Abstract
The organization and allelic recombination of the merozoite surface protein-1 gene of Plasmodium vivax (PvMsp-1), the most widely prevalent human malaria parasite, were evaluated in complete nucleotide sequences of 40 isolates from various geographic areas. Alignment of 31 distinct alleles revealed the mosaic organization of PvMsp-1, consisting of seven interallele conserved blocks flanked by six variable blocks. The variable blocks showed extensive variation in repeats and nonrepeat unique sequences. Numerous recombination sites were distributed throughout PvMsp-1, in both conserved blocks and variable block unique sequences, and the distribution was not uniform. Heterozygosity of PvMsp-1 alleles was higher in Asia (0.953 ± 0.009) than in Brazil (0.813 ± 0.047). No identical alleles were shared between Asia and Brazil, whereas all but one variable block nonrepeat sequence found in Brazil occurred in Asia. These observations suggest that P. vivax populations in Asia are ancestral to Brazilian populations, and that PvMsp-1 has heterogeneity in frequency of allelic recombination events. Recurrent origins of new PvMsp-1 alleles by repeated recombination events were supported by a rapid decline in linkage disequilibrium between pairs of synonymous sites with increasing nucleotide distance, with little linkage disequilibrium at a distance of over 3 kb in a P. vivax population from Thailand, evidence for an effectively high recombination rate of the parasite. Meanwhile, highly reduced nucleotide diversity was noted in a region encoding the 19-kDa C-terminal epidermal growth factor-like domain of merozoite surface protein-1, a vaccine candidate.
The human malaria parasite Plasmodium vivax is prevalent worldwide, and accounts for 70–80 million cases annually, mostly in Asia and Latin America (1). Growing resistance of P. vivax strains to chloroquine is spurring the development of a vaccine against P. vivax malaria. One current vaccine candidate is merozoite surface protein-1 (MSP-1), a 200-kDa protein expressed on the surface of the P. vivax merozoite (2). MSP-1 of Plasmodium species is synthesized as a high-molecular-weight precursor and then processed into several fragments (3). At the time of red cell invasion by the merozoite, only the 19-kDa C-terminal fragment (MSP-119), which contains two epidermal growth factor-like domains, remains on the surface. Antibodies against MSP-119 inhibit merozoite entry into red cells (4), and immunization with MSP-119 protects monkeys from challenging infections (5, 6). Hence, MSP-119 is considered a promising vaccine candidate.
Importantly, there is extensive allelic diversity of MSP-1 among isolates (7), and this polymorphism may hamper development of effective vaccines. In Plasmodium falciparum, the most virulent malaria parasite, polymorphism in PfMsp-1 is well characterized. PfMsp-1 consists of several interallele variable blocks flanked by conserved or semiconserved blocks. Variation in this gene is basically dimorphic; i.e., one or the other of two different residues (8, 9). Meiotic recombination in the anopheline mosquito is the major mechanism for allelic variation of PfMsp-1 (8); thus, intragenic recombination between unlike alleles generates new alleles in the progeny (10). Recombination sites are confined to the 5′ and 3′ regions of the gene. Limited sequences suggest similar patterns of allelic recombination of P. vivax Msp-1 (PvMsp-1) (11–13). However, to date, only two complete sequences from monkey-adapted strains of P. vivax have been available (14, 15). Consequently, the overall picture of allelic recombination of PvMsp-l, as well as its structural organization, remains unclear.
P. vivax and P. falciparum are distantly related in phylogeny, with a divergence time of 75–165 million years (Myr) ago (16), greatly predating the origin of hominids. P. vivax and the related monkey malaria parasite species Plasmodium cynomogi and Plasmodium knowlesi diverged 11.7–30.5 Myr ago, whereas P. falciparum and the closely related chimpanzee malaria parasite Plasmodium reichenowi diverged 5–8 Myr ago (16). The genomic compositions of these species also differ greatly: the A+T content in the protein-coding region is ≈75% in P. falciparum/P. reichenowi and ≈60% in P. vivax/P. cynomolgi/P. knowlesi. These facts suggest that the evolutionary history of polymorphism of Msp-1 differs greatly between P. falciparum and P. vivax. Estimating frequency of allelic recombination in Msp-1 has important implications for understanding the evolution of its polymorphism. Because this locus encodes a surface antigen targeted by host immune responses (17), selective pressure is likely to operate to maintain polymorphism in Msp-1. Under selective pressure, the level of polymorphism would be affected by frequency of allelic recombination. Within a locus with a high recombination rate, polymorphic sites separated by a relatively short distance segregate independently at meiosis and therefore may have different evolutionary histories. In the present study, we examined the organization of PvMsp-1 and the frequency of its allelic recombination by analyzing complete nucleotide sequences of 40 isolates of diverse geographic origin. Here, we show that PvMsp-1 has mosaic organization with heterogeneity in frequency of allelic recombination. Further, we present evidence for an effectively high recombination rate of P. vivax.
Materials and Methods
Isolates.
Initially, 61 clinical isolates of P. vivax, diagnosed by microscopy on Giemsa-stained blood smears, were included. After informed consent was obtained, blood was sampled from patients as 1-ml samples in EDTA or as 75-μl samples spotted on filter paper. P. vivax DNA was extracted by using the Wizard Genomic DNA Purification kit (Promega) or a standard chloroform/phenol method. Of 61 isolates, 40 had a single PvMsp-1 allele, as determined by preliminary PCR-based haplotyping (not shown); these 40 isolates were sequenced. The sources of these 40 samples were as follows: 19 isolates from Tak Province, northern Thailand (1997 and 1998); one isolate (TV400) from a hospital in Bangkok (1997) (all 20 Thai isolates were assigned a “T” prefix before their isolate number); eight isolates from Rondonia, northwestern Brazil (1995 and 1997) (a “BR” or “BP” prefix); five isolates from Cox Bazai village in Bangladesh (1994) (a “BD” prefix); four isolates (SK-1 to SK-4) from Kyongi-do, South Korea (1998); two isolates (VM278 and VM55) from Malakula, Vanuatu (1996 and 1998); and one isolate (IN1) from a Japanese patient who traveled to India in 1999. The genomic DNA of monkey-adapted strains of P. vivax (Sal-1 and Belem strains) were used as reference sequences.
Sequencing.
The entire PvMsp-1 (5.1–5.3 kb) was amplified by PCR (18) by using primers PVF0 and PVR0 (which are published as Table 2 in the supporting information on the PNAS web site, www.pnas.org). PCR products were purified by using a Pharmacia Micro Spin S400 HR. DNA sequencing was performed from both directions by using the dRhodamine Terminator Cycle Sequencing kit (Applied Biosystems) in an ABI 310 DNA sequencer (Applied Biosystems). The sequencing primers used are published in Table 2. Two independent PCR products were sequenced for each isolate. Wherever singleton polymorphism occurred, sequencing was repeated after another independent PCR. The rate of sequence error was 1.4 × 10−5 per base; thus, duplication of sequencing reduced the rate to 2.0 × 10−10. We confirmed the Sal-1 PvMsp-1 sequence (15) but found multiple sites in the Belem sequence inconsistent with the reported sequence (14). The reported sequence most likely contains artifacts, as indicated by others (19). Hence, we used our Belem sequence for analysis.
Sequence Alignment.
Sequences were aligned by using the clustal w program (20) and GENETYX MAC 10.1.2. Nucleotide diversity (π) was calculated by using a sliding window of 100 bases, with a step size of 20 bases. Boundaries of conserved and variable blocks were determined from amino acid sequence homology. Subsequently, π and its standard deviation were computed for each block, excluding alignment gaps, by using DnaSP 3.52 (21). Due to size polymorphism in variable blocks, nucleotide distance was referred to after multiple sequence alignment. To localize interspecies conserved residues, PvMsp-1 alleles were aligned with Msp-1 sequences from four other Plasmodium species: three rodent species (Plasmodium yoelii, Plasmodium berghei, and Plasmodium chabaudi; GenBank accession nos. J04668, U43521, and M34947) and two alleles of P. falciparum (K1 and MAD20 strains; X03371, X05624). Partial Msp-1 sequences from P. cynomolgi (U25743) and P. knowlesi (AF298219) were also compared with PvMsp-1.
Statistical Analysis.
The minimum number of recombination events (Rm) was estimated by the four-gamete test (22) by using DnaSP 3.52. Informative polymorphic sites were used to compute Rm after excluding singleton sites and those sites segregating for more than three different nucleotides. Alignment in variable blocks was not completely reliable, because sequence variation was extensive due to insertions/deletions and varying numbers of degenerating short tandem repeats, in addition to point mutations. Therefore, we did not calculate Rm in variable blocks. Evidence for recombination in variable blocks was obtained by visually inspecting sequence comparisons. Linkage disequilibrium between polymorphic sites within PvMsp-1 was tested by the R2 index (23), by using DnaSP 3.52; variable blocks and noninformative sites were excluded from analysis. Statistical significance was set at P < 0.05 for the R2 index for each pair of polymorphic sites by the two-tailed Fisher's exact test. Heterozygosity (h) was calculated as described (24). The number of synonymous substitutions per synonymous site, and that of nonsynonymous substitutions per nonsynonymous site were computed by using Nei and Gojobori's method (25), with the modification of Zhang et al. (26), with MEGA 2.1 (27).
Results
Mosaic Organization of PvMsp-1.
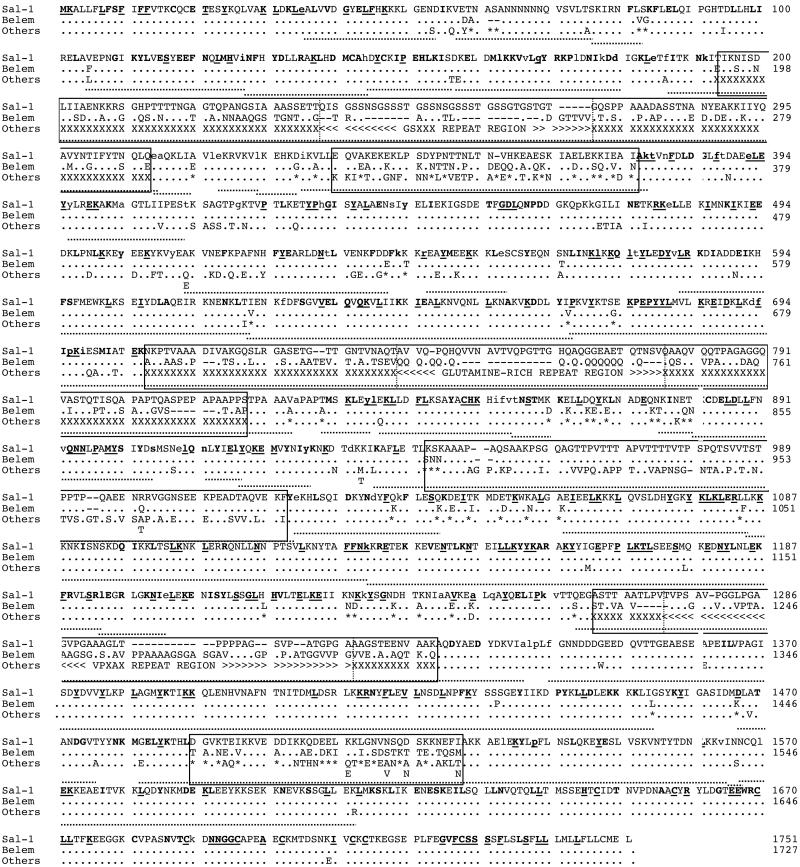
We obtained 31 distinct PvMsp-1 alleles from 40 isolates. Alignment of their deduced amino acid sequences, together with the Sal-1 and Belem sequences, revealed that PvMsp-1 consists of six interallele variable blocks (445 aa) flanked by seven conserved blocks (1,306 aa) (Fig. 1). Nucleotide diversity (π) varied from 0.0023 ± 0.0003 to 0.0495 ± 0.0025 in conserved blocks and from 0.1193 ± 0.0178 to 0.2055 ± 0.0064 in variable blocks (which are published as Fig. 5 in the supporting information on the PNAS web site). Conserved blocks contain 211 nucleotide substitutions with 86 synonymous changes, with the lowest value of π in conserved block 13. As in PfMsp-1, nucleotide substitutions are principally dimorphic (i.e., one or the other of two different nucleotides), with four exceptions (trimorphic). In addition, we confirmed by Southern blot hybridization that each haploid genome contains a single copy of PvMsp-1 (not shown), as previously shown for PfMsp-1 (8). Variable blocks show extensive sequence variations consisting of a number of substitutions, insertions/deletions and varying numbers of short tandem repeats (which are published as Table 3 in the supporting information on the PNAS web site). Of the six variable blocks, four possess apparent tandem repeats: block 2, degenerating 5-mer (GSXXX)n=1–9 (X = any residue, n = the number of repeats); block 6, poly(Q)n=10–21; block 8, degenerating repeats of (PVTTTX)n=2 or (PXVAPXX)n=2; block 10, degenerating 5-mer (VPXXX)n=5–8. Both sides of each repeat are flanked by nonrepeat unique sequences in which variation is rather polymorphic, with two to four basic sequence types (Fig. 2). Blocks 4 and 12 do not have apparent repeats.
Fig 1.
Sequence variation in PvMsp-1 alleles. Amino acid sequences of 40 isolates and two monkey-adapted strains (Sal-1 and Belem strains) of P. vivax are aligned. Dots and dashes represent residues identical to Sal-1 and deletions, respectively. Asterisks denote residues identical to either the Sal-1 or Belem sequence. Other types of substitutions are listed under “Others.” Interallele variable blocks are boxed; repeats and polymorphic residues are indicated by <, >, and X, respectively; and boundaries for subblocks are demarcated by dotted vertical lines. Codons with synonymous nucleotide substitutions are indicated by lowercase letters. Bold letters with and without underlining denote residues conserved perfectly among P. vivax, P. falciparum (K1 allele and MAD20 allele), P. chabaudi, P. berghei, and P. yoelii, and residues conserved among P. vivax and three of the other four species, respectively. Dotted underlining indicates the sites of Rm (22).
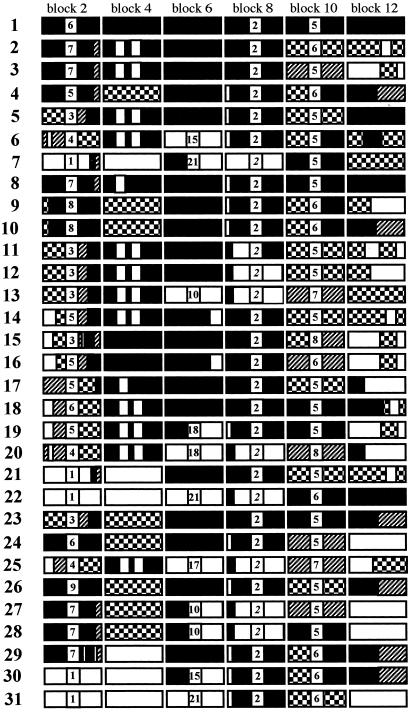
Fig 2.
Allelic variation in PvMsp-1 represented by associations of sequences of six variable blocks. Variable blocks 2, 6, 8, and 10 contain short tandem repeats: block 2, degenerating 5-mer (GSXXX)n=1–9 (X stands for any residue); block 6, poly (Q)n=10–21; block 8, degenerating repeats of (PVTTTX)n=2 or (PXVAPXX)n=2 (italicized); block 10, degenerating 5-mer (VPXXX)n=5–8. Variable blocks 4 and 12 and subblocks flanking these repeats have unique sequences, with each indicated by filled, unfilled, diagonal bricked, or checkerboard patterns. To simplify presentation, patterns are used to indicate different sequences within each block; i.e., a given pattern does not represent the same sequence in different blocks. In block 12, a small conserved region is excluded. Assignment of 42 isolates into 31 allelic types is as follows: 1 = Sal-1; 2 = BP29; 3 = BP1; 4 = BP13, BP63; 5 = BP30; 6 = BR07; 7 = BP39, BR44; 8 = T064, TF14, TF127; 9 = TV400; 10 = TG57; 11 = TC22; 12 = TG44; 13 = T107; 14 = T124; 15 = TFF18; 16 = T131, TG40; 17 = TC28, TC103; 18 = TG55; 19 = TD29; 20 = TG46; 21 = T077, TG48; 22 = TE26; 23 = BD4; 24 = BD1, BD2, BD9; 25 = BD6; 26 = IN1; 27 = SK1, SK2, SK4; 28 = SK3; 29 = VM55; 30 = VM278; 31 = Belem. Complete nucleotide identity occurs within each association type, except type 8, in which three isolates show substitutions at codons 1,564 and 1,705 (Fig. 1).
Alignments of PvMsp-1 alleles with Msp-1 from four other Plasmodium species (described above) revealed that those amino acids that are perfectly conserved among all five species, and those that are conserved in P. vivax and three of the other four species are distributed exclusively in conserved blocks. These include the N-terminal signal peptide region, the cysteine-rich C-terminal epidermal growth factor-like domains and regions of as yet unknown function. In all conserved blocks, the number of synonymous substitutions per synonymous site was greater than nonsynonymous substitutions per nonsynonymous site, with statistically significant differences in blocks 1, 3, 5, and 7 (Table 1), suggesting that structure-related functions are conserved in MSP-1 in nature, and that therefore purifying selection due to functional constraints is operating in conserved blocks.
Table 1.
Synonymous (dS) and nonsynonymous (dN) nucleotide substitutions in conserved blocks of PvMsp-1
| Block | Base pairs | dS ± SE | dN ± SE | P |
|---|---|---|---|---|
| 1 | 570 | 3.57 ± 1.04 | 1.34 ± 0.40 | <0.01 |
| 3 | 78 | 13.76 ± 5.94 | 2.53 ± 1.47 | <0.05 |
| 5 | 993 | 4.48 ± 0.72 | 1.91 ± 0.31 | <0.001 |
| 7 | 372 | 7.66 ± 1.82 | 3.13 ± 0.76 | <0.05 |
| 9 | 729 | 3.22 ± 0.90 | 1.87 ± 0.44 | NS |
| 11 | 498 | 1.77 ± 0.77 | 0.68 ± 0.26 | NS |
| 13 | 672 | 0.59 ± 0.32 | 0.18 ± 0.11 | NS |
dS and dN are per 100 sites. NS, not significant.
Recombination Sites.
The 31 PvMsp-1 alleles show unique combinations of variable block sequences (Fig. 2 and Table 3). Numerous potential recombination sites are readily identifiable within and between variable blocks, suggesting that allelic variation is generated by intragenic recombination events. Computation of Rm identified a minimum of 37 recombination sites: 31 sites within conserved blocks and six sites spanning each variable block (Fig. 1). Many recombination sites, demarcated by different patterns in each variable block, can be identified by visual inspection in their nonrepeat unique sequences. Notably, recombination sites are distributed across PvMsp-1, and the distribution is not uniform, with the lowest number of Rm in block 13 (Fig. 1).
Geographic Distribution.
We compared geographic distribution of PvMsp-1 alleles to infer the age of the alleles and frequency of recombination events. We first observed a very high allelic diversity in Asia (h = 0.953 ± 0.009), which is significantly higher than diversity in Brazil (h = 0.813 ± 0.047) (P < 0.01). Consistently, the number of nonrepeat unique sequence types in each variable block was higher in Asia than in Brazil. In Asia and Brazil, in blocks 2, 4, 6, 8, 10, and 12, the numbers of nonrepeat unique sequence types were 11, 5, 4, 4, 5, and 11 (allelic types nos. 8–28 in Fig. 2) and 4, 3, 3, 3, 4, and 6 (allelic types nos. 2–7), respectively. Importantly, all but one nonrepeat sequence found in Brazil also occurred in Asia, whereas no identical PvMsp-1 alleles were shared between Asia and Brazil or among different areas in Asia: Thailand (nos. 8–22), Bangladesh (nos. 23–25), India (no. 26), and South Korea (nos. 27 and 28). Further, unique associations were also apparent between all neighboring variable blocks; e.g., an association of nonrepeat sequences of blocks 2 and 4 in type no. 2 was unique to Brazil, whereas association of nonrepeat sequences in type no. 4 of Brazil occurred in Asia as well. Together, these findings suggest that (i) P. vivax populations in Asia are ancestral to populations in Brazil; (ii) PvMsp-1 has heterogeneity in frequency of allelic recombination events; and (iii) the history of recombination events in these unique sequences predates subdivision of P. vivax populations in the New World.
Linkage Disequilibrium.
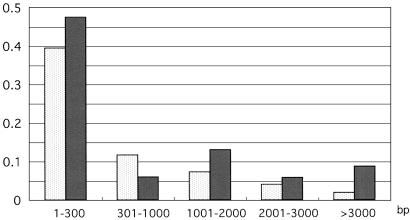
To estimate the frequency of recombination events in PvMsp-1, we analyzed linkage disequilibrium between polymorphic sites of the 20 Thai sequences (h = 0.935), which contained 175 informative polymorphic sites with 69 synonymous and 109 nonsynonymous substitutions (which are published as Fig. 6 in the supporting information on the PNAS web site). A total of 285/2,346 pairs of synonymous sites (12.1%) had significant linkage disequilibrium. Proportions of pairs with significant linkage disequilibrium declined rapidly in a curvilinear fashion with increasing nucleotide distance (Fig. 3): 39.4% and 11.7% of pairs were statistically significant at a distance of <0.3 and 0.3–1.0 kb, respectively; only 1.9% of pairs were significant at >3.0 kb. Meanwhile, a total of 830/5,265 pairs of nonsynonymous sites (15.8%) had significant linkage disequilibrium. A substantial proportion of nonsynonymous sites (8.7%) had significant linkage disequilibrium at a distance of >3 kb (Fig. 3). The overall proportions of pairs with significant linkage disequilibrium were significantly higher for nonsynonymous sites than for synonymous sites (P < 0.001). These results indicate that P. vivax has an effectively high recombination rate, with limited linkage disequilibria between nonsynonymous sites.
Fig 3.
Proportions of linkage disequilibrium between polymorphic sites within PvMsp-1 in a population from Thailand. Percentages of synonymous sites (2,346 pairs, stippled bar) and nonsynonymous sites (5,265 pairs, filled bar) that had statistically significant linkage disequilibrium (P < 0.05) are shown according to the nucleotide distance between the sites.
Discussion
The present study revealed that the organization of PvMsp-1 is basically similar to that of PfMsp-1: (i) mosaic organization of several interallele variable blocks flanked by conserved blocks; (ii) dimorphic substitutions in conserved blocks and rather polymorphic substitutions in variable blocks; and (iii) allelic recombination as a mechanism for the generation of new alleles. However, PvMsp-1 clearly differs from PfMsp-1 in that numerous recombination sites occur throughout PvMsp-1 in both conserved blocks and variable blocks. We identified at least 31 Rm in conserved blocks of PvMsp-1; this is most likely an underestimate, because we excluded variable blocks from estimation of Rm. Thus, the localization of recombination sites in PvMsp-1 is very different from that of PfMsp-1, in which recombination sites are confined to the 5′ and 3′ regions with no recombination events in the central 3.7-kb region (8).
The mosaic organization of PvMsp-1 presents a highly informative pattern of intragenic recombination at the level of population sequence diversity. In the present study, we obtained evidence for heterogeneity in frequency of allelic recombination events. It is generally agreed that malaria was first introduced into the New World ≈500 years ago by Europeans, followed by subsequent migrations from Europe and Africa (28), suggesting that P. vivax populations subdivided in the New World within the past few hundred years. During that time period, recombination events within variable block nonrepeat sequences have been rare, whereas the events have occurred repeatedly in conserved blocks. The chance of recombination is considerably reduced with increasing sequence divergence. In a bacterial system, sequence divergence of 10% results in 1,000-fold decrease in recombination frequency (29). In variable blocks of PvMsp-1, there are two to four basic nonrepeat sequence types, and their homologies are less than 40%. Therefore, mismatching duplexes would suppress the chance of meiotic recombination to a level several orders of magnitude below that of homologous recombination.
We believe that the high number of recombination sites in conserved blocks is related to the high frequency of recombination events. This hypothesis is consistent with the present analysis of linkage disequilibrium within PvMsp-1 in a P. vivax population from Thailand: linkage disequilibrium between pairs of synonymous sites declines rapidly with increasing nucleotide distance, with little linkage disequilibrium at a distance of >3 kb. We were able to confirm linkage “equilibrium” at relatively short distances in the reported sequences of other P. vivax genes, i.e., polymorphic sites separated by 350 bp in Ama-1 and by 840 bp in Csp, in P. vivax populations from the Philippines and Papua New Guinea (19, 30). Thus, not only PvMsp-1 but other loci of P. vivax show a high recombination rate. The recombination rate of P. falciparum is ≈6 × 10−7 Morgans/base (31), which is much higher than that of Drosophila (2 × 10−8 Morgans/base) (32). It has been reported that linkage disequilibrium within PfMsp-1 rapidly declines with distance, with little linkage disequilibrium at distances of over 1 kb in P. falciparum populations from Africa (33). Although direct estimates of the recombination rate of P. vivax from experimental crosses are not available, the present data show that allelic recombination events in PvMsp-1 are common, and that P. vivax has a considerably higher effective recombination rate than Drosophila.
The high recombination rate of P. vivax has important implications for understanding the population structure of the parasite. A recent study demonstrated that P. falciparum has a spectrum of population structures: strong linkage disequilibrium and low genetic diversity in areas with low levels of transmission; and linkage “equilibrium” and high diversity in areas with high levels of transmission (28). Such a spectrum may also exist for P. vivax populations. However, in the present study, we observed linkage “equilibrium” within a few kilobases of PvMsp-1 and a high level of heterozygosity (h = 0.935) of PvMsp-1 alleles in a P. vivax population from Thailand, where malaria transmission is low. High frequency of mixed-allele infections is common in natural populations of P. vivax: 34% of the present samples had mixed PvMsp-1 alleles, and it has been reported that 40% of infections in Brazil have mixed PvMsp-1 alleles (34). Long-lasting and recurrent parasitemia often associated with relapse, a characteristic of P. vivax infection, may contribute to an increased rate of heterozygote formation and subsequent interallele recombination in the anopheline mosquito vector. Therefore, new PvMsp-1 alleles could be generated by recombination events even in areas with low levels of malaria transmission.
The present population sequence analysis highlights the evolution of polymorphism of PvMsp-1. Alignments of PvMsp-1 alleles with reported partial Msp-1 sequences of P. cynomolgi and P. knowlesi for variable block 12 revealed striking conservation of sequence segments (Fig. 4): VNAQI and OLN sequences of P. cynomolgi/P. knowlesi are identical to types 4 and 2 alleles of P. vivax, respectively. The rest of the P. cynomolgi sequence is mostly a mosaic, composed of segments of types 1–4 alleles. These findings indicate that allelic sequences in variable block 12 nonrepeat regions predate the origin of P. vivax, and that P. vivax has maintained this ancestral polymorphism for 11.7–30.5 million years. Ancestral origins of polymorphic sequences shared by phylogenetically distant species have also been suggested for P. falciparum: alleles of Msp-2 and Msp-3 of P. falciparum are closer to those of P. reichenowi than to different alleles of P. falciparum (35, 36). Balancing selection appears to be a likely mechanism for maintaining polymorphism of variable block nonrepeat allelic sequences of PvMsp-1 (37, 38).
Fig 4.
Alignment of block 12 of PvMsp-1 alleles and Msp-1 of P. cynomolgi and P. knowlesi. P. vivax basic types 1–4 form mosaic mixtures of recombinant types 5–10. Note that P. cynomolgi sequence mostly consists of the four basic types of P. vivax. Residues conserved among P. vivax alleles are boxed, and those unique to P. cynomolgi and/or P. knowlesi are shaded. Symbols are: b = E or K, x = D or A, and z = K or T.
In conserved blocks, the number of nonsynonymous substitutions per nonsynonymous site was not higher than that of synonymous substitutions per synonymous site. However, this does not necessarily mean that positive selection is not operating in these blocks. Linkage disequilibrium between nonsynonymous sites and alterations in T cell epitope sequences deserve attention in this context. First, we observed that proportions of linkage disequilibrium in PvMsp-1 were significantly higher for pairs of nonsynonymous sites than for pairs of synonymous sites. At the merozoite surface, MSP-1 fragments, which are formed by processing from a precursor protein, are weakly associated with each other. Conformation-dependent B cell epitopes, some of which are formed by association between these fragments, are allotype-specific, and allotype-specific antibodies occasionally compete with other antibodies for binding, probably by steric hindrance (39). Hence, epistatic selection at the protein level might lead to linkage disequilibrium between nonsynonymous sites in PvMsp-1. Second, a single amino acid change or clustered replacements in polymorphic T cell epitopes may potentially cause reduction in binding of MSP-1 to a peptide-binding pocket of HLA. Using a T cell epitope prediction algorithm (40), we were able to identify potential T cell epitopes in polymorphic sites in conserved blocks. Notably, binding of a predicted T cell epitope peptide to a particular HLA allotype is markedly affected by single or multiple amino acid substitution. For example, change of T to A and/or L to V at positions 75 and 82, respectively, of the predicted T cell epitope of VSVLTSKIRNFLSKF (Fig. 1) results in reduction of binding to HLA DRB1*0301, with scores declining from 27 to 18 or 19. A substantial percentage of replacements occurring in conserved blocks result in alterations in binding to HLA class II allotypes, despite the fact that algorithms available to date predict only limited numbers of class II allotypes. Polymorphism in T cell epitope regions could enable parasites to escape host immune responses. Therefore, we consider that host immune pressure plays a crucial role in the evolution and maintenance of polymorphism in PvMsp-1.
Finally, we observed very low nucleotide diversity in block 13, which contains a region encoding the 19-kDa C-terminal epidermal growth factor-like domain of PvMsp-1, a vaccine candidate. Interestingly, this block had the lowest Rm value and lowest nucleotide diversity of all blocks (Fig. 1). Recombination-associated hitchhiking (41) could have reduced the level of polymorphism in this region. The low polymorphism in the 19-kDa domain among geographically diverse P. vivax populations suggests that this portion of MSP-1 has promise as a vaccine against P. vivax.
Supplementary Material
Acknowledgments
We thank A. Lal and I. Goldman (Center for Disease Control, Chamblee, GA) for Sal-1 DNA, P. David (Institut Pasteur, Paris) for Belem DNA, and D. J. Conway for advice. We are grateful to all patients who participated in this study. This work was supported by a Grant-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (no. 14021125 to K.T.) and from the Japan Society for the Promotion of Science (13576030 and 14570224 to A.K.). C.P. was supported by the Hitachi Scholarship Foundation.
Abbreviations
Rm, the minimum number of recombination events
MSP-1, merozoite surface protein-1
Pv, Plasmodium vivax
Pf, Plasmodium falciparum
h, heterozygosity
References
- 1.Mendis K., Sina, B. J., Marchesini, P. & Carter, R. (2001) Am. J. Trop. Med. Hyg. 64, 97-106. [DOI] [PubMed] [Google Scholar]
- 2.Udagama P. V., Gamage-Mendis, A. C., David, P. H., Peiris, J. S. M., Perera, K. L. R. L., Mendis, K. N. & Carter, R. (1990) Am. J. Trop. Med. Hyg. 42, 104-110. [DOI] [PubMed] [Google Scholar]
- 3.Holder A. A. (1988) Prog. Allergy 41, 72-97. [PubMed] [Google Scholar]
- 4.Blackman M. J., Heidrich, H. G., Donachie, S., McBride, J. S. & Holder, A. A. (1990) J. Exp. Med. 172, 379-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar S., Yadava, A., Keister, D. B., Tian, J. H., Ohl, M., Perdue-Greenfield, K. A., Miller, L. H. & Kaslow, D. C. (1995) Mol. Med. 1, 325-333. [PMC free article] [PubMed] [Google Scholar]
- 6.Collins W. E., Kaslow, D. C., Sullivan, J. S., Morris, C. L., Galland, G., Yang, C., Saekhou, A. E., Xiao, L. & Lal, A. (1999) Am. J. Trop. Med. Hyg. 60, 350-356. [DOI] [PubMed] [Google Scholar]
- 7.McBride J. S., Newbold, C. I. & Anand, R. (1985) J. Exp. Med. 161, 160-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanabe K., Mackay, M., Goman, M. & Scaife, J. (1987) J. Mol. Biol. 195, 273-287. [DOI] [PubMed] [Google Scholar]
- 9.Miller L. H., Roberts, T., Shahabuddin, M. & McCutchan, T. F. (1993) Mol. Biochem. Parasitol. 59, 1-14. [DOI] [PubMed] [Google Scholar]
- 10.Kerr P. J., Ranford-Cartwright, L. C. & Walliker, D. (1994) Mol. Biochem. Parasitol. 66, 241-248. [DOI] [PubMed] [Google Scholar]
- 11.Kolakovich K. A., Ssengoba, A., Wojcik, K., Tsuboi, T., Al-Yaman, F., Alpers, M. & Adams, J. H. (1996) Exp. Parasitol. 83, 11-18. [DOI] [PubMed] [Google Scholar]
- 12.Premawansa S., Snewin, V. A., Khouri, E., Mendis, K. & David, P. H. (1993) Exp. Parasitol. 76, 192-199. [DOI] [PubMed] [Google Scholar]
- 13.Putaporntip C., Jongwutiwes, S., Seethamchai, S., Kanbara, H. & Tanabe, K. (2000) Mol. Biochem. Parasitol. 109, 111-119. [DOI] [PubMed] [Google Scholar]
- 14.del Portillo H. A., Longacre, S., Khouri, E. & David, P. H. (1991) Proc. Natl. Acad. Sci. USA 88, 4030-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson H., Tucker, J. E., Kaslow, D. C., Krettli, A. U., Collins, W. E., Kiefer, M. C., Bathurst, I. C. & Barr, P. J. (1992) Mol. Biochem. Parasitol. 50, 325-334. [DOI] [PubMed] [Google Scholar]
- 16.Escalante A. A., Barrio, E. & Ayala, F. J. (1995) Mol. Biol. Evol. 12, 616-626. [DOI] [PubMed] [Google Scholar]
- 17.Riley E. M., Allen, S. J., Wheeler, J. G., Blackman, M. J., Bennett, S., Takacs, B., Schonfeld, H.-J., Holder, A. A. & Greenwood, B. M. (1992) Parasite Immunol. 14, 321-337. [DOI] [PubMed] [Google Scholar]
- 18.Sakihama N., Mitamura, T., Kaneko, A., Horii, T. & Tanabe, K. (2001) Exp. Parasitol. 97, 50-54. [DOI] [PubMed] [Google Scholar]
- 19.Figtree M., Pasay, C., Slade, R., Cheng, Q., Cloonan, N., Walker, J. & Saul, A. (2000) Mol. Biochem. Parasitol. 108, 53-66. [DOI] [PubMed] [Google Scholar]
- 20.Higgins D. G., Bleasby, A. J. & Fuchs, R. (1992) Comput. Appl. Biosci. 8, 189-191. [DOI] [PubMed] [Google Scholar]
- 21.Rozas J. & Rozas, R. (1999) Bioinformatics 15, 174-175. [DOI] [PubMed] [Google Scholar]
- 22.Hudson R. R. & Kaplan, N. L. (1985) Genetics 111, 147-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill W. G. & Robertson, A. (1968) Theor. Appl. Genet. 38, 226-231. [DOI] [PubMed] [Google Scholar]
- 24.Sakihama N., Kaneko, A., Hattori, T. & Tanabe, K. (2001) Gene 279, 41-48. [DOI] [PubMed] [Google Scholar]
- 25.Nei M. & Gojobori, T. (1986) Mol. Biol. Evol. 3, 418-426. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J., Rosenberg, H. F. & Nei, M. (1998) Proc. Natl. Acad. Sci. USA 95, 3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244-1245. [DOI] [PubMed] [Google Scholar]
- 28.Anderson T. J. C., Haubold, B., Williams, J. T., Estrada-Franco, J. G., Richardson, L., Mollinedo, R., Bockarie, M., Mokili, J., Mharakurwa, S., French, N., et al. (2000) Mol. Biol. Evol. 17, 1467-1482. [DOI] [PubMed] [Google Scholar]
- 29.Rayssiguier C., Thaler, D. S. & Radman, M. (1989) Nature 342, 396-401. [DOI] [PubMed] [Google Scholar]
- 30.Qari S. H., Goldman, I. F., Povoa, M. M., di Santi, S., Alpers, M. P. & Lal, A. A. (1992) Mol. Biochem. Parasitol. 55, 105-114. [DOI] [PubMed] [Google Scholar]
- 31.Su X.-Z., Ferdig, M., Huang, Y., Huynh, C. Q., Liu, A., You, J., Wootton, J. C. & Wellems, T. E. (1999) Science 286, 1351-1353. [DOI] [PubMed] [Google Scholar]
- 32.Chakravarti A., Buetow, K. H., Antonarakis, S. E., Waber, P. G., Boehm, C. D. & Kazazian, H. H. (1984) Am. J. Hum. Genet. 36, 1239-1258. [PMC free article] [PubMed] [Google Scholar]
- 33.Conway D. J., Roper, C., Oduola, A. M. J., Arnot, D. E., Kremsner, P., Grobusch, M. P., Curtis, C. & Greenwood, B. M. (1999) Proc. Natl. Acad. Sci. USA 96, 4506-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soares I. S., Barnwell, J. W., Ferreira, M. U., Gomes Da Cunha, M., Laurino, J. P., Castilho, B. A. & Rodrigues, M. M. (1999) Mol. Med. 5, 459-470. [PMC free article] [PubMed] [Google Scholar]
- 35.Dubbeld M. A., Kochen, C. H. & Thomas, A. W. (1998) Mol. Biochem. Parasitol. 92, 187-192. [DOI] [PubMed] [Google Scholar]
- 36.Okenu D. M. N., Thomas, A. W. & Conway, D. J. (2000) Mol. Biochem. Parasitol. 109, 185-188. [DOI] [PubMed] [Google Scholar]
- 37.Hughes M. K. & Hughes, A. L. (1995) Mol. Biochem. Parasitol. 71, 99-113. [DOI] [PubMed] [Google Scholar]
- 38.Conway D. J. & Baum, J. (2002) Trends Parasitol. 18, 351-355. [DOI] [PubMed] [Google Scholar]
- 39.Wilson C. F., Anand, R., Clark, J. T. & McBride, J. S. (1987) Parasite Immunol. 9, 737-746. [DOI] [PubMed] [Google Scholar]
- 40.Rammensee H.-G., Bachmann, J., Emmmerich, N. P. N., Bachor, O. A. & Stevanvic, S. (1999) Immunogenetics 50, 213-219. [DOI] [PubMed] [Google Scholar]
- 41.Maynard Smith J. & Haigh, J. (1976) Genet. Res. 23, 23-35. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.