Abstract
Helicobacter pylori genomes contain about 30 different hop genes, which encode outer membrane proteins. In this study, we analyzed genetic diversity in the H. pylori hopQ (omp27) locus, which corresponds to HP1177 in the genome of H. pylori reference strain 26695. hopQ and its flanking genes were PCR amplified from multiple H. pylori strains, and the nucleotide sequences were determined. This analysis revealed the existence of two different families of hopQ alleles. Type I hopQ alleles are present in the genomes of two fully sequenced H. pylori strains, whereas the existence of type II hopQ alleles has not previously been recognized. Type I and type II hopQ alleles are 75 to 80% identical in nucleotide sequences and encode predicted outer membrane proteins that are 68 to 72% identical in amino acid sequences. PCR-based methods were developed to enable rapid differentiation between type I and type II hopQ alleles. Type I hopQ alleles were found significantly more commonly in cag+/type s1-vacA strains from patients with peptic ulcer disease than in cag-negative/s2-vacA strains from patients without ulcer disease (P < 0.001). Determination of hopQ allelic types provides a new method for classification of H. pylori strains. Further studies in multiple populations of patients are indicated to evaluate the usefulness of this approach for distinguishing potentially ulcerogenic H. pylori strains from less virulent strains.
Helicobacter pylori is a gram-negative bacterium that persistently colonizes the stomachs of more than half of the world's human population. Colonization of the stomach by H. pylori consistently induces gastric inflammation, known as superficial chronic gastritis, and is a risk factor for development of peptic ulcer disease and gastric malignancies (16, 20).
H. pylori exhibits a very high level of intraspecies genetic diversity (11, 31, 55). When H. pylori strains isolated from unrelated humans are compared, each isolate is genetically unique, with a level of relatedness among most orthologous sequences ranging from about 90 to 99% nucleotide identity (1, 4, 21, 24, 42, 49). This diversity is the consequence of numerous point mutations, combined with a very high rate of intraspecies genetic recombination (11, 24, 49, 55). In addition, certain genes are present in some H. pylori strains but absent from others (3, 6, 13, 45). Insight into genetic variation among H. pylori strains has been gained by comparing the complete genome sequences of two different H. pylori strains (26695 and J99) (6, 51). Overall, the nucleotide sequences of these two genomes are about 94% identical. However, 206 different genes have been identified that are present in only one strain but not the other (117 genes present only in strain 26695 and 89 present only in strain J99) (6).
The majority of H. pylori-infected persons remain asymptomatic for decades, but a subset develop peptic ulcer disease, distal gastric adenocarcinoma, or gastric lymphomas (16, 20). Genetic variation among H. pylori strains could be a factor that helps to explain these diverse clinical outcomes of H. pylori infection. Thus far, two candidate markers for distinguishing virulent H. pylori strains (i.e., those associated with peptic ulceration or gastric carcinoma) from less virulent strains have been studied extensively: presence or absence of the cag pathogenicity island and polymorphisms in vacA alleles.
The cag pathogenicity island is a ∼40-kb DNA segment that encodes a highly antigenic protein (CagA) and a type IV secretion pathway utilized for translocating CagA into host cells (2, 7, 14, 15, 35, 46, 47, 52). Upon entry into eukaryotic cells, CagA undergoes phosphorylation of tyrosine residues and induces cytoskeletal alterations in cells (7, 15, 35, 46, 47). The cag pathogenicity island also encodes factors that activate proinflammatory signal transduction pathways in epithelial cells and that probably contribute to an enhanced inflammatory response in the gastric mucosa (2, 14, 52).
The vacA gene encodes a secreted “vacuolating toxin” (9, 34, 39). Effects of VacA on cells include the formation of intracellular vacuoles, formation of anion-selective channels in the plasma membrane, induction of apoptosis, and alterations in the permeability of epithelial monolayers. In contrast to the cag island, which is present in some H. pylori strains but absent from others, all H. pylori strains contain a vacA allele. Two families of vacA alleles (designated type s1 and type s2) can be differentiated by analysis of the 5′ end of vacA (8). Type s1 and type s2-VacA protoxins undergo cleavage of amino-terminal signal sequences at two different sites, and as a result, secreted type s2-VacA toxins contain a 12-amino-acid amino-terminal hydrophilic segment that is absent from secreted type s1 toxins (8, 30, 32). The presence of this segment renders type s2-VacA toxins defective in vacuolating toxin activity.
Type s1-vacA alleles are found predominantly in H. pylori strains that contain the cag pathogenicity island, and type s2-vacA alleles are found predominantly in strains that lack the cag pathogenicity island (8, 22, 44, 54). In many different studies, infection with cag+/s1-vacA strains has been associated with a higher risk of peptic ulceration than has infection with cag-negative/s2-vacA strains (8, 18, 22, 25, 29, 43, 44, 48, 53, 54). A role for the cag pathogenicity island and VacA in the pathogenesis of peptic ulcer disease also has been demonstrated in animal models (36, 50). In several studies, strains classified as cag+/s1-vacA have been associated with an increased risk of distal gastric carcinoma (12, 33). Notably, associations between cag+/s1-vacA strains and severe clinical illness have not been detected as readily in Asian populations as in Western populations (38).
In addition to the cag pathogenicity island and vacA, several other candidate markers for distinguishing virulent H. pylori strains from less virulent strains have been proposed. These include iceA, babA2, and oipA (22, 41, 54, 56). In an effort to identify new markers of virulence, a recent study used whole-genome DNA microarray methodology to analyze gene content in 15 different H. pylori strains (45). The cag pathogenicity island was present in 10 strains and was absent or present in an incomplete form in 5 strains. Ten genes located elsewhere in the H. pylori genome were reported to be “coinherited” with the cag island (i.e., present more frequently in cag+ strains than in cag-negative strains). This group included six genes predicted to encode proteins of unknown function (hypothetical proteins), two genes predicted to encode type II DNA methyltransferases, and two genes predicted to encode outer membrane proteins (babA and omp27). It was suggested that these genes coinherited with the cag island might contribute to the virulence of cag+ strains (45).
In the present study, we sought to examine in further detail the diversity that is reported to exist in the omp27 locus. H. pylori omp27, corresponding to HP1177 in the genome of H. pylori 26695 and JHP1103 in the genome of H. pylori J99 (6, 51), belongs to the hop family of paralogous genes and has recently been renamed hopQ (5). We report here that hopQ alleles can be classified into two highly divergent families and report the use of PCR-based methodology for classifying H. pylori strains based on hopQ genotypes. We report that type I hopQ alleles are found significantly more commonly in cag+/s1-vacA strains from patients with peptic ulcer disease than in cag-negative/s2-vacA strains from patients without ulcer disease. These data confirm and extend the microarray results reported by Salama et al. (45) and suggest that variation in hopQ genotypes among H. pylori strains may be a factor that influences the clinical outcome of H. pylori infections.
MATERIALS AND METHODS
Bacterial strains.
H. pylori strains 26695 and J99 are reference strains for which the entire genome sequences are known (6, 51), and H. pylori Tx30a is an additional reference strain (ATCC 51932). The other 30 H. pylori strains utilized in this study were isolated from patients in Denver, Colorado, or Nashville, Tennessee, who underwent routine upper gastrointestinal endoscopy for a variety of indications. Fifteen of these patients had peptic ulcer disease; the other 15 patients had no ulcer detected at the time of endoscopy and no previous history of peptic ulceration. The cagA and vacA genotypes of these strains have been reported previously (8, 17, 19). H. pylori strains were cultured at 37°C on trypticase soy agar plates containing 5% sheep blood in ambient air containing 5% CO2.
Nucleotide sequence analysis of hopQ alleles.
DNA was extracted from H. pylori as described previously (8). In initial experiments, 3-kb amplicons containing hopQ (corresponding to HP1177 in the genome of H. pylori 26695) were PCR amplified by using primers BA7676 and BA7674, derived from the sequences of two flanking genes (HP1178 and HP1175, respectively) (Table 1). The nucleotide sequences of 3-kb amplicons were then determined in the Vanderbilt University Core Nucleotide Sequencing Laboratory, using the primer-walking method. The relatedness of hopQ alleles or deduced hopQ products from different strains was analyzed by aligning sequences using the ClustalW algorithm in a Macvector software package.
TABLE 1.
Oligonucleotide primers used for analysis of H. pylori hopQ alleles
| Region amplified | Primer designation | Primer sequence |
|---|---|---|
| hopQ and flank- ing genes | BA7676a | 5′AACGCTAAGGCTTTATGCTTATGCTC |
| BA7674b | 5′GTCGTTAAACCCGCTCTAAACTCGG | |
| Type I hopQ | OP5136c | 5′CAACGATAATGGCACAAACT |
| OP4829d | 5′GTCGTATCAATAACAGAAGTTG | |
| Type II hopQ | BA8363e | 5′TCCAATCCAGAAGCGATTAA |
| BA8364f | 5′GTTTTAATGGTTACTTCCACC |
Primer derived from HP1178 (deoD).
Primer derived from HP1175.
Forward primer corresponding to nucleotides 186 to 205 in hopQ from H. pylori 26695.
Reverse primer corresponding to nucleotides 689 to 710 in hopQ from H. pylori 26695.
Forward primer corresponding to nucleotides 199 to 218 in hopQ from H. pylori J262.
Reverse primer corresponding to nucleotides 609 to 629 in hopQ from H. pylori J262.
The hopQ (omp27 or HP1177) sequence from H. pylori strain 26695 corresponds to GenBank accession number AE000623, and the hopQ (JHP1103) sequence from H. pylori strain J99 corresponds to GenBank accession number AE001538 (6, 51).
PCR-based methodology for typing hopQ alleles.
Primers used for PCR-based typing of hopQ alleles are shown in Table 1. Primers OP5136 and OP4829 were designed to specifically amplify type I hopQ fragments, and primers BA8363 and BA8364 were designed to specifically amplify type II hopQ fragments. Thermal cycling parameters for each pair of primers were set at 95°C for 1 min, 50 or 55°C for 1 min, and 72°C for 2 min, for a total of 30 cycles.
Nucleotide sequence accession number.
The hopQ nucleotide sequences described in this study have been assigned GenBank accession numbers AY147193 to AY147201.
RESULTS
Genetic diversity in the hopQ locus.
As a first step in investigating genetic diversity present in the hopQ locus (corresponding to HP1177 in the genome of H. pylori 26695), we designed PCR primers based on the nucleotide sequences of two flanking genes (HP1175 [predicted to encode a conserved hypothetical integral membrane protein] and HP1178 [designated deoD, predicted to encode purine-nucleoside phosphorylase]) (Table 1). After use of these primers (designated BA7674 and BA7676, respectively) in PCR, a 3-kb product was successfully amplified from two strains for which the entire genome sequence is known (strains 26695 and J99), as well as from nine additional H. pylori strains (J104, J258, J262, Tx30a, J154, J190, J63, 86-313, and 87-230). Thus, we did not detect any evidence of large deletions in the hopQ loci of any of these strains. As a next step, we analyzed the nucleotide sequences of these 3-kb PCR products. Each PCR product contained a hopQ orthologue (corresponding to HP1177 in the genome of H. pylori 26695), flanked by sequences orthologous to HP1175 and HP1178 from H. pylori strain 26695.
The hopQ alleles in these strains ranged from 1,890 to 1,926 nucleotides in length. The nucleotide sequences of hopQ alleles amplified from H. pylori strains J104 and J258 and the hopQ sequences from reference strains 26695 and J99 were all relatively closely related to each other (ranging from 89 to 95% nucleotide identity) (Table 2). Similarly, the nucleotide sequences of hopQ alleles amplified from H. pylori strains J262, Tx30a, J154, J190, J63, 86-313, and 87-230 were all closely related to each other (93 to 99% nucleotide identity). In contrast, the sequences of hopQ alleles amplified from strains J262, Tx30a, J154, J190, J63, 86-313, and 87-230 were only 75 to 80% identical to hopQ sequences from strains 26695, J99, J104, and J258. Thus, phylogenetic analysis indicated the existence of two separate lineages of hopQ alleles (Table 2). Hereafter, we define the family of hopQ alleles similar to those of the fully sequenced reference strains (26695 and J99) as type I hopQ alleles and define the newly recognized family of hopQ alleles present in strains J262, Tx30a, J154, J190, J63, 86-313, and 87-230 as type II hopQ alleles.
TABLE 2.
Analysis of relatedness among hopQ alleles and deduced hopQ products from different H. pylori strainsa
| Strain | Nucleotide or amino acid identity (%)b |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 26695 | J99 | J104 | J258 | J262 | Tx30a | J154 | J190 | J63 | 86-313 | 87-230 | |
| 26695 | 89 | 95 | 95 | 76 | 79 | 79 | 77 | 77 | 79 | 75 | |
| J99 | 87 | 91 | 90 | 77 | 79 | 79 | 78 | 78 | 77 | 77 | |
| J104 | 95 | 89 | 93 | 75 | 77 | 77 | 76 | 77 | 77 | 75 | |
| J258 | 94 | 88 | 93 | 78 | 80 | 80 | 80 | 78 | 79 | 78 | |
| J262 | 70 | 70 | 68 | 70 | 94 | 94 | 95 | 95 | 94 | 93 | |
| Tx30a | 70 | 70 | 69 | 71 | 94 | 99 | 97 | 96 | 94 | 93 | |
| J154 | 71 | 70 | 69 | 71 | 94 | 99 | 97 | 96 | 94 | 93 | |
| J190 | 71 | 71 | 69 | 72 | 95 | 96 | 96 | 95 | 94 | 93 | |
| J63 | 71 | 71 | 70 | 72 | 94 | 96 | 96 | 95 | 93 | 93 | |
| 86-313 | 72 | 70 | 70 | 70 | 94 | 93 | 93 | 93 | 92 | 93 | |
| 87-230 | 69 | 70 | 68 | 70 | 93 | 93 | 93 | 94 | 93 | 93 | |
The hopQ alleles in strains 26695, J99, J104, and J258 are classified as type I, and the hopQ alleles in strains J262, Tx30a, J154, J190, J63, 86-313, and 87-230 are classified as type II. Levels of relatedness shown in boldface indicate the considerable divergence between type I and type II hopQ alleles.
Values in the top half of the table represent nucleotide identity, and values in the bottom half represent amino acid identity.
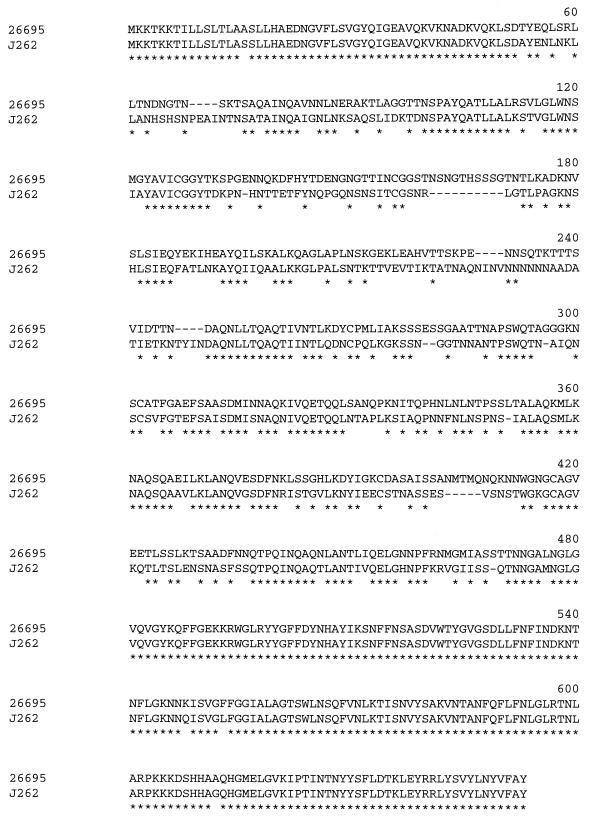
All of the hopQ alleles analyzed encoded predicted proteins with molecular masses ranging from 68 to 70 kDa. The deduced type I HopQ protein sequences encoded by strains 26695, J99, J104, and J258 were all relatively closely related to each other (ranging from 88 to 95% amino acid identity) (Table 2). Similarly, the deduced type II HopQ protein sequences encoded by strains J262, Tx30a, J154, J190, J63, 86-313, and 87-230 were closely related to each other (92 to 99% amino acid identity). In contrast, the deduced type II HopQ sequences encoded by strains J262, Tx30a, J154, J190, J63, 86-313, and 87-230 were highly divergent (68 to 71% amino acid identity) when compared with HopQ sequences from strains 26695, J99, J104, and J258 (Table 2). Type I and type II HopQ protein sequences were closely related within a region comprising 55 amino acids at the amino terminus (which includes a predicted 21-amino-acid amino-terminal signal sequence) and within a region comprising about 183 amino acids at the carboxy terminus (Fig. 1). In contrast, the midregions of type I and type II HopQ proteins were highly divergent (Fig. 1).
FIG. 1.
Alignment of the deduced HopQ amino acid sequence of H. pylori strain 26695 (the prototype type I HopQ sequence) with that of HopQ from strain J262 (the prototype type II HopQ sequence). Stars indicate identical amino acid residues. The amino-terminal and carboxy-terminal regions of the two proteins are closely related, but the midregions are highly divergent.
As reported previously (5), the paralogous H. pylori genes to which type I hopQ alleles are most closely related (based on phylogenic analysis of the deduced protein sequences) include hopA, hopZ, hopD, hopS (babA), hopU, hopT (babB), hopM, hopN, hopO, and hopP. Analysis of type II hopQ alleles indicated that they were related to the same group of paralogous genes. When type II hopQ alleles were compared with the known genome sequences of H. pylori strains 26695 and J99, using the BLAST network service of the National Center for Biotechnology Information, the highest level of relatedness was with the type I hopQ alleles (HP1177 and JHP1103) in these two genomes (76 to 79% nucleotide identity). Sequences closely related (>80% nucleotide identity) to type II hopQ alleles were not identified in either of the two fully sequenced H. pylori genomes. Thus, each of the fully sequenced H. pylori genomes contains a single type I hopQ allele.
Distribution of type I and type II hopQ alleles among H. pylori strains.
To investigate further the prevalence and distribution of type I and type II hopQ alleles among H. pylori strains, we next examined a larger panel of well-characterized strains. We selected 15 H. pylori strains (each of which possessed the cag pathogenicity island and type s1-vacA alleles) that had been isolated from patients with peptic ulcer disease and 15 strains (each of which lacked the cag pathogenicity island and possessed type s2-vacA alleles) that had been isolated from patients with no history of peptic ulcer disease (Table 3). We reasoned that these two groups of strains should represent two ends of a spectrum of virulence. DNA was extracted from the 30 different strains, and, when used as a template for PCR amplification of a fragment of the 16S rRNA gene (40), each of the DNA preparations yielded a product of the expected size (data not shown). Using primers BA7676 and BA7674, we attempted to amplify 3-kb products containing hopQ sequences from each of the 30 DNA preparations, as described above. Amplicons of the expected size (3 to 3.5 kb) were obtained from 28 of the 30 strains, a 5-kb amplicon was obtained from one strain (H. pylori strain J166), and no amplicon was obtained from one strain (H. pylori strain 87-75). The complete hopQ sequences of eight of these strains were determined as described above (Table 2). Partial nucleotide sequences of hopQ alleles from the remaining strains then were determined by sequencing the PCR products, using a single primer (OP4702, 5′ATGAAAAAAACGAAAAAAAC), derived from the nucleotide sequence of the 5′ end of hopQ. This approach yielded about 400 nucleotides of hopQ sequence from 20 different strains. Each of these 20 hopQ sequences could be classified into either the type I or type II family, based on alignments with the two families of hopQ alleles shown in Table 2. Thus, among the 28 H. pylori strains from which hopQ sequence data were successfully obtained, 14 strains contained type I hopQ alleles and 14 strains contained type II hopQ alleles (Table 3). Thirteen (87%) of 15 cag+ H. pylori strains from patients with peptic ulcer disease contained type I hopQ alleles, and 1 (8%) of 13 evaluable cag-negative H. pylori strains from patients with gastritis alone contained type I hopQ alleles (P < 0.001, Fisher's exact test). Conversely, two (13%) of 15 cag+ H. pylori strains from patients with peptic ulcer disease contained type II hopQ alleles, and 12 (92%) of 13 evaluable cag-negative H. pylori strains from patients with gastritis alone contained type II hopQ alleles (P < 0.001).
TABLE 3.
Genotypes of H. pylori strains analyzed in this studya
| Strain | cagA type | vacA type | Type of hopQ allele in HP1177 locus based on nucleotide sequence analysisb | Type of hopQ allele detected using PCR-based typing methodsc |
|---|---|---|---|---|
| J104 | + | sla/m1 | I∗ | I |
| J166 | + | slb/m1 | I | I, II |
| J258 | + | sla/m1 | I∗ | I |
| J116 | + | sla/m2 | I | I, II |
| J123 | + | sla/m2 | I | I |
| J87 | + | sla/m2 | I | I, II |
| J223 | + | sla/m2 | I | I, II |
| 87-29 | + | slb/m1 | I | I |
| 87-199 | + | sla/m1 | I | I |
| 87-33 | + | slb/m1 | I | I |
| 92-21 | + | sla/m1 | II | I, II |
| J128 | + | sla/m2 | I | I |
| 92-25 | + | slb/m1 | I | I |
| J178 | + | sla/m1 | I | I |
| J133 | + | slb/m2 | II | II |
| J262 | − | s2/m2 | II∗ | II |
| J154 | − | s2/m2 | II∗ | II |
| J190 | − | s2/m2 | II∗ | II |
| J195 | − | s2/m2 | II | II |
| 87-203 | − | s2/m2 | I | I, II |
| 87-75 | − | s2/m2 | NAd | I, II |
| 86-313 | − | s2/m2 | II∗ | II |
| 87-225 | − | s2/m2 | II | II |
| 87-90 | − | s2/m2 | NAd | None |
| 87-230 | − | s2/m2 | II∗ | II |
| 92-20 | − | s2/m2 | II | II |
| 92-23 | − | s2/m2 | II | II |
| J63 | − | s2/m2 | II∗ | II |
| 86-338 | − | s2/m2 | II | II |
| 92-28 | − | s2/m2 | II | II |
All 15 cagA+ strains were isolated from patients with peptic ulcer disease, whereas all 15 cagA-negative strains were isolated from patients without peptic ulcer disease.
The HP1177 locus was PCR amplified using primers BA7676 and BA7674 (derived from flanking genes). Subsequently, either complete nucleotide sequence analysis of the hopQ allele was performed (indicated by asterisks), or sequence analysis of the 5′ end of hopQ was performed using primer OP4702.
Data are based on results of PCRs using primers that selectively amplified type I hopQ alleles or primers that selectively amplified type II hopQ alleles.
NA, not applicable, due to failure to obtain the desired PCR product (strain 87-75) or failure to obtain hopQ sequence data (strain 87-90).
PCR-based methodology for detecting type I and type II hopQ alleles.
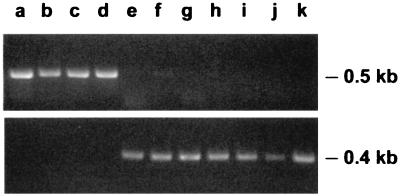
Based on analysis of the hopQ nucleotide sequences described above, we designed PCR primers that could be used to selectively amplify type I hopQ sequences but not type II hopQ sequences (Table 1). When tested on a panel of 11 strains for which the entire hopQ nucleotide sequences had been determined (Table 2), these primers functioned effectively to amplify products of the expected size from strains known to contain type I hopQ alleles but not from strains known to contain type II hopQ alleles (Fig. 2). We also designed primers that could be used to selectively amplify only type II hopQ sequences but not type I hopQ sequences (Table 1). When tested on the same group of strains, these primers functioned as predicted and amplified products of the expected size only from strains known to contain type II hopQ alleles (Fig. 2).
FIG. 2.
PCR-based methodology for identifying two families of hopQ alleles. Top panel: primers OP5136 and OP4829 amplified a product of the expected size from strains J99, 26695, J104, and J258 (lanes a to d, respectively) but not from the other strains tested (lanes e to k). Bottom panel: primers BA8363 and BA8364 amplified a product of the expected size from strains J262, Tx30a, J154, J190, J63, 86-313, and 87-230 (lanes e to k, respectively) but not from the other strains tested (lanes a to d).
We then used these primers to detect the presence of type I and type II hopQ alleles in the panel of 30 different well-characterized H. pylori strains. By use of PCR primers specific for the amplification of type I hopQ fragments, PCR products of the expected size were successfully amplified from 16 of the 30 strains (Tables 3 and 4). By use of PCR primers specific for amplifying type II hopQ fragments, PCR products of the expected size were successfully amplified from 20 of the 30 strains (Tables 3 and 4). As described above, based on partial nucleotide sequence analysis of 3-kb amplicons, 14 strains were known to contain type I hopQ alleles and 14 strains were known to contain type II hopQ alleles. These allelic types were successfully identified in all 28 strains by using PCR with type-specific hopQ primers (Table 3). Thus, there was a high level of agreement between results obtained using two different experimental approaches for analyzing hopQ genotypes. The only apparent discrepancy involved the amplification of both type I and type II products from seven different H. pylori strains, whereas sequence analysis of 3-kb amplicons from these strains revealed the presence of only one type of hopQ allele in each strain (Table 3).
TABLE 4.
Detection of hopQ alleles in H. pylori strains using PCR-based methodologya
| Type of H. pylori strain and associated clinical condition (n) | No. of strains from which the indicated hopQ product was amplified |
|||
|---|---|---|---|---|
| Type I only | Type II only | Type I and type II | Neither type | |
| cag+/s1-vacA, peptic ulcer (15) | 9 | 1 | 5 | 0 |
| cag-negative/s2-vacA, no peptic ulcer (15) | 0 | 12 | 2 | 1 |
Type I hopQ fragments were amplified from cag+ strains significantly more frequently than from cag-negative strains (P < 0.001). Type II hopQ fragments were amplified significantly more frequently from cag mutant strains than from cag+ strains (P = 0.005).
To investigate the basis for amplification of both type I and type II hopQ fragments from seven strains, two strains (J116 and J166) were selected for more detailed analysis. Nucleotide sequence analysis of the ∼0.5-kb PCR products amplified from these strains using type I-specific and type II-specific hopQ primers confirmed that both type I and type II hopQ products had been amplified from each strain. To rule out the possibility that this phenomenon reflected the presence of a mixed population of bacteria, single colonies of strain J116 and strain J166 were isolated and then reanalyzed by PCR. By using type I-specific and type II-specific hopQ primers, both type I and type II hopQ fragments were amplified from single colonies of each strain. Therefore, it seems likely that these strains contain two different hopQ alleles (one in a locus corresponding to HP1177 and one in a different chromosomal locus). The amplification of only one hopQ allelic type from these strains using primers derived from HP1178 and HP1175 suggests that the second hopQ locus is flanked by genes unrelated to HP1178 and HP1175.
Based on PCR-based detection of hopQ alleles, type I hopQ alleles were detected in 14 (93%) of 15 cag+ H. pylori strains cultured from patients with peptic ulcer disease. In contrast, type I hopQ alleles were detected in only 2 (13%) of 15 cag-negative H. pylori strains cultured from patients with no history of peptic ulcer disease (P < 0.001). By use of PCR primers specific for the amplification of type II hopQ fragments, PCR products of the expected size were successfully amplified from 6 (40%) of 15 cag+ H. pylori strains cultured from patients with peptic ulcer disease and from 14 (93%) of 15 cag-negative H. pylori strains cultured from patients with no history of peptic ulcer disease (P = 0.005) (Table 4). These data indicate that type I hopQ sequences are found significantly more frequently in cag+ H. pylori strains from patients with peptic ulcer disease (putative “ulcerogenic” strains) than in cag-negative strains from patients with no history of ulcer disease (putative nonulcerogenic strains).
DISCUSSION
A comparison of the complete genome sequences of two different H. pylori strains has provided considerable insight into intraspecies genetic diversity in H. pylori (6). Most orthologous sequences in these two genomes show levels of relatedness ranging from 90 to 99% nucleotide identity. Analyses of larger collections of strains have reported similar levels of relatedness for most H. pylori genes analyzed thus far, including flaA, ureA, babA, and cysS (1, 21, 42, 49).
In the present study, we analyzed genetic diversity among hopQ alleles of H. pylori. The two previously known hopQ alleles (from the two fully sequenced reference strains, 26695 and J99) are 89% identical in nucleotide sequences. Among the nine completely sequenced hopQ alleles described in the present study, we report that hopQ alleles from two different H. pylori strains (J104 and J258) are closely related to these previously characterized hopQ alleles. In contrast, hopQ sequences from seven strains are considerably different from the previously characterized hopQ alleles. Based on phylogenetic analysis (Table 2), it seems appropriate to classify H. pylori hopQ alleles into two different families, designated type I and type II. The hopQ alleles in the two fully sequenced H. pylori genomes (from strains 26695 and J99) represent prototypes for the type I family of hopQ alleles. In contrast, type II hopQ alleles have not been previously recognized. These data indicate that, despite the availability of two complete H. pylori genome sequences, the full range of genetic diversity among H. pylori strains is by no means completely characterized.
Each of the two fully sequenced H. pylori genomes contains a single copy of a type I hopQ allele and no sequences closely related (i.e., >80% nucleotide identity) to type II hopQ alleles. Interestingly, in the present study, we identified several strains that apparently contain both a type I hopQ allele and a type II hopQ allele. Nucleotide sequence analysis indicates that one allele is located in a locus corresponding to HP1177 in the genome of H. pylori 26695, and the second hopQ allele is presumed to be in a different, not-yet-identified locus. Duplication of H. pylori genes encoding outer membrane proteins has been reported previously for babA, HopJ/K, and hopM/N, and in the case of babA, such duplication can be strain specific (5, 26). Our data indicating the presence of strain-specific variation in the number of hopQ alleles per genome are therefore not entirely surprising.
Intraspecies genetic recombination occurs commonly in H. pylori, and evidence of such recombination has been readily detected by analysis of several different H. pylori genes (1, 8, 10, 28, 49). It seems likely that evidence of recombination between type I and type II hopQ alleles could be detected if further analysis were undertaken. However, the striking finding presented here is that the phylogenetic structure of these two families of hopQ alleles seems to have remained relatively intact despite frequent intraspecies genetic recombination. Based on the substantial amino acid sequence diversity that exists between type I and type II HopQ proteins, we think that it is likely that these two families of proteins possess differing functional properties. For example, if HopQ proteins function as adhesins, there might be differences in the binding specificities of type I versus type II HopQ proteins. Similar to the classification of hopQ alleles into two highly divergent families, H. pylori vacA alleles (encoding secreted vacuolating toxins) have been classified into highly divergent families, designated type m1 and type m2 (8, 10). The nucleotide sequences of type m1- and type m2-vacA alleles are only about 70% identical within a 0.7-kb midregion of the gene. As predicted, based on the existence of two families of vacA allelic types, the products of type m1- and type m2-vacA alleles have indeed been reported to differ in functional properties (27, 37).
In a recent study, the gene content of 15 different H. pylori strains was analyzed by using DNA microarray methodology (45). A group of 1,281 different genes was detected in all 15 strains, whereas 362 genes were nonconserved (and hence presumed to be nonessential). Based on the detection of hopQ sequences in 80% of the 15 strains studied, hopQ (omp27) was classified as a nonconserved gene. The region of hopQ utilized on these microarray chips corresponded to a 1,308-nucleotide fragment (nucleotides 70 to 1377 of hopQ from strain 26695) (N. Salama, personal communication), i.e., a type I hopQ allele. It seems likely that type II hopQ sequences would not hybridize efficiently to this DNA under high-stringency conditions, and we speculate that type II hopQ alleles were not detected using the previously described microarray methodology. In the present study, by using two different sets of PCR primers designed to specifically amplify only type I and type II hopQ alleles, we were able to detect the presence of hopQ sequences in 29 of 30 different H. pylori strains (Table 3). These results suggest that hopQ alleles are present in all or nearly all H. pylori strains.
Interestingly, the microarray study by Salama et al. reported that hopQ alleles were detected in cag+ strains more commonly than in cag-negative strains (10 of 10 cag+ strains versus 2 of 5 cag-negative strains) (45). In the present study, we found that type I hopQ alleles were present in cag+ strains significantly more commonly than in cag-negative strains. Based on the presumption that the previous microarray methodology only could detect type I hopQ alleles, our present results seem consistent with the previous microarray results (45).
Previous studies have concluded that type s1-vacA alleles are found predominantly in H. pylori strains that contain the cag pathogenicity island, whereas type s2-vacA alleles are found predominantly in H. pylori strains that lack the cag pathogenicity island (8, 22, 44, 54). Other genes found more commonly in cag+ strains than in cag-negative strains include babA2 alleles (encoding a functional Lewis b binding adhesin) (22, 26), several additional genes identified in a microarray analysis (45), and type I hopQ alleles. Because H. pylori has a recombinational population structure (23, 49), these groups of strains probably do not represent distinct phylogenetic lineages. Instead, these examples of linkage disequilibrium presumably indicate that a selective advantage is conferred to strains with certain genotypes.
A genotype-based classification of H. pylori strains is potentially useful for predicting the clinical outcome of infections. For example, previous studies have concluded that H. pylori strains carrying the cag pathogenicity island and type s1-vacA alleles are associated with a higher risk of peptic ulcer disease than are strains that lack these genetic elements (8, 18, 22, 25, 29, 43, 44, 48, 53, 54). One recent study concluded that “triple-positive” strains (i.e., possessing the cag pathogenicity island, type s1-vacA alleles, and babA2 alleles) are associated with a particularly high risk of peptic ulceration (22). The results of the present study provide further evidence indicating that putative ulcerogenic strains may contain genetic elements different from those found in nonulcerogenic strains. In future studies, it will be important to analyze H. pylori strains from multiple populations of patients and from various parts of the world in order to gain further insight into the patterns of linkage disequilibrium that exist in this species. In addition, multivariate analysis of large collections of H. pylori strains from humans with well-defined clinical diagnoses may permit identification of the H. pylori factors that are most relevant for predicting clinical outcome.
Acknowledgments
This work was supported in part by the National Institutes of Health (R01 DK53623 and AI 39657) and the Medical Research Service of the Department of Veterans Affairs.
REFERENCES
- 1.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. [DOI] [PubMed] [Google Scholar]
- 2.Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol. 28:37-53. [DOI] [PubMed] [Google Scholar]
- 3.Akopyants, N. S., A. Fradkov, L. Diatchenko, J. E. Hill, P. D. Siebert, S. A. Lukyanov, E. D. Sverdlov, and D. E. Berg. 1998. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akopyanz, N., N. O. Bukanov, T. U. Westblom, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alm, R. A., J. Bina, B. M. Andrews, P. Doig, R. E. Hancock, and T. J. Trust. 2000. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect. Immun. 68:4155-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 7.Asahi, M., T. Azuma, S. Ito, Y. Ito, H. Suto, Y. Nagai, M. Tsubokawa, Y. Tohyama, S. Maeda, M. Omata, T. Suzuki, and C. Sasakawa. 2000. Helicobacter pylori CagA protein can be tyrosine phosphorylated in gastric epithelial cells. J. Exp. Med. 191:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atherton, J. C., P. Cao, R. M. Peek, Jr., M. K. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270:17771-17777. [DOI] [PubMed] [Google Scholar]
- 9.Atherton, J. C., T. L. Cover, E. Papini, and J. L. Telford. 2001. Vacuolating cytotoxin. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C.
- 10.Atherton, J. C., P. M. Sharp, T. L. Cover, G. Gonzalez-Valencia, R. M. Peek, Jr., S. A. Thompson, C. J. Hawkey, and M. J. Blaser. 1999. Vacuolating cytotoxin (vacA) alleles of Helicobacter pylori comprise two geographically widespread types, m1 and m2, and have evolved through limited recombination. Curr. Microbiol. 39:211-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaser, M. J., and D. E. Berg. 2001. Helicobacter pylori genetic diversity and risk of human disease. J. Clin. Investig. 107:767-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blaser, M. J., G. I. Perez-Perez, H. Kleanthous, T. L. Cover, R. M. Peek, P. H. Chyou, G. N. Stemmermann, and A. Nomura. 1995. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 55:2111-2115. [PubMed] [Google Scholar]
- 13.Cao, P., and T. L. Cover. 1997. High-level genetic diversity in the vapD chromosomal region of Helicobacter pylori. J. Bacteriol. 179:2852-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Covacci, A., and R. Rappuoli. 2000. Tyrosine-phosphorylated bacterial proteins: Trojan horses for the host cell. J. Exp. Med. 191:587-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cover, T. L., D. E. Berg, M. J. Blaser, and H. L. T. Mobley. 2001. Helicobacter pylori pathogenesis, p. 510-558. In E. A. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, San Diego, Calif.
- 17.Cover, T. L., P. Cao, C. D. Lind, K. T. Tham, and M. J. Blaser. 1993. Correlation between vacuolating cytotoxin production by Helicobacter pylori isolates in vitro and in vivo. Infect. Immun. 61:5008-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cover, T. L., Y. Glupczynski, A. P. Lage, A. Burette, M. K. R. Tummuru, G. I. Perez-Perez, and M. J. Blaser. 1995. Serologic detection of infection with cagA+ Helicobacter pylori strains. J. Clin. Microbiol. 33:1496-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cover, T. L., M. K. R. Tummuru, P. Cao, S. A. Thompson, and M. J. Blaser. 1994. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J. Biol. Chem. 269:10566-10573. [PubMed] [Google Scholar]
- 20.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garner, J. A., and T. L. Cover. 1995. Analysis of genetic diversity in cytotoxin-producing and non-cytotoxin-producing Helicobacter pylori strains. J. Infect. Dis. 172:290-293. [DOI] [PubMed] [Google Scholar]
- 22.Gerhard, M., N. Lehn, N. Neumayer, T. Boren, R. Rad, W. Schepp, S. Miehlke, M. Classen, and C. Prinz. 1999. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc. Natl. Acad. Sci. USA 96:12778-12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Go, M. F., V. Kapur, D. Y. Graham, and J. M. Musser. 1996. Population genetic analysis of Helicobacter pylori by multilocus enzyme electrophoresis: extensive allelic diversity and recombinational population structure. J. Bacteriol. 178:3934-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottke, M. U., C. A. Fallone, A. N. Barkun, K. Vogt, V. Loo, M. Trautmann, J. Z. Tong, T. N. Nguyen, T. Fainsilber, H. H. Hahn, J. Korber, A. Lowe, and R. N. Beech. 2000. Genetic variability determinants of Helicobacter pylori: influence of clinical background and geographic origin of isolates. J. Infect. Dis. 181:1674-1681. [DOI] [PubMed] [Google Scholar]
- 25.Han, S. R., H. J. Schreiber, S. Bhakdi, M. Loos, and M. J. Maeurer. 1998. vacA genotypes and genetic diversity in clinical isolates of Helicobacter pylori. Clin. Diagn. Lab. Immunol. 5:139-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ilver, D., A. Arnqvist, J. Ogren, I. M. Frick, D. Kersulyte, E. T. Incecik, D. E. Berg, A. Covacci, L. Engstrand, and T. Boren. 1998. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373-377. [DOI] [PubMed] [Google Scholar]
- 27.Ji, X., T. Fernandez, D. Burroni, C. Pagliaccia, J. C. Atherton, J. M. Reyrat, R. Rappuoli, and J. L. Telford. 2000. Cell specificity of Helicobacter pylori cytotoxin is determined by a short region in the polymorphic midregion. Infect. Immun. 68:3754-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kersulyte, D., H. Chalkauskas, and D. E. Berg. 1999. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol. Microbiol. 31:31-43. [DOI] [PubMed] [Google Scholar]
- 29.Kidd, M., A. J. Lastovica, J. C. Atherton, and J. A. Louw. 1999. Heterogeneity in the Helicobacter pylori vacA and cagA genes: association with gastroduodenal disease in South Africa? Gut 45:499-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letley, D. P., and J. C. Atherton. 2000. Natural diversity in the N terminus of the mature vacuolating cytotoxin of Helicobacter pylori determines cytotoxin activity. J. Bacteriol. 182:3278-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marshall, D. G., W. G. Dundon, S. M. Beesley, and C. J. Smyth. 1998. Helicobacter pylori—a conundrum of genetic diversity. Microbiology 144:2925-2939. [DOI] [PubMed] [Google Scholar]
- 32.McClain, M. S., P. Cao, H. Iwamoto, A. D. Vinion-Dubiel, G. Szabo, Z. Shao, and T. L. Cover. 2001. A 12-amino-acid segment, present in type s2 but not type s1 Helicobacter pylori VacA proteins, abolishes cytotoxin activity and alters membrane channel formation. J. Bacteriol. 183:6499-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miehlke, S., C. Kirsch, K. Agha-Amiri, T. Gunther, N. Lehn, P. Malfertheiner, M. Stolte, G. Ehninger, and E. Bayerdorffer. 2000. The Helicobacter pylori vacA s1, m1 genotype and cagA is associated with gastric carcinoma in Germany. Int. J. Cancer 87:322-327. [PubMed] [Google Scholar]
- 34.Montecucco, C., E. Papini, M. de Bernard, J. L. Telford, and R. Rappuoli. 1999. Helicobacter pylori vacuolating cytotoxin and associated pathogenic factors, p. 264-286. In J. E. Alouf and J. H. Freer (ed.), The comprehensive sourcebook of bacterial protein toxins. Academic Press, San Diego, Calif.
- 35.Odenbreit, S., J. Puls, B. Sedlmaier, E. Gerland, W. Fischer, and R. Haas. 2000. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287:1497-1500. [DOI] [PubMed] [Google Scholar]
- 36.Ogura, K., S. Maeda, M. Nakao, T. Watanabe, M. Tada, T. Kyutoku, H. Yoshida, Y. Shiratori, and M. Omata. 2000. Virulence factors of Helicobacter pylori responsible for gastric diseases in Mongolian gerbil. J. Exp. Med. 192:1601-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pagliaccia, C., M. de Bernard, P. Lupetti, X. Ji, D. Burroni, T. L. Cover, E. Papini, R. Rappuoli, J. L. Telford, and J. M. Reyrat. 1998. The m2 form of the Helicobacter pylori cytotoxin has cell type-specific vacuolating activity. Proc. Natl. Acad. Sci. USA 95:10212-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan, Z. J., D. E. Berg, R. W. van der Hulst, W. W. Su, A. Raudonikiene, S. D. Xiao, J. Dankert, G. N. Tytgat, and A. van der Ende. 1998. Prevalence of vacuolating cytotoxin production and distribution of distinct vacA alleles in Helicobacter pylori from China. J. Infect. Dis. 178:220-226. [DOI] [PubMed] [Google Scholar]
- 39.Papini, E., M. Zoratti, and T. L. Cover. 2001. In search of the Helicobacter pylori VacA mechanism of action. Toxicon 39:1757-1767. [DOI] [PubMed] [Google Scholar]
- 40.Peek, R. M., Jr., G. G. Miller, K. T. Tham, G. I. Perez-Perez, T. L. Cover, J. C. Atherton, G. D. Dunn, and M. J. Blaser. 1995. Detection of Helicobacter pylori gene expression in human gastric mucosa. J. Clin. Microbiol. 33:28-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peek, R. M., Jr., S. A. Thompson, J. P. Donahue, K. T. Tham, J. C. Atherton, M. J. Blaser, and G. G. Miller. 1998. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc. Assoc. Am. Physicians 110:531-544. [PubMed] [Google Scholar]
- 42.Pride, D. T., R. J. Meinersmann, and M. J. Blaser. 2001. Allelic variation within Helicobacter pylori babA and babB. Infect. Immun. 69:1160-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ribeiro De Gusmão, V., E. Nogueira Mendes, D. M. De Magalhães Queiroz, G. Aguiar Rocha, A. M. Camargos Rocha, A. A. R. Ashour, and A. S. T. Carvalho. 2000. vacA genotypes in Helicobacter pylori strains isolated from children with and without duodenal ulcer in Brazil. J. Clin. Microbiol. 38:2853-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rudi, J., C. Kolb, M. Maiwald, D. Kuck, A. Sieg, P. R. Galle, and W. Stremmel. 1998. Diversity of Helicobacter pylori vacA and cagA genes and relationship to VacA and CagA protein expression, cytotoxin production, and associated diseases. J. Clin. Microbiol. 36:944-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segal, E. D., J. Cha, J. Lo, S. Falkow, and L. S. Tompkins. 1999. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. USA 96:14559-14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stein, M., R. Rappuoli, and A. Covacci. 2000. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc. Natl. Acad. Sci. USA 97:1263-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strobel, S., S. Bereswill, P. Balig, P. Allgaier, H. G. Sonntag, and M. Kist. 1998. Identification and analysis of a new vacA genotype variant of Helicobacter pylori in different patient groups in Germany. J. Clin. Microbiol. 36:1285-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Telford, J. L., P. Ghiara, M. Dell'Orco, M. Comanducci, D. Burroni, M. Bugnoli, M. F. Tecce, S. Censini, A. Covacci, Z. Xiang, et al. 1994. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. J. Exp. Med. 179:1653-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 52.Tummuru, M. K., S. A. Sharma, and M. J. Blaser. 1995. Helicobacter pylori picB, a homologue of the Bordetella pertussis toxin secretion protein, is required for induction of IL-8 in gastric epithelial cells. Mol. Microbiol. 18:867-876. [DOI] [PubMed] [Google Scholar]
- 53.Van Doorn, L. J., C. Figueiredo, F. Megraud, S. Pena, P. Midolo, D. M. Queiroz, F. Carneiro, B. Vanderborght, M. D. Pegado, R. Sanna, W. De Boer, P. M. Schneeberger, P. Correa, E. K. Ng, J. Atherton, M. J. Blaser, and W. G. Quint. 1999. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology 116:823-830. [DOI] [PubMed] [Google Scholar]
- 54.van Doorn, L. J., C. Figueiredo, R. Sanna, A. Plaisier, P. Schneeberger, W. de Boer, and W. Quint. 1998. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology 115:58-66. [DOI] [PubMed] [Google Scholar]
- 55.Wang, G., M. Z. Humayun, and D. E. Taylor. 1999. Mutation as an origin of genetic variability in Helicobacter pylori. Trends Microbiol. 7:488-493. [DOI] [PubMed] [Google Scholar]
- 56.Yamaoka, Y., D. H. Kwon, and D. Y. Graham. 2000. A M(r) 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 97:7533-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]