Abstract
Some insight into human evolution has been gained from the sequencing of four Y chromosome genes. Primary genomic sequencing determined gene SMCY to be composed of 27 exons that comprise 4,620 bp of coding sequence. The unfinished sequencing of the 5′ portion of gene UTY1 was completed by primer walking, and a total of 20 exons were found. By using denaturing HPLC, these two genes, as well as DBY and DFFRY, were screened for polymorphic sites in 53–72 representatives of the five continents. A total of 98 variants were found, yielding nucleotide diversity estimates of 2.45 × 10−5, 5.07 × 10−5, and 8.54 × 10−5 for the coding regions of SMCY, DFFRY, and UTY1, respectively, with no variant having been observed in DBY. In agreement with most autosomal genes, diversity estimates for the noncoding regions were about 2- to 3-fold higher and ranged from 9.16 × 10−5 to 14.2 × 10−5 for the four genes. Analysis of the frequencies of derived alleles for all four genes showed that they more closely fit the expectation of a Luria–Delbrück distribution than a distribution expected under a constant population size model, providing evidence for exponential population growth. Pairwise nucleotide mismatch distributions date the occurrence of population expansion to ≈28,000 years ago. This estimate is in accord with the spread of Aurignacian technology and the disappearance of the Neanderthals.
The human Y chromosome consists of a nonrecombining region (NRY), which makes up 95% of its length, flanked by pseudoautosomal regions (1). Aside from the absence of recombination, NRY differs from all other nuclear human chromosomes by its presence in males only, its common ancestry and persistent meiotic relationship with the X chromosome, and the tendency of its genes to degenerate during evolution. Other than for its role in male sex determination, mediated by the SRY gene (2), the Y chromosome has been perceived as functionally desolate. Recently, 20 genes or gene families have been identified in the NRY (3). They fall into two classes. One class of nine genes is expressed in many organs and has X homologs that escape X inactivation. Among them are the single-copy genes DBY, DFFRY, and UTY, all of which have been mapped into the deletion interval 5C, and SMCY, located in the interval 5O (3). Most of the other NRY genes are highly duplicated, including the 11 NRY genes that are expressed specifically in testes. By deletion mapping, putative roles in sex determination, germ-cell tumorigenesis, determination of stature, and spermatogenesis have been established for most of the NRY genes (3). In particular, deletion of the AZFa region that contains DBY, DFFRY, and UTY has been shown to disrupt spermatogenesis, causing infertility in otherwise healthy men (4). SMCY, on the other hand, has been found to contain the human H-Y epitope, H-Y/HLA-B7, which alone or in part accounts for the male-specific transplantation antigen (5). SMCY's 65% homology to the retinoblastoma 2 gene suggests that the gene may code for a transcription factor (5).
The NRY has been also considered devoid of sequence variation (6). However, recently, biallelic variation has been revealed, including three variants in an 18.3-kilobase (kb) stretch of the SRY sequence (7), two variants within 949 bp of the RPS4Y gene (8), and five variants in the DFFRY gene (9). In other studies that used sequenced-tagged sites dispersed throughout the NRY (10) as templates for PCR, direct sequencing alone or in combination with DHPLC led to the discovery of more than 30 biallelic sites in a wide range of ethnicities (11, 12).
The present study reports a total of 98 simple sequence polymorphisms that were discovered by DHPLC in the coding and intronic regions of the DBY, DFFRY, SMCY, and UTY1 genes screened in a set of 53–72 males representing as many as 46 diverse populations from the five continents. We also provide evidence for exponential population growth and its onset with the spread of Aurignacian technology.
Materials and Methods
DNA Samples.
The continental and ethnic affiliations of the 53–72 males used for screening SMCY, DBY, DFFRY, and UTY1 are given in Tables 1 and 3. All samples were collected according to approved human subject protocols.
Table 1.
Nucleotide position, defined with respect to the GenBank entry U52195, and geographic distribution of the 47 SMCY polymorphisms detected in 53 representatives from Africa, Asia, Europe, America, and Oceania
| Population | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 3 | 4 | 5 | 5 | 5 | 5 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H | 4 | 4 | 5 | 6 | 9 | 9 | 2 | 2 | 3 | 3 | 3 | 3 | 4 | 1 | 6 | 6 | 6 | 6 | 9 | 9 | 9 | 3 | 7 | 7 | 7 | 3 | 4 | 4 | 3 | 4 | 4 | 4 | 4 | 4 | ||||||||||||||
| a | 2 | 2 | 0 | 2 | 3 | 3 | 0 | 0 | 6 | 6 | 6 | 6 | 8 | 5 | 4 | 4 | 4 | 4 | 9 | 9 | 9 | 0 | 6 | 6 | 6 | 5 | 0 | 2 | 4 | 9 | 6 | 6 | 6 | 6 | ||||||||||||||
| P | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 5 | 5 | 4 | 7 | 2 | 3 | 8 | 8 | 7 | 7 | 8 | 8 | 7 | 5 | 6 | 6 | 6 | 6 | 1 | 1 | 2 | 6 | 2 | 2 | 2 | 6 | 4 | 9 | 4 | 8 | 7 | 7 | 7 | 7 | |
| l | - | - | - | - | - | - | - | - | - | - | - | - | - | + | + | - | - | + | - | + | + | + | + | - | - | + | 1 | + | + | + | + | + | + | - | + | - | - | - | + | 3 | 8 | + | 9 | + | + | + | + | |
| o | 3 | 3 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 8 | 4 | 1 | 3 | 2 | 9 | 5 | 7 | 9 | 1 | 2 | 6 | 1 | 7 | 5 | * | 1 | 3 | 3 | 4 | 7 | 1 | 2 | 9 | 9 | 5 | 3 | 1 | 8 | U | 4 | 8 | 8 | 1 | |||
| t | 1 | 1 | 9 | 6 | 4 | 3 | 3 | 9 | 8 | 7 | 0 | 4 | 0 | 6 | 5 | 8 | 6 | 2 | 7 | 0 | 3 | 3 | 2 | 1 | 0 | 4 | 8 | 7 | 9 | 1 | 0 | 3 | 3 | 2 | 8 | 2 | T | 6 | 1 | 8 | 4 | |||||||
| y | 7 | 4 | 4 | 2 | 9 | 6 | 5 | 4 | 8 | 6 | 6 | 2 | 1 | 2 | 4 | 0 | 5 | 6 | 7 | 2 | 4 | 8 | 8 | 9 | 8 | 2 | 8 | 7 | 0 | 8 | 0 | 3 | R | 8 | 7 | 4 | 8 | |||||||||||
| p | 6 | 7 | 5 | 5 | 5 | 0 | 8 | 2 | 1 | 4 | 2 | 2 | 6 | 1 | 1 | 3 | 5 | 7 | ||||||||||||||||||||||||||||||
| e | 0 | 4 | 4 | 1 | 4 | 3 | 0 | 9 | 7 | 6 | 0 | |||||||||||||||||||||||||||||||||||||
| Orangutan | - | G | - | A | - | A | 5bp | - | - | G | G | 9A | A | - | - | A | T | - | - | - | - | G | T | - | A | T | G | - | C | C | - | - | T | T | O | - | T | A | C | C | A | A | C | T | G | T | A | |
| Gorilla | - | G | G | A | T | A | 5bp | C | A | G | G | 7A | A | A | A | A | T | - | 4A | - | - | G | T | AT | A | T | G | G | T | C | AC | 7T | T | T | O | G | T | G | C | C | A | A | C | T | - | - | C | |
| Chimpanzee | G | G | G | A | T | A | 5bp | C | A | G | G | 9A | A | A | A | A | T | T | 4A+C+4A | C | G | G | T | AT | A | T | G | G | C | C | AC | TA+7T | T | T | O | G | T | A | T | C | A | A | C | T | G | T | C | |
| Human, ancestral | G | G | G | A | T | A | 5bp | C | A | G | G | 9A | A | A | A | A | T | T | 9A | C | G | G | T | AT | A | T | G | G | C | C | AC | 8T | T | T | O | G | T | A | C | C | A | A | C | T | G | T | C | |
| AF09 Khoisan | 1 | . | . | . | . | . | . | . | . | . | . | . | 8A | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . |
| AF11 Surma | 2 | . | . | . | . | . | . | . | . | G | . | . | 8A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . |
| AF01, AF08, OC02† | 3 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . |
| AF02, AF03, AF05‡ | 4 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | A | . | . | . |
| AF07 Lisongo | 5 | . | T | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | A | . | . | . |
| AF04 Mbuti Zaire Pygmy | 6 | . | T | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | T | . | . | . | A | . | . | . |
| AF06 Lisongo | 7 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | A | . | . | . |
| AF12 Bozo | 8 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . |
| AF13 Tuareg | 9 | . | T | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . |
| AS18 Sindhi | 10 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 8A | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . |
| AS21 Kazak | 11 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . |
| AS06 Japanese | 12 | . | T | . | . | . | G | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | A | . | . | . |
| AS07 Japanese | 13 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | A | . | . |
| AF10 and others§ | 14 | . | T | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . |
| AS10 and others¶ | 15 | . | T | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | T | A | . | . | . |
| AS19 Arab, Central Asia | 16 | . | T | . | . | . | . | . | . | . | . | A | . | . | G | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | T | A | . | . | . |
| AS20 Arab, Central Asia | 17 | . | T | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | O | . | . | . | . | . | . | . | . | . | . | . | T | A | . | . | . |
| AS22 Uzbek | 18 | . | T | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | A | . | . | . | . | . | T | A | . | . | . |
| EU04 Italian | 19 | . | T | . | . | . | . | O | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | T | A | . | . | . |
| AS02, AS11, AS17‖ | 20 | . | T | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | O | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . |
| OC03, 04 Melanessian | 21 | . | T | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | C | . |
| OC01 New Guinean | 22 | . | T | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | A | . | C | . |
| EU05, 06 Basque | 23 | . | T | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | T |
| AS01 Cambodian | 24 | A | T | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . |
| AS08 Han Chinese | 25 | A | T | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . |
| AS15 Brahul | 26 | . | T | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . |
| EU01 Sardinian | 27 | . | T | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 9T | . | A | . | . | . | . | . | . | . | . | . | A | . | . | . |
| AS14 Baloochi | 28 | . | T | . | . | . | . | . | . | . | . | A | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | A | . | . | . |
| EU02 Italian | 29 | . | T | . | . | . | . | . | G | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . |
| EU03 German | 30 | . | T | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . |
| AS03 Atayal | 31 | . | T | A | . | . | . | . | . | . | . | A | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | A | . | . | . |
AF, Africa; AS, Asia; EU, Europe; AM, America; OC, Oceania; -, sequence not available.
Nonsynonymous substitution.
AF01, Biaka Pygmy; AF08, Khoisan; OC02, New Guinean.
AF02, 03, Biaka Pygmy; AF05, Mbuti Zaire Pygmy.
AF10, Berta; AS04, Ami; AS05, Japanese; AS09, Han Chinese; AS12, 13, Pathan; AS16, Sindhi; AS23, Druze; OC06, Australian Aborigine.
AS10, Brushaski; EU07, Basque; OC05, Australian Aborigine; AM01, Karitiana; AM02, Surui; AM03, Colombian Indian; AM04, Mayan Indian.
AS02, Cambodian; AS11, Brushaski; AS17, Sindhi.
Table 3.
Variable sites in 72 individuals for DFFRY, DBY, and UTY1, defining 18 major haplotypes, not including polymorphic sites observed once
| Population |
DFFRY†
|
DBY†
|
UTY1†
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 2 | 3 | 5 | 5 | 6 | 8 | 9 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 3 | 3 | 3 | 3 | 3 | |||||||||||||||||||
| H | 1 | 5 | 2 | 4 | 9 | 3 | 0 | 4 | 4 | 4 | 9 | 0 | 6 | 6 | 7 | 8 | 1 | 1 | 6 | 7 | 0 | 9 | 3 | 3 | 3 | 3 | 3 | 1 | 1 | 2 | 2 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 6 | 9 | 2 | 6 | 6 | 6 | 6 | 6 | 7 | |||||
| a | 2 | 4 | 2 | 3 | 8 | 0 | 8 | 2 | 8 | 7 | 0 | 5 | 1 | 7 | 9 | 8 | 9 | 7 | 0 | 4 | 9 | 7 | 1 | 1 | 1 | 1 | 1 | 0 | 5 | 2 | 2 | 3 | 3 | 3 | 3 | 8 | 8 | 8 | 1 | 7 | 3 | 7 | 7 | 7 | 7 | 7 | 8 | |||||
| p | 7 | 4 | 9 | 7 | 1 | 7 | 7 | 9 | 4 | 8 | 3 | 0 | 5 | 7 | 9 | 6 | 4 | 1 | 1 | 1 | 1 | 5 | 9 | 5 | 1 | 4 | 2 | 2 | 2 | 2 | 2 | 1 | 8 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 5 | 1 | 5 | 8 | 8 | 9 | 9 | 9 | 4 | |
| l | 7 | + | 5 | + | + | 7 | - | 2 | 2 | - | - | + | 5 | 7 | 1 | - | + | - | - | - | - | + | - | - | - | + | + | + | + | + | + | 4 | - | + | + | + | + | + | + | + | - | - | + | - | 8 | + | + | - | - | - | + | |
| o | U | 7 | * | 1 | 9 | 4 | * | 1 | 1 | 9 | * | 3 | 4 | 2 | 2 | 2 | 2 | 1 | 3 | 5 | 1 | 3 | 3 | 4 | 8 | 1 | 1 | 4 | 3 | 3 | 1 | 5 | 6 | 6 | 1 | 2 | 7 | 8 | 6 | 5 | 7 | 2 | 2 | 3 | 1 | |||||||
| t | T | 3 | 8 | 6 | 0 | 0 | 1 | 8 | 5 | 2 | 2 | 4 | 2 | 2 | 2 | 3 | 8 | 9 | 6 | 1 | 0 | 1 | 6 | 6 | 8 | 7 | 2 | 2 | 6 | 2 | 8 | 8 | 0 | 3 | 6 | 8 | 9 | 2 | 6 | |||||||||||||
| y | R | 1 | 6 | 9 | 1 | 7 | 2 | 0 | 0 | 0 | 0 | 9 | 9 | 3 | 2 | 7 | 9 | 1 | 2 | 2 | 7 | 8 | 1 | 4 | 9 | 5 | ||||||||||||||||||||||||||
| p | 5 | 2 | 7 | 9 | 0 | 4 | 1 | 8 | 1 | 1 | 8 | |||||||||||||||||||||||||||||||||||||||||
| e | 3 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Orangutan | - | - | C | - | - | T | - | A | G | T | - | - | - | G | T | - | T | C | O | T | G | T | T | - | G | - | - | - | - | A | G | C | A | - | - | T | - | - | - | - | - | - | 5bp | - | T | G | T | G | - | - | - | |
| Gorilla | - | - | C | - | - | - | - | - | G | T | - | - | - | - | T | - | - | - | - | - | - | - | - | - | - | T | - | - | - | - | C | - | - | - | T | - | - | - | - | G | - | - | - | - | - | T | - | - | - | C | ||
| Chimpanzee | T | C | C | - | T | T | C | A | G | T | - | - | C | G | T | A | T | C | O | T | A | T | T | A | G | T | C | C | O | A | G | C | A | A | A | T | G | T | T | G | G | A | 5bp | A | T | G | T | G | T | A | C | |
| Human, ancestral | T | C | C | A | T | T | C | A | G | C | G | O | C | G | T | A | T | C | O | T | A | T | T | A | G | T | C | C | O | A | G | C | A | A | A | T | G | T | T | G | G | A | 5bp | A | T | G | T | G | T | A | C | |
| AF09 Khoisan | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | C | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . |
| AF15 Falasha, Ethiopia | 1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | G | . |
| AF14 Nuba | 2 | . | . | . | . | . | C | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . |
| AF02 Biaka Pygmy | 3 | . | . | . | . | . | C | T | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| AF03 Biaka Pygmy | 3 | . | . | . | . | . | C | T | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . |
| AF05 Mbuti Zaire Pygmy | 3 | C | . | . | . | . | C | T | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| AF07 Lissongo | 3 | . | . | . | . | . | C | T | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| AF04 Mbuti Zaire Pygmy | 4 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| AS37 Bedouine | 4 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| AF01 Biaka Pygmy | 5 | . | T | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| AF06 Lissongo | 5 | . | T | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| AF16 Nuba‡ | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| AS06 Japanese | 6 | . | T | T | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| AS33 Hmong, Laos | 6 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| AS18 Sindhi | 7 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . |
| AS38 Bedouine | 7 | . | T | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . |
| AS07 Japanese | 8 | . | T | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . |
| OC02 New Guinean | 8 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | A | . | . | . | . | . |
| OC07 Australian Aborigine | 8 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | A | . | . | . | . | . |
| AS28, AS30, AM06§ | 9 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | G | . | . | . | . |
| AS02 and others¶ | 10 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . |
| AM02 Surul | 10 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . |
| AM04 Mayan | 10 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . |
| AS11 Brushaski | 10 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . |
| AS12 Pathan | 10 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . |
| AS15 Brahui | 10 | . | T | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | 4bp | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . |
| EU03 and others‖ | 11 | . | T | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . |
| AS16, AS25** | 12 | . | T | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . |
| OC01 New Guinean | 13 | . | T | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . |
| OC03, OC04 Melanessian | 13 | . | T | . | . | . | . | . | . | . | . | O | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . |
| AS14, AS29, AS35, EU08‡‡ | 14 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . |
| AS40 Yemenite | 15 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | T |
| EU02 Italian | 15 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | T |
| AS01 and others†† | 16 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | O | G | . | . | . | . | . | . | . |
| AS09 Han Chinese | 16 | . | T | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | G | O | G | . | . | . | . | . | . | . |
| EU04 and others§§ | 17 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . |
| EU13 Italian‡ | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | . | . | . | . | C | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . |
| AS10, AS24¶¶ | 18 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | T | . | . | . | . | . | . | . | C | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . |
| AS39 Arab, Central Asia | 18 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | T | . | . | . | . | C | . | . | C | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . |
AF, Africa; AS, Asia; EU, Europe; AM, America; OC, Oceania; -, sequence not available.
Nonsynonymous substitution.
Variant nucleotide positions are defined with respect to the cDNA sequences in GenBank for DFFRY (AF000986), DBY (AF000985), and UTY1 (AF000996).
Screened for UTY1 only.
AS28, Japanese; AS30, Hazara; AM06, Navajo Indian.
AS02, Cambodian; AS17, Sindhi; AS23, Druze; AS32, Tamil; AS34, 36, Bedouine; EU12, Ashkenazi; AM01, Karitiana; AM03, Colombian Indian.
EU03, German; EU05, 06, Basque; EU09, Finn; OC06, Australian Aborigine; AM05, Muskogee.
AS16, Sindhi; AS25, Makrani.
AS14, Baloochi; AS29, Hazara; AS35; Sepharadim; EU08, Ashkenazi.
AS01, Cambodian; AS03, Atayal; AS04, Ami; AS05, 27; Japanese; AS08, 26, Han Chinese; AS13, Pathan.
EU04, Italian; EU07, Basque; EU10, Finn; EU11, Georgia; AS31, Haraza; OC05, Australian Aborigine.
AS10, Brushaski; AS24, Makrani.
Reference DNA Sequences.
A contiguous 75,850-bp reference sequence containing the SMCY gene was obtained by joining two cosmid sequences (GenBank accession nos. AC003031 and AC003094), which were shotgun sequenced at the Stanford DNA Sequencing and Technology Center. The repeatmasker2 program (http://ftp.genome.washington.edu) was used to identify human repeat DNA sequences. The respective reference genomic and cDNA sequences of DFFRY, AC002531, and AF000986 and of DBY, AC004474, and AF000985 were downloaded from GenBank, as was the genomic sequence of the first 16 exons of UTY1 (AC006376). A 9-kb sequence gap containing exons 17–20 was closed by long-range PCR (PCR SuperMix High Fidelity kit, Life Technologies, Rockville, MD) by using exon 16 and the 3′ untranslated region (AF000996) as priming sites.
PCR.
Primers designed for SMCY covered all unique sequences and repeat elements other than long interspersed elements (LINEs), yielding amplicons 300–500 bp in length (http://www.ncbi.nlm.nih.gov/SNP). For DBY, DFFRY, and UTY1, only the coding and flanking noncoding sequences were amplified. The PCR protocol comprised an initial denaturation at 95°C for 10 min to activate AmpliTaq Gold, 14 cycles of denaturation at 94°C for 20 s, primer annealing at 63–56°C with 0.5°C decrements, and extension at 72°C for 1 min, followed by 20 cycles at 94°C for 20 s, 56°C for 1 min, and 72°C for 1 min, and a final 5-min extension at 72°C. Each 50-μl PCR contained 1 unit of AmpliTaq Gold polymerase, 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 0.1 mM each of the four deoxyribonucleotide triphosphates, 0.2 μM each of forward/reverse primers, and 50 ng of genomic DNA. PCR yields were determined semiquantitatively on ethidium bromide-stained agarose gels.
DHPLC Analysis.
Unpurified PCR products were mixed at an equimolar ratio with a reference Y chromosome and subjected to a 3-min 95°C denaturing step followed by gradual reannealing from 95°C to 65°C over 30 min. Each mixture (10 μl) was loaded onto a DNASep column (Transgenomic, San Jose, CA), and the amplicons were eluted in 0.1 M triethylammonium acetate (pH 7) with a linear acetonitrile gradient at a flow rate of 0.9 ml/min (11). Under appropriate temperature conditions, which were optimized by computer simulation (available at http://insertion.stanford.edu/melt.html), mismatches were recognized by the appearance of two or more peaks in the elution profiles.
DNA Sequencing.
Polymorphic and reference PCR samples were purified with Qiagen (Valencia, CA) QIAquick spin columns and sequenced with the PE Biosystems (Foster City, CA) Dye Terminator Cycle Sequencing Kit and a model 373A DNA sequencer. Chimpanzee, gorilla, and orangutan samples were sequenced for the entire 4.62-kb coding region of SMCY and, whenever possible, for those NRY segments that were polymorphic in humans.
Statistical Analysis.
The program dnaml in phylip (version 3.57c) was used to construct phylogenetic networks. Insertions and deletions were treated as single-nucleotide substitutions. The estimated nucleotide diversity, π, in a sample of n chromosomes was calculated from the equation
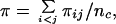 |
1 |
where πij is the number of nucleotide differences between the ith and jth DNA sequences and nc = n(n − 1)/2 (13).
Tajima's D statistic (14) is defined by the equation
 |
2 |
S is the number of segregating nucleotides, and V̂(d) an estimator of the variance of d. The value of D is 0 for selectively neutral mutations in a constant population infinite sites model. A negative value of D indicates either purifying selection or population expansion (15).
The expected Luria–Delbrück/Lea–Coulson (16, 17) distribution of the number of mutants for each gene was fitted by maximum likelihood, treating each nucleotide of the screened sequence as analogous to a parallel, independent bacterial culture. The distributions under the expectation of constant population size were calculated according to the method of Watterson (18).
For the mismatch distribution, the mean number of differences (m) between pairs of chromosomes and variance (ν) of this number were computed for each gene. The corresponding parameters were θ (θ = 2μN0, where N0 is the effective population size before a single instantaneous expansion), μ (the mutation rate for each gene), and τ (time in mutational units since the expansion of the population, assuming a generation length of 25 years). For estimation of θ and τ, we used the following equations (see ref. 19)
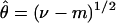 |
3 |
and
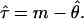 |
4 |
The standard errors for τ̂ were estimated by simulation (20), averaging the mean number of differences in 1,000 coalescent trees, each of which was constructed randomly with a sample size and an estimate of θ matched to the observed distribution. Errors from estimating θ itself and any uncertainties from the estimation of mutation rates are not included. The actual time (T) since the expansion was estimated by τ̂/2 μl, where μ is the mutation rate (based on the number of sequence differences observed between human and chimpanzee and assuming a divergence time of 4.9 million years) and l is the number of base pairs tested. The mutation rate per nucleotide for SMCY was 1.53 × 10−9, calculated on the basis of 597 observed differences between human and chimpanzee over 39,931 bp, whereas that of DBY, DFFRY, and UTY1 was 1.25 × 10−9 (470 differences over 38,468 bp).
Results and Discussion
SMCY.
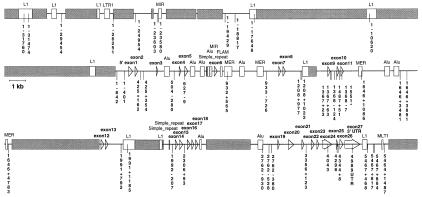
The genomic structure of SMCY is shown in Fig. 1. The gene consists of 27 exons. The shadowed areas (35.9 kb) represent LINE elements that have been excluded from DHPLC analysis. A total of 39,931 bp (4,620 bp coding) was screened in 53 human male samples from the five continents, and 47 polymorphic sites (Table 1) were detected: 30 (64%) transitions, 10 (21%) transversions, and 7 (15%) deletions or insertions including two runs of As or Ts, respectively. Three variants in the coding region (exon 11, G1,551A, Val426Met; exon 24, C4,043T, Pro1,256Pro; exon 25, A4,298G, Gly1,341Gly) were observed only once in a Sindhi, Zaire Pygmy, and Khoisan, respectively. One site was recurrent, namely T627-9C in intron 4. Of the 47 biallelic sites, 15, 19, 5, and 2 were observed exclusively in Africa, Asia, Europe, and Oceania, respectively. Three polymorphisms were found in all five continents. Two additional sites were detected everywhere except in Africa, and one was present in both a Sardinian and a Baloochi.
Figure 1.
Genomic structure of SMCY and the nucleotide positions of 47 sequence variants detected in 53 individuals. SMCY consists of 27 exons spaced over 39.5 kb, 11.6% of which are coding. The AUG start codon is located in exon 2. Shadowed areas (35.9 kb) represent LINE elements that were excluded from the variation search. The numbers indicate the nucleotide positions of the polymorphic sites with respect to the published cDNA sequence (GenBank accession no. U52195).
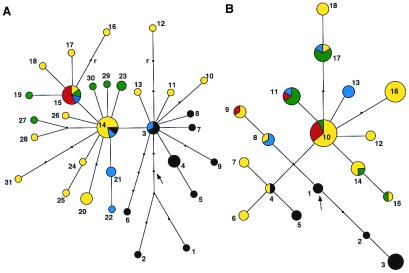
The 47 polymorphisms were used to infer a phylogenetic network comprising 31 distinct haplotypes (Table 1 and Fig. 2A). Asia and Africa have the most haplotypes with 15 and 10, respectively, followed by Europe, Oceania, and America with 6, 5, and 1, respectively. The root of the network, based on primate sequence data, is located in Africa, with the most ancestral patrilineages represented today by a Khoisan and an East African Surma. As suggested by the three clades centered at haplotypes 3, 14 and 15, respectively, anatomically modern humans migrated in, at least, two waves from Africa into Asia and New Guinea, followed by a later dispersal probably out of Western Asia into Central Asia, Europe, and the Americas. Independent corroborating evidence for the occurrence of these three major prehistoric migratory episodes has been derived from the geographic distribution of haplotypes from a chromosome 21 region (21). A detailed coalescence analysis and dating of the key mutations in the SMCY phylogeny is provided in the companion paper by Thomson et al. (22).
Figure 2.
Maximum likelihood networks of the SMCY haplotypes (A) and the major haplotypes constructed from the combined variant sites in DFFRY, DBY, and UTY1 (B). Haplotype numbers are from Tables 1 and 3. The representations of color are black, Africa; yellow, Asia; green, Europe; red, America; and blue, Oceania. The areas of the circles represent the number of individuals carrying each haplotype. The arrows indicate the location of the most likely root in the phylogenies. The occurrence of a single recurrent mutation (r) did not generate any ambiguity in the parsimonious mutational pathway when considered in the context of other polymorphisms.
The three primates (chimpanzee, gorilla, and orangutan) were sequenced over the entire coding region of SMCY (4,620 bp) and 4,758 bp of flanking noncoding sequences. In the coding region, humans differ in a total of 125 positions (available on request) from apes, 80% of which are transitions. Nucleotide position 3,917 is triallelic (C in human and chimpanzee, A in gorilla, and G in orangutan). There are four nonsynonymous differences separating the human from all great apes, only one of which resulted in a nonconservative amino acid change (Val1,186Ala). The two transitions and two transversions were found in exon 3 (A480G, Val69Ile), exon 7 (A963C, His230Asn), exon 10 (C1,423G, Ser383Thr), and exon 24 (C3,832T, Val1,186Ala). Generally, nonsynonymous differences are more frequent in the C-terminal part of the gene. No nonsynonymous change was detected in the H-Y epitope region (nucleotides 3,160–3,200). Table 2 summarizes the number of sequence differences found among the four species. Assuming 4.9 million years for the divergence time of human and chimpanzee, the divergence time of human and gorilla was estimated from the log proportion of identical nucleotides as 7.0 and 7.4 million years for coding and noncoding segments, respectively. The corresponding divergence times for human and orangutan were 16.2 and 16.9 million years. These numbers are in fairly good agreement with published divergence times of 6.6 and 13.0 million years, respectively, for gorilla and orangutan based on mitochondrial DNA substitution rates (23).
Table 2.
Number of sequence differences between humans and apes in coding and flanking noncoding sequences of SMCY
| Species | Coding (4,620 bp)* | Noncoding (4,758 bp) |
|---|---|---|
| Human/chimpanzee | 29 (51.7) | 79 |
| Human/gorilla | 41 (70.7) | 109 |
| Human/orangutan | 89 (69.7) | 258 |
| Chimpanzee/gorilla | 40 (55.0) | 130 |
| Chimpanzee/orangutan | 94 (64.9) | 275 |
| Gorilla/orangutan | 95 (67.4) | 282 |
The percentage of silent mutations is given in parentheses.
DFFRY, DBY, and UTY1.
Table 3 lists the polymorphic sites identified in DFFRY, DBY, and UTY1 in 70 (72 for UTY1) Y chromosomes representing 41 ethnic groups. For DFFRY, 17 polymorphisms were discovered over 16,699 bp (7,668 bp coding). Seven of the variant sites are located in exons 6, 11, 18, 22, 30, 33, and 37. Three are nonsynonymous, resulting in two nonconservative (exon 6, C2,295T, Arg211Cys; exon 22, G4,842A, Ala1,060Thr) and one conservative amino acid change (exon 33, G6,777T, Ala1,705Ser). G4,842A had been already observed three times in 576 infertile men and once in 96 fertile men (9). In the present study, G4,842A was detected only once in an individual from Pakistan. The other three polymorphisms reported by Sun et al. (9) were not observed in our populations.
For DBY, a total of 14 polymorphic sites were found over 9,000 bp (1,983 bp coding), and they are all noncoding. This observation is in concordance with the study of Sun et al. (9), who also failed to detect a single coding variant. For UTY1, 20 polymorphic sites were discovered over 15,317 bp (3,240 bp coding). Two sites, both synonymous, are located in exons 1 and 14. By combining the polymorphisms observed in DFFRY, DBY, and UTY1 and excluding those observed only once, 18 major haplotypes could be inferred, most of them found in Asia (n = 10) and Africa (n = 5). The similarity of the phylogeny of the three genes (Fig. 2B) that are located within the same deletion interval on the NRY (5C) to that of SMCY (deletion interval 5O) attests to the coherence of the NRY, which, therefore, can be viewed evolutionarily as if it were a single locus. For instance, among the individuals present in both ascertainment sets, the same Asian individuals (As06, As07, and As18) and the New Guinean (OC02) cluster with the Africans, supporting the hypothesis of at least two distinct migratory waves out of Africa. A more detailed analysis of DBY and DFFRY including coalescence time estimates is provided in the companion paper by Thomson et al. (22).
Nucleotide Diversity and Tajima's D Values.
The following nucleotide diversity estimates (± SEM) were obtained for the coding and noncoding segments, respectively, of the four NRY genes investigated: DBY, 0 and 10.5 ± 8.75 × 10−5; DFFRY, 5.07 ± 5.34 × 10−5 and 9.85 ± 7.72 × 10−5; SMCY, 2.45 ± 4.44 × 10−5 and 9.16 ± 5.33 × 10−5; and UTY1, 8.54 ± 10.3 × 10−5 and 14.2 ± 9.29 × 10−5. Both the higher than expected percentage of synonymous mutations (67%) and the approximately 2-fold lower nucleotide diversity in coding versus noncoding segments imply selection against deleterious alleles. However, the ratios of nucleotide diversity estimates for coding and noncoding segments on the NRY are very similar to those observed for 16 autosomal genes, with an arithmetic mean of 2.6 and a range of 0.63 to 6.67. Further, the average nucleotide diversity for the coding regions of the four NRY genes (0.51 × 10−4) is about five times lower than the corresponding estimate of 2.62 ± 0.51 × 10−4 (based on 223 polymorphic sites over 58,093 bp) for the 16 autosomal genes AMPD1, ATM, BRCA1, BRCA2, COL, COX2, CYP1A1, EA2, FBN1, IL2, IL4, MTE, STR, Tubulin M40, Werner, and XRCC1. A similar ratio was obtained comparing the noncoding regions of the same 16 autosomal genes (5.07 ± 0.55 × 10−4, based on 479 polymorphic sites over 79, 445 bp) to the 4 NRY genes (0.99 × 10−4). The ratio is somewhat lower than expected given that the effective population size of autosomes relative to the Y chromosome is only 4-fold greater. To test for reduced variability on the NRY, a pairwise comparison of loci linked to noncoding segments of the NRY and the autosomal genes was performed with the Hudson–Kreitman–Aguadé test (24) and chimpanzee sequences for homologous genes (data not shown). The NRY clearly shows evidence of reduced variability to the autosomes (χ2[1] =4.0; P = 0.045), confirming similar comparisons of smaller numbers of polymorphisms on previously reported NRY sequences with X-linked human genes (25, 26). Possible explanations include not only selection on NRY but also differences between male and female effective population sizes and/or in the dispersal of males and females.
The Tajima's D values for the combined coding and noncoding segments of DBY, DFFRY, SMCY, and UTY1 were −2.15 (P < 0.05), −1.89, −2.31 (P < 0.05), and −1.57, respectively. At first glance, these values suggest purifying selection on the human Y chromosome. However, unpublished Tajima's D values obtained for mtDNA (−2.11; P < 0.05), AMPD1 (−2.07; P < 0.05), ATM (−2.25; P < 0.05), BRCA2 (−2.10; P < 0.05), COL (−1.19), COX2 (−2.16; P < 0.05), CYP1A1 (−2.20, P < 0.05), EA2 (−1.72), FBN1 (−1.80), IL4 (−1.19), MTE (−1.71), STR (−1.09), Tubulin M40 (−1.21), XRCC1 (−1.45), DIAPH2 (−1.36), and MCM (−0.96) are also all negative. Other than being characteristic of selection, negative D values are also characteristic of growing populations where initially rare alleles have a stronger effect on S than on π (15). The relative homogeneity of values obtained for 20 genes of four different genetic systems is therefore more in agreement with population growth, which will affect all genes equally. Signatures of population expansion were also reported for microsatellite repeat data (27).
An independent observation points unequivocally to population growth. We analyzed the distribution of the number of mutants at independent sites in an exponentially growing population by fitting the Luria–Delbrück distribution (16, 17) to the observed frequency spectra in the four NRY genes (Fig. 3). Although far from perfect, the marginal fits obtained for the former (SMCY, χ2[4] = 9.99, P = 0.041; DBY, χ2[3] = 6.08, P = 0.108; DFFRY, χ2[3] = 5.96, P = 0.114; and UTY1, χ2[4] = 10.38, P = 0.034) are strikingly better than those observed with distributions generated under the hypothesis of constant population size (18). The respective χ2 values for the latter distributions were 66.96 (df = 4; P ≈ 10−13), 20.51 (df = 3; P ≈ 0.0001), 13.73 (df = 3; P ≈ 0.003), and 18.03 (df = 4; P ≈ 0.001) for SMCY, DBY, DFFRY, and UTY1, respectively. Deviations from the Luria–Delbrück distribution may result from population growth rates not exactly constant in time or space, variable mutation rates, random genetic drift, and the difficulty of obtaining a truly random sample of the world population.
Figure 3.
Allele frequency spectra of the four Y chromosome genes. Shaded columns show the observed distribution of allele frequencies. Black columns depict the expected numbers of derived alleles under the expectation of the Luria–Delbrück/Lea–Coulson theory (16, 17), assuming that each nucleotide in the screened regions is analogous to a parallel, independent bacterial culture. The concordance of the observed and expected distributions is consistent with a significant population expansion. In contrast, there is strong incongruity with the expectation of constant population size (blank columns) with the distribution of alleles estimated by Watterson (18).
Mismatch Distributions.
To investigate further the hypothesis of population expansion and the time of its occurrence, we computed the distribution of pairwise differences from the segregating sites of both the 70 individuals studied for all of DBY, DFFRY, and UTY1, and the 53 SMCY sequences (Fig. 4). In the two cases, nearly unimodal and smooth mismatch distributions were observed with a raggedness (r) of 0.0133 and 0.0231, respectively (20). A similar distribution has been reported for segregating sites in the hypervariable segments of human mitochondrial DNAs, with an estimated time of population expansion of 40,000 years ago. Although based on a simple model of a sudden expansion to a very large population size (19), mismatch distributions provide a means of comparing different genetic systems. In this regard, it is interesting that our NRY data suggest a more recent expansion time of ≈28,000 years ago. This time falls within the range of 7,000–41,000 years, with a mean of 18,000 years, that has been obtained with a rejection algorithm for eight NRY microsatellites surveyed in 445 individuals (28). The discrepancy in expansion time estimates between the NRY genes on the one hand and mtDNA on the other hand is most likely due to the reduced variability on the former. Nevertheless, the NRY expansion time estimates seem to be correlated with the spread of Aurignacian technology (29) and the disappearance of Neanderthals, who lived in Europe as recently as 28,000 years ago (30).
Figure 4.
Histograms display the numbers of sequence differences among all possible pairs of sequences in the set of 70 individuals for 51 segregating sites in DFFRY, DBY, and UTY1, over 41,016 bp (A) and 53 individuals for 47 segregating sites in SMCY over 39,931 bp (B). The mean values and standard errors (SE) for human expansion times were estimated as described in the text.
In conclusion, polymorphic genes on the NRY are evolutionarily neutral or nearly so. Largely immune to recurrent mutation, they provide a valuable system for the reconstruction of the history of our species.
Acknowledgments
We thank the DNA donors and the investigators who provided the samples: M. E. Ibrahim, T. Jenkins, J. Kidd, S. Q. Mehdi, P. Parham, M. T. Seielstad, and R. S. Wells. Marc Feldman and Steve Sherry kindly provided constructive criticism. This work was supported by National Institutes of Health Grants GM55273 and HG01707.
Abbreviation
- NRY
nonrecombining region of the Y chromosome
- DHPLC
denaturing HPLC
- kb
kilobase
Footnotes
References
- 1.Cooke H J, Brown W R, Rappold G A. Nature (London) 1985;317:687–692. doi: 10.1038/317687a0. [DOI] [PubMed] [Google Scholar]
- 2.Sinclair A H, Berta P, Palmer M S, Hawkins J R, Griffiths B L, Smith M J, Foster J W, Frischauf A M, Lovell-Badge R, Goodfellow P N. Nature (London) 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 3.Lahn B T, Page D C. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- 4.Vogt P H, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, Kohn F M, Schill W B, Farah S, Ramos C, et al. Hum Mol Genet. 1996;5:933–943. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- 5.Kent-First M G, Maffitt M, Muallem A, Brisco P, Shultz J, Ekenberg S, Agulnik A I, Agulnik I, Shramm D, Bavister B, et al. Nat Genet. 1996;14:128–129. doi: 10.1038/ng1096-128. [DOI] [PubMed] [Google Scholar]
- 6.Dorit R L, Akashi H, Gilbert W. Science. 1995;268:1183–1185. doi: 10.1126/science.7761836. [DOI] [PubMed] [Google Scholar]
- 7.Whitfield L S, Sulston J E, Goodfellow P N. Nature (London) 1995;368:379–380. doi: 10.1038/378379a0. [DOI] [PubMed] [Google Scholar]
- 8.Bergen A W, Wang C Y, Tsai J, Jefferson K, Dey C, Smith K D, Pear S C, Tsai S J, Goldman D. Ann Hum Genet. 1999;63:62–80. doi: 10.1046/j.1469-1809.1999.6310063.x. [DOI] [PubMed] [Google Scholar]
- 9.Sun C, Skaletsky H, Birren B, Devon K, Tang Z, Silber S, Oates R, Page D. Nat Genet. 1999;23:429–432. doi: 10.1038/70539. [DOI] [PubMed] [Google Scholar]
- 10.Vollrath D, Foote S, Hilton A, Brown L G, Beer-Romero P, Bogan J S, Page D C. Science. 1992;258:52–59. doi: 10.1126/science.1439769. [DOI] [PubMed] [Google Scholar]
- 11.Underhill P A, Jin L, Lin A A, Mehdi S Q, Jenkins T, Vollrath D, Davis R W, Cavalli-Sforza L L, Oefner P J. Genome Res. 1997;7:996–1005. doi: 10.1101/gr.7.10.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su B, Xiao J, Underhill P, Deka R, Zhang W, Akey J, Huang W, Shen D, Lu D, Luo J, et al. Am J Hum Genet. 1999;65:1718–1724. doi: 10.1086/302680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ. Press; 1987. pp. 256–258. [Google Scholar]
- 14.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tajima F. Genetics. 1989;123:597–601. doi: 10.1093/genetics/123.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luria S E, Delbrück M. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lea D E, Coulson A C. Genetics. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 18.Watterson G A. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 19.Sherry S T, Rogers A R, Harpending H, Soodyall H, Jenkins T, Stoneking M. Hum Biol. 1994;66:761–775. [PubMed] [Google Scholar]
- 20.Harpending H C, Sherry S T, Rogers A R, Stoneking M. Curr Anthropol. 1993;34:483–496. [Google Scholar]
- 21.Jin L, Underhill P A, Doctor V, Davis R W, Shen P, Cavalli-Sforza L L, Oefner P J. Proc Natl Acad Sci USA. 1999;96:3796–3800. doi: 10.1073/pnas.96.7.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomson R, Pritchard J K, Shen P, Oefner P J, Feldman M W. Proc Natl Acad Sci USA. 2000;97:7360–7365. doi: 10.1073/pnas.97.13.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horai S, Hayasaka K, Kondo R, Tsugane K, Takahata N. Proc Natl Acad Sci USA. 1995;92:532–536. doi: 10.1073/pnas.92.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudson R R, Kreitman M, Aguadé M. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nachman M W. Mol Biol Evol. 1998;15:1744–1750. doi: 10.1093/oxfordjournals.molbev.a025900. [DOI] [PubMed] [Google Scholar]
- 26.Jaruzelska J, Zietkiewicz E, Labuda D. Mol Biol Evol. 1999;16:1633–1640. doi: 10.1093/oxfordjournals.molbev.a026076. [DOI] [PubMed] [Google Scholar]
- 27.Kimmel M, Chakraborty R, King J P, Bamshad M, Watkins W S, Jorde L B. Genetics. 1998;148:1921–1930. doi: 10.1093/genetics/148.4.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard J K, Seielstad M T, Perez-Lezaun A, Feldman M W. Mol Biol Evol. 1999;16:1791–1798. doi: 10.1093/oxfordjournals.molbev.a026091. [DOI] [PubMed] [Google Scholar]
- 29.Klein R G. The Human Career: Human Biological and Cultural Origins. 2nd Ed. Chicago: Univ. Chicago Press; 1999. [Google Scholar]
- 30.Smith F H, Trinkaus E, Pettitt P B, Karavanic I, Paunovic M. Proc Natl Acad Sci USA. 1999;96:12281–12286. doi: 10.1073/pnas.96.22.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]