Abstract
To shed light on the origin of adaptive immunity, a cDNA library was prepared from purified lymphocyte-like cells of a jawless vertebrate, the sea lamprey (Petromyzon marinus). Randomly selected cDNA clones were sequenced, and their homologies to proteins in the databases were determined. Of the sequences homologous to proteins involved in immune responses, five were selected for further characterization. Their encoding genes corresponded to loci that in jawed vertebrates are essential for activities of lymphocytes. These activities include regulation of T and B cell stimulation and proliferation (CD45); stabilization of molecular complexes involved in lymphocyte activation, adhesion, migration, and differentiation (CD9/CD81); adaptor functions in signaling leading to the activation of B lymphocytes (BCAP) and T lymphocytes (CAST); and amino acid transport associated with cell activation (CD98). The presence of these genes in the lamprey genome and their expression in lymphocyte-like cells support the notion that these cells perform many of the functions of gnathostome lymphocytes. It reopens the question of the stage jawless fishes reached in the evolution of their immune system.
Vertebrates are divided into two main groups—those without jaws and those with jaws, the agnathans and the gnathostomes, respectively (1). Jawless vertebrates dominated the seas of the Early Paleozoic 450–500 million years ago but are now represented by only two orders and fewer than 100 species: the Petromyzontiformes (the lampreys) and the Myxiniformes (the hagfishes) (2). The gnathostomes are a conspicuous component of present-day life, both aquatic and terrestrial. The transition from jawless to jawed vertebrates was occasioned by a number of evolutionary innovations, among which the appearance of the jaws and a full-blown adaptive (anticipatory) immune system may have been the most consequential (3). All animals are capable of protecting themselves against pathogens and parasites by mechanisms collectively referred to as a nonadaptive (nonanticipatory, innate) immune system (4). Each of these mechanisms involves different sets of molecules with recognition sites for components of a particular group of parasites. Jawed vertebrates added to the nonadaptive immune system an adaptive immune system that has the ability to anticipate any non-self-challenge by diversifying a limited set of specialized molecules. A central cell in the adaptive immune system is the lymphocyte. Gnathostome lymphocytes are derived from hemopoietic stem cells, which they share with other blood cells (5). They are distinguished by their morphology and behavior, which includes the ability to respond to stimuli by activation, transformation into blast cells, proliferation, migration, and differentiation into effector cells (4). These activities are effected by molecules, most of which are also expressed in other cells but in a different combination.
The definition of lymphocytes in jawless vertebrates is an unresolved issue (3). In an accompanying paper (6), we describe the isolation from the lamprey of a cell population that morphologically resembles mammalian lymphocytes. To characterize the expression profile of the purified lamprey lymphocyte-like cells, we isolated their RNA and sequenced a large number of cDNA clones obtained from it (6). From this collection of sequences we chose several for further characterization. Here we describe five lamprey genes, the mammalian counterparts of which play important roles in lymphocyte signaling and activation.
Materials and Methods
Animals and Cells.
Lymphocyte-like cells were isolated from the intestine of ammocoete larvae of the sea lamprey Petromyzon marinus by sorting in a flow cytometer on the basis of light-scatter characteristics (6). Light- and electron-microscopic evaluation revealed the isolated cells to resemble small lymphocytes of humans and mice (6).
Construction of cDNA Library.
Total RNA was extracted from 1 × 106 sorted cells with the help of the RNeasy mini kit (Qiagen, Hilden, Germany). Approximately 200–300 ng of the total RNA was reverse-transcribed by Power Script reverse transcriptase (BD Biosciences/CLONTECH, Heidelberg, Germany) and then subjected to 25 cycles of cDNA amplification by long-distance PCR using the SMART cDNA library-construction kit (BD Biosciences/CLONTECH). The cDNA was size-fractionated by chromatography on Chromaspin-400 columns, and fractions containing cDNA larger than 500 bp were pooled and ligated to the λTriplEx2 vector (BD Biosciences/CLONTECH). Ligated DNA was packaged into phage particles by using the Gigapack Gold in vitro packaging kit (Stratagene, Amsterdam), transfected into the Escherichia coli strain XL1-blue bacteria (BD Biosciences/CLONTECH), and plated. A total of 2 × 106 independent plaque-forming units was obtained.
Isolation and Analysis of cDNA Clones.
The unamplified library was plated at a dilution yielding single, well separated plaques. A total of 9,000 clones were picked manually and suspended in 500 μl of Lambda dilution buffer (100 mM NaCl/10 mM MgCl2/35 mM Tris⋅HCl, pH 7.5/0.01% gelatin). Insert DNA was PCR-amplified from 1 μl of the phage suspension in a 25-μl reaction volume by using the primer pair TriplEx2LD5 (5′-CTC GGG AAG CGC GCC ATT GTG TTG GT-3′) and TriplEx2LD3 (5′-ATA CGA CTC ACT ATA GGG CGA ATT GGC C-3′) and either the HotStar Taq polymerase (Qiagen) or the Expand long-template PCR system (Roche Diagnostics). Cycling conditions for short PCR in a PTC-200 Peltier thermal cycler (MJ Research and Biozym, Hessisch Oldendorf, Germany) were as follows: initial denaturation at 94°C for 2 min (Expand long-template PCR system) or at 95°C for 15 min (HotStar TaqDNA polymerase) followed by 10 cycles of denaturation at 94°C for 10 sec, primer annealing at 65°C for 30 sec, and elongation at 68°C (Expand long-template PCR system) or 72°C (HotStar TaqDNA polymerase) for 6 min. An additional 20 PCR cycles then were performed under the same conditions except that each elongation step was 20 sec longer than in the preceding cycle. The PCR was completed by a final elongation for 7 min. Long-PCR conditions were as follows: denaturation for 2 min at 92°C followed by 10 cycles of 10 sec at 92°C, 30 sec at 64°C, and 25 min at 68°C, and then by 20 cycles of 10 sec at 92°C, 30 sec at 64°C, and 25 min at 68°C (20-sec cycle elongation for each successive cycle) followed by 7 min at 68°C. The DNA was amplified in the GeneAmp PCR system 9700. The PCR products were purified and sequenced by the custom-sequencing service of MediGenomix by using the primer pTripl5Seq2 (5′-GAA GCG CGC CAT TGT GTT-3′) annealing next to the 5′ end of the insert cDNA. Rapid amplification of cDNA ends PCR was used to extend the cDNA clones.
Construction and Screening of Genomic Library.
Genomic DNA was isolated from the sea lamprey and digested partially with the Sau3A enzyme. The 5′-overhanging DNA ends were partially filled in with dATP and dGTP, and the fragments were ligated to the arms of a λ phage vector by using the lambda FIX II/XhoI partial fill-in vector kit (Stratagene). One million plaque-forming units of the library were plated and nitrocellulose replica filters (Schleicher & Schüll) were hybridized with specific probes by using the AlkPhos Direct hybridization system (Amersham Biosciences). The inserts were isolated and digested with appropriate restriction enzymes, and the fragments were ligated to the pUC18 and pGEM-11Zf(+) vectors (Promega). Long subcloned inserts were subcloned further as PCR fragments and sequenced by using the Thermo Sequenase primer cycle-sequencing kit (Amersham Biosciences) and the Li-Cor 4200 DNA sequencer (MWG Biotech, Ebersberg, Germany).
Southern Blot Hybridization.
Genomic DNA (5 μg) was digested with HindIII restriction endonuclease for 18 h under the conditions recommended by the supplier (Roche Diagnostics), and fragments were separated by agarose-gel electrophoresis and blotted onto Hybond N+ nylon filters (Amersham Biosciences). Prehybridization, hybridization, and probe labeling were carried out by using the AlkPhos Direct kit (Amersham Biosciences). DNA (100 ng) was used for the labeling of the probe. After overnight hybridization, the filters were washed according to the AlkPhos Direct protocol. After the application of the chemiluminescent detection reagent CDP-Star of the Hyperfilm ECL kit (Amersham Biosciences) was exposed to the blot for 6 h and developed.
Phylogenetic Analysis.
Sequences were aligned by using SEQPUP 0.6f software for Macintosh (ref. 7, http://iubio.bio.indiana.edu/soft/molbio). Variability of the sequences was assessed with the help of the MEGA 2.1 program (8) by using synonymous and nonsynonymous substitutions estimated by the Nei and Gojobori method (9) for exons and Kimura's two-parameter distances (10) for introns. Phylogenetic trees drawn by the neighbor-joining method (11) using the PAUP* 4.0b10 program (12) were based on P distances for amino acid sequences and Kimura's two-parameter distances for nucleotide sequences. Trees were rooted at midpoint. Maximum-parsimony trees were drawn by the same program using the heuristic search algorithm. Gaps were treated as missing data. The topological stability of the trees was assessed by 500 bootstrap replications.
Results
Total RNA isolated from purified lamprey lymphocyte-like cells was converted into cDNA, which was used to prepare a cDNA library. Clones randomly picked from the library were sequenced partially, and the nucleotide sequences were translated into amino acid sequences and then subjected to blastp searches to ascertain matches to proteins in the databases. Interesting clones identified in this manner were selected for further analysis. The sequences were extended by rapid amplification of cDNA ends PCR, isolation and sequencing of overlapping cDNA clones, and long PCR of genomic DNA templates or of cloned genomic fragments identified by Southern blotting and hybridization. Exon–intron borders were determined by PCR amplification of genomic DNA with primers placed in exons expected to flank an intron or by long PCRs designed to amplify either the entire gene or overlapping segments of a gene. The amino acid sequences then were subjected to phylogenetic analysis aimed at establishing their homologies to known gnathostome proteins. Numerous orthologies to proteins/genes expressed in lymphocytes of jawed vertebrates were identified in this manner. The description of five homologs of immunologically relevant genes follows. The lamprey genes/proteins are referred to by the names of their human counterparts.
CD45 (PTPRC).
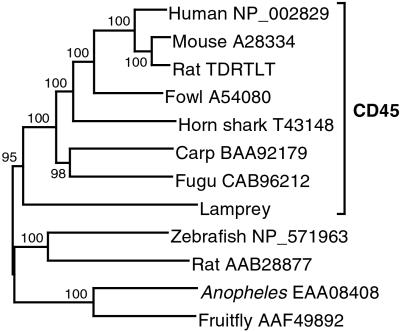
Human CD45, a member of the receptor-type protein tyrosine phosphatase (PTP) family, is a large protein composed of 1,281 amino acid residues in its mature form (13, 14). A single transmembrane (TM) segment divides the molecule into two large parts, the N-terminal extracellular (EC) domain (576 residues) and the C-terminal cytoplasmic part (683 residues). The translated sequence of the original lamprey cDNA clone identified as being homologous to human CD45 covered only a short segment of the C-terminal part, but the sequence was extended later by the analysis of six overlapping clones. The continuity of this sequence was confirmed by analysis of lamprey genomic DNA (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). Although the sequence of the lamprey cytoplasmic part aligns well with the known CD45 proteins, that of the EC part is virtually unalignable, and the exon–intron organization of the corresponding part of the gene also deviates from that of the CD45-encoding human PTPRC gene (ref. 15, and data to be published elsewhere). Nevertheless, phylogenetic analysis based on the well alignable C-terminal part clearly indicates that the lamprey sequence is orthologous to the mammalian CD45 sequence (Fig. 1) rather than to some other member of the PTP family. This conclusion is supported also by the similarity in the overall structural organization of the lamprey and human proteins: the length of the molecules, the organization and size of the domains, the placement of the putative TM segment, and the high number of cysteine residues (16 in human and 13 in lamprey) as well as the abundance of putative N-linked glycosylation sites (9 in human and 6 in lamprey) in the EC part of both molecules. These similarities also argue against the possibility that the lamprey sequence is an evolutionary chimera produced by coupling the C-terminal part of CD45 to an unrelated protein. More likely, the two parts of the molecule evolved under different selection pressures. Although the need to retain constancy of the catalytic sites constrains variation in the C-terminal part, the N-terminal part may be free of such restrictions. On the contrary, the divergence of the N-terminal part may be promoted by the necessity to coevolve with other proteins with which it might interact. The documented occurrence of human CD45 isoforms differing in the length of their EC regions and produced by alternative splicing (16) supports the contention that the function of this domain requires structural flexibility. Such a form of differential evolution, whereby one part of a molecule remains highly conserved while another part has diverged beyond recognition in distant taxa, has also been documented for other proteins (17, 18). Furthermore, the divergence of the CD45 N-terminal domain is not restricted to comparisons between agnathan and gnathostome proteins; the shark CD45 sequence (see Fig. 6) is similarly unalignable with the mammalian proteins in this part of the molecule. The high degree of sequence conservation in the catalytic part indicates that the enzymatic activity of the CD45 protein and hence its principal function is similar in the lamprey and humans.
Fig 1.
Neighbor-joining tree of CD45 sequences and related PTPs. The tree is based on the amino acid residues 906–972 and 1,104–1,661 of the alignment in Fig. 6. Numbers on the branches indicate bootstrap values. The tree is rooted at midpoint. The sequences are designated by the species name followed by the GenBank accession number. The highest scoring non-CD45 PTPs in a blastp search were chosen as outgroup sequences. Zebrafish NP_571963, PTP, receptor type A; rat AAB28877, receptor PTP-σ; fruit fly AAF49892 and Anopheles EAA08408 are translated from genes obtained from the genome-sequencing projects.
B Cell Adaptor for Phosphoinositide 3-Kinase (BCAP).
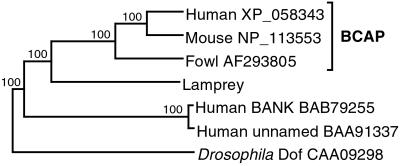
BCAP is a protein with a sequence that has been deduced from cDNA clones recently identified in the domestic fowl and house mouse (19). We obtained the corresponding human sequence from the database of the human genome sequencing project (GenBank accession nos. AL358235 and NT_029394) and used it to infer the exon–intron organization of the encoding gene. The three sequences are distantly related to the Drosophila Dof protein, which is essential for signal transmission by the fibroblast growth factor receptor (20). The human genome, however, also contains two other, more distantly related sequences: B cell scaffold protein with ankyrin repeats (BANK; ref. 21) and an as-yet-unnamed sequence BAA91337 (Fig. 2). The lamprey cDNA library yielded a clone with a sequence that, when translated into amino acids, was 37% identical with that of the fowl protein (Fig. 7, which is published as supporting information on the PNAS web site). The lamprey BCAP protein displays all the motifs that characterize the fowl, mouse, and human sequences (Fig. 7): the ankyrin repeat region extending between residues 336 and 404 (in the fowl-sequence numbering), the coiled-coil region (residues 636–666), the proline-rich segment (residues 436–453), and at least three of the six potential tyrosine phosphorylation sites found in the fowl sequence—one YXXN site (Tyr-378) for GRB2 and two YXXM sites (Tyr-423 and Tyr-448) for phosphoinositide 3-kinase. On the phylogenetic tree of available BCAP and related sequences, the lamprey sequence assumes the position in the BCAP clade expected on the basis of its taxonomical origin (Fig. 2).
Fig 2.
Neighbor-joining tree of BCAP protein sequences. The tree is based on the amino acid alignment in Fig. 7. Numbers on the branches indicate bootstrap values. The tree is rooted at midpoint. The sequences are designated by the species name followed by the GenBank accession number.
CD3ɛ-Associated Signal Transducer (CAST).
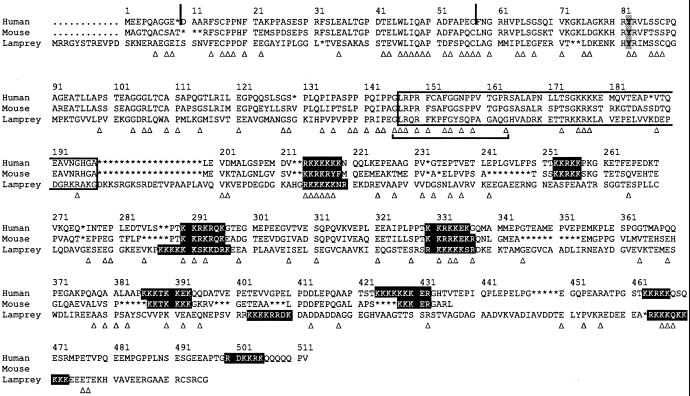
CAST (22) was identified originally as an ORF positioned anti-sense to the ERCC1 gene in humans and yeast (23) and hence is also named ASE1 (24), and independently as nucleolin (25), a ubiquitous nucleolar protein. The protein sequence translated from the lamprey cDNA sequence shows 21% identity with the human CAST protein (Fig. 3). The first in-frame methionine codon at the 5′ end of the lamprey sequence is 13 positions upstream from that of the human sequence. The lamprey and human CASTs share a tyrosine at position 82 (human sequence numbering), which in the human protein is believed to be part of a half-immunoreceptor tyrosine-based activation motif (ITAM) and the signal for the interaction with the μ2 chain of the adaptor AP-2 complex. In the lamprey, however, the motif is YRIMSS rather than YRVLSS as it is in the human protein, and thus it does not conform to the YXXL form thought to be required for the motif's functionality. The region of the lamprey sequence corresponding to that identified by Yamazaki et al. (22) as being the minimum required for binding with CD3ɛ (residues 147–198) is conserved only in its 5′ part. The conserved part (residues 146164) is rich in glycine, arginine, and phenylalanine (= GRFrich region), which forms β-turns and is believed to be necessary for interaction with nucleic acids. Of the eight lysine/arginine-rich segments in the human sequence (22), five are also present in the lamprey sequence. The alternating basic and acidic residues are thought to provide a nucleolar-localization signal for the protein.
Fig 3.
Alignment of the human, mouse, and lamprey CAST protein sequences. Amino acid residues are designated in the International Union of Pure and Applied Chemistry single-letter code. Exon–intron borders are indicated by vertical lines. Asterisks indicate indels introduced to optimize the alignment. The CD3ɛ-binding region is enclosed in an open box, and the lysine/arginine-rich segments enclosed in filled boxes. The conserved Tyr-82 is shaded. The GRF-rich region is indicated by a bracket, and shared residues between human/mouse and lamprey are indicated by open triangles. Accession numbers: NP_036231, human CAST; NP_665821, mouse CAST.
CD9.
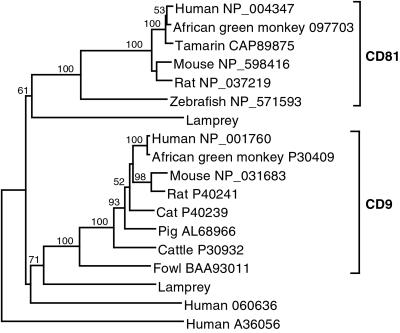
The mammalian CD9 is a member of a large tetraspanin protein family, which in humans has 28 members (26). The proteins share four conserved membrane-spanning regions and a set of sequence motifs. The different members of the family function by associating with various proteins and thus influencing their functions in signal transduction, activation, adhesion, migration, and differentiation. We found two clones in the lamprey cDNA library, which a blastx search identified as members of the tetraspanin family (Fig. 8, which is published as supporting information on the PNAS web site). Phylogenetic analysis of the clones identified them as members of the subfamily that includes mammalian CD9, CD81, and Tspan2 sequences, although the low bootstrap values make the assignment of the lamprey sequences ambiguous. The two lamprey proteins translated from the cDNA sequences share some residues with gnathostome CD81 but more residues are shared with the CD9 group (Fig. 8). The analysis thus assigns them to the CD9 cluster and suggests that they diverged from each other within the lamprey lineage after its divergence from the gnathostome lineage (Fig. 4). The proteins are 228 and 222 residues long and 36% identical in their sequence. An alignment with other members of the CD9/CD81 subfamily (Fig. 8) as well as secondary structure predictions (not shown) suggest that the proteins contain four TM regions (TM1–TM4), two EC segments (short EC1 between TM1 and TM2 and long EC2 between TM3 and TM4), a very short intracellular segment (between TM2 and TM3), and short intracellular N and C termini. The lamprey EC2 is 84 (79) residues long and contains four cysteine residues, which, in analogy with the structure of the human CD81 EC2 domain (27), in all probability form two disulfide bonds. It is presumably the EC2 domain via which the CD9 molecules interact with other proteins and thus exert their influence on cellular functions. Except for the conserved cysteines and a few other widely shared residues, the sequences of the lamprey EC2 domains are quite different from those of their mammalian counterparts. This difference perhaps is not surprising, because it presumably reflects the requirement to interact with other lamprey proteins that can be expected to be very different from their mammalian homologs.
Fig 4.
Neighbor-joining tree of CD9 and CD81 proteins. The tree is based on the amino acid alignment in Fig. 8. Numbers on the branches indicate bootstrap values. The tree is rooted at midpoint. The sequences are designated by the species name followed by the GenBank accession number. O60636 and A36056 are the human Tspan2 and CO-029 sequences, respectively. Both are tetraspanins related to CD9 and CD81, and they were used as outgroups in the phylogenetic analysis.
CD98 (SLC3A2).
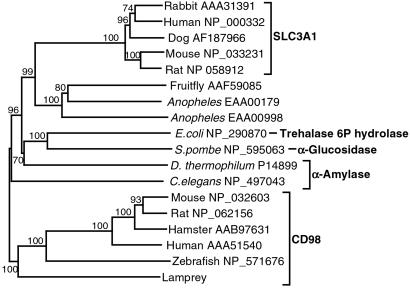
In the human nomenclature, CD98 is a designation used for the heavy chain of amino acid transporters; the gene encoding the heavy chain is denoted solute carrier family 3, member 2 or SLC3A2 (28). The ORF of the lamprey cDNA sequence translates into 524 amino acid residues, as compared with 529 residues of the human protein (29). The lamprey protein is identical with its human counterpart at 32.7% of the positions (Fig. 9, which is published as supporting information on the PNAS web site). The shared residues are scattered mostly along the entire sequence, but some are clustered into largely invariant blocks. Particularly conspicuous is the block encompassing positions 102–124, which in the human sequence has been identified as the TM domain. This location of the domain suggests that the N terminus of the protein is located intracellularly and the C terminus extracellularly, as has been demonstrated for the human CD98 heavy chain (30). Hence the chain is a type II protein. The invariant Cys-129 is involved in the formation of a disulfide bond that associates the heavy chain with one of several light chains encoded in separate genes. By contrast, Cys-474, which is shared by all known mammalian C98 heavy chains, is replaced in the lamprey by isoleucine (and in the zebrafish by leucine). The lamprey sequence contains nine potential N-glycosylation sites, of which one is shared with one of the sites found in the human sequence. A phylogenetic tree drawn for the available protein sequences confirms the orthology of the lamprey and human CD98 heavy chain sequences (Fig. 5).
Fig 5.
Neighbor-joining tree of CD98 proteins from different species. The tree is based on positions 136–800 of the amino acid alignment in Fig. 9. Numbers on the branches indicate bootstrap values. The tree is rooted at midpoint. The sequences are designated by the species name followed by the GenBank accession number. E. coli, Escherichia coli; S. pombe, Schizosaccharomyces pombe; D. thermophilum, Dictyoglomus thermophilum; C. elegans, Caenorhabditis elegans.
Discussion
The five kinds of lamprey sequences described in this study have two features in common. First, they all are expressed in cells that resemble mammalian lymphocytes morphologically (6). Second, all their mammalian counterparts are involved in lymphocyte activation. CD45, BCAP, and CAST participate directly in the transduction of signals generated by lymphocyte stimulation, whereas CD9 and CD98 participate in processes such as proliferation, adhesion, and migration, which follow the activation. Mammalian CD45 is a PTP located in a lipid raft that is mobilized after the engagement of the T cell receptor (TCR) by the major histocompatibility complex-peptide assemblies (31, 32). The coalescence of CD45- and TCR/CD3-bearing rafts enables CD45 to dephosphorylate TCR/CD3-associated Srk-family protein tyrosine kinases and thus activate them. The activated protein tyrosine kinases then phosphorylate a host of proteins and thus initiate the signaling pathway. Mammalian BCAP is one of the adaptor molecules (19, 33)—proteins that lack enzymatic or transcriptional activity but express a variety of binding sites enabling them to mediate interactions between proteins of the signaling network. In a B lymphocyte, protein tyrosine kinases activated by the interaction of the B cell receptor with an antigen phosphorylate a tyrosine residue of cytosolic BCAP molecules and thus unmask on them binding sites for phosphatidylinositol 3-kinase. Binding of BCAP to phosphatidylinositol 3-kinase up-regulates the activity of this enzyme and directs it to CD19-bearing lipid rafts rich in phosphatidylinositol (4,5)-bisphosphate [PtdIns (4,5)P2], the enzyme's target, thus initiating another signaling pathway. CAST is also an adaptor protein, constitutively associated with the ɛ chain of the TCR–CD3 complex (22). The engagement of the TCR–CD3 by the major histocompatibility complex-peptide assembly leads to phosphorylation of the CAST molecule, enabling it to bind to proteins in the pathway leading to the activation of the interleukin-2 and other genes. The mammalian CD9 molecule (26, 34) has been shown to associate with integrins, enzymes involved in signal transduction, and a variety of molecules of the immune system (major histocompatibility complex, CD3, CD4, CD19, and others). These associations enable it to influence the function of the activated cells. Finally, CD98 was described originally as a T cell-activation antigen (35) and later shown to possess at least two functions. First, by associating with different light chains it regulates, via its EC domain, the expression and cellular localization of amino acid transport activities and thus of cell proliferation (36, 37). Second, it interacts physically and functionally, via its TM and intracellular domains, with integrin adhesion receptors and through them influences stimulation of T lymphocytes and other cells (38).
The expression of the five kind of proteins is not restricted to lymphocytes. CD45 is synthesized in all hemopoietic cells except erythrocytes and platelets (31). BCAP has been demonstrated in B lymphocytes and macrophages of a variety of tissues (19). CAST is expressed in a wide variety of tissues, in both the nucleus (nucleolus) and cytoplasm of the cells (22, 39). CD9 and CD98 are also ubiquitously expressed (28, 34). Nevertheless, the expression of these molecules in lymphocytes is essential for the normal function of the immune system. Thus, for example, in humans the failure to express CD45 results in a life-threatening immunodeficiency (40). These observations do not necessarily imply that the molecules carry out the same immunological functions in the lamprey lymphocyte-like cells as they do in mammalian lymphocytes. They do support, however, the notion that the lamprey cells indeed are lymphocyte-like. In addition to the five proteins, the lamprey lymphocyte-like cells also express Spi (18) and Ikaros-family proteins (41), PSMB7 (42), ABCB9 (T.U.-o., W.E.M., M.D.C., and J.K., unpublished results), MIF (A.S., T.U.-o., N. Kuroda, W.E.M., N. Takezaki, R.D., F. Figueroa, M.D.C., and J.K., unpublished results), receptors for small inducible cytokines (N. Kuroda, T.U.-o., A.S., I. E. Samonte, F. Figueroa, W.E.M., and J.K., unpublished results), and other proteins (6), all of which also are expressed by mammalian lymphocytes. Thus lamprey cells are not only morphologically but also genetically and evolutionarily closely related to gnathostome lymphocytes.
Note Added in Proof.
While the manuscript was processed for publication, an article describing a CD45 sequence of another agnathan, the Pacific hagfish, appeared (43).
Supplementary Material
Acknowledgments
We thank Ms. Jane Kraushaar for editorial assistance and Ms. Heike Hausmann and Ms. Sabine Rosner for technical assistance; Dr. James G. Seeley for supplying us with lampreys; and Dr. Herbert Tichy for his help and advice.
Abbreviations
PTP, protein tyrosine phosphatase
TM, transmembrane
EC, extracellular
BCAP, B cell adaptor for phosphoinositide 3-kinase
CAST, CD3ɛ-associated signal transducer
TCR, T cell receptor
References
- 1.Pough F. H., Janis, C. M. & Heiser, J. B., (1998) Vertebrate Life (Prentice–Hall, Upper Saddle River, NJ).
- 2.Paxton J. R. & Eschmeyer, W. N., (1994) Encyclopedia of Fishes (Univ. of New South Wales Press, Sydney).
- 3.Klein J., Sato, A. & Mayer, W. E. (2000) in Major Histocompatibility Complex: Evolution, Structure, and Function, ed. Kasahara, M. (Springer, Tokyo), pp. 3–26.
- 4.Klein J. & Horejsi, V., (1997) Immunology (Blackwell Scientific, Oxford).
- 5.Kondo M., Scherer, D. C., King, A. G., Manz, M. G. & Weissman, I. L. (2001) Curr. Opin. Genet. Dev. 11, 520-526. [DOI] [PubMed] [Google Scholar]
- 6.Mayer W. E., Uinuk-ool, T., Tichy, H., Gartland, L. A., Klein, J. & Cooper, M. D. (2002) Proc. Natl. Acad. Sci. USA 99, 14350-14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert, D. G. (1996) SEQPUP, A Biosequence Editor and Analysis Application, Version 0.6f, http://iubio.bio.indiana.edu/soft/molbiol.
- 8.Kumar S., Tamura, K., Jakobsen, I. B. & Nei, M., (2001) mega 2, Molecular Evolutionary Genetic Analysis Software (Arizona State Univ., Tempe).
- 9.Nei M. & Gojobori, T. (1986) Mol. Biol. Evol. 3, 418-426. [DOI] [PubMed] [Google Scholar]
- 10.Kimura M. (1980) J. Mol. Evol. 16, 111-120. [DOI] [PubMed] [Google Scholar]
- 11.Saitou N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406-425. [DOI] [PubMed] [Google Scholar]
- 12.Swofford D. L., (2002) PAUP*, Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 4.
- 13.Thomas M. L. (1989) Annu. Rev. Immunol. 7, 339-369. [DOI] [PubMed] [Google Scholar]
- 14.Fischer E. H., Charbonneau, H. & Tonks, N. K. (1991) Science 253, 401-406. [DOI] [PubMed] [Google Scholar]
- 15.Hall L. R., Streuli, M., Schlossman, S. F. & Saito, H. (1988) J. Immunol. 141, 2781-2787. [PubMed] [Google Scholar]
- 16.Ralph S. J., Thomas, M. L., Morton, C. C. & Trowbridge, I. S. (1987) EMBO J. 6, 1251-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toyosawa S., O'hUigin, C., Tichy, H. & Klein, J. (1999) Gene 234, 307-314. [DOI] [PubMed] [Google Scholar]
- 18.Shintani S., Sato, A., O'hUigin, C. & Klein, J. (2000) J. Mol. Evol. 51, 363-373. [DOI] [PubMed] [Google Scholar]
- 19.Okada T., Maeda, A., Iwamatsu, A., Gotoh, K. & Kurosaki, T. (2000) Immunity 13, 817-827. [DOI] [PubMed] [Google Scholar]
- 20.Vincent S., Wilson, R., Coelho, C., Affolter, M. & Leptin, M. (1998) Mol. Cell 2, 515-525. [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama K., Su, I., Tezuka, T., Yasuda, T., Mikoshiba, K., Tarakhovsky, A. & Yamamoto, T. (2002) EMBO J. 21, 83-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamazaki T., Hamano, Y., Tashiro, H., Itoh, K., Nakano, H., Miyatake, S. & Saito, T. (1999) J. Biol. Chem. 274, 18173-18180. [DOI] [PubMed] [Google Scholar]
- 23.Van Duin M., Van Den Tol, J., Hoeijmakers, H. H. J., Bootsma, D., Rupp, I. P., Reynolds, P., Prakash, L. & Prakash, S. (1989) Mol. Cell. Biol. 9, 1794-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin-Gallardo A., McCombie, W. R., Gocayne, J. D., FitzGerald, M. G., Wallace, S., Lee, B. M. B., Lamerdin, J., Trapp, S., Kelly, J. M. & Liu, L. (1992) Nat. Genet. 1, 34-39. [DOI] [PubMed] [Google Scholar]
- 25.Lapeyre B., Caizergues-Ferrer, M., Bouche, G. & Amalric, F. (1985) Nucleic Acids Res. 13, 5805-5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boucheix C. & Rubinstein, E. (2001) Cell. Mol. Life Sci. 58, 1189-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitadokoro K., Bordo, D., Galli, G., Petracca, R., Falugi, F., Abrignani, S., Grandi, G. & Bolognesi, M. (2001) EMBO J. 20, 12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devés R. & Boyd, C. A. R. (2000) J. Membr. Biol. 173, 165-177. [DOI] [PubMed] [Google Scholar]
- 29.Lumadue J. A., Glick, A. B. & Ruddle, F. H. (1987) Proc. Natl. Acad. Sci. USA 84, 9204-9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fenczik C. A., Zent, R., Dellos, M., Calderwood, D. A., Satriano, J., Kelly, C. & Ginsberg, M. H. (2001) J. Biol. Chem. 276, 8746-8752. [DOI] [PubMed] [Google Scholar]
- 31.Justement L. B. (1997) Adv. Immunol. 66, 1-65. [DOI] [PubMed] [Google Scholar]
- 32.Alexander D. R. (2000) Semin. Immunol. 12, 349-359. [DOI] [PubMed] [Google Scholar]
- 33.Yamazaki T., Takeda, K., Gotoh, K., Takeshima, H., Akira, S. & Kurosaki, T. (2002) J. Exp. Med. 195, 535-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berditchevski F. (2001) J. Cell Sci. 114, 4143-4151. [DOI] [PubMed] [Google Scholar]
- 35.Haynes B. F., Hemler, M. E., Mann, D. L., Eisenbarth, G. S., Shelhamer, J., Mostowski, H. S., Thomas, C. A., Strominger, J. L. & Fauci, A. S. (1981) J. Immunol. 126, 1409-1414. [PubMed] [Google Scholar]
- 36.Mastroberardino L., Spindler, B., Pfeiffer, R., Skelly, P. J., Loffing, J., Shoemaker, C. B. & Verrey, F. (1998) Nature 395, 288-291. [DOI] [PubMed] [Google Scholar]
- 37.Torrents D., Estevez, R., Pineda, M., Fernandez, E., Lloberas, J., Shi, Y. B., Zorzano, A. & Palacin, M. (1998) J. Biol. Chem. 273, 32437-32445. [DOI] [PubMed] [Google Scholar]
- 38.Fenczik C. A., Sethi, T., Ramos, J. W., Hughes, P. E. & Ginsberg, M. H. (1997) Nature 390, 81-85. [DOI] [PubMed] [Google Scholar]
- 39.Ginisty H., Sicard, H., Roger, B. & Bouvet, P. (1999) J. Cell Sci. 112, 761-772. [DOI] [PubMed] [Google Scholar]
- 40.Kung C., Pingel, J. T., Heikinheimo, M., Klemola, T., Varkila, K., Yoo, L. I., Vuopala, K., Poyhonen, M., Uhari, M., Rogers, M., et al. (2000) Nat. Med. 6, 343-345. [DOI] [PubMed] [Google Scholar]
- 41.Mayer W. E., O'hUigin, C., Tichy, H., Terzic, J. & Saraga-Babic, M. (2002) Scand. J. Immunol. 55, 162-170. [DOI] [PubMed] [Google Scholar]
- 42.Takezaki N., Zaleska-Rutczynska, Z. & Figueroa, F. (2002) Gene 282, 179-187. [DOI] [PubMed] [Google Scholar]
- 43.Nagata T., Suzuki, T., Ohta, Y., Flajnik, M. F. & Kasahara, M. (2002) Immunogenetics 54, 286-291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.