Abstract
Forward genetic screens for mutations that rescue the paralysis of ric-8 (Synembryn) reduction-of-function mutations frequently reveal mutations that cause hyperactivation of one or more components of the Gαs pathway. Here, we report that one of these mutations strongly reduces the function of the Dunce cAMP phosphodiesterase PDE-4 by disrupting a conserved active site residue. Loss of function and neural overexpression of PDE-4 have profound and opposite effects on locomotion rate, but drug-response assays suggest that loss of PDE-4 function does not affect steady-state acetylcholine release or reception. Our genetic analysis suggests that PDE-4 regulates both Gαs-dependent and Gαs-independent cAMP pools in the neurons controlling locomotion rate. By immunostaining, PDE-4 is strongly expressed throughout the nervous system, where it localizes to small regions at the outside boundaries of synaptic vesicle clusters as well as intersynaptic regions. The synaptic subregions containing PDE-4 are distinct from those containing active zones, as indicated by costaining with an antibody against the long form of UNC-13. This highly focal subsynaptic localization suggests that PDE-4 may exert its effects by spatially regulating intrasynaptic cAMP pools.
THE presynaptic function of cAMP and its role in the generation and execution of behaviors is poorly understood. Recent Caenorhabditis elegans genetics studies have shown that presynaptic cAMP plays a critical role in regulating locomotion rate (Reynolds et al. 2005; Schade et al. 2005). Mutants specifically lacking the neuronal Gαs pathway that generates cAMP are nearly paralyzed and yet, paradoxically, they seem to have normal levels of steady-state neurotransmitter release, as indicated by live animal drug-response assays (Reynolds et al. 2005; Charlie et al. 2006). Electrophysiological studies of a Drosophila Gαs null and reduction-of-function mutants also found normal (Hou et al. 2003; Wolfgang et al. 2004) or increased (Renden and Broadie 2003) neurotransmitter release in response to low-frequency nerve stimulation, although these studies also found that Gαs mutants, unlike wild type, showed no potentiation of neurotransmitter release after tetanic stimulation. Fly Gαs nulls, like worm mutants lacking a neuronal Gαs pathway, show little or no movement (Wolfgang et al. 2001). The strong contrast between the behavioral and physiological effects of decreased synaptic cAMP is puzzling and suggests that we do not adequately understand what cAMP is doing at the synapse.
Studies using Drosophila have investigated the neuronal function of cAMP using the learning and memory mutants dunce and rutabaga. dunce encodes a cAMP phosphodiesterase that normally functions to reduce cAMP levels, whereas rutabaga encodes a Ca2+-calmodulin-stimulated adenylyl cyclase that represents one, but probably not the only, source of cAMP in Drosophila neurons. These studies, and others using the Aplysia model system, have established that cAMP plays a central role in learning and memory formation (Davis et al. 1995; Kandel 2001). Some of these studies have focused on long-term facilitation mediated by cAMP response element binding (CREB) protein and map kinase-induced transcriptional changes (Bailey et al. 1996; Kandel and Pittenger 1999; Kandel 2001). While such gene expression changes appear highly relevant to long-term memory, the available evidence suggests that cAMP has another conserved function that is CREB independent and yet plays a central role in the execution of all behaviors, learned or otherwise. For example, the C. elegans CREB ortholog CRH-1 does not appear to be expressed in most neurons and, in strong contrast to the neuronal Gαs pathway, is not required for normal locomotion (Kimura et al. 2002).
Researchers have also directly investigated the effects of dunce and rubabaga mutations on synaptic physiology and ultrastructure. These studies found impaired synaptic facilitation at both the whole-cell and individual synapse levels in both dunce and rutabaga mutants (Zhong and Wu 1991; Kuromi and Kidokoro 2000; Renger et al. 2000). Synaptic recordings from individual synapses found an ∼50% reduction in the frequency of spontaneous release of individual vesicles in both dunce and rutabaga mutant larvae (Renger et al. 2000), although whole-cell recordings of spontaneous release from dunce mutant embryos showed no difference from wild type (Suzuki et al. 2002). Nerve-evoked release from individual synapses is reduced or unchanged in rutabaga mutants (depending on the study), but not significantly different in dunce mutants (Cheung et al. 1999; Renger et al. 2000). Renger et al. (2000) found substantial variation in decay times of spontaneous and evoked currents from individual dunce and rutabaga synapses and increased variability in the responses of both kinds of mutant synapses during tetanic stimulation. Other intriguing studies have found that dunce and rutabaga mutations affect the mobilization of synaptic vesicles between different defined vesicle pools (Kuromi and Kidokoro 2000; Suzuki et al. 2002; Kidokoro et al. 2004) and, ultrastructurally, the ratios of docked/undocked synaptic vesicles (Renger et al. 2000).
Despite the many important insights provided by the Drosophila dunce and rutabaga studies, pressing questions remain that must be answered to understand the role of synaptic cAMP in the generation and execution of behaviors. For example, what controls the activation of the Gαs pathway at specific synapses or populations of synapses? How does the Gαs pathway interact with other Gα-signaling pathways that regulate neurotransmitter release? Does the Gαs pathway make all of the cAMP that regulates synaptic function, or is there also a Gαs-independent pool? Is the Dunce cAMP phosphodiesterase targeted to specific synaptic subregions to spatially regulate cAMP within synapses?
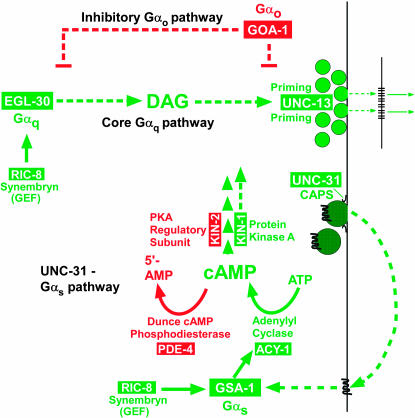
Using a collection of synaptic signaling mutants that is unique in its scope among model organisms, genetic studies in C. elegans have begun to address some of these questions by showing that presynaptic UNC-31 (a dense core vesicle priming protein) controls activation of the Gαs pathway (Charlie et al. 2006) and that the UNC-31–Gαs pathway is part of a larger synaptic signaling network composed of at least three major Gα-signaling pathways that drive the locomotion behavior and may control neurotransmitter release at all synapses (Figure 3 and references in legend; see also Introduction of Reynolds et al. 2005). In addition to the Gαs pathway, this synaptic signaling network includes an essential Gαq pathway, which appears to function upstream of the synaptic vesicle priming mechanism, and a powerful inhibitory Gαo pathway. The Gαs pathway is largely dependent on the Gαq pathway to exert its effects on locomotion and is required for a proper response to the diacyclglycerol that the Gαq pathway produces (Reynolds et al. 2005). C. elegans researchers have identified many of the components of this network through large forward genetic screens centered around easily recognizable phenotypes that affect locomotion, egg laying, and growth on the pesticide aldicarb. With respect to the locomotion behavior, loss-of-function mutations in positive regulators cause paralysis or strongly decreased rates of locomotion, while loss-of-function mutations in negative regulators cause hyperactive locomotion.
Figure 3.
Schematic of the C. elegans synaptic signaling network with emphasis on the Gαs pathway. Shown are the three major Gα-signaling pathways of the network with the Gαo and Gαq branches of the network in skeletal form (most components not shown). Solid lines indicate that direct interactions are known or likely, while dashed lines indicate predicted interactions, unknown components, or known components that are not shown. Proteins that promote locomotion and/or neurotransmitter release are shown in green, while proteins that inhibit locomotion and/or neurotransmitter release are shown in red. The model is based on findings from this study (for PDE-4) and the following previous studies: Maruyama and Brenner (1991), Mendel et al. (1995), Segalat et al. (1995), Brundage et al. (1996), Korswagen et al. (1997), Berger et al. (1998), Hajdu-Cronin et al. (1999), Lackner et al. (1999), Miller et al. (1999, 2000), Nurrish et al. (1999), Richmond et al. (1999, 2001), Moorman and Plasterk (2002), Tall et al. (2003), Reynolds et al. (2005), Schade et al. (2005), and Charlie et al. (2006).
Previously reported forward genetic screens identified a group of mutants that strongly rescue the nearly paralyzed phenotype caused by the strong reduction-of-function ric-8(md303) mutation (Schade et al. 2005). RIC-8 (Synembryn) is a guanine nucleotide exchange factor (Tall et al. 2003) that is required to maintain both the Gαq and the Gαs pathways of the synaptic signaling network in a functional state (Miller et al. 2000; Tall et al. 2003; Reynolds et al. 2005). The ric-8(md303) rescuing mutations restore coordinated locomotion to the ric-8 mutant and improve its locomotion rate to wild-type levels or greater. Mapping and molecular characterization of some of the ric-8(md303) suppressor mutations revealed that they are often mutations that cause hyperactivation of the Gαs pathway (Schade et al. 2005). The Gαs pathway mutations uncovered by these studies include dominant gain-of-function alleles of GSA-1 (Gαs) and ACY-1 (a highly conserved type IX adenylyl cyclase) and reduction-of-function mutations in KIN-2 (the regulatory subunit of protein kinase A) (Schade et al. 2005). Although the first genetic studies of the C. elegans Gαs pathway showed that transgenic hyperactivation of the Gαs pathway causes paralysis via necrotic death of neurons (Korswagen et al. 1997; Berger et al. 1998), the native mutations that activate the Gαs pathway confer only minimal cell death and instead cause highly coordinated, strongly hyperactive locomotion as single mutants (Schade et al. 2005).
Other mutations from the ric-8(md303) rescue screen remain to be characterized and could reveal other components of the neuronal Gαs pathway or an interacting pathway of the synaptic signaling network. Here, we report that one of these mutations strongly reduces the function of the Dunce cAMP phosphodiesterase PDE-4 by disrupting a conserved active site residue. Loss of function and neuronal overexpression of PDE-4 have profound and opposite effects on locomotion rate, but our live animal drug-response assays suggest that loss of PDE-4 function does not affect steady-state acetylcholine release or reception. Our genetic analysis suggests that PDE-4 regulates both Gαs-dependent and Gαs-independent cAMP pools in the neurons controlling locomotion rate. By immunostaining, PDE-4 is strongly expressed throughout the nervous system, where it localizes to small regions at the outside boundaries of synaptic vesicle clusters as well as intersynaptic regions. The synaptic subregions containing PDE-4 are distinct from those containing active zones, as indicated by costaining with an antibody against the long form of UNC-13. This highly focal subsynaptic localization suggests that PDE-4 may exert its effects by spatially regulating intrasynaptic cAMP pools or gradients.
MATERIALS AND METHODS
Strains and worm culture:
References and descriptions of all of the strains used in this study are cited in the text and/or figure legends. Methods used to make transgenic strains and double and triple mutants are further described below. All culture media were made with Sigma (St. Louis) A-7002 agar. Worm culture followed previously described methods (Brenner 1974). We observed and manipulated live animals under Olympus SZX-12 stereomicroscopes equipped with ×1.2, 0.13 numerical aperture (NA) plan apochromatic objectives. Unless otherwise specified, wild-type worms were C. elegans variety Bristol, strain N2. acy-1(pk1279) was maintained over the closely linked mutation dpy-17(e164) as the strain NL1999, which was kindly provided by Celine Moorman and Ron Plasterk. NL1999 cultures were maintained by cloning six wild-type larvae to individual culture plates, growing them 4 days at 20°, and saving plates that segregated paralyzed larvae and Dpy-17 animals. We used SP754 unc-4(e120) mnDf88/mnC1 dpy-10(e128) unc-52(e444) in the genetic test for haplo-insufficiency of the pde-4 locus. mnDf88 deletes multiple genes, including markers known to physically flank pde-4.
Isolation and outcrossing pde-4(ce268):
We isolated the ce268 mutation in a previously described suppressor screen for mutations that could rescue the nearly paralyzed phenotype of ric-8(md303) (Schade et al. 2005). We outcrossed this semidominant mutation in a ric-8(md303) background by crossing ce268/+; md303/+ males to md303 hermaphrodites. From the virgin progeny of this cross, we cloned partially rescued (non-wild-type) animals [putative genotype ce268/+; ric-8(md303)]. In the next generation, we then cloned candidate ce268; ric-8(md303) homozygotes on the basis of the strong rescue of ric-8(md303)'s paralysis. In the next generation we confirmed the homozygosity of the strain by the absence of paralyzed ric-8(md303) single mutants. We then repeated this process to produce a 4× outcrossed ce268; ric-8(md303) strain. After mapping the mutation (see below), we isolated 5× outcrossed ce268 single mutants by producing the strain ce268/dpy-10(e128); ric-8(md303)/+ and then relying on the close linkage of dpy-10 and ce268 to positively identify ce268 homozygotes in the next generation and then choosing a line that did not segregate ce268; ric-8(md303) nonhyperactive double mutants.
Single-nucleotide-polymorphism-based mapping of ce268 to a physical interval:
We mapped ce268 by its ric-8 suppressor phenotype using the ric-8(md303) mapping strain ric-8(md303) [CB4856] that we had outcrossed 12× into a CB4856 background (Schade et al. 2005). To map ce268, we crossed md303 [CB4856]/+[CB4856] males to ce268; ric-8(md303) double mutants. From the virgin progeny of this cross, we cloned partially rescued (non-wild-type) animals [putative genotype ce268/+; ric-8(md303) containing the CB4856 markers]. In the next generation, we cloned candidate ce268; ric-8(md303) homozygotes on the basis of the strong rescue of the ric-8 mutant's paralysis. We then checked the progeny of these animals for homozygosity [absence of wild-type animals or ric-8(md303) single mutants], and, upon starvation, we checked the homozygous cultures for various CB4856 single nucleotide polymorphisms (SNPs) as described (Schade et al. 2005). We used SNPs identified by Wicks et al. (2001) to map the ce268 to a chromosome and subregion in a manner similar to that previously described (Schade et al. 2005). After determining that the mutation mapped to the recombination-poor center of chromosome II, we repeated the crosses in 13 weekly cycles to produce the ∼1900 mapping lineages necessary to narrow the mutation to a somewhat tractable interval of 474 kb. The SNPs that we used to narrow ce268 to this interval have been previously reported as locations on specific genomic DNA clones (Wicks et al. 2001; Swan et al. 2002). The specific locations of the SNPs used in this study are as follows (genomic DNA clone/location on clone): ceP136 (E04F6/1447); ceP126 (C30G12/37,286); ceP124 (F43E2/25,033); ceP105 (ZK1290/15,120); ceP128 (F18A1/17,419).
Identification of the ce268 mutation through large-scale sequence analysis:
After narrowing the ce268 mutation to as small a region as possible (474 kb) using SNP-based mapping, we chose an ∼400-kb region of highest likelihood for detailed sequence analysis to find the mutation. To sequence this region, we used Expand 20 Kb+ (Roche) to amplify overlapping 11-kb PCR fragments from purified ce268 genomic DNA. We then designed primers to completely sequence each product. In the 400-kb interval, there were three regions of sizes 13.8, 0.5, and 0.15 kb that we could not amplify and/or sequence despite repeated attempts. We also excluded another 4.2 kb of sequencing gaps after finding the mutation near the end of the project.
RT–PCR analysis of pde-4(ok1290):
To analyze the pde-4 mRNA produced in the ok1290 mutant, we isolated total RNA from N2 (wild type) and pde-4(ok1290) using the RNeasy mini kit (QIAGEN, Chatsworth, CA) followed by reverse transcription and PCR amplification using Stratascript RT polymerase (Stratagene, La Jolla, CA) and Accuprime Pfx DNA polymerase, respectively. We then compared N2 and pde-4(ok1290) RT–PCR products by gel analysis and partial sequencing.
Plasmids:
To make KG#203 [rab-3∷pde-4d(+) cDNA], we used StrataScript reverse transcriptase (Stratagene) and a primer engineered with a restriction site to transcribe a pde-4d (+) DNA template from purified wild-type C. elegans mRNA. For reasons not clear, the reverse transcription product did not serve as an adequate template for subsequent amplification, despite repeated attempts with different primer combinations and PCR conditions. We therefore used RNAse H to nick the duplexed RNA/DNA hybrid molecule and synthesized the second strand of DNA using Escherichia coli DNA polymerase I. We then used Accuprime Pfx and primers engineered with restriction sites to amplify and clone the cDNA into NheI/AgeI-cut KG#59 (a neuronal-specific C. elegans expression vector described in Schade et al. 2005). To make KG#205 [rab-3∷pde-4d (D448N) cDNA], we used the QuikChange site-directed mutagenesis method (Stratagene) to introduce the D448N mutation into the pde-4d(+) cDNA on the KG#203 template. The mutation changes codon 448 of pde-4d from GAT (Asp) to AAT (Asn) (HDVDH to HNVDH). After sequencing the entire insert and identifying a clone with only this mutation, we recloned the insert into a fresh KG#59 expression vector background with the same restriction sites used to make KG#203. To make KG#213 [rab-3∷pde-4d (298–end) cDNA], we used Accuprime Pfx and primers engineered with restriction sites to amplify a fragment containing codons 298–764 [the end of the pde-4d(+) coding region] using KG#203 as a template and cloned the fragment into NheI/AgeI-cut KG#59. The coding region of this construct starts with a native methionine (M298). To make KG#200 [glutathione-S-transferase (GST)–UNC-13L (422–602)], we used Accuprime Pfx and primers engineered with restriction sites to amplify codons S422–A602 of the unc-13a isoform, as defined by the WormBase WS 140 freeze, and cloned the fragment into NcoI/XhoI-cut pGex-KG, which is a bacterial GST expression vector (Guan and Dixon 1991). An artificial stop codon follows A602. The region S422–A602 is the last ∼1/3 of the L region of UNC-13LR as defined by Kohn et al. (2000) and does not include any part of the R region. To make KG#193 [GST–PDE-4d (151–278)] and KG#194 (GST–PDE-4d (279–405)], we used Accuprime Pfx and primers engineered with restriction sites to amplify the desired codons from pde-4d(+) in KG#203 and cloned them into BamHI/XhoI- or BamHI/NcoI-cut pGex-KG, respectively. Both of these PDE-4 regions are common to all known products of the pde-4 locus. In all bacterial expression constructs, the protein of interest follows the C terminus of GST. In all constructs involving the cloning of PCR fragments, we sequenced the inserts and used clones containing no mutations in the fragment of interest to establish the final plasmid stock.
Transgenes:
We produced transgenic strains bearing extrachromosomal arrays by the method of Mello et al. (1991). We used pBluescript carrier DNA to bring the final concentration of DNA in the injection mixture to 175 ng/μl. All injection mixtures used in this study included the cotransformation marker plasmids KG#68 [rab-3∷GFP] and KG#67 [ttx-3∷GFP] at 15 ng/μl each. We made pde-4(ce268); ceEx166 [rab-3∷pde-4d (+) cDNA] by injecting pde-4(ce268) mutants with KG#203 [rab-3∷pde-4d (+) cDNA] at 40 ng/μl. We made ceEx148 [rab-3∷pde-4d (298–end) cDNA] by injecting N2 (wild type) with KG#213 at 20 ng/μl. We made ceIs33 [rab-3∷pde-4d (+) cDNA] by injecting N2 (wild type) with KG#203 [rab-3∷pde-4d (+) cDNA] at 40 ng/μl and integrating the resulting transgene. We made ceIs35 [rab-3∷pde-4d (D448N) cDNA] by injecting N2 (wild-type) animals with KG#205 [rab-3∷pde-4 (D448N) cDNA] at 40 ng/μl and integrating the resulting transgene. We performed the integrations as described, except that cultures were screened for 100% transmittance of the GFP, not the pha-1 marker (Reynolds et al. 2005).
Double mutants:
Unless otherwise specified, we constructed double mutants using standard genetic methods without additional marker mutations, and we confirmed homozygosity of both mutations by double-amplification PCR with nested primers [for strains containing the deletion mutations acy-1(pk1279) or unc-31(e928)] or by PCR and sequencing (for single-base-change mutations). Reynolds et al. (2005) describes examples of strain constructions involving acy-1(pk1279).
Standard locomotion assays:
Unless otherwise specified, we performed standard locomotion assays as described, using standardized plates and assay procedures (Miller et al. 1999; Reynolds et al. 2005). We chose animals for the assays from growing cultures that had not been previously starved. We collected 6- to 30-hr-old larval-arrested pde-4(ce268); acy-1(pk1279) double mutants and identical stage controls of N2 (wild type) and pde-4(ce268) single mutants for assays, documentation, and genotype verification as described for other larval arrested strains (Reynolds et al. 2005). For all locomotion assays on larval stage animals, we counted body bends for a single 6-min period for each of 10 animals that had been freshly plated at standard-room-temperature-equilibrated locomotion plates and stabilized for 30 min–2 hr before assaying. For the two strains containing extrachromosomal arrays [pde-4(ce268); ceEx166 and ceEx148], we handpicked young adult animals with uniformly green nervous systems (due to the stable presence of the rab-3∷GFP cotransformation marker). During the locomotion assay, we used predetermined specific light power and filter settings to control for possible light effects and an external light source to prevent stage warming.
Stimulated locomotion assays:
For stimulated locomotion assays, we prepicked 30 young adult animals from the strain to be assayed to a standard culture plate. We chose animals for the assays from growing cultures that had not been previously starved. We then plated one healthy representative adult on each of 10 labeled locomotion assay plates, taking care to handle the plates gently without jarring. The remaining 20 animals were stored on the culture plate for later loading on the locomotion plates. After this plating, we waited at least 10 min before starting the assay to allow animals to settle. For the stimulus, we made a standard weight consisting of two steel washers (o.d., 2 in.; i.d., 1 and 5/32 in.; thickness, 1/8 in.) inside an empty 60-mm petri plate and secured the plate lid and base to each other with adhesive tape. The total weight was 79.57 g. To set up for stimulation, the experimental locomotion assay plate containing the animal to be assayed was placed lid side up and aligned with marks on the glass stage of the scope that precisely line up with the objective above. We produced the stage marks by placing a stack of four 60-mm petri plates lid side up under the scope objective, aligning the stack of plates so they are exactly centered below the objective, and marking the location of the bottom plate on the stage with a fine Sharpie pen. We place two three-channel timers (Fisher #06-662-46) on the back part of the scope stage and set the timers at 5, 25, 45, 65, 85, and 105 sec. We then arranged five standard lab counting devices and placed them in order next to the scope. To initiate the stimulus, we held the weighted plate lid side up and aligned it precisely against the objective of the scope (the protective casing around the objective prevents the plate from touching the glass part of the objective). We then dropped the weighted plate on the aligned experimental plate three times in rapid succession. We did not touch or hold the experimental plate during this process. The falling distance is 38 mm from the bottom of the weighted plate to the top of the experimental plate. As soon as the weighted plate hit the experimental plate for the last time, we started all of the channels on both timers by hitting both start buttons simultaneously (there was an ∼0.5-sec delay as we reached for the timers after dropping the plate). The next step must be completed within 5 sec. We quickly removed the weighted plate from the scope and found the animal on the experimental plate. We zoomed into the desired magnification and, when we heard the timer beep at 5 sec, we began counting body bends, recording each bend on the first counter. When the second channel of the timer began beeping, we switched counting to the second counter, etc., until the fourth channel started beeping at which time we stopped counting and recorded the raw data from each counter. We then removed the animal from the plate and repeated the above steps for the other nine experimental plates. We then repeated the assay twice to get a sample size of 30.
Video production of living worms on culture plates:
We captured and converted videos of worms on agar plates containing OP-50 bacterial lawns as described (Schade et al. 2005).
Drug-sensitivity assays:
We performed drug paralysis assays on solid media as described (Lackner et al. 1999; Nurrish et al. 1999) with the modifications noted below. We added levamisole to 55° molten media to a final concentration of 800 μm from a 200-mm stock in ddH2O. We added aldicarb to a final concentration of 2 mm from a 10-mm stock solution in ddH2O (allowing ∼2–3 hr for dissolving the aldicarb in water before adding to the 55° molten media), and we made the media with 20% less water than normal to compensate for the large drug volume. We seeded aldicarb- and levamisole-containing plates with OP-50 on the day they were poured and stored them at room temperature for 2 days, lid side up, before using. We used 20 animals for each strain/condition per trial. We chose animals for the assays from growing cultures that had not been previously starved.
Antibodies:
To produce the UNC-13L-specific antibody KM21E-3.1, we prepared a recombinant UNC-13 fragment by first transforming KG#200 [GST–UNC-13 (422–602); see Plasmids] into the bacterial expression host BL21-DE3. We grew a 5-liter culture to an A600 of 0.4, lowering the incubation temperature gradually during growth such that the 0.4-OD culture was at 19°. We then added IPTG to 30 μm and continued growth at 19° for ∼17 hr before harvesting the cells. Cell pellets were frozen in liquid nitrogen and rapidly thawed before resuspending in 67-ml ice cold lysis buffer [50 mm Tris–HCl, pH 8.0, 10% glycerol, 2 mm β-mercaptoethanol, 0.1 mm PMSF, 1 μg/ml pepstatin, 1 μg/ml leupeptin, 0.255 mg/ml lysozyme (Sigma L-6876)] per liter of culture. All steps thereafter were at 4°. To lyse, we stirred the suspension for 30 min and then added 5 mg of DNAse (Sigma D-4527) and 400 μl of 1 m MgSO4 and incubated the suspension 30 min more with stirring. After clearing the lysate with a 30 min × 33,000 × g spin, we added NaCl to 300 mm and loaded the lysate at 0.5 ml/min onto a 5-ml glutathione–agarose (Sigma #G4510) column hooked to a Biologic (Bio-Rad, Hercules, CA) low-pressure chromatography system. We washed the column with 50 ml of GST buffer B [45 mm Tris–HCl (pH 8.0 at 4°), 150 mm NaCl, 10% glycerol, 1 mm DTT, 11 mm MgCl2, 2 mm ATP], followed by 25 ml of GST buffer C [50 mm Tris (pH 8.0 at 4°), 50 mm NaCl, 10% glycerol, 2 mm βme]. We eluted the protein with 25 ml of GST buffer D (GST buffer C plus 10 mm reduced glutathione, adjusted to pH 8.0 at 4°) while collecting 1-ml fractions. We analyzed peak fractions by SDS–PAGE and determined that a band of the correct size was ∼50% pure. We further purified the protein on an SDS–PAGE gel and homogenized the Coomassie-stained gel band in a 10-ml Potter-Elvehjem tissue grinder (Kontes), driven by a drill press, such that the final concentration was 600 μg/ml of gel suspension. We sent the suspension to Cocalico (Cocalico, PA) for injection into a goat (1200-μg initial injection plus 2 × 600-μg boosts at 3- and 4-week intervals, respectively). We affinity purified UNC-13-specific antibodies by incubating 12 ml of serum with nitrocellulose strips containing 2000 μg of SDS–PAGE-purified GST–UNC-13 (422–602) using a previously described method (Miller et al. 2000). We produced the PDE-4-specific antibody KM25B-3.1 using KG#193 [GST–PDE-4 (151–278); see Plasmids] and a second PDE-4-specific antibody KM24B-3.1 using KG#194 [GST–PDE-4 (279–405)]. We produced these antibodies using the same methods as for the KM21E-3.1 antibody; however, the GST–PDE-4 (151–278) protein was ∼85% pure after purification, and we did not further purify it before submitting it for injection into the host animal. We produced both PDE-4 antibodies in rabbits. Janet Duerr provided the UNC-17-specific antibody mAb1403 (as mouse monoclonal ascites) (Duerr et al. 2001), and Mike Nonet provided the UNC-10-specific antibody RIM5431 (as rabbit serum) (Koushika et al. 2001).
Immunostaining:
We developed a new immunostaining procedure for a recent study (described in detail in Charlie et al. 2006), and we also used it for this study. To carry out the preadsorption control staining in supplemental Figure S2 at http://www.genetics.org/supplemental/, we purified GST–PDE-4D (151–278) by SDS–PAGE and transferred the protein to nitrocellulose. After staining with Ponceau S, we cut out the strip containing the purified protein (estimated at close to 100% purity) and destained and blocked nonspecific sites on the strip by incubation with 10 mm Tris, pH 8.0, 150 mm NaCl, 0.05% Tween-20 plus 3% nonfat dry milk. We then inserted a 5-mm strip of the blotted protein (containing ∼30 μg of protein) into the primary antibody mixture and incubated for 1 hr × RT, before removing the strip and adding the worm suspension. We used the primary antibodies at the following dilutions: KM25B-3.1 [α-PDE-4 (151–278), 1/200]; KM24B-3.1 [α-PDE-4 (279–405), 1/150]; mAb1403 (α-UNC-17; 1/5000 from ascites), RIM5431 (α-UNC-10; 1/12,000). We produced secondary antibodies by coupling whole IgG (Jackson Immunoresearch, West Grove, PA; “ML” quality for multi-labeling) to Alexa 555 or Alexa 647 (Molecular Probes, Eugene, OR) according to the manufacturer's instructions. We purchased Cy2-coupled secondary antibodies directly from Jackson Immunoresearch as whole IgG “ML” quality antibodies. We used all of the secondary antibodies at 1.8 μg/ml final concentration.
Imaging:
We collected fluorescent images using a Nikon Eclipse TE2000-E inverted microscope equipped with a ×60 1.4 NA oil planapochromat objective (CF160-type), a ×1.5 tube lens, a motorized linear-encoded z-drive, and a motorized filter turret containing the GFP, Cy3, and Cy5 image-registered filter cubes (Semrock). Our illumination source was an X-Cite 120 illuminator (EXFO), and we captured 12-bit images with an ORCA-AG camera (Hamamatsu, Bridgewater, NJ) controlled by Metamorph Premier software. To preserve quantitative information, we chose exposure times in which the maximum pixel intensity did not exceed 90% of the dynamic range of the camera, and we recorded light source power output before and after each image, using a radiometer (EXFO). We collected all images from animals in which the nerve cord of interest was oriented up and close to the surface of the coverslip, and all images are maximum projections of a z-series taken through the entire cord, including most of the out-of-focus light, using a step size of 140 nm. We used AutoDeblur Gold CWF (Autoquant) to deconvolve image stacks using the Adaptive PSF blind method and 10 iterations. After deconvolving, we used AutoDeblur Gold CWF to double the XY size of the images to reduce pixel size, using the suggested ideal linear interpolation algorithm to maintain quantitative accuracy. We applied pixel shifts of ∼54 nm in the X and Y directions to the final overlayed images to reflect a small amount of chromatic aberration inherent in the system (in the center of the field of view), which we determined experimentally by triple-staining UNC-10 active zones with three different color secondary antibodies. We collected images for negative controls using exposure times identical to positive controls, and we recorded light source power output as above. We processed nondeconvolved images used for negative control staining similarly, but without applying deconvolution and with the following modifications. Metamorph automatically assigns the brightest pixel a value of 255 for display of 24-bit three-color files. Thus even background pixels with low absolute values get boosted up in the absence of a real signal. To make the displayed pixel intensities of the control images comparable to the experimental images, we used Metamorph to analyze the original 12-bit maximum projection image of the color used for the control staining and determined the mean value of 10 of the brightest areas along the image. We did this for both the positive and the negative control. We then determined the ratio of the mean negative control value and the mean N2 (wild-type) value and used this ratio to adjust color intensity of the negative control in the final 24-bit display image using Paintshop Pro for this adjustment.
RESULTS
The ce268 mutation confers phenotypes similar to mutants with a hyperactivated Gαs pathway:
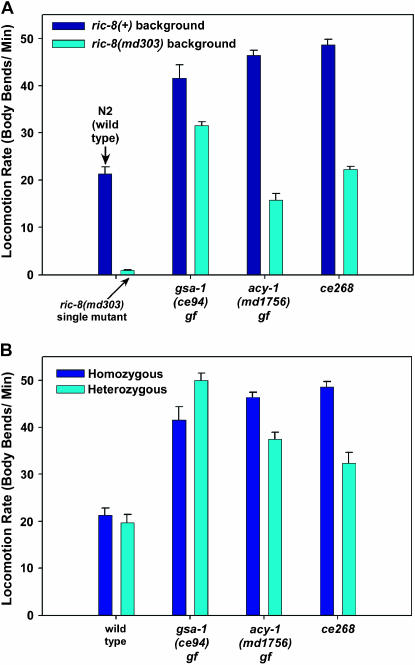
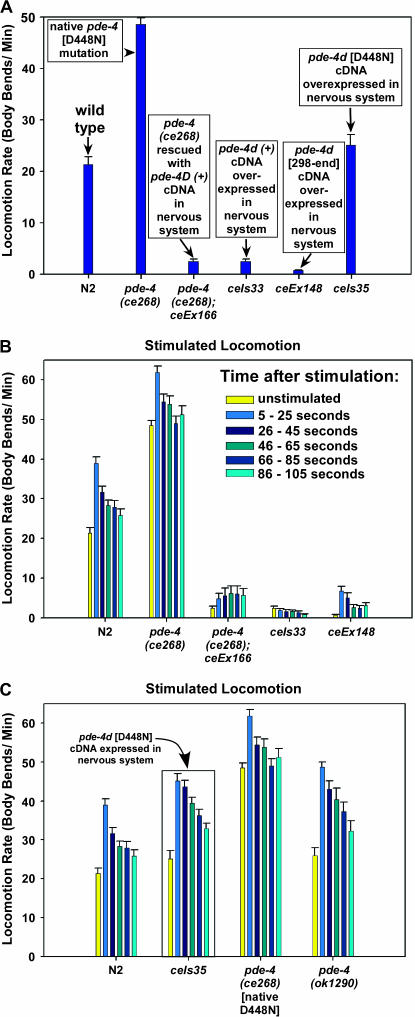
The ce268 mutant is one of the remaining uncharacterized mutants from the ric-8 suppressor screen that closely resembles mutants with a hyperactivated Gαs pathway and yet, as described below, it maps to a location distinct from the Gαs pathway components identified in the previous study (Schade et al. 2005). Similar to other mutations that hyperactivate the Gαs pathway, ce268 restores coordinated locomotion to ric-8(md303) and dramatically improves its locomotion rate (∼31-fold) to a level not significantly different from that of wild type (Figure 1A and supplemental Figure 1 movies at http://www.genetics.org/supplemental/). In comparison to the known Gαs-pathway-activating mutations, ce268 improves ric-8(md303)'s locomotion rate slightly more than the strongest dominant mutation in acy-1 (adenylyl cyclase) does, but less than the strongest dominant mutation in gsa-1 (Gαs) does (Figure 1A). As a single mutant, ce268 confers strongly hyperactive locomotion (∼2.3-fold greater than wild type), similar to the gsa-1 and acy-1 gain-of-function mutants (Figure 1A). Like the gsa-1 and acy-1 gain-of-function mutants, ce268/+ heterozygotes showed significantly hyperactive locomotion (∼50% greater than wild type). However, this dominant effect was not as strong as the acy-1 and gsa-1 gain-of-function mutations, which had locomotion rates that were 80 and 130% greater than that of wild type as heterozygotes (Figure 1B).
Figure 1.
The ce268 mutation confers phenotypes similar to mutants with a hyperactivated Gαs pathway. (A) The mean locomotion rates, expressed as body bends per minute, of strains carrying gsa-1(ce94) gf (Gαs gain of function), acy-1(md1756) gf (adenylyl cyclase gain of function), or ce268. Dark blue bars represent the mutants in a ric-8(+) (wild type for ric-8) background, while cyan bars represent double mutants carrying the indicated mutations in a ric-8(md303) (strong reduction-of-function) background. The first set of two bars shows wild-type animals and ric-8(md303) single mutants. Error bars represent the standard error of the mean for 10 animals. Values for strains carrying gsa-1(ce94) and acy-1(md1756) are reprinted with permission from Schade et al. (2005) for comparison. See also supplemental QuickTime movies for Figure 1 at http://www.genetics.org/supplemental/. (B) The ce268 mutation is semidominant. Shown are the mean locomotion rates, expressed as body bends per minute, of strains carrying gsa-1(ce94) gf (Gαs gain of function), acy-1(md1756) gf (adenylyl cyclase gain of function), or ce268. Dark blue and cyan bars represent animals homozygous or heterozygous, respectively, for the indicated mutations. All heterozygotes are also heterozygous for dpy-5(e61), which we used as a recessive marker mutation to identify heterozygotes. The first set of two bars shows wild type and dpy-5(e61)/+. Similar to the dominant mutations gsa-1(ce94) and acy-1(md1756), the ce268 mutation confers significantly hyperactive locomotion even as a heterozygote (P-value for ce268/+ compared to dpy-5(e61)/+ is 0.0009 using the unpaired t-test with Welch correction). Error bars represent the standard error of the mean for 10 animals. Values for strains carrying gsa-1(ce94) and acy-1(md1756) are reprinted with permission from Schade et al. (2005) for comparison.
The ce268 mutation maps to the pde-4 gene and disrupts the cAMP phosphodiesterase catalytic domain:
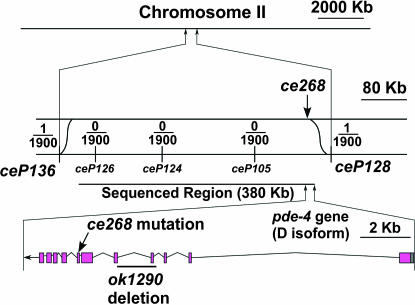
We mapped ce268 by its ric-8(md303) suppressor phenotype using the mapping strain ric-8(md303) [CB4856] that we had outcrossed 12× into a CB4856 background (Schade et al. 2005). CB4856 is a wild-type strain that contains thousands of SNPs relative to the N2 wild-type strain (Wicks et al. 2001; Swan et al. 2002). In a previous study, we described a method for high-resolution mapping of both dominant and recessive mutations entirely with respect to SNPs (Schade et al. 2005). Our initial attempt to map the ce268 mutation with a small number of mapping lines showed that it mapped to the recombination-poor center of chromosome II. The low probability of recombination in this region then forced us to scale up the mapping procedure considerably. Ultimately, we had to generate ∼1900 mapping lines to narrow the mutation to a somewhat tractable interval of 474 kb (Figure 2).
Figure 2.
Mapping the ce268 mutation relative to SNPs. Shown is a drawing of the region on chromosome II that contains the ce268 mutation. Low-, medium-, and high-resolution views are layered from top to bottom, accompanied by scale bars. (Top) The entire chromosome. (Middle) The region between the SNPs ceP136 and ceP128, which is the smallest region to which we were able to map the mutation by SNP analysis of 1900 lineages. The top strand represents the mutant chromosome, and the bottom strand represents the CB4856 chromosome containing the indicated SNP markers. This is the expected arrangement of the two chromosomes during the crossing-over stage of meiosis when recombination could occur. Sinusoidal lines represent recombination events that could place the mutation on the same chromosome as a SNP marker. Fractions represent the number of actual recombination events, inferred from SNP mapping data over the total number of homozygous mutant lines tested (1900). See materials and methods for the specific location of each SNP. The vertical arrow points to the actual location of the mutation, based on sequence analysis of ce268 genomic DNA. The 380-kb region we chose for sequence analysis is indicated. (Bottom) Just the region containing the pde-4 gene wherein lies the ce268 mutation, as indicated by a diagonal arrow. A line indicates the location of the 1554-bp ok1290 deletion isolated by the C. elegans Gene Knockout Consortium. The missing sequence begins at [AC]TGTTTACCTG and ends at AAGATGCTCA[AC]. Since the deletion begins and ends with the same two nucleotides, the exact breakpoint is ambiguous. All of exon 4 and the beginning of exon 5 (i.e., including the splice acceptor) are missing. ok1290 also duplicates a 203-bp segment of genomic DNA that immediately follows the deletion.
Proceeding on the assumption that the dominance of ce268 was due to a gain-of-function mutation similar to the other dominant Gαs-pathway-activating mutations, we analyzed this interval for candidate genes whose gain of function could cause a Gαs pathway activation phenotype, but we found none. Our mapping data were insufficient to make rational inferences about where within this large interval the mutation might lie, since we found only one recombinant relative to each SNP that bounded the interval and zero recombinants in three other SNPs distributed equally throughout the interval. Furthermore, we decided against trying to narrow the interval further by transformation rescue experiments with large genomic fragments due to uncertainty about our chances of rescuing the dominant phenotype with wild-type sequences. Therefore, to find the ce268 mutation, we chose an ∼400-kb region of highest likelihood and sequenced this region from ce268 genomic DNA (Figure 2). We found 23 homozygous differences between our ce268 genomic sequence and the curated N2 sequence stored in WormBase (Chen et al. 2005). Twenty-one of these differences amounted to changes unlikely to be caused by the EMS mutagen that we used to produce the ce268 mutant. These changes included single-base-pair insertions or deletions (18) and a smaller number of multiple-base-pair insertions or deletions (3). We found only two homozygous single-base-pair changes, both of which had the chemical signature of EMS (GC-to-AT changes). One of these changes fell in an intergenic region not close to the beginning or end of any predicted gene, and the other fell within the pde-4 coding region. The pde-4 gene lay at the extreme end of the 400-kb region that we sequenced and, since we sequenced from the center out in both directions, it was the last gene that we sequenced (Figure 2).
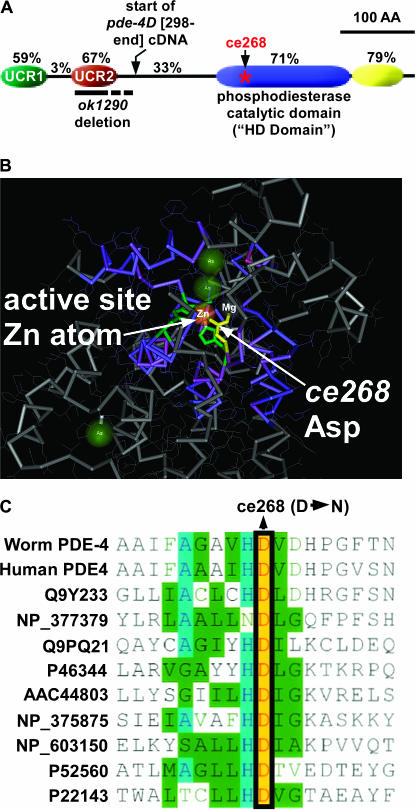
pde-4 encodes a cAMP-specific phosphodiesterase that is the closest C. elegans homolog of human cAMP phosphodiesterase 4D and the Drosophila Dunce protein. cAMP phosphodiesterases, which convert cAMP to 5′-AMP, normally function to lower cAMP levels generated by adenylyl cyclase activation in response to Gαs pathway activation (Sunahara et al. 1996) (Figure 3). Like its human and fly counterparts, the worm pde-4 gene locus generates a diverse array of pde-4 gene products. An analysis of pde-4 cDNA sequences in WormBase WS140 found evidence of at least five isoforms, designated pde-4a–e (Chen et al. 2005). These isoforms differ from each other exclusively at the N terminus and appear to be generated by alternative promoters and alternative transcription start sites. We compared the core region of the worm and human homologs (the region common to all worm isoforms) and found that they are similar in length (623 amino acids in human vs. 558 in worm) and share 52% overall identity. Within this shared region, the subregions with the highest conservation are the 183-amino-acid phosphodiesterase catalytic domain (71% identical) and an immediately adjacent 70-amino-acid domain of unknown function near the C terminus of the protein (79% identical) (Figure 4A). The shared region of PDE-4 also contains two other highly conserved subregions of 63 and 64 amino acids near its N terminus (Figure 4A). A previous study identified and named these upstream conserved regions as UCR1 and UCR2 after comparing fly and mammalian forms of the protein (Bolger et al. 1993).
Figure 4.
Conserved PDE-4 domains and mutation locations. (A) A drawing of C. elegans PDE-4 highlighting four conserved domains. Percentages indicate the percentage identity to human phosphodiesterase 4D (NP_006194). Locations of the ce268 mutation and the ok1290 deletion are indicated. The dashed line at the right half of the ok1290 deletion indicates uncertainty about which cryptic splice site(s) are used at the ok1290 mutant locus (see text). The drawing begins with Arg 117 of the PDE-4D isoform (this is the beginning of exon 2 of the D isoform, sequence context RRESFLYR) and does not include N-terminal sequences that vary between the five predicted products the pde-4 locus. A previous comparison of Drosophila and human PDE-4D identified and named the upstream conserved regions (UCR1 and UCR2) (Bolger et al. 1993). Domain boundaries relative to the PDE-4D isoform are as follows: UCR1 (R117–N179), UCR2 (A211–K274), catalytic domain (Y406–W588), and C-terminal conserved domain (R589–P658). The boundaries of the catalytic HD domain were identified using the National Center for Biotechnology Information Conserved Domain Search tool (Marchler-Bauer et al. 2005) during a BLASTP search using the PDE-4D isoform. The PDE-4D protein sequence used for this analysis is taken from WormBase, WS 149 (Chen et al. 2005). (B) The crystal structure of the catalytic domain of human PDE4 as determined by Hatley et al. (2000). The Asp that is mutated in pde-4(ce268) is yellow. Note that this residue participates in coordinating a zinc atom as well as a second metal ion that is probably a magnesium atom (Hatley et al. 2000). Other residues that coordinate the zinc atom are partially visible and green. This image was exported from Cn3D version 4.1 (National Center for Biotechnology Information). (C) An amino acid alignment of various metal-dependent phosphodiesterases in the region around the ce268 mutation. The alignment includes human phosphodiesterase 4, other cyclic nucleotide phosphodiesterases, and distantly related bacterial phosphodiesterases that are not involved in cyclic nucleotide metabolism. Accession numbers (left column) can be used for details on the source of each sequence.
The ce268 mutation is a D448N change relative to the PDE-4D isoform. It disrupts the catalytic domain by changing one of the four active site residues that together chelate an active-site zinc atom (Figure 4B). This Asp at position 448 is conserved in all metal-dependent phosphohydrolases from bacteria to humans (Figure 4C). ce268 is likely to strongly decrease the function of PDE-4, because mutating the same Asp to an Ala in bovine PDE5 (cGMP-specific phosphodiesterase) decreases the rate of catalysis more than eightfold (Turko et al. 1998). By immunostaining, using the anti-PDE-4 antibody described below, we found no difference in the levels of PDE-4 protein produced by wild type and the pde-4(ce268) mutant or in its localization pattern (N. K. Charlie and K. G. Miller, unpublished results), which is consistent with the mutation affecting the protein's function rather than its production or localization.
Other than ce268, we isolated no other alleles of pde-4 in either the hyperactive mutant or the ric-8(md303) suppressor screens described in Schade et al. (2005), and there are no reports of pde-4 alleles in previous C. elegans studies; however, the C. elegans Gene Knockout Consortium has produced the deletion allele pde-4(ok1290). ok1290 removes 1554 bp from an intron-rich region of the gene that includes 95 bp of coding sequence from two exons (see Figure 2 and legend for exact deletion boundaries). All predicted products of the pde-4 locus share these exons. Splicing around the deletion to go to the next unaffected exon would disrupt the reading frame well before the catalytic domain and produce a nonfunctional protein. We verified that this splicing pattern does in fact occur by comparing pde-4(+) and pde-4(ok1290) mRNA products in the region of the deletion using RT–PCR followed by gel and sequence analysis (N. K. Charlie and K. G. Miller, unpublished results). However, we also found that the pde-4(ok1290) uses at least three other cryptic splice sites between the end of the deletion and the first unaffected exon that are not used by the pde-4(+) locus (N. K. Charlie and K. G. Miller, unpublished results). This observation, combined with pde-4(ok1290) mutant phenotype data, presented below, suggests that pde-4(ok1290) can produce an mRNA that maintains reading frame downstream of the deletion. The amino acids that are missing in ok1290 are almost exclusively from the UCR2 subregion and are well upstream of the catalytic domain (Figure 4A shows the boundaries of the deletion).
Loss of function and overexpression of PDE-4 in neurons cause opposite locomotion phenotypes:
Our molecular analysis of the ce268 mutation suggests that it strongly reduces PDE-4 function. The ce268 mutant phenotypes support this conclusion. Reducing PDE-4 function should increase cAMP levels and, as we found, cause a phenotype similar to mutants with a hyperactivated Gαs pathway. Although dominant/semidominant patterns of inheritance, such as we observed for ce268, are sometimes associated with gain-of-function mutations, our molecular and phenotypic analyses are more consistent with dominance caused by a dominant negative effect and/or haplo-insufficiency. If, as our analysis of pde-4(ce268) suggests, PDE-4 normally functions to inhibit locomotion, then overexpression of PDE-4 might confer sluggish locomotion. To test this, we produced transgenic animals containing the pde-4d(+) cDNA driven by the rab-3 nervous-system-specific promoter. We chose the pde-4d isoform for this study only because it seemed to be a relatively abundant splice product on the basis of its representation in the Kohara mixed-stage cDNA library. We found that the neuronally overexpressed pde-4d(+) transgene not only rescued the hyperactive locomotion of the pde-4(ce268) mutant, but also further reduced its locomotion rate to 11% of the wild-type rate (Figure 5A). When we injected the rab-3∷pde-4d(+) cDNA into a wild-type background and integrated the resulting array, it caused a similarly sluggish locomotion rate (Figure 5A). We also produced transgenic strains carrying a truncated version of the cDNA [pde-4d (298–end)] that lacks the UCR1 and UCR2 domains, but includes the catalytic and C-terminal domains (Figure 4A shows where this cDNA starts). Neuronal overexpression of pde-4d (298–end) conferred a locomotion rate ∼3% of the wild-type rate (Figure 5A). These strongly sluggish phenotypes are caused by too much PDE-4 cAMP phosphodiesterase activity rather than indirect strong toxic effects of overproducing the PDE-4 protein, because neuronal overexpression of a pde-4d cDNA containing the inactivating ce268 [D448N] mutation resulted in wild-type or slightly hyperactive locomotion (17% higher locomotion rate than that of wild type, but not statistically different; P = 0.16) (Figure 5A). These results suggest that PDE-4 normally functions to inhibit the locomotion rate by lowering cAMP levels via its cAMP phosphodiesterase activity.
Figure 5.
Loss of function and overexpression of PDE-4 in neurons cause opposite locomotion phenotypes. (A) The mean locomotion rates, expressed as body bends per minute, of strains carrying various dosages and forms of pde-4. Note that transgenic neuron-specific overexpression of the pde-4d(+) cDNA in pde-4(ce268) or pde-4(+) backgrounds results in strong sluggish locomotion, as does a truncated version of the cDNA pde-4d (298–end) that lacks the N-terminal conserved domains UCR1 and UCR2, but includes the catalytic and C-terminal conserved domains (see Figure 4A for start site of the truncated cDNA). The locomotion rate of ceIs35 is not significantly different from wild type (P = 0.16 using the unpaired t-test with Welch correction). The transgenic pde-4 cDNAs are driven by the neuron-specific rab-3 promoter (Schade et al. 2005). Error bars represent the standard error of the mean for 10 animals. (B) The mean locomotion rates of the indicated strains at various time points before and after delivery of a strong standard stimulus (see materials and methods). Each group of bars represents the stimulated locomotion response of the indicated strain over a 100-sec period, divided into 5- × 20-sec intervals as indicated. The first bar in each group shows the unstimulated locomotion rate. ceIs33, the transgenic strain that overexpresses PDE-4 in the nervous system, decreases its locomotion rate after the strong stimulus. ceIs35, which expresses the D448N mutation in a neuronally driven pde-4d cDNA, shows hyperactive locomotion relative to stimulated wild type at each time point after the stimulus, but not prior to stimulation (P ≤ 0.018 for each time point after stimulation compared to the corresponding wild-type time points). The native ce268 mutant (which also has the D448N mutation) is more hyperactive than the ceIs35 transgenic strain before and after stimulation. The pde-4(ok1290) mutant is significantly hyperactive at all time points after stimulation compared to stimulated wild type (when compared to the wild-type time points, P < 0.0006 at all time points except the last, where P = 0.047), but pde-4(ok1290) is not quite significantly hyperactive in the unstimulated locomotion assay (P = 0.084 compared to wild type). Error bars represent the standard error of the mean for 30 animals at each time point and 10 animals for the unstimulated assay. Statistical significance tests used the unpaired t-test with Welch correction.
A transgenic D448N mutation has a dominant negative effect on stimulated locomotion rate:
The lack of a significantly hyperactive locomotion rate in the transgenic strain overexpressing the pde-4d [D448N] cDNA suggests that the ce268 [D448N] mutation is not a strong dominant negative mutation or that transgenic overproduction of PDE-4 may have slight toxic effects that are enough to counteract the hyperactivity. Our observations of the D448N transgenic strains on culture plates indicated that they were hypersensitive to stimuli. To further investigate the extent to which the ce268 [D448N] mutation exerts a dominant negative effect, we developed a stimulated locomotion assay for quantifying hypersensitivity to a standard stimulus. Our assay, which we describe in detail in materials and methods, consists of dropping a standard weight from a fixed height onto the lid of a culture plate containing the animal to be assayed. We then quantify the animal's locomotion rate for 20-sec intervals, starting 5 sec after application of the stimulus and ending five intervals later at 105 sec. Under these conditions, wild-type animals increase their locomotion rate ∼83% in the first 20-sec interval; however, by the third 20-sec interval they have returned to a level that is only ∼25% greater than the unstimulated state, and this does not change significantly during the last three intervals (Figure 5B). pde-4(ce268) mutants, which are strongly hyperactive even in the unstimulated state, increase their locomotion rate only 26% in the first interval after the stimulus and are not significantly different from the unstimulated rate by the fourth interval (Figure 5B). Interestingly, the strain carrying an integrated version of the neuronally driven pde-4d(+) cDNA decreased its locomotion rate at the first interval after stimulation, and the rate continued to decrease steadily throughout the 105-sec assay period, ending at only 31% of the unstimulated rate (see ceIs33 in Figure 5B). In contrast, the strain containing the D448N mutation in a neuronally overexpressed pde-4d cDNA showed significantly hyperactive locomotion relative to stimulated wild type at each time point after the stimulus (Figure 5B). Consistent with our observations that this strain seemed hypersensitive to stimuli on culture plates, the rate at which its locomotion rate slowed down after stimulation was less than the rate at which the wild-type rate slowed down. For example, its locomotion rate during the second 20-sec interval was not significantly different from the first interval, in contrast to wild type, which showed a strongly significant decrease between the first and second intervals (compare N2 and ceIs35 in Figure 5C). Although these data show that the transgenic D448N mutation can exert a significant dominant negative effect in response to stimulation, the native homozygous ce268 [D448N] mutation still has significantly stronger effects on locomotion rate, both unstimulated and stimulated. One possibility is that haplo-insufficiency may contribute to ce268's semidominant pattern of inheritance; however, in testing the locomotion rate of a strain carrying a heterozygous deficiency that includes pde-4 and other genes, we found no evidence of this using quantitative locomotion assays (data not shown). Alternatively, the native mutation solely may cause a dominant negative effect, whereas transgenic overproduction of the D448N mutant protein may cause a combination of dominant negative effects that cause hyperactivity and indirect toxic effects that reduce locomotion rate.
Deleting the UCR2 domain reduces, but does not eliminate, the function of PDE-4:
To investigate the importance of the UCR2 domain to the function of PDE-4, we quantified the extent to which the ok1290 deletion disrupts the ability of PDE-4 to control locomotion rate using both unstimulated and stimulated locomotion assays. We found that ok1290 is a relatively weak allele of pde-4. Its unstimulated locomotion rate is only 22% greater than that of unstimulated wild type, and this difference is not quite statistically significant; however, its stimulated locomotion rate was significantly hyperactive at every interval after stimulation when compared to stimulated wild type at the same intervals (Figure 5B). These results suggest that PDE-4 function is mildly or moderately reduced in the absence of the UCR2 domain.
Genetic analysis suggests the presence of a Gαs-independent pool of cAMP in neurons:
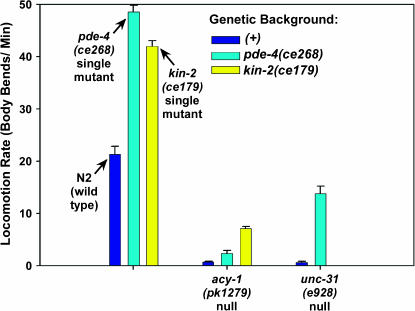
Since cAMP phosphodiesterases lower cAMP levels by converting cAMP to 5′-AMP, and since the pde-4(ce268) mutation specifically disrupts the catalytic site that is necessary for cAMP phosphodiesterase activity and causes phenotypes similar to mutants with a hyperactivated Gαs pathway, we infer that high cAMP levels cause the hyperactive locomotion phenotype of pde-4(ce268). What is the source of this cAMP? Or is there more than one source? One candidate for the cAMP source is ACY-1 (adenylyl cyclase). Moorman and Plasterk (2002) produced the acy-1 null mutation pk1279 and showed that it causes larval growth arrest and paralysis. A previous study used native mutations that hyperactivate the GSA-1 (Gαs) pathway to show that GSA-1 is completely dependent on ACY-1 to exert its effects on locomotion rate and larval growth (Schade et al. 2005). In other words, strongly hyperactivating the Gαs pathway has no detectable effect on the locomotion rate or larval arrest phenotype of animals lacking ACY-1. This shows that there are no other adenylyl cyclases that GSA-1 can activate to stimulate locomotion rate and larval growth; however, it does not rule out the possibility that there is a pool of cAMP in the cells controlling locomotion rate and larval growth that is produced independently of GSA-1–ACY-1 pathway activation. If there is such a pool, then strongly reducing the function of PDE-4 might partially rescue the paralysis and larval growth defects caused by lack of ACY-1. If there is no other source of cAMP, then reducing the function of PDE-4 should have no further effect in a mutant lacking ACY-1 (just as hyperactivating Gαs has no effect on an acy-1 null). To test this, we constructed a double mutant containing an acy-1 null mutation in combination with pde-4(ce268). We found that the pde-4(ce268) mutation improved the locomotion rate of the acy-1 null more than threefold to a level that is ∼10% that of wild type (Figure 6). pde-4(ce268) also partially rescued the larval growth arrest phenotype of the acy-1 null. Whereas 0/24 acy-1 null single mutants reached adulthood over a 25-day period (Schade et al. 2005), 8/42 (19%) of pde-4(ce268); acy-1(pk1279) double mutants became egg-laying adults within 5 days of plating 6- to 30-hr-old larvae, and we were able to establish a viable homozygous culture of this strain. These results, taken with our previous findings that strongly hyperactivating GSA-1 (Gαs) has no effect on the locomotion rate or larval growth of the acy-1 null, suggest that there is a cAMP pool produced independently of the Gαs pathway and regulated by PDE-4.
Figure 6.
Loss of PDE-4 partially rescues the paralysis of animals lacking a functional neuronal Gαs pathway. Shown are the mean locomotion rates, expressed as body bends per minute, of strains carrying acy-1(pk1279) (acy-1 null in muscle and nervous system) or unc-31(e928) (unc-31 null). Dark blue bars represent the mutants in a pde-4(+) (wild type for pde-4) background, cyan bars in the second and third sets represent double mutants carrying the indicated mutations in the pde-4(ce268) (strong reduction-of-function) background, and the yellow bar in the second set represents the acy-1(pk1279); kin-2(ce179) double mutant. Wild-type animals and single-mutant controls are shown in the first set of three bars. Error bars represent the standard error of the mean for 10 animals. Values for acy-1(pk1279) and kin-2(ce179) are reprinted with permission from Schade et al. (2005); values for unc-31(e928) single mutants are reprinted with permission from Charlie et al. (2006).
Given that there is a pool of cAMP produced independently of the Gαs pathway, it should be possible to partially bypass the acy-1 null state via a mutation that makes the cAMP effector KIN-2 (a regulatory subunit of protein kinase A) hypersensitive to low concentrations of cAMP. The kin-2(ce179) mutation changes a conserved Arg in the four-amino-acid inhibitory pseudosubstrate domain that normally functions to keep protein kinase A turned off in the absence of cAMP (Schade et al. 2005). Biochemical studies in vertebrates have shown that mutating this Arg produces a holoenzyme that is hypersensitive to, but still dependent on, cAMP for protein kinase A activation (Buechler et al. 1993). We found that the kin-2(ce179) mutation improves the locomotion rate of the acy-1 null more than ninefold to 7.1 ± 0.37 body bends/min, a level that is ∼33% that of wild type (Figure 6). Like pde-4(ce268), the kin-2 mutation also rescues the larval growth arrest of the acy-1 null. Specifically, 28/48 (58%) of acy-1(pk1279); kin-2(ce179) double mutants became egg-laying adults within 5 days of plating 6- to 30-hr-old larvae. Since the stronger Gαs pathway activating the mutation gsa-1(ce81) has no effect on the larval growth arrest or locomotion rate of the acy-1 null, the ability of kin-2(ce179) to significantly rescue the acy-1 null again suggests that there is a Gαs-independent cAMP pool.
ACY-1 functions in both the nervous system and the body-wall muscle (Reynolds et al. 2005). Lack of ACY-1 in either tissue causes sluggish locomotion, although animals lacking ACY-1 specifically in the nervous system are significantly more paralyzed (Reynolds et al. 2005). Data presented below suggest that PDE-4 functions specifically in the nervous system. Therefore, pde-4(ce268)'s effect on the locomotion rate of the acy-1 null is likely to be limited by having to overcome low cAMP levels in body-wall muscle that would inhibit locomotion. To specifically investigate the effect of PDE-4 on the neuronal pool of Gαs-independent cAMP, we transferred the pde-4(ce268) mutation into an unc-31 null background. In a recent study, we found that unc-31 null mutants specifically lack the ability to activate their neuronal Gαs pathway for controlling locomotion rate (Charlie et al. 2006). unc-31 is expressed only in the nervous system (not in body-wall muscle) and encodes the C. elegans ortholog of the dense-core vesicle priming protein CAPS. On the basis of this study and the findings of Charlie et al. (2006), the pde-4(ce268) mutation should improve the locomotion rate of the unc-31 null mutant by making the Gαs-independent pool of neuronal cAMP available to drive locomotion, and that is indeed the case. pde-4(ce268) improves the locomotion rate of the unc-31 null >20-fold from ∼3 to 64% of the wild-type rate (Figure 6). This strong effect suggests that the Gαs-independent pool of cAMP in neurons is capable of supporting substantial locomotion when PDE-4's function is reduced or downregulated. The strong rescue of the unc-31 null further suggests that pde-4(ce268)'s weaker effect on the locomotion rate of the similarly paralyzed acy-1 null is likely due to lack of PDE-4 expression in body-wall muscle (and thus an inability of the PDE-4 reduction-of-function mutation to increase the low cAMP levels in the body-wall muscle of acy-1 null mutants).
Loss of PDE-4 function does not significantly affect levamisole or aldicarb sensitivity:
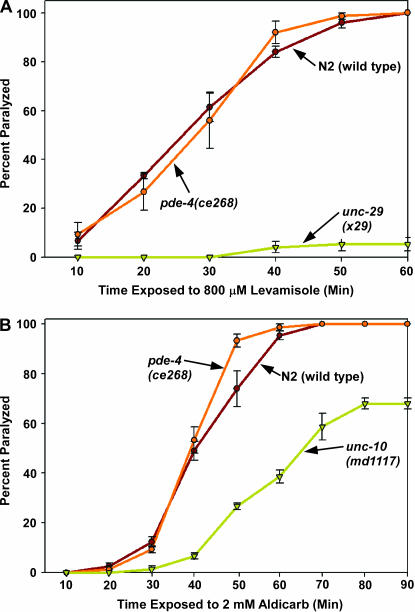
We next explored the possible causes of the hyperactive locomotion of pde-4(ce268) mutants. One possible cause is an increased response of the body-wall muscle to acetylcholine, the major neurotransmitter that drives the locomotion rate at the C. elegans neuromuscular junction. To test this, we compared the paralysis response of wild-type animals and pde-4(ce268) mutants to the ACh receptor agonist levamisole. When placed on plates containing levamisole, we found that wild-type animals and pde-4(ce268) mutants were indistinguishable from one another in their paralysis response over three trials, whereas an unc-29 null mutant that lacks functional levamisole-type ACh receptors remained unparalyzed during the assay (Figure 7A). This suggests that pde-4(ce268) mutants have a normal body-wall muscle ACh receptor response.
Figure 7.
Loss of PDE-4 function does not significantly affect levamisole or aldicarb sensitivity. (A) Loss of PDE-4 function does not affect the postsynaptic body-wall muscle response to the acetylcholine receptor agonist levamisole. The percentage of animals that are paralyzed over a 60-min time course of exposure to 800 μm levamisole on solid media plates is shown. unc-29(x29), included as a negative control, lacks functional levamisole ACh receptors (Gally et al. 2004). Error bars represent the standard errors of the mean for populations of 20 animals at each time point. (B) Loss of PDE-4 function does not significantly affect sensitivity to the acetylcholinesterase inhibitor aldicarb. The percentage of animals that are paralyzed over a 90-min time course of exposure to 2 mm aldicarb on solid media plates is shown. pde-4(ce268) appears slightly hypersensitive to aldicarb only at the 50-min time point, but this is not statistically significant (P = 0.13 using the unpaired t-test with Welch correction). The unc-10 null mutant md1117 is shown as a positive control for aldicarb resistance (Koushika et al. 2001). Error bars represent the standard errors of the mean for populations of 20 animals at each time point.
We next asked if increased acetycholine release contributes significantly to pde-4(ce268)'s hyperactive locomotion by quantifying its sensitivity to the acetylcholinesterase inhibitor aldicarb. Since the secreted acetylcholine that accumulates in the presence of aldicarb is toxic, mutations that decrease or increase the steady-state rate of acetylcholine release confer resistance or hypersensitivity to aldicarb, respectively (Rand and Nonet 1997). We found that wild-type animals and pde-4(ce268) mutants were largely indistinguishable from one another in their paralysis response to aldicarb over three trials, whereas an unc-10 (RIM) null mutant, previously shown to have decreased neurotransmitter release (Koushika et al. 2001), showed strong resistance to paralysis during the assay (Figure 7B). This suggests that pde-4(ce268) mutants have normal or near-normal levels of steady-state ACh release despite their hyperactive locomotion; however, the resolution of this whole-animal aldicarb assay may be insufficient to detect increased ACh release if it were occurring only at a subset of cholinergic synapses.
In a previous study we found that native mutations that hyperactivate the Gαs pathway in both body-wall muscle and neurons confer significant levamisole resistance and aldicarb hypersensitivity, but that transgenically hyperactivating the Gαs pathway solely in the nervous system or solely in body-wall muscle resulted in normal sensitivity to both drugs (Schade et al. 2005). In light of these previous findings, and combined with our experiments showing rescue of the pde-4(ce268) mutant phenotypes with a neuron-specific promoter, the normal levamisole and aldicarb sensitivity of pde-4(ce268) suggests that PDE-4 functions primarily or solely in neurons, and not in both body-wall muscle and neurons.
PDE-4 localizes to cholinergic and noncholinergic synapses in the ventral nerve cord:
To further investigate where PDE-4 acts, we produced an affinity-purified antibody to a 128-amino-acid region of PDE-4 that is shared among all known products of the pde-4 locus and used it to immunostain whole adult animals. At the whole-animal level, we observed concentrated staining throughout the nervous system, primarily localized to the ganglia and nerve cords, including the nerve ring, ventral, dorsal, and sublateral nerve cords (data not shown for nerve ring and dorsal nerve cord; ventral and sublateral cord images are described and shown below). To test the specificity of our antibody, we produced a second affinity-purified antibody against a 126-amino-acid region of PDE-4 that is completely nonoverlapping with the region used for the first antibody, but is again shared by all known products of the pde-4 locus. Both antigen regions are poorly conserved among the different phosphodiesterase proteins in C. elegans. The second antibody gave an identical staining pattern at both the whole-animal level (data not shown) and in high-resolution images of individual nerve cords. Supplemental Figure S1A at http://www.genetics.org/supplemental/ compares the staining pattern of the two antibodies along a 36-μm length of the ventral nerve cord. Both antibodies produced the unique pattern of staining described below. The staining pattern completely disappears after preadsorption of the antibody against gel-purified and nitrocellulose-immobilized recombinant PDE-4 (supplemental Figure S1B at http://www.genetics.org/supplemental/; materials and methods).
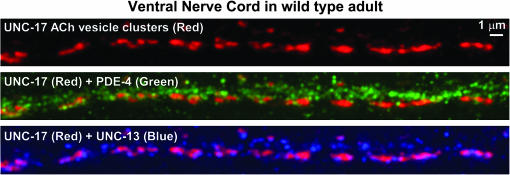
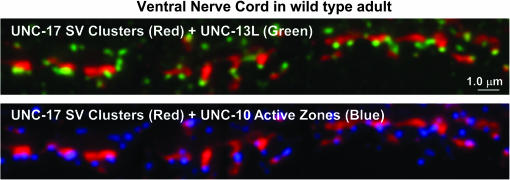
Within the ventral nerve cord, which contains a high density of the cholinergic synapses that drive the locomotion behavior as well as other noncholinergic synapses, we observed PDE-4 staining in small, but variable, size spots that appeared to be distributed throughout the thickness of the cord (Figure 8). The size of these spots was close to the theoretical limit of resolution for light microscopy (∼180 nm). Some of these spots appeared to lie within and/or just outside of the boundaries of cholinergic synaptic vesicle clusters, as visualized by costaining with an antibody to the synaptic vesicle ACh transporter UNC-17 (Figure 8). To investigate the location of the PDE-4 spots relative to cholinergic and noncholinergic regions of the ventral cord, we included a third antibody in the staining reaction that we produced using the N-terminal “L” region of the synaptic vesicle priming protein UNC-13. Below we provide evidence that our UNC-13L antibody specifically recognizes UNC-13L. We observed noncholinergic regions of the ventral nerve cord (regions devoid of UNC-17 staining) that contained abundant UNC-13L spots together with PDE-4 spots. This triple-staining experiment therefore suggests that PDE-4 does not exclusively associate with cholinergic synapses, but is also found at or near noncholinergic synapses (Figure 8).
Figure 8.
PDE-4 localizes to cholinergic and noncholinergic synapses in the ventral nerve cord. Maximum projection of a z-series through the cholinergic region of the ventral nerve cord. Shown are different color combinations from the same triple-stained image. (Top) UNC-17 cholinergic vesicle clusters (red blobs). (Middle) Cholinergic vesicle clusters (red blobs) + PDE-4 immunostaining (green spots). (Bottom) Cholinergic vesicle clusters (red blobs) + UNC-13L active zones (blue spots). Green and blue spots not lining up with cholinergic synapses likely represent noncholinergic synapses in the ventral cord (e.g., GABAergic or glutamatergic). Note the widespread occurrence of PDE-4 immunostaining in both cholinergic and noncholinergic regions of the nerve cord and the much larger number of PDE-4 spots when compared to UNC-13L active zones. We used the KM25B-3.1 PDE-4 antibody for this experiment (produced against the PDE-4D [151–278] protein region).
UNC-13L localizes to the active zone subregion of synapses:
In both worms and vertebrates, UNC-13 and its vertebrate ortholog are found in two major sizes: a long form and a short form, plus splice variants of each (Kohn et al. 2000; Betz et al. 2001). In worms, a GFP-tagged version of the UNC-13 short form (UNC-13S) is, for the most part, diffusely distributed in the nerve cords of wild-type animals, but is also localized to puncta that correspond to a subset of synapses (Nurrish et al. 1999). The UNC-13 long form (UNC-13L) is almost exclusively localized to synaptic puncta by immunostaining (Kohn et al. 2000); however, no previous study in any organism has analyzed the localization of UNC-13L or its vertebrate ortholog relative to a known active zone protein. By costaining with our antibody produced against the L region of UNC-13 and an antibody that recognizes the active zone protein UNC-10 (RIM) (Koushika et al. 2001), we found that UNC-13L localizes to the active zone subregion of synapses (Figure 9). This staining pattern is completely specific for UNC-13L, because it is absent in the mutant unc-13(md1072) (supplemental Figure S2 at http://www.genetics.org/supplemental/), which lacks the L region used to produce the antibody (Kohn et al. 2000).
Figure 9.
The UNC-13L protein colocalizes with the synaptic active zone protein UNC-10 (RIM). Maximum projection of a z-series through the cholinergic region of the ventral nerve cord. Shown are two color combinations from the same triple-stained image. (Top) UNC-17 cholinergic vesicle clusters (red blobs) + UNC-13L active zones (green spots). (Bottom) The same image in the red (UNC-17) and blue (UNC-10) channels. Note the strong correspondence in the positions of the UNC-13 and UNC-10 spots relative to each cholinergic synapse. Green and blue spots not lining up with cholinergic synapses likely represent noncholinergic synapses in the ventral cord (e.g., GABAergic or glutamatergic).
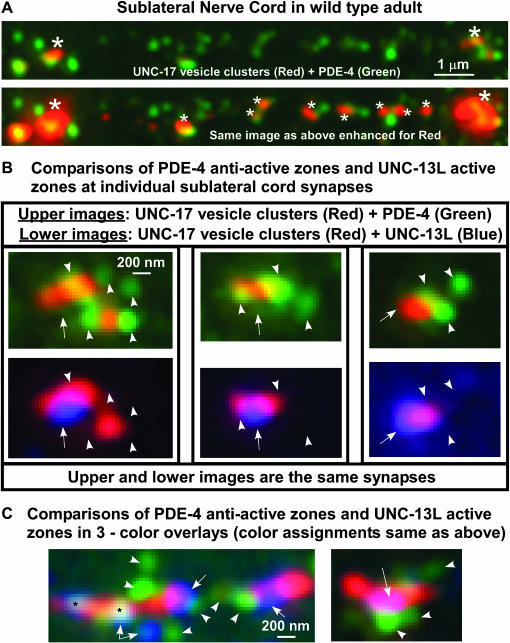
PDE-4 associates with the outside boundaries of synaptic vesicle clusters in regions that do not overlap with synaptic active zones:
To further investigate the subcellular localization of PDE-4 and its relationship to active zones, we focused our imaging efforts on the sublateral nerve cords. These nerve cords are simpler than the ventral and dorsal nerve cords for synaptic image analysis, because they contain only cholinergic neuromuscular synapses, and the synapses are at a much lower density than those in the ventral and dorsal nerve cords. Our images of the sublateral nerve cord revealed that PDE-4 is often localized to spots at or near the outer edges of synaptic vesicle clusters. We also observed numerous PDE-4 spots in apparent intersynaptic regions between major synaptic vesicle clusters; however, when we further enhanced the intensity of the channel that represents UNC-17-positive synaptic vesicles, we found that many, but not all, of the “intersynaptic” PDE-4 spots were associated with small clusters of synaptic vesicles that are difficult to see when the display is optimized for the major synapses (Figure 10A). To determine the precise relationship of PDE-4 to synaptic active zones, we analyzed single synapses triple stained for UNC-17 vesicle clusters, UNC-13L active zones, and PDE-4. We found that the PDE-4 spots were rarely, if ever, concentrated in regions overlapping with synaptic active zones. Instead, the PDE-4 spots were either adjacent to the active zone region or on the opposite side of the synapse (Figure 10, B and C). This highly localized PDE-4 pattern suggests that PDE-4, instead of acting from a uniformly distributed cytosolic pool, may exert its effects by reducing cAMP at specific locations within and around nonactive zone regions of synapses.
Figure 10.
PDE-4 associates with the outside boundaries of synaptic vesicle clusters in regions away from active zones. (A) Maximum projection of a z-series through the purely cholinergic sublateral nerve cord of a wild-type adult. UNC-17 cholinergic vesicle clusters (red) and PDE-4 (green). (Top) A region containing two major synapses (large asterisks; synaptic density is low in the sublateral cords). Note that PDE-4 tends to concentrate, sometimes asymmetrically, near the edges of the synaptic vesicle clusters as well as in intersynaptic intervals. (Bottom) The same image, enhanced in the red channel, to show that much of the “intersynaptic” PDE-4 is associated with the edges of small ACh vesicle clusters (the small clusters are indicated with small asterisks). (B) The PDE-4 associated with synaptic vesicle clusters tends to concentrate in areas away from the UNC-13L active zones. Shown are three individual sublateral cord synapses triple-stained for UNC-17 (red), PDE-4 (green), and UNC-13L (blue). The top and bottom image in each pair is the same synapse showing different color channels for comparison. Note that the blue active zone regions tend to occur in regions largely devoid of PDE-4 staining. Also note that the majority of PDE-4 staining appears to be just outside each cluster of synaptic vesicles, although there may be some overlap. Regions of PDE-4 staining are indicated with arrowheads in the top image of each pair, while UNC-13 active zones are indicated with arrows in the bottom image of each pair. The arrows and arrowheads are duplicated at the same sites on the paired images to facilitate comparing the locations of PDE-4 and UNC-13 active zones. (C) Triple-color images showing the relationship of UNC-17 (red), PDE-4 (green), and UNC-13L (blue) at sublateral cholinergic synapses. Note the tight association of PDE-4 with the outside boundaries of the vesicle clusters in regions away from the active zone region. Arrowheads indicate regions of PDE-4 staining, while arrows indicate UNC-13 active zones. The two areas marked by black asterisks indicate regions of white where all three markers (UNC-17, PDE-4, and UNC-13L) are apparently closely juxtaposed; however, since the image was produced by compressing a z-series, there is no way to determine the relative positions of each marker on the z-axis in these images. (Right) The UNC-13L active zone shows up as purple due to z-axis compression of the red UNC-17 marker onto the blue UNC-13L marker. In A–C we used the KM25B-3.1 PDE-4 antibody (produced against the PDE-4D [151–278] protein region).
DISCUSSION
Strongly reducing the function of the Dunce cAMP phosphodiesterase PDE-4 confers phenotypes similar to that of mutants with a hyperactivated Gαs pathway:
Here we report the first isolation of a mutant in the C. elegans pde-4 (Dunce cAMP phosphodiesterase) gene. Our genetic and molecular analyses of the ce268 mutation demonstrate that it strongly reduces the function of PDE-4. However, the frequency with which we isolated pde-4 alleles suggests that ce268 is not a null mutation. We estimate that we have screened ∼100,000 EMS-mutagenized genomes in genetic screens where a ce268-like mutant phenotype would have been a clear target and goal of the screen (Miller et al. 1999; Schade et al. 2005). Since the frequency with which EMS eliminates the function of an average-sized C. elegans gene is ∼1/2500 mutagenized genomes (Greenwald and Horvitz 1980), our screens should have identified ∼40 pde-4 loss-of-function mutations if the null phenotype were similar to pde-4(ce268). The fact that we found only one allele suggests that total elimination of PDE-4's function results in a phenotype that would mask its role in regulating locomotion rate. For example, the Drosophila PDE-4 ortholog dunce is required for oogenesis and embryogenesis (Bellen et al. 1987). However, since our pde-4(ce268) mutant has wild-type fertility and embryogenesis (K. G. Miller, unpublished results), it seems likely that the pde-4's null phenotype will be confined to the disruption of neurons. Since overproduction of cAMP can kill neurons and cause paralysis in C. elegans (Korswagen et al. 1997; Berger et al. 1998), pde-4's null phenotype may be paralysis or lethality caused by dead neurons.
The fact that we found only one allele also suggests that we had to hit the pde-4 gene just right to confer a strong hyperactive phenotype without eliminating its function. Our molecular analysis of ce268 demonstrates the surgical precision of the mutation, which affects an active-site residue conserved in all metal-dependent phosphohydrolases from bacteria to humans. Mutating the Asp that is mutated in ce268 to an Ala in bovine PDE5 (cGMP-specific phosphodiesterase) decreases the rate of catalysis >8-fold (Turko et al. 1998). This is a relatively small effect compared with mutating the adjacent conserved His to an Ala, which decreases the catalysis rate 267-fold (Turko et al. 1998). However, ce268 changes Asp to a polar Asn residue, not Asp to Ala, and it is unclear how different substitutions at this site could perturb the configuration of the active site and the function of the adjacent critical His.
Our phenotypic comparison of pde-4(ce268) to other Gαs pathway mutants suggests that it promotes pathway activation similarly to some of the strongest Gαs-pathway-activating mutations on the basis of its ability to rescue the paralysis of ric-8(md303). By this criterion, ce268 is stronger than our acy-1 gain-of-function mutants, and comparable to, but clearly not as strong as, the strongest gsa-1 gain-of-function mutants. The strong mutant phenotypes of pde-4(ce268) suggest that there are no other phosphodiesterases in C. elegans that can substitute for the specific function of PDE-4. As shown in Table 1, PDE-4 is the most highly conserved of the six known phosphodiesterases in C. elegans. There have been no reports of mutant phenotypes or expression patterns for the other five forms.
TABLE 1.
C. elegans cyclic nucleotide phosphodiesterases
| C. elegans protein name | Closest human homolog | BLASTP smallest sum probablility relative to PDE-4 | BLASTP smallest sum probability relative to closest human homolog and (% identity) |
|---|---|---|---|
| PDE-4 (ortholog of fly Dunce) | cAMP-specific 3′,5′-cyclic phosphodiesterase 4D | — | 7.1e-169 (47) |
| PDE-1 | Calcium/calmodulin-dependent 3′,5′-cyclic nucleotide phosphodiesterases1C (Cam-PDE 1C) | 8.9e-42 | 4.4e-136 (40) |
| PDE-6 | Splice isoform 1 of high-affinity cAMP-specific and IBMX-insensitive 3′,5′-cyclic phosphodiesterase 8A | 1.2e-36 | 5.5e-88 (26) |
| PDE-2 | Splice isoform PDE2A3 of cGMP-dependent 3′,5′-cyclic phosphodiesterase | 1.9e-21 | 8.8e-94 (24) |
| PDE-5 | cAMP and cAMP-inhibited cGMP 3′,5′-cyclic phosphodiesterase 10A | 2.0e-14 | 7.7e-98 (48) |
| PDE-3 | cGMP-inhibited 3′,5′-cyclic phosphodiesterase | 0.00082 | 3.2e-89 (34) |
Column 1 lists all C. elegans proteins producing “high-scoring segment pairs” in a TBLASTN 2.0MP search of the C. elegans genome using PDE-4D as the query. The proteins are listed in order of their similarity to PDE-4 on the basis of BLASTP smallest sum probabilities. Closest human homolog descriptions and the corresponding smallest sum probabilities are taken from WormBase Release WS148 (http://www.wormbase.org). Percentage identities were calculated using Vector NTI AlignX and do not include nonaligning N- or C- terminal ends of each protein pair. Swissprot accession numbers for the C. elegans/human proteins are: PDE-1 (O18696/ Q14123); PDE-2 (P30645-1/ O00408); PDE-3 (Q810P7-2/ Q13370); PDE-4 (Q22000-1/ Q08499); PDE-5 (P91119/ Q9Y233); PDE-6 (Q810P7-2/ O60658).
By comparison to ce268, the pde-4(ok1290) deletion, isolated by targeted reverse genetics, is a relatively weak allele. Its lesion is largely confined to the UCR2 domain and a short section of poorly conserved sequence downstream of UCR2. By immunostaining we found no difference in the levels of protein produced by wild type and the pde-4(ok1290) mutant or in its localization (N. K. Charlie and K. G. Miller, unpublished results). A previous study suggested that this domain may act as an inhibitory domain, because deletion of part of this domain from a vertebrate PDE4 resulted in a sixfold increase in PDE activity after expression in bacteria and purification of the recombinant enzyme (Kovala et al. 1997). However, two other studies using yeast and COS-cell-produced recombinant enzymes found no effect of removing this domain on catalytic activity (Jacobitz et al. 1996; Owens et al. 1997), suggesting that the inhibitory effect may be specific to the way in which the recombinant protein is produced in bacteria. Our study did not find evidence for an inhibitory function for this domain, because deleting it in the ok1290 mutant reduces PDE-4 function, albeit mildly.
A Gαs-independent pool of cAMP in neurons:
Reducing PDE-4 function should result in abnormally high levels of cAMP in neurons. We used genetic pathway analysis to investigate the source(s) of the cAMP that cause hyperactive locomotion in mutants with reduced PDE-4 function. Our analysis suggests the presence of a Gαs-independent pool of cAMP in neurons, because pde-4(ce268) significantly rescued the paralysis and larval growth arrest of an acy-1 null mutant, and our previous study showed that GSA-1 (Gαs) exerts all of its effects on locomotion and larval growth through ACY-1 (Schade et al. 2005). The fact that a strong gsa-1 gain-of-function mutation has no effect on the paralysis or the larval growth of the acy-1 null, but the weaker pde-4(ce268) mutation significantly rescues both, suggests that neurons have a pool of cAMP that is produced independently of Gαs and that is normally regulated by PDE-4. We also found evidence of the Gαs-independent pool of cAMP using the kin-2(ce179) mutant, which contains a mutation that makes protein kinase A hypersensitive to cAMP. This mutation also conferred substantial rescue of the larval growth arrest and paralysis of an acy-1 null.
To specifically investigate the neuronal pool of Gαs-independent cAMP, we analyzed the effect of reducing PDE-4 function on an unc-31 null mutant, since a previous study suggested that it lacks the ability to activate its neuronal Gαs pathway (Charlie et al. 2006). The strong rescue of the unc-31 null's paralysis by pde-4(ce268) suggests that the Gαs-independent pool of cAMP in neurons is capable of supporting substantial locomotion when PDE-4's function is reduced or downregulated. This result may also shed light on ce268's strong rescue of ric-8(md303)'s paralysis. RIC-8 appears to be required to maintain both the EGL-30 (Gαq) and the GSA-1 (Gαs) pathways in a functional state (Reynolds et al. 2005). ric-8(md303) reduction-of-function mutants are likely to have a decreased ability to activate their Gαs pathway similarly to unc-31 null mutants. Reducing the function of PDE-4 would help overcome this defect by potentiating the Gαs-independent cAMP as well as whatever amount of Gαs-dependent cAMP that the ric-8(md303) mutant manages to produce.
What is the source of the Gαs-independent pool of cAMP in C. elegans? Interestingly, biochemical studies in vertebrates have found that Ca2+ and Ca2+–CaM can stimulate rodent adenylyl cyclase isoforms I, III, and VIII in the absence of Gαs activation, although these isoforms can also be activated by Gαs or synergistically by both activators (Sunahara et al. 1996). Type VIII adenylyl cyclase is the ortholog of the rutabaga protein that is required for learning and memory in Drosophila. The C. elegans homolog of this protein is ACY-3, although it is the least conserved of the four worm adenylyl cyclase proteins (Table 2). Of the two remaining worm adenylyl cyclase genes, the restricted expression pattern of ACY-2 appears to rule out a strong role for it in the ventral cord motor neurons that control locomotion rate (Korswagen et al. 1998). ACY-4 is almost as highly conserved as ACY-1 (Table 2), but nothing is known about its expression pattern, and no mutations have been reported. Future studies will require null mutants of acy-3 and acy-4 to investigate possible roles for ACY-3 and/or ACY-4 in synthesizing the Gαs-independent pool of cAMP. Double or triple mutants with acy-1 nulls could then reveal the cAMP null phenotype for the locomotion behavior, which, in light of the current findings, is unknown.
TABLE 2.
C. elegans adenylyl cyclases
| C. elegans protein name | Closest human homolog | BLASTP smallest sum probablility relative to ACY-1 | BLASTP smallest sum probability relative to closest human homolog and (% identity) |
|---|---|---|---|
| ACY-1 | Adenylyl cyclase, type IX | — | 3.3e-166 (32) |
| ACY-4 | Adenylyl cyclase, type VI Ca(2+)-inhibitable adenylyl cyclase | 6.6e-60 | 3.0e-163 (32) |
| ACY-2 | Adenylyl cyclase, type II (=mouse type IV) | 6.0e-37 | 8.2e-131 (28) |
| ACY-3 (ortholog of fly Rutabaga) | Adenylyl cyclase, type VIII Ca2+/calmodulin activated adenylyl cyclase | 0.058 | 4.4e-44 (9) |
Column 1 lists all C. elegans proteins producing “high-scoring segment pairs” in a TBLASTN 2.0MP search of the C. elegans genome using ACY-1 as the query. Closely related guanylyl cyclases, of which there are many in C. elegans, are not included. The proteins are listed in order of their similarity to ACY-1 on the basis of BLASTP smallest sum probabilities. Closest human homolog descriptions and the corresponding smallest sum probabilities are taken from WormBase Release WS148 (http://www.wormbase.org). Percentage identities were calculated using Vector NTI AlignX and do not include nonaligning N- or C- terminal ends of each protein pair. Accession numbers for the C. elegans/human proteins are: ACY-1 (NP_497970/ O60503); ACY-2 (NP_504553/ Q08462); ACY-3 (NP_497765/ P40145); ACY-4 (NP_504486/ O43306).
How do PDE-4 reduction-of-function mutations affect neurotransmitter release?
We found a striking discrepancy between the strong behavioral phenotypes of pde-4(ce268) and its near wild-type sensitivity to levamisole and aldicarb. This suggests that pde-4(ce268) mutants have near wild-type levels of steady-state ACh release. In a previous study, we found that native mutations that hyperactivate GSA-1 (Gαs) or ACY-1 cause significant hypersensitivity to aldicarb, which indicates increased neurotransmitter release (Schade et al. 2005). However, GSA-1 and ACY-1 are expressed in both the nervous system and the body-wall muscle (Korswagen et al. 1997, 1998), and PDE-4 appears confined to the nervous system (this study). Schade et al. (2005) demonstrated that transgenic expression of an acy-1 gain-of-function mutation only in the nervous system or only in body-wall muscle does not affect aldicarb sensitivity, despite causing hyperactive locomotion when expressed from either tissue and rescuing the paralysis of ric-8(md303) when expressed from the nervous system. Electrophysiological studies of dunce mutants in Drosophila suggest that our live animal aldicarb assays accurately reflect the state of neurotransmitter release in populations of pde-4 mutant synapses. Synaptic recordings from dunce mutant embryos found no difference from wild type in either the resting spontaneous synaptic current frequency or the nerve-evoked synaptic currents (Suzuki et al. 2002), although a previous study reported a ∼50% reduction in the frequency of spontaneous release events when recording from individual larval synapses (Renger et al. 2000). Reducing the function of dunce does cause profound defects in synaptic physiology, however, such as a substantial variation in the decay times of spontaneous and evoked currents and a severe reduction in synaptic facilitation (the ability of synapses to increase evoked neurotransmitter release after tetanic stimulation) (Zhong and Wu 1991; Kuromi and Kidokoro 2000; Renger et al. 2000). Given the reduced facilitation, it is surprising but very interesting that dunce mutations increase mobilization of synaptic vesicles from the reserve pool to the readily releasable pool during tetanic stimulation (Kuromi and Kidokoro 2000; Kidokoro et al. 2004). Furthermore, dunce mutant embryos differ substantially from wild type in having increased hypertonicity-induced neurotransmitter release (Suzuki et al. 2002). Remarkably, hypertonicity-induced release occurs independently of Ca2+ influx or release of Ca2+ from internal stores (Tanabe and Kijima 1988; Rosenmund and Stevens 1996). It seems to be related to stretch-induced neurotransmitter release, which also occurs independently of Ca2+ (Chen and Grinnell 1997; Kashani et al. 2001). Hypertonicity-induced release is conserved in C. elegans (Richmond et al. 1999) and is frequently used to define the size of the readily releasable primed pool of synaptic vesicles (Rosenmund and Stevens 1996; Aravamudan et al. 1999; Richmond et al. 1999). Because cAMP appears to play an important role in this Ca2+-independent release mechanism, an understanding of how pde-4 mutations cause hyperactive locomotion will likely require a more detailed knowledge of the signal transduction molecules that control Ca2+-independent release phenomena such as hypertonic and stretch-induced release.
Focal localization of PDE-4 suggests intrasynaptic spatial regulation of cAMP:
Using triple-label immmunostaining combined with wide-field deconvolution microscopy, we showed that PDE-4 is concentrated in the C. elegans nervous system and is associated with both cholinergic and noncholinergic synapses. Although we cannot rule out the presence of PDE-4 in muscle cells below the detection level of our method, the neuronal-specific rescue of the pde-4(ce268) mutant and the absence of levamisole and aldicarb sensitivity phenotypes (which, as discussed above, seem to require combined muscle and nervous system activation of the Gαs pathway) argue against a requirement for PDE-4 in muscle cells during the locomotion behavior. Consistent with this, a previous Drosophila study found that reduction or overexpression of dunce strongly affected cAMP levels in nervous system tissue, but had no effect on muscle cAMP levels (Cheung et al. 1999).
Our study is the first to examine the high-resolution subsynaptic localization of PDE-4 or any of its orthologs in other organisms. We found that PDE-4 associates with the outside boundaries of synaptic vesicle clusters in regions that do not overlap with synaptic active zones. The size of the PDE-4 spots (or aggregates) is at or below the level resolvable by visible light (i.e., ≤ ∼180 nm), and there are generally one to several per synapse. PDE-4's highly focal subsynaptic localization suggests that it may exert its negative regulatory effects on locomotion rate by spatially regulating intrasynaptic cAMP pools or gradients. Several important questions remain. Is PDE-4 targeted to these specific locations? Does it self-associate/aggregate, as suggested by biochemical studies (Kovala et al. 1997)? What keeps it away from active zones? What controls the enzymatic activity of PDE-4 at these sites, or is it always active? Does PDE-4 activity from these highly focal sites produce intrasynaptic cAMP gradients?
Conclusion:
Although our study provides some potentially important new insights into the Gαs pathway of the synaptic signaling network, it also further highlights the puzzling nature of this pathway. Bridging the gap between synaptic function and behavior will require a clear understanding of the function of the neuronal Gαs pathway, which is currently lacking. Our apparent discovery of a Gαs-independent pool of cAMP in neurons points to the need for future studies to determine the phenotype of animals that completely lack cAMP at synapses. Because cAMP appears to play an important role in Ca2+-independent release (Suzuki et al. 2002), an understanding of why pde-4 and other Gαs-pathway-activating mutations cause hyperactive locomotion, and why mutants lacking the neuronal Gαs pathway are paralyzed, will likely require identification of the signal transduction molecules that control Ca2+-independent release phenomena. Whether these signaling molecules overlap with the hypothesized signaling proteins that control activation of the UNC-31–neuronal Gαs pathway (Charlie et al. 2006) remains an open question.
Acknowledgments
The authors thank Celine Moorman and Ronald Plasterk for the acy-1(pk1279) mutant, Leon Avery for providing the unc-31(e928) mutant, and the C. elegans Gene Knockout group at the Oklahoma Medical Research Foundation for providing the pde-4(ok1290) mutant. We also thank Janet Duerr and Jim Rand for their gift of the UNC-17 antibody and Mike Nonet for his gift of the UNC-10 antibody. We obtained all DNA sequences from the Oklahoma Medical Research Foundation's Core DNA Sequencing Facility. Some of the strains used here were provided by the C. elegans Genetics Center. Grants to K.G.M. from the National Institute of Mental Health (MH62400) and the Oklahoma Center for the Advancement of Science and Technology (HR02-093) supported this work.
References
- Aravamudan, B., T. Fergestad, W. S. Davis, C. K. Rodesch and K. Broadie, 1999. Drosophila Unc-13 is essential for synaptic transmission. Nat. Neurosci. 2(11): 965–971. [DOI] [PubMed] [Google Scholar]
- Bailey, C. H., D. Bartsch and E. R. Kandel, 1996. Toward a molecular definition of long-term memory storage. Proc. Natl. Acad. Sci. USA 93: 13445–13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen, H. J., B. K. Gregory, C. L. Olsson and J. A. Kiger, Jr., 1987. Two Drosophila learning mutants, dunce and rutabaga, provide evidence of a maternal role for cAMP on embryogenesis. Dev. Biol. 121: 432–444. [DOI] [PubMed] [Google Scholar]
- Berger, A. J., A. C. Hart and J. M. Kaplan, 1998. Gαs-induced neurodegeneration in Caenorhabditis elegans. J. Neurosci. 18: 2871–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz, A., P. Thakur, H. J. Junge, U. Ashery, J. S. Rhee et al., 2001. Functional interaction of the active zone proteins Munc13–1 and RIM1 in synaptic vesicle priming. Neuron 30: 183–196. [DOI] [PubMed] [Google Scholar]
- Bolger, G., T. Michaeli, T. Martins, T. St. John, B. Steiner et al., 1993. A family of human phosphodiesterases homologous to the dunce learning and memory gene product of Drosophila melanogaster are potential targets for antidepressant drugs. Mol. Cell. Biol. 13: 6558–6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of C. elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundage, L., L. Avery, A. Katz, U. Kim, J. E. Mendel et al., 1996. Mutations in a C. elegans Gqα gene disrupt movement, egg laying, and viability. Neuron 16: 999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechler, Y. J., F. W. Herberg and S. S. Taylor, 1993. Regulation-defective mutants of type I cAMP-dependent protein kinase. J. Biol. Chem. 268: 16495–16503. [PubMed] [Google Scholar]
- Charlie, N. K., M. A. Schade, A. M. Thomure and K. G. Miller, 2006. Presynaptic UNC-31 (CAPS) is required to activate the Gαs pathway of the Caenorhabditis elegans synaptic signaling network. Genetics 172: 943–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B. M., and A. D. Grinnell, 1997. Kinetics, Ca2+ dependence, and biophysical properties of integrin-mediated mechanical modulation of transmitter release from frog motor nerve terminals. J. Neurosci. 17: 904–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, N., T. W. Harris, I. Antoshechkin, C. Bastiani, T. Bieri et al., 2005. WormBase: a comprehensive data resource for Caenorhabditis biology and genomics. Nucleic Acids Res. 33: D383–D389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, U. S., A. J. Shayan, G. L. Boulianne and H. L. Atwood, 1999. Drosophila larval neuromuscular junction's responses to reduction of cAMP in the nervous system. J. Neurobiol. 40: 1–13. [DOI] [PubMed] [Google Scholar]
- Davis, R. L., J. Cherry, B. Dauwalder, P. L. Han and E. Skoulakis, 1995. The cyclic AMP system and Drosophila learning. Mol. Cell. Biochem. 149–150: 271–278. [DOI] [PubMed] [Google Scholar]
- Duerr, J. S., J. Gaskin and J. B. Rand, 2001. Identified neurons in C. elegans coexpress vesicular transporters for acetylcholine and monoamines. Am. J. Physiol. Cell Physiol. 280: C1616–C1622. [DOI] [PubMed] [Google Scholar]
- Gally, C., S. Eimer, J. E. Richmond and J. L. Bessereau, 2004. A transmembrane protein required for acetylcholine receptor clustering in Caenorhabditis elegans. Nature 431: 578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald, I. S., and H. R. Horvitz, 1980. unc-93 (e1500): a behavioral mutant of Caenorhabditis elegans that defines a gene with a wild-type null phenotype. Genetics 96: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, K. L., and J. E. Dixon, 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192: 262–267. [DOI] [PubMed] [Google Scholar]
- Hajdu-Cronin, Y. M., W. J. Chen, G. Patikoglou, M. R. Koelle and P. W. Sternberg, 1999. Antagonism between Goα and Gqα in C. elegans: the RGS protein EAT-16 is necessary for Goα signaling and regulates Gqα activity. Genes Dev. 13: 1780–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatley, M. E., B. K. Benton, J. Xu, J. P. Manfredi, A. G. Gillman et al., 2000. Isolation and characterization of constitutively active mutants of mammalian adenylyl cyclase. J. Biol. Chem. 275: 38626–38632. [DOI] [PubMed] [Google Scholar]
- Hou, D., K. Suzuki, W. J. Wolfgang, C. Clay, M. Forte et al., 2003. Presynaptic impairment of synaptic transmission in Drosophila embryos lacking Gsα. J. Neurosci. 23: 5897–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobitz, S., M. M. McLaughlin, G. P. Livi, M. Burman and T. J. Torphy, 1996. Mapping the functional domains of human recombinant phosphodiesterase 4A: structural requirements for catalytic activity and rolipram binding. Mol. Pharmacol. 50: 891–899. [PubMed] [Google Scholar]
- Kandel, E. R., 2001. The molecular biology of memory storage: a dialogue between genes and synapses. Science 294: 1030–1038. [DOI] [PubMed] [Google Scholar]
- Kandel, E. R., and C. Pittenger, 1999. The past, the future and the biology of memory storage. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354: 2027–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashani, A. H., B. M. Chen and A. D. Grinnell, 2001. Hypertonic enhancement of transmitter release from frog motor nerve terminals: Ca2+ independence and role of integrins. J. Physiol. 530: 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro, Y., H. Kuromi, R. Delgado, C. Maureira, C. Oliva et al., 2004. Synaptic vesicle pools and plasticity of synaptic transmission at the Drosophila synapse. Brain Res. Brain Res. Rev. 47: 18–32. [DOI] [PubMed] [Google Scholar]
- Kimura, Y., E. E. Corcoran, K. Eto, K. Gengyo-Ando, M. A. Muramatsu et al., 2002. A CaMK cascade activates CRE-mediated transcription in neurons of Caenorhabditis elegans. EMBO Rep. 3: 962–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn, R. E., J. S. Duerr, J. McManus, A. Duke, T. L. Rakow et al., 2000. Expression of multiple UNC-13 proteins in the Caenorhabditis elegans nervous system. Mol. Biol. Cell 11: 3441–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korswagen, H. C., J. Park, Y. Ohshima and R. H. Plasterk, 1997. An activating mutation in a Caenorhabditis elegans Gs protein induces neural degeneration. Genes Dev. 11: 1493–1503. [DOI] [PubMed] [Google Scholar]
- Korswagen, H. C., A. M. van der Linden and R. H. Plasterk, 1998. G protein hyperactivation of the Caenorhabditis elegans adenylyl cyclase SGS-1 induces neuronal degeneration. EMBO J. 17: 5059–5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika, S. P., J. E. Richmond, G. Hadwiger, R. M. Weimer, E. M. Jorgensen et al., 2001. A post-docking role for active zone protein Rim. Nat. Neurosci. 4: 997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovala, T., B. D. Sanwal and E. H. Ball, 1997. Recombinant expression of a type IV, cAMP-specific phosphodiesterase: characterization and structure-function studies of deletion mutants. Biochemistry 36: 2968–2976. [DOI] [PubMed] [Google Scholar]
- Kuromi, H., and Y. Kidokoro, 2000. Tetanic stimulation recruits vesicles from reserve pool via a cAMP-mediated process in Drosophila synapses. Neuron 27: 133–143. [DOI] [PubMed] [Google Scholar]
- Lackner, M. R., S. J. Nurrish and J. M. Kaplan, 1999. Facilitation of synaptic transmission by EGL-30 Gqα and EGL-8 PLCβ: DAG binding to UNC-13 is required to stimulate acetylcholine release. Neuron 24: 335–346. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer et al., 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33: D192–D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama, I. N., and S. Brenner, 1991. A phorbol ester/diacylglycerol-binding protein encoded by the unc-13 gene of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 88: 5729–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello, C. C., J. M. Kramer, D. Stinchcomb and V. Ambros, 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10(12): 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel, J. E., H. C. Korswagen, K. S. Liu, Y. M. Hajdu-Cronin, M. I. Simon et al., 1995. Participation of the protein Go in multiple aspects of behavior in C. elegans. Nature 267: 1652–1655. [DOI] [PubMed] [Google Scholar]
- Miller, K. G., M. D. Emerson and J. B. Rand, 1999. Goα and diacylglycerol kinase negatively regulate the Gqα pathway in C. elegans. Neuron 24: 323–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, K. G., M. D. Emerson, J. McManus and J. B. Rand, 2000. RIC-8 (synembryn): a novel conserved protein that is required for Gqα signaling in the C. elegans nervous system. Neuron 27: 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman, C., and R. H. Plasterk, 2002. Functional characterization of the adenylyl cyclase gene sgs-1 by analysis of a mutational spectrum in Caenorhabditis elegans. Genetics 161: 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurrish, S., L. Segalat and J. M. Kaplan, 1999. Serotonin inhibition of synaptic transmission: Gαo decreases the abundance of UNC-13 at release sites. Neuron 24: 231–242. [DOI] [PubMed] [Google Scholar]
- Owens, R. J., C. Catterall, D. Batty, J. Jappy, A. Russell et al., 1997. Human phosphodiesterase 4A: characterization of full-length and truncated enzymes expressed in COS cells. Biochem. J. 326(Pt. 1): 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand, J. B., and M. L. Nonet, 1997. Synaptic transmission, pp. 611–643 in C. elegans II, edited by D. H. Riddle, T. Blumenthal, B. J. Meyer and J. R. Priess. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Renden, R. B., and K. Broadie, 2003. Mutation and activation of Gαs similarly alters pre- and postsynaptic mechanisms modulating neurotransmission. J. Neurophysiol. 89: 2620–2638. [DOI] [PubMed] [Google Scholar]
- Renger, J. J., A. Ueda, H. L. Atwood, C. K. Govind and C. F. Wu, 2000. Role of cAMP cascade in synaptic stability and plasticity: ultrastructural and physiological analyses of individual synaptic boutons in Drosophila memory mutants. J. Neurosci. 20: 3980–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, N. K., M. A. Schade and K. G. Miller, 2005. Convergent, RIC-8-dependent Gα signaling pathways in the Caenorhabditis elegans synaptic signaling network. Genetics 169: 650–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond, J. E., W. S. Davis and E. M. Jorgensen, 1999. UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat. Neurosci. 2(11): 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond, J. E., R. M. Weimer and E. M. Jorgensen, 2001. An open form of syntaxin bypasses the requirement for UNC-13 in vesicle priming. Nature 412: 338–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund, C., and C. F. Stevens, 1996. Definition of the readily releasable pool of vesicles at hippocampal synapses. Neuron 16: 1197–1207. [DOI] [PubMed] [Google Scholar]
- Schade, M. A., N. K. Reynolds, C. M. Dollins and K. G. Miller, 2005. Mutations that rescue the paralysis of Caenhorabditis elegans ric-8 (synembryn) mutants activate the Gαs pathway and define a third major branch of the synaptic signaling network. Genetics 169: 631–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segalat, L., D. A. Elkes and J. M. Kaplan, 1995. Modulation of serotonin-controlled behaviors by Go in Caenhorabditis elegans. Nature 267: 1648–1651. [DOI] [PubMed] [Google Scholar]
- Sunahara, R. K., C. W. Dessauer and A. G. Gilman, 1996. Complexity and diversity of mammalian adenylyl cyclases. Annu. Rev. Pharmacol. Toxicol. 36: 461–480. [DOI] [PubMed] [Google Scholar]
- Suzuki, K., A. D. Grinnell and Y. Kidokoro, 2002. Hypertonicity-induced transmitter release at Drosophila neuromuscular junctions is partly mediated by integrins and cAMP/protein kinase A. J. Physiol. 538: 103–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan, K. A., D. E. Curtis, K. B. McKusick, A. V. Voinov, F. A. Mapa et al., 2002. High-throughput gene mapping in Caenorhabditis elegans. Genome Res. 12: 1100–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall, G. G., A. M. Krumins and A. G. Gilman, 2003. Mammalian Ric-8A (synembryn) is a heterotrimeric Galpha protein guanine nucleotide exchange factor. J. Biol. Chem. 278: 8356–8362. [DOI] [PubMed] [Google Scholar]
- Tanabe, N., and H. Kijima, 1988. Transmitter release at frog end-plate loaded with a Ca2+-chelator, BAPTA: hypertonicity and erythrosin B augment the release independently of internal Ca2+. Neurosci. Lett. 92: 52–57. [DOI] [PubMed] [Google Scholar]
- Turko, I. V., S. H. Francis and J. D. Corbin, 1998. Potential roles of conserved amino acids in the catalytic domain of the cGMP-binding cGMP-specific phosphodiesterase. J. Biol. Chem. 273: 6460–6466. [DOI] [PubMed] [Google Scholar]
- Wolfgang, W. J., C. Clay, J. Parker, R. Delgado, P. Labarca et al., 2004. Signaling through Gs alpha is required for the growth and function of neuromuscular synapses in Drosophila. Dev. Biol. 268: 295–311. [DOI] [PubMed] [Google Scholar]
- Wolfgang, W. J., A. Hoskote, I. J. Roberts, S. Jackson and M. Forte, 2001. Genetic analysis of the Drosophila Gsα gene. Genetics 158: 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston and R. H. Plasterk, 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164. [DOI] [PubMed] [Google Scholar]
- Zhong, Y., and C. F. Wu, 1991. Altered synaptic plasticity in Drosophila memory mutants with a defective cyclic AMP cascade. Science 251: 198–201. [DOI] [PubMed] [Google Scholar]