Abstract
Microarrays are an effective tool for monitoring genome-wide gene expression levels. In current microarray analyses, the majority of genes on arrays are frequently eliminated for further analysis because the changes in their expression levels (ratios) are considered to be not significant. This strategy risks failure to discover whole sets of genes related to a quantitative trait of interest, which is generally controlled by several loci that make various contributions. Here, we describe a high-throughput gene discovery method based on correspondence analysis with a new index for expression ratios [arctan (1/ratio)] and three artificial marker genes. This method allows us to quickly analyze the whole microarray dataset and discover up-/down-regulated genes related to a trait of interest. We employed an example dataset to show the theoretical advantage of this method. We then used the method to identify 88 cancer-related genes from a published microarray data from patients with breast cancer. This method also allows us to predict the phenotype of a given sample from the gene expression profile. This method can be easily performed and the result is also visible in 3D viewing software that we have developed.
INTRODUCTION
Microarray experiments are widely used to simultaneously monitor the expression levels of thousands to tens of thousands of genes in many organisms (1–3). In microarray data analyses, genes showing 2-fold relative expression levels at least or >2 SDs away from the mean among expression levels are often considered to show precise measurement or significantly different expression from the control. These genes are selected for further analysis (4–6). This approach usually eliminates the majority of genes on the array for further analysis.
The expression levels of many genes show wide natural variation (7,8). There is no firm theoretical basis for defining a significant expression level (9). The considerable elimination of microarray data poses a serious problem for the analyses of quantitative traits. The quantitative traits are affected by several or more loci. The effects of each loci on the phenotype are different (10). The current approach with threshold values for expression levels could eliminate genes which affect the phenotypes with small changes of expression levels.
We suspected that there is a practical reason for the tendency to over-reduce the number of genes for further analysis. Analyses of microarray data are commonly performed by hierarchical clustering methods (11–13). However, the hierarchical clustering for a large microarray dataset with >10 000 genes is too much time consuming and not practical. The detection of a clear cut-off point in large dendrograms is also difficult. Eliminating genes with small fold changes in expression levels for further analysis allows us to perform hierarchical clustering analyses in a short time and easily detect clusters with differerent gene expression profiles.
Recently, principal component analysis (PCA) has been used to process microarray data (14–16). PCA calculations for the whole microarray dataset are not time consuming. PCA reduces the high dimensionality of a large microarray dataset (matrix). The scores (coordinates) of the first three principal components allow visual assessment of associations between genes and phenotypes in a 3D subspace. However, correspondence analysis (CA) (17) is more effective than PCA for discovering genes related to phenotypes of interest.
As with PCA, CA allows us to summarize an originally high-dimensional data matrix (row [gene] and column [sample]) with a low-dimensional projection. In CA, genes and samples (phenotypes) are projected into a 2- or more-dimensional subspace at the same time (bi-plot). This bi-plot reveals associations across genes and samples. Kishino and Waddell (18) applied CA to a matrix of normalized fluorescence intensities. To apply CA for log-transformed intensity ratios, Fellenberg et al. (19) additively shifted the log-ratios to a positive range. However, minimum values of log-ratios are not same among experiments. Differences of logarithmic bases (e.g. 2, 10 and e) also provide different values of log-ratios. Consequently, even though many expression datasets are available from public databases, care must be taken in the logarithmic bases prior to analyses. Ranking is another transformation method for expression data (20), but it causes a loss of information in gene expression levels. These two current indices, additively shifted log-ratios and ranks, for gene expression profiles are unsuitable in CA.
We describe here a high-throughput gene discovery method based on CA with a new index for expression ratios and three artificial marker genes. This method also allows prediction of phenotypes from the gene expression profiles.
MATERIALS AND METHODS
CA and PCA for microarray data
CA and PCA were performed using the statistical software package R (http://cran.r-project.org/) and its library ‘multiv’ on a 2.60 GHz Intel Pentium4 personal computer with 2 GB of random access memory. CA with a new index was also performed with our developed software mentioned below. PCA and CA provide scores (coordinates) for genes and samples.
Preparation of an example dataset
An example dataset contains gene expression ratios for 516 genes and 100 samples (Supplementary Table 1). Of the 100 samples, 50 were phenotype A and 50 were phenotype B. There are five down-regulated genes (D1 to D5) and five up-regulated genes (U1 to U5) in phenotype A. Among the same phenotype, these 10 genes have the same expression ratios. In addition, three housekeeping genes (HK1, HK2 and HK3) have the same expression ratios among all samples. There are 500 genes unrelated to phenotypes (Unrelated1 to Unrelated 500). The expression ratios for the unrelated genes were randomly selected from the published microarray data (21). This example dataset includes three artificial marker genes.
Preparation of breast cancer expression data
The published microarray data (21) includes 24 481 genes and 117 samples. Two samples and 457 genes that had more than two missing values were eliminated from the dataset. For the remainder, the missing values, at most two, were replaced by the average expression ratio for the same phenotype. Out of 115 samples, 62 samples were from patients that developed metastases within 5 years after their initial diagnosis (poor prognoses), and 53 samples were from patients that remained free of disease for at least 5 years after diagnosis (good prognoses). The dataset for 24 024 genes and 115 samples is shown in Supplementary Table 2.
Significant distances in a low-dimensional projection
We used a confidence area for a location of a point (plot) of genes in a low-dimensional projection obtained by CA to identify up-/down-regulated and housekeeping genes. For an ith row (gene) in a contingency table, a 95% confidence area is defined as a confidence circle centered at the location of the gene in the 2D subspace (22), where the radius is , the value of the statistic χ2 with two degrees of freedom (d.f.) is 5.99 at a 0.05 significance level, and Ki• is the total of the elements in the i-th row. The d.f. of χ2 are equal to the number of dimensions in the subspace.
Detection of gene ontology terms
We performed statistical analyses of gene ontology (GO) terms for the candidate cancer-related genes with the web-based tool GOTM (http://genereg.ornl.gov/gotm/) (23). The GOTM provides GO terms and their significant probabilities (P-values). GO terms with P-values <0.05 were retrieved.
Identification of MeSH terms for genes
We identified ‘Disease’ MeSH terms related to genes using BioCompass (NEC Corporation), which searches for MeSH terms significantly related to genes using a supervised classification method. The reliability of the assignment is shown as a score. MeSH terms with scores over 0.05 were selected as highly significant.
Hierarchical clustering
Hierarchical clustering (complete linkage clustering with Spearman rank correlation) was performed using Cluster (Stanford University). Dendrograms and expression maps were generated by Treeview (Stanford University).
Prediction ratio of phenotypes in CA
A Monte-Carlo simulation with 10 000 runs was performed to investigate the distribution of the prediction ratios. In each run, the 115 samples were randomly divided into 95 ‘supervised’ and 20 ‘query’ samples. Although the phenotypes of the supervised samples were available, the phenotypes of the query samples were not. The 7D distances between each query sample and each supervised sample were calculated. The reciprocal distance was used as the score to weight the distance to a close supervised sample.
We compared the two total scores and for each query sample, where DPi is the distance to ith supervised poor prognosis sample and DGj is the distance to jth supervised good prognosis sample. When , the query sample was predicted to be from a patient with a poor prognosis and vice versa. The prediction ratio in each run was computed from 20 query samples.
RESULTS
A new index for gene expression ratios
The new index for gene expression ratios is the arctangent (inverse tangent) of the reciprocal intensity ratio (arctan[1/ratio]). The reciprocal ratio is equivalent to the differential coefficient of the natural logarithm of the ratio. Consequently, this index ranges from 0 to 90°. When the ratio is equal to one, the index is 45°. As the gene is repressed or induced, this index increases or decreases from 45°, respectively. This index changes more substantially than conventional indices (log[ratio]) when the ratio is between 0.1 and 10 (Supplementary Figure 1). When the ratio is <0.1 or >10, the new index changes less than the current indices. Nonetheless, the power of gene discovery is maintained because the new index still allows heavily repressed or induced genes to be easily identified.
Three artificial marker genes to identify genes associated with a trait
We added three artificial genes (ExtraGenes) to the dataset to classify all genes on the array. Assuming that there are two phenotypes (A and B) of a quantitative trait, genes on the array can be classified into the following four categories: (i) genes specifically expressed in either phenotype, (ii) genes up- or down-regulated between the two phenotypes, (iii) genes up- or down-regulated that are unrelated to the phenotypes, and (iv) housekeeping genes that show constant levels of expression in all samples. The genes related to the phenotypes are included in categories (i) and (ii). To classify all genes, we used three ExtraGenes (ExtraGene1, ExtraGene2 and ExtraGene3) to the dataset. The expression ratio of ExtraGene1 is zero in phenotype A samples, and the maximum expression ratio is given to phenotype B samples. ExtraGene2 has the inverse gene expression pattern as ExtraGene1. ExtraGene1 and ExtraGene2 assist in the discovery of genes related to the phenotypes and housekeeping genes [categories (i), (ii) and (iv)] as described below. ExtraGene3 shows the same ratio (1.0) in all samples and also aids in the identification of housekeeping genes [category (iv)]. Consequently, genes related to the phenotypes can be obtained. The ExtraGenes introduced here are different from the ‘virtual genes’ employed by Fellenberg et al. (19) to directly interpret a distance between a gene and a sample.
A line segment to identify up-/down-regulated genes
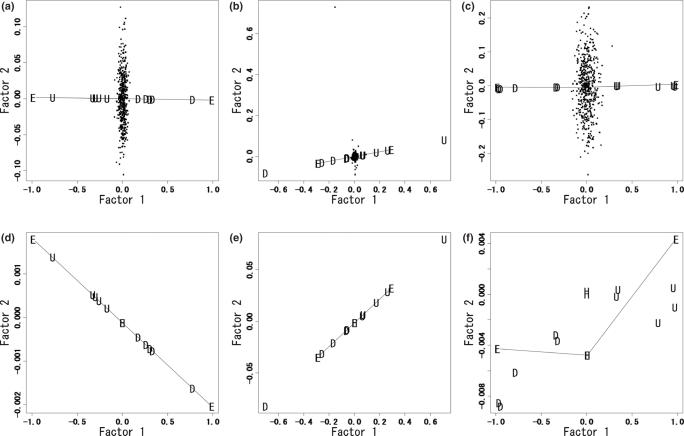
We performed CA and PCA using the new index (arctan[1/ratio]) and current indices (additively shifted log2[ratio] and rank) using an example dataset. This example dataset includes three ExtraGenes (ExtraGene1 to ExtraGene3). In ExtraGene1, the ratios in phenotypes A and B are 0 and 100, respectively. ExtraGene2 has the inverse profile as ExtraGene1, and ExtraGene3 has the same ratio (1.0) for all samples. As expected, regardless of the index, CA separates the samples into positive and negative scores along the first axis (Factor1) according to phenotypes A and B (data not shown). However, projections of genes into the first 2D subspaces show different patterns among the indices (Figure 1). The cumulative contribution ratios in Figure 1a–c are 60.0, 63.9 and 40.3%, respectively. From CA using the new index (Figure 1a and d), up- and down-regulated genes have negative and positive scores in Factor1, respectively, and lie on a line segment between ExtraGene1 and ExtraGene2. We call this line segment the UDL (up/down line). As expected from CA, all housekeeping genes and ExtraGene3 lie in the center of the UDL, which is the origin of the subspace. Locations of genes unrelated to phenotypes are random and independent of the UDL.
Figure 1.
CA with three indices. Factor1 and Factor2, the first two axes obtained from CA, respectively; U, genes up-regulated (U1 to U5) in phenotype A samples; D, down-regulated genes (D1 to D5) in phenotype A samples; H, housekeeping genes (HK1 to HK3); E, ExtraGenes (ExtraGene1 to ExtraGene3); dots, unrelated genes (Unrelated1 to Unrelated500). (a) CA with the new index. (b) CA with an additively shifted logarithmic ratio. (c) CA with a rank index. (d–f) Plots of only U, D, H and E for (a–c), respectively.
CA with an additively shifted log2(ratio) index does not give a UDL (Figure 1b and e) because genes D1 and U1 lie outside of the line segment between ExtraGene1 and ExtraGene2. This is due to the fact that the expression ratios between the two phenotypes are the largest for these two genes. As in Figure 1d, housekeeping genes and ExtraGene3 are plotted in the middle between ExtraGene1 and ExtraGene2. CA with a rank index cannot create a UDL (Figure 1c and f). Both the up-/down-regulated and housekeeping genes are randomly placed away from the line segments obtained from the ExtraGenes. Only CA with the new index can define a UDL that allow us to predict up-/down-regulated genes among phenotypes. The UDL defined here is not identical to the line to ‘standard coordinates’ with mean 0 and variance 1 (24). Fellenberg et al. (19) used standard coordinates to classify genes and samples in a bi-plot.
The results of PCA with the three indices are shown in Supplementary Figure 2. The cumulative contribution ratios in Supplementary Figure 2a–c are 71.1, 85.1 and 43.5%, respectively. Regardless of the index, PCA did not generate a UDL. The genes were all randomly located with the ExtraGenes in the subspace. This result shows that, regardless of the index, PCA is not appropriate for the clustering of genes according to their expression patterns.
Analysis of breast cancer data
We next applied CA with the new index to published human breast cancer microarray data (21). This available data contains 24 024 gene expression ratios from 115 samples (Supplementary Table 2). The three ExtraGenes were also added to the dataset. We calculated 7D scores to 24 027 genes and the 115 samples. This process takes only ∼10 s. Thus, like PCA, CA requires considerably less time for calculation than hierarchical clustering.
Up-/down-regulated genes in significant regions
As shown in Figure 1d, a UDL was determined as a line segment between ExtraGene1 and ExtraGene2 (Figure 2a and Supplementary Figure 3a). Genes up- or down-regulated between good and poor prognosis samples and housekeeping genes are expected to lie on the UDL. However, the locations of these genes can statistically deviate from the UDL as well as biometric data generally deviate from the expected value.
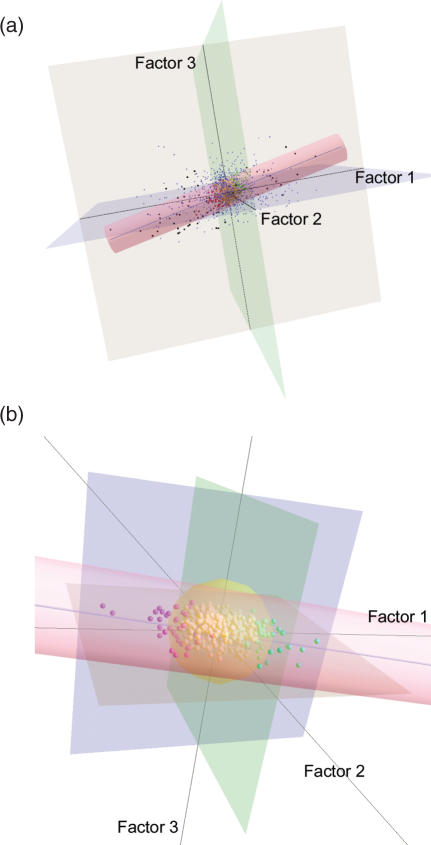
Figure 2.
CA plots for 24 024 genes in the first 3D subspace. Factor1, Factor2 and Factor3 show the first three axes obtained from CA, respectively. (a) CA plot for all of the analyzed 24 024 genes. The cylinder indicates the UDR. The blue line inside the UDR is the UDL. The 23 480 genes (small blue dots) unrelated to cancer are outside the UDR. The black dots out of the UDR correspond to 70 candidate genes identified by van't Veer et al. (21). (b) CA plot for the 544 genes inside the UDR. The 456 yellow spheres indicate significant housekeeping genes. Of the remaining 88 genes, the 43 red and 45 green spheres indicate statistically significant up- and down-regulated genes, respectively.
For the breast cancer data, we applied the confidence areas to a 7D space. The value of the statistic χ2 with seven d.f. at a significance level of 0.05 is 14.0671. The significant distance from an ExtraGene becomes , where n is the number of samples and fi is the new index (arctan[1/ratio]) for the ExtraGene of the ith sample. Consequently, the significant distance from ExtraGene1 is 0.0502 because . Similarly, the significant distances from ExtraGene2 and ExtraGene3 are 0.0543 and 0.0521, respectively.
The significant distance from ExtraGene1 was used as the significant distance from the UDL because they are nearly equal. Up-/down-regulated and housekeeping genes were located inside the confidence area of UDL with a 95% probability. We call this confidence area the up/down region (UDR). In the first 3D subspace, the UDR is visualized as a cylindrical shape (Figure 2a and Supplementary Figure 3a). Using this UDR, we estimated that 544 genes are up-/down-regulated or housekeeping genes.
Detection of 88 genes related to breast cancer
It is expected that housekeeping genes cluster around the position of ExtraGene3. Housekeeping genes were defined as those that have less than the significant distance from ExtraGene3. A statistical test with a 95% significant distance from ExtraGene3 is also available. This significant region forms as a spherical space in the 3D subspace (Figure 2b and Supplementary Figure 3b). Here, we call this region the HKR (housekeeping region). Consequently, we detected 88 genes associated with the diagnosis of breast cancer (Supplementary Table 3). Out of the 88 genes, 45 and 43 genes had positive and negative first-axis coordinates, respectively.
Functions of the detected genes
The set of 88 genes does not include any marker genes identified in the previous report (21). To compare the biological functions of the two gene sets, we investigated GO annotations. The result shows that GO terms related to cell cycle (e.g. cell cycle and division) and apoptosis (e.g. I-kappaB kinase/NF-kappaB cascade and induction of programmed cell death) are highly significant for the 88 genes detected here (Supplementary Table 4a). These biological processes are well known to be affected by the activities of oncogenes. As shown in Supplementary Table 4b, the majority of GO terms for the previously reported 70 genes are related to cellular development (e.g. cellular growth and morphogenesis) and DNA metabolism (e.g. DNA metabolism and strand elongation). We also investigated MeSH terms assigned for the 88 genes shown here (Supplementary Table 5). The 14 MeSH terms are significantly related to neoplasms. Together with the GO terms, these MeSH terms suggested that the 88 genes detected here are related to cancer.
The biological functions of 35 of the 88 genes identified here have been investigated in previous studies. There is no detailed information on the function of the other 53 genes in public databases or published reports. Based on the published reports on the 35 previously studied genes, 18 of them are oncogenes, candidate target genes for tumor therapy, or genes with known carcinogenic functions (Table 1).
Table 1.
Biological functions of representative genes detected in this work
| Gene symbol | Aliases | Regulation | Description | Reference |
|---|---|---|---|---|
| ALK | – | Up | Having oncogenetic roles of haematopoietic and non-haematopoietic tumors | Pulford et al. (25) |
| BCL10 | Bcl10 | Up | Activation of NF-κB cascade through ubiquitination of NEMO | Zhou et al. (26) |
| ERN1 | IRE1 | Up | Mediating endoplasmic reticulum stress-induced NF-κB activation | Kaneko et al. (27) |
| MTCP1 | MTCP-1 | Up | A candidate gene potentially involved in the leukemogenic process of mature T cell proliferations | Stern et al. (28) |
| SAFB | SAFB1 | Up | A repressor of ERα activity via indirect association with histone deacetylation | Townson et al. (29) |
| ASRGL1 | hALP | Up | A transactivator of telomerase activity | Lv et al. (30) |
| DHX9 | RHA | Up | A component of the transactivation complex for the transcriptional activity of NF-κB | Tetsuka et al. (31) |
| STAG1 | – | Up | A transcriptional target for p53 and a mediator of p53-dependent apoptosis | Anazawa et al. (32) |
| CDK5R1 | p35, p25 | Up | A mediator of apoptosis in digoxin-triggered prostate cancer cell | Lin et al. (33) |
| RASSF1 | RASSF1A | Down | DNA methylation of RASSF1 promortor is associated with poor outcome of breast cancer | Müller et al. (34) |
| SRA1 | SRA | Down | A coactivator of ERα transcriptional activity | Cavarretta et al. (35) |
| TNFSF12 | TWEAK | Down | Inducing multiple pathways of cell death | Nakayama et al. (36) |
| CST3 | CystC | Down | Inhibiting the invasion of breast cancer cell | Sokol et al. (37) |
| EGR3 | – | Down | A target for transcriptional factor ERα | Inoue et al. (38) |
| CCNL2 | Cyclin L2 | Down | A regulator of the transcription and RNA processing of certain apoptosis-related factors | Yang et al. (39) |
| SEPW1 | – | Down | Allelic loss of the chromosome 19q arm is a frequent event in human diffuse gliomas | Smith et al. (40) |
| SYNPO2 | Myopodin | Down | A tumor suppressor gene to limit the growth and to inhibit the metastasis of cancer cells | Jing et al. (41) |
| ZDHHC13 | FLJ10852 | Down | Forced expression of ZDHHC13 activates the NF-κB signaling pathway | Matsuda et al. (42) |
Gene symbol, representative gene symbol of the candidate gene; aliases, other names of the gene or its product in references; regulation, ‘up’ or ‘down’ indicates the gene regulation detected in poor prognosis patient group by our method.
Sample and gene classification in CA
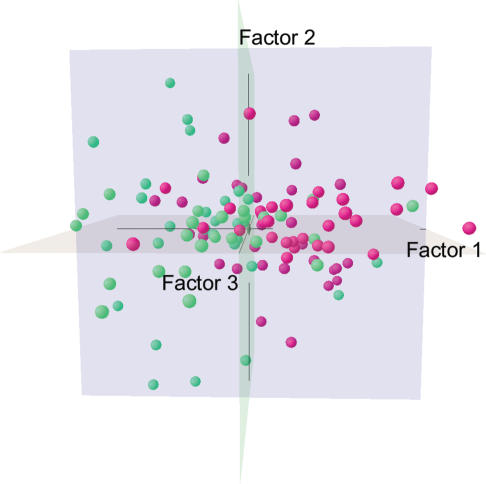
We used the new method on the 88 detected genes from the 115 samples to evaluate the power of sample and gene classification. The majority of poor and good prognosis samples separate into positive and negative first-axis scores, respectively (Figure 3). This incomplete classification is due to the low cumulative explained percentage (63.1%) in the first 3D subspace. However, even if the cumulative explained percentage is not low, the classification of the phenotypes of a quantitative trait (disease outcome) would be still difficult because heritability of a quantitative trait is generally not high. Quantitative traits are influenced not only by gene expression (genetic) but also by environmental (non-genetic) factors. This raises the possibility that gene expression patterns alone cannot adequately account for differences between phenotypes of a quantitative trait.
Figure 3.
CA plot for the 115 samples. Factor1, Factor2 and Factor3 show the first three axes obtained from CA, respectively. Green and red spheres indicate samples from patients with poor and good prognoses, respectively.
We performed hierarchical clustering for the 115 samples and 88 genes detected here to verify the gene and sample classifications obtained from CA. Both genes and samples divided into two subclusters (Supplementary Figure 4). Most of the samples were correctly classified into poor and good prognosis groups. The incomplete classification of samples again is likely due to the same reasons as in the CA results (Figure 3). In the two gene subclusters, 41 and 47 genes are up- or down-regulated in the poor prognosis samples. Except for only four genes, the two gene sets in the subclusters are consistent with the two gene sets separated by the positive and negative scores of the first axis in CA (Figure 2b and Supplementary Table 3).
Predictions of phenotypes by the new method
van't Veer et al. (21) suggested that gene expression profiles could correctly predict phenotypes of samples (83% prediction rate). This conclusion was based on a single population. The predictability is expected to change according to the population sets used. The predictability of phenotypes from gene expression profile alone is one of the most important issues. A Monte-Carlo simulation with 10 000 runs was performed to obtain the distribution of prediction rates. The average of the prediction rates across all runs was ∼73%. The SD was 9%. The range was from 35 to 100%, and 7325 runs showed prediction rates over 70%.
Development of tools for the new method
Finally, as part of the current studies, we developed a software GuCAL that can easily carry out our method of analysis (Supplementary Data). The results can be visualized as a 3D image using Java3D software, which was developed with the J2SE Software Development Kit (SDK). This viewing software allows rotation, zooming in and out, and panning of the image. The 3D subspace for any analyzed data can be created using GuCAL and another Perl script, CAView (Supplementary Data).
DISCUSSION
We describe here a method for gene discovery from microarray data. Our method, CA with a new index for expression ratios coupled with the inclusion of ExtraGenes, allows us to define a UDL, UDR and HKR, which assist in the detection of genes related to the phenotype of interest. Although the confidence regions used here for UDR and HKR are defined for a contingency table, the application shows good classifications of genes. Our method also dramatically reduces the calculation time, and it is effective at predicting the phenotype based on the gene expression profile.
Using this method, we detected 88 prognostic marker genes from a published human breast cancer dataset (21). van't Veer et al. (21) selected 4968 genes from the 24 481 genes on this array, from which 70 marker genes were identified using a three-step supervised classification method. Both the 70 candidate genes identified in this previous report and the 88 genes detected here show up-/down-regulation between poor and good prognosis samples. The 88 genes identified here do not include any of 70 previously identified genes, but it does include known cancer-related genes (Table 1). Especially, gene associated with breast cancer, such as tumor suppressers, NF-κB activators and genes associated with estrogen receptor-α (ERα) were identified.
RASSF1 and CST3 are tumor suppresser genes in breast cancer (Table 1). RASSF1 regulates cell cycle progression and apoptosis (43). Müller et al. (34) suggested that the aberrant DNA methylation of RASSF1 is a powerful prognostic factor in breast cancer. CST3 is an antagonist of oncogenic TGF-β signaling, which promotes invasion in malignant human breast cancer cells (37). Our result shows the down-regulations of the above two tumor suppresser genes in poor prognosis samples (Table 1). It supports the previous reports.
NF-κB activators, BCL10, ERN1 and DHX9, were also identified (Table 1). NF-κB, which is general carcinogenesis including breast cancer, functions as a cancer-related transcription factor involved in cell proliferation and anti-apoptosis (44). For example, NF-κB cascade induces the proliferation of mammary epithelial cells through cyclin D1 expression in healthy subjects (45). In breast cancer cell, the constitutive activation of NF-κB was observed prior to malignant transformation (46). It raises the possibility of NF-κB as a candidate prognostic factor. Moreover, the NF-κB activators, BCL10, ERN1 and DHX9, are up-regulated in poor prognosis samples (Table 1). This is consistent with the previous reports on breast cancer.
Strong correlation between down-regulation of ERα-related genes and poor prognosis in breast cancer was reported by van't Veer et al. (21). Fujita et al. (47) reported that the probability of invasion and metastasis of breast cancer is increased by aberrant regulation of cell adhesion-related pathway including MTA3, Snail and E-cadherin in ER-negative breast epithelial cells. Our results also indicate that ERα can be down-regulated by up-regulation of SAFB and down-regulation of SRA1 and EGR3 (Table 1).
We also identified other genes which are involved in cancer-related biological processes such as cell cycle and apoptosis. Uncontrolled cell cycle, or abnormal cell proliferation is closely related with general carcinogenesis (48). Our detected MTCP1 (Table 1) plays a key role in T-cell prolymphocytic leukaemia (28) and its higher expression is correlated with T-cell malignancies (49). Up-regulation of this oncogene can be correlated with malignancy of other cancer including breast cancer. Apoptosis is also an important mechanism to control normal cell proliferation and anti-apoptosis is a hallmark of various carcinogenesis (50). In our experiment two apoptosis-related factors were detected as TNFSF12 and CCNL2 (Table 1). TNFSF12 induces cell death in several cancer cells (36). Overexpression of CCNL2 suppresses the growth of human hepatocellular carcinoma cell (39). Down-regulation of these apoptosis regulators might be involved in breast cancer development. Finally, detected SYNPO2 is a homolog of myopodin which suppresses tumor growth and metastasis in prostate cancer (41). In our result, the down-regulation of SYNPO2 in poor prognosis was observed. It suggests that the regulation of SYNPO2 is also involved in breast cancer.
We suspect that the discrepancy between this work and the previous report (21) arises from the current tendency to over-reduce the number of genes for further analyses. Genes that do not have >2-fold differences in expression ratios and P-values <0.01 are commonly excluded. The threshold in the previous work would have eliminated 84 of the 88 genes detected here. Furthermore, the candidate genes identified in the previous report are located outside the UDR, although some are close. This result implies that the candidate genes that were identified from the 4968 threshold-selected genes include those that are the closest to the UDR (Figure 2a). Generally, the expression ratios show greater variation at lower expression levels. Yang et al. (51) suggested that even <2-fold expression levels can be significant. We also believe that a method that assesses the expression ratios and/or P-values of detected genes from the whole microarray dataset would be better than the current method, which detects candidate genes from those selected using the threshold. Differences in the selection of genes using the UDR, which can vary according to the significant distance (significant level), may also explain the difference between the two sets of candidate genes.
The two phenotypes used here (poor and good prognoses) may be not sufficient as supervisors. Inclusion of more useful environmental (e.g. age) and diagnostic information the phenotyping could facilitate gene discovery using our method.
As the information for samples increases, the number of phenotypes may increase to more than two. Our method can be extended to more than two phenotypes. In the current study, we prepared two ExtraGenes for the two phenotypes, wherein the ExtraGene is specifically expressed in either phenotype. When there are more than two phenotypes, ExtraGenes specific to each phenotype would be created to detect the genes related to the trait.
Our method also makes it possible to perform an accurate supervised prediction of phenotypes. This supervised classification is based on our detected genes. This provides further supports that our method can correctly select genes associated with prognosis of cancer.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Dr Ken-ichi Tanno for many detailed discussions. The authors also acknowledge Naomi Ishida and Hironori Mizuguchi (BioInformatics Business Promotion Center, NEC Corporation) for the large-scale analyses of MeSH and GO terms using the NEC product ‘BioCompass’. Funding to pay the Open Access publication charges for this article was provided by the authors' private funds.
Conflict of interest statement. The authors declare that they have no competing financial interests.
REFERENCES
- 1.Tanaka T.S., Kunath T., Kimber W.L., Jaradat S.A., Stagg C.A., Usuda M., Yokota T., Niwa H., Rossant J., Ko M.S. Gene expression profiling of embryo-derived stem cells reveals candidate genes associated with pluripotency and lineage specificity. Genome Res. 2002;12:1921–1928. doi: 10.1101/gr.670002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hughes T.R., Mao M., Jones A.R., Burchard J., Marton M.J., Shannon K.W., Lefkowitz S.M., Ziman M., Schelter J.M., Meyer M.R., et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 3.Whitney A.R., Diehn M., Popper S.J., Alizadeh A.A., Boldrick J.C., Relman D.A., Brown P.O. Individuality and variation in gene expression patterns in human blood. Proc. Natl Acad. Sci. USA. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mueller A., O'Rourke J., Grimm J., Guillemin K., Dixon M.F., Lee A., Falkow S. Distinct gene expression profiles characterize the histopathological stages of disease in Helicobacter-induced mucosa-associated lymphoid tissue lymphoma. Proc. Natl Acad. Sci. USA. 2003;100:1292–1297. doi: 10.1073/pnas.242741699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sperger J.M., Chen X., Draper J.S., Antosiewicz J.E., Chon C.H., Jones S.B., Brooks J.D., Andrews P.W., Brown P.O., Thomson J.A. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc. Natl Acad. Sci. USA. 2003;100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y., Ma C., Delohery T., Nasipak B., Foat B.C., Bounoutas A., Bussemaker H.J., Kim S.K., Chalfie M. Identification of genes expressed in C.elegans touch receptor neurons. Nature. 2002;418:331–335. doi: 10.1038/nature00891. [DOI] [PubMed] [Google Scholar]
- 7.Cheung V.G., Conlin L.K., Weber T.M., Arcaro M., Jen K.Y., Morley M., Spielman R.S. Natural variation in human gene expression assessed in lymphoblastoid cells. Nature Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 8.Kuo W.P., Jenssen T.K., Butte A.J., Ohno-Machado L., Kohane I.S. Analysis of matched mRNA measurements from two different microarray technologies. Bioinformatics. 2002;18:405–412. doi: 10.1093/bioinformatics/18.3.405. [DOI] [PubMed] [Google Scholar]
- 9.Quackenbush J. Computational analysis of microarray data. Nature Rev. Genet. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- 10.Falconer D.S., Mackay T.F.C. Introduction to Quantitative Genetics. Longman, Essex; 1996. [Google Scholar]
- 11.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caceres M., Lachuer J., Zapala M.A., Redmond J.C., Kudo L., Geschwind D.H., Lockhart D.J., Preuss T.M., Barlow C. Elevated gene expression levels distinguish human from non-human primate brains. Proc. Natl Acad. Sci. USA. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomson J.M., Parker J., Perou C.M., Hammond S.M. A custom microarray platform for analysis of microRNA gene expression. Nature Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 14.Pomeroy S.L., Tamayo P., Gaasenbeek M., Sturla L.M., Angelo M., McLaughlin M.E., Kim J.Y., Goumnerova L.C., Black P.M., Lau C., et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 15.Cline E.I., Bicciato S., DiBello C., Lingen M.W. Prediction of in vivo synergistic activity of antiangiogenic compounds by gene expression profiling. Cancer Res. 2002;62:7143–7148. [PubMed] [Google Scholar]
- 16.Hirai M.Y., Yano M., Goodenowe D.B., Kanaya S., Kimura T., Awazuhara M., Arita M., Fujiwara T., Saito K. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2004;101:10205–10210. doi: 10.1073/pnas.0403218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishisato S. Analysis of Categorical Data: Dual Scaling and its Applications. Toronto: University of Toronto Press; 1980. [Google Scholar]
- 18.Kishino H., Waddell P.J. Correspondence analysis of genes and tissue types and finding genetic links from microarray data. Genome Inform. Ser. Workshop Genome Inform. 2000;11:83–95. [PubMed] [Google Scholar]
- 19.Fellenberg K., Hauser N.C., Brors B., Neutzner A., Hoheisel J.D., Vingron M. Correspondence analysis applied to microarray data. Proc. Natl Acad. Sci. USA. 2001;98:10781–10786. doi: 10.1073/pnas.181597298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perelman S., Mazzella M.A., Muschietti J., Zhu T., Casal J.J. Finding unexpected patterns in microarray data. Plant Physiol. 2003;133:1717–1725. doi: 10.1104/pp.103.028753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van't Veer L.J., Dai H., van de Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 22.Lebart L., Morineau A., Warwick K.M. Multivariate descriptive statistical analysis: correspondence analysis and related techniques for large matrices. 1984. Translated by Berry, E.M. Wiley, NY. [Google Scholar]
- 23.Zhang B., Schmoyer D., Kirov S., Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenacre M.J. Correspondence Analysis in Practice. London: Academic Press; 1993. [Google Scholar]
- 25.Pulford K., Lamant L., Espinos E., Jiang Q., Xue L., Turturro F., Delsol G., Morris S.W. The emerging normal and disease-related roles of anaplastic lymphoma kinase. Cell Mol. Life Sci. 2004;61:2939–2953. doi: 10.1007/s00018-004-4275-9. [DOI] [PubMed] [Google Scholar]
- 26.Zhou H., Wertz I., O'Rourke K., Ultsch M., Seshagiri S., Eby M., Xiao W., Dixit V.M. Bcl10 activates the NF-kappaB pathway through ubiquitination of NEMO. Nature. 2004;427:167–171. doi: 10.1038/nature02273. [DOI] [PubMed] [Google Scholar]
- 27.Kaneko M., Niinuma Y., Nomura Y. Activation signal of nuclear factor-kappa B in response to endoplasmic reticulum stress is transduced via IRE1 and tumor necrosis factor receptor-associated factor 2. Biol. Pharm. Bull. 2003;26:931–935. doi: 10.1248/bpb.26.931. [DOI] [PubMed] [Google Scholar]
- 28.Stern M.H., Soulier J., Rosenzwajg M., Nakahara K., Canki-Klain N., Aurias A., Sigaux F., Kirsch I.R. MTCP-1: a novel gene on the human chromosome Xq28 translocated to the T cell receptor alpha/delta locus in mature T cell proliferations. Oncogene. 1993;8:2475–2483. [PubMed] [Google Scholar]
- 29.Townson S.M., Kang K., Lee A.V., Oesterreich S. Structure-function analysis of the estrogen receptor alpha corepressor scaffold attachment factor-B1: identification of a potent transcriptional repression domain. J. Biol. Chem. 2004;279:26074–26081. doi: 10.1074/jbc.M313726200. [DOI] [PubMed] [Google Scholar]
- 30.Lv J., Liu H., Wang Q., Tang Z., Hou L., Zhang B. Molecular cloning of a novel human gene encoding histone acetyltransferase-like protein involved in transcriptional activation of hTERT. Biochem. Biophys. Res. Commun. 2003;311:506–513. doi: 10.1016/j.bbrc.2003.09.235. [DOI] [PubMed] [Google Scholar]
- 31.Tetsuka T., Uranishi H., Sanda T., Asamitsu K., Yang J.P., Wong-Staal F., Okamoto T. RNA helicase A interacts with nuclear factor kappaB p65 and functions as a transcriptional coactivator. Eur. J. Biochem. 2004;271:3741–3751. doi: 10.1111/j.1432-1033.2004.04314.x. [DOI] [PubMed] [Google Scholar]
- 32.Anazawa Y., Arakawa H., Nakagawa H., Nakamura Y. Identification of STAG1 as a key mediator of a p53-dependent apoptotic pathway. Oncogene. 2004;23:7621–7627. doi: 10.1038/sj.onc.1207270. [DOI] [PubMed] [Google Scholar]
- 33.Lin H., Juang J.L., Wang P.S. Involvement of Cdk5/p25 in digoxin-triggered prostate cancer cell apoptosis. J. Biol. Chem. 2004;279:29302–29307. doi: 10.1074/jbc.M403664200. [DOI] [PubMed] [Google Scholar]
- 34.Müller H.M., Widschwendter A., Fiegl H., Ivarsson L., Goebel G., Perkmann E., Marth C., Widschwendter M. DNA methylation in serum of breast cancer patients: an independent prognostic marker. Cancer Res. 2003;63:7641–7645. [PubMed] [Google Scholar]
- 35.Cavarretta I.T., Mukopadhyay R., Lonard D.M., Cowsert L.M., Bennett C.F., O'Malley B.W., Smith C.L. Reduction of coactivator expression by antisense oligodeoxynucleotides inhibits ERalpha transcriptional activity and MCF-7 proliferation. Mol. Endocrinol. 2002;16:253–270. doi: 10.1210/mend.16.2.0770. [DOI] [PubMed] [Google Scholar]
- 36.Nakayama M., Ishidoh K., Kayagaki N., Kojima Y., Yamaguchi N., Nakano H., Kominami E., Okumura K., Yagita H. Multiple pathways of TWEAK-induced cell death. J. Immunol. 2002;168:734–743. doi: 10.4049/jimmunol.168.2.734. [DOI] [PubMed] [Google Scholar]
- 37.Sokol J.P., Neil J.R., Schiemann B.J., Schiemann W.P. The use of cystatin C to inhibit epithelial–mesenchymal transition and morphological transformation stimulated by transforming growth factor-beta. Breast Cancer Res. 2005;7:R844–R853. doi: 10.1186/bcr1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inoue A., Omoto Y., Yamaguchi Y., Kiyama R., Hayashi S.I. Transcription factor EGR3 is involved in the estrogen-signaling pathway in breast cancer cells. J. Mol. Endocrinol. 2004;32:649–661. doi: 10.1677/jme.0.0320649. [DOI] [PubMed] [Google Scholar]
- 39.Yang L., Li N., Wang C., Yu Y., Yuan L., Zhang M., Cao X. Cyclin L2, a novel RNA polymerase II-associated cyclin, is involved in pre-mRNA splicing and induces apoptosis of human hepatocellular carcinoma cells. J. Biol. Chem. 2004;279:11639–11648. doi: 10.1074/jbc.M312895200. [DOI] [PubMed] [Google Scholar]
- 40.Smith J.S., Tachibana I., Pohl U., Lee H.K., Thanarajasingam U., Portier B.P., Ueki K., Ramaswamy S., Billings S.J., Mohrenweiser H.W., et al. A transcript map of the chromosome 19q-arm glioma tumor suppressor region. Genomics. 2000;64:44–50. doi: 10.1006/geno.1999.6101. [DOI] [PubMed] [Google Scholar]
- 41.Jing L., Liu L., Yu Y.P., Dhir R., Acquafondada M., Landsittel D., Cieply K., Wells A., Luo J.H. Expression of myopodin induces suppression of tumor growth and metastasis. Am. J. Pathol. 2004;164:1799–1806. doi: 10.1016/S0002-9440(10)63738-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuda A., Suzuki Y., Honda G., Muramatsu S., Matsuzaki O., Nagano Y., Doi T., Shimotohno K., Harada T., Nishida E., et al. Large-scale identification and characterization of human genes that activate NF-kappaB and MAPK signaling pathways. Oncogene. 2003;22:3307–3318. doi: 10.1038/sj.onc.1206406. [DOI] [PubMed] [Google Scholar]
- 43.Agathanggelou A., Cooper W.N., Latif F. Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 2005;65:3497–3508. doi: 10.1158/0008-5472.CAN-04-4088. [DOI] [PubMed] [Google Scholar]
- 44.Karin M., Cao Y., Greten F.R., Li Z.W. NF-kappaB in cancer: from innocent bystander to major culprit. Nature Rev. Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 45.Cao Y., Bonizzi G., Seagroves T.N., Greten F.R., Johnson R., Schmidt E.V., Karin M. IKKalpha provides an essential link between RANK signaling and cyclin D1 expression during mammary gland development. Cell. 2001;107:763–775. doi: 10.1016/s0092-8674(01)00599-2. [DOI] [PubMed] [Google Scholar]
- 46.Kim D.W., Sovak M.A., Zanieski G., Nonet G., Romieu-Mourez R., Lau A.W., Hafer L.J., Yaswen P., Stampfer M., Rogers A.E., et al. Activation of NF-kappaB/Rel occurs early during neoplastic transformation of mammary cells. Carcinogenesis. 2000;21:871–879. doi: 10.1093/carcin/21.5.871. [DOI] [PubMed] [Google Scholar]
- 47.Fujita N., Jaye D.L., Kajita M., Geigerman C., Moreno C.S., Wade P.A. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell. 2003;113:207–219. doi: 10.1016/s0092-8674(03)00234-4. [DOI] [PubMed] [Google Scholar]
- 48.Sherr C.J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 49.Gritti C., Dastot H., Soulier J., Janin A., Daniel M.T., Madani A., Grimber G., Briand P., Sigaux F., Stern M.H. Transgenic mice for MTCP1 develop T-cell prolymphocytic leukemia. Blood. 1998;92:368–373. [PubMed] [Google Scholar]
- 50.Schmitt C.A. Senescence, apoptosis and therapy—cutting the lifelines of cancer. Nature Rev. Cancer. 2003;3:286–295. doi: 10.1038/nrc1044. [DOI] [PubMed] [Google Scholar]
- 51.Yang I.V., Chen E., Hasseman J.P., Liang W., Frank B.C., Wang S., Sharov V., Saeed A.I., White J., Li J., et al. Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol. 2002;24:research0062. doi: 10.1186/gb-2002-3-11-research0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.