Abstract
Anaplasma phagocytophilum, the etiologic agent of human granulocytic anaplasmosis, has a large paralog cluster (approximate 90 members) that encodes the 44-kDa major outer membrane proteins (P44s). Gene conversion at a single p44 expression locus leads to P44 antigenic variation. Homologs of genes for the RecA-dependent RecF pathway, but not the RecBCD or RecE pathways, of recombination were detected in the A. phagocytophilum genome. In the present study, we examined whether the RecF pathway is involved in p44 gene conversion. The recombination intermediate structure between a donor p44 and the p44 expression locus of A. phagocytophilum was detected in an HL-60 cell culture by Southern blot analysis followed by sequencing the band and in blood samples from infected SCID mice by PCR, followed by sequencing. The sequences were consistent with the RecF pathway recombination: a half-crossover structure, consisting of the donor p44 locus connected to the 3′ conserved region of the recipient p44 in the p44 expression locus in direct orientation. To determine whether the p44 recombination intermediate structure can be generated in a RecF-active Escherichia coli strain, we constructed a double-origin plasmid carrying the p44 expression locus and a donor p44 locus and introduced the plasmid into various E. coli strains. The recombination intermediate was recovered in an E. coli strain with active RecF recombination pathway but not in strains with deficient RecF pathway. Our results support the view that the p44 gene conversion in A. phagocytophilum occurs through the RecF pathway.
Human granulocytic anaplasmosis (HGA; formerly human granulocytic ehrlichiosis or HGE) is a significant, emerging tick-borne infectious disease, first reported in 1994 (9). The disease had been increasingly recognized in the United States and Europe, and HGA was designated as a nationally notifiable disease for the United States in 1998 (14). HGA is a potentially fatal systemic disease characterized by fever, headache, myalgia, anorexia, and chills and is frequently accompanied by leukopenia, thrombocytopenia, anemia, and elevations in serum hepatic aminotransferases (2). The etiologic agent, isolated from HGA patients in 1995 (15), is an obligate intracellular rickettsial pathogen that was recently reclassified with other related Ehrlichia spp. as Anaplasma phagocytophilum (10).
The p44 multigene family of A. phagocytophilum encodes immunodominant 44-kDa major outer membrane proteins, P44s (4, 11, 25-27, 39, 42-44). P44 plays critical roles in infection. For example, anti-P44 antibodies can prevent A. phagocytophilum infection in cell culture (39) and partially protect mice from experimental infection with A. phagocytophilum (20), and a recombinant P44 protein induces proinflammatory cytokines in human leukocytes in vitro (21). The p44 gene family has a central hypervariable region of approximately 280 bp. This region is flanked by 50- to 500-bp sequences from each of 5′ and 3′ conserved regions (see Fig. 1A). To date, 88 individual p44 paralogs or orthologs had been identified by their signature hypervariable nucleotide sequences. Many of p44s are considered functional pseudogenes (silent storage copies) rather than nonfunctional pseudogenes on the way to elimination since, despite truncation of 5′- and/or 3′-terminal sequences, some express the full-length p44 transcripts and P44 proteins (26, 42).
FIG. 1.
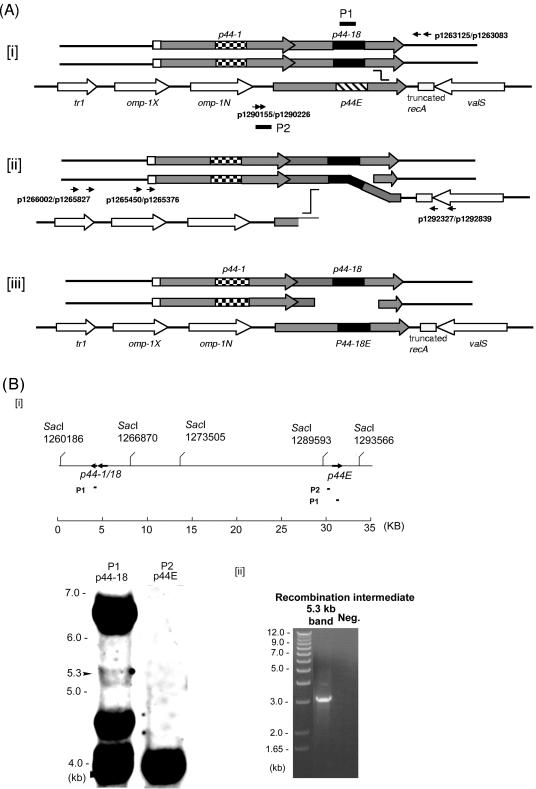
Analysis of the p44 recombination intermediate in A. phagocytophilum. (A) Successive half-crossover model for p44-18 conversion to the p44E and the experimental design. [i] The donor locus was expected to be a part of replicated chromosome; the p44-1/18 locus in a donor chromosome (p44-1/18D) synapses with the 3′ conserved region of p44E in the recipient chromosome. [ii] A half crossover between p44E and p44-1/18D generates one recombination duplex and two ends. The upstream of the synapsed region of p44E is presumably degraded to generate a single-stranded tail at the 5′ conserved region by an exonuclease. [iii] The putative final products of the successive crossover are one duplex with two double strands exchanged in the recipient p44 expression locus and two ends in the donor locus. The gray boxes are p44 conserved regions, and the large checkerboard, black, and wide downward diagonal are hypervariable regions of p44-1, p44-18, and the recipient p44E, respectively. The arrows of boxes indicate transcriptional orientation. Horizontal arrows with numbers indicate primers used to isolate the intermediate structure by PCR and primers used to demonstrate the absence of a reciprocal recombination product. Black short bars indicate the locations of two probes (P1 and P2) used for Southern blot analysis in Fig. 1B. The “stairstep” symbol indicates the potential pair of the recombination sites. (B) Southern blot analysis of the half-crossover intermediate structure of A. phagocytophilum cultured in HL-60 cells. [i] The probe positions and SacI digestion sites within the 35-kb region containing p44-1/18 and p44E are indicated. The numbers under the SacI sites indicate the positions of the cleavage sites in the genome. The genomic DNA was purified from A. phagocytophilum cultivated in HL-60 cells. Southern blot analysis of genomic DNA digested with the restriction enzyme SacI with two probes (P1 and P2) showed the ∼6.6-kb band containing the donor p44-1/18 locus and a 4-kb band containing the recipient p44E. An ∼5.3-kb band contained duplicated p44-1/18. [ii] PCR was performed using the primer pairs p1265450-p1292839 and p1265376-p1292327 (Fig. 1Aii) and the DNA isolated from the 5.3-kb region of the gel as a template. The numbers on the left indicate the molecular sizes. A band of approximately 3.2 kb was amplified and sequenced GenBank accession no. DQ011270. “Neg.” refers to a negative control with water as a template.
Several studies reported that diverse p44 paralogs are expressed in patients, in animal models of infection (mouse and horse), and in ticks (4, 11, 25, 26, 40, 42, 44). This antigenic variation system is expected to allow A. phagocytophilum to avoid and escape host immune recognition and to allow adaptation to new environments, especially during tick transmission of A. phagocytophilum (11, 33, 34, 44). A single polymorphic p44 expression locus that consists of four tandem genes—tr1, omp-1X, omp-1N (corresponding to p44ESup1 described by Barbet et al. [4]), and p44E (any p44 species at the p44 expression locus, corresponding to msp2 described by Barbet et al. [4])—was identified in several A. phagocytophilum strains (4, 26). Recently, the successful development of an isogenic clone from A. phagocytophilum HZ strain allowed us to demonstrate the nonreciprocal recombination (gene conversion) of paralogous p44s at this p44 expression locus (27). Although expression locus is expected to be the primary locus for diverse p44 gene expression in A. phagocytophilum (4, 26), the molecular mechanisms of p44 gene conversion, and thus antigenic variation, are largely unknown.
Several bacterial pathogens such as Borrelia burgdorferi and Neisseria gonorrhoeae exhibit antigenic variation by gene conversion (the nonreciprocal transfer of DNA sequences between homologous genes) within their hosts (5). Only a few studies, however, have described recombination mechanisms responsible for antigenic switching in bacteria. RecA-dependent RecF-mediated recombination was suggested to mediate N. gonorrhoeae pilin antigenic variation (32). Three RecA-dependent homologous recombination pathways, RecF, RecE, and RecBCD, have been identified in Escherichia coli (22, 23). Several studies using E. coli mutant strains and plasmids showed that RecF pathway recombination is nonreciprocal without crossover, but RecBCD and RecE pathway recombination can be reciprocal and associated with crossover (36). We previously reported that A. phagocytophilum lacks homologs of genes required for RecBCD or RecE recombination pathways but has homologs of most of the genes involved in RecF recombination (26, 27). The p44 recombination is apparently nonreciprocal (i.e., gene conversion), in that the donor p44 is copied at the p44 expression locus and previous resident p44E vanishes after conversion (26, 27); this gene conversion occurs without crossover, preserving the entire donor region and noncoding regions flanking p44E; this gene conversion is nonsegmental, in that the p44 hypervariable region is identical between donor p44 and p44E (11, 26, 27), although Barbet et al. proposed that p44 gene conversion is segmental (4). Our analysis using cloned A. phagocytophilum population is consistent with our previous observation that both 5′ and 3′ conserved regions of p44E flanking the hypervariable region contain nucleotide sequence variations (11, 26, 27). This analysis is also in agreement with our previous prediction that instead of using the entire conserved regions for gene conversion, various lengths (50 to 200 bp) of partial 5′ and 3′ conserved region sequences at the border of the hypervariable region are used for gene conversion in the p44 expression locus (27). Kobayashi (22) proposed a novel successive two-half-crossover (no crossover) model as the mechanism of RecF pathway gene conversion since two types of half-crossover intermediates could be isolated as only one product of possible crossover using a double-origin plasmid in the E. coli RecF active strain (22, 38, 41). A similar mode of gene conversion without crossover during yeast meiosis, called the synthesis-dependent strand annealing model, can be also explained by the successive two-half-crossover model (1). For obligatory intracellular bacteria including A. phagocytophilum, there is no useful genetic system and no naturally isolated mutant, making the genetic analysis of p44 recombination mechanisms impossible. In the present study, therefore, we first examined whether the half-crossover intermediate structure is formed between the donor p44 and recipient p44E in A. phagocytophilum. Second, we constructed a double-origin plasmid carrying a donor p44 locus and a recipient p44E locus with the antibiotic selection markers to investigate whether the p44 half-crossover occurs in the RecF pathway active E. coli strain. This study is the first analysis of the recombination pathway and intermediate structure involved in the A. phagocytophilum p44 antigenic variation.
MATERIALS AND METHODS
Southern blot analysis of A. phagocytophilum genomic DNA digested by SacI and sequencing of the 5.3-kb DNA fragment.
The genomic DNA of A. phagocytophilum HZ was extracted from organisms purified from HL-60 culture by Sephacryl S1000 chromatography (26). Total DNA (8 μg) was digested by restriction enzyme SacI and loaded into each well of a 0.7% agarose gel. The p44-18 (P1) probe was amplified by the primer pair p1263960 (5′-CGTGGAGATTTCTAATTCCGG-3′) and p1263637 (5′-TTCAGGGGTGAGCTTCTTAG-3′). The probe (P2) p44E upstream intergenic region was amplified by using the primer pair p1289877 (5′-TGGACGAGAAGAATGGGATC-3′) and p1290352 (5′-TCTTCGTCTCCTCACTTCAG-3′). PCR for preparing Southern blot probes was performed in a 50-μl reaction mixture, containing 10 ng of genomic DNA of A. phagocytophilum HZ, 10 pmol of each primer, a 0.2 mM concentration of each deoxynucleoside triphosphate, 1.5 mM MgCl2, and 5 U of Taq DNA polymerase. PCR was performed with 2 min of denaturation at 94°C, followed by 35 cycles of 30 s of denaturation at 94°C, 30 s of annealing at 58°C, and 1 min of extension at 72°C. The probes were labeled by PCR amplification using a biotinylation kit according to the manufacturer's instructions (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD). DNA transfer, hybridization, and detection were carried out by using an AP Chemiluminescent Blotting Kit (Kirkegaard & Perry).
The 5.3-kb region DNA fragments digested by SacI were recovered from 0.7% agarose gel by using QIAEX II gel extraction kit (QIAGEN, Valencia, CA). The intermediate sequence was PCR amplified with extracted DNA as a template with the primer pair p1266002 (5′-CAATCTTACCCCTAGTGAGCAGT-3′) and p1292839 (5′-TGTACAGCTTGTAGCCGGTAAT-3′) and the primer pair p1265827 (5′-TTACCTATTACAGTATCACCATGC-3′) and p1292327 (5′-AACAAGATATGCATCCGCAAATC-3′). The first PCR product was 1:100 diluted, and 5 μl was used for the second PCR. The products of the second PCR were sequenced. Sequence assembling, alignments, and analysis were performed by using the SeqMan, MegAlign, and MapDraw programs (DNAStar, Inc., Madison, WI).
Analysis of A. phagocytophilum recombination intermediate structure in an infected SCID mouse.
Ten 4-week-old ICR SCID male mice (Taconic Farm, Inc., Germantown, NY) were each inoculated intraperitoneally with the cloned A. phagocytophilum HZ strain (27). The blood sample of SCID mouse 9 was used to investigate recombination based on the previous temporal p44E sequence population analysis study (27). The nested PCR was performed with primers specific to the upstream sequence of the donor p44-1/18 locus and the downstream sequence of the p44E and was performed with DNA isolated from peripheral blood leukocytes from the SCID mouse. The primer pairs used were primers p1265450 (5′-GCTATGGGAGATTACTATTC-3′) and p1292839 and primers p1265376 (5′-CATTTTCTTTAAAAGGCAGAC-3′) and p1292327 (Fig. 1A).
PCR with Pfu DNA polymerase was performed in a 50-μl reaction mixture containing 5 μl of the DNA product, 10 pmol of each primer, a 0.2 mM concentration of each deoxynucleoside triphosphate, and 5 U of Pfu Ultra High-Fidelity DNA polymerase (Stratagene, La Jolla, CA). PCR was performed with 2 min of denaturation at 94°C, followed by 35 cycles of 20 s of denaturation at 94°C, 20 s of annealing at 58°C, and 5 min of extension at 70°C. PCR products were purified from a gel and cloned into the pCR-Blunt vector (Invitrogen, Carlsbad, CA). DNA clones were randomly selected from the transformants and sequenced on an ABI 373XL Stretch DNA sequencer by using the ABI Prism BigDye terminator cycle sequencing reaction kit (ABI, Foster City, CA).
To examine the possibility of PCR artifacts formed during amplification, p44-1/18 and p44E were cloned into the TA cloning vector pCRII (Invitrogen), and 20 ng of plasmid mixture was spiked in genomic DNA from an uninfected SCID mouse and used as the PCR template. The PCR was performed with the primer pairs p1265450-p1292839 and p1265376-p1292327. The first PCR product was 1:100 diluted, and 5 μl was used for the second PCR. To investigate possibility of the reciprocal crossing over between the upstream sequence of the p44E and the downstream sequence of donor p44-1/18 locus, nested PCR was performed by using the same SCID mouse DNA sample. For this reaction, the two forward primers were p1290155 (5′-CGTTATTTGTTCTAGAGAAAG-3′) and p1290226 (5′-ATTGGACTTTTGAGCTGTCTT-3′), and the reverse primers were p1263125 (5′-CACCACGCAGGAATATCGATCT-3′) and p1263083 (5′-GCTTTTGCCACTAGAGACAGG-3′) (Fig. 1A).
The p44-18 donor locus, p44-1/18, was PCR amplified by using the primer pair p1265450 and p1263125 from the SCID mice DNA and sequenced. The PCR conditions used were as described above.
Construction of a plasmid encoding p44E expression and p44 donor loci and analysis of their recombination.
Plasmid pEKD30, a double-origin (ColE1 and p15A) plasmid of 9.3 kb in length (see Fig. 3Ai) was constructed by using the plasmid pKEN33 (generously provided by Ichizo Kobayashi [38]) as the backbone. A recipient locus and a donor locus derived from A. phagocytophilum were inserted into the plasmid pKEN33. The recipient locus consisted of the 213 bp upstream of p44E (p44E IR in Fig. 3Ai), a kanamycin-resistant gene km, an E. coli ribosome binding site (TAAGGAG), and p44-18E and its 446 bp downstream sequence (p44E DS in Fig. 3Ai). The km gene was PCR amplified from plasmid pCR-XL-TOPO (Invitrogen). The 213-bp intergenic upstream sequence, p44-18E and its 466-bp downstream sequence were PCR amplified from A. phagocytophilum genomic DNA. The intergenic region was amplified by the primer pair p1291699 (5′-GGTCGACGGGCTAAGGGCTCCCCTTTT-3′) and p1292129 (5′-ATCTAGAGCAATAGACCCAGTAG-3′) to have XbaI and SalI sites at each end, respectively. The p44-18E and the downstream sequence was amplified by primer pair p1290170 (5′-GGGCATGCATGCGACGTCAGAAAGATGTGCGTAAGAGGTAA-3′) and p1291698 (5′-CCTGCAGCCCTCTTTAGATAAGCAAGCTTA-3′) to have AatII and PstI sites at each end, respectively. km was amplified by using the primer pair pkmF (5′-GCTGCAGTAAGGAGGTTTCGC ATGATTGAACAAGATG-3′) and pkmR (5′-CGTCGACTCAGAAGAACTCGTCAAGAAGG-3′), including a ribosomal binding site sequence (5′-TAAGGAG-3′) immediately upstream of the km gene and SalI and PstI site at each end, respectively. Restriction enzyme sites are underlined. These three fragments were digested by restriction enzymes AatII, PstI, SalI, and XbaI and were then ligated in direct orientation into pKEN33 that had been digested by XbaI and AatII to give a plasmid pKEN33-EKD.
FIG.3.
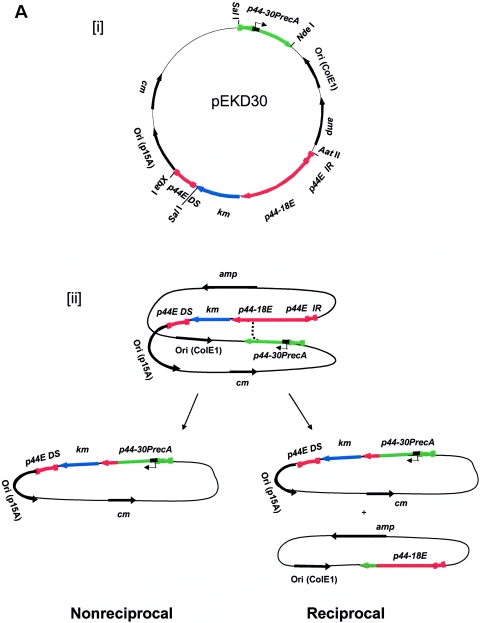
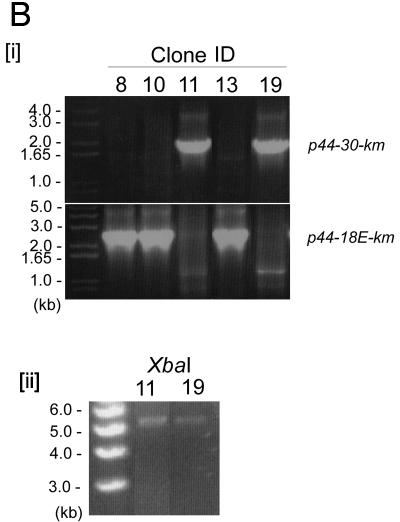
Analysis of a p44 recombination intermediate in E. coli using plasmid encoding p44 expression and donor loci. (A) Double-origin plasmid pEKD30 and E. coli system design. [i] Plasmid pEKD30 carries the recipient site of the p44 expression locus p44E upstream region (p44E IR)- p44-18E-km-p44E downstream sequence (p44E DS) and the donor site p44-30PrecA in a direct orientation. The donor p44-30PrecA has an E. coli recA gene promoter in the hypervariable region (bent arrow). The plasmid carries two more antibiotic-resistant genes (cm and amp) and two compatible replication origins (P15A and ColE). The restriction enzyme cleavage sites are shown. If p44-30PrecA recombines to p44-18E, km is transcribed from the recA promoter, allowing isolation of the recombination intermediates in the presence of Km. [ii] Experimental design. If nonreciprocal recombination occurs between p44-30PrecA and p44-18E, only one type of plasmid (5.7 kb) that carries km with upstream p44-30PrecA is expected to be isolated in the presence of Km. If reciprocal recombination occurs between the donor and the recipient sequences, two plasmids should be generated: one identical to the plasmid generated by the nonreciprocal recombination (5.7 kb) and another that carries p44-18E and amp (3.6 kb). Thus, in nonreciprocal recombination, Kmr E. coli strains are Ams, while in reciprocal recombination Kmr E. coli strains are Amr. [iii] Sequence alignment of 5′- and 3′-end conserved regions of donor p44-30D and recipient p44-18E in the plasmid pEKD30. The different nucleotides between p44-30D and p44-18E conserved regions are shaded in light gray. (B) Analysis of the Kmr Ams plasmids isolated from E. coli strain JC7623 with an active RecF pathway. [i] PCR amplification using the primer pair located upstream of p44-30PrecA and downstream of km, respectively, showed a band of the expected size for the recombined p44-30PrecA-km structure in Kmr Ams clones 11 and 19. The remaining three Kmr Ams clones had the original p44-18-km structure. [ii] The two PCR positive plasmids from clones 11 and 19 were analyzed by digestion with XbaI to determine the size of the recombined plasmid. As predicted from the restriction sites shown in Fig. 3Ai, XbaI digestion generated a single 5.7-kb band from the recombined p44-30-km plasmid. [iii] The sequence between the two SalI sites in clones 11 and 19, which confirmed the recombination between p44-30PrecA and p44-18E.
As a donor p44 sequence, p44-30, which is a truncated p44, 897 bp in length (as a SalI and NdeI fragment), was selected. In order to insert an E. coli recA promoter (5′-CAAAACACTTGATA-35CTGTATGAGCATACAGTATAAT-10TGCTTCA-3′) (28) into the p44-30 hypervariable region, two primer pairs were designed to amplify the 5′ and 3′ fragments of p44-30 that were overlapped at the RecA promoter sequence: p1416475 (5′-GGGTCGACGTCGAC GTATACCAAAAGCCTATGCAATAA-3′)-p1416768 (5′-TGAAGCAATTATACTGTATGCTCATACAGTATCAAGTGTTTTGGTATTCCGTTACGCTTCCTCC-3′) and p1416771 (5′-CAAAACACTTGATACTGTATGAGCATACAGTATAATTGCTTCAGGCAGAGCCGGATGAAAACAC-3′)-p1417466 (5′-GGCATATGCATATGAGAATTAAAGTAGAAAAGGGGAG-3′). Then, using these two PCR fragments as a template, the p44-30 with the E. coli recA gene promoter in the hypervariable region (p44-30PrecA) was PCR amplified by primers p1416475 and p1417466. The PCR product was digested by restriction enzymes SalI and NdeI and was ligated into pKEN33-EKD that had been digested by SalI and NdeI (this also removed the original km gene in pKEN33) to give pEKD30. All inserted fragments were sequenced and the plasmid pEKD30 was kept in a ΔrecA strain, INVαF′, with minimal generation numbers.
Detection and analysis of recombination.
The isogenic E. coli K-12 strains JC7623 (recB21 recC22 sbcB15 sbcC201) (24, 29), JC8111 (recB21 recC22 sbcB15 sbcC201 recF143), and JC8679 (recB21 recC22 sbcA23) were from E. coli genetic stock center (Yale University, New Haven, CT). JC12190 (recB21 recC22 sbcB15 sbcC201 recJ153) (17, 30) was obtained from Ichizo Kobayashi (University of Tokyo, Tokyo, Japan). Competent cells were prepared as described previously (35). Approximately 200 ng of the intact plasmid DNA was delivered into 1010 bacterial cells at a field strength of 12.5 kV/cm for 5.2 to 5.5 ms. Even when recombination generates two plasmids, they may be lost during growth in the absence of selection; therefore, we only let the bacteria recover at 37°C, with rotation at 220 rpm for 1 h after electroporation. The bacteria were then applied to SOB plates containing different combinations of chloramphenicol (Cm), kanamycin (Km), and amphotericin B (Am), each at 50 μg/ml, and colony numbers on plates were determined to assess the recombination frequency. Km-resistant colonies were randomly selected and analyzed by site-specific PCR. The sites for binding of primers p1416475 and pkmR (sequence shown above) were located upstream of the donor p44-30 and in the 3′ end of the km gene, respectively. The binding sites for the other primer pair, p1290226-pkmR, were located upstream of p44E and 3′ end of the km gene, respectively. The plasmid was extracted from the Km-resistant colonies and digested by XbaI or by XbaI and SalI to confirm the fragment sizes. The fragments were cloned and sequenced.
RESULTS
Analysis of the p44 half-crossover structure in A. phagocytophilum-infected HL-60 cells by Southern blot analysis followed by sequencing.
The majority (80 to 90%) of noncloned A. phagocytophilum HZ isolated after many passages in cell culture (>100 passages, with each passage after 3 to 4 days of culture) has p44-18 at its p44 expression locus (p44-18E) (39, 40, 44). Lower percentages are sometimes observed due to presently undefined reasons, but during subsequent passages the percentage eventually returns to the original 80 to 90% of p44-18E in the population. Therefore, we hypothesized that the recombination intermediate structure linking the donor p44-18 sequence to the recipient p44 expression locus is detected in A. phagocytophilum in culture. The donor p44-1/18 locus consists of tandem p44-1 (1,242-bp full-length p44 gene) and p44-18 (762-bp pseudogene) (Fig. 1A). We isolated genomic DNA from this high-passage A. phagocytophilum HZ to detect the recombination intermediate by Southern blot analysis. For Southern blot analysis we chose restriction enzyme SacI. As shown in our previous sequencing data (26) and the map of SacI restriction sites (Fig. 1Bi), there is no SacI site within the p44 expression locus and the donor p44-1/18 locus, and the p44 expression locus is expected to be in an ∼4.0-kb fragment and the p44-1/18 locus is expected be in an ∼6.6-kb fragment. The recombination intermediate structure is predicted to be approximately 5.3 kb (Fig. 1Aii and Bi).
Probes for Southern blotting were designed as shown in Fig. 1A and Bi. The P1 probe was designed to be specific to p44-18 hypervariable region, and the P2 probe was designed to be specific to a region upstream of p44E in the expression locus. By using P1 probe, the ∼6.6-kb band corresponding to the p44-1/18 locus, and the ∼4.0-kb band corresponding p44-18E in the expression locus were detected (Fig. 1Bi). In addition, a new and weak band of ∼5.3 kb was consistently detected by using P1 probe (Fig. 1Bi). An ∼4.7-kb band hybridized with P1 probe. p44-18 was not detected in this band region; however, the sequence which has 134/171 bp (78%) identity with P1 probe was detected in the SacI fragment of A. phagocytophilum genomic DNA of this size.
The P2 probe was expected to hybridize to the expression locus 4.0-kb band. As shown Fig. 1Bi, the P2 probe hybridized to the 4.0-kb and did not hybridize to the 5.3-kb band, suggesting that if this band is the recombination intermediate, this is a nonreciprocal event (Fig. 1Bi). The 5.3 kb-band region which did not include 6.6- and 4.0-kb bands (therefore, there is no chance of strand jumping between these two fragments) was recovered from the agarose gel and used as a template for PCR using the primer pairs p1266002-p1292839 and p1265827-p1292327 (Fig. 1Aii). The PCR generated the 3.2-kb band (Fig. 1Bii). The sequencing result showed a half-crossover structure between the p44-1/18 and the p44 expression loci as shown in Fig. 1Aii and Fig. 2B. The GenBank accession number for the intermediate sequence of the cell culture is DQ011270. Southern blot analyses were independently repeated by two investigators more than four times using different batch of A. phagocytophilum cultures and probe preparations of different sizes, as well as restriction enzymes purchased from different sources. The results with the P1 and P2 probes were reproducible. These results suggest that p44E recombination occurs via the RecF pathway successive half-crossover mechanism in the cell culture system.
FIG.2.
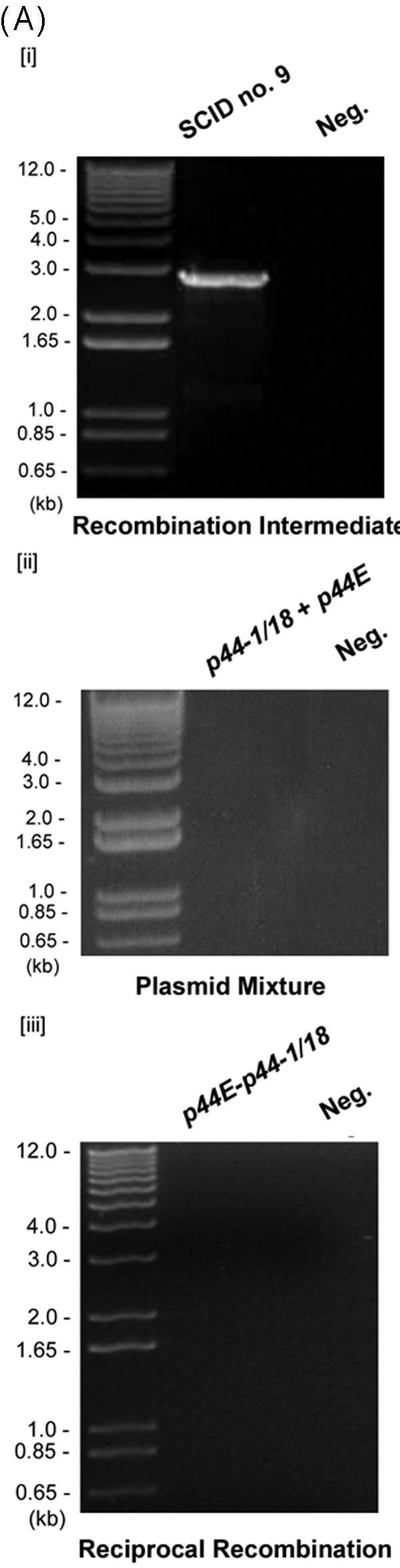
Analysis of the p44 recombination intermediate in A. phagocytophilum from an infected SCID mouse. (A) The half-crossover recombination intermediate was detected by PCR of peripheral blood leukocytes from an A. phagocytophilum-infected SCID mouse. [i] The ∼2.9-kb band was amplified by the primer pairs p1265450-p1292839 and p1265376-p1292327 (Fig. 1Aii) using Pfu DNA polymerase. [ii] No PCR product was detected with the same primer pairs from uninfected SCID mouse DNA spiked with plasmids containing p44-1/18 and p44E as a template. [iii] No PCR product was detected in peripheral blood leukocytes of infected mice with forward primers located in the intergenic region of p44E and omp-1N and with reverse primers located downstream of donor p44-1/18 locus (Fig. 1Ai). “Neg.” refers to a negative control with water as a template. (B) The nucleotide sequence of the half-crossover recombination duplex between p44E and p44-1/18D. The second set of primers used for the nested PCR are indicated by horizontal arrows and labeled with primer ID. p44-1/18DE INT is the intermediate structure containing donor site p44-1/18 and the recipient site p44E. The sequence is identical to that expected for a half-crossover recombination intermediate as illustrated in Fig. 1Aii. The light-shaded areas indicate sequence identity between the crossover structure and the putative donor p44-1/18 locus, and the dark shaded areas indicate identical sequence between the hybrid structure and the recipient p44E, or among all three structures. The boxed nucleotides are start codons for p44-1 and for p44-18. The stop codons are underlined. Dashes indicate sequence gaps that helped to determine the origins of conserved sequences in the recombination intermediate.
Analysis of p44 recombination intermediate structure from A. phagocytophilum infecting a SCID mouse.
To confirm that the RecF-pathway recombination intermediate structure in A. phagocytophilum in infected animals, we analyzed peripheral blood leukocytes from a A. phagocytophilum-infected severe combined immunodeficiency (SCID) mouse 50 days after inoculation with A. phagocytophilum. This time point was used for analysis because populations of p44-18E appeared to increase at this time point in a previous study (27) and should provide a high probability of detecting the recombination intermediate structure. However, we could not obtain A. phagocytophilum DNA from infected SCID mice that were sufficiently enriched for Southern blot analysis due to the low infection rate of granulocytes and small amount of blood specimens. Therefore, we developed a nested PCR method as shown in Fig. 1A. The forward primers p1265450 and p1265376 are complementary to regions starting 136 and 60 bp upstream of p44-1 of the p44-1/18 locus, respectively; the reverse primers p1292839 and p1292327 are complementary to regions starting 619 and 107 bp from the 3′ end of the valS in the p44 expression locus (Fig. 1A). Three elements of our PCR procedures were designed to prevent artifactual, recombination-like events that might occur during PCR. Pfu DNA polymerase was used for PCR, since it has been demonstrated that Pfu DNA polymerase does not have discernible recombination activity, in contrast to Taq and Vent DNA polymerases, due to in part high proofreading activity of Pfu DNA polymerase (37). We used prolonged polymerase extension times (4 min) in each cycle, since an incompletely extended segment of DNA and the partially amplified sequence act as a primer during subsequent amplification. In addition, rapid cooling between the denaturation step and the annealing step (20 s) was used to discourage annealing of incomplete products over the more abundant primer.
The PCR of DNA from peripheral blood leukocytes from the infected mouse amplified a band of ∼2.9 kb (Fig. 2Ai). The size was smaller than the 3.2-kb band observed in the cell culture specimen since the 5′ primers used here anneal to a region further downstream of the donor locus than those used to analyze the cell culture samples (Fig. 1A). DNA sequencing (Fig. 2B) revealed that the 2.9-kb band had a sequence expected for the half-crossover structure consisting of the p44-1/18 sequence upstream and a p44E sequence downstream as illustrated in Fig. 1Aii and almost identical to the sequence DQ011270 detected in cell culture. The next half-crossover event in the 5′ p44 conserved region of p44-18 of the intermediate is expected to create the structure illustrated in Fig. 1Aiii, which was previously demonstrated by PCR and sequencing (27).
As a control to rule out the possibility of a PCR shuffling between the p44 expression locus and donor p44-1/18, we performed the same PCR using as a template a mixture of two p44 plasmid clones each carrying donor p44-1/18 and p44E sequences of the A. phagocytophilum HZ strain. Each of the DNAs at 20 ng was spiked into genomic DNA from an uninfected SCID mouse. The concentration of each p44 in the plasmid present in this PCR was estimated to be >4,000-fold the concentration of total p44 genes present in the mouse blood. No bands between 2 and 3 kb were amplified in this PCR control (Fig. 2Aii). As another control, the nested PCR was performed with forward primers complementary to the intergenic region of p44E and omp-1N and reverse primers that would anneal downstream of the p44-1/18 locus (Fig. 1A). This PCR did not generate a band (Fig. 2Aiii). This result also confirmed the absence of reciprocal recombination between the recipient p44 expression locus and the donor p44-1/18 locus (Fig. 1 and 2Aiii). To confirm absence of crossover at the donor locus, we sequenced the p44-1/18 locus by using DNA sample extracted from the blood of the same mouse. The sequences of randomly selected 10 clones of the PCR product were identical and did not show any sequence variation (data not shown). These results support that the first half-crossover structure in the successive half-crossover model in the RecF pathway recombination is also present in A. phagocytophilum in the SCID mouse (22).
Construction of a plasmid encoding p44E expression and p44 donor loci and their recombination through the RecF pathway.
Kobayashi (22) could isolate half-crossover intermediates using a double-origin plasmid in the E. coli RecF active strain (22, 38, 41). A stable plasmid expression system or gene knockout system is not currently available for any obligate intracellular bacteria. It may be, however, possible to demonstrate the p44 recombination intermediate by constructing an E. coli plasmid and by using E. coli mutants that are deficient in one or more of the recombination pathways. Therefore, in order to analyze the p44 recombination mechanism, we developed an E. coli plasmid system. We constructed the pEKD30 plasmid from the double-origin plasmid pKEN33 (38) by replacing the two original Km resistance genes (km) with the recipient p44 expression locus sequence and a donor p44 sequence, respectively, in direct orientation (Fig. 3Ai). The double-origin plasmid was used, since intraplasmid recombination causes segregation of the plasmid into two plasmids and each origin allows recovery of two plasmids (Fig. 3Aii) under permissive conditions. The recipient p44E consists of p44-18E hypervariable region flanked by 5′ and 3′ conserved sequences (Fig. 3Aiii). A km gene was inserted downstream of p44E to create an operon fusion with an E. coli ribosome-binding site sequence (5′-TAAGGAG-3′) upstream of the ATG start codon of km gene. The p44E-km fragment contained a 213-bp sequence upstream of p44-18E (p44E IR) and a 446 bp downstream sequence (p44E DS). The donor was derived from the p44-30 pseudogene (897 bp). An E. coli recA promoter (5′-CAAAACAC TTGATA−35 CTGTATGAGCATACAG TATAAT−10 TGCTTCA-3′) was inserted into the hypervariable region of the p44-30 (Fig. 3Ai) to create p44-30PrecA. When the donor p44-30PrecA recombines to the p44-18E, km will be expressed from the RecA promoter in the p44-30PrecA and bacteria become resistant to Km; therefore, we could recover the recombined clones in the presence of Km. Various E. coli strains were cultured at 37°C for 1 h after introduction of the plasmid. The bacteria were then applied to SOB plates containing different combinations of Cm, Km, and Am. If recombination is nonreciprocal, the cells become Am sensitive, since nonreciprocal recombination causes loss of the Am resistance gene (amp) due to breakage in p44E (Fig. 3Aii). However, if recombination is reciprocal as catalyzed by the RecE pathway, the E. coli clone remains Am resistant, since p44E receives p44-30 and the segregated plasmid with amp is ligated and thus survives (Fig. 3Aii).
In the RecBCD-deficient, RecF-active E. coli strain JC7623 (recB21 recC22 sbcB15 sbcC201), nonreciprocal recombination occurred as the Km-resistant, Cm-resistant, and Am-sensitive (Kmr Cmr Ams) transformants were detected. The ratio of Kmr/Amr transformants was approximately 0.01 as determined by colony counting in the presence of Km and Am, respectively. DNA extracted from transformant clones were examined by two types of PCR: one specific for the expected product of recombination, using primers located downstream of the km gene and upstream of the donor p44-30 (Fig. 3A), and another specific for the original p44-18-km locus using primers located downstream of the km gene and upstream of the donor p44-18. Two transformants of five Kmr clones examined had plasmids with the recombined p44-30-km locus and had lost the original p44-18-km locus (Fig. 3Bi). The isolated plasmids from the two recombined Kmr transformants were analyzed by restriction enzyme digestion with XbaI (restriction sites in the pEKD30 plasmid are shown in Fig. 3Ai). By using the restriction enzyme XbaI, a linear plasmid of ∼5.7 kb was detected in the two Kmr transformants (Fig. 3Bii) rather than the original pEKD30 plasmid (9.3 kb). Sequencing of the amplicons (1,836 bp) obtained by using primers located upstream of the donor p44-30 and in the 3′ end of the km gene, respectively, from the two clones demonstrated that these were indeed the half-crossover structure expected from the intraplasmid recombination (Fig. 3Biii). Thus, the p44 gene recombination can occur in the nonreciprocal RecF pathway of E. coli.
recF and recJ are essential genes involved in RecF pathway homologous recombination (22, 23). In the recF-deficient strain JC8111 (recB21 recC22 sbcB15 sbcC201 recF143), no transformants were recovered from the plate with Km. Using this strain, the transformants on Am plate were 4 × 104 to 6 × 104. In the recJ-deficient strain JC12190 (recB21 recC22 sbcB15 sbcC201 recJ153), no transformants were recovered from the Km plate. The transformants in Am plate were 4 × 103 to 5 × 103. These results confirmed that the p44 gene conversion in the pEKD30 plasmid was RecF dependent and that recF and recJ were essential for p44 gene conversion in E. coli.
A. phagocytophilum lacks genes in the RecE pathway (26, 27). However, to test whether p44 recombination could occur through the RecE pathway, we examined the recombination in the RecE-active strain JC8679 (recB21recC22sbcA23). The ratio of Kmr/Amr transformants was less than 0.001, suggesting that RecE-dependent recombination (crossover) can also occur, albeit at a lower frequency than that observed through the RecF pathway.
DISCUSSION
The present experimental data generated using cell culture and infected animal specimens and the E. coli plasmid construct are consistent with our prediction based on the genome sequence data analysis (26, 27) and the characteristics of p44 gene conversion (4, 11, 26, 27) that the p44 gene conversion in A. phagocytophilum occurs through the RecF pathway. Isolation of recombination intermediates has been extremely difficult and possible thus far in limited systems, such as the yeast (16), E. coli (22, 41), and N. gonorrhoeae (18, 19). The recombined half-crossover-like structure in A. phagocytophilum in HL-60 cell culture and in the blood of an infected SCID mouse was very similar to those previously described in RecF-dependent intraplasmid recombination in an E. coli strain (22, 41). The weak 5.3-kb band in Southern blot analysis suggests that this structure is unstable or exists at very low frequency in A. phagocytophilum in cell culture under the conditions of the present study. Although a large amount of bacterial DNA can be isolated, p44E recombination rates appear to be much lower in cell culture than in animals (27). On the other hand, in infected animals a very small amount of A. phagocytophilum DNA is present in the overwhelming background of host cell DNA. These facts make the Southern blot detection extremely difficult. Further experiments and analysis would help find conditions or factors that enhance the p44 recombination rates to facilitate the detection of recombination intermediates in Southern blot analyses. The intermediate structure observed by sequencing is probably not a PCR shuffling artifact between a donor p44 and a recipient p44 expression locus DNA for reasons. (i) The intermediate structure sequence was obtained from the 5.3 kb-band region that did not include donor and recipient sequences. (ii) The existence of this structure was demonstrated by PCR with a high-fidelity Pfu DNA polymerase with no discernible recombination activity (37). (iii) Extremely low concentrations of p44 DNAs in the total DNA from infected blood specimens, since most of it is host cell derived in our assay, prevents the PCR shuffling artifact. (iv) Even when the PCR was performed with a high concentration of two plasmids, one carrying p44E and another the p44 donor sequence, a recombined sequence was not generated. (v) The PCR detected only a single unidirectional half-crossover structure in which 3′ end of donor p44 recombined with the recipient p44E but not the structure in which the 5′ end of p44E recombined with the p44 donor. (vi) Similar recombination intermediates using a double-origin plasmid carrying the p44 expression locus and a donor p44 locus were recovered in an E. coli strain with an active RecF recombination pathway.
A stable gene knockout method has not been developed for obligate intracellular organisms, but the E. coli system may provide an alternative strategy for characterization of factors involved in the p44 recombination in A. phagocytophilum. We inserted a donor p44 and the recipient p44E in the double-origin plasmid. Using this plasmid, pEKD30, we isolated the half-crossover intermediate in E. coli strain JC7623 with an active RecF-pathway (recBCsbcBC). The demonstrated fidelity of the E. coli system for p44 recombination should encourage the use of E. coli enzymes and mutant systems for the analysis of antigenic variation in obligatory intracellular bacteria. Using a BLAST search, genes involved in the RecF recombination pathway were found in the A. phagocytophilum genome sequence database (26, 27). RecJ is a single-strand specific 5′ to 3′ exonuclease (23); the homolog in A. phagocytophilum may digest the 5′ tail end of p44. RecFOR and RecA homologs were also found that may help the 3′-tailed single strand invade the donor p44 duplex. In addition, the A. phagocytophilum genome contains the Holliday junction DNA helicase genes, ruvA and ruvB, and the crossover junction endodeoxyribonuclease gene, ruvC. RuvA and RuvB denature and renature duplex DNA at the cruciform structure to promote its migration, while the RuvC protein acts to resolve the Holliday junction structure. Genes of the recBCD-dependent and recE-dependent pathways were absent from our bacterium, although homologs of AddAB have been found in Bacillus (8) and Rhizobium etli (45). AddAB was detected in the genome of A. phagocytophilum (APH0258 and APH0849 with E-values of 6e-79 and 9e-07, respectively) by BLAST search using AddAB of R. etli. This new class of enzymes have both helicase and nuclease activity and function like the RecBCD enzyme (8). In the future, we may be able to express these genes in our pEKD30 and E. coli system and define their roles in p44 recombination.
Further experiments and analysis would help optimize the present E. coli system for molecular analysis of p44 recombination. An experiment should be performed to determine how much noncoding sequence upstream and downstream of p44E is required for recombination. Three of five Kmr clones did not have the recombined p44-30-km locus but retained the original p44-18-km locus. These transformants were not investigated in detail. It is possible that this background transformation may be due to the insertion of the whole intergenic region between omp-1N and p44E into the plasmid. This region contains the A. phagocytophilum promoter sequence upstream of p44E (26) that may have been recognized by E. coli transcription and translation system. Thus, the downstream km gene could have been expressed without recombination. By removing or changing this promoter region, we may be able to reduce the Kmr background colonies. However, our speculation may not be correct, since we did not obtain any Kmr colonies when the recF mutant strain JC8111 or the recJ mutant strain JC12190 was used. An experiment that compares the lengths and sequences of donor p44s for their efficiency of recombination will be also informative. The functional pseudogene p44-30 used for pEKD30 plasmid has approximately 200 bp of the 5′conserved region and 410 bp of the 3′ conserved region, and thus far all p44s found to recombine to p44E have at least 50 bp from each of these regions (26, 27, 40). According to Fujitani et al. (12), 50 bp is the minimum length required for the efficient homologous recombination. By systematically modifying the pEKD30 construct, we may be able to define the sequence that influences efficiency of p44 recombination.
In the bovine intraerythrocytic agent, Anaplasma marginale, the major surface protein 2 (msp2) multigene family, homologous to the p44 gene family, also shows variation in gene expression in a similar gene expression locus (3, 6, 7, 13). Recently, Futse et al. proposed an “anchoring” model of segmental gene conversion for annealing donor and recipient msp2 sequences (13). however, a recombination intermediate structure or a recombination mechanism has not been described. The RecF pathway recombination may also involve gene conversion of the msp2 pseudogene to the msp2 expression locus, because A. marginale also lacks the RecBCD and RecE pathway genes (31) and because msp2s have 5′ and 3′ conserved regions and a central hypervariable region similar to the p44 locus suitable for homologous recombination.
In summary, the present study supports that the sequential half-crossover model accounts for the gene conversion in the p44 expression locus. The present study opened the possibility of many mechanistic experiments to determine the molecular mechanisms of p44 recombination and thus P44 antigenic variation.
Acknowledgments
This research was supported by a grant R01AI47407 from the National Institutes of Health.
We thank Ichizo Kobayashi for plasmid pKEN33 and E. coli strains. We also thank the E. coli Genetic Stock Center at Yale University, New Haven, Conn., for E. coli strains. We also appreciate the helpful discussions with Yumi Kumagai.
Editor: J. T. Barbieri
REFERENCES
- 1.Allers, T., and M. Lichten. 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106:47-57. [DOI] [PubMed] [Google Scholar]
- 2.Bakken, J. S., J. Krueth, C. Wilson-Nordskog, R. L. Tilden, K. Asanovich, and J. S. Dumler. 1996. Clinical and laboratory characteristics of human granulocytic ehrlichiosis. JAMA 275:199-205. [PubMed] [Google Scholar]
- 3.Barbet, A. F., A. Lundgren, J. Yi, F. R. Rurangirwa, and G. H. Palmer. 2000. Antigenic variation of Anaplasma marginale by expression of MSP2 mosaics. Infect. Immun. 68:6133-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbet, A. F., P. F. M. Meeus, M. Belanger, M. V. Bowie, J. Yi, A. M. Lundgren, A. R. Alleman, S. J. Wong, F. K. Chu, U. G. Munderloh, and S. D. Jauron. 2003. Expression of multiple outer membrane protein sequence variants from a single genomic locus of Anaplasma phagocytophilum. Infect. Immun. 71:1706-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbour, A. G., and B. I. Restrepo. 2000. Antigenic variation in vector-borne pathogens. Emerg. Infect. Dis. 6:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brayton, K. A., D. P. Knowles, T. C. McGuire, and G. H. Palmer. 2001. Efficient use of a small genome to generate antigenic diversity in tick-borne ehrlichial pathogens. Proc. Natl. Acad. Sci. USA 98:4130-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brayton, K. A., G. H. Palmer, A. Lundgren, J. Yi, and A. F. Barbet. 2002. Antigenic variation of Anaplasma marginale msp2 occurs by combinatorial gene conversion. Mol. Microbiol. 43:1151-1159. [DOI] [PubMed] [Google Scholar]
- 8.Chedin, F., and S. C. Kowalczykowski. 2002. A novel family of regulated helicases/nucleases from gram-positive bacteria: insights into the initiation of DNA recombination. Mol. Microbiol. 43:823-834. [DOI] [PubMed] [Google Scholar]
- 9.Chen, S. M., J. S. Dumler, J. S. Bakken, and D. H. Walker. 1994. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J. Clin. Microbiol. 32:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumler, J. S., A. F. Barbet, C. P. Bekker, G. A. Dasch, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. R. Rurangirwa. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145-2165. [DOI] [PubMed] [Google Scholar]
- 11.Felek, S., S. Telford III, R. C. Falco, and Y. Rikihisa. 2004. Sequence analysis of p44 homologs expressed by Anaplasma phagocytophilum in infected ticks feeding on naive hosts and in mice infected by tick attachment. Infect. Immun. 72:659-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujitani, Y., K. Yamamoto, and I. Kobayashi. 1995. Dependence of frequency of homologous recombination on the homology length. Genetics 140:797-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Futse, J. E., K. Y. Brayton, D. P. Knowles, and G. H. Palmer. 2005. Structural basis for segmental gene conversion in generation of Anaplasma marginale outer membrane protein variants. Mol. Microbiol. 57:212-221. [DOI] [PubMed] [Google Scholar]
- 14.Gardner, S. L., R. C. Holman, J. W. Krebs, R. Berkelman, and J. E. Childs. 2003. National surveillance for the human ehrlichioses in the United States, 1997-2001, and proposed methods for evaluation of data quality. Ann. N. Y. Acad. Sci. 990:80-89. [DOI] [PubMed] [Google Scholar]
- 15.Goodman, J. L., C. Nelson, B. Vitale, J. E. Madigan, J. S. Dumler, T. J. Kurtti, and U. G. Munderloh. 1996. Direct cultivation of the causative agent of human granulocytic ehrlichiosis. N. Engl. J. Med. 334:209-215. [DOI] [PubMed] [Google Scholar]
- 16.Haber, J. E. 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32:561-599. [DOI] [PubMed] [Google Scholar]
- 17.Horii, Z., and A. J. Clark. 1973. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K-12: isolation and characterization of mutants. J. Mol. Biol. 80:327-344. [DOI] [PubMed] [Google Scholar]
- 18.Howell-Adams, B., L. A. Wainwright, and H. S. Seifert. 1996. The size and position of heterologous insertions in a silent locus differentially affect pilin recombination in Neisseria gonorrhoeae. Mol. Microbiol. 22:509-522. [DOI] [PubMed] [Google Scholar]
- 19.Howell-Adams, B., and H. S. Seifert. 2000. Molecular models accounting for the gene conversion reactions mediating gonococcal pilin antigenic variation. Mol. Microbiol. 37:1146-1158. [DOI] [PubMed] [Google Scholar]
- 20.Kim, H. Y., and Y. Rikihisa. 1998. Characterization of monoclonal antibodies to the 44-kilodalton major outer membrane protein of the human granulocytic ehrlichiosis agent. J. Clin. Microbiol. 36:3278-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, H. Y., and Y. Rikihisa. 2000. Expression of interleukin-1β, tumor necrosis factor alpha, and interleukin-6 in human peripheral blood leukocytes exposed to human granulocytic ehrlichiosis agent or recombinant major surface protein P44. Infect. Immun. 68:3394-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi, I. 1992. Mechanisms for gene conversion and homologous recombination: the double-strand break repair model and the successive half crossing-over model. Adv. Biophys. 28:81-133. [DOI] [PubMed] [Google Scholar]
- 23.Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58:401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kushner, S. R., H. Nagaishi, A. Templin, and A. J. Clark. 1971. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc. Natl. Acad. Sci. USA 68:824-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, Q., N. Zhi, N. Ohashi, H. W. Horowitz, M. E. Aguero-Rosenfeld, J. Raffalli, G. P. Wormser, and Y. Rikihisa. 2002. Analysis of sequences and loci of p44 homologs expressed by Anaplasma phagocytophila in acutely infected patients. J. Clin. Microbiol. 40:2981-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, Q., Y. Rikihisa, N. Ohashi, and N. Zhi. 2003. Mechanisms of variable p44 expression by Anaplasma phagocytophilum. Infect. Immun. 71:5650-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, Q., and Y. Rikihisa. 2005. Establishment of cloned Anaplasma phagocytophilum and analysis of p44 gene conversion within infected horse and SCID mice. Infect. Immun. 73:5106-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lisser, S., and H. Margalit. 1993. Compilation of Escherichia coli mRNA promoter sequences. Nucleic Acids Res. 21:1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lloyd, R. G., and C. Buckman. 1985. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J. Bacteriol. 164:836-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovett, S. T., and A. J. Clark. 1984. Genetic analysis of the recJ gene of Escherichia coli K-12. J. Bacteriol. 157:190-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meeus, P. F., K. A. Brayton, G. H. Palmer, and A. F. Barbet. 2003. Comparison of the major surface protein 3 pseudogene loci in two different isolates of Anaplasma marginale. Abstr. 37. Am. Soc. Rickettsiol. 18th Meet. American Society for Rickettsiology, Cumberland, Md.
- 32.Mehr, I. J., and H. S. Seifert. 1998. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol. Microbiol. 30:697-710. [DOI] [PubMed] [Google Scholar]
- 33.Rikihisa, Y. 2000. Ehrlichial strategy for survival and proliferation in leukocytes. Subcell. Biochem. 33:517-538. [DOI] [PubMed] [Google Scholar]
- 34.Rikihisa, Y. 2003. Mechanisms to create a safe haven by members of the family Anaplasmataceae. Ann. N. Y. Acad. Sci. 990:548-555. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 2nd ed., p. 119-122. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Santoyo, G., and D. Romero. 2005. Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol. Rev. 29:169-183. [DOI] [PubMed] [Google Scholar]
- 37.Shafikhani, S. 2002. Factors affecting PCR-mediated recombination. Environ. Microbiol. 4:482-486. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi, N. K., K. Yamamoto, Y. Kitamura, S. Q. Luo, H. Yoshikura, and I. Kobayashi. 1992. Nonconservative recombination in Escherichia coli. Proc. Natl. Acad. Sci. USA 89:5912-5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, X., T. Kikuchi, and Y. Rikihisa. 2006. Two neutralization sites defined by epitope-mapping and monoclonal antibody inhibition analysis on P44 major surface proteins of Anaplasma phagocytophilum. Infect. Immun. 74:1873-1882. [DOI] [PMC free article] [PubMed]
- 40.Wang, X., Y. Rikihisa, T. H. Lai, Y. Kumagai, N. Zhi, and S. M. Reed. 2004. Rapid sequential changeover of expressed p44 genes during the acute phase of Anaplasma phagocytophilum infection in horses. Infect. Immun. 72:6852-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamoto, K., K. Kusano, N. K. Takahashi, H. Yoshikura, and I. Kobayashi. 1992. Gene conversion in the Escherichia coli RecF pathway: a successive half crossing-over model. Mol. Gen. Genet. 234:1-13. [DOI] [PubMed] [Google Scholar]
- 42.Zhi, N., N. Ohashi, and Y. Rikihisa. 1999. Multiple p44 genes encoding major outer membrane proteins are expressed in the human granulocytic ehrlichiosis agent. J. Biol. Chem. 274:17828-17836. [DOI] [PubMed] [Google Scholar]
- 43.Zhi, N., N. Ohashi, and Y. Rikihisa. 2002. Activation of a p44 pseudogene in Anaplasma phagocytophila by bacterial RNA splicing: a novel mechanism for posttranscriptional regulation of a multigene family encoding immunodominant major outer membrane proteins. Mol. Microbiol. 46:135-145. [DOI] [PubMed] [Google Scholar]
- 44.Zhi, N., N. Ohashi, T. Tajima, J. Mott, R. W. Stich, D. Grover, S. R. Telford III, Q. Lin, and Y. Rikihisa. 2002. Transcript heterogeneity of the p44 multigene family in a human granulocytic ehrlichiosis agent transmitted by ticks. Infect. Immun. 70:1175-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuniga-Castillo, J., D. Romero, and J. M. Martinez-Salazar. 2004. The recombination genes addAB are not restricted to gram-positive bacteria: genetic analysis of the recombination initiation enzymes RecF and AddAB in Rhizobium etli. J. Bacteriol. 186:7905-7913. [DOI] [PMC free article] [PubMed] [Google Scholar]