi-limiting conditions, roots of mycorrhizal plants acquired 2.31 (±0.5) ppm/g dry weight (dw) P, whereas roots of control plants had only 1.23 (±0.4) ppm/g dw P (p < 0.022). A similar result was obtained for leaf tissue; leaves of mycorrhizal plants contained 2.89 (±0.59) ppm/g dw P, and controls contained 1.84 (±0.28) ppm/g dw P (p < 0.023). These results illustrate the full functionality of the experimentally established rice symbiosis system.
Expression of Pi Transporters in Mycorrhizal Rice Roots.
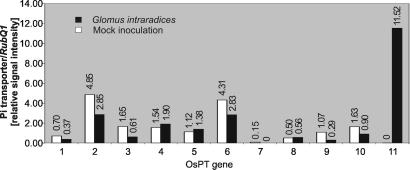
To determine which of the Pi transporters are transcriptionally induced upon mycorrhizal colonization, gene-specific primers and probes were designed, and quantitative reverse transcription-PCR (RT-PCR) and TaqMan chemistry (24) were used to monitor transcript levels for every Pi transporter gene. To ensure specificity, the primer and probe sequences were aligned with the rice genome. To obtain representative results and at the same time reduce the size and the costs of the expression analysis, three independent experiments were carried out, each on 30 plants for the respective treatment. After the isolation of total RNA from corresponding tissue samples, the RNA was pooled for examining the expression of the Pi transporter genes. Expression analysis revealed that 10 of the 13 Pi transporter genes are expressed in roots; only transcripts of OsPT12 and OsPT13 were not detected in roots, and the mRNA of OsPT7 was barely detectable (Fig. 1). This result could be due to inadequate primers and probes for transcript detection, or it could reflect either pseudogenes or genes that are expressed in other tissues but which are silent in roots under the applied conditions. Because the rhizosphere was the focus of this study, we concentrated on genes transcriptionally active in roots. Messenger RNA of OsPT1, OsPT2, OsPT3, OsPT6, OsPT9, and OsPT10 was present in both mock- and G. intraradices-inoculated roots but displayed somewhat lower transcript levels upon mycorrhizal colonization (Fig. 1). Expression levels of OsPT4, OsPT5, and OsPT8 appeared similar in the presence or absence of G. intraradices (Fig. 1). To confirm the observed expression pattern on a smaller sample number, we selected the two genes, OsPT2 and OsPT6, and examined their expression in three additional equivalent experiments. Messenger RNA of both genes was strongly reduced in response to colonization by G. intraradices (data not shown). Remarkably, however, OsPT11 was observed to be transcriptionally silent in mock-inoculated roots but was strongly induced upon mycorrhizal colonization (Fig. 1). This finding was observed repeatedly in four independent experiments.
Figure 1.
Levels of mRNA of rice high-affinity Pi transporters in roots of mock- and G. intraradices-inoculated plants determined by TaqMan real-time PCR. The bars show the expression level of each individual Pi transporter normalized to the expression level of RubQ1.
OsPT2 and OsPT11 Expression at Different Pi Availability.
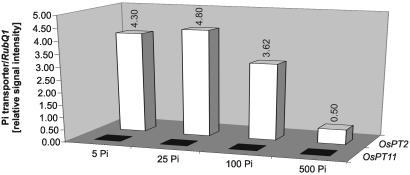
Although the drastic accumulation of OsPT11 transcripts upon mycorrhizal colonization could be symbiosis specific, it also may be a secondary effect of the enhanced Pi supply, in this case, provided by the fungus. To distinguish between OsPT11 regulation by nutritional or fungus-specific signals, we examined OsPT11 mRNA levels in plants grown at various levels of Pi. To define plant perception of Pi-limiting vs. Pi-sufficient conditions, we explored a high-affinity Pi transporter gene as a molecular marker of Pi starvation. The regulation of OsPT2 resembled that of the two Medicago truncatula Pi transporter genes MtPT1 and MtPT2, which have been shown to be transcriptionally induced upon Pi starvation and down-regulated during AM symbiosis (25). In fact, transcript levels of OsPT2 were high at 5, 25, and 100 μM Pi but were strongly reduced at 500 μM Pi (Fig. 2), which allowed the monitoring of restricted Pi availability by the induction of OsPT2 transcription. Importantly, transcript levels of OsPT11 were unchanged, remaining below the limit of detection at all Pi concentrations (Fig. 2). Thus, activation of this gene is not mediated by Pi availability or the nutritional P status of the plant.
Figure 2.
Expression levels of OsPT2 and OsPT11 in roots of plants grown at different levels of Pi. The bars show the expression levels of each individual Pi transporter gene relative to the expression levels of RubQ1. The same pattern of expression was observed in two additional experiments.
Developmental and Organ-Specific Regulation of OsPT2 and OsPT11 Expression.
To determine the tissue and developmental specificity of OsPT2 and OsPT11 expression, mRNA was examined at various stages in the maturation of roots, leafs, stems, panicles, and seeds as well as of embryos and endosperm tissues. To decrease the size of the experiment, tissue was collected from three independent experiments (in each case from at least 15 plants) for isolation of total RNA. OsPT2 was low in all tissues except roots, where high levels of transcripts were observed at different developmental stages, the highest being in roots of mature plants (Fig. 7). This elevated expression in mature roots could reflect specific developmental regulation, or it may be that the Pi depletion zone around older roots results in Pi starvation and, hence, high activation of this gene. Expression patterns of OsPT2 were corroborated by using the Affymetrix rice GeneChip (data not shown). Importantly, expression of OsPT11 was not detected in plants grown in the absence of mycorrhizal colonization in any tissues or at any developmental stage (Fig. 7).
Temporal Expression of OsPT11.
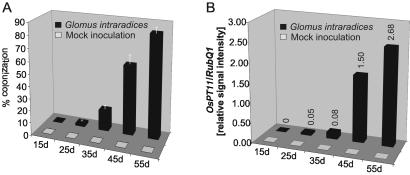
To study correlation of mycorrhizal colonization with the specific accumulation of OsPT11 mRNA further, we examined the kinetics of mycorrhizal colonization and OsPT11 expression at intervals of 15, 25, 35, 45, and 55 dpi. Microscopic determination of the degree of colonization revealed rare intraradical structures (2 ± 1.4%) at 25 dpi but more clearly at 35 dpi (25 ± 8.5%). A rapid increase in colonization to 56 ± 7.1% occurred between 35 and 45 dpi, reaching 83 (±5.7%) at 55 dpi (Fig. 3A). OsPT11 mRNA was not detectable at early time points (15 dpi) or in mock-inoculated roots. A low signal was detected first at 25 dpi, and a slight signal increase was seen at 35 dpi, followed by a rapid induction between 35 and 45 dpi, and a further enhancement between 45 and 55 dpi (Fig. 3B). We observed considerable variations in OsPT11 transcript level by comparing individual time points of different experiments but found a consistent increase in expression level accompanying the progress of mycorrhizal colonization. Thus, the kinetics of OsPT11 gene expression mimicked the kinetics of fungal spread within the mycorrhizal roots.
Figure 3.
Progressive colonization of rice roots by G. intraradices and levels of OsPT11 mRNA at the corresponding stages of the symbiosis. (A) Abundance of intraradical fungal structures displayed as the percentage of the root system colonized. No uncontrolled infection was observed in mock-inoculated plants. (B) The bars show the expression levels of OsPT11 relative to the expression levels of RubQ1.
Absence of OsPT11 Expression in Roots Infected with Pathogenic Fungi.
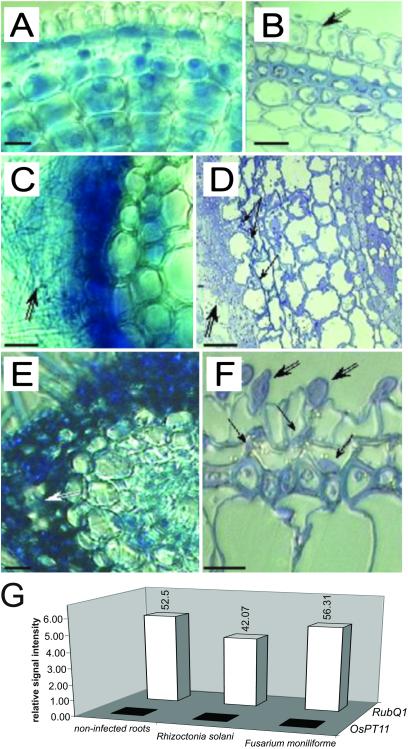
To examine in more detail the specificity of the OsPT11 expression in response to colonization by G. intraradices, we challenged the plants with pathogenic fungi on the assumption that fungal elicitors can provide signals for OsPT11 activation. For this purpose, rice roots were inoculated with R. solani and F. moniliforme. To our knowledge, there are no specific marker genes described for rice roots infected by R. solani and F. moniliforme. Instead, we have used microscopic inspection to determine when efficient penetration had occurred before collapsing of the plants. Microscopic inspection of root cross sections at 8 dpi confirmed that both R. solani and F. moniliforme had penetrated into the cortex tissue and had spread inside and around the roots (Fig. 4 C–F). Although pathogen-infected roots showed high amounts of constitutive RubQ1 mRNA, there were no detectable levels of OsPT11 message (Fig. 4G). Thus, fungal infection per se was not able to trigger transcriptional activation of the gene, which suggests that not fungus- but mycorrhiza-specific signals are responsible for OsPT11 activation.
Figure 4.
Rice root infection by Fusarium moniliforme or Rhizoctonia solani and its influence on OsPT11 expression. (A–F) Hand cuttings (A, C, and E) and semifine sections (B, D, and F) of control roots (A and B) or roots infected by F. moniliforme (C and D) or R. solani (E and F) at 8 dpi. Control rhizoderm cells appear joined (B, double black arrow). A dense mycelium of F. moniliforme has formed around the root (C and D, double black arrow) and the fungus has penetrated intercellularly (D, black arrow) disorganizing the rhizodermal root layer. A dense mycelium of R. solani has formed around the root (E, double white arrow), disorganizing the rhizodermal root cells. (F) External hyphae of R. solani have attached to the rhizoderm (double black arrows) and have invaginated the root cells (black arrows). (Bars = 10 μm.) (G) Expression of OsPT11. The expression levels of RubQ1 itself and of OsPT11 relative to the expression levels of RubQ1 are shown. The same pattern of expression was observed in an additional experiment.
Cloning of the OsPT11 cDNA and Genomic DNA.
OsPT11 cDNA was obtained after reverse transcription of total RNA isolated from mycorrhizal rice roots, followed by PCR amplification using primers predicted from the gene sequence. The same primers were used for amplification of the gene region. The cDNA was 1,668 bp and the genomic clone contained an intron of 92 bp. The predicted protein of 555 amino acids had 11 or 13 potential transmembrane domains, depending on topology prediction programs [TMHMM: 11 (26), HMMTOP: 13 (27), DAS: 11 (28)]. However, according to TMPRED (www.ch.embnet.org/software/TMPRED_form.html), the preferred topology for the OsPT11 protein consists of 12 membrane-spanning domains with intracellular N and C termini and an intracellular central loop.
Phylogenetic Relationship of OsPT11 and Other Rice and Dicotyledonous Pi Transporters.
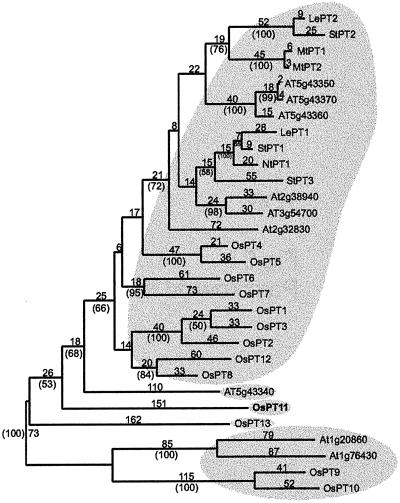
To estimate the phylogenetic relationship between OsPT11 and other rice and dicotyledonous high-affinity Pi transporter proteins, we used maximum parsimony to generate an unrooted phylogram and performed a bootstrap analysis on the data set to determine the robustness of the phylogram's topology (Fig. 5). Two main groups of Pi transporters are inferred, as they are supported by bootstrap values ≥90%. Genes from mono- and dicotylenous plants are found in both main groups, indicating their evolutionary divergence before the common ancestor of monocots and dicots.
Figure 5.
Maximum parsimony tree of rice and other plant high-affinity Pi transporter proteins. The phylogenetic analysis was performed on the T-COFFEE amino acid alignment with maximum parsimony and bootstrapping. The numbers above the lines refer to the number of sequence changes, the numbers in parentheses refer to bootstrap values.
The first group contains the majority of proteins from the mono- and the dicotyledonous plant species, in which they are subgrouped by phylogeny. For example, one group of solanaceous Pi transporters, including the mycorrhizae-induced StPT3 gene, forms a cluster together with the two brassicaceous Arabidopsis proteins At2g38940 and AT3g54700. These proteins are part of a larger group that includes solanaceous, leguminous, and brassicaceous transporters but not a single rice protein. For rice, 9 of the 13 Pi transporters form one larger group with other Pi transporters. The second group of Pi transporters is distantly related to the first group and includes a pair of proteins from rice (OsPT9 and OsPT10) and a pair from Arabidopsis (Atg20860 and At1g76430). Three orphans fall outside of the two main groups: OsPT11, OsPT13, and AT5g43340. These proteins are only distantly related to other rice and dicotyledonous plant Pi transporter proteins, which is particularly surprising for OsPT11. This protein has no close homologue among the known Pi transporters of dicotyledonous species including StPT3, the Pi transporter induced by mycorrhizal symbiosis in potato (Fig. 5). Therefore, rice and potato seem to have symbiotically induced Pi transporters that are functionally homologous but phylogenetically divergent. Furthermore, OsPT11 is distant to known fungal Pi transporters and displays levels of SI lower than 36% (data not shown).
Analysis of the OsPT11 Promoter Sequence.
Previously (6), Miniature Inverted repeat Transposable Element (MITE)-like and mycorrhiza and resistant-related (MRR) elements were identified within the promoter region of the StPT3 gene, and their possible regulatory functions were discussed. The promoter region (2,149 bp 5′ proximal to the ATG) of the OsPT11 gene was examined for the presence of inverted repeats of 10-bp length or longer, indicative of MITE- or other transposon-like elements, and was also searched for the occurrence of the conserved MRR element by BLASTN (29). The OsPT11 promoter neither contained MITE- nor any MRR-related sequences (data not shown).
Functional Analysis in pho84.
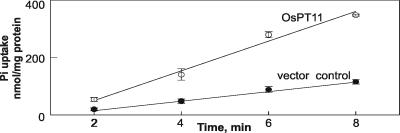
The function of the OsPT11 protein as a Pi transporter was assayed by 32P-uptake experiments by using the yeast deletion-mutant pho84 (www-sequence.stanford.edu/group/yeast_deletion_project/), which is affected in Pi uptake at reduced Pi supply (30). OsPT11 cDNA regulated by the constitutive promoter of alcohol dehydrogenase was introduced into pho84. Despite the high background of cells carrying the empty vector control, the mutant cells expressing the OsPT11 protein displayed a 3.25-fold higher Pi acquisition than control cells (Fig. 6). This increased Pi uptake showed that the OsPT11 gene encodes a functional Pi transporter.
Figure 6.
Kinetics of Pi uptake. Phosphate uptake by yeast pho84 cells transformed with either the empty vector or vector with the OsPT11 construct. Cells, buffered to pH 5.0 with KOH, were supplied with 0.2 mM Pi (18.5 MBq [32P]/μmol) at t = 0 min. Values represent mean and SD of five replicates.
Discussion
Here, we describe a rice gene set encoding proteins that are homologous to high-affinity Pi transporters. We have examined the expression of each gene in relation to the AM symbiosis. Although the mRNA levels of OsPT1, OsPT2, OsPT3, OsPT6, OsPT9, and OsPT10 expressed in rice roots (hereafter called REPT for root-expressed phosphate transporters) declined during symbiosis, the mRNA of OsPT11 accumulated synchronously with the progression of mycorrhizal colonization. This observation is interesting for several reasons. First, it suggests basic differences in the coordination of activation of this gene family, which is shown here to be regulated in response to mycorrhizal colonization. Second, although expression of the REPT genes was not suppressed to an undetectable level, the general tendency was an “either-or” balance between OsPT11 and the REPT genes: in the absence of mycorrhizae, OsPT11 is silent but mRNA levels of the REPT genes are high. This situation is reversed upon formation of the AM symbiosis: transcription of OsPT11 is enhanced strongly while the expression of the REPT genes is suppressed. A similar observation was made about a subset of Pi transporter genes from potato and M. truncatula. Potato StPT1 and StPT2 mRNA levels were reduced upon interaction with AM fungi and StPT3 expression was strongly induced, exhibiting mRNA in cells containing arbuscules (6). Also, transcript levels of the M. truncatula Pi transporter genes MtPT1 and MtPT2 were reduced in roots of mycorrhizal plants compared with nonmycorrhizal controls (25). Thus, the coordinated and concurrent suppression and induction of gene activation seems to be a common feature reflecting a switch in Pi supply from direct root uptake to symbiotic Pi delivery, which agrees with an earlier observation that fungal Pi uptake in mycorrhizal roots accounts for most of the total Pi acquisition (31). Thus, the expression of transporters involved in direct Pi uptake may be superfluous. The observation that direct supply to roots of Pi at high concentration also suppressed expression of OsPT2, one of the REPT genes, supports their rather direct regulation by Pi availability independent of the source of supply. However, results obtained for Mt4 (a Pi-starvation inducible and mycorrhizae-suppressed gene of unknown function from M. truncatula) suggested that a signal released by the fungus and not Pi availability accounted for the reduction in Mt4 mRNA levels. M. truncatula myc− mutant plants blocked at fungal penetration and not able to develop a functional symbiotic relationship, including fungal Pi delivery, still show repression of the Mt4 gene (32). Thus, the regulatory network of REPT gene expression may explore a combination of fungal and nutritional signals. In contrast, OsPT11 encodes a functional Pi transporter but is neither regulated in response to Pi availability nor is it influenced by the Pi demand of the plant; in the absence of mycorrhizae, it is simply silent. Thus OsPT11 seems to be linked exclusively to mycorrhizal colonization, which is consistent with AM fungus-controlled regulation.
Within the soil, Pi occurs at concentrations of <10 μM. The Km values of high-affinity plant Pi transporters, therefore, could have been predicted to be below 10 μM. Using heterologous yeast Pi uptake mutants, however, revealed Km values ranging from 31 μM (8) to 280 μM (33). In contrast, the expression of AtPT1 in tobacco tissue culture cells yielded a Km of 3.1 μM, thus matching the expected value of <10 μM (34) and indicating that these Pi transporters either require plant specific modifications or plant specific interactors (4, 23, 30, 34). Complementation of the high-affinity Pi-uptake deficiency of the yeast mutants by plant Pi transporter genes, though confirmed that they encode functional Pi transporter proteins (5, 6), which, as we show, also applies to the monocotyledonous OsPT11 protein.
The AM symbiosis dates back to at least Devonian times and coincides with the occurrence of early land plants (1, 11, 12). Within the flowering plants, AM fungi have shown no taxonomic specificity for host plants, and most members of both dicotyledonous and monocotyledonous species readily enter into this association. Independently of the type of host plant, the morphological and cytological consequences of the AM symbiosis are homologous, and enhanced Pi nutrition is a common characteristic among distantly related mycorrhizal plants. Therefore, factors involved in the core steps of AM symbiosis formation could have been conserved during evolution, beyond the division of angiosperms into mono- and dicotyledonous plants. It is, therefore, surprising that the mycorrhizae-induced Pi transporters from rice and potato (6) are phylogenetically so divergent. Furthermore, although StPT3 is closely related to other solanaceous Pi transporters, the OsPT11 protein is only distantly related to all other rice Pi transporters. It may still be the case that potato has another gene in addition to StPT3 that is orthologous to OsPT11. There is no OsPT11 ortholog among the full set of Arabidopsis Pi transporter genes; however, Arabidopsis (and all Brassicaceae) belongs to the minority of plant species that are nonhosts for AM fungi and, thus, may be an exception with respect to the absence of an OsPT11 ortholog.
Because OsPT11 and StPT3 both encode functional Pi transporters, it is tempting to speculate that various genes for symbiosis-mediated Pi uptake into plants have arisen during plant evolution. Because the basic requirement for Pi transporter function could have been satisfied by more than one of these proteins, additional adaptive steps, which probably occurred independently in different plant evolutionary lineages, have determined their functional specialization and symbiosis-specific regulation. This hypothesis is supported by the analysis of OsPT11 and StPT3 promoter regions, which have no obvious common structural characteristics. Rausch et al. (6) suggested that the regulatory impact of an MITE-like element and its internally enclosed and conserved sequence motifs (MRR1 and MRR2) could determine symbiosis-induced gene transcription, which may be true for potato, but the rice OsPT11 promoter has neither inverted repeat sequences reminiscent of transposable elements nor any of the MRR motifs. Therefore, the MITE/MRR elements as acquired signals directing Pi transporter regulation toward AM symbiosis may be not generally relevant. It is, however, intriguing that MRR motifs are present in the regulatory regions of genes encoding resistance determinants and are indicative of promoters responding to plant pathogen attack. Thus, the regulation of genes by promoter-containing MRR-like elements may or may not differentiate between pathogens and AM fungi. In infection studies with the rice pathogens R. solani and F. moniliforme described here, we found that pathogen invasion of roots did not result in OsPT11 gene induction. Thus, the observed gene activation was specific to mycorrhizal colonization and was not triggered by other fungal root infections.
This report opens up the characterization of Pi transporters from a monocotyledonous plant and provides a survey of the complete assortment of this group of proteins. Rice is the only member of the collection of plants used for studying AM symbiosis and Pi nutrition suitable for a genome-wide approach, and further exploitation of this system should substantially contribute to a better understanding of both these processes not only in rice but—because of the syntenic relationship between the grasses—in cereals in general.
Supplementary Material
Acknowledgments
We thank Bret Cooper and Bram Estes for assisting with the TaqMan real-time PCR; Natalie Catlett and Julie Ann Bick for help with yeast studies; Georg Felix for the Sigmaplots; Jeff Harper and coworkers for the P analyses; Sonia Guimil and Paul Budworth for providing RNA samples; RongLin Wang and Gernot Presting for computational promoter analysis; and Pat King and Jerzy Paszkowski for critically reading the manuscript.
Abbreviations
- AM
arbuscular mycorrhizal
- dpi
days post inoculation
- MRR
mycorrhiza and resistant-related element
- REPT
root-expressed phosphate transporters
Note Added in Proof.
A gene from M. truncatula (MtPT4) encoding a Pi transporter has been shown to be exclusively expressed in roots colonized by AM fungi. It was further determined that the MtPT4 protein is located on the peri-arbuscular membrane (35).
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF536961, OsPT1; AF536962, OsPT2; AF536963, OsPT3; AF536964, OsPT4; AF536965, OsPT5; AF536966, OsPT6; AF536967, OsPT7; AF536968, OsPT8; AF536969, OsPT9; AF536970, OsPT10; AF536971, OsPT11; AF5369672, OsPT12; and AF536973, OsPT13).
References
- 1.Smith S E, Read D J. Mycorrhizal Symbiosis. San Diego: Academic; 1997. [Google Scholar]
- 2.Smith S E, Dickson S, Smith F A. Aust J Plant Physiol. 2001;28:683–694. [Google Scholar]
- 3.Schachtman D P, Reid R J, Ayling S M. Plant Physiol. 1998;116:447–453. doi: 10.1104/pp.116.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chrispeels M J, Crawford N M, Schroeder J I. Plant Cell. 1999;11:661–676. doi: 10.1105/tpc.11.4.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raghothama K G. Curr Opin Plant Biol. 2000;3:182–187. [PubMed] [Google Scholar]
- 6.Rausch C, Daram P, Brunner S, Jansa J, Laloi M, Leggewie G, Amrhein N, Bucher M. Nature (London) 2001;414:462–470. doi: 10.1038/35106601. [DOI] [PubMed] [Google Scholar]
- 7.Rosewarne G M, Barker S J, Smith S E, Smith F A, Schachtman D P. New Phytol. 1999;144:507–516. doi: 10.1046/j.1469-8137.1999.00536.x. [DOI] [PubMed] [Google Scholar]
- 8.Daram P, Brunner S, Persson B L, Amrhein N, Bucher M. Planta. 1998;206:225–233. doi: 10.1007/s004250050394. [DOI] [PubMed] [Google Scholar]
- 9.Goff S A, Ricke D, Lan T H, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H. Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, Hu S, Wang J, Wong G K, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X. Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 11.Redeker D, Kodner R, Graham L. Science. 2000;289:1920–1921. doi: 10.1126/science.289.5486.1920. [DOI] [PubMed] [Google Scholar]
- 12.Remy W, Taylor T N, Hass H, Kerp H. Proc Natl Acad Sci USA. 1994;91:11841–11843. doi: 10.1073/pnas.91.25.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bécard G, Fortin J A. New Phytol. 1988;108:211–218. doi: 10.1111/j.1469-8137.1988.tb03698.x. [DOI] [PubMed] [Google Scholar]
- 14.Brundrett M C, Piché Y, Peterson R L. Can J Bot. 1984;62:2128–2134. [Google Scholar]
- 15.Giovanetti M, Mosse B. New Phytol. 1980;84:489–500. [Google Scholar]
- 16.Jauneau A, Quentin M, Driouich A. Protoplasma. 1997;198:9–19. [Google Scholar]
- 17.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Notredame C, Higgins D G, Heringa J. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 19.Swofford D L. paup*, Phylogenetic Analysis Using Parsimony. Sunderland, MA: Sinauer; 2001. , Version 4.0b8. [Google Scholar]
- 20.Wang J, Jiang J, Oard J H. Plant Science. 2000;156:201–211. doi: 10.1016/s0168-9452(00)00255-7. [DOI] [PubMed] [Google Scholar]
- 21.Elble R. BioTechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- 22.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 23.Smith F W, Rae A L, Hawkesford M J. Biochim Biophys Acta. 2000;1465:236–245. doi: 10.1016/s0005-2736(00)00141-3. [DOI] [PubMed] [Google Scholar]
- 24.Holland P M, Abramson R D, Watson R, Gelfand D H. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Trieu A T, Blaylock L A, Harrison M J. Mol Plant–Microbe Interact. 1998;11:14–22. doi: 10.1094/MPMI.1998.11.1.14. [DOI] [PubMed] [Google Scholar]
- 26.Krogh A, Larsson B, von Heijne G, Sonnhammer E L. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 27.Tusnady G E, Simon I. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 28.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 29.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bun-Ya M, Nishimura M, Harashima S, Oshima Y. Mol Cell Biol. 1991;11:3229–3238. doi: 10.1128/mcb.11.6.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearson J N, Jakobsen I. New Phytol. 1993;124:489–494. [Google Scholar]
- 32.Burleigh S H, Harrison M J. Plant Mol Biol. 1997;34:199–208. doi: 10.1023/a:1005841119665. [DOI] [PubMed] [Google Scholar]
- 33.Leggewie G, Willmitzer L, Riesmeier J W. Plant Cell. 1997;9:381–392. doi: 10.1105/tpc.9.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitsukawa N, Okumura S, Shirano Y, Sato S, Kato T, Harashima S, Shibata D. Proc Natl Acad Sci USA. 1997;94:7098–7102. doi: 10.1073/pnas.94.13.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harrison, M. J., Dewbre, G. R. & Liu, J. (2002) Plant Cell, in press. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.