Abstract
Anaplasma phagocytophilum is the causative agent of an emerging tick-borne zoonosis in the United States and Europe. The organism causes a febrile illness accompanied by other nonspecific symptoms and can be fatal, especially if treatment is delayed. Persistence of A. phagocytophilum within mammalian reservoir hosts is important for ensuring continued disease transmission. In the related organism Anaplasma marginale, persistence is associated with antigenic variation of the immunoprotective outer membrane protein MSP2. Extensive diversity of MSP2 is achieved by combinatorial gene conversion of a genomic expression site by truncated pseudogenes. The major outer membrane protein of A. phagocytophilum, MSP2(P44), is homologous to MSP2 of A. marginale, has a similar organization of conserved and variable regions, and is also encoded by a multigene family containing some truncated gene copies. This suggests that the two organisms could use similar mechanisms to generate diversity in outer membrane proteins from their small genomes. We define here a genomic expression site for MSP2(P44) in A. phagocytophilum. As in A. marginale, the msp2(p44) gene in this expression site is polymorphic in all populations of organisms we have examined, whether organisms are obtained from in vitro culture in human HL-60 cells, from culture in the tick cell line ISE6, or from infected human blood. Changes in culture conditions were found to favor the growth and predominance of certain msp2(p44) variants. Insertions, deletions, and substitutions in the region of the genomic expression site encoding the central hypervariable region matched sequence polymorphisms in msp2(p44) mRNA. These data suggest that, similarly to A. marginale, A. phagocytophilum uses combinatorial mechanisms to generate a large array of outer membrane protein variants. Such gene polymorphism has profound implications for the design of vaccines, diagnostic tests, and therapy.
Anaplasma (Ehrlichia) phagocytophilum (8, 10) is a causative agent of granulocytic anaplasmosis. The organism infects humans as well as animals including dogs, cattle, sheep, deer, horses, and rodents and is transmitted by Ixodes species ticks (31, 36). Clinical disease associated with human infections is usually acute, although patients may have long-term adverse health outcomes such as recurrent fevers even after antibiotic treatment (34). Infections in ruminants and rodents can be persistent. A. phagocytophilum typically causes transient microscopically detectable infections in laboratory mice or in the white-footed mouse, Peromyscus leucopus. However, following this initial rickettsemia and even when infection is not microscopically apparent, blood from infected animals was still infective to naive animals (35). Splenectomy of mice with inapparent infections also caused reappearance of the rickettsiae in granulocytes within 1 week of the procedure (35). Persistence of rickettsiae within mammalian hosts may be an important factor allowing continuous disease transmission. The closely related pathogen Anaplasma marginale can persist in ruminant hosts for the lifetime of the animal (32). In cattle persistently infected with A. marginale, there are cyclic peaks of rickettsemia every 2 to 6 weeks (23) containing different variants of the immunoprotective major outer membrane protein MSP2 (11-13). These variants contain MSP2 epitopes recognized by antibodies appearing subsequently, but not prior, to the respective peak rickettsemia, suggesting that antigenic variation is responsible for persistence. MSP2 is encoded by a multigene family (11, 33), and sequence variation is achieved by segmental gene conversion of a single polycistronic expression site by different pseudogenes (3, 5, 32). These pseudogenes contain a hypervariable region and portions of flanking 5′ and 3′ conserved sequences but are otherwise truncated and cannot encode full-length MSP2 (4).
There are similarities at the molecular level between A. marginale and A. phagocytophilum. As in A. marginale infections, a dominant antibody response in patients infected with A. phagocytophilum is expressed against a variable ∼40-kDa outer membrane protein (20) [termed MSP2(P44) here]. This protein has different apparent molecular weights, reactivities with infection sera, and reactivities with MSP2(P44)-specific monoclonal antibodies in different strains (2, 24, 40). The gene encoding MSP2(P44) has been cloned from genomic DNAs of several strains of A. phagocytophilum (18, 30, 41). This gene, like msp2, is a member of a cross-hybridizing multigene family and is homologous to A. marginale msp2 (60 to 66% similarity and 40 to 53% identity, depending on the gene and the strain). Importantly, sequence alignment of different msp2(p44) variants and A. marginale msp2 reveals significant variation in the same central hypervariable region (CVR) (12). As in A. marginale, the A. phagocytophilum genome contains incomplete msp2(p44) genes with a unique CVR and conserved 5′ and 3′ flanking sequences (39) that could be a source of diversity for combinatorial recombination mechanisms. mRNA encoding MSP2(P44) is heterogeneous in populations of A. phagocytophilum, containing diverse hypervariable regions (7, 42).
Despite these similarities between the two organisms, a concept that is widely favored currently is that variation in msp2(p44) arises from the differential transcription of multiple paralogous genes interspersed in the A. phagocytophilum genome (7, 39, 43). Here we present evidence that this concept may not be correct and describe a polymorphic genomic expression site for MSP2(P44). This expression site transcribes the majority of different MSP2(P44) mRNAs observed in organisms grown in vitro in cultured HL-60 cells. The genomic expression site in A. phagocytophilum has several features similar to those of the genomic expression site in A. marginale. Taken together with the similar structures of the respective outer membrane proteins and the gene families encoding them, this suggests that comparable mechanisms are employed for the generation of outer membrane protein diversity in the two species.
MATERIALS AND METHODS
Growth of A. phagocytophilum in HL-60 cells.
The promyelocytic leukemia cell line HL-60 (CCL-240; American Type Culture Collection) was infected with A. phagocytophilum strain NY-18 (1, 17), graciously supplied by M. E. Aguero-Rosenfeld (New York Medical College, Valhalla, N.Y.), or strain Webster (2), generously provided by J. S. Dumler (The Johns Hopkins Medical Institutions, Baltimore, Md.). The cell line was propagated in RPMI 1640 medium containing 2 mM l-glutamine (Cellgro; Mediatech, Inc., Herndon, Va.) and 10% fetal bovine serum (Cellgro) at 37°C in the presence of 5% CO2, as described previously (37). Briefly, cultured cells were maintained at approximately 2 × 105 cells/ml. When cells were 70 to 100% infected, cell cultures were split at a 1:3 ratio (infected to uninfected cells). The percentage of infected cells was determined by microscopic examination of cytospin preparations stained with Wright-Giemsa stain. Cells were harvested when they were 90 to 100% infected.
RT-PCR and PCR.
Confluent HL-60 cells infected with A. phagocytophilum were centrifuged at 4°C and 150 × g for 10 min, washed in phosphate-buffered saline, and then resuspended in 5 to 6 volumes of RNAlater (Ambion, Austin, Tex.) for extraction of RNA. Cells were incubated overnight at 4°C in RNAlater and then at −20°C for about 8 h before long-term storage at −80°C. RNA was isolated from stored aliquots of ∼3 × 106 infected HL-60 cells by using the RNAqueous kit (Ambion), which yielded 7 to 15 μg of total RNA/aliquot. RNA was digested with DNase I (DNA-free; Ambion), followed by removal of DNase I with DNase Inactivation Reagent (Ambion). For reverse transcription (RT) reactions, 1 to 2 μg of RNA template was used per reaction with the Retroscript kit (Ambion). The complete msp2(p44) gene was RT-PCR amplified from RNA with RT primer AB1001 and PCR primers AB1000 and AB1005 by using Taq DNA polymerase (Perkin-Elmer, Wellesley, Mass.). These primers are located in the mRNA and in the genomic expression site in regions immediately flanking the 5′ and 3′ ends of the msp2(p44) coding sequence. For RT-PCR amplification of the CVR, primer AB943 (RT primer) and primers AB970 and AB976 (PCR primers), located in the 5′ and 3′ conserved sequences flanking the CVR, were used. RT-PCR amplification of the intercistronic region between p44ESup1 and msp2(p44) utilized AB974 as the RT primer, followed by nested PCRs with oligonucleotides AB1043 and AB1046 in the primary PCR and AB1045 and AB1047 in the secondary PCR. The locations of RT-PCR products are shown in Fig. 1. Controls always included reactions without reverse transcriptase and without template. PCRs were conducted similarly on A. phagocytophilum genomic DNA prepared from in vitro cultures (37) by using the Nucleospin nucleic acid purification kit (Clontech, Palo Alto, Calif.). The msp2(p44) gene in the expression site was PCR amplified with primers AB1000 and AB1001.
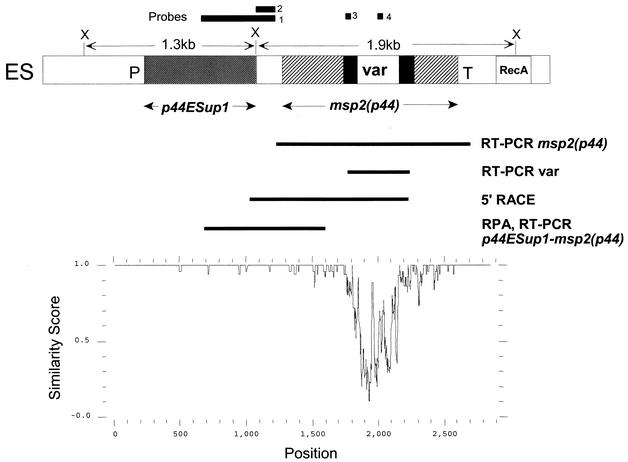
FIG. 1.
Structure and variability of a genomic expression site for msp2(p44) in A. phagocytophilum. (Top) Diagram indicating the location of msp2(p44) and an upstream gene (p44ESup1) within the expression site. Solid areas immediately flanking the variable region (var) represent sequence present in the expression site and in most genomic pseudogenes. The locations of RT-PCR and 5′-RACE products and of an RPA probe used to establish the structure of mRNA carrying msp2(p44) are indicated below the diagram. DNA probes used in Southern blots to verify locus structure are indicated above the diagram. ES, expression site; P, promoter sequence; T, terminator; RecA, a downstream sequence homologous to the recA gene; X, XbaI cleavage site. (Bottom) PLOTSIMILARITY graph, drawn to the same scale as the diagram above, demonstrating the variability of this expression locus in five different populations of A. phagocytophilum. These populations are the NY18 strain cultured in HL-60 cells, the Webster strain cultured in HL-60 cells, the HGE2 strain cultured in ISE6 tick cells (population II in Fig. 7), and two populations of A. phagocytophilum from infected human blood (patient 2, day 3, and patient 2, day 27 [Fig. 8]). A similarity score of 1.0 indicates identical sequence in a sliding window of 10 nucleotides, and a score decreasing from 1.0 to 0.0 indicates increasing variation.
5′-RACE.
For 5′ rapid analysis of cDNA ends (5′-RACE) (14), mRNA was reverse transcribed into cDNA by using primer AB943. After first-strand cDNA synthesis, the original mRNA template was removed by treatment with an RNase mix, and the cDNA was purified using a GLASSMAX Spin Cartridge according to the manufacturer's instructions (Invitrogen, Carlsbad, Calif.). A homopolymeric tail was then added to the 5′ end of the cDNA by using terminal deoxynucleotidyl transferase and dCTP (Invitrogen). An aliquot of the reaction product was directly amplified by PCR using SuperTaq (Ambion), a nested gene-specific primer (AB970), and a deoxyinosine-containing anchor primer (Invitrogen). Both the RT primer and the nested gene-specific primer are located in the 3′ conserved region flanking the CVR; therefore, 5′-RACE products should contain the CVR and the 5′ end of expressed msp2(p44). The PCR conditions used were as follows: an initial denaturation cycle of 2 min at 93°C, followed by 35 cycles of 30 s at 93°C, 1 min at 55°C, and 5 min at 72°C, and a final extension of 25 min at 72°C. Two control reactions were conducted similarly, one without reverse transcriptase in the initial RT reaction and another with no template in the PCR. The products of 5′-RACE were resolved by agarose gel electrophoresis, transferred to nylon membranes, and hybridized with the fluoresceinated oligonucleotide probe AB945. This probe is located in the 5′ conserved region flanking the CVR. Specific hybridization was obtained only in reactions containing reverse transcriptase and template. The 5′-RACE products were cloned into plasmid vector pCR-XL-TOPO (Invitrogen), and clones containing the 5′ conserved region of msp2(p44) were identified with probe AB945 and sequenced. 5′-RACE clones were of variable size and contained sequence extending up to approximately 1,200 bp from the 3′ conserved region flanking the CVR.
RNase protection assay (RPA).
An RNA probe was prepared from the cloned RT-PCR product p44ESup1-msp2(p44) encompassing the N-terminal coding region of msp2(p44), a C-terminal region of the upstream gene p44ESup1, and the intercistronic sequence (Fig. 1). The RT-PCR product was cloned into plasmid vector pCR4-TOPO (Invitrogen) and linearized prior to probe preparation with PvuII, which cuts within the plasmid vector sequence. Antisense, biotin-labeled RNA probes were prepared by in vitro transcription with T3 RNA polymerase. Probes were hybridized with varying quantities of A. phagocytophilum RNA, treated with RNase to degrade unprotected probe, and analyzed by electrophoresis on 5% polyacrylamide gels containing 8 M urea to determine the sizes of protected probe fragments (SuperSignal RPA III Chemiluminescent Detection Kit; Pierce, Rockford, Ill.). Controls included probe that was not digested with RNase and probe that was hybridized with equivalent amounts of yeast RNA and then digested. The sizes of protected probe fragments were determined by reference to molecular weight standards (biotinylated RNA Century Plus; Ambion). The PvuII-linearized RNA probe contained 907 bp of A. phagocytophilum sequence (Fig. 1) including 302 bp of N-terminal coding sequence from msp2(p44), 213 bp of intercistronic sequence, and 392 bp of C-terminal coding sequence of p44ESup1. The location of the major protected RNA fragment was confirmed by using a shorter probe terminating at the XbaI site at the C terminus of p44ESup1.
RFLP analysis of msp2(p44) genomic expression site clones.
RT-PCR and PCR products containing the complete msp2(p44) sequence amplified from the genomic expression site were cloned into the pCR4-TOPO vector and Escherichia coli TOP10 cells (Invitrogen). Individual colonies were grown overnight in 96-well deep blocks containing 1.5 ml of Luria-Bertani medium and kanamycin (50 μg/ml). Plasmid DNA was prepared from cultures by centrifuging the blocks at 1,100 × g for 15 min, resuspending cells in 400 μl of 50 mM Tris-HCl (pH 8.0)-10 mM EDTA-100 μg of RNase A/ml, adding 400 μl of 200 mM NaOH-1% sodium dodecyl sulfate, and inverting five times to lyse cells. Four hundred microliters of 3 M sodium acetate, pH 4.5, was added, and the plates were covered with sealing tape and inverted five times. The blocks were placed at −80°C for 1 to 2 h, then thawed and centrifuged for 30 min at 2,830 × g and 4°C. Nine hundred microliters of cleared supernatant was transferred to a 96-well filter plate (Unifilter; Whatman, Clifton, N.J.) on a new 96-well block and centrifuged for 3 to 4 min at 1,140 × g. An equal volume of isopropanol was added to each well, and the block was covered with sealing tape and inverted once or twice before being centrifuged for 45 to 60 min at 2,830 × g and 4°C. The supernatant was carefully decanted, and pellets of DNA were washed with 1 ml of 70% ethanol and air dried before resuspension in TE buffer (10 mM Tris-HCl-1 mM EDTA [pH 8.0]). All samples were digested with EcoRI to release insert DNA and with EcoRI and RsaI to analyze restriction fragment length polymorphism (RFLP) patterns. Digested DNA was resolved by electrophoresis on 1.5% agarose gels and visualized by staining with ethidium bromide. Clones were selected for DNA sequencing based on the RFLP patterns obtained.
Southern blotting.
A. phagocytophilum genomic DNA prepared from organisms cultured in vitro (37) was digested with XbaI, which cleaves on either side of the msp2(p44) gene in the expression site (Fig. 1), to release an approximately 1.9 kb fragment containing the gene. Digested DNA was separated by electrophoresis on a 1% agarose gel and transferred to nylon membranes for hybridization. Probes were either oligonucleotides synthesized with a 5′ fluorescein end label (Genosys Biotechnologies, The Woodlands, Tex.) or PCR-amplified products labeled with fluorescein-dUTP by using the Prime-It Fluor labeling kit (Stratagene, La Jolla, Calif.). Hybridization and detection were performed by methods described previously (3), by using a rabbit anti-fluorescein antibody conjugated to alkaline phosphatase and chemiluminescence detection of bound probe. Four probes were used in the hybridization studies: probe 1 (554 bp) was derived from amplification of NY18 genomic DNA with primers AB990 and AB1015, probe 2 (154 bp) was derived from amplification with primers AB990 and AB1044, probe 3 was fluorescein-labeled oligonucleotide AB945, and probe 4 was fluorescein-labeled oligonucleotide AB1016 (see Fig. 1 for locations of probes).
DNA sequencing.
Sequencing was performed at the University of Florida DNA Sequencing Core Laboratory (Gainesville) by using ABI Prism dye terminator cycle sequencing protocols developed by Applied Biosystems (Foster City, Calif.). The fluorescently labeled extension products were analyzed on an Applied Biosystems model 373 Stretch DNA Sequencer. Oligonucleotide primers were designed by using OLIGO 5.0 (Molecular Biology Insights, Cascade, Colo.) software and synthesized by Genosys Biotechnologies. Nucleotide sequences were analyzed by using the Wisconsin Package, version 10.3 (Accelrys Inc., San Diego, Calif.), available through the Biological Computing Core facilities of the Interdisciplinary Center for Biotechnology Research at the University of Florida. Sequence alignments were made by using PILEUP and GAP, and similarities were displayed by using PLOTSIMILARITY. Prokaryotic factor-independent RNA polymerase terminator sequences were predicted by using TERMINATOR. To obtain the sequence of an msp2(p44) expression site and flanking regions, the sequence present in mRNA was first obtained from 5′-RACE and RT-PCR clones from both strain Webster and strain NY18 RNA. This sequence was then extended (22, 38) in both 5′ and 3′ directions from strain NY18 genomic DNA by using unique 5′ flanking and CVR sequences to identify genomic loci capable of transcribing the observed mRNA. The sequence of a single genomic region matching the observed mRNA was obtained. This sequence was confirmed on both strands following PCR amplification of the entire locus with primers AB1041 and AB1042. For comparison, the sequence of this genomic locus was obtained from strains NY18 and Webster cultivated in HL-60 mammalian cells, from the HGE2 strain cultivated in ISE6 tick cells (29), and from organisms present in infected human blood.
Preliminary genome sequence data was obtained from The Institute for Genomic Research through the website at http://www.tigr.org.
A. phagocytophilum transferred from HL-60 to tick cells.
The growth of organisms in both the HL-60 human promyelocytic cell line and the ISE6 cell line from Ixodes scapularis has been described previously (21). The HGE2 strain (15) was passaged in HL-60 cells for 6 months and then was grown alternately in human and tick cells. HH represents the population of organisms grown continuously in HL-60 cells, and II represents the population of organisms grown continuously in tick cells. HI represents organisms that had been transferred from HL-60 to tick cells and had been established in tick cell culture, while HIH represents HI organisms transferred back to growth in HL-60 cells (21). A. phagocytophilum DNA was isolated from infected cultures passaged in the above four ways. The msp2(p44) gene from the genomic expression site was PCR amplified with oligonucleotide primers AB1000 and AB1001. PCR products were cloned into the pCR4-TOPO vector, and individual clones were analyzed by RFLP mapping and sequencing.
A. phagocytophilum from human blood.
DNA was extracted from samples of human blood that were submitted to the New York State Department of Health and were PCR positive for A. phagocytophilum, as defined by specific amplification of a 920-bp fragment of the 16S rRNA gene by primers HGE1F and HGE3R, as described elsewhere (9). Samples were collected from a total of six patients. For patient 1, we had three PCR-positive samples taken on days 0, 8, and 12 after the onset of clinical symptoms. For patient 2, we had two PCR-positive samples taken on days 3 and 27 postonset. Patient 2 was congenitally asplenic. This patient initially received a short course of doxycycline consisting of eight doses over 4 days (days 3 to 6 postonset), improved, and was discharged from the hospital for 19 days. The patient was readmitted to the hospital on day 27 with clinical symptoms consistent with a relapsing A. phagocytophilum infection. Only one sample each was available for patients 3 to 6, and these were taken on days 1, 3, 5, and 6 after the onset of clinical symptoms, respectively. Patients 1 and 3 to 6 appeared to be normal immunocompetent individuals. The msp2(p44) expression site locus was PCR amplified from all samples, and individual clones of the locus were analyzed by RFLP mapping and sequencing as described previously.
Oligonucleotides.
Oligonucleotides used as primers and probes were as follows (5′ to 3′; “F” stands for 5′ fluorescein): AB943, AAGAAGATCATAACAAGCATT; AB945, FGCTAAGGAGTTAGCTTATGATGTTGTTACTGGRCAGACTGATAA; AB970, CCTTCAATAGTYTTAGCTAGTAACCC; AB974, TCATAAGCTAACTCCTTAGC; AB976, GGAGTTAGCTTATGATGTTGTTA; AB990, GGCTAACCCCCTCTAACATCT; AB1000, CCGGCTGAAGTGAGGAGACGA; AB1001, AAGTACCGCAGGAAGTAGAAT; AB1005, TTAAAGTAGAAAAGGGGAGCC; AB1015, FTTCACTGCCGGAAAGAGTGGGGCTAAAGGAGAAGT; AB1016, FCCCGCGGGCCAAACGATACCACAGGTGCTAAAGGA; AB1041, ATGTCAGTACCGGCATATCTTGAAATC; AB1042, AACTGCTCAACAATAGACATTGAAGCC; AB1043, TGGGTATAGAGATAGAGGGAAGTGAG; AB1044, TCTAGAGAAAGATGTGCGTAAGAGG; AB1045, AGAGTGGGGCTAAAGGAGAAGTG; AB1046, CCACCAATACCATAACCAACACTAC; AB1047, ATGTTGTCCTTAAACCCAATCC.
Nucleotide sequence accession numbers.
The sequences reported here have been assigned GenBank accession numbers AY164490 to AY164513. AY164490 and AY164491 are the msp2(p44) genomic expression loci from strains NY18 and Webster cultured in HL-60 cells. AY164492 is the locus from the HGE2 strain cultured in ISE6 cells. AY164493 and AY164494 are the loci from patient-2 blood samples collected on day 3 and day 27 postonset, respectively. AY164495 to AY164508 are variable-region sequences from the genomic expression locus in blood samples from patients 1 to 6. AY164509 to AY164512 are variable-region sequences from the genomic expression locus in the HGE2 strain, variants HH1, HH2, HI1, and II1. AY164513 is a genomic pseudogene from the NY18 strain.
RESULTS
Structure of a genomic expression site for MSP2(P44).
Like msp2 of A. marginale, msp2(p44) genomic copies and transcribed mRNA contain a CVR flanked by 5′ and 3′ conserved sequences (Fig. 2). This was confirmed for strains NY18 and Webster of A. phagocytophilum by using oligonucleotide primers in the conserved regions to amplify the CVR from mRNA by RT-PCR. As expected, different clones of the RT-PCR products contained diverse CVR sequences. 5′-RACE was then used to obtain sequence from mRNA extending 5′ from the CVR. The locations of RT-PCR and 5′-RACE products are shown in Fig. 1. Six different 5′-RACE clones from strain NY18 and two from strain Webster were sequenced. From the sequenced clones we obtained mRNA sequence extending 5′ to the CVR sequence and indeed, by comparison with known gene sequences, 5′ to the msp2(p44) gene initiation codon. Interestingly, the two longest 5′-RACE clones from NY18 mRNA contained, at their 5′ ends, sequence identical to that of the two longest clones from Webster mRNA, even though the 5′-RACE clones contained different CVR sequences. These data suggested that different msp2(p44) mRNAs were possibly derived from the same genomic locus.
FIG. 2.
Conservation of msp2 sequence between A. marginale and A. phagocytophilum. The msp2E sequence is the predominant variant sequence encoded in the msp2 expression site of an acute bloodstream population of A. marginale strain Florida (GenBank accession number AF200925), and msp2pseud is encoded by a pseudogene present in genomic DNA of the same strain of A. marginale (accession number U60780). The msp2(p44)E sequence is the predominant variant sequence encoded in the msp2(p44) expression site of A. phagocytophilum strain NY18 (this study) grown in HL-60 cells, and msp2(p44)pseud is encoded by a pseudogene present in genomic DNA of the same strain of A. phagocytophilum (this study). Amino acids that are identical in all four sequences are capitalized. The N-terminal amino acid sequences of native MSP2 and MSP2(P44) are underlined.
Probing of genomic DNA with sequence present in mRNA 5′ to the initiation codon (Fig. 1 and 3, probe 2) indicated that this genomic region exists as a single copy in the genome. This was in contrast to results obtained with a probe from the 5′ conserved region immediately flanking the CVR (Fig. 1 and 3, probe 3), which hybridized with multiple members of the msp2(p44) multigene family. We sequenced genomic DNA from the potential expression site locus in strains NY18 and Webster, using sequence information from the 5′ end of mRNA to identify the locus. The sequence obtained encoded a complete msp2(p44) gene, including the CVR, flanking conserved sequences, and complete N and C termini. At the N terminus was a potential signal peptide (Fig. 2) followed by the sequence HDDVSAL(D/E)TG (HDDVSALDTG in strain NY18 and HDDVSALETG in strain Webster [data not shown]), corresponding to that obtained previously by Zhi et al. (39, 41) by N-terminal sequencing of the native protein. Upstream of the msp2(p44) gene (Fig. 1) was a second open reading frame encoding a polypeptide with a molecular weight of 29,329, p44ESup1, most closely related (BLASTP E value, 9e−23) to a polypeptide (OpAG3) in the analogous msp2 expression site (3) of A. marginale (Fig. 4A). Whereas opag3 is separated from msp2 by two open reading frames in A. marginale, p44ESup1 was immediately upstream of msp2(p44) in A. phagocytophilum. Approximately 80 bp upstream of p44ESup1 was a potential promoter region that closely resembled the promoter identified by primer extension analysis (3) of msp2 in A. marginale (Fig. 4B). Thirty of 31 bases were identical in the −35 and −10 predicted promoter regions in A. marginale and A. phagocytophilum, despite little similarity in regions immediately flanking the promoter. Downstream of msp2(p44) in the expression site (Fig. 1) was a predicted prokaryotic terminator sequence followed by sequence homologous to numerous bacterial RecA polypeptides (e.g., BLASTP E value with Brucella melitensis RecA, 3e−25). Comparison of the sequence of the complete expression site locus in strains NY18 and Webster revealed that, with the exception of the CVR, the sequence was largely conserved. Only one amino acid substitution was predicted in p44ESup1 between strains NY18 and Webster.
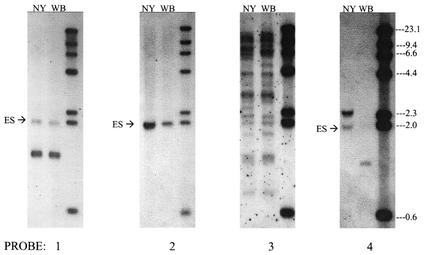
FIG. 3.
Verification of expression site structure by Southern blotting of genomic DNA. A. phagocytophilum DNA from either strain NY18 (NY) or strain Webster (WB) was digested with the enzyme XbaI to release the 1.9-kb fragment containing the expressed msp2(p44) gene (ES) and the 1.3-kb fragment containing p44ESup1. Digested and separated DNA was hybridized with probes 1 to 4 against different regions of the expression site (see Fig. 1 for locations of probes). Molecular weight markers are in the far right lane of each blot.
FIG. 4.
Comparison of outer membrane protein expression site structures in A. marginale and A. phagocytophilum. (A) Diagram showing the locations of expressed msp2 and msp2(p44) genes in the two organisms. Nomenclature for opag1 to opag3 of A. marginale is as in reference 27. P, predicted promoter; T, predicted prokaryotic terminator sequence; var, variable region. (B) Comparison of predicted promoter sequences in A. marginale and A. phagocytophilum expression sites.
Polymorphisms in the msp2(p44) expression site reflect similar polymorphisms in msp2(p44) mRNA.
An important question is whether expression from this genomic locus could yield the diverse species of msp2(p44) mRNA that are observed in A. phagocytophilum (7, 42). In A. marginale, different mRNAs are encoded from a single msp2 expression locus because the msp2 gene in this locus is continually undergoing segmental gene conversion by pseudogenes containing diverse CVR sequences (5). To answer this question, we compared the structures of multiple independent clones of the locus from genomic DNA with the structures of multiple independent clones reverse transcribed from msp2(p44) mRNA (Fig. 5). As expected, RFLP patterns of the genomic clones were diverse (Fig. 5A), showing that msp2(p44) in the expression site is very polymorphic. Similar RFLP patterns were obtained from clones derived from msp2(p44) mRNA, at corresponding frequencies. The sequences of the CVR region in the genomic clones closely matched CVR sequences in mRNA-derived clones (Fig. 5B), which were generated by three independent methods [RT-PCR amplifying the complete msp2(p44) gene, RT-PCR amplifying the CVR alone, and 5′-RACE from the 3′ conserved region (see Fig. 1)]. As in A. marginale (3), CVR sequences differed by multiple substitutions, insertions, and deletions, with some sequence blocks shared by otherwise different variants. For example, KDLVQELTPE at positions 124 to 133 (Fig. 5B) is shared by sequence variants of the expression locus in both strains Webster and NY18, but further upstream the major Webster strain variant (WebESDNAseqA [Fig. 5B]) diverges from the others. These types of changes would be consistent with a mechanism of segmental gene conversion.
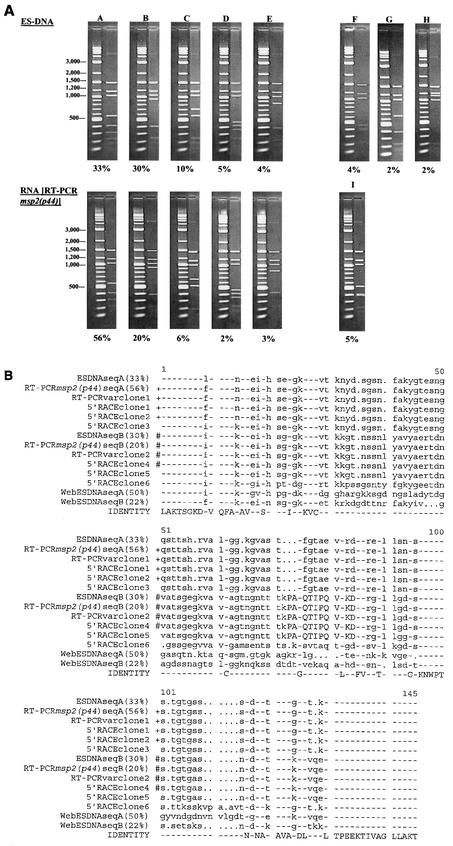
FIG.5.
Sequence diversity in mRNA encoding msp2(p44) reflects polymorphisms in the msp2(p44) copy within the genomic expression site. (A) RFLP analysis of expression site clones compared to RFLP analysis of msp2(p44) mRNA. Plasmid DNAs from 135 independent strain NY18 genomic expression site clones (ES-DNA) and 95 independent clones derived by RT-PCR from NY18 msp2(p44) mRNA were digested with EcoRI and RsaI and analyzed by agarose gel electrophoresis. Clones with identical digestion patterns were grouped together, and the frequencies of the major patterns were determined. Each digestion pattern (on the right) is shown next to a marker lane of molecular weight standards. The percentage of clones with each digestion pattern is shown below. Minor patterns representing <2% of the population are not shown; therefore, the sums of the percentages shown are <100%. The predominant variants, represented by clones A through E, have the same digestion patterns whether they are derived from DNA or from RNA. Patterns F through I, representing minor variants, were unique to clones derived from either DNA or RNA. (B) Sequence comparison of genomic expression site clones with clones derived from msp2(p44) mRNA. Individual expression site clones representing the predominant RFLP patterns A and B (Fig. 5A) were sequenced and aligned with the sequences of six 5′-RACE clones and two RT-PCRvar clones, also derived from strain NY18 msp2(p44) mRNA. For comparison, the two predominant genomic expression site sequence variants present in strain Webster (WebESDNAseqA and WebESDNAseqB) are included at the bottom of the alignment. The amino acids encoded by probe 4 (see Fig. 1 and 3), specific for the CVR sequence expressed in strain NY18, are capitalized (amino acids 73 to 84). Identical sequences are indicated by identical symbols (+ or #) to the left of the aligned sequences. Although not identical, the sequences of ESDNAseqA and 5′-RACE clone 5 each differed by only a single amino acid from the two groups of identical sequence variants. These changes could represent actual minor variation in mRNA species or a mutation occurring during in vitro amplification and cloning.
An oligonucleotide probe was designed based on sequence differences in the expression site encoding the CVR in strains NY18 and Webster (Fig. 5B). This probe should be specific for sequence found in the expression site in about 30% of the NY18 population. On Southern blots, the 1.9-kb XbaI band containing the expression site was detected only in the NY18 population (Fig. 3, probe 4), as expected, even though the 1.9-kb fragment from the locus was clearly present in strain Webster and could be revealed by using probes to conserved expression site sequence (Fig. 3, probes 1 and 2).
This expression site encodes the majority of full-length msp2(p44) mRNA.
RT-PCR using oligonucleotide primers located in the N-terminal coding region of msp2(p44) and the C terminus of p44ESup1 revealed a polycistronic mRNA of ∼900 bases encompassing the intergenic region (Fig. 6, RT-PCR). However, when this fragment was used to synthesize an RNA probe for RPA, only a minor proportion of the probe was fully protected at 900 bases (Fig. 6, RPA). The major protected probe band was ∼600 to 650 bases, containing the N-terminal coding sequence of msp2(p44) and the intergenic region between msp2(p44) and p44ESup1. These data suggested that, although there is polycistronic mRNA detectable by RT-PCR carrying both msp2(p44) and p44ESup1, the major species of mRNA carrying msp2(p44) present at steady-state levels in infected HL-60 cells extends through the intergenic region and XbaI site (Fig. 1) and no more than 150 bases into the coding region of p44ESup1. This could possibly be explained by differential stability of a polycistronic mRNA to endonucleolytic degradation during cellular processing (16) and agrees with the previous observation that 5′-RACE clones frequently terminate in the same sequence region (Fig. 1).
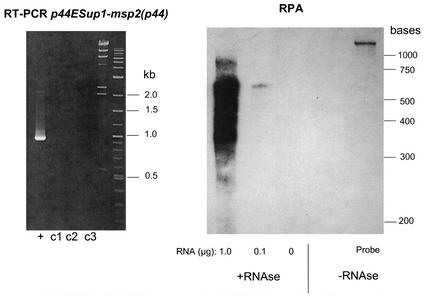
FIG. 6.
The major mRNA species encoding the N terminus of msp2(p44) in A. phagocytophilum-infected HL-60 cells also contains the msp2(p44)-p44ESup1 intergenic region. (Left) Ethidium bromide-stained gel of RT-PCRs amplifying the C-terminal region encoding p44ESup1, the N-terminal region encoding msp2(p44), and the intergenic region between them. +, RT-PCR; c1 to c3, control reactions containing either no reverse transcriptase enzyme, no template in primary PCR, or no template in secondary PCR, respectively. (Right) RPA using the cloned and sequenced 907-bp RT-PCR product to generate an antisense RNA probe. The complete RNA probe is ∼1,100 bases, as it also contains vector sequence that should not be protected by hybridization with A. phagocytophilum RNA. The quantity of protecting A. phagocytophilum RNA is indicated below the gel. Positions of molecular size standards are indicated on the right for both panels.
Variability of the expressed msp2(p44)gene during in vitro passage of A. phagocytophilum between human and tick cells.
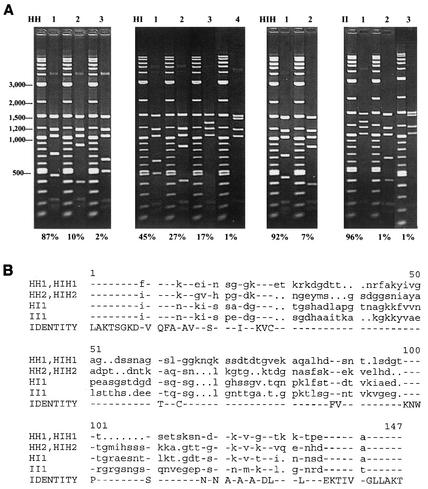
Jauron et al. (21) have maintained the HGE2 strain of A. phagocytophilum in parallel in the HL-60 human promyelocytic cell line and the ISE6 tick cell line from I. scapularis. They have also documented antigenic changes in MSP2(P44) when the organism was transferred between these cell lines. Importantly, expression of an MSP2(P44) epitope recognized by a monoclonal antibody was lost within 3 weeks of transfer of organisms from human to tick cells, whereas organisms continually grown in human cells continued to express the epitope (21). Moreover, when organisms were returned from tick to human cells, they regained expression of the same MSP2(P44) epitope. We analyzed the genomic expression site in these samples, by RFLP mapping and sequencing, to determine if there were sequence changes in msp2(p44) variants expressed from this locus that may correlate with the antigenic data (Fig. 7). As with previous populations of A. phagocytophilum analyzed, RFLP mapping showed multiple variant sequences present in all four samples. Growth in specific host cells and passage between host cells clearly influenced which variants were found and the proportions of each (Fig. 7A). Interestingly, the predominant variants present in organisms grown continuously in HL-60 cells (HH1 and HH2 [Fig. 7]), which represented approximately 97% of this population, were not detected after organisms had been transferred to tick cells but reappeared when organisms were transferred back to HL-60 cells and again became the predominant variants (HIH1 and HIH2 [Fig. 7]). Although all populations in both HL-60 and ISE6 cultures were diverse, the effects of these environmental changes were different for different variants. Growth of variants HI1 and II1 appeared to be favored in tick cells, similar to the increased representation of HH1 and HH2 in mammalian cells. These data correlate with the previous data showing changes in the expressed MSP2(P44) epitopes induced by transfer from HL-60 to tick cells (21).
FIG. 7.
The predominant msp2(p44) sequence variants in the expression site are different during in vitro growth in HL-60 and ISE6 cells. A. phagocytophilum was grown continuously in HL-60 (HH) or ISE6 (II) cells or transferred between them (HI, HIH). A total of 70 to 92 independent clones of the genomic expression site were prepared and analyzed from each of the four populations of A. phagocytophilum, as in Fig. 5. (A) RFLP analysis of expression site clones from each population. The percentage of clones with each digestion pattern is given below each gel. (B) Alignment of the CVRs of the msp2(p44) expression sites from the predominant sequence variants determined in panel A.
A large array of variant sequences is present in the expression site in human infections.
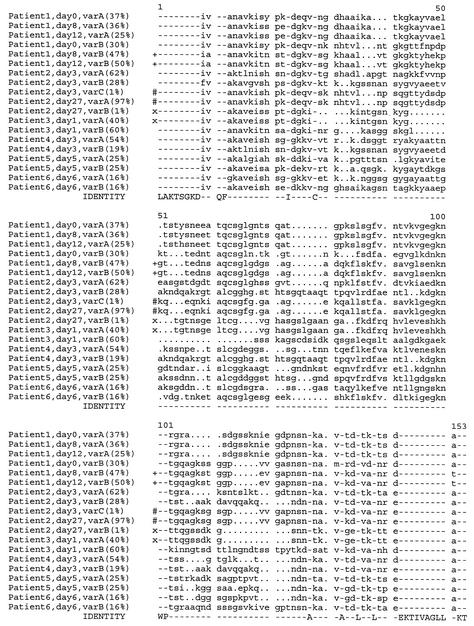
We compared the sequences of this genomic locus of A. phagocytophilum in blood samples from six infected persons. The variability of the locus was primarily localized to the CVR and was similar to that observed in comparisons of strains NY18 and Webster from in vitro culture (Fig. 1, PLOTSIMILARITY graph). Generally, multiple and diverse sequences of the CVR were obtained from each patient (Fig. 8). The predominant variants in a single patient were different from each other and from the predominant sequence variants in other patients, suggesting a vast capacity for variation. Paired blood samples that were all PCR positive for A. phagocytophilum were available to us from two patients and were taken either at days 0, 8, and 12 (patient 1) or at days 3 and 27 (patient 2) after onset of clinical symptoms. The clinical history and symptoms (see Materials and Methods) of patient 2 were consistent with the day-27 blood sample representing a relapse of the initial infection. The paired blood samples from each of the two patients contained multiple msp2(p44) variants. Similar predominant variants were obtained in the three samples from the first patient taken at days 0, 8, and 12, with the exception of day 0, varB. Interestingly, in patient 2, with a potential relapse infection, a minor variant present on day 3 (varC) became the predominant variant on day 27 (varA), and the initial predominant variants were not detected (Fig. 8).
FIG. 8.
A diverse repertoire of msp2(p44) sequences is encoded in the genomic expression site in individual patients infected with A. phagocytophilum. Genomic DNA was prepared from infected human blood of each patient. For two patients (patients 1 and 2), samples were available at differing times following the first onset of clinical symptoms. Independent clones of the genomic expression site were prepared and analyzed by RFLP mapping as in Fig. 5 and 7. The predominant variants (A and B) in each sample were sequenced and aligned by using PILEUP. In one case a minor variant in one population (patient 2, day 3, varC) that was identical to a predominant variant in a subsequent A. phagocytophilum population from the same patient (patient 2, day 27, varA) is also shown. The percentage of each sequence variant within a population, as determined by RFLP mapping, is indicated next to the variant designation. Identical sequences are indicated by identical symbols (+, #, or x) to the left of aligned sequences.
DISCUSSION
The data presented here reveal a genomic expression site for msp2(p44) that is used for expression of multiple mRNAs with diverse CVRs in strains NY18 and Webster of A. phagocytophilum. Polymorphisms in mRNA transcribed in infected mammalian cells are accompanied by similar polymorphisms in the expressed msp2(p44) gene. RNase protection data and comparison of clones derived from DNA and RNA suggest that this expression site is used for transcription of the majority, if not all, of msp2(p44) polymorphic mRNAs. Lin et al. (26) have recently described conservation of amino acid residues at five positions within the CVR encoded in msp2(p44) mRNA transcribed during human infections. These five positions are also conserved in the different variant sequences present in this expression site in infected patients (positions 28, 63, 101, 102, and 132, encoding amino acids C, C, W, P, and A, respectively [Fig. 8]). Therefore, both variable and conserved sequences present in transcribed mRNA correspond to variable and conserved sequences present in this expression locus. This further supports designation of this locus as the primary site for expression of polymorphic msp2(p44) mRNA.
Zhi et al. (43) have described an alternative mechanism for expression of one truncated msp2(p44) copy, p44-18, involving RNA splicing with the transcript from an adjacent msp2(p44) gene. However, this mechanism could not explain the diversity of transcripts observed, since other pseudogenes, not similarly juxtaposed (39), could not be expressed in this way. It was proposed that this unusual splicing mechanism may contribute toward the dominant expression of p44-18 in mammals. Interestingly, the HH1 variant (Fig. 7) is also a dominant variant in HL-60 cells and has a sequence almost identical (1 amino acid difference) to that encoded by p44-18. However, the HH1 variable region is present in the same expression site as the other expressed variants that we have characterized. Therefore, it appears unnecessary to postulate a different mechanism for expression of this dominant variant.
The structure of the expression site has similarities to that described previously for msp2 of A. marginale (3). These similarities include the expressed msp2(p44) gene, the coding sequence for an upstream gene, and a predicted promoter sequence. Taken together with previous information that shows the presence of homologous multigene families in both A. phagocytophilum and A. marginale, the presence of truncated genes in both organisms, and a similar organization of variable and conserved regions in the expressed proteins (3, 4, 12, 18, 30, 33, 39, 41), it is highly likely that the two organisms use similar mechanisms for the generation of outer membrane protein diversity.
The close juxtaposition of the expression site with a sequence homologous to RecA is intriguing, given the pivotal role of RecA in homologous recombination. In Neisseria gonorrhoeae antigenic variation of the pilus proceeds by a RecA-dependent process that involves unidirectional recombination of portions of incomplete silent pilin genes, pilS, into the expressed pilE gene (25, 28). This is thought to involve a pilE-pilS hybrid intermediate, with a crossover in small regions of conserved sequence. There are clear analogies between this mechanism and the use of segments of incomplete, silent gene copies to recombine unidirectionally into the msp2 expression site in A. marginale (5). However, as yet it is unknown whether the mechanism in A. marginale is RecA dependent or why the recA sequence is immediately downstream from the expression site in A. phagocytophilum but not in A. marginale. Also, the recA gene at this position in A. phagocytophilum is not complete, but encodes a segment of 87 amino acids of the more conserved RecA N terminus, followed by unrelated sequence and termination codons in all three reading frames.
There were multiple variant sequences present in the expression site in all populations of A. phagocytophilum that we examined, including organisms from in vitro culture in human or tick cells, and organisms in blood samples. The proportions of different variants and the detection of new variants were affected by environmental conditions such as growth in tick or human cells and by the time from disease onset in humans. We hypothesize that this is due to high-frequency segmental gene conversion of the expression site by different msp2(p44) genes, including incomplete copies, followed by selection of different variants. This hypothesis is supported by Southern blotting and genome sequence data. Figure 3 shows that there are numerous potential donor copies existing elsewhere in the genome for expression site variable-region sequence. Also, we compared 20 different variable-region sequences found in the expression site in this study with the unfinished A. phagocytophilum genome database (http://www.tigr.org). Fourteen variable-region amino acid sequences encoded in the expression site are identical (10 sequences) or nearly identical (4 sequences with one or two amino acid changes) to sequence encoded by genomic copies found elsewhere in the genome. Four expression site variants (AY164495, AY164498, AY164499, and AY164511) each appear to be a composite of sequence from two to three different genomic copies, and two expression site variants (AY164505 and AY164510) did not clearly match copies in this database. Therefore, there appear to be potential donor copies for the majority of different sequences that we have observed here in the expression site.
Selection of different variants could be mediated in infected hosts by the host's immune responses, as there is evidence that antibodies to MSP2(P44) have protective capacity, at least against homologous variants (19, 24). This would explain the observation of different predominant variants found in paired blood samples taken 24 days apart in the case of the congenitally asplenic patient with symptoms indicative of a relapse infection. The data from human-tick cell transfers suggest that selection of variants can also involve host cell environment or growth temperature.
The large number of msp2(p44) variants indicates that it could be difficult to use this antigen reliably in diagnostic tests or vaccines. The possibility of using conserved regions outside the CVR remains, although there are data showing that antibodies to MSP2(P44) in human infections primarily recognize variable epitopes (2). One should also exercise caution in the use of msp2(p44) genes for molecular typing and epidemiological analyses of different strains, as has been suggested (6), until more data are available on the stability of the different multigene loci. It may be difficult to distinguish between sequence polymorphisms caused by evolutionary divergence and rapid recombination mechanisms.
Clinically, the ability of the organism to express large numbers of different variants of an immunoprotective outer membrane protein has important implications for therapy and patient monitoring. The possibility of relapse infections should be considered, especially for immunosuppressed patients. Such patients should be closely monitored, even after apparent initial clearance of organisms following therapy.
In summary, these data show that a single genomic expression site is capable of expressing multiple msp2(p44) mRNAs with diverse CVRs. All populations of A. phagocytophilum examined were polymorphic with respect to the CVR, although there were few changes in regions flanking msp2(p44) in this genomic locus. The observed diversity of sequence variants was influenced both by the host cell culture environment and by the duration of infection in humans. The overall similarities between these data in A. phagocytophilum and A. marginale suggest that similar mechanisms for generating outer membrane protein diversity and establishing persistent infections are available to the two organisms.
Acknowledgments
We thank Annie Moreland and Hyun Yi for excellent technical assistance, the Molecular Genetics core facility of the Wadsworth Center, New York State Department of Health, for synthesis of oligonucleotide primers for confirmation of disease in patient samples by PCR, and Guy Palmer for critical review of the manuscript.
This investigation was supported by NIH grant AI45580 and by grant 814-2346A from the Centers for Disease Control and Prevention of the U.S. Public Health Service for laboratory surveillance of human granulocytic anaplasmosis as a part of the Emerging Infections Program. Preliminary sequence data were obtained from The Institute for Genomic Research through the website at http://www.tigr.org. Sequencing of A. phagocytophilum was accomplished with support from NIAID/Ohio State University (grant RO1 AI47885 to Y. Rikihisa).
Editor: J. T. Barbieri
REFERENCES
- 1.Aguero, R. M., H. W. Horowitz, G. P. Wormser, D. F. McKenna, J. Nowakowski, J. Munoz, and J. S. Dumler. 1996. Human granulocytic ehrlichiosis: a case series from a medical center in New York State. Ann. Intern. Med. 125:904-908. [DOI] [PubMed] [Google Scholar]
- 2.Asanovich, K. M., J. S. Bakken, J. E. Madigan, R. M. Aguero, G. P. Wormser, and J. S. Dumler. 1997. Antigenic diversity of granulocytic Ehrlichia isolates from humans in Wisconsin and New York and a horse in California. J. Infect. Dis. 176:1029-1034. [DOI] [PubMed] [Google Scholar]
- 3.Barbet, A. F., A. Lundgren, J. Yi, F. R. Rurangirwa, and G. H. Palmer. 2000. Antigenic variation of Anaplasma marginale by expression of MSP2 mosaics. Infect. Immun. 68:6133-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brayton, K. A., D. P. Knowles, T. C. McGuire, and G. H. Palmer. 2001. Efficient use of a small genome to generate diversity in tick-borne ehrlichial pathogens. Proc. Natl. Acad. Sci. USA 98:4130-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brayton, K. A., G. H. Palmer, A. Lundgren, J. Yi, and A. F. Barbet. 2002. Antigenic variation of Anaplasma marginale msp2 occurs by combinatorial gene conversion. Mol. Microbiol. 43:1151-1159. [DOI] [PubMed] [Google Scholar]
- 6.Carter, S. E., M. D. Ravyn, Y. Xu, and R. C. Johnson. 2001. Molecular typing of the etiologic agent of human granulocytic ehrlichiosis. J. Clin. Microbiol. 39:3398-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caspersen, K., J. H. Park, S. Patil, and J. S. Dumler. 2002. Genetic variability and stability of Anaplasma phagocytophila msp2 (p44). Infect. Immun. 70:1230-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, S. M., J. S. Dumler, J. S. Bakken, and D. H. Walker. 1994. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J. Clin. Microbiol. 32:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu, F. K. 1998. Rapid and sensitive PCR-based detection and differentiation of aetiologic agents of human granulocytotropic and monocytotropic ehrlichiosis. Mol. Cell. Probes 12:93-99. [DOI] [PubMed] [Google Scholar]
- 10.Dumler, J. S., A. F. Barbet, C. P. Bekker, G. A. Dasch, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. R. Rurangirwa. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ′HGE agent' as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145-2165. (Corrigendum, 52:5-6, 2002.) [DOI] [PubMed]
- 11.Eid, G., D. M. French, A. M. Lundgren, A. F. Barbet, T. F. McElwain, and G. H. Palmer. 1996. Expression of major surface protein 2 antigenic variants during acute Anaplasma marginale rickettsemia. Infect. Immun. 64:836-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.French, D. M., W. C. Brown, and G. H. Palmer. 1999. Emergence of Anaplasma marginale antigenic variants during persistent rickettsemia. Infect. Immun. 67:5834-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.French, D. M., T. F. McElwain, T. C. McGuire, and G. H. Palmer. 1998. Expression of Anaplasma marginale major surface protein 2 variants during persistent cyclic rickettsemia. Infect. Immun. 66:1200-1207. (Erratum, 66:2400.) [DOI] [PMC free article] [PubMed]
- 14.Frohman, M. A. 1993. Rapid amplification of complementary DNA ends for generation of full-length complementary DNAs: thermal RACE. Methods Enzymol. 218:340-356. [DOI] [PubMed] [Google Scholar]
- 15.Goodman, J. L., C. M. Nelson, B. Vitale, J. E. Madigan, J. S. Dumler, T. J. Kurtti, and U. G. Munderloh. 1996. Direct cultivation of the causative agent from patients with human granulocytic ehrlichiosis. N. Engl. J. Med. 334:209-215. [DOI] [PubMed] [Google Scholar]
- 16.Grunberg-Manago, M. 1999. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33:193-227. [DOI] [PubMed] [Google Scholar]
- 17.Horowitz, H. W., T. C. Hsieh, M. E. Aguero-Rosenfeld, F. Kalantarpour, I. Chowdhury, G. P. Wormser, and J. M. Wu. 2001. Antimicrobial susceptibility of Ehrlichia phagocytophila. Antimicrob. Agents Chemother. 45:786-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ijdo, J. W., W. Sun, Y. Zhang, L. A. Magnarelli, and E. Fikrig. 1998. Cloning of the gene encoding the 44-kilodalton antigen of the agent of human granulocytic ehrlichiosis and characterization of the humoral response. Infect. Immun. 66:3264-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ijdo, J. W., C. Wu, S. R. Telford III, and E. Fikrig. 2002. Differential expression of the p44 gene family in the agent of human granulocytic ehrlichiosis. Infect. Immun. 70:5295-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ijdo, J. W., Y. Zhang, E. Hodzic, L. A. Magnarelli, M. L. Wilson, S. R. Telford, S. W. Barthold, and E. Fikrig. 1997. The early humoral response in human granulocytic ehrlichiosis. J. Infect. Dis. 176:687-692. [DOI] [PubMed] [Google Scholar]
- 21.Jauron, S. D., C. M. Nelson, V. Fingerle, M. D. Ravyn, J. L. Goodman, R. C. Johnson, R. Lobentanzer, B. Wilske, and U. G. Munderloh. 2001. Host cell-specific expression of a p44 epitope by the human granulocytic ehrlichiosis agent. J. Infect. Dis. 184:1445-1450. [DOI] [PubMed] [Google Scholar]
- 22.Johnston, S. L., M. Strausbauch, G. Sarkar, and P. J. Wettstein. 1995. A novel method for sequencing members of multi-gene families. Nucleic Acids Res. 23:3074-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kieser, S. T., I. S. Eriks, and G. H. Palmer. 1990. Cyclic rickettsemia during persistent Anaplasma marginale infection of cattle. Infect. Immun. 58:1117-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, H. Y., and Y. Rikihisa. 1998. Characterization of monoclonal antibodies to the 44-kilodalton major outer membrane protein of the human granulocytic ehrlichiosis agent. J. Clin. Microbiol. 36:3278-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koomey, J. M., and S. Falkow. 1987. Cloning of the recA gene of Neisseria gonorrhoeae and construction of gonococcal recA mutants. J. Bacteriol. 169:790-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, Q., N. Zhi, N. Ohashi, H. W. Horowitz, M. E. Aguero-Rosenfeld, J. Raffalli, G. P. Wormser, and Y. Rikihisa. 2002. Analysis of sequences and loci of p44 homologs expressed by Anaplasma phagocytophila in acutely infected patients. J. Clin. Microbiol. 40:2981-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohr, C. V., K. A. Brayton, V. Shkap, T. Molad, A. F. Barbet, W. C. Brown, and G. H. Palmer. 2002. Expression of Anaplasma marginale msp2 operon-associated proteins during mammalian and arthropod infection. Infect. Immun. 70:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehr, I. J., and H. S. Seifert. 1998. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol. Microbiol. 30:697-710. [DOI] [PubMed] [Google Scholar]
- 29.Munderloh, U. G., S. D. Jauron, V. Fingerle, L. Leitritz, S. F. Hayes, J. M. Hautman, C. M. Nelson, B. W. Huberty, T. J. Kurtti, G. G. Ahlstrand, B. Greig, M. A. Mellencamp, and J. L. Goodman. 1999. Invasion and intracellular development of the human granulocytic ehrlichiosis agent in tick cell culture. J. Clin. Microbiol. 37:2518-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy, C. I., J. R. Storey, J. Recchia, L. A. Doros-Richert, C. Gingrich-Baker, K. Munroe, J. S. Bakken, R. T. Coughlin, and G. A. Beltz. 1998. Major antigenic proteins of the agent of human granulocytic ehrlichiosis are encoded by members of a multigene family. Infect. Immun. 66:3711-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogden, N. H., Z. Woldehiwet, and C. A. Hart. 1998. Granulocytic ehrlichiosis: an emerging or rediscovered tick-borne disease? J. Med. Microbiol. 47:475-482. [DOI] [PubMed] [Google Scholar]
- 32.Palmer, G. H., W. C. Brown, and F. R. Rurangirwa. 2000. Antigenic variation in the persistence and transmission of the ehrlichia Anaplasma marginale. Microbes Infect. 2:167-176. [DOI] [PubMed] [Google Scholar]
- 33.Palmer, G. H., G. Eid, A. F. Barbet, T. C. McGuire, and T. F. McElwain. 1994. The immunoprotective Anaplasma marginale major surface protein 2 is encoded by a polymorphic multigene family. Infect. Immun. 62:3808-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramsey, A. H., E. A. Belongia, C. M. Gale, and J. P. Davis. 2002. Outcomes of treated human granulocytic ehrlichiosis cases. Emerg. Infect. Dis. 8:398-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Telford, S. R., J. E. Dawson, P. Katavolos, C. K. Warner, C. P. Kolbert, and D. H. Persing. 1996. Perpetuation of the agent of human granulocytic ehrlichiosis in a deer tick-rodent cycle. Proc. Natl. Acad. Sci. USA 93:6209-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker, D. H., and J. S. Dumler. 1996. Emergence of the ehrlichioses as human health problems. Emerg. Infect. Dis. 2:18-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walls, J. J., M. Aguero-Rosenfeld, J. S. Bakken, J. L. Goodman, D. Hossain, R. C. Johnson, and J. S. Dumler. 1999. Inter- and intralaboratory comparison of Ehrlichia equi and human granulocytic ehrlichiosis (HGE) agent strains for serodiagnosis of HGE by the immunofluorescent-antibody test. J. Clin. Microbiol. 37:2968-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weber, K. L., M. E. Bolander, and G. Sarkar. 1998. Rapid acquisition of unknown DNA sequence adjacent to a known segment by multiplex restriction site PCR. BioTechniques 25:415-419. [DOI] [PubMed] [Google Scholar]
- 39.Zhi, N., N. Ohashi, and Y. Rikihisa. 1999. Multiple p44 genes encoding major outer membrane proteins are expressed in the human granulocytic ehrlichiosis agent. J. Biol. Chem. 274:17828-17836. [DOI] [PubMed] [Google Scholar]
- 40.Zhi, N., Y. Rikihisa, H. Y. Kim, G. P. Wormser, and H. W. Horowitz. 1997. Comparison of major antigenic proteins of six strains of the human granulocytic ehrlichiosis agent by Western immunoblot analysis. J. Clin. Microbiol. 35:2606-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhi, N., N. Ohashi, Y. Rikihisa, H. W. Horowitz, G. P. Wormser, and K. Hechemy. 1998. Cloning and expression of the 44-kilodalton major outer membrane protein gene of the human granulocytic ehrlichiosis agent and application of the recombinant protein to serodiagnosis. J. Clin. Microbiol. 36:1666-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhi, N., N. Ohashi, T. Tajima, J. Mott, R. W. Stich, D. Grover, S. R. Telford III, Q. Lin, and Y. Rikihisa. 2002. Transcript heterogeneity of the p44 multigene family in a human granulocytic ehrlichiosis agent transmitted by ticks. Infect. Immun. 70:1175-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhi, N., N. Ohashi, and Y. Rikihisa. 2002. Activation of a p44 pseudogene in Anaplasma phagocytophila by bacterial RNA splicing: a novel mechanism for post-transcriptional regulation of a multigene family encoding immunodominant major outer membrane proteins. Mol. Microbiol. 46:135-145. [DOI] [PubMed] [Google Scholar]