Abstract
Many plant, animal, and fungal genomes contain cytosine DNA methylation in asymmetric sequence contexts (CpHpH, H = A, T, C). Although the enzymes responsible for this methylation are unknown, it has been assumed that asymmetric methylation is maintained by the persistent activity of de novo methyltransferases (enzymes capable of methylating previously unmodified DNA). We recently reported that the DOMAINS REARRANGED METHYLASE (DRM) genes are required for de novo DNA methylation in Arabidopsis thaliana because drm1 drm2 double mutants lack the de novo methylation normally associated with transgene silencing. In this study, we have used bisulfite sequencing and Southern blot analysis to examine the role of the DRM loci in the maintenance of asymmetric methylation. At some loci, drm1 drm2 double mutants eliminated all asymmetric methylation. However, at the SUPERMAN locus, asymmetric methylation was only completely abolished in drm1 drm2 chromomethylase 3 (cmt3) triple mutant plants. drm1 drm2 double mutants also showed a strong reduction of CpNpG (n = A, T, C, or G) methylation at some loci, but not at others. The drm1 drm2 cmt3 triple mutant plants did not affect CpG methylation at any locus tested, suggesting that the primary CpG methylases are encoded by the MET1 class of genes. Although neither the drm1 drm2 double mutants nor the cmt3 single mutants show morphological defects, drm1 drm2 cmt3 triple mutant plants show pleiotropic effects on plant development. Our results suggest that the DRM and CMT3 genes act in a partially redundant and locus-specific manner to control asymmetric and CpNpG methylation.
Cytosine DNA methylation plays a major role in gene silencing and heterochromatin formation (1). In mammals, methylation is largely restricted to CpG dinucleotides, but low levels of methylation at asymmetric sites are found in some cell types (2). Asymmetric DNA methylation is also found in some fungal genomes. For instance, Neurospora crassa shows dense asymmetric methylation associated with a phenomenon called Repeat-Induced Point mutation (RIP) (3), and Ascobolus immersus shows asymmetric methylation associated with the Methylation Induced Premeiotically (MIP) phenomenon (4). Plant genomes contain high levels of asymmetric methylation, and also contain abundant CpNpG methylation (5–8).
The symmetry of the CpG site was proposed to be important for stable maintenance of methylation patterns after DNA replication (9, 10). Replication of symmetrical sites would produce hemimethylated sequences, which were proposed to be preferred targets for a maintenance methyltransferase that would methylate cytosines in the newly synthesized DNA strand (11). Consistent with these early ideas, Dnmt1, the major mammalian CpG methyltransferase (12), is known to prefer DNA substrates containing hemimethylated CpG dinucleotides (13). Dnmt1 also localizes to DNA replication foci (14) consistent with the notion that maintenance methylation and DNA replication are tightly coupled. The CpNpG site methylated in plants is also symmetric. Although it is attractive to imagine that maintenance of CpNpG methylation is similar to that of CpG methylation, essentially nothing of the mechanism is known. Methylation of asymmetric or nonpalindromic sequences, on the other hand, could well be maintained by a mechanism different from that of symmetric sites. Asymmetric methylation must be reestablished after each DNA replication cycle, because there is no complementary sequence to serve as a guide for remethylation of particular cytosines (2, 15, 16).
The function of asymmetric methylation remains unclear. In one study, all symmetric CpG and CpNpG sites were removed from a 35S transgene that is normally subject to transsilencing by a second transgene which contains the 35S promotor present in a repetitive array (17). The symmetric site-free transgene became heavily methylated at the asymmetric sites. Furthermore, the removal of symmetric sites did not prevent transcriptional silencing of this transgene, but did prevent the maintenance of silencing in the absence of the repetitive array. In vitro methylation of the asymmetric cytosines also reduced the transcriptional activity of this 35S promotor in protoplasts (18). These studies show that asymmetric methylation can exist in the absence of symmetric methylation and may contribute to gene silencing.
Arabidopsis thaliana has at least three classes of DNA methyltransferase genes that are possible candidates for controlling asymmetric methylation, the MET1 class, the CMT3 (CHROMOMETHYLASE 3) class, and the DOMAINS REARRANGED METHYLASE (DRM) class (19). Although it seems likely that these genes encode enzymes that are active methyltransferases, this has not yet been directly demonstrated by studying the in vitro enzymatic properties of the proteins.
The MET1 class of genes (20) is most similar to Dnmt1 in both sequence and function. Loss-of-function met1 mutants, (also called ddm2 mutants) and antisense-MET1 transgenic plants lack the majority of CpG methylation (20–25). In contrast to mutant Dnmt1 mice, which die after 9 days of development, antisense-MET1 or met1 mutant plants are viable, but display a number of developmental abnormalities that become progressively more extreme as the mutants are inbred (22, 23). Surprisingly, some of these abnormalities are caused by ectopic hypermethylation of particular genes, such as SUPERMAN (SUP) and AGAMOUS (8, 26), a phenomenon that superficially resembles the hypermethylation of human tumor suppressor genes in otherwise hypomethylated genomic backgrounds (27).
The CMT3 class of genes are specific to the plant kingdom and encode methyltransferase proteins containing a chromo domain (28). CMT3 loss-of-function mutants were isolated in three independent studies, and show a genome-wide loss of CpNpG methylation and a reduction of asymmetric methylation at some loci (24, 29, 30). CpNpG methylation is also affected by histone methylation, because loss-of-function mutations in the KRYPTONITE (KYP) histone H3 lysine 9 methyltransferase gene show a loss of DNA methylation in CpNpG contexts (31).
The third class of genes, composed of DRM1 and DRM2, contain catalytic domains showing sequence similarity to those of the mammalian Dnmt3 methyltransferases (32). Dnmt3a and Dnmt3b encode important de novo methyltransferases (33–36). For instance, mice harboring mutations in both enzymes lack the ability to de novo methylate retroviruses when introduced into embryonic stem cells (35). Furthermore, in humans, Dnmt3b mutations reduce CpG methylation at some pericentromeric sequences and are the cause of a rare recessive disorder called ICF syndrome, for immunodeficiency, centromeric instability, and facial anomalies (35, 37, 38). We recently reported that the DRM genes are required for the initial establishment of methylation of cytosines in all known sequence contexts: CpG, CpNpG, and asymmetric (39). We found that drm1 drm2 double mutants lacked de novo methylation of the direct repeats of the FWA locus, which normally occurs when FWA is transformed into wild-type plants (39). drm1 drm2 double mutants also blocked de novo methylation of the SUP locus, which occurs in the presence of a SUP inverted repeat. However, the drm mutants did not show reactivation of previously methylated and silenced FWA or SUP epigenetic alleles, suggesting that the DRM genes are required for the establishment but not the maintenance of gene silencing (39).
In this report, we show that the DRM loci are required for the maintenance of asymmetric DNA methylation. However, at some loci such as SUP, we find that DRMs act redundantly with CMT3, so that only in drm1 drm2 cmt3 triple mutants is all asymmetric methylation lost. Furthermore, we find that at some loci the DRMs are more important for the maintenance of CpNpG methylation than CMT3. Our data suggest that the DRM and CMT3 genes encode partially redundant methyltransferases, and that different sequence or chromatin contexts can modulate their function.
Materials and Methods
Genetic Analysis.
The drm1 drm2, clk-st, drm1 drm2 clk-st, and cmt3-7 clk-st strains used in these studies, and the PCR-based molecular markers used to follow each gene, were recently described (24, 39). We constructed drm1 drm2 cmt3-7 triple mutants in a homozygous clk-st background by crossing a cmt3-7 clk-st plant to a drm1 drm2 clk-st plant and PCR genotyping the F2 progeny. We identified five independent drm1 drm2 cmt3-7 clk-st plants. Seeds of the met1 (ddm2) mutant were a kind gift of Eric Richards (Department of Biology, Washington University, St. Louis).
Genomic Bisulfite Sequencing.
Two micrograms of genomic DNA from the appropriate genotype was digested with restriction enzymes that cut just outside of the region of interest. Bisulfite treatment was performed as described (26). Because our previous studies found similar patterns of methylation on the two strands of DNA (8, 40), only one strand was analyzed for each locus. The bottom strand of MEA-ISR analyzed corresponds to positions 68067–68320 of bacterial artificial chromosome (BAC) clone T14P4 (GenBank accession no. AC022521). Primers for MEA-ISR are: JP1026 5′-AAA GTG GTT GTA GTT TAT GAA AGG TTT TAT and JP1027 5′-CTT AAA AAA TTT TCA ACT CAT TTT TTT TAA AAA A. PCR products were cloned by using the TOPO TA cloning kit (Invitrogen), and 15 individual clones were sequenced. Bisulfite sequencing on the top strand of SUP was performed as described (24). Bisulfite sequencing of the top strand of FWA was as described (40).
Southern Blot Analysis.
MEA-ISR probe was amplified from genomic DNA with following primers: JP980 5′-AAA CCT TTC GTA AGC TAC AGC CAC TTT GTT-3′; JP981 5′-TCG GAT TGG TTC TTC CTA CCT CTT TAC CTT-3′, corresponding to positions 68348–69438 on BAC clone T14P4 (GenBank accession no. AC022521). FWA probe was a 1.7-kb PCR-generated DNA fragment corresponding to positions 498-2281 in GenBank accession no. AF178688. PCR primers were JP 1084 5′-CTT CGC CTT TCT CTT CCT CAT CTG CGC TT-3′ and JP1085 5′-GAG TTT GAT AAG CAG TGC GCC TAT GGG TT-3′. Ta3 probe was as described (41) and amplified by JP1154 5′-GAT CTA TCT GGC CCC AGA CGT AGA TCT AA-3′ and JP1155 5′-CCG GCA ATC TAC TAT ATG AGA TCT TTA CAA-3′; 180-bp centromere repeat probes were described in ref. 21.
Results
DRMs Control Asymmetric and CpNpG Methylation at FWA and MEA-ISR.
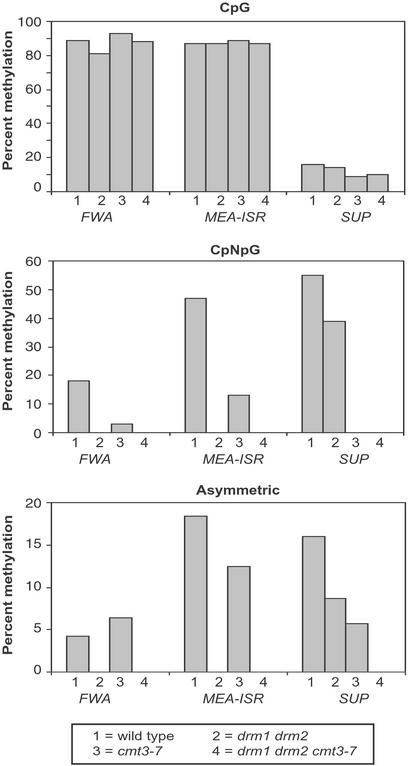
To determine the effect of the DRM loci in the maintenance of asymmetric methylation, we used bisulfite genomic sequencing to study the levels of methylation at two endogenous sequences that are methylated at asymmetric sites, FWA and MEA-ISR. The FWA locus encodes a homeodomain-containing protein whose expression is silenced in the vegetative tissues of wild-type plants. This silencing is associated with methylation of two direct repeats in the 5′ region of the gene (40). When this methylation is lost, either in spontaneous hypomethylated epigenetic mutants such as fwa-1, or in the methylation mutants ddm1 and met1, the FWA gene is overexpressed causing a dominant late flowering phenotype (24, 40, 42). In wild type, the FWA direct repeats contain 89% CpG, 18% CpNpG, and 4% asymmetric methylation (Fig. 1 and Table 2, which is published as supporting information on the PNAS web site, www.pnas.org) (40). MEA-ISR is an approximately 183-bp sequence present in seven direct repeats lying in an intergenic region between the imprinted medea (MEA) gene (43, 44) and the aldehyde oxidase gene on BAC clone T14P4 near the upper end of chromosome 1, approximately 500,000 bp from the end. These repeats are also found in 12 other genomic locations, all of which are also subtelomeric. For this reason, we named this sequence MEA-ISR for Intergenic Subtelomeric Repeat. We found that this repeat shows high levels of DNA methylation in wild-type strains, namely 87% CpG, 47% CpNpG, and 18% asymmetric (Fig. 1).
Figure 1.
Diagram represents bisulfite sequencing of a 500-nt region of the top strand of the FWA gene, 219-nt region of the bottom strand of the MEA-ISR region, and 1,028-nt region of the top strand of the SUP gene in different mutant backgrounds. Detailed data supporting the graphical presentation can be found in Table 2 and Fig. 6. For the analysis of FWA and MEA-ISR, the wild-type strain is WS. For the SUP gene, the wild-type strain is clk-st in the Landsberg erecta background. drm1 drm2, cmt3-7, and drm1 drm2 cmt3 mutants are homozygous for the clk-st inverted repeat SUP transgene. Height of the bars represents the percentage of methylation at each site of 15 clones analyzed by bisulfite sequencing.
We compared the parental Wassilewskija (WS) strain with a drm1 drm2 double mutant strain by using bisulfite genomic sequencing and found that the drm1 drm2 mutations eliminated all asymmetric methylation of both the FWA and MEA-ISR sequences. Therefore, the DRM loci are important for the methylation of asymmetric cytosines.
The drm1 drm2 mutations also eliminated the CpNpG methylation of both FWA and MEA-ISR (Fig. 1). Because cmt3 mutants were previously found to strongly reduce methylation of CpNpG sites of all loci tested (24, 30), we next analyzed FWA and MEA-ISR in the cmt3-7 mutant, a null cmt3 allele (24). We found that cmt3-7 reduced, but did not eliminate, the CpNpG methylation (Fig. 1). Therefore, both DRM and CMT3 are required for proper maintenance of CpNpG methylation patterns, and at some loci, such as FWA and MEA-ISR, the drm1 drm2 double mutant is more effective than cmt3-7 at reducing CpNpG methylation.
We also analyzed drm1 drm2 cmt3-7 triple mutants by bisulfite sequencing (Fig. 1) and found that, like drm1 drm2 double mutants, the triple mutants lacked all traces of asymmetric and CpNpG methylation at FWA and MEA-ISR. However, CpG methylation levels were similar to the wild type.
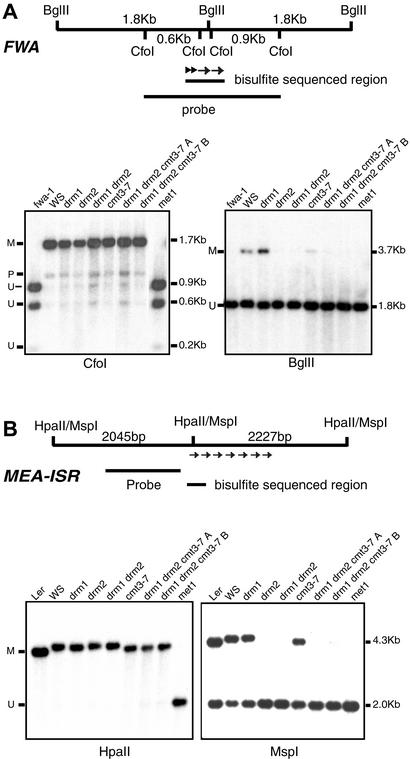
We used Southern blot analysis with methylation-sensitive restriction enzymes to confirm some of the bisulfite sequencing results at FWA and MEA-ISR. In these analyses, we also included the drm1 and drm2 single mutants as well as the met1 mutant. For the FWA blots, we included the fwa-1 epigenetic mutant (40) as a completely unmethylated control. Methylation at the FWA gene was assayed with two methylation-sensitive restriction enzymes, CfoI and BglII, which are found within the methylated direct repeats (Fig. 2A). CfoI recognizes the sequence GCGC and is inhibited by CpG methylation of the site. Previous Southern blot results showed that drm1 drm2 double mutants (39) and the cmt3-7 single mutant (31) did not affect CpG methylation at FWA. Here we compared two independently isolated drm1 drm2 cmt3 triple mutants (named A and B) with all of the single mutants, and found that all mutants or combinations of mutants showed the same level of CpG methylation as the wild-type control WS (Fig. 2A). This finding is consistent with the bisulfite sequence data indicating that the drm1 drm2 cmt3 triple mutants do not affect preexisting CpG methylation at FWA. CpG methylation was completely lost in a met1 mutant line that had developed an fwa late flowering phenotype (Fig. 2A). This finding further confirms that MET1 maintains CpG methylation (20, 22–25).
Figure 2.
Southern blot analysis of FWA and MEA-ISR. (A Upper) A diagram of the CfoI and BglII restriction fragments present in the FWA gene. The inner two CfoI sites and the BglII site are within the methylated direct repeats of the FWA promoter, shown as arrows above. The region analyzed by bisulfite sequencing is also noted. (Lower) DNA blots of equivalent amounts of CfoI (Left) and BglII (Right) digested genomic DNA probed with a 1.74-kb fragment corresponding to the diagram on top. The positions of the methylated (M), unmethylated (U), and partially methylated (P) bands are shown, as well as the size of the bands in kilobases. drm1 drm2 cmt3-7 A and B are two independently isolated triple mutant strains. (B Upper) A diagram of the HpaII/MspI restriction fragments present in the MEA-ISR region. The inner HpaII/MspI site lies in the first of seven direct repeats (shown as arrows) and the probe lies in the unique sequence between these repeats and the MEA locus. (Lower) DNA blots of equivalent amounts of HpaII (Left) and MspI (Right) digested genomic DNA probed with a 1.1-kb fragment corresponding to the diagram on top.
The restriction enzyme BglII is inhibited by cytosine methylation within its recognition site AGATCT (45), and we have used this enzyme to detect CpNpG methylation of FWA. Because of the sequence context at FWA (AAGATCTG), BglII will detect CpTpG methylation at a site that we previously found to be methylated by bisulfite sequencing (40). BglII would also be inhibited by methylation of the asymmetric CpTpT site on the other strand, but because we have not found methylation at this site by bisulfite sequencing, this is unlikely to complicate the analysis. A 3.7-kb BglII fragment caused by cytosine methylation at the BglII site appears in wild-type WS but not in fwa-1 (Fig. 2A). A faint 3.7-kb methylated band is detected in cmt3-7, consistent with bisulfite sequence data showing that CpNpG methylation was reduced but not completely lost in cmt3-7 (Fig. 1). drm1 drm2 double mutants and drm1 drm2 cmt3-7 triple mutants eliminated methylation of the BglII site, consistent with the bisulfite data showing a lack of CpNpG methylation in these strains (Fig. 1).
Methylation of the BglII site was also lost in the drm2 single mutant, but not in the drm1 single mutant (Fig. 2A). This finding fits with our previous observations that drm2 but not drm1 blocked FWA transgene de novo methylation (39), and with the fact that DRM2 RNA is expressed at a much higher level than DRM1 RNA (32). Thus DRM2 is likely to encode the predominant enzyme in the DRM family.
For Southern blot analysis of the MEA-ISR region, we used a single HpaII/MspI site (CCGG) present within the first direct repeat. We used a probe within unique sequence adjacent to these repeats to assay methylation of this site (Fig. 2B). HpaII is inhibited by methylation of either cytosine of its recognition site allowing detection of CpG and CpNpG methylation, whereas MspI is only inhibited by methylation of the outer cytosine allowing detection of CpNpG methylation. Similar to the wild-type strains WS and Ler, drm1 drm2, cmt3-7 and drm1 drm2 cmt3-7 triple mutant plants were not cut by HpaII, confirming the bisulfite data showing that these mutants do not affect CpG methylation (Fig. 1). However, the met1 mutant showed complete digestion with HpaII suggesting a loss of methylation of the site (Fig. 2B).
The MspI Southern blot of the MEA-ISR sequence revealed about 50% cleavage of wild-type Ler and WS DNAs (Fig. 2B), consistent with the 47% CpNpG methylation determined by bisulfite sequence (Fig. 1). Similar to results with the FWA gene, drm2 but not drm1 caused a loss of all CpNpG methylation at MEA-ISR. cmt3-7 retained a fair amount of CpNpG methylation, again consistent with the bisulfite data (Fig. 1). These results confirm that, at the MEA-ISR locus, cmt3-7 single mutants and drm1 drm2 double mutants both reduce methylation at CpNpG sites, but drm1 drm2 does so much more efficiently.
In conclusion, drm1 drm2 double mutants showed a complete loss of non-CpG methylation at both FWA and MEA-ISR. Interestingly, drm1 drm2 plants do not show a late flowering phenotype typical of plants in which expression of the FWA gene has been reactivated (39). This suggests that non-CpG methylation does not play major role in the silencing of FWA.
Both DRM and CMT3 Control Asymmetric Methylation at SUP.
Epigenetic silenced alleles of the SUP locus (the clark kent or clk alleles) are densely methylated in all sequence contexts (8, 24, 25). Whereas the originally isolated clk alleles spontaneously revert to an unmethylated state ≈3% of the time (8), a transgenic allele called clk-st shows a stable nonreverting phenotype, making it more suitable for genetic studies (24). clk-st contains a single inverted repeat of the SUP locus, which can induce de novo methylation of itself as well as of the endogenous SUP locus (described in detail in reference 39). The methylation consists of 16% CpG, 55% CpNpG and 16% asymmetric (Fig. 1) (24). We previously found that cmt3-7 eliminated the CpNpG methylation, and reduced the asymmetric methylation by approximately 60% (Fig. 1) (24). To test the effect of the drm mutations on SUP methylation, we compared clk-st with a drm1 drm2 clk-st strain (39). Contrary to what was found with FWA and MEA-ISR, we observed that drm1 drm2 double mutants reduced but did not eliminate asymmetric methylation (Fig. 1 and Fig. 6 and Table 2, which are published as supporting information on the PNAS web site) (24, 39). However, this residual asymmetric methylation was not detected in drm1 drm2 cmt3-7 clk-st strains (Fig. 1). Therefore, DRM and CMT3 act redundantly to control SUP asymmetric methylation.
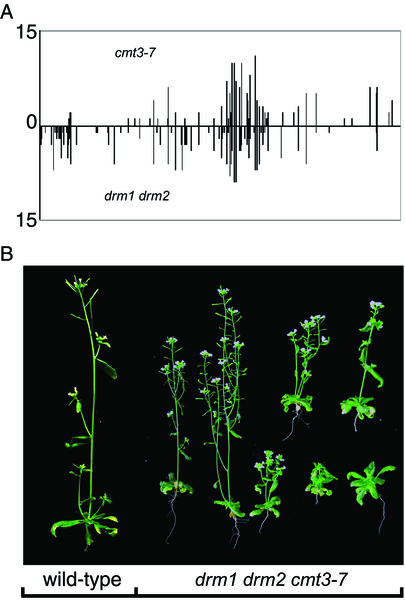
Because drm1 drm2 cmt3-7 triple mutants eliminate all asymmetric methylation at SUP, an analysis of the methylation remaining in the cmt3-7 mutants should reflect the residual activity of DRM, and that remaining in drm1 drm2 should reflect the residual activity of CMT3. Fig. 4A shows that the positions of the asymmetric methylation of SUP remaining in either cmt3-7 plants or drm1 drm2 plants were largely overlapping (also see Fig. 6). An analysis of the sequence context of the asymmetric methylation in these mutants suggests that both CMT3 and DRM prefer to methylate sites that follow cytosine residues (Table 1). However, both CMT3 and DRM showed a bias against sites that immediately precede cytosines; CpA and CpT methylation were much more frequent than CpC methylation (Table 1). These results suggest that CMT3 and DRM can methylate the same asymmetric sites, and both show roughly the same preference for particular DNA sequence contexts.
Figure 4.
Genetic redundancy of the DRM and CMT3 genes. (A) Graph showing the level and positions of asymmetric methylation of SUP in the cmt3-7 mutant or the drm1 drm2 double mutant, both in the clk-st background. Height of the bars represents the frequency of methylation at each site of 15 clones analyzed by bisulfite sequencing. The x axis represents the 1,028-nt methylated region on the top strand of SUP. (B) Photographs of a four-week-old wild-type Ler plant (Left) and several drm1 drm2 cmt3-7 triple mutant plants (Right).
Table 1.
Sequence context of the asymmetric methylation found at the SUP locus within 15 cloned PCR products of bisulfite-treated DNA
| Site | % methylation of site
|
||||
|---|---|---|---|---|---|
| Wild type | cmt3-7 | drm1 drm2 | drm1 drm2 cmt3-7 | No. of sites | |
| All sites* | 16.2 | 5.7 | 8.7 | 0 | 204 |
| CpA | 17.1 | 4.6 | 8.7 | 0 | 70 |
| CpT | 22 | 9.5 | 13.0 | 0 | 86 |
| CpC | 4.7 | 0.7 | 1.0 | 0 | 48 |
| ApC | 8.6 | 0.6 | 4.5 | 0 | 46 |
| TpC | 18.3 | 7.1 | 8.9 | 0 | 83 |
| GpC | 13.3 | 4.7 | 5.8 | 0 | 31 |
| CpC | 22.4 | 9.1 | 14.7 | 0 | 44 |
| CpNpC† | 22.5 | 8.4 | 12.7 | 0 | 61 |
| CpCpC | 40.0 | 15.6 | 30.4 | 0 | 9 |
The genotypes analyzed were either the unmutagenized clk-st line (wild-type), or clk-st containing the cmt3-7 and/or drm1 drm2 mutations.
Asymmetric methylation (All sites) are cytosines within the context CpHpH, where H = A, T, or C. In each context shown, the methylated cytosine is in bold.
N = A, T, C, or G.
The effects of the drm mutations on CpNpG methylation at SUP were also different from that seen at FWA and MEA-ISR. drm1 drm2 only reduced CpNpG methylation at the SUP locus by about 30%, whereas cmt3-7 completely abolished this methylation. Thus, the effect of drm1 drm2 on CpNpG methylation is locus-specific, completely eliminating the CpNpG methylation of FWA and MEA-ISR, but only reducing it moderately at SUP.
CMT3 Controls CpNpG Methylation at the Ta3 Retrotransposon and at Centromeric Repeats.
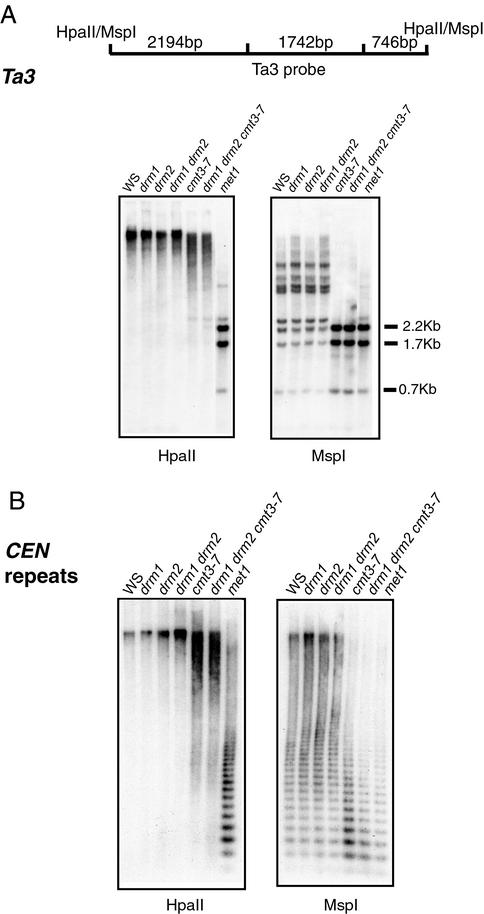
We tested the effect of the various single, double, and triple methyltransferase mutants on two pericentromeric sequences, the Ta3 retrotransposon, and the 180-bp centromeric repeat sequence, using Southern blot analysis with HpaII and MspI (Fig. 3). Neither the drm single mutants nor the drm1 drm2 double mutants affected the pattern of enzyme digestion, suggesting that the DRM genes do not play a role in maintaining CpG or CpNpG methylation at these sequences. In contrast, cmt3-7 and drm1 drm2 cmt3-7 triple mutants showed nearly complete digestion by MspI, but not HpaII, showing that that CMT3 is responsible for the maintenance of CpNpG methylation at both Ta3 and the centromeric repeats (24, 31).
Figure 3.
Southern blot analysis of the Ta3 retrotransposon and 180-bp centromeric repeat sequences. Each panel shows lanes that contain equivalent amounts of genomic DNA of the indicated genotype digested with HpaII (Left) and MspI (Right). (A Upper) A diagram of the HpaII/MspI restriction fragments present in the Ta3 probe. (Lower) A DNA blot probed with Ta3 probe. (B) Blot probed with a 180-bp centromeric repeat probe.
We also found that, in contrast to cmt3 mutants (24) and met1 mutants (X.C, L. Johnson, and S.E.J. unpublished observation), drm1 drm2 double mutants did not show reactivation of RNA expression of the Ta3 sequence (data not shown). Therefore, the DRM genes do not appear to play a role the maintenance of gene silencing of Ta3.
MET1 Likely Affects CpNpG and Asymmetric Methylation Indirectly.
Bisulfite sequencing and Southern blot data show that methylation of CpG sites are unaffected in drm1 drm2 cmt3-7 triple mutant strains at all sequences tested (Figs. 1–3). In contrast, at every sequence tested, the met1 mutant showed a strong reduction in CpG methylation (Figs. 2 and 3). Therefore, like its mammalian counterpart Dnmt1, MET1 appears to be the primary methyltransferase for the maintenance of CpG methylation. However, consistent with earlier studies (22, 23, 46), we observed that met1 mutants greatly reduced CpNpG methylation at FWA, MEA-ISR, Ta3, and the centromeric repeat sequences (Figs. 2 and 3), and eliminated the asymmetric methylation of FWA and MEA-ISR as detected by bisulfite sequencing (data not shown). Because MET1 cannot substitute for the maintenance of CpNpG and asymmetric methylation in drm1 drm2 cmt3-7 strains, it seems most likely that the losses of CpNpG and asymmetric methylation in met1 are not directly caused by promiscuous enzymatic activity of the MET1 enzyme, but are instead a secondary effect caused by the primary loss of CpG methylation.
A Pleiotropic Morphological Phenotype Marks DRM and CMT3 Redundancy.
Although neither the cmt3 mutants (24) nor the drm1 drm2 double mutants (39) show morphological differences from wild type, drm1 drm2 cmt3-7 plants showed a pleiotropic phenotype including developmental retardation, reduced plant size, and partial sterility (Fig. 4B). We did not observe, however, several defects commonly seen in the ddm1 and met1 methylation mutants, such as clavata-like flowers, apetala2-like flowers, agamous-like flowers, sup-like flowers, or extreme late flowering (22, 23, 26, 42). Thus, DRM and CMT3 act in a redundant fashion to control some aspects of plant growth and development, which may be different from those affected in ddm1 and met1 mutants.
Discussion
Locus-Specific Action of the DRM and CMT3 Methyltransferase Genes.
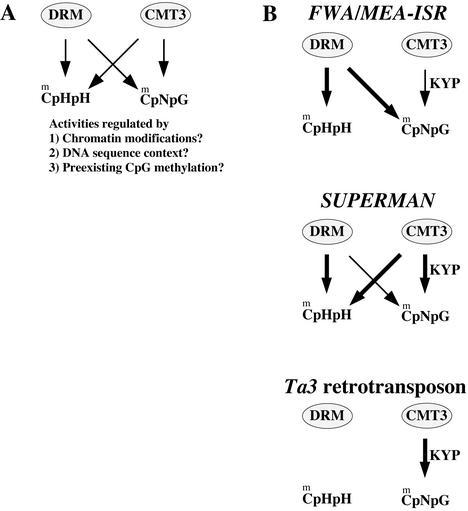
Our results suggest that the DRM and CMT3 genes encode methyltransferase enzymes that show overlapping roles in the control of asymmetric and CpNpG methylation (Fig. 5A). However, the activities of these methyltransferases are highly dependent on the locus under study, giving a surprising number of different patterns of dependence on either DRM, CMT3, or both. Fig. 5B summarizes the results of this study, showing the dependence of each methylated gene on DRM and/or CMT3 for both asymmetric and CpNpG methylation. What are the signals that target these methylases to vary their function? It seems likely that several factors could be involved, including chromatin modifications present at specific loci, the DNA sequences involved, and cross talk between CpG methylation and non-CpG methylation.
Figure 5.
Model illustrating the complex relationship between DRM and CMT3 activities. (A) DRM and CMT3 both have the capacity to methylate asymmetric and CpNpG sites, but these activities are locus-specific, and therefore may be regulated by various factors as discussed in the text. (B) Summary of the activities of DRM and CMT3 inferred from the analysis of the types of methylation lost at each locus in the drm1 drm2, cmt3-7, or drm1 drm2 cmt3-7 mutants. Thicker arrows denote more activity, which is inferred from the greater loss of methylation in a particular mutant.
Chromatin modifications can be considered at two levels: overall chromatin structure, and specific histone modifications. Examples of overall chromatin structure are provided by the Ta3 retrotransposon and centromeric repeats, which are nested within the highly condensed, pericentromeric, constitutive heterochromatin. The higher order chromatin structures present near centromeres may modulate methyltransferase function in such a way that CMT3 is solely responsible for the methylation of CpNpG sites (Fig. 5B). In contrast, MEA-ISR is subtelomeric, and SUP and FWA are in the midst of single copy sequence, which is presumably euchromatic in nature. An example of a specific chromatin modification is histone H3 Lys-9 methylation. Mutations in the KYP gene, which encodes a histone H3 Lys-9-specific methyltransferase, reduce CpNpG methylation (31). This observation suggests that CpNpG DNA methylation is at least partially controlled by histone H3 Lys-9 methylation, through an interaction of CMT3 with methylated chromatin. One possibility is that H3 Lys-9 methylation simply acts to more efficiently recruit CMT3 to particular loci, possibly through an interaction with a plant homolog of the H3 Lys-9 binding protein HP1 (31). A second possibility is that H3 Lys-9 methylation and/or HP1 binding acts to modulate CMT3′s biochemical properties, stimulating its CpNpG methylating activity or modulating its preference for unmethylated or hemimethylated CpNpG sites.
At least two DNA sequence parameters can be imagined that could modulate methyltransferase function; overall sequence architecture and sequence composition. An example of the overall sequence architecture could be the presence of direct or inverted repeats, which are in fact found in all of the genes in this study. FWA, MEA-ISR, and the 180-bp centromeric repeat sequences all contain direct repeats that are immediately adjacent to one another. The Ta3 retrotransposon contains long terminal repeat sequences at each end of the element, which are direct repeats separated by several kilobases. Finally, the clk allele used in this study, clk-st, contains an inverted repeat of the SUP gene in addition to the endogenous SUP locus (39). With respect to sequence composition, it is possible that the density of particular target sites is important. In particular, we note that SUP, a locus whose methylation and silencing is highly dependent on CMT3 function, is rich in CpNpG sites but has few CpG sites. Within the methylated 1,028-bp region there are 9 CpGs and 27 CpNpG sites. Conversely, the FWA locus, whose methylation and silencing is mostly dependent on MET1 function, is rich in CpG. The 500-bp methylated region of FWA contains 20 CpG sites (more than four times the density at SUP), and 14 CpNpG sites.
Another possibility to be considered is that CpG methylation could target non-CpG methylation. The results of this and other studies suggest that MET1 is specific for CpG methylation, and yet met1 mutants show a strong reduction in non-CpG methylation at most loci examined (20, 22–25). The strongest evidence that MET1 does not directly methylate non-CpG sites, is that all traces of non-CpG methylation are lost in drm1 drm2 cmt3-7 triple mutants, which contain a wild-type MET1 gene. In addition, at the SUP locus met1 mutants cause a large reduction of CpG methylation without major losses of CpNpG and asymmetric methylation (24, 25). These results suggest that MET1 is specific for CpG methylation, and that the losses of CpNpG and asymmetric methylation in met1 mutants are indirect. One interpretation of these findings is that preexisting CpG methylation generates signals for the further methylation of cytosines in other contexts. An example is FWA, which is methylated primarily at CpG sites (89%), and less so at CpNpG sites (18%) and asymmetric sites (4%). The DRM loci are mainly responsible for maintaining this non-CpG methylation, because it is lost in the drm1 drm2 double mutants. However, in fwa mutants induced in a met1 background, all traces of both CpG and non-CpG methylation are lost (24, 40). Thus, DRM may methylate CpNpG and asymmetric sites at FWA, either because of the preexisting CpG methylation itself, or because of other chromatin modifications that result from the presence of CpG methylation. A second possibility is that the loss of CpG methylation of FWA is sufficient to reactivate gene expression, and that transcription produces a chromatin structure that is repressive for non-CpG methylation.
An interesting aspect of this study is that CMT3 appears to methylate asymmetric cytosines at the SUP locus, but not at the other tested loci. One possible explanation is that the inverted repeat structure of the SUP locus present in the transgenic clk-st line activates CMT3's ability to methylate asymmetric sites. However, we found residual asymmetric methylation in drm1 drm2 clk-3 triple mutant plants (ref. 39 and data not shown), suggesting that CMT3 can methylate asymmetric sites on the single SUP endogene present in this strain. Therefore, it seems that some aspect of the chromatin or DNA structure of SUP activates CMT3's asymmetric methylation activity. We previously found that the most densely methylated region of SUP contains a pyrimidine-rich sequence, which is predicted to form a small hairpin (26). Similar sequences were also found in both hypermethylated regions of the agamous locus. It seems possible that the unusual DNA structure of the SUP hairpin, or the potential of this hairpin to be transcribed into a small RNA, activates dense non-CpG methylation of SUP. One might predict that the multiple copy PAI loci (47) would show a relationship with DNA methyltransferases similar to SUP, with asymmetric methylation depending on both DRM and CMT3 activities. This is because previous observations of the methylation patterns of the PAI genes showed high levels of asymmetric methylation that were significantly but not completely reduced in cmt3 mutants (30).
In summary, several chromatin or DNA sequence parameters may regulate non-CpG methylation, either alone in combination, and further insight into this problem may only come by studying additional genetic modifiers of non-CpG methylation.
Asymmetric Methylation Sequence Specificity.
An analysis of the sequence context of the asymmetric sites methylated by DRM and CMT3 suggests that both enzymes prefer to methylate CpA and CpT sites relative to CpC sites (Table 1). These results are consistent with those of other studies of asymmetric methylation in plants, where CpA and CpT have been found in more abundance than CpC methylation (6, 17, 48). These results are also consistent with nearest neighbor analysis of plant DNA (5), and with studies of the in vitro substrate preference of purified plant methyltransferases (49, 50). Interestingly, mouse embryonic stem cells also contain a significant amount CpA and CpT methylation and lower amounts of CpC methylation, and indirect evidence suggests an important role for Dnmt3 genes (the closest mammalian DRM homologs) in maintaining this methylation (2). For example, mouse embryonic stem cells mutant for Dnmt1 still contain a significant level of CpA and CpT methylation, and the high level of expression of Dnmt3a implicates it as the methyltransferase responsible (2). Furthermore, Dnmt3a can methylate non-CpG sites in vitro, or when ectopically expressed in Drosophila cells (2, 16) and in both cases CpA and CpT sites are methylated more frequently than CpC sites. Finally, the site CmeC(A/T)GG is also found to be methylated in mammals (51–55). Thus a preference for the methylation of CpA and CpT sites may be a conserved property of asymmetric methyltransferases.
De Novo Methylation vs. the Maintenance of Non-CpG Methylation.
Previous characterization of the drm mutants suggested that the DRMs encode the major de novo methyltransferases in Arabidopsis (39). For instance, drm1 drm2 double mutants completely blocked de novo CpG methylation and transgene silencing of FWA, which normally occurs when FWA is transformed into plants. Furthermore, drm1 drm2 double mutants blocked de novo CpNpG and asymmetric methylation and gene silencing of the endogenous SUP locus, which normally occurs in the presence of a SUP inverted repeat transgene locus. These experiments suggest that the DRM genes encode enzymes capable of methylating previously unmodified DNA, and that CMT3 cannot substitute for this function. However, in this study we show that, with respect to genes whose silencing is already established, the DRM and CMT3 loci act redundantly to maintain the proper patterns of asymmetric and CpNpG methylation. Thus, a distinction must be made between the initial methylation and silencing of a gene (normally termed de novo methylation) and the maintenance of overall patterns of non-CpG methylation once a gene has been initially methylated. DRM appears to participate in both of these processes, and CMT3 only in the latter.
Further, although the DRM and CMT3 loci are functionally redundant in terms of maintenance of overall patterns of non-CpG methylation, it is possible, particularly in the case of CpNpG methylation that the two act by different mechanisms. For instance, it is possible that DRM2 is more efficient at methylating completely unmodified CpNpG sites, whereas CMT3 may show a preference for hemimethylated CpNpG sites.
The Biological Function of Non-CpG Methylation.
We found that although neither the drm1 drm2 double mutants nor the cmt3-7 single mutant showed morphological differences from the wild type, drm1 drm2 cmt3-7 plants showed pleiotropic developmental defects. These drm1 drm2 cmt3-7 plants retained CpG methylation at all of the sequences tested (Figs. 1–3), suggesting that the pleiotropic phenotype is caused by a reduction of non-CpG methylation, which is important for some endogenous plant process(es). In plants, high levels of non-CpG methylation are associated with the RNA-directed DNA methylation that is observed during transcriptional and posttranscriptional gene silencing, and this methylation may contribute to the silencing of viral and transposon sequences (15, 56–62). Thus, one possibility is that the pleiotropic phenotype of drm1 drm2 cmt3-7 plants is caused by defects in genome defense/gene silencing processes. Further study of these triple mutant plants may help to reveal possible biological functions of non-CpG methylation.
Supplementary Material
Acknowledgments
We thank Mike Huang and Yoo Lee for technical assistance, Eric Richards for providing seeds of the met1 (ddm2) mutant, and Daniel Zilberman, Jim Jackson, Anders Lindroth, Lianna Johnson, Nathan Springer, and Shawn Kaeppler for helpful discussions. This work was supported by National Institutes of Health Grant GM60398 (to S.E.J.).
Abbreviations
- CMT3
chromomethylase 3
- DRM
domains rearranged methylase
- WS
Wassilewskija
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Self-Perpetuating Structural States in Biology, Disease, and Genetics,” held March 22–24, 2002, at the National Academy of Sciences in Washington, DC.
References
- 1.Richards E J, Elgin S C. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- 2.Ramsahoye B H, Biniszkiewicz D, Lyko F, Clark V, Bird A P, Jaenisch R. Proc Natl Acad Sci USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selker E U, Fritz D Y, Singer M J. Science. 1993;262:1724–1728. doi: 10.1126/science.8259516. [DOI] [PubMed] [Google Scholar]
- 4.Goyon C, Nogueira T I, Faugeron G. J Mol Biol. 1994;240:42–51. doi: 10.1006/jmbi.1994.1416. [DOI] [PubMed] [Google Scholar]
- 5.Gruenbaum Y, Naveh-Many T, Cedar H, Razin A. Nature (London) 1981;292:860–862. doi: 10.1038/292860a0. [DOI] [PubMed] [Google Scholar]
- 6.Meyer P, Niedenhof I, ten Lohuis M. EMBO J. 1994;13:2084–2088. doi: 10.1002/j.1460-2075.1994.tb06483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingelbrecht I, Van Houdt H, Van Montagu M, Depicker A. Proc Natl Acad Sci USA. 1994;91:10502–10526. doi: 10.1073/pnas.91.22.10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobsen S E, Meyerowitz E M. Science. 1997;277:1100–1103. doi: 10.1126/science.277.5329.1100. [DOI] [PubMed] [Google Scholar]
- 9.Riggs A D. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 10.Holliday R, Pugh J E. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 11.Bird A P. J Mol Biol. 1978;118:49–60. doi: 10.1016/0022-2836(78)90243-7. [DOI] [PubMed] [Google Scholar]
- 12.Bestor T, Laudano A, Mattaliano R, Ingram V. J Mol Biol. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- 13.Bestor T H. EMBO J. 1992;11:2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leonhardt H, Page A W, Weier H U, Bestor T H. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 15.Pelissier T, Thalmeir S, Kempe D, Sanger H L, Wassenegger M. Nucleic Acids Res. 1999;27:1625–1634. doi: 10.1093/nar/27.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gowher H, Jeltsch A. J Mol Biol. 2001;309:1201–1208. doi: 10.1006/jmbi.2001.4710. [DOI] [PubMed] [Google Scholar]
- 17.Dieguez M J, Vaucheret H, Paszkowski J, Mittelsten Scheid O. Mol Gen Genet. 1998;259:207–215. doi: 10.1007/s004380050806. [DOI] [PubMed] [Google Scholar]
- 18.Dieguez M J, Bellotto M, Afsar K, Mittelsten Scheid O, Paszkowski J. Mol Gen Genet. 1997;253:581–588. doi: 10.1007/s004380050360. [DOI] [PubMed] [Google Scholar]
- 19.Finnegan E J, Kovac K A. Plant Mol Biol. 2000;43:189–201. doi: 10.1023/a:1006427226972. [DOI] [PubMed] [Google Scholar]
- 20.Finnegan E J, Dennis E S. Nucleic Acids Res. 1993;21:2383–2388. doi: 10.1093/nar/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vongs A, Kakutani T, Martienssen R A, Richards E J. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 22.Finnegan E J, Peacock W J, Dennis E S. Proc Natl Acad Sci USA. 1996;93:8449–8454. doi: 10.1073/pnas.93.16.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronemus M J, Galbiati M, Ticknor C, Chen J, Dellaporta S L. Science. 1996;273:654–657. doi: 10.1126/science.273.5275.654. [DOI] [PubMed] [Google Scholar]
- 24.Lindroth A M, Cao X, Jackson J P, Zilberman D, McCallum C M, Henikoff S, Jacobsen S E. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 25.Kishimoto N, Sakai H, Jackson J, Jacobsen S E, Meyerowitz E M, Dennis E S, Finnegan E J. Plant Mol Biol. 2001;46:171–183. doi: 10.1023/a:1010636222327. [DOI] [PubMed] [Google Scholar]
- 26.Jacobsen S E, Sakai H, Finnegan E J, Cao X, Meyerowitz E M. Curr Biol. 2000;10:179–186. doi: 10.1016/s0960-9822(00)00324-9. [DOI] [PubMed] [Google Scholar]
- 27.Baylin S B, Herman J G. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 28.Henikoff S, Comai L. Genetics. 1998;149:307–318. doi: 10.1093/genetics/149.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCallum C M, Comai L, Greene E A, Henikoff S. Nat Biotechnol. 2000;18:455–457. doi: 10.1038/74542. [DOI] [PubMed] [Google Scholar]
- 30.Bartee L, Malagnac F, Bender J. Genes Dev. 2001;15:1753–1758. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson J P, Lindroth A M, Cao X, Jacobsen S E. Nature (London) 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 32.Cao X, Springer N M, Muszynski M G, Phillips R L, Kaeppler S, Jacobsen S E. Proc Natl Acad Sci USA. 2000;97:4979–4984. doi: 10.1073/pnas.97.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okano M, Xie S, Li E. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 34.Hsieh C L. Mol Cell Biol. 1999;19:8211–8218. doi: 10.1128/mcb.19.12.8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okano M, Bell D W, Haber D A, Li E. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 36.Lyko F, Ramsahoye B H, Kashevsky H, Tudor M, Mastrangelo M A, Orr-Weaver T L, Jaenisch R. Nat Genet. 1999;23:363–366. doi: 10.1038/15551. [DOI] [PubMed] [Google Scholar]
- 37.Xu G L, Bestor T H, Bourc'his D, Hsieh C L, Tommerup N, Bugge M, Hulten M, Qu X Y, Russo J J, Viegas-Pequignot E. Nature (London) 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 38.Hansen R S, Wijmenga C, Luo P, Stanek A M, Canfield T K, Weemaes C M, Gartler S M. Proc Natl Acad Sci USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao X, Jacobsen S E. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- 40.Soppe W J, Jacobsen S E, Alonso-Blanco C, Jackson J P, Kakutani T, Koornneef M, Peeters A J. Mol Cell. 2000;6:791–802. doi: 10.1016/s1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- 41.Kakutani T, Munakata K, Richards E J, Hirochika H. Genetics. 1999;151:831–838. doi: 10.1093/genetics/151.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kakutani T, Jeddeloh J A, Flowers S K, Munakata K, Richards E J. Proc Natl Acad Sci USA. 1996;93:12406–12411. doi: 10.1073/pnas.93.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grossniklaus U, Vielle-Calzada J P, Hoeppner M A, Gagliano W B. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- 44.Kiyosue T, Ohad N, Yadegari R, Hannon M, Dinneny J, Wells D, Katz A, Margossian L, Harada J J, Goldberg R B, Fischer R L. Proc Natl Acad Sci USA. 1999;96:4186–4191. doi: 10.1073/pnas.96.7.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McClelland M, Nelson M, Raschke E. Nucleic Acids Res. 1994;22:3640–3659. doi: 10.1093/nar/22.17.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartee L, Bender J. Nucleic Acids Res. 2001;29:2127–2134. doi: 10.1093/nar/29.10.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bender J, Fink G R. Cell. 1995;83:725–734. doi: 10.1016/0092-8674(95)90185-x. [DOI] [PubMed] [Google Scholar]
- 48.Luff B, Pawlowski L, Bender J. Mol Cell. 1999;3:505–511. doi: 10.1016/s1097-2765(00)80478-5. [DOI] [PubMed] [Google Scholar]
- 49.Theiss G, Schleicher R, Schimpff-Weiland G, Follmann H. Eur J Biochem. 1987;167:89–96. doi: 10.1111/j.1432-1033.1987.tb13307.x. [DOI] [PubMed] [Google Scholar]
- 50.Adams R L, Lindsay H, Reale A, Seivwright C, Kass S, Cummings M, Houlston C. EXS. 1993;64:120–144. doi: 10.1007/978-3-0348-9118-9_6. [DOI] [PubMed] [Google Scholar]
- 51.Clark S J, Harrison J, Frommer M. Nat Genet. 1995;10:20–27. doi: 10.1038/ng0595-20. [DOI] [PubMed] [Google Scholar]
- 52.Franchina M, Kay P H. DNA Cell Biol. 2000;19:521–526. doi: 10.1089/104454900439755. [DOI] [PubMed] [Google Scholar]
- 53.Lorincz M C, Schubeler D, Goeke S C, Walters M, Groudine M, Martin D I. Mol Cell Biol. 2000;20:842–850. doi: 10.1128/mcb.20.3.842-850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malone C S, Miner M D, Doerr J R, Jackson J P, Jacobsen S E, Wall R, Teitell M. Proc Natl Acad Sci USA. 2001;98:10404–10409. doi: 10.1073/pnas.181206898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lorincz M C, Groudine M. Proc Natl Acad Sci USA. 2001;98:10034–10036. doi: 10.1073/pnas.201392598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wassenegger M, Heimes S, Riedel L, Sanger H L. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 57.Jones A L, Thomas C L, Maule A J. EMBO J. 1998;17:6385–6393. doi: 10.1093/emboj/17.21.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mette M F, van der Winden J, Matzke M A, Matzke A J. EMBO J. 1999;18:241–248. doi: 10.1093/emboj/18.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mette M F, Aufsatz W, van der Winden J, Matzke M A, Matzke A J. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wassenegger M. Plant Mol Biol. 2000;43:203–220. doi: 10.1023/a:1006479327881. [DOI] [PubMed] [Google Scholar]
- 61.Matzke M, Matzke A J M, Kooter J M. Science. 2001;293:1080–1083. doi: 10.1126/science.1063051. [DOI] [PubMed] [Google Scholar]
- 62.Jones L, Ratcliff F, Baulcombe D C. Curr Biol. 2001;11:747–757. doi: 10.1016/s0960-9822(01)00226-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.