Abstract
The global pattern of growth-phase-dependent gene expression of Helicobacter pylori during in vitro culture was analyzed by using a high-density DNA microarray. To detect consistent coordinated gene expression in this bacterium, temporal changes in transcription were assessed in two independent time courses. Cluster analysis of the expression profiles highlighted a major switch in gene expression during the late log-to-stationary phase transition that we have termed the Log-Stat switch. Statistical analysis of the genes that were significantly induced or repressed during the Log-Stat switch revealed that many of these genes were related to virulence. Among these, expression of the genes for the neutrophil activating protein (napA) and the major flagellin subunit (flaA) were significantly induced. Additionally, the expression of a number of genes involved in iron homeostasis changed dramatically at this switch; the gene for the iron-storage protein, pfr, was induced, while the genes for two putative iron uptake proteins, fecA and frpB, were significantly repressed. These data suggest that the late log phase may correspond to the most virulent phase of growth in H. pylori and may be intimately related to its pathogenesis. The use of microarrays to analyze the kinetics of the transcriptional response of a bacterial pathogen to a changing environment has enabled the discovery of previously unappreciated relationships between genes by elucidation of coordinated gene expression profiles.
Helicobacter pylori is a human pathogen that inhabits the harsh environmental niche of the stomach. Its colonization causes a number of outcomes, including gastric and duodenal ulcer, gastric cancer, and B-cell mucosa-associated lymphoid tissue lymphoma (18, 42). The ability of H. pylori to live in this acidic environment makes its physiology unique, and much research has focused on understanding the factors that enable this bacterium to survive in the stomach. A number of factors known to be involved in virulence, such as the cag pathogenicity island (cag PAI), motility, and the urease enzyme, have been extensively studied, and significant advances regarding the regulation of expression of these factors have been made (1, 31). However, little is known about the global mechanisms of gene expression regulation in H. pylori and how this expression is modified to cope with changes in the environment or to facilitate chronic infections in the stomach.
Transcriptional regulation in H. pylori is unique compared to that of other pathogens, as it possesses relatively few genes encoding transcriptional regulators. This may be due, in part, to the relatively small size of the H. pylori genome, which has only ∼1,500 predicted open reading frames (ORFs), compared to the ∼1,740 ORFs predicted for Haemophilus influenzae and ∼4,290 ORFs in Escherichia coli. Only four genes in H. pylori code for proteins with helix-turn-helix motifs compared to 34 such proteins in H. influenzae and 148 proteins in E. coli (54). In addition, only one-third of the number of two-component regulatory systems of E. coli are present in H. pylori (54). This apparent lack of regulation may reflect the fact that H. pylori is exposed to few different environments, the stomachs of humans and primates being the only known reservoirs of the bacterium (37). There is, however, evidence that H. pylori uses other mechanisms of regulation. These include slipped-strand mispairing within genes (28) and in putative promoter regions (4) and methylation by its nine type II methyltransferases (37, 54). To date, little is known regarding posttranscriptional or translational control in H. pylori, but evidence from two-dimensional gel electrophoresis analysis suggests that these exist (32). Finally, the H. pylori genome does not have extensive operon structure. For example, the flagellar regulon is not contained in operons in this organism, which further confounds the apparent lack of regulation (54).
Whole-genome expression profiling is now possible with the use of DNA microarrays. This technique has recently been used to profile the global gene expression of numerous model microbial organisms, such as E. coli (44), Caulobacter crescentus (33), Bacillus subtilis (24), and Streptomyces coelicolor (26). However, with the exception of two studies with Streptococcus pneumoniae investigating competence development (45) and the response to an autoinducer peptide (16), few comprehensive expression profiling experiments of pathogenic microorganisms have been performed. Those studies that have been conducted have concentrated on bimodal gene expression such as iron limitation in Pasteurella multocida (41) and low oxygen tension in Mycobacterium tuberculosis (51). No extensive time course (TC) gene expression profiling of fastidious pathogenic organisms has been previously reported.
Herein, we describe comprehensive gene expression profiling of H. pylori grown in vitro. This TC analysis highlights a major switch in gene expression at the late log phase-to-stationary phase transition. The corresponding up-regulation of many of the known virulence factors and the possibility that this switch in expression profiles is triggered by the changing availability of iron in the cytoplasm is discussed. Previously unappreciated coregulated genes were discovered, highlighting the power of TC analysis to simultaneously investigate expression patterns of known virulence factors and to infer the function of unknown genes from their pattern of gene expression. This approach can provide a better understanding of the gene expression regulation of virulence factors and how this may affect H. pylori's ability to cause disease.
MATERIALS AND METHODS
Bacterial strains and growth.
The H. pylori strain Sydney strain 1 (SS1) (34) was revived from frozen stocks and grown on solid media on horse blood plates containing 4% (wt/vol) Columbia agar base (Oxoid, Basingstoke, Hampshire, United Kingdom), 5% (vol/vol) defibrinated horse blood (HemoStat Labs, Dixon, Calif.), 0.2% (wt/vol) β-cyclodextrin (Sigma Chemical Company, St. Louis, Mo.), 10 μg of vancomycin (Sigma)/ml, 5 μg of cefsulodin (Sigma)/ml, 2.5 U of polymyxin B (Sigma)/ml, 50 μg of cycloheximide (Sigma)/ml, 5 μg of trimethoprim (Sigma)/ml, and 8 μg of amphotericin B (Sigma)/ml under microaerobic conditions obtained by using a CampyGen sachet (Oxoid) and an anaerobe jar at 37°C for 48 to 72 h.
TCs.
Two individual TC experiments of H. pylori growth in broth culture were performed. These were done on different days and used individual cultures for each time point. Plate-grown H. pylori was used to inoculate brucella broth (BD, Franklin Lakes, N.J.) liquid media supplemented with 10% (vol/vol) fetal calf serum (GIBCO-Invitrogen, Carlsbad, Calif.) (BB) and grown in microaerobic conditions with shaking at 37°C for 24 h (starter culture). For the first TC, BB media was inoculated with the starter culture to an optical density at 600 nm (OD600) of 0.05 and 5-ml aliquots were distributed into 9- by 50-ml conical tubes (BD), one tube for each time point taken at 6, 12, 18, 24, 30, 36, 42, 48, and 60 h. For the second TC, 20-ml aliquots were distributed into 8- by 125-ml conical flasks, one for each time point taken at 6, 12, 18, 22, 28, 35, 42, and 50 h. RNA was extracted from the remaining time zero hour (T0 h) culture (as described below). For each time point, an aliquot was removed from the sample for OD600 measurement, CFU counts, and microscopic visualization of the culture for assessment of motility and morphology, and the remaining culture was passed through a 0.45-μm-pore-size cellulose acetate filter by vacuum (Millipore, Bedford, Mass.) to remove the bacteria from the media. The filter was immediately placed into a 50-ml conical tube, frozen in liquid N2, and stored at −80°C.
RNA isolation.
Each 50-ml conical tube was thawed on ice, and an appropriate volume of Trizol (Gibco) was added directly to the membrane for lysis and fixation of RNA (1 ml/1 × 107 to 5 × 107 CFU). Total RNA was purified as described previously (39) with the exception that, rather than precipitating the RNA after Trizol extraction, the aqueous phase was used directly in the RNeasy clean-up protocol (Qiagen, Chatsworth, Calif.). Also, an on-column DNase (Qiagen) digestion for 40 min was performed during the RNeasy clean-up. RNA was eluted in RNase-free water and quantified by OD260.
Preparation and hybridization of cDNA probes.
Each time point sample was labeled and hybridized on separate H. pylori microarrays (which have been previously described [46]) together with reference RNA. The reference for both experiments was the T0 h RNA. cDNA was synthesized from 2 μg of total RNA in a standard reverse transcriptase reaction by using Superscript II (−) (Invitrogen) with 1 μg of Panorama H. pylori cDNA labeling primers (SigmaGenosys, The Woodlands, Tex.). Aminoallyl dUTP was then incorporated into the purified cDNA by addition of 5 μl of 10× buffer (400 μg of random octamers/ml, 0.5 M Tris-HCl, 100 mM MgSO4, 10 mM dithiothreitol), 5 μl of dNTP-dUTP mix (0.5 mM [each] dGTP, dATP, dCTP; 0.2 mM aminoallyl dUTP; and 0.3 mM dTTP), and 2 μl of Klenow (New England Biolabs, Beverly, Mass.), and the reaction mixture was incubated for 16 h at 37°C. Free amines were removed by using Microcon YM 30 (Millipore) columns as instructed by the manufacturer, and the concentrated samples were dried in a Speed Vac (Savant). The probe was labeled by the addition of 1/16 of one reaction vial of FluoroLink Cy5 (emission at 635 nm, red) or Cy3 (emission at 532 nm, green) monofunctional dye (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.) in 0.05 M sodium bicarbonate (pH 9.0) and incubated for 1 h at room temperature in the dark. The reaction was quenched by the addition of 1.3 M hydroxylamine and incubated for 15 min at room temperature in the dark. The Cy3 (reference RNA) and Cy5 (time point RNA) reaction mixtures were combined, and unincorporated dye was removed by using a Qia-Quick PCR purification column according to the manufacturer's instructions (Qiagen). The probe was dried in a Speed Vac and resuspended in Tris-EDTA (10 mM Tris [pH 8.0], 1 mM EDTA). For hybridization, 25 μg of yeast tRNA, 3× SSC (3 M NaCl, 0.3 M trisodium citrate [pH 7]) (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and 0.3% (wt/vol) sodium dodecyl sulfate were added to the labeled probe and then heat denatured for 2 min at 99°C. The probe was cooled briefly and then applied to the H. pylori microarray for hybridization in chambers for 16 to 20 h at 55°C. Stringency washes were then performed as previously described (20). The hybridized slides were scanned and analyzed by using a Gene Pix 4000A scanner and the GENEPIX 3.0 software (Axon Instruments, Redwood City, Calif.).
Data analysis.
Data were collated by using the Stanford University Microarray Database (50). Spots were excluded from analysis due to obvious spot abnormalities, low signal (if the sum of the median intensities for the two channels was ≤500), or uneven distribution of pixel intensities in the spot (the standard deviation of pixel intensity ratios was >3.5). The data obtained for the net pixel intensity in each channel of each microarray were normalized by using the default-computed normalization described on the Stanford Microarray Database web site (http://genome-www.stanford.edu/MicroArray/help/results_normalization.shtml). The ratio of the Red (time point sample) to Green (reference) channels for each spot were expressed as log2 (R/G). The data within each TC were normalized by mathematical transformation such that the abundance of each gene's transcript represented by a given spot was relative to the level of that transcript at the time point at the end of the lag phase (the 6-h time point). Only spots which contained data for >80% of the arrays were used, and duplicate spots for each ORF on the microarray were averaged for analysis. There were 1,590 values representing unique genes used for analysis. The data from all arrays used in this study are available at http://genome-www.stanford.edu/MicroArray.
Visualization and statistical analysis of the data.
The log-transformed data were analyzed with the CLUSTER program (version 2.11.01) by performing self-organizing map (SOM) analysis, and the results were displayed by using the TREEVIEW program (version 1.50.1.1) (21) (http://www.microarrays.org /software.html). Genes whose expression level varied by ≥2-fold over the course of both TCs were extracted for visualization. Genes whose average net intensity (above background) across each entire TC was ≥500 and whose expression level changed by less than 1.3-fold in both TCs were deemed constitutively expressed. The statistical significance of the major changes observed by using the clustering analysis was assessed by using the significance analysis of microarrays (SAM) program (described in references 17 and 55) (http://www-stat-class.stanford.edu/SAM/servlet/SAMServlet). In brief, SAM performs iterative t tests between the data for two groups of arrays (assigned by the user) and reports genes whose levels are significantly different between them. For these analyses, the missing data points were first estimated by using a K-nearest neighbor imputation with 10 neighbors (55). Two sets of unpaired two-class SAM analyses were performed on the imputed data, where two time points prior to the major changes in gene expression in each TC were assigned to the first group while two time points after the switch were assigned to the second group. In each case, the data for the first TC were analyzed separately from those of the second TC and only the genes found to be significant in both data sets are reported. For the first analyses to assess changes between mid-log and stationary phases, the time points used were as follows: first TC, mid-log (T18 and T24 h) and stationary phase (T42 and T60 h); second TC, mid-log (T12 and T18 h) and stationary phase (T42 and T50 h). For the second analyses, to assess dramatic changes during the transition from the log to stationary phase, the time points used were as follows: first TC, mid-late log (T30 and T36 h) and early stationary phase (T42 and T60 h); second TC, mid-late log (T18 and T22 h) and early stationary phase (T28 and T35 h). The SAM program calculated a list of genes whose transcript levels were significantly increased or decreased between the two groups and produced a false discovery rate, which is an estimate of the percentage of false positives called. In both analyses, a calculated false discovery rate of <1% was used to assign significance and a twofold cutoff in the change in expression level was imposed. The relative level of significance calculated by the program is also reported (score values are correlated with significance) (17, 55).
The expression pattern of genes of interest were plotted over time with the Microsoft Excel program. Patterns are representative of both TCs.
RPAs.
RNase protection assays (RPAs) were conducted as previously described (40). For each gene, 1 μg of total RNA from the 18-, 22-, 42-, and 50-h time points from the second TC was hybridized to its respective antisense riboprobe. Riboprobe templates were generated by using the primer pairs listed in Table 1, and these produced the following sizes of templates: 302 bp for flaA, 330 bp for pfr, 293 bp for fecA, 359 bp for frpB, and 261 bp for amiE. In each case, the templates were generated by PCR with Taq polymerase and the amplification products were ligated to pGemT (Promega, Madison, Wis.), proper orientation was confirmed, and riboprobes were synthesized by using the Maxiscript kit (Ambion, Austin, Tex.), the appropriate RNA polymerase, and 50 μCi of [32P]UTP (NEN, Boston, Mass.), as previously described (40). The products of RPAs were separated on 5% denaturing polyacrylamide gels and exposed to phosphor-screens (Kodak, Rochester, N.Y.). Quantification and peak analysis of bands were conducted by using a PhosphorImager and the ImageQuant program (Molecular Dynamics, Sunnyvale, Calif.).
TABLE 1.
Primers used in this study
| Primer | DNA sequence (5′-3′) |
|---|---|
| amiE-RPA-F | GGTTTGCCTGGGTTGGAT |
| amiE-RPA-R | GATTTTGCGGTATTTTTG |
| fecA-RPA-F | AAGCGCCAATCAGAGCAT |
| fecA-RPA-R | TCACACCGCCAAAAACAT |
| flaA-RPA-F | CTGACATCGTTCGTTTGA |
| flaA-RPA-R | AATCCCTGTGCCTGCTGA |
| frpB-RPA-F | CTAACCCTGATGTGAATG |
| frpB-RPA-R | ATGCGCGTTTTGATAAGC |
| pfr-RPA-F | GCGGCTGAAGAATACGAG |
| pfr-RPA-R | CTGATCAGCCAAATACAA |
Motility measurements.
A third TC of H. pylori growth in batch culture was performed in order to quantitatively measure changes in motility concurrent with gene expression changes. This TC was performed as described for the first two TCs except that a single 70-ml BB culture was inoculated with an overnight starter culture of H. pylori to an OD600 of 0.06. Samples were taken (3 to 20 ml depending on culture density) at 0-, 6-, 12-, 18-, 24-, 30-, and 36-h time points for OD measurement, RNA extraction, and motility tracings.
The motility of 1-μl samples was monitored by live phase-contrast microscopy with glass slides and coverslips prewarmed to 37°C. A Hammamatsu C2400 video charge-coupled device camera was used to record movement in the field of view via an Argus-20 image processor (by using the TRACE function) onto S-VHS video. Movement was traced over a 5-s period. Two sets of selected video frames for each time point were digitized for the generation of time-lapse films with the National Institutes of Health ObjectImage program. The percentage of motile bacteria at each time was estimated by using these films. In addition, the lengths of 5 to 10 individual motility traces were measured for each time point, and the curvilinear velocity (CLV) of each of these bacteria was calculated in micrometers/second. The average percent motility and CLV for each time point was plotted over time by using Microsoft Excel.
The gene expression profiles from this TC were assessed by microarray as described for the first two TCs. The data for all of the genes involved in flagellar structure, biosynthesis, regulation, and function were extracted, and these data were visualized by using the CLUSTER and TREEVIEW programs. The Excel plot containing the motility data was then compared with the transcriptional profile of the flagellar regulon for this TC.
Supplementary material.
The following material is available at the web site http://falkow.stanford.edu/whatwedo/supplementarydata/. Table S1 is a full list of the genes from the induced set indicated in Fig. 1B that vary by at least twofold over time in both TCs. Table S2 is a full list of the genes from the repressed set indicated in Fig. 1B that vary by at least twofold over time in both TCs.
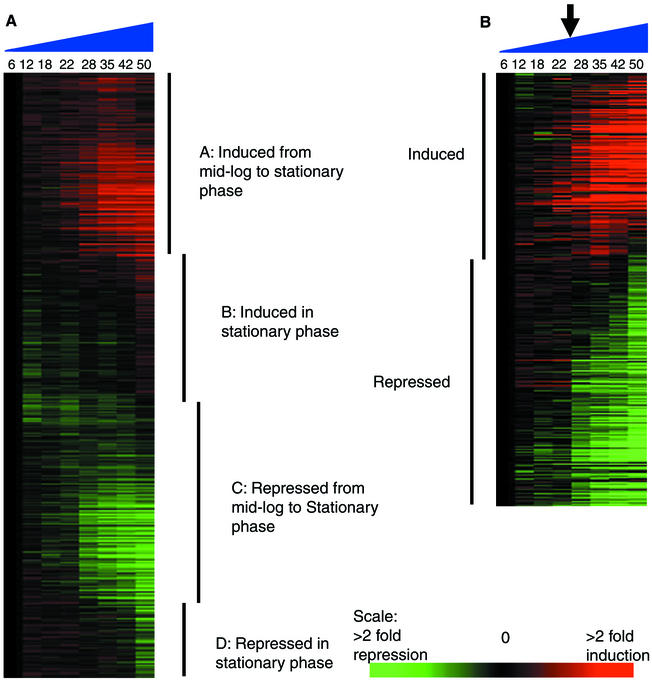
FIG. 1.
Self-organizing maps showing the temporal dependence of gene expression patterns in a TC of H. pylori growth. (A) All 1,590 genes that passed the filtering criteria; (B) genes that were induced or repressed by at least twofold in both TC experiments (325 genes). The major classes of expression patterns are indicated in panel A, and these are reduced to the induced and repressed genes shown in panel B. The progression of time in the TC is shown from 6 to 50 h (blue triangles). The arrow shows the position of the Log-Stat switch. The scale indicates the relative level of expression of each gene, where red indicates induced expression and green indicates repression.
RESULTS AND DISCUSSION
Reliability of array data.
To assess whether the gene expression of H. pylori varied according to the phase of growth and whether this corresponded with coordinated gene expression regulation, two independent TC experiments were performed. RNA samples were collected at time points covering the entire growth cycle. The level of RNA transcript at each time point (time point RNA, labeled with Cy-5 [red]) was compared with the level of transcript at the T0 h time point (reference, labeled with Cy-3 [green]) by using an H. pylori DNA microarray (46). The normalized log ratio of the red to green intensities [log2 (R/G)] for each spot were collated and expressed as the level relative to the time point at the end of the lag phase, the 6-h time point.
The H. pylori microarray used in this study contained duplicate spots representing each ORF designated in the two sequenced strains, 26695 and J99 (46). Duplicate spots provided an internal estimation of array quality and ensured a greater coverage of represented ORFs. Using this approach, reliable data for 96% (1,590 of 1,660) of the represented ORFs on the array were obtained. A pooled estimate of variance calculation showed that the median variance between the log2 (R/G) values for duplicate spots was 0.02 (0.089, 95th percentile), indicating a high correlation in the values for each duplicate measurement. This result also indicated that the quality of data obtained from the H. pylori arrays varied little across each array and thus, the duplicate measurements for each gene were averaged for further analysis.
The gene expression patterns obtained for the two independent TCs were very similar. To ascertain reproducible growth-phase-dependent changes, the data obtained from each TC were assessed separately and only the genes which showed consistent expression patterns between experiments were reported.
Gene expression is temporally regulated during H. pylori growth.
SOM analysis was used to order genes such that those with similar patterns of expression were grouped together and the resultant order of these groups approximated the time of first induction or repression during the TCs (14). Similar, coordinated gene expression patterns were detected in both TCs. The SOM analysis (Fig. 1A) showed that gene expression patterns varied in a time- and growth-phase-dependent manner. Four major expression patterns were observed and are indicated in Fig. 1A. In this SOM analysis, all genes which passed the filtering criteria (described in Materials and Methods) were included (1,590 genes). It is evident that the expression level of many genes did not vary significantly over time (80% of spots varied by <2-fold). This suggests that a large number of genes were either constitutively expressed or were not expressed at all during batch culture. A gene was considered to be expressed only if the net intensity value in the red channel was ≥500. Using this criterion, it was found that the average number of genes expressed at any one time point in these TCs was ∼40% (data not shown). The genes which were expressed and had the least variance in expression over time in both TCs were considered constitutively expressed genes (Table 2). This set of 15 genes includes those that are likely to be involved in homeostasis during culture, such as the central intermediary metabolism genes (hypE and ppk) and the transport and binding genes (narK and proWX). Others are involved in the maintenance of cell structure (dgkA and neuB). Interestingly, a number of genes of unknown function were also constitutively expressed, suggesting an important as-yet-unidentified role for these gene products.
TABLE 2.
Constitutively expressed genes
| UNIQIDb | Symbol | Name | Category |
|---|---|---|---|
| HP0178 | neuB | Spore coat polysaccharide biosynthesis protein E, sialic acid synthase | Cell envelope |
| HP0289 | Putative | Toxin-like OMP, putative (VacA paralog) | Cell envelope |
| HP0047 | hypE | Hydrogenase expression/formation protein | Central intermediary metabolism |
| HP1010a | ppk | Polyphosphate kinase | Central intermediary metabolism |
| HP0821 | uvrC | Excinuclease ABC subunit C | DNA metabolism |
| HP0700 | dgkA | Diacylglycerol kinase | Fatty acid and phospholipid metabolism |
| HP0313a | narK | Nitrite extrusion protein, putative | Transport and binding |
| HP0818a | proWX | Osmoprotection binding protein | Transport and binding |
| HP0639 | Conserved | Conserved hypothetical | |
| HP0810 | Conserved | Conserved hypothetical | |
| HP0952a | Conserved | Integral membrane protein | Conserved hypothetical |
| HP1066 | Conserved | Putative OMP | Conserved hypothetical |
| HP1221a | Conserved | Conserved hypothetical | |
| HP0287 | Unknown | H. pylori specific | |
| HP0608 | Unknown | H. pylori specific |
Genes with the highest expression level in this group (based on an average net intensity in the red channel of >1,000).
UNIQID, unique identifier for each gene assigned by The Institute for Genomic Research (TIGR) (54).
Those genes whose expression level was increased or decreased by at least twofold over both TCs are depicted in Fig. 1B (325 of 1,590, 20%). Only 40 of these genes changed by ≥4-fold over time. The subtle expression patterns observed in the first SOM in Fig. 1A were reduced to two prominent patterns of expression in Fig. 1B: genes whose level of expression was reduced (repressed set) and genes whose expression was increased (induced set) during the transition into the stationary phase. More genes comprised the repressed set (64%) than the induced set (36%). Interestingly, this is a similar result to the TC analysis of S. coelicolor growth, where 80% of the genes analyzed did not change substantially over time and the remaining 20% were equally divided into up- and down-regulated genes (26). The full set of genes which change by at least twofold are listed in the supplementary material in Tables S1 (induced set) and S2 (repressed set).
The repressed set of genes was composed primarily of ribosomal genes; genes involved in DNA synthesis, transcription, and translation; genes encoding transport and binding proteins; and genes involved in energy metabolism. The gene showing the greatest reduction in expression over the TC was that for aliphatic amidase (amiE), which was reduced by ≥5-fold in both TCs.
The induced set of genes was more heterogeneous in nature but included many of the genes known to be involved in virulence. These include the cytotoxin-associated gene (cagA), the neutrophil-activating protein (napA), a large number of genes coding for outer membrane proteins (OMPs), some regulatory protein genes, and many of the genes encoding stress-related proteins, such as the chaperone and heat shock protein genes, clpB and dnaK. The gene with the highest induction was the non-heme iron-containing ferritin (pfr) that was induced by ≥10-fold in both TCs.
A major switch in gene expression profiles occurs during the late log phase.
The gene expression pattern observed in Fig. 1B indicated that there was a switch in gene expression during the growth curve, which corresponds with the transition from late log phase to stationary phase. We termed this dramatic shift in gene expression the Log-Stat switch. Prior to this point, the expression levels of the affected genes changed little, while after the switch, levels of many genes began to increase or decrease dramatically. This switch also directly followed a change from maximum motility and spiral shape to a decline in these characteristics (data not shown).
The majority of previous studies have assessed the mRNA level of a given gene at just one time point in the growth cycle, comparing different growth conditions or mutants (9, 10). Two recent microarray studies investigated the transcriptional response of H. pylori to acid. These studies used a single time point and showed no overlap between the genes identified (2, 5). This emphasizes the difficulty in comparing only a single time point for analysis of global transcriptional changes, particularly when the conditions being compared may cause the bacterium to grow at different rates.
The data presented in the present study show that gene expression patterns in H. pylori can vary dramatically within short periods of time, particularly at the transition between the log and stationary phases. Thus, our data highlight the importance of examining a number of time points during the growth cycle to investigate the kinetic response of transcription to environmental changes or mutations. This study represents the first use of TC experiments for microarray analysis of global transcriptional coordination in H. pylori.
Significance analysis of the Log-Stat switch.
To assess the significance of the observed changes in gene expression levels during the Log-Stat switch, we performed SAM. The SAM program identifies genes which have significant changes in expression between the assigned groups of arrays by using a series of iterative t tests. A two-class, unpaired SAM analysis was performed on each TC to determine genes showing significant changes between the mid-log and stationary phases. A group of 75 genes were found to be significantly changed in this analysis, and these included genes representative of the trends observed in Fig. 1B (Table 3). genes that included genes involved in virulence: the cag PAI genes cagA and cag1, the neutrophil-activating protein napA, the major flagellin flaA, and a number of OMP genes, omp5, omp29, omp11, and hopA. Included in the repressed set of 52 genes were a large number of transcription and translation genes as well as the urease structural subunit gene ureA and the regulatory genes gppA and spoT. Perhaps not surprisingly, it appears that the transition from log to stationary phase was characterized by the repression of many of the genes required for bacterial growth and replication. In contrast, apparently non-growth-related genes were up-regulated at this time. This suggests that other cellular processes are important in the stationary phase. The up-regulation of many key virulence genes may indicate an increase in the organism's ability to cause disease in this growth phase.
TABLE 3.
Genes whose expression is significantly induced or repressed between the mid-log and stationary phases as assessed by a two-class unpaired SAM analysis
| Gene set | Category and TIGR no.a | Symbol | Putative function | First TC
|
Second TC
|
||
|---|---|---|---|---|---|---|---|
| Scoreb | Fold changec | Scoreb | Fold changec | ||||
| Induced genes | Cell envelope | ||||||
| HP0227 | omp5 | OMP | 1.7 | 2.2 | 2.5 | 5.0 | |
| HP0229 | hopA | OMP/porin | 1.4 | 2.5 | 2.3 | 3.9 | |
| HP0472 | omp11 | OMP | 1.9 | 2.5 | 1.7 | 3.1 | |
| HP1342 | omp29 | OMP | 1.6 | 2.1 | 2.5 | 5.1 | |
| Cellular processes | |||||||
| HP0109d | dnaK | Chaperone and heat shock protein, 70 kDa | 2.7 | 4.2 | 2.9 | 6.8 | |
| HP0110d | grpE | Cochaperone and heat shock protein, 24 kDa | 1.7 | 3.5 | 3.8 | 6.2 | |
| HP0111d | hrcA | Heat shock regulator | 1.3 | 2.9 | 3.2 | 4.7 | |
| HP0243 | napA | Neutrophil-activating protein | 2.3 | 4.5 | 4.3 | 9.3 | |
| HP0520 | cag1 | cag PAI protein | 1.0 | 2.1 | 1.9 | 3.9 | |
| HP0547 | cagA | Cytotoxiticity-associated gene A | 2.3 | 2.6 | 2.3 | 6.2 | |
| HP1006 | traG | Conjugal transfer protein | 2.9 | 3.2 | 1.6 | 4.8 | |
| Degradation of proteins | |||||||
| HP0264 | clpB | ATP-dependent protease binding subunit, heat shock | 2.1 | 2.6 | 2.2 | 3.3 | |
| Energy metabolism | |||||||
| HP0631d | hydA | Quinine-reactive Ni/Fe hydrogenase, small subunit | 3.8 | 4.1 | 4.4 | 8.6 | |
| HP0632d,e | hydB | Quinine-reactive Ni/Fe hydrogenase, large subunit | 3.0 | 3.3 | 3.8 | 5.9 | |
| HP0633d,e | hydC | Quinine-reactive Ni/Fe hydrogenase, cytochrome b subunit | 2.6 | 2.7 | 3.8 | 6.0 | |
| HP1458 | Thioredoxin | Thioredoxin, putative | 1.7 | 4.0 | 2.8 | 8.8 | |
| Motility | |||||||
| HP0601 | flaA | Flagellin A | 2.2 | 2.7 | 2.3 | 5.4 | |
| Transport and binding | |||||||
| HP0653a | pfr | Non-heme iron-containing ferritin | 6.7 | 13.7 | 6.5 | 22.2 | |
| Unknown function | |||||||
| HP0023d | 3.5 | 3.5 | 2.3 | 3.0 | |||
| HP0964d | 2.4 | 2.9 | 2.3 | 3.9 | |||
| HP0965d | 2.5 | 2.8 | 2.2 | 3.3 | |||
| HP0966d | 1.8 | 2.1 | 1.7 | 2.6 | |||
| HP1588 | 1.5 | 2.4 | 3.1 | 4.9 | |||
| Repressed genes | Amino acid biosynthesis | ||||||
| HP0649e | aspA | Aspartate ammonia-lyase | −2.2 | 2.1 | −1.5 | 2.4 | |
| HP0672 | aspB | Aspartate aminotransferase | −3.2 | 3.3 | −1.4 | 2.1 | |
| Biosynthesis of cofactors | |||||||
| HP1582d,e | pdxJ | Pyridoxal phosphate biosynthetic protein J | −2.3 | 2.9 | −2.6 | 4.3 | |
| HP1583d | pdxA | Pyridoxal phosphate biosynthetic protein A | −2.3 | 2.5 | −2.1 | 3.2 | |
| Cell envelope | |||||||
| HP1373 | mreB | Rod shape-determining protein | −1.7 | 3.1 | −1.2 | 2.2 | |
| HP1429 | kpsF | Polysialic acid capsule expression protein | −2.4 | 2.8 | −1.4 | 2.2 | |
| Cellular processes | |||||||
| HP0630 | mda66 | Modulator of drug activity | −1.6 | 2.5 | −1.6 | 3.0 | |
| Central intermediary metabolism | |||||||
| HP0004 | icfA | Carbonic anhydrase | −1.9 | 2.3 | −1.3 | 2.1 | |
| HP0073 | ureA | Urease alpha subunit (urea amidohydrolase) | −1.3 | 2.1 | −1.3 | 2.2 | |
| HP1532 | glmS | Glucosamine fructose-6-phosphate aminotransferase | −1.6 | 2.3 | −1.6 | 2.1 | |
| DNA metabolism | |||||||
| HP0213 | gidA | Glucose-inhibited division protein | −1.6 | 2.0 | −2.0 | 3.3 | |
| HP0259 | xseA | Exonuclease VII, large subunit | −2.1 | 2.4 | −1.2 | 2.0 | |
| Energy metabolism | |||||||
| HP0294 | amiE | Aliphatic amidase | −6.8 | 16.8 | −5.1 | 14.4 | |
| HP1133 | atpG | ATP synthase F1, subunit gamma | −1.1 | 2.3 | −1.3 | 2.6 | |
| Fatty acid and phospholipid metabolism | |||||||
| HP0201d | plsX | Fatty acid/phospholipid synthesis protein | −1.6 | 2.8 | −1.8 | 2.7 | |
| HP0202d | fabH | Beta-ketoacyl-acyl carrier protein synthase III | −1.8 | 2.6 | −1.8 | 2.2 | |
| Regulatory functions | |||||||
| HP0278e | gppA | Guanosine pentaphosphate phosphohydrolase | −1.6 | 2.8 | −1.8 | 2.4 | |
| HP0775 | spoT | Penta-phosphate guanosine-3′-pyrophosphohydrolase | −1.6 | 2.0 | −1.4 | 2.2/PICK> | |
| Transcription and translation | |||||||
| HP0077f | prfA | Peptide chain release factor RF-1 | −2.3 | 2.1 | −4.7 | 2.2 | |
| HP0297 | rpl27 | Ribosomal protein L27, 50S | −1.6 | 2.6 | −1.3 | 2.9 | |
| HP1195d | fusA | Translation elongation factor EF-G | −1.7 | 4.2 | −2.0 | 5.5 | |
| HP1196d | rps7 | Ribosomal protein S7, 30S | −1.5 | 2.7 | −1.5 | 3.5 | |
| HP1198d | rpoB | DNA-directed RNA polymerase, beta subunit | −1.5 | 2.9 | −2.1 | 3.6 | |
| HP1199d | rpl7/l12 | Ribosomal protein L7/L12, 50S | −1.4 | 2.2 | −2.0 | 3.5 | |
| HP1200d | rpl10 | Ribosomal protein L10, 50S | −1.9 | 2.3 | −2.5 | 4.3 | |
| HP1292d | rpl17 | Ribosomal protein L17, 50S | −1.8 | 3.1 | −2.1 | 3.5 | |
| HP1293d | rpoA | DNA-directed RNA polymerase, alpha subunit | −1.8 | 4.2 | −2.5 | 5.5 | |
| HP1294d | rps4 | Ribosomal protein S4, 30S | −1.6 | 3.6 | −1.7 | 4.0 | |
| HP1295d | rps11 | Ribosomal protein S11, 30S | −1.3 | 3.1 | −1.3 | 2.8 | |
| HP1296d | rps13 | Ribosomal protein S13, 30S | −1.6 | 2.5 | −1.5 | 2.7 | |
| HP1299d | map | Methionine amino peptidase | −2.1 | 2.9 | −2.3 | 4.1 | |
| HP1300d | secY | Preprotein translocase subunit | −2.1 | 4.0 | −3.1 | 6.4 | |
| HP1302d | rps5 | Ribosomal protein S5, 30S | −1.7 | 2.5 | −2.3 | 4.0 | |
| HP1304d | rpl6 | Ribosomal protein L6, 50S | −1.3 | 2.0 | −1.7 | 3.6 | |
| HP1307d | rpl5 | Ribosomal protein L5, 50S | −1.4 | 2.0 | −2.0 | 4.0 | |
| HP1309d | rpl14 | Ribosomal protein L14, 50S | −1.8 | 2.8 | −2.2 | 4.3 | |
| HP1312d | rpl16 | Ribosomal protein L16, 50S | −1.6 | 2.8 | −2.9 | 4.8 | |
| HP1313d | rps3 | Ribosomal protein S3, 30S | −1.9 | 3.0 | −2.6 | 7.1 | |
| HP1319d | rpl3 | Ribosomal protein L3, 50S | −2.2 | 2.5 | −2.2 | 4.4 | |
| HP1555 | tfs | Translation elongation factor EF-Ts | −1.1 | 2.0 | −1.5 | 2.6 | |
| Transport and binding | |||||||
| HP0140 | lctP | l-Lactate permease | −1.6 | 2.1 | −2.1 | 2.9 | |
| HP0686e | fecA | Iron(III) dicitrate transport protein | −3.9 | 4.7 | −3.3 | 5.3 | |
| HP0876e | frpB | Iron-regulated OMP | −4.6 | 6.5 | −4.7 | 9.6 | |
| HP1170d | glnP | Glutamine ABC transporter, permease protein | −1.8 | 2.3 | −1.4 | 2.0 | |
| HP1172d,e | glnH | Glutamine ABC transporter, periplasmic protein | −2.4 | 2.9 | −2.1 | 3.9 | |
| HP1400 | fecA | Iron(III) dicitrate transport protein | −1.2 | 2.6 | −2.0 | 4.2 | |
| Unknown function | |||||||
| HP0184 | −1.1 | 2.2 | −1.1 | 2.3 | |||
| HP0719 | −1.1 | 2.4 | −1.9 | 3.2 | |||
| HP1124 | −1.3 | 2.6 | −1.2 | 2.7 | |||
| HP1173e | −2.0 | 3.6 | −1.3 | 2.3 | |||
| HP0318e | −2.1 | 2.7 | −1.0 | 2.1 | |||
| HP1490 | −1.5 | 2.6 | −1.2 | 2.1 | |||
Gene number assigned by The Institute for Genomic Research (TIGR) (54).
Relative level of significance assigned by SAM.
Change (n-fold) between mid-log and stationary phase.
Part of putative operon.
Significantly changed in level at the Log-Stat switch and between the mid-log and stationary phases.
Level significantly changed only at the Log-Stat switch.
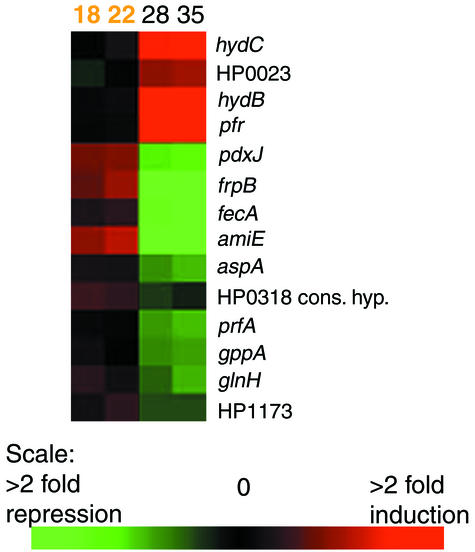
Since the expression levels of some genes changed precisely at the Log-Stat switch, a second two-class, unpaired SAM analysis was performed by using the two time points immediately prior to the switch and the two time points directly following the switch. A small number of genes (14 genes) were found to change significantly at this time (Fig. 2). Four of these genes were significantly increased in level while the levels of 10 genes were significantly decreased. All of these genes were also found to change significantly in the first SAM analysis, except for one gene, prfA. These genes also represent those whose expression changes the most over the entire TC.
FIG. 2.
A hierarchical cluster of the genes found to be significantly induced or repressed at the Log-Stat switch in both TCs by SAM analysis. The data for one TC is shown at the time points used for this analysis comparing expression at T18 and T22 h (shown in orange at the top) with expression at T28 and T35 h (black). The gene names are indicated on the right side (details are shown in Table 3). The scale indicates the relative level of expression of each gene, where red indicates induced expression and green indicates repression. Cons. hyp., conserved hypothetical protein.
The significantly repressed set includes three genes involved in biosynthesis and translation, some key transport and binding protein genes (including two iron uptake genes), and the regulatory gene, gppA. Those genes that had significantly increased expression levels included the iron storage protein gene pfr and two genes involved in energy metabolism (hydA and hydB). These results suggest that the Log-Stat switch comprises a specific change in physiology that, considering the inclusion of iron homeostasis genes, may be driven by changing levels of iron in the cytoplasm.
Validation of microarray results.
To validate the ability of this H. pylori microarray to determine significant changes in gene expression, RPAs were performed. Five representative genes from those found to change significantly between the mid-log and stationary phases of growth were chosen for further analysis (Table 3). As shown in Fig. 3, transcript levels of flaA and pfr were greatly induced in the stationary phase while transcript levels of amiE, fecA (HP0686), and frpB (HP0876) were repressed. Thus, RPA analysis confirms the ability of the H. pylori microarray to detect changes in gene expression.
FIG. 3.
Independent validation of microarray results by RPA. Relative levels of transcript for each of the indicated genes were assessed by using antisense riboprobes as described in Materials and Methods. Clear patterns of expression for the log phase (T18 and T22 h) and stationary phase (T42 and T50 h) are evident and support data obtained via DNA microarray.
The H. pylori regulatory genes.
The apparent coordinated temporal regulation of gene expression found in these TCs suggests that transcriptional regulators may be involved. Little is known of the mechanism of action of the predicted regulatory proteins of H. pylori, with the exception of the HspR, HrcA, and Fur repressor proteins, which are well described (15, 25, 52). In the present study, SAM analysis revealed that hrcA expression was significantly induced while expression of gppA and spoT, encoding known regulatory proteins, were significantly reduced during the transition from log to stationary phase.
The spoT gene of H. pylori has a high degree of similarity to other spoT genes involved in the production of guanosine-3′-diphosphate-5′-diphosphate (ppGpp), whereas the gppA gene controls the synthesis of pppGpp. These nucleotides are known to mediate the stringent response in other bacterial species (11, 13). In these systems, the stringent response has been shown to be involved in diverse cellular processes, such as sporulation and virulence. These functions include both positive and negative regulation of various factors involved in adaptation to metabolic signals (13). It was previously believed that H. pylori exhibits a relaxed phenotype indicating no classical stringent response (48). However, the presence of both the spoT and gppA genes and their significant regulation during the growth curve suggests that H. pylori at least is able to produce the ppGpp and pppGpp nucleotides, and thus, they are likely to be involved in some kind of metabolic regulatory response. Future investigation of the growth-phase regulation of the levels of ppGpp and pppGpp in H. pylori may help elucidate the function of these nucleotides and should reveal whether H. pylori is in fact able to undergo a stringent response.
Operon structure and gene regulation.
In most bacteria, the expression of genes carried by multicistronic units is coordinately regulated. There are relatively few of these operonic structures in H. pylori in comparison to other bacteria, which further confounds the relative scarcity of regulatory proteins in H. pylori (54). Those genes found to be significantly regulated in this study, which are likely to be contained in operons, are indicated in Table 3. Among these are those coding for the stress-related genes dnaK, grpE, and hrcA (DnaK operon) and a set of H. pylori-specific genes of unknown function, HP0963 to HP0966.
The expression pattern of the genes in the DnaK operon and another stress-related operon, HspR, consisting of the genes cbpA, hspR, and orf (HP1026) are shown in Fig. 4A. As expected, this analysis shows that genes within each of these operons have highly related expression profiles. The DnaK operon showed a biphasic pattern of expression, where levels increased following the Log-Stat switch, subsided briefly, and then increased again in the stationary phase. In contrast, the expression levels of the three genes in the HspR operon showed only the spike in expression level after the Log-Stat switch.
FIG. 4.
Line graphs showing the change in expression level [log2 (R/G)] of selected genes over time (in hours). (A) The heat shock operons: DnaK (dnaK, grpE, and hrcA) and HspR (cbpA, hspR, and orf [HP1026]). (B) An operon containing H. pylori-specific genes of unknown function. The legend on the right in each case indicates the names of the genes plotted.
The transcriptional profiles of the two operons are supported by their proposed transcriptional regulation. Both the HspR and HrcA proteins are transcriptional repressors that act on σ80-dependent promoters (25). The promoter of the HspR operon is negatively autoregulated by the HspR protein (25). In contrast, the promoter of the DnaK operon is negatively regulated by both the HrcA and the HspR proteins (25). This difference in promoter activity may explain the monophasic (HspR operon) versus biphasic (DnaK operon) expression profiles of these two operons (Fig. 4A). Thus, from these examples it can be seen that the transcriptional profile data from the present study may provide some insight into the control of operon structures in relation to growth phase.
Interestingly, three of the four genes in the putative operon HP0964 to HP0966 are significantly induced in the stationary phase, and the expression profiles of all the genes in the operon are shown in Fig. 4B. All of these genes are induced two- to fourfold in the stationary phase, which suggests a growth-phase-specific function for these gene products. Since this operon appears to be coregulated with known virulence genes, it is possible that these gene products may also be important in this process.
Expression of virulence factors and the Log-Stat switch.
The microarray data in this study revealed that many of the known virulence factors of H. pylori, such as napA, cagA, flaA, and pfr exhibit peak expression levels in the late log or stationary phase of growth. To date, little is known about the coordinated transcriptional expression of these virulence factors and how this may relate to infection and pathogenicity.
The expression levels of two of these, the neutrophil-activating protein napA and the cytotoxin-associated gene cagA, were both significantly induced over time in these TC studies. The level of gene expression of the cagA gene began to increase at the Log-Stat switch and continued to increase over the entire period of growth sampled (Fig. 5A). The CagA protein is a major effector molecule of the cag PAI which encodes a type IV secretion apparatus and is considered one of the most important virulence factors in H. pylori (6, 12). Despite this, little is known about the functions of the individual proteins encoded by the cag PAI or the transcriptional control of these genes. In the present study, the majority of the genes in the cag PAI did not change significantly during the growth curve. Only one other cag PAI gene, cag1, was found to be significantly regulated during the transition from log to stationary phase (Table 3). The cag1 gene has been shown to be unnecessary for the function of the type IV secretion apparatus (24). Interestingly, another gene, traG, thought to encode part of a different type IV secretion apparatus (47), is also significantly induced after the Log-Stat switch, possibly suggesting a redundant function for this gene product in CagA secretion (Table 3).
FIG. 5.
Line graphs showing the change in expression level [log2 (R/G)] of selected genes over time (in hours). (A) Coexpression of the cagA and omp5/29 genes; (B) some key iron homeostasis genes; (C) selected iron-cofactored genes. The legend on the right in each case indicates the names of the genes plotted.
It was also observed that the expression levels of two genes previously unrelated to the cag PAI, omp5 and omp29, closely followed the expression profile of the cagA gene (Fig. 5A) (correlation, 0.97). omp5 and omp29 are duplicated genes encoding OMPs, (also known as hopM/N) and are predicted to be porins (3). The close coexpression of the cagA gene with the omp5/29 genes may suggest that these OMPs are involved in CagA secretion and/or activation.
The expression of napA begins to increase after the Log-Stat switch and then levels out in the late-stationary phase (Fig. 5B). Dundon et al. (19) have previously demonstrated that the HP-NAP protein accumulates in the stationary phase under normal growth conditions. HP-NAP is important for pathogenesis as H. pylori-induced gastritis is characterized by infiltration of neutrophils and monocytes into the gastric mucosa (38). The HP-NAP protein induces neutrophil adhesion to endothelial cells, directing these cells to the gastric mucosa, and stimulates NADPH-oxidase, which in turn induces the release of oxygen radicals (22). This results in tissue damage, causing the release of nutrients, which promotes H. pylori survival (23). H. pylori can protect itself from the toxic effects of the released oxygen radicals by producing superoxide dismutase and catalase enzymes (8). Interestingly, in the present study, both the sodB gene and the catalase-like gene, HP0485 (data not shown), are shown to be induced at the same time or directly following the induction of napA. As discussed earlier, the stress-related DnaK operon was also found to be up-regulated at this time and may also be necessary for protection of the organism from oxidative stress.
Some pathogenic bacteria such as Salmonella enterica serovar Typhimurium have been shown to be most virulent in the late log phase of growth (35). In S. enterica serovar Typhimurium this has been attributed to the peak in expression of the type III secretion apparatus, one of the major aspects of the virulence determinants in Salmonella sp. (35). Based on the results of the present study, we would predict that H. pylori may be most virulent in the late log phase of growth. Based on our microarray findings, M. Amieva (personal communication) has established that H. pylori in the late log phase of growth is most efficient in delivering CagA protein and inducing cell elongation in AGS cells. This is considered to be one measure of virulence in this organism (49). These observations may suggest that the Log-Stat switch does indeed correspond with an increase in virulence attributes.
Iron homeostasis regulation.
The regulation of iron homeostasis is very important in bacterial pathogens, as the host sequesters available iron from the tissues as a defense mechanism (43). H. pylori has an extensive ability to scavenge iron that may contribute significantly to its virulence, as infection has been linked with iron deficiency anemia (7). A large proportion of the genes whose expression changed dramatically at the Log-Stat switch have previously been shown to be involved in iron uptake and storage (22, 57) or encode proteins that are iron cofactored (19, 36). During the log phase, the putative iron uptake genes, fecA (HP0686), an iron (III) dicitrate transport protein, and frpB (HP0876), an iron-regulated OMP, were expressed at a maximal level (Fig. 5C). The expression of these genes was significantly repressed during the Log-Stat switch (Fig. 3 and Table 3). Interestingly, another gene previously unrelated to iron uptake, amiE (HP0294), had a very similar pattern of expression to these two iron uptake genes (correlation coefficient of 0.95) (Fig. 5C). In contrast, the non-heme iron-containing ferritin, pfr, which codes for the major iron storage protein had the opposite pattern of expression at the Log-Stat switch (Fig. 5C). The expression of this ferritin in H. pylori has previously been shown to accumulate in the stationary phase during normal growth, and our expression data would support this finding (19). A number of the known iron-cofactored protein genes also had an increased level of expression at this time, such as the quinone-reactive Ni-Fe hydrogenase subunit genes hydA-C and napA (which has been shown to bind iron) (19) (Fig. 5B). Interestingly, the expression level of the major iron-dependent regulator gene, fur, did not change significantly over time (data not shown). The net result of this switch appears to be the cessation of iron uptake and the storage of excess iron in the cytoplasm in order to prevent iron toxicity as well as the expression of proteins which require iron as a cofactor. This tight relationship between the expression levels of these particular iron uptake genes and the pfr gene during the growth cycle have not been previously reported and indicates the utility of microarray expression studies in discovering new relationships between genes.
Motility and the corresponding expression of the flagellar regulon.
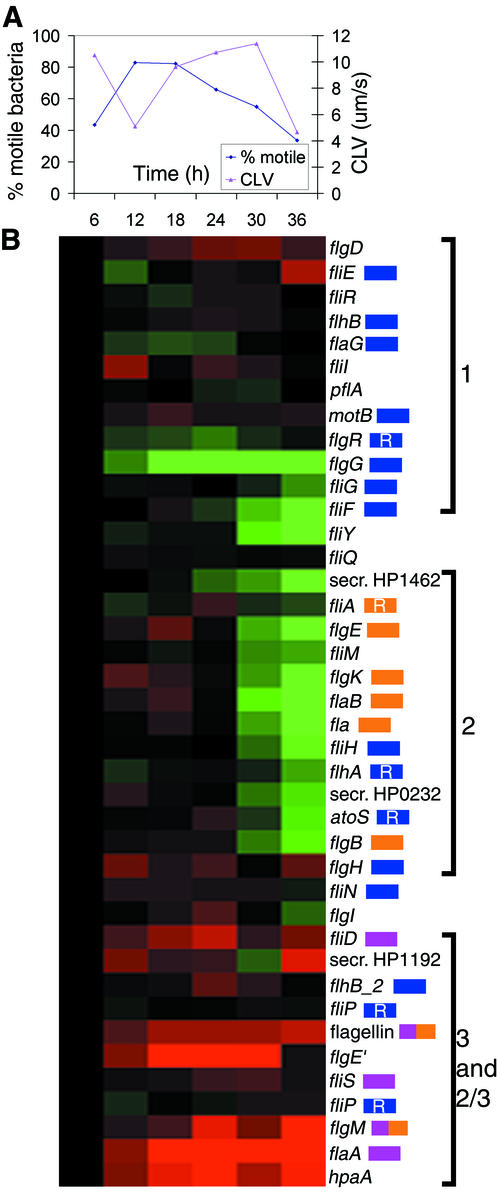
Since it was observed that motility appeared to be regulated in relation to the Log-Stat switch in the first two TCs, a third TC was conducted and used to assess gene expression and to quantitate motility. As was observed for the first two TCs, the percentage of motile bacteria in the culture peaked just prior to the transition from log- to stationary-phase growth (Fig. 6A). In contrast, the average CLV (in micrometers/second) of the bacteria was found to peak in early to mid-stationary phase and then to drop dramatically in late-stationary phase (Fig. 6A). These data are in agreement with a similar study where motility was measured over the growth curve (56).
FIG. 6.
Changes in H. pylori motility and flagellar gene expression over time. (A) Plot showing the changes in the percentage of motile bacteria in the culture and the changes in CLV (in micrometers/second) of the motile bacteria over time (in hours). (B) A hierarchical cluster showing the expression of the flagellar regulon over the same time points indicated in panel A. The gene names are shown on the right side along with color coding showing the predicted class of each gene (blue, class 1; orange, class 2; purple, class 3; orange and purple, classes 2 and 3). R indicates genes which are predicted to be regula-tors. The three clusters containing the majority of the genes in each class (1, 2, 3, and 2 and 3) are shown on the far right. Secr., secreted protein involved in motility.
To understand the role of individual genes in the observed motility, we correlated the expression of the genes of the flagellar regulon with motility. The hierarchical cluster of all of these genes (Fig. 6B) indicates that there were temporal changes in flagellar gene expression and that these were clustered into groups of genes which roughly approximate their proposed flagellar gene class. In S. enterica serovar Typhimurium, the transcriptional regulation of bacterial flagella has been shown to be composed of an intricate network of temporally regulated genes, which are organized into three classes of genes. These are class 1, the early gene complex, class 2, and class 3, the late flagellar gene complex (30). A similar network of flagellar gene transcription appears to be present in H. pylori. However, since the flagellar regulon in this organism is not organized in an operon structure, many of the components of this system are yet to be elucidated (30). The predicted class for some of the flagellar genes is indicated in Fig. 6B (30, 53). The predicted class 1 genes show quite diverse patterns of expression in this study, suggesting that there may be more than one means of regulation for these genes. In contrast, the class 2 genes, including the minor flagellin, flaB, are all down-regulated after transition into the stationary phase. The class 3 genes, including the major structural component of the flagella, flaA, are expressed at a high level from log into stationary phase. The expression of this class of genes appears to be correlated with the highest CLV for this TC (Fig. 6A). Finally, two genes which are predicted to belong to both class 2 and 3, flgM and a flagellin gene (flaG), were expressed at an intermediate level between these classes.
Using these temporal data, it may be possible to predict the class of other genes in the flagellar regulon which have not yet been assigned. For example, the expression of flaA and the hpaA gene were closely correlated (Fig. 6B). The H. pylori flagellum is covered by a flagellar sheath encoded by the hpaA gene, which is thought to be necessary to protect it from gastric acid (37). This close correlation of expression between the flaA and the hpaA genes detected in the present study has not been previously reported. The promoter region of hpaA has been shown to contain a putative σ70 sequence but no apparent σ28 sequence (27). Thus, regulation of the expression of these two genes may not be directly related, especially considering that in flaA-flaB knockout mutants, the flagellar sheath is still produced (29). Another example is the expression of the flagellar hook homolog flgE′ (HP0908), which appears to be regulated in a fashion similar to that of the class 3 genes (Fig. 6B).
This global expression profiling experiment has highlighted the particular advantage of TC analysis for illuminating previously unknown programmed physiological processes in H. pylori. Through the investigation of coordinated expression profiles, the importance of a number of genes of unknown function have been inferred, including a number of constitutively expressed genes. In addition, we have shown that the transition from log- to stationary-phase growth in H. pylori is particularly important in the regulation of iron homeostasis, motility, and virulence gene expression. Although a somewhat simplistic view, these data may suggest that the late log phase corresponds to the most virulent phase of growth and thus may be intimately related to its pathogenesis. It also suggests that the ability of H. pylori to withstand conditions of stress, such as iron limitation, may vary depending on growth phase, and this possibility is currently being investigated.
Acknowledgments
This work was funded by the National Institutes of Health, grant R01 AI38459, and the National Health and Medical Research Council of Australia, and by a special grant provided by the Vice Chancellor of the University of New South Wales, J. Niland, for L.J.T. to complete this work at Stanford University. D.S.M. was supported by the Damon Runyon Cancer Foundation and Digestive Disease Center DK56339.
We are grateful to N. Salama, K. Guillemin, D. Baldwin, and C. Detweiler for assistance with microarray experiments and data analysis, to M. Amieva for the growth-phase-dependent virulence experiments, to I. Dawes for critically reading the manuscript, to N. Saunders and C. Kim for help in programming, and to L. Satkamp for technical assistance.
Editor: B. B. Finlay
REFERENCES
- 1.Akada, J. K., M. Shirai, H. Takeuchi, M. Tsuda, and T. Nakazawa. 2000. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol. Microbiol. 36:1071-1084. [DOI] [PubMed] [Google Scholar]
- 2.Allan, E., C. L. Clayton, A. McLaren, D. M. Wallace, and B. W. Wren. 2001. Characterization of the low-pH responses of Helicobacter pylori using genomic DNA arrays. Microbiology 147:2285-2292. [DOI] [PubMed] [Google Scholar]
- 3.Alm, R. A., J. Bina, B. M. Andrews, P. Doig, R. E. Hancock, and T. J. Trust. 2000. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect. Immun. 68:4155-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 5.Ang, S., C. Z. Lee, K. Peck, M. Sindici, U. Matrubutham, M. A. Gleeson, and J. T. Wang. 2001. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect. Immun. 69:1679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Backert, S., E. Ziska, V. Brinkmann, U. Zimny-Arndt, A. Fauconnier, P. R. Jungblut, M. Naumann, and T. F. Meyer. 2000. Translocation of the Helicobacter pylori CagA protein in gastric epithelial cells by a type IV secretion apparatus. Cell. Microbiol. 2:155-164. [DOI] [PubMed] [Google Scholar]
- 7.Barabino, A. 2002. Helicobacter pylori-related iron deficiency anemia: a review. Helicobacter 7:71-75. [DOI] [PubMed] [Google Scholar]
- 8.Bereswill, S., O. Neuner, S. Strobel, and M. Kist. 2000. Identification and molecular analysis of superoxide dismutase isoforms in Helicobacter pylori. FEMS Microbiol Lett. 183:241-245. [DOI] [PubMed] [Google Scholar]
- 9.Bereswill, S., U. Waidner, S. Odenbreit, F. Lichte, F. Fassbinder, G. Bode, and M. Kist. 1998. Structural, functional and mutational analysis of the pfr gene encoding a ferritin from Helicobacter pylori. Microbiology 144:2505-2516. [DOI] [PubMed] [Google Scholar]
- 10.Bijlsma, J. J. E., B. Waidner, A. H. M. van Vliet, N. J. Hughes, S. Hag, S. Bereswill, D. J. Kelly, C. M. J. E. Vandenbroucke-Grauls, M. Kist, and J. G. Kusters. 2002. The Helicobacter pylori homologue of the ferric uptake regulator is involved in acid resistance. Infect. Immun. 70:606-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cassels, R., B. Oliva, and D. Knowles. 1995. Occurrence of the regulatory nucleotides ppGpp and pppGpp following induction of the stringent response in staphylococci. J. Bacteriol. 177:5161-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterji, D., and A. K. Ojha. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4:160-165. [DOI] [PubMed] [Google Scholar]
- 14.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 15.Delany, I., G. Spohn, R. Rappuoli, and V. Scarlato. 2001. The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol. Microbiol. 42:1297-1309. [DOI] [PubMed] [Google Scholar]
- 16.de Saizieu, A., C. Gardes, N. Flint, C. Wagner, M. Kamber, T. J. Mitchell, W. Keck, K. E. Amrein, and R. Lange. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 182:4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Detweiler, C. S., D. B. Cunanan, and S. Falkow. 2001. Host microarray analysis reveals a role for the Salmonella response regulator phoP in human macrophage cell death. Proc. Natl. Acad. Sci. USA 98:5850-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du, M. Q., and P. G. Isaccson. 2002. Gastric MALT lymphoma: from aetiology to treatment. Lancet Oncol. 3:97-104. [DOI] [PubMed] [Google Scholar]
- 19.Dundon, W. G., A. Polenghi, G. Del Guidice, R. Rappuoli, and C. Montecucco. 2001. Neutrophil-activating protein (HP-NAP) versus ferritin (Pfr): comparison of synthesis in Helicobacter pylori. FEMS Microbiol. Lett. 199:143-149. [DOI] [PubMed] [Google Scholar]
- 20.Eisen, M. B., and P. O. Brown. 1999. DNA arrays for analysis of gene expression. Methods Enzymol. 303:179-205. [DOI] [PubMed] [Google Scholar]
- 21.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans, D. J., Jr., D. G. Evans, H. C. Lampert, and H. Nakano. 1995. Identification of four new prokaryotic bacterioferritins, from Helicobacter pylori, Anabaena variabilis, Bacillus subtilis and Treponema pallidum, by analysis of gene sequences. Gene 153:123-127. [DOI] [PubMed] [Google Scholar]
- 23.Fiocca, R., O. Luinetti, L. Villani, A. M. Chiaravalli, C. Capella, and E. Solcia. 1994. Epithelial cytotoxicity, immune responses, and inflammatory components of Helicobacter pylori gastritis. Scand. J. Gastroenterol. Suppl. 205:11-21. [PubMed] [Google Scholar]
- 24.Fischer, W., J. Puls, R. Buhrdorf, B. Gebert, S. Odenbreit, and R. Haas. 2001. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 42:1337-1348. [DOI] [PubMed] [Google Scholar]
- 25.Homuth, G., S. Domm, D. Kleiner, and W. Schumann. 2000. Transcriptional analysis of major heat shock genes of Helicobacter pylori. J. Bacteriol. 182:4257-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, J., C. J. Lih, K. H. Pan, and S. N. Cohen. 2001. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 15:3183-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, A. C., R. P. Logan, S. Foynes, A. Cockayne, B. W. Wren, and C. W. Penn. 1997. A flagellar sheath protein of Helicobacter pylori is identical to HpaA, a putative N-acetylneuraminyllactose-binding hemagglutinin, but is not an adhesin for AGS cells. J. Bacteriol. 179:5643-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Josenhans, C., K. A. Eaton, T. Thevenot, and S. Suerbaum. 2000. Switching of flagellar motility in Helicobacter pylori by reversible length variation of a short homopolymeric sequence repeat in fliP, a gene encoding a basal body protein. Infect. Immun. 68:4598-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Josenhans, C., A. Labigne, and S. Suerbaum. 1995. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J. Bacteriol. 177:3010-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Josenhans, C., E. Niehus, S. Amersbach, A. Horster, C. Betz, B. Drescher, K. T. Hughes, and S. Suerbaum. 2002. Functional characterization of the antagonistic flagellar late regulators FliA and FlgM of Helicobacter pylori and their effects on the H. pylori transcriptome. Mol. Microbiol. 43:307-322. [DOI] [PubMed] [Google Scholar]
- 31.Joyce, E. A., J. V. Gilbert, K. A. Eaton, A. Plaut, and A. Wright. 2001. Differential gene expression from two transcriptional units in the cag pathogenicity island of Helicobacter pylori. Infect. Immun. 69:4202-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jungblut, P. R., D. Bumann, G. Haas, U. Zimny-Arndt, P. Holland, S. Lamer, F. Siejak, A. Aebischer, and T. F. Meyer. 2000. Comparative proteome analysis of Helicobacter pylori. Mol. Microbiol. 36:710-725. [DOI] [PubMed] [Google Scholar]
- 33.Laub, M. T., H. H. McAdams, T. Feldblyum, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290:2144-2148. [DOI] [PubMed] [Google Scholar]
- 34.Lee, A., J. O'Rourke, M. C. De Ungria, B. Robertson, G. Daskalopoulos, and M. F. Dixon. 1997. A standardized mouse model of Helicobacter pylori infection-introducing the Sydney strain. Gastroenterology 112:1386-1397. [DOI] [PubMed] [Google Scholar]
- 35.Lundberg, U., U. Vinatzer, D. Berdnik, A. von Gabain, and M. Baccarini. 1999. Growth phase-regulated induction of Salmonella-induced macrophage apoptosis correlates with transient expression of SPI-1 genes. J. Bacteriol. 181:3433-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maier, R. J., C. Fu, J. Gilbert, F. Moshiri, J. Olson, and A. G. Plaut. 1996. Hydrogen uptake hydrogenase in Helicobacter pylori. FEMS Microbiol. Lett. 141:71-76. [DOI] [PubMed] [Google Scholar]
- 37.Marais, A., G. L. Mendz, S. L. Hazell, and F. Megraud. 1999. Metabolism and genetics of Helicobacter pylori: the genome era. Microbiol. Mol. Biol. Rev. 63:642-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311-1315. [DOI] [PubMed]
- 39.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schoolnik, and A. Camilli. 2002. Host-induced epidemic spread of the Cholera bacterium. Nature 417:642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merrell, D. S., and A. Camilli. 1999. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol. Microbiol. 34:836-849. [DOI] [PubMed] [Google Scholar]
- 41.Paustian, M. L., B. J. May, and V. Kapur. 2001. Pasteurella multocida gene expression in response to iron limitation. Infect. Immun. 69:4109-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peek, R. M., Jr., and M. J. Blaser. 2002. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2:28-37. [DOI] [PubMed] [Google Scholar]
- 43.Perez-Perez, G. I., and D. A. Israel. 2000. Role of iron in Helicobacter pylori: its influence in outer membrane protein expression and in pathogenicity. Eur. J. Gastroenterol. Hepatol. 12:1263-1265. [DOI] [PubMed] [Google Scholar]
- 44.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rimini, R., B. Jansson, G. Feger, T. C. Roberts, M. de Francesco, A. Gozzi, F. Faggioni, E. Domenici, D. M. Wallace, N. Frandsen, and A. Polissi. 2000. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol. Microbiol. 36:1279-1292. [DOI] [PubMed] [Google Scholar]
- 46.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schroder, G., S. Krause, E. L. Zechner, B. Traxler, H. J. Yeo, R. Lurz, G. Waksman, and E. Lanka. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184:2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scoarughi, G. L., C. Cimmino, and P. Donini. 1999. Helicobacter pylori: a eubacterium lacking the stringent response. J. Bacteriol. 181:552-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Segal, E. D., S. Falkow, and L. S. Tompkins. 1996. Helicobacter pylori attachment to gastric cells induces cytoskeletal rearrangements and tyrosine phosphorylation of host cell proteins. Proc. Natl. Acad. Sci. USA 93:1259-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sherlock, G., T. Hernandez-Boussard, A. Kasarskis, G. Binkley, J. C. Matese, S. S. Dwight, M. Kaloper, S. Weng, H. Jin, C. A. Ball, M. B. Eisen, P. T. Spellman, P. O. Brown, D. Botstein, and J. M. Cherry. 2001. The Stanford Microarray Database. Nucleic Acids Res. 29:152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. USA 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spohn, G., and V. Scarlato. 1999. The autoregulatory HspR repressor protein governs chaperone gene transcription in Helicobacter pylori. Mol. Microbiol. 34:663-674. [DOI] [PubMed] [Google Scholar]
- 53.Spohn, G., and V. Scarlato. 1999. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J. Bacteriol. 181:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 55.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Worku, M. L., R. L. Sidebotham, M. M. Walker, T. Keshavarz, and Q. N. Karim. 1999. The relationship between Helicobacter pylori motility, morphology and phase of growth: implications for gastric colonization and pathology. Microbiology 145:2803-2811. [DOI] [PubMed] [Google Scholar]
- 57.Worst, D. J., B. R. Otto, and J. de Graaff. 1995. Iron-repressible outer membrane proteins of Helicobacter pylori involved in heme uptake. Infect. Immun. 63:4161-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]