Abstract
The genome of Borrelia burgdorferi encodes a large number of lipoproteins, many of which are expressed only at certain stages of the spirochete's life cycle. In the current study we describe the B. burgdorferi population structure with respect to the production of two lipoproteins [outer surface protein A (OspA) and outer surface protein C (OspC)] during transmission from the tick vector to the mammalian host. Before the blood meal, the bacteria in the tick were a homogeneous population that mainly produced OspA only. During the blood meal, the population became more heterogeneous; many bacteria produced both OspA and OspC, whereas others produced only a single Osp and a few produced neither Osp. From the heterogeneous spirochetal population in the gut, a subset depleted of OspA entered the salivary glands and stably infected the host at time points >53 hr into the blood meal. We also examined genetic heterogeneity at the B. burgdorferi vlsE locus before and during the blood meal. In unfed ticks, the vlsE locus was stable and one predominant and two minor alleles were detected. During the blood meal, multiple vlsE alleles were observed in the tick. Tick feeding may increase recombination at the vlsE locus or selectively amplify rare vlsE alleles present in unfed ticks. On the basis of our data we propose a model, which is different from the established model for B. burgdorferi transmission. Implicit in our model is the concept that tick transmission converts a homogeneous spirochete population into a heterogeneous population that is poised to infect the mammalian host.
Here we present the results of a study on the antigenic and genetic changes experienced by Lyme disease spirochetes (Borrelia burgdorferi) as they move from a tick to a vertebrate host. Ticks acquire B. burgdorferi infection when larval or nymphal ticks feed on an infected host (1, 2). The ingested spirochetes colonize the tick gut and persist through the molt. After the molt, when ticks feed again, the bacteria multiply in the gut, after which some of the spirochetes escape from the gut into the hemolymph, and invade salivary glands and enter the host dermis (3–7).
The B. burgdorferi genome codes for a large number of lipoproteins, many of which localize to the outer membrane and are likely to play a role in transmission. Outer surface protein A (OspA) and outer surface protein C (OspC) are among the most intensely studied lipoproteins of B. burgdorferi. Several studies point to the production of OspA and -C changing during tick transmission (4, 8–11). Within an unfed tick, the spirochetes mostly produce OspA, whereas OspC is rarely detected (8, 11). Once the tick starts to feed, many spirochetes no longer produce OspA, whereas large numbers now produce OspC, most likely through increased transcription of the ospC gene (8, 10–12). The down-regulation of OspA during tick feeding and the up-regulation of OspC support the hypothesis that OspA may function within the tick gut, possibly as a receptor that mediates attachment to the gut epithelium (13). OspC may be involved in spirochetes escaping the gut, invading the salivary glands, and establishing an infection in the mammal. One prediction of this hypothesis is that OspA-producing bacteria may be confined to the gut, whereas OspC-producing bacteria may selectively escape the gut and invade the salivary glands and the host dermis. In the current study we have followed the Osp phenotype of spirochete populations within feeding ticks to test the above hypothesis. The results reveal that the population dynamics that occur during transmission are complex and contradict the established hypothesis.
In addition to transcriptional activation, recombination also leads to the expression of novel molecules on the surface of spirochetes. The best-studied recombination site in B. burgdorferi is the vlsE locus, which consists of an active telomeric expression site flanked on the 5′ side by multiple silent vls cassettes (14). Unidirectional recombination events between segments of the cassettes and the vlsE gene at the expression site lead to the generation of new vlsE antigenic variants in the mammalian host (14–16). The genetic stability of the vlsE locus in the tick vector has not been studied. Here we describe genetic changes that take place at the vlsE locus during ticks' feeding. On the basis of genetic (vlsE) and antigenic (OspA and -C) changes, we propose a model for spirochete transmission by ticks. The model has important implications for transmission and vaccine development.
Materials and Methods
Mice, Bacteria, and Ticks.
Mice used in the study were 6- to 8-week-old C3H, HenJ mice purchased from the National Institutes of Health (Bethesda, MD). A low-passage B31 strain of B. burgdorferi (Centers for Disease Control and prevention, Fort Collins, CO) was grown on solid BSK-II medium (38) and a single clone designated B31-C1 was isolated and used in the current study. B. burgdorferi B31-C1 was grown in liquid BSK-II medium (17). The ticks used in this study originated from female Ixodes scapularis collected in Bridgeport, CT. The larvae were F1 generation of the wild ticks. Mice were infected by injecting 1 × 107 B31-C1 cells per mouse. Larvae were infected with B31-C1 by feeding on the infected mice as previously described (18). Ninety-seven percent of larvae cultured were infected. The larval ticks were kept in humid chambers at 21°C until they molted to the nymphal stage.
Infection of Mice.
Two to three weeks after tick removal, the mice were tested for B. burgdorferi infection by Western blotting and culture as previously described (19, 20).
Direct Fluorescent Antibody (DFA) Staining of Spirochetes Within Ticks.
Tick salivary glands and guts (homogenized and whole mount) were prepared for double-labeling DFA as previously described (4). Monoclonal antibodies (mAb) directed to OspA (C3.78) (4) and OspC (B5) (21) were conjugated with fluorescent dyes, Alexa 488 (Alexa) and Texas red-X (TR), respectively, as described in the manufacturer's manual (Molecular Probes). The antibody combinations used were (i) FITC-labeled goat anti-Borrelia (FITC-Bb; KPL, Gaithersburg, MD) and TR-labeled mouse C3.78 mAb against OspA; (ii) FITC-labeled goat anti-Borrelia and TR-labeled mouse B5 mAb against OspC; and (iii) Alexa-labeled mouse C3.78 mAb against OspA and TR-labeled mouse B5 mAb against OspC. After the antibody incubations, the slides were mounted and observed by epifluorescence microscopy (Eclipse E 600, Nikon) with a digital camera (SPOT II, Diagnostic Instruments, Sterling Heights, MI). Images were captured with spot software version 2.2 (Diagnostic Instruments). Some specimens were also observed with a Leica TCS-NT confocal microscope system (Leica Microsystems, Wetzlar, Germany).
We determined the proportion of spirochetes producing only OspA, only OspC, both OspA and C, and neither protein as described below:
Combination i, TR-OspA/FITC-Bb = X (proportion of total population producing OspA).
Combination ii, TR-OspC/FITC-Bb = Y (proportion of total population producing OspC).
Combination iii, Alexa-OspC/TR-OspA = Z (proportion of OspA-producing spirochetes that also produce OspC).
Proportion of the Borrelia producing both OspA and OspC (A+/C+) = X × Z.
Proportion of Borrelia producing only OspA (A+/C−) = X × (1 − Z).
Proportion of Borrelia producing only OspC (A−/C+) = Y − (X × Z).
Proportion of Borrelia producing neither OspA nor OspC (A−/C−) = 1 − X − Y + (X × Y).
DFA Staining of Spirochetes in Mouse Dermis at the Site of Tick Attachment.
Often, when ticks were forcibly removed from mice, a small piece of the mouse dermis remained attached to the tick hypostome. A total of 233 skin samples remaining on the hypostome were examined for spirochetes and Osp phenotype. The skin samples were stained with antibody as described above except that antibody incubations were performed at 4°C for 2 days.
PCR Cloning of the vlsE Locus and Restriction Fragment Length Polymorphism (RFLP) Analysis.
Total DNA was purified from infected ticks by using the QIAamp Tissue kit (Qiagen, Valencia, CA). PCR primers (vlsE-F, 5′-GTAGTACGACGGGGAAACCAGATAGTAC-3′; vlsE-R, 5′-CCTAAAACTTTGCGAACTGCAGACTCAGC-3′) were designed to amplify the variable region of the vlsE locus (14) from total infected tick DNA. PCR amplifications were performed with the following program: 1 cycle for 5 min at 94°C; 35 cycles at 94°C for 30 sec, 60°C for 30 sec, 72°C for 40 sec, and concluding with 72°C for 5 min. PCR products were purified and cloned into the vector pCR2.1-TOPO (Invitrogen, Carlsbad, CA) and then used to transform Escherichia coli (TOP10 One Shot Cell). Twenty-five to 30 clones were randomly selected and the inserts from each individual clone were PCR amplified. AluI and MboI were used to digest the PCR-amplified inserts. The digested DNA was separated on agarose gels and stained with ethidium bromide to compare the RFLP patterns at the vlsE locus in fed and unfed ticks.
Results
Timing of B. burgdorferi Transmission from Ticks to Mice.
Previous studies have documented that ticks transmit B. burgdorferi to hosts 48–60 hr after attachment (5, 22). Experiments were performed to better define the timing of tick transmission. Groups of 20–25 nymphal ticks infected with a clonal isolate of B. burgdorferi strain B31 were placed on individual mice, and all of the ticks were removed from individual mice at 2-h intervals starting at 37 hr and ending at 72 hr into the blood meal (Fig. 1B). Mice were tested for spirochete infection by serology and culture of selected organs. None of the mice exposed to ticks for 52 hr or less were infected, whereas all of the mice exposed for at least 53 hr were infected with B. burgdorferi (Fig. 2).
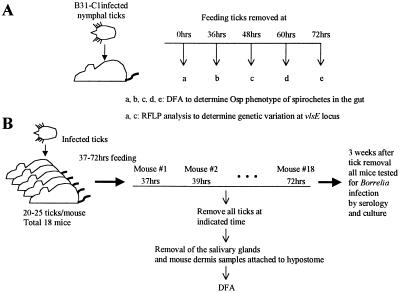
Figure 1.
Outline of experimental approach. (A) B. burgdorferi Osp phenotype in the tick gut. Infected ticks were placed on mice and removed at the indicated times. Twelve unfed and 93 fed nymphal ticks (12 ticks from 36 hr, 39 ticks from 48 hr, 36 ticks from 60 hr, and 6 ticks from 72 hr) were removed. The guts were dissected and examined by DFA to determine the OspA and OspC phenotype within the gut. DNA was purified from gut homogenates prepared from unfed and partially engorged (48-hr) ticks to determine the extent of variation at the B. burgdorferi vlsE locus. (B) B. burgdorferi infection of tick salivary glands and mouse dermis. A total of 18 mice were infested with 20–25 infected nymphal ticks. All of the ticks were removed from individual mice at 2-hr intervals starting at 37 hr and ending at 72 hr into the blood meal. Mouse and tick samples were analyzed for B. burgdorferi infection and Osp phenotypes.
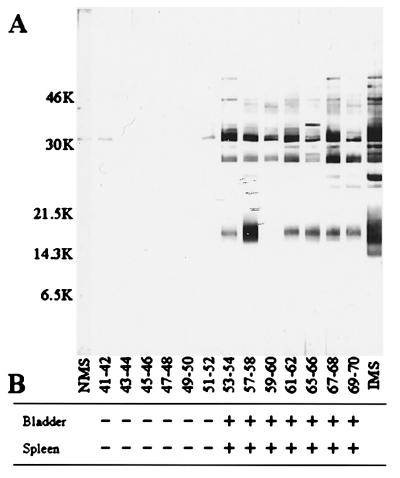
Figure 2.
Timing of tick transmission of B. burgdorferi to mice. Mice exposed to infected ticks for various time periods (Fig. 1B) were tested for infection by Western blotting (A) or culture (B). Numbers on the left in A are molecular masses of markers in kDa. In a few cases (hr 37–38, 39–40, 55–56) all of the ticks were not recovered from the mice. These animals were not tested for infection because we could not accurately determine the length of tick exposure. IMS, infected mouse serum; NMS, normal (uninfected) mouse serum.
The Osp Phenotype of the B. burgdorferi Population Within the Gut of Feeding Ticks.
During tick feeding, spirochetes multiply within the gut (23) before some of the bacteria disseminate to the salivary glands. Experiments were next performed to characterize the Osp phenotype of spirochetes within the gut of feeding ticks. DFA staining was used to determine the OspA and/or OspC phenotype of the population of B. burgdorferi in the gut of unfed and feeding nymphal I. scapularis (Fig. 1A). The majority of spirochetes within unfed ticks produced only OspA (85.6% ± 7.3%) and a minority (14.1% ± 7.5%) did not produce either protein (Fig. 3). None of the spirochetes within the gut of unfed ticks produced OspC (Fig. 3; see also Fig. 8, which is published as supplemental data on the PNAS web site, www.pnas.org).
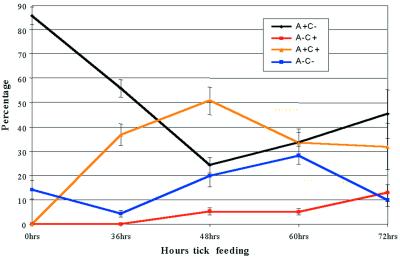
Figure 3.
Population dynamics of B. burgdorferi producing OspA and OspC in the tick gut during the blood meal. Infected ticks were allowed to feed on mice for various time periods as described for Fig. 1A. The guts were removed from ticks and the proportion of bacteria displaying each Osp phenotype was determined as described in the text.
Dramatic changes in the spirochete population in the gut were observed in response to tick feeding (Figs. 3 and 4). Two novel phenotypes of spirochetes producing OspC (A+/C+ and A−/C+) were observed during the blood meal. Forty-eight hours into the blood meal the major (50.6% ± 11.3%) population in the gut were spirochetes that produced both OspA and OspC (Figs. 3 and 4). By 60 hr into the blood meal the double-positive spirochetes had decreased to 33.4% ± 8.8% (Fig. 3). Although spirochetes that produced solely OspA were a major population within unfed ticks, after 48 and 60 hr of feeding, this population decreased to 24.3% ± 6.3% and 33.7% ± 11.0%, respectively (Fig. 3). Minor populations that produced solely OspC as well as neither Osp were also observed within the gut of feeding ticks (Fig. 3). These results demonstrated that before feeding the gut was colonized by a relatively homogeneous population of spirochetes that mainly produced only OspA. As the infected tick began to feed, novel OspC-positive phenotypes were generated in the tick gut and the gut was populated by an antigenically heterogeneous population of B. burgdorferi (Figs. 3 and 4).
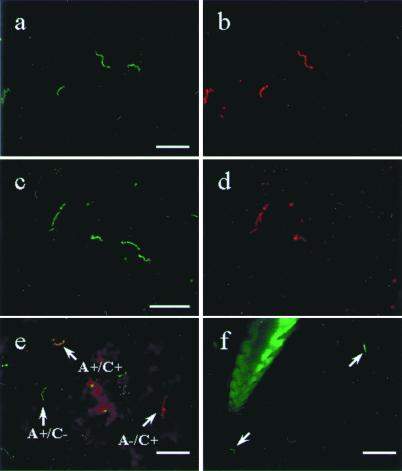
Figure 4.
Direct dual immunofluorescence of B. burgdorferi in the gut of partially engorged (48 hr) ticks and in the mouse dermis. (a and b) The same field double-labeled with a FITC-conjugated polyclonal B. burgdorferi antibody (a) and a TR-conjugated mAb against OspA (b). (c and d) The same field labeled with FITC-conjugated polyclonal Borrelia antibody (c) and a TR-conjugated mAb against OspC (d). (e) Merged image of spirochetes stained with Alexa 488-conjugated OspA (green) and TR-conjugated OspC (red). Three phenotypes were observed in e: arrows, Borrelia producing only OspA in green, only OspC in red, and both OspA and OspC in orange. (f) Mouse skin sample attached to the hypostome of a tick that had fed for 40 hr is labeled with the FITC-conjugated polyclonal Borrelia antibody. The hypostome itself autofluoresces, and bacteria staining with the antibody are indicated by the arrowhead. (Bars in a, c, and e represent 20 μm, and the bar in f represents 50 μm.)
Timing of Salivary Gland Invasion.
Many studies point to B. burgdorferi moving from the gut to the salivary glands and then entering the host through the salivary ducts of the tick (6, 7, 22). Spirochetes are first seen in salivary glands as early as 36 hr into the blood meal, whereas transmission has been noted to occur at some point between 48 and 60 hr into the blood meal (22, 23). Experiments were performed to better define the relationship between timing of salivary gland invasion and host infection. Salivary glands were removed from unfed ticks as well as from feeding ticks at 2-hr intervals between 37 and 72 hr into the blood meal (Fig. 1B). Salivary glands were examined for the presence of spirochetes and their Osp phenotype by using DFA. No spirochetes were detected in the salivary glands of the unfed ticks (12 pairs examined). Among ticks that were removed before the mice became infected (37–52 hr of feeding), 18.45% ± 18.71% of the salivary glands were infected. When ticks had fed for long enough (≥53 hr) to stably infect the mice (Fig. 2), the proportion of infected salivary glands (65.67% ± 14.2%) was significantly greater (t test, t = 8.3, P < 0.001) than in ticks that had fed for <53 hr (also see Fig. 9, published as supplemental data).
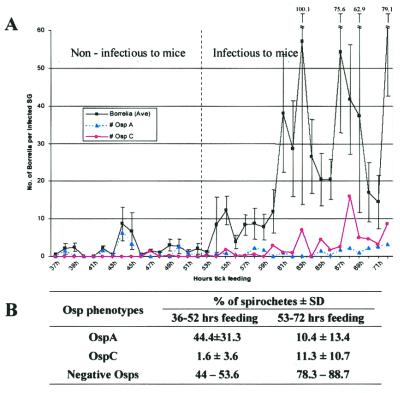
Many of the ticks that fed for between 36 and 52 hr had bacteria in the salivary glands, yet they did not infect the mice (Fig. 2). Experiments were performed to determine the mean number of bacteria within each infected salivary gland at various times during the blood meal. Ticks that had fed for <53 hr had very few bacteria in the infected glands (2.92 ± 2.67 bacteria per gland) (Fig. 5). In contrast, the number of bacteria per gland steadily increased after 53 hr and reached a maximum of 61 bacteria per gland at 72 hr into the blood meal (Fig. 5). The mean number of bacteria per salivary gland (23.66 ± 18.04) between 53 and 72 hr into the blood meal was significantly higher than the number observed before 53 hr (t test, t = 5.07, P < 0.001). Ticks that had attached for long enough to infect mice had a greater number of infected salivary glands and more spirochetes within individual salivary glands than ticks that were attached for a shorter period and failed to infect mice.
Figure 5.
B. burgdorferi infection of tick salivary glands. (A) Salivary glands were removed from ticks attached to mice for various time periods as described for Fig. 1B. At each time point the samples were analyzed to determine the mean number of total spirochetes per infected salivary gland (SG) pair (♦) and the mean number of OspA- (▴) and OspC- (●) producing spirochetes per infected pair of glands. The vertical dashed line separates ticks that had attached to the host for too short a time to infect mice (<53 hr) from those that had attached long enough to infect mice (≥53 hr) (see Fig. 2 for mouse transmission data). (B) Summary of Osp phenotypes in the salivary glands before and after mouse infection during tick feeding. The salivary gland data are based on A.
Osp Phenotype of Bacteria in Tick Salivary Glands.
Both OspA- and OspC-positive spirochetes have been observed in the salivary glands of ticks (9, 10). DFA was used to determine the proportion of OspA- and OspC-producing bacteria in the salivary glands of feeding ticks (Fig. 5). The small number of bacteria that infected salivary glands in ticks that had fed for <53 hr consisted mainly of spirochetes that produced only OspA (44.4%) and neither Osp (44–53.6%) (Fig. 5). At times <53 hr very few bacteria (1.6%) in the salivary glands produced OspC. At ≥53 hr into the blood meal when mice were infected, the proportion of OspC-positive bacteria increased to 11.3%, whereas the proportion of OspA-positive bacteria declined from 44.4% to 10.4%. The major population of bacteria in the salivary glands of ticks that had attached for long enough to transmit Borrelia did not produce either OspA or OspC (78.3–88.7%) (Fig. 5). These observations established that as the blood meal progressed the Osp phenotype of the bacteria entering the salivary glands changed (also see Fig. 10 in the supplemental data).
B. burgdorferi in the Mouse Dermis at the Site of Tick Attachment.
Mice infected by tick bite readily seroconvert to OspC and rarely seroconvert to OspA, suggesting that the bacteria entering the host produce OspC and little or no OspA (8, 24–26). It has proven difficult to directly examine the Osp phenotype of spirochetes in mouse skin during tick transmission because of the difficulty of finding bacteria in the skin. However, in one study OspC-positive spirochetes have been observed in the skin of mice infected by tick bite (27). The hypostome is a tick mouth part armed with numerous, sharp, curved denticles and is inserted into the host dermis during tick feeding (28). The food canal, which is dorsally located on the hypostome surface, is the route through which blood is drawn into the tick as well as the route through which saliva is excreted into the host. Because of the hypostome's characteristic barbed structure, often a piece of the host's dermis remains attached to the hypostome after the forced removal of feeding ticks.
We examined the mouse dermis attached to the hypostome for spirochetes as well as their Osp phenotype. Two hundred and thirty-three skin samples were examined by DFA with the antibody combinations i and ii (see Materials and Methods). Sixty-one of the 233 skin samples contained spirochetes (see Fig. 4F, for example). Much to our surprise, spirochetes were observed in 23.6% ± 24.3% of skin samples from mice that did not become infected because of early (<53 hr) removal of ticks. Dermis samples collected from mice that became stably infected (53- to 72-hr attachment) had a prevalence of 30.2% ± 31.8%. At both early (37–52 hr) and late (53–72 hr) time points into the blood meal, bacteria migrated to the salivary glands and entered the host dermis. However, the bacteria that entered early were unable to establish an infection, unlike the organisms that were introduced into the host later (Fig. 2). Double-labeling DFA experiments were performed to determine the Osp phenotype of bacteria that entered the host dermis. Before stable infection of mice, the bacteria that entered the skin at the site of tick attachment primarily produced OspA (71.2%), while a few (18.0%) produced OspC. This pattern was reversed in bacteria present in skin samples that led to stable infections, with fewer OspA- (12.5%) and more OspC- (29.9%) producing spirochetes in the dermis. As in the case of salivary glands at late stages of tick feeding (Fig. 5), the majority of bacteria (58–70%) in the dermis produced neither OspA nor OspC.
Genetic Variation of B. burgdorferi vlsE During Tick Transmission.
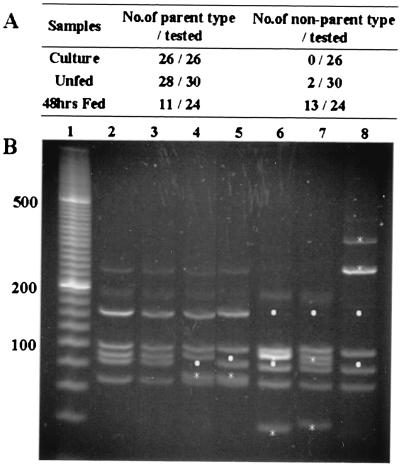
In addition to ospA and ospC genes, the B. burgdorferi genome codes for many other lipoproteins. The vlsE gene of B. burgdorferi is especially interesting because unidirectional recombination events between silent partial gene fragments and the vlsE gene lead to the generation of new antigenic variants in the mammalian host (14–16). RFLP analysis was used to determine the vlsE genotype of spirochetes within unfed and feeding ticks. The variable region of the vlsE locus of B. burgdorferi was PCR amplified and cloned into a vector. RFLP analysis was performed by digesting the vlsE inserts with AluI and MboI. The vlsE locus of 26 individual clones from cultured B. burgdorferi were tested. All 26 clones had the same RFLP pattern (Fig. 6). Thirty clones from unfed ticks and 24 clones from 48-hr fed ticks were analyzed. Of the 30 clones examined from unfed ticks, 28 had a RFLP pattern that was identical to the parental clonal population of culture-grown B31 used in this study (Fig. 6). Two of the 30 clones from unfed ticks had patterns that differed from the parental strain. Surprisingly, 24 clones derived from feeding ticks had 13 different RFLP patterns, all of which were different from that of the parental strain (Fig. 6).
Figure 6.
RFLP analysis of the vlsE locus of B. burgdorferi in culture, unfed, and partially engorged (48-hr) ticks. (A) Summary of the vlsE variants detected by RFLP analysis. The single vlsE allele in cultured bacteria was designated as the parental allele. In partially fed ticks, the 24 clones belonged to 13 different RFLP types, all of which were different from the parent type. (B) Lane 1, 20-bp ladder; lane 2, RFLP pattern of parental B31-C1; lanes 3 and 4, RFLP patterns of 2 of the 3 alleles detected in unfed ticks; lanes 5–9, RFLP patterns of 4 of the 13 alleles detected in fed ticks. The asterisks mark new bands in the clones that are absent from the parental strain. The dots mark bands present in parental strain that are absent from the clone tested.
Discussion
Previous studies on tick transmission have been based on a few time points or total tick homogenates (4, 8, 10). These studies demonstrated that during the blood meal many spirochetes up-regulate OspC and down-regulate OspA. This simple inverse relationship between OspA and OspC led to the hypothesis that bacteria producing OspC invaded the salivary glands and entered the host. The current study reveals that transmission is more complex than expected from previous studies and involves an antigenically and genetically polymorphic population of spirochetes entering the host. Furthermore, our data point to OspC-producing bacteria being a minor population entering the host. On the basis of our data and the observations of others we propose a model for B. burgdorferi transmission from ticks to mammals (Fig. 7) and discuss the implications of the model for transmission and vaccine development.
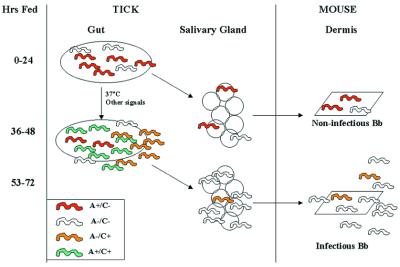
Figure 7.
Model for population dynamics of OspA and OspC during B. burgdorferi (Bb) transmission from ticks to mice. The model is based on the predominant Osp phenotypes observed in the different organs. See text for details of model. In brief, we propose that at early times of feeding (<53 hr) noninfectious bacteria that mainly produce OspA and others that produce neither Osp invade the salivary glands and host dermis in small numbers, but they fail to stably infect the mice. Stable transmission is initiated later (≥53 hr) when bacteria producing only OspC leave the gut. These infectious organisms clear OspC en route to the salivary glands and then enter the host dermis. In our model, from the heterogeneous population in the gut, only a subpopulation that is depleted of OspA is successfully transmitted to mice.
Spirochetes within unfed ticks were homogeneous with most producing only OspA and also displayed little genetic heterogeneity at the vlsE locus. During tick feeding many spirochetes in the gut started to produce OspC and the gut became populated with a heterogeneous population with respect to OspA and OspC production. The induction of OspC that occurred during the blood meal did not require the clearance of OspA from the surface. Rather, OspA-producing spirochetes present within the tick appeared to produce OspC also so that initially both proteins were present on the surface of the spirochetes in the gut. Tick feeding also increased heterogeneity at the vlsE locus of B. burgdorferi, possibly by stimulating recombination at the vlsE gene. Alternately, the observed increase in the number of vlsE alleles may be due to selection within the feeding tick of rare vlsE variants already present in the population. A major feature of our model is that tick feeding converts a relatively homogeneous population of bacteria in the gut to a highly heterogeneous population with both antigenic and genetic variants (Fig. 7).
Mice were infected only by ticks that had fed for longer than 53 hr. At times greater than 53 hr when mice were stably infected, large numbers of tick salivary glands were infected and individual salivary glands had large numbers of bacteria. Furthermore, at time points >53 hr the gut had a mix of bacteria with all four phenotypes well represented (Fig. 3). In contrast, most of the bacteria in the salivary glands and host dermis produced neither OspA or OspC and a significant minority produced only OspC. From the heterogeneous population in the gut, only a subset of the bacteria invaded the salivary glands and infected the host. These observations indicate that the tick gut epithelium acts as a selective barrier that allows only certain phenotypes to preferentially escape into the hemocele and enter the host during productive infection.
Much to our surprise, a few spirochetes were regularly observed in tick salivary glands and host skin samples at 37–52 hr into the blood meal when mice failed to become infected. Even though some spirochetes move to the host dermis as early as 37 hr into the blood meal, why do the ticks have to remain attached for at least 53 hr for stable transmission? Our results point to several differences between noninfectious spirochetes that prematurely invade the host and infectious spirochetes that enter the host at >53 hr after attachment. Bacteria that invaded the salivary glands and the host dermis prematurely mostly produced OspA only; infectious bacteria that invaded the host later consisted of bacteria that produced neither OspA nor OspC and others that produced only OspC. These phenotypic differences between early and late arrivals in the salivary glands and host dermis may contribute to the differences in infectivity. At later times (>53 hr) a greater number of salivary glands were infected and individual salivary glands contained more bacteria than salivary glands that were infected between 37 and 53 hr. Therefore, in addition to phenotypic differences, an infectious inoculum may be delivered only at time points >53 hr.
Role of OspA and OspC in B. burgdorferi Transmission.
OspA is an adhesin that selectively binds to tick gut epithelial cells (13). The reason OspA-producing bacteria rarely entered the salivary glands and the host may be that OspA-producing bacteria are selectively retained in the tick gut. Especially striking was the sudden burst of OspC synthesis by spirochetes in the tick gut, which peaked at 48 hr, just before infectious spirochetes entered the host. Previous studies pointed to OspC playing an important role in early infection in the mammal because infected animals readily seroconvert to OspC (22). However, this study has demonstrated that the protein was not produced by most spirochetes that actually entered the salivary glands and moved into the host. On the basis of this pattern of expression, we propose that OspC may play a role in allowing the bacteria to escape from the gut into the hemocele. Once they exit the gut, the bacteria may begin to down-regulate OspC production (Fig. 7). The recent report by Gilmore and Piesman (29) that OspC antibodies block the movement of spirochetes from the tick gut to the salivary glands supports this hypothesis. Even though most bacteria invading the salivary glands and entering the host did not produce OspC, small numbers of bacteria did have the protein on their surface, and this may be sufficient to stimulate the early OspC antibody response that is regularly observed. The OspC immune response in mice wanes over time (30, 31), possibly because the rodent immune response is due to a small number of OspC-producing bacteria being carried over from the tick to the host.
Our working model that OspA serves to retain bacteria in the tick gut and that OspC facilitates transfer from the gut lumen to the hemocele is a simplification and does not explain all our observations. Small numbers of OspA-producing bacteria were observed in the salivary glands, and many OspC-producing bacteria remained confined to the gut. Many individual bacteria within the feeding tick produced both OspA and OspC, and the location of these double-positives may depend on the actual level of OspA and -C on individual bacteria. Furthermore, in addition to OspA and -C, other bacterial proteins may control transmission, and their expression pattern may explain the apparent mislocalization of some of the spirochetes.
Genetic and Antigenic Diversity Generated During Transmission.
In addition to the role of OspC in the movement of B. burgdorferi from the gut to the hemocele, another key feature of our model is that the population of spirochetes that enter the host is heterogeneous. The genome of B. burgdorferi contains close to 150 genes encoding lipoproteins (32, 33). In this study we have examined the production of just two lipoproteins and find that the population that infects the host consists of bacteria that produce neither Osp as well as some that produce only OspC. The true complexity of the population that is transmitted is undoubtedly greater because many of the other surface proteins may also be differentially produced within the population.
Recent studies on paralogous gene families of B. burgdorferi such as the ospE-related genes (34), the mlp gene family (35), and individual loci such as vlsE (15, 16) and ospC (36) indicate that genetic recombination occurs frequently within the spirochete genome. Spirochetes cultured in BSK-II medium do not display high levels of recombination, whereas both vlsE- and ospE-related genes have been observed to recombine during murine infection (14, 34, 37). The data we present here point to another potential site of genetic recombination—the tick gut. RFLP patterns of the vlsE locus from unfed ticks revealed a homogeneous profile consisting of 1 predominant allele and 2 minor alleles. During the blood meal 13 vlsE alleles, all of which were different from the parental strain, were observed. Therefore, tick feeding may stimulate recombination at the vlsE locus. Alternately, rare vlsE alleles present in the spirochete population may be preferentially amplified during tick feeding. The population of bacteria that the tick delivers into the host's dermis consists of individual bacteria producing different Osps as well as different alleles of lipoproteins.
Implications for Vaccine Development.
We have previously reported that tick-transmitted B. burgdorferi are better able to evade host immunity than are culture-grown organisms (19). Our data provide an explanation for the greater ability of tick-borne organisms to evade host immunity. Culture-grown bacteria predominantly produce OspA and do not show evidence of active recombination, and they resemble the homogeneous spirochetes within unfed nymphal ticks. This uniform population is susceptible to immune serum. In contrast, the genetic and antigenic heterogeneity that is generated during tick transmission may allow some of the bacteria delivered by tick bite to evade antibodies and establish an infection within hosts despite the circulating antibodies. The differences in the antigenic and genetic structure of the population of tick-transmitted and culture-grown spirochetes have profound implications for vaccine testing and development. To begin with, it is essential that vaccines be tested with tick challenge because cultured organisms are not representative of organisms introduced by ticks. Simply identifying antigens produced early during infection in the host may not be sufficient for identifying vaccine candidates because large numbers of bacteria that enter the host may not produce this antigen. The search for additional Lyme disease vaccine candidates must focus on antigens produced within the tick at early stages of blood meal that may lead to transmission-blocking immunity or antigens that are produced by most if not all bacteria entering the host.
Supplementary Material
Acknowledgments
We thank Dr. Janne Cannon for her comments on the manuscript. This work was supported by grants from the National Institutes of Health (AR-02061) and The Arthritis Foundation.
Abbreviations
- OspA
outer surface protein A
- OspC
outer surface protein C
- DFA
direct fluorescent antibody
- TR
Texas red-X
- RFLP
restriction fragment length polymorphism
References
- 1.Piesman J. Exp Appl Acarol. 1989;7:71–80. doi: 10.1007/BF01200454. [DOI] [PubMed] [Google Scholar]
- 2.Burgdorfer W, Lane R S, Barbour A G, Gresbrink R A, Anderson J R. Am J Trop Med Hyg. 1985;34:925–930. doi: 10.4269/ajtmh.1985.34.925. [DOI] [PubMed] [Google Scholar]
- 3.Benach J L, Coleman J L, Skinner R A, Bosler E M. J Infect Dis. 1987;155:1300–1306. doi: 10.1093/infdis/155.6.1300. [DOI] [PubMed] [Google Scholar]
- 4.de Silva A M, Telford S R, 3rd, Brunet L R, Barthold S W, Fikrig E. J Exp Med. 1996;183:271–275. doi: 10.1084/jem.183.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piesman J. J Infect Dis. 1993;167:1082–1085. doi: 10.1093/infdis/167.5.1082. [DOI] [PubMed] [Google Scholar]
- 6.Ribeiro J M, Mather T N, Piesman J, Spielman A. J Med Entomol. 1987;24:201–205. doi: 10.1093/jmedent/24.2.201. [DOI] [PubMed] [Google Scholar]
- 7.Zung J L, Lewengrub S, Rudzibnska M A, Spielman A, Telford S R, Piesman J. Can J Zool. 1989;67:1737–1748. [Google Scholar]
- 8.Schwan T G, Piesman J, Golde W T, Dolan M C, Rosa P A. Proc Natl Acad Sci USA. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fingerle V, Liegl G, Munderloh U, Wilske B. Med Microbiol Immunol (Berlin) 1998;187:121–126. doi: 10.1007/s004300050083. [DOI] [PubMed] [Google Scholar]
- 10.Coleman J L, Gebbia J A, Piesman J, Degen J L, Bugge T H, Benach J L. Cell. 1997;89:1111–1119. doi: 10.1016/s0092-8674(00)80298-6. [DOI] [PubMed] [Google Scholar]
- 11.Schwan T G, Piesman J. J Clin Microbiol. 2000;38:382–388. doi: 10.1128/jcm.38.1.382-388.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Silva A M, Zeidner N S, Zhang Y, Dolan M C, Piesman J, Fikrig E. Infect Immun. 1999;67:30–35. doi: 10.1128/iai.67.1.30-35.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pal U, de Silva A M, Montgomery R R, Fish D, Anguita J, Anderson J F, Lobet Y, Fikrig E. J Clin Invest. 2000;106:561–569. doi: 10.1172/JCI9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J R, Hardham J M, Barbour A G, Norris S J. Cell. 1997;89:275–285. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J R, Norris S J. Infect Immun. 1998;66:3698–3704. doi: 10.1128/iai.66.8.3698-3704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J R, Norris S J. Infect Immun. 1998;66:3689–3697. doi: 10.1128/iai.66.8.3689-3697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbour A G. Yale J Biol Med. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 18.Piesman J. J Med Entomol. 1993;30:199–203. doi: 10.1093/jmedent/30.1.199. [DOI] [PubMed] [Google Scholar]
- 19.de Silva A M, Fikrig E, Hodzic E, Kantor F S, Telford S R, 3rd, Barthold S W. J Infect Dis. 1998;177:395–400. doi: 10.1086/514200. [DOI] [PubMed] [Google Scholar]
- 20.Fikrig E, Barthold S W, Kantor F S, Flavell R A. Science. 1990;250:553–556. doi: 10.1126/science.2237407. [DOI] [PubMed] [Google Scholar]
- 21.Mbow M L, Gilmore R D, Jr, Titus R G. Infect Immun. 1999;67:5470–5472. doi: 10.1128/iai.67.10.5470-5472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piesman J. J Med Entomol. 1995;32:519–521. doi: 10.1093/jmedent/32.4.519. [DOI] [PubMed] [Google Scholar]
- 23.de Silva A M, Fikrig E. Am J Trop Med Hyg. 1995;53:397–404. doi: 10.4269/ajtmh.1995.53.397. [DOI] [PubMed] [Google Scholar]
- 24.Golde W T, Burkot T R, Sviat S, Keen M G, Mayer L W, Johnson B J, Piesman J. J Exp Med. 1993;177:9–17. doi: 10.1084/jem.177.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gern L, Schaible U E, Simon M M. J Infect Dis. 1993;167:971–975. doi: 10.1093/infdis/167.4.971. [DOI] [PubMed] [Google Scholar]
- 26.Brunet L R, Sellitto C, Spielman A, Telford S R., 3rd Infect Immun. 1995;63:3030–3036. doi: 10.1128/iai.63.8.3030-3036.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leuba-Garcia S, Martinez R, Gern L. Zentralbl Bakteriol. 1998;287:475–484. doi: 10.1016/s0934-8840(98)80187-4. [DOI] [PubMed] [Google Scholar]
- 28.Sonenshine D E. Biology of Ticks. Vol. 1. New York: Oxford Univ. Press; 1991. pp. 119–158. [Google Scholar]
- 29.Gilmore R D, Jr, Piesman J. Infect Immun. 2000;68:411–414. doi: 10.1128/iai.68.1.411-414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwan T G, Simpson W J. Scand J Infect Dis Suppl. 1991;77:94–101. [PubMed] [Google Scholar]
- 31.Schwan T G. Infect Agents Dis. 1996;5:167–181. [PubMed] [Google Scholar]
- 32.Casjens S, Palmer N, van Vugt R, Huang W M, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson R J, et al. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 33.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, et al. Nature (London) 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 34.Akins D R, Caimano M J, Yang X, Cerna F, Norgard M V, Radolf J D. Infect Immun. 1999;67:1526–1532. doi: 10.1128/iai.67.3.1526-1532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X, Popova T G, Hagman K E, Wikel S K, Schoeler G B, Caimano M J, Radolf J D, Norgard M V. Infect Immun. 1999;67:6008–6018. doi: 10.1128/iai.67.11.6008-6018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryan J R, Levine J F, Apperson C S, Lubke L, Wirtz R A, Spears P A, Orndorff P E. Mol Microbiol. 1998;30:365–379. doi: 10.1046/j.1365-2958.1998.01071.x. [DOI] [PubMed] [Google Scholar]
- 37.Sung S Y, Lavoie C P, Carlyon J A, Marconi R T. Infect Immun. 1998;66:4656–4668. doi: 10.1128/iai.66.10.4656-4668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurtti T J, Munderloh U G, Johnson R C, Ahlstrand G C. J Clin Microbiol. 1987;25:2054–2058. doi: 10.1128/jcm.25.11.2054-2058.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.