Abstract
The forces stabilizing the three-dimensional structures of membrane proteins are currently not well understood. Previously, it was shown that a single Asn side chain in a transmembrane segment can mediate the dimerization and trimerization of a variety of hydrophobic helices. Here, we examine the tendencies of a representative set of amino acids (Asn, Gln, Asp, Glu, Lys, Ala, Val, Leu, Ser, Thr) to direct the oligomerization of a model transmembrane helix. The model peptide is entirely hydrophobic throughout a 20-residue segment and contains a single central site for the introduction of various amino acid “guests.” Analytical ultracentrifugation and gel electrophoresis were used to determine the stoichiometry and free energy of association of the entire set of peptides within micelles. Variants with two polar atoms at the guest site—Asn, Gln, Asp, and Glu—formed stable trimers, whereas residues with one or fewer polar atoms showed a much weaker tendency to associate. The data are examined in light of the frequencies of occurrence of various amino acid side chains in membrane proteins and provide insight into the role of polar interactions in directing transmembrane helix association. These data also suggest an approach to the design of variants of natural single-span transmembrane proteins with various potentials to associate in the bilayer.
The forces that drive membrane protein folding are not well understood. A reasonable model for membrane protein folding (1) posits that folding occurs in two kinetically separate steps. The first involves insertion of the helical regions of the protein in the bilayer, whereas the second involves the formation of specific interactions between these helices, to form a tightly packed native structure. The second step can occur in an intermolecular process as in the folding/assembly of multimeric ion channel proteins or in an intramolecular process, as in the folding of monomolecular proteins. The features required for the insertion of peptides into bilayers are largely hydrophobic in nature and have been quantified through various model systems (reviewed in ref. 2). However, the features required for the subsequent association of inserted helices are controversial and less well understood (3). One early study (4) suggested that the composition of the interiors of membrane proteins are similar to those of water-soluble proteins; both predominantly consist of well-packed apolar residues. Buried polar side chains occur less frequently but may be important for function, conformational specificity, and thermodynamic stability. A more recent study suggests that small residues such as Gly and Ala may additionally play an important role in the association of some transmembrane helix pairs (5–7), although this is not a universal phenomenon (8). Finally, the role of amino acid sequence in defining the orientation and topology of transmembrane helical segments has been defined by using a glycosylation mapping method (9–11).
To determine experimentally the features required for folding membrane proteins, we built a transmembrane peptide beginning with a water-soluble two-stranded coiled coil from GCN4 (GCN4-P1; ref. 12). This dimer is stabilized in aqueous solution by a series of hydrophobic interactions. Only one polar interaction occurs between side chains that are buried in the center of the structure, involving a hydrogen bond between the carboxamide groups of two Asn side chains on neighboring chains. Mutational studies indicate that this interaction is actually destabilizing relative to a hydrophobic interaction. However, it is important for specifying a dimeric state, relative to other aggregation states that are observed when the buried Asn is changed to hydrophobic residues (13, 14). Crystal structures of several variants of GCN4-P1 in trimeric and tetrameric aggregation states have been determined, providing an excellent system for comparing the effects of substitutions in water-soluble versus membrane-soluble proteins. Previously (15), we converted GCN4-P1 to a membrane-soluble peptide, MS1, by changing the exterior polar side chains of the water-soluble peptide to a combination of apolar side chains while maintaining its buried side chains invariant. The resulting peptide, MS1, was found to associate in nonionic detergent micelles and bilayers. In micelles, the association was a thermodynamically reversible monomer–dimer–trimer equilibrium, which favored formation of trimers at high peptide/detergent ratios. The Asn appeared critical for association: when this residue was replaced by Val, the corresponding peptide failed to associate. By contrast, the same mutation strongly favored the formation of trimers in the water-soluble parent peptide (14). Thus, polar interactions involving the transmembrane Asn residue appear essential for association of the peptide in micelles. An independent study by using transmembrane peptides fused to water-soluble protein domains came to a similar conclusion and additionally provided evidence for hydrogen bonding between the buried Asn side chain (16). Both studies also found that interactions between Asn side chains were the primary driving force for the interaction, with the packing of apolar side chains playing a less important energetic role in bilayers and micelles.
Here, we ask whether other polar side chains are able to similarly mediate the association of transmembrane helices. The transmembrane Asn-14 is therefore changed to Ser, Thr, and Gln as examples of neutral polar side chains. Asp Glu, and Lys were also examined as examples of ionizable residues. Finally, Ala, Val, and Leu were investigated as examples of small and large apolar side chains.
Materials and Methods
Peptide Synthesis and Purification.
MS1 (Fig. 1) was synthesized and purified according to Choma et al. (15). For five of the peptides, the synthesis was split into five portions before coupling the guest amino acid, sealed in “tea bags” (a gift from Enrique Perez Paya, University of Valencia, Valencia, Spain) (17) and the desired amino acid coupled manually. The bags were then returned to the reaction vessel, and the synthesis was completed on the peptide synthesizer. The N termini were labeled with 4-fluoro-7-nitrobenz-2-oxa-1,3-diazole [(NBD) fluoride; Molecular Probes] by stirring resin-bound peptide for 24 hours with a 3-fold molar excess of label in dimethylformamide with sufficient N-methylmorpholine to make the reaction mix slightly basic.
Figure 1.
Peptide sequences.
All SDS electrophoresis was performed by using 4% acrylamide in the stacking gel and 20% acrylamide in the resolving gel. The gel was stained with Colloidal blue (NOVEX, San Diego) to show all protein bands. Samples contained ≈5 μg peptide in 4% SDS in Tris pH 6.8 buffer. Running buffer was 1% SDS Tris/tricine, pH 8.25, buffer. The NBD-labeled peptides were used because they stained better than the unlabeled peptides.
Analytical Ultracentrifugation.
The concentration of the NBD-labeled peptide was determined by the absorbance of ethanolic solutions (NBD: ɛ458 = 21,000). Ethanolic solutions of the NBD-labeled peptide plus C14-betaine detergent were dried under reduced pressure, then dissolved in 100 mM sodium phosphate buffer, pH 7.0, with 13% D2O present to adjust for the detergent density (18). Sedimentation equilibrium experiments were performed at 25°C by using a Beckman (Beckman Coulter) XL-I analytical ultracentrifuge, as described (19). The samples were centrifuged in three-compartment carbon–epoxy centerpieces with sapphire windows for lengths of time sufficient to achieve equilibrium. Data obtained by absorbance at 470 nm were analyzed by nonlinear least-squares curve fitting of radial concentration profiles by using the Marquardt–Levenberg algorithm implemented in Igor Pro (WaveMetrics, Lake Oswego, OR) with a user-defined function coding the equations describing reversible association in centrifugation. The monomeric molecular masses and partial specific volumes were calculated by using the program sedinterp (20), modified to include revised values for individual amino acid residues (21) and corrected for hydrogen–deuterium exchange by using averaged H-D exchanged amino acid residue weights. The calculated values of these parameters were held constant in fitting the absorbance versus radius profiles to various equilibrium models.
A monomer–trimer equilibrium model provides a very good fit to the sedimentation curves for MS1 (15) and most of the variants. However, the curves for MS1-N14E and MS1-N14D indicated that they were forming higher-order aggregates as well as trimers. The correction associated with the higher-order aggregate is considerable for MS1-N14E and even more substantial for MS1-N14D. Inclusion of an electrolyte (0.25 M NaCl) reduced the formation of higher-order aggregates in both cases, while having very little effect on the trimerization constant for MS1. Also, as the pH was lowered from 8.0 to 6.0, the higher-order aggregation of the MS1-N14E and MS1-N14D peptides reached nearly undetectable levels (published as supplemental data, Figs. 8 and 9, on the PNAS web site, www.pnas.org). Thus, it is likely that the trimers involve a neutral protonated Asp or Glu side chain, whereas the higher-order aggregates arise from surface-absorbed peptide with the Glu or Asp side chains in an ionized state. The negatively charged side chain might associate with the positively charged residues of neighboring helices, leading to the formation of aggregates. Here, we report sedimentation of MS1-N14E and MS1-N14D at neutral pH in the presence of 0.25 M NaCl. Under these conditions, the curves for MS1-N14E conformed well to a monomer–trimer equilibrium, whereas MS1-N14D required a monomer–trimer–hexamer scheme to describe adequately the data (supplemental data). At a peptide/detergent ratio of 1:1,000, equal amounts of trimer and hexamer were present in MS1-N14D including only 10% monomer.
The degree of association of MS1-N14A, MS1-N14V, MS1-N14L, MS1-N14T, and MS1-N14S was relatively small under accessible peptide/detergent ratios, making it difficult to determine uniquely the association state of the aggregated form. Therefore, for sake of uniformity of analysis, these curves were also analyzed by using a monomer–trimer equilibrium, giving a very good fit to the data. A monomer–tetramer association scheme also fit the data for these mutants, but the difference in ΔΔG (reported in Fig. 5), when expressed on a per-monomer basis was the same within experimental error.
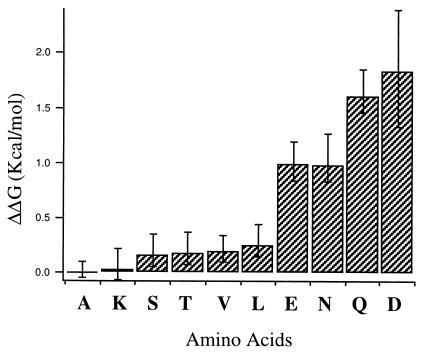
Figure 5.
Histogram showing the differences in free energies of trimerization (−ΔΔG in kcal/mol per monomer) for each of the variants relative to the MS1-N14A variant. Error bars reflect the uncertainties in free energies as obtained by a sensitivity analysis of the analytical ultracentrifugation data. Because the ΔΔG values are calculated as differences from alanine, the alanine variant errors were added to each of the others.
Structural Analysis of Transmembrane Helices.
A total of 1,118 residues distributed over 62 transmembrane helices in 7 membrane proteins were examined. The dataset of Stevens and Arkin (3) was used with one exception; helix I of 1occ was reported to span from residues 21 to 53, but examination of the structure suggested a more typical length of 18 residues (34 to 52). To characterize the exposure of the side chains in the structures, a probe-accessible surface was calculated by using a probe radius of 1.5 Å and compared with an appropriate standard for the “unfolded state.” In the two-state model, the unfolded state is considered an isolated helix. Therefore, the measured area was compared with the corresponding value for the same residue type in a polyAla helix [the most frequent rotamer for the side chain in a helical conformation (22, 23) was used in this latter calculation]. If the solvent accessibility for the residue in the transmembrane segments was less than 20% of the exposed surface area of that residue in the polyalanine helix, the side chain was considered buried. In each case, probe accessibility was calculated by using the biologically relevant association state of the protein, although only one subunit of each different chain was included for compiling the statistical frequencies of occurrence of the various amino acid types.
Results
SDS/PAGE.
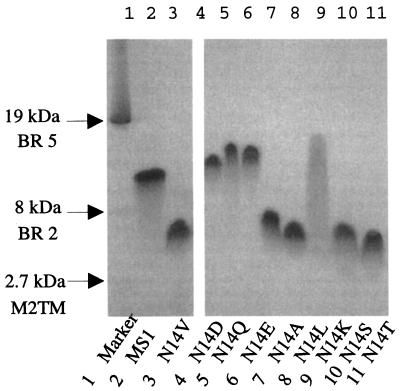
SDS/PAGE has been used extensively to assess the degree of association of various membrane peptides, such as the transmembrane domain of phospholamban (24) and glycophorin (25). Therefore, this method was initially used to determine the association of the different MS1 variants. MS1 migrates with an apparent molecular weight approximately equal to that of the trimeric state of the peptide, whereas MS1-N14V migrates with a mobility consistent with a monomeric state (Fig. 2). The variants in which the central Asn was replaced with Asp (MS1-N14D), Gln (MS1-N14Q), and Glu (MS1-N14E) show mobilities similar to MS1. The peptides containing a central Ala (MS1-N14A), Leu (MS1-N14L), Ser (MS1-N14S), and Thr (MS1-N14T) run in a similar fashion to MS1-N14V, indicating that these peptides are monomeric in SDS. The peptide N14K smears on the gel, possibly because of interactions between the charged Lys and the SDS micelles.
Figure 2.
SDS/PAGE of MS1 and variants. Lane 1 is a membrane standard consisting of the 19- and 8-kDa fragments of bacteriorhodopsin and a 2.7-kDa fragment from the M2 proton channel from influenza A virus (15). Lanes 2 and 3 contain MS1 and MS1-N14V, which migrate with molecular weights similar to that expected for a trimer and monomer, respectively. Lane 4 (MS1-N14D), Lane 5 (MS1-N14Q), and Lane 6 (MS1-N14E) all migrate similarly to MS1 at approximately the trimeric MW. Lanes 7, 8, 10, and 11 (MS1-N14A, MS1-N14L, MS1-N14S, MS1-N14T) all migrate as apparent monomers. Lane 9 contains the N14K peptide.
Analytical Ultracentrifugation.
All of the peptides except N14K run as single bands in SDS/PAGE, denoting a single association state. In nonionic detergent micelles, however, MS1 exists in a monomer–n-mer equilibrium (15). The association of peptides in SDS micelles requires days to equilibrate (26). Therefore, a more quantitative experimental method was necessary to determine the association states of the MS1 variants.
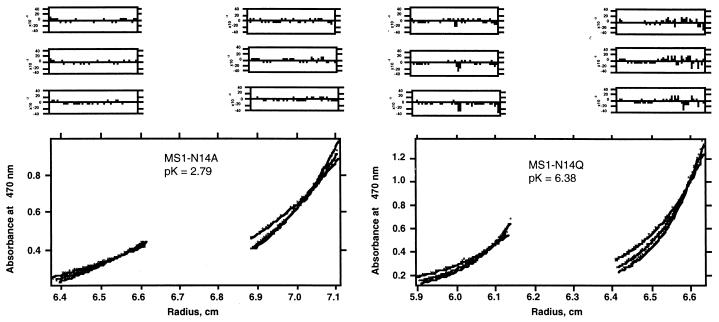
Analytical ultracentrifugation and fluorescence quenching experiments have shown that MS1 associates to form trimers in a variety of micellar systems including C12E8, DPC, SDS, and C14-betaine micelles (15). Therefore, analytical ultracentrifugation was used to examine the association of the variants in micelles of the detergent C14-betaine, chosen for its chemical stability and moderately low critical micelle concentration (≈0.1 mM). To eliminate the contributions of the micelles to the sedimentation of the peptides, solvent density was adjusted by using D2O to match the density of the detergent (18). Analysis of the sedimentation curves under these conditions provides the buoyant molecular weight of only the peptide component. The equilibrium sedimentation curves were well described by a monomer–trimer equilibrium (Fig. 3). The variant MS1-N14A is primarily monomeric at all accessible peptide/detergent ratios, whereas MS1-N14Q shows an even greater tendency to trimerize than MS1.
Figure 3.
Analytical ultracentrifugation of NBD-MS1-N14A and NBD-MS1-N14Q in C14-betaine micelles. The peptides at different peptide-to-detergent ratios (1:150 and 1:300 molar ratios) in 100 mM sodium phosphate buffer, pH 7.0, 13% D2O, were centrifuged at 40, 45, and 48 K. A monomer–trimer equilibrium scheme was used to fit curves to the experimental data. Residuals of curve fits for each individual data set are shown above the data.
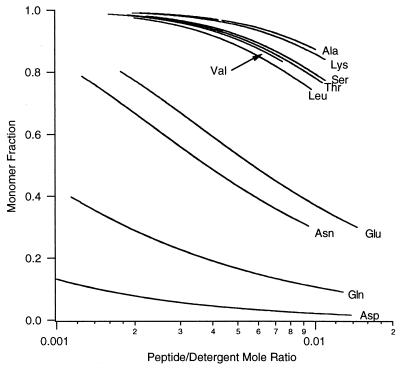
Fig. 4 illustrates the computed fraction of monomer as a function of the mol fraction of the peptide in the micellar phase. The differences in the free energy of trimerization (on a per-peptide basis) of the variants relative to MS1-N14A are presented in Fig. 5. Variants with carboxamide- or carboxylic acid-containing side chains at position 14 (Asp, Asn, Glu, Gln) have a high tendency to associate, whereas the remaining peptides show much weaker association. N14K, which was not well behaved in SDS/PAGE, was clearly demonstrated to be predominantly monomeric in the zwitterionic C14-betaine detergent micelles.
Figure 4.
The variation in monomer fraction with peptide/detergent mol ratio for each of the peptide variants studied in this work. Curves were calculated by using dissociation constants determined by curve-fitting analytical ultracentrifugation data.
Amino Acid Distribution in Membrane Proteins.
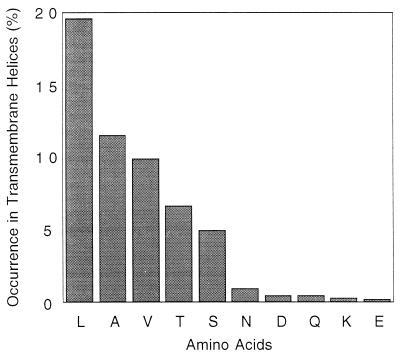
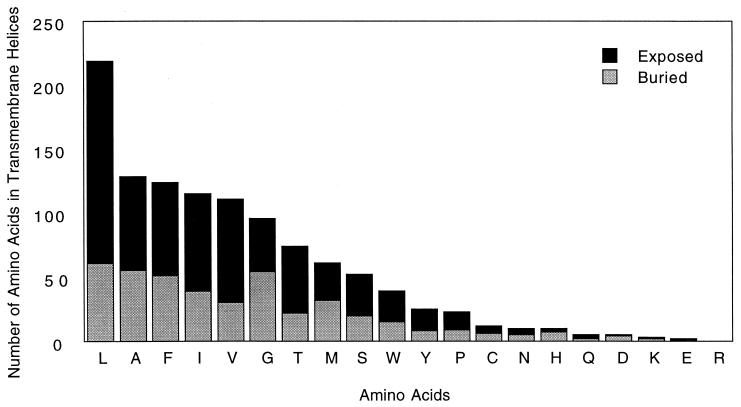
To determine whether the trends measured in our model system accurately reflect trends in natural helical membrane proteins, we measured the frequency of occurrence of the amino acid side chains in the transmembrane segments of membrane proteins available from the protein data bank (27). Asn, Asp, Gln, and Glu would be expected to occur infrequently on the surface of a membrane protein; otherwise, they would cause oligomerization (15, 16). In a sample of 1,118 transmembrane helical residues, these 4 side chains occur only 22 times. As expected, those that occur at buried positions inevitably hydrogen bond to the backbone of neighboring helices and/or polar side chains. Visual inspection of the 10 exposed side chains from this group shows that they are all located near the ends of the helices and that their side chains tend to project away from the hydrophobic portion of the transmembrane segment (i.e., toward the headgroup region of the bilayer; Table 1; Figs. 6 and 7). For example, in the recent crystal structure of rhodopsin, there is a highly conserved Asn whose side chain is involved in a hydrogen bond with an Asp and a backbone carbonyl, as well as another Asn in which both polar atoms are involved in a hydrogen-bonding network (28).
Table 1.
Summary of occurrence for all 20 amino acids in the transmembrane segments of the 7 helical proteins examined
| Amino acid | Buried | Exposed | Total | Fraction buried |
|---|---|---|---|---|
| Ala | 55 | 74 | 129 | 0.43 |
| Arg | 0 | 0 | 0 | |
| Asn | 5 | 5 | 10 | |
| Asp | 4 | 1 | 5 | |
| Cys | 6 | 6 | 12 | 0.50 |
| Gln | 2 | 3 | 5 | |
| Glu | 1 | 1 | 2 | |
| Gly | 54 | 42 | 96 | 0.56 |
| His | 7 | 3 | 10 | |
| Ile | 39 | 76 | 115 | 0.34 |
| Leu | 61 | 158 | 219 | 0.28 |
| Lys | 2 | 1 | 3 | |
| Met | 32 | 29 | 61 | 0.52 |
| Phe | 51 | 73 | 124 | 0.41 |
| Pro | 9 | 14 | 23 | 0.39 |
| Ser | 20 | 32 | 55 | 0.36 |
| Thr | 22 | 52 | 74 | 0.30 |
| Trp | 15 | 24 | 39 | 0.38 |
| Tyr | 8 | 17 | 25 | 0.32 |
| Val | 30 | 81 | 111 | 0.27 |
When at least 12 side chains were present, the fraction buried was calculated and is shown along with the total number of occurrences.
Figure 6.
Frequency of side chains in transmembrane regions of protein structures deposited in the Protein Data Bank. The proteins included bacteriorhodopsin, the photosynthetic reaction center, the KCSA K channel, glycophorin, the light-harvesting complex, cytochrome C oxidase, and the cytochrome bc1 complex. Transmembrane regions were defined according to Stevens and Arkin (3).
Figure 7.
Frequency of side chains exposed to lipid and buried within the protein structure for all 20 amino acids in 7 helical transmembrane protein structures deposited in the Protein Data Bank.
Ser and Thr are also small polar side chains, but they failed to lead to transmembrane peptide association in variants of MS1. Therefore, it was of considerable interest that they occur in much greater frequency than Asn, Asp, Gln, and Glu in both buried as well as exposed locations. At exposed positions, Ser and Thr taken together occur 8.4-fold more frequently than the sum of the frequencies of Asn, Asp, Gln, and Glu. The corresponding value for buried locations is 3.8. Also, Leu Ala, and Val, which failed to direct strongly oligomerization of variants of MS1, were among the most frequently occurring of amino acids. Lys occurs only three times in the population.
Discussion
Previously, it has been shown that a single Asn in a transmembrane helix provides a strong driving force for dimer or trimer formation (15, 16). Here we extend these studies to examine the potential of other commonly occurring amino acids to direct the oligomerization of transmembrane helices. Within the context of the MS1 peptide, Gln is shown to have an even higher potential than Asn to induce the formation of trimers. The side chain of Gln is one methylene longer than Asn, possibly providing additional flexibility for forming optimal hydrogen bonds within the interior of the trimer. Both Asn and Gln often occur in buried locations of water-soluble three-stranded coiled coils, where they form hydrogen bonds to one another, a buried water molecule, and/or chloride ions (29–33).
Because trimerization occurs in a region of low dielectric, it is likely that the carboxyl-containing side chains Asp and Glu are protonated. Consistent with this observation, the fit of a monomer–trimer equilibrium becomes increasingly poor as the pH is increased (supplemental data). Similarly, in a variant of GCN4-P1, in which a buried Asn was replaced by Asp, association was linked to protonation of the carboxyl group (33). It has long been known that carboxylic acids form strong hydrogen bonds, particularly in solvents of low dielectric (34, 35). Finally, it has previously been shown that the introduction of a Glu into the transmembrane helix of the neu oncogene protein product leads to the formation of oligomers (36, 37). Again, the formation of the transmembrane oligomeric form appears to be linked with protonation of the Glu side chain (36).
Although Ser and Thr are also small polar amino acids, they failed to direct trimer formation in variants of MS1. Their side chains contain a single polar hydroxyl group, whose proton can hydrogen bond back to a carbonyl oxygen of a residue preceding it by one helical turn (38). An analogous hydrogen bond does not occur frequently for Asn or Gln residues when located within an α helix (38). Thus, Ser and Thr behave more like apolar amino acids when in a transmembrane helix (39).
The incorporation of Lys into MS1 led to a peptide that failed to show a strong tendency to form trimers. Like Asp and Glu, its pKa is approximately 3 logs removed from the experimental pH of 7.0 (the pKas of Asp and Glu are near 4, whereas the pKa of Lys is approximately 10). Thus, it would appear to be thermodynamically feasible to deprotonate the Lys side chain in the low dielectric of the membrane in a manner analogous to Asp and Glu. However, the failure of N14K to form oligomers suggests that the deprotonated Lys side chain has a lower tendency to form stabilizing intermolecular interactions, possibly because its side chain has only one polar atom with which to form such interactions, whereas Asp and Glu have two. Thus acetic acid is a liquid, whereas methylamine is a gas. Further, the formation of specific hydrogen-bonded interactions that involve a Lys side chain would require freezing five torsional angles of its long side chain, resulting in an unfavorable entropy of interaction. By contrast, only two and three torsional angles are required to specify the conformation of Asp and Glu, respectively.
The aliphatic variants of MS1 with Ala, Val, and Leu failed to associate strongly in the membrane. Modeling and crystallographic studies of the corresponding water-soluble structures suggest that MS1-N12V and MS1-N12L could form densely packed trimers. In an aqueous environment, this interaction is highly favored, but the lack of a hydrophobic effect in a membrane-like environment substantially decreases the driving force for trimerization. Presumably, any improvement in van der Waals interactions that occurs in trimerization is largely balanced by an unfavorable decrease in entropy of the amino acid side chains. Of these variants, MS1-N14A showed the lowest tendency to associate, possibly because its small side chain would lead to the formation of voids in the helix–helix interface. However, high-resolution structural information will be necessary to confirm this conclusion. Interestingly, small side chains have been shown to mediate the interaction of some transmembrane helices (5). Our findings indicate that they need to be placed in the proper context to be effective.
The frequencies with which different types of amino acids occur in the transmembrane helical portions of membrane proteins of known structure can be rationalized largely on the basis of our experimental measurements. Two features need to be considered that contribute to the observed frequencies: first, a transmembrane helix must partition into the bilayer, which tends to maximize the number of hydrophobic side chains and minimize polar residues, particularly those bearing side chains that are charged at neutral pH. However, the requirement for function and folding dictates the occasional need for polar side chains. This tradeoff between partitioning versus folding and function is apparent in a comparison of the frequencies of occurrence of Asn and Gln in the population of type I single-span membrane proteins (0.2%; ref. 40) versus the multispan membrane proteins (1.3%) studied in this work. The corresponding values for Asp plus Glu are 0.2% in single-span proteins versus 0.7% in multispan proteins. Thus the requirement for folding and function leads to an expansion of the number of these polar side chains in the transmembrane region. Indeed, when Asp, Asn, Glu, and Gln occur in multispan proteins, they are found either in the headgroup region or in the interior of the protein, where they appear to be essential for folding, proton translocation activity, or other functional roles. By contrast, the presence of even a single Asn or Gln in a single-span membrane protein might potentially lead to deleterious oligomerization, as discussed previously (16).
In conclusion, we have determined the potential of a representative collection of amino acids to mediate association of transmembrane helices in membranes. The results show a clear distinction between side chains with a single polar heavy atom (Ser, Thr, Lys) versus side chains with two polar atoms (Asp, Asn, Glu, Gln). An important issue for future studies will be to determine the extent to which differences between Asn, Gln, Glu, and Asp are specific to the MS1 peptide or instead reflect intrinsic differences between these side chains. Our experimental data also help explain the observed frequencies of occurrence of side chains in transmembrane segments, which encourages one to believe that both measures reflect simple physicochemical properties of the side chains. Finally, the availability of these data should allow one to design variants of single-span transmembrane proteins with defined tendencies to associate. Such experiments may prove useful in the study of the mechanisms of signal transduction.
Supplementary Material
Acknowledgments
We are grateful for the initial studies and insights of Christin Choma. Kim A. Sharp and Christopher M. Summa were of great help in the analysis of the membrane protein structures. This work was supported by National Institutes of Health Grant 56423 and by the Materials Research Science and Engineering Centers program of the National Science Foundation, award no. DMR 96–32598.
Abbreviation
- NBD
4-fluoro-7-nitrobenz-2-oxa-1,3-diazole
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Popot J L, Gerchman S E, Engelman D M. J Mol Biol. 1987;198:655–676. doi: 10.1016/0022-2836(87)90208-7. [DOI] [PubMed] [Google Scholar]
- 2.White S H, Wimley W C. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 3.Stevens T J, Arkin I T. Proteins. 1999;36:135–143. doi: 10.1002/(sici)1097-0134(19990701)36:1<135::aid-prot11>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 4.Rees D C, DeAntonio L, Eisenberg D. Science. 1989;245:510–513. doi: 10.1126/science.2667138. [DOI] [PubMed] [Google Scholar]
- 5.Eilers M, Shekar S C, Shieh T, Smith S O, Fleming P J. Proc Natl Acad Sci USA. 2000;97:5796–5801. doi: 10.1073/pnas.97.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Javadpour M M, Eilers M, Groesbeek M, Smith S O. Biophys J. 1999;77:1609–1618. doi: 10.1016/S0006-3495(99)77009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russ W P, Engelman D M. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 8.Bowie J U. J Mol Biol. 1997;272:780–789. doi: 10.1006/jmbi.1997.1279. [DOI] [PubMed] [Google Scholar]
- 9.Braun P, von Heijne G. Biochemistry. 1999;38:9778–9782. doi: 10.1021/bi990923a. [DOI] [PubMed] [Google Scholar]
- 10.Monne M, Nilsson I, Johansson M, Elmhed N, von Heijne G. J Mol Biol. 1998;284:1177–1183. doi: 10.1006/jmbi.1998.2218. [DOI] [PubMed] [Google Scholar]
- 11.Monne M, Nilsson I, Elofsson A, von Heijne G. J Mol Biol. 1999;293:807–814. doi: 10.1006/jmbi.1999.3183. [DOI] [PubMed] [Google Scholar]
- 12.O'Shea E K, Klemm J D, Kim P S, Alber T. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 13.Harbury P B, Zhang T, Kim P S, Alber T. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 14.Harbury P B, Kim P S, Alber T. Nature (London) 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- 15.Choma C, Gratkowski H, Lear J D, DeGrado W F. Nat Struct Biol. 2000;7:161–166. doi: 10.1038/72440. [DOI] [PubMed] [Google Scholar]
- 16.Zhou F X, Cocco M J, Russ W P, Brunger A T, Engelman D M. Nat Struct Biol. 2000;7:154–160. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
- 17.Pinilla C, Appel J R, Houghten R A. Methods Mol Biol. 1996;66:171–179. doi: 10.1385/0-89603-375-9:171. [DOI] [PubMed] [Google Scholar]
- 18.Tanford C, Nozaki Y, Reynolds J A, Makino S. Biochemistry. 1974;13:2369–2376. doi: 10.1021/bi00708a021. [DOI] [PubMed] [Google Scholar]
- 19.Kochendoerfer G G, Salom D, Lear J D, Wilk-Orescan R, Kent S B, DeGrado W F. Biochemistry. 1999;38:11905–11913. doi: 10.1021/bi990720m. [DOI] [PubMed] [Google Scholar]
- 20.Laue T, Shaw B D, Ridgeway T M, Pelletier S L. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding S E, Rowe A J, Horton J C, editors. Cambridge, U.K.: R. Soc. Chem.; 1992. pp. 90–125. [Google Scholar]
- 21.Kharakoz D P. Biochemistry. 1997;36:10276–10285. doi: 10.1021/bi961781c. [DOI] [PubMed] [Google Scholar]
- 22.Dunbrack R L, Jr, Karplus M. Nat Struct Biol. 1994;1:334–340. doi: 10.1038/nsb0594-334. [DOI] [PubMed] [Google Scholar]
- 23.McGregor M J, Islam S A, Sternberg M J. J Mol Biol. 1987;198:295–310. doi: 10.1016/0022-2836(87)90314-7. [DOI] [PubMed] [Google Scholar]
- 24.Simmerman H K, Kobayashi Y M, Autry J M, Jones L R. J Biol Chem. 1996;271:5941–5946. doi: 10.1074/jbc.271.10.5941. [DOI] [PubMed] [Google Scholar]
- 25.Lemmon M A, Flanagan J M, Hunt J F, Adair B D, Bormann B J, Dempsey C E, Engelman D M. J Biol Chem. 1992;267:7683–7689. [PubMed] [Google Scholar]
- 26.Reddy L G, Jones L R, Thomas D D. Biochemistry. 1999;38:3954–3962. doi: 10.1021/bi981795d. [DOI] [PubMed] [Google Scholar]
- 27.Berman H M, Westbrook J, Feng Z, Gilliland G, Bhat T N, Weissig H, Shindyalov I N, Bourne P E. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palczewski K, Kumasaka T, Hori T, Behnke C A, Motoshima H, Fox B A, Le Trong I, Teller D C, Okada T, Stenkamp R E, et al. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 29.Baker K A, Dutch R E, Lamb R A, Jardetzky T S. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 30.Eckert D M, Malashkevich V N, Kim P S. J Mol Biol. 1998;284:859–865. doi: 10.1006/jmbi.1998.2214. [DOI] [PubMed] [Google Scholar]
- 31.Fass D, Harrison S C, Kim P S. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 32.Ji H, Bracken C, Lu M. Biochemistry. 2000;39:676–685. doi: 10.1021/bi991893e. [DOI] [PubMed] [Google Scholar]
- 33.Schneider J P, Lombardi A, DeGrado W F. Folding Des. 1998;3:R29–R40. doi: 10.1016/S1359-0278(98)00011-X. [DOI] [PubMed] [Google Scholar]
- 34.Pauling L, Brockway L O. Proc Natl Acad Sci USA. 1934;20:336–340. doi: 10.1073/pnas.20.6.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pauling L, Sherman J. Proc Natl Acad Sci USA. 1934;20:340–345. doi: 10.1073/pnas.20.6.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith S O, Smith C S, Bormann B J. Nat Struct Biol. 1996;3:252–258. doi: 10.1038/nsb0396-252. [DOI] [PubMed] [Google Scholar]
- 37.Weiner D B, Liu J, Cohen J A, Williams W V, Greene M I. Nature (London) 1989;339:230–231. doi: 10.1038/339230a0. [DOI] [PubMed] [Google Scholar]
- 38.Baker E N, Hubbard R E. Prog Biophys Mol Biol. 1984;44:97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- 39.Lemmon M A, Engelman D M. Q Rev Biophys. 1994;27:157–218. doi: 10.1017/s0033583500004522. [DOI] [PubMed] [Google Scholar]
- 40.Landolt-Marticorena C, Williams K A, Deber C M, Reithmeier R A. J Mol Biol. 1993;229:602–608. doi: 10.1006/jmbi.1993.1066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.