Abstract
The combined observations of elevated DNA repair gene expression, high uracil-DNA glycosylase-initiated base excision repair, and a low spontaneous mutant frequency for a lacI transgene in spermatogenic cells from young mice suggest that base excision repair activity is high in spermatogenic cell types. Notably, the spontaneous mutant frequency of the lacI transgene is greater in spermatogenic cells obtained from old mice, suggesting that germ line DNA repair activity may decline with age. A paternal age effect in spermatogenic cells is recognized for the human population as well. To determine if male germ cell base excision repair activity changes with age, uracil-DNA glycosylase-initiated base excision repair activity was measured in mixed germ cell (i.e., all spermatogenic cell types in adult testis) nuclear extracts prepared from young, middle-aged, and old mice. Base excision repair activity was also assessed in nuclear extracts from premeiotic, meiotic, and postmeiotic spermatogenic cell types obtained from young mice. Mixed germ cell nuclear extracts exhibited an age-related decrease in base excision repair activity that was restored by addition of apurinic/apyrimidinic (AP) endonuclease. Uracil-DNA glycosylase and DNA ligase were determined to be limiting in mixed germ cell nuclear extracts prepared from young animals. Base excision repair activity was only modestly elevated in pachytene spermatocytes and round spermatids relative to other spermatogenic cells. Thus, germ line short-patch base excision repair activity appears to be relatively constant throughout spermatogenesis in young animals, limited by uracil-DNA glycosylase and DNA ligase in young animals, and limited by AP endonuclease in old animals.
Germ line genomic stability is an important factor in reproductive health, with approximately 20% of genetic diseases attributed to new germ line mutations (12, 13, 46). Several autosomal dominant genetic diseases due to de novo male germ line mutations are associated with increased paternal age (18, 21, 45, 49), suggesting that male germ line genomic stability is compromised with age. Analysis of chromosomes in spermatozoa revealed an increased frequency of aberrations with age (24, 34, 41, 54), providing additional evidence that male germ line genomic stability decreases with age. Similarly in mice, the spontaneous mutant frequency in a lacI transgene recovered from pachytene spermatocytes, round spermatids, and epididymal spermatozoa obtained from old animals was approximately 10-fold higher than the mutant frequency observed for young mice (60).
In contrast to the decline in germ line genetic integrity with old age, the germ line DNA of young mice appears to be well maintained. Compared to somatic tissues, a lower spontaneous mutant frequency in the male germ line for a lacI transgene has been reported for fish (65), mice (32, 60), and rats (16). The murine mitotically proliferating male germ cell pool is comprised of primitive type A spermatogonia, which sequentially give rise to mitotically active, type A spermatogonia, intermediate spermatogonia, and type B spermatogonia (3). Type B spermatogonia generate a population of primary spermatocytes that proceed through meiosis and become haploid round spermatids (5) and then undergo complex morphological and molecular transformations to generate spermatozoa (3).
The spontaneous mutant frequency in a lacI transgene was analyzed by using specific spermatogenic cell types and found to decline between primitive type A spermatogonia and type B spermatogonia (60). The lower mutant frequency was maintained throughout the remainder of spermatogenesis. These observations suggest that mechanisms operate to provide spermatozoa with a low mutant frequency.
The specific-locus test and analysis of embryonic malformations have been used to indirectly study germ line mutagenesis in mice (19, 44, 47, 52). These studies revealed a differential susceptibility to induced mutagenesis. For example, many chemicals have been shown to exert their most potent mutagenic germ line effects on postmeiotic cell types (reviewed in reference 53). The mechanism(s) mediating the apparent differential mutability has not been described.
DNA base excision repair (BER) may be one of the mechanisms regulating germ line genomic stability because this repair pathway is largely responsible for ameliorating spontaneous base damage (33). Consistent with this are the observations that high levels of DNA BER gene transcripts are detected in the testis (1, 11, 17, 25, 36, 59, 61, 66). Specifically, male germ cells appear to be the source of the highest observed expression of several base excision repair genes, including those for Xrcc-1 (59, 61), DNA polymerase β (β-pol) (1, 25), and DNA ligase III (11, 36). Peak expression of these repair genes generally occurs in pachytene spermatocytes and round spermatids.
Western blot analyses have supported the RNA data showing greater levels of DNA ligases I and III, Xrcc-1, Ape/Ref-1, and β-pol proteins in mixed germ cell (MGC, i.e., all spermatogenic cell types in adult testis) nuclear extracts than in adult somatic tissues (26, 48). Furthermore, we have reported higher levels of uracil-DNA glycosylase-initiated base excision repair (UDG-BER) activity in MGC nuclear extracts obtained from young adult mice compared to extracts from brain, liver, enriched preparations of prepubertal Sertoli cells, thymocytes, and small intestine (26). Together, these data support the hypothesis that UDG-BER activity is greater in spermatogenic cells than in somatic tissues.
Two major BER pathways, based on the number of nucleotides incorporated during repair, are recognized. Short-patch BER is characterized by incorporation of a single nucleotide into the DNA strand undergoing repair in a β-pol-dependent manner (55, 56). During long-patch BER, 2 to 6 nucleotides are typically incorporated into the DNA strand undergoing repair in a PCNA-dependent manner (20, 31, 40). While substantial biochemical characterization of these pathways has been realized, there are few reports documenting the repair activities in mammalian tissues or primary cell types.
The studies described herein were performed to determine if DNA repair activity varies among spermatogenic cell types and whether DNA repair activity in spermatogenic cells obtained from old animals is diminished. Such changes could at least partially account for the mutagenesis observations described above. Modest variation in UDG-BER was detected among enriched populations of spermatogenic cell types obtained from young mice. In vitro UDG-BER activity in MGC nuclear extracts prepared from old mice was significantly lower than in MGC nuclear extracts from young adult mice. A significant reduction in the relative abundance of Ape/Ref-1 in MGC nuclear extracts obtained from old mice coincided with the decreased UDG-BER activity, and addition of purified recombinant APE/REF-1 restored the activity in germ cell nuclear extracts prepared from old mice. In contrast, addition of purified uracil-DNA glycosylase and DNA ligase III each elevated the UDG-BER activity in nuclear extracts prepared from MGCs obtained from young animals. Finally, short-patch and long-patch BER pathways were found to function in murine spermatogenic cell nuclear extracts.
MATERIALS AND METHODS
Animals.
Male CD1 mice (6, 8, 30, and 60 days old) were obtained from Charles River, and B6D2F1 male mice (3, 16, and 28 months old) were obtained from the National Institute on Aging (NIA). Mice heterozygous for Apex were obtained from in-house breedings. Apex heterozygous knockout mice were originally developed using 129-derived embryonic stem cells (35), but were backcrossed into C57BL/6J mice for at least five generations before the studies described herein. All animals were housed in an American Association for the Accreditation of Laboratory Animal Care-accredited animal facility and fed standard mouse lab chow and water ad libitum. Mice were specific pathogen free.
At the appropriate ages, mice were humanely euthanized, and then testes were rapidly removed and used immediately for MGC preparations or enriched spermatogenic cell type and Sertoli cell isolations. Thymuses were removed from 6-day-old mice and used immediately for thymocyte isolation. All procedures were performed in accordance with federal and institutional guidelines for animal welfare.
MGC preparations, spermatogenic cell types, Sertoli cells, and thymocyte isolations.
Testes were collected from three to four adult male B6D2F1 or Apex heterozygous knockout mice and used to prepare MGCs as described previously (4, 5, 51). The composition of the MGC preparations from adult mice was approximately 4% spermatogonia, 22% spermatocytes, 71% spermatids, and <3% Sertoli cells (3). Specific populations of spermatogenic cell types and Sertoli cells were prepared from male CD1 mice of the appropriate ages using a Sta Put gradient system as described previously (4, 51).
Primitive type A spermatogonia and prepubertal Sertoli cells were recovered from mice 6 days old, type A and type B spermatogonia were from mice 8 days old, and pachytene spermatocytes and round spermatids were from mice 30 and >60 days old. Purities of recovered spermatogonia and Sertoli cells were ≥85% by morphological criteria. Purities of pachytene spermatocytes and round spermatids were ≥95%. Thymuses were obtained from sets of 50 6-day-old male CD1 mice and were teased apart in homogenization buffer as previously described (26). Cells were dissociated by gentle passage through a 16-gauge needle, and thymocytes were enriched by passage through Nytex mesh.
Nuclear extracts.
Nuclear extracts were prepared from MGCs, enriched spermatogenic cell types, Sertoli cells, and thymocytes as described by Widen and Wilson (63) with modification, as described previously (26). Briefly, a known amount of luciferase protein (Roche Molecular Biochemicals, Indianapolis, Ind.) was added by including it in the buffer used to resuspend pelleted nuclei for homogenization. Protein concentrations were determined using the Bradford assay (7) with immunoglobulin (Ig) as a standard (Bio-Rad, Hercules, Calif.). Nuclear extracts were routinely diluted to 10 mg/ml, separated into single-use aliquots, and stored at −80°C until use.
Prior to use in repair assays or Western analyses, luciferase assays were performed by adding 2 μl of nuclear extract to 400 μl of luciferase buffer (60 mM Tris-acetate [pH 7.5], 2.5 mM EDTA, 12 mM magnesium acetate, 60 mM dithiothreitol, 5 mM ATP, 0.075% bovine serum albumin, and 150 μM luciferin). Relative light units were measured on a Lumat LB 9501 (Berthold) luminometer and compared to a luciferase standard curve (14, 15). The amount of luciferase recovered was used to correct for differences in protein recovered between samples (26).
Short-patch BER assay.
The UDG-BER assay was performed as previously described (26, 55). Briefly, 3 pmol of a 51-mer oligonucleotide containing a single G:U mismatch and a 5′ fluorescein label on the U-containing strand (Integrated DNA Technologies) was combined with nuclear extract and incubated at 37°C for 10 min in reaction buffer (100 mM Tris-HCl [pH 7.5], 0.1 mM EDTA, 5 mM MgCl2, 1 mM dithiothreitol, 2 mM ATP, 0.5 mM NAD, 20 μM each dATP, dGTP, and dTTP, 5 mM ditrisphosphocreatine, 10 U of creatine phosphokinase, 20 nM unlabeled dCTP, and 20 μCi of [α-33P]dCTP [3,000 Ci/mmol]). Ten micrograms of nuclear extract prepared from MGCs and purified populations of spermatogenic cell types and 40 μg of nuclear extract prepared from somatic cell types were used for UDG-BER assays. Reactions were stopped by placing the mixes on ice and the addition of 4.5 μl of stop solution (50 mM EDTA, 0.3 M NaCl, and 80% formamide) and then subjected to denaturing polyacrylamide gel electrophoresis (PAGE).
Oligonucleotide standards encompassing the linear range of fluorescent quantification were run simultaneously on each gel. Fluorescent visualization and quantification of oligonucleotide recovery were performed on a ChemiImage 4400 (Alpha Innotech), and radionucleotide incorporation was measured using a GS-363 Molecular Imager System (Bio-Rad). Various purified BER proteins, Escherichia coli UDG (Life Technologies, Grand Island, N.Y.), human β-pol, and human APE/REF-1 (Trevigen, Gaithersburg, Md.) and murine DNA ligases I and IIIβ (A. Tomkinson, University of Texas Health Science Center Institute of Biotechnology, San Antonio, Tex.) were added independently to nuclear extracts in a volume of 1 μl to determine which activity could enhance overall UDG-BER activity.
Long-patch BER assay.
Three different long-patch BER substrates were prepared containing a G:U mismatch, a natural apurinic/apyrimidinic (AP) site, and a reduced AP site. Covalently closed circular DNA (cccDNA) was prepared as previously described (39, 40). cccDNA containing a reduced AP site was prepared by treating uracil-containing cccDNA with uracil-DNA glycosylase (10 U/μg of DNA) for 1 h at 37°C in the presence of NaBH4. Substrate DNA was 32P labeled as previously described (39).
Long-patch BER was conducted as previously described (6) with a few modifications. Briefly, 100 ng of cccDNA (50 fmol of reduced AP:G) was incubated with 5 μg of nuclear extract for 5, 10, 20, and 40 min at 25°C in a reaction buffer (20 mM HEPES, 100 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, 20 μM each of the four deoxynucleoside triphosphates [dNTPs], 2 mM ATP, 2 mM NAD, 40 mM phosphocreatine, and 1 U of creatine phophokinase) in a total reaction volume of 20 μl. Repair reactions were stopped by addition of sodium dodecyl sulfate (SDS) to a final concentration of 0.4%. Samples were incubated with 2 μg of proteinase K for 20 min at 37°C, and the DNA was recovered after phenol-chloroform extraction by ethanol precipitation. DNA was digested with HinfI as previously described (40), and repair products were analyzed by gel electrophoresis through 20% polyacrylamide and 8 M urea and detected by autoradiography. Quantitative analysis was performed with a Fuji BAS1000.
Western blot analysis.
Nuclear extracts prepared from enriched populations of spermatogenic cell types (50 μg) and MGCs (50 μg) obtained from young (3-month-old), middle-aged (16-month-old), and old (28-month-old) mice were fractionated on SDS-10% PAGE gels (acrylamide-bisacrylamide, 29:1), followed by electroblotting onto Trans-Blot transfer medium (Bio-Rad). To facilitate detection of specific antigens, blots were cut into three sections based on molecular mass.
Rabbit polyclonal anti-ligase I and -ligase III antibodies (A. Tomkinson, University of Texas Health Science Center at San Antonio, San Antonio, Tex.) were used to detect DNA ligases I and III, respectively. Xrcc1 was detected with rabbit anti-human XRCC1 (hXRCC1) polyclonal antibody (Serotec, Raleigh, N.C.). Rabbit anti-human APE/REF-1 (hAPE/REF-1; Novus Biologicals, Littleton, Colo.) and rabbit polyclonal anti-β-pol (S. Wilson, NIEHS, Research Triangle Park, N.C.) were used to detect Ape/Ref-1 and β-pol, respectively. Purified β-pol and APE/REF-1 (Trevigen, Gaithersburg, Md.) and DNA ligases I and III (A. Tomkinson) were included as standards and controls.
Primary antibody was followed by horseradish peroxidase-conjugated goat anti-rabbit Ig antibody (Pierce, Rockford, Ill.), and signal was generated with enhanced chemiluminescence (ECL; Pierce, Rockford, Ill.). Intensity of visualized bands was measured using a ChemiImager 4400 (Alpha Innotech, San Leandro, Calif.) as an integrated density value.
Statistical analysis.
The data were analyzed using analysis of variance. For spermatogenic cell types for which all pairs of means were compared, the Duncan's new multiple range test was used. For analyses in which fewer than all pairs of means were compared, the Bonferroni adjustment was used. P values are presented from analysis of log-transformed data, whereas means and standard errors computed from untransformed data are presented. P values of <0.05 were considered significant.
RESULTS
In vitro BER activity in spermatogenic cells.
To assess UDG-BER activity for specific spermatogenic cell types, nuclear extracts were prepared from premeiotic cell types (enriched populations each of primitive type A, type A, and type B spermatogonia), a meiotic cell type (pachytene spermatocytes), and a postmeiotic cell type (round spermatids) obtained from appropriately aged CD1 mice (a strain commonly used for obtaining defined populations of spermatogenic cell types). Two products can potentially be detected in the UDG-BER assay. One product, a 51-mer, represents complete repair, while the second product, a 23-mer, represents repair through incorporation of [33P]dCTP without ligation. In every assay, the 23-mer represented less than 5% of the counts. Consequently, the data are presented as completely repaired oligonucleotide based on counts detected for the 51-mer.
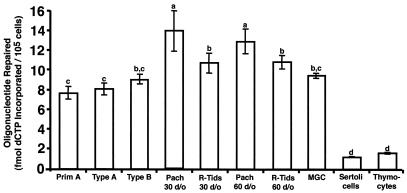
No significant differences were detected in UDG-BER activity between nuclear extracts prepared from primitive type A spermatogonia, type A spermatogonia, and type B spermatogonia (Fig. 1). Nuclear extracts prepared from pachytene spermatocytes (obtained from 30- and >60-day-old mice) displayed a very modest 1.4-fold-higher UDG-BER activity than nuclear extracts prepared from MGCs obtained from 3- to 6-month-old CD1 mice and 1.6-fold-higher UDG-BER activity than spermatogonia. Pachytene spermatocytes exhibited a modest but significant 1.2-fold-higher UDG-BER activity than nuclear extracts prepared from round spermatids. Round spermatid nuclear extracts prepared from 30- and >60-day-old mice exhibited a modest 1.3-fold-higher UDG-BER activity than nuclear extracts prepared from spermatogonia. While these differences are small, statistical analyses revealed that the differences were significant, and thus they are reported here. The UDG-BER activity of nuclear extracts prepared from prepubertal Sertoli cells, a somatic cell type present in the testis, and thymocytes was similar (P < 0.05) and lower than that in nuclear extracts prepared from enriched spermatogenic cell types (P < 0.05; Fig. 1).
FIG. 1.
UDG-BER activities for nuclear extracts prepared from enriched populations of spermatogenic cell types obtained from CD1 mice. Results are presented as mean ± standard error of the mean (SEM) of three replicate assays for each of three independent nuclear extract preparations. Cell types exhibiting similar UDG-BER activity are indicated by having the same letter. Prim A, primitive type A spermatogonia; Type A, type A spermatogonia; Type B, type B spermatogonia; Pach., pachytene spermatocytes; R-Tids, round spermatids.
In vitro BER activity in MGCs.
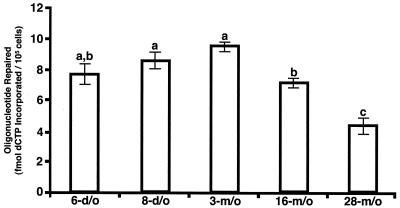
Nuclear extracts were prepared from MGCs isolated from neonatal (6- and 8-day-old), young adult (3-month-old), middle-aged (16-month-old), and old (28-month-old) male B6D2F1 mice and used to assess UDG-BER activity relative to age (Fig. 2). We have previously demonstrated that UDG-BER is similar in C57BL/6J, B6D2F1, and CD1 mice (26). B6D2F1 mice were used to assess the relationship between age and UDG-BER because they are maintained by the NIA for such studies, while CD1 mice are not maintained for aging studies.
FIG. 2.
UDG-BER activities for MGC nuclear extracts prepared from 6-day-old (6-d/o), 8-day-old, 3-month-old (3-m/o), 16-month-old, and 28-month-old male B6D2F1 mice. Results are presented as means ± SEM of three replicate assays for each of three independent nuclear extract preparations. MGC nuclear extracts from different-aged animals with similar UDG-BER activity are indicated by having the same letter.
UDG-BER activity was reduced 25% (P < 0.05) in nuclear extracts prepared from middle-aged mice and 53% (P < 0.05) in old mice compared to nuclear extracts prepared from neonatal and young adult mice (Fig. 2). While UDG-BER activity was significantly reduced in nuclear extracts prepared from 16- and 28-month-old mice, the activity of MGC nuclear extracts from 16-month-old mice was 5.8-fold and 4.3-fold higher than that in Sertoli cell and thymocyte nuclear extracts, respectively. Activity in MGC nuclear extracts from 28-month-old mice was 3.6-fold and 2.7-fold higher than that in Sertoli cell and thymocyte nuclear extracts, respectively (Fig. 1 and 2).
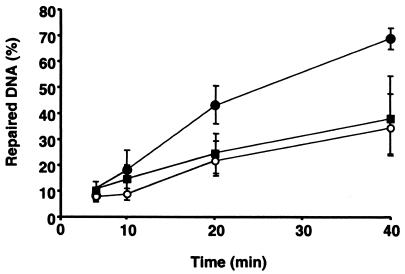
Nuclear extracts prepared from MGCs isolated from young adult, middle-aged, and old male B6D2F1 mice were also used to assess long-patch BER activity. Repair of G:U mismatch on a cccDNA substrate was not detected using MGC nuclear extracts obtained from young mice (data not shown), but an AP site generated by preincubation of cccDNA containing a G:U mismatch with uracil-DNA glycosylase was repaired (data not shown). Long-patch BER activity of cccDNA containing a reduced AP site was 50% lower (P < 0.05) in MGC nuclear extracts isolated from middle-aged and old mice compared to MGC nuclear extracts prepared from young mice (Fig. 3).
FIG. 3.
Long-patch BER activities on reduced AP site cccDNA for MGC nuclear extracts prepared from 3-month-old, 16-month-old, and 28-month-old male B6D2F1 mice. Results are presented as means ± SEM of assays for each of three independent nuclear extract preparations. Solid circles, 3-month-old mice; open circles, 16-month-old mice; solid squares, 28-month-old mice.
BER protein composition in nuclear extracts.
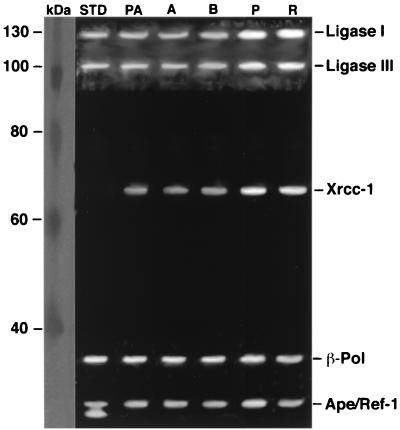
Relative proportions of BER proteins in nuclear extracts prepared from enriched populations of spermatogenic cells were determined by Western blot analyses (Fig. 4). The relative proportions of DNA ligase III and Xrcc-1 were 1.7-fold and 2.5-fold higher, respectively, in pachytene spermatocytes and round spermatids than in spermatogonial cell types (Table 1).
FIG. 4.
Western blot analysis of BER proteins in nuclear extracts prepared from enriched populations of primitive type A spermatogonia (PA), type A spermatogonia (A), type B spermatogonia (B), pachytene spermatocytes (P), and round spermatids (R). Bands corresponding to DNA ligase I (130 kDa), DNA ligase III (93 kDa), Xrcc-1 (69 kDa), β-pol (39 kDa), and Ape/Ref-1 (37 kDa) proteins were visualized. Triplicate assays for each of three independent nuclear extract preparations were performed. Some variation between replicates was noted. A summary of all data is shown in Table 1. Molecular mass protein standards and purified DNA ligases I and III, β-pol, and Ape/Ref-1 (lane STD) are shown for comparison.
TABLE 1.
Abundance of BER proteins in nuclear extracts prepared from enriched spermatogenic cell types
| Protein | IDV/103 cells ± SEM (% of total)a
|
||||
|---|---|---|---|---|---|
| Primitive type A spermatogonia | Type A spermatogonia | Type B spermatogonia | Pachytene spermatocytes | Round spermatids | |
| DNA ligase I | 21.0 ± 3.1 (26.6) | 20.7 ± 2.1 (26.0) | 20.8 ± 0.7 (24.9) | 22.6 ± 1.8 (20.4) | 22.1 ± 0.7 (20.2) |
| DNA ligase III | 15.3 ± 2.6 (19.3) | 15.2 ± 1.9 (19.1) | 16.3 ± 1.1 (19.6) | 26.5 ± 0.4 (24.3) | 26.3 ± 1.0 (24.1) |
| Xrcc-1 | 9.3 ± 0.2 (11.7) | 9.0 ± 0.8 (11.2) | 8.6 ± 0.2 (10.3) | 22.1 ± 1.8 (20.3) | 22.8 ± 0.2 (20.9) |
| β-Pol | 19.0 ± 2.5 (18.4) | 19.0 ± 0.6 (20.0) | 18.9 ± 1.8 (22.5) | 19.0 ± 0.7 (17.5) | 19.1 ± 0.3 (17.3) |
| Ape/Ref-1 | 14.6 ± 2.7 (24.0) | 15.9 ± 1.3 (23.7) | 18.8 ± 0.6 (22.7) | 19.0 ± 2.8 (17.5) | 18.9 ± 1.3 (17.5) |
Values are expressed as integrated density value (IDV) per 103 cells ± SEM, with the percentage of total chemiluminescence for a tissue, calculated by (protein IDV/total IDV for 103 cells) × 100. Values in boldface type are significantly different from values for proteins in other cell types.
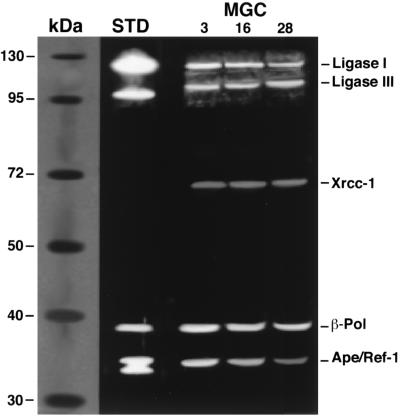
The relative proportions of BER proteins in MGC nuclear extracts prepared from young, middle-aged, and old mice were also determined by Western blot analyses (Fig. 5). Quantification of chemiluminescence from Western blots revealed no significant changes in the abundances of DNA ligase I, DNA ligase III, Xrcc-1, and β-pol among the age groups (Table 2). In contrast, MGC nuclear extracts prepared from 16-month-old mice exhibited a 38% reduction in Ape/Ref-1 compared to nuclear extracts from 3-month-old mice (Table 2). Abundance of Ape/Ref-1 in MGC nuclear extracts prepared from 28-month-old mice was reduced by 62% compared to nuclear extracts from 3-month-old mice (P < 0.05; Table 2).
FIG. 5.
Western blot analysis of BER proteins in MGC nuclear extracts prepared from 3-month-old, 16-month-old, and 28-month-old mice. Bands corresponding to DNA ligase I (130 kDa), DNA ligase III (93 kDa), Xrcc-1 (69 kDa), β-pol (39 kDa), and Ape/Ref-1 (37 kDa) proteins were visualized. Triplicate assays were performed for each of three independent nuclear extract preparations. Some variation in signal intensities was observed among Western blots. A summary of all data is shown in Table 2. Molecular mass protein standards and purified DNA ligases I and III, β-pol, and APE/REF-1 (lane STD) are shown for comparison.
TABLE 2.
Abundance of BER proteins in nuclear extracts prepared from MGCs obtained from different-aged mice
| Protein | Mean IDV/103 cells ± SEM (% of total)a
|
||
|---|---|---|---|
| 3-month-old mice | 16-month-old mice | 28-month-old mice | |
| DNA ligase I | 12.0 ± 1.3 (24.2) | 12.2 ± 0.3 (26.5) | 11.9 ± 0.4 (26.5) |
| DNA ligase III | 11.7 ± 0.8 (23.6) | 11.4 ± 0.3 (24.8) | 12.8 ± 0.4 (28.5) |
| Xrcc-1 | 5.0 ± 0.6 (10.1) | 5.3 ± 0.3 (11.4) | 5.1 ± 0.1 (11.3) |
| β-Pol | 13.3 ± 0.6 (26.8) | 12.3 ± 0.4 (27.4) | 12.4 ± 0.4 (27.7) |
| Ape/Ref-1 | 7.6 ± 0.9 (15.3) | 4.6 ± 0.3 (9.9) | 2.7 ± 0.9 (6.0) |
See Table 1, footnote a.
Identification of limiting BER enzyme activities in MGC nuclear extracts.
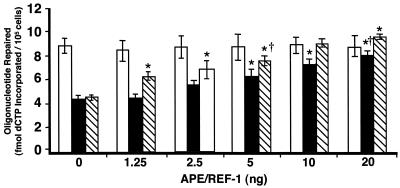
Due to the concomitant reduction in UDG-BER activity and decreased Ape/Ref-1 levels in MGCs obtained from old mice, the ability of purified Ape/Ref-1 to restore UDG-BER activity in nuclear extracts from old mice was tested. Young heterozygous Apex knockout mice (35) were used as controls for reduced Ape/Ref-1 activity. Addition of purified APE/REF-1 to MGC nuclear extracts prepared from 3-month-old mice had no effect on UDG-BER activity (Fig. 6). MGC nuclear extracts prepared from heterozygous Apex knockout mice exhibited 50% of the UDG-BER activity of nuclear extracts from MGCs obtained from young wild-type mice (Fig. 6). Purified APE/REF-1 (5 ng) added to MGC nuclear extracts prepared from Apex heterozygous knockout mice was sufficient to restore the UDG-BER activity to that of young wild-type animals. Addition of 5 ng of APE/REF-1 resulted in a significant increase in UDG-BER activity for nuclear extracts from MGCs obtained from 28-month-old mice (Fig. 6), and supplementation with 20 ng of purified APE/REF-1 resulted in UDG-BER activity similar to that of MGCs obtained from young mice.
FIG. 6.
UDG-BER activities for MGC nuclear extracts prepared from 3-month-old B6D2F1 (open), 28-month-old B6D2F1 (solid), and 3-month-old Apex heterozygous knockout mice (striped). Results are presented as means ± SEM of three replicate assays for each of three independent nuclear extract preparations. ∗, significantly (P < 0.05) different from addition of 0 ng of protein within a specific group. †, amount of purified APE/REF-1 required to restore activity to that of 3-month-old mice with 0 ng of protein added.
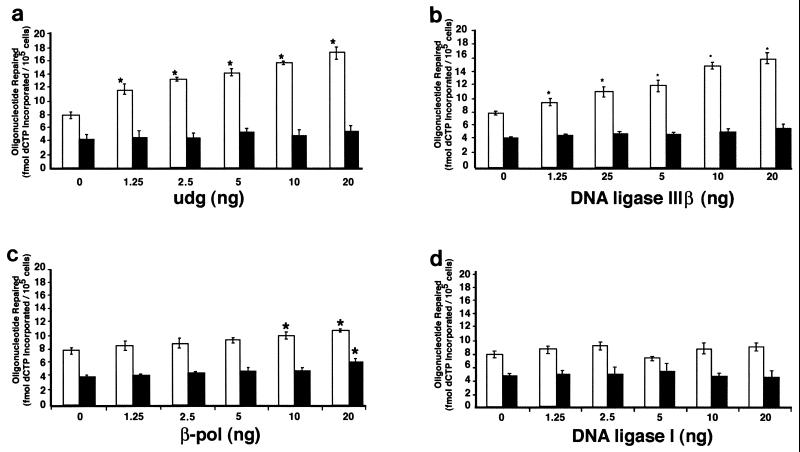
The addition of 1.25 ng of purified uracil-DNA glycosylase significantly increased UDG-BER activity in nuclear extracts prepared from MGCs obtained from 3-month-old mice (Fig. 7a). Furthermore, UDG-BER activity was increased approximately 2-fold after addition of 20 ng of purified uracil-DNA glycosylase to MGC nuclear extracts prepared from 3-month-old mice. In comparison, addition of purified uracil-DNA glycosylase to MGC nuclear extracts prepared from 28-month-old mice did not alter UDG-BER activity. Purified DNA ligase IIIβ also significantly increased UDG-BER activity in nuclear extracts prepared from 3-month-old mice. However, no significant change in UDG-BER activity was observed after addition of purified DNA ligase IIIβ to MGC nuclear extracts prepared from 28-month-old mice (Fig. 7b).
FIG. 7.
UDG-BER activities for MGC nuclear extracts prepared from 3-month-old (open) and 28-month-old (solid) male B6D2F1 mice. Results are presented as means ± SEM of three replicate assays for each of three independent nuclear extract preparations. Results are for UDG-BER assays in which purified uracil-DNA glycosylase (a), DNA ligase III (b), β-pol (c), and DNA ligase I (d) were added to MGC nuclear extracts. ∗, significantly (P < 0.05) different from addition of 0 ng of protein within a specific age group.
Addition of 10 or 20 ng of purified β-pol modestly but significantly increased UDG-BER activity in nuclear extracts prepared from MGCs obtained from 3-month-old (1.35-fold) and 28-month-old (1.5-fold) mice (Fig. 7c). No significant changes in UDG-BER activity were detected with the addition of DNA ligase I to nuclear extracts from young or old mice (Fig. 7d).
DISCUSSION
Based on previous observations identifying variable DNA repair gene expression among defined populations of murine spermatogenic cell types (1, 11, 25, 36, 59, 61, 64), differences in spontaneous mutation frequencies among spermatogenic cell types (60), and differences in induced mutagenesis among murine spermatogenic cell types (reviewed in reference 53) nuclear extracts prepared from defined populations of spermatogenic cell types were examined for short-patch UDG-BER activity to determine if there was cell type variation in repair activity.
The modest increase in activity observed for nuclear extracts prepared from pachytene spermatocytes and round spermatids compared to earlier spermatogenic cell types is generally consistent with elevated DNA repair gene expression and inversely correlated with decreased spontaneous mutant frequencies (1, 11, 25, 36, 59, 61, 64, 66). These results suggest that high UDG-BER activity measured in spermatogenic cell types may contribute to the overall reduced mutation frequency observed in male germ cells. Consistent with Olsen et al. (48), we detected no variation between the relative abundances of Ape/Ref-1, DNA ligase I, and β-pol in premeiotic, meiotic, and postmeiotic male germ cells from young mice. However, increases in Xrcc-1 and DNA ligase III were detected in meiotic and postmeiotic mouse male germ cells, although these increases in protein are modest compared to the 10-fold increase reported for Xrcc-1 (59) and LigIII (11) transcript levels. Transcription ceases in condensing spermatids, and several genes required in later stages of spermatogenesis are expressed and accumulated in meiotic and postmeiotic spermatogenic cell types (23, 29, 30, 37, 38, 43, 58). Such a phenomenon may explain the discrepancy between transcript and protein levels observed for these BER genes.
Pachytene spermatocyte and round spermatid nuclear extracts were prepared from 30-day-old and 60-day-old animals to determine whether there were differences in activity between cells in the first wave of spermatogenesis (30-day-old) and later (60-day-old). No significant differences were detected. These results were reinforced by the comparisons among MGC nuclear extracts prepared from 6-day-old (composed principally of primitive type A spermatogonia), 8-day-old (types A and B spermatogonia), and 3-month-old mice (all stages of spermatogenesis and after the first wave of spermatogenesis). Accordingly, no significant difference was detected among nuclear extracts prepared from different-aged mice reflecting the first wave of spermatogenesis (6-day-old and 8-day-old) compared to animals that have progressed beyond the first wave of spermatogenesis (3-month-old).
A paternal age effect has long been recognized in humans due to the corresponding impact on paternally derived de novo mutations associated with certain autosomal dominant disorders. A similar age-related increase in spontaneous mutant frequencies for murine spermatogenic cells has been observed using lacI transgenic mice (60). Because DNA repair pathways contribute to maintaining genetic integrity and because BER in particular is largely responsible for ameliorating spontaneous base damage (22, 33), short-patch and long-patch UDG-BER assays were employed to assess repair activity in relation to age. A decrease in short-patch UDG-BER activity measured in MGC nuclear extracts from 16-month-old mice compared to young mice and an additional reduction in nuclear extracts from 28-month-old mice were observed.
Despite the decline in UDG-BER activity in MGCs from old animals, the activity is greater than that found in thymocytes and Sertoli cells obtained from 6-day-old animals that display relatively high UDG-BER activity for somatic cell types. Analogous to short-patch BER, a decline in long-patch BER was detected in nuclear extracts prepared from 16- and 28-month-old mice with the reduced AP site-containing substrate. Together, these data indicate that an overall decline in BER activity may contribute to the increased mutant frequency observed in spermatogenic cells from old mice.
The limiting enzyme activity in the short-patch BER pathway has not been conclusively identified, although it has been reported that in some cell culture lines Ape/Ref-1 is limiting (27, 50). However, the lyase activity of β-pol appears to be rate limiting using a reconstituted BER pathway (2, 57). The apparent discrepancies may be explained by the different assay systems used; the limiting activity may vary among cell types and tissues and may vary for different DNA lesions.
In this study we show that uracil-DNA glycosylase appears to be a major limiting activity for short-patch BER in MGC nuclear extracts prepared from young mice. The cccDNA substrate containing a G:U mismatch was not repaired at a detectable level by MGC nuclear extracts prepared from young male mice. Due to the smaller amount of cccDNA used for the long-patch assay compared to the short-patch assay, it is possible that the same relative proportion of G:U-containing substrate was repaired via the long-patch pathway, but the actual amount of product was too small to be detected. However, cccDNA substrate containing a natural AP site was repaired efficiently. This suggests that uracil-DNA glycosylase is also one of the rate-limiting factors in long-patch BER.
Because there are many DNA glycosylases (22), it is reasonable to speculate that the BER proteins functioning downstream of these enzymes are present at greater activity levels than any one DNA glycosylase, so that the BER pathway can be completed. In this scenario, the initiating DNA glycosylase may be expected to be rate limiting for any specific base lesion.
Therefore, it was somewhat surprising that DNA ligase IIIβ was nearly as effective as uracil-DNA glycosylase in elevating the overall UDG-BER activity of MGC nuclear extracts prepared from young mice. Two forms of DNA ligase III are present in the testis (36): DNA ligase IIIα (103 kDa), which is ubiquitously expressed, and DNA ligase IIIβ (96 kDa), which is expressed in meiotic and postmeiotic cell types. However, peak expression of DNA ligases IIIα and IIIβ occurs in pachytene spermatocytes (11, 36).
The relative contributions of DNA ligase IIIα and IIIβ in BER have not been clearly delineated. Although our results suggest that DNA ligase IIIβ can participate in short-patch BER, the data do not exclude a contribution from DNA ligase IIIα. Notably, DNA ligase IIIα has been shown to interact with Xrcc-1 to facilitate ligation of DNA single-strand breaks during BER (8, 9, 10, 36, 62). The addition of APE/REF-1, β-pol, and DNA ligase I to MGC nuclear extracts obtained from young animals had little to no effect on UDG-BER activity, suggesting that uracil-DNA glycosylase and DNA ligase III are the major limiting activities in these nuclear extracts.
In contrast, the major limiting activity in MGC nuclear extracts prepared from old mice appears to be Ape/Ref-1. Decreased signal for Ape/Ref-1 in Western blots was observed for MGC nuclear extracts obtained from 28-month-old mice compared to young mice, and addition of APE/REF-1 to extracts from old mice restored short-patch BER activity to the levels observed for extracts obtained from young mice. Recently, cell cultures derived from Apex heterozygous knockout mice were shown to be more susceptible to oxidative stress than wild-type cells (42). Together, these data reveal biological consequences of reduced Ape/Ref-1 levels. In agreement with earlier reports, no changes were detected in the relative abundance of Xrcc-1 with increased age (61).
In conclusion, there is only modest variation in short-patch UDG-BER activity between spermatogenic cell types. However, there are differences between BER activity in spermatogenic cells from young and old mice. First, short-patch and long-patch BER activities are substantially lower in germ cell nuclear extracts prepared from old mice compared to extracts prepared from young mice. Second, uracil-DNA glycosylase and DNA ligase III appear to be the principal limiting activities in short-patch UDG-BER for MGC nuclear extracts obtained from young mice, while Ape/Ref-1 is the major limiting activity in MGC nuclear extracts obtained from old mice.
Considered in conjunction with the recently reported low level of nucleotide excision repair in rat spermatogenic cell types (28), our data suggest that BER is an important pathway for maintaining genome integrity in male germ cells and that a reduction in BER with age may contribute to the increased mutant frequency observed for male germ cells obtained from old mice.
Acknowledgments
This publication was made possible by grants ESO9136, AG13560, AG14674, AG00205, CA50519, and CA75137 from the NIEHS, NIA, and NCI, NIH, the Environmental Hazards Center at the South Texas Veteran's Health Care System (STVHCS), the STVHCS, the Nutritional and Interventional Gerontology Training Program, DOE grant AC03-76SF00098, and dissertation research support from the NIA.
We thank Sam Wilson for supplying antibody against β-pol, Alan Tomkinson for supplying purified DNA ligase I and IIIβ and antibodies against DNA ligase I and III, and Allison McKenna for conducting the long-patch BER assays.
REFERENCES
- 1.Alcivar, A. A., L. E. Hake, and N. B. Hecht. 1992. DNA polymerase-beta and poly(ADP)ribose polymerase mRNAs are differentially expressed during the development of male germinal cells. Biol. Reprod. 46:201-207. [DOI] [PubMed] [Google Scholar]
- 2.Beard, W. A., and S. H. Wilson. 2000. Structural design of eukaryotic DNA repair polymerase: DNA polymerase β. Mutat. Res. 460:231-244. [DOI] [PubMed] [Google Scholar]
- 3.Bellve, A. R. 1993. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 225:84-113. [DOI] [PubMed] [Google Scholar]
- 4.Bellve, A. R., C. F. Millette, Y. M. Bhatnagar, and D. A. O'Brien. 1977. Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J. Histochem. Cytochem. 25:480-494. [DOI] [PubMed] [Google Scholar]
- 5.Bellve, A. R., J. C. Cavicchia, C. F. Millette, D. A. O'Brien, Y. M. Bhatnagar, and M. Dym. 1977. Spermatogenic cells of the prepuberal mouse. J. Cell Biol. 74:68-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biade, S., R. W. Sobol, S. H. Wilson, and Y. Matsumoto. 1998. Impairment of proliferating cell nuclear antigen-dependent apurinic/apyrimidinic site repair on linear DNA. J. Biol. Chem. 273:898-902. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Caldecott, K. W., C. K. McKeown, J. D. Tucker, S. Ljungquist, and L. H. Thompson. 1994. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol. 14:68-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caldecott, K. W., J. D. Tucker, L. H. Stanker, and L. H. Thompson. 1995. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 23:4836-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cappelli, E., R. Taylor, M. Cevasco, A. Abbondandolo, K. Caldecott, and G. Frosina. 1997. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J. Biol. Chem. 272:23970-23975. [DOI] [PubMed] [Google Scholar]
- 11.Chen, J., A. E. Tomkinson, W. Ramos, Z. B. Mackey, S. Danehower, C. A. Walter, R. A. Shultz, J. M. Besterman, and I. Husain. 1995. Mammalian DNA ligase III: molecular cloning, chromosomal location and expression in spermatocytes undergoing meiotic recombination. Mol. Cell. Biol. 15:5412-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crow, J. F. 1997. The high spontaneous mutation rate: is it a health risk? Proc. Natl. Acad. Sci. USA 94:8380-8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crow, J. F., and C. Denniston. 1981. The mutation component of genetic damage. Science 212:888-893. [DOI] [PubMed] [Google Scholar]
- 14.De Wet, J. R., K. V. Wood, D. R. Helinski, and M. DeLuca. 1985. Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proc. Natl. Acad. Sci. USA 82:7870-7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Wet, J. R., K. V. Wood, M. DeLuca, D. R. Helinski, and S. Subramani. 1987. Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol. 7:725-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dycaico, M. J., G. S. Provost, P. L. Kretz, S. L. Ransom, J. C. Moores, and J. M. Short. 1994. The use of shuttle vectors for mutation analysis in transgenic mice and rats. Mutat. Res. 307:461-478. [DOI] [PubMed] [Google Scholar]
- 17.Engelward, B. P., M. S. Boosalis, B. J. Chen, Z. Deng, M. J. Siciliano, and L. D. Samson. 1993. Cloning and characterization of a mouse 3-methyladenine/7-methylguanine/3-methylguanine DNA glycosylase cDNA whose gene maps to chromosome 11. Carcinogenesis 14:175-181. [DOI] [PubMed] [Google Scholar]
- 18.Erickson, J. D., and M. M. Cohen, Jr. 1974. A study of parental age effects on the occurrence of fresh mutations for the Apert syndrome. Ann. Hum. Genet. 38:89-96. [DOI] [PubMed] [Google Scholar]
- 19.Favor, J. 1990. Multiple endpoint mutational analysis in the mouse. Prog. Clin. Biol. Res. 340C:115-124. [PubMed]
- 20.Frosina, G., P. Fortini, O. Rossi, F. Carrozzino, G. Raspaglio, L. S. Cox, D. P. Lane, A. Abbondandolo, and E. Dogliotti. 1996. Two pathways for base excision repair in mammalian cells. J. Biol. Chem. 271:9573-9578. [DOI] [PubMed] [Google Scholar]
- 21.Glaser, L. R., W. Jiang, S. A. Boyadjiev, A. K. Tran, A. A. Zachary, L. V. Maldergem, D. Johnson, S. Walsh, M. Oldridge, S. A. Wall, A. O. M. Wilkie, and E. W. Jabs. 2000. Paternal origin of FGFR2 mutations in sporadic cases of Crouzon syndrome and Pfeiffer syndrome. Am. J. Hum. Genet. 66:768-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glassner, B. J., L. M. Posnick, and L. D. Samson. 1998. The influence of DNA glycosylases on spontaneous mutation. Mutat. Res. 400:33-44. [DOI] [PubMed] [Google Scholar]
- 23.Gold, B., H. Fugimoto, J. M. Kramer, R. P. Erickson, and N. B. Hecht. 1983. Haploid accumulation and translational control of phosphoglycerate kinase 2 messenger RNA during mouse spermatogenesis. Dev. Biol. 98:392-399. [DOI] [PubMed] [Google Scholar]
- 24.Griffin, D. K., M. A. Abruzzo, E. A. Millie, L. A. Sheean, E. Feingold, S. L. Sherman, and T. J. Hassold. 1995. Non-disjunction in human sperm: evidence for an effect of increasing paternal age. Hum. Mol. Genet. 4:2227-2232. [DOI] [PubMed] [Google Scholar]
- 25.Hirose, F., Y. Hotta, M. Yamaguchi, and A. Matsukage. 1989. Difference in the expression level of DNA polymerase beta among mouse tissues: high expression in the pachytene spermatocytes. Exp. Cell Res. 181:169-180. [DOI] [PubMed] [Google Scholar]
- 26.Intano, G. W., C. A. McMahan, R. B. Walter, J. R. McCarrey, and C. A. Walter. 2001. Mixed spermatogenic germ cell nuclear extracts exhibit high base excision repair activity. Nucleic Acids Res. 29:1366-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izumi, T., T. K. Hazra, I. Boldogh, A. E. Tomkinson, M. S. Park, S. Ikeda, and S. Mitra. 2000. Requirement for human AP endonuclease 1 for repair of 3′-blocking damage at DNA single-strand breaks induced by reactive oxygen species. Carcinogenesis 21:1329-1334. [PubMed] [Google Scholar]
- 28.Jansen, J., A. K. Olsen, R. Wiger, H. Naegeli, P. deBoer, F. van der Hoeven, J. A. Holme, G. Brunborg, and L. Mullenders. 2001. Nucleotide excision repair in rat male germ cells: low level of repair in intact cells contrasts with high dual incision activity in vitro. Nucleic Acids Res. 29:1791-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleene, C. K., R. J. Distel, and N. B. Hecht. 1984. Translational regulation and deadenylation of a protamine mRNA during spermiogenesis in the mouse. Dev. Biol. 105:71-79. [DOI] [PubMed] [Google Scholar]
- 30.Kleene, C. K. 1996. Patterns of translational regulation in the mammalian testis. Mol. Reprod. Dev. 43:268-281. [DOI] [PubMed] [Google Scholar]
- 31.Klungland, A., and T. Lindahl. 1997. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J. 16:3341-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohler, S. W., G. S. Provost, A. Fieck, P. L. Kretz, W. O. Bullock, J. A. Sorge, D. L. Putman, and J. M. Short. 1991. Spectra of spontaneous and mutagen-induced mutations in the lacI gene in transgenic mice. Proc. Natl. Acad. Sci. USA 88:7958-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindahl, T., P. Karran, and R. D. Wood. 1997. DNA excision repair pathways. Curr. Opin. Genet. Dev. 7:158-169. [DOI] [PubMed] [Google Scholar]
- 34.Lowe, X., B. Collins, J. Allen, N. Titenko-Holland, J. Breneman, M. van Beek, J. Bishop, and A. J. Wyrobeck. 1995. Aneuploidies and micronuclei in germ cells of male mice of advanced age. Mutat. Res. 338:59-76. [DOI] [PubMed] [Google Scholar]
- 35.Ludwig, D. L., M. A. MacInnes, Y. Takiguchi, P. E. Purtymun, M. Henrie, M. Flanery, J. Meneses, R. A. Pedersen, and D. J. Chen. 1998. A murine AP-endonuclease gene-targeted deficiency with postimplantation embryonic progression and ionizing radiation sensitivity. Mutat. Res. 409:17-29. [DOI] [PubMed] [Google Scholar]
- 36.Mackey, Z. B., W. Ramos, D. S. Levin, C. A. Walter, J. R. McCarrey, and A. E. Tomkinson. 1997. An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Mol. Cell. Biol. 17:989-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsui, M., H. Horiguchi, H. Kamma, M. Fujiwara, R. Ohtsubo, and T. Ogata. 2000. Testis- and developmental stage-specific expression of hnRNP A2/B1 splicing isoforms, B0a/b. Biochim. Biophys. Acta 1493:33-40. [DOI] [PubMed] [Google Scholar]
- 38.Matsumoto, M., S.-I. Kurata, H. Fujimoto, and M. Hoshi. 1993. Haploid specific activations of protamine 1 and hsc70t genes in mouse spermatogenesis. Biochem. Biophys. Acta 1174:274-278. [DOI] [PubMed] [Google Scholar]
- 39.Matsumoto, Y. 1999. Base excision repair assay using Xenopus laevis oocyte extracts, p. 289-300. In D.S. Henderson (ed.), Methods in molecular biology, vol. 113: DNA repair protocols: eukaryotic systems. Humana, Totowa, N.J. [DOI] [PubMed] [Google Scholar]
- 40.Matsumoto, Y., K. Kyung, and D. F. Bogenhagen. 1994. Proliferating cell nuclear antigen-dependent abasic site repair in Xenopus laevis oocytes: an alternative pathway of base excision DNA repair. Mol. Cell. Biol. 14:6187-6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McInnes, B., A. Rademaker, and R. Martin. 1998. Donor age and the frequency of disomy for chromosomes 1, 13, 21 and structural fluorescence in situ hybridization. Hum. Reprod. 13:2489-2494. [DOI] [PubMed] [Google Scholar]
- 42.Meira, L. B., S. Devaraj, G. E. Kisby, D. K. Burns, R. L. Daniel, R. E. Hammer, S. Grundy, I. Jiaial, and E. C. Friedberg. 2001. Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Res. 61:5552-5557. [PubMed] [Google Scholar]
- 43.Morales, C. R., Y. K. Kwon, and N. B. Hecht. 1991. Cytoplasmic localization during storage and translation of the mRNAs of transition protein 1 and protamine 1, two translationally regulated transcripts of the mammalian testis. J. Cell Sci. 100:119-131. [DOI] [PubMed] [Google Scholar]
- 44.Müller, W.-U., C. Streffer, A. Wojcik, and F. Niedereichholz. 1999. Radiation-induced malformations after exposure of murine germ cells in various stages of spermatogenesis. Mutat. Res. 425:99-106. [DOI] [PubMed] [Google Scholar]
- 45.Murdoch, J. L., B. A. Walker, and V. A. McKusick. 1972. Parental age effects on the occurrence of new mutations for the Marfan syndrome. Ann. Hum. Genet. 35:331-336. [DOI] [PubMed] [Google Scholar]
- 46.Nelson, K., and L. B. Holmes. 1989. Malformations due to presumed spontaneous mutations in newborn infants. N. Engl. J. Med. 320:19-23. [DOI] [PubMed] [Google Scholar]
- 47.Nomura, T. 1988. X-ray-and chemically induced germ-line mutation causing phenotypical anomalies in mice. Mutat. Res. 198:309-320. [DOI] [PubMed] [Google Scholar]
- 48.Olsen, A.-K., H. Bjørtuft, R. Wiger, J. A. Holme, E. C. Seeberg, M. Bjørås, and G. Brunborg. 2001. Highly efficient base excision repair (BER) in human and rat male germ cells. Nucleic Acids Res. 8:1781-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olshan, F. F., P. G. Schnitzer, and P. A. Baird. 1994. Paternal age and the risk of congenital heart defects. Teratology 50:80-84. [DOI] [PubMed] [Google Scholar]
- 50.Ramana, C. V., I. Boldogh, T. Izumi, and S. Mitra. 1998. Activation of apurinic/apyrimidinic endonuclease in human cells by reactive oxygen species and its correlation with their adaptive response to genotoxicity of free radicals. Proc. Natl. Acad. Sci. USA 95:5061-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romrell, L. J., A. R. Bellve, and D. W. Fawcett. 1976. Separation of mouse spermatogenic cells by sedimentation velocity. Dev. Biol. 49:119-131. [DOI] [PubMed] [Google Scholar]
- 52.Russell, L. B., P. R. Hunsicker, D. K. Johnson, and M. D. Shelby. 1998. Unlike other chemicals, etoposide (a topoisomerase-II inhibitor) produces peak mutagenicity in primary spermatocytes of the mouse. Mutat. Res. 400:279-286. [DOI] [PubMed]
- 53.Shelby, M. D., J. B. Bishop, J. M. Mason, and K. R. Tindall. 1993. Fertility, reproduction, and genetic disease: studies on the mutagenic effects of environmental agents on mammalian germ cells. Environ. Health Perspect. 100:283-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi, Q., and R. H. Martin. 2000. Aneuploidy in human sperm: a review of the frequency and distribution of aneuploidy, effects of donor age lifestyle factors. Cytogenet. Cell Genet. 90:219-226. [DOI] [PubMed] [Google Scholar]
- 55.Singhal, R. K., R. Prasad, and S. H. Wilson. 1995. DNA polymerase β conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J. Biol. Chem. 270:949-957. [DOI] [PubMed] [Google Scholar]
- 56.Sobol, R. W., J. K. Horton, R. Kühn, H. Gu, R. K. Singhal, R. Prasad, K. Rajewsky, and S. H. Wilson. 1996. Requirement of mammalian DNA polymerase-β in base-excision repair. Nature 379:183-186. [DOI] [PubMed] [Google Scholar]
- 57.Srivastava, D. K., B. J. Vande Berg, R. Prasad, J. T. Molina, W. A. Beard, A. E. Romkinson, and S. H. Wilson. 1998. Mammalian abasic site base excision repair: identification of the reaction sequence and rate-determining steps. J. Biol. Chem. 273:21203-21209. [DOI] [PubMed] [Google Scholar]
- 58.Stern, L., K. C. Kleene, B. Gold, and N. B. Hecht. 1983. Gene expression during mammalian spermatogenesis. III. Changes in populations of mRNA during spermiogenesis. Exp. Cell Res. 143:247-255. [DOI] [PubMed] [Google Scholar]
- 59.Walter, C. A., D. A. Trolian, M. B. McFarland, K. A. Street, G. R. Gurram, and J. R. McCarrey. 1996. Xrcc-1 expression during male meiosis. Biol. Reprod. 55:630-635. [DOI] [PubMed] [Google Scholar]
- 60.Walter, C. A., G. W. Intano, J. R. McCarrey, C. A. McMahan, and R. B. Walter. 1998. Mutation frequency declines during spermatogenesis in young mice but increases in old mice. Proc. Natl. Acad. Sci. USA 95:10015-10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walter, C. A., J. Lu, M. Bhakta, Z. Q. Zhou, L. H. Thompson, and J. R. McCarrey. 1994. Testis and somatic Xrcc-1 DNA repair gene expression. Somatic Cell Mol. Gen. 20:451-461. [DOI] [PubMed] [Google Scholar]
- 62.Wei, Y.-F., P. Robins, K. Carter, K. Caldecott, D. J. C. Pappin, G.-L. Yu, R.-P. Wang, B. K. Shell, R. A. Nash, P. Schar, D. E. Barnes, W. A. Haseltine, and T. Lindahl. 1995. Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and recombination. Mol. Cell. Biol. 15:3206-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Widen, S. G., and S. H. Wilson. 1991. Mammalian β-polymerase promoter: large-scale purification and properties of ATF/CREB palindrome binding protein from bovine testis. Biochemistry 30:6296-6305. [DOI] [PubMed] [Google Scholar]
- 64.Wilson, T. M., S. A. Rivkees, W. A. Deutsch, and M. R. Kelley. 1996. Differential expression of the apurinic/apyrimidinic endonuclease (Ape/Ref-1/ref-1) multifunctional DNA base excision repair gene during fetal development and in adult rat brain and testis. Mutat. Res. 362:237-248. [DOI] [PubMed] [Google Scholar]
- 65.Winn, R. N., M. B. Norris, K. J. Brayer, C. Torres, and S. L. Muller. 2000. Detection of mutations in transgenic fish carrying a bacteriophage λ cII transgene target. Proc. Natl. Acad. Sci. USA 97:12655-12660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou, Z. Q., and C. A. Walter. 1995. Expression of the DNA repair gene XRCC1 in baboon tissues. Mutat. Res. 348:111-116. [DOI] [PubMed] [Google Scholar]