Abstract
The number of telomeric DNA repeats at chromosome ends is maintained around a mean value by a dynamic balance between elongation and shortening. In particular, proteins binding along the duplex part of telomeric DNA set the number of repeats by progressively limiting telomere growth. The paradigm of this counting mechanism is the Rap1 protein in Saccharomyces cerevisiae. We demonstrate here that a Rap1-independent mechanism regulates the number of yeast telomeric repeats (TG1–3) and of vertebrate repeats (T2AG3) when TEL1, a yeast ortholog of the human gene encoding the ATM kinase, is inactivated. In addition, we show that a T2AG3-only telomere can be formed and maintained in humanized yeast cells carrying a template mutation of the gene encoding the telomerase RNA, which leads to the synthesis of vertebrate instead of yeast repeats. Genetic and biochemical evidences indicate that this telomere is regulated in a Rap1-independent manner, both in TEL1 and in tel1Δ humanized yeast cells. Altogether, these findings shed light on multiple repeat-counting mechanisms, which may share critical features between lower and higher eukaryotes.
Keywords: evolution/TEL1/telomerase/telomere/yeast
Introduction
Telomeres are nucleoprotein structures found at the ends of all eukaryotic linear chromosomes (reviewed in Blackburn and Greider, 1995; Zakian, 1995). They play essential roles in the maintenance of genome integrity and in the control of cell proliferation. They specifically cap the ends of chromosomes such that they are not recognized by DNA damage checkpoints and repair systems (Sandell and Zakian, 1993). They are also involved in the control of gene expression and in the spatial and functional organization of chromosomes (Gottschling et al., 1990; reviewed in Gilson et al., 1993).
In most eukaryotes, the sequence of telomeric DNA consists of a tandem array of short guanine-rich repeats. It is an oriented structure with the G-rich strand running 5′ to 3′ towards the distal end of the chromosome and ending as a short single-stranded 3′ overhang. Telomeric DNA decreases in length as cells divide. This is due to a replication defect in the most distal telomeric sequence, which cannot be achieved by the standard polymerase machinery, and also to nuclease activities coupled to replication (reviewed in Lingner et al., 1995). The dynamics of telomere length control cell proliferation and genome integrity, triggering senescence, apoptosis and chromosomal instability (reviewed in Blackburn, 2001). In many organisms, telomere shortening is compensated by telomerase, a unique reverse transcriptase, which specifically extends the 3′ G-rich telomeric strand (reviewed in Blackburn, 2000). This enzyme appears to be constitutively expressed in many unicellular organisms, including the budding and fission yeasts, and to be highly regulated during the development of metazoans.
Usually, the telomere length of telomerase-positive cells is maintained at a constant mean value (reviewed in McEachern et al., 2000). Remarkably, the mean telomere length exhibits variations between yeast strains or human individuals. This cellular determination of the mean number of telomeric repeats is best represented by the so-called counting mechanism, that is the ability of the cell to retain the ‘memory’ of the number of telomeric repeats corresponding to a constant mean value. This mechanism is involved in the progressive return to equilibrium of too short telomeres either during the de novo formation of telomeres (Szostak and Blackburn, 1982; Barnett et al., 1993) or when a single telomere is abruptly shortened (Marcand et al., 1999). Another demonstration of repeat counting is the ability of telomeric repeats or telomeric protein-binding sites inserted at an internal position from the telomere, even when misorientated, to be taken as part of the total telomere length (Marcand et al., 1997; Ray and Runge, 1999b; Grossi et al., 2001). Consequently, the number of telomeric repeats distal to this internal sequence decreases. In budding yeast, the counting mechanism appears to be based on a progressive decline in the rate of telomere elongation when the number of repeats increases until elongation is balanced by degradation (Marcand et al., 1999).
In budding yeast, an important counting factor is Rap1. An excess of Rap1 bound to a telomere causes repression of telomerase activity in cis, thereby creating a negative feedback loop that contributes to the setting of telomere length (Kyrion et al., 1992; Krauskopf and Blackburn, 1996; Marcand et al., 1997, 1999; Ray and Runge, 1999b). Two factors interacting with Rap1, Rif1 and Rif2, are also involved in this Rap1-dependent regulation (Wotton and Shore, 1997). There is now increasing evidence that mechanisms involving protein binding to telomeric DNA sequences and limiting telomere elongation are conserved in distantly related organisms. For instance, in fission yeast, a deletion of TAZ1, a gene encoding a telomeric DNA-binding protein, dramatically increases telomere length (Cooper et al., 1997). Interestingly, although the fission yeast SpRap1 negatively regulates telomere elongation like its budding yeast ortholog, it is not directly bound to telomeric DNA, but associates to the telomere through an interaction with Taz1 (Chikashige and Hiraoka, 2001; Kanoh and Ishikawa, 2001; Park et al., 2002). In human cells, the known telomeric DNA-binding proteins are TRF1 and TRF2 (Chong et al., 1995; Bilaud et al., 1996, 1997; Broccoli et al., 1997). Reminiscent of the properties of the yeast Rap1, an excess of TRF1 and TRF2 inhibits telomere elongation (van Steensel and de Lange, 1997; Smogorzewska et al., 2000). Moreover, their targeting to a single telomere leads to telomere shortening, indicating that they act in cis to limit telomere elongation (Ancelin et al., 2002). As in fission yeast, the human ortholog of Rap1 does not bind to telomeric DNA, but is likely to be targeted to telomeres via its interaction with TRF2 (Li et al., 2000). Interestingly, TRF2 can shorten telomeres in the absence of telomerase (Ancelin et al., 2002; Karlseder et al., 2002).
A fascinating connection between the counting mechanism of telomere length and chromosome end stability has been revealed by the functions of the PI3-like kinase ATM, which plays an important role in the checkpoint of double-strand break in human cells, and of its ortholog in budding yeast TEL1 (reviewed in Pandita, 2002). Loss of function of these proteins leads to a new steady-state number of telomeric repeats, shorter than the wild type. In tel1Δ yeast cells, in contrast to wild-type cells, non-telomeric Rap1-binding sites are not counted (Ray and Runge, 1999a). Moreover, in the absence of TEL1, a deletion of the C-terminal part of Rap1 as well as the disruption of RIF1 or RIF2 have no effect on mean telomere length (Craven and Petes, 1999; Ray and Runge, 1999a). This led us to investigate Rap1-independent mechanisms of telomere length regulation in budding yeast.
Results
Yeast repeats are counted both in TEL1 and in tel1Δ cells while the vertebrate repeats are counted only in tel1Δ cells
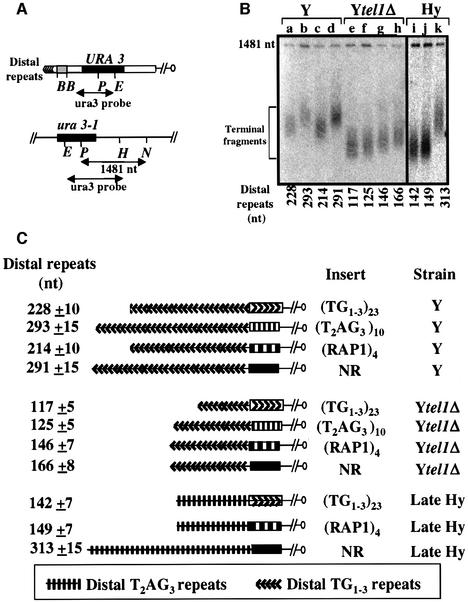
In order to characterize the counting factor acting in tel1Δ cells, we asked whether internal stretchs of the budding yeast TG1–3 repeats or of the vertebrate T2AG3 repeats could be taken into account for telomere length regulation in the absence of TEL1. To this end, the left extremity of chromosome VII beyond the ADH4 gene was replaced by a newly formed URA3-tagged telomere containing the sequences (TG1–3)23 or (T2AG3)10 inserted at the vicinity of the distal TG1–3 repeats. We also inserted in the same location a sequence containing four non-telomeric Rap1-binding sites [named (RAP1)4] and a portion of the RAD52 gene (named NR). The counting effect of the inserted sequences was evaluated by measuring the size of a terminal restriction fragment containing part of the URA3 gene (Marcand et al., 1997; Ray and Runge, 1999b; Figure 1).
Fig. 1. In tel1Δ cells TG1–3 and T2AG3 repeats are counted by Rap1-independent pathways. (A) Schematic representation of the modified VII-L telomere used to determine the counting capacity of the internal sequence. The positions of the EcoRV (E), SapI (P), NaeI (N), HindIII (H) and BamHI (B) restriction sites are indicated. The gray box bracketed by BamHI sites indicates the position of the inserted sequences (see Materials and methods). (B) Genomic DNA of the Y, Ytel1Δ and Hy strains was digested with SapI and NaeI and hybridized with the ura3 probe. The median length of the telomeric restriction fragment was calculated using a set of molecular weight markers (not shown) and the non-telomeric 1481 nt fragment as an internal control. In order to obtain the length of the distal telomeric repeats, 378 nt was subtracted from the size of the measured terminal fragment, as shown below each lane. The inserted sequence is (TG1–3)23 in lanes a, e and i; (T2AG3)10 in lanes b and f; (RAP1)4 in lanes c, g and j; and NR in lanes d, h and k. (C) Schematic representation of the results.
When the control sequence NR is inserted close to the telomere, the length of the distal TG1–3 repeats, is identical to the mean length of the strains without insert, i.e. ∼300 nucleotides (nt) for TEL1 and ∼160 nt for tel1Δ cells (Figure 1B, lanes d and h, and C; data not shown). This validates the use of the NR sequence as a non-counted control. As expected, in TEL1 cells, both (TG1–3)23 and (RAP1)4 shorten the distal repeats as compared with NR, indicating that they are taken into account for length regulation (Figure 1B, lanes a and c, and C). In tel1Δ cells, the shortening was more pronounced for (TG1–3)23 than for (RAP1)4 (∼45 nt versus 20 nt) (Figure 1B, lanes e and g, and C). Thus, the yeast telomeric repeats mediate a more potent counting mechanism than non-telomeric Rap1 sites when TEL1 is inactivated. The counting effect of yeast repeats was confirmed in another tel1Δ background using an internal stretch of 270 nt of TG1–3 repeats (see Supplementary figure 1A, available at The EMBO Journal Online). In this case, the internal stretch of TG1–3 repeats is flanked on both sides by a FRT site, which can be excised upon induction of the expression of the FLP1 gene in the presence of galactose. Using this experimental design, we demonstrated previously that in vivo telomerase activity is gradually repressed upon telomere elongation, suggesting that the accumulation of counting factors leads to a progressive inhibition of telomerase activity (Marcand et al., 1999). As in wild-type cells, we observed a progressive decline in the elongation rate as a function of telomere length in tel1Δ cells, suggesting a seemingly similar type of counting mechanism in TEL1 and in tel1Δ yeast cells (see Supplementary figure 1B and C).
Strikingly, the (T2AG3)10 sequence is correctly counted in tel1Δ cells, leading to a shortening of 49 nt as compared with NR (Figure 1B, lane f, and C). Importantly, the (T2AG3)10 insert is not taken into consideration for telomere length regulation in the presence of TEL1 (Figure 1B, lane b, and C). We conclude that a cryptic Rap1-independent counting mechanism, which is revealed in the absence of TEL1, can accommodate both T2AG3 and TG1–3 repeats, while the Rap1-dependent mechanism is specific for TG1–3 repeats.
Progressive replacement of TG1–3 by T2AG3 repeats in humanized yeast cells
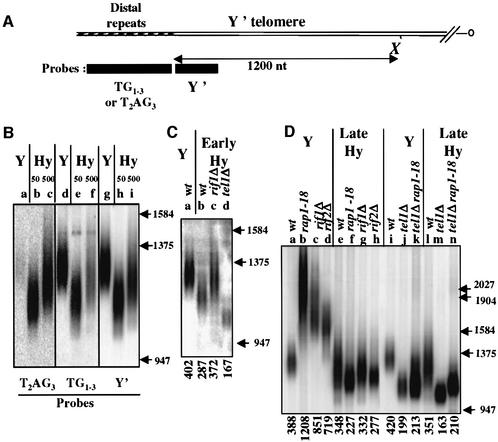
The ability of tel1Δ cells to count subtelomeric T2AG3 repeats (see above) prompted us to investigate whether the number of T2AG3 repeats can be regulated over the entire length of a yeast telomere. To this end, we made use of a humanized yeast strain, which carries the tlc1-human (tlc1-h) allele of the RNA telomerase TLC1 gene. The presence of the tlc1-h allele results in the incorporation of T2AG3 instead of TG1–3 repeats at the end of yeast chromosomes (Henning et al., 1998). Shortly after the growth of haploid spores carrying tlc1-h instead of TLC1, the mean length of the Y′ telomeres appears stable but shorter than telomeres of cells originating from wild-type TLC1 spores (Henning et al., 1998). We repeated this experiment starting from the heterozygote TLC1/tlc1-h (designated C6.1 in Henning et al., 1998). We measured the Y′ telomere length in several tlc1-h haploid cells after ∼50 generations post-germination and confirmed that telomeres were shortened of ∼100–150 nt as compared with TLC1 cells (Figure 2A, B, lanes b, e and h, and C, lane b; data not shown). One TLC1 haploid and one tlc1-h haploid were chosen for further analysis, and were named Y and Hy cells, respectively. After ∼500 generations of Hy cells, we noticed a decrease in the intensity of the Y′ telomere hybridization signal with a TG1–3 probe (Figure 2B, lanes e and f) as compared with a T2AG3 probe or with a Y′ probe (Figure 2B, lanes b and c, and h and i, respectively). Remarkably, the long-term growth of Hy cells also led to a slight increase in the length of the Y′ telomeres (Figure 2B, lane i, and D, lanes e and l). This elongation stopped after ∼500 generations at a new mean length, which is still shorter than wild-type yeast telomeres. This new length setting appears to be stable over an extended period of growth, up to 800 generations after germination (data not shown). It does not seem to result from a selection of cells that grow faster since a slight increase in generation time is observed for late cultures of Hy cells and since it occurs both when cells are serially diluted from liquid culture or upon successive restreaking on agar plates (data not shown). Because of these observations, we will refer hereafter to early Hy cells as those that had grown for <100 generations after germination, and to late Hy cells as those that had grown for >300 generations.
Fig. 2. Maintenance of Y′ telomere in humanized yeast. (A) Schematic representation of yeast telomeres associated with a Y′ subtelomeric element, the position of the invariable XhoI (X) site is reported. Digestion of genomic DNA by XhoI allows to measure the size of the terminal fragment of all Y′ telomeres in the cells. The different probes used for Southern blot analysis are shown. The length of the distal repeats was estimated by subtracting 1200 nt from the measured length of the terminal XhoI fragment. (B) Example of Y′ telomere Southern blot hybridized successively to a T2AG3, TG1–3 or Y′ probe. The number of generation after germination is indicated for the Hy cells. Lanes a, d and g: Y cells; lanes b, e and h: Hy cells after 50 generations (early passages); lanes c, f and i: Hy cells after 500 generations (late passages). (C and D) The XhoI blots were hybridized with the Y′ probe. wt: wild type for the RAP1, RIF1, RIF2 and TEL1 genes. In (B–D), the molecular weight markers, used to measure the mean telomere lengths, are indicated in nt. In (C) and (D), the length of distal repeats is indicated at the bottom of each lane.
Formation and maintenance of a telomere made exclusively of T2AG3 repeats
In order to investigate whether a telomere made solely of T2AG3 repeats can be maintained in Hy cells, the left extremity of chromosome VII was replaced in Y and Hy cells by an URA3-containing DNA ending with a stretch of 10 T2AG3 repeats. In Y cells, such a construct led to the addition of TG1–3 repeats distally to the T2AG3 repeats (data not shown). This is in agreement with previous work showing that various types of telomeric repeats can be used as a seeding sequence (Szostak and Blackburn, 1982; Lustig, 1992). In late Hy cells, the majority of randomly selected URA+ clones exhibited a restriction pattern indicative of a successful seeding event, i.e. a smeary signal for the VII-L terminal restriction fragment probed with an ura3 probe (Figure 3A and B, lanes g and k; data not shown).
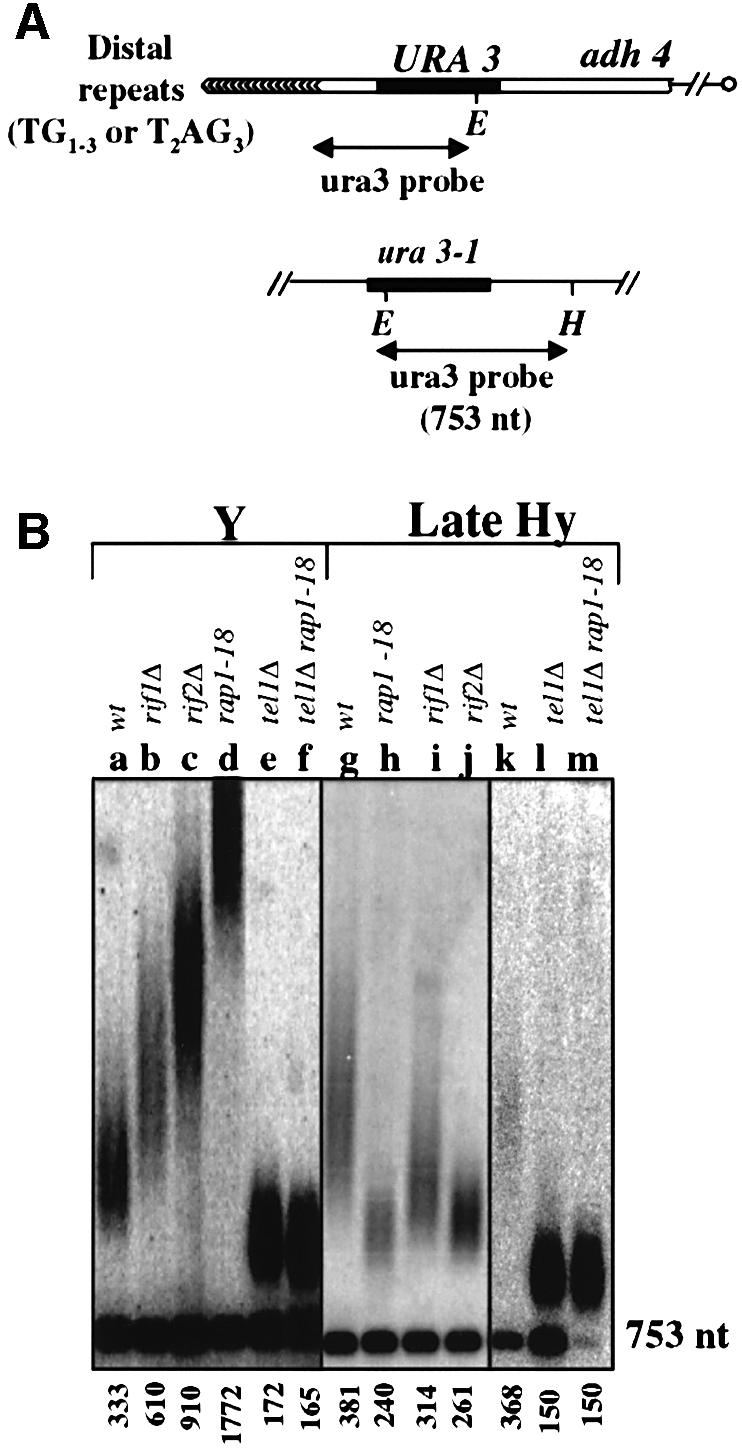
Fig. 3. Maintenance of the T2AG3-only telomere. (A) Schematic representation of the modified VII-L telomere used to monitor the length of the wild type and of T2AG3-only telomere. A representation of the ura3-1 allele is shown to indicate the existence of a 753 nt fragment hybridizing with the ura3 probe. (B) Genomic DNA was digested with EcoRV and HindIII and hybridized with the ura3 probe. The median length of the telomeric restriction fragment was calculated using a set of molecular weight markers (not shown) and using the ura3-1 753 nt fragment as an internal control. 720 bp was subtracted from this value to give the size of the distal TG1–3 repeats (Y strains) or the distal T2AG3 repeats (Hy strains). wt: wild type for the RAP1, RIF1, RIF2 and TEL1 genes.
The URA3-tagged telomeric DNA fragment was PCR-amplified from the genomic DNA of one of these clones, and the recovered plasmids contained an insert in the size range of 400–650 nt that hybridizes to a T2AG3 probe (data not shown). The sequencing of four randomly chosen plasmid DNAs shows the presence of an uninterrupted array of T2AG3 repeats over the entire length of the cloned telomere, demonstrating the de novo formation of a telomere composed solely of T2AG3 repeats. This T2AG3-only telomere can be formed and propagated normally when the RAD52 gene is disrupted (data not shown). Thus, the T2AG3-only telomere is unlikely to be maintained by homologous recombination. We have been unable to disrupt the EST2 gene in Hy cells (data not shown). Based on the RAD52-independence and the failure to introduce an EST2 disruption, we conclude that telomerase plays a critical role in the maintenance of the T2AG3-only telomere.
The length profile of the T2AG3-only telomere exhibits an increased heterogeneity as compared with a URA3-tagged telomere in Y cells (Figure 3B, compare lane a with g and k). During 40 restreaks that correspond to ∼800 generations, the length and heterogeneity of the T2AG3-only telomere appears relatively well preserved, although some fluctuations can be observed, with a mean length ranging from 350 to 500 nt (data not shown). We conclude that the length of the T2AG3-only telomere can be regulated in budding yeast, although the mechanism involved appears less efficient than the one acting in TLC1 cells.
The Rap1 pathway does not limit elongation of telomeres made principally or exclusively of T2AG3 repeats
We then investigated genetically whether the Rap1 pathway plays a role in telomere length regulation in Hy cells. To this end, we introduced in tlc1-h cells carrying an URA3-tagged telomere the rif1Δ, rif2Δ and rap1-18 mutations, which overelongate telomeres in TLC1 cells by disrupting the Rap1-mediated cis-repression of telomerase activity (Kyrion et al, 1992; Wotton and Shore, 1997) and the tel1Δ mutation, which shortens telomeres by inhibiting telomerase activity (Craven and Petes, 1999). Since the sporulation efficiency of tlc1-h/tlc1-h homozygote was very poor, probably due to their telomeric defects, the production of mutated haploid cells from the diploids was very inefficient. Consequently, the mutations were introduced by gene replacement in haploid cells of the Y and Hy strains. Several independent transformants were analyzed and in each case the telomeric phenotypes were similar. In addition, each mutant was backcrossed to tlc1-h cells of opposite mating type and was shown to be complemented by the wild-type allele (data not shown). For the sake of clarity, we will present in Figures 2 and 3 the results obtained for a single transformant of each construction.
As expected, the Y′ telomeres and the URA3-tagged telomere were overelongated in Y cells deleted for RIF1 (Figures 2D, lane c, and 3B, lane b). When the RIF1 disruption is introduced into early Hy cells, this also leads to overelongation of Y′ telomeres as compared with RIF1 Hy cells (Figure 2C, lane c). However, and strikingly, when the same RIF1 disruption was introduced into late Hy cells, a faint shortening of Y′ telomeres was observed (Figure 2D, lane g). Similarly, the rap1-18 allele or the deletion of RIF2 leads to Y′ telomere shortening in late Hy cells (Figure 2D, lanes f and h). The rap1-18, rif1Δ and rif2Δ mutations were also found to decrease the length of the T2AG3-only telomere in late Hy cells (Figure 3B, lanes h–j). In contrast to what has been reported (Wotton and Shore, 1997), the RIF1 disruption has a less pronounced deregulation effect than the RIF2 disruption at the URA3-tagged telomere of Y cells (Figure 3B, lanes b and c). The reason for this unexpected hierarchy between the mutation phenotypes is unknown, but might be related to strain differences. It is noteworthy that the relative strength between rif1Δ and rif2Δ is opposite at the Y′ telomeres (Figure 2D, lanes c and d).
The fact that the length of Y′ telomere slightly increases in early but not in late rif1Δ Hy cells raises the possibility that the repressive effect of the Rap1–Rif complex is lost in late Hy cells because of the progressive replacement of TG1–3 repeats by T2AG3 (see above and Figure 2B). This hypothesis predicts that TG1–3 repeats in late Hy cells are still able to negatively regulate telomere length. This was directly tested by forming in late Hy cells composite telomeres containing an internal (TG1–3)23 or (RAP1)4 sequence and distal T2AG3 repeats. The lengths of these telomeres are maintained at a mean value, which is twice as short as one of the T2AG3-only telomere (Figure 1B, lanes i and j, and C). This reveals a very potent counting effect in late Hy cells exerted by the internal TG1–3 repeats and the non-telomeric Rap1 sites. This counting is even stronger than the one observed with an identical amount of internal repeats in Y cells (Figure 1C). This could be explained by a greater sensitivity of the humanized telomerase and/or of the T2AG3-enriched telomeres to Rap1-mediated repression.
In TLC1 cells, the absence of TEL1 revealed a cryptic length regulation pathway recognizing internal T2AG3 repeats (see above and Figure 1). Therefore, the TEL1 gene product might be inoperative at the telomeres of Hy cells, explaining the ability of these cells to regulate telomere length. However, in Hy cells, the deletion of TEL1 leads to a pronounced shortening of the Y′ telomere (Figure 2C, lane d, and D, lane m) and of the T2AG3-only telomere (Figure 3B, lane l). These data indicate that TEL1 is required for a proper length regulation in Hy cells. The double mutation tel1Δ rap1-18 does not lead to an additional shortening of the T2AG3-only telomere in late Hy cells (Figure 3B, lane m), suggesting that a Rap1-dependent elongation in Hy cells also requires TEL1 activity. We believe that the very short Y′ telomeres in Hy tel1Δ cells (Figure 2D, lane m) might be attributed to the potent ‘counting capacities’ of TG1–3 repeats in Hy cells (Figure 1C).
The T2AG3-only telomeric DNA appears devoid of Rap1
In order to assay the binding of Rap1 to the T2AG3-only telomere in Hy cells, we performed chromatin immunoprecipitation experiments (ChIP) with formaldehyde-mediated cross-linked cells expressing a myc-tagged version of Rap1. The RAP1-MYC Hy cells behave as RAP1 Hy cells with respect to viability, growth rate and telomere length (data not shown). However, we noticed that the telomeres of RAP1-MYC Y cells were slightly longer than those of RAP1 cells (+30 nt). This, together with the absence of telomere length change due to Rap1-myc expression in Y tel1Δ cells, suggests that the addition of the myc tag to the C-terminus of Rap1 induces a minor loss of function of the C-terminus of Rap1. This is not expected to affect the binding of Rap1 to its natural targets, both because the cells expressing Rap1-myc do not show any growth defect and because the DNA-binding properties of Rap1 depend mainly on the central domain. Overall, we considered that the Rap1-myc fusion protein is largely functional.
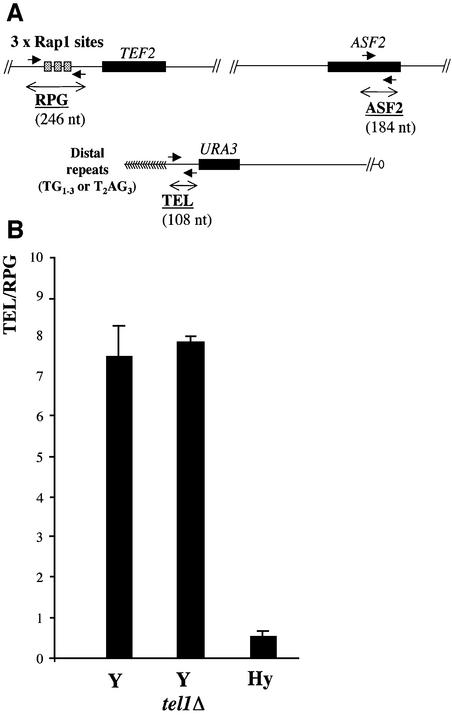
The DNA recovered from the immunoprecipitates was accurately quantified by real-time PCR (Ladenburger et al., 2002). The RPG PCR fragment of the TEF2 promoter, which is bound in vivo by three Rap1 proteins simultaneously (De Sanctis et al., 2002), was used as a positive non-telomeric control for Rap1 binding (Figure 4A). We used as a negative control the ASF2 open reading frame, which is known to be devoid of Rap1 sites (Liu et al., 2002) (Figure 4A). The specificity of our ChIP procedure was estimated by the ratio of the immunoprecipitated RPG to ASF2 DNA. This ratio fluctuated between 8 and 12 in all the experiments performed with Y, Y tel1Δ and Hy cells (data not shown). When the same ChIP experiments were performed but with cells expressing the wild-type Rap1 protein or in the absence of formaldehyde cross-linking, we were unable to detect any specific amplification of the RPG fragment. We conclude that our ChIP experiments were reliable since our conditions led to a expected specific enrichment in Rap1-bound DNA at the TEF2 promoter in all types of yeast cells used in this study.
Fig. 4. The binding of Rap1 to the T2AG3-only telomere is impaired. A myc tag epitope was inserted at the C-terminus of the RAP1 endogenous gene in Y, Y tel1Δ and Hy strains carrying an URA3-tagged telomere. (A) Schematic representation of the three DNA fragments analyzed for Rap1 binding by ChIP and by real-time PCR. (B) Results of the immunoprecipitation are expressed as the ratio of the immunoprecipitated TEL DNA to that of the RPG DNA. The average value for at least three experiments is given in the diagram as well as the estimated standard deviation.
The TEL PCR fragment lies adjacent to an URA3-tagged telomere (Figure 4A). It does not contain a Rap1-binding site, but its immediate juxtaposition to the telomeric DNA guarantees its preferential coprecipitation with telomeric DNA. In order to compare telomeric versus non-telomeric binding of Rap1, we measured the ratio between the quantities of TEL to RPG DNA in the immunoprecipitates (TEL/RPG in Figure 4B). As expected, we found a 7.5-fold enrichment of TEL DNA as compared with RPG DNA in ChIP with Y cells. This value could be a slight underestimation of the amount of telomeric Rap1 since the TEL fragment does not overlap with the telomeric repeats (see above). In any case, the enrichment is compatible with the number of Rap1 proteins bound at a wild-type yeast telomere, which was estimated to be ∼18 (Gilson et al., 1993). Remarkably, there is no enrichment of TEL over RPG in experiments performed with Hy cells (Figure 4B). To the contrary, less TEL than RPG DNA was immunoprecipitated, leading to a TEL/RPG ratio of 0.3. Since RPG is expected to be bound by three Rap1 molecules, one can infer that fewer than three Rap1 molecules are bound to the TEL fragment in Hy cells. This absence or quasi-absence of Rap1 from the T2AG3-only telomeric DNA cannot be explained by telomere shortening, since the mean length of the URA3-tagged telomere in Y and Hy cells are roughly similar (Figure 3B, lanes a, g and k). Moreover, ChIP experiments performed with Y tel1Δ cells that exhibit a mean length much shorter than wild type (Figure 3B, lane e) leads to an enrichment of TEL over RPG similar to that observed in Y cells. The recovery of a seemingly equal amount of TEL DNA from Y and Y tel1Δ cells could be explained if the Rap1 molecules in the cross-linked telomeric chromatin are more exposed to antibodies in tel1Δ cells and/or if the density of telomeric and subtelomeric Rap1 molecules is higher in tel1Δ than in wild-type cells. Nevertheless, these data clearly show that in Y cells, a large number of Rap1 molecules are bound to telomeres in the absence of TEL1. In conclusion, the telomerase-repressive functions of Rap1 appear inoperative at telomeres made principally or exclusively of T2AG3 repeats because of the absence or quasi-absence of Rap1 associated in vivo to vertebrate repeats.
Discussion
Rap1-independent counting of vertebrate repeats in budding yeast
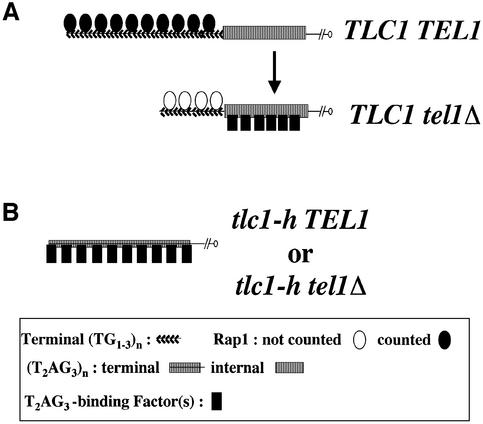
The major finding of this work is to have revealed Rap1-independent mechanisms capable of counting the number of yeast telomeric repeats (TG1–3) and of vertebrate telomeric repeats (T2AG3) at the telomeres of budding yeast. This is based upon two distinct experimental situations (depicted in Figure 5). In the absence of TEL1, vertebrate repeats inserted at the internal side of a single telomere are taken into account for telomere length regulation (Figure 5A). In an identical setting, authentic yeast repeats (TG1–3) are also counted in a seemingly Rap1-independant manner (Craven and Petes, 1999; Ray and Runge, 1999a; this work). In tlc1-h cells, also known as humanized yeast (Henning et al., 1998), a T2AG3-only telomere formed only by vertebrate repeats can be maintained at a regulated mean length (Figure 5B). In agreement with these data, Alexander and Zakian (2003) also show that a fully vertebrate telomere is maintained at a regulated telomere length (Alexander and Zakian, 2003). The disruption of TEL1 in humanized yeast leads to shorter but stable telomeres, like in wild-type telomerase yeast. Thus, while the presence of TEL1 does not allow the counting of internal vertebrate repeats in a wild-type telomerase context, the mean length of a T2AG3-only telomere appears to be regulated both in TEL1 and in tel1Δ humanized yeast. Overall, these differential effects of TEL1 between TLC1 and tlc1-h yeast on the regulation of vertebrate repeats raise the interesting possibility of multiple, possibly overlapping, mechanisms able to regulate the number vertebrate repeats at budding yeast telomeres.
Fig. 5. Multiple pathways regulate the number of vertebrate telomeric repeats at chromosome ends of budding yeast. The symbols are explained at the bottom of the figure. (A) In tel1Δ cells, the Rap1-counting pathway is no more operative although Rap1 stills binds to telomeres, as shown by ChIP assays. Our data indicate that unknown counting factors able to recognize T2AG3 repeats set the mean telomere length in tel1Δ cells. Whether the T2AG3-binding factor is associated to vertebrate repeats in TEL1 cells is unknown. (B) In humanized yeast, a T2AG3-only telomere can be formed and maintained at a regulated mean length. Noteworthy, this regulation can occur both in TEL1 and in tel1Δ cells and can co-habit with a Rap1-dependent regulation pathway when TG1–3 repeats or Rap1 sites are inserted internally. Whether the T2AG3-binding factor(s) involved in length regulation is (are) the same in TLC1 tel1Δ and in tlc1-h cells is not known.
Telomere length regulation in tel1Δ cells
The cryptic pathway(s) revealed in tel1Δ cells does (do) not seem to be active in TEL1 cells, since the deletion of the C-terminus of Rap1 in the presence of TEL1 leads to a fully deregulated telomere (Kyrion et al., 1992). One explanation would be that TEL1 has a dual effect, being both required for the yeast repeat-specific pathway and suppressing the cryptic pathway(s).
What is the nature of the factors able to regulate telomere length and to recognize yeast and vertebrate repeats in tel1Δ cells? Our data and those of others (Craven and Petes, 1999; Ray and Runge, 1999a) clearly eliminate Rap1, and to our knowledge there are no known Saccharomyces cerevisiae factors able to specifically bind both types of repeats. Alternatively, yeast and vertebrate repeats might be bound by distinct counting factors. The product of the TBF1 gene might be such a candidate because it binds T2AG3 repeats with high affinity, both in vitro (Liu and Tye, 1991; Brigati et al., 1993) and in vivo (Koering et al., 2000; Alexander and Zakian, 2003).
Humanized yeast as a model to study telomere functions
Long-term cultures of humanized yeast resulted in a change in telomere length regulation, switching from a partially Rap1-dependent mode with short telomeres in early passages to a Rap1-independant mode with longer telomeres in late cultures. Although less pronounced, the elongation of telomeres in late tlc1-h cells is reminiscent of the runaway phenotype described previously for certain template mutations of the Kluyveromyces lactis telomerase (McEachern and Blackburn, 1995). This is likely to be explained by a progressive replacement of TG1–3 by T2AG3 repeats, in agreement with the existence of a turnover mechanism that continuously degrades and resynthesizes telomeric repeats (Krauskopf and Blackburn, 1998). In agreement with this hypothesis, we show that an internal stretch of 23 TG1–3 repeats or an array of non-telomeric Rap1 sites can play a role in length regulation of telomeres in late humanized yeast. This provides an explanation of the lengthening of Y′ telomeres in late Hy cells that correlates with a progressive decline in the number of residual yeast repeats.
A yeast telomere without Rap1
One provocative finding of this work is the demonstration that humanized yeast can seed and regulate a telomere formed only of an array of vertebrate repeats. This T2AG3-only telomere can be maintained in long-term culture for >200 generations without provoking massive cell cycle arrest or growth defects. It does not appear to be associated to Rap1 by ChIP experiments. In addition, its length is not elongated by mutations in RAP1, RIF1 or RIF2 genes. We conclude that the T2AG3-only telomere is largely, if not totally, devoid of Rap1. This absence of in vivo Rap1 binding to T2AG3 repeats is in agreement with various studies showing that Rap1 is a very poor binder of T2AG3 repeats in vitro (Liu and Tye, 1991; Pollice et al., 2000). These results are also consistent with data presented by Alexander and Zakian (2003) showing that the loss rate of a yeast chromosome is not affected by having a fully vertebrate telomere, and that Rap1 and Rif2 are not binding to a T2AG3-only telomere (Alexander and Zakian, 2003). Our work shows that a yeast telomere is not strictly dependent on the specific binding of Rap1. This might be related to the capacity of the Ku and Cdc13 proteins to efficiently bind T2AG3 DNA extremities (Lin and Zakian, 1996; Bianchi and de Lange, 1999; Alexander and Zakian, 2003).
Unexpectedly, the rap1-18, rif1Δ or rif2Δ mutations, instead of overelongating telomeres as in wild-type cells, provoke a slight telomere shortening in humanized cells. Thus, the genes involved in the Rap1-dependent mechanism that limits telomere elongation in wild-type telomerase cells slightly stimulate telomere elongation in humanized cells. Since this occurs at the T2AG3-only telomere, seemingly devoid of Rap1, one can evoke indirect effects, like an increase in telomerase-activating components or a decrease in telomerase-repressive components when the number of telomeric Rap1 complexes is low. An alternative explanation is that a small number of Rap1 complexes at the T2AG3-only telomere stimulates telomere elongation in a way similar to the activation of telomerase by the C-terminal part of Rap1, described previously for short telomeres in TLC1 cells (Ray and Runge, 1998). Finally, it is worth noting that a yeast strain maintaining a T2AG3-only telomere constitutes a valuable tool to reconstitute and to study the functions of vertebrate telomere proteins in a highly tractable organism. A similar strategy could be applied to different telomeric sequences, like those from plants or ciliates.
Multiple counting pathways: evolutionary and functional considerations
The data presented here raise the possibility that natural subtelomeric regions, containing vertebrate-like repeats, participate in the regulation of yeast telomere length. This would provide an explanation for the differential length regulation of natural telomeres (Craven and Petes, 1999). Furthermore, physiological variations in TEL1 activity might link the pathway of natural telomere length regulation to DNA-damage checkpoints.
The fact that budding yeast can recognize and use vertebrate repeats suggests that the essential features required for telomere function have been highly conserved in evolution. Indeed, human cells appear to exhibit multiple counting pathways, dependent or not dependent on hRAP1 (Smogorzewska et al., 2000; Ancelin et al., 2002).
In conclusion, our work demonstrates the existence of multiple pathways of the counting mechanism in yeast that appear to share key components with those of the higher eukaryotes. The Rap1-independent pathway revealed in tel1Δ cells might well share common features with the shorter length setting that occurs in cells of ataxia telangiectasia patients. These findings show the importance for the organisms to properly control telomere length and telomerase activity at the level of the telomere itself.
Materials and methods
Plasmid and yeast strains
pHumaTel is a derivative of pURTel (Gottschling et al., 1990) where the 80 nt sequence of TG1–3 repeats is replaced by a sequence containing 12 T2AG3 repeats. Double-stranded oligonucleotides with BamHI-compatible ends were inserted into the BamHI site of pURTel or of pHumaTel. An AscI site has been included at one side of the oligonucleotides in order to determine their orientation once inserted into the plasmid DNA. The integrity of the cloned sequences has been checked by sequencing. The sequences of the inserted DNA between the two BamHI sites are the following (the BamHI sites are in bold and the AscI sites are underlined): (i) NR: 5′-GGATCCGGCGCGCCTCTT CAATTCTTGCCTTCTTTCTATAATAATTCTTCAGCTTGATCTCG ATTACTTTACTAGGATCC; (ii) (TG1–3)23: 5′-GGATCCGGCGCG CCGTGTGTGGGTGTGTGGGTGTGGTGTGTGGGTGTGGTGTGT GGGTGTGGGTGTGTGGGTGTGGATCC; (iii) (RAP1)4: 5′-GGA TCCAATGTGTGGGTGCAATGTGTGGGTGCAATGTGTGGGTGC AATGTGTGGGTGCAATGTGTGGGCGCGCCGGATCC; and (iv) (T2AG3)10: 5′-GGATCCGGCGCGCCTTAGGGTTAGGGTTAGGGT TAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGG GATCC.
The strains Y (MATa trp1 his3 ade2 lys2 ura3) and Hy (MATα trp1 his5 ade2 lys2 ura3 tlc1-h) derive from spores of the diploid strain C6.1, a generous gift of P.P.Liu (Henning et al., 1998). The Y and Hy strains were used to fragment VII-L in order to insert a linearized pURTel or pHumaTel DNA derivatives containing the oligonucleotide NR, (RAP1)4, (T2AG3)10 and (TG1–3)23. Subsequently, tel1Δ derivatives were obtained.
Null mutations of TEL1, RIF1 and RIF2 genes were created using a PCR product as described in Wach et al. (1994). All gene disruptions or replacements were confirmed by Southern blot analysis. At least five restreaks (∼100 generations) were systematically carried out before measuring telomere length, so that sufficient divisions had occurred to reach a steady-state telomere length.
A myc tag epitope was inserted at the C-terminus of the RAP1 endogenous gene in fragmented Y, Ytel1Δ and Hy strains. Plasmid pFA6a-13Myc-TRP1 was used for the PCR-based epitope tagging technique, which was performed as described by Knop et al. (1999).
The rap1-18 mutation was obtained as follows: the last 435 nucleotides of the RAP1 gene, missing in the rap1-18 mutation (Kyrion et al., 1992), were replaced by a cassette containing six HA tags in-frame with the RAP1 gene as well as the Sp TRP1 selection gene. Plasmid pYM4 was used for the PCR-based epitope tagging technique (Knop et al., 1999).
Assays for telomeric structure and composition
Telomere blotting and telomere length measurement were performed as described in Marcand et al. (1999). By analyzing the same gels by two different experimenters or the same samples on two different gels, we estimated that the standard error in telomere length measurements was <10%. For each strain, the length was measured from two or three independent cultures. The difference between cultures does not usually exceed 10%, i.e. the standard error due to the procedure of length measurement.
The URA3-tagged telomeric DNA was amplified as described in Tzfati et al. (2000) using the following primers: anchor, 5′(phosphate)-AGGGTTAATAAGCGGCCGCGTCG(amine)-3′; hyb-anchor, 5′-CGA CGCGGCCGCTTATTAACCCT; pTUF, 5′-CCATCGATGGCATA TTTGAGAAGATGAGGCCAGC. The anchor primer was ligated to genomic DNA using RNA ligase (Promega); subsequently, the DNA was digested with EcoRI and amplified with hyb-anchor and pTUF primers. The PCR product was controlled by gel electrophoresis, which revealed the characteristic smear of telomeric fragments. Subsequently, after purification, it was cloned into the pGEMT vector (Promega) and sequenced.
ChIP and quantitative PCR using the LightCycler instrument (Roche Biochemicals) were performed as described previously (Strahl-Bolsinger et al., 1997; Schramke et al., 2001). We used three pairs of primers: primers in the ASF2 gene as a negative control (ORF, primers: GCGCCTCTTTCTACTCTTAATCC, TCAGGTTGGCCCTTATATGT GGTG); primers surrounding three Rap1-binding sites inside the promoter of TEF2 (RPG, primers: GCCGGCTGTAGGGGGGC GCCAT, CTTTCCAGCATAGTCGAAGAGA); and primers located between the telomere and the subtelomeric URA3 gene (TEL, primers: GCAATATTTGAGAAGATGCGGCCAGC, TAATAACTGATATAA TTAAATTGAA). The quantitative analyses by real-time PCR were performed mainly as described in Ladenburger et al. (2002). Briefly, genomic DNA of known concentration was amplified by using the relevant primers to produce a standard curve. Identical PCRs were performed with the DNA from immunoprecipitated chromatin as a template. The concentration of a given DNA fragment (either RPG, TEL or ASF2) was determined by extrapolation to the relevant standard curve.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We would like to thank P.P.Liu, D.Gottschling and D.Shore for various strains, plasmids and advice. We would like also to thank M.Brunori for critical reading of the manuscript and M.K.Alexander and V.A.Zakian for communicating unpublished results. The work in E.G.’s laboratory was supported by ‘La Ligue Nationale contre le Cancer’. Work in the laboratory of V.G. was supported by the ‘Association pour la Recherche contre le Cancer’ and by the ‘Fondation pour la Recherche Medicale’. Work in F.A.’s laboratory was supported by University of Rome ‘La Sapienza’ (La cellula di lievito come modello in biologia cellulare e per lo sviluppo di sistemi biotecnologici).
References
- Alexander M.K. and Zakian,V.A. (2003) Rap1p telomere association is not required for mitotic stability of a C3TA2 telomere in yeast. EMBO J., 22, 1688–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancelin K., Brunori,M., Bauwens,S., Koering,C.E., Brun,C., Ricoul,M., Pommier,J.P., Sabatier,L. and Gilson,E. (2002) Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell. Biol., 22, 3474–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett M.A., Buckle,V.J., Evans,E.P., Porter,A.C.G., Rout,D., Smith,A.G. and Brown,W.R.A. (1993) Telomere directed fragmentation of mammalian chromosomes. Nucleic Acids Res., 21, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A. and de Lange,T. (1999) Ku binds telomeric DNA in vitro. J. Biol. Chem., 274, 21223–21227. [DOI] [PubMed] [Google Scholar]
- Bilaud T., Koering,C.E., Binet-Brasselet,E., Ancelin,K., Pollice,A., Gasser,S.M. and Gilson,E. (1996) The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res., 24, 1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T., Brun,C., Ancelin,K., Koering,C.E., Laroche,T. and Gilson,E. (1997) Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet., 17, 236–239. [DOI] [PubMed] [Google Scholar]
- Blackburn E.H. (2000) The end of the (DNA) line. Nat. Struct. Biol., 7, 847–850. [DOI] [PubMed] [Google Scholar]
- Blackburn E.H. (2001) Switching and signaling at the telomere. Cell, 106, 661–673. [DOI] [PubMed] [Google Scholar]
- Blackburn E.H. and Greider,C.W. (1995) Telomeres. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Brigati C., Kurtz,S., Balderes,D., Vidali,G. and Shore,D. (1993) An essential yeast gene encoding a TTAGGG repeat-binding protein. Mol. Cell. Biol., 13, 1306–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccoli D., Smogorzewska,A., Chong,L. and de Lange,T. (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet., 17, 231–235. [DOI] [PubMed] [Google Scholar]
- Chikashige Y. and Hiraoka,Y. (2001) Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr. Biol., 11, 1618–1623. [DOI] [PubMed] [Google Scholar]
- Chong L., van Steensel,B., Broccoli,D., Erdjument-Bromage,H., Hanish,J., Tempst,P. and de Lange,T. (1995) A human telomeric protein. Science, 270, 1663–1667. [DOI] [PubMed] [Google Scholar]
- Cooper J.P., Nimmo,E.R., Allshire,R.C. and Cech,T.R. (1997) Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature, 385, 744–747. [DOI] [PubMed] [Google Scholar]
- Craven R.J. and Petes,T.D. (1999) Dependence of the regulation of telomere length on the type of subtelomeric repeat in the yeast Saccharomyces cerevisiae. Genetics, 152, 1531–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sanctis V., La Terra,S., Bianchi,A., Shore,D., Burderi,L., Di Mauro,E. and Negri,R. (2002) In vivo topography of Rap1p-DNA complex at Saccharomyces cerevisiae TEF2 UAS(RPG) during transcriptional regulation. J. Mol. Biol., 318, 333–349. [DOI] [PubMed] [Google Scholar]
- Gilson E., Laroche,T. and Gasser,S. (1993) Telomeres and the functional architecture of the nucleus. Trends Cell Biol., 3, 128–134. [DOI] [PubMed] [Google Scholar]
- Gottschling D.E., Aparicio,O.M., Billington,B.L. and Zakian,V.A. (1990) Position effect at S. cerevisiae telomeres: reversible represssion of Pol II transcription. Cell, 63, 751–762. [DOI] [PubMed] [Google Scholar]
- Grossi S., Bianchi,A., Damay,P. and Shore,D. (2001) Telomere formation by rap1p binding site arrays reveals end-specific length regulation requirements and active telomeric recombination. Mol. Cell. Biol., 21, 8117–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning K.A., Moskowitz,N., Ashlock,M.A. and Liu,P.P. (1998) Humanizing the yeast telomerase template. Proc. Natl Acad. Sci. USA, 95, 5667–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J. and Ishikawa,F. (2001) spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol., 11, 1624–1630. [DOI] [PubMed] [Google Scholar]
- Karlseder J., Smogorzewska,A. and de Lange,T. (2002) Senescence induced by altered telomere state, not telomere loss. Science, 295, 2446–2449. [DOI] [PubMed] [Google Scholar]
- Knop M., Siegers,K., Pereira,G., Zachariae,W., Winsor,B., Nasmyth,K. and Schiebel,E. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Koering C.E., Fourel,G., Binet-Brasselet,E., Laroche,T., Klein,F. and Gilson,E. (2000) Identification of high affinity Tbf1p-binding sites within the budding yeast genome. Nucleic Acids Res., 28, 2519–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauskopf A. and Blackburn,E.H. (1996) Control of telomere growth by interaction of RAP1 with the most distal telomeric repeats. Nature, 383, 354–357. [DOI] [PubMed] [Google Scholar]
- Krauskopf A. and Blackburn,E.H. (1998) Rap1 protein regulates telomere turnover in yeast. Proc. Natl Acad. Sci. USA, 95, 12486–12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrion G., Boakye,K.A. and Lustig,A.J. (1992) C-terminal truncation of RAP1 results in the deregulation of telomere size, stability and function in Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 5159–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladenburger E.M., Keller,C. and Knippers,R. (2002) Identification of a binding region for human origin recognition complex proteins 1 and 2 that coincides with an origin of DNA replication. Mol. Cell. Biol., 22, 1036–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Oestreich,S. and de Lange,T. (2000) Identification of human Rap1: implications for telomere evolution. Cell, 101, 471–483. [DOI] [PubMed] [Google Scholar]
- Lin J.J. and Zakian,V.A. (1996) The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl Acad. Sci. USA, 93, 13760–13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Cooper,J.P. and Cech,T.R. (1995) Telomerase and DNA end replication: no longer a lagging strand problem? Science, 269, 1533–1534. [DOI] [PubMed] [Google Scholar]
- Liu X.S., Brutlag,D.L. and Liu,J.S. (2002) An algorithm for finding protein DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol., 20, 835–839. [DOI] [PubMed] [Google Scholar]
- Liu Z. and Tye,B.K. (1991) A yeast protein that binds to vertebrate telomeres and conserved yeast telomeric junctions. Genes Dev., 5, 49–59. [DOI] [PubMed] [Google Scholar]
- Lustig A.J. (1992) Hoogsteen G–G base pairing is dispensable for telomere healing in yeast. Nucleic Acids Res., 20, 3021–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S., Gilson,E. and Shore,D. (1997) A protein-counting mechanism for telomere length regulation in yeast. Science, 275, 986–990. [DOI] [PubMed] [Google Scholar]
- Marcand S., Brevet,V. and Gilson,E. (1999) Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J., 18, 3509–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern M.J. and Blackburn,E.H. (1995) Runaway telomere elongation caused by telomerase RNA gene mutations. Nature, 376, 403–409. [DOI] [PubMed] [Google Scholar]
- McEachern M.J., Krauskopf,A. and Blackburn,E.H. (2000) Telomeres and their control. Annu. Rev. Genet., 34, 331–358. [DOI] [PubMed] [Google Scholar]
- Pandita T.K. (2002) ATM function and telomere stability. Oncogene, 21, 611–618. [DOI] [PubMed] [Google Scholar]
- Park M.J., Jang,Y.K., Choi,E.S., Kim,H.S. and Park,S.D. (2002) Fission yeast Rap1 homolog is a telomere-specific silencing factor and interacts with Taz1p. Mol. Cells, 13, 327–333. [PubMed] [Google Scholar]
- Pollice A., Zibella,M.P., Bilaud,T., Laroche,T., Pulitzer,J.F. and Gilson,E. (2000) In vitro binding of nucleolin to double-stranded telomeric DNA. Biochem. Biophys. Res. Commun., 268, 909–915. [DOI] [PubMed] [Google Scholar]
- Ray A. and Runge,K.W. (1998) The C terminus of the major yeast telomere binding protein Rap1p enhances telomere formation. Mol. Cell. Biol., 18, 1284–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A. and Runge,K.W. (1999a) Varying the number of telomere-bound proteins does not alter telomere length in tel1Δ cells. Proc. Natl Acad. Sci. USA, 96, 15044–15049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A. and Runge,K.W. (1999b) The yeast telomere length counting machinery is sensitive to sequences at the telomere-nontelomere junction. Mol. Cell. Biol., 19, 31–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell L.L. and Zakian,V.A. (1993) Loss of a yeast telomere: arrest, recovery and chromosome loss. Cell, 75, 729–739. [DOI] [PubMed] [Google Scholar]
- Schramke V., Neecke,H., Brevet,V., Corda,Y., Lucchini,G., Longhese,M.P., Gilson,E. and Geli,V. (2001) The set1Δ mutation unveils a novel signaling pathway relayed by the Rad53-dependent hyperphosphorylation of replication protein A that leads to transcriptional activation of repair genes. Genes Dev., 15, 1845–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A., van Steensel,B., Bianchi,A., Oelmann,S., Schaefer,M.R., Schnapp,G. and de Lange,T. (2000) Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol., 20, 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Hecht,A., Luo,K. and Grunstein,M. (1997) SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev., 11, 83–93. [DOI] [PubMed] [Google Scholar]
- Szostak J.W. and Blackburn,E.H. (1982) Cloning yeast telomeres on linear plasmid vectors. Cell, 29, 245–255. [DOI] [PubMed] [Google Scholar]
- Tzfati Y., Fulton,T.B., Roy,J. and Blackburn,E.H. (2000) Template boundary in a yeast telomerase specified by RNA structure. Science, 288, 863–867. [DOI] [PubMed] [Google Scholar]
- van Steensel B. and de Lange,T. (1997) Control of telomere length by the human telomeric protein TRF1. Nature, 385, 740–743. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat,A., Pohlmann,R. and Philippsen,P. (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wotton D. and Shore,D. (1997) A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in S. cerevisiae. Genes Dev., 11, 748–760. [DOI] [PubMed] [Google Scholar]
- Zakian V.A. (1995) Telomeres: beginning to understand the end. Science, 270, 1601–1607. [DOI] [PubMed] [Google Scholar]