Abstract
The role of DNA barcoding as a tool to accelerate the inventory and analysis of diversity for hyperdiverse arthropods is tested using ants in Madagascar. We demonstrate how DNA barcoding helps address the failure of current inventory methods to rapidly respond to pressing biodiversity needs, specifically in the assessment of richness and turnover across landscapes with hyperdiverse taxa. In a comparison of inventories at four localities in northern Madagascar, patterns of richness were not significantly different when richness was determined using morphological taxonomy (morphospecies) or sequence divergence thresholds (Molecular Operational Taxonomic Unit(s); MOTU). However, sequence-based methods tended to yield greater richness and significantly lower indices of similarity than morphological taxonomy. MOTU determined using our molecular technique were a remarkably local phenomenon—indicative of highly restricted dispersal and/or long-term isolation. In cases where molecular and morphological methods differed in their assignment of individuals to categories, the morphological estimate was always more conservative than the molecular estimate. In those cases where morphospecies descriptions collapsed distinct molecular groups, sequence divergences of 16% (on average) were contained within the same morphospecies. Such high divergences highlight taxa for further detailed genetic, morphological, life history, and behavioral studies.
Keywords: cox1, CO1, Madagascar, collaborative taxonomy, DNA barcode, biodiversity
1. Introduction
The increasing loss of biodiversity presents a daunting challenge to taxonomists and requires the discovery and analysis of biodiversity at a greatly accelerated pace. If we are really serious about ‘zero biodiversity loss’ in Madagascar and elsewhere, then conservation planning needs to be based more fundamentally on biodiversity data, and this requires taxonomic knowledge (Brooks et al. 2004a). However, if nothing is done to change the slow pace of current taxonomic efforts and practice, it will take centuries to complete even a preliminary ‘Encyclopedia of life’ on Earth (Wilson 2003). It is clear that if systematics is going to play a practical role in directing the preservation and development of natural systems (Gotelli 2004), changes need to occur throughout the entire taxonomic process, from collecting to description, from publication to dissemination, and from public outreach to advocacy.
In this paper, we show how DNA barcoding (using cytochrome oxidase 1 (cox1 or CO1)—Hebert et al. 2003a,b), enables taxonomic data on hyperdiverse arthropods to be gathered, analysed, and synthesized into useful products in a timeframe that meets the challenge presented by the rate of biodiversity loss. We test a model for accelerating the taxonomic process with the aims of providing the necessary data for effective taxonomy, and—most importantly—the tools for making data accessible and applicable to the conservation agenda. The model is tested on a key taxonomic group, ants, and in an especially threatened area, Madagascar. We describe how cox1 DNA barcoding enables rapid identification of Molecular Operational Taxonomic Units (MOTU—Floyd et al. 2002; Blaxter 2004) for the assessment of richness and turnover across landscapes.
Madagascar has been identified as one of the world's outstanding biodiversity hotspots, harbouring a unique and threatened biota, whose composition and origins are linked to the breakup of Gondwana (Battistini & Richard-Vindard 1972; Jolly et al. 1984; Storey et al. 1995; Lourenço 1996; Goodman & Patterson 1997; Goodman & Benstead 2003). As in many island environments (Gillespie & Roderick 2002), Madagascar's indigenous terrestrial arthropods are in severe danger of extinction due to habitat deterioration and invasion of exotic species. Since humans colonized Madagascar approximately 1500–2000 years ago (Burney 1997), it is estimated that as much as 80% of Madagascar's original habitat has been destroyed (Sussman et al. 1996). Much of the island is now species-poor secondary grassland, which is annually burned and highly eroded.
Never has there been a more supportive political environment in which to address these threats in Madagascar. Over the next five years, the Malagasy government plans to more than triple the number of protected areas and is committed to sustainable conservation planning. To accomplish these goals, areas of conservation importance must be determined. One major obstacle in the identification of areas for protection in Madagascar is incomplete knowledge of the island's patterns of species richness, turnover, and endemism (Schatz 2002). It is unclear which of the remaining patches of natural vegetation should be of highest priority for conservation. Existing data are often at an inappropriate spatial scale for conservation implementation, not standardized across sites, and focused on vertebrates—which represent only a small proportion of the biota.
There is strong and growing evidence that the investment in collection and compilation of species data yields great value in conservation planning (Brooks et al. 2004a,b). Species data are a precondition of good, efficient conservation (Ferrier et al. 2004). Current knowledge of species distributions, however, is often limited to select vertebrate taxa. Vertebrates, however, are frequently inadequate indicators of biodiversity as a whole; they fail to represent the bulk of diversity, especially the fine-scale patterns of diversity shown by arthropods (Fisher 2000; Ferrier et al. 2004). Arthropods, such as ants, often exhibit high rates of spatial turnover (replacement of species) and therefore provide the essential fine-scale maps for assessing biodiversity at a scale at which conservation decisions are typically made on the ground.
The ant fauna of Madagascar is currently estimated to include approximately 1000 species, of which 96% are endemic (Fisher 1996b; Fisher & Girman 2000). An estimated 75% of the ant fauna in Madagascar, however, remain undescribed (Fisher 1996a). For example, of the 71 species of the genus Strumigenys described in a recent revision, 70 were endemic and newly described (Fisher 2000). Although ants dominate the biomass of most terrestrial communities (Davidson 1997; Davidson et al. 2003), are critical pollinators and seed dispersers (Beattie 1985; Ness et al. 2004), and are critical to nutrient cycling and ecosystem function (Thorp 1949; Majer 1983; Andersen 1993; Andersen 1997; Diehl et al. 2004), there is a global lack of studies of ant diversity or community structure. This may be largely because of the difficulty of species-level identifications (Bolton 2003).
A DNA-based system of species identification using a single gene (cytochrome oxidase 1) was proposed by Hebert et al. (2003a,b) who coined the term DNA barcoding. Since then, the utility of DNA barcodes for species identification has been successfully demonstrated with several taxonomic groups (Hebert et al. 2004b; Powers 2004; Hajibabaei et al. 2005; Lambert et al. 2005; Ward et al. 2005). The potential for such a system is evident to many who study biodiversity, especially, in smaller (Floyd et al. 2002; Blaxter 2004; Blaxter et al. 2004), understudied, or hyperdiverse (Hebert et al. 2004a) groups or in areas where the estimates of diversity lag well behind what is actually there.
There have been great improvements in overcoming other obstacles to including arthropods in biodiversity assessment. Efficient methods exist for their collection (Longino & Colwell 1997; Fisher 1999; Longino et al. 2002) and processing (Fisher 2005). The enormous amount of material collected and processed, however, presents new challenges: how to accelerate their identification and description? In this study, we test if rapid DNA barcoding can be used to provide a surrogate to species diversity patterns. In our analysis we test whether a diversity estimate based on a DNA barcode MOTU is significantly different from estimates based on traditional morphological taxonomy using an understudied taxa (ants) from a part of the world where established taxonomic frameworks are only now emerging (Madagascar). We demonstrate that DNA barcode functional units are an effective surrogate for traditional morphological species and discover the same relative patterns of diversity within and between collection sites. This is important as it allows the rapid and scalable identification of diversity from widespread, numerous and thorough inventories. A sequence-based assessment of diversity can scale to many more sites and specimens in the same amount of time possible for traditional analyses. MOTU will not always be coincident with species (determined using whatever concept one chooses), but rather are categories whose membership is a hypothesis open to re-testing. Thus, this method is not one of strictly DNA-based taxonomy. Our goal is to test whether MOTU perform as an estimate, or surrogate, of species richness patterns derived from morphological based determination of species (morphospecies).
We compare richness and turnover of MOTU from sequence data with species richness and turnover patterns from morphological determinations for ant specimens from four sites in northern Madagascar. Using an operational, tree-based approach to the identification of MOTU, we identified clusters of specimens beneath a threshold of similarity (Hebert et al. 2003a). The identification of MOTU using a sequence divergence threshold represents just one possible means to determine MOTU (for other examples see Floyd et al. 2002; Sites & Marshall 2003; DeSalle & Amato 2004) and is used here only as an example.
2. Methods
(a) Field sites
We surveyed ants at four localities in November and December 2004 in northeastern Madagascar in the Province of Antsiranana:
Antsahabe: Forêt Antsahabe, 11.4 km 275° Daraina, 550 m, 13°13.7′S, 49°33.4′E 15–18.xi.2004, tropical dry forest.
Binara: Forêt Binara, 9.1 km 233° Daraina, 650–800 m, 13°15.8′S, 49°36.2′E 19–22.xi.2004, rainforest.
Marojejy: Parc National de Marojejy, 28.0 km 38° Andapa, 450 m, 14°26.2′S, 49°46.5′E 23–25.xi.2004, rainforest.
Ambato: Forêt Classée Ambato, 26.6 km 33° Ambanja, 150 m, 13°27.87′S, 48°33.10′E, 8–11.xii.2004, rainforest.
At each of the sites, we spent two days searching by hand for ants. This technique differs from standardized inventory protocols for collecting ants (Fisher 1999) but provides a manageable case study for testing the use of DNA barcoding for assessing patterns of richness and turnover. Specimens from every collection from each locality were used for cox1 sequencing and morphological study. All specimen data for material examined in this study are available on AntWeb (www.antweb.org). Material was deposited at California Academy of Sciences, San Francisco, USA (CAS).
(b) Genetic analysis
Specimens were identified to genera immediately upon collection by parataxonomists in Madagascar (Wheeler 1995; Basset et al. 2000) and preserved in 95% ethanol. Upon return to California, specimens were loaded into ScrewTop TrakMates® boxes (Matrix Technologies) and shipped to the University of Guelph, Canada. Here, DNA was extracted from tissues rich in mitochondria (e.g. legs), employing primers with high universality, and amplifying a relatively long PCR product (>600 base pairs; bp). Total genomic DNA extracts were prepared from small pieces (≤1 mm) of leg using the NucleoSpin® 96 Tissue kit (Macherey–Nagel Duren, Germany), following the manufacturer's protocols. Extracts were resuspended in 30 μl of dH2O, and a 650-base bp region near the 5′ terminus of the cox1 gene was amplified following standard protocol (Hebert et al. 2003a). Briefly, full length sequences were amplified using primers (LF1—ATTCAACCAATCATAAAGATATTGG and LR1—TGATTTTTTGGACATCCAGAAGTTTA (Hebert et al. 2004a)). In cases where a 650 bp product was not successfully generated, internal primer pairs (LF1-ANTMR1D–ATGMWGGNKYMGGWACWGGWTG) and (MLF1—GCTTTCCCACGAATAAATAATA (Hajibabaei et al. 2005)—LR) were employed to generate shorter overlapping sequences that allowed the creation of a composite sequence (contig). PCR reactions were carried out in 96-well plates in 12.5 μl reaction volumes containing: 2.5 mM MgCl2, 5 pmol of each primer, 20 μM dNTPs, 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 10–20 ng (1–2 μl) of genomic DNA, and 1 unit of TaqDNA polymerase using a thermocycling profile of one cycle of 2 min at 94 °C, five cycles of 40 s at 94 °C, 40 s at 45 °C, and 1 min at 72 °C, followed by 36 cycles of 40 sec at 94 °C, 40 sec at 51 °C, and 1 min at 72 °C, with a final step of 5 min at 72 °C. Products were visualized on a 2% agarose E-Gel® 96-well system (Invitrogen) and samples containing clean single bands were bidirectionally sequenced using BigDye v3.1 on an ABI 3730 DNA Analyzer (Applied Biosystems). Contigs were made using Sequencher v.4.0.5 (Gene Codes) and were subsequently aligned by eye in Bioedit (Hall 1999). Sequence divergences were calculated using the K2P distance model (Kimura 1980) and a NJ tree of distances (Saitou & Nei 1987) was created to provide a graphic representation of the among-species divergences using MEGA2 (Kumar et al. 2001), and BOLD (www.barcodinglife.org). Unless otherwise stated, all genetic distances we report are corrected. Sequence neutrality (Tajima's D—Tajima 1989) and rates of substitution were calculated with DnaSP v.3 (Rozas & Rozas 1999). Sequences and other specimen information are available in the project Ant Diversity in Northern Madagascar file in the Completed Projects section of the Guelph Barcode of Life website (www.barcodinglife.org) with complete collection information deposited at www.antweb.org. Sequences have also been deposited in GenBank (DQ176049–DQ176316).
To infer the biodiversity at each collection locality from cox1 sequences, we used two classes of sequence divergence threshold (Hebert et al. 2004b). In this approach, clusters that grouped at sequence divergence values less than the applied threshold were grouped together into a single MOTU. Using a sequence divergence threshold represents just one possible means to determine MOTU (for other examples see Sites & Marshall 2003; DeSalle & Amato 2004).
In our analysis, we tested threshold values of 2 and 3%. Previous studies have shown that a 3% threshold revealed approximately 98% of lepidopteran species identified through conventional morphological taxonomy (Hebert et al. 2003a), while a 2.7% threshold revealed 90% of 260 bird species from North America (Hebert et al. 2004b). Within the Hymenoptera, preliminary work with the ant species in North America found the average conspecific pairwise divergence to be 1.9% (377 sequences from 104 species—Smith et al. 2005), suggesting that 2–3% threshold are appropriate hypotheses to test ant species identity against. Importantly, a sequence threshold set too high will not identify closely related, or new, species (e.g. a 2% threshold would not identify the 10 cryptic species of Astraptes fulgerator—Hebert et al. 2004a). We explore this restriction further in §4.
The diversity and species turnover of MOTU, and morphospecies (m) within and between sites were compared using the Jaccard Index of similarity (Magurran 2004) calculated using the program EstimateS v.7.5 (Colwell 2004). Here, where for any two sites 1 and 2: A represents the species present in both site 1 and 2, B those species only at site 1, and C those species only at site 2.
3. Results
PCR products were easily produced and aligned as no insertions; deletions or stop codons were observed. Additionally, no visualized PCR product contained double bands. These observations support the conclusion that the sequences we analysed were mitochondrial DNA and not nuclear pseudogenes (Bensasson et al. 2001).
PCR products were generated from 268 of 280 specimens (95.7%) from 28 genera from four collection sites in northern Madagascar (Electronic Appendix, figure 5). Of these 268, 224 were generated using primers LF_LR (84.6%), 26 were contigs generated using two overlapping primer sites (9.7%), and 18 were shorter sequences generated using primers MLF_LR (6.7%). The average congeneric pairwise divergence was 8.51%. Sequences were heavily AT biased, as expected in insect mtDNA (Crozier & Crozier 1993) (table 1). The observed mutation patterns in the sequence data are consistent with a neutral model of molecular evolution (Tajima's D: 0.05643, p>0.10). Each individual was assigned to a morphospecies using morphological characteristics and to a MOTU using sequence thresholds of 2 and 3%.
Table 1.
Sequence statistics for the specimens listed in the Electronic Appendix, table 4.
| average | ||||||||
|---|---|---|---|---|---|---|---|---|
| domain | identical pairs | transitional pairs | transversional pairs | R=si/sv | T | C | A | G |
| avg | 473 | 62 | 71 | 0.9 | 39 | 18.7 | 30.4 | 11.9 |
| 1st position | 168 | 18 | 16 | 1.2 | 28.7 | 17.5 | 34.6 | 19.2 |
| 2nd position | 193 | 5 | 4 | 1.2 | 44.8 | 24 | 17.1 | 14.1 |
| 3rd position | 113 | 38 | 51 | 0.8 | 43.5 | 14.5 | 39.5 | 2.5 |
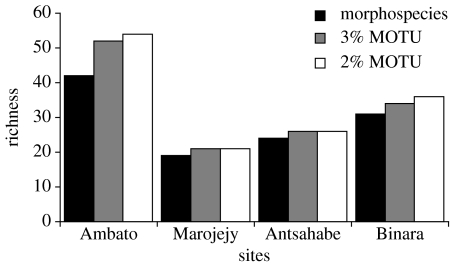
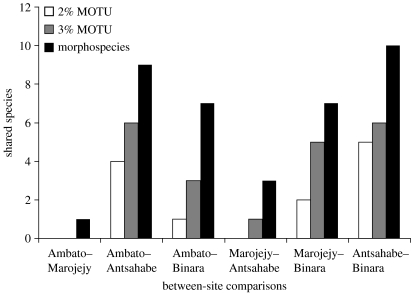
There was no significant difference between richness (number of morphospecies or MOTU) using the threshold approach or morphological taxonomy (ANOVA, F=0.216, df=2, p=0.811) (figure 1). There was a significant difference in turnover between sites using the different methodologies to assign individuals to morphospecies or MOTU (Shared species or MOTUs—ANOVA F=4.420, df=2, p=0.033, Jaccard Index— F=7.552, df=2, p=0.006) (figure 2, table 2). Molecular similarity thresholds tended to emphasis the uniqueness of each site while morphological taxonomy tended to find more overlap between sites.
Figure 1.
Estimates of richness (number of MOTU or morphospecies) and sample size for each site surveyed in 2004 using a 2% and 3% threshold for recognition and morphological taxonomic estimates of richness.
Figure 2.
Estimates of turnover (shared species) for pairwise comparisons between each of four sites surveyed in 2004 using three methods for diversity estimation.
Table 2.
Jaccard indices of similarity of four collection sites in Madagascar.
| 2% | 3% | morphospecies | |
|---|---|---|---|
| Ambato–Marojejy | 0 | 0 | 0.017 |
| Ambato–Antsahabe | 0.053 | 0.083 | 0.158 |
| Ambato–Binara | 0.011 | 0.036 | 0.106 |
| Marojejy–Antsahabe | 0 | 0.021 | 0.075 |
| Marojejy–Binara | 0.036 | 0.098 | 0.163 |
| Antsahabe–Binara | 0.089 | 0.111 | 0.222 |
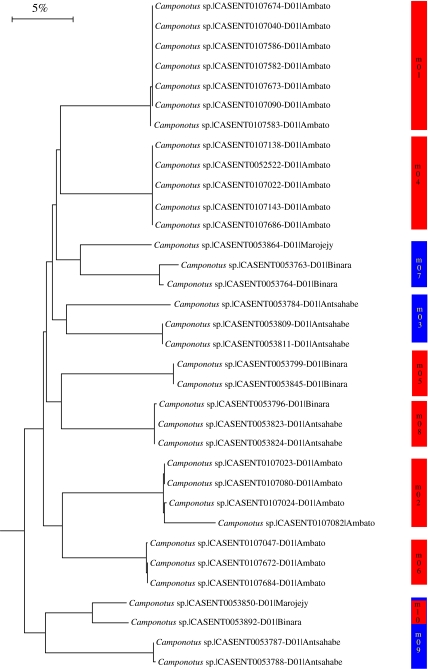
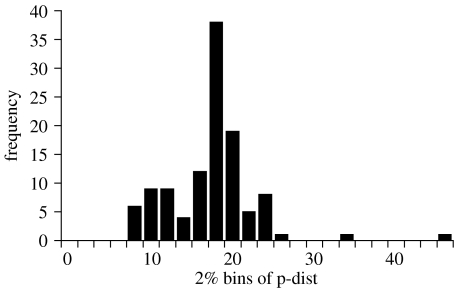
In general, the comparisons between 2% MOTU with morphology and 3% MOTU with morphology are the same (compare richness patterns in table 3, figure 1). Therefore, for brevity, only the results for the comparison between 3% MOTU and morphology are presented here. There was a strong correlation between 2% (∼13 substitutions/657 bp) and 3% (∼19 substitutions/657 bp) MOTU with the species estimates using morphological taxonomy. For example, in Camponotus, individuals were allocated nearly identically using morphology or a 3% threshold (figure 3). Exceptions occurred where deep sequence divergences were apparent between individuals identified as the same morphospecies but were collected at different sites (morphospecies m07, m09), or the same collection sites (m03). There were assignment differences between the 3% MOTU with morphology that are detailed in table 3 and table 4 in the Electronic Appendix. Generally, morphological taxonomy ‘lumped’ MOTU separated using the threshold approach. The average molecular divergence of morphospecies which contained multiple MOTU was 16.27% (n=113 comparisons, average=16.27, s.e.=0.51). On average, there were 2.3 cases per genera where morphology failed to recognize MOTU erected with a 3% threshold (50 times total). Pairwise sequence divergences for only those 50 cases where morphospecies contained multiple 3% MOTU are shown in a frequency histogram in figure 4.
Table 3.
Comparison of the number of morphospecies and MOTU contained within each of the 28 genera examined in this analysis.
| genera | morphospecies | 3% MOTU | 2% MOTU |
|---|---|---|---|
| Anochetus | 3 | 3 | 4 |
| Aphaenogaster | 2 | 3 | 3 |
| Camponotus | 10 | 12 | 12 |
| Cataulcus | 2 | 2 | 5 |
| Cerapachys | 4 | 5 | 5 |
| Crematogaster | 3 | 5 | 5 |
| Eutetramorium | 1 | 2 | 2 |
| Hypoponera | 4 | 4 | 4 |
| Leptogenys | 5 | 6 | 6 |
| Leptothorax | 3 | 6 | 6 |
| Melissotarsus | 1 | 1 | 1 |
| Monomorium | 2 | 4 | 4 |
| Mystrium | 1 | 2 | 2 |
| Odontomachus | 1 | 1 | 1 |
| Pachycondyla | 5 | 7 | 7 |
| Paratrechina | 2 | 2 | 2 |
| Pheidole | 9 | 12 | 12 |
| Plagiolepis | 3 | 4 | 5 |
| Platythyrea | 2 | 3 | 3 |
| Prionopelta | 1 | 1 | 1 |
| Pyramica | 1 | 1 | 1 |
| Simopone | 1 | 1 | 1 |
| Strumigenys | 3 | 3 | 4 |
| Tapinoma | 1 | 1 | 1 |
| Technomyrmex | 2 | 2 | 2 |
| Terataner | 2 | 3 | 4 |
| Tetramorium | 9 | 11 | 13 |
| Tetraponera | 7 | 10 | 10 |
| totals | 90 | 117 | 126 |
Figure 3.
Camponotus taxon NJ tree of K2P distance. Coloured bars with names indicate the samples described as a species using morphological taxonomy. Red indicates perfect correlation between 3% MOTU and morphological estimate of species identity. Blue indicates cases where a morphospecies contained multiple 3% MOTU. Accession numbers and collection sites are indicated.
Figure 4.
Frequency histogram of the genetic variation (uncorrected p-dist) for only those cases where morphospecies contains multiple 3% MOTU. (n=113 comparisons, average=16.27, s.e.=0.51).
4. Discussion
(a) Biodiversity assessment
Our analysis is unique in that it has compared diversity measures at four sites in Madagascar using both the morphologically defined species units and MOTU based on two different threshold values for DNA barcode sequence divergence. Patterns of richness and turnover of MOTU and morphospecies were not significantly different. The take-home message is not that the values are the same (although for the most case they are remarkably similar), but rather that the patterns of richness within sites and turnover between sites were so similar. Thus, richness and turnover assessments determined using DNA barcode variability, accrued with less than three weeks of preparatory analysis, provided an effective surrogate for species determined through time intensive detailed morphological analyses. It is important to recognize that our analyses do not use the DNA barcode to define an ontological species concept, but rather, to recognize classes of diversity. We test the epistemological hypothesis that the barcode MOTU can be used as a surrogate for the identification of diversity within and between collection localities. Species are spatio-temporally bounded individuals, not a class of items (Baum 1998). However, our recognition of these individuals defined ontologically must frequently come after using epistemological methodology, including category membership. Different epistemological methods of recognizing diversity frequently arrive at different answers (e.g. the Ensatina eschsholtzii complex salamanders of California may include from 1 to 11 different species—Frost & Hillis 1990; Graybeal 1995; Highton 1998; Wake & Schneider 1998)
For many inventories of hyperdiverse taxa, the lack of taxonomic expertise, or concentration of expertise to only a few individuals, inhibits morphological assessment of large numbers of specimens. Thus, an ambitious arthropod inventory can quickly overwhelm taxonomists with too many specimens, and thus are often unable to provide fine scale data for conservation for many groups. As an example, the NSF-funded arthropod inventory of Madagascar has shipped over a third of a million specimens to over 100 participating taxonomic collaborators (Fisher 2005). Major taxonomic products from these inventories, that will take decades to produce, represent only a fraction of the diversity collected, and provide no short-term return of biodiversity data to Madagascar. Inventories are essential for documenting global diversity and generating necessary material for taxonomic study. However, for inventories to be relevant in the short term, the inventory process must reduce the bottlenecks in returning relevant data for conservation.
Although in this study morphological determination of species only took a matter of months, the morphological protocol would not easily scale to include additional inventories across a wider geographic region. With each new site and each new species added, the documentation and analyses of morphological variation becomes increasingly more difficult. A sequence-based approach to the analysis of diversity, backed by a database of single gene barcodes, allows the exploration of diversity to scale to a rate that is not currently feasible using morphology alone. The DNA barcode provides a surrogate method for identifying units of diversity—a surrogate that will later serve as an additional character set for taxonomic assessment. If taxonomy is to provide a necessary tool to ecology and conservation science in hotspots it must be done at a much faster rate than in the past—especially with small, hyperdiverse or as yet undescribed fauna.
Our analysis provides an example of the speed and complementarity with which DNA barcoding can work in concert with a more conventional morphological taxonomic approach—neither competing nor replacing (Hebert & Gregory 2005). This result has important ramifications for the argument to include hyperdiverse taxa in conservation planning. Hyperdiverse groups can be included, knowing that in the short term, richness and turnover assessments can be provided and are valuable surrogates of diversity. Most importantly, with each inventory, additional data will be accrued to assist in taxonomic analysis.
(b) What do these results suggest about tropical ant diversity?
Discrepancies between estimates of morphospecies and MOTU identified with cox1 DNA barcodes occur in restricted geographic areas and usually involve sharp ecological and elevational differentiation (Fisher 1999). For instance, within Anochetus madagascarensis, we see subtle phylogeographic structure within the DNA barcode region. Individual A. madagascarensis were collected from the Binara (800 m elevation) on the east coast and Ambato (150 m elevation) on the west coast. These two localities are separated by the highest mountain in Madagascar (2876 m). Individuals from these populations are separated by, on average 1.5% sequence divergence. Are these populations operating as separate species? Are these populations members of the same species but highly divergent? Our data alone cannot answer this question. However, of critical import, our data have identified the surprising level of within-species divergence and lays bare these differences to further study. A standard arthropod molecular clock for cox1 is 1.2–1.5% per million years (Caccone & Sbordoni 2001; Farrell 2001; Dick et al. 2004). Hymenopterans exhibit rate acceleration for this gene (Hebert et al. 2003b), and therefore average estimates should be interpreted with caution. However, accelerating these rates suggests that populations have been isolated for several hundred thousand years. The opportunity now exists to employ a suite of approaches (behavioural observations, tests of interbreeding, and phylogeographic resolution of more quickly evolving genetic markers) to test species membership. We recognize that a molecular approach to biodiversity estimation may underestimate diversity when collections include quickly evolving species-pairs (Hebert et al. 2003a). Using the DNA barcode based MOTU approach, these pairs would be coded as one MOTU—as perhaps we have seen in A. madagascarensis. Indeed, ecological demarcation can make significant contributions to species formation and eventual morphological divergence. Distinct and extremely local divergence has been seen in the tropics in Costa Rican salamanders (Garcia-Paris et al. 2000), and the Amazonian poison-dart frog (Lougheed et al. 1999). As in these examples, the allopatric Anocheus MOTU documented here may represent cryptic species that owe their formation to orogeny: isolative and habitat effects of the development of mountain ranges. Mountains could produce unique lineages and species through the production of alternative refugial habitats through climatic changes (Hewitt 2004). Palynological studies throughout Madagascar have indicated a dynamic environmental history in response to periods of glaciation and interglaciation (Burney 1997; Gasse & Van Campo 1998). These climatic shifts have had a considerable impact on forest vegetation structure and therefore ant habitat.
Our sampling methodology in this preliminary study has resulted in divergent singleton specimens which were summarily coded as 2 or 3% MOTU in our analysis. However, it will only be by sampling multiple individuals from a provisional species, or MOTU, that interspecific variation will be properly assessed, allowing the hypothesis of species-level monophyly to be tested (Funk & Omland 2003—and references within are a thorough review). This is a valid concern in an analysis of MOTU from inventories of hyperdiverse groups such as ants, which often include many taxa known only from single individuals (Fisher 1999; Longino et al. 2002). With additional inventories in the future, many of these singletons will be represented in collections by more specimens. Caution must be expressed regarding any paraphyletic relationship discovered with a MOTU represented by one specimen (Funk & Omland 2003).
In cases where the 2–3% MOTU and the morphological estimation of a species differed, the molecular variants were either isolated geographically or were specimens contained by a species epithet whose actual status is, as yet, undetermined. This discrepancy does not compromise the use of a DNA barcode for species identification; just the opposite. In this case, DNA barcoding has identified regional lineages that can be further tested for species status or phylogeographic structure.
(c) Future
DNA barcoding proved an effective surrogate for morphospecies diversity patterns across localities in northern Madagascar. This study demonstrates how inventories of hyperdiverse taxa such as ants can provide rapid analysis of diversity for conservation assessment. Sequence data generated during the inventory process will also provide an alternative set of characters to assist in inferring species boundaries during future taxonomic studies. Thus, the application of DNA tools during diversity assessment will facilitate and complement taxonomic study. The combination of DNA sequencing data coupled with inventory and traditional taxonomy is a model that can be applied across disciplines and will allow analytical needs to scale to the enormity of the biodiversity crisis (DeSalle & Amato 2004). It will help in the identification and conservation of the evolutionary processes that generate and preserve biodiversity.
Of course, species boundaries are too complex and fuzzy (Green 1996) to be only described by DNA barcode sequence divergence. Rather a suite of informative characters (multi-gene analyses, behavioural studies and taxonomic expertise) is required for the delineation of a species. However, we show here that DNA barcodes allow the rapid identification of functional units of diversity that can scale to the magnitude of hyperdiverse arthropods at a timeframe needed by conservation groups responding to habitat destruction and degradation. Ant diversity, measured using MOTU in collaboration with taxonomists should provide the essential fine-scale maps for assessing biodiversity at a scale at which conservation decisions are made.
Little time remains for the documentation of global biodiversity. Taxonomists, equipped with modern tools and collaborations, have a chance to move systematics to the forefront of conservation and the public's attention. With increased taxonomic output and improved public access and visibility, public support for the discovery of life on this planet should follow.
Acknowledgments
The authors wish to thank the Consortium for Barcoding Life for organizing, the British Natural History Museum for hosting, and the Alfred P. Sloan Foundation for funding the First International Conference on Barcoding Life. Funding for this research was provided by the National Science Foundation awards to BLF and CE Griswold (DEB-0072713) and to BLF and PS Ward (DEB-0344731); the Gordon and Betty Moore Foundation, Natural Sciences and Engineering Research Council (Canada) and Canada Research Chairs program to PDNH, and a Fonds Nature et Technologies B3 postdoctoral fellowship (Quebec) to MAS. We are grateful to April Nobile for image creation, Dylan Burge for specimen preparation and Michele Esposito for specimen data entry. Thanks to Jeremy DeWaard and Angela Holliss for sequencing assistance, and Sujeevan Ratnasingham and Rob Dooh for analytical assistance. Fieldwork that provided the basis for this work could not have been completed without the gracious support of the Malagasy people, and the Arthropod Inventory Team (Balsama Rajemison, Helian Ratsirarson, Jean Claude Rakotonirina, Jean-Jacques Rafanomezantsoa, Chrislain Ranaivo, Coco Randriambololona, Hanitriniana Rasoazanamavo, Nicole Rasoamanana, Clavier Randrianandrasana, Pascal Rabeson, Valerie Rakotomalala, Dimby Raharinjanahary and Dylan Burge). John Dumbacher and Gary Umphrey and two anonymous reviewers provided valuable comments on an earlier version of this manuscript.
One contribution of 18 to a Theme Issue ‘DNA barcoding of life’.
MAS and BLF have contributed in equal part to this paper and each author reserves the right to be considered first author.
Supplementary Material
References
- Andersen A.N. Ants as indicators of restoration success at a uranium mine in tropical Australia. Restoration Ecol. 1993;1:156–167. [Google Scholar]
- Andersen A.N. Using ants as bioindicators: multiscale issues in ant community ecology. Cons. Ecol. 1997;1 [Google Scholar]
- Basset Y, Novotny V, Miller S.E, Pyle R. Quantifying biodiversity: experience with parataxonomists and digital photography in Papua New Guinea and Guyana. Bioscience. 2000;50:899–908. [Google Scholar]
- Battistini R, Richard-Vindard G. Junk; The Hague: 1972. Biogeography and ecology of Madagascar. [Google Scholar]
- Baum D.A. Individuality and the existence of species through time. Syst. Zool. 1998;47:641–653. doi: 10.1080/106351598260644. [DOI] [PubMed] [Google Scholar]
- Beattie A.J. Cambridge University Press; Cambridge, MA: 1985. The evolutionary ecology of ant–plant mutualisms. [Google Scholar]
- Bensasson D, Zhang D, Hartl D.L, Hewitt G.M. Mitochondrial pseudogenes: evolution's misplaced witnesses. Trends Ecol. Evol. 2001;16:314–321. doi: 10.1016/s0169-5347(01)02151-6. 10.1016/S0169-5347(01)02151-6 [DOI] [PubMed] [Google Scholar]
- Blaxter M.L. The promise of a DNA taxonomy. Phil. Trans. R. Soc. B. 2004;359:669–679. doi: 10.1098/rstb.2003.1447. 10.1098/rstb.2003.1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M, Elsworth B, Daub J. DNA taxonomy of a neglected animal phylum: an unexpected diversity of tardigrades. Proc. R. Soc. B. 2004;271(Suppl 4):S189–S192. doi: 10.1098/rsbl.2003.0130. 10.1098/rsbl.2003.0130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton B. Harvard University Press; Cambridge, MA: 2003. Identification guide to the ant genera of the World. [Google Scholar]
- Brooks T.M, da Fonseca G.A.B, Rodrigues A.S.L. Protected areas and species. Cons. Biol. 2004a;18:616–618. 10.1111/j.1523-1739.2004.01836.x [Google Scholar]
- Brooks T.M, da Fonseca G.A.B, Rodrigues A.S.L. Species, data, and conservation planning. Cons. Biol. 2004b;18:1682–1688. 10.1111/j.1523-1739.2004.00457.x [Google Scholar]
- Burney D.A. Theories and facts regarding Holocene environmental change before and after human colonization. In: Goodman S.M, Patterson B.D, editors. Natural change and human impact in Madagascar. Smithsonian Institution Press; Washington, DC: 1997. pp. 75–79. [Google Scholar]
- Caccone A, Sbordoni V. Molecular biogeography, evolutionary rates, and morphological adaptations to cave life: a case study using Bathysciine beetles and sequence data from the mitochondria CO1 gene. Evolution. 2001;55:122–130. doi: 10.1111/j.0014-3820.2001.tb01278.x. [DOI] [PubMed] [Google Scholar]
- Colwell, R.K. 2004 EstimateS: Statistical estimation of species richness and shared species from samples: available at http://viceroy.eeb.uconn.edu/estimates Persistent URL http://purl.oclc.org/estimates
- Crozier R.H, Crozier Y.C. The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization. Genetics. 1993;133:97–117. doi: 10.1093/genetics/133.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson D.W. The role of resource imbalances in the evolutionary ecology of tropical arboreal ants. Biol. J. Linn. Soc. 1997;61:153–181. 10.1006/bijl.1996.0128 [Google Scholar]
- Davidson D.W, Cook S.C, Snelling R.R, Chua T.H. Explaining the abundance of ants in lowland tropical rainforest canopies. Science. 2003;300:969–972. doi: 10.1126/science.1082074. 10.1126/science.1082074 [DOI] [PubMed] [Google Scholar]
- DeSalle R, Amato G. The expansion of conservation genetics. Nat. Rev. Gen. 2004;5:702–712. doi: 10.1038/nrg1425. 10.1038/nrg1425 [DOI] [PubMed] [Google Scholar]
- Dick C.W, Roubik D.W, Gruber K.F, Bermingham E. Long-distance gene flow and cross-Andean dispersal of lowland rainforest bees (Apidae: Euglossini) revealed by comparative mitochondrial DNA phylogeography. Mol. Ecol. 2004;13:3775–3785. doi: 10.1111/j.1365-294X.2004.02374.x. 10.1111/j.1365-294X.2004.02374.x [DOI] [PubMed] [Google Scholar]
- Diehl E, Sanhudo C.E, Diehl-Fleig E. Ground-dwelling ant fauna of sites with high levels of copper. Braz. J. Biol. 2004;64:33–39. doi: 10.1590/s1519-69842004000100005. 10.1590/S1519-69842004000100005 [DOI] [PubMed] [Google Scholar]
- Farrell B.D. Evolutionary assembly of the milkweed fauna: cytochrome oxidase 1 and the age of Tetraopes beetles. Mol. Phylogenet. Evol. 2001;18:469–478. doi: 10.1006/mpev.2000.0888. [DOI] [PubMed] [Google Scholar]
- Ferrier S.G, et al. Mapping more of terrestrial biodiversity for global conservation assessment. BioScience. 2004;54:1101–1109. [Google Scholar]
- Fisher B.L. Ant diversity patterns along an elevational gradient in the Reserve Naturelle Integrale d'Andringitra, Madagascar. Fieldiana Zool. 1996a;85:93–108. [Google Scholar]
- Fisher B.L. Origins and affinities of the ant fauna of Madagascar. In: Lourenco W.R, editor. Biogeographie de Madagascar. Editions de L'Orstrom; Paris: 1996b. pp. 457–465. [Google Scholar]
- Fisher B.L. Improving inventory efficiency: a case study of leaf litter ant diversity in Madagascar. Ecol. Appl. 1999;9:714–731. [Google Scholar]
- Fisher B.L. The Malagasy fauna of Strumigenys. In: Bolton B, editor. The ant tribe Dacetini. vol. 65. 2000. Memoirs of the American Entomological Institute. [Google Scholar]
- Fisher B.L. A model for a global inventory of ants: a case study in Madagascar. In: Jablonski N.G, editor. Biodiversity: a symposium held on the occasion of the 150th anniversary of the California Academy of Sciences. Proceedings of the California Academy of Sciences June 17–18. vol. 56. 2005. pp. 78–89. [Google Scholar]
- Fisher B.L, Girman D.J. Biogeography of ants in eastern Madagascar. In: Lourenco W.R, Goodman S.M, editors. Diversite et endemism a Madagascar. 2000. pp. 331–344. [Google Scholar]
- Floyd R, Abebe E, Papert A, Blaxter M. Molecular barcodes for soil nematode identification. Mol. Ecol. 2002;11:839–850. doi: 10.1046/j.1365-294x.2002.01485.x. 10.1046/j.1365-294X.2002.01485.x [DOI] [PubMed] [Google Scholar]
- Frost D.R, Hillis D.M. Species in concept and practice: herpetological applications. Herpetologica. 1990;46:87–104. [Google Scholar]
- Funk D.J, Omland K.E. Species-level paraphyly and polyphyly: frequency, causes and consequences, with insights from animal mitochondrial DNA. Ann. Rev. Ecol. Syst. 2003;34:397–423. 10.1146/annurev.ecolsys.34.011802.132421 [Google Scholar]
- Garcia-Paris M, Good D.A, Parra-Olea G, Wake D.B. Biodiversity of Costa Rican salamanders: implications of high levels of genetic differentiation and phylogeographic structure for species formation. Proc. Natl Acad. Sci. USA. 2000;97:1640–1647. doi: 10.1073/pnas.97.4.1640. 10.1073/pnas.97.4.1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasse F, Van Campo E. A 40 000 year pollen and diatom record from Lake Tritrivakely, Madagascar, in the southern Tropics. Quat. Res. (Orlando) 1998;49:299–311. 10.1006/qres.1998.1967 [Google Scholar]
- Gillespie R.G, Roderick G.K. Arthropods on islands: colonization, speciation, and conservation. Ann. Rev. Entomol. 2002;47:595–632. doi: 10.1146/annurev.ento.47.091201.145244. 10.1146/annurev.ento.47.091201.145244 [DOI] [PubMed] [Google Scholar]
- Goodman S.M, Benstead J. University of Chicago Press; Chicago: 2003. Natural history of Madagascar. [Google Scholar]
- Goodman S.M, Patterson B.D. Smithsonian Institution Press; Washington DC.: 1997. Natural change and human impact in Madagascar. [Google Scholar]
- Gotelli N.J. A taxonomic wish-list for community ecology. Phil. Trans. R. Soc. B. 2004;359:585–597. doi: 10.1098/rstb.2003.1443. 10.1098/rstb.2003.1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybeal A. Naming species. Syst. Biol. 1995;44:237–250. [Google Scholar]
- Green D.M. The bounds of species: hybridization in the Bufo americanus group of North American toads. Israel J. Zool. 1996;42:95–109. [Google Scholar]
- Hajibabaei M, de Waard J.R, Ivanova N.V, Ratnasingham S, Dooh R.T, Kirk S.L, Mackie P.M, Hebert P.D.N. Critical factors for assembling a high volume of DNA barcodes. Phil. Trans. R. Soc. B. 2005;360:1959–1967. doi: 10.1098/rstb.2005.1727. 10.1098/rstb.2005.1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hebert, P. D. N. & Gregory, T. R. In press. The promise of DNA barcoding for taxonomy. Syst. Biol. [DOI] [PubMed]
- Hebert P.D, Cywinska A, Ball S.L, deWaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B. 2003a;270:313–322. doi: 10.1098/rspb.2002.2218. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N, Ratnasingham S, deWaard J.R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B. 2003b;270:S96–S99. doi: 10.1098/rsbl.2003.0025. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D, Penton E.H, Burns J.M, Janzen D.H, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl Acad. Sci. USA. 2004;101:14 812–14 817. doi: 10.1073/pnas.0406166101. 10.1073/pnas.0406166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D, Stoeckle M.Y, Zemlak T.S, Francis C.M. Identification of birds through DNA barcodes. PLoS Biol. 2004b;2:e312. doi: 10.1371/journal.pbio.0020312. 10.1371/journal.pbio.0020312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt G.M. The structure of biodiversity—insights from molecular phylogeography. Front. Zool. 2004;1:4. doi: 10.1186/1742-9994-1-4. 10.1186/1742-9994-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highton R. Is Ensatina eschscholtzii a ring-species? Herpetologica. 1998;54:254–278. [Google Scholar]
- Jolly A, Oberlé P, Albignac R. Pergamon Press; Oxford: 1984. Key environments: Madagascar. [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Kumar S.K, Tamura I, Jakobsen B, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. 10.1093/bioinformatics/17.12.1244 [DOI] [PubMed] [Google Scholar]
- Lambert D.M, Baker A, Huynen L, Haddrath O, Hebert P.D, Millar C.D. Is a large-scale DNA-based inventory of ancient life possible? J. Hered. 2005;96:279–284. doi: 10.1093/jhered/esi035. 10.1093/jhered/esi035 [DOI] [PubMed] [Google Scholar]
- Longino J.T, Colwell R.K. Biodiversity assessment using structured inventory: capturing the ant fauna of a tropical rain forest. Ecol. Appl. 1997;7:1263–1277. [Google Scholar]
- Longino J.T, Coddington J, Colwell R.K. The ant fauna of a tropical rain forest: estimating species richness three different ways. Ecology. 2002;83:689–702. [Google Scholar]
- Lougheed S.C, Gascon C, Jones D.A, Bogart J.P, Boag P.T. Ridges and rivers: a test of competing hypotheses of Amazonian diversification using a dart-poison frog (Epipedobates femoralis) Proc. R. Soc. B. 1999;266:1829–1835. doi: 10.1098/rspb.1999.0853. 10.1098/rspb.1999.0853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenço W.R. ORSTOM; Paris: 1996. Biogéographie de Madagascar. [Google Scholar]
- Magurran A.E. Blackwell; Oxford, UK: 2004. Measuring biological diversity. [Google Scholar]
- Majer J.D. Ants: bio-indicators of minesite rehabilitation, land-use, and land conservation. Envir. Manag. 1983;7:375–383. 10.1007/BF01866920 [Google Scholar]
- Ness J.H, Bronstein J.L, Andersen A.N, Holland J.N. Ant body size predicts dispersal distance of ant-adapted seeds: Implications of small-ant invasions. Ecology. 2004;85:1244–1250. [Google Scholar]
- Powers T. Nematode molecular diagnostics: from bands to barcodes. Annu. Rev. Phytopathol. 2004;42:367–383. doi: 10.1146/annurev.phyto.42.040803.140348. 10.1146/annurev.phyto.42.040803.140348 [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. 10.1093/bioinformatics/15.2.174 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schatz G.E. Taxonomy and herbaria in service of plant conservation: Lessons from Madagascar's endemic families. Ann. Missouri Bot. Gard. 2002;89:145–152. [Google Scholar]
- Sites J, Marshall J.C. Delimiting species: a renaissance issue in systematic biology. Trends Ecol. Evol. 2003;18:461–471. 10.1016/S0169-5347(03)00184-8 [Google Scholar]
- Smith M.A, Umphrey G.J, Fisher B.L, Hebert P.D.N. The Natural History Museum; London, UK: 2005. Rapid assessment of ant diversity in a northern world heritage site using DNA barcodes. p. Poster. [Google Scholar]
- Storey M, Mahoney J.J, Saunders A.D, Duncan R.A, Kelley S.P, Coffin M.F. Timing of hot spot-related volcanism and the breakup of Madagascar and India. Science. 1995;267:852–855. doi: 10.1126/science.267.5199.852. [DOI] [PubMed] [Google Scholar]
- Sussman R.W, Green G.M, Sussman L.K. The use of satellite imagery and anthropology to assess the causes of deforestation in Madagascar. In: Sponsel L.E, Headland T.N, Bailey R.C, editors. Tropical deforestation: the human dimension. Columbia University Press; New York, NY: 1996. pp. 296–315. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorp J. Effects of certain animals that live in soils. In: Drew J.V, editor. Selected papers in soil formation and classification. vol. 1. SSSA Special Publication Series; Madison, WI: 1949. [Google Scholar]
- Wake D.B, Schneider C.J. Taxonomy of the plethodontid salamander genus Ensatina. Herpetologica. 1998;54:279–298. [Google Scholar]
- Ward R.D, Zemlak T.S, Innes B.H, Last P.R, Hebert P.D.N. DNA barcoding Australia's fish species. Phil. Trans. R. Soc. B. 2005;360:1847–1857. doi: 10.1098/rstb.2005.1716. 10.1098/rstb.2005.1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler Q. Systematics, the scientific basis for inventories of biodiversity. Biodiv. Cons. 1995;4:476–489. 10.1007/BF00056338 [Google Scholar]
- Wilson E.O. The encyclopedia of life. Trends Ecol. Evol. 2003;18:77–80. 10.1016/S0169-5347(02)00040-X [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.